
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 March 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.832669
This article is part of the Research TopicImprovement for quality and safety traits in horticultural plantsView all 25 articles
 Jinfen Wen1†
Jinfen Wen1† Junheng Lv2†
Junheng Lv2† Kai Zhao2†
Kai Zhao2† Xiang Zhang2†
Xiang Zhang2† Zuosen Li2†
Zuosen Li2† Hong Zhang2†
Hong Zhang2† Jinlong Huo2†
Jinlong Huo2† Hongjian Wan3
Hongjian Wan3 Ziran Wang2
Ziran Wang2 Haishan Zhu2
Haishan Zhu2 Minghua Deng2*
Minghua Deng2*Ethylene is very important in the process of plant development and regulates the biosynthesis of many secondary metabolites. In these regulatory mechanisms, transcription factors (TFs) that mediate ethylene signals play a very important role. Capsaicinoids (CAPs) are only synthesized and accumulated in Capsicum species, causing their fruit to have a special pungent taste, which can protect against attack from herbivores and pathogens. In this study, we identified the TF CcERF2, which is induced by ethylene, and demonstrated its regulatory effect on CAPs biosynthesis. Transcriptome sequencing analysis revealed that the expression patterns of CcERF2 and multiple genes associated with CAPs biosynthesis were basically the same. The spatiotemporal expression results showed CcERF2 was preferentially expressed in the placenta of the spicy fruit. Ethylene can induce the expression of CcERF2 and CAPs biosynthesis genes (CBGs). CcERF2 gene silencing and 1-methylcyclopropene (1-MCP) and pyrazinamide (PZA) treatments caused a decrease in expression of CBGs and a sharp decrease in content of CAPs. The results indicated that CcERF2 was indeed involved in the regulation of structural genes of the CAPs biosynthetic pathway.
Peppers (Capsicum spp.) are important vegetables worldwide. Pepper fruits are diverse in color, rich in nutrients, have a special pungency and aroma, and so are widely used as food additives (Ou et al., 2018). The pungency of pepper fruit is derived from capsaicinoids (CAPs), which are only biosynthesized in Capsicum plants (Iwai et al., 1979; Liu et al., 2019). The CAPs have the effect of curbing attack by herbivores and microorganisms. At the same time, CAPs are also widely used in many fields such as food, medicine, cosmetics, and agricultural pest control; CAPs are also widely used in riot prevention, personal defense, and in the military and national defense (Lejeune et al., 2007; Tewksbury et al., 2008; Ludy et al., 2012). The CAPs are synthesized and accumulated in fruit placental tissues (Iwai et al., 1979). So far, more than 22 CAPs have been identified in peppers. The most important components of CAPs are capsaicin (CaP) and dihydrocapsaicin (DhCaP), and these represent approximately 90% of the total content of CAPs. The accumulation of CAPs is affected by many factors, including endogenous (for example, variety and developmental stage of the fruit) and exogenous factors (for example, light, temperature, water conditions, and biological stress; Phimchan and Techawongstien, 2012; Garruña-Hernández et al., 2013). Genotype is the most important determinant of CAPs content (Bosland et al., 2012; Liu et al., 2017); among the five domesticated species of Capsicum, Capsicum chinense, and Capsicum frutescens accumulate significantly higher contents of CAPs (Deng et al., 2009; Bosland et al., 2012). It is reported that factors such as plant growth regulators, chemicals, temperature, light, and drought stress can change the content of CAPs (Gurung et al., 2011).
The CAPs are synthesized by the fusion of phenylpropane and branched-chain fatty acid pathways in the placenta (Aluru et al., 2003; Blum et al., 2003; Mazourek et al., 2009). The CAPs biosynthesis genes (CBGs) that have been determined to be involved in biosynthesis of CAPs include PAL, Ca4H, 4CL, BCAT, Kas, FatA, ACL, ACS, CoMT, pAMT, and CS (Abraham-Juárez et al., 2008; Liu et al., 2013). The biosynthetic mechanism of CAPs has been widely elucidated using bioinformatics analysis and multi-omics (Qin et al., 2014). As CAPs have considerable application value and commercial use, and much effort has gone into enhancing their content. However, their biosynthesis greatly changes spatiotemporally, and expression of CBGs is precisely regulated at the transcriptional level. Manipulating the expression level of crucial CBGs seems to determine increasing the content of CAPs (Abraham-Juárez et al., 2008; Sun et al., 2019; Zhu et al., 2019). In highly spicy peppers, the transcription level of CBGs (such as pAMT, Kas, and CS) is always higher than that of less spicy varieties (Abraham-Juárez et al., 2008). It is very important that manipulating the expression levels of some transcription factors (TFs) usually change the transcription levels of all genes in the metabolic pathway, thereby affecting the final content of the compound.
For example, the TFs CaMYB31, CaMYB108, CaMYB48, Erf, and Jerf are associated with regulating the content of CAPs (Keyhaninejad et al., 2014; Phimchan et al., 2014; Arce-Rodríguez and Ochoa-Alejo, 2017; Sun et al., 2019; Zhu et al., 2019). It was found that the Solanaceae-specific TF MYB31 affects the accumulation of CAPs by directly targeting CBGs, resulting in changing the expression level of CBGs (Zhu et al., 2019). The TF CaMYB108 activates the CBG promoters, especially those of COMT, pAMT, and KasI, and promotes enhanced expression of CBGs, thereby promoting the biosynthesis of CAPs (Sun et al., 2019). By directly binding and regulating the expression of CBGs, TF CaMYB48 participates in the biosynthesis of CAPs, but the transcriptional regulation of CAPs biosynthesis has not been fully clarified (Sun et al., 2020). Therefore, it is necessary to identify TFs associated with the biosynthesis of CAPs.
Ethylene plays a significant role in the plant life process (Hu et al., 2020). After synthesis of ethylene, it binds to the receptor ETR and transmits the signal to the nucleus through MAPKK and EIN2 (Gray, 2004). The EIN2 binds to EIN3/EIL1 and EIN3/EIL1 binds to the ERF1 promoter. The ERF1 binds to the GCC-box-containing genes in the downstream promoter region specifically and their expression can promote secondary metabolism product synthesis (Fujimoto et al., 2000; Brown et al., 2003; Paul et al., 2020). For example, in tobacco, ERF189 and ERF163 can bind to the GCC-box in the promoter region of the tobacco nicotine synthesis-related gene PMT2 specifically, and directly promote synthesis of tobacco alkaloids (Shoji et al., 2010). The B3 subfamily proteins ORCA3 and ORA59 of the ERF family are two typical TFs associated with the regulation of secondary metabolites. The ORCA3 can upregulate the expression level of indole alkaloid synthesis-related genes in terpenoids, and promote the terpenoid indole biosynthesis (van der Fits and Memelink, 2000, 2001). Lithospermum erythrophyllum LeERF-1 affects the secondary metabolites of shikonin positively through a mechanism similar to that of ORCA3 in affecting secondary metabolites (Zhang et al., 2011). In Catharanthus roseus, CrERF5 upregulates the biosynthesis and accumulation of bisindole alkaloids (Pan et al., 2019).
The TFs and key enzymes in ethylene synthesis and its signal transduction can promote the biosynthesis of secondary metabolites. In many cases (plants, cells, and hairy roots), the addition of ethylene, addition of 1-aminocyclopropane 1-carboxylic acid (ACC), and overexpression of related TFs can increase the biosynthesis of secondary metabolites (Pan et al., 2000; Gantet and Memelink, 2002; Zhao et al., 2004; Buer et al., 2006; De Boer et al., 2011). In previous studies, PAL genes possessed a homolog of the GCC-box in their promoters and ERF genes could combine with their cis-acting element (Ohme-Takagi and Shinshi, 1995; Keyhaninejad et al., 2014). Both Erf and Jerf in pepper have been proposed to be involved in accumulation of pungency (Keyhaninejad et al., 2014). Many of the ERF family are TFs that are candidates for regulating CAP biosynthesis (Song et al., 2020). However, how CAP biosynthesis is regulated at the transcription level is still unknown in peppers.
In this paper, the ethylene-induced AP2/ERF TF CcERF2, which was particularly expressed in the placenta was identified. We confirmed that CcERF2 was associated with CAPs biosynthesis.
The C. chinense inbred line SL08 has a high CAPs content and was derived from Shuan La, the hottest pepper genotype in China (Deng et al., 2009). The Capsicum annuum inbred line H19 has a low CAPs content, and was derived from Xiangtan Chi Ban Jiao. Pepper seeds germinated in the soil of cell plastic flats in complete darkness at 28°C. The seedlings were grown in a greenhouse of Yunnan Agricultural University on campus under normal conditions.
The CaP and DhCaP were extracted and detected according to Deng et al. (2009). The total CAPs content was calculated as (CaP + DhCaP)/0.91 (Deng et al., 2009). Fruits in different development stages of SL08 and H19 were used as material for extraction and detection of CaP and DhCaP. All experiments were repeated three times.
RNA-seq was performed according to Liu et al. (2012). Fruits of SL08 at 4, 14, 24, 34, 44, and 54 days after pollination (DAP) were used to isolate total RNA. All of RNA-seq data generated in this study are available from the NCBI Short Read Archive (SRA, BioProject ID: PRJNA789050), and the raw RNA-seq data are freely available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789050.
