- State Key Laboratory of Crop Genetics and Germplasm Enhancement, Cotton Germplasm Enhancement and Application Engineering Research Center, Ministry of Education, Nanjing Agricultural University, Nanjing, China
Cotton fiber is the most important natural textile material in the world. Identification and functional characterization of genes regulating fiber development are fundamental for improving fiber quality and yield. However, stable cotton transformation is time-consuming, low in efficiency, and technically complex. Moreover, heterologous systems, such as Arabidopsis and tobacco, did not always work to elucidate the function of cotton fiber specifically expressed genes or their promoters. For these reasons, constructing a rapid transformation system using cotton fibers is necessary to study fiber’s specifically expressed genes. In this study, we developed an easy and rapid Agrobacterium-mediated method for the transient transformation of genes and promoters in cotton fibers. First, we found that exogenous genes could be expressed in cotton fibers via using β-glucuronidase (GUS) and green fluorescence protein (GFP) as reporters. Second, parameters affecting transformation efficiency, including LBA4404 Agrobacterium strain, 3 h infection time, and 2-day incubation time, were determined. Third, four different cotton genes that are specifically expressed in fibers were transiently transformed in cotton fibers, and the transcripts of these genes were detected ten to thousand times increase over the control. Fourth, GUS staining and activity analysis demonstrated that the activity profiles of GhMYB212 and GhFSN1 promoters in transformed fibers are similar to their native activity in developmental fibers. Furthermore, the transient transformation method was confirmed to be suitable for subcellular localization studies. In summary, the presented Agrobacterium-mediated transient transformation method is a fast, simple, and effective system for promoter characterization and protein expression in cotton fibers.
Introduction
Cotton is an important industrial crop in the world. Cotton fiber, a highly elongated and thickened single cell of the seed epidermis, is the world’s most important natural textile raw material (Kohel et al., 1970). Cotton fiber development can be divided into four distinct but overlapping stages, namely, differentiation, elongation, secondary cell wall (SCW) thickening, and maturation (Graves and Stewart, 1988). On the day of anthesis, approximately one-third of the epidermal cells of the cotton ovule will differentiate into unbranched fiber cells (Stewart, 1975). During the elongation stage, cotton fiber cells elongate to ∼3 cm in 20 days before they switch to intensive secondary cell wall cellulose synthesis (Ruan, 2007). After maturation, more than 95% of fiber weight is cellulose. Due to these cellular and developmental characteristics, in addition to its important industrial value, the cotton fiber represents an outstanding single-cell model to study the control of cell differentiation, rapid elongation, and cellulose synthesis. Mining key genes, investigating their expression pattern and biological functions in fiber cell development will not only contribute to improving the yield and quality of cotton fibers but also will benefit the plant science communities.
Stable cotton transformation is the most revealing method for characterizing the gene functions and promoter activities in fiber cells. A few genes and promoters have been stably transformed into cotton to detect their functions in cotton fiber development (Deng et al., 2012; Lv et al., 2015; Zhang et al., 2018; Sun et al., 2019). However, cotton transformation is time-consuming, low in efficiency, and technically complex (Zhang, 2013). Transformation of cotton genes or promoters into model plants, such as Arabidopsis thaliana and tobacco, was an alternative way to investigate biological function. A previous study showed that an R2R3-MYB transcription factor that was specifically expressed in cotton fibers was transformed into Arabidopsis and affected the secondary cell wall biosynthesis and deposition in the transgenic plants (Sun et al., 2015). In addition, a cotton fiber-preferential promoter, pGbEXPA2, was verified to be regulated by gibberellins and abscisic acid via transformation in Arabidopsis (Li et al., 2015). The promoter of a cotton lipid transfer protein gene, FSltp4, showed strong activity in all types of trichomes in transgenic tobacco plants (Delaney et al., 2007). Despite these advances, promoters of many genes specifically expressed in cotton fibers have very low activities or are not active in these model plants, maybe due to the heterologous system of different species. Also, the functions of cotton fiber-specific genes cannot be fully demonstrated via ectopic expression in model plants. Establishing a simple, fast, and efficient transformation method in cotton fiber would greatly facilitate the studies on cotton fiber development.
Agrobacterium-mediated transient transformation, which is much more versatile, quick, and efficient in comparison with stable transformation, has been enormously applied for gene functional studies or biological material production in plants, including gene expression studies (Merle et al., 2002), promoter characterization (Yang et al., 2000; Rancé et al., 2002), gene silencing (Dubey et al., 2017), elicitor identification (Palanichelvam et al., 2000), vaccine production (D’Aoust et al., 2008), and CRISPR/Cas9-based genome editing (Kaur et al., 2021). Agrobacterium-mediated transient transformation is implemented via agroinfiltration. Generally, syringe and vacuum infiltration, which are applicable for a variety of plant species, are the mainly used methods for agroinfiltration. Genes are successfully transiently expressed in various tissues of different plants via syringe infiltration, such as Arabidopsis, soybean leaves (Kim et al., 2009; King et al., 2015), and melon fruit (Han et al., 2015). Vacuum infiltration has also been utilized for transient transformation in different plant tissues, including tobacco, black pepper leaves (Mani and Manjula, 2011; Fujiuchi et al., 2016), and lisianthus pollen (Sung, 2004). Although specialized equipment is required, vacuum infiltration exhibits improved transformation potential or yield (Zhao et al., 2017).
In this study, we developed an easy and rapid Agrobacterium-mediated transient transformation method in cotton fibers that can be used for promoter characterization and protein subcellular localization. By using the optimized vacuum infiltration method, we successfully overexpressed four previously reported cotton genes, analyzed the activities of two promoters, and performed the subcellular localization assay. The established method will provide an easy and fast way to perform functional studies on cotton fiber preferentially or specifically expressed genes.
Materials and Methods
Plant Materials
The allotetraploid cotton cultivar Gossypium hirsutum (accession TM-1) was used in this study. Cotton plants were cultivated in a glasshouse with a photoperiod of 14 h/10 h (light/dark) and a temperature of 28°C of day and 20°C of night. Cotton boll age was determined by tagging each pedicel on the day of flowering. Cotton bolls were collected on selected days post-anthesis (DPA). Fresh cotton fibers attached to seeds were used for Agrobacterium infiltration.
Plasmid Construction
To test whether genes can be expressed in cotton fibers via Agrobacterium-mediated transient transformation, binary overexpressing vectors were constructed. Coding sequences (CDS) of GhMYB212 (Gh_D11G3078) (Sun et al., 2019), GhCFE1 (Gh_A05G1404) (Lv et al., 2015), GhSusC (Gh_D06G0825) (Brill et al., 2011), and GhFSN1 (Gh_A12G1049) (Zhang et al., 2018) were cloned from G. hirsutum TM-1 and inserted downstream of the constitutive CaMV35S promoter in pBI121 vector, respectively. Additionally, the 1,944 bp promoter sequences of GhMYB212 and 2,035 bp promoter sequences of GhFSN1 were replaced by the CaMV35S promoter in the pBI121 vector, respectively, to construct the recombinant plasmids of pGhMYB212::GUS and pGhFSN1::GUS.
