
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 March 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.825209
Scientists have long been captivated by biogeographic disjunctions, and disjunctions between East Asia and North America have been particularly well-studied at the genus and family levels. By contrast, disjunctions between eastern and western Asia have received less attention. Euonymus L. is taxonomically divided into two sections based on the number of cells in anthers as follows: E. sect. Uniloculares has one-celled anthers and occurs mainly in Asia, whereas E. sect. Biloculares has two-celled anthers and is distributed globally. We used Illumina sequencing to investigate the genomes of four species in sect. Uniloculares. The chloroplast (cp) genomes are highly conserved (157,290–158,094 bp). Pseudogenisation of ndhF and intron loss in rps16 was detected. Based on the cp genomes of the four species of E. sect. Uniloculares, we propose a novel hypothesis of disjunction between eastern and western Asia. Biogeographic reconstruction and molecular dating revealed that sect. Uniloculares separated from its sect. Biloculares forebears 4.0 Mya during the Pliocene era. The radial diversification of sect. Uniloculares from East Asia and the establishment of the western Asian clade during the Pleistocene era (1.9 Mya) were the results of both dispersal and vicariance, making the section the youngest diverged clade conforming to age estimation. The centre of origin of sect. Uniloculares was determined to be in East Asia. Disjunctions and diversification between eastern and western Asia in sect. Uniloculares are thought to have been caused by changes in monsoon patterns, temperature variations, and the emergence of the Gobi Desert.
Understanding intercontinental disjunction patterns is among the main objectives of biogeography (Cox et al., 2016; Costa et al., 2020; Namgung et al., 2021). Such disjunctions and fragmentations have resulted from continental drift, the formation of the Bering land bridge, the formation of deserts and lakes, and global climatic oscillations during the Cenozoic era (Ickert-Bond and Wen, 2006; Liao et al., 2007; Nie et al., 2008; Kim et al., 2015; Kim et al., 2017). Disjunctions between the temperate regions of eastern North America and East Asia (Wen, 1999; Xiang et al., 2000; Wen, 2001), western North America and southern Europe (Wen and Ickert-Bond, 2009; Mao et al., 2010), and eastern and western North America (Xiang et al., 1998) have received much attention (Xiang et al., 1998; Wen, 1999; Deng et al., 2015; Kim et al., 2017; Song et al., 2020). However, less emphasis has been placed on understanding the disjunction between East and West Asia (Song et al., 2020).
Euonymus L. is among the largest genera in the Celastraceae, comprising 129–200 species (Leonova, 1974; Ma, 2001; Savinov and Baikov, 2007; Gavrilova et al., 2018) of deciduous and evergreen shrubs and small trees (Gavrilova et al., 2018). The genus is widely distributed throughout tropical and temperate regions (Blakelock, 1951; Ma, 2001; Xia et al., 2018), but it is concentrated within east, south, and southeast Asia and the Himalayas (Ma, 2001; Yao et al., 2018). Blakelock (1951) also noted its occurrence in Madagascar, North Africa, and Australia and concluded that Euonymus is a cosmopolitan genus (Blakelock, 1951).
With respect to classification, Ma (2001) divided Euonymus into five sections, such as sects. Uniloculares, Echinococcus, Illicifolia, Melanocarya, and Euonymus, based on capsule texture (angular, echinate, or smooth). In contrast, Rouy and Foucaud, in Flora of France (Fl. De France 4, 158-9, 1897), recognised the following two sections: the Biloculares, with bilocular anthers (e.g., Euonymus europeaus), and the Uniloculares, with unilocular anthers (e.g., Euonymus latifolius) (Blakelock, 1951).
Species in E. sect. Uniloculares are described as deciduous, evergreen shrubs, or small trees and are characterised by tetra- or pentamerous flowers (Ma and Funston, 2008), winged capsules, and one-celled anthers. Sect. Uniloculares has been alternatively classified as subgenus Kalonymus; however, the key characteristics of subgenus Kalonymus (one-celled anthers and winged capsules) are similar to those of sect. Uniloculares, and as such subgenus Kalonymus was declared illegitimate (McNeill et al., 2012; Du et al., 2016). Of the 16 species discussed in this section, only E. latifolius occurs in western Asia, Europe, and North Africa. The other 15 species, including Euonymus macropterus, Euonymus sachalinensis, and Euonymus oxyphyllus, are distributed throughout East Asia and the Russian Far East. China is a hotspot for species discussed in this section and hosts eight endemic species, including Euonymus schensianus and Euonymus szechuanensis (included in this study) (Ma and Funston, 2008).
We used the members of sect. Uniloculares as a model to understand vicariance and disjunction, as the section exhibits a disjunction between eastern and western Asia. We used the whole chloroplast (cp) genomes of six species in sect. Uniloculares, of which we sequenced the following four species: E. macropterus, E. sachalinensis, E. oxyphyllus, and E. latifolius. The genomes of E. schensianus and E. szechuanensis were obtained from the National Centre for Biotechnology Information (NCBI) GenBank. E. macropterus, E. sachalinensis, and E. oxyphyllus are distributed in East Asia and the Russian Far East, whereas E. latifolius is distributed in western Asia, Europe, and North Africa, making this section a suitable model for exploring disjunction and evolution based on the whole cp genome.
Analysis of cp genomes is widely used in phylogenetic studies. In angiosperms, the cp genome is transferred from the female parent (Palmer et al., 1988). It is a type of plastid with a double-layered membrane and thylakoid structures with a high concentration of chlorophyll, which plays a vital role in photosynthesis and other biochemical processes (Neuhaus and Emes, 2000; Cheng et al., 2020; Liang et al., 2020). Similar to mitochondria, cp is among the organelles that have their genome (Cheng et al., 2020). Gene deletions, duplications, mutations, and rearrangements have been observed in the cp genomes of angiosperms (Lee et al., 2007). Phylogenetic studies use cp genomes as they are more highly conserved among taxa than nuclear and mitochondrial genomes. Genomes may provide markers for phylogenetic analyses and may be used to estimate the time of divergence of higher taxonomic ranks (Moore et al., 2010; Lee et al., 2019).
The insights obtained from phylogenetic, age estimation, and biogeographic studies rely on analysis of the whole cp genome, which is considered essential to infer backbone phylogeny and phylogenetic trees. Analysis of the cp genome may also help to resolve the relationships among complex groups of plants, which remain unclear due to taxonomic gaps.
Samples of E. macropterus, E. oxyphyllus, and E. sachalinensis were collected from different sites in South Korea, whereas E. latifolius samples were collected from the Tsytsin Main Botanical Garden, Russian Academy of Sciences (RAS), Moscow, Russia (refer to Table 1). All samples were dried using silica gel, and voucher specimens for the Korean samples were prepared and submitted to the Gachon University Herbarium (GCU). Dried samples were used for DNA extraction using the common 2 × cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). Concentrations of extracted DNA were measured using a spectrophotometer (BioSpec-nano Shimadzu, Kyoto, Japan). The DNA bands were observed using gel electrophoresis, and samples with good-quality bands were subjected to quality tests.
DNA samples with concentrations of >20 ng/μl were transferred to the next-generation sequencing using the Illumina MiSeq sequencing system (Illumina, Seoul, South Korea). Sequencing libraries were prepared using an Illumina TruSeq Nano DNA Library Preparation Kit. Raw reads (300-bp paired-end reads) were trimmed to remove regions with error probabilities > 3% per base using Geneious version 7.1.9 (Kearse et al., 2012). Total reads, assembled reads, and coverage are summarised in Supplementary Table 5. Paired-end reads were assembled by performing a “map to reference” using the cp genome of E. schensianus (GenBank Acc. no: KY511610), which is accessible in NCBI. Gene content was annotated in Geneious version 7.1.9 using E. schensianus as a reference and applying a 75% similarity index to identify genes. Following the generation of quadripartite structures, further steps were carried out. The start and end of transfer RNAs (tRNAs) were confirmed using tRNAScan with the default settings (Chan and Lowe, 2019). Circular and linear cp maps were then converted into graphical maps using OrgaellarGenomeDRAW (Greiner et al., 2019).
We compared the cp genomes of six species in sect. Unilocualres using the Shuffle-LGAN mode in mVISTA (Frazer et al., 2004). Euonymus hamiltonianus (KY926695) was used as a reference genome. To evaluate differences among the six species, whole cp genomes, including coding genes, tRNAs, ribosomal RNAs (rRNAs), and intergenic spacers, were aligned using MAFFT version 7.017 embedded in Geneious version 7.1.9 (Katoh et al., 2002). Nucleotide diversity (π) was calculated using DnaSP version 6.12.03 (Rozas, 2009). The structure and junctions between inverted repeats (IRs), large single copies (LSCs), and small single copies (SSCs) were investigated using IRscope (Amiryousefi et al., 2018).
A total of 11 Euonymus species were used to construct a phylogenetic tree; four were sequenced as part of this study, whereas the data for the remaining seven species were obtained from NCBI GenBank (Table 2 and Supplementary Table 1). We extracted and aligned 80 protein-coding genes using MAFFT version 7.017 embedded in Geneious version 7.1.9 (Katoh et al., 2002). Maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) were used to infer relationships among the 11 Euonymus L. species, while six other genera in the Celastraceae were used as outgroups. Maximum parsimony was conducted using PAUP version 4.0 (Wilgenbusch and Swofford, 2003), using equal character weighting and treating gaps as missing data. Searches of 1,000 random additions used tree bisection and reconnection branch swapping, and MulTree permitted 10 trees to be held at each step. Bootstrap analysis, with the same parameters, was used to find internal support. J-Model test version2.1.10 was used to identify the best model based on Akaike’s Information Criterion (AIC). AIC values indicated that TVM + I + G was the best model, and we used this model for the ML and BI analyses. IQ-TREE was also used for the ML analysis (Trifinopoulos et al., 2016). A support value was calculated with 1,000 ultrafast bootstrap replicates. We used MrBayes version 3.2.6 for the BI analysis (Ronquist et al., 2012). Two runs were conducted from random trees for a minimum of 1,000,000 generations. One tree was sampled every 1,000 generations. The first 25% of trees were discarded as burn-in, and the remaining trees were used to construct a 50% majority-rule consensus tree, with the proportion of bifurcations in the consensus tree provided as a posterior probability (PP) to estimate the robustness of half of the BI tree. Effective sample size (ESS) values were then examined for model parameters (≥200). Trees were edited using FigTree version 1.4.4 (Rambaut, 2020).
