
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 March 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.819360
This article is part of the Research Topic Plant Cell Wall Polysaccharides as Biofuels and Biomaterials View all 19 articles
 Yoshimi Nakano1†‡
Yoshimi Nakano1†‡ Hitoshi Endo1
Hitoshi Endo1 Lorenz Gerber2
Lorenz Gerber2 Chiaki Hori3‡
Chiaki Hori3‡ Ayumi Ihara1,3,4
Ayumi Ihara1,3,4 Masayo Sekimoto3
Masayo Sekimoto3 Tomoko Matsumoto3
Tomoko Matsumoto3 Jun Kikuchi3
Jun Kikuchi3 Misato Ohtani1,3,4*†
Misato Ohtani1,3,4*† Taku Demura1,3*
Taku Demura1,3*The secondary cell wall (SCW) in the xylem is one of the largest sink organs of carbon in woody plants, and is considered a promising sustainable bioresource for biofuels and biomaterials. To enhance SCW formation in poplar (Populus sp.) xylem, we developed a self-reinforced system of SCW-related transcription factors from Arabidopsis thaliana, involving VASCULAR-RELATED NAC-DOMAIN7 (VND7), SECONDARY WALL-ASSOCIATED NAC-DOMAIN PROTEIN 1/NAC SECONDARY WALL THICKENING-PROMOTING FACTOR3 (SND1/NST3), and MYB46. In this system, these transcription factors were fused with the transactivation domain VP16 and expressed under the control of the Populus trichocarpa CesA18 (PtCesA18) gene promoter, creating the chimeric genes PtCesA18pro::AtVND7:VP16, PtCesA18pro::AtSND1:VP16, and PtCesA18pro::AtMYB46:VP16. The PtCesA18 promoter is active in tissues generating SCWs, and can be regulated by AtVND7, AtSND1, and AtMYB46; thus, the expression levels of PtCesA18pro::AtVND7:VP16, PtCesA18pro::AtSND1:VP16, and PtCesA18pro::AtMYB46:VP16 are expected to be boosted in SCW-generating tissues. In the transgenic hybrid aspens (Populus tremula × tremuloides T89) expressing PtCesA18pro::AtSND1:VP16 or PtCesA18pro::AtMYB46:VP16 grown in sterile half-strength Murashige and Skoog growth medium, SCW thickening was significantly enhanced in the secondary xylem cells, while the PtCesA18pro::AtVND7:VP16 plants showed stunted xylem formation, possibly because of the enhanced programmed cell death (PCD) in the xylem regions. After acclimation, the transgenic plants were transferred from the sterile growth medium to pots of soil in the greenhouse, where only the PtCesA18pro::AtMYB46:VP16 aspens survived. A nuclear magnetic resonance footprinting cell wall analysis and enzymatic saccharification analysis demonstrated that PtCesA18pro::AtMYB46:VP16 influences cell wall properties such as the ratio of syringyl (S) and guaiacyl (G) units of lignin, the abundance of the lignin β-aryl ether and resinol bonds, and hemicellulose acetylation levels. Together, these data indicate that we have created a self-reinforced system using SCW-related transcription factors to enhance SCW accumulation.
In recent years, mounting environmental problems, such as global warming from fossil fuels, have increased the importance of sustainable and carbon-neutral bioresources. Lignocellulosic biomass, the most abundant above-ground bioresource, is found in the secondary cell walls (SCWs) of xylem tissues. To improve the availability and use of lignocellulosic biomass, the molecular mechanisms of SCW formation have been actively studied. In the last 2 decades especially, our understanding of the key transcriptional regulators for SCW formation has greatly expanded (Zhong et al., 2010a; Nakano et al., 2015; Ohtani and Demura, 2019; Kamon and Ohtani, 2021). One of the most important findings is the identification of key transcriptional factors of SCW formation; in vascular plants, a specific group of NAC (NAM/ATAF/CUC) family transcription factors, called VNS [VASCULAR-RELATED NAC-DOMAIN (VND), NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NST)/SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND), and SOMBRERO (SMB)-related] proteins, were shown to function as master regulators of SCW formation, activating all of the events required for SCW formation in A. thaliana (Arabidopsis; Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2010b; Yamaguchi et al., 2008, 2011; Ohashi-Ito et al., 2010; Ohtani et al., 2011; Xu et al., 2014; Akiyoshi et al., 2020). Additionally, downstream of the VNS proteins, the MYB transcription factors, such as Arabidopsis MYB46 and MYB83, function to upregulate the expression of genes encoding SCW-related enzymes as secondary master regulators of xylem cell formation (Zhong et al., 2007; Ko et al., 2009, 2014; McCarthy et al., 2009, 2010; Nakano et al., 2010). The NAC–MYB-based transcriptional network of SCW formation is widely conserved among land plants (Zhong et al., 2010a; Xu et al., 2014; Nakano et al., 2015; Bowman et al., 2017; Ohtani et al., 2017a), suggesting that these NAC and MYB transcription factors would be effective targets for modifying the quantity and quality of lignocellulosic biomass.
Phylogenetic tree analysis indicates that three subgroups can be recognized within VNS proteins; VND, SND/NST, and SMB subgroups (Nakano et al., 2015; Bowman et al., 2017; Ohtani et al., 2017a; Akiyoshi et al., 2020). In Arabidopsis, VND and SND/NST subgroups regulate the differentiation of xylem vessels and xylem fibers, respectively (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2010b; Yamaguchi et al., 2008, 2011). It has been shown that cell wall characteristics are different between xylem vessel cells and fiber cells, such as syringyl (S)/guaiacyl (G) ratio of lignin subunits (Saito et al., 2012). Therefore, the regulatory targets of SCW-related genes should be different between VND and SND/NST subgroups; indeed, the transcriptome analysis of Arabidopsis cells carrying the inducible overexpression system of VND or SND/NST revealed that the genes for lignin monomer biosynthesis could be more strongly induced by SND/NST proteins than by VND proteins (Ohashi-Ito et al., 2010; Zhong et al., 2010b; Yamaguchi et al., 2011). In addition, a part of SCW-biosynthetic genes, for example SCW-specific cellulose synthase genes CesA4 and CesA8, are common targets of VNS and MYB proteins (Zhong et al., 2007, 2010b; Ko et al., 2009, 2014; McCarthy et al., 2009, 2010; Nakano et al., 2010; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Interestingly, in Arabidopsis, most of SCW-biosynthetic genes would be mainly regulated by the MYB46 and MYB83 proteins, since the SCW deposition in xylem cells is severely defective in the myb46 myb83 double mutant (McCarthy et al., 2009), and the cesa mutant phenotype could not be rescued when the MYB46-regulatory cis element was mutated in the promoter controlling CesA genes (Kim et al., 2013). These findings collectively suggest that each SCW-related transcription factor would differently regulate the characteristics of SCW in xylem tissues.
The effects of VNS and MYB overexpression have already been reported in several plant species, including Arabidopsis, rice (Oryza sativa), and a hybrid aspen (Populus tremula × tremuloides; Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007, 2011; McCarthy et al., 2010; Ohtani et al., 2011; Valdivia et al., 2013; Yoshida et al., 2013; Xu et al., 2014; Endo et al., 2015; Akiyoshi et al., 2020). These reports showed that the continuous overexpression of VNS or MYB under the control of the cauliflower mosaic virus 35S promoter resulted in the ectopic deposition of SCWs, resulting in the inhibition of plant growth. Moreover, the transgenic expression of VNS genes from other species showed a higher induction activity of ectopic SCW deposition than the overexpression of the gene from the same species (Kubo et al., 2005; Ohtani et al., 2011; Sakamoto et al., 2016). These observations led us to develop a new hypothesis that the enhanced xylem tissue–specific expression of heterologous VNS and MYB genes would be an effective strategy for enhancing SCW deposition in the xylem without negatively impacting plant growth.
In order to test the hypothesis described above, we designed a self-reinforced system of SCW-related transcription factors from Arabidopsis: AtVND7, AtSND1/AtNST3, and AtMYB46. In this system, to make an artificial positive feedback loop (Yan et al., 2012), the transcription factors were fused with the VP16 transcription activation domain to enhance their function (Yamaguchi et al., 2010). In addition, they were expressed under the control of the Populus trichocarpa CesA18 (PtCesA18) promoter, which is active and the gene is expressed in xylem tissue (Suzuki et al., 2006). The resultant chimeric genes, PtCesA18pro::AtVND7:VP16, PtCesA18pro::AtSND1:VP16, and PtCesA18pro::AtMYB46:VP16, were introduced into the hybrid aspen (P. tremula × tremuloides T89). The phenotypic analysis showed that our strategy could work to enhance SCW deposition in xylem tissues in PtCesA18pro::AtSND1:VP16 and PtCesA18::AtMYB46:VP16. We also found that only transgenic plants expressing PtCesA18pro::AtMYB46:VP16 were able to grow in pots of soil in the greenhouse, while PtCesA18pro::AtVND7:VP16 and PtCesA18pro::AtSND1:VP16 could grow only in a sterile half-strength Murashige and Skoog (MS) medium. The lignocellulosic property in the xylem tissues was altered in the PtCesA18pro::AtMYB46:VP16 line 17 with the highest expression of AtMYB46. Together, these data collectively demonstrate the practicality of using a self-reinforced system of SCW-related transcription factors to enhance lignocellulose accumulation and SCW deposition, especially for the second master regulator of xylem cell formation (AtMYB46) in woody plants.
The promoter sequence of the PtCesA18 gene was amplified from the genome of black cottonwood (Populus trichocarpa). Hybrid aspen (P. tremula × P. tremuloides, T89) was used for the transformation, according to the methods described by Ohtani et al. (2011). The plants were maintained and propagated aseptically on a medium containing a half-strength Murashige and Skoog (MS) salt mixture (Duchefa Biochemie, Haarlem, The Netherlands; pH 5.8) under a photoperiod of 16 h light/8 h dark at 23°C (Ohtani et al., 2011; Hori et al., 2020). For the analysis of growth and xylem lignocellulose, the transgenic plants were acclimated to the pots of soil in the greenhouse and grown under a 16-h light/8-h dark photoperiod at 23°C (Ohtani et al., 2011; Hori et al., 2020). To clone the transcription factor genes, total RNAs were extracted from 7-day-old A. thaliana (ecotype Columbia) seedlings grown in half-strength MS medium containing 0.5% (w/v) sucrose, as described by Nakano et al. (2010).
