- 1National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3CAS-JIC Centre of Excellence for Plant and Microbial Science, Chinese Academy of Sciences, Shanghai, China
Transposable elements (TEs, transposons) are mobile DNAs that are prevalent in most eukaryotic genomes. In plants, their mobility has vastly contributed to genetic diversity which is essential for adaptive changes and evolution of a species. Such mobile nature of transposon has been also actively exploited in plant science research by generating genetic mutants in non-model plant systems. On the other hand, transposon mobilization can bring about detrimental effects to host genomes and they are therefore mostly silenced by the epigenetic mechanisms. TEs have been studied as major silencing targets and acted a main feature in the remarkable growth of the plant epigenetics field. Despite the importance of transposon in plant biology and biotechnology, their mobilization and the underlying mechanisms are largely left unanswered. This is mainly because of the sequence repetitiveness of transposons, which makes their detection and analyses difficult and complicated. Recently, some attempts have been made to develop new experimental methods detecting active transposons and their mobilization behavior. These techniques reveal TE mobility in various levels, including the molecular, cellular, organismal and population scales. In this review, we will highlight the novel technical approaches in the study of mobile genetic elements and discuss how these techniques impacted on the advancement of transposon research and broadened our understanding of plant genome plasticity.
Introduction
Transposable elements (TEs or transposons) are stretches of DNA that move around the genomes and are ubiquitous in most eukaryotic genomes (Feschotte, 2008; Lisch, 2012; Chuong et al., 2017). Particularly, the genomes of major food crops such as barley, wheat and maize contain myriads of transposons making up more than 80% of their genomes (Tenaillon et al., 2010). Among the diverse types of transposons, the long terminal repeat (LTR) retrotransposon is the predominant type of TEs in most plant genomes (Casacuberta and Santiago, 2003; Grandbastien, 2015; Cho, 2018; Satheesh et al., 2021) and thus will be the main focus of this review. The mobilization of an LTR retrotransposon is mediated by the reverse transcription of TE mRNAs to cDNAs (also referred to as extrachromosomal DNA, ecDNA), which happens in virus-like particles (VLPs) and is followed by the insertion to new genomic positions by the integrase (Cho et al., 2019; Satheesh et al., 2021). Due to the mobile nature of transposons and thereby potential danger of genomic instability, they are subject to the host genomes’ epigenetic silencing pathways, including chromatin modification and DNA methylation (Slotkin and Martienssen, 2007; Matzke and Mosher, 2014). On the other hand, transposon is one of the major sources of genetic diversity, which is critical for evolution and adaptive changes of plants (Lisch, 2012; Dubin et al., 2018). Besides, TEs have been actively exploited in the plant science field as useful mutagenic reagents. For example, Tos17 in rice is specifically activated by in vitro tissue culture and the resulting random insertional mutants tagged with Tos17 are important genetic resources in the rice functional genomics (Hirochika et al., 1996; Hirochika, 2010). Similarly, Tnt1 was used to generate genetic mutants in Medicago truncatula, Brachypodium distachyon, and Glycine max (D’Erfurth et al., 2003; Tadege et al., 2008; Revalska et al., 2011; Cui et al., 2012; Nandety et al., 2020), and the maize Ac/Ds DNA transposon system was used as a functional genomics tool in Arabidopsis, Oryza sativa, and Glycine max (Long et al., 1993; Mathieu et al., 2009; Wang et al., 2013). Despite the vast importance of transposons, little is known about the regulatory mechanisms of their mobilization, which is largely because of the lack of experimental methods that can detect the transposition events with sufficient sensitivity and precision.
It is well documented that transposons can be transcriptionally activated by the environmental challenges and at specific cell types and developmental stages (Martínez and Slotkin, 2012; Cho, 2018; Cho et al., 2019). However, the mobilization of activated transposons hardly happens likely because of complex regulation at the post-transcriptional steps (Hung and Slotkin, 2021; Kim et al., 2021b). Owing to the scarcity of transposition events and technical difficulty to detect it, it has been challenging to study transposon mobilization. In the past, transposon insertion was inferred by phenotypic abnormalities caused by deleterious mutations of a gene disrupted by TE integration. For example, some of the epigenetic recombinant inbred lines (epiRILs) generated from the met1 mutant in Arabidopsis exhibited various abnormal phenotypes, which were associated with gene disruption caused by the transposition of Evade retroelement (Mirouze et al., 2009; Reinders et al., 2009; Reinders and Paszkowski, 2009). A PCR-based technique called transposon display (TD) and its derivative methods are usually the experimental approaches of choice to detect and locate new insertions of a transposon of interest (Kim et al., 2021a). Briefly, the adapter with known sequence is ligated to the restriction enzyme-digested DNA ends. PCR amplification by the specific sequences of the adapter and transposon ends yields amplicons containing the genomic regions flanking the transposon of interest. Although TD is an efficient and versatile method to study transposition events, it has certain fundamental limitations; for instance, transposon of high copy number is difficult to be amplified and hardly detected for new insertions. In addition, TD requires prior knowledge of TE sequences and thus relies on the quality of TE annotation. Most importantly, TD can only reveal the insertions that are meiotically inherited and fixed in the genomes, thus is not able to detect transpositions in real time and those happened in somatic cells (Figure 1). Over the last several years, there have been significant efforts to unveil the landscape of transpositions in the plant genomes by developing novel experimental methods. These innovative approaches reveal the mobilomes at varying scales from molecular to population levels. In this review, we will introduce and discuss the up-to-date experimental techniques tracing mobile DNAs in the plant genomes.
