
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 June 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.820767
This article is part of the Research Topic Necrotrophic Fungal Plant Pathogens, Volume II View all 7 articles
Botrytis cinerea is a necrotrophic microbe that causes gray mold disease in a broad range of hosts. In the present study, we conducted molecular microbiology and transcriptomic analyses of the host–B. cinerea interaction to investigate the plant defense response and fungal pathogenicity. Upon B. cinerea infection, plant defense responses changed from activation to repression; thus, the expression of many defense genes decreased in Arabidopsis thaliana. B. cinerea Zn(II)2Cys6 transcription factor BcSpd1 was involved in the suppression of plant defense as ΔBcSpd1 altered wild-type B05.10 virulence by recovering part of the defense responses at the early infection stage. BcSpd1 affected genes involved in the fungal sclerotium development, infection cushion formation, biosynthesis of melanin, and change in environmental pH values, which were reported to influence fungal virulence. Specifically, BcSpd1 bound to the promoter of the gene encoding quercetin dioxygenase (BcQdo) and positively affected the gene expression, which was involved in catalyzing antifungal flavonoid degradation. This study indicates BcSpd1 plays a key role in the necrotrophic microbe B. cinerea virulence toward plants by regulating pathogenicity-related compounds and thereby suppressing early plant defense.
Plant pathogens are classified as biotrophs, necrotrophs, or hemibiotrophs based on their relationship with host plants. Fungal pathogens that feed on living host tissues are known as biotrophs, those that kill and feed on dead host tissues are known as necrotrophs, while the hemibiotrophs exhibit a biphasic feeding strategy—an early biotrophic stage for colonizing and a late necrotrophic stage for feeding (Newman and Derbyshire, 2020). Necrotrophic fungi are reported to be far more economically damaging than biotrophs (Dean et al., 2012; Lorang, 2019; Newman and Derbyshire, 2020). Many necrotrophic fungi are host-specific and only inflict disease on a narrow range of plants, while others have a broad host range. Botrytis cinerea is a typical broad host-range necrotrophic fungal pathogen that can affect more than 1,400 plant species (Fillinger and Elad, 2016). The underlying molecular mechanisms facilitating broad host-range necrotrophy have not been well defined to date.
Molecular studies on B. cinerea, including whole-genome sequencing and analysis of B. cinerea isolates B05.10 and T4, indicated that the fungi navigate a delicate balance of suppressing and inducing several events, such as hormone-regulated defense, cell wall degradation enzymes, fungal effectors, detoxification of plant defense compounds, and activation of transcription regulators (Castillo et al., 2017; Zhang et al., 2018; Cheung et al., 2020; Newman and Derbyshire, 2020; John et al., 2021; Shao et al., 2021; Westrick et al., 2021). Each factor may have its own unique role, or several factors together, causing B. cinerea to successfully invade the host plant and to inflict disease (Fillinger and Elad, 2016). For example, transcription factors (TFs) are sequence-specific DNA-binding proteins required to modulate gene expression (Charoensawan et al., 2010; Hughes, 2011; Caramori et al., 2019). Consequently, an organism relies on a set of suitably operating TFs to orchestrate the expression of genes involved in phytopathogenicity. The characterization of such regulators provides an avenue to identify virulence factors, informing research strategies aimed at building durable resistance in plants (Nejat et al., 2017; Zhang et al., 2018; Jones et al., 2019; Keller, 2019). In addition, their direct inhibition is considered an effective method for targeted disease control (Tietjen and Schreier, 2013; Bahn, 2015; Cho, 2015; Sang and Kim, 2020). One of the transcriptional regulator families is Zn(II)2Cys6, which has only been identified in the fungal kingdom (MacPherson et al., 2006; Campbell et al., 2008). B. cinerea Zn(II)2Cys6 BcBot6 positively regulates botrydial biosynthetic genes and is required for fungal virulence (Porquier et al., 2016, 2019). Since Zn(II)2Cys6 is a transcription regulator, it is interesting to determine if it is involved in the regulation of target gene expression in B. cinerea for modulating host defenses.
Plant immunity contains two interconnected levels based on the recognition of molecular patterns, microbe-associated molecular patterns (MAMPs) or damage-associated molecular patterns (DAMPs) by pattern recognition receptors (PRRs), termed as MAMP-triggered immunity (MTI), or DAMP-triggered immunity (DTI), and on the recognition of effectors from adapted pathogens with plant resistance genes (R-genes) termed as effector-triggered immunity (ETI) (Jones and Dangl, 2006; Couto and Zipfel, 2016; Boutrot and Zipfel, 2017; Gust et al., 2017; Ngou et al., 2021; Yuan et al., 2021). The interaction between effectors and R proteins results in a classical arms race and is described as the “zigzag” model of plant parasite interactions (Cook et al., 2015; Kanyuka and Rudd, 2019). However, the necrotrophic pathogens have evolved to elicit ETI in plants since ETI-derived program cell death (PCD), which restricts the growth of biotrophs but provides a metabolizable substrate for necrotrophs (Lorang, 2019; Shao et al., 2021). This interaction between necrotrophic effectors and R proteins has been described as “inverse” gene-for-gene model of coevolution (Faris and Friesen, 2020; Shao et al., 2021). Thus, a new model to explain plant immunity called “spatial invasion model (SIM),” integrates MTI/DTI and ETI in a common framework, which describes plant detecting and preventing invasion of general pathogenic species, including broad-range host necrotrophic fungi (Kanyuka and Rudd, 2019).
Upon pathogen recognition, a set of downstream responses in plants are induced, such as alteration in hormone levels, transcriptional reprogramming, and changes in plant metabolites. The defense-related genes like transcription factors (TFs) and hormones are repeatedly mentioned (Glazebrook, 2005; Tsuda et al., 2009; Birkenbihl et al., 2017). In addition, plants produce antimicrobial metabolites to suppress fungal growth. Thus, in response to B. cinerea, plant TFs such as WRKYs, ERFs, and NACs; the phytohormones such as JA, ET, SA, and ABA; and metabolites such as camalexin, flavonoids, and glucosinolates (GS) are involved, which either positively or negatively affect plant defenses. Plant resistance and its susceptibility to pathogens are two sides of one coin (Vidhyasekaran, 2014). When plant defense response is suppressed, pathogens succeed in infecting plants. When fungal pathogenicity is repressed, plants succeed in resisting pathogens. It is still a question of how fungi regulate the growth, development, and pathogenicity during the fungus–plant interaction.
In this study, we identified that a pathogenesis-related gene, BcSpd1, encoding Zn(II)2Cys6 transcriptional regulator is involved in the regulation of fungal growth, development, and virulence in the necrotrophic B. cinerea. BcSpd1 is involved in plant–B. cinerea interaction and impairs host defense responses by targeting many virulence genes. We compared the transcripts change from B05.10- and ΔBcSpd1-infected plants to obtain an integrated understanding of B. cinerea’s promotion of disease development by suppressing defense-related gene expression. The results of this study will increase our understanding of the complexity of the plant–B. cinerea interaction and will enhance efforts to identify pathogenicity- or toxicity-related genes in necrotrophic microbes.
For the experiments, 4-week-old A. thaliana plants were used. The plants were grown under a microbe-free climate chamber with 10-h light and 14-h dark cycles. B. cinerea B05.10 and the indicated mutants were grown in the PDA plate to produce the conidia spores. The spores were harvested as previously reported (Liu et al., 2017).
For incubation, the leaves were spray-infected with 5 × 105 spores ml–1. The buffer without any spore was sprayed as untreated control (CK). All leaves were harvested at 14 h and frozen at −80°C for RNA sequencing. For qPCR assay, the plants were infected with B. cinerea B0510 and ΔBcSpd1 and collected at different time points. All the samples were repeated three times.
Total RNA samples were prepared for Illumina sequencing. RNA isolation, purification, and monitoring, and cDNA library construction and sequencing were performed as described previously (Liu et al., 2015). All clean data with high-quality reads were used for analyses. Reference genome and gene model annotation files were downloaded from the website.1 The index of the reference genome was built, and paired-end clean reads were aligned to the reference by using the HISAT package (Kim et al., 2015). Finally, the FPKM of each gene was calculated based on the length and read counts mapped to the gene (Trapnell et al., 2010).
Differential expression analysis of all samples (CK, Bc) was performed as described in a previous study (Liu et al., 2015). The differentially expressed genes (DEGs) were selected with log2 (fold change) > 1 or log2 (fold change) < −1 and with statistical significance (p-value < 0.05) by R package.
Gene Ontology (GO) enrichment analysis of DEGs was implemented by the GOseq R package, and GO terms with corrected p-values less than 0.05 were considered significantly enriched by DEGs (Kanehisa et al., 2008). Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level information, based on large-scale molecular datasets generated by genome sequencing and other high throughput experimental technologies (Young et al., 2010).
Real-time quantitative PCR was performed as previously described (He et al., 2020). BcActin was used for normalization. All analyses were repeated three times using biological replicates. Primer sequences are listed in Supplementary Table 1.
To construct the BcSpd1 (suppression of plant defense 1, Bcin06g05230) gene replacement vectors, flanking sequences of the gene were PCR-amplified from the B05.10 genomic DNA and inserted into the PXEH vector, respectively (Feng et al., 2017). The final replacement vector was generated and then transformed with B05.10 spores using an Agrobacterium tumefaciens AGL-1 strain. Knockout resistant transformants were initially screened on a selective medium (PDA containing 50 μg ml–1 HygB) and then confirmed by PCR and qPCR with indicated primers (Supplementary Table 1). To investigate BcSpd1 complement lines, PCR fragments encoding the full-length open reading frames (ORFs) of genes were isolated and cloned in-frame into the modified pCAMBIA1303 (Yellisetty et al., 2015). Then, the vectors were transformed into ΔBcSpd1 spores to obtain the complement strains (ΔBcSpd1-C). The strain showing wild-type gene expression levels was used for further analysis.
The fungal growth of the tested B. cinerea strains was determined by measuring the radial diameter of colonies on solid CM (1.0% glucose, 0.2% peptone, 0.1% yeast extract, 0.1% casamino acids, nitrate salts, trace elements, 0.01% vitamins, 1.2% agar, pH 6.5) or PDA. Other mediums used in the assays included liquid CM (CM without agar) and PDB (PDA without agar). Fungal growth, infection structure formation, sclerotia formation, melanin biosynthesis, and organic acid production were determined as follows.
For conidium germination assays, fresh conidia of the WT, ΔBcSpd1, and ΔBcSpd1-C strains were harvested from CM plates with ddH2O, and the conidial suspension was adjusted to the concentration of 1 × 105 conidia mL–1 in CM buffer. To examine melanin biosynthesis, the spores of each strain were inoculated on the CM liquid medium in a bottle for 4 days, and melanin production was examined for each bottle. The production of organic acid was determined by a pH indicator assay using CM agar with 0.1% bromophenol blue (CM, pH5.5) or 0.1% bromothymol blue (CM, pH7.5). The acidification of the medium caused by the fungus resulted in a color change. For pH value examination, the indicated fungal strains were incubated on CM liquid medium with the original pH of 6.0. For infection cushion formation assays, the conidial suspension droplets (1 × 105 conidia mL–1, 20 μL) or mycelial plugs were added to the liquid medium (CM) on the glass, and the inoculated fungi were incubated in a moistened chamber at 25°C. The formation of infection cushions was observed and photographically documented at the indicated time points post-incubation. For sclerotia formation assays, strains were cultivated on CM plates at 25°C in darkness; the production of sclerotia by the test strains was observed and photographed 30 days post-incubation.
