
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 July 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.833326
This article is part of the Research Topic Abiotic Stress Signaling in Plants: Functional Genomic Intervention, Volume II View all 18 articles
 Zhenzhen Han1,2,3,4,5†
Zhenzhen Han1,2,3,4,5† Jinyu Wang1,2,3,4,5†
Jinyu Wang1,2,3,4,5† Xinxin Wang1,2,3,4,5
Xinxin Wang1,2,3,4,5 Xijia Zhang1,2,3,4,5
Xijia Zhang1,2,3,4,5 Yanbo Cheng1,2,3,4,5
Yanbo Cheng1,2,3,4,5 Zhandong Cai1,2,3,4,5
Zhandong Cai1,2,3,4,5 Hai Nian1,2,3,4,5*
Hai Nian1,2,3,4,5* Qibin Ma1,2,3,4,5*
Qibin Ma1,2,3,4,5*The WRKY transcription factors (TFs) are one of the largest families of TFs in plants and play multiple roles in plant growth and development and stress response. In this study, GmWRKY21 encoding a WRKY transcription factor was functionally characterized in Arabidopsis and soybean. The GmWRKY21 protein containing a highly conserved WRKY domain and a C2H2 zinc-finger structure is located in the nucleus and has the characteristics of transcriptional activation ability. The GmWRKY21 gene presented a constitutive expression pattern rich in the roots, leaves, and flowers of soybean with over 6-fold of relative expression levels and could be substantially induced by aluminum stress. As compared to the control, overexpression of GmWRKY21 in Arabidopsis increased the root growth of seedlings in transgenic lines under the AlCl3 concentrations of 25, 50, and 100 μM with higher proline and lower MDA accumulation. The results of quantitative real-time polymerase chain reaction (qRT-PCR) showed that the marker genes relative to aluminum stress including ALMT, ALS3, MATE, and STOP1 were induced in GmWRKY21 transgenic plants under AlCl3 treatment. The stress-related genes, such as KIN1, COR15A, COR15B, COR47, GLOS3, and RD29A, were also upregulated in GmWRKY21 transgenic Arabidopsis under aluminum stress. Similarly, stress-related genes, such as GmCOR47, GmDREB2A, GmMYB84, GmKIN1, GmGST1, and GmLEA, were upregulated in hair roots of GmWRKY21 transgenic plants. In summary, these results suggested that the GmWRKY21 transcription factor may promote the tolerance to aluminum stress mediated by the pathways regulating the expression of the acidic aluminum stress-responsive genes and abiotic stress-responsive genes.
Aluminum (Al) mainly existing in the forms of harmless oxides and aluminosilicates is the third most abundant metal in the earth's crust next to oxygen and silicon (Kochian et al., 2004). Acidic soils are widespread with ~30% of the world's land area and about 50% of the potential of the world's potentially arable lands (Silambarasan et al., 2019). In acidic soil (pH <5), Al3+ ions are activated and dissolved into the soil solution from clay minerals, which can quickly inhibit root growth resulting in reduced crop yield (Kochian et al., 2015). Therefore, Al toxicity is the main factor restricting plant growth in acidic soil (Bojorquez-Quintal et al., 2017). The solubility of aluminum increases with the decrease in pH value (Brautigan et al., 2012). In an acidic solution, Al3+ exists as the octahedral hexahydrate commonly known as Al3+ (Kochian et al., 2015), while the Al toxicity in acidic soil is mainly caused by Al3+(Singh et al., 2017). The most apparent feature of Al toxicity is to inhibit the root tip cell elongation and cell division. It was generally believed that the root tip is the central part of Al toxicity (Kochian, 1995). The transition zone (DTZ) of the root tip 1–2 mm between the meristematic zone and the elongation zone is the most sensitive area for aluminum poisoning (Kollmeier et al., 2000). Al3+ is electrostatically bound to the pectin of the negatively charged root tip cell wall (Kochian, 1995), which reduces the rate of cell division and extension and affects root elongation.
Previous studies showed that the higher concentration of Al3+ will cause severe toxicity to plants because it will inhibit cell elongation and cell division in the roots, which will lead to enlarged root tips, reduced root hairs, or no root hair development, thus hindering the absorption of nutrients and water (Kochian et al., 2015). Al3+ toxicity increases the formation of reactive oxygen species (ROS) in plants by leading to lipid membrane peroxidation, ion leakage, protein oxidation, callose accumulation, and cell death (Ezaki et al., 2000; Yamamoto et al., 2002; Boscolo et al., 2003).
In adaptation to Al stress, plants have evolved two resistance mechanisms: external and internal tolerance mechanisms (Liu et al., 2014; Kochian et al., 2015; Wang N. et al., 2017). Several main components exist in the external mechanism including the cell wall of Al3+ fixation, induced pH increase, organic acid (OA) anions chelate Al3+ ions, and Al3+ transmembrane outflow, while some main components are involved in the internal tolerance mechanism including plants chelate aluminum ions through organic acids and proteins in cells and transporting aluminum ions from sensitive cytoplasmic areas to non-sensitive areas, such as vacuoles (Feng, 2000; Ryan et al., 2001; Kochian et al., 2015; Silambarasan et al., 2019). Aluminum-activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) identified in plants are two transporter families that confer Al tolerance through the secretion of the organic acids. TaALMT1 was the first identified malate efflux gene in wheat (Sasaki et al., 2004). Another organic acid efflux transporter family MATEs as a plasma membrane efflux transporter are responsible for the Al-activated citrate release (Liu et al., 2009, 2012).
Transcription factors are proteins that regulate gene transcription to adapt to the environmental stimulus (Finatto et al., 2018). WRKY proteins have one or two conserved WRKY domains of about 60 amino acids with an invariable sequence motif WRKYGQK at the N-terminus and a zinc-finger domain-like (CX4−5CX22−23HXH or CX7CX23HXC) (Eulgem et al., 2000; Rushton et al., 2010). The WRKY TFs can be classified into three main groups (I–III) based on the number of WRKY domains and the type of their zinc-finger motifs. The WRKY TFs in group I have two WRKY domains and a C2H2-type zinc-finger structure. The TFs in group II have a single WRKY domain and contain the zinc-finger motif C2H2, which is divided into five subgroups (IIa–e). The TFs in group III hold a WRKY domain and contain a C2-HC motif (Eulgem et al., 2000; Zhang and Wang, 2005; Song et al., 2018; Xie et al., 2018).
WRKY is a superfamily of transcription factors in plants that play an essential role in many life processes, especially in biotic and abiotic stresses (Eulgem et al., 2000; Zhang and Wang, 2005; Rushton et al., 2010; Chen et al., 2012; Li et al., 2013). SPF1 was the first WRKY gene isolated from sweet potato in 1994 (Ishiguro and Nakamura, 1994), and then, the ABF1 and ABF2 genes were discovered in wild oats (Rushton et al., 1995). The genes of WRKY1, WRKY2, and WRKY3 from parsley were confirmed for the first time that WRKY protein could regulate the response of plants to pathogens (Rushton et al., 1996).
In Arabidopsis, overexpression of AtWRKY25 and AtWRKY33 increased salt tolerance in transgenic plants (Jiang and Deyholos, 2009). Overexpression of AtWRKY57, ABO3, AtWRKY70, AtWRKY46, and AtWRKY54 in Arabidopsis conferred the tolerance of transgenic plants to drought stress (Ren et al., 2010; Jiang et al., 2012; Chen et al., 2017). Overexpression of GhWRKY34 enhanced the tolerance of transgenic Arabidopsis to salt stress by selectively taking up Na+ and K+ (Zhou et al., 2015). GhWRKY1-like encodes a WRKY TF-mediated drought tolerance in Arabidopsis via modulating ABA biosynthesis (Hu et al., 2021). Besides these, AtWRKY3 and AtWRKY4 increased salt stress tolerance in transgenic Arabidopsis (Li C. et al., 2021). In rice, overexpression of OsWRKY11 controlled by the HSP101 promoter resulted in the enhancement of drought resistance, which was characterized by slower leaf wilting and a higher survival rate (Wu et al., 2009). OsWRKY30 in transgenic rice enhanced the tolerance to drought stress (Shen et al., 2012). Overexpression of OsWRKY45 and OsWRKY72 can change the drought resistance of transgenic Arabidopsis, which may be related to the induction of genes related to abscisic acid stress (Qiu and Yu, 2009; Song et al., 2010). Recent studies have shown that OsWRKY108 was an integrative regulator of P homeostasis and leaf inclination (Wang et al., 2021). The inactivated OsWRKY5 enhanced drought tolerance through abscisic acid signaling pathways (Lim et al., 2021). At 4°C, overexpression of OsWRKY76 improved the tolerance to low-temperature stress (Babitha et al., 2013). OsWRKY50 enhanced salt stress tolerance via an ABA-independent pathway (Huang et al., 2021). In wheat, TaWRKY2 overexpression improved salt and drought tolerance by reducing the expression of STZ and RD29B (Niu et al., 2012). TaWRKY146 enhanced drought tolerance by inducing stomatal closure in Arabidopsis (Ma et al., 2017). TaWRKY1 and TaWRKY33 made transgenic Arabidopsis resistant to drought and high-temperature stress (He et al., 2016). The heterologous transgenic plants of the TaWRKY19 gene had salt and drought tolerance and showed tolerance to low temperatures (Niu et al., 2012). TaWRKY74 participated in copper tolerance through regulation of TaGST1 expression and GSH content (Li G. Z. et al., 2021). Ectopic expression of TaWRKY75-A improved drought and salt resistance in transgenic Arabidopsis (Ye et al., 2021). Among the WRKY genes in soybean, overexpression of GmWRKY21 enhanced the tolerance to cold stress in Arabidopsis (Zhou et al., 2008). GmWRKY54 and GmWRKY49 enhanced the tolerance to salt and drought stress in transgenic Arabidopsis (Zhou et al., 2008; Xu et al., 2018), whereas GmWRKY16 conferred salt and drought tolerance (Ma et al., 2019). Overexpression of GmWRKY12 increased the tolerance of transgenic soybean seedlings to drought and salt stress by proline accumulation and malondialdehyde decrease. GmWRKY46 overexpression showed that it was involved in hairy root development and subsequently affected plant growth and Pi uptake (Li C. et al., 2021). It was found that the research progresses on the functions of WRKY family genes in soybean mostly focused on the abiotic stress response. However, there are few reports on the functions of WRKY family genes in acidic aluminum stress in soybean. In this study, GmWRKY21, an abiotic stress-responsive gene induced by Al stress, was investigated for its functional characterization of Al stress tolerance both in Arabidopsis and in soybean. The systems of Arabidopsis genetic transformation and soybean hairy root transformation were used to study the resistance and molecular mechanism of GmWRKY21 responding to acidic aluminum stress.