Fruit at 24 DAP was used to investigate the effect of different exogenous substances on expression level of CcERF2 and CBGs after 0.5, 1.0, 1.5, 3.5, 5.5, 8.5, and 11.5 h. Treated with exogenous substances, the 24-DAP fruits of SL08 were soaked in sterile water with 0.01 g/L 6-benzylaminopurine (6-BA), 20 g/L PEG6000, 30% H2O2, 0.17 g/L gibberellic acid (GA3), 0.14 g/L salicylic acid (SA), 17.53 g/L NaCl, 90 g/L glutamate (Glu), or 0.1 mmol/L ethephon (with sterile water as control) under normal conditions (16 h of light at 30 ± 2°C and 8 h of darkness at 20 ± 2°C). After treatment, all fruits were frozen in liquid nitrogen and immediately stored at −80°C for gene expression analysis. All experiments were repeated three times.
The open reading frame of CcERF2 was cloned according to Deng et al. (2012). The primers used in the experiment are shown in Supplementary Table S1. The BLAST-protein-nucleic acid (BLASTP) analysis was performed on CcERF2 through a database1; PSORT Prediction was performed for subcellular location prediction; and MEME was used to analyze the amino acid sequence motif. The Portparam tool was used to predict the physical and chemical properties of the protein. SignalP-5.0, TMHMM, and Nepos were used for signal peptide, transmembrane structure, and phosphorylation site of the protein prediction, respectively; SOPMA was used to predict its secondary structure; and ClustalX was used for sequence alignment and phylogenetic analysis. The MEGA 6.0 software was used to construct a phylogenetic tree based on the amino acid sequence through the neighbor joining method, and the bootstrap method was used to evaluate the reliability of each node in the tree, repeated 1,000 times.
Subcellular localization and transcriptional activation analysis were performed according to Zhu et al. (2019) and Sun et al. (2020). The full-length coding sequences of CcERF2 was cloned into the pAN580 (green fluorescent protein, GFP) vector and fused to the N-terminus of GFP under the control of the CaMV 35S promoter. The constructs were separately introduced into tobacco protoplasts for transient expression, and the GFP fluorescence signals were detected using a Zeiss lsm710 confocal laser scanning microscope (Carl Zeiss Inc., Jena, Germany).
The RNAiso Plus (Takara, Dalian, P. R. China) was used to extract total RNA from pepper fruit. The synthesis of cDNA first strand refers to the instructions of the High Fidelity PrimeScript® RT-PCR Kit (Takara). According to the sequence of related genes revealed by transcriptome sequencing data, specific primers were respectively designed, and the internal reference was the β-ACTIN gene. Quantitative real-time PCR (qRT-PCR) was performed according to the method provided by the SYBR®Premix Ex TaqTM II (Tli RNaseH Plus) kit (Takara). The primers used in this study are shown in Supplementary Table S1.
A fragment of the CcERF2 coding sequence (CDS) with low similarity to other genes was cloned into pTRV2 and generated the silencing vector pTRV2–CcERF2. The VIGS were carried as reported (Zhu et al., 2019). In short, the pTRV2–CcERF2 and pTVR1 vectors were co-injected into the cotyledon stage seedlings of line SL08. The empty vectors pTRV2 and pTVR1 were co-infiltrated as a control, and pTRV2–PDS and pTVR1 were co-infiltrated as a technical control. The RNA isolated from 24-DAP fruits was used for expression analysis, and 34-DAP fruits were used for CaP and DhCaP measurement. The primer information used in this study is shown in Supplementary Table S1.
The SL08 pepper fruit of 24 DAP was used to study the effects of 1-methylcyclopropene (1-MCP) and pyrazinamide (PZA) on expression of CBGs and content of CAPs. The 1-MCP treatment follows: 1 μl L−1 1-MCP fumigated for 12 h and kept in darkness at a temperature of 20 ± 2°C and humidity of 85 ± 5%. The PZA treatment follows: 100 mM PZA was sprayed onto the surface of the fruit until there were droplets, and kept in darkness at a temperature of 20 ± 2°C and humidity of 85 ± 5%. The fruits after 5 days of treatment were stored in liquid nitrogen and used for subsequent analysis. All experiments were repeated three times.
Ethylene release rate was determined according to Xu et al. (2018).
Each experiment contained three biological replicates and three technical replicates. The error bars indicate SEs. One-way ANOVA was performed to identify significant differences. The relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Control expression was without induction at the 0 h time point.
Accumulation of CaP and DhCaP was detected in both lines, but the content of CaP was much higher (about 3–4 times) than that of DhCaP. The CaP and DhCaP contents were much higher in the placenta of SL08 than in H19 (Supplementary Figure S1). Both CaP and DhCaP began to accumulate at 9 DAP in the placenta of SL08, reached a peak at 44 DAP, and then began to decline. Both CaP and DhCaP began to accumulate at 11 DAP in the placenta of H19, and began to decrease at 49 DAP. The CaP, DhCaP, and total CAPs contents in the placenta of SL08 were 44.4, 63.1, and 48.0 times those in H19, respectively.
Expression patterns of CBGs during development stages of the placenta were analyzed using transcriptome sequencing data of the placenta and pericarp of line SL08 during the early stage of the laboratory study (Figure 1). Expressions of CcPAL, CcCa4H, Cc4CL, CcCoMT, CcpAMT, CcCS, CcBCAT, CcKAS, CcACL, CcFAT, and CcACS genes were positively associated with accumulation of CAPs in SL08. The expression of these genes (except CcACL) showed a low expression level in fruit at 4 DAP, increased, and then decreased. Gene CcCS (Capana05g000531) was not expressed in 4-DAP fruit, and reached a maximum in 24-DAP fruit. Expression of CcCoMT (Capana03g001811), CATCcBCAT (Capana04g000751), CcKAS (Capana01g000111), and CcFATA (Capana06g000197) reached their highest values in 14-DAP fruit. Expression of CcCa4H (Capana06g000272) and CcpAMT (Capana10g001341) genes reached their maximum in 34-DAP fruit. Expression of CcPAL (Capana09g002199) reached its highest value in 44-DAP fruit.
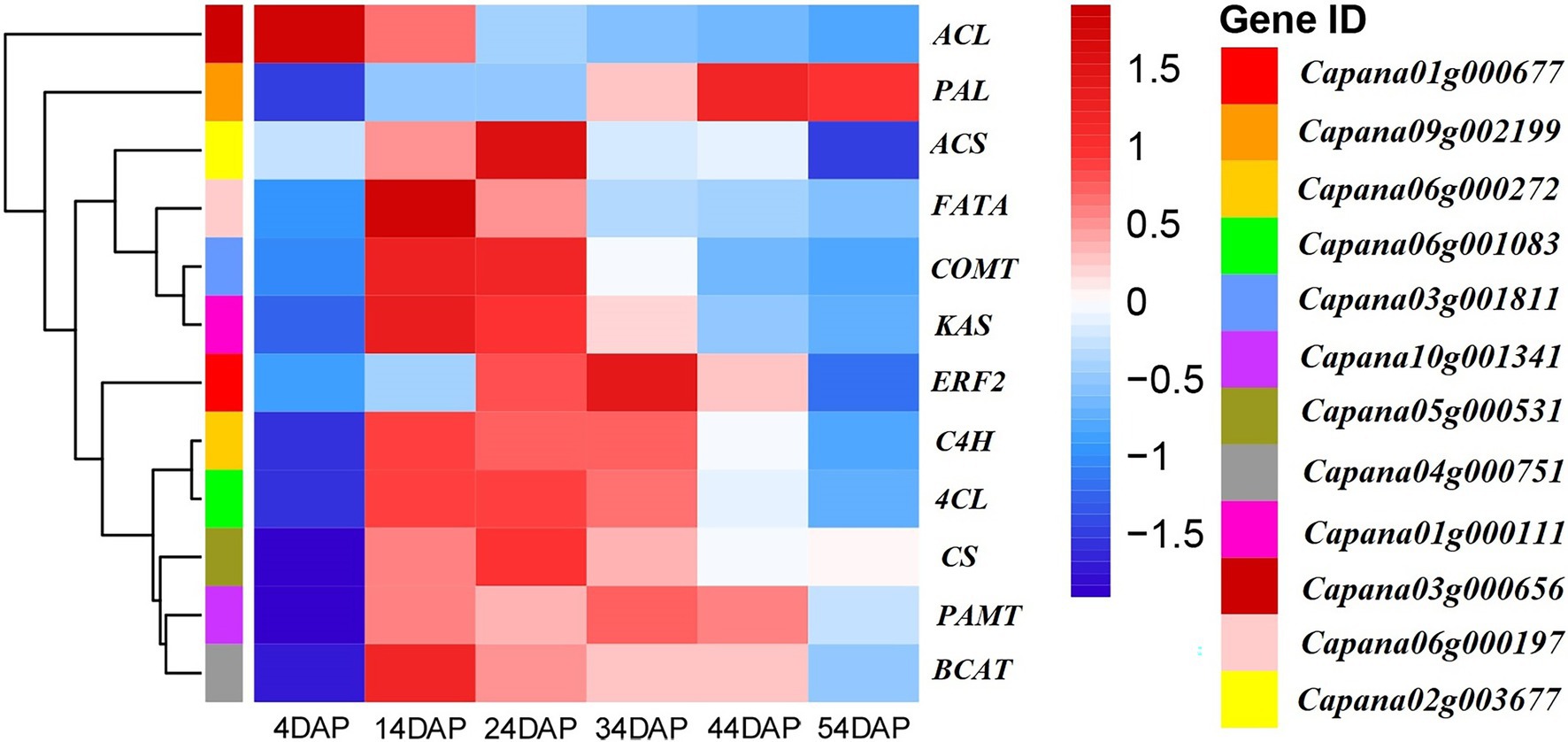
Figure 1. Heat map indicating the capsaicinoids (CAPs) biosynthesis genes (CBGs) and CcERF2 expression patterns in fruit of inbred line SL08. The heatmap was generated by transcriptome sequencing data with R software packages. The data were normalized in each row, and the numbers on the right side of the figure indicate the gene expression level corresponding to color.