An approximately 3-kbp promoter sequence of PtCesA18 (also known as PtrCesA1A/PtCesA3-2/PtiCesA8-B or eugene3.00040363; Taylor et al., 2003; Suzuki et al., 2006; Kumar et al., 2009), which was most homologous to a Populus tremuloides CesA that was previously characterized as a xylem-specific cellulose synthase gene (Wu et al., 2000), was amplified using PCR from the genomic DNA isolated from Populus trichocarpa shoots as described by Ohtani et al. (2011), and then cloned into pENTR™/D-TOPO™ (Thermo Fisher Scientific, Waltham, MA, United States). The coding regions (excluding the stop codon) of AtVND7 (AT1G71930), AtSND1 (AT1G32770), and AtMYB46 (AT5G12870) were also PCR-amplified from the cDNAs prepared from the total RNAs of the Arabidopsis seedlings, as described by Nakano et al. (2010), and then cloned into pENTR™/D-TOPO™ (Thermo Fisher Scientific, Waltham, MA, United States). The HYGROMYCIN PHOSPHOTRANSFERASE gene and NOS terminator of the pSMAH621 vector (Kubo et al., 2005) were replaced with the NEOMYCIN PHOSPHOTRANSFERASE II (NPTII) gene and the Arabidopsis heat shock protein terminator (AtHSP ter), respectively, to increase mRNA expression (Nagaya et al., 2010). To generate the construction of PtCesA18pro::GUS, PtCesA18 promoter sequence was inserted into the Sma I site located the upstream of GUS gene in the modified pSMAH621 containing NPTII and AtHSP ter. In addition, the 35S promoter and the GUS gene were removed from pSMAH621 using restriction enzymes, and the sequence of the human herpes virus–derived VP16 transcriptional activation domain was inserted, to obtain the pSMAH-VP16-Thsp vector.
To generate the sequence corresponding to PtCesA18pro::AtVND7, PtCesA18pro::AtSND1, or PtCesA18pro::AtMYB46, the sequences of PtCesA18pro and the coding regions of AtVND7, AtSND1, and AtMYB46 were first independently amplified by PCR with a set of primers containing the overlapped sequences between the PtCesA18pro sequence and the coding regions of AtVND7, AtSND1, or AtMYB46, and a primer containing the recognition site of a restriction enzyme. The PCR products were used as the templates for a second PCR to amplify the sequences of PtCesA18pro::AtVND7, PtCesA18pro::AtSND1, or PtCesA18pro::AtMYB46, using the primer sets comprising PtCesA18p-EV_F and VND7(−stop)_Avr_R, SND1(−stop)_Avr_R, or MYB46(−stop)_Avr_R. The resultant PCR products were cloned into the pGEM T-easy cloning vector (Promega, Madison, WI, United States), and the sequences of PtCesA18pro::AtVND7, PtCesA18pro::AtSND1, or PtCesA18pro::AtMYB46 were cut out with EcoRV (TaKaRa Bio, Kusatsu, Japan) and AvrII (TaKaRa Bio). The pSMAH-VP16-Thsp vector was treated with BamHI (TaKaRa Bio), followed by a treatment with Klenow Fragment (TaKaRa Bio) to generate the blunt ends. The resultant sample was subsequently treated with XbaI, and then used for the ligation with the PtCesA18pro::AtVND7, PtCesA18pro::AtSND1, or PtCesA18pro::AtMYB46 fragments, to obtain the plant expression vector for the expression of PtCesA18pro::AtVND7:VP16, PtCesA18pro::AtSND1:VP16, or PtCesA18pro::AtMYB46:VP16 (Figure 1E). The plasmid was electroporated into the Agrobacterium tumefaciens (GV3101::pMP90), and each individual clone was used for the transformation of hybrid aspen (Ohtani et al., 2011). The successful insertion of the transgene in each transgenic aspen was confirmed by PCR with the extracted genomic DNA samples using specific primer sets. The primer sequence information is indicated in Supplementary Table 1.
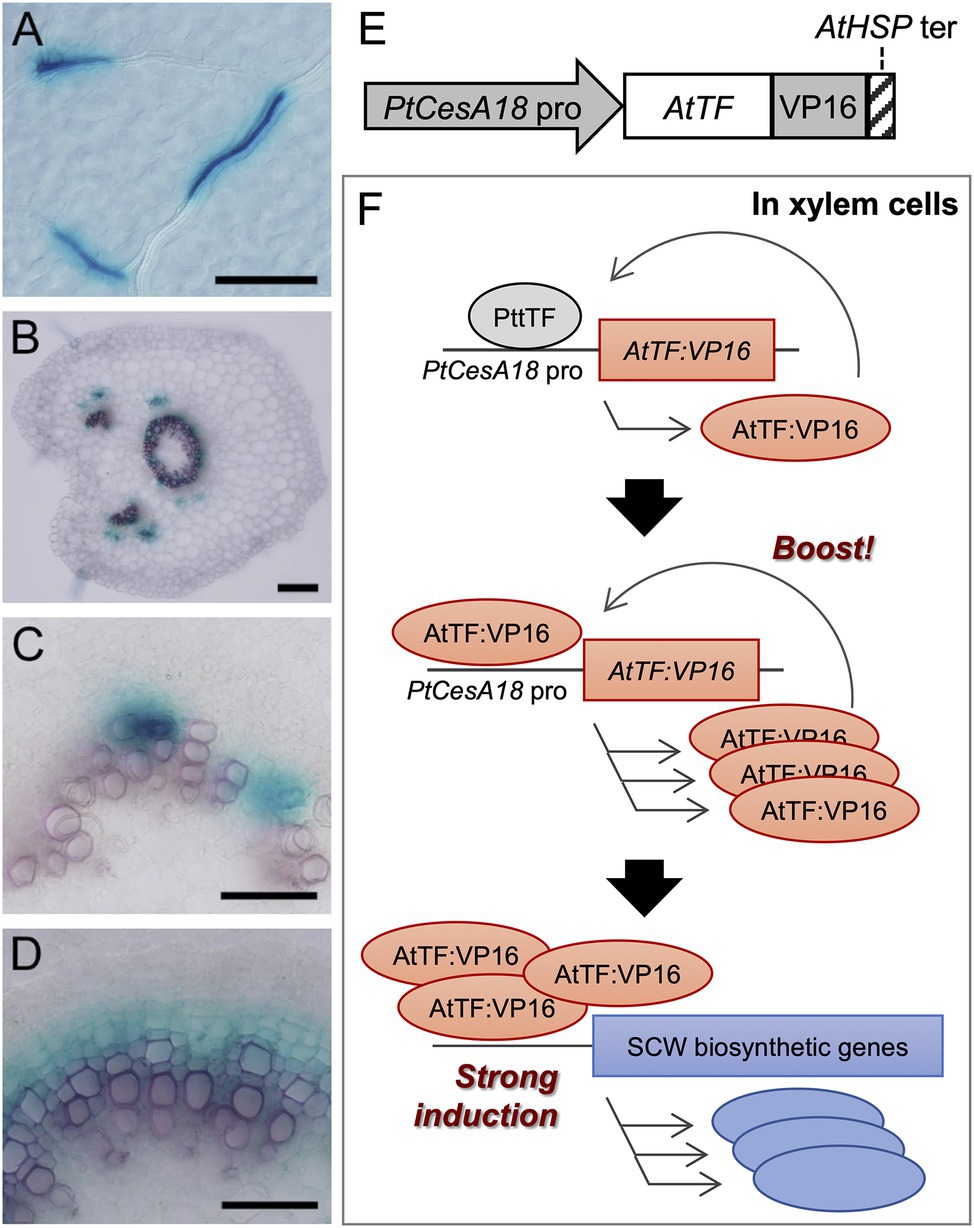
Figure 1. The concept of the self-reinforced system with secondary cell wall (SCW)-related transcription factors. (A–D) The promoter activity of the Populus trichocarpa CesA18 (PtCesA18) gene in the hybrid aspen (Populus tremula × tremuloides T89). The activity was determined using the PtCesA18pro::GUS reporter in the leaf (A), the petiole (B), the elongating third internode (C), and the elongating fifth internode (D). GUS signals were observed in the developing leaf veins (A), xylem and phloem fibers in the petiole (B), and developing xylem tissues (C,D). Bars = 100 µm in (A,B), 50 µm in (C,D). (E) Schematic diagram of the construct used for the self-reinforced system. In this system, a gene encoding an Arabidopsis thaliana transcription factor (TF) is designed to be expressed under the control of the PtCesA18 promoter, forming a fusion protein with VP16, a transcriptional activation domain derived from the human herpes virus. The transcription of transgene is terminated by the Arabidopsis heat shock protein terminator (AtHSP ter; Nagaya et al., 2010). (F) Mode of action of the self-reinforced system with SCW-related transcription factors. (Upper) In the xylem cells of hybrid aspen (Populus tremula × tremuloides), the PtCesA18 promoter is activated by an endogenous transcription factor (PtTF), resulting in the generation of AtTF:VP16, which is introduced by the construct described in (E). (Middle) The generated AtTF:VP16 can strongly activate the PtCesA18 promoter activity to boost the synthesis of AtTF:VP16 in a self-reinforcing manner. (Lower) The generated AtTF:VP16 fusion protein subsequently induces endogenous SCW-related genes. TF, transcription factor; At, Arabidopsis thaliana; Pt, Populus trichocarpa; and Ptt, Populus tremula × tremuloides.
Stem tissues of transgenic aspens were collected and frozen in liquid nitrogen, then ground with a mortar and pestle. A 600-μl aliquot of extraction buffer [2% (w/v) cetyltrimethylammonium bromide, 2.5% (w/v) polyvinylpolypyrrolidone 40, 2 M NaCl, 100 mM Tris–HCl (pH 8.0), 25 mM EDTA (pH 8.0), and 2% (v/v) 2-mercaptoethanol] was added to each 100–150-mg sample, and the mixture was homogenized by vortexing. After incubating the samples at 65°C for 10 min, an equal volume of chloroform:isoamylalcohol (24:1) was added to the mixture, followed by vigorous shaking. Subsequently, the samples were centrifuged at 11,000 g for 10 min at 4°C, after which the aqueous phase was transferred into a new tube. The extraction using chloroform:isoamylalcohol and the centrifugation were repeated two times in total. LiCl solution (to a final concentration of 3 M) was added to the collected aqueous parts, and the mixture was incubated overnight at 4°C. The samples were centrifuged at 15,000 g for 20 min at 4°C to precipitate the RNA fractions, and the precipitated RNA fractions were purified using RNeasy Mini Kit (QIAGEN, Venlo, The Netherlands) to obtain the total RNA samples. The RNA samples were kept at −80°C until required.
The total RNA samples from transgenic aspens were treated with RQ1 RNase-Free DNase (Promega) following the manufacturer’s instructions. A 2-μg aliquot of DNase-treated RNA was used to synthesize cDNA with the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Basel, Switzerland) using oligo dT(18) primers (Promega). The cDNA samples were kept at −20°C until required. The amounts of mRNAs were quantified using LightCycler 480 SYBR Green I Master (Roche), gene-specific primer sets (Supplementary Table 1), and LightCycler 480 Real-Time PCR System (Roche). The expression levels of the analyzed genes were normalized against the expression level of ELONGATION INITIATION FACTOR 4a (ELF4A; Ohtani et al., 2011; Hori et al., 2020; Akiyoshi et al., 2021).
