- Department of Life Sciences, Korea University, Seoul, South Korea
RNA polymerase II–associated factor 1 complex (PAF1C) regulates the transition from the vegetative to the reproductive phase primarily by modulating the expression of FLOWERING LOCUS C (FLC) and FLOWERING LOCUS M [FLM, also known as MADS AFFECTING FLOWERING1 (MAF1)] at standard growth temperatures. However, the role of PAF1C in the regulation of flowering time at chilling temperatures (i.e., cold temperatures that are above freezing) and whether PAF1C affects other FLC-clade genes (MAF2–MAF5) remains unknown. Here, we showed that Arabidopsis thaliana mutants of any of the six known genes that encode components of PAF1C [CELL DIVISION CYCLE73/PLANT HOMOLOGOUS TO PARAFIBROMIN, VERNALIZATION INDEPENDENCE2 (VIP2)/EARLY FLOWERING7 (ELF7), VIP3, VIP4, VIP5, and VIP6/ELF8] showed temperature-insensitive early flowering across a broad temperature range (10°C–27°C). Flowering of PAF1C-deficient mutants at 10°C was even earlier than that in flc, flm, and flc flm mutants, suggesting that PAF1C regulates additional factors. Indeed, RNA sequencing (RNA-Seq) of PAF1C-deficient mutants revealed downregulation of MAF2–MAF5 in addition to FLC and FLM at both 10 and 23°C. Consistent with the reduced expression of FLC and the FLC-clade members FLM/MAF1 and MAF2–MAF5, chromatin immunoprecipitation (ChIP)-quantitative PCR assays showed reduced levels of the permissive epigenetic modification H3K4me3/H3K36me3 and increased levels of the repressive modification H3K27me3 at their chromatin. Knocking down MAF2–MAF5 using artificial microRNAs (amiRNAs) in the flc flm background (35S::amiR-MAF2–5 flc flm) resulted in significantly earlier flowering than flc flm mutants and even earlier than short vegetative phase (svp) mutants at 10°C. Wild-type seedlings showed higher accumulation of FLC and FLC-clade gene transcripts at 10°C compared to 23°C. Our yeast two-hybrid assays and in vivo co-immunoprecipitation (Co-IP) analyses revealed that MAF2–MAF5 directly interact with the prominent floral repressor SVP. Late flowering caused by SVP overexpression was almost completely suppressed by the elf7 and vip4 mutations, suggesting that SVP-mediated floral repression required a functional PAF1C. Taken together, our results showed that PAF1C regulates the transcription of FLC and FLC-clade genes to modulate temperature-responsive flowering at a broad range of temperatures and that the interaction between SVP and these FLC-clade proteins is important for floral repression.
Introduction
Plant survival and fitness depends on timely seed production through precise control of flowering time. Flowering time is modulated by a number of endogenous and environmental cues, such as daylength, age, and prolonged exposure to cold and ambient temperatures (Amasino, 2010; Srikanth and Schmid, 2011). To successfully survive a range of varying environmental conditions, plants have evolved a complex regulatory network that integrates these cues to control flowering time (Srikanth and Schmid, 2011). In Arabidopsis (Arabidopsis thaliana), nearly 400 flowering genes are known to regulate flowering time in genetically distinct pathways, e.g., the photoperiod, ambient temperature, aging, vernalization, hormonal, and sugar pathways (Bernier and Périlleux, 2005; Bouché et al., 2016). These pathways modulate flowering in response to different endogenous and environmental signals to optimize reproductive success.
Among the flowering time genes, FLOWERING LOCUS C (FLC), encoding a MADS-box transcription factor, is an important repressor of flowering in various plant species, including Arabidopsis (Michaels and Amasino, 1999; Sheldon et al., 1999; Ruelens et al., 2013). In Arabidopsis winter accessions, FRIGIDA (FRI) induces FLC transcription, whereas vernalization (prolonged exposure to cold) epigenetically represses FLC transcription (Johanson et al., 2000; Choi et al., 2011). By contrast, in rapid-cycling accessions lacking functional FRI, members of the autonomous pathway regulate FLC transcription (Amasino, 2010). FLC negatively regulates flowering by directly repressing the transcription of two important genes that promote flowering, FLOWERING LOCUS T (FT), and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1; Helliwell et al., 2006; Searle et al., 2006; Li et al., 2008).
In addition to FLC, the Arabidopsis genome contains five more members of the FLC clade, e.g., MADS AFFECTING FLOWERING1 [MAF1, also known as FLOWERING LOCUS M (FLM)] and MAF2–MAF5 (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001). The MAF2–MAF5 genes occur in a tandem repeat within a 22-kb region. The role of FLM as a floral repressor has been well-studied (Scortecci et al., 2003; Balasubramanian et al., 2006), and its loss of function causes temperature-insensitive flowering (Lee et al., 2013). Like FLM, MAF2 acts as a floral repressor, and the loss of MAF2 function results in strong acceleration of flowering upon vernalization, whereas plants overexpressing MAF2 flowered significantly later than wild-type plants (Ratcliffe et al., 2003). Similarly, MAF3 also represses flowering by directly binding to the promoter sequences of FT and SOC1 and repressing their transcription. Interestingly, the effect of the loss of MAF3 function was more evident at lower temperatures than at normal growth temperatures (Gu et al., 2013). Overexpression of MAF3 produced a stronger floral delay in the Landsberg erecta (Ler) accession compared to Columbia (Col-0; Ratcliffe et al., 2003). T-DNA mutants of MAF4 exhibited accelerated flowering (Gu et al., 2009), and MAF4 overexpression in Ler resulted in a strong delay of flowering (Scortecci et al., 2003). Unlike MAF1–MAF4, the floral repressive effect of MAF5 is not strong, and its overexpression only delayed flowering under non-inductive short-day conditions (Kim and Sung, 2010). These FLC-clade transcription factors physically interact with each other, and some of them interact with SHORT VEGETATIVE PHASE (SVP) to make repressor complexes for efficient floral repression (Gu et al., 2013).
Expression of FLC-clade genes is epigenetically regulated by a number of histone modifiers that are recruited to these loci by Polymerase II-Associated Factor 1 Complex (PAF1C; Zhang and Van Nocker, 2002; Zhang et al., 2003; Oh et al., 2004). In Arabidopsis, PAF1C components include CELL DIVISION CYCLE73 (CDC73)/PLANT HOMOLOGOUS TO PARAFIBROMIN (PHP), VERNALIZATION INDEPENDENCE2 (VIP2)/EARLY FLOWERING7 (ELF7), VIP3, VIP4, VIP5, and VIP6/ELF8 (Zhang and Van Nocker, 2002; Zhang et al., 2003; He, 2009; Yu and Michaels, 2010). Components of PAF1C interact with RNA Polymerase II (Betz et al., 2002) and recruit H3K4 methyltransferase to their target genes, primarily in the 5′ transcribed regions, thereby leading to the active expression of their target genes (Ng et al., 2003b). In addition to methyltransferases, PAF1C interacts with the splicing factor SKI-INTERACTING PROTEIN (SKIP) to modulate expression of FLC and FLM (Cao et al., 2015; Li et al., 2019).
Genetic studies revealed that lesions in these PAF1C components resulted in nearly identical early flowering phenotypes under standard growth conditions (He et al., 2004; Oh et al., 2004). PAF1C is required for the enrichment of active epigenetic marks, primarily H3K4me3, at the chromatin of FLC and its homologs to maintain their expression; PAF1C deficiency results in reduced expression of these floral repressors, which eventually accelerates flowering (Zhang et al., 2003; He et al., 2004; Yu and Michaels, 2010). For instance, loss of function of ELF7 and VIP6 results in the reduced expression of FLC, FLM/MAF1, and MAF2 (He et al., 2004). Mutations in VIP3 strongly reduced FLC expression and strongly accelerated flowering. In particular, vip3 mutants flowered significantly earlier than flc mutants, suggesting that additional genes are involved in the early flowering of vip3 mutants (Zhang et al., 2003). Loss of function of VIP4 and VIP5 also resulted in strong flowering acceleration that was comparable to that seen in vip3 single mutants. However, after vernalization, the H3K27 methyltransferase complex Polycomb repressive complex 2 (PRC2) is involved in silencing FLC expression (and probably MAF expression) by depositing repressive H3K27me3 marks on the FLC chromatin (Wood et al., 2006; De Lucia et al., 2008). Although the regulation of FLC (and some MAF genes) is well-studied under standard growth temperature conditions, whether PAF1C-mediated regulation involves the entire FLC clade and the functional importance of this clade in regulating flowering at chilling temperatures remain unclear.
Here, we showed that PAF1C epigenetically regulates the entire set of FLC-clade genes. PAF1C-defective mutants showed ambient temperature-insensitive early flowering due to the downregulation of FLC-clade genes. The epigenetic status of the chromatin of FLC and FLC-clade genes was altered in PAF1C-defective mutants. Expression of FLC and FLC-clade genes was upregulated in response to low temperature in wild-type plants, and these genes play an important role in floral repression at chilling temperatures. Furthermore, MAF2–MAF5 physically interacted with SVP, and SVP-mediated floral repression requires PAF1C, suggesting the possibility that larger repressive complexes form to prevent precocious flowering, especially at chilling temperatures.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutant lines for ELF7 (elf7-2; SALK_ 046605 and SALK_070632; hereafter elf7-4), CDC73 (cdc73-1; SALK_150644 and cdc73-2; SALK_008357), VIP3 (vip3-2; SALK_083364 and vip3-6; SALK_060207), VIP4 (vip4-1; SALK_122755 and vip4-3; SALK_006392), VIP5 (vip5-2; SALK_062223 and SALK_055889; hereafter vip5-3), and VIP6 (vip6-2; SALK_065364 and SALK_119910, hereafter vip6-5) were obtained from the ABRC. Genotyping of the mutant lines was performed using the primers described in Supplementary Table 1. The effect of the T-DNA insertion on gene expression was confirmed via RT-PCR for the uncharacterized mutant lines (elf7-4, vip5-3, and vip6-5). The 35S::SVP:HA lines were previously generated (Cho et al., 2012). For expression analyses at different temperatures, seedlings from the identical developmental stage of 1.02 (8-day-old seedlings at 23°C and 22-day-old seedlings at 10°C; Boyes et al., 2001) grown under standard long-day (LD) conditions (16:8 h light:dark) were used. LED lights with a light intensity of 120 μmol m−2 s−1 were used in this study. Flowering time was measured as the total leaf number, and the data are presented as box plots generated using the “PlotsOfData” app in the R package shiny (Postma and Goedhart, 2019), with customized settings.
RNA Sequencing
RNA sequencing (RNA-Seq) was performed using 8-day-old seedlings grown at 23°C and 23-day-old seedlings grown at 10°C under standard LD conditions in two biological replicates for each sample. About 60–80 seedlings were harvested at Zeitgeber Time 16 (ZT16) and pooled for RNA extraction using Invitrogen’s Plant RNA Purification Reagent. For RNA sequencing, library preparation was performed with an Illumina TruSeq Stranded Total RNA Sample Prep kit (Illumina), according to the manufacturer’s protocols, and paired-end reads were produced using an Illumina HiSeq2000 sequencer. The raw RNA-seq data of PAF1C-deficient mutants generated in this study were deposited at the Gene Expression Omnibus (GEO) NCBI database and are available under the accession number GSE171778. Transcriptome data for sdg8 mutants (GEO accession number GSE8528) were previously published (Pajoro et al., 2017).
RNA-Seq Data Analyses
The raw sequence reads were processed by adapter trimming, followed by qualitative analysis of raw reads using FastQC.1 The resulting good quality reads were aligned to the TAIR10 reference genome using CLC Genomics Workbench v.11. Differentially expressed genes (DEGs) were defined as genes with at least a 1.5-fold change, unless mentioned otherwise. Heatmaps were generated using the built-in function of CLC Genomics Workbench. Gene Ontology (GO) analysis was performed with DAVID (Ashburner et al., 2000), and GO enrichment data plotting was performed using the R package ggplot2 (Wickham, 2011). For the identification of common targets, the intersection of the gene lists was identified using the R package UpSetR (Lex et al., 2014) and the Java-based program VennDis (Ignatchenko et al., 2015).
Reverse Transcription Quantitative PCR Analyses
Reverse transcription quantitative PCR (RT-qPCR) was used to validate the RNA-seq data obtained from the PAF1C-deficient mutants. Total RNA was extracted from seedlings at the identical developmental stage at ZT16 at different temperatures. Plant RNA purification reagent (Invitrogen) was used for RNA extraction. The DNase I–treated RNA (~2 μg) was reverse transcribed into cDNA using MMLV enzyme (ELPIS Biotech). qPCR analyses of cDNA or immunoprecipitated DNA (see below) were performed using ×2 A-Star Real Time PCR Master Mix (BioFACT) in a Thermo Fisher QuantStudio 5 real-time PCR machine. All qPCR experiments were conducted in three biological replicates, each with three technical replicates.