A soybean cultivar, “Huaxia 3” (HX3), bred by the Guangdong Subcenter of the National Center for Soybean Improvement in South China Agricultural University, was used to investigate the tissue expression pattern of GmWRKY21 and its responses to Al stress. Soybean seeds of HX3 were germinated in vermiculite with room temperature set at 28/26°C and the light time set as 14-h light/10-h dark under a light intensity of 100 μmol/(m2·s) (Zeng et al., 2012). For the Al dose–response experiment, seedlings were subjected to a modified 0.5 mM CaCl2 solution containing 0, 25, 50, 75, or 100 μM of AlCl3 for 24 h. During the time-course experiment, seedlings were subjected to a modified 0.5 mM CaCl2 solution with 50 μM AlCl3 for 0, 6, 9, 12, 24, 36, or 48 h.
The cDNA sequences of GmWRKY21 gene were isolated using specific primers (Supplementary Table 1) and inserted into the multicloning sites of the pLB vector to form the GmWRKY21-pLB construct. Then, the full-length coding sequence of GmWRKY21 amplified from the GmWRKY21-pLB vector was inserted into the SacI and XbaI sites of a pTF101.1 vector. The non-conserved region sequence 200 bp of GmWRKY21 amplified by PCR using the specific primers was inserted into the AvrII and AscI sites, and then, the 200-bp sequence was reversely inserted into the SacI and SpeI sites of a pMU103 vector using the ClonExpressR II One Step Cloning Kit (C112, Vazyme, Nanjing, China). The GmWRKY21-pTF101 and GmWRKY21-pMU103 plasmids were then transformed into Agrobacterium strains, GV3101 and K599 competent cells by heat shock, respectively. The subsequent transformations were carried out by the methods of the Agrobacterium-mediated floral dip (Clough and Bent, 2010) and Agrobacterium rhizogenes-based transformation (Matthews and Youssef, 2016).
The sequence information of the GmWRKY21 gene was obtained from the database of the National Center for Biotechnology Information (NCBI) with the accession number NP_001237327.2 (Wang et al., 2019). The full-length sequence of GmWRKY21 was amplified by RT-PCR using specific primers (Supplementary Table 1) and Super-Fidelity DNA polymerase (Phanta Max, Vazyme Biotech Co., Ltd., Nanjing, China). The seedlings of soybean cultivar HX3 under the treatment of 50 μM AlCl3 (1 mM CaCl2, pH 4.5) were used to extract total RNA by TRIzol reagent (Invitrogen). The methods of generated cDNA, RT-PCR, and agarose gel electrophoresis were described in detail previously (Lu et al., 2015). The purified PCR product was then inserted into the multiple cloning sites of the pLB vector (Tiangen Rapid DNA Ligation Kit, Beijing, China). The positive clones in Escherichia coli were used to obtain the full cDNA sequence of GmWRKY21 identified by PCR, enzyme digestion, and sequencing (Sangon Biotech Co., Ltd., Shanghai, China) (Lu et al., 2015; Ma et al., 2018).
The nucleotide sequence of GmWRKY21 and the amino acid sequence of GmWRKY21 protein were used to search its homologous genes and proteins using the database of NCBI. The alignment of nucleotide sequences was performed with the software of DNAMAN. The structure prediction of the GmWRKY21 protein was carried out using the NCBI blast results. The phylogenetic analysis was enforced by using the software of MEGA X (Kumar et al., 2016). The alignment was adjusted manually, while the unrooted phylogenetic trees were constructed by the neighbor-joining method (Sievers et al., 2011).
Localization of the ‘protein was carried out using the method described previously (Lu et al., 2015). The complete coding sequence of GmWRKY21 without a stop codon amplified by PCR using the specific primers was inserted into the NcoI and SpeI sites to generate a fusion construct of the GmWRKY21-pCAMBIA1302 vector under the control of CaMV 35S promoter. The plasmids of pCAMBIA1302 and GmWRKY21-pCAMBIA1302 were transformed into Agrobacterium tumefaciens strain GV3101 by the heat shock method. The young leaves of 4-week-old tobacco plants were contaminated by the recombinant Agrobacterium tumefaciens using the method described previously in detail (Kokkirala et al., 2010). The agro-infiltration leaves of tobacco plants cultured for 48h to 72 h were photographed with a confocal laser scanning microscope (Leica, Germany).
The ORF sequence of GmWRKY21 amplified by PCR using the specific primers was inserted into the EcoRI and BamHI sites to create a fusion construct of GmWRKY21-pGBKT7 (Supplementary Table 1). The GmWRKY21-pGBKT7 and pGBKT7 plasmids were transformed into the cells of yeast strain Y2H. Transcriptional activation of GmWRKY21 protein was analyzed according to the methods described previously (Yang et al., 2010).
Total RNA was extracted from the seedlings of soybean or Arabidopsis using the Plant Total RNA Kit (GeneMark, Taiwan, China). Reversing transcription for the first-strand cDNA synthesis was performed with 2 μg of total RNA using a PrimeScriptTM RT reagent Kit (Takara, Beijing, China). The qRT-PCR analysis was performed on a CFX96TM Real-Time System (Bio-Rad, Hercules, CA, USA) with SYBR Premix Ex Taq II (Takara, Beijing, China). The inner reference gene Actin 3 was used to normalize the data. The quantitative variations of gene expression between the examined replicates were evaluated by the 2−ΔΔCt method described previously (Willems et al., 2008; Lu et al., 2015). Three independent biological repeats were performed to ensure accurate statistical analysis. The specific primers are listed in Supplementary Table 1.
Two homozygous lines of T3 generation of GmWRKY21 transgenic plants were used for Al sensitivity assay by measuring relative root elongation according to the method described previously (Yokosho et al., 2016). After seed germination in the MS medium, 5-day-old seedlings of wild-type and GmWRKY21 transgenic plants were transferred to a solid agar medium supplied with 1 mM CaCl2 and 1% sucrose containing different concentrations of AlCl3 (0, 25, 50, and 100 μM; pH 4.5). To prepare 100 mM of AlCl3 stock solution, 2.414 g of AlCl3·6H2O was dissolved in 100 ml of distilled water and sterile-filtered. The root lengths were measured before and after 3 days of different treatments. The relative root elongation (RRE) was computed as the formula, that is, RRE = (root elongation with different treatments/root elongation without treatments) ×100 (Cai et al., 2020).
The plasmids of the empty vector (EV), GmWRKY21-pTF101 (OE), and GmWRKY21-pMU103 (RNAi) constructs were transformed into the competent cells of Agrobacterium strains K599 by heat shock. Hairy root transformation was performed according to previous reports (Kereszt et al., 2007; Ren et al., 2019). Hypocotyls of 5-day-old soybean seedlings were injected with Agrobacterium rhizogenes K599 and kept at high humidity (>90%) with plastic lids until hairy roots at the injection sites were developed to 5–10 cm long. Then, the main roots of the original seedlings were cut, and the hairy roots were immersed in water for ~5 days before the plants with hairy roots were transferred into the solutions of 0 and 50 μM AlCl3 (0.5 mM CaCl2, pH 4.3) for 24 h.
All data were presented as the mean of three biological replicates ± SEM. Student's t-test at P = 0.01 or P = 0.05 was used to identify the difference between observation values (Lu et al., 2015).
Based on recent reports of QTL mapping for the aluminum tolerance of soybean (Wang et al., 2019), an aluminum stress-induced gene encoding a transcription factor of WRKY protein was obtained from the database under the gene locus GLYMA_04G218700 and the protein accession number of NP_001237327.2. The WRKY gene designed as GmWRKY21 is located on chromosome 4 of soybean. The GmWRKY21 open reading frame (ORF) was isolated from the soybean variety Huaxia 3 based on putative sequence information available from the Phytozome database. The sequence analysis showed that the GmWRKY21 gene contained three exons and two introns, which encoded a 196-amino-acid protein with an estimated molecular mass of 22.521 KDa and an isoelectric point of 6.143 (data not shown).
The protein BLAST analysis using the complete amino acid sequences indicated that the GmWRKY21 protein had a WRKY DNA-binding domain at the location of the peptide chain between 111 aa and 168 aa. The relationship analysis between GmWRKY21 and other WRKY proteins in soybean and Arabidopsis by the phylogenetic tree showed that Glyma.06g147100 was 91.3% sequence similar to its homology of the GmWRKY21 gene in soybean (Figure 1A), while AT5G64810 with the highest homology in Arabidopsis was 40.8% sequence similar to that of the GmWRKY21 gene. The results of multiple sequence alignment indicated there were three groups for all the WRKY proteins with the largest subgroup of group II. The GmWRKY21 protein was a member of group IIc containing a typical WRKY domain and a C2H2 zinc-finger motif (Eulgem et al., 2000; Figure 1B). In group IIc, some genes had higher sequence similarity to that of GmWRKY21. Among them, GmWRKY6 held the tolerance to salt and drought stress (Zhou et al., 2008). The results suggested that GmWRKY21 protein was a WRKY TF in soybean which may play certain roles in responding to abiotic stress.
