- 1Key Laboratory of Germplasm Innovation and Utilization of Triticeae Crops at Universities of Inner Mongolia Autonomous Region, College of Life Sciences, Inner Mongolia Agricultural University, Hohhot, China
- 2Institute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot, China
- 3Henan Key Laboratory of Wheat Biology, National Engineering Laboratory for Wheat, Key Laboratory of Wheat Biology and Genetic Breeding in Central Huang-Huai Region, Ministry of Agriculture and Rural Affairs, Wheat Research Institute, Henan Academy of Agricultural Sciences, Zhengzhou, China
Background: Wheat (Triticum aestivum L.) ABA insensitive five (ABI5) binding protein gene (TaAFP) is a homologue of the ABI5 binding protein (AFP) gene in Arabidopsis thaliana. It is well documented that AtAFP is a negative regulator of ABA signaling that regulates embryo germination and seed dormancy. TaABI5 was earlier shown to be expressed specifically in seed and its transcript accumulated during wheat grain maturation and acquisition of dormancy. It plays an important role in seed dormancy. In a previous study, we identified two allelic variants TaAFP-B1a and TaAFP-B1b of TaAFP on chromosome arm 2BS in common wheat, designated as, respectively. Sequence analysis revealed a 4 bp insertion in the promoter of TaAFP-B1a compared with TaAFP-B1b that affected mRNA transcription level, mRNA stability, GUS and tdTomatoER translation level, and GUS activity determining seed dormancy.
Results: The transcription and translation levels of TaAFP-B were significantly reduced in TaAFP-Ba and TaAFP-Ba-GFP transgenic plants compared with TaAFP-Bb and TaAFP-Bb-GFP. The average GI (germination index) values of TaAFP-Ba and TaAFP-Ba-GFP were significantly lower than those of TaAFP-Bb and TaAFP-Bb-GFP in T1 and T2 transgenic rice seeds, whereas mature TaAFP-Ba and TaAFP-Ba-GFP transgenic seeds exhibited increased ABA sensitivity and content of endogenous ABA compared with TaAFP-Bb and TaAFP-Bb-GFP.
Conclusion: The 4 bp insertion in the promoter of TaAFP-Ba decreased transcript abundance and translation level in transgenic rice. This insertion increased sensitivity to ABA and content of endogenous ABA in mature seeds, leading to a higher seed dormancy and pre-harvest sprouting tolerance in transgenic rice.
Background
Pre-harvest sprouting (PHS) is a worldwide problem in wheat production. It causes significant losses in grain weight and reduced end-use quality (Groos et al., 2002; Humphreys and Noil, 2002). Generally, PHS decreases grain yield by 6–10%, and the value sprouted wheat can be reduced by 20–50% (Zhang et al., 2017). Severely sprouted wheat cannot be used for flour production or other applications in the food industry and therefore can only be used for animal feed (Simsek et al., 2014). Seed dormancy is the major factor reducing the risk of PHS under wet weather conditions (Li et al., 2004). Therefore, it is important to understand the genetic mechanism of seed dormancy in wheat.
The balance of abscisic acid (ABA) and gibberellin (GA) levels is a major regulator of seed dormancy in plants. ABA regulates germination and promotes mature seed dormancy by inhibiting α-amylase synthesis (Hoffmann, 1997). The mechanism of ABA sensitivity in seeds has been extensively studied in Arabidopsis. Several genes associated with seed dormancy have been identified as factors involved in ABA signaling and ABA biosynthesis (Lee and Kende, 2001; Oikawa et al., 2004; Liu et al., 2007; Sugimoto et al., 2010). The ABA signaling pathway includes transcription factors that serve as both positive and negative regulators. The positive regulatory factors are viviparous-1 (Vp-1), ABI3, ABI4, and ABI5 (Finkelstein, 1994; Finkelstein and Lynch, 2000). In ABA signaling, ABI3 and ABI5 act as intermediates, regulating the maturation and germination of seeds; expression of these genes facilitates desiccation tolerance of seeds at later stages of maturity (Lopez-Molina et al., 2001, 2002; Carles et al., 2002). ABI3, ABI4, and ABI5 also affect seed development and ABA sensitivity. A null mutation of abi3 conferred a more severe effect than that of abi4 or abi5 (Parcy et al., 1994; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Negative regulators include ABI1, ABI2, AIP2 E3 ligase, RING E3 ligase, and ABA insensitive five binding protein (AFP) that regulates ABI5 (Gosti et al., 1999; Lopez-Molina et al., 2003; Stone et al., 2006). ABI3 is required for appropriate expression of ABI5 (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002; Sugimoto et al., 2010). ABI3 is an ortholog of Viviparous-1 (Vp-1) in wheat, and Vp-1 homologs play an important role in seed maturation processes such as seed dormancy and seed desiccation (McCarty et al., 1989, 1991; Giraudat et al., 1992). Mis-spliced transcripts of TaVp-1 were identified in the A and B genomes of wheat, and different expression levels of the properly spliced transcripts of TaVp-1A and TaVp-1B were associated with different levels of seed dormancy in white-grained wheat genotypes (McKibbin et al., 2002; Yang et al., 2007a,b, 2014; Sun et al., 2012). ABI3 encodes a transcription factor and interacts with ABI5 to condition embryonic gene expression and seed sensitivity to ABA (Koornneef et al., 1984; Finkelstein, 1994; Lopez-Molina and Chua, 2000). ABI5 functions as a critical factor in seed maturation and dormancy, and in dehydration tolerance of young Arabidopsis seedlings (Lopez-Molina et al., 2001; Sugimoto et al., 2010).
ABI5 binding protein (AFP) acts as a negative regulator in the ABA signaling pathway by facilitating degradation of ABI5 (Lopez-Molina et al., 2003). AFP transcription and translation increases during seed development and desiccation, ultimately reaching a plateau in mature seeds (Lopez-Molina et al., 2003; Garcia et al., 2008). Three wheat AFP genes (TaAFPs), designated TaAFP-A, TaAFP-B, and TaAFP-D, were isolated and localized to the short arms of chromosomes 2A, 2B, and 2D, respectively (Ohnishi et al., 2008). In our previous study, two allelic variants of TaAFP-B were identified and designated as TaAFP-Ba and TaAFP-Bb; TaAFP-Ba contained a 4 bp insertion in the promoter region compared with TaAFP-Bb (Feng et al., 2019). The 4 bp insertion affected mRNA stability, mRNA levels, accumulation of tdTomatoER and GUS, and GUS activity in TaAFP-B promoter fusions. AFPB, a co-dominant functional marker of TaAFP-B, was developed based on this 4 bp InDel. Mature seeds of genotype TaAFP-B1b generating a 203 bp fragment had stronger dormancy than the TaAFP-B1a genotype that produced a 207 bp fragment. The average germination indices (GI) of plants homozygous for TaAFP-Ba and TaAFP-Bb were 45.18 and 30.72%, respectively, as determined by a test of 91 white-grained Chinese wheat cultivars and advanced lines. The difference was highly significant (P < 0.001). Thus, TaAFP-B was significantly associated with PHS tolerance in common wheat (Feng et al., 2019).
In order to further elucidate the functions of TaAFP-Ba and TaAFP-Bb and investigate the mechanisms underlying seed dormancy or PHS tolerance in wheat, both alleles were transformed and investigated in rice.
Results
Identification and Determination of Copy Number of the TaAFP-B Alleles in Transgenic Rice
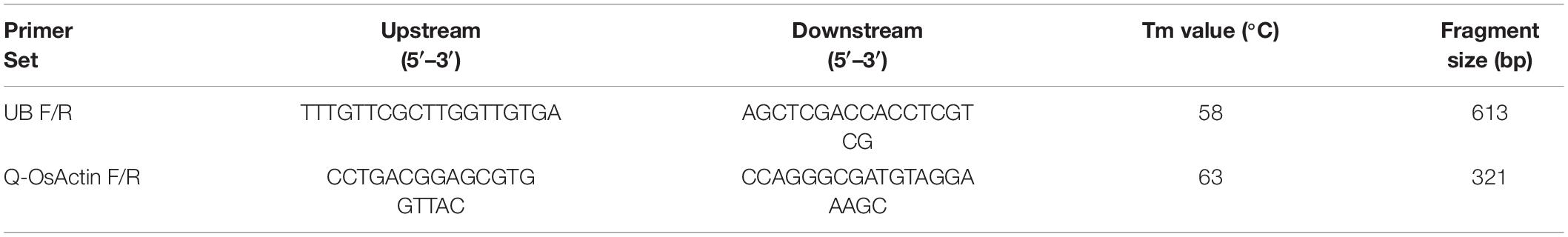
Twenty transgenic rice lines (T1) for each of TaAFP-Ba, TaAFP-Bb, TaAFP-Ba-GFP and TaAFP-Bb-GFP constructs were selected using hygromycin. UBF/R primers were used with the genomic DNA template from candidate lines to validate transgenic events. The frequencies of PCR positive TaAFP-Ba, TaAFP-Bb, TaAFP-Ba-GFP and TaAFP-Bb-GFP transgenic rice lines were 50.0, 70.0, 55.0, and 75.0%, respectively, for their respective transgenes (Table 1).
