
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 March 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.814015
This article is part of the Research TopicEngagement of Transcription Cofactors in the Developmental Processes and Response to Environmental CuesView all 4 articles
 Hua Chen1,2
Hua Chen1,2 Qiang Yang1,2
Qiang Yang1,2 Huiwen Fu1,2
Huiwen Fu1,2 Kun Chen1,2
Kun Chen1,2 Shanshan Zhao1,2
Shanshan Zhao1,2 Chong Zhang1,2
Chong Zhang1,2 Tiecheng Cai1,2
Tiecheng Cai1,2 Lihui Wang1,2
Lihui Wang1,2 Wenzhi Lu1,2
Wenzhi Lu1,2 Hao Dang1,2
Hao Dang1,2 Meijia Gao1,3
Meijia Gao1,3 Huaqi Li1,3
Huaqi Li1,3 Xinyi Yuan1,2
Xinyi Yuan1,2 Rajeev K. Varshney1,3
Rajeev K. Varshney1,3 Weijian Zhuang1,2*
Weijian Zhuang1,2*Peanut embryo development is easily affected by a variety of nutrient elements in the soil, especially the calcium level. Peanut produces abortive embryos in calcium-deficient soil, but underlying mechanism remains unclear. Thus, identifying key transcriptional regulators and their associated regulatory networks promises to contribute to a better understanding of this process. In this study, cellular biology and gene expression analyses were performed to investigate peanut embryo development with the aim to discern the global architecture of gene regulatory networks underlying peanut embryo abortion under calcium deficiency conditions. The endomembrane systems tended to disintegrate, impairing cell growth and starch, protein and lipid body accumulation, resulting in aborted seeds. RNA-seq analysis showed that the gene expression profile in peanut embryos was significantly changed under calcium deficiency. Further analysis indicated that multiple signal pathways were involved in the peanut embryo abortion. Differential expressed genes (DEGs) related to cytoplasmic free Ca2+ were significantly altered. DEGs in plant hormone signaling pathways tended to be associated with increased IAA and ethylene but with decreased ABA, gibberellin, cytokinin, and brassinosteroid levels. Certain vital genes, including apoptosis-inducing factor, WRKYs and ethylene-responsive transcription factors, were up-regulated, while key regulators of embryo development, such as TCP4, WRI1, FUS3, ABI3, and GLK1 were down-regulated. Weighted gene co-expression network analysis (WGCNA) identified 16 significant modules associated with the plant hormone signaling, MAPK signaling, ubiquitin mediated proteolysis, reserve substance biosynthesis and metabolism pathways to decipher regulatory network. The most significant module was darkolivegreen2 and FUS3 (AH06G23930) had the highest connectivity among this module. Importantly, key transcription factors involved in embryogenesis or ovule development including TCP4, GLK1, ABI3, bHLH115, MYC2, etc., were also present in this module and down regulated under calcium deficiency. This study presents the first global view of the gene regulatory network involved in peanut embryo abortion under calcium deficiency conditions and lays foundation for improving peanut tolerances to calcium deficiency by a targeted manipulation of molecular breeding.
Peanut (Arachis hypogaea L.) is a major agronomic crop providing important sources of plant oil and proteins worldwide. Peanut productivity has been adversely challenged by various biotic and abiotic stresses, leading to a significant reduction (Pandey et al., 2012).
Peanut embryo development is easily affected by a variety of nutrient elements in the soil, especially the calcium level. Calcium is a universal second messenger and essential mineral nutrient for plant growth and development. Peanut is a calcium-addicted crop. Calcium deficiency can seriously affect peanut growth and development and causes a series of physiological changes in peanut. Young leaves turn yellow in older tissues owing to the immobility of Ca2+. The number of flowers increases during anthesis, but most flowers are aborted. The mature leaves in the bottom outer layer form atrophic lesions in the late growth stage, vacuolar membrane of mesophyll cells breaks, chloroplast expands loosely, capsule breaks, and grana lamella structure is destroyed, which accelerates leaf senescence (Lin, 1996). Necrosis occurred at bud tip and root tip under the extreme calcium deficiency condition. The cell wall is loose and distorted and plasmolysis occurred. More than 90% calcium is directly absorbed from the soil by the peanut pod during development (Beringer and Taha, 1976). Peanuts growing in calcium-deficient soil produce a high rate of abortive embryos with empty or defectively filled pods, resulting in great economic losses (Jain et al., 2011) which poses a serious problem for global peanut production, especially in tropical and sub-tropical areas. Fortunately, application of calcium fertilizer could significantly increase the pod yield of peanut in acid soil (Zhang et al., 2015). Zhang has reported that peanut yields can increase by 26.92% with 210 kg ha–1 fused CaO treatment, which might be related to higher pod numbers per plant, higher double kernel rate, and higher plumpness of kernel under CaO treatment (Zhang et al., 2015). Thilakarathna also reported that the addition of 250 kg ha–1 of gypsum can increase the pod dry weight of peanuts by 39% (Thilakarathna et al., 2015). In addition to poor peanut yield and quality (Murata et al., 2008), calcium deficiency deteriorates seed viability and germination.
Different cultivars showed diverse sensitivities to calcium (Wu H. et al., 2017). Large pod varieties tend to produce a much higher proportion of empty pods in calcium-deficient soil. Tropical and subtropical areas thus use only peanut cultivars of small pod with yield performance reductions by approximately half compared with the large pod varieties. In China, more than 1.1 million hectares were planted with peanut in the south of peanut production regions. However, the yield per unit area of peanut is much lower than the national average, which is mainly due to the lack of calcium in the soil. It is of great significance to elucidate the mechanism controlling peanut embryo abortion for molecular breeding of large pod varieties with higher tolerance to low calcium. To address this important biological problem, different methods such as DDRT-PCR (Zhang et al., 2008), SSHaLL (Chen et al., 2016), proteomics (Zhang et al., 2007) and microRNA analysis (Chen et al., 2019) have been employed and various essential genes identified (Jain et al., 2011), providing initial insights into the mechanism of low calcium-induced embryo abortion.
With the rapid development of next-generation sequencing technologies, transcriptome profiling is a powerful approach for generating a transcriptional map at the whole-genome scale to discern regulatory networks and has been widely applied to plants such as peanut for screening candidate genes involved in plant growth and development as well as response to abiotic and biotic stresses (Xia et al., 2013). Weighted gene correlation network analysis (WGCNA) is an effective and robust network modeling method, which can identify co-expression modules regulating plant development and responsive to various stresses (Zhang and Horvath, 2005).
In this study, we firstly investigated the cell structure and morphology changes of peanut embryos under calcium deficiency conditions at the stage of 15, 20, and 30 DAP (days after pegging) using transmission electronic microscopy (TEM). Then global gene expression atlas showing genome-wide expression of genes in peanut embryos under calcium deficiency were developed from RNA-seq data to identify the key regulated genes in embryo abortion. Furthermore, WGCNA analysis was employed to comprehensively identify important regulators or pathways contributing to peanut abortion under calcium deficiency conditions. The results will further our understanding of dissecting the networks regulating calcium deficiency-induced embryo abortion and provide insights into mechanisms responsible for embryo development, especially its abortion under calcium deficiency. It will also lay the foundation for peanut molecular breeding of large pod varieties with higher tolerance to low calcium.
Plant material was collected from peanut cultivar MH6 grown in Ca2+-deficient soil in Pingtan, Fujian Province of China. The exchangeable Ca2+ content in the soil was 0.6 cmol/kg soil. Peanuts grown in this soil were used as the Ca2+ deficiency-treated material, and peanuts grown in the same soil fertilized with 75 kg/667 m2 plaster (CaO) were used as the Ca2+ sufficiency-treated material. The exchangeable Ca2+ content after fertilization was 4.2 cmol/kg soil, and generally the critical value of Ca2+ content in soil that could result in peanut embryo abortion was < 3.0 cmol/kg soil (Zhang, 2004). Embryos (15, 20, and 30 DAP) were manually dissected, frozen in liquid nitrogen and stored at −80°C for RNA-seq and qRT-PCR. Three biological replicates were prepared for all treatments.
Peanut embryos (15, 20, and 30 DAP) were fixed in 5% (v/v) glutaric dialdehyde in 0.1 M phosphate buffer (pH 7.3) for 24 h at 4°C and postfixed in 1% (w/v) osmic acid for 1–2 h. The embryos were dehydrated using a graded ethanol series and embedded in ERL-4206. The embedded embryos were then placed in an incubator chamber at 70°C for polymerization for 8–10 h. The specimens were sectioned to a thickness of 50–70 nm, and the sections were moved to a copper screen coated with a thin film. The sections were stained with both uranyl acetate and lead citrate to observe the ultrastructure of peanut embryos using TEM.
Total RNA was extracted from calcium deficiency-treated and control peanut embryos with TRIzol reagent (Invitrogen, Carlsbad, CA, United States). The RNAs were treated with RNase-free DNase I (Takara, Dalian, China) to eliminate contaminating genomic DNA. cDNA libraries were prepared using Illumina Paired End Sample Prep Kit. Three biological replicates were performed for each library. The quality of the libraries was evaluated using a Qubit2.0 Fluorometer (Life Technologies, CA, United States) and Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). RNA-seq libraries were sequenced via Illumina HiSeq™ 2500 platform (BIOMARKER, China). Raw reads were filtered to discard adapter sequences, low-quality reads (including reads containing more than > 10% poly-N and reads with more than 50% bases with a Q value ≤ 10). Filtered reads were mapped to the cultivated peanut reference genome1 (Zhuang et al., 2019) using the Bowtie tools software v.1.0.0 (Langmead et al., 2009) and TopHat2 (Kim et al., 2013) for reporting all mapping locations. The mapped reads were assembled by Cufflinks (Trapnell et al., 2010). Fragments per Kilobase of exon per Million Fragments (FPKM) was used to measure Cuffdiff to describe the transcript sufficiency. DESeq software (version 1.18.0) was applied with a criterion of a | log2fold change| ≥ 1 and FDR (false discovery rate) < 0.01 (Anders and Huber, 2010) to detect differentially expressed genes (DEGs).
The differentially expressed genes were subjected to NR, Swiss-Prot, GO, and COG to predict their biological functions. In addition, DEGs were subjected to the KEGG, KOG and Pfam databases to perceive their biological roles. KOBAS software (version 2.0) was used to test the statistical enrichment of differentially expressed genes in KEGG pathways. GO enrichment analysis was implemented with a p ≤ 0.05 as the threshold, and topGO was used to generate the acyclic graph.
Weighted gene co-correlation network analysis (WGCNA) was performed to construct gene co-expression networks using WGCNA R (version 4.0.3) package (Langfelder and Horvath, 2008). Genes with low abundance (FPKM value < 0.5) were filtered to eliminate noise. All genes were used as input to the signed WGCNA network construction. The co-expression modules were built using the automatic network construction function block wise Modules with default settings, except that the soft power was set to 30, merge Cut Height value was 0.25 and min Module Size was 30. The network of modules of co-expressed genes (edge weight > 0.15) was visualized by Cytoscape 3.9.0 (Saito et al., 2012).
