
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 22 February 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.825341
This article is part of the Research Topic Growth Patterns Underlying Plant Development View all 20 articles
Post-mitotic cell growth is a key process in plant growth and development. Cell expansion drives major growth during morphogenesis and is influenced by both endogenous factors and environmental stimuli. Though both isotropic and anisotropic cell growth can contribute to organ size and shape at different degrees, anisotropic cell growth is more likely to contribute to shape change. While much is known about the mechanisms that increase cellular turgor and cell-wall biomass during expansion, the genetic factors that regulate these processes are less studied. In the past quarter of a century, the role of the CINCINNATA-like TCP (CIN-TCP) transcription factors has been well documented in regulating diverse aspects of plant growth and development including flower asymmetry, plant architecture, leaf morphogenesis, and plant maturation. The molecular activity of the CIN-TCP proteins common to these biological processes has been identified as their ability to suppress cell proliferation. However, reports on their role regulating post-mitotic cell growth have been scanty, partly because of functional redundancy among them. In addition, it is difficult to tease out the effect of gene activity on cell division and expansion since these two processes are linked by compensation, a phenomenon where perturbation in proliferation is compensated by an opposite effect on cell growth to keep the final organ size relatively unaltered. Despite these technical limitations, recent genetic and growth kinematic studies have shown a distinct role of CIN-TCPs in promoting cellular growth in cotyledons and hypocotyls, the embryonic organs that grow solely by cell expansion. In this review, we highlight these recent advances in our understanding of how CIN-TCPs promote cell growth.
Plant growth and development are remarkably plastic, which helps them overcome the constraints posed by their sessile existence and enables them to thrive in diverse environmental conditions. Developmental plasticity is achieved partly by preserving the self-renewing stem cells within the meristem niches and by post-embryonic initiation of lateral organs from the flanks of the meristem (Powell and Lenhard, 2012). Organ growth is a resultant outcome of two key cellular processes, proliferation, and post-mitotic expansion, the former preceding the latter. Proliferation produces an adequate cell reserve at an early stage of organ growth, and expansion increases cell volume under the force of turgor pressure (Donnelly et al., 1999; Cosgrove, 2005; Gonzalez et al., 2012). For example, mitotic cycle in Arabidopsis and tobacco leaves ceases when their blades reach ∼10% of their final area, triggering endoreduplication-mediated cell expansion that drives the remainder of the blade growth (Poethig and Sussex, 1985; Donnelly et al., 1999).
Though the physical basis of cell expansion through turgor pressure and cell-wall extensibility has been studied in detail (Ray et al., 1972; Braidwood et al., 2014), its genetic regulation remains less clear. This is primarily because of an overlapping occurrence of cell division and expansion in the lateral organs of post-embryonic origin. For example, within the growing primordium of most angiosperm leaves, cell division and expansion coexist in the proximal and distal domains, respectively (Nath et al., 2003; Das Gupta and Nath, 2015). Cells transiting from an actively dividing state to differentiation characterize a dynamic transition zone, flanked by mitotic cells to its proximal end and differentiated cells to its distal end. Perturbations in rate, duration, or distribution of cell proliferation and expansion often affect the final size and shape of organs (Donnelly et al., 1999; Efroni et al., 2008; Andriankaja et al., 2012). Because proliferation and differentiation are linked by a compensatory mechanism (Gonzalez et al., 2012) in post-embryonic organs, it is difficult to uncouple the contribution of cell growth from that of cell division to the final organ morphology. However, the organs of embryonic origin such as hypocotyl and cotyledon serve as an ideal model for cell growth studies since cell division is absent or insignificant during their growth after germination (Tsukaya et al., 1994; Stoynova-Bakalova et al., 2004; Boron and Vissenberg, 2014). Besides, these embryonic organs possess simpler and uniform epidermal cell morphology, making their analysis more tractable. Utilization of these simplifying advantages of hypocotyl and cotyledon has shed light on the molecular players involved in unidirectional and planar cell expansion, respectively. In this review, we highlight recent advances in our understanding of the genetic regulation of post-mitotic cell growth, with an emphasis on CIN-TCP transcription factors and their new-found role in cell expansion.
Because the rigid wall precludes migration of plant cells, organ morphology is greatly influenced by the direction and duration of cell enlargement (Braidwood et al., 2014). Plant cells predominantly show anisotropic expansion, where the rate and direction of growth vary across the cell surface, allowing cells to acquire a variety of forms to limit the magnitude of mechanical stress and simultaneously enhance tissue strength (Johnson and Lenhard, 2011; Braidwood et al., 2014; Sapala et al., 2018). Anisotropic cell growth can be either diffused type where expansion is dispersed over the cell surface, or tip-growth type where expansion occurs only at localized tips (Martin et al., 2001). For example, cells in hypocotyl, stem, and root show growth along the longitudinal direction at their side walls over the entire surface. By contrast, root hairs and pollen tubes exhibit restricted growth only at a single site at the cell tip (Braidwood et al., 2014). In yet another growth pattern, the pavement cells in leaves and cotyledons show multiple growth polarities on segments of their surface, ultimately producing puzzle piece-shaped cells with lobes and indentations. We refer to this type of cell growth as planar growth since the expansion is primarily in the X–Y plane, with little expansion in the orthogonal direction. Distinct cytological properties such as differential deposition of cell-wall material, orientation of cellulose microfibrils, and polarized accumulation of cytoplasmic content determine the mode of cell growth and organ morphology (Braidwood et al., 2014). However, certain cells such as parenchyma of potato tuber or apple fruit display a diffused isotropic mode of growth, forming isodiametric cells compacted together where the cellulose microfilaments are randomly oriented (Cosgrove, 2014).
In most tissue types, rapid increase in cell volume is directly associated with endoreplication cycle where DNA is replicated without cytokinesis or chromosome segregation, leading to enhanced nuclear ploidy level (Melaragno et al., 1993; Edgar and Orr-Weaver, 2001; Breuer et al., 2010). Polyploidization is known to promote ribosome biogenesis, resulting in an increase in protein synthesis and in total cytoplasmic content, which ultimately leads to cell growth (Larkins et al., 2001; Sugimoto-Shirasu and Roberts, 2003). Thus, modulating the onset of endocycle and the duration of its progression substantially influence organ size with a strong correlation with cellular ploidy level. FASCIATA1 (FAS1), a chromatin assembly subunit protein, and E2F TARGET GENE 1 (ETG1), a replisome factor, interfere in the chromatin assembly that triggers endoreplication in plants. Consequently, fas1 and etg1 mutants perform additional rounds of endocycle, leading to larger cells and organs (Ramirez-Parra and Gutierrez, 2007; Takahashi et al., 2010). Similarly, cell cycle inhibitory KIP-RELATED PROTEIN (KRP) genes controlling endoreplication and chromatin structure regulate expression of genes required for cell expansion (Jegu et al., 2013). The quintuple krp mutant exhibits leaves with smaller epidermal cells and KRP-overexpression lines form larger cells, suggesting a direct correlation of KRP abundance and endoreplication-mediated cell growth (Verkest et al., 2005; Cheng et al., 2013). Additional rounds of endoreplication in dark-grown hypocotyl cells serve as a major determinant of de-etiolated seedling growth in contrast to light-grown seedlings (Gendreau et al., 1997). In floral organs, suppressing the transition to endoreduplication in the frill1 mutant, which has a reduced sterol methyltransferase activity, results in expanded cells with enlarged nuclei during the late stage of petal development (Hase et al., 2000). It is also suggested that the combination of phytohormones, nutrients, and environmental signals trigger endoreplication and cell growth (Kondorosi et al., 2000; Sugimoto-Shirasu and Roberts, 2003). For instance, gibberellic acid (GA) and ethylene coordinate with DNA synthesis and endoreplication-mediated hypocotyl cell elongation (Gendreau et al., 1999). Many reports in multiple plant species strongly indicate the presence of endocycle-independent mechanism of cell growth affecting organ size (Sugimoto-Shirasu and Roberts, 2003).
