- State Key Laboratory of Crop Biology, Agronomy College, Shandong Agricultural University, Tai’an, China
Water uptake facilitates the initiation of seed germination. It is presumed that aquaporin (AQP)-mediated water inflow contributes to seed germination, but the genetic evidence is still lacking. This study aimed at genome-wide identification of ZmAQPs and further determined the physiological functions. Following a comprehensive search, a total of 41 ZmAQPs were identified according to the latest genome database. Through bioinformatic approaches, the physicochemical characteristics, phylogenetic relationships, and structural features of ZmAQPs were analyzed. The gene expression analysis of 20 high-resolution and multi-tissues samples showed that ZmAQPs had distinct spatiotemporal and tissue-specific expression profiles during seed germination and early seedling development. We then focused on the aquaporin of maize tonoplast intrinsic protein 3 (ZmTIP3), which is specifically expressed in germinating seed. A mutant zmtip3-1 with disruption of the ZmTIP3-1 gene showed shorter shoot and root length, and decreased seedling dry weight compared with the control (W22). The result revealed that ZmTIP3-1 improved the absolute content of seed protein and promoted storage reserves mobilization, suggesting that ZmTIP3 may be a positive regulator of seed vigor. This work provides valuable clues for understanding the function and possible regulatory mechanism of ZmAQPs in seed germination and seedling growth.
Introduction
Maize (Zea mays) is an important staple food and economic crop worldwide. Seed germination and seedling establishment are crucial initial stages for plant growth and are closely related to the rate and uniformity of field crop establishment, which ultimately determines the yield, especially for maize, in the absence of tillering (Rajjou et al., 2012; Finch-Savage and Bassel, 2016). Germination begins with water uptake by quiescent seed and ends with radicle protrusion, and the subsequent post-germination seedling growth. Seed hydration during germination is displayed in a typical triphasic pattern (Nonogaki et al., 2010; Bewley et al., 2013). Meanwhile, there are substantial physical and metabolic events that occur, including cell expansion, cell division, and reserve mobilization (Nonogaki et al., 2010). However, fundamental research on maize seed germination and early seedling growth is limited.
Water is a prerequisite for germination and is the medium of life activities. Aquaporins (AQPs) are integral membrane proteins that facilitate the movement of water across biological membranes (Chaumont and Tyerman, 2017). Aquaporins exist in membranes with homologous or heterologous tetramer forms (Daniels et al., 1999), and a typical structure of an aquaporin monomer has two conserved NPA (Asn-Pro-Ala) motifs, six membrane-spanning α-helixes (H1-H6), and two short highly hydrophobic α-helixes (HB and HE), with the N- and C-termini exposed to the cytosol (Maurel, 1997; Murata et al., 2000; Chaumont et al., 2001). Based on the subcellular localization and sequence homology, the aquaporin family in higher plants was mainly classified into four subfamilies, including the plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), and small basic intrinsic proteins (SIPs) (Chaumont and Tyerman, 2017).
The plant AQP family comprises many members, and their cellular localization diversification and regulatory complexity enables AQPs to function in plant growth and in the response to environmental signals. Therefore, plant AQPs have received increasing attention. With the availability of genomic sequences of plant species, the identification of AQPs provides basic information to better understand the AQPs gene family. Numerous studies have confirmed that plant AQPs can be ubiquitously expressed in all tissues and exhibit clear spatiotemporal-specific patterns at the transcription level (Barrieu et al., 1998; Chaumont et al., 1998; Takahashi et al., 2004; Guo et al., 2006; Vander Willigen et al., 2006; Hunter et al., 2007; Soto et al., 2008; Gattolin et al., 2009; Novikova et al., 2014; Utsugi et al., 2015). Moreover, AQPs are highly regulated by posttranslational modifications, including phosphorylation and heteromerization, that influence the amount and activity of AQPs, trafficking, gating and re-localization, to ultimately control plant water relations (Zelazny et al., 2007; Chaumont and Tyerman, 2014; Maurel et al., 2015; Berny et al., 2016). Few studies have investigated the biological function of AQPs in plant development and growth. The expression of TIP3-1 was detected exclusively in developing seeds and early seed germination and ceased after radicle emergence at the transcriptional level (Johnson et al., 1989; Melroy and Herman, 1991; Vander Willigen et al., 2006; Novikova et al., 2014). In contrast, other AQPs exhibited relatively low expression in dry and germinating seeds. Extensive research has shown that TIP1 and TIP2 substitute for TIP3 after seed germination and retain high expression levels in elongating and dividing tissues (Maurel et al., 1997a; Chaumont et al., 1998; Vander Willigen et al., 2006; Novikova et al., 2014). According to our knowledge, only two previous reports provided genetic evidence that AQPs function in seed germination, that is, Arabidopsis AtTIP3 (Footitt et al., 2019) and rice OsPIP1 (Liu et al., 2007) influence seed germination under stress. Water inflow is essential for the initiation of embryonic cell elongation during early germination. Water imbibed by seed maintains cell turgor pressure and causes tissue expansion and subsequent seedling growth; AQPs may be involved in the finely tuned regulation of water transport (Obroucheva et al., 2017). Despite the predicted significance of TIPs in seed germination and seedling emergence based on expression analysis, genetic evidence is currently lacking in maize, and the mechanism of the spatial and temporal fine-tuning of ZmTIPs remains largely unknown.
Here, we performed a genome-wide characterization of maize aquaporins (ZmAQPs) and a comprehensive analysis of the phylogenetic relationships and physicochemical and structural properties of 41 identified genes. We also investigated the expression profile of AQPs during the seed germination process and then focused on seed-specific ZmTIP3-1. ZmTIP3-1 acts in the regulation of seedling growth and the protein content of the seed. Furthermore, the regulation mechanism of the spatiotemporal-specific pattern of ZmTIPs during seed germination is discussed in depth.
Materials and Methods
Materials
The maize inbred lines B73 and Z58 (Zheng58), with different genetic backgrounds, were used to investigate the expression patterns of ZmAQPs during seed germination and seedling emergence. For the physiological function analysis, the UniformMu insertion mutant zmtip3-1 (UFMu-13193) was obtained from the Maize Genetics Cooperation Stock Center. The zmtip3-1 mutant was backcrossed five generations with W22 and then selfed to generate BC5F2, from which homozygous zmtip3-1 was identified for the phenotype analysis of seed germination and seedling growth. All the maize materials were grown at the Experimental Station of Shandong Agricultural University.
Identification of Maize Aquaporins and Bioinformatic Analysis
Two methods were used to identify the members of the ZmAQP gene family. The Hidden Markov Model (HMM) file corresponding to the MIP domain (PF00230) was downloaded from the PFAM database.1 HMMER (3.3.1) was used to search for AQP proteins from the maize genome database. The AQP protein sequences of Arabidopsis and rice reported in previous research (Quigley et al., 2001; Sakurai et al., 2005) were downloaded from the UniProt database.2 These protein sequences were used as queries in BLASTP searches with BLASTP suite (blast 2.5.0). The latest maize genome information, Zm-B73-REFERENCE-NAM-5.0 (GCF_902167145.1), was retrieved from NCBI.3 Multiple sequence alignment and phylogenetic trees were analyzed by MEGA-X (Sudhir et al., 2018), and the trees were visualized by FigTree (v1.4.4). The specific chromosomal positions and collinearity of AQPs were processed with MapInspect software and TBtools (MCScanX) (Chen et al., 2020). Basic physical and chemical characteristics, the subcellular localization, transmembrane domains, gene structures, motifs, cis-acting regulatory elements and the phosphorylation site of AQPs were determined by online websites, as follows: ProtParam tool,4 WoLF PSORT5 and TMHMM Server v.2.0,6 Gene Structure Display Server (GSDS 2.0) (Hu B. et al., 2015), MEME web server (Bailey et al., 2009), plantCARE database (Lescot et al., 2002) and NetPhos 3.1,7 respectively. The 3D structure was predicted by the phyre2.0 website8 and visualized by VMD software (Kelley et al., 2015). The interaction network for ZmAQPs was searched from the string site9 and displayed through CytoScope software (Shannon et al., 2003).
Subcellular Localization Assays
The full-length cDNAs of ZmAQPs were subcloned into the pSAT6-EYFP_N1 vector to create the ZmAQP-YFP constructs. The protoplast isolation and transient expression were conducted as described previously (Tu et al., 2020). Mesophyll protoplasts were isolated using the 7-d-old seedling leaves under dark condition. The ZmAQP-YFP fusion plasmid was transfected into protoplasts using the PEG-calcium solution (0.4 g/mL PEG 4000, 0.8 M mannitol, 0.1 M CaCl2). After being washed and resuspendeding with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES), mesophyll protoplasts were incubated under dark for 12–18 h. The YFP fluorescence was examined and imaged with a confocal microscope.
Seed Germination Test and Phytohormone Treatment
The standard germination test was performed as described previously (Zhao et al., 2020). The trials were conducted in sand with three 50-seed replicates for each material. Radicle emergence characteristic was recorded every 4 h after imbibition. Radicle emergence is defined as radicle > 2 mm long. The seedlings were allowed to grow for 7 days and the number of germinated seeds was counted at 24 h-intervals. Germination index (GI) and seed vigor index were calculated, GI = Σ (Gt/Dt); VI = GI × W, where Gt is the number of germinated seeds on day, t and Dt is time in days, W is the dry weight per seedling (g). For recording dry weight, all normal seedlings were dried at 105°C for 30 min and then transferred to 80°C and dried until constant weight.
The samples used for gene expression profiles analysis were obtained from the process of germination and post-germination, including the imbibed seeds, root and shoot at different stages, and 4 days seedlings (mesocotyl, plumule, and coleoptile included). Different zones of primary maize root were collected according to previous studies (Kozlova et al., 2012; Yu et al., 2016). To determine the effects of phytohormones on ZmAQPs, we set three groups of treatments: dry seeds were imbibed in the solutions of 10 μM ABA (Solarbio, A8060) and 10 μM GA (Solarbio, G8040); 10 μM IAA (Aladdin, I101074), and 10 μM ethephon (Solarbio, E8020) were spared on the germinated seeds (just after radicle emergence) and seedlings (just growing out of the soil), respectively. Water treatment was used as a control. All the samples for analysis were collected 3 h after treatment.
Gene Expression Analysis
Total RNA was extracted using the TIANGEN RNA Extraction Kit (DP445) according to the manufacture’s protocol. The first-strand cDNA was synthesized using the PrimeScript™ RT reagent Kit (Takara, RR047A). RT-qPCR analysis was performed using the TB Green® Premix Ex Taq™ (Takara, RR420A) on QuantStudio™ 12K Flex Software (Thermo Fisher Scientific). Each sample was quantified at least in triplicate and normalized using ACTIN 1 as a reference gene. The primer sequences of ACTIN 1 used were described previously (Zhang Z. et al., 2015). The gene-specific primer pairs for RT-qPCR are listed in Supplementary Table 5.
Western Blot
Protein expression levels of zmtip3-1 mutant and W22 were determined by western blotting. Seeds imbibed for 0, 12, and 24 h of both zmtip3-1 and W22 were harvested for analysis. Total proteins were extracted in an extraction buffer (50-mM Tris-HCl at pH 7.5, 150-mM NaCl, 1 mM EDTA, 10 mM NAF, 25 mM β-glycerophosphate, 2 mM sodium orthovanadate, 10% triglyceride, 0.1% tween-20, 0.6% SDS, 1 mM DTT, and 1 mM PMSF) containing protease inhibitor cocktail (Roche). The proteins were detected by SDS-PAGE gel with an anti-TIP3 antibody, which was produced using a synthetic peptide specific for maize aquaporin TIP3 (Abmart). Anti-Actin antibody (CW0096, Abmart) was used as a control.
