
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 April 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.832981
 Diaa Abd El-Moneim*†‡
Diaa Abd El-Moneim*†‡ Roberto Contreras‡
Roberto Contreras‡ Javier Silva-Navas
Javier Silva-Navas Francisco Javier Gallego
Francisco Javier Gallego Ana M. Figueiras†
Ana M. Figueiras† Cesar Benito†
Cesar Benito†Aluminum (Al) toxicity in acid soils influences plant development and yield. Almost 50% of arable land is acidic. Plants have evolved a variety of tolerance mechanisms for Al. In response to the presence of Al, various species exudate citrate from their roots. Rye (Secale cereale L.) secretes both citrate and malate, making it one of the most Al-tolerant cereal crops. However, no research has been done on the role of the mitochondrial citrate synthase (mCS) gene in Al-induced stress in the rye. We have isolated an mCS gene, encoding a mitochondrial CS isozyme, in two S. cereale cultivars (Al-tolerant cv. Ailés and Al-sensitive inbred rye line Riodeva; ScCS4 gene) and in two Brachypodium distachyon lines (Al-tolerant ABR8 line and Al-sensitive ABR1 line; BdCS4 gene). Both mCS4 genes have 19 exons and 18 introns. The ScCS4 gene was located on the 6RL rye chromosome arm. Phylogenetic studies using cDNA and protein sequences have shown that the ScCS4 gene and their ScCS protein are orthologous to mCS genes and CS proteins of different Poaceae plants. Expression studies of the ScCS4 and BdSC4 genes show that the amount of their corresponding mRNAs in the roots is higher than that in the leaves and that the amounts of mRNAs in plants treated and not treated with Al were higher in the Al-tolerant lines than that in the Al-sensitive lines of both species. In addition, the levels of ScCS4 and BdCS4 mRNAs were reduced in response to Al (repressive behavior) in the roots of the tolerant and sensitive lines of S. cereale and B. distachyon.
The most common metal in the Earth’s crust is aluminum (Al). In non-acid soils, Al is harmless; but in acidic soils, Al is solubilized into the toxic Al3+ cation (Ritchie, 1995), which inhibits root elongation (Panda et al., 2009). Internal and external mechanisms have developed in plants to deal with Al3+. The exudation of organic anions from root apices is an exclusion mechanism identified for many species with strong genetic support (e.g., malate, citrate, and, in some species, oxalate) (Mariano and Keltjens, 2001). Two distinct patterns of Al-induced organic acid secretion have been identified. The release of organic acids starts immediately after the addition of Al in pattern I (Hede et al., 2001; Ma et al., 2001). The activation of the anion channel was involved in this pattern. In pattern II, the release of organic acids begins after a marked lag phase (Hede et al., 2001; Ma et al., 2001). The genes of organic acids metabolism are implicated in this pattern. The ability of different plants to withstand toxic concentrations of Al3+ varies greatly. In response to Al stress, the rye S. cereale, one of the most Al-tolerant cereal crops, secretes citrate and malate from its roots (Li et al., 2000). Al-stress tolerant cultivars (Ailés and Petkus) and the sensitive Riodeva inbred line show pattern II (inducible) of malic exudation (Abd El-Moneim et al., 2014, 2015; Sánchez-Parra et al., 2015). For citrate, the tolerant Petkus also showed an inducible exudation pattern (Abd El-Moneim et al., 2014). According to the results of Ma et al. (2001), however, the sensitive Riodeva does not show such an inducible pattern (Abd El-Moneim et al., 2014). The model grass B. distachyon also secretes both organic acids following an inducible exudation pattern (pattern II) (Contreras et al., 2014). Tolerance genes for Al-activated malate transporter (ALMT) and Al-activated citrate transporter (AACT or MATE) promote organic anion efflux from roots. These genes have been identified in various species for Al tolerance. In many plants, the activities of enzymes involved in citrate metabolism have been investigated (Mariano et al., 2005). One of them is citrate synthase (CS) activity encoded by several genes. In S. cereale (Li et al., 2000), Phaseolus vulgaris (Mugai et al., 2000), and Cassia tora (Yang et al., 2004), exposure to Al increased CS root activities, but not in wheat (Triticum aestivum) or triticale (Yang et al., 2004). In soybean (Glycine max), Al treatment enhanced mitochondrial CS enzyme activity and their gene expression (Eticha et al., 2010). On the contrary, in Coffea arabica, CS activities decrease after Al shock (Ramírez-Benítez et al., 2008). Finally, CS activities in the tolerant and sensitive triticale lines and their response to pH in the presence or absence of Al were similar (Hayes and Ma, 2003). In this work, the authors propose that the exudation of organic acids is not very related to their metabolism. In addition to soybean, variations in CS gene expression have also been examined in P. vulgaris and Arabidopsis thaliana. In P. vulgaris, no major differences in CS expression were found (Eticha et al., 2010). Transcriptomic analysis of A. thaliana root responses to Al revealed no significant enhancement in CS transcript abundance (Kumari et al., 2008). In cultured carrot (Daucus carota) cells (Koyama et al., 1999), A. thaliana (Koyama et al., 2000), and canola (Brassica napus) (Anoop et al., 2003) plants, overexpression of mitochondrial CS genes (mCS) resulted in enhanced citrate efflux. There is an essential controversy over whether CS overexpression results in the increase in citrate exudation and greater tolerance to Al stress in tobacco and alfalfa transgenic plants (de la Fuente et al., 1997; Delhaize et al., 2001; Osawa and Kojima, 2006; Barone et al., 2008; Han et al., 2009). The overexpression of a bacterial CS gene in tobacco and papaya transgenic plants improved Al tolerance and citrate overproduction (de la Fuente et al., 1997). Delhaize et al. (2003) concluded that internal citrate concentrations and citrate efflux in tobacco are highly insensitive to large changes in either mitochondrial CS activity or cytosolic isocitrate dehydrogenase (IDH) activity and suggested other factors such as transport out of the roots or control citrate efflux. The findings obtained by Osawa and Kojima (2006) indicated that Al-inducible expression of mCS coupled with enhanced citrate release mediates Al resistance in the tree Paraserianthes falcataria. Despite the wealth of data on mCS gene activity reported in other plant species, less is known on their responsiveness to Al stress in S. cereale and B. distachyon.
This study aimed to isolate the gene, identify the chromosomal location, analyze the molecular variability, and observe the changes in the expression of mitochondrial citrate synthase gene (mCS) by Al stress in S. cereale and B. distachyon.
Six different cultivars were used in the experiments (four S. cereale cultivars and two B. distachyon cultivars). The tolerance or sensitivity of the S. cereale cultivars and of the lines of B. distachyon to Al was established by root growth tests in the absence of and in the presence of Al at different concentrations in previous studies carried out by our research group (Contreras et al., 2014; Abd El-Moneim et al., 2015). Table 1 shows the Al tolerance for the studied cultivars. In these previous works, we also estimated the exudation by the malate and citrate roots in the presence and absence of Al. Chromosomal location of the ScCS gene was identified in rye (Imperial cultivar), hexaploid wheat T. aestivum (Chinese Spring cultivar), their corresponding wheat-rye amphiploid (Chinese Spring-Imperial), seven wheat-rye disomic addition lines (1R to 7R), and the ditelosomic wheat-rye addition line 6RS.
Seven-day-old seedlings were subjected to a nutrient solution containing Al in the form of AlK (SO4)2⋅12H2O (pH 4.0) at a concentration of 150 μM for 24 h to isolate the ScCS4 gene of rye. Root samples were frozen in liquid nitrogen and held at −80°C until required. Using TRIzol Kit (Invitrogen), RNA was obtained from the roots and then converted to cDNA using High-Capacity cDNA Reverse Transcription Package (Applied Biosystems). From the sequence of the HvCS4 gene (reference AK248736.1) in barley, one pair of primers was designed to amplify ScCS4 cDNA in the rye by PCR (Table 2, ScCS4-cDNA–F and ScCS4-cDNA-R).
