
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 March 2022
Sec. Aquatic Photosynthetic Organisms
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.817852
Heat shock protein 20 (Hsp20) genes play important roles in plant growth, development, and response to environmental stress. However, the Hsp20 gene family has not yet been systematically investigated, and its function in red algae (Rhodophyta) remains poorly understood. Herein, we characterized Hsp20 gene families in red algae by studying gene structure, conserved motifs, phylogenetic relationships, chromosome location, gene duplication, cis-regulatory elements, and expression profiles. In this study, 97 Hsp20 genes were identified using bioinformatic methods and classified into 13 subfamilies based on phylogenetic relationships. Phylogenetic analysis revealed that Hsp20 genes might have a polyphyletic origin and a complex evolutionary pattern. Gene structure analysis revealed that most Hsp20 genes possessed no introns, and all Hsp20 genes contained a conserved α-crystalline domain in the C-terminal region. Conserved motif analysis revealed that Hsp20 genes belonging to the same subfamily shared similar motifs. Gene duplication analysis demonstrated that tandem and segmental duplication events occurred in these gene families. Additionally, these gene families in red algae might have experienced strong purifying selection pressure during evolution, and Hsp20 genes in Pyropia yezoensis, Pyropia haitanensis, and Porphyra umbilicalis were highly evolutionarily conserved. The cis-elements of phytohormone-, light-, stress-responsive, and development-related were identified in the red algal Hsp20 gene promoter sequences. Finally, using Py. yezoensis, as a representative of red algae, the Hsp20 gene expression profile was explored. Based on the RNA-seq data, Py. yezoensis Hsp20 (PyyHsp20) genes were found to be involved in Py. yezoensis responses against abiotic and biotic stresses and exhibited diverse expression patterns. Moreover, PyyHsp20 is involved in Py. yezoensis growth and development and revealed spatial and temporal expression patterns. These results provide comprehensive and valuable information on Hsp20 gene families in red algae and lay a foundation for their functional characterization. In addition, our study provides new insights into the evolution of Hsp20 gene families in red algae and will help understand the adaptability of red algae to diverse environments.
In recent years, heat has become a prominent abiotic stress factor owing to global warming. All organisms synthesize a set of proteins called heat shock proteins (HSPs) to combat heat stress (Lindquist and Craig, 1988). According to protein molecular weight and sequence homology, HSPs can be divided into five families: Hsp100s, Hsp90s, Hsp70s, Hsp60s, and Hsp20s, among which, Hsp20s are considered the most abundant and complex members (Vierling, 1991). In addition, the molecular size of Hsp20s ranges from 12 to 42 kDa, and thus they are called small HSPs. Hsp20s function as ATP-independent molecular chaperones to prevent proteins from irreversible aggregation and play a fundamental role in plant-acquired thermotolerance processes (Waters, 2013). Hsp20s mainly possess a highly conserved sequence of approximately 90 amino acid residues, called the α-crystalline domain (ACD) or the Hsp20 domain, in the C-terminal region, which is involved in substrate interactions (Waters and Vierling, 2020). In addition, the ACD is flanked by a variable N-terminal domain and a short C-terminal extension, which performs two different functions (Waters and Vierling, 2020).
Analyses of genome sequences from eukaryotic organisms have shown that Hsp20 genes belong to a group of diverse and complex families. Plant Hsp20s can be divided into several subfamilies (CI-CVI, MTI, MTII, ER, CP, and PX) based on cellular location, sequence homology, and function (Waters and Vierling, 2020). CI-CVI subfamilies are localized in the cytoplasm and nucleus; MTI and MTII subfamilies are localized in the mitochondria; and the other three subfamilies (ER, CP, and PX) are localized in the endoplasmic reticulum, chloroplast, and peroxisome, respectively (Waters and Vierling, 2020). Previous studies have suggested that plant Hsp20s are not only induced by heat stress but also by a wide variety of other stresses, such as drought, salinity, osmotic stress, oxidative stress, and UV-B radiation (Wang et al., 2004; Waters, 2013). In general, stress-related Hsp20s are tightly repressed at low temperatures, whereas their overexpression might induce deleterious effects on plant growth and development (Sun et al., 2016; Bourgine and Guihur, 2021). Hsp20s plays an important role in plant immunity (Park and Seo, 2015). Currently, an increasing amount of data reveals a close correlation between Hsp20s accumulation and plant stress tolerance. In addition, Hsp20s are involved in plant growth and development, such as embryo development, somatic embryogenesis, seed germination, pollen development, and fruit maturation (Waters et al., 1996).
Red algae (Rhodophyta) are one of the most ancient groups of eukaryotic algae, with over 7,000 known species.1 They form a distinct photosynthetic eukaryotic lineage with primitive features, such as the absence of centrioles, flagella, and parenchyma; the presence of unstacked thylakoids; and phycobilisomes in the chloroplast (Lopez-Bautista, 2010). Red algae are classified into seven classes (Cyanidiophyceae, Bangiophyceae, Florideophyceae, Compsopogonophyceae, Porphyridiophyceae, Rhodellophyceae, and Stylonematophyceae) and exhibit a remarkable diversity in their habitats (e.g., hot springs, acidic sulfur fumes, fresh water, deep ocean, and intertidal zone) and morphology (e.g., unicells, filaments, leaf-shaped thallus, parenchymatous blades, single-cell-thick tubular form) (Yoon et al., 2010). Red algae comprise many commercially valuable species that can be used as food, pharmaceuticals, nutraceuticals, cosmetics, phycocolloids, and phyco supplements (e.g., soil additives, fertilizers, and animal feed). For example, Pyropia spp. are consumed worldwide. Chondrus, Eucheuma, and Kappaphycus serve as raw materials for carrageenan production. Gelidium and Gracilaria are agarophytes used in the agar industry (Reddy et al., 2010). In addition, red algae are important for eukaryotic evolution (Yoon et al., 2010). Over the last 20 years, research on the physiological ecology, evolution, and commercial importance of red algae has surged.
To date, the Hsp20 gene family has been investigated in many plant species; for instance,19 Hsp20 genes have been identified in Arabidopsis (Scharf et al., 2001), 39 in rice (Ouyang et al., 2009), 48 in potato (Zhao et al., 2018), 35 in pepper (Guo et al., 2015), 42 in tomato (Yu et al., 2016), and 51 in soybean (Lopes-Caitar et al., 2013). However, only a few studies have investigated the Hsp20 gene families in algae. Currently, only six Hsp20 genes have been identified in the green alga Chlamydomonas reinhardtii (Waters and Rioflorido, 2007), two in the unicellular red alga Cyanidioschyzon merolae (Kobayashi et al., 2014), and five in the multicellular red alga Py. yezoensis (Uji et al., 2019). Overall, systematic and comprehensive studies on the Hsp20 gene family in algae are lacking. In recent years, with the development of high-throughput sequencing technology, high-quality and complete reference genomes of eight red algae have been assembled, annotated, and made available through public databases (Matsuzaki et al., 2004; Bhattacharya et al., 2013; Collen et al., 2013; Schoenknecht et al., 2013; Brawley et al., 2017; Lee et al., 2018; Cao et al., 2020; Wang et al., 2020). In addition, some red algal protein data without reference genomes are available on EukProt (Richter et al., 2020). These studies provide convenient and useful resources for further understanding the Hsp20 gene families in red algae.
In the present study, we performed a comprehensive and systematic analysis of Hsp20 gene families in red algae. We identified the members of the red algal Hsp20 gene families using bioinformatic methods and analyzed the gene structure, conserved motifs, phylogenetic relationships, evolutionary origin, chromosome location, gene duplication, cis-regulatory elements, and expression profiles. These results provide valuable information for further functional characterization of red algal Hsp20 genes and elucidation of the evolutionary history of red algal Hsp20 gene families.
To identify Hsp20 genes throughout the red algae, 17 kinds of red algae were chosen depending on the availability of genome or protein data (Supplementary Table 1). High-quality Py. yezoensis and Pp. haitanensis reference genome assemblies and protein sequences were obtained from our laboratory (Cao et al., 2020; Wang et al., 2020). Chondrus crispus, Cy. merolae, Gracilariopsis chorda, Galdieria sulphuraria, Porphyridium purpureum, and Ph. umbilicalis reference genome assemblies and protein sequences were downloaded from the National Center for Biotechnology Information (NCBI2). Other red algal protein sequences were downloaded from the EukProt database3. The hidden Markov model (HMM) profile of the Hsp20 domain (PF00011) was downloaded from Pfam4 to identify Hsp20 genes in red algae. HMMER software version 3.0 (Finn et al., 2011) was used to search for Hsp20s from the red algal protein sequences with an E-value cutoff value of 0.001. According to the HMMER results, all candidate Hsp20s that may contain the Hsp20 domain were submitted to Pfam and CDD5 to confirm the Hsp20 domain. The red algal Hsp20 genes were named according to their molecular weights. The molecular weight, instability index, and theoretical isoelectric points (pI) of the red algal Hsp20s were calculated using the ProtParam tool (Gasteiger et al., 2003). Subcellular protein localization was predicted using WoLF PSORT, CELLO, Yloc, BUSCA, MULocDeep, and SeqNLS (Yu et al., 2006; Horton et al., 2007; Briesemeister et al., 2010; Lin and Hu, 2013; Savojardo et al., 2018; Jiang et al., 2021).
First, we constructed a maximum likelihood (ML) phylogenetic tree to classify Hsp20 genes in red algae. All highly conserved ACD sequences of the predicted red algal Hsp20s were aligned using the MEGA X-ClustalW program with default parameters. An ML tree was inferred using IQ-TREE 2.1.3 (Minh et al., 2020), and the best-fit substitution model was automatically selected using ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQ-TREE. Branch support was calculated using ultrafast bootstrap approximation with 1,000 replicates (Hoang et al., 2018). Finally, the phylogenetic trees were edited using EvolView (Subramanian et al., 2019).
