
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 29 April 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.837563
This article is part of the Research Topic Insights in Plant Biotechnology: 2021 View all 22 articles
Orchids have a huge reservoir of secondary metabolites making these plants of immense therapeutic importance. Their potential as curatives has been realized since times immemorial and are extensively studied for their medicinal properties. Secondary metabolism is under stringent genetic control in plants and several molecular factors are involved in regulating the production of the metabolites. However, due to the complex molecular networks, a complete understanding of the specific molecular cues is lacking. High-throughput omics technologies have the potential to fill up this lacuna. The present study deals with comparative analysis of high-throughput transcript data involving gene identification, functional annotation, and differential expression in more than 30 orchid transcriptome data sets, with a focus to elucidate the role of various factors in alkaloid and flavonoid biosynthesis. Comprehensive analysis of the mevalonate (MVA) pathway, methyl-D-erythritol 4-phosphate (MEP) pathway, and phenylpropanoid pathway provide specific insights to the potential gene targets for drug discovery. It is envisaged that a positive stimulation of these pathways through regulation of pivotal genes and alteration of specific gene expression, could facilitate the production of secondary metabolites and enable efficient tapping of the therapeutic potential of orchids. This further would lay the foundation for developing strategies for genetic and epigenetic improvement of these plants for development of therapeutic products.
Orchids are members of one of the most advanced plant families, the Orchidaceae with their unique morphology (labellum, gynostemium), functional characteristics, and ecological adaptations (mycorrhizal association, and velamen) that are not found in model plants. Though popular as affluent ornamentals, orchids were first discovered for their therapeutic properties. The restorative properties of orchids have been well documented since times immemorial, Theophrastus in his book named “Enquiry into Plants” reported the use of orchids as therapeutics. These plants have also found reference in Indian and Chinese traditional pharmacopeia. In Indian Ayurvedic system of medicine, “Ashtavarga” is an important formulation, consisting of eight herbs, out of which four are orchids, that is, Habenaria edgeworthii (vriddhi), Habenaria intermedia (riddhi), Malaxis acuminata (jeevaka), and Malaxis muscifera (rishibhak). Similarly, in Chinese medicine, Anoectochilus roxburghii has been promoted as “King medicine” to treat snake bites, lung and liver disease, and hypertension (He et al., 2006). “Shi-Hu,” an orchid-based therapeutic formulation, prepared from Dendrobium nobile and allied species, is prized as a tonic because of its efficiency in treating lung, kidney, and stomach diseases, hyperglycemia, and diabetes (Bulpitt et al., 2007). “Tian-Ma” derived from tubers of Gastrodia elata is effectively used in the treatment of headaches, migraines, epilepsy, high blood pressure, rheumatism, fever, and nervous problems (Kong et al., 2003). In addition to their use as therapeutics, these plants have also been widely used as tonics and restoratives. The most important example is Dactylorhiza hatagirea which is used as an aphrodisiac (Lawler, 1984). Several orchids, such as Shwethuli (Zeuxine strateumatica) and Salabmisri (Eulophia dabia), Vanda testacea, and Rhynchostylis retusa, are used as aphrodisiacs, blood purifiers, general restorative tonics, and for treating rheumatism, piles, bronchitis, and inflammations (Chauhan, 1990; Vij et al., 2013; Hossain et al., 2020; Figure 1). These healing and restorative properties are due to the presence of a rich diversity of phytochemicals which are bioactive and are responsible for the pharmacognostic potential of these plants (Teoh, 2016).

Figure 1. Some therapeutically important orchid species. (A), Eulophia dabia (D.Don) Hochr.; (B), Zeuxine strateumatica (L.) Schltr.; (C), Dactylorhiza hatagirea (D.Don) Soó; (D), Malaxis muscifera (Lindl.) Kuntze; (E), Habenaria intermedia D.Don; (F), Habenaria pectinata D.Don; (G), Vanda testacea (Lindl.) Rchb.f.; (H), Platanthera edgeworthii (Hook.f. ex Collett) R.K.Gupta; (I), Crepidium acuminatum (D.Don) Szlach.
The integration of traditional knowledge with modern research can pave a way as promising leads for the discovery of novel drugs with greater therapeutic potential than synthetic medicine offering new horizons in the field of therapeutics and drug discovery. However, the studies in this direction are not commensurate with the immense potential of these plants. This is mainly due to lack of complete understanding of the spectrum of molecular networks of secondary metabolism. Even though there have been a number of studies on the phytochemical profiling and the biological activity, there is limited information about the regulating molecular cues and the alternate biosynthetic routes which are utilized in these plants as a survival strategy in harsh and dynamic climatic conditions. Various omics approaches can be instrumental to understand and elucidate these complex mazes and help in utilization of these plants as therapeutics to their fullest potential.
Recent times have revolutionized the process of deciphering the genetic identity of the germplasm by using minimal amount of tissues to generate humongous volume of data using transcriptomic approach. Genome editing with the help of transcriptomic sequencing provide extra choices for genetic improvement in orchids. For techniques like CRISPR/Cas9, the sequence of the genome of the host can ascertain the specific and accurate target sites to increase the efficiency of the genome editing process (Kui et al., 2017) and can be highly beneficial for overall improvement of the germplasm. Transcriptomic sequencing has also helped in increasing the pace for the development of Simple Sequence Repeats (SSR), which are the microsatellite markers with random tandem repeats of 2–6 nucleotides. These markers are widely used because of their reproducibility, co-dominant nature, extreme polymorphism, simplicity, abundance, and easy amplification. The development of SSR markers in these medicinally important orchids can help in germplasm breeding, marker-assisted selection, parentage analysis, and genetic diversity studies. The SSR markers identified can help in evaluating and understanding genetic relationships quantitatively and qualitatively (Li et al., 2014) and help in constructing genetic maps of these plants which will further help in taxonomy, genetics, and genomic studies.
The reference genome of many medicinal non-model plants is not available. Transcriptomic approach provides an alternative way for collecting high-throughput data for gene identification, expression analysis, and putative functional characterization using metabolic profiling data (Góngora-Castillo and Buell, 2013). Whole transcriptome shotgun sequencing (WTSS) makes it possible to probe the genes of various metabolite biosynthesis processes and the relationship between the genes and plant metabolites. Another approach, termed as the Phytochemical genomics approach, involves sequence data sets combined with metabolomic data sets to elucidate the complete profile of secondary metabolites. In Digital gene expression analysis, differential expression of genes which are involved in secondary metabolism is studied to decipher the genetic variability and help in the drug discovery. The development of single-cell transcriptomics will aid in identifying networks and pathways and further facilitate drug discovery and development. The present study is an exhaustive review of the omics research on secondary metabolism in orchids, primarily focusing on the use of transcriptomic data for the analysis of genes and pathways associated with the synthesis of secondary metabolites and could be further be used for establishing the therapeutic potential of the orchids.
The therapeutic potential of orchids has been reported since times immemorial. In 1579, Langham (1579) reported the antipyretic and anti-diarrheal properties of orchids. A Caribbean folklore mentions the use of Vanilla claviculata for treating wounds and syphilis (Griffith, 1847) while the flowers of Vanilla griffithii (Burkill, 1935) and leaf paste of Vanda roxburghii were used in treating fever (Chawla et al., 1992). Dendrobium huoshanense stems are reported to be beneficial for the eye, stomach, and liver ailments (Hsieh et al., 2008; Luo et al., 2008) while those of Dendrobium monoliforme are reported to be antipyretic (Zhao et al., 2003). Oil-based extracts of stems and leaves of Anoectochilus formosanus are effective for the treatment of hypertension, impotency, liver spleen disorders, and chest and abdominal pains (Satish et al., 2003). Leaf decoction of Dendrobium candidum is used for treating diabetes (Wu et al., 2004). Traditional usage of orchids as restoratives and tonics have been widely and commonly reported. The tubers of Dactylorhiza hatagirea have been used for the preparation of “Salep” which possess healing qualities (Lawler, 1984). Similar preparations like “dbang lag” have been used to provide sustenance for Tibetan monks practicing in remote caves (Teoh, 2019). Such studies coupled with ethnobotanical knowledge formed basis of many systematic reviews on utilization of orchids as therapeutic agents (Lawler, 1984).