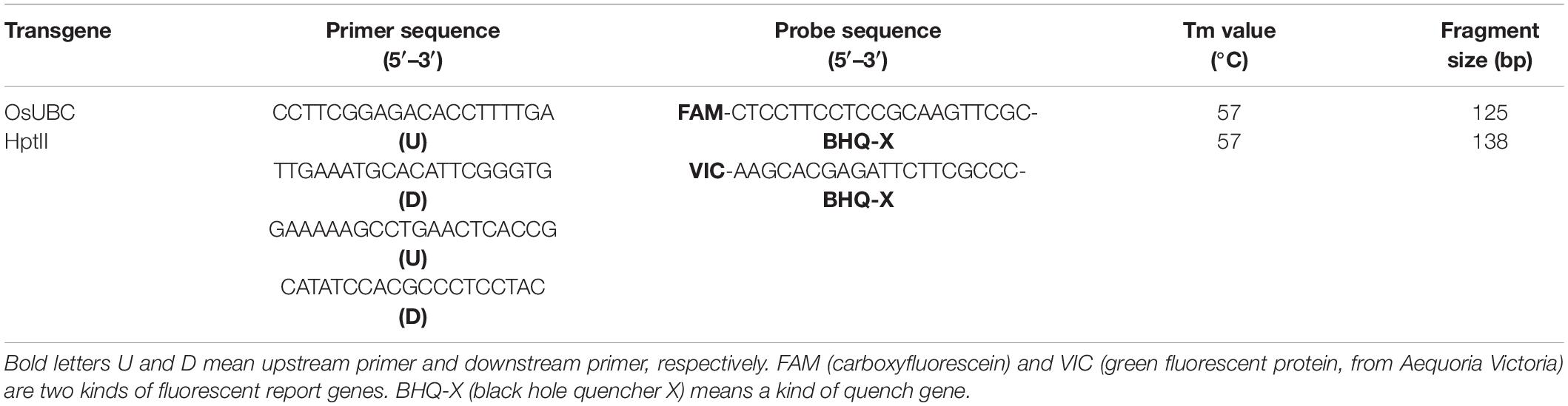
The hptII (hygromycin phosphotransferase II) transgene copy numbers were determined in transgenic plants using droplet digital PCR (ddPCR) and OsUBC reference gene with probe labels of FAM™ and VIC™ (Tables 1, 2). The ddPCR data quality was good as indicated by the significantly different fluorescence values from negative and positive droplets, and no positive droplets were found in the negative control samples (Supplementary Figure 1). The lines had one, two, or three copies of the hptII transgene, and the proportions of single copy lines were 77.8, 66.7, 75.0, and 55.6% in transgenic lines with TaAFP-Ba and TaAFP-Bb, TaAFP-Ba-GFP and TaAFP-Bb-GFP, respectively. The single-copy T1 plants were grown to maturity and T2 seeds were harvested.
Expression of the TaAFP-B Gene in Transgenic Rice
Because transgene copy numbers can influence expression levels, we selected only single-copy transgenic lines for further analysis. Transcript expression levels of TaAFP-B in seeds were measured by qPCR using an empty vector transformant as a control (Figure 1). The transcript levels of TaAFP-Ba and TaAFP-Ba-GFP were lower than those of TaAFP-Bb and TaAFP-Bb-GFP in mature transgenic rice seeds. The expression patterns and transcript levels in different tissues (roots, stems, leaves) of T2 plants 30 DAG (days after germination) and in mature seeds from T2 plants were quantified by qPCR with Q-TaAFP-B-F/R primers (Table 1). The pattern of TaAFP-B expression levels for all four genotypes was similar, in the order of seeds > roots > stems > leaves, and all values for transgenic plants were significantly higher than the empty vector control (Figure 2). Transcript expression levels of TaAFP-Ba-GFP in all four tissues were significantly lower than those of TaAFP-Bb-GFP (Figure 2), whereas the expression levels of TaAFP-B in TaAFP-Ba lines were slightly lower than in TaAFP-Bb lines (Figure 2), suggesting expression level was influenced by presence of the GFP reporter gene.
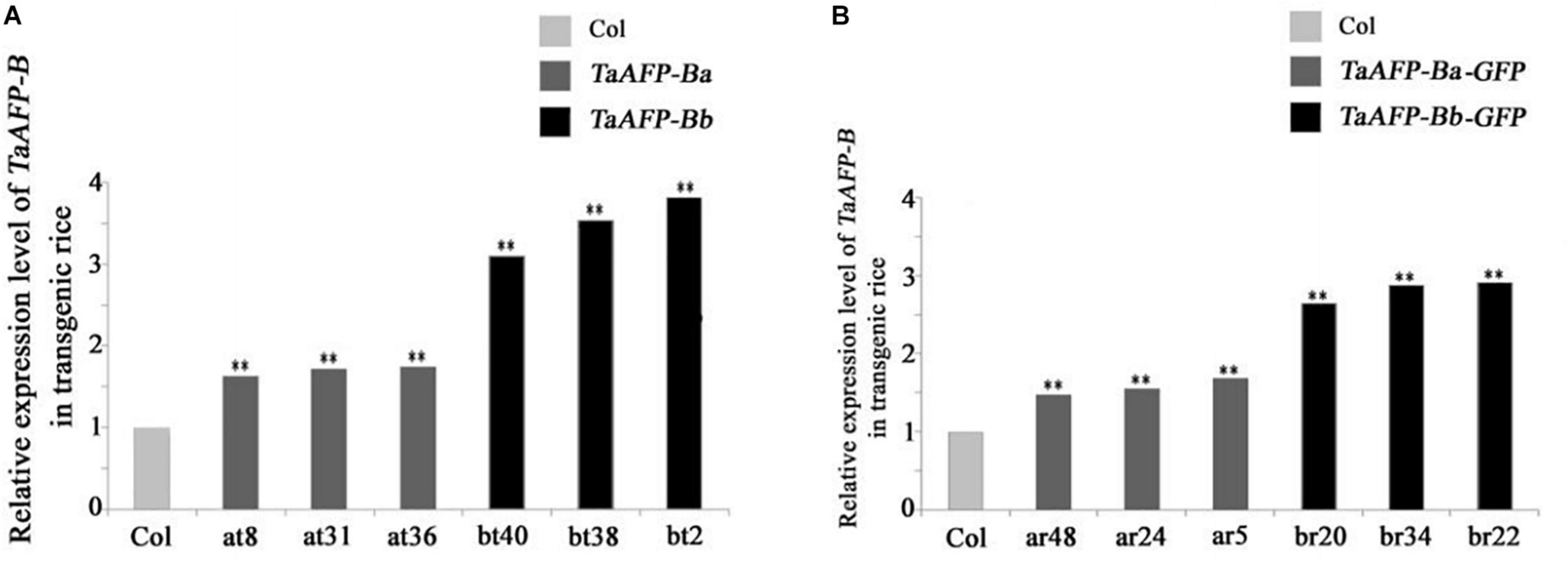
Figure 1. Expression levels of TaAFP-B in seeds of transgenic rice lines with single copies seeds of TaAFP-Ba (A), TaAFP-Bb (A), TaAFP-Ba-GFP (B) and TaAFP-Bb-GFP (B). **Significant differences between transgenic plants and controls at P < 0.01; Col, transgenic lines with an empty vector.
