
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 06 April 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.836244
This article is part of the Research TopicPlant Circadian RhythmsView all 7 articles
Plants require an endogenous regulatory network and mechanism to cope with diurnal environmental changes and compensate for their sessile nature. Plants use the circadian clock to anticipate diurnal changes. Circadian rhythm predicts a 24-h cycle with 16 h of light and 8 h of darkness in response to abiotic and biotic factors as well as the appropriate temperature. For a plant’s fitness, proper growth, and development, these rhythms synchronize the diurnal photoperiodic changes. Input pathway, central oscillator, and output pathway are the three components that make up the endogenous clock. There are also transcriptional and translational feedback loops (TTFLs) in the clock, which are dependent on the results of gene expression. Several physiological processes, such as stress acclimatization, hormone signaling, morphogenesis, carbon metabolism, and defense response, are currently being investigated for their interactions with the circadian clock using phenotypic, genomic, and metabolic studies. This review examines the role of circadian rhythms in the regulation of plant metabolic pathways, such as photosynthesis and carbon metabolism, as well as developmental and degenerative processes, such as flowering and senescence. Furthermore, we summarized signaling pathways related to circadian rhythms, such as defense response and gene regulatory pathways.
Living organisms, such as animals, cyanobacteria, and plants, have an internal endogenous oscillator known as the circadian clock, which predicts the alteration during the light and dark cycles. The term “circadian” was first coined in 1959 by Franz Halberg, originated from two different Latin words “circa” means “around” and “diem or dies” means “day” (McClung, 2006; Srivastava et al., 2019; Man et al., 2020). It permits organisms to synchronize and assemble for daily or seasonal changes depending on the surrounding environmental conditions. The circadian clock is an internal biological timekeeper that helps the plants to attain fitness, proper growth, and development (Inoue et al., 2017). This endogenous oscillator is generally comprised of three different modules: (i) input pathway, which gives information about the surrounding environment; (ii) central oscillator that consists of “canonical clock gene,” which composes the elite clock design; and (iii) the output pathway that constitutes the clock-driven downstream processes. The central oscillator includes complex TTFLs (de Dios et al., 2018) that blend with the post-transcriptional and post-translational modifications (Dalchau et al., 2011; Yan et al., 2021). The circadian clock has a self-reliant mechanism and their metabolic processes can also be administered by circadian rhythm, which was previously studied with the model plant Arabidopsis, also in the potato, and rice crops (Kim et al., 2017; Inoue et al., 2018).
The circadian rhythm oscillates daily for about 24 h in the period of light and dark cycles in response to biotic and abiotic factors (Kiełbowicz-Matuk et al., 2014). From the studies of Harmer (2009), it was detected that there are observed three feedback loops, such as morning, central, and evening loops, from the central oscillator, in the model organism Arabidopsis thaliana. The central loop consists of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) (Kim et al., 2016) encode MYB-related transcription factors that are classified as the morning expressed genes. CCA1 modulates clock-independent and clock-dependent responses (Lei et al., 2019). TIMING OF CAB EXPRESSION (TOC1) is an evening expressed gene from the core or central loop, which is from the family of Pseudo-Response Regulator (PRR) (Shogo et al., 2008). Thus, these core loops combine with the morning and evening loops and help in the primary construction of circadian rhythm in plants (Wang and Ma, 2013).
From the previous studies, it was perceived that in the morning loop, CCA1 or LHY gene forms a negative feedback loop by combining with a PRR7 or PRR9 and represses the expression of CCA1 and LHY (Farré et al., 2005; Nakamichi et al., 2010). Likewise, in the evening loop, the TOC1 gene forms a negative feedback loop along with GIGANTEA (GI); also it represses an unknown Y factor, which further activates the expression of the TOC1 gene (Huq et al., 2000; Mizoguchi et al., 2005; Chen et al., 2013; Saini et al., 2019). GI activates ZEITLUPE (ZLT) protein and then acts along with it and targets TOC1 protein for degradation (Figure 1). In addition, LUX ARRHYTHMO (LUX) is a gene and EARLY FLOWERING (ELF), such as ELF 3 and ELF 4, that is the protein of the evening complex (EC), which forms various sets of interlocked negative feedback loops (Nohales and Kay, 2016). In the previous studies (Rawat et al., 2011; Rugnone et al., 2013; Wu et al., 2016), it is mentioned that REVEILLE (RVE) genes, LIGHT-REGULATED WD (LWD) 1 and 2 genes (transcriptional coactivators) (McClung, 2019), and NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED (LNK) genes were the positive regulators found in the loops of the circadian oscillator.
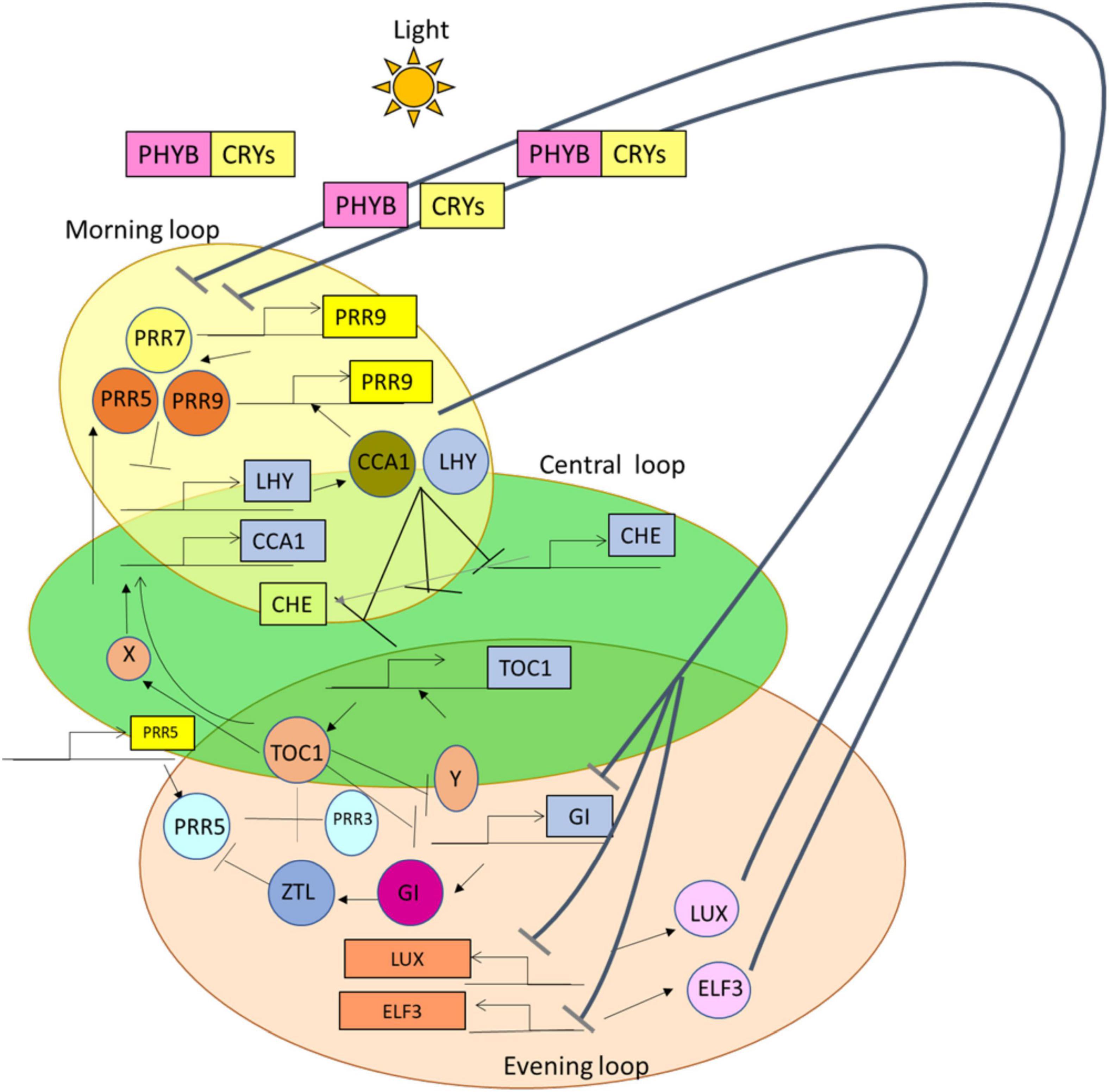
Figure 1. A simplified representation of the suppression of genes having the proteins and photoreceptors present during the functioning of 24 h circadian rhythm. In the presence of light, these photoreceptors Cryptochromes (CRYs) and Phytochrome B (PHY B), which are represented in yellow and pink squares, help in the functioning of genes and proteins; along with the formation of two different negative loops. That is, these morning loop genes [CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) or LATE ELONGATED HYPOCOTYL (LHY)] combine with Pseudo-Response Regulators (PRR7 or PRR9) and suppress the action of CCA1 or LHY (represented with black lines with arrows). On the other hand, TIMING OF CAB EXPRESSION (TOC1) combines with GI, which then leads to the activation of the TOC1 gene. ZEITLUPE (ZTL) is activated with the help of GI, and then GI itself combines with ZTL and suppresses the function of TOC1 (indicated with the black lines along with arrows).