Due to the significant role of orchids in the traditional medicine system, it has become imperative that these traditional remedies should be utilized for the discovery of new therapeutics. A plethora of studies has been reported ever since, to investigate the role of orchids as promising source of bioactive agents. A number of reports on the antioxidant and anti-inflammatory potential of various orchids like Phalaenopsis hybrids (Minh et al., 2016) and Dendrobium officinale (Zhang et al., 2017) have come up. Cytotoxic and apoptotic effects have also been reported in Dendrobium crepidatum and D. chrysanthum (Prasad and Koch, 2016). Antimicrobial activity has also been documented in Dendrobium monoliforme (Paudel et al., 2018). Antihyperglycemic (Dactylorhiza hatagirea; Choukarya et al., 2019), anti-diabetic and hepatoprotective activity (Calanthe fimbriata; Peng et al., 2019) have been reported.
To provide a sound scientific scaffolding for development of potential therapeutic products, efforts have been also directed to isolate and profile the phytochemicals from plant extracts. Various classes of secondary metabolites have been isolated from different plant parts and evaluated for biological activity. Phenanthrenes, like denbinobin, from Dendrobium nobile, showed potential cytotoxic activity (Lee et al., 1995), prevented metastatic gastric cancer, and showed potent therapeutic activity against hepatic fibrosis (Yang et al., 2007; Song et al., 2012). Similarly, kinsenoside from Anoectochilus roxburghii showed antihyperglycemic activity (Zhang et al., 2007). Cymbidine A from Cymbidium goeringii is responsible for the hypotensive and diuretic activity (Watanabe et al., 2007). Flavones C-glycosides and anthocyanins from red Phalaenopsis hybrids exhibited antioxidant activity (Kuo et al., 2010). Polysaccharides from Dendrobium officinale (Liu et al., 2011) and Gastrodia elata (Bao et al., 2017) have exhibited immune-enhancing potential. Galactoxyloglucan (GXG), a purified polysaccharide from Dendrobium huoshanense, improved insulin sensitivity, thus preventing hyperglycemia (Wang et al., 2019). Role of flavonoids especially rutin, in imparting antioxidant potential have also been highlighted in Dendrobium officinale (Zhang et al., 2017). Flavonoids of Dactylorhiza hatagirea also exhibited antihyperglycemic activity (Choukarya et al., 2019). Sesquiterpenoids from Dendrobium nobile exhibited neuroprotective activity (Ma et al., 2019b), while bibenzyl compounds from Dendrobium officinale showed cytotoxic activity (Ren et al., 2020). A group of compounds (phenanthrenes, bibenzyls, glucosyloxybenzyl succinate derivatives, flavonoids, lignans, terpenoids, etc.) isolated from Pleione, showed anti-tumor, anti-neurodegenerative, and anti-inflammatory biological activities (Wu et al., 2019). Despite a large number of reports on the phytochemical profiling in orchids, the studies are not commensurate with the immense potential of orchids as therapeutic agents. Omics techniques offer a great opportunity to provide an alternate and efficient method to study and characterize specific phytochemicals. Transcriptomic approaches can generate insights to the secondary metabolite biosynthetic pathways and can aid in functional characterization of their key regulatory genes.
Undeterred by the peculiarity in their unique characteristics, orchids are depreciated with respect to understanding their molecular complexities. A complete understanding of the spectrum of the molecular networks by isolated analyses of gene families is not plausible due to the limited availability of orchid genomes. On the other hand, transcriptome-wide analyses can help resolve complex metabolic pathways which are at play in these plants. Transcriptome is a complete set of mRNA and non-coding RNA produced by a cell or organism at a particular point of time. It generates large-scale transcripts that could help in analyzing different gene families all at once and could also guide toward understanding cross-links in mechanisms involved. The analysis begins with the collection of the desired tissue and subsequent isolation of RNA from the collected sample. The isolated RNA is used for the synthesis of complementary DNA which is eventually utilized for the construction of libraries after sequencing. There are large numbers of sequencing techniques that are prevalent nowadays, such as Roche/454, Illumina, Applied Biosystems SOLiD, and Helicos HeliScope (Magi et al., 2010). Even though these techniques produce abundant short reads at a much higher throughput than any Sanger sequencer but data presented after such analysis is a set of short reads composed of several hundred base pairs. The reads, thus, obtained are curated as raw reads. These read are first filtered and adjusted based on the quality control measures. Then the filtered reads are first either reconstructed using de novo assembly in absence of reference genome or assembled by alignment to the reference genome (Wolf, 2013). The assembly of the reads can be performed with tools like Trinity (Grabherr et al., 2011), Velvet (Zerbino and Birney, 2008), SPAdes (Bankevich et al., 2012), or SOAPdenovo-Trans (Xie et al., 2014). The assembled reads form contigs or singletons; both of these are part of unigenes. The functional annotation of the unigenes or transcripts is completed using various databases, such as NCBI,1 KEGG,2 and SwissProt.3 Additionally, the number of reads for a transcript provides the level of its abundance, thus serving as the starting point for biological inference of spatiotemporal gene expression (Wolf, 2013; Ma et al., 2019a). Transcriptome helps in identification of transcripts involved in primary and secondary metabolism and their splice variants (Wang et al., 2009). Comparing the levels of differentially expressed genes at different developmental stages or environmental conditions, provide insights into the physiological status of the tissue at a specific time. These data sets also contain information of small RNAs, long non-coding RNAs, and molecular repeats etc., and provide a tentative framework for functional assertion for putative annotations. These data can serve as an important lead for modern pharmaceutical industry toward development of herbal-based medicines.
High-throughput transcriptomic approaches produce extensive data sets that can be applied to identify candidate key genes in specific physiological processes using co-expression networks analysis (Carrera et al., 2009; Windram et al., 2014). On the other hand, targeted sequencing using degenerate primers proves to be economical and enables exhaustive analysis of specific genes. Specific genes exhibiting significant sequence similarity with gens involved in similar biological processes can be amplified by degenerate primers in related organisms (Wei et al., 2003). Functional validation of putative genes using metabolic profiling of flavonoids using gene-insertion mutants and transgenic plants with overexpressing genes could be used to understand the role genes in secondary metabolism. Further, recombinant proteins and in vitro biochemical assays could be used to decipher catalytic activity of the proteins. This “reverse genetics” approach for gene identification is very promising where bioinformatic prediction of candidate genes preceded the experimental analysis.
There have been a limited number of transcriptome-wide studies in orchids to explore and elucidate different aspects of orchid development (Table 1), however, the efforts are not in line with the immense advantage of using transcriptomic techniques to decipher various molecular networks. The therapeutic potential of orchids is closely associated with the intricate maze of secondary metabolism pathways and their by-products is mainly responsible for their diverse therapeutic properties. These pathways are, in turn, under strict control of an array of molecular factors which regulate the synthesis of phytochemicals. A large number of gene families are specifically associated with various biosynthetic pathways. Transcriptomic data emerging from various studies conducted in orchids have been tabulated in Table 1 and it is evident that Illumina sequencing was the most commonly used sequencing method and Trinity was the most common assembler software used. A maximum number of final reads were obtained in Dendrobium officinale (81,284,898; Yuan et al., 2020) and highest number of unigenes were identified in Dendrobium huoshanense (499,190, Zhou et al., 2020). A huge variation was noticed in the total number of unigenes as reported in different plant parts using different techniques. In Dendrobium officinale, the range in the number of unigenes was observed from 2,99,107 (Shen et al., 2017) to 23,131 (Adejobi et al., 2021) as reported from various tissues. Similarly, in Dendrobium catenatum, 23,139 unigenes were reported from stem tissue (Lei et al., 2018) and the number drastically increased to 478,361 in Dendrobium huoshanense when roots and leaves were also included for analysis (Yuan et al., 2018). This can be attributed to specific gene expression in tissues at various stages of growth and development and environmental conditions. In Phalaenopsis amabilis, a comparative number of unigenes were reported, 37,723 and 34,020, from petals and labellum, respectively (Yang et al., 2014), indicating that a similar genetic profile can be seen in tissues at comparable physiological stages. In Anoectochilus roxburghii, 186,865 unigenes were reported from root, stem, and leaves (Chen et al., 2020). Interestingly, different techniques and platforms used for sequencing analysis can also play a role in this variation. Root, stem, and leaf tissues of Dendrobium huoshanense reported 4,99,190 unigenes when the Illumina HiSeq2000 platform was used (Zhou et al., 2020) while 4,78,361 unigenes were identified when Illumina Hiseq 2500 platform was used (Yuan et al., 2018). Hence, it can be concluded that a lot of variation is observed in the transcriptomic data, and hence, the analysis needs to be supported with substantial functional studies.