The observation of conidial germination, infection structure formation, etc., was performed under a Nikon Eclipse 80i fluorescence microscope system. At least three independent experiments with triplicated replicates per experiment were performed.
cDNA of the BcSpd1 gene was PCR-amplified and cloned into the expression vector pET28a to produce the plasmid pET28a-BcSpd1 with His-tag. The recombinant vector was transformed to competent bacterial cells of E. coli BL21. The recombinant protein was induced and expressed in E. coli BL21 cells.
The BcSpd1 protein with His-Ttag was run through 12% SDS-PAGE and transferred to a PVDF Western blotting membrane (Roche Diagnostics GmbH) with a Bio-Rad electroblotting apparatus. The recombinant protein was detected as following the instructions of a previous study (Liu et al., 2015).
For Electrophoretic Mobility Shift Assay (EMSA), about 50-bp upstream and downstream primers labeled with biotin were synthesized using the primers, as shown in Supplementary Table 1. EMSA detection was performed following the LightShift Chemiluminescent EMSA procedure (Thermo-Scientific) as described previously (Liu et al., 2015).
The data were analyzed by an analysis of variance (ANOVA) using SPSS 18 software (IBM). The differences were considered significant at *P < 0.05, **P < 0.01, and ***P < 0.001, respectively. All the data are represented as the mean ± SEM of at least three independent experiments.
Our previous study indicated that B. cinerea B05.10 is a virulent strain and suppresses WRKY33-mediated plant early defense (Liu et al., 2017). Our recent study revealed that B. cinerea B05.10 promotes disease development in the medicinal plant Panax ginseng by suppressing plant early defense signals and antifungal metabolite biosynthesis (Chen et al., 2022). To identify the genes involved in B. cinerea, we screened the transcripts during the plant–Botrytis interaction. A qPCR analysis indicated that a gene Bcin06g05230 is highly expressed in B. cinerea at 14 h after interaction (Figure 1A). The expression of the gene decreased at 24 and 48 h. The earlier expression of this gene reveals that it might play a role in the host–Botrytis interaction.
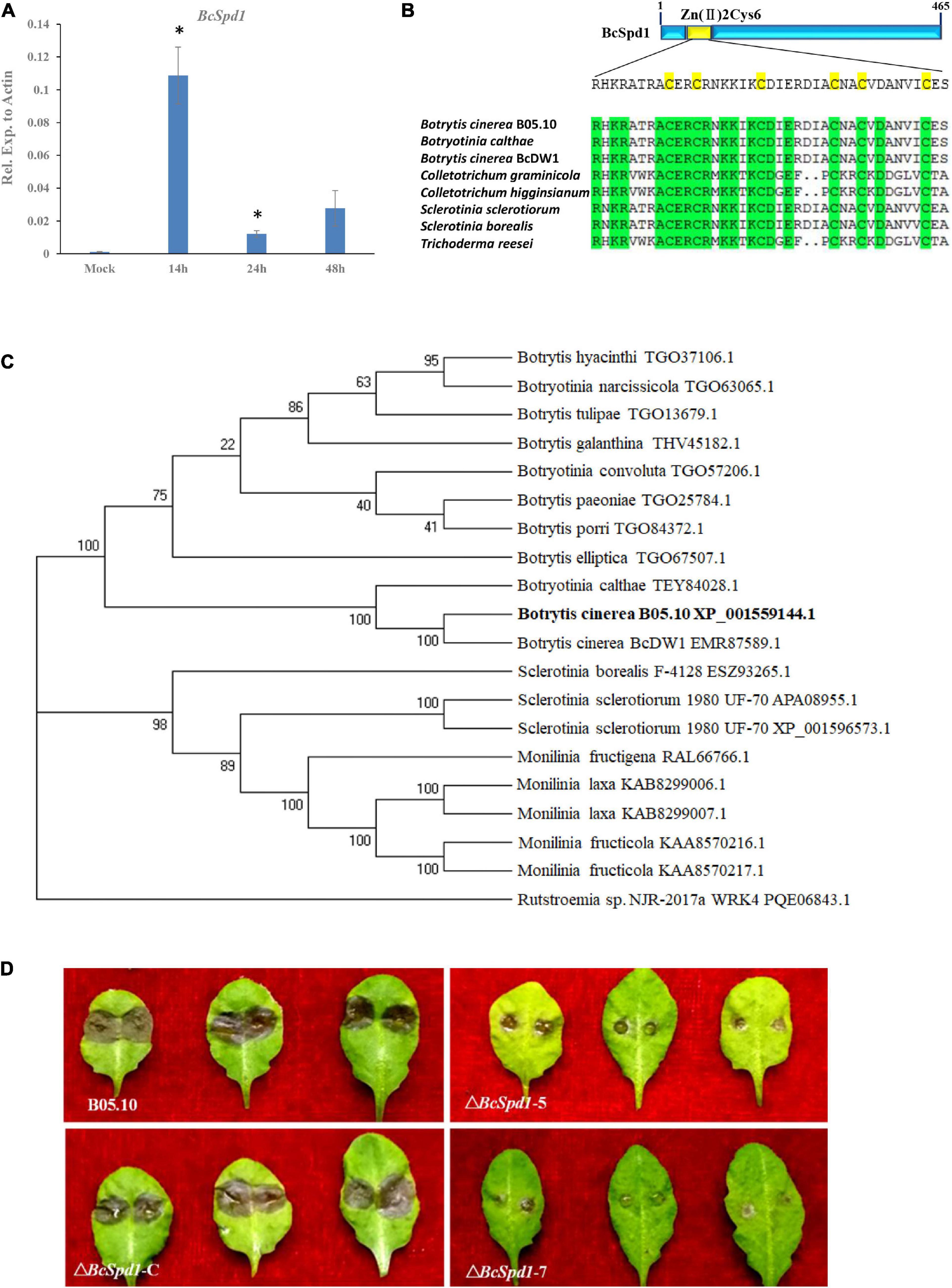
Figure 1. Identification and characterization of BcSpd1 in B. cinerea. (A) qRT-PCR analysis of BcSpd1 gene expression in B. cinerea during fungus infection of the ginseng plant at different timepoints. (B) Zn2Cys6 domain alignment of the BcSpd1 and the indicated species. (C) Phylogenetic tree of BcSpd1 proteins and the indicated proteins including Botrytis cinerea BcDW1 (EMR87589.1), Botrytis elliptica (TGO67507.1), Botrytis porri (TGO84372.1), Botrytis paeoniae (TGO25784.1), Botrytis galanthina (THV45182.1), Botrytis tulipae (TGO13679.1), Botrytis hyacinthi (TGO37106.1), Botryotinia calthae (TEY84028.1), Botryotinia convoluta (TGO57206.1), Botryotinia narcissicola (TGO63065.1), Sclerotinia borealis F-4128 (ESZ93265.1), Sclerotinia sclerotiorum 1980 UF-70 (APA08955.1), Sclerotinia sclerotiorum 1980 UF-70 (XP_001596573.1), Monilinia fructigena (RAL66766.1), Monilinia laxa (KAB8299006.1), Monilinia laxa (KAB8299007.1), Monilinia fructicola (KAA8570216.1), Monilinia fructicola (KAA8570217.1), and Rutstroemia sp. NJR-2017a WRK4 (PQE06843.1). (D) ΔBcSpd1 shows reduced disease symptoms on Arabidopsis thaliana Col-0 leaves. Photographs were taken 3 days post-infection. Wild-type strain B05.10 presents large lesions compared with ΔBcSpd1-5- and ΔBcSpd1-7-infected leaves that show small lesions or no necrotic symptoms on leaves. The complement line ΔBcSpd1-C recovered from wild-type pathogenicity.
Protein sequence analysis revealed several conserved domains (Supplementary Figure 1). The phylogenetic analysis indicated the protein had a higher similarity with the C6 transcription factors (Figures 1B,C). Previous research indicated the genes encoding Zn(II)2Cys6 transcription activator (Todd and Andrianopoulos, 1997). Since the gene encoding transcription activator positively affected downstream gene expression, we hypothesized that the gene and its targets might be involved in fungal virulence.
In order to determine whether Bcin06g05230 is involved in B. cinerea pathogenicity, four mutants were obtained (Supplementary Figure 2). Genome PCR and mRNA qPCR amplification of these mutants showed no expected band and no expression of the gene (Supplementary Figures 2B,C). The gene was cloned, and the complement lines were then generated (Supplementary Figure 3). The virulence of the two mutant strains (ΔBcSpd1-5 and ΔBcSpd1-7) and one complement line (ΔBcSpd1-C) was tested on A. thaliana. Both the mutants showed small lesion sizes compared with the wild-type strains (Figure 1D). Visual inspection of infections in different plants such as tomato and bean also showed a general trend of decreased virulence for the mutants, in comparison with the wild-type (Supplementary Figure 4). Since the wild-type fungus suppressed plant defense and the gene mutants reduced fungal virulence, we named the gene BcSpd1 (suppression of plant defense gene 1). The reintroduction of the BcSpd1 gene into the mutant strains resulted in restoration of the virulence to the levels of the wild-type strain B0510, unequivocally assigning the decrease in virulence to the mutation of ΔBcSpd1 (Figure 1D). These results indicated that BcSpd1 is involved in B. cinerea virulence.
B. cinerea ΔBcSpd1 mutants grew similar to the wild-type B05.10 and the complement strain ΔBcSpd1-C in CM medium. The germination rate of ΔBcSpd1 also has no significant difference compared to the wild-type and complemented strains. Because the sclerotia formation within dying host tissues represented an important survival mechanism of B. cinerea, we were interested in investigating the effects of BcSpd1 gene deletion on sclerotia development. After 1 month of incubation in the dark, ΔBcSpd1 was unable to develop any sclerotia on the CM plate, while the wild-type and ΔBcSpd1-C produced numerous sclerotia, indicating that BcSpd1 is essential for sclerotia formation in B. cinerea (Figure 2A).