Fossil data for Maytenus spp. (17.4–7.1 Mya), Salacia spp. (7.1–2.8 Mya), and Celastrus madagascariensis (21.7–11.7 Mya), all from Africa and Madagascar, were used as minimum age constraints for three nodes (Bacon et al., 2016). To infer time of divergence, we used the uncorrelated relaxed molecular clock model for Markov Chain Monte Carlo (MCMC) simulation in BEAST version 1.8.3, featured in Beauti version 1.8.3. The Yule speciation tree prior (Gernhard, 2008) was used with an uncorrelated relaxed molecular clock. The stem nodes C1–C3 were calibrated using a normal distribution because of the availability of means for the fossil data used in age estimation (Figure 7). C1 and C3 had mean values of 12.2 and 16.5 (SD = 2.9), respectively, whereas C2 had a mean value of 4.7 (SD = 1.2). MCMC length was limited to 100 million generations, and parameter sampling was conducted every 1,000 generations. Tracer version 1.65 was used for convergence (Rambaut and Drummond, 2003), and ESS was assessed. A maximum clade credibility tree was generated using TreeAnnotator version 1.8.3, following the removal of the first 25% of trees as burn-in and to display the mean age and 95% of highest posterior density (HPD) interval for each node (Drummond and Rambaut, 2007). The tree was annotated using FigTree version 1.4.4 (Rambaut, 2020).
Eight areas were defined within the geographic ranges of the 11 focal species (five species from sect. Biloculares and six species from sect. Uniloculares) based on available literature (Agea et al., 2021) and web sources, particularly Plants of the World Online (POWO, 2019). These areas included East Asia; the Russian Far East; western Asia; south, southeast, and central Europe; North Africa; south and southeast Asia and the eastern and western Himalayas; North America; and South Africa. Ancestral area reconstruction was conducted using Reconstruct Ancestral State in Phylogenies (RASP) version 2.1b (formerly S-DIVA) (Yu et al., 2010; Yu et al., 2013).
The whole cp genomes of four species of sect. Uniloculares, including E. latifolius, E. sachalinensis, E. macropterus, and E. oxyphyllus, were constructed. The results indicated that all four species had typical cp genomes consisting of LSC and SSC regions and two IR copies, which are typically referred to as IRa and IRb. The genomes of two additional species in sect. Uniloculares (E. schensianus and E. szechuanensis) were obtained from GenBank. The four newly assembled cp genomes were compared with those of the aforementioned species. No discernible differences were observed among the six species. However, E. latifolius had a large genome (158,094 bp), whereas that of E. sachalinensis was small relative to the other species (157,290 bp). The LSC in E. oxyphyllus had more base pairs (85,924 bp). E. latifolius also had a large number of base pairs in the SSC (18,581 bp), whereas the IR in E. macropterus comprised 26,709 bp (Figure 1).
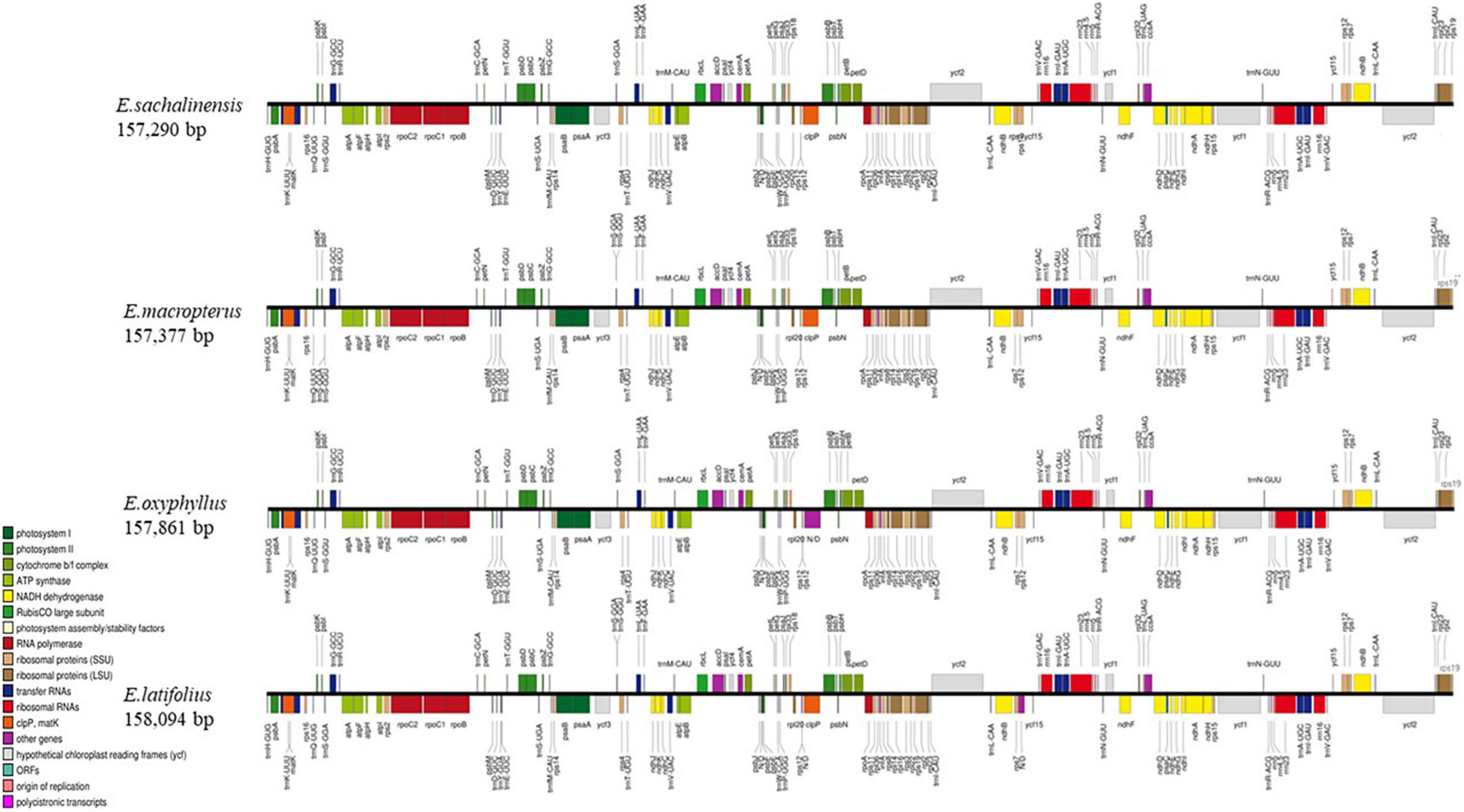
Figure 1. Linear gene map of four species of Euonymus sect. Uniloculares. Genes shown above are transcribed clockwise, and genes shown below are transcribed counterclockwise. The functional group of genes is shown by the colour code on the left.
The numbers of coding genes and rRNAs were similar among species. E. latifolius had 38 rRNAs compared with 39 for E. macropterus and 37 for the other four species. Pseudogenisation was detected in rps16 (LSC), ycf1 (IRb), and ndhF (SSC) (Table 2), and rps16 was observed without non-coding genes (introns) in all the taxa studied. Pairwise determination of divergent regions was conducted using mVISTA, in which the six species were compared with E. hamiltonianus (KY926695) as a reference. Introns exhibited variation, whereas coding regions demonstrated a higher level of conservation (Figure 2).
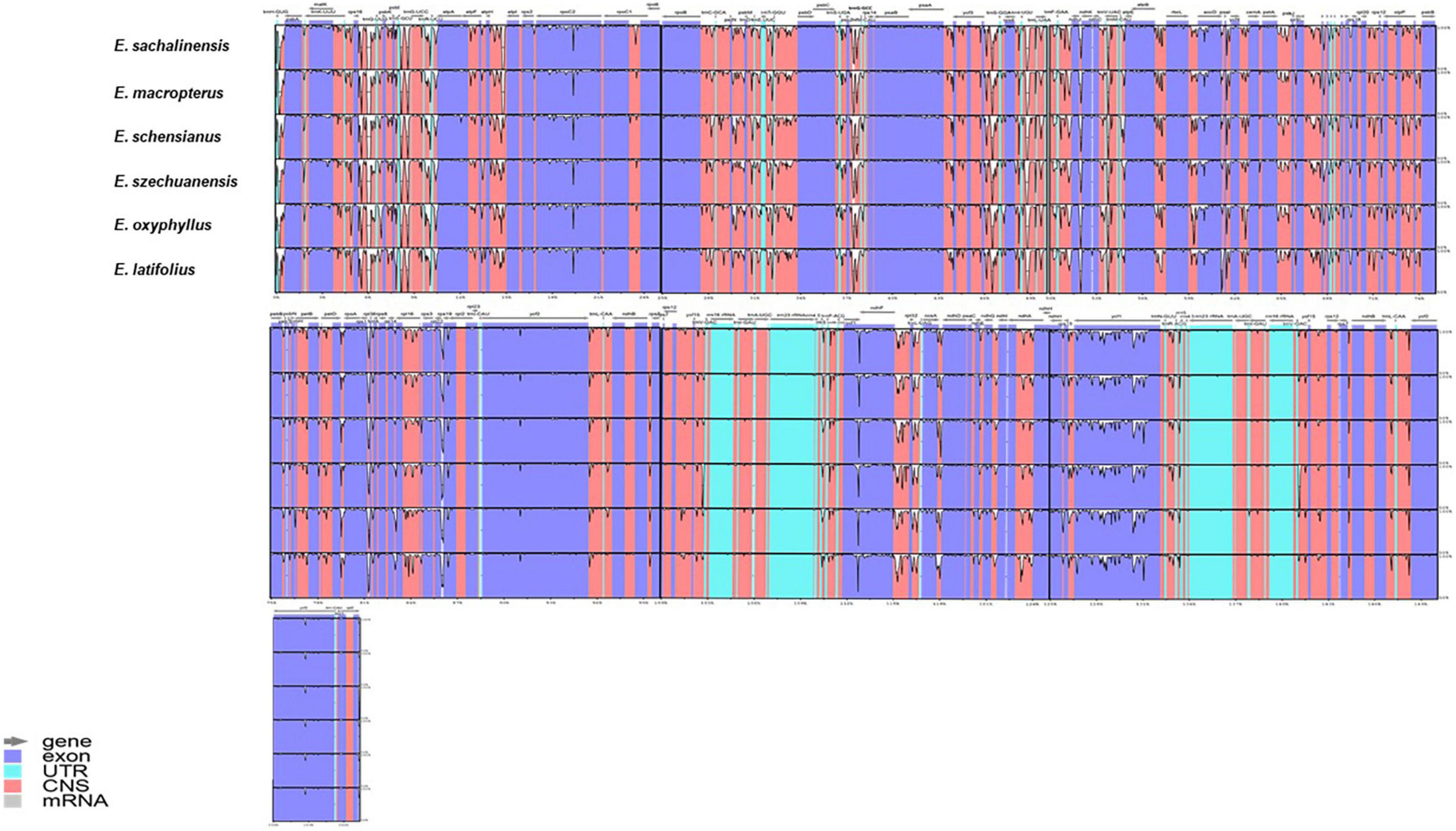
Figure 2. Using mVISTA, six chloroplast genomes of Euonymus sect. Uniloculares were aligned in parallel using Euonymus hamiltonianus as a reference. The transcriptional directional order of genes is indicated by arrows above. The identity on Y-scale axis ranges from 50 to 100%. Colour-coded regions include exon, intron, and non-coding regions.