For RT-qPCR, data analyses were performed according to the previously published ΔΔCT method (Livak and Schmittgen, 2001), with the modification of using two reference genes, PP2AA3 (AT1G13320) and a SAND family gene (AT2G28390) to normalize the data (Hong et al., 2010). Data normalization was performed using the geometric mean of the two reference genes. Sequences of primers used in RT-qPCR analyses are shown in Supplementary Table 1. The statistical significance of differences in gene expression levels among samples was assessed using one-way ANOVA with a 0.05 level of significance (95% CI).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays were performed using wild-type and vip3 mutant seedlings, as described previously (Susila et al., 2021). Briefly, seedlings of each genotype were harvested at the 1.02 stage (Boyes et al., 2001) and crosslinked using fixation buffer (1% formaldehyde). Immunoprecipitation was performed with polyclonal anti-H3K4me3 (Millipore, 04-745) or anti-H3K27me3 (Millipore, 07-449) antibodies bound to Dynabead Protein A (Thermo Scientific). The ChIPed DNA was extracted using the ChIP DNA Clean & Concentrator kit (Zymo Research). Relative enrichment of histone modifications was analyzed using qPCR as described earlier (Susila et al., 2021). The primers used in ChIP-qPCR are shown in Supplementary Table 1. All ChIP experiments were performed with three biological replicates and three technical replicates for each genotype.
Designing and Cloning amiRNAs That Target MAF Genes
To posttranscriptionally knock down the MAF2–MAF5 genes, artificial microRNAs (amiRNAs) were designed using the WMD3 webtool (Schwab et al., 2006). Predominantly expressed splice variants of MAF2–MAF5 (MAF2.3, MAF3.1, MAF4.3, and MAF5.2) were selected from the Araport11 cDNA collection and used as target genes for subsequent studies. Two independent amiRNAs were selected and amplified using pRS300 as a template with four amiRNA-specific primers (Supplementary Table 1), as previously described (Schwab et al., 2006). The amplified amiRNAs were cloned into the pENTR2B vector and subsequently into the pEG100 vector containing the 35S promoter.
Protein–Protein Interaction Analyses Using Deep Learning Algorithms
To test whether MAF2–MAF5 interact with SVP, we used two recently developed artificial intelligence (AI)–based deep learning programs, D-SCRIPT (Sledzieski et al., 2021) and PPI-Detect (Romero-Molina et al., 2019). Protein sequences were provided in fasta format as an input for both programs. Protein phosphatase 2A A3 (PP2AA3) was used as a negative interactor control.
Yeast Two-Hybrid Assays
For confirmation of the physical interaction between MAF2–MAF5 and SVP, the full-length coding sequences of the predominantly expressed splice variants of MAF2–MAF5 (MAF2.3, MAF3.1, MAF4.3, and MAF5.2) were fused in-frame to the DNA binding domain (BD) in the pGBKT7 vector. For the activation domain (AD)–fused SVP, full-length coding sequences of SVP were fused to the GAL4 AD in the pGADT7 vector. The double transformation was performed by introducing a combination of SVP with the MAF2–MAF5 vectors into the yeast strain AH109.
Co-immunoprecipitation Assays
Co-immunoprecipitation (Co-IP) experiments were performed in Arabidopsis mesophyll protoplasts as described earlier (Wu et al., 2009). The coding sequences of MAF2–MAF5 were fused with GFP (35S::MAF-GFP) in the 326-GFP vector (Lee et al., 2001) and co-transfected with 35S::SVP-2HA in the protoplasts isolated from wild-type plants. The transfected protoplasts were incubated at 23°C for 3 h to allow production of these proteins in sufficient quantities before overnight incubation at 10°C. After incubation, the protoplasts were pelleted and lysed with lysis buffer (10 mM Tris–HCl, pH 7.5, with.1% Triton X-100, and ×1 Roche Protease Inhibitor Cocktail). The lysate was then incubated overnight with GFP-Trap magnetic beads (Chromotek). Anti-GFP monoclonal antibodies (Roche) and anti-HA high-affinity monoclonal antibody clone 3F10 (Sigma-Aldrich) were used as primary antibodies for the western blots.
Results
PAF1C-Deficient Mutants Flower Early at a Broad Range of Temperatures
To test the effect of a lesion in PAF1C on flowering time, we measured flowering time of PAF1C-deficient mutants at different temperatures ranging from chilling (i.e., cold but not freezing, 10°C) to high temperature (27°C). To show that any observed flowering time change was not specific to a single allele, we used two independent homozygous mutant lines for each PAF1C gene (CDC73, ELF7, VIP3, VIP4, VIP5, and VIP6; Figures 1A,B). For the previously uncharacterized T-DNA lines elf7-4 (SALK_070632), vip5-3 (SALK_055889), and vip6-5 (SALK_119910), we performed conventional RT-PCR to examine their transcript levels; indeed, all three lines were found to be RNA-null alleles (Supplementary Figure 1).
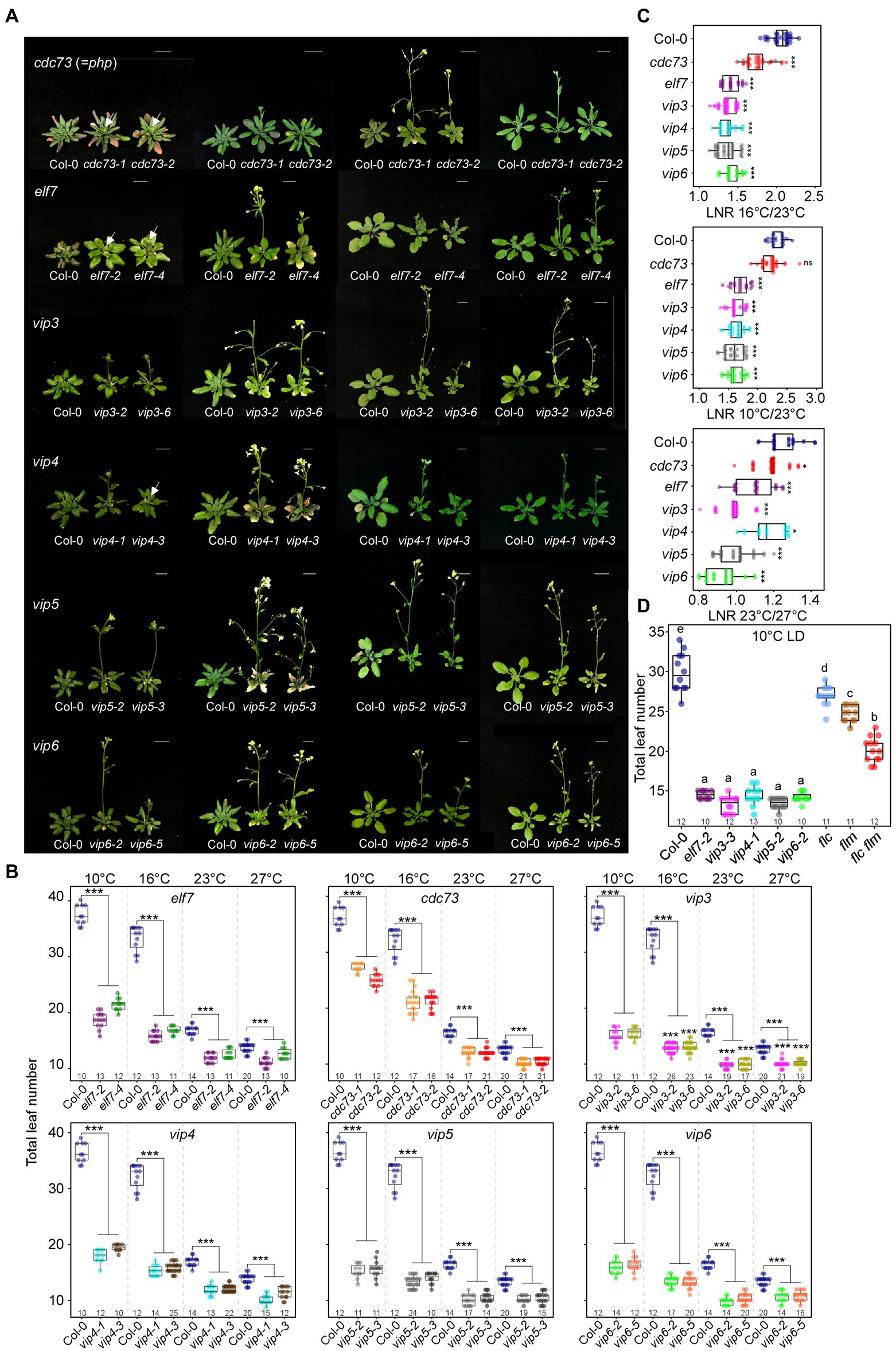
Figure 1. Early flowering of polymerase II–associated factor 1 complex (PAF1C)-deficient mutants at all tested temperatures under long-day (LD) conditions. (A,B) Phenotype (A) and flowering time quantified as total leaf number (B) of PAF1C-deficient mutant lines at 10, 16, 23, and 27°C. Arrows indicate inflorescences. Note that the same leaf number data from Col-0 plants were used in each panel (B). One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance. Scale bar = 1 cm. (C) Leaf number ratio (LNR) of Col-0 and PAF1C-deficient mutants. (D) Flowering time comparison of PAF1C-deficient mutants with flc, flm, and flc flm mutants at 10°C. One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance (p < 0.05). *p < 0.05 and ***p ≤ 0.001. Letters in (D) indicate significant difference by one-way ANOVA followed by Tukey’s range tests Numbers above the x-axis represent n.
Flowering time measurement showed that wild-type plants flowered with a mean total leaf number (TLN) of 36.3 ± 1.8, 32.0 ± 2.3, 15.6 ± 0.9, and 12.5 ± 0.5 at 10, 16, 23, and 27°C, respectively (Figures 1A,B; Supplementary Table 2). At all tested temperatures, the PAF1C-deficient mutants flowered significantly earlier than the wild-type plants, indicating that a lesion in PAF1C caused early flowering at a broad range of temperatures. In particular, PAF1C-deficient mutants showed very early flowering, compared with the wild type, as temperature decreased. We selected an allele that showed a strong early flowering phenotype from each gene (cdc73-2, elf7-2, vip3-2, vip4-1, vip5-2, and vip6-2) and used these mutants for further analyses.
To assess the temperature sensitivity of PAF1C-deficient mutants, we calculated the leaf number ratio (LNR) of cdc73-2, elf7-2, vip3-2, vip4-1, vip5-2, and vip6-2 mutants, using the TLN values at different temperatures. A LNR close to 1 indicates that temperature has little effect on flowering. All PAF1C-deficient mutants showed significantly lower LNRs compared with wild-type plants (Figure 1C; Supplementary Table 3). These results indicated that a lesion in PAF1C caused ambient temperature-insensitive flowering, especially at lower temperatures.
We then compared flowering time of PAF1C mutants with that of flc, flm, and flc flm mutants at 10°C. The flc, flm, and flc flm mutants flowered with 27.0 ± 1.3, 24.8 ± 1.1, and 20.2 ± 1.6 leaves, respectively, at 10°C (Figure 1D). Interestingly, their flowering times (measured as TLN) were later than elf7, vip3, vip4, vip5, and vip6 single mutants. This indicated that a lesion in both FLC and FLM was insufficient to phenocopy the early flowering time seen in PAF1C-deficient mutants. Therefore, considering that the PAF1C regulates FLC and FLM transcription (Zhang and Van Nocker, 2002; Zhang et al., 2003; Oh et al., 2004), these results suggested the possibility that PAF1C regulates other factors, in addition to FLC and FLM, in modulating flowering time at 10°C.