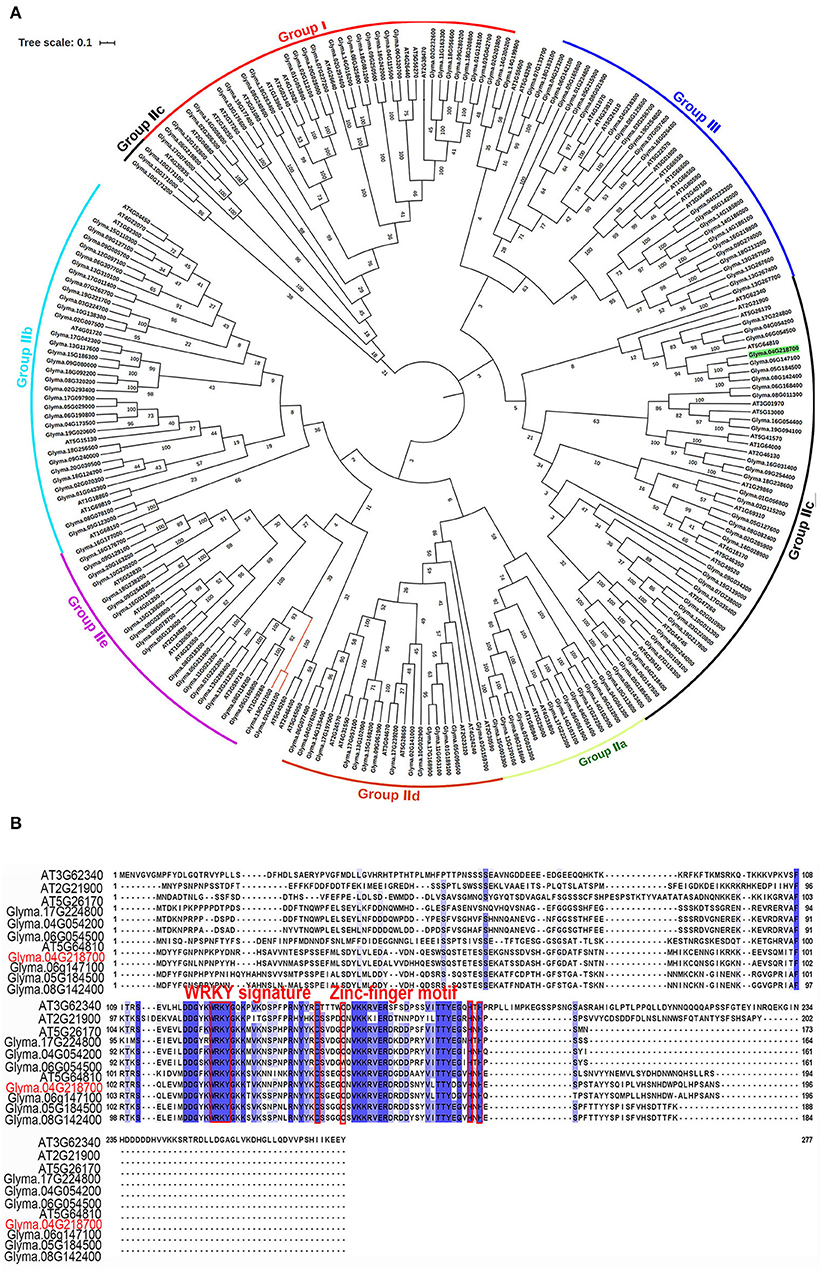
Figure 1. Bioinformatics analysis of the GmWRKY21 gene. (A) Phylogenetic tree was constructed to analyze GmWRKY21 and WRKY proteins in soybean and Arabidopsis by the neighbor-joining method using the software of MEGA X. We selected 177 and 71 WRKT family genes in soybean and Arabidopsis, respectively (Supplementary Table 1). The amino acid sequences and the accession numbers of WRKY proteins from soybean and Arabidopsis were obtained from the databases of NCBI (https://www.ncbi.nlm.nih.gov/). (B) Multiple sequence alignment of genes belonged to the same branch of GmWRKY21 from the phylogenetic tree in (A). Red box highlights the WRKY domains and the zinc-finger motifs.
To determine the transcriptional activity of GmWRKY21 protein, the complete GmWRKY21 ORF was inserted into the pGBKT7 vector. The results showed that the cell deposits transformed by the GmWRKY21-pGBKT7 construct turned blue on the SD/-Trp/X-α-Gal medium. However, the yeast cells transformed by the pGBKT7 vector did not turn blue (Figure 2A). These results indicated that the GmWRKY21 protein had transcriptional activity in yeast cells.
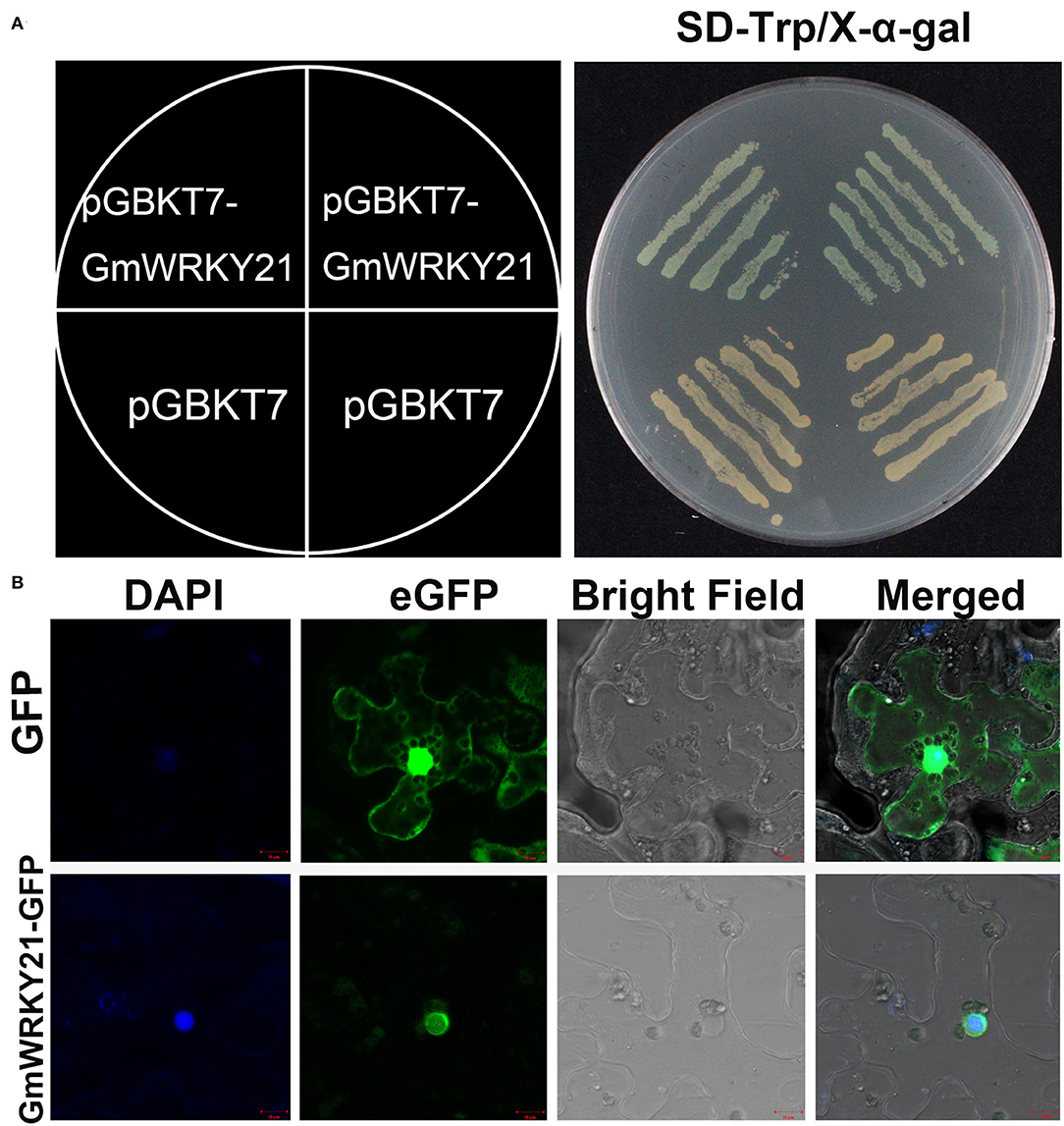
Figure 2. Transcriptional activation and subcellular localization of GmWRKY21 protein. (A) Transcriptional activation analysis of GmWRKY21 protein. (B) Subcellular localization of GmWRKY21 protein in leaf epidermal cells of tobacco. The ORF sequence of GmWRKY21 was inserted into the sites of EcoRI and BamHI of pGBKT7 vector to form the fusion carrier of GmWRKY21-pGBKT7. The fusion plasmid of GmWRKY21-pGBKT7 and pGBKT7 alone (negative control) were transformed into the cells of yeast strain Y2H. The cell deposits were colored with a chromogenic substrate of X-gal. The full coding sequence of GmWRKY21 (without TGA) was inserted into the NcoI and SpeI sites of pCAMBIA1302 vector to obtain the GmWRKY21-GFP fusion construct. The pCAMBIA1302-GFP vector was transformed as a control. Scale bar (A,B): 10 μm.
To investigate the subcellular localization of GmWRKY21 protein, the GmWRKY21-GFP (green fluorescent protein) recombinant was transformed into the cells of tobacco leaves. The results showed that the green fluorescence was observed in both cytoplasm and nucleus in the cells transformed with the pCAMBIA1302 vector, while the green fluorescence was only observed in the nucleus in the cells transformed with the GmWRKY21-pCAMBIA1302 vector (Figure 2B). The results indicated that the GmWRKY21 protein is localized in the nucleus.
The qRT-PCR was used to investigate the expression patterns of GmWRKY21. The results showed that GmWRKY21 presented a constitutive expression pattern, which was rich in soybean roots with about a 10-fold expression value, and more transcripts in leaves and flowers than those in stems or pods (Figure 3A). The GmWRKY21 gene was upregulated under the dose treatments of AlCl3 with more than 160-fold relative expression values, and the GmWRKY21 transcripts reached the maximum value under the treatment of 50 μM AlCl3 with about 350-fold relative expression value compared with that under the control treatment (Figure 3B). While under the time-course treatments of 50 μM AlCl3, the GmWRKY21 gene was promptly upregulated with over 50-fold relative expression value and reached its much higher value after the treatment of 50 μM AlCl3 for 24 h (Figure 3C). After 50 μM AlCl3 treatment for 48 h, the expression of GmWRKY21 was still at a relatively high level with over 250-fold relative expression (Figure 3C).