Nucleotide diversity was analysed for both coding and non-coding regions of the six species. We aligned 114 genes from the LSC, SSC, and IR and calculated π values. Of these, 53 genes, including psbI, rpoC1, rps14, and rps4, exhibited no nucleotide diversity (π = 0). The other 61 genes exhibited nucleotide diversity; the highest overall value was recorded for psbM (π = 0.04); the highest value observed among tRNAs was for trnI-CAU (π = 0.2). The π-value was 0.006 for ycf1, whereas psbZ, atpE, and rpl14 had π-values of 0.005. We observed π-values of 0.003 in rpoA, accD, petl, pasJ, and rpl22 (Figure 3). Detailed values are shown in Supplementary Table 2.
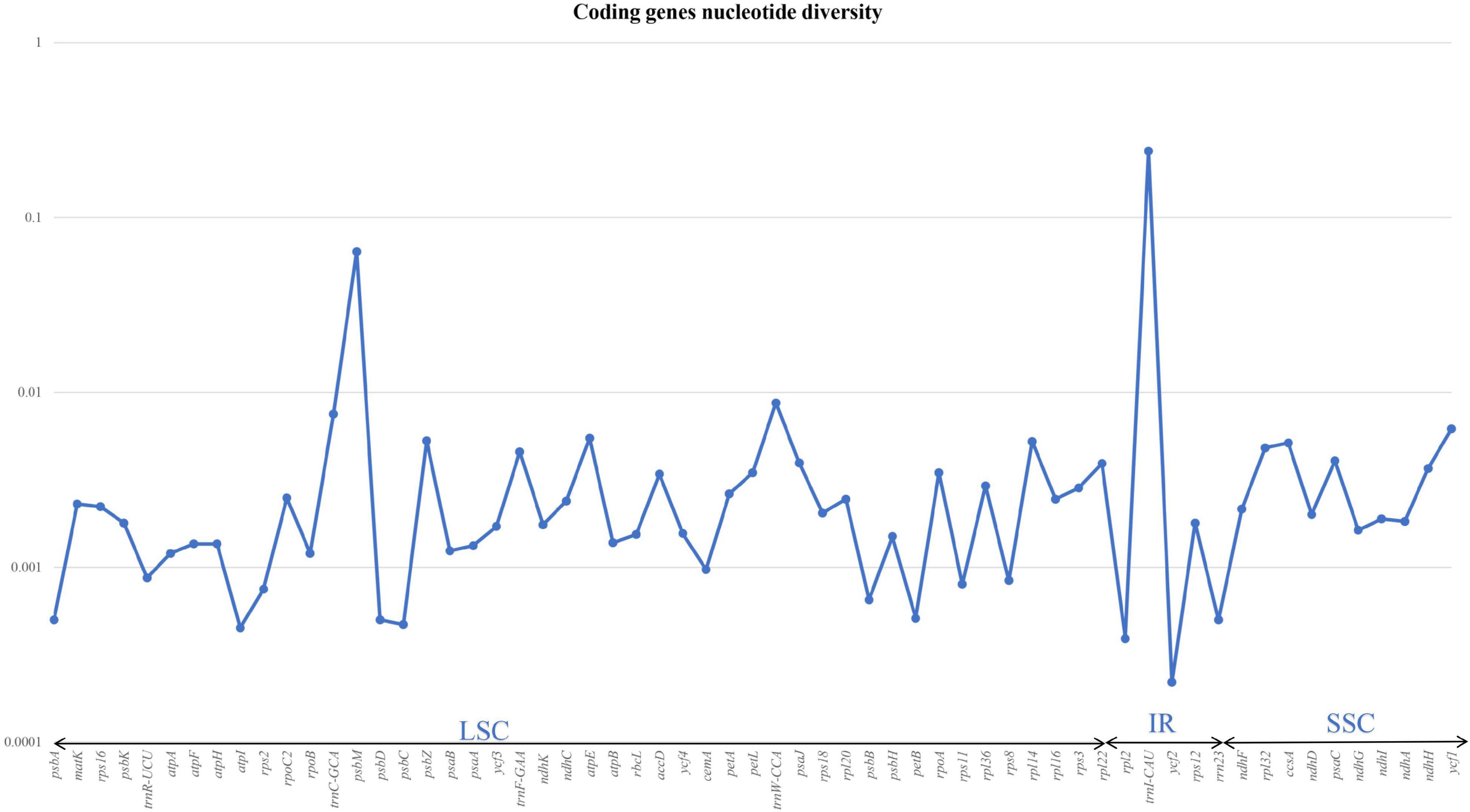
Figure 3. Graph illustrating the nucleotide variations in the coding genes of six sect. Uniloculares species. The graph does not include genes with zero variability.
The nucleotide diversity of non-coding regions was also analysed. We analysed 130 regions and introns between the genes. Of these, only 21 exhibited no diversity, including rpoC1_rpoB, psaB_psaA, and ndhJ_ndhK. The highest π-value (0.04) observed was for trnM-CAU_atpE, rpl22_rps19, and rps19_rpl2. Other introns had π-values of 0.01 (Figure 4 and Supplementary Table 3).
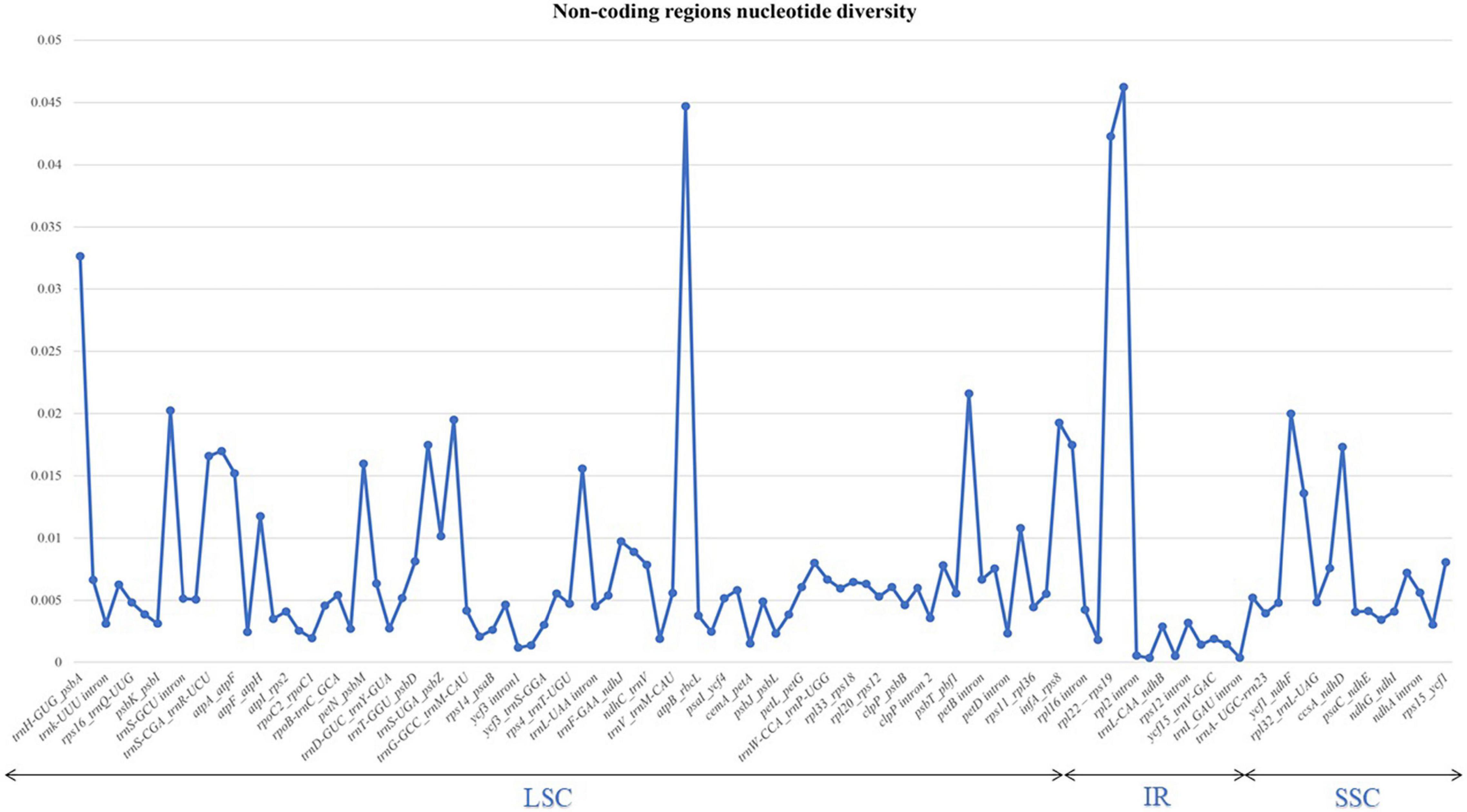
Figure 4. Graph illustrating the variations in nucleotide sequences in the non-coding regions (introns) of six sect. Uniloculares species. The graph does not include genes with zero variability.