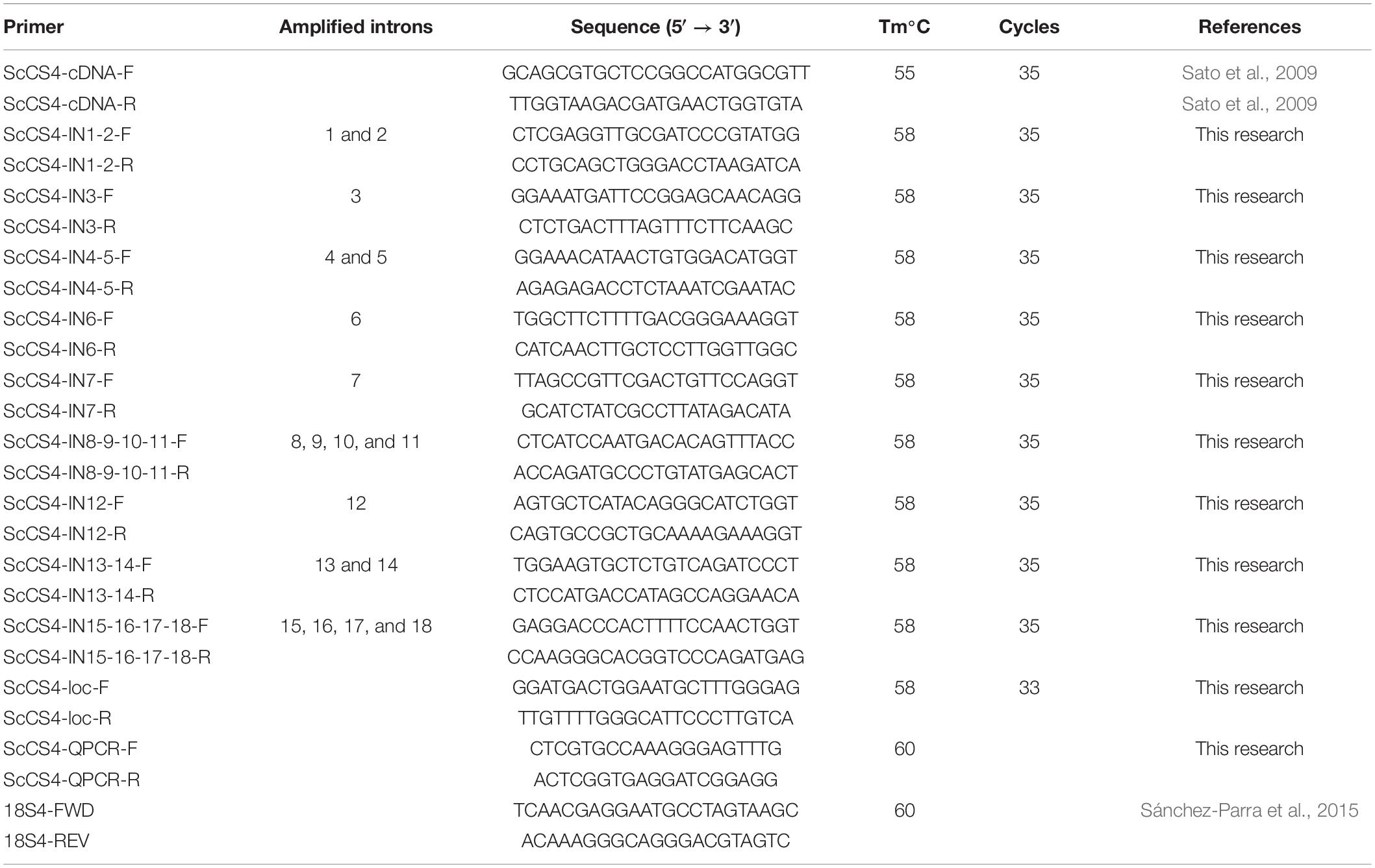
Table 2. Sequences of the primers used in this work, annealing temperature (Tm) and number of cycles used in the PCR.
The PCR was performed using 20 μl of the reaction mixture containing 30 ng of DNA, 5 pmols of each primer, and 10 μl of Taq PCR Master Mix (Qiagen). Amplifications were carried out in the thermal cyclers PTC-100 (MJ Research) with the following program: an initial step of 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C followed by a final extension step of 7 min at 72°C. PCR products were visualized on 1.5–2% TAE agarose gels.
The PCR product was cloned into the TOPO TA Cloning Kit according to the conditions specified by the supplier (Invitrogen). The sequences were identified via an ABI PRISM 3700 Genetic Analyzer. Nine pairs of primers (from ScCS4-IN1-2-F to ScCS4-IN15-16-17-18-R) were designed on successive exons (Figure 1A) to amplify different introns to obtain the genomic sequence of the ScCS4 gene (Table 2). The PCR mixture was the same as described previously but nine pairs of primers were designed to amplify the introns. The PCR program used was also the same as described previously but the annealing temperature was 58°C (Table 2).
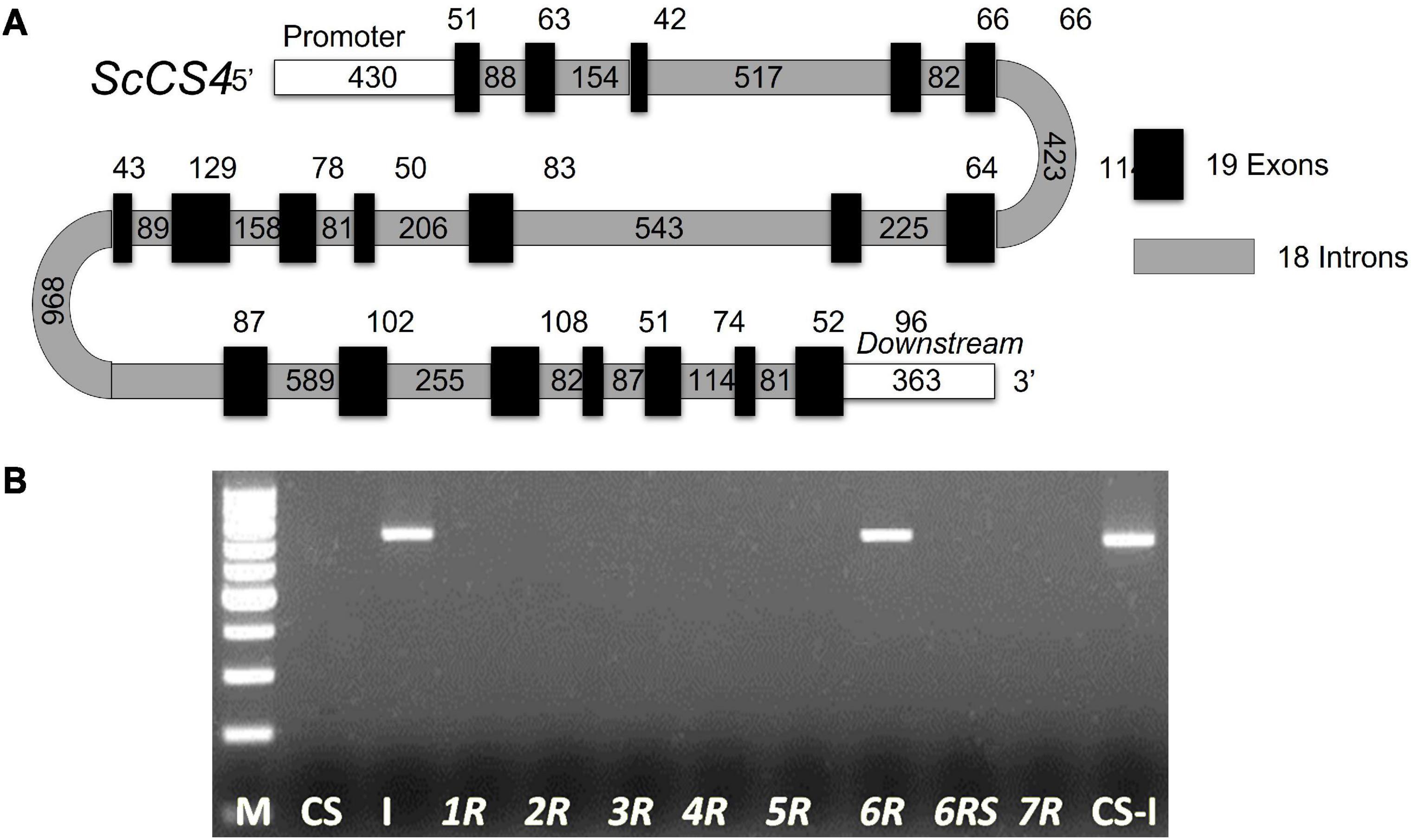
Figure 1. (A) Schematic representation of the ScCS4 gene. This gene has 19 exons and 18 introns. The exons are depicted as black boxes, and the introns are depicted as gray boxes. The numbers indicate their respective sizes in bp. A 430-bp fragment corresponding to the upstream (promoter) region and a 363-bp fragment corresponding to the downstream region were isolated. (B) Chromosomal location of the ScCS4 gene. CS: Chinese Spring (T. aestivum). I: Imperial (S. cereale). CS-I: Wheat–rye amphiploid. 1R, 2R, 3R, 4R, 5R, 6R, and 7R: Wheat-rye disomic addition lines (Chinese Spring–Imperial). 6RS: rye chromosome arm 6RS ditelosomic addition line. M: molecular marker.
The secondary structures of the hypothetical rye proteins were obtained using the PSIPRED protein structure prediction server,1 while the SWISS-MODEL program was used to create a three-dimensional model of the hypothetical rye ScCS protein. In this regard, Jmol software2 was used to generate a protein model for ScCS protein (cultivar Petkus).
Two pairs of primers (Table 2, ScCS4-loc-F, and ScCS4-loc-F) were designed based on the sequences of ScCS4 of Imperial rye cultivar and TaCS4 of T. aestivum to locate the ScCS4 gene. This pair of primers was developed just to amplify in Imperial rye but not in wheat. The PCR conditions (reaction mix and program) were the same as described previously but with 33 cycles and 58°C annealing temperature (Table 2). To locate the ScCS4 gene, gDNAs of wheat-rye disomic and ditelosomic lines were extracted from young leaves with the DNeasy Plant Mini Kit (Qiagen).
Different parameters (NS: number of sites, INDEL: insertion-deletion polymorphism, IMS: invariable monomorphic sites, VPS: variable polymorphic sites, SVS: singleton variable sites, PIS: parsimony-informative sites, CS: sequence conservation, SC: synonymous changes, NSC: non-synonymous changes; Supplementary Table 1) were computed to describe the genetic diversity for ScCS4 among rye cultivars and some Poaceae species using DnaSP software v5.
In addition, using MEGA X software (Tamura et al., 2013), phylogenetic relationships between several CS proteins from Poaceae and other eudicot species were studied using different approaches, such as the amino acid substitution model and the neighbor-joining clustering method. The dendrogram’s robustness was examined using bootstraps with 10,000 replicates.