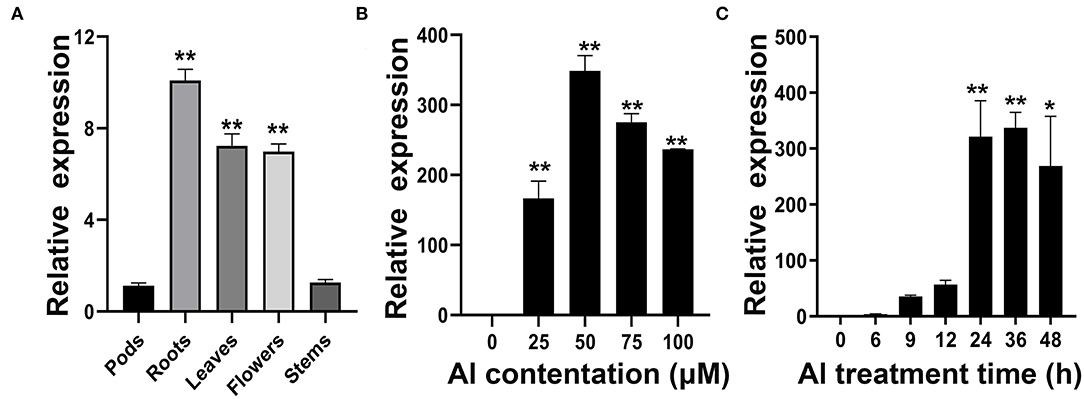
Figure 3. Analysis of GmWRKY21 expression patterns in different tissues and in response to Al stress. (A) qRT-PCR analysis of the GmWRKY21 transcript in different tissues of the soybean variety Huaxia 3. The total RNA was isolated from the samples of soybean roots, stems, leaves, flowers, and pods. (B) Dose-dependent GmWRKY21 expression pattern in the roots of soybean seedlings. The root samples were taken from the seedlings exposed to different treatments of AlCl3 concentrations (0, 25, 50, 75, and 100 μM) for 24 h. (C) The time-course GmWRKY21 expression pattern in the roots of soybean seedlings. The soybean seedlings were exposed to 50 μM AlCl3 for 0, 6, 9, 12, 24, 36, or 48 h. The root samples were separately harvested for qRT-PCR analysis. The expressing values are designed as the means ± SEM (n = 3). The experiments were performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *P < 0.05 and **P < 0.01.
To investigate GmWRKY21 response to Al stress, three homozygous lines (OE-2, OE-5, and OE-10) and wild type were used to observe the phenotypes under the treatments of AlCl3 in Arabidopsis. As shown in Figure 4A, no statistical difference was observed in root growth between WT and GmWRKY21 transgenic lines in the absence of AlCl3. However, in the presence of 25 and 50 μM AlCl3, the root elongation of WT was inhibited by 15 and 16% after 5 days, but the root elongation of GmWRKY21 transgenic lines was not significantly influenced. The root elongation of both WT and transgenic lines was inhibited with the increase in Al concentrations. At 100 μM AlCl3, the root elongation of WT was inhibited by 38% and that of the transgenic lines was inhibited by 12% (Figure 4B). The determination results showed that MDA was accumulated in both WT and GmWRKY21 transgenic lines with 0.2733 and 0.1643 μg/g fresh weight, respectively (Figure 4C). In contrast, proline contents in both wild-type plants and GmWRKY21 transgenic lines were 24 and 38–51 μg/g fresh weight, respectively, under the treatment of 100 μM AlCl3 (Figure 4D).
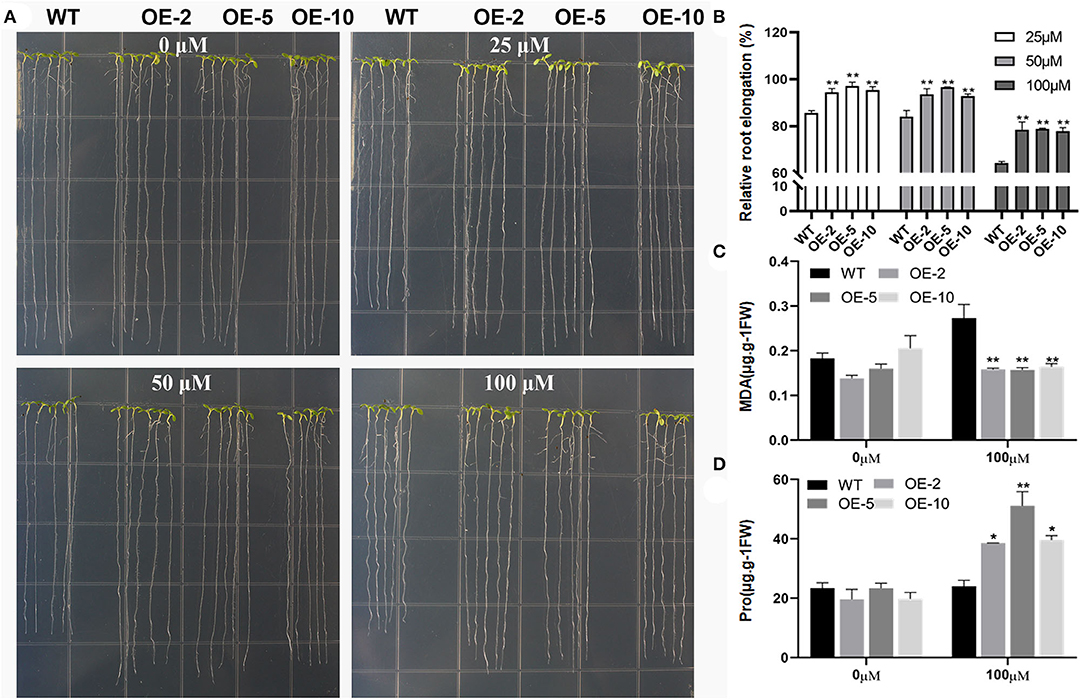
Figure 4. Overexpression of GmWRKY21 conferred enhanced Al tolerance in transgenic Arabidopsis. (A) The phenotypes of GmWRKY21 transgenic lines (OE-2, 5, and 10) and wild type (WT) under different treatments of AlCl3 solutions. Five-day-old seedlings were transferred to solid agar medium supplied with 1 mM CaCl2 and 1% sucrose containing different concentrations of AlCl3 (0, 25, 50, or 100 μM; pH 4.5) for 5 days. (B) The analysis of relative root elongation for the GmWRKY21 transgenic lines and WT. (C) The determination of MDA content. (D) The determination of free proline content. All data are presented as means ± SEM. A significant difference according to the one-way analysis of variance is denoted as follows: *P < 0.05, **P < 0.01.
To assess the effect of GmWRKY21 overexpression on Al tolerance in soybean, transgenic hairy roots were produced using the A. rhizogenes-mediated transformation system (Figure 5A). Transcript analysis revealed that GmWRKY21 was upregulated in the overexpression lines at an average fold of 16 and the expression of GmWRKY21 was downregulated in the RNAi transgenic lines at an average fold of 0.5 (Figure 5B). Hematoxylin staining analysis of root tips showed that the GmWRKY21-silenced transgenic lines accumulated more Al3+ than those of the overexpression lines. The determination in root tips showed that the Al3+ contents were 88.1, 162.4, and 120.7 μg/g in the tips of OE, RNAi, and WT plants, respectively (Figure 5C). The results indicated that overexpression of GmWRKY21 improves the tolerance of the hairy roots to Al stress by reducing Al3+ accumulation in soybean roots.
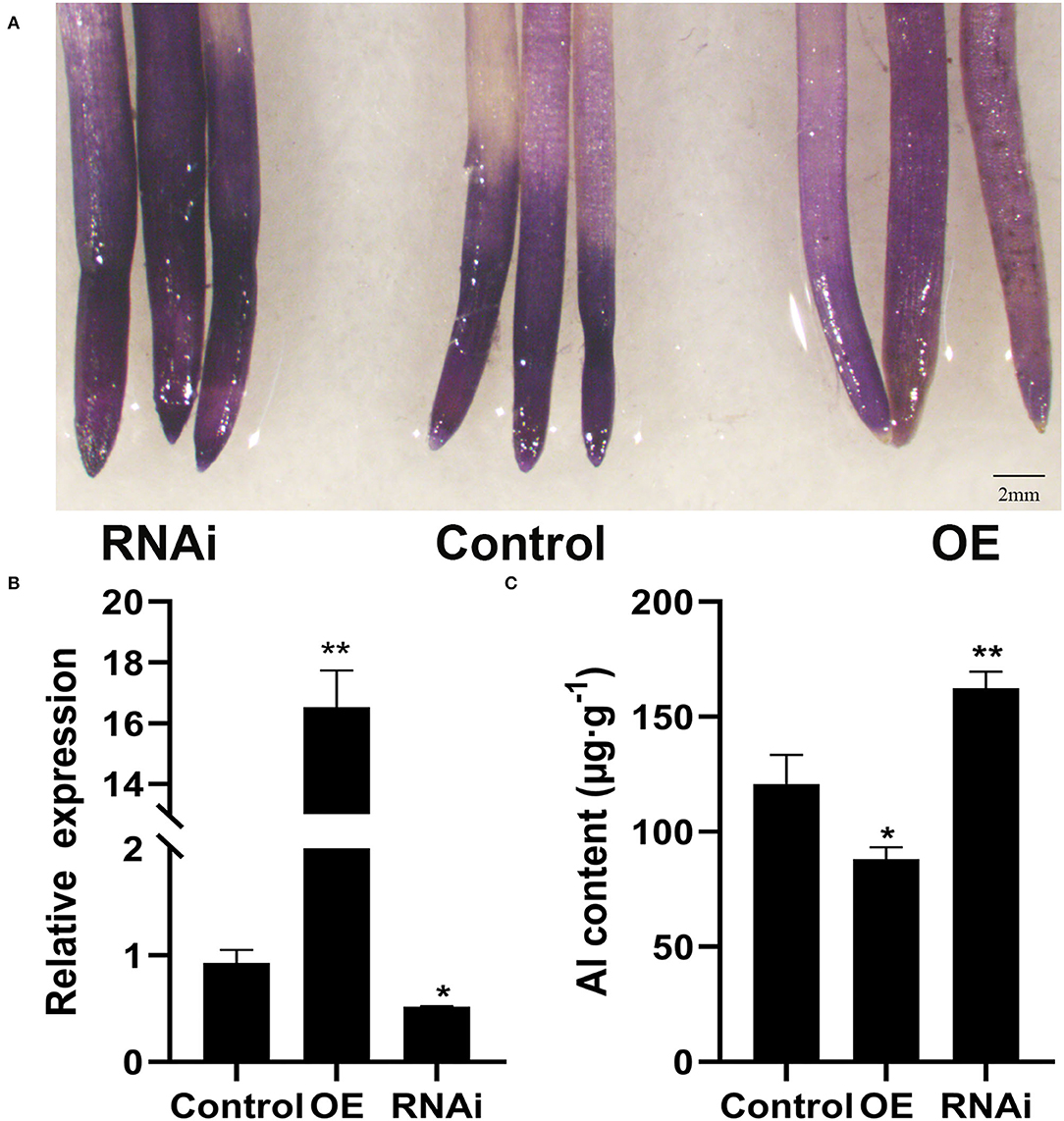
Figure 5. Hematoxylin staining in soybean hairy roots. (A) Hematoxylin staining in soybean hairy roots. (B) Detection of RNA level in soybean hairy roots. (C) Determination of Al3+ content in root tips. OE: the hairy roots of GmWRKY21-overexpressing transgenic soybean; RNAi: the hairy roots of GmWRKY21-RNAi transgenic soybean; Control: the hairy roots of Agrobacterium rhizogenes pathogenic strain K599 in soybean. The experiments were performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *P < 0.05 and **P < 0.01.
