- Shandong Peanut Research Institute, Qingdao, China
Lipoxygenases (LOXs) are a gene family of nonheme iron-containing dioxygenases that play important roles in plant development and defense responses. To date, a comprehensive analysis of LOX genes and their biological functions in response to abiotic stresses in peanut has not been performed. In this study, a total of 72 putative LOX genes were identified in cultivated (Arachis hypogaea) and wild-type peanut (Arachis duranensis and Arachis ipaensis) and classified into three subfamilies: 9-LOX, type I 13-LOX and type II 13-LOX. The gene structures and protein motifs of these peanut LOX genes were highly conserved among most LOXs. We found that the chromosomal distribution of peanut LOXs was not random and that gene duplication played a crucial role in the expansion of the LOX gene family. Cis-acting elements related to development, hormones, and biotic and abiotic stresses were identified in the promoters of peanut LOX genes. The expression patterns of peanut LOX genes were tissue-specific and stress-inducible. Quantitative real-time PCR results further confirmed that peanut LOX gene expression could be induced by drought, salt, methyl jasmonate and abscisic acid treatments, and these genes exhibited diverse expression patterns. Furthermore, overexpression of AhLOX29 in Arabidopsis enhanced the resistance to drought stress. Compared with wide-type, AhLOX29-overexpressing plants showed significantly decreased malondialdehyde contents, as well as increased chlorophyll degradation, proline accumulation and superoxide dismutase activity, suggesting that the transgenic plants exhibit strengthened capacity to scavenge reactive oxygen species and prevent membrane damage. This systematic study provides valuable information about the functional characteristics of AhLOXs in the regulation of abiotic stress responses of peanut.
Introduction
Peanut is one of the most important economic crops producing healthy oil and high-quality protein for global human diets (Bertioli et al., 2016). The cultivated peanut (AABB genome) originated from two diploid progenitor species: Arachis duranensis and Arachis ipaensis (Bertioli et al., 2016; Zhuang et al., 2019). However, substantial peanut crop losses occur worldwide each year due to a variety of abiotic and biotic stresses, such as drought, salt, herbivorous insects and viruses. In plants, the synthesis of oxylipins and their derivatives, such as jasmonic acid (JA), green leaf volatiles, divinyl ethers and traumatic acid, is catalyzed by LOXs in the LOX pathway (Silke and Baldwin, 2010; Christensen et al., 2015). These oxylipins have been evidenced to be involved in plant responses to various stresses (Porta and Rocha-Sosa, 2002; Zhou et al., 2009).
Lipoxygenases are nonheme and iron-containing dioxygenases that compose multiple subfamilies in plants, fungi and animals (Brash, 1999). On the basis of specific LOX localization during the process of oxygenating substrates, including linoleic acid, α-linolenic acid (α-LeA), and arachidonic acid, plant LOXs have been divided into two classes: 9-LOXs and 13-LOXs. Furthermore, 13-LOXs can be classified into two subfamilies: type I 13-LOXs and type II 13-LOXs. Type I 13-LOX genes exhibit high sequence similarity (75%) and have no transit peptides, while type II 13-LOX genes exhibit low similarity with each other (up to 35%) and possess a chloroplast transit peptide (Veldink et al., 1977; Brash, 1999). The LOX pathway in plants contains four major metabolic routes: the peroxygenase pathway, the allene oxide synthase pathway, the hydroperoxide lyase pathway and the divinyl ether synthase pathway (Feussner and Wasternack, 2002; Porta and Rocha-Sosa, 2002). In the LOX metabolic pathway, LOXs catalyze polyunsaturated fatty acids, such as LA, α-LeA and arachidonic acid, to produce either 13S- or 9S-hydroperoxy derivatives (Feussner and Wasternack, 2002). These hydroperoxides are then metabolized via several secondary reactions and converted into diverse oxylipins. The different oxylipins generated by these metabolic routes participate in plant development (tendril coiling, cell death, etc.) and defense responses (responses to wounds, pathogens, fungi, herbivory, and other stresses; Reymond et al., 2000). In addition, LOXs have been proved to be key factors regulating the growth, lipid metabolism, seed storage and vigor, maturation and senescence, as well as stress resistance in soybean, rice, maize, and peanut (Carrera et al., 2007; Ying and Yz, 2020). More recent studies have focused on the functions of plant LOXs, especially with regard to growth, development, and stress responses.
Due to the rapid development of structural and functional genomics, many LOX genes were identified in a wide range of plants, and their functions in development (Liu and Chervin, 1997; Kolomiets et al., 2001; Yan et al., 2017) and defense responses (Heitz et al., 1997; Hui et al., 2016a) have also been well studied. A total of six LOXs (AtLOX1 to AtLOX6) were characterized in Arabidopsis (Umate, 2011). With the applications of high-performance liquid chromatography and gas chromatography–mass spectrometry analysis, AtLOX1 and AtLOX5 were classified as 9-LOXs, and AtLOX2, AtLOX3, AtLOX4 and AtLOX6 were classified as 13-LOXs (Bannenberg et al., 2009). AtLOX1 and AtLOX5 are reportedly involved in lateral root development and defense against pathogens (Tamara et al., 2007). Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade (Tamara et al., 2007). AtLOX3 and AtLOX4 were proved not only to be essential for male fertility but also to be involved in global proliferation (Caldelari et al., 2011). Besides, Arabidopsis lox3 lox4 double mutants are male sterile, and the double mutation results in defective global proliferative arrest; these genes perform distinct functions in resistance to plant-parasitic nematodes (Ozalvo et al., 2014). AtLOX2 and AtLOX6 participate in the JA biosynthesis pathway, and can be induced by different stresses (Bell et al., 1995; Grebner et al., 2013). In addition, rice OsHI-LOX, maize ZmLOX10 and tobacco NaLOX3 mediate herbivore-induced defense, and they are involved in the JA biosynthesis (Zhou et al., 2009; Rayko and Baldwin, 2010; Christensen et al., 2013). In maize, the LOX8 localizes to chloroplasts and participates in the wound-induced JA biosynthesis pathway, during which the production of LOX10-derived oxylipins is essential (Christensen et al., 2013). Furthermore, the ZmLOX10 localizes to organelles and acts to modulate both direct and indirect defenses against herbivores (Christensen et al., 2013).
The LOX gene family has been systematically studied in several plants, such as Arabidopsis, tomato, and maize (Bannenberg et al., 2009; Mariutto, 2011; Ogunola et al., 2017), but data for peanut was sparse. In peanut, scientists casted most attention on the potential roles of LOX genes in resistance to Aspergillus flavus. The phylogenetic relationships and molecular functions (Aspergillus flavus infection) of wild-type peanut LOX genes have been investigated (Hui et al., 2016b). A body of evidence has demonstrated that LOX and its products 9S- and 13S-hydroperoxy fatty acids (9S- and 13S- HPODE) play a significant role in the Aspergillus/seed interaction (Burow et al., 2000; Willis, 2005; Müller et al., 2014; Korani et al., 2018; Khan et al., 2020). However, the genome-wide identification and functional characterization of the LOX gene family under abiotic stresses in cultivated peanut has not been conducted. The availability of peanut genome annotation information made it possible to systematically and comprehensively characterize the LOX gene family. Here, 72 putative peanut LOX genes were identified by genome-wide searches of the wild-type and cultivated peanut genomes. The phylogenetic relationships, gene structures, conserved protein motifs and structures, subcellular localization, chromosome localization, gene duplication as well as cis-acting elements of LOX genes were characterized in peanut. To investigate the expression patterns of peanut LOXs in different tissues and under various stress challenges, the RNA-seq results were integratedly studied. Moreover, the expression patterns of five LOXs from three subfamilies in response to two stress treatments (drought and salt) and two hormone treatments (MeJA and ABA) were determined. These results will contribute to the selection of candidate genes for further characterization of peanut LOX genes which function in growth, development and defenses against various abiotic stresses.
Materials and Methods
Identification of LOXs in Peanut
The protein sequences of the allotetraploid Arachis hypogaea (AABB) and its two wild diploid progenitor species A. duranensis (AA) and A. ipaensis (BB; Milligan et al., 1998) were obtained from the PeanutBase database.1 The GmLOX amino acid sequences were downloaded at https://phytozome.jgi.doe.gov/pz/portal.html, and the MtLOX protein sequences were downloaded at http://jcvi.org/medicago/display.php?pa-geName=General§ion=Download.
A local BLAST search was performed to identify putative peanut LOX genes. First, the HMM profile of lipoxygenase (accession: PF00305) was obtained from PFAM,2 and the peanut local protein database was downloaded from PeanutBase. The versions of the peanut genomes from PeanutBase used in this study were as follows: Cultivated peanut (Version 2: A. hypogaea cv. Tifrunner) and A. ipaensis (Version 2). The HMM profile was then utilized to identify putative peanut LOXs in the peanut local protein database via the hmmsearch tool in HMMER3.0 software. All the putative LOX genes were further confirmed to contain LOX domains using the PFAM databases3 (Finn et al., 2006), InterPro4 (Quevillon et al., 2005) and the NCBI Batch CD-search database5 (Marchler-Bauer et al., 2005). In addition, the physical and chemical properties of the putative LOX proteins, including the number of amino acids (NA), molecular weight (Liu et al., 2016) and isoelectric point (theoretical pI), were calculated using the online tool ExPASy6 (Gasteiger et al., 1999). Detailed information on these physical and chemical properties of the peanut LOXs was presented in Supplementary Table S1.
Phylogenetic Analysis
Multiple alignments of LOX proteins from four species, including peanut (A. duranensis, A. ipaensis and A. hypogaea), Glycine max, Medicago truncatula and Arabidopsis thaliana, were conducted using ClustalW. The Gblocks Server7 was used to select conserved protein blocks for the above multiple alignment. A maximum likelihood phylogenetic tree with 1,000 bootstrap replicates was constructed via Molecular Evolutionary Genetics Analysis (MEGA X) software (Sudhir et al., 2018). The tree was further edited with specific colors indiciating the different subfamilies by EvolView.8
Analysis of Gene Structures, Conserved Motifs and Promoters
The exon–intron structures were visualized with the online tool Gene Structure Display Server (GSDS) using the CDS and genomic sequences of peanut LOXs9 (Guo et al., 2007). The conserved motifs of the peanut LOX proteins were identified using Multiple Expectation Maximization for Motif Elicitation (MEME Suite; Timothy et al., 2006).10 The promoter sequences (1.5 kb upstream of the peanut LOX transcription start site) were downloaded from PeanutBase, and were used to predict cis-acting regulatory elements with PlantCARE.11 Detailed information on the cis-acting elements in each LOX promoter was shown in Supplementary Table S4.
Prediction of the 3D Structures of LOX Proteins
The 3D protein structures of the LOXs were predicted using SWISS-MODEL12 (Andrew et al., 2018). We randomly selected genes from each subfamily and from different species for modeling. As shown in Figure 1, 9-LOX (AtLOX1, GmLOX1, MtLOX1, AiLOX9, AdLOX11, and AhLOX20), type I 13-LOX (GmLOX6, MtLOX10, AiLOX4, AdLOX4, and AhLOX8) and type II 13-LOX (AtLOX3, GmLOX12, MtLOX18, AiLOX15, AdLOX17, and AhLOX30) were displayed.