For expression analyses, 30 seeds were always germinated and grown per treatment. The seeds were sterilized with HgCl2 (0.1%) for 10 min, washed several times with de-ionized water, and incubated overnight on moist filter paper in the dark at 4°C in Petri dishes. Later, the seeds were incubated for two more days in the dark at 20°C in Petri dishes. Germinated seeds were placed on a nylon mesh floating on a continuously aerated nutrient solution containing 0.4 mm CaCl2, 0.65 mm KNO3, 0.25 mm MgCl2⋅6H2O, 0.01 mm (NH4)2SO4, and 0.04 mm NH4NO3 (pH 4.0). Containers with this aerated nutrient solution were placed in a growth chamber at 20°C under 16-h day illumination, a light intensity of 12 W/m2, and 60% of humidity (Abd El-Moneim et al., 2015). The seedlings of the cv. Petkus and the Riodeva inbred line of S. cereale were exposed to five different nutrient solutions containing 0, 25, 50, 100, and 300 μM AlK (SO4)2, respectively. Meanwhile, the same number of seedlings were exposed to 300 μM AlK (SO4)2 at different times (0, 4, 8, 12, and 24 h). In the case of ABR1 and ABR8 cultivars of B. distachyon, the seedlings were exposed to a nutrient solution containing 0 and 20 μM AlK (SO4)2 for 24 h. Following that, the root apices (15 mm) and leaves were excised and frozen in liquid nitrogen. TissueLyser II (Qiagen) and 5-mm Stainless Steel Beads were used to homogenize the samples. Total RNA was extracted from leaves and roots from 30 plants per genotype and exposure time using TRIzol and a PureLink RNA Mini Kit. A NanoDrop ND-1000 spectrophotometer was used to determine the quality of the RNA. A High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to reverse transcript (2 μg) the total RNA. Depending on cDNA sequences from the previously isolated ScCS4 gene and the DOE-JGI genome sequence for BdCS4 gene of B. distachyon accession Bd21, specific primers were designed with Oligo software (Table 2, ScCS4-QPCR-F, and ScCS4-QPCR-R). Furthermore, the 18S gene’s mRNA (Table 2, 18S-FWD, and 18S-Rev) was used as a housekeeping gene. The qRT-PCR was performed on an Applied Biosystems 7900 HT fast real-time PCR device under the following conditions: one stage at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All PCR samples and controls were prepared in duplicates. All 96-well plates contained two standard curves (target gene and endogenous control) for the use of the “relative standard curve approach” for data analysis. Unknown sample values are interpolated from the standard curves in this method. To assure that PCR products were not attributable to amplification of contaminant gDNA, duplicate control reactions without reverse transcription were included for each sample.
A 1,419-bp cDNA fragment (including the stop codon) was amplified from the roots of Ailés, Imperial, Petkus, and Riodeva mRNA treated with Al for 24 h using primers of the HvCS4 gene (Table 2, ScCS4-cDNA–F, and ScCS4-cDNA-R). The rye fragments from Ailés, Imperial, Petkus, and Riodeva were of the same size as that found in barley HvCS4 gene. Eight samples of rye cDNA fragments were cloned (one from Ailés, four from Imperial, two from Petkus, and one from Riodeva), sequenced, and compared among them (Supplementary Data 1) and with other plant species’ sequences (Supplementary Data 2). These eight cDNA fragments were coded for putative CS enzymes with 472 amino acids (Supplementary Data 1A). These sequences are in the NCBI (GenBank accession numbers from OM718863 to OM718870).
The ScCS4 rye cDNA sequence of Ailés shared 99% identity with T. aestivum TaCS4 (Accession No. AK455004.1), 98% identity with Aegilops tauschii AetCS4 (Accession No. XM020301802.1), 97% identity with barley HvCS4 cDNA (Accession No. AK248736.1), 94% with B. distachyon (Accession No. XM003571152.3), and 88% of Oryza sativa OsCS4 cDNA (Accession No. XM015762564.1).
The genomic sequence of the ScCS4 gene from the Ailés cultivar was obtained using nine primer pairs designed in successive exons (Table 2, from ScCS4-IN1-2-F to ScCS4-IN15-16-17-18-R) in order to amplify the corresponding introns. The predicted location of the different rye ScCS4 introns was described by comparison of the rye cDNA sequence with the rye genomic sequence and with the orthologous B. distachyon gene. The ScCS4 gene displayed a total of 19 exons and 18 introns. Exon and intron sizes are shown in Figure 1A.
We analyzed the variability among 19 exons in the ScCS gene’s cDNAs using the eight different sequences (different clones) obtained: one from Ailés, four from Imperial, two from Petkus, and one from Riodeva (Supplementary Data 1, 1A). Moreover, we carried out a diversity analysis in each of the 19 exons of CS4 in rye, comparing to several orthologous exons from other Poaceae and eudicots species (exon sequences of six species of Poaceae) (Supplementary Data 2).
In rye cDNA sequences, only exons 4, 5, 11, 14, 15, 17, and 19 showed SNPs, being exon 17 the most variable (Supplementary Table 1). Twelve SNPs of the ScCS4 gene were detected; four and two were exclusive for Imperial (clone Imperial 5) and sensitive inbred line (Riodeva), respectively, while one SNP was exclusive for Imperial (clone Imperial 1), Imperial (clone Imperial 4), Petkus (clone Petkus 2), and Petkus (clone Petkus 9) (Supplementary Table 1). Two non-synonymous changes (H62Y and K343R) were observed in the ScCS4 gene of Riodeva (Supplementary Data 1A). We used the PROVEAN software3 to predict whether these changes have an impact on the biological function of the ScCS4 protein. The results obtained indicate that both changes are neutral (prediction cutoff −2.5) (Supplementary Figure 1).
In the comparisons between Poaceae (Supplementary Table 2), exon 9 revealed the highest average SNPs per 100 nucleotides (30). Furthermore, all the exons were variable, the total number of SNPs was 329 (Supplementary Tables 2, 3), and the nucleotide diversity was 0.10097 (Supplementary Table 3). Regarding the changes in the hypothetical proteins encoded by the ScCS gene, the number of synonymous changes was higher than that of non-synonymous changes in comparing Poaceae species. However, the situation was reversed in the case of rye sequences, where the number of non-synonymous changes was higher than that of synonymous changes (Supplementary Table 3). Likewise, the degree of conservation between the rye sequences (0.992) was much higher than the degree of conservation between the Poaceae sequences (0.768), as expected for different species. However, the nucleotide diversity among studied rye sequences was 0.00269, lower than the nucleotide diversity (0.10097) among different Poaceae sequences (Supplementary Table 3). It was noted that no insertions or deletions (Indels) were detected among all the studied cDNA sequences of rye as well as among cDNA sequences of Poaceae species.
A specific band of Imperial (I) rye cultivar was amplified using a CS4 specific pair of primers (Table 2, ScCS4-loc-F and ScCS4-loc-F) in this cultivar and in the addition line Chinese Spring-Imperial (CS-I) with the chromosome 6R. However, this band was not observed in the addition line (CS-I) with the chromosome arm 6RS. Therefore, this gene was located by disomic and ditelosomic wheat-rye addition lines on the chromosome arm 6RL (Figure 1B) of the Imperial rye.
The secondary and tertiary structures of the hypothetical rye proteins (Supplementary Data 3) were obtained using PSIPRED v3.0 and SWISS-MODEL programs, respectively. These programs indicated that ScCS4 is a mitochondrial rye protein. The CS domain has 23 α-helix regions with high predictive confidence and 3 β-sheet regions (Supplementary Figure 1); the first observed β-sheet region (in yellow color and without assigned number) has a very low predictive confidence. Only four amino acid substitutions were detected after comparing the four hypothetical rye proteins obtained in this study. However, when comparing them with the barley sequence (AK248736) and rice sequences (OsCSY5 and OsCS), we found 5, 30, and 33 amino acid substitutions, respectively. The SWISS-MODEL software identified (with a resolution of 2.00 Å) an A. thaliana CS (6k5v.1.A) as a template for all rye mitochondrial CS sequences (Nishio and Mizushima, 2020). In all cases, the percentage of identity in the amino acid sequence was 83.48%. The predicted hypothetical quaternary structure is a homo-dimer. The generated protein model for ScCS protein (cultivars Ailés and Petkus) is shown in Supplementary Figure 1. Nishio and Mizushima (2020) have described three functionally important residues (His308, His354, and Asp409) in A. thaliana. In our rye sequences, the equivalent residues are His307, His353, and Asp408. These residues are conserved in all rye cultivars and Riodeva line. Several disulfide bridge-forming residues and other cysteine residues are conserved in all rye cultivars (Cys 108, Cys209, Cys 365, and Cys 467). The two non-synonymous changes (H62Y and K343R) observed in the Riodeva ScCS4 protein (Supplementary Data 1A) do not alter the secondary and tertiary structures of the protein. Therefore, these changes probably do not affect the correct functioning of the enzyme.
We used two methods to confirm that the isolated rye gene is the ortholog of the mitochondrial CS genes of rice and barley (which were employed to design the primers used to amplify rye sequence). First, we studied the phylogenetic relationships among the cDNAs of ScCS4 gene and other CS4 Poaceae genes. Second, we studied the phylogenetic relationships among the amino acids of the putative ScCS4 protein and the amino acid sequences of other CS4 Poaceae proteins that are available in the NCBI databases (Supplementary Data 2). To ensure that the dendrogram structure and the different clusters obtained were consistent, different models of nucleotide substitution genetic distances for cDNAs, amino acid substitutions for proteins, and clustering methods were used. All the phylogenetic trees obtained always gave dendrograms with the same structure and very similar bootstrap values. Figure 2 showed the dendrogram obtained using cDNA sequences. The rye sequences are grouped in the same subcluster with those of other Triticines, such as wheat and barley. Maize and sorghum are grouped into another subcluster.
