- Genetics and Biotechnology Lab, Plant and AgriBiosciences Research Centre, Ryan Institute, National University of Ireland Galway, Galway, Ireland
Parent-of-origin effects arise when a phenotype depends on whether it is inherited maternally or paternally. Parent-of-origin effects can exert a strong influence on F1 seed size in flowering plants, an important agronomic and life-history trait that can contribute to biomass heterosis. Here we investigate the natural variation in the relative contributions of the maternal and paternal genomes to F1 seed size across 71 reciprocal pairs of F1 hybrid diploids and the parental effect on F1 seed size heterosis. We demonstrate that the paternally derived genome influences F1 seed size more significantly than previously appreciated. We further demonstrate (by disruption of parental genome dosage balance in F1 triploid seeds) that hybridity acts as an enhancer of genome dosage effects on F1 seed size, beyond that observed from hybridity or genome dosage effects on their own. Our findings indicate that interactions between genetic hybridity and parental genome dosage can enhance heterosis effects in plants, opening new avenues for boosting heterosis breeding in crop plants.
Introduction
Hybridisation is widely used in crop breeding to harness hybrid vigour effects and for generation of improved phenotypes (Goulet et al., 2017). Improvement of F1 hybrid performance relative to parental lines is known as heterosis and can affect yield, biomass and many other characters (Birchler et al., 2010; McKeown et al., 2013a; Schnable and Springer, 2013; Miyaji and Fujimoto, 2018; Liu et al., 2020). Heterosis breeding schemes underpin higher-yielding F1 hybrid varieties of many crops, including maize, rice, sugar beet, oilseed rape, and sorghum (Cheng et al., 2007; Ma and Yuan, 2015; Goulet et al., 2017; Dimitrijevic and Horn, 2018; McGrath and Panella, 2018; Wolko et al., 2019). Both positive and negative heterosis can be useful, increasing or decreasing a trait value relative to the parents, respectively (Ryder et al., 2014). While positive heterosis is preferable for yield, negative heterosis for growth duration is also useful for earliness in rainfed crops (Kumar et al., 2015).
Seed/grain size is a prime breeding target and a major contributor to seed weight, ultimately influencing yield (Shomura et al., 2008; Kesavan et al., 2013; Makino et al., 2020). Despite the traditional yield gain attributed to seed/grain number, modern crop varieties have demonstrated that seed/grain weight, and therefore seed size, can also be associated with grain yield, driven by artificial selection which can alter the constraints on the trade-off between seed size and number (Griffiths et al., 2015; Golan et al., 2019; Calderini et al., 2021; Martin, 2021). The non-additive mode of inheritance that characterises heterosis prevents the prediction of F1 hybrid phenotypes from performance in the parental generation, which necessitates laborious field testing of hybrids developed from complex breeding designs to identify germplasm pools with good combining ability (Fridman, 2015; Seymour et al., 2016; Vasseur et al., 2019). In this context, the model plant Arabidopsis thaliana (hereafter Arabidopsis) provides an opportunity for studying parental genotype combinations that generate heterosis effects (Stokes et al., 2007; Schnable and Springer, 2013) due to the number of genotyped accessions (Weigel, 2012). Indeed, seed size heterosis has been reported in some Arabidopsis intraspecific hybrids (Alonso-Blanco et al., 1999; Stokes et al., 2007; House et al., 2010; Groszmann et al., 2014; van Hulten et al., 2018; Wang et al., 2020), making Arabidopsis a suitable system for improving understanding of the control of seed size heterosis.
Seed size is a life-history trait that is also influenced by parent-of-origin effects, which can occur when the phenotype differs according to whether it is transmitted maternally or paternally (Spillane et al., 2002; Vielle-Calzada et al., 2002; Lawson et al., 2013). Analysis of reciprocal crosses in several species have demonstrated strong maternal effects on F1 seed size, while paternal effects are only rarely reported (de Jong and Scott, 2007; Diggle et al., 2010; House et al., 2010; de Jong et al., 2011; Li and Li, 2015; Singh et al., 2017; Li et al., 2019). The mechanisms underlying maternal effects on seed size include (1) uniparental inheritance of cytoplasmic organelles, usually via the female gametes (although paternal inheritance/leakage of organellar genomes has been reported in certain species), (2) gametophytic effects involving the egg cell and/or central cell, (3) sporophytic effects (mediated by e.g., maternal integuments, the seed coat), and (4) genomic imprinting in the seed endosperm (Grossniklaus et al., 2001; Donohue, 2009; Diggle et al., 2010; Greiner et al., 2015; Armenta-Medina and Gillmor, 2019; Kirkbride et al., 2019).
Paternal effects can also influence seed development in a range of ways, although these are more limited than for the maternal parent. Paternal effects can occur through male gametogenesis or post-fertilisation, due to factors deposited by the sperm cells in the egg and/or central cells, as well as by genomic imprinting (Spillane et al., 2002; Wolff et al., 2015; Pires et al., 2016; Batista and Köhler, 2020). Indeed, the role of sperm-accumulated transcripts (including coding and non-coding RNAs) is apparent in early development in both animals and plants (Gòdia et al., 2018). For example, interruption of the delivery of small RNAs by the sperm causes arrested zygotic divisions in mouse, leading to embryonic lethality (Yuan et al., 2016). In Arabidopsis, the sperm-transmitted microRNA, miR159, triggers endosperm nuclear divisions (Zhao et al., 2018), while the sperm-delivered SHORT SUSPENSOR (SSP) mRNA is necessary for the first zygotic division (Bayer et al., 2009). Epigenetic modifications present in sperm and/or semen can influence offspring development in many taxa, including insects, fish, and nematodes (Macartney et al., 2018) and include inheritance of DNA methylation patterns and chromatin structure, potentially affecting the embryo’s transcriptional programs (Skinner et al., 2015; Klosin et al., 2017; Macartney et al., 2018).
Studies in Arabidopsis demonstrate that altering the relative dosages of the parental genomes, typically in inter-ploidy crosses, causes particularly strong parent-of-origin effects on seed endosperm development, resulting in dramatic seed size changes even in the absence of genetic hybridity (Scott et al., 1998; Fort et al., 2016; Lafon-Placette and Köhler, 2016). While doubling the paternal genome dosage in crosses between diploid (2x) and tetraploid (4x) parents can result in prolonged endosperm proliferation and abnormally large seeds which frequently abort, the reciprocal cross between a 4x maternal parent and a 2x paternal parent (generating a maternal genome excess in the F1 seed), causes precocious endosperm cellularisation leading to small F1 seeds (Scott et al., 1998; Fort et al., 2016; Lafon-Placette and Köhler, 2016). We previously demonstrated that paternal excess F1 triploids stably demonstrated heterosis for biomass in a panel of ten accession combinations, in addition to any heterosis seen in crosses between diploids of the same accessions. We could not however associate this with any increase in relative growth rate and concluded that the effect is likely to be due to paternal genomes driving an increase in seed size (Fort et al., 2016).
In this study, we test this hypothesis across Arabidopsis genetic diversity by reciprocally crossing 71 accessions to tetraploid tester lines. We quantify the contribution of both the maternal and paternal genomes on F1 seed size and investigate associations with the extent of seed size heterosis. Finally, using reciprocal pairs of maternal and paternal genome excess F1 triploids, we investigate whether parental genome dosage can enhance hybridity-associated parent-of-origin effects on F1 seed size in pairs of equivalent reciprocal F1 hybrid diploids beyond that achieved by polyploidy alone. This analysis allows us to quantify the impact of natural variation on parent-of-origin effects for F1 seed size in crosses between plants of equal or unequal ploidy, and to determine whether there are modifier effects on seed size heterosis arising from the interactions between hybridity and parental genome dosage.
Materials and Methods
Plant Growth Conditions and Crossing
Arabidopsis seeds were surfaced-sterilised with chlorine gas for 1 h by mixing 30 ml of sodium hypochlorite (10%) and 3 ml of hydrochloric acid (HCl) (37%) in a bell chamber. Sterile seeds were stratified for 3–4 days at 4°C in the dark and germinated directly in 5:1:1 mixture of compost:vermiculite:perlite (National Agrochemical Distributors Ltd., Dublin, Ireland). Plants were grown in a randomised block design under long-day conditions (16 h light, 21 °C /8 h dark, 18 °C) with light intensity 120–150 μE m–2 s–1 and humidity 70%. Manual crosses were performed on a Leica MZ6 stereo microscope; only flowers on the primary stem were emasculated to reduce possible alterations in seed size due to natural variation between branches. Sepals, petals and stamens were removed using #5 Forceps (Dumont, Montignez, Switzerland) and stigmas allowed to mature for two days before being pollinated.
Plant Material and Crossing Design
The accessions used in this study and the geolocation of where they were collected (Supplementary Table 1) were provided by Arthur Korte from University Würzburg. The 71 accessions (plus the Ler-0 tester line) used to generate the reciprocal F1 hybrids are part of an original panel of 400 accessions designed by Arthur Korte which comprised 200 Swedish accessions (Long et al., 2013) and an additional random set of 200 accessions derived from a wide geographic range including Central and Western Europe, Asia, and North America. Tetraploid Ler-0 seeds (4x Ler-0) were the kind gift of Brian Dilkes (Purdue University, West Lafayette, IN, United States), generated by colchicine doubling (Blakeslee, 1941). A diploid tester line in Ler-0 background (2x Ler-0) was used to generate the set of 142 Ler-0/Accession hybrids (including reciprocals). To minimise any possible environmental effects, three replicates of every cross combination were generated (for a total of 426 manual crosses) and grown in a randomised block design. The seed size value of parental lines was obtained from three biological replicates of self-fertilised plants.
Seed Size Analysis
Approximately 20 DAP (Days After Pollination) siliques were harvested once they had turned brown before dropping seeds and stored in paper envelopes before seed size analysis. Dried silique material and aborted seeds were removed using forceps, seeds were spread onto an EPSON Perfection V600 Photo Scanner and evenly separated using a painting brush to ensure further single-seed measurements. Images were taken at a resolution of 900 dpi in a black and white format. ImageJ was used to measure seed area (Abramoff et al., 2004), with a range of 0.08–0.3 mm2 for diploid and paternal-excess triploid seeds and 0.03–0.2 mm2 for maternal-excess triploid seeds to exclude any non-seed material.
Calculation of Heterosis, Genetic Distance, and Heritability
The mean size (mm2) of exclusively plump seeds was used for seed size heterosis calculations following Falconer and Mackay (1996) to evaluate F1 performance over their mid-parent (MP), best-parent (BP), or worst-parent (WP) values (Falconer and Mackay, 1996). Mid-Parent Heterosis (MPH) levels were determined as: %MPH = (F1 – MP)/MP × 100, where the MP value consists of the mean value of both parents. MPH is a relative measure to assess whether F1 hybrids have a greater performance (bigger seed size) than the average performance of both parents. In contrast, Best-Parent Heterosis (BPH) levels evaluate the F1 hybrid value compared only to the best performing parent (BP), determining offspring with increased seed size over the largest parent. BPH levels were calculated as: %BPH = (F1 – BP)/BP × 100. Worst-Parent Heterosis (WPH) was calculated as %WPH = (F1 – WP)/WP × 100 to determine F1 hybrids with a smaller seed size than the worst performing parent (WP). The relative genetic distance was calculated as the proportion of alleles in an accession of a different genotype than that of the tester line Ler-0, given the assumption of complete homozygosity across all accessions. Calculations were made using a subset of 3,866,657 SNPs acquired from 1001 genomes rather than an absolute distance using all 10,709,949 available SNPs, with a SNP excluded when all 71 accessions as well as Ler-0 were found to share a genotype at that locus. For each of the cross directions, heritability was calculated by taking the proportion of variance explained by genotype as a fraction of overall variance. Variance were calculated using a random effects model such that:
where Yij is the jth seed of the ith accession cross with Ler-0, gi is a factor accounting for the effect of the ith accession and ?ij is the error associated with each measurement. The above equation was used to calculate the variance components of the 2x Ler-0 X 2x Accession cross progenies and the 2x Accession X 2x Ler-0 cross progenies independent of one another. Heritability in this case is assumed to be broad-sense rather than narrow-sense as the inbred nature of the parental accessions means that all F1 offspring generated by a given cross should be genetically identical.
