
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 February 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.824358
This article is part of the Research TopicStructure and Function of Chloroplasts, Volume IIIView all 13 articles
Chloroplast thylakoid protein rubredoxin 1 (RBD1) in Chlamydomonas and its cyanobacterial homolog RubA contain a rubredoxin domain. These proteins have been proposed to participate in the assembly of photosystem II (PSII) at early stages. However, the effects of inactivation of RBD1 on PSII assembly in higher plants are largely unclear. Here, we characterized an Arabidopsis rbd1 mutant in detail. A drastic reduction of intact PSII complex but relatively higher levels of assembly intermediates including PSII RC, pre-CP47, and pre-CP43 were found in rbd1. Polysome association and ribosome profiling revealed that ribosome recruitment of psbA mRNA is specifically reduced. Consistently, in vivo protein pulse-chase labeling showed that the rate of D1/pD1 synthesis is significantly reduced in rbd1 compared with WT. Moreover, newly synthesized mature D1 and pD1/D2 can assemble into the PSII reaction center (RC) complex but further formation of larger PSII complexes is nearly totally blocked in rbd1. Our data imply that RBD1 is not only required for the formation of a functional PSII core complex during the early stages of PSII assembly but may also be involved in the translation of D1 in higher plants.
Photosystem II (PSII) is the primary pigment-protein complex in the photosynthetic electron transport chain in oxygenic organisms. PSII harvests light energy to split water resulting in the release of oxygen and electrons that are transferred from water to plastoquinone in cyanobacteria, algae, and plants. PSII drives oxygenic photosynthesis together with PSI (Photosystem I), Cyt b6f (Cytochrome b6f), and ATP synthase, and provides energy and oxygen for most living organisms on earth (Nelson and Junge, 2015; Shen, 2015). In higher plants, PSII is composed of the PSII reaction center (RC), CP43 and CP47, the oxygen-evolving complex (OEC), and LHCII as well as several small subunits with a low molecular mass. PSII RC is the minimum unit for the primary photochemical reaction and is composed of the subunits D1, D2, PsbI, and two subunits of cytochrome b559 (PsbE and PsbF) (Nelson and Yocum, 2006). The D1 and D2 proteins form a heterodimer and bind the basal redox components required for PSII electron transport such as P680, pheophytin, non-heme iron and quinone (Umena et al., 2011). CP43 and CP47 are peripheral core antenna subunits attached to the PSII RC. They function in energy transfer from light harvesting complex (LHCII) to the PSII reaction center. As the catalytic site for water splitting, OEC is composed of one Ca atom, four Mn atoms, and five oxygen atoms (Mn4CaO5 cluster). Seven amino acid residues from D1 and CP43 provide ligands to OEC (Umena et al., 2011; Shen, 2015). PsbO, PsbP, and PsbQ are attached to the lumen side of PSII. Together with CP43, these proteins provide protection and contribute to access channels for substrate and products during water oxidation. LHCII proteins bind to PSII forming the PSII-LHCII complex which in turn forms PSII-LHCII supercomplexes in the thylakoid membrane.
Assembly of PSII in chloroplasts occurs in a stepwise manner (Baena-González and Aro, 2002; Rokka et al., 2005; Mulo et al., 2008; Nixon et al., 2010; Komenda et al., 2012; Lyska et al., 2013; Nickelsen and Rengstl, 2013). First, the precursor of D1 (pD1) is co-translationally assembled into the D2-Cyt b559 receptor complex to form the PSII RC with the PsbI subunit. The C-terminal 9–13 residues of pD1 are subsequently removed by the CtpA enzyme to form functional D1 protein (Che et al., 2013). Second, CP47 is integrated into the RC to form the CP47-RC complex. The chloroplast-encoded proteins PsbH, PsbM, PsbTc, and the nucleus-encoded protein PsbR are assembled into the CP47-RC complex to form CP43-less PSII. In the next step, CP43 and other subunits are integrated to form the PSII monomer. Finally, the PsbO, PsbP, and PsbQ subunits associate with the PSII monomer and form supercomplexes with LHCII eventually with the help of PsbJ and PsbZ (Rokka et al., 2005).
More than a dozen PSII assembly factors are required for the PSII assembly at various steps in higher plants. For the biogenesis of the PSII RC complex, several facilitating factors are involved in particular steps such as psbA translation, pD1 processing, and assembly of the complex. HCF173 is a protein weakly related to short-chain dehydrogenases/reductases. It forms a high molecular weight complex that both facilitates translational initiation of D1 and stabilizes the psbA mRNA (Schult et al., 2007). After translation, the nine residue C-terminal tail of pD1 is cleaved by the lumen-localized C-terminal processing protease CtpA (Che et al., 2013). Two One-Helix-Proteins OHP1 and OHP2 as well as the HCF244 protein form a complex essential for the formation of the PSII RC complex. Formation of the OHP1/OHP2/HCF244 complex is likely required for the efficient recruitment of ribosomes with psbA mRNA encoding D1 and also for the delivery of chlorophyll and/or other cofactors to the D1/D2 heterodimer (Beck et al., 2017; Hey and Grimm, 2018; Li et al., 2018; Myouga et al., 2018; Chotewutmontri et al., 2020). In addition, thylakoid lumen protein HCF136 is essential for PSII RC formation (Meurer et al., 1998; Plücken et al., 2002). YCF48, the ortholog of HCF136 in cyanobacteria and red algae, is a seven-bladed beta-propeller that promotes efficient incorporation of D1 into the PSII RC complex by interacting with newly synthesized pD1 on the thylakoid lumen side (Komenda et al., 2008; Yu et al., 2018). Chloroplast-encoded PsbN is not a PSII subunit but rather acts as a molecular chaperone required in the early phase of PSII RC formation (Torabi et al., 2014).
Rubredoxins are iron-containing proteins in which one iron atom is coordinated by four cysteine residues, and they participate in electron transfer reactions (Lovenberg and Sobel, 1965; Adman et al., 1975). RBD1 (Rubredoxin 1) from Chlamydomonas and its ortholog from cyanobacteria RubA contain one rubredoxin domain and one transmembrane domain. Loss of RBD1 in photosynthetic eukaryotes impacts the functional accumulation of PSII (Calderon et al., 2013). Further studies demonstrated that they may participate in PSII RC formation by participating together with cyt b559 to protect PSII assembly intermediates from photooxidation or maintaining PSII co-factors in a suitable redox state to prevent photodamage (García-Cerdán et al., 2019; Kiss et al., 2019).
In this study, we characterized the rbd1 mutant of Arabidopsis. We demonstrate that ribosome recruitment of psbA mRNA encoding the D1 protein is specifically reduced in rbd1, suggesting that RBD1 may act as a regulatory factor involving D1 translation. The PSII reaction center accumulates in higher amounts in rbd1 but further association with other PSII assembly intermediates to form the larger PSII complex and supercomplexes is nearly totally blocked. These data imply that Arabidopsis RBD1, similarly to the orthologs found in Chlamydomonas and cyanobacteria, exerts a role in the formation of a functional PSII core complex during the early stages of PSII assembly.
The T-DNA insertion mutant rbd1 (WiscDsLoxHs187_05C) was obtained from Nottingham Arabidopsis Stock Center (NASC). Position of the T-DNA insertion in rbd1 was confirmed by PCR using primers in Supplementary Table 1 and subsequent sequencing of the PCR products. Wild type (Columbia, Col-0), rbd1, and rbd1-com plants were grown on Murashige and Skoog (MS) basal medium containing 3% sucrose and 0.05% (w/v) MES (2-morpholinoethanesulfonic acid) (pH 5.8) under a 16-h photoperiod with 50 μmol photons m–2 s–1 at 23°C in a greenhouse.
For complementation analysis, a genomic DNA fragment containing RBD1 and its promoter (750 bp upstream of transcription initiation site) was amplified by PCR using primers RBD1-1301-F and RBD1-1301-R (Supplementary Table 1). PCR products were then cloned into the pCAMBIA1301 vector. The resulting vector was then transferred into Agrobacterium tumefaciens C58C and transformed into heterozygous rbd1 plants by the floral dipping method (Clough and Bent, 1998).
Images of chlorophyll a fluorescence were captured with a MAXI-IMAGING-PAM chlorophyll fluorometer (Walz, Germany) with default parameters. Plants were first kept in darkness for 20 min and the minimum fluorescence (Fo) was measured by turning on the measuring light (ML). Maximum fluorescence (Fm) was then determined with a saturating pulse (SP). The maximum quantum yield of PSII was calculated as Fv/Fm = (Fm-Fo)/Fm. Images of Fo and Fv/Fm were displayed using a false color scale from 0 to 1.