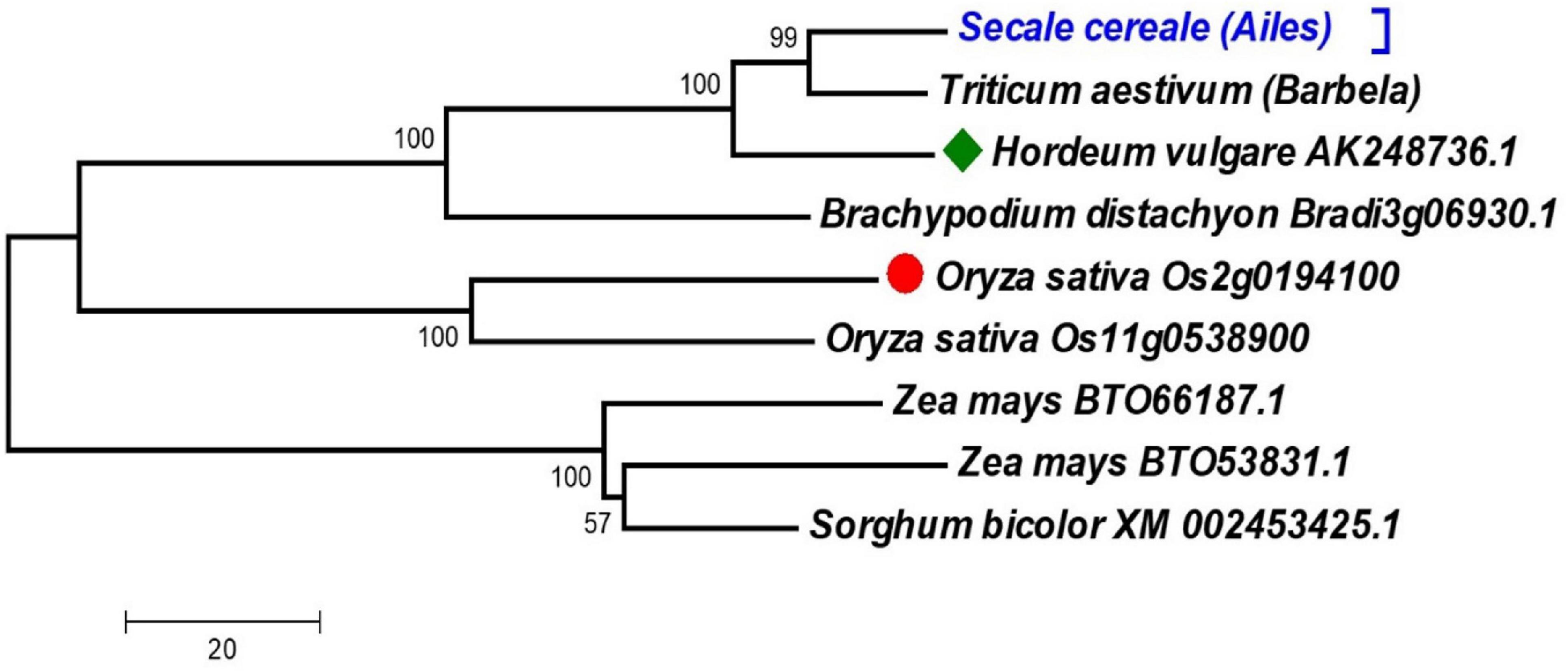
Figure 2. The dendrogram was obtained from the sequences of cDNAs of ScCS4 gene and CS genes of seven different Poaceae species. The studied sequences of cDNAs and proteins were extracted from NCBI and Phytozome. Bootstraps with 10,000 replicates were calculated to test the robustness of the dendrogram. The red circle indicates the starting rice sequence and the green diamond indicates the barley sequence. The ScCS4 sequence obtained has been indicated with light blue text.
In the same context, Supplementary Figure 2 showed the phylogenetic tree obtained by various CS4 proteins belonging to rye and other Poaceae and eudicots species (Supplementary Data 4). The CS4 proteins were assembled in two clusters, one with the monocot species and the other with the eudicot species. The monocot cluster has two subclusters, one of them containing the rye ScCS4 protein and the CS4 proteins of T. aestivum (from the genomes A, B, and D), T. urartu, Ae. tauschii, H. vulgare, B. distachyon, and B. stacei. The other subcluster groups the CS4 proteins of O. sativa, O. brachyantha, Sorghum bicolor, Zea mays, Panicum virgatum, P. hallii, Setaria italica, and S. viridis. The eudicots cluster also has two subclusters. One of them contains the Brassicaceae species (B. napus, B. oleracea, B. rapa, A. thaliana, A. lyrata, Raphanus sativus, Eutrema salsugineum, Camelina sativa, and Capsella rubella). The other subcluster contains a Malvaceae species (Theobroma cacao), three Rosaceae and Rhamnaceae species (Prunus sibirica, P. persica, and Ziziphus jujuba), and two Cucurbitoidae species (Cucumis sativus and C. melo).
Both the tolerant cultivar Petkus and the sensitive inbred line Riodeva had higher levels of ScCS4 transcripts in their roots than that in their leaves with Al (+) and without Al (−) after 24 h of treatment. This disparity was most noticeable and significant in the roots of plants that had not been exposed to Al (Figure 3A). Petkus roots have 50 times more ScCS4 transcripts than leaves in the absence of Al during 24 h (−). However, Petkus roots have about 7.1-fold more ScCS4 transcripts than the leaves after being exposed to Al for 24 h (+) (Figure 3A). Similar findings have been observed for the roots of Riodeva inbred line. Without Al, the roots of Riodeva have almost 4.7-fold more ScCS4 transcripts than the leaves. Instead, Riodeva roots have 2.2-fold more ScCS4 transcripts than leaves in the presence of Al (+) (Figure 3A). Moreover, ScCS4 expression was seven times higher in the roots of Petkus than those of Riodeva without Al (−) (Figure 3B). Roots treated with Al for 24 h (+) obtained similar findings: the ScCS4 expression was 3.4-fold higher in Petkus than in Riodeva (Figure 3B). This higher amount of mitochondrial CS transcripts in Petkus than in Riodeva could explain part of the Al tolerance of Petkus. However, without Al, the ScCS4 expression levels in leaves were similar in Petkus and Riodeva (Figure 3B). With Al, the expression level in Riodeva leaves is slightly higher than that in Petkus (Figure 3B). ScCS4 transcription in roots was diminished after exposure to Al in both Petkus and Riodeva, with higher gene repression in Petkus than Riodeva, according to time-course studies (Figures 4A,B). Furthermore, ScCS4 transcription in roots decreased following 24 h of exposure to various Al doses (25, 50, 100, and 300 μM) (Figures 4C,D).
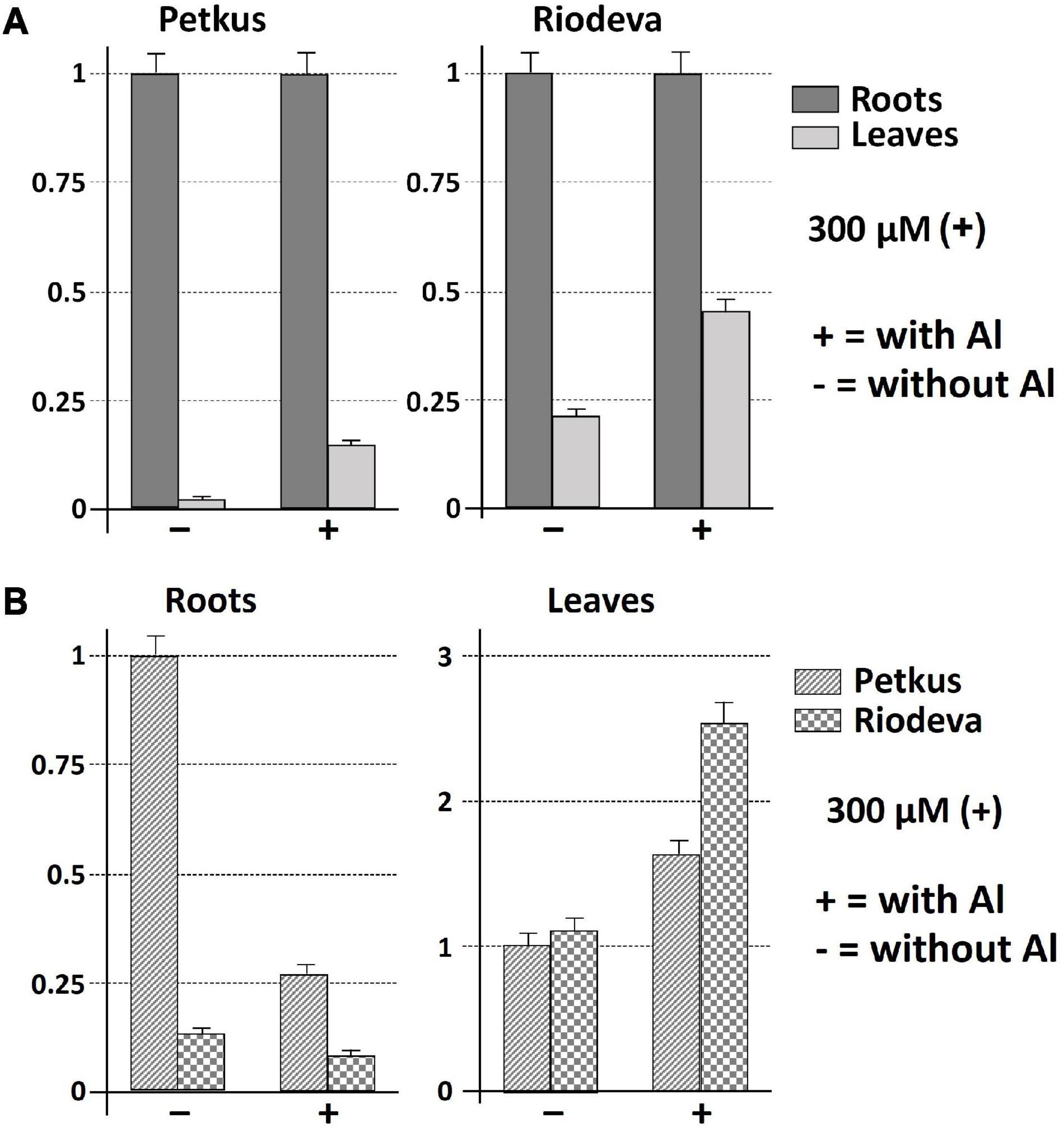
Figure 3. Expression patterns (qRT-PCR) of ScCS4 under 300 μM AlK (SO4)2 at 24 h of treatment with AlK (SO4)2 (+) and without AlK (SO4)2 (−) using root apices and leafs of Riodeva (sensitive inbred line) and Petkus (tolerant cultivar). The expression level of 18S rRNA was used as a loading control. Error bars represent standard deviations (SDs). All data are the means of three independent biological and technical replicates. (A) ScCS4 expression level of leaves compared with roots in Petkus and Riodeva. The relative expression level 1 for Petkus roots with and without aluminum and the relative expression level 1 for Riodeva roots with and without aluminum do not mean that there is the same amount of citrate synthase messenger RNA in the different situations. In all four cases, the roots have been selected as a reference. (B) ScCS4 expression level of Riodeva compared with Petkus in roots and leaves. Change at each time point is expressed as the relative difference in expression compared with roots of the Petkus without Al or with leaves of the Petkus without Al. Y-axis: change (fold difference) at each time point is expressed as the relative difference in expression compared with the case selected as reference (relative expression level 1).
