- Molecular Plant Pathology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
Plant pathogens employ secreted proteins, among which are effectors, to manipulate and colonize their hosts. A large fraction of effectors is translocated into host cells, where they can suppress defense signaling. Bacterial pathogens directly inject effectors into host cells via the type three secretion system, but it is little understood how eukaryotic pathogens, such as fungi, accomplish this critical process and how their secreted effectors enter host cells. The root-infecting fungus Fusarium oxysporum (Fo) secrets numerous effectors into the extracellular space. Some of these, such as Foa3, function inside the plant cell to suppress host defenses. Here, we show that Foa3 suppresses pattern-triggered defense responses to the same extent when it is produced in planta irrespective of whether the protein carries the PR1 secretory signal peptide or not. When a GFP-tagged Foa3 was targeted for secretion it localized, among other locations, to mobile subcellular structures of unknown identity. Furthermore, like the well-known cell penetrating peptide Arginine 9, Foa3 was found to deliver an orthotospovirus avirulence protein-derived peptide into the cytosol, resulting in the activation of the matching resistance protein. Finally, we show that infiltrating Foa3 into the apoplast results in strong suppression of the pattern-triggered immune responses, potentially indicating its uptake by the host cells in absence of a pathogen.
Introduction
Plant pathogens are a major threat to agriculture causing significant yield losses. For successful colonization, pathogens need to manipulate their host, for example, by evading or suppressing its defense responses. Microbial pathogens, such as bacteria, fungi, or oomycetes, generally do this via a cocktail of secreted proteins, called effectors (Lo Presti et al., 2015; Uhse and Djamei, 2018; He et al., 2020). Plants on the other hand detect the presence of potential pathogens through a sophisticated surveillance system comprised of both cell-surface and intracellular receptors (Bentham et al., 2020; Zhou and Zhang, 2020). Detection of conserved structures that are typical for microbes, called pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs) is mediated by transmembrane receptors. A well characterized example is flagellin (or its derived peptide flg22) that is recognized via FLS2, leading to a series of immune responses such as increased calcium influx, production of reactive oxygen species (ROS) and activation of mitogen-activated protein kinases (MAPKs; Defalco and Zipfel, 2021). Intracellularly localized immune receptors generally belong to the nucleotide binding and leucin-rich repeat containing proteins (NLRs) and detect race-specific effectors. Activation of NLRs results in a strong defense response, often culminating in localized cell death (referred to as hypersensitive response, HR; Yuan et al., 2021).
A significant portion of effectors is translocated into host cells, where they act as virulence or avirulence factors (Lo Presti et al., 2015; He et al., 2020). Whereas translocation of bacterial effectors via the type three secretion system is well established, it is little understood how fungal- or oomycete effectors reach the cytoplasm of host cells (Petre and Kamoun, 2014). Intracellular-acting oomycete effectors are frequently characterized by well-defined N-terminal amino acid motifs such as the RXLR or CRN sequence (Rehmany et al., 2005; Schornack et al., 2010). Initially, it was proposed that the RXLR motif mediates effector uptake by binding to specific phospholipids in the host cell membrane; however, recent data suggest that it rather plays a role during secretion into the extracellular space (Kale et al., 2010; Wawra et al., 2017). An intracellular function is shown for many fungal effectors; however, a shared amino acid motif is not apparent among them (Lo Presti et al., 2015). Prior to their internalization, Magnaporthe oryzae effectors localize to the biotrophic interfacial complex, a membrane-rich structure between the fungal hyphae and the plant cell membrane (Khang et al., 2010). Recently, a protein complex identified in Ustilago maydis was proposed to mediate translocation of effectors into the host cytosol (Ludwig et al., 2021). However, these structures have not been described in other plant pathogenic fungi. Thus, it remains a key question whether filamentous plant pathogens use special machineries to deliver effectors into host cells and/or whether certain effectors possess properties that allow them to pass the host cell membrane independent of a pathogen-encoded structure.
Fusarium oxysporum (Fo) is a widespread root-colonizing fungus, that can cause disease in more than 120 different plant species, many of which are agronomically important (Edel-Hermann and Lecomte, 2019). Individual strains carry partially overlapping effector sets, potentially determining their host range (Van Dam et al., 2016). Several Fo effectors were shown to act intracellularly as (a)virulence factors (Houterman et al., 2009; Di et al., 2017; Tintor et al., 2020). However, it remains unknown how they enter plant cells, since they do not share obvious motifs, or have common physio-chemical properties such as size or charge. Here we focus on Foa3, an effector of the Arabidopsis-infecting strain Fo5176 that inhibits both the flg22- and chitin-triggered ROS burst when expressed in planta without its endogenous signal peptide, indicating that it acts via an intracellular mechanism (Tintor et al., 2020). However, expression of full length Foa3 (e.g. including its signal peptide) resulted in a comparable suppression of both flg22- and chitin-triggered ROS accumulation, suggesting that the effector is taken up by the host cells. In this study we provide several lines of evidence pointing to translocation of Foa3 into the host cytosol when it is targeted for secretion or infiltrated into the apoplast.
Materials and Methods
Plasmid Construction
The constructs pCAMBIA-35S::Sw-5b and pCAMBIA-35S::NS21-YFP were described previously (Zhu et al., 2017). The signal peptide coding sequence of Nicotiana tabacum PR1 was amplified from PVX::ECP1 (Laugé et al., 2000) and subcloned into pDONR201 and pGWB451. To generate SPpr1-NS24-YFP, the signal peptide sequence was PCR-amplified using primer FP8256/FP8257 and as template pDONR-SPpr1 (primers are listed in Table 1). Likewise, the NS24-YFP cassette was amplified using primer FP8258/FP8259 and the template pCAMBIA-NS21-YFP (Zhu et al., 2017). Primer FP8258 N-terminally introduced three additional amino acids to NS21-YFP, resulting in NS24-YFP. This was done to ensure that a stretch of hydrophilic amino acids follows the signal peptide and thus ensures proper cleavage by the plant secretion machinery. The amino acids were the same as in the full length NSm protein at that position. The SPpr1 and NS24-YFP fragments were fused by overlapping PCR with primers FP8260/FP8259, thereby introducing an XbaI and a BamHI site. The obtained product was cloned into the XbaI/BamHI sites of pBIN61 (SLDB3104; (Tameling et al., 2010) and named pBIN-SPpr1-NS24-YFP (Supplementary Figure 1). To create SPpr1-NS24-YFP-Arg9, the SPpr1-NS24-YFP cassette was amplified by PCR using primers FP8260/FP8592 and pBIN-SPpr1-NS24-YFP as template. In a second PCR the restriction sites XbaI/XmaI were added by PCR using primers FP8260/FP8593. Finally, the obtained fragment was cloned into the XbaI/XmaI sites of pBIN61, resulting in pBIN-SPpr1-NS24-YFP-Arg9. To generate SPpr1-NS24-Foa3, the backbone of pBIN-SPpr1-NS24-YFP was amplified by PCR using primers FP8847/FP8848. The Foa3 coding sequence (genbank ID: FOXB_16928) lacking the signal peptide sequence (dsp for deleted sp) was amplified from pBIN-dspFoa3-HB (Tintor et al., 2020) with primers FP8849/FP8850, thereby introducing overlaps with NS24 and the vector backbone. Finally, these two fragments were fused using the In-Fusion technology, following the manufacturer’s instructions (New England Biolabs). The obtained construct was named pBIN-SPpr1-NS24-Foa3.
