
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 February 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.823019
This article is part of the Research Topic Environmental and molecular control of bud dormancy and bud break in woody perennials: An integrative approach View all 23 articles
SHORT VEGETATIVE PHASE (SVP) is an important regulator of FLOWERING LOCUS T (FT) in the thermosensory pathway of Arabidopsis. It is a negative regulator of flowering and represses FT transcription. In poplar trees, FT2 is central for the photoperiodic control of growth cessation, which also requires the decrease of bioactive gibberellins (GAs). In angiosperm trees, genes similar to SVP, sometimes named DORMANCY-ASSOCIATED MADS-BOX genes, control temperature-mediated bud dormancy. Here we show that SVL, an SVP ortholog in aspen trees, besides its role in controlling dormancy through its expression in buds, is also contributing to the regulation of short day induced growth cessation and bud set through its expression in leaves. SVL is upregulated during short days in leaves and binds to the FT2 promoter to repress its transcription. It furthermore decreases the amount of active GAs, whose downregulation is essential for growth cessation, by repressing the transcription of GA20 oxidase. Finally, the SVL protein is more stable in colder temperatures, thus integrating the temperature signal into the response. We conclude that the molecular function of SVL in the photoperiodic pathway has been conserved between Arabidopsis and poplar trees, albeit the physiological process it controls has changed. SVL is thus both involved in regulating the photoperiod response in leaves, modulating the timing of growth cessation and bud set, and in the subsequent temperature regulation of dormancy in the buds.
Photoperiod is an important environmental cue that controls diverse developmental processes in plants, for example, flowering in Arabidopsis and timing of growth cessation in Populus trees (Pin and Nilsson, 2012). At the center of the mechanism, with which plants sense day length, is the CONSTANS/FLOWERING LOCUS T module. This module is partially conserved between Populus and Arabidopsis, but best understood in the latter. In Arabidopsis, FT expression is tightly regulated by many factors and becomes a hub for the integration of different signals, which fine-tunes the response. In addition to photoperiod (Kobayashi et al., 1999), it is regulated by age (Wang, 2014), vernalization (Searle et al., 2006), and ambient temperature (Lee et al., 2007). SHORT VEGETATIVE PHASE (SVP) is part of the latter pathway and represses FT expression by binding to its promoter (Lee et al., 2007). In Populus two FT orthologs have been identified, called FT1 and FT2. Only FT2 has a comparable expression pattern to the Arabidopsis FT, being expressed in leaves under long photoperiods, while FT1 is only expressed in buds during winter (Hsu et al., 2011).
For trees in boreal forests, fine-tuning of the photoperiod response is critical for survival; they need to adapt to the rapidly changing seasons. Especially during the autumn months, temperature and day length are decreasing quickly. Once the day length falls under the critical day length, a threshold for growth permitting conditions, the trees stop their growth and set terminal buds, which protect the enclosed leaf primordia and shoot apical meristems from the subsequent low temperatures (Rohde and Bhalerao, 2007). These short days (SDs) are a reliable signal, with which the trees can anticipate the onset of winter. The signal is transmitted through FT2, which is downregulated within a few days after shifts to SDs (Böhlenius et al., 2006; Hsu et al., 2006). Trees failing to downregulate FT2 are unable to respond to the SD signal and continue growth indefinitely, while plants with reduced FT2 expression respond more quickly (Böhlenius et al., 2006), leading to early growth cessation and bud set.
CO and GI have been identified as positive regulators of FT2 in long days (LDs). However, their expression profiles do not dramatically change upon shift to SDs (Ding et al., 2018). Arabidopsis CO is rapidly degraded in the dark, thus unable to induce FT in SDs (Valverde et al., 2004), and it is so far unclear if the same is true for poplar CO. However, the lack of induction by CO is not enough to explain the rapid downregulation of FT2 in SDs, especially since GI is still expressed and of higher relative importance for FT2 expression (Ding et al., 2018). GI might contribute to the release of repressive activity on FT2 expression, as has been shown for poplar CYCLING DOF FACORS (Ding et al., 2018). Such repressors might therefore contribute to the downregulation of FT2 expression in response to shorter photoperiods. Another possible candidate for such a repressor would be SVP, a MADS domain-containing gene and a strong repressor of FT expression in Arabidopsis (Hartmann et al., 2000).
SVP homologs have been found in other tree species. For example, in peach trees, six DAM (dormancy-associated MADS-box) genes have been associated with the non-dormant phenotype of the evergrowing mutant (Bielenberg et al., 2008). DAM1 and DAM4 peak in their expression at the end of summer and are hypothesized to be involved in the regulation of growth cessation (Li et al., 2009). Also, in apple, DAM- and SVP-like genes have been suggested to control bud set and dormancy (Wu et al., 2017, 2021; Falavigna et al., 2019, 2021; Moser et al., 2020). Recently, a Populus SVP ortholog named SVL has been shown to be expressed in buds where it is involved in dormancy establishment and maintenance (Singh et al., 2018, 2019). However, all analysis so far has been focused on the role of SVP/DAM genes in the buds, and their role in regulating the photoperiodic response in leaves is still unclear.
Besides FT2, another important factor of the short-day response is gibberellins (GAs, Eriksson et al., 2000). GAs are growth-promoting hormones and work both through and independently of FT2 (Eriksson et al., 2015). A decrease in the levels of active GAs is essential for growth cessation and bud set (Eriksson et al., 2000). So far it is poorly understood how the levels of active GAs are regulated upon shift to SD. In short-day grown Arabidopsis, SVP represses the expression of GA20 oxidase, a gene encoding a rate-limiting enzyme in the GA biosynthesis pathway, thereby keeping the amount of bioactive GAs low (Rieu et al., 2008; Andrés et al., 2014). If this function of SVP was conserved in trees, it could be another mechanism through which SVL could potentially control growth cessation.
The ability of SVP to control FT expression and GA biosynthesis in Arabidopsis as well as the involvement of MADS genes in the phenology of other tree species prompted us to investigate the role of SVL in the regulation of growth cessation and bud set in Populus. Our data show that SVL expression in the leaves modulates the timing of SD-induced growth cessation and is able to repress both FT2 and GA20 oxidase by binding to their promoters. Thus, SVPs mode of action has been conserved between Arabidopsis and Populus, even though the biological process it is involved in has changed.
Hybrid aspen (Populus tremula × tremuloides) clone T89 was used as WT control and all genetic modifications were done in this background. Plants were cultivated on ½ Murashige and Skoog medium under sterile conditions for 4 weeks or until they had rooted (max. 8 weeks). After transfer to soil, plants were grown in growth chambers in LD (18 h light, 20°C/6 h dark, 18°C) and with weekly fertilization (10 ml NPK-Rika S/plant). To induce growth cessation, plants were moved to SD (14-h light, 20°C/ 10-h dark, 18°C) and fertilization was stopped. For dormancy release, plants were treated with cold (8 h light, 6°C/16 h dark, 6°C). In both SD and LD, previously published bud scores (Ibáñez et al., 2010) were used to assess effects on bud development (set/flush). For year-around gene expression analysis, a ca. 40-year-old local (Umeå, Sweden) aspen tree was sampled once a month around midday (May to August leaves, buds from September to April).
Protein sequences of SVP homologues were aligned in CLC main workbench (Qiagen) and a Maximum Likelihood Phylogeny was constructed with neighbor-joining method and 1,000 bootstrap replicates.