Statistical Analysis
The data were analyzed by Student’s t-test using SPSS 19.0 software (SPSS, Inc., Chicago, United States).
Results
Genome-Wide Identification of Aquaporins in Maize
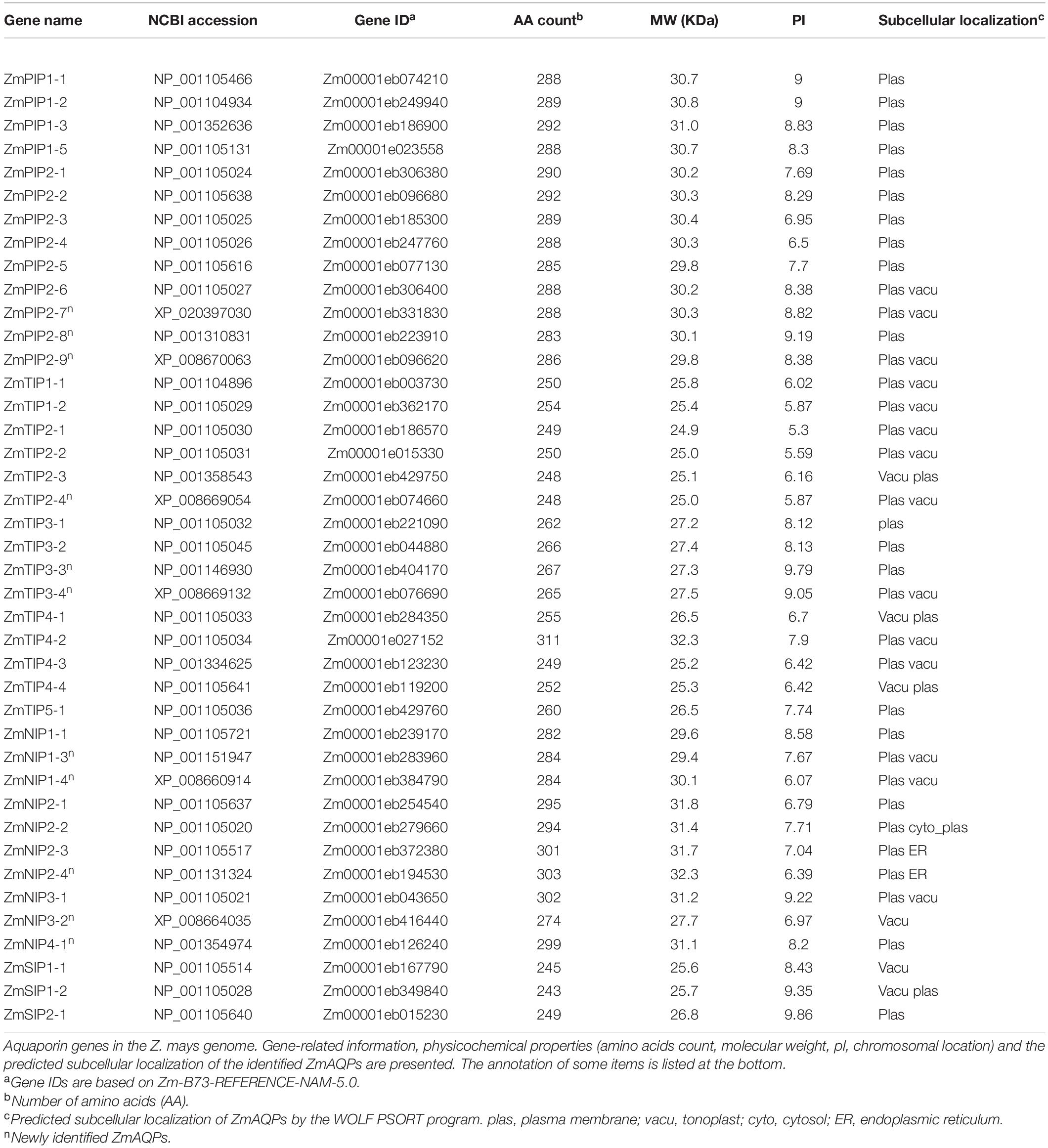
A comparison of HMMER and BLASTP searches of the Z. mays genome identified 43 putative ZmAQP sequence items. Based on the predicted conserved domains and annotation of the sequences, duplicate sequences were discarded. We finally obtained 41 complete and unique genes encoding ZmAQPs. Despite 41 ZmAQPs being identified in the work of Bari et al. (2018), there were errors, duplicate genes, and confusing nomenclature. For example, the ZmTIP4;1 and ZmTIP4;1a were in the same gene ID (Zm00001d037779) by searching the maize genome B73 RefGen_v4. Besides, ZmTIP4; 1a was named as ZmTIP4-1 by Chaumont et al. (2001). Here, ZmAQP genes were named with reference to homologous genes of Arabidopsis and rice, and of which 30 genes were annotated by Chaumont et al. (2001). The ZmAQP names, gene ID, protein length, molecular weight (MW), and theoretical isoelectric point (PI) are shown in Table 1. Compared with the work of Chaumont et al. (2001), two genes, zmpip1-4 and zmpip1-6, were removed, while eleven novel identified genes were added (Table 1). Initially, Chaumont et al. (2001) noted that zmpip1-3 and zmpip1-4 present in the B73 inbred line encoded the same protein with different nucleotide sequence. We confirmed that the zmpip1-3 and zmpip1-4 had the same accession (NP_001352636) and the same gene ID (Zm00001eb186900) in Zm-B73-REFERENCE-NAM-5.0 genome database. For zmpip1-6, was not in the Zm-B73-REFERENCE-NAM-5.0 but was in B73 RefGen_v4 with the gene ID Zm00001d020383. Multiple protein sequence alignments of the 41 identified ZmAQPs are presented in Supplementary Figure 1. All of the ZmAQPs contained two conserved NPA motifs and some highly conserved domains or sites (Supplementary Table 1). The NPA motifs were well conserved, by contrast, ZmNIP3-1, ZmNIP4-1, ZmSIP1s, and ZmSIP-1 encoded motifs with a variable third residue in which A was replaced by either I, L, T, or V (Supplementary Table 1).
Protein domain prediction analysis demonstrated that all of the sequences contained putative transmembrane α-helices but showed differences in the number of transmembrane domains. Most AQP proteins have a typical structure with six membrane-spanning domains, and five ZmAQPs contain five or seven (Supplementary Table 1). As a highly reliable means, the 3D structure prediction was further conducted for the prediction of the tertiary structure of the five ZmAQPs maps. 3D structural modeling revealed that each aquaporin monomer was formed by six long helices (namely TM1-TM6, with two on the surface and four at the bottom of the picture) and two re-entrant short helices (HB and HE, facing the readers) (Supplementary Figure 2). Hence, we considered that all 41 ZmAQPs had six transmembrane domains.
To characterize the phylogenetic relationships and to predicate the putative function of the ZmAQPs, a phylogenetic tree of 109 AQP sequences was constructed among Arabidopsis, rice, and maize. Phylogenetic analysis showed that ZmAQPs clustered into four different subfamilies (PIPs, TIPs, NIPs, and SIPs), the same as OsAQPs and AtAQPs (Figure 1). Among the 41 identified ZmAQPs proteins, 13 belong to PIPs, 15 to TIPs, 10 to NIPs, and 3 to SIPs. The phylogenetic analysis and subcellular localization of ZmAQPs contribute to further understanding or predicting their functional properties. According to the predicted subcellular localization, ZmAQP subfamilies exhibited distinct subcellular localizations and certain preferences. The PIPs are predominately localized to the plasma membrane, while most of the TIPs members (TIP1s, TIP2s, and TIP4s) are located at the plasma membrane and vacuolar membrane (tonoplast) (Table 1). Dual localization of plant AQPs was found in previous studies, including ZmPIP1-2 and AtTIP3s (Chaumont et al., 2000; Gattolin et al., 2011), suggesting versatile roles of AQPs. ZmTIP3-1 and TIP3-2 were predicted to localize to the plasma membrane (Table 1). In accordance with the prediction analysis, subcellular localization of ZmTIP3s and some other AQPs was mainly on the plasma membrane (Supplementary Figure 3). All observations showed that the complexity of AQP subcellular localization, which is compatible with the AQP functional diversity.
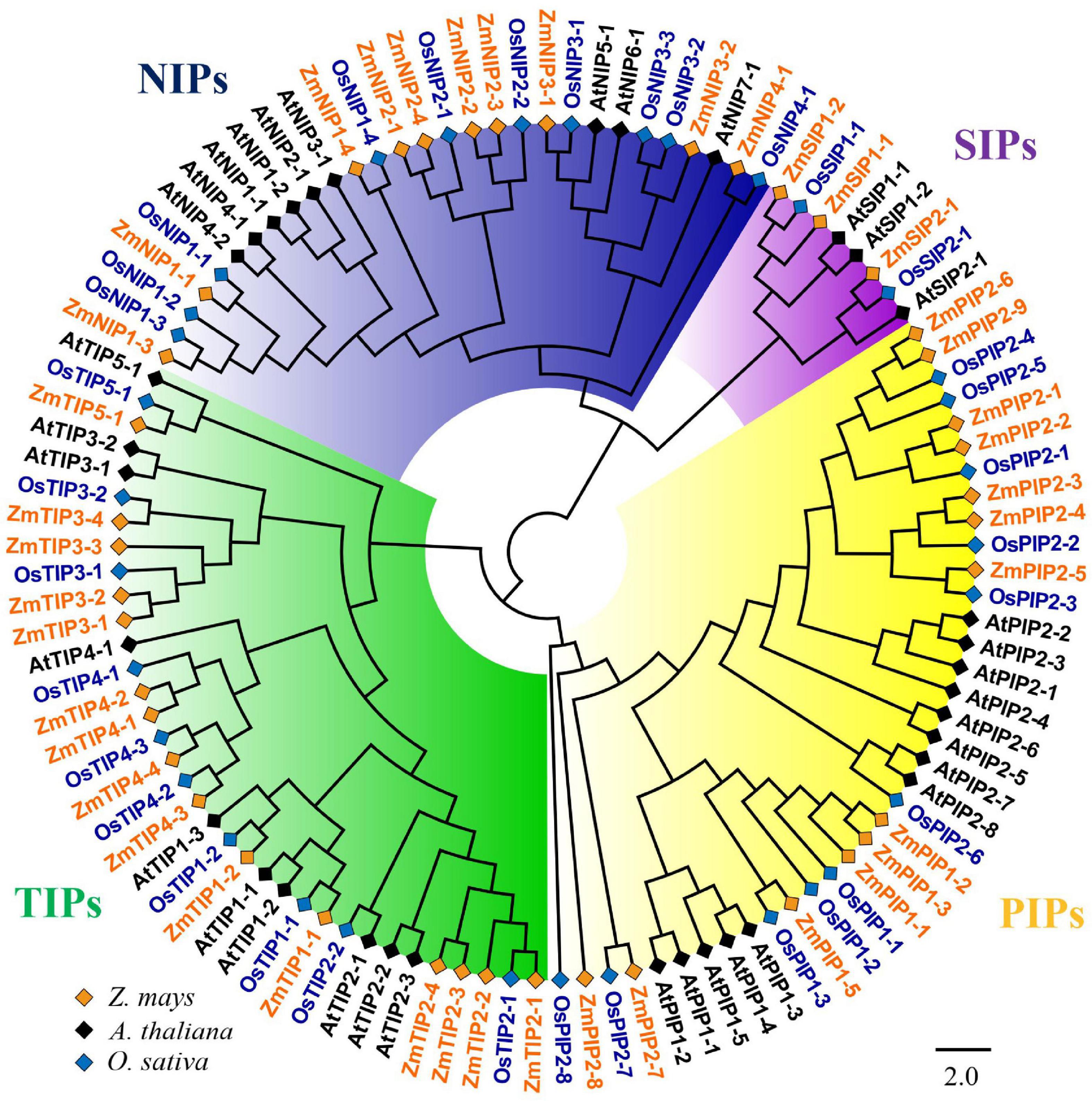
Figure 1. Phylogenetic analysis of 109 aquaporin proteins from Z. mays (Zm, indicated in orange), A. thaliana (At, indicated in black), and O. sativa (Os, indicated in blue). The ZmAQPs clustered into four different subfamilies (PIPs, TIPs, NIPs, and SIPs), with the corresponding OsAQP and AtAQP subfamilies. Each AQP subfamily is highlighted with a specific background color. PIPs, TIPs, NIPs, and SIPs are indicated in yellow, green, blue and purple, respectively. Multi-species protein sequence alignment was performed using Clustal W software. The phylogenetic tree was constructed using MEGA X based on the NJ method and further improved using the FigTree (v1.4.4) software.