To C-terminally fuse them to GFP, dspFoa3 and SPpr1-Foa3 were first subcloned into pDONR207. dspFoa3 was amplified with primers FP9156/FP9157, and the Gateway adapters were added with primers FP872/FP873. The obtained fragment was subcloned into pDONR207 following the manufacturer’s instructions. The start codon was added by Quick Change using primers FP9627/FP9628. SPpr1-Foa3 was amplified from a pBIN-SPpr1-Foa3-NS24 construct by using primers FP9155/FP9157 and introduced into pDONR207. pGWB451-dspFoa3-GFP and pGWB451-SPpr1-Foa3-GFP were generated by Gateway recombination.
Transient Transformation of Nicotiana benthamiana
Agrobacterium tumefaciens strain GV3101 was transformed with binary vectors and used to transiently transform 4–5-week-old N. benthamiana plants as described previously (Tintor et al., 2020). Plants were kept in a greenhouse at 20°C without artificial illumination. For Western blots, apoplastic fluid isolation, ROS and MAPK assays, Agrobacteria were infiltrated at an OD600 of 0.3. A strain containing the silencing suppressor P19 was co-infiltrated at the same OD600 (Tintor et al., 2020). For cell death assays and confocal microscopy, all Agrobacterium strains, including P19, were infiltrated at an OD600 of 0.1.
Apoplastic Fluid Collection
Apoplastic fluid was isolated from N. benthamiana leaves 2–3 days after infiltration with Agrobacteria. To this end, deionized water (MQ) was infiltrated into leaves using a needleless syringe. Excess water was removed from the surface with filter paper. Next, the leaves were rolled up and placed into the outer unit of a 20-ml syringe, which was inserted into a 50 ml falcon tube. Apoplastic fluid was released by centrifugation at 1,000 g for 10 min at 4°C. Next, the apoplastic fluid was filtered through a PVDF membrane (0.45 μm pore size, low protein binding) to remove any Agrobacteria, and either used directly or stored at −20°C.
ROS and MAPK Assays
Flg22-triggered ROS generation was monitored as described earlier (Felix et al., 1999). ROS production was induced with 200 nM flg22 and measured with help of a luminol/peroxidase solution [250 μM luminol (Sigma), 10 μg/ml peroxidase (Sigma)]. Apoplastic fluid derived from N. benthamiana plants that produced dspFo3, spFoa3 or spGFP was infiltrated into leaves of 4–5-week-old N. benthamiana plants and 3 h later leaf discs were excised and kept overnight on MQ. Flg22-triggered ROS production was measured the next morning.
Flg22-triggered MAPK activation was performed as described previously, with a few modifications (Flury et al., 2013). Three days after Agro-infiltration, N. benthamiana leaves were infiltrated with 0.5 μM flg22 or MQ and 10 min later leaf discs corresponding to 15 mg leaf material were collected and immediately frozen in liquid nitrogen. Total protein was isolated as described earlier and MAPK activation was monitored by Western blot using an anti-p44/p42 MAPK antibody (D13.14.4E; Cell Signaling Technology). Apoplastic fluid derived from N. benthamiana plants that produced dspFo3, spFoa3 or spGFP was infiltrated into leaves of 4–5-week-old N. benthamiana plants and 3 h later 1 μM flg22 or MQ was infiltrated. Ten minutes after flg22 treatment, leaf discs were collected, frozen in liquid nitrogen, and processed as described above.
Protein Isolation and Western Blotting
Protein isolation and western blotting was carried out largely as reported before (Tintor et al., 2020). Briefly, soluble proteins were isolated from N. benthamiana by grinding 20 mg leaf tissue in 150 μl lysis buffer [50 mM Tris–HCl pH 7.5, 2% SDS, 10 mM DTT, 1x protease inhibitor cocktail (Roche), 1 mM PMSF] and centrifuging at 16,000 g for 10 min. Proteins were separated by SDS-PAGE, using 12% gels. As primary antibody monoclonal anti-GFP antibodies (ChromoTek) were used at 1:6,000 dilution. The secondary goat-anti-rat antibody (Pierce) was used at 1:8,000. The signal was visualized using the ECL Plus kit (GE Healthcare) according to the manufacturer’s instructions.
Cell Death and Confocal Imaging
Cell death in N. benthamiana leaves was visualized similarly as described (Landeo Villanueva et al., 2021). Four days after Agrobacterium infiltration red fluorescence was visualized using a ChemiDoc imaging system (Universal Hood III, BioRad), using Qdot 605 filters and 30 s exposure time. Live cell imaging was carried out 2–3 days after Agro-infiltration using a Nikon A1 confocal microscope. GFP was excited at 488 nm with an Ar-ion laser, and emission was detected at 500–550 nm. RFP was excited at 561 nm using a dpss-laser and emission was detected at 570–620 nm. Images were captured using a 20x water-immersion objective or a 40x oil-immersion objective. Images were composed using Fiji software.
Results
Both dspFoa3 and SPpr1-Foa3 Inhibit PTI
We previously reported that a Foa3 version lacking its endogenous signal peptide (dspFoa3) suppressed flg22- and chitin-triggered ROS production, pointing to an intracellular virulence function (Tintor et al., 2020). Interestingly, expressing the full-length gene, thus encoding the signal peptide, resulted in a comparable level of ROS suppression. To exclude the possibility that the fungal Foa3 signal peptide is not efficiently recognized by the plant secretory system, we generated a construct where the signal peptide sequence derived from Nicotiana tabacum PR1 was fused to Foa3. This PR1 signal peptide was shown in previous studies to effectively target proteins for secretion (Hammond-Kosack et al., 1994; Laugé et al., 2000; Ziegler et al., 2000). In addition, both dspFoa3 and SPpr1-Foa3 were C-terminally tagged with GFP to allow their detection via Western blot and live cell imaging. To test whether the fusion proteins were produced to similar amounts in planta, both constructs were expressed in N. benthamiana using Agrobacterium mediated transient transformation. A construct encoding GFP fused to the PR1 signal peptide sequence was included as a control. Western blotting with anti-GFP antibodies revealed that dspFoa3-GFP migrated at around 50 kDa, which corresponds to the expected size of the dspFoa3-GFP fusion protein (Figure 1A). In contrast, for SPpr1-Foa3-GFP two bands were detected at roughly 25 and 50 kDa, indicating that the GFP tag was frequently cleaved off, but also that a significant amount of the GFP tag remained fused to the effector. Next, apoplastic fluid was collected to test for presence of secreted Foa3. The apoplastic fluid of leaves expressing dspFoa3-GFP revealed only a very weak GFP signal, indicating that in the absence of a signal peptide, this protein remains largely intracellular (Figure 1A). The residual amount detected in the apoplast could potentially originate from damaged cells. In contrast, both free GFP as well as the Foa3-GFP fusion protein were readily detected in the apoplastic fluid of SPpr1-Foa3-GFP expressing leaves (Figure 1A). These results show that dspFoa3 remains intracellular, but that adding the PR1 signal peptide results in its secretion into the apoplast.