The 10th elongated internodes of the stems were harvested and fixed in 0.05 M sodium phosphate buffer (pH 7.2) containing 1.25% (v/v) glutaraldehyde and 2% (w/v) paraformaldehyde, then fixed with 1% (w/v) osmium tetroxide. The fixed samples were washed with 8% (w/v) sucrose solution and dehydrated using an ethanol series [25, 60, 80, 99, and 100% (v/v)]. The ethanol was replaced with propylene oxide, and thereafter the propylene oxide was replaced with Spurr’s resin (Polysciences, Warrington, PA, United States). Subsequently, the samples were fully embedded in Spurr’s resin. Sections (80 µm thickness) were cut and post-stained with lead stain of Sato (1968) and observed using a transmission electron microscopy (TEM; HITACHI H-7100, HITACHI High-Tech, Tokyo, Japan). The area of the cell walls was calculated by subtracting the area of the cell region surrounded by the plasma membrane from the area of entire cell including the cell wall, using ImageJ.1 The statistical analysis was performed using Tukey–Kramer test (p < 0.05) in R.2
The method for nuclear magnetic resonance (NMR) footprint analysis was described by Hori et al. (2020) and Akiyoshi et al. (2021). The stem samples (about 10-cm in length) were collected from transgenic aspens grown in the greenhouse, freeze-dried, and debarked, then ground in an automill (TK-AM7; Tokken, Kashiwa, Japan) for 10 min. The samples were then further ground in a Pulverisette 5 ball mill (Fritsch, Idar-Oberstein, Germany) for 12 h. Next, 30 mg of powdered sample was dissolved in dimethyl sulfoxide (DMSO)-d6:pyridine-d5 (4:1) and heated at 50°C for 30 min with shaking at 14,000 rpm in a Thermomixer Comfort (Eppendorf, Hamburg, Germany). After centrifugation at 15,000 rpm for 5 min, the supernatant was transferred into NMR tubes for analysis. Same supernatant samples were used for Pyrolysis-GC/MS analysis (section “Pyrolysis-GC/MS Analysis”).
Nuclear magnetic resonance spectra were collected on an Avance II HD-700 instrument (Bruker, Billerica, MA, United States) with a 5-mm cryoTCI probe. For the 1H and 13C analyses, 700.130 and 176.06 MHz resonance frequencies were used, respectively. The temperature for all NMR samples was kept at 318 K. Chemical shifts were referenced to the methyl group of DMSO-d6 at 1H = 2.49 ppm and 13C = 39.5 ppm. The echo/antiecho gradient selections were used for the collection of 2D 1H-13C hetero-nuclear single quantum coherence (HSQC) spectra. A total of 51 regions of interest (ROI) were identified based on previously assigned chemical shifts (Komatsu and Kikuchi, 2013; Watanabe et al., 2014; Mori et al., 2015). The protocols described in Mansfield et al. (2012) and in Tsuji et al. (2015) were used to quantify HSQC. The peak intensity of pyridine-d5 used for the normalization. The statistical analysis was performed using Tukey–Kramer test (p < 0.05) in R.
The 8-μl of the cell wall extraction sample described in the section “Nuclear magnetic resonance footprinting analysis of transgenic aspen stem tissues” was dropped on pyrofoil F500 (500°C; Japan Analytical Industry Co., Japan). Pyrolysis GC–MS analysis was performed using a Curie Point Pyrolyzer JPS-900 (Curie Point Pyrolyzer, Automated Model, Japan Analytical Industry Co., Ltd., Japan) connected directly to a gas chromatograph system 7890B GC System (Agilent, United States). Pyrolysis was performed using the following conditions; 280°C oven temperature, 280°C needle temperature, and 500°C pyrolysis for 5 s. The pyrolysates were then transferred to the gas chromatograph (5977A MSD, Agilent, United States) equipped with Agilent J&W DB-1 MS column (60 m × 0.25 mm × 0.25 μm, Agilent, United States), and analyzed under the following conditions; carrier, helium; injector temperature, 280°C; split ratio, 100:1; mass range, 50–300; column temperature, heat up from 40 to 280°C at a rate of 4°C/min, and then keep 280°C for 5 min.
Pyrolysis GC–MS data were analyzed based on the methods described by Gerber et al. (2012), Pinto et al. (2012), and Gerber et al. (2016). Briefly, the raw pyrolysis GC–MS data were converted to the NetCDF format by the software Agilent Chemstation Data Analysis, and then the smoothing and alignment of chromatograms were performed (Gerber et al., 2012). After the background subtraction, the Multivariate Curve-Resolution by Alternate Regression (MCR-AR) analysis was performed, to deconvolute the chromatographic and mass spectral profiles. Based on the annotation information of each peak by Gerber et al. (2012), we computed the values for carbohydrate:lignin (C/L) ratio and lignin S/G ratio.
The powdered stem samples were treated with a mixture of cellulase from Trichoderma reesei ATCC 26921 (Sigma-Aldrich, Merck KgaA, Darmstadt, Germany) and cellobiase from Aspergillus niger (Sigma-Aldrich, Merck KgaA) for 24 h, according to the methods of Okubo-Kurihara et al. (2016) and Ohtani et al. (2017b). The supernatant was collected after centrifugation, after which 0.1 M NaOH solution was added to stop the reaction. The released glucose and xylose were measured with a “Glucose CII-Test” (FUJIFILM Wako Pure Chemical Corporation, Osak, Japan) and a “D-Xylose Assay Kit” (Megazyme, Bray, Ireland), respectively.
First, we examined the promoter activity of P. trichocarpa CesA18 (PtCesA18), by generating the transgenic PtCesA18pro:GUSline of hybrid aspen (P. tremula × tremuloides T89; Figures 1A–D). GUS signals were detected in developing xylem vessel cells in the leaf veins (Figure 1A), developing xylem and phloem fiber cells in the petiole (Figure 1B), developing vessel cells in the primary xylem of young internodes (Figure 1C), and developing xylem and fiber cells in the secondary xylem of the older internodes (Figure 1D). These observations indicate that PtCesA18 promoter activity is tightly associated with SCW formation, as expected; thus, we decided to use the PtCesA18 promoter for the self-reinforced system of SCW-related transcription factors (Figures 1E,F).
In our self-reinforced system, the transcription factors associated with SCW formation are expressed under the control of the PtCesA18 promoter, as a fusion protein with the human herpes virus–derived transcriptional activation domain VP16 (Figure 1E). The transcription factor genes used in the present work were AtSND1, AtVND7, and AtMYB46, which are master regulators of SCW formation in A. thaliana (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007; Ko et al., 2009, 2014; Nakano et al., 2015; Ohtani and Demura, 2019; Kamon and Ohtani, 2021). The key feature of this self-reinforced system is that the PtCesA18 promoter can be upregulated by AtSND1, AtVND7, and AtMYB46, since these transcription factors can recognize and bind their cis-regulatory elements, which are highly conserved among each ortholog (Zhong et al., 2010a; Ohtani et al., 2011; Kim et al., 2012; Zhong and Ye, 2012). First, in the xylem cells, the PtCesA18 promoter activity is upregulated by endogenous VNS and MYB functions, resulting in the induction of AtSND1:VP16, AtVND7:VP16, or AtMYB46:VP16 expression. Subsequently, these induced AtSND1:VP16, AtVND7:VP16, or AtMYB46:VP16 proteins can bind and upregulate the PtCesA18 promoter to generate large amounts of AtSND1:VP16, AtVND7:VP16, and AtMYB46:VP16 mRNAs. We can expect that our constructs will result in the high production of AtSND1:VP16, AtVND7:VP16, and AtMYB46:VP16 proteins, facilitating the strong induction of the SCW biosynthetic genes, as the combined effects of PtCesA18 promoter and transcriptional activation domain VP16 (Figure 1F).
We introduced the chimeric genes PtCesA18pro::AtSND1:VP16, PtCesA18pro::AtVND7:VP16, and PtCesA18pro::AtMYB46:VP16, or the vector control PtCesA18pro::GUS, into the hybrid aspen P. tremula × tremuloides line T89 (Figures 2A–D, Supplementary Figure 1). As the results, 13, 11, 16, and 14 independent lines of PtCesA18pro::AtSND1:VP16, PtCesA18pro::AtVND7:VP16, PtCesA18pro::AtMYB46:VP16, and PtCesA18pro::GUS, respectively, were established. All PtCesA18pro::AtMYB46:VP16 or PtCesA18pro::AtSND1:VP16 were indistinguishable from the non-transgenic aspen and control transgenic line PtCesA18pro::GUS in terms of their size and morphology when grown in the sterile half-strength MS medium (Figures 2A–D); however, all transgenic PtCesA18pro::AtVND7:VP16 plants showed severe growth defects (Supplementary Figure 1). The stem sections of the PtCesA18pro::AtVND7:VP16 lines indicated no clear secondary growth in the xylem and collapsed primary xylem cells (Supplementary Figure 1). It is possible that the decrease of water transport ability and mechanical strength of the stem tissues by the reduction in xylem cells would lead to growth defects in the PtCesA18pro::AtVND7:VP16 plants. There is a clear difference in transcriptional activation activity between AtVND7 and AtSND1; AtVND7 can strongly induce programmed cell death (PCD)-related genes as a master regulator of xylem vessel cell differentiation, which include PCD process, while AtSND1 possesses a relatively low ability to do so since AtSND1 functions in fiber cell differentiation (Kubo et al., 2005; Ohashi-Ito et al., 2010; Zhong et al., 2010b; Yamaguchi et al., 2011) Thus, the results of the PtCesA18pro::AtVND7:VP16 lines might reflect this strong activity of AtVND7 to induce PCD.