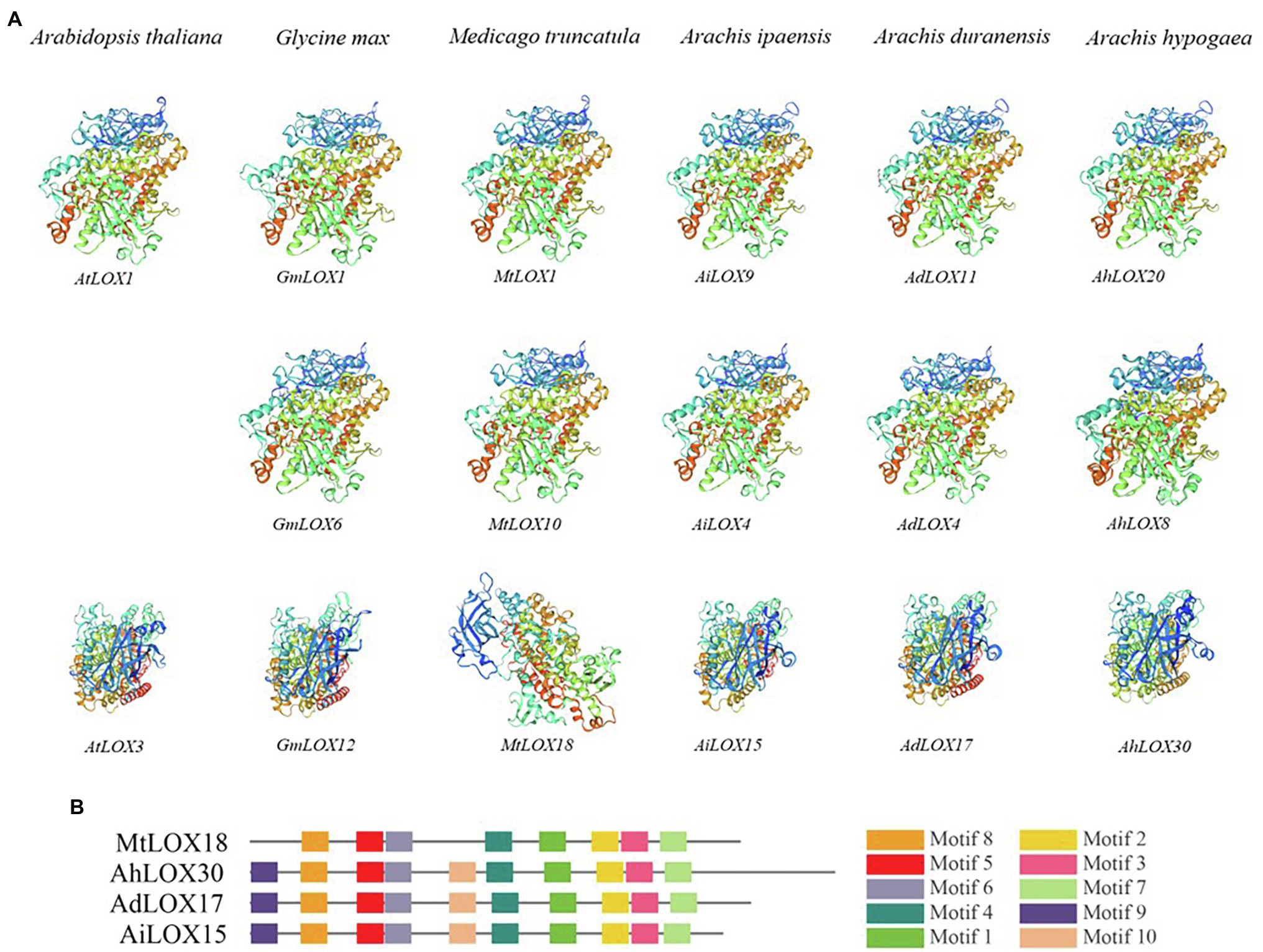
Figure 1. Predicted structures of LOX proteins. The modeled structures of LOX genes from different species are displayed. 9-LOX (AtLOX1, GmLOX1, MtLOX1, AiLOX9, AdLOX11, and AhLOX20), type I 13-LOX (GmLOX6, MtLOX10, AiLOX4, AdLOX4, and AhLOX8) and type II 13-LOX (AtLOX3, GmLOX12, MtLOX18, AiLOX15, AdLOX17, and AhLOX30).
Chromosomal Location and Gene Duplication
The chromosomal locations of the putative LOX genes in peanut were obtained from the PeanutBase database.13 Gene duplication events of the peanut LOX genes were analyzed (E-value<1e-10) using the Multiple Collinearity Scan toolkit (MCScanX).14 The segmental duplicated genes were characterized as homologous genes between different chromosomes. To visualize the segmentally duplicated pairs of peanut LOX genes, we draw a map of the chromosomal distribution and duplication of peanut LOX genes using Circos software.15
Analysis of the Gene Expression Patterns of Peanut LOX Using RNA-Seq Values
To study the expression patterns of peanut LOX genes in different tissues and under different stresses, the fragments per kilobase of transcript per million fragments (FPKM) values of 22 tissues were obtained from the PeanutBase database (Josh et al., 2016),16 and the FPKM values of LOX genes in response to drought and salt treatments were obtained from our previous work. The LOX accession number and FPKM values of different tissues and under different stresses were provided in Supplementary Tables S6 and S7. A total of 72 LOX gene expression data points were log2-transformed and visualized by Heml software (Wankun et al., 2014).
Plant Materials and Treatments
Seedlings of the peanut cultivar Huayu71 (a cultivated peanut bred by our team) were grown until the three-leaf stage and used for the gene expression analysis. All the materials were grown in a climate chamber under controlled conditions: 20°C /16 h light and 20°C/8 h dark, with 60% relative humidity. For the drought and salt stresses, the seedlings were immersed in Hoagland liquid medium supplemented with 20% PEG6000, 200 mM NaCl, 100 μM MeJA or 100 μM ABA. The roots of the seedlings were harvested after 0, 6, 12, 24, and 48 h and rapidly frozen in liquid nitrogen. Three biological replicates were performed for the stress analysis in this study.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was extracted using the Takara RNA Extraction kit (Code No. 9767, TaKaRa, Dalian) according to the manufacturer’s instructions. First-strand cDNA was synthesized using the Takara PrimeScript RT Reagent kit (Code No. RR037, Takara, Dalian). The quantitative real-time PCR (qRT–PCR) was carried out in a 20 μl reaction system containing 1 μl (100 ng) of cDNA, 0.4 μl of forward primer (10.0 μmol/l), 0.4 μl of reverse primer (10.0 μmol/l), 10 μl of 2 × qPCR mixture (2× Takara TB Premix Ex Taq Mix, No. RR820, Takara, Dalian) and 8.2 μl of RNase-free water using the ABI7500 Fast System (Applied Biosystems, CA, United States). The PCR procedure was set as follows: 95°C for 5 min; 40 cycles of 95°C for 10 s and 60°C for 30 s. The relative expression levels of all the genes were analyzed using 2–∆∆CT. The primers specific for the AhLOX genes and actin used in this work were shown in Supplementary Table S8.
Vector Construction and Transformation Into Arabidopsis
The coding DNA sequence (CDS) of AhLOX29 was amplified from “Huayu71” cDNA. The 2,754-bp CDS was then ligated between a 35S promoter and the nopaline synthase (Nos) terminator in the pCambia2300EC vector to construct the overexpression vector (35S. AhLOX29. Nos). The constructed overexpression plasmid was transformed into Arabidopsis according to an established transformation procedure. After being cultured on MS media supplemented with 50 mg/l kanamycin, positive seedlings were obtained from the selective media. Subsequently, the surviving seedlings were transplanted to nutritional soil for further confirmation by PCR using the vector-specific primers: VSP-F and VSP-R (Supplementary Table S8).
Determination of Drought-Related Physiological Parameters
Drought stress treatment was applied to both WT and transgenic plants while they grew to the seedling stage in a climate chamber. The WT and transgenic plants were grown in the same amount of nutrient soil, and the same amount of water was poured at each watering. The total chlorophyll contents, proline contents, malondialdehyde (MDA) contents and superoxide dismutase (SOD) enzyme activity were measured before drought treatment. The four physiological parameters were measured after water was stopped for 10 days. The total chlorophyll contents in leaves were determined using the chlorophyll assay kit (Comin, CPL-2-G, Suzhou). Leaves of Arabidopsis plants were measured for three replicates, and the average values were calculated as total chlorophyll levels. For quantification of the proline contents, MDA contents and SOD enzyme activity, the assay Kit (Comin, PRO-2-Y, MDA-2-Y and SOD-2-W, Suzhou) was used following previously described procedures (Bates et al., 1973; Hao et al., 2013; Yu et al., 2015).
Results
Identification of Peanut LOX Genes and Phylogenetic Analysis of LOXs From Different Species
To identify LOX genes in peanut, the HMM profile of LOX (accession: PF00305) was used to search the peanut local protein database. A total of 89 putative LOX genes were identified in peanut. After confirming the presence of the LOX domain using the PFAM, InterPro and NCBI Batch CD-search databases, 72 peanut LOX genes were determined to have the whole lipoxygenase domain and were used for further analysis. Based on the analysis of physical and chemical properties, we found that the length of peanut LOX proteins ranged between 509 (AdLOX5) and 1,491 amino acids (AhLOX25), and the molecular weight of LOX proteins ranged from 58.35 kDa (AdLOX5) to 170.64 kDa (AhLOX25). The pI was predicted to range from 4.98 (AhLOX5) to 9.15 (AiLOX17), which indicated that various microenvironments were needed for each LOX protein to perform optimal function.
The classification and phylogenetics of LOX proteins have been analyzed in several plants, such as Arabidopsis, Glycine max, and Medicago truncatula (Jin et al., 2008; Umate, 2011; Ogunola et al., 2017), helping us to classify the evolutionary relationships of the peanut LOX proteins. Multiple sequence alignments of 117 LOX conserved protein blocks, including 72 peanut LOXs, 6 AtLOXs, 19 GmLOXs and 20 MtLOXs, were performed using ClustalW, and their GenBank accession IDs were presented in Supplementary Table S2. A maximum likelihood phylogenetic tree with 1,000 bootstraps was constructed using the LOX proteins of these four species. On the basis of the phylogenetic relationships and the classifications of LOX proteins from other species, the peanut LOX gene family could be classified into three clades: 9-LOX, type I 13-LOX and type II 13-LOX (Figure 2). As shown in Figure 2, the peanut LOXs were randomly distributed among the LOXs of the other three species. In the 9-LOX subfamily, AdLOX9-12, AiLOX8-10, and AhLOX15-22 are closely related to AtLOX1, AtLOX5, GmLOX1-5, and MtLOX1-3. AdLOX1-8, AiLOX1-7, and AhLOX1-14 are type I 13-LOXs and are closely related to GmLOX6-10 and MtLOX4-13. In the type II 13-LOX clade, AdLOX13-18, AiLOX11-18, and AhLOX23-36 are closely related to AtLOX2-4, AtLOX6, GmLOX11-19, and MtLOX14-20. The bootstrap values for some parts of the phylogenetic tree are low, which may have occurred due to the low similarity of protein sequences.