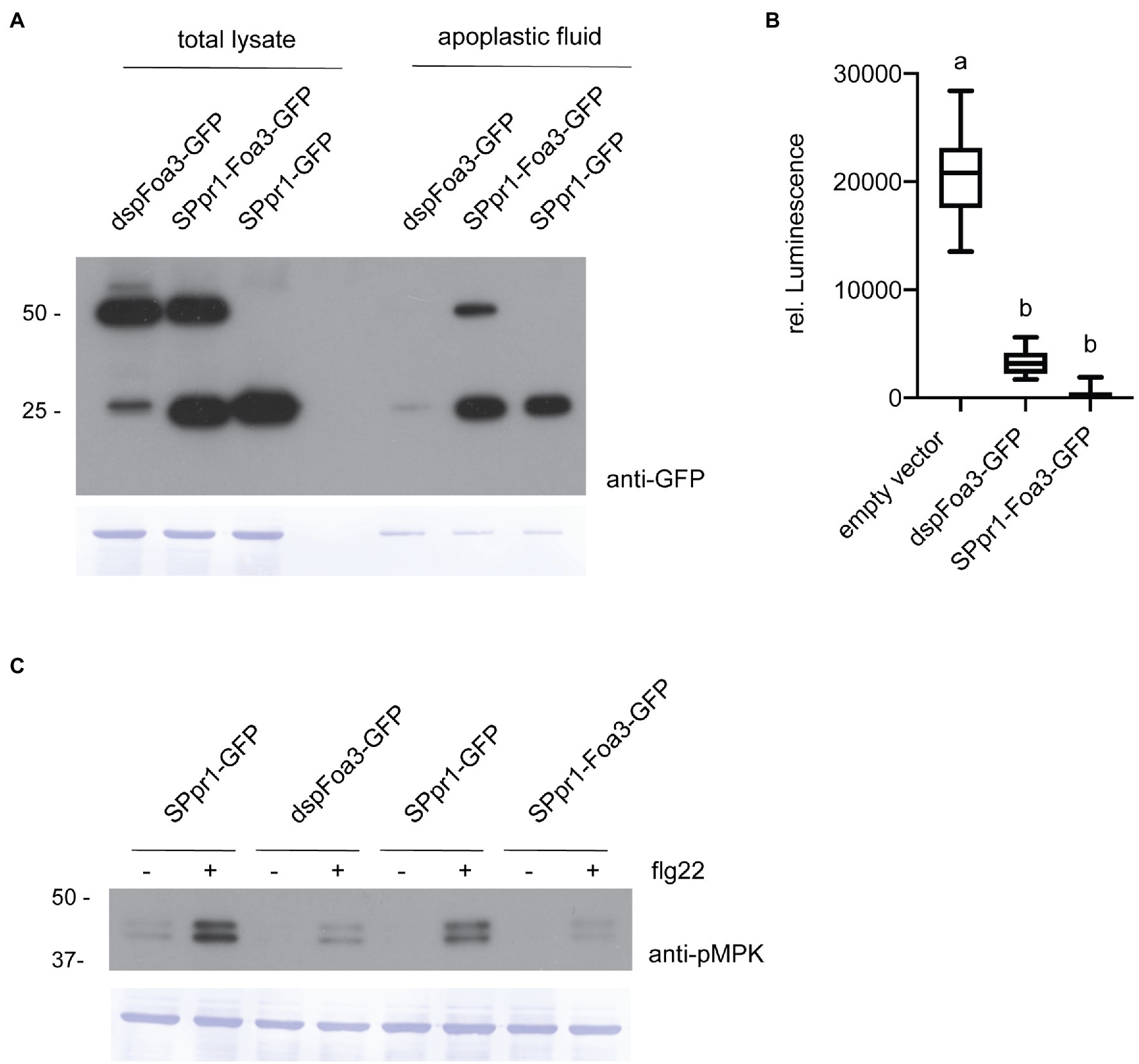
Figure 1. Expression of Foa3 with or without the PR1 signal peptide sequence suppresses flg22-triggered defense responses. (A) Anti-GFP western blots showing accumulation of dspFoa3-GFP, SPpr1-Foa3-GFP and SPpr1-GFP in total protein lysates and in apoplastic fluid of Agrobacterium infiltrated Nicotiana benthamiana leaves. Molecular weight (kDa) markers are shown on the left. Equal loading is verified by Coomassie staining of the blots. (B) Flg22-triggered ROS burst in Nicotiana benthamiana plants expressing the indicated constructs. Boxes extend from the 25th to the 75th percentile, whiskers from the lowest to highest values, bar indicates the median, n = 12 leaf discs. Significant differences are indicated by the different letters (p < 0.05, Kruskal-Wallace test). (C) Western blot showing flg22-triggered activation of MAP kinases in Nicotiana benthamiana leaves expressing the indicated constructs. Leaf discs were exposed to water (−) or flg22 (+) for 10 min. Molecular weight (kDa) markers are shown on the left. Equal loading is verified by Coomassie staining of the blots. Experiments were repeated three times with similar results.
Next, we wanted to test whether Foa3 could still exert its (intracellular) defense-suppressing activity after being targeted to the apoplast by the PR1 signal peptide. To test this, N. benthamiana leaves infiltrated with Agrobacteria strains containing, respectively, an empty vector, dspFoa3-GFP or SPpr1-Foa3-GFP were treated with the bacterial elicitor flg22. Subsequently, the production of ROS was measured over time. As expected, the empty vector-treated leaves showed high levels of ROS induction, whereas the presence of dspFoa3-GFP resulted in a strong suppression of this defense response upon flg22 application (Figure 1B). Importantly, the SPpr1-Foa3-GFP producing leaves showed a similar or even slightly stronger ROS suppression as compared to the dspFoa3-GFP leaves (Figure 1B). To corroborate this finding, a second defense output was tested, namely activation of MAP kinases by flg22. For this N. benthamiana leaves expressing either spGFP as control or dspFoa3-GFP or SPpr1-Foa3-GFP were mock-treated or infiltrated with flg22. The activation of MAP kinases was monitored by Western blot using an antibody that recognizes phosphorylated MAP kinases. Whereas the spGFP-expressing leaves showed a strong accumulation of phosphorylated MAPKs, presence of dspFoa3-GFP or SPpr1-Foa3-GFP resulted in an almost complete suppression of this defense response (Figure 1C). These findings show that even though the effector was targeted for secretion, Foa3 still efficiently suppressed PTI responses.
Subcellular Localization of Foa3
Since both dspFoa3 and SPpr1-Foa3 efficiently suppressed defense responses despite being targeted to different locations, we set out to investigate their subcellular localization. First, N. benthamiana leaves expressing dspFoa3-GFP were subjected to laser scanning confocal microscopy. A GFP signal was revealed that is typical for a nuclear-cytosolic localization (Figure 2A). Indeed, co-expression with a nuclear marker (REP-NLS-RFP; Maio et al., 2019) confirmed accumulation in the nucleus, whereas only very little overlap with an endoplasmic reticulum (ER) marker was observed (Cao et al., 2018). In conclusion, expression of Foa3-GFP without a secretory signal peptide leads to its accumulation mainly in the cell nucleus and cytosol. This indicates that Foa3 exerts its PTI-suppressing activity at these subcellular locations. In contrast, expression of SPpr1-Foa3-GFP showed a very different localization pattern for the fluorescent effector protein. The GFP signal was predominantly observed in bright spots, most of which were mobile (Figure 2). A somewhat weaker signal was outlining the cell and was also visible around the nucleus and in cytosolic strands (Figure 2A). In addition, the GFP signal was detected in a net-like structure typical for the ER which indeed co-localized with the ER marker (Figure 2A). The bright punctate structures localized in the proximity of, but did not show a clear overlap with, the ER marker suggesting that they are not part of the ER. We did not detect a strong accumulation of SPpr1-Foa3-GFP in the cytosol or nucleus, even though it suppresses the PTI responses to an extent similar as dspFoa3-GFP. Since in a large fraction of the tagged effector the GFP was cleaved off, it is possible that mainly untagged Foa3 enters the cytosol and nucleus and is responsible for the observed PTI suppression. Altogether, SPpr1-Foa3-GFP seems to partially localize to the ER, but in addition it strongly accumulates in subcellular structures of unknown identity that can localize in physical proximity to the ER, but also move along cytosolic strands.
