
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 May 2022
Sec. Plant Nutrition
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.816475
 Maria Oszvald1
Maria Oszvald1 Kirsty L. Hassall2
Kirsty L. Hassall2 David Hughes2
David Hughes2 Adriana Torres-Ballesteros3
Adriana Torres-Ballesteros3 Ian Clark3
Ian Clark3 Andrew B. Riche1
Andrew B. Riche1 Sigrid Heuer1,4*
Sigrid Heuer1,4*
Crops assimilate nitrogen (N) as ammonium via the glutamine synthetase/glutamate synthase (GS/GOGAT) pathway which is of central importance for N uptake and potentially represents a bottle neck for N fertiliser-use efficiency. The aim of this study was to assess whether genetic diversity for N-assimilation capacity exists in wheat and could be exploited for breeding. Wheat plants rapidly, within 6 h, responded to N application with an increase in GS activity. This was not accompanied by an increase in GS gene transcript abundance and a comparison of GS1 and GS2 protein models revealed a high degree of sequence conservation. N responsiveness amongst ten wheat varieties was assessed by measuring GS enzyme activity, leaf tissue ammonium, and by a leaf-disc assay as a proxy for apoplastic ammonia. Based on these data, a high-GS group showing an overall positive response to N could be distinguished from an inefficient, low-GS group. Subsequent gas emission measurements confirmed plant ammonia emission in response to N application and also revealed emission of N2O when N was provided as nitrate, which is in agreement with our current understanding that N2O is a by-product of nitrate reduction. Taken together, the data suggest that there is scope for improving N assimilation capacity in wheat and that further investigations into the regulation and role of GS-GOGAT in NH3 emission is justified. Likewise, emission of the climate gas N2O needs to be reduced, and future research should focus on assessing the nitrate reductase pathway in wheat and explore fertiliser management options.
Nitrogen (N) is essential for plant growth and is a primary driver of crop yield and grain quality. One of the main challenges for modern agriculture is to reduce the use of N-fertiliser, whilst maintaining or increasing grain yield (Hirel et al., 2011; Hawkesford, 2014). Currently, the amount of applied fertiliser required to reach the potential yield of wheat in the United Kingdom, which is estimated at 20 t ha–1, ranges from 150 kg to 350 kg ha–1 (Mitchell and Sheehy, 2018). However, generally less than half of the applied N-fertiliser is assimilated by crops (Masclaux-Daubresse et al., 2010) and the runoff of excess N contaminates water ways and causes emission of gaseous ammonia (NH3) and the potent greenhouse gas nitrous oxide (N2O) from soil microbial activities (Linquist et al., 2012; Gao et al., 2019).
Plants have developed sophisticated and highly regulated N-uptake systems. Gene families coding for low- and high-affinity NO3– and NH4+ transporters, respectively, have been identified across different plant species and are located in roots, for primary N acquisition, and other organs for internal distribution and storage (for a review see Li et al., 2017). Once in the plant, NO3– is reduced to NH4+ by nitrate reductase (NR) and nitrite reductase (NiR) (Campbell, 1988) which is then assimilated by the glutamine synthetase-glutamate synthase (GS-GOGAT) cycle, generating glutamine (Balotf et al., 2016). The GS-GOGAT cycle is thus central to the ability of plants to assimilate and use N (e.g., Bernard and Habash, 2009; Thomsen et al., 2014).
Genes in the GS-GOGAT pathway have been studied in some detail, revealing that glutamine synthetase occurs in two major forms, one localised to the cytosol (GS1) and the other localised to the chloroplast (GS2) (García-Gutiérrez et al., 2018). In diploid plants, GS1 is usually encoded by three to five genes, predominantly expressed in the vascular tissues and involved in generating glutamine for intercellular N transport (Brugiére et al., 1999; Tabuchi et al., 2005; Cánovas et al., 2007; Bernard et al., 2008). In contrast, GS2 is generally encoded by a single gene and is located in chloroplasts and mitochondria in green tissues (Tingey et al., 1988; Taira et al., 2004). The chloroplastic GS2 isoform assimilates fertiliser NH4+ and NH4+ generated by nitrate reduction and photorespiration in leaves, whereas the cytosolic GS1 isoform is the major form present in roots for primary N assimilation from soil (Lam et al., 1996; Ishiyama et al., 2004). GS1 is also the isoform mainly responsible for N remobilisation during leaf senescence and grain filling (Zhang Z. et al., 2017). In wheat, an initial study had identified ten GS cDNA sequences classified into four subfamilies (GS2, GS1, GSr, and GSe; Bernard et al., 2008). Gene names for the GS1 genes were subsequently updated to GS1;1, GS1;2, and GS1;3 (Wei et al., 2021a) and detailed gene expression and immunolocalisation studies of the proteins showed tissue-specific expression and differential N responsiveness of the individual genes (Wei et al., 2020, 2021b).
A range of plant species have been modified to overexpress cytosolic GS1, following different strategies with regards to the promoter used and transgene selection (for review see Thomsen et al., 2014; Li et al., 2017). The outcomes of these studies have been variable but underscored the importance of cytosolic GS1 for efficient N assimilation, plant growth, and biomass accumulation. For instance, in barley it has been shown that cis-genic overexpression of HvGS1-1 improved grain yield and nitrogen use efficiency (NUE), which is encouraging (Gao et al., 2019). In light of the importance of this pathway, there are surprisingly few studies that have investigated whether natural variation in GS enzyme activity exists. Evidence for this has been provided in studies in rice (Kumagai et al., 2011a,b) and tomato (Sánchez-Rodríguez et al., 2011), and a recent study in durum wheat showed differences in GS activity and GS gene expression in genotypes with contrasting grain protein content (Nigro et al., 2016). In the present study, we are complementing these findings with a comparative study of GS enzyme activity and gene expression in response to N fertiliser in ten bread wheat genotypes, providing further evidence that natural variation does exist that can be exploited for breeding.
Whilst NH4+ is essential for plant growth and development, at high micromolar concentrations it is toxic (Britto and Kronzucker, 2002) and it has been suggested that tolerance is related to N assimilation capacity (Cruz et al., 2006). Ammonium toxicity can lead to the inhibition of root and shoot growth and to leaf chlorosis, and it has been related to ionic imbalances, disturbance of pH gradients across plasma membranes and oxidative stress (Gerendás et al., 1997; Britto and Kronzucker, 2002; Bittsánszky et al., 2015; Esteban et al., 2016; Li et al., 2017). High levels of intracellular NH4+ can also build up via plant-internal processes, such as lignin biosynthesis, protein and nucleic acid degradation, and photorespiration (Taira et al., 2004; Carvalho et al., 2011; Kumagai et al., 2011b; Bittsánszky et al., 2015; Melino et al., 2018), with the latter two especially relevant under senescence and stress, such as drought or high light. Importantly, it has already been shown that GS activity is compromised under drought stress, for example, and it is therefore important to identify genotypes that maintain GS-GOGAT activity under stress to prevent cellular damage and maintain growth (Sánchez-Rodríguez et al., 2011).
Whilst the importance of emission of N gas from intensive agricultural soils is well known and recently re-gained attention in relation to the evolving field of microbiome studies, it is less widely known that N-gas emission also occurs from plants. Apoplastic NH4+ and NH3 emission was first shown more than 20 years ago in studies in barley (Mattsson and Schjoerring, 1996; Mattsson et al., 1997; Pearson et al., 1998), wheat (Parton et al., 1988a,b; Morgan and Parton, 1989), oilseed rape (Husted and Schjoerring, 1995) and grasses (Heckathorn and DeLucia, 1995; Hanstein and Felle, 1999; Mattsson and Schjoerring, 2002; van Hove et al., 2002; Loubet et al., 2012), and more recently also in rice (Kumagai et al., 2011a). First reports on N2O emission from plants came from studies in tobacco (Goshima et al., 1999; Hakata et al., 2003) and wheat (Smart and Bloom, 2001) and this was confirmed in more recent studies in wheat (Baruah et al., 2012), as well as in rice (Gogoi and Baruah, 2012) and grasses (Bowatte et al., 2014). Furthermore, an extensive study conducted by Lenhart et al. (2019) measured N2O emission from thirty-two plant species and the significance of this was discussed in relation to climate change.
Taken together, there is sufficient justification for further investigation into diversity of N assimilation capacity in crops and this has gained importance with regards to emission of gasses involved in climate change. In this study we therefore assessed allelic variation in GS genes, GS enzyme activity and N-gas emission within United Kingdom bread wheat to explore, within an already elite germplasm pool, whether genetic diversity exists that can be exploited for improving NUE and reducing N-gas emission. The data presented suggest that there are constitutive differences in N assimilation capacity and GS activity within elite modern wheat. The data also confirm that wheat does emit N gasses in response to ammonia and nitrate fertiliser application, and that N2O emission is linked to nitrate.
Ten United Kingdom wheat (Triticum aestivum L.) varieties were used in this study, including two spring-type bread wheat varieties (Cadenza and Paragon) and eight winter wheat varieties (Alchemy, Brompton, Claire, Hereward, Rialto, Robigus, Soissons, and XL-19). The winter wheat varieties were chosen because they were used as parents for the development of an “elite MAGIC” population (Mackay et al., 2014) and seeds were provided by NIAB, Cambridge, United Kingdom. Seeds of the spring wheat varieties are available at Rothamsted Research, Harpenden, United Kingdom.
Seeds were surface sterilised with 100% EtOH for 2 min and 5% bleach for 15 min. After 5× washing with sterile water, seeds were germinated on wet filter paper at room temperature (RT) for 7 days before transfer into plastic pots (9 × 9 × 10 cm for the genetic diversity study, 13 × 10 × 11 cm for the gas emission experiment, (see below) containing potting mix composed of 75% medium grade peat, 12% screened sterilised loam, 3% medium grade vermiculite, 10% grit (5 mm screened, lime free), 3.5 kg m–3 Osmocote Exact (total N 16%) (Supplier: Scotts UK Professional, Ipswich, Suffolk, United Kingdom) and 0.5 kg m–3 PG compound fertiliser (Supplier: Yara UK Ltd., Harvest House, Europarc, Grimsby, North East Lincolnshire). For the N-gas emission the same potting mix was used but without Osmocote (see below). Pots were placed on benches in a controlled environment glasshouse and plants were grown at 22°C/16°C (day/night) with 16 h light. To keep the light intensity above 400 μmol m–2 s–1, supplementary light was supplied by SON-T lighting as required. Plants were watered daily from the bottom with tap water.
To assess temporal GS enzyme activity in response to N-fertiliser application, an initial time course experiment was conducted with the two spring wheat varieties Cadenza and Paragon. Plants were grown as described above in a completely randomised design. At the 2–3 tiller vegetative stage each pot was supplied with NH4NO3 equivalent to 150 kg N ha–1 (0.345 g NH4NO3) and 250 kg N ha–1 (0.575 g NH4NO3), respectively, dissolved in 50 ml distilled water.
The calculation was based on pot area (cm2)/ha * amount of N fertiliser * % of the N content of the fertiliser.
N was applied at 10 a.m. and the youngest fully expanded leaf from six replicate plants for each treatment were harvested at 2, 6, 24, and 30 h after N application and immediately frozen in liquid N and stored at −80°C for further analysis. Water was supplied to control plants which were always sampled in parallel.
To assess the effect of water-stress on N responses, a dry-down pot experiment was carried out with Cadenza and Paragon. Plants were grown as described above and watered daily from the bottom with tap water. At the 3–4 tiller stage watering was discontinued for a subset of the plants to induce water stress. In a pilot study, leaf water content was measured during the dry down period as an indicator of water stress. For this, sampled leaves were weighted (W) before incubation in tap water for 3–4 h in the light at RT. After tapping the leaves dry leaves were weighed (TW) and samples were oven dried 80°C for 24 h and dry weight (DW) determined. Relative water content was calculated as RWC (%) = [(W-DW)/ (TW-DW)] × 100. For subsequent experiments, samples at 72 h after end of watering were collected, when RWC was reduced by about 20% compared to control plants.
Eight winter wheat varieties (see above), in addition to Paragon and Cadenza, were grown in a randomised complete block design with six replicates for each treatment in the greenhouse as described above. Plants at the vegetative state (4–5 weeks old plants) were treated with the equivalent of 250 kg N ha–1 supplied as NH4NO3 (0.575 g per pot) and urea (0.264 g per pot), respectively, dissolved in 50 ml de-ionised water. De-ionised water was supplied to control plants. Plants were treated at 10 a.m. and the youngest fully expanded leaf of individual plants was harvested before (0 h), and at 6 and 30 h after N application. Control plants were always sampled in parallel. Samples for the leaf-disc ammonium assay (see below) were taken and directly transferred into buffer. The remainder of the leaf was frozen in liquid N and stored at −80°C for analyses of GS activity and quantification of tissue ammonium (see below).
The field site was located at the Rothamsted Experimental Farm (Harpenden, United Kingdom) and utilised the long-term Wheat Genetic Improvement Network (WGIN) Diversity Trial, which was in its 16th year at the time of this experiment (2019). The experiment was set up as a fully randomised split plot design, with three replicates. Plots (1.8 × 9 m) were sown on 09/10/2018 and standard farm protocols were used for crop protection. Three levels of N fertilisation were applied in three splits, amounting to a total seasonal N doses equivalent to 100 (N1), 200 (N2), and 350 (N3) kg N ha–1, respectively. For the present study, two selected varieties (Cadenza and Paragon) were sampled from the three replicate N1 and N3 plots, directly before (T0) and at 48 h after application of the second N split (50 kg N ha–1 for N1; 250 kg N ha–1 for N3). Both plots had previously received 50 kg N in the first split application (February) and received a final, third split application of 50 kg N in May. Leaf samples of six randomly selected plants from each plot were immediately frozen in liquid N and stored at −80°C for further analysis.
