
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 March 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.838421
This article is part of the Research TopicZygotic and Non-Zygotic Embryogenesis: Evolutionary, Developmental and Practical AspectsView all 13 articles
Although full sequence data of several embryogenesis-related genes are available in conifers, their functions are still poorly understood. In this study, we focused on the transcription factor WUSCHEL-related HOMEOBOX 2 (WOX2), which is involved in determination of the apical domain during early embryogenesis, and is required for initiation of the stem cell program in the embryogenic shoot meristem of Arabidopsis. We studied the effects of constitutive overexpression of Pinus pinaster WOX2 (PpWOX2) by Agrobacterium-mediated transformation of P. pinaster somatic embryos and Arabidopsis seedlings. Overexpression of PpWOX2 during proliferation and maturation of somatic embryos of P. pinaster led to alterations in the quantity and quality of cotyledonary embryos. In addition, transgenic somatic seedlings of P. pinaster showed non-embryogenic callus formation in the region of roots and subsequently inhibited root growth. Overexpression of PpWOX2 in Arabidopsis promoted somatic embryogenesis and organogenesis in a part of the transgenic seedlings of the first and second generations. A concomitant increased expression of endogenous embryogenesis-related genes such as AtLEC1 was detected in transgenic plants of the first generation. Various plant phenotypes observed from single overexpressing transgenic lines of the second generation suggest some significant interactions between PpWOX2 and AtWOX2. As an explanation, functional redundancy in the WOX family is suggested for seed plants. Our results demonstrate that the constitutive high expression of PpWOX2 in Arabidopsis and P. pinaster affected embryogenesis-related traits. These findings further support some evolutionary conserved roles of this gene in embryo development of seed plants and have practical implications toward somatic embryogenesis induction in conifers.
With respect to the divergence of angiosperms and gymnosperms, many morphological differences of embryogenesis and embryo development have to be considered (Bowe and Coat, 2000; Cairney et al., 2006; Cairney and Pullman, 2007). Many studies using Arabidopsis as a model plant have increased our knowledge of the function of embryogenesis-related genes in angiosperms (Laux et al., 1996; Lotan et al., 1998; Boutilier et al., 2002; Haecker et al., 2004; Breuninger et al., 2008; Jeong et al., 2011; Chung et al., 2016; Horstman et al., 2017). In contrast, little is known about the function of genes that regulate embryogenesis in conifers (Cairney and Pullman, 2007; De Silva et al., 2008; Trontin et al., 2016a). This is because of particular challenges of conifers. These species have long breeding cycles, large physical size, and slow growth. As a result, a number of powerful genetic approaches used in model plants, such as identification of zygotic embryo-defective mutants and T-DNA insertional mutagenesis, are impracticable. Moreover, recalcitrance to vegetative propagation through conventional or tissue culture methods (mostly aging/phase-change effects) often results in lack of an efficient system for direct plant regeneration from selected superior trees. Available methods are, therefore, mostly “retroactive” and based on somatic embryogenesis initiation from immature seeds coupled with cryopreservation to preserve the juvenility of embryogenic tissue. Selected genotypes after field evaluation of somatic seedlings in clonal tests can be propagated from a cryopreserved juvenile stock. This is currently the preferred and powerful strategy in conifers to overcome recalcitrance of mature trees (Klimaszewska and Cyr, 2002; Bonga et al., 2010) to achieve clonal propagation of superior genotypes and for gene functional studies by genetic transformation of embryogenic cultures (Klimaszewska et al., 2007, 2009, 2016).
The first step for assignment of gene functions in conifers is usually based on sequence similarities between conifers and model angiosperms. Most embryogenesis-related genes identified in Arabidopsis (a global network of ca. 300–450 genes) (Tzafrir et al., 2004; De Smet et al., 2010) have homologous candidate sequences in conifers (Cairney and Pullman, 2007; Zhang et al., 2012). The second one is analysis of transgenic plants following overexpression and/or knock-down of the candidate genes. Such reverse genetics approach for functional gene studies remains challenging in most conifers, because significant genomic resources are needed (genome sequence, expressed sequence tags, etc.), as well as efficient genetic transformation and plant regeneration systems (Trontin et al., 2007). Significant progress has been achieved in several conifer species of high economical interest (Pinus, Picea, and Larix) based on Agrobacterium-mediated genetic transformation of embryogenic cultures and plant regeneration by somatic embryogenesis (Trontin et al., 2007; Klimaszewska et al., 2009, 2016; Bonga et al., 2010). Such a system has been developed for reverse genetics in maritime pine (Pinus pinaster Ait.) during the past 15 years (Trontin et al., 2002, 2007, 2016b; Lelu-Walter et al., 2016) and was implemented by multinational EU consortia (e.g., Sustainpine, 2010–20131; ProCoGen, 2012–20152) for functional gene studies (Trontin et al., 2013; El-Azaz et al., 2020).
An embryogenesis-related gene that is crucial for early embryo development in angiosperms is WUSCHEL-related HOMEOBOX 2 (WOX2). In Arabidopsis, WOX2 is required for cell fate decisions and domain delineation in the apical domain during embryo development, including initiation of shoot meristem stem cells (Haecker et al., 2004; Xiao et al., 2006; Breuninger et al., 2008; Zhang et al., 2017). In post-embryonic development, WOX2 seems to be involved in lateral organ formation and separation (Chung et al., 2016). The WOX2 gene encodes a member of the WOX family of homeodomain transcription factors. WOX proteins share a homeobox that encodes a WUS-type homeodomain. In addition to the homeodomain, many WUS/WOX proteins contain a conserved WUS-box (TLPLFPMH) located downstream of the homeodomain in both angiosperms and gymnosperms (Haecker et al., 2004; Nardmann and Werr, 2006; see also Figure 1). Dolzblasz et al. (2016) reported that the canonical WUS-box is essential for stem cell maintenance within shoot apical meristem in Arabidopsis. Moreover, several overexpression studies indicate that WOX family members containing the conserved WUS-box are involved in plant growth and development. For example, induced overexpression of WOX5 in Arabidopsis produces extra layers of stem cells in columella root cap (Zhang et al., 2010). Overexpression of WOX1 in Arabidopsis leads to defects in meristem development (Zhang et al., 2011). Transgenic plants overexpressing WUSCHEL (WUS, founding member of the WOX family) in Arabidopsis led to high frequency of somatic embryo (SE) formation in all tissues without any need for exogenous plant hormone supply (Laux et al., 1996; Zuo et al., 2002). Furthermore, in angiosperms other than Arabidopsis, the overexpression of AtWUS induced somatic embryogenesis and callus formation (Arroyo-Herrera et al., 2008; Solís-Ramos et al., 2009; Bouchabke-Coussa et al., 2013).
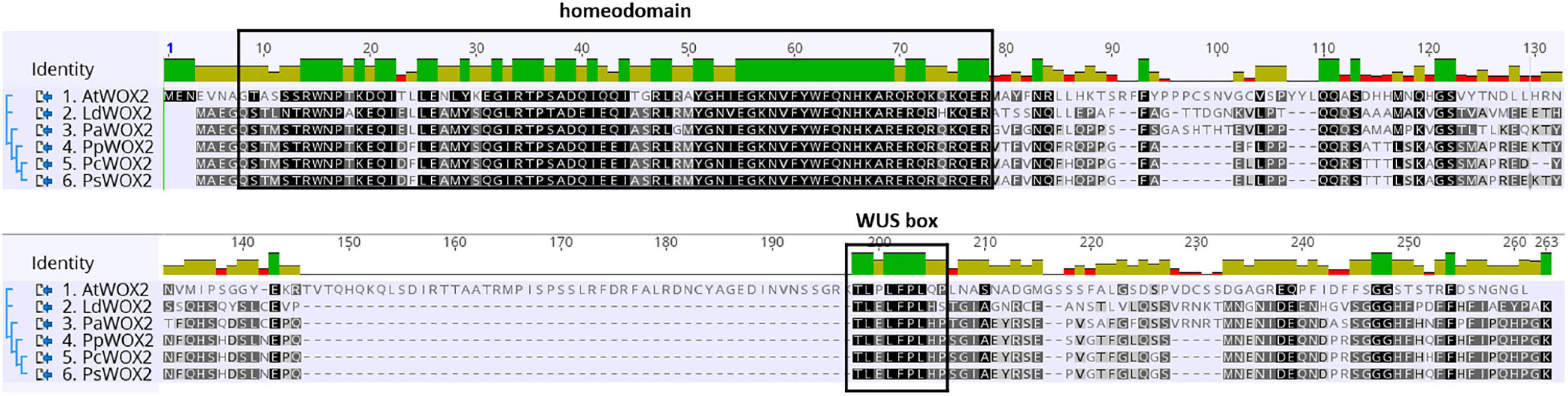
Figure 1. Comparative WOX2 amino acid sequences in the angiosperm Arabidopsis thaliana (AtWOX2) and in 5 conifers, namely, Larix decidua (LdWOX2), Picea abies (PaWOX2), Pinus pinaster (PpWOX2), Pinus contorta (PcWOX2), and Pinus sylvestris (PsWOX2). Introduced gaps in the amino acid sequences are indicated by dashes. Boxes show the putative WOX homeodomain and WUS box. Amino acid identity is shown by a color code.
In conifers, the transcription factor WOX2 is considered as an important developmental regulator during early somatic embryogenesis (Klimaszewska et al., 2010; Trontin et al., 2016a) and has been proposed as a putative marker for effective initiation of somatic embryogenesis and to predict embryogenic potential (Palovaara and Hakman, 2008; Klimaszewska et al., 2010, 2011; Miguel et al., 2016; Alvarez et al., 2018). Zhu et al. (2016) revealed that the WOX2 from Picea abies (PaWOX2) plays more crucial roles during early embryogenesis (i.e., protoderm formation and suspensor expansion) than during late embryogenesis.
Genomic resources are now accumulating in model conifers, especially for Picea and Pinus species (Birol et al., 2013; Nystedt et al., 2013; Neale et al., 2014; Plomion et al., 2016). For this study, we focused on P. pinaster, as the production of similar genomic sequence data was initiated during the European ProCoGen project (2011–20152). Moreover, a reference transcriptome has been made available for this species (SustainpineDB database1) (Canales et al., 2014).
To our knowledge, there has been no report up to now on the overexpression of WOX2 isolated from conifers in both gymnosperms and, more specifically, angiosperms. The genetic transformation of Pinus is a long and difficult process typically achieved in about 12 months. Deregulating WOX2 with same binary vector in both P. pinaster and Arabidopsis would provide additional insights into evolutionary conserved roles of this gene in gymnosperms and angiosperms, particularly during somatic embryogenesis. In this study, we analyzed the constitutive overexpression of WOX2 isolated from P. pinaster (PpWOX2) in both P. pinaster SE and Arabidopsis seedlings. Our results showed that the endogenous expression of PpWOX2 is enhanced in early proliferating embryos and significantly decreased during SE development in P. pinaster. Constitutive overexpression of PpWOX2 in P. pinaster embryogenic cultures affected SE development and promoted callus formation in the root region of regenerated somatic seedling. In Arabidopsis seedlings, PpWOX2 overexpression led to increased propensity to somatic embryogenesis and organogenesis, likely by alteration of expression patterns of embryogenesis-related genes.
Somatic embryos (SEs) of a P. pinaster embryogenic line (PN519) were used for genetic transformation and transgenic plant production. PN519 was initiated at FCBA in 1999 from an immature zygotic embryo (full-sib progeny, Breton et al., 2006) according to the method of Bercetche and Pâques (1995). This line has good abilities for both genetic transformation and plant regeneration by somatic embryogenesis and has been, therefore, extensively characterized for the past 15 years (Trontin et al., 2002, 2007, 2013, 2016b; Breton et al., 2006; Lelu-Walter et al., 2016).
Seeds of Arabidopsis thaliana (L.) Heynh. (ecotype Columbia-0) were obtained from the Nottingham Arabidopsis Stock Center3. For germination, the Arabidopsis seeds were soaked first in tap water at 4°C for 2 days and thereafter transferred to potting soil (Stender, Schermbeck, Germany) and grown for 4–6 weeks at 21–23°C under short day conditions (10-h light/14-h dark cycle) with illumination at ∼120 μE m–2 s–1 provided by HQI Powerstar 400W/D (Osram Licht AG, Munich, Germany). For flower induction, plants were cultivated under the same but longer day conditions (16-h light).
Both SEs and non-embryogenic calluses were obtained from wild-type (WT) Arabidopsis plant seedlings after induction following the method reported by Gaj (2001). WT SE and non-embryogenic WT calluses were used as positive (embryogenic) and negative (non-embryogenic) controls, respectively.
In silico search for WOX2 sequences in the P. pinaster expressional database (see Canales et al., 2014) was performed by Philippe Label (INRAE, France). The cDNA sequence of a putative P. pinaster homolog to WOX2 (PpWOX2) was identified in SustainpineDB (nine unigenes assigned to conifer WOX2) and submitted under accession number (Acc.) KY773924.1 at the National Center for Biotechnology Information (NCBI) GenBank, and its cloning was performed. PpWOX2 was concurrently sequenced by Alvarez et al. (2018) and deposited in GenBank under accession number KU962991. The two published sequences are identical (570 bp).
Alignment of the deduced amino acid sequence of PpWOX2 [Acc. ARS01278 in this study; concurrently deposited by Alvarez et al. (2018), Acc. ANC94872] with homologous WOX2 protein sequences from Pinus sylvestris (PsWOX2, Acc. CAT02937.2; Nardmann et al., unpublished), Pinus contorta (PcWOX2, Acc. ADR10436.1; Park et al., 2010), Picea abies (PaWOX2, Acc. CAL18267.1; Palovaara and Hakman, 2008), Larix decidua (LdWOX2, Acc. AEF56564.2; Rupps et al., 2016), and A. thaliana (AtWOX2, Acc. NP_200742.2; Tabata et al., 2000) was carried out using the Geneious R10.2.3 software (Biomatters Ltd., Auckland, New Zealand). Gaps were included in the sequences to optimize the alignment.
To generate the 35S::PpWOX2 vector, the GUSPlus gene of the 35S::GUSPlus cassette from the binary vector pCAMBIA1305.2 (GenBank Acc. AF354046)4 was replaced with the PpWOX2 gene isolated from P. pinaster (Acc. KY773924). The GUSPlus gene was excised from pCAMBIA1305.2 by restriction with NcoI and PmlI. The entire open reading frame of PpWOX2 was amplified from cDNA (obtained from total RNA extract) using specific primers (Table 1). The PCR fragment (570 bp) was then ligated into the NcoI and PmlI sites of the digested pCAMBIA1305.2. The resulting 35S::PpWOX2 binary vector was checked by sequencing and introduced into the disarmed Agrobacterium tumefaciens strain C58/C1 using the freeze-thaw method (Nishiguchi et al., 1987). The pCAMBIA1305.2 vector harbors a constitutive expression cassette (35S::HPTII) for selection of transgenic events with hygromycin. This antibiotic is effective in maritime pine (Trontin et al., 2002, 2007).