Results
Parent-of-Origin Effects on Size of F1 Hybrid Arabidopsis Seeds
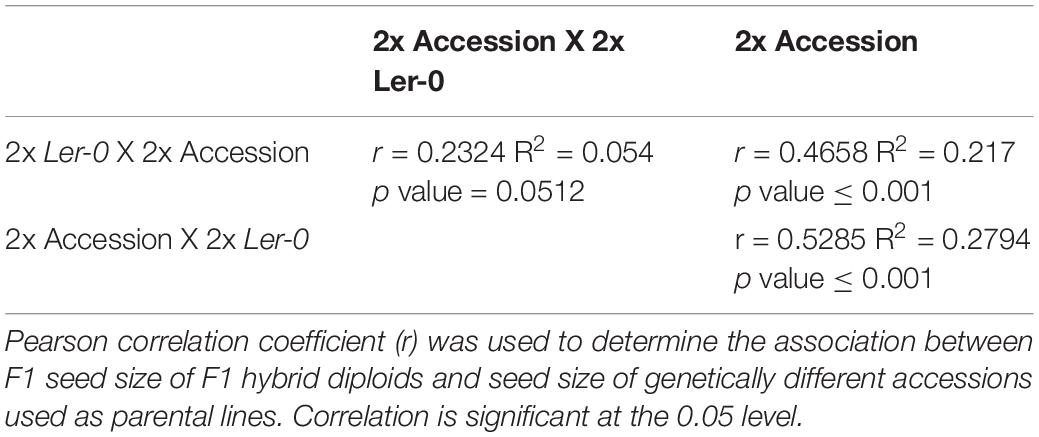
To determine the extent of parent-of-origin effects on F1 seed size heterosis from crosses between different Arabidopsis accessions, a panel of 71 genetically different diploid (2x) accessions (Supplementary Table 1) and a 2x tester accession in Ler-0 background were used as crossing parents to generate reciprocal Ler-0/Accession F1 hybrids. As a baseline, we first determined the F1 seed size of each parental line when grown under controlled, randomised conditions. As expected, we observed significant natural variation for seed size across the accessions when selfed (Figure 1A), ranging from the small-seeded accessions IP-Pro-0 (0.1179 mm2) and the Ler-0 tester accession (0.1332 mm2), to the considerably larger seeds of Pt-0 (0.1672 mm2) and Ta-0 (0.1781 mm2).
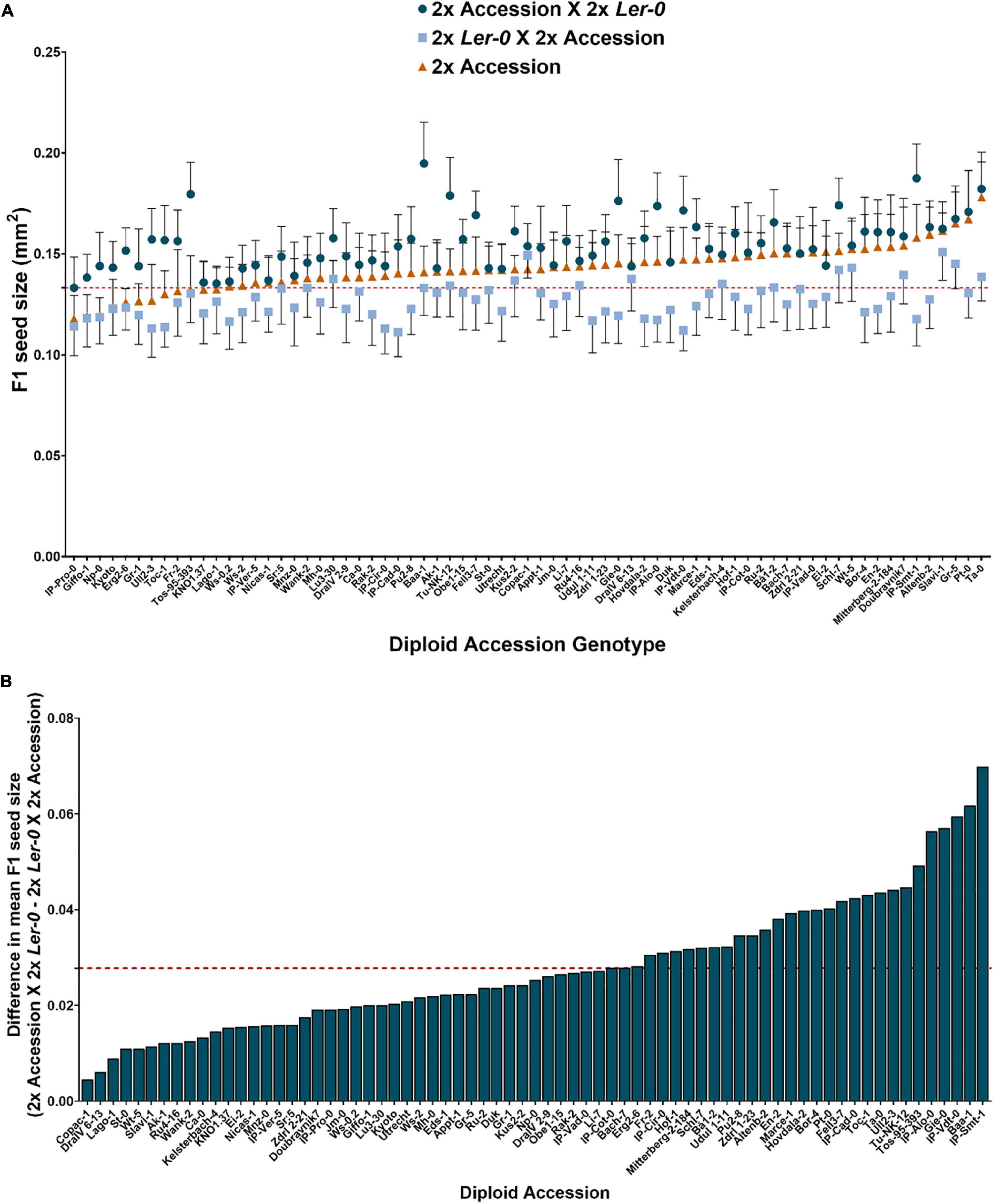
Figure 1. Heterosis effects on F1 seed size observed in reciprocal F1 hybrid diploid seeds depends on parent-of-origin effects. (A) F1 hybrid diploid seed size of reciprocal crosses using 2x Ler-0 as pollen donor (represented by blue circles) or as maternal parent (light blue squares). Seed size of selfed isogenic parental lines is also represented (orange triangles) for comparison with their reciprocal F1 offspring. The horizontal red dashed line corresponds to the mean seed size value of the accession 2x Ler-0 (0.1332 mm2). Error bars represent SD. N ≥ 40 seeds/genotype. (B) Natural variation for parent-of-origin effects on seed size across 71 reciprocal F1 hybrid diploids. The red dashed line represents the average difference in mean F1 seed area for the entire set of 71 reciprocal hybrids.
To systematically quantify the extent of parental effects, each cross was performed in both directions (to generate a set of reciprocal F1 hybrid seeds (2x Ler-0 X 2x Accession vs 2x Accession X 2x Ler-0). We found that F1 seed size was strongly influenced by parent-of-origin effects, as many pairs of reciprocal crosses displayed large parental effect differences in F1 seed size. Generally, Ler-0 when used as maternal parent produced the smallest seeds (0.82 times smaller on average than their reciprocal counterparts), whereas Ler-0 used as pollen donor led to substantially larger F1 seeds (Figure 1A). These results are consistent with previous findings of parent-of-origin effects on seed size in crosses involving Ler-0 (see section “Discussion”). To quantify the relationship between maternal or paternal genotype and F1 seed size, we performed a correlation analysis (Table 1). A strong correlation (r = 0.5285, p-value ≤ 0.0001) was found between the seed size of F1 hybrid diploid seeds derived from 2x Accession X 2x Ler-0 crosses and the seed size of that of the accessions used as maternal parent, confirming that F1 seed size is highly influenced by maternal effects. Notably, we also found a strong positive correlation (r = 0.4658, p-value ≤ 0.0001) between the seed size of F1 hybrid seeds from 2x Ler-0 X 2x Accession crosses and the parental seed size of the 2x accessions used as pollen donors, suggesting that F1 hybrid seed size can also be significantly influenced by the paternal genome (Figure 1A). Such “paternal effects” have been reported before from small numbers of inter-accession crosses (House et al., 2010), but have typically been found to be quite small.
To quantify the extent of parent-of-origin effects observed in F1 seed size between pairs of reciprocal F1 hybrid diploids, the mean seed size area of F1 seeds from 2x Ler-0 X 2x Accession crosses was subtracted from the mean F1 seed size of the reciprocal cross 2x Accession X 2x Ler-0. The positive values obtained (Figure 1B) confirmed the consistently bigger seed size of 2x Accession X 2x Ler-0 F1 hybrids. Not all genetically different F1 hybrids displayed the same magnitude in seed size differences between pairs of reciprocal F1 hybrid diploids (Figure 1B). For instance, the parental accessions Copac-1, DraIV 6–13, and Lago-1 generated the three F1 hybrids with the weakest parent-of-origin effects (Δ = 0.0045 mm2, Δ = 0.0061 mm2, and Δ = 0.0089 mm2, respectively), while IP-Vdt-0, Baa-1 and IP-Smt-1 were the three parental accessions with the strongest parent-of-origin effects on F1 seed size (Δ = 0.0595 mm2, Δ = 0.0617 mm2, and Δ = 0.0698 mm2, respectively). As shown in Figures 1B and 2, weak parent-of-origin effects led to similar F1 seed size between reciprocal F1 hybrid diploids, whereas strong parent-of-origin effects led to dramatic differences in F1 seed size.
