- 1Plant Gene Engineering Center, Chinese Academy of Sciences Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement, South China Botanical Garden, Chinese Academy of Sciences, Guangdong Key Laboratory of Applied Botany, Guangzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
Transgene integration typically takes place in an easy-to-transform laboratory variety before the transformation event is introgressed through backcrosses to elite cultivars. As new traits are added to existing transgenic lines, site-specific integration can stack new transgenes into a previously created transgenic locus. In planta site-specific integration minimizes the number of segregating loci to assemble into a breeding line, but cannot break genetic linkage between the transgenic locus and nearby undesirable traits. In this study, we describe an additional feature of an in planta gene-stacking scheme, in which the Cre (control of recombination) recombinase not only deletes transgenic DNA no longer needed after transformation but also mediates recombination between homologous or non-homologous chromosomes. Although the target site must first be introgressed through conventional breeding, subsequent transgenes inserted into the same locus would be able to use Cre-mediated translocation to expedite a linkage drag-free introgression to field cultivars.
Introduction
Crop development via transgenesis is typically done by inserting DNA into an easily transformable variety and then introgressing the transgene out to many different locale-specific cultivars. As new traits are developed, it becomes a challenge of where to integrate new trait genes. If inserted into a new locus, breeders will have more loci to reassemble back into a breeding line. Efforts to cluster multiple transgenes at a single integration locus can be achieved through prior stacking of the many genes in vitro into a single plant transformation construct (Goderis et al., 2002; Chen et al., 2006; Shih et al., 2016; Zhu et al., 2017; Collier et al., 2018). However, relying solely on this approach means that further addition of transgenes would require combining new genes with previously introduced transgenes into a larger transformation vector. Though this is not a technical limitation, it could have legal ramifications. Previously introduced traits could then require a new round of de-regulation for being a new transformation event. Adding more transgenes to an existing transgene locus is possible via in planta site-specific integration through the use of site-specific nucleases (Puchta et al., 1993; Wright et al., 2005; Fauser et al., 2012; Zhang et al., 2012; Dong and Ronald, 2021) or site-specific recombinases (Albert et al., 1995; Day et al., 2000; Li et al., 2009; Hou et al., 2014; Nandy et al., 2015; Chen and Ow, 2017; Chen et al., 2019). This would insure co-introgression to field cultivars without additional loci to impede downstream breeding.
We had described an in planta gene-stacking system that uses mycobacteriophage Bxb1 integrase (recombinase) for site-specific integration (Hou et al., 2014). In this system, a target site is first created by the insertion of a first trait gene linked to an attP (phage attachment) sequence that serves as a “target site” (Figure 1A). New DNA is introduced through a donor construct, such as a second trait gene plasmid that also carries two complementary attB (bacterial attachment) sequences (Figure 1B). The recombination of one plasmid-encoded attB with the genomic attP places the incoming DNA precisely into the genomic target (Figure 1C). Since the donor DNA can carry two attB sites, two configurations are possible depending on which attB site recombines. Figure 1C shows the preferred configuration that can be screened by polymerase chain reaction (PCR), and the attB not used in the first round of integration can serve as a target site for the next round of integration by a donor plasmid with two attP sites (not shown; refer to Hou et al., 2014). In theory, this permits serial gene stacking by alternating between the uses of attB and attP donor plasmids. Cre (control of recombination) recombinase is then introduced to delete away transgenic DNA flanked by lox (locus of x-over) sites that is no longer needed after transformation (Figure 1E).
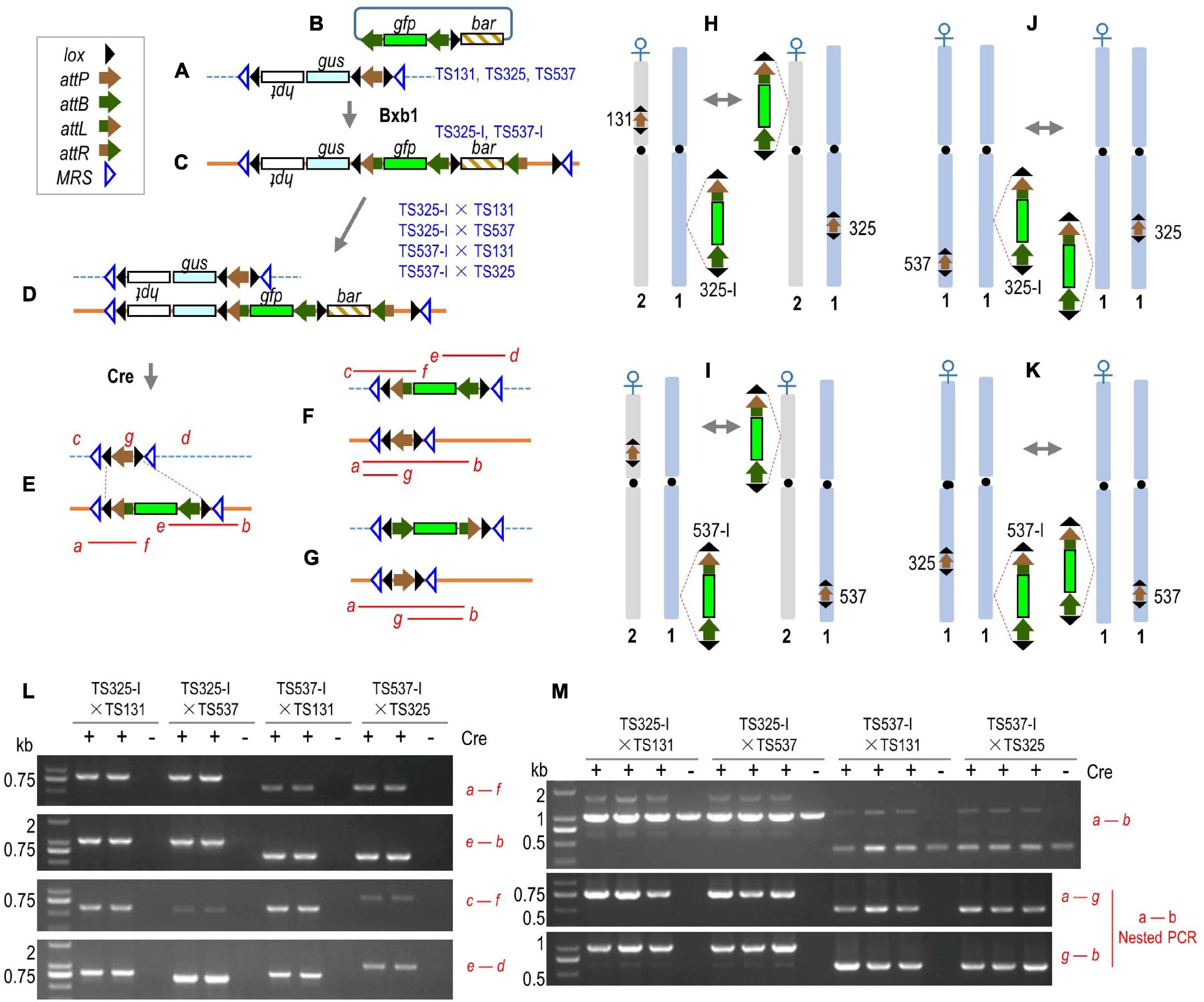