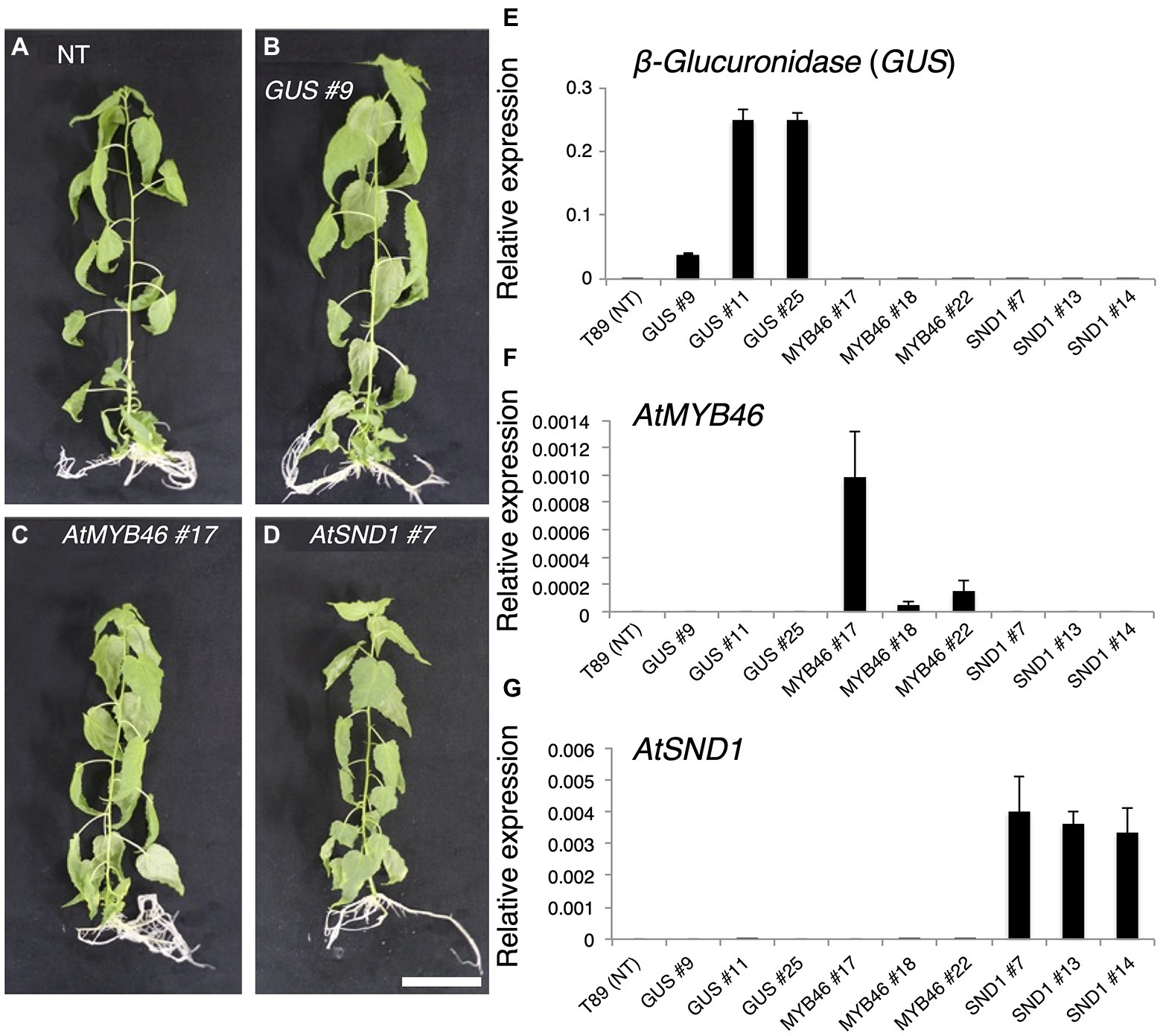
Figure 2. Generation of transgenic aspens carrying PtCesA18pro::AtMYB46:VP16, PtCesA18pro::AtSND1:VP16, or PtCesA18pro::AtVND7:VP16. (A–D) Transgenic aspens grown in a sterile half-strength Murashige and Skoog growth medium. Typical views of the non-transgenic (NT) plant (T89; A) and plants carrying the PtCesA18pro::GUS (GUS) line 9 (B), PtCesA18pro::AtMYB46:VP16 (AtMYB46) line 17 (C), or PtCesA18pro::AtSND1:VP16 (AtSND1) line 7 (D). Bar = 5 cm. (E–G) Expression levels of the introduced genes, β-Glucuronidase (GUS; E), AtMYB46 (F), and AtSND1 (G) in the transgenic lines, as quantified using Reverse Transcription-Quantitative PCR (RT-qPCR) analysis. The data are displayed as means ± SD (n = 3).
Based on the expression levels of the transgenes (Figures 2E–G), we selected three lines of each transgenic plant for further detailed analysis. First, the expression levels of Populus tremula × tremuloides XYLEM CYSTEINE PROTEASE 1 (PttXCP1), encoding a cysteine protease involved in PCD (Avci et al., 2008); PttMYB003, encoding a poplar AtMYB46 homolog (Ohtani et al., 2011; Zhong et al., 2013); PttCesA18, encoding a xylem-specific cellulose synthase (Wu et al., 2000; Suzuki et al., 2006); PttGT47A, encoding an enzyme for hemicellulose biosynthesis; and CONIFERALDEHYDE 5-HYDROXYLASE (PttCAld5H), encoding an enzyme for monolignol biosynthesis (Hori et al., 2020) were examined using Reverse Transcription-Quantitative PCR (RT-qPCR) with the stem-derived total RNAs (Figure 3). PttXCP1 and PtMYB0003 were upregulated only in the PtCesA18pro::AtSND1:VP16 plants (Figure 3), while the other SCW biosynthesis genes were upregulated in both the PtCesA18pro::AtMYB46:VP16 and PtCesA18pro::AtSND1:VP16 lines, relative to their expression in the non-transgenic plants and the vector control PtCesA18pro::GUS plants (Figure 3). This is consistent with the known transcriptional regulatory hierarchy, in which the VNS proteins, including AtSND1, can induce the expression of the secondary master switch MYB genes such as AtMYB46 and AtMYB83, as well as the PCD-related and SCW-related genes, while the secondary master switch MYB proteins mainly upregulate SCW-related genes (Zhong et al., 2010a; Schuetz et al., 2013; Nakano et al., 2015; Ohtani and Demura, 2019; Kamon and Ohtani, 2021). We could not confirm the increased abundance of the proteins of AtVND7:VP16, AtSND1:VP16, nor AtMYB46:VP16 proteins, due to the absence of specific antibodies for these transcription factors. However, based on the increased expression of the downstream genes of AtSND1 and AtMYB46 in transgenic plants (Figure 3), we concluded that our system could successfully function as expected (Figure 1F).
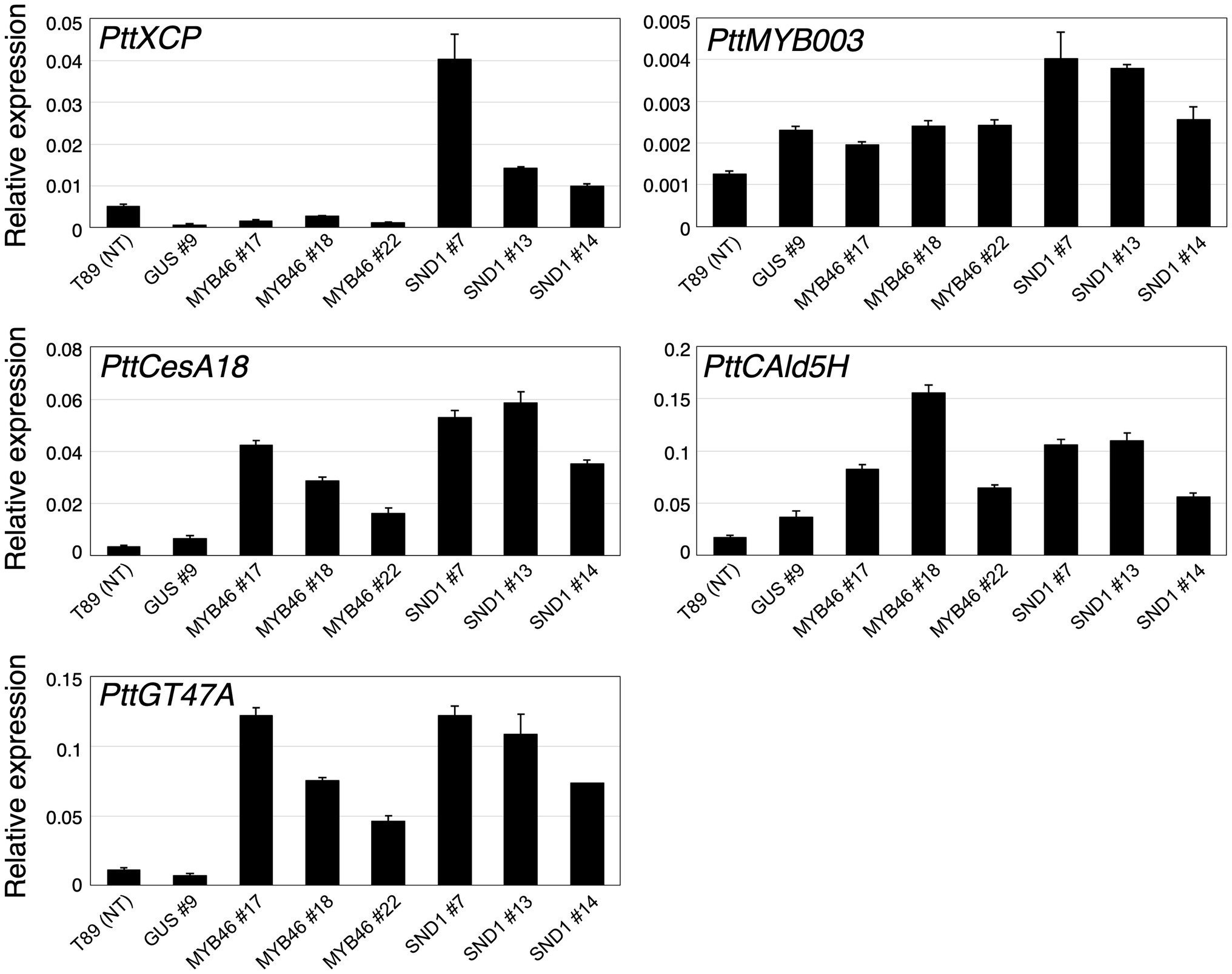
Figure 3. Expression levels of xylem-related genes in transgenic aspens. The PttXCP1, PttMYB003, PttCesA18, PttCAld5H, and PttGT47A mRNAs were quantified in non-transgenic (NT) aspen (T89), the PtCesA18pro::GUS (GUS) lines, the PtCesA18pro::AtMYB46:VP16 (MYB46) lines, and the PtCesA18pro::AtSND1:VP16 (SND1) lines using RT-qPCR. The expression levels of the analyzed genes were normalized against the expression level of ELFA. The data are displayed as means ± SD (n = 3).
Next, we examined the cell arrangement and SCW deposition in the secondary xylem of the PtCesA18pro::AtMYB46:VP16 and PtCesA18pro::AtSND1:VP16 transgenic plants (Figure 4). Toluidine blue staining showed that the size and cell arrangement of the xylem tissues of PtCesA18pro::AtMYB46:VP16 lines 18 and 22 and PtCesA18pro::AtSND1:VP16 line 13 were not significantly different from that of the control PtCesA18pro::GUS plants, whereas the width of the xylem was smaller in PtCesA18pro::AtMYB46:VP16 line 17, which had a high AtMYB46 expression level (Figure 2F), and in PtCesA18pro::AtSND1:VP16 lines 7 and 14 (Figure 4A). In the xylems of PtCesA18pro::AtMYB46:VP16 line 17 and PtCesA18pro::AtSND1:VP16 lines 7 and 14, bright blue–stained cells were observed in the inner part of the secondary xylem and phloem fiber cells (Figure 4A). The bright blue signal by toluidine blue staining indicates lignin deposition (Pradhan Mitra and Loqué, 2014), thus we next used phloroglucinol, which stains lignin deposits red, to make clear lignin deposition additionally. The results showed strong red signals in the phloem fibers of PtCesA18pro::AtMYB46:VP16 line 17 and PtCesA18pro::AtSND1:VP16 lines 7 and 14 (Figure 4B). Lacking of bright blue toluidine blue signals and red phloroglucinol signals in PtCesA18pro::AtSND1:VP16 line 13 indicated that no changes in lignification in the line 13. In addition, intense red signals and bright pink to white signals were observed in the xylem cells of PtCesA18pro::AtMYB46:VP16 line 17 and PtCesA18pro::AtSND1:VP16 lines 7 and 14, respectively, in comparison with the control plants (Figure 4B). The different coloration of phloroglucinol signals has been shown to reflect the different thickness of SCW and/or lignin S/G ratio (Pradhan Mitra and Loqué, 2014; Blaschek et al., 2020), and the bright pink to white phloroglucinol signals are observed when the SCW are thick or S-lignin rich SCWs are present (Blaschek et al., 2020). Thus, these observations suggested that SCW deposition, including lignification, was enhanced in these three lines, and that the acceleration of SCW deposition occurs simultaneously with a decrease in the size of the xylem regions.