Table 1. List of gene-specific primers used for transgene analyses and reverse transcription quantitative (RT-q) PCR in Pinus pinaster and Arabidopsis.
Agrobacterium-mediated genetic transformation of P. pinaster proliferating SE was performed with proliferating SE, 7 days after sub-culture, i.e., during the phase of active SE growth on proliferation medium.
The transformation procedure was a combination of a reference FCBA/INRAE protocol designed during the Sustainpine project for P. pinaster [Trontin and Lelu-Walter, 2010; partially published in Trontin et al. (2002, 2007)] and the “droplet method” adapted for Larix decidua (Taryono, 2000). The co-cultivation procedure was performed according to the “droplet method”. One colony of Agrobacterium C58/C1 was inoculated in a liquid YEB medium with appropriate antibiotics at 28°C for approximately 16 h while shaking (200 rpm). The optical density at 600 nm (OD600) of Agrobacterium suspension was adjusted to about 0.5. Each clump of immature proliferating SEs was inoculated with one droplet (30 μl) of the Agrobacterium suspension. The SEs and agrobacteria were co-cultivated for 2 days in an MSG medium (Klimaszewska, 1989) with 0.1 M acetosyringone in darkness at 21–23°C. For elimination of agrobacteria, the co-cultivated SEs were washed with a 200-ml liquid MSG medium supplemented with 300 mg l–1 timentin (MSG-T) for 30 min. Afterward, the SEs were collected on filter paper in a Büchner funnel using a vacuum pump and a short low-pressure pulse. The filter paper was subsequently placed into a solidified MSG-T medium for 10 days. Selection for transgenic embryo clumps (transgenic lines) was performed on an MSG-T medium supplemented with 10 mg l–1 hygromycin for 2 weeks. The SEs were then sub-cultured every 2 weeks into the MSG-T medium plus 15 mg l–1 hygromycin for 6 weeks. After the selection procedure, transformation efficiency was calculated as the number of hygromycin-resistant lines confirmed by PCR analysis (PCR+ lines) per gram fresh mass (FM) of co-cultivated proliferating SEs.
All transformation experiments were performed twice, and up to 10 PCR+ lines were cryopreserved.
After preliminary SE maturation experiments, we selected two independent PCR+ lines that showed quite stable PpWOX2 overexpression during SE proliferation (lines OE_#11 and OE_#15). The maturation experiments were performed at the same time for both 35S::PpWOX2 lines, as well as for one empty vector transgenic control line, pCAMBIA 1305.2 (EV-pC05), and the non-transgenic WT PN519 control line. All transgenic and control lines were managed simultaneously and at the same subculture frequency to avoid any differences related with aging effect.
The whole process of SE development since reactivation from the cryopreserved stock involved three developmental stages: proliferation, maturation, and conversion (Ramarosandratana et al., 2001). In proliferating embryogenic tissues, an immature SE consists of an embryonic region of small, densely cytoplasmic cells subtended by long, highly vacuolated suspensor cells (Figures 2A,B). During maturation, the embryonic region became more prominent and opaque. Finally, a cotyledonary SE shows a shoot apex and a root pole containing primary meristems, a well-defined cotyledon ring, and a hypocotyl (Figure 2C). Cotyledonary SEs have a potential to germinate and develop into rooted somatic seedlings (Figure 2D).
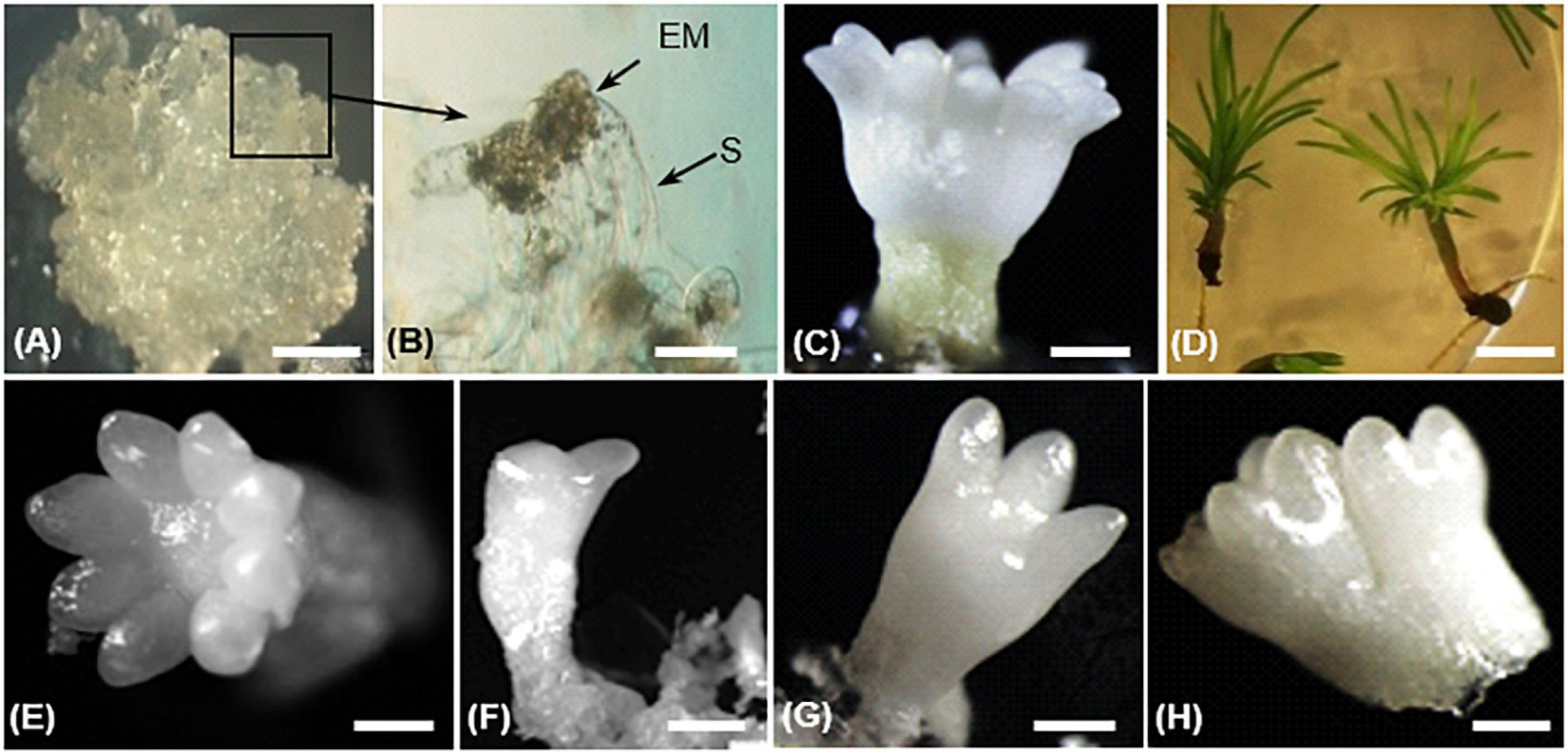
Figure 2. Different steps of somatic embryo (SE) development in Pinus pinaster (A–D) and morphology of harvested cotyledonary SEs (E–H) in transgenic 35S::PpWOX2 lines and control lines (transgenic EV-pC05 line and the non-transgenic WT).(A) Embryogenic culture containing proliferating immature SE; (B) proliferating SEs showing embryo proper (EP) and suspensor cells (S); (C) cotyledonary SE after 14 weeks in maturation medium; (D) somatic seedlings after 8 weeks in conversion medium. Cotyledonary SEs from PpWOX2 and control lines were classified according to different categories such as (E) normal (cotyledons with symmetrical radial development), (F,G) deformed (cotyledons with asymmetrical development), and (H) not separated, fused embryos. Scale bars: (A) 3 mm, (B) 0.1 mm, (B,C,E–H) 2 mm, and (D) 5 mm.
The PN519 embryogenic line was initially reactivated from the FCBA cryopreserved stock following the method described by Harvengt (2005) but using a modified Litvay et al. (1985) basal formulation (mLV) as reported in Klimaszewska et al. (2001, 2007). Proliferating immature SEs were sub-cultured every 10–14 days on Petri dishes in an mLV basal medium (Klimaszewska et al., 2001, 2007) supplemented with 2 μM 2,4-D (2,4-dichlorophenoxyacetic) acid (Sigma-Aldrich, Germany) and 1 μM BAP (6-benzylaminopurine; Sigma-Aldrich, Germany). Embryogenic cultures were maintained in darkness at 22 ± 2°C. Re-cryopreservation of PN519 transgenic and non-transgenic control WT lines was performed using a protocol previously established for Abies and Larix species (Zoglauer et al., 2003). Any thawed SE culture batch was reactivated within 1–2 months prior to maturation.
Development of SEs to reach the cotyledonary stage was performed on mLV-based maturation medium as the best option reported for maritime pine (Lelu-Walter et al., 2016; Trontin et al., 2016b). The method described by Morel et al. (2014) was used with slight modifications. Briefly, 1 g of proliferating SEs (3–5 days after sub-culture in mLV proliferation medium) was washed first and then suspended in 10 ml of the mLV liquid medium without plant growth regulators (PGRs). An aliquot of a suspension (1 ml containing ∼100 mg proliferating SEs) was spread on a sterile filter paper (Whatman No. 2, diameter 11 cm), air-dried at ambient temperature, and placed on a Petri dish in an mLV medium supplemented with 80 μM abscisic acid (ABA; Duchefa, Haarlem, The Netherlands) and 0.2 M sucrose, and solidified with 1% (w/v) gellan gum (Gelrite; Duchefa, Haarlem, The Netherlands). All maturation cultures were maintained in darkness and at 22 ± 2°C. Each maturation experiment was performed on 10 Petri dishes and repeated 2–3 times.
After 12–14 weeks, SE development reached the cotyledonary stage (Morel et al., 2014). Embryogenic potential was estimated as the number of cotyledonary SEs per g FM of embryogenic tissue. Cotyledonary SEs of PpWOX2 transgenic lines and controls (transgenic EV-pC05 line and non-transgenic WT) were classified according to their morphological characteristics as “normal” (cotyledons with symmetrical radial development, Figure 2E), “deformed” (cotyledons with asymmetric development, e.g., one cotyledon, two cotyledons, etc., Figures 2F,G), or “not separated,” fused embryos (two embryos not clearly separated from each other, Figure 2H; Marum et al., 2009).
To convert cotyledonary SEs into plantlets, fully developed embryos with well-shaped cotyledons (normal SEs) were placed horizontally onto Petri dishes on the surface of an mLV medium supplemented with 3% sucrose (w/v) and solidified with 0.4% (w/v) Gelrite and 0.06% (w/v) plant agar (Duchefa, Haarlem, The Netherlands). The cultures were maintained in darkness at 22 ± 2°C for 10 days. Afterward, the seedlings were transferred to a long day photoperiod (14-h light, 120 μE m–2 s–1, OSRAM Lumilux Cool White light) at 22 ± 2°C for 6–8 more weeks. Seven embryos were placed in each plate. Three replicates were used for each treatment, and experiments for SE conversion were repeated three times. After 6 weeks, the SEs were scored as converted into viable somatic seedlings if an elongated epicotyl, green cotyledons, and primary needles as well as an elongating root apex were observed. The percentage of somatic seedlings with root elongation was determined after 8 weeks of culture.
The ability of different explants from P. pinaster somatic seedlings to initiate embryogenic tissue and/or non-embryogenic callus was tested for PpWOX2 transgenic lines, the empty vector (EV-pC05), and non-transgenic WT control lines. Needle/hypocotyl and root explants of somatic seedlings were collected 6–8 weeks after germination of cotyledonary SEs and cultivated in a conversion medium, i.e., mLV basal medium (Klimaszewska et al., 2001, 2007) without PGRs. The cultures were incubated in darkness for 8 weeks at 21–23°C.
Agrobacterium-mediated transformation of Arabidopsis seedling plants using the 35S::PpWOX2 vector was performed with a modified floral-dip method to obtain T0 seeds (Weigel and Glazebrook, 2002). To select transgenic lines and for regeneration of first-generation (T1) plants, water-soaked and cold-treated (2 days at 4°C) T0 seeds were surface-sterilized by immersion in 1.5% NaOCl (active chlorine) for 15 min, followed by three rinses in autoclaved Aqua purificata (aqua pur.). Then, the T0 seeds were cultured in vitro in an MS medium (Murashige and Skoog, 1962) supplemented with 20 mg l–1 hygromycin and incubated for 4–6 weeks under short day conditions (10-h light, 120 μE m–2 s–1). Hygromycin-resistant transgenic T1 plants obtained from treated T0 seeds were transferred to potting soil for 3–4 weeks under long day conditions to promote flowering. Plants from two independent T1 transgenic lines (PpWOX2-line #1 and PpWOX2-line #2) were self-pollinated. Resulting seeds of the T1 generation were harvested and subsequently cultured in an MS medium supplemented with 20 mg l–1 hygromycin to regenerate T2 transgenic plants. The two selected T1 transgenic lines were shown to be homozygotes for the PpWOX2 transgene based on both hygromycin-resistance assay and PCR analysis of their T2 progenies (data not shown).
For preparation of histological sections, Arabidopsis tissues (green callus containing embryo-like or leaf-like structures) were fixed in 3% (v/v) glutaraldehyde for 24 h at 4°C. Samples were rinsed in 1 × PBS and dehydrated in an ethanol series (30, 50, 70, 90, and 100%) at 4°C. The samples were then infiltrated in an ethanol:historesin solution (glycol methacrylate, GMA-Leica) ratio series (4:1, 4:2, 4:3, 4:4, 3:4, 2:4, 1:4, and finally 100% historesin) and embedded in historesin (15 ml infiltration solution + 1 ml hardener; Leica, Wetzlar, Germany). Five-μm thick slices were obtained with a rotary microtome (HM 355S; Microm, Walldorf, Germany). The sections were floated on aqua pur. at room temperature, picked up in clean glass slides, and air-dried at 42°C. The sections were stained with 0.5% (w/v) ponceau S for 6 min, rinsed in aqua pur., and stained again with 0.5% (w/v) methylene blue for 15 s followed by rapid rinse in aqua pur. The stained sections were examined using an Olympus SZ X12 microscope.
Plant genomic DNA from both P. pinaster and Arabidopsis materials (transgenic line, embryo, and seedlings) was isolated from a 100-mg FM tissue according to a modified Doyle (1990) protocol (Dumolin et al., 1995).