Based on the transcriptome sequencing data of lines SL08 and H19 at different developmental stages of placenta completed in the laboratory, combined with the changes in the CaP and DhCaP contents of the placenta of both lines (Supplementary Figure S1), the AP2/ERF TF CcERF2 (Capana01g000677) was identified. Its expression pattern was similar to that of CBGs (Figure 1). Expression of CcERF2 was almost undetectable in the early stage of placenta (4 DAP), then rose rapidly, reached a maximum at 34 DAP, and then decreased.
We measured the expression level of CcERF2 in some tissues of line SL08 and found that it was mainly expressed in the placenta (Figure 2A). It also showed considerable level of expression in seeds and pericarp, meaning that it has a role in these tissues. We also investigated CcERF2 expression in the placenta of both lines, and found significantly higher expression in SL08 than in H19 (Figure 2B). Expression of CcERF2 in the placenta had the same pattern as the contents of CaP and DhCaP. Thus, our results indicated that CcERF2 may play a vital role in CAPs biosynthesis.
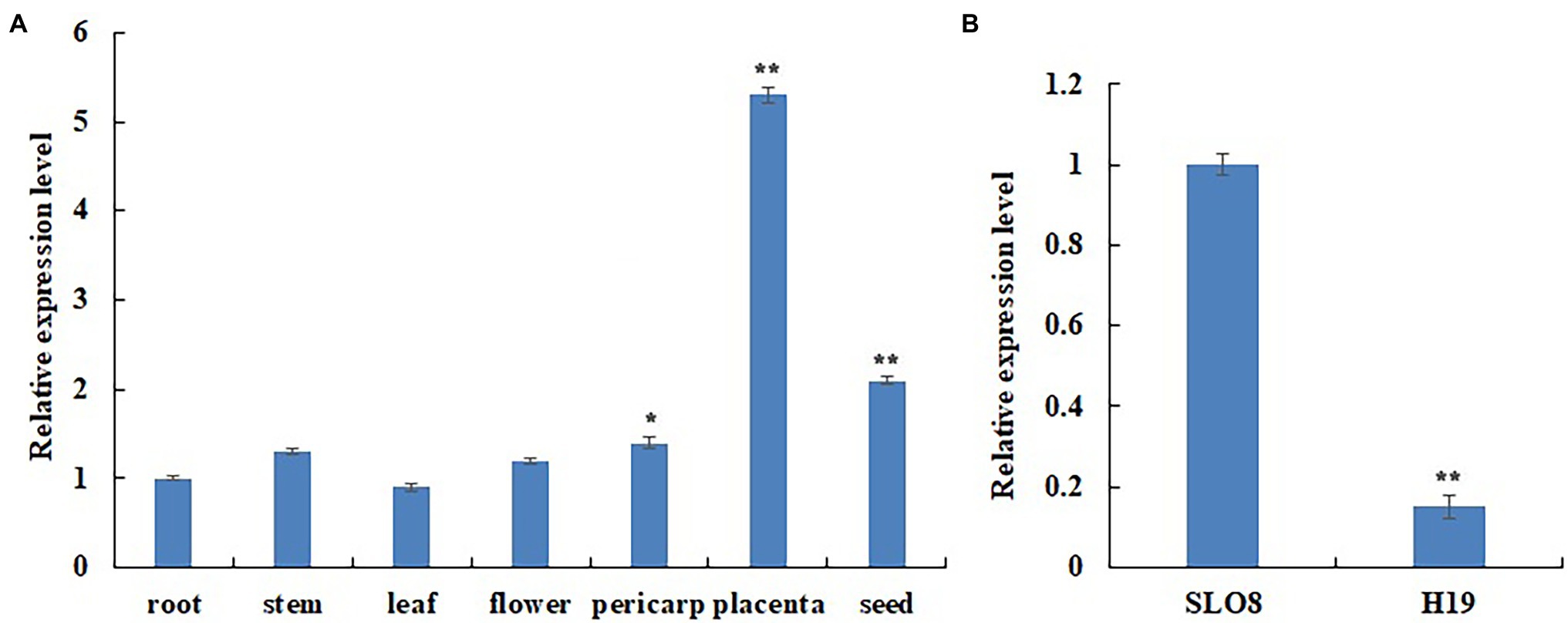
Figure 2. Differential expression assays of CcERF2. (A) Differential expression assays of CcERF2 in different tissues of pepper inbred line SL08; Roots, stems, leaves and flowers were collected from 30-day-old seedlings. Fruits at 24 days after pollination (DAP) was collected and was divided into pericarps, placentas and seeds. The relative expression of the root was set to 1, and that of all the other tissues was measured relative to that of the root. (B) Differential expression assays of CcERF2 in pepper inbred line SL08 and H19 fruits. Fruits at 24 DAP was collected to investigate the CcERF2 expression. The relative expression of the SL08 was set to 1, and that of H19 was measured relative to that of the SL08. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9). Student’s t-test was used to identify significant differences compared to the control (*p < 0.05, **p < 0.01).
Based on our transcriptome sequencing data of SL08 and the pepper genome data in the public database, specific primers were designed to clone the full-length CDS of CcERF2 from SL08. Nucleotide sequence analysis showed that CcERF2 length was 795 bp. The molecular formula of CcERF2 is C1328H2050N376O409S9, molecular weight is 30,115.70 D, and theoretical isoelectric point is 5.74. It is a fat-soluble, hydrophilic, and unstable protein. CcERF2 had no signal peptide and no transmembrane structure, and is located in the nucleus (88.8% probability).
The protein domain prediction results showed that CcERF2 belonged to the AP2 superfamily, and the conservative amino acid sequence position was 74–131 (LYRGIRQRPWGKWAAEIRDPRKGVRVWLGTFNTAEEAARAYDKEARKIRGEKAKVNFP; Figure 3A). This domain specifically bound to the 11-bp GCC-box of the ethylene response element and was essential for the ethylene response. Based on BLAST, 48 amino acid sequences including CcERF2 were obtained, and 43 motifs were obtained after motif significance test and analysis. On the whole, the predicted motifs differed within the same family, but the conserved motifs in the same subgroup were almost the same; that is, the closer the related species, the more similar were the motifs. For peppers, they all contained 13 conserved motifs: 1–8, 11, 12, 17, 19, and 22. Of these, motifs 1, 3, and 5 covered the AP2 conserved structure domain (Figure 3B). The secondary structure of CcERF2 protein contained 73 alpha helices (27.56%), 22 extension chains (8.42%), and nine beta turns (3.41%). There were 160 random coils (60.61%). The evolutionary relationship of amino acid sequences showed that the selected ERF sequences were grouped together in different families. This meant that they were conserved in the evolutionary relationship. In genetic relationship, CcERF2 protein was the closest to Solanaceae. In the Solanaceae, it was closest to Capsicum and Solanum, and most distant to Petunia (Figure 3C). The phosphorylation sites of CcERF2 protein were predicted, and 19 amino acid residue types were screened: six types of T, three types of Y, and 10 types of S. The CcERF2 was localized to the cell nucleus (Figure 4A) and the CcERF2 protein had strong activation activity (Figure 4B).
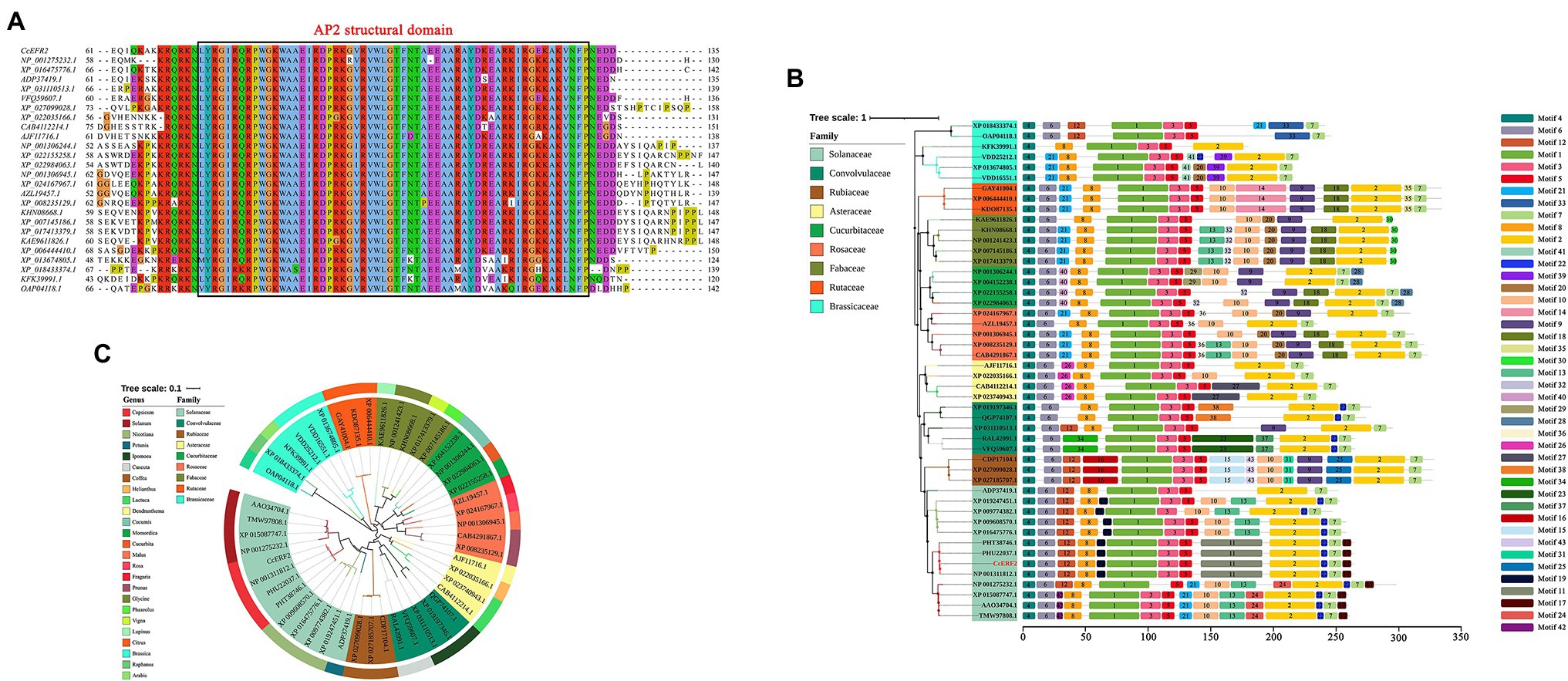
Figure 3. Phylogenic tree of ERF transcription factors (TFs), analysis of promoter cis-acting elements and multiple amino acid sequence alignment of CcERF2 TFs. (A) Multiple amino acid sequence alignment of ERF2 TFs. (B) Promoter cis-acting elements comprising CcERF2 in Capsicum chinense and 47 ERF2 promoters in plant. (C) Phylogenetic tree comprising CcERF2 in C. chinense and 47 ERF2 TFs in plant.