In model organism A. thaliana, it was found that circadian cycle duration can be extended or lengthened significantly using the overexpressed transcription factors BBX18, BBX19, and BBX32 (B-Box); also BBX18 increases the speed of this biological cycle and BBX32 overexpression may even lead to late flowering (Yuan et al., 2021). Circadian gating is the process where the circadian rhythm balances the plant response with different environmental cues; thus, the response depends upon the daytime (Belbin et al., 2019). The regulation of the circadian clock is intrinsically connected with the responses toward decreasing in temperature. Adaptation of cold has proteins involved, such as C-REPEAT/DRE BINDING FACTOR (CBF), which has a role as a key regulator (Panter et al., 2019).
The input pathway reveals to extend some of the environmental variations or signals, recognized by the photoreceptors, such as Phytochrome B (PHY B) and Cryptochromes (CRYs), which develops the components of the negative loops and temperature to entrain the stage and waveform of the circadian oscillator (Aguilar-Arnal and Sassone-Corsi, 2015). The output pathway controls several processes, such as reproductive development, hormonal production, defense responses, and the minimal percent of expression of the genome (Shor et al., 2017). Various physiological processes take place, such as stress acclimatization, hormone signaling, morphogenesis, carbon metabolism, and defense response including phenotypic, genomic, and metabolic studies in the later stages, along with the interaction with this circadian clock (Sanchez et al., 2011).
Circadian research has been extremely interdisciplinary, attracting researchers from a wide range of scientific disciplines. The existence of circadian rhythms was demonstrated by astronomer Jean Jacques d’Ortous deMairan in 1729. Jean Jacques d’Ortous deMairan discovered that the daily leaf movements of the heliotrope plant, Mimosa pudica, continued in complete darkness that implies the existence of an endogenous time-generating mechanism in accordance with geophysical time. Bünning, however, provided the first evidence for a genetic basis of circadian rhythms two centuries later. Bünning reported that in common beans, the offspring’s period lengths ranged between the extremes of the parent generation’s period lengths.
Circadian rhythms generate a biological time of day measurement. Circadian regulation is an important adaptation in plants to their changing environment. The majority of our understanding of the molecular aspects of circadian regulation in plants comes from controlled laboratory experiments. However, it is becoming clear that the circadian clock plays complex roles in transcriptome coordination under natural conditions, in both naturally occurring plant populations and crop species. This review has a work scheme based on the role of circadian rhythms in the regulation of plant metabolic pathways, such as photosynthesis and carbon metabolism, the regulation of developmental processes, such as flowering and degenerative processes, i.e., senescence, the regulation of plant signaling pathways, such as defense response, and the gene regulatory pathways interrelated with the circadian rhythms. Further, the review will also be highlighted with the research gaps that identify new domains and suggest recommendations for future investigation (Table 1).
The continual pattern of the circadian rhythm in many organisms, mainly in plants, leads to the opening of stomata during daytime and the closing of stomata during the nighttime (Webb, 2003). The circadian rhythm plays an essential role in the interaction between photosynthesis activities and the diurnal variation based upon the availability of light (Hennessey and Field, 1991). The minute opening in the leaves is said to be stomata; thus, the opening and closing of stomata that concern the balanced amount of carbon dioxide taken inside along with the proportion of water and oxygen are released out. Photosynthesis itself is a salient circadian rhythm in the plant, which is composed of various molecular and physiological processes, such as stomatal opening, chlorophyll contents, chlorophyll fluorescence, and net carbon assimilation rate (Rascher et al., 2001; Pan et al., 2015).
The opening of stomata is an essential process for net carbon assimilation. It is also essential for the carbon dioxide diffusion into mesophyll cells from the atmosphere, based on the cost of water. Naturally, photosynthesis production gets affected by the action of circadian responsiveness or closing of stomata (Youngsung et al., 2017). Furthermore, the rhythm produced in stomatal conductance could affect the carbon assimilation rate by restricting the flow of carbon dioxide into the leaves. The process of photosynthesis influences plant metabolism; thus, it synchronizes the Photosystem II activity or stomatal opening and movement of the chloroplast. From the previous study on A. thaliana (Dodd et al., 2015), the overexpression of CCA1 in the plant could lead to the reduction of net carbon assimilation, which may also lead to the reduction of net carbon assimilation maximal plant fitness.
There is a bidirectional relationship between the biological clock and photosynthesis because photosynthesis gets affected by circadian regulation. In addition, the circadian clock has a core structure due to which it gets affected by photosynthesis (Haydon et al., 2013). The regulation of this oscillator affects the diurnal fluctuation in the stomatal conductance and photosynthesis process and various processes, such as respiration and growth. Even the behavior of stomata gets affected by circadian regulation (de Dios et al., 2020). The fraction of the diurnal pattern of variation in the stomatal conductance, which is allocated to the clock, is higher in amount during the changes in the daytime than in the changes allocated to the clock by photosynthesis. Thus, this indicates that the stomatal conductance has a vigorous circadian regulation than photosynthesis (de Dios et al., 2018).
Several studies on A. thaliana found that the endogenous rhythm controls photosynthesis and physiology, and the plant attains a peak of fitness (Green et al., 2002; Dodd et al., 2005; Müller et al., 2014). The regulation of the circadian oscillator raises the plant productivity, viability of seed, and plant survival. From de Dios et al. (2018), it is known that, in the Calvin cycle, the rate of photosynthesis depends upon the stomatal conductance and the mesophyll conductance and the biochemical process. The adjustment made in the accumulation of carbon dioxide present within the intercellular spaces by the stomatal conductance helps in modulating the process of photosynthesis. It is also known that there is a negative interaction or lack of interaction between photosynthesis and stomatal conductance. This suggests that the circadian regulation in stomatal conductance is implausible to guide the regulation of photosynthesis by circadian oscillation (Males and Howard, 2017). There is a primary role for endogenous rhythm in mesophyll conductance in balancing the accumulation of carbon dioxide in the chloroplast.