The qRT-PCR was carried out to analyze the molecular regulation mechanism of the GmWRKY21 gene tolerant to acidic aluminum. The results showed that the expression of AtALMT1, AtALS3, AtMATE, and AtSTOP1 was all upregulated under Al stress in transgenic Arabidopsis (Figure 6). The transcripts of AtALMT1 and AtALS3 induced by Al stress were much higher than those of them under the control; however, the expression of AtMATE and AtSTOP1 was not significantly higher compared with that of the control (Figure 6). The results showed that GmWRKY21 enhances the transgenic plants tolerant to Al stress mediated by the multiple genes through the pathway responding to aluminum toxicity.
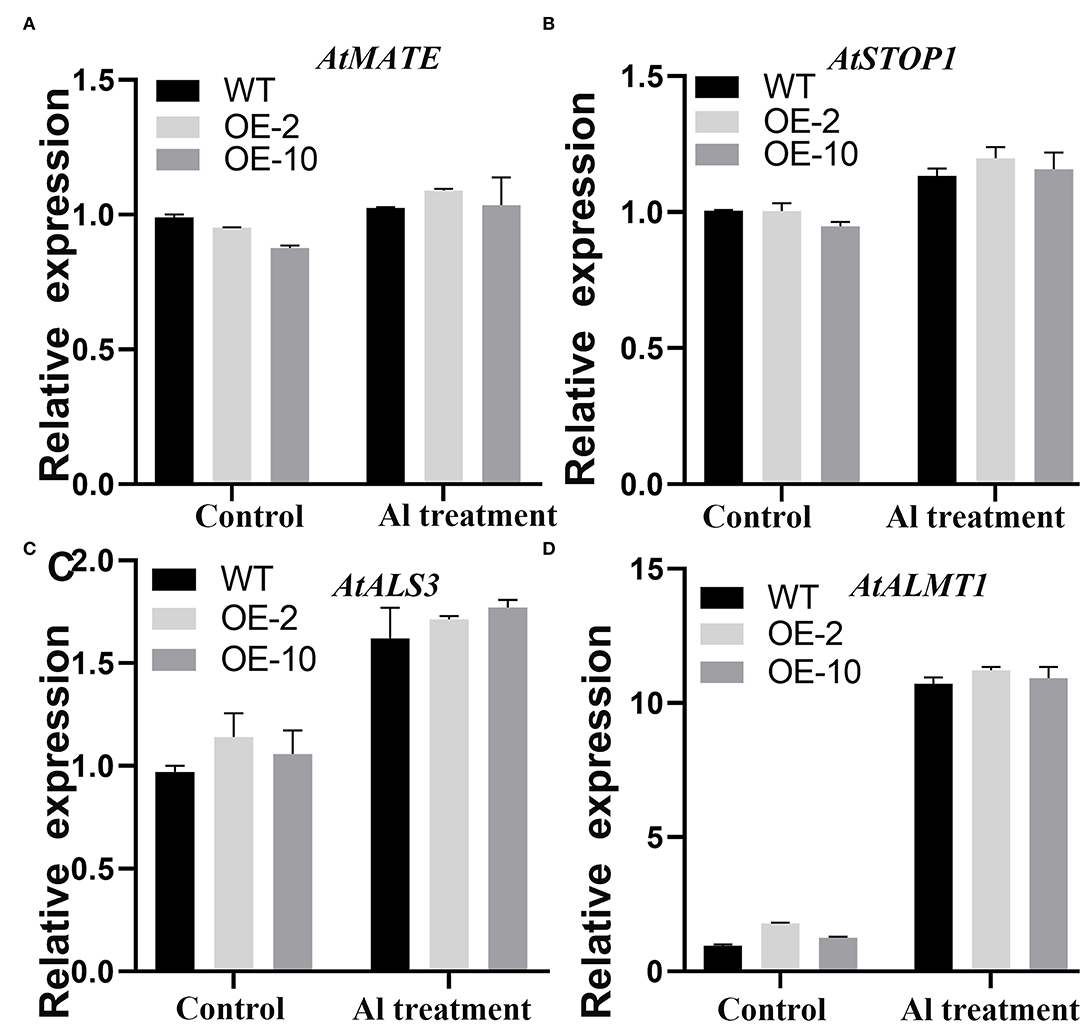
Figure 6. (A–D) Expression patterns of genes responsive to Al stress. WT: wild-type Arabidopsis; OE-2, OE-10: GmWRKY21 overexpression transgenic lines. WT and transgenic Arabidopsis were grown on 0 or 100 μM AlCl3 (pH 4.3) 1/2 MS medium for about 10 days. All data are presented as means ± SEM (n = 3). Significant differences according to the one-way analysis of variance are denoted.
To further investigate the pathways regulated by GmWRKY21 under Al stress, the expression patterns of stress-responsive genes were performed by qRT-PCR. The results showed that the stress-responsive genes, such as KIN1, COR15A, COR15B, COR47, GLOS3, and RD29A, were upregulated by the treatment of 100 μM AlCl3 (Figure 7). Among them, the transcripts of KIN1, GLOS3, COR15A, and COR15B genes were significantly higher in the OE-10 line than those of WT under Al stress. However, only the expression levels of KIN1, GLOS3, and RD29A were significantly different from those in the OE-2 lines. The results showed that the GmWRKY21 TF enhances the tolerance of transgenic Arabidopsis to acidic aluminum stress mediated by regulating the expression of the stress-responsive genes.
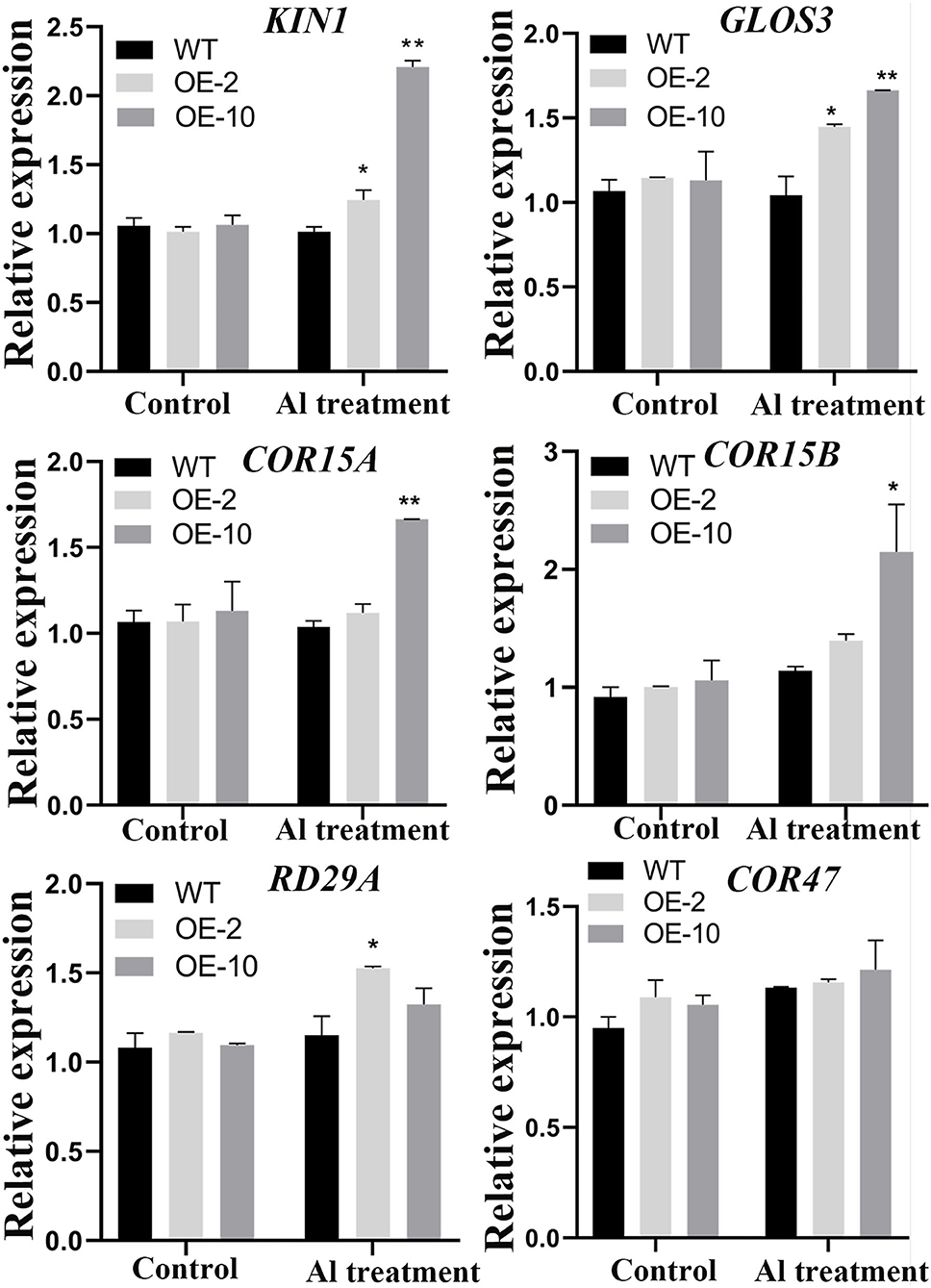
Figure 7. Expression patterns of stress-responsive genes regulated by GmWRKY21in Arabidopsis. WT, wild-type Arabidopsis; OE-2, OE-10: GmWRKY21 overexpression transgenic lines. WT and transgenic Arabidopsis were grown on 0 or 100 μM AlCl3 (pH 4.3) 1/2 MS medium for about 10 days. All data are presented as means ± SEM (n = 3). Significant differences according to the one-way analysis of variance are denoted as follows: **P < 0.01 and *P < 0.05.
Several genes such as GmCOR47 (Guo et al., 1992), GmDREB2A (Agarwal et al., 2007), GmMYB84 (Wang Y. Q. et al., 2017), GmKIN1 (Wang et al., 1995), GmGST1 (Qi et al., 2018), and GmLEA (Magwanga et al., 2018) play significant roles in abiotic stress. The expression levels of these marker genes were analyzed between EV and GmWRKY21 transgenic (OE or RNAi) hairy roots under normal and Al stress conditions by qRT-PCR analysis. There was no significant difference between the EV roots and either of the GmWRKY21 transgenic soybean hairy roots under normal growth conditions. The expression levels of GmCOR47, GmDREB2A, GmMYB84, GmKIN1, GmGST1, and GmLEA in the OE soybean hairy roots were significantly higher than those in the EV roots. Conversely, the expression levels of these stress-responsive genes in the RNAi soybean hairy roots were significantly lower than those in the EV-control soybean hairy roots (Figure 8).