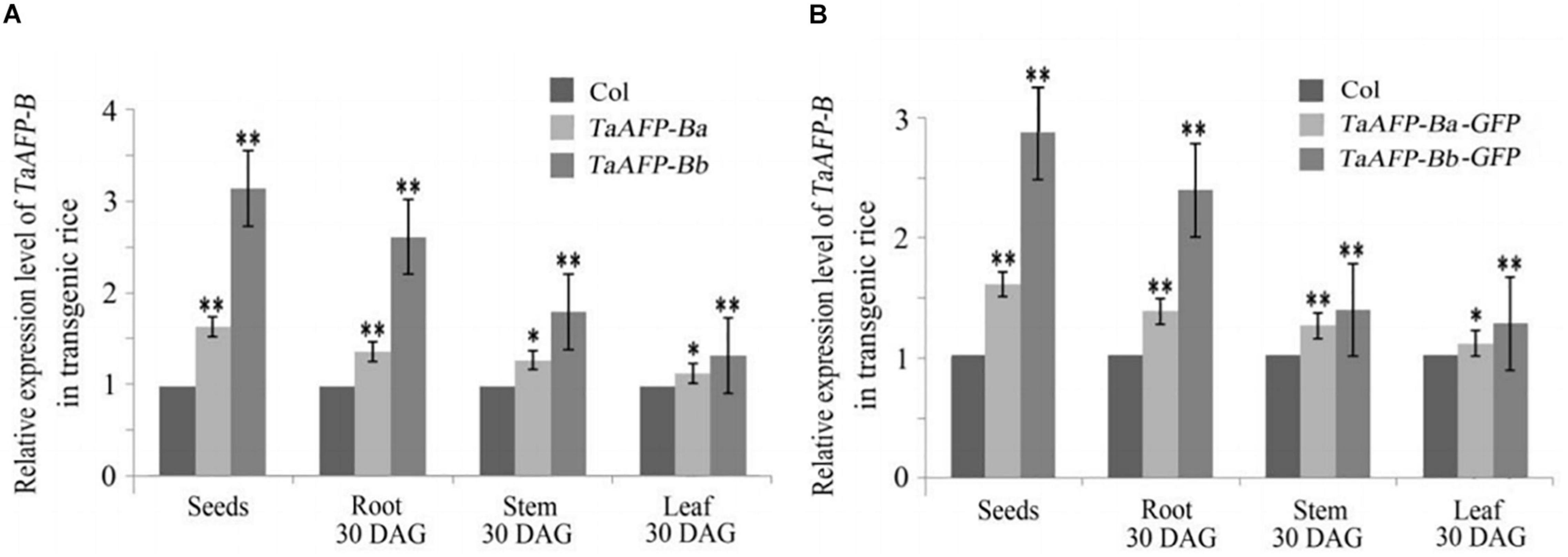
Figure 2. Expression levels of TaAFP-B in different tissues of single-copy transgenic rice of TaAFP-Ba, (A) TaAFP-Bb, (A) TaAFP-Ba-GFP and (B) TaAFP-Bb-GFP. (B) ** and * significant differences between transgenic plants and controls at P < 0.01 and P < 0.05, respectively; Col, transgenic lines with an empty vector.
TaAFP-B Is Induced by Abiotic Stress
To understand how transgenic rice lines behave during abiotic stress response we examined the expression of TaAFP-B in the TaAFP-Ba and TaAFP-Bb lines under different stress conditions. Mature seeds from T2 plants were treated with ABA (3 mM), salt (150 mM NaCl), and mannitol (300 mM to mimic osmotic stress and dehydration) for 1–5 days. Expression levels of TaAFP-B in mature rice seeds with TaAFP-Ba were lower than those with TaAFP-Bb after 1 or 2 days treatment with NaCl, mannitol, and the control treated with water, whereas expression levels of TaAFP-B in the TaAFP-Ba line were higher than those in the TaAFP-Bb line after 1–5 days treatment with ABA, indicating that seeds of the TaAFP-Ba line were more sensitive to exogenous ABA than those of the TaAFP-Bb line. After treatment for 3 d, 4 d, and 5 d, the expression levels of TaAFP-B in TaAFP-Ba line were all higher than in the TaAFP-Bb line after treatment with ABA, NaCl, and mannitol, respectively (Figure 3).
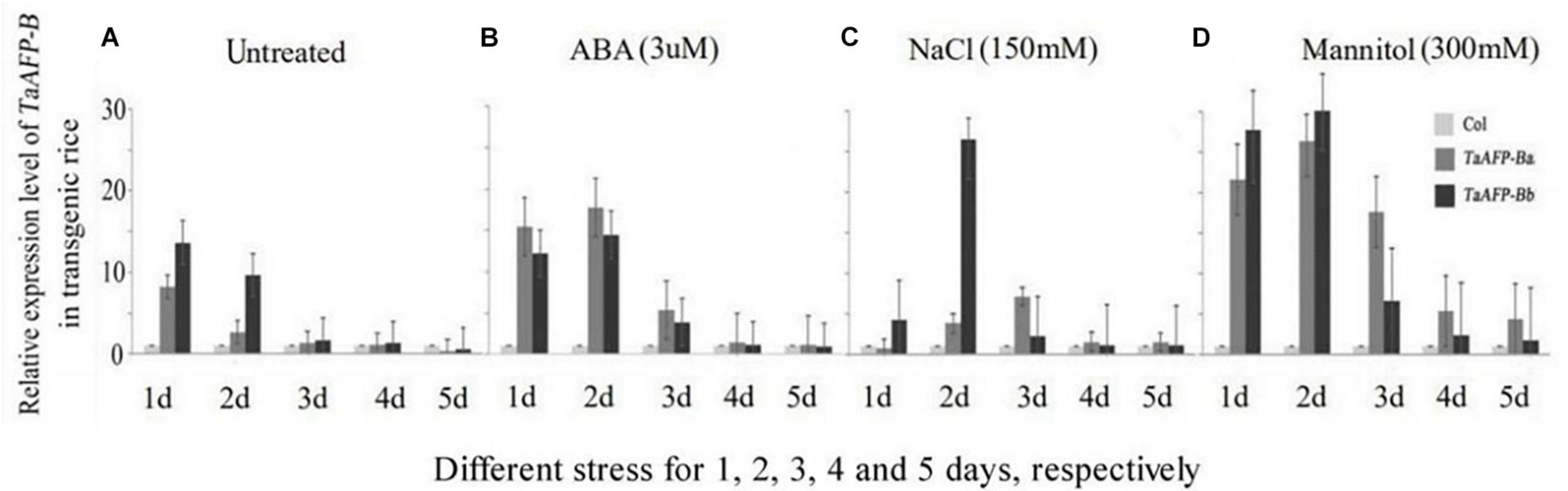
Figure 3. Expression profiles of TaAFP-B by qPCR upon exposure to different stress for 1, 2, 3, 4, and 5 days, respectively, in mature seeds of transgenic rice lines Col, transgenic lines with an empty vector. (A) Untreated; (B) ABA stress treatment; (C) NaCl stress treatment; (D) Mannitol stress treatment.
Green Fluorescent Protein Localization and Western Blotting in Transgenic Rice Carrying TaAFP-B
Green fluorescent protein fluorescence in leaves of 30-day-old transgenic rice plants carrying TaAFP-Ba-GFP and TaAFP-Bb-GFP was visualized using confocal laser scanning microscopy. Intensity and density of GFP fluorescence was weakest in the control with the empty vector and strongest in plants with TaAFP-Bb-GFP. The overall trend of GFP intensity was: TaAFP-Bb-GFP > TaAFP-Ba-GFP > control (Figure 4). Western blotting analysis was performed using an Anti-GFP monoclonal antibody to detect the expression level of TaAFP-B in transgenic rice. The level of TaAFP-B-GFP fusion protein was higher than in TaAFP-Bb-GFP seeds than TaAFP-Ba-GFP seeds (Figure 5) indicating that the 4 bp insertion in the TaAFP-Ba promoter decreased both GFP fluorescence and TaAFP-B-GFP protein levels in mature transgenic rice seeds.