Transcriptomic data can provide a basic lead for functional studies if a unified, systematic, and statistically significant approach is adopted for its assembly and characterization. To scrutinize the functionality of the unigenes identified from the transcriptomic data set, their assessment was carried out against different databases like KEGG, Swissprot, and non-redundant database (Nr; Table 2). The highest similarity of the unigenes was found against the Nr database except in the case of Dendrobium huoshanense where the SwissProt similarity of unigenes was the highest (Zhou et al., 2020). Out of 186,865 unigenes identified in Anoectochilus roxburghii, approximately 35, 32, and 47% were annotated using KEGG, SwissProt, and Nr database (Chen et al., 2020). However, only 9,946 out of 73,528 unigenes were annotated by KEGG in Calanthe tsoongiana (Jiang et al., 2021a). In Dendrobium officinale, the unigenes characterized using SwissProt varied from 13,418 (Guo et al., 2013) to 62,695 unigenes (Wang et al., 2021). The variation could be due to the use of different platforms used for sequencing or assembly and due to the type of tissue used in different studies.
The annotation of genes or transcripts obtained using various servers helped in the characterization of genes based on their functional roles. The KEGG analysis of different studies in association with pathways of secondary metabolism has been summarized in Table 3. KEGG analysis of stem, leaves, and roots revealed the presence of cyanoamino acid metabolism, phenylpropanoid biosynthesis, diterpenoid biosynthesis, flavonoid and flavonol biosynthesis, steroid biosynthesis, and isoflavonoid biosynthesis pathways in Anoectochilus roxburghii (Chen et al., 2020). 65,286 unigenes in Dendrobium officinale (Shen et al., 2017), 10,723 unigenes in Cymbidium longibracteatum (Jiang et al., 2018), and 9,130 unigenes in Dactylorhiza hatagirea (Dhiman et al., 2019) were annotated by KEGG analysis. Differential gene expression (DEG) of the different colored buds of Phalaenopsis sp. suggested that most DEGs were of phenylpropanoid biosynthesis which suggests the role of anthocyanins for variable colors (Gao et al., 2016). The number of unigenes annotated to phenylpropanoid varies from 49 unigenes in Phalaenopsis sp. (Gao et al., 2016) to 466 in Cymbidium goeringii (Ramya et al., 2019). In a transcriptomic study of Pleione limprichtii, 11,067 genes were mapped to 131 KEGG pathways and 1,294 unigenes were associated with secondary metabolite synthesis (Zhang et al., 2020b).
Even though large numbers of secondary metabolites are produced by plants, only a selected compounds have important medicinal properties. These secondary metabolites can be grouped into various classes like alkaloids, terpenoids, polyphenols, phenanthrene, bibenzyl derivatives, etc. Therapeutic effects of different alkaloids especially terpenoid alkaloids have been widely reported in orchids (Sut et al., 2017; Gantait et al., 2021; Ghai et al., 2021). These terpenes alkaloids are formed through the mevalonate (MVA) pathway and methyl-D-erythritol 4-phosphate (MEP) pathway (Figure 2). MVA pathway initiates with acetyl-CoA as a precursor. Acetyl-CoA undergoes a series of catalyzation reactions to produce isopentyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). These IPP units are further processed to form sesquiterpenes. Meanwhile, the MEP pathway begins with the condensation of pyruvate and D-glyceraldehyde-3-phosphate by 1-Deoxy-D-xylulose-5-phosphate synthase (DXS). The regulatory mechanisms and biochemistry of the mevalonate (MVA) and methyl-D-erythritol 4-phosphate (MEP) pathway are well characterized. In MVA, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) lays an important role in controlling the metabolic flux. The regulation of mevalonate kinase (MK) is regulated by feedback mechanism at both transcriptional and post-translational levels (Hinson et al., 1997). The isopentyl pyrophosphate (IPP) units are isomerized into dimethylallyl pyrophosphate (DMAPP) by IPP isomerase which is the initiating molecule in terpenoid biosynthesis. The enzyme, 1-deoxy-D-xylulose-5-phosphate synthase (DXS), is key player in controlling influx into the MEP pathway through decarboxylation reaction and is regulated by feedback mechanism through IPP and DMAPP (Banerjee et al., 2013). This can be corroborated by the higher expression of DXS and the terpenoid levels in the inflorescences in Arabidopsis (Carretero-Paulet et al., 2002). This process proceeds to form IPP and DMAPP via multistep reactions catalyzed by a series of enzymes. The MEP and MVA pathways are both linked by an intermediary precursor isopentenyl pyrophosphate. Subsequently, the pathways result in the formation of monoterpenoids, diterpenoids, carotenoids, sesquiterpenoids, and some other metabolites. Sesquiterpene alkaloids are the most abundant types of alkaloids of Dendrobium (Chen et al., 2019). Hsiao et al. (2011) reported the identification of 50 unigenes of the MEP and MVA pathways in Phalaenopsis while in Cymbidium goeringii, 32 unigenes of MVA and 38 unigenes of MEP pathway were identified (Ramya et al., 2019). Forty-six unigenes in Dendrobium huoshanense (Yuan et al., 2018) and 36 in Dendrobium officinale (Shen et al., 2017) related to the MEP and MVA pathway were identified. According to Li et al. (2017), isoprene units obtained through the MEP pathway were responsible for the biosynthesis of dendrobine in Dendrobium nobile. The expression of acetyl-CoA acetyltransferase (AACT), mevalonate diphosphosphate decarboxylase (MVD), phosphomevalonate kinase (PMK), and Alpha-humulene synthase (TPS21) changes upon inoculation of the orchid with MF23 (Mycena sp.) which results in induction of pathway leading to dendrobine biosynthesis (Li et al., 2017). Besides fungal stimulation, methyl jasmonate (MeJA) treatment of D. officinale also results in increased expression of genes associated with MEP and MVA pathway (Chen et al., 2019). Toh et al. (2017) also studied the fragrant sites in Vanda Mimi Palmer which indirectly points toward the sites of high monoterpenoid production. Higher expression of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) and 1-deoxyxylulose-5-phosphate synthetase (DXS) was observed in root than in leaf but DXS and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) were abundant mainly in stems of Dendrobium huoshanense (Yuan et al., 2018). Yuan et al. (2018) suggested stem-specific accumulation of alkaloids in D. huoshanense but leaf-specific accumulation is observed in D. officinale (Shen et al., 2017). A series of enzymes associated with strictosidine were also identified in a study on Dendrobium officinale (Shen et al., 2017). Accumulation of dendrobine, a sesquiterpene alkaloid, was consistently more when the expression of PMK and MVD was high but got reduced as the expression of the aforementioned genes decreased in MF23 infected Dendrobium nobile orchid plant (Li et al., 2017). In the same study, the dendrobine pathway was negatively correlated with the expression of TPS21 but no relation with genes of MEP was observed (Li et al., 2017). Secologanin synthase (SCS) which is essential for the synthesis of secologanin has also been reported to be involved in alkaloid biosynthesis (Guo et al., 2013). Different terpenes are synthesized from isopentenyl diphosphate through two pathways mevalonate pathway and methylerythritol phosphate pathway. Hsiao et al. (2006) analyzed transcriptomes of Phalaenopsis bellina and Phalaenopsis equestris where genes related to the DXP-geraniol linalool pathway were identified by data mining. In another study, regulation of monoterpene biosynthesis by PbbHLH4 in Phalaenopsis orchid was provided (Chuang et al., 2018). Terpene synthases (TPSs) are responsible for the structure diversity of terpene while cytochromes P450 (CYPs) further modifies the products from TPSs which provide further diversification of terpenes (Tsai et al., 2017). In D. huoshanense, 229 unigenes of the P450 superfamily were identified (Yuan et al., 2018) but in D. officinale, 236 unigenes associated with P450 were mined (Shen et al., 2017). Strictosidine synthase had higher expression levels in protocorm like bodies (PLBs) than in leaves suggesting the higher content of total alkaloid is related to the higher amount of precursor strictosidine produced in D. officinale (Wang et al., 2021). The positive stimulation of either MEP or MVA pathway could eventually lead to an increase in the production of alkaloids which could eventually increase the therapeutic potential of the orchid plant.