To generate SVL RNAi plants, the RNAi fragment was amplified by PCR using PtSVLRNAiF and PtSVLRNAiR primers, which contain attB1 and attB2 sites, respectively. The fragment was introduced into the pDONOR 201 vector (Invitrogen) by BP recombination. The PttSVLRNAi fragment was then transferred to the final destination vector pK7GWIGWI (Karimi et al., 2002) with Invitrogen LR recombinase, creating a double-stranded RNAi molecule driven by the constitutive Cauliflower Mosaic Virus 35S promoter. For construction of PttSVLoe—(35S::PttSVL:Myc), full-length PttSVL CDS was amplified from hybrid aspen mRNA with oxPttSVLF and oxPttSVLR primers and cloned into pDONOR 201 with BP clonase (Invitrogen). The fragment was then transferred to the destination vector pGWB18 (Karimi et al., 2002). Primers used for construct generation are listed in Supplementary Table S1. All cloning reactions were performed according to the manufacturer’s instructions. Hybrid aspen was transformed as previously described (Nilsson et al., 1992). Arabidopsis thaliana was transformed by using the floral dip method (Clough and Bent, 1998). The svp-32 (Salk_072930) mutant seeds were ordered from Nottingham Arabidopsis Stock Centre (NASC).
Arabidopsis thaliana WT Col-0, svp-32, and PttSVLoe plants were grown on soil in LD (16 h light/8 h dark, 22°C). To measure flowering time, rosette leaves and cauline leaves of 10 plants per line were counted until first flowers were visible.
Poplar leaves (youngest fully expanded leaves) were ground to fine powder, of which 100 mg were used for RNA extraction with CTAB extraction buffer (Chang et al., 1993; 2% CTAB, 100 mM Tris-HCl (pH 8.0), 25 mM EDTA, 2 M NaCl, 2% PVP). The samples were incubated at 65°C for 2 min and extracted twice with an equal volume of chloroform–isoamyl alcohol (24:1). Nucleic acids were precipitated at −20°C for 3 h with ¼ volumes 10 M LiCl. Precipitate was collected by centrifugation (13,000 rpm, 4°C, 20 min) and washed with 70% EtOH. After drying it was dissolved in 60 μl H2O (DEPC treated). Contamination of genomic DNA was removed from 2,5 μg total nucleic acid by DNase treatment (TURBO DNA-free™ Kit, Ambion®), and 1,000 ng RNA were used for cDNA synthesis with iScript™ cDNA Synthesis Kit (Bio-Rad). The cDNA was diluted 50 times for downstream applications. Quantitative real-time PCR (qPCR) was run on a LightCycler® 480 with SYBR Green I Master (Roche). All kits and machines were used according to the manufacturer’s instructions. The reaction protocol started with 5 min pre-incubation at 95°C, followed by 50 cycles of amplification consisting of 10 s denaturation at 95°C, 15 s annealing at 60°C and 20 s elongation at 72°C. For the acquisition of a melting curve, fluorescence was measured during the step-wise increase in temperature from 65°C to 97°C. Relative expression levels were obtained using the 2-ΔΔCq method (Livak and Schmittgen, 2001). GeNorm (Vandesompele et al., 2002) identified UBQ and 18S as most stable reference genes. All used primers had an efficiency of >1,8 and their correct product was confirmed by sequencing. A complete list of primer sequences can be found in Supplementary Table S1.
Material (about 150 mg fresh weight of the youngest fully expanded leaves) was suspended in 80% methanol–1% acetic acid containing internal standards and mixed by shaking during 1 hour at 4°C. The extract was kept a − 20°C overnight and then centrifuged and the supernatant dried in a vacuum evaporator. The dry residue was dissolved in 1% acetic acid and passed through a Oasis HLB (reverse-phase) column as described in (Seo et al., 2011). The dried eluate was dissolved in 5% acetonitrile–1% acetic acid, and the GAs were separated using an autosampler and reverse-phase UHPLC chromatography (2.6 μm Accucore RP-MS column, 100 mm length x 2.1 mm i.d.; Thermo Fisher Scientific) with a 5 to 50% acetonitrile gradient containing 0.05% acetic acid, at 400 μl/min over 21 min.
The hormones were analyzed with a Q-Exactive mass spectrometer (Orbitrap detector; Thermo Fisher Scientific) by targeted Selected Ion Monitoring. The concentrations of GAs in the extracts were determined using embedded calibration curves and the Xcalibur 4.0 and TraceFinder 4.1 SP1 programs. The internal standards for quantification were the deuterium-labeled hormones.
For RNA sequencing experiments RNA was isolated as described above and purified with RNeasy kit (Qiagen) according to the manufacturer’s instructions. DNAse treatment was performed on column (Qiagen). Concentration and quality of RNA were assessed using Qubit™ RNA BR Assay Kit (Invitrogen) and Bioanalyzer (Agilent), respectively. 3 μg total RNA with RIN ≧8 were sent for sequencing to SciLife Lab, Stockholm. Library preparation was carried out with an Agilent NGS Bravo workstation in 96-well plates with TruSeq Stranded mRNA kit (Illumina) according to the manufacturer’s instructions. mRNA was purified through selective binding to poly dT-coated beads and fragmented using divalent cations under elevated temperature. cDNA was synthesized using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific), cleaned with AMPure XP solution (Thermo Fisher Scientific), 3′ adenylated, and ligated to adapters. Fragments were cleaned with AMPure XP beads (Thermo Fisher Scientific), amplified by PCR, and purified with AMPure XP beads (Thermo Fisher Scientific). After washing with 80% ethanol, they were eluted in EB (Qiagen). The quality and concentration of the adapter-ligated libraries were checked on the LabChip GX/HT DNA high sensitivity kit and by Quant-iT, respectively. The libraries were then sequenced using the Illumina NovaSeq-6,000 platform, generating from 20 to 110 million paired-end reads (2 × 150 bp) per sample.
The data pre-processing was performed as described here: http://www.epigenesys.eu/en/protocols/bio-informatics/1283-guidelines-for-rna-seq-data-analysis. The quality of the raw sequence data was assessed using FastQC.1 Residual ribosomal RNA (rRNA) contamination was assessed and filtered using SortMeRNA [v2.1 (Kopylova et al., 2012); settings--log --paired_in --fastx--sam --num_alignments 1] using the rRNA sequences provided with SortMeRNA (rfam-5 s-database-id98.fasta, rfam-5.8 s-database-id98.fasta, silva-arc-16 s-database-id95.fasta, silva-bac-16 s-database-id85.fasta, silva-euk-18 s-database-id95.fasta, silva-arc-23 s-database-id98.fasta, silva-bac-23 s-database-id98.fasta and silva-euk-28 s-database-id98.fasta). Data were then filtered to remove adapters and trimmed for quality using Trimmomatic [v0.39 (Bolger et al., 2014); settings TruSeq3-PE-2.fa:2:30:10 LEADING:3 SLIDINGWINDOW:5:20 MINLEN:50]. After both filtering steps, FastQC was run again to ensure that no technical artefacts were introduced. Filtered reads were pseudo-aligned to v1.1 of the P. tremula transcripts {retrieved from the PopGenIE resource (Sundell et al., 2015) using salmon [v1.1.0 (Patro et al., 2017)], with non-default parameters --gcBias--seqBias --validateMappings} against an index containing the P. tremula v1.1 genome sequence as decoy. Statistical analysis of single-gene differential expression between conditions was performed in R (v4.0.0; R Core Team 2020) using the Bioconductor [v3.10 (Huber et al., 2015)] DESeq2 package [v1.28.1 (Love et al., 2014)]. FDR adjusted values of p were used to assess significance; a common threshold of 1% was used throughout. For the data quality assessment (QA) and visualization, the read counts were normalized using a variance stabilizing transformation as implemented in DESeq2. The biological relevance of the data—for example, biological replicates similarity—was assessed by principal component analysis and other visualizations (e.g., heatmaps), using custom R scripts, available from https://github.com/DomeniqueA/SVL. The raw data are available from the European Nucleotide Archive2 under the accession number PRJEB46749.