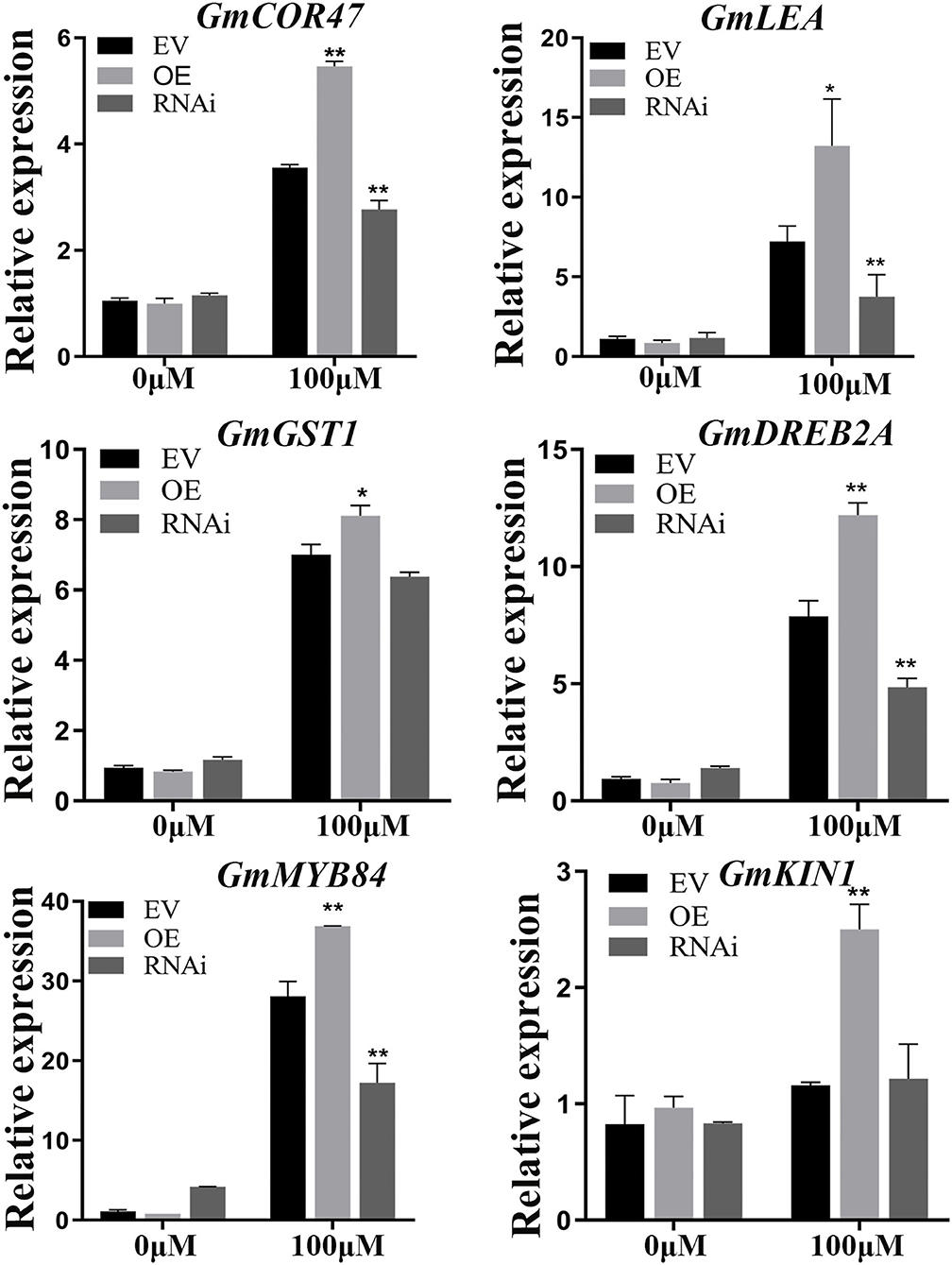
Figure 8. Expression patterns of the stress-responsive genes including GmCOR47, GmLEA, GmGST1, GmDREB2A, GmMYB84, and GmKIN1. EV, empty vector; OE, overexpressing GmWRKY21 soybean hair roots; RNAi: the hairy roots of GmWRKY21-RNAi transgenic soybean. The EV, OE, and RNAi soybean hair roots under normal and Al stress conditions. The hair roots were exposed to 0 and 100 μM AlCl3 (pH 4.3) for 24 h. All data are presented as means ± SEM (n = 3). Significant differences according to the one-way analysis of variance are denoted as follows: **P < 0.01 and *P < 0.05.
Since the first WRKY transcription factor gene SPF1 was found in sweet potato in 1994 (Ishiguro and Nakamura, 1994), the WRKY family genes have been broadly investigated for the diverse biological roles in plant growth and development and responses to biotic and abiotic stress (Dai et al., 2016; Kiranmai et al., 2018). However, few genes are known about the WRKYs' roles and their mechanisms responsive to Al stress in soybean. In this study, the candidate gene GmWRKY21 derived from the RIL (recombinant inbred line, F12) population of Zhonghuang 24 and Huaxia 3 (160 lines) was investigated for its functional characterization of tolerance to aluminum stress in Arabidopsis (Wang et al., 2019). The GmWRKY21 protein located in subgroup IIc contained the WRKY domain and C2H2-type zinc-finger structure with the classical characteristic of WRKY proteins from soybean and Arabidopsis (Figures 1A,B). The GmWRKY21 protein had more similarities to other WRKY proteins, such as GmWRKY6, GmWRKY25, GmWRKY45, GmWRKY53, GmWRKY8, GmWRKY54, and GmWRKY45, in soybean and AtWRKY57, AtWRKY45, AtWRKY71, AtWRKY28, AtWRKY75, and AtWRKY12 in Arabidopsis (Supplementary Figure 1). Previous reports indicated that these genes were all related to biotic and abiotic stress. For example, GmWRKY6, GmWRKY25, GmWRKY53, and GmWRKY8 all respond to salt and drought stress (Zhou et al., 2008). GmWRKY45 enhanced the adaptation to phosphate deficiency and salt tolerance in transgenic Arabidopsis (Zhou et al., 2008; Li et al., 2019; Wei et al., 2019). Overexpression of GmWRKY54 resulted in the tolerance to salt and drought stress in Arabidopsis and the resistance to drought stress through activating genes in Ca2+ signaling pathways and abscisic acid in transgenic soybean (Wei et al., 2019). In addition, the WRKY family genes also play multiple roles in regulating plant growth and development, responding to biotic and abiotic stresses. Previous reports indicated that AtWRKY57 conferred drought tolerance in Arabidopsis (Jiang et al., 2012). AtWRKY45 transcription factor activated PHT1; 1 expression in response to phosphate starvation (Wang et al., 2014). AtWRKY71 accelerated flowering in Arabidopsis in the presence of salt stress (Yu Z. Y. et al., 2018). Co-expression of AtWRKY28 and AtbHLH17 conferred resistance to abiotic stress in Arabidopsis (Babitha et al., 2013). AtWRKY75 interacted with DELLA protein to positively regulate Arabidopsis flowering (Zhang et al., 2018), while AtWRKY12 negatively regulated cadmium tolerance in Arabidopsis by directly targeting GSH1 (Han et al., 2019). Furthermore, the phylogenetic tree indicated that the GmWRKY21 protein responding to salt and drought stress had more similarities to the GmWRKY6 protein (Supplementary Figure 1). Previous reports showed that overexpression of GmWRKY21 responding to various abiotic stresses, such as salt and drought, enhanced the tolerance of low-temperature stress in transgenic Arabidopsis (Zhou et al., 2008). Therefore, the GmWRKY21 protein was identified to be a WRKY transcription factor to participate in abiotic stress and/or development in plants.
Previous studies revealed that WRKY TFs were widely investigated for key roles in acidic aluminum stress (Ding et al., 2013; Li et al., 2018, 2020). In this study, the GmWRKY21 gene was rich in soybean root and quickly upregulated under AlCl3 treatment with the differences in Al3+ concentrations and temporal differences (Figure 3). Aluminum stress increased the formation of reactive oxygen species (ROS) in plants, leading to lipid membrane peroxidation, ion leakage, and protein oxidation (Ezaki et al., 2000; Boscolo et al., 2003). Malondialdehyde (MDA) was one of the most commonly used indicators for the destruction of ROS under stress conditions (Shi et al., 2013), while proline was a multifunctional amino acid to protect plant growth by accumulating in large quantities when plants were subjected to adversity stress (Natarajan et al., 2012). In this study, overexpression of GmWRKY21 enhanced the tolerance of transgenic Arabidopsis and soybean hairy roots to acid–aluminum stress with the corresponding changes of MDA and proline (Figures 4, 6). Hematoxylin was easy to combine with the root aluminum to form a purplish red complex, and the root tip aluminum ion content can be indirectly judged by the depth of root tip color so as to obtain the sensitivity of plants to aluminum toxicity (Rincon and Gonzales, 1992). In this study, the lines of GmWRKY21 overexpression were dyed shallower by hematoxylin staining with less Al3+ content in the root tips compared to those of WT and RNAi plants (Figure 5). Recent reports indicated that other WRKY TFs also have the function of coping with acidic aluminum stress. For example, AtWRKY46 was a transcriptional suppressor of ALMT1 that regulates alumina-induced Arabidopsis malic acid secretion. The mutation of AtWRKY46 led to increased malic acid secretion and reduced aluminum accumulation in the root tip resulting in higher aluminum tolerance in Arabidopsis (Ding et al., 2013). OsWRKY22 enhanced the tolerance to aluminum stress by activating the expression of OsFRDL4 and enhancing the secretion of citric acid in rice (Li et al., 2018). AtWRKY47 made Arabidopsis aluminum tolerant by regulating cell wall modification genes (Li et al., 2020). Therefore, the GmWRKY21 gene may play an important role in the response of plants to acidic aluminum stress.