Transients of Chlorophyll a fluorescence were monitored using a PAM-2500 chlorophyll fluorometer (Walz, Germany). Plants were dark adapted for 20 min. Then Fo and Fm were determined with the measuring light (ML, red light, intensity 2) or a saturating pulse (SP, red light, intensity 10, 0.6 s), respectively. Then, the leaves were illuminated with actinic light (AL, red light, 64 μmol photons m–2 s–1) for 4 min and the steady-state fluorescence (Ft) was recorded.
The sequence encoding the soluble part of RBD1 protein (amino acid residues 13–174) was amplified by PCR from cDNA with primers of RBD1-ab-F and RBD1-ab-R (Supplementary Table 1). The PCR products were cloned into the pET28a expression vector (Novagen, United States) by double enzyme digestion and the reconstructed vector was transformed into E. coli Rosetta competent cells for expression of the recombinant protein. After induction with 0.5 mM IPTG for 4 h at 37°C, the cells were harvested and recombinant protein was purified from the supernatant on a Ni-NTA agarose column (Qiagen). Polyclonal antibody against RBD1 was raised in rabbits.
Antibodies of D1 (PHY0057, 1:2,000), D2 (PHY0060, 1:2,000), CP43 (PHY0318, 1:5,000), CP47 (PHY0319, 1:2,000), PsbE (PHY0506A, 1:5,000), PsbI (PHY0132A, 1:1,000), PsbO (PHY0344, 1:2,000), PsaA (PHY0342, 1:2,000), PsaD (PHY0343, 1:5,000), Cyt b6 (PHY0020S, 1:1,000), CF1γ (PHY0313, 1:1,000), CF1ε (PHY0314, 1:1,000), and SBPase (PHY0410S, 1:1,000) were obtained from PhytoAB.
Chloroplast thylakoids were isolated according to Zhang et al. (2016). Briefly, leaves of 4-week-old plants grown on MS medium were homogenized in isolation buffer I (0.33 M sorbitol, 30 mM Tricine/KOH, pH 8.4, 10 mM NaHCO3, 5 mM EGTA, 5 mM EDTA), and the homogenate was filtered through three layers of Miracloth (Calbiochem, United States). After centrifugation at 4,200 g for 5 min at 4°C and resuspension in isolation buffer II (0.3 M sorbitol, 20 mM Hepes-KOH, pH 7.6, 5 mM MgCl2, 2.5 mM EDTA), intact chloroplasts were osmotically ruptured in lysis buffer (5 mM MgCl2, 2.5 mM EDTA, 20 mM Hepes-KOH, pH 7.6). Thylakoid membranes were separated by centrifugation at 10,000 g for 2 min at 4°C.
BN-PAGE, SDS-urea-PAGE, and immunoblot analysis were performed as previously described (Zhang et al., 2016). For 2D/SDS-PAGE, excised BN-PAGE strips were incubated with 2 × SDS sample buffer (50 mM Tris–HCl, pH 6.8, 8 M urea, 5% SDS, 20% glycerol, 5% b-mercaptoethanol, 1% bromophenol blue) for 30 min at 25°C and layered onto denaturing 15% SDS-urea-PAGE gels. For Tricine-SDS-PAGE, samples were solubilized in 4 × Tricine sample buffer (12% SDS, 6% β-mercaptoethanol, 30% glycerol, 150 mM Tris–HCl, pH 7.0, 0.05% Coomassie Brilliant Blue G250) and separated on 10/16% step-gradient Tricine-SDS gels. For immunoblot analysis, proteins separated by SDS-urea-PAGE or Tricine-SDS-PAGE were transferred to nitrocellulose membranes (GE Healthcare, United States) and incubated with various antibodies. Signals were captured with a WSE-6200 LuminoGraph II Chemiluminescence Imaging system (ATTO Technology, Japan).
Arabidopsis crude DNA was extracted from leaves through shock crushing with DNA extraction buffer (0.2 M Tris–HCl, pH 8.0; 12.5 mM EDTA, 0.25 M NaCl, 0.5% SDS), and the DNA was precipitated with ethanol and centrifuged at 13,500 g for 10 min at room temperature. The DNA was solubilized and subjected to PCR analysis. RNA gel blot analysis was performed according to Zhang et al. (2018). Total RNA was extracted from the leaves of 4-week-old plants with liquid nitrogen using TRlzol regent (Invitrogen, United States). Equal amounts of total RNA were separated by formamide denaturing agarose gel electrophoresis and transferred to a nylon membrane (GE Healthcare, United States). RNA was detected using nucleic acid probes labeled with digoxigenin (DIG, Roche, Switzerland).
Polysome association analysis was performed according to our previous studies (Zhang et al., 2018). Leaves of 4-week-old plants were ground in liquid nitrogen, and polysomes were extracted with polysome extraction buffer (0.2 M Tris–HCl, pH 9.0, 0.2 M Sucrose, 0.2 M KCl, 35 mM MgCl2, 25 mM EGTA, 1% [v/v] Triton X-100, 2% [v/v] polyoxyethylene-10-tridecyl ether, 0.5 mg mL–1 heparin, 0.1 M 2-mercaproethanol, 0.1 mg mL–1 chloramphenicol, 25 μg mL–1 cycloheximide). After centrifugation at 20,000 g for 5 min at 4°C, the supernatants were further solubilized with 0.5% (w/v) sodium deoxycholate. Unresolved membrane was removed by centrifugation at 20,000 g for 15 min at 4°C, and the supernatants were subsequently layered on 15–55% sucrose gradients. After centrifugation at 237,000 g in the SW55 rotor for 65 min at 4°C, each gradient was divided into 10 0.5-mL fractions and proteins associated with RNA were denatured with 0.5% (w/v) SDS and 20 mM EDTA, and then RNA was extracted with acid phenol/chloroform/isoamyl alcohol (25:24:1). The RNA associated with polysomes was precipitated with 95% ethanol at room temperature and solubilized in DEPC-treated water. The samples were subjected to RNA gel blot analysis.
Chloroplast proteins labeling and chasing analysis was performed as previously described (Zhang et al., 2018). Labeled proteins were fractionated by SDS-urea-PAGE or 2D BN/SDS-PAGE, and signals were detected by autoradiography.
The ribosomal profiling analysis was performed by Gene Denovo Biotechnology Co., (Guangzhou, China) according to Ingolia et al. (2012) with modifications. Leaves of 4-week-old mutants and WT plants were ground to powder in liquid nitrogen and then dissolved in lysis buffer (20 mM Tris HCl pH 8.0, 1.5 mM MgCl2, 140 mM KCl, 100 μg mL–1 chloramphenicol, 100 mg mL–1 cycloheximide, 1% Triton-X-100). After centrifugation at 20,000 g for 10 min at 4°C, the supernatant was treated with RNase I (NEB, United States) and DNase I (NEB, United States) at room temperature for 45 min to degrade DNA and unprotected RNA. Nuclease digestion was stopped by adding RNase inhibitor (SUPERase•In; Ambion, United States). Then, digested RNAs were separated by size exclusion columns (illustra MicroSpin S-400 HR Columns; GE Healthcare) and SDS was added to the elution at a final concentration of 1% to denature RNA-associated proteins. RNAs with a size greater than 17 nt were isolated with RNA Clean and Concentrator-25 kit (Zymo Research, United States). rRNA was removed as reported previously (Morlan et al., 2012), and RNAs were further purified using magnetic beads (Vazyme, China).
Ribo-seq libraries were prepared with NEBNext Multiple Small RNA Library Prep Set for Illumina (NEB, United States) according to the user manual. PCR products with DNA fragments at 140–160 bp (representing insert sizes of 18–35 bp) were enriched to generate a cDNA library and sequenced with Illumina HiSeqTM X10 (Gene Denovo Biotechnology Co., China).
Read processing, alignment, and analysis were carried out according to Chotewutmontri and Barkan (2018). Reads counts in the open reading frame (ORF) of coding genes were calculated by software RSEM, and gene expression level was normalized by using RPKM (Reads Per Kilobase of transcript per Million mapped reads) method. Read counts and RPKM values for chloroplast genes are summarized in Supplementary Table 2. Distribution of ribosome footprints along the ORF was normalized by the sum of the total read counts.