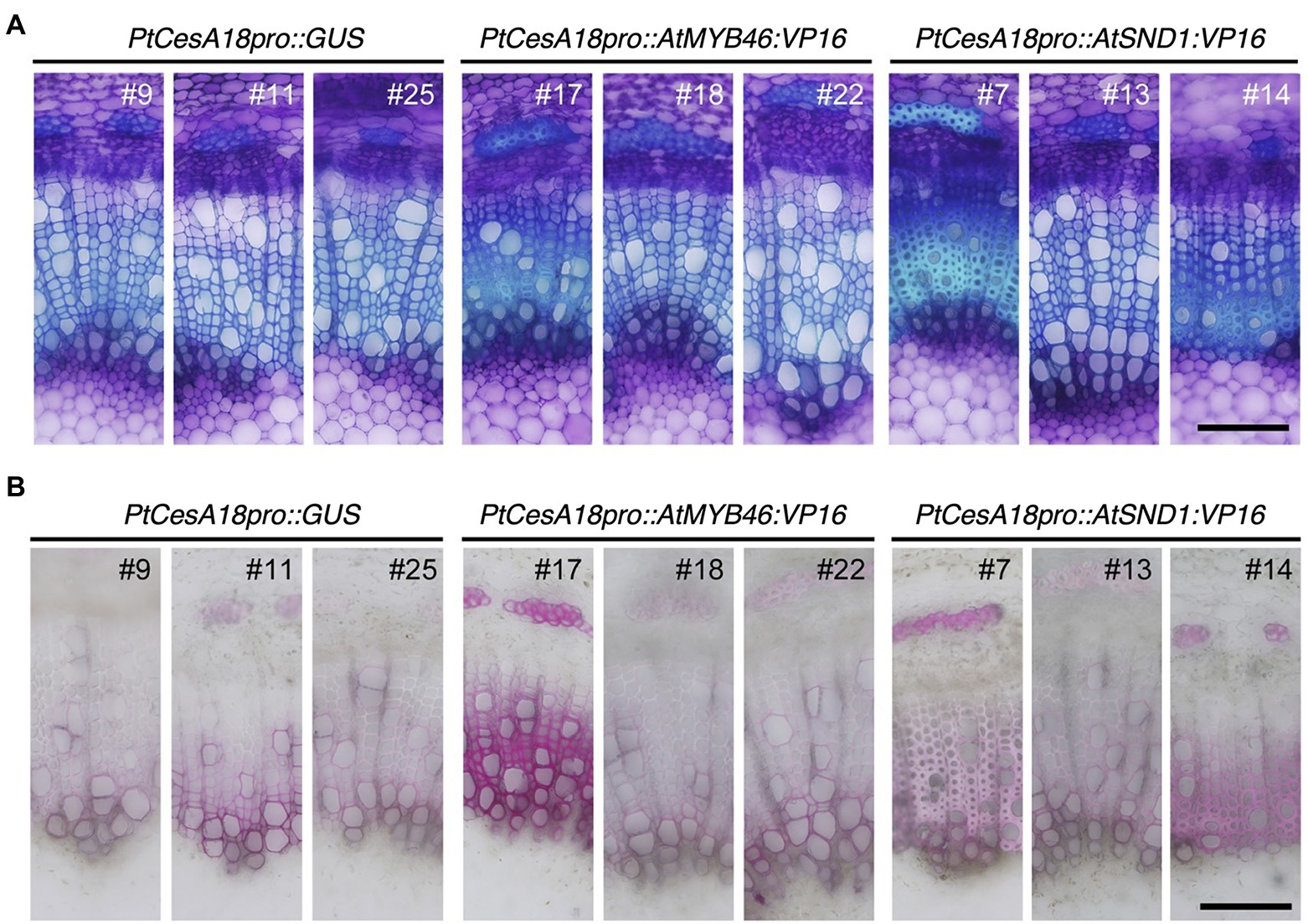
Figure 4. Histological observations of xylem tissues in transgenic aspens. (A) Stem sections of PtCesA18pro::GUS, PtCesA18pro::AtMYB46:VP16, and PtCesA18pro::AtSND1:VP16 stained with toluidine blue. (B) Stem sections of PtCesA18pro::GUS, PtCesA18pro::AtMYB46:VP16, and PtCesA18pro::AtSND1:VP16 stained with phloroglucinol-HCl for the detection of lignin. Bars = 100 μm.
To examine the cell wall thickness of the secondary xylem cells, a TEM analysis was performed for both the xylem vessel cells (Figures 5A–C) and the xylem fiber cells (Figures 5D–F). In the PtCesA18pro::AtMYB46:VP16 plants, the cell wall thickness of both the vessel and fiber cells was only statistically increased in line 7 compared with the controls (Figures 5G,H). By contrast, all three lines of PtCesA18pro::AtSND1:VP16 showed a significant increase in the cell wall thickness of both the xylem vessel and fiber cells relative to the control plants (Figures 5G,H), despite the lack of clear changes of fiber cells in the line 13 by the histological observation (Figure 4). PtCesA18pro::AtSND1:VP16 line 13 has a larger variation in cell wall thickness of fiber cells than the other two lines, line 7 and 14 (Figure 5H), thus the effects of cell wall thickness could be unstable in the line 13. Notably, the effect of the high transgenic AtSND1 expression on cell wall thickness was greater than that of AtMYB46 transgenic expression (Figures 5G,H), and in the case of the PtCesA18pro::AtMYB46:VP16 plants, the increase in cell wall thickness was detected only in line 17, which had the highest AtMYB46 expression level (Figure 2F). Such differences in overexpression effects might reflect the difference of molecular function between AtSND1 and AtMYB46; AtSND1 activates the entire molecular program of fiber cell differentiation, including not only SCW biosynthesis enzymes but also the other regulatory factors, such as secretion machinery (Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2010b; Ohashi-Ito et al., 2010), while the regulatory targets of AtMYB46 are mainly SCW-related enzymatic genes (Zhong et al., 2007; Ko et al., 2009; McCarthy et al., 2009).
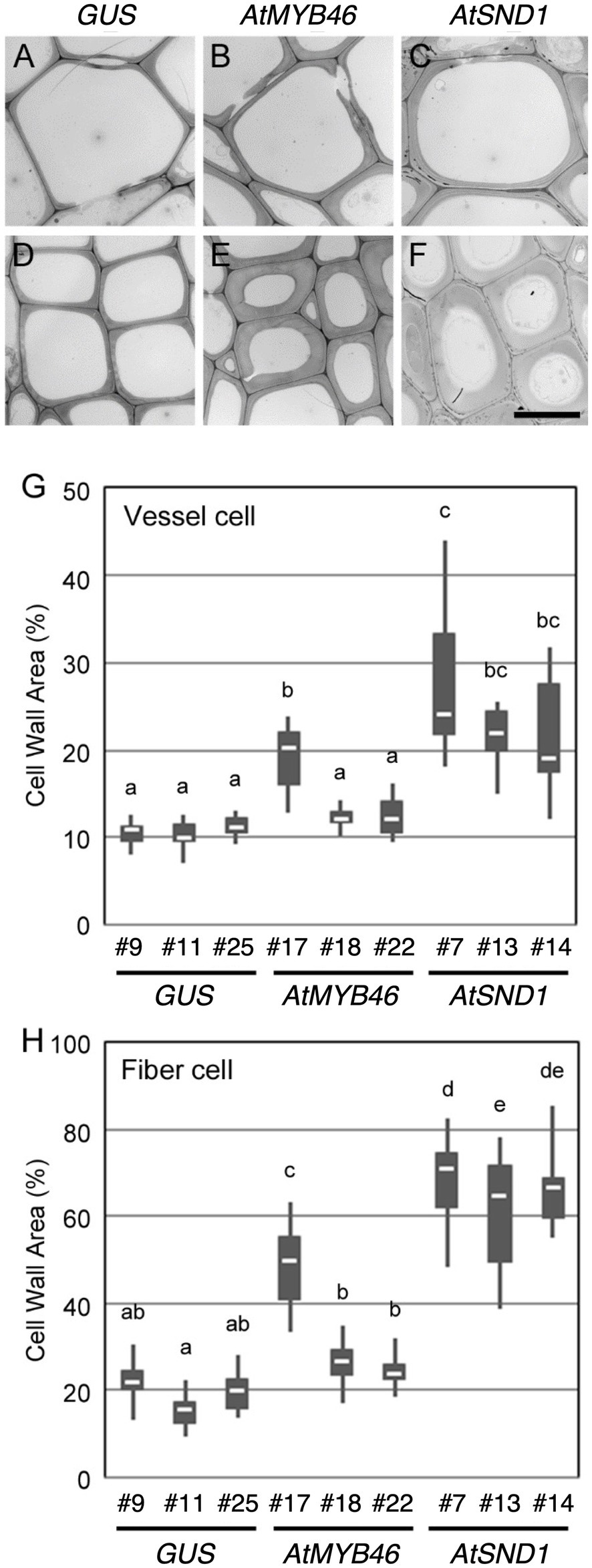
Figure 5. Cell wall thickness in the transgenic aspens. (A–F) Typical transmission electron microscope (TEM) images of xylem vessel cells (A–C) and xylem fibers cells (D–F) in the 10th elongating internodes of PtCesA18pro::GUS line 9 (A,D), PtCesA18pro::AtMYB46:VP16 line 17 (B,E), and PtCesA18pro::AtSND1:VP16 line 7 (C,F). Bar = 10 μm. (G,H) Box plots of the cell wall area of the xylem vessel cells (G) and xylem fiber cells (H). Cell wall area is shown as a ratio of the entire cell area. Different letters indicate statistically significant differences (Tukey–Kramer test: p < 0.05; n = 7–12 for xylem vessel cells, n = 14–29 for xylem fiber cells).
The transgenic aspens grown in the sterile MS growth medium showed enhanced SCW deposition in their secondary xylem (Figures 4, 5). To further assess the wood structure and wood cell wall properties, we tried to grow the transgenic plants in pots of soil in the greenhouse (Figure 6). Five plants of each transgenic line (PtCesA18pro::AtMYB46:VP16 and PtCesA18pro::AtSND1:VP16) were prepared and transferred into the soil. Unfortunately, no PtCesA18pro::AtSND1:VP16 plants were able to survive being transplanted into the pots of soil. It was previously reported that the transgenic aspen expressing OsSWN1:VP16, a chimeric gene comprising a rice homolog of AtSND1 and VP16, under the control of the AtSND1 promoter showed no growth defects in the pots of soil (Sakamoto et al., 2016); thus, the lack of viability of our self-reinforced system with PtCesA18pro::AtSND1:VP16 plants expressing AtSND1 is likely caused by some factors that would be affected only in our self-reinforced system, but not in the traditional overexpression. Previously, we observed that cell wall modification actively occurred during the stress response in poplar (Chen and Polle, 2010; Janz et al., 2012; Polle et al., 2019; Hori et al., 2020), and the expression levels of the VNS genes, especially the poplar SND1 homologs, were decreased in response to stresses in poplar (Hori et al., 2020), suggesting that the regulation of VNS activity could be crucial for the plant stress response. Based on this idea, we hypothesize that the PtCesA18pro::AtSND1:VP16 plants cannot perform the proper acclimatization processes required for the transfer from humid aseptic growth medium containers to relatively dry soils in the greenhouse.