To confirm genetic transformation, putative independent transgenic events (hygromycin-resistant lines) were tested by PCR using specific primers (Table 1) targeting PpWOX2 (PpWOX2-fw and T35S_rev, 592-bp fragment; PpWOX2-rev and CaMV35S, 315-bp fragment) and HPTII (HPTII-fw, HPTII-rev, 412-bp fragment).
For detection of persistent Agrobacterium in the putative transgenic events (false positive amplification), PCR was also performed with NPTII-specific primers (Table 1, 557-bp fragment).
Total RNA from proliferating SEs and cotyledonary SEs of P. pinaster was isolated with RNeasy® Plant Mini Kit (Qiagen, Venlo, The Netherlands) using an RLC lysis buffer containing guanidine hydrochloride according to manufacturer’s instructions. Total RNA from somatic seedlings of P. pinaster (germinated for 6–8 weeks) was isolated according to Chang et al. (1993). Total RNA from Arabidopsis seedlings was extracted using TRIzol (Invitrogen) following manufacturer’s instructions. cDNA was synthesized from 1 μg total RNA using QuantiTect® Reverse Transcription Kit (Qiagen, Venlo, The Netherlands) as described in the manufacturer’s instructions.
Reverse transcription-quantitative (RT-q) PCR reactions were performed with a CFX96™ Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States). Reactions were performed with 1 μl template (10 ng cDNA), 5 μl 2 × SYBR Green Master mix (SensiMix™ SYBR No-ROX Kit; Bioline, Memphis, TN, USA), 0.3 μl forward and reverse primers (10 μmol μl–1), and aqua pur. to a final volume of 10 μl.
Measurements were carried out on three reactions per cDNA sample (technical replicates) and three cDNA samples that were synthesized from independent RNA samples (biological replicates). Negative controls, i.e., either autoclaved aqua pur. or RNA without reverse transcriptase as template, were included in each run. Three reference genes encoding Helicase (TC151784 from P. pinaster), expressed protein At2g32170 (TC125990 from Pinus taeda), and mitosis protein YLS8 (TC136697 from P. pinaster) were used in combination for normalization of the relative expression of PpWOX2 in P. pinaster (Table 1). The expression of these genes was shown to be constant during SE development (Arndt, 2013). The relative expression of PpWOX2 (both transgene and endogenous genes), AtWOX2, AtWUSCHEL (AtWUS), and AtLEC1 in Arabidopsis was normalized to the expression levels of three reference genes from Arabidopsis (SAND: At2g28390, PP2A: At1g13320, and PEX4: At5g25760, Table 1) validated by Czechowski et al. (2005).
Real-time RT-qPCR data were analyzed using Bio-Rad CFX Manager Version 1.6.541.1028 and qbase PLUS. Accurate normalization based on multiple internal control genes was achieved following the method of Vandesompele et al. (2002).
The cDNA sequence (570 bp) of a putative P. pinaster homolog to WOX2 (PpWOX2) was independently identified in SustainpineDB [(Acc. KY773924); in this study: Acc. KU962991, Alvarez et al., 2018].
The deduced PpWOX2 protein sequence (Acc. ARS01278; in this work: Acc. ANC94872, Alvarez et al., 2018) shares conserved characteristic features of the plant WOX family (Alvarez et al., 2018). In addition to the conserved homeodomain, many plant WUS/WOX proteins contain a conserved WUS-box (TLPLFP) located downstream of the homeodomain (Figure 1, Haecker et al., 2004; Nardmann and Werr, 2006), including in Pinus pinaster (Alvarez et al., 2018). Such a putative WUS-box was identified in the PpWOX2 protein sequence and shows the same differences in amino acid sequence compared to Arabidopsis (TLPLFPLQP) as other conifers from genera Pinus, Picea and Larix (TLELFPLHP).
Pinus pinaster WOX2 expression (both transgene and endogenous genes) was investigated during normal development of P. pinaster SE by RT-qPCR (wild-type samples). Significant variation was detected by ANOVA [F(8,18) = 23.479, P < 0.05; Figure 3A]. Relative expression was highest in proliferating immature SEs and then significantly decreased in cotyledonary SEs. A very low level of PpWOX2 expression was detected in 6–8-week–old somatic seedlings although not significantly different from that of cotyledonary SEs.
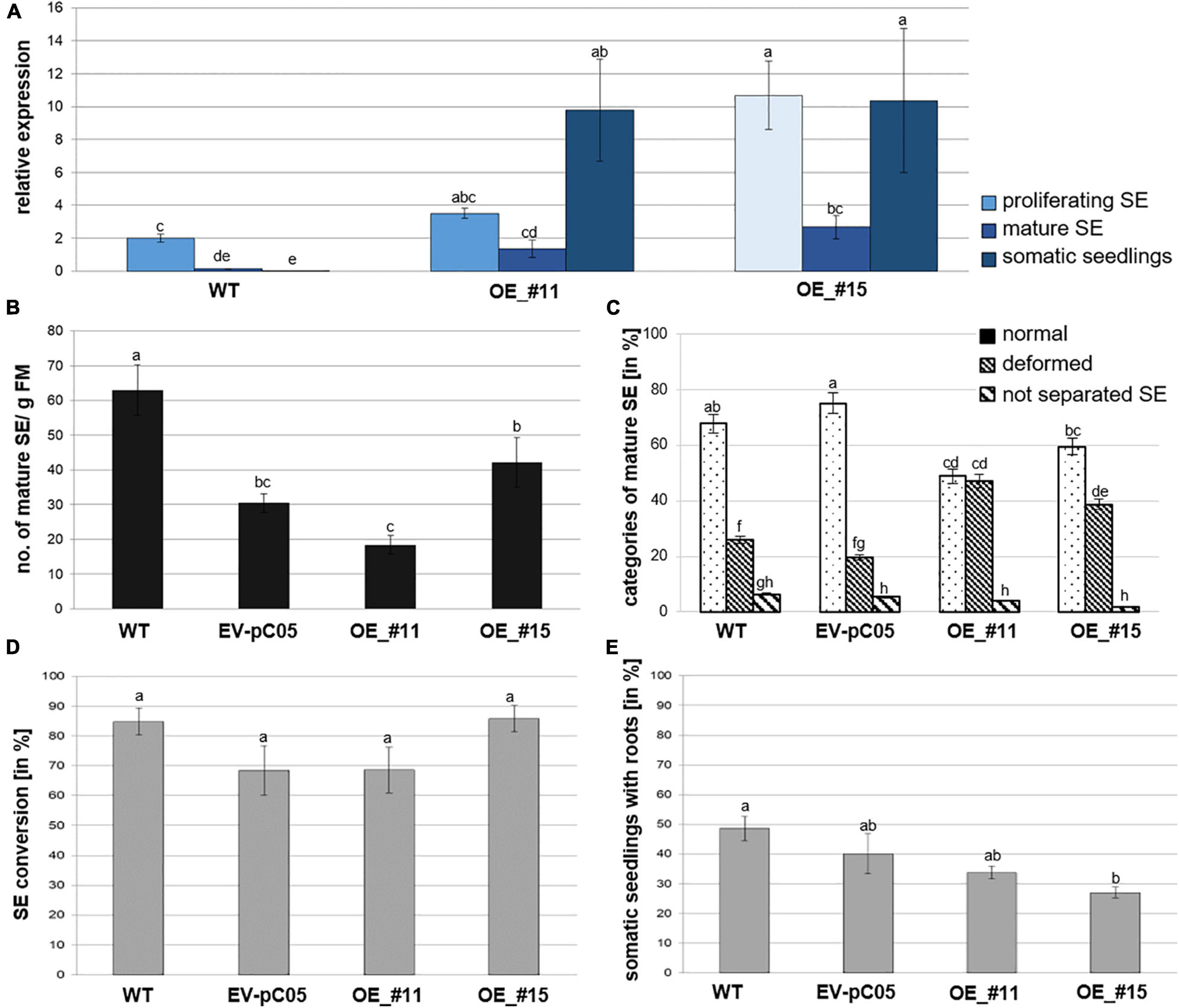
Figure 3. Analysis of 35S::PpWOX2 overexpressing P. pinaster lines #11 and #15 (OE_#11 and OE_#15) and controls (WT, EV-pC05) with regard to PpWOX2 transgene expression following (A) RT-qPCR analysis, (B) SE maturation, (C) cotyledonary SE morphology, and (D) conversion (D) as well as (E) root development. (A) Relative expression of PpWOX2 in proliferating SEs, cotyledonary SEs, and somatic seedlings by RT-qPCR. (B) Number of cotyledonary SEs per g FM after 12–14 weeks in maturation medium. (C) Percentage of cotyledonary SEs (n = 70) with different morphological phenotypes classified as normal, deformed, or not separated (fused) after 12–14 weeks in maturation medium. (D) Percentage of SEs (n = 21) converted into somatic seedlings after 6 weeks in germination medium. (E) Percentage of somatic seedlings (n = 21) with active root growth after 8 weeks in conversion medium. Embryos of control transgenic line EV-pC05 were transformed with pCAMBIA1305.2 (empty vector). WT line was the original PN519 line used for transformation experiment. Each value represents mean ± standard error obtained from three biological replicates and (A) 3, (D,E) 4, or (B,C) 7–10 technical replicates. Bars with different letters are significantly different (ANOVA, P < 0.05).
The transformation efficiency of proliferating SEs with the 35S::PpWOX2 construct or the control empty vector pCAMBIA1305.2 was 11.9 and 26.0 PCR+ lines g–1 FM, respectively. The presence of T-DNA (HPTII and PpWOX2 PCR detection) was confirmed in hygromycin-resistant 35S::PpWOX2 lines in different developmental steps (proliferating SEs, cotyledonary SEs, and somatic seedlings; Figure 4). The NPTII bacterial selectable marker was not detected in these PCR+ lines during SE development (Figure 4), supporting that PCR+ lines are free from contaminating agrobacteria. Ten 35S::PpWOX2 PCR+ lines and five transgenic control lines (EV-pC05) could be cryopreserved.

Figure 4. PCR analyses to confirm the presence and stability of the transgene in 35S::PpWOX2 P. pinaster lines in various developmental steps from proliferating immature SEs to cotyledonary SEs and somatic seedlings. Lines #11 and #15 are PCR-positive (PCR+) throughout SE development. PCR amplification of HPTII (412 bp), PpWOX2 (315 bp), and NPTII (557 bp). Empty vector (EV): embryos of control line EV-pC05 transformed with pCAMBIA1305.2, P = 35S::PpWOX2-isolated plasmid (positive PCR control). WT, wild-type, non-transgenic PN519 control line. H2O, negative PCR control (water as template).
Following RT-qPCR, two independent 35S::PpWOX2 lines (#11 and #15) showed higher PpWOX2 relative expression compared to the wild type (WT) in the three analyzed developmental steps [F(8,23) = 7.27, P < 0.05; Figure 3A]. Observed differences were only significant for line #15 in the proliferating and mature stages but highly significant for both transgenic lines (#11 and #15) in the seedling stage. Similar results were obtained for any of the eight additional 35S::PpWOX2 lines that were generated and cryopreserved (data not shown). Similar to WT, relative expression was decreased in cotyledonary SEs compared to proliferating SEs (although only significant for line #15), suggesting some phase change effect on transition between early and late embryogenesis. Although some differences may exist, in our experiments, PpWOX2 expression was similar between proliferating SEs and somatic seedlings (no significant differences) in both lines #11 and #15.
The effect of high constitutive overexpression of PpWOX2 on SE development in P. pinaster was analyzed by evaluating SE maturation and conversion into plantlets for the 35S::PpWOX2 lines (#11 and #15) and controls (WT and EV-pC05).
PpWOX2 over-expressing lines #11 and #15 produced similar and significantly reduced yields in cotyledonary SEs compared to WT (Figure 3B). However, embryos of the control transgenic line EV-pC05 showed a similar significant trend [F(4,84) = 17.046, P < 0.05; Figure 3B], suggesting some technical issue related to the genetic transformation procedure (see Discussion and Supplementary Table 1). In maritime pine, it has been reported that hygromycin selection could result in reduced yields in cotyledonary SEs (Trontin et al., 2007).
No significant morphological differences were observed between WT and transgenic control line EV-pC05. In contrast, the ANOVA revealed significant variation in normal and deformed embryos among PpWOX2 transgenic and control lines [F(11,213) = 30.16, P < 0.05; Figure 3C]. The transgenic lines showed significantly reduced yields in normal embryos. Accordingly, a significant, higher percentage of proliferating SEs from lines #11 (47%) and #15 (39%) developed into deformed embryos, mostly with asymmetric development of cotyledons when compared to the WT (25%) and control line EV-pC05 (18%).
The overexpression of PpWOX2 had no detectable early effect on conversion of cotyledonary SEs into somatic seedlings for both 35S::PpWOX2 lines #11 and #15 as compared with the WT and EV-pC05 (Figure 3D). High conversion rates were observed for all lines (68–85%). Accordingly, no significant difference in the frequency of somatic seedlings showing active root growth could be detected between 35S::PpWOX2 lines and the EV-pC05 control (Figure 3E). As some reduced frequency is observed for all transgenic lines compared to the WT, especially in the case of line #15 F(4,19) = 5.002, P = 0.006; Figure 3E, (as for SE yield), some adverse effects of the genetic transformation procedure have again been suggested.
Somatic embryo (SE) viability was significantly reduced compared to controls after 8–10 weeks of culture in a conversion medium (Figure 5). Interestingly, line #15, which had the lowest ability for rooting, showed callus formation in the root region, and further root development was prevented in 73% of the somatic seedlings (Figures 3E, 5D,E).