Transcriptome Analyses of PAF1C-Deficient Mutants
Mutants in all PAF1C components (except CDC73) flowered significantly earlier than flc flm double mutants at 10°C (Figure 1D), suggesting that additional factors are involved in this early flowering. To identify these factors, we performed RNA-seq using PAF1C-deficient mutants grown at 10 and 23°C. Euclidean distance clustering associated with complete linkage classified the transcriptome profiles into two major clades (Figure 2A): clade I, containing Col-0 and cdc73 mutants grown at 23°C; and clade II, containing the remaining mutants grown at 10 and 23°C. This indicates that the Col-0 plants and cdc73 mutants grown at 23°C have similar transcriptome profiles. Clade II was further divided into plants grown at 23°C and plants grown at 10°C. Among plants grown at 10°C, Col-0 plants and cdc73 mutants grouped together, whereas elf7, vip3, vip4, vip5, and vip6 mutants grouped together. This expression profile-based classification was consistent with the flowering time changes of PAF1C-deficient mutants at low temperature. As both cdc73 mutants flowered later than other PAF1C-deficient mutants (Figure 1) and cdc73 mutants were grouped in the same clade with Col-0 plants based on RNA-seq data (Figure 2A), we excluded CDC73 from further analyses.
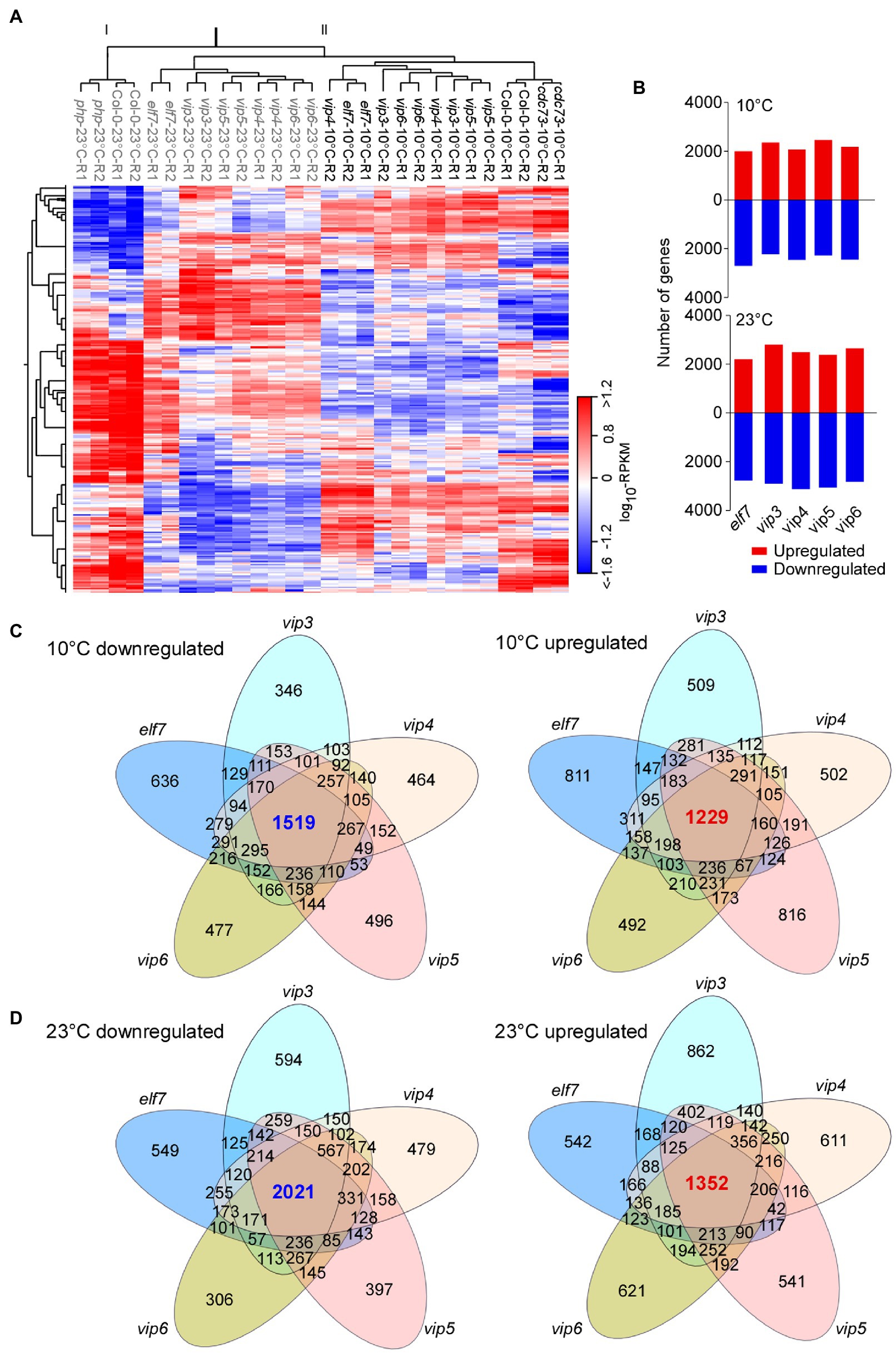
Figure 2. Transcriptome analyses of PAF1C-deficient mutants. (A) Heatmap showing the global expression differences between the wild type (Col-0) and PAF1C-deficient mutants. The genotypes of plants grown at 23°C are shown in grey, whereas plants grown at 10°C are shown in black. (B) Differentially expressed genes (DEGs) in PAF1C-deficient mutants compared to wild-type (Col-0) seedlings at 10°C (upper panel) and 23°C (lower panel). (C,D) Venn diagrams representing the commonly downregulated and upregulated genes at 10°C (C) and 23°C (D) in PAF1C-deficient mutants.
We selected DEGs that showed increased or decreased transcript levels (>1.5-fold). Transcriptome analyses revealed large numbers of DEGs in PAF1C-deficient mutants at both 10 and 23°C (Figure 2B). We then analyzed DEGs that were commonly upregulated and downregulated in different mutants (Figures 2C,D). At 10°C, 1,519 genes were commonly downregulated in elf7, vip3, vip4, vip5, and vip6 mutants, whereas 1,229 genes were commonly upregulated in these mutants (Figure 2C). At 23°C, 2,021 genes were commonly downregulated in elf7, vip3, vip4, vip5, and vip6 mutants, and 1,352 genes were upregulated in these mutants (Figure 2D). These analyses indicated that the largest number of intersecting DEGs was in the set containing the elf7, vip3, vip4, vip5, and vip6 mutants, suggesting that a large number of genes were commonly altered in these mutants at both temperatures. This observation was also consistent with the similar early flowering phenotypes of elf7, vip3, vip4, vip5, and vip6 mutants at 10 and 23°C.
To understand the biological significance of the common DEGs in PAF1C-deficient mutants, we performed Gene Ontology (GO) analyses using the webtool DAVID (Ashburner et al., 2000). At 10°C, the downregulated genes showed significant enrichment for GO terms related to the response to transcription, microtubule-based movement, different metabolic processes, response to jasmonic acid, and stomatal complex development. The upregulated genes were enriched in GO terms related to different metabolic processes and response to different stimuli (Supplementary Figure 2A). At 23°C, the downregulated genes were enriched in GO terms related to response to Karrikin, different metabolic processes, MAPK cascade, and photosynthesis, whereas the upregulated genes were enriched with GO terms related to different metabolic processes, response to oxidative stress, and cell wall organization (Supplementary Figure 2B).
PAF1C Regulates the Expression of FLC and the Other FLC-Clade Genes
The Flowering Interactive Database (FLOR-ID; Bouché et al., 2016) contains known genes involved in regulating flowering time. To check whether PAF1C deficiency affects the expression of known flowering time genes, we analyzed which genes in FLOR-ID were included among the common DEGs in PAF1C-deficient mutants at 10 and 23°C (Figures 2C,D). As PAF1C is involved in maintaining the active transcription of its target genes (He et al., 2004; Oh et al., 2004; Yu and Michaels, 2010), we expect that the direct targets of PAF1C will be downregulated in the PAF1C-deficient plants. Interestingly, our analyses showed that 26 and 27 flowering time genes were downregulated at 10 and 23°C, respectively, and both sets included FLC and MAF1, 2, 3, 4, and 5 (Figures 3A,B).
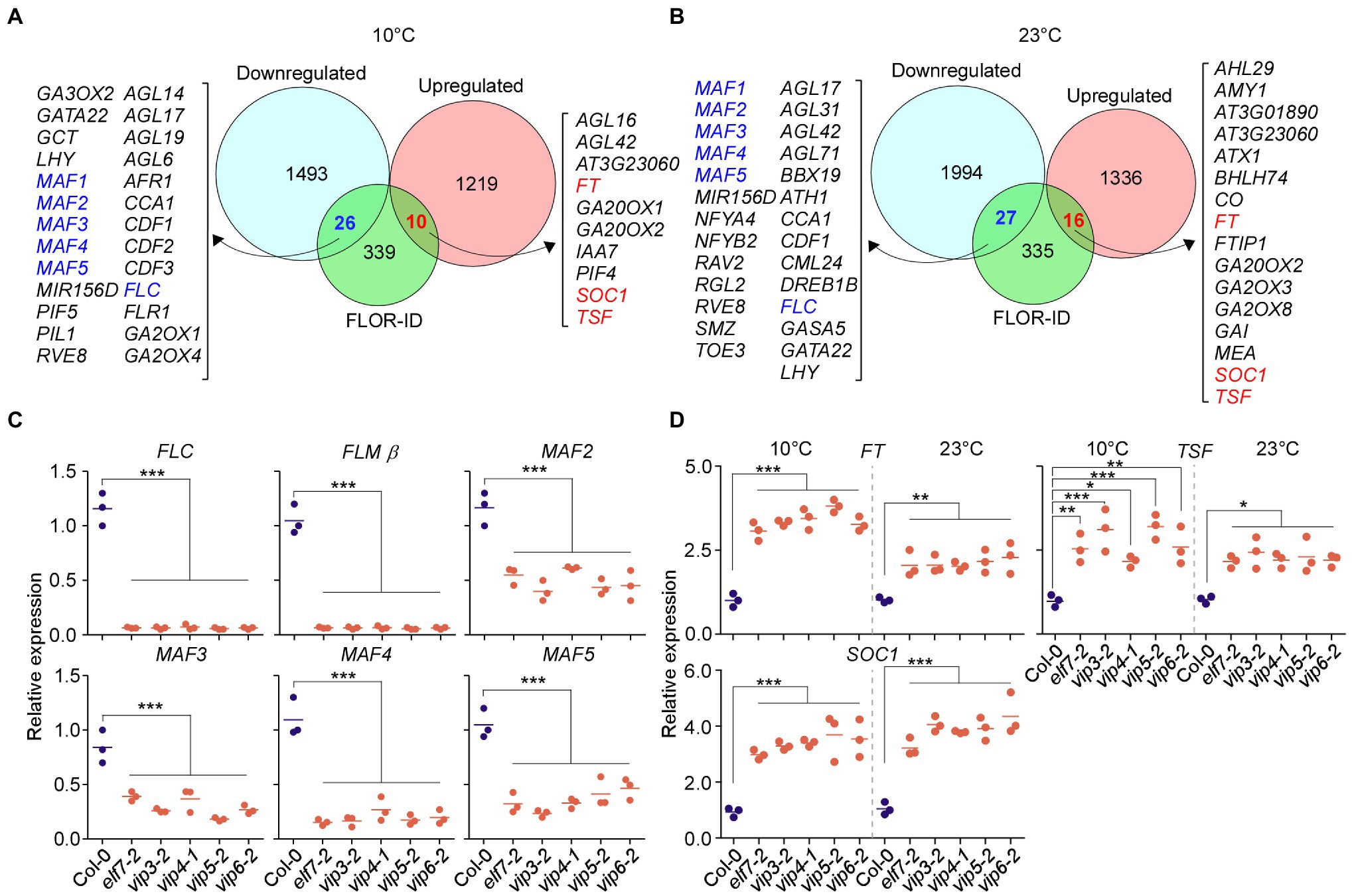
Figure 3. Polymerase II–associated factor 1 complex regulates expression of FLOWERING LOCUS C (FLC) and the FLC-clade genes MADS AFFECTING FLOWERING1 (MAF1)/FLOWERING LOCUS M (FLM) and MAF2–MAF5. (A,B) Commonly downregulated and upregulated flowering genes in PAF1C-deficient mutants at 10°C (A) and 23°C (B). (C) qPCR confirmation of downregulation of FLC and FLC-clade genes in PAF1C-deficient mutants at 10°C. (D) mRNA levels of FLOWERING LOCUS T (FT), TSF, and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) in PAF1C-deficient mutants at 10 and 23°C. One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance. *p < 0.05; **p < 0.01; and ***p ≤ 0.001.