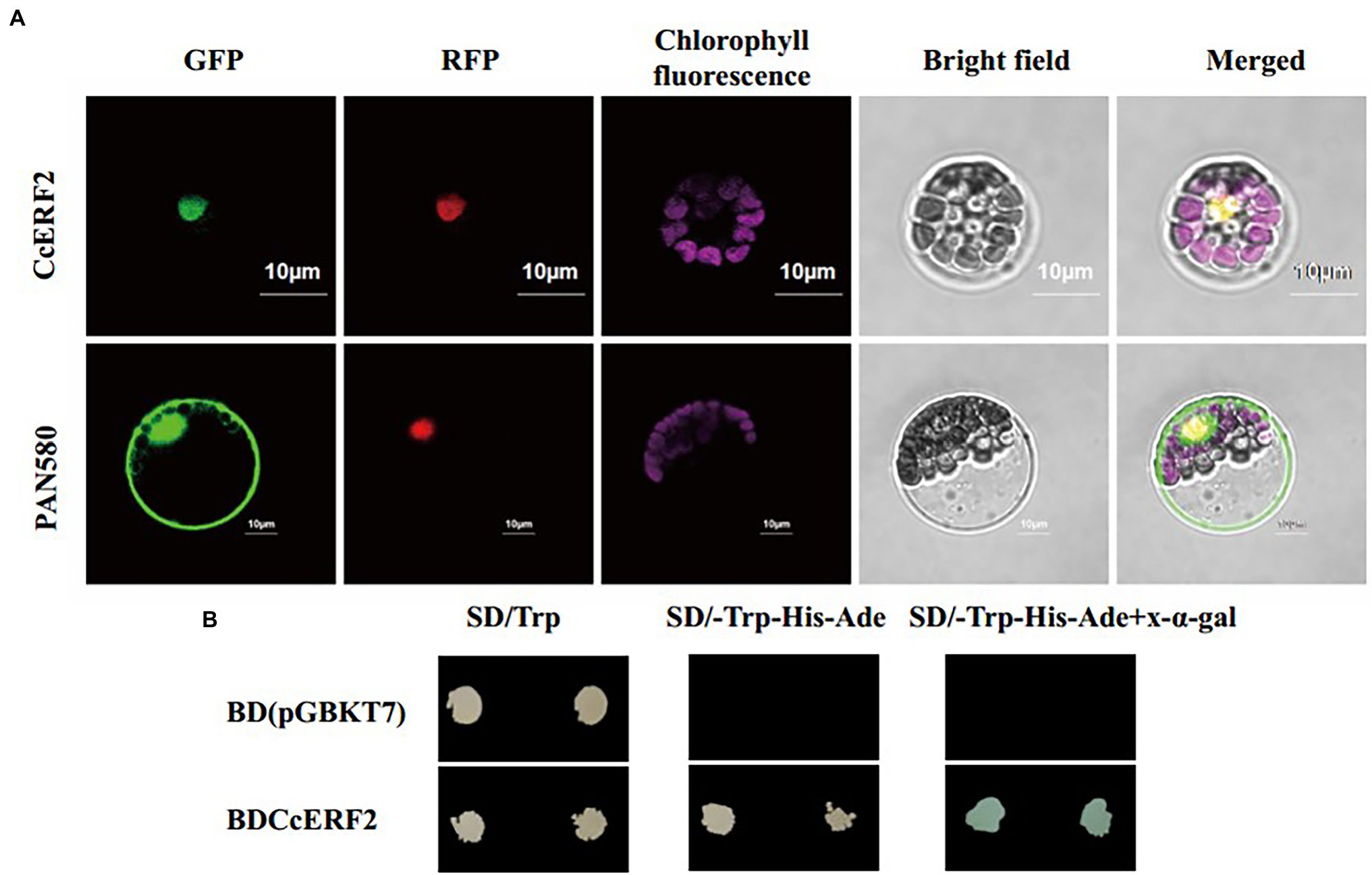
Figure 4. Subcellular localization and transcriptional activation analysis. (A) Green fluorescent protein (GFP) signals indicated that CcERF2 was localized to nucleus. GFP fluorescence signals were detected using a Zeiss lsm710 confocal laser scanning microscope (Carl Zeiss, Inc., Jena, Germany). Scale bars: 10 μm. (B) Auxotroph plates of SD/–Leu–His–Ade (middle) and SD/–Leu–His–Ade–x-α-gal (right) showing transcriptional activation of protein. SD/Trp, medium lacking tryptophan; SD/-Trp-His-Ade, medium lacking tryptophan, histidine, and adenine.
Exogenous substances were applied to the 24-DAP fruit of SL08 to test the regulation model. Expression of CcERF2 in placental tissues was analyzed using qRT-PCR (Figure 5).
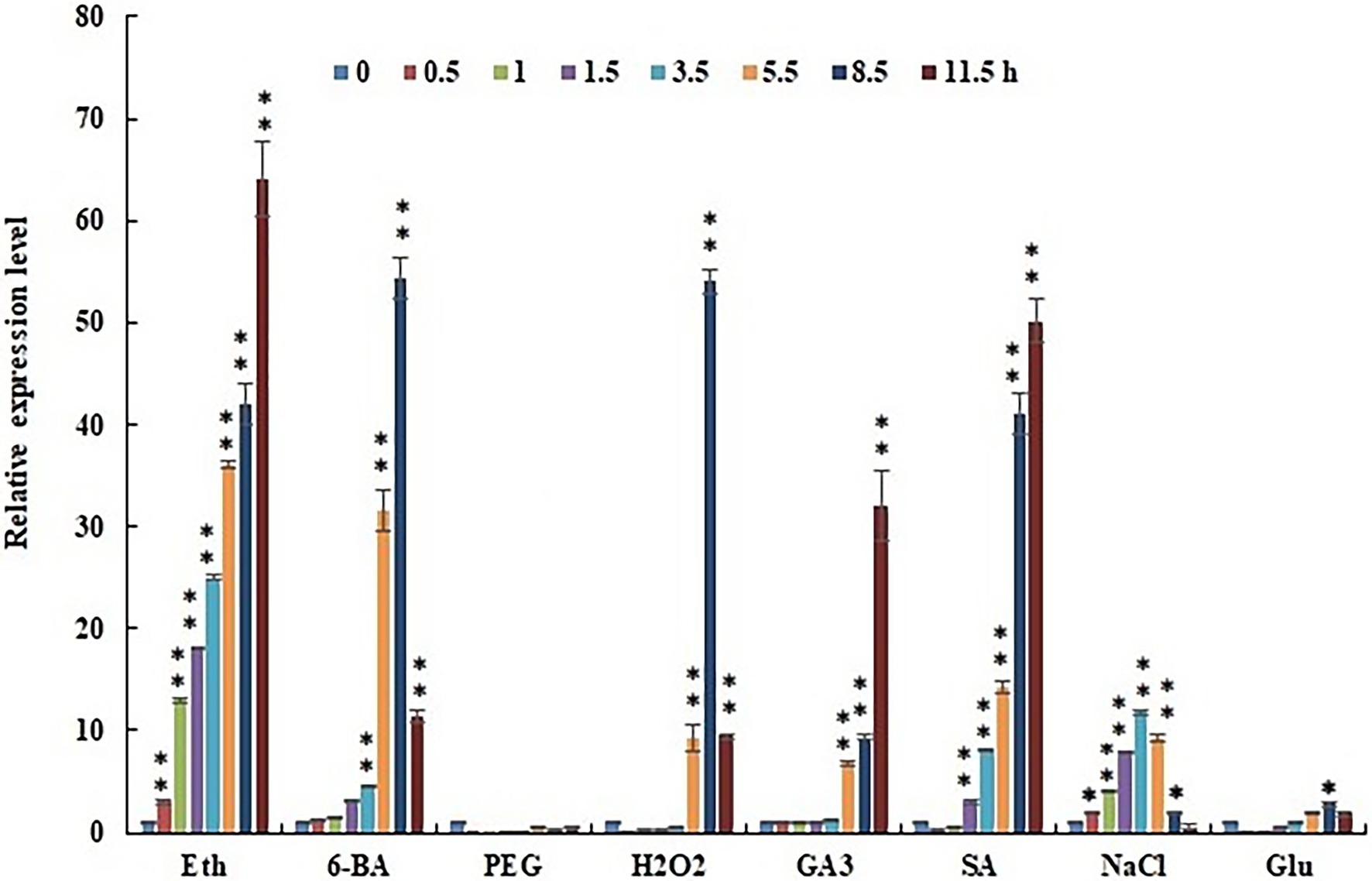
Figure 5. Effect of elicitors (Eth, 6-BA, PEG, H2O2, GA3, SA, NaCl, and Glu) on CcERF2 expression. The 24 DAP pepper fruits were treated with elicitors (Eth, 6-BA, PEG, H2O2, GA3, SA, NaCl, or Glu), and the fruit were sampled after elicitation for 0.5, 1.0, 1.5, 3.5, 5.5, 8.5, and 11.5 h. Without induction at the 0 h time point was used as control expression. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9). Student’s t-test was used to identify significant differences compared to the control (*p < 0.05, **p < 0.01).