Three selected genotypes (Cadenza, Paragon, and Claire) were grown under controlled conditions as describe above in a randomised complete block design to measure N gas emission using a Picarro G2508 Greenhouse Gas Analyser (Picarro Inc., Santa Clara, CA, United States). One-week-old seedlings were transplanted into pots (13 × 10 × 11 cm) with potting mix (see above) without additional nutrients. After 24 h, plants were supplied with a modified Letcombe’s solution according to Masters-Clark et al. (2020). For the low-N treatment, 0.325 ml of KNO3 (1.6 M) was supplied to each pot and 3.25 ml were supplied for the high-N treatment. Each pot received the same amount of P (3.25 ml, 0.17 M KH2PO4) and micronutrients (Masters-Clark et al., 2020). Plants were kept well-watered using tap water. For the gas measurement, plants at the 3–4 tiller stage were treated with the equivalent of 250 kg ha–1 N supplied either as KNO3 or NH4NO3. Water was supplied to controls. The soil surface of each pot was covered with aluminium foil and cling film before the Picarro clear plexiglass chambers were mounted and sealed with tape. As a soil control, pots were used from which the above-ground plant biomass was removed to account for any emission coming from roots and/or the soil microbiome. These pots were treated the same way as pots with plants. The experiment was carried out in six replicates for each genotype and treatment. The Picarro instrument was set up to measures sixteen chambers for 3:49 min each consecutively and repeatedly over 24 h. Data output from the G2508 was on average every 2 s as dry mole fraction in ppb for NH3 gas, and ppm for N2O. The flow rate between the G2508 and incubation vessel and chamber were 250 mL min–1 using a low-leak external vacuum pump. At the end of the experiment, above-ground biomass was oven dried at 80°C for 24 h to determine the dry weight, which was used to normalize maximum NH3 and N2O gas emissions.
A second experiment was carried out to compare Cadenza and Paragon with an elite wheat from a comparably low-N environment and for this the variety Mace (AGT, Australia) was used. The experiment was conducted as described above except that plants were grown under high-N conditions in potting mix supplemented with Osmocote (see above). Plants were treated with the equivalent of 250 kg N ha–1 supplied as NH4NO3. Water was supplied to controls.
A third experiment was carried out to investigate if N2O gas emission is derived from nitrate reduction and/or ammonia assimilation. For this, Cadenza plants were grown under low N conditions as described above and treated with the equivalent of 250 kg N ha–1 supplied as urea and KNO3 respectively with and without 150 μM of the nitrate reductase inhibitor (NI) tungsten sulphate. Water was supplied to controls.
Total (combined GS1 and GS2) extractable GS activity was determined in 100 mg ground fresh leaf material extracted in a 1 mL buffer solution containing 25 mM TRIS–HCl, 1 mM EDTA, 1 mM DTT, 1 mM MgCl2, 0.2 g polyvenyl pyrrolidone and 2 mM leupeptin, pH 7.6. GS activity was measured according to O’Neal and Joy (1974) by incubating 50 μl leaf extracts in an assay medium containing 600 mM glutamate, 45 mM hydroxylamine hydrochloride, 150 mM MgSO4*7H2O, 30 mM EDTA-Na2, and 60 mM ATP for 30 min at 30°C in a shaker incubator. The enzyme reaction was terminated with 300 μl stopping solution (0.67M FeCl3, 0.2M TCA, 0.7M HCl). After centrifugation (5 min, 13,000 × g) the optical density of supernatants was measured at 540 nm using a Jenway 6715 UV/Vis spectrophotometer.
For the present study, a protocol for the extraction of apoplastic ammonium (Husted and Schjoerring, 1995) was modified and we refer to this method as “leaf-disc” ammonium assay.
For this, four leaf-discs (5 mm diameter) from the middle of a fully expanded leaf were cut with a hole punch and immediately submerged in 1 ml isotonic sorbitol solution (0.28M) in an Eppendorf tube. Samples were incubated at 4°C for 24 h before vacuum infiltration in a desiccator (DURAN, DN 150) for 2 min at a pressure of 4 atm. In contrast to the published protocol, leaf-discs were centrifuged within the sorbitol solution (2,000 × g, 5 min, 4°C) and aliquots (10 μl) of the supernatant were used to measure NH4+ concentration using the Megazyme rapid Ammonia kit (K-AMIAR 04/18). This assay is based on reductive amination of 2-oxoglutarate with glutamate dehydrogenase (GDH) and NADPH. Assays were always performed with two technical replicates and ammonium concentration was normalised to leaf-disc weight.
The absence of cytosolic contamination of the leaf-disc extract with cytoplasm was confirmed by measuring GS enzyme activity (see below) and activity of the cytosolic glycolysis enzyme G6PDH according to Lohr and Waller (1974). Briefly, G6PDH activity is measured in a spectrophotometer following the reduction of NADP in a solution containing 66 mM KH2PO4/K2HPO4 (pH 7.6), 10 mM MgCl2, 300 mM NADP, 2 mM glucose-6-phosphate (G6P) and 10 μl of leaf tissue extracts in a final volume of 300 μl. G6PDH activity in the samples was calculated using a standard curve for NADPH at 340 nm (Spectra 340PC plate reader). All assays were performed in two technical replicates.
To determine the NH4+ concentration in leaf tissue, 100 mg of fully expanded leaf samples were ground in liquid nitrogen and incubated in 1 ml 0.05M H2SO4 at RT for 1 h. The homogenate was centrifuged at 13,000 × g for 15 min at RT and the supernatant was used to quantify NH4+ using the Megazyme rapid ammonia kit (K-AMIAR 04/18) according to the manufacturer’s instructions.
The nitrate content in leaves was determined in water extracts of 100 mg ground leaf tissue according to Zhao and Wang (2017). Samples were mixed with 100 μl of deionised water and boiled at 100°C for at least 20 min. Deionised water (0.1 ml) was used as a control. After centrifugation at 15,871 × g for 10 min, the supernatant was transferred into a new tube and 40 μl salicylic acid-sulphuric acid was added. The reaction was incubated at RT for 20 min before 0.95 ml of 8% (w/v) NaOH solution was added and tubes were allowed to cool to RT for 20–30 min. The OD410 value of each sample was measured in a spectrophotometer and water was used as a blank. The nitrate concentration was calculated as Y = CV/W [Y: nitrate content (μg/g); C: nitrate concentration calculated with OD410 using a regression equation from the standard curve (μg/ml); V: total volume of extracted sample (ml); W: weight of sample (g)].
Glutamine was extracted from 50 mg frozen, ground leaf samples with 0.5 ml 80% EtOH for 3 h at 40°C in a heating block and centrifuged at 10,000 × g for 15 min at RT. The supernatant (10 μl) was used to determine leaf L-glutamine content using the L-glutamine/ammonia rapid kit from Megazyme (K-GLNAM 04/17). Samples were normalised to 100 mg FW.
The total soluble protein concentration in leaves was measured using the Bradford method with BSA as standard (Bradford, 1976).
Genomic sequences of the wheat GS1 and GS2 genes were initially retrieved from the Chinese Spring reference genome (IWGSC, INSDC Assembly GCA_900519105.1; July 2018) available in Ensembl Plants.1 Accession numbers are provided in Table 1. Representatives of the different GS1 and GS2 homeologs were then used for a BLASTp search in Ensembl Plants to ensure all GS genes and homeologs had been identified. This search revealed an additional low confidence (LC) GS1 homeolog on chromosome 6D. The chromosomal position of the individual sequences was derived from Ensembl Plants. The genomic sequences of representative Chinese Spring GS1 and GS2 homeologs were then used for a BLASTn search of the available genomic scaffolds of four wheat varieties (Cadenza, Paragon, Robigus, and Claire)2 and a tetraploid durum wheat (Kronos) (sequence versions 2018-02-19).3 The identified GS-gene containing scaffolds (Supplementary Table 1) were aligned to the genomic sequence of the respective GS Chinese Spring gene sequences using the LASTZ algorithm of the Geneious 10.2.3 software and the aligned regions with some 5′ and 3′ flanking regions were extracted. Gene models were predicted by aligning the extracted scaffold sequences with the respective Chinese Spring genomic sequence, the longest cDNA, and the corresponding coding regions (CDR) retrieved from Ensembl Plants. Extraction of the putative CDRs, translation into protein, and sequence alignments using the MAFFT algorithm were conducted using Geneious 10.2.3. A comparison of the predicted protein sequences to create a circular tree was done in Geneious. Additional gene expression data were derived from the wheat expression database.4
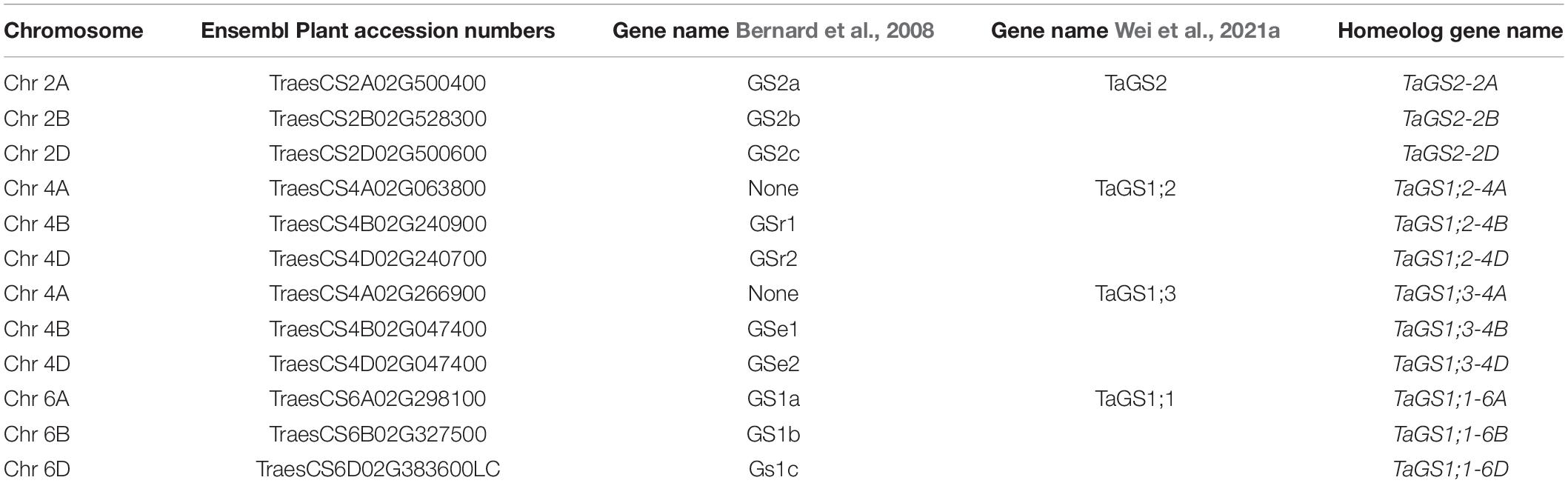
Table 1. Putative glutamine synthetase (GS1 and GS2) genes from the Chinese Spring reference genome.
Total RNA was extracted from 50 mg frozen, ground leaf samples using TRIzol reagent (Invitrogen Cat No. 15596-026) (Chomczynski and Sacchi, 1987). Integrity of the RNA was confirmed in a 2% (w/v) agarose gel and RNA concentration was quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, United States). cDNA synthesis was performed with 2 μg of total RNA per sample using Invitrogen Superscript III reverse transcriptase with dT-adapter primers (Invitrogen, United Kingdom) in a 20 μl reaction according to the manufacturer’s protocol. cDNA quality was verified by PCR amplification of the actin gene (for primer sequences see Table 2).
For the quantification of gene transcripts, the SYBR® Green Jump Start™ Taq Ready Mix™ (Sigma-Aldrich, United Kingdom) was used in the Applied Biosystems 7500 Real Time PCR System. Each 25 μl reaction contained 0.5 μl cDNA and 10 μM forward and reverse primers, respectively.
For primer design, detailed comparative nucleotide sequence analyses of the above-described sequences were conducted to identify gene- and homeolog-specific regions and primer pairs were positioned manually in selected regions (data not shown). For the purpose of this study, primers were designed targeting regions highly conserved within GS2 and the GS1 genes, respectively, specifically annealing to and amplifying the three GS2 homeologs together, as well as the three GS1 genes and their corresponding homeologs combined. Efficiency of at least three primer pairs were tested in cDNA dilution series and the mean primer efficiency was estimated using the linear phase of all individual reaction amplification curves (Ramakers et al., 2003), calculated by using the LinRegPCR package (Tuomi et al., 2010). Three house-keeping reference genes (Table 2) were used to normalise relative quantification (NRQ) of expression (NE). The NRQ of NE was calculated in relation to the CT values, the primer efficiency (E) of the target gene (X), and the normalising reference gene (N) as Normalised Relative Expression NRE using the formula: NRE = (EX)-CT,X/(EN)CT,N according to Rieu and Powers (2009).