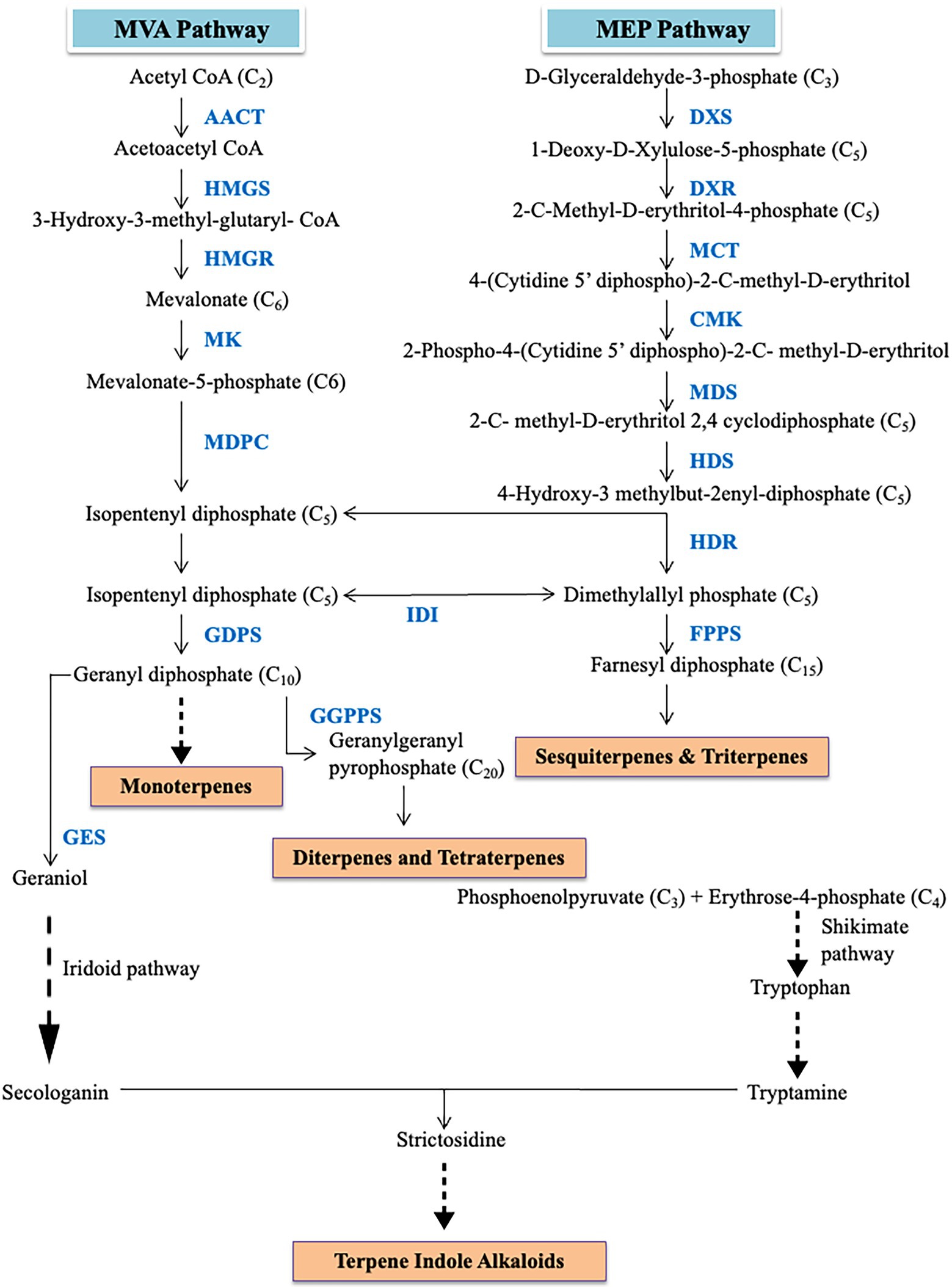
Figure 2. Diagrammatic representation of methyl-D-erythritol 4-phosphate (MEP) and mevalonate (MVA) pathway.
Besides alkaloids, the role of flavonoids as antioxidant, anti-cancer, and anti-aging agents has also been highlighted (Middleton et al., 2000). The flavonoids are compounds with bridged phenyl rings which are synthesized through the phenylpropanoid pathway. Flavonoid also provides resistance against disease and insects in plants and enable the plant for adapting to adverse environmental conditions with the help of increased production in secondary metabolites (Campos and Hamdan, 2000; Yuan et al., 2020). Anoectochilus roxburghii is rich in flavonoid compounds, such as dihydroquercetin, quercetin, kaempferol, and myricetin (Ye et al., 2017), which are responsible for the drug activity of this orchid plant (Chen et al., 2020). Lei et al. (2018) reported about C-glycosides type flavonoids are more abundant than O-glycosides in Dendrobium. The metabolic analysis of Anoectochilus roxburghii revealed an abundance of flavonoids in leaves than in roots or stems (Chen et al., 2020). The by-products of the shikimate pathway are the precursor for a large assortment of secondary metabolites (Tzin et al., 2012; Takayuki et al., 2013). It is a multistep process that starts with the condensation of phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P; Figure 3). The intermediate chorismite after further processing leads to the independent formation of aromatic amino acids, tryptophan, tyrosine, phenylalanine. Phenylalanine is the precursor for the Phenylpropanoid pathway which ultimately results in the synthesis of flavonoids. PAL is the most important rate limiting fulcrum enzyme that links primary metabolism with secondary metabolism (Vogt, 2010; Fraser and Chapple, 2011). A positive correlation between the PAL enzyme activity and accumulation of phenylpropanoid compounds has been widely reported (Bate et al., 1994; Vogt, 2010). Carbon flux into different branches of flavonoid synthesis is regulated by flavonol synthase (FLS; Davies et al., 2003). In Arabidopsis, activity of hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) led to maneuvering of the metabolic flux into flavonoids through Chalcone synthase (CHS) activity (Besseau et al., 2007). Additionally, there are several transcription factors that regulate the gene expression which ultimately controls the metabolic flux. The expression of regulatory molecules like MYB is inversely proportional to lignin production, thus facilitating the metabolic flux toward flavonoid production (Fornale et al., 2010). Similarly, elicitors like salicylic acid and methyl jasmonate positively diverts the metabolic flux toward increased production of secondary metabolites (Creelman and Mullet, 1997; Kessler and Baldwin, 2002). Phenylalanine is catalyzed by phenylalanine ammonia lyase (PAL) to form cinnamate which is converted to p-coumaroyl-CoA by trans-cinnamate 4-monooxygenase (C4H) and 4-coumaroyl-CoA synthase (4CL). p-coumaroyl-CoA is further processed by series of different enzymes to form flavonoids, flavonols, flavanones, and anthocyanins.