The fruits were treated with ethephon, which significantly induced CcERF2 expression. Compared with the control, CcERF2 expression increased by 64 times within 11.5 h of applying ethephon. At the same time, the effects of exogenous 6-BA, PEG, H2O2, GA3, SA, NaCl, and Glu on CcERF2 expression were also tested. Of these, 6-BA, H2O2, GA3, SA, and NaCl significantly induced CcERF2 expression. Compared with the control, these exogenous substances increased CcERF2 expression; however, the modes of induced expression were not the same. In the test time range, under the 6-BA, H2O2, and NaCl treatments, the CcERF2 expression showed a trend of initial increase and then decrease; however, under GA3 and SA treatment, CcERF2 expression showed an increasing trend. Treatment with Glu had no significant effect on CcERF2 expression and PEG treatment was inhibitory.
Ethephon treatment could significantly increase the expression level of CBGs, but the induction effect was inconsistent. Compared to controls, within 11.5 h of applying ethephon, gene expression levels of CcPAL, CcCoMT, and CcFAT were increased about 2.3–5.0 times; of CcCa4H, CcpAmt, CcBCAT, CcACL, and CcACS were increased about 5.8–8.3 times; of CcKAS was increased about 11.1 times; and of CcCS was increased more than 100 times. Considering that ethephon significantly induced the expression of CcERF2, our results showed that CcERF2 was a key factor in ethylene-mediated biosynthesis of CAPs (Figure 6).
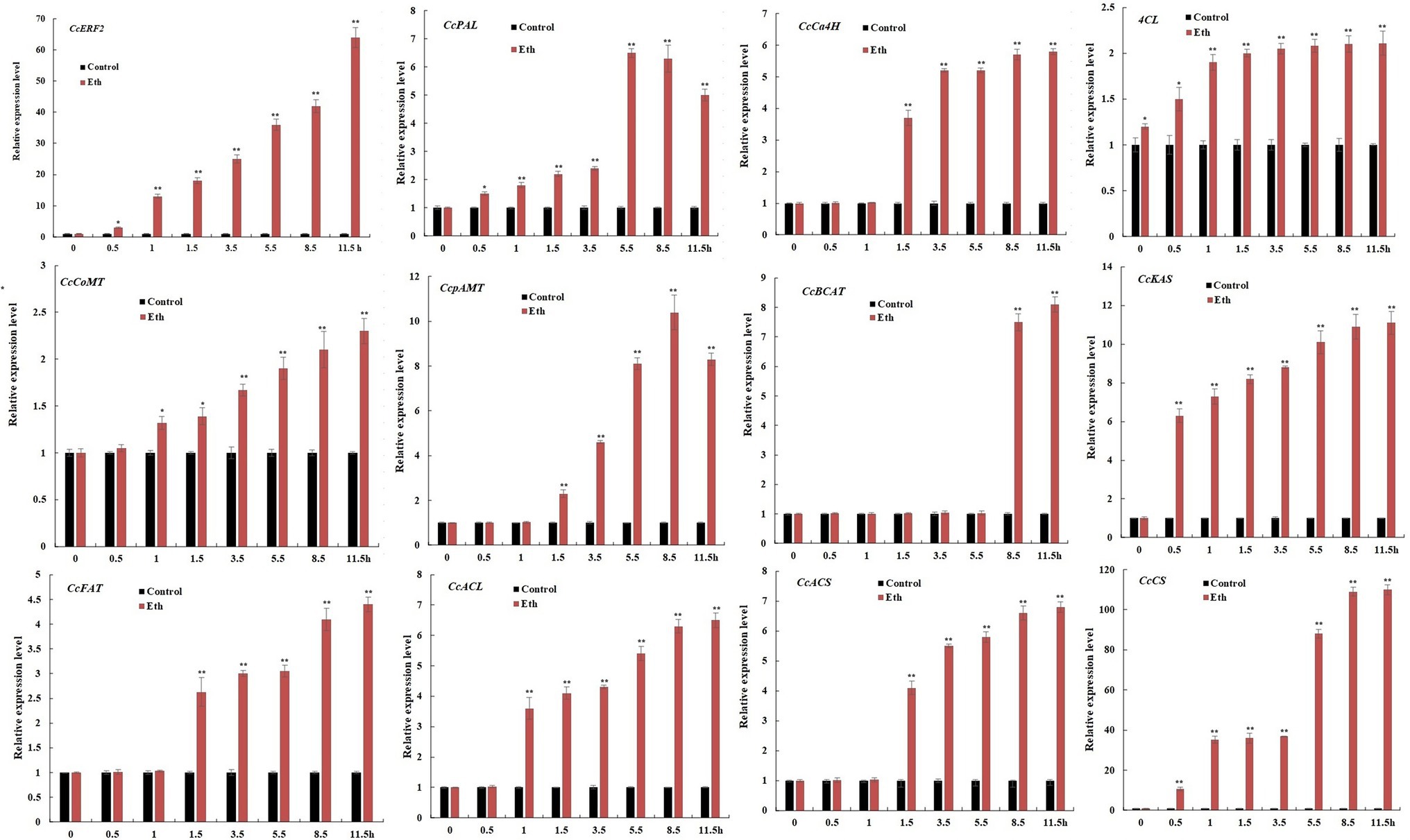
Figure 6. Expression profiles of CBGs after treatment with Eth. The 24 DAP pepper fruits were treated with Eth, and the fruit were sampled after elicitation for 0.5, 1.0, 1.5, 3.5, 5.5, 8.5, and 11.5 h. Without induction at the 0 h time point was used as control expression. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9). Student’s t-test was used to identify significant differences compared to the control (*p < 0.05, **p < 0.01).
The effect of blocking ethylene signal transmission by 1-MCP on CBG expression and CAP contents was studied. The 1-MCP treatment remarkably reduced the expression of PAL, C4H, COMT, pAMT, and CS genes (Figure 7A). The 1-MCP treatment significantly reduced the content of CAPs (Figure 7B). The results indicated that ethylene signal transduction was involved in the regulation of CAP biosynthesis.
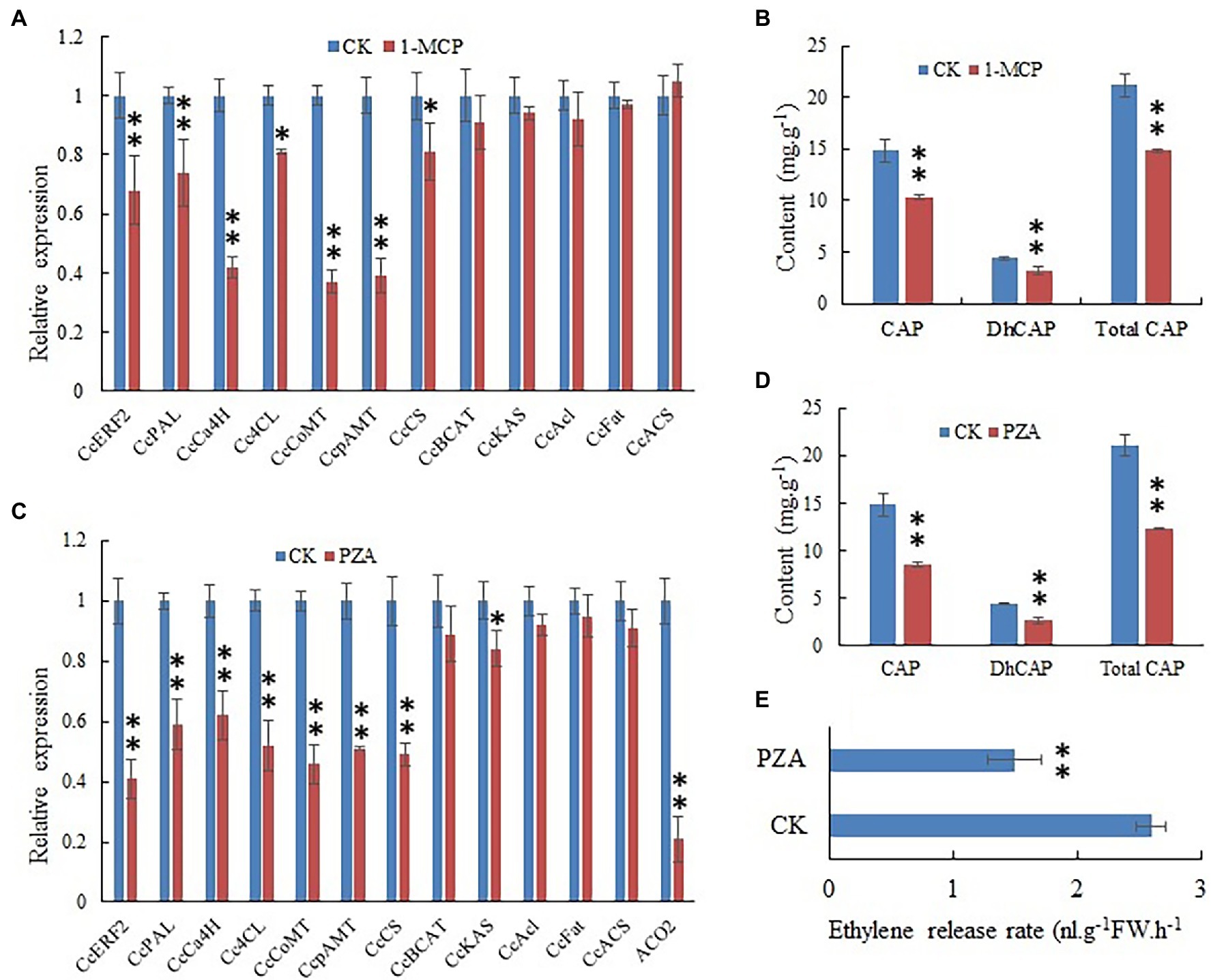
Figure 7. Expression level of CBGs and CcERF2, relative contents of capsaicin (CaP), dihydrocapsaicin (DhCaP), and CAPs in 1-methylcyclopropene (1-MCP) and pyrazinamide (PZA) treatment fruit. (A) Expression level of CBGs and CcERF2 in the 1-MCP treatment fruit. Fruits at 24 DAP was sampled to perform the expression level of CBGs and CcERF2. The relative expression level of the CBGs and CcERF2 in the control were set to 1, and that of in 1-MCP treatment fruit were measured relative to that in the control. (B) Relative contents of CaP, DhCaP, and CAPs in the 1-MCP treatment fruit at 29 DAP. (C) Expression level of CBGs, CcERF2, and ACC oxidase-2 (ACO2) in the PZA treatment fruit. Fruits at 24 DAP was sampled to perform the expression level of CBGs and CcERF2. The relative expression level of the CBGs and CcERF2 in the control were set to 1, and that of in PZA treatment fruit were measured relative to that in the control. (D) Relative contents of CaP, DhCaP, and CAPs in the PZA treatment fruit at 29 DAP. (E) Ethylene release rate in the PZA treatment fruit at 29 DAP. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9). Student’s t-test was used to identify significant differences compared to the control (*p < 0.05, **p < 0.01).