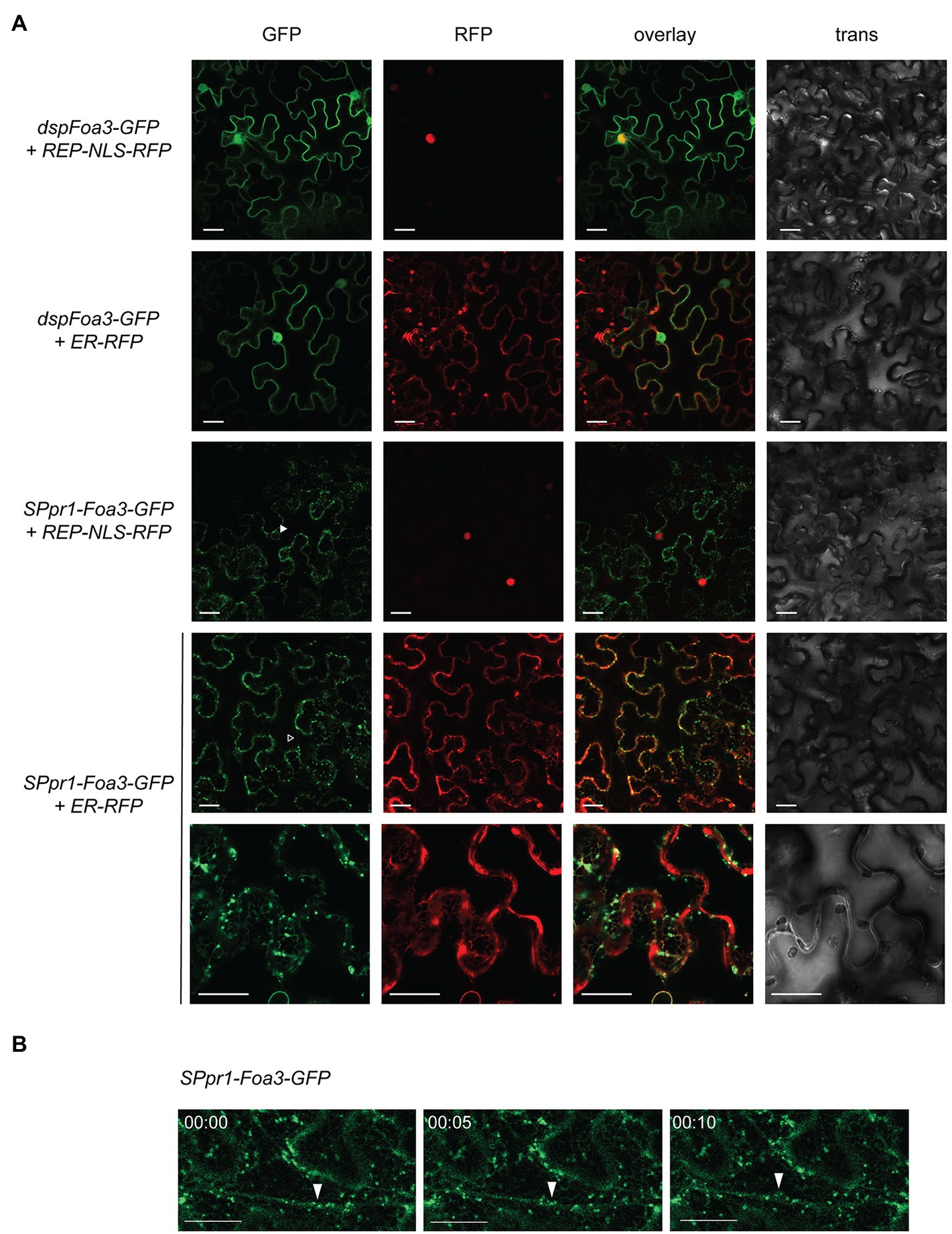
Figure 2. Subcellular localization of Foa3 produced with (sp) or without (dsp) a secretory signal peptide. (A) Confocal microscopy of Agro-infiltrated Nicotiana benthamiana leaves expressing the indicated constructs. Closed arrowhead indicates SPpr1-Foa3-GFP accumulating around a nucleus. Open arrowhead indicates SPpr1-Foa3-GFP at cytosolic strands. Size bars = 20 μm. (B) Time-lapse confocal images showing accumulation of SPpr1-Foa3-GFP in mobile subcellular structures. Closed arrows indicate Foa3-GFP labelled puncta moving along cytosolic strands.
Foa3 Can Deliver NS24 Into Host Cells
To provide additional evidence for intracellular translocation of SPpr1-Foa3, we monitored its ability to deliver an Avirulence protein to the cytosol. The non-structural movement protein (NSm) of tomato spotted wilt virus is recognized by the resistance protein Sw-5b, leading to an HR. Furthermore, it was shown that attaching a 21 amino acid peptide derived from NSm (NS21) to YFP is sufficient to physically bind to and activate Sw-5b (Zhu et al., 2017). As the interaction between NS21 and Sw-5b occurs intracellularly and produces an easily detectable cell death response, we predicted that attaching NS21 to Foa3 could serve as a proxy for its intracellular localization.
First, we investigated whether attaching the PR1 signal peptide to NS21-YFP would re-direct it to the secretory path and thus prevent intracellular accumulation and subsequent recognition by Sw-5b. To create a typical cleavage site in which the signal peptide sequence is followed by a set of polar amino acids, the NS21 unit was N-terminally extended by three amino acids, resulting in SPpr1-NS24-YFP (Figure 3A; section Materials and Methods). Zhu et al. established that N- or C-terminally extending the NS21 minimal unit does not interfere with its recognition by Sw-5b (Zhu et al., 2017). As reported earlier, co-expression of NS21-YFP with Sw-5b in N. benthamiana induced a strong cell death response (Figure 3C). In contrast, co-expression of SPpr1-NS24-YFP and Sw-5b did not produce a visible cell death response, indicating that secretion of NS24-YFP prevented its intracellular accumulation and thus the activation of the intracellularly localized Sw-5b resistance protein. Importantly, Western blotting showed that both NS21-YFP and SPpr1-NS24-YFP accumulated to similar levels, thus the lack of an HR was not due to poor stability or accumulation of the secreted NS24-YFP protein (Figure 3B). Instead, this result suggests that NS24-YFP lacks the ability to efficiently re-localize to the cytosol after being secreted.