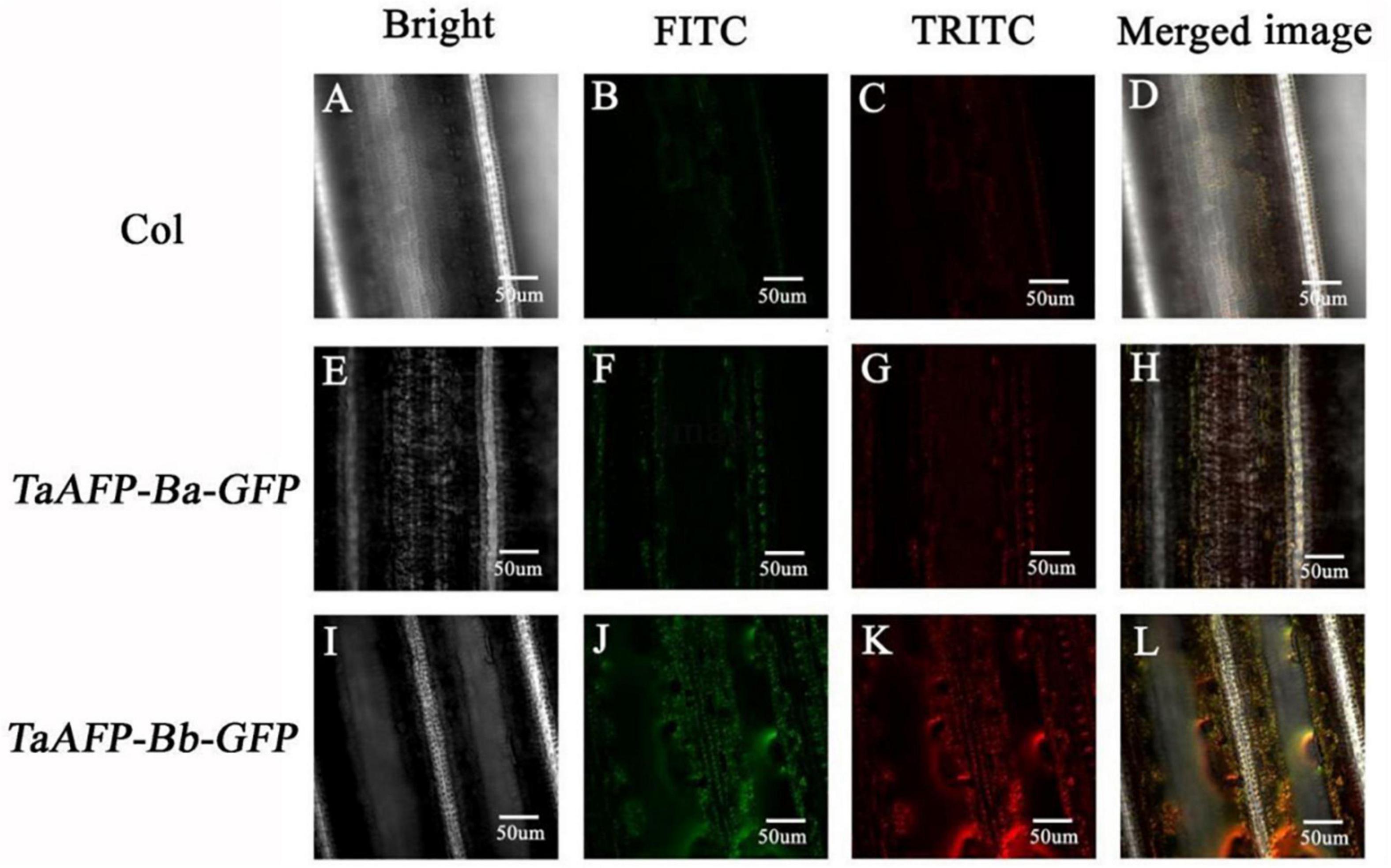
Figure 4. Detection of green (FITC) and red (TRITC) fluorescence in leaves of transgenic rice containing the empty vector (Col), TaAFP-Ba-GFP and TaAFP-Bb-GFP by confocal laser scanning microscopy. Magnification 40X. (A) Brightfield in empty vector transgenic rice leaves. (B) Green fluorescence in empty vector transgenic rice leaves. (C) Red fluorescence in empty vector transgenic rice leaves. (D) Merged images of A–C. (E) Brightfield in TaAFP-BaS-GFP leaves. (F) Green fluorescence in TaAFP-BaS-GFP leaves. (G) Red fluorescence in TaAFP-BaS-GFP leaves. (H) Merged images of E–G. (I) Brightfield in TaAFP-BbS-GFP leaves. (J) Green fluorescence in TaAFP-BbS-GFP leaves. (K) Red fluorescence in TaAFP-BbS-GFP leaves. (L) Merged images of I–K.
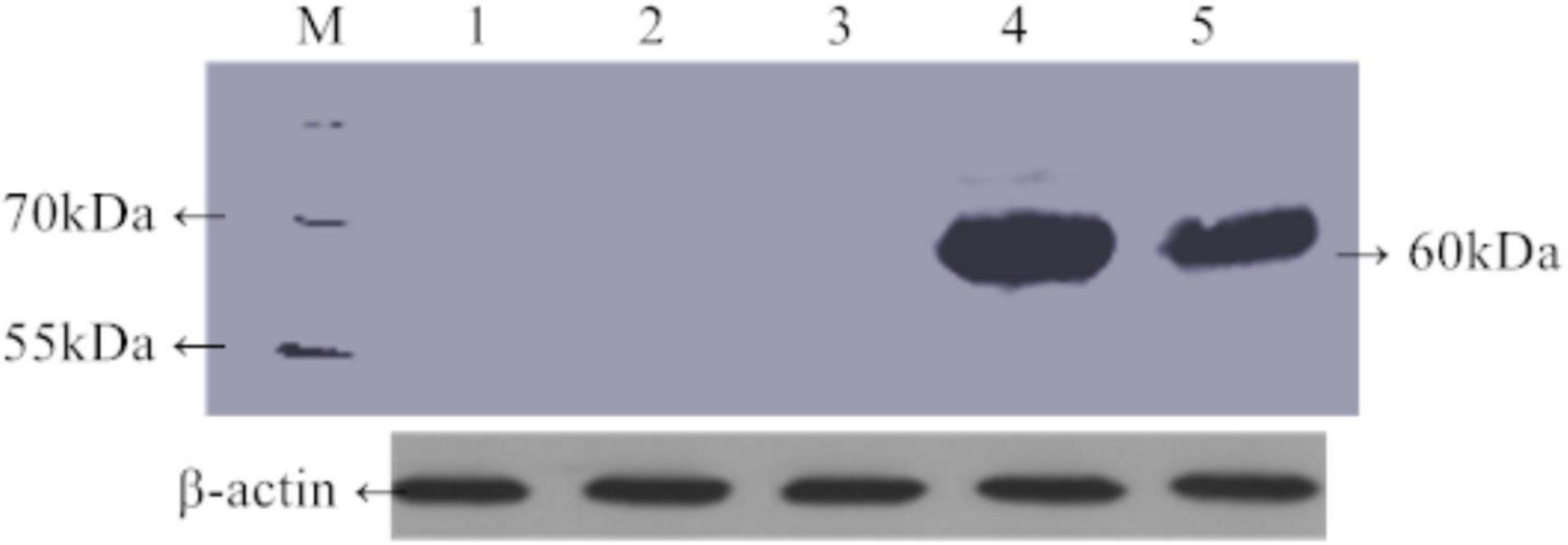
Figure 5. Expression level of TaAFP-B in mature seeds of transgenic rice of TaAFP-Ba-GFP and TaAFP-Bb-GFP using western blotting. M, Prestained protein marker (product#26616). 1–4: Mature seeds of 1, wild type; 2, TaAFP-Bb transformant; 3, TaAFP-Ba transformant; 4, TaAFP-Bb-GFP transformant; 5, TaAFP-Ba-GFP transformant.
Phenotypes of Transgenic Rice Lines
Germination index, plant height, tiller number, 100-seed weight, and length and diameter of stem internodes) of single copy T2 lines were phenotyped and analyzed (Figure 6 and Table 3). Average GI values of TaAFP-Ba and TaAFP-Bb plants were 58.5 and 65.5%, respectively, a significant difference (P < 0.05), compared with the control group (61.7%). The plant heights of 51.6, 59.0 and 59.8 cm for the TaAFP-BaS, TaAFP-BbS and control lines, respectively, showed that TaAFP-Bb lines were significantly shorter (P < 0.05). The lengths of the stem second internodes of the TaAFP-Ba, TaAFP-Bb and control genotypes were 5.2, 3.8 and 5.8 cm, respectively, with the difference between TaAFP-Bb genotype and control being significant (P < 0.05). The average GI values of TaAFP-Ba-GFP and TaAFP-Bb-GFP were 66.1 and 69.1%, respectively, both significantly different from the control (61.7%) (P < 0.05). The lengths of the stem second internodes of TaAFP-Ba-GFP and TaAFP-Bb-GFP were 50.5 and 57.3 cm, respectively, a significant difference (P < 0.01), whereas the length for the control was 59.0 cm. The diameters of the stem first, second and third internodes of TaAFP-Bb-GFP were 2.3, 2.5, and 2.5 mm, respectively, significantly different (P < 0.05) from those of the control (2.9, 3.1, and 3.1 mm, respectively). Analysis of variance indicated that differences in average GI values between TaAFP-Ba and TaAFP-Bb and between TaAFP-Ba-GFP and TaAFP-Bb-GFP genotypes were the most highly significant (P < 0.01) among all measured traits. These data showed that the 4 bp InDel in the TaAFP-B promoter affected not only average GI values in transgenic rice lines, but also internode length and diameters of all three internodes.