The rigid wall of plant cells – comprised of complex meshwork of cellulose, hemicellulose, pectin, and glycoproteins – accommodates two seemingly contradictory biological functions - providing a strong structural support to the cell and an adjustable elasticity necessary for its growth (Bashline et al., 2014). The turgor pressure triggered by osmotic activity leads to cell-wall loosening and produces enlarged cells with balanced water potential without losing their integrity (Cosgrove, 2005; Seifert and Blaukopf, 2010; Bashline et al., 2014). The physical properties of cell-wall are altered to promote expansion by the activity of several cell-wall-modifying enzymes such as glycosyltransferases, methylesterases, hydroxylases, and hydrolases. These enzymes target major components of the cell wall (Braidwood et al., 2014). For example, the EXPANSIN (EXP) family proteins promote cell-wall loosening by breaking the non-covalent bonds between cellulose microfibrils in an acidic environment and correspondingly produce bigger leaves with larger cells when overexpressed (Cho and Cosgrove, 2000; Marowa et al., 2016). Similarly, pectin methyl esterases hydrolyze ester bond and enhance negative electrostatic charge within the cell wall to provide flexibility and mobility to homo-galacturonan in the cell wall. Mutant analysis of multiple cellulose synthase (CESA) genes has demonstrated the redundant function of these cell-wall synthesis promoting genes in cell expansion in multiple organs (Burn et al., 2002).
Phytohormones play a critical role in promoting cell growth in association with either cell-wall-remodeling enzymes or by enhancing orientated deposition of cellular microfibrils (Wolters and Jurgens, 2009). In addition, auxin, brassinosteroid (BR), and GA promote cell-wall biosynthesis and expansion by activating several regulatory transcription factors. Auxin induces rapid cell expansion in stem, hypocotyl, leaf, and coleoptile by activating a proton (H+) pump ATPase in the plasma membrane (PM), resulting in extracellular acidification, an environment favoring the cell-wall loosening enzymes (Rayle, 1973; Rayle and Cleland, 1977, 1992; Perrot-Rechenmann, 2010). The early auxin-responsive SMALL AUXIN UPREGULATED RNA (SAUR) gene family members participate in modulating PM H+-ATPase activity through direct interaction and inhibition of PP2C-D protein phosphatases (Spartz et al., 2014). Recent reports have shed light on tissue-specific expression pattern of SAUR genes and their involvement in distinct morphological processes under the regulation of developmental and environmental factors (Ren and Gray, 2015; Stortenbeker and Bemer, 2019). BR, GA, jasmonic acid (JA), and abscisic acid (ABA) also regulate SAUR-mediated cell growth (Ren and Gray, 2015). Auxin, BR, and GA integrate environmental signals through the DELLA-ARF6-BZR1-PIF4 complex to form a central regulatory network and maintain cell elongation by regulating an overlapping set of SAUR genes (Oh et al., 2014; Van Mourik et al., 2017). Mutants deficient in AUXIN RESPONSE FACTORS (ARFs) form shorter hypocotyl compared to the wild type, further supporting the role of auxin response in cell expansion (Reed et al., 2018). Another homeodomain transcription factor HAT2 is also known to promote cell elongation in hypocotyl in response to auxin signaling (Sawa et al., 2002). A recent report suggests that increased endogenous auxin, triggered by a low-sugar state in the cotyledons following seed germination, promotes compensation-mediated cell enlargement (Tabeta et al., 2021). Another recent study demonstrates that constitutively activated salicylic acid (SA) signaling suppresses compensation-induced cell expansion in leaves, thus assigning a role for SA in organ size regulation (Fujikura et al., 2020).
The regulators of the cell-wall extensibility factors described above are studied in less detail. The ARGOS-LIKE (ARL) gene is preferentially expressed in cotyledon, mature root, and leaves and promotes organ size as a result of increased cell expansion (Hu et al., 2006). Similarly, overexpression of ZINC-FINGER HOMEODOMAIN 5 (ZHD5) produces bigger leaves with enlarged epidermal cells by acting on yet unidentified target genes required for cell expansion (Hong et al., 2011). Few more examples of cell expansion regulators include TARGET OF RAPAMYCIN (TOR), CELL CYCLE SWITCH PROTEIN 52 (CCS27A), CYTOCHROME P450, FAMILY 78, SUBFAMILY A (CYP78A6), and ERBB-3 BINDING PROTEIN 1 (EBP1), overexpression of which results in larger organs due to increased cell expansion (Gonzalez et al., 2012; Vanhaeren et al., 2015). The list of the transcription factors that directly or indirectly regulate cell-wall expansion is still growing, with the recent addition of the TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR (TCP) family proteins.
Teosinte branched1, cycloidea, proliferating cell factor proteins participate in sequence-specific DNA-binding, transcriptional activation, and protein–protein interaction to regulate diverse developmental processes including flower symmetry, leaf and petal morphogenesis, trichome development, axillary branching, and pathogen defense (Martin-Trillo and Cubas, 2010; Sugio et al., 2014; Danisman, 2016; Sarvepalli and Nath, 2018; Challa et al., 2019). They are distinguished by their non-canonical basic helix-loop-helix (bHLH) domain known as the TCP domain and are present from chlorophyte algae to higher land plants (Navaud et al., 2007; Dhaka et al., 2017). Based on sequence diversity in amino acid residues within the TCP-domain, the 24 TCP proteins encoded by the Arabidopsis genome are divided into the class I subfamily with thirteen members and the class II subfamily with eleven members (Kosugi and Ohashi, 2002; Li, 2015). Eight class II TCPs are collectively called CINCINNATA-like TCPs (CIN-TCPs) that form a subclade and are expressed in the transition zone in leaf primordia where they redundantly suppress proliferation and promote differentiation in dividing cells (Nath et al., 2003; Das Gupta et al., 2014; Sarvepalli and Nath, 2018; Challa et al., 2019). Transcripts of five CIN-TCP genes – TCP2, 3, 4, 10, and 24 – are degraded by the microRNA miR319 (Palatnik et al., 2003), adding an additional level of complexity in CIN-TCP-mediated regulation of leaf morphogenesis. In addition to leaf maturation, CIN-TCPs also regulate leaflet initiation, biotic and abiotic stress tolerance, photomorphogenic seedling growth, phytohormone signaling, flowering time control, cell-wall thickening, etc (Koyama et al., 2007, 2010a, 2017; Navaud et al., 2007; Efroni et al., 2008; Schommer et al., 2008; Sarvepalli and Nath, 2011a; Aguilar-Martinez and Sinha, 2013; Das Gupta et al., 2014; Wang et al., 2014, 2015; Challa et al., 2016, 2019; Kubota et al., 2017; Lei et al., 2017; Zhou et al., 2018, 2019; Fan et al., 2020; Fang et al., 2021). Most developmental processes regulated by CIN-TCPs are linked to their involvement in suppressing proliferation and promoting differentiation of cells. Effects of CIN-TCPs on the exit from division cycle within leaf primordia have been reviewed elsewhere (Sarvepalli and Nath, 2018).
Accumulating evidence in recent years has revealed a direct participation of CIN-TCPs in post-mitotic cell expansion, which could not be elucidated earlier due to extensive functional redundancy among the members, making the loss-of-function analysis less effective (Efroni et al., 2010; Sarvepalli and Nath, 2011a; Koyama et al., 2017). On the other hand, overexpression of miR319-resistant CIN-TCPs in their endogenous expression domain causes embryonic and seedling lethality, making gain-of-function analysis challenging (Palatnik et al., 2003; Koyama et al., 2007). Moreover, a change in cell proliferation in leaf primordia is often accompanied by an opposite change in cell size by a phenomenon called compensation, making the analysis of gene function in cellular regulation difficult to interpret (Horiguchi and Tsukaya, 2011; Hisanaga et al., 2015). Considering the contribution of CIN-TCPs in repressing cell division potential (Challa et al., 2019), it is rather a daunting task to uncouple TCP-dependent cell expansion from compensatory cellular growth in post-embryonic organs such as leaves.