An endogenous rhythm in the framework of the light-harvesting compounds is an essential feature for the circadian rhythm in photosynthesis. The chlorophyll synthesis rate is controlled by the circadian rhythm and diurnal variations in chlorophyll a/b, which is also connected with an endogenous rhythm in the process of photosynthesis. The gene light-harvesting complex A/B protein (LHCB) activation occurs when the CCA1 and LHY moderate; it also directly gets binding with the promoters. The circadian regulation synchronizes the gene transcription linked with the Calvin cycle. Photosynthesis interacts with the PRR7 gene to manage the maintenance and entrainment of vigorous circadian oscillators (Haydon et al., 2013). In addition to this, the expression level of LHY/CCA1 and PRR7/PRR9 has the capability to induce the performance of plant growth and development (Müller et al., 2014).
In the C3 photosynthesis pathway or photosynthetic carbon reduction (PCR) cycle (Furbank and Taylor, 1995), plants consist of a single chloroplast that conducts many reactions; mainly, it helps in the conversion of light energy into chemical energy. Ribulose-1, 5-bisphosphate carboxylase/oxygenase (RuBisCO) is the protein used to activate the initial fixation of carbon called ribulose-1, 5-bisphosphate (RuBP), which is a five-carbon sugar-phosphate, where carbon dioxide is getting converted as two molecules in a three-carbon compound called 3-phosphoglyceric acid (PGA) or 3-carbon acid (Locke et al., 2018). It is also found that the RuBisCO protein dispersed inside the chloroplast changes according to the rhythmic duration; additionally, this dispersal of RuBisCO protein interacts with the rhythm of carbon dioxide fixation (Nassoury et al., 2001).
During the daytime, nicotinamide adenine dinucleotide phosphate (NADPH) and adenosine triphosphate (ATP) are produced with the help of a light reaction to charge the assimilation of carbon. The carbon present, in addition, is deposited as starch inside the chloroplast, which ends up as the diurnal variations of starch concentration and reduction (Locke et al., 2018). Photorespiration is the process where the oxygen fixation gets activated, and then it directly takes place in the carbon dioxide fixation. The coherence of C3 photosynthesis in the air can be estimated based on the conflict between carbon dioxide and oxygen and the cost of energyrelated by reclaiming phosphoglycolate. In C4 plants, photosynthesis occurs where NADP-malic enzyme (ME) helps transfer the malate from mesophyll to bundle sheath chloroplast.
Jones (2017) described that crassulacean acid metabolism (CAM) photosynthetic pathway allows enhanced productivity of water usage by isolating the carbon dioxide fixation for the opening of stomata during the daytime, and it closes the stomata and controls the water loss. Thus, these CAM plants open their stomata and fix the carbon dioxide with the aid of phosphoenolpyruvate carboxylase (PPC) and malate dehydrogenase activity during nighttime (Boxall et al., 2020). The activity of PPC has synchronized the mixture of PPC with circadian-regulated expression along with the PPC phosphorylation, which further ends up changing the activity of carboxylase. The circadian oscillator-related PPC activity initiates malate, then deposited into vacuoles as the malic acid (Hartwell et al., 2016). During the night, malate comes out of vacuoles. It enters the cytosol, where the decarboxylation takes place with the help of an enzyme Phosphopyruvate Carboxykinase (PCK) or by NAD-malic enzyme (NAD-ME) on CAM variants. This could further lead to an increase in the level of carbon dioxide in leaves, which influences the stomatal closure authorizing RuBisCO to return the carbon dioxide into the Calvin Benson cycle in the presence of light.
The circadian oscillation has been intimated for modulating many enzymes included in plant carbon (primary) metabolism (Harmer et al., 2000). The photosynthate inflation notifies the advancement of this biological system. The providence of carbon plays an essential role in plants while scrutinizing the different methods for improving crop productivity. Many different enzymes are involved in isolation during carbon metabolisms, such as chlorophyll a/b binding protein (CAB), RuBisCO (RBCS), phosphoenopyruvate carboxykinase (PEPC-K), phosphoenolpyruvate carboxylase (PEPC), ME, pyruvate kinase (PPDK), sucrose phosphate synthase (SPS), pyruvate kinase (PK), sucrose-non-fermentation1-related protein kinase1 (SnRK1), and the DNA-binding with one finger (Dof) co-expression; these were the genes looked over during the diurnal conditions (Kanwal et al., 2014).
Generally, in carbon metabolism, the efficacy of light is considered a driving force, and an entraining signal of the Calvin-Benson-Bassham (CBB) cycle or C3 cycle tries to isolate their parts. These spotted genes have recognized an endogenous rhythm present in the genotypes and the various patterns during the high range of expression. At the time of dawn, the compulsion of photosynthetic carbon dioxide directs the sucrose synthesis and the accumulation of starch; thus, it assists for the pursued production of starch during the time of dusk (Graf and Smith, 2011; Stitt and Zeeman, 2012; Pokhilko and Ebenhoh, 2015; Kim et al., 2017). At dusk, the starch stockpiled throughout the day before night will be deteriorated and devoured. Starch yield disruption occurs due to an unpredicted rapid arrival of dusk, which leads to the early consumption of starch and malnourishment of carbon. This malnourishment of carbon can end up in the swift alternate in gene expression; metabolism can lead to the minute difference in the rate of growth, which could further result in a higher difference in biomass production within a few weeks. It is feasible to directly or indirectly manage these mechanisms of endogenous clock genes (Lu et al., 2005; Graf et al., 2010; Kotting et al., 2010). From previous (Sulpice et al., 2014) studies, it is known that the clock incidentally regulates the synthesis of starch by the modulation of starch degradation.
Mostly, in plants, the amount of carbon secured along with starch is higher in level when compared to wild type depending upon the decreasing range of circadian rhythm period (K€olling et al., 2015; Jones, 2017). They reveal the transcriptional alternation, which is an indication for the starvation of carbon compounds before night. The plants were usually grown in a 12-h light and 12-h dark period for 3 continual weeks. It is known from Graf et al. (2010), when the plants are further relocated into dark conditions only after the 8 h of light exhibited a high range of starch degradation, and also there was no indication of carbon starvation found throughout the next 16 h of dark condition. In well-sustained plants, when the plant growth is strictly restricted by the carbon supply then the growth is managed either by CCA1 or LHY (Yazdanbakhsh et al., 2011; Müller et al., 2014).
The oilseed and vegetable varieties have various morphologies and different harvestable sections. This oilseed type of crop has a minimal period of circadian rhythm and maximal range of net carbon assimilation when compared to the vegetable varieties (Yarkhunova et al., 2016). From Smith and Stitt (2007), it is known that the rate of photosynthesis becomes limited under the condition of a shorter duration of photoperiods (or low irradiance). In such a case, it could lead to a limited source phenotype, which is the condition of insufficient or lack of carbon and minimal plant growth. Whereas, whenever more carbon is obtained from the atmosphere for the better growth of plants, having higher irradiance or long timing of photoperiod and the plants are of limited sink phenotype.
The clock maintains a certain level of carbon supply rate with the help of clock component CCA1/LHY, which could be further stored and used until the next day. During the nighttime, the instability of starch can be managed by the clock component CCA1/LHY in A. thaliana. This regulation plays a major role in avoiding either the lack or breakdown of sucrose and decreased growth level during nighttime (Müller et al., 2014). The rhythmic clock component TOC1 provides a specific idea for the biological system in the maintenance of cellular mechanisms (Cervela-Cardona et al., 2021).