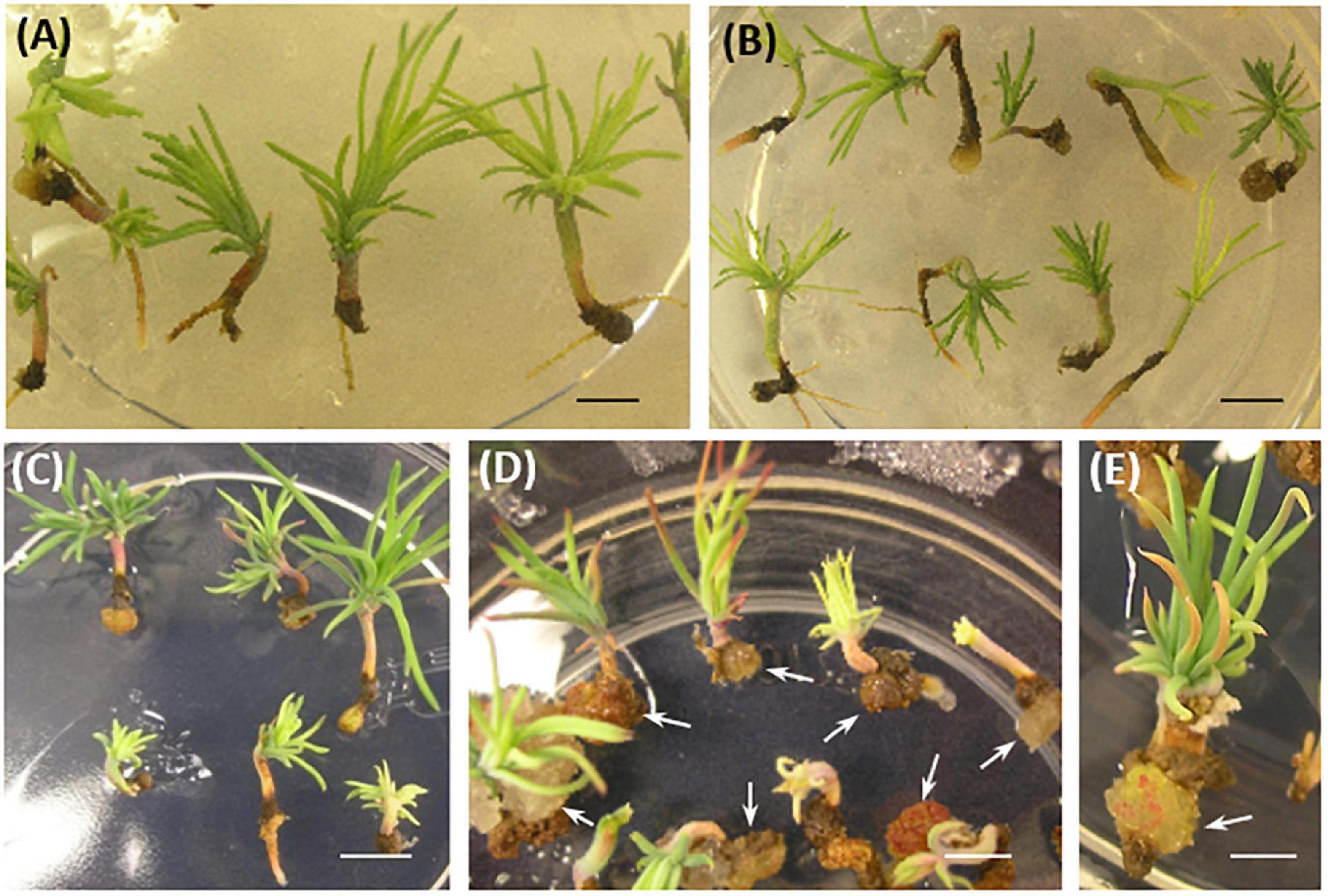
Figure 5. P. pinaster somatic seedlings from 35S::PpWOX2 transgenic lines and controls (EV-pC05, WT) after 8–10 weeks in conversion medium. (A) WT and (B) EV-pC05 somatic seedlings with similar and normal phenotype. The root growth of somatic seedlings from the 35S::PpWOX2 lines was reduced (C,D) and ultimately affected SE viability and behavior. Note the formation of callus in the root region of line #15 [(D,E), arrows] compared to the WT and EV-pC05 controls (A,B). Scale bars: (A) 2 mm, (B–D) 3 mm, and (E) 1 mm.
To further examine the ability of transgenic plants overexpressing PpWOX2 to initiate callus production, primary needle/hypocotyls and root explants from the 35S::PpWOX2, EV-pC05, and WT somatic seedlings were cultured in an mLV medium without PGRs for 8 weeks. No direct somatic embryogenesis initiation could be detected under such experimental conditions. The calli obtained from the different types of explants from all lines were compact and white yellow, and did not differentiate into proliferating SEs (non-embryogenic calli).
The percentage of non-embryogenic callus formation from root explants was significantly higher in line #15 (100%) than in the WT (16%) and EV-pC05 (48%) controls [F(7,16) = 6.407, P = 0; Figure 6]. The observed increase in EV-pC05 compared to the WT explants was also significant and could indicate some habituation phenomenon of transgenic lines (following repeated subculture) to auxin supplemented in the proliferation medium. Nevertheless, this result shows that constitutive overexpression of PpWOX2 in line #15 resulted in increased propensity to callus formation in the root pole, which, in turn, could be detrimental to root growth and subsequent plant viability (Figures 3E, 5). In the case of line #11, non-embryogenic callus formation (45%) was significantly increased compared to WT, but was not when compared to EV-pC05 [F(4,84) = 17.046, P = 0; Figure 6].
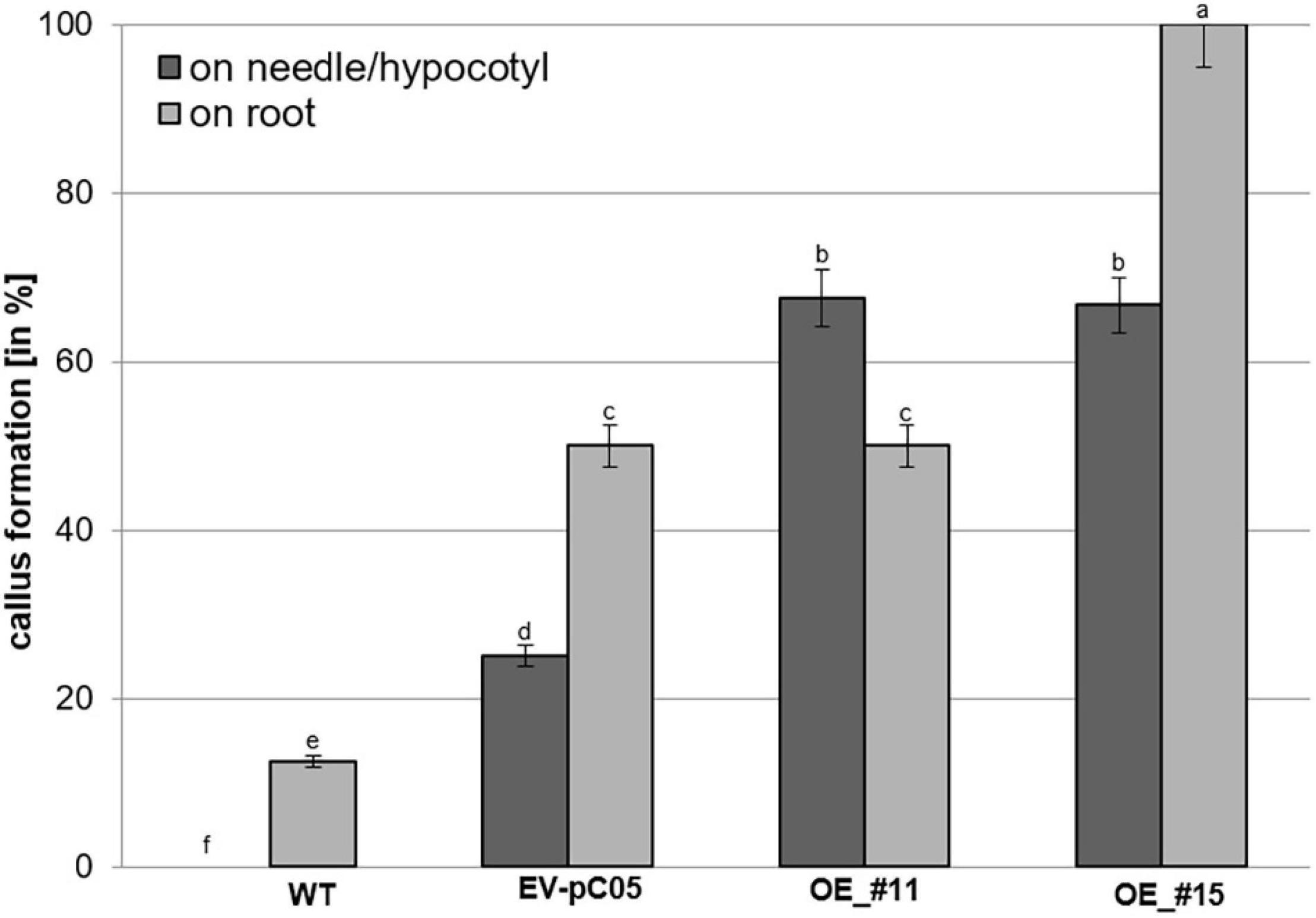
Figure 6. Percentage of non-embryogenic callus formation from P. pinaster primary needle/hypocotyl or root explants (N ≈ 20) of the 35S::PpWOX2 lines #11 and #15, transgenic EV-pC05, and non-transgenic WT controls in mLV medium without PGR (conversion medium). Each value represents mean ± standard error from three biological replicates and five Petri dishes as technical replicates. Bars with different letters indicate significant differences (ANOVA, P < 0.05).
Nearly 65% of the needle/hypocotyl explants of both 35S::PpWOX2 lines #11 and #15 showed callus formation (Figure 6), and this was a significantly higher rate than that of the EV-pC05 (24%) and WT controls (0%). Similar to the root explants, EV-pC05 produced significantly more callus from needle/hypocotyl explants than the WT, suggesting, as mentioned earlier, some impact of the genetic transformation procedure.
Overall, the data showed that somatic seedlings of the 35S::PpWOX2 lines have an increased ability to initiate non-embryogenic callus on needle/hypocotyl only (line #11) or both needle/hypocotyl and root explants (line #15) compared to the non-transgenic WT and transgenic EV-pC05 controls.
The overexpression of PpWOX2 in Arabidopsis had different effects on the phenotype of transgenic (PCR+) seedlings from the T1 and T2 generations.
Considering PpWOX2-expressing plants from the T1 generation, significant variation in the frequency of four classes of phenotypes were detected [F(3,8) = 9.74, P = 0.005, Figure 7A]. Only 16% of the PpWOX2-T1 transgenic seedlings showed the WT normal phenotype (Figures 8A,F), and the remaining 84% of the plants showed alterations such as stunted growth (34%, Figure 8G), embryo-like structures (35%, Figure 8H), and leaf-like structures (14%, Figure 8J). Histological examinations of the embryo-like structures (Figure 8H) revealed the presence of globular SEs with actively dividing cells (Figure 8I). Histochemical observation of the leaf-like structures (Figure 8J) only showed leaf anatomical characteristics without any evidence for embryo formation (Figure 8K).
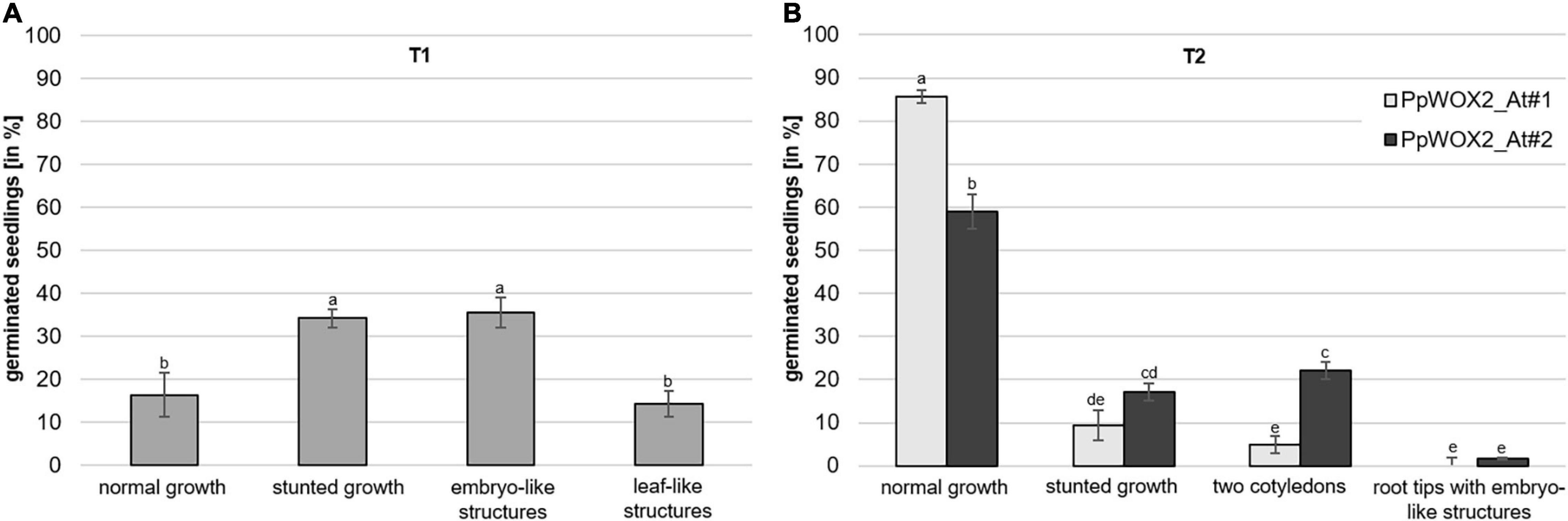
Figure 7. Frequency of different phenotypes among PpWOX2 overexpressing Arabidopsis plants from the T1 or T2 generation. (A) T0 seeds transformed with the 35S::PpWOX2 gene cassette were germinated and resulting T1 plants (N = 70–80) developed different phenotypes (normal or stunted growth, embryo-like and leaf-like structures) in various frequencies. (B) T1 seeds transformed with the 35S::PpWOX2 gene cassette (N = 70–80) were germinated and resulting T2 plants from 2 independent transgenic lines (PpWOX2_At#1 and PpWOX2_At#2) developed either some phenotypes similar to those of T1 plants (normal growth and stunted growth) or new phenotypes not previously detected in the T1 plants (only two cotyledons and plants with root tips that formed embryo-like structures). Error bars indicate standard error for means of three biological replicates and 7–8 technical replicates. Bars with different letters show significant statistical differences (P < 0.05).