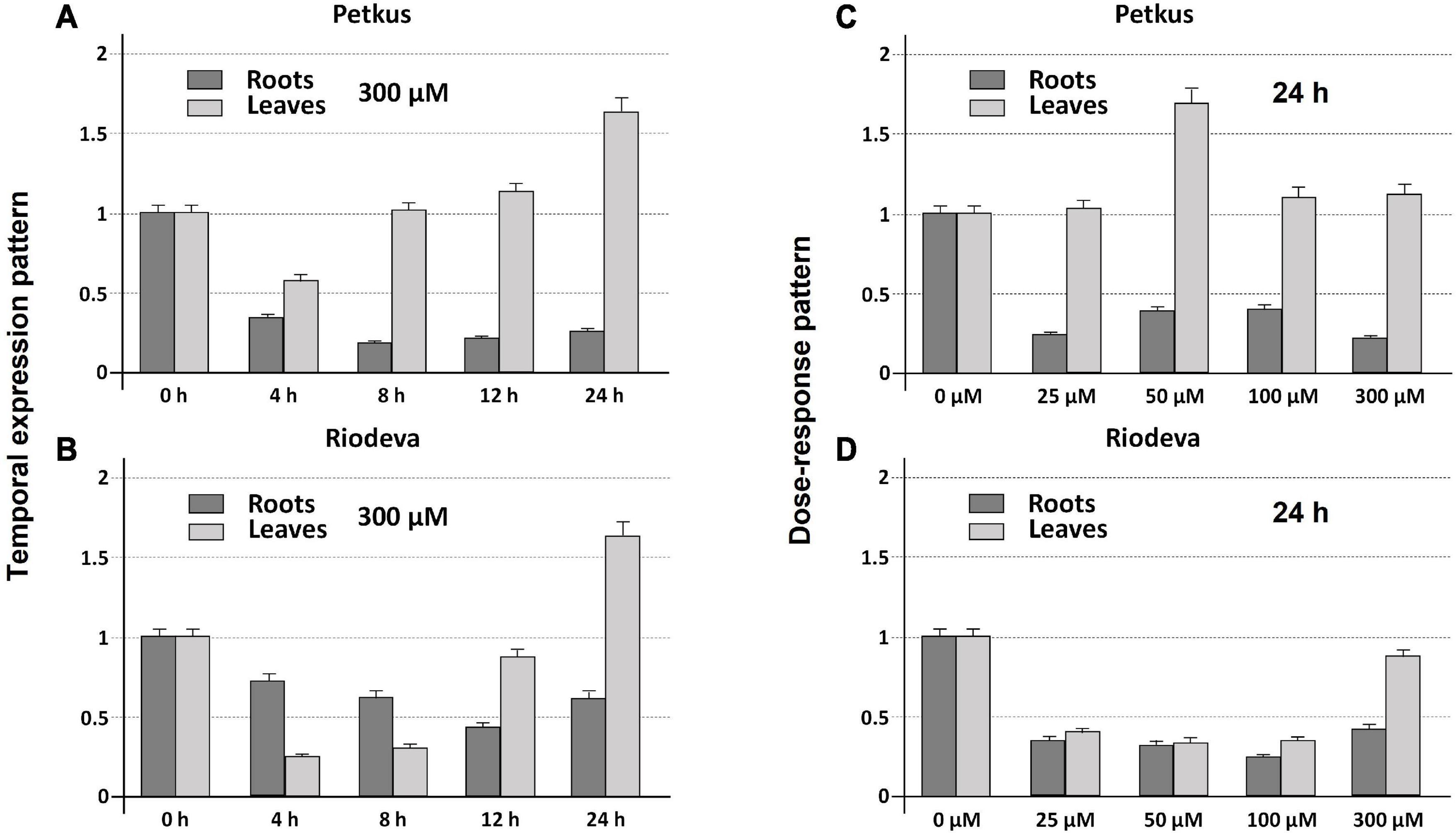
Figure 4. Temporal expression (A,B) and dose-response patterns (C,D) (qRT-PCR) of root apices and leaf cDNA transcripts of ScCS4 gene from the Petkus (tolerant cultivar) (A,C) and Riodeva (sensitive inbred line) (B,D) in the absence of Al (0 h) or with 300 μM of AlK (SO4)2 at 4, 8, 12, and 24 h (temporal expression patterns) or with Al at 0, 25, 50, 100, and 300 μM for 24 h (dose–response patterns). Change is expressed as the relative difference in expression without Al (0 h or 0 μM) compared with each time point. The expression level of 18S rRNA was used as a loading control. Error bars represent standard deviations (SDs). All data are the means of three independent biological and technical replicates. Y-axis: Change (fold difference) at each time point is expressed as the relative difference in expression compared with the case selected as reference (relative expression level 1).
On the contrary, the levels of BdCS4 transcripts were consistently higher in roots than leaves of both the resistant (ABR8) and sensitive (ABR1) Brachypodium lines, in the absence of Al (−) and in the presence of Al for 24 h (+) (Figure 5). The BdCS4 expression level in the sensitive ABR1 roots without Al was slightly higher in comparison with that of the tolerant ABR8 (Figure 5). mRNA transcript levels of ScCS4 in Petkus roots with Al were like that found for BdCS4 in both Brachypodium lines (Figure 5). The BdCS4 transcription level in roots reduced upon exposure to Al in both the tolerant ABR8 and sensitive ABR1 lines, with higher gene repression in ABR1 than in ABR8 (Figure 5). However, both lines with and without Al have similar levels of transcription in their leaves.
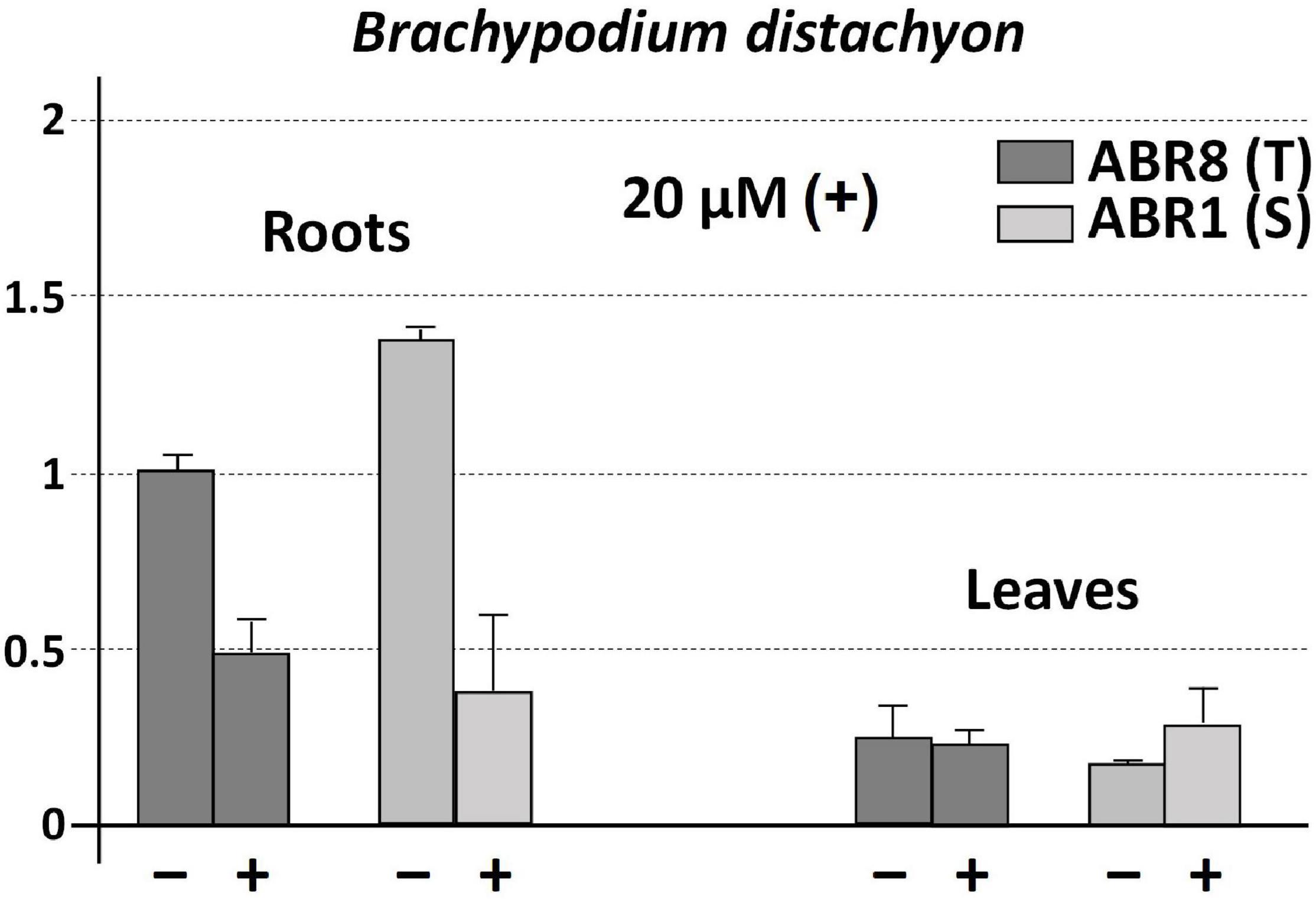
Figure 5. Expression patterns (qRT-PCR) of BdCS4 at 24 h with 20 μM AlK (SO4)2 (+) and without AlK (SO4)2 (−) treatments using root apices and leaf of ABR1 (sensitive line) and ABR8 (tolerant line) of B. distachyon. The expression level of 18S rRNA was used as a loading control. The change at each treatment is expressed as the relative difference in expression compared with the tolerant genotype diploid (ABR8) without exposure to Al (−). Error bars represent standard deviations (SDs). All data are the means of three independent biological and technical replicates.