The results indicated that after pepper fruits were treated with ethylene biosynthesis inhibitor PZA for 12 h, the expression of ACC oxidase-2 (ACO2) gene was extremely reduced (Figure 7C); expression of CBGs such as PAL, C4H, COMT, pAMT, and CS was inhibited (Figure 7C); CAPs contents were also obviously reduced (Figure 7D); and the endogenous ethylene release rate was conspicuously reduced (Figure 7E). This indicated that the content of endogenous ethylene significantly affected biosynthesis of CAPs.
The VIGS vector pTRV2–CcERF2 was constructed using a vector derived from tobacco rattle virus (TRV), and a CcERF2-silenced experiment was performed. The 24-DAP fruits of SL08 were used for the study. It was demonstrated that Agrobacterium infection with an empty pTRV2 vector resulted in distinctive changes in expression level of CBGs in pepper fruits (Abraham-Juárez et al., 2008). Compared with empty vector plants, the fruits infected with the pTRV2–CcERF2 construct showed a distinctive reduction in CcERF2 expression (22.4% of the empty vector; Figure 8A). Consistent with the expression level of CcERF2, CBGs in silent plants were also significantly downregulated. In the CcERF2-silent placenta, the expression of genes derived from fatty acid metabolism pathways (such as CcKAS, CcACL, CcFAT, and CcACS) only slightly changed. However, expression of genes derived from the phenylpropane pathway (such as CcPAL, CcCa4H, Cc4CL, CcComt, and CcpAmt) and CcCS underwent significant changes, and the genes whose expression was altered were consistent with the transcription level of CcERF2 (Figure 8A).
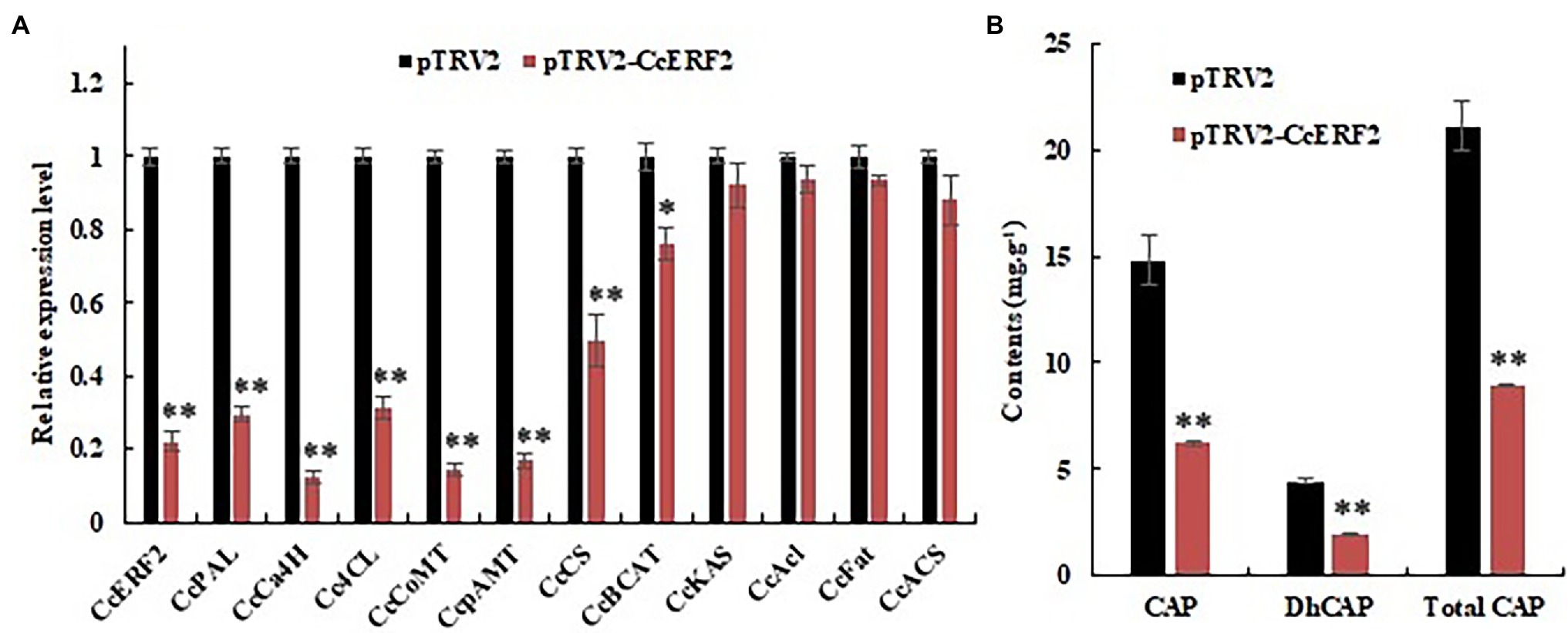
Figure 8. Expression level of CBGs and CcERF2, relative contents of CaP, DhCaP, and CAPs in the control (empty vector) and CcERF2-silenced fruit. (A) Expression level of CBGs and CcERF2 in the control (empty vector) and CcERF2-silenced fruit. Fruits at 24 DAP was sampled to perform the expression level of CBGs and CcERF2. The relative expression level of the CBGs and CcERF2 in the control were set to 1, and that of in the CcERF2-silenced fruit were measured relative to that in the control. (B) Relative contents of CaP, DhCaP, and CAPs in the control (empty vector) and CcERF2-silenced fruit at 34 DAP. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9). Student’s t-test was used to identify significant differences compared to the control (*p < 0.05, **p < 0.01).
The CAPs contents of the pepper fruits infected with the empty pTRV2 vector were similar to those of uninfected plants (Abraham-Juárez et al., 2008). The 34-DAP fruits were used to study the effect of CcERF2 silencing on the CaP and DhCaP contents. Compared with fruits infected with empty pTRV2, the CaP and DhCaP contents in fruits infected with pTRV2–CcERF2 were significantly reduced by 74.2 and 73.0%, respectively (Figure 8B). The above results strongly supported that CcERF2 regulated certain CBGs to control CAPs biosynthesis.
Plant secondary metabolism comprises the life activity, formed in the long-term evolutionary process. The secondary metabolites are not only associated with the regulation of plant growth, but also allow plants to effectively deal with all kinds of stresses, such as fungi, pests, and herbivores (Dangl and Jones, 2001). The distribution of secondary metabolites is usually specific to species, organs, and tissues; CAPs are uniquely synthesized in the fruit placenta of Capsicum and have obvious specificity (Mazourek et al., 2009). A variety of stress conditions (such as high temperature, hydropenia, and herbivore invasion) can induce the production of secondary metabolites, and this process is affected by a lot of plant endogenous hormones (such as ethylene). Studies have found that low temperature promotes the accumulation of CAPs in pepper fruits (Keyhaninejad et al., 2014; Arce-Rodríguez and Ochoa-Alejo, 2017); drought induces biosynthesis of CAPs (Phimchan et al., 2014); and injury significantly increases the CAPs content of pepper fruits (Arce-Rodríguez and Ochoa-Alejo, 2017). Plants under stress (such as high temperature, hydropenia, and herbivore attack) usually show an increase in ethylene content, which can increase the formation of secondary metabolites. Ethylene plays a vital factor in response to all kinds of stresses (Fang et al., 2014; Hu et al., 2020). However, the effect of CAPs biosynthesis induced by ethylene needs further study.