Due to the fluctuation in the photoperiod, the rise in sugars changes, that is, the increase or decrease of sugars depends upon the presence of light and photosynthesis rate. Thus, it indicates sugar does not increase in the early morning or in late evening; also clock loses the sensitivity at the end of the light period, as it is not phase advanced (Haydon et al., 2013). The early rise of sugar takes place on sunny days, which helps the circadian clock in showing the appropriate phase for driving processes that are linked with sugar metabolism. Sugar accumulation in the morning helps in the activation of the photosynthesis process on all days of the cycle.
Due to sugars, the activation of the CCA1 promoter increases; also simultaneously, it also suppresses the PRR7 (Haydon et al., 2013; Dodd et al., 2015). The regulation of sugar with the circadian clock has the capability to develop the action of the oscillator for dealing with the clock timing; this may also form a mechanism where biological processes, which are considered as the outputs of biological oscillators, are getting affected by sugars. These all are because sugars can manage the outputs of the circadian clock directly, even without performing any role in the core region of the clock. The circadian clock may get affected by sugars in long-term response pathway, which strengthens the rhythmicity in leaves of A. thaliana during the nighttime, after which there will be a need for GI (Dalchau et al., 2010).
With the help of PHY and CRY photoreceptors, the entrainment cues, including signals of light, are obtained. It also comprises the regulation of FLOWERING LOCUS T (FT) through the sugar signaling of Trehalose-6-Phosphate (T6P); even though the photoperiod synchronization of FT is not totally dependent upon this T6P (Dodd et al., 2015). In the late daytime, during the process of photosynthesis, sugars are synthesized and these sugars are then conserved and further exploited in the process of respiration during the nighttime. On the basis of different carbohydrate transports along with the various stages of cycle rhythmicity, it develops the diel oscillation of sugars (Haydon et al., 2011).
The development of flowers is an important stage in plant development. The formation of flower organs occurs when the gene expressions are accurately controlled during the flower development stage (Schaffer et al., 1998). This process is activated by the circadian rhythm (Schaffer et al., 1998). The flowering genotype depends upon the merged reactions of external and internal features, such as photoperiod and temperature (Min, 2005). The clock recognizes the length of photoperiod and regulates the phloem companion cells FT and CONSTANS (CO); where CO is a cue regulator for flowering when there is a conflict between the length of photoperiod and the circadian rhythm based on external coincidence theory (Sawa et al., 2007). The leaf senescence regulators ORESARA 1 (ORE 1) are controlled by endogenous rhythm and activated by various other components, such as CCA1, PRR9, ELF3, ELF4, and LUX.
The ORE 1 gene is present downstream of ETHYLENE INSENSITIVE 2 (EIN 2), which activates the ethylene signaling pathway. This ORE 1 gene encodes with protein-specific transcriptional factor (TF) NAM, ATAF, and CUC (NAC), which then positively activates the senescence in leaves and moderates various pathways of leaf senescence by activating SENESCENCE ASSOCIATED GENES (SAGs) expression, such as SAG 29 and BIFUNCTIONAL NUCLEASE 1 (BFN 1) (Matallana-Ramirez et al., 2013). The ORE 1 expression increases during cell aging, leading to age-induced cell apoptosis. The microRNA164 (miR164) negatively activates the expression of ORE 1. The trifurcate feed-forward loop is formed with the combination of EIN 2, miR164, and ORE 1, which changes the senescence of the leaf (Kim et al., 2009). During leaf aging, the circadian rhythm gets shortened, which is found in the model organism Arabidopsis (Kim et al., 2016). From the previous study, it is known that the duration of flowering in Arabidopsis takes long days (LDs) and also its mechanism is well explained (Corbesier and Coupland, 2005).
In A. thaliana, the GI, which is the component of the clock, activates the photoperiodic flowering. This GI binds along with the CO promoter to activate the expression of CO (Figure 2). GI controls leaf senescence as the time of flowering, where it depends on the location. GI often acts as the mediator to make the interaction between leaf senescence and flowering and develops the productivity and fitness of plants. The ELF 4 links directly with the GI and suppresses its interaction with the ORE 1 promoter (Kim et al., 2020). The result of the regulatory network controls the transcription of CO, and CO mRNA gets collected after the night in a short-time duration. Then CO stops to assemble in the dusk due to degradation of protein by the combination of CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP 1) and SUPPRESSOR OF PHYA-105 1 (SPA 1) (Laubinger et al., 2006; Jang et al., 2008). The CRYPTOCHROME 2 (CRY 2), which is photoactivated, combines with COP 1 and SPA 1 to make a complex compound to inhibit the degradation of CO (Zuo et al., 2011).
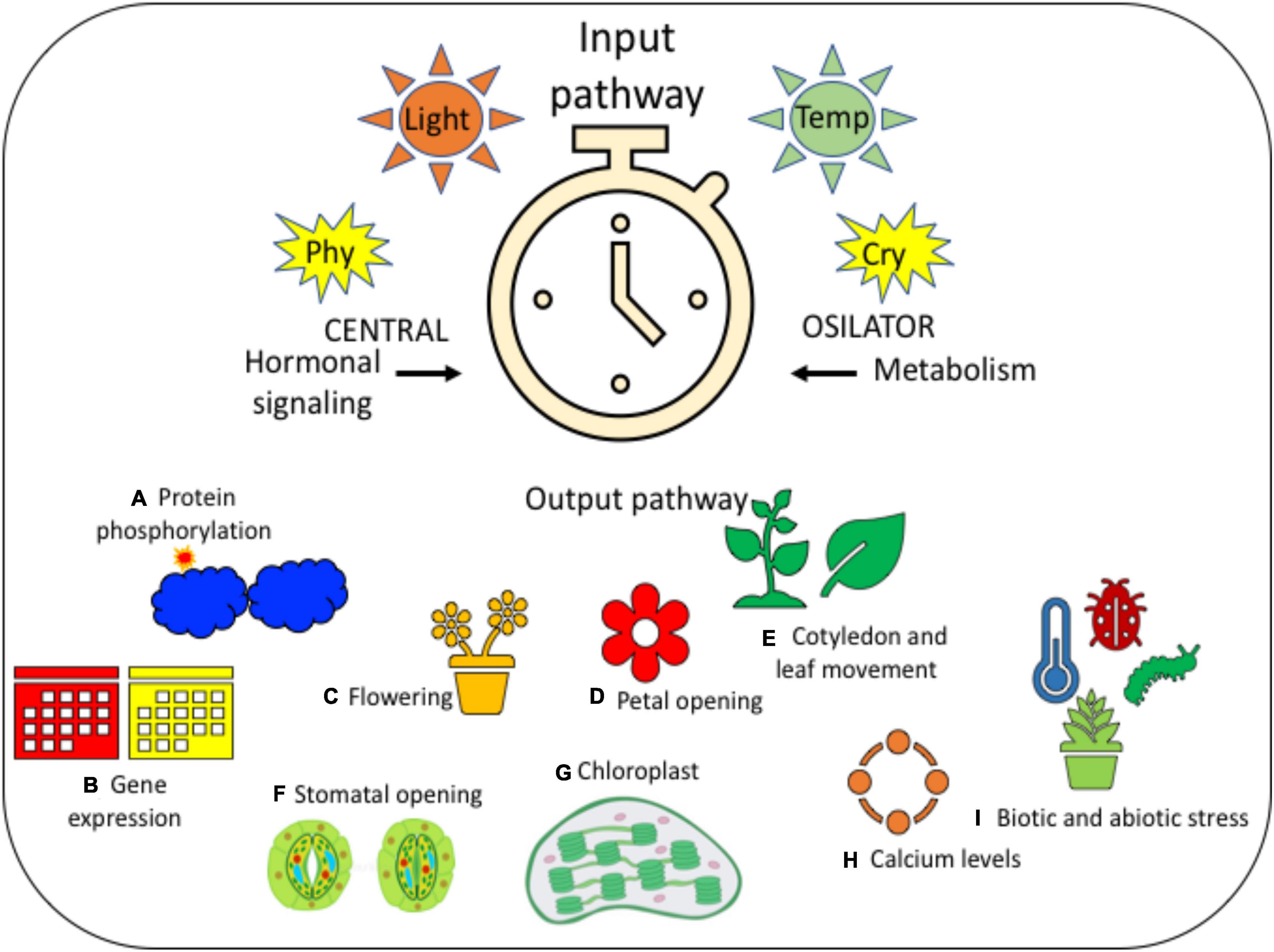
Figure 2. It is a diagrammatic representation of the circadian rhythm for providing a clear idea on the same. It has three different stages, such as (i) input pathway that contains light and temperature, which is then followed by (ii) central oscillator, in the presence of photoreceptors, such as PHY and CRY; further leading to the (iii) output pathway, which consists of many different physiological and developmental processes, such as (A) protein phosphorylation, (B) gene expression, (C) flowering, (D) petal opening, (E) cotyledon and leaf movement, (F) stomatal opening, (G) chloroplast, (H) calcium levels, and (I) biotic and abiotic stress. All these processes indicate the step-by-step variations that take place in every plant during the changes in photoperiodic rhythm.