The study of identity and similarity of cDNA sequences of the hypothetical protein encoded by them (Supplementary Data 1, 1A, 2) and phylogenetic relationships indicated that the sequences isolated and characterized in rye are orthologous to the barley sequence used to design the primers. Furthermore, the isolated exon and intron structures (Figure 1A) match the structure expected in most cases for the orthologous gene of B. distachyon. Moreover, ScCS4 was predicted to be a mitochondrial protein. This result is consistent with that expected for mitochondrial CS proteins of the Poaceae.
Recently, advances in rye genomics have achieved the sequencing of two genomes of S. cereale L. (Bauer et al., 2017; Li et al., 2021). In the rye genome sequence database,4 we have found a cDNA and a protein sequence (SECCE6Rv1G0388260.1) that have 100 and 99.789% identity with our Imperial 2 and Ailes 1 sequences, respectively. Therefore, the sequences isolated in our different rye cultivars match the sequence of the rye genome database.
Our ScCS4 chromosomal location on chromosome arm 6RL (Figure 1B) agrees with the previously mentioned syntenic relationships between wheat, barley, rice, Brachypodium, and rye (Naranjo et al., 1987; Devos, 2005; The International Brachypodium Initiative, 2010; Martis et al., 2013). As a result, our data support the idea that the ScCS4 gene isolated from rye is orthologous to the T. aestivum genes TaCS4 (TaCS4-A, TaCS4-B, and Ta CS4-D), HvCS4 (H. vulgare), OsCS4 (O. sativa), and BdCS4 (B. distachyon) (Figure 2 and Supplementary Figure 2). In addition, the Al tolerance gene Alt1 has been located on chromosome 6R in S. cereale L. (Gallego and Benito, 1997), and the ScMATE3 gene, probably involved in Al detoxification mechanisms, is located on the same chromosome (Santos et al., 2020).
The ScCS4 gene is located on a different chromosome (6R) than the two tolerance genes that have been detected in our crosses between sensitive and tolerant rye plants in previous publications (Fontecha et al., 2007; Silva-Navas et al., 2012). These two candidate tolerance genes, namely, ScALMT1 and ScMATE1, are on the chromosome arm 7RS (Fontecha et al., 2007; Collins et al., 2008; Silva-Navas et al., 2012).
Genetic diversity studies (Supplementary Tables 1, 2) in the Secale genus are essential to better characterize its genome for breeding purposes and even within S. cereale. This genus is one of the most tolerant to Al stress (Santos et al., 2018). All the estimated diversity parameters, including nucleotide diversity, were always higher between Poaceae species than between S. cereale. Our results indicated that the studied gene is highly conserved during evolution. The same is observed for other genes that encode the tricarboxylic acid cycle enzymes (Abd El-Moneim et al., 2015). This could explain the high degree of conservation detected among Poaceae species, which was even higher among rye samples. However, Santos et al. (2016, 2018) found significant genetic variability among ScMATE1 cDNA sequences within the subspecies of S. cereale and even higher in the whole Secale genus. The obtained dendrograms using cDNA nucleotide (Figure 2) and amino acid sequences (Supplementary Figure 2) from rye and B. distachyon are grouped in the same cluster as the HvCS4 cDNA and the CS4 protein from barley (this last cDNA sequence was used to design the primers). These findings support the hypothesis that both the ScCS4 gene and protein of rye isolated in this research are orthologous to the CS4 genes and proteins of T. aestivum, T. urartu, A. tauschii, H. vulgare, B. distachyon, and B. stacei. Moreover, these findings agree with the chromosomal location results and coincide quite well with the phylogeny of the species included in our study.
In response to Al stress, many Al-tolerant species secrete organic acids from their roots. Triticale, B. napus, Avena sativa, R. sativus, and S. cereale and some tolerant varieties of T. aestivum, B. distachyon, and B. hybridum exudate citrate and malate. Malate is released by Al-tolerant cultivars of P. vulgaris, Z. mays, C. tora, A. thaliana, and G. max. Citrate is exudated by the Al-tolerant cultivars of H. vulgare. Finally, Fagopyrum esculentum and Colocasia esculenta exudate oxalate. However, this classification is in permanent change due to the new findings in different plant species. For instance, the roots of P. vulgaris and G. max also exudate citrate in response to Al treatment (Yang et al., 2000; Rangel et al., 2010). Moreover, another essential factor that should not be forgotten is intraspecific variability. The roots of several varieties of T. aestivum release only malate in response to Al stress, others only citrate, and some both organic acids simultaneously (Ryan et al., 2009; Garcia-Oliveira et al., 2016, 2018). A similar situation can be expected in other plant species. In allogamous species, the cultivars are constituted by plants with different genotypes. Therefore, our ability to detect the different organic acids exuded by the roots depends largely on the quantity of varieties and/or lines analyzed of a given species. So far, the S. cereale cultivars and lines studied exude malate and citrate from the roots, although in different amounts. However, because rye is an allogamous species, internal variability in the cultivars is possible since the plants would have different genotypes. Therefore, there could be plants that exude only malic or citric acids and others that exude both organic acids simultaneously. Considering that, in most of the studies on organic acid exudation in rye, pools of at least 15 or 20 plants were analyzed, it may be that some of them exude only malate or only citrate and others, both organic acids. To solve this problem, it would be necessary to analyze the exudation of each plant separately.
Consequently, the possible relationship between citrate exudation by the roots in response to Al treatment and changes in the CS enzymatic activity or CS messengers’ expression could be affected.
As already indicated in the introduction of this work, the changes in the enzymatic activity of CS produced by Al stress are different in the plant species studied. The Al treatment increases CS activity in S. cereale (Li et al., 2000), P. vulgaris (Mugai et al., 2000), and C. tora (Yang et al., 2004), and the roots of these three species release citrate. However, the CS activity does not increase in T. aestivum and triticale in their sensitive and tolerant lines (Li et al., 2000; Hayes and Ma, 2003). The Al-tolerant varieties of T. aestivum usually exudate only malate, and some of them release citrate. Since the wheat variety used by Hayes and Ma (2003) only releases malate, it is expected that an increase in the CS enzymatic activity will not be found. In soybean, Al stress enhances mCS gene expression and enzyme activity of CS (Eticha et al., 2010), and their roots release citrate and, in addition, malate. Instead, the CS activity decreases after Al treatment in C. arabica (Ramírez-Benítez et al., 2008). Another critical factor is that all these plant species have different genes codifying CS enzymes, some of them expressed in the cytosol and one in the mitochondria. Thereby, the enzymatic activity estimated in the previously cited works is due to the sum of activities of these different CS enzymes.
A crucial issue is the citric acid exudation pattern. There are plant species whose roots are always exuding citric acid, in the absence and in the presence of Al, showing a constitutive pattern. However, in other species, citric acid’s exudation is induced after treatment with Al, showing an inducible pattern. In rye, the inbred lines Riodeva and P105 show a constitutive citrate exudation pattern (Santos et al., 2018). However, the tolerant cultivars Imperial and Petkus have an inducible pattern of citrate exudation (Abd El-Moneim et al., 2014, 2015; Sánchez-Parra et al., 2015). In addition, different species of the Secale genus show different citric acid exudation patterns (Santos et al., 2018). The intraspecific variability of the exudation pattern has been less studied in plant species. The allogamous cultivars of S. cereale have plants with different genotypes; some could show a constitutive exudation pattern, and others could show an inducible pattern. Considering this, when analyzing the citric acid exudation pattern using groups of 15–20 plants, the expected result, if there are plants of both types, could be an inducible pattern. For B. distachyon, roots of sensitive line ABR1 show low citrate exudation without Al, and no changes were observed by Al treatment. However, roots of the tolerant line ABR8 show a clearly Al-inducible citrate exudation pattern (Contreras et al., 2014).
Changes in CS gene expression due to Al treatment are also variable depending on the species or whether plants are sensitive or tolerant to Al. CS expression does not change significantly due to Al treatment in both P. vulgaris (Eticha et al., 2010) and A. thaliana (Kumari et al., 2008). However, overexpression of mCS genes led to increased citrate efflux in carrot, A. thaliana, and canola cells (Koyama et al., 1999, 2000; Anoop et al., 2003), respectively.
As mentioned in the introduction, overexpression of a bacterial CS gene in transgenic tobacco and papaya plants resulted in improved Al tolerance and citrate overproduction (de la Fuente et al., 1997). However, Delhaize et al. (2001), using transgenic tobacco plants, did not observe these results and reported that two transgenic alfalfa plants expressing CS did not show an improvement in Al tolerance. On the contrary, Barone et al. (2008) detected transgenic alfalfa plants with more Al tolerance than non-transgenic control. The rice mitochondrial gene OsCS1 was induced by Al treatment, and several tobacco transgenic lines expressing OsCS1 showed increased citrate efflux and high Al tolerance (Han et al., 2009). Finally, the Al-inducible expression of mitochondrial CS together with enhanced citrate release mediates Al resistance in the tree P. falcataria (Osawa and Kojima, 2006).