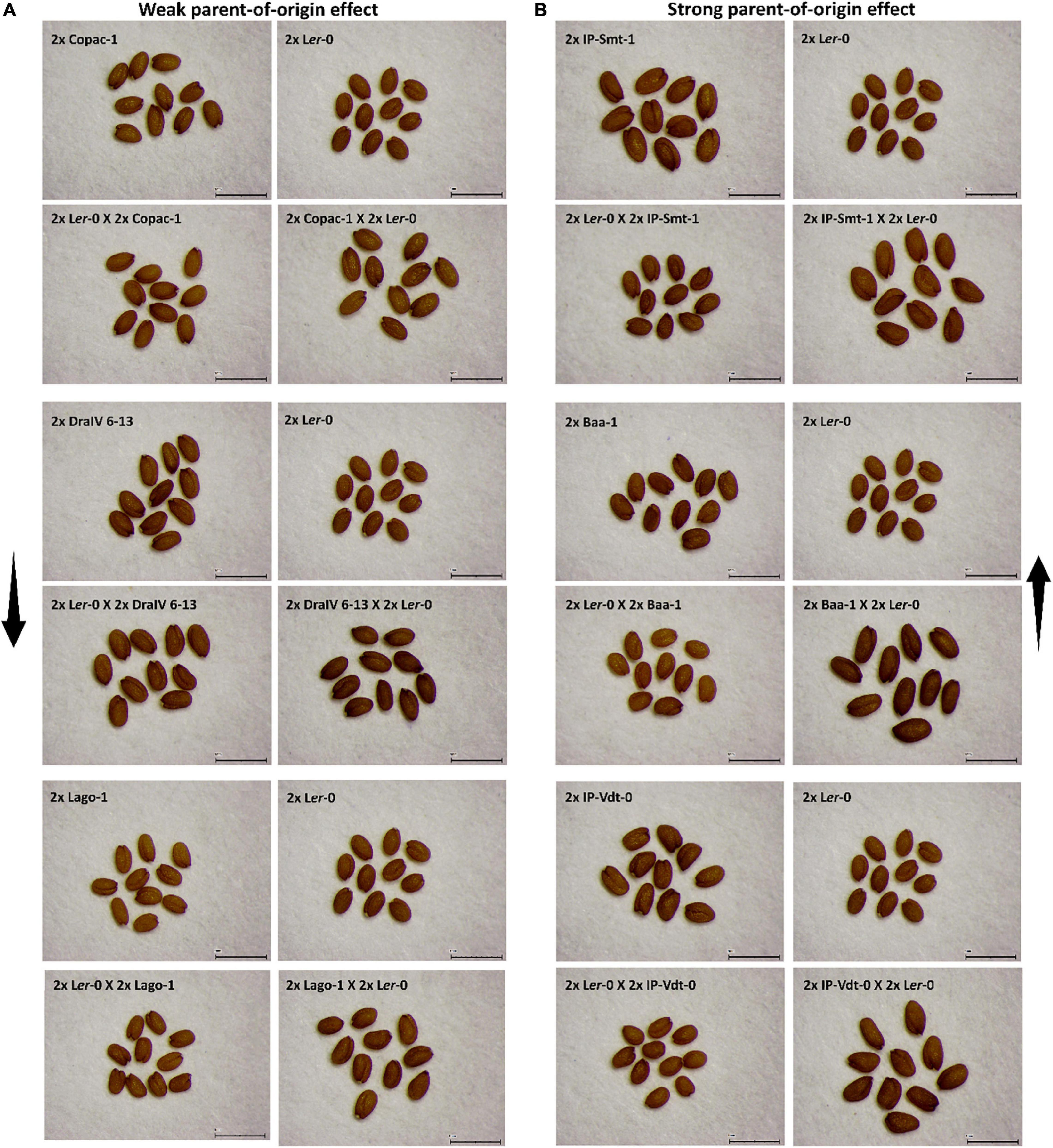
Figure 2. Seed size phenotypes of reciprocal F1 hybrid diploid seeds and their parental lines. (A) The three F1 hybrid diploids with the weakest parent-of-origin effects on F1 seed size in this study and the accessions used as parental lines. The black arrow represents the intensity of parent-of-origin effects in ascending order. (B) The three F1 hybrid diploids with the strongest parent-of-origin effects in this study and their parental lines. The black arrow represents the intensity of parent-of-origin effects, from lowest to highest. Scale bar = 1 mm.
Extent of Heterosis Effects on F1 Seed Size Strongly Influenced by Parent-of-Origin Effects
Seed size in Arabidopsis inter-accession hybrids frequently demonstrates heterosis when compared to the parental values. To quantify the extent of heterosis on size of F1 seeds and of any impact of the parent-of-origin effects described above, we calculated Best-Parent (%BPH), Mid-Parent (%MPH) and Worst-Parent Heterosis (%WPH) for F1 seed size for all 71 reciprocal Ler-0/Accession hybrids (Figure 3 and Table 2). Best-Parent Heterosis was found for F1 seed size in all 2x Accession X 2x Ler-0 cross direction F1 hybrids, except those derived from the parental accessions DraIV-6–13 (BPH = –1.17%), Duk (BPH = –0.68%), Zdrl 2–21 (BPH = –0.4%), Ei-2 (BPH = –4.57%), and IP-Pro-0 (BPH = 0%) (where a negative value for %BPH indicates that the F1 hybrid does not exceed the seed size of the BP value). Notably, IP-Pro-0 when used as maternal parent produced F1 hybrid seeds that were the same size as the BP value (Figure 3B and Supplementary Table 2). All four accessions did, however, display Mid-Parent heterosis, meaning that the F1 hybrids exceeded the seed size of the average value of the two parental lines. The highest levels of BPH were found in Baa-1/Ler-0 F1 hybrids (following the convention the maternal parent is always mentioned first), whose F1 seeds were nearly 40% larger than the BP value, whereas the lowest levels of BPH were found in the Utrecht/Ler-0 F1 hybrids, with seeds only 0.4% larger than the BP value.
Figure 3. The heterotic effect on F1 diploid seed size depends on parent-of-origin effects. (A) Levels of Best-Parent (%BPH), Mid-Parent (%MPH) and Worst-Parent Heterosis (%WPH) across reciprocal F1 hybrid diploid seed size. (B) F1 hybrids that do not follow the trend observed among 71 reciprocal Ler-0/Accession F1 hybrids for %BPH, %MPH or %WPH. Exceptions are indicated in red. The horizontal dotted line represents the critical value 0.
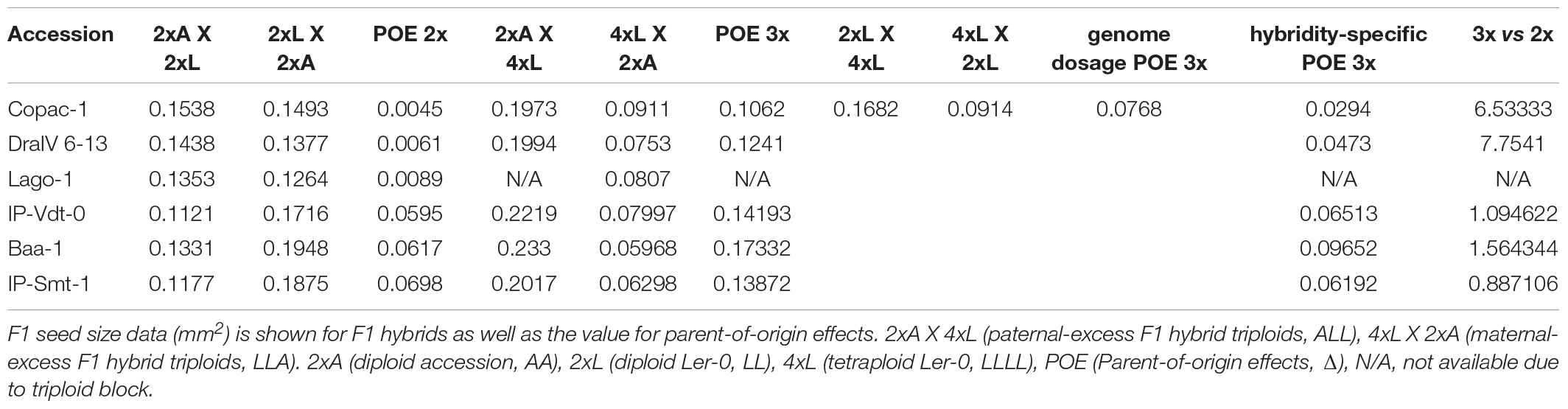
Table 2. Hybridity-associated parent-of-origin effects on seed size in F1 hybrid triploids compared with their diploid counterparts.
Conversely, the F1 hybrids generated from the 2x Ler-0 X 2x Accession cross direction showed a tendency toward WPH for seed size (Figure 3B and Supplementary Table 2). The highest levels of WPH were found in the Ler-0/IP-Cad-0 and Ler-0/IP-Vdt-0 F1 hybrids, with F1 seeds 16% smaller than the WP value. However, this pattern was more likely to be disrupted by the genome of the pollen donors, with numerous accessions such as Wank-2, Lu3-30, Tu-NK-12, Kus2-2, Copac-1, Ru4-16, DraIV 6–13, Kelsterbach-4, Bå1-2, Schl-7, Wt-5, Doubravnik7, Slavi-1, Gr-5, and Ta-0 producing Best-Parent and/or Mid-Parent Heterosis instead. For example, Ler-0/Copac-1 and Ler-0/Slavi-1 crosses produced F1 seeds that were more than 10% larger than their respective WP value. Ler-0/Copac-1 F1 hybrids displayed BPH levels (BPH = 4.8%), while Ler-0/Slavi-1 showed MPH levels (MPH = 2.4%). These parental effect results indicate that in these genotype combinations, the pollen donor genotype alters the heterotic response and suppresses the usual small-seeded “Ler-0 mother effect.”
We also investigated whether there was any correlation between the genetic distance (or geolocation) between the tester Ler-0 genotype and the accession genotypes used as either maternal or paternal parents in crosses with Ler-0. No correlation was observed between genetic distance (or geolocation) and heterosis effects on F1 seed size (Supplementary Figure 2 and Supplementary Table 6). We further investigated the broad-sense heritability of hybrid F1 seed size in relation to maternal and paternal parent of origin, where the maternal genotype explains 41.87% of the phenotypic variance in F1 hybrid seed size, whereas the paternal genotype explains 24.8% (Supplementary Table 7).
Overall, our results indicate that while F1 seed size heterosis is subject to natural variation within species, the extent of the heterosis effects are strongly influenced by parent-of-origin effects. While positive F1 seed size heterosis was typically associated with a Ler-0 father effect, negative heterosis was generally induced by Ler-0 maternal inheritance. However, there are inter-accession crosses which generate F1 seeds that do not conform with the general trends.
Hybridity Enhances Parent-of-Origin Effects on F1 Seed Size in Inter-Ploidy Crosses Beyond Those Observed in Diploid Crosses
To investigate whether parental genome dosage modifies the “Ler-0 effect” observed across our reciprocal F1 hybrid diploid population, we repeated our crosses using a subset of the 71 accessions, but this time crossed to a tetraploid (4x) tester line, again in the Ler-0 background. These genotypes were denoted “AA” for the diploid accession, “LLLL” for the tetraploid Ler-0 tester, and “ALL” or “LLA” for the two sets of reciprocal F1 hybrid triploid seeds (using the genetically identical genotype of the embryo to indicate the two sets of offspring) (Supplementary Figure 1). We performed these crosses using the three accessions that displayed the weakest parent-of-origin effects on F1 seed size in diploid-diploid crosses (Copac-1, DraIV 6-13 and Lago-1) and those which displayed the strongest parental effects (IP-Vdt-0, Baa-1 and IP-Smt-1). Furthermore, reciprocal isogenic (genetically identical) inter-ploidy crosses between 2x and 4x Ler-0 plants were also conducted to disaggregate heterosis effects due to genetic hybridity vs genome dosage (Figure 4 and Table 2). We have previously shown that these effects can interact in complex ways in control of plant biomass (Fort et al., 2016), and had hypothesised that similar interactions in control of seed size could help to explain later biomass accumulation.
