- 1Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, China
- 2Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Mengla, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 5Plant Germplasm and Genomics Centre, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 6Department of Biological Sciences, Sam Houston State University, Huntsville, TX, United States
- 7Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, Myanmar
Morphological approaches often fail to delimit species in recently derived species complexes. This can be exacerbated in historical collections which may have lost key features in specimen preparation and preservation. Here, we examine the Pedicularis siphonantha complex, endemic to the Mountains of Southwest China. This complex is characterized by its red/purple/pink and long-tubular corolla, and twisted, beaked galea. However, herbarium specimens are often difficult to identify to species. Molecular approaches using nrITS or nuclear ribosomal internal transcribed spacer (nrITS) + plastid DNA (ptDNA) have been successfully used for species identification in Pedicularis. To resolve taxonomic confusion in the Pedicularis siphonantha complex, we reconstructed phylogenies of the complex using nrITS and four plastid DNA loci (matK, rbcL, trnH-psbA, and trnL-F). To recover as much of the phylogenetic history as possible, we sampled individuals at the population level. Topological incongruence between the nrITS and ptDNA datasets was recovered in clades including two widely distributed species, Pedicularis milliana and Pedicularis tenuituba. Based on morphological, geographical, and genetic evidence, we suggest that hybridization/introgression has occurred between P. milliana and Pedicularis sigmoidea/Pedicularis sp. 1 in the Yulong Snow Mountain of Lijiang, northwest Yunnan, and between P. tenuituba and Pedicularis leptosiphon in Ninglang, northwest Yunnan. After removing conflicting DNA regions in Pedicularis dolichosiphon (nrITS) and P. milliana (ptDNA), the concatenated nrITS and ptDNA phylogenies distinguish 11 species in the P. siphonantha complex, including two undescribed species, from the Jiaozi and Yulong Snow Mountains, respectively. Phylogeographical analyses indicate that the P. siponantha complex originated from south of the Hengduan Mountains, expanding north to the Himalayas and the Yunnan-Guizhou Plateau. Moreover, the uplift of the Qinghai-Tibet Plateau and climate oscillations may have driven further diversification in the complex.
Introduction
The Mountains of Southwest China host one of the richest temperate floras, with a high proportion of endemic species (Boufford, 2014). Mountain uplifts and the monsoon climate create geographically and ecologically isolated habitats, which have driven plant diversification in this region (Hoorn et al., 2013; Xing and Ree, 2017; Ding et al., 2020). Rapid diversification and frequent introgression compound taxonomic confusion, as documented in megadiverse genera of the region, such as Meconopsis Vig. (Papaveraceae), Primula L. (Primulaceae), and Rhododendron L. (Ericaceae) (Zha et al., 2010; Yang et al., 2012; Favre et al., 2016). Species delimitation is traditionally based on morphological characters. However, morphological approaches often fail to delimit recently diverged species, resulting in cryptic species complexes (Bickford et al., 2007; Struck et al., 2018). The study of cryptic species, therefore, offers a window into the diversification and the maintenance of recently divergent species groups.