Figure 1. Cre-mediated recombination between different target sites. (A) Rice target lines TS131, TS325, and TS537 structure. (B) Integrating vector pZH210B was inserted into TS325 and TS537 to form TS325-I and TS537-I, respectively (C). F1 hybrids from crosses (D) and expected structures from Cre-mediated deletion of DNA flanked by directly oriented lox sites (E); inversion from recombination of oppositely oriented lox sites not shown. Translocation of gfp-containing segment to another chromosome (F) and after inversion (G). Red italic letters, PCR primers; red lines, PCR products. (H–K) Chromosome location of target sites in TS131, TS325, and TS537 with relevant lox sites and the direction of attP. Crosses with gfp-stacked lines (i.e., 325-I and 537-I) leads to Cre-mediated reciprocal translocation of the lox-flanked DNA. (L,M) PCR results from deletion and chromosome recombination in F1 + cre plants. Genes described in the Methods section and were transcribed in direction of lettering, promoters, and terminators not shown. Recombination sites indicated in inset legend. DNA size is mentioned in kb.
Of particular relevance is that the target construct has been designed with a set of lox sites in the opposing orientations (Figure 1A), in which they are retained after Cre-mediated deletion regardless of the number of transgenes stacked into the target site (Figure 1E). This pair of inverted lox sites can serve as recombination substrates for Cre-mediated intra-chromosomal inversion, as well as inter-chromosomal recombination. Prior studies have shown that Cre is capable of causing recombination between different chromosomes (Qin et al., 1994; Smith et al., 1995; Koshinsky et al., 2000; Vergunst et al., 2000; Zong et al., 2005; Titen et al., 2020). Inter-chromosomal recombination would break genetic linkage that could potentially expedite transgene introgression from a laboratory-transformed line to field cultivars (Figures 1F,G). In this study, we show that inter-chromosomal recombination of lox sites can relocate a transgene to a different chromosome, whether to the same location of a homologous chromosome or to another location in a non-homologous chromosome. Although the original target construct must first be introgressed through conventional breeding, subsequent transgenes appended to that locus would be able to use site-specific translocation for linkage drag-free introgression to field cultivars.
Materials and Methods
Site-Specific Integration and Rice Transformation
Biolistic-mediated site-specific integration of rice (Oryza sativa cv. Zhonghua 11) target line TS131 (Figure 1A) using integrating vector pZH201B has been described (Li et al., 2016) and greater details are available including lines TS325 and TS537 (Figure 1A; Li et al., submitted). Each of these three target lines has a full-length T-DNA construct-expressing reporter gene gus (encoding β-glucuronidase), with correct sequence recombination sites, and is located at a non-gene coding DNA >2.9 kb and >0.8 kb from nearest start and stop codons, respectively. Line TS*537* was generated from TS537 through CRISPR/Cas9-mediated mutagenesis (Ma et al., 2015) using oligonucleotides listed in Supplementary Table 1. The primer pair was connected to vector pYLCRISPR/CasPubi-B after annealing. Agrobacterium-mediated transformation of rice calluses with a cre-expressing construct was conducted as described (Li and Li, 2003). Other genes shown in Figure 1A are hpt, gfp, and bar that encode, respectively, hygromycin phosphotransferase, green fluorescence protein, and bialaphos resistance enzyme.
Mutated PCR and Restriction Endonuclease Digestion
Restriction endonuclease analysis was carried out to distinguish between TS537 and TS*537*. However, because the CRISPR-mediated changes in TS*537* did not destroy an existing restriction site, CRS-PCR (created restriction site PCR) (Qiao et al., 2013; Wang et al., 2016; Avanus and Altınel, 2017; Ding et al., 2017) and overlapping PCR were used to create a restriction site for the TS537 PCR product, but not for the TS*537* PCR product. Specific steps are outlined in Figure 2H as follows: primers h + e, h + f, e + k, and f + k can only be amplified from F1 + cre; one or two bases were changed near the 3′-end of the oligonucleotide m or o, respectively (Supplementary Figure 1); primers h + n and m + i created the overlapping PCR product h-i, and similarly, primers j + p and o + k created the overlapping PCR product j–k. The product h–i or j–k would be cleaved by AseI or AgeI (NEB Beijing, China), respectively, if amplified from TS537, but not from TS*537* (Figure 2H).