It is the first time that repression of the mCS gene following Al treatment was observed in S. cereale and B. distachyon, although repression of other genes related to organic acid metabolism (Abd El-Moneim et al., 2015) or related to Al stress (Sánchez-Parra et al., 2015) has been previously detected in rye.
This gene repression could be indirectly associated with malate and citrate exudation. When the mitochondrial genes MDH (malate dehydrogenase) and FUM (fumarase) were repressed in transgenic Solanum lycopersicum plants, a decrease in both root length and exuded pH was observed, and the exudation of malate and citrate was activated (van der Merwe et al., 2009). Thus, increased root exudation of organic acids may be associated with decreased pH. In yeast and transgenic B. napus, Anoop et al. (2003) found that the regulation of several enzymes involved in citrate synthesis and turnover, such as MDH, CS, ACO (aconitase), and IDH (isocitrate dehydrogenase), could be recognized as potential targets for gene manipulation to understand better the role of citrate metabolism in mediating Al tolerance. Another vital aspect that could be related to mitochondrial CS expression is the exudation of citrate by the roots carried out by citrate-transporting membrane proteins that may or may not be activated by aluminum treatment.
At least two different genes, probably involved in root citrate exudation and Al tolerance, namely, ScMATE1 (ScFRLD1 or ACCT1) and ScMATE2 (ScFRLD2), have been reported in rye (Yokosho et al., 2010; Silva-Navas et al., 2012). The ScMATE1 is induced by Al treatment in the sensitive Riodeva inbred line (Silva-Navas et al., 2012), but not in the IR51 rye inbred line (Yokosho et al., 2010). The transcription factors ScSTOP1 and TaSTOP1 seem to be necessary for the expression of the genes ScALMT1, ScMATE1 (from rye), TaALMT1, and TaMATE1 (from wheat) (Garcia-Oliveira et al., 2013; Silva-Navas et al., 2021). The ScMATE1 gene is Al-induced in other tolerant rye cultivars like Imperial, Petkus, and the inbred line 2,672/4. However, Al stress does not induce this gene in Ailés tolerant rye cultivar. The tolerance of the Ailés cultivar is mainly due to the presence of an allele of the ScALMT1 gene that codes for a malate transporter activated by Al (Fontecha et al., 2007; Collins et al., 2008). The ScMATE1 allele present in the sensitive inbred line Riodeva is a tolerant allele (Silva-Navas et al., 2012). This contradictory finding is explained because the roots of the inbred line Riodeva can grow at concentrations between 100 and 150 μM. Therefore, inbred line Riodeva compared with other rye cultivars and lines whose roots grow at concentrations higher than 300 μM is a sensitive rye. However, Riodeva is tolerant compared with other plant species such as H. vulgare, T. aestivum, T. turgidum, A. thaliana, and B. distachyon whose roots do not grow at concentrations higher than 100 μM.
Repression of ScCS4 and BdCS4 mRNA transcription in response to Al in S. cereale and B. distachyon roots indicates that manipulation of mitochondrial CS gene expression can enhance Al tolerance in rye and B. distachyon and that overexpression of these genes in sensitive lines can also improve their tolerance.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
CB designed and performed this research. DAE-M and RC obtained the rye CS sequences and the expression data. FG and JS-N analyzed the sequences and carried out the phylogenetic analyses. AF revised this draft by rewriting the discussion and commenting. All authors commented on the manuscript.
This study was funded by the Spanish Ministry of Education and Science (AGL2008-03049/AGR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank A.J. Lukaszewski for providing the wheat-rye addition lines.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.832981/full#supplementary-material
Supplementary Figure 1 | On the left, the hypothetical ScCS protein secondary structure was obtained with PSIPRED v3.0 (http://bioinf.cs.ucl.ac.uk/psipred/). The secondary structure is made up of 23 α-helix regions and 3 β-sheet regions. On the right, the hypothetical tertiary structure of ScCS from the cultivars Ailés and Petkus. The tertiary structure was obtained with the SWISS-MODEL program. The protein from the Protein DataBase (PDB) showed that the greatest similarity to ours (83.3%) was from Arabidopsis thaliana (with a resolution of 2.00 Å).
Supplementary Figure 2 | The dendrogram was obtained from the sequences of amino acids of the hypothetical ScCS4 protein and CS proteins of various species of Poaceae and other eudicots. The studied sequences of proteins were extracted from NCBI and Phytozome. Bootstraps with 10,000 replicates were calculated to test the robustness of the dendrogram. Secale cereale L. is in red color, the blue lines indicate the Poaceae species, and the red lines indicate some eudicot species.
Supplementary Data 1 | Alignment of ScCS4 cDNA sequences corresponding to eight different sequences (different clones) of S. cereale, one from cv. Ailés, four from cv. Imperial, two from cv. Petkus, and the last from inbred line Riodeva.
Supplementary Data 1A | Alignment of ScCS4 amino acid sequences corresponding to eight different sequences (different clones) of S. cereale, one from cv. Ailés, four from cv. Imperial, two from cv. Petkus, and the last from inbred line Riodeva.
Supplementary Data 2 | Alignment of ScCS4 cDNA sequence corresponding to S. cereale cv. Ailés with those found in other six species of Poaceae.
Supplementary Data 3 | Alignment of ScCS4 putative protein sequences corresponding to the rye cultivars Ailés, Imperial, and Petkus, and the inbred line Riodeva.
Supplementary Data 4 | Alignment of ScCS4 putative protein sequences corresponding to S. cereale cv. Imperial, several Poaceae species, and other plant species.
Abd El-Moneim, D., Contreras, R., Silva-Navas, J., Gallego, F. J., Figueiras, A. M., and Benito, C. (2015). On the consequences of aluminium stress in rye: repression of two mitochondrial malate dehydrogenase mRNAs. Plant Biol. 17, 123–133. doi: 10.1111/plb.12219
Abd El-Moneim, D., Contreras, R., Silva-Navas, J., Gallego, F. J., Figueiras, A. M., and Benito, C. (2014). Pectin methylesterase gene and aluminum tolerance in Secale cereale. Environ. Exp. Bot. 107, 125–133. doi: 10.1016/j.envexpbot.2014.06.006
Anoop, V. M., Basu, U., McCammon, M. T., McAlister-Henn, L., and Taylor, G. J. (2003). Modulation of citrate metabolism alters Aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol. 132, 2205–2217. doi: 10.1104/pp.103.023903
Barone, P., Rosellini, D., Lafayette, P., Bouton, J., Veronesi, F., and Parrott, W. (2008). Bacterial citrate synthase expression and soil Aluminum tolerance in transgenic alfalfa. Plant Cell Rep. 27, 893–901. doi: 10.1007/s00299-008-0517-x
Bauer, E., Schmutzer, T., Barilar, I., Mascher, M., Gundlach, H., Martis, M. M., et al. (2017). Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 89, 853–869. doi: 10.1111/tpj.13436
Collins, N. C., Shirley, N. J., Saeed, M., Pallotta, M., and Gustafson, J. P. (2008). An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 179, 669–682. doi: 10.1534/genetics.107.083451
Contreras, R., Figueiras, A. M., Gallego, F. J., and Benito, C. (2014). Brachypodium distachyon: a model species for aluminium tolerance in Poaceae. Funct. Plant Biol. 41, 1270–1283. doi: 10.1071/FP13362
de la Fuente, J. M., Ramírez-Rodríguez, V., Cabrera-Ponce, J. L., and Herrera-Estrella, L. (1997). Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 276, 1566–1568. doi: 10.1126/science.276.5318.1566
Delhaize, E., Hebb, D. M., and Ryan, P. R. (2001). Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiol. 125, 2059–2067. doi: 10.1104/pp.125.4.2059
Delhaize, E., Kataoka, T., Hebb, D. M., White, R. G., and Ryan, P. R. (2003). Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15, 1131–1142. doi: 10.1105/tpc.009134
Devos, K. M. (2005). Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8, 155–162. doi: 10.1016/j.pbi.2005.01.005
Eticha, D., Zahn, M., Bremer, M., Yang, Z., Rangel, A. F., Rao, I. M., et al. (2010). Transcriptomic analysis reveals differential gene expression in response to Aluminum in common bean (Phaseolus vulgaris) genotypes. Ann. Bot. 105, 1119–1128. doi: 10.1093/aob/mcq049
Fontecha, G., Silva-Navas, J., Benito, C., Mestres, M. A., Espino, F. J., Hernández-Riquer, M. V., et al. (2007). Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.). Theor. Appl. Genet. 114, 249–260. doi: 10.1007/s00122-006-0427-7
Gallego, F. J., and Benito, C. (1997). Genetic control of aluminium tolerance in rye (Secale cereale L.). Theor. Appl. Genet. 95, 393–399. doi: 10.1007/s001220050575
Garcia-Oliveira, A., Martin-Lopes, P., Tolrá, R., Poschendrieder, C., Guedes-Pinto, H., and Benito, C. (2016). Differential physiological responses of portuguese bread wheat (Triticum aestivum L.) genotypes under aluminium stress. Diversity 8:26. doi: 10.3390/d8040026
Garcia-Oliveira, A. L., Benito, C., Guedes-Pinto, H., and Martins-Lopes, P. (2018). Molecular cloning of TaMATE2 homoeologues potentially related to aluminium tolerance in bread wheat (Triticum aestivum L.). Plant Biol. 20, 817–824. doi: 10.1111/plb.12864
Garcia-Oliveira, A. L., Benito, C., Prieto, P., Andrade Menezes, R., Rodrigues-Pousada, C., Guedes-Pinto, H., et al. (2013). Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton (H(+)) toxicity in bread wheat (Triticum aestivum L.). BMC Plant Biol. 13:134. doi: 10.1186/1471-2229-13-134
Han, Y., Zhang, W., Zhang, B., Zhang, S., Wan, W., and Ming, F. (2009). One novel mitochondrial citrate synthase from Oryza sativa l. can enhance Aluminum tolerance in transgenic tobacco. Mol. Biotechnol. 42, 299–305. doi: 10.1007/s12033-009-9162-z
Hayes, J. E., and Ma, J. F. (2003). Al-induced efflux of organic acid anions is poorly associated with internal organic acid metabolism in triticale roots. J. Exp. Bot. 54, 1753–1759. doi: 10.1093/jxb/erg188
Hede, A. R., Skovmand, B., and Lopez-Cesati, J. (2001). “Acid soils and aluminum toxicity,” in Application of physiology in Wheat Breeding, eds M. P. Reynolds, J. I. Ortiz-Monasterio, and A. MacNab (Mexico, DF: CYMMYT), 172–182.