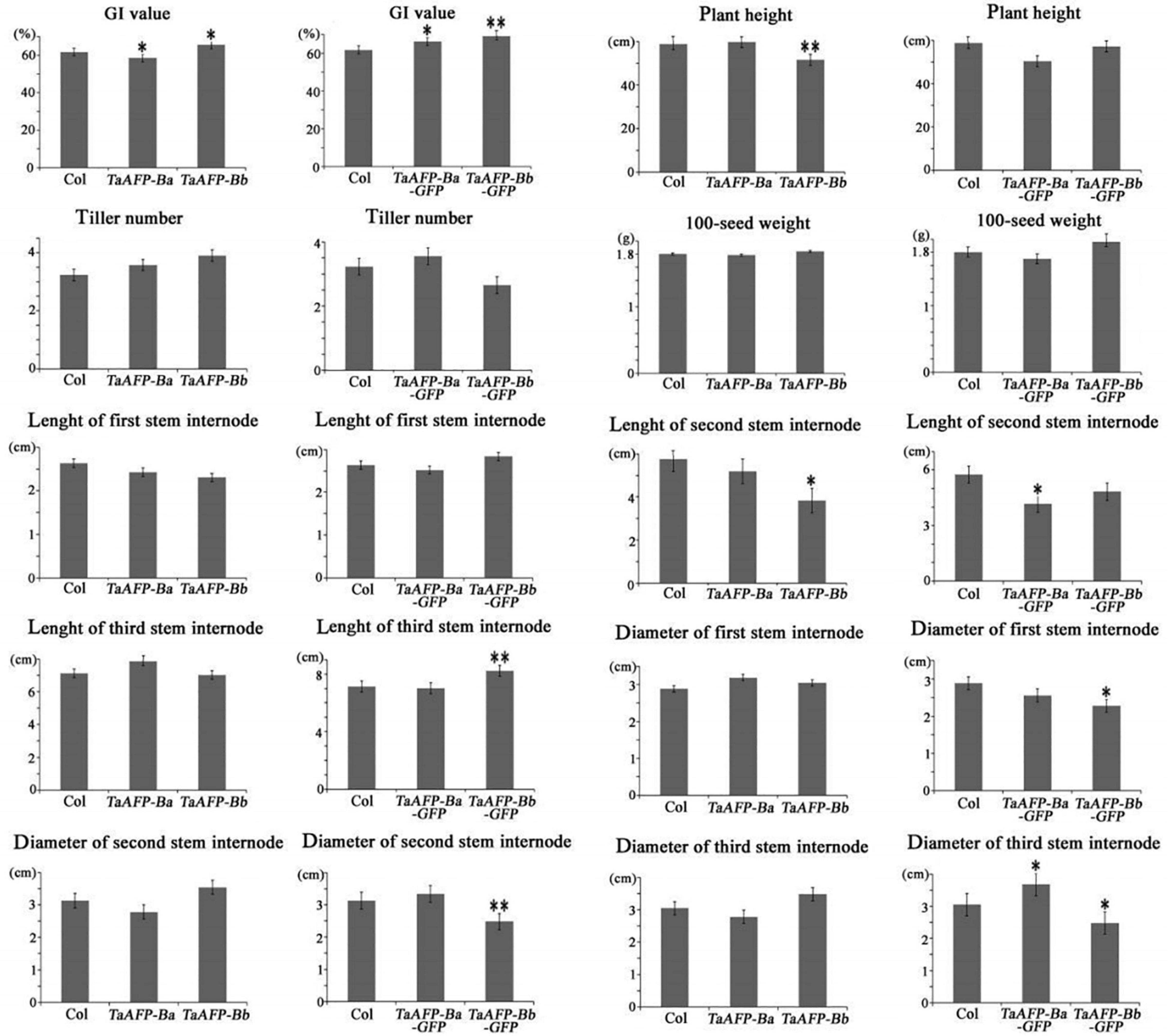
Figure 6. Phenotypic traits from single-copy between TaAFP-Ba and TaAFP-Bb, and between TaAFP-Ba-GFP and TaAFP-Bb-GFP transgenic rice lines. ** and * mean significant differences between transgenic plants and controls at P < 0.01 and P < 0.05, respectively.
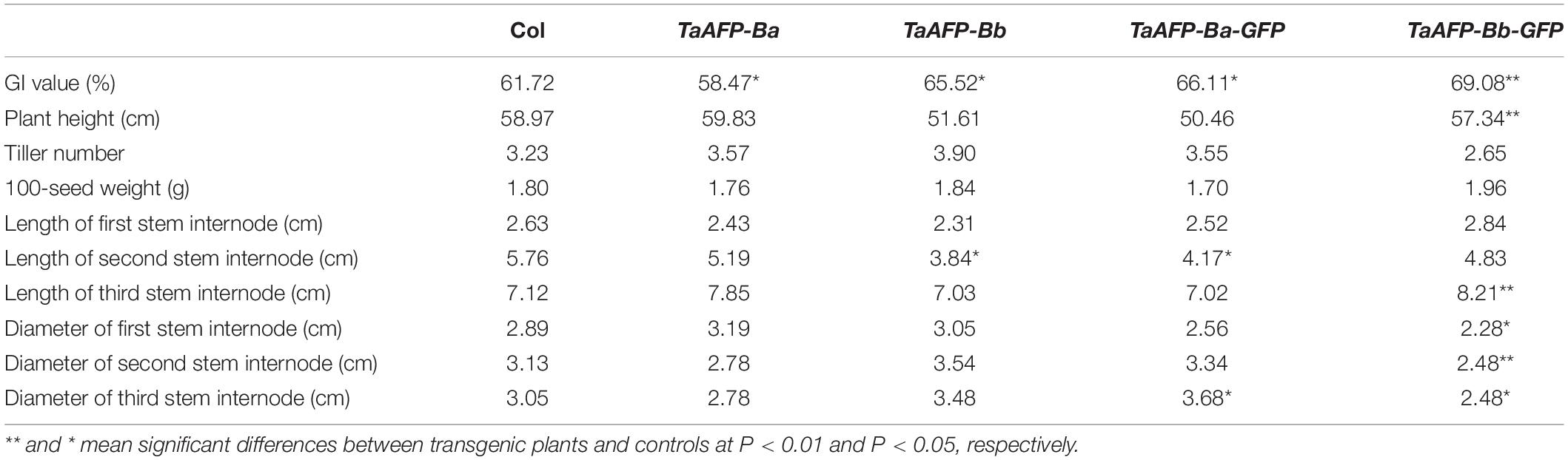
Table 3. Dates of phenotypic traits from single-copy between TaAFP-Ba and TaAFP-Bb, and between TaAFP-Ba-GFP and TaAFP-Bb-GFP transgenic rice lines.
Endogenous ABA content was examined in mature seeds of the TaAFP-Ba, TaAFP-Bb, TaAFP-Ba-GFP and TaAFP-Bb-GFP lines by HPLC (high performance liquid chromatography). The average values of endogenous ABA in seeds of the TaAFP-Ba and TaAFP-Bb lines at 0.8 and 0.5 μg/g, respectively, were significantly different (P < 0.05), and the value for the TaAFP-Ba-GFP line (0.6 μg/g) was also higher than that of the TaAFP-Bb-GFP line (0.5 μg/g) (Figure 7). Thus, the 4 bp insertion in the TaAFP-Ba promoter led to a higher endogenous ABA content in mature seeds of transgenic rice.