Real-time PCR for the relative expression level of DEGs was performed using ChamQ SYBR qPCR Master Mix (High ROX Premixed) (Vazyme, Nanjing, China) with specific primers (Supplementary Table 11), and Ahactin was used as an internal reference gene. All reactions were performed on an ABI7500 system in triplicate. The relative expression levels of the DEGs were calculated using the comparative CT method (2–△ △ CT method) (Schmittgen and Livak, 2008), followed by normalization to the PCR threshold cycle number (Ct value) of the DEGs to that of the reference gene. The Student’s t-test was employed to compare differences between the control and experimental values.
Our previous study indicated that peanut pods at 15, 20 and 30 days after pegging (DAP) tended to be aborted and reduced the peanut yield and quality (Chen et al., 2019). Here, the embryo cells were investigated using transmission electronic microscopy (TEM) at 15, 20 and 30 DAP under calcium deficiency and sufficiency conditions. No obvious differences in cell morphology at 15 DAP were observed, except that the calcium-deficient cells contained fewer and smaller starch granules in the cytoplasm. The embryo cells contained large central vacuoles and thin cell walls at this stage (Figures 1C,D). At 20 DAP, the cell wall was clearly thickening under calcium sufficiency condition, while the calcium deficiency embryo cells remained thinner (Figures 1E,F). Moreover, the central vacuoles were compartmented into smaller ones surrounded by cytoplasm under calcium sufficiency. More vacuole proteins with different sizes were developed inside the vacuoles. Small lipid bodies began to form between the vacuoles and inside the cell walls (Figure 1E). Nevertheless, no clear developmental events occurred in calcium deficiency embryo cells. Their endomembrane systems appeared somewhat disintegrated, with a large reduction of lipid bodies inside the cells (Figure 1F). At 30 DAP, the embryo cell morphology with deficient calcium stress was further distorted as the cell wall was inclined to degrade (Figure 1H). Both protein and lipid bodies exhibited damage without new formation under calcium-deficient conditions. Moreover, as the endomembrane systems tended to decompose, the organelles showed a disordered arrangement inside the embryo cells. By contrast, massive lipid bodies continued to form around many vacuole protein bodies in an orderly manner in embryo cells under calcium-sufficient conditions (Figure 1G). Therefore, calcium deficiency caused irregular embryo development at early stages, especially the membrane systems, which impaired the accumulation of reserve substances including starch, proteins and lipids, leading to the abortion of seeds.
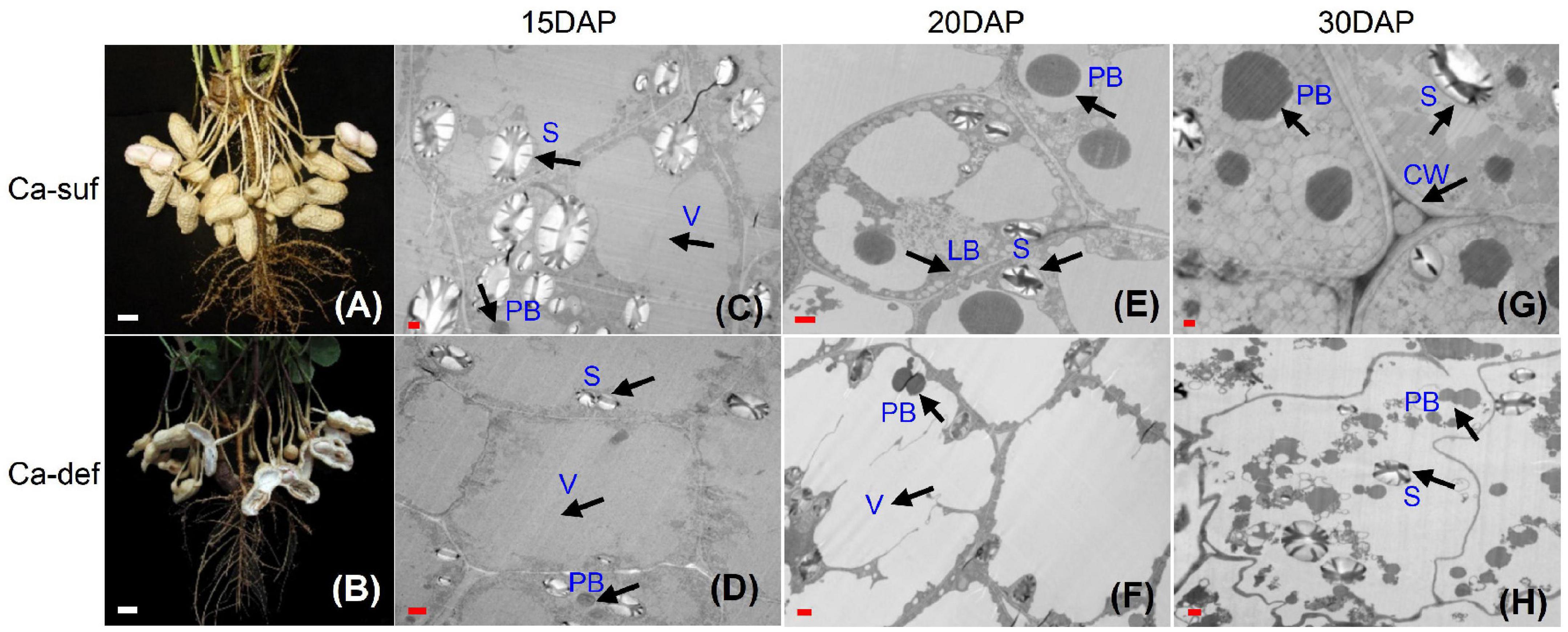
Figure 1. Effects of calcium on peanut pod development and embryo cell characteristics. (A) Peanut pods grown under calcium sufficiency conditions at 30 DAP. (B) Peanut pods grown under calcium deficiency conditions at 30 DAP. (C) Embryo cell of 15 DAP with calcium sufficiency. (D) Embryo cell of 15 DAP with calcium deficiency. (E) Embryo cell of 20 DAP with calcium sufficiency. (F) Embryo cell of 20 DAP with calcium deficiency. (G) Embryo cell of 30 DAP with calcium sufficiency; (H) Embryo cell of 30 DAP with calcium deficiency. LB, lipid body; PB, protein body; N, nucleus; S, starch; V, vacuole; CW, cell wall. Bars indicated in panels (A,B) are 10 mm; Bars in panels (C,D) are 1 μm and Bars in panels (E,F) are 0.1 μm.
To investigate responsive genes to calcium deficiency in peanut embryos, RNA-seq libraries for calcium deficiency and sufficiency at 15, 20 and 30 DAP were constructed and the global gene expression profiles surveyed using the Illumina HiSeq™ 2500 platform. After removal of adaptor sequences and low-quality reads, a total of ∼20–26 million clean reads for each sample were produced, comprising 100,743,125,153 nucleotides (100.74 Gb) with ∼90% Q30 and ∼46% GC content (Supplementary Table 1). After alignment with the peanut reference genomes (Zhuang et al., 2019), a high proportion of reads (∼86–93%) were mapped to the genomes. Among them, ∼69–82% were mapped uniquely to one location and more than 80% to the exons (Supplementary Figure 1). The transcriptional sufficiency of the genes was quantified using Cufflinks and measured as Fragments Per Kilobase per Million mapped reads (FPKM). As a result, 60,011 transcripts were identified. Accordingly, we were convinced that the obtained sequencing qualities were sufficiently high to sustain further analysis.
To identify calcium deficiency-responsive genes that regulate embryo development in peanut, the normalized expression levels of all genes were subsequently analyzed for their expression patterns. A total of 4,540 DEGs were identified, of which 638, 4,023, and 3,764 DEGs were obtained at 15, 20 and 30 DAP (Figure 2A and Supplementary Table 2). Among them, a total of 337, 2,028 and 1,898 DEGs were up-regulated and 301, 1,995 and 1,866 DEGs down-regulated at 15, 20 and 30 DAP, respectively (Figures 2C–E and Supplementary Table 2). Additionally, 570 DEGs showed differential expression at the three stages. Among these DEGs, 319 were up-regulated and 251 down-regulated (Figures 2B, 4E and Supplementary Table 2). The differentially expressed genes reflected the broad molecular changes associated with peanut embryo abortion under calcium deficiency conditions.
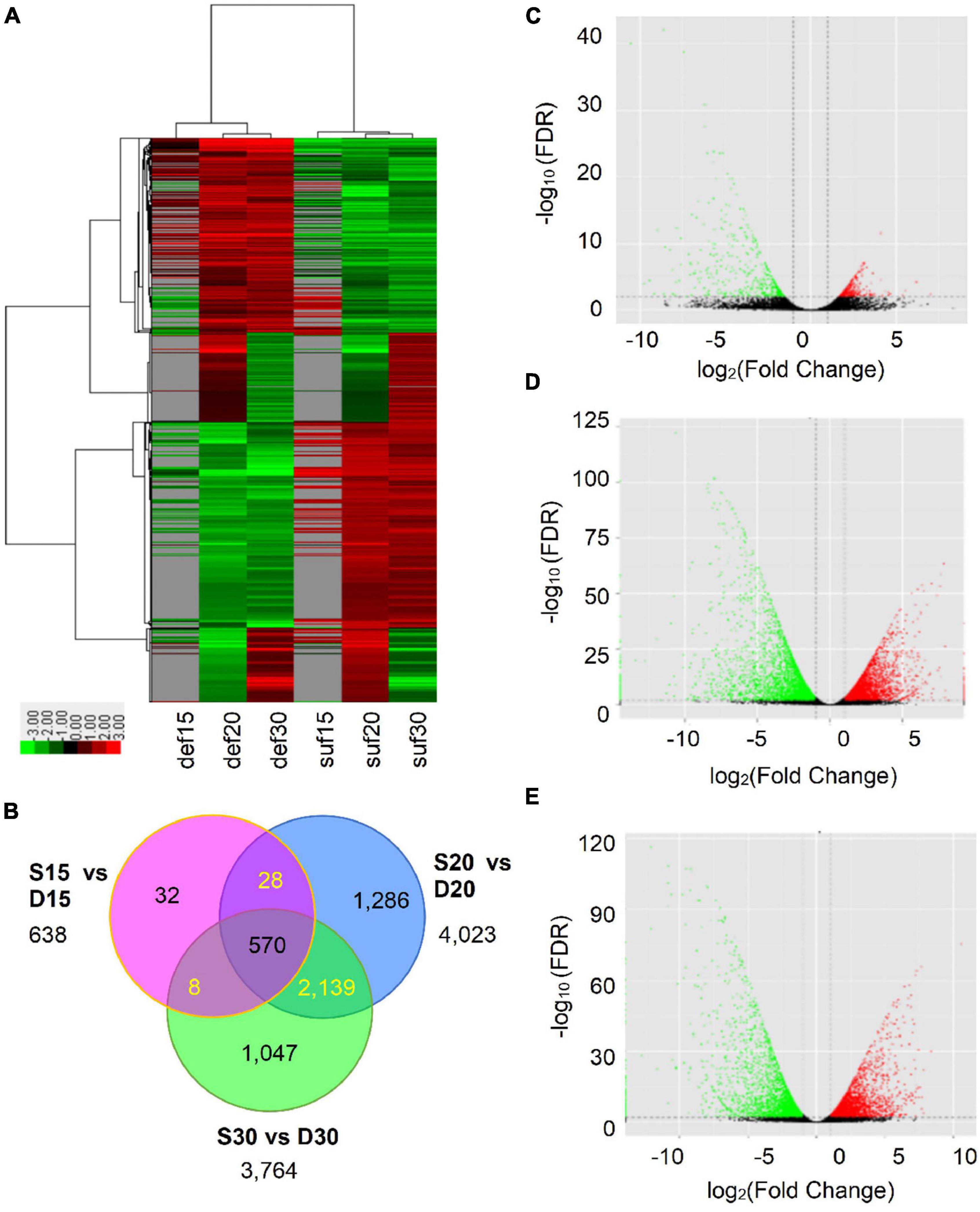
Figure 2. Transcriptional changes in peanut embryos responded to calcium deficiency. (A) Expression profiles of the DEGs response to calcium deficiency were analyzed and clustered by average linkage method. Expression level was calculated using log2 (FPKM + 1). Red indicates up-regulated genes, green indicates down-regulated genes and gray indicates no expression data. (B) The distribution of differentially expressed transcripts. (C) The volcano plot of DEGs at 15 DAP. (D) The volcano plot of DEGs at 20 DAP. (E) The volcano plot of DEGs at 30 DAP. FDR in panels (C–E) means false discovery rate.