WT and SVLoe plants were grown in LD (18 h light, 20°C/6 h dark, 18°C) for 4 weeks before the experiment started. Upon shift to SD, half of the plants were transferred to regular SD (14 h light, 20°C/ 10 h dark, 18°C), while the other half were transferred to cold SD (14 h light, 15°C/10 h dark, 10°C). Proteins were isolated and visualized on a Western blot using anti-myc antibodies (Agrisera).
WT and SVPoe plants were grown in LD. Per genotype, one fully expanded leaf was harvested from each of four biological replicates at ZT 18 and cut into small pieces. These were cross-linked in 50 ml PBS buffer +1% formaldehyde and vacuum (4 times 5 min). The reaction was stopped with addition of glycine to a final concentration of 100 mM. The pieces of leaves were frozen in liquid N2 and ground to fine powder. Nuclei were extracted, lysed in nuclei isolation buffer [50 mM HEPES pH 7.4, 5 mM MgCl2, 25 mM NaCl, 5% sucrose, 30% glycerol, 0.25% Triton X-100, 0.1% ß-mercaptoethanol, 0.1% proteinase inhibitor cocktail (Sigma)] and sonicated, resulting in DNA fragments of 500–1,000 base pair length. For immunoprecipitation, 300 μl of the nuclear extract were homogenized with 200 μl IP buffer (80 mM Tris-HCl pH 7.4, 230 mM NaCl, 1.7% NP40, 0.17% DOC) followed by 1 μl 1 M DTT, 1 μl protease inhibitor cocktail, 1 μl 10 mg/ml RNase A and 5 μl of a monoclonal myc antibody (ab32, Abcam). The mixture was incubated under soft agitation at 4°C over night and centrifuged at full speed for 15 min at 4°C. 40 μl Protein A beads were added into the supernatant and incubation was continued for another 2 hours with soft agitation at 4°C. Protein beads were first washed two times with ice-cold low salt buffer (20 mM Tris-HCl pH 8, 150 mM NaCl, 0.1%SDS, 1% Triton X-100, and 2 mM EDTA), and two times with high salt buffer (20 mM Tris-HCl pH 8, 500 mM NaCl, 0.1%SDS, 1% Triton X-100, and 2 mM EDTA). Then, beads were washed two times with ice-cold LiCl buffer (10 mM Tris-HCl pH 8, 250 mM LiCl, 1% Igepal Ca-630, 1% DOC, and 1 mM EDTA). Chromatins were eluted from the beads with elution buffer (100 mM NaHCO3, 1% SDS) at 65°C for 20 min. To de-crosslink the extract, it was incubated with proteinase K (10 ng/ml) for 1 hour at 55°C. Afterward, DNA was extracted by Chromatin immunoprecipitation (ChIP) DNA Clean & Concentrator Kits (Biosite D5205). Quantities of immunoprecipitations were quantified using SYBR green (Roche) and the iQ5 light cycler (Bio-Rad). A similarly treated extract from WT without tagged protein was used as control. Primers used for amplification of genomic fragments are listed in Supplementary Table S1.
Populus has one orthologous gene to Arabidopsis SVP called SVL (Singh et al., 2018; Supplementary Figure S1A). AtSVP and PtSVL share 66% identity on the amino acid level, making SVL the only likely SVP ortholog compared to other MADS domain-containing genes in Populus (Supplementary Figure S1A; Singh et al., 2018). Because of the high similarity to SVP (Supplementary Figure S1B), we hypothesized that SVL could act like SVP in Arabidopsis by having a function in the photoperiodic response in leaves. The svp mutant is early flowering (Hartmann et al., 2000) and we tested whether SVL could rescue this phenotype. For this we expressed SVL cDNA under the control of the 35S promoter in svp-32 plants. Flowering time was determined by counting rosette and cauline leaves. These plants produced significantly more leaves than svp-32 mutants and wild-type (WT) Arabidopsis plants before developing the first flowers (Supplementary Figure S2). These results imply that the functionality of has been conserved between Arabidopsis SVP and Populus SVL.
Populus SVL function has previously been described in the shoot apex in relationship to the regulation of bud dormancy (Singh et al., 2018, 2019). We wanted to investigate to what extent leaf-expressed SVL also contributes to the regulation of growth cessation and bud set. Investigation of the SVL annual expression pattern in local adult aspen trees (Umeå, Sweden) sampled in the middle of the day showed that it is highly expressed in leaves during the short days (SDs) of late summer and early autumn (Figure 1A), after FT2 expression declined (Supplementary Figure S3), and to higher levels than what can be detected in buds. To test whether this expression pattern is consistent in juvenile trees grown in controlled growth conditions, we checked the diurnal expression pattern of SVL in leaves first in long days (LD) and after 2 weeks of SD treatment (Figure 1B). In these conditions, one of the first genes to respond is FT2 which shows a clear downregulation after 2 weeks in 14 h SD (Ding et al., 2018). In long days, SVL displayed a minor peak of expression at ZT 6–8 (Figure 1B). The SVL expression increased after shift to SD and showed a prominent morning peak at around 4 hours after dawn, suggesting a role for SVL in the photoperiodic response in leaves. We then also wanted to know if a decrease in ambient temperature could increase the stability of the SVL protein, as has been shown for Arabidopsis SVP (Lee et al., 2013). When plants expressing myc-tagged SVL from a constitutive promoter were exposed to lower temperatures than our standard SD treatment, the accumulation of SVL protein was increased (Figures 1C,D). This indicates that there could be a role for leaf-expressed SVL in response to both short photoperiods and lower temperatures.