We performed RT-qPCR analyses to confirm the downregulation of FLC and FLC-clade genes in PAF1C-deficient mutants at 10°C. Among the alternatively spliced forms produced from the FLM locus, FLM-ß is the functional form (Scortecci et al., 2001; Pose et al., 2013); therefore, we measured FLM-ß transcript levels for FLM. The RT-qPCR showed that that FLC, FLM, and MAF2–MAF5 showed significant downregulation in PAF1C-deficient mutants (Figure 3C). FLC showed 17.6-fold downregulation in elf7-2 mutants, 18-fold in vip3-2 mutants, 16-fold in vip4-1 mutants, 20.1-fold in vip5-2 mutants, and 18.3-fold in vip6-2 mutants (Figure 3C), consistent with a previous study (Oh et al., 2004). Similarly, FLM-ß showed 15.6- to 18.2-fold downregulation in PAF1C-deficient mutants. MAF2 showed 2.1-fold downregulation in elf7-2 mutants, 2.9-fold in vip3-2 mutants, 1.9-fold in vip4-1 mutants, 2.7-fold in vip5-2 mutants, and 2.6-fold in vip6-2 mutants at 10°C. Similarly, MAF3 was downregulated by 2.1- to 4.6-fold in the PAF1C-deficient mutants at 10°C. MAF4 mRNA levels were downregulated by 4.1- to 7.1-fold and MAF5 mRNA levels were downregulated 2.2- to 4.4-fold in the PAF1C-deficient mutants at 10°C.
We then measured the mRNA levels of FT, TSF, and SOC1 by RT-qPCR and found that the mRNA levels of FT, TSF, and SOC1 were significantly higher in the PAF1C-deficient mutants at 10 and 23°C compared with the wild type. FT mRNA levels were increased by 3.1- to 3.8-fold in the PAF1C-deficient mutants at 10°C (Figure 3D). At 23°C, FT was upregulated by 1.8- to 2.3-fold. TSF was also significantly upregulated in the PAF1C-deficient mutants at both temperatures. SOC1 transcript levels showed a similar pattern, with a fold increase of 3.2–4.0 and 3.1–4.1 in PAF1C-deficient mutants at 10 and 23°C, respectively, consistent with the downregulation of FLC and FLC-clade genes in PAF1C-deficient mutants (Figure 3C). These results suggested that functional PAF1C is required for the expression of FLC and FLC-clade genes and that a lesion in one of its components results in the downregulation of FLC and FLC-clade genes, which leads to the derepression of FT, TSF, and SOC1, and early flowering. Furthermore, considering that the expression of all MAF genes was affected in PAF1C-deficient mutants and PAF1C-deficient mutants flowered significantly earlier than flc flm double mutants (Figure 1D), it is likely that the MAF2–MAF5 genes play a significant role in floral repression at chilling temperatures. Although FLC and FLC-clade genes are known to undergo alternative splicing and their splice variants might have differential effects on flowering time (Caicedo et al., 2004; Lee et al., 2013; Pose et al., 2013; Rosloski et al., 2013), our RNA-seq data showed no significant difference in the levels of the splice variants of FLC and FLC-clade genes (Supplementary Figure 3), except for their overall downregulation, suggesting that the flowering time change seen in PAF1C-decificient mutants was not associated with the differential splicing patterns of FLC and FLC-clade genes.
PAF1C Deficiency Alters the Epigenetic Status of the FLC-Clade Genes
Our expression analyses showed that in addition to FLC and FLM, MAF2–MAF5 were downregulated in PAF1C-deficient mutants (Figure 3), suggesting that this downregulation may be due to the altered epigenetic status at these loci in PAF1C-deficient mutants. To test this possibility, we analyzed the levels of the repressive H3K27me3 marks and permissive H3K4me3/H3K36me3 marks of FLC and the FLC-clade genes in vip3-2 mutants (as a representative PAF1C-deficient mutant line) and wild-type plants grown at 10°C. Four different qPCR primer sets (P1–P4), spanning the entire gene bodies of the target genes, were used to assess the enrichment of the repressive/permissive marks (Figure 4A).
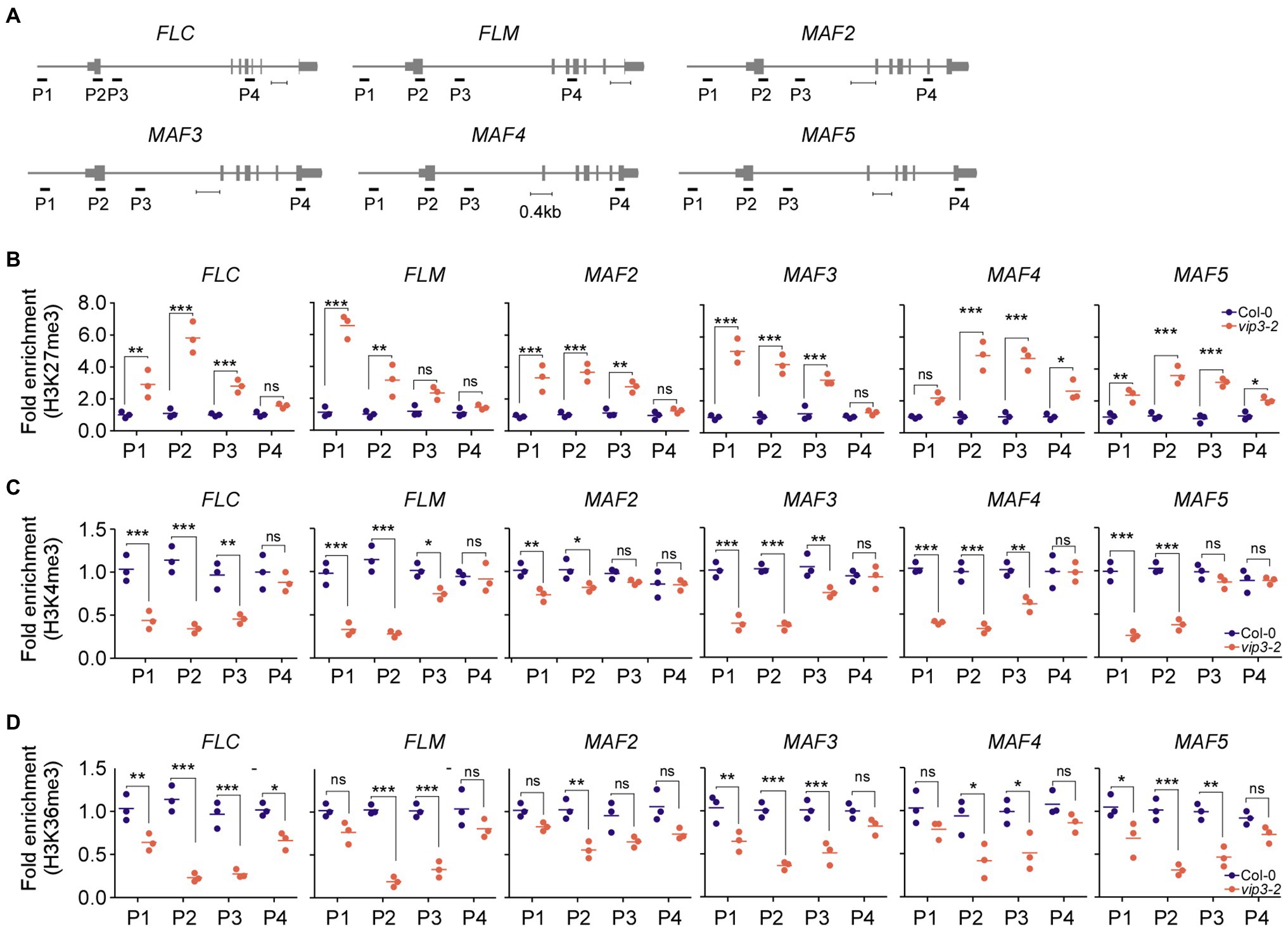
Figure 4. Polymerase II–associated factor 1 complex deficiency alters the epigenetic status of chromatin of the FLC and the FLC-clade genes MAF1/FLM and MAF2–MAF5. (A) Schematic representation of the genomic regions of FLC and FLC-clade genes and the regions amplified for chromatin immunoprecipitation (ChIP) assays. Scale bar = 0.4 kb. Note that our P2 region in FLC partially overlaps with the region that was used in a previous study (He et al., 2004). In the case of FLM, our P2 region is included in a region that was previously shown to be affected (He et al., 2004). (B–D) Fold enrichment of the repressive epigenetic H3K27me3 marks (B), permissive H3K4me3 marks (C), and permissive H3K36me3 marks (D) at the genomic regions of FLC and FLC-clade genes in wild-type (Col-0) and PAF1C-deficient vip3 mutant seedlings. For each primer pair, the enrichment was normalized to the wild-type control (Col-0). One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance. *p < 0.05; **p < 0.01; ***p ≤ 0.001; and ns: not significant.
Consistent with their downregulation, the FLC and FLC-clade genes showed significantly increased enrichment of the repressive H3K27me3 marks throughout their gene bodies in vip3 mutants (Figure 4B). In FLC, the highest enrichment (5.8-fold higher enrichment compared to wild-type plants) of repressive H3K27me3 marks was observed in the P2 region, which contains the transcription start site (Figure 4B), consistent with a previous finding (He et al., 2004). Significantly higher enrichment was also observed in the P1 (2.9-fold) and P3 (2.8-fold) regions of FLC. In FLM chromatin, the H3K27me3 enrichment was highest in the P1 region (5.8-fold) followed by P2 (3.1-fold) and P3 (2.0-fold) regions. MAF2 and MAF3 showed similar H3K27me3 patterns with the highest enrichment in the P1 region (3.7- and 5.3-fold, respectively), followed by P2 (3.6- and 4.4-fold, respectively) and P3 regions (2.4- and 2.7-fold, respectively). The enrichment of H3K27me3 in the MAF5 chromatin was highest in the P3 and P2 regions (3.5- and 3.4-fold, respectively) followed by P1 with 2.4-fold higher enrichment of H3K27me3 in vip3 mutants compared to wild-type plants at 10°C. H3K27me3 enrichment in the P4 regions of FLC, FLM, MAF2, and MAF3 of vip3 mutants was comparable with wild-type samples, whereas MAF4 and MAF5 showed slightly higher enrichment in the P4 regions (1.3- and 2.5-fold, respectively) in vip3 mutants (Figure 4B).
By contrast, enrichment of the permissive H3K4me3 marks was significantly reduced in the chromatin of FLC and FLC-clade genes in vip3 mutants, primarily in the P1 and P2 regions (Figure 4C). Enrichment of H3K4me3 in the FLC chromatin in vip3 mutants was reduced 3.3-fold in the P2 region, 2.4-fold in P1, and 2.1-fold in P3 compared to wild-type plants. FLM also showed reduced enrichment of H3K4me3 marks in the P2 (4.1-fold) and P1 (2.9-fold) regions. In the MAF2 chromatin, reduced H3K4me3 enrichment was seen in the P1 (1.4-fold) and P2 (1.3-fold) regions. In addition, in the MAF3-5 chromatin, H3K4me3 enrichment was significantly lower in the P2 region (2.8-, 3.0-, and 2.7-fold, respectively) and the P1 region (2.6-, 2.5-, and 3.9-fold, respectively) in vip3 mutants (Figure 4C). Similar reduction patterns were found for the H3K36me3 mark at the gene bodies of these genes, with significantly reduced enrichment at the P2 and P3 regions (Figure 4D). Taken together, these results suggest that PAF1C is required to maintain permissive epigenetic marks and prevent deposition of repressive marks at the FLC and FLC-clade genes, thereby maintaining their active transcription.
FLC-Clade Genes Are Upregulated in Wild-Type Plants at 10°C
The early flowering of PAF1C-deficient mutants at 10°C is likely due to the combinatorial effect of FLC, FLM, and the other MAFs, suggesting the functional importance of MAF2–MAF5 at low temperature (10°C). To test whether expression of these genes is upregulated in wild-type plants at 10°C, we compared the transcript levels of these genes in wild-type plants at 10 and 23°C using our RNA-seq data. This analysis revealed upregulation of FLC, FLM, and MAF2–MAF5 in wild-type plants at 10°C compared to 23°C by at least 1.5-fold (Figure 5A). RT-qPCR analyses also showed statistically significant induction of FLC, FLM, and MAF2–MAF5 in wild-type plants at 10°C compared to 23°C (Figure 5B). Consistent with the upregulation of FLC and FLC-clade genes, transcript levels of FT and TSF, their downstream targets, were significantly reduced (>3-fold) in wild-type plants at 10°C in comparison to 23°C (Figure 5C). This suggests that these MAFs might play important roles in modulating flowering time at chilling temperatures by regulating FT and TSF.