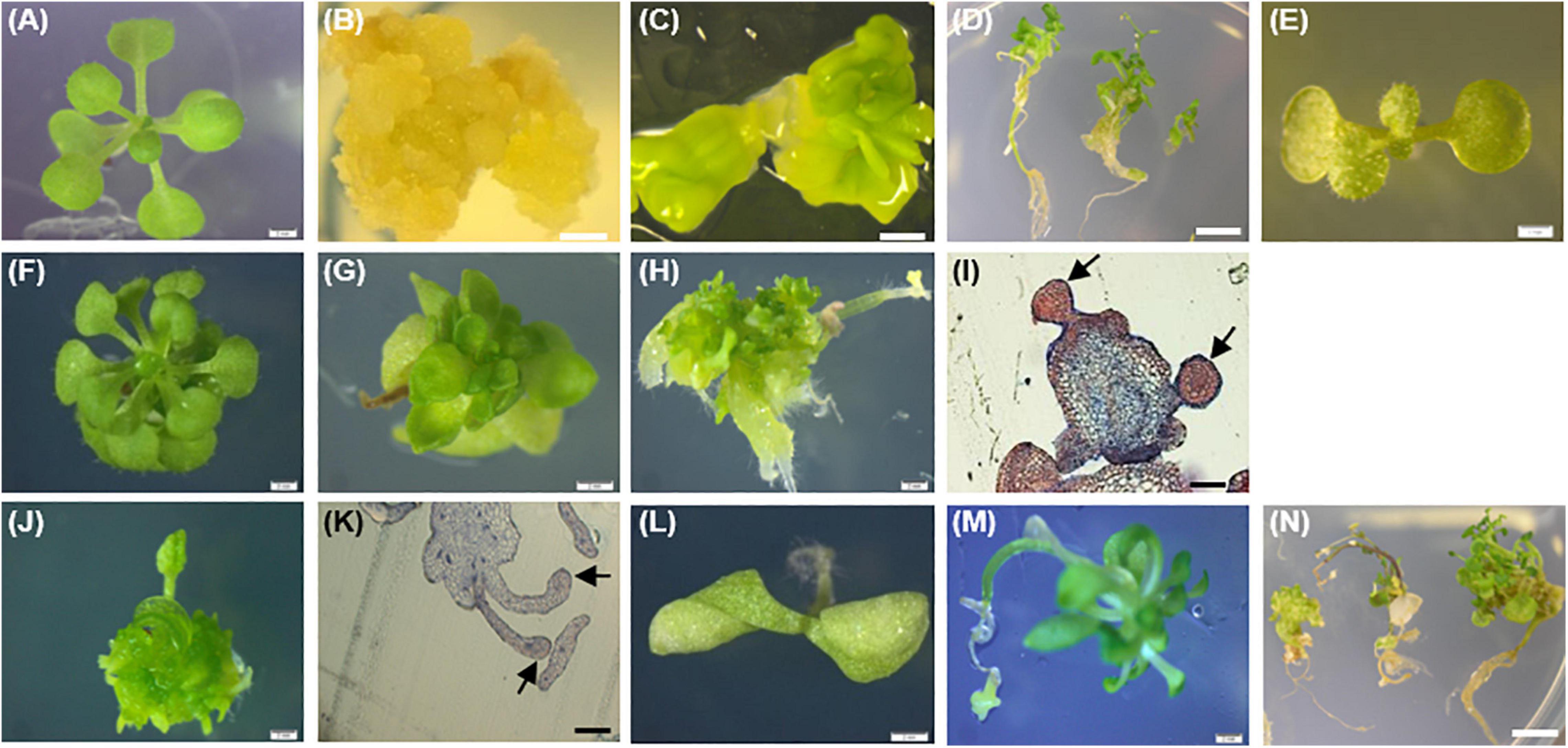
Figure 8. Phenotypes of the 35S::PpWOX2 transgenic plants, transgenic EV-pC05, and non-transgenic WT control plants of Arabidopsis. (A) WT seedling from first (T1) or second (T2) generation grown in MS medium without PGRs for 7 weeks. (B) WT non-embryogenic callus. (C) WT SEs obtained from explants of immature zygotic embryos cultivated in an induction medium (B5 medium supplemented with 5 μM 2,4-D) after 4 weeks. (D) WT SEs germinated and developed into somatic plants in MSR medium after 2 weeks. (E) EV-pC05 transgenic plant (transformed with the pCAMBIA1305.2 empty vector) grown in MS medium supplemented with 20 mg l–1 hygromycin. These control plants exhibited a normal phenotype similar to that of WT seedlings (A) in both T1 and T2 generations. PpWOX2-transformed seeds of the T1 generation germinated in MS medium supplemented with 20 mg l–1 hygromycin and exhibiting (F) normal growth, (G) stunted growth, (H) embryo-like structures, or (J) leaf-like structures. Histological aspects of (I) embryo-like structures and (K) leaf-like structures (arrows) were analyzed after staining with Ponceau-methylene blue. The T2 generation of PpWOX2 transgenic seeds developed into plants with (F) normal growth, (G) stunted growth, (L) two cotyledons, or (M) root tips forming embryo-like structures. (N) Embryo-like structure from the PpWOX2 T1 plant generation converted into somatic plants in MSR medium. Scale bars: (B,C,H,J), and (L) 0.5 mm; (A,E,F,G) 1 mm; (D,N) 1.2 mm; and (I,K) 0.2 mm.
In contrast to the T1 generation, a high percentage of the PpWOX2-expressing T2 plants showed normal (WT) growth with significant differences between the two investigated lines, PpWOX2_At#1 (85%) and PpWOX2_At#2 [60%, F(7,24) = 21.635, P = 0; Figure 7B]. Stunted growth was observed in only 9–15% of the plants without significant difference between the lines (Figure 7B). In addition, two new phenotypes were observed that did not appear previously in the T1 generation. First, a number of T2 seedlings (5–22%) were arrested in the two-cotyledon stage (Figure 7B), and no further leaf formation was observed (Figure 8L). This phenotype was found more frequently in PpWOX2_At#2 (22%) than in PpWOX2_At#1 (5%) lines [F(7,24) = 21.63, P = 0, Figure 7B]. Second, embryo-like structures suggesting direct somatic embryogenesis was observed at root tips of some plants from both PpWOX2 transgenic lines (Figure 8M) but in quite low frequency (≤1.7%, Figure 7B).
The morphology and regeneration ability of embryo-like structures obtained from PpWOX2 T1 plants (Figure 8H) were compared with that of SEs (Figure 8C) and non-embryogenic calluses (Figure 8B) initiated from the WT seedlings. Embryo-like structures and SEs were similar in their germination and regeneration abilities and phenotypes, as illustrated in Figure 8D (WT SEs) and 8N (PpWOX2 plants derived from embryo-like structures), respectively. No plant regeneration could be obtained from non-embryogenic calluses.
The PpWOX2 transgenic plants initiated flower formation earlier (12–15 days after germination) than the WT and EV-pC05 control plants (22–28 days). Control transgenic line EV-pC05 exhibited normal growth (a phenotype similar to that of WT-plants) in both the T1 and T2 generations (Figure 8E).
In order to investigate whether the constitutive overexpression of PpWOX2 in Arabidopsis affects the expression of selected embryogenesis-related endogenous genes, the expression of AtWOX2, AtWUS, and AtLEC1 was estimated by RT-qPCR in 35S::PpWOX2 T1 and T2 transgenic lines exhibiting different phenotypes (Figure 9). Controls were the empty vector transgenic line (EV-pC05_At) and various non-transgenic lines with unknown (WT seedlings), demonstrated (WT SE), or no (non-embryogenic callus) embryogenic ability. All analyzed tissues (excluding non-embryogenic callus) were sampled from plants of the same age and developmental stage.
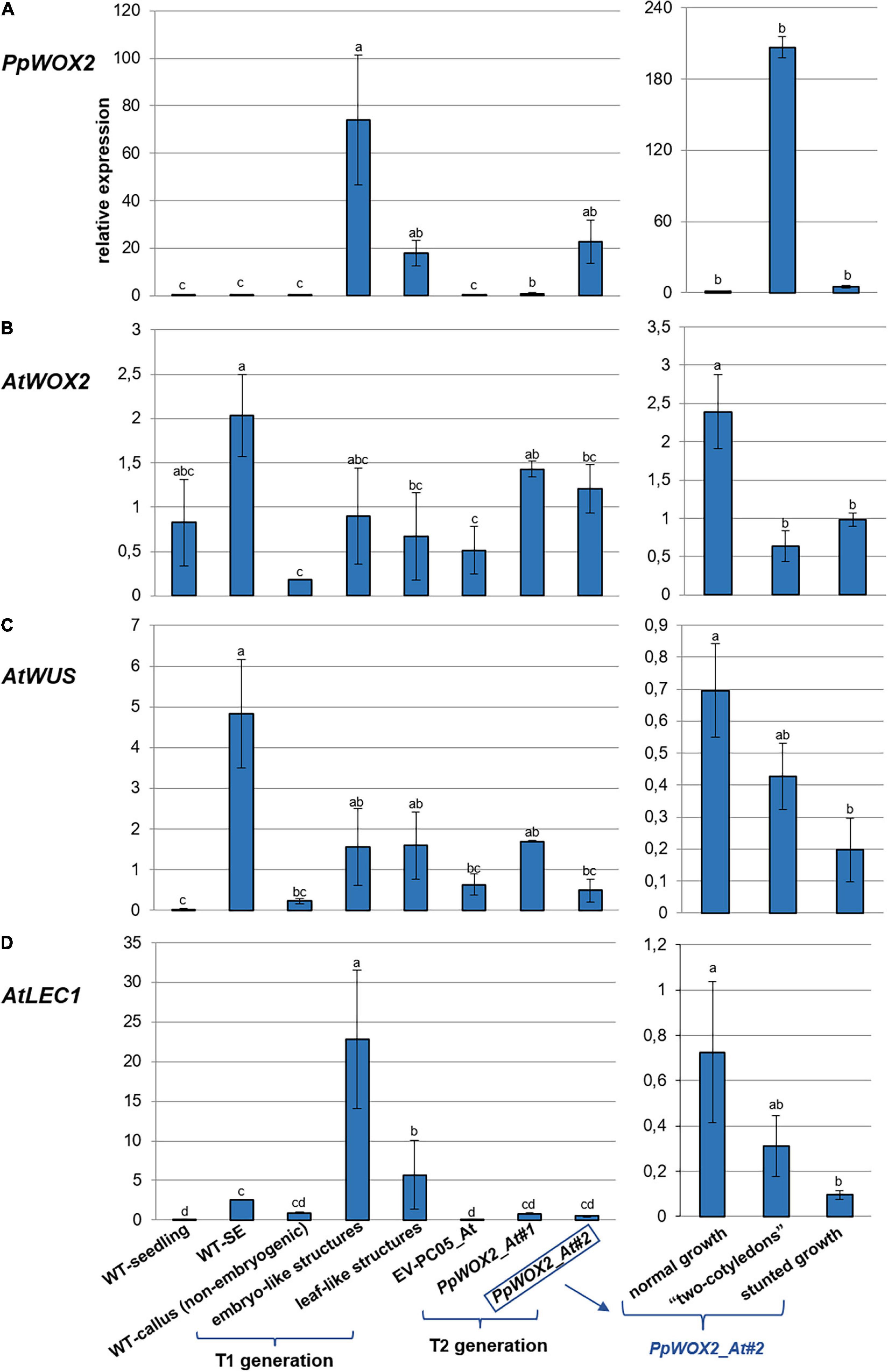
Figure 9. Relative expression of (A) PpWOX2, (B) AtWOX2, (C) AtWUS, and (D) AtLEC1 in Arabidopsis plants obtained from PpWOX2 overexpressing lines (T1 and T2 generations), transgenic EV-pC05_At, and non-transgenic WT controls (seedlings and somatic embryos/SE) at the same age and developmental stage by RT-qPCR. Each value represents mean ± standard error of three biological replicates and three technical replicates. Bars with different letters indicate significant differences (ANOVA, P < 0.05). The different phenotypes are illustrated in Figure 8. Samples PpWOX2_At#1 and PpWOX2_At#2 from the T2 generation are a mixture of plants with the “normal,” “two-cotyledon,” and “stunted growth” phenotypes. Gene expression is presented separately for these three phenotypes for line PpWOX2_At#2 (right part of the figure).
The relative expression of PpWOX2 was confirmed to be significantly higher in transgenic Arabidopsis plants of both T1 and T2 generations than in transgenic (EV-pC05_At) and non-transgenic (WT) controls [F(7,16) = 3.882, P = 0.012; Figure 9A]. Among the different phenotypes observed for the T1 and T2 plants, expression was similar and lowest for plants showing normal and stunted growth as illustrated for the PpWOX2_At#2 line (Figure 9A, right graph). PpWOX2 expression was much higher in T1 plants showing embryo- or leaf-like structures than in transgenic plants with normal growth (10- to 35-fold ratio, data not shown). Similarly, PpWOX2 expression was significantly higher in T2 plants with the “two-cotyledon” phenotype for both lines PpWOX2_At#1 (data not shown) and PpWOX2_At#2 (128-fold ratio, Figure 9A, right graph). The reduced behavior of T2 transgenic plants with root tips showing an embryo-like structure did not allow for gene expression analysis.
In contrast to PpWOX2, the expression of AtWOX2 (Figure 9B) was mostly similar in the T1 plants and controls (Figure 9B). Only plants with the “leaf-like structure” phenotype showed significantly reduced AtWOX2 expression compared to the non-transgenic SE control. Considering the T2 plants, some significant differences were observed for the PpWOX2_At#1 line, which showed higher AtWOX2 expression than the EV-pC05_At and non-embryogenic callus controls. However, the expression was similar to that of control non-transgenic seedlings and SE (WT). Similarly, no significant difference compared to controls could be detected in the case of line PpWOX2_At#2 when all plant phenotypes were considered together as a mixture. Analyzing separately the different plant phenotypes for this line revealed that AtWOX2 expression is similarly and significantly reduced in plants with two cotyledons or stunted growth compared to plants with normal growth (Figure 9B, right graph). We concluded that PpWOX2 overexpression had apparently little effect on the expression of endogenous WOX2. The PpWOX2_At#2 data for plants with normal, stunted, or two-cotyledon phenotypes, however, suggest some inverse relationship between PpWOX2 (Figure 9A) and AtWOX2 expressions (Figure 9B).
In the case of AtWUS expression (Figure 9C), some significant differences with non-transgenic control seedlings were detected for T1 plants and T2 plants from the PpWOX2_At#1 line (higher expression than in WT seedlings). AtWUS expression in T2 plants from the PpWOX2_At#2 line was also significantly lower than in non-transgenic control SEs. However, AtWUS expression was found to be similar in both T1 and T2 PpWOX2 transgenic plants and EV-pC05_At transgenic control, suggesting that PpWOX2 overexpression had no drastic effect on the expression of this gene. AtWUS expression was found to be higher in T2 plants from line PpWOX2_At#2 exhibiting normal growth (significant) and “2 cotyledon” phenotype (non-significant) compared to plants with “stunted” phenotype (Figure 9C, right graph). However, there is no clear correlation with PpWOX2 transgene expression.
Expression of AtLEC1 is significantly higher in T1 plants with embryo-like and leaf-like structures than in all the controls (Figure 9D). In contrast, no significant difference could be detected in the case of T2 plants. However, considering line PpWOX2_At#2, AtLEC1 expression (Figure 9D, right graph) showed a pattern similar to that for AtWUS (Figure 9C, right graph), i.e., higher expression in plants with normal growth (significant) and “two-cotyledon” phenotype (non-significant) compared to plants with stunted growth.