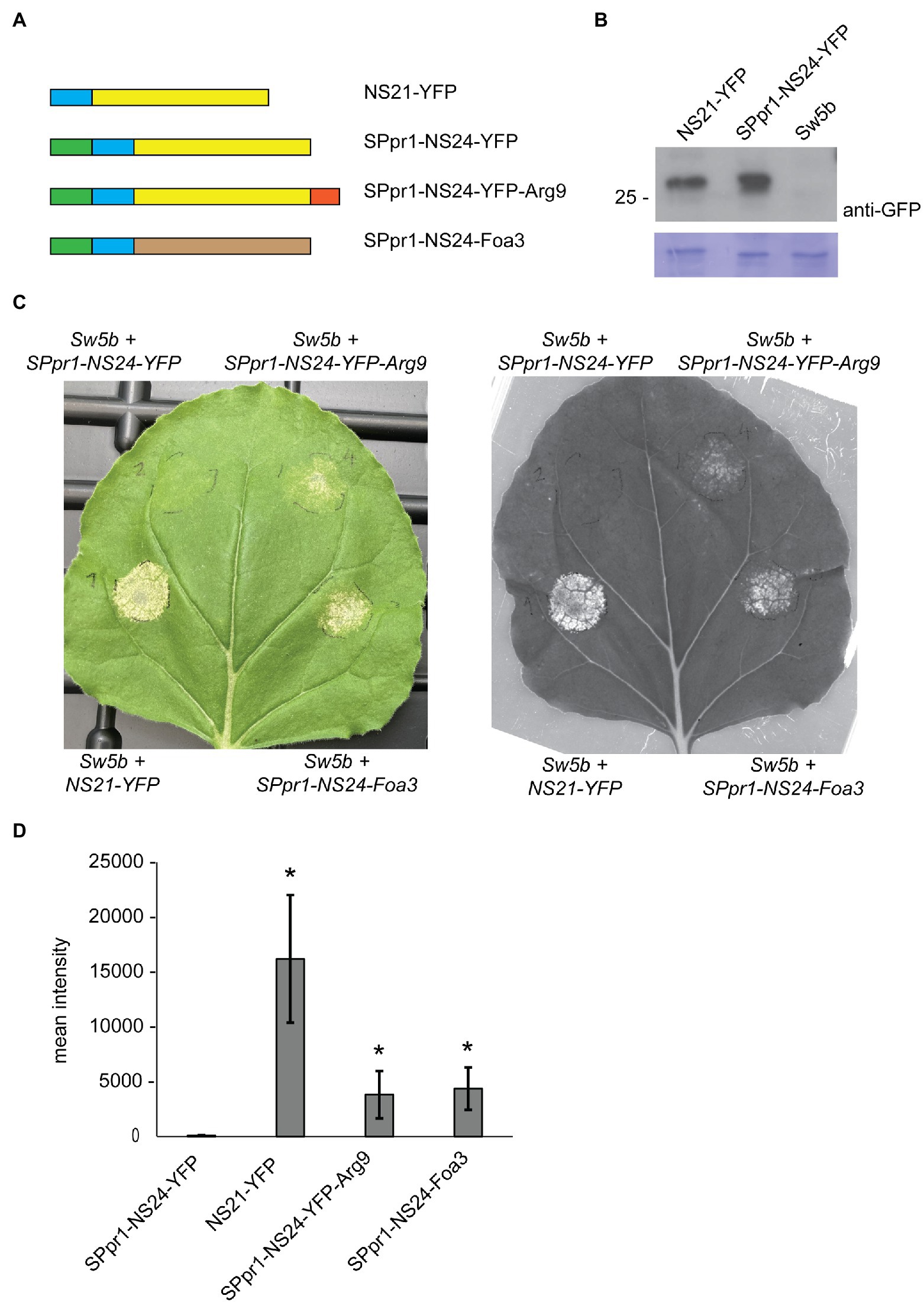
Figure 3. Foa3 can deliver an avirulence protein of tomato spotted wilt virus into host cells. (A) Schematic representation of the constructs used in Nicotiana benthamiana Agro-infiltration experiments. NS21/24 (blue), YFP (yellow), PR1 signal peptide (green), Arginine 9 (red) and Foa3 (brown) are shown. (B) Anti-YFP Western blot showing similar accumulation levels of NS21-YFP and SPpr1-NS24-YFP in Agro-infiltrated Nicotiana benthamiana leaves. Molecular weight (kDa) markers are shown on the left. Equal loading is verified by Coomassie staining of the blots. (C) Sw-5b triggered cell death in Nicotiana benthamiana leaves expressing the indicated constructs. A representative leaf is shown upon regular photography (left) or following red fluorescence imaging using a Chemidoc imager (right). Images were taken 5 days after Agro-infiltration. (D) Signal quantification of red fluorescence imaging of Nicotiana benthamiana leaves producing the indicated constructs. Statistically significant differences to the SPpr1-NS24-YFP construct are indicated (p < 0.05, t-test, n = 6 leaves). Experiments were repeated at least three times with similar results.
Next, we tested whether attaching a cell penetrating peptide of nine consecutive arginine residues (Arg9) to NS24-YFP would result in its uptake, thereby triggering Sw-5b dependent cell death. Arg9-mediated uptake of target proteins has been demonstrated in different systems, among others in soybean and tomato (Dou et al., 2008; Numata et al., 2018). When the SPpr1-NS24-YFP-Arg9 construct was co-expressed with Sw-5b, a cell death response was observed, indicating that NS24-YFP-Arg9 was successfully taken up by the plant cells. The cell death response was weaker than the one induced by cytoplasmic NS21-YFP but was readily detectable both by visual inspection as well as by monitoring red fluorescent emission as a proxy for cell death (Landeo Villanueva et al., 2021; Figure 3C). Next, we tested whether Foa3 could replace the function of Arg9 and mediate cellular entry of NS24. Thereto, SPpr1-NS24 was N-terminally fused to Foa3, and when this SPpr1-NS24-Foa3 construct was co-expressed with Sw-5b, a clear cell death response was observed (Figure 3C). Quantification of red fluorescence showed that both SPpr1-NS24-YFP-Arg9 and SPpr1-NS24-Foa3 induced comparable levels of Sw-5b activation (Figure 3D). In conclusion, both Foa3 and Arg9 seem to enable cellular uptake of NS24 and thus activation of Sw-5b.
Foa3 Can Inhibit PTI Upon Its Infiltration to the Apoplast
Intracellular accumulation of SPpr1-Foa3-GFP could result from its uptake from the apoplast or from re-translocation (e.g., escape) out of the secretory pathway. To distinguish between these scenarios, we investigated whether infiltration of Foa3 into the apoplast would result in PTI suppression. Since SPpr1-Foa3-GFP expressing N. benthamiana accumulates Foa3 in the apoplast (Figure 1A), apoplastic fluid from these plants was collected and infiltrated into untreated plants. In addition, several controls were infiltrated: (1) apoplastic fluid from dspFoa3-GFP expressing plants, containing only trace amounts of Foa3-GFP; (2) apoplastic fluid of SPpr1-GFP expressing plants, containing high levels of GFP; and (3) MQ water. Subsequently, leaf discs obtained from the infiltrated plants were exposed to flg22 and ROS generation was measured. Plants that were infiltrated with either water or the apoplastic fluid from dspFoa3-GFP or SPpr1-GFP expressing plants showed a strong accumulation of flg22-triggered ROS (Figure 4A). In contrast, infiltration of the apoplastic fluid that contained high amounts of Foa3-GFP resulted in an almost complete suppression of the flg22-triggered ROS response (Figure 4A). These results could imply that Foa3 is taken up from the apoplast, reaching its intracellular localization where it suppresses flg22-triggered ROS generation.