A total of 15 unigenes encoding seven enzymes of the flavonoid pathway were identified from D. huoshanense (Zhou et al., 2020) while 31 and 19 unigenes in D. catenatum (Lei et al., 2018) and Pleione limprichtii (Zhang et al., 2020b). In a study on Anoectochilus roxburghii, inoculation with Ceratobasidium sp. AR2 increases the flavonoid content of the plant by upregulating PAL, chalcone synthase (CHS), 4CL and downregulating of cinnamate 4-hydroxylase (C4H), and chalcone isomerase (CHI) genes (Zhang et al., 2020a). In a new cultivar of Cymbidium longibracteatum with yellow leaves and tubers, seven unigenes related to flavonoid biosynthesis were upregulated (Jiang et al., 2018). Similarly, expression levels of CHS, CHI, dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS1), and UDP-glucose: flavonoid-3-O-glucosyltransferase (UFGT) were comparatively higher in red, corroborating with higher anthocyanin content in the red stems of D. candidum (Jia et al., 2021). Similarly, most of the genes involved in anthocyanin biosynthesis were upregulated during floral development of Dendrobium nestor (Cui et al., 2021). Expression of PAL and HMG-CoA reductase was upregulated in the abaxial surface of the tissue of Vanda Mimi Palmer (Toh et al., 2017). The rate of flavonoid production in plants was reported to be controlled by CHS with associated CHI. The higher expression levels of CHS, CHI, flavonol synthase (FLS), DFR, and Anthocyanidin reductase (ANR) in roots than in stems and leaves of A. roxburghii were reported as well (Chen et al., 2020). Upregulation of LAR1, DFR3, flavanone 3-hydroxylase (F3H), CHS1, CHS2, and CHS3 in leaves facilitates the copious accumulation of flavonoids in leaves of Dedrobium officinale (Yuan et al., 2020). In the same study, Dihydroflavonol reductase (DFR), which is responsible for the conversion of flavonoids into anthocyanin biosynthesis, has higher expression in stems and leaves. MeJA treatment in D. officinale lead to the accumulation of bibenzyl (erianin and gigantol) increased due to upregulation of PAL, 4CL, C4H, and CYP450 (Adejobi et al., 2021). During explant browning in Phalaenopsis sp., higher expression of PhPAL, PhCHS, and Ph4CL was observed which suggest the role of anthocyanin in the early stages of tissue browning (Xu et al., 2015). Similarly, upregulated expression of Pa4CL, PaANS, PaF3H, and PaDFR was detected in purple petal cultivar of Phalaenopsis amabilis (Meng et al., 2019). The study on Phalaenopsis did not identify any DEGs related to CHS, ANS, DFR, and flavonoid-3′-hydroxylase (F3′H) in white petals which could be due to either technical limitations or due to absence of anthocyanin pathway (Yang et al., 2014). Similarly, no transcript of flavonoid-3′,5′-hydroxylase (F3′5′H) was identified from the transcriptome of Ophrys even though 61 transcripts of anthocyanins pathway were mined (Sedeek et al., 2013). Expression of PlCHS, PlCHI, and PlFLS was upregulated in white petals but colored petals had higher expression of PlF3’H, PlDFR, and PlANS in Pleione limprichtii (Zhang et al., 2020b). PAL, 4CL, and C4H were upregulated in 8 and 10 weeks old seeds of Vanilla planifolia (Rao et al., 2014). Expression of trans-resveratrol-di-O-methyltransferase-like (ROMT) encoding gene, responsible for resveratrol biosynthesis, was high in tubers of Dactylorhiza hatagirea (Dhiman et al., 2019). It positively correlates with the fact that tubers of this plant are used as anti-inflammatory, anticarcinogenic, and as a cardioprotective agent. Higher expression of ROMT correlated with the abundant quantity of resveratrol and stilbenes (Dhiman et al., 2019). The role of caffeic acid, coumaric acid, and Caffeoyl CoA in the synthesis of resveratrol and stilbenes has also been pointed out in the same study. Genes associated with flavonoid pathways were reported to be regulated by UDP-glycosyltransferase and cytochrome P450 (Liu et al., 2013). DcTT8, a bHLH transcription factor in D. candidum, regulated the anthocyanin production by binding to the promoter region of DcF3′H and DcUFGT (Jia et al., 2021). The above review asserts that transcriptomic approaches can serve as a boon for gene discovery, functional annotation, and expression profiling in non-model organisms.
Orchids grow in a variety of habits and habitats mainly owing to the presence of an array of unique secondary metabolites which help these plants sustain the stressful conditions. Therefore, these plants have emerged as important source for bioprospecting following traditional approaches. Omics technology, on the other hand, offer great potential for analysis of the complete metabolic pathways and provides detailed insights to gene function for drug discovery and other therapeutic interventions. The present study is a comprehensive analysis of transcriptomes more than 30 orchids mainly focusing on the alkaloids and flavonoids pathways. It can form the basis of an effective resource for the functional studies on tapping the immense potential of unique orchid secondary metabolites to facilitate development of novel therapeutic products from these plants.
JS conceptualized the work. DG and AK performed the analysis and prepared the original draft. PK, SP, and JS critically reviewed and edited the draft. All the authors have read and approved the final version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
DG is grateful to Council of Scientific and Industrial Research for Senior Research Fellowship (File No. 09/135(0809)/2018-EMR-I). AK is thankful to Department of Science and Technology (DST) for INSPIRE Fellowship for Research Students (File No. DST/INSPIRE/03/2021/002638). JS is thankful for partial financial support received from Department of Science and Technology, Government of India under Promotion of University Research and Scientific Excellence (PURSE) grant scheme. Authors are thankful to Jagdeep Verma for the photographs of orchid plants.
Adejobi, O. I. I., Guan, J., Yang, L., Hu, J. M., Yu, A., Muraguri, S., et al. (2021). Transcriptomic analyses shed light on critical genes associated with bibenzyl biosynthesis in Dendrobium officinale. Plan. Theory 10:633. doi: 10.3390/plants10040633
Banerjee, A., Wu, Y., Banerjee, R., Li, Y., Yan, H., and Sharkey, T. D. (2013). Feedback inhibition of deoxy-d-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 288, 16926–16936. doi: 10.1074/jbc.M113.464636
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bao, Q., Qian, L., Gong, C., and Shen, X. (2017). Immune-enhancing activity of polysaccharides from Gastrodia elata. J. Food Process. Preserv. 41:e13016. doi: 10.1111/jfpp.13016
Bate, N. J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P. W., et al. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. U. S. A. 91, 7608–7612. doi: 10.1073/pnas.91.16.7608
Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162. doi: 10.1105/tpc.106.044495
Bulpitt, C. J., Li, Y., Bulpitt, P. F., and Wang, J. J. (2007). The use of orchids in Chinese medicine. J. R. Soc. Med. 100, 558–563. doi: 10.1177/0141076807100012014
Burkill, I. H. (1935). A Dictionary of the Economic Products of the Malay Peninsula, Vol. II. London: Crown Agents.
Campos, E. G., and Hamdan, F. F. (2000). Cloning of the chaperonin t-complex polypeptide 1 gene from Schistosoma mansoni and studies of its expression levels under heat shock and oxidative stress. Parasitol. Res. 86, 253–258. doi: 10.1007/s004360050039
Carrera, J., Rodrigo, G., Jaramillo, A., and Elena, S. F. (2009). Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 10, R96–R15. doi: 10.1186/gb-2009-10-9-r96
Carretero-Paulet, L., Ahumada, I., Cunillera, N., Rodriguez-Concepcion, M., Ferrer, A., Boronat, A., et al. (2002). Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol. 129, 1581–1591. doi: 10.1104/pp.003798
Chawla, A. S., Sharma, A. K., Handa, S. S., and Dhar, K. L. (1992). Chemical studies and anti-inflammatory activity of Vanda roxburghii roots. Indian J. Pharm. Sci. 54, 159–161.