Furthermore, by forming a blue light-dependent complex with FKF1 (FLAVIN-BINDING, KELCH REPEAT, F-BOX 1), GI regulates the expression of floral activators CO and FT (Imaizumi et al., 2003, 2005; Sawa et al., 2007; Sawa and Kay, 2011). CYCLING DOF FACTOR 1 (CDF1), a protein that represses CO and FT transcription, is degraded by the GI-FKF1 complex (Imaizumi et al., 2005; Fornara et al., 2009; Song et al., 2012). CO protein levels, which are stabilized by FKF1 at the end of the long photoperiod, control the activation of FT expression in LDs (Suárez-López et al., 2001; Song et al., 2012). Furthermore, light regulates CO protein, so CO levels are low in red light and high in blue light. While blue and far-red light help to keep CO stable (Valverde et al., 2004). This control increases the floral signal by reinforcing the accumulation of CO protein levels in the evening of LDs. These molecular interactions theoretically result in a double external coincidence mechanism that involves multiple clock outputs, but the combined effects of this rhythmic mechanism have not been quantitatively tested or incorporated into previous mathematical models (Song et al., 2012).
Similarly, the senescence in plants requires a functional endogenous system; also, many different senescence-related genes encode the TFs in which NAC and WRKY show a clear circadian rhythm pattern (Kim and Hong, 2019). The clock rhythm has combined the function of both flowering and leaf senescence, due to which the early leaf senescence takes place because of the ELF period as per the causes of embryonal carcinoma (EC) mutants (Kim et al., 2018). The analysis of transcriptional profiling exhibits the senescence regulatory genes; not only the ORE 1, NAP, WRKY 53, and WRKY 70 but also the phytohormone jasmonate (JA) responsive and signaling genes are getting activated at the stage of EC-mediated senescence of leaf (Zhang et al., 2018). The salicylic acid (SA) pathway has a connection with the PHY-associated leaf senescence mechanism. The regulation of senescence on the basis of circadian oscillator and light has been explained in a detailed manner in a recent study (Lee et al., 2021).
The circadian rhythm can also be expressed with the help of the plant defense system, also in the absence of the pathogen. The glycine-rich RNA binding protein is a pathogen responsive gene, and it was already seen in several crops that exhibit the regulation of the clock (Lu et al., 2017). Defense response plays a significant role in plants for the normal functioning of cells against the infection caused by the pathogen (Spoel and Dong, 2008). In the case of a pathogen attack, the cuticle supply, which is a physical plant barrier, is considered the first line of plant defense. This form of defense system blocks the pest and pathogen invasion. Whenever a swap takes place in the cuticle, it is immediately realized by the plant and instantly, it begins the defense response during the time of pathogen attack (Malik et al., 2020). The instigated interaction, which occurs between pattern recognition receptors and pathogen-associated molecular patterns (PAMPs), activates a considerable amount of plant defense systems, such as stomatal closure, to keep away from the invasion of the pathogen (Jones and Dangl, 2006; Miller et al., 2017; Butt et al., 2020).
Phytohormones, such as JA and SA, play an essential role in defense responses (Roden and Ingle, 2009; Tamaoki et al., 2013). Depending upon the mode of pathogen attack, the phytohormones are synthesized by the plants (Loake and Grant, 2007). If the pathogen moves inside the plant cells, the polymorphic Nucleotide Binding and Leucine-Rich Repeat (NB-LRR) proteins available within the host cell interconnect with specifically effecting molecules of the pathogen (Jones and Dangl, 2006; Miller et al., 2017). The growth by the addition of SA influences the strengthening of the cell wall, phenolic accumulation, and production. It also triggers resistance (R) and other different defense response genes (activated of clock gene LUX). The PRRs recognize the PAMPs that stop the formation of pathogen colonies, which end up in the PAMP triggered immunity (PTI). This PTI restricts the released action of the pathogen inside the host cell, and also it switches on the defense system, which functions in the way of non-host-specific R. The four stages of plant immunity have become a well-known process from a recent study (Butt et al., 2020).
The prompt in reactive oxygen species (ROS) production is considered the initial defense response state in plants. The interaction between the clock system and redox state in the cellular system confirms the stability between the immunity of plants and their proper growth (Brody, 2019; Zhang J. et al., 2019). The LUX gets bonded with EDS 1 and JASMONATE ZIM DOMAIN (JAZ) 5 promoters, which could affect the signaling of JA and SA (Zhang C. et al., 2019). The formation of co-receptor complex takes place with the association of transcriptional repressor proteins, such as JASMONOYL-ISOLEUCINE (JA-Ile), JAZ, and CORONATINE-INSENSITIVE1 (COI1), which is an F-Box protein in the existence of vital appearance of JA (Nitschke et al., 2016). This leads to the JAZ repressor degradation due to the ubiquitin-proteasome system (Thines et al., 2007), and then it rescues the TFs, such as MYC 2. Thus, the transcription for response genes of JA is enabled, such as JAZ genes, also MYC 2, and all the biosynthesis genes of JA, i.e., LIPOXYGENASE (LOX) and OXO PHYTODIENOATE REDUCTASE 3 (OPR3) (Chung et al., 2008; Wasternack and Hause, 2013).
The clock entrainment with the light is conciliated by CRY and PHY; they are translocated to the nucleus concerning the light where the gene expression is activated. Several defense genes are moderated by the function of PHY and the circadian clock (Roden and Ingle, 2009). Plants having the deficiency of PHY A/B show the complete reduction in the expression of PRR; thus, this stipulates the importance of the defense signaling pathway (Genoud et al., 2002; Butt et al., 2020). From Griebel and Zeier (2008), it is known that the plant defense has an increased function during the time of dawn rather than dusk; also, they have used the pressure inoculum in the leaves of the organism Pseudomonas syringae (P. syringae) along with the diversion of defense response in stomata. The rhythmicity of the clock blocks the susceptibility of the plants toward many different pathogens and insects, where the plant susceptibility will have the most little part at the time of interaction of endogenous clock with the defense hormones, such as JA and SA (Goodspeed et al., 2012; Korneli et al., 2014; Ingle et al., 2015).