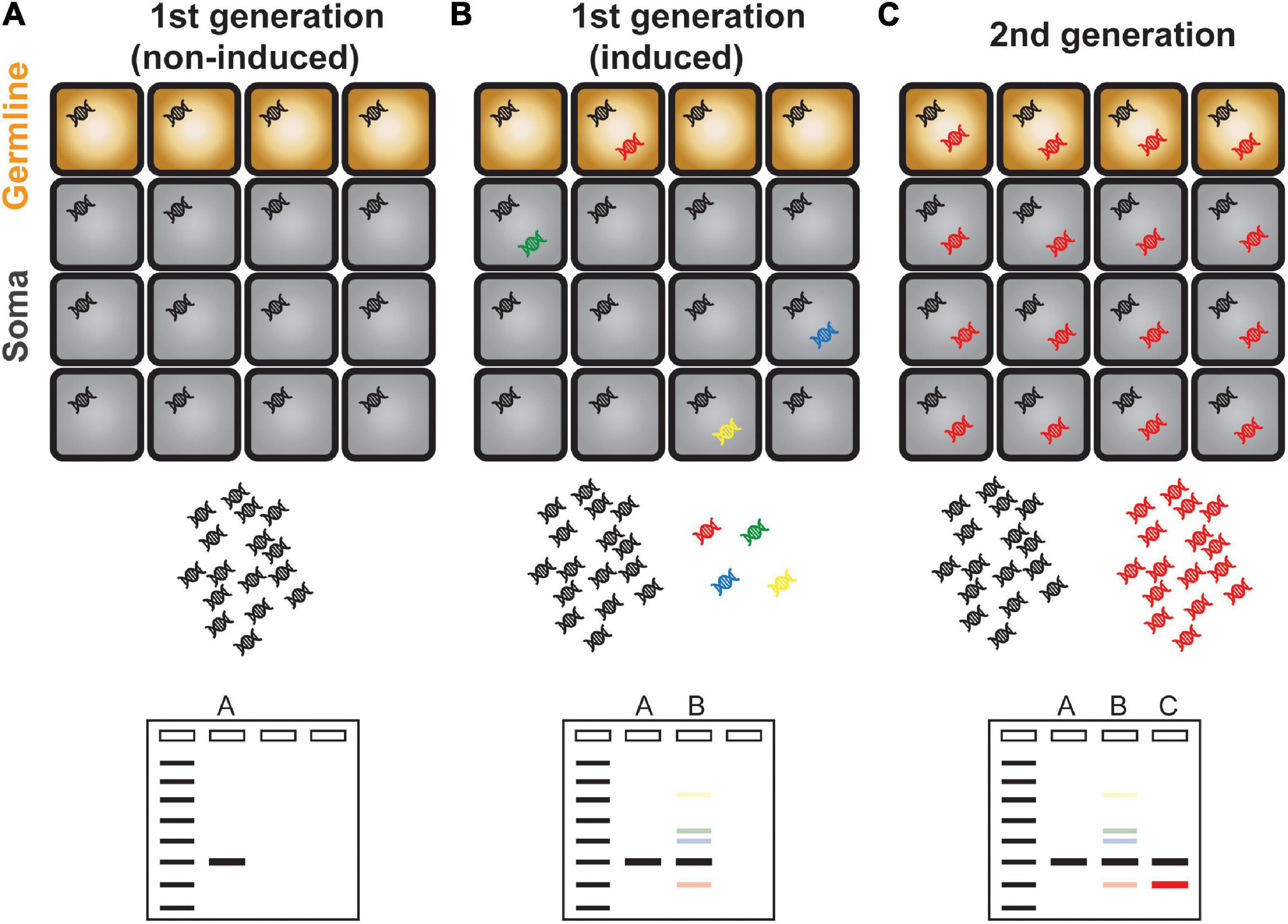
Figure 1. Schematic illustration of transposon display. (A) A single copy TE is present evenly in all cells and is represented as a single band in a transposon display experiment. (B) Activation and mobilization of a TE gave rise to new and additional copies inserted in different genomic positions as represented in different colors. Because of the scarcity of newly copied DNA, transposon display method is unable to amplify these DNAs which are illustrated as faint bands. (C) The new TE copy that mobilized in germline cells is inherited to the next generation. The transgenerationally maintained new TE DNA can be amplified efficiently and is visible as a discrete band in a gel electrophoresis.
Molecular Level
The mobilization cycle of an LTR retrotransposon consists of transcription, reverse transcription, and integration to new genomic positions. Since the direct detection of transposon integration is relatively more challenging, the DNA intermediate which is the final product of reverse transcription reaction and the direct target of integration has been studied to infer the transposon mobility. In this section, the cutting edge methods detecting the DNA intermediates of LTR retrotransposons will be highlighted (Figures 2A,B).