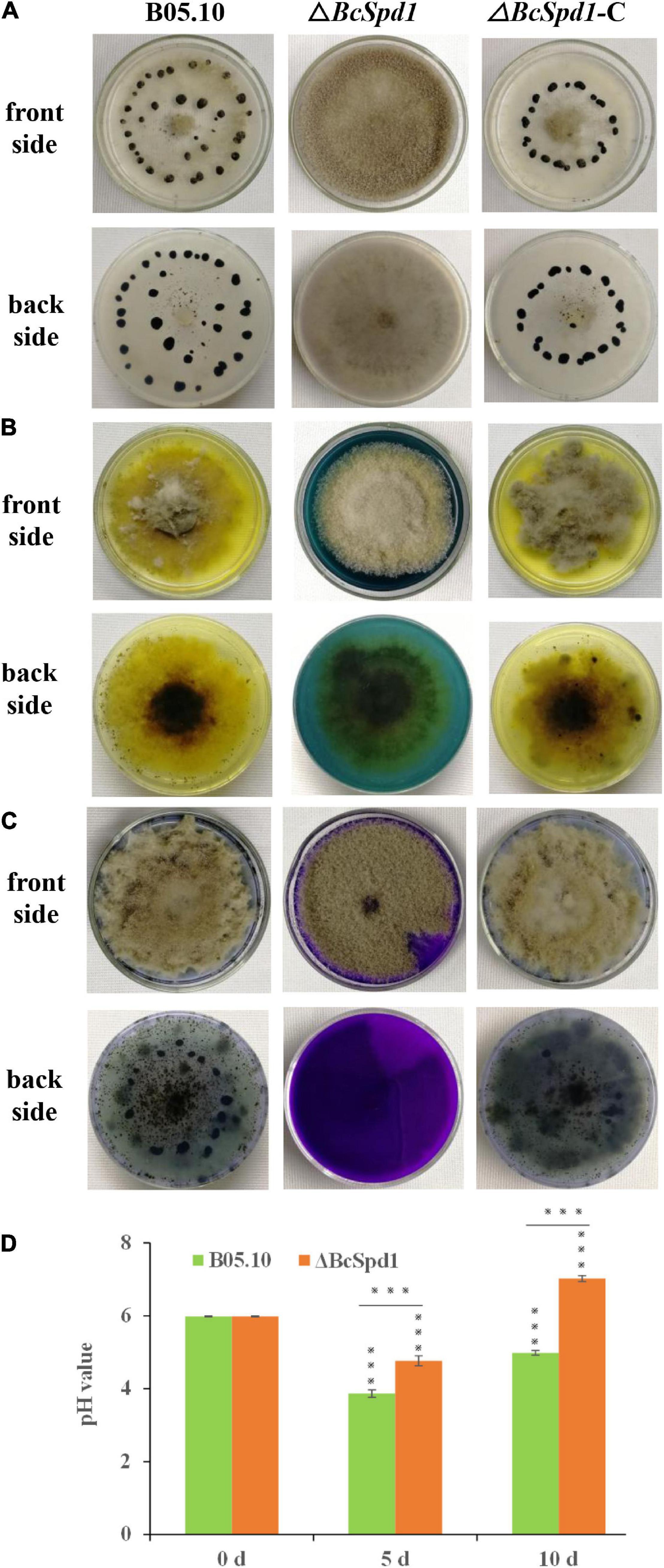
Figure 2. Impact of BcSpd1 deletion on sclerotia development and organic acid biosynthesis. (A) Wild-type strain B05.10, ΔBcSpd1, and the complemented strain ΔBcSpd1-C were incubated on CM plates at 25°C for 4 weeks in darkness. Both B05.10 and ΔBcSpd1-C produce sclerotia on the plates, while ΔBcSpd1 does not. (B) Indicated strains were incubated on CM plates with 0.1% bromophenol blue for 4 days. Both B05.10- and ΔBcSpd1-C-incubated plates show yellow color, while ΔBcSpd1 shows blue. (C) The indicated strains were incubated on CM plates with 0.1% bromothymol blue for 4 days. Both B05.10- and ΔBcSpd1-C-incubated plates show weak blue color, while ΔBcSpd1 show bluish violet. (D) pH changes in wild-type strain B05.10- and ΔBcSpd1 mutant-incubated CM liquid mediums at different times (0 d, 5 d, or 10 d). The unincubated CM medium was used as control (CK). Asterisks indicate significant differences between treatments (***P < 0.001, two-tailed t-test).
The production of organic acid is thought to contribute to B. cinerea virulence by affecting its pH environment and thus involve in the production and activity of secreted enzymes. By performing a CM plate assay using the pH indicator bromophenol blue or bromothymol blue, we compared and examined the ability of the wild-type B05.10, the ΔBcSpd1 mutant, and the ΔBcSpd1-C complement mutant to acidify the agar medium by secreting organic acid. If no fungus was present on the CM plates, the color was blue when bromophenol blue was added, or the color was bluish violet when bromothymol blue was added. After the indicated fungal strains were incubated, ΔBcSpd1 mutants affected the organic acid forming as the pH changed in both the wild-type B0510 and complement line ΔBcSpd1-C (Figures 2B,C). We next tested the changes in pH values in the CM medium after incubation with B05.10 and ΔBcSpd1 mutant, respectively. The original pH of the CM medium was adjusted around pH 6.0 before incubation; then, 5 days after incubation at 25°C under shaking, the pH value decreased to 3.8 in B05.10-incubated CM medium, while the pH value decreased to 4.8 in the ΔBcSpd1-incubated medium (Figure 2D). After 10 days, the pH value was about 5.0 in the B05.10-incubated medium, while the pH value was around 7.0 in the ΔBcSpd1-incubated medium (Figure 2D). The pH value is higher in the ΔBcSpd1-incubated medium than in B05.10, suggesting that BcSpd1 is involved in pH changes during incubation. These results confirmed that BcSpd1 contributed to lower pH values in both plates and the medium.
The infection structures, like infection cushions, play a critical role in B. cinerea host penetration and virulence (Feng et al., 2017; Cao et al., 2018; Liu et al., 2018). To evaluate the effect of BcSpd1 on infection structure formation, we compared the infection cushion formation among B0510, ΔBcSpd1, and ΔBcSpd1-C strains. As demonstrated in Figure 3A, much less infection cushion was produced in ΔBcSpd1 than in B0510 and ΔBcSpd1-C strains at 36, 48, and 72 h after the colony was incubated on hydrophobic glass. Similar results were observed in fungal spores incubated on the hydrophobic glass as observed at 22 hpi (Figure 3B). These data indicated that BcSpd1 involved and positively regulated fungal infection cushion formation.
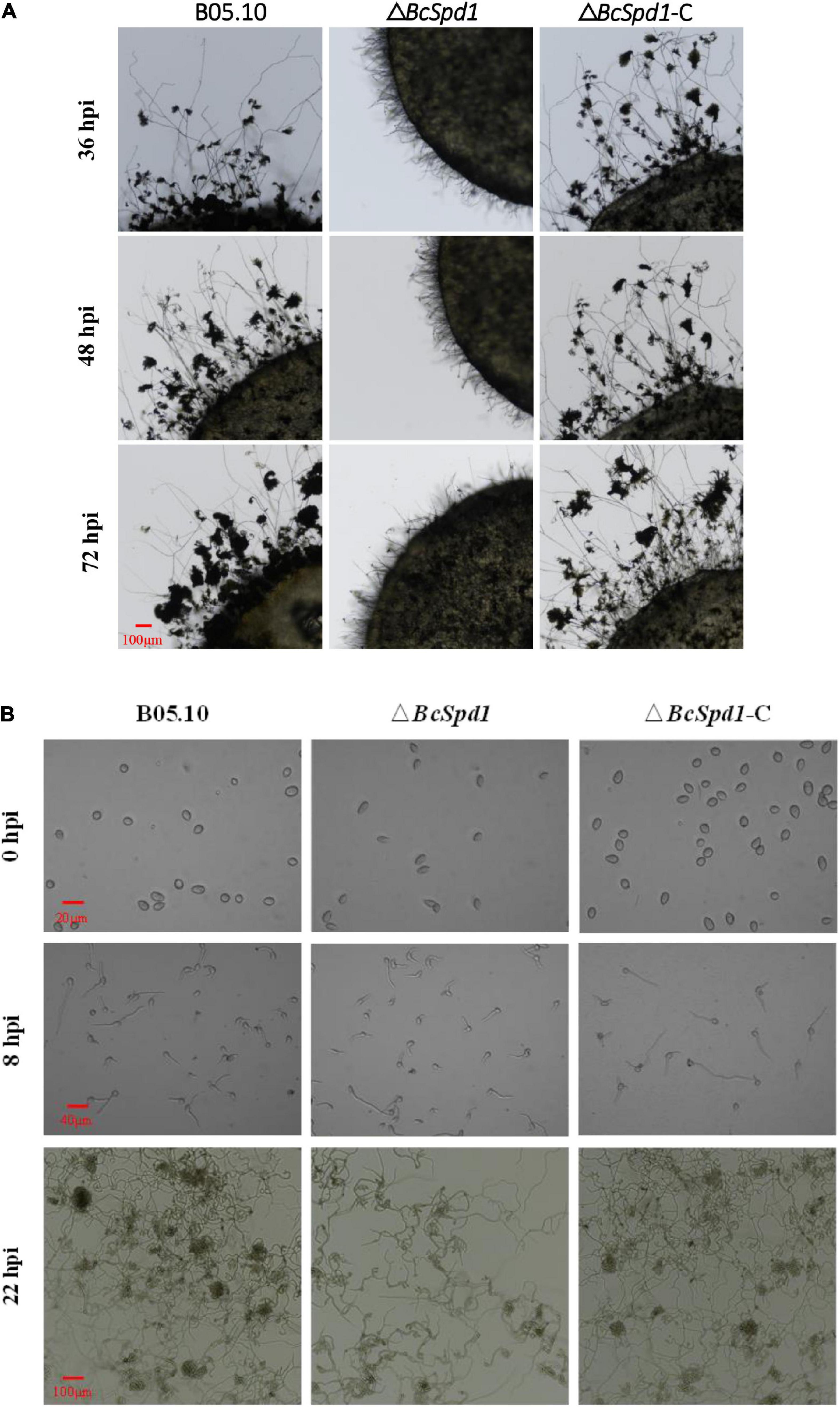
Figure 3. Impact of BcSpd1 deletion on infection cushion formation. (A) Mycelial plugs of wild-type strain B05.10, ΔBcSpd1, and the complemented strain ΔBcSpd1-C were incubated on a hydrophobic glass. The photos were taken at 36, 48, and 72 h, respectively. (B) Formation of infection cushion when fungal spores were incubated on the hydrophobic glass. Photos were taken at 0, 8, and 22 h, respectively.
In addition, after growing on CM for 4 days, ΔBcSpd1 mutants produced significantly more dark pigments than the wild-type B05.10 or the complemented strain (Figure 4A). It has been well accepted that melanin is primarily responsible for dark pigmentation in many filamentous fungi. In ΔBcSpd1, both the culture (Figure 4B, left) and the mycelium (Figure 4B, right) showed more dark pigments after growing in the CM medium. Furthermore, when 0.01% tricyclazole, a melanin biosynthesis inhibitor, was added to the CM medium, the production of melanin disappeared both in ΔBcSpd1 mycelium and the incubation culture, which confirmed the overproduction of melanin by deletion of BcSpd1 (Figure 4C).