To further explore the biological function of DEGs during peanut embryo abortion under calcium deficiency, the DEGs were annotated based on the different databases. A total of 244, 130 and 176 enriched GO terms in biological process, cellular component and molecular function, respectively, were identified among the DEGs. GO term enrichment analysis indicated that signal transduction, biosynthetic processes of plant hormones, and responses to biotic and abiotic stimulus were significantly enriched (Supplementary Figures 2, 4 and Supplementary Tables 3, 5). Furthermore, KEGG pathway analysis indicated DEGs were assigned to 68, 113, and 113 pathways involved in peanut embryo abortion under calcium deficiency at 15, 20, and 30 DAP, respectively. It is worth noting that plant hormone signal transduction and plant-pathogen interaction pathways were also identified as significantly enriched (Figure 3A). Most DEGs participating metabolism, plant hormone levels and the plant-pathogen interaction presumably play vital roles in peanut embryo abortion under calcium deficiency. Among them, 21 pathways were significantly enriched (Q value < 0.05), including photosynthesis-antenna proteins, photosynthesis, flavonoid biosynthesis and fatty acid biosynthesis (Figure 3B and Supplementary Table 6). These pathways clearly conformed to the morphological variances of embryo development and differences in reverse synthesis and accumulation, as observed based on the number and size of starches, protein bodies, and lipid bodies between calcium sufficiency and deficiency conditions (Figure 1).
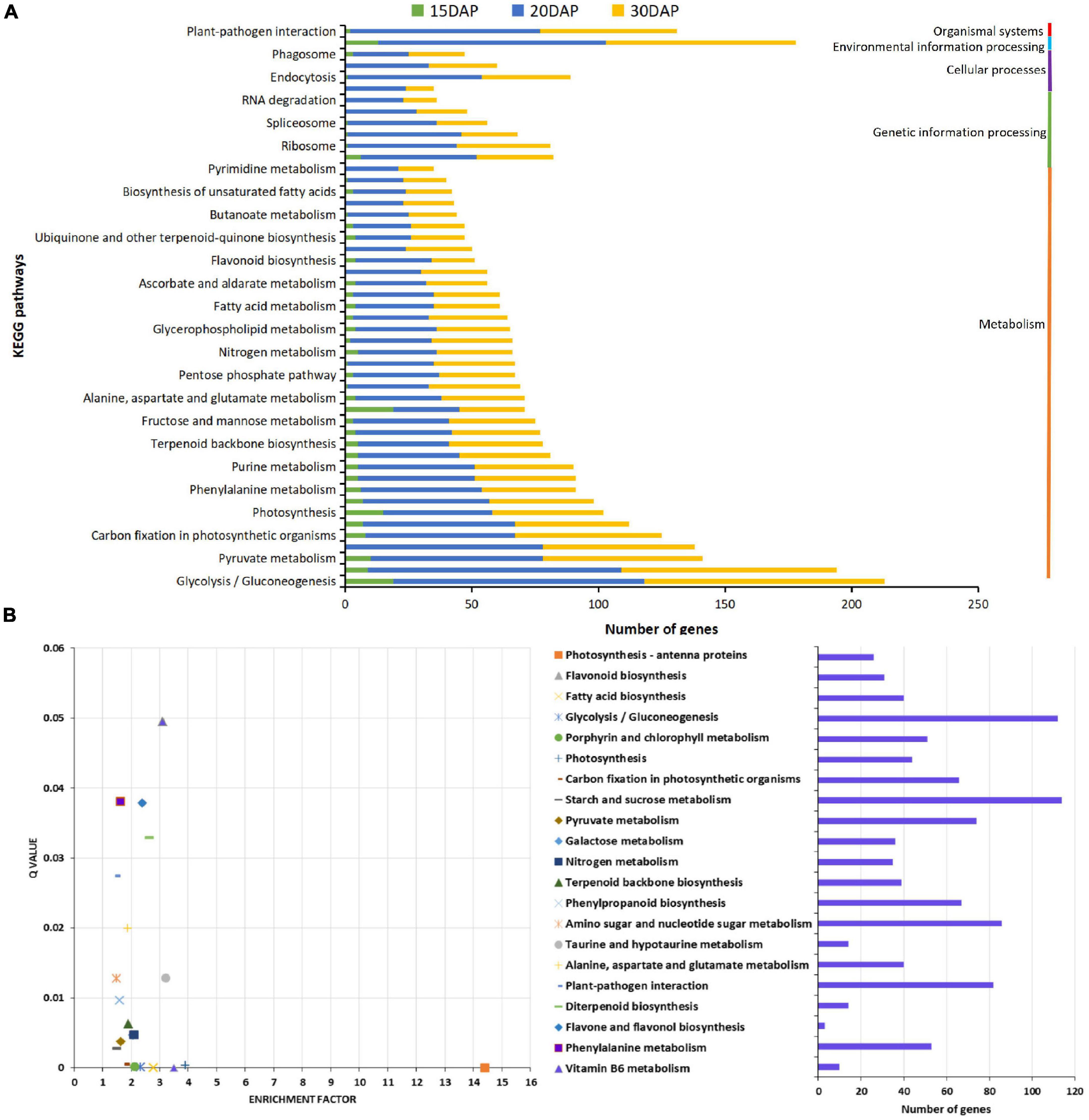
Figure 3. KEGG pathways analysis for differentially expressed genes. (A) Statistical analysis of KEGG pathways for DEGs; (B) KEGG pathways enrichment analysis of DEGs.
Given the importance of calcium in peanut growth and development, we were first prompted to focus on calcium signaling pathway-related genes for revealing the relationship between calcium and embryo development. The expression levels of many genes involved in calcium signaling pathway were significantly altered, presumably in association with embryo abortion under calcium deficiency. A group of genes controlling Ca2+ efflux and influx presented expression changes (Figure 4A and Supplementary Table 7). Among them, eight DEGs encoding Ca2+ transporting ATPases (ACA13, ACA12, ACA4 and ECA4) and one DEG encoding the mitochondrial calcium uniporter protein (MCU2) were up-regulated under calcium deficiency. Three DEGs encoding vacuolar cation/proton exchanger 3 (CAX3) showed an apparent down-regulation. Additionally, there was a difference in the accumulation of transcript levels of four DEGs encoding the cyclic nucleotide-gated ion channel (CNGC), of which CNGC15 and CNGC20 were up-regulated and CNGC1 was down-regulated. In addition, epidermal growth factor receptor (EGFR) and phosphatidylinositol phospholipase C (PI-PLC, PLC2 and PLC4) were down-regulated. Accordingly, it was predicted that calcium deficiency might induce embryo abortion through expression changes in these genes regulating cytoplasmic Ca2+ homeostasis. In contrast, calcium signal components, such as calcium-dependent protein kinases (CPK28, CPK10), calmodulin-binding receptor-like cytoplasmic kinases (CRCK2, CRCK3) and calmodulin-like proteins (CML11, CML39), were up-regulated. It is suggested the Ca2+ homeostasis was altered in the cytoplasm under calcium deficiency, resulting in defects in embryo development as a consequence of embryo abortion.
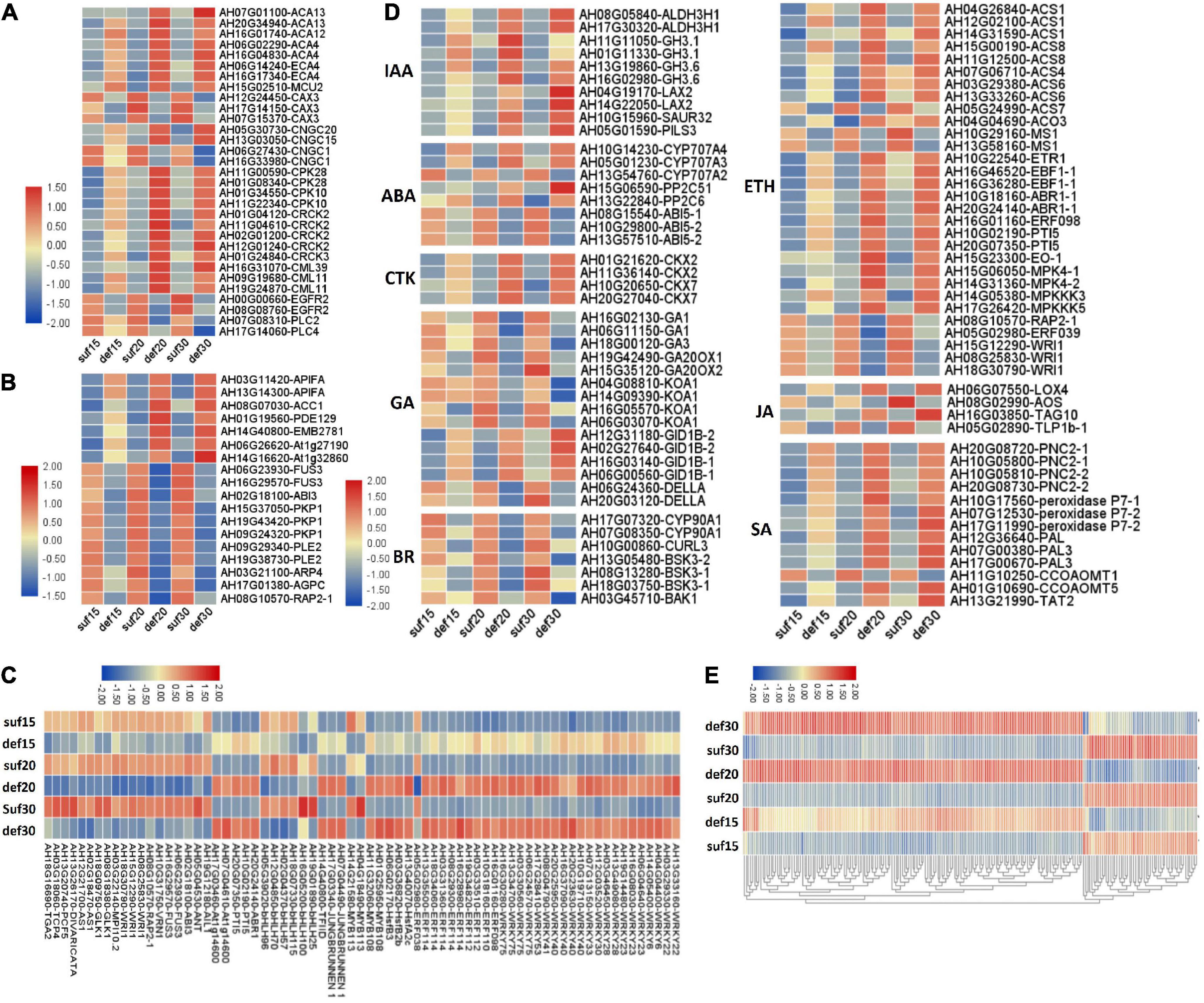
Figure 4. Relative expression changes of important differential expression genes and microarray validation of expression profiles of identified DEGs under deficiency calcium. (A) Calcium signaling pathway-related genes. (B) Embryo development related genes. (C) Transcription factors. (D) Plant hormone related genes. IAA, indole-3-acetic acid; ABA, abscisic acid; CTK, cytokinin; GA, gibberellin; BR, brassinosteroid; ETH, ethylene; JA, jasmonic acid; SA, salicylic acid. (E) Parts of common DEGs at the three stages. The expression level was calculated using log2 (FPKM + 1).