We constructed a phylogenetic tree based on 80 protein-coding genes in the cp genome using MP, ML, and BI. Only the ML tree is discussed further. A total of 11 Euonymus species were aligned with six Celastraceae species for which the whole cp genome data were available were used as outgroups, which includes Catha edulis (KT861471), Celastrus orbiculatus (MW316708), Maytenus guangxiensis (MN707924), Monimopetalum chinense (MK450440), Parnassia palustris (MH544205), and Salacia amplifolia (MK799641). We aligned 69,975 bps using MAFFT (Katoh et al., 2002). The six sect. Uniloculares species were grouped together in a clade with a bootstrap value of 100. This clade was monophyletic to sect. Biloculares, whereas the Euonymus japonicus and Euonymus fortunei clades were sister taxa to the sect. Uniloculares clade. Within the Uniloculares clade, E. macropterus and E. sachalinensis were sister taxa with 100% resolution. Sect. Uniloculares is closely related to E. japonicus and E. fortunei. E. latifolius, the representative taxon from western Asia, was located in a separate branch of sect. Uniloculares clade with 100% resolution. E. japonicus and E. fortunei were grouped in a clade and exhibited a close association, with a bootstrap value of 100%. E. hamiltonianus, Euonymus yunnanensis, and Euonymus phellomanus were grouped in a separate clade with 100% resolution within the sister group, whereas E. hamiltonianus and E. yunnanensis exhibited a 100% relationship to one another. Values < 50 are not shown in the tree. Within the Euonymus ingroup, ndhF was recorded as a true gene in E. hamiltonianus, E. yunnanensis, and E. phellomanus. In contrast, ndhF was a pseudogene in all members of sect. Uniloculares included in our analysis, along with E. japonicus and E. fortunei. The length of the gene was reduced above the clade. Pseudogenisation was also observed in rps16 (LSC) and ycf1 (IRb) in both the ingroups and outgroups (Figure 5).
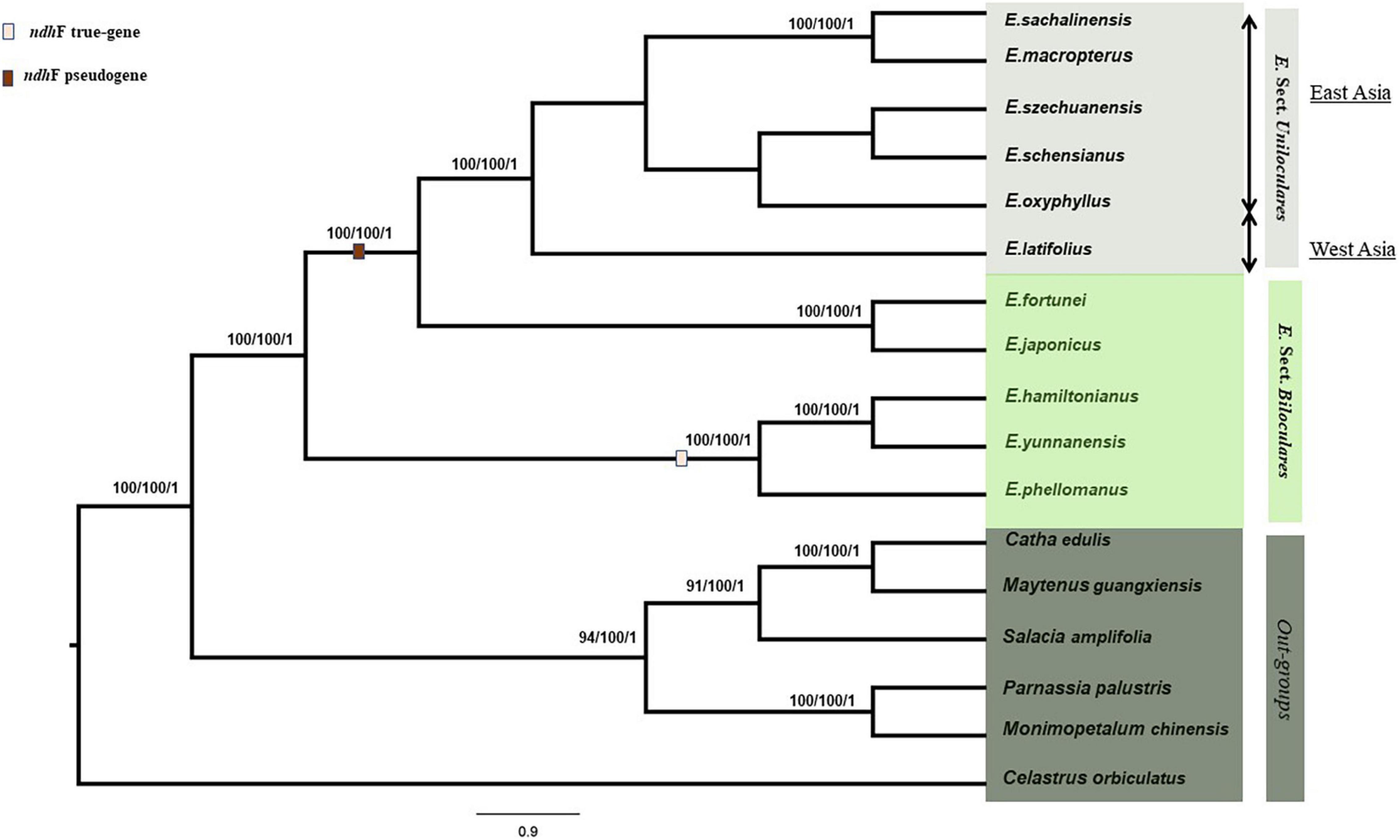
Figure 5. Maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) were used to generate a phylogenetic tree based on whole chloroplast genomes of 11 Euonymus L. species (6 species of sect. Uniloculares and 5 of sect. Biloculares). Bootstrap values were 100, and the posterior probability values were 1. Values < 50 are ignored.
The IRs in the cp genomes of flowering plants differ only slightly among species (Goulding et al., 1996). We compared four plastid genomes along with two previously available genomes from sect. Uniloculares to investigate the expansion or contraction of the IR and SSC regions. An expansion of the IR junction is evident in Figure 6, where rps19 is present in both IRa and IRb. Except E. szechuanensis, the IR region in the other five species extended from 7 to 66 bp in IRb. The pseudogene ycf1 (IRb) was adjusted to the boundary between IRb and the SSC. Both the JLA and JLS appeared to be unstable, as trnH was observed to move toward the IR in all six species. Slight variations in ycf1, from 4,639 to 4,720 bp, were observed in the SSC. Pseudogenisation of ndhF was observed in all members of sect. Uniloculares, as its length was short compared with the true gene. As a pseudogene, ndhF is 792–879 bp from the junction JSB.
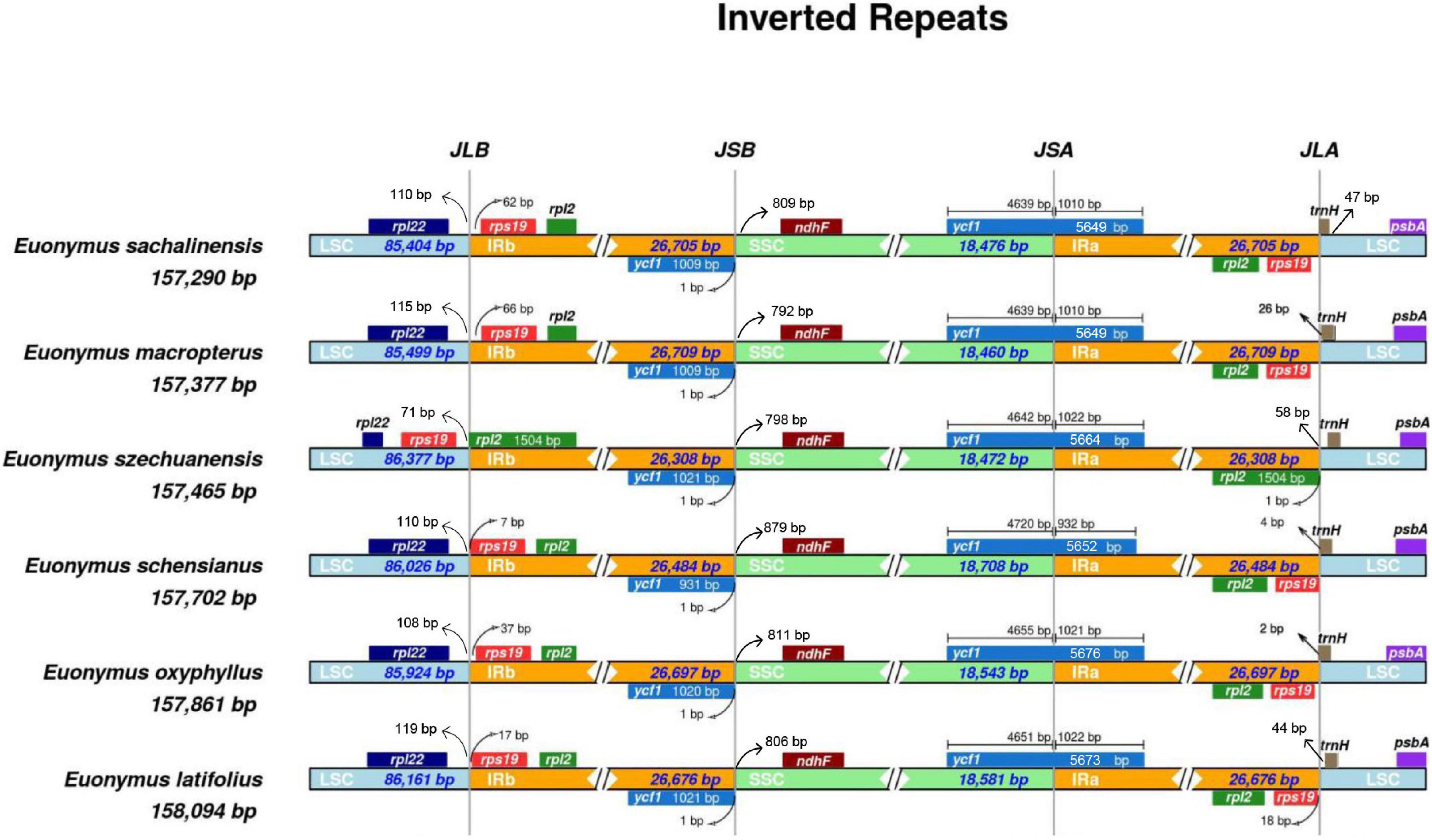
Figure 6. Large single copy (LSC), inverted repeat (IR), and small single copy (SSC) junction comparison among chloroplast genomes of six species of Euonymus sect. Uniloculares.