Structural Characterization and Evolutionary Analysis of the Maize Aquaporin Gene Family
The exon-intron organizations of all identified ZmAQP genes were examined. The gene structure analysis showed that the exon-intron counts and distribution in the ZmAQP genes, varied from one to five exons per gene (Figure 2B and Supplementary Table 1). The exon-intron distribution and length were closely coincident with the alignment clusters of ZmAQP genes (Figure 2). The variation in the ZmAQP gene structure was also observed in the four subfamily members. Overall, the gene structure was diverse in the ZmAQP gene family but was relatively conserved in each subfamily. The differences in gene structure might confer functional variations. We further identified 15 conserved motifs (designated as motif 1 to motif 15, Supplementary Table 2) in the ZmAQP family to gain more insight into the ZmAQP diversity. As shown in Figure 2C, ZmAQP members within the same subfamily were usually found to share a similar motif composition. The differences among ZmAQP subfamilies were reflected in the motif number and their distribution (Figure 2C). The PIPs possess the most motifs and the motif arrangements are regular with specific ordering and similar positions. In summary, the similar gene structure and highly conserved protein motifs within the same subfamily were in agreement with the phylogenetic analysis, indicating the reliability of the above analysis. As shown in Supplementary Figure 4, the identified 41 ZmAQP genes were unevenly distributed among 10 chromosomes according to the number of genes on each chromosome. The distribution of ZmAQP genes in the maize genome could be affected by some genetic events such as segmental duplication (Lynch and Conery, 2000). The result indicated that 11 segmental duplication events within 15 ZmAQP genes were identified. Furthermore, the duplication events between genes usually occurred on chr2, chr4, and chr5 (Supplementary Figure 4). The tandem duplication events were also attributed to gene family evolution. Duplications of individual genes or chromosomal segments may provide an idea for research directions regarding new gene functions and expression patterns.
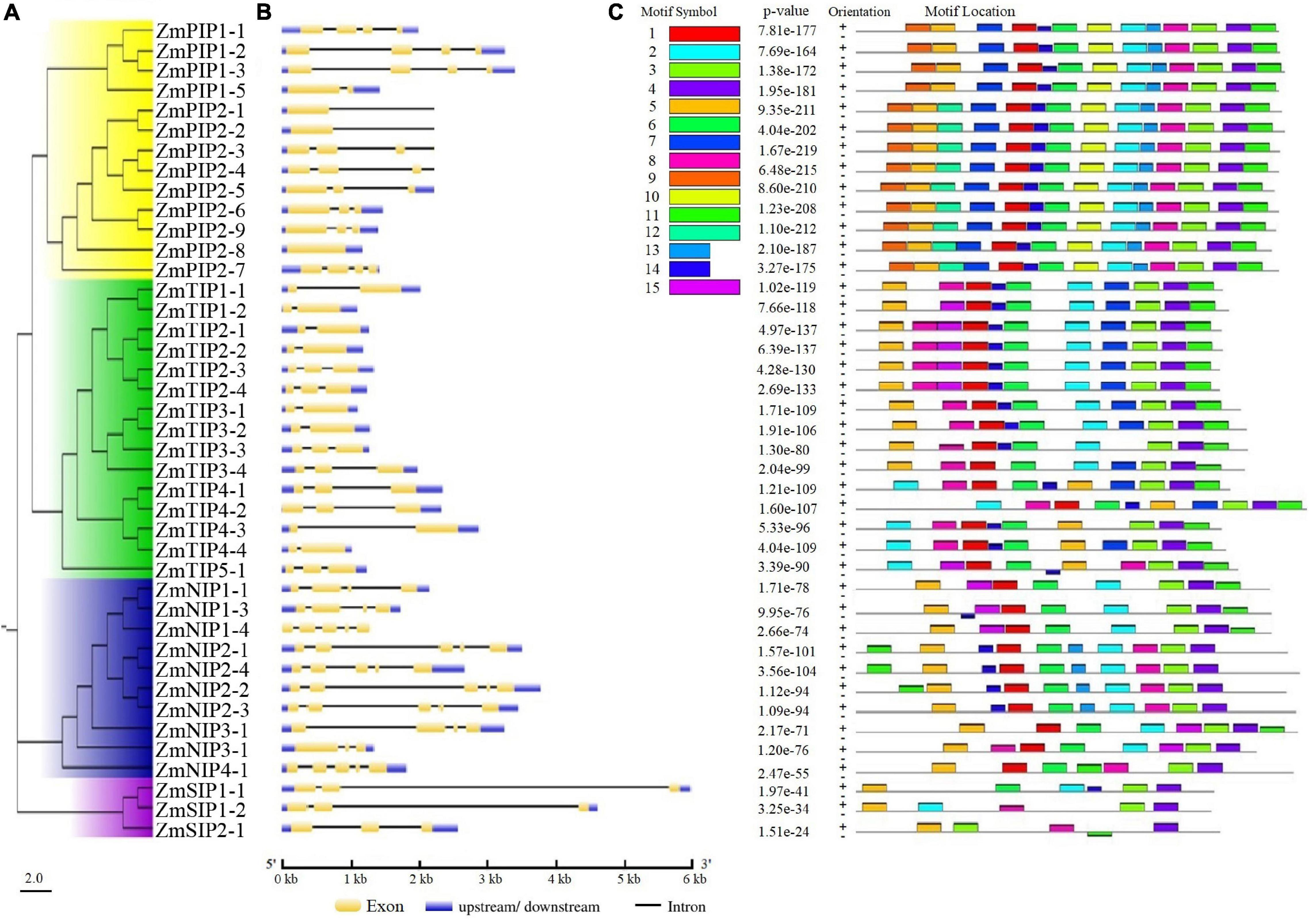
Figure 2. Phylogenetic relationships, gene structure and motif compositions of ZmAQPs. (A) The neighbor-joining evolutionary tree of 41 ZmAQP protein sequences constructed using MEGA X software. The details of four subfamilies are shown in different background colors. (B) Exon-intron gene structure of ZmAQP genes. Exons (CDS) and introns are represented by yellow boxes and black lines, respectively. Blue boxes indicate the upstream or downstream regions of genes. Gene length is shown in kb on the x-axis. (C) Distribution of conserved motifs in ZmAQPs. The conserved motifs were identified using the MEME online program. Different motifs, numbers 1–15, are highlighted with different colors. The sequence information for each motif is provided in Supplementary Table 2.
Analysis of the Spatial-Temporal Expression Patterns of Maize Aquaporins
To elucidate the spatial-temporal pattern of the ZmAQPs, we reanalyzed the publicly available RNA-seq data of 79 samples in different developmental stages, including roots, internodes, leaves, young stems, reproductive organs, and seeds (Stelpflug et al., 2015). As shown in Supplementary Table 3, most of the PIPs had high expression in all the samples examined, indicating a constitutive expression profile during plant development. Notably, PIPs showed relatively higher expression levels in roots, with relatively low expression levels in the other tissues especially the developing seeds. Indeed, it is not difficult to understand the results, as root is the primary organ for water uptake and the general role of aquaporins is to regulate transmembrane water transport (Peret et al., 2012; Chaumont and Tyerman, 2017; Ding et al., 2020).
Meanwhile, ZmTIP1-2 showed higher expression in the root and anthers, ZmTIP2s mainly in the roots, and ZmTIP4s in the leaves. The result also proved that ZmTIP3s are seed-specific and are exclusively expressed in developing seed and early germination stages. Overall, the RNA-seq analysis suggests that the expression profile of ZmAQPs varies with tissues and developmental stages.
Despite the vital roles in plant development and stress responses, the function of AQPs in the process of seed germination and seedling morphogenesis remains largely unclear. Next, we explored the expression patterns of ZmPIPs and ZmTIPs during seed germination and early seedling growth using qRT-PCR. Here, we applied a high temporal/spatial-resolution analysis using 20 samples collected within the first 96 h after B73 seed imbibition, including the embryo and endosperm at different times after seed imbibition and the root and shoot (mesocotyl, plumule, and coleoptile) at different stages of seedling growth. In the embryo of imbibed seeds, there was a clear trend toward increasing PIP transcript accumulation, peaking in the seeds imbibed for 36 h (EM-36 h, radicle protrusion) (Figure 3A). In post-germination seedlings, all the PIPs genes were ubiquitously expressed in the root, mesocotyl, or coleoptile, with significantly higher expression levels in seedlings than in imbibed seeds (Figure 3A). Because of the function of roots in water uptake and because most ZmAQPs showed the highest transcript abundances in the root (Figure 3), the transcriptional levels of some selected PIP genes in different zones of the root were also investigated. Almost all of the PIPs examined showed the highest expression in the rapid elongation zones of roots at 4-d-old seedlings (EZ-4 d), as opposed to 7-d-old seedlings (Figure 4A).
