
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 March 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.821647
This article is part of the Research TopicIdentification and Characterization of Contrasting Genotypes/Cultivars to Discover Novel Players in Crop Responses to Abiotic/Biotic Stresses, Volume IIView all 15 articles
 Zhihong Hou1,2†
Zhihong Hou1,2† Yongli Li2†
Yongli Li2† Yuhan Cheng3†
Yuhan Cheng3† Weiwei Li4†
Weiwei Li4† Tai Li2
Tai Li2 Hao Du2
Hao Du2 Fanjiang Kong2
Fanjiang Kong2 Lidong Dong2
Lidong Dong2 Dianfeng Zheng1,5,6*
Dianfeng Zheng1,5,6* Naijie Feng1,5,6*
Naijie Feng1,5,6* Baohui Liu2*
Baohui Liu2* Qun Cheng2*
Qun Cheng2*
Salt stress is a major factor limiting the growth and yield of soybean (Glycine max). Wild soybeans (Glycine soja) contain high allelic diversity and beneficial alleles that can be re-introduced into domesticated soybeans to improve adaption to the environment. However, very few beneficial alleles have been identified from wild soybean. Here, we demonstrate that wild soybean is more salt tolerant than cultivated soybean and examine dehydration responsive element-binding (DREB) family transcription factor genes to look for advantageous alleles that might improve drought tolerance in cultivated soybean. Our genome-wide analysis identified 103 DREB genes from the Glycine max genome. By combined RNA-sequencing and population genetics of wild, landrace, and cultivated soybean accessions, we show that the natural variation in DREB3a and DREB3b is related to differences in salt tolerance in soybean accessions. Interestingly, DREB3b, but not DREB3a, appears to have undergone artificial selection. Soybean plants carrying the wild soybean DREB3b allele (DREB3b39Del) are more salt tolerant than those containing the reference genome allele (DREB3bRef). Together, our results suggest that the loss of the DREB3b39Del allele through domestication of cultivated soybean may be associated with a reduction in salt tolerance. Our findings provide crucial information for improving salt tolerance in soybean through molecular breeding.
High salt levels in soil cause stress that affects plant growth and salinization of farmland restricts global agricultural production (Flowers, 2004; Munns and Tester, 2008; Ondrasek et al., 2011). Soil salinization is exacerbated by natural and human activities, including irrigation (Munns et al., 2020). Under salt stress, plants experience dehydration, metabolic toxicity, nutrient deficiencies, and membrane dysfunction, leading to tissue damage and early senescence (Essah et al., 2003). To adapt to various environmental stresses, plants have evolved many complexes signaling networks, and the molecular basis of these pathways is currently being studied (Zhu, 2001).
Soybean [Glycine max (L.) Merr.] is an economically important crop for oil and protein (Graham and Vance, 2003). It is classified as a moderately salt-sensitive crop; under salt stress, soybean yields may be reduced by up to 40% (Papiernik et al., 2005; Munns and Tester, 2008). Moreover, soybean cultivars show variation in their salt sensitivity: under saline conditions, salt-sensitive cultivars have a 37% decrease in yield compared with salt-tolerant cultivars (Parker et al., 1983). This natural variation in salt tolerance provides opportunities to increase soybean production under saline conditions (Guan et al., 2014; Qi et al., 2014). However, few salt-tolerant variants have been identified (Guan et al., 2014; Qi et al., 2014). This lack of knowledge has greatly inhibited attempts to improve salt tolerance in crops.
Cultivated soybean was domesticated from its wild counterpart Glycine soja Sieb. & Zucc. 6,000–9,000 years ago (Hymowitz, 1970; Carter et al., 2004). Due to artificial selection and population/genetic bottlenecks, cultivated soybeans have much lower genetic diversity than their wild relative (Hyten et al., 2006; Lam et al., 2010). This reduced variation may have led to the loss of some genes or alleles that are important for adapting to different environments. Therefore, wild soybean, with its high allelic diversity, may be a good source for beneficial alleles that can be re-introduced into domesticated soybean cultivars through breeding.
The dehydration responsive element-binding (DREB) family is a subgroup of the APETALA2/ethylene-responsive factor (AP2/ERF) transcription factor (TF) superfamily; in multiple plants, DREB genes respond to a wide variety of abiotic stresses, such as drought, high salt, cold, and heat (Xu et al., 2011; Kudo et al., 2017; Ali and Hadi, 2018; Eckardt, 2019). Some DREBs are involved in the salt stress response in soybean (Gao et al., 2005; Li et al., 2005; Chen et al., 2007). However, the variation in the DREB family in wild soybean remains to be explored. To fill this gap, we combined genome-wide analysis, association studies, population genetics, and RNA-sequencing to identify candidate causal DREB genes for salt tolerance. Our results show that DREB3b contributes to salt tolerance and may have undergone natural and artificial selection. In addition, the DREB3b39Del allele from wild soybean improves salt tolerance compared to the DREB3bRef allele from cultivated soybean. The identification of these DREB3b alleles provides important information for improving the yield of soybean and other crops.
The soybean cultivar Williams 82 (W82) and 424 soybean varieties (Lu et al., 2020) were used in this study. 424 soybean accessions were grown in a greenhouse using vermiculite as the growth medium, and maintained at 25°C and 70% relative humidity with a 16 h light/8 h dark. Fifteen days after planting, seedlings at the first-node stage (V1; Fehr et al., 1971) were subjected to 200 mM NaCl treatment. After being treated for 2 weeks, salt tolerance index was determined with two independent experiments. Briefly, five to eight plants of each accession in each replication were scored for salt tolerance. A salt-tolerance rating for each of the accession was assigned by the respective level of leaf chlorosis (Liu et al., 2020). The salt-tolerance ratings ranged on a scale from 1 (normal green leaves) to 5 (complete death). All primers used for vector construction, PCR, and quantitative reverse transcription (qRT)-PCR assays for all target genes are listed in Supplementary Table 3.
The soybean genome in Phytozome1 was used to identify all members of DREB family genes. A database search was carried out against all the genome-coded soybean proteins with the BLASTP program (E-value < 1e–20 and coverage >75%) in TBtools (Chen et al., 2020) using 56 full-length amino acid sequences of DREB homologs from A. thaliana (Sakuma et al., 2002). Multiple alignment of the protein sequences of 159 DREB genes (56 from A. thaliana and 103 from soybean) were aligned by ClustalW. A neighbor-joining phylogenetic tree was generated using MEGA_X with bootstrap repeated 1,000 times. The expression data of 103 DREB genes in different tissues (leaf, stem, root, flower, seed, pod, and cotyledon) of soybean were obtained from RNA-seq database (Machado et al., 2020).2 Location of DREB genes was visualized using the soybean genome generic feature format (.gff) using TBtools.
For RNA-sequencing, the W82 seedlings were grown in a growth chamber maintained at 25°C and 70% relative humidity with a 16-h light/8-h dark. Fifteen days after planting, seedlings at the first-node stage (V1; Fehr et al., 1971) were subjected to 200 mM NaCl treatment. The control plants were treated with water. After being treated for 2 weeks, total RNA was isolated using TRIzol reagent kit (Invitrogen, Carlsbad, CA, United States). cDNA synthesis was conducted using an M-MLV reverse transcriptase kit (Takara, Dalian, China) according to the manufacturer’s protocol. The cDNA fragments were purified using a QiaQuick PCR extraction kit, and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR amplified, and sequenced using an Illumina HiSeq 2500 system by Gene Denovo Biotechnology Co. (Guangzhou, China).
The full-length coding sequence of DREB3a and DREB3b in 1295 soybean accessions were retrieved from the 1295 resequencing data (Lu et al., 2020). The resequencing data of 1295 accessions were deposited into the NCBI database under accession number PRJNA394629, and the GSA database in BIG Data Center under accession numbers PRJCA000205 and PRJCA001691. Only haplotypes found in ≥3 soybean accessions were recorded. A haplotype network was constructed based on polymorphic sites of the whole coding sequences of DREB3a and DREB3b using the Median-Joining method in the NETWORK version 4.6.1.2 software (Fluxus Technology Ltd., Sudbury, Suffolk, United Kingdom) with data preparation using DNASP version 5 (Librado and Rozas, 2009).
A 20-kb sequence set (Chr01:48411780.48452989 and Chr15:1392382…1433465) centered on the candidate genes (DREB3a and DREB3b) were obtained from the 1295 soybean accessions whole-genome dataset. Nucleotide diversity (π) of each accession was estimated using the script vcftools with parameters–window-pi 20000–window-pi-step 10000.