Second, highly conserved ACD sequences were used to construct an ML phylogenetic tree to study the origin of red algal Hsp20 genes. This conserved domain has proven to be useful in evolutionary studies of Hsp20 genes (Caspers et al., 1995; de Jong et al., 1998; Huang et al., 2008; Kobayashi et al., 2014). The phylogenetic tree contained not only ACD sequences of the previously predicted red algal Hsp20s but also the other 154 ACD sequences of Hsp20s. Hsp20 protein sequences of Cyanophora paradoxa were downloaded from the Cyanophora paradoxa Genome Project (Price et al., 2019). Other Hsp20 protein sequences are shown in Supplementary Table 2. Multiple sequence alignment was performed using the MEGA X-ClustalW program with the default parameters. An ML tree was inferred using IQ-TREE 2.1.3 (Minh et al., 2020), and the best-fit substitution model was automatically selected using ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQ-TREE. Branch support was calculated using ultrafast bootstrap approximation with 1,000 replicates (Hoang et al., 2018). The ML tree was compiled and visualized using FigTree 1.4.4.6
Exon-intron structures of the red algal Hsp20 genes were identified using the TBtools software (Chen et al., 2020). Furthermore, conserved motifs of red algal Hsp20s were identified using the MEME program (version 5.4.1)7 with the following parameters: number of motifs 6 and optimum motif widths from 6 to 50 amino acid residues (Bailey et al., 2015). The conserved ACD of red algal Hsp20 protein sequences was aligned using the MEGA X-ClustalW program with default parameters. GeneDoc software8 was used for homology shading and scoring of the aligned sequences. Sequence logos were generated using WebLogo (Crooks et al., 2004).
Chromosomal locations of Hsp20 genes were visualized using TBtools with eight red algae genomic sequences and annotation files. In addition, using the One-Step MCScanX program of TBtools, we analyzed tandem duplication events and segmental duplication events of the red algal Hsp20 gene family (Wang et al., 2012; Chen et al., 2020). Similarly, the One-Step MCScanX program of TBtools was used to analyze the collinearity relationship of Hsp20 genes between different red algae. Furthermore, to determine the selection pressure, the rates of non-synonymous (Ka) and synonymous (Ks) substitutions were calculated using TBtools.
To identify the cis-acting regulatory elements in the promoter sequences of red algal Hsp20 genes, the upstream 1.5 kb promoter sequences of red algal Hsp20 genes were submitted to PlantCARE9 (Lescot et al., 2002). The detected cis-acting regulatory elements were classified into different response types based on their annotated functions, and the number of detected cis-acting regulatory elements was displayed using a heatmap. In addition, we followed manual inspection and the motif-based sequence analysis tool MEME to search for heat-shock elements (HSEs).
To investigate the expression pattern of PyyHsp20 (Py. yezoensis Hsp20) genes, Illumina RNA-seq data were collected from previous studies conducted in our laboratory (Tang et al., 2019; Wang et al., 2020). The RNA-seq data were deposited in NCBI under BioProject PRJNA589917 and PRJNA560692. These data included different developmental stages and abiotic and biotic stresses. RNA-seq data analysis, including experimental design, quality control, read alignment, and quantification of gene and transcript levels, was performed as previously described (Tang et al., 2019; Wang et al., 2020). Furthermore, gene expression levels were quantified by fragments per kilobase of transcript per million fragments mapped (FPKM) values, and expression heat maps were created using TBtools software based on log2 transformed FPKM values. In addition, RNA-seq data including different parts of the blade were collected from previous studies conducted in our laboratory, and the FPKM values are given in Supplementary Table 3. RNA-seq data and gene expression analyses were performed as described previously.
In this study, we analyzed 17 representative species of red algae with available genomic or protein information. A total of 97 putative Hsp20 genes in red algae were identified and named based on their molecular weights. Two members were identified in Cy. merolae, three in Ga. sulphuraria, two in Cyanidium caldarium, five in Ch. crispus, six in Gr. chorda, nine in Agarophyton chilense, seven in Madagascaria erythrocladioides, eight in Compsopogon caeruleus, three in Erythrolobus australicus, six in Porphyridium aerugineum, eight in Pr. purpureum, seven in Py. yezoensis, eight in Pp. haitanensis, nine in Ph. umbilicalis, three in Rhodella violacea, six in Rhodosorus marinus, and five in Stylonematophyceae sp. Among the 97 putative Hsp20 genes, eight candidate sequences (AcHsp20-14.9, AcHsp20-13.0, CocHsp20-7.6, PyyHsp20-20.3, PphHsp20-15.6, PphHsp20-20.8, PhuHsp20-20.5, and RomHsp20-13.4) contained incomplete Hsp20 domains according to Pfam and CDD. Detailed information on the red algal Hsp20 genes is shown in Supplementary Table 4, including names, chromosome locations, intron numbers, protein lengths, molecular weights, pI values, and instability index. The corresponding predicted molecular weights and length of encoded proteins varied from 6.61 to 49.27 kDa and 60 to 434 amino acids. The pI of Hsp20s ranged from 3.97 to 10.88. The instability index of Hsp20s ranged from 21.76 to 73.59. The protein sequences of the red algal Hsp20 genes are provided in Supplementary Data Sheet 1. Protein subcellular localization is closely related to protein function, and their prediction is helpful in understanding protein function. In this study, we used six protein subcellular localization prediction tools to predict the subcellular location of red algal proteins. The prediction of the subcellular localization of red algal Hsp20s is shown in Supplementary Table 5. However, the results were not entirely consistent. Finally, we accepted the majority consensus.
An unrooted ML phylogenetic tree was constructed based on the alignment of the conserved ACD sequences of the red algal Hsp20 protein sequences (Figure 1). Based on the phylogenetic tree of the conserved ACD sequences, all identified 97 Hsp20 genes were classified into 13 distinct subfamilies. There were two types of subfamilies in the red algal Hsp20 gene families. One type was a broadly distributed subfamily, including the II, IV, XI, and XII subfamilies, which included members from different classes. For instance, in the II subfamily, eleven members were from the class Compsopogonophyceae, five members were from the class Florideophyceae, four members were from the class Porphyridiophyceae, and one member were from the class Cyanidiophyceae. The other type was a class-specific subfamily, such as the I, III, V, VI, VII, VIII, IX, X, and XIII subfamilies, which included members from the same classes. For instance, in subfamily I, all 13 members came from the same class, Florideophyceae. It is worth noting that only 4 of 97 red algal Hsp20s (RomHsp20-31.6, RomHsp20-34.8, RomHsp20-34.0, and StHsp20-35.5) belonging to the class Stylonematophyceae possessed three ACDs, and the other red algal Hsp20s possessed only one ACD. Most strikingly, in these 12 ACDs, ACDs from different Hsp20s clustered together rather than those from the same Hsp20. In addition, classification did not correlate with predicted subcellular localization.
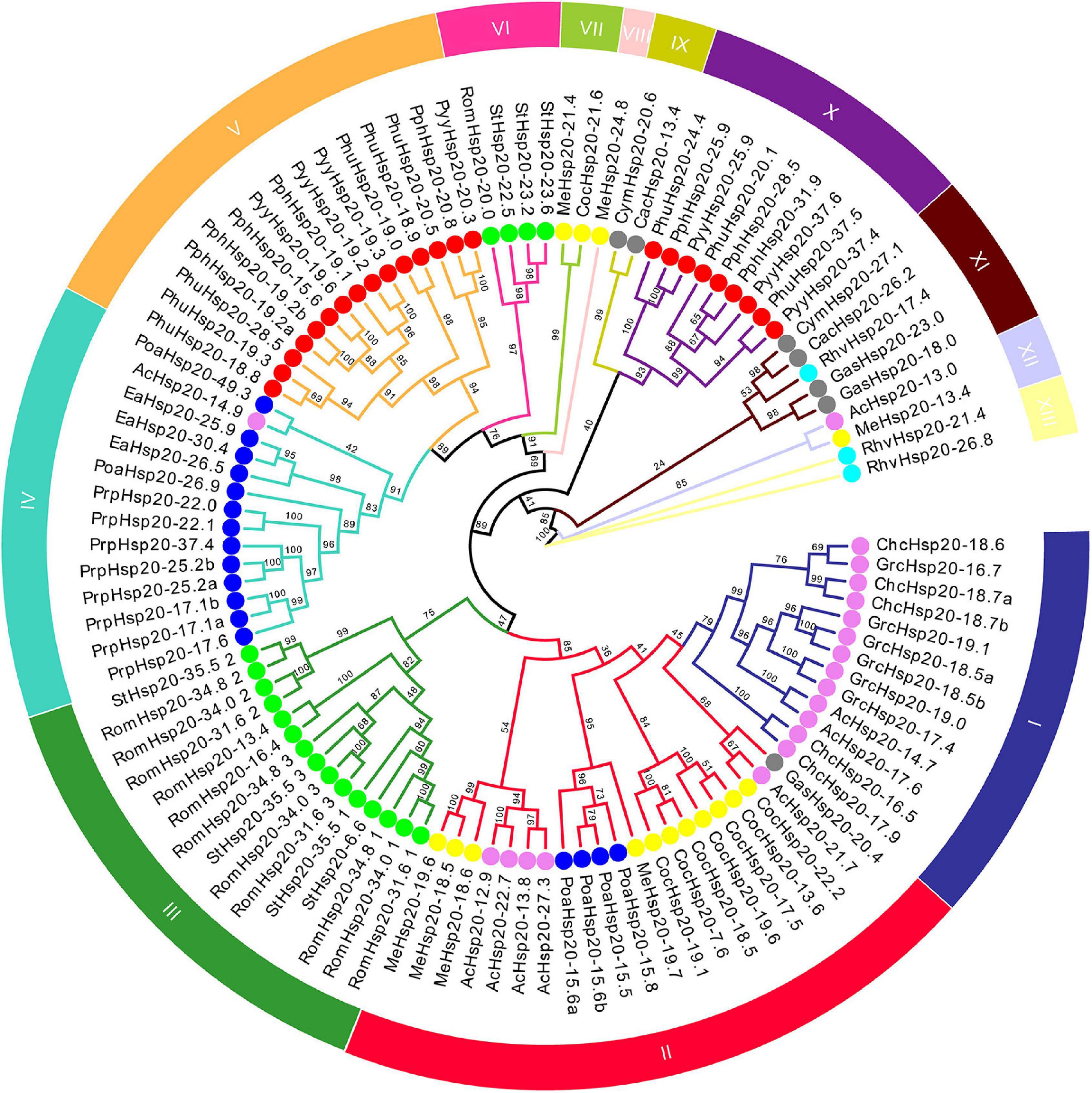
Figure 1. Phylogenetic tree of conserved ACD sequences of red algal Hsp20 proteins. The maximum likelihood (ML) phylogenetic tree of conserved ACD sequences of red algal Hsp20 proteins was constructed using IQ-TREE 2.1.3. The different subfamilies, numbered from I to XIII, were marked using different colors. Red, gray, violet, yellow, blue, cyan, and lime circles represent Bangiophyceae, Cyanidiophyceae, Florideophyceae, Compsopogonophyceae, Porphyridiophyceae, Rhodellophyceae, and Stylonematophyceae, respectively.