The raw data of ribosome profiling reported in this paper have been deposited in the GenBank (NCBI) Sequence Read Archive (SRA) with accession number PRJNA7856661.
The sequences data in this assay obtain from the Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: AtRBD1 (AT1G54500, Arabidopsis thaliana), CrRBD1 (Cre07.g315150.t1.2, Chlamydomonas reinhardtii), and SyRubA (slr2033, Synechocystis sp. PCC 6803).
To investigate the function of RBD1 in higher plants, the T-DNA insertion Arabidopsis mutant rbd1 was obtained from NASC (Nottingham Arabidopsis Stock Center). As reported previously, the rbd1 mutant cannot grow photoautotrophically on soil and exhibits a yellow-green leaf phenotype when grown on Murashige and Skoog (MS) medium (Figure 1A, Calderon et al., 2013). Chlorophyll a fluorescence imaging and fluorescence transients showed that the minimum chlorophyll a fluorescence (Fo) is increased and the variable fluorescence (Fv) decreased in rbd1, resulting in a reduced maximum quantum yield of PSII (Fv/Fm) (Figures 1A,B). These results indicate that the PSII activity is reduced in the rbd1 mutant.
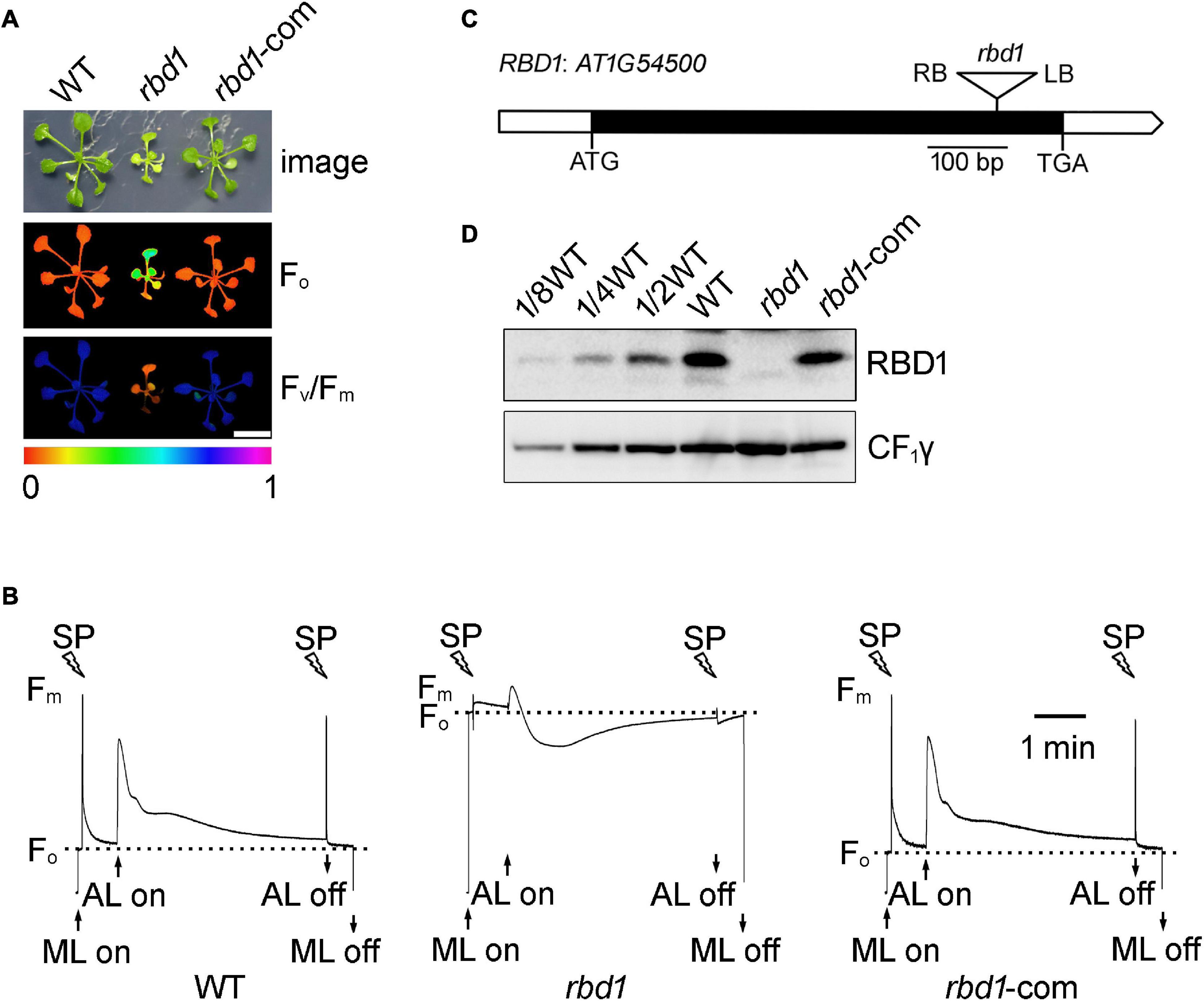
Figure 1. Characterization of the rbd1 mutant. (A) Phenotypes and complementation of the rbd1 mutant. Growth (upper panel) and chlorophyll a fluorescence (Fo, middle panel; Fv/Fm, lower panel) phenotype of wild type (WT), rbd1, and rbd1-com plants. Values for Fo and Fv/Fm are indicated using a false color scale at the bottom. rbd1-com, rbd1 mutant complemented using the RBD1 genomic DNA from WT. (B) Chlorophyll a fluorescence transients of the rbd1 mutant. After 20-min dark adaption, minimal fluorescence (Fo) was measured with measuring light (ML) and a saturation pulse (SP) was applied to record maximal fluorescence (Fm). Subsequently, leaves were exposed to actinic light (AL, 64 μmol photons m–2 s–1) for 4 min. (C) Structure of the RBD1 gene. White and black boxes represent the untranslated and coding regions, respectively. The T-DNA insertion is represented by a triangle. (D) Immunoblot analysis of RBD1 protein. Thylakoid proteins extracted from WT, rbd1, and rbd1-com plants were separated by SDS-urea-PAGE and detected with an antibody against RBD1. CF1γ is shown as a loading control.
The RBD1 gene of Arabidopsis does not contain introns (Figure 1C). Sequencing of the PCR products showed that the T-DNA was inserted into the coding region of RBD1, resulting in a complete loss of RBD1 protein in thylakoids (Figure 1D). Introduction of the genomic RBD1 gene driven by its own promoter in the rbd1 mutant completely rescued growth and wild-type chlorophyll a fluorescence patterns (Figures 1A,B). Consistently, similar levels of RBD1 protein were detected in WT and complemented plants (Figure 1D). These results demonstrate that loss of the RBD1 protein is responsible for the severe mutant phenotype of rbd1.
To examine the effects of RBD1 on the abundance of thylakoid complexes, thylakoids freshly isolated from wild type and rbd1 mutant were solubilized with 1% n-dodecyl β-D-maltoside (β-DM), and protein complexes were separated by blue native polyacrylamide gel electrophoresis (BN-PAGE) (Figure 2A). The results revealed a drastic reduction of PSII complexes including PSII supercomplexes, PSII dimer, PSII monomer, and CP43-less PSII, whereas accumulation of NDH-PSI supercomplex was identical in wild type and rbd1 mutant (Figure 2A). To quantitate the abundance of PSII subunits in rbd1, immunoblotting was performed using WT and rbd1 thylakoids. Accumulation of the PSII core subunits D1, D2, CP43, CP47, PsbE, and PsbI is significantly reduced to less than 1/8 of WT in rbd1 (Figure 2B). The level of PSII oxygen-evolving complex subunit PsbO and PSI subunits (PsaA and PsaD) in the rbd1 mutant is reduced to about 1/2 compared with WT. However, the amounts of LHCII complex (Lhcb1), cytochrome b6f complex (Cyt b6), and ATP synthase (CF1ε) are slightly increased in rbd1 relative to WT (Figure 2B). Similar levels of these thylakoid membrane proteins were observed in the Chlamydomonas RBD1-deficient mutant 2apc. The decrease of PSI in rbd1 is likely a secondary effect of the drastic reduction of PSII (Calderon et al., 2013). These data indicate that, similar to its Chlamydomonas ortholog, Arabidopsis RBD1 is required for the accumulation of PSII complexes (García-Cerdán et al., 2019).