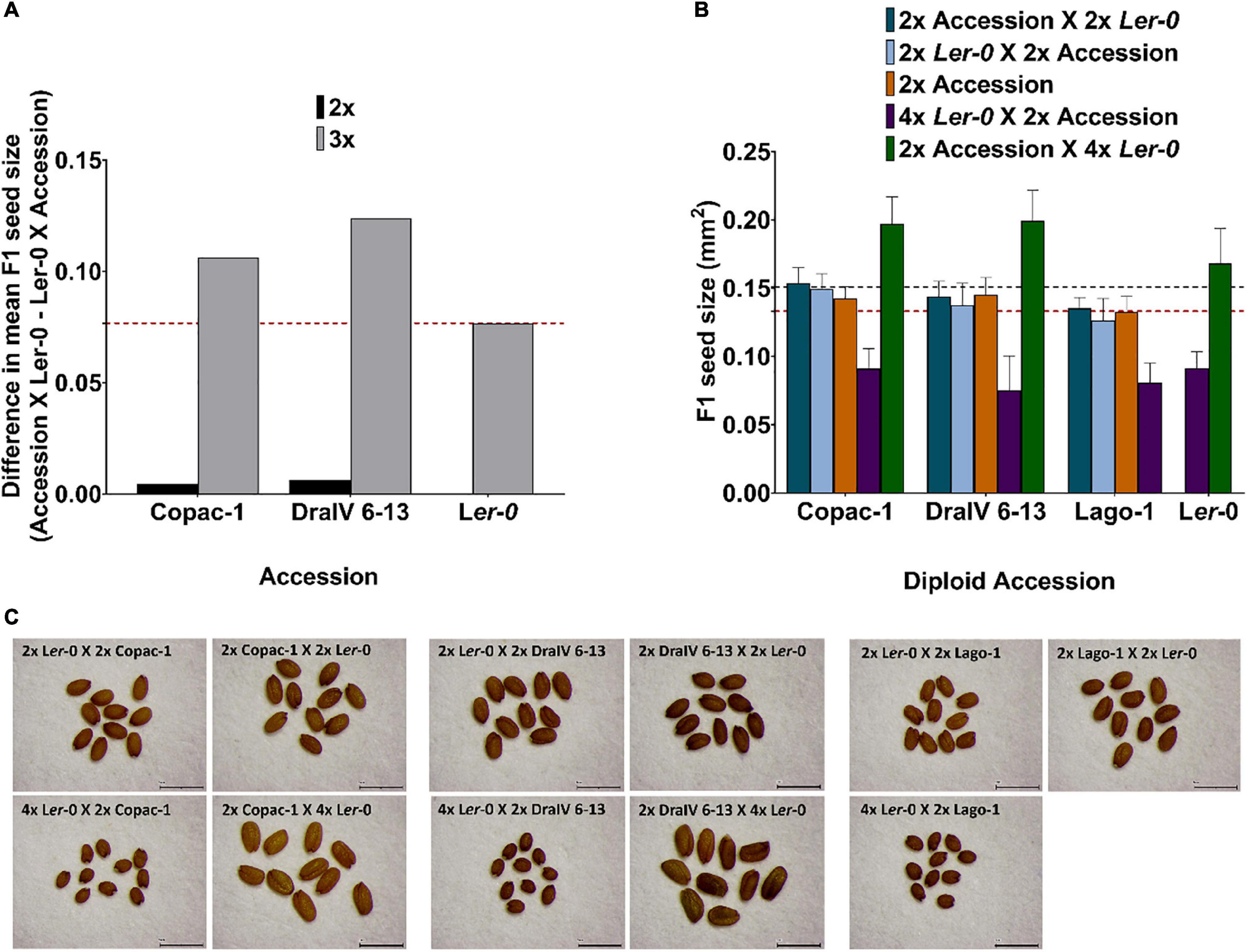
Figure 4. Hybridity enhances parent-of-origin effects in F1 triploids beyond that achieved at the diploid level. (A) Difference in F1 mean seed size for reciprocal isogenic (Ler-0) and hybrid crosses at the diploid and triploid level. The red dashed line represents the parent-of-origin effects for Ler-0 F1 isogenic triploids. (B) F1 hybrid seed size of balanced-ploidy crosses (2x Accession X 2x Ler-0 and 2x Ler-0 X 2x Accession) and inter-ploidy crosses (2x Accession X 4x Ler-0 and 4x Ler-0 X 2x Accession). Seed size of parental lines is also represented (orange for the natural accessions, a horizontal red dashed line for 2x Ler-0 and black dashed line for 4x Ler-0) for comparison with ploidy levels and their reciprocal F1 offspring. Error bars represent SD. (C) Seed size phenotypes of reciprocal F1 seeds in balanced and inter-ploidy crosses. Scale bar = 1mm.
Notably, we found that the “Ler-0 effect” can be enhanced in pairs of reciprocal F1 hybrid triploid crosses of the accessions from the weakest parent-of-origin effects group. At the triploid level (3x), parent-of-origin effects in Copac-1/Ler-0 hybrids increased up to Δ = 0.1062 mm2, in contrast to those of 2x level (Δ = 0.0045 mm2) (Table 2 and Figure 4A). To calculate parent-of-origin effects strictly associated with genetic hybridity, we subtracted the parent-of-origin effects caused by genome dosage observed in isogenic inter-ploidy reciprocal crosses of Ler-0 (Δ = 0.0768 mm2) from that of Copac-1/Ler-0 hybrid triploids (Δ = 0.1062 mm2). Strikingly, we found that genetic hybridity alone induced parent-of-origin effects that were 6.5 times higher in Copac-1/Ler-0 hybrid triploids than in diploids (Table 2 and Figure 4A). Similarly, we detected parent-of-origin effects in DraIV 6-13/Ler-0 hybrids triploids that were 7.75 times higher than those at the 2x level, enhancing the usual “Ler-0 effect.” It was not possible to determine parent-of-origin effects at the 3x level in Lago-1/Ler-0 reciprocal crosses, as paternal genome excess in this genotype cross combination leads to the postzygotic reproductive barrier known as the triploid block (Köhler et al., 2010, 2021). In contrast to these results, we observed that such “Ler-0 effects” remained similar at both the 2x and 3x level in F1 hybrids of the accessions from the strongest parent-of-origin group (Table 2).
Overall, our results indicate that crosses between plants of unequal ploidy can drive additional hybridity-associated parent-of-origin effects in certain genotype combinations, even where no such parent-of-origin effects are seen in equivalent inter-accession crosses between diploids.
Hybridity Has a Greater Effect on F1 Seed Size Heterosis at Triploid Level Compared to Diploid Level
To investigate whether the additional hybridity-associated parent-of-origin effects found in F1 hybrid triploids of Ler-0 and the accessions Copac-1, DraIV 6-13 and Lago-1 modify the extent of heterosis, we calculated MPH levels at the 2x and 3x level. MPH ensures that both parental lines are considered in determining the level of heterosis in the F1 hybrid. For instance, a positive value for MPH may indicate that the F1 hybrid outperforms the average value of the parental lines, while a negative value indicates the hybrid falls below it. We therefore quantified the hybridity-specific contribution to deviations from the Mid-Parent value (MPV), beyond that achieved by genome dosage effects alone on 3x seed size.
On average, we found that hybridity alone increased F1 seed size by extra 17.9% in paternal-excess F1 triploids (3xp) and decreased it by 9.9% in maternal-excess F1 triploids (3xm) when compared to the Ler-0 F1 isogenic 3x control (Supplementary Table 3). Despite the occurrence of MPH at the 2x level for certain cross combinations (Supplementary Table 4), we detected a greater deviation from the MPV in reciprocal inter-ploidy crosses (Supplementary Tables 3, 4), which exceeded both the relative levels of MPH achieved by polyploidy alone in Ler-0 F1 isogenic triploids and those in F1 hybrid diploids (Supplementary Table 3). On one hand, F1 hybrid 3xp seeds were on average 16.1% larger than the MPV due to hybridity alone (after subtracting the MPH levels observed in Ler-0 F1 isogenic 3x control crosses due to genome dosage), unlike their 2x counterparts which were only 5.5% larger. On the other hand, F1 hybrid 3xm seeds were 7.7% smaller than the MPV without considering genome dosage effects, in contrast to their hybrid diploid counterparts which were 0.8% larger (Figure 5 and Supplementary Table 3).
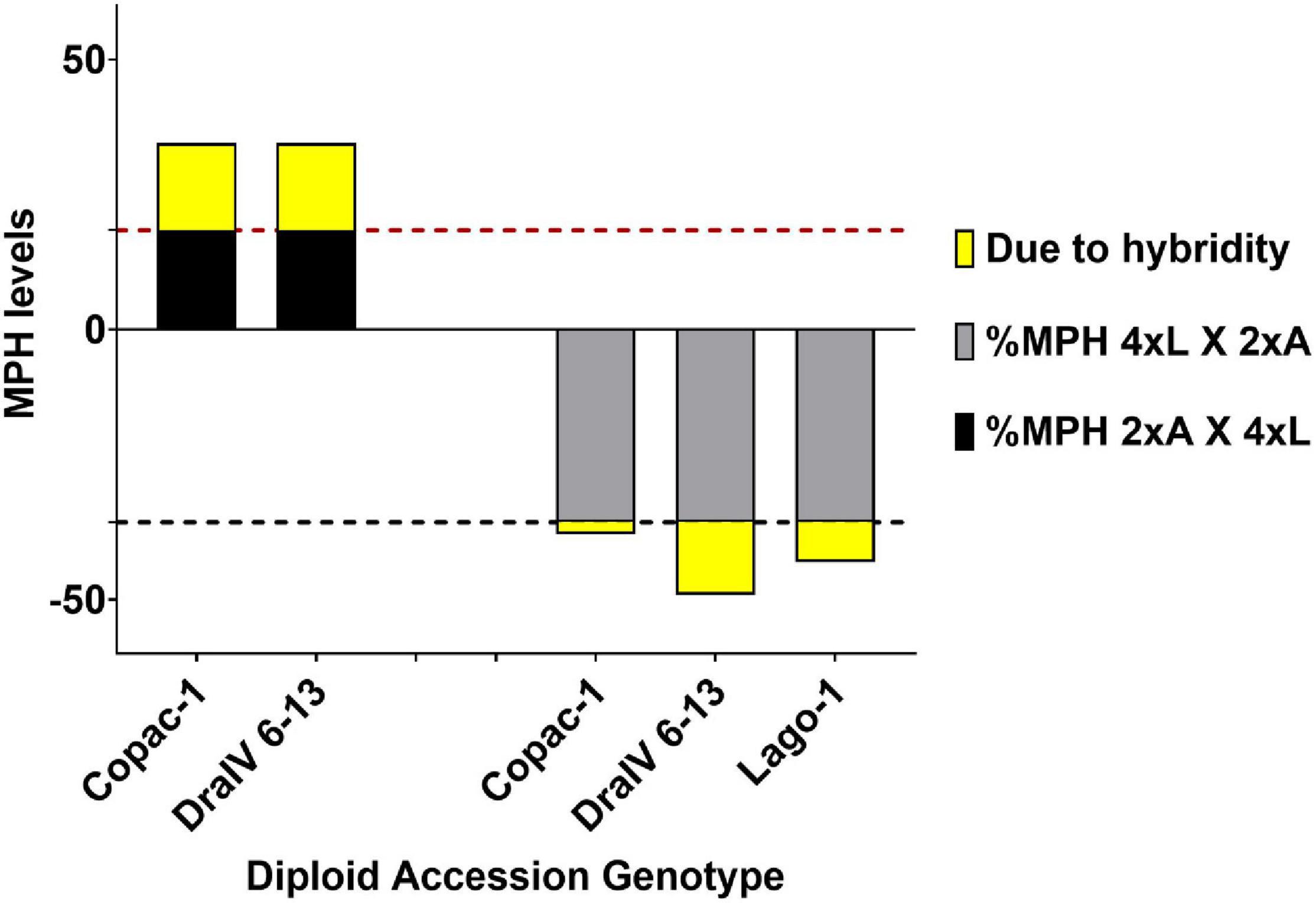
Figure 5. Hybridity has a greater effect on F1 seed size heterosis at the triploid level (3x). Mid-Parent Heterosis (MPH) levels in F1 3x seeds from reciprocal inter-ploidy crosses are shown. 2xA X 4xL (2x Accession X 4x Ler-0), 4xL X 2xA (4x Ler-0 X 2x Accession). The proportion due to hybridity is highlighted in yellow and it was calculated by subtracting the %MPH of reciprocal Ler-0 F1 isogenic inter-ploidy crosses (18.4% for 2x4 crosses, represented by a horizontal red dashed line, and –35.7% for 4x2 crosses, represented by a horizontal black dashed line) from the MPH levels of F1 hybrid triploids.