Previous studies have found that the ALMT protein family can transport malic acid in the root system to the rhizosphere conferring plants tolerant to aluminum toxicity (Sasaki et al., 2004). The MATE protein family was also involved in the aluminum rejection mechanism by transporting citric acid from the root system to the rhizosphere (Ryan et al., 2011; Kochian et al., 2015). In Arabidopsis, ALMT1, MATE, and ALS3 were upregulated by the STOP1 transcription factor (Sawaki et al., 2009). In this study, the STOP1, ALMT1, ALS3, and MATE genes were upregulated under aluminum acid stress, while the STOP1, ALMT1, and ALS3 genes were dramatically induced by GmWRKY21 in transgenic lines. However, the transcripts of the four genes in transgenic Arabidopsis under Al stress were statistically non-significant differences compared to those under the control (Figure 6). The results suggested that GmWRKY21 enhances the tolerance to Al stress in transgenic plants by the comprehensive effect of multiple genes. Previous research found that GmWRKY21 was involved in a variety of abiotic stresses, such as low-temperature, salt, and drought stress (Zhou et al., 2008). In this study, the stress-related genes of KIN1, GLOS3, and RD29A were significantly upregulated in transgenic Arabidopsis plants under the AlCl3 treatment (Figure 7; Yu et al., 2016; Chu et al., 2018; Wang et al., 2018; Ma et al., 2019). The LEA genes of COR15A and COR15B were upregulated with more transcripts in transgenic Arabidopsis plants (Figure 7), which were abundantly present in vegetative plant tissues under drought, low-temperature or salt, and other environmental stresses (Hundertmark and Hincha, 2008). Overexpression of COR15A or COR15B significantly improved the freezing resistance of Arabidopsis, while silencing these two genes at the same time reduced the freezing resistance of Arabidopsis (Artus et al., 1996; Thalhammer et al., 2014). The results indicated that GmWRKY21 enhanced the tolerance to aluminum stress in Arabidopsis by several genes mediated by certain stress-responsive pathways.
A previous study indicated that some genes such as GmCOR47, GmMYB84, GmDREB2A, GmGST1, GmKIN1, and GmLEA play certain roles in regulating plant response to abiotic stress by potential mechanism. GmGST1 helps in maintaining redox homeostasis of cells, thus enhancing plant ability to resist abiotic stress (Qi et al., 2018). GmMYB84 can improve the expression levels of GmRBOHB-1 and GmRBOHB-2 genes by binding to the MBS cis-elements in their promoter, enhancing stress tolerance in soybean (Wang N. et al., 2017). The COR47 and LEA as group 2 LEA (LEA II) proteins encode dehydrins, which are cold-inducible proteins that are supposed to sustain membrane stabilization and prevent protein aggregation (Guo et al., 1992; Yu Y. C. et al., 2018; Yu Z. Y. et al., 2018; Li P. et al., 2021). The DREB protein plays an important role in regulating abiotic stress-related genes and thereby imparting stress tolerance to the plant system (Dubouzet et al., 2003). KIN1 was induced by low temperature, ABA, osmoticum, and dehydration (Wang et al., 1995). These stress-responsive genes were significantly upregulated in the hairy root of transgenic GmWRKY21 soybean (Figure 8). This indicates that GmWRKY21 improved soybean resistance to aluminum stress by regulating the expression of several stress-responsive genes.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
QM, HN, and ZH conceived and designed the study. ZH, JW, XW, XZ, YC, and ZC conducted the experiments. ZH, JW, and QM performed the data and statistical analysis. ZH wrote the manuscript which was reviewed and edited by HN and QM. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (31971965 and 31771816), the Major Project of New Varieties Cultivation of Genetically Modified Organisms (2016ZX08004002-007), the Special Supervision on Quality and Safety of Agricultural Products of the Ministry of Agriculture and Rural Areas (4100-C17106 and 21301091702101), the Key Projects of International Scientific and Technological Innovation Cooperation among Governments under National Key R&D Plan (2018YFE0116900-06), the Key Area Research and Development Program of Guangdong Province (2020B020220008), the China Agricultural Research System (CARS-04-PS09), and the Project of Science and Technology of Guangzhou (201804020015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.833326/full#supplementary-material
Agarwal, P., Agarwal, P. K., Nair, S., Sopory, S. K., and Reddy, M. K. (2007). Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol. Genet. Genomics 277, 189–198. doi: 10.1007/s00438-006-0183-z
Artus, N. N., Uemura, M., Steponkus, P. L., Gilmour, S. J., Lin, C., and Thomashow, M. F. (1996). Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. U.S.A. 93, 13404–13409. doi: 10.1073/pnas.93.23.13404
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Bojorquez-Quintal, E., Escalante-Magana, C., Echevarria-Machado, I., and Martinez-Estevez, M. (2017). Aluminum, a friend or foe of higher plants in acid soils. Front. Plant Sci. 8, 1767. doi: 10.3389/fpls.2017.01767
Boscolo, P., Menossi, M., and Jorge, R. A. (2003). Aluminum-induced oxidative stress in maize. Phytochemistry 62, 181–189. doi: 10.1016/S0031-9422(02)00491-0
Brautigan, D. J., Rengasamy, P., and Chittleborough, D. J. (2012). Aluminium speciation and phytotoxicity in alkaline soils. Plant Soil 360, 187–196. doi: 10.1007/s11104-012-1232-5
Cai, Z. D., Xian, P. Q., Lin, R. B., Cheng, Y. B., Lian, T. X., Ma, Q. B., et al. (2020). Characterization of the soybean GmIREG family genes and the function of GmIREG3 in conferring tolerance to aluminum stress. Int. J. Mol. Sci. 21, 497. doi: 10.3390/ijms21020497
Chen, J. N., Nolan, T. M., Ye, H. X., Zhang, M. C., Tong, H. N., Xin, P. Y., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, L. G., Song, Y., Li, S. J., Zhang, L. P., Zou, C. S., and Yu, D. Q. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chu, M. X., Li, J. J., Zhang, J. Y., Shen, S. F., Li, C. N., Gao, Y. J., et al. (2018). AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 69, 5241–5253. doi: 10.1093/jxb/ery278
Clough, S. J., and Bent, A. F. (2010). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dai, X. Y., Wang, Y. Y., and Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Ezaki, B., Gardner, R. C., Ezaki, Y., and Matsumoto, H. (2000). Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 122, 657–665. doi: 10.1104/pp.122.3.657
Feng, M. J. (2000). Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 41, 383–390. doi: 10.1093/pcp/41.4.383
Finatto, T., Viana, V. E., Woyann, L. G., Busanello, C., Maia, L. C. D., and Oliveira, A. C. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 41, 533–544. doi: 10.1590/1678-4685-gmb-2017-0232
Guo, W., Ward, R. W., and Thomashow, M. F. (1992). Characterization of a cold-regulated wheat gene related to Arabidopsis cor47. Plant Physiol. 100, 915–922. doi: 10.1104/pp.100.2.915
Han, Y. Y., Fan, T. T., Zhu, X. Y., Wu, X., Ouyang, J., Jiang, L., et al. (2019). WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol. Biol. 99, 149–159. doi: 10.1007/s11103-018-0809-7
He, G. H., Xu, J. Y., Wang, Y. X., Liu, J. M., Li, P. S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16, 116. doi: 10.1186/s12870-016-0806-4
Hu, Q., Ao, C. W., Wang, X. R., Wu, Y. F., and Du, X. Z. (2021). GhWRKY1-like, a WRKY transcription factor, mediates drought tolerance in Arabidopsis via modulating ABA biosynthesis. BMC Plant Biol. 21, 458. doi: 10.1186/s12870-021-03238-5
Huang, S. Z., Hu, L. J., Zhang, S. H., Zhang, M. X., Jiang, W. Z., Wu, T., et al. (2021). Rice OsWRKY50 mediates ABA-dependent seed germination and seedling growth, and ABA-independent salt stress tolerance. Int. J. Mol. Sci. 22, 8625. doi: 10.3390/ijms22168625
Hundertmark, M., and Hincha, D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118. doi: 10.1186/1471-2164-9-118
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jiang, Y. J., Liang, G., and Yu, D. Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Jiang, Y. Q., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Kereszt, A., Li, D. X., Indrasumunar, A., Nguyen, C. D. T., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kiranmai, K., Rao, G. L., Pandurangaiah, M., Nareshkumar, A., Reddy, V. A., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9, 346. doi: 10.3389/fpls.2018.00346
Kochian, L. V. (1995). Cellular mechanism of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. 46, 675–681. doi: 10.1146/annurev.pp.46.060195.001321
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soil? Mechanisms of aluminum tolerance and phosphorous efficiency. Ann. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kochian, L. V., Pineros, M. A., Liu, J., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571. doi: 10.1146/annurev-arplant-043014-114822
Kokkirala, V. R., Yonggang, P., Abbagani, S., Zhu, Z., and Umate, P. (2010). Subcellular localization of proteins of Oryza sativa L. In the model tobacco and tomato plants. Plant Signal. Behav. 5, 1336–1341. doi: 10.4161/psb.5.11.13318
Kollmeier, M., Felle, H. H., and Horst, W. J. (2000). Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. Is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 122, 945–956. doi: 10.1104/pp.122.3.945
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, C., Li, K. N., Liu, X. Y., Ruan, H., Zheng, M. M., Yu, Z. J., et al. (2021). Transcription factor GmWRKY46 enhanced phosphate starvation tolerance and root development in transgenic plants. Front. Plant Sci. 12, 700651. doi: 10.3389/fpls.2021.700651
Li, C., Liu, X. Y., Ruan, H., Zhang, J. Y., Xie, F. B., Gai, J. Y., et al. (2019). GmWRKY45 enhances tolerance to phosphate starvation and salt stress, and changes fertility in transgenic Arabidopsis. Front. Plant Sci. 10, 1714. doi: 10.3389/fpls.2019.01714
Li, C. X., Yan, J. Y., Ren, J. Y., Sun, L., Xu, C., Li, G. X., et al. (2020). A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 62, 1176–1192. doi: 10.1111/jipb.12888
Li, G. Z., Wang, Z. Q., Yokosho, K., Ding, B., Fan, W., Gong, Q. Q., et al. (2018). Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 219, 149–162. doi: 10.1111/nph.15143