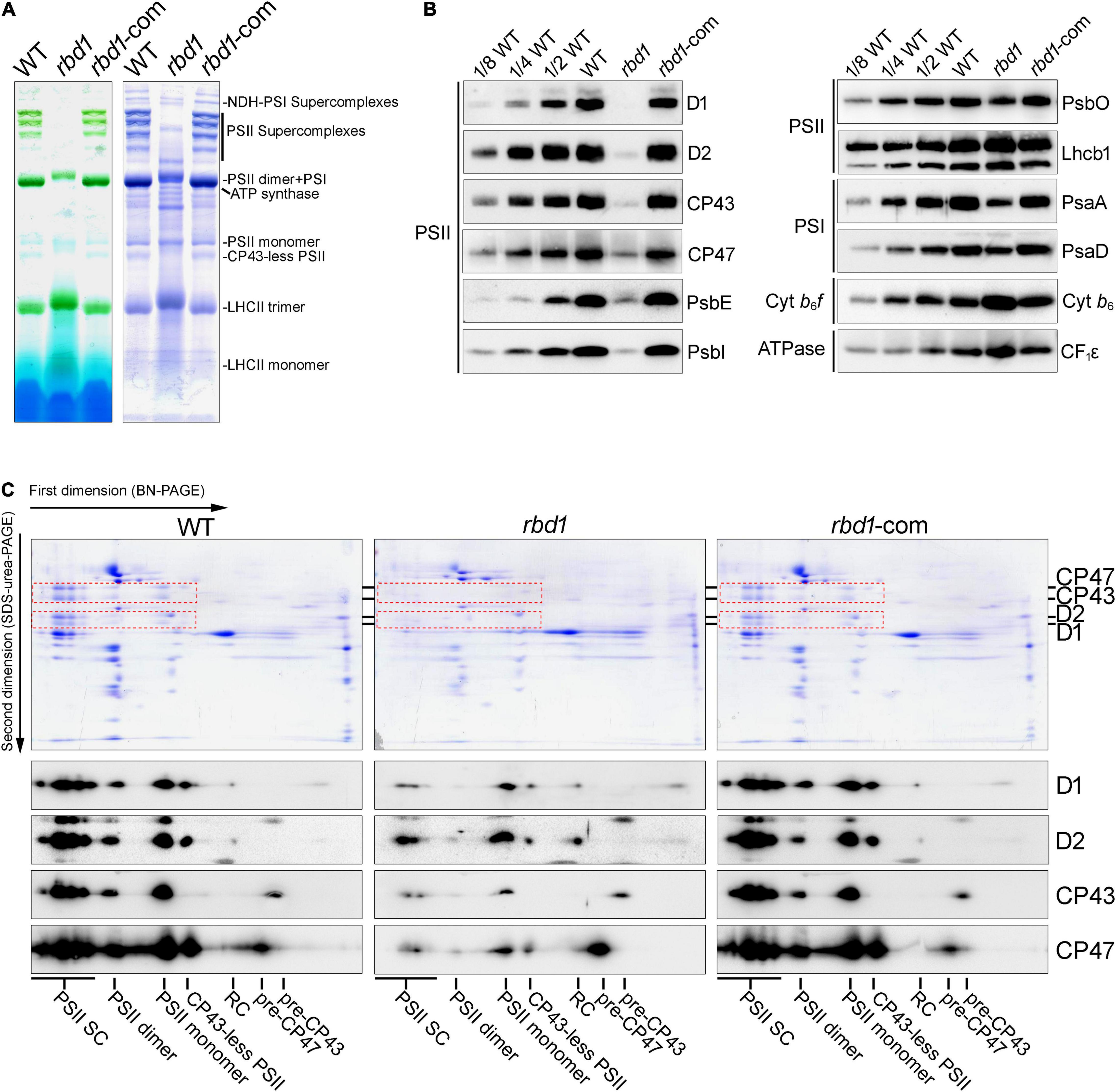
Figure 2. Analysis of thylakoid protein complexes from WT, rbd1, and rbd1-com plants. (A) BN-PAGE analysis of thylakoid membrane protein complexes. Thylakoid protein complexes corresponding to equal amounts of chlorophyll were solubilized with 1% dodecyl-β-D-maltoside (β-DM) and separated by BN-PAGE (left). The gels were stained with Coomassie Brilliant Blue (CBB) (right). (B) Immunoblot analysis of representative subunits of thylakoid protein complexes in the rbd1 mutant. Equal amounts of thylakoid proteins isolated from WT, rbd1, and rbd1-com plants were separated by SDS-urea-PAGE or Tricine-SDS-PAGE. Immunoblot analysis was performed with specific antibodies indicated on the right side of each panel. A dilution series of the WT sample (1/2, 1/4, and 1/8) was loaded to quantitatively assess protein accumulation in the mutants. (C) Two-dimensional BN/SDS-PAGE analysis. For each genotype, a total of five excised BN-PAGE strips (A) were incubated with SDS sample buffer for 30 min and layered onto denaturing 15% SDS-urea-PAGE gels. Proteins were stained with CBB (top part) or transferred to nitrocellulose membranes to blot with D1, D2, CP43, and CP47 antibodies, respectively (bottom part). Signals with the same exposure time are shown for each antibody. The spots of D1, D2, CP43, and CP47 on the CBB-stained gels are framed by red dot boxes. PSII-SC, PSII supercomplexes; RC, reaction center.
To further investigate the accumulation of the PSII assembly intermediates in rbd1, thylakoid protein complexes separated in the first dimension by (1D) BN-PAGE were then resolved into their subunits in the second dimension by (2D) electrophoresis on 15% SDS-urea-PAGE (Figure 2C). Immunoblot analysis showed that the PSII core subunits D1, D2, CP43, and CP47 are present as expected in the PSII supercomplexes (PSII-SC), PSII dimer, PSII monomer, and CP43-less PSII in WT and complemented plants (Figure 2C). However, in the rbd1 mutant, D1 and D2 are mainly present in the PSII monomer and CP43 and CP47 are mainly present in the PSII monomer and their assembly intermediates (pre-CP43 and pre-CP47) (Figure 2C). Moreover, only trace amounts of D1 and D2 were detected in the PSII reaction center (RC) in WT and complemented plants but a relatively higher level of RC assembly intermediate was found in the rbd1 mutant (Figure 2C). These results show that rbd1 accumulates a high amount of PSII assembly intermediates including PSII RC, pre-CP43, and pre-CP47.
To study the assembly kinetics of PSII in the rbd1 mutant, chloroplast-encoded proteins were labeled with [35S]-Met for 20 min using primary leaves of 12-day-old plants in the presence of cyclohexmide which specifically inhibits cytosolic protein synthesis. After labeling, thylakoids were extracted and solubilized in the SDS sample buffer. Equal amounts of protein were separated by 15% SDS-PAGE and the labeled proteins were visualized by autoradiography. As shown in Figure 3A, the most striking difference is that the rate of synthesis of mature D1 protein is drastically reduced in rbd1 compared with WT (Figure 3A). In contrast, proteins corresponding to pD1 and D2 were heavily labeled in rbd1 (Figure 3A). As mature D1 is produced from pD1 by removing the C-terminal 9-AA with the CtpA enzyme, it is possible that processing of pD1 protein is retarded in rbd1, resulting in a higher accumulation of pD1. Since this band also contains D2 protein, it is possible that the rate of synthesis of D2 is increased in rbd1 compared with WT.