We also found that Copac-1 suppressed the “Ler-0 mother effect,” as 2x Ler-0 X 2x Copac-1 F1 seeds exceeded the MPV by 8.3% (Supplementary Table 3), while Ler-0 mothers tended to generate smaller seeds (Figure 3). Paternal inheritance of Copac-1 increased F1 seed size independently of the ploidy level: i.e., F1 3xm derived from 4x Ler-0 X 2x Copac-1 crosses did not decrease F1 seed size beyond the genome dosage effect, unlike the other genotype combinations. Similarly, maternal contribution of Copac-1 also led to a F1 seed size increase in both 2x and 3x levels, although triploidy enhanced this positive effect on F1 seed size (Figure 5 and Supplementary Table 3). Finally, we sought to quantify the positive impact of paternal genome excess on F1 triploid seed size across 48 genetically distinct hybrids (Supplementary Figure 3 and Supplementary Table 5). On average, the introduction of hybridity at the 2x level increased seed size by 12.6% over the MPV. However, altering the paternal genome dosage of these F1 hybrids led to a further F1 seed size increase of 38.8%, where 20.4% of such enhancement was due to hybridity alone.
Overall, these findings indicate that hybridity has a greater effect than parental genome dosage effects on F1 seed size in a triploid context, and that genetic hybridity acts as a ‘heterosis enhancer’ under parental genome imbalance.
Discussion
Heterosis effects on F1 seed size can occur in inter-accession crosses of Arabidopsis. Indeed, we and others have demonstrated pervasive parent-of-origin effects during plant reproduction (House et al., 2010). In Fort et al. (2016) we proposed such parental effects as causal to increased rosette biomass accumulation in polyploid F1 hybrids. In this study, we use a diversity panel of Arabidopsis accessions to (1) investigate natural variation in parent-of-origin effects on F1 seed size; (2) how parent-of-origin effects modify the heterosis response for F1 seed size; and (3) to identify “modifier” accessions which ablate or enhance parent-of-origin effects on F1 seed size. Here, we discuss our findings in relation to seed size control and heterosis effects in plant reproduction, breeding and biotechnology.
Parent-of-Origin Effects Modify Paternal and Maternal Control of F1 Seed Size Heterosis in Hybrid Diploids
The Arabidopsis accession Ler-0 generates small F1 seeds when used as a maternal (ovule) parent in inter-accession crosses, and larger ones when used as the paternal (pollen) parent. In this study we used Ler-0 as a tester line on the basis of this property, as we have done previously (Duszynska et al., 2013, 2019), reasoning that, if this effect is robust across hybrid genotypes, it provides a basis for identifying those “modifier” accessions which enhance or suppress this heterosis effect, as well as for estimating the range of variation which this trait may display. We did indeed confirm that the expected pattern was generally robust in reciprocal crosses of 71 accessions, with F1 seeds produced using Ler-0 as ovule parent being on average 0.82 times smaller their reciprocals (Figure 1A). This indicates that the trends reported from reciprocal crosses between Ler-0/Accession such as Cvi by Alonso-Blanco et al. (1999), C24 by Zhu et al. (2016), and with groups of three or four accessions by House et al. (2010) and Groszmann et al. (2014) can be generalised across European and North American Arabidopsis accessions. Whether this generalised pattern holds true for crosses with more recently identified glacial relictual populations (Lee et al., 2017; Fulgione and Hancock, 2018; Toledo et al., 2020) or the poorly-understood African diversity of the species (Durvasula et al., 2017) remains to be determined.
Most studies on parental control of F1 seed size report pivotal roles for the maternal parent, especially in crops, where paternal effects are rarely reported (Singh et al., 2017; Baskin and Baskin, 2019). However, our data indicates significant paternal effects on F1 seed size heterosis in balanced-ploidy crosses. Indeed, we find that the maternal genotype explains 28% of the variation in F1 hybrid diploid seed size, similar to that of House et al. (2010), who found that maternal genotype explained 29.3% of the variation in F1 seed size in reciprocal hybrid diploid crosses. In contrast, we found that a further 22% of variation is contributed by the paternal genotype (r = 0.4658, p < 0.0001), considerably higher than the 10.4% estimated by House et al. and supporting our earlier supposition that paternal effects on seed size in Arabidopsis thaliana could lead to significant changes in biomass in mature plants (Fort et al., 2016). Furthermore, parent-of-origin effects due to genetic hybridity alone were on average 7.14 times higher in F1 hybrid triploids than those found in the diploid counterparts for the accessions from the weakest parent-of-origin effects group (Figure 4 and Table 2), while it remained similar at both the 2x and 3x level in F1 hybrids of the accessions from the strongest parent-of-origin group. These results suggest a hidden phenotypic plasticity in the “Ler-0 effect” that can be modified by genome dosage.
Differences in Mechanisms of Maternal and Paternal Control of F1 Seed Size
Notably, we could not find any significant correlation between the F1 seed size of pairs of reciprocal F1 hybrid diploids (Table 1), suggesting that the mechanisms for increases in F1 offspring seed size differ depending on whether the genome is inherited maternally or paternally. It is well-established that natural hybrid populations in both plants and animals tend to establish with one species consistently being the maternal parent, i.e., monodirectionally (Trier et al., 2014; Nieto Feliner et al., 2017). This may be due to ploidy differences that are only tolerated in the endosperm when in maternal excess (Donoghue et al., 2014; Köhler et al., 2021; Städler et al., 2021) or more rarely in paternal excess (Vallejo-Marín et al., 2016). However, in homoploid F1 hybrids, the basis of this bias remains unknown (except where maternal size is critical for gestation, as in mammals (Abbott et al., 2013). As F1 seed size is strongly associated with successful offspring establishment, differences in the genetic basis of seed size by parent-of-origin could provide a further explanation for such phenomena in plants.
Despite the critical importance of maternal control of seed size, we could not attribute the observed increase in size of F1 hybrid diploid seeds from 2x accession X 2x Ler-0 crosses entirely to maternal effects. On average, the parental accessions used for reciprocal crosses had larger seed sizes than the tester line 2x Ler-0, except for IP-Pro-0, Giffo-1, Np-0, Kyoto, Erg2-6, Gr-1, UII2-3, Toc-1, Fr-2, Tos-95-393, KNO1.37 and Lago-1. Despite the relatively smaller seed size of these exceptions, they produced larger F1 hybrid diploid seeds when used as maternal parent (Figure 1A). These results reveal a significant paternal effect of Ler-0 that is independent of any effects derived as a female parent, as Ler-0 tends to generate the biggest F1 seeds when used as paternal parent, irrespective of the maternal genotype, with only some exceptions (Supplementary Table 2 and Figure 3). The mechanisms for this “Ler-effect” remain unknown but likely involve increased seed sink strength: this has been argued as a likely role for imprinted Paternally Expressed Genes (iPEGs) (Baroux et al., 2002; McKeown et al., 2013b), which we have shown to be preferentially subject to positive selection in Arabidopsis (Tuteja et al., 2019), although direct evidence for iPEGs controlling sink strength in this species is lacking.
We also found that parent-of-origin effects on F1 hybrid seed size are widely influenced by natural variation (Figure 1B). We detect genotype combinations that lead to both strong and weak parent-of-origin effects on F1 seeds in homoploid crosses. Such reciprocal F1 hybrid diploid seeds are composed of embryos who are genetically identical (but epigenetically different), but which also differ in their seed coat composition which is of maternal origin. Similarly, the endosperm genome in F1 seeds may act as a source of variation, as it differs in the genome dosage composition (2m:1p maternal:paternal genome ratio), potentially resulting in unequal parental genome contribution effects that may explain the phenotypic F1 seed size differences we observe (Lafon-Placette and Köhler, 2014; Pignatta et al., 2014; Lafon Placette, 2020; Picard et al., 2021). Another possibility is that parent-of-origin effects in F1 hybrid seeds can alter cyto-nuclear interactions that contribute to F1 seed size heterosis (Flood et al., 2020), ultimately altering the F1 hybrid’s transcriptional program (Botet and Keurentjes, 2020). Flood et al. (2020) have revealed how organellar genomes affect seed size through new plasmotype–nucleotype combinations (cybrids) across five genetically different reciprocal cybrids of Arabidopsis. In particular, a seed size increment (1.6× larger), in comparison with its nuclear parent, was found in Ler-0/Sha cybrids (Ler-0 nucleotype, Sha plasmotype), suggesting that cyto-nuclear interactions are important for seed size heterosis and may be altered in F1 hybrids (Flood et al., 2020).
We did not detect any significant correlation between differences in mean F1 seed size of parental lines and that of reciprocal F1 hybrid diploids. Hence, similar seed size in the parental lines does not necessarily lead to similar F1 size in reciprocal F1 hybrid diploids. This was particularly apparent in reciprocal crosses of 2x Ler-0 with some accessions (e.g., Toc-1, Fr-2, UII2-3, Tu-NK-12, Tos-95-393, IP-Vdt-0, and Baa-1), where the parental seed size was very similar to diploid Ler-0, but the F1 hybrid seeds differed considerably (Figures 1, 2).
Seed Size Heterosis Depends on Parent-of-Origin Effects
We detected significant natural variation in the extent of the heterosis effects on F1 seed size when diploid accessions were reciprocally crossed, which depended on the cross direction (Figure 3 and Table 1). BPH compares the F1 hybrid trait values only to the best performing parent, in contrast to MPH, which is a relative measure that ensures that both parental lines are considered in determining the level of heterosis in the F1 hybrid. As the F1 hybrid populations share a common parent (2x Ler-0), we could attribute the differences in MPH levels to the differences across the different natural accessions used as either maternal or paternal parent. Consistent with our previous observations on Ler-0 fathers (pollen) generating bigger F1 hybrid seeds, high levels of BPH were found across 2x Accession X 2x Ler-0 F1 hybrid population except for the F1 hybrids derived from the accessions DraIV-6-13, Duk, Zdrl 2-21, Ei-2 and IP-Pro-0, whose F1 seeds instead displayed MPH.