All the plasmids were introduced into the Agrobacterium cells as described previously (Chen et al., 1994). The primers used for vector construction are listed in Supplementary Table 1.
Agrobacterium-Mediated Transient Transformation of Cotton Fibers
Transformed Agrobacterium was cultured at 28°C in liquid Luria-Bertani (LB) medium (supplemented with 50 μg/mL rifampicin and 50 μg/mL kanamycin) until the OD600 reached 1.5. Then, the Agrobacterium cells were collected by centrifugation (4,000 rpm, 10 min) and resuspended with transformation solution [1/2 Murashige and Skoog (MS) medium supplemented with 165 μM acetosyringone and 3% sucrose (w/v), pH = 5.8] to an OD600 between 0.8 and 0.9. Before the resuspension procedure, the transformation solution was degassed with a vacuum pump (Sanplatec Inc., Satoshi Kato, Japan) under 0.07 MPa for 3 min.
Cotton bolls at selected DPA were collected, sterilized with 70% ethanol, and carefully stripped off to avoid damaging to the fibers. Then, fibers attached to the seeds were placed into a conical flask containing the resuspended Agrobacterium solution. After being treated with a vacuum pump at a pressure of 0.07 MPa for 2 min, 0.01% (v/v) Tween 20 was added into the transformation solution. The cotton fibers were infected by Agrobacterium cells under dark (25°C and 50 rpm/min) conditions for a determined period of time. Later, cotton fibers were taken out and rinsed with sterile water more than five times until the water is clear. Then, the fibers were transferred to 1/2 MS medium [supplemented with 0.3% (w/v) plant gel, 50 μg/mL rifampicin, and 50 μg/mL kanamycin, pH = 5.8] and placed in an incubator with a temperature of 26°C/20°C (day/night) and a photoperiod of 16 h of light and 8 h of dark. After co-incubation, fibers were used for gene expression analysis, enzyme activity determination, and fluorescence signal observation. Figure 1 summarizes the procedure to generate transient transgenic cotton fibers.

Figure 1. Schematic procedures for Agrobacterium-mediated transient transformation of cotton fibers.
β-Glucuronidase Activity Detection
Cotton fibers transformed with Agrobacterium containing p35S::GUS, pGhMYB212::GUS, or pGhFSN1::GUS constructs were subjected to GUS histochemical staining and enzyme activity determination, with fibers transformed with empty Agrobacterium as the negative control. The GUS staining and activity assay were performed according to Jefferson et al. (1987). Fibers on each seed were collected and used as one biological replicate. Three biological replicates were used for each reaction.
Green Fluorescence Protein Fluorescence Observation
Coding sequences of enhanced GFP gene (eGFP) under the control of CaMV35S promoter were transiently transformed into cotton fibers. Fluorescence of eGFP was observed using a confocal laser scanning microscope (TCS SP8, Leica, Germany) with the 488 nm excitation and 495–530 nm emission wavelengths. Fibers transformed with empty Agrobacterium were used as the negative control.
RNA Isolation, Quantitative Real-Time PCR, and Expression Analysis
Total RNA was isolated from transformed fibers using the Biospin Plant Total RNA Extraction Kit (Hangzhou Bioer Technology Co., Ltd., Hangzhou, China). The cDNA was synthesized from RNA using the HiScript III RT SuperMix (+gDNA wiper) (Vazyme, Inc., China) according to the manufacturer’s instructions. Gene-specific primers for quantitative reverse transcript PCR (qRT-PCR) analysis were designed using Primer Premier 5.0 software. The qRT-PCR reaction was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) using AceQ SYBR Green Master (Vazyme, Inc., China). The cotton ubiquitin 7 (GhUBQ7, accession number: DQ116441) was used as a reference for normalization (Li et al., 2018). Relative expression levels were calculated according to the method described by Livak and Schmittgen (2001). Three biological replicates were used for each sample. Detailed primer information is shown in Supplementary Table 1.
Subcellular Localization of Proteins in Fiber Cells
To test the application of subcellular localization in cotton fiber cells via the developed Agrobacterium-mediated transient transformation, a nuclear-localized protein histone B2, AtHTB2 (Zhang et al., 2020), and a plasma membrane-localized aquaporin, AtPIP2A (Qiu et al., 2020), were selected. Open reading frames of these two proteins without stop codon were fused with mCherry to generate 35S::AtH2B-mCherry and 35S::AtPIP2A-mCherry constructs, respectively. The recombinant vectors were transiently expressed in fiber cells by using the Agrobacterium-mediated method as described above. Subcellular localization was observed using a confocal laser scanning microscope at the 587 nm excitation and 610 nm emission wavelengths (TCS SP8, Leica, Germany).
Results
Construction of the Transient Expression Method in Cotton Fiber Cells
As shown in Figure 1, procedures of transient expression of the protein in cotton fiber cells are involved in the screening of the target gene, Agrobacterium strain, fiber developmental stage, infection time, and co-incubation time. In the beginning, we selected eGFP as the target gene due to its ready observation. The plasmid containing 35S::eGFP was introduced into two commonly used Agrobacterium tumefaciens strains GV3101 and LBA4404, respectively. The cotton fibers at 20 DPA were selected to perform Agrobacterium infection. Fiber cells were infected with Agrobacterium for 3 h, followed by 3-day incubation. Strong eGFP fluorescence signals were observed in cotton fiber cells that were infected with GV3101 or LBA4404 (Figures 2A,B), indicating that the eGFP protein was successfully expressed in fiber cells. However, no fluorescence signals were observed in the control fibers, which were not infected with Agrobacterium (Figure 2C). At 10× magnification, the eGFP signals were similar between fiber cells infected with GV3101 and LBA4404; however, at 40× magnification, strong fluorescence signals were observed in the cytoplasm of fibers infected with LBA4404, while many weak signals in fibers with GV3101 (Figures 2A,B). Then, we used the GUS report system to further verify the transient expression of proteins in cotton fiber cells. GUS staining showed that the control fibers that were not infected with Agrobacterium exhibited weak signals (Figure 2D), which is consistent with previous reports that the enzyme β-glucuronidase is present in developing cotton fibers (Sudan et al., 2006). Further analysis demonstrated that fiber cells infected with LBA4404 or GV3101 harbors 35S::GUS construct showed much stronger GUS expression than that in the control fibers (Figure 2D). Additionally, LBA4404-inoculated fiber cells exhibited deeper staining than that in GV3101-inoculated fiber cells. GUS activity showed that fibers transformed with LBA4404 or GV3101 had higher GUS activity than the control (Figure 2D). Furthermore, LBA4404-inoculated fiber cells showed higher GUS activities than those in GV3101-inoculated fiber cells. These results suggest that the target gene could be transiently expressed in cotton fiber cells via the Agrobacterium-mediated transient transformation method, and the LBA4404 strain is more efficient to perform the fiber cell infection process. Thus, the LBA4404 strain was used in the following experiments.