In brief, high PpWOX2 expression in T1 transgenic plants with embryo-like or leaf-like structures is correlated with increased expression of AtLEC1 but not AtWOX2 or AtWUS. High PpWOX2 expression in T2 transgenic plants with the “2-cotyledon” phenotype is correlated with decreased expression of AtWOX2.
In angiosperms, the transcription factor WOX2 is necessary for cell fate and delineation of the apical embryo domain during embryogenesis (Haecker et al., 2004; Xiao et al., 2006). Zhang et al. (2017) demonstrated that WOX2 contributes to the initiation of shoot meristem stem cells in the embryo of Arabidopsis. Several reverse genetic studies have been reported on knockout and overexpression of WOX2 in angiosperms, mainly in Arabidopsis (Haecker et al., 2004; Jeong et al., 2011; Elhiti et al., 2013; Chung et al., 2016; Zhang et al., 2017). For example, loss of AtWOX2 function in zygotic embryos of Arabidopsis has only relatively mild consequences and results in aberrant divisions at the apex of the globular pro-embryo and, occasionally, seedlings with a single cotyledon (Haecker et al., 2004; Jeong et al., 2011). In contrast, overexpression of AtWOX2 in Arabidopsis causes severe growth defects and further morphological phenotypes by impairing plant organ formation (Chung et al., 2016).
In conifers, much attention has been paid to identify genes that are important during embryogenesis [Park et al., 2010; Rupps et al., 2016; reviewed in Cairney and Pullman (2007), Miguel et al. (2016), Trontin et al. (2016a)] but a considerable number of studies are still needed to unravel the precise spatiotemporal function of each gene. Zhu et al. (2016) reported that downregulation of WOX2 of Picea abies (PaWOX2) during early embryogenesis resulted in significant decrease in the yield of mature embryos. In contrast, downregulation of PaWOX2 after late embryo formation had no effect on further embryo development and maturation. In this study, we focused on the functional study on a WOX2 gene isolated from P. pinaster (PpWOX2) by constitutive overexpression of PpWOX2 in both P. pinaster SEs and Arabidopsis seedlings.
The deduced WOX2 protein shares the WUS-type homeodomain and the conserved WUS-box of the WOX family proteins (Figure 1). Based on phylogenetic analyses, the 15 members of the WOX family are divided into three clades: the ancient clade, the intermediate clade, and the WUS clade (modern clade) (van der Graaff et al., 2009; Hedman et al., 2013; Lian et al., 2014; Alvarez et al., 2018). The modern clade contains WUS and WOX1-7 members that are found in seed plants, confirming an evolutionary relationship among WOX genes of this group (Lian et al., 2014; Alvarez et al., 2018). PpWOX2 shares high identity to its homolog in Pinus sylvestris (97%) and Pinus contorta (95%), low global identity to the WOX2 of Picea abies (76%) and Larix decidua (63%), and very low identity to the WOX2 of Arabidopsis (35%) (Hedman et al., 2013; Zhu et al., 2016).
The expression of endogenous PpWOX2 in P. pinaster is high in the SE proliferation step but significantly decreases after SE maturation. No PpWOX2 expression could be detected later in somatic seedlings. These data confirmed (SE development) and extend (somatic plant development) previous results obtained by Alvarez et al. (2018) from a different P. pinaster embryogenic line and following a different protocol for somatic embryogenesis. A similar WOX2 expression pattern has been found in other conifers such as Larix decidua (Rupps et al., 2016), Picea abies (Palovaara et al., 2010a; Hedman et al., 2013), and Picea glauca (Klimaszewska et al., 2011). These differences in WOX2 expression during SE development may be explained by an internal control of PpWOX2 by other genes that are differentially expressed during development, such as polar auxin transport-related genes (PIN genes) (Palovaara and Hakman, 2009). Considering these expression data, it would be interesting to confirm that PpWOX2 can be used in P. pinaster to distinguish in the proliferation stage embryogenic from non-embryogenic tissues. In conifers, the expression of WOX2 is indeed known as a putative marker for effective initiation of somatic embryogenesis and to predict the embryogenic potential (embryogenecity) of a culture (Palovaara and Hakman, 2008; Park et al., 2010; Klimaszewska et al., 2011; Miguel et al., 2016).
Over-expression of PpWOX2 in proliferating SEs of P. pinaster had, apparently, a negative effect on maturation (Figure 3B). Both lines, OE_#11 and OE_#15, were found to have a significant lower maturation ability than the non-transgenic WT control. These results are consistent with those reported by Klimaszewska et al. (2010). In this report, it was shown that induced overexpression of AtWUS in SEs of Picea glauca had a striking effect on SE maturation by disrupting embryo development. According to this result, it may be postulated that WOX genes of (at least) the modern clade influence embryo development in conifers. However, embryos of the empty vector control line (EV-pC05) also showed significant decrease in maturation yield in our experiments compared to non-transgenic WT embryos, suggesting that reduced yield could also result from transgene positional effects and/or technical issues such as differences in physiological aging between transgenic lines and WT controls (discussed in Supplementary Table 1). In maritime pine, physiological aging of embryogenic lines results in reduced maturation yield (Breton et al., 2006; Lelu-Walter et al., 2016; Trontin et al., 2016b). Aging effects could, therefore, result from unperceived differences in management of transgenic and WTs during the long process of genetic transformation. The development of a simplified, rapid, and improved genetic transformation protocol of embryogenic tissues would be profitable to reduce possible protocol-related effects on maturation yield. Furthermore, the choice of a selective agent may influence maturation results. Hygromycin is known to affect negatively maturation yield in P. pinaster when used at 20 mg l–1 (Trontin et al., 2007). Although hygromycin concentration was reduced to 10–15 mg l–1 in our experiments, hygromycin could have affected the regeneration of transgenic embryos of the PpWOX2 and control EV-pC05 lines.
The overexpression of PpWOX2 in P. pinaster negatively affected SE development with i) significantly increased frequency of deformed SE, especially asymmetrical cotyledon development (lines OE_#11 and OE_#15, Figure 3C) and ii) non-embryogenic callus formation in the root pole of germinating somatic seedlings, with subsequent inhibition of root growth (especially in the highly overexpressing line OE_#15, Figures 3E, 5D,E). Similarly, Klimaszewska et al. (2010) reported the inhibition of root growth in somatic seedlings of Picea glauca after induced overexpression of AtWUS during SE conversion. Following downregulation of WOX2 during early embryogenesis in Picea abies, a unique function of WOX2 was suggested in conifers during protoderm development and suspensor expansion, which are important steps for proper early embryo development (Zhu et al., 2016).
To further investigate the propensity of young P. pinaster somatic seedlings transformed with PpWOX2 by callus formation, needle/hypocotyl and root explants from two transgenic lines were cultured in the mLV medium (Litvay et al., 1985) without PGRs. No formation of embryogenic tissue could be detected from both transgenic lines and controls, suggesting that PpWOX2 constitutive overexpression had no major and direct effect that could stimulate embryogenic potential in young somatic seedlings without PGRs. mLV is the currently preferred basal medium for P. pinaster to initiate somatic embryogenesis from immature zygotic embryos (with or without PGRs, Trontin et al., 2016b) and secondary embryogenesis from cotyledonary SEs and somatic seedlings (with PGRs, Klimaszewska et al., 2009). Furthermore, mLV is useful to induce non-embryogenic callus formation from young to more mature materials with or without PGRs (Trontin et al., 2016b,c). Therefore, we initially considered that the overexpression of PpWOX2 could be sufficient to stimulate the formation of embryogenic tissues from somatic seedling explants. However, as in most conifer species (see Bonga et al., 2010), initiation of somatic embryogenesis in P. pinaster materials older than immature zygotic embryos (Trontin et al., 2016c) or cotyledonary or young germinating SEs (Klimaszewska et al., 2009) is still difficult to achieve and may require the additional use of PGRs such as auxin and/or cytokinins. Recently, it was shown that abscisic acid (ABA) supplementation can also lead to initiation of somatic embryogenesis in the Douglas fir (Pseudotsuga menziesii) (Walther et al., 2021) and has to be taken into consideration as a signal for SE activation. Induction of secondary somatic embryogenesis from germinated SEs could be obtained but at low rate with a combination of 2,4-D (9.5 μM) and BA (4.5 μM) in P. pinaster (Klimaszewska et al., 2009). In Capsicum annuum, ectopic BABY BOOM expression is not sufficient to induce embryogenesis, and exogenous cytokinin is required for SE formation (Heidmann et al., 2011). However, ectopic expression of BABY BOOM in Arabidopsis and Brassica led to spontaneous formation of SEs and cotyledon-like structures in seedlings without the use of exogenous PGRs (Boutilier et al., 2002). The enhanced embryogenic potential of young somatic seedlings in P. pinaster may require a combination of ectopic expression of embryogenesis-related gene(s) and adequate PGR treatment and/or other environmental conditions.
Instead of embryogenic callus, non-embryogenic callus formation was determined with high frequency in needle/hypocotyl (significant in both lines #11 and #15) and/or root explants (significant in line #15) of PpWOX2 somatic seedlings (Figure 6) cultivated in mLV deprived of PGRs. Non-embryogenic callus formation was similarly significantly enhanced in the transgenic control EV-pC05 line compared to the WT, suggesting some technical issues related with genetic transformation. However, this increase was lower than that observed for the 35S::PpWOX2 lines except in the case of line #11 for the root explants. These results may be explained by alteration in levels of endogenous hormones, particularly auxin, in response to PpWOX2 overexpression in somatic seedlings. Recent studies showed that at the time of stem cell initiation, WOX2 affects the auxin pathway by increase in expression of the auxin transporter PIN1 gene (Zhang et al., 2017). In conifers, Palovaara and Hakman (2009) suggested that polar auxin transport is involved in regulation of the expression of both the auxin efflux carrier (encoded by PIN1) and WOX2. N-1-naphthylphthalamic acid (NPA) treatment of embryos before cotyledon initiation, indeed, disrupted the endogenous auxin pattern and expression of both PIN1-like and WOX2 (Hakman et al., 2009). Interestingly, the expression of PIN homologs was associated with the auxin immunolocalization pattern during cotyledon formation in Picea abies (Palovaara et al., 2010b). It was suggested that correct auxin transport is crucial during transition from early to pre-cotyledonary embryos and that it is involved in the coordinated regulation of WOX2 and PIN1 (Trontin et al., 2016a), which, in turn, could affect both cotyledonary embryo development (especially cotyledon formation) and normal root formation. Further analysis is needed to consider the interplay of PpWOX2 and auxin in P. pinaster and other conifers.
Furthermore, the heterologous overexpression of PpWOX2 in Arabidopsis resulted in transgenic seedlings (T1 and T2 generations) with different phenotypes such as normal growth, stunted growth, and embryo-like and leaf-like structures. One-third (35%) of the first generation of PpWOX2-At plants showed embryo-like structures. In addition, a similarly high number of T1 plants exhibited stunted growth (35%) and leaf-like structures (15%). PpWOX2 expression was comparatively low in T1 plants exhibiting a normal growth phenotype (similar data to “normal growth”-type of T2 plants, Figure 9A, right graph). In different phenotypes of T2 plants, reduced PpWOX2 expression in the normal phenotype compared to stunted- and even more (significant) two-cotyledon phenotypes (Figure 9A) apparently resulted in increase in frequency of the normal phenotype in T2 plants compared to the other phenotypes (Figure 7B), while a positive link between the expression of PpWOX2 and the frequency of abnormal phenotypes was observed in the T2 plants. Similarly, the overexpression of PpWOX2 in immature SEs of P. pinaster (Figure 3A) is correlated to decrease in the frequency of normal, well-developed, cotyledonary embryos compared to the WT (Figure 3B), although not significant in our experiments when compared to the EV-PC05 transgenic control. Overall, these results suggest that the high, constitutive expression of PpWOX2 negatively affects the frequency of the normal phenotype in both Arabidopsis and P. pinaster.
PpWOX2 expression is higher in the transgenic T1 and T2 plants than in the WT control material (seedlings, SEs, non-embryogenic callus) and transgenic control EV-pC05 (Figure 9A and Supplementary Table 1). However, the different phenotypes observed in transgenic T1 and T2 plants (Figures 7, 8) are only weakly supported by gene expression data (e.g., lower PpWOX2 expression in the T2 plants with normal and stunted growth than in the “two-cotyledon” plants, Figure 9). Some post-transformation silencing effects related to 35S promoter inactivation (a well-known effect during transgenesis) may be partially responsible for such discrepancy. It can be suspected that transgene inactivation (and associated phenotype reversion) could be high in the T2 generation as the frequency of plants with normal phenotype is significantly higher (60–85%) than in T1 (15–20% only). Different PpWOX2 expression levels revealed by RT-qPCR may, therefore, not reflect the actual levels of PpWOX2 protein production. Our data do not allow for a study on whether transcriptional or post-transcriptional silencing mechanisms could be involved. However, considering the high levels of PpWOX2 expression observed in some plants with abnormal phenotypes, such as the two-cotyledon T2 plants (At#2), and embryo-like and leaf-like T1 structures (Figure 9A), it could largely be an effect of post-transcriptional or even translational regulation of PpWOX2 expression. There is generally low support in our experiments for an effect of PpWOX2 overexpression (Figure 9A) on the germination rate of T1 and T2 plants (Figure 7). Observed data may be more related with various degrees of silencing, as only T2 plants with normal phenotype germinated with excellent rates (similar to those of WT seedlings).
Moreover, in Arabidopsis, the embryo-like and leaf-like phenotypes in T1 plants, persistence of the stunted growth phenotype in T1 (35%) and T2 plants (up to 15%), and production of T2 plants with two cotyledons (5–20%) or formation of embryo-like structures on the root tip (1.7%) directed us to analyze the endogenous expression of several major embryogenesis-related genes (AtWOX2, AtLEC1, and AtWUS) to investigate a putative association with observed phenotypes. As hundreds of genes are probably involved in Arabidopsis embryogenesis, an extension of this expression study and additional transcriptomic and/or proteomic approaches are required to gain further insight into intrinsic processes (Tzafrir et al., 2004; De Smet et al., 2010; Elhiti et al., 2013).
PpWOX2 constitutive over-expression in Arabidopsis had a globally low effect on endogenous AtWOX2 and AtWUS expressions, which are roughly similar in transgenic lines and controls from the T1 and T2 generations.