Statistical significance was assessed using ANOVA and Student’s t-test in Genstat 20th Edition (VSNi) and in Microsoft Excel. For both, the drought and genetic diversity experiments no blocking terms were included. For the field trial the block structure block/whole plot/plot was used to reflect the split-plot structure of N application to whole plots. The genetic diversity and field trials consisted of the three-way factorial treatment structure genotype×treatment×time. The drought experiment consisted of the treatment structure [baseline/(time×treatment)]×genotype, where baseline is indicative of the first time point sampled before any treatments were applied. Where appropriate, response variables were transformed to ensure homogeneity of variance, specifically concentration of leaf-disc ammonium was analysed on the square root scale and tissue ammonium was analysed on the scale of the natural logarithm. Small levels of missingness in these three variables were present across the three experiments and were estimated iteratively through the ANOVA. Student’s t-test was applied to test the significance of differences between the data from Cadenza and Paragon in the time course experiment and in the gene expression analysis. For the leaf NO3– and glutamine measurements, a regression analysis was performed, and the F-statistics reported are the sequential F-statistics associated with adding terms in the order time, treatment, genotype and associated higher order interactions. Different orders of fitting were tested and although quantitative results deviated marginally, conclusions remained qualitatively the same. For the comparative analysis of the N response in ten genotypes, mean values were based on sqrt (raw data leaf-disc ammonia) and log2 transformation of data [LFoldChange = log2(treatment/control Mean)]. For the Picarro experiment, ANOVA was used to compare emissions from plant vs. soil using the raw data. To compare emissions between genotypes and treatment effects, ANOVA was carried out on the normalised data with treatment structure comprised of: Genotype*(Trt_type/Treatment)*N.level. To ensure homogeneity of variance, all response variables of the maximum emission data were first log transformed.
To assess the response of glutamine synthetase (GS) to high N application a time-course experiment was conducted using the United Kingdom spring wheat varieties Cadenza and Paragon at the vegetative stage grown in fully fertilised potting mix (Figures 1A,B). Significant differences in GS enzyme activity were observed over time (F3,66 = 16.65, p < 0.001), with this temporal trend varying both with N treatment (F6,66 = 2.72, p = 0.02) and with genotype (F3,66 = 3.13, p = 0.031) Under control conditions (zero additional N), GS activity was similar in both genotypes at all time points with a GS activity around 60 μmol GHA mg FW–1 h–1 NH4+ (Figures 1A,B). The application of the equivalent of 150 kg N ha–1 (as NH4NO3) significantly increased GS activity at 2 h and at 30 h in Paragon (Figure 1A) whereas there was no significant GS response in Cadenza to this N level (Figure 1B). Application of the higher N250 doses had a significant positive effect on GS activity in Paragon at 6 h and in Cadenza at 30 h, increasing GS activity to about 90 and 100 μmol GHA mg FW–1 h–1, respectively.
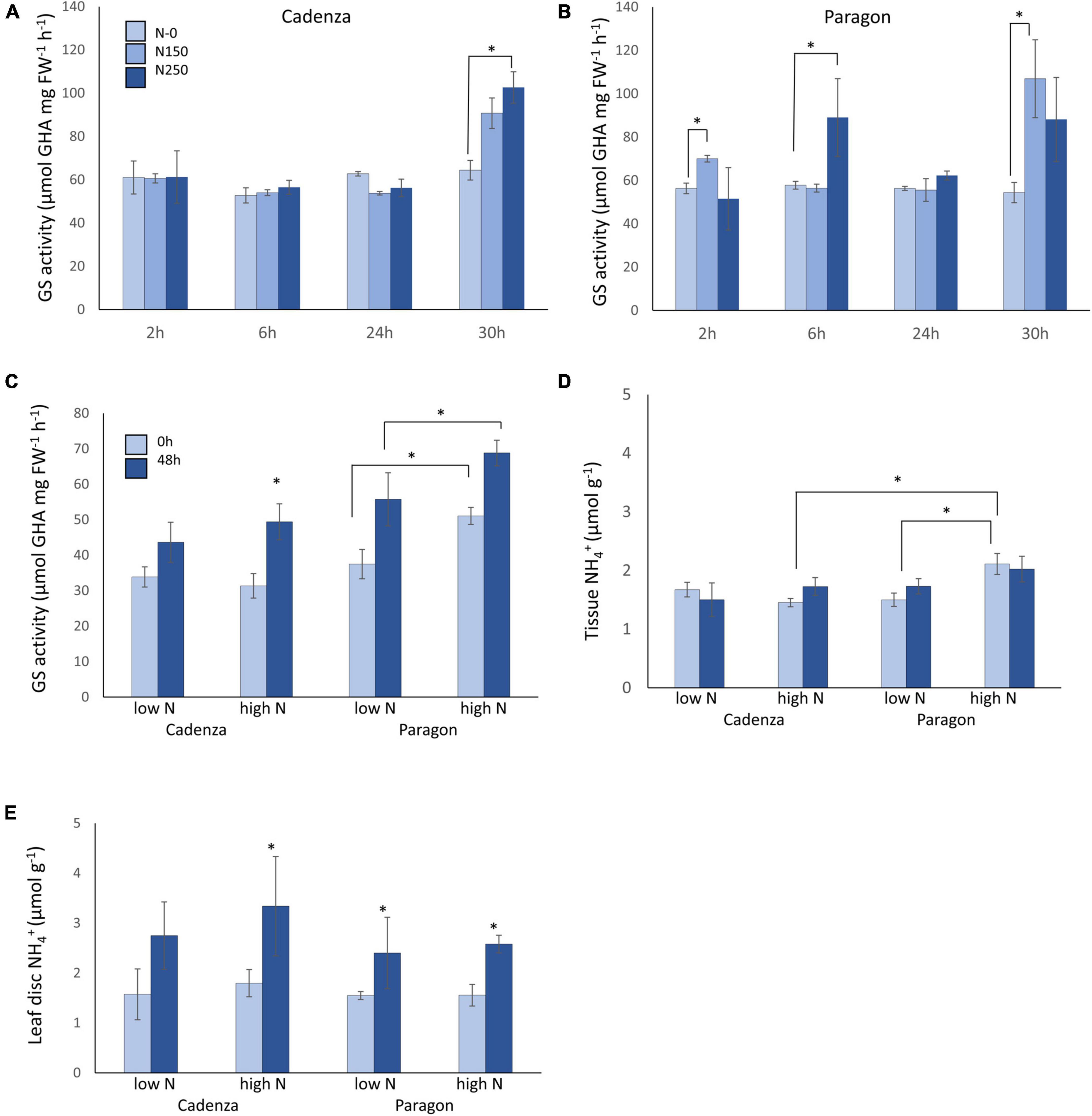
Figure 1. Temporal response of wheat glutamine synthetase to N fertiliser under controlled and field conditions. The response of glutamine synthetase (GS) enzyme activity was measured in two bread wheat genotypes grown in fully fertilised (Osmocote slow-release fertiliser) potting mix. GS activity in leaf samples of Cadenza (A) and Paragon (B) was measured at four timepoints after application of NH4NO3, equivalent to 150 kg N ha–1 (N150) and 250 kg ha–1 (N250), respectively. Water was applied to control plants (N-0) and measured in parallel. Each value represents the mean ± SE of four biological replicates. The same genotypes were sampled from a long-term field experiment with plots adjusted to low (100 kg N ha–1) and high (350 kg N ha–1) total seasonal N application (see text for details). GS activity (C), tissue ammonium (D) and leaf-disc assay ammonium (E) were measured in leaf samples collected before (0 h) and at 48 h after the second split application of NH4SO4 fertiliser equivalent to 50 kg N ha–1 (low N plot) and 250 kg N ha–1 (high-N plot), respectively. Each value represents the mean ± SE of six biological replicates. Asterisks indicate significant differences between means (p < 0.05).
To validate the results from the pot experiment, the response to different levels of N-fertiliser application was analysed in vegetative stage Cadenza and Paragon plants grown in a long-term field experiment with low and high N-plots (see section “Materials and Methods” for details). To assess N responses, plants were sampled before and at 48 h after N application and analysed for GS enzyme activity and tissue NH4+, as well as for leaf-disc NH4+ (Figures 1C–E). A significant change in GS activity was seen over time (F1,117 = 43.62, p < 0.001). In Cadenza, GS activity prior to N application was similar in the low-N and the high-N plots and a significant increase in GS activity after fertiliser application was observed only in plants grown in the high-N plot (Figure 1C). In contrast, GS activity in Paragon before N application was already significantly higher in the high-N plots and significantly increased further after N application in both, the low-N and high-N plots (up to 68.81 μmol GHA g FW–1 h–1; Figure 1C).
In contrast to the observed changes in GS enzyme activity, tissue NH4+ concentration did not respond to N-fertiliser application (F1,115 = 2.39, p = 0.125). However, whilst, tissue NH4+ was similar (between 1.49 and 1.72 μmol g–1) in both genotypes under low-N conditions, Paragon had a significantly higher tissue ammonium concentration under high N conditions (2.11 μmol g–1 FW) compared to plants grown in the low N plots (1.49 μmol g–1) (Figure 1D). Interestingly, leaf-disc extracted ammonium, used here as a proxy for potential volatile NH3 losses, increased significantly (F1,128 = 31.85, p < 0.001) in response to N application in the high N plot in Cadenza (from 1.82 to 3.10 μmol g–1) and in both, the high and low N plots in Paragon, although it remained comparably lower in the latter (max 2.73 μmol g–1) (Figure 1E).
Glutamine synthetases constitute a multi-gene family in wheat (Figure 2A) and for the purpose of this study the sequence information has been updated based on recent versions of the Chinese Spring reference genome (IWGSC RefSeq v1.0; International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018) and complemented with protein models based on available genomic scaffold sequences of five wheat varieties (Figure 2 and Supplementary Table 1). In Chinese Spring, the GS2 gene is located on chromosome group 2, two GS1 genes (GS1;2 and GS1;3) are located on chromosome group 4, and a third GS1 gene (GS1;1) is located on chromosome group 6 (Figure 2B).
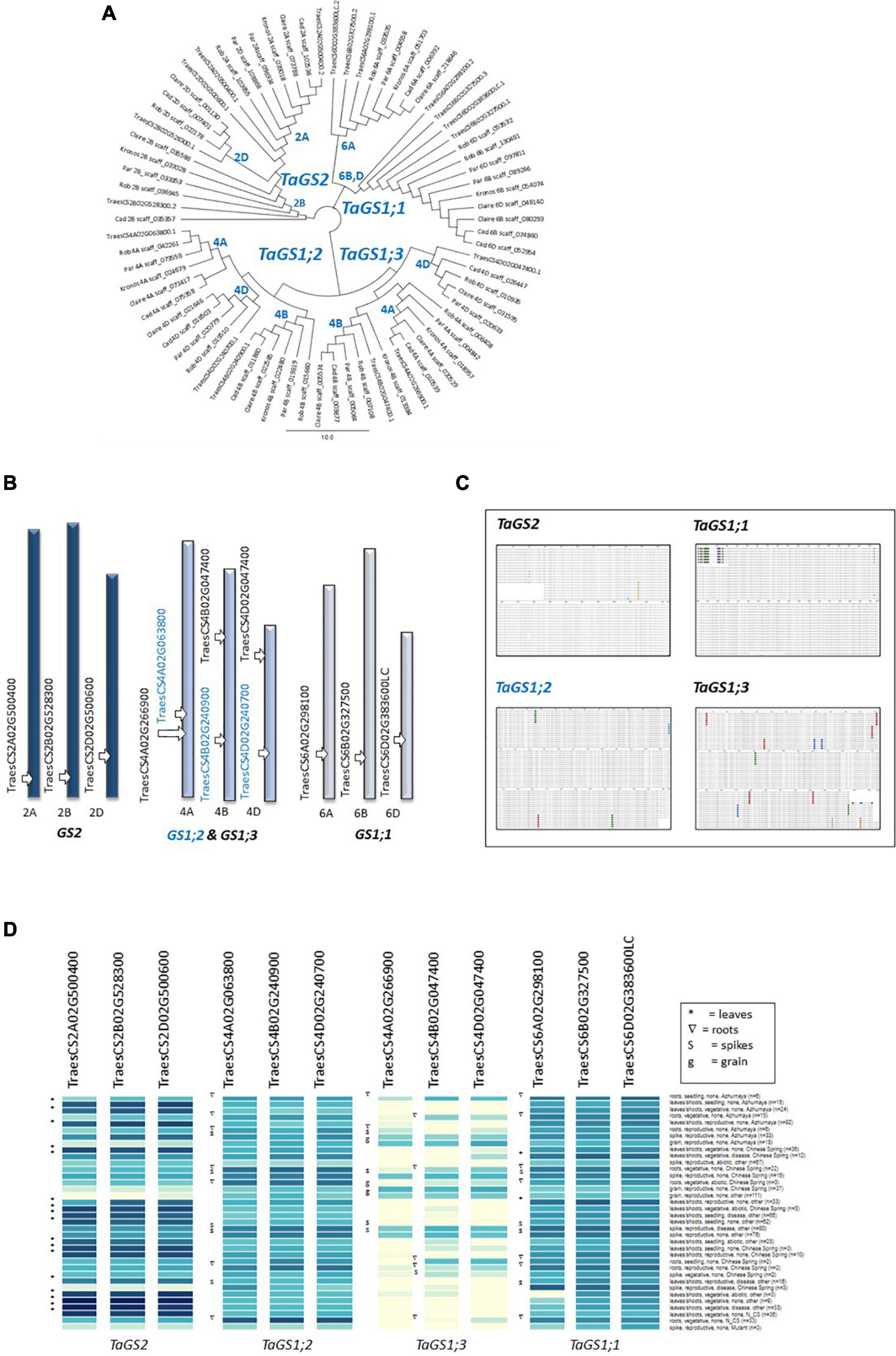
Figure 2. Comparison of glutamine synthetase genes in six wheat varieties. Sequences of the wheat GS1 and GS2 genes of the reference Chinese Spring reference genome were extracted from Ensembl Plants (https://plants.ensembl.org/Triticum_aestivum) and used as a query and template to extract genomic sequences from publicly available scaffolds and to predict GS1 and GS2 protein models in four bread wheat varieties (Cadenza, Paragon, Claire, and Robigus) and one durum wheat (Kronos) (see text for details; Supplementary Table 1). A comparison of the protein models is shown in the tree generated in Geneious (A). The gene IDs and chromosomal positions of the genes in the Chinese Spring reference genome are shown in (B). The high degree of conservation of the genes across the six analysed varieties is shown in the alignment in (C) (see Supplementary Figures 1–3 for details). Available gene expression data (http://www.wheatexpression.com) show a preferential expression of GS2 in leaves, and the additional expression of GS1 genes in roots, spikes and grain (D). Note that the GS1;3 genes show a distinct expression pattern and had the highest number of amino acid changes (C) (http://www.wheatexpression.com; Ramírez-González and Borrill, 2018).