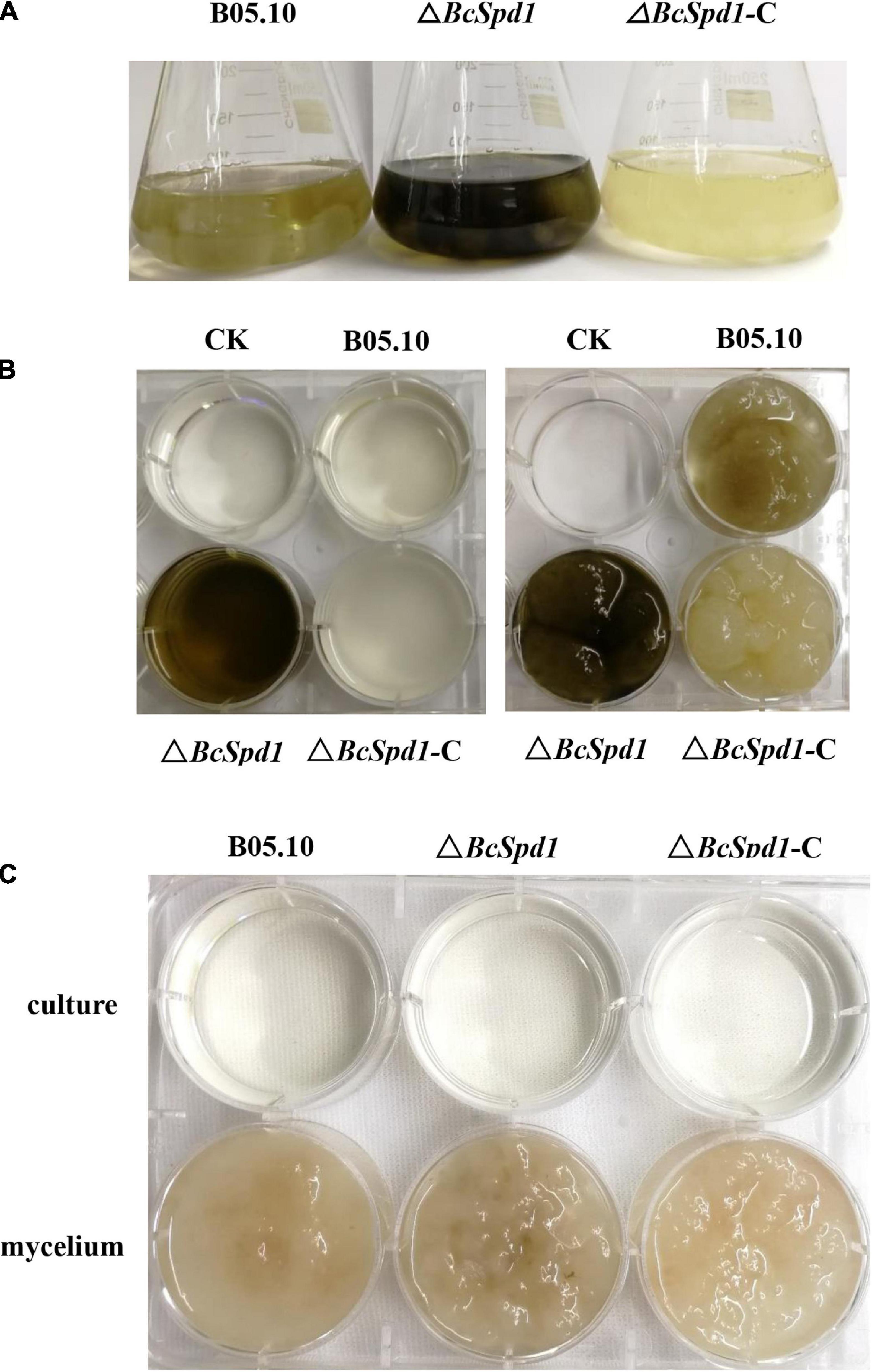
Figure 4. Impact of BcSpd1 deletion on melanin production. (A) Comparisons of mycelial pigmentation among the wild-type strain B05.10, ΔBcSpd1, and the complemented strain ΔBcSpd1-C after 4 days of incubation on CM liquid medium. (B) Mycelia (right) and the CM buffer (left) from (A) were photographed, respectively. (C) CM liquid medium with 0.01% tricyclazole inhibited the production of melanin both in the ΔBcSpd1 mycelium and in the incubation culture.
Since BcSpd1 is involved in B. cinerea sclerotia development, infection structure formation, pH value changes, melanin biosynthesis, and fungal virulence, we next aimed to identify how BcSpd1 was involved. Several genes were reported to affect B. cinerea sclerotia formation, including NOP53, PDE2, BMP1, LTF1, LTF2, FRQ, SAK1, and BcG3 (Zheng et al., 2000; Gronover et al., 2001; Doehlemann et al., 2006; Segmüller et al., 2007; Liu et al., 2011; Harren et al., 2013; Hevia et al., 2015; Cohrs et al., 2016; Cao et al., 2018; Cheung et al., 2020). Lost function of these genes delayed or was unable to form sclerotia, indicating that the genes were involved in or regulated sclerotia production. The expression of a Bcpks12 gene was observed at the sclerotia formation stage, suggesting the involvement of the gene (Zhang et al., 2015). Further study indicated that Bcpks12 was exclusively required for the melanization of sclerotia that are specifically expressed during sclerotia development (Zhu et al., 2017). We performed qPCR to test their expression in ΔBcSpd1 and B05.10. As indicated in the heatmap (Figure 5), the expression of nop53, ltf1, pde2, bmp1, and Bcg3 was decreased in ΔBcSpd1 compared with that in B05.10, indicating that BcSpd1 positively affected their expression. The expression of ltf2, frq, and sak1 was increased in ΔBcSpd1, indicating a negative role of these genes in sclerotium formation (Segmüller et al., 2007; Liu et al., 2011; Harren et al., 2013; Cohrs et al., 2016). NOP53 was also involved in infection cushion formation as the indicated structure formed late in NOP53 mutants compared with B05.10 (Cao et al., 2018). Oxaloacetate hydrolase (BcOAH1) was reported to synthesize oxalate, and ΔBcoah1 mutants did not produce oxalate in vitro (Han et al., 2007). As seen in Figure 5, the expression of Bcoah1 decreased in ΔBcSpd1, indicating that BcSpd1 positively regulated the expression of Bcoah1. Here, the downregulation of Bcoah1 in ΔBcSpd1 might partly lead to the increase in the pH value, as seen in Figure 2. All the data observed before revealed that BcSpd1 regulated genes in the formation of sclerotia, the production of infection cushion, and the decrease in environmental pH value, which played a role in B05.10 growth, development, and virulence. However, BcSpd1 negatively affected the melanin biosynthesis as more dark pigments were observed in the ΔBcSpd1 mutant. Interestingly, the expression of brn1, scd1, cmr1, and bos1 was increased in ΔBcSpd1 mutant compared with B05.10, indicating that BcSpd1 negatively affected the gene expression. BRN1, SCD1, CMR1, and BOS1 were involved in fungal melanin biosynthesis (Liu et al., 2011; Yang et al., 2013; Cheung et al., 2020). The induction of these genes in ΔBcSpd1 mutants might contribute to the accumulation of melanin, as seen in Figure 4. In addition, BcSpd1 positively affected the expression of thr1, chk1, and pks1, which were also involved in melanin biosynthesis (Yang et al., 2013). In this condition, BcSpd1 might play a complex role in regulating B. cinerea melanin biosynthesis.
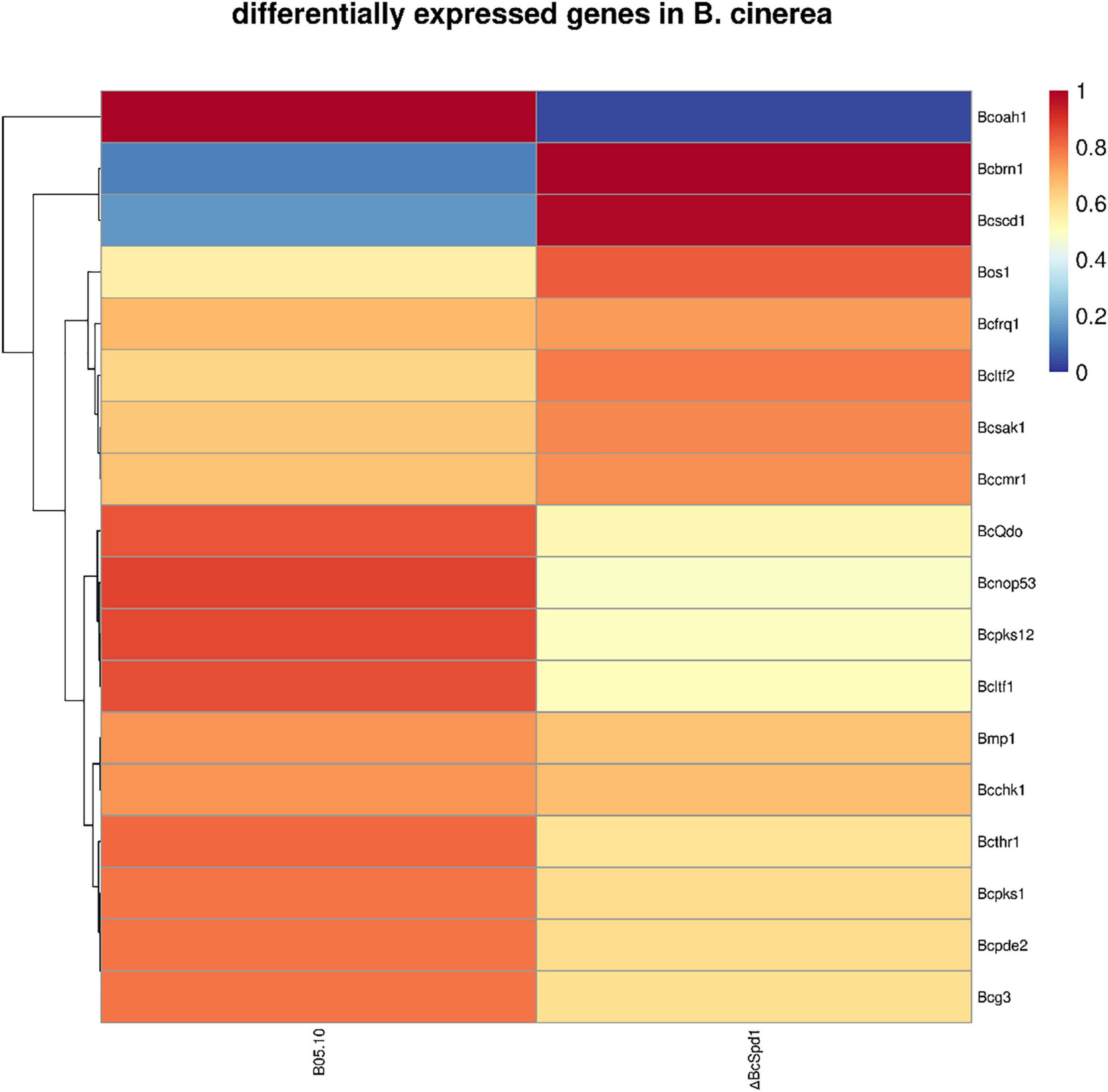
Figure 5. A heatmap analysis of differentially expressed genes involved in B. cinerea development and virulence between the wild-type strain B05.10 and ΔBcSpd1.