The coding sequence of DREB3b was obtained from W82 and GDW013 (wild soybean variety) by KOD-Plus-Neo (TOYOBO) with the primer in Supplementary Table 3. The coding sequence of DREB3b was cloned into plant expression vector pCAMBIA3301 with gene-specific primers. Transgenic soybean hairy roots were generated by Agrobacterium rhizogenes-mediated transformation as described by Graham et al. (2007) and Kereszt et al. (2007) with some modifications. The cotyledons were cut into rough triangles and immediately placed in Petri dishes containing 0.6% agar medium to keep them moist. The cut surface was treated with 20 μl A. rhizogenes suspension. The dishes were sealed with Parafilm and placed in an incubator at 25°C. Transformed hairy roots were abundant along a callus ridge on the inoculated cotyledons after approximately 3 weeks. Overexpression of the target gene in transgenic hairy roots was tested via quantitative PCR (qPCR) and GUS staining.
The sequences of the DREB3b39Del allele and DREB3bRef allele were obtained by sequencing. Primers were designed using Primer Premier 5.0, with a product size < 200 bp. The InDel marker was developed on the basis of the variation in DREB3b. Supplementary Table 3 lists the InDel markers that were used in this study.
In this study, for 424 accessions phenotypic evaluation, at least 6 individual plants were analyzed. all values were presented as mean ± s.e.m. and numbers (n) of samples or replicates are indicated in figure legends. Data were analyzed with GraphPad Prism 8 (ver. 8.0.1). Significance levels of differences were calculated by one-tailed, two-sample Student’s t-tests or one-way ANOVA with GraphPad Prism 8 (ver. 8.0.1).
Owing to “domestication syndrome” through artificial selection for traits to increase yield, cultivated soybean has lost substantial genetic diversity (Qiu et al., 2013; Li et al., 2014). Here, we examined a panel of 424 previously described accessions (Lu et al., 2020), which comprised 85 wild soybeans, 153 landraces, and 186 cultivars. To assess the genetic diversity in this panel, we compared the shapes of the wild, landrace, and cultivated soybean seeds (Figure 1A) and estimated the number of single-nucleotide polymorphisms (SNPs) and the nucleotide diversity across chromosome 1 in the 424 accessions. The number of SNPs and the nucleotide diversity are lowest in cultivated soybean and highest in wild soybean (Figures 1B,C). These results are consistent with a previous preliminary report (Lam et al., 2010).
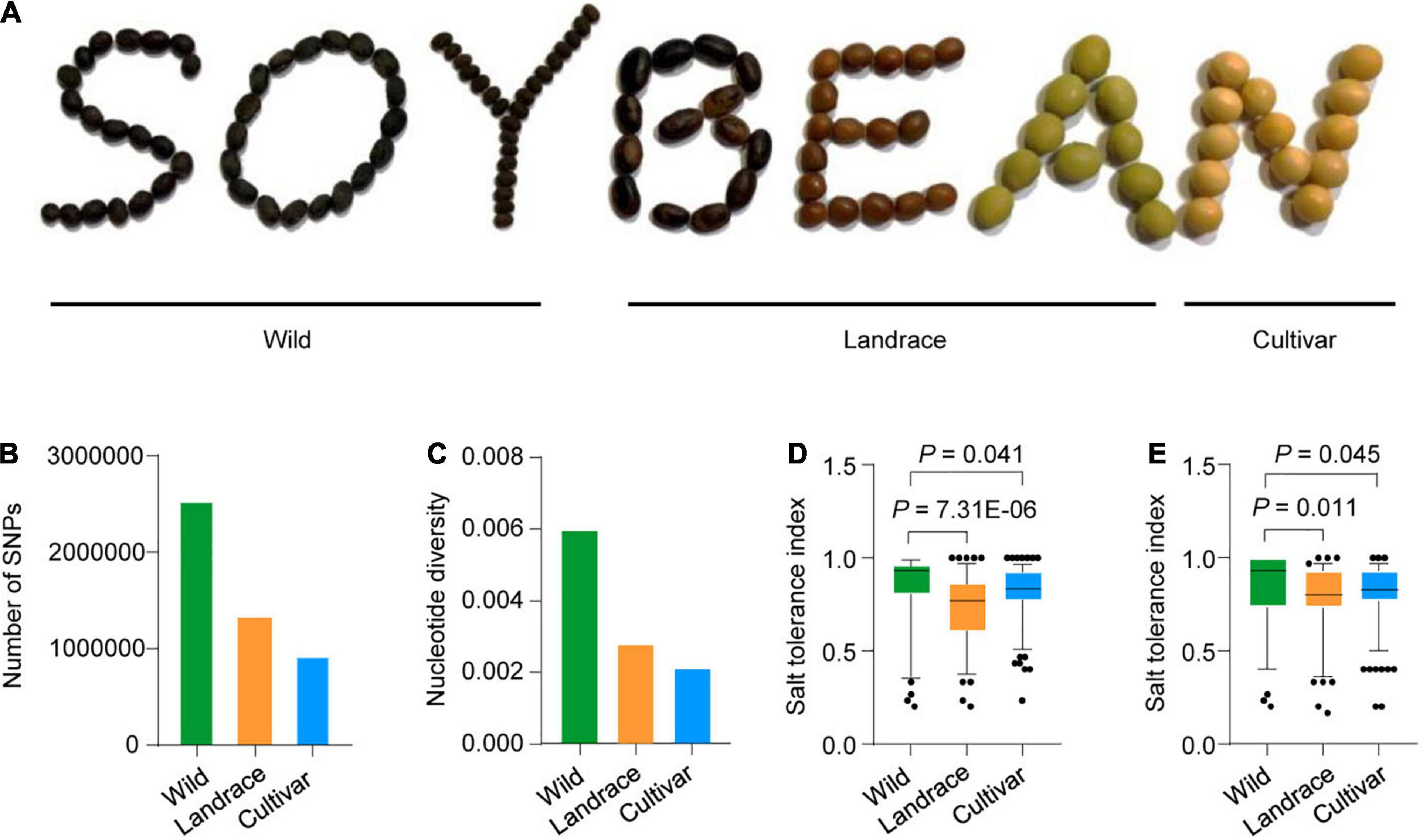
Figure 1. The morphologic characteristics and salt tolerance of wild, landrace and cultivated soybean. (A) The morphologic characteristics of wild, landrace, and cultivated soybean. (B) Number of SNPs of 424 accessions in chromosome 1 genomic region. (C) Nucleotide diversity of 424 accessions in chromosome 1 genomic region. (D,E) Comparison of the salt tolerance index of 424 accessions (85 wild soybeans, 153 landraces and 186 improved cultivars); two biological replicates in greenhouse.
We also measured the salt tolerance indexes of the 424-accession panel. Phenotype was evaluated in a growth chamber at 25°C, revealing significantly reduced salt tolerance in landrace and cultivated soybean compared to wild accessions in two biological replicates (Figures 1D,E). These results suggest that artificial selection may have had a strong effect on the genetic diversity in the cultivated soybeans.
The DREB family plays an important role in plant responses to abiotic stresses (Gao et al., 2005; Chen et al., 2007; Ali and Hadi, 2018; Eckardt, 2019), so it would be meaningful to identify the genes encoding DREB TFs in soybean. In this study, 56 Arabidopsis thaliana DREB sequences (Sakuma et al., 2002) were used as a query to search the soybean database. This identified 103 DREB genes from the Glycine max genome (Supplementary Table 1), 73 of which have been reported previously (Zhou et al., 2020b). The DREB genes were distributed on all chromosomes of soybean, with 8, 3, 4, 5, 6, 8, 4, 3, 4, 5, 7, 7, 8, 9, 3, 2, 8, 2, 5, and 2 DREB genes on chromosome 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, and 20, respectively (Supplementary Figure 1). Phylogenetic analyses based on previously reported Arabidopsis DREB sequences (Sakuma et al., 2002) classified these genes into six subgroups: 14 genes in A-1 group, 35 genes in A-2 group, 19 genes in A-3 group, 2 genes in A-4 group, 21 genes in A-5 group, and 12 genes in A-6 group (Figure 2A and Supplementary Table 1).