The plant circadian rhythm is cell-independent and self-assisting under the seasonal and diurnal input sources from the environmental cue for encountering the stimulant-navigated response. The endogenous clock components will be synchronized at either transcriptional or post-transcriptional extent. The role and the function of CCA1 and LHY (the two main clock components in light signaling) in circadian timing and control of the flowering plant could be explained with the separation and depiction of clock genes. It has been found that after dawn, the genes help much in controlling the chlorophyll; it was also known that these clock components help maintain the fitness of plants with improved immunity for the best performance.
In the present review, we have tried to explain the different biological or developmental processes of plants through the circadian clock, which will also help the researchers understand how the genes present in the circadian system, are expressed, and how they are functioning. The circadian rhythm has been analyzed with the model organism A. thaliana and in other plants, such as Nicotiana benthamiana (N. benthamiana), Chrysanthemum, and Petunia plant. Yet, there are many vegetable crops or flowering plants, which could be readily available. In addition, it can be used to analyze circadian biology to provide a clear vision about the same in a very detailed manner. Both in vitro and in vivo studies for the analysis of this photoperiodic rhythmicity along with the abiotic stress conditions using molecular work can be performed as a brief future study. The study can be accomplished to increase the growth and yield of edible crops that are highly tolerant toward the abiotic stresses, for example, salinity, heat, and cold stress conditions, based on the functioning of their circadian rhythm.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Horticultural and Molecular Physiology Lab was funded by DST-SERB (project no. SRG/2020/000170), Govt. of India, and Timac Agro International (Roulliers group, France), India. HMP Lab also acknowledges VIT seed grant for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar-Arnal, L., and Sassone-Corsi, P. (2015). Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proc. Nat. Acad. Sci. U S A. 112, 6863–6870. doi: 10.1073/pnas.1411264111
Belbin, F. E., Hall, G. J., Jackson, A. B., Schanschieff, F. E., Archibald, G., Formstone, C., et al. (2019). Plant circadian rhythms regulate the effectiveness of a glyphosate-based herbicide. Nature Commun. 10, 1–1. doi: 10.1038/s41467-019-11709-5
Boxall, S. F., Kadu, N., Dever, L. V., Kneřová, J., Waller, J. L., Gould, P. J. D., et al. (2020). Kalanchoë PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell 32, 1136–1160. doi: 10.1105/tpc.19.00481
Brody, S. (2019). Circadian Rhythms in Fungi: Structure/Function/Evolution of Some Clock Components. J. Biol. Rhythm 34, 364–379. doi: 10.1177/0748730419852832
Butt, G. R., Qayyum, Z. A., and Jones, M. A. (2020). Plant Defence Mechanisms Are Modulated by the Circadian System. Biology 9:454. doi: 10.3390/biology9120454
Cervela-Cardona, L., Alary, B., and Mas, P. (2021). The Arabidopsis circadian clock and metabolic energy: A question of time. Front. Plant Sci. 12:804468. doi: 10.3389/fpls.2021.804468
Chen, Y. Y., Wang, Y., Shin, L. J., Wu, J. F., Shanmugam, V., Tsednee, M., et al. (2013). Iron is involved in the maintenance of circadian period length in Arabidopsis. Plant Physiol. 161, 1409–1420. doi: 10.1104/pp.112.212068
Chung, H. S., Koo, A. J., Gao, X., Jayanty, S., Thines, B., Jones, A. D., et al. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. doi: 10.1104/pp.107.115691
Corbesier, L., and Coupland, G. (2005). Photoperiodic flowering of Arabidopsis: integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 28, 54–66. doi: 10.1111/j.1365-3040.2005.01283.x
Dalchau, N., Baek, S. J., Briggs, H. M., Robertson, F. C., Dodd, A. N., Gardner, M. J., et al. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Nat. Acad. Sci. U S A. 108, 5104–5109. doi: 10.1073/pnas.1015452108
Dalchau, N., Hubbard, K. E., Robertson, F. C., Hotta, C. T., Briggs, H. M., Stan, G. B., et al. (2010). Correct biological timing in Arabidopsis requires multiple light-signaling pathways. Proc. Nat. Acad. Sci. U S A. 107, 13171–13176. doi: 10.1073/pnas.1001429107
de Dios, V. R., Anderegg, W. R. L., Li, X., Tissue, D. T., Bahn, M., Damien, L., et al. (2020). Circadian regulation does not optimize stomatal behaviour. Plants 9:1091. doi: 10.3390/plants9091091
de Dios, V.R., Anderegg, W. R. L., Li, X., Tissue, D. T. et al. (2018). Circadian regulation of photosynthesis and transpiration from genes to ecosystems. Environ. Exp. Bot. 152, 37–48. doi: 10.1016/j.envexpbot.2017.09.010
Dodd, A. N., Belbin, F. E., Frank, A., and Webb, A. A. R. (2015). Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 6:245. doi: 10.3389/fpls.2015.00245
Dodd, A. N., Salathia, N., Hall, A., Ke’vei, E., To’th, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Farré, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., and Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/j.cub.2004.12.067
Feke, A., Vanderwall, M., Liu, W., Gendron, J. M., et al. (2021). Functional domain studies uncover novel roles for the ZTL Kelch repeat domain in clock function. PLoS One 16:e0235938. doi: 10.1371/journal.pone.0235938
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Franklin, K. A. (2020). PRR proteins of the circadian clock call time on shade avoidance. Proc. Nat. Acad. Sci. U S A. 117, 5095–5096. doi: 10.1073/pnas.2000716117
Fukuda, N., Suenaga, T., Miura, E., Tsukamoto, A., et al. (2020). The Expression of ELF4-Like Genes Is Influenced by Light Quality in Petunia. Agronomy 10:1800. doi: 10.3390/agronomy10111800
Furbank, R. T., and Taylor, W. C. (1995). Regulation of photosynthesis in C3 and C4 plants: a molecular approach. Plant Cell 7:797. doi: 10.1105/tpc.7.7.797
Genoud, T., Buchala, A. J., Chua, N. H., and Métraux, J. P. (2002). Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J. 31, 87–95. doi: 10.1046/j.1365-313x.2002.01338.x
Goodspeed, D., Chehab, E. W., Min-Venditti, A., Braam, J., and Covington, M. F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Nat. Acad. Sci. U S A. 109, 4674–4677. doi: 10.1073/pnas.1116368109
Graf, A., and Smith, A. M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16, 169–175. doi: 10.1016/j.tplants.2010.12.003
Graf, A., Schlereth, A., Stitt, M., and Smith, A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Nat. Acad. Sci. U S A. 107, 9458–9463. doi: 10.1073/pnas.0914299107
Green, R. M., and Elaine, M. T. (2002). The role of CCA1 and LHY in the plant circadian clock. Dev. Cell 2, 516–518. doi: 10.1016/s1534-5807(02)00184-3
Green, R. M., Tingay, S., Wang, Z. Y., and Tobin, E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584. doi: 10.1104/pp.004374
Griebel, T., and Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147, 790–801. doi: 10.1104/pp.108.119503
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Hartwell, J., Dever, L. V., and Boxall, S. F. (2016). Emerging model systems for functional genomics analysis of Crassulacean acid metabolism. Curr. Opin. Plant Biol. 31, 100–108. doi: 10.1016/j.pbi.2016.03.019