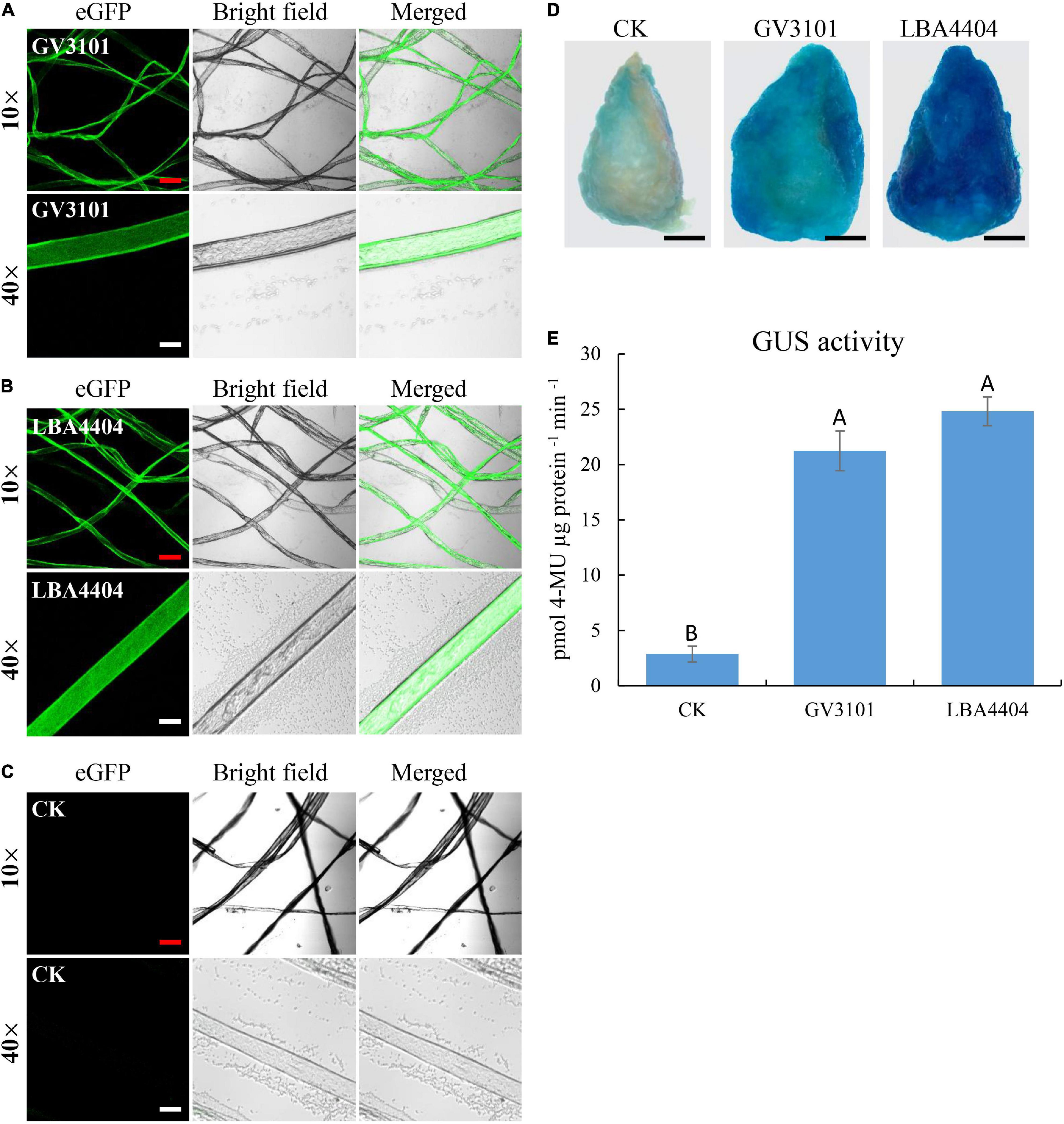
Figure 2. Transient expression of enhanced green fluorescence protein (GFP) gene (eGFP) and β-glucuronidase (GUS) protein in cotton fibers incubated with different Agrobacterium strains. (A,B) eGFP signal observation in cotton fibers transiently expressing 35S::eGFP construct using GV3101 (A) or LBA4404 (B). (C) Fibers without Agrobacterium infection were used as control. The red line and white line indicate 100 and 25 μm, respectively. (D,E) GUS staining (D) and activity determination (E) of cotton fibers transiently expressing 35S::GUS construct using GV3101, LBA4404, or fibers without Agrobacterium infection. Fibers on one seed were used as one biological replicate, and at least, three independent biological replicates were detected. The black line indicates 2 mm. Different upper case letters in (E) indicate significant differences at P < 0.01 (Turkey’s test). Error bars indicate the SD of three biological replicates. Fibers at 20-day post-anthesis (DPA) were used for transformation.
To determine the optimal parameters for transient expression, the infection time was investigated using the GUS report system. Cotton fibers were infected with Agrobacterium carrying the 35S::GUS plasmid for 0, 2, 3, and 4 h, respectively. After incubation for 1 day, GUS staining was performed on these fibers. Compared with the weak GUS staining signal in cotton fibers without Agrobacterium infection (0 h), 2, 3, and 4 h infection resulted in a strong GUS signal in fiber cells (Figure 3A). GUS activity further showed that the activities in 3 and 4 h infected fibers were significantly higher than that in 2 h fibers, and there was no significant difference between 3 and 4 h infected fibers (Figure 3B). Therefore, we chose 3 h infection as the optimal parameter for our transient transformation system.