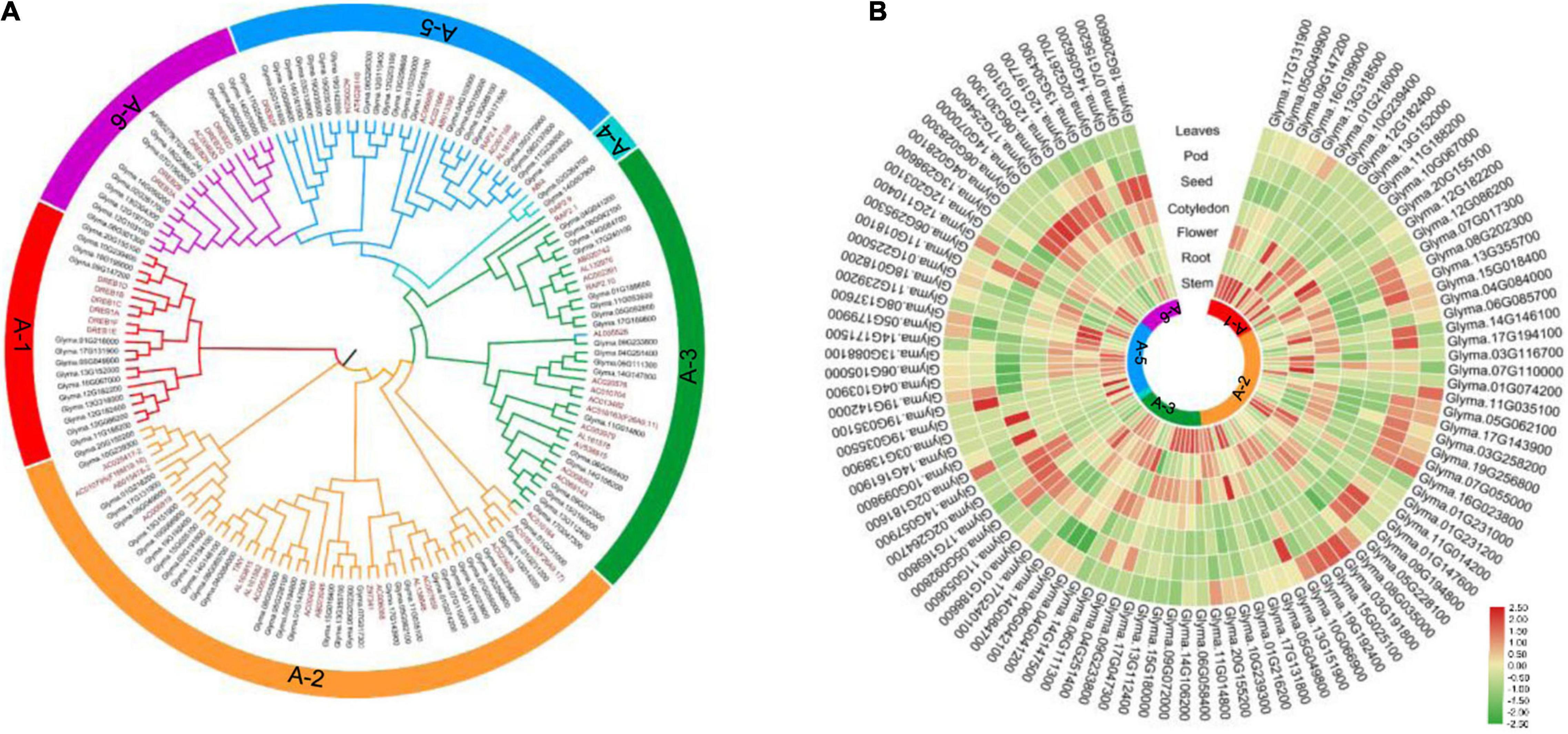
Figure 2. Phylogenetic tree of the DREB genes of Arabidopsis thaliana and soybean, and gene expression profile of soybean DREB genes in different tissues. (A) The NJ tree was constructed using MEGA_X based on alignment of a total of 159 DREB proteins, including 56 Arabidopsis DREBs and 103 Glycine max DREBs. DREB subfamilies are divided into six groups (A-1 to A-6). (B) The normalized microarray expression data of 103 Glycine max DREBs was downloaded from PLEXdb (http://www.plexdb.org/). The heatmap was produced using TBtools.
The transcript levels of 103 DREB genes in different soybean tissues were analyzed using the Phytozome database.3 The A-1 subgroup was highly expressed in stem and flower; the A-2 subgroup was highly expressed in root, flower, pod, and leaves; the A-3 subgroup was highly expressed in stem, root, and flower; the A-4 subgroup and the A-6 subgroup were highly expressed in seed and cotyledon; the A-5 subgroup was highly expressed in stem (Figure 2B). These data suggest that DREB genes of the same subgroup may have similar functions, whereas DREB genes of different subgroups may have functional differentiation.
As a few DREB genes have been shown to be induced by salinity stress (Chen et al., 2007, 2009; Zhou et al., 2020a), we asked whether other DREB family members also control salt tolerance and whether these homologs could be used in agricultural applications. We thus performed RNA-sequencing experiments with three biological replicates, comparing the cultivated soybean Williams 82 (W82) grown under control conditions (W82-Water) with W82 treated with 200 mM NaCl for 15 days (W82-NaCl). The differentially expressed genes (DEGs) were identified by a pairwise comparison of the transcriptome datasets (W82-Water vs. W82-NaCl). As expected, the gene expression patterns were similar between three biological replicates but differed significantly between the control and salt treatments (Figure 3A).
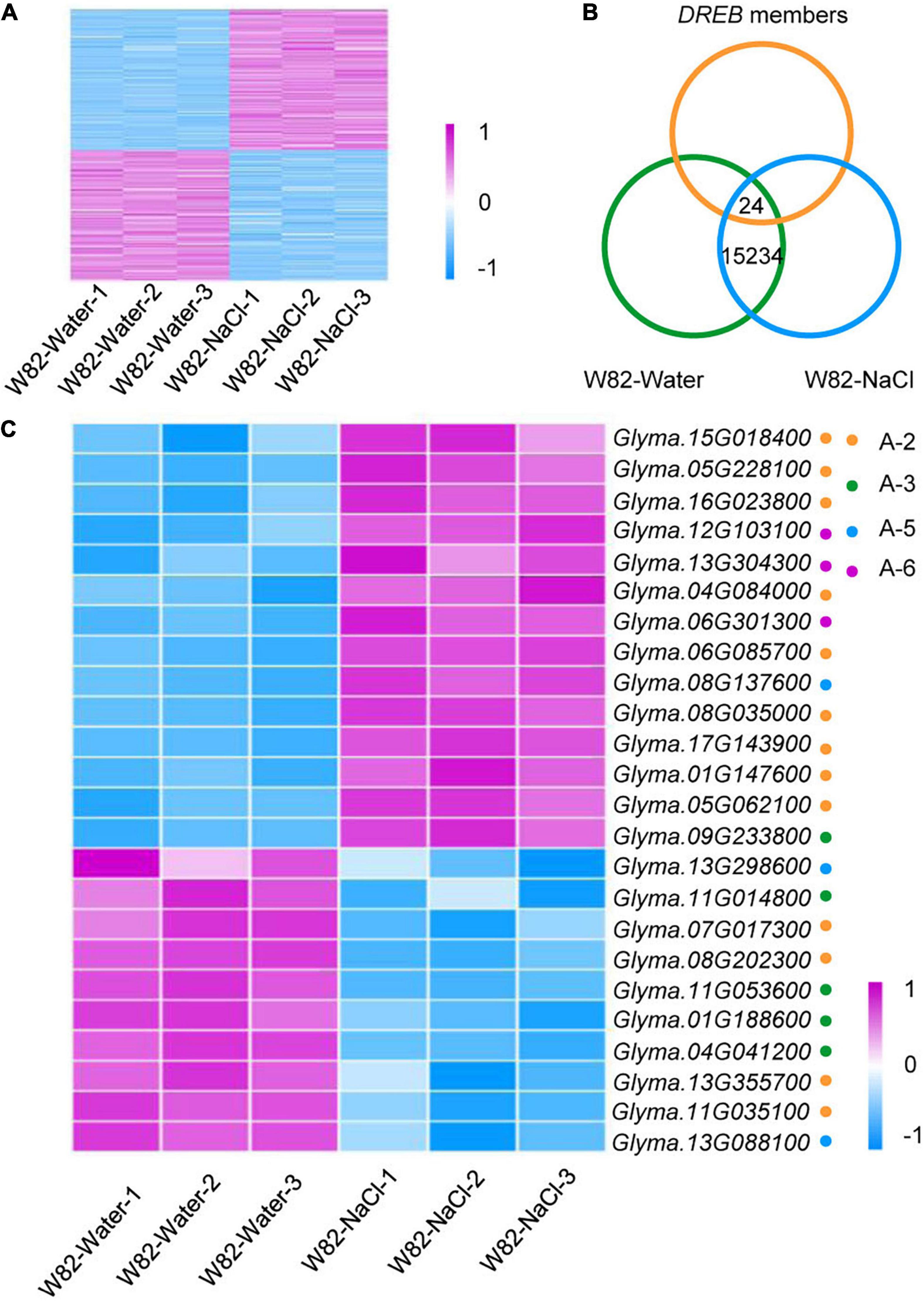
Figure 3. Identification of differentially expressed DREB genes in NaCl-treated W82 vs. mock treated. (A) Heat map of differentially expressed genes in W82-Water and W82-NaCl; n = 3 for each group. Relative transcript level is indicated on a color scale from purple (high) to blue (low). (B) Overlap between W82-Water, W82-NaCl and DREB genes. (C) Heat map of differentially expressed 24 DREB genes under NaCl treatment.