Using the Chinese Spring genomic sequences as a query, GS1 and GS2 genomic sequences available via Decypher were identified in four bread wheat varieties (Cadenza, Paragon, Robigus, and Claire), as well as in the tetraploid durum wheat variety Kronos. For the prediction of the protein models, Chinese Spring genomic intron-exon structures were then superimposed onto the assembled genomic sequences and exons were extracted and translated into putative protein sequences using Geneious (see section “Materials and Methods” for details). Details on the wheat scaffolds from which sequences were retrieved and gene names are provided in Table 1 and Supplementary Table 1.
A comparison of the predicted protein models across the six analysed wheat varieties revealed a very high degree of conservation with 98–100% sequence identity over the entire length of the protein (Figure 2C and see Supplementary Figures 1–3 for full size alignments). The highest degree of sequence variation was observed for the TaGS1;3 gene on chromosome 4 with thirteen amino acid substitutions (Figures 2A,C). Based on available expression data5 GS2 is predominantly expressed in leaves (Figure 2D) in agreement with its localisation in the chloroplast and role in photorespiratory N re-assimilation. The cytoplasmic GS1 genes are predominantly expressed in roots and spikes/grain consistent with their role in primary N assimilation in the roots and N remobilisation during senescence and grain filling (Figure 2D). Whereas the TaGS1;1 homeologs on chromosome 6 and TaGS1;2 homeologs on chromosome 4 are also expressed in leaves, the more variable chromosome 4 TaGS1;3 gene appears not to be expressed in leaves (Figure 2D).
To enable gene expression analysis, detailed DNA sequence comparisons between the six genotypes were conducted and, for the purpose of this study, primers were designed that specifically bind to the three GS2 homeologs, as well as GS1-specific primers that bind to all homeologs of the three GS1 genes (for primer sequences see Table 2). Based on the respond in the GS activity in the different genotypes, N-responsiveness of the GS genes was assessed in three selected genotypes (Cadenza, Paragon, Claire), grown under N replete conditions, at the vegetative stage before and at 6 and 30 h after N application (NH4NO3; equivalent of 250 kg N ha–1). In the analysed leaf samples, combined expression of the GS1 genes was much lower than expression of the GS2 homeologs and showed no significant N response (Figures 3A,B). Contrary to our expectation, increases in GS2 gene expression after N application were also largely not significant (Figure 3B) although there was a weak (r = 0.4459) correlation between GS activity and GS2 gene expression (see inlay in Figure 3B).
Figure 3. GS1 and GS2 gene expression analysis in response to N and water deficit. For gene expression analyses, specific primers were designed that specifically amplify the GS1 genes and GS2 genes, respectively (see Figure 2). Expression of the three GS1 genes and their corresponding homeologs (A) and the GS2 gene with its three homeologs (B) was quantified in leaf samples from the four indicated wheat genotypes at 6 h and 30 after NH4NO3 application equivalent of 250 kg N ha–1 (N250) and in water-treated control plants (N-0). Each value represents the mean ± SE of four biological replicates. The inlay in (B) shows the correlation of GS gene expression and GS enzyme activity (corresponding GS activity data are shown in Supplementary Figure 4). The response of GS1 (C) and GS2 (D) gene expression to a dry-down treatment was analysed at 72 h after withholding water. Well-watered (WW) plants were sampled in parallel. Each value represents the mean ± SE of four biological replicates. In the dry-down experiment, GS enzyme activity (E) and leaf tissue ammonium content (F) were measured in parallel at 0 h and at 72 h after withholding water. The inlay shows a negative correlation of tissue ammonium and GS activity. Each value represents the mean ± SE of six biological replicates. Asterisks indicate significant differences between means (p < 0.05) of each treatment.
In order to corroborate lack of transcriptional regulation of GS genes, gene expression in Cadenza and Paragon was assessed in response to water stress. As had been overserved in the previous experiment, under water stress, GS1 transcript abundance was about 10-times lower compared to GS2 in both genotypes, however, GS1 transcript abundance significantly increased in response to water stress (F11 = 7.921, p < 0.02) whilst GS2 transcript abundance was significantly reduced (F11 = 17.660, p < 0.001) (Figures 3C,D). The parallel analysis of GS enzyme activity showed a significant difference between genotypes (F1,67 = 9.13 p = 0.004) with Paragon having higher GS activity on average (92.8 μmol GHA g FW–1 h–1) than Cadenza (76.9 μmol GHA g FW–1 h–1) (Figure 3E). In response to the dry-down treatment, GS enzyme activity significantly decreased in both genotypes compared to the well-watered controls by the end of the experiment, at 72 h after withholding water (Figure 3F). In agreement with that, leaf tissue NH4+ and GS activity were negatively correlated (r = 0.486) and increased under water stress (see inlay in Figure 3F).
Based on the observed positive response of GS activity and increase in leaf-disc NH4+ in the two analysed spring wheat genotypes Cadenza and Paragon, this study was extended to winter wheat. As mentioned above, the target environment for this study is the United Kingdom intensive system and we therefore chose eight founders of an elite MAGIC population (Alchemy, Brompton, Claire, Hereward, Rialto, Robigus, Soissons, and Xl-19; NIAB Elite MAGIC, Mackay et al., 2014). Plants were grown under N-replete conditions and at the 3–4 tiller vegetative stage were supplied with the equivalent of 250 kg N ha–1, either as solubilised NH4NO3 or as urea and GS enzyme activity, leaf tissue and leaf-disc NH4+were measured at 6 and 30 h after N application. Control plants were supplied with water and analysed in parallel (Figure 4 and Supplementary Table 2). NH4NO3 was included in this study to assess if N assimilation might be higher when plants employ both, the plant inherent ammonium and nitrate uptake systems. However, no significant differences between N sources were detected under the experimental conditions applied in this study. In contrast. a strong significant effect of time was seen in GS activity (F1,296 = 34.46, p < 0.001) with this effect differing between treatments (F2,296 = 10.59, p < 0.001). Both leaf-disc and leaf tissue NH4+ concentration differed significantly across times, treatment and genotype with significant three-way interactions (F18,295 = 1.80, p = 0.024 and F18,288 = 4.11, p < 0.001).
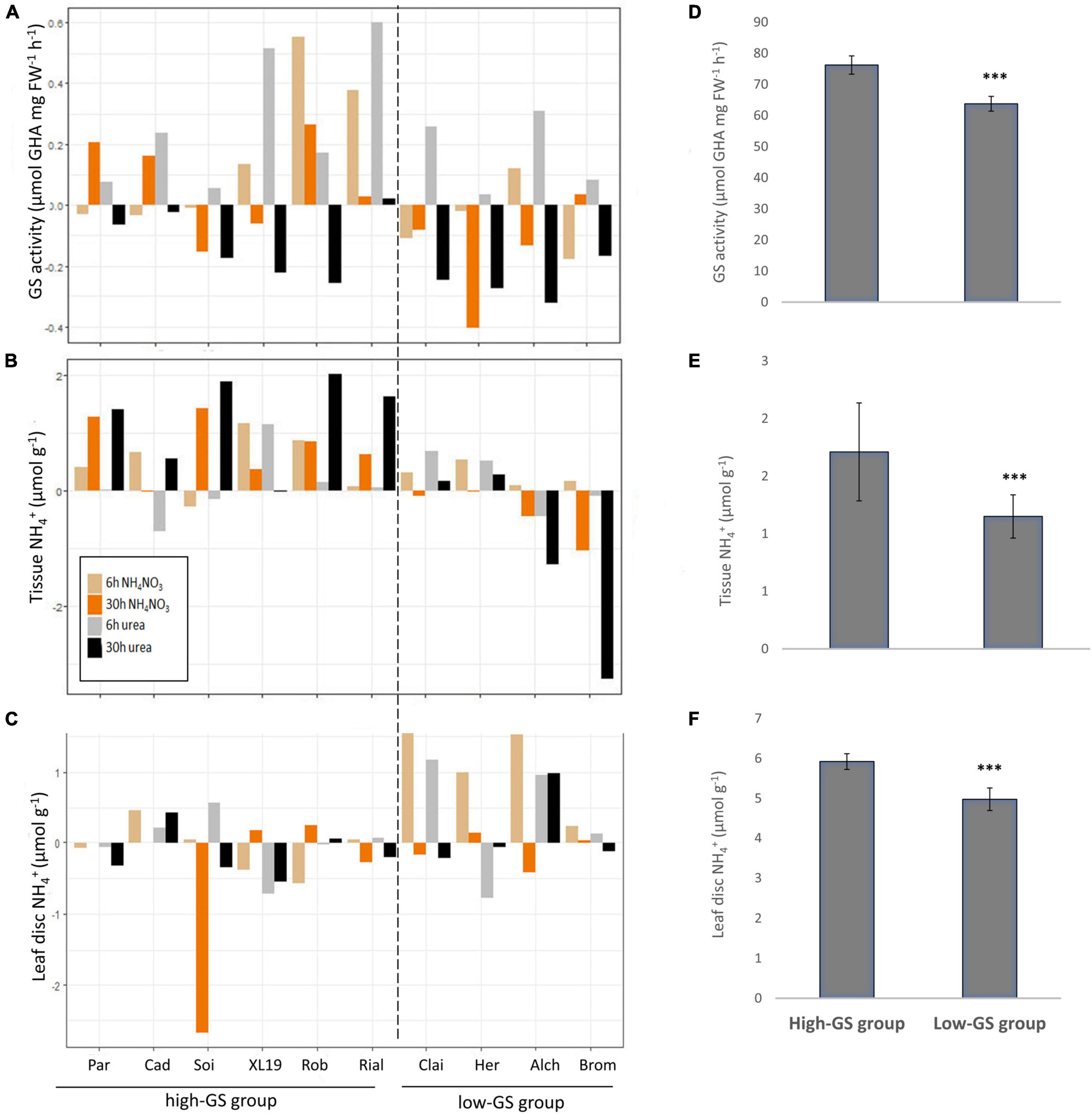
Figure 4. Differential response of ten wheat genotypes to N fertilisers. Ten elite United Kingdom wheat genotypes were treated with the equivalent of 250 kg N ha–1 applied either as NH4NO3 or urea and the response of GS activity (A), leaf tissue ammonium (B) and leaf-disc ammonium (C) were measured at 6 and 30 h after N application. The data shown represent log2 transformed relative changes compared to water-treated control plants (average values of six replicates). Data were calculated as LFold_Change = log2(treatment/control_mean). Average values of the actual data for the high-GS and the low-GS group, respectively, are given in (D–F). Error bars represent standard errors. Differences between groups were significant at p < 0.001. Cad, Cadenza; Par, Paragon; Soi, Soissons; Bro, Brompton; Alc, Alchemy; Her, Hereward; Cl, Claire; XL-19, XL-19; Rob, Robigus; Ria, Rialto. Differences between groups were significant at ***p < 0.001.
Overall, the data were variable and difficult to interpret, however, two significantly distinct groups could be distinguished, both in absolute GS activity (F1,296 = 52.85; p < 0.001) and in the relative change in GS activity [raw data/F1,296 = 11,22 (log2 data); p < 0.001] based on an ANOVA of log2 transformed change data and actual data (Figures 4A,D and Supplementary Table 2). This differentiation is mainly based on the higher GS activity present in the “high-GS group” (mean 76.2 μmol GHA h–1 FW–1) compared to the “low-GS group” (mean 63.8 μmol GHA h–1 FW–1) (Figure 4D). Likewise, the log2-fold change in response to N application was significantly different between the high-GS group (mean 0.09375 log2 fold change) and the low-GS group (mean −0.08875 log2 fold change) (Figure 4A). This grouping was also supported by the tissue NH4+ concentration [F1,296 = 13.01 (raw data)/F1,296 = 45.29 (log 2 data); p < 0.001] and leaf-disc NH4+ data [F1,296 = 12.97 (sqrt raw data)/F1,296 = 14.9 (log 2 data); p < 0.001], which were both significantly higher in the high-GS group (Figures 4B,C,E,F).
The pattern that emerged from the analysis of this complex data set revealed an overall positive N response of the high-GS group, i.e., generally the six genotypes within that group (Paragon, Cadenza, Soissons, XL-19, Robigus, and Rialto) had a constitutively higher GS activity which tended to respond positively to N application (Figures 4A,D). In contrast, the data suggest a possible inhibitory effect of N application on GS activity in the low-GS group (Claire, Hereward, Alchemy, Brompton; Figures 4A,D). Likewise, tissue NH4+ concentration in the high-GS group increased after N application and was constitutively higher (mean 1.1 μmol g–1 NH4+) compared to the low-GS group (mean 0.89 μmol g–1 NH4+) (Figures 5B,E). Contrary to this, leaf-disc NH4+, as a proxy for potential N losses via volatile NH3 gas, increased in the low-GS group (Figures 4C,F). However, although in the high-GS group leaf-disc NH4+ did not increase in response to N, it was overall higher in this group (mean 5.5 μmol g–1 NH4+) compared to the low GS-group (4.5 μmol g–1 NH4+) (Figure 4F).