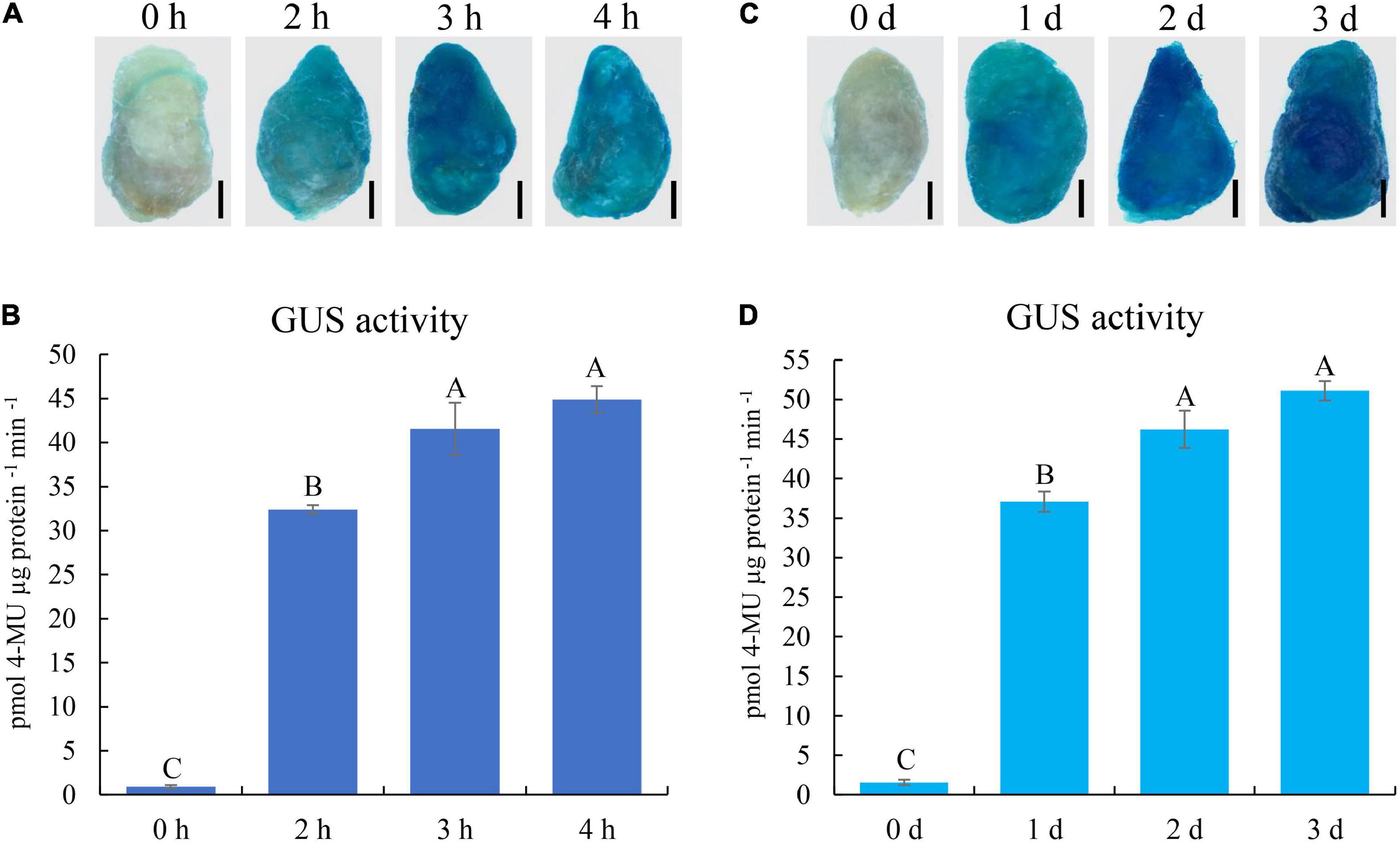
Figure 3. GUS expression analyses of 35S::GUS construct in cotton fibers under different transformation conditions. (A) GUS staining of fibers under different infection times with Agrobacterium. (B) GUS activity of cotton fibers infected with Agrobacterium containing 35S::GUS construct under different infection times. (C) GUS staining of fibers under different incubation times after Agrobacterium infection. (D) GUS activity of cotton fibers under different incubation times following Agrobacterium infection. The black line indicates 2 mm in (A,C). Error bars indicate SD. Different letters indicate significant differences (Turkey’s test; P < 0.01). h, hour; d, day. Fibers at 20 DPA were used for transformation.
We also optimized the incubation time using the GUS report system. Cotton fibers infected with Agrobacterium for 3 h were harvested immediately (0 day) or were further incubated on medium for 1, 2, or 3 days. GUS staining and activity determination showed that the GUS enzyme activities in cotton fibers without incubation were very weak, whereas they were much higher in 1-, 2-, or 3-day incubated fibers (Figures 3C,D). GUS activities in 2 and 3-day incubated fibers were significantly higher than that in 1-d fibers, and there was no significant difference between 2 and 3 days incubated fibers (Figure 3D). Thus, 2-day incubation was selected in our transient transformation system.
Taken together, three parameters, including LBA4404 Agrobacterium strain, 3 h infection time, and 2 days incubation time, were determined to study the transient expression in cotton fibers.
Transient Overexpression of Four Cotton Fiber Preferentially Expressed Genes via Agrobacterium-Mediated Method
We selected four previously characterized cotton genes that were preferentially expressed in fibers, including GhMYB212 (Sun et al., 2019), GhCFE1 (Lv et al., 2015), GhSusC (Brill et al., 2011), and GhFSN1 (Zhang et al., 2018) to study the expression levels by using the transient transformation method. Expression analyses in G. hirsutum TM-1 showed that GhCFE1 and GhMYB212 were preferentially expressed in fibers undergoing rapid elongation (5–15 DPA), while GhSusC and GhFSN1 were mainly expressed in fibers at the secondary cell wall thickening stage (15–25 DPA) (Figure 4A), similar to previous reports. The coding regions of these four genes were cloned and inserted downstream of the 35S promoter in the pBI121 binary expression vector to obtain 35S::GhCFE1, 35S::GhMYB212, 35S::GhSusC, and 35S::GhFSN1 constructs, respectively. Furthermore, the four constructs were individually transformed into Agrobacterium and used for subsequent transient transformation. Fibers at 20 DPA were subjected to transformation and were collected for gene expression analysis. Quantitative RT-PCR assay showed that the transcript levels of GhMYB212, GhCFE1, GhSusC, and GhFSN1 increased by 44.19, 1,535.33, 11.59, and 205.74 times in transiently transformed fibers in comparison to the control that transformed with empty pBI121 binary expression vector, respectively (Figure 4B). These results confirmed the success of our method in transient overexpressing target genes in cotton fibers.