Several class I TCP proteins have also been implicated in cell division and cell growth. For example, TCP20 is enriched in the chromatin regions that encode cyclin CYCB1;1 and several ribosomal proteins, suggesting its role in coupling division and growth of cells (Li et al., 2005). However, we focus on the class II TCP proteins in this review and set aside the discussion on the class I proteins for future.
Hypocotyl initials are formed by patterned cell division during embryo development. Upon seed germination and seedling establishment, hypocotyl grows in length solely by directional cell expansion (Nemhauser and Chory, 2002; Boron and Vissenberg, 2014). Several studies on hypocotyl growth at the cellular level suggest an extreme elongation of the under-differentiated epidermal cells (up to 100-fold elongation compared to embryonic cell stage) as a result of rounds of endoduplication under darkness as opposed to the light-grown seedlings (Gendreau et al., 1997). The epidermal cells in hypocotyl follow a steep acropetal mode of growth where elongation starts at the basal cells and gradually moves upward (Miséra et al., 1994; Wei et al., 1994; Gendreau et al., 1997).
Both miR319-targeted and non-targeted CIN-TCP proteins promote hypocotyl elongation under the influence of environmental signals (Figure 1). Analysis of loss and gain-of-function mutants has demonstrated that miR319-regulated CIN-TCPs promote hypocotyl elongation by directly activating YUCCA5 (YUC5)-mediated auxin biosynthesis, which in turn activates the central ARF6/8-BZR1 circuit and downstream cell expansion genes (Challa et al., 2016). TCP1, a non-CIN-TCP class-II TCP protein, also enhances cell elongation in hypocotyl by directly activating the BR biosynthetic gene DWARF4 (Guo et al., 2010), suggesting that the class II TCPs promote hypocotyl cell growth by acting on multiple phytohormone pathways. Interestingly, TCP3, another miR319-regulated CIN-TCP member, is reported to activate INDOLE-3-ACETIC ACID3/SHORT HYPOCOTYL2 (IAA3/SHY2), a negative regulator of auxin signaling, though overexpression of a miR319-resistant form of TCP3 causes elongation of hypocotyl cells (Koyama et al., 2010a). These observations imply distinct functions of redundant CIN-TCP partners in promoting cell growth, perhaps to maintain homeostasis in hormone response.
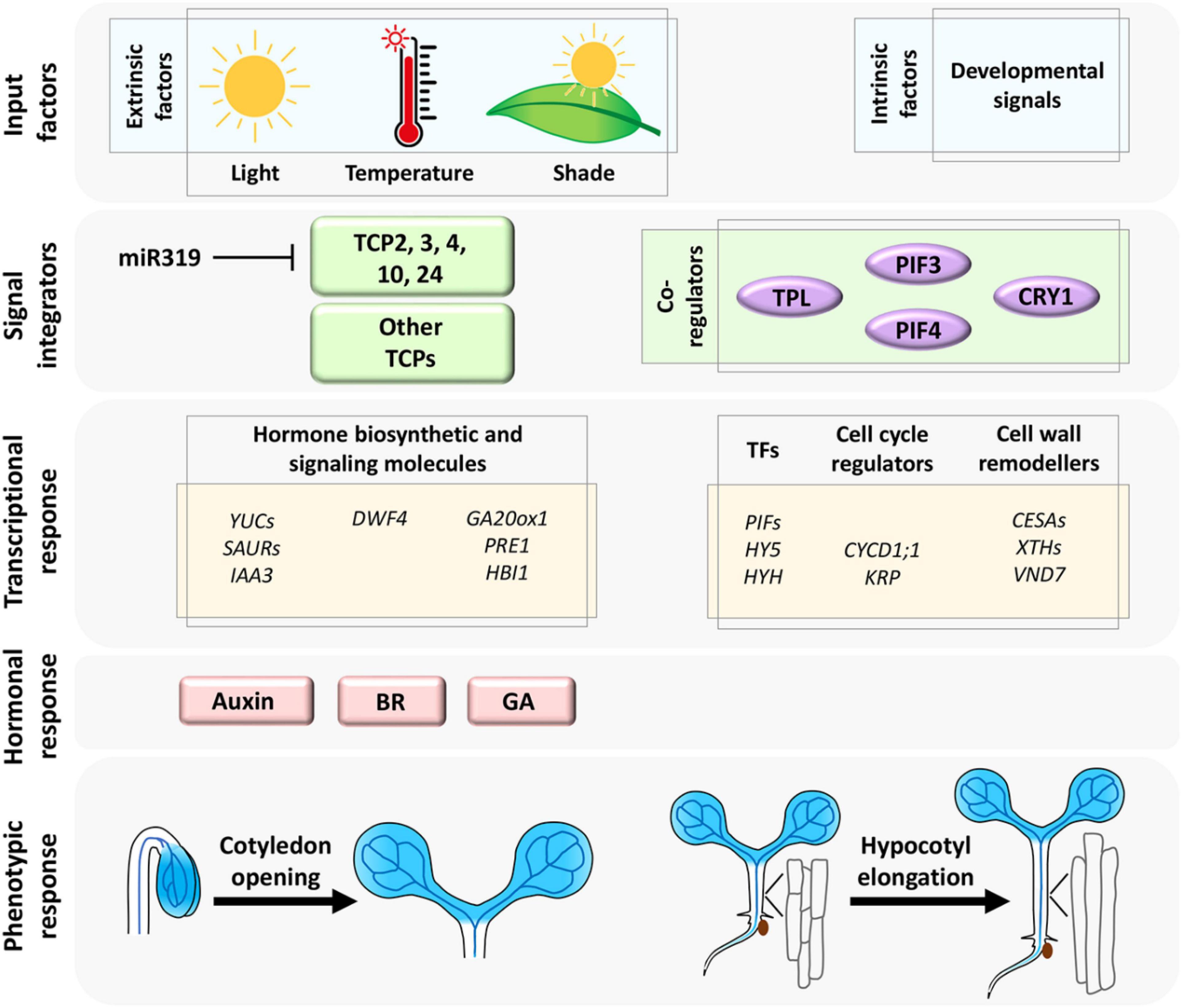
Figure 1. The extrinsic (environmental) and intrinsic (developmental) signals are consolidated by miR319-regulated CIN-TCP transcription factors and their co-regulators (Dong et al., 2019; Ferrero et al., 2019; Zhang et al., 2019; Zhou et al., 2019) to form a regulatory network that activates downstream target molecules and phytohormones to promote cell growth linked to hypocotyl elongation and cotyledon opening. Shaded blue color represents CIN-TCP gene expression pattern.
Though CIN-TCPs promote hypocotyl elongation, their promoters are primarily active in the cotyledons (Koyama et al., 2007; Challa et al., 2016; Dong et al., 2019). It is possible that these proteins exert a non-cell-autonomous effect on hypocotyl elongation through systemic enhancement of cotyledon-derived auxin response. This is supported by the fact that polar auxin transport is required to promote hypocotyl elongation in light-grown seedlings, though not in dark-grown seedlings (Jensen et al., 1998). Much of the environmental cues are sensed by the cotyledons where most auxin biosynthesis, response, and transport genes are expressed, and this cotyledon-derived mobile auxin travels to hypocotyl where it promotes cell elongation, as identified by chemical intervention and surgical experiments (Tao et al., 2008; Keuskamp et al., 2010; Procko et al., 2014). Effects of mobile auxin is seen primarily in the epidermal tissue where auxin interacts with the BR pathway to induce hypocotyl growth (Procko et al., 2016). In addition to CIN-TCPs, several class-I TCP members also promote hypocotyl elongation (listed in Table 1) in a context and condition-dependent manner.