Embryo developmental genes among the DEGs according to the annotation results was then investigated. It was reported that 481 genes related to seed development in Arabidopsis were disrupted by a loss-of-function mutation exhibiting a seed phenotype (Meinke et al., 2008). To identify peanut embryo developmental genes, a blast analysis of our DEGs against these 481 genes was performed. Among all the DEGs, four were predicted to encode plastidial pyruvate kinase 2 (pPK2), which showed a high similarity to PKP1 in Arabidopsis and were down-regulated in peanut embryos under calcium deficiency (Figure 4B and Supplementary Table 7). Two DEGs encoding the B3 domain-containing transcription factor FUS3 and one DEG encoding ABI3 displayed significant down-regulation in the three developmental stages (Figure 4B and Supplementary Table 7). Two DEGs were predicted to encode dihydrolipoyllysine-residue acetyltransferase component 4 of the pyruvate dehydrogenase complex (LTA2), which shared high similarity to PLE2 in Arabidopsis, as well as a down-regulation in peanut embryos under calcium-deficient conditions (Figure 4B and Supplementary Table 7). One DEG was predicted to encode the ethylene-responsive transcription factor RAP2-1, which was significantly down-regulated under calcium deficiency (Figure 4B and Supplementary Table 7). Furthermore, eight DEGs were significantly up-regulated under calcium-deficient conditions (Figure 4B and Supplementary Table 7). Among the up-regulated DEGs, AH14G40800 was suggested to encode the transcription initiation factor TFIID subunit 6, which exhibits high similarity to EMB2781 in Arabidopsis. Two DEGs encoding the leucine-rich repeat protein kinase family protein and one for the glycosyl hydrolase superfamily protein showed a remarkable up-regulation. These DEGs might also play important roles in peanut embryo abortion under calcium deficiency conditions.
A series of genes related to plant hormones biosynthesis and signal transduction were differentially expressed in calcium deficiency-treated peanut embryos (Figure 4C and Supplementary Table 7). Aldehyde dehydrogenase family 3 member H1 (ALDH3H1) participating in tryptophan metabolism and in indole-3-acetic acid (IAA) biosynthesis were up regulated. The indole-3-acetic acid-amido synthetase GH3.1 and GH3.6 were also significantly up-regulated. Auxin transporter protein1 (AUX1), protein PIN-LIKES 3 (PILS3) and auxin-responsive protein SAUR32 were significantly up-regulated under deficient calcium stress. Cytokinin dehydrogenase (CRX), including CKX2 and CKX7, were significantly up-regulated under calcium deficiency. For gibberellin, the ent-copalyl diphosphate synthases GA1 and GA3, gibberellin 20 oxidases GA20OX1 and GA20OX2, and ent-kaurenoic acid oxidase 1 (KAO1) were down-regulated. Surprisingly, gibberellin receptor GID1B was up-regulated and DELLA protein GAI down-regulated. In terms of the brassinosteroid signaling transduction pathway, the brassinosteroid LRR receptor kinase CURL3 and protein kinase protein with the tetratricopeptide repeat domain (BSK3) and BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 (BAK1) were all significantly down-regulated.
The abscisic acid 8′-hydroxylase genes (CYP707A1, CYP707A3, CYP707A4) were up-regulated in calcium deficiency peanut embryos (Figure 5). Moreover, the xanthoxin dehydrogenase gene ABA2 and zeaxanthin epoxidase gene ABA1 involved in the biosynthesis of abscisic acid were down-regulated. The ABA receptor PYL showed expression changes with PYL9 down-regulation and PYL4 and PYL11 up-regulation. Additionally, two protein phosphatase 2C (PP2C) were also up-regulated. ABSCISIC ACID-INSENSITIVE 5 (ABI5) was down regulated. The expression changes in ABA signal transduction pathway-related genes should be responsible for embryo abortion under calcium deficiency conditions. A series of genes involved in ethylene biosynthesis and signal transduction exhibited altered expression. For example, 1-aminocyclopropane-1-carboxylate synthase 1 (ACS1) and 1-aminocyclopropane-1-carboxylate oxidase 3 (ACO3) related to ethylene biosynthesis were up-regulated. Ethylene receptor 1 ETR1, ethylene-responsive transcription factor ABR1 and ERF098 and the pathogenesis-related gene transcriptional activator PTI5 were all up-regulated. Mitogen-activated protein kinase 4 (MPK4), mitogen-activated protein kinase kinase kinase 3 (MAPKK3) and mitogen-activated protein kinase kinase kinase 5 (MAPKK5) were also up-regulated. However, several other ethylene-responsive transcription factors, including RAP2-1, ERF039 and WRI1, were down-regulated. Moreover, the TGACG-sequence-specific DNA-binding protein TGA10 and thaumatin-like protein TLP1b were up-regulated and down-regulated, respectively, in the jasmonic acid signaling pathway. Both peroxidases, including cationic peroxidase 2 (PNC2) and peroxidase P7-1, as well as phenylalanine ammonia-lyase (PAL), were up-regulated in the salicylic acid signaling pathway. Briefly, the altered transcript levels of the above genes in peanut embryos under calcium deficiency condition should change the plant hormone levels and affect signal transduction.
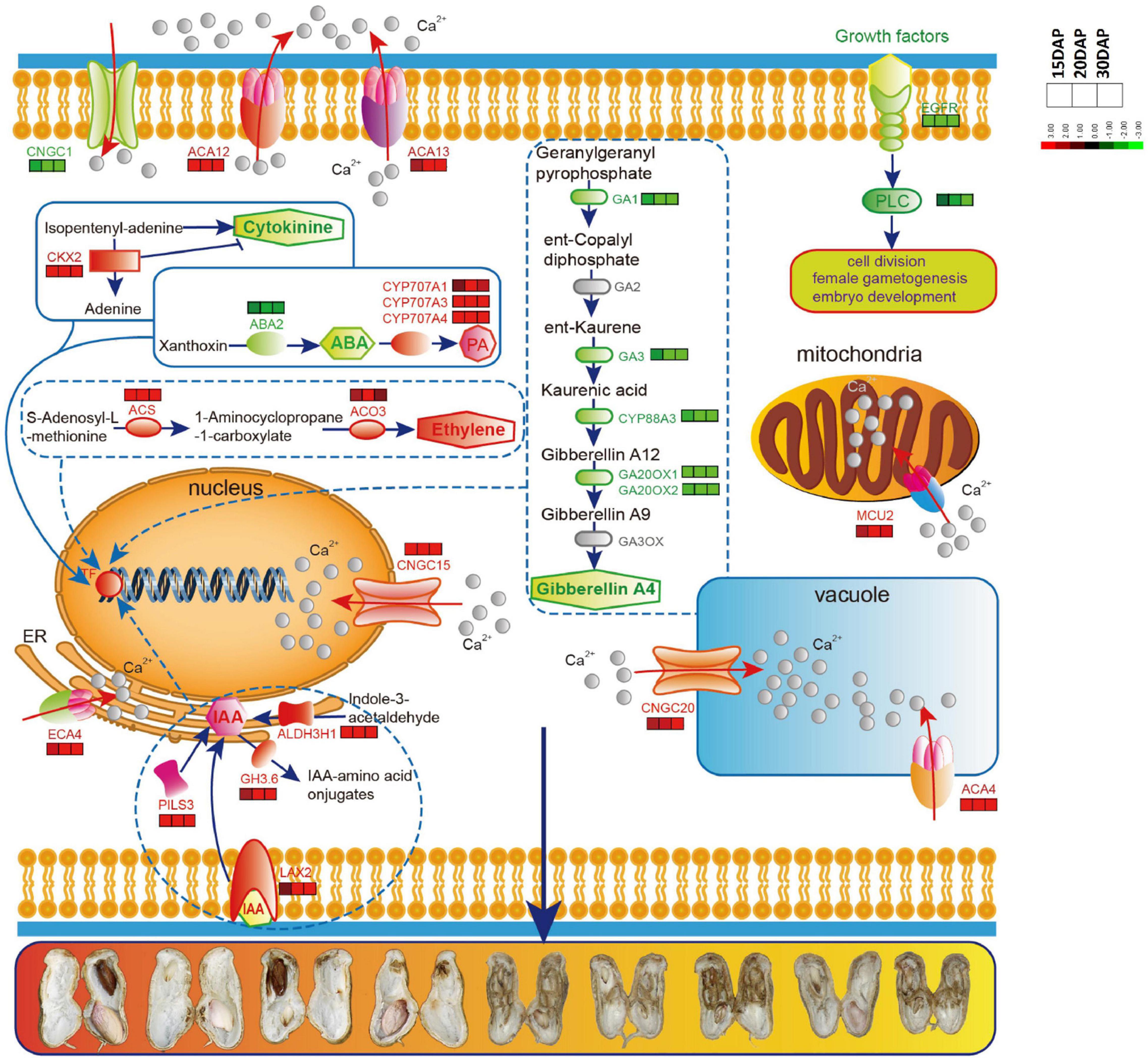
Figure 5. The predicted schematic diagram of calcium signaling pathway and hormone biosynthesis related genes in peanut embryo cells and their subcellular localization. Red color indicates up-regulated genes, green indicates down-regulated genes. Calcium-transporting ATPases and Cyclic nucleotide-gated ion channels (CNGC) are localized in different intracellular organelles and have high capacity Ca2+ flow across membranes. ACA12 and ACA13 are present in the plasma membrane, ACA4 in the vacuole and ECA4 in endoplasmic reticulum (ER). CNGC1 is in the plasma membrane, CNGC15 in nucleus and CNGC20 in vacuole membrane. EGFR, epidermal growth factor receptor; PLC, phosphoinositide phospholipase C; MCU2, calcium uniporter protein 2, mitochondrial; GA1, Ent-copalyl diphosphate synthase; GA3, Ent-kaurene oxidase; GA20OX1, Gibberellin 20 oxidase 1; GA20OX2, Gibberellin 20 oxidase 2; CKX2, Cytokinin dehydrogenase 2; ABA2, Xanthoxin dehydrogenase; CYP707A1, Abscisic acid 8′-hydroxylase 1; CYP707A3, Abscisic acid 8′-hydroxylase3; CYP707A4, Abscisic acid 8′-hydroxylase 4, ACS, 1-aminocyclopropane-1-carboxylate synthase; ACO3, PILS3, protein PIN-LIKES 3; GH3.6, Indole-3-acetic acid-amido synthetase GH3.6; ALDH3H1, Aldehyde dehydrogenase family 3 member H1.