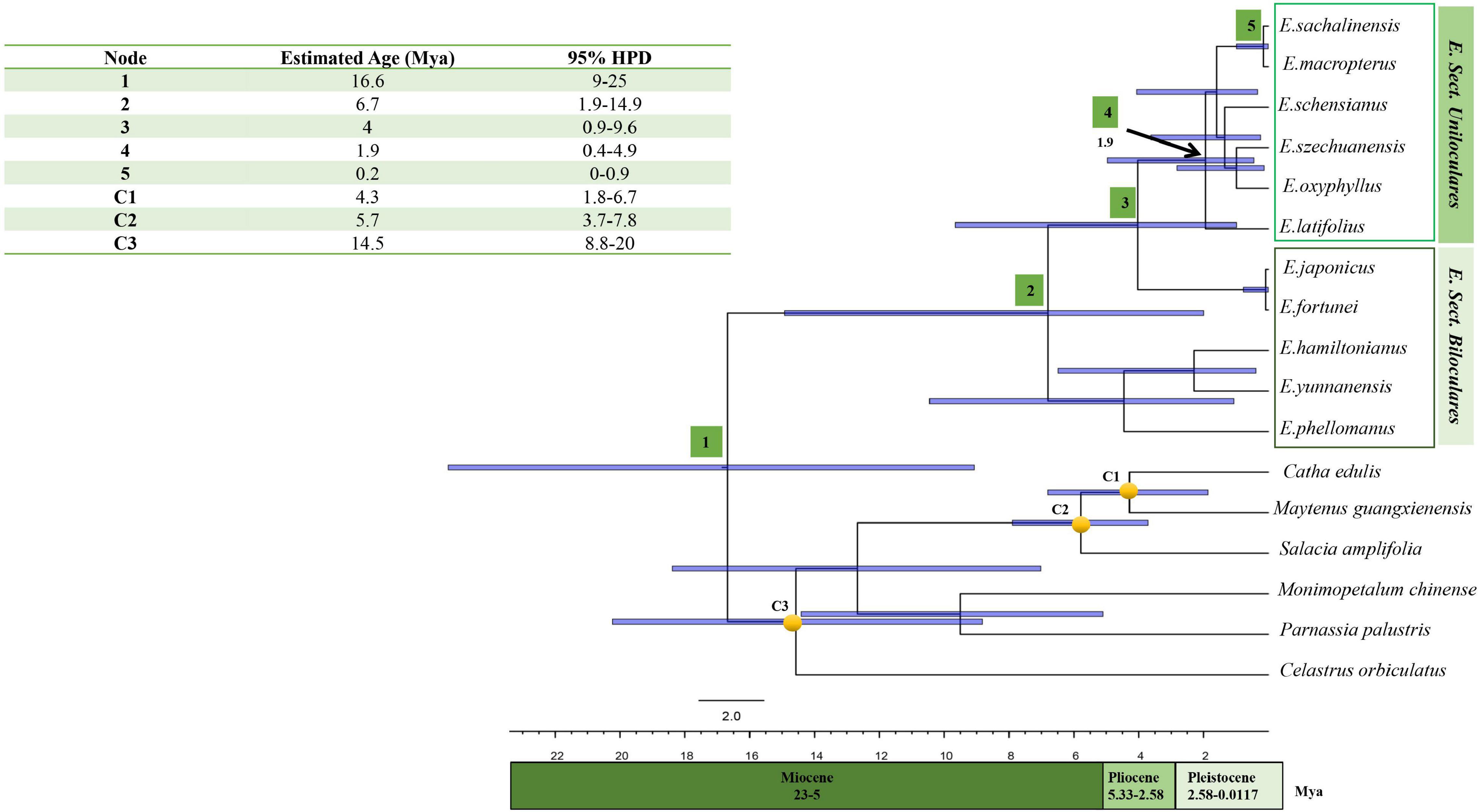
Figure 7. Chronogram depicting the divergence time estimated in BEAST using chloroplast coding sequences. Approximately 95% posterior probability is represented by the bar on each node. Nodes 1–5 are the nodes of interests. The calibration points utilised in the analysis were represented by nodes labelled as C1, C2, and C3.
Our time-of-divergence estimate indicated that extant members of Euonymus diverged from other members of the Celastraceae approximately 16.6 Mya (Node 1; 95% HPD = 9–25 Mya), locating it within the Middle Miocene era. The crown age of the clade comprising E. japonicus, E. fortunei, and members of sect. Uniloculares was estimated to have diverged approximately 6.7 Mya (Node 2; 95% HPD = 1.9–14.4 Mya). The crown age of sect. Uniloculares was estimated to have occurred approximately 4.0 Mya (Node 3; 95% HPD = 0.9–9.6 Mya) during the Pliocene era. With respect to the divergence of E. latifolius, the western Asian representative of sect. Uniloculares, the estimated crown age of E. sect. Uniloculares was approximately 1.9 Mya (Node 4; 95% HPD = 0.4–4.9 Mya). Species in sect. Uniloculares diverged during the Pleistocene era. The clade was estimated to be young and to have diverged from ancestral taxa with two-celled anthers (Biloculares) (Figure 7).
A biogeographical evaluation was conducted in RASP using the Bayesian binary method analysis. This assessment indicated that sect. Uniloculares originated in East Asia (Node 29, relative probability = 0.6). A vicariance event was also observed at Node 29. Euonymus schenisanus and E. szechuanensis are endemic to China (East Asia; A), whereas E. macropeterus, E. sachalinensis, and E. oxyphyllus also occur in the Russian Far East (East Asia–Russian Far East; AB). Only E. latifolius occurs in western Asia, south, southeast, and Central Europe, and North Africa (=CDE), making its distribution opposite to the East Asian sister clade (Figure 8).
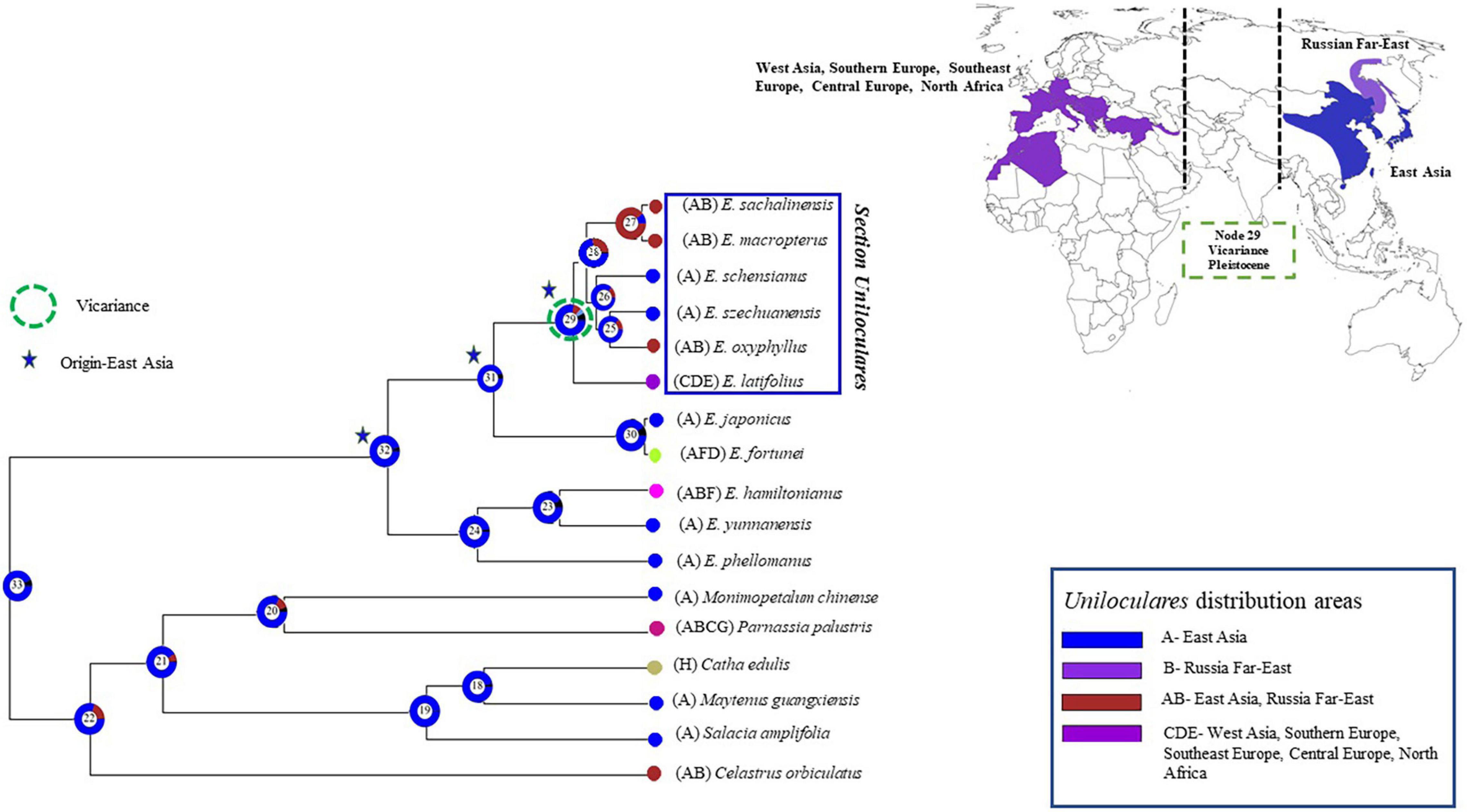
Figure 8. The Bayesian binary method (BBM) was used to analyse the biogeography of Euonymus sect. Uniloculares. The vicariance between East and West Asia is highlighted on the map to the right. The figure highlights four primary areas of interest. The Supplementary Table 4 contains the detail of each node’s events.