A phylogenetic tree based on the conserved ACD sequences of 251 Hsp20s from various species of archaea, cyanobacteria, bacteria, fungi, algae, and plants was constructed using the ML method (Figure 2). The goal of our analysis was to elucidate the origin of red algal Hsp20 genes. Phylogenetic analysis showed that archaea Hsp20s clustered at the base of the tree, followed by bacteria, cyanobacteria, glaucophyta, and fungi clades. Plant Hsp20s clustered into two major clades, and clustered according to the cellular localization. In green algae, apart from two Hsp20s, all green algal Hsp20s clustered together in one clade. In red algae, Hsp20s were always grouped together. It is worth noting that some red algal Hsp20s clustered with bacterial Hsp20s. This indicated that these red algal Hsp20s may originate from bacteria via horizontal gene transfer (HGT). In addition, none of the red algal Hsp20s belonged to land plant organelle-localized Hsp20s.
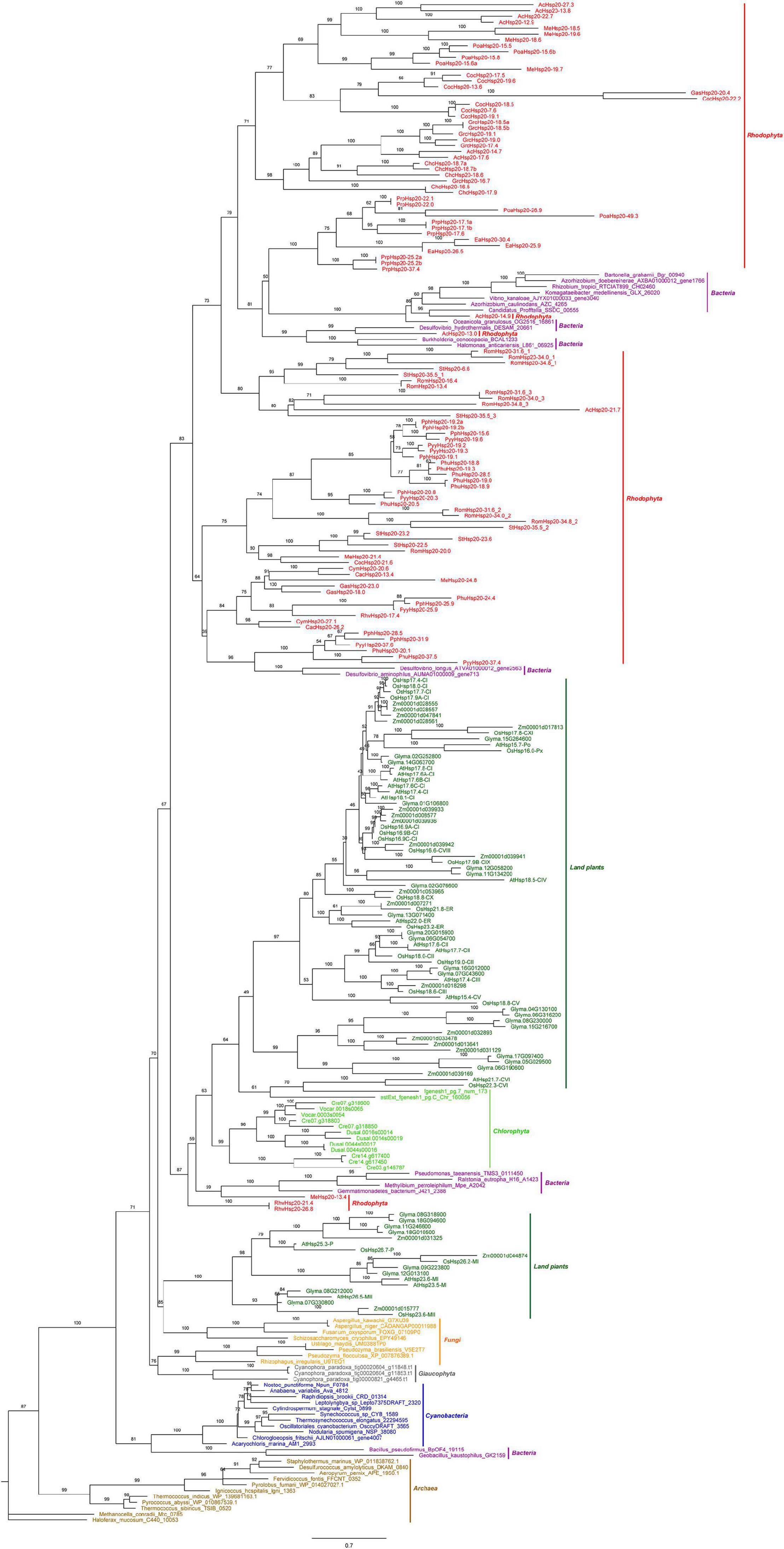
Figure 2. The maximum likelihood (ML) phylogenetic tree based on the conserved ACD sequences of Hsp20s. The tree was rooted by the ACD sequences of Haloferax mucosum Hsp20 protein. We applied different colors for species in different phyla, such as saddle brown for the archaea, purple for the bacteria, blue for the cyanobacteria, orange for the fungi, green for the land plants, gray for the Glaucophyta, lime for the Chlorophyta, and red for the Rhodophyta. Detailed information for the proteins included in the analysis can be found in the Supplementary Material.
The gene structure plays a crucial role in the evolution of multiple gene families. Our results showed that most of the red algal Hsp20 genes contained no introns and only three contained one intron (Figure 3D). In some studies, genes with few or no introns were considered to be rapidly activated in response to various stresses (Jeffares et al., 2008).
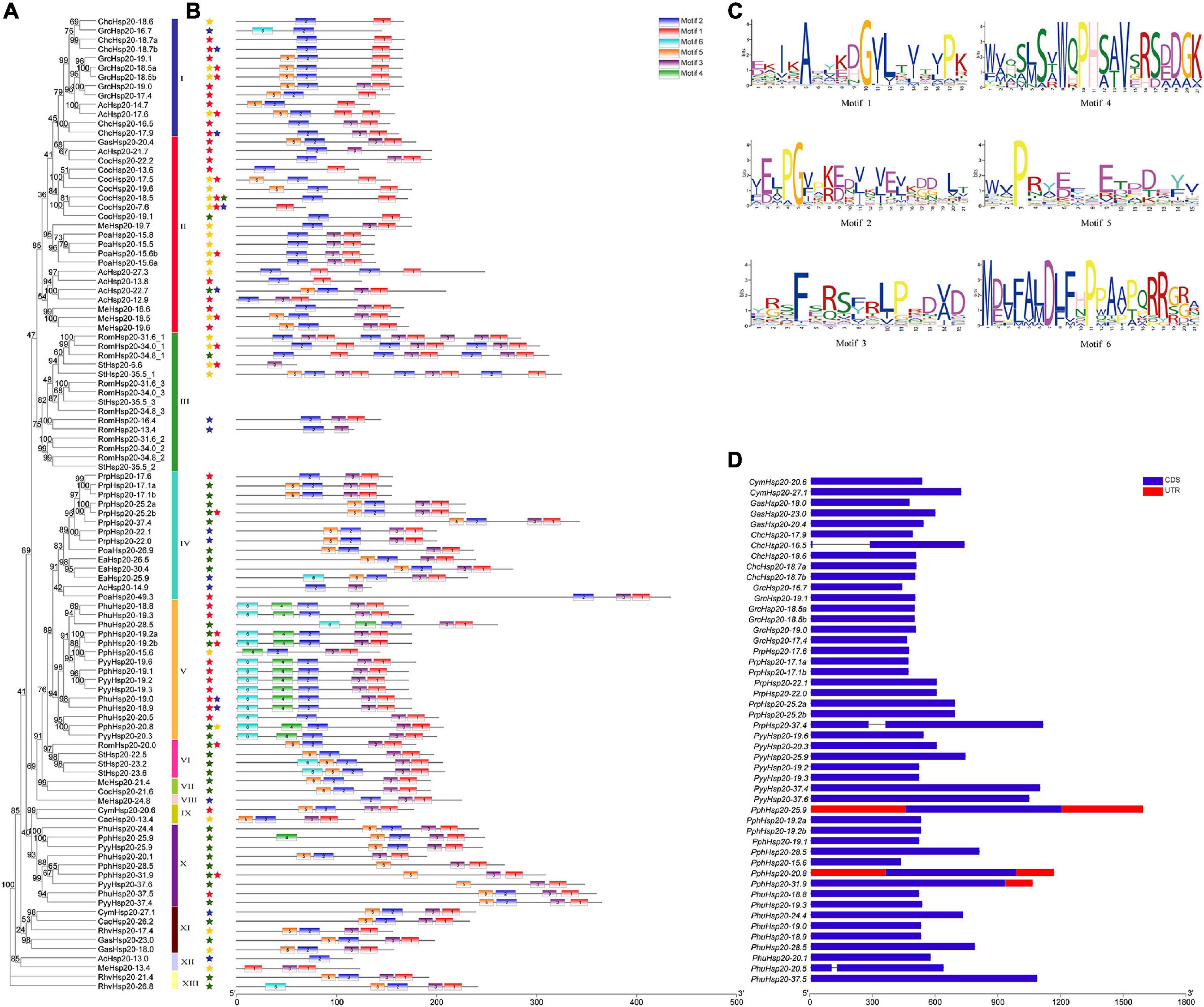
Figure 3. Gene structure and motif compositions of Hsp20 genes in red algae. (A) Maximum likelihood (ML) phylogenetic tree and subcellular localization prediction. Red, yellow, blue, and green stars represent nucleus, cytoplasm, mitochondrion, and plastid, respectively. (B) Motif analysis was performed using the MEME software. Different colored boxes represent different motifs in the corresponding positions of each red algal Hsp20 protein. The scale at the bottom represents the length of the protein and motif. (C) Motif logos of the conserved motifs. (D) Gene structure was analyzed using the TBtools software. Boxes filled with blue represent exons, and solid black lines represent introns. The untranslated regions are indicated using red boxes. The scale at the bottom is in bp.