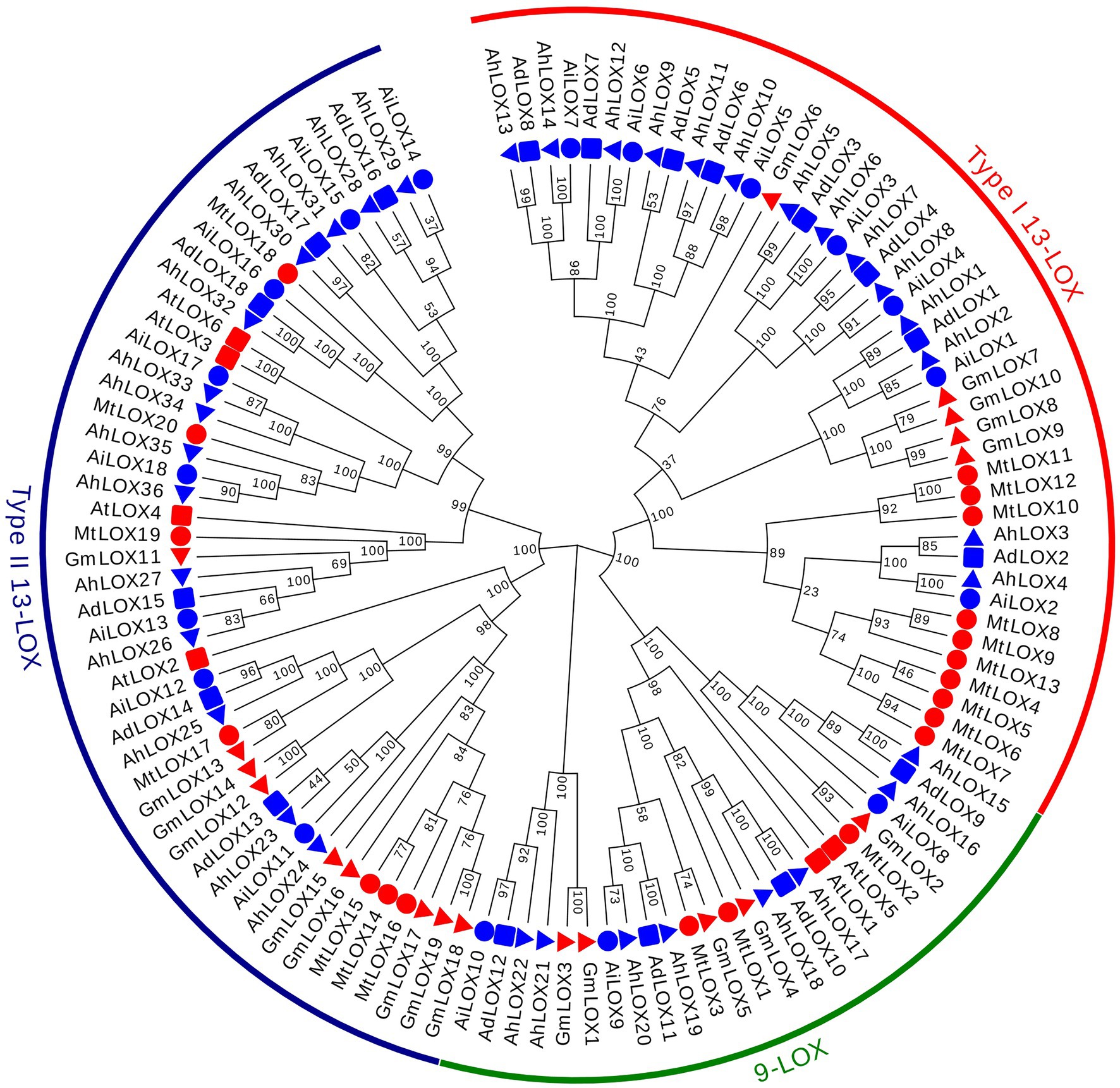
Figure 2. Phylogenetic tree of the peanut LOX gene family. Phylogenetic tree of 117 LOX proteins from peanut (72), Arabidopsis (6), Glycine max (19) and Medicago truncatula (20). MEGA X software was used to construct the maximum-likelihood (ML) phylogenetic tree with 1,000 replications.
Phylogenetic, Gene Structure and Conserved-Domain Analysis of Peanut LOX Genes
To gain a further understanding of the gene structure and conserved protein motifs of LOX genes, we constructed a maximum likelihood phylogenetic tree with the peanut LOX proteins, and compared the protein motifs (Figure 3A) and exon/intron structures (Figure 3B) of each LOX gene. The exon/intron structures of the LOXs were analyzed using GSDS. As shown in Figure 3B, most LOXs were found to have similar gene structures in terms of both intron numbers (7–9) and exon lengths. However, variations in the intron numbers of some LOXs were observed. The maximum number of introns (17 introns) was observed in AdLOX14. In addition, we found that AhLOX25 had 15 introns while AiLOX15 and AhLOX30-31 contained 10 introns.
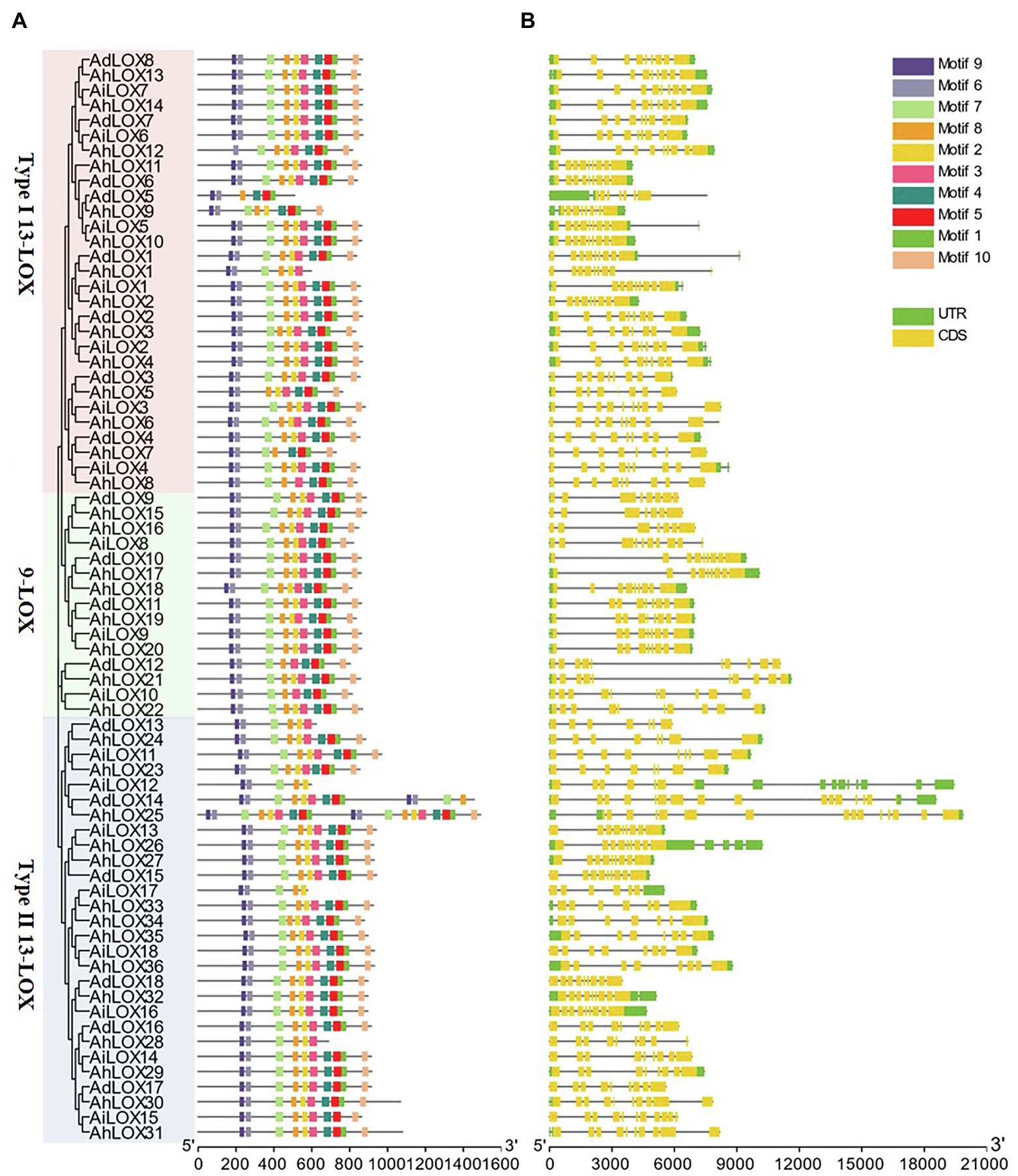
Figure 3. Phylogenetic relationship, gene structure and conserved-motif analysis of the LOX genes in peanut. The maximum-likelihood phylogenetic tree was constructed using MEGA X with 1,000 replicates. (A) Conserved motifs of LOX proteins. Ten conserved motifs are shown in different-colored boxes, and detailed information on the motifs is provided in Supplementary Table S3. (B) Exon-intron structures of LOX genes. The yellow boxes represent exons, and the gray lines represent introns.
The LOX protein motifs were identified by searching the MEME Suite databases to explore the similarity and diversity of motifs within subfamilies. We identified 10 distinct motifs in the peanut LOX proteins, named as motifs 1–10 (Figure 3A and Supplementary Table S3). As expected, all peanut LOX proteins contained highly representative motifs, including 38-residue (motif 1), substrate-binding (motif 3), and oxygen-binding (motif 2) motifs. Among them, motif 1 was essential for the stability of LOX protein, and the AhLOX1, AhLOX28, AdLOX13, AiLOX12, and AiLOX17 lacked this motif, indicating that these genes might exhibit altered enzymatic activity. In addition, motif 5 (except AhLOX1, AhLOX28, AdLOX13, AiLOX12, and AiLOX17), motif 6, motif 7 (except AdLOX5 and AhLOX5), motif 8, motif 9, and motif 10 (except AdLOX5, AhLOX1, AdLOX13, AiLOX12, AiLOX17, and AhLOX28) were conserved in all peanut LOXs. The AhLOX1, AdLOX13, AiLOX12, AiLOX17, and AhLOX28 lacked motif 4. Whether the absence of these motifs confers unique functions to these proteins requires further investigation. In any case, the highly conserved motifs in peanut LOX proteins would contribute to the functional analysis of the peanut LOX proteins.
Analysis of the Predicted Structures of LOX Proteins
We randomly selected certain LOX proteins from each subfamily in the six species and modeled their structures using SWISS-MODEL. The 9-LOX proteins had similar structures among different species, whereas the 13-LOX proteins seemed to have different structures among these species. As shown in Figure 1, the modeled structures of the type I 13-LOX proteins in peanut and the other two species were similar. However, the structure of type II 13-LOX proteins was not conserved; there were two types, and we found that the modeled structure of type II 13-LOX in Medicago truncatula was different from those of the other species. The MtLOX18 lacked motifs 9 and 10, thereby resulting in the difference of modeled structure with other subfamily members (Figure 1). In general, the modeled structures of 9-LOX and type I 13-LOX subfamily proteins in different species were similar, whereas the type II 13-LOX protein had diverse structures among different species.
Analysis of Cis-Acting Elements in Peanut LOX Genes
Cis-acting elements in the promoter are crucial for initiating transcription and regulating gene expression. To analyze the cis-acting elements in peanut LOX gene promoters, the sequence 1.5 kb upstream from the initiation codon was predicted by searching the online PlantCARE database. The identified cis-acting elements were related to transcription, cell cycle, development, hormones as well as stresses, and most of these elements were hormone-related and stress-related elements (Figure 4). It was prediceted that the expression of peanut LOXs could be induced by hormones, such as MeJA, ABA, salicylic acid (SA), gibberellin (GA), auxin (IAA), and ethylene (ET). Moreover, many elements were involved in various stresses, such as anoxic induction, drought, zein metabolism, wounding, and low temperature. All peanut LOX genes contained many light-responsive elements. In addition, elements associated with growth and development, including meristem and endosperm expression, were found in most LOXs. Detailed information about the elements and functions in each gene were presented in Supplementary Table S4.