BPH and WPH for F1 seed size are indicative of extreme or transgressive phenotypes (Rieseberg et al., 1999; Mackay et al., 2021) and can be of agricultural importance when genotype combinations lead to larger (BPH) or smaller seeds (WPH), depending on the desired seed size for certain species. WPH can be considered an inverse of conventional heterosis and is typically hypothesised to be due to outbreeding depression, potentially derived from negative epistatic interactions (Bomblies et al., 2007; Oakley et al., 2015). In this case, hybridisation would disrupt favourable allele interactions (Burke and Arnold, 2001; Bomblies et al., 2007; Alcázar et al., 2010; Chae et al., 2014; Chen et al., 2016; Boeven et al., 2020). We found genotype combinations that led to transgressive phenotypes in both directions, which confirm the non-additive mode of inheritance typical of heterosis.
The Arabidopsis accessions used in this study covered a wide geographic distribution (Supplementary Table 1) and showed wide natural variation for heterosis effects on F1 seed size. It is unclear whether the extent of heterosis can increase with genetic distance between parents, with most studies failing to support such correlations (Schnable and Springer, 2013; Seymour et al., 2016; Labroo et al., 2021), consistent with our results in this study (Supplementary Figure 2). Furthermore, we find no significant association between geolocation (latitude and longitude) and phenotypic variance for parent-of-origin effects, reciprocal F1 hybrid diploid seed size or seed size of selfed isogenic parental lines (Supplementary Table 6). Our results are in agreement with Ren et al. (2019), where no significant association between geographical distribution of 191 accessions and seed size from selfed plants was found. Our heritability analysis indicates that the maternal genotype explains 41.87% of the phenotypic variance in F1 hybrid seed size, whereas the paternal genotype explains 24.8% (Supplementary Table 7). These results are consistent with our findings in Table 1 that show a higher correlation between maternal seed size and its F1 hybrid seed offspring in a 2x Accession X 2x Ler-0 cross than the opposite 2x Ler-0 X 2x Accession cross.
Hybridity Enhances the “Ler-0 Effect” on F1 Hybrid Triploids
Inter-ploidy crosses alter the genome dosage balance ratio in the endosperm (and the timing of endosperm cellularisation), leading to developmental effects on F1 seed size. Many genes can be dosage-sensitive, due to genomic imprinting (Batista and Köhler, 2020), stoichiometric imbalances in macromolecular complexes (Veitia and Birchler, 2015), or other dosage sensitivities (Birchler et al., 2005), as well as possible maternal-zygotic crosstalk mechanisms (Doughty et al., 2014; Lafon-Placette and Köhler, 2016; Robert et al., 2018). Several studies suggest dosage-dependent phenotypes can also contribute to heterosis (Birchler and Veitia, 2012; Birchler et al., 2016). For instance, the major Quantitative Trait Loci (QTL) in tomato fruit weight 2.2 (fw2.2) displays a dosage-dependent heterotic response, as higher levels of gene expression correlated negatively with smaller fruit size (Cong et al., 2002; Liu et al., 2003). Furthermore, SINGLE FLOWER TRUSS (SFT) gene in tomato generates yield heterosis only when the loss-of-function allele is heterozygous, consistent with dosage-sensitivity (Krieger et al., 2010; McKeown et al., 2013a). Our results indicate that effects such as these may be revealed by parental genome dosage imbalance, and that these effects can enhance hybridity-associated parent-of-origin effects. Triploidy and hybridity can therefore generate unique phenotypes and potentially reveal cryptic genetic variation and/or interactions that are hidden in euploids and/or isogenics (Pires et al., 2016; Tabib et al., 2016).
Conclusion
In this study we have quantified natural variation in parent-of-origin effects on F1 seed size in the model plant, Arabidopsis, in both homoploid diploid and inter-ploidy (tetraploid X diploid) crosses. Our results indicates that parental effects can strongly influence F1 seed size, with a much larger influence of the paternal genome on F1 seed size than previously appreciated. Parent-of-origin effects on seed phenotypes in angiosperms can be sporophytic or gametophytic (Spillane et al., 2002). While maternal effects on seed size can have a sporophytic (e.g., seed coat tissues) and/or gametophytic (e.g., egg cell, central cell) basis, paternal effects are most likely to be gametophytic effects. The underlying molecular mechanisms responsible for the significant maternal and paternal effects on F1 seed size we identify in this study remain to be identified. Our findings have implications for hybrid seed biology and may open new avenues for the study of paternal contribution to F1 seed size, potentially generating new applications in plant breeding programs. The lack of correlation between maternal and paternal effects indicates that both the genetic pathways mediated, and the evolutionary pressures likely to act on them, differ between pathways for maternal and paternal control of F1 seed size. We also demonstrate that genetic hybridity alone can enhance parent-of-origin effects over seven-fold in F1 hybrids generated from inter-ploidy crosses, indicating that genetic hybridity effects can be genetically enhanced by parental genome dosage effects. We conclude that the interaction between the effects of hybridity and parental genome dosage need to be considered for more complete understanding of the mechanisms underpinning heterosis effects.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
CS and RC-B planned and designed the research. RC-B and AF performed the experiments. RC-B and RC analysed the data. CS, RC-B, PM, and GB drafted the manuscript, which was reviewed and revised by all authors.
Funding
This work was supported by a Science Foundation Ireland Principal Investigator Grant (13/IA/1820) to CS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.835219/full#supplementary-material
Supplementary Figure 1 | Illustration of the crossing scheme used and schematic diagram of the ploidy level of the different F1 seed compartments from inter-ploidy (unbalanced) crosses. Chromosomes of the tetraploid Ler-0 tester line (genotype LLLL) are represented in black, while those of Copac-1, DraIV 6-13, Lago-1, IP-Vdt-0, Baa-1 and IP-Smt-1 (genotype AA) are shown in gray.
Supplementary Figure 2 | Correlation between the parent-of-origin effects of the 71 accessions on F1 hybrid seed size when crossed with the tester Line Ler-0, and the genetic distance between these accessions and the reference accession Ler-0. The red line shows the regression of parent-of-origin effects onto the genetic distance, to which the R2 value and p-value correspond.
Supplementary Figure 3 | Seed size (mm2) of paternal-excess F1 hybrid triploids using 48 genetically different maternal genotypes that are part of the accession panel used in this study. The horizontal red dashed line represents the mean seed size value for the Ler-0 F1 isogenic triploid control (0.1682 mm2). Error bars represent SD, N > 20 seeds/genotype.
Supplementary Table 1 | List of Arabidopsis thaliana natural diploid accessions used in this study and their geolocation.
Supplementary Table 2 | Calculations of Best-Parent (%BPH), Mid-Parent (%MPH) and Worst-Parent Heterosis (%WPH) for F1 hybrid diploid seed size. Values that do not follow the observed trend are highlighted in red.
Supplementary Table 3 | MPH levels of reciprocal balanced (2x X 2x) and inter-ploidy hybrid crosses (2x X 4x, 4x X 2x). 2xA (diploid accession), 2xL (2x Ler-0), 4xL (4x Ler-0). N/A, not available due to triploid block.
Supplementary Table 4 | Deviations from the mid-parent value (MPV) in F1 hybrid diploids and triploids. Data represents the mean F1 seed size. Asterisks represent significant differences between the F1 hybrids and the MPV (‘****’ and ‘*’ indicate statistical significance at p-value < 0.0001 and p-value < 0.05, respectively, determined by Student’s t-test. Ns, not significant). N/A, not available due to triploid block.
Supplementary Table 5 | MPH levels across 48 genetically different F1 hybrids at the diploid and paternal-excess triploid level. 2xA (diploid accession), 2xL (2x Ler-0), 4xL (4x Ler-0).
Supplementary Table 6 | Influence of geolocation on phenotypic variation. Pearson correlation coefficient (r) was used to determine the latitudinal and longitudinal association of the 71 genetically different accessions of this study with reciprocal F1 hybrid diploid seed size, seed size of selfed isogenic diploid parental lines and parent-of-origin effects. Correlation is significant at the 0.05 level.
Supplementary Table 7 | Broad-sense heritability of hybrid F1 seed size in terms of the parent of origin.
Abbreviations
Arabidopsis, Arabidopsis thaliana; POE, parent-of-origin effects; 2x, diploid; 4x, tetraploid; 3xm, maternal-excess F1 triploids; 3xp, paternal-excess F1 triploids; BPH, Best-Parent Heterosis levels; MPH, Mid-Parent Heterosis levels; WPH, Worst-Parent Heterosis levels; MPV, mid-parent value.
References
Abbott, R., Albach, D., Ansell, S., Arntzen, J. W., Baird, S. J., Bierne, N., et al. (2013). Hybridization and speciation. J. Evol. Biol. 26, 229–246.
Abramoff, M. D., Magalhaes, P. J., and Ram, S. J. (2004). Image processing with imageJ. Biophoton. Int. 11, 36–42.
Alcázar, R., García, A. V., Kronholm, I., De Meaux, J., Koornneef, M., Parker, J. E., et al. (2010). Natural variation at strubbelig receptor kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat. Genet. 42, 1135–1139. doi: 10.1038/ng.704
Alonso-Blanco, C., Blankestijn-De Vries, H., Hanhart, C. J., and Koornneef, M. (1999). Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Nat. Acad. Sci. 96, 4710–4717. doi: 10.1073/pnas.96.8.4710
Armenta-Medina, A., and Gillmor, C. S. (2019). “Chapter Eighteen - Genetic, molecular and parent-of-origin regulation of early embryogenesis in flowering plants,” in Current Topics in Developmental Biology, ed. U. Grossniklaus (Cambridge: Academic Press), 497–543. doi: 10.1016/bs.ctdb.2018.11.008
Baroux, C., Spillane, C., and Grossniklaus, U. (2002). Genomic imprinting during seed development. Adv. Genet. 46, 165–214. doi: 10.1016/s0065-2660(02)46007-5
Baskin, J. M., and Baskin, C. C. (2019). How much influence does the paternal parent have on seed germination? Seed Sci. Res. 29, 1–11. doi: 10.1038/sj.hdy.6800131
Batista, R. A., and Köhler, C. (2020). Genomic imprinting in plants-revisiting existing models. Genes Dev. 34, 24–36. doi: 10.1101/gad.332924.119
Bayer, M., Nawy, T., Giglione, C., Galli, M., Meinnel, T., and Lukowitz, W. (2009). Paternal control of embryonic patterning in arabidopsis thaliana. Science 323, 1485–1488. doi: 10.1126/science.1167784
Birchler, J. A., and Veitia, R. A. (2012). Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc. Nat. Acad. Sci. 109, 14746–14753. doi: 10.1073/pnas.1207726109
Birchler, J. A., Johnson, A. F., and Veitia, R. A. (2016). Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 245, 128–134. doi: 10.1016/j.plantsci.2016.02.002
Birchler, J. A., Riddle, N. C., Auger, D. L., and Veitia, R. A. (2005). Dosage balance in gene regulation: biological implications. Trends Genet. 21, 219–226. doi: 10.1016/j.tig.2005.02.010
Birchler, J. A., Yao, H., Chudalayandi, S., Vaiman, D., and Veitia, R. A. (2010). Heterosis. Plant Cell 22, 2105–2112.