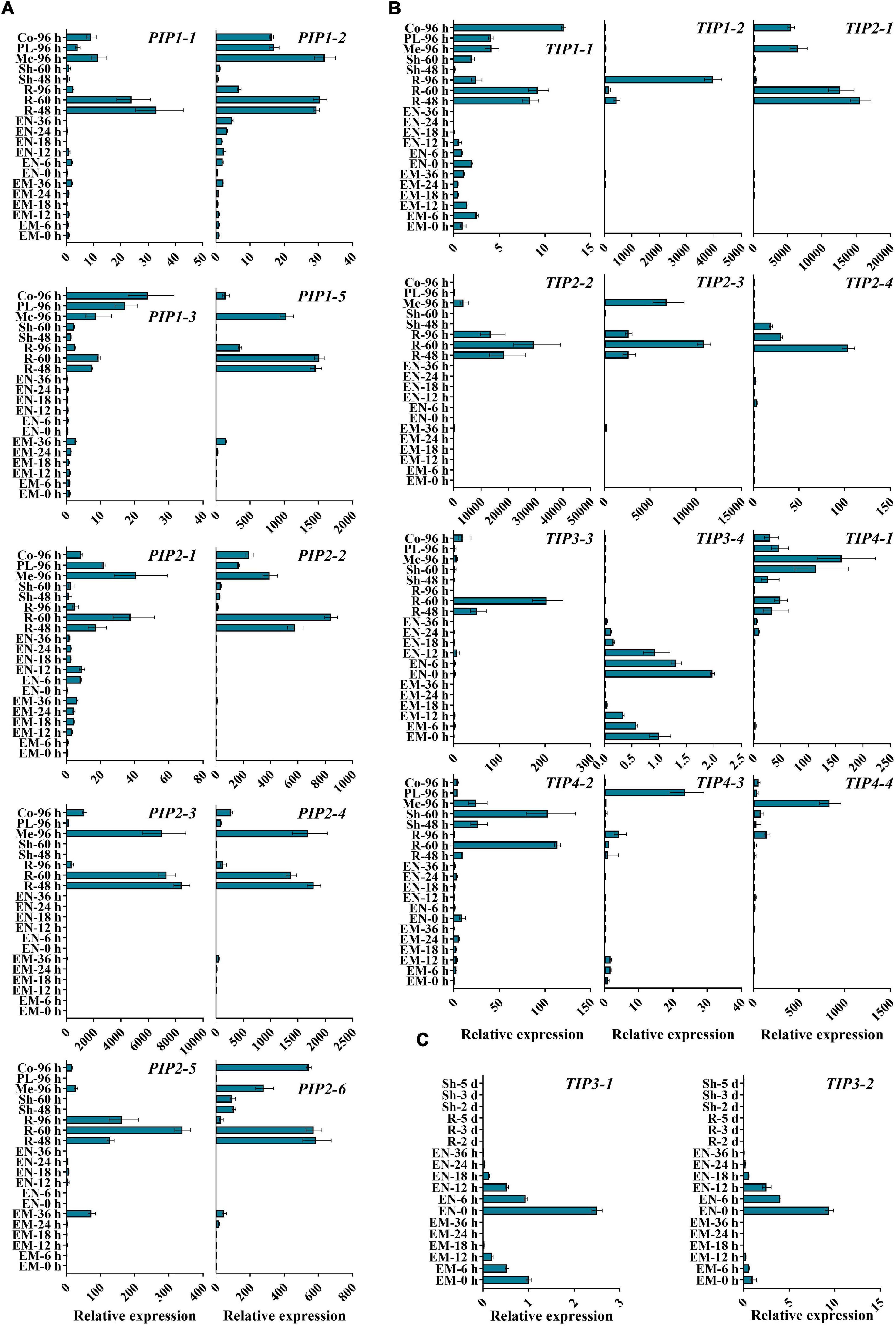
Figure 3. qRT-PCR analysis of the expression levels of ZmAOPs during early seed germination. The relative transcript levels of ZmPIPs (A) and ZmTIPs (B,C) in B73 during the seed germination process. Twenty samples from seeds and seedlings at different times after seed imbibition were used in the assay. ACTIN 1 was used as an internal control. Data are means ± SD (n = 3). Three independent experiments produced similar results. EM, embryo; EN, endosperm; R, root; Sh, shoot; Me, mesocotyl; PL, plumule; Co, coleoptile.
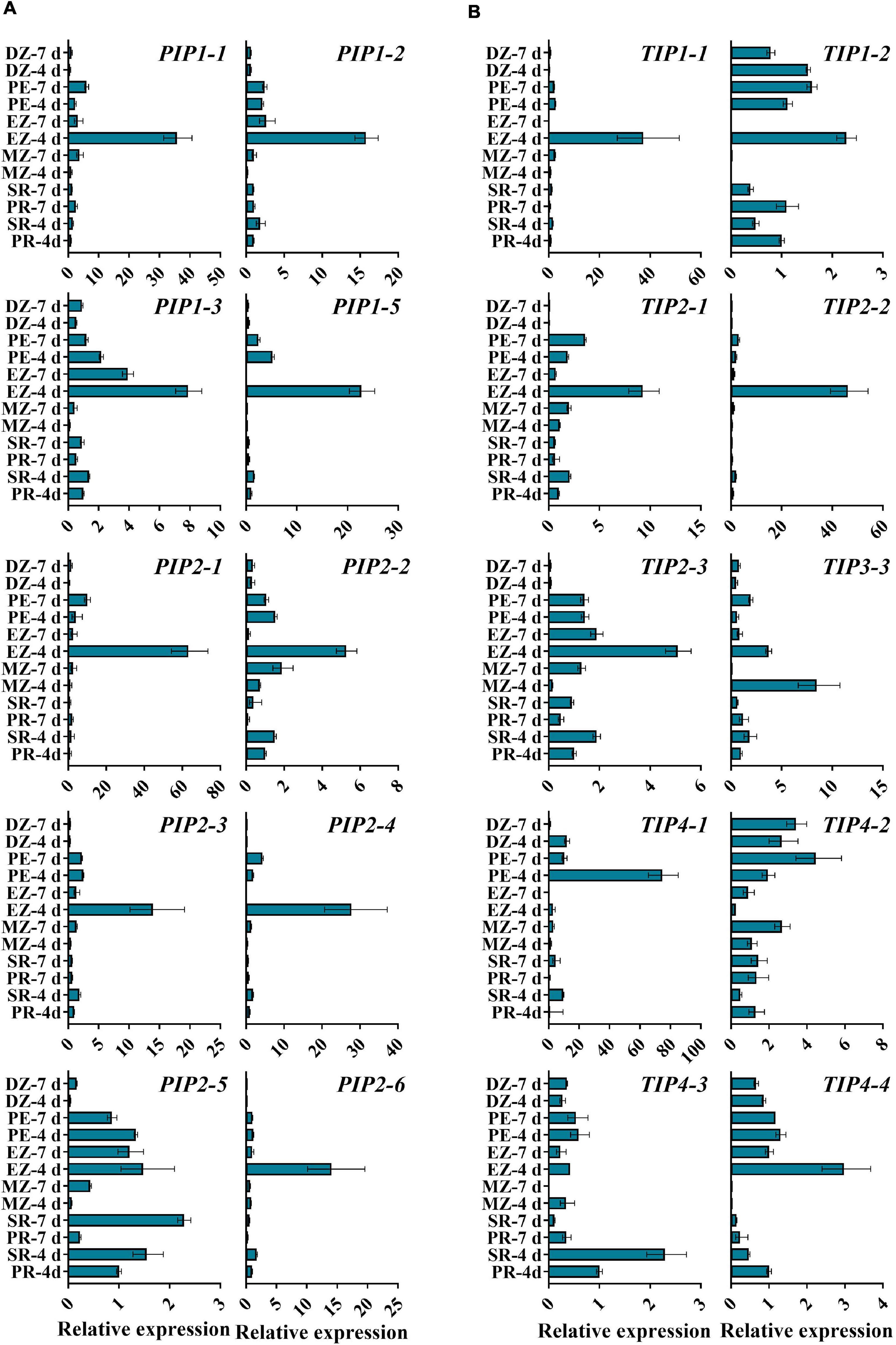
Figure 4. qRT-PCR analysis of the expression levels of ZmAOPs in different zones of the seedling root. The relative transcript levels of ZmPIPs (A) and ZmTIPs (B) in B73. Samples from roots at 4 or 7 days after seed imbibition were used in the assay. ACTIN 1 was used as an internal control. Data are means ± SD (n = 3). PR, primary root; SR, secondary root; MZ-root cap and meristem zone of the primary root; EZ-elongation zone of the primary root; PZ-post elongation zones of the primary root; DZ-differentiation zone of the primary root.
The transcripts of TIP genes exhibited tissue- and stage-specific profiles of accumulation (Figures 3B,C). High expression levels of TIP3s (TIP3-1, TIP3-2, and TIP3-4) were observed in dry and imbibed seeds, but the transcripts of TIP3s disappeared upon completion of germination (Figure 3C). In contrast, prevalent expression of TIP1, TIP2, and TIP4 subgroup members was induced hundreds and thousands of times after radicle emergence (Figure 3B). The expression patterns of TIPs in different root zones are in general agreement with those of PIPs (Figure 4). Several genes displayed different expression patterns. PIP2-5 and TIP1-2 were detected in several zones of roots at different stages, and there were few significant overall differences among the samples (Figure 4). Similar to TIP4-1, the expression levels were higher in the PE (post-elongation zones) compared with the other root zones (Figure 4). The results of the ZmAQP expression profiles remained largely similar to those observed in the Z58 line (Supplementary Figures 5, 6). Our qRT-PCR analysis indicated that TIP1/TIP2 progressively substituted for TIP3 after radicle protrusion and remained at relatively high expressed levels during early seedling development, consistent with previous research (Maurel et al., 1997a; Novikova et al., 2014). Together, in the maize seed germination and post-germination processes, our results revealed a strong link between ZmAQP expression and tissue expansion (Figures 3, 4).
The elongation of the radicle and mesocotyl is the key event for maize seed germination and seedlings growing out of the soil (Kutschera and Wang, 2016). These processes are closely related to AQP-mediated water uptake and transport, which maintain cell turgor and cause cell elongation (Maurel et al., 1997a; Obroucheva et al., 2017; Wang et al., 2020). Considering the seed-specific TIP3s during early germination, we first validated the biological function of TIP3s in subsequent experiments.
Maize Tonoplast Intrinsic Protein 3 Enhances Maize Seed Vigor and Improves Seedling Growth
To determine the role of ZmTIP3-1 in seed germination, a Mu-inserted zmtip3-1 mutant line was obtained from the Maize Genetics Stock Center. The uniform Mu was inserted into the 5′UTR (43 bp upstream from ATG) of the ZmTIP3-1 gene (Figure 5A). We first crossed the zmtip3-1 mutant into the W22 background and backcrossed 5 generations with W22 to generate BC5F1, followed by selfing, from which a homozygous zmtip3-1 line with the Mu locus was identified by PCR analysis for subsequent studies (Figure 5B). Furthermore, the qRT-PCR analysis demonstrated that Mu insertion resulted in complete inhibition of ZmTIP3-1 expression in the zmtip3-1 mutant at different times after imbibition (Figure 5C). With the western blot analysis, the protein accumulation level of ZmTIP3-1 was significantly lower in the zmtip3-1 mutant compared with W22 before radicle protrusion (Figure 5D). A standard seed germination test disclosed that relative to the control, mutation of ZmTIP3-1 was essentially unchanged in terms of radicle emergence or germination index (GI, which reflects germination speed and uniformity) (Figures 5E,F). There were no differences in the 100-grain weight between the mutant and W22 (Figure 5H). Based on these results, ZmTIP3-1 minimally influences the seed germination rate and does not regulate seed size and weight. However, as shown in Figure 5G, zmtip3-1 showed a significant decrease in the vigor index (VI), which is an important index used to evaluate seed quality; VI determines the rate and uniformity of seed germination, and seedling growth (ISTA, 2014). Phenotypic analysis indicated that disruption of ZmTIP3-1 resulted in weaker seedlings (Figure 5I). The growth of 7-d-old mutant seedlings was significantly reduced compared with that of W22 according to the length and dry weight of the seedlings (Figures 5J–L). Accordingly, our results demonstrated that disruption of ZmTIP3-1 significantly lowered the vigor index mainly by reducing the seedling growth.