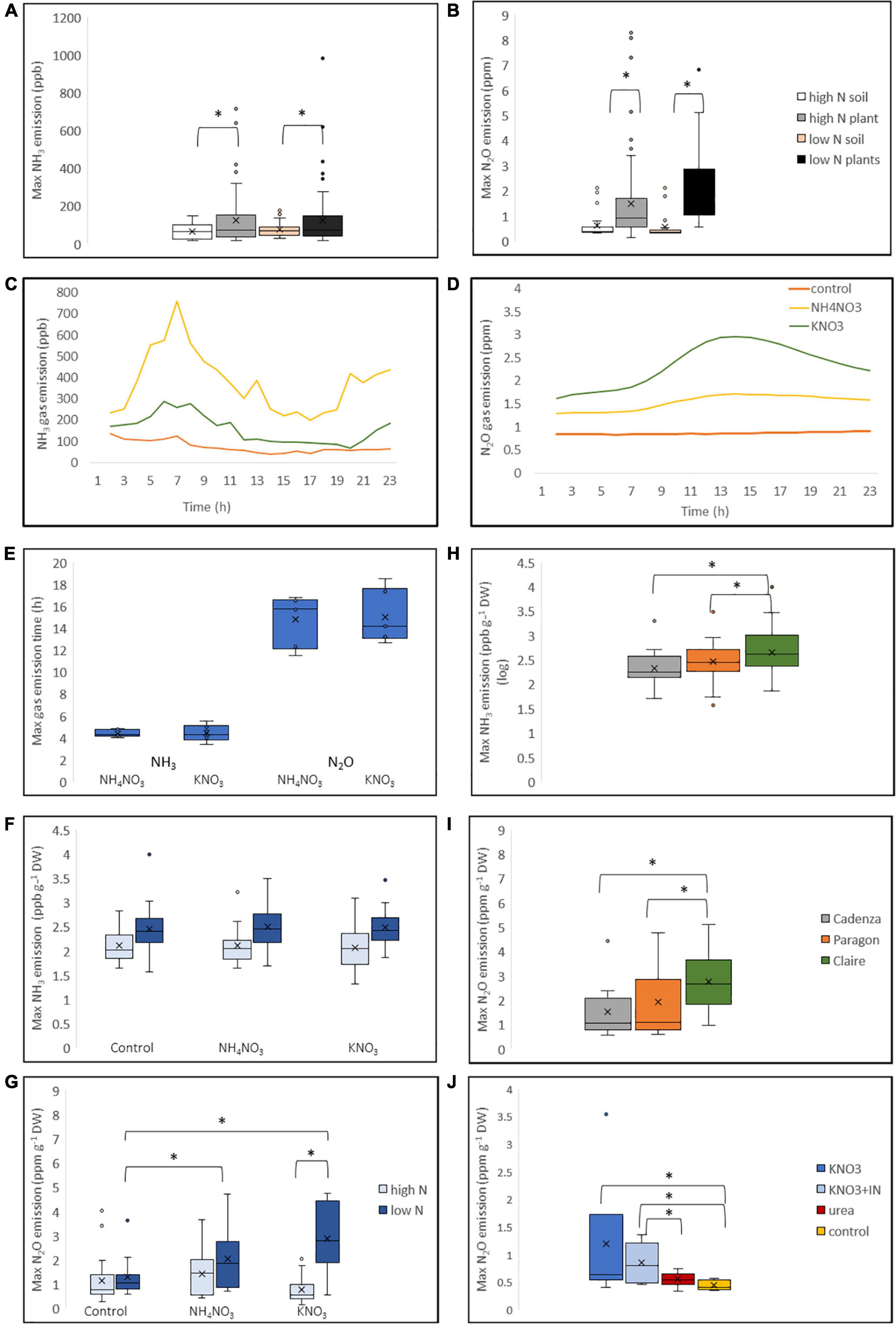
Figure 5. Ammonia and N2O gas emission in response to N application in wheat. Three wheat varieties representative of the high-GS group (Cadenza, Paragon) and low GS-group (Claire) were grown under low and high N conditions, respectively, and treated with the equivalent of 250 kg N ha–1 applied either as NH4NO3 or KNO3 immediately before gas measurements. Water was applied to control plants measured in parallel. For the soil control, above-ground plant tissue was cut off, but roots were retained (see text for details). Gas emission was measured using a gas analyser (Picarro G2508; United States) over a 24 h period. Maximum NH3 (A) and N2O (B) emission from plants was significantly higher than the background emission from soil. Representative processed Picarro data for NH3 (C) and N2O (D) emission are shown, with maximum NH3 emission occurring earlier than maximum N2O emission (E). The effect of the growth conditions and N treatment on maximum NH3 and N2O emission was calculated using the combined data of the three genotypes showing higher emission from N-starved plants (F,G). Genotypic differences between plants grown under low-N conditions are shown in (H,I). Cadenza plants grown under low-N conditions were treated with urea and KNO3, with and with nitrate reductase inhibitor (NI), showing highest N2O emission in the KNO3 treatment (J). Data are derived from six replicates for each genotype and treatment. Asterisks indicate significant differences between means (p < 0.05) of each treatment.
To further investigate the observed genotypic differences, three genotypes were selected to additionally determine changes in leaf nitrate and L-glutamine content in response to NH4NO3 application. Claire was selected as a representative of the low-GS group and Cadenza and Paragon as representatives of the high-GS group. The combined data of three selected genotypes, showed a significant increase in leaf tissue L-glutamine, NO3– and NH4+ content, as well as in GS activity, in response to N application (Supplementary Figures 4A–D). Of the three genotypes, Claire had the lowest GS activity and leaf-disc NH4+, as well as the lowest L-glutamine and tissue NH4+, although the latter differences were not significant (Supplementary Figures 4F–J). Overall, there was a positive correlation of GS activity with leaf L-glutamine content (r = 0.58), as well as tissue NH4+ and leaf-disc NH4+ (r = 0.54; r = 0.56). Total soluble protein content of samples was also analysed but did not show any changes in response to N application at the two timepoints (6 and 30 h) analysed (data not shown).
To corroborate the leaf-disc NH4+ data and confirm that wheat plants do in fact emit NH3 gas, genotypes from the high-GS group (Cadenza and Paragon) and low-GS group (Claire) were analysed using a Picarro real-time gas analyser. For this experiment, plants were grown under high-N and low-N conditions, with the latter treatment significantly reducing plant dry weight in all genotypes by about 50% (Supplementary Figure 5). Plants were supplied with the equivalent of 250 kg N ha–1 either as NH4NO3 or KNO3 in the morning, and gas emission was measured over a period of 24 h. Emission of NH3, as well as N2O, showed significant differences (F181 = 5.803, p < 0.05; F150 = 17.6975, p < 0.001, respectively) between plants and soil controls, with higher maximum emission from plants under high-N and low-N conditions (Figures 5A,B). Maximum NH3 gas emission occurred at about 5–6 h after N application, whereas maximum N2O emission occurred later, at about 15 h after N application (Figures 5C–E). Representative NH3 and N2O emission data generated from processed Picarro data (Paragon grown under low N conditions) are shown in Figures 5C,D. N2O emission was overall higher and within the ppm range, whereas NH3 gas emission was in the ppb range.
Regardless of N treatment, max NH3 emission was significantly different between plants grown under high-N and low-N conditions, with low-N plants showing higher NH3 emission (F1.0289 = 26.4939, p < 0.001; Figure 5F). Similarly, maximum N2O emission was significantly higher from plants grown under low-N conditions (F0.7262 = 32.8576, p < 0.001; Figure 5G). In the combined data of the three analysed genotypes, a significant effect of the N treatment was observed on N2O emission in plants grown under low-N conditions, with a significantly higher N2O emission after both, NH4NO3 and KNO3 application compared to water-treated control plants (Figure 5G). N2O emission was highest when KNO3 (F0.7262 = 6.0957, p < 0.05) was applied, in agreement with our current understanding that N2O is a by-product of nitrate reductase. In the combined data, there was no significant difference in NH3 emission between plants that received N and water control plants (Figure 5F). However, the log transformed NH3 emission data showed genotypic differences, with a significantly higher emission from the low GS-group representative variety Claire, compared to Cadenza and Paragon (Figure 5H). Likewise, N2O emission was significantly higher in Claire (Figure 5I). This was observed only in plants grown under low-N conditions but not in plants grown under high-N conditions (data not shown). To further investigate the effect of the N source on gas emission, Cadenza plants grown under low-N conditions were treated with urea and KNO3, with and without addition of the nitrate-reductase inhibitor Tungsten, respectively. In agreement with the expectation, the data show a significantly higher N2O when N was supplied as KNO3 compared with urea, and N2O emission was significantly reduced in the presence of Tungsten (Figure 5J).
To assess N-uptake and assimilation of the three genotypes under low-N and high-N growth conditions, leaf samples of the previous experiment were collected at the end of the gas measurement (at 24 h after N application) and analysed for GS activity, and tissue and leaf-disc NH4+ (Figure 6). The analysis of the combined data showed a significantly (F0.59 = 0.16, p < 0.01) higher tissue NH4+ content in plants grown under low-N conditions, though in the combined data there was no significant difference in GS activity or leaf-disc NH4+ (Figures 6A,C,E). However, when comparing the genotypes individually, GS activity in Claire was variable in response to NH4NO3 and was significantly lower compared to Cadenza and Paragon when N was supplied as KNO3 (Figure 6B). All genotypes showed a significant increase in tissue NH4+ after N application, and this was most pronounced when N was supplied as NH4NO3 (Figure 6D). Under the experimental conditions applied, there was no significant difference between the genotypes in leaf tissue NH4+ (Figure 6D), however, in contrast leaf-disc NH4+ was significantly lower in Claire compared to the other two genotypes, after both, NH4NO3 and KNO3 application (Figure 6F). This is in agreement with the data from the diversity study (Figure 4F) showing an overall lower leaf-disc NH4+ in the low-GS group.
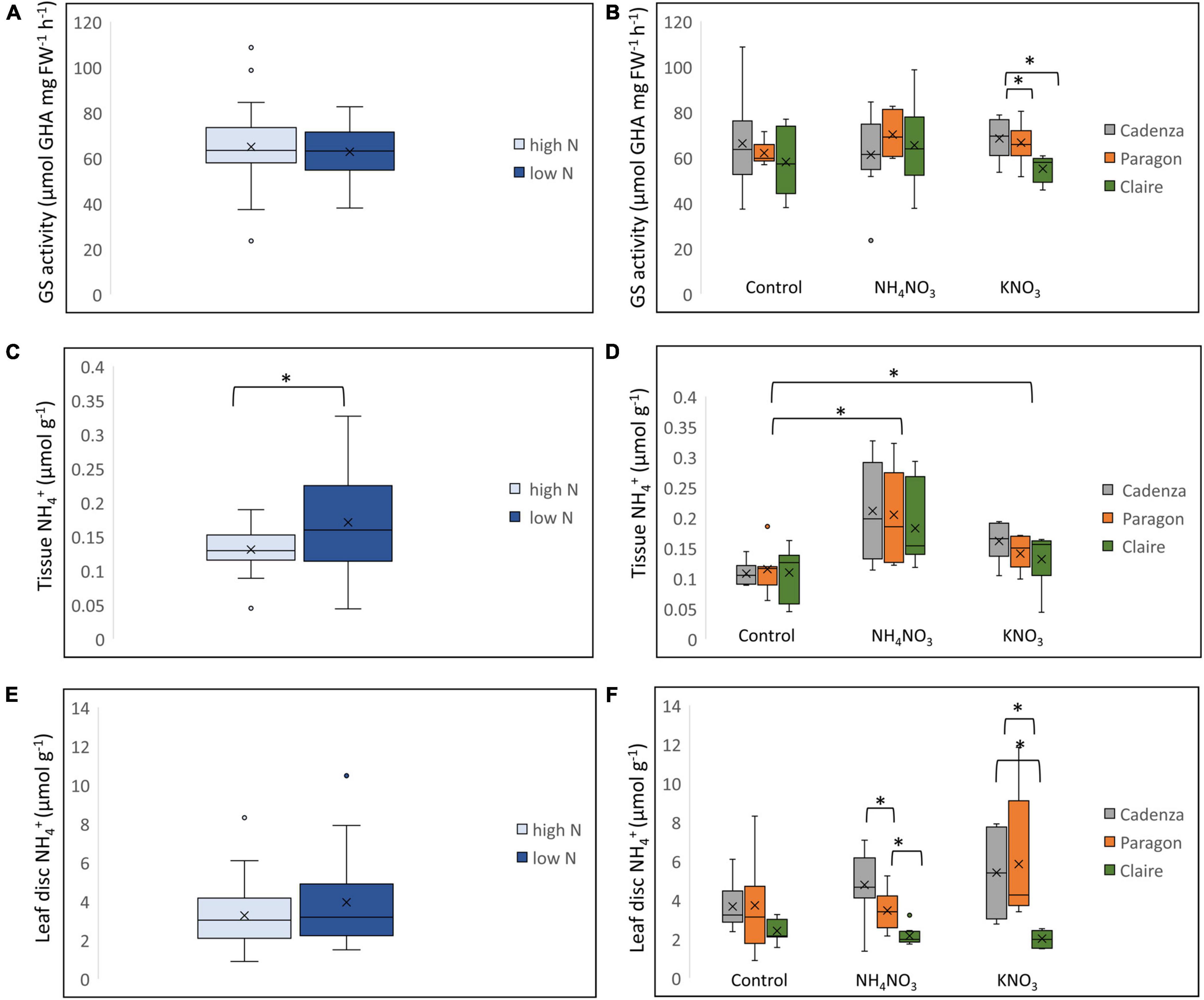
Figure 6. Differential N response in plants grown under high and low N conditions. Selected genotypes, Cadenza, Paragon and Claire, were grown under low-N and high-N conditions until the 3–4 tiller stage and then treated with the equivalent of 250 kg N ha–1 as NH4NO3 and KNO3, respectively. Water was used for control plants. GS activity (A), leaf tissue NH4+ (C) and leaf-disc NH4+ (E) were measured and compared between low-N and high-N plants. The effect of different treatments on GS enzyme activity (B), leaf tissue NH4+ (D) and leaf-disc NH4+ (F) was compared between the three genotypes. Asterisks indicate significant differences between means (p < 0.05) of each treatment.