The MEME online tool was employed to identify conserved motifs in red algal Hsp20s, and six types of consensus motifs were identified (Figure 3B). The length of these conserved motifs varied from 15 to 21 amino acids (Figure 3C). The locations of the six motifs matched the conserved regions, as revealed through multiple sequence alignment analysis. As shown in Figure 3B, motifs 1, 2, and 3 were found in most members of the red algal Hsp20 family. Motifs 4 and 6 were mainly found in the Hsp20s of Py. yezoensis, Pp. haitanensis, and Ph. umbilicalis belonging to the same class, Bangiophyceae. The type, order, and number of motifs were similar in proteins within the same subfamily but differed from those of other subfamilies, indicating that these genes may be highly conserved (Figure 3A). However, the functions of these highly conserved amino acid motifs remain elusive.
Multiple sequence alignments of the conserved ACD domains among the red algal Hsp20s are shown in Figure 4. In previous studies, the ACD domain was divided into consensus I and II domains separated by a hydrophilic domain of variable length in plants. These two conserved regions are separated by a hydrophilic domain of variable length and characterized by residues Pro-X (14)-Gly-Val-Leu and Pro-X (14)-X-Val/Leu/Ile-Val/Leu/Ile, respectively (Waters et al., 1996). These conserved regions are known to play important roles in the chaperone function of Hsp20s. To date, the structure of the ACD region in red algae remains relatively unknown. In our study, the ACD domain of red algal Hsp20s could also be divided into two parts (consensus I and consensus II) separated by a hydrophilic domain of variable length. Moreover, most motifs 1 and 3 were located in consensus region I, and most motifs 2 were located in consensus region II.

Figure 4. Multiple sequence alignment of α-crystalline domains of Hsp20s in red algae. (A) Sequence logo of the consensus II region. The height of individual symbols within a stack indicates the relative frequency of an amino acid at that position. (B) Sequence logo of the consensus I region. The height of individual symbols within a stack indicates the relative frequency of an amino acid at that position. (C) Multiple sequence alignment of the red algal Hsp20s containing consensus I and II regions. Conserved amino acid residues are indicated using color shading. Consensus I and II regions are underlined at the bottom and the typical amino acid residues within these regions are indicated by asterisks.
To better understand the chromosomal locations of red algal Hsp20 genes, their positions on each chromosome were marked. As shown in Figure 5A, 48 Hsp20 genes were mapped to 31 chromosomes. The number of Hsp20 genes on each chromosome was between one and four. Except for Cy. merolae and Ga. sulphuraria, Hsp20 genes were unevenly distributed on 27 chromosomes. In Cy. merolae, both Hsp20 genes were located on one chromosome. In Ga. sulphuraria, the three Hsp20 genes were evenly distributed on the three chromosomes. Interestingly, the chromosomal locations of Ch. crispus and Gr. chordas exhibited similar patterns. It might be related to their close relationship.
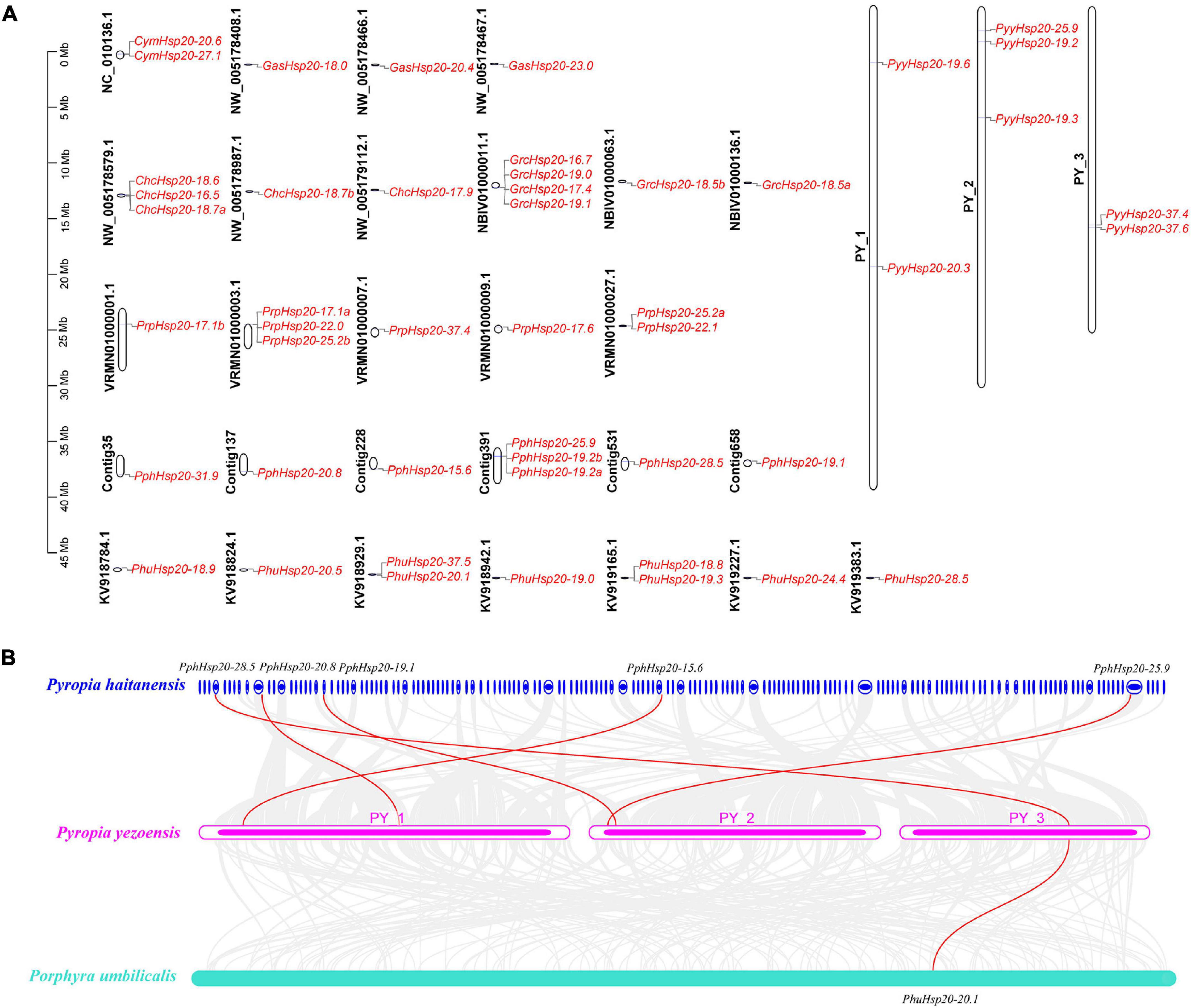
Figure 5. Chromosome location and synteny analysis of red algal Hsp20 genes. (A) Chromosome location of Hsp20 genes in eight red algae. (B) Synteny analysis of the Hsp20 genes between three red algae. Gray lines in the background indicate collinear blocks within the three red algal genomes. The red lines highlight syntenic Hsp20 gene pairs.
Subsequently, using the One-Step MCScanX program of TBtools, we analyzed tandem duplication and segmental duplication events. The results are shown in Supplementary Table 6. We identified five gene pairs with tandem duplication events and two gene pairs with segmental duplication events. Notably, these events occurred mainly in Pr. purpureum. These results suggest that these events are the main driving forces for the diversity of Hsp20 genes in Pr. purpureum.
Furthermore, we explored the collinearity relationships among the eight red algae. As shown in Figure 5B (Supplementary Table 7), five Py. yezoensis Hsp20 genes had collinearity relationships with Pp. haitanensis Hsp20 genes and one Py. yezoensis Hsp20 gene had collinearity relationships with Ph. umbilicalis Hsp20 genes. The other five red algae species showed no collinearity. These results indicate that the Hsp20 genes in Py. yezoensis, Pp. haitanensis, and Ph. umbilicalis are highly evolutionarily conserved.
TBtools was used to calculate the non-synonymous (Ka)/synonymous (Ks) ratios for each gene pair (Supplementary Tables 6, 7). When the Ka/Ks ratio is equal to 1, it shows a neutral selection; when it is >1, it denotes positive selection; when it is <1, it is used for purifying selection. The Ka/Ks ratios of tandem duplication, segmental duplication, and collinearity gene pairs were <1, indicating that the red algal Hsp20 gene families might have experienced strong purifying selection pressures during evolution.
Cis-regulatory elements are important molecular switches involved in the transcriptional regulation of gene expression and control various biological processes, including stress responses, hormone responses, and developmental processes (Yamaguchi-Shinozaki and Shinozaki, 2005). To further explore the regulatory mechanisms of Hsp20 genes in red algal growth, development, and stress response, the cis-elements in the promoter region (1.5 kb upstream sequences from the translation start sites) of the 48 red algal Hsp20 genes were further analyzed. Four categories of cis-elements, phytohormone-, stress-, light-responsive, and development-related, were identified (Figure 6A). Among the four cis-element categories, the phytohormone-responsive category accounted for the highest proportion, including auxin, gibberellin, MeJA, abscisic acid, and salicylic acid-responsive elements. Among these elements, MeJA-responsive and abscisic acid-responsive cis-elements accounted for the largest proportion of the phytohormone-responsive category. In the stress-responsive category, stress response-related cis-elements, such as the GC-motif (anoxic specific inducibility), LTR (low temperature-responsive), ARE (anaerobic induction), MBS (drought-inducibility), TC-rich repeats (defense and stress responsiveness), and WUN motifs (wound-responsiveness) were detected. However, we did not find HSEs in the promoters of the red algal Hsp20 genes using the PlantCARE tool. In general, the expression of Hsp genes was regulated at the transcriptional level by binding of heat shock transcription factors (HSFs) to HSEs. Eukaryotic HSEs are categorized into three types: perfect (P), gap (G), and step (S). P-type HSEs have three inverted repeats in a contiguous array (nGAAnnTTCnnGAAn or TTCnnGAAnnTTC). G-type HSEs have two consecutive inverted sequences, with the third sequence separated by 5 bp [nTTCnnGAAn (5 bp) nGAAn]. S-type HSEs have 5 bp gaps separating all three modules [nTTCn (5 bp) nTTCn (5 bp) nTTCn] (Mittal et al., 2011). We subsequently used manual inspection and the motif-based sequence analysis tool MEME to search for HSEs. In total, six, two, and six red algal Hsp20 promoters contained only P-, G-, and S-type HSEs, respectively, and one red algal Hsp20 promoter showed both P- and G-type HSEs. One red algal Hsp20 promoter showed two P-type HSEs, and one red algal Hsp20 promoter showed two S-type HSEs (Figure 6B and Supplementary Data Sheet 2). In addition, one motif showed consensus sequences similar to the known perfect HSE consensus sequence (Figure 6C). The main components of the light-responsive elements were G-box and Sp1. In the last category, plant development-related elements, including meristem-specific activation (NON-box), meristem expression (CAT-box), circadian, and cell cycle regulation (MSA-like) were identified. These results indicate that Hsp20 genes play crucial roles in the growth, development, and stress response of red algae. In addition, there are some differences in cis-acting elements between different red algae species. For example, Sp1 was found in Pp. haitanensis but not in Ga. sulphuraria. These results suggest that Hsp20 genes not only play common roles in different red algae species but also play specific roles in certain red algae species.