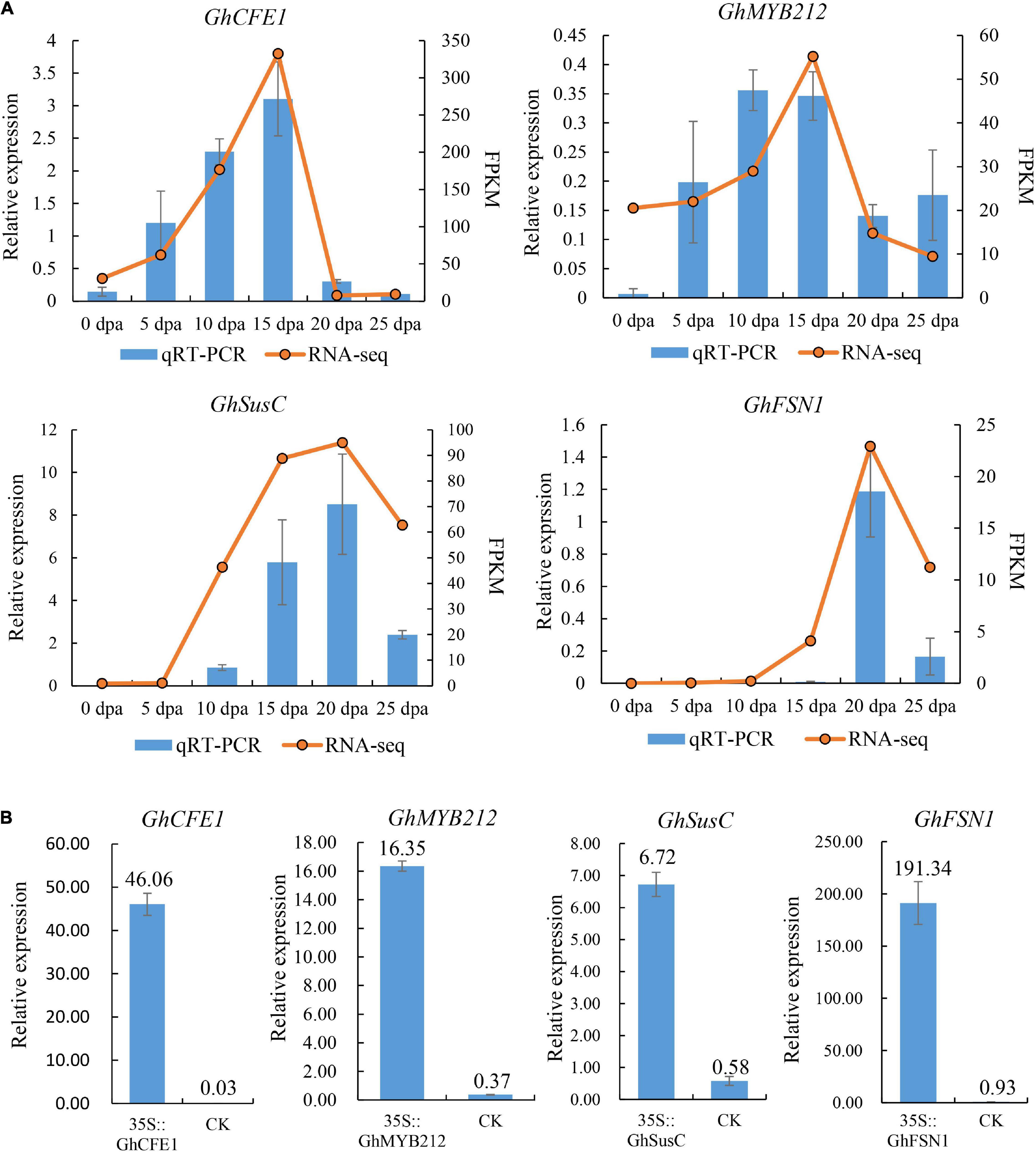
Figure 4. Transient overexpression of four cotton genes in fibers. (A) The expression pattern of four genes used for transient overexpression experiment was investigated in Gossypium hirsutum TM-1, including two genes, GhCFE and GhMYB212, which are preferentially expressed in elongating fibers, and two genes, GhSusC and GhFSN1, which are preferentially expressed in secondary cell wall depositing fibers. (B) The expression levels of GhCFE1, GhMYB212, GhSusC, and GhFSN1 were significantly higher in transiently transformed fibers than that in the control, which was transformed with empty pBI121 binary expression vector, respectively. Data above each column indicate the relative expression of each gene compared with the internal reference gene, GhUBQ7. Fibers at 20 DPA were used for transformation. Error bars indicate SD.
Promoter Activity Analysis in Fiber Cells Using Agrobacterium-Mediated Transient Transformation
To further test the efficiency of the method, we cloned the promoter regions (∼2 kb) of GhFSN1 and GhMYB212 into pBI121 vector upstream of GUS to make the recombinant plasmids, pGhFSN1::GUS and pGhMYB212::GUS, respectively. Cotton fibers at 10 and 20 DPA were transiently transformed with Agrobacterium and then were subjected to GUS staining and activity determination. The staining results revealed that weak signals were observed both in 10 and 20 DPA fibers without Agrobacterium infection (Figure 5A). The signal of pGhFSN1::GUS was higher in 20 DPA fibers than that in 10 DPA fibers; on the contrary, the signal of pGhMYB212::GUS was higher in 10 DPA fibers than that in 20 DPA fibers. GUS activity assays confirmed that the control fibers had low β-glucuronidase activities (Figure 5B). Again, GUS activities in 20 DPA fibers were significantly higher than that in 10 DPA fibers transformed with pGhFSN1::GUS, while GUS activities in 10 DPA fibers were significantly higher than that in 20 DPA fibers transformed with pGhMYB212::GUS. These results are consistent with the GhFSN1 and GhMYB212 endogenous transcript levels quantified by qRT-PCR in cotton fibers (Figure 4A), indicating that the Agrobacterium-mediated transient transformation method was suitable for promoter characterization in fiber cells.