Chen, Y., Pan, W., Jin, S., and Lin, S. (2020). Combined metabolomic and transcriptomic analysis reveals key candidate genes involved in the regulation of flavonoid accumulation in Anoectochilus roxburghii. Process Biochem. 91, 339–351. doi: 10.1016/j.procbio.2020.01.004
Chen, Y., Wang, Y., Lyu, P., Chen, L., Shen, C., and Sun, C. (2019). Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 132, 419–429. doi: 10.1007/s10265-019-01099-6
Chen, J., Zhu, F., Liu, L., Yi, L., Dai, Y., Chen, S., et al. (2021). Integrative analyses of transcriptome and metabolome shed light on the regulation of secondary metabolites in pseudobulbs of two Bletilla striata (Thunb.) Reichb. f. varieties. J. Appl. Res. Med. Aromat. Plants 293:110738. doi: 10.1016/j.scienta.2021.110738
Choukarya, R., Choursia, A., and Rathi, J. (2019). In vivo and in vitro antidiabetic activity of hydroalcoholic extract of Dactylorhiza hatagirea roots: an evaluation of possible phytoconstituents. J. Drug Deliv. Ther. 9, 76–81. doi: 10.22270/jddt.v9i6-s.3752
Chuang, Y., Hung, Y., Tsai, W., Chen, W., and Chen, H. (2018). PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 69, 4363–4377. doi: 10.1093/jxb/ery246
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Cui, X., Deng, J., Huang, C., Tang, X., Li, X., Li, X., et al. (2021). Transcriptomic analysis of the anthocyanin biosynthetic pathway reveals the molecular mechanism associated with purple color formation in Dendrobium Nestor. Life 11:113. doi: 10.3390/life11020113
Davies, K. M., Schwinn, K. E., Deroles, S. C., Manson, D. G., Lewis, D. H., Bloor, S. J., et al. (2003). Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131, 259–268. doi: 10.1023/A:1024018729349
Dhiman, N., Sharma, N. K., Thapa, P., Sharma, I., Swarnkar, M. K., Chawla, A., et al. (2019). De novo transcriptome provides insights into the growth behaviour and resveratrol and trans-stilbenes biosynthesis in Dactylorhiza hatagirea:– an endangered alpine terrestrial orchid of western Himalaya. Sci. Rep. 9, 13133–13113. doi: 10.1038/s41598-019-49446-w
Fang, L., Xu, X., Li, J., Zheng, F., Li, M., Yan, J., et al. (2020). Transcriptome analysis provides insights into the non-methylated lignin synthesis in Paphiopedilum armeniacum seed. BMC Genomics 21:524. doi: 10.1186/s12864-020-06931-1
Fornale, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644. doi: 10.1111/j.1365-313X.2010.04363.x
Fraser, C. M., and Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9:e0152. doi: 10.1199/tab.0152
Gantait, S., Das, A., Mitra, M., and Chen, J. T. (2021). Secondary metabolites in orchids: biosynthesis, medicinal uses, and biotechnology. S. Afr. J. Bot. 139, 338–351. doi: 10.1016/j.sajb.2021.03.015
Gao, L. W., Jiang, D. H., Yang, Y. X., Li, Y. X., Sun, G. S., Ma, Z. H., et al. (2016). De novo sequencing and comparative analysis of two Phalaenopsis orchid tissue-specific transcriptomes. Russ. J. Plant Physiol. 63, 391–400. doi: 10.1134/S1021443716020072
Ghai, D., Verma, J., Kaur, A., Thakur, K., Pawar, S. V., and Sembi, J. K. (2021). “Bioprospection of orchids and appraisal of their therapeutic indications,” in Bioprospecting of Plant Biodiversity for Industrial Molecules. eds. S. K. Upadhyay and S. P. Singh (Chichester, West Sussex, UK: John Wiley & Sons Ltd), 401–424.
Góngora-Castillo, E., and Buell, C. R. (2013). Bioinformatics challenges in de novo transcriptome assembly using short read sequences in the absence of a reference genome sequence. Nat. Prod. Rep. 30, 490–500. doi: 10.1039/c3np20099j
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Griffith, R. E. (1847). Medicinal Botany or Descriptions of the More Important Plants Used in Medicine With Their History, Properties and Mode of Administration. Philadelphia: Lea and Blanchard.
Guo, X., Li, Y., Li, C., Luo, H., Wang, L., Qian, J., et al. (2013). Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 527, 131–138. doi: 10.1016/j.gene.2013.05.073
He, C., Wang, C., Guo, S., Yang, J., and Xiao, P. (2006). A novel flavonoid Glucoside from Anoectochilus roxburghii (Wall.) Lindl. J. Integr. Plant Biol. 48, 359–363. doi: 10.1111/j.1744-7909.2006.00179.x
Hinson, D. D., Chambliss, K. L., Toth, M. J., Tanaka, R. D., and Gibson, K. M. (1997). Post-translational regulation of mevalonate kinase by intermediates of the cholesterol and nonsterol isoprene biosynthetic pathways. J. Lipid Res. 38, 2216–2223. doi: 10.1016/S0022-2275(20)34935-X
Hossain, M. M., Akter, S., and Uddin, S. B. (2020). “Screening of bioactive phytochemicals in some indigenous epiphytic orchids of Bangladesh” in Orchid Biology: Recent Trends and Challenges (Singapore: Springer Nature), 481–506.
Hsiao, Y., Chen, Y., Huang, S., Pan, Z., Fu, C., Chen, W., et al. (2011). Gene discovery using next-generation pyrosequencing to develop ESTs for Phalaenopsis orchids. BMC Genomics 12:360. doi: 10.1186/1471-2164-12-360
Hsiao, Y., Tsai, W., Kuoh, C., Huang, T., Wang, H., Wu, T., et al. (2006). Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biol. 6:14. doi: 10.1186/1471-2229-6-14
Hsieh, Y. S., Chien, C., Liao, S. K., Liao, S. F., Hung, W. T., Yang, W. B., et al. (2008). Structure and bioactivity of the polysaccharides in medicinal plant Dendrobium huoshanense. Bioorg. Med. Chem. 16, 6054–6068. doi: 10.1016/j.bmc.2008.04.042
Jia, N., Wang, J. J., Liu, J., Jiang, J., Sun, J., Yan, P., et al. (2021). DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum. Plant Physiol. Biochem. 162, 603–612. doi: 10.1016/j.plaphy.2021.03.006
Jiang, Y., Liu, Y., Song, H., and He, J. (2021b). Integrated transcriptomics and metabolomics uncover the molecular basis of flavonoid accumulation in the rhizomes of two Cymbidium tortisepalum var. longibracteatum cultivars. Sci. Hortic. 293:110738. doi: 10.1016/j.scienta.2021.110738
Jiang, Y., Song, H. Y., He, J. R., Wang, Q., and Liu, J. (2018). Comparative transcriptome analysis provides global insight into gene expression differences between two orchid cultivars. PLoS One 13:e0200155. doi: 10.1371/journal.pone.0200155
Jiang, Y., Tian, M., Wang, C., and Zhang, Y. (2021a). Transcriptome sequencing and differential gene expression analysis reveal the mechanisms involved in seed germination and protocorm development of Calanthe tsoongiana. Gene 772:145355. doi: 10.1016/j.gene.2020.145355
Kessler, A., and Baldwin, I. T. (2002). Plant responses to insect herbivory: the emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. doi: 10.1146/annurev.arplant.53.100301.135207
Kong, J. M., Goh, N. K., Chia, L. S., and Chia, T. F. (2003). Recent advances in traditional plant drugs and orchids. Acta Pharmacol. Sin. 24, 7–21.
Kui, L., Chen, H., Zhang, W., He, S., Xiong, Z., Zhang, Y., et al. (2017). Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 7:2036. doi: 10.3389/fpls.2016.02036
Kuo, P. C., Chen, G. F., Yang, M. L., and Wu, T. S. (2010). High-performance liquid chromatography profiling of pigments from Phalaenopsis hybrids and their contribution to antioxidant and antityrosinase activities. Acta Hortic. 878, 89–95. doi: 10.17660/ActaHortic.2010.878.9
Lawler, L. (1984). “Ethnobotany of the Orchidaceae,” in Orchid Biology, Reviews & Perspectives III. ed. J. Arditti (Ithaca: Cornell University), 27–149.