Our recent study indicated that flavonoids such as quercetin, kaempferol, and luteolin play a role in plant defense against B. cinerea, but the biosynthesis of such compounds was suppressed in ginseng plants upon fungal early infection (Chen et al., 2022). A gene encoding quercetin dioxygenase was reported to degrade flavonoids, and the ΔBcQdo mutants showed less virulence and partly lost their function in degrading certain flavonoids (Chen et al., 2019, 2022). Next, we aimed to investigate how BcQdo was regulated by B. cinerea. Interestingly, the expression of BcQdo was decreased in the ΔBcSpd1 mutant compared with B05.10 (Figure 5), indicating that BcSpd1 is involved in the regulation of BcQdo expression. We further identified several conserved DNA motifs in the promoter of BcQdo, which could be recognized by Zn(II)2Cys6 transcription factors (Figure 6A). Next, we performed an EMSA experiment to examine if BcSpd1 can bind to these sequences. The BcSpd1 gene was cloned into the pET28a expression vector, and the protein was overexpressed in Escherichia coli BL21 (Figure 6B). The recombinant BcSpd1 was used to bind the DNA fragment in vitro. Totally, four biotin-labeled fragments from the promoter of BcQdo were designed and synthesized (Figure 6A). As seen in Figure 6C, BcSpd1 clearly interacted with fragment 3 in the promoter sequence of BcQdo genes by EMSA. Fragment 3 contains the conserved domain CGGN8CCG, which is typical binding sites for Zn(II)2Cys6 proteins (Todd and Andrianopoulos, 1997). The excess of unlabeled DNA fragments blocked its interaction with BcSpd1, underlining the specificity of the protein/DNA interaction. Since BcSpd1 belongs to the C6 transcription factors, the highly expressed BcSpd1 mutant in B05.10 at the early infection stage might target and positively regulate BcQdo expression, thereby reducing the antifungal flavonoid concentration.
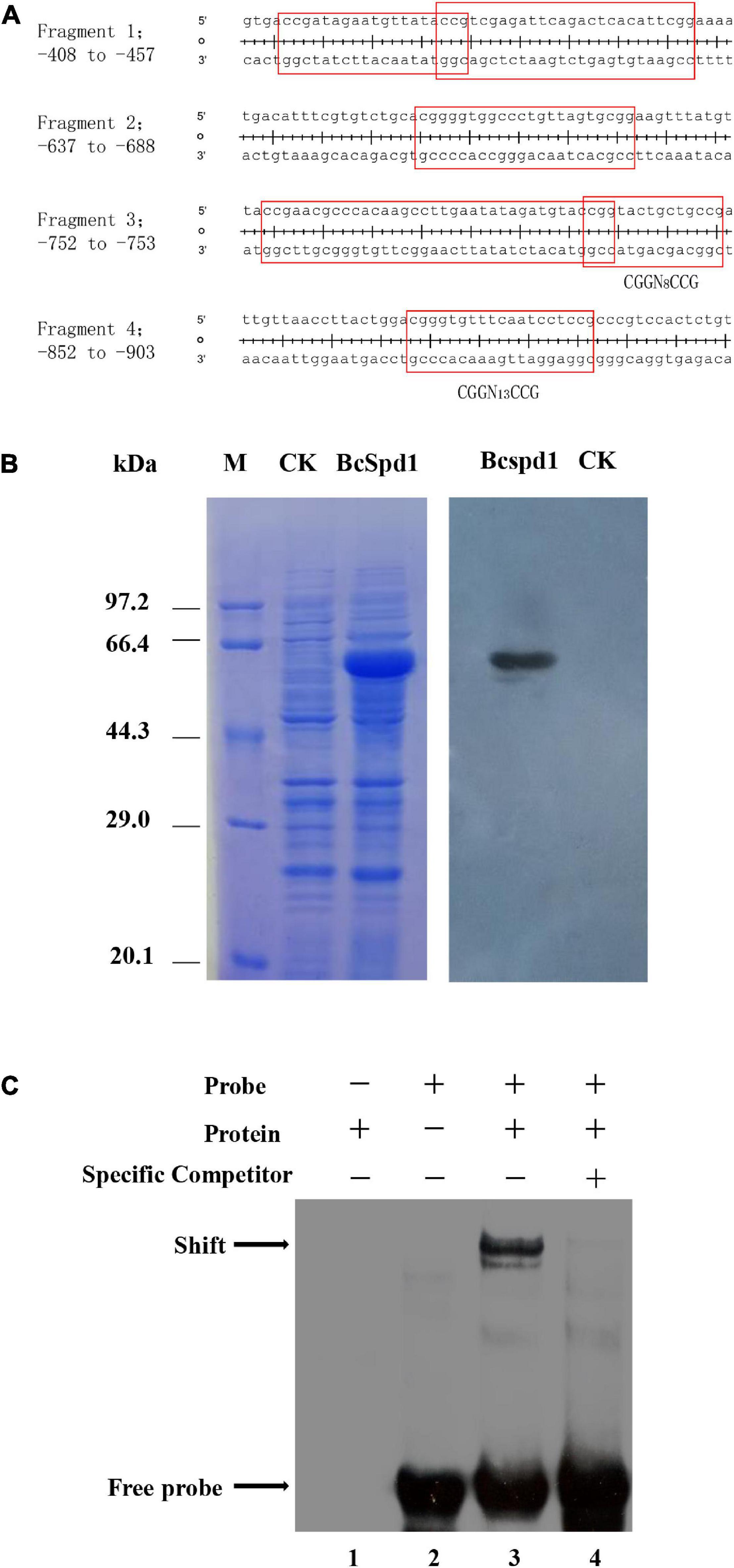
Figure 6. BcSpd1 regulates BcQdo-mediated B. cinerea virulence. (A) Conserved DNA fragments in the promoter of BcQdo. The predicted transcription factor-binding sites were framed. (B) SDS-PAGE and Western blot analysis of recombinant BcSpd1 in Escherichia coli. Left: The proteins were stained with Coomassie brilliant blue R-250. M, Positions of marker proteins. CK, SDS-PAGE of the extracts from empty pET28a-transformed bacterial cultures. BcSpd1, SDS-PAGE of the recombinant BcSpd1 extracted from pET28a-BcSpd1-transformed bacterial culture. Right: Western blot analysis of BcSpd1 protein in the indicated bacterial cultures (anti-His). (C) EMSA analysis of BcSpd1 binding to biotin-labeled promoter fragments of BcQdo. Line 1, the reaction only with recombinant BcSpd1 protein; line 2, the reaction with only biotin-labeled probes; line 3, the reaction with both recombinant BcSpd1 protein and biotin-labeled probes, which showed binding shift; line 4, the unlabeled probes were added to the reaction and competed the binding.
Since B. cinerea B05.10 is virulent toward A. thaliana and the Col-0 and ΔBcSpd1 mutants altered the wild-type fungal virulence, we next aimed to determine how B05.10 affects plant defenses. We performed RNA sequencing to test A. thaliana transcript changes. B. cinerea B05.10 spray-inoculated leaves (14 h post-infection, Col-0 B0510), B. cinerea ΔBcSpd1 spray-inoculated leaves (14 h post-infection, Col-0 ΔBcSpd1), and mock-treated leaves (control, Col-0 CK) were used. The raw sequence data were submitted to the NCBI (GSE186842). The reads were aligned with the A. thaliana genome.
To identify genes involved in A. thaliana response to B. cinerea on a genome-wide level, we compared statistically significantly differentially changed genes (altered two-fold or more, P ≤ 0.05, SSTF) between B. cinerea B05.10-treated, B. cinerea ΔBcSpd1-treated, and un-treated (CK) Col-0 plants (Figure 7A). About 3,940 SSTF genes were identified in ΔBcSpd1-treated plants, while around 3,840 SSTF genes were observed in B05.10-treated plants when compared with untreated plants, respectively. The significantly differentially expressed genes between ΔBcSpd1- and B05.10-treated plants were about 340. When compared to CK, 2992 SSTF genes were both observed in ΔBcSpd1- and B05.10-treated plants, while 949 SSTF genes were only observed in ΔBcSpd1-infected plants and 848 SSTF genes were only observed in B05.10-treated plants (Figure 7B and Supplementary File 1).
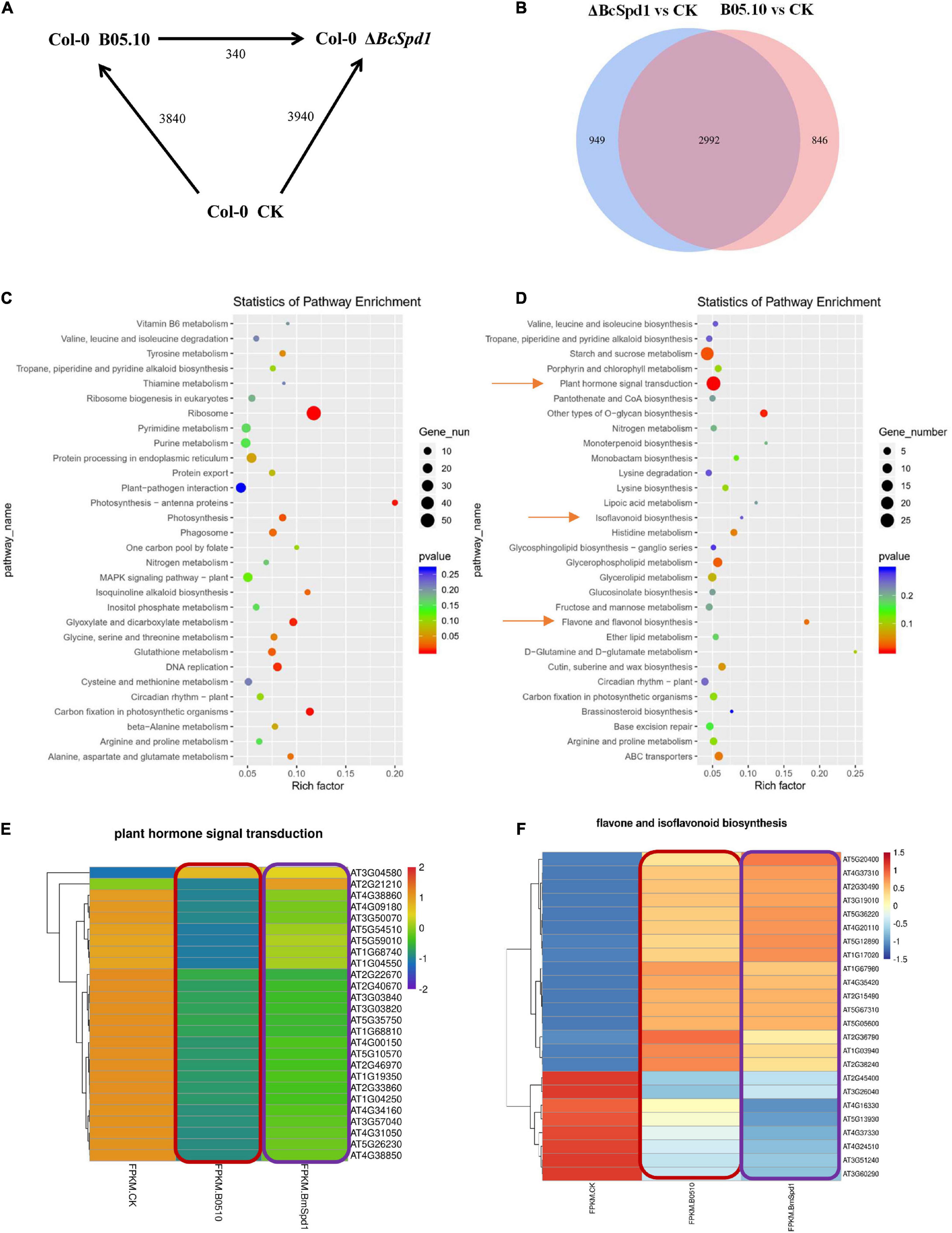
Figure 7. Transcription analysis of B. cinerea B05.10- and ΔBcSpd1-infected A. thaliana Col-0 plants. (A) Number of differentially expressed genes (≥ 2-fold; P ≤ 0.05) in Col-0 at 14 h after mock treatment (CK) or B05.10 or ΔBcSpd1 spray inoculation. The total number of genes between treatments or fungal strains is indicated. (B) Gene analysis of the overlap genes between B05.10-affected and ΔBcSpd1-affected differentially regulated genes. (C) KEGG analysis of differentially expressed genes only observed in B05.10-affected plants. (D) KEGG analysis of differentially expressed genes only observed in ΔBcSpd1-affected plants. (E) A heatmap analysis of differentially expressed genes in the plant hormone transduction pathway among CK, B05.10-, and ΔBcSpd1-treated Col-0 plants. (F) A heatmap analysis of differentially expressed genes in the flavone and isoflavonoid biosynthesis pathways among CK, B05.10-, and ΔBcSpd1-treated Col-0 plants.