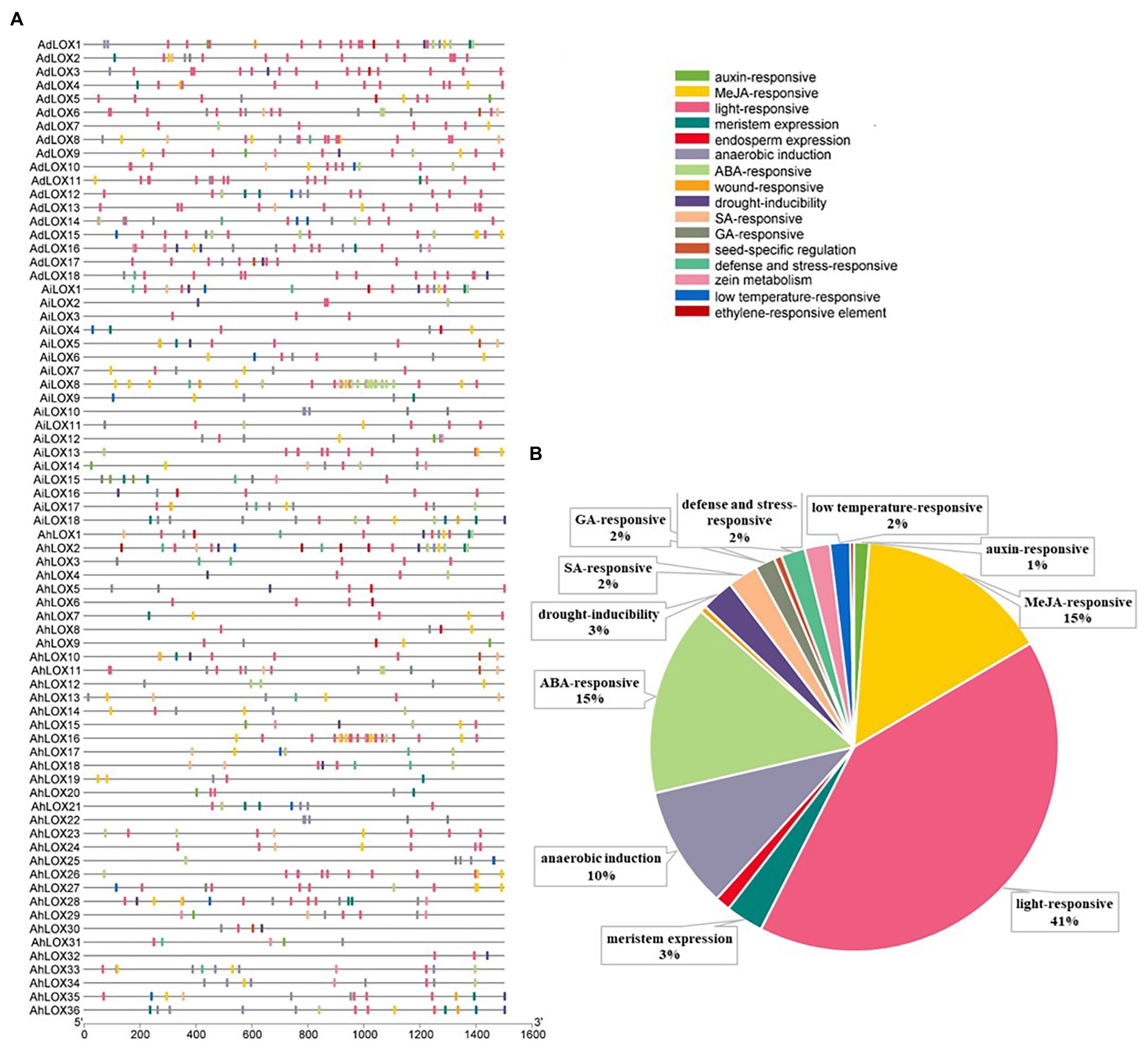
Figure 4. Distribution of cis-acting regulatory elements in the peanut LOX gene promoters. (A) The distribution of cis-acting elements in the promoter of each peanut LOX. (B) The ratios of different functional cis-acting elements are shown in the form of pie graphs.
Chromosomal Location and Gene Duplication of Peanut LOXs
As shown in Figure 5, the distribution of peanut LOXs on the chromosomes were not random. AdLOXs and AiLOXs were centrally distributed on chromosomes A9 and B9, respectively. Similarly, almost half of the AhLOX genes (17 out of 36) were located on chromosomes 9 and 19. The chromosomal locations of AdLOX were mapped onto 6 of the 10 A. duranensis (A genome) chromosomes, and AiLOX was also distributed on 6 of the 10 A. ipaensis (B genome) chromosomes. Chromosomes A3, A8, B3, B6, 3, 6, 8, and 16 each had 3 LOXs. There was only one LOX gene on chromosomes A2, A10, B2, B10, 2, 10, 12 13, and 20, and two LOX genes on chromosomes A6, B8, and 18. Chromosomes A1, A4, A5, A7, B1, B4, B5, B7, 1, 4, 5, 7, 11, 14, 15, and 17 had no LOXs. These results indicated that gene duplication events might have occurred on chromosomes A9, B9, 9, and 19 during the evolutionary process of the LOX gene family.
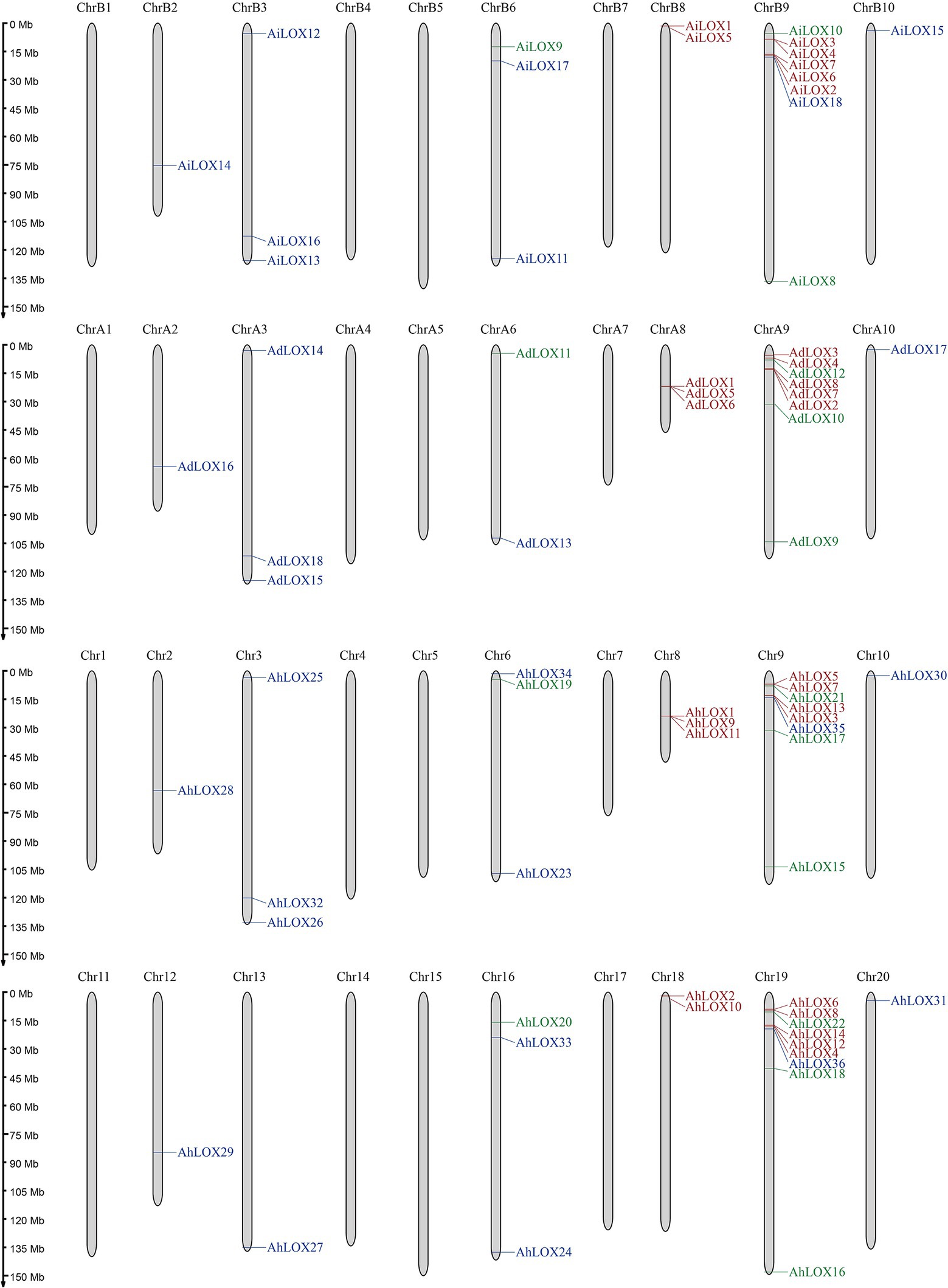
Figure 5. Chromosomal locations of LOX genes. The approximate locations of the 72 peanut LOX genes are displayed on the right side of the chromosomes. Genes from different subfamilies are shown in different colors (9-LOX: green, type I 13-LOX: red, and type II 13-LOX: blue).
Gene duplication is widespread in the peanut genome. We analyzed the segmental duplication events of the LOX genes in peanut using MCScanX software. As shown in Figure 6, among 72 peanut LOXs, 48 pairs of segmental duplication genes were identified (Figure 6, Supplementary Table S5), and 14 pairs were located on chromosomes A9, B9, 9, and 19. The largest number of gene duplication events occurred in type II 13-LOX (24 pairs), followed by type I 13-LOX (14 pairs) and 9-LOX (10 pairs). These results suggested that segmental duplication played a vital role in the expansion of peanut LOX gene family.
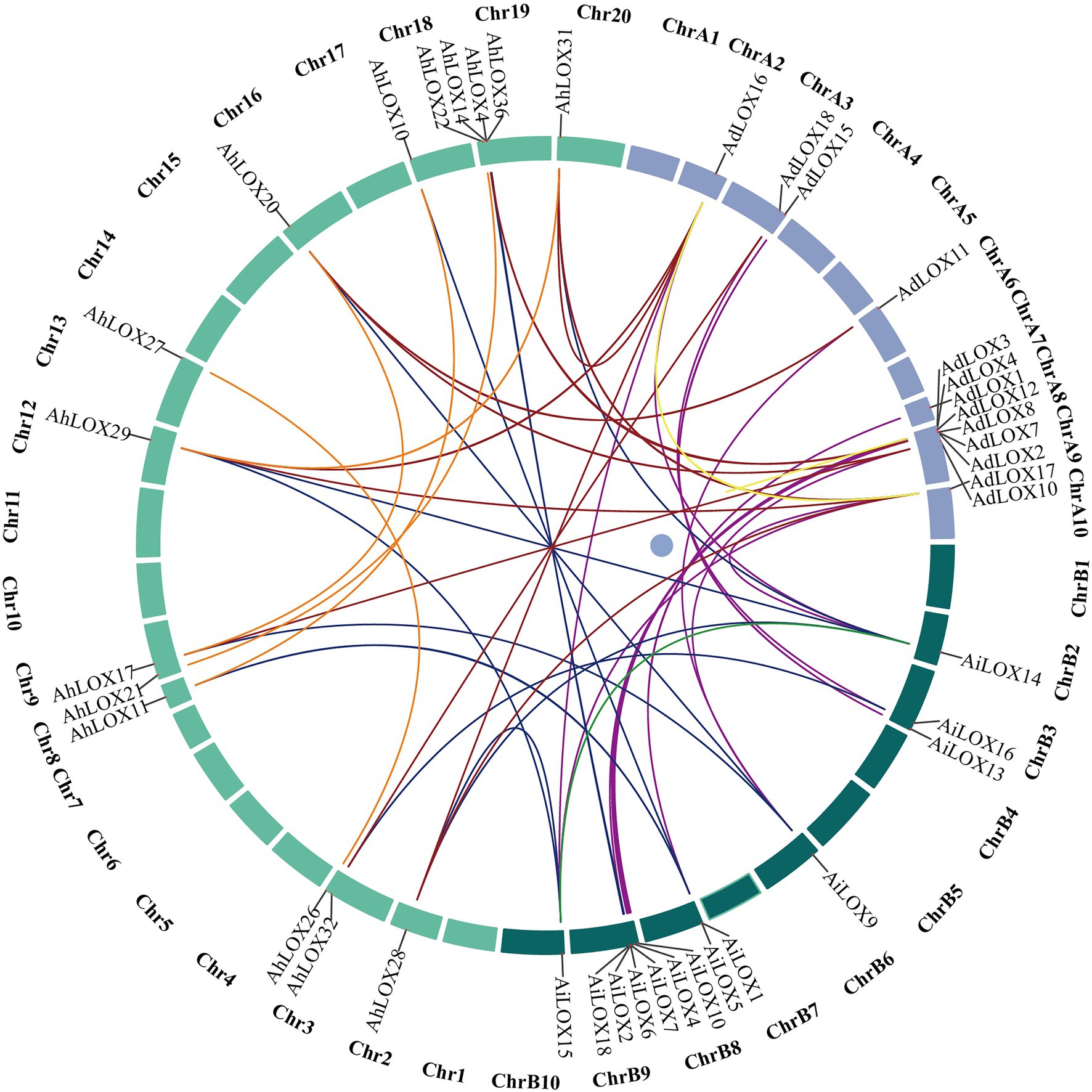
Figure 6. Segmentally duplicated gene pairs of LOXs in peanut. The lines highlighted in different colors indicate duplicated LOX gene pairs.