Figure 6. Growth and morphology of the PtCesA18pro::AtMYB46:VP16 plants grown in pots of soil in the greenhouse. Phenotypes of the transgenic plants PtCesA18pro::GUS (vector control; A–D) and PtCesA18pro::AtMYB46:VP16 (E–I) 3 months after being transferred from the sterile half-strength Murashige and Skoog growth medium to pots of soil. Scale = 1 m.
In contrast, the PtCesA18pro::AtMYB46:VP16 plants and the vector control plants continuously grew in the greenhouse pots of soil, reaching ~1.5 m in height during the 3-month culture (Figure 6). The height and morphology of the PtCesA18pro::AtMYB46:VP16 plants were comparable to the vector control plants, suggesting no significant growth effect of the high transgenic AtMYB46 expression in our self-reinforced system. These results indicated that the current version of the self-reinforced system with the VNS genes is not suitable for directly obtaining plant biomass for practical use, but that the self-reinforced system using the AtMYB46 gene has potential for further practical use.
Next, the cell walls of the PtCesA18pro::AtMYB46:VP16 stems (line 17, 18, and 22) were assessed by NMR footprinting analysis (Kim and Ralph, 2010; Mansfield et al., 2012; Hori et al., 2020; Akiyoshi et al., 2021). The cell wall fractions were extracted from the stem samples of transgenic plants using a DMSO-d6/pyridine-d5 solution, then subjected to the HSQC NMR analysis, which has proven to be an effective method to examine the polysaccharides and lignin components in unfractionated plant cell wall materials (Kim and Ralph, 2010; Mansfield et al., 2012; Tobimatsu et al., 2013; Kim et al., 2017; Zhang et al., 2019; Figure 7A). We performed the principal component analysis (PCA) of the signal intensities of the NMR peaks. The PCA plot demonstrated that the cell wall properties of PtCesA18pro::AtMYB46:VP16 woods could be differentiated from that of the vector control plants (Figure 7B); the PtCesA18pro::AtMYB46:VP16 samples were generally located in the regions where PC2 takes a negative value (see pink circle in Figure 7B). The samples of PtCesA18pro::AtMYB46:VP16 line 17 in particular were clustered in the region with positive PC1 values and negative PC2 values (see red circle in Figure 7B), suggesting relatively large changes in cell wall composition in this line.
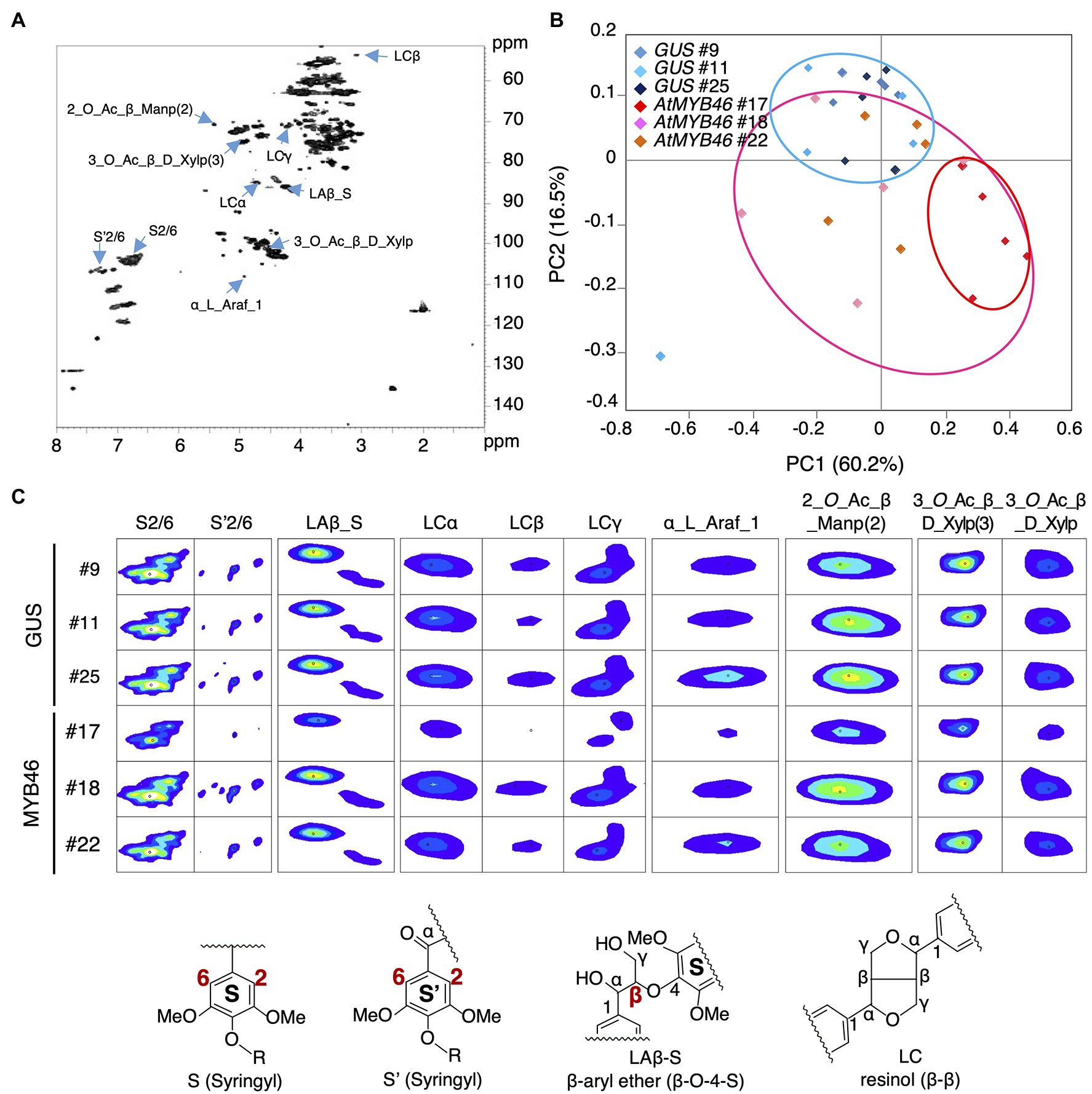
Figure 7. Nuclear magnetic resonance (NMR) footprint analysis of stem samples of the transgenic aspens. (A) Typical hetero-nuclear single quantum coherence (HSQC) image of the stem samples. The peaks whose signals were decreased in PtCesA18pro::AtMYB46:VP16 line 17 are indicated with right blue arrows. (B) Principal component analysis (PCA) plots of the PtCesA18pro::GUS (vector control) and PtCesA18pro::AtMYB46:VP16 stem samples. Blue, pink, and red ovals indicate the distribution ranges of the PtCesA18pro::GUS samples, all PtCesA18pro::AtMYB46:VP16 samples, and the samples specifically from PtCesA18pro::AtMYB46:VP16 line 17, respectively. (C) NMR signal peaks that were significantly decreased in PtCesA18pro::AtMYB46:VP16 line 17. Representative structures for lignin-related NMR peaks are shown at the bottom. For each line, five individuals were examined as biological replicates.
We further examined the peaks that contributed to the PC1 score in Figure 7B, and found that such peaks contained many lignin-related NMR peaks (Figures 7A,C). In PtCesA18pro::AtMYB46:VP16 line 17, the signal intensities of the NMR peaks corresponding to syringyl (S) lignin (S2/6 and S’2/6), β-aryl ether (Laβ_S), and resinol (LCα, LCβ, and LCγ) were significantly reduced (Figure 7C), suggesting that the S-lignin monomer and related structures would be decreased in line 17. Indeed, the S/G ratio of lignin was about 1.0 in this line (Figure 8A), which is lower than the typical S/G ratio of poplar wood (1.8–2.3; Davison et al., 2006). In Arabidopsis mature stems, the S/G ratio is around 0.4 (Sibout et al., 2005); thus, the high transgenic expression of Arabidopsis MYB46 might influence the lignin biosynthesis process in the transgenic hybrid aspen, enabling the biosynthesis of lignin with intermediate characters between poplar and Arabidopsis. In addition to lignin-related NMR peaks, sugar-related peaks also showed decreased signal intensities in the PtCesA18pro::AtMYB46:VP16 line 17 (Figure 7C). The intensity of the signal related to arabinose (α_L_Araf_1), which is largely contained in the primary cell wall, was decreased, possibly due to the enhancement of SCW formation in line 17 (Figure 5). Interestingly, the signals related to acetylated hemicellulosic components, such as acetylated mannan [2-O-Ac-β-D-Manp (2)] and acetylated xylan [3-O-Ac-β-D-Xylp and 3-O-Ac-β-D-Xylp (3)], were also reduced in line 17 (Figure 7C); therefore, in line 17, the inhibition of hemicellulose acetylation and/or the deacetylation of hemicellulose is enhanced.
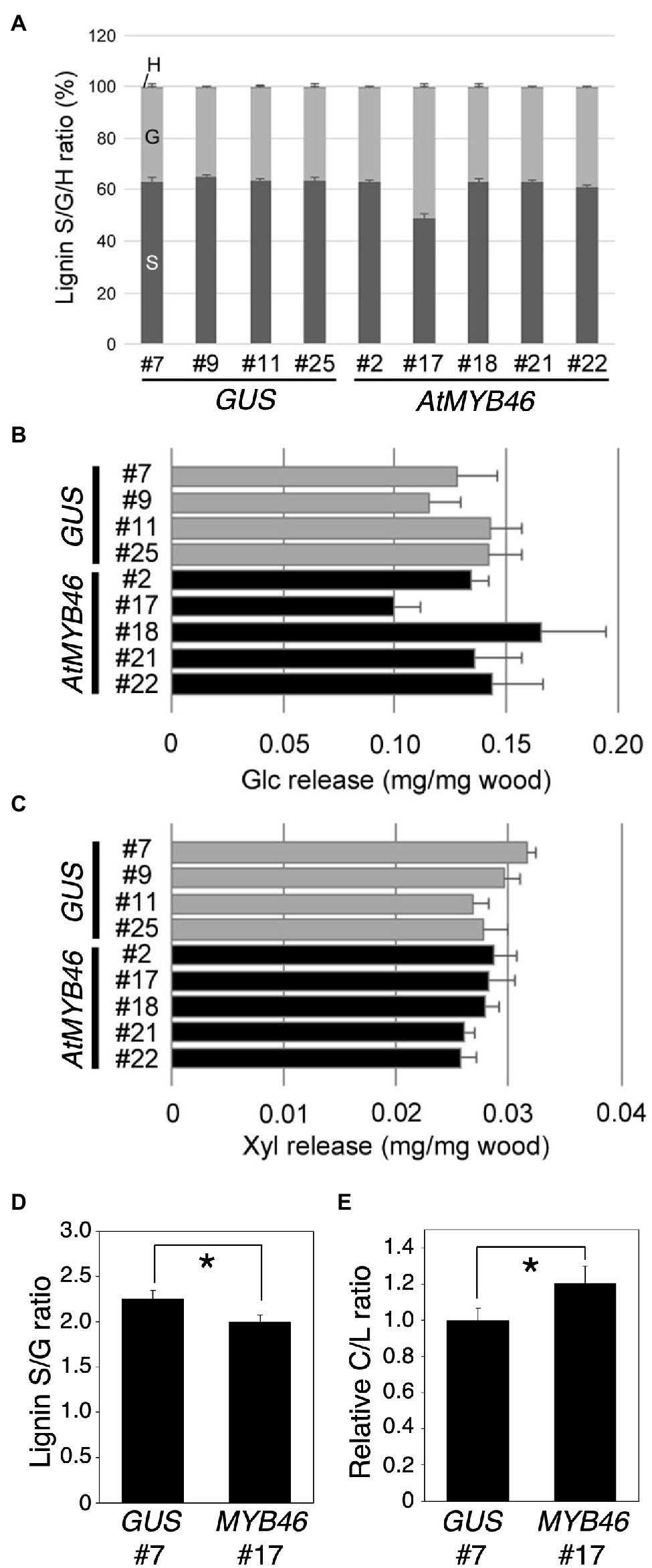
Figure 8. Lignin S/G ratio and enzymatic saccharification efficiency of transgenic aspens. (A) Lignin S/G ratio of the PtCesA18pro::GUS (vector control) and PtCesA18pro::AtMYB46:VP16 stem samples. The data are shown as means ± SD (n = 5). (B,C) Sugar release from the PtCesA18pro::GUS (vector control) and PtCesA18pro::AtMYB46:VP16 stem samples after enzymatic treatment. The amounts of glucose (Glc; B) and xylose (Xyl; C) obtained from the treated wood samples are shown (means ± SD; n = 5). (D,E) Pyrolysis-GC/MS analysis of PtCesA18pro::GUS line 7 and PtCesA18pro::AtMYB46:VP16 line 17. The lignin S/G ratio (D) and carbohydrate:lignin (C/L) ratio (E) were shown (n = 3). For the C/L ratio, the average value of PtCesA18pro::GUS line 7 data was set as 1.0, and all results were shown as relative values. Asterisk indicates statistically significant difference (Student’s t test, p < 0.05).