Lee, Y. H., Park, J. D., Baek, N. I., Kim, S. I., and Ahn, B. Z. (1995). In vitro and in vivo antitumoral phenanthrenes from the aerial parts of Dendrobium nobile. Planta Med. 61, 178–180. doi: 10.1055/s-2006-958043
Lei, Z., Zhou, C., Ji, X., Wei, G., Huang, Y., Yu, W., et al. (2018). Transcriptome analysis reveals genes involved in flavonoid biosynthesis and accumulation in Dendrobium catenatum from different locations. Sci. Rep. 8, 1–16. doi: 10.1038/s41598-018-24751-y
Li, Q., Ding, G., Li, B., and Guo, S. X. (2017). Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. Infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 7, 1–16. doi: 10.1038/s41598-017-00445-9
Li, N., Dong, Y., Lv, M., Qian, L., Sun, X., Liu, L., et al. (2021a). Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 12:661296. doi: 10.3389/fgene.2021.661296
Li, X., Fan, J., Luo, S., Yin, L., Liao, H., Cui, X., et al. (2021b). Comparative transcriptome analysis identified important genes and regulatory pathways for flower color variation in Paphiopedilum hirsutissimum. BMC Plant Biol. 21:495. doi: 10.1186/s12870-021-03256-3
Li, X., Jin, F., Jin, L., Jackson, A., Huang, C., Li, K., et al. (2014). Development of Cymbidium ensifolium genic-SSR markers and their utility in genetic diversity and population structure analysis in cymbidiums. BMC Genet. 15, 1–14. doi: 10.1186/s12863-014-0124-5
Liu, X. J., Lu, Y., Yuan, Y., Liu, S., Guan, C., Chen, S., et al. (2013). De novo transcriptome of Brassica juncea seed coat and identification of genes for the biosynthesis of flavonoids. PLoS One 8:e71110. doi: 10.1371/journal.pone.0071110
Liu, X. F., Zhu, J., Ge, S. Y., Xia, L. J., Yang, H. Y., Qian, Y. T., et al. (2011). Orally administered Dendrobium officinale and its polysaccharides enhance immune functions in BALB/c mice. Nat. Prod. Commun. 6, 867–870.
Luo, J. P., Deng, Y. Y., and Zha, X. Q. (2008). Mechanism of polysaccharides from Dendrobium huoshanense on streptozotocin-induced diabetic cataract. Pharm. Biol. 46, 243–249. doi: 10.1080/13880200701739397
Ma, X., Meng, Y., Wang, P., Tang, Z., Wang, H., and Tian, X. (2019a). Bioinformatics-assisted, integrated omics studies on medicinal plants. Brief. Bioinform. 21, 1857–1874. doi: 10.1093/bib/bbz132
Ma, C., Meng, C. W., Zhou, Q. M., Peng, C., Liu, F., Zhang, J. W., et al. (2019b). New sesquiterpenoids from the stems of Dendrobium nobile and their neuroprotective activities. Fitoterapia 138:104351. doi: 10.1016/j.fitote.2019.104351
Ma, X., Tang, K., Tang, Z., Dong, A., Meng, Y., and Wang, P. (2021). Organ-specific, integrated omics data-based study on the metabolic pathways of the medicinal plant Bletilla striata (Orchidaceae). BMC Plant Biol. 21:504. doi: 10.1186/s12870-021-03288-9
Magi, A., Benelli, M., Gozzini, A., Girolami, F., Torricelli, F., and Brandi, M. L. (2010). Bioinformatics for next generation sequencing data. Genes 1, 294–307. doi: 10.3390/genes1020294
Meng, X., Li, G., Gu, L., Sun, Y., Li, Z., Liu, J., et al. (2019). Comparative metabolomic and transcriptome analysis reveal distinct flavonoid biosynthesis regulation between petals of white and purple Phalaenopsis amabilis. J. Plant Growth Regul. 39, 823–840. doi: 10.1007/s00344-019-10025-y
Middleton, E. J., Kandaswami, C., and Theoharides, T. C. (2000). The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 52, 673–751.
Minh, T. N., do Khang, T., Tuyen, P. T., Minh, L. T., Anh, L. H., Quan, L. V., et al. (2016). Phenolic compounds and antioxidant activity of Phalaenopsis orchid hybrids. Antioxidants 5:31. doi: 10.3390/antiox5030031
Mohd-Hairul, A. R., Ong Abdullah, J., Song, A. A. L., Foo, H. L., Namasivayam, P., and Rahim, R. A. (2020). Volatile constituents and floral transcriptome analyses provide new insights into fragrance biosynthesis of Vanda ‘Tan Chay Yan’. Plant Biosyst. 1–8. doi: 10.1080/11263504.2020.1829733 [Epub ahead of print].
Paudel, M. R., Rajbanshi, N., Sah, A. K., Acharya, S., and Pant, B. (2018). Antibacterial activity of selected Dendrobium species against clinically isolated multiple drug resistant bacteria. Afr. J. Microbiol. Res. 12, 426–432. doi: 10.5897/AJMR2018.8846
Peng, Y., Gao, Y., Zhang, X., Zhang, C., Wang, X., Zhang, H., et al. (2019). Antidiabetic and hepatoprotective activity of the roots of Calanthe fimbriata Franch. Biomed. Pharmacother. 111, 60–67. doi: 10.1016/j.biopha.2018.12.066
Prasad, R., and Koch, B. (2016). In vitro anticancer activities of Ethanolic extracts of Dendrobium crepidatum and Dendrobium chrysanthum against T-cell lymphoma. J. Cytol. Histol. 07:432. doi: 10.4172/2157-7099.1000432
Ramya, M., Park, P. H., Chuang, Y., Kwon, O. K., An, H. R., Park, P. M., et al. (2019). RNA sequencing analysis of cymbidium goeringii identifies floral scent biosynthesis related genes. BMC Plant Biol. 19:337. doi: 10.1186/s12870-019-1940-6
Rao, X., Krom, N., Tang, Y., Widiez, T., Havkin-Frenkel, D., Belanger, F. C., et al. (2014). A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia). BMC Genomics 15:964. doi: 10.1186/1471-2164-15-964
Ren, G., Deng, W. Z., Xie, Y. F., Wu, C. H., Li, W. Y., Xiao, C. Y., et al. (2020). Bibenzyl derivatives from leaves of Dendrobium officinale. Nat. Prod. Commun. 15, 1934578X2090867–1934578X2090865. doi: 10.1177/1934578X20908678
Satish, M. N., Abhay, P. S., Chen-Yue, L., Chao-Lin, K., and Hsin-Sheng, T. (2003). Studies on tissue culture of Chinese medicinal plant resources in Taiwan and their sustainable utilization. Bot. Bull. Acad. Sin. 44, 79–98.
Sedeek, K. E. M., Qi, W., Schauer, M. A., Gupta, A. K., Poveda, L., Xu, S., et al. (2013). Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS One 8:e64621. doi: 10.1371/journal.pone.0064621
Shan, T., Yin, M., Wu, J., Yu, H., Liu, M., Xu, R., et al. (2021). Comparative transcriptome analysis of tubers, stems, and flowers of Gastrodia elata Blume reveals potential genes involved in the biosynthesis of phenolics. Fitoterapia 153:104988. doi: 10.1016/j.fitote.2021.104988
Shen, C., Guo, H., Chen, H., Shi, Y., Meng, Y., Lu, J., et al. (2017). Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNASeq. Sci. Rep. 7:187. doi: 10.1038/s41598-017-00292-8
Song, J. I., Kang, Y. J., Yong, H., Kim, Y. C., and Moon, A. (2012). Denbinobin, a phenanthrene from Dendrobium nobile, inhibits invasion and induces apoptosis in SNU-484 human gastric cancer cells. Oncol. Rep. 27, 813–818. doi: 10.3892/or.2011.1551
Sut, S., Maggi, F., and Dall’Acqua, S. (2017). Bioactive secondary metabolites from orchids (Orchidaceae). Chem. Biodivers. 14:e1700172. doi: 10.1002/cbdv.201700172
Takayuki, T., Mutsumi, W., Rainer, H., and Fernie, A. R. (2013). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4:62. doi: 10.3389/fpls.2013.00062
Toh, C., Mohd-Hairul, A. R., Ain, N. M., Namasivayam, P., Go, R., Abdullah, N. A. P., et al. (2017). Floral micromorphology and transcriptome analyses of a fragrant Vandaceous orchid, Vanda Mimi palmer, for its fragrance production sites. BMC. Res. Notes 10:554. doi: 10.1186/s13104-017-2872-6
Tsai, W. C., Dievart, A., Hsu, C. C., Hsiao, Y. Y., Chiou, S. Y., Huang, H., et al. (2017). Post genomics era for orchid research. Bot. Stud. 22, 1–22. doi: 10.3390/ijms22136947
Tzin, V., Malitsky, S., Zvi, M. M. B., Bedair, M., Sumner, L., Aharoni, A., et al. (2012). Expression of a bacterial feedback-insensitive 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase of the shikimate pathway in Arabidopsis elucidates potential metabolic bottlenecks between primary and secondary metabolism. New Phytol. 194, 430–439. doi: 10.1111/j.1469-8137.2012.04052.x
Vij, S. P., Verma, J., and Kumar, S. C. (2013). Orchids of Himachal Pradesh. Bishen Singh Mahendra Pal Singh, Dehradun.