Tissue-Specific Expression Patterns of Peanut LOXs
To confirm the tissue expression patterns and potential functions of LOX genes in peanut growth and development, the expression levels of the genes in 22 tissues (leaves, shoot tips, roots, nodules, perianths, flowers, pistils, stamens, gynophore tips, fruits, pericarps, and seeds in nearly all developmental stages) were investigated using RNA-seq data published by Josh et al. (2016). Based on the log2-transformed FPKM values, we found that the expression levels of peanut LOXs were notably different among the 22 tissues (Figure 7). Most LOXs were expressed in all 22 tissues, except for AdLOX4, AdLOX7, AiLOX3, AiLOX4, AiLOX6, AhLOX6-8, AhLOX12, AhLOX15, AhLOX23-25, and AhLOX28. Interestingly, certain genes, AdLOX2, AiLOX2, AiLOX8, AiLOX13, AiLOX18, AhLOX3, AhLOX4, AhLOX17, AhLOX26, and AhLOX33-36, were expressed at extremely high levels in all the investigated tissues. Besides, a large number of genes with tissue-specific expression patterns were identified. AdLOX1, AdLOX6, AiLOX1, AiLOX5, AhLOX1-2, and AhLOX9-11 were specifically highly expressed in seeds, while AdLOX2-3, AdLOX8, AiLOX2, AiLOX7-8, AhLOX3-4, and AhLOX13-14 were found to be specifically expressed in the shoot tips, roots, gynophore tips, fruits, and pericarps. As shown in Figure 7 and Supplementary Table S6, most homoeologous genes, such as AdLOX8 and AiLOX7, AdLOX1 and AiLOX1, and AhLOX26 and AhLOX27, exhibited similar expression patterns. Nevertheless, some peanut LOXs showed diverse expression patterns; for example, AhLOX2 was specifically expressed in seeds, while its homoeologous gene AhLOX4 was highly expressed in leaves, shoot tips, roots, nodules, perianths, flowers, pistils, stamens, gynophore tips, fruits, and pericarps. The different expression patterns of homoeologous genes may be due to the polyploidization and duplication events that occurred in peanut evolution.
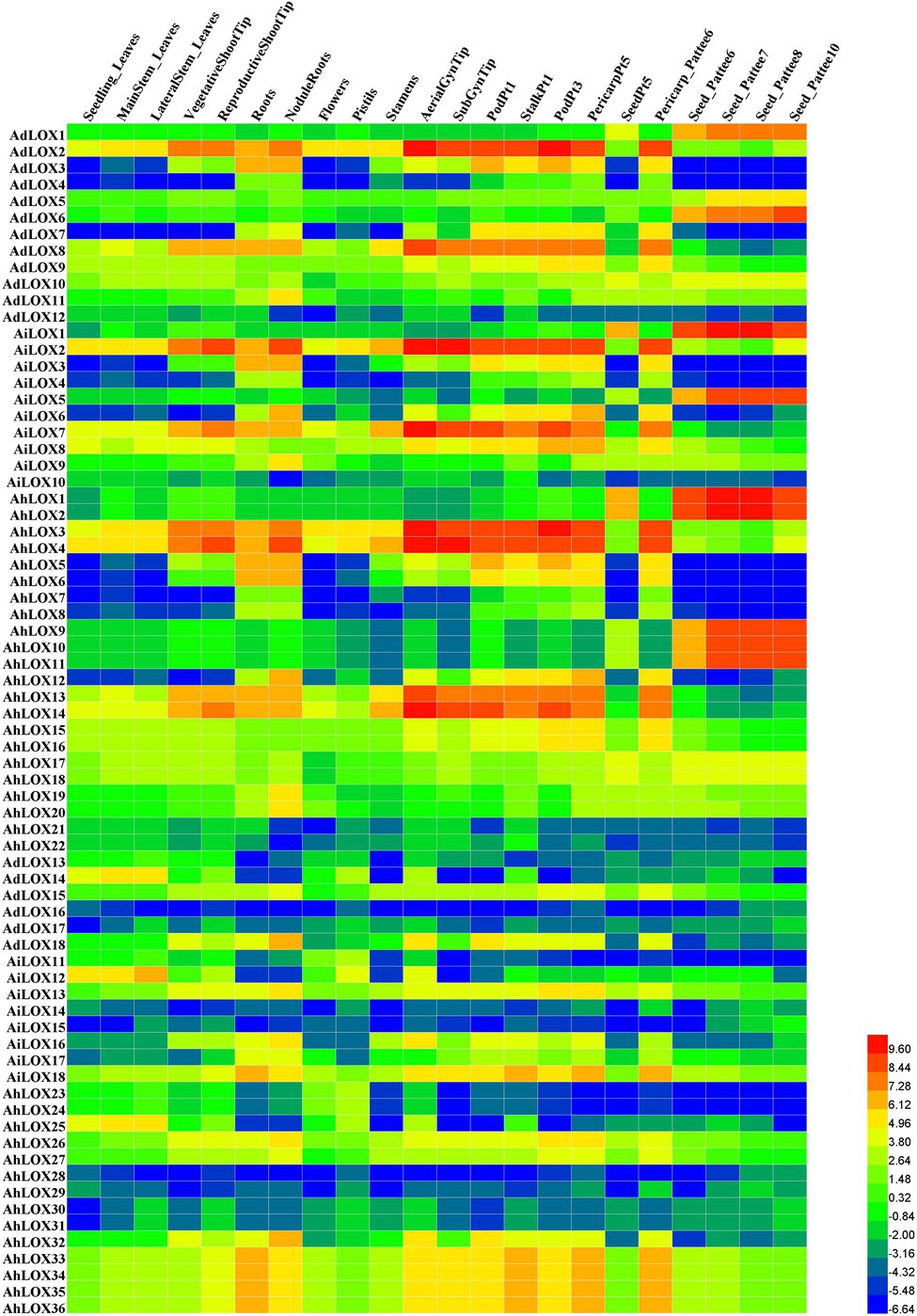
Figure 7. Expression profiles of LOX genes in 22 different peanut tissues. The heatmap was generated by Heml software, and the fragments per kilobase of transcript per million fragments (FPMK) values of peanut LOX genes were log2-transformed. The red and blue colors represent the maximum and minimum values, respectively.
Peanut LOX Expression Patterns Under Abiotic Stresses
Previous studies have revealed that LOX genes play an important role in plant defenses against abiotic and biotic stresses. In this study, the expression profiles of peanut LOXs were analyzed under salt and drought stresses using published transcriptome sequencing results. The peanut LOX genes presented diverse expression patterns under salt and drought stress. In Figure 8, the expression of 35 peanut LOXs was upregulated more than 2-fold under salt stress, and that of 19 LOXs was upregulated more than 5-fold. Moreover, the greatest increase was up to 30-fold (AiLOX9 and AhLOX20). The expression of eight peanut LOXs was downregulated nearly or more than 2-fold. Unlike the salt treatment, only a few peanut LOXs were upregulated under the drought treatment. Similarly, there were only 11 genes that were downregulated more than 2-fold after the drought treatment. It is worth noting that most of the homoeologous LOX genes presented similar expression patterns under salt and drought stress. In particular, the expression of AdLOX10 decreased while the expression of AhLOX20 stay unchanged in response to drought stress.
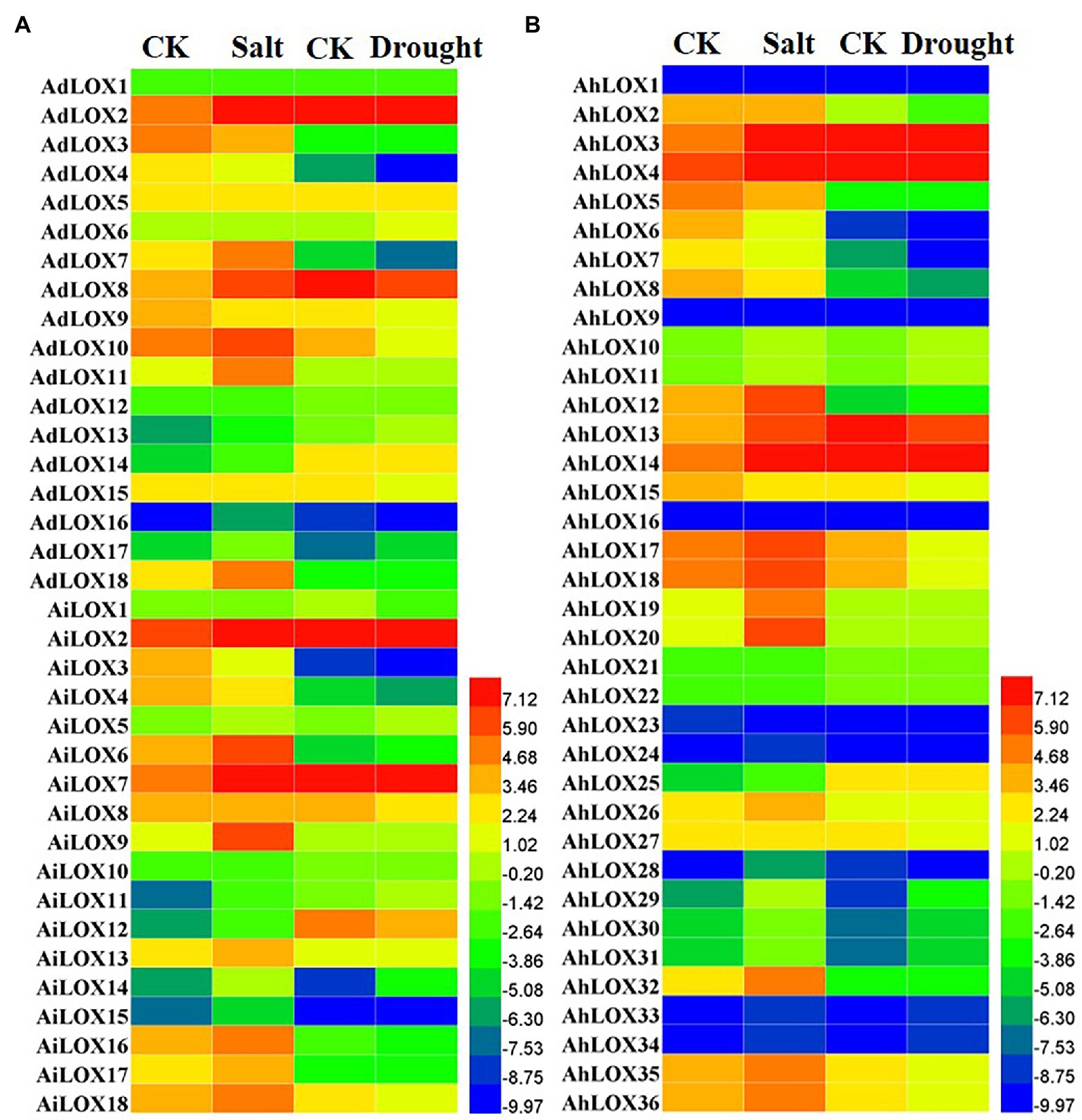
Figure 8. Heatmap of the expression patterns of LOX genes under salt and drought stress treatments in peanut (A and B). The FPKM values of the peanut LOX genes were log2-transformed to create the heatmap using Heml software. The red and blue colors represent the higher or lower relative abundance of each LOX gene, respectively.
Analysis of the Expression Patterns of Peanut LOXs in Response to Four Treatments by qRT-PCR
The qRT-PCR was used to validate the peanut LOX expression patterns under abiotic stresses (salt and drought) and hormone treatment (MeJA and ABA). As shown in Figure 9, five genes from different subfamilies, AhLOX20 (9-LOX), AhLOX8 (type I 13-LOX), AhLOX12 (type I 13-LOX), AhLOX29 (type II 13-LOX), and AhLOX30 (type II 13-LOX), were investigated at six time points (0, 6, 12, 24, and 48 h) using peanut leaves (Figure 9A) and roots (Figure 9B) in the three-leaf stage.