The lignin S/G ratio and degree of hemicellulose acetylation are critical factors for the regulation of SCW properties and the enzymatic saccharification of wood biomass (Chen and Dixon, 2007; Studer et al., 2011; Pawar et al., 2013, 2017), and the NMR data showed a decreased S/G ratio in PtCesA18pro::AtMYB46:VP16 line 17 (Figure 8A). Thus, the enzymatic saccharification efficiency of PtCesA18pro::AtMYB46:VP16 was examined (Figures 8B,C). No statistically significant differences in the release of glucose or xylose were detected between PtCesA18pro::AtMYB46:VP16 and the vector control PtCesA18pro::GUS plants (Figures 8B,C). Since the SCW accumulation was enhanced in PtCesA18pro::AtMYB46:VP16 line 17 (Figure 5), this finding of saccharification efficiency seemed to be a discrepancy; however, in the PtCesA18pro::AtMYB46:VP16 line 17, lignin deposition may have been increased (Figure 4B) and the S/G ratio was decreased (Figure 8A). High amounts of lignin with a low S/G ratio can decrease the saccharification efficiency (Studer et al., 2011); therefore, it is possible that the accumulation of SCW material in PtCesA18pro::AtMYB46:VP16 line 17 increased its recalcitrance for sugar release, offsetting the effects of enhanced SCW accumulation to result in no difference in enzymatic saccharification efficiency (Figure 8C). To make clear this point, we performed the pyrolysis-GC/MS analysis of cell wall extracts (Figures 8D,E). The pyrolysis-GC/MS analysis confirmed the lower S/G ratio in the PtCesA18pro::AtMYB46:VP16 line 17 (Figure 8D). It was also indicated that the carbohydrate portion to lignin was increased in the PtCesA18pro::AtMYB46:VP16 line 17 (Figure 8D), suggesting that the MYB46 overexpression would not increase the lignin contents, but possibly change the balancing between carbohydrates and lignin in the enhanced SCW. Taken together, our data demonstrated the basic effectiveness of our SCW-related transcription factor–based self-reinforced system for enhanced SCW accumulation (Figures 4, 5) and the modification of SCW properties (Figures 7, 8). However, the current system could not increase sugar yields through enzymatic saccharification (Figures 8B,C), suggesting that we must further improve the design of this self-reinforced system.
In this work, we tested our idea of a SCW-related transcription factor–based self-reinforced system in the useful woody plant Populus. The results indicated that this system can work to enhance SCW accumulation in hybrid aspen without visible difference of plant growth, clearly indicating the effectiveness of this system for increasing biomass production per space; however, we also recognized that the selection of transcription factors is critical for this system. The strong boosting of VNS protein activity is not suitable for the self-reinforced system, since the expression of VNS genes would be tightly regulated to generate proper xylem cells in response to environmental conditions. Indeed, our results demonstrated that the transgenic aspens carrying PtCesA18pro::AtVND7:VP16 or PtCesA18pro::AtSND1:VP16 cannot survive well. By contrast, PtCesA18pro::AtMYB46:VP16 successfully functioned to enhance SCW accumulation in the xylem cells of the transgenic aspens, as expected (Figures 4, 5). Enhanced SCW accumulation was observed only in PtCesA18pro::AtMYB46:VP16 line 17, which showed the highest expression of the transgene AtMYB46 (Figure 2F), and even in this line, the yields of glucose and xylose following enzymatic saccharification were no different to the control lines (Figures 8B,C). These results suggest that there is still much room for improvement in the selection of transcription factors for this self-reinforced system.
Interestingly, the RT-qPCR results of the PtCesA18pro::AtSND1:VP16 and PtCesA18pro::AtMYB46:VP16 expression levels showed that the degree of enhancement of SCW deposition did not correlate well with the expression levels of the tested SCW-related genes, which are all well-known SCW-related factors working downstream of the VNS module (Figures 3–5). Considering that the heterologous VNS genes induce the accumulation and/or modification of the SCW more effectively than endogenous VNS genes (Ohtani et al., 2011; Sakamoto et al., 2016; Akiyoshi et al., 2021), genes unexpectedly related to SCW accumulation could be affected in PtCesA18pro::AtMYB46:VP16 line 17. These differences in target genes may be related to the differences in the cis-sequence binding properties of the VNS proteins from each plant species, as well as the differences in their interactivity with other proteins. Thus, we have to consider the endogenous molecular mechanisms of SCW formation in hybrid aspen more carefully, and identify the critical points for further engineering of woody biomass. To improve the design of the SCW-related transcription factor–based self-reinforced system for the further enhancement of woody biomass utilization, we should further analyze the characteristics of the VNS and MYB proteins as transcription factors, especially focusing on their functionality in target plant species for the engineering.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
YN, MO, and TD designed the experiments. YN, AI, TM, and MO grew, collected, and sampled the material. YN, HE, CH, and MO performed the microscope and TEM observations, gene expression analysis, and saccharification test. LG and MS performed pyrolysis-GC-MS analysis. TM and JK designed and performed the NMR analyses. YN and MO wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was in part supported by the RIKEN Center for Sustainable Sciences, MEXT KAKENHI (JP18H05484 and JP18H05489 to MO and TD, and JP20H05405 and JP21H05652 to MO, Grants-in-Aid from the NC-CARP project to YN, HE, and TD), JSPS KAKENHI (JP20H03271 to MO and JP18H02466 to TD), and ERATO JST (JPMJER1602 to MO).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Masatoshi Yamaguchi, Arata Yoneda, Ko Kato, Minoru Kubo, Seiji Takayama, Masaaki Umeda, and Akiho Yokota (NAIST, Japan) for their fruitful discussions. We are also grateful to Nobuko Shizawa, Shizuka Nishida, Eriko Tanaka, Yuki Mitsubayasi, Hanae Sugita, Aiko Morita, and Hitomi Ichikawa (NAIST, Japan); Arika Takebayashi, Ryoko Hiroyama, Kayo Kitaura, and Akiko Sato (RIKEN, Japan); and Megumi Takahashi (University of Tokyo, Japan) for their technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.819360/full#supplementary-material
Akiyoshi, N., Ihara, A., Matsumoto, T., Takebayashi, A., Hiroyama, R., Kikuchi, J., et al. (2021). Functional analysis of poplar SOMBRERO-type NAC transcription factors yields a strategy to modify woody cell wall properties. Plant Cell Physiol. 62, 1963–1974. doi: 10.1093/pcp/pcab102
Akiyoshi, N., Nakano, Y., Sano, R., Kunigita, Y., Ohtani, M., and Demura, T. (2020). Involvement of VNS NAC-domain transcription factors in tracheid formation in Pinus taeda. Tree Physiol. 40, 704–716. doi: 10.1093/treephys/tpz106
Avci, U., Earl, P. H., Ismail, I. O., Beers, E. P., and Haigler, C. H. (2008). Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 56, 303–315. doi: 10.1111/j.1365-313X.2008.03592.x
Blaschek, L., Champagne, A., Dimotakis, C., Nuoendagula, D. R., Hishiyama, S., Kratzer, S., et al. (2020). Cellular and genetic regulation of coniferaldehyde incorporation in lignin of herbaceous and woody plants by quantitative Wiesner staining. Front. Plant Sci. 11:109. doi: 10.3389/fpls.2020.00109
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
Chen, S., and Polle, A. (2010). Salinity tolerance of Populus. Plant Biol. 12, 317–333. doi: 10.1111/j.1438-8677.2009.00301.x
Davison, B. H., Drescher, S. R., Tuskan, G. A., Davis, M. F., and Nghiem, N. P. (2006). Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl. Biochem. Biotechnol. 130, 427–435. doi: 10.1385/ABAB:130:1:427
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATEDNAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Gerber, L., Eliasson, M., Trygg, J., Moritz, T., and Sundberg, B. (2012). Multivariate curve resolution provides a high-throughput data processing pipeline for pyrolysis-gas chromatography/mass spectrometry. J. Anal. Appl. Pyrolysis 95, 95–100. doi: 10.1016/j.jaap.2012.01.011
Gerber, L., Öhman, D., Kumar, M., Ranocha, P., Goffner, D., and Sundberg, B. (2016). High-throughput microanalysis of large lignocellulosic sample sets by pyrolysis-gas chromatography/mass spectrometry. Physiol. Plant. 156, 127–138. doi: 10.1111/ppl.12397
Hori, C., Yu, X., Mortimer, J. C., Sano, R., Matsumoto, T., Kikuchi, J., et al. (2020). Impact of abiotic stress on the regulation of cell wall biosynthesis in Populus trichocarpa. Plant Biotechnol. 37, 273–283. doi: 10.5511/plantbiotechnology.20.0326a
Janz, D., Lautner, S., Wildhagen, H., Behnke, K., Schnitzler, J. P., Rennenberg, H., et al. (2012). Salt stress induces the formation of a novel type of ‘pressure wood’ in two Populus species. New Phytol. 194, 129–141. doi: 10.1111/j.1469-8137.2011.03975.x
Kamon, E., and Ohtani, M. (2021). Xylem vessel cell differentiation: a best model for new integrative cell biology? Curr. Opin. Plant Biol. 64:102135. doi: 10.1016/j.pbi.2021.102135
Kim, W. C., Kim, J. Y., Ko, J. H., Kim, J., and Han, K. H. (2013). Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J. Plant Physiol. 170, 1374–1378. doi: 10.1016/j.jplph.2013.04.012
Kim, W. C., Ko, J. H., and Han, K. H. (2012). Identification of a cis-acting regulatory motif recognized by MYB46, a master regulator of secondary wall biosynthesis. Plant Mol. Biol. 78, 489–501. doi: 10.1007/s11103-012-9880-7
Kim, H., Padmakshan, D., Li, Y., Rencoret, J., Hatfield, R. D., and Ralph, J. (2017). Characterization and elimination of undesirable protein residues in plant cell wall materials for enhancing lignin analysis by solution-state nuclear magnetic resonance spectroscopy. Biomacromolecules 18, 4184–4195. doi: 10.1021/acs.biomac.7b01223