Haydon, M. J., Bell, L. J., and Webb, A. A. (2011). Interactions between plant circadian clocks and solute transport. J. Exp. Bot. 62, 2333–2348. doi: 10.1093/jxb/err040
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E., and Webb, A. A. R. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. doi: 10.1038/nature12603
Hennessey, T. L., and Field, C. B. (1991). Circadian rhythms in photosynthesis: oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol. 96, 831–836. doi: 10.1104/pp.96.3.831
Hicks, K. A., Tina, M. A., and Wagner, D. R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13, 1281–1292. doi: 10.1105/tpc.13.6.1281
Huq, E., Tepperman, J. M., and Quail, P. H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Nat. Acad. Sci. U S A. 97, 9789–9794. doi: 10.1073/pnas.170283997
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Imaizumi, T., Tran, H. G., Swartz, T. E., Briggs, W. R., and Kay, S. A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. doi: 10.1038/nature02090
Ingle, R. A., Stoker, C., Stone, W., Adams, N., Smith, R., Grant, M., et al. (2015). Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 84, 937–948. doi: 10.1111/tpj.13050
Jang, S., Marchal, V., Panigrahi, K. C., Wenkel, S., Soppe, W., Deng, X. W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288. doi: 10.1038/emboj.2008.68
Jones, M. A. (2017). Interplay of circadian rhythms and light in the regulation of photosynthesis-derived metabolism. Progress Bot. 79, 147–171. doi: 10.1007/124_2017_2
K€olling, K., Thalmann, M., Müller, A., Jenny, C., and Zeeman, S. C. (2015). Carbon partitioning in Arabidopsis thaliana is a dynamic process controlled by the plants metabolic status and its circadian clock. Plant Cell Environ. 38, 1965–1979. doi: 10.1111/pce.12512
Kanwal, P., Gupta, S., Arora, S., and Kumar, A. (2014). Identification of genes involved in carbon metabolism from Eleusine coracana (L.) for understanding their light-mediated entrainment and regulation. Plant Cell Rep. 33, 1403–1411. doi: 10.1007/s00299-014-1625-4
Inoue, K., Araki, T., and Endo, M. (2017). Integration of input signals into the gene network in the plant circadian clock. Plant Cell Physiol. 58, 977–982. doi: 10.1093/pcp/pcx066
Inoue, K., Araki, T., and Endo, M. (2018). Circadian clock during plant development. J. Plant Res. 131, 59–66. doi: 10.1007/s10265-017-0991-8
Kiełbowicz-Matuk, A., Rey, P., and Rorat, T. (2014). Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot. 113, 831–842. doi: 10.1093/aob/mct303
Kim, C., Kim, S. J., Jeong, J., Park, E., Oh, E., Park, Y. I., et al. (2020). High ambient temperature accelerates leaf senescence via PHYTOCHROME-INTERACTING FACTOR 4 and 5 in Arabidopsis. Mol. Cells 43:645. doi: 10.14348/molcells.2020.0117
Kim, H., and Hong, S. (2019). Role of the circadian clock in fine-tuning the process of leaf senescence in plants. Translat. Med. Aging 3, 26–30. doi: 10.1016/j.tma.2018.12.001
Kim, H., Kim, H. J., Vu, Q. T., Jung, S., McClung, C. R., Hong, S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Nat. Acad. Sci. U S A. 115, 8448–8453. doi: 10.1073/pnas.1722407115
Kim, H., Kim, Y., Yeom, M., Lim, J., and Nam, H. G. (2016). Age-associated circadian period changes in Arabidopsis leaves. J. Exp. Bot. 67, 2665–2673. doi: 10.1093/jxb/erw097
Kim, J. A., Kim, H. S., Choi, H. S., Jang, J. Y., Jeong, M. J., and Lee, S. I. (2017). The importance of the circadian clock in regulating plant metabolism. Int. J. Mol. Sci. 18:2680. doi: 10.3390/ijms18122680
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323:1053e1057. doi: 10.1126/science.1166386
Korneli, C., Danisman, S., and Staiger, D. (2014). Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock. Plant Cell Physiol. 55, 1613–1622. doi: 10.1093/pcp/pcu092
Kotting, O., Kossmann, J., Zeeman, S. C., and Lloyd, J. R. (2010). Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 13, 321–329. doi: 10.1016/j.pbi.2010.01.003
Laubinger, S., Marchal, V., Le Gourrierec, J., Wenkel, S., Adrian, J., Jang, S., et al. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213–3222. doi: 10.1242/dev.02481
Lee, J., Kang, M. H., Kim, J. Y., and Lim, P. O. (2021). The role of light and circadian clock in regulation of leaf senescence. Front. Plant Sci. 12:669170. doi: 10.3389/fpls.2021.669170
Lei, J., Jayaprakasha, G. K., Singh, J., Uckoo, R., Borrego, E. J., Finlayson, S., et al. (2019). CIRCADIAN CLOCK-ASSOCIATED1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol. 181, 1344–1359. doi: 10.1104/pp.19.00676
Liew, L. C., Mohan, B. S., and Prem, L. B. (2017). A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development. Sci. Rep. 7, 1–16. doi: 10.1038/s41598-017-10823-y
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
Locke, A. M., Slattery, R. A., and Ort, D. R. (2018). Field-grown soybean transcriptome shows diurnal patterns in photosynthesis-related processes. Plant Direct 2:e00099. doi: 10.1002/pld3.99
Lopez, L., Fasano, C., Perrella, G., Facella, P., et al. (2021). Cryptochromes and the Circadian Clock: The Story of a Very Complex Relationship in a Spinning World. Genes 12:672. doi: 10.3390/genes12050672
Lu, H., McClung, C. R., and Zhang, C. (2017). Tick Tock: Circadian Regulation of Plant Innate Immunity. Annu. Rev. Phytopathol. 55, 287–311. doi: 10.1146/annurev-phyto-080516-035451
Lu, Y., Gehan, J. P., and Sharkey, T. D. (2005). Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 138, 2280–2291. doi: 10.1104/pp.105.061903
Males, J., and Howard, G. (2017). Stomatal biology of CAM plants. Plant Physiol. 174, 550–560. doi: 10.1104/pp.17.00114
Malik, N. A. A., Kumar, I. S., and Nadarajah, K. (2020). Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 21:963. doi: 10.3390/ijms21030963
Man, A. W. C., Xia, N., and Li, H. (2020). Circadian rhythm in adipose tissue: novel antioxidant target for metabolic and cardiovascular diseases. Antioxidants 9:968. doi: 10.3390/antiox9100968
Más, P., Alabadí, D., Yanovsky, M. J., Oyama, T., and Kay, S. A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236. doi: 10.1105/tpc.006734
Matallana-Ramirez, L. P., Rauf, M., Farage-Barhom, S., Dortay, H., Xue, G. P., Dröge-Laser, W., et al. (2013). NAC transcription factor ORE1 and senescence induced bifunctional nuclease1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6:1438e1452. doi: 10.1093/mp/sst012
McWatters, H. G., Kolmos, E., Hall, A., Doyle, M. R., Amasino, R. M., Gyula, P., et al. (2007). ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 144, 391–401. doi: 10.1104/pp.107.096206
Miller, R. N. G., Alves, G. S. C., and Van Sluys, M. A. (2017). Plant immunity: Unravelling the complexity of plant responses to biotic stresses. Ann. Bot. 119, 681–687. doi: 10.1093/aob/mcw284
Min, N. I. (2005). Integration of light signaling with photoperiodic flowering and circadian rhythm. Cell Res. 15, 559–566. doi: 10.1038/sj.cr.7290325
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Müller, L. M., von Korff, M., and Davis, S. J. (2014). Connections between circadian clocks and carbon metabolism reveal species-specific effects on growth control. J. Exp. Bot. 65, 2915–2923. doi: 10.1093/jxb/eru117