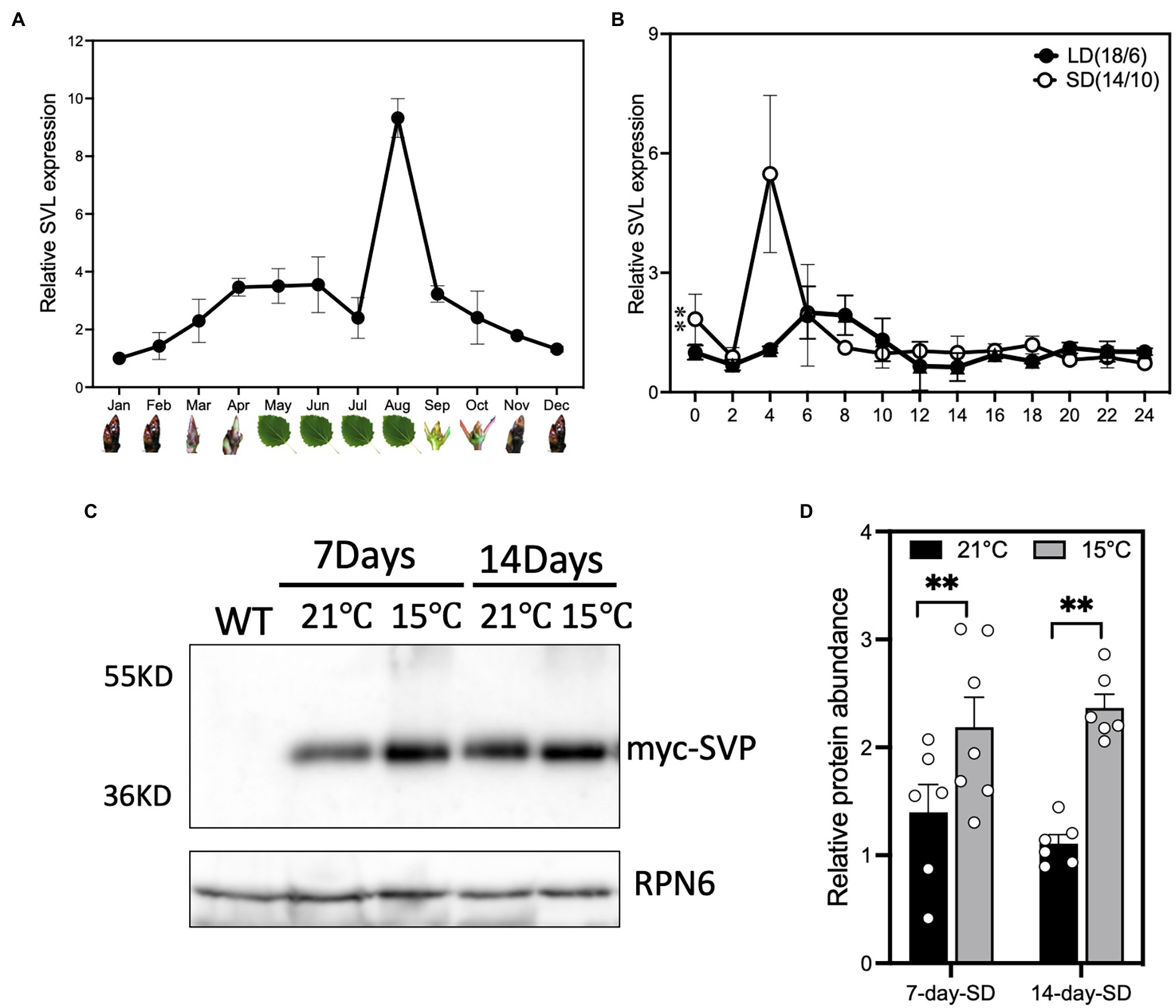
Figure 1. SVL is expressed during the autumn. (A) SVL expression in field-grown mature Populus tremula over the course of 1 year. Samples were taken at 2 p.m. in the middle of each month. May–August leaves, September–April terminal buds. Values are relative to the expression in January samples. Error bars indicate standard error of biological replicates, n = 3. (B) Relative SVL expression in WT during long days (18 h light/6 h dark) and after 2 weeks of short days (14-h light/10-h dark) treatment. Lowest expression in LD was set as 1. The asterisks indicate a statistically significant difference between LD and SD samples by two-way ANOVA Fisher’s test. Error bars indicate standard error of biological replicates, n = 3. **Indicate p < 0.01. (C) Western blot showing that the myc-SVL protein accumulates in leaves of 35S::myc:SVL-expressing plants during SD treatment at both 21°C and 15°C day temperature. (D) Relative protein abundance of myc-SVL in leaves of 35S::myc:SVL-expressing plants after 1 week and 2 weeks of SD treatment at both 21°C and 15°C day temperature. All values are relative to the protein amount at 14-day SD at 21°C. The asterisks indicate a statistically significant difference between two temperature samples by Welch’s test. Error bars indicate standard error of biological replicates, n ≥ 6. **Indicate p < 0.01.
To test the role of SVL, we generated SVL RNAi and SVL over-expressing (SVLoe) trees. Downregulation was up to 80% effective, while overexpression resulted in a six-fold increase of SVL expression at ZT17 compared to wild-type T89 (WT; Supplementary Figure S4). Neither downregulation nor overexpression of SVL had a striking effect on vegetative growth; all transgenic lines were indistinguishable from WT controls after 3 weeks in LD (Figure 2A). After shift to SD, poplars respond with growth cessation and bud set. We used previously described bud scores (Ibáñez et al., 2010) to test the speed of SD response in three independent transgenic lines per construct. SVL RNAi plants showed a small but consistent delay of bud set compared to WT (Figure 2B). Both growth cessation (score 2) and bud set (score 1) were delayed by ca. 1 week. SVLoe plants on the other hand ceased growth several weeks earlier than WT (Figure 2C). This indicates that SVL is a repressor of vegetative growth and promoter of SD-induced growth cessation.
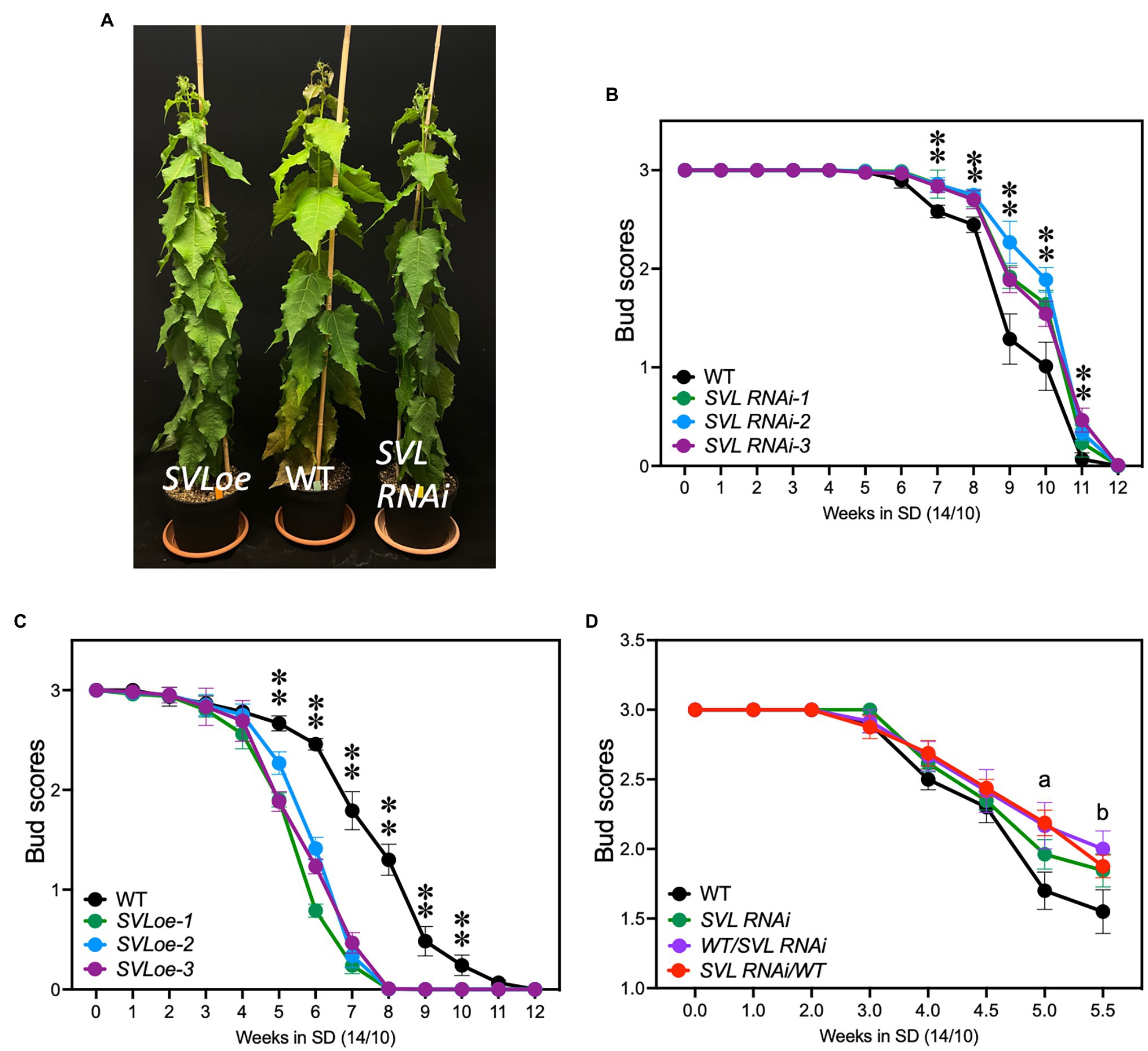
Figure 2. SVL controls growth cessation and bud set. (A) Photograph of SVLoe, WT, and SVL RNAi plants after 3 weeks in LD. (B) Plot of bud scores showing that growth cessation and bud set of three independent SVL RNAi lines were delayed compared to WT. Error bars indicate standard error of biological replicates, n = 9. (C) Plot of bud scores describing growth cessation and bud set of three independent SVLoe lines compared to WT. SVLoe plants stopped growth earlier than WT. The asterisks indicate statistically significant difference of each transgenic line from WT by two-way ANOVA Fisher’s test. Error bars indicate standard error of biological replicates, n = 9. **Indicate p < 0.01. (D) Plot of bud scores showing growth cessation of WT, SVL RNAi, and grafts thereof. a, significant differences were observed from multiple comparisons of WT vs. SVLRNAi, WT vs. WT/SVLRNAi, WT vs. SVLRNAi/WT, and SVLRNAi vs. SVLRNAi/WT. b, significant differences were observed from multiple comparisons of WT vs. SVLRNAi, WT vs. WT/SVLRNAi, and WT vs. SVLRNAi/WT. Statistic significance was determined by two-way ANOVA Fisher’s test (p < 0.05). Error bars indicate standard error of biological replicates, n ≥ 6.