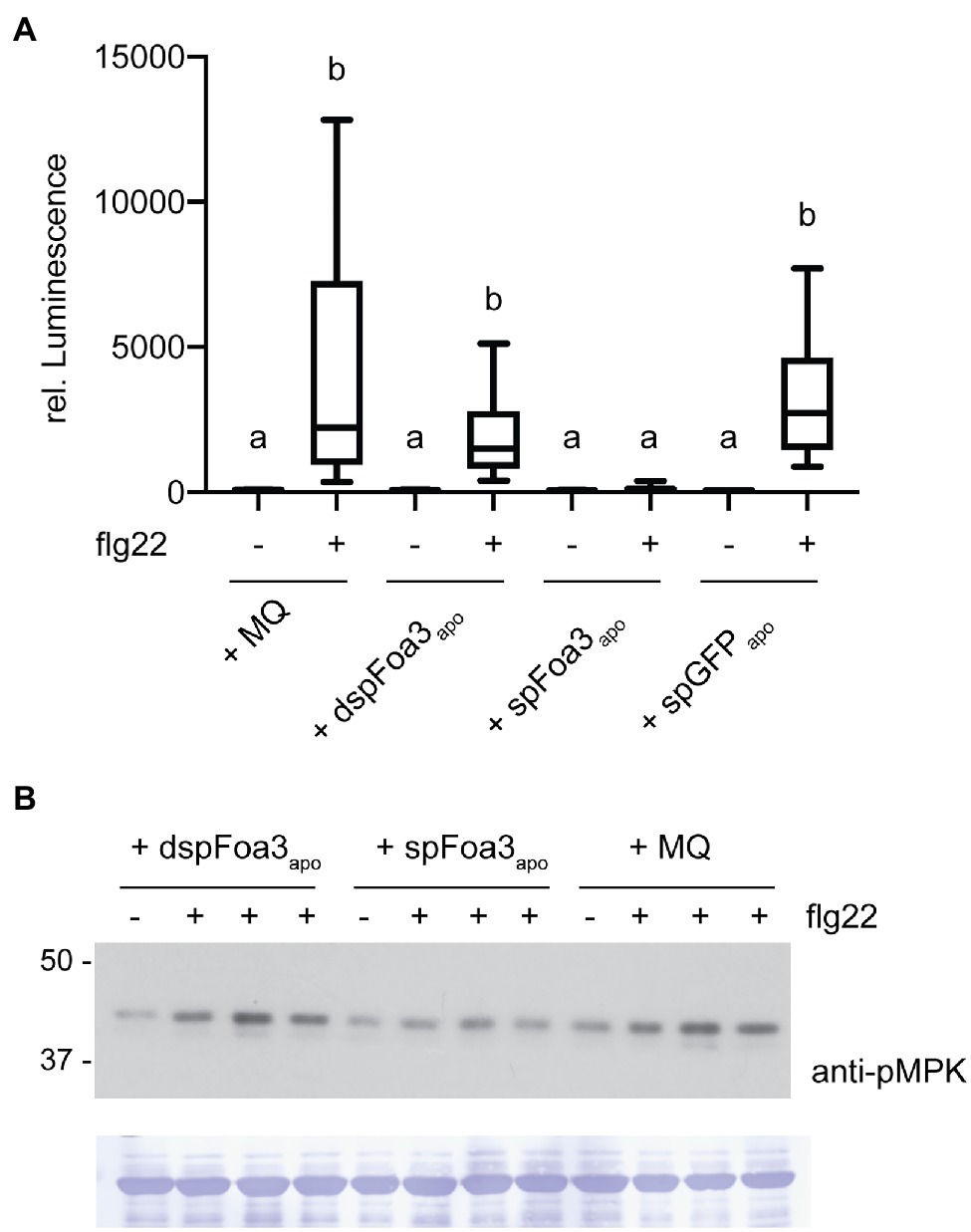
Figure 4. Infiltration of Foa3-containing apopastic fluid into Nicotiana benthamiana leaves results in suppression of flg22-triggered defense responses. (A) Flg22-triggered ROS burst in Nicotiana benthamiana leaves that were treated as follows: Infiltrated with water (MQ), infiltrated with apoplastic fluid isolated from Nicotiana benthamiana plants expressing either dspFoa3-GFP (dspFoa3-GFPapo) or SPpr1-Foa3-GFP (SPpr1-Foa3-GFPapo) or SPpr1-GFP (SPpr1-GFPapo), respectively. Boxes extend from the 25th to the 75th percentile, whiskers from the lowest to highest values, bar indicates the median, n = 16 leaf discs. Significant differences are indicated (p < 0.05, Kruskal-Wallace test). (B) Western blot showing flg22-triggered activation of MAP kinases in Nicotiana benthamiana leaves infiltrated with water (MQ) or apoplastic fluid isolated from Nicotiana benthamiana plants expressing either dspFoa3-GFP (dspFoa3-GFPapo) or SPpr1-Foa3-GFP (SPpr1-Foa3-GFPapo). Subsequently, leaves were treated with water (−) or flg22 (+) for 10 min. Three independent flg22-treated samples were loaded on blots next to each other. Molecular weight (kDa) markers are shown on the left. Equal loading is verified by Coomassie staining of the blots. Experiments were repeated three times with similar results.
To further validate these findings, we monitored flg22-triggered activation of MAPKs in N. benthamiana plants that were treated similarly as for the ROS assay. Infiltration of water or apoplastic fluid from a dspFoa3-GFP expressing plant resulted in MAPK activation as revealed by detection of phosphorylated MAPKs by Western blotting (Figure 4B). In contrast, the infiltration of SPpr1-Foa3-GFP containing apoplastic fluid resulted in much lower levels of activated MAPKs (Figure 4B). In this experiment, predominantly the upper MAPK band was detected, which corresponds to SIPK (SALICYLIC ACID-INDUCED PROTEIN KINASE; Segonzac et al., 2011). This difference may be due to the presence of Agrobacteria in the experimental setup of Figure 1, and absence thereof in Figure 4. Nonetheless, only the treatment with Foa3-containing apoplastic fluid led to suppression of the flg22-triggered accumulation of activated MAPKs. Together with the observed suppression of ROS generation by apoplastically delivered Foa3, these findings point to effector uptake by plant cells from the apoplast.
Discussion
In this study we demonstrate that, when produced in planta without a secretory signal peptide, Foa3 efficiently suppresses flg22-triggered ROS accumulation and -MAPK activation, indicating that it targets an intracellular host factor required for early flg22 signaling. Since chitin-triggered ROS accumulation was suppressed as well (Tintor et al., 2020), Foa3 apparently intercepts signaling triggered by multiple pattern recognition receptors. As dspFoa3-GFP shows a nucleo-cytosolic accumulation, it is likely that its PTI suppressing activity is carried out at these subcellular locations. Interestingly, suppression of the same PTI responses was observed when Foa3 was targeted for secretion via the PR1 signal peptide or when the effector was directly infiltrated into the apoplast. Two scenarios could explain this finding. In the first one, Foa3 is taken up from the apoplast and subsequently translocated to the nucleo-cytoplasmic compartment where it exerts a virulence activity. In the second scenario, Foa3 would have to act on two different targets, resulting in both cases in almost complete inhibition of flg22 and chitin-signaling. Even though effectors can have multiple targets, these are generally present at the same location and/or affect different processes. To our knowledge, an effector targeting the same process via separate intracellular and extracellular functions has not yet been described, making the latter possibility less likely.