We identified 24 differentially expressed DREB genes in the W82-water vs. W82-NaCl comparison. There were nine up-regulated genes in the A-2 subgroup, one up-regulated gene each in the A-3 and A-5 subgroups, three up-regulated genes in the A-6 subgroup, four down-regulated genes in the A-2 and A-3 subgroups, and two down-regulated genes in the A-5 subgroup (Figures 3B,C and Supplementary Table 2).
The gene structures of the 24 differentially expressed DREB genes were diagrammed, including the untranslated regions (UTRs) and the locations of exons and introns (Supplementary Figure 2). Among the up-regulated DREB genes, nine genes had no introns and five genes contained one intron (Supplementary Figure 2A). Among the down-regulated DREB genes, nine genes had no introns and only one gene contained one intron (Supplementary Figure 2B). This observation is consistent with the structural characteristics described by Zhou et al. (2020b).
Some DREB genes positively regulate tolerance to abiotic stress in soybean (Chen et al., 2007, 2009; Zhou et al., 2020a); therefore, we focused on the 14 up-regulated DREB genes. To systematically study natural variation of DREB genes, we investigated the variation of all 14 up-regulated DREB genes in the 424-accession panel and found that only nine DREB genes arose through natural variation: Glyma.01G147600, Glyma.15G018400, Glyma.04G084000, Glyma.08G035000, Glyma.16G023800, Glyma.09G233800, Glyma.08G137600, Glyma.12G103100, Glyma.06G301300 (Figures 4A,B and Supplementary Figures 3–6).
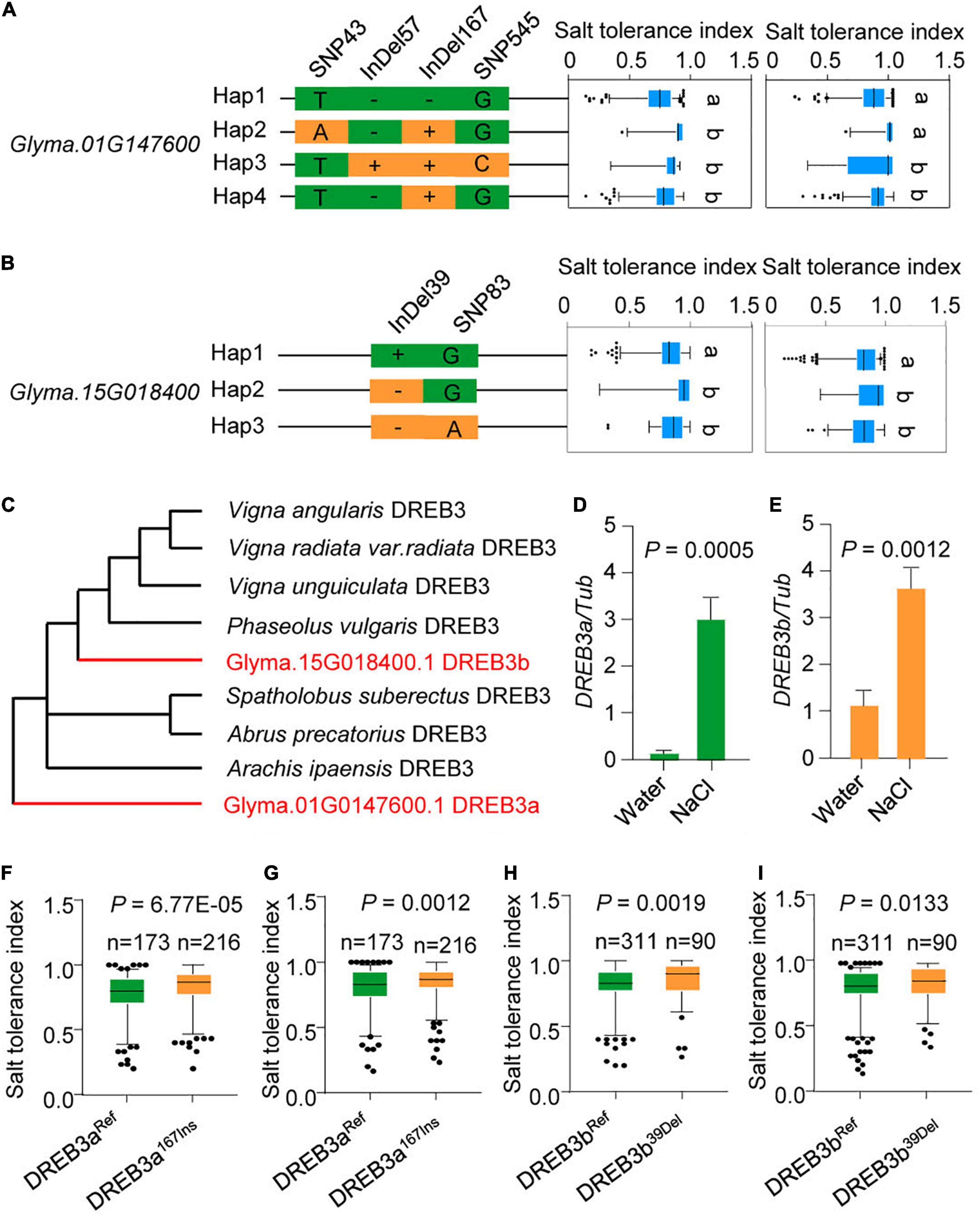
Figure 4. Natural variation of DREB3a and DREB3b are significantly associated with the salt tolerance. (A) Boxplots for salt tolerance index based on the haplotypes of Glyma.01G147600 among 424 soybean accessions. The Glyma.01G147600 haplotypes (Hap1–Hap4) were categorized by the four significant variants. (B) Boxplots for salt tolerance index based on the haplotypes of Glyma.15G018400 among 424 soybean accessions. The Glyma.15G018400 haplotypes (Hap1–Hap3) were categorized by the two significant variants. The experiment was performed using two biological replicates in (A,B). The same lowercase letters above the histogram bars in (A,B) denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values. (C) Phylogenetic tree of DREB proteins in plants. Red indicates DREB3a and DREB3b protein. (D) The transcription levels of DREB3a under salt stress (200 mM NaCl). (E) The transcript levels of DREB3b under salt stress (200 mM NaCl). All data were given as mean ± s.e.m. (n = 3 biological replicates). (F,G) GmDREB3aRef andDREB3a167Ins alleles associated with salt tolerance index. (H,I) DREB3bRef andDREB3b39Del alleles associated with salt tolerance index. One-tailed Student’s t-test was used to generate the P-values in (D–I).
We then measured the salt tolerance indexes of the population to determine if salt tolerance is associated with any of the nine genes. After correcting for population structure (at least ten individual plants were analyzed per accession), two out of the nine DREB genes were significantly correlated with the salt tolerance index (p < 0.05): Glyma.01G147600 and Glyma.15G018400 (Figures 4A,B).
Phylogenetic investigation indicated that Glyma.01G147600 and Glyma.15G018400 are orthologs of DREB3 in other plants. The two orthologs were thus named DREB3a and DREB3b, respectively (Figure 4C). To determine whether DREB3a and DREB3b respond to salt stress, we performed qRT-PCR to examine their expression profiles under salt stress. The results showed that DREB3a and DREB3b were up-regulated under salt stress (Figures 4D,E), consistent with RNA-sequencing data (Figure 3C).
Interestingly, Hap. 2-4 of DREB3a harbored a 15-bp InDel predicted to insert 5 amino acids, and Hap. 2–3 of DREB3b lacked a 6-bp InDel predicted to lack 2 amino acids (hereafter referred to as DREB3a167Ins and DREB3b39Del, respectively). We then compared the phenotypic differences of the accessions carrying either the reference genome DREB3a allele (DREB3aRef) or DREB3a167Ins allele, and the two accessions carrying either the reference genome DREB3b allele (DREB3bRef) or DREB3b39Del allele. The salt tolerance indexes of DREB3a167Ins and DREB3b39Del allele accessions were significantly higher than DREB3aRef and DREB3bRef accessions in two biological replicates (Figures 4F–I). These results strongly suggest that DREB3a and DREB3b are excellent candidate genes that respond to salt stress.