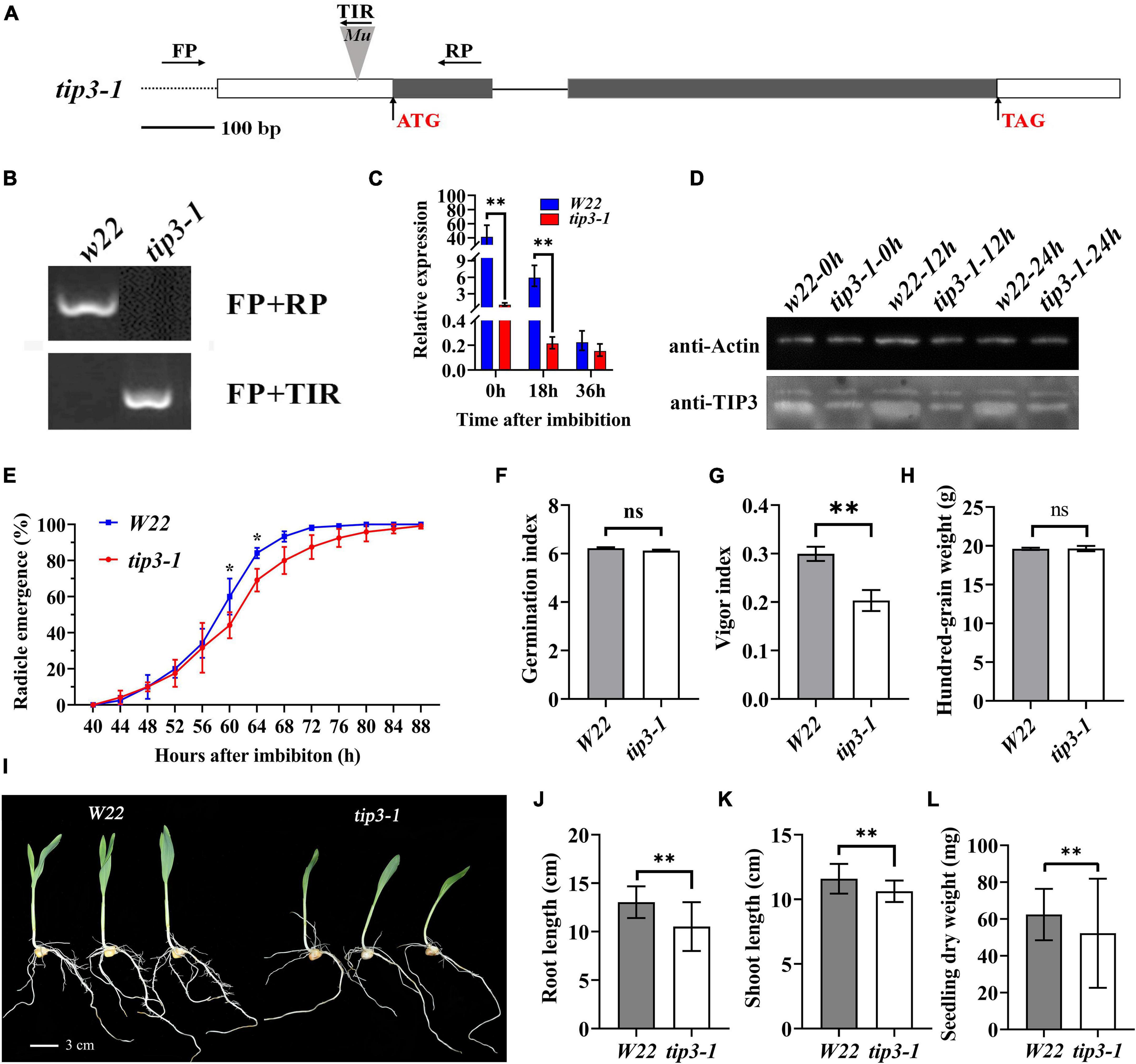
Figure 5. Phenotypic analysis of the zmtip3-1 mutant. (A) Gene structure of the ZmTIP3-1 and Mu insertion sites. Exons are shown as black boxes and introns as lines. The 5’ UTR and 3’ UTR are shown as white boxes. The Mu insertion site and primer sites are indicated. (B) Identification of the mutant by PCR at the genome level. (C) qRT–PCR analysis of ZmTIP3-1 transcript abundance in W22 and zmtip3-1 seeds. The samples were collected at 0, 18, and 36 h after imbibition. ACTIN 1 was used as an internal control. Data are means ± SD. (D) Immunoblot analysis of anti-TIP3 in zmtip3-1 and W22. Anti-actin antibody was used as the internal control. Total proteins were extracted from whole seeds at 0, 12, and 24 h after imbibition. (E) Comparison of the germination rate between zmtip3-1 and W22. Radicle emergence characteristics were recorded every 4 h after imbibition. Radicle emergence is defined as radicle length > 2 mm. Data are means ± SD (n = 3). There were three biological replicates and each included 100 grains. Comparison of the germination index (GI, F), vigor index (VI, G), and 100-grain weight (H) between zmtip3-1 and W22. Seedling growth (I), root length (J), shoot length (K) and seedling dry weight (L) in zmtip3-1 and W22 at 7 days after imbibition. * and ** indicate the significant differences at 5 and 1% levels, respectively, according to Student’s t-test, ns indicates non-significant.
Based on the temporal and spatial expression patterns of TIP3-1, there was an observation argues this it could be related to preparation for germination (Obroucheva and Sin’kevich, 2010). Our results corroborated this idea. The study further attempted to investigate the underlying mechanisms.
Maize Tonoplast Intrinsic Protein 3-1 Influences the Absolute Content of Protein in Maize Seeds and Storage Reserves Mobilization
In maize, the accumulation of ZmTIP3-1 transcripts increased with maize kernel development and was highest at the mature stage (Supplementary Figure 7). Notably, the expression of ZmTIP3-1 in the dry seed was higher than that of other AQPs (Supplementary Figure 8). TIP3-1, as a representative aquaporin in PSV, is highly conserved in higher plants (Johnson et al., 1989; Chaumont and Tyerman, 2017). Previous research determined that the abundance of TIP3-1 was not correlated with the content of storage protein in the cotyledon of protein-rich soybean seed (Johnson et al., 1989; Melroy and Herman, 1991). Our previous study verified that the absolute content of protein was significantly correlated with the seedling dry weight of maize and wheat seeds (Wen et al., 2018). However, the correlation between TIP3 accumulation and the protein content in starch-rich cereal seeds remains unclear. To this end, the impact of ZmTIP3-1 on the grain protein content needs to be evaluated. In the present study, we detected a significant decrease in the absolute content of protein in zmtip3-1 mutant seeds compared with the W22 (Figures 6A,B). Collectively, we considered that ZmTIP3-1 has a desirable effect on the absolute content of protein of maize seed, which might result in reduced seedling growth of the zmtip3-1 mutant.
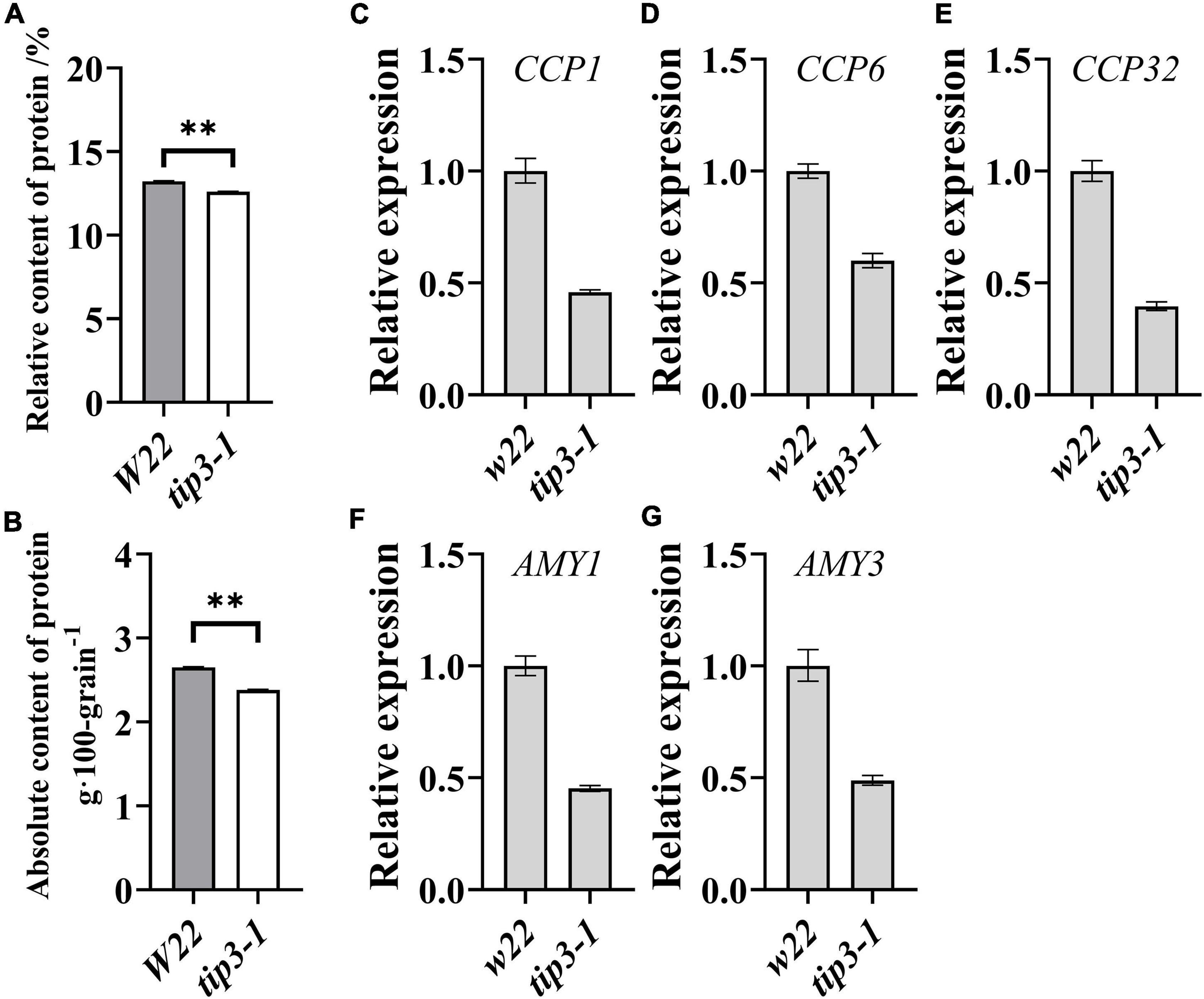
Figure 6. Protein content and mobilization-related genes expression in seeds. Determination of the relative protein content (A) and the absolute protein content per 100 grains (B) in tip3-1 mutants and W22. All the seeds were harvested at maturity. We multiplied the relative content of the components by the 100-grain weight to calculate the absolute content of the components (g⋅100 grain–1). Data are means ± SD (n = 3). qRT-PCR analysis of the expression levels of mobilization-related genes of cysteine protease1 (CCP1, C), cysteine protease 6 (CCP6, D), cysteine protease 32 (CCP32, E), alpha amylase 1 (AMY1, F), alpha amylase 3 (AMY3, G). The samples were collected at 36 h after imbibition. ACTIN 1 was used as an internal control. Data are means ± SD. ** indicates the significant differences at 1% level, according to Student’s t-test, ns indicates non-significant.