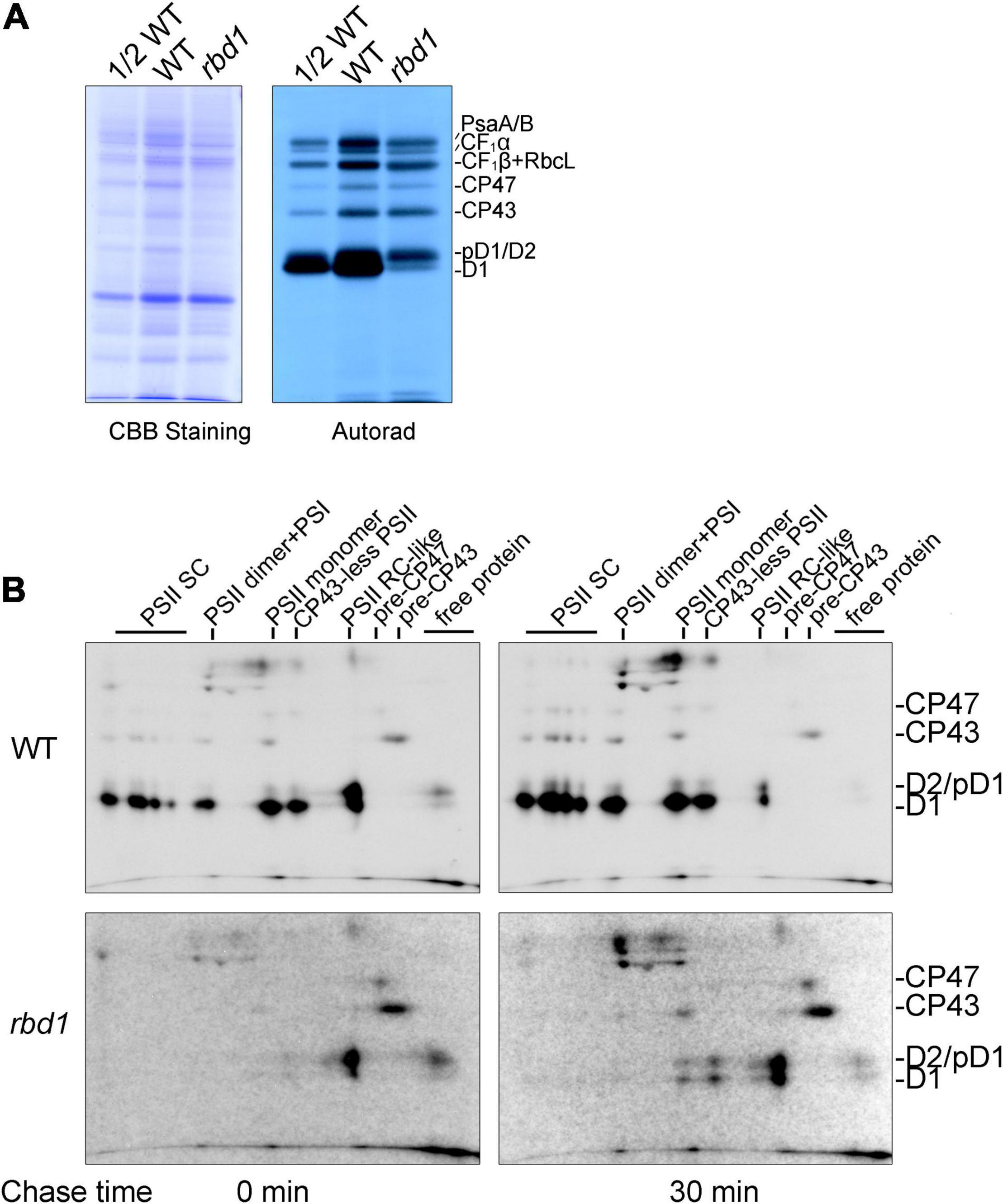
Figure 3. PSII assembly is impaired in the rbd1 mutant. (A) Pulse labeling of thylakoid proteins in the rbd1 mutant. Primary leaves of 12-day-old plants were labeled with [35S]-Met for 20 min in the presence of cycloheximide. Thylakoid proteins were extracted and separated by SDS-urea-PAGE. The gels were stained with Coomassie Brilliant Blue (CBB, left), and labeled proteins were visualized by autoradiography (right). (B) Analysis of PSII assembly in rbd1 mutant. Leaves were labeled as in A (indicated as 0 min) and chased with cold Met for 30 min (indicated as 30 min). Thylakoid proteins were isolated and separated by 2D BN/SDS-urea-PAGE. PSII-SC, PSII supercomplexes. Positions of CP47, CP43, D2/pD1, and D1 are shown on the left. The exposure times of autoradiography for WT and rbd1 are 3 and 15 days, respectively.
To distinguish between these two possibilities, we performed pulse-chase labeling experiment. After labeling for 20 min (indicated as 0 in Figure 3B), the labeled proteins were chased with unlabeled Met for 30 min (indicated as 30 in Figure 3B). To investigate the dynamic fates of newly synthesized thylakoid proteins, thylakoid protein complexes were separated by 2D BN/SDS-PAGE and individual subunits were visualized by autoradiography. As shown in Figure 3B, in the WT, mature D1, D2, and CP43 proteins were readily detected in the PSII supercomplexes (PSII SC), PSII dimer, PSII monomer, CP43-less PSII, and PSII reaction center (PSII RC) after labeling for 20 min (indicated as 0 in Figure 3B), indicating efficient PSII assembly. At the PSII RC position, the signal of the protein spot corresponding to pD1/D2 is comparable with mature D1 (Figure 3B). 2D BN/SDS-PAGE of labeled proteins in WT showed that the level of labeled D2 protein is significantly lower than that of D1 in the different PSII complexes. Thus, it is reasonable to speculate that the protein spot corresponding to pD1/D2 in the PSII RC complex represents mainly pD1. As expected, during the 30 min chase, pD1 was rapidly processed to mature D1 protein in the PSII RC complex which was further assembled into various PSII complexes in WT (Figure 3B).
However, after labeling for 20 min, the majority of mature D1 and pD1/D2 was present in the PSII RC and trace amounts of these proteins were in the free form in rbd1 (Figure 3B). The level of pD1/D2 was significantly higher than mature D1 protein in the PSII RC complex. Even after chasing for 30 min, the majority of D1/pD1 was still present in the PSII RC complex and the level of pD1/D2 was higher than mature D1 in PSII RC (Figure 3B). In addition, CP43 and CP47 were mainly present in the pre-CP43 and pre-CP47 complex, respectively (Figure 3B). These results indicate that PSII assembly of PSII RC complex to CP43-less PSII and other intact PSII complexes is severely impaired in rbd1. Since PSII RC contains a high level of mature D1 after chasing for 30 min, it is unlikely that processing of pD1 into D1 represents the main limitation for assembly of PSII in rbd1.
Our protein labeling experiments also showed that the rate of synthesis of the PSI core subunits PsaA/B, ATP synthase subunits CF1α, as well as the PSII core subunits CP43 and CP47, are slightly reduced in rbd1 compared with WT (Figure 3A). A similar reduction was also reported for other PSII mutants such as hcf136, hcf244, hcf173, ohp1, and ohp2 (Meurer et al., 1998; Schult et al., 2007; Link et al., 2012; Li et al., 2018) and it is most likely a secondary effect of PSII deficiency.
To investigate why the rate of synthesis of D1 is reduced in rbd1, RNA gel blot and polysome association analysis were performed. As shown in Figure 4A, there was no obvious defect in the accumulation and expression pattern of psbA, psbB, psbC, psbD, psbEFJL, and psbKI in rbd1 compared with WT indicating that RBD1 is not involved in transcription and post-transcriptional processes of the chloroplast psb genes. Polysome association analysis which reflects translation initiation of transcripts showed that the distribution of psbB, psbC, psbD, psbEFJL, and psbKI transcripts was almost identical between rbd1 and WT (Figure 4B). In contrast, the distribution of psbA transcripts was significantly shifted toward the lower molecular weight polysomal fractions in the rbd1 mutant compared with WT (Figure 4B) indicating that translation initiation of psbA transcript is impaired in rbd1, resulting in a reduced synthesis rate of D1 in vivo. To confirm that the reduction of psbA mRNA in the higher molecular weight fractions in rbd1 was caused by the dissociation with polysomes we performed a control experiment by treating the extract with EDTA before ultracentrifugation. It is known to cause ribosome dissociation from mRNA. As shown in Figure 4C, EDTA treatment induced shifts of psbA transcripts towards lower molecular weight fractions in both the rbd1 mutant and WT, indicating that the amount of polysomes associated with psbA transcripts was reduced.

Figure 4. RNA blot analysis of plastid psb transcripts encoding PSII subunits and their polysome association. (A) RNA blot analysis of the plastid psb transcripts encoding PSII subunits. Equal amounts of total RNA extracted with Trizol reagent were separated by electrophoresis on denaturation agarose gels. RNA gel-blot hybridization was performed with DIG-labeled probes specific for psbA, psbB, psbC, psbD, psbEFJL, psbKI, and ACTIN7. ACTIN7 was used as a loading control. (B) Polysome analysis. mRNA-polysome complexes isolated from WT and rbd1 plants were fractionated by 15–55% sucrose density gradient centrifugation. Each gradient was divided into ten 0.5-mL fractions and total RNA was extracted. RNA gel-blot hybridization was then performed with DIG-labeled probes specific for psbA, psbB, psbC, psbD, psbEFJL, and psbKI. RNA staining is shown as a loading control. (C) Dissociation of polysomes monitored with a psbA probe. Samples were prepared as in B except for the addition of EDTA (10 mM) in the polysome extraction buffer. EDTA was used to dissociate polysomes from mRNA.