Expression of both FT2 and GA20oxidase in rootstocks of grafted trees is sufficient to significantly delay growth cessation and bud set (Miskolczi et al., 2019). We then asked if the role of SVL in modulating the timing of growth cessation is due to SVL activity in the leaf or shoot apex or both. To investigate this we performed reciprocal graftings of SVL RNAi and wild-type trees and compared the timing of growth cessation to trees where scions had been grafted to their own stock. In both types of heterografts growth cessation was delayed to the same extent as in SVL RNAi homografts suggesting that SVL modulates the timing of growth cessation trough activity both in the leaf and in the shoot apex (Figure 2D).
We then tested whether the different speeds of response in transgenic lines were due to altered expression of FT2. After 2 weeks of SD treatment, FT2 expression had ceased in WT and SVLoe, while it was still strongly expressed in the SVL RNAi lines (Figure 3A). In addition to FT2, gibberellins are also known to affect growth cessation and bud set. We therefore analyzed the expression of a GA20 oxidase2, a key enzyme in gibberellin biosynthesis and found that it was increased in the leaves of SVL RNAi lines, while being reduced in SVLoe (Figure 3B). We focused on GA20 oxidase2 because we have found that it is the predominantly expressed GA20 oxidase gene in leaves (not shown). Consequently, the amount of the active gibberellin GA1 was increased in leaves of SVL RNAi lines in both LD and SD (Supplementary Figure S5) compared to wild type. This suggests that SVL can influence the timing of growth cessation through a repression of both the expression of FT2 and the biosynthesis of gibberellins.
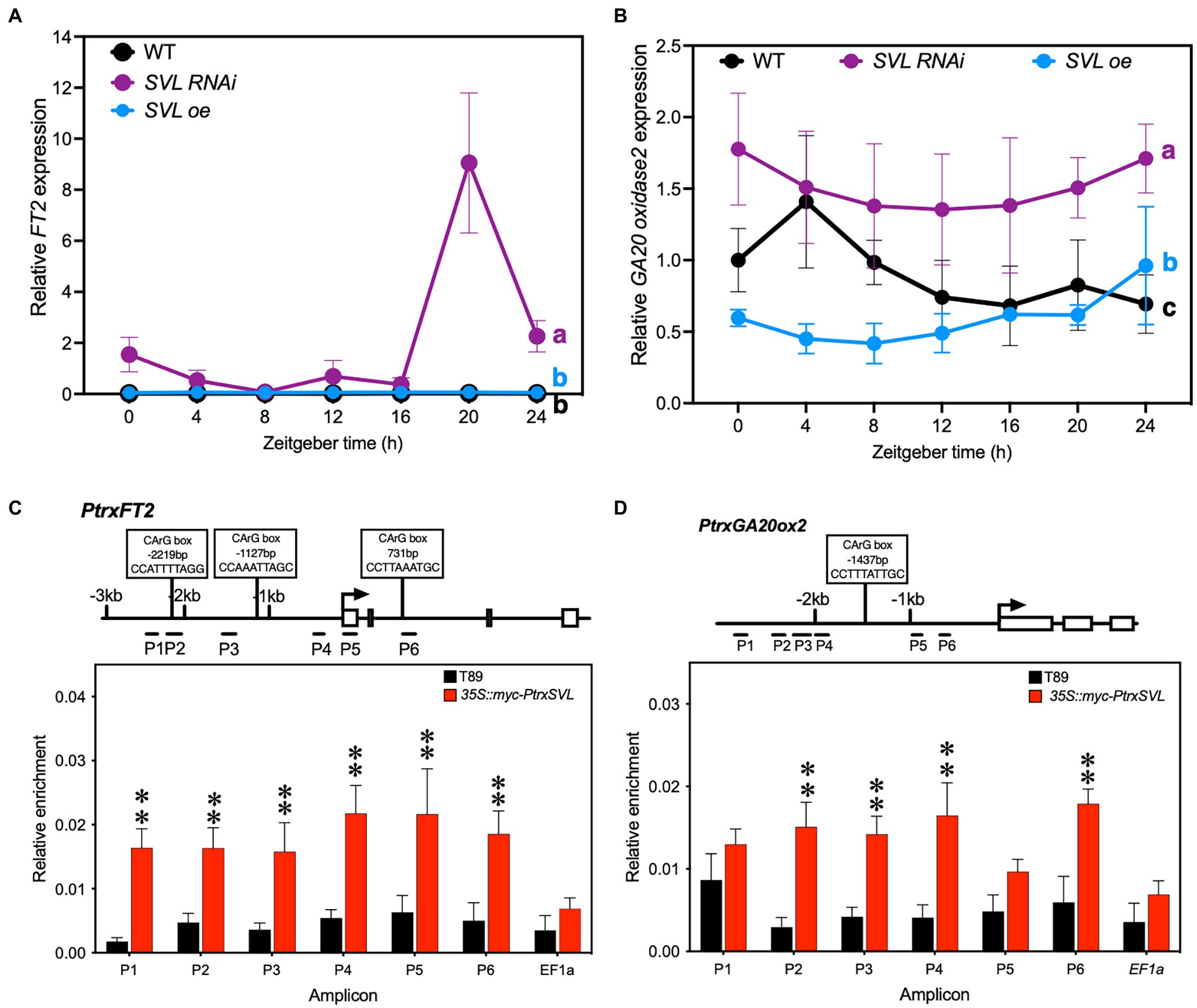
Figure 3. SVL is a transcriptional repressor and is associated with the promoters of its targets. (A,B) Gene expression of FT2 (A) and GA20 oxidase2 (B) after 2 weeks of SD (14 h light/10 h dark) treatment. In both cases expression was increased in SVL RNAi and decreased in SVLoe lines. Lines that do not share letters are significantly different from each other according to post-two-way ANOVA Fisher’s test (p < 0.05). Error bars indicate standard error of biological replicates, n = 3. (C,D) Relative enrichment of promoter fragments after ChIP quantified by qPCR of FT2 (C) and GA20 oxidase2 (D). Values are normalized against input. Position of putative SVL binding sites, CArG boxes, shown to bind to Arabidopsis SVP in leaves (Gregis et al., 2013), are indicated. The asterisks indicate a statistically significant difference from WT by Welch’s test. Error bars indicate standard error of biological replicates, n = 4, **indicate p < 0.01.