The expression of AtLEC1 was much more affected, especially in the T1 generation (significant overexpression). Indeed, transgenic PpWOX2 plants from the T1 generation with embryo-like and leaf-like structures showed very high expression of AtLEC1 compared to the WT non-transgenic (seedlings, SEs, and non-embryogenic callus) and transgenic (EV-pC05_At) controls. These strong results suggest that overexpression of PpWOX2 in Arabidopsis may stimulate embryogenesis and organogenesis through coordinated overexpression of AtLEC1. Consistent with this outcome, previous reports support that the over-expression of AtLEC1 in Arabidopsis induced somatic embryogenesis in vegetative cells (Lotan et al., 1998).
In addition, the expression of PpWOX2 in transgenic plants with embryo-like and leaf-like structures was much higher (10 − 35-fold) than in transgenic plants with normal growth. Therefore, a plausible explanation is that ectopic expression of PpWOX2 in Arabidopsis might act redundant with AtWOX2 in inducing embryogenesis and/or organogenesis. Another possible explanation is that the overexpression of PpWOX2 in Arabidopsis might result in the over-expression of AtWUS, which activates the expression of LEC1 and promotes somatic embryogenesis. It was reported that AtWUS promoted somatic embryogenesis and activated the expression of LEC1, LEC2, and FUS3 in cotton (Zheng et al., 2014). The authors suggested that AtWUS may alter PIN expression, which could lead to establishment of new auxin gradients. Subsequently, a new auxin response was formed and stimulated the expression of LEC1, LEC2, and FUS3. In addition, the overexpression of GgWUS from Gnetum gnemon (Coniferopsida) in Arabidopsis was observed to induce somatic embryogenesis and organogenesis (Nardmann et al., 2009). In fact, WUS is involved in regulation of both meristematic stem cells (pluripotent) and embryogenic stem cells (totipotent) (Elhiti et al., 2013). Moreover, WUS is known as an embryogenesis marker in embryonic cells, and its overexpression in Arabidopsis resulted in somatic embryogenesis and repeated formation of adventitious shoots in the absence of auxin (Zuo et al., 2002; Elhiti, 2010). We did not obtain clear results of AtWUS expression during our experiments. The data showed some high expression of AtWUS compared to the non-transgenic WT seedling control, but the differences were not significant compared with the other non-transgenic (WT-SE, WT-callus) and transgenic (EV-pC05_At) controls. The heterologous expression of PpWOX2 in Arabidopsis (which only has 35% similarity with AtWOX2, Figure 1) cannot apparently direct increased expression of either AtWUS (Figure 9C) or AtWOX2 (Figure 9B), which, in turn, could affect the expression of AtWUS and/or AtLEC1. Specific experiments are, therefore, needed to confirm this hypothesis.
Considering the emergence of various phenotypes in the T2 generation following PpWOX2 overexpression, our data showed that the normal/abnormal plant phenotype is associated with altered AtWOX2, AtWUS, and AtLEC1 expression (PpWOX2_At#2). It is suggested that variable levels of ectopic overexpression of PpWOX2 could affect the expression of these three embryogenesis-related genes.
One possible further explanation on how overexpression of PpWOX2 in Arabidopsis can stimulate somatic embryogenesis and organogenesis is the high similarity of the WUS family-specific homeodomain and the WUS-box from PpWOX2 and AtWOX2 proteins. In plants, the WOX homeobox is assigned to a subfamily of homeobox transcription factors that are involved in plant embryonic patterning. However, the function of the WOX homeobox has not yet been analyzed in plants. In animals, it is confirmed that a homeobox gene family (HOX), which is expressed in specific embryo domains, has a major regulatory role during early pattern formation, similar to that of the WOX homeobox family (Haecker et al., 2004). In this study, the complete protein sequences of PpWOX2, AtWOX2, and AtWUS show low similarity (35%), but they share high similarity (78%) in the WUS-type homeodomain and WUS-box. Wu et al. (2007) suggested that functional redundancy in the WOX family was not solely determined by overall protein sequence similarity but by high similarities of the homeodomains.
Thus, it can be concluded that high similarities of the WUS-type homeodomain and WUS-box among PpWOX2, AtWOX2, and AtWUS might be sufficient for functional redundancy in these WOX proteins. This speculation is supported by several lines of evidence:
First, the homology of WOX homeodomains among the WOX genes supports three major clades: the ancient, intermediate, and modern clades. According to phylogenetic analysis, the WOX1-7 and WUS genes are grouped in the modern clade. Some subgroups from the modern clade have been lost in several species (e.g., lack of WOX1/6 subgroup in rice). These studies imply that WOX members in the modern clade may have a conserved and redundant function (Hedman et al., 2013; Lian et al., 2014; Alvarez et al., 2018).
Second, in angiosperms, the expression of WUS and WOX5 is specific to the shoot and root regions, respectively. Furthermore, Sarkar et al. (2007) demonstrated that WOX5 and WUS are interchangeable for stem cell control in Arabidopsis. In contrast, Nardmann et al. (2009) reported that Ginkgo biloba WUS (GbWUS) and Pinus sylvestris WUS (PsWUS) are expressed in both the shoots and the roots, and suggested that the WUS and WOX5 genes are the result of an angiosperm-specific gene duplication.
Our results suggest that constitutive overexpression of PpWOX2 in Arabidopsis seedlings alters the expression of embryogenesis-related genes (AtLEC1 and/or AtWUS), which, in turn, could promote the formation of SEs and organs. Further reverse genetics studies through inducible overexpression of combinations of embryogenesis-related genes and possibly an additional (hormonal) stimulus might be helpful to overcome recalcitrance to somatic embryogenesis of already differentiated tissues in Pinus pinaster. These findings might be helpful to gain insights into conifer embryogenesis and, in best case, to develop strategies for induction of somatic embryogenesis from adult conifer trees (Lelu-Walter et al., 2016; Trontin et al., 2016c).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, KY773924.1.
AR, KZ, JR, and SH conceived and designed the research project with expert guidance from J-FT for the maritime pine work. J-FT provided PN519 as starting material for maritime pine. J-FT and AR provided the plasmids and methods for the plant transformation experiments. SH and AR conducted the experiments, analysed the data and wrote the manuscript. KZ and AR supervised the lab work. All authors reviewed and approved the final version of the manuscript.
This study was supported by a Yousef Jameel Scholarship Fund. This study also received specific financial support from Bundes Ministerium für Bildung und Forschung (Germany, BMBF-FKZ-AZ0315706B) and French National Research Agency (ANR-09-KBBE-0007) through a transnational KBBE project (SUSTAINPINE 2010-2013, PLE2009-0016, Coord. F. Cánovas/University of Málaga) as well as a MULTIFOREVER transnational project (since 2019, Coord. J-FT/FCBA, Pierroton, and/or AR/HUB, Berlin) supported within the framework of the European Research Area Network Cofund “ForestValue – Innovating forest-based bioeconomy” (773324) jointly funded by national organizations (in France: ANR-19-SUM2-0002-01); in Germany: Fachagentur Nachwachsende Rohstoffe e.V. (FKZ 22040218). Handling of the PN519 embryogenic line at FCBA (cryopreservation, reactivation, and multiplication) benefited from the technical support of the XYLOBIOTECH facility (XYLOFOREST platform, ANR-10-EQPX-16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Philippe Label (INRAE, France) for the identification of the PpWOX2 sequence in Pinus pinaster databases.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.838421/full#supplementary-material
Alvarez, J. M., Bueno, N., Cañas, R. A., Avila, C., Cánovas, F. M., and Ordás, R. J. (2018). Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: new insights into the gene family evolution. Plant Physiol. Biochem. 123, 304–318. doi: 10.1016/j.plaphy.2017.12.031
Arndt, N. (2013). Establishing of New Reference Genes and Quantitative PCR Analysis During Somatic Embryogenesis of P. pinaster Ait. M.Sc. thesis. Berlin: Humboldt-Universität zu Berlin.
Arroyo-Herrera, A., Gonzalez, A. K., Moo, R. C., Quiroz-Figueroa, F. R., Loyola-Vargas, V. M., Rodriguez-Zapata, L. C., et al. (2008). Expression of WUSCHEL in Coffeacanephoracauses ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Cult. 94, 171–180. doi: 10.1007/s11240-008-9401-1
Bercetche, J., and Pâques, M. (1995). “Somatic embryogenesis in maritime pine (Pinus pinaster),” in Protocol for Somatic Embryogenesis in Woody Plants, eds S. Jain, P. Gupta, and R. Newton (Dordrecht: Springer), 221–242.
Birol, I., Raymond, A., Jackman, S. D., Pleasance, S., Coope, R., Taylor, G. A., et al. (2013). Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29, 1492–1497. doi: 10.1093/bioinformatics/btt178
Bonga, J. M., Klimaszewska, K. K., and Aderkas, P. (2010). Recalcitrance in clonal propagation, in particular of conifers. Plant Cell Tissue Organ Cult. 100, 241–254. doi: 10.1007/s11240-009-9647-2
Bouchabke-Coussa, O., Obellianne, M., Linderme, D., Montes, E., Maia-Grondard, A., Vilaine, F., et al. (2013). WUSCHEL over-expression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 32, 675–686. doi: 10.1007/s00299-013-1402-9
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Bowe, L. M., and Coat, G. (2000). Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales’ closet relatives are conifers. Proc. Natl. Acad. Sci. U.S.A. 97, 4092–4097. doi: 10.1073/pnas.97.8.4092
Breton, D., Harvengt, L., Trontin, J.-F., Bouvet, A., and Favre, J.-M. (2006). Long-term sub-culture randomly affects morphology and subsequent maturation of early somatic embryos in maritime pine. Plant Cell Tissue Organ Cult. 87, 95–108. doi: 10.1007/s11240-006-9144-9
Breuninger, H., Rikirsch, E., Hermann, M., Ueda, M., and Laux, T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876. doi: 10.1016/j.devcel.2008.03.008
Cairney, J., and Pullman, G. S. (2007). The cellular and molecular biology of conifer embryogenesis. New Phytol. 176, 511–536. doi: 10.1111/j.1469-8137.2007.02239.x
Cairney, J., Zheng, L., Cowels, A., Hsioa, J., Zismann, V., Liu, J., et al. (2006). Expressed sequence tags from loblolly pine embryos reveal similarities with angiosperm embryogenesis. Plant Mol. Biol. 62, 485–501. doi: 10.1007/s11103-006-9035-9
Canales, J., Bautista, R., Label, P., Gómez-Maldonado, J., Lesur, I., Fernández-Pozo, N., et al. (2014). De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology. Plant Biotechnol. J. 12, 286–299. doi: 10.1111/pbi.12136
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1385/MB:19:2:201
Chung, K., Sakamoto, S., Mitsuda, N., Suzuki, K., Ohme-Takagi, M., and Fujiwara, S. (2016). WUSCHEL-RELATED HOMEOBOX 2 is a transcriptional repressor involved in lateral organ formation and separation in Arabidopsis. Plant Biotechnol. 33, 245–253. doi: 10.5511/plantbiotechnology.16.0202a
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
De Silva, V., Bostwick, D., Burns, K. L., Oldham, C. D., Skryabina, A., Sullards, M. C., et al. (2008). Isolation and characterization of a molecule stimulatory to growth of somatic embryos from early stage female gametophyte tissue of loblolly pine. Plant Cell Rep. 27, 633–646. doi: 10.1007/s00299-007-0484-7
De Smet, I., Lau, S., Mayer, U., and Jürgen, G. (2010). Embryogenesis – the humble beginnings of plant life. Plant J. 61, 959–970. doi: 10.1111/j.1365-313X.2010.04143.x
Dolzblasz, A., Nardmann, J., Clerici, E., Causier, B., van der Graaff, E., Chen, J., et al. (2016). Stem cell regulation by Arabidopsis WOX genes. Mol. Plant 9, 1028–1039. doi: 10.1016/j.molp.2016.04.007
Dumolin, S., Demesure, B., and Petit, R. J. (1995). Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 91, 1253–1256. doi: 10.1007/BF00220937
El-Azaz, J., de la Torre, F., Pascual, M. B., Debille, S., Canlet, F., Harvengt, L., et al. (2020). Transcriptional analysis of arogenate dehydratase genes identifies a link between phenylalanine biosynthesis and lignin biosynthesis. J. Exp. Bot. 71, 3080–3093. doi: 10.1093/jxb/eraa099
Elhiti, M. A. (2010). Molecular Characterization of Several Brassica Shoot Apical Meristem Genes and the Effect of Their Altered Expression During in Vitro Morphogenesis. Dissertation. Winnipeg, MB: University of Manitoba.
Elhiti, M., Stasolla, C., and Wang, A. (2013). Molecular regulation of plant somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 49, 631-642. doi: 10.1007/s11627-013-9547-3
Gaj, M. D. (2001). Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 64, 39–46. doi: 10.1023/A:1010679614721
Haecker, A., Gross-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., et al. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. doi: 10.1242/dev.00963
Hakman, I., Hallberg, H., and Palovaara, J. (2009). The polar auxin transport inhibitor NPA impairs embryo morphology and increases the expression of an auxin efflux facilitator protein PIN during Piceaabies somatic embryo development. Tree Physiol. 29, 483–496. doi: 10.1093/treephys/tpn048
Harvengt, L. (2005). “Somatic embryogenesis in Maritime pine (Pinus pinasterAit.),” in Protocols of Somatic Embryogenesis in Woody Plants, Forestry Sciences, Vol. 77, eds S. M. Jain and P. K. Gupta (Berlin: Springer Verlag), 107–120. doi: 10.1007/1-4020-2985-3_10
Hedman, H., Zhu, T., von Arnold, S., and Sohlberg, J. J. (2013). Analysis of the WUSCHEL RELATED HOMEOBOX gene family in the conifer Piceaabies reveals extensive conservation as well as dynamic patterns. BMC Plant Biol. 13:89. doi: 10.1186/1471-2229-13-89
Heidmann, I., De Lange, B., Lambalk, J., Angenent, G. C., and Boutilier, K. (2011). Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 30, 1107–1115. doi: 10.1007/s00299-011-1018-x
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857. doi: 10.1104/pp.17.00232
Jeong, S., Bayer, M., and Lukowitz, W. (2011). Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. J. Exp. Bot. 62, 1687–1697. doi: 10.1093/jxb/erq398
Klimaszewska, K. (1989). Plantlet development from immature zygotic embryos of hybrid larch through somatic embryogenesis. Plant Sci. 63, 95–103. doi: 10.1016/0168-9452(89)90105-2
Klimaszewska, K., and Cyr, D. R. (2002). Conifer somatic embryogenesis: I. Development. Dendrobiologia 48, 31–39.