Besides its PTI-suppressing activity, Foa3 successfully delivered a secreted NSm24 avirulence peptide to the matching intracellular Sw-5b resistance protein, resulting in a cell death response. In contrast, when an NSm24-YFP fusion protein was targeted for secretion, it did not induce a Sw-5b-triggered cell death, indicating that YFP itself was not able to translocate to the cytosol. To exclude whether the observed Sw-5-specific cell death could be due to escape of the NSm24-Foa3, but not the NSm24-YFP protein, from the secretory pathway upon Agro-infiltration, apoplastic fluid isolated from wildtype N. benthamiana leaves Agro-infiltrated with NSm21-YFP, SPpr1-NSm24-YFP, SPpr1-NSm24-YFP-Arg9 and SPpr1-NSm24-Foa3 was infiltrated in Sw5b expressing N. benthamiana leaves (data not shown). No Sw5b-specific cell death was observed following infiltration of either the NSm24-Foa3 fusion or the positive control NSm24-YFP-Arg9. The same constructs induced cell death when expressed in Sw5b plants, showing their functionality (Figure 3). The lack of Sw5b-induced cell death following apoplastic fluid infiltration is likely due to proteolytic activity in the extracellular spaces resulting in cleavage of the NSm24 peptide from the Foa3 effector protein and from the Arg9 peptide. It is well established that the extracellular spaces from Solanaceous species contain potent proteases that remove (affinity) tags from effector proteins secreted to the apoplast (van Esse et al., 2006) as also can be seen in Figure 1A. Cleavage could have occurred at any step between agroinfiltration, harvesting of the apoplastic fluid or following infiltration, making the latter experiment inconclusive. Potential uptake of the Foa3 protein from the apoplastic space could occur directly from the apoplast into the cytosol, or via retrograde transport from the ER following its internalisation by endocytosis and/or following escape from endosomal vesicles. Future experiments using in situ immune labeling could reveal whether there is a preferred uptake mechanism, or whether these processes occur simultaneously. Notably, when expressed in planta with a secretory signal peptide, numerous fungal and oomycete effector also appear to localize intracellularly, mostly demonstrated by an intracellular avirulence activity (Rehmany et al., 2005; Bos et al., 2006; Rafiqi et al., 2010; Blondeau et al., 2015). However, besides the P. nodorum effector ToxA (Liu et al., 2009), in these cases it was not tested whether the avirulence activity was also observed after infiltration of these effectors into the apoplast, as shown here.
When expressed without a signal peptide Foa3 accumulates in the cytosol and nucleus, strongly suggesting that it acts at these locations to inhibit PTI signaling. In contrast, the GFP fluorescence of secreted Foa3 showed a relatively weak signal in the cytosol and ER but was strongly detected in punctate structures that localized close to the ER, but also moved along cytosolic strands. It is possible that these structures represent vesicles containing proteins targeted for secretion. However, expression of secreted GFP did not produce similar bright fluorescent structures (Supplementary Figure 2). This could indicate that Foa3 accumulates at an intermediate step of the secretory path, even though it eventually reaches the apoplast, since it is present in the apoplastic fluid (Figure 1A). Interestingly, several host-translocated effectors showed similar concentrated accumulation when expressed in planta with a signal peptide (Kloppholz et al., 2011; Petre et al., 2016). In contrast, other intracellular effectors showed a more uniform distribution, indicating that this localization pattern is effector-specific (Rafiqi et al., 2010). It is unlikely that this localization is merely a consequence of overexpression since dspFoa3-GFP accumulates to a similar level but does not exert the same localization pattern. However, whether the punctate localization structures in which Foa3 accumulate are functionally correlated with its ability to translocate into host cells is at present unclear. Additional cell biological studies are required to elucidate the identity and role of these subcellular structures.
Whether a potential cell entry of Foa3 is mediated via binding to a cell surface structure such as (glycol) lipids is currently unknown and would require experimental testing. Our data indicate that Foa3 mediates uptake of NS24 as efficiently as the known cell entry peptide Arginine 9. The precise working mechanism of Arginine 9 is not understood, but it is thought that positively charged arginine residues electrostatically bind to negatively charged cell-surface molecules, possibly inducing endocytosis of the target protein (Duchardt et al., 2007; Schmidt et al., 2010). Foa3 itself carries six arginine and two lysine residues, which are not clustered in a protein structure fold prediction by AlphaFold, arguing against localized strong positive surface charges. However, several positively charged amino acids are predicted to be surface-localized, theoretically enabling an Arginine 9-like, charge-based uptake mechanism of Foa3. The supposed uptake of Foa3 and its strong PTI-suppressing activity provide an ideal tool to investigate the underlying cell biological processes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
NT, IB, and GN performed the experiments. NT and FT designed the project. NT wrote the paper with help of FT. All authors contributed to the article and approved the submitted version.
Funding
FT and NT obtained support from the NWO-Earth and Life Sciences funded VICI Project No. 865.14.003 and from ENZA Zaden.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the van Leeuwenhoek Center for Advanced Microscopy, and there especially Ronald Breedijk for instruction at the usage of the confocal microscope, and we would like to thank Reinofke van de Vis for help with the Chemidoc Imager. In addition, we are grateful to Bas Beerens and Margarita Simkovicova for help with experiments, Lisong Ma for plasmids as well as to Harold Lemereis and Ludek Tikovsky for plant care.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.813181/full#supplementary-material
Supplementary Figure 1 | Plasmid maps of pBIN61 containing the cassettes SPpr1-NSm24-YFP, SPpr1-NSm24-YFP-Arg9 and SPpr1-NSm24-FOA3 (A), as well as pGWB-dspFOA3-GFP and pGWB451-SPpr1-FOA3-GFP (B).
Supplementary Figure 2 | Confocal microscopy of Agro-infiltrated N. benthamiana leaves expressing SPpr1-GFP, hence producing secreted GFP. The bright punctate structures observed with SPpr1-Foa3-GFP were not observed upon SPpr1-GFP expression.