We next analyzed the significantly differentially expressed genes by using GO and KEGG methods, respectively. As shown in Supplementary Figure 5, GO analysis of the 2992 SSTF genes observed both in ΔBcSpd1- and B05.10-treated plants indicated the enrichment of response to hormones (SA, JA, ABA), response to chitin, response to pathogens (fungus, bacterium, oomycetes), response to wounding, response to heat, defense response, defense response to pathogens (fungus, bacterium, oomycetes), etc. (Supplementary Figure 5A and Supplementary File 2). KEGG analysis of 2992 SSTF genes indicated the enrichment of plant–pathogen interaction, plant hormone signal transduction, plant MAPK signaling pathway, linoleic acid metabolism, fatty acid degradation, alpha-linolenic acid metabolism, etc. (Supplementary Figure 5B). When analyzing the SSTF genes only in ΔBcSpd1-infected plants or B05.10-treated plants by using the GO method, we observed the genes in DNA replication, DNA binding, mRNA binding, rRNA processing, ribosome, etc. were enriched in ΔBcSpd1-infected plants (Supplementary Figure 5C); however, all the genes mentioned before were not significantly enriched in B05.10-treated plants (Supplementary Figure 5D), suggesting that these genes played a role in plant defense. GO analysis of SSTF genes only in B05.10-treated plants was associated with microtubules and chloroplasts, response to ROS, response to hydrogen peroxide, etc. (Supplementary Figure 5D). These data indicated different genes were enriched in different treatments. When analyzing these SSTF genes by using the KEGG method, plant hormone signal transduction, isoflavonoid biosynthesis, and flavone biosynthesis were enriched in B05.10-treated plants (Figure 7D) compared with ΔBcSpd1-infected plants (Figure 7C).
We further analyzed the heatmap of differentially regulated genes isolated by using the KEGG method. As indicated in Figure 7E, the expression of the genes in plant hormone signal transduction was decreased in B05.10-infected plants compared with CK, while the expression of the same genes increased in ΔBcSpd1-infected plants compared with that in B05.10-treated plants (Figure 7E and Supplementary File 3). It indicated plant defense-related genes were downregulated by B05.10, and these genes were partly upregulated by ΔBcSpd1. The genes in the flavone and isoflavonoid biosynthesis pathway also changed their expression in B05.10- and ΔBcSpd1-treated plants (Figure 7F and Supplementary File 4), indicating the biosynthesis of the compounds were affected. Since transcription factors were reported to involve in plant defense, either positively or negatively, we next compared the differentially expressed genes encoding transcription factors. As shown in Supplementary Figure 6 and Supplementary File 5, the transcription factors altered their expression in ΔBcSpd1-treated plants compared with B05.10-treated plants. The expression of certain TFs is higher in ΔBcSpd1-treated plants than in B05.10-treated plants (red frame labeled), suggesting their potential role in ΔBcSpd1-mediated defense. Similar results were observed in gene response to ABA, a plant hormone involved in both biotic stress and abiotic stress (Supplementary Figure 7 and Supplementary File 6). The expression of certain genes was higher in B05.10-treated plants than in ΔBcSpd1-treated plants, suggesting these genes may involve in fungal virulence. Thus, BcSpd1 was involved in B. cinerea suppression of host defense responses.
The broad host range necrotrophy B. cinerea feed on many hosts even without any relationships. The molecular bases of broad host range necrotrophy in plant pathogens are not well defined and form an ongoing area of research (Newman and Derbyshire, 2020). Specific recognition of B. cinerea by the gene-for-gene resistant mechanism has not been well studied so far (Lorang, 2019). No major R-gene has been identified with resistance to B. cinerea. Plant immunity toward this fungus appears to be under complex poorly understood genetic control (Rowe and Kliebenstein, 2008). The broad host range necrotrophic plant pathogens have evolved diverse, and sometimes convergent, responses to similar selective regime governed by interactions with a highly heterogeneous host landscape (Newman and Derbyshire, 2020). Here, we reported that B. cinerea BcSpd1 played a key role in regulating fungal growth, development, and virulence, and the gene presented as the pathogenic factor which repressed plant defense responses (Figure 8).
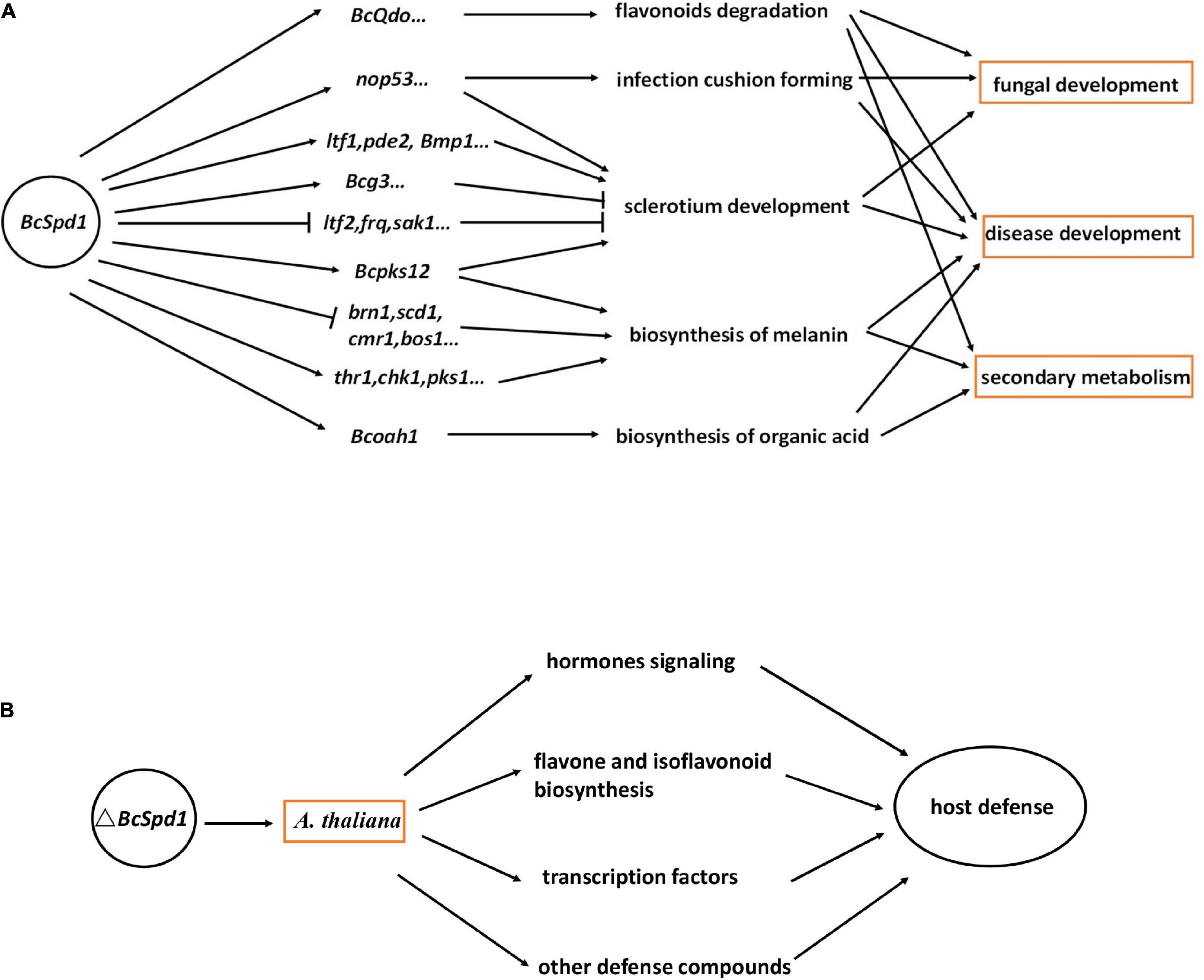
Figure 8. Models showing the role of BcSpd1 in plant–B. cinerea interaction. (A) B. cinerea BcSpd1 is involved in regulating genes in fungal growth, development, secondary metabolism, pathogenicity, and anti-microbe plant metabolism degradation. (B) ΔBcSpd1 recovered from host defense by upregulating many genes in the hormone pathway, transcription factors, and defense-related secondary metabolism biosynthesis.