To investigate whether RBD1 is involved in the translation of other transcripts encoding PSII subunits and other thylakoid complexes, ribosome profiling was performed using WT and rbd1 plants. In this experiment, ribosome-protected mRNA fragments were deep-sequenced and the reads of each transcript can be calculated by normalizing to total chloroplast reads (cpRPKM). We obtained 63,216,962 and 57,604,852 high-quality clean reads in two replicates for WT and rbd1, respectively, after the removal of rRNA, tRNAs, snoRNA, snRNA, miRNA, and low-quality reads (Supplementary Table 2). The proportion of total reads that mapped to the reference genome ranged from 70.43% to 89.39%, and 22,719 genes were identified. As shown in Figure 5A, psbA mRNA abundance of ribosome occupancy was drastically reduced to about 14% of WT in rbd1, consistent with the lower abundance of polysome association of psbA mRNA in the mutant (Figure 4B and Supplementary Table 2). The abundance of ribosome footprints for psbN, and psaA and psaB mRNAs encoding PSI subunits were comparable in rbd1 and WT. The ribosome footprint abundance of other chloroplast transcripts varied between 1.5- and 4-fold increase compared to WT (Figure 5A). Similar results were also reported for the PSII mutants of hcf244, ohp1, and ohp2 (Chotewutmontri et al., 2020). They can be explained by the fact that psbA ribosome footprints represent a high fraction of the total footprints in WT and the drastic decrease in psbA reads in the mutant therefore increases the cpRPKM values for other genes. If one takes into account this compensation effect, the relatively low rbd1/WT cpRPKM ratios for psbN, psaA, and psaB suggest that translation of these proteins may be slightly affected. PsbN is not a PSII subunit but it is involved in the PSII RC formation (Torabi et al., 2014). These facts imply that RBD1 and PsbN share some biochemical or genetic interaction during PSII RC biogenesis.
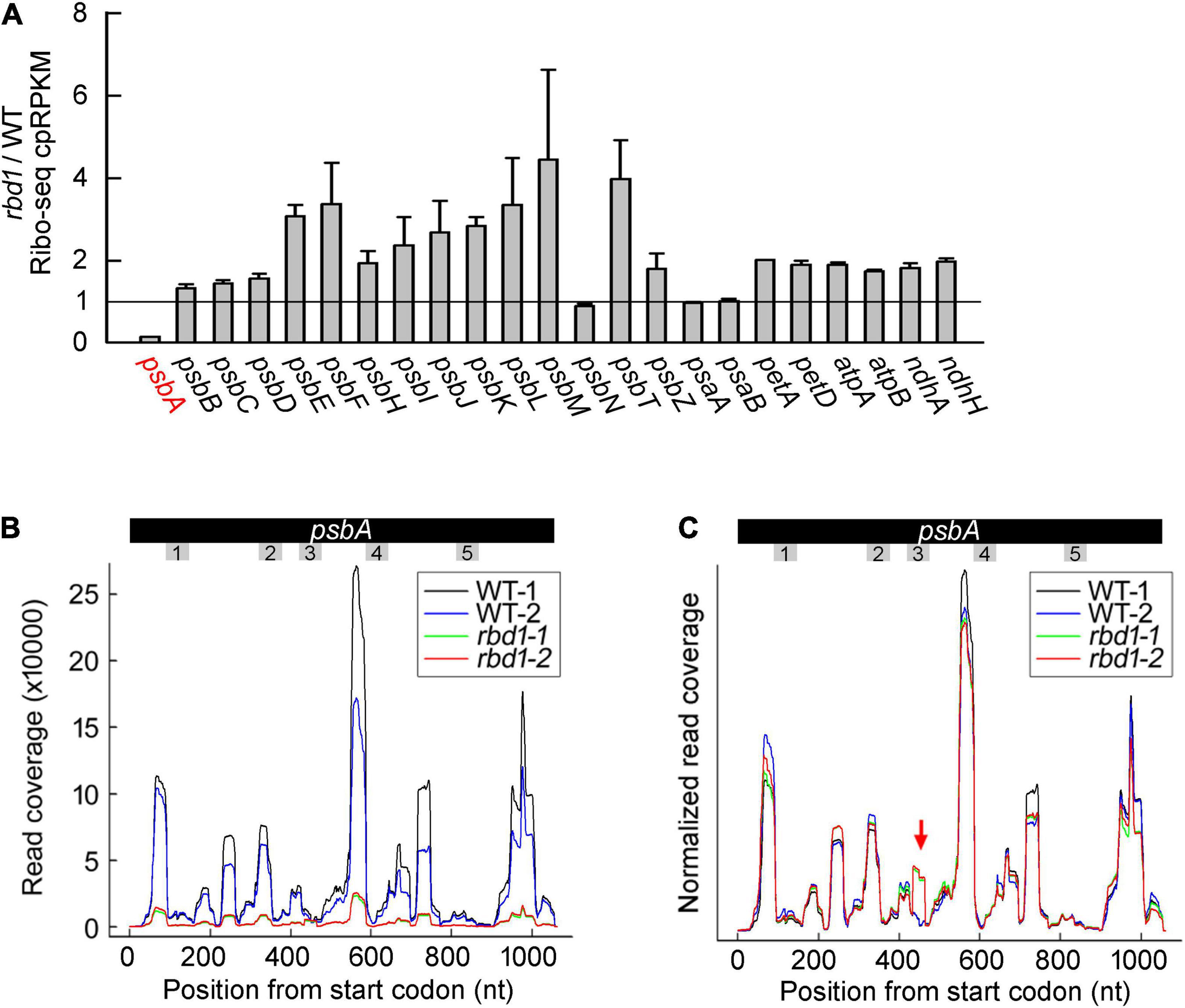
Figure 5. Ribo-seq analysis of the expression of chloroplast gene in the rbd1 mutant. (A) Ribosome footprint abundance for chloroplast genes in rbd1 relative to wild type. Average of two replicates (mean ± SD) is shown. cpRPKM, reads per kilobase in the ORF per million reads mapped to chloroplast ORFs. (B) Distribution of ribosome footprints along the psbA ORF in the two replicates of the rbd1 mutant (rbd1-1 and rbd1-2) and WT (WT-1 and WT-2). The regions of psbA mRNA encoding the transmembrane domains (TMD) are labelled from 1 to 5. (C) Normalized reads of (B). The Y-axis was normalized to the sum of the read coverage. The red arrow points to a new pausing site in rbd1.
To further investigate the role of RBD1 in psbA mRNA translation, we analyzed the distribution of ribosome footprints along the psbA mRNA. Several major peaks were detected both in the mutant and wild type that represent the positions of ribosomes paused temporarily along the mRNA during translation. As expected, the signals of rbd1 detected in the distribution of psbA mRNA were much lower compared to WT because of the lower abundance of ribosome association in rbd1 (Figure 5B). To facilitate comparison of the ribosome distribution along the psbA mRNA in WT and rbd1, the Y-axis was adjusted as previously reported (Chotewutmontri et al., 2020). As shown in Figure 5C, the pattern of the ribosome distribution along the psbA mRNA in rbd1 was similar to wild type (Figure 5C). However, a peak was specifically detected in rbd1 in the region of psbA mRNA encoding the latter part of the third transmembrane domain (TMD) (Figure 5C). These results suggest that ribosomes pause during psbA translation in rbd1 when the third TMD of D1 emerges from the ribosomal tunnel. It is thus likely that integration of the first two, probably also the third, TMDs of D1 is retarded during PSII biogenesis in rbd1. RBD1 may, directly or indirectly, facilitate the insertion of D1 into thylakoids to form the functional PSII RC complex during assembly and repair of PSII.
Our results show that the levels of the PSII core subunits D1, D2, CP43, and CP47 are reduced to less than 1/8 of WT resulting in a drastic decrease of the various intact PSII complexes as well as PSII activity (Figures 1, 2). Similar phenotypes were also found for the Chlamydomonas rbd1 and Synechocystis rubA deletion mutant (García-Cerdán et al., 2019; Kiss et al., 2019). However, in the Arabidopsis rbd1 mutant, the steady-state levels of PSII assembly intermediates including PSII RC, pre-CP43, and pre-CP47 complexes are relatively higher compared to WT and the complemented plants (Figure 2C). Consistent with these results, pulse labeling and pulse-chase experiments showed that newly synthesized PSII core subunits are mainly present in the PSII RC, pre-CP43, and pre-CP47 intermediates after labeling for 20 min and an additional 30-min chase (Figure 3B). These results confirm that RBD1 is required for PSII assembly in vivo (García-Cerdán et al., 2019; Kiss et al., 2019).