To gain further insight into the evolutionary history of DREB3a and DREB3b, we examined the variation in the DREB3a and DREB3b coding sequences in our previously described collection of 1295 resequenced accessions (Supplementary Figure 7A), of which four (Hap. 1, 2, 3, and 4) are described above (Figure 4A), and identified five haplotypes in DREB3b (Supplementary Figure 7B), of which three (Hap. 1, 2, 3) are described above (Figure 4B). The residues of Hap. 2 for both DREB3a and DREB3b are identical to all other homologs in legume (Supplementary Figures 8A,B), suggesting that Hap. 2 is the original haplotype in the soybean species. Median-joining network analysis of DREB3a showed that Hap. 5 is derived from Hap. 2, Hap. 1, and Hap. 3 are derived from Hap. 5, and Hap. 4 is derived from Hap. 3 (Supplementary Figure 9A). Median-joining network analysis of DREB3b showed that Hap. 1, 3, and 5 are derived from Hap. 2, and Hap. 4 is derived from Hap. 3 (Supplementary Figure 9B).
We then analyzed the percentage of DREB3a167Ins and DREB3b39Del alleles in 1295 resequenced soybean accessions. Surprisingly, the most common allele DREB3bRef was present in the vast majority of improved cultivars (507/515) and in most landraces (418/485) (Figure 5A), indicating that the DREB3bRef allele is nearly fixed in the landrace population. This dramatic increase of the DREB3bRef allele in the soybean population indicates that the DREB3bRef allele might have been under strong artificial selection during domestication.
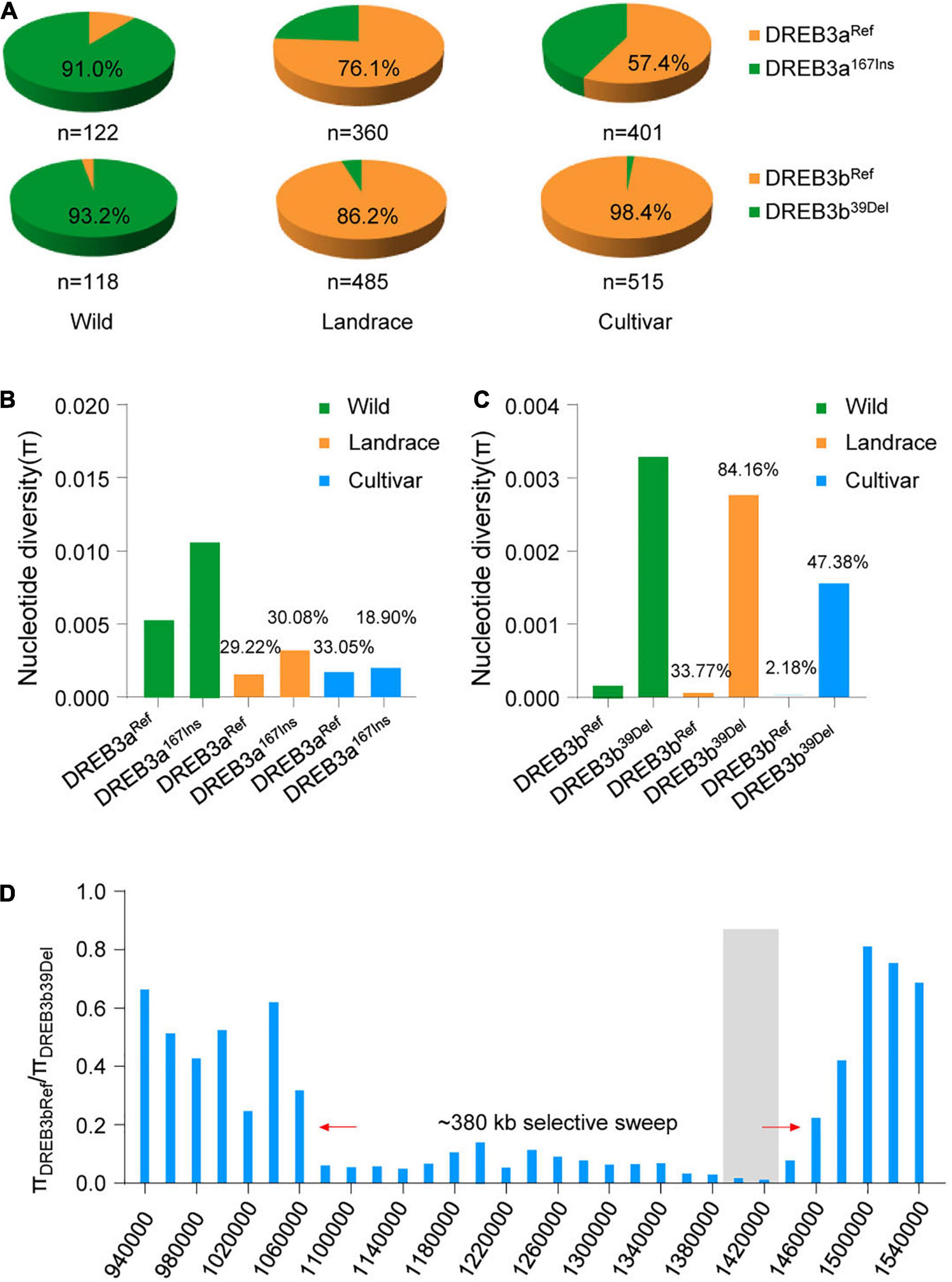
Figure 5. DREB3b underwent artificial selection but DREB3a did not. (A) Proportions of DREB3a and DREB3b alleles and their co-occurrence with each of the three germplasm groups (wild, landrace, and cultivar). (B) Nucleotide diversity analysis of the region surrounding DREB3a in wild, landrace, and cultivated soybean. (C) Nucleotide diversity analysis of the region surrounding DREB3b in wild, landrace, and cultivated soybean. (D) Selective sweep in the 380-kb DREB3b genomic region.
To examine whether selection has acted on DREB3bRef, we analyzed the nucleotide diversity of the 20-kb region flanking DREB3bRef in domesticated and wild soybean lines. Landraces carrying the DREB3b39Del allele retained 84.16% of the nucleotide diversity of their wild counterparts, and improved cultivars retained 47.38% of the nucleotide diversity of landraces (Figure 5C). Whereas, landraces carrying the DREB3bRef allele retained only 33.77% of the nucleotide diversity of wild lines, and improved cultivars retained only 2.18% of the nucleotide diversity of landraces (Figure 5C). We identified strong evidence of selection in a region of 380 kb around the DREB3b gene (Figure 5D). These results suggest that the DREB3bRef allele was targeted by selection, thereby causing its rapid accumulation in domesticated soybeans. This is also supported by similar negative values of Tajima’s D statistic for DREB3b in domesticated soybeans (Supplementary Table 5), indicating that DREB3b (and in particular the DREB3bRef allele) experienced selection during the transition from wild soybean to landrace.
Given that salt tolerance is not considered a soybean domestication trait, it is not clear why DREB3bRef was selected during early domestication. We propose that the DREB3b region is a hotspot for many domestication and agronomic traits. Consistent with expectation, the DREB3bRef allele improves flowering, increases seed size, and decreases plant height relative to the DREB3b39Del allele (Supplementary Figures 10A–C). Therefore, DREB3bRef may have been selected for an effect on flowering time, seed size and plant height, and the change in salt tolerance was a correlated response. Considering the enrichment of the DREB3b39Del allele in wild soybeans, and the higher salt tolerance index of the DREB3b39Del allele compared to that of the DREB3bRef allele, the DREB3b39Del allele may be undergoing strong natural selection to facilitate the response of wild soybeans to salt stress.
For DREB3a, the DREB3aRef allele was the most abundant in landraces (274/360) and in improved cultivars (230/401) (Figure 5A). However, landraces carrying the DREB3a167Ins allele retained 30.08% of the nucleotide diversity of their wild counterparts, and improved cultivars retained 18.90% of the nucleotide diversity of landraces (Figure 5B). Whereas, landraces carrying DREB3aRef allele retained 29.22% of the nucleotide diversity of wild, and cultivated retained 33.05% of the nucleotide diversity of landraces (Figure 5B). We identified no strong evidence of selection in a region of the DREB3a gene (Figure 5D, Supplementary Figure 11, and Supplementary Table 5). This implies that DREB3b, but not DREB3a, has undergone artificial selection during in the domestication process.
We next focused on the function of DREB3b. To further characterize the potential contribution of DREB3b to salt tolerance in different soybean varieties, we generated DREB3bRef-overexpressing and DREB3b39Del-overexpressing transgenic soybean hairy roots by high-efficiency A. rhizogenes-mediated transformation (Graham et al., 2007; Kereszt et al., 2007). We examined the DREB3bRef-overexpressing and DREB3b39Del-overexpressing transgenic hairy roots by GUS staining and qRT-PCR (Figures 6A,B). Under normal conditions (0 mM NaCl), all soybean hairy roots grew well, with no noticeable difference (Figure 6C). Under 100 mM NaCl stress, the average root length of DREB3b39Del-overexpressing soybean hairy roots was much longer than that of DREB3bRef-overexpressing transgenic hairy roots and much longer than that of non-transgenic hairy roots (Figures 6C,D).