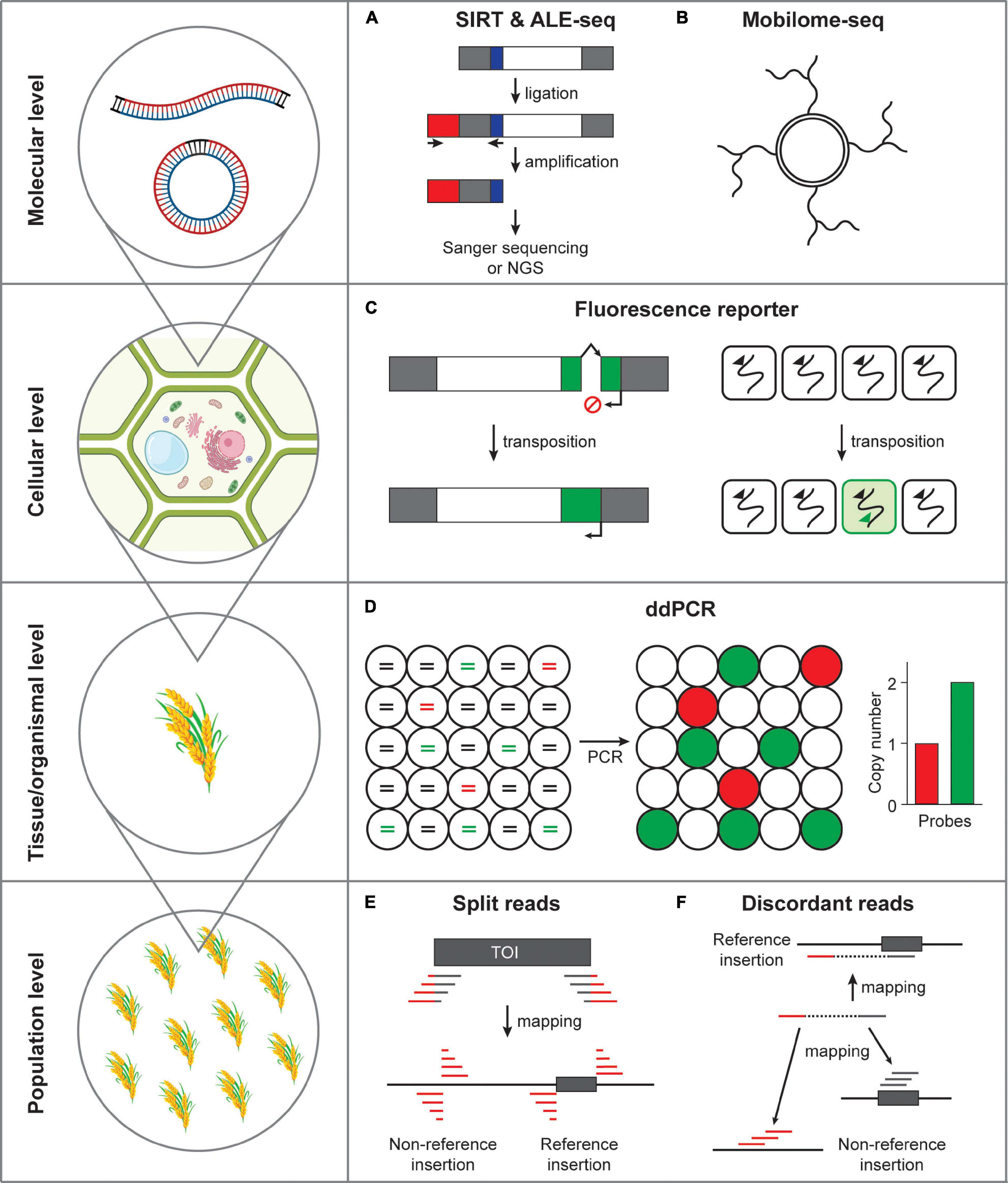
Figure 2. Methods detecting transposon mobility at varying scales. (A) Activated retrotransposon gives rise to extrachromosomal DNAs in both linear and circular forms. SIRT and ALE-seq are the methods that can detect linear DNA intermediates of retroelements. Gray box, long terminal repeats; white box, open reading frame; red box, adapter; blue box, PBS. (B) Extrachromosomal circular DNAs can be amplified by the isothermal strand displacement DNA polymerization. (C) Fluorescence reporter retrotransposition system can specifically label the cells in which transposition occurred. Green box, fluorescence reporter gene; arrows, transcriptional start sites; broken arrow, intron. (D) Transposon mobilization at tissue or organismal level can be assessed by ddPCR. Fragmented DNAs are randomly distributed in separate droplets and independently amplified. Lines in circles represent fragmented genomic DNA colored for different PCR templates. Red, a single-copy reference gene; green, transposon of interest; black, non-templated DNA. (E) The split reads method uses the reads tagged with transposon sequences that map to the insertion positions. TOI, transposon of interest; gray box, transposon; gray line, transposon sequence; red line, flanking sequence of transposon. (F) The discordant reads method uses the paired-end reads, which map to transposon from one side and to a distant genomic site from the other side of a read pair.
Detection of Linear ecDNA
The reverse transcription reaction of transposon gives rise to linear extrachromosomal DNAs (eclDNAs) and it is the linear form of ecDNAs that is capable of integrating to genomic DNA (Cho et al., 2019; Wang et al., 2021a,b). As an attempt to detect eclDNA, Griffiths et al. (2018) established a method named sequence-independent retrotransposon trapping (SIRT). SIRT employs the adapter ligation to the end of eclDNAs and specific amplification targeted to the conserved primer-binding site (PBS) sequence, which is located immediately after the upstream LTR. Using this method a novel family of LTR retrotransposon named DODGER was identified in the Landsberg erecta ecotype of Arabidopsis mutated with MET1 (Griffiths et al., 2018). Unfortunately, SIRT exhibited limited robustness when tested in crop genomes, presumably because of the large size of the crop genomes and abundance of transposon-related sequences. An improved method was then developed named amplification of LTR extrachromosomal DNA followed by sequencing (ALE-seq), which is able to detect the LTRs of crop genomes with larger size (Cho et al., 2019). ALE-seq uses two primers specific to sequences of the adapter and PBS in two separate reactions: in vitro transcription and reverse transcription. Using this novel method, Cho et al. (2019) identified a new Copia-family LTR retrotransposon Go-on in the heat-stressed rice plants. Importantly, the ALE-seq method is particularly useful in non-reference crop species because the final amplicon product can reveal the full-length sequences of the LTR region. Such reference- and annotation-free approach was successfully tested in tomato pericarp samples and identified a novel Gypsy-family retroelement Fruit-Induced RetroElement (FIRE) (Cho et al., 2019). Although ALE-seq is sensitive enough to identify eclDNAs from crop genomes, it can only sequence the 5′ LTR regions and it is desired to further improve this method to cover full range of a TE. Altogether, ALE-seq is a versatile, efficient and high-throughput method identifying active LTR retroelements in crop genomes.