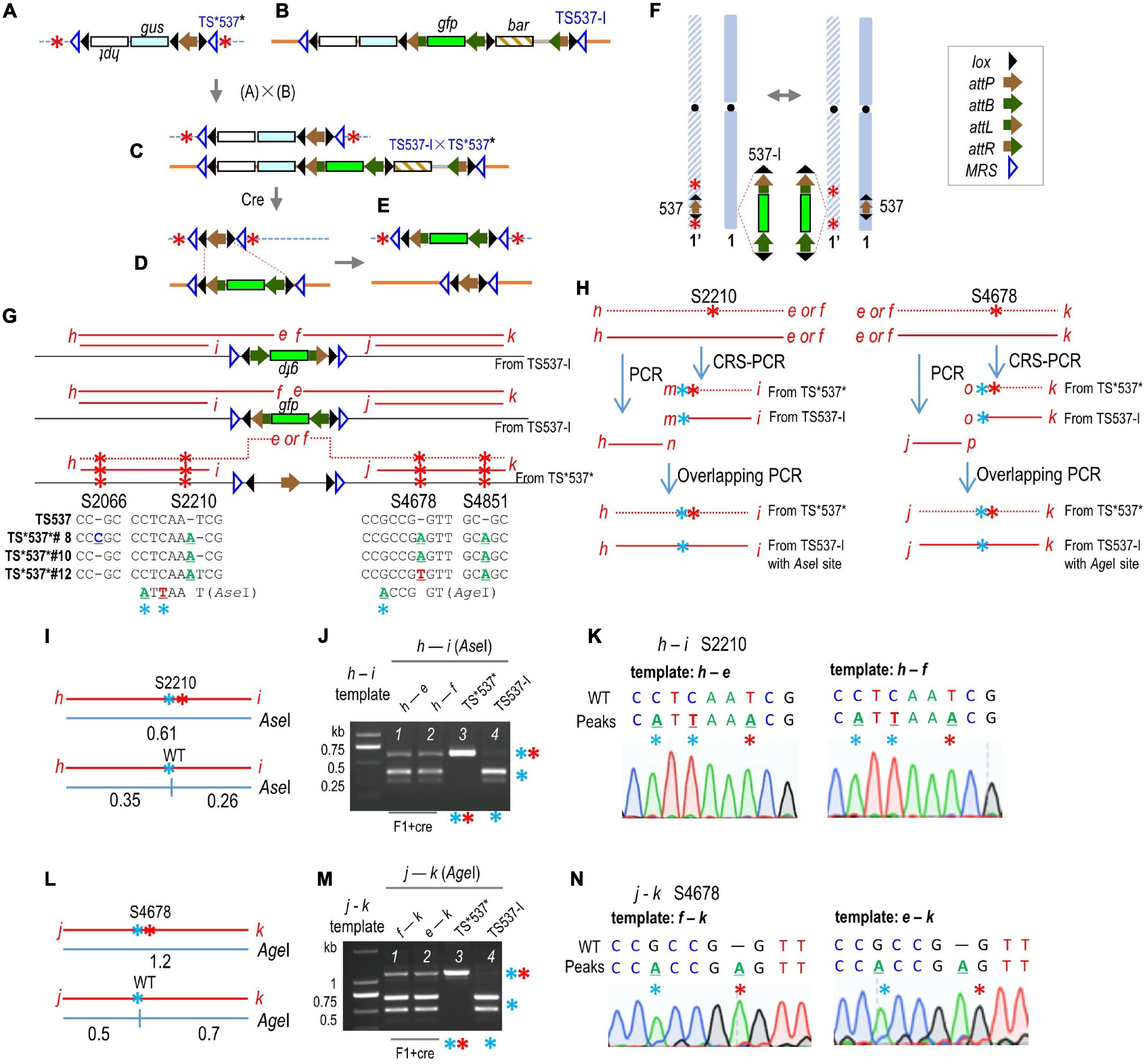
Figure 2. Gene translocation from a target site to an allelic target site. (A) TS*537* structure mutated by CRISPR. (B) Structure of gfp-stacking line TS537-I. (C) Structure of F1 hybrid from TS537-I × TS*537*. (D) Resolved structure from Cre-mediated deletion; inversion not shown. (E) Structure after Cre-mediation reciprocal translocation from recombination between lox sites (red dashed lines) from a TS537-I × TS*537* cross (F). (G) CRISPR generated mutations in TS*537* (S2066, S2210, S4678, and S4851). Red letters, PCR primers; red lines, PCR products. (H) CRS-PCR and overlapping PCR to create AseI or AgeI restriction site for PCR products derived from TS537, but not from TS*537*. Blue * indicates mutation in PCR primer to form restriction site, red * indicates CRISPR mutations that abolish restriction site. Expected fragment sizes (in kb) and data from AseI (I,J) or AgeI (L,M) cleavage of CRS-PCR/overlapping PCR products of F1 + cre DNA. Sequencing results of h–i fragment resistant to AseI (K) or AgeI (N).
PCR and DNA Sequencing
DNA was isolated from ∼100 mg of a 60-day-old rice leaf tissue and ground in liquid nitrogen as described (Lu and Zheng, 1992). PCR was conducted under standard conditions using 1.1 × T3 super PCR mix (Tsingke Biotechnology, Beijing, China) and KOD-FX High-Fidelity DNA Polymerase (TOYOBO, Osaka, Japan) with primers listed in Supplementary Table 2. PCR products were isolated by using HiPure Gel Pure DNA Mini Kit (Magen, Guangzhou, China) and sequenced by Sangon Biotech (Shanghai, China).
Results and Discussion
Recombination Between Non-homologous Chromosomes
We sought to test Cre recombinase-mediated recombination of lox sites between chromosomes and whether the recombination between sets of flanking inverted lox sites could translocate a transgene to another chromosome. Target lines TS131, TS325, and TS537 served as receptor lines, and each harbors a single copy of the target construct (Figure 1A). TS131 is located in the short arm of chromosome 2, TS325 and TS537 are both located in the long arm of chromosome 1, and their chromosome orientation is indicated by the direction of attP flanked by the relevant lox sites (Figures 1H–K). From Bxb1-mediated integration of the gfp-containing plasmid pZH210B (Figure 1B) into TS325 and TS537 (Li et al., submitted), integrant lines TS325-I and TS537-I, respectively, were generated to serve as donor lines (Figure 1C).