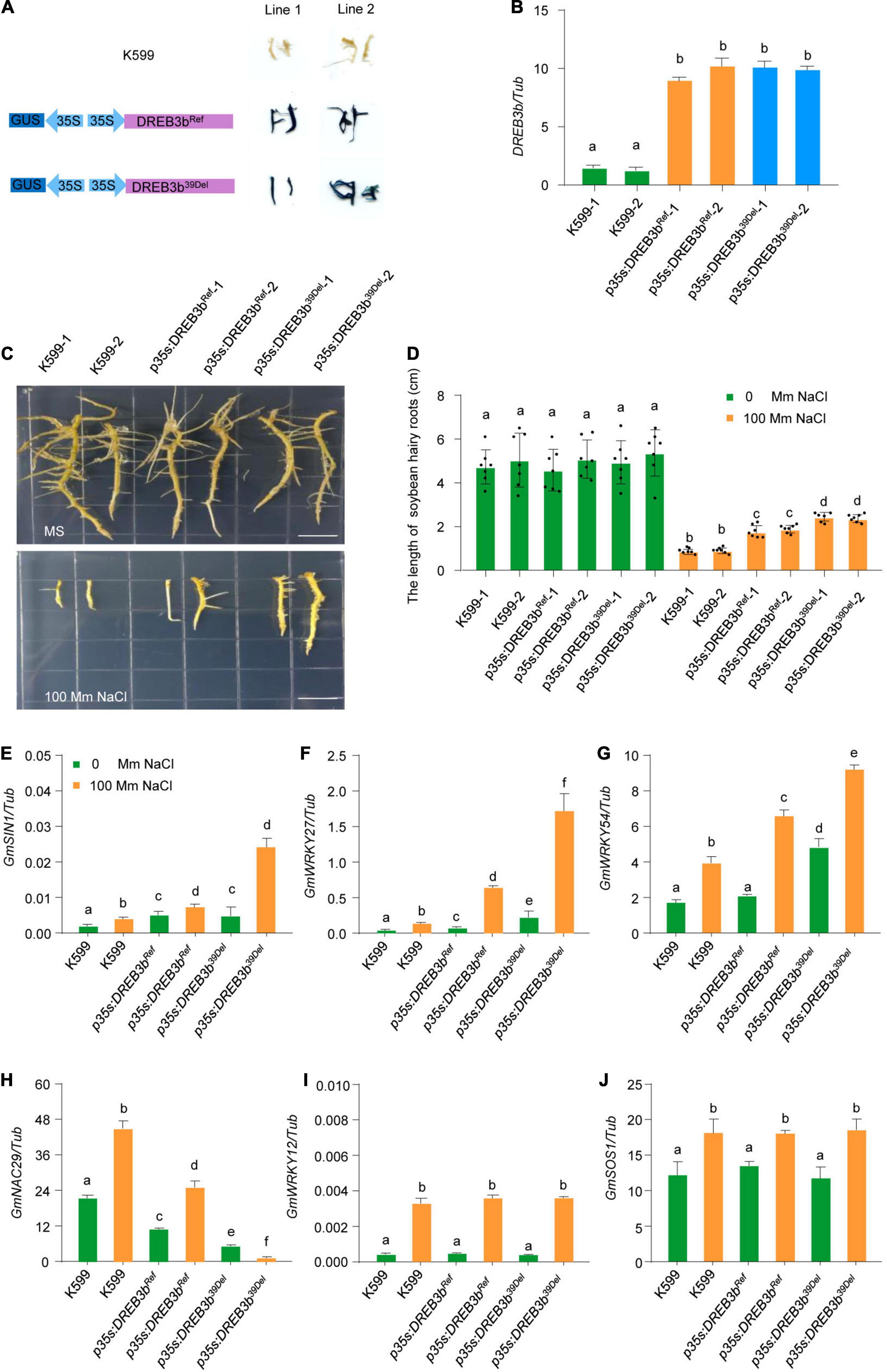
Figure 6. DREB3b39Del improves salt tolerance more than DREB3bRef in transgenic soybean hairy roots. (A) Schematic diagram of overexpression vector and GUS staining of overexpressing DREB3b39Ref andDREB3b39Del in soybean hairy roots. (B) The transcript levels of DREB3b in W82 and two independent DREB3b39Ref and DREB3b39Del transgenic lines grown under normal growth conditions. (C,D) Phenotype of the transgenic lines overexpressing DREB3b39Ref and DREB3b39Del with or without 100 mM NaCl treatment. The lengths of soybean hairy roots were obtained from at least seven roots. (E–J) DREB3b regulates the expression of salt stress-tolerant gene in soybean with or without 100 mM NaCl treatment. Experiment was performed using three biological replicates. The same lowercase letters above the histogram bars denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values.
Many positive and negative regulators of salt tolerance in soybean have been identified (Zhou et al., 2008; Wang et al., 2015; Shi et al., 2018; Li et al., 2019). To test whether DREB3b can regulate the expression of these genes, we performed qRT-PCR analysis in DREB3bRef-overexpressing and DREB3b39Del-overexpressing soybean hairy roots under mock and NaCl treatment (100 mM). DREB3bRef and DREB3b39Del up-regulated the expression of the positive regulators of salt tolerance SIN1, WRKY27, and WRKY54, and down-regulated the expression of negative regulator of salt tolerance NAC29, but could regulate the expression of WRKY12 and SOS1 under mock and NaCl treatment (Figures 6E–J). Interestingly, the transcript levels of SIN1, WRKY27 and WRKY54 were higher in DREB3b39Del soybean hairy roots than in DREB3bRef soybean hairy roots, and the transcript level of NAC29 was lower in DREB3b39Del soybean hairy roots than in DREB3bRef soybean hairy roots under NaCl treatment (Figures 6E–J). These results show that the DREB3b39Del allele enhances salt tolerance more than the DREB3bRef allele in soybean.
Genetic markers are an important and effective tool for identifying mutant alleles in molecular-assisted studies, and could accelerate the genotyping procedure in future generations. We thus developed an InDel marker to identify the DREB3b39Del allele. After PCR amplification using DREB3b-specific primers (Supplementary Table 3), the DREB3bRef allele produced 106-bp amplicons, whereas the DREB3b39Del allele produced 100-bp amplicons, and the differences could be detected by electrophoresis (Supplementary Figure 12). We used this InDel marker to detect the genotypes of soybean accessions at random, and the results were consistent with the sequencing results (Supplementary Figure 12 and Supplementary Table 4). Therefore, this InDel marker can be used to improve the breeding efficiency of salt tolerant soybean varieties in molecular breeding.
Soybean is a leguminous plant that is capable of closed flower fertilization (closed flower pollination). This feature may be helpful for maintaining genomic homogeneity and reducing genomic variation, which may have been further intensified by the domestication process (Lam et al., 2010). Due to bottlenecks and artificial selection, some genes that play important roles for adaptation to different environments have been lost during the soybean domestication process. Wild soybean (Glycine soja) may have high allelic diversity at certain loci, including advantageous alleles that can be re-introduced into domesticated soybeans via breeding to improve their adaption to the environment (Zhang et al., 2017). Consistent with a previous report (Lam et al., 2010), we show that the SNPs number of both landrace and improved cultivars were significantly lower than that of wild soybeans (Figure 1B), and wild soybeans have retained higher genetic diversity, but have been lost in cultivated soybeans (Figure 1C). We also show that the salt tolerance index of wild soybeans was much higher than that of improved cultivars (Figures 1D,E). These data indicate that a large number of useful alleles and genes from wild soybeans may be great significance for improving the salt tolerance of soybean. Importantly, to explore the variation in excellent alleles in wild soybeans, we combined genome-wide analysis and RNA-sequencing and identified an excellent allele from wild soybean, which could be useful in breeding for improved salt tolerance in soybean.