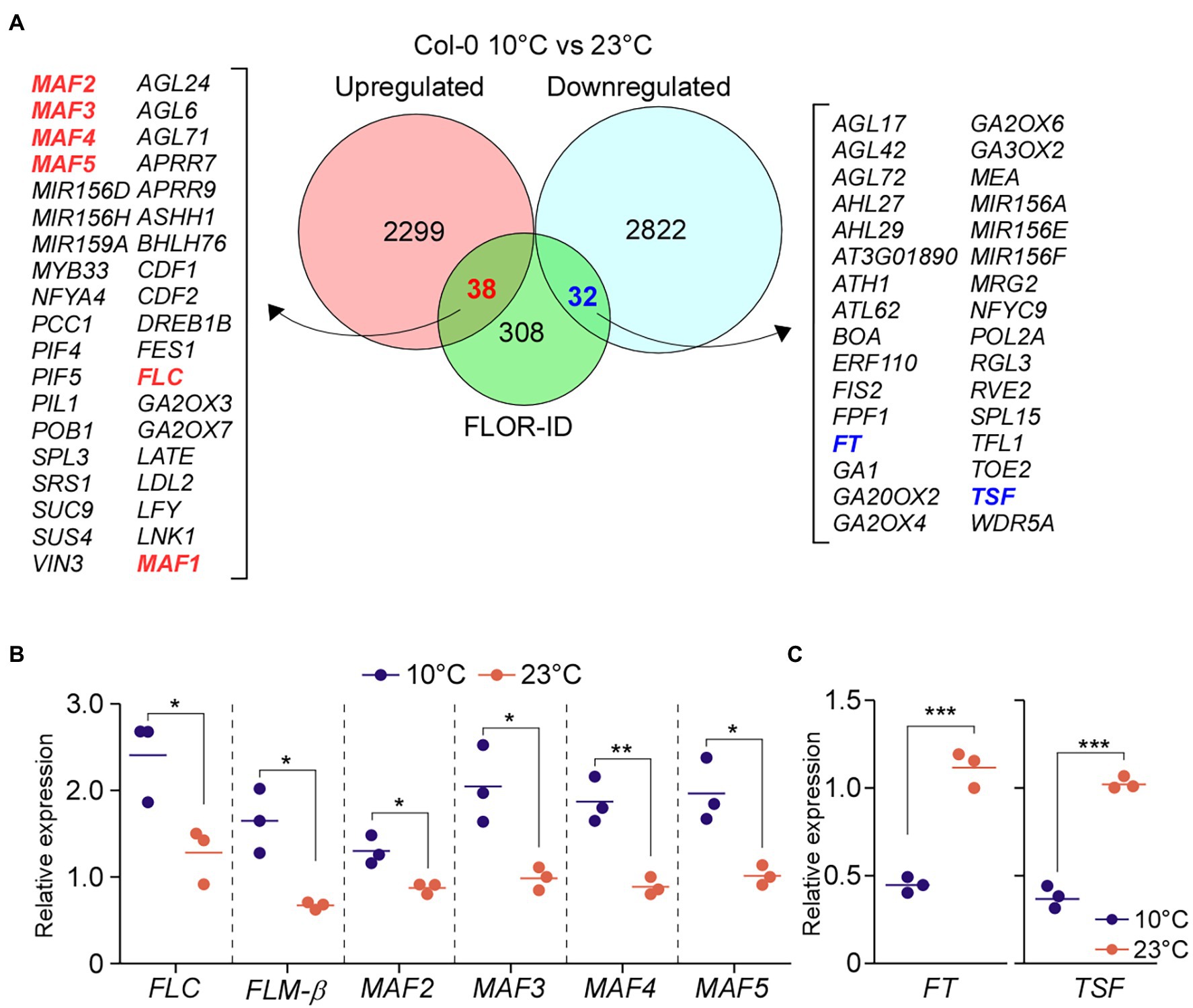
Figure 5. Low temperature induces the expression of FLC and FLC-clade genes. (A) Differentially expressed genes (DEGs) between wild-type (Col-0) seedlings grown at 10 and 23°C under LD conditions. (B,C) Relative mRNA levels of FLC and FLC-clade genes (B) and FT (C) at 10°C under LD conditions. One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance. *p < 0.05; **p < 0.01; and ***p ≤ 0.001.
FLC-Clade Genes Are Important for Floral Repression at Chilling Temperatures
Polymerase II–associated factor 1 complex modulates the expression of downstream genes by recruiting a number of histone modifiers, including SET DOMAIN GROUP8 (SDG8; Wood et al., 2003; Ng et al., 2003a; Kim et al., 2005). SDG8 recruited by PAF1C then regulates the expression of FLC and FLM; therefore, a mutation in SDG8 causes early flowering at normal temperatures (Kim et al., 2005). Because the flowering response of sdg8 mutants under chilling-stress temperatures is not known, we analyzed the flowering time of sdg8 mutants at 10 and 23°C. We found that the sdg8 mutants flowered significantly earlier than flc, flm, and flc flm mutants at 10°C (Figures 6A,B). The sdg8 mutants flowered with 18.1 ± 1.0 leaves, which was significantly earlier than flc mutants (32.6 ± 1.6 leaves), flm mutants (26.5 ± 1.7 leaves), and flc flm double mutants (23.0 ± 1.1 leaves) at 10°C. However, at 23°C, the flowering time of sdg8 mutants (8.5 ± .6 leaves) was only slightly earlier than flc and flm mutants (10.0 ± 0.7 and 9.7 ± 0.7 leaves, respectively) and was comparable to flc flm double mutants (8.4 ± .5 leaves; Figure 6B). LNR analyses revealed significantly decreased LNR values of sdg8 mutants to low temperature (Figure 6C), indicating that the temperature responsiveness of sdg8 mutants was reduced.
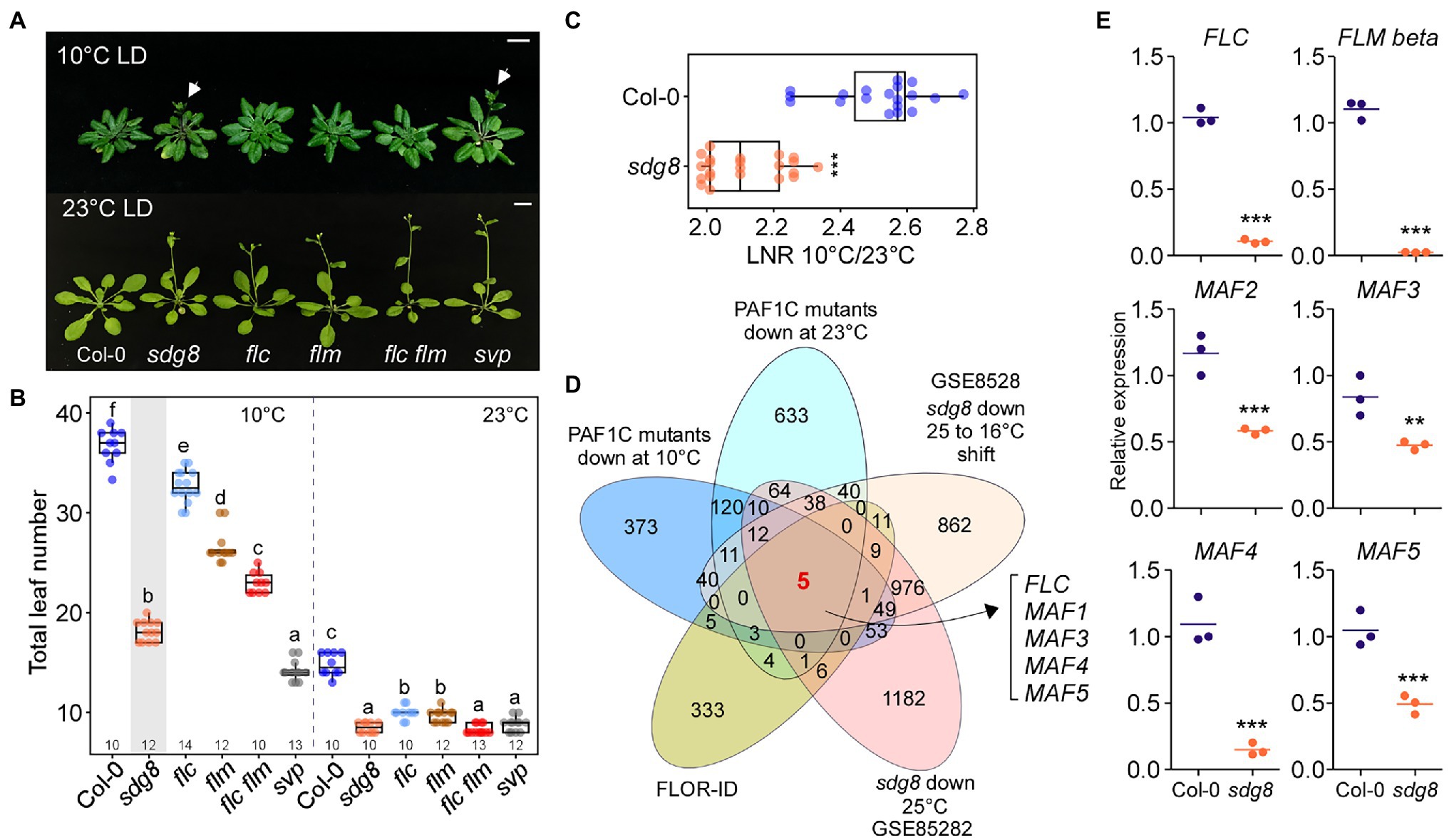
Figure 6. MAF genes are important for floral repression at low temperature. (A,B) Flowering phenotype (A) and flowering time (B) of sdg8, flc, flm, flc flm, and short vegetative phase (svp) mutants at 10 and 23°C under LD conditions. Arrows indicate inflorescences. (C) Leaf number ratio (LNR; 10°C/23°C) of sdg8 mutants. (D) Venn diagram of the genes that were commonly downregulated in sdg8 mutants and PAF1C-deficient mutants at different temperatures. (E) Reverse transcription quantitative PCR (RT-qPCR) of FLC and FLC-clade genes in the wild type (Col-0) and sdg8 mutants at 10°C under LD conditions. One-way ANOVA followed by Dunnett’s multiple comparison tests were performed to test the statistical significance. *p < 0.05; **p < 0.01; ***p ≤ 0.001; and ns: not significant. Letters indicate significant differences by one-way ANOVA followed by Tukey’s range tests (p < 0.05). Numbers above the x-axis in (B) represent n. Scale bar = 1 cm.
To determine whether MAF genes play a role in the regulation of flowering time in sdg8 mutants, we first analyzed publicly available RNA-seq data for sdg8 mutants grown at 16°C with or without shifting to 25°C (GSE85282; Pajoro et al., 2017) and then found the intersection of (1) the set of DEGs in sdg8 mutants, (2) the genes that were commonly downregulated in PAF1C-deficient mutants at 10°C (Figure 2C) and 23°C (Figure 2D), and (3) the list of flowering time genes from FLOR-ID (Bouché et al., 2016). From this comparison, we found that FLC, FLM, and MAF3–MAF5 were commonly downregulated in sdg8 mutants (Figure 6D; Supplementary Figure 4), like in PAF1C-deficient mutants (Figure 3). MAF2, which showed 1.8-fold downregulation in sdg8 mutants, was not identified here, due to the criteria for selecting DEGs (2-fold change).
We then performed RT-qPCR to confirm the downregulation of these genes at 10°C. The RT-qPCR results showed statistically significant downregulation of FLC, FLM-ß, and MAF2–MAF5 mRNA levels in sdg8 mutants at 10°C (Figure 6E). FLC mRNA levels were decreased by 13.2-fold, whereas FLM-ß showed 15.9-fold downregulation in sdg8 mutants at 10°C. In addition, expression of MAF2–MAF5 was downregulated by 2.0-, 1.7-, 7.3-, and 1.9-fold, respectively, in sdg8 mutants at 10°C (Figure 6E). Taken together, these results suggested that the early flowering phenotype of sdg8 mutants at chilling temperature is mediated by the downregulation of FLC and MAF genes. However, it should be noted that sdg8 mutants flowered slightly later than PAF1C-dificient mutants at 10°C (Supplementary Figure 5), suggesting a possibility that the PAF1C recruits an additional histone modifier(s), besides SDG8, to regulate the expression of the FLC-clade genes.