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kita, M., Ito, S., Yamashino, T., Mizuno, T., et al. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698. doi: 10.1093/pcp/pci086
Nassoury, N., Fritz, L., and Morse, D. (2001). Circadian changes in ribulose-1,5-bisphosphate carboxylase/oxygenase distribution inside individual chloroplasts can account for the rhythm in dinoflagellate carbon fixation. Plant Cell 13, 923–934. doi: 10.1105/tpc.13.4.923
Nitschke, S., Cortleven, A., Iven, T., Feussner, I., Havaux, M., Riefler, M., et al. (2016). Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28, 1616–1639. doi: 10.1105/tpc.16.00016
Nohales, M. A., and Kay, S. A. (2016). Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 23:1061. doi: 10.1038/nsmb.3327
Pan, W. J., Wang, X., Deng, Y. R., Li, J. H., Chen, W., Chiang, J. Y., et al. (2015). Nondestructive and intuitive determination of circadian chlorophyll rhythms in soybean leaves using multispectral imaging. Sci. Rep. 5, 1–13. doi: 10.1038/srep11108
Panter, P. E., Muranaka, T., Cuitun-Coronado, D., Graham, C. A., Yochikawa, A., Kudoh, H., et al. (2019). Circadian regulation of the plant transcriptome under natural conditions. Front. Genet. 10:1239. doi: 10.3389/fgene.2019.01239
Pokhilko, A., and Ebenhoh, O. (2015). Mathematical modelling of diurnal regulation of carbohydrate allocation by osmo-related processes in plants. J. R. Soc. Interface 12, 2014–1357. doi: 10.1098/rsif.2014.1357
Rascher, U., Hütt, M. T., Siebke, K., Osmond, B., Beck, F., and Lüttge, U. (2001). Spatiotemporal variation of metabolism in a plant circadian rhythm: the biological clock as an assembly of coupled individual oscillators. Proc. Nat. Acad. Sci. U S A. 98, 11801–11805. doi: 10.1073/pnas.191169598
Rawat, R., Takahashi, N., Hsu, P. Y., Matthew, A., Jacob Schwartz, J., Salemi, M. R., et al. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7:e1001350. doi: 10.1371/journal.pgen.1001350
Roden, L. C., and Ingle, R. A. (2009). Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. Plant Cell 21, 2546–2552. doi: 10.1105/tpc.109.069922
Rugnone, M. L., Soverna, A. F., Sanchez, S. E., Schlaen, R. G., Hernando, C. E., Seymour, D. K., et al. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Nat. Acad. Sci. U S A. 110, 12120–12125. doi: 10.1073/pnas.1302170110
Saini, R., Jaskolski, M., and Davis, S. J. (2019). Circadian oscillator proteins across the kingdoms of life: structural aspects. BMC Biol. 17, 1–39. doi: 10.1186/s12915-018-0623-3
Sanchez, A., Shin, J., and Davis, S. J. (2011). Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6, 223–231. doi: 10.4161/psb.6.2.14893
Sawa, M., and Kay, S. A. (2011). GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc. Nat. Acad. Sci. U S A. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I. A., et al. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 1219–1229. doi: 10.1016/s0092-8674(00)81465-8
Shogo, I., Niwa, Y., Nakamichi, N., Kawamura, H., Yamashino, T., and Mizuno, T. (2008). Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 49, 201–213. doi: 10.1093/pcp/pcm178
Shor, E., Paik, I., Kangisser, S., Green, R., and Huq, E. (2017). PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 215, 217–228. doi: 10.1111/nph.14579
Smith, A. M., and Stitt, M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30, 1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Spoel, S. H., and Dong, X. (2008). Making Sense of Hormone Crosstalk during Plant Immune Responses. Cell Host Microbe 3, 348–351. doi: 10.1016/j.chom.2008.05.009
Srivastava, D., Shamim, M. D., Kumar, M., Mishra, A., Maurya, R., Sharma, D., et al. (2019). Role of circadian rhythm in plant system: An update from development to stress response. Environ. Exp. Bot. 162, 256–271. doi: 10.1186/s13054-016-1208-6
Stitt, M., and Zeeman, S. C. (2012). Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292. doi: 10.1016/j.pbi.2012.03.016
Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Sulpice, R., Flis, A., Ivakov, A. A., Apelt, F., Krohn, N., Encke, B., et al. (2014). Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol. Plant 7, 137–155. doi: 10.1093/mp/sst127
Tamaoki, D., Seo, S., Yamada, S., Kano, A., Miyamoto, A., Shishido, H., et al. (2013). Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 8:e24260. doi: 10.4161/psb.24260
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Wang, X., and Ma, L. (2013). Unraveling the circadian clock in Arabidopsis. Plant Signal Behav. 8:e23014. doi: 10.4161/psb.23014
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Webb, A. A. R. (2003). The physiology of circadian rhythms in plants. New Phytol. 160, 281–303. doi: 10.1046/j.1469-8137.2003.00895.x
Wu, J. F., Tsai, H. L., Joanito, I., Wu, Y. C., Chang, C. E., Li, Y. H., et al. (2016). LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7, 1–10. doi: 10.1038/ncomms13181
Yan, J., Kim, Y. J., and Somers, D. E. (2021). Post-Translational Mechanisms of Plant Circadian Regulation. Genes 12:325. doi: 10.3390/genes12030325
Yarkhunova, Y., Edwards, C. E., Ewers, B. E., Baker, R. L., Aston, T. L., McClung, C. R., et al. (2016). Selection during crop diversification involves correlated evolution of the circadian clock and ecophysiological traits in Brassica rapa. New Phytol. 210, 133–144. doi: 10.1111/nph.13758
Yazdanbakhsh, N., Sulpice, R., Graf, A., Stitt, M., and Fisahn, J. (2011). Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ. 34, 877–894. doi: 10.1111/j.1365-3040.2011.02286.x
Yeom, M., Kim, H., Lim, J., Shin, A. Y., Hong, S., Kim, J. I., et al. (2014). How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol. Plant 7, 1701–1704. doi: 10.1093/mp/ssu086
Youngsung, J., Fragoso, V., Yon, F., Baldwin, I. T., and Kim, S. G. (2017). Circadian clock component, LHY, tells a plant when to respond photosynthetically to light in nature. J. Int. Plant Biol. 59, 572–587. doi: 10.1111/jipb.12547
Yuan, L., Yu, Y., Liu, M., Song, Y., Li, H., Sun, J., et al. (2021). BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell. 33, 2602–2617. doi: 10.1093/plcell/koab133
Zhang, C., Min, G., Seitz, N. C., Angel, W., Hallworth, A., Wiratan, L., et al. (2019). LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-019-10485-6
Zhang, J., Ren, Z., Zhou, Y., Ma, Z., Ma, Y., Hou, D., et al. (2019). NPR1 and Redox Rhythms: Connections, between Circadian Clock and Plant Immunity. Int. J. Mol. Sci. 20:1211. doi: 10.3390/ijms20051211
Zhang, Y., Wang, Y., Wei, H., Li, N., Tian, W., Chong, K., et al. (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11, 326–337. doi: 10.1016/j.molp.2017.12.017
Keywords: circadian rhythms, carbon metabolism, gene regulatory pathways, plant metabolism, signaling pathways, photosynthesis, flowering and senescence, defense response
Citation: Venkat A and Muneer S (2022) Role of Circadian Rhythms in Major Plant Metabolic and Signaling Pathways. Front. Plant Sci. 13:836244. doi: 10.3389/fpls.2022.836244
Received: 15 December 2021; Accepted: 23 February 2022;
Published: 06 April 2022.
Edited by:
Xiaodong Xu, Henan University, ChinaReviewed by:
Norihito Nakamichi, Nagoya University, JapanCopyright © 2022 Venkat and Muneer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sowbiya Muneer, c293Yml5YS5tdW5lZXJAdml0LmFjLmlu, c29iaXlha2hhbjEyNkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.