A total of 24 WRKY transcription factors, 10 ethylene-responsive transcription factors (ERFs) containing 5 ERF families, 3 heat stress transcription factors and 4 MYB transcription factors, were all up-regulated in response to low calcium (Figure 4D and Supplementary Table 7). Additionally, two pathogenesis-related gene transcriptional activators (PTI5) and two transcription factors JUNGBRUNNEN 1 were also up-regulated under calcium-deficient conditions.
The down-regulated transcription factors under deficient calcium conditions included 6 bHLH transcription factors, 5 ethylene-responsive transcription factors, 5 B3 domain-containing transcription factors and 3 AP2-like ethylene-responsive transcription factors. Importantly, two transcription factors related to embryogenesis or ovule development were significantly down-regulated, including the transcription factor TCP4 encoding protein MATERNAL EFFECT EMBRYO ARREST 35 (MEE35) and AP2-like ethylene-responsive transcription factor ANT. In addition, several transcription factors related to cell differentiation were also down-regulated, such as MYB113, AS1 and GATA transcription factor 21. Several other transcription factors that participate in development or defense responses, such as GLK1 (related to plastid development and resistance), DIVARICATA, PCF5 and TGA2, were also down-regulated.
A total of 12 important candidate DEGs were then selected for the quantitative RT-PCR assays to confirm the differential expression of the important DEGs screened from the RNA-Seq analysis. The expression levels of the DEGs fit well with those determined by RNA-Seq analysis (Figure 6). Two DEGs encoding apoptosis-inducing factor homolog A (APIFA) showed sharp increases in calcium deficiency-treated embryos at 15, 20 and 30 DAP (Figures 4B, 7). APIFA presumably might play a pivotal role in peanut embryo abortion under calcium deficiency. The relative expression changes in two transcription factors (TCP4 and ANT) related to embryogenesis or ovule development were also validated by qRT-PCR suggesting that they were down-regulated in the calcium deficiency-treated embryos, consistent with the RNA-seq (Figures 6, 4D). The down-regulation of these two transcription factors might play a key role in peanut embryo development barriers under conditions of calcium deficiency.

Figure 6. Quantitative RT-PCR validation of selected DEGs under calcium deficiency and sufficiency in peanut embryo at 15, 20 and 30 DAP. Blue bar represents relative expression level changes calculated by the FPKM using RNA-seq. Orange line with indicates relative expression level changes determined by qRT-PCR analysis.
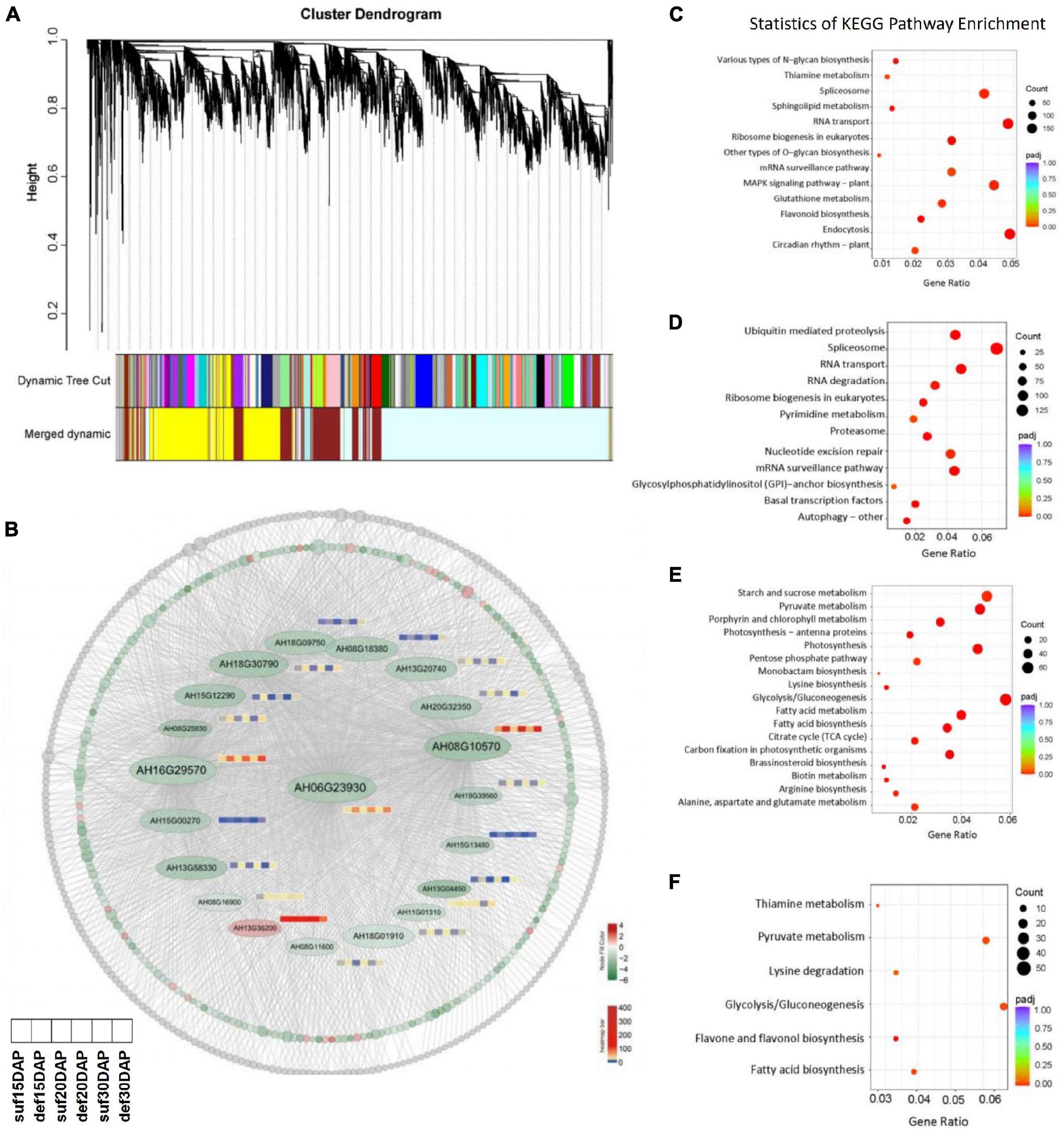
Figure 7. WGCNA of the genes under calcium deficiency and sufficiency in peanut embryo at 15, 20 and 30 DAP. (A) Hierarchical clustering tree indicates 16 modules of co-expressed genes identified by WGCNA. (B) Network of the hub gene AhFUS3 (AH06G23930) and related genes with an edge weight of > 0.15 from the navajowhite2 module. Each circle represents one gene and red color indicates up-regulated genes, green indicates down-regulated genes, gray color represents the genes with no significant differences under calcium deficiency and sufficiency conditions. The expression profiles for important transcription factors in the navajowhite2 module under calcium deficiency and sufficiency conditions at the stage of 15, 20 and 30 DAP were analyzed. (C–F) KEGG analysis of genes in the darkolivegreen2, firebrick3, navajowhite, navajowhite2 modules.
WGCNA was performed to construct the co-expression networks across all samples and 16 distinct gene co-expression modules defined using different color codes were identified (Figure 7A). The genes with high correlation were clustered into one module, and genes within the same module are co-expressed. KEGG pathway enrichment analysis showed that the pathways related to MAPK signaling pathway was significantly enriched in the darkolivegreen2 module, ubiquitin mediated proteolysis pathway was significantly enriched in the firebrick3 module, reserve substance biosynthesis and metabolism such as fatty acid, starch and sucrose, lysine and arginine was significantly enriched in the navajowhite and navajowhite2 modules (Figures 7A,C–F and Supplementary Table 8). Cytoscape 3.9.0 was used to construct the co-expression network of hub gene FUS3 (AH06G23930), which had the highest connectivity among the navajowhite2 module and plays important roles in embryo development (Figure 7B). Interestingly, several other transcription factors such as TCP4, GLK1, ABI3, bHLH115, MYC2, etc., were also included in this module and down regulated under calcium deficiency (Figures 4C, 6 and Supplementary Tables 9, 10), which had been identified to be involved in embryogenesis or ovule development. Thus these genes in navajowhite2 module may play important roles in peanut embryo development.
Peanut grown in calcium-deficient soil generates a high degree of empty pods, showing special sensitivity to calcium level in the soil (Jain et al., 2011). Deficiency calcium severely impaired the normal development of embryo cells, causing the endomembrane system disintegrated, the cell wall to remain thin and later degrade, and the vacuoles to fail compartmentation in early stages and thus leading to reserve accumulation (Figure 1). The abnormal embryo cell growth was clearly the first step toward embryo abortion. As a result, the accumulation of reserve substances, including starch granules, protein and lipid bodies, were all seriously impaired and dissolved (Figure 1). Thereby, deficient calcium weakened early embryo cell development, abrogating reverse synthesis and accumulation and thus leading to embryo abortion.
To elucidate the cellular events, a large number of DEGs were identified involved in various pathways (Figure 3). Diverse important biological processes, such as signal transduction, biosynthetic processes of plant hormones, and responses to biotic and abiotic stimuli were significantly enriched (Supplementary Figures 2, 4 and Supplementary Tables 3–5). Specifically, many genes involved in carbohydrate metabolic pathways such as starch and sugar metabolism, glycolysis and gluconeogenesis, and citrate cycle pathways, for example, the alpha-amylase gene AMY1.1, were up-regulated in response to calcium deficiency (Figure 3; Yu et al., 2018). These phenomena explain the reduced number of plastids in early embryo cells and greatly reduced starch accumulation size (Figure 1). Genes related to fatty acid biosynthesis and metabolism and glycerolipid and glycerophospholipid metabolisms pathways, such as TGL1 (Fan et al., 2017), WRI1 (Chen et al., 2018), LOX1.5 (Lõpez et al., 2011), lipid-transfer protein 1 (LTP1) and peroxygenase (SOP1), regulating fatty acid biosynthesis, were down-regulated. These results were consistent with the decreased accumulation and distorted morphology of lipid bodies (Figure 1). Many genes, such as amino acid permease 7 (AAP7), an amino acid-proton symporter and a stereospecific transporter with broad specificity for neutral amino acids in amino acid metabolism and protein synthesis and processing, such as glycine, alanine, aspartate, glutamate, and phenylalanine metabolism, processing, and amino sugar and nucleotide sugar metabolism pathways, were down-regulated, impairing protein synthesis, as observed in the protein bodies behavior in calcium-deficient embryo cells (Figure 1).