Knockout/Knockdown of FLC and FLC-Clade Genes Results in Extremely Early Flowering at 10°C
To genetically confirm the importance of MAF genes in repressing flowering at chilling temperatures, we used amiRNAs to repress the MAF genes. To this end, we designed two amiRNAs that simultaneously target MAF2–MAF5 (Figure 7A) and overexpressed these amiRNAs in flc flm double mutants. Two independent transgenic lines overexpressing amiRNAs against MAF2–MAF5 (35S::amiR-MAF2-5 flc flm #1 and #2) flowered significantly earlier than flc flm double mutants (Figures 7B,C). At 10°C, The 35S::amiR-MAF2-5 flc flm #1 and #2 plants flowered with an average TLN of 12.1 to 12.9 leaves (c.f., wild-type plants: 32.8 ± 3.8), indicating that knockout/down of FLC and all five FLC-clade genes caused extremely early flowering at 10°C. Furthermore, these transgenic lines flowered earlier than flc flm double mutants (21.6 ± 1.6 leaves; Figure 7C). Interestingly, both 35S::amiR-MAF2-5 flc flm lines flowered earlier than svp mutants (14.6 ± 1.4 leaves). However, at 23°C, The 35S::amiR-MAF2-5 flc flm #1 and #2 plants flowered with an average TLN of 11.1–11.4 leaves (c.f., wild-type plants: 16.0 ± 1.1), which was similar to the TLN of flc flm double (11.7 ± 0.6 leaves) and svp single mutants (11.1 ± 0.7 leaves; Figure 7C). These results highlighted the importance of the MAF2–MAF5 transcription factors in repressing flowering at chilling temperatures.
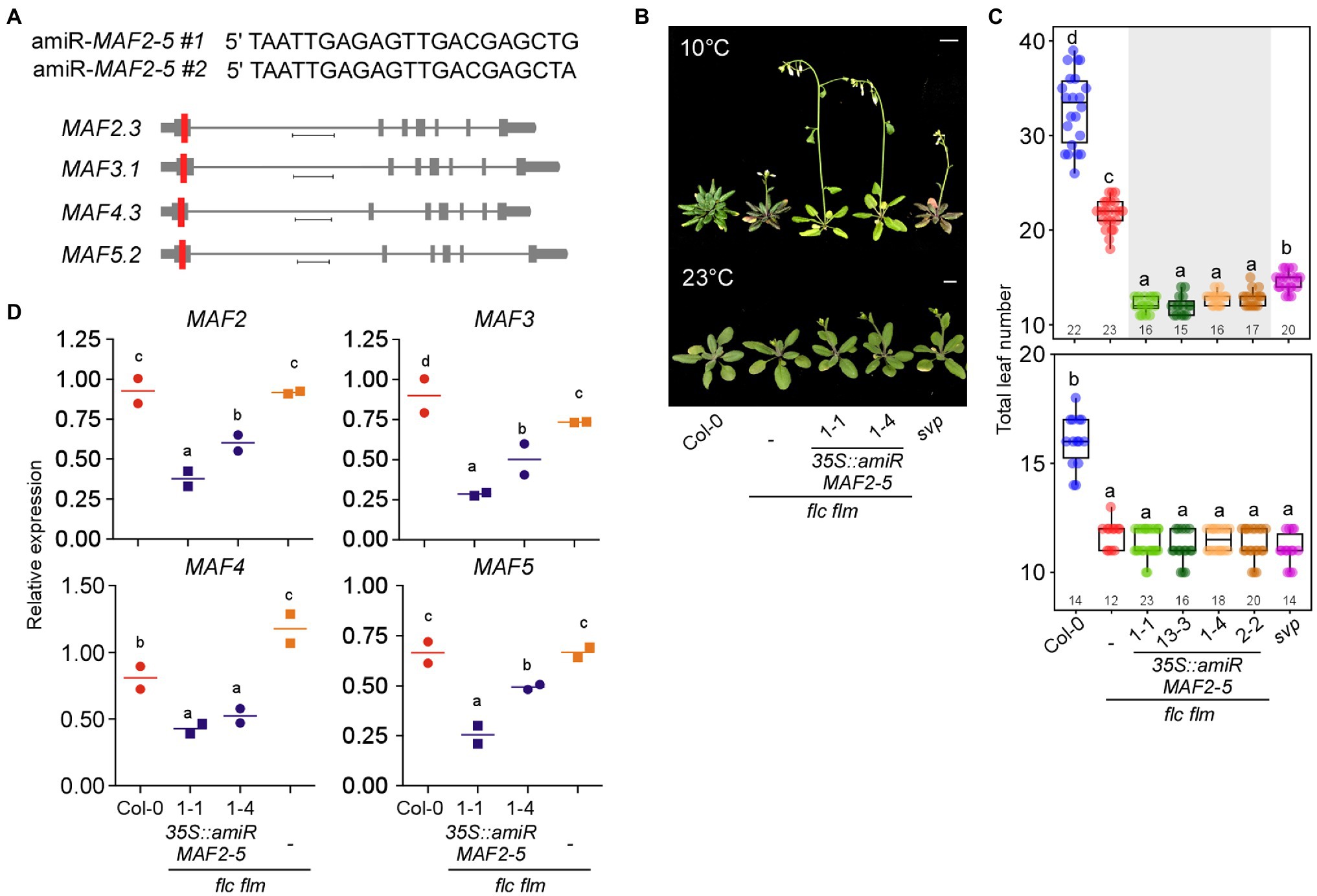
Figure 7. Knockout/down of FLC and MAFs together results in precocious flowering at chilling temperatures. (A) Sequences of artificial microRNAs (amiRNAs) designed to knock down MAF2–MAF5 genes (top) and schematic representation of their location (red vertical line) in a predominantly expressed spliced variant of MAF2–MAF5 genes (bottom). Scale bar: 0.4 kb (B,C) Flowering phenotype (B) and flowering time (C) of 35S::amiR-MAF2-5 flc flm transgenic seedlings at 10 and 23°C under LD conditions. Scale bar = 1 cm. (D) RT-qPCR confirmation of knockdown of MAF2–MAF5 in 35S::amiR-MAF2-5 flc flm transgenic lines. Letters indicate significant differences by one-way ANOVA followed by Tukey’s range tests (p < 0.05). Numbers above the x-axis in (C) represent n.
To confirm that this early flowering of 35S::amiR-MAF2-5 flc flm #1 (1–1 and 13–3) and #2 (1–4 and 2–2) plants was indeed due to the downregulation of MAF2–MAF5, we performed RT-qPCR analyses. These analyses confirmed the transgenic seedlings showed significantly lower MAF mRNA levels compared with the wild-type plants and flc flm double mutants (Figure 7D). Furthermore, MAF2–MAF5 mRNA levels were significantly lower in 35S::amiR-MAF2-5 flc flm #1 plants than in 35S::amiR-MAF2-5 flc flm #2 plants, except MAF4. The stronger reduction of MAF transcript levels in the 35S::amiR-MAF2-5 flc flm #1 plants was consistent with their earlier flowering time phenotype compared with 35S::amiR-MAF2-5 flc flm #2 plants (Figure 7C). Taken together, these data suggest that ablation of function of FLC and all FLC-clade members resulted in earlier flowering than flc flm mutants at chilling temperatures; therefore, MAF2–MAF5 also play a role in repressing flowering at chilling temperatures.
MAFs Physically Interact With SVP to Form Repressor Complexes
Loss of SVP function results in early flowering across a broad range of temperatures (10°C–27°C; Lee et al., 2013). SVP interacts with FLC (Fujiwara et al., 2008; Li et al., 2008) and with FLM (Lee et al., 2013; Pose et al., 2013). In vivo and yeast two-hybrid (Y2H) analyses showed that SVP interacts with MAF2 and MAF4 (Gu et al., 2013). Since the 35S::amiR-MAF2-5 flc flm (#1 and #2) plants showed significantly earlier flowering at 10°C compared with svp mutants (Figure 7C), one possible scenario is that SVP alone is insufficient to repress flowering at 10°C and may require FLC-clade proteins to repress flowering.
To test whether FLC-clade proteins physically interact with SVP, we first used two recently developed artificial intelligence (AI)–based deep learning programs that were designed to predict protein–protein interactions: D-SCRIPT (Sledzieski et al., 2021) and PPI-Detect (Romero-Molina et al., 2019). These programs produce an interaction score between 0 (no interaction predicted) and 1 (interaction strongly predicted). In these analyses, FLC was used as a known interacting partner of SVP (Li et al., 2008), and PP2AA3 was used a negative control. The FLC–SVP interaction scores were 0.977 (D-SCRIPT) and 0.705 (PPI-Detect), whereas the PP2AA3–SVP interaction scores were 0.004 (D-SCRIPT) and 0.278 (PPI-Detect; Figure 8A). From D-SCRIPT analyses, the MAF2–SVP, MAF3–SVP, MAF4–SVP, and MAF5–SVP interaction scores were 0.789, 0.740, 0.586, and 0.779, respectively. From PPI-Detect analyses, the MAF2–SVP, MAF3–SVP, MAF4–SVP, and MAF5–SVP interaction scores were 0.584, 0.512, 0.915, and 0.822, respectively. All of these interaction scores were above the cut-off value of 0.5, suggesting that SVP interacts with MAF2–MAF5 in vivo.
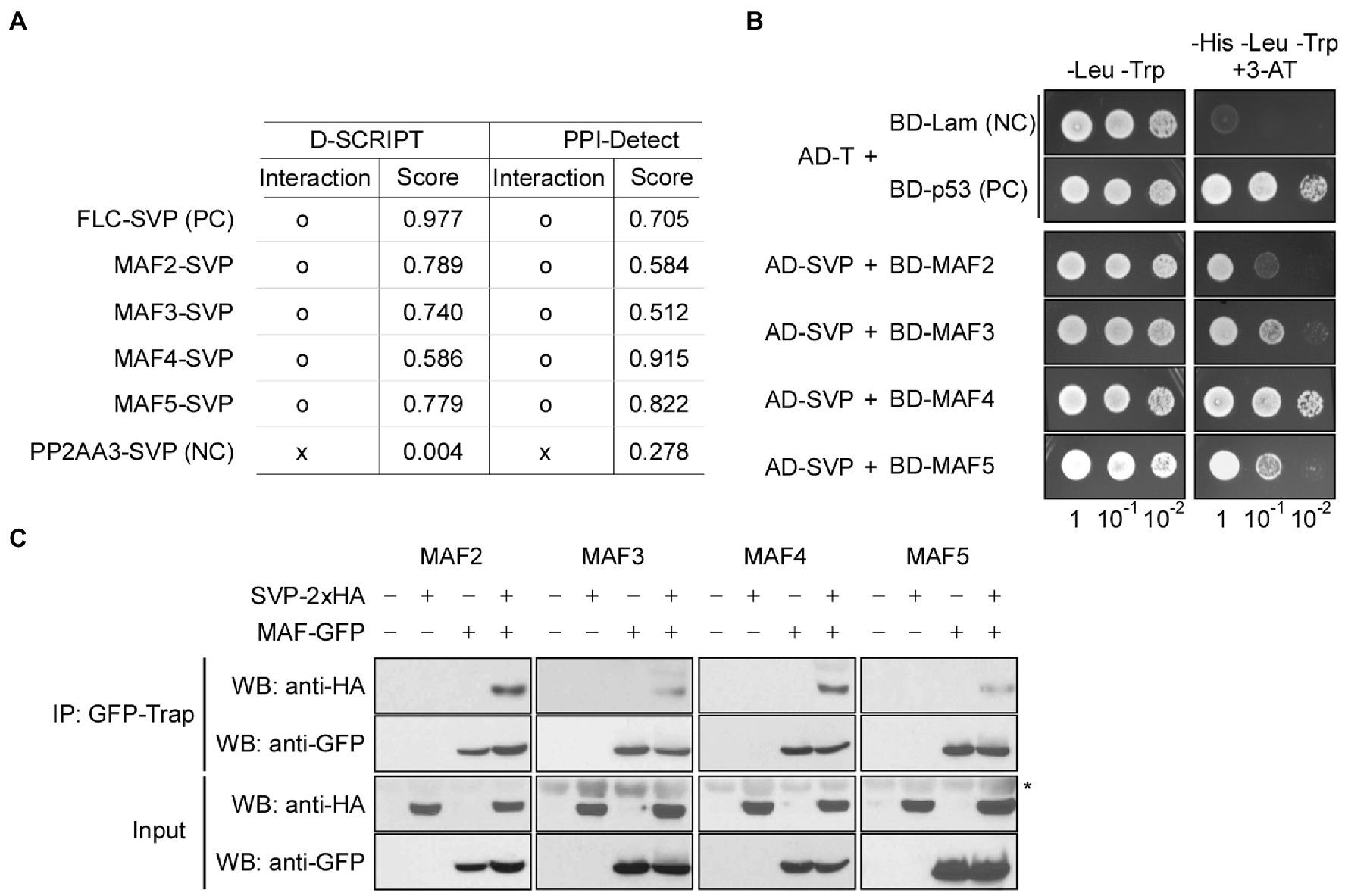
Figure 8. MAFs physically interact with SVP. (A) Artificial intelligence (AI)–based prediction of interaction between MAFs and SVP. o: interaction, x: no interaction. Score (ranging from 0 to 1) represents the likelihood of interaction between two input proteins, where 0 represents no predicted interaction and 1 represents a high-confidence prediction that the proteins interact. (B) Yeast two-hybrid assays showing the interaction between MAFs and SVP. Positive control (PC): AD-T (SV40 large T-antigen) and BD-p53 (murine p53); negative control (NC): AD-T and BD-Lam (Lamin). (C) In vivo confirmation of MAF–SVP interactions via co-immunoprecipitation assays using Arabidopsis protoplasts at 10°C. PC; positive control, NC; negative control. The asterisk (*) represents a nonspecific signal on the anti-HA blot.