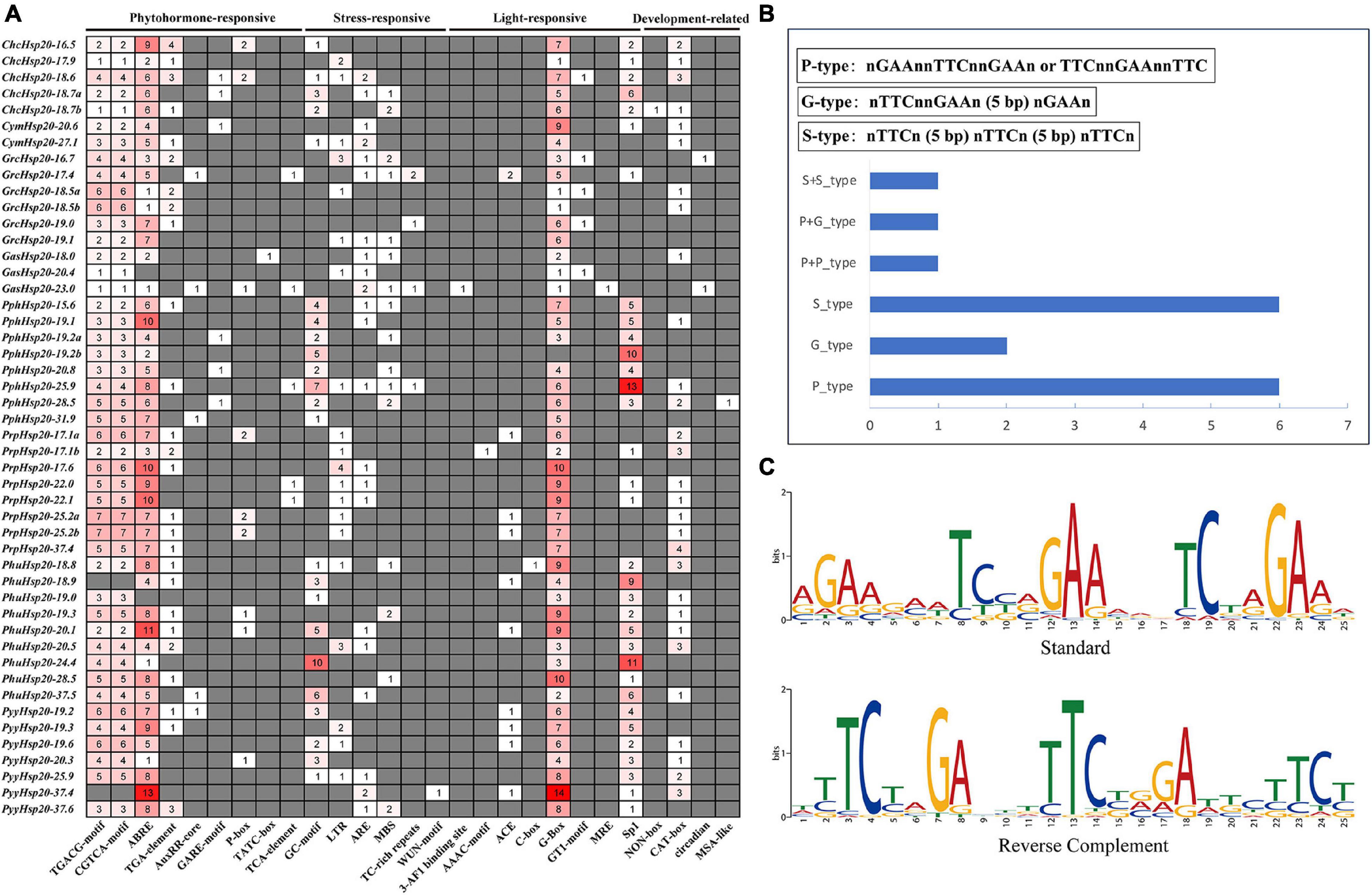
Figure 6. Predicted cis-elements in red algal Hsp20 promoters. (A) Analysis of 1.5-kb upstream cis-acting elements found in red algal Hsp20 genes. The different colors and numbers of the grid indicated the numbers of different promoter elements. (B) Distribution of HSEs in red algal Hsp20 promoters. (C) Motif logos of the putative heat shock elements (HSEs) in red algae Hsp20 promoters. The motifs were obtained using MEME analysis based on promoter sequences of red algal Hsp20 genes.
To explore the functions of red algal Hsp20 genes in response to biotic and abiotic stress, we performed a comprehensive expression analysis using the available RNA-seq data of Py. yezoensis from our laboratory to investigate the expression patterns of PyyHsp20 genes in response to dehydration/rehydration and red rot disease. Different expression patterns of PyyHsp20 genes were observed in response to dehydration/rehydration and red rot disease (Figure 7). As shown in Figure 7A, under dehydrated and rehydrated stresses, except for PyyHsp20-37.4 and PyyHsp20-25.9, the expression of all the other PyyHsp20 genes was significantly downregulated under dehydrated conditions and significantly upregulated under rehydrated conditions. Moreover, PyyHsp20-19.3, PyyHsp20-19.6, PyyHsp20-20.3, and PyyHsp20-37.6 were up-regulated under 50% water loss conditions. As shown in Figure 7B, the expression of all PyyHsp20 genes was significantly upregulated when Py. yezoensis was infected with the oomycete pathogen Pythium porphyrae. Among these genes, PyyHsp20-37.6 showed the highest expression level. These results indicated that most PyyHsp20 genes responded to biotic and abiotic stresses, and the response mechanisms of different PyyHsp20 genes to biotic and abiotic stresses were different. In conclusion, these results suggest that PyyHsp20 plays a role in mediating the response of Py. yezoensis under environmental stress conditions.
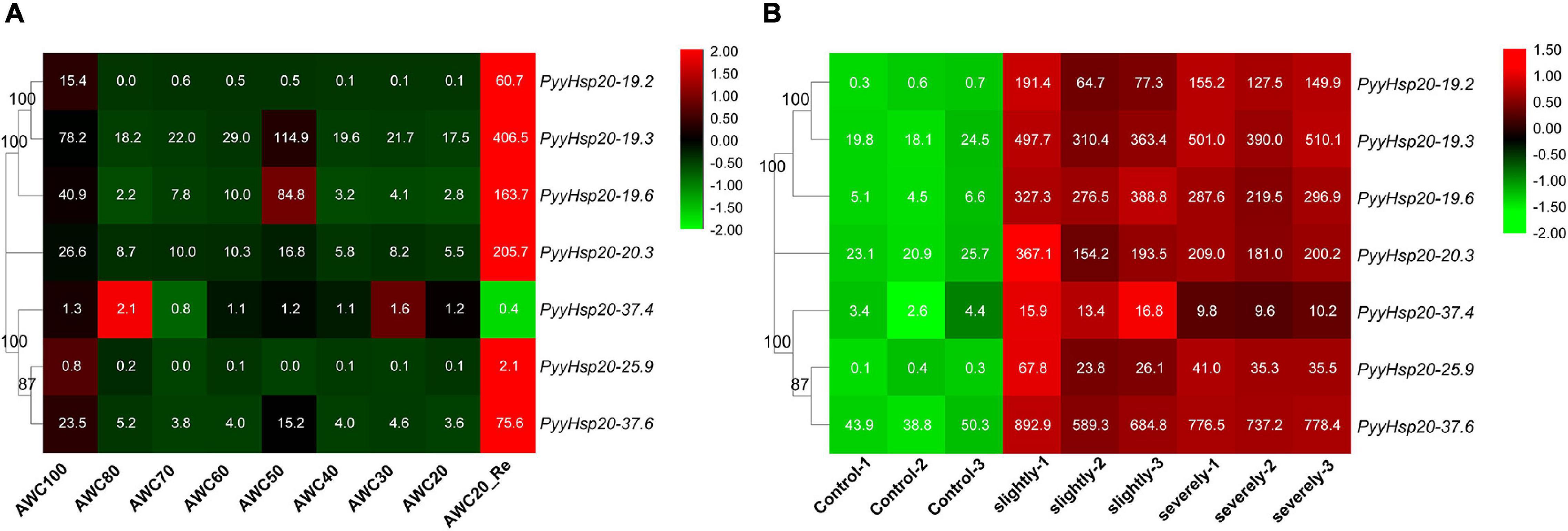
Figure 7. Heat map of the expression profiles of PyyHsp20 genes under various biotic and abiotic stress conditions. (A) Heat map of the expression profiles of PyyHsp20 genes under the dehydrated and rehydrated stresses. AWC100, AWC80, AWC70, AWC60, AWC50, AWC40, AWC30, and AWC20 represent water loss at 0, 20, 30, 40, 50, 60, 70, and 80%, respectively. AWC20_Re represents rehydration 30 min after 80% water loss. Differential expression is shown in different colors. The numbers represent relative expression quantity. Each experimental condition contained four biological repetitions. (B) Heat map of the expression profiles of PyyHsp20 genes in Py. yezoensis during infection. Control represents a healthy blade of Py. yezoensis. Slightly represents a slightly infected stage. Severely represents severely infected stage. Differential expression is shown in different colors. The number represents relative expression quantity.