We investigated the mechanism of ethylene on the expression of CcERF2 and CBGs. After fruits of line SL08 were treated with ethephon, expressions of CcERF2 and CBGs were significantly increased, equivalent to 2.1–110.3 times that of the control. However, the response pattern of each gene induced by ethephon slightly differed (Figure 5). Many studies have indicated that ethephon can upregulate the expression of multiple genes in the phenylpropane metabolic pathway, thereby promoting biosynthesis of flavonoids, anthocyanins, rutin, lignin, and procyanidins (El-Kereamy et al., 2003; Buer et al., 2006; Hossain et al., 2009; Pan et al., 2019; Hu et al., 2020; Paul et al., 2020; Zhao et al., 2020). Our experimental results are similar to those reports.
Most AP2/ERF family proteins have conserved AP2 domains, and it was reported that this type of TF can regulate the synthesis of plant secondary metabolites (Paul et al., 2020). These AP2/ERF factors seem to play a similar role in regulation metabolic genes (Hu et al., 2020). In this paper, the AP2/ERF TF CcERF2, which can be used as a transcription activator to regulate CAPs biosynthesis was identified. There were 43 motifs obtained by analyzing the motif significance of the CcERF2 protein. On the whole, the predicted motifs differed within the same family, but the conserved motifs in the same subgroups were almost the same. That is, the closer the related species, the more similar were the motifs. The peppers all contained 13 conserved motifs (1–8, 11, 12, 17, 19, and 22), of which motifs 1, 3, and 5 covered the conserved AP2 structural domain. Our results showed that the function of CcERF2 was convergent and divergent among different plant species.
Various genes related to CAPs biosynthesis have been studied (Aluru et al., 2003). However, very few TFs associated with CAPs biosynthesis have been isolated and characterized, except for some MYB TFs (Arce-Rodríguez and Ochoa-Alejo, 2017; Sun et al., 2019, 2020; Zhu et al., 2019). We need to identify some TFs, especially AP2/ERF family TFs, which can be used as positive regulators to promote accumulation of CAPs. In the current study, based on transcription data, the AP2/ERF TF CcERF2 was selected for further analysis, because CcERF2 had a similar expression pattern to the CBGs in the transcriptome data. Silencing CcERF2 downregulated the expression of CBGs, especially the expression level of genes associated with the phenylpropane pathway, and therefore reduced its CAPs content. The results showed that CcERF2 was involved in regulating this metabolic process.
The 1-MCP is an ethylene receptor inhibitor (Sisler and Serek, 1997; Huber, 2008). It can effectively inhibit ethylene signal transduction (Du et al., 2021). A large number of studies have shown that 1-MCP can affect the maturation, senescence, and secondary metabolism of non-climacteric fruits by blocking ethylene signal transduction (Gómez-Lobato et al., 2012; Li et al., 2016; Zhang et al., 2018). Our research results showed that 1-MCP significantly reduced the expression of some capsaicin biosynthesis genes, and obviously reduced the content of CAPs in fruits.
The PZA is a new type of ethylene biosynthesis inhibitor that acts by inhibiting expression of ACO2 (Sun et al., 2017). Our research showed that PZA obviously reduced ACO2 expression in pepper fruits and remarkably reduced ethylene production. At the same time, PZA treatment greatly reduced the expression of some genes related to capsaicin biosynthesis, thereby reducing the content of CAPs.
This study showed strong evidence that ethylene induced the expression of CcERF2 and CBGs, and 1-MCP and PZA treatments caused a decrease in expression of CBGs and a sharp decrease in content of CAPs in fruit. It also demonstrated that CcERF2 could promote CAP biosynthesis in Capsicum. It is necessary to further study whether CcERF2 acts directly on the promoter of CBGs of the CAP pathway or through formation of a complex with other TFs.
The original contributions presented in the study are publicly available. This data can be found at: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA789050.
MD and KZ conceived and designed the study. JL, XZ, ZL, HZ, JH, and HW performed the research. ZW, HZ, XZ, and JW analyzed the data. MD, XZ, and JW prepared the paper. All authors contributed to the article and approved the submitted version.
This study was supported by the Natural Science Foundation of China (Grant Nos. 31560556 and 31160394) and the Major Science and Technology Projects in Yunnan Province (2018BB020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.832669/full#supplementary-material
Supplementary Figure S1 | Capsaicin (CaP), dihydrocapsaicin (DhCaP), and total capsaicinoids (CAPs) contents determination in inbred line SL08 and H19 fruits. Fifteen fruits were sampled for the measurement of the contents of CAP, DhCAP, and total CAPD at each developmental stage. The experiments were replicated three biological times and three technical times. Data are expressed as the mean ± SD (n = 9).
Abraham-Juárez, M. R., Rocha-Granados, M. C., López, M. G., Rivera-Bustamante, R. F., and Ochoa-Alejo, N. (2008). Virus-induced silencing of Comt, pAmt and Kas genes results in a reduction of capsaicinoid accumulation in chili pepper fruits. Planta 227, 681–695. doi: 10.1007/s00425-007-0651-7
Aluru, M. R., Mazourek, M., Landry, L. G., Curry, J., Jahn, M., and O'Connell, M. A. (2003). Differential expression of fatty acid synthase genes, Acl, Fat and Kas, in Capsicum fruit. J. Exp. Bot. 54, 1655–1664. doi: 10.1093/jxb/erg176
Arce-Rodríguez, M. L., and Ochoa-Alejo, N. (2017). An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 174, 1359–1370. doi: 10.1104/pp.17.00506
Blum, E., Mazourek, M., O'Connell, M. A., Curry, J., Thorup, T., Liu, K., et al. (2003). Molecular mapping of capsaicinoid biosynthesis genes and quantitative trait loci analysis for capsaicinoid content in Capsicum. Theor. Appl. Genet. 108, 79–86. doi: 10.1007/s00122-003-1405-y
Bosland, P. W., Coon, D., and Reeves, G. (2012). Trinidad moruga scorpion pepper is the world‘s hottest measured chile pepper at more than two million scoville heat units. HortTechnology 22, 534–538. doi: 10.21273/HORTTECH.22.4.534
Brown, R. L., Kazan, K., McGrath, K. C., Maclean, D. J., and Manners, J. M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Buer, C. S., Sukumar, P., and Muday, G. K. (2006). Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140, 1384–1396. doi: 10.1104/pp.105.075671
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
De Boer, K., Tilleman, S., Pauwels, L., Vanden Bossche, R., De Sutter, V., Vanderhaeghen, R., et al. (2011). APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66, 1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x
Deng, M. H., Wen, J. F., Huo, J. L., Zhu, H. S., Wang, P., Dai, X. Z., et al. (2012). Molecular characterization and prokaryotic expression of orf507 sterility-associated gene in chilli pepper (Capsicum annum L.) cytoplasmic male sterility. Pak. J. Bot. 44, 1497–1502.