An additional Picarro experiment was carried out to compare United Kingdom varieties with wheat from a different agro-ecological zone and for this, the Australian elite variety Mace (Wyalkatchem/Stylet), which has been shown to yield well across different Australian environments (SARDI Crop Harvest Report, 2015), was included in this study. It is noteworthy that compared to the United Kingdom, wheat systems in Australia are relatively low-N input and low-yielding, suggesting that N responses in Mace might be different compared to Cadenza and Paragon, which are adapted to high N levels. In agreement with that, the data show that at 24 h after N application, Mace had a significantly lower GS activity, as well as lower leaf-tissue and leaf-disc NH4+ content compared to the United Kingdom varieties (Supplementary Figures 6A–C). Likewise, NH3 and N2O gas emission were also significantly lower in Mace (Supplementary Figures 6D,E) suggesting that the N assimilation system in wheat is indeed plastic and subject to breeders’ selections.
The main objective of this study was to assess the immediate and short-term responses within wheat to N-fertiliser application at the vegetative stage and to see if there is variation within the N-assimilation capacity that can be used for improving N-fertiliser assimilation.
An initial time course experiment was conducted to assess whether and how rapidly GS activity in wheat responds to N-fertiliser application and if genotypic differences could be observed. The data showed that the spring wheat variety Paragon was overall more responsive to N compared with Cadenza, i.e., GS activity increased as early as 6 h after N application and also responded to lower N doses. The subsequent extended study, including the two spring wheat varieties and eight winter wheat varieties, confirmed that genotypic differences do exist, though there was considerable variation within the data. However, in response to N application, two significantly distinct groups, a high-GS group and low-GS group, could be distinguished. Members of the high-GS group (including both, Cadenza and Paragon) showed a constitutively higher and more N-responsive GS activity and glutamine content in response to N compared to the low GS-group, but also an increased tissue NH4+ level. An increase in tissue NH4+ concentration when GS activity is high may seem counterintuitive, however, this has also been described in rice by Kumagai et al. (2011a) suggesting that plants store excess NH4+ in the leaf when N uptake exceeds the N-assimilation capacity, likely in the vacuoles (Howitt and Udvardi, 2000). The response to N-fertiliser application was markedly different in genotypes constituting the low-GS group, compared with the high-GS group, with the former showing negative regulation of GS activity, no or little increase in tissue NH4+ and low glutamine content. This, in conjunction with the observed increase in leaf-disc NH4+, suggests overall poor N assimilation capacity and potentially high N losses in this group.
A negative response of leaf GS activity to N-fertiliser application has previously been reported from durum wheat (Nigro et al., 2016) and bread wheat (Zhang M. et al., 2017). This was only observed in high-grain protein genotypes; however, the high-grain protein genotypes had an overall higher GS activity, similar to the constitutive differences between bread wheat genotypes reported here.
Recent detailed studies in wheat (Wei et al., 2020) showed an increase in GS2 protein in leaves in response to both, NH4+ and NO3– application and this was confirmed by an in-gel GS activity assay showing increased GS2 activity in response to both N sources. In agreement with that, there was no significant differences between N sources in the genotypes analysed in the present study. Interestingly, Wei et al. showed that, in contrast to GS2, GS1 expression and activity showed differential N responses with a positive NH4+ response in roots and a positive NO3+ in shoots (Wei et al., 2020).
It has been shown that the response of GS activity to high leaf tissue NH4+ differs among plant species. Studies in lupin and mustard have found that high NH4+ tissue concentration was inhibitory (Ratajczak et al., 1981; Schmidt and Mohr, 1989), whilst it was stimulatory in sugar beet, mustard and pine (Vollbrecht et al., 1989; Mäck and Tischner, 1990). It was suggested that one of the underlying reasons for these conflicting reports may be related to the plant carbon (C) to N ratio, e.g., plants with low C:N ratios might lead to a deficiency in N-acceptor carbon skeletons. The complex interplay and potential limitation of C in N metabolism has been studied intensively in relation to the alanine aminotransferase (AlaAT) pathway in various crops (e.g., McAllister et al., 2012; Tiong et al., 2021).
In order to study the expression of the GS genes, we have extracted the sequences of the wheat GS1 and GS2 genes from available, but as yet unannotated, genome sequences from five wheat varieties, in addition to the available Chinese Spring reference genome. The models are therefore putative and will need to be experimentally validated. However, comparison of the predicted protein sequences revealed a near 100% sequence conservation between genotypes across the entire protein and there was no non-synonymous change that could explain differences in GS activities between the high-GS and low-GS activity group. GS enzymes are composed of eight monomers assembled into a complex, high-molecular holoenzyme and given its essential function, this high degree of conservation within wheat and across species is not surprising (Kumada et al., 1993; Bernard et al., 2008; Thomsen et al., 2014; Yang et al., 2016).
Analysis of GS gene expression showed an approximately six-fold higher expression of GS2 in leaves, which is in agreement with other studies in wheat (Bernard et al., 2008; Zhang M. et al., 2017; Wei et al., 2020) and rice (Zhao and Shi, 2006). Although GS2 expression was higher than GS1 overall in Cadenza and Paragon, which also had the highest GS activity, observed positive transcriptional changes after N application were largely not significant; suggesting that constitutive differences might be more relevant than N-triggered changes. Other studies report similar findings, with expression of GS genes being higher in an N-efficient compared to an N-inefficient genotype regardless of N treatment (Tian et al., 2015) and an overall higher expression was also observed in high grain-N genotypes compared with low grain-N genotypes (Nigro et al., 2016; Zhang M. et al., 2017).
There are very few studies in cereals that have assessed dynamic changes in GS gene expression that allow comparison with the short-term responses assessed here. The earliest analysis of N responses in wheat we could find was conducted at 10 days post fertiliser application showing an increase in GS transcript abundance (Nigro et al., 2019). Zhang Z. et al. (2017) monitored TaGS1 and TaGS2 transcript level in two wheat genotypes, grown under low- and high-N conditions, throughout development and in different tissues. The data showed a steep increase in leaf GS2 expression at the jointing stage and this was higher under high N, whereas GS1 expression was highest at 14 days post-anthesis, in agreement with their roles in N-assimilation and N-remobilisation, respectively. The detailed studies in wheat reported by Wei et al. (2020) were conducted with plants grown for 12 days under a range of different steady-stated N concentrations but nevertheless revealed interesting details on the differential regulation of GS1 and GS2 by different N sources, as mentioned above. Strikingly, the authors showed that GS2 is highly expressed in roots whilst they were unable to detect the corresponding protein, suggesting negative post-transcriptional regulation of TaGS2 in roots.
A study in rice assessed GS1 and GS2 gene expression at 6 h after N application to N-starved plants and the authors report an increase in the low-abundance OsGln1;1 gene, however, in agreement with our data the highly expressed OsGln1;2 gene, as well as OsGln2 were not responsive to N application at 6 h (Zhao and Shi, 2006). In another study in rice, GS gene expression in roots in response N-fertiliser application was analysed, showing a significant increase in transcripts of OsGln1;2, which is the most abundant GS gene in roots, in response to NH4+ but not to NO3–. In contrast, expression of another GS1 isoform (OsGln1;1) decreased in response to both N sources, whilst GS2 remained unchanged (low expression in roots). The analysed GOGAT genes were all significantly suppressed (Zhao and Shi, 2006). In a time-course experiment in wheat conducted with plants grown at two different N levels revealed little differences in GS1 and GS2 expression though in flag leaves, GS2 expression was higher under high N conditions (Wei et al., 2021b). In an RNA-seq study in durum wheat GS1 and GS2 expression in leaves did not respond to long-term N starvation, however, GS1 expression in roots increased consistent with its role in N uptake (Curci et al., 2017). Taken together, data on the transcriptional regulation of GS genes in response to N are inconsistent, and other factors need to be considered, such as post-transcriptional regulation and homeolog-specific expression (Wei et al., 2020), as well as post-translational regulation (Finnemann and Schjoerring, 2000; Swarbreck et al., 2008).
In contrast to the inconclusive role of transcriptional regulation in response to N, our dry-down experiment showed a significant upregulation of GS1 and downregulation of GS2 demonstrating that GS genes, in principle, can be regulated at the transcriptional level. Downregulation of GS2 was also observed in durum wheat exposed to osmotic stress, however, GS1 expression and GS activity remained unchanged in the three analysed genotypes (Jallouli et al., 2019). In fact, high GS expression has been implicated with a beneficial role under drought stress in a recent study with simultaneous overexpression of GS1;1 and GS2 (James et al., 2018), and had previously been observed in naturally drought-tolerant rice (Singh and Ghosh, 2013). GS activity was also responsive to salinity stress and specifically increased in the oldest of three analysed leaves, possibly related to N remobilisation under stress (Carillo et al., 2008).
Volatilisation of NH3 occurs when NH4+-generating processes exceed the re-assimilation capacity (Sutton et al., 1995) and was first described in relation to photorespiration in soybean (Weiland and Stutte, 1985) and spring wheat (Morgan and Parton, 1989). Since then it has been reported in barley (Mattsson et al., 1998), rice (Kumagai et al., 2011a,b), and many other species (see introduction), including Lolium and Bromus grass (Mattsson and Schjoerring, 2002). In the latter study, a significant correlation between high leaf tissue NH4+ and apoplastic NH4+ concentration was observed. From the apoplast, NH4+ can be emitted as NH3 gas via stomata (Husted and Schjoerring, 1995; Pearson et al., 1998; Mattsson and Schjoerring, 2002). Likewise, in rice high NH3 emission was observed at high tissue NH4+ concentrations and this was associated with genotypic differences observed between japonica rice and an aus-type landrace, which had low GS activity (Kumagai et al., 2011a).
These studies prompted us to assess if the observed differences in GS activity and leaf tissue NH4+ between the high GS-group and the low-GS group was associated with emission of NH3.
Due to the physico-chemical properties of NH3 gas it is difficult to measure emission directly from plants, and specialised equipment is required (Schjoerring et al., 1992). However, it has been suggested that tissue NH4+ concentration can be used to estimate the NH3 compensation point because it often increases proportionally to apoplastic NH4+ Mattsson and Schjoerring (2002) and Sutton et al. (2009) found that NH3 emission was closely correlated with NH4+ concentration in leaf tissue. Therefore, we used the leaf-disc assay where NH4+ is extracted from fresh, intact leaf-discs as a proxy for apoplastic NH4+ (see section “Materials and Methods” for details). In agreement with the above-mentioned studies, we found that leaf-disc NH4+ was significantly higher in the high-GS group, which also had a significantly higher leaf tissue NH4+. However, contrary to the expectation, there was no obvious increase in leaf-disc NH4+ after N application, although tissue NH4+ increased in this group. This suggests that genotypes in the high-GS group are indeed better able to assimilate and store excess NH4+ in contrast to the low-GS group. In the latter group, the N storage and assimilation capacity was possibly exceeded as indicated by lack of increase in leaf tissue NH4+ and low glutamine content. The increase in leaf-disc NH4+ might thus be a way to prevent accumulation of toxic cellular NH4+ levels.
Based on these indicative data, a follow-up study was conducted using a Picarro gas analyser to quantify NH3 as well as N2O gas emission. Cadenza and Paragon were selected as representatives of the high-GS group and Claire as a representative of the low-GS group.
Our current understanding of the roles of plants in N2O emission is still controversial with field-based studies suggesting that N2O is in fact produced by soil microorganisms and simply diffuses through plants (Chang et al., 1998; Rusch and Rennenberg, 1998; Baruah et al., 2010; Machacova et al., 2013; Bowatte et al., 2014; Wen et al., 2017). However, other studies using a range of different plant species and conducted under sterile conditions provided convincing evidence that the N2O gas indeed is derived from a plant-inherent process, and not from the soil microbiome or microbes present on leaves (Goshima et al., 1999; Hakata et al., 2003; Lenhart et al., 2019). Although the underlying mechanisms remain to be elucidated in detail, there is evidence that N2O can be produced from nitric oxide (NO) in the mitochondria of plants under hypoxic conditions (for a review see Timilsina et al., 2020) and this is also relevant in the context that NO acts as a second messenger in plants (for recent reviews see Astier et al., 2018; Gupta et al., 2020). However, more relevant in relation to this study is the finding that N2O emission was specifically measured only when NO3– was applied as the N source, but not with NH4+ or glycine (Lenhart et al., 2019).