In the Uniloculares clade, four dispersal events occurred at Node 29, two at Nodes 27 and 25, and one at Node 28. No extinctions were observed (Figure 9).
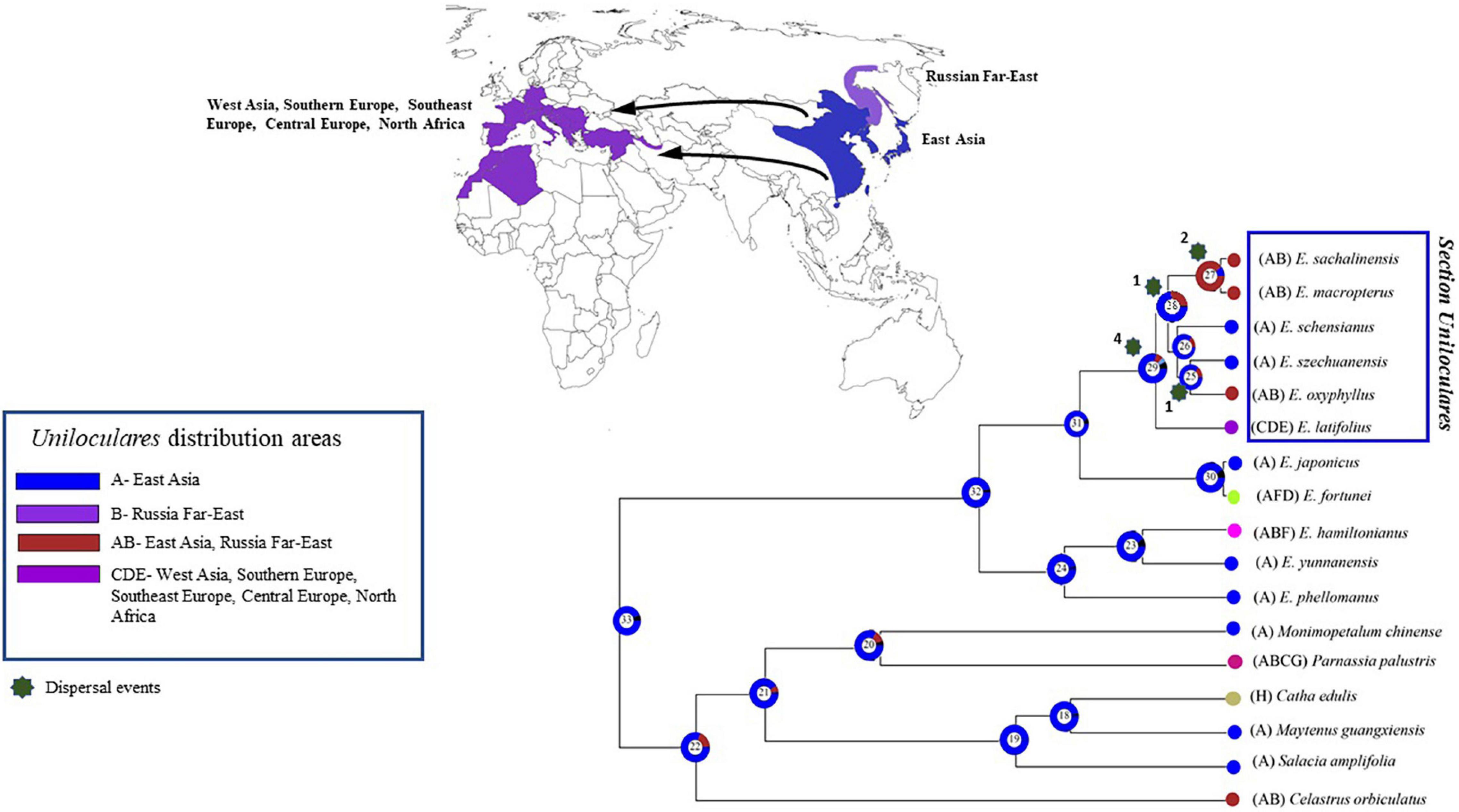
Figure 9. Figure illustrating the dispersal events. Number above star indicates the number of times of dispersal in the sect. Uniloculares clade. Node 29 is the main node of interest. Arrows on the map point the dispersal from East to West Asia. The Supplementary Table 4 contains the detail of each node’s events.
Pseudogenisation of the ycf1 gene in IRb is a common phenomenon in most cp genomes, but the loss of function in other coding genes is more intriguing. The first whole cp genome sequencing in Euonymus was the sequencing of E. japonicus according to the study by Choi and Park (2016), who observed pseudogenisation of ndhF. Pseudogenisation of ndhF has also been reported in the genomes of several other plant species, including Saniculiphyllum sp. (Saxifragaceae) (Folk et al., 2020), Zoysia sp., Sporobolus sp. (Poaceae) (Cheon et al., 2021), and some members of the Orchidaceae (Zavala-Páez et al., 2020). Intron loss in rps16 has been confirmed in eight members of the Celastraceae and was successfully detected by primers designed to identify such losses (Gu et al., 2018). Pseudogenenisation of rps16 has been reported in Gentiana straminea (Ni et al., 2016), Gentiana crassicaulis, and Gentiana robusta (Ni et al., 2017), as well as in Hepatica sp. (Li et al., 2021), Populus sp., Veratrum sp., non-parasitic members of the Fabaceae, and members of the Orchidaceae (Nagano et al., 1991; Saski et al., 2005; Tuskan et al., 2006; Ni et al., 2016).
Euonymus is difficult to identify at the species level because the flowers are not showy and shed shortly after blooming, complicating the task of identifying specimens based on reproductive structures (Du et al., 2016). As a result, species identifications are disputed, and taxonomists are currently attempting to resolve the debate using genetic data. Wang et al. (2020) constructed phylogenetic trees based on whole cp genomes, in which E. schensianus and E. szechuanensis were grouped together in a clade with 100% resolution. E. japonicus was grouped in a subclade with the latter, closely approximating our results. According to the study by Simmons et al. (2012), who analysed matK, trnL-F, ITS, and 26S rDNA, sect. Uniloculares (=subgenus Kalonymus) was placed in a subclade with sect. Illicifolia, which includes E. japonicus and E. fortunei.
We propose that the diversification of the sect. Uniloculares clade occurred in East Asia in the late Pliocene, approximately 4 Mya (Node 3) (Figure 7). In their study, Wang et al. (2019) confirmed that several climatic changes occurred in East Asia throughout the Pliocene epoch, including an unanticipated change in monsoon cycles approximately 4.2 Mya due to high-latitude cooling.
The mid-Pliocene warm period occurs between 3.264 and 3.025 Mya, during which global temperatures are considered warmer than current temperatures, whereas paleogeography appears to be modern. These warm ages are assumed to share many parallels with the global warming and carbon dioxide levels (Meng and Kubatko, 2009) observed throughout the past century, with global temperature estimates ranging from 1.8 to 3.6°C (Dowsett et al., 2010; Haywood et al., 2013; Zhang et al., 2013).
The evolution and diversification of the Asian flora can be traced back to the formation of the Gobi Desert, which is situated in an arid region of Asia. This change is considered to have occurred 2.6 Mya and may be attributable to changes in geography and climate. Other potential causal factors include geological deformation and changes in the Asian monsoon due to global warming (Lu et al., 2019). The desert could potentially limit gene flow. Global climatic oscillations occurred approximately 2.0 Mya, during the Pleistocene epoch, causing environmental changes that led to the development and migration of numerous species in the Northern Hemisphere (Hewitt, 2000; Hewitt, 2004; Qiu et al., 2011).
The diversification of species in sect. Uniloculares is estimated to have occurred between 0.9 and 1.9 Mya and appears to have originated in East Asia. E. latifolius, the western Asian representative, diverged approximately 1.9 Mya, indicating that the clade diverged recently. Species that diverged around the same time as the East Asian clade of sect. Uniloculares due to phylogeographic segregation in East Asia include Platycrater arguta (Hydrangeaceae; 0.89 Mya) (Qiu et al., 2009b), Kirengeshoma koreana and Kirengeshoma palmata (Hydrangeaceae; 0.45 and 0.16 Mya, respectively) (Qiu et al., 2009c), and Dysosma versipellis (Berberidaceae; 0.4 Mya) (Qiu et al., 2009a). According to the study by Wadia (1955), available evidence implies that desertification occurred after glacial events. There were seven or eight well-established oscillations during the Pleistocene ice age. It is thought that such fluctuations, e.g., from cold to warm temperatures, occurred approximately 10,000–15,000 years ago.