Blakeslee, A. F. (1941). Effect of Induced Polyploidy in Plants. Am. Natural. 75, 117–135. doi: 10.1086/280940
Boeven, P. H. G., Zhao, Y., Thorwarth, P., Liu, F., Maurer, H. P., Gils, M., et al. (2020). Negative dominance and dominance-by-dominance epistatic effects reduce grain-yield heterosis in wide crosses in wheat. Sci. Adv. 6:eaay4897. doi: 10.1126/sciadv.aay4897
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5:e236. doi: 10.1371/journal.pbio.0050236
Botet, R., and Keurentjes, J. J. B. (2020). The role of transcriptional regulation in hybrid Vigor. Front. Plant Sci. 11:410. doi: 10.3389/fpls.2020.00410
Burke, J. M., and Arnold, M. L. (2001). Genetics and the fitness of hybrids. Annu. Rev. Genet. 35, 31–52. doi: 10.1146/annurev.genet.35.102401.085719
Calderini, D. F., Castillo, F. M., Arenas-M, A., Molero, G., Reynolds, M. P., Craze, M., et al. (2021). Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential. New Phytol. 230, 629–640. doi: 10.1111/nph.17048
Chae, E., Bomblies, K., Kim, S. T., Karelina, D., Zaidem, M., Ossowski, S., et al. (2014). Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159, 1341–1351. doi: 10.1016/j.cell.2014.10.049
Chen, C., Ea, Z., and Lin, H. X. (2016). Evolution and molecular control of hybrid incompatibility in Plants. Front. Plant Sci. 7:1208. doi: 10.3389/fpls.2016.01208
Cheng, S.-H., Cao, L.-Y., Zhuang, J.-Y., Chen, S.-G., Zhan, X.-D., Fan, Y.-Y., et al. (2007). Super hybrid rice breeding in china: achievements and prospects. J. Integ. Plant Biol. 49, 805–810.
Cong, B., Liu, J., and Tanksley, S. D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Nat. Acad. Sci. 99, 13606–13611. doi: 10.1073/pnas.172520999
de Jong, T. J., and Scott, R. J. (2007). Parental conflict does not necessarily lead to the evolution of imprinting. Trends Plant Sci. 12, 439–443. doi: 10.1016/j.tplants.2007.07.003
de Jong, T. J., Hermans, C. M., and Van Der Veen-Van Wijk, K. C. (2011). Paternal effects on seed mass in Arabidopsis thaliana. Plant Biol. 13, (Suppl. 1), 71–77. doi: 10.1111/j.1438-8677.2009.00287.x
Diggle, P. K., Abrahamson, N. J., Baker, R. L., Barnes, M. G., Koontz, T. L., Lay, C. R., et al. (2010). Dynamics of maternal and paternal effects on embryo and seed development in wild radish (Raphanus sativus). Ann. Bot. 106, 309–319. doi: 10.1093/aob/mcq110
Dimitrijevic, A., and Horn, R. (2018). Sunflower hybrid breeding: from markers to genomic selection. Front. Plant Sci. 8:2238. doi: 10.3389/fpls.2017.02238
Donoghue, M. T., Fort, A., Clifton, R., Zhang, X., Mckeown, P. C., Voigt-Zielinksi, M. L., et al. (2014). C(m)CGG methylation-independent parent-of-origin effects on genome-wide transcript levels in isogenic reciprocal F1 triploid plants. DNA Res. 21, 141–151. doi: 10.1093/dnares/dst046
Donohue, K. (2009). Completing the cycle: maternal effects as the missing link in plant life histories. Philos. Trans. R.Soc. London. Ser. B, Biol. Sci. 364, 1059–1074. doi: 10.1098/rstb.2008.0291
Doughty, J., Aljabri, M., and Scott, R. J. (2014). Flavonoids and the regulation of seed size in Arabidopsis. Biochem. Soc. Trans. 42, 364–369. doi: 10.1042/BST20140040
Durvasula, A., Fulgione, A., Gutaker, R. M., Alacakaptan, S. I., Flood, P. J., Neto, C., et al. (2017). African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Nat. Acad. Sci. 114, 5213–5218. doi: 10.1073/pnas.1616736114
Duszynska, D., Mckeown, P. C., Juenger, T. E., Pietraszewska-Bogiel, A., Geelen, D., and Spillane, C. (2013). Gamete fertility and ovule number variation in selfed reciprocal F1 hybrid triploid plants are heritable and display epigenetic parent-of-origin effects. New Phytol. 198, 71–81. doi: 10.1111/nph.12147
Duszynska, D., Vilhjalmsson, B., Castillo Bravo, R., Swamidatta, S., Juenger, T. E., Donoghue, M. T. A., et al. (2019). Transgenerational effects of inter-ploidy cross direction on reproduction and F2 seed development of Arabidopsis thaliana F1 hybrid triploids. Plant Reprod. 32, 275–289. doi: 10.1007/s00497-019-00369-6
Flood, P. J., Theeuwen, T. P. J. M., Schneeberger, K., Keizer, P., Kruijer, W., Severing, E., et al. (2020). Reciprocal cybrids reveal how organellar genomes affect plant phenotypes. Nat. Plants 6, 13–21. doi: 10.1038/s41477-019-0575-9
Fort, A., Ryder, P., Mckeown, P. C., Wijnen, C., Aarts, M. G., Sulpice, R., et al. (2016). Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol. 209, 590–599. doi: 10.1111/nph.13650
Fridman, E. (2015). Consequences of hybridization and heterozygosity on plant vigor and phenotypic stability. Plant Sci. 232, 35–40. doi: 10.1016/j.plantsci.2014.11.014
Fulgione, A., and Hancock, A. M. (2018). Archaic lineages broaden our view on the history of Arabidopsis thaliana. New Phytol. 219, 1194–1198. doi: 10.1111/nph.15244
Gòdia, M., Swanson, G., and Krawetz, S. A. (2018). A history of why fathers’ RNA matters. Biol. Reprod. 99, 147–159. doi: 10.1093/biolre/ioy007
Golan, G., Ayalon, I., Perry, A., Zimran, G., Ade-Ajayi, T., Mosquna, A., et al. (2019). GNI-A1 mediates trade-off between grain number and grain weight in tetraploid wheat. Theor. Appl. Genet. 132, 2353–2365. doi: 10.1007/s00122-019-03358-5
Goulet, B. E., Roda, F., and Hopkins, R. (2017). Hybridization in Plants: Old Ideas. New Techn. Plant Physiol. 173, 65–78. doi: 10.1104/pp.16.01340
Greiner, S., Sobanski, J., and Bock, R. (2015). Why are most organelle genomes transmitted maternally? BioEssays 37, 80–94. doi: 10.1002/bies.201400110
Griffiths, S., Wingen, L., Pietragalla, J., Garcia, G., Hasan, A., Miralles, D., et al. (2015). Genetic Dissection of Grain Size and Grain Number Trade-Offs in CIMMYT Wheat Germplasm. PLoS One 10:e0118847. doi: 10.1371/journal.pone.0118847
Grossniklaus, U., Spillane, C., Page, D. R., and Köhler, C. (2001). Genomic imprinting and seed development: endosperm formation with and without sex. Curr. Opin. Plant Biol. 4, 21–27. doi: 10.1016/s1369-5266(00)00130-8
Groszmann, M., Gonzalez-Bayon, R., Greaves, I. K., Wang, L., Huen, A. K., Peacock, W. J., et al. (2014). Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 166, 265–280. doi: 10.1104/pp.114.243998
House, C., Roth, C., Hunt, J., and Kover, P. X. (2010). Paternal effects in Arabidopsis indicate that offspring can influence their own size. Proc. Biol. Sci. 277, 2885–2893. doi: 10.1098/rspb.2010.0572
Kesavan, M., Song, J. T., and Seo, H. S. (2013). Seed size: a priority trait in cereal crops. Physiol. Plant 147, 113–120. doi: 10.1111/j.1399-3054.2012.01664.x
Kirkbride, R. C., Lu, J., Zhang, C., Mosher, R. A., Baulcombe, D. C., and Chen, Z. J. (2019). Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Nat. Acad. Sci. 116, 2761–2766. doi: 10.1073/pnas.1807621116
Klosin, A., Casas, E., Hidalgo-Carcedo, C., Vavouri, T., and Lehner, B. (2017). Transgenerational transmission of environmental information in C. elegans. Science 356, 320–323. doi: 10.1126/science.aah6412
Köhler, C., Dziasek, K., and Del Toro-De León, G. (2021). Postzygotic reproductive isolation established in the endosperm: mechanisms, drivers and relevance. Philos. Trans. R Soc. B: Biol. Sci. 376:20200118. doi: 10.1098/rstb.2020.0118
Köhler, C., Mittelsten Scheid, O., and Erilova, A. (2010). The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 26, 142–148. doi: 10.1016/j.tig.2009.12.006
Krieger, U., Lippman, Z. B., and Zamir, D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. doi: 10.1038/ng.550
Kumar, N., Tikka, S., Ram, B., and Dagla, M. C. (2015). Heterosis studies for agronomic trait under different environmental conditions in sesame (Sesamum indicum L.). Elect. J. Plant Breed. 6, 130–140.
Labroo, M. R., Studer, A. J., and Rutkoski, J. E. (2021). Heterosis and Hybrid Crop Breeding: a multidisciplinary review. Front. Genet. 12:643761. doi: 10.3389/fgene.2021.643761
Lafon Placette, C. (2020). Endosperm genome dosage, hybrid seed failure, and parental imprinting: sexual selection as an alternative to parental conflict. Am. J. Bot. 107, 17–19. doi: 10.1002/ajb2.1401
Lafon-Placette, C., and Köhler, C. (2014). Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 17, 64–69. doi: 10.1016/j.pbi.2013.11.008
Lafon-Placette, C., and Köhler, C. (2016). Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol. Ecol. 25, 2620–2629. doi: 10.1111/mec.13552
Lawson, H. A., Cheverud, J. M., and Wolf, J. B. (2013). Genomic imprinting and parent-of-origin effects on complex traits. Nat. Rev. Genet. 14, 609–617. doi: 10.1038/nrg3543
Lee, C. R., Svardal, H., Farlow, A., Exposito-Alonso, M., Ding, W., Novikova, P., et al. (2017). On the post-glacial spread of human commensal Arabidopsis thaliana. Nat. Commun. 8:14458. doi: 10.1038/ncomms14458
Li, N., and Li, Y. (2015). Maternal control of seed size in plants. J. Exp. Bot. 66, 1087–1097. doi: 10.1093/jxb/eru549
Li, N., Xu, R., and Li, Y. (2019). Molecular networks of seed size control in plants. Ann. Rev. Plant Biol. 70, 435–463. doi: 10.1146/annurev-arplant-050718-095851
Liu, J., Cong, B., and Tanksley, S. D. (2003). Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiol. 132, 292–299. doi: 10.1104/pp.102.018143
Liu, J., Li, M., Zhang, Q., Wei, X., and Huang, X. (2020). Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 62, 287–298. doi: 10.1111/jipb.12804
Long, Q., Rabanal, F. A., Meng, D., Huber, C. D., Farlow, A., Platzer, A., et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45, 884–890. doi: 10.1038/ng.2678
Ma, G.-H., and Yuan, L.-P. (2015). Hybrid rice achievements, development and prospect in China. J. Integ. Agricult. 14, 197–205. doi: 10.16288/j.yczz.18-213
Macartney, E. L., Crean, A. J., and Bonduriansky, R. (2018). Epigenetic paternal effects as costly, condition-dependent traits. Heredity 121, 248–256. doi: 10.1038/s41437-018-0096-8
Mackay, I. J., Cockram, J., Howell, P., and Powell, W. (2021). Understanding the classics: the unifying concepts of transgressive segregation, inbreeding depression and heterosis and their central relevance for crop breeding. Plant Biotechnol. J. 19, 26–34. doi: 10.1111/pbi.13481
Makino, A., Kaneta, Y., Obara, M., Ishiyama, K., Kanno, K., Kondo, E., et al. (2020). High yielding ability of a large-grain rice cultivar, Akita 63. Sci. Rep. 10:12231. doi: 10.1038/s41598-020-69289-0
Martin, A. R. (2021). Crops and the seed mass–seed output trade-off in plants. Int. J. Plant Sci. 182, 84–90.