Detection of Circular ecDNA
Two LTRs of eclDNAs are bound by the integrases and homodimerization of integrases place two ends of an eclDNA close next to each other, which is then recognized as a DNA double-strand break by the cellular DNA damage response pathways (Møller et al., 2015, 2016; Lanciano et al., 2017). The homologous recombination and non-homologous end joining pathways repair the LTR-LTR gap, resulting in single-LTR and double-LTR extrachromosomal circular DNAs (eccDNAs), respectively (Lanciano et al., 2017). As a by-product of an activated LTR retrotransposon (albeit incapable of integration) eccDNA is considered to represent active TE mobility. Lanciano et al. (2017) established an experimental method called mobilome-seq that specifically sequences circular DNAs including retrotransposon-derived eccDNAs. The mobilome-seq procedure first initiates with digestion of linear DNA (mostly derived from genomic DNA) and randomly amplifying the remaining circular DNA by the isothermal stand displacement amplification (i.e., rolling circle amplification, Figure 2B). Unlike ALE-seq, mobilome-seq has additional advantage that can sequence full-length retroelement; however, it is also important to note that it reads sequences derived from organellar circular DNAs requiring additional filtering steps to remove them, which compromises the sequencing efficiency (Lanciano et al., 2017; Satheesh et al., 2021). Nonetheless, mobilome-seq can be a useful approach to investigate active retroelements because it requires relatively low sequence coverage, which can be particularly useful to studies using rare plant materials and samples with limited availability. For example, Lanciano et al. (2017) discovered a PopRice retrotransposon family that becomes active in the rice endosperm. In addition, Thieme et al. (2017) identified Houba, a Copia-like retrotransposon in rice, that was activated by the treatment of chemical inhibitors of RNA Polymerase II and DNA methylation. Moreover, Esposito et al. (2019) found that nightshade, a Copia/Ale retrotransposon in potatoes, produces large amount of eccDNAs in non-stressed plants, while under the cold stress condition is no longer active, presumably because of the hypermethylation induced by cold stress. More recently, mobilome-seq revealed that Onsen, a Copia-like retrotransposon specifically activated in the heat-stressed Arabidopsis plants, produces eccDNAs mostly from two copies, AT1G11265 and AT5G13205 (Roquis et al., 2021). In summary, mobilome-seq is a useful method detecting the retrotransposon mobility by sequencing eccDNAs.
Long-Read Sequencing
One of the challenges in the study of transposon is that TE sequences are repetitive in genomes and thus cause serious ambiguity in their analysis. This is particularly more troublesome when analyzing short-read sequencing data. Recently, the long-read sequencing technologies advanced remarkably and greatly improved the accuracy of transposon sequence analysis. For example, in the recent work of Panda and Slotkin, Oxford Nanopore Technology (ONT) sequencing was tested in the DNA methylation-deficient mutants of Arabidopsis, which significantly improved the quality of TE annotation (Panda and Slotkin, 2020). In an independent work by Lee et al. (2020) the ONT method was tested in the VLP fraction collected from the epigenetic mutants of Arabidopsis. This allowed direct identification of active transposable elements in their full lengths and also revealed diverse forms of DNA intermediates (Lee et al., 2020). Overall, the long-read sequencing technology is apparently a game-changer in the field of transposon research and highly expected to unveil the hidden aspects of transposon mobilization which was previously unable to be studied.
Cellular Level
In the previous section, we focused on the methods detecting the DNA intermediates produced from active LTR retrotransposons which could be used as a proxy of TE mobility. It is important to note that the presence of DNA intermediates can be a good indication of TE activation; however, it does not necessarily represent transposition events directly. While in plants there has not been any robust methods detecting transposition events at the cellular level so far, the transposition reporter system used in humans and yeast has served as a standard method assessing transposon mobility. In this section, the transposition reporter assay systems revealing transposon insertion at the cellular level will be introduced (Figure 2C).