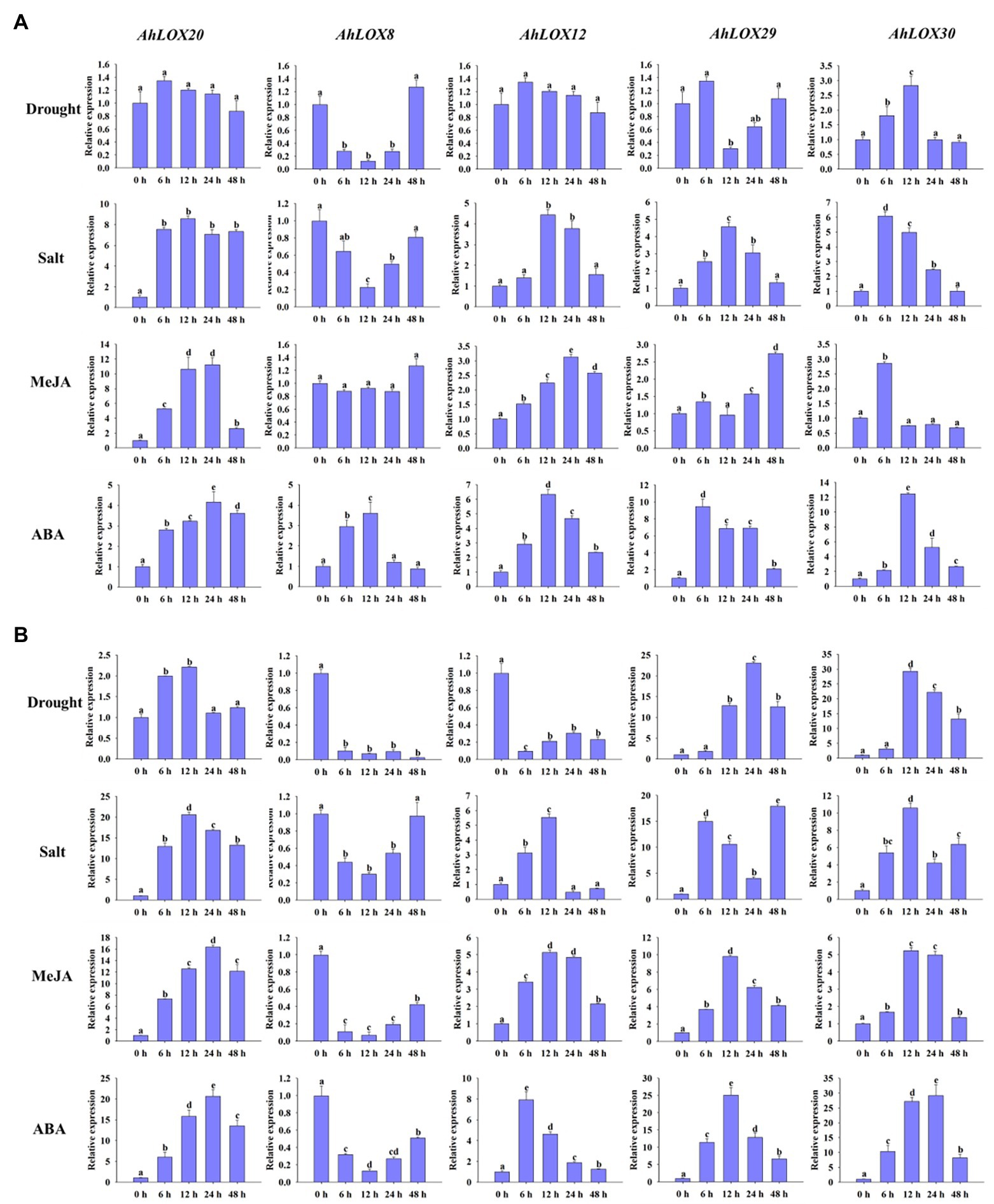
Figure 9. Analysis of the relative expression levels of peanut LOX genes by qRT-PCR. The expression profiles of five peanut LOX genes (AhLOX20, AhLOX8, AhLOX12, AhLOX29, and AhLOX30) under two stresses (drought and salt) and two hormone treatments (MeJA and ABA) were validated. (A) Leaves; (B) Roots. The data are presented as the mean ± SD (n = 3), and the values differed significantly at p < 0.05. The different letters indicate significant differences.
The five selected peanut LOX genes exhibited different expression patterns in response to the salt, drought, MeJA and ABA treatments. After drought treatment, the expression profiles of most LOX genes were similar in both leaves and roots, while AhLOX12 presented opposite expression patterns in leaves and roots. The expression of two genes (AhLOX12 and AhLOX30) in leaves and that of three genes (AhLOX20, AhLOX29, and AhLOX30) in roots were upregulated. Remarkably, the relative expression of AhLOX29 and AhLOX30 was upregulated more than 20-fold in roots. A significant decrease of the relative expression of AhLOX8 at 6 h in leaves and roots, and AhLOX29 at 12 h in leaves after drought treatment were observed. After salt treatment, only the AhLOX8 was downregulated in both leaves and roots. The expression levels of selected LOX genes (AhLOX12, 20, 29, and 30) were increased by 4- to 8-fold in leaves. The greatest changes were found for the AhLOX8 and AhLOX29 in roots, which were increased up to 20-fold (Figure 9). The above-described results show that only AhLOX12 in roots exhibited different expression patterns in response to drought and salt stress. In addition, the relative expression level of AhLOX29 in roots was increased by approximately 20-fold in response to both drought and salt treatments. As for the MeJA and ABA treatments, the expression of all the genes except AhLOX8 was upregulated, and the maximum change was nearly 30-fold (AhLOX30 after ABA treatment) in roots. Both MeJA and ABA stress led to the downregulation of AhLOX8 in roots, and ABA treatment resulted in upregulation of AhLOX8 in leaves. Together, the relative expression of three genes, AhLOX20 (9-LOX), AhLOX29 (type II 13-LOX), and AhLOX30 (type II 13-LOX), increased in response to the four treatments (drought, salt, MeJA, and ABA). The expression of other AhLOX gene members, such as AhLOX8 (type I 13-LOX), was decreased after exposure to these four stresses. The expression of AhLOX12 (type II 13-LOX) was upregulated by salt, MeJA, and ABA treatment and downregulated by exposure to drought.
Overexpression of the AhLOX29 Gene in Arabidopsis Enhances Resistance to Drought
WT and transgenic Arabidopsis were exposed to drought stress during the seedling stage, and the drought tolerance of the AhLOX29 transgenic lines was significantly improved (Figure 10A). The relative expression levels of AhLOX29 in the transgenic and WT lines were measured by qRT-PCR. Results showed that the relative expression levels of the two overexpression lines were significantly higher than that of the WT lines (Figure 10B), indicating that the AhLOX29 was successfully transcribed in Arabidopsis. After being treated with 20% PEG6000 for 1 h, the expression of AhLOX29 in the AhOL and WT lines was obviously increased, and the transcript levels of AhLOX29 in the AtOL lines were significantly higher than those in the WT (Figure 10B). Compared with 1 h, the expression of the AhLOX29 in the WT was unchanged after 6 h of 20% PEG6000 treatment, but the expression of AtOL1 and AtOL2 was decreased at 6 h (Figure 10B). To briefly sum up, the AhLOX29 is a stress-responsive gene, and the rapid accumulation of AhLOX29 at 1 h suggests its positive role in drought resistance.
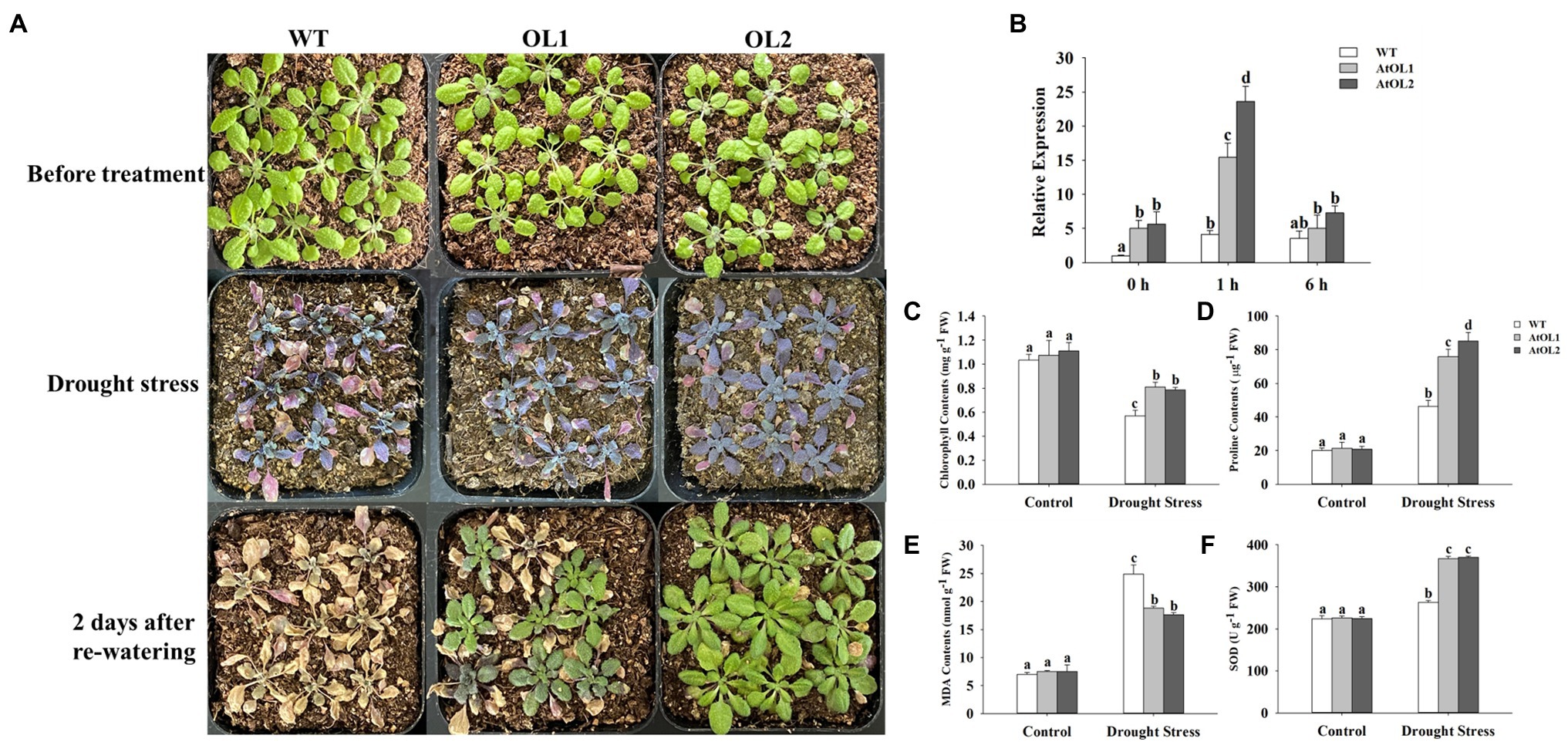
Figure 10. Overexpression of AhLOX29 improved tolerance to drought stress in transgenic Arabidopsis. (A) Phenotype of control and transgenic plants before and after drought stress treatment; (B) the relative expression level of AhLOX29 in WT and transgenic plants; (C) chlorophyll contents; (D) proline contents; (E) MDA contents and (F) SOD activity.
The total chlorophyll, proline and MDA contents as well as SOD activity in the leaves on day 0 and day 10 after drought stress were measured (Figures 10C–F). After 10 days of drought treatment, the chlorophyll content in leaves decreased, and the chlorophyll contents in the gene-overexpressed plants were significantly higher than those in the WT plants (Figure 10C). The proline content in plants increased under drought challenges, thereby functioning as an osmotic protective agent to alleviate or resist the damage caused by drought stress. After drought stress, the proline content in Arabidopsis leaves increased by approximately 2-fold in the WT plants and 4-fold in the transgenic plants, and the gene-overexpressed lines accumulated higher levels of proline than the WT (Figure 10D). The MDA contents were remarkably increased by drought stress, and higher MDA resulted in greater damage to the membrane and plants. The MDA contents in the WT and transgenic Arabidopsis lines were equal before drought treatment, and drought treatment significantly increased the MDA contents from 6.99 to 24.85 nmol/g in the WT plants and to 18.8 and 17.6 nmol/g in the AtOL1 and AtOL2 lines, respectively (Figure 10E). Under normal conditions, no difference in SOD activity were found between the WT and transgenic plants (Figure 10F). However, after drought challenge for 10 days, the SOD activity was obviously increased, and gene-overexpressed Arabidopsis displayed a notable increase in SOD activity compared with WT Arabidopsis.