The data presented here are in support of the findings that N gas emission originates from plants since there was a significant difference between pots with plants and control pots, from which the above-ground plant tissue was removed immediately before the gas measurement. Since the roots were retained in the controls and similar microbiomes should be present in all pots, differences in gas emissions can be ascribed to activities from leaves. Overall, after N application, emission was higher in plants that had been grown under N-starvation conditions, suggesting that, within the duration of the experiments, the N-assimilation and -storage capacity in N-starved plants was insufficient, leading to higher N-gas emission. This is in agreement with the study from Schjoerring and Mattsson (2001) who reported higher NH3 losses from winter wheat that had received 75% of a standard N dose, compared to fully fertilised plants.
Likewise, differences between the genotypes included in this study were only observed in plants grown under N deficiency where NH3 and N2O emission was significantly higher in the low-GS genotype Claire compared to Paragon and Cadenza. However, analysis of the Australian variety Mace, which had low GS activity and also low gas emission, suggests that N gas emission is not simply a matter of GS activity.
Thus, more detailed comparative studies on N-gas emission in plants is clearly needed to establish the underlying mechanisms. However, the presented data are in support of studies by others that N-gas emission does occur from plants and that this is a response to N fertiliser application. Importantly, in this study we have found that emission of N2O is related to nitrate fertiliser application since it was not observed when N was applied as urea and was significantly reduced in the presence a nitrate reductase inhibitor, which is in agreement with the study by Lenhart et al. (2019) and our current understanding that N2O originates from plant NO3– reduction. The role of N2O emission from plants in climate change has been realised (e.g., Smart and Bloom, 2001; Gogoi and Baruah, 2012; Lenhart et al., 2019) and needs to be addressed as a matter of urgency. The data presented here suggest that genotypic differences do exist that can be exploited for breeding and that N management and fertiliser choices would provide opportunities to reduce N2O emission from cropping systems.
The data presented here are showing that natural variation of GS activity exists in wheat and that genotypes respond differently to N-fertiliser application. There is thus an opportunity to exploit this natural variation for breeding. However, for this it will be important to better understand the underlying mechanisms of post-transcriptional and post-translational regulation of GS enzyme activity. It will also be important to establish to what extend GS activity is directly relevant for preventing N-gas emission and if other factors, such as e.g., NH4+ and NO3– storage capacity play important roles. This is not only relevant in relation to developing novel approaches towards enhancing crop N-fertiliser use efficiency, but even more so for the reduction of emission of the potent greenhouse gas N2O. Our data indicate that N2O emission is higher when N is applied to N-starved plants which warrants further investigations into the NO3– uptake and assimilation pathways in plants, however, it also opens opportunities for addressing this by fertiliser management options and fertiliser choices.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SH was leading the project. MO conducted the glasshouse experiments. KH carried out the statistical analysis. AR conducted the field experiments. IC provided access to the Picarro instrument. DH and AT-B analysed the Picarro data. SH and MO wrote the manuscript. All authors contributed to the article and approved the submitted version.
Rothamsted Research receives strategic funding from the Biotechnology and Biological Sciences Research Council of the United Kingdom. We acknowledge support through the Designing Future Wheat (DFW) Strategic Programme (BB/P016855/1). The field experiment was supported by the United Kingdom Department for Environment, Food and Rural Affairs funding of the Wheat Genetic Improvement Network (CH1090). We also acknowledge support of MO and SH through the project “Novel Fertiliser Formulations and Management for African Agriculture” in collaboration with the Mohammed VI Polytechnic University, Morocco, funded by the Office Chérifien des Phosphates (OCP group).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank our RRes colleagues Rowan Mitchell and Andy Phillips for their help retrieving the scaffold GS gene sequences.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.816475/full#supplementary-material
Astier, J., Gross, I., and Durner, J. (2018). Nitric oxide production in plants: an update. J. Exp. Bot. 69:420. doi: 10.1093/jxb/erx420
Balotf, S., Kavoosi, G., and Kholdebarin, B. (2016). Nitrate reductase, nitrite reductase, glutamine synthetase, and glutamate synthase expression and activity in response to different nitrogen sources in nitrogen-starved wheat seedlings. Biotechnol. Appl. Biochem. 63:1362. doi: 10.1002/bab.1362
Baruah, K. K., Gogoi, B., Borah, L., Gogoi, M., and Boruah, R. (2012). Plant morphophysiological and anatomical factors associated with nitrous oxide flux from wheat (Triticum aestivum). J. Plant Res. 125, 507–516. doi: 10.1007/s10265-011-0464-4
Baruah, K. K., Gogoi, B., and Gogoi, P. (2010). Plant physiological and soil characteristics associated with methane and nitrous oxide emission from rice paddy. Physiol. Mol. Biol. Plants 16, 79–91. doi: 10.1007/s12298-010-0010-1
Bernard, S. M., and Habash, D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182:2823. doi: 10.1111/j.1469-8137.2009.02823.x
Bernard, S. M., Møller, A. L. B., Dionisio, G., Kichey, T., Jahn, T. P., Dubois, F., et al. (2008). Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol. Biol. 67:9303. doi: 10.1007/s11103-008-9303-y
Bittsánszky, A., Pilinszky, K., Gyulai, G., and Komives, T. (2015). Overcoming ammonium toxicity. Plant Sci. 231:e5. doi: 10.1016/j.plantsci.2014.12.005
Brugiére, N., Dubois, F., Limami, A. M., Lelandais, M., Roux, Y., Sangwan, R. S., et al. (1999). Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11, 1995–2011. doi: 10.1105/tpc.11.10.1995
Bowatte, S., Newton, P. C. D., Theobald, P., Brock, S., Hunt, C., Lieffering, M., et al. (2014). Emissions of nitrous oxide from the leaves of grasses. Plant Soil 374, 275–283. doi: 10.1007/s11104-013-1879-6
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Britto, D. T., and Kronzucker, H. J. (2002). NH4+ toxicity in higher plants: a critical review. J. Plant Physiol. 159, 567–584. doi: 10.1078/0176-1617-0774
Campbell, W. H. (1988). Nitrate reductase and its role in nitrate assimilation in plants. Physiol. Plant. 74, doi: 10.1111/j.1399-3054.1988.tb04965.x
Cánovas, F. M., Avila, C., Cantón, F. R., Cañas, R. A., and de la Torre, F. (2007). Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 58, 2307–2318. doi: 10.1093/jxb/erm051
Carillo, P., Mastrolonardo, G., Nacca, F., Parisi, D., Verlotta, A., and Fuggi, A. (2008). Nitrogen metabolism in durum wheat under salinity: accumulation of proline and glycine betaine. Funct. Plant Biol. FPB 35, 412–426. doi: 10.1071/FP08108
Carvalho, J., de, F. C., Madgwick, P. J., Powers, S. J., Keys, A. J., Lea, P. J., et al. (2011). An engineered pathway for glyoxylate metabolism in tobacco plants aimed to avoid the release of ammonia in photorespiration. BMC Biotechnol. 11:111. doi: 10.1186/1472-6750-11-111
Chang, C., Janzen, H. H., Nakonechny, E. M., and Cho, C. M. (1998). Nitrous oxide emission through plants. Soil Sci. Soc. Am. J. 62, 35–38. doi: 10.2136/sssaj1998.03615995006200010005x
Chomczynski, P., and Sacchi, N. (1987). Single-step method of rna isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162: 9999. doi: 10.1006/abio.1987.9999
Cruz, C., Bio, A. F. M., Domínguez-Valdivia, M. D., Aparicio-Tejo, P. M., Lamsfus, C., and Martins-Loução, M. A. (2006). How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223:155. doi: 10.1007/s00425-005-0155-2
Curci, P. L., Aiese Cigliano, R., Zuluaga, D. L., Janni, M., Sanseverino, W., and Sonnante, G. (2017). Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 7:1176. doi: 10.1038/s41598-017-01377-0
Esteban, R., Ariz, I., Cruz, C., and Moran, J. F. (2016). Review: mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248:e8. doi: 10.1016/j.plantsci.2016.04.008
Finnemann, J., and Schjoerring, J. K. (2000). Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J. 24, 171–181. doi: 10.1046/j.1365-313X.2000.00863
Gao, Y., de Bang, T. C., and Schjoerring, J. K. (2019). Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 17, 1209–1221. doi: 10.1111/pbi.13046
García-Gutiérrez, Á, Cánovas, F. M., and Ávila, C. (2018). Glutamate synthases from conifers: gene structure and phylogenetic studiesNo title. BMC Genom. 19:65. doi: 10.1186/s12864-018-4454-y
Gerendás, J., Zhu, Z., Bendixen, R., Ratcliffe, R. G., and Sattelmacher, B. (1997). Physiological and biochemical processes related to ammonium toxicity in higher plants. Pflanzenernaehr. Bodenk 1997, 239–251. doi: 10.1002/jpln.19971600218
Gogoi, B., and Baruah, K. K. (2012). Nitrous oxide emissions from fields with different wheat and rice varietiesno title. Pedosphere 22, 112–121. doi: 10.1016/s1002-0160(11)60197-5
Goshima, N., Mukai, T., Suemori, M., Takahashi, M., and Caboche, M. (1999). Short communication: emission of nitrous oxide (N2O) from transgenic tobacco expressing antisense NiR mRNA. plant JNo title. Plant J. 19, 75–80. doi: 10.1046/j.1365-313x.1999.00494.x
Gupta, K. J. L., Mur, L. A. J., Wany, A., Kumari, A., Fernie, A. R., and Ratcliffe, R. G. (2020). The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol. 225, 1143–1151. doi: 10.1111/nph.15969
Hakata, M., Takahashi, M., Zumft, W. G., Ssakamoto, A. M. H., and Morikawa, H. (2003). Conversion of the nitrate nitrogen and nitrogen dioxide to nitrous oxides in plants. Acta Biotechnol. 23, 249–257. doi: 10.1016/j.copbio.2018.01.014
Hanstein, S., and Felle, H. H. (1999). The influence of atmospheric NH 3 on the apoplastic pH of green leaves: a non-invasive approach with pH-sensitive microelectrodes. New Phytol. 143, 333–338. doi: 10.1046/j.1469-8137.1999.00453.x
Hawkesford, M. J. (2014). Reducing the reliance on nitrogen fertilizer for wheat production. J. Cereal Sci. 59, 276–283. doi: 10.1016/j.jcs.2013.12.001
Heckathorn, S. A., and DeLucia, E. H. (1995). Ammonia volatilization during drought in perennial C4 grasses of tallgrass prairie. Oecologia 101, 361–365. doi: 10.1007/BF00328823
Hirel, B., Tétu, T., Lea, P. J., and Frédéric, D. (2011). Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 3, 1452–1485. doi: 10.3390/su3091452
Howitt, S. M., and Udvardi, M. K. (2000). Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta Biomembr. 1465, 152–170. doi: 10.1016/s0005-2736(00)00136-x
Husted, S., and Schjoerring, J. K. (1995). Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol. 109, 1453–1460. doi: 10.1104/pp.109.4.1453
International Wheat Genome Sequencing Consortium (IWGSC), Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Ishiyama, K., Inoue, E., Tabuchi, M., and Yamaya, T. (2004). Biochemical background and compartmentalized functions of cytosolic glutamine synthetase for active ammonium assimilation in rice roots. Plant Cell Physiol. 45, 1640–1647. doi: 10.1093/pcp/pch190
Jallouli, S., Ayadi, S., Landi, S., Capasso, G., Santini, G., Chamekh, Z., et al. (2019). Physiological and molecular osmotic stress responses in three durum wheat (triticum turgidum ssp durum) genotypes. Agronomy 9:550. doi: 10.3390/agronomy9090550
James, D., Borphukan, B., Fartyal, D., Ram, B., Singh, J., Manna, M., et al. (2018). Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic rice (Oryza sativa L.): impact on tolerance to abiotic stresses. Front. Plant Sci. 9:786. doi: 10.3389/fpls.2018.00786
Kumada, Y., Hillemann, D., Hosted, T. J., Rochefort, D. A., Thompson, C. J., Wohlleben, W., et al. (1993). Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. PNAS 90, 3009–3013. doi: 10.1073/pnas.90.7.3009
Kumagai, E., Araki, T., Hamaoka, N., and Ueno, O. (2011a). Ammonia emission from rice leaves in relation to photorespiration and genotypic differences in glutamine synthetase activity. Ann. Bot. 108, 1381–1386. doi: 10.1093/aob/mcr245
Kumagai, E., Araki, T., and Ueno, O. (2011b). Ammonia emission from leaves of different ice (Oryza sativa L.) cultivars. Plant Prod. Sci. 14, 249–253. doi: 10.1626/pps.14.249
Lam, H.-M., Coschigano, K. T., Oliveira, I. C., Melo-Oliveira, R., and Coruzzi, G. M. (1996). The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Lenhart, K., Behrendt, T., Greiner, S., Steinkamp, J., Well, R., Giesemann, A., et al. (2019). Nitrous oxide effluxes from plants as a potentially important source to the atmosphere. New Phytol. 221, 1398–1408. doi: 10.1111/nph.15455
Li, H., Hu, B., and Chu, C. (2017). Nitrogen use efficiency in crops: lessons from arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Linquist, B. A., Adviento-Borbe, M. A., Pittelkow, C. M., van Kessel, C., and van Groenigen, K. J. (2012). Fertilizer management practices and greenhouse gas emissions from rice systems: a quantitative review and analysis. Front. Crop. Res. 135:10–21. doi: 10.1016/j.fcr.2012.06.007
Lohr, G. W., and Waller, H. D. (1974). “Methods in enzymatic analysis,” in Verlag Chemie, Weinheim, ed. H. U. Bergmeyer (New York: Academic Press), 636–643.