Retrotransposition reporter system was first suggested in yeast using the Ty retroelement TyH3 that includes an intron fragment (Boeke et al., 1985). Heidmann et al. (1988) had later developed an improved version using the neo (neomycin phosphotransferase) gene cassette (neoRT). In this system, the neo gene is disrupted by an artificial intron containing polyadenylation signals, thereby the functional neo proteins can be produced only from the transposed intron-free DNA (Heidmann et al., 1988). This method allows for determination of transposition efficiency when cells are grown in the selective G418-containing media (Heidmann et al., 1988). Similar methods have been developed to study mobilization of other types of TEs including the intracisternal A-type particles (IAPs) in mice and the long interspersed elements (LINEs) in Drosophila and human cells (Heldmann and Heidmann, 1991; Jensen and Heidmann, 1991; Tchenio et al., 1993; Maestre et al., 1995; Esnault et al., 2000). Further improvement of retrotransposition assay system was attempted by Moran et al. (1996) by developing the reporter cassette consisting of an antisense copy of neo gene incorporated in two human L1 elements (L1.2 and LRE2) in a cultured human cell line (Rangwala and Kazazian, 2009). In addition, other alternative methods have also been developed by employing blasticidin S deaminase, his3 auxotrophic marker and a lacZ colorimetric indicator (Curcio and Garfinkel, 1991; Tchenio and Heidmann, 1992; Goodier et al., 2007). However, such intron-containing reporter systems had some drawbacks that retrotransposition assay is dependent on antibiotics resistance and assessed by counting colonies, which usually takes long time and has relatively low throughput. Recently, innovations to this classical method have been made by replacing the antibiotics resistance genes to visual fluorescence (Ostertag et al., 2000) and bioluminescence genes (Xie et al., 2011), which dramatically increases the sensitivity and throughput, enabling large-scale screening experiments. In summary, the retrotransposition assay systems have been widely used to determine the transposition rate of a retroelement mostly in non-plant systems. Introducing such system to the plant systems will enable single-cell detection of transposition and greatly improve our understanding of transposon mobilization.
Tissue/Organism Level
Droplet Digital PCR
The retrotransposition reporter assay system described in the previous section can be potentially useful for cell- and tissue-level detection of transposition events. The synthetic artificial retrotransposon mobility assay is powerful because it enables direct visualization of transposition; however, such transgenic approach can be challenging in many non-reference plant species. Determination of copy number changes of an endogenous TE can be one of the easiest alternative methods to assess transpositional activity. It is worth noting, however, that the logarithmic quantitative real-time PCR analysis is difficult to measure subtle differences of copy number (Bubner and Baldwin, 2004; Bubner et al., 2004; Fan and Cho, 2021). Droplet digital PCR (ddPCR) is a far more accurate and sensitive technique that allows for digital measurement of DNA copy number (Hindson et al., 2013; Doi et al., 2015; Campomenosi et al., 2016; Głowacka et al., 2016; Fan and Cho, 2021). The ddPCR experiment performs DNA amplification in thousands of nanoliter-scale droplets that read-out positive or negative fluorescence signals (Figure 2D). The resulting digital data is then processed by a Poisson probability distribution to derive copy numbers. In fact, we previously showed that ddPCR can be a robust method that accurately detects the copy number of a retrotransposon (Fan and Cho, 2021). Importantly, the ddPCR technique only requires a trace amount of DNA and is therefore possible to be performed in DNAs extracted from small amount of tissues and rare samples.
Population Level
Next-Generation Sequencing-Based Transposable Element Mapping
Arabidopsis 1,001 genome project produced massive paired-end short-read whole-genome sequencing data from 1,135 accessions from a worldwide collection (Weigel and Mott, 2009; Cao et al., 2011; Alonso-Blanco et al., 2016). Similar attempt has been made in rice generating sequencing data from a total of more than 3,000 germplasm accessions (Li J.-Y. et al., 2014; Li Z. et al., 2014). Equipped with relatively well-assembled and annotated reference genomes available for both plant species, TE insertion polymorphisms have been intensively profiled at population level. Several softwares have been developed so far to systematically identify transposon insertions. These tools take advantage of diverse sequencing read information; for instance, split reads in Transposon Insertion Finder (TIF) (Nakagome et al., 2014), SPLITREADER (Baduel et al., 2021b), and RTRIP (Liu et al., 2020), discordant read pair alignment in TRACKPOSON (Carpentier et al., 2019), and combination of these two as demonstrated in TEPID (Stuart et al., 2016). Additionally, in a recent work of Baduel et al. (2021a) SPLITREADER and TEPID pipelines were integrated, building an intensive map of TE landscape in Arabidopsis.
The split-reads method first searches for reads containing the end sequences of a TE and target site duplications (TSDs), which are identical sequences flanking a TE and created as a result of transposition (Figure 2E). In TIF, the read sequences tagged with transposon end sequences are mapped to the reference genome to identify the locations of de novo insertions (Nakagome et al., 2014). Similarly, SPLITREADER extracts reads that do not properly map to the reference genome and forcedly map to 5′ and 3′ TE sequence extremities (within a range of 300 bp) by soft clipping (Baduel et al., 2021b). Then, the bona fide insertions and their locations are identified by mapping the clipped reads to the reference genome. Recently, Liu et al. (2020) tested a similar method in rice and generated the RTRIP database, which contains the comprehensive profile of transposon insertion polymorphisms in the rice 3K genome project.
The discordant read pair method employs mapping of reads from one side to the target TE and the other side to a distant genomic region (Figure 2F). TRACKPOSON, for instance, first maps all reads of a given accession onto each TE family represented by a single consensus sequence, and then maps the unmapped paired reads to the rice reference genome to determine its location (Carpentier et al., 2019). The transposition landscape revealed by these methods uncovered that transposon proliferation is most strongly associated with the presence of a transposon at a specific location, which was somehow activated during the evolutionary process (Carpentier et al., 2019).