Deng, M. H., Wen, J. F., Zhu, H. S., and Zou, X. X. (2009). The hottest pepepr variety in China. Genet. Resour. Crop. Evol. 56, 605–608. doi: 10.1007/s10722-009-9445-z
Du, Y., Jin, T., Zhao, H., Han, C., Sun, F., Chen, Q., et al. (2021). Synergistic inhibitory effect of 1-methylcyclopropene (1-MCP) and chlorine dioxide (ClO2) treatment on chlorophyll degradation of green pepper fruit during storage. Postharvest Biol. Technol. 171:111363. doi: 10.1016/j.postharvbio.2020.111363
El-Kereamy, A., Chervin, C., Roustan, J. P., Cheynier, V., Souquet, J. M., Moutounet, M., et al. (2003). Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol. Plant. 119, 175–182. doi: 10.1034/j.1399-3054.2003.00165.x
Fang, R. J., Zhao, H., Liao, R. H., Tang, C. Y., Wu, F. Y., Zhu, Y., et al. (2014). Dual regulating effects of ethylene on the formation of plant secondary metabolites. Chin. Bull. Bot. 49, 626–639. doi: 10.3724/SP.J.1259.2014.00626
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Gantet, P., and Memelink, J. (2002). Transcription factors: tools to engineer the production of pharmacologically active plant metabolites. Trends Pharmacol. Sci. 23, 563–569. doi: 10.1016/S0165-6147(02)02098-9
Garruña-Hernández, R., Monforte-González, M., Canto-Aguilar, A., Flota, F. V., and Orellana, R. (2013). Enrichment of carbon dioxide in the atmosphere increases the capsaicinoids content in Habanero peppers (Capsicum chinense Jacq.). J. Agric. Food Chem. 93, 1385–1388. doi: 10.1002/jsfa.5904
Gómez-Lobato, M. E., Hasperue, J. H., Civello, P. M., Chaves, A. R., and Martínez, G. A. (2012). Effect of 1-MCP on the expression of chlorophyll degrading genes during senescence of broccoli (Brassica oleracea L.). Sci. Hortic. 144, 208–211. doi: 10.1016/j.scienta.2012.07.017
Gray, W. M. (2004). Hormonal regulation of plant growth and development. PLoS Biol. 2:E311. doi: 10.1016/j.scienta.2012.07.017
Gurung, T., Techawongstien, S., Suriharn, B., and Techawongstien, S. (2011). Impact of environments on the accumulation of capsaicinoids in Capsicum spp. HortScience 46, 1–6. doi: 10.2514/1.16041
Hossain, M. A., Kim, S., Kim, K. H., Lee, S. J., and Lee, H. (2009). Flavonoid compounds are enriched in lemon balm (Melissa officinalis) leaves by a high level of sucrose and confer increased antioxidant activity. HortScience 44, 1907–1913. doi: 10.21273/HORTSCI.44.7.1907
Hu, Y.-T., Xu, Z.-C., Tian, Y., Gao, R.-R., Ji, A.-J., Pu, X.-D., et al. (2020). Genome-wide identification and analysis of AP2/ERF transcription factors related to camptothecin biosynthesis in Camptotheca acuminate. Chin. J. Nat. Med. 18, 582–593. doi: 10.1016/S1875-5364(20)30070-4
Huber, D. J. (2008). Suppression of ethylene responses through application of 1-methylcyclopropene: a powerful tool for elucidating ripening and senescence mechanisms in climacteric and nonclimacteric fruits and vegetables. HortScience 43, 106–111. doi: 10.21273/HORTSCI.43.1.106
Iwai, K., Suzuki, T., and Fujiwake, H. (1979). Formation and accumulation of pungent principle of hot pepper fruits, capsaicin and its analogues, in Capsicum annuun var. annuun cv. karayatsubusa at different growth stages after flowering. Agric. Biol. Chem. 43, 2493–2498. doi: 10.1080/00021369.1979.10863843
Keyhaninejad, N., Curry, J., Romero, J., and O’Connell, M. A. (2014). Fruit specific variability in capsaicinoid accumulation and transcription of structural and regulatory genes in Capsicum fruit. Plant Sci. 215–216, 59–68. doi: 10.1016/j.plantsci.2013.10.013
Lejeune, M. P. G. M., Kovacs, E. M. R., and Westerterp-Plantenga, M. S. (2007). Effect of capsaicin on substrate oxidation and weight maintenance after modest bodyweight loss in human subjects. Brit. J. Nutr. 90, 651–659. doi: 10.1079/BJN2003938
Li, L., Lichter, A., Chalupowicz, D., Gamrasni, D., Goldberg, T., Nerya, O., et al. (2016). Effects of the ethylene-action inhibitor 1-methylcyclopropene on postharvest quality of non-climacteric fruit crops. Postharvest Biol. Technol. 111, 322–329. doi: 10.1016/j.postharvbio.2015.09.031
Liu, S. Q., Chen, C. M., Chen, G. J., Cao, B. H., Chen, Q. H., and Lei, J. J. (2012). RNA-sequencing tag profiling of the placenta and pericarp of pungent pepper provides robust candidates, contributing to capsaicinoid biosynthesis. Plant Cell Tissue Organ Cult. 110, 111–121. doi: 10.1007/s11240-012-0135-8
Liu, F., Deng, Y., Zheng, J., Liu, M., Ou, L., Yang, B., et al. (2017). PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132. doi: 10.1016/j.molp.2017.03.005
Liu, S., Li, W., Wu, Y., Chen, C., and Lei, J. (2013). De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLoS One 8:e48156. doi: 10.1371/journal.pone.0048156
Liu, Y., Lv, J., Liu, Z., Wang, J., Yang, B., Chen, W., et al. (2019). Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 306:125629. doi: 10.1016/j.foodchem.2019.125629
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ludy, M. J., Moore, G. E., and Mattes, R. D. (2012). The effects of capsaicin and capsiate on energy balance: critical review and meta-analyses of studies in humans. Chem. Senses 37, 103–121. doi: 10.1093/chemse/bjr100
Mazourek, M., Pujar, A., Borovsky, Y., Paran, I., Mueller, L., and Jahn, M. M. (2009). A dynamic interface for capsaicinoid symstems biology. Plant Physiol. 150, 1806–1821. doi: 10.1104/pp.109.136549
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Ou, L., Li, D., Lv, J., Chen, W., Zhang, Z., Li, X., et al. (2018). Pan-genome of cultivated pepper (Capsicum) and its use in gene presence-absence variation analyses. New Phytol. 220:360. doi: 10.1111/nph.15413
Pan, Q., Wang, C., Xiong, Z., Wang, H., Fu, X., Shen, Q., et al. (2019). CrERF5, an AP2/ERF transcription factor, positively regulates the biosynthesis of bisindole alkaloids and their precursors in Catharanthus roseus. Front. Plant Sci. 10:931. doi: 10.3389/fpls.2019.00931
Pan, Z. W., Wang, H. Q., and Zhong, J. J. (2000). Scale-up study on suspension cultures of Taxus chinensis cells for production of taxane diterpene. Enzym. Microb. Technol. 27, 714–723. doi: 10.1016/S0141-0229(00)00276-3
Paul, P., Singh, S. K., Patra, B., Liu, X., Pattanaik, S., and Yuan, L. (2020). Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants. Plant Physiol. 182, 840–856. doi: 10.1104/pp.19.00772
Phimchan, P., Chanthai, S., Bosland, P. W., and Techawongstien, S. (2014). Enzymatic changes in phenylalanine ammonia-lyase, cinnamic-4-hydroxylase, capsaicin synthase, and peroxidase activities in Capsicum under drought stress. J. Agric. Food Chem. 62, 7057–7062. doi: 10.1021/jf4051717
Phimchan, P., and Techawongstien, S. (2012). Impact of drought stress on the accumulation of capsaicinoids in Capsicum cultivars with different initial capsaicinoid levels. HortScience 47, 1204–1209. doi: 10.21273/HORTSCI.47.9.1204
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U. S. A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Shoji, T., Kajikawa, M., and Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22, 3390–3409. doi: 10.1105/tpc.110.078543
Sisler, E. C., and Serek, M. (1997). Inhibitors of ethylene responses in plants at the receptor level: recent developments. Physiol. Plant. 100, 577–582. doi: 10.1111/j.1399-3054.1997.tb03063.x
Song, J., Chen, C., Zhang, S., Wang, J., Huang, Z., Chen, M., et al. (2020). Systematic analysis of the Capsicum ERF transcription factor family: identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genomics 21:573. doi: 10.1186/s12864-020-06983-3
Sun, X., Li, Y., He, W., Ji, C., Xia, P., Wang, Y., et al. (2017). Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nat. Commun. 8:15758. doi: 10.1038/ncomms15758
Sun, B., Zhou, X., Chen, C., Chen, C., Chen, K., Chen, M., et al. (2020). Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic. Res. 7:162. doi: 10.1038/s41438-020-00381-2
Sun, B., Zhu, Z., Chen, C., Chen, G., Cao, B., Chen, C., et al. (2019). Jasmonate-inducible R2R3-MYB transcription factor regulates capsaicinoid biosynthesis and stamen development in Capsicum. J. Agric. Food Chem. 67, 10891–10903. doi: 10.1021/acs.jafc.9b04978
Tewksbury, J. J., Reagan, K. M., Machnicki, N. J., Carlo, T. A., Haak, D. C., Peñaloza, L. C., et al. (2008). Evolutionary ecology of pungency in wild chilies. Proc. Natl. Acad. Sci. U. S. A. 105, 11808–11811. doi: 10.1073/pnas.0802691105
van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonateresponsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. doi: 10.1126/science.289.5477.295
van der Fits, L., and Memelink, J. (2001). The jasmonateinducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25, 43–53. doi: 10.1046/j.1365-313x.2001.00932.x
Xu, L., Yue, Q., Xiang, G., Bian, F., and Yao, Y. (2018). Melatonin promotes ripening of grape berry via increasing the levels of ABA, H2O2, and particularly ethylene. Hortic. Res. 5, 1–11. doi: 10.1038/s41438-018-0045-y
Zhang, X. M., Li, J. P., Fu, M. R., Jin, T., Yang, X. Y., Han, C., et al. (2018). Effects of the combination of 1-MCP and ClO2 on shelf life quality of green pepper. Sci. Technol. Food Ind. 39, 275–280. doi: 10.13386/j.issn1002-0306.2018.13.050
Zhang, W., Zou, A., Miao, J., Yin, Y., Tian, R., Pang, Y., et al. (2011). LeERF-1, a novel AP2/ERF family gene within the B3 subcluster, is down-regulated by light signals in Lithospermum erythrorhizon. Plant Biol. 13, 343–348. doi: 10.1111/j.1438-8677.2010.00375.x
Zhao, C., Liu, X., Gong, Q., Cao, J., Shen, W., Yin, X., et al. (2020). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhao, J., Zheng, S. H., Fujita, K., and Sakai, K. (2004). Jasmonate and ethylene signaling and their interaction are integral parts of the elicitor signaling pathway leading to β-thujaplicin biosynthesis in Cupressus lusitanica cell cultures. J. Exp. Bot. 55, 1003–1012. doi: 10.1093/jxb/erh127
Keywords: CcERF2, pepper, capsaicinoids, ethylene, VIGS
Citation: Wen J, Lv J, Zhao K, Zhang X, Li Z, Zhang H, Huo J, Wan H, Wang Z, Zhu H and Deng M (2022) Ethylene-Inducible AP2/ERF Transcription Factor Involved in the Capsaicinoid Biosynthesis in Capsicum. Front. Plant Sci. 13:832669. doi: 10.3389/fpls.2022.832669
Received: 10 December 2021; Accepted: 14 February 2022;
Published: 03 March 2022.
Edited by:
Xinchao Wang, Tea Research Institute (CAAS), ChinaReviewed by:
Changming Chen, South China Agricultural University, ChinaCopyright © 2022 Wen, Lv, Zhao, Zhang, Li, Zhang, Huo, Wan, Wang, Zhu and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minghua Deng, ZGVuZ21pbmdodWEyMDEzQHNpbmEuY29t, orcid.org/0000-0001-8293-9035
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.