Kim, H., and Ralph, J. (2010). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5). Org. Biomol. Chem. 8, 576–591. doi: 10.1039/b916070a
Ko, J. H., Jeon, H. W., Kim, W. C., and Han, K. H. (2014). The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 114, 1099–1107. doi: 10.1093/aob/mcu126
Ko, J. H., Kim, W. C., and Han, K. H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665. doi: 10.1111/j.1365-313X.2009.03989.x
Komatsu, T., and Kikuchi, J. (2013). Comprehensive signal assignment of 13C-labeled lignocellulose using multidimensional solution NMR and 13C chemical shift comparison with solid-state NMR. Anal. Chem. 85, 8857–8865. doi: 10.1021/ac402197h
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Kumar, M., Thammannagowda, S., Bulone, V., Chiang, V., Han, K. H., Joshi, C. P., et al. (2009). An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci. 14, 248–254. doi: 10.1016/j.tplants.2009.02.004
Mansfield, S. D., Kim, H., Lu, F., and Ralph, J. (2012). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7, 1579–1589. doi: 10.1038/nprot.2012.064
McCarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. doi: 10.1093/pcp/pcq064
McCarthy, R. L., Zhong, R., and Ye, Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mori, T., Tsuboi, Y., Ishida, N., Nishikubo, N., Demura, T., and Kikuchi, J. (2015). Multidimensional high-resolution magic angle spinning and solution-state NMR characterization of 13C-labeled plant metabolites and lignocellulose. Sci. Rep. 5:11848. doi: 10.1038/srep11848
Nagaya, S., Kawamura, K., Shinmyo, A., and Kato, K. (2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332. doi: 10.1093/pcp/pcp188
Nakano, Y., Nishikubo, N., Goué, N., Ohtani, M., Yamaguchi, M., Katayama, Y., et al. (2010). MYB transcription factors orchestrating the developmental program of xylem vessels in Arabidopsis roots. Plant Biotechnol. 27, 267–272. doi: 10.5511/plantbiotechnology.27.267
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Ohtani, M., Akiyoshi, N., Takenaka, Y., Sano, R., and Demura, T. (2017a). Evolution of plant conducting cells: perspectives from key regulators of vascular cell differentiation. J. Exp. Bot. 68, 17–26. doi: 10.1093/jxb/erw473
Ohtani, M., and Demura, T. (2019). The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 56, 82–87. doi: 10.1016/j.copbio.2018.10.002
Ohtani, M., Nishikubo, N., Xu, B., Yamaguchi, M., Mitsuda, N., Goue, N., et al. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67, 499–512. doi: 10.1111/j.1365-313X.2011.04614.x
Ohtani, M., Ramachandran, V., Tokumoto, T., Takebayashi, A., Ihara, A., Matsumoto, T., et al. (2017b). Identification of novel factors that increase enzymatic saccharification efficiency in Arabidopsis wood cells. Plant Biotechnol. 34, 203–206. doi: 10.5511/plantbiotechnology
Okubo-Kurihara, E., Ohtani, M., Kurihara, Y., Kakegawa, K., Kobayashi, M., Nagata, N., et al. (2016). Modification of plant cell wall structure accompanied by enhancement of saccharification efficiency using a chemical, lasalocid sodium. Sci. Rep. 6:34602. doi: 10.1038/srep34602
Pawar, P. M., Derba-Maceluch, M., Chong, S. L., Gandla, M. L., Bashar, S. S., Sparrman, T., et al. (2017). In muro deacetylation of xylan affects lignin properties and improves saccharification of aspen wood. Biotechnol. Biofuels 10:98. doi: 10.1186/s13068-017-0782-4
Pawar, P. M., Koutaniemi, S., Tenkanen, M., and Mellerowicz, E. J. (2013). Acetylation of woody lignocellulose: significance and regulation. Front. Plant Sci. 4:118. doi: 10.3389/fpls.2013.00118
Pinto, R. C., Gerber, L., Eliasson, M., Sundberg, B., and Trygg, J. (2012). Strategy for minimizing between-study variation of large-scale phenotypic experiments using multivariate analysis. Anal. Chem. 84, 8675–8681. doi: 10.1021/ac301869p
Polle, A., Chen, S. L., Eckert, C., and Harfouche, A. (2019). Engineering drought resistance in forest trees. Front. Plant Sci. 9:1875. doi: 10.3389/fpls.2018.01875
Pradhan Mitra, P., and Loqué, D. (2014). Histochemical staining of Arabidopsis thaliana secondary cell wall elements. JoVE 87:51381. doi: 10.3791/51381
Saito, K., Watanabe, Y., Shirakawa, M., Matsushita, Y., Imai, T., Koike, T., et al. (2012). Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant. J. 69, 542–552. doi: 10.1111/j.1365-313X.2011.04811.x
Sakamoto, S., Takata, N., Oshima, Y., Yoshida, K., Taniguchi, T., and Mitsuda, N. (2016). Wood reinforcement of poplar by rice NAC transcription factor. Sci. Rep. 6:19925. doi: 10.1038/srep19925
Sato, T. A. (1968). Modified method for lead staining of thin sections. J. Electron. Microsc. 17, 158–9.
Schuetz, M., Smith, R., and Ellis, B. (2013). Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 64, 11–31. doi: 10.1093/jxb/ers287
Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L., et al. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17, 2059–2076. doi: 10.1105/tpc.105.030767
Studer, M. H., DeMartini, J. D., Davis, M. F., Sykes, R. W., Davison, B., Keller, M., et al. (2011). Lignin content in natural Populus variants affects sugar release. Proc. Natl. Acad. Sci. U. S. A. 108, 6300–6305. doi: 10.1073/pnas.1009252108
Suzuki, S., Li, L., Sun, Y. H., and Chiang, V. L. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. doi: 10.1104/pp.106.086678
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U. S. A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Tobimatsu, Y., Chen, F., Nakashima, J., Escamilla-Treviño, L. L., Jackson, L., Dixon, R. A., et al. (2013). Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25, 2587–2600. doi: 10.1105/tpc.113.113142
Tsuji, Y., Vanholme, R., Tobimatsu, Y., Ishikawa, Y., Foster, C. E., Kamimura, N., et al. (2015). Introduction of chemically labile substructures into Arabidopsis lignin through the use of LigD, the Cα-dehydrogenase from Sphingobium sp. strain SYK-6. Plant Biotechnol. J. 13, 821–832. doi: 10.1111/pbi.12316
Valdivia, E. R., Herrera, M. T., Gianzo, C., Fidalgo, J., Revilla, G., Zarra, I., et al. (2013). Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64, 1333–1343. doi: 10.1093/jxb/ers394
Watanabe, T., Shino, A., Akashi, K., and Kikuchi, J. (2014). Chemical profiling of Jatropha tissues under different torrefaction conditions: application to biomass waste recovery. PLoS One 9:e106893. doi: 10.1371/journal.pone.0106893
Wu, L., Joshi, C. P., and Chiang, V. L. (2000). A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J. 22, 495–502. doi: 10.1046/j.1365-313x.2000.00758.x
Xu, B., Ohtani, M., Yamaguchi, M., Toyooka, K., Wakazaki, M., Sato, M., et al. (2014). Contribution of NAC transcription factors of plant adaptation to land. Science 343, 1505–1508. doi: 10.1126/science.1248417
Yamaguchi, M., Goué, N., Igarashi, H., Ohtani, M., Nakano, Y., Mortimer, J. C., et al. (2010). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant. Physiol. 153, 906–914. doi: 10.1104/pp.110.154013
Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. doi: 10.1111/j.1365-313X.2008.03533.x
Yamaguchi, M., Mitsuda, N., Ohtani, M., Ohme-Takagi, M., and Demura, T. (2011). VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of broad range of genes for xylem vessel formation. Plant J. 66, 579–590. doi: 10.1111/j.1365-313X.2011.04514.x
Yan, F., Mitra, P., Zhang, L., Prak, L., Verhertbruggen, Y., Kim, J. S., et al. (2012). Engineering secondary cell wall deposition in plants. Plant. Biotechnol. J. 11, 325–335. doi: 10.1111/pbi.12016
Yoshida, K., Sakamoto, S., Kawai, T., Kobayashi, Y., Sato, K., Ichinose, Y., et al. (2013). Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4:383. doi: 10.3389/fpls.2013.00383
Zhang, J., Li, M., Bryan, A. C., Yoo, C. G., Rottmann, W., Winkeler, K. A., et al. (2019). Overexpression of a serine hydroxymethyltransferase increases biomass production and reduces recalcitrance in the bioenergy crop Populus. Sustain. Energy Fuels 3, 195–207. doi: 10.1039/C8SE00471D
Zhong, R., Demura, T., and Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, R., Lee, C., and Ye, Z. H. (2010a). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15, 625–632. doi: 10.1016/j.tplants.2010.08.007
Zhong, R., Lee, C., and Ye, Z. H. (2010b). Functional characterization of poplar wood associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., McCarthy, R. L., Haghighat, M., and Ye, Z. H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8:e69219. doi: 10.1371/journal.pone.0069219
Zhong, R., Richardson, E. A., and Ye, Z. H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary cell wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. doi: 10.1105/tpc.107.053678
Keywords: secondary cell wall, xylem, transcription factor, AtVND7, AtSND1, AtMYB46, hybrid aspen
Citation: Nakano Y, Endo H, Gerber L, Hori C, Ihara A, Sekimoto M, Matsumoto T, Kikuchi J, Ohtani M and Demura T (2022) Enhancement of Secondary Cell Wall Formation in Poplar Xylem Using a Self-Reinforced System of Secondary Cell Wall-Related Transcription Factors. Front. Plant Sci. 13:819360. doi: 10.3389/fpls.2022.819360
Received: 21 November 2021; Accepted: 17 February 2022;
Published: 14 March 2022.
Edited by:
Berit Ebert, Ruhr University Bochum, GermanyReviewed by:
Aimin Wu, South China Agricultural University, ChinaCopyright © 2022 Nakano, Endo, Gerber, Hori, Ihara, Sekimoto, Matsumoto, Kikuchi, Ohtani and Demura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Misato Ohtani, bWlzYXRvQGVkdS5rLnUtdG9reW8uYWMuanA=; Taku Demura, ZGVtdXJhQGJzLm5haXN0Lmpw
‡Present addresses: Yoshimi Nakano, Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan
Chiaki Hori, Research faculty of Engineering, Hokkaido University, Sapporo, Japan
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.