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wang, Z., Jiang, W., Liu, Y., Meng, X., Su, X., Cao, M., et al. (2021). Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between Protocorm-like bodies and leaves. BMC Genomics 22:579. doi: 10.1186/s12864-021-07887-6
Wang, H. Y., Li, Q. M., Yu, N. J., Chen, W. D., Zha, X. Q., Wu, D. L., et al. (2019). Dendrobium huoshanense polysaccharide regulates hepatic glucose homeostasis and pancreatic β-cell function in type 2 diabetic mice. Carbohydr. Polym. 211, 39–48. doi: 10.1016/j.carbpol.2019.01.101
Wang, Y., Shahid, M. Q., Ghouri, F., and Baloch, F. S. (2020). De novo assembly and annotation of the juvenile tuber transcriptome of a Gastrodia elata hybrid by RNA sequencing: detection of SSR markers. Biochem. Genet. 58, 914–934. doi: 10.1007/s10528-020-09983-w
Watanabe, K., Tanaka, R., Sakurai, H., Iguchi, K., Yamada, Y., Hsu, C. S., et al. (2007). Structure of Cymbidine A, a monomeric peptidoglycan-related compound with hypotensive and diuretic activities, isolated from a higher plant, cymbidium goeringii (Orchidaceae). Chem. Pharm. Bull. 55, 780–783. doi: 10.1248/cpb.55.780
Wei, X., Kuhn, D. N., and Narasimhan, G. (2003). “Degenerate primer design via clustering.” in Computational Systems Bioinformatics. CSB2003. Proceedings of the 2003 IEEE Bioinformatics Conference, 75–83.
Windram, O., Penfold, C. A., and Denby, K. J. (2014). Network modeling to understand plant immunity. Annu. Rev. Phytopathol. 52, 93–111. doi: 10.1146/annurev-phyto-102313-050103
Wolf, J. B. W. (2013). Principles of transcriptome analysis and gene expression quantification: an RNA-seq tutorial. Mol. Ecol. Resour. 13, 559–572. doi: 10.1111/1755-0998.12109
Wu, X. Q., Li, W., Chen, J. X., Zhai, J. W., Xu, H. Y., Ni, L., et al. (2019). Chemical constituents and biological activity profiles on Pleione (Orchidaceae). Molecules 24:3195. doi: 10.3390/molecules24173195
Wu, H. S., Xu, J. H., Chen, L. Z., and Sun, J. J. (2004). Studies on anti-hyperglycemic effect and its mechanism of Dendrobium candidum. Zhongguo ZhongYao Za Zhi 29, 160–163.
Xie, Y., Wu, G., Tang, J., Luo, R., Patterson, J., Liu, S., et al. (2014). SOAPdenovo-trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. doi: 10.1093/bioinformatics/btu077
Xu, C., Zeng, B., Huang, J., Huang, W., and Liu, Y. (2015). Genome-wide transcriptome and expression profile analysis of Phalaenopsis during explant browning. PLoS One 10:e0123356. doi: 10.1371/journal.pone.0123356
Yang, H., Sung, S. H., and Kim, Y. C. (2007). Antifibrotic phenanthrenes of Dendrobium nobile stems. J. Nat. Prod. 70, 1925–1929. doi: 10.1021/np070423f
Yang, Y., Wang, J., Ma, Z., Sun, G., and Zhang, C. (2014). De novo sequencing and comparative transcriptome analysis of white petals and red labella in Phalaenopsis for discovery of genes related to flower color and floral differentiation. Acta Soc. Bot. Pol. 83, 191–199. doi: 10.5586/asbp.2014.023
Ye, S., Zhao, Q., and Zhang, A. (2017). Anoectochilus roxburghii: a review of its phytochemistry, pharmacology, and clinical applications. J. Ethnopharmacol. 209, 184–202. doi: 10.1016/j.jep.2017.07.032
Yuan, Y., Yu, M., Jia, Z., Song, X., Liang, Y., and Zhang, J. (2018). Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genomics 19:978. doi: 10.1186/s12864-018-5305-6
Yuan, Y., Zhang, J., Liu, X., Meng, M., Wang, J., and Lin, J. (2020). Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics 112, 1781–1794. doi: 10.1016/j.ygeno.2019.10.010
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Zhang, Y., Cai, J., Ruan, H., Pi, H., and Wu, J. (2007). Antihyperglycemic activity of kinsenoside, a high yielding constituent from Anoectochilus roxburghii in streptozotocin diabetic rats. J. Ethnopharmacol. 114, 141–145. doi: 10.1016/j.jep.2007.05.022
Zhang, C., Chen, J., Huang, W., Song, X., and Niu, J. (2021a). Transcriptomics and metabolomics reveal purine and phenylpropanoid metabolism response to drought stress in Dendrobium sinense, an endemic orchid species in Hainan Island. Front. Physiol. 12:692702. doi: 10.3389/fgene.2021.692702
Zhang, Y., Li, Y., Chen, X., Meng, Z., and Guo, S. (2020a). Combined metabolome and transcriptome analyses reveal the effects of mycorrhizal fungus Ceratobasidium sp. AR2 on the flavonoid accumulation in Anoectochilus roxburghii during different growth stages. Int. J. Mol. Sci. 21:564. doi: 10.3390/ijms21020564
Zhang, M., Yu, Z., Zeng, D., Si, C., Zhao, C., Wang, H., et al. (2021b). Transcriptome and metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomol. Ther. 11:736. doi: 10.3390/biom11050736
Zhang, Y., Zhang, L., Liu, J., Liang, J., Si, J., and Wu, S. (2017). Dendrobium officinale leaves as a new antioxidant source. J. Funct. Foods 37, 400–415. doi: 10.1016/j.jff.2017.08.006
Zhang, Y., Zhou, T., Dai, Z., Dai, X., Li, W., Cao, M., et al. (2020b). Comparative transcriptomics provides insight into floral color polymorphism in a Pleione limprichtii orchid population. Int. J. Mol. Sci. 21:247. doi: 10.3390/ijms21010247
Zhao, C., Liu, Q., Halaweish, F., Shao, B., Ye, Y., and Zhao, W. (2003). Copacamphane, picrotoxane, and alloaromadendrane sesquiterpene glycosides and phenolic glycosides from Dendrobium moniliforme. J. Nat. Prod. 66, 1140–1143. doi: 10.1021/np0301801
Zhou, P., Pu, T., Gui, C., Zhang, X., and Gong, L. (2020). Transcriptome analysis reveals biosynthesis of important bioactive constituents and mechanism of stem formation of Dendrobium huoshanense. Sci. Rep. 10:2857. doi: 10.1038/s41598-020-59737-2
Keywords: secondary metabolism, transcriptome, orchids, alkaloids, flavonoids
Citation: Ghai D, Kaur A, Kahlon PS, Pawar SV and Sembi JK (2022) A Walk Through the Maze of Secondary Metabolism in Orchids: A Transcriptomic Approach. Front. Plant Sci. 13:837563. doi: 10.3389/fpls.2022.837563
Received: 16 December 2021; Accepted: 11 April 2022;
Published: 29 April 2022.
Edited by:
Manoj K. Sharma, Jawaharlal Nehru University, IndiaReviewed by:
Pardeep Kumar Bhardwaj, Institute of Bio-Resources and Sustainable Development (IBSD), IndiaCopyright © 2022 Ghai, Kaur, Kahlon, Pawar and Sembi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaspreet K. Sembi, amFzcHJlZXQuc2VtYmlAcHUuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.