The F1 hybrid from an integrant and target line would be expected to harbor two chromosomes with lox sites that can recombine with each other if Cre is introduced (Figure 1D). Cre-mediated intra-molecular recombination of lox sites is expected to produce a resolved structure (Figure 1E), while inter-chromosomal recombination can generate various intermediates, including one final outcome being the translocation of gfp to another chromosome in either orientation (Figures 1F,G). To test this possibility, we conducted four pairs of crosses between homozygous plants, namely, TS325-I♂ × TS131♀(Figure 1H), TS537-I♂ × TS131♀(Figure 1I), TS325-I♂ × TS325♀(Figure 1K). The F1 seeds, hemizygous for two different transgenic loci, were used to induce embryogenic calluses. The cre gene was then transformed into these calluses by Agrobacterium-mediated gene transfer.
After regeneration of F1 + cre plants from calluses, the non-recombined structures were tested by PCR using primers a + f and e + b (Figures 1E,L), whereas the translocation of the gfp-containing fragment to a different chromosome was also detected by primers c + f and e + d (Figures 1F,L). Note that the primers a, b, c, and d lie outside the target construct and are, therefore, unique for each chromosome location. Detection by location-specific primers c + f and e + d, however, could not distinguish between a double recombination events in the same cell vs. separate recombination events in different cells. Location-specific primers c + d also failed to amplify a contiguous fragment containing gfp that would be ∼4.5, ∼4.9, or ∼4.3 kb from TS131, TS325, or TS537 chromosomes, respectively, likely due to competing reactions of the smaller ∼0.6, ∼1.0, or ∼0.4 kb fragment from the corresponding WT chromosomes. In contrast, the reciprocal product from a translocation, the replacement of the gfp-containing fragment by an attP fragment, was only 0.8 kb larger than the WT chromosome-derived product. Indeed, location-specific primers a + b amplified the WT ∼1 kb and a larger ∼1.8 kb band from the TS325-I × TS131- and TS325-I × TS537-derived F1 plants, and a ∼1.2-kb product was detected along with the WT ∼0.4 kb product from the TS537-I × TS131 and TS537-I × TS325 F1 plants (Figure 1M). A contiguous fragment from location-specific primers a + b indicated a double recombination event from at least some cells.
Since Cre-mediated recombination could also invert lox-flanked DNA, the attP could be in the opposite orientation in the chromosome (Figure 1G). Using nested PCR of the location-specific a–b PCR product, where primer g corresponds to within the attP sequence (Figures 1E–G), both attP orientations were found as amplified products were detected with primers a + g as well as by primers g + b (Figure 1M).
Recombination Between Homologous Chromosomes
To test for potential transgene cassette exchange between homologous chromosomes, it was necessary to have sequence differences in the flanking regions. Therefore, we used CRISPR/Cas9 technology to mutate both sides of TS537 to generate TS*537* (Figure 2A). Out of 96 transgenic plants, 43 had segregated away cas9, and 15 of those were sequenced for PCR products from primers h + i and j + k (Figure 2G). Three independent TS*537* lines were found with mutations on both sides of the target construct and without heterozygosity, which suggested the same mutations generated in both homolog chromosomes or that the mutations were copied onto its homologous chromosome. Four mutations were found at chromosome 1 positions 35,912,066, 35,912,210, 35,914,678, and 35,914,851, hereafter named sites S2066, S2210, S4678, and S4851, respectively (Figure 2G). TS*537*#8 has mutations at all four sites, whereas TS*537*#10 and TS*537*#12 lack a mutation at S2066. These three lines (i.e., TS*537*#8, TS*537*#10, and TS*537*#12) were crossed with homozygous TS537-I♀ (Figure 2B) to generate F1 heterozygotes (Figure 2C). Cre-lox intramolecular recombination is expected to produce a resolved structure (Figure 2D), but inter-chromosomal recombination can also generate various intermediates including the translocation of gfp to homologous chromosome (Figures 2E,F). Embryogenic calluses of the F1 heterozygotes were then transformed with a cre-expressing construct through Agrobacterium infection.
To detect possible chromosome recombination, CRS-PCR and overlapping PCR were used to create a restriction site for the PCR product from TS537-I, but not from TS*537* (Figure 2H and Supplementary Figure 1). Beginning with a template from primers h + e or h + f, primers m + i were then used to change the WT sequence to create an AseI site near S2210 (Figure 2H and Supplementary Figure 1A). Primers n and m overlap by 23 bp, and the h–n and m–i fragments were templates for primers h + i, which would, therefore, have an AseI site if copied from TS537-I, but not from TS*537*. Indeed, the ∼0.61 kb h–i fragment (Figure 2I) was cleaved by AseI into 0.35 and 0.26 kb products if from TS537-I DNA, but not from TS*537* DNA (Figure 2J, lanes 3 and 4). Likewise, primers o + k were used to create an AgeI site near S4678 (Figure 2L, Supplementary Figure 1B) from template e–k or f–k (Figure 2H). Primers p and o overlap by 20 bp, and the j–p and o-k fragments were templates for primers j + k. The j–k fragment would have an AgeI site if copied from TS537-I, but not from TS*537*. As shown in Figure 2M, the ∼1.2 kb j–k fragments (Figure 2L) were cleaved by AgeI into 0.5 and 0.7 kb fragments from TS537-I DNA, but not from that of TS*537* DNA (lanes 3 and 4).