Klimaszewska, K., Hargreaves, C., Lelu-Walter, M.-A., and Trontin, J.-F. (2016). “Advances in conifer somatic embryogenesis since year 2000,” in In Vitro Embryogenesis in Higher Plants, eds M. A. Germanà and M. Lambardi (New York, NY: Springer), 131–166. doi: 10.1007/978-1-4939-3061-6_7
Klimaszewska, K., Noceda, C., Pelletier, G., Label, P., Rodriguez, R., and Lelu-Walter, M. (2009). Biological characterization of young and aged embryogenic cultures of Pinus pinaster (Ait.). In Vitro Cell. Dev. Biol. Plant 45, 20–33. doi: 10.1007/s11627-008-9158-6
Klimaszewska, K., Overton, C., Stewart, D., and Rutledge, R. G. (2011). Initiation of somatic embryos and regeneration of plants from primordial shoots of 10-year-old somatic white spruce and expression profiles of 11 genes followed during the tissue culture process. Planta 233, 635–647. doi: 10.1007/s00425-010-1325-4
Klimaszewska, K., Park, Y. S., Overton, C., MacEacheron, I., and Bonga, J. M. (2001). Optimized somatic embryogenesis in Pinus strobus L. In Vitro Cell. Dev. Biol. Plant 37, 392–399. doi: 10.1007/s11627-001-0069-z
Klimaszewska, K., Pelletier, G., Overton, C., Stewart, D., and Rutledge, R. G. (2010). Hormonally regulated overexpression of Arabidopsis WUS and conifer LEC1 (CHAP3A) in transgenic white spruce: implications for somatic embryo development and somatic seedling growth. Plant Cell Rep. 29, 723–734. doi: 10.1007/s00299-010-0859-z
Klimaszewska, K., Trontin, J.-F., Becwar, M. R., Devillard, C., Park, Y. S., and Lelu-Walter, M. A. (2007). Recent progress in somatic embryogenesis of four Pinus spp. Tree For. Sci. Biotechnol. 1, 11–25.
Laux, T., Mayer, K. F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. doi: 10.1242/dev.122.1.87
Lelu-Walter, M.-A., Klimaszewska, K., Miguel, C., Aronen, T., Hargreaves, C., Teyssier, C., et al. (2016). “Somatic embryogenesis for more effective breeding and deployment of improved varieties in Pinus sp.: bottlenecks and recent advances,” in Somatic Embryogenesis – Fundamental Aspects and Applications, eds V. M. Loyola-Vargas and N. Ochoa-Alejo (Cham: Springer Verlag), 319–365. doi: 10.1007/978-3-319-33705-0_19
Lian, G., Ding, Z., Wang, Q., Zhang, D., and Xu, J. (2014). Origins and evolution of WUSCHELRELATEDHOMEOBOX protein family in plant kingdom. Sci. World J. 2014:534140. doi: 10.1155/2014/534140
Litvay, J. D., Verma, D. C., and Johnson, M. A. (1985). Influence of a loblolly pine (Pinus taeda L.). Culture medium and its components on growth and somatic embryogenesis of the wild carrot (Daucus carota L.). Plant Cell Rep. 4, 325–328. doi: 10.1007/BF00269890
Lotan, T., Ohto, M. A., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195-1205. doi: 10.1016/s0092-8674(00)81463-4
Marum, L., Loureiro, J., Rodriguez, E., Santos, C., Oliveira, M. M., and Miguel, C. (2009). Flow cytometric and morphological analyses of Pinus pinaster somatic embryogenesis. J. Biotechnol. 143, 288–295. doi: 10.1016/j.jbiotec.2009.08.001
Miguel, C. M., Rupps, A., Raschke, J., Rodrigues, A. S., and Trontin, J.-F. (2016). “Impact of molecular studies on somatic embryogenesis development for implementation in conifer multi-varietal forestry,” in Vegetative Propagation of Forest trees, eds Y.-S. Park, J. M. Bonga, and H.-K. Moon (Seoul: Korea Forest Research Institute), 373–421.
Morel, A., Trontin, J.-F., Corbineau, F., Lomenech, A.-M., Beaufour, M., Reymond, I., et al. (2014). Cotyledonary somatic embryos of Pinus pinasterAit. most closely resemble fresh, maturing cotyledonary zygotic embryos: biological, carbohydrate and proteomic analyses. Planta 240, 1075–1095. doi: 10.1007/s00425-014-2125-z
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
Nardmann, J., and Werr, W. (2006). The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono-and dicot evolution. Mol. Biol. Evol. 23, 2492–2504. doi: 10.1093/molbev/msl125
Nardmann, J., Reisewitz, P., and Werr, W. (2009). Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 26, 1745–1755. doi: 10.1093/molbev/msp084
Neale, D., Wegrzyn, J., Stevens, K., Zimin, A. V., Puiu, D., Crepeau, M. W., et al. (2014). Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15:R59. doi: 10.1186/gb-2014-15-3-r59
Nishiguchi, R., Takanami, M., and Oka, A. (1987). Characterization and sequence determination of the replicator region in the hairy-root-inducing plasmid pRiA 4b. Mol. Gen. Genet. 206, 1–8. doi: 10.1007/BF00326529
Nystedt, B., Street, N. R., Wetterbom, A., Zuccolo, A., Lin, Y. C., Scofield, D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. doi: 10.1038/nature12211
Palovaara, J., and Hakman, I. (2008). Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Piceaabies somatic embryogenesis. Plant Mol. Biol. 66, 533–549. doi: 10.1007/s11103-008-9289-5
Palovaara, J., and Hakman, I. (2009). WOX2 and polar auxin transport during spruce embryo pattern formation. Plant Signal. Behav. 4, 153–155. doi: 10.4161/psb.4.2.7684
Palovaara, J., Hallberg, H., Stasolla, C., and Hakman, I. (2010a). Comparative expression pattern analysis of WUSCHEL-RELATEDHOMEOBOX 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Piceaabies. New Phytol. 188, 122–135. doi: 10.1111/j.1469-8137.2010.03336.x
Palovaara, J., Hallberg, H., Stasolla, C., Luit, B., and Hakman, I. (2010b). Expression of a gymnosperm PIN homologous gene correlates with auxin immunolocalization pattern at cotyledon formation and in demarcation of the procambium during Piceaabies somatic embryo development and in seedling tissues. Tree Physiol. 30, 479–489. doi: 10.1093/treephys/tpp126
Park, S. Y., Klimaszewska, K., Park, J. Y., and Mansfield, S. D. (2010). Lodgepole pine: the first evidence of seed-based somatic embryogenesis and the expression of embryogenesis marker genes in shoot bud cultures of adult trees. Tree Physiol. 30, 1469–1478. doi: 10.1093/treephys/tpq081
Plomion, C., Bastien, C., Bogeat-Triboulot, M.-B., Bouffier, L., Déjardin, A., Duplessis, S., et al. (2016). Forest tree genomics: 10 achievements from the past 10 years and future prospects. Ann. For. Sci. 73, 77–103. doi: 10.1007/s13595-015-0488-3
Ramarosandratana, A., Harvengt, L., Bouvet, A., Calvayrac, R., and Pâques, M. (2001). Influence of the embryonal-suspensor mass (ESM) sampling on development and proliferation of maritime pine somatic embryos. Plant Sci. 160, 473–479. doi: 10.1016/s0168-9452(00)00410-6
Rupps, A., Raschke, J., Rümmler, M., Linke, B., and Zoglauer, K. (2016). Identification of putative homologs of Larix decidua to BABYBOOM (BBM), LEAFY COTYLEDON1 (LEC1), WUSCHEL-related HOMEOBOX2 (WOX2) and SOMATIC EMBRYOGENESIS RECEPTOR-like KINASE (SERK) during somatic embryogenesis. Planta 243, 473–488. doi: 10.1007/s00425-015-2409-y
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Solís-Ramos, L. Y., González-Estrada, T., Nahuath-Dzib, S., Zapata-Rodriguez, L. C., and Castano, E. (2009). Over-expression of WUSCHEL in C. Chinense causes ectopic morphogenesis. Plant Cell Tissue Organ Cult. 96, 279–287. doi: 10.1007/s11240-008-9485-7
Tabata, S., Kaneko, T., Nakamura, Y., Kotani, H., Kato, T., Asamizu, E., et al. (2000). Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826. doi: 10.1038/35048507
Taryono, M. (2000). Somatische Embryogenese and Transformation bei der Europäischen Lärche (Larix decidua Mill.). Dissertation. Berlin: Humboldt-Universität zu Berlin.
Trontin, J.-F., and Lelu-Walter, M. A. (2010). Agrobacterium-Mediated Genetic Transformation of Maritime Pine (Embryogenic Line PN519) Using pCbar as Standard, Reference Binary Vector. Sustainpine WP3 Common Protocol. N°1/V01(10/06/2010). Protocol Disseminated to Sustainpine WP3 Partners. SUSTAINPINE Multinational Project. Plant KBBE, PL2-2009-0016.
Trontin, J.-F., Debille, S., Canlet, F., Harvengt, L., Lelu-Walter, M. A., Label, P., et al. (2013). “Somatic embryogenesis as an effective regeneration support for reverse genetics in maritime pine: the Sustainpine collaborative project as a case study,” in Proceeding of the IUFRO Working Party 2.09.02 Conference on “Integrating Vegetative Propagation, Biotechnology and Genetic Improvement for Tree Production and Sustainable Forest Management”, 25-28/06/2012, Brno, eds Y. S. Park and J. M. Bonga, 184–187. Available online at: http://www.iufro.org/science/divisions/division-2/20000/20900/20902/publications (accessed March 20, 2013).
Trontin, J.-F., Harvengt, L., Garin, E., Lopez-Vernaza, M. A., Arancio, L., Hoebeke, J., et al. (2002). Towards genetic engineering of maritime pine (Pinus pinasterAit.). Ann. For. Sci. 59, 687-697. doi: 10.1051/forest:2002057
Trontin, J.-F., Klimaszewska, K., Morel, A., Hargreaves, C., and Lelu-Walter, M. A. (2016a). “Molecular aspects of conifer zygotic and somatic embryo development: a review of genome-wide approaches and recent insights,” in In Vitro Embryogenesis in Higher Plants, eds M. A. Germanà and M. Lambardi (New York, NY: Springer Science+Business Media), 167–207. doi: 10.1007/978-1-4939-3061-6_8
Trontin, J.-F., Teyssier, C., Morel, A., Harvengt, L., and Lelu-Walter, M. A. (2016b). “Prospects for new variety deployment through somatic embryogenesis in maritime pine,” in Vegetative Propagation of Forest Trees, eds Y.-S. Park, J. M. Bonga, and H.-K. Moon (Seoul: Korea Forest Research Institute), 572–606.
Trontin, J.-F., Aronen, T., Hargreaves, C., Montalbán, I. A., Moncaleán, P., Reeves, C., et al. (2016c). “International effort to induce somatic embryogenesis in adult pine trees,” in Vegetative Propagation of Forest Trees, eds Y.-S. Park, J. M. Bonga, and H.-K. Moon (Seoul: Korea Forest Research Institute), 211–260.
Trontin, J.-F., Walter, C., Klimaszewska, K., Park, Y. S., and Lelu-Walter, M. A. (2007). Recent progress in genetic transformation of four Pinus spp. Transgenic Plant J. 1, 314–329.
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
van der Graaff, E., Laux, T., and Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10:248. doi: 10.1186/gb-2009-10-12-248
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research34.1. doi: 10.1186/gb-2002-3-7-research0034
Walther, M., Wagner, I., Raschke, J., Zoglauer, K., and Rupps, A. (2021). Abscisic acid induces somatic embryogenesis and enables the capture of high-value genotypes in Douglas fir (Pseudotsuga menziesii [MIRB.] Franco). Plant Cell Tissue Organ Cult. 148, 45–59. doi: 10.1007/s11240-021-02159-3
Weigel, D., and Glazebrook, J. (2002). Arabidopsis. A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press, 165.
Wu, X., Chory, J., and Weigel, D. (2007). Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev. Biol. 309, 306–316. doi: 10.1016/j.ydbio.2007.07.019
Xiao, W., Custard, K. D., Brown, R. C., Lemmon, B. E., Harada, J. J., Goldberg, R. B., et al. (2006). DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18, 805–814. doi: 10.1105/tpc.105.038836
Zhang, H., Han, W., De Smet, I., Talboys, P., Loya, R., Hassan, A., et al. (2010). ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64, 764–774. doi: 10.1111/j.1365-313X.2010.04367.x
Zhang, Y., Wu, R., Qin, G., Chen, Z., Gu, H., and Qu, L. J. (2011). Over-expression of WOX1leads to defects in meristem development and polyamine homeostasis in Arabidopsis. J. Integr. Plant Biol. 53, 493–506. doi: 10.1111/j.1744-7909.2011.01054.x
Zhang, Y., Zhang, S., Han, S., Li, X., and Qi, L. (2012). Transcriptome profiling and in silico analysis of somatic embryos in Japanese larch (Larix leptolepis). Plant Cell Rep. 31, 1637–1657. doi: 10.1007/s00299-012-1277-1
Zhang, Z., Tucker, E., Hermann, M., and Laux, T. (2017). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40, 264–277. doi: 10.1016/j.devcel.2017.01.002
Zheng, W., Zhang, X., Yang, Z., Wu, J., Li, F., Duan, L., et al. (2014). AtWUSCHEL promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS One 9:e87502. doi: 10.1371/journal.pone.0087502
Zhu, T., Moschou, P. N., Alvarez, J. M., Sohlberg, J. J., and Arnold, S. (2016). WUSCHEL-RELATED HOMEOBOX 2 is important for protoderm and suspensor development in the gymnosperm Norway spruce. BMC Plant Biol. 16:19. doi: 10.1186/s12870-016-0706-7
Zoglauer, K., Behrendt, U., Rahmat, A., and Ross, H. (2003). Somatic Embryogenesis—the Gate to Biotechnology in Conifers. Vienna: Springer, 175–202. doi: 10.1007/978-3-7091-6040-4_11
Keywords: homologous genes, homologous and heterologous overexpression, genetic transformation, WOX2, WUSCHEL, LEC1
Citation: Hassani SB, Trontin J-F, Raschke J, Zoglauer K and Rupps A (2022) Constitutive Overexpression of a Conifer WOX2 Homolog Affects Somatic Embryo Development in Pinus pinaster and Promotes Somatic Embryogenesis and Organogenesis in Arabidopsis Seedlings. Front. Plant Sci. 13:838421. doi: 10.3389/fpls.2022.838421
Received: 17 December 2021; Accepted: 24 January 2022;
Published: 10 March 2022.
Edited by:
Víctor M. Loyola-Vargas, Scientific Research Center of Yucatán (CICY), MexicoReviewed by:
Neftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoCopyright © 2022 Hassani, Trontin, Raschke, Zoglauer and Rupps. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Rupps, YW5kcmVhLnJ1cHBzQGJpb2xvZ2llLmh1LWJlcmxpbi5kZQ==
†Present address: Seyedeh Batool Hassani, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University, Tehran, Iran
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.