Koyama, H., Kawamura, A., Kihara, T., Hara, T., Takita, E., and Shibata, D. (2000). Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus limited soil. Plant Cell Physiol. 41, 1030–1037. doi: 10.1093/pcp/pcd029
Koyama, H., Takita, E., Kawamura, A., Hara, T., and Shibata, D. (1999). Over expression of mitochondrial citrate synthase gene improves the growth of carrot cells in Aluminum-phosphate medium. Plant Cell Physiol. 40, 482–488. doi: 10.1093/oxfordjournals.pcp.a029568
Kumari, M., Taylor, G. J., and Deyholos, M. K. (2008). Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol. Genet. Genomics 279, 339–357.
Li, G., Wang, L., Yang, J., He, H., Jin, H., Li, X., et al. (2021). A high-quality genome assembly highlights rye genomic characteristic and agronomically important genes. Nat. Genet. 53, 574–584. doi: 10.1038/s41588-021-00808-z
Li, X. F., Ma, J. F., and Matsumoto, H. (2000). Pattern of Aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol. 123, 1537–1543. doi: 10.1104/pp.123.4.1537
Ma, J. F., Ryan, P. R., and Delhaize, E. (2001). Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/s1360-1385(01)01961-6
Mariano, E. D., Jorge, R. A., Keltjens, W. G., and Menossi, M. (2005). Metabolism and root exudation of organic acid anions under aluminium stress. Braz. J. Plant Physiol. 17, 157–172. doi: 10.1590/s1677-04202005000100013
Mariano, E. D., and Keltjens, W. G. (2001). “Exudation of organic acid anions from root apices as an aluminium resistance mechanism in maize,” in Plant Nutrition, eds W. J. Horst, M. K. Schenk, A. Bürkert, N. Claassen, H. Flessa, W. B. Frommer, et al. (Dordrecht: Springer), 494–495. doi: 10.1007/0-306-47624-x_239
Martis, M. M., Zhou, R., Haseneyer, G., Schmutzer, T., Vrána, J., Kubaláková, M., et al. (2013). Reticulate evolution of the rye genome. Plant Cell 25, 3685–3698. doi: 10.1105/tpc.113.114553
Mugai, E. N., Agong, S. G., and Matsumoto, H. (2000). Aluminium tolerance mechanisms in Phaseolus vulgaris L.: citrate synthase activity and TTC reduction are well correlated with citrate secretion. Soil Sci. Plant Nutr. 46, 939–950. doi: 10.1080/00380768.2000.10409159
Naranjo, T., Roca, A., Goicoechea, P. G., and Giráldez, R. (1987). Arm homoeology of wheat and rye chromosomes. Genome 29, 873–882. doi: 10.1139/g87-149
Nishio, K., and Mizushima, T. (2020). Structural and biochemical characterization of mitochondrial citrate synthase 4 from Arabidopsis thaliana. Acta Crystallogr. F: Struct. Biol. Commun. 76, 109–115. doi: 10.1107/S2053230X20001521
Osawa, H., and Kojima, K. (2006). Citrate-release-mediated Aluminum resistance is coupled to the inducible expression of mitochondrial citrate synthase gene in Paraserianthes falcataria. Tree Physiol. 26, 565–574. doi: 10.1093/treephys/26.5.565
Panda, S. K., Baluska, F., and Matsumoto, H. (2009). Aluminum stress signaling in plants. Plant Signal. Behav. 4, 592–597. doi: 10.4161/psb.4.7.8903
Ramírez-Benítez, J. E., Chee-González, L., and Hernández-Sotomayor, S. M. (2008). Aluminium induces changes in organic acids metabolism in Coffea arabica suspension cells with differential Al-tolerance. J. Inorg. Biochem. 102, 1631–1637. doi: 10.1016/j.jinorgbio.2008.03.002
Rangel, A. F., Rao, I. M., Braun, H. P., and Horst, W. J. (2010). Aluminum resistance in common bean (Phaseolus vulgaris) involves induction and maintenance of citrate exudation from root apices. Physiol. Plant. 138, 176–190. doi: 10.1111/j.1399-3054.2009.01303.x
Ritchie, G. S. P. (1995). “Soluble aluminium in acidics soils: principles and practicalities,” in Plant-Soil Interactions at Low pH: Principles and Management, eds R. A. Date, N. J. Grundon, G. E. Rayment, and M. E. Probert (Alphen aan den Rijn: Kluwer Academic Publisher), 23–33. doi: 10.1016/j.scitotenv.2014.03.064
Ryan, P. R., Raman, H., Gupta, S., Horst, W. J., and Delhaize, E. (2009). A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol. 149, 340–351. doi: 10.1104/pp.108.129155
Sánchez-Parra, B., Figueiras, A. M., Abd El-Moneim, D., Contreras, R., Rouco, R., Gallego, F. J., et al. (2015). The role of two superoxide dismutase mRNAs in rye aluminium tolerance. Plant Biol. 17, 694–702. doi: 10.1111/plb.12281
Santos, E., Benito, C., Silva-Navas, J., Gallego, F. J., Figueiras, A. M., Pinto-Carnide, O., et al. (2018). Characterization, genetic diversity, phylogenetic relationships, and expression of the aluminum tolerance MATE1 gene in Secale species. Biol. Plant. 62, 109–120. doi: 10.1007/s10535-017-0749-0
Santos, E., Matos, M., and Benito, C. (2020). Isolation and characterization of a new MATE gene located in the same chromosome arm of the aluminium tolerance (Alt1) rye locus. Plant Biol. 22, 691–700. https://doi.org/10.1111/plb.13107, doi: 10.1111/plb.13107
Santos, E., Matos, M., Silva, P., Figueiras, A.M., Benito, C., and Pinto-Carnide, O. (2016). Molecular diversity and genetic relationships in Secale. J. Genet. 95, 273–281. doi: 10.1007/s12041-016-0632-3
Sato, K., Shin-I, T., Seki, M., Shinozak, K., Yoshida, H., Takeda, K., et al. (2009). Development of 5006 full-length CDNAs in Barley: a tool for accessing cereal genomics resources. DNA Res. 16, 81–89. doi: 10.1093/dnares/dsn034
Silva-Navas, J., Benito, C., Téllez-Robledo, B., El-Moneim, D. A., and Gallego, F. J. (2012). The ScAACT1 gene at the Qalt5 locus as a candidate for increased aluminum tolerance in rye (Secale cereale L.). Mol. Breed. 30, 845–856. doi: 10.1007/s11032-011-9668-5
Silva-Navas, J., Salvador, N., Del Pozo, J. C., Benito, C., and Gallego, F. J. (2021). The rye transcription factor ScSTOP1 regulates the tolerance to Aluminum by activating the ALMT1 transporter. Plant Sci. 310:110951. doi: 10.1016/j.plantsci.2021.110951
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA 6: molecular evolutionary genetics analysis version 6. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
The International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
van der Merwe, M. J., Osorio, S., Moritz, T., Nunes-Nesi, A., and Fernie, A. R. (2009). Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol. 149, 653–669. doi: 10.1104/pp.108.130518
Yang, Y. Y., Jung, J. Y., Song, W. J., Suh, H. S., and Lee, Y. (2000). Identification of rice varieties with high tolerance or sensitivity to lead and characterization of the mechanism of tolerance. Plant Physiol. 124, 1019–1026.
Yang, Z. M., Yang, H., Wang, J., and Wang, Y. S. (2004). Aluminum regulation of citrate metabolism for Al-responsive citrate efflux in the roots of (Cassia tora L.). Plant Sci. 166, 1589–1594. doi: 10.1016/j.plantsci.2004.02.012
Keywords: aluminum tolerance, mitochondrial citrate synthase, expression changes, Secale cereale, Brachypodium distachyon
Citation: Abd El-Moneim D, Contreras R, Silva-Navas J, Gallego FJ, Figueiras AM and Benito C (2022) Repression of Mitochondrial Citrate Synthase Genes by Aluminum Stress in Roots of Secale cereale and Brachypodium distachyon. Front. Plant Sci. 13:832981. doi: 10.3389/fpls.2022.832981
Received: 10 December 2021; Accepted: 02 March 2022;
Published: 07 April 2022.
Edited by:
Ahmad M. Alqudah, Aarhus University, DenmarkReviewed by:
Ertugrul Filiz, Duzce University, TurkeyCopyright © 2022 Abd El-Moneim, Contreras, Silva-Navas, Gallego, Figueiras and Benito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diaa Abd El-Moneim, ZGFiZGVsbW9uaWVtQEFydS5lZHUuZWc=
†ORCID: Diaa Abd El-Moneim, orcid.org/0000-0003-3285-0563; Cesar Benito, orcid.org/0000-0003-1074-1294; Ana M. Figueiras, orcid.org/0000-0002-0944-1668
‡Present addresses: Diaa Abd El-Moneim, Department of Plant Production, (Genetic Branch), Faculty of Agricultural Environmental Sciences, Arish University, El-Arish, Egypt; Roberto Contreras, University of Atacama, CRIDESAT, Copiapó, Chile
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.