To investigate the function of PyyHsp20 in Py. yezoensis growth and development, we analyzed the expression profiles of PyyHsp20 genes in different parts and developmental stages of Py. yezoensis. Significant differences were observed in the expression profiles of PyyHsp20 genes at different developmental stages and different parts (Figure 8). First, RNA-seq data were used to explore the expression levels of PyyHsp20 genes in two different parts of the blade: the base and middle parts. Cell morphology and size differed significantly between the two parts of the blade. However, this finding has not yet been reported. As illustrated in Figure 8A, most PyyHsp20 genes were expressed in two parts, except for PyyHsp20-19.2 and PyyHsp20-25.9, which were barely expressed in any part. The expression levels of three PyyHsp20 genes (PyyHsp20-19.3, PyyHsp20-19.6, and PyyHsp20-37.6) in the base part were relatively higher than those in the middle part. Second, RNA-seq data were used to explore the expression levels of PyyHsp20 at two different developmental stages, including leafy gametophytes and filamentous sporophytes (Figure 8B). All PyyHsp20 genes were expressed in the sporophyte, and most PyyHsp20 genes were expressed in the gametophyte, except for PyyHsp20-19.2 and PyyHsp20-25.9, which were present at almost undetectable levels. The expression levels of PyyHsp20-19.2, PyyHsp20-19.3, PyyHsp20-37.4, and PyyHsp20-25.9 were higher in sporophytes than in gametophytes. Conversely, the expression levels of PyyHsp20-19.6, PyyHsp20-20.3, and PyyHsp20-37.6 in sporophytes were lower than those in gametophytes. These results indicate that PyyHsp20 genes showed spatial and temporal expression patterns during different developmental stages and in different parts of Py. yezoensis. In conclusion, these results suggested that PyyHsp20 plays a crucial role in the growth and development of Py. yezoensis.
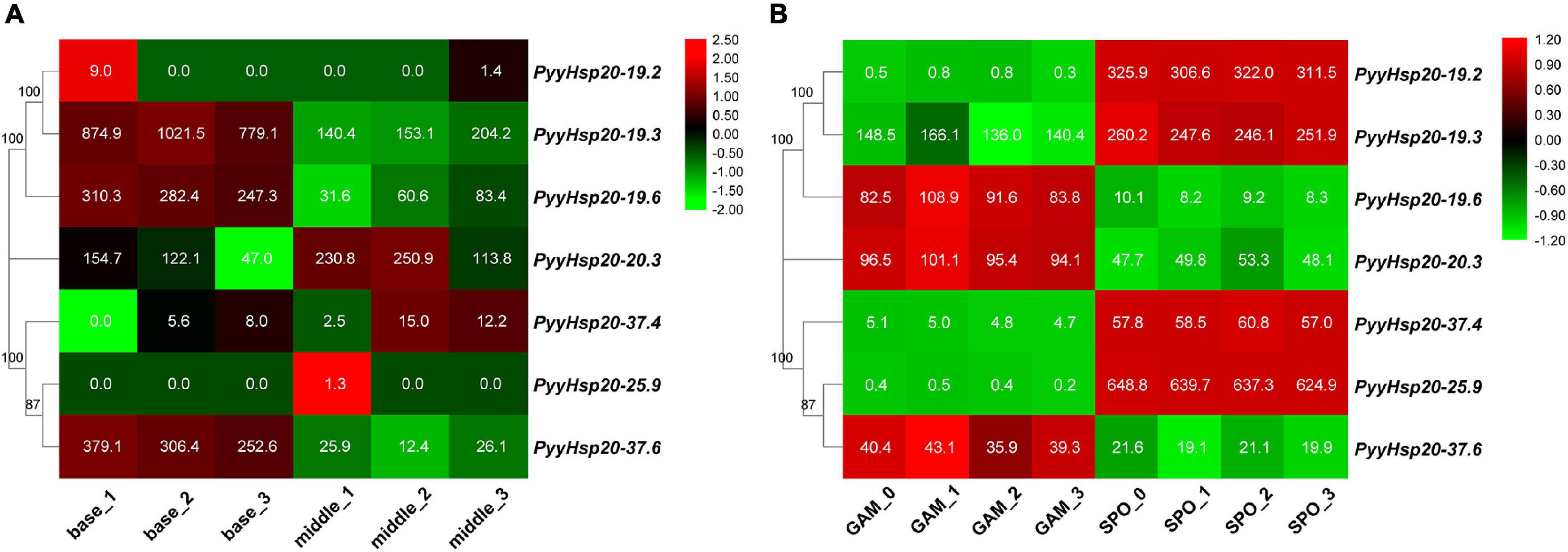
Figure 8. Heat map of the expression profiles of PyyHsp20 genes in various parts and developmental stages of Py. yezoensis. (A) Heat map of the expression profiles of PyyHsp20 genes in different parts of the blade in Py. yezoensis. Differential expression is shown in different colors. The number represents relative expression quantity. (B) Heat map of the expression profiles of PyyHsp20 genes in different developmental stages of Py. yezoensis. Differential expression is shown in different colors. The number represents relative expression quantity.
Although Hsp20 gene families have been investigated in many plant species, only a few Hsp20 genes have been identified in red algae. Moreover, a comprehensive analysis of the Hsp20 gene family in red algae is lacking. Therefore, we analyzed 17 representative species covering all seven classes of Rhodophyta with available genomic or transcriptomic information. Detailed information on the red algal Hsp20 genes is shown in Supplementary Table 4, including names, chromosome locations, intron numbers, protein lengths, molecular weights, pI values, and instability index. However, some information, such as chromosome locations and intron numbers, may be incomplete owing to the lack of a relevant reference genome. To date, only eight high-quality and complete reference genomes of red algae have been assembled, annotated, and made available through public databases (Matsuzaki et al., 2004; Bhattacharya et al., 2013; Collen et al., 2013; Schoenknecht et al., 2013; Brawley et al., 2017; Lee et al., 2018; Cao et al., 2020; Wang et al., 2020). In addition, some red algal protein data without reference genomes are available through EukProt (Richter et al., 2020). Although some of the red algal protein sequences in EukProt are incomplete, such as StHsp20-6.6, their Hsp20 domains are complete. Therefore, we chose these red algal Hsp20s to study the classification and origin of the red algal Hsp20 gene families. We believe that a unified classification for Rhodophyta would be beneficial for future studies, when genomic data are available for these taxa. In addition, these genomic data will soon be available owing to rapidly developing sequencing technologies and declining costs, which will be of great help in the study of the red algal Hsp20 gene families.
Protein subcellular localization is closely related to protein function, and its prediction is helpful for understanding protein function. In general, proteins can only perform their functions at specific subcellular positions (Wang et al., 2014). Currently, numerous programs or websites are available for the prediction of protein subcellular localization, such as WoLF PSORT, CELLO, Yloc, BUSCA, MULocDeep, and SeqNLS (Yu et al., 2006; Horton et al., 2007; Briesemeister et al., 2010; Lin and Hu, 2013; Savojardo et al., 2018; Jiang et al., 2021). However, no software has been specifically designed to predict the subcellular localization of red algae proteins. An earlier study reported that a subcellular localization prediction tool called PredAlgo is dedicated to green algae (Tardif et al., 2012). Tardif et al. (2012) also pointed out that currently available predictors are unreliable when used to predict the localization of algal proteins. However, the PredAlgo website is currently unavailable. In addition, the use of PredAlgo is inappropriate for red algal proteins (Mori et al., 2016). To accurately predict the subcellular location of red algal proteins, we used six protein subcellular localization prediction tools to predict the subcellular location of red algal proteins. The prediction of the subcellular localization of red algal Hsp20s is shown in Supplementary Table 5. Unfortunately, the results of the six tools are not entirely consistent. The reasons for this may be as follows: First, existing algorithms for protein subcellular localization prediction are not suitable for red algal proteins because of their unique cell structure, such as the presence of unstacked thylakoids and phycobilisomes in the chloroplast. Second, there is little experimental evidence of the subcellular localization of red algal proteins. Finally, we accepted the majority consensus. Therefore, it is necessary to develop a new algorithm with the specific aim of predicting red algae targeting. This will be helpful to study protein function in red algae. In addition, more experimental evidence about the subcellular localization of red algal proteins is required.
The gene structure plays a crucial role in the evolution of multiple gene families. The gene structure of Hsp20 proteins were investigated where genomic data were available. In addition, gene duplication, chromosome location, collinearity analysis, and cis-regulatory element analysis were explored in the Hsp20 proteins of eight red algae. Our results showed that most of the red algal Hsp20 genes contained no introns, and only three contained one intron, suggesting relatively simple gene structures. This is consistent with previous results in higher plants (Zhao et al., 2018). In some studies, genes with few or no introns were considered to be rapidly activated in response to various stresses (Jeffares et al., 2008). The instability index provides an estimate of protein stability in a test tube, which can be predicted, as described by Gasteiger et al. (2005). A protein whose instability index is smaller than 40 is predicted to be stable, and a value above 40 predicts that the protein may be unstable. In our study, the instability index of most red algae Hsp20s was greater than 40, indicating that most of them were unstable proteins (Supplementary Table 4). Instability is also considered to be a common trait of stress-responsive proteins. Hsp20 genes are considered as one of the rapidly expressed genes under various stress conditions. The absence of introns and the presence of instability may be in accordance with the need for rapid induction of Hsp20 genes in response to various stresses and rapid disposal of proteins after the stress response.