To protect against the disease caused by B. cinerea, we must understand the mechanisms by which the pathogen causes disease. Virulence factors and pathogenicity genes have been identified for the availability of fungal genomes, but in many cases, their roles remain elusive. It is becoming increasingly clear that gene regulation is vital to enable plant infection and TFs play an essential role. The significance of TFs as regulatory elements in plant-pathogenic fungi has been functionally characterized (John et al., 2021). The TFs are involved in controlling various aspects of fungal development, stress tolerance, and the biosynthesis of virulence factors such as effectors and secondary metabolites. There are a significant number of Zn(II)2Cys6 TF encoding genes, whose activation or functional products have not been resolved (Deepika et al., 2016; Keller, 2019; Romsdahl and Wang, 2019; Graham-Taylor et al., 2020). Here, we reported a new Zn(II)2Cys6 TF, BcSpd1, that played a key role in B. cinerea. Loss of BcSpd1 function in ΔBcSpd1 reduced B. cinerea virulence, and the fungal structures or secondary metabolites associated with BcSpd1 would contribute to fungal pathogenicity. BcSpd1 positively regulated genes involved in the sclerotium development and the infection cushion formation and decreased the environment pH values, while BcSpd1 negatively affected the melanin biosynthesis (Figure 8A). Similar results were reported in several recent studies. For example, the cucurbit pathogen Colletotrichum orbiculare Zn(II)2Cys6 TF Mtf4 was shown to control the development of the appressorium, which played a role in host penetration (Kodama et al., 2019). In Verticillium dahliae, VdFtf1 was identified as a Zn(II)2Cys6 TF that is required for full virulence on cotton (Zhang et al., 2018). Zn(II)2Cys6 TPC1 is involved in the early stage of plant infection by M. oryzae. TPC1 is required for polarized growth and virulence in M. oryzae by regulated synthesis of reactive oxygen species and the MAPK pathway during host invasion (Galhano et al., 2017). In Alternaria brassicicola, AbPf2 was dispensable for normal growth but crucial for virulence (Cho et al., 2013). Gene deletion of Pf2 orthologs in Parastagonospora nodorum and Pyrenophora triticirepentis resulted in downregulation of key effector genes including ToxA and Tox3, leading to the loss of host-specific virulence in wheat (Rybak et al., 2017). In Zymoseptoria tritici, the putative Pf2 ortholog Zt107320 was reported to mediate virulence and sporulation during infection (Habig et al., 2020). Fundamental knowledge of TF regulation provides avenues to identify novel virulence factors in plant–fungus interaction and improves the understanding of the regulatory networks linked to pathogen evolution, while TFs (e.g., BcSpd1) can themselves be specifically targeted for disease management. The development of inhibitors or fungicides, which could suppress BcSpd1 function in B. cinerea, would help to control this pathogen in the future.
From the comparative analysis of host response toward the B. cinerea virulent strain B05.10 and the ΔBcSpd1 mutant in this study, it appears that the outcome of the interaction between the hosts and the two fungal pathogens is much different. The difference is mainly determined by qualitative and quantitative differences in the ΔBcSpd1-dependent activation of similar defense response in the same plant (Figure 8B). In one aspect, the plants inoculated with strain B05.10 were impaired in the accumulation of hormone pathway genes compared with plants infected with the mutant strain at the early stage, and this impairment seemed to be causal to disease development. The strain B05.10 actively suppressed host hormones and signaling was the consequence of early expression of BcSpd1. Similar results were reported in the A. thaliana wrky33 mutant, which was highly susceptible to B. cinerea 2100, while the wrky33nced3nced5 triple mutants were deficient in ABA biosynthesis restored Col-0 resistance by recovering from the expression of plant defense-related genes such as TFs and hormone signaling (Liu et al., 2015). The ABA signaling acted as the susceptible factor in plants not only toward B. cinerea isolate 2100 but also to the isolate B05.10 (Liu et al., 2015, 2017). Quite interestingly, the gene responses to ABA were partly reduced in ΔBcSpd1-infected Col-0 plants and additionally indicated that the susceptibility of plants toward B0510 was very likely by BcSpd1-modulated host ABA signaling. In other aspects, our data suggested that the transcripts of some TFs were affected by B05.10 infection. Interestingly, compared with B05.10, the ΔBcSpd1-infected plants partly recovered from the expression of these genes at an early stage, suggesting their potential role in plant defense. Here, BcSpd1 is involved in such repression and contributes to B. cinerea virulence. Our study further indicates that B. cinerea BcSpd1 is involved in suppressing certain flavones as the compounds play a role in host defense (Chen et al., 2022). On the one hand, BcSpd1 positively regulated BcQdo expression, which is involved in the degradation of certain flavones. On the other hand, many genes in the flavone and isoflavonoid biosynthetic pathway are activated by the ΔBcSpd1 mutants, suggesting that BcSpd1 repressed their expression levels at early infection stages, which could also explain the reduction of antifungal metabolisms in ginseng plants upon B05.10 infection (Chen et al., 2022). Our study proves that B. cinerea BcSpd1 plays a key role in pathogenicity by suppression of plant early defense responses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number GSE186842.
SL and HC designed the research plan and wrote the manuscript. SL, HC, SZ, SH, WL, and RA performed the research. All authors contributed to the article and approved the submitted version.
This work was partly supported by the National Natural Science Foundation of China (NSFC grant no. 32171801 to SL) and the Cross-Disciplinary Innovation Founding of Jilin University (No. JLUXKJC2020313 to SL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Paul Tudzynski from the University of Münster, for kindly providing Botrytis cinerea strain B05.10.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.820767/full#supplementary-material
Supplementary Figure 1 | Protein blast analysis revealed conserved domains in BcSpd1. The amino acid of BcSpd1 was submitted to NCBI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the conserved domain was indicated.
Supplementary Figure 2 | Construction of the BcSpd1 gene deletion mutants (ΔBcSpd1). Replacement of BcSpd1 with a hygromycin gene (Hyg) cassette by homologous recombination (A) and PCR verification of transformants (B). The agarose gels depict (in order): PCR amplification of Hyg genes, PCR amplification of BcSpd1 genes (one heterozygotic line, four transformed homozygotic lines). (C) qPCR analysis of BcSpd1 genes in wild-type B0510 and the indicated ΔBcSpd1 mutants.
Supplementary Figure 3 | Construction of the complemented lines of BcSpd1 (ΔBcSpd1-C).
Supplementary Figure 4 | Phenotype of BcSpd1 deletion mutants showed less pathogenicity than wild-type B. cinerea B0510.
Supplementary Figure 5 | GO and KEGG analysis of differentially expressed genes. (A) GO analysis of differentially expressed genes observed in both B05.10 and ΔBcSpd1 affected Col-0 plants compared with CK. (B) KEGG analysis of differentially expressed genes observed in both B05.10 and ΔBcSpd1 affected Col-0 plants compared with CK. (C) GO analysis of differentially expressed genes observed only in ΔBcSpd1 affected Col-0 plants compared with CK. (D) GO analysis of differentially expressed genes observed only in B05.10 affected Col-0 plants compared with CK.
Supplementary Figure 6 | Heatmap analysis of differentially expressed genes encoding transcription factors among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Supplementary Figure 7 | Heatmap analysis of differentially expressed genes response to ABA among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Supplementary File 1 | The top 100 differentially expressed genes in both B05.10- and ΔBcSpd1-treated plants compared with CK.
Supplementary File 2 | GO enrichment pathway analysis of the differentially expressed genes in both B05.10 and ΔBcSpd1 treated Col-0 plants compared with CK.
Supplementary File 3 | Differentially expressed genes involved in plant hormones signal transduction among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Supplementary File 4 | Differentially expressed genes involved in flavone and isoflavonoid biosynthesis pathway among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Supplementary File 5 | Differentially expression genes encoding transcription factors among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Supplementary File 6 | Differentially expressed genes response to ABA among CK, B05.10, and ΔBcSpd1 treated Col-0 plants.
Bahn, Y. S. (2015). Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets. PLoS Pathog. 11:e1004936. doi: 10.1371/journal.ppat.1004936
Birkenbihl, R. P., Liu, S., and Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Boutrot, F., and Zipfel, C. (2017). Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Campbell, R. N., Leverentz, M. K., Ryan, L. A., and Reece, R. J. (2008). Metabolic control of transcription: paradigms and lessons from Saccharomyces cerevisiae. Biochem. J. 414, 177–187. doi: 10.1042/BJ20080923
Cao, S. N., Yuan, Y., Qin, Y. H., Zhang, M. Z., de Figueiredo, P., Li, G. H., et al. (2018). The pre-rRNA processing factor Nop53 regulates fungal development and pathogenesis via mediating production of reactive oxygen species. Environ. Microbiol. 20, 1531–1549. doi: 10.1111/1462-2920.14082
Caramori, G., Ruggeri, P., Mumby, S., Atzeni, F., and Adcock, I. M. (2019). Transcription factors. eLS 153, 1398–1412.
Castillo, L., Plaza, V., Larrondo, L. F., and Canessa, P. (2017). Recent Advances in the Study of the Plant Pathogenic Fungus Botrytis cinerea and its Interaction with the Environment. Curr. Protein Pept. Sci. 18, 976–989.
Charoensawan, V., Wilson, D., and Teichmann, S. A. (2010). Genomic repertoires of DNA-binding transcription factors across the tree of life. Nucleic Acids Res. 38, 7364–7377. doi: 10.1093/nar/gkq617
Chen, H., Zhang, S., He, S. A. R., Wang, M., and Liu, S. (2022). The necrotroph Botrytis cinerea promotes disease development in Panax ginseng by manipulating plant defense signals and antifungal metabolites degradation. J. Ginseng Res. doi: 10.1016/j.jgr.2022.03.005
Chen, J., Ullah, C., Reichelt, M., Gershenzon, J., and Hammerbacher, A. (2019). Sclerotinia sclerotiorum Circumvents Flavonoid Defenses by Catabolizing Flavonol Glycosides and Aglycones. Plant Physiol. 180, 1975–1987. doi: 10.1104/pp.19.00461
Cheung, N., Tian, L., Liu, X., and Li, X. (2020). The Destructive Fungal Pathogen Botrytis cinerea-Insights from Genes Studied with Mutant Analysis. Pathogens 9:923. doi: 10.3390/pathogens9110923
Cho, Y. (2015). How the necrotrophic fungus Alternaria brassicicola kills plant cells remains an enigma. Eukaryot. Cell 14, 335–344. doi: 10.1128/EC.00226-14
Cho, Y., Ohm, R. A., Grigoriev, I. V., and Srivastava, A. (2013). Fungal-specific transcription factor AbPf2 activates pathogenicity in Alternaria brassicicola. Plant J. 75, 498–514. doi: 10.1111/tpj.12217
Cohrs, K. C., Simon, A., Viaud, M., and Schumacher, J. (2016). Light governs asexual differentiation in the grey mould fungus Botrytis cinerea via the putative transcription factor BcLTF2. Environ. Microbiol. 18, 4068–4086. doi: 10.1111/1462-2920.13431
Cook, D. E., Mesarich, C. H., and Thomma, B. P. (2015). Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. doi: 10.1146/annurev-phyto-080614-120114
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430.
Deepika, V. B., Murali, T. S., and Satyamoorthy, K. (2016). Modulation of genetic clusters for synthesis of bioactive molecules in fungal endophytes: a review. Microbiol. Res. 182, 125–140. doi: 10.1016/j.micres.2015.10.009
Doehlemann, G., Berndt, P., and Hahn, M. (2006). Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59, 821–835. doi: 10.1111/j.1365-2958.2005.04991.x
Faris, J. D., and Friesen, T. L. (2020). Plant genes hijacked by necrotrophic fungal pathogens. Curr. Opin. Plant Biol. 56, 74–80. doi: 10.1016/j.pbi.2020.04.003
Feng, H. Q., Li, G. H., Du, S. W., Yang, S., Li, X. Q., de Figueiredo, P., et al. (2017). The septin protein Sep4 facilitates host infection by plant fungal pathogens via mediating initiation of infection structure formation. Environ. Microbiol. 19, 1730–1749. doi: 10.1111/1462-2920.13613
Fillinger, S., and Elad, Y. (2016). Botrytis – The Fungus, the Pathogen and its Management in Agricultural Systems. Cham: Spinger.