McGrath, M., and Panella, L. (2018). Sugar Beet Breeding in Plant Breeding Reviews. Hoboken: Wiley, 167–218.
McKeown, P. C., Fort, A., and Spillane, C. (2013b). “Genomic Imprinting: Parental Control of Gene Expression in Higher Plants,” in Polyploid and Hybrid Genomics, eds Z. J Chen and J. A. Birchler Hobeken: Wiley. 257–270. doi: 10.1002/9781118552872.ch16
McKeown, P. C., Fort, A., Duszynska, D., Sulpice, R., and Spillane, C. (2013a). Emerging molecular mechanisms for biotechnological harnessing of heterosis in crops. Trends Biotechnol. 31, 549–551. doi: 10.1016/j.tibtech.2013.06.008
Miyaji, N., and Fujimoto, R. (2018). “Chapter Eight - Hybrid Vigor: Importance of Epigenetic Processes and Consequences for Breeding,” in Advances in Botanical Research, eds M. Mirouze, E. Bucher, and P. Gallusci (Cambridge: Academic Press), 247–275.
Nieto Feliner, G., Álvarez, I., Fuertes-Aguilar, J., Heuertz, M., Marques, I., Moharrek, F., et al. (2017). Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity 118, 513–516. doi: 10.1038/hdy.2017.7
Oakley, C. G., Ågren, J., and Schemske, D. W. (2015). Heterosis and outbreeding depression in crosses between natural populations of Arabidopsis thaliana. Heredity 115, 73–82. doi: 10.1038/hdy.2015.18
Picard, C. L., Povilus, R. A., Williams, B. P., and Gehring, M. (2021). Transcriptional and imprinting complexity in Arabidopsis seeds at single-nucleus resolution. Nat. Plants 7, 730–738. doi: 10.1038/s41477-021-00922-0
Pignatta, D., Erdmann, R. M., Scheer, E., Picard, C. L., Bell, G. W., and Gehring, M. (2014). Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 3:e03198.
Pires, N. D., Bemer, M., Müller, L. M., Baroux, C., Spillane, C., and Grossniklaus, U. (2016). Quantitative genetics identifies cryptic genetic variation involved in the paternal regulation of seed development. PLoS Genet. 12:e1005806. doi: 10.1371/journal.pgen.1005806
Ren, D., Wang, X., Yang, M., Yang, L., He, G., and Deng, X. W. (2019). A new regulator of seed size control in Arabidopsis identified by a genome-wide association study. New Phytol. 222, 895–906. doi: 10.1111/nph.15642
Rieseberg, L. H., Archer, M. A., and Wayne, R. K. (1999). Transgressive segregation, adaptation and speciation. Heredity 83, 363–372. doi: 10.1038/sj.hdy.6886170
Robert, H. S., Park, C., Gutièrrez, C. L., Wójcikowska, B., Pěnčík, A., Novák, O., et al. (2018). Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4, 548–553. doi: 10.1038/s41477-018-0204-z
Ryder, P., Mckeown, P. C., Fort, A., and Spillane, C. (2014). “Epigenetics and Heterosis in Crop Plants,” in Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications: Transcriptional Regulation and Chromatin Remodelling in Plants, eds R. Alvarez-Venegas, C. De-La-Peña, and J. A. Casas-Mollano (Cham: Springer International Publishing), 129–147. doi: 10.1007/978-3-030-14760-0_4
Schnable, P. S., and Springer, N. M. (2013). Progress Toward Understanding Heterosis in Crop Plants. Ann. Rev. Plant Biol. 64, 71–88. doi: 10.1146/annurev-arplant-042110-103827
Scott, R. J., Spielman, M., Bailey, J., and Dickinson, H. G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341.
Seymour, D. K., Chae, E., Grimm, D. G., Martín Pizarro, C., Habring-Müller, A., Vasseur, F., et al. (2016). Genetic architecture of nonadditive inheritance in Arabidopsis thaliana hybrids. Proc. Nat. Acad. Sci. 113, E7317–E7326. doi: 10.1073/pnas.1615268113
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Singh, J., Clavijo Michelangeli, J. A., Gezan, S. A., Lee, H., and Vallejos, C. E. (2017). Maternal effects on seed and seedling phenotypes in reciprocal f1 hybrids of the common Bean (Phaseolus vulgaris L.). Front. Plant Sci. 8:42. doi: 10.3389/fpls.2017.00042
Skinner, M. K., Guerrero-Bosagna, C., and Haque, M. M. (2015). Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 10, 762–771. doi: 10.1080/15592294.2015.1062207
Spillane, C., Vielle-Calzada, J., and Grossniklaus, U. (2002). “Parent-of-origin effects and seed development: Genetics and epigenetics,” in Transgenic Plants and Crops, eds G. C. Y. Khachatourians, H. Hui, R. Scorza, W.-K. Nip, and Y. H. Hui Boca Raton: CRC Press. 109–135.
Städler, T., Florez-Rueda, A. M., and Roth, M. (2021). A revival of effective ploidy: the asymmetry of parental roles in endosperm-based hybridization barriers. Curr. Opin. Plant Biol. 61:102015. doi: 10.1016/j.pbi.2021.102015
Stokes, D., Morgan, C., O’neill, C., and Bancroft, I. (2007). Evaluating the utility of Arabidopsis thaliana as a model for understanding heterosis in hybrid crops. Euphytica 156, 157–171. doi: 10.1007/s10681-007-9362-1
Tabib, A., Vishwanathan, S., Seleznev, A., Mckeown, P. C., Downing, T., Dent, C., et al. (2016). A Polynucleotide repeat expansion causing temperature-sensitivity persists in wild irish accessions of arabidopsis thaliana. Front. Plant Sci. 7:1311. doi: 10.3389/fpls.2016.01311
Toledo, B., Marcer, A., Méndez-Vigo, B., Alonso-Blanco, C., and Picó, F. X. (2020). An ecological history of the relict genetic lineage of Arabidopsis thaliana. Environ. Exp. Bot. 170:103800. doi: 10.1016/j.envexpbot.2019.103800
Trier, C. N., Hermansen, J. S., Sætre, G.-P., and Bailey, R. I. (2014). Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid italian sparrow and its parent species. PLoS Genet. 10:e1004075. doi: 10.1371/journal.pgen.1004075
Tuteja, R., Mckeown, P. C., Ryan, P., Morgan, C. C., Donoghue, M. T. A., Downing, T., et al. (2019). Paternally expressed imprinted genes under positive darwinian selection in arabidopsis thaliana. Mol. Biol. Evol. 36, 1239–1253. doi: 10.1093/molbev/msz063
Vallejo-Marín, M., Cooley, A. M., Lee, M. Y., Folmer, M., Mckain, M. R., and Puzey, J. R. (2016). Strongly asymmetric hybridization barriers shape the origin of a new polyploid species and its hybrid ancestor. Am. J. Bot. 103, 1272–1288. doi: 10.3732/ajb.1500471
van Hulten, M. H. A., Paulo, M.-J., Kruijer, W., Blankestijn-De Vries, H., Kemperman, B., Becker, F. F. M., et al. (2018). Assessment of heterosis in two Arabidopsis thaliana common-reference mapping populations. PLoS One 13:e0205564. doi: 10.1371/journal.pone.0205564
Vasseur, F., Fouqueau, L., De Vienne, D., Nidelet, T., Violle, C., and Weigel, D. (2019). Nonlinear phenotypic variation uncovers the emergence of heterosis in Arabidopsis thaliana. PLoS Biol. 17:e3000214. doi: 10.1371/journal.pbio.3000214
Veitia, R. A., and Birchler, J. A. (2015). Models of buffering of dosage imbalances in protein complexes. Biol. Direct 10:42. doi: 10.1186/s13062-015-0063-8
Vielle-Calzada, J.-P., Grossniklaus, U., and Spillane, C. (2002). “Parent-of-Origin Effects and Seed Development: Genetics and Epigenetics,” in Transgenic plants and crops,, eds G. C. Y. Khachatourians, H. Hui, R. Scorza, W.-K. Nip, and Y. H. Hui Boca Raton: CRC Press. 109–136.
Wang, L., Yamashita, M., Greaves, I. K., Peacock, W. J., and Dennis, E. S. (2020). Arabidopsis Col/Ler and Ws/Ler hybrids and Hybrid Mimics produce seed yield heterosis through increased height, inflorescence branch and silique number. Planta 252:40. doi: 10.1007/s00425-020-03444-9
Weigel, D. (2012). Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158, 2–22. doi: 10.1104/pp.111.189845
Wolff, P., Jiang, H., Wang, G., Santos-González, J., and Köhler, C. (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. ELife 4:e10074. doi: 10.7554/eLife.10074
Wolko, J., Dobrzycka, A., Bocianowski, J., and Bartkowiak-Broda, I. (2019). Estimation of heterosis for yield-related traits for single cross and three-way cross hybrids of oilseed rape (Brassica napus L.). Euphytica 215:56.
Yuan, S., Schuster, A., Tang, C., Yu, T., Ortogero, N., Bao, J., et al. (2016). Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143, 635–647. doi: 10.1242/dev.131755
Zhao, Y., Wang, S., Wu, W., Li, L., Jiang, T., and Zheng, B. (2018). Clearance of maternal barriers by paternal miR159 to initiate endosperm nuclear division in Arabidopsis. Nat. Commun. 9:5011. doi: 10.1038/s41467-018-07429-x
Keywords: Parent-of-origin effects, seed size, hybridity, heterosis, parental genome dosage, ploidy, paternal contribution, Arabidopsis
Citation: Castillo-Bravo R, Fort A, Cashell R, Brychkova G, McKeown PC and Spillane C (2022) Parent-of-Origin Effects on Seed Size Modify Heterosis Responses in Arabidopsis thaliana. Front. Plant Sci. 13:835219. doi: 10.3389/fpls.2022.835219
Received: 14 December 2021; Accepted: 19 January 2022;
Published: 07 March 2022.
Edited by:
Koen Geuten, KU Leuven, BelgiumReviewed by:
James A. Birchler, University of Missouri, United StatesTakashi Tsuchimatsu, The University of Tokyo, Japan
Copyright © 2022 Castillo-Bravo, Fort, Cashell, Brychkova, McKeown and Spillane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles Spillane, Y2hhcmxlcy5zcGlsbGFuZUBudWlnYWx3YXkuaWU=
†ORCID: Rosa Castillo-Bravo, orcid.org/0000-0002-0002-6097; Antoine Fort, orcid.org/0000-0002-2210-7234; Ronan Cashell, orcid.org/0000-0003-1003-1292; Galina Brychkova, orcid.org/0000-0003-2452-7635; Peter C. McKeown, orcid.org/0000-0002-7255-6062; Charles Spillane, orcid.org/0000-0003-3318-323X
 Rosa Castillo-Bravo
Rosa Castillo-Bravo Antoine Fort†
Antoine Fort† Peter C. McKeown
Peter C. McKeown Charles Spillane
Charles Spillane