From the F1 + cre genome, primers h + f and e + k should reveal whether gfp is linked to WT or mutated DNA (Figure 2G) as primers e and f were anchored to gfp. However, since gfp could also be inverted, primers h + e and f + k were also tested. These PCR products were then used as templates for nested PCR to amplify h–i and j–k as described above, followed by AseI or AgeI treatment to detect mutated sites at S2210 and S4678, respectively (Figure 2H). As shown in the representative data of a 60-day-old F1 + cre plant from TS537-I♂ × TS*537*#10♀, some h–i products derived from h–e and h–f templates were immune to AseI cleavage (Figure 2J, lanes 1 and 2). Likewise, some j–k products derived from e–k and f–k templates resisted AgeI (Figure 2M, lanes 1 and 2). Assuming that the gfp-anchored primer sets amplified TS537-I and TS*537-I* equally, and that what translocated across can translocate back, at most 50% of gfp DNA would be linked to mutated sites. Based on band intensity, ∼20% of the gfp DNA was linked to mutations of the TS*537* genome. The h–i and j–k fragments that were immune to AseI and AgeI cleavage (upper bands from Figures 2J,M, lanes 1 and 2) were gel-purified for DNA sequencing. As shown in Figures 2K,N, the predominant peaks show the TS*537*#10 sequence. As these h–i and j–k fragments were derived from h–f, h–e, f–k, and e–k templates, with primers e and f anchored to gfp, this demonstrates that the S2210 and S4678 mutations in TS*537*#10 were each linked to gfp.
Since recombination could generate at 8 genotypes, namely, TS537-I and TS*537* parental types, TS537 and TS*537-I* from cassette exchange, and TS*537-I, TS537*, TS537-I*, and TS*537 from single crossovers, primers h + k were used to preferentially amplify the smaller ∼4.4 kb size products from TS*537* × TS537-I F1 + cre genomic DNA (Figure 3B). Although lox-flanked DNA could invert, the regions corresponding to mutated sites should remain constant. In competing reactions, primers h + k amplified only the ∼4.4 kb h-k band with the attP site, but not the longer ∼7.5 kb fragment containing gfp (Figure 3), and the segments corresponding to the four CRISPR mutations were sequenced. The major peaks were consistent with the TS*537* sequence (CRISPR-mutated sequence) before translocation. Minor peaks were also found that correspond to WT sequence in TS537 or TS537-I (Figure 3C). This would be consistent with translocation of the attP site from TS*537* to its homologous chromosome from TS537-I.
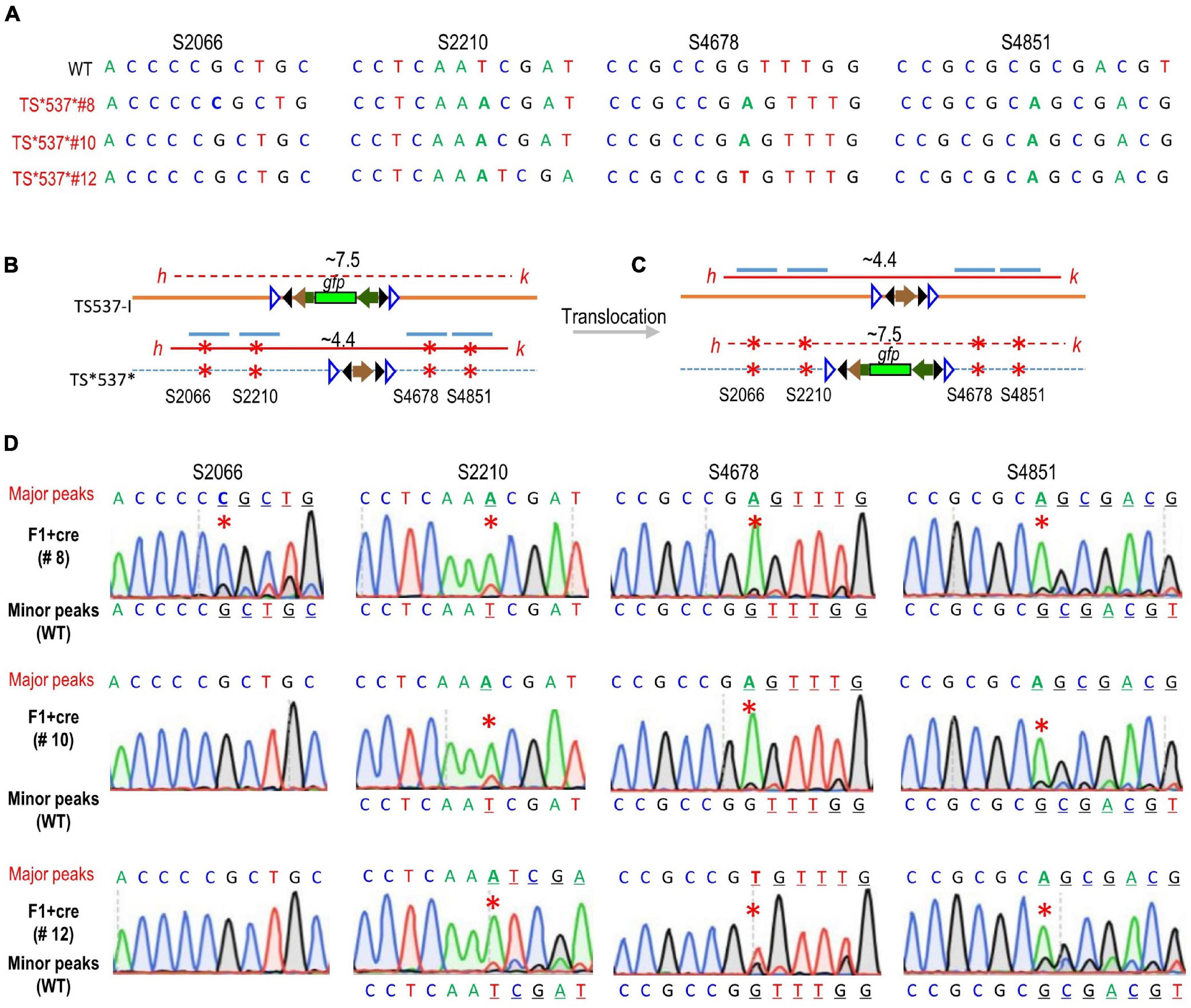
Figure 3. Sequencing results after gene exchange between allelic target sites. (A) Sequences of regions corresponding to CRISPR mutation sites. PCR product h–k containing gfp before (B) or after translocation (C) was too long (∼7.5 kb) for amplification (dashed red line) in competing reaction with shorter (∼4.4 kb) attP fragment (solid red line) without gfp. Blue lines depict the four regions sequenced, respectively. (D) Sequencing data from the ∼4.4 kb h–k fragments from different F1 + cre genomes. Major peaks consistent with the TS*537*-derived sequence before translocation (B); minor peaks consistent with the TS537-derived sequence after translocation (C). Red * indicates CRISPR mutations.