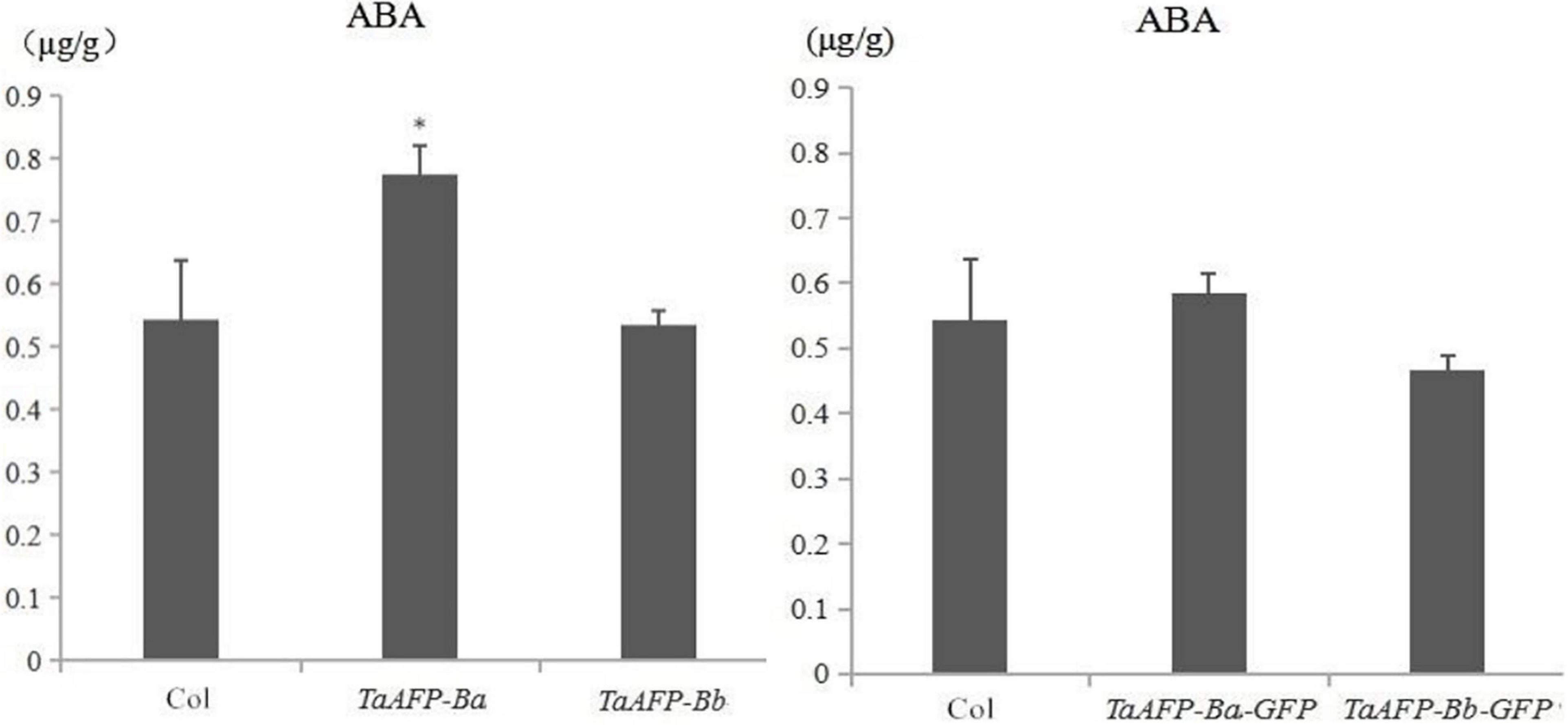
Figure 7. Endogenous ABA contents in mature seeds of transgenic rice lines containing TaAFP-Ba/b, TaAFP-Ba/b, TaAFP-Ba-GFP and TaAFP-Bb-GFP determined by HLPC. *Significant difference between transgenic lines containing TaAFP-Ba and TaAFP-Bb at P < 0.05.
Discussion
Transcript levels of the TaAFP-Ba allele were markedly higher than those of the TaAFP-Bb in different tissues and were correlated with the observed GUS activity in ProTaAFP-Ba:GUS transgenic rice (Feng et al., 2019). Those results suggested that the 4 bp InDel in the 5′UTR of TaAFP-B affected gene expression and function. The present analysis showed (1) that transcript levels of TaAFP-B were lower in transgenic rice lines containing TaAFP-Ba and TaAFP-Ba-GFP than in lines with TaAFP-Bb and TaAFP-Bb-GFP (Figure 2); (2) that the GFP fusion protein levels in seeds of lines containing TaAFP-Ba-GFP was lower than in those with TaAFP-Bb-GFP (Figures 4, 5); and (3) that average GI values of lines with TaAFP-Ba and TaAFP-Ba-GFP lines were lower than those of lines containing TaAFP-Bb and TaAFP-Bb-GFP (Figure 6). Taken together, these results showed that the higher transcript expression and protein levels of TaAFP-Bb led to higher GI values in mature transgenic rice seed. Thus, the 4 bp InDel in the 5′UTR of TaAFP-B had a significant effect on expression level of TaAFP-B which, in turn, influenced the GI value of mature seeds and several other traits.
ABA plays an important role in seed maturation and dormancy (Gubler et al., 2005; Nakashima et al., 2006). Accumulation and activity of AFP in Arabidopsis are induced by ABA during seed germination (Lopez-Molina et al., 2002). It was shown that a negative correlation is present between the germination ability of embryos and the sensitivity of embryos to ABA. Moreover, there is evidence that ABA levels in mature wheat embryos are similar in both PHS-sensitive and -resistant cultivars, and that sprouting behavior is related more to the extent of ABA sensitivity of embryos than to actual endogenous ABA content (Walker-Simmons and Sesing, 1990). In this study, the transcript expression levels of TaAFP-B in mature seeds of transgenic lines carrying TaAFP-Ba and TaAFP-Bb were determined following treatment with ABA. Our results indicated that TaAFP-B transcript expression levels in the transgenic lines had different sensitivities to ABA (Figure 3) that not only affected dormancy but also plant height and length and diameter stem internodes. The 4 bp InDel in the promoter of TaAFP-B not only affected the mRNA transcription level, mRNA decay, translation levels of GUS, tdTomatoER, and GUS activity in wheat (Feng et al., 2019), but also transcript and translation levels of TaAFP-B, sensitivity to ABA, content of endogenous ABA, and average GI values in mature seeds of transgenic rice. The allelic variation of TaAFP-B was markedly associated with seed dormancy and therefore pre-harvest sprouting tolerance in wheat. Furthermore, the 4-bp InDel in the promoter of TaAFP-B also affected markedly the plant height, lengths of the second and third internodes, diameters of the first to third internodes, but these values of phenotypes have not a same trend between transgenic rice plants of TaAFP-Ba and TaAFP-Bb, and between TaAFP-Ba-GFP and TaAFP-Bb-GFP (Figure 6), which may be a reason of GFP fusion protein affecting these phenotypes. In addition, the change in sensitivity to ABA in TaAFP-Ba transgenic rice might also be responsible for changes of lengths and diameters of the second and third stem internodes and plant height in transgenic rice, because ABI5 is a key positive factor in ABA signaling pathway in plants, which has the capability of binding the SbGA2ox3 5′-regulatory region and promote SbGA2ox3 protein accumulation, resulting in degradation of GA4 in sorghum (Renata et al., 2013); in addition, during phyB-dependent inhibition of germination in Arabidopsis, AtABI5 interacts with AtGA3ox1 and AtGA3ox2, but suppresses their expression instead, which results in lower GA levels in the seed (Lee et al., 2012). These proofs of ABI5 function showed a cross-talk between function of ABI5, ABA signaling and GA metabolism, corroborating the fact that TaAFPs were orthologs of AtAFP (Ohnishi et al., 2008), so it is deduced that there is also indirect relationship between function of TaAFP, ABA signaling and GA metabolism. The change in sensitivity to ABA in TaAFP-Ba transgenic rice might lead to change of GA content and further affect the lengths and diameters of the second and third stem internodes and plant height in transgenic rice, but these requires further investigation.