Planar expansion of pavement cells with interdigitation has been widely studied using cotyledon as a model organ due to their simplicity and lack of concomitant cell division. Hyperactivated TCP4 promotes cotyledon cell expansion (Sarvepalli and Nath, 2011a). The organ-specific activity of the early auxin response genes SAUR16 and SAUR50 contribute to differential cellular response in cotyledon opening, which is driven by cell expansion and hypocotyl elongation upon enhancement in TCP4 level during seedling de-etiolation (Dong et al., 2019). SAUR16 and SAUR50 act as potential direct targets of TCP4 and this transcriptional activity of TCP4 is inhibited by PHYTOCHROME INTERACTING FACTOR3 (PIF3), a canonical bHLH transcription factor abundantly expressed in etiolated seedlings (Dong et al., 2019). PIF3 is recruited to the SAUR promoter and interferes with DNA-binding by TCP4 to retain closed unexpanded cotyledons under darkness. TCP4 does not seem to physically interact with PIF3 in this process, and thus the exact mechanism of this competitive binding to SAUR promoter is still an open question. By acting on the ARF-BZR1 signaling node and SAUR genes, CIN-TCPs promote cotyledon cell expansion perhaps by cell-wall remodeling, though evidence in support of this proposition has remained elusive. Distinct phytohormones such as GA, ethylene, and ABA also regulate cotyledon cell expansion independently (Collett et al., 2000; Stoynova-Bakalova et al., 2004). Crosstalk between CIN-TCPs and these hormone response pathways are not well studied. Based on the involvement of CIN-TCPs in transcriptional regulation of multiple phytohormones (Guo et al., 2010; Sarvepalli and Nath, 2011b; Challa et al., 2016; Gonzalez-Grandio et al., 2017), one can speculate their participation as signal integrators, which can be further explored (Figure 1).
Like cotyledons, post-embryonic lateral organs such as leaves also display planar growth driven by cell expansion in both transverse and longitudinal axes. Phytohormones such as ethylene and cytokinin promote cell growth along the transverse axes, whereas auxin, BR, and GA promote expansion along longitudinal axes (Mizukami, 2001). These hormones, individually or in combination, affect the organization of cellular and cortical cytoskeleton elements leading to differential expansion rate along growth axes (Blume et al., 2012). In addition, hormones also impact tubulin and actin transcript abundance (Kandasamy et al., 2001; Blume et al., 2012).
As mentioned earlier, analysis of gene function on cell growth in leaf primordium is compounded by compensation wherein an alteration in cell number is compensated by an opposite effect on cell area, resulting in a tendency of the final leaf size to remain unaffected. Genetic analysis coupled with growth kinematic studies have revealed the effect of CIN-TCP proteins on cell growth (Challa et al., 2019). Loss-of-function mutants of CIN-TCPs form bigger leaves with excess cells, and their overexpression results in smaller leaves with fewer cells (Figure 2A; Efroni et al., 2008; Schommer et al., 2014; Challa et al., 2019). Increased cell number in cin-tcp loss-of-function mutants is accompanied by smaller cells, suggesting a compensatory effect. Though the cell-size defect is corrected to the wild-type level when a dominant form of TCP4 is activated, the cell size does not increase beyond the wild-type level, indicating that CIN-TCPs are required but not sufficient for pavement cell growth. These cell kinematics results have been interpreted as CIN-TCPs promoting the division-to-differentiation switch in the proliferating leaf pavement cells (Challa et al., 2019). According to this interpretation, reduced CIN-TCP function causes fewer cells to depart the division cycle and enter the expansion phase, resulting in excess cells at maturity with a higher proportion of smaller cells. On the contrary, gain of CIN-TCP function causes more cells to depart cell cycle and enter differentiation, leaving fewer cells to divide. This interpretation appears to explain both the loss and gain-of-function phenotypes of CIN-TCP mutants without ascribing a cell growth function to them. It also partly explains the cellular basis of compensation in mature leaves: prolonging the proliferation phase accumulates more cells of smaller size, measuring the average cell area at maturity smaller than that in wild type. CIN-TCP proteins possibly promote the division-to-differentiation transition by increasing the nuclear ploidy level via endoreplication (Challa et al., 2019; Zhang et al., 2019). Transition to cell differentiation by CIN-TCPs is redundant with the NGATHA and class II KNOTTED1-LIKE transcription factors, and higher order mutants of these genes produce persistent leaf growth with delayed differentiation status (Alvarez et al., 2016; Challa et al., 2021). The inability of CIN-TCPs to increase cell size over and above the wildtype level can also be interpreted as a threshold-dependent activity below which they are unable to promote cell expansion. Alternatively, it is also possible that CIN-TCPs depend on other factors for driving cell expansion in leaf. Cell-size analysis using transgenic lines expressing a higher level of CIN-TCP would test these possibilities.
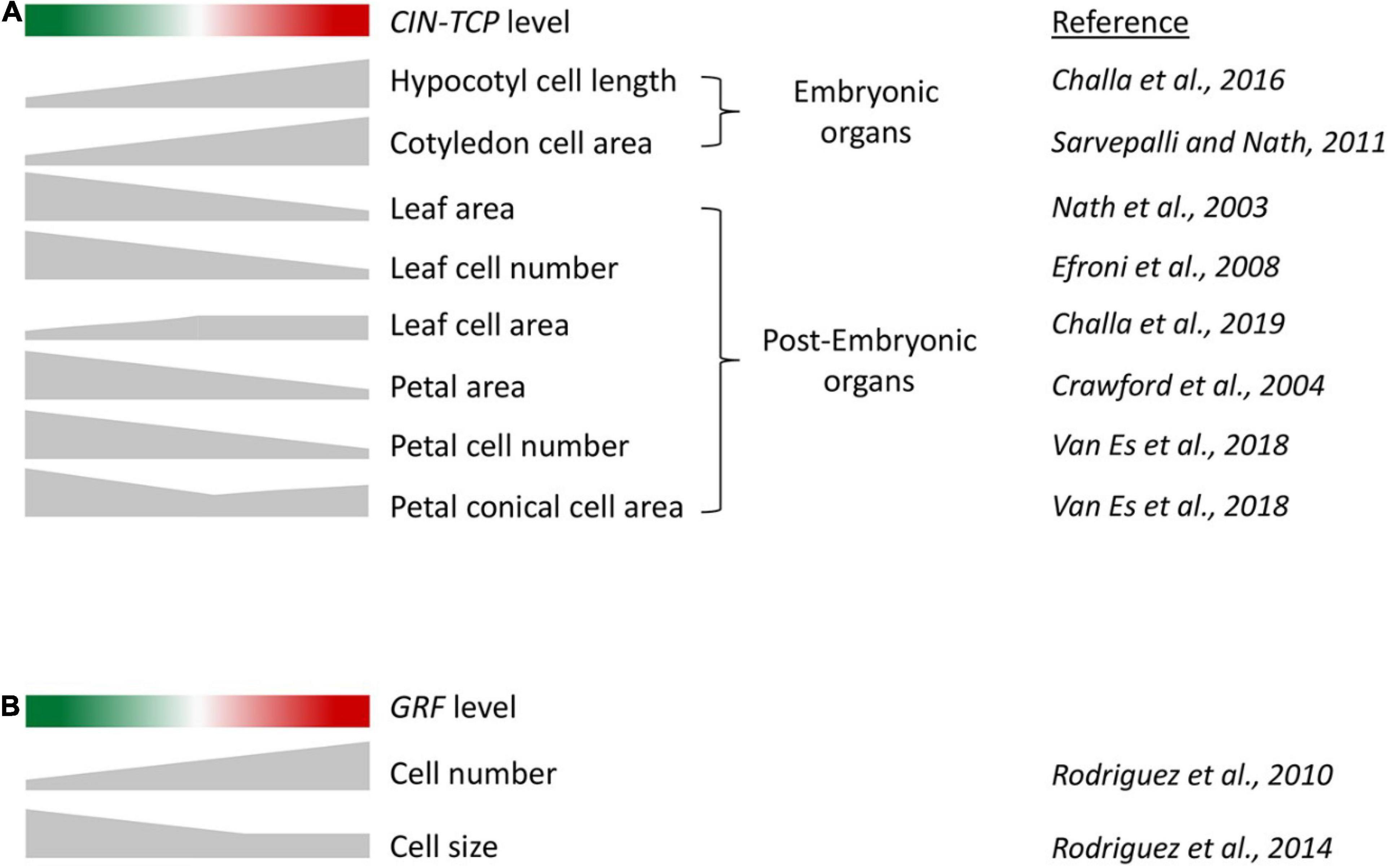
Figure 2. (A) A schematic to describe the correlation between CIN-TCP activity and cell growth in embryonic and post-embryonic organs. Red and green shades indicate progressively higher and lower CIN-TCP levels, respectively, compared to wild-type level in the middle. Height of the gray-shaded objects corresponds to the extent of phenotype indicated on the right. Organs with embryonic origin such as cotyledons and hypocotyl where cell division and expansion are temporally exclusive, exhibit cell expansion directly proportional to CIN-TCP level. In post-embryonic lateral organs such as leaves and petals where cell division and expansion are contemporary, the correlation of cell growth and CIN-TCP abundance is more complex due to combined action of proliferation and expansion. (B) A schematic describing the correlation of GRF activity and cell number and size in leaf.