Farmlands worldwide are increasingly affected by soil salinization, and most crops are sensitive to salt stress (Ren et al., 2005; Munns et al., 2012; An et al., 2017). Therefore, salt tolerance is an ecologically significant trait for crop breeding. Previous studies have confirmed that genetic variation in salt tolerance exists widely within and among major crops, providing an opportunity to use salt tolerance variation to improve crop salt tolerance (Wu et al., 2012; Qi et al., 2014; Zhang et al., 2018). For example, natural variation in SALT3/CHX1, a cation H+ exchanger gene, could modulate salt tolerance in soybean (Guan et al., 2014; Qi et al., 2014). Moreover, the DREB family is one of the most important TF families in vascular plants, and DREB TFs are involved in salt stress responses (Li et al., 2005; Xu et al., 2011; Kudo et al., 2017; Ali and Hadi, 2018; Eckardt, 2019). For example, DREBa, DREBb, and DREBc are up-regulated under salt stress conditions (Li et al., 2005); GmDREB2, a soybean DRE-binding transcription factor, confers drought- and salt-tolerance in transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants (Chen et al., 2007). However, the natural variation of DREB genes has not been examined in soybean. Here, we have identified one DREB gene in soybean, DREB3b, which conferred salt tolerance in soybean (Figure 4B). The molecular footprint showed strong selection at DREB3b during domestication (Figure 5D). The DREB3b39Del allele was highly abundant in wild soybeans (about 95%), and the DREB3bRef allele was highly abundant in improve cultivar (about 98%, Figure 5A). Together, the data suggest that the DREB3bRef allele may have undergone artificial selection during domestication. We also demonstrate that, compared to DREB3bRef overexpression, DREB3b39Del overexpression led to (i) better salt tolerance in soybean hairy root, and (ii) stronger up-regulation of positive regulators of salt tolerance and stronger down-regulation of a negative regulator of salt tolerance. Therefore, the loss of the DREB3b39Del allele in the domestication of improved cultivar may be a reason for its reduced salt tolerance.
Based on our data, we propose a model (Figure 7) describing how the artificial selection of the DREB3bRef allele occurred in cultivated soybean, thereby reducing salt tolerance. According to this model, wild soybeans contain a large number of alleles in DREB3b but the DREB3bRef allele underwent artificial selection in the domestication of cultivated soybean, resulting in the reduction of salt tolerance. Overexpression of DREB3b could improve salt tolerance by up-regulating positive regulators of salt tolerance and down-regulating negative regulators of salt tolerance.
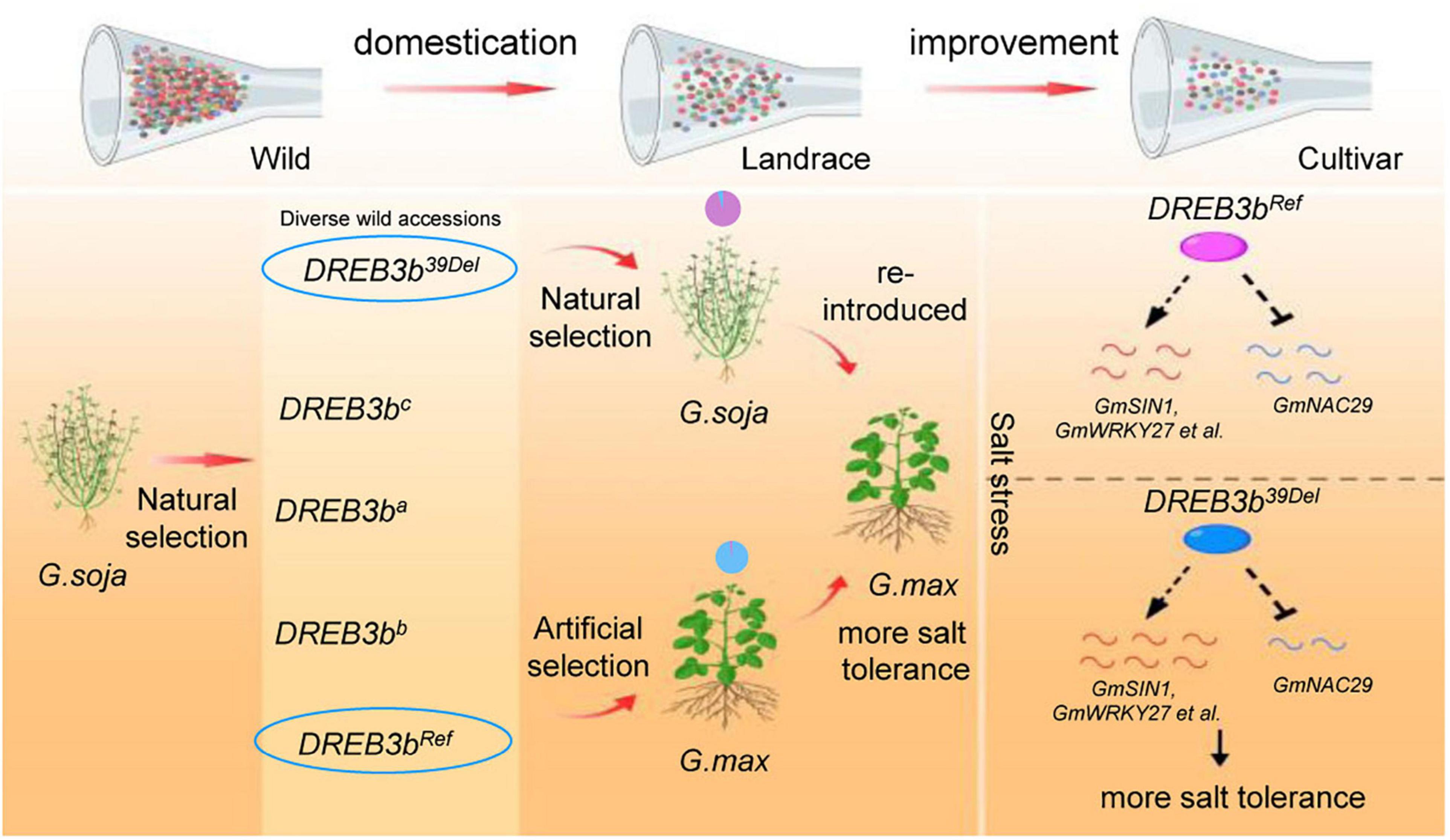
Figure 7. Model of artificial selection in DREB3b in soybean and the underlying mechanism. A large number of potentially advantageous alleles were lost during domestication and cultivar improvement, resulting in lower salt tolerance of cultivated soybean than wild soybean. The DREB3bRef allele underwent artificial selection in cultivated soybean. The DREB3bRef allele reduced the salt tolerance of cultivated soybean. DREB3b up-regulates positive regulators of salt tolerance and down-regulates negative regulators of salt tolerance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI BioProject, PRJNA797227.
QC and BL designed and interpreted the results. QC, BL, DZ, and NF coordinated the projects. QC, LD, FK, HD, TL, YC, YL, and ZH performed the experiments. LD, QC, YL, and ZH performed the data analysis. QC and ZH wrote the manuscript. WL revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32090065 and 32001508 to LD, 32090064 and 31725021 to FK, 31930083 to BL, and 31901568 to QC) and the Major Program of Guangdong Basic and Applied Research (Grant No. 2019B030302006 to FK and BL).
YC is employed by Beijing Zhongnong Futong Horticulture Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.821647/full#supplementary-material
Supplementary Figure 1 | Distribution of 103 DREB genes on 20 chromosomes in soybean.
Supplementary Figure 2 | Gene structures of 24 DREB genes. (A) Gene structures of up-regulated DREB genes. (B) Gene structures of down-regulated DREB genes. The exon-intron organization and UTRs of DREB genes were analyzed by aligning the DNA sequences using the GSDS 2.0 server (http://gsds.gao-lab.org/index.php). Introns and exons are shown as black lines and orange boxes, respectively. Green boxes at 5’ and 3’ ends represent untranslated regions (UTRs).
Supplementary Figure 3 | Boxplots for salt tolerance index based on the haplotypes of salt-induced DREB genes from the A-2 subgroup. The experiment was performed using two biological replicates. The same lowercase letters above the histogram bars denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values.
Supplementary Figure 4 | Boxplots for salt tolerance index based on the haplotypes of salt-induced DREB gene from the A-3 subgroup. The experiment was performed using two biological replicates. The same lowercase letters above the histogram bars denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values.
Supplementary Figure 5 | Boxplots for salt tolerance index based on the haplotypes of salt-induced DREB gene from the A-5 subgroup. The experiment was performed using two biological replicates. The same lowercase letters above the histogram bars denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values.
Supplementary Figure 6 | Boxplots for salt tolerance index based on the haplotypes of salt-induced DREB genes from the A-6 subgroup. The experiment was performed using two biological replicates. The same lowercase letters above the histogram bars denote non-significant differences across the two panels (P > 0.05). One-way ANOVA was used to generate the P-values.
Supplementary Figure 7 | Haplotypes of DREB3a and DREB3b. (A) Haplotypes of DREB3a. (B) Haplotypes of DREB3b. Haplotype was extracted from the 1,295 panel of 146 wild soybeans, 575 landraces and 574 improved cultivars.