There was a difference in transcript levels of TaAFP-Ba and TaAFP-Bb in common wheat and transgenic rice. In a previous study in common wheat (Feng et al., 2019), trend of transcript levels of TaAFP-B in seeds at different days after pollination was always TaAFP-Ba > TaAFP-Bb compared with TaAFP-Bb > TaAFP-Ba in the present work with transgenic rice. The reasons for the different trend of transcript expression levels of TaAFP-Ba and TaAFP-Bb between wheat and transgenic rice might be associated with different genetic backgrounds of these two species or result of TaAFP-B overexpression. Moreover, the transcript levels of TaAFP-B in TaAFP-Ba transgenic rice seeds treated with ABA were more sensitive than those in the TaAFP-Bb genotype (Figure 3). Endogenous ABA content in mature seeds of the transgenic lines measured by atomic absorption showed that the endogenous ABA content was higher in mature seeds containing TaAFP-Ba and TaAFP-Ba-GFP than those containing TaAFP-Bb and TaAFP-Bb-GFP (Figure 7). It can therefore be deduced that the level of endogenous ABA in mature seeds of transgenic rice had been altered by overexpression of TaAFP-B. ABI5 is a positive regulatory factor in the ABA signaling pathway (Lopez-Molina et al., 2001) and regulates the maturation and germination of seeds at later stages of maturity (Lopez-Molina et al., 2001, 2002; Carles et al., 2002), and AFP is a novel negative regulator of ABA signaling that attenuates ABA signaling by targeting ABI5 for ubiquitin-mediated degradation in nuclear bodies (Lopez-Molina et al., 2003). The higher level of endogenous ABA in mature seed of the transgenic line with TaAFP-Ba is attributed to less TaAFP-B transcript and less TaAFP-B protein that lead to higher seed dormancy.
Conclusion
The 4 bp insertion in the promoter of TaAFP-Ba decreased the transcript expression and translation level in transgenic rice. Increased endogenous ABA content in mature seeds and affected their sensitivity to ABA leading to higher seed dormancy and PHS tolerance, and also effected the diameter and length of the second and third internodes and plant height. These results provide evidence for potential application of the allelic variation in TaAFP-B for improvement of PHS and lodging tolerance in wheat.
Materials and Methods
Plasmid Construction
The pCAMBIA1390-Ubi-GFP vector was kindly supplied by Professor Lanqin Xia, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (CAAS). The sequences of TaAFP-Ba and TaAFP-Bb were 1,058 bp (105 bp fragment of 5′UTR and 953 bp CDS) and 1,054 (101 bp fragment of 5′UTR and 953 bp CDS) bp, respectively (the difference due to a 4 bp InDel in the 5′UTR). The sequence of the TaAFP-B alleles were amplified and cloned into vectors pCAMBIA1390-Ubi and pCAMBIA1390-Ubi-GFP by General Biosystems (Anhui) Co., Ltd., respectively.
Transformation of Rice Plants
Agrobacterium tumefaciens strain EHA105 was used to genetically transform embryonic calli of rice (Oryza sativa L. ssp. japonica cultivar Nipponbare, kindly supplied by Professor Lanqin Xia, CAAS) with previously described recombinant vectors (Feng et al., 2019). Transformed calli were screened and planted on regeneration medium under hygromycin selection (50 mg/L) to acquire T1 plants.
Identification of Transgenic Plants and Copy Number Determination
All transgenic rice lines were planted in a greenhouse and genomic DNA was isolated from bulked young leaf samples from 5 plants of each T1 line according to the guidelines for the DNA Secure Plant Kit (TIANGEN Biotech). PCR was performed as described previously to identify positive transgenic lines (Feng et al., 2019). Amplified PCR fragments and primer sets are listed in Table 1. The quality of genomic DNA was determined using a BioDrop spectrophotometer, and copy numbers were detected using ddPCR (Collier et al., 2017, Shanghai Biotechnology Co., Ltd.). A primer pair and probe were designed to detect a unique single-copy insertion for target and endogenous reference genes, respectively. The target and endogenous reference genes were hptII and OsUBC, respectively (Table 2). Reference gene amplification was detected using a FAM™-labeled probe whereas target gene amplification was detected with the VIC™-labeled probe. The ddPCR reaction mixture consisted of 2 μL DNA, 10 μL of 1X QX200 EvaGreen Supermix (Bio-Rad), 450 nM of each primer pair, and 250 nM of each probe. Thermal cycling conditions were 95°C for 10 min, followed by 40 cycles of 94°C for 30s and 60°C for 30s, 98°C for 10 min, and a final hold at 4°C. The detailed experimental procedures were referenced from McCord (2016) and Collier et al. (2017). We planted and identified at least three positive independent single-copy lines for each transgenic type. Plants for subsequent analysis were grown at 25°C under 16/8 h light/darkness in a greenhouse at the Anhui Academy of Agricultural Sciences Base for transgenic rice.
Transgenic Plant Phenotyping
The single-copy positive plants of each transgenic rice line were selected for phenotypic analysis after qPCR. The T2 generations, 42 plants from three single-copy positive lines of transgenic rice of TaAFP-Ba, TaAFP-Bb, TaAFP-Ba-GFP and TaAFP-Bb-GFP, respectively, were analyzed for germination index, plant height, tiller number, 100-seed weight, and stem length diameter.
Gene Expression Analysis
Total RNA was extracted from 5 g samples of roots, stems, leaves and seeds, respectively, with the TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa). First-strand cDNA was synthesized and qPCR was carried out using a LightCycler®480 System Real-Time PCR as described by Feng et al. (2019). All primers for the target genes are listed in Table 1. Relative expression of target genes was evaluated by the 2–ΔΔCt method (Li et al., 2018). The experiments were conducted in three biological replications.
Germination Assays and Stress Treatments
The seeds from single-copy T2 lines were sterilized in 75% ethyl alcohol and were sown in a growth chamber with a 16/8 h light/darkness cycle at 28°C. For germination assays, 100 seeds were germinated and grown on solid 1/2-MS medium for 7 days. The GI value for each transgenic line were calculated according to the number of germinated seeds on a daily basis (Walker-Simmons, 1988). For stress treatments, the seeds were incubated on solid 1/2-MS medium including 3 μM ABA, 150 mM NaCl (for salt stress), or 300 mM mannitol (for dehydration stress), respectively, for 1, 2, 3, 4, and 5 days. Transgenic seeds from empty vector transformants was used as controls and each experiment included three biological replicates. All samples were collected after each treatment and frozen in liquid nitrogen and stored at −80°C.
Fluorescent Protein Expression and Western Blotting
FITC (fluorescein isothiocyanate) and TRITC (tetramethylrho damine) filters were used to assess green fluorescence for GFP and red fluorescence for chlorophyll autofluorescence from chloroplasts. The fluorescence intensity of merged images with orange color revealed the expression of GFP. We selected young leaves of single copy positive plants at 30 days after germination (DAG) to analyze the expression of GFP in transgenic rice with TaAFP-Ba-GFP and TaAFP-Bb-GFP constructs mounted on covered glass slides by an Olympus BX-60 of Confocal Laser Scanning Microscope. We used excitation lasers at 488 and 568 nm to excitate GFP and chlorophyll, respectively; images were collected by FITC and TRITC filters, and single-channel images were superimposed to observe the expression of GFP in leaves. At the same time, Anti-GFP (TransGen Biotech) was selected as a probe to detect the TaAFPB fusion protein in transgenic rice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YF and YH performed the experiments and wrote the manuscript. BH assisted in performing experiments. YZ planted the experimental materials and determined the GI values. YY and YX designed the experiments and assisted in writing the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31760382), the Inner Mongolia Natural Science Foundation (Grant Nos. 2019MS03016 and 2019MS03067), and the Science and Technology Project of Henan Province (Grant Nos. 152102110136 and 172102110076). The funding source had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to R. A. McIntosh, Plant Breeding Institute, University of Sydney, for critical review of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.837805/full#supplementary-material
Abbreviations
PHS, pre-harvest sprouting; ABA, abscisic acid; GA, gibberellins; Vp-1, viviparous-1; LEA, late embryogenesis abundant; InDel, insertion-deletion; UTR, untranslated region; GFP, green fluorescent protein; FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine; HPLC, high performance liquid chromatography.