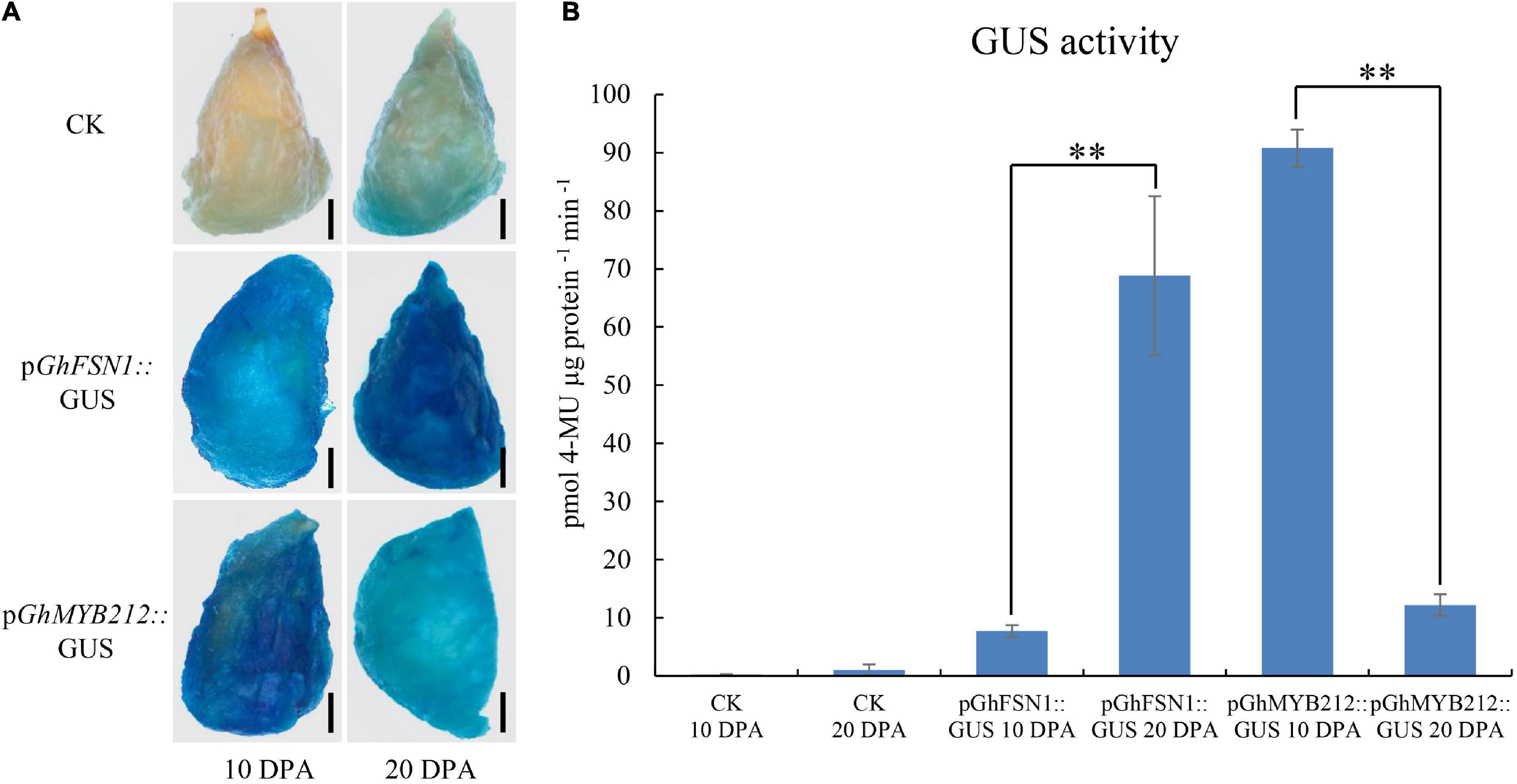
Figure 5. Activities of promoters of GhFSN1 and GhMYB212 at different fiber development stages were analyzed via Agrobacterium-mediated transient transformation. (A) GUS staining results showed that the GhFSN1 promoter had high activity at 20 DPA fibers, while the GhMYB212 promoter had high activity at 10 DPA fibers. Black line indicates 2 mm. (B) GUS activity analysis of cotton fibers infected with Agrobacterium containing pGhFSN1::GUS or pGhMYB212::GUS constructs, respectively. Error bars indicate SD. ** indicates a significant difference using the t-test with a P-value of 0.01.
The Developed Transient Expression Method in Cotton Fiber Can Be Used for Subcellular Localization Assay
Having validated that proteins could be transiently expressed in cotton fibers, we further investigated whether the expressed proteins were correctly subcellular localized in fiber cells. A nuclear-localized protein, histone B2 (AtHTB2) (Zhang et al., 2020), and a plasma membrane-localized aquaporin, AtPIP2A (Qiu et al., 2020), were ligated with mCherry protein at the N-terminal, respectively, to form 35S::AtHTB2-mCherry and 35S::AtPIP2A-mCherry constructs. Agrobacterium-mediated transient transformation with the 35S::AtHTB2-mCherry construct produced a strong fluorescence signal confined to nuclear (Figure 6A), while fiber cell expressing AtPIP2A-mCherry showed strong fluorescence at the plasma membrane (Figure 6B). These results suggested that the transient transformation method was suitable for subcellular localization studies in cotton fiber cells.
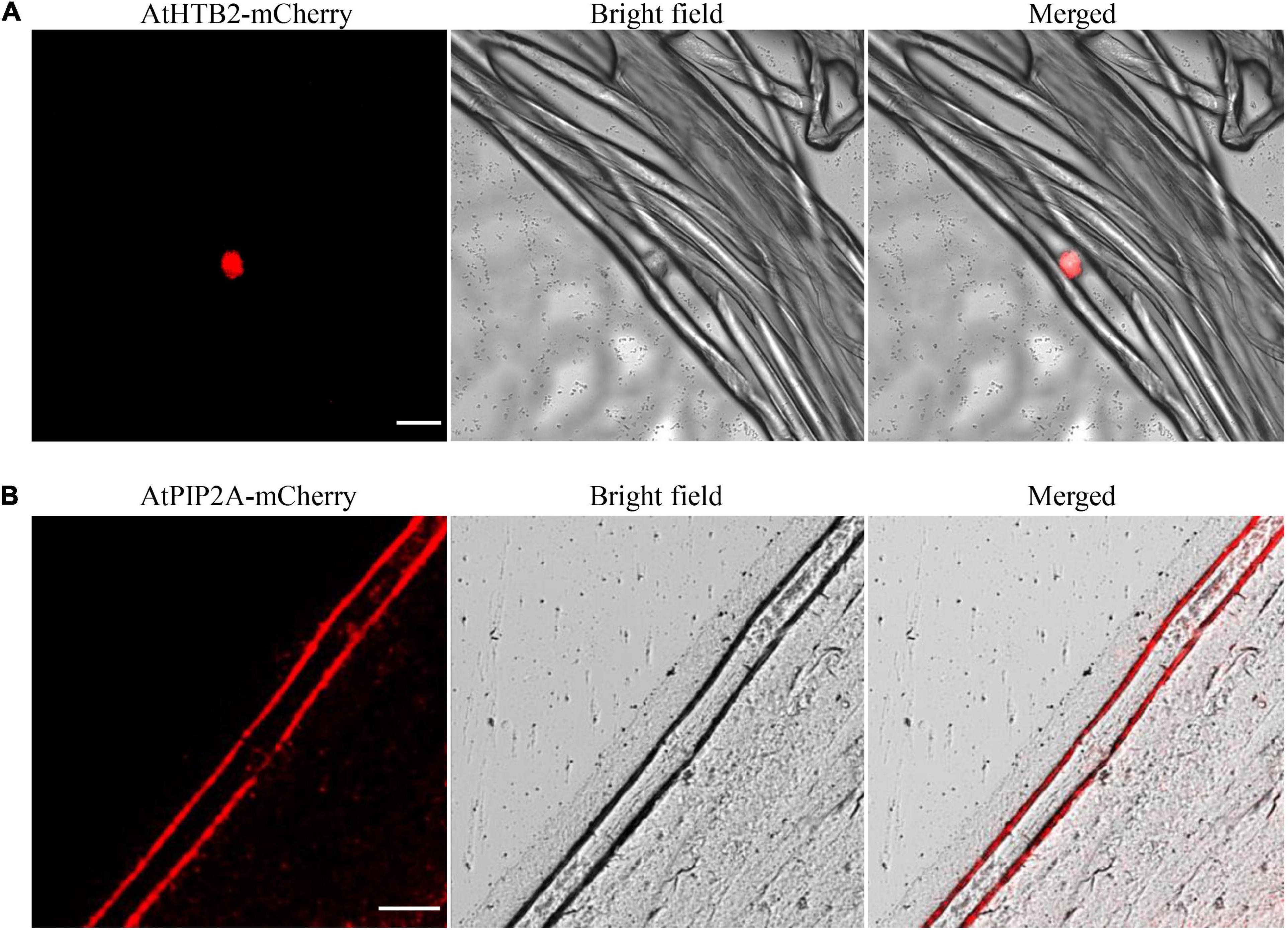
Figure 6. Subcellular localization of mCherry-tagged proteins in fiber cells. (A) A strong fluorescence signal was observed in the nucleus of fiber cells transformed with Agrobacterium harboring 35S::AtHTB2-mCherry construct. (B) A strong fluorescence signal was observed in the plasma membrane of fiber cells transformed with Agrobacterium harboring 35S::AtPIP2A-mCherry construct. White lines indicate 50 μm. AtHTB2, Arabidopsis thaliana histone B2; AtPIP2A, an aquaporin, short for Arabidopsis thaliana plasma membrane intrinsic protein. Fibers at 20 DPA were used for transformation.