Supplementary Figure 8 | Comparison of the amino acid sequences of DREB3a, DREB3b and its homologs in plants. (A) Protein sequence comparisons of DREB3a alleles and its homologs in plants. (B) Protein sequence comparisons of DREB3b alleles and its homologs in plants.
Supplementary Figure 9 | Haplotype origins of DREB3a and DREB3b.
Supplementary Figure 10 | Boxplots for yield-related traits based on the haplotypes of DREB3b gene. (A) Flowering time. (B) Plant height. (C) 100-grain weight. One-tailed Student’s t-test was used to generate the P-values.
Supplementary Figure 11 | Selective sweep in the DREB3a genomic region.
Supplementary Figure 12 | Development of functional genetic markers for DREB3bRef and DREB3b39Del alleles. Lines 1–7, soybean plants harbor DREB3bRef allele. Lines 8–13, soybean plants harbor DREB3bRef allele.
Ali, N., and Hadi, F. (2018). CBF/DREB transcription factor genes play role in cadmium tolerance and phytoaccumulation in Ricinus communis under molybdenum treatments. Chemosphere 208, 425–432. doi: 10.1016/j.chemosphere.2018.05.165
An, D., Chen, J. G., Gao, Y. Q., Li, X., Chao, Z. F., Chen, Z. R., et al. (2017). AtHKT1 drives adaptation of Arabidopsis thaliana to salinity by reducing floral sodium content. PLoS Genet. 13:e1007086. doi: 10.1371/journal.pgen.1007086
Carter, T. E., Nelson, R., Sneller, C. H., and Cui, Z. (2004). Soybeans: Improvement, Production and Uses, 3rd Edn. Madison, WI: American Society of Agronomy. doi: 10.1016/j.agsy.2004.06.006
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, M., Wang, Q. Y., Cheng, X. G., Xu, Z. S., Li, L. C., Ye, X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. doi: 10.1016/j.bbrc.2006.12.027
Chen, M., Xu, Z., Xia, L., Li, L., Cheng, X., Dong, J., et al. (2009). Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 60, 121–135. doi: 10.1093/jxb/ern269
Eckardt, N. A. (2019). DREB duo defines distinct drought and cold response pathways. Plant Cell 31, 1196–1197. doi: 10.1105/tpc.19.00331
Essah, P. A., Davenport, R., and Tester, M. (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol. 133, 307–318. doi: 10.1104/pp.103.022178
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. S. (1971). Stage of development descriptions for soybeans. Crop Sci. 11, 929–931. doi: 10.2135/cropsci1971.0011183x001100060051x
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55, 307–319. doi: 10.1093/jxb/erh003
Gao, S. Q., Xu, H. J., Cheng, X. G., Chen, M., Xu, Z. S., Li, L. C., et al. (2005). Improvement of wheat drought and salt tolerance by expression of a stress-inducible transcription factor GmDREB of soybean (Glycine max). Chin. Sci. Bull. 50, 2714–2723. doi: 10.1360/982005-1234
Graham, P. H., and Vance, C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Graham, T. L., Graham, M. Y., Subramanian, S., and Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144, 728–740. doi: 10.1104/pp.107.097865
Guan, R., Qu, Y., Guo, Y., Yu, L., Liu, Y., Jiang, J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80, 937–950. doi: 10.1111/tpj.12695
Hymowitz, T. (1970). On the domestication of the soybean. Econ. Bot. 24, 408–421. doi: 10.1007/BF02860745
Hyten, D. L., Song, Q., Zhu, Y., Choi, I. Y., Nelson, R. L., Costa, J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. U.S.A. 103, 16666–16671. doi: 10.1073/pnas.0604379103
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kudo, M., Kidokoro, S., Yoshida, T., Mizoi, J., Todaka, D., Fernie, A. R., et al. (2017). Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 15, 458–471. doi: 10.1111/pbi.12644
Lam, H. M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F. L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Li, S., Wang, N., Ji, D., Zhang, W., Wang, Y., Yu, Y., et al. (2019). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31, 2107–2130. doi: 10.1105/tpc.18.00662
Li, X. P., Tian, A. G., Luo, G. Z., Gong, Z. Z., Zhang, J. S., and Chen, S. Y. (2005). Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor. Appl. Genet. 110, 1355–1362. doi: 10.1007/s00122-004-1867-6
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi: 10.1038/nbt.2979
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, X. X., Chang, R. Z., Guan, R. X., and Qiu, L. J. (2020). Establishment of screening method for salt tolerant soybean at emergence stage and screening of tolerant germplasm. Acta Agron. Sin. 46, 1–8. doi: 10.3724/SP.J.1006.2020.94062
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Machado, F. B., Moharana, K. C., Almeida-Silva, F., Gazara, R. K., Pedrosa-Silva, F., Coelho, F. S., et al. (2020). Systematic analysis of 1298 RNA-Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas. Plant J. 103, 1894–1909. doi: 10.1111/tpj.14850
Munns, R., Day, D. A., Fricke, W., Watt, M., Arsova, B., Barkla, B. J., et al. (2020). Energy costs of salt tolerance in crop plants. New Phytol. 225, 1072–1090. doi: 10.1111/nph.15864
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., and Jordans, C. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Papiernik, S. K., Grieve, C. M., Lesch, S. M., and Yates, S. R. (2005). Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil Sci. Plant Anal. 36, 951–967. doi: 10.1081/CSS-200050280
Parker, M. B., Gascho, G. J., and Gaines, T. P. (1983). Chloride toxicity of soybeans grown on Atlantic coast flatwoods soils. Agron. J. 75, 439–443. doi: 10.2134/agronj1983.00021962007500030
Qi, X., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340. doi: 10.1038/ncomms5340
Qiu, L. J., Xing, L. L., Guo, Y., Wang, J., Jackson, S. A., and Chang, R. Z. (2013). A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol. Biol. 83, 41–50. doi: 10.1007/s11103-013-0076-6
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., and Chao, D. Y. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Ondrasek, G., Rengel, Z., and Veres, S. (2011). “Soil salinisation and salt stress in crop production,” in Abiotic Stress in Plants: Mechanisms and Adaptations, eds A. K. Shanker and B. Venkateswarlu (Rijeka: INTECH Press), 171–190. doi: 10.5772/22248
Shi, W. Y., Du, Y. T., Ma, J., Min, D. H., Jin, L. G., Chen, J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19:4087. doi: 10.3390/ijms19124087
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wu, H. J., Zhang, Z., Wang, J. Y., Oh, D. H., Dassanayake, M., Liu, B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224. doi: 10.1073/pnas.1209954109
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Zhang, H., Li, Y., and Zhu, J. K. (2018). Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. doi: 10.1038/s41477-018-0309-4
Zhang, H., Song, Q., Griffin, J. D., and Song, B. H. (2017). Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol. Genet. Genomics 292, 1257–1265. doi: 10.1007/s00438-017-1345-x
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhou, Y., Chen, M., Guo, J., Wang, Y., Min, D., Jiang, Q., et al. (2020a). Overexpression of soybean DREB1 enhances drought stress tolerance of transgenic wheat in the field. J. Exp. Bot. 71, 1842–1857. doi: 10.1093/jxb/erz569
Zhou, Y. X., Zhou, W., Liu, H., Liu, P., and Li, Z. G. (2020b). Genome-wide analysis of the soybean DREB gene family: identification, genomic organization and expression profiles in response to drought stress. Plant Breed. 139, 1158–1167. doi: 10.1111/pbr.12867
Keywords: artificial selection, DREB, natural variation, salt tolerance, cultivated soybean, wild soybean
Citation: Hou Z, Li Y, Cheng Y, Li W, Li T, Du H, Kong F, Dong L, Zheng D, Feng N, Liu B and Cheng Q (2022) Genome-Wide Analysis of DREB Genes Identifies a Novel Salt Tolerance Gene in Wild Soybean (Glycine soja). Front. Plant Sci. 13:821647. doi: 10.3389/fpls.2022.821647
Received: 24 November 2021; Accepted: 17 January 2022;
Published: 04 March 2022.
Edited by:
Guihua Bai, Independent Researcher, New York, NY, United StatesReviewed by:
Milad Eskandari, University of Guelph, CanadaCopyright © 2022 Hou, Li, Cheng, Li, Li, Du, Kong, Dong, Zheng, Feng, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dianfeng Zheng, emRmZm5qQDI2My5uZXQ=; Naijie Feng, ZmVuZ25qQGdkb3UuZWR1LmNu; Baohui Liu, bGl1YmhAZ3podS5lZHUuY25y; Qun Cheng, Y2hlbmdxdW4wMTE4QGd6aHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.