Pedicularis L. (Orobanchaceae) consists of approximately 600–800 species, of which two-thirds are endemic to the Mountains of Southwest China (Li, 1948, 1949). Pedicularis exhibits dramatic variations in corolla structure and galea form, beak length and shape, and corolla tube length, which are key characters for species delimitation. Four general corolla types are recognized: (A) short-tubular corolla with a beakless, toothless galea (upper lip), (B) short-tubular corolla with a toothed galea, (C) short-tubular corolla with a beaked galea, and (D) long-tubular corolla with a beaked galea (Maximowicz, 1888; Li, 1948, 1949; Tsoong, 1955, 1956, 1963). Long-tubular corollas always bear a beaked galea, which has been considered as a derived corolla type. Ree (2005) and Yu et al. (2015) have demonstrated that long-tubular corollas appear to have been derived from short-tubular corollas several times, resulting in taxonomic confusion. The Pedicularis siphonantha complex includes only long-tubular species with the purple-red corolla and an S-shaped, beaked galea (Figure 1). Pedicularis siphonantha D. Don from the Himalayas was the first described species. To date, at least 11 species of this complex are recognized from the western Himalayas to the Mountains of Southwest China (Li, 1949; Yang et al., 1998; Yu et al., 2015, 2018). The P. siphonantha complex was supported as monophyletic by Yu et al. (2015). However, species within this complex are difficult to distinguish, especially as herbarium specimens, which lose three-dimensional corolla structure and color and often lack field photos and descriptions of key diagnostic characters. This results in uncertainty about the number of species and their geographical distributions. For example, pressed specimens of Pedicularis delavayi Franch. ex Maxim. resemble P. siphonantha, though the three-dimensional structure of the middle lobe of the lower lip and corolla throat color allows easy distinction. Because of the loss of structural and color characteristics in preserved specimens, Tsoong (1963) inferred that the distribution of P. siphonantha extended to the Mountains of Southwest China, and included the species as a variety of P. siphonantha, i.e., P. siphonantha var. delavayi (Franch. ex Maxim.) P. C. Tsoong. In contrast, Li (1949) considered P. delavayi as a separated species. Li (1949) and Tsoong (1963), as well as Yang et al. (1998) and other botanists, misidentified most herbarium specimens of Pedicularis tenuituba H. L. Li and Pedicularis milliana W. B. Yu et al. as P. delavayi (Yu et al., 2018). In addition, the infraspecific taxonomy of P. siphonantha is not fully resolved yet in the Himalaya region (Yu et al., 2018).

Figure 1. Field photos of 11 species of the Pedicularis siphonantha complex and Pedicularis delavayi, (A) Pedicularis tenuituba H. L. Li; (B) Pedicularis leptosiphon H. L. Li; (C) Pedicularis dolichosiphon (Hand.-Mazz.) H. L. Li; (D) P. siphonantha D. Don; (E) Pedicularis sp. 2 from Jiaozi snow mountain; (F) Pedicularis milliana W. B. Yu, D. Z. Li, and H. Wang; (G) Pedicularis sp. 1 from Ganheba, Lijiang; (H) Pedicularis sigmoidea Franch. ex Maxim; (I) Pedicularis variegata H. L. Li; (J) Pedicularis dolichantha Bonati; (K) Pedicularis humilis Bonati. (L) Pedicularis delavayi Franch. ex Maxim.
Molecular approaches have been widely applied for species identification (Hebert et al., 2003; Hebert and Gregory, 2005; Hollingsworth et al., 2016; Kress, 2017). Four candidate DNA barcodes [nuclear ribosomal internal transcribed spacer (nrITS), and three plastid matK, rbcL, and trnH-psbA regions] can be used to discriminate more than 89.0% of Pedicularis species (Yu et al., 2011; Liu et al., 2013). Based on nrITS and four plastid loci (matK, rbcL, trnH-psbA, and trnL-F), no samples of P. delavayi cluster with other species of the P. siphonanta complex. Yu et al. (2018) have reinstated P. delavayi as a separate species and discovered an undescribed species, P. milliana, which was previously misidentified as P. siphonantha var. delavayi or P. delavayi. To date, species delimitation of the P. siphonantha complex is not fully resolved, due to limited sampling and poorly known species distributions (Li, 1949; Yu et al., 2015, 2018). For example, though P. milliana occurs widely in northwest Yunnan and Pedicularis sigmoidea Franch. ex Maxim. occurs in the south margin of P. milliana (Figure 2), specimens (see Figure 1G) from the Yulong Snow Mountain (G1 in Figure 2) bear a remarkable S-shaped beak similar to that of P. sigmoidea. Other specimens (see Figure 1E), from the Jiaozi Snow Mountain (E1-3 in Figure 2), appear to be distinct from known species (Yu et al., 2015, 2018). Morphological ambiguity is supported by topological incongruence between nrITS and plastid gene datasets (Yu et al., 2015), which might be caused by hybridization or introgression.