Concluding Remarks and Future Perspectives
We reviewed the recent technical advances in transposon research by highlighting several new methods identifying active TEs and detecting transposition events (Figure 2). These novel experimental methods and improved transposon annotation aided by the long-read sequencing technologies and population-scale genome resequencing databases will open-up a new window to unveil a long-lasting mystery of jumping genes. Although the experimental techniques described above has greatly improved our ability to observe transposition events, there are still several issues left to be dealt with. Firstly, detection of transposition events at single-cell level will be obviously the next task to be accomplished. To this end, a novel approach for the single-cell detection of transposon mobilization is highly desired. Secondly, the single-cell genomics will vastly benefit the transposon biology. The transposition reporter systems introduced above rely on the artificially engineered TE sequences. The investigation of the native TEs and their transposition at high resolution will only be possible when the single-cell genomics technologies become more available. Thirdly, the detection sensitivity of transposon research tools will have to be improved further. The new experimental tools to study transposon such as ALE-seq and mobilome-seq are mostly tested in the epigenetic mutants where transposons become unusually active in mobility. Although these methods were sensitive enough to discover novel active retroelements (Go-on and PopRice), the moderately active TEs were difficult to be identified (Lanciano et al., 2017; Cho et al., 2019). Considering the rarity of DNAs representing activated TE intermediates or derived from transposed copy, the improvement of detection sensitivity of these methods will help identify new transposons that are present in small niches of cells or activated only to a moderate level. Altogether, the technical advances of transposon research at varying scales have greatly contributed to our understanding of TE life cycle and will broaden the breadth of knowledge on mobile genetic elements and genome plasticity.
Author Contributions
WF, LW, JiC, HL, and EK drafted the manuscript. EK and JuC edited the manuscript. JuC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the grants from the National Natural Science Foundation of China (31970518, 32150610473, and 32111540256), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030209), and the General Program of Natural Science Foundation of Shanghai (21ZR1470700). EK was the recipient of a President’s International Fellowship Initiative (PIFI) young staff fellowship (2021FYB0001) from CAS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alonso-Blanco, C., Andrade, J., Becker, C., Bemm, F., Bergelson, J., Borgwardt, K. M., et al. (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491. doi: 10.1016/j.cell.2016.05.063
Baduel, P., Quadrana, L., and Colot, V. (2021b). Efficient detection of transposable element insertion polymorphisms between genomes using short-read sequencing data. Methods Mol. Biol. 2250, 157–169. doi: 10.1007/978-1-0716-1134-0_15
Baduel, P., Leduque, B., Ignace, A., Gy, I., Gil, J., Loudet, O., et al. (2021a). Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biol. 22, 138. doi: 10.1186/s13059-021-02348-5
Boeke, J. D., Garfinkel, D. J., Styles, C. A., and Fink, G. R. (1985). Ty elements transpose through an RNA intermediate. Cell 40, 491–500. doi: 10.1016/0092-8674(85)90197-7
Bubner, B., and Baldwin, I. T. (2004). Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 23, 263–271. doi: 10.1007/s00299-004-0859-y
Bubner, B., Gase, K., and Baldwin, I. T. (2004). Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnol. 4:14. doi: 10.1186/1472-6750-4-14
Campomenosi, P., Gini, E., Noonan, D. M., Poli, A., D’Antona, P., Rotolo, N., et al. (2016). A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 16:60. doi: 10.1186/s12896-016-0292-7
Cao, J., Schneeberger, K., Ossowski, S., Günther, T., Bender, S., Fitz, J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43, 956–963. doi: 10.1038/ng.911
Carpentier, M.-C., Manfroi, E., Wei, F.-J., Wu, H.-P., Lasserre, E., Llauro, C., et al. (2019). Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat. Commun. 10:24. doi: 10.1038/s41467-018-07974-5
Casacuberta, J. M., and Santiago, N. (2003). Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311, 1–11. doi: 10.1016/S0378-1119(03)00557-2
Cho, J. (2018). Transposon-derived Non-coding RNAs and their function in plants. Front. Plant Sci. 9:600. doi: 10.3389/fpls.2018.00600
Cho, J., Benoit, M., Catoni, M., Drost, H.-G., Brestovitsky, A., Oosterbeek, M., et al. (2019). Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nat. Plants 5, 26–33. doi: 10.1038/s41477-018-0320-9
Chuong, E. B., Elde, N. C., and Feschotte, C. (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. doi: 10.1038/nrg.2016.139
Cui, Y., Barampuram, S., Stacey, M. G., Hancock, C. N., Findley, S., Mathieu, M., et al. (2012). Tnt1 retrotransposon mutagenesis: a tool for soybean functional genomics. Plant Physiol. 161, 36–47. doi: 10.1104/pp.112.205369
Curcio, M. J., and Garfinkel, D. J. (1991). Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 88, 936–940. doi: 10.1073/pnas.88.3.936
D’Erfurth, I., Cosson, V., Eschstruth, A., Lucas, H., Kondorosi, A., and Ratet, P. (2003). Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J. 34, 95–106. doi: 10.1046/j.1365-313X.2003.01701.x
Doi, H., Takahara, T., Minamoto, T., Matsuhashi, S., Uchii, K., and Yamanaka, H. (2015). Droplet digital polymerase chain reaction (PCR) outperforms real-time PCR in the detection of environmental DNA from an invasive fish species. Environ. Sci. Technol. 49, 5601–5608. doi: 10.1021/acs.est.5b00253