It was also detected several up-regulation for cell wall degradation-related genes but down-regulation of cell wall biogenesis and organization-related genes (Supplementary Table 2). XTH, participating in cell wall biogenesis and construction (Miedes et al., 2013), was up-regulated under calcium deficiency. PME, related to the modification of cell walls via demethylesterification of cell wall pectin, was also up-regulated. The up-regulation of XTH and PME could probably lead to a loss of the cell wall and a distorted cell morphology and structure, consistent with the TEM observation (Figure 1). Therefore, the transcriptome changes under calcium deficiency were likely responsible for the distorted cell morphology and structure, abated embryonic cell growth and hindered accumulation of lipid and protein bodies, ultimately leading to embryo abortion.
Cytoplasmic Ca2+ signals have been reported to play vital roles in the activation of programmed cell death (PCD) (Ma and Berkowitz, 2007). Cytoplasmic Ca2+ homeostasis is essential to mediate plant growth and development, as well as adaptive responses to a broad range of abiotic and biotic stresses (Clapham, 2007). In the present study, many calcium-related genes were changed under calcium deficiency. Genes related to Ca2+ influx, such as CNGC1 and CNGC17, were down-regulated, and genes controlling Ca2+ efflux, such as ACA4, ACA12, ACA13, ECA4 and MCU2, were up-regulated (Figure 5). CNGC1 has been confirmed to relate Ca2+ uptake and root development in Arabidopsis (Ma et al., 2006). CNGC17 plays important roles in phytosulfokine (PSK) peptide-induced cell expansion (Ladwig et al., 2015). Additionally, the expression level of CNGC15 in the present study was increased, resulting in a flow of Ca2+ flow from the cytoplasm to the nucleus (Figure 5). Mt-CNGC15a/b/c have been found to be responsible for nuclear Ca2+ spiking in Medicago truncatula (Charpentier et al., 2016). CNGC20 is also up-regulated and involved in Ca2+ oscillations in the nucleus, playing essential roles in the establishment of both rhizobial and mycorrhizal symbioses in roots (Capoen et al., 2011). Obviously, the above genes control cytoplasmic Ca2+ homeostasis and are intimately involved in plant development. Notwithstanding, little is currently known about their effects on embryo development. In addition, EGFR, PLC2 and PLC4 were down-regulated. Accumulating evidence places PI-PLC enzymes at the center of plant innate immunity, which can regulate the increased cytoplasmic levels of free Ca2+, lowering the intercellular pH and the oxidative burst (Abd-El-Haliem and Joosten, 2017). Hence, reduced expression levels of PI-PLC lead to a decrease in cytoplasmic levels of free Ca2+. Importantly, PLC2 is essentially involved in reproductive and embryonic development, presumably by regulating mitosis and/or formation of the cell division plane (Di Fino et al., 2017). Disruption of PLC2 leads to sterility, indicating a significant role of PLC2 in reproductive development (Li et al., 2015). Especially important is the observation of elevated auxin levels in plc2 floral tissues, suggesting that the infertility of plc2 plants may be associated with increased auxin concentrations in reproductive organs (Li et al., 2015). Therefore, calcium deficiency induced embryo abortion associated with expression changes in these genes by regulating cytoplasmic Ca2+ homeostasis.
During calcium signal transduction, Ca2+-binding proteins or Ca2+ sensors decode the stimulus-specific Ca2+ signals into downstream responses. In the present study, many calcium signal components, such as CPK28, CPK10, CRCK2, CRCK3, CML11 and CML39, were up-regulated. As calcium sensors, calcium-dependent protein kinases (CDPKs) play important roles in the regulation of plant growth and development, as well as responses to biotic and abiotic stresses. In Arabidopsis CPK28 controls stem elongation and vascular development (Matschi et al., 2013). CPK28 targets methionine adenosyltransferase (MAT), an essential enzyme in ethylene biosynthesis. cpk28 mutants display short hypocotyls and ectopic lignification caused by ethylene overproduction (Jin et al., 2017). Furthermore, CPK28 also regulates development by balancing JA and GA levels (Matschi et al., 2015). When plants are exposed to microbial infection, the primary responses mediated by surface-localized immune receptors are to increase cytosolic calcium (Ca2+) levels followed by a burst of apoplastic reactive oxygen species (ROS). CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst (Monaghan et al., 2015). Hence, CPK28 is an important regulatory component related to plant growth and development, associated with changes in phytohormones and cytosolic Ca2+ levels. In the present study, CPK28 was significantly up-regulated under conditions of calcium deficiency, concurring with aborted embryos and decreases in cytosolic Ca2+ levels. CPK10 participates in ABA- and Ca2+-mediated regulation of stomatal movements in response to drought stress (Zou et al., 2010). Notwithstanding, the functions of CPK10 in seed/embryo development have remained unclear. Here, CPK10 was up-regulated under calcium deficiency, implying its important roles in peanut embryo development. CML39 belonging to extended families of unique Ca2+ sensors, is involved in light signal transduction and early seedling establishment (Bender et al., 2013). CML11 participated in the defense response against Verticillium dahliae infection in upland cotton (Cheng et al., 2016). However, the functions of CML in seed/embryo development have not been clarified. In addition, CRCK2 and CRCK3 are also up-regulated. Disruption of CRCK2 induces defects in male gametophyte development (Boavida et al., 2009). The up-regulation of CRCKs might suggest that peanut plants have an adaptive reaction to conditions of calcium deficiency. De novo transcriptome sequencing has revealed potential mechanisms of seed abortion in dove tree and shown that calcium might be an important second messenger in the regulation of abortion (Li et al., 2016). A number of calcium-related genes, including CML and ACA, are up-regulated in aborted seeds (Li et al., 2016). The expression of three Ca2+-transporting ATPase genes were lower in peanut kernels under the Ca2+ sufficient treatment (Yang et al., 2020). All the above results are consistent with ours.
Accordingly, a model was proposed to generalize Ca2+ homeostasis in embryo cells under calcium deficiency conditions (Figure 5). Calcium deficiency first decreased the Ca2+ level in the apoplast and tissue. Subsequently, it altered the transcriptome through the expression of genes encoding Ca2+ influx and efflux transporters, allowing free Ca2+ in the cytoplasm and extracellular space to enter intracellular organelles, including the vacuole, mitochondria, endoplasmic reticulum and nucleus, leading to a decline in cytoplasmic free Ca2+ levels. These changes clearly disrupt cytoplasmic Ca2+ homeostasis, mediate the hormone metabolic balance, alter gene expression, and ultimately lead to peanut embryo abortion under calcium deficiency conditions through hormone signaling and transcriptome changes (Figure 5).
A series of genes related to plant hormone biosynthesis and signal transduction were differentially expressed in calcium-deficient peanut embryos (Figure 3). The endogenous IAA content is significantly increased in aerial pods with embryo abortion (Chen et al., 2013). IAA content increased in peanut kernels under Ca2+ deficiency (Yang et al., 2020). In our study, ALDH3H1, participating in tryptophan metabolism and the key enzyme in IAA biosynthesis, was up-regulated. IAA synthetase GH3.1 and GH3.6 were also up-regulated, which catalyze the synthesis of IAA-amino acid conjugates, providing a mechanism for plants to cope with the presence of excess auxin (Staswick et al., 2005) in embryo cells and this result was consistent with the previous reports (Yang et al., 2020). Auxin transporter protein (AUX1/LAX2) was significantly up-regulated under deficient calcium stress, presumably leading to an increase in IAA content. PILS3, required for auxin-dependent regulation of plant growth by determining the cellular sensitivity to auxin, was also up-regulated. In Arabidopsis, ectopic expression of PILS3 resulted in dwarfed and/or bushy plants showing severe defects in flower development leading to sterility (Barbez et al., 2012). The related genes in the auxin signaling transduction pathway, including the auxin-responsive protein SAUR32, were also markedly up-regulated. Over-expression of SAUR32 leads to reduce hypocotyl elongation in Arabidopsis (Park et al., 2007). It is possible that the up-regulation of SAUR32 should result in some defects in peanut embryo cell growth.
Cytokinin dehydrogenase 2 (CKX3) and CKX7 were significantly up-regulated in peanut embryo under calcium deficiency. CKX is a key negative regulator controlling endogenous cytokinin in plants (Zhao et al., 2015). Thereby, this change could lead to a decrease in endogenous cytokinin, which might affect seed development and ultimately lead to embryo abortion. However, the previous study showed that two genes encoding cytokinin dehydrogenase were down-regulated in peanut kernels under Ca2+ deficient conditions (Yang et al., 2020), which is contrary to our findings. Therefore, cytokinin content in peanut seeds under calcium deficiency and sufficiency conditions should be assayed in the future study.
The level of endogenous abscisic acid has been reported to function as a positive regulator during plant embryo development (González-Guzmán et al., 2002). CYP707As encode abscisic acid (ABA) 8′-hydroxylases, key enzymes in ABA catabolism. CYP707A1, CYP707A3, and CYP707A4 were up-regulated in calcium-deficient peanut embryos, leading to a decrease in the endogenous ABA level. Previous studies have demonstrated that overexpression of AhCYP707A4 in Nicotiana benthamiana decreased ABA content with high numbers of aborted embryos, small pods and fewer seeds, confirming role in the regulation of Ca2+ deficiency-induced embryo abortion via ABA-mediated apoptosis (Chen et al., 2016). Additionally, ABA2 and ABA1, involving in the biosynthesis of ABA, were down-regulated. ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde, a key step in ABA biosynthesis (González-Guzmán et al., 2002). The up-regulation of CYP707As and down-regulation of ABA2 and ABA1 might thus decrease the ABA level under calcium deficiency conditions, affecting embryo development. It was reported that abscisic acid 8′-hydroxylase gene was up-regulated in peanut kernels under Ca2+ deficiency, which is also consistent with our results (Yang et al., 2020).
Several key enzymes involved in gibberellin biosynthesis were down-regulated under deficient calcium conditions: GA1 and GA3 (Sun and Kamiya, 1994), GA20OX1 and GA20OX2 (Rieu et al., 2008), and KAO1 (Helliwell et al., 2001). Gibberellin 20 oxidase gene were down-regulated in peanut kernels under Ca2+ deficiency, which is also consistent with our results (Yang et al., 2020). The down-regulation of these genes decrease the GA level in calcium deficiency-treated embryos, affecting embryo development. Surprisingly, the gibberellin receptor GID1B was up-regulated and the DELLA protein GAI down-regulated. GID1B is expressed in ovules as a dominant GA receptor at low GA concentrations during germination (Gallego-Giraldo et al., 2014). GID1B targets DELLA proteins, the repressors of GA-induced growth, for degradation via the proteasome as the feedback mechanism caused by low levels of GAs in calcium-deficient embryos.