The mobilization of seed storage reserves is one of the major events in germination and seedling growth. We determined the expression levels of genes related to storage reserves mobilization in the zmtip3-1 mutant. Compared with the W22, the expression levels of starch metabolism-related genes, alpha amylase 1 (AMY1) and alpha amylase 3 (AMY3), and protein degradation related genes, cysteine protease1 (CCP1), cysteine protease 6 (CCP6), cysteine protease 32 (CCP32), are significantly decreased in the zmtip3-1 mutant (Figures 6C–G), suggesting that ZmTIP3-1 enhances seedling growth may be by promoting storage reserves mobilization.
Discussion
Spatiotemporal Expression of Maize Aquaporins During Seed Germination and Seedling Emergence
Water uptake mediates the initiation of germination and sequential metabolic activation (Nonogaki et al., 2010), in which AQPs accumulate and are actively involved (Obroucheva et al., 2017; Hoai et al., 2020). For maize, immediately after radicle emergence, the expressions of large members of ZmAQPs are activated in rapid expansion tissues, such as the root elongation zone and elongated mesocotyl, indicating the roles of AQPs in seedling emergence (Figures 3, 4 and Supplementary Figures 5, 6). The growing tissues require a constant water supply for ensuring the increased turgor pressure upon cell enlargement (Obroucheva and Sin’kevich, 2010). Based on the functions of AQPs, PIPs probably mainly act on water inflow and cell-to-cell water transport, whereas TIPs function in maintaining turgor pressure (Trofimova et al., 2001; Javot and Maurel, 2002). Consistent with previous findings of dual localization of AtTIP3s (Gattolin et al., 2011), our results suggest that ZmTIPs can localize in the plasma membrane (Table 1 and Supplementary Figure 3). Despite the significantly higher expression level of ZmTIP3s compared with other ZmAQPs in dry and imbibed seed (Figure 3 and Supplementary Figure 8), plasma membrane localization of ZmTIP3s may ensure effective water absorption during early germination. Compared with the dry seed, most of ZmTIP1s and TIP2s had up to 10,000-fold greater expression post-germination, which was significantly higher than the magnitude of the increase in ZmPIPs (Figures 3A,B and Supplementary Figures 5A,B). In addition, in vitro experiments showed water permeability and aquaporin activity of the tonoplast were higher than those of the plasma membrane (Maurel et al., 1997b; Niemietz and Tyerman, 1997; Trofimova et al., 2001). This would help to explain the dramatic changes in TIP expression after germination. Arabidopsis AtTIP1 and AtTIP2 have been confirmed to promote cell elongation (Reinhardt et al., 2016). Given the roles of the vacuole in plant cells and dual localization, ZmTIPs could participate in water inflow into the cells and water exchange between the vacuole and the cytoplasm for maintaining turgor to meet early seedling growth.
In contrast to full-scale activation of other ZmTIPs post-germination, ZmTIP3s (including TIP3-1, TIP3-2, and newly characterized TIP3-4) were distinctive (Figure 3C and Supplementary Figure 5C). Since the first identification of α-TIP (TIP3) in plants, the expression profile had been unequivocally determined, to be seed-specific with high expression in maturing and germinating seed, and almost completely abolished after germination (Johnson et al., 1989). Our results (Figure 3C and Supplementary Figure 9) aligned with previous experiments and confirmed this finding. Although it was presumed that TIP3 contributed to seed germination, this hypothesis was not validated by functional analysis until 2019, i.e., that AtTIP3 influences seed germination under water stress (Footitt et al., 2019). We established that ZmTIP3-1 had no desirable effects on germination rate but did affect maize seedling growth (Figure 5). Additionally, the accumulation of TIP3 was opposed to that of TIP1s (TIP2s) at the transcript level, which raised great concern (Maurel et al., 2008; Chaumont and Tyerman, 2017). There was a direct correlation between the disappearance of TIP3 and the coalescence of protein storage vacuoles (PSVs) (Lee et al., 2015, 2020), which involved in protein mobilization during seed germination (Bewley et al., 2013). TIP1 and TIP2 are preferentially localized in the lytic vacuoles formed by PSV and vegetative storage proteins vacuoles, respectively (Maurel et al., 2008). It has also been observed that ZmTIP1 was highly expressed in dividing and expanding cells of seedling roots (Barrieu et al., 1998; Chaumont et al., 1998). Here, we further clarified the expression trend of ZmTIP1s, i.e., that they appeared in only the protruding radicle, increased gradually with the germination process, and increased to substantial levels in the roots of seedlings (Figure 3B). Numerous studies confirmed the spatiotemporal expression of TIP aquaporins. However, the mechanisms governing the shift of TIPs remain unclear. The study also observed the disappearance of ZmTIP3-1 after radicle emergence was only at transcription levels, not at the protein level (Figures 5C,D), suggesting that ZmTIP3-1 was still functioning somehow. Considering the high accumulation of ZmTIP3-1 in mature kernels and dry seeds (Supplementary Figures 7, 8), we further speculated that the transcription of ZmTIP3-1 did not occur while the stored ZmTIP3-1 mRNA in dry seed did work during seed germination. Arabidopsis AtTIP1-1 and AtTIP3-1 were both found in the central vacuole of seedlings (Gattolin et al., 2011). In barley seed development, HvTIP3-1 interacted with HvTIP1-2 to form a heterotetramer, which facilitates the water transport capacity (Utsugi et al., 2015). In maize germination, further investigation is required to determine whether there is a physical interaction between ZmTIP3 and ZmTIP1 or TIP2 to supply considerable water for rapid seedling growth.
Seed germination and seedling emergence, beginning from a quiescent state to a metabolically active state in a short period, involve a series of physiological, biochemical, and morphological changes, and various tissues or organs such as the radicle, coleoptile, plumule, and mesocotyl in cereal seeds (Bewley et al., 2013). These activities cannot be separated from AQP-facilitated water uptake and transport in seed and seedlings (Obroucheva and Sin’kevich, 2010; Obroucheva, 2013; Obroucheva et al., 2017). AQPs exhibited exquisite spatiotemporal-specific expression patterns in the germination process, which is complementary to the versatile roles of AQPs in response to the establishment of new tissues/organs and various signals such as hormones or environmental stresses (Maurel et al., 2008; Li et al., 2014; Kapilan et al., 2018). Therefore, we suggest that the germinating seed may be an excellent model to examine the regulation mechanisms of the shift of the different AQPs during these sequential developmental stages.
Regulation of Maize Aquaporins
The finely tuned biological functions of plant AQPs can be regulated in multiple ways, including transcriptional and posttranslational modifications (Maurel et al., 2008, 2015; Chaumont and Tyerman, 2014). The transcriptional response to various signals and stimuli was mainly mediated by cis-acting regulatory elements. A bioinformatic promoter cis-element analysis of ZmAQP was performed to understand its versatile roles and dynamic changes during different development stages. The cis-elements of ZmAQP promoters were identified and classified into four major groups: stress, phytohormone, and light response, as well as plant growth-related cis-elements (Supplementary Figure 11).
Here, the results indicated that most ZmAQPs contain rich cis-elements in response to phytohormones and light. ABRE, as a major cis-acting element in ABA-responsive gene expression, and G-box (involved in light response) were the most abundant cis-acting elements (Supplementary Figures 11B–D). The ABA-signaling pathway is central to stress-responses in plants (Zhu, 2016). At the same time, we found that most of the ZmAQP genes had multiple stress-responsive elements, such as ARE (essential for the anaerobic induction), LTR (involved in low-temperature stress), and MBS (MYB binding site involved in drought-stress) (Supplementary Figure 11A). Numerous studies have implicated plant AQPs in abiotic stress regulation (Vera-Estrella et al., 2004; Xu et al., 2013; Ge et al., 2014; Zhang D. et al., 2015; Rios et al., 2017; Footitt et al., 2019). ABA has an essential role in seed development and germination. For seed-specific ZmTIP3s, an abundance of ABRE elements was identified (Supplementary Figure 11A), suggesting a potential role of ZmTIPs in ABA responses. The ABA content gradually increased as the grain developed, and the ZmTIP3 expression also exhibited a steadily rising trend (Supplementary Figure 7). Studies of Arabidopsis and barley have shown that the transcripts of TIP3s are regulated by GA and ABA during germination (Lee et al., 2015; Footitt et al., 2019). Furthermore, the transcription factor ABSCISIC ACID INSENSITIVE 3 (ABI3) directly binds to the RY motif of AtTIP3 promoters, which in turn induces AtTIP3 expression (Mao and Sun, 2015). However, the seed-specific RY-motif was found only in ZmTIP3-2, not in ZmTIP3-1 (Supplementary Figure 11C). Our data are in agreement with the findings of Footitt et al. (2019) for the different responses of TIP3-1 and TIP3-2 to hormones (Supplementary Figure 9A), which indicates they perform different functions, despite that their sequences shared high similarity. Light-responsive G-box element was the most enriched cis-acting element in the promoters of ZmAQP (Supplementary Figure 11D). PIF3 (Phytochrome interacting factor 3) and HY5 (Elongated hypocotyl 5), as two critical transcription factors in the light signaling pathway, can bind to G-box elements of light-responsive genes to regulate seedling morphogenesis (Liu et al., 2013; Zhao et al., 2019). Promoter cis-element analysis showed ZmTIP1s and ZmTIP2s contained abundant G-box elements (Supplementary Figure 11), which was in line with the extremely high expression of ZmTIPs in the mesocotyl of 4-d-old seedlings (Figure 3B). Additionally, ethylene, facilitating seedling emergence through the soil (Zhong et al., 2014), significantly induced the expression of ZmTIP1-1 (Supplementary Figure 9C). These results suggest that ZmTIPs may contribute to seedling establishment. The pivotal role of gibberellin (GA) in seed germination is self-evident. ZmPIP genes contain a relatively large number of GA-responsive elements, including P-box, GARE-box, and TATC-box (Supplementary Figure 11B). On the contrary, ZmTIP1, TIP2, and TIP3 contain few GA-related elements, except for ZmTIP1-2 (Supplementary Figure 11), and its expression was markedly increased under GA treatment during seedling emergence (Supplementary Figure 9C). How the environmental signal-related trans-acting factors (or transcription factors) recognize and interact with the cis-acting elements, thereby positively or negatively regulating ZmAQPs at the transcriptional level, should be further investigated.