The function of Hsp20 genes has been systematically investigated in many higher plants (Scharf et al., 2001; Ouyang et al., 2009; Lopes-Caitar et al., 2013; Guo et al., 2015; Yu et al., 2016; Zhao et al., 2018). Hsp20 genes not only play important roles in plant responses to various stresses but are also involved in plant developmental processes (Yu et al., 2016). However, the function of Hsp20 in red algae is relatively unknown. Py. yezoensis is one of the most economically important marine red algae worldwide, and is recognized as an ideal model for studying the molecular mechanisms of stress resistance (Wang et al., 2020). In this study, Py. yezoensis was used as a representative of red algae to explore the expression profiles of Hsp20 genes under abiotic (dehydration/rehydration) and biotic (red rot disease) stresses. The data demonstrated that numerous PyyHsp20 genes were significantly induced to a larger extent under dehydration/rehydration stress and showed differential expression patterns, indicating that different PyyHsp20 genes may play different roles in response to dehydration/rehydration stress. Previous studies have shown that PyyHsp20 genes are induced by other abiotic stresses, such as heat, oxidative, and copper stress (Uji et al., 2019). Furthermore, the Hsp20 gene in Cy. merolae, and Pp. haitanensis are also induced by heat stress (Kobayashi et al., 2014; Chang et al., 2021). Several Hsp20 genes also participate in the interactions between plants and pathogens, such as viruses, bacteria, and fungi (Park and Seo, 2015). Tang et al. (2019) previously found that PyyHsp20 genes showed increased expression during the slight and severe stages of the oomycete pathogen Pythium porphyrae infection. In the present study, we found that all PyyHsp20 genes are induced by pathogen infection. These results were inconsistent with the report that biotic stress can induce the expression of some, but not all Hsp20 genes (Li and Liu, 2019). In higher plants, spatiotemporal regulation of the Hsp20 gene family has been observed in various tissues and developmental stages (Yu et al., 2016). In our study, spatiotemporal regulation of the Hsp20 gene family was also observed in different parts and developmental stages of Py. yezoensis. Py. yezoensis exhibits a haploid-diploid heteromorphic life cycle with a haploid macroscopic blade-forming gametophyte and a diploid microscopic filamentous sporophyte. In addition, Py. yezoensis require fertilization and meiosis for the transition from gametophytes to sporophytes and from sporophytes to gametophytes, respectively. In our study, we found that some PyyHsp20 genes were highly expressed in sporophytes, whereas some PyyHsp20 genes were highly expressed in gametophytes. In the two different parts of the blade, PyyHsp20 genes also displayed differential expression. These results indicate that Py. yezoensis seems to have established a sophisticated mechanism to tightly regulate the expression of PyyHsp20 genes, where and when required.
Cis-regulatory elements are important molecular switches involved in the transcriptional regulation of gene expression and control various biological processes, including stress responses, hormone responses, and developmental processes (Yamaguchi-Shinozaki and Shinozaki, 2005). Cis-regulatory element predictions have been widely used to explore the functions of Hsp20s in several species (Wang et al., 2021). In our study, various stress-responsive, phytohormone-responsive, light-responsive, and plant development-related cis-elements were found in the promoter regions of red algal Hsp20 genes (Figure 6). Among the four cis-element categories, the phytohormone-responsive category accounted for the highest proportion. Phytohormones are a large category of small endogenous, low-molecular-weight molecules that not only regulate plant growth and development at low concentrations, but can also act as signaling molecules that participate in plant responses to environmental stresses. Phytohormones such as auxin, ethylene, jasmonic acid, salicylic acid (SA), abscisic acid (ABA), cytokinin (CK), and gibberellins (GAs) can play important roles in plant development and stress responses (Saidi and Hajibarat, 2019). Emerging studies suggest that red algae contain phytohormones such as auxin, ethylene, ABA, CK, GAs, jasmonic acid, SA, and methyl jasmonate (MeJA) (Lu and Xu, 2015; Song et al., 2017). In higher plants, phytohormones have been shown to be linked to heat stress signaling and modulate the expression of Hsp under heat stress (Zhang and Wang, 2011; Nazar et al., 2017). Previous studies have reported that Hsp20 genes can be induced by the exogenous application of MeJA, MeJA/SA, and the ethylene precursor 1-aminocylopropane-1-carboxylic acid (ACC) in the red algae Ch. crispus, Gracilariopsis lemaneiformis, and Py. yezoensis (Collen et al., 2006; Wang et al., 2017; Uji et al., 2019). In addition, cis-regulatory elements in the promoters are involved in the cross-talk of different stress signals in gene expression and constitute gene expression cascades during abiotic stress responses and control the molecular processes of stress responses and stress tolerance (Yamaguchi-Shinozaki and Shinozaki, 2005). Thus, we may assume a crosstalk between Hsp20s and phytohormones in the response of red algae to environmental stresses. Taken together, these genes may play important roles in regulating the growth, development, and stress responses of red algae. However, further analysis is needed to investigate how red algal Hsp20 genes perform their functions.
Hsp20s is a ubiquitous protein family found in archaea, bacteria, and eukaryotes. All organisms possess Hsp20s, indicating that these proteins evolved very early, prior to the divergence of the three domains of life (Archaea, Bacteria, and Eukarya) (Waters, 2013). A previous study reported that Hsp20s was the first in the last universal common ancestor (LUCA) (Rebeaud et al., 2021). Plant Hsp20s are particularly diverse and numerous in number. In evolutionary terms, they have exhibited lineage-specific expansion patterns (Waters and Vierling, 2020). According to their subcellular localization, plant Hsp20s are classified into several subfamilies (CI-CVI, MTI, MTII, ER, CP, and PX) (Waters and Vierling, 2020). Previous studies based on the limited number of algal Hsp20s sequences suggested that none of the algal Hsp20s sequences were found within the land plant lineage of cytosolic and organelle-localized proteins (Waters and Rioflorido, 2007). However, it is important to note that although algae do not possess close homologs of land plant CP Hsp20s, chloroplast-targeted Hsp20s are present in Cl. reinhardtii (Waters and Vierling, 2020). Further analysis indicated that algal Hsp20s are closely related only to Hsp20s from the same species (Waters and Rioflorido, 2007), which is different from the higher plant Hsp20s. Higher plant Hsp20s are more closely related to members of the same subfamily from divergent species than to other Hsp20s from the same species (Waters et al., 1996). In our study, red algal Hsp20s were classified as not based on predicted subcellular localizations. This might be because the lack of experimental data for the red algae organisms studied and the existing tools used to predict subcellular localization are inaccurate for red algae, or because the red algal Hsp20 gene families might have distinct but currently unclear classification mechanisms. In addition, we identified two types of subfamilies in the red algal Hsp20 gene families. One type was a class-specific subfamily that included members from the same class. The other type is a broadly distributed subfamily that includes members from different classes (Figure 1). In the broadly distributed subfamily, red algal Hsp20 genes were always closely related to Hsp20 genes from the same species. This suggests that a significant expansion of red algal Hsp20 genes occurred after the divergence of the red algae (Rhodophyta). Furthermore, we found that only four of 97 red algal Hsp20s belonging to the class Stylonematophyceae possessed three ACDs, and the other red algal Hsp20s possessed only one ACD. Moreover, among these 12 ACDs, ACDs were more closely related to ACDs from different Hsp20s than to ACDs from the same Hsp20s. This implies that Hsp20 genes in the class Stylonematophyceae might have experienced a specific evolutionary process. In our study, red algal Hsp20s were distinct from those of land plants and green algae, indicating that Hsp20s probably evolved independently in red algae and plants (green algae) (Figure 2). Furthermore, some red algal Hsp20s clustered with bacterial Hsp20s, indicating that these red algal Hsp20s may originate from bacteria via HGT, while some red algal Hsp20s were not clustered with bacterial Hsp20s. Taken together, Hsp20 genes may have a polyphyletic origin and a complex evolutionary pattern. In addition, plant Hsp20s clustered according to cellular localization, which was consistent with the findings of a previous study (Huang et al., 2008; Waters and Vierling, 2020). Our study provides new insights into the evolution of Hsp20 gene families in red algae.
In this study, 97 putative Hsp20 genes were identified in red algae. A comprehensive analysis of red algal Hsp20 genes using gene structures, conserved motifs, phylogenetic relationships, chromosome location, gene duplication, and cis-regulatory elements was performed. Finally, Py. yezoensis was selected as a representative of red algae to explore the expression profiles of Hsp20 genes under different stresses, at different developmental stages, and in different parts of the blade using RNA-seq data. This study provides comprehensive information on Hsp20 gene families in red algae and lays a foundation for their functional characterization. In addition, our study provides new insights into the evolution of Hsp20 gene families in red algae and can help understand the adaptability of red algae to diverse environments.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
TG collected the public dataset, analyzed the data, and performed the experiments. TG and ZM drafted the manuscript. LT and XY contributed to bioinformatics analysis and gene expression profile analysis. ZM and YM conceived and designed the study. GD and YM reviewed the manuscript. All authors have read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant No.32060829), the National Key R&D Program of China (2020YFD0901101), the Fundamental Research Funds for the Central Universities (202064006), the 2020 Research Program of Sanya Yazhou Bay Science and Technology City (No. SKJC-2020-02-009), Hainan Provincial Natural Science Foundation of China (320QN292), China Postdoctoral Science Foundation (2021M703033), Special Project of Central Government Guiding Local Science and Technology Development (ZY2020HN02) and the Innovation Platform for Academicians of Hainan Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.817852/full#supplementary-material
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bhattacharya, D., Price, D. C., Chan, C. X., Qiu, H., Rose, N., Ball, S., et al. (2013). Genome of the red alga Porphyridium purpureum. Nat. Commun. 4:1941. doi: 10.1038/ncomms2931
Bourgine, B., and Guihur, A. (2021). Heat shock signaling in land plants: from plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 12:710801. doi: 10.3389/fpls.2021.710801
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Wheeler, G. L., Lohr, M., Goodson, H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. U.S.A. 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Briesemeister, S., Rahnenfuehrer, J., and Kohlbacher, O. (2010). YLoc-an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38, W497–W502. doi: 10.1093/nar/gkq477
Cao, M., Xu, K., Yu, X., Bi, G., Liu, Y., Kong, F., et al. (2020). A chromosome-level genome assembly of Pyropia haitanensis (Bangiales. Rhodophyta). Mol. Ecol. Resour. 20, 216–227. doi: 10.1111/1755-0998.13102
Caspers, G. J., Leunissen, J. A. M., and Dejong, W. W. (1995). The expanding small heat-shock protein family, and structure predictions of the conserved alpha-crystallin domain. J. Mol. Evol. 40, 238–248. doi: 10.1007/bf00163229
Chang, J., Shi, J., Lin, J., Ji, D., Xu, Y., Chen, C., et al. (2021). Molecular mechanism underlying Pyropia haitanensis PhHsp22-mediated increase in the high-temperature tolerance of Chlamydomonas reinhardtii. J. Appl. Phycol. 33, 1137–1148. doi: 10.1007/s10811-020-02351-6
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Collen, J., Herve, C., Guisle-Marsollier, I., Leger, J. J., and Boyen, C. (2006). Expression profiling of Chondrus crispus (Rhodophyta) after exposure to methyl jasmonate. J. Exp. Bot. 57, 3869–3881. doi: 10.1093/jxb/erl171
Collen, J., Porcel, B., Carre, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
de Jong, W. W., Caspers, G.-J., and Leunissen, J. A. (1998). Genealogy of the α-crystallin—small heat-shock protein superfamily. Int. J. Biol. Macromol. 22, 151–162. doi: 10.1016/s0141-8130(98)00013-0
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the expasy server,” in Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Guo, M., Liu, J.-H., Lu, J.-P., Zhai, Y.-F., Wang, H., Gong, Z.-H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6:806. doi: 10.3389/fpls.2015.00806
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522.