References
Bentham, A. R., De La Concepcion, J. C., Mukhi, N., Zdrzalek, R., Draeger, M., Gorenkin, D., et al. (2020). A molecular roadmap to the plant immune system. J. Biol. Chem. 295, 14916–14935. doi: 10.1074/jbc.REV120.010852
Blondeau, K., Blaise, F., Graille, M., Kale, S. D., Linglin, J., Ollivier, B., et al. (2015). Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins. Plant J. 83, 610–624. doi: 10.1111/tpj.12913
Bos, J. I., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., and Takken, F. L. W. (2018). The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 11, 691–705. doi: 10.1016/j.molp.2018.02.011
Defalco, T. A., and Zipfel, C. (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 81, 3449–3467. doi: 10.1016/j.molcel.2021.07.029
Di, X., Cao, L., Hughes, R. K., Tintor, N., Banfield, M. J., and Takken, F. L. W. (2017). Structure-function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 216, 897–914. doi: 10.1111/nph.14733
Dou, D., Kale, S. D., Wang, X., Jiang, R. H., Bruce, N. A., Arredondo, F. D., et al. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Duchardt, F., Fotin-Mleczek, M., Schwarz, H., Fischer, R., and Brock, R. (2007). A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic 8, 848–866. doi: 10.1111/j.1600-0854.2007.00572.x
Edel-Hermann, V., and Lecomte, C. (2019). Current status of Fusarium oxysporum Formae speciales and races. Phytopathology 109, 512–530. doi: 10.1094/PHYTO-08-18-0320-RVW
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313x.1999.00265.x
Flury, P., Klauser, D., Schulze, B., Boller, T., and Bartels, S. (2013). The anticipation of danger: microbe-associated molecular pattern perception enhances AtPep-triggered oxidative burst. Plant Physiol. 161, 2023–2035. doi: 10.1104/pp.113.216077
Hammond-Kosack, K. E., Harrison, K., and Jones, J. D. G. (1994). Developmentally regulated cell death on expression of the fungal avirulence gene Avr 9 in tomato seedlings carrying the disease-resistance gene Cf-9. Proc. Natl. Acad. Sci. U. S. A. 91, 10445–10449. doi: 10.1073/pnas.91.22.10445
He, Q., Mclellan, H., Boevink, P. C., and Birch, P. R. J. (2020). All roads lead to susceptibility: The many modes of action of fungal and oomycete intracellular effectors. Plant Commun. 1:100050. doi: 10.1016/j.xplc.2020.100050
Houterman, P. M., Ma, L., Van Ooijen, G., De Vroomen, M. J., Cornelissen, B. J. C., Takken, F. L. W., et al. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58, 970–978. doi: 10.1111/j.1365-313X.2009.03838.x
Kale, S. D., Gu, B., Capelluto, D. G., Dou, D., Feldman, E., Rumore, A., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295. doi: 10.1016/j.cell.2010.06.008
Khang, C. H., Berruyer, R., Giraldo, M. C., Kankanala, P., Park, S. Y., Czymmek, K., et al. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22, 1388–1403. doi: 10.1105/tpc.109.069666
Kloppholz, S., Kuhn, H., and Requena, N. (2011). A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. doi: 10.1016/j.cub.2011.06.044
Landeo Villanueva, S., Malvestiti, M. C., Van Ieperen, W., Joosten, M. H. A. J., and Van Kan, J. A. L. (2021). Red light imaging for programmed cell death visualization and quantification in plant-pathogen interactions. Mol. Plant Pathol. 22, 361–372. doi: 10.1111/mpp.13027
Laugé, R., Goodwin, P. H., De Wit, P. J. G. M., and Joosten, M. H. A. J. (2000). Specific HR-associated recognition of secreted proteins from Cladosporium fulvum occurs in both host and non-host plants. Plant J. 23, 735–745. doi: 10.1046/j.1365-313x.2000.00843.x
Liu, Z., Faris, J. D., Oliver, R. P., Tan, K.-C., Solomon, P. S., McDonald, M. C., et al. (2009). SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn 3 gene. PLoS Pathogens 5:e1000581. doi: 10.1371/journal.ppat.1000581.Epub
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Ludwig, N., Reissmann, S., Schipper, K., Gonzalez, C., Assmann, D., Glatter, T., et al. (2021). A cell surface-exposed protein complex with an essential virulence function in Ustilago maydis. Nat. Microbiol. 6, 722–730. doi: 10.1038/s41564-021-00896-x
Maio, F., Arroyo-Mateos, M., Bobay, B. G., Bejarano, E. R., Prins, M., and Van Den Burg, H. A. (2019). A lysine residue essential for geminivirus replication also controls nuclear localization of the Tomato yellow leaf curl virus Rep protein. J. Virol. 93, e01910–e01918. doi: 10.1128/JVI.01910-18
Numata, K., Horii, Y., Oikawa, K., Miyagi, Y., Demura, T., and Ohtani, M. (2018). Library screening of cell-penetrating peptide for BY-2 cells, leaves of Arabidopsis, tobacco, tomato, poplar, and rice callus. Sci. Rep. 8:10966. doi: 10.1038/s41598-018-29298-6
Petre, B., and Kamoun, S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12:e1001801. doi: 10.1371/journal.pbio.1001801
Petre, B., Kopischke, M., Evrard, A., Robatzek, S., and Kamoun, S. (2016). Cell re-entry assays do not support models of pathogen-independent translocation of AvrM and AVR3a effectors into plant cells. Biorxiv [Preprint]. doi: 10.1101/038232
Rafiqi, M., Gan, P. H., Ravensdale, M., Lawrence, G. J., Ellis, J. G., Jones, D. A., et al. (2010). Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22, 2017–2032. doi: 10.1105/tpc.109.072983
Rehmany, A. P., Gordon, A., Rose, L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., et al. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850. doi: 10.1105/tpc.105.031807
Schmidt, N., Mishra, A., Lai, G. H., and Wong, G. C. (2010). Arginine-rich cell-penetrating peptides. FEBS Lett. 584, 1806–1813. doi: 10.1016/j.febslet.2009.11.046
Schornack, S., Van Damme, M., Bozkurt, T. O., Cano, L. M., Smoker, M., Thines, M., et al. (2010). Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. U. S. A. 107, 17421–17426. doi: 10.1073/pnas.1008491107
Segonzac, C., Feike, D., Gimenez-Ibanez, S., Hann, D. R., Zipfel, C., and Rathjen, J. P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 156, 687–699. doi: 10.1104/pp.110.171249
Tameling, W. I. L., Nooijen, C., Ludwig, N., Boter, M., Slootweg, E., Goverse, A., et al. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22, 4176–4194. doi: 10.1105/tpc.110.077461
Tintor, N., Paauw, M., Rep, M., and Takken, F. L. W. (2020). The root-invading pathogen Fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytol. 227, 1479–1492. doi: 10.1111/nph.16618
Uhse, S., and Djamei, A. (2018). Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 14:e1006992. doi: 10.1371/journal.ppat.1006992
Van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H., Kistler, H. C., Ma, L. J., et al. (2016). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18, 4087–4102. doi: 10.1111/1462-2920.13445
van Esse, H. P., Thomma, B. P. H. J., van 't Klooster, J. W., and de Wit, P. J. G. M. (2006). Affinity-tags are removed from Cladosporium fulvum effector proteins expressed in the tomato leaf apoplast. J. Exp. Bot. 57, 599–608. doi: 10.1093/jxb/erj044
Wawra, S., Trusch, F., Matena, A., Apostolakis, K., Linne, U., Zhukov, I., et al. (2017). The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell 29, 1184–1195. doi: 10.1105/tpc.16.00552
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zhou, J. M., and Zhang, Y. (2020). Plant immunity: danger perception and signaling. Cell 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Zhu, M., Jiang, L., Bai, B., Zhao, W., Chen, X., Li, J., et al. (2017). The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 29, 2214–2232. doi: 10.1105/tpc.17.00180
Keywords: PAMP, microbe, effector-triggered immunity, wilt disease, flg22, endocytosis, Nicotiana benthamiana, haustorium
Citation: Tintor N, Nieuweboer GAM, Bakker IAW and Takken FLW (2022) The Intracellularly Acting Effector Foa3 Suppresses Defense Responses When Infiltrated Into the Apoplast. Front. Plant Sci. 13:813181. doi: 10.3389/fpls.2022.813181
Edited by:
Maryam Rafiqi, Mohammed VI Polytechnic University, MoroccoReviewed by:
Miaoying Tian, University of Hawaii at Manoa, United StatesPeter Henry, United States Department of Agriculture, United States
Copyright © 2022 Tintor, Nieuweboer, Bakker and Takken. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frank L. W. Takken, Zi5sLncudGFra2VuQHV2YS5ubA==
†ORCID: Nico Tintor, orcid.org/0000-0001-5368-7668
Gea A. M. Nieuweboer, orcid.org/0000-0001-8062-8938
Ilse A. W. Bakker, orcid.org/0000-0003-2922-7412
Frank L. W. Takken, orcid.org/0000-0003-2655-3108
 Nico Tintor
Nico Tintor Gea A. M. Nieuweboer
Gea A. M. Nieuweboer Ilse A. W. Bakker†
Ilse A. W. Bakker† Frank L. W. Takken
Frank L. W. Takken