References
Carles, C., Bies-Etheve, N., Aspart, L., Léon-Kloosterziel, K. M., Koornneef, M., Echeverria, M., and Delseny, M. (2002). Regulation of Arabidopsis thaliana Em genes role of ABI5. Plant J. 30, 373–383. doi: 10.1046/j.1365-313x.2002.01295.x
Collier, R., Dasgupta, K., Xing, Y. P., Hernandez, B. T., Shao, M., Rohozinski, D, et al. (2017). Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 90, 1014–1025. doi: 10.1111/tpj.13517
Feng, Y. M., Liu, M., Wang, Z., Zhao, X. L., Han, B., Xing, Y. P., et al. (2019). A 4-bp deletion in the 5′UTR of TaAFP-B is associated with seed dormancy in common wheat (Triticum aestivum L.). BMC Plant Biol. 19:349. doi: 10.1186/s12870-019-1950-4
Finkelstein, R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutation. Plant J. 5, 765–71. doi: 10.1046/j.1365-313X.1994.5060765.x
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.2307/3871072
Finkelstein, R. R., Wang, M. L., Lynch, T. J., Rao, S., and Good-man, H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. doi: 10.1105/tpc.10.6.10
Garcia, M.E., Lynch, T., Peters, J., and Snowden, C, Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658. doi: 10.1007/s11103-008-9344-2
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., and Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1910. doi: 10.2307/3871085
Groos, C., Gay, G., Perretant, M. R., Gervais, L., Bernard, M., Dedryver, F., and Charmet, G. (2002). Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor. Appl. Genet. 104, 39–47. doi: 10.1007/s001220200004
Gubler, F., Millar, A. A., and Jacobsen, J. V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8, 183–187. doi: 10.1016/j.pbi.2005.01.011
Hoffmann, F. (1997). Plant dormancy: physiology, biochemistry and molecular biology. Plant Sci. 125, 231–232. doi: 10.1016/S0168-9452(97)00062-9
Humphreys, D. G., and Noil, J. (2002). Methods for characterization of pre-harvest sprouting resistance in a wheat breeding program. Euphytica 126, 61–65. doi: 10.1023/A:1019671622356
Koornneef, M., Reuling, G., and Karssen, C. M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61, 377–83. doi: 10.1111/j.1399-3054.1984.tb06343.x
Lee, Y., and Kende, H. (2001). Expression of beta-expansins is correlated with internodal elongation in deep water rice. Plant Physiol. 127, 645–654. doi: 10.1104/pp.108.125518
Lee, K. P., Piskurewicz, U., Turecková, V., Carat, S., Chappuis, R., Strnad, M., et al. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26, 1984–1996. doi: 10.1101/gad.194266.112
Li, C. D., Ni, P. X., Francki, M., Hunter, A., Zhang, Y., Schibed, D., et al. (2004). Genes controlling seed dormancy and preharvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genom. 4, 84–93. doi: 10.1007/s10142-004-0104-3
Li, H., Li, J., Xu, R. F., Qin, R. Y., Song, F. S., Li, L., et al. (2018). Isolation of five rice non-endosperm tissue-expressed promoters and evaluation of their activities in transgenic rice. Plant Biotech. J. 16, 1138–1147. doi: 10.1111/pbi.12858
Liu, H. Y., Yu, X., Cui, D. Y., Sun, M. H., Sun, W. N., Tang, Z. C., et al. (2007). The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 17, 638–49. doi: 10.1038/cr.2007.34
Lopez-Molina, L., and Chua, N. H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. doi: 10.1093/pcp/41.5.541
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A post germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–7. doi: 10.1073/pnas.081594298
Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N. H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. doi: 10.1101/gad.1055803
Lopez-Molina, L., Mongrand, S., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–323. doi: 10.1046/j.1365-313x.2002.01430.x
McCarty, D. R., Carson, C. B., Stinard, P. S., and Robertson, D. S. (1989). Molecular analysis of VIVIPAROUS-1: an abscisic acid insensitive mutant of maize. Plant Cell 1, 523–532. doi: 10.1105/tpc.1.5.523
McCarty, D. R., Hattori, T., Carson, C. B., Vasil, V., Lazar, M., and Vasil, I. K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. doi: 10.1016/0092-8674(91)90436-3
McCord, P.H. (2016). Using droplet digital PCR (ddPCR) to detect copy number variation in sugarcane, a high-level polyploid. Euphytica 209, 439–448. doi: 10.1007/s10681-016-1657-7
McKibbin, R. S., Wilkinson, M. D., Bailey, P. C., Flintham, J. E., Andrew, L. M., Lazzeri, P. A., et al. (2002). Transcripts of Vp-1 homologues are mis spliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. U.S.A. 99, 10203–10208. doi: 10.1073/pnas.152318599
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes, including RD29B and RD29A in seeds, germinating embryos and seedlings of Arabidopsis. Plant Mol. Biol. 60, 50–68. doi: 10.1007/s11103-005-2418-5
Ohnishi, N., Himi, E., Yamasaki, Y., and Noda, K. (2008). Differential expression of three ABA insensitive five binding protein (AFP)-like genes in wheat. Genes Genet. Syst. 83, 167–177. doi: 10.1266/ggs.83.167
Oikawa, T., Koshioka, M., Kojima, K., Yoshida, H., and Kawata, M. (2004). A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 55, 687–700. doi: 10.1007/s11103-004-1692-y
Parcy, F., Valon, C., and Raynal, M. (1994). Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic Acid. Plant Cell 6, 1567–1582. doi: 10.2307/3869944
Renata, C., Daniel, C. C., Luis, B. A. R., and Verónica, R.M. (2013). In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 64, 5721–5735. doi: 10.1093/jxb/ert347
Simsek, S., Ohm, J. B., Lu, H., Rugg, M., Berzonsky, W., Alamri, M. S., and Mergoum, M. (2014). Effect of pre-harvest sprouting on physicochemical changes of proteins in wheat. J. Sci. Food Agr. 94, 205–212. doi: 10.1002/jsfa.6229
Stone, S. L., Williams, L. A., and Farmer, L. (2006). Keep on going a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Sugimoto, K., Takeuchi, Y., Ebana, K., Miyao, A., Hirochika, H., Hara, N., et al. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. U.S.A. 107(13), 5792–5797. doi: 10.1073/pnas.0911965107
Sun, Y. W., Yang, Y., Shewry, P. R., Jones, H. D., and Xia, L. Q. (2012). Isolation and characterization of Viviparous-1 haplotypes in wheat related species. Euphytica 188, 71–84. doi: 10.1007/s10681-012-0659-3
Walker-Simmons, M. K. (1988). Enhancement of ABA responsiveness in wheat embryos at higher temperature. Plant Cell Environ. 11, 769–775.
Walker-Simmons, M., and Sesing, J. (1990). Temperature effects on embryonic abscisic acid levels during development of wheat grain dormancy. Plant Growth Regul. 9, 51–56. doi: 10.1007/BF02041941
Yang, Y., Ma, Y. Z., Xu, Z. S., Chen, X. M., He, Z. H., Yu, Z., et al. (2007b). Isolation and characterization of Vp-1 genes in wheat varieties with distinct pre-harvest sprouting tolerance and ABA sensitivity. J. Exp. Bot. 58, 2863–2871. doi: 10.1093/jxb/erm073
Yang, Y., Zhang, C. L., Liu, S. X., Sun, Y. Q., Meng, J. Y., and Xia, L. Q. (2014). Characterization of the rich haplotypes of Viviparous-1A in Chinese wheats and development of a novel sequence-tagged site marker for pre-harvest sprouting resistance. Mol. Breed. 33, 75–88. doi: 10.1007/s11032-013-9935-8
Yang, Y., Zhao, X. L., Xia, L. Q., Chen, X. M., Xia, X. C., Yu, Z., and He, Z. H. (2007a). Development and validation of a Viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor. Appl. Genet. 115, 971–980. doi: 10.1007/s00122-007-0624-z
Keywords: agrobacterium-mediated transformation, allelic variation, InDel, pre-harvest sprouting, wheat
Citation: Feng Y, Han Y, Han B, Zhao Y, Yang Y and Xing Y (2022) A 4 bp InDel in the Promoter of Wheat Gene TaAFP-B Affecting Seed Dormancy Confirmed in Transgenic Rice. Front. Plant Sci. 13:837805. doi: 10.3389/fpls.2022.837805
Received: 17 December 2021; Accepted: 21 February 2022;
Published: 31 March 2022.
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Shingo Nakamura, Institute of Crop Science (NARO), JapanSven Krishnan Nelson, Heliponix, LLC, United States
Copyright © 2022 Feng, Han, Han, Zhao, Yang and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Yang, eWFuZ3lhbmNodXRhb0AxMjYuY29t; Yanping Xing, eHlwaW5nODMxNUAxNjMuY29t
†These authors have contributed equally to this work
 Yumei Feng1,2†
Yumei Feng1,2† Yan Yang
Yan Yang Yanping Xing
Yanping Xing