Loubet, B., Decuq, C., Personne, E., Massad, R. S., Flechard, C., Fanucci, O., et al. (2012). Investigating the stomatal, cuticular and soil ammonia fluxes over a growing tritical crop under high acidic loads. Biogeosciences 9, 1537–1552. doi: 10.5194/bg-9-1537-2012
Machacova, K., Papen, H., Kreuzwieser, J., and Rennenberg, H. (2013). Inundation strongly stimulates nitrous oxide emissions from stems of the upland tree Fagus sylvatica and the riparian tree Alnus glutinosa. Plant Soil 364, 287–301. doi: 10.1007/s11104-012-1359-4
Mäck, G., and Tischner, R. (1990). Glutamine synthetase oligomers and isoforms in sugarbeet (Beta vulgaris L.). Planta 181, 10–17. doi: 10.1007/BF00202319
Mackay, I. J., Bansept-Basler, P., Barber, T., Bentley, A. R., Cockram, J., Gosman, N., et al. (2014). An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 Genes Gen. Genet. 4, 1603–1610. doi: 10.1534/g3.114.012963
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Masters-Clark, E., Shone, E., Paradelo, M., Hirsch, P. R., Clark, I. M., Otten, W., et al. (2020). Development of a defined compost system for the study of plant-microbe interactions. Sci. Rep. 10:7521. doi: 10.1038/s41598-020-64249-0
Mattsson, M., Hausler, R. E., Leegood, R. C., Lea, P. J., and Schjoerring, J. K. (1997). Leaf-atmosphere NH3 exchange in barley mutants with reduced activities of glutamine synthetase. Plant Physiol. 114, 1307–1312. doi: 10.1104/pp.114.4.1307
Mattsson, M., Husted, S., and Schjørring, J. K. (1998). Influence of nitrogen nutrition and metabolism on ammonia volatilization in plants. Nutr. Cyc. Agroecosys 51, 35–40.
Mattsson, M., and Schjoerring, J. K. (1996). Ammonia emission from young barley plants: influence of N source, light/dark cycles and inhibition of glutamine synthetase. J. Exp. Bot. 47, 477–484. doi: 10.1093/jxb/47.4.477
Mattsson, M., and Schjoerring, J. K. (2002). Dynamic and steady-state responses of inorganic nitrogen pools and NH3 exchange in leaves of lolium perenne and bromus erectus to changes in root nitrogen supply. Plant Physiol. 128, 742–750. doi: 10.1104/pp.010602
McAllister, C. H., Beatty, P. H., and Good, A. G. (2012). Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol. J. 10, 1011–1025. doi: 10.1111/j.1467-7652.2012.00700.x
Melino, V. J., Casartelli, A., George, J., Rupasinghe, T., Roessner, U., Okamoto, M., et al. (2018). RNA catabolites contribute to the nitrogen pool and support growth recovery of wheat. Front. Plant Sci. 9:e01539. doi: 10.3389/fpls.2018.01539
Mitchell, P. L., and Sheehy, J. E. (2018). Potential yield of wheat in the united kingdom: how to reach 20 t ha-1. Front. Crop. Res. 224, 115–125. doi: 10.1016/j.fcr.2018.05.008
Morgan, J. A., and Parton, W. J. (1989). Characteristics of ammonia volatilization from spring wheat. Crop Sci. 29, 726–731. doi: 10.2135/cropsci1989.0011183X002900030038x
Nigro, D., Fortunato, S., Giove, S. L., Paradiso, A., Gu, Y. Q., Blanco, A., et al. (2016). Glutamine synthetase in durum wheat: genotypic variation and relationship with grain protein content. Front. Plant Sci. 7:971. doi: 10.3389/fpls.2016.00971
Nigro, D., Gadaleta, A., Mangini, G., Colasuonno, P., Marcotuli, I., Giancaspro, A., et al. (2019). Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta 249, 1157–1175. doi: 10.1007/s00425-018-03075-1
O’Neal, D., and Joy, K. W. (1974). Glutamine synthetase of pea leaves. Plant Physiol. 54, 773–779. doi: 10.1104/pp.54.5.773
Parton, W. J., Morgan, J. A., Altenhofen, J. M., and Harper, L. A. (1988a). Ammonia Volatilization from Spring Wheat Plants Account for Losses of from 20 to of the Applied N in Agricultural Systems.
Parton, W. J., Morgan, J. A., Altenhofen, J. M., and Harper, L. A. (1988b). Ammonia volatilization from spring wheat plants. Agron. J. 80, 419–425. doi: 10.2134/agronj1988.00021962008000030008x
Pearson, J., Clough, E. C. M., Woodall, J., Havill, D. C., and Zhang, X. H. (1998). Ammonia emissions to the atmosphere from leaves of wild plants and hordeum vulgare treated with methionine sulphoximine. New Phytol. 138, 37–48. doi: 10.1046/j.1469-8137.1998.00880.x
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., and Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of hexaploid wheat across tissues and cultivars. Science 361:eaar6089. doi: 10.1126/science.aar6089
Ratajczak, L., Ratajczak, W., and Mazurowa, H. (1981). The effect of different carbon and nitrogen sources on the activity of glutamine synthetase and glutamate dehydrogenase in lupine embryonic axes. Physiol. Plant. 51, 277–280. doi: 10.1111/j.1399-3054.1981.tb04477.x
Rieu, I., and Powers, S. J. (2009). Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21, 1031–1033. doi: 10.1105/tpc.109.066001
Rusch, H., and Rennenberg, H. (1998). Black alder (alnus glutinosa (L.) gaertn.) trees mediate methane and nitrous oxide emission from the soil to the atmosphere. Plant Soil 201, 1–7. doi: 10.1023/A:1004331521059
SARDI Crop Harvest Report (2015). Trojan Delivers a Surprise in 2014 Wheat Trials. Available online at: https://pir.sa.gov.au/__data/assets/pdf_file/0020/243452/all_trials_2014.pdf
Sánchez-Rodríguez, E., Rubio-Wilhelmi, M., del, M., Ríos, J. J., Blasco, B., Rosales, M. Á, et al. (2011). Ammonia production and assimilation: Its importance as a tolerance mechanism during moderate water deficit in tomato plants. J. Plant Physiol. 168, 816–823. doi: 10.1016/j.jplph.2010.11.018
Schjoerring, J. K., and Mattsson, M. (2001). Quantification of ammonia exchange between agricultural cropland and the atmosphere: measurements over two complete growth cycles of oilseed rape, wheat, barley and pea. Plant Soil 228, 105–115. doi: 10.1023/A:1004851001342
Schjoerring, J. K., Sommer, S. G., and Ferm, M. (1992). A simple passive sampler for measuring ammonia emission in the field. Water. Air. Soil Pollut. 62, 13–24. doi: 10.1007/BF00478450
Schmidt, S., and Mohr, H. (1989). Regulation of the appearance of glutamine synthetase in mustard (Sinapis alba L.) cotyledons by light, nitrate and ammonium. Planta 177, 526–534. doi: 10.1007/BF00392621
Singh, K. K., and Ghosh, S. (2013). Regulation of glutamine synthetase isoforms in two differentially drought-tolerant rice (Oryza sativa L.) cultivars under water deficit conditions. Plant Cell Rep. 32, 183–193. doi: 10.1007/s00299-012-1353-6
Smart, D. R., and Bloom, A. J. (2001). Wheat leaves emit nitrous oxide during nitrate assimilation. Proc. Natl. Acad. Sci. U.S.A. 98, 7875–7878. doi: 10.1073/pnas.131572798
Sutton, M. A., Nemitz, E., Theobald, M. R., Milford, C., Dorsey, J. R., Gallagher, M. W., et al. (2009). Dynamics of ammonia exchange with cut grassland: strategy and implementation of the GRAMINAE integrated experiment. Biogeosciences 6, 309–331. doi: 10.5194/bg-6-309-2009
Sutton, M. A., Schjorring, K. J., Wyers, G. P., Duyzer, J. H., Ineson, P., and Powlson, D. S. (1995). Plant-atmosphere exchange of ammonia [and discussion]. Philos. Trans. Phys. Sci. Eng. 351, 261–278.
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T. Z., Garcia-Hernandez, M., Foerster, H., et al. (2008). The arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014. doi: 10.1093/nar/gkm965
Tabuchi, M., Sugiyama, K., Ishiyama, K., Inoue, E., Sato, T., Takahashi, H., et al. (2005). Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 42, 641–651. doi: 10.1111/j.1365-313X.2005.02406.x
Taira, M., Valtersson, U., Burkhardt, B., and Ludwig, R. A. (2004). Arabidopsis thaliana GLN2 -encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16, 2048–2058. doi: 10.1105/tpc.104.022046
Thomsen, H. C., Eriksson, D., Møller, I. S., and Schjoerring, J. K. (2014). Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 19, 656–663. doi: 10.1016/j.tplants.2014.06.002
Tian, Y.-S., Xu, J., Zhao, W., Xing, X.-J., Fu, X.-Y., Peng, R.-H., et al. (2015). Identification of a phosphinothricin-resistant mutant of rice glutamine synthetase using DNA shuffling. Sci. Rep. 5:15495. doi: 10.1038/srep15495
Timilsina, A., Bizimana, F., Pandey, B., Yadav, R. K. P., Dong, W., and Hu, C. (2020). Nitrous oxide emissions from paddies: understanding the role of rice plants. Plants 9:180. doi: 10.3390/plants9020180
Tingey, S. V., Tsai, F. Y., Edwards, J. W., Walker, E. L., and Coruzzi, G. M. (1988). Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J. Biol. Chem. 263, 9651–9657. doi: 10.1016/S0021-9258(19)81566-1
Tiong, J., Sharma, N., Sampath, R., MacKenzie, N., Watanabe, S., Metot, C., et al. (2021). Improving nitrogen use efficiency through overexpression of alanine aminotransferase in rice, wheat, and barley. Front. Plant Sci. 12:29. doi: 10.3389/fpls.2021.628521
Tuomi, J. M., Voorbraak, F., Jones, D. L., and Ruijter, J. M. (2010). Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50, 313–322. doi: 10.1016/j.ymeth.2010.02.003
van Hove, L. W. A., Heeres, P., and Bossen, M. E. (2002). The annual variation in stomatal ammonia compensation point of rye grass (Lolium perenne L.) leaves in an intensively managed grassland. Atmos. Environ. 36, 2965–2977. doi: 10.1016/S1352-2310(02)00242-X
Vollbrecht, P., Klein, E., and Kasemir, H. (1989). Different effects of supplied ammonium on glutamine synthetase activity in mustard (Sinapis alba) and pine (Pinus sylvestris) seedlings. Physiol. Plant. 77, 129–135. doi: 10.1111/j.1399-3054.1989.tb05988.x
Wei, Y., Xiong, S., Zhang, Z., Meng, X., Wang, L., Zhang, X., et al. (2021a). Localization, gene expression, and functions of glutamine synthetase isozymes in wheat grain (Triticum aestivum L.). Front. Plant Sci. 77:129–135.
Wei, Y., Wang, L., Qin, B., Li, H., Wang, X., Zhang, Z., et al. (2021b). A New perspective on the role of glutamine synthetase in nitrogen remobilization in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 22:11083. doi: 10.3390/ijms222011083
Wei, Y., Wang, X., Zhang, Z., Xiong, S., Meng, X., Zhang, J., et al. (2020). Nitrogen regulating the expression and localization of four glutamine synthetase isoforms in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 21:6299.
Weiland, R. T., and Stutte, C. A. (1985). Oxygen influence on foliar nitrogen loss from soyabean and sorghum plants. Ann. Bot. 55, 279–282. doi: 10.1093/oxfordjournals.aob.a086902
Wen, Y., Corre, M. D., Rachow, C., Chen, L., and Veldkamp, E. (2017). Nitrous oxide emissions from stems of alder, beech and spruce in a temperate forest. Plant Soil 420, 423–434. doi: 10.1007/s11104-017-3416-5
Yang, X., Nian, J., Xie, Q., Feng, J., Zhang, F., Jing, H., et al. (2016). Rice ferredoxin-dependent glutamate synthase regulates nitrogen–carbon metabolomes and is genetically differentiated between japonica and indica subspecies. Mol. Plant 9, 1520–1534. doi: 10.1016/j.molp.2016.09.004
Zhang, M., Ma, D., Ma, G., Wang, C., Xie, X., and Kang, G. (2017). Responses of glutamine synthetase activity and gene expression to nitrogen levels in winter wheat cultivars with different grain protein content. J. Cereal Sci. 74, 187–193. doi: 10.1016/j.jcs.2017.01.012
Zhang, Z., Xiong, S., Wei, Y., Meng, X., Wang, X., and Ma, X. (2017). The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci. Rep. 7:1000. doi: 10.1038/s41598-017-01071-1
Zhao, L., and Wang, Y. (2017). Nitrate assay for plant tissues. Bio Prot. 7:e2029. doi: 10.21769/BioProtoc.2029
Keywords: Triticum aestivum L, nitrogen fertiliser, N2O greenhouse gas, genetic diversity, glutamine synthetase, drought
Citation: Oszvald M, Hassall KL, Hughes D, Torres-Ballesteros A, Clark I, Riche AB and Heuer S (2022) Genetic Diversity in Nitrogen Fertiliser Responses and N Gas Emission in Modern Wheat. Front. Plant Sci. 13:816475. doi: 10.3389/fpls.2022.816475
Received: 17 November 2021; Accepted: 20 January 2022;
Published: 04 May 2022.
Edited by:
Guillermo Esteban Santa-María, National University of General San Martín, ArgentinaReviewed by:
Yihao Wei, Henan Agricultural University, ChinaCopyright © 2022 Oszvald, Hassall, Hughes, Torres-Ballesteros, Clark, Riche and Heuer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sigrid Heuer, c2lncmlkLmhldWVyQG5pYWIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.