Li, G. Z., Zheng, Y. X., Chen, S. J., Liu, J., Wang, P. F., Wang, Y. H., et al. (2021). TaWRKY74 participates copper tolerance through regulation of TaGST1 expression and GSH content in wheat. Ecotoxicol. Environ. Saf. 221, 112469. doi: 10.1016/j.ecoenv.2021.112469
Li, J., Besseau, S., Trnen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, P., Li, X., and Jiang, M. (2021). CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 48, 5821–5832. doi: 10.1007/s11033-021-06541-4
Lim, C., Kang, K., Shim, Y., Yoo, S. C., and Paek, N. C. (2021). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188, 1900–1916. doi: 10.1093/plphys/kiab492
Liu, J. P., Luo, X. Y., Shaff, J., Liang, C. Y., Jia, X. M., Li, Z. Y., et al. (2012). A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J. 71, 327–373. doi: 10.1111/j.1365-313X.2012.04994.x
Liu, J. P., Magalhaes, J. V., Shaff, J., and Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Liu, J. P., Pineros, M. A., and Kochian, L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56, 221–230. doi: 10.1111/jipb.12162
Lu, J., Suo, H. C., Yi, R., Ma, Q. B., and Nian, H. (2015). Glyma11g13220, a homolog of the vernalization pathway gene VERNALIZATION 1 from soybean [Glycine max (L.) Merr.], Promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 15, 232. doi: 10.1186/s12870-015-0602-6
Ma, J. H., Gao, X. L., Liu, Q., Shao, Y., Zhang, D. J., Jiang, L. N., et al. (2017). Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 8, 2036. doi: 10.3389/fpls.2017.02036
Ma, Q. B., Xia, Z. L., Cai, Z. D., Li, L., Cheng, Y. B., Liu, J., et al. (2019). GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 9, 1979. doi: 10.3389/fpls.2018.01979
Ma, Q. B., Yi, R., Li, L., Liang, Z. Y., Zeng, T. T., Zhang, Y., et al. (2018). GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 18, 212. doi: 10.1186/s12870-018-1397-z
Magwanga, R. O., Lu, P., Kirungu, J. N., Lu, H. J., Wang, X. X., Cai, X. Y., et al. (2018). Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 19, 6. doi: 10.1186/s12863-017-0596-1
Matthews, B., and Youssef, R. (2016). Agrobacterium rhizogenes-based transformation of soybean roots to form composite plants. Bio-Protocol 6, e1708. doi: 10.21769/BioProtoc.1708
Natarajan, S. K., Zhu, W. D., Liang, X. W., Zhang, L., Demers, A. J., Zimmerman, M. C., et al. (2012). Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free Radic. Biol. Med. 53, 1181–1191. doi: 10.1016/j.freeradbiomed.2012.07.002
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Qi, J. S., Song, C. P., Wang, B. S., Zhou, J. M., Kangasjarvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qiu, Y. P., and Yu, D. Q. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Ren, B., Wang, X. T., Duan, J. B., and Ma, J. X. (2019). Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922. doi: 10.1126/science.aav8907
Ren, X. Z., Chen, Z. Z., Liu, Y., Zhang, H. R., Zhang, M., Liu, Q. A., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Rincon, M., and Gonzales, R. A. (1992). Aluminum partitioning in intact roots of aluminum-tolerant and aluminum-sensitive wheat (Triticum aestivum L.) cultivars. Plant Physiol. 99, 1021–1028. doi: 10.1104/pp.99.3.1021
Rushton, P. J., Macdonald, H., Huttly, A. K., Lazarus, C. M., and Hooley, R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 29, 691–702. doi: 10.1007/BF00041160
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, P. J., Torres, J. T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I. E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. Embo J. 15, 5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x
Ryan, P. R., Delhaize, E., and Jones, D. L. (2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 527–560. doi: 10.1146/annurev.arplant.52.1.527
Ryan, P. R., Tyerman, S. D., Sasaki, T., Furuichi, T., Yamamoto, Y., Zhang, W. H., et al. (2011). The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 62, 9–20. doi: 10.1093/jxb/erq272
Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313X.2003.01991.x
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. doi: 10.1104/pp.108.134700
Shen, H. S., Liu, C. T., Zhang, Y., Meng, X. P., Zhou, X., Chu, C. C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80, 241–253. doi: 10.1007/s11103-012-9941-y
Shi, H. T., Ye, T. T., and Chan, Z. L. (2013). Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass [Cynodon dactylon (L). Pers.]. Plant Physiol. Biochem. 71, 226–234. doi: 10.1016/j.plaphy.2013.07.021
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W. Z., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Silambarasan, S., Logeswari, P., Valentine, A., and Cornejo, P. (2019). Role of Curtobacterium herbarum strain CAH5 on aluminum bioaccumulation and enhancement of Lactuca sativa growth under aluminum and drought stresses. Ecotoxicol. Environ. Saf. 183, 109571–109573. doi: 10.1016/j.ecoenv.2019.109573
Singh, S., Tripathi, D. K., Singh, S., Sharma, S., Dubey, N. K., Chauhan, D. K., et al. (2017). Toxicity of aluminium on various levels of plant cells and organism. Environ. Exp. Bot. 137, 177–193. doi: 10.1016/j.envexpbot.2017.01.005
Song, H., Sun, W. H., Yang, G. F., and Sun, J. (2018). WRKY transcription factors in legumes. BMC Plant Biol. 18, 243. doi: 10.1186/s12870-018-1467-2
Song, Y., Chen, L. G., Zhang, L. P., and Yu, D. Q. (2010). Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35, 459–471. doi: 10.1007/s12038-010-0051-1
Thalhammer, A., Bryant, G., Sulpice, R., and Hincha, D. K. (2014). Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 166, 190–201. doi: 10.1104/pp.114.245399
Wang, H., Datla, R., Georges, F., Loewen, M., and Cutler, A. J. (1995). Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 28, 605–617. doi: 10.1007/BF00021187
Wang, H., Xu, Q., Kong, Y. H., Chen, Y., Duan, J. Y., Wu, W. H., et al. (2014). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077
Wang, N., Zhang, W. X., Qin, M. Y., Li, S., Qiao, M., Liu, Z. H., et al. (2017). Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol 58, 1764–1776. doi: 10.1093/pcp/pcx111
Wang, S. C., Zhang, J., Gu, M., and Xu, G. H. (2021). OsWRKY108 is an integrative regulator of phosphorus homeostasis and leaf inclination in rice. Plant Signal. Behav. 16, 1976545. doi: 10.1080/15592324.2021.1976545
Wang, X. L., Wang, Y., Wang, L., Liu, H., Zhang, B., Cao, Q. J., et al. (2018). Arabidopsis PCaP2 functions as a linker between ABA and SA signals in plant water deficit tolerance. Front. Plant Sci. 9, 578. doi: 10.3389/fpls.2018.01062
Wang, X. X., Cheng, Y. B., Yang, C., Yang, C. Y., Mu, Y. H., Xia, Q. J., et al. (2019). QTL mapping for aluminum tolerance in RIL population of soybean (Glycine max L.) by RAD sequencing. PLoS ONE 14, e223674. doi: 10.1371/journal.pone.0223674
Wang, Y. Q., Li, R. H., Li, D. M., Jia, X. M., Zhou, D. W., Li, J. Y., et al. (2017). NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. U.S.A. 114, 5047. doi: 10.1073/pnas.1618557114
Wei, W., Liang, D. W., Bian, X. H., Shen, M., Xiao, J. H., Zhang, W. K., et al. (2019). GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 100, 384–398. doi: 10.1111/tpj.14449
Willems, E., Leyns, L., and Vandesompele, J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379, 127–129. doi: 10.1016/j.ab.2008.04.036
Wu, X. L., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xie, T., Chen, C. J., Li, C. H., Liu, J. R., Liu, C. Y., and He, Y. H. (2018). Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genomics 19, 490. doi: 10.1186/s12864-018-4880-x
Xu, Z. L., Raza, Q., Xu, L., He, X. L., Huang, Y. H., Yi, J. X., et al. (2018). GmWRKY49, a salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 809. doi: 10.3389/fpls.2018.00809
Yamamoto, Y., Kobayashi, Y., Devi, S. R., Rikiishi, S., and Matsumoto, H. (2002). Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 128, 63–72. doi: 10.1104/pp.010417
Yang, L. A., Ji, W., Zhu, Y. M., Gao, P., Li, Y., Cai, H., et al. (2010). GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 61, 2519–2533. doi: 10.1093/jxb/erq084
Ye, H., Qiao, L. Y., Guo, H. Y., Guo, L. P., Ren, F., Bai, J. F., et al. (2021). Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 12, 663118. doi: 10.3389/fpls.2021.663118
Yokosho, K., Yamaji, N., Mitani-Ueno, N., Shen, R. F., and Ma, J. F. (2016). An aluminum-inducible IREG gene is required for internal detoxification of aluminum in buckwheat. Plant Cell Physiol. 57, 1169–1178. doi: 10.1093/pcp/pcw065
Yu, X. W., Liu, Y. M., Wang, S., Tao, Y., Wang, Z. K., Shu, Y. J., et al. (2016). CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 35, 613–627. doi: 10.1007/s00299-015-1907-5
Yu, Y. C., Wang, L., Chen, J. C., Liu, Z. H., Park, C. M., and Xiang, F. N. (2018). WRKY71 acts antagonistically against salt-delayed flowering in Arabidopsis thaliana. Plant Cell Physiol. 59, 414–422. doi: 10.1093/pcp/pcx201
Yu, Z. Y., Wang, X., and Zhang, L. S. (2018). Structural and functional dynamics of dehydrins: A plant protector protein under abiotic stress. Int. J. Mol. Sci. 19, 3420. doi: 10.3390/ijms19113420
Zeng, Q. Y., Yang, C. Y., Ma, Q. B., Li, X. P., Dong, W. W., and Nian, H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12, 182. doi: 10.1186/1471-2229-12-182
Zhang, L. P., Chen, L. G., and Yu, D. Q. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, Y. J., and Wang, L. J. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhou, L., Wang, N. N., Gong, S. Y., Lu, R., Li, Y., and Li, X. B. (2015). Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 96, 311–320. doi: 10.1016/j.plaphy.2015.08.016
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: transcription factor, GmWRKY21, soybean, Al stress, Arabidopsis thaliana
Citation: Han Z, Wang J, Wang X, Zhang X, Cheng Y, Cai Z, Nian H and Ma Q (2022) GmWRKY21, a Soybean WRKY Transcription Factor Gene, Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. Front. Plant Sci. 13:833326. doi: 10.3389/fpls.2022.833326
Received: 11 December 2021; Accepted: 15 June 2022;
Published: 25 July 2022.
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Branka Salopek Sondi, Rudjer Boskovic Institute, CroatiaCopyright © 2022 Han, Wang, Wang, Zhang, Cheng, Cai, Nian and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qibin Ma, bWFxaWJpbkBzY2F1LmVkdS5jbg==; Hai Nian, aG5pYW5Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.