The miR396-GRF module regulates leaf size and influences cell proliferation in a manner opposite to that of miR319-CIN-TCP. The GRF transcription factors, which are repressed by miR396, promote cell proliferation in the leaf primordia (Rodriguez et al., 2010; Schommer et al., 2014). Leaves overexpressing GRF accumulate excess pavement cells, whereas GRF down-regulation leads to fewer cells (Rodriguez et al., 2010; Challa et al., 2019). These results are in agreement with a molecular link between these two modules where the miR319-CIN-TCP module acts as an upstream regulator of miR396-GRF, with CIN-TCPs directly activating MIR396 transcription (Schommer et al., 2014). However, contrary to the effects of CIN-TCPs on cell size, loss of GRFs leads to bigger pavement cells whereas their gain-of-function fails to alter cell size (Kim and Lee, 2006; Rodriguez et al., 2010, 2014; Horiguchi and Tsukaya, 2011). These results suggest a diverging effect of CIN-TCPs and GRFs on cell size regulation (Figure 2B).
At the molecular level, only a few cell-expansion genes have been identified as targets of TCP proteins, which control cell growth in lateral organs (listed in Table 2). For instance, TCP4 induces pavement cell differentiation by elevating auxin response as well as by directly activating the HOMEODOMAIN ARABIDOPSIS THALIANA 1 (HAT2)-encoding gene in an auxin-independent manner (Challa et al., 2019). A recent report suggests that the miR319-non-targeted CIN-TCPs such as TCP5, 13, and 17 reduces cell expansion; TCP13 directly associates with the upstream regulatory region of Arabidopsis thaliana homeobox 12 (ATHB12), a known cell expansion gene, and reduces its transcription (Hur et al., 2019). Mutation in the class-I TCP member TCP20 results in enlarged pavement cell during early leaf development, which is antagonistic to the effect shown by their class-II counterparts (Danisman et al., 2012).
Loss of miR319-targeted CIN-TCPs in the MIR319A-overexpressing leaves, in addition to causing smaller cell size, also reduces the typical jigsaw-puzzle morphology, making the pavement cells relatively rounder (Efroni et al., 2008). The fact that CIN-TCPs transcriptionally regulate several phytohormones which are necessary for intercalated expansion of pavement cells might provide a mechanistic insight to this observation. Small non-lobed pavement cells also appear in the mutant of the chromatin remodeler SWI/SNF ATPase protein BRAHMA, which is a well-established protein interactor of TCP4, indicating an epigenetic regulation of cell expansion (Efroni et al., 2013; Vercruyssen et al., 2014; Thouly et al., 2020).
The petals of the cin mutant in snapdragon are made of fewer epidermal cells that are larger in size compared to wild type, suggesting that CIN-TCPs exert an opposite effect on cell expansion in petals to their effect on leaf (Crawford et al., 2004). TCP5, along with TCP13 and TCP17, also suppresses the area of the conical cells in Arabidopsis petals by inhibiting ethylene biosynthesis through direct binding to the 1-aminocyclopropane-1-carboxylate (ACC) synthase 2 (ACS2) locus (Van Es et al., 2018). The tcp5; tcp13; tcp17 triple mutant forms bigger and broader petals with excess and larger conical cells, suggesting an inhibitory effect of these genes on both cell division and expansion, as opposed to the compensatory effect observed in leaves. Conversely, TCP5-overexpressing lines form smaller petals with fewer cells (Van Es et al., 2018). However, the conical cells in these TCP5 gain-of-function petals are slightly larger than wild type, perhaps due to compensation.
CINCINNATA-like TCP members such as TCP4 are recruited to the genomic regions of several genes involved in cell-wall biogenesis and lignin deposition and promote secondary cell-wall thickening in diverse species. For instance, activation of miR319-resistant TCP4 triggers transcription of a xylem differentiation-promoting gene VASCULAR-RELATED NAC-DOMAIN7 (VND7), and several cellulose (CESA4, 7, and 8) and lignin (LAC4 and LAC17) biosynthetic genes (Sun et al., 2017). Overexpression of GhTCP4 promotes cell-wall thickening by binding to the cis elements of secondary cell-wall biosynthetic genes such as GhCESA7 and GhFSN1 in cotton (Cao et al., 2020). On the contrary, TCP24 negatively regulates genes involved in secondary cell-wall biogenesis and thickening, indicating a possible involvement in cell-wall remodeling and extensibility (Wang et al., 2015). In summary, these results point toward a new field of study which can connect CIN-TCPs directly to the cell-wall biochemistry and provide a mechanistic basis of TCP-mediated cell expansion.
Recent findings implicate a role for the CIN-TCP genes in modulating hypocotyl elongation in response to environmental cues such as light and temperature. Multiple alleles of the jaw-D dominant mutation, where overexpression of miR319 downregulates five cognate CIN-TCP targets (TCP2, 3, 4, 10, and 24), show light-hypersensitive effect, where TCP4 and TCP24 suppress photomorphogenesis and promote hypocotyl elongation (Tsai et al., 2014). By contrast to TCP4 and TCP24, TCP2 inhibits hypocotyl elongation specifically under blue light condition by interacting with the blue-light receptor CRYPTOCHROME 1 and activating the transcription of ELONGATED HYPOCOTYL5 (HY5) and HY5-HOMOLOG, genes that promote photomorphogenesis (He et al., 2016). The non-miR319-targeted CIN-TCP members TCP5, TCP13, and TCP17 also promote hypocotyl elongation in response to canopy-shade by directly activating the auxin biosynthetic YUC genes and PIF4/5 (Zhou et al., 2018). TCP5/13/17 promote seedling growth also in response to high temperature by targeting PIF4 activity at both transcriptional and post-transcriptional level (Han et al., 2019; Zhou et al., 2019). Interestingly, TCP5 is expressed in both cotyledon and hypocotyl where it directly enhances local increase in auxin biosynthesis and BR response upon elevated temperature (Bellstaedt et al., 2019; Han et al., 2019). Heat treatment shifts TCP5 expression from leaf blade to petiole, supporting its role in differential cell expansion of petiole and reduced leaf size (Han et al., 2019). Thus, there appears to be a positive correlation between the level of CIN-TCP expression and the extent of cell elongation in embryonic organs such as hypocotyl (Figure 2A). Environmental challenges such as low light intensity and rise in ambient temperature induce unequal growth rates between adaxial and abaxial regions of the petiole cells, leading to hyponastic growth in association with major phytohormones (Millenaar et al., 2009). Chimeric repression of the CIN-TCP member TCP3 shows irregularly differentiated petiole formation, whereas overexpression of TCP5/13/17 displays longer petiole in response to high temperature, consistent with their petiole-specific expression (Koyama et al., 2007; Han et al., 2019). Further, enhanced and reduced activity of TCP1 leads to elongated and shortened petiole, respectively, which is mediated by auxin and BR response (Guo et al., 2010; Koyama et al., 2010b).