We then asked if FT2 and GA20 oxidase2 are direct targets of SVL. Since SVL is a MADS-box transcription factor, we tested the ability of the SVL protein to bind to the promoter region of these genes. For that we performed a chromatin immunoprecipitation assay in leaves of WT and our myc-tagged SVLoe lines. Quantification by qPCR showed significant enrichments of six fragments surrounding the FT2 transcriptional start site, up to 2.5 kb upstream and 1 kb downstream (Figure 3C). Enrichment at the GA20 oxidase2 promoter was significant for four of six fragments (Figure 3D). No enrichment could be detected at a control locus (Figures 3C,D). These results show that SVL can associate with the promoters of FT2 and GA20 oxidase2 and potentially repress their expression through direct binding.
To better understand what role SVL plays in leaves during SD treatment, we performed RNA sequencing analysis on leaves of wild-type and SVL RNAi plants. Samples were harvested at ZT17 during LD and after one, two, three and 10 weeks of SD treatment, respectively. Major transcriptional changes happened after the shift from LD to SD in both WT and SVL RNAi lines with more than 10,000 genes being differentially expressed between the two time points (Figure 4A; Supplementary Figure S6). However, no further significant changes were detectable after two and 3 weeks of SD. At the individual time points, only a small number of differentially expressed genes (DEG) could be detected between the two lines (Figure 4B; Supplementary Figure S6) and gene ontology (GO) enrichment resulted in no specific terms. These results indicate that the role of SVL in leaves might be limited to the regulation of a very limited set of genes.
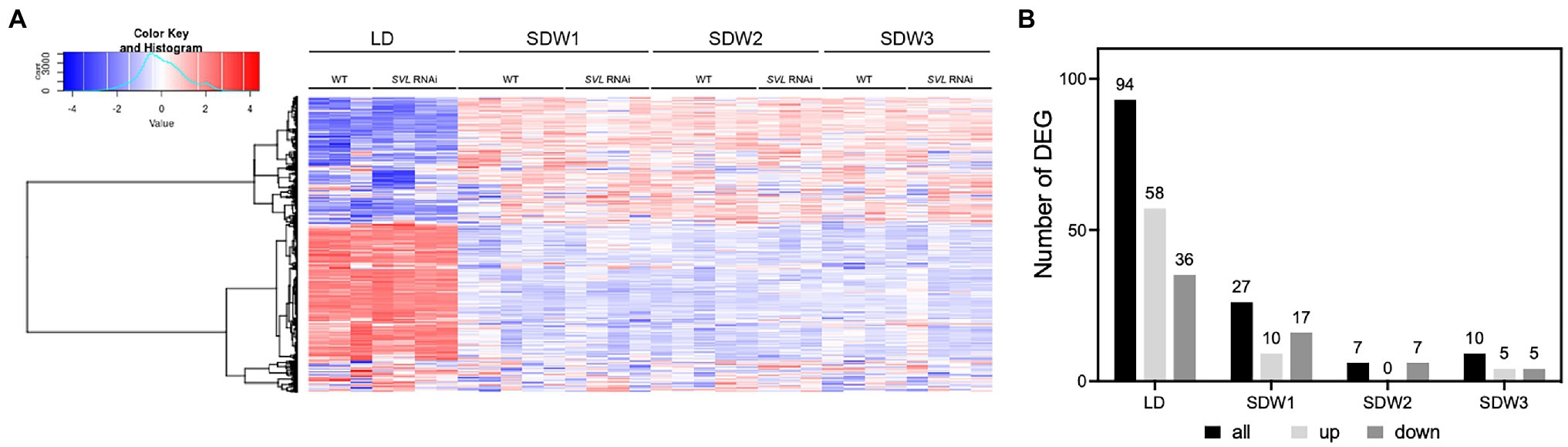
Figure 4. Global transcriptional changes in response to SD treatment. (A) Heatmap of gene expression in WT and SVL RNAi in LD and SD (week 1 to week 3). (B) Number of differentially expressed genes between WT and SVL RNAi over time.
AtSVP is a known floral repressor (Hartmann et al., 2000), acting on the FT promoter (Lee et al., 2007). Here we show that PtSVL (hereafter SVL) is functionally conserved as was recently shown for apple (Falavigna et al., 2021) and previously also, for instance, Kiwi SVP-like genes, (Wu et al., 2012). Both SVP and SVL proteins share 66% identity and SVL overexpression was sufficient to complement the Arabidopsis svp-32 mutant phenotype. ChIP analysis showed that SVL could bind to both FT2 and GA20 oxidase promoters, repressing their transcription. This suggests that SVL expression in the leaf can fulfil a similar role in poplar as in Arabidopsis, modulating the photoperiodic regulation of FT and GA20 oxidase expression. To repress downstream targets, AtSVP forms heterodimers with MADS-box proteins like FLM and FLC (Lee et al., 2007, 2013; Posé et al., 2013). In apple, it has recently been shown that the SVP-like protein MdSVPa can form transcriptional complexes with various MADS-box proteins that are expressed in buds at different dormancy-specific phases (Falavigna et al., 2021). It remains to be shown if the SVL activity in the Populus leaf is also dependent on one or more MADS-box proteins.
In Arabidopsis, SVP is part of the thermosensory pathway, which inhibits flowering in cold conditions (Lee et al., 2007), partially mediated by a stabilization of the SVP protein at colder temperatures (Lee et al., 2013). Temperatures drop during autumn and we could show that SVL protein was more abundant in a combination of short-day treatment and cold (Figure 1C), mimicking autumn conditions. While the timing of growth cessation is mainly regulated by photoperiod, integration of temperature signals through SVL could give trees more flexibility to fine-tune their SD response, it could also be an important factor contributing to SVLs function in buds during the induction of dormancy.
There is no strict correlation between the downregulation of FT2 and the upregulation of SVL in the year-around samples (Figure 1A; Supplementary Figure S3) confirming that SVL is not the primary regulator of the photoperiodic response and the regulation of FT2. However, alteration of SVL expression levels influenced the timing of growth cessation and bud set upon short-day (SD) treatment. We could show that this was caused by an effect on FT2 expression and levels of GAs. Consistently, SVL RNAi lines had increased FT2 expression and a delayed growth cessation, while SVLoe plants had strongly reduced FT2 expression and early growth cessation. Furthermore, levels of the bioactive gibberellin GA1 were increased in SVL RNAi plants. This suggests that the role of SVL is to modulate the timing of the photoperiodic response.
The interest for the role of SVP-like genes in the regulation of dormancy originally stemmed from studies of the evergrowing mutant (evg) in peach (Prunus persica), which fails to form terminal vegetative buds and continues to grow indeterminately under dormancy-inducing conditions. The evg locus was mapped and found to contain a large deletion of six tandemly repeated SVP-like genes called DAM genes (Bielenberg et al., 2008). The simultaneous downregulation of the expression of three SVP/DAM genes in apple through RNAi also caused an evergrowing phenotype (Wu et al., 2021). Because of the focus on the expression and function of these genes during dormancy-induction in the bud, to our knowledge, there appears to be no characterization of the photoperiodic regulation of expression of FT-like genes in leaves of the evergrowing mutants. It is an interesting possibility that the loss of SVP/DAM-like expression in the leaves of peach and apple could contribute to an inability to keep FT expression down in response to the short-day signal. Since downregulation of FT expression appears to be a prerequisite for growth cessation and bud set (Böhlenius et al., 2006), this could at least partially explain the fact that the evergrowing mutants are not only failing to establish dormancy, but are also not able to enter into growth cessation and bud set—which is also a prerequisite for the entry into dormancy.