The aquaporin activity, trafficking, and interaction with other partners are influenced by post-translational modifications (Chaumont and Tyerman, 2017). Phosphorylation is a major posttranslational modification of aquaporins. Hence, we performed the prediction of maize aquaporin phosphorylation sites. According to the prediction results, serine was the most dominant phosphorylated residue in ZmAQPs, and the sites were relatively conserved within each subfamily (Supplementary Figure 1). The results showed that ZmPIPs had N-terminal Ser- and Thr-phosphorylated residues. In addition, ZmPIP2s had highly conserved Ser-phosphorylated sites in the C-terminal (Supplementary Figure 1). The identified sites in the previous study are annotated. These results prove the same in accordance with the previous analysis based on mass spectrometry and phosphoproteomic analysis (Van Wilder et al., 2008; Hu X. et al., 2015), thus confirming the prediction analysis. In ZmTIPs, the most highly conserved phosphorylation site was the Ser-residue located in the α-helixes of HE (Supplementary Figure 1). However, the site was not present in seed-specific ZmTIP3s. Calcium-dependent protein kinases acting in aquaporin phosphorylation have been identified (Harvengt et al., 2000; Sjövall-Larsen et al., 2006). Phosphorylation of AQPs confers plants the ability to gate aquaporins, which strongly involved in water channel activity (Chaumont and Tyerman, 2017; Daniels and Yeager, 2019). Moreover, the identification of interaction partners of AQPs may facilitate the discovery of novel protein kinases for phosphorylating AQPs, as well as new heterotetramers. There were studies indicating that plants had the capacity to regulate water transport activity and subcellular localization through heterotetramers (Zelazny et al., 2007; Utsugi et al., 2015; Bellati et al., 2016; Berny et al., 2016). The AQP protein-interacting networks were constructed based on the STRING. We identified a total of 104 ZmAQP-interacting proteins, mainly involved in sugar transport, transcription factors, and protein kinases (Supplementary Figure 10). The detailed information of the interacting proteins is shown in Supplementary Table 4. For example, the protein kinases will help to provide a better understanding of the phosphorylation of ZmAQPs. More rigorous experimental approaches are required to validate these predictive results. Further identification and characterization of cis-regulatory elements and posttranslational modifications may help to reveal the regulation mechanism of plant AQPs.
ZmTIPs: Are They Markers of Seed and Seedling Vigor?
Seed vigor, a comprehensive trait for evaluating seed quality, can be demonstrated by germination characteristics, such as the rate and uniformity of seed germination and seedling growth (ISTA, 2014). First, our study clarified the shift in ZmTIP3s and TIP1s/2s at the transcriptional level during seed germination (Figure 3B and Supplementary Figure 5B). Decreased the expression of TIP3s was relative to the coalescence of PSVs to central vacuoles (Novikova et al., 2014; Lee et al., 2015). Generally, TIP3s and TIP1s are considered to be characteristic of PSVs and vegetative vacuoles (Maurel et al., 2008). Early studies have proposed hypotheses about the roles of TIP3 in preparation for germination (Maurel, 1997). Aquaporins ZmTIP3s could be biomarkers of seed vigor based on proteomics analysis in our research for the first time (Li et al., 2018). The study demonstrated that a peak in ZmTIP3s expression was observed at maturity and decreased during germination (Figure 3C and Supplementary Figure 7). In parallel, we confirmed that ZmTIP3-1 improves the seed protein content and enhances seedling growth (Figures 5, 6). Because of the evaluation of seed vigor by the protein content of seed in cereals (Wen et al., 2018), we thus consider whether the ZmTIP3 expression levels can be used as a marker of the protein content in maize. Unfortunately, no significant positive correlation between the ZmTIP3 transcript accumulation and the stored protein content of seeds in about 270 maize accessions was observed, which may be attributed to the differences in the genetic background of the materials used. The finding is in alignment with previous experiments on soybean (Melroy and Herman, 1991). We suggest that ZmTIP3-1 might promote stored protein mobilization and thereby increase seedling growth. After radicle emergence, ZmTIP3s were replaced with ZmTIP1s/2s to perform functions in the tonoplast. Given the importance of mesocotyl elongation in seedling establishment and the extremely high expression of ZmTIP1s in the mesocotyl, A more detailed analysis is needed to elucidate whether and how ZmTIPs regulate seedlings growing out of the soil. Further, is the high accumulation of ZmTIP3 related to water uptake during seed imbibition? How does the dual localization of ZmTIP3 occur? Does ZmTIP3-2 have a different function in regulating seed germination? Answers to the above questions of both fundamental and practical importance remain to be revealed.
Conclusion
In conclusion, based on the latest maize genome database, we identified 41 ZmAQPs and performed a prediction analysis of their basic information involved in the physicochemical characteristics and structural properties. We focused on the expression profiles of ZmAQPs during the seed germination and seedling emergence, and further confirmed the positive roles of seed-specific ZmTIP3-1 in early seedling growth. The results provide direct genetic evidence of the functions of ZmAQPs in seed germination.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
LZ and CZ conceived the research. YS, ZL, and JS performed most of the experiments. CZ, LZ, and YS supervised the experiments and analyzed the data. CW and YL provided technical assistance and suggestions for writing. LZ and YS wrote the manuscript. All authors commented on the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31801450), the Modern Agricultural Industry Technology System Innovation Team of Shandong Province (SDAIT-02-02), and the Good Seed Project of Shandong province (2019LZGC002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.831916/full#supplementary-material
Footnotes
- ^ http://pfam.xfam.org/
- ^ http://www.uniprot.org/
- ^ https://www.ncbi.nlm.nih.gov/assembly/GCF_902167145.1
- ^ https://web.expasy.org/protparam/
- ^ https://wolfpsort.hgc.jp/
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ https://services.healthtech.dtu.dk/service.php?NetPhos-3.1
- ^ http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index
- ^ https://string-db.org/
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bari, A., Farooq, M., Hussain, A., Muhammad, T., Abbas, M., Mustafa, G., et al. (2018). Genome-wide bioinformatics analysis of aquaporin gene family in maize (Zea mays L.). J. Phylogenet. Evol. Biol. 6:197. doi: 10.4172/2329-9002.1000197
Barrieu, F., Chaumont, F., and Chrispeels, M. J. (1998). High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol. 117, 1153–1163. doi: 10.1104/pp.117.4.1153
Bellati, J., Champeyroux, C., Hem, S., Rofidal, V., Krouk, G., Maurel, C., et al. (2016). Novel aquaporin regulatory mechanisms revealed by interactomics. Mol. Cell Proteom. 15, 3473–3487. doi: 10.1074/mcp.M116.060087
Berny, M. C., Gilis, D., Rooman, M., and Chaumont, F. (2016). Single mutations in the transmembrane domains of maize plasma membrane aquaporins affect the activity of monomers within a heterotetramer. Mol. Plant 9, 986–1003. doi: 10.1016/j.molp.2016.04.006
Bewley, J., Bradford, K., Hilhorst, H., and Nonogaki, H. (2013). Seeds: physiology of development, germination and dormancy. New York, NY: Springer. doi: 10.1007/978-1-4614-4693-4
Chaumont, F., Barrieu, F., Herman, E. M., and Chrispeels, M. J. (1998). Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 117, 1143–1152. doi: 10.1104/pp.117.4.1143
Chaumont, F., Barrieu, F., Jung, R., and Chrispeels, M. J. (2000). Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol. 122, 1025–1034. doi: 10.1104/pp.122.4.1025
Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M. J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. doi: 10.1104/pp.125.3.1206
Chaumont, F., and Tyerman, S. (2017). Plant Aquaporins. New York, NY: Springer. doi: 10.1007/978-3-319-49395-4
Chaumont, F., and Tyerman, S. D. (2014). Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164, 1600–1618. doi: 10.1104/pp.113.233791
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194-1202. doi: 10.1016/j.molp.2020.06.009
Daniels, M. J., Chrispeels, M. J., and Yeager, M. (1999). Projection structure of a plant vacuole membrane aquaporin by electron cryo-crystallography1 1Edited by W. Baumeister. J. Mol. Biol. 294, 1337–1349. doi: 10.1006/jmbi.1999.3293
Daniels, M. J., and Yeager, M. (2019). Phosphorylation of TIP3 aquaporins during phaseolus vulgaris embryo development. Cells 8:1362. doi: 10.3390/cells8111362
Ding, L., Milhiet, T., Couvreur, V., Nelissen, H., Meziane, A., Parent, B., et al. (2020). Modification of the expression of the aquaporin ZmPIP2; 5 affects water relations and plant growth. Plant Physiol. 182, 2154–2165. doi: 10.1104/pp.19.01183
Finch-Savage, W. E., and Bassel, G. W. (2016). Seed vigour and crop establishment: extending performance beyond adaptation. J. Exp. Bot. 67, 567–591. doi: 10.1093/jxb/erv490
Footitt, S., Clewes, R., Feeney, M., Finch-Savage, W. E., and Frigerio, L. (2019). Aquaporins influence seed dormancy and germination in response to stress. Plant Cell Environ. 42, 2325–2339. doi: 10.1111/pce.13561
Gattolin, S., Sorieul, M., and Frigerio, L. (2011). Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Mol. Plant 4, 180–189. doi: 10.1093/mp/ssq051
Gattolin, S., Sorieul, M., Hunter, P. R., Khonsari, R. H., and Frigerio, L. (2009). In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biol. 9:133. doi: 10.1186/1471-2229-9-133
Ge, F. W., Tao, P., Zhang, Y., and Wang, J. B. (2014). Characterization of AQP gene expressions in Brassica napus during seed germination and in response to abiotic stresses. Biol. Plant. 58, 274–282. doi: 10.1007/s10535-013-0386-1
Guo, L., Wang, Z. Y., Lin, H., Cui, W. E., Chen, J., Liu, M., et al. (2006). Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 16, 277–286. doi: 10.1038/sj.cr.7310035
Harvengt, P., Vlerick, A., Fuks, B., Wattiez, R., Ruysschaert, J. M., and Homble, F. (2000). Lentil seed aquaporins form a hetero-oligomer which is phosphorylated by a Mg2+-dependent and Ca2+-regulated kinase. Biochem. J. 352, 183–190. doi: 10.1042/bj3520183
Hoai, P. T. T., Tyerman, S. D., Schnell, N., Tucker, M., McGaughey, S. A., Qiu, J., et al. (2020). Deciphering aquaporin regulation and roles in seed biology. J. Exp. Bot. 71, 1763–1773. doi: 10.1093/jxb/erz555
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, X., Wu, L., Zhao, F., Zhang, D., Li, N., Zhu, G., et al. (2015). Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 6:298. doi: 10.3389/fpls.2015.00298
Hunter, P. R., Craddock, C. P., Di Benedetto, S., Roberts, L. M., and Frigerio, L. (2007). Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 145, 1371–1382. doi: 10.1104/pp.107.103945