The apparent divergent effects of CIN-TCP transcription factors on the morphogenesis of various primordia including flower, axillary meristem, leaf, and petal converge at their inhibitory effect on cell proliferation. This effect is at least in part mediated by directly activating the transcription of KRP cell-cycle inhibitor genes. By advancing the cessation of cell proliferation and consequently triggering differentiation in various primordia, these proteins regulate the shape and size that are manifested in the mature organs. This rather simplified effect of these proteins has now started to expand with the identification of several target genes, interacting partners, and upstream regulatory proteins, presenting a complex regulatory network involving these transcription factors. One such effect is on cell growth of embryonic organs, which grow solely by cell expansion post-germination. Elongation of hypocotyl cells by CIN-TCPs can be explained by YUCCA-mediated auxin biosynthesis at the shoot apex that is transported to the growing hypocotyl (Challa et al., 2016). Though activation of dominant CIN-TCPs within the cell proliferation phase of leaf primordia reduces cell number, their activation beyond the proliferation phase has no effect on the cell or leaf size (Challa et al., 2019). This is perhaps not surprising, considering that the CIN-TCP genes are primarily expressed in developing leaves. However, the failure of the CIN-TCPs to increase cell expansion in leaves is rather intriguing.
Studies emerging over the past decade have elucidated the function of the CIN-TCP proteins as central integrators of environmental, developmental, and hormonal signals to perform several key developmental processes. By participating at multiple stages of organ growth including the duration and extent of cell proliferation, cell-fate transition from division to differentiation, onset of post-mitotic cell growth and scope of cell expansion, CIN-TCPs program diverse organs to achieve final size and shape over a wide range of plant species. To overcome the intertwined relationship between cell division and differentiation, and to study the exact contribution of CIN-TCP genes in cell expansion per se, the embryonic organs such as hypocotyl and cotyledons are useful model organs. However, the sequence of molecular events leading to cell enlargement and the exact kinetics of cell elongation in association with TCP genes awaits detailed elucidation. Finally, understanding the conservation of this CIN-TCP-dependent modulation in post-mitotic cellular growth across species is a challenge for future studies, considering organ size is a major contributor to crop productivity.
MR wrote the first draft of the manuscript and progressively modified it to the final version. KC and KS contributed to writing and corrections. UN corrected the manuscript and guided the other authors to produce the final version. All authors contributed to the article and approved the submitted version.
This work was supported by the Ministry of Human Resource Development, Government of India (fellowships to MR and KC), Department of Science and Technology for Improvement of S&T Infrastructure (DST-FIST), and Department of Biotechnology (DBT)-IISc Partnership Program Phase-II at IISc (Sanction No. BT/PR27952/INF/22/212/2018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar-Martinez, J. A., and Sinha, N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4:406. doi: 10.3389/fpls.2013.00406
Alvarez, J. P., Furumizu, C., Efroni, I., Eshed, Y., and Bowman, J. L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5:e15023. doi: 10.7554/eLife.15023
Andriankaja, M., Dhondt, S., De Bodt, S., Vanhaeren, H., Coppens, F., De Milde, L., et al. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78. doi: 10.1016/j.devcel.2011.11.011
Bashline, L., Lei, L., Li, S., and Gu, Y. (2014). Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 7, 586–600. doi: 10.1093/mp/ssu018
Bellstaedt, J., Trenner, J., Lippmann, R., Poeschl, Y., Zhang, X., Friml, J., et al. (2019). A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 180, 757–766. doi: 10.1104/pp.18.01377
Blume, Y. B., Krasylenko, Y. A., and Yemets, A. I. (2012). Effects of phytohormones on the cytoskeleton of the plant cell. Russian J. Plant Physiol. 59, 515–529.
Boron, A. K., and Vissenberg, K. (2014). The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep. 33, 697–706. doi: 10.1007/s00299-014-1591-x
Braidwood, L., Breuer, C., and Sugimoto, K. (2014). My body is a cage: mechanisms and modulation of plant cell growth. New Phytol. 201, 388–402. doi: 10.1111/nph.12473
Breuer, C., Ishida, T., and Sugimoto, K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13, 654–660. doi: 10.1016/j.pbi.2010.10.006
Burn, J. E., Hocart, C. H., Birch, R. J., Cork, A. C., and Williamson, R. E. (2002). Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 129, 797–807. doi: 10.1104/pp.010931
Cao, J. F., Zhao, B., Huang, C. C., Chen, Z. W., Zhao, T., Liu, H. R., et al. (2020). The miR319-targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol. Plant 13, 1063–1077. doi: 10.1016/j.molp.2020.05.006
Challa, K. R., Aggarwal, P., and Nath, U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28, 2117–2130. doi: 10.1105/tpc.16.00360
Challa, K. R., Rath, M., and Nath, U. (2019). The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 15:e1007988. doi: 10.1371/journal.pgen.1007988
Challa, K. R., Rath, M., Sharma, A. N., Bajpai, A. K., Davuluri, S., Acharya, K. K., et al. (2021). Active suppression of leaflet emergence as a mechanism of simple leaf development. Nat. Plants 7, 1264–1275. doi: 10.1038/s41477-021-00965-3
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., Liu, H., et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Cho, H. T., and Cosgrove, D. J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 97, 9783–9788. doi: 10.1073/pnas.160276997
Collett, C. E., Harberd, N. P., and Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124, 553–562. doi: 10.1104/pp.124.2.553
Cosgrove, D.J. (2014). “Plant cell growth and elongation,” in eLS, ed. John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0001688.pub2
Crawford, B. C., Nath, U., Carpenter, R., and Coen, E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135, 244–253. doi: 10.1104/pp.103.036368
Danisman, S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930. doi: 10.3389/fpls.2016.01930
Danisman, S., Van Der Wal, F., Dhondt, S., Waites, R., De Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Das Gupta, M., Aggarwal, P., and Nath, U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204, 901–912. doi: 10.1111/nph.12963
Das Gupta, M., and Nath, U. (2015). Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell 27, 2785–2799.
Dhaka, N., Bhardwaj, V., Sharma, M. K., and Sharma, R. (2017). Evolving tale of TCPs: new paradigms and old lacunae. Front. Plant Sci. 8:479. doi: 10.3389/fpls.2017.00479
Dong, J., Sun, N., Yang, J., Deng, Z., Lan, J., Qin, G., et al. (2019). The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31, 1155–1170. doi: 10.1105/tpc.18.00803
Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E., and Dengler, N. G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. doi: 10.1006/dbio.1999.9443
Edgar, B. A., and Orr-Weaver, T. L. (2001). Endoreplication cell cycles: more for less. Cell 105, 297–306. doi: 10.1016/s0092-8674(01)00334-8
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Efroni, I., Eshed, Y., and Lifschitz, E. (2010). Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22, 1019–1032.
Efroni, I., Han, S. K., Kim, H. J., Wu, M. F., Steiner, E., Birnbaum, K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24, 438–445. doi: 10.1016/j.devcel.2013.01.019
Fan, D., Ran, L., Hu, J., Ye, X., Xu, D., Li, J., et al. (2020). miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 227, 867–883. doi: 10.1111/nph.16585
Fang, Y., Zheng, Y., Lu, W., Li, J., Duan, Y., Zhang, S., et al. (2021). Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 9, 17–28.