There are several other SVP-like and DAM-like genes in Populus (Supplementary Figure S1). In order to fully understand the role of the individual genes, it will be important to know if complete knockouts of SVL expression through CRISPR-Cas9-mediated gene editing, or combinations of the deactivation of several SVP/DAM-like genes, also leads to an evergrowing phenotype in poplar trees.
Previous studies showed the role of SVL in the shoot apex, both during the establishment of dormancy during SD (Singh et al., 2019) and its release during winter (Singh et al., 2018). Both pathways work through differential regulation of gibberellin biosynthesis with SVL inducing GA2 oxidase in the developing bud, presumably in order to reduce the amount of active GAs that can reach the shoot apex, while, at the same time, repressing the expression of GA20 oxidase to downregulate GA biosynthesis. We show that SVL functions through a similar pathway in leaves. Grafting studies in Populus have shown that not only FT2 but also GA20 oxidase expression in leaves, clearly affect growth cessation and bud set (Miskolczi et al., 2019), suggesting that both FT2 and GAs are moving from leaf to shoot apex in order to modulate this process. Our data show that SVL affects the expression of both FT2 and GA20 oxidase in leaves and that SVL associates with both the FT2 and GA20 oxidase2 loci that both contain putative SVL binding sites in the form of CArG boxes shown to bind to Arabidopsis SVP (Gregis et al., 2013). However, in contrast to our findings here, Singh et al. (2019) could not detect an interaction between SVL and the GA20 oxidase loci. This discrepancy could be attributed to the use of different protocols and primers for the ChIP assay, but could also be related to the fact that we looked for interactions in leaf samples rather than in shoot apex samples as used by Singh et al. (2019). SVP-like proteins acts in complexes with other MADS-box proteins and co-transcription factors. These other factors are likely to differ between leaf and shoot apex samples and might affect the SVL binding. They might also contribute to an indirect binding to the GA20 oxidase locus in the ChIP assay.
One of the direct targets of FT2 in the shoot apex is the gene Like-AP1 (LAP1) which in turn mediates the regulation of cell cycle-related genes that are downregulated during growth cessation. (Azeez et al., 2014). Is it possible that LAP1 is also a SVL target? Although our grafting experiments did not allow us to collect enough material to analyze the expression of target genes in the shoot apex, we did check for LAP1 expression in leaves of SVL RNAi plants. LAP1 is normally expressed to very low levels in leaves, but is dramatically upregulated in leaves of SVL RNAi trees (Supplementary Figure S7). The LAP1 locus also contain several potential SVL binding sites (Supplementary Figure S7). Although the relevance of the LAP1 regulation in the leaf is unclear, it shows a potential for SVL to also control growth cessation through a repression of LAP1 expression in the shoot apex that could be part of the explanation to the contribution of SVL expression for growth cessation in both leaf and shoot apex (Figure 2D).
Our data suggest that SVL has only minor effects on the leaf transcriptome, suggesting that it might have relative few targets in the leaf compared to in the shoot apex. When the dataset was specifically quired both FT2 and GA20 oxidase2 were not found to be higher expressed in the SVL RNAi dataset compared to wild type after the shift to SD (not shown). The reason for these genes not appearing in the DEG list is probably due to relatively low expression levels and/or the fact that this was samples from a single time-point at the end of the day when expression was not significantly different.
Additionally, SVL has been shown to affect FT1 expression in buds. Thus, SVL is involved in three similar pathways regulating different aspects of the annual growth cycle of Populus trees (Figure 5); First through a regulation of the photoperiod response in leaves, contributing to the downregulation of FT2 and GA20 oxidase, leading to growth cessation and bud set (this study); Then as an inducer of GA2 oxidase and CALLOSE SYNTHASE 1 in the buds to prevent growth-inducing signals to reach the shoot apex to establish dormancy (Singh et al., 2019); And finally, its expression is reduced in response to low temperatures in winter, leading to a relieved repression of FT1 and reduced expression of TCP18/BRC1, hypothesized to lead to bud break (Singh et al., 2018). Consequently, SVL serves as an important regulator of both the beginning and end of the growing season as well as the establishment of winter dormancy. Interestingly, these three different phases of SVL regulation corresponds to three different clusters of expression profiles for DAM and SVP-like genes from different fruit tree species (Falavigna et al., 2019, 2021). In Rosaceae, DAM and SVP-like genes have evolved into different clades. In apple, the SVP-like protein SVPa provides DNA-binding activity to different complexes with DAM proteins that are specifically expressed during different phases of the dormancy cycle (Falavigna et al., 2021). Our data from growth cessation together with the previous data from bud set and bud break (Singh et al., 2018, 2019) suggest that this could also be true in Populus trees, with SVL serving as the central DNA-binding hub.
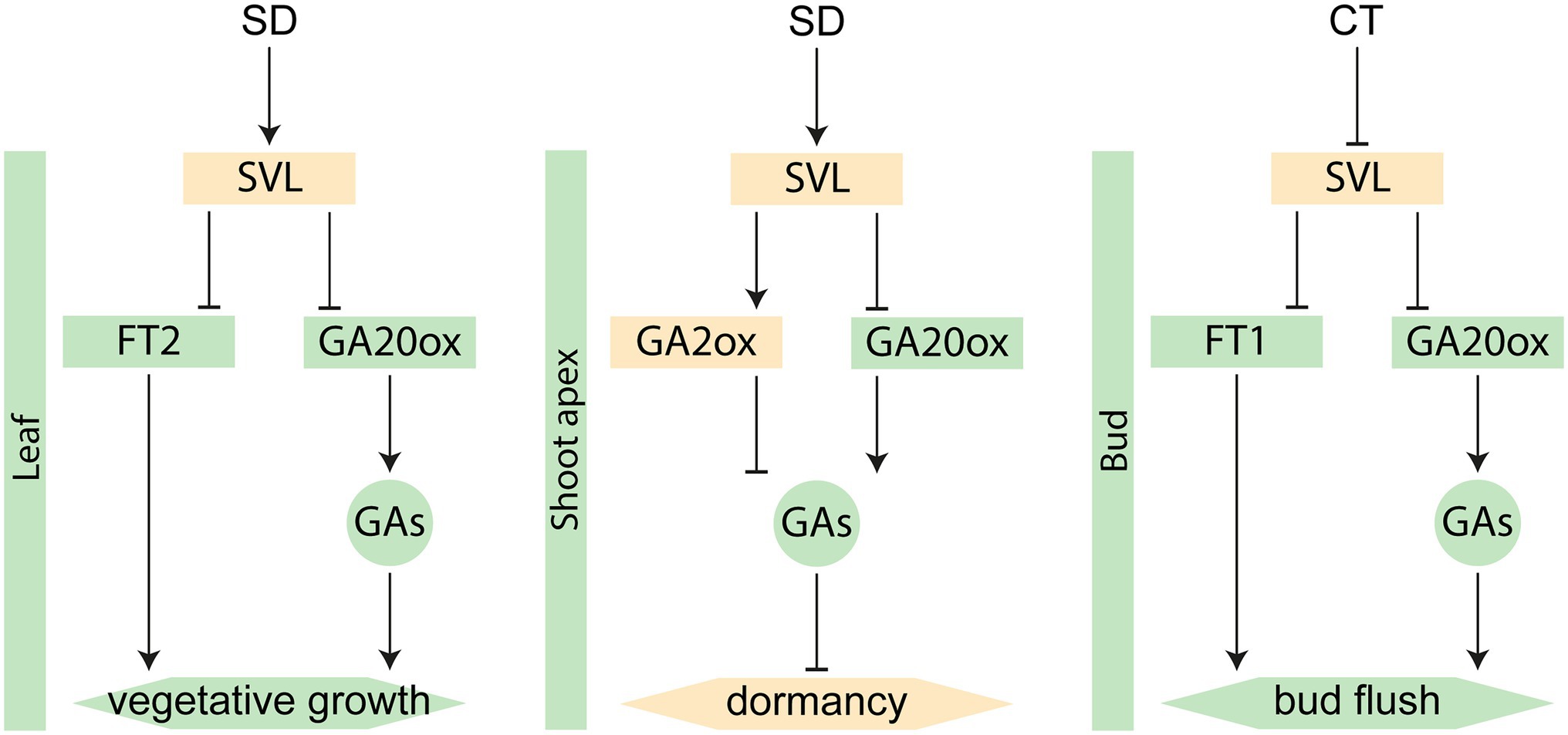
Figure 5. Different roles of SVL in the annual growth cycle. Green color indicates growth-promoting factors, while orange color indicates repressors. Boxes represent genes and circles represent hormones. SD = short day, CT = cold treatment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB46749.