Although the rate of synthesis of pD1 and D1 is reduced in rbd1, newly synthesized pD1/D1/D2 proteins are able to assemble into the PSII RC complex in this mutant (Figure 3). However, formation of the larger PSII complexes is severely decreased (Figure 3B). In higher plants, after the formation of the PSII RC complex, CP47 and several low molecular mass subunits (LMM) attach to RC forming CP43-less PSII. Subsequently, CP43 and other LMM PSII subunits are assembled to form the PSII monomer (Rokka et al., 2005). In rbd1, newly synthesized CP47 and CP43 are mainly present in the pre-CP47 and pre-CP43 assembly intermediates, respectively (Figure 3B). Pulse labeling showed that the rate of synthesis of CP47 and CP43 is slightly reduced in rbd1 comparable with WT (Figure 3A). Also, ribosome occupancy of psbB and psbC mRNA is not reduced in rbd1 (Figure 5A). These results suggest that newly synthesized CP47 and CP43 are functional and available for further association with the PSII RC complex. However, they are unable to sequentially associate with PSII RC during the labeling and chase period of 50 min (Figure 3B). Hence, it is reasonable to propose that the newly formed PSII RC complex is largely non-functional and therefore cannot bind antennas CP47 and CP43 to form functional PSII. However, we could not exclude the possibility that PSII RC is functional, but the CP47 and CP43 antennas do not have access to PSII RC in the rbd1 mutant.
In the Synechocystis rubA deletion mutant, however, the rates of synthesis of D1 as well as those of the other core subunits D2, CP43, and CP47 are as in WT (Kiss et al., 2019). Moreover, the majority of newly synthesized D1 and D2 remained in an unassembled state in thylakoids and could not assemble into PSII RC complexes as in the Arabidopsis rbd1 mutant (Figure 3B; Kiss et al., 2019). Since RBD1 in chloroplasts and its cyanobacterial ortholog RubA have conserved rubredoxin and transmembrane domains, it is reasonable to assume that they exert a similar physiological function during PSII assembly. How can one explain the differences between these two species? One explanation could be that the PSII assembly process in cyanobacteria is slightly different. The Arabidopsis pam68-2 mutant accumulates ∼1/10 level of PSII complex compared with WT but in the Synechocystis sll0933, in which the PAM68 cyanobacterial ortholog Sll0933 is disrupted, the PSII complex accumulates almost normally (Armbruster et al., 2010). Further analysis showed that a high level of PSII RC complex is present in pam68-2 but almost undetectable in the sll0933 mutant (Armbruster et al., 2010). Similar phenotypes with different levels of PSII assembly intermediates were also found between the hcf136 Arabidopsis and ycf48 cyanobacterial mutant (Meurer et al., 1998; Komenda et al., 2008). Moreover, several specific chloroplast PSII assembly factors are absent in cyanobacteria (Nickelsen and Rengstl, 2013). Reciprocally, cyanobacteria also possess specific PSII assembly factors that are missing in chloroplasts (Nickelsen and Rengstl, 2013). These differences in the PSII assembly process and assembly factors may be the result of different environmental living pressures during evolution. As discussed by Nixon et al. (2010), it is also possible that a more effective protein quality system in chloroplasts may lead to differences in accumulation of PSII assembly intermediates in the mutants mentioned above.
How does RBD1 facilitate formation of the functional PSII RC complex? Our protein labeling experiments show that PSII RC containing mature D1 is produced implying that newly synthesized pD1 protein can be properly inserted into thylakoids in the correct topology for processing by the CtpA enzyme in the lumen. RBD1 contains a redox-active rubredoxin domain with a redox midpoint potential of +114 mV that is capable to transfer electrons to cytochrome c in vitro (Calderon et al., 2013). Thus, formation of inactive PSII RC in rbd1 is most likely due to deficient redox control. It is known that D1 integrates co-translationally into the PSII complex during its assembly (Zhang et al., 1999). Addition of DTT at a low concentration partially activated translation elongation of D1 protein pointing to the existence of a redox-sensitive regulatory component (Zhang et al., 2000). It has also been reported that two Cys residues of the cyanobacterial ortholog RubA are reversibly oxidized upon a light-to-dark transition (Guo et al., 2014). It is interesting that one of the two Cys residues (Cys-125) in D1 is positioned at the end of the second transmembrane domain (another one is in the middle of the first loop of D1). Therefore, RBD1 might be the redox regulatory component during the biogenesis of pD1, especially at early stages, when ribosomes pause after the first two transmembrane domains have emerged from the ribosomal tunnel in rbd1 (Figure 5C).
An alternative possibility for the function of RBD1 is delivery co-factors into PSII RC, as discussed previously (Kiss et al., 2019). The D1/D2 heterodimer binds several redox components including six chlorophyll a, two pheophytin (Pheno) a, one non-heme iron, as well as quinones QA and QB (Umena et al., 2011). Non-pigmented forms of most chlorophyll-binding proteins are rapidly degraded, suggesting that these co-factors are incorporated into PSII RC during the biogenesis of D1/D2 (Funk et al., 1995). Thus, RBD1 might also facilitate the binding of redox components to D1, such as pheophytin which binds to the residue Glu-130 of D1 at the end of the second transmembrane segment (Giorgi et al., 1996; Sharma et al., 1997). Moreover, it has been shown that iron can be released from the active site of rubredoxin protein under certain conditions (Zheng et al., 2013). Hence, another possibility could be that RBD1 directly delivers the non-heme Fe to the D1/D2 heterodimer.
Our polysome association and ribosome profiling analysis revealed that ribosome occupancy on psbA mRNA is drastically reduced in the absence of RBD1 (Figure 5A). A similar decrease was also found in the hcf244, ohp1, and ohp2 mutants (Chotewutmontri et al., 2020). These results imply that RBD1 and HCF244/OHP1/OHP2 are involved in the translation initiation of D1. However, a peak downstream of the region encoding the latter part of the third transmembrane domain of D1 is relatively enhanced in rbd1 compared to WT (Figures 5B,C), indicating that ribosomes pause in rbd1 after the second transmembrane domain of D1 is released from the ribosomal tunnel. It is possible that, in the absence of RBD1, the first two transmembrane domains cannot properly insert in the D2-Cyt b559 precomplex because of unavailability for the redox-active cofactors, resulting in ribosome pausing on psbA mRNA, and subsequently in the degradation of this ribosome-nascent chain complex, ultimately leading to a decrease of psbA mRNA translation (Figures 3, 5). A similar scenario may be also valid for the HCF244/OHP1/OHP2 proteins, which were proposed to deliver chlorophyll to PSII RC (Hey and Grimm, 2018; Li et al., 2018; Myouga et al., 2018; Chotewutmontri et al., 2020). In this case, reduction of polysome occupancy on the psbA mRNA is likely a secondary effect of membrane integration deficiency of D1.
In summary, we provide evidence that RBD1 is required for the formation of the functional PSII core complex and probably also required for the translation of D1 during the assembly of PSII at early stages in Arabidopsis. Together with the reports on RBD1 from Chlamydomonas and its ortholog from cyanobacteria RubA (García-Cerdán et al., 2019; Kiss et al., 2019), it is likely that RBD1 orthologs exert a similar role for forming a functional PSII among various photosynthetic organisms.
Thylakoid intrinsic protein RBD1 is required for assembly of PSII core complex at early stages.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author is: LZ (emhhbmdsaW4yMDE3QHNobnUuZWR1LmNu).
The original contributions presented in the study are publicly available. These data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA785666.
LC, HM, and LZ conceived the study and designed the experiments. LC, HM, and JR performed the experiments. HM and LZ produced the figures. LC, HM, LP, and LZ wrote the manuscript. LZ supervised the whole study. All authors analyzed the data.