Several genes in the brassinosteroid signaling pathway were down-regulated, including CYP90A1, CURL3, BSK3 and BAK1. CYP90A1 (CPD), a cytochrome P450 gene encoding steroid hydroxylase, is a BR-specific biosynthesis gene and controls cell elongation (Tanaka et al., 2005). Brassinosteroids could rescue the phenotype of the cpd mutant (Szekeres et al., 1996). CURL3 responds to BR, and the curl-3 mutant shows reduced fertility, strikingly similar to the Arabidopsis mutant brassinosteroid insensitive 1 (bri1) (Koka et al., 2000). BSK3 is a positive regulator of BR signaling (Zhang et al., 2016). BAK1 and BKK1 regulate BR-dependent growth and BR-independent cell-death pathways, and bak1 bkk1 double mutants exhibit a seedling-lethality phenotype (He et al., 2007). It was also found that CYP90A1 was down-regulated under calcium deficiency, which should decrease BR levels in the embryo and affect the expression of downstream gene, such as CURL3, BSK3 and BAK1, ultimately affecting normal development.
In addition, many genes involved in ethylene biosynthesis and signal transduction showed different expression levels under low calcium conditions. For example, ACS1 and ACO3, which are related to ethylene biosynthesis, were up-regulated, possibly leading to an increase in ethylene levels. Accordingly, ethylene receptor (ETR), EIN3-binding F-box protein 1 (EBF1), and ethylene-responsive transcription factors (ABR1 and ERF098) were also up-regulated. The increased expression of these genes might promote advanced embryo maturity and halt continuing development, as supported by the observation that peanuts growing in calcium deficiency soil produced many more geminated seeds.
In addition, some important genes in the jasmonic acid and salicylic acid signal transduction pathways were also significantly changed. Thus, peanuts growing in calcium-deficient conditions activated a series of gene expression changes and cross-talk among various hormones.
Overall, peanut embryo development is associated and regulated by various hormones. However, the mechanisms that regulate the crosstalk among these plant hormones are still not well understood. Furthermore, changes in the cytoplasmic calcium concentration have been demonstrated to function as a secondary signal messenger for most plant hormone pathways (Fortes et al., 2015). Recently, a growing number of studies have shown that the calcium signal transduction pathway is involved in plant hormone pathways. CPK28 plays a vital role in balancing JA and GA in Arabidopsis development (Matschi et al., 2015) and affects ethylene biosynthesis (Jin et al., 2017). CPK4 is related to the modulation of ABA signaling (Wang et al., 2017). CML20 negatively regulates ABA-induced stomatal movement in Arabidopsis (Wu X. et al., 2017). StCDPK3 is involved in the cross-talk between ABA and GA signaling at the onset of potato tuber development (Grandellis et al., 2016). In brief, peanut seed development may be regulated by the collaboration of the Ca2+ signal transduction pathway and its involvement in hormone regulation pathways. In a word, a predicted schematic diagram was proposed to generalize calcium signaling pathway and hormone biosynthesis related genes in peanut embryo cells according to our and previous study (Figure 5). The predicted schematic diagram of calcium signaling pathway and hormone biosynthesis related genes in peanut embryo cells and their subcellular localization. More research is needed to delve into the underlying molecular mechanisms for peanut abortion under calcium deficiency conditions.
Previous study indicated that Ca2+ deficiency can affect several other ions transport (zinc, aluminum, sodium, potassium, iron, and magnesium) and nutrient absorption (nitrate and phosphate) (Yang et al., 2020). Here we found that zinc transporter 5 was up-regulated under calcium deficiency (Supplementary Table 2). Sodium/hydrogen exchanger 4 (NHX4), sodium/metabolite cotransporter BASS1 (T30F21.11) and sodium-dependent phosphate transport protein 1 (T27A16.25) were down regulated (Supplementary Table 2). Two DEGs encoding potassium transporter 8 (T9L3_180) were down regulated (Supplementary Table 2). However, DEGs encoding potassium transporter 5 (T9E8.160), potassium transporter 12 (T13D8.5) and potassium channel SKOR (SKOR) were up regulated (Supplementary Table 2). Several DEGs related to magnesium transport including magnesium transporter MRS2-1, MRS2-3, MRS2-4, MRS2-11 were also found (Supplementary Table 2). Nitrogen, phosphorus and potassium are essential elements for plant growth and development. In this study, the expression levels of fourteen NRT1/PTR family protein genes were changed with four genes down-regulated and ten genes up-regulated under Ca2+ deficiency conditions (Supplementary Table 2). In addition, one DEG encoding high-affinity nitrate transporter 3.1 (At5g50200) and one DEG encoding peptide/nitrate transporter (ZIFL2) were up regulated under Ca2+ deficiency conditions, which is consistent with previous study (Yang et al., 2020). It was reported that SPX domain proteins are essential for maintaining phosphorus homeostasis in plants (Secco et al., 2012; Wang et al., 2021). Two DEGs encoding SPX domain-containing protein 1 and two DEGs encoding SPX domain-containing membrane protein were down regulated under Ca2+ deficiency conditions (Supplementary Table 2). Obviously, Ca2+ deficiency not only affects intracellular Ca2+ homeostasis but also other ions transport and homeostasis such as Zn2+, Na+, K+, Mg2+, etc., and hinders the absorption of important peanut development needed nutrients. But the underlying mechanism for the complicated regulatory relationships among these ions is not clear. Our results poses an interesting research direction for further study.
Several putative important regulatory genes were obtained that might play critical roles in peanut embryo abortion under conditions of calcium deficiency. Two APIFAs showed a sharp increase in calcium deficiency-treated embryos (Figures 4B, 6). These results implied that APIFA played a pivotal role in peanut embryo abortion under calcium deficiency. Apoptosis-inducing factor (AIF) is a phylogenetically conserved mitochondrial intermembrane flavoprotein that induces apoptosis via a caspase-independent pathway (Lorenzo et al., 1999). AIF is involved in the induction of nuclear chromatin condensation as well as large-scale DNA fragmentation (approximately 50 kb), and it is essential for programmed cell death during the cavitation of embryoid bodies (Lü et al., 2003). The up-regulation of APIFA indicated that apoptosis occurred in the early stages of peanut development under calcium-deficient conditions, an essential cause of the embryo abortion.
Several transcription factors (TCP4, ANT, WRI1, and FUS3) are related to embryogenesis or ovule development, and they were down-regulated in the calcium deficiency-treated embryos. TCP4 is essential for post-zygotic development (Pagnussat et al., 2005). ANT plays a critical role in regulating ovule and female gametophyte development (Klucher et al., 1996). FUS3 is an essential regulator of embryogenesis (Tsuchiya et al., 2004). WRI1 is involved in the regulation of seed storage substance metabolism in Arabidopsis, especially seed oil accumulation (Cernac and Benning, 2004). The down-regulation of these transcription factors yields embryo development barriers under calcium deficiency.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA470988.
WZ and HC conceived the project, designed the research and conducted the experiments. HC performed most of the experiments, analyzed the data, and drafted the manuscript. QY carried out the data analysis and participated in drafting the manuscript. HF assisted in data collection and analysis. KC and SZ performed qRT-PCR analysis. CZ, TC, and LW participated in data collection and analysis. HD, MG, HL, and XY helped to prepare and take the samples. WZ and RV revised the manuscript. All authors have read, edited, and approved the current version of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (31601337) and the Science and Technology Foundation of Fujian Province of China (2017N0006 and 2021N5007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Ensheng Zhou and Shoucheng Liu for their help in the experimental field selection and plant management. We would also like to thank Zhiyong Wang (Carnegie Institution for Science, Stanford) and Ronglong Pan (National Tsing Hua University) for a critical review of the article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.814015/full#supplementary-material
Supplementary Figure 1 | The percentage of mapped reads distribution in different sites (intro, intergenic and exon) of gene in peanut embryos under calcium deficiency and sufficiency conditions. The X axis represented three repetitions of peanut embryos under calcium deficiency and sufficiency conditions at 15, 20, and 30 DAP, respectively. The Y axis is represented the percentage of mapped reads distributed in different sites of gene. Blue column: exon; Orange: intergenic; Gray: intron. Three biological replicates were carried out for each sample.
Abd-El-Haliem, A. M., and Joosten, M. H. A. J. (2017). Plant phosphatidylinositol-specific phospholipase C at the center of plant innate immunity. J. Integr. Plant Biol. 59, 164–179. doi: 10.1111/jipb.12520
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Barbez, E., Kubeš, M., Rolčík, J., Béziat, C., Pěnčík, A., Wang, B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122. doi: 10.1038/nature11001
Bender, K. W., Rosenbaum, D. M., Vanderbeld, B., Ubaid, M., and Snedden, W. A. (2013). The Arabidopsis calmodulin-like protein, CML39, functions during early seedling establishment. Plant J. 76, 634–647. doi: 10.1111/tpj.12323
Beringer, H., and Taha, M. A. (1976). 45Calcium absorption by two cultivars of groundnut (Arachis hypogea). Exp. Agric. 12, 1–7. doi: 10.1017/S0014479700006992
Boavida, L. C., Shuai, B., Yu, H. J., Pagnussat, G. C., Sundaresan, V., and McCormick, S. (2009). A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181, 1369–1385. doi: 10.1534/genetics.108.090852
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Charpentier, M., Sun, J., Martins, T. V., Radhakrishnan, G. V., Findlay, K., Soumpourou, E., et al. (2016). Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105. doi: 10.1126/science.aae0109
Chen, H., Yang, Q., Chen, K., Zhao, S., Zhang, C., Pan, R., et al. (2019). Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genomics 20:392. doi: 10.1186/s12864-019-5770-6
Chen, H., Zhang, C., Cai, T. C., Deng, Y., Zhou, S., Zheng, Y., et al. (2016). Identification of low Ca2+ stress-induced embryo apoptosis response genes in Arachis hypogaea by SSH-associated library lift (SSHaLL). Plant Biotechnol. J. 14, 682–698. doi: 10.1111/pbi.12415
Chen, L., Zheng, Y., Dong, Z., Meng, F., Sun, X., Fan, X., et al. (2018). Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genomics 293, 401–415. doi: 10.1007/s00438-017-1393-2
Chen, X., Zhu, W., Azam, S., Li, H., Zhu, F., Li, H., et al. (2013). Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant Biotechnol. J. 11, 115–127. doi: 10.1111/pbi.12018
Cheng, H. Q., Han, L. B., Yang, C. L., Wu, X. M., Zhong, N. Q., Wu, J. H., et al. (2016). The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67, 1935–1950. doi: 10.1093/jxb/erw016
Di Fino, L. M., D’Ambrosio, J. M., Tejos, R., van Wijk, R., Lamattina, L., Munnik, T., et al. (2017). Arabidopsis phosphatidylinositol-phospholipase C2 (PLC2) is required for female gametogenesis and embryo development. Planta 245, 717–728. doi: 10.1007/s00425-016-2634-z
Fan, J., Yu, L., and Xu, C. (2017). A central role for triacylglycerol in membrane lipid breakdown, fatty acid β-oxidation, and plant survival under extended darkness. Plant Physiol. 174, 1517–1530. doi: 10.1104/pp.17.00653
Fortes, A. M., Teixeira, R. T., and Agudelo-Romero, P. (2015). Complex interplay of hormonal signals during grape berry ripening. Molecules 20, 9326–9343. doi: 10.3390/molecules20059326
Gallego-Giraldo, C., Hu, J., Urbez, C., Gomez, M. D., Sun, T. P., and Perez-Amador, M. A. (2014). Role of the gibberellin receptors GID1 during fruit-set in Arabidopsis. Plant J. 79, 1020–1032. doi: 10.1111/tpj.12603
González-Guzmán, M., Apostolova, N., Bellés, J. M., Barrero, J. M., Piqueras, P., Ponce, M. R., et al. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. doi: 10.1105/tpc.002477
Grandellis, C., Fantino, E., García, M. N. M., Bialer, M. G., Santin, F., Capiati, D. A., et al. (2016). StCDPK3 phosphorylates in vitro two transcription factors involved in GA and ABA signaling in potato: StRSG1 and StABF1. PLoS One 11:e0167389. doi: 10.1371/journal.pone.0167389
He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., et al. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115. doi: 10.1016/j.cub.2007.05.036
Helliwell, C. A., Chandler, P. M., Poole, A., Dennis, E. S., and Peacock, W. J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 98, 2065–2070. doi: 10.1073/pnas.98.4.2065
Jain, M., Pathak, B. P., Harmon, A. C., Tillman, B. L., and Gallo, M. (2011). Calcium dependent protein kinase (CDPK) expression during fruit development in cultivated peanut (Arachis hypogaea) under Ca2+-sufficient and -deficient growth regimens. J. Plant Physiol. 168, 2272–2277. doi: 10.1016/j.jplph.2011.07.005
Jin, Y., Ye, N., Zhu, F., Li, H., Wang, J., Jiang, L., et al. (2017). Calcium-dependent protein kinase CPK28 targets the methionine adenosyltransferases for degradation by the 26S proteasome and affects ethylene biosynthesis and lignin deposition in Arabidopsis. Plant J. 90, 304–318. doi: 10.1111/tpj.13493
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Klucher, K. M., Chow, H., Reiser, L., and Fischer, R. L. (1996). The AINTEGUMENTA gene of arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. doi: 10.1105/tpc.8.2.137
Koka, C. V., Cerny, R. E., Gardner, R. G., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122, 85–98. doi: 10.1104/pp.122.1.85
Ladwig, F., Dahlke, R. I., Stührwohldt, N., Hartmann, J., Harter, K., and Sauter, M. (2015). Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes cyclic nucleotide-gated channel17, H+-ATPase, and BAK1. Plant Cell 27, 1718–1729. doi: 10.1105/tpc.15.00306
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, 1–13. doi: 10.1186/gb-2009-10-3-r25
Li, L., He, Y., Wang, Y., Zhao, S., Chen, X., Ye, T., et al. (2015). Arabidopsis PLC2 is involved in auxin-modulated reproductive development. Plant J. 84, 504–515. doi: 10.1111/tpj.13016
Li, M., Dong, X., Peng, J., Xu, W., Ren, R., Liu, J., et al. (2016). De novo transcriptome sequencing and gene expression analysis reveal potential mechanisms of seed abortion in dove tree (Davidia involucrata Baill.). BMC Plant Biol. 16:82. doi: 10.1186/s12870-016-0772-x
Lin, W. Z. (1996). A study on the symptom and ultrastructure of calcium deficiency for peanut. Sci. Agric. Sin. 29, 53–60.