Dubin, M. J., Mittelsten Scheid, O., and Becker, C. (2018). Transposons: a blessing curse. Curr. Opin. Plant Biol. 42, 23–29. doi: 10.1016/j.pbi.2018.01.003
Esnault, C., Maestre, J., and Heidmann, T. (2000). Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. doi: 10.1038/74184
Esposito, S., Barteri, F., Casacuberta, J., Mirouze, M., Carputo, D., and Aversano, R. (2019). LTR-TEs abundance, timing and mobility in Solanum commersonii and S. tuberosum genomes following cold-stress conditions. Planta 250, 1781–1787. doi: 10.1007/s00425-019-03283-3
Fan, W., and Cho, J. (2021). “Quantitative measurement of transposon copy number using the droplet digital PCR,” in Plant Transposable Elements. Methods in Molecular Biology, Vol. 2250, ed. J. Cho (New York, NY: Humana), 171–176. doi: 10.1007/978-1-0716-1134-0_16
Feschotte, C. (2008). Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405. doi: 10.1038/nrg2337
Głowacka, K., Kromdijk, J., Leonelli, L., Niyogi, K. K., Clemente, T. E., and Long, S. P. (2016). An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 39, 908–917. doi: 10.1111/pce.12693
Goodier, J. L., Zhang, L., Vetter, M. R., and Kazazian, H. H. (2007). LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483. doi: 10.1128/mcb.00332-07
Grandbastien, M. A. (2015). LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta Gene Regul. Mech. 1849, 403–416. doi: 10.1016/j.bbagrm.2014.07.017
Griffiths, J., Catoni, M., Iwasaki, M., and Paszkowski, J. (2018). Sequence-independent identification of active LTR Retrotransposons in Arabidopsis. Mol. Plant 11, 508–511. doi: 10.1016/j.molp.2017.10.012
Heidmann, T., Heidmann, O., and Nicolas, J. F. (1988). An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl. Acad. Sci. U.S.A. 85, 2219–2223. doi: 10.1073/pnas.85.7.2219
Heldmann, O., and Heidmann, T. (1991). Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64, 159–170. doi: 10.1016/0092-8674(91)90217-M
Hindson, C. M., Chevillet, J. R., Briggs, H. A., Gallichotte, E. N., Ruf, I. K., Hindson, B. J., et al. (2013). Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10, 1003–1005. doi: 10.1038/nmeth.2633
Hirochika, H. (2010). Insertional mutagenesis with Tos17 for functional analysis of rice genes. Breed. Sci. 60, 486–492. doi: 10.1270/jsbbs.60.486
Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A. 93, 7783–7788. doi: 10.1073/pnas.93.15.7783
Hung, Y.-H., and Slotkin, R. K. (2021). The initiation of RNA interference (RNAi) in plants. Curr. Opin. Plant Biol. 61, 102014. doi: 10.1016/j.pbi.2021.102014
Jensen, S. A., and Heidmann, T. (1991). An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 10, 1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x
Kim, E. Y., Wang, L., Lei, Z., Li, H., Fan, W., and Cho, J. (2021b). Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat. Plants 7, 303–309. doi: 10.1038/s41477-021-00867-4
Kim, E. Y., Fan, W., and Cho, J. (2021a). Determination of TE insertion positions using transposon display. Methods Mol. Biol. 2250, 115–121. doi: 10.1007/978-1-0716-1134-0_11
Lanciano, S., Carpentier, M.-C., Llauro, C., Jobet, E., Robakowska-Hyzorek, D., Lasserre, E., et al. (2017). Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. 13:e1006630. doi: 10.1371/journal.pgen.1006630
Lee, S. C., Ernst, E., Berube, B., Borges, F., Parent, J.-S., Ledon, P., et al. (2020). Arabidopsis retrotransposon virus-like particles and their regulation by epigenetically activated small RNA. Genome Res. 30, 576–588. doi: 10.1101/gr.259044.119
Li, J.-Y., Wang, J., and Zeigler, R. S. (2014). The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience 3:8. doi: 10.1186/2047-217X-3-8
Li, Z., Fu, B. Y., Gao, Y. M., Wang, W. S., Xu, J. L., Zhang, F., et al. (2014). The 3,000 rice genomes project. Gigascience 3:7. doi: 10.1186/2047-217X-3-7
Lisch, D. (2012). How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49–61. doi: 10.1038/nrg3374
Liu, Z., Wang, T., Wang, L., Zhao, H., Yue, E., Yan, Y., et al. (2020). RTRIP: a comprehensive profile of transposon insertion polymorphisms in rice. Plant Biotechnol. J. 18, 2379–2381. doi: 10.1111/pbi.13425
Long, D., Martin, M., Sundberg, E., Swinburne, J., Puangsomlee, P., and Coupland, G. (1993). The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc. Natl. Acad. Sci. U.S.A. 90, 10370–10374. doi: 10.1073/pnas.90.21.10370
Maestre, J., Tchénio, T., Dhellin, O., and Heidmann, T. (1995). mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 14, 6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x
Martínez, G., and Slotkin, R. K. (2012). Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr. Opin. Plant Biol. 15, 496–502. doi: 10.1016/j.pbi.2012.09.001
Mathieu, M., Winters, E. K., Kong, F., Wan, J., Wang, S., Eckert, H., et al. (2009). Establishment of a soybean (Glycine max Merr. L) transposon-based mutagenesis repository. Planta 229, 279–289. doi: 10.1007/s00425-008-0827-9
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Mirouze, M., Reinders, J., Bucher, E., Nishimura, T., Schneeberger, K., Ossowski, S., et al. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430. doi: 10.1038/nature08328
Møller, H. D., Larsen, C. E., Parsons, L., Hansen, A. J., Regenberg, B., and Mourier, T. (2016). Formation of extrachromosomal circular DNA from long terminal repeats of retrotransposons in Saccharomyces cerevisiae. G3 6, 453–462. doi: 10.1534/g3.115.025858
Møller, H. D., Parsons, L., Jørgensen, T. S., Botstein, D., and Regenberg, B. (2015). Extrachromosomal circular DNA is common in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 3114–3122. doi: 10.1073/pnas.1508825112
Moran, J. V., Holmes, S. E., Naas, T. P., Deberardinis, R. J., Boeke, J. D., and Kazazian, H. H. (1996). High frequency retrotransposition inin cultured mammalian cells. Cell 87, 917–927.