This work was supported by the Fund of Shanghai Engineering Research Center of Plant Germplasm Resources (Grant No. 17DZ2252700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.824358/full#supplementary-material
Adman, E., Watenpaugh, K. D., and Jensen, L. H. (1975). NH-S hydrogen bonds in Peptococcus aerogenes ferredoxin, Clostridium pasteurianum rubredoxin, and Chromatium high potential iron protein. Proc. Natl. Acad. Sci. U.S.A. 72, 4854–4858. doi: 10.1073/pnas.72.12.4854
Armbruster, U., Zühlke, J., Rengstl, B., Kreller, R., Makarenko, E., Ruhle, T., et al. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22, 3439–3460. doi: 10.1105/tpc.110.077453
Baena-González, E., and Aro, E.-M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1451–1460. doi: 10.1098/rstb.2002.1141
Beck, J., Lohscheider, J. N., Albert, S., Andersson, U., Mendgen, K. W., Rojas-Stütz, M. C., et al. (2017). Small one-helix proteins are essential for photosynthesis in Arabidopsis. Front. Plant Sci. 8:7. doi: 10.3389/fpls.2017.00007
Calderon, R. H., García-Cerdán, J. G., Alizée, M., Cook, R., and Niyogi, K. K. (2013). A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 288, 26688–26696. doi: 10.1074/jbc.M113.487629
Che, Y., Fu, A., Hou, X., McDonald, K., Buchanan, B. B., Huang, W., et al. (2013). C-terminal processing of reaction center protein D1 is essential for the function and assembly of Photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 16247–16252. doi: 10.1073/pnas.1313894110
Chotewutmontri, P., and Barkan, A. (2018). Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 14:e1007555. doi: 10.1371/journal.pgen.1007555
Chotewutmontri, P., Williams-Carrier, R., and Barkan, A. (2020). Exploring the link between photosystem II assembly and translation of the Chloroplast psbA mRNA. Plants 9:152. doi: 10.3390/plants9020152
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Funk, C., Adamska, I., Green, B. R., Andersson, B., and Renger, G. (1995). The nuclear-encoded chlorophyll-binding Photosystem II-S protein is stable in the absence of pigments. J. Biol. Chem. 270, 30141–30147. doi: 10.1074/jbc.270.50.30141
García-Cerdán, J. G., Furst, A. L., Mcdonald, K. L., Schünemann, D., and Niyogi, K. K. (2019). A thylakoid membrane-bound and redox-active rubredoxin (RBD1) functions in de novo assembly and repair of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 33, 116–126. doi: 10.1073/pnas.1903314116
Giorgi, L. B., Nixon, P. J., Merry, S. A. P., Joseph, D. M., Durrant, J. R., Rivas, J. D., et al. (1996). Comparison of primary charge separation in the photosystem II reaction center complex isolated from wild-type and D1-130 mutants of the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 271, 2093–2101. doi: 10.1074/jbc.271.4.2093
Guo, J., Nguyen, A. Y., Dai, Z., Su, D., Gaffrey, M. J., Moore, R. J., et al. (2014). Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specifific redox proteomics. Mol. Cell Proteomics 13, 3270–3285. doi: 10.1074/mcp.M114.041160
Hey, D., and Grimm, B. (2018). ONE-HELIX PROTEIN2 (OHP2) is required for the stability of OHP1 and assembly factor HCF244 and is functionally linked to PSII biogenesis. Plant Physiol. 177, 1453–1472. doi: 10.1104/pp.18.00540
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M., and Weissman, J. S. (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550. doi: 10.1038/nprot.2012.086
Kiss, E., Knoppova, J., Aznar, G. P., Pilny, J., and Komenda, J. (2019). A photosynthesis-specific rubredoxin-like protein is required for efficient association of the D1 and D2 proteins during the initial steps of photosystem ii assembly. Plant Cell 31, 2241–2258. doi: 10.1105/tpc.19.00155
Komenda, J., Nickelsen, J., Tichý, M., Prásil, O., Eichacker, L. A., and Nixon, P. J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 22390–22399. doi: 10.1074/jbc.M801917200
Komenda, J., Sobotka, R., and Nixon, P. J. (2012). Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251. doi: 10.1016/j.pbi.2012.01.017
Li, Y., Liu, B., Zhang, J., Kong, F., Zhang, L., Meng, H., et al. (2018). OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol. 179, 195–208. doi: 10.1104/pp.18.01231
Link, S., Engelmann, K., Meierhoff, K., and Westhoff, P. (2012). The atypical shortchain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol. 160, 2202–2218. doi: 10.1104/pp.112.205104
Lovenberg, W., and Sobel, B. E. (1965). Rubredoxin: a new electron transfer protein from Clostridium pasteurianum. Proc. Natl. Acad. Sci. U.S.A. 54, 193–199. doi: 10.1073/pnas.54.1.193
Lyska, D., Meierhoff, K., and Westhoff, P. (2013). How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237, 413–428. doi: 10.1007/s00425-012-1752-5
Meurer, J., Plücken, H., Kowallik, K. V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. doi: 10.1093/emboj/17.18.5286
Morlan, J. D., Qu, K., and Sinicropi, D. V. (2012). Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One 7:e42882. doi: 10.1371/journal.pone.0042882
Mulo, P., Sirpiö, S., Suorsa, M., and Aro, E.-M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98, 489–501. doi: 10.1007/s11120-008-9320-3
Myouga, F., Takahashi, K., Tanaka, R., Nagata, N., Kiss, A. Z., Funk, C., et al. (2018). Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol. 176, 2277–2291. doi: 10.1104/pp.17.01782
Nelson, N., and Junge, W. (2015). Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683. doi: 10.1146/annurev-biochem-092914-041942
Nelson, N., and Yocum, C. F. (2006). Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57, 521–565. doi: 10.1146/annurev.arplant.57.032905.105350
Nickelsen, J., and Rengstl, R. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 605–639. doi: 10.1146/annurev-arplant-050312-120124
Nixon, P. J., Michoux, F., Yu, J., Boehm, M., and Komenda, J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16. doi: 10.1093/aob/mcq059
Plücken, H., Müller, B., Grohmann, D., Westhoff, P., and Eichacker, L. A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532, 85–90. doi: 10.1016/s0014-5793(02)03634-7
Rokka, A., Suora, M., Saleem, A., Battchikova, N., and Aro, E. M. (2005). Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem. J. 388, 159–168. doi: 10.1042/BJ20042098
Schult, K., Meierhoff, K., Paradies, S., Töller, T., Wolff, P., and Westhoff, P. (2007). The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19, 1329–1346. doi: 10.1105/tpc.106.042895
Sharma, J., Panico, M., Shipton, C. A., Nilsson, F., Morris, H. R., and Barber, J. (1997). Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272, 33158–33166. doi: 10.1074/jbc.272.52.33158
Shen, J. R. (2015). The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48. doi: 10.1146/annurev-arplant-050312-120129
Torabi, S., Umate, P., Manavski, N., Plöchinger, M., Kleinknecht, L., Bogireddi, H., et al. (2014). PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26, 1183–1199. doi: 10.1105/tpc.113.120444
Umena, Y., Kawakami, K., Shen, J. R., and Kamiya, N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473:55. doi: 10.1038/nature09913
Yu, J., Knoppová, J., Michoux, F., Bialek, W., Cota, E., Shukla, M. K., et al. (2018). Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein. Proc. Natl. Acad. Sci. U.S.A. 115, E7824–E7833. doi: 10.1073/pnas.1800609115
Zhang, L., Duan, Z., Zhang, J., and Peng, L. (2016). Biogenesis factor required for ATP synthase 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol. 171, 1291–1306. doi: 10.1104/pp.16.00248
Zhang, L., Paakkarinen, V., van Wijk, K. J., and Aro, E.-M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274, 16062–16067. doi: 10.1074/jbc.274.23.16062
Zhang, L., Paakkarinen, V., van Wijk, K. J., and Aro, E.-M. (2000). Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12, 1769–1781. doi: 10.1105/tpc.12.9.1769
Zhang, L., Pu, H., Duan, Z., Li, Y., Liu, B., Zhang, Q., et al. (2018). Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell 30, 1770–1788. doi: 10.1105/tpc.18.00075
Keywords: rubredoxin, photosystem II, reaction center, psbA, ribosome profiling, Arabidopsis thaliana
Citation: Che L, Meng H, Ruan J, Peng L and Zhang L (2022) Rubredoxin 1 Is Required for Formation of the Functional Photosystem II Core Complex in Arabidopsis thaliana. Front. Plant Sci. 13:824358. doi: 10.3389/fpls.2022.824358
Received: 29 November 2021; Accepted: 13 January 2022;
Published: 23 February 2022.
Edited by:
Alistair McCormick, University of Edinburgh, United KingdomReviewed by:
Julian Eaton-Rye, University of Otago, New ZealandCopyright © 2022 Che, Meng, Ruan, Peng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Zhang, emhhbmdsaW4yMDE3QHNobnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.