Ferrero, L. V., Gastaldi, V., Ariel, F. D., Viola, I. L., and Gonzalez, D. H. (2021). Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 105, 147–159. doi: 10.1007/s11103-020-01075-y
Ferrero, V., Viola, I. L., Ariel, F. D., and Gonzalez, D. H. (2019). Class I TCP transcription factors target the gibberellin biosynthesis gene GA20ox1 and the growth-promoting genes HBI1 and PRE6 during thermomorphogenic growth in Arabidopsis. Plant Cell Physiol. 60, 1633–1645. doi: 10.1093/pcp/pcz137
Fujikura, U., Ezaki, K., Horiguchi, G., Seo, M., Kanno, Y., Kamiya, Y., et al. (2020). Suppression of class I compensated cell enlargement by xs2 mutation is mediated by salicylic acid signaling. PLoS Genet. 16:e1008873. doi: 10.1371/journal.pgen.1008873
Gastaldi, V., Lucero, L. E., Ferrero, L. V., Ariel, F. D., and Gonzalez, D. H. (2020). Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 182, 2096–2110. doi: 10.1104/pp.19.01501
Gendreau, E., Orbovic, V., Höfte, H., and Traas, J. (1999). Gibberellin and ethylene control endoreduplication levels in the Arabidopsis thaliana hypocotyl. Planta 209, 513–516. doi: 10.1007/PL00008123
Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. doi: 10.1104/pp.114.1.295
Gonzalez, N., Vanhaeren, H., and Inze, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Gonzalez-Grandio, E., Pajoro, A., Franco-Zorrilla, J. M., Tarancon, C., Immink, R. G., and Cubas, P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U.S.A. 114, E245–E254. doi: 10.1073/pnas.1613199114
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., and Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
Han, X., Yu, H., Yuan, R., Yang, Y., An, F., and Qin, G. (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15, 611–622. doi: 10.1016/j.isci.2019.04.005
Hase, Y., Tanaka, A., Baba, T., and Watanabe, H. (2000). FRL1 is required for petal and sepal development in Arabidopsis. Plant J. 24, 21–32. doi: 10.1046/j.1365-313x.2000.00851.x
He, Z., Zhao, X., Kong, F., Zuo, Z., and Liu, X. (2016). TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J. Exp. Bot. 67, 775–785. doi: 10.1093/jxb/erv495
He, Z., Zhou, X., Chen, J., Yin, L., Zeng, Z., Xiang, J., et al. (2021). Identification of a consensus DNA-binding site for the TCP domain transcription factor TCP2 and its important roles in the growth and development of Arabidopsis. Mol. Biol. Rep. 48, 2223–2233. doi: 10.1007/s11033-021-06233-z
Herve, C., Dabos, P., Bardet, C., Jauneau, A., Auriac, M. C., Ramboer, A., et al. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149, 1462–1477. doi: 10.1104/pp.108.126136
Hisanaga, T., Kawade, K., and Tsukaya, H. (2015). Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 66, 1055–1063. doi: 10.1093/jxb/erv028
Hong, S. Y., Kim, O. K., Kim, S. G., Yang, M. S., and Park, C. M. (2011). Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286, 1659–1668. doi: 10.1074/jbc.M110.167692
Horiguchi, G., and Tsukaya, H. (2011). Organ size regulation in plants: insights from compensation. Front. Plant Sci. 2:24. doi: 10.3389/fpls.2011.00024
Hu, Y., Poh, H. M., and Chua, N. H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47, 1–9.
Hur, Y. S., Kim, J., Kim, S., Son, O., Kim, W. Y., Kim, G. T., et al. (2019). Identification of TCP13 as an upstream regulator of ATHB12 during leaf development. Genes 10:644. doi: 10.3390/genes10090644
Jegu, T., Latrasse, D., Delarue, M., Mazubert, C., Bourge, M., Hudik, E., et al. (2013). Multiple functions of Kip-related protein5 connect endoreduplication and cell elongation. Plant Physiol. 161, 1694–1705. doi: 10.1104/pp.112.212357
Jensen, P. J., Hangarter, R. P., and Estelle, M. (1998). Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116, 455–462.
Johnson, K., and Lenhard, M. (2011). Genetic control of plant organ growth. New Phytol. 191, 319–333.
Kandasamy, M. K., Gilliland, L. U., Mckinney, E. C., and Meagher, R. B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13, 1541–1554. doi: 10.1105/tpc.010026
Keuskamp, D. H., Pollmann, S., Voesenek, L. A., Peeters, A. J., and Pierik, R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. U.S.A. 107, 22740–22744. doi: 10.1073/pnas.1013457108
Kim, J. H., and Lee, B. H. (2006). GROWTH-REGULATING FACTOR4 ofArabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468.
Kondorosi, E., Roudier, F., and Gendreau, E. (2000). Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3, 488–492. doi: 10.1016/s1369-5266(00)00118-7
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Koyama, T., Furutani, M., Tasaka, M., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2010a). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Koyama, T., Sato, F., and Ohme-Takagi, M. (2010b). A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 74, 2145–2147. doi: 10.1271/bbb.100442
Koyama, T., Sato, F., and Ohme-Takagi, M. (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 175, 874–885. doi: 10.1104/pp.17.00732
Kubota, A., Ito, S., Shim, J. S., Johnson, R. S., Song, Y. H., Breton, G., et al. (2017). TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 13:e1006856. doi: 10.1371/journal.pgen.1006856
Larkins, B. A., Dilkes, B. P., Dante, R. A., Coelho, C. M., Woo, Y. M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52, 183–192.
Lei, N., Yu, X., Li, S., Zeng, C., Zou, L., Liao, W., et al. (2017). Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 7:10016. doi: 10.1038/s41598-017-09398-5
Li, C., Potuschak, T., Colon-Carmona, A., Gutierrez, R. A., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, S. (2015). The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 10:e1044192. doi: 10.1080/15592324.2015.1044192
Marowa, P., Ding, A., and Kong, Y. (2016). Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 35, 949–965. doi: 10.1007/s00299-016-1948-4
Martin, C., Bhatt, K., and Baumann, K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4, 540–549.
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Melaragno, J. E., Mehrotra, B., and Coleman, A. W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668. doi: 10.1105/tpc.5.11.1661
Millenaar, F. F., Van Zanten, M., Cox, M. C. H., Pierik, R., Voesenek, L., and Peeters, A. J. M. (2009). Differential petiole growth in Arabidopsis thaliana: photocontrol and hormonal regulation. New Phytol. 184, 141–152. doi: 10.1111/j.1469-8137.2009.02921.x
Miséra, S., Müller, A. J., Weiland-Heidecker, U., and Jürgens, G. (1994). The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol. Gen. Genet. MGG 244, 242–252. doi: 10.1007/BF00285451
Mizukami, Y. (2001). A matter of size: developmental control of organ size in plants. Curr. Opin. Plant Biol. 4, 533–539. doi: 10.1016/s1369-5266(00)00212-0
Nath, U., Crawford, B. C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Herve, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., and Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3:e03031. doi: 10.7554/eLife.03031
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Perrot-Rechenmann, C. (2010). Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2:a001446. doi: 10.1101/cshperspect.a001446
Poethig, R. S., and Sussex, I. M. (1985). The developmental morphology and growth dynamics of the tobacco leaf. Planta 165, 158–169. doi: 10.1007/BF00395038
Procko, C., Burko, Y., Jaillais, Y., Ljung, K., Long, J. A., and Chory, J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30, 1529–1541.
Procko, C., Crenshaw, C. M., Ljung, K., Noel, J. P., and Chory, J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165, 1285–1301. doi: 10.1104/pp.114.241844
Ramirez-Parra, E., and Gutierrez, C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144, 105–120. doi: 10.1104/pp.106.094979
Ray, P. M., Green, P. B., and Cleland, R. (1972). Role of turgor in plant cell growth. Nature 239, 163–164.
Rayle, D. L. (1973). Auxin-induced hydrogen-ion secretion in Avena coleoptiles and its implications. Planta 114, 63–73.