In recent years, phylogeographic studies have investigated the connection between the evolution of organisms and geological uplift. Mountainous uplifts result in the formation of geographical barriers, leading to habitat fragmentation, loss of dispersal corridors, and restricted gene flow (Luo et al., 2016). Due to previous climatic changes, eastern and western Asia became colonised by numerous trees species (Wolfe, 1975; Qiu et al., 2011; Gratzfeld, 2013). Song et al. (2020) discussed disjunction and vicariance between eastern and western Asia using Petrocarya spp. (Juglandaceae) as a model species and posited the formation of the Gobi Desert as a factor in the disjunction between eastern and western Asia. Woody taxa such as Fagus (Fagaceae), Acer (Sapindaceae), Buxus (Buxaceae), Sorbus (Rosaceae), and Parrotia (Hamamelidaceae) are reported to have similar distribution patterns in eastern and western Asia (Ickert-Bond et al., 2005; Li and Tredici, 2008; Jia and Bartish, 2018; Kozlowski et al., 2018).
Our study provides an evolutionary framework for evaluating the biogeographic history of Euonymus sect. Uniloculares. The monophyly of sect. Uniloculares was supported based on analyses of whole cp genomes. We posit that East Asia is the centre of origin of sect. Uniloculares. Temperature fluctuations, desertification, and changes in rainfall cycles may have played a role in the disjunction by disrupting gene flow between regions. Scientific exploration and experimentation are ongoing, and more studies should be conducted on the phylogenomics, evolution, demographic history, and biogeography of Euonymus, as many questions about the genus remain unanswered.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession numbers OL770076-OL770079.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This study was supported by grants received from the Korean National Arboretum (Grant no. KNA1-1-13,14-1) and the National Research Foundation of Korea (NRF) (NRF-2017R1D1A1B06029326).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The leaf samples and herbarium record scans for E. latifolius were contributed by Nikolay Aleksandrovich Trusov, a senior researcher in the Laboratory of Dendrology, and Igor Olegovich Yatsenko, a researcher in the Laboratory of Dendrology, Tsytsin Main Botanical Garden, RAS, Moscow, Russia.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.825209/full#supplementary-material
BI, Bayesian inference; BBM, Bayesian binary method; Bp, base pairs; Cp, chloroplast; HPD, highest posterior density; IR, inverted repeat; LSC, large single copy; MCMC, Markov Chain Monte Carlo; Mya, million years ago; ML, maximum likelihood; MP, maximum parsimony; NCBI, National Centre for Biotechnology Information; sect., section; st. dev., standard deviation; sp., species; SSC, small single copy.
Agea, D., García De Lucas, S., and Lorite, J. (2021). Regeneration of submediterranean species Euonymus latifolius (L.) Mill. at its southernmost limit in Europe. Mediterr. Bot. 42:e68137. doi: 10.5209/mbot.68137
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Bacon, C. D., Simmons, M. P., Archer, R. H., Zhao, L. C., and Andriantiana, J. (2016). Biogeography of the Malagasy Celastraceae: multiple independent origins followed by widespread dispersal of genera from Madagascar. Mol. Phylogenet. Evol. 94, 365–382. doi: 10.1016/j.ympev.2015.09.013
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Meth. Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Cheng, Y., Zhang, L., Qi, J., and Zhang, L. (2020). Complete chloroplast genome sequence of Hibiscus cannabinus and comparative analysis of the Malvaceae family. Front. Genet. 11:227. doi: 10.3389/fgene.2020.00227
Cheon, S. H., Woo, M. A., Jo, S., Kim, Y. K., and Kim, K. J. (2021). The Chloroplast Phylogenomics and Systematics of Zoysia (Poaceae). Plants 10:1517. doi: 10.3390/plants10081517
Choi, K. S., and Park, S. (2016). The complete chloroplast genome sequence of Euonymus japonicus (Celastraceae). Mitochondrial DNA A 27, 3577–3578. doi: 10.3109/19401736.2015.1075127
Costa, L., Jimenez, H., Carvalho, R., Carvalho-Sobrinho, J., Escobar, I., and Souza, G. (2020). Divide to conquer: evolutionary history of Allioideae Tribes (Amaryllidaceae) is linked to distinct trends of karyotype evolution. Front. Plant Sci. 11:320. doi: 10.3389/fpls.2020.00320
Cox, C. B., Moore, P. D., and Ladle, R. J. (2016). Biogeography: An Ecological and Evolutionary Approach. New York: John Wiley & Sons.
Deng, T., Nie, Z. L., Drew, B. T., Volis, S., Kim, C., Xiang, C. L., et al. (2015). Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of Northern Hemisphere herbaceous plants? The case of Meehania (Lamiaceae). PLoS One 10:e0117171. doi: 10.1371/journal.pone.0117171
Dowsett, H., Robinson, M., Haywood, A., Salzmann, U., Hill, D., Sohl, L., et al. (2010). The PRISM3D paleoenvironmental reconstruction. Stratigraphy 7, 123–139.
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Du, C., Zuo, Y., Wang, Y., Liu, Q., and Ma, J. (2016). Revision of seven species of Euonymus (Celastraceae) from East and South Asia. Phytotaxa 288, 19–31. doi: 10.11646/phytotaxa.288.1.2
Folk, R. A., Sewnath, N., Xiang, C. L., Sinn, B. T., and Guralnick, R. P. (2020). Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae). BMC Plant Biol. 20:324. doi: 10.1186/s12870-020-02533-x
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Gavrilova, O. A., Britski, D. A., Grigorieva, V. V., Tarasevich, V. F., Pozhidaev, A. E., and Leunova, V. M. (2018). Pollen morphology of the genus Euonymus (Celastraceae). Turczaninowia 21, 188–206.
Gernhard, T. (2008). The conditioned reconstructed process. J. Theor. Biol. 253, 769–778. doi: 10.1016/j.jtbi.2008.04.005
Goulding, S. E., Wolfe, K., Olmstead, R., and Morden, C. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 252, 195–206. doi: 10.1007/BF02173220
Gratzfeld, G. K. J. (2013). Zelkova–an Ancient Tree. Global Status and Conservation Action. Fribourg: Natural History Museum.
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Gu, C., Tembrock, L. R., Zheng, S., and Wu, Z. (2018). The complete chloroplast genome of Catha edulis: a comparative analysis of genome features with related species. Int. J. Mol. Sci. 19:525. doi: 10.3390/ijms19020525
Haywood, A. M., Hill, D., Dolan, A., Otto-Bliesner, B. L., Bragg, F., Chan, W. L., et al. (2013). Large-scale features of Pliocene climate: results from the Pliocene Model Intercomparison Project. Clim. Past 9, 191–209.
Hewitt, G. (2000). The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. Ser. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
Ickert-Bond, S. M., and Wen, J. (2006). Phylogeny and biogeography of Altingiaceae: evidence from combined analysis of five non-coding chloroplast regions. Mol. Phylogenet. Evol. 39, 512–528. doi: 10.1016/j.ympev.2005.12.003
Ickert-Bond, S. M., Pigg, K. B., and Wen, J. (2005). Comparative infructescence morphology in Liquidambar (Altingiaceae) and its evolutionary significance. Am. J. Bot. 92, 1234–1255. doi: 10.3732/ajb.92.8.1234
Jia, D. R., and Bartish, I. V. (2018). Climatic changes and Orogeneses in the late Miocene of Eurasia: the main triggers of an expansion at a continental scale? Front. Plant Sci. 9:1400. doi: 10.3389/fpls.2018.01400
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, C., Cameron, K. M., and Kim, J. H. (2017). Molecular systematics and historical biogeography of Maianthemum ss. Am. J. Bot. 104, 939–952. doi: 10.3732/ajb.1600454
Kim, C., Deng, T., Wen, J., Nie, Z. L., and Sun, H. (2015). Systematics, biogeography, and character evolution of Deutzia (Hydrangeaceae) inferred from nuclear and chloroplast DNA sequences. Mol. Phylogenet. Evol. 87, 91–104. doi: 10.1016/j.ympev.2015.03.002
Kozlowski, G., Song, Y. G., Bétrisey, S., Alvarado, E. V., and Bétrisey, S. (2018). Wingnuts (” Pterocarya”) & Walnut Family: Relict Trees: Linking the Past, Present and Future. Fribourg: Natural History Museum.
Lee, H. L., Jansen, R. K., Chumley, T. W., and Kim, K. J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Lee, S. R., Kim, K., Lee, B. Y., and Lim, C. E. (2019). Complete chloroplast genomes of all six Hosta species occurring in Korea: molecular structures, comparative, and phylogenetic analyses. BMC Genom. 20:833. doi: 10.1186/s12864-019-6215-y
Leonova, T. G. (1974). The Genus Euonymus L. of the USSR and Neighbouring Countries. Leningrad: Nauka.
Li, J., and Tredici, P. (2008). The Chinese Parrotia: a sibling species of the Persian Parrotia. Arnoldia 66, 2–9.
Li, Y., Dong, Y., Liu, Y., Yu, X., Yang, M., and Huang, Y. (2021). Comparative analyses of Euonymus chloroplast genomes: genetic structure, screening for loci with suitable polymorphism, positive selection genes, and phylogenetic relationships within Celastrineae. Front. Plant Sci. 11:2307. doi: 10.3389/fpls.2020.593984
Liang, H., Zhang, Y., Deng, J., Gao, G., Ding, C., Zhang, L., et al. (2020). The complete chloroplast genome sequences of 14 Curcuma species: insights into genome evolution and phylogenetic relationships within zingiberales. Front. Genet. 11:802. doi: 10.3389/fgene.2020.00802
Liao, W. J., Yuan, Y. M., and Zhang, D. Y. (2007). Biogeography and evolution of flower color in Veratrum (Melanthiaceae) through inference of a phylogeny based on multiple DNA markers. Plant Syst. Evol. 267, 177–190.