Galhano, R., Illana, A., Ryder, L. S., Rodríguez-Romero, J., Demuez, M., Badaruddin, M., et al. (2017). Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 13:e1006516. doi: 10.1371/journal.ppat.1006516
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Graham-Taylor, C., Kamphuis, L. G., and Derbyshire, M. C. (2020). A detailed in silico analysis of secondary metabolite biosynthesis clusters in the genome of the broad host range plant pathogenic fungus Sclerotinia sclerotiorum. BMC Genomics 21:7. doi: 10.1186/s12864-019-6424-4
Gronover, C. S., Kasulke, D., Tudzynski, P., and Tudzynski, B. (2001). The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 14, 1293–1302. doi: 10.1094/MPMI.2001.14.11.1293
Gust, A. A., Pruitt, R., and Nurnberger, T. (2017). Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791.
Habig, M., Bahena-Garrido, S. M., Barkmann, F., Haueisen, J., and Stukenbrock, E. H. (2020). The transcription factor Zt107320 affects the dimorphic switch, growth and virulence of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 21, 124–138. doi: 10.1111/mpp.12886
Han, Y., Joosten, H. J., Niu, W., Zhao, Z., Mariano, P. S., McCalman, M., et al. (2007). Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi. J. Biol. Chem. 282, 9581–9590. doi: 10.1074/jbc.M608961200
Harren, K., Brandhoff, B., Knodler, M., and Tudzynski, B. (2013). The High-Affinity Phosphodiesterase BcPde2 Has Impact on Growth, Differentiation and Virulence of the Phytopathogenic Ascomycete Botrytis cinerea. PLoS One 8:e78525. doi: 10.1371/journal.pone.0078525
He, S., Chen, H., Wei, Y., An, T., and Liu, S. (2020). Development of a DNA-based real-time PCR assay for the quantification of Colletotrichum camelliae growth in tea (Camellia sinensis). Plant Methods 16:17. doi: 10.1186/s13007-020-00564-x
Hevia, M. A., Canessa, P., Müller-Esparza, H., and Larrondo, L. F. (2015). A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. PNAS 112, 8744–8749. doi: 10.1073/pnas.1508432112
Hughes, T. R. (2011). Introduction to “a handbook of transcription factors. Subcell Biochem. 52, 1–6. doi: 10.1007/978-90-481-9069-0_1
John, E., Singh, K. B., Oliver, R. P., and Tan, K. C. (2021). Transcription factor control of virulence in phytopathogenic fungi. Mol. Plant Pathol. 22, 858–881. doi: 10.1111/mpp.13056
Jones, D. A. B., John, E., Rybak, K., Phan, H. T. T., Singh, K. B., Lin, S. Y., et al. (2019). A specific fungal transcription factor controls effector gene expression and orchestrates the establishment of the necrotrophic pathogen lifestyle on wheat. Sci. Rep. 9:15884. doi: 10.1038/s41598-019-52444-7
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kanyuka, K., and Rudd, J. J. (2019). Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8. doi: 10.1016/j.pbi.2019.02.005
Keller, N. P. (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180. doi: 10.1038/s41579-018-0121-1
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kodama, S., Nishiuchi, T., and Kubo, Y. (2019). Colletotrichum orbiculare MTF4 Is a Key Transcription Factor Downstream of MOR Essential for Plant Signal-Dependent Appressorium Development and Pathogenesis. Mol. Plant Microbe Interact. 32, 313–324. doi: 10.1094/MPMI-05-18-0118-R
Liu, J. K., Chang, H. W., Liu, Y., Qin, Y. H., Ding, Y. H., Wang, L., et al. (2018). The key gluconeogenic gene PCK1 is crucial for virulence of Botrytis cinerea via initiating its conidial germination and host penetration. Environ. Microbiol. 20, 1794–1814. doi: 10.1111/1462-2920.14112
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., and Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4:e07295. doi: 10.7554/eLife.07295
Liu, S., Ziegler, J., Zeier, J., Birkenbihl, R. P., and Somssich, I. E. (2017). Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ. 40, 2189–2206. doi: 10.1111/pce.13022
Liu, W., Soulié, M. C., Perrino, C., and Fillinger, S. (2011). The osmosensing signal transduction pathway from Botrytis cinerea regulates cell wall integrity and MAP kinase pathways control melanin biosynthesis with influence of light. Fungal Genet. Biol. 48, 377–387. doi: 10.1016/j.fgb.2010.12.004
Lorang, J. (2019). Necrotrophic Exploitation and Subversion of Plant Defense: a Lifestyle or Just a Phase, and Implications in Breeding Resistance. Phytopathology 109, 332–346. doi: 10.1094/PHYTO-09-18-0334-IA
MacPherson, S., Larochelle, M., and Turcotte, B. (2006). A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604.
Nejat, N., Rookes, J., Mantri, N. L., and Cahill, D. M. (2017). Plant-pathogen interactions: toward development of next-generation disease-resistant plants. Crit. Rev. Biotechnol. 37, 229–237. doi: 10.3109/07388551.2015.1134437
Newman, T. E., and Derbyshire, M. C. (2020). The Evolutionary and Molecular Features of Broad Host-Range Necrotrophy in Plant Pathogenic Fungi. Front. Plant Sci. 11:591733. doi: 10.3389/fpls.2020.591733
Ngou, B. P. M., Ahn, H. K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Porquier, A., Moraga, J., Morgant, G., Dalmais, B., Simon, A., Sghyer, H., et al. (2019). Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn(2)Cys(6) transcription factor BcBoa13. Curr. Genet. 65, 965–980. doi: 10.1007/s00294-019-00952-4
Porquier, A., Morgant, G., Moraga, J., Dalmais, B., Luyten, I., Simon, A., et al. (2016). The botrydial biosynthetic gene cluster of Botrytis cinerea displays a bipartite genomic structure and is positively regulated by the putative Zn(II)(2)Cys(6) transcription factor BcBot6. Fungal Genet. Biol. 96, 33–46. doi: 10.1016/j.fgb.2016.10.003
Romsdahl, J., and Wang, C. C. C. (2019). Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012-2018). Medchemcomm 10, 840–866.
Rowe, H. C., and Kliebenstein, D. J. (2008). Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180, 2237–2250. doi: 10.1534/genetics.108.091439
Rybak, K., See, P. T., Phan, H. T. T., Syme, R. A., Moffat, C. S., Oliver, R. P., et al. (2017). A functionally conserved Zn2Cys6 binuclear cluster transcription factor class regulates necrotrophic effector gene expression and host-specific virulence of two major Pleosporales fungal pathogens of wheat. Mol. Plant Pathol. 18, 420–434.
Sang, H., and Kim, J. I. (2020). Advanced strategies to control plant pathogenic fungi by host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS). Plant Biotechnol. Rep. 14, 1–8. doi: 10.3390/ijms22157956
Segmüller, N., Ellendorf, U., Tudzynski, B., and Tudzynski, P. (2007). BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryotic. Cell 6, 211–221. doi: 10.1128/EC.00153-06
Shao, D., Smith, D. L., Kabbage, M., and Roth, M. G. (2021). Effectors of Plant Necrotrophic Fungi. Front. Plant Sci. 12:687713. doi: 10.3389/fpls.2021.687713
Tietjen, K., and Schreier, P. H. (2013). “New targets for fungicides,” in Modern Methods in Crop Protection Research, eds P. Jeschke, W. Krämer, U. Schirmer, and M. Witschel (Hoboken, NJ: John Wiley & Sons Ltd), 197–216.
Todd, R. B., and Andrianopoulos, A. (1997). Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21, 388–405. doi: 10.1006/fgbi.1997.0993
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J., and Katagiri, F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5:e1000772. doi: 10.1371/journal.pgen.1000772
Vidhyasekaran, P. (2014). “PAMP signaling in plant innate immunity,” in PAMP Signals in Plant Innate Immunity. Signaling and Communication in Plants, Vol. 21. (Dordrecht: Springer). doi: 10.1007/978-94-007-7426-1_2
Westrick, N. M., Smith, D. L., and Kabbage, M. (2021). Disarming the Host: detoxification of Plant Defense Compounds During Fungal Necrotrophy. Front. Plant Sci. 12:651716. doi: 10.3389/fpls.2021.651716
Yang, Q., Chen, Y., and Ma, Z. (2013). Involvement of BcVeA and BcVelB in regulating conidiation, pigmentation and virulence in Botrytis cinerea. Fungal Genet. Biol. 50, 63–71. doi: 10.1016/j.fgb.2012.10.003
Yellisetty, V., Reddy, L. A., and Mandapaka, M. (2015). In planta transformation of sorghum (Sorghum bicolor (L.) Moench) using TPS1 gene for enhancing tolerance to abiotic stresses. J. Genet. 94, 425–434. doi: 10.1007/s12041-015-0540-y
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zhang, C., He, Y., Zhu, P., Chen, L., Wang, Y., Ni, B., et al. (2015). Loss of bcbrn1 and bcpks13 in Botrytis cinerea Not Only Blocks Melanization But Also Increases Vegetative Growth and Virulence. Mol. Plant Microbe Interact. 28, 1091–1101. doi: 10.1094/MPMI-04-15-0085-R
Zhang, W. Q., Gui, Y. J., Short, D. P. G., Li, T. G., Zhang, D. D., Zhou, L., et al. (2018). Verticillium dahliae transcription factor VdFTF1 regulates the expression of multiple secreted virulence factors and is required for full virulence in cotton. Mol. Plant Pathol. 19, 841–857. doi: 10.1111/mpp.12569
Zheng, L., Campbell, M., Murphy, J., Lam, S., and Xu, J. R. (2000). The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 13, 724–732. doi: 10.1094/MPMI.2000.13.7.724
Keywords: Botrytis cinerea, Zn(II)2Cys6 transcription factor, the necrotrophic microbe, pathogenicity, host defense
Citation: Chen H, He S, Zhang S, A R, Li W and Liu S (2022) The Necrotroph Botrytis cinerea BcSpd1 Plays a Key Role in Modulating Both Fungal Pathogenic Factors and Plant Disease Development. Front. Plant Sci. 13:820767. doi: 10.3389/fpls.2022.820767
Received: 23 November 2021; Accepted: 01 June 2022;
Published: 30 June 2022.
Edited by:
Paloma Melgarejo, Ministerio de Agricultura, Alimentación y Medio Ambiente, SpainReviewed by:
Wenjun Zhu, Wuhan Polytechnic University, ChinaCopyright © 2022 Chen, He, Zhang, A, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouan Liu, c2hvdWFuQGpsdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.