We then performed Y2H analyses to experimentally validate the predicted interactions. Indeed, Y2H analyses showed that SVP interacts with MAF2–MAF5 in yeast cells (Figure 8B). To further test these interactions in vivo, we performed Co-IP experiments using Arabidopsis mesophyll protoplasts. To this end, 35S::2 × HA:SVP and 35S::GFP:MAF vectors were transiently co-expressed in protoplasts to produce HA-tagged SVP and GFP-tagged MAF2–MAF5 proteins and then the transfected protoplasts were shifted to 10°C to test the protein–protein interaction at 10°C. We precipitated protein extracts using GFP-Trap and probed the resulting precipitates with anti-HA antibodies. SVP-2 × HA successfully co-immunoprecipitated with each MAF transcription factor (Figure 8C; asterisk), confirming the interactions between SVP and MAFs. Taken together, these results suggest that MAF2–MAF5, like FLC and FLM, physically interact with SVP, further implying that SVP forms a repressor complex including FLC and MAFs. It is therefore likely that the formation of the complex leads to efficient floral repression, thus allowing the plant to acclimate to chilling temperatures.
SVP-Mediated Floral Repression Likely Requires FLC and FLC-Clade Genes
Since the PAF1C-deficient mutants showed strong early flowering at 10°C (Figure 1), we tested whether SVP transcript levels were affected in PAF1C-deficient mutants, as svp mutants flower early across a range of temperatures (10°C–27°C; Lee et al., 2013). RT-qPCR analyses showed that SVP mRNA levels in PAF1C-deficient mutants were similar to those of Col-0 plants at both 10 and 23°C (Figure 9A; He et al., 2004). This suggested that the flowering time change seen in PAF1C-deficient mutants at both 10 and 23°C was independent of SVP transcript levels.
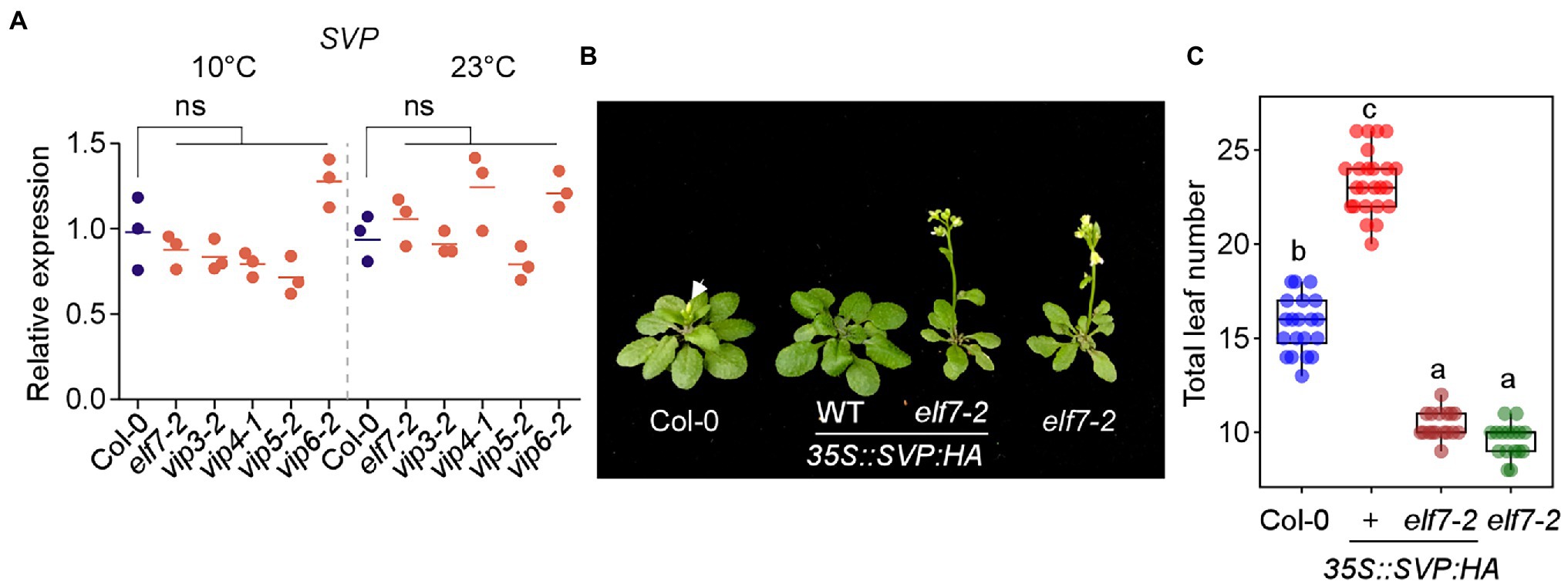
Figure 9. SVP-mediated floral repression is dependent on FLC and FLC-clade genes. (A) SVP mRNA levels in PAF1C-deficient mutants at 10 and 23°C under LD conditions. ns; not significant. (B,C) Phenotype (B) and total leaf number (C) of 35S::SVP-HA and 35S::SVP-HA elf7 plants. Note that these plants were first grown on medium containing phosphinothricin for 6 days at 23°C, transferred to soil, and then grown at 16°C. Letters indicate significant differences by one-way ANOVA followed by Tukey’s range tests (p < 0.05).
We then tested whether SVP requires PAF1C to delay flowering time. For this experiment, we used elf7-2 and vip4-1 mutants as representative PAF1C-deficient mutants and crossed them with 35S::SVP:HA plants. The 35S::SVP:HA plants showed delayed flowering (23.3 ± 1.7 leaves) at 23°C, but the 35S::SVP:HA elf7-2 plants flowered with 10.4 ± .7 leaves (Figures 9B,C). Considering that elf7-2 mutants flowered with 9.6 ± 0.9 leaves at 23°C, this genetic interaction study showed that the late flowering caused by SVP overexpression was almost completely suppressed by elf7-2 mutation. Similarly, the late flowering of 35S::SVP:HA plants was strongly suppressed by vip4-1 mutation (Supplementary Figure 6). These results suggested that SVP overexpression was unable to delay flowering in the absence of a functional PAF1C.
Short vegetative phase was unable to repress flowering in elf7-2 mutants, which have dramatically decreased mRNA levels of FLC and FLC-clade genes (Figure 3B), suggesting the possibility that SVP binding to its targets requires functional FLC and FLC-clade transcription factors. To test this hypothesis, we took advantage of a publicly available ChIP-seq dataset (GSE54881) for SVP-GFP in the presence/absence of FLC (FRI FLC and FRI flc; Mateos et al., 2015). Consistent with a previous study (Mateos et al., 2015), the number of targets bound by SVP-GFP was substantially reduced in plants without functional FLC (FRI flc), compared to the plants with functional FLC (FRI FLC; Supplementary Figure 7). In terms of the number of bound targets, SVP-GFP was only able to bind to 39.2% of its target genes in the absence of functional FLC (the number of targets bound by SVP-GFP in FRI FLC was set to 100%; Supplementary Figure 7A). Furthermore, SVP-GFP was able to bind to 553 additional target genes in the presence of FLC (Supplementary Figure 7B; Mateos et al., 2015), indicating the importance of FLC for SVP binding ability. Since other FLC-clade transcription factors also interact with SVP (Figure 8), these results suggest that the FLC-clade transcription factors play a similar role, especially at low temperatures.
Discussion
In Arabidopsis, PAF1C regulates flowering primarily through epigenetic modulation of FLC and FLM expression under standard growth conditions (Kim et al., 2005; Yu and Michaels, 2010). However, the role of PAF1C in regulating flowering time at chilling temperatures remains unknown. In this study, we show that PAF1C not only regulates FLC and FLM, but also regulates the entire FLC clade of genes (FLM/MAF1 and MAF2–MAF5), which play important roles in repressing flowering at low temperatures.
Several environmental factors, including temperature, affect flowering. At lower temperatures, Arabidopsis plants flower late compared to plants at elevated temperatures (Lee et al., 2013). Several MADS-box transcription factors, including FLC, FLM, and SVP, play an important role in delaying flowering (Lee et al., 2007, 2013). FLC and FLM are well-known to be epigenetically regulated by a number of histone modifiers, including SET domain-containing histone methyltransferases (He et al., 2003; Zhang et al., 2003; Oh et al., 2004; Nasim et al., 2021). PAF1C recruits these histone modifiers to modulate expression of its target genes, including FLC, FLM, and MAF2 (Zhang and Van Nocker, 2002; He et al., 2003; Zhang et al., 2003; Oh et al., 2004).
Polymerase II–associated factor 1 complex function is critical for proper plant growth and development, as lesions in PAF1C components result in strong defects in the vegetative and reproductive stages, such as severely stunted growth, stem cell maintenance defects, floral structure defects, and male sterility (Zhang et al., 2003; He et al., 2004; Oh et al., 2004; Fal et al., 2019). Flowering time analyses across a range of ambient temperatures showed that all PAF1C mutants flowered early in a temperature-independent manner with reduced expression of FLC and FLC-clade genes (Figures 1, 3). This observation validates the previous findings that mutations in VIP3, VIP5, and VIP6 result in reduced FLC and FLM expression and, hence, accelerated flowering (Zhang et al., 2003; Oh et al., 2004). However, flowering of flc, flm, and flc flm mutants was delayed at a chilling temperature of 10°C, indicating that these mutants showed temperature-sensitive flowering at 10°C. This further indicated that FLC and FLM function primarily from 16 to 27°C (Lee et al., 2013), suggesting that other genes may play important roles at lower temperatures. In this study, we observed that PAF1C-deficient mutants had lower transcript levels of FLC and the other FLC-clade genes at 10°C (Figure 3), indicating that FLC-clade genes are involved in repressing flowering at 10°C. Our ChIP-qPCR assays showed that downregulation of FLC and FLC-clade genes in PAF1C-deficient mutants is associated with higher enrichment of the repressive mark H3K27me3 and reduced levels of the permissive marks H3K4me3 and H3K36me3 in the chromatin of these genes (Figure 4). This is consistent with previous findings that PAF1C-deficient mutants had reduced FLC and FLM expression due to the reduced H3K4me3 levels at these loci (He et al., 2004; Oh et al., 2004; Xu et al., 2008). Moreover, we showed that PAF1C-mediated epigenetic regulation is not limited to FLC and FLM, but also affects the entire FLC clade.
A previous study showed that MAF3 function is more important at lower temperatures than higher temperatures, as the flc flm maf3 triple mutants flowered earlier than flc flm double mutants at 16°C, compared to 23°C (Gu et al., 2013). This supports our hypothesis that FLC-clade genes are important to repress flowering at low temperatures, as the 35S::amiR-MAF2-5 flc flm plants flowered significantly earlier than the flc flm double mutants. Furthermore, we observed induction of FLC and FLC-clade genes in wild-type plants at 10°C (Figure 5), supporting previous findings that the mRNA levels of FLC and FLM-ß increased at low temperatures (16°C compared to 23°C; Lee et al., 2007; Pose et al., 2013). This might mean that plants increase transcription of these FLC family genes in response to low temperatures to ensure efficient floral repression, and this increase in transcription likely requires PAF1C. Since PAF1C components and functions are conserved from unicellular yeast to complex eukaryotic organisms (Tomson and Arndt, 2013), this regulatory mechanism might be conserved and have important functions in other plant species.