To examine whether PCR template switching had been a factor in our analysis, we tested a simulated experiment mixing 1:1 the genomic DNA of TS*537*#8 and TS537-I before PCR with primers h + x (Supplementary Figure 2). Primer x lies within the ampicillin resistance gene in donor vector pZH210B, and the h–x band should be ∼3.3 kb, as primer h is separated from TS537 by ∼2 kb of genomic DNA. If template switching occurred at a significant rate, S2066 and S2210 mutations should appear as minor peaks. Despite conducting this test under various PCR conditions, minor peaks corresponding to mutations were below detection (Supplementary Figure 2).
Future Prospects
In practice, the F1 plant in this exercise would represent a hybrid between a lab cultivar with a newly integrated transgene and a field cultivar previously introgressed with a target site containing already inserted transgenes. To use the Cre-mediated recombination to break genetic linkage on one or both sides of the transgene, the field cultivar must also have a target site. This can be done by conventional breeding (Figures 4A–C) of the target locus with or without a first transgene. Subsequent stacking of additional transgenes would be into the laboratory line (Figure 4D), which can then be crossed with the transgenic field cultivar (Figures 4C,D). Introduction of Cre recombinase would most likely be through a genetic cross, and preferably by a cre line already introgressed into elite genotype. Cre would then translocate the new transgenic locus from the lab line to the field line at the same chromosome position (Figure 4E), or optionally, to a different chromosome with a target locus introgressed into a field line (Figures 4F,G). This may be necessary if non-elite traits on either side of the transgene are too close to the target locus to obtain a suitable field target line by conventional breeding. In short, the first introgression is by conventional breeding, while subsequent introgression is facilitated by Cre-lox-mediated recombination. This linkage breakage strategy may not be universally applicable, but is suitable for the particular in planta gene-stacking method we described (Hou et al., 2014), and this principle can be adapted for other in planta gene-stacking schemes.
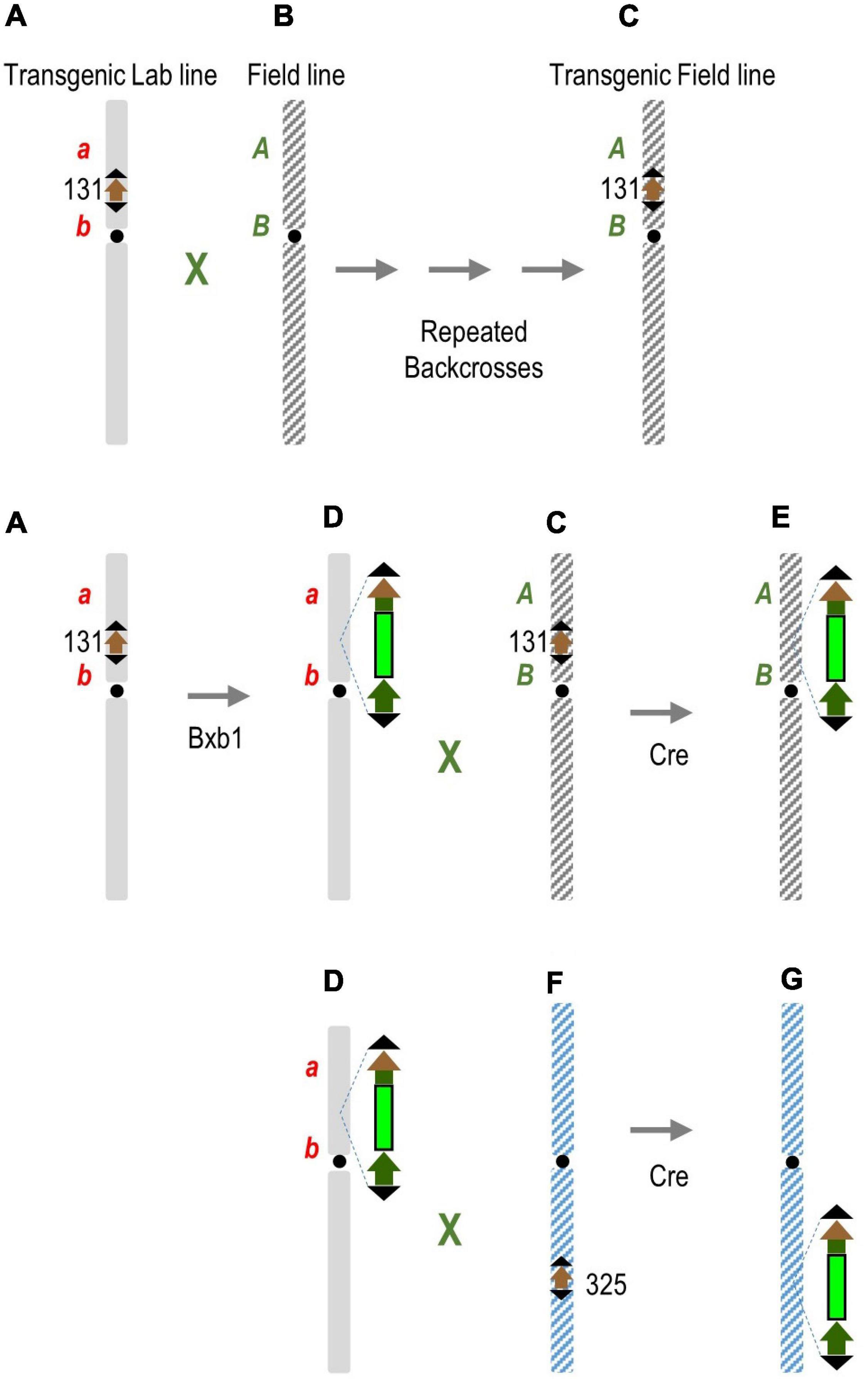
Figure 4. Cre-mediated transgene translocation to break linkage drag. (A) Transgenic lab line created by gene transfer, with or without a first transgene (not shown), is crossed repeatedly to a field line (B) to introgress the transgenic locus into transgenic field line (C) with the combination of elite traits. Subsequent stacking of additional transgenes to the same target site (D) can be crossed with the transgenic elite line (C). Introduction of Cre recombinase translocates the new transgenic locus from lab line to field line at the same chromosome position (E). Optionally, Cre translocates the transgenic locus in line (D) to a different chromosome (F) with a target locus introgressed into a field line (G), which might be necessary if the non-elite traits a and/or b are too close to the target locus and fail to segregate away. A, B represent elite traits; a, b non-elite traits.