Discussion
To date, the LOX multigene family has been identified and functionally characterized in various plant species, such as Arabidopsis (Umate, 2011), rice (Umate, 2011), cotton (Shaban et al., 2018), tomato (Upadhyay et al., 2019), and pepper (Sarde et al., 2018). Furthermore, previous studies in these species have focused mainly on its roles in protein structure, evolution, growth and development, improvement of seed quality as well as defense responses to biotic and abiotic stresses (Feussner and Wasternack, 2002; Porta and Rocha-Sosa, 2002; Roy Chowdhury et al., 2016). Although several genes related to the molecular characterization of peanut LOXs have been reported, a genome-wide identification of the LOX gene family in cultivated peanut and the functional analysis of these genes under abiotic stress conditions have not been performed. The fully annotated reference genomes of A. duranensis and A. ipaensis have been released (Bertioli et al., 2016), and whole-genome sequences of cultivated peanut (Tifrunner) were recently available as well (Bertioli et al., 2019). Accordingly, it becomes possible to identify all the putative LOX genes in peanut and analyze potential functions.
Unlike the other plant species investigated in this study, the number of AhLOXs in peanut (36) was significantly higher than that of AtLOXs (6), AiLOXs (18), and AdLOXs (18). The high numbers of AhLOXs might be attributed to the allotetraploid genome of peanut, which experienced whole-genome duplication and gene duplication events (Bertioli et al., 2011). Gene duplication and whole genome duplication events contributed to the expansion of gene families in plant evolution (Cannon et al., 2004). In addition, a collinearity analysis showed that there were three groups of tandem duplications, and 48 pairs of segmental duplication genes were identified in peanut. Our results regarding gene duplication events further elucidated the mechanism underlying the expansion of the LOX gene family. New genes might have been generated during such duplication events, thereby resulting in the generation of new biological functions (Conant and Wolfe, 2008).
Based on the protein structure, sequence similarity and presence of a chloroplast transit peptide (Feussner and Wasternack, 2002), peanut LOXs were classified into three subfamilies via phylogenetic analysis, 9-LOX, type I 13-LOX, and type II 13-LOX. The gene structure analysis indicated that most of these peanut genes had similar structures. Intron gain or loss was observed in several genes in the 9-LOX, type I 13-LOX, and type II 13-LOX subfamilies, which might be due to the selection pressures during peanut evolution (Zhang, 2003; Babenko et al., 2004). Accordingly, the diverse exon–intron structure contributed to their functional diversification (Mattick, 1994). Moreover, we gained additional insights into protein motifs in peanut (Figure 3). As expected, the motifs of the peanut LOXs were conserved, demonstrating that these conserved motifs might play key roles in the conservation of LOX gene functions. The predicted 3D structures of proteins were also determined to compare protein structures within the three subfamilies in different species (Figure 1). The modeled structures of four genes (AhLOX1, AhLOX28, AdLOX13, and AiLOX17), which lacked motifs 1, 4, 5, and 10, were looser than those possessing all of the 10 motifs (Supplementary Table S3). In addition, the structures of these four genes were different from each other as well, and the variation was likely to be the main factor leading to a variety of protein functions. Taken together, the structure and phylogeny of the LOX gene family showed diversity and complexity, thereby contributing to functional diversification.
The results for cis-acting elements revealed that peanut LOXs might participate in regulating various biological and molecular processes, such as transcription, cell cycle, development, hormones, and biotic/abiotic stresses. A large number of cis-acting regulatory elements related to hormones (MeJA, ABA, SA, GA, IAA, ET, etc.) and various stresses (anoxic conditions, drought, zein metabolism, wounding, low temperature, etc.) were identified in each peanut LOX promoter. Our findings were in consistent with previous studies which proved that LOX played multiple roles in cotton (Shaban et al., 2018) and tomato (Upadhyay et al., 2019). A recent study demonstrated that the cis-acting elements of LOX genes could regulate the response to abiotic stresses in tomato (Upadhyay et al., 2019). In addition, two previous studies revealed that the LOX gene played an important role in resistance to root-knot nematodes (Gao et al., 2008) and phloem feeders (Zhou et al., 2009). Therefore, this analysis further proved that the LOXs belonged to a multifunctional gene family, and they were involved in defense responses to various environmental stresses as well as in plant growth and development.
The expression patterns of peanut LOX genes were analyzed using public RNA-seq data for 22 tissues. The 72 candidate LOX genes exhibited prominently different expression profiles in 22 tissues, and even genes in the same subfamily were expressed differently. Thus it could be summarized that the LOX genes performed diverse and crucial functions in different growth and developmental stages. For instance, LOXs played a dual role in the fruit development of tomato; the three tomato LOX genes were separately regulated during fruit ripening, and the expressions could be influenced by ethylene and developmental factors (Griffiths et al., 1999). In Arabidopsis, LOX genes have been evidenced to participate in the developmental transition from vegetative to flowering, lateral root development and stress-induced senescence (Feng et al., 2012). Hence, different LOXs could regulate various physiological processes in different tissues by specific expression patterns.
Lipoxygenases were a multifunctional gene family, which was confirmed via the RNA-seq data and qRT-PCR results in this study. We found that peanut LOXs exhibited different expression patterns in response to abiotic stresses and hormone treatments, suggesting that they might be involved in various plant stress response pathways and closely related to hormonal regulation. As for rice, OsLOX-silenced plants were more susceptible to striped stem borers, which were chewing herbivores, but more resistant to brown planthoppers, which were phloem feeders; thus, the LOX played opposite roles with regard to resistance to chewing and phloem-feeding herbivores (Zhou et al., 2009). Additionally, the LOX participated in herbivore-induced JA biosynthesis, and the modulation of JA activity was essential for the plant defense responses (Zhou et al., 2009). It was also corroborated that the rice LOX3 gene could function in response to drought and pathogens (Liu et al., 2008). In peanut, a number of LOX genes were functionally characterized according to the response to Aspergillus-seed interactions. For instance, PnLOX1 and PnLOX2/PnLOX3 were either positively or negatively regulated during Aspergillus infection, and these genes were proved to be one of the host genetic factors influencing the aflatoxin contamination (Korani et al., 2018). In mature peanut seeds, AhLOX1 (PnLOX1) expression could be highly induced by Aspergillus spp., or by MeJA and wounding (Burow et al., 2000). Although multifunctional LOX gene families have been found in many plants, the functional characterization of LOXs in peanut was rarely investigated.
Drought is one of the key stresses that has negative impact on crop yield (Dietz et al., 2021). Above results from the analysis of cis-acting elements in promoters, gene expression profiles, and transgenic plants suggested the potential role of AhLOX genes in drought resistance. Overexpression of AhLOX29 significantly enhanced the resistance to drought in Arabidopsis (Figure 10). Supporting our findings, previous studides revealed that LOX6 was essential for the production of 12-oxo-phytodienoic acid in leaves and roots in response to osmotic stress and drought (Grebner et al., 2013; Savchenko et al., 2014). Furthermore, jasmonates produced in roots were independent in leaves, which was demonstrated by grafting experiments with jasmonate-deficient mutants (Grebner et al., 2013). Osmotic and salt stresses could trigger the overproduction of reactive oxygen species (ROS) in plants, and the scavenging of ROS was an important mechanism for the resistance to abiotic stresses (Ullah et al., 2017). MDA is widely used as an indicator of drought exposure to evaluate the degree of plasma membrane damage and the drought tolerance capacity of plants (Zhang et al., 2021). The decrease of chlorophyll contents in transgenic Arabidopsis was obviously lower than that in WT, which indicated that the overexpression of the AhLOX29 gene could reduce the negative effect of drought stress on Arabidopsis photosynthesis. Compared with the WT, the OE-AhLOX29 lines had higher SOD activities and lower MDA contents, suggesting the improved capacity to scavenge ROS, thereby protected the membrane from being damaged in transgenic plants. Hence, the AhLOX29 gene could enhance the resistance to drought stress in Arabidopsis. Peanut LOX genes are of critical significance for peanut growth and development as well as the response to various stresses.
Conclusion
In this study, 72 LOX genes in the whole peanut genome were identified and classified into 9-LOX, type I 13-LOX and type II 13-LOX subfamilies. The investigation of phylogenetic relationships, gene structures and protein motifs revealed that the structure and function of most LOXs in peanut were relatively conserved during evolution. The LOX genes are nonrandomly located on chromosomes, and most of them are located on chromosomes A9, B9, 9, and 19. The segmentally duplicated genes contribute significantly to the expansion of the LOX gene family. Peanut LOX genes are involved in various biological processes, including growth and development, hormones (MeJA, ABA, SA, and JA) and responses to multiple stresses (drought, salt, anoxic induction, wounding, and low temperature). Overexpression of AhLOX29 in Arabidopsis increased the tolerance to drought stress possibly by scavenging ROS and alleviating membrane damage. The systematic and comprehensive data from this study not only provide a foundation for revealing the functional roles of peanut LOX genes but also contribute to the selection of appropriate candidate LOX genes for further characterization with regard to plant development and stress responses.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YM and SS conceived and designed the study. YM performed the experiments and wrote the manuscript. QS helped the bioinformatics analysis. CY provided the plant materials. JW, XZ, CY, and CL revised the paper. All authors have read and approved the final version of the manuscript.