One important question raised by our observations is how these MAFs play such a critical role in floral repression at 10°C. One possible answer is their effect on SVP function. MADS-box transcription factors physically interact to form larger complexes that synergistically enhance their abilities to regulate transcription. Consistent with this, the MADS-box transcription factors FLC (Fujiwara et al., 2008; Li et al., 2008) and FLM (Lee et al., 2013) form floral repressor complexes with SVP and enhance their repression of flowering. Our data revealed that SVP interacts with all five MAF transcription factors in vitro and in vivo (Figure 8). This finding is consistent with our observation that in plants that lack (or have downregulated expression of) FLC and FLC-clade transcription factor genes, such as the PAF1C-deficient mutants and 35S::amiR-MAF2-5 flc flm plants, SVP alone is not sufficient to repress flowering, especially at lower temperatures. Consistent with this notion, SVP overexpression was unable to delay flowering in PAF1C-deficient elf7 and vip4 mutants (Figure 9; Supplementary Figure 6), with significantly low levels of FLC and FLC-clade transcripts, suggesting that SVP function depends on FLC and FLC-clade transcription factors, and thus providing novel insight into SVP protein function at low temperatures. However, it should be noted that further genetic interaction analyses, such as analyses of plants overexpressing SVP in the 35S::amiR-MAF2-5 flc flm background, will provide more direct genetic evidence.
Furthermore, our analysis of a previously published ChIP-seq dataset suggested that the presence/absence of FLC influences SVP binding to its targets, as the number of SVP-bound targets nearly doubled in the presence of functional FLC, implying that SVP function depends on FLC, as previously reported (Mateos et al., 2015). It is likely that FLC-clade transcription factors play similar roles, enhancing SVP binding to its targets and/or enhancing its ability to repress transcription; however, further experiments are required to confirm this hypothesis. It would be interesting to perform a genome-wide analysis of whether SVP can bind and repress its target genes in plants with reduced expression of FLC and FLC-clade genes, such as PAF1-deficient mutants or the 35S::amiR-MAF2-5 flc flm plants.
In conclusion, our findings showed that PAF1C epigenetically regulates all the FLC-clade genes and that these genes play an important role in repressing flowering at chilling temperatures by forming floral repressor complexes with SVP. Wild-type plants accumulate higher levels of FLC-clade transcripts in response to chilling temperature to prevent precocious flowering. Our work uncovers the functional importance of MAF transcription factors in repressing flowering at chilling temperatures and increases the current understanding of how flowering is regulated in response to temperature.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/, under the accession number GSE171778.
Author Contributions
ZN and JA designed the research. ZN performed the bioinformatic analyses and conducted experimental work. HS helped with ChIP-qPCR experiments. SJ and GY provided technical assistance. JA supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (NRF-2017R1A2B3009624 to JA) and Samsung Science and Technology Foundation under Project Number SSTF-BA1602-12.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Elizabeth S. Haswell for providing research materials, and Y. J. Kim for her technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.817356/full#supplementary-material
Footnotes
References
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Balasubramanian, S., Sureshkumar, S., Lempe, J., and Weigel, D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2:e106. doi: 10.1371/journal.pgen.0020106
Bernier, G., and Périlleux, C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3, 3–16. doi: 10.1111/j.1467-7652.2004.00114.x
Betz, J., Chang, M., Washburn, T., Porter, S., Mueller, C., and Jaehning, J. (2002). Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Gen. Genomics. 268, 272–285. doi: 10.1007/s00438-002-0752-8
Bouché, F., Lobet, G., Tocquin, P., and Périlleux, C. (2016). FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, D1167–D1171. doi: 10.1093/nar/gkv1054
Boyes, D. C., Zayed, A. M., Ascenzi, R., McCaskill, A. J., Hoffman, N. E., Davis, K. R., et al. (2001). Growth stage–based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510. doi: 10.1105/TPC.010011
Caicedo, A. L., Stinchcombe, J. R., Olsen, K. M., Schmitt, J., and Purugganan, M. D. (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. 101, 15670–15675. doi: 10.1073/pnas.0406232101
Cao, Y., Wen, L., Wang, Z., and Ma, L. (2015). SKIP interacts with the Paf1 complex to regulate flowering via the activation of FLC transcription in Arabidopsis. Mol. Plant 8, 1816–1819. doi: 10.1016/j.molp.2015.09.004
Cho, H. J., Kim, J. J., Lee, J. H., Kim, W., Jung, J.-H., Park, C.-M., et al. (2012). SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett. 586, 2332–2337. doi: 10.1016/j.febslet.2012.05.035
Choi, K., Kim, J., Hwang, H.-J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
De Lucia, F., Crevillen, P., Jones, A. M., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. 105, 16831–16836. doi: 10.1073/pnas.0808687105
Fal, K., Cortes, M., Liu, M., Collaudin, S., Das, P., Hamant, O., et al. (2019). Paf1c defects challenge the robustness of flower meristem termination in Arabidopsis thaliana. Development 146:dev173377. doi: 10.1242/dev.173377
Fujiwara, S., Oda, A., Yoshida, R., Niinuma, K., Miyata, K., Tomozoe, Y., et al. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20, 2960–2971. doi: 10.1105/tpc.108.061531
Gu, X., Jiang, D., Wang, Y., Bachmair, A., and He, Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57, 522–533. doi: 10.1111/j.1365-313X.2008.03709.x
Gu, X., Le, C., Wang, Y., Li, Z., Jiang, D., Wang, Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4:1947. doi: 10.1038/ncomms2947
He, Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564. doi: 10.1093/mp/ssp005
He, Y., Doyle, M. R., and Amasino, R. M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18, 2774–2784. doi: 10.1101/gad.1244504
He, Y., Michaels, S. D., and Amasino, R. M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. doi: 10.1126/science.1091109
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P., and Minghim, R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 16:169. doi: 10.1186/s12859-015-0611-3
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W., and Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Hong, S. M., Bahn, S. C., Lyu, A., Jung, H. S., and Ahn, J. H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51, 1694–1706. doi: 10.1093/pcp/pcq128
Ignatchenko, V., Ignatchenko, A., Sinha, A., Boutros, P. C., and Kislinger, T. (2015). VennDIS: A JavaFX-based Venn and Euler diagram software to generate publication quality figures. Proteomics 15, 1239–1244. doi: 10.1002/pmic.201400320
Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. doi: 10.1126/science.290.5490.344
Kim, S. Y., He, Y., Jacob, Y., Noh, Y.-S., Michaels, S., and Amasino, R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17, 3301–3310. doi: 10.1105/tpc.105.034645
Kim, D.-H., and Sung, S. (2010). The plant Homeo domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc. Natl. Acad. Sci. 107, 17029–17034. doi: 10.1073/pnas.1010834107
Lee, Y. J., Kim, D. H., Kim, Y.-W., and Hwang, I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190. doi: 10.1105/tpc.010232
Lee, J. H., Ryu, H.-S., Chung, K. S., Posé, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., and Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Lex, A., Gehlenborg, N., Strobelt, H., Vuillemot, R., and Pfister, H. (2014). UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992. doi: 10.1109/TVCG.2014.2346248
Li, D., Liu, C., Shen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Li, Y., Yang, J., Shang, X., Lv, W., Xia, C., Wang, C., et al. (2019). SKIP regulates environmental fitness and floral transition by forming two distinct complexes in Arabidopsis. New Phytol. 224, 321–335. doi: 10.1111/nph.15990
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mateos, J. L., Madrigal, P., Tsuda, K., Rawat, V., Richter, R., Romera-Branchat, M., et al. (2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16:31. doi: 10.1186/s13059-015-0597-1
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Nasim, Z., Fahim, M., Hwang, H., Susila, H., Jin, S., Youn, G., et al. (2021). Nonsense-mediated mRNA decay modulates Arabidopsis flowering time via the SET DOMAIN GROUP 40–FLOWERING LOCUS C module. J. Exp. Bot. 72, 7049–7066. doi: 10.1093/jxb/erab331
Ng, H. H., Dole, S., and Struhl, K. (2003a). The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625–33628. doi: 10.1074/jbc.C300270200
Ng, H. H., Robert, F., Young, R. A., and Struhl, K. (2003b). Targeted recruitment of Set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719. doi: 10.1016/S1097-2765(03)00092-3
Oh, S., Zhang, H., Ludwig, P., and van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16, 2940–2953. doi: 10.1105/tpc.104.026062
Pajoro, A., Severing, E., Angenent, G., and Immink, R. (2017). Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 18:102. doi: 10.1186/s13059-017-1235-x
Pose, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Postma, M., and Goedhart, J. (2019). PlotsOfData—A web app for visualizing data together with their summaries. PLoS Biol. 17:e3000202. doi: 10.1371/journal.pbio.3000202
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J., and Riechmann, J. L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. doi: 10.1105/tpc.009506
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L., and Riechmann, J. L. (2001). Regulation of flowering in Arabidopsis by an FLCHomologue. Plant Physiol. 126, 122–132. doi: 10.1104/pp.126.1.122
Romero-Molina, S., Ruiz-Blanco, Y. B., Harms, M., Münch, J., and Sanchez-Garcia, E. (2019). PPI-detect: a support vector machine model for sequence-based prediction of protein–protein interactions. J. Comput. Chem. 40, 1233–1242. doi: 10.1002/jcc.25780
Rosloski, S. M., Singh, A., Jali, S. S., Balasubramanian, S., Weigel, D., and Grbic, V. (2013). Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Mol. Biol. 81, 57–69. doi: 10.1007/s11103-012-9982-2
Ruelens, P., De Maagd, R. A., Proost, S., Theißen, G., Geuten, K., and Kaufmann, K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280. doi: 10.1038/ncomms3280
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Scortecci, K. C., Michaels, S. D., and Amasino, R. M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M that represses flowering. Plant J. 26, 229–236. doi: 10.1046/j.1365-313x.2001.01024.x
Scortecci, K., Michaels, S. D., and Amasino, R. M. (2003). Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 52, 915–922. doi: 10.1023/A:1025426920923
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. doi: 10.1105/tpc.11.3.445
Sledzieski, S., Singh, R., Cowen, L., and Berger, B. (2021). Sequence-based prediction of protein-protein interactions: a structure-aware interpretable deep learning model. bioRxiv [Preprint]. doi: 10.1101/2021.01.22.427866
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Susila, H., Nasim, Z., Jin, S., Youn, G., Jeong, H., Jung, J.-Y., et al. (2021). “Profiling protein–DNA interactions by chromatin immunoprecipitation in Arabidopsis,” in Proteomic Profiling. ed. A. Posch (New York City, US: Springer), 345–356.
Tomson, B. N., and Arndt, K. M. (2013). The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829, 116–126. doi: 10.1016/j.bbagrm.2012.08.011
Wickham, H. (2011). ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185. doi: 10.1002/wics.147
Wood, C. C., Robertson, M., Tanner, G., Peacock, W. J., Dennis, E. S., and Helliwell, C. A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. 103, 14631–14636. doi: 10.1073/pnas.0606385103
Wood, A., Schneider, J., Dover, J., Johnston, M., and Shilatifard, A. (2003). The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742. doi: 10.1074/jbc.C300269200
Wu, F.-H., Shen, S.-C., Lee, L.-Y., Lee, S.-H., Chan, M.-T., and Lin, C.-S. (2009). Tape-Arabidopsis sandwich-a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. doi: 10.1186/1746-4811-5-16
Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J.-P., Steinmetz, A., et al. (2008). Di-and tri-but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28, 1348–1360. doi: 10.1128/MCB.01607-07
Yu, X., and Michaels, S. D. (2010). The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol. 153, 1074–1084. doi: 10.1104/pp.110.158386
Zhang, H., Ransom, C., Ludwig, P., and Van Nocker, S. (2003). Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch flowering locus C. Genetics 164, 347–358. doi: 10.1093/genetics/164.1.347
Keywords: Arabidopsis, FLC, MAFs, PAF1C, flowering, epigenetics, temperature
Citation: Nasim Z, Susila H, Jin S, Youn G and Ahn JH (2022) Polymerase II–Associated Factor 1 Complex-Regulated FLOWERING LOCUS C-Clade Genes Repress Flowering in Response to Chilling. Front. Plant Sci. 13:817356. doi: 10.3389/fpls.2022.817356
Edited by:
Marta Adelina Mendes, University of Milan, ItalyReviewed by:
Alice Pajoro, Max Planck Institute for Plant Breeding Research, GermanyMartina Cerise, Max Planck Institute for Plant Breeding Research, Germany
Copyright © 2022 Nasim, Susila, Jin, Youn and Ahn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji Hoon Ahn, amFobkBrb3JlYS5hYy5rcg==
 Zeeshan Nasim
Zeeshan Nasim Hendry Susila
Hendry Susila Suhyun Jin
Suhyun Jin Geummin Youn
Geummin Youn Ji Hoon Ahn
Ji Hoon Ahn