Lu, H., Wang, X., Wang, X., Chang, X., Zhang, H., Xu, Z., et al. (2019). Formation and evolution of Gobi Desert in central and eastern Asia. Earth Sci. Rev. 194, 251–263.
Luo, D., Yue, J. P., Sun, W. G., Xu, B., Li, Z. M., Comes, H. P., et al. (2016). Evolutionary history of the subnival flora of the Himalaya-Hengduan Mountains: first insights from comparative phylogeography of four perennial herbs. J. Biogeogr. 43, 31–43.
Ma, J., and Funston, A. (2008). “Euonymus,” in Flora of China 11, eds Z. Y. Wu, P. H. Raven, and D. Y. Hong (St. Louis: Science Press, Beijing & Missouri Botanical Garden Press), 440–463.
Mao, K., Hao, G., Liu, J., Adams, R. P., and Milne, R. I. (2010). Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytol. 188, 254–272. doi: 10.1111/j.1469-8137.2010.03351.x
McNeill, J., Barrie, F., Buck, W., Demoulin, V., Greuter, W., Hawksworth, D., et al. (2012). International Code of Nomenclature for Algae, Fungi and Plants (Melbourne Code). Königstein: Koeltz Scientific Books.
Meng, C., and Kubatko, L. S. (2009). Detecting hybrid speciation in the presence of incomplete lineage sorting using gene tree incongruence: a model. Theor. Popul. Biol. 75, 35–45. doi: 10.1016/j.tpb.2008.10.004
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G., and Soltis, D. E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A. 107, 4623–4628. doi: 10.1073/pnas.0907801107
Nagano, Y., Matsuno, R., and Sasaki, Y. (1991). Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr. Genet. 20, 431–436. doi: 10.1007/BF00317074
Namgung, J., Do, H. D. K., Kim, C., Choi, H. J., and Kim, J. H. (2021). Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae). Sci. Rep. 11:3262. doi: 10.1038/s41598-021-82692-5
Neuhaus, H., and Emes, M. (2000). Nonphotosynthetic metabolism in plastids. Ann. Rev. Plant Biol. 51, 111–140. doi: 10.1146/annurev.arplant.51.1.111
Ni, L., Zhao, Z., Xu, H., Chen, S., and Dorje, G. (2016). The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 577, 281–288. doi: 10.1016/j.gene.2015.12.005
Ni, L., Zhao, Z., Xu, H., Chen, S., and Dorje, G. (2017). Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr. Genet. 63, 241–252. doi: 10.1007/s00294-016-0631-1
Nie, Z. L., Wen, J., Azuma, H., Qiu, Y. L., Sun, H., Meng, Y., et al. (2008). Phylogenetic and biogeographic complexity of Magnoliaceae in the Northern Hemisphere inferred from three nuclear data sets. Mol. Phylogenet. Evol. 48, 1027–1040. doi: 10.1016/j.ympev.2008.06.004
Palmer, J. D., Jansen, R. K., Michaels, H. J., Chase, M. W., andManhart, J. R. (1988). Chloroplast DNA variation and plant phylogeny. Ann. Missouri Bot. Gard. 75, 1180–1206. doi: 10.2307/2399279
POWO. (2019). Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Richmond: Royal Botanic Gardens, Kew.
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Qiu, Y. X., Qi, X. S., Jin, X. F., Tao, X. Y., Fu, C. X., Naiki, A., et al. (2009b). Population genetic structure, phylogeography, and demographic history of Platycrater arguta (Hydrangeaceae) endemic to East China and South Japan, inferred from chloroplast DNA sequence variation. Taxon 58, 1226–1241.
Qiu, Y. X., Sun, Y., Zhang, X. P., Lee, J., Fu, C. X., and Comes, H. P. (2009c). Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to Quaternary climate change and landbridge configurations. New Phytol. 183, 480–495. doi: 10.1111/j.1469-8137.2009.02876.x
Qiu, Y. X., Guan, B. C., Fu, C. X., and Comes, H. P. (2009a). Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol. Phylogenet. Evol. 51, 281–293. doi: 10.1016/j.ympev.2009.01.016
Rambaut, A., and Drummond, A. (2003). Tracer: MCMC Trace Analysis Tool. Oxford: University of Oxford.
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saski, C., Lee, S. B., Daniell, H., Wood, T. C., Tomkins, J., Kim, H. G., et al. (2005). Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59, 309–322. doi: 10.1007/s11103-005-8882-0
Savinov, I., and Baikov, K. (2007). The analysis of phylogenetic relations in the genus Euonymus (Celastraceae R. Br.) using SYNAP method. Turczaninowia 10, 3–4.
Simmons, M. P., Mckenna, M. J., Bacon, C. D., Yakobson, K., Cappa, J. J., Archer, R. H., et al. (2012). Phylogeny of Celastraceae tribe Euonymeae inferred from morphological characters and nuclear and plastid genes. Mol. Phylogenet. Evol. 62, 9–20. doi: 10.1016/j.ympev.2011.08.022
Song, Y. G., Li, Y., Meng, H. H., Fragnière, Y., Ge, B. J., Sakio, H., et al. (2020). Phylogeny, taxonomy, and biogeography of Pterocarya (Juglandaceae). Plants 9:1524. doi: 10.3390/plants9111524
Trifinopoulos, J., Nguyen, L. T., Von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604.
Wadia, D. N. (1955). Deserts of Asia-Their Origin and Growth in the Late Pleistocene Time. Lucknow: Birbal Sahni Institute Of Palaeobotany.
Wang, H., Lu, H., Zhao, L., Zhang, H., Lei, F., and Wang, Y. (2019). Asian monsoon rainfall variation during the Pliocene forced by global temperature change. Nat. Commun. 10:5272. doi: 10.1038/s41467-019-13338-4
Wang, W. C., Chen, S. Y., and Zhang, X. Z. (2017). Characterization of the complete chloroplast genome of the golden crane butterfly, Euonymus schensianus (Celastraceae). Conserv. Genet. Resour. 9, 545–547. doi: 10.1007/s12686-017-0719-z
Wang, X., Li, H., Zheng, M., and Jiang, J. (2020). The complete chloroplast genome of Euonymus szechuanensis. Mitochondrial DNA B 5, 1130–1131. doi: 10.1080/23802359.2019.1698345
Wen, J. (1999). Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Ann. Rev. Ecol. Syst. 30, 421–455. doi: 10.1146/annurev.ecolsys.30.1.421
Wen, J. (2001). Evolution of eastern Asian–eastern North American biogeographic disjunctions: a few additional issues. Int. J. Plant Sci. 162, S117–S122.
Wen, J., and Ickert-Bond, S. M. (2009). Evolution of the Madrean–Tethyan disjunctions and the North and South American amphitropical disjunctions in plants. J. Syst. Evol. 47, 331–348. doi: 10.1111/j.1759-6831.2009.00054.x
Wilgenbusch, J. C., and Swofford, D. (2003). Inferring evolutionary trees with PAUP. Curr. Protoc. Bioinform. 6.4, 1–6.4.28. doi: 10.1002/0471250953.bi0604s00
Wolfe, J. A. (1975). Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Ann. Missouri Bot. Gard. 62, 264–279. doi: 10.2307/2395198
Xia, M., Zhang, F., Rao, H., Chi, X., Khan, G., Zhang, Y., et al. (2018). Complete chloroplast genome sequence of Parnassia brevistyla (Celastraceae) and phylogenetic analysis with related species. Mitochondrial DNA B 3, 1187–1188. doi: 10.1080/23802359.2018.1524725
Xiang, Q. Y., Soltis, D. E., and Soltis, P. S. (1998). The eastern Asian and eastern and western North American floristic disjunction: congruent phylogenetic patterns in seven diverse genera. Mol. Phylogenet. Evol. 10, 178–190. doi: 10.1006/mpev.1998.0524
Xiang, Q. Y., Soltis, D. E., Soltis, P. S., Manchester, S. R., and Crawford, D. J. (2000). Timing the eastern Asian–eastern North American floristic disjunction: molecular clock corroborates paleontological estimates. Mol. Phylogenet. Evol. 15, 462–472. doi: 10.1006/mpev.2000.0766
Yao, C., Zuo, Y., Du, C., and Ma, J. (2018). Morphological study of floral nectaries in Euonymus and the probable origin of the echinate fruit surface. Plant Divers. 40, 35–40. doi: 10.1016/j.pld.2017.12.004
Yu, Y., Harris, A., and He, X. (2010). S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. doi: 10.1016/j.ympev.2010.04.011
Yu, Y., Harris, A., and He, X. (2013). RASP (Reconstruct Ancestral State in Phylogenies), version 2.0. Availabe online at: http://mnh.scu.edu.cn/soft/blog/RASP (accessed September 16, 2021).
Zavala-Páez, M., Vieira, L. D. N., Baura, V. A. D., Balsanelli, E., Souza, E. M. D., Cevallos, M. C., et al. (2020). Comparative Plastid Genomics of Neotropical Bulbophyllum (Orchidaceae; Epidendroideae). Front. Plant Sci. 11:799. doi: 10.3389/fpls.2020.00799
Keywords: chloroplast genome, divergence, dispersal, disjunction, East Asia, E. sect. Uniloculares, vicariance, West Asia
Citation: Jamshed S and Kim J-H (2022) Disjunction and Vicariance Between East and West Asia: A Case Study on Euonymus sect. Uniloculares Based on Plastid Genome Analysis. Front. Plant Sci. 13:825209. doi: 10.3389/fpls.2022.825209
Received: 30 November 2021; Accepted: 11 February 2022;
Published: 11 March 2022.
Edited by:
Robert Philipp Wagensommer, University of Bari Aldo Moro, ItalyCopyright © 2022 Jamshed and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joo-Hwan Kim, a2ltamgyMDA5QGdhY2hvbi5hYy5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.