It is interesting that inter-chromosomal recombination was detected in the F1 generation that had not gone through gamogenesis. This could mean that homologous chromosome synapsis was not a factor, and whether it could further increase the translocation rate remains to be tested. We admit that the sequence data were derived from a population of PCR products; hence, we could not exclude the possibility that some or all fragments had undergone intermolecular recombination on only one side of the donor target site. Nonetheless, breaking linkage drag does not require a cassette exchange reaction if it were between the same homologous chromosome locations, but merely the inter-chromosomal recombination between the transgene and nearby DNA. Beying et al. (2020) reported chromosome arm exchange frequencies at ∼0.01% in Arabidopsis somatic cells through the use of CRISPR/Cas9. It is not clear whether the CRISPR/Cas9 reaction is reversible, but Cre definitely catalyzes reversible recombination. Since what translocates across can also translocate back, 100% recombination efficiency would translate to 50% transgene translocation. In this study, transgene translocation reached ∼20%, which ought to be a sufficient rate for recovering progeny with genetic linkages broken. This study did not proceed to the stage of recovering progeny, but since transmission of recombination events and segregation of the cre locus have been documented in many previous studies, there is no a prior reason to think these would not be possible.
In summary, this study demonstrates in principle that introducing Cre recombinase into a F1 hybrid serves not only to remove unnecessary DNA such as marker genes and plasmid backbone as previously shown in many studies but could also break genetic linkage on either or both sides of the transgenic locus. Naturally, Cre activity has to be sufficiently efficient in germline cells to insure transmission of recombination events, which means practical implementation of this method would still require the tedious task of testing many different germline-specific promoters for any given crop species.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
DO designed the experiments. QY and RL performed the experiments. QY and DO analyzed the data, discussed results, and wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Key R&D Project (Grant No. 2016YFD0101904) from the Ministry of Science and Technology of China and by the Key Research Program of Frontier Sciences CAS (Grant No. QYZDY-SSW-SMC010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank X. Liang for assistance in modifying the CRISPR vector provided by Y. Liu of South China Agricultural University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.828960/full#supplementary-material
References
Albert, H., Dale, E. C., Lee, E., and Ow, D. W. (1995). Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7, 649–659. doi: 10.1046/j.1365-313X.1995.7040649.x
Avanus, K., and Altınel, A. (2017). Comparison of allele-specific PCR, created restriction-site PCR, and PCR with primer-introduced restriction analysis methods used for screening complex vertebral malformation carriers in Holstein cattle. J. Vet. Sci. 18, 465–470. doi: 10.4142/jvs.2017.18.4.465
Beying, N., Schmidt, C., Pacher, M., Houben, A., and Puchta, H. (2020). CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 6, 638–645. doi: 10.1038/s41477-020-0663-x
Chen, Q. J., Zhou, H. M., Chen, J., and Wang, X. C. (2006). A gateway-based platform for multigene plant transformation. Plant Mol. Biol. 62, 927–936. doi: 10.1007/s11103-006-9065-3
Chen, W., and Ow, D. W. (2017). Precise, flexible and affordable gene stacking for crop improvement. Bioengineered 8, 451–456. doi: 10.1080/21655979.2016.1276679
Chen, W., Kaur, G., Hou, L., Li, R., and Ow, D. W. (2019). Replacement of stacked transgenes in planta. Plant Biotechnol. J. 17, 2029–2031. doi: 10.1111/pbi.13172
Collier, R., Thomson, J. G., and Thilmony, R. (2018). A versatile and robust Agrobacterium-based gene stacking system generates high-quality transgenic Arabidopsis plants. Plant J. 95, 573–583. doi: 10.1111/tpj.13992
Day, C. D., Lee, E., Kobayashi, J., Holappa, L. D., Albert, H., and Ow, D. W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14, 2869–2880. doi: 10.1101/gad.849600
Ding, M., Duan, X., Feng, X., Wang, P., and Wang, W. (2017). Application of CRS-PCR-RFLP to identify CYP1A1 gene polymorphism. J. Clin. Lab. Anal. 31:e22149. doi: 10.1002/jcla.22149
Dong, O. X., and Ronald, P. C. (2021). Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. U.S.A. 118:e2004834117. doi: 10.1073/pnas.2004834117
Fauser, F., Roth, N., Pacher, M., Ilg, G., Sánchez-Fernández, R., Biesgen, C., et al. (2012). In planta gene targeting. Proc. Natl. Acad. Sci. U.S.A. 109, 7535–7540. doi: 10.1073/pnas.1202191109
Goderis, I., De Bolle, M. F. C., Francois, I., Wouters, P. F. J., Broekaert, W. F., and Cammue, B. P. A. (2002). A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol. Biol. 50, 17–27. doi: 10.1023/a:1016052416053
Hou, L., Yau, Y.-Y., Wei, J., Han, Z., Dong, Z., and Ow, D. W. (2014). An open-source system for in planta gene stacking by Bxb1 and Cre recombinases. Mol. Plant 7, 1756–1765. doi: 10.1093/mp/ssu107
Koshinsky, H. A., Lee, E., and Ow, D. W. (2000). Cre-lox site-specific recombination between Arabidopsis and tobacco chromosomes. Plant J. 23, 715–722. doi: 10.1046/j.1365-313x.2000.00839.x
Li, M., and Li, H. (2003). A simple and highly efficient Agrobacterium-mediated rice transformation system. Chin. J. Exp. Biol. 36, 289–294.