Discussion
In this study, we developed an Agrobacterium-mediated transient transformation system with cotton fiber as the receptor aiming to accelerate the functional research on cotton fiber development. As far as we know, this is the first report in which the target gene was transiently expressed in cotton fiber via the Agrobacterium-mediated method. In plant research, model plants, such as Arabidopsis thaliana or tobacco, are generally used as transgenic receptors for gene function investigation if the species itself is difficult to make the transgenic plant, such as cotton. However, as cotton fiber has its own unique developmental characteristics, some genes or promoters specifically function in cotton fiber are expressed at very low or no expression at all in the model plants after transformation. Moreover, although different methods have been developed in cotton transformation, it is still time-consuming, labor-intensive, costly, and technically complex to investigate functions of genes and activities of promoters via stable genetic transformation, which has seriously hindered the research progress on the function of genes in cotton fiber development. The rapid and technically easy method described herein is supposed to partially solve this problem. In our studies, we first showed that four exogenous proteins, including GFP, GUS, AtHTB2-mCherry, and AtPIP2A-mCherry, and four cotton endogenous proteins, including GhCFE1, GhMYB212, GhSusC, and GhFSN1, were all successfully expressed in cotton fibers via the developed methods within a few days (Figures 2, 4, 6). Second, the promoter activities of GhFSN1 and GhMYB212 determined using the transient transformation method in this study were well consistent with their promoting activities in normal developing cotton fibers (Figures 4, 5), indicating that the promoter activity can be quickly and accurately determined in cotton fiber via the transient transformation method in this study. Additionally, AtHTB2 protein was successfully localized at nuclear and AtPIP2A at the plasma membrane in the subcellular localization assay (Figure 6), indicating that cotton fiber could be used for subcellular localization studies of the cotton endogenous proteins. In summary, these findings implied that the method developed in this study can be used for protein expression and promoter activity investigation studies.
We also optimized the transformation system by determining the affecting parameters, including the Agrobacterium strain, infection time, and incubation time. Through comparison of GFP fluorescence signals, GUS staining, and GUS activity determination, we showed that Agrobacterium strain LBA4404 had higher transformation efficiency than GV3101 (Figure 2). Furthermore, based on comprehensive consideration of expression levels of the GUS gene and operation time, infection time of 3 h, and incubation time of 2 days were suggested for performing transformation in the method (Figure 3). Based on these, we successfully developed the transient transformation system in cotton fiber with the detected infiltration parameters.
As discussed above, the method reported in this study is quite applicable for promoter characterization and in situ subcellular localization of proteins that are preferentially or specifically expressed in cotton fibers. Hopefully, the transient transformation of cotton fiber can be used to test the regulation of transcription factors on downstream genes and interaction studies of different proteins in fiber cells. In the future study, we will further optimize the parameters to enhance the transformation efficiency and extend its application.
Each cotton fiber is a single cell that is highly elongated and thickened from the seed epidermis. To our best knowledge, this is the first report that proteins are transiently expressed using plant single cell as the receptor via the Agrobacterium-mediated method. The results in this study imply that single intact plant cells can be transformed with Agrobacterium, providing important clues for future plant transformation studies.
Conclusion
In this study, we developed an easy and fast Agrobacterium-mediated transient transformation method using cotton fiber as the receptor. This method can be used for in situ promoter characterization and subcellular localization of proteins that are preferentially or specifically expressed in cotton fibers. Furthermore, the method shows a promising application in gene functional studies, such as protein–protein interaction and regulatory network detection, after further optimization.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
WG conceived the study and revised the manuscript. XS and WG designed the experiments. XS, LZ, YD, MZ, and YY contributed to the cotton fiber infiltration, observation, and detection. QH performed the quantitative RT-PCR analysis. XS interpreted the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This program was financially supported in part by the Guidance Foundation, Sanya Institute of Nanjing Agricultural University (NAUSY-MS01), National Natural Science Foundation of China (31701472), and Collaborative Innovation Center for Modern Crop Production cosponsored by Province and Ministry (CIC-MCP) (No. 10). The funders were not involved in the experimental design of the study, data collection, analysis, and interpretation, and writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.837994/full#supplementary-material
References
Brill, E., van Thournout, M., White, R. G., Llewellyn, D., Campbell, P. M., Engelen, S., et al. (2011). A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol. 157, 40–54. doi: 10.1104/pp.111.178574
Chen, H., Nelson, R. S., and Sherwood, J. L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16:670.
D’Aoust, M. A., Lavoie, P. O., Couture, M. M. J., Trépanier, S., Guay, J.-M., Dargis, M., et al. (2008). Influenza virus-like particles produced by transient expression in nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6, 930–940. doi: 10.1111/j.1467-7652.2008.00384
Delaney, S. K., Orford, S. J., Martin-Harris, M., and Timmis, J. N. (2007). The fiber specificity of the cotton FSltp4 gene promoter is regulated by an AT-rich promoter region and the AT-hook transcription factor GhAT1. Plant Cell Physiol. 48, 1426–1437. doi: 10.1093/pcp/pcm111
Deng, F., Tu, L., Tan, J., Li, Y., Nie, Y., and Zhang, X. (2012). GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. doi: 10.1104/pp.111.186742
Dubey, V. K., Lee, U. G., Kwon, D. H., and Lee, S. H. (2017). Agroinfiltration-based expression of hairpin RNA in soybean plants for RNA interference against tetranychus urticae. Pestic. Biochem. Physiol. 142, 53–58. doi: 10.1016/j.pestbp.2017.01.004
Fujiuchi, N., Matsuda, R., Matoba, N., and Fujiwara, K. (2016). Removal of bacterial suspension water occupying the intercellular space of detached leaves after agroinfiltration improves the yield of recombinant hemagglutinin in a nicotiana benthamiana transient gene expression system. Biotechnol. Bioeng. 113, 901–906. doi: 10.1002/bit.25854
Graves, D. A., and Stewart, J. M. (1988). Chronology of the differentiation of cotton (Gossypium hirsutum L.) fiber cells. Planta 175, 254–258. doi: 10.1007/BF00392435
Han, L., Zhang, L., Liu, J., Li, H., Wang, Y., and Hasi, A. (2015). Transient expression of optimized and synthesized natto kinase gene in melon (Cucumis melo L.) fruit by agroinfiltration. Plant Biotechnol. 15:0430a. doi: 10.5511/plantbiotechnology.15.0430a
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 6, 3901–3907.