Rayle, D. L., and Cleland, R. (1977). “Chapter 6 control of plant cell enlargement by hydrogen ions,” in Current Topics in Developmental Biology, eds A. A. Moscona and A. Monroy (Cambridge, MA: Academic Press), 187–214. doi: 10.1016/s0070-2153(08)60746-2
Rayle, D. L., and Cleland, R. E. (1992). The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99, 1271–1274. doi: 10.1104/pp.99.4.1271
Reed, J. W., Wu, M. F., Reeves, P. H., Hodgens, C., Yadav, V., Hayes, S., et al. (2018). Three auxin response factors promote hypocotyl elongation. Plant Physiol. 178, 864–875. doi: 10.1104/pp.18.00718
Ren, H., and Gray, W. M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8, 1153–1164. doi: 10.1016/j.molp.2015.05.003
Rodriguez, R. E., Debernardi, J. M., and Palatnik, J. F. (2014). Morphogenesis of simple leaves: regulation of leaf size and shape. Wiley Interdiscip. Rev. Dev. Biol. 3, 41–57. doi: 10.1002/wdev.115
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Sapala, A., Runions, A., Routier-Kierzkowska, A. L., Das Gupta, M., Hong, L., Hofhuis, H., et al. (2018). Why plants make puzzle cells, and how their shape emerges. eLife 7:e32794. doi: 10.7554/eLife.32794
Sarvepalli, K., and Nath, U. (2011a). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67, 595–607. doi: 10.1111/j.1365-313X.2011.04616.x
Sarvepalli, K., and Nath, U. (2011b). Interaction of TCP4-mediated growth module with phytohormones. Plant Signal Behav. 6, 1440–1443. doi: 10.4161/psb.6.10.17097
Sarvepalli, K., and Nath, U. (2018). CIN-TCP transcription factors: Transiting cell proliferation in plants. IUBMB Life 70, 718–731. doi: 10.1002/iub.1874
Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S., et al. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32, 1011–1022. doi: 10.1046/j.1365-313x.2002.01488.x
Schommer, C., Debernardi, J. M., Bresso, E. G., Rodriguez, R. E., and Palatnik, J. F. (2014). Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 7, 1533–1544. doi: 10.1093/mp/ssu084
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Seifert, G. J., and Blaukopf, C. (2010). Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 153, 467–478. doi: 10.1104/pp.110.153940
Shen, J., Ge, D., Song, X., Xiao, J., Liu, X., Che, G., et al. (2021). Roles of CsBRC1-like in leaf and lateral branch development in cucumber. Plant Sci. 302:110681. doi: 10.1016/j.plantsci.2020.110681
Spartz, A. K., Ren, H., Park, M. Y., Grandt, K. N., Lee, S. H., Murphy, A. S., et al. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26, 2129–2142. doi: 10.1105/tpc.114.126037
Stortenbeker, N., and Bemer, M. (2019). The SAUR gene family: the plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 70, 17–27. doi: 10.1093/jxb/ery332
Stoynova-Bakalova, E., Karanov, E., Petrov, P., and Hall, M. A. (2004). Cell division and cell expansion in cotyledons of Arabidopsis seedlings. New Phytol. 162, 471–479.
Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553. doi: 10.1016/j.pbi.2003.09.009
Sugio, A., Maclean, A. M., and Hogenhout, S. A. (2014). The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 202, 838–848. doi: 10.1111/nph.12721
Sun, X., Wang, C., Xiang, N., Li, X., Yang, S., Du, J., et al. (2017). Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 15, 1284–1294. doi: 10.1111/pbi.12715
Tabeta, H., Watanabe, S., Fukuda, K., Gunji, S., Asaoka, M., Hirai, M. Y., et al. (2021). An auxin signaling network translates low-sugar-state input into compensated cell enlargement in the fugu5 cotyledon. PLoS Genet. 17:e1009674. doi: 10.1371/journal.pgen.1009674
Takahashi, N., Quimbaya, M., Schubert, V., Lammens, T., Vandepoele, K., Schubert, I., et al. (2010). The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 6:e1000817. doi: 10.1371/journal.pgen.1000817
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Thouly, C., Le Masson, M., Lai, X., Carles, C. C., and Vachon, G. (2020). Unwinding BRAHMA functions in plants. Genes 11:90. doi: 10.3390/genes11010090
Tsai, H. L., Li, Y. H., Hsieh, W. P., Lin, M. C., Ahn, J. H., and Wu, S. H. (2014). HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26, 2858–2872. doi: 10.1105/tpc.114.126722
Tsukaya, H., Tsuge, T., and Uchimiya, H. (1994). The cotyledon: a superior system for studies of leaf development. Planta 195, 309–312.
Van Es, S. W., Silveira, S. R., Rocha, D. I., Bimbo, A., Martinelli, A. P., Dornelas, M. C., et al. (2018). Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 94, 867–879. doi: 10.1111/tpj.13904
Van Mourik, H., Van Dijk, A. D. J., Stortenbeker, N., Angenent, G. C., and Bemer, M. (2017). Divergent regulation of Arabidopsis SAUR genes: a focus on the SAUR10-clade. BMC Plant Biol. 17:245. doi: 10.1186/s12870-017-1210-4
Vanhaeren, H., Gonzalez, N., and Inze, D. (2015). A journey through a leaf: phenomics analysis of leaf growth in Arabidopsis thaliana. Arabidopsis Book 13:e0181. doi: 10.1199/tab.0181
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S. K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Verkest, A., Manes, C. L., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., et al. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736. doi: 10.1105/tpc.105.032383
Wang, H., Mao, Y., Yang, J., and He, Y. (2015). TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 6:436. doi: 10.3389/fpls.2015.00436
Wang, M. Y., Zhao, P. M., Cheng, H. Q., Han, L. B., Wu, X. M., Gao, P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. doi: 10.1104/pp.113.215673
Wang, S. T., Sun, X. L., Hoshino, Y., Yu, Y., Jia, B., Sun, Z. W., et al. (2014). MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS One 9:e91357. doi: 10.1371/journal.pone.0091357
Wei, N., Kwok, S. F., Von Arnim, A. G., Lee, A., Mcnellis, T. W., Piekos, B., et al. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6, 629–643. doi: 10.1105/tpc.6.5.629
Wolters, H., and Jurgens, G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. doi: 10.1038/nrg2558
Zhang, G., Zhao, H., Zhang, C., Li, X., Lyu, Y., Qi, D., et al. (2019). TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis. J. Integr. Plant Biol. 61, 1151–1170. doi: 10.1111/jipb.12749
Zhang, T., Qu, Y., Wang, H., Wang, J., Song, A., Hu, Y., et al. (2017). The heterologous expression of a chrysanthemum TCP-P transcription factor CmTCP14 suppresses organ size and delays senescence in Arabidopsis thaliana. Plant Physiol. Biochem. 115, 239–248. doi: 10.1016/j.plaphy.2017.03.026
Zhou, Y., Xun, Q., Zhang, D., Lv, M., Ou, Y., and Li, J. (2019). TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate Thermomorphogenesis in Arabidopsis thaliana. iScience 15, 600–610. doi: 10.1016/j.isci.2019.04.002
Keywords: cell expansion and differentiation, cell proliferation, CIN-TCP, Arabidopsis, hypocotyl, cotyledon, leaf
Citation: Rath M, Challa KR, Sarvepalli K and Nath U (2022) CINCINNATA-Like TCP Transcription Factors in Cell Growth – An Expanding Portfolio. Front. Plant Sci. 13:825341. doi: 10.3389/fpls.2022.825341
Received: 30 November 2021; Accepted: 13 January 2022;
Published: 22 February 2022.
Edited by:
Daniel Kierzkowski, Université de Montréal, CanadaReviewed by:
Marie Monniaux, UMR 5667 Laboratoire Reproduction et Développement des Plantes (RDP), FranceCopyright © 2022 Rath, Challa, Sarvepalli and Nath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Utpal Nath, dXRwYWxuYXRoQGlpc2MuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.