Nakagome, M., Solovieva, E., Takahashi, A., Yasue, H., Hirochika, H., and Miyao, A. (2014). Transposon insertion finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics 15:71. doi: 10.1186/1471-2105-15-71
Nandety, R. S., Serrani-Yarce, J. C., Gill, U. S., Oh, S., Lee, H., Zhang, X., et al. (2020). Insertional mutagenesis of Brachypodium distachyon using the Tnt1 retrotransposable element. Plant J. 103, 1924–1936. doi: 10.1111/tpj.14813
Ostertag, E. M., Luning Prak, E. T., DeBerardinis, R. J., Moran, J. V., and Kazazian, H. H. (2000). Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423. doi: 10.1093/nar/28.6.1418
Panda, K., and Slotkin, R. K. (2020). Long-Read cDNA sequencing enables a “Gene-Like” transcript annotation of transposable elements. Plant Cell 32, 2687–2698. doi: 10.1105/tpc.20.00115
Rangwala, S. H., and Kazazian, H. H. (2009). The L1 retrotransposition assay: a retrospective and toolkit. Methods 49, 219–226. doi: 10.1016/j.ymeth.2009.04.012
Reinders, J., and Paszkowski, J. (2009). Unlocking the Arabidopsis epigenome. Epigenetics 4, 557–563. doi: 10.4161/epi.4.8.10347
Reinders, J., Wulff, B. B. H., Mirouze, M., Mari-Ordonez, A., Dapp, M., Rozhon, W., et al. (2009). Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23, 939–950. doi: 10.1101/gad.524609
Revalska, M., Vassileva, V., Goormachtig, S., Van Hautegem, T., Ratet, P., and Iantcheva, A. (2011). Recent progress in development of Tnt1 functional genomics platform for Medicago truncatula and Lotus japonicus in Bulgaria. Curr. Genomics 12, 147–152. doi: 10.2174/138920211795564313
Roquis, D., Robertson, M., Yu, L., Thieme, M., Julkowska, M., and Bucher, E. (2021). Genomic impact of stress-induced transposable element mobility in Arabidopsis. Nucleic Acids Res. 49, 10431–10447. doi: 10.1093/nar/gkab828
Satheesh, V., Fan, W., Chu, J., and Cho, J. (2021). Recent advancement of NGS technologies to detect active transposable elements in plants. Genes Genomics 43, 289–294. doi: 10.1007/s13258-021-01040-z
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Stuart, T., Eichten, S. R., Cahn, J., Karpievitch, Y. V., Borevitz, J. O., and Lister, R. (2016). Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife 5:e20777. doi: 10.7554/eLife.20777
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Tchenio, T., and Heidmann, T. (1992). High-frequency intracellular transposition of a defective mammalian provirus detected by an in situ colorimetric assay. J. Virol. 66, 1571–1578. doi: 10.1128/jvi.66.3.1571-1578.1992
Tchenio, T., Segal-Bendirdjian, E., and Heidmann, T. (1993). Generation of processed pseudogenes in murine cells. EMBO J. 12, 1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x
Tenaillon, M. I., Hollister, J. D., and Gaut, B. S. (2010). A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15, 471–478. doi: 10.1016/j.tplants.2010.05.003
Thieme, M., Lanciano, S., Balzergue, S., Daccord, N., Mirouze, M., and Bucher, E. (2017). Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 18, 134. doi: 10.1186/s13059-017-1265-4
Wang, L., Cho, J., and Satheesh, V. (2021a). Bioinformatics analysis guides to LTR retrotransposon-derived extrachromosomal linear DNAs identified by ALE-seq. Methods Mol. Biol. 2250, 111–114. doi: 10.1007/978-1-0716-1134-0_10
Wang, L., Kim, E. Y., and Cho, J. (2021b). High-throughput profiling of extrachromosomal linear DNAs of long terminal repeat retrotransposons by ALE-seq. Methods Mol. Biol. 2250, 103–110. doi: 10.1007/978-1-0716-1134-0_9
Wang, N., Long, T., Yao, W., Xiong, L., Zhang, Q., and Wu, C. (2013). Mutant resources for the functional analysis of the rice genome. Mol. Plant 6, 596–604. doi: 10.1093/mp/sss142
Weigel, D., and Mott, R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10:107. doi: 10.1186/gb-2009-10-5-107
Keywords: transposon, long terminal repeat (LTR) retrotransposon, retrotransposition, ALE-seq, mobilome-seq, long-read sequencing, droplet digital PCR (ddPCR)
Citation: Fan W, Wang L, Chu J, Li H, Kim EY and Cho J (2022) Tracing Mobile DNAs: From Molecular to Population Scales. Front. Plant Sci. 13:837378. doi: 10.3389/fpls.2022.837378
Received: 16 December 2021; Accepted: 11 January 2022;
Published: 01 February 2022.
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Stefan Grob, University of Zurich, SwitzerlandCopyright © 2022 Fan, Wang, Chu, Li, Kim and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun Yu Kim, ZXVueXVraW1AY2VtcHMuYWMuY24=; Jungnam Cho, anVuZ25hbWNob0BjZW1wcy5hYy5jbg==
†These authors have contributed equally to this work
 Wenwen Fan1,2†
Wenwen Fan1,2† Ling Wang
Ling Wang Hui Li
Hui Li Eun Yu Kim
Eun Yu Kim Jungnam Cho
Jungnam Cho