Javot, H., and Maurel, C. (2002). The role of aquaporins in root water uptake. Ann. Bot. 90, 301–313. doi: 10.1093/aob/mcf199
Johnson, K. D., Herman, E. M., and Chrispeels, M. J. (1989). An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 91, 1006–1013. doi: 10.1104/pp.91.3.1006
Kapilan, R., Vaziri, M., and Zwiazek, J. J. (2018). Regulation of aquaporins in plants under stress. Biol. Res. 51:4. doi: 10.1186/s40659-018-0152-0
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kozlova, L. V., Snegireva, A. V., and Gorshkova, T. A. (2012). Distribution and structure of mixed linkage glucan at different stages of elongation of maize root cells. Rus. J. Plant Physiol. 59, 339–347. doi: 10.1134/s1021443712030090
Kutschera, U., and Wang, Z. Y. (2016). Growth-limiting proteins in maize coleoptiles and the auxin-brassinosteroid hypothesis of mesocotyl elongation. Protoplasma 253, 3–14. doi: 10.1007/s00709-015-0787-4
Lee, S. E., Yim, H. K., Lim, M. N., Yoon, I., Kim, J., and Hwang, Y. S. (2015). Abscisic acid prevents the coalescence of protein storage vacuoles by upregulating expression of a tonoplast intrinsic protein gene in barley aleurone. J. Exp. Bot. 66, 1191–1203. doi: 10.1093/jxb/eru467
Lee, S. E., Yoon, I. S., and Hwang, Y. S. (2020). Aquaporin activity of barley tonoplast intrinsic proteins is involved in the delay of the coalescence of protein storage vacuoles in aleurone cells. J. Plant Physiol. 251:153186. doi: 10.1016/j.jplph.2020.153186
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, G., Santoni, V., and Maurel, C. (2014). Plant aquaporins: roles in plant physiology. Biochim. Biophys. Acta 1840, 1574–1582. doi: 10.1016/j.bbagen.2013.11.004
Li, Y., Qu, H., Zhu, P., Su, K., and Zhang, C. (2018). Comparative proteomics reveals the mechanisms underlying variations in seed vigor based on maize (Zea mays L.) ear positions. Plant Mol. Biol. Rep. 36, 738–749. doi: 10.1007/s11105-018-1115-x
Liu, H. Y., Yu, X., Cui, D. Y., Sun, M. H., Sun, W. N., Tang, Z. C., et al. (2007). The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 17, 638–649. doi: 10.1038/cr.2007.34
Liu, X., Chen, C.-Y., Wang, K.-C., Luo, M., Tai, R., Yuan, L., et al. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25, 1258–1273. doi: 10.1105/tpc.113.109710
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Mao, Z., and Sun, W. (2015). Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. J. Exp. Bot. 66, 4781–4794. doi: 10.1093/jxb/erv244
Maurel, C. (1997). Aquaporins and water permeability of plant membranes. Ann. Rev. Plant Biol. 48, 399–429. doi: 10.1146/annurev.arplant.48.1.399
Maurel, C., Boursiac, Y., Luu, D. T., Santoni, V., Shahzad, Z., and Verdoucq, L. (2015). Aquaporins in Plants. Physiol. Rev. 95, 1321–1358. doi: 10.1152/physrev.00008.2015
Maurel, C., Chrispeels, M., Lurin, C., Tacnet, F., Geelen, D., Ripoche, P., et al. (1997a). Function and regulation of seed aquaporins. J. Exp. Bot. 48, 421–430. doi: 10.1093/jxb/48.Special_Issue.421
Maurel, C., Tacnet, F., Güclü, J., Guern, J., and Ripoche, P. (1997b). Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc. Nat. Acad. Sci 94, 7103–7108. doi: 10.1073/pnas.94.13.7103
Maurel, C., Verdoucq, L., Luu, D. T., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
Melroy, D. L., and Herman, E. M. (1991). TIP, an integral membrane protein of the protein-storage vacuoles of the soybean cotyledon undergoes developmentally regulated membrane accumulation and removal. Planta 184, 113–122. doi: 10.1007/BF00208244
Murata, K., Mitsuoka, K., Hirai, T., Walz, T., and Fujiyoshi, Y. (2000). Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605. doi: 10.1038/35036519
Niemietz, C. M., and Tyerman, S. D. (1997). Characterization of water channels in wheat root membrane vesicles. Plant Physiol. 115, 561–567. doi: 10.1104/pp.115.2.561
Nonogaki, H., Bassel, G. W., and Bewley, J. D. (2010). Germination—Still a mystery. Plant Sci. 179, 574–581. doi: 10.1016/j.plantsci.2010.02.010
Novikova, G. V., Tournaire-Roux, C., Sinkevich, I. A., Lityagina, S. V., Maurel, C., and Obroucheva, N. (2014). Vacuolar biogenesis and aquaporin expression at early germination of broad bean seeds. Plant Physiol. Biochem. 82, 123–132. doi: 10.1016/j.plaphy.2014.05.014
Obroucheva, N. V. (2013). Aquaporins in seeds. Seed Sci. Res. 23, 213–216. doi: 10.1017/s0960258513000275
Obroucheva, N. V., and Sin’kevich, I. A. (2010). Aquaporins and cell growth. Rus. J. Plant Physiol. 57, 153–165. doi: 10.1134/s1021443710020019
Obroucheva, N. V., Sinkevich, I. A., Lityagina, S. V., and Novikova, G. V. (2017). Water relations in germinating seeds. Rus. J. Plant Physiol. 64, 625–633. doi: 10.1134/s102144371703013x
Peret, B., Li, G., Zhao, J., Band, L. R., Voss, U., Postaire, O., et al. (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991–998. doi: 10.1038/ncb2573
Quigley, F., Rosenberg, J. M., Shachar-Hill, Y., and Bohnert, H. J. (2001). From genome to function: the Arabidopsis aquaporins. Genome Biol. 3, 1–17. doi: 10.1186/gb-2001-3-1-research0001
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi: 10.1146/annurev-arplant-042811-105550
Reinhardt, H., Hachez, C., Bienert, M. D., Beebo, A., Swarup, K., Voss, U., et al. (2016). Tonoplast aquaporins facilitate lateral root emergence. Plant Physiol. 170, 1640–1654. doi: 10.1104/pp.15.01635
Rios, J. J., Martinez-Ballesta, M. C., Ruiz, J. M., Blasco, B., and Carvajal, M. (2017). Silicon-mediated Improvement in plant salinity tolerance: the role of aquaporins. Front. Plant Sci. 8:948. doi: 10.3389/fpls.2017.00948
Sakurai, J., Ishikawa, F., Yamaguchi, T., Uemura, M., and Maeshima, M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. doi: 10.1093/pcp/pci172
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sjövall-Larsen, S., Alexandersson, E., Johansson, I., Karlsson, M., Johanson, U., and Kjellbom, P. (2006). Purification and characterization of two protein kinases acting on the aquaporin SoPIP2; 1. Biochimica et Biophysica Acta (BBA)-Biomembranes 1758, 1157–1164.
Soto, G., Alleva, K., Mazzella, M. A., Amodeo, G., and Muschietti, J. P. (2008). AtTIP1; 3 and AtTIP5; 1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582, 4077–4082. doi: 10.1016/j.febslet.2008.11.002
Stelpflug, S. C., Sekhon, R. S., Vaillancourt, B., Hirsch, C. N., Buell, C. R., de Leon, N., et al. (2015). An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9:25. doi: 10.3835/plantgenome2015.04.0025
Sudhir, K., Glen, S., Michael, L., Christina, K., and Koichiro, T. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 6:96.
Takahashi, H., Rai, M., Kitagawa, T., Morita, S., Masumura, T., and Tanaka, K. (2004). Differential localization of tonoplast intrinsic proteins on the membrane of protein body type II and aleurone grain in rice seeds. Biosci. Biotechnol. Biochem. 68, 1728–1736. doi: 10.1271/bbb.68.1728
Trofimova, M., Zhestkova, I., Andreev, I., Svinov, M., Bobylev, Y. S., and Sorokin, E. (2001). Osmotic water permeability of vacuolar and plasma membranes isolated from maize roots. Rus. J. Plant Physiol. 48, 287–293. doi: 10.1023/A:1016697813072
Tu, X. Y., Mejia-Guerra, M. K., Franco, J. A. V., Tzeng, D., Chu, P. Y., Shen, W., et al. (2020). Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat. Commun. 11:18832. doi: 10.1038/s41467-020-18832-8
Utsugi, S., Shibasaka, M., Maekawa, M., and Katsuhara, M. (2015). Control of the water transport activity of barley HvTIP3;1 specifically expressed in seeds. Plant Cell Physiol. 56, 1831–1840. doi: 10.1093/pcp/pcv104
Van Wilder, V., Miecielica, U., Degand, H., Derua, R., Waelkens, E., and Chaumont, F. (2008). Maize plasma membrane aquaporins belonging to the PIP1 and PIP2 subgroups are in vivo phosphorylated. Plant Cell Physiol. 49, 1364–1377. doi: 10.1093/pcp/pcn112
Vander Willigen, C., Postaire, O., Tournaire-Roux, C., Boursiac, Y., and Maurel, C. (2006). Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant Cell Physiol. 47, 1241–1250. doi: 10.1093/pcp/pcj094
Vera-Estrella, R., Barkla, B. J., Bohnert, H. J., and Pantoja, O. (2004). Novel regulation of aquaporins during osmotic stress. Plant Physiol. 135, 2318–2329. doi: 10.1104/pp.104.044891
Wang, Y., Zhao, Z., Liu, F., Sun, L., and Hao, F. (2020). Versatile roles of aquaporins in plant growth and development. Int. J. Mol. Sci. 21:21249485. doi: 10.3390/ijms21249485
Wen, D., Hou, H., Meng, A., Meng, J., Xie, L., and Zhang, C. (2018). Rapid evaluation of seed vigor by the absolute content of protein in seed within the same crop. Sci. Rep. 8:5569. doi: 10.1038/s41598-018-23909-y
Xu, C., Wang, M., Zhou, L., Quan, T., and Xia, G. (2013). Heterologous expression of the wheat aquaporin gene TaTIP2;2 compromises the abiotic stress tolerance of Arabidopsis thaliana. PLoS One 8:e79618. doi: 10.1371/journal.pone.0079618
Yu, P., Gutjahr, C., Li, C., and Hochholdinger, F. (2016). Genetic control of lateral root formation in cereals. Trends Plant Sci. 21, 951–961. doi: 10.1016/j.tplants.2016.07.011
Zelazny, E., Borst, J. W., Muylaert, M., Batoko, H., Hemminga, M. A., and Chaumont, F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Nat. Acad. Sci. 104, 12359–12364. doi: 10.1073/pnas.0701180104
Zhang, D., Tong, J., He, X., Xu, Z., Xu, L., Wei, P., et al. (2015). A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic stress. Front. Plant Sci. 6:1237. doi: 10.3389/fpls.2015.01237
Zhang, Z., Yang, J., and Wu, Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 Heterodimerizing Proteins. Plant Cell 27, 1162–1172. doi: 10.1105/tpc.15.00035
Zhao, L., Peng, T., Chen, C.-Y., Ji, R., Gu, D., Li, T., et al. (2019). HY5 interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis. Plant Physiol. 180, 1450–1466. doi: 10.1104/pp.19.00055
Zhao, L., Xie, L., Huang, J., Su, Y., and Zhang, C. (2020). Proper Glyphosate application at post-anthesis lowers grain moisture content at harvest and reallocates non-structural carbohydrates in maize. Front. Plant Sci. 11:2018. doi: 10.3389/fpls.2020.580883
Zhong, S., Shi, H., Xue, C., Wei, N., Guo, H., and Deng, X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Nat. Acad. Sci. 111, 3913–3920. doi: 10.1073/pnas.1402491111
Keywords: aquaporin, tonoplast intrinsic protein 3, seed germination and seedling emergence, seed vigor, gene family analysis, maize
Citation: Su Y, Liu Z, Sun J, Wu C, Li Y, Zhang C and Zhao L (2022) Genome-Wide Identification of Maize Aquaporin and Functional Analysis During Seed Germination and Seedling Establishment. Front. Plant Sci. 13:831916. doi: 10.3389/fpls.2022.831916
Received: 10 December 2021; Accepted: 07 January 2022;
Published: 27 January 2022.
Edited by:
Abidur Rahman, Iwate University, JapanReviewed by:
Louai Rishmawi, UMR 5004 Biochimie et Physiologie Moléculaire des Plantes (BPMP), FranceSteven Footitt, Boǧaziçi University, Turkey
Copyright © 2022 Su, Liu, Sun, Wu, Li, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunqing Zhang, Y3F6aGFuZ0BzZGF1LmVkdS5jbg==; Linmao Zhao, bG16aGFvQHNkYXUuZWR1LmNu
 Yingchun Su
Yingchun Su Zhonghao Liu
Zhonghao Liu Linmao Zhao
Linmao Zhao