Funding
This research was supported by the innovation Project of SAAS (CXGC2021B33) and (CXGC2021A30), the National Natural Science Foundation of China (31901506), the Taishan Scholar Project of Shandong Province (ts201712080), the Agro-industry Technology Research System of Shandong Province (SDAIT-04\u201302), the Natural Science Foundation of Shandong Province (ZR2021MC128) and Shandong Elite Variety Project (2020LZGC001). The funders had no role in the design and data analysis of this study, and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to all the colleagues who provided helpful databases and useful assistance in our laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.832785/full#supplementary-material
Footnotes
1. ^https://www.peanutbase.org/
4. ^http://www.ebi.ac.uk/interpro/scan.html
5. ^https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
6. ^https://web.expasy.org/protparam/
7. ^http://molevol.cmima.csic.es/castresana/Gblocks_server.html
8. ^http://www.evolgenius.info/evolview/
9. ^http://gsds.cbi.pku.edu.cn/
11. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
12. ^https://beta.swissmodel.expasy.org/
13. ^https://www.peanutbase.org/
References
Andrew, W., Martino, B., Stefan, B., Gabriel, S., Gerardo, T., Rafal, G., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Babenko, V. N., Rogozin, I. B., Mekhedov, S. L., and Koonin, E. V. (2004). Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 32, 3724–3733. doi: 10.1093/nar/gkh686
Bannenberg, G., Martinez, M., Hamberg, M., and Castresana, C. (2009). Diversity of the enzymatic activity in the Lipoxygenase gene family of Arabidopsis thaliana. Lipids 44, 85–95. doi: 10.1007/s11745-008-3245-7
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bell, E., Creelman, R. A., and Mullet, J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 92, 8675–8679. doi: 10.1073/pnas.92.19.8675
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., and Schmutz, J. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi: 10.1038/s41588-019-0405-z
Bertioli, D. J., Seijo, G., Freitas, F. O., Valls, J., Leal-Bertioli, S., and Moretzsohn, M. C. (2011). An overview of peanut and its wild relatives. Plant Genet. Res. 9, 134–149. doi: 10.1017/S1479262110000444
Brash, A. R. (1999). Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 274, 23679–23682. doi: 10.1074/jbc.274.34.23679
Burow, G. B., Gardner, H. W., and Keller, N. P. (2000). A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant Mol. Biol. 42, 689–701. doi: 10.1023/A:1006361305703
Caldelari, D., Wang, G., Farmer, E. E., and Dong, X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75, 25–33. doi: 10.1007/s11103-010-9701-9
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Carrera, A., Echenique, V., Zhang, W., Helguera, M., Manthey, F., Schrager, A., et al. (2007). A deletion at the Lpx-B1 locus is associated with low lipoxygenase activity and improved pasta color in durum wheat (Triticum turgidum ssp. durum). J. Cereal Sci. 45, 67–77. doi: 10.1016/j.jcs.2006.07.001
Christensen, S. A., Huffaker, A., Kaplan, F., Sims, J., Ziemann, S., Doehlemann, G., et al. (2015). Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. U. S. A. 112, 11407–11412. doi: 10.1073/pnas.1511131112
Christensen, S. A., Nemchenko, A., Borrego, E., Murray, I., Sobhy, I. S., Bosak, L., et al. (2013). The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 74, 59–73. doi: 10.1111/tpj.12101
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Dietz, K.-J., Zörb, C., and Geilfus, C.-M. (2021). Drought and crop yield. Plant Biol. 23, 881–893. doi: 10.1111/plb.13304
Feng, B., Dong, Z., Xu, Z., Wang, D., and Wang, T. (2012). Molecular characterization of a novel type of lipoxygenase (LOX) gene from common wheat (Triticum aestivum L.). Mol. Breed. 30, 113–124. doi: 10.1007/s11032-011-9603-9
Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi: 10.1146/annurev.arplant.53.100301.135248
Finn, R. D., Jaina, M., Benjamin, S. B. C., Sam, G. J., Volker, H., Timo, L., et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–D251. doi: 10.1093/nar/gkj149
Gao, X., Starr, J., Goebel, C., Engelberth, J., Feussner, I., Tumlinson, J., et al. (2008). Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant-Microbe Interact. 21, 98–109. doi: 10.1094/MPMI-21-1-0098
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. E., Wilkins, M. R., Appel, R. D., et al. (1999). Protein identification and analysis tools on the ExPASy server. Methods Mol. Biol. 112:531. doi: 10.1385/1.59259-584-7:531
Grebner, W., Stingl, N. E., Oenel, A., Mueller, M. J., and Berger, S. (2013). Lipoxygenase6-dependent Oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 161, 2159–2170. doi: 10.1104/pp.113.214544
Griffiths, A., Barry, C., Alpuche-Solis, A. G., and Grierson, D. (1999). Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. Front. Plant Sci. 50:793. doi: 10.1093/jexbot/50.335.793
Guo, A.-Y., Zhu, Q.-H., Chen, X., and Luo, J.-C. (2007). GSDS: a gene structure display server. Yichuan 29, 1023–1026.
Hao, L., Wang, Y., Zhang, J., Xie, Y., Zhang, M., Duan, L., et al. (2013). Coronatine enhances drought tolerance via improving antioxidative capacity to maintaining higher photosynthetic performance in soybean. Plant Sci. 210, 1–9. doi: 10.1016/j.plantsci.2013.05.006
Heitz, T., Bergey, D. R., and Ryan, C. A. (1997). A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114, 1085–1093. doi: 10.1104/pp.114.3.1085
Hui, D., Lai, J., Qian, W., Zhang, S., Liang, C., Dai, Y. S., et al. (2016a). Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 244, 1–7. doi: 10.1016/j.plantsci.2015.11.009
Hui, S., Wang, P., Li, C., Han, S., and Wang, X. (2016b). Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 6:35245. doi: 10.1038/srep35245
Jin, H. S., Van, K., Dong, H. K., Kim, K. D., Jang, Y. E., Choi, B. S., et al. (2008). The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 8:133. doi: 10.1186/1471-2229-8-133
Josh, C., Chu, Y., Brian, S., and Peggy, O. A. (2016). A developmental Transcriptome map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 7:1446. doi: 10.3389/fpls.2016.01446
Khan, S. A., Zhang, C., Ali, N., and Gandeka, M. (2020). Highdensity SNP map facilitates fine mapping of QTLs and candidate genes discovery for Aspergillus favus resistance in peanut (Arachis hypogaea). Theor. Appl. Genet. 133, 2239–2257. doi: 10.1007/s00122-020-03594-0
Kolomiets, M. V., Hannapel, D. J., Chen, H., Tymeson, M., and Gladon, R. J. (2001). Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13, 613–626. doi: 10.1105/tpc.13.3.613
Korani, W., Chu, Y., Holbrook, C. C., and Ozias-Akins, P. (2018). Insight into genes regulating post-harvest Aflatoxin contamination of Tetraploid Peanut from transcriptional profiling. Genetics 209, 143–156. doi: 10.1534/genetics.118.300478
Liu, M., and Chervin, C. (1997). Ethylene and fruit ripening. Physiol. Plant. 101, 727–739. doi: 10.1111/j.1399-3054.1997.tb01057.x
Liu, N., Jiang, L., Zhang, W., Liu, L., Zhai, H., and Wan, J. (2008). Role of LOX3 gene in alleviating adverse effects of drought and pathogens in Rice. Rice Sci. 15, 276–282. doi: 10.1016/S1672-6308(09)60004-4
Liu, L., Sonbol, F.-M., Huot, B., Gu, Y., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7. doi: 10.1038/ncomms13099
Marchler-Bauer, A., Anderson, J., Cherukuri, P., Dewweese-Scott, C., Geer, L., Gwadz, M., et al. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196. doi: 10.1093/nar/gki069
Mariutto, M. (2011). The elicitation of a systemic resistance by pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 11, 29–29. doi: 10.1186/1471-2229-11-29
Mattick, J. S. (1994). Introns: evolution and function. Curr. Opin. Genet. Dev. 4, 823–831. doi: 10.1016/0959-437X(94)90066-3
Milligan, S. J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V. M. (1998). The root knot nematode resistance gene mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319. doi: 10.1105/tpc.10.8.1307
Müller, V., Amé, M., Carrari, F., Gieco, J. O., and Asis, R. (2014). Lipoxygenase activation in Peanut seed cultivars resistant and susceptible to Aspergillus parasiticus colonization. Phytopathology 104, 1340–1348. doi: 10.1094/PHYTO-12-13-0338-R
Ogunola, O. F., Hawkins, L. K., Mylroie, E., Kolomiets, M. V., Borrego, E., Tang, J. D., et al. (2017). Characterization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS One 12, e0181265. doi: 10.1371/journal.pone.0181265
Ozalvo, R., Cabrera, J., Escobar, C., Christensen, S. A., Borrego, E. J., Kolomiets, M. V., et al. (2014). Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol. Plant Pathol. 15, 319–332. doi: 10.1111/mpp.12094
Porta, H., and Rocha-Sosa, M. (2002). Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 130, 15–21. doi: 10.1104/pp.010787
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Rayko, H., and Baldwin, I. T. (2010). Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. Cell Mol. Biol. 36, 794–807. doi: 10.1046/j.1365-313X.2003.01921.x
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. doi: 10.1105/tpc.12.5.707
Roy Chowdhury, M., Li, X., Qi, H., Li, W., Sun, J., Huang, C., et al. (2016). Functional characterization of 9−/13-LOXs in Rice and silencing their expressions to improve grain qualities. Biomed. Res. Int. 2016, 1–8. doi: 10.1155/2016/4275904
Sarde, S. J., Kumar, A., Remme, R. N., and Dicke, M. (2018). Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Mol. Biol. 98, 375–387. doi: 10.1007/s11103-018-0785-y
Savchenko, T., Kolla, V. A., Wang, C.-Q., Nasafi, Z., Hicks, D. R., Phadungchob, B., et al. (2014). Functional convergence of Oxylipin and Abscisic acid pathways controls Stomatal closure in response to drought. Plant Physiol. 164, 1151–1160. doi: 10.1104/pp.113.234310
Shaban, M., Ahmed, M. M., Sun, H., Ullah, A., and Zhu, L. (2018). Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics 19, 599. doi: 10.1186/s12864-018-4985-2
Silke, A., and Baldwin, I. T. (2010). Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles. Science 329, 1075–1078. doi: 10.1126/science.1191634
Sudhir, K., Glen, S., Li, M., Christina, K., and Koichiro, T. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Tamara, V., Marta, M., Miguel Angel, L., Jorge, V., Tomas, C., Liam, D., et al. (2007). Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19, 831–846. doi: 10.1105/tpc.106.046052
Timothy, B. L., Nadya, W., Chris, M., and Wilfred, W. L. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Ullah, A., Sun, H., Hakim, X., and Yang, and Zhang, X., (2017). A novel cotton WRKY-gene, GhWRKY6-like, improves salt tolerance by activating the ABA signalling pathway and scavenging of reactive oxygen species. Physiol. Plant. 162, 439–454. doi: 10.1111/ppl.12651
Umate, P. (2011). Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 6, 335–338. doi: 10.4161/psb.6.3.13546
Upadhyay, R. K., Handa, A. K., and Mattoo, A. K. (2019). Transcript abundance patterns of 9- and 13-Lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to Each stress. Genes 10. doi: 10.3390/genes10090683
Veldink, G. A., Vliegenthart, J. F., and Boldingh, J. (1977). Plant lipoxygenases. Prog. Chem. Fats Other Lipids 15, 131–166. doi: 10.1016/0079-6832(77)90014-3
Wankun, D., Yongbo, W., Zexian, L., Han, C., and Yu, X. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Willis, D. (2005). Aspergillus infection inhibits the expression of peanut 13S-HPODE-forming seed lipoxygenases. Mol. Plant Microbe Interact. 18, 1081–1089. doi: 10.1094/MPMI-18-1081
Yan, J., Chia, J. C., Sheng, H., Jung, H. I., Zavodna, T. O., Lu, Z., et al. (2017). Arabidopsis pollen fertility requires the transcription factors CIT1 and SPL7 that regulate copper delivery to anthers and Jasmonic acid synthesis. Plant Cell 29, 3012–3029. doi: 10.1105/tpc.17.00363
Ying, Z. A., and Yz, B. (2020). Effect of lipoxygenase-3 on storage characteristics of peanut seeds - ScienceDirect. J. Stored Prod. Res. 87:101589. doi: 10.1016/j.jspr.2020.101589
Yu, L. H., Wu, S. J., Peng, Y. S., Liu, R. N., and Xiang, C. B. (2015). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. doi: 10.1111/pbi.12358
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhang, Y., Luan, Q., Jiang, J., and Li, Y. (2021). Prediction and utilization of malondialdehyde in exotic pine Under drought stress using near-infrared spectroscopy. Frontiers. Plant Sci. 12. doi: 10.3389/fpls.2021.735275
Zhou, G., Qi, J., Ren, N., Cheng, J., Erb, M., Mao, B., et al. (2009). Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 60, 638–648. doi: 10.1111/j.1365-313X.2009.03988.x
Keywords: peanut, lipoxygenase, gene family, expression pattern, drought stress
Citation: Mou Y, Sun Q, Yuan C, Zhao X, Wang J, Yan C, Li C and Shan S (2022) Identification of the LOX Gene Family in Peanut and Functional Characterization of AhLOX29 in Drought Tolerance. Front. Plant Sci. 13:832785. doi: 10.3389/fpls.2022.832785
Edited by:
Swarup Roy Choudhury, Indian Institute of Science Education and Research, Tirupati, IndiaReviewed by:
Gongke Zhou, Qingdao Agricultural University, ChinaAutar Krishen Mattoo, Agricultural Research Service (USDA), United States
Jay Prakash Awasthi, Assam University, India
Copyright © 2022 Mou, Sun, Yuan, Zhao, Wang, Yan, Li and Shan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifei Mou, eWlmZWltb3UxMTIzQDE2My5jb20=; Shihua Shan, c2hhbnNoX3NwcmlAMTYzLmNvbQ==
 Yifei Mou
Yifei Mou Quanxi Sun
Quanxi Sun Cuiling Yuan
Cuiling Yuan Xiaobo Zhao
Xiaobo Zhao Juan Wang
Juan Wang Caixia Yan
Caixia Yan Chunjuan Li
Chunjuan Li Shihua Shan*
Shihua Shan*