DA, JZ, and ON planned the research. DA, JZ, BZ, MR, KL, and AM performed experiments and analyzed data. DA and ON wrote the manuscript and all authors reviewed and approved its final version.
VR supported PhD positions for AM and DA. Kempe foundation supported a post-doctoral stipend for JZ. VINNOVA and KAW supported positions, consumables, and platform support for BZ, MR, and DA. This work was supported by grants from the Swedish Research Council, the Knut and Alice Wallenberg Foundation, Kempe Foundation and the Swedish Governmental Agency for Innovation Systems (VINNOVA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by the Science for Life Laboratory. We thank SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We also thank Esther Carrera for the GA quantifications carried out at the Plant Hormone Quantification Service, IBMCP, Valencia, Spain.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.823019/full#supplementary-material
Andrés, F., Porri, A., Torti, S., Mateos, J., Romera-Branchat, M., García-Martínez, J. L., et al. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. U. S. A. 111, E2760–E2769. doi: 10.1073/pnas.1409567111
Azeez, A., Miskolczi, P., Tylewicz, S., and Bhalerao, R. P. (2014). A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr. Biol. 24, 717–724. doi: 10.1016/j.cub.2014.02.037
Bielenberg, D. G., Wang, Y. E., Li, Z., Zhebentyayeva, T., Fan, S., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genom. 4, 495–507. doi: 10.1007/s11295-007-0126-9
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 11, 113–116. doi: 10.1007/BF02670468
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ding, J., Böhlenius, H., Rühl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). GIGANTEA-like genes control seasonal growth cessation in Populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Eriksson, M. E., Hoffman, D., Kaduk, M., Mauriat, M., and Moritz, T. (2015). Transgenic hybrid aspen trees with increased gibberellin (GA) concentrations suggest that GA acts in parallel with FLOWERING LOCUS T 2 to control shoot elongation. New Phytol. 205, 1288–1295. doi: 10.1111/nph.13144
Eriksson, M. E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Falavigna, V. D. S., Guitton, B., Costes, E., and Andrés, F. (2019). I want to (bud) break free: the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9:1990. doi: 10.3389/fpls.2018.01990
Falavigna, V. D. S., Severing, E., Lai, X., Estevan, J., Farrera, I., Hugouvieux, V., et al. (2021). Unraveling the role of MADS transcription factor complexes in apple tree dormancy. New Phytol. 232, 2071–2088. doi: 10.1111/nph.17710
Gregis, V., Andrés, F., Sessa, A., Guerra, R. F., Simonini, S., Mateos, J. L., et al. (2013). Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14:R56. doi: 10.1186/gb-2013-14-6-r56
Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., and Huijser, P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Hsu, C.-Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U. S. A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Hsu, C.-Y., Liu, Y., Luthe, D. S., and Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18, 1846–1861. doi: 10.1105/tpc.106.041038
Huber, W., Carey, V. J., Gentleman, R., Anders, S., Carlson, M., Carvalho, B. S., et al. (2015). Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods 12, 115–121. doi: 10.1038/nmeth.3252
Ibáñez, C., Kozarewa, I., Johansson, M., Ögren, E., Rohde, A., and Eriksson, M. E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153, 1823–1833. doi: 10.1104/pp.110.158220
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY™ vectors for agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Lee, J. H., Ryu, H.-S., Chung, K. S., Posé, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., and Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Li, Z., Reighard, G. L., Abbott, A. G., and Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530. doi: 10.1093/jxb/erp195
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Miskolczi, P., Singh, R. K., Tylewicz, S., Azeez, A., Maurya, J. P., Tarkowská, D., et al. (2019). Long-range mobile signals mediate seasonal control of shoot growth. Proc. Natl. Acad. Sci. U. S. A. 116, 10852–10857. doi: 10.1073/pnas.1902199116
Moser, M., Asquini, E., Miolli, G. V., Weigl, K., Hanke, M.-V., Flachowsky, H., et al. (2020). The MADS-box gene MdDAM1 controls growth cessation and bud dormancy in apple. Front. Plant Sci. 11:1003. doi: 10.3389/fpls.2020.01003
Nilsson, O., Aldén, T., Sitbon, F., Little, C. A., Chalupa, V., Sandberg, G., et al. (1992). Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1, 209–220. doi: 10.1007/BF02524751
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pin, P., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S. J., Gong, F., et al. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504. doi: 10.1111/j.1365-313X.2007.03356.x
Rohde, A., and Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Seo, M., Jikumaru, Y., and Kamiya, Y. (2011). Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods Mol. Biol. 773, 99–111. doi: 10.1007/978-1-61779-231-1_7
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkovič, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Miskolczi, P., Maurya, J. P., and Bhalerao, R. P. (2019). A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 29, 128–133. doi: 10.1016/j.cub.2018.11.006
Sundell, D., Mannapperuma, C., Netotea, S., Delhomme, N., Lin, Y.-C., Sjödin, A., et al. (2015). The plant genome integrative explorer resource: PlantGenIE. Org. New Phytol. 208, 1149–1156. doi: 10.1111/nph.13557
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–12. doi: 10.1186/gb-2002-3-7-research0034
Wang, J.-W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65, 4723–4730. doi: 10.1093/jxb/eru246
Wu, R., Cooney, J., Tomes, S., Rebstock, R., Karunairetnam, S., Allan, A. C., et al. (2021). RNAi-mediated repression of dormancy-related genes results in evergrowing apple trees. Tree Physiol. 41, 1510–1523. doi: 10.1093/treephys/tpab007
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8:477. doi: 10.3389/fpls.2017.00477
Keywords: poplar, FLOWERING LOCUS T, phenology, dormancy, SHORT VEGETATIVE PHASE
Citation: André D, Zambrano JA, Zhang B, Lee KC, Rühl M, Marcon A and Nilsson O (2022) Populus SVL Acts in Leaves to Modulate the Timing of Growth Cessation and Bud Set. Front. Plant Sci. 13:823019. doi: 10.3389/fpls.2022.823019
Received: 26 November 2021; Accepted: 27 January 2022;
Published: 17 February 2022.
Edited by:
Fernando Andrés, INRA UMR Amélioration Génétique et Adaptation des Plantes Méditerranéennes et Tropicales, FranceReviewed by:
Timo Hytonen, University of Helsinki, FinlandCopyright © 2022 André, Zambrano, Zhang, Lee, Rühl, Marcon and Nilsson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ove Nilsson, b3ZlLm5pbHNzb25Ac2x1LnNl
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.