Kaur, M., Manchanda, P., Kalia, A., Ahmed, F. K., Nepovimova, E., Kuca, K., et al. (2021). Agroinfiltration mediated scalable transient gene expression in genome edited crop plants. Int. J. Mol. Sci. 22:10882. doi: 10.3390/ijms221910882
Kim, M. J., Baek, K., and Park, C. M. (2009). Optimization of conditions for transient agrobacterium mediated gene expression assays in arabidopsis. Plant Cell Rep. 2009, 1159–1167. doi: 10.1007/s00299-009-0717-z
King, J. L., Finer, J. J., and McHale, L. K. (2015). Development and optimization of agroinfiltration for soybean. Plant Cell Rep. 34, 133–140. doi: 10.1007/s00299-014-1694-4
Kohel, R. J., Richmond, T. R., and Lewis, C. F. (1970). Texas marker-1. description of a genetic standard for Gossypium hirsutum L. Crop Sci. 10, 670–671. doi: 10.2135/cropsci1970.0011183X001000060019x
Li, H., Li, K., Guo, Y., Guo, J., Miao, K., Botella, J. R., et al. (2018). A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum). Plant Methods 14:50. doi: 10.1186/s13007-018-0319-2
Li, Y., Tu, L., Ye, Z., Wang, M., Gao, W., and Zhang, X. (2015). A cotton fiber-preferential promoter. PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep. 34, 1539–1549. doi: 10.1007/s00299-015-1805-x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, F. N., Wang, H. H., Wang, X. Y., Han, L. B., Ma, Y. P., Wang, S., et al. (2015). GhCFE1A, a dynamic linker between the ER network and actin cytoskeleton, plays an important role in cotton fibre cell initiation and elongation. J. Exp. Bot. 66, 1877–1889. doi: 10.1093/jxb/eru530
Mani, T., and Manjula, S. (2011). Optimization of agrobacterium mediated transient gene expression and endogenous gene silencing in piper colubrinum link. by vacuum infiltration. Plant Cell Tiss. Organ Cult. 105, 113–119. doi: 10.1007/s11240-010-9836-z
Merle, C., Perret, S., Lacour, T., Jonval, V., Hudaverdian, S., Garrone, R., et al. (2002). Hydroxylated human homotrimeric collagen I in Agrobacterium tumefaciens-mediated transient expression and in transgenic tobacco plant. FEBS Lett. 515, 114–118. doi: 10.1016/s0014-5793(02)02452-3
Palanichelvam, K., Cole, A. B., Shababi, M., and Schoelz, J. E. (2000). Agroinfiltration of. cauliflower mosaic virus gene vi elicits hypersensitive response in nicotiana species. Mol. Plant Microbe Interact. 13, 1275–1279. doi: 10.1094/MPMI.2000.13.11.1275
Qiu, J., McGaughey, S. A., Groszmann, M., Tyerman, S. D., and Byrt, C. S. (2020). Phosphorylation influences water and ion channel function of AtPIP2;1. Plant Cell Environ. 43, 2428–2442. doi: 10.1111/pce.13851
Rancé, I., Norre, F., Gruber, V., and Theisen, M. (2002). Combination of viral promoter sequences to generate highly active promoters for heterologous therapeutic protein over-expression in plants. Plant Sci. 162, 833–842. doi: 10.1016/S0168-9452(02)00031-6
Ruan, Y. L. (2007). Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct. Plant Biol. 34, 1–10. doi: 10.1071/fp06234
Stewart, J. M. (1975). Fiber initiation on the cotton ovule (Gossypium hirsutum). Am. J. Bot. 62, 723–730. doi: 10.1002/j.1537-2197.1975.tb14105.x
Sudan, C., Prakash, S., Bhomkar, P., Jain, S., and Bhalla-Sarin, N. (2006). Ubiquitous presence of β-glucuronidase (GUS) in plants and its regulation in some model plants. Planta 224, 853–864. doi: 10.1007/s00425-006-0276-2
Sun, W., Gao, Z., Wang, J., Huang, Y., Chen, Y., Li, J., et al. (2019). Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 222, 864–881. doi: 10.1111/nph.15620
Sun, X., Gong, S. Y., Nie, X. Y., Li, Y., Li, W., Huang, G. Q., et al. (2015). A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic arabidopsis. Physiol. Plant 154, 420–432. doi: 10.1111/ppl.12317
Sung, H. P. (2004). In Vitro culture and transformation by agroinfiltration of lisianthus (Eustoma russellianus) pollen. J. Life Sci. 14, 1018–1022. doi: 10.5352/JLS.2004.14.6.101
Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi: 10.5352/JLS.2004.14.6.1018
Zhang, B. (2013). Transgenic cotton: from biotransformation methods to agricultural application. Methods Mol. Biol. 958, 3–15. doi: 10.1007/978-1-62703-212-4_1
Zhang, J., Huang, G. Q., Zou, D., Yan, J. Q., Li, Y., Hu, S., et al. (2018). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217, 625–640. doi: 10.1111/nph.14864
Zhang, L., Zhao, Y., Liang, H., Li, X., Gallagher, K. L., and Wu, S. (2020). Gateway-compatible vectors for functional analysis of proteins in cell type specific manner. Plant Methods 16:635. doi: 10.1186/s13007-020-00635-z
Keywords: cotton fiber, Agrobacterium, transient transformation, promoter activity, subcellular localization
Citation: Shang X, Zhu L, Duan Y, He Q, Zhao M, Yu Y and Guo W (2022) An Easy and Rapid Transformation Protocol for Transient Expression in Cotton Fiber. Front. Plant Sci. 13:837994. doi: 10.3389/fpls.2022.837994
Received: 17 December 2021; Accepted: 08 February 2022;
Published: 22 March 2022.
Edited by:
Vladimir Orbovic, University of Florida, United StatesCopyright © 2022 Shang, Zhu, Duan, He, Zhao, Yu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wangzhen Guo, bW9lbGFiQG5qYXUuZWR1LmNu
 Xiaoguang Shang
Xiaoguang Shang Lijie Zhu
Lijie Zhu Wangzhen Guo
Wangzhen Guo