Li, R., Han, Z., Hou, L., Kaur, G., Yin, Q., and Ow, D. W. (2016). “Method for biolistic site-specific integration in plants catalyzed by Bxb1 integrase,” in Chromosome and Genomic Engineering in Plants: Methods and Protocols, ed. M. Murata (New York, NY: Springer New York), 15–30.
Li, Z., Xing, A., Moon, B. P., McCardell, R. P., Mills, K., and Falco, S. C. (2009). Site-specific integration of transgenes in soybean via recombinase-mediated DNA cassette exchange. Plant Physiol. 151, 1087–1095. doi: 10.1104/pp.109.137612
Lu, Y., and Zheng, K. (1992). A simple method for isolation of rice DNA. Chin. J. Rice Sci. 6, 47–48.
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A Robust CRISPR/Cas9 System for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Nandy, S., Zhao, S., Pathak, B. P., Manoharan, M., and Srivastava, V. (2015). Gene stacking in plant cell using recombinases for gene integration and nucleases for marker gene deletion. BMC Biotechnol. 15:93. doi: 10.1186/s12896-015-0212-2
Puchta, H., Dujon, B., and Hohn, B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21, 5034–5040. doi: 10.1093/nar/21.22.5034
Qiao, W., Wang, T., Zhang, L., Tang, Q., Wang, D., and Sun, H. (2013). Association study of single nucleotide polymorphisms in XRCC1 gene with the risk of gastric cancer in Chinese population. Int. J. Biol. Sci. 9, 753–758. doi: 10.7150/ijbs.6783
Qin, M., Bayley, C., Stockton, T., and Ow, D. W. (1994). Cre recombinase-mediated site-specific recombination between plant chromosomes. Proc. Natl. Acad. Sci. U.S.A. 91, 1706–1710. doi: 10.1073/pnas.91.5.1706
Shih, P. M., Vuu, K., Mansoori, N., Ayad, L., Louie, K. B., Bowen, B. P., et al. (2016). A robust gene-stacking method utilizing yeast assembly for plant synthetic biology. Nat. Commun. 7:8. doi: 10.1038/ncomms13215
Smith, A. J., De Sousa, M. A., Kwabi-Addo, B., Heppell-Parton, A., Impey, H., and Rabbitts, P. (1995). A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination. Nat. Genet. 9, 376–385. doi: 10.1038/ng0495-376
Titen, S. W. A., Johnson, M. T. B., Capecchi, M., and Golic, K. G. (2020). Site-specific recombination with inverted target sites: a cautionary tale of dicentric and acentric chromosomes. Genetics 215, 923–930. doi: 10.1534/genetics.120.303394
Vergunst, A. C., Jansen, L. E., Fransz, P. F., de Jong, J. H., and Hooykaas, P. J. (2000). Cre/lox-mediated recombination in Arabidopsis: evidence for transmission of a translocation and a deletion event. Chromosoma 109, 287–297. doi: 10.1007/s004120000079
Wang, S., Duan, X., Wang, T., Feng, X., Wang, P., Yao, W., et al. (2016). Detection of the rs10250202 polymorphism in protection of telomeres 1 gene through introducing a new restriction enzyme site for PCR-RFLP assay. Springerplus 5:592. doi: 10.1186/s40064-016-2214-5
Wright, D. A., Townsend, J. A., Winfrey, R. J. Jr., Irwin, P. A., Rajagopal, J., Lonosky, P. M., et al. (2005). High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44, 693–705. doi: 10.1111/j.1365-313X.2005.02551.x
Zhang, Y., Zhang, F., Li, X., Baller, J. A., Qi, Y., Starker, C. G., et al. (2012). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161, 20–27. doi: 10.1104/pp.112.205179
Zhu, Q. L., Yu, S. Z., Zeng, D. C., Liu, H. M., Wang, H. C., Yang, Z. F., et al. (2017). Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant 10, 918–929. doi: 10.1016/j.molp.2017.05.008
Keywords: gene stacking, transgene replacement, transgene translocation, GMO, recombinase, Cre
Citation: Yin Q, Li R and Ow DW (2022) Site-Specific Sequence Exchange Between Homologous and Non-homologous Chromosomes. Front. Plant Sci. 13:828960. doi: 10.3389/fpls.2022.828960
Received: 04 December 2021; Accepted: 05 January 2022;
Published: 03 February 2022.
Edited by:
James A. Birchler, University of Missouri, United StatesReviewed by:
Yiping Qi, University of Maryland, College Park, United StatesHolger Puchta, Karlsruhe Institute of Technology (KIT), Germany
Copyright © 2022 Yin, Li and Ow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruyu Li, bGlydXl1QHNjYmcuYWMuY24=; David W. Ow, ZG93QGJlcmtlbGV5LmVkdQ==
 Qian Yin
Qian Yin Ruyu Li
Ruyu Li David W. Ow
David W. Ow