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Huang, L.-H., Wang, H.-S., and Kang, L. (2008). Different evolutionary lineages of large and small heat shock proteins in eukaryotes. Cell Res. 18, 1074–1076. doi: 10.1038/cr.2008.283
Jeffares, D. C., Penkett, C. J., and Baehler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jiang, Y., Wang, D., Yao, Y., Eubel, H., Kunzler, P., Moller, I. M., et al. (2021). MULocDeep: a deep-learning framework for protein subcellular and suborganellar localization prediction with residue-level interpretation. Comput. Struct. Biotechnol. J. 19, 4825–4839. doi: 10.1016/j.csbj.2021.08.027
Kalyaanamoorthy, S., Bui Quang, M., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kobayashi, Y., Harada, N., Nishimura, Y., Saito, T., Nakamura, M., Fujiwara, T., et al. (2014). Algae sense exact temperatures: small heat shock proteins are expressed at the survival threshold temperature in Cyanidioschyzon merolae and Chlamydomonas reinhardtii. Genome Biol. Evol. 6, 2731–2740. doi: 10.1093/gbe/evu216
Lee, J., Yang, E. C., Graf, L., Yang, J. H., Qiu, H., Zelzion, U., et al. (2018). Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in Rhodophyta. Mol. Biolo. Evol. 35, 1869–1886. doi: 10.1093/molbev/msy081
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., and Liu, X. (2019). Genome-wide identification and expression profile analysis of the Hsp20 gene family in Barley (Hordeum vulgare L.). PeerJ 7:e6832. doi: 10.7717/peerj.6832
Lin, J.-R., and Hu, J. (2013). SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One 8:e0076864. doi: 10.1371/journal.pone.0076864
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Lopes-Caitar, V. S., de Carvalho, M. C. C. G., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14:577. doi: 10.1186/1471-2164-14-577
Lopez-Bautista, J. M. (2010). “Red algal genomics: a synopsis,” in Red Algae in the Genomic Age, eds J. Seckbach, & D.J. Chapman (Dordrecht: Springer) 227, 229–240.
Lu, Y., and Xu, J. (2015). Phytohormones in microalgae: a new opportunity for microalgal biotechnology? Trends Plant Sci. 20, 273–282. doi: 10.1016/j.tplants.2015.01.006
Matsuzaki, M., Misumi, O., Shin-I, T., Maruyama, S., Takahara, M., Miyagishima, S. Y., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mittal, D., Enoki, Y., Lavania, D., Singh, A., Sakurai, H., and Grover, A. (2011). Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L.). FEBS J. 278, 3076–3085. doi: 10.1111/j.1742-4658.2011.08229.x
Mori, N., Moriyama, T., Toyoshima, M., and Sato, N. (2016). Construction of global acyl lipid metabolic map by comparative genomics and subcellular localization analysis in the red alga Cyanidioschyzon merolae. Front. Plant Sci. 7:958. doi: 10.3389/fpls.2016.00958
Nazar, R., Iqbal, N., and Umar, S. (2017). “Heat stress tolerance in plants: action of salicylic acid,” in Salicylic Acid: A Multifaceted Hormone, eds R. Nazar, N. Iqbal, and N. Khan (Berlin: Springer), 145–161.
Ouyang, Y., Chen, J., Xie, W., Wang, L., and Zhang, Q. (2009). Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol. Biol. 70, 341–357. doi: 10.1007/s11103-009-9477-y
Park, C.-J., and Seo, Y.-S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323–333. doi: 10.5423/ppj.Rw.08.2015.0150
Price, D. C., Goodenough, U. W., Roth, R., Lee, J.-H., Kariyawasam, T., Mutwil, M., et al. (2019). Analysis of an improved Cyanophora paradoxa genome assembly. DNA Res. 26, 287–299. doi: 10.1093/dnares/dsz009
Rebeaud, M. E., Mallik, S., Goloubinoff, P., and Tawfik, D. S. (2021). On the evolution of chaperones and cochaperones and the expansion of proteomes across the tree of life. Proc. Natl. Acad. Sci. U.S.A. 118:e2020885118. doi: 10.1073/pnas.2020885118
Reddy, C. R. K., Gupta, V., and Jha, B. (2010). “Developments in biotechnology of red algae,” in Red Algae in the Genomic Age, eds J. Seckbach and D. J. Chapman (Dordrecht: Springer), 307–341.
Richter, D., Berney, C., Strassert, J., Burki, F., and de Vargas, C. (2020). EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotic life. bioRxiv [Preprint] doi: 10.1101/2020.06.30.180687
Saidi, A., and Hajibarat, Z. (2019). Characterization of cis-elements in hormonal stress-responsive genes in Oryza sativa. AsPac. J. Mol. Biol. Biotechnol. 27, 95–102.
Savojardo, C., Martelli, P. L., Fariselli, P., Profiti, G., and Casadio, R. (2018). BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 46, W459–W466. doi: 10.1093/nar/gky320
Scharf, K. D., Siddique, M., and Vierling, E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6, 225–237.
Schoenknecht, G., Chen, W.-H., Ternes, C. M., Barbier, G. G., Shrestha, R. P., Stanke, M., et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210. doi: 10.1126/science.1231707
Song, Y., Cui, X., Chen, J., Yang, R., and Yan, X. (2017). The profiling of eleven phytohormones in Pyropia haitanensis under different high-temperature environments. J. Fish. China 41, 1578–1587.
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sun, X., Sun, C., Li, Z., Hu, Q., Han, L., and Luo, H. (2016). AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 39, 1320–1337. doi: 10.1111/pce.12683
Tang, L., Qiu, L., Liu, C., Du, G., Mo, Z., Tang, X., et al. (2019). Transcriptomic insights into innate immunity responding to red rot disease in red alga Pyropia yezoensis. Int. J. Mol. Sci. 20:5970. doi: 10.3390/ijms20235970
Tardif, M., Atteia, A., Specht, M., Cogne, G., Rolland, N., Brugiere, S., et al. (2012). PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29, 3625–3639. doi: 10.1093/molbev/mss178
Uji, T., Gondaira, Y., Fukuda, S., Mizuta, H., and Saga, N. (2019). Characterization and expression profiles of small heat shock proteins in the marine red alga Pyropia yezoensis. Cell Stress Chaperones 24, 223–233. doi: 10.1007/s12192-018-00959-9
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 42, 579–620.
Wang, D., Yu, X., Xu, K., Bi, G., Cao, M., Zelzion, E., et al. (2020). Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 11:4028. doi: 10.1038/s41467s-020-17689-1
Wang, F., Wang, C., Zou, T., Xu, N., and Sun, X. (2017). Comparative transcriptional profiling of Gracilariopsis lemaneiformis in response to salicylic acid- and methyl jasmonate-mediated heat resistance. PLoS One 12:e0176531. doi: 10.1371/journal.pone.0176531
Wang, W. X., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, X. A., Zheng, Y., Chen, B., Zhi, C., Qiao, L., Liu, C., et al. (2021). Genome-wide identification of small heat shock protein (HSP20) homologs in three cucurbit species and the expression profiles of CsHSP20s under several abiotic stresses. Int. J. Biol. Macromol. 190, 827–836. doi: 10.1016/j.ijbiomac.2021.08.222
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z., Zou, Q., Jiang, Y., Ju, Y., and Zeng, X. (2014). Review of protein subcellular localization prediction. Curr. Bioinform. 9, 331–342. doi: 10.2174/1574893609666140212000304
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Waters, E. R., Lee, G. J., and Vierling, E. (1996). Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 47, 325–338. doi: 10.1093/jxb/47.3.325
Waters, E. R., and Rioflorido, I. (2007). Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J. Mol. Evol. 65, 162–174. doi: 10.1007/s00239-006-0223-7
Waters, E. R., and Vierling, E. (2020). Plant small heat shock proteins – evolutionary and functional diversity. New Phytol. 227, 24–37. doi: 10.1111/nph.16536
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94.
Yoon, H. S., Zuccarello, G. C., and Bhattacharya, D. (2010). “Evolutionary history and taxonomy of red algae,” in Red Algae in the Genomic Age, eds J. Seckbach and D. J. Chapman (Dordrecht: Springer), 25–42.
Yu, C.-S., Chen, Y.-C., Lu, C.-H., and Hwang, J.-K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yu, J., Cheng, Y., Feng, K., Ruan, M., Ye, Q., Wang, R., et al. (2016). Genome-wide identification and expression profiling of tomato hsp20 gene family in response to biotic and abiotic stresses. Front. Plant Sci. 7:1215. doi: 10.3389/fpls.2016.01215
Zhang, S., and Wang, X. (2011). Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 168, 2093–2101. doi: 10.1016/j.jplph.2011.06.010
Keywords: heat shock protein 20, red algae, phylogenetic analysis, expression profile, abiotic and biotic stresses, growth and development
Citation: Gao T, Mo Z, Tang L, Yu X, Du G and Mao Y (2022) Heat Shock Protein 20 Gene Superfamilies in Red Algae: Evolutionary and Functional Diversities. Front. Plant Sci. 13:817852. doi: 10.3389/fpls.2022.817852
Received: 18 November 2021; Accepted: 11 February 2022;
Published: 16 March 2022.
Edited by:
Eric Marechal, UMR 5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Anthony Guihur, University of Lausanne, SwitzerlandCopyright © 2022 Gao, Mo, Tang, Yu, Du and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunxiang Mao, eXhtYW9AaG50b3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.