Lõpez, M. A., Vicente, J., Kulasekaran, S., Vellosillo, T., Martínez, M., Irigoyen, M. L., et al. (2011). Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 67, 447–458. doi: 10.1111/j.1365-313X.2011.04608.x
Lorenzo, H. K., Susin, S. A., Penninger, J., and Kroemer, G. (1999). Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 6, 516–524. doi: 10.1038/sj.cdd.4400527
Lü, C. X., Fan, T. J., Hu, G. B., and Cong, R. S. (2003). Apoptosis-inducing factor and apoptosis. Acta Biochim. Biophys. Sin. (Shanghai) 35, 881–885.
Ma, W., Ali, R., and Berkowitz, G. A. (2006). Characterization of plant phenotypes associated with loss-of-function of AtCNGC1, a plant cyclic nucleotide gated cation channel. Plant Physiol. Biochem. 44, 494–505. doi: 10.1016/j.plaphy.2006.08.007
Ma, W., and Berkowitz, G. A. (2007). The grateful dead: calcium and cell death in plant innate immunity. Cell. Microbiol. 9, 2571–2585. doi: 10.1111/j.1462-5822.2007.01031.x
Matschi, S., Hake, K., Herde, M., Hause, B., and Romeis, T. (2015). The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27, 591–606. doi: 10.1105/tpc.15.00024
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., and Romeis, T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. doi: 10.1111/tpj.12090
Meinke, D., Muralla, R., Sweeney, C., and Dickerman, A. (2008). Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 13, 483–491. doi: 10.1016/j.tplants.2008.06.003
Miedes, E., Suslov, D., Vandenbussche, F., Kenobi, K., Ivakov, A., Van Der Straeten, D., et al. (2013). Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 64, 2481–2497. doi: 10.1093/jxb/ert107
Monaghan, J., Matschi, S., Romeis, T., and Zipfel, C. (2015). The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP induced calcium burst. Plant Signal. Behav. 10, 1–5. doi: 10.1080/15592324.2015.1018497
Murata, M. R., Zharare, G. E., and Hammes, P. S. (2008). pH of the pod-zone affects reproductive growth of groundnut. J. Plant Nutr. 31, 69–79. doi: 10.1080/01904160701741859
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Pandey, M. K., Monyo, E., Ozias-Akins, P., Liang, X., Guimarães, P., Nigam, S. N., et al. (2012). Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 30, 639–651. doi: 10.1016/j.biotechadv.2011.11.001
Park, J. E., Kim, Y. S., Yoon, H. K., and Park, C. M. (2007). Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 172, 150–157. doi: 10.1016/j.plantsci.2006.08.005
Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S. J., Gong, F., et al. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504. doi: 10.1111/j.1365-313X.2007.03356.x
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076. doi: 10.1038/nmeth.2212
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Sun, T. P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase a of gibberellin biosynthesis. Plant Cell 6, 1509–1518. doi: 10.2307/3869986
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Tanaka, K., Asami, T., Yoshida, S., Nakamura, Y., Matsuo, T., and Okamoto, S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125. doi: 10.1104/pp.104.058040
Thilakarathna, S. M. C. R., Kirthisinghe, J. P., Gunathilaka, B. L., and Dissanayaka, D. M. P. V. (2015). Influence of gypsum application on yield and visual quality of groundnut (Arachis hypogaea L.) grown in Maspotha in Kurunegala district of Sri Lanka. Trop. Agric. Res. 25:432. doi: 10.4038/tar.v25i3.8050
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsuchiya, Y., Nambara, E., Naito, S., and McCourt, P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37, 73–81. doi: 10.1046/j.1365-313X.2003.01939.x
Wang, L., Jia, X., Zhang, Y., Xu, L., Menand, B., Zhao, H., et al. (2021). Loss of two families of SPX domain-containing proteins required for vacuolar polyphosphate accumulation coincides with the transition to phosphate storage in green plants. Mol. Plant 14, 838–846. doi: 10.1016/j.molp.2021.01.015
Wang, P., Yang, Q., Sang, S., Chen, Y., Zhong, Y., and Wei, Z. (2017). Arabidopsis inositol polyphosphate kinase AtIpk2β is phosphorylated by CPK4 and positively modulates ABA signaling. Biochem. Biophys. Res. Commun. 490, 441–446. doi: 10.1016/j.bbrc.2017.06.060
Wu, H., Li, C., Huang, Z., Tang, X., Han, Z., Zhong, R., et al. (2017). Comparision on acid and low calcium resistance ability of different peanut varieties. J. Anhui Agric. Sci. 11, 27–30.
Wu, X., Qiao, Z., Liu, H., Acharya, B. R., Li, C., and Zhang, W. (2017). CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 8:824. doi: 10.3389/fpls.2017.00824
Xia, H., Zhao, C., Hou, L., Li, A., Zhao, S., Bi, Y., et al. (2013). Transcriptome profiling of peanut gynophores revealed global reprogramming of gene expression during early pod development in darkness. BMC Genomics 14:517. doi: 10.1186/1471-2164-14-517
Yang, S., Wang, J., Tang, Z., Guo, F., Zhang, Y., Zhang, J., et al. (2020). Transcriptome of peanut kernel and shell reveals the mechanism of calcium on peanut pod development. Sci. Rep. 10:15723. doi: 10.1038/s41598-020-72893-9
Yu, W., Zou, W., Dhital, S., Wu, P., Gidley, M. J., Fox, G. P., et al. (2018). The adsorption of α-amylase on barley proteins affects the in vitro digestion of starch in barley flour. Food Chem. 241, 493–501. doi: 10.1016/j.foodchem.2017.09.021
Zhang, B., and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4:Article17. doi: 10.2202/1544-6115.1128
Zhang, B., Wang, X., Zhao, Z., Wang, R., Huang, X., Zhu, Y., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170, 1149–1161. doi: 10.1104/pp.15.01668
Zhang, J. (2004). Molecular Mechanism of Embryo Abortion of Peanut Affected by Calcium Deficiency. Available online at: http://d.wanfangdata.com.cn/Thesis_Y629120.aspx (accessed February 28, 2005).
Zhang, J. C., Cai, N. B., Zhang, X. W., and Zhuang, W. J. (2007). Isolation and identification of specific expressed proteins from peanut (Arachis hypogaea) development/abortion embryo mediated by Ca2 +. Acta Agron. Sin. 33, 814–819.
Zhang, J. C., Zhang, X. W., and Song, Y. H. (2008). A MADS box gene in peanut embryo was involved in embryo development regulated by Ca2 + level. Acta Agron. Sin. 24, 180–185.
Zhang, J. L., Feng, G., Jing-Jing, M., Xiao-Xia, Y., Sha, Y., Si-Bin, Z., et al. (2015). Effects of calcium fertilizer on yield, quality and related enzyme activities of peanut in acidic soil. Chin. J. Plant Ecol. 39, 1101–1109. doi: 10.17521/cjpe.2015.0107
Zhao, J., Bai, W., Zeng, Q., Song, S., Zhang, M., Li, X., et al. (2015). Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 35:60. doi: 10.1007/s11032-015-0232-6
Zhuang, W., Chen, H., Yang, M., Wang, J., Pandey, M. K., Zhang, C., et al. (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51, 865–876. doi: 10.1038/s41588-019-0402-2
Keywords: embryo abortion, calcium, hormone, WGCNA, Arachis hypogaea L.
Citation: Chen H, Yang Q, Fu H, Chen K, Zhao S, Zhang C, Cai T, Wang L, Lu W, Dang H, Gao M, Li H, Yuan X, Varshney RK and Zhuang W (2022) Identification of Key Gene Networks and Deciphering Transcriptional Regulators Associated With Peanut Embryo Abortion Mediated by Calcium Deficiency. Front. Plant Sci. 13:814015. doi: 10.3389/fpls.2022.814015
Received: 12 November 2021; Accepted: 14 February 2022;
Published: 21 March 2022.
Edited by:
Michael Nicolas, Wageningen University and Research, NetherlandsReviewed by:
Ashutosh Pandey, National Institute of Plant Genome Research (NIPGR), IndiaCopyright © 2022 Chen, Yang, Fu, Chen, Zhao, Zhang, Cai, Wang, Lu, Dang, Gao, Li, Yuan, Varshney and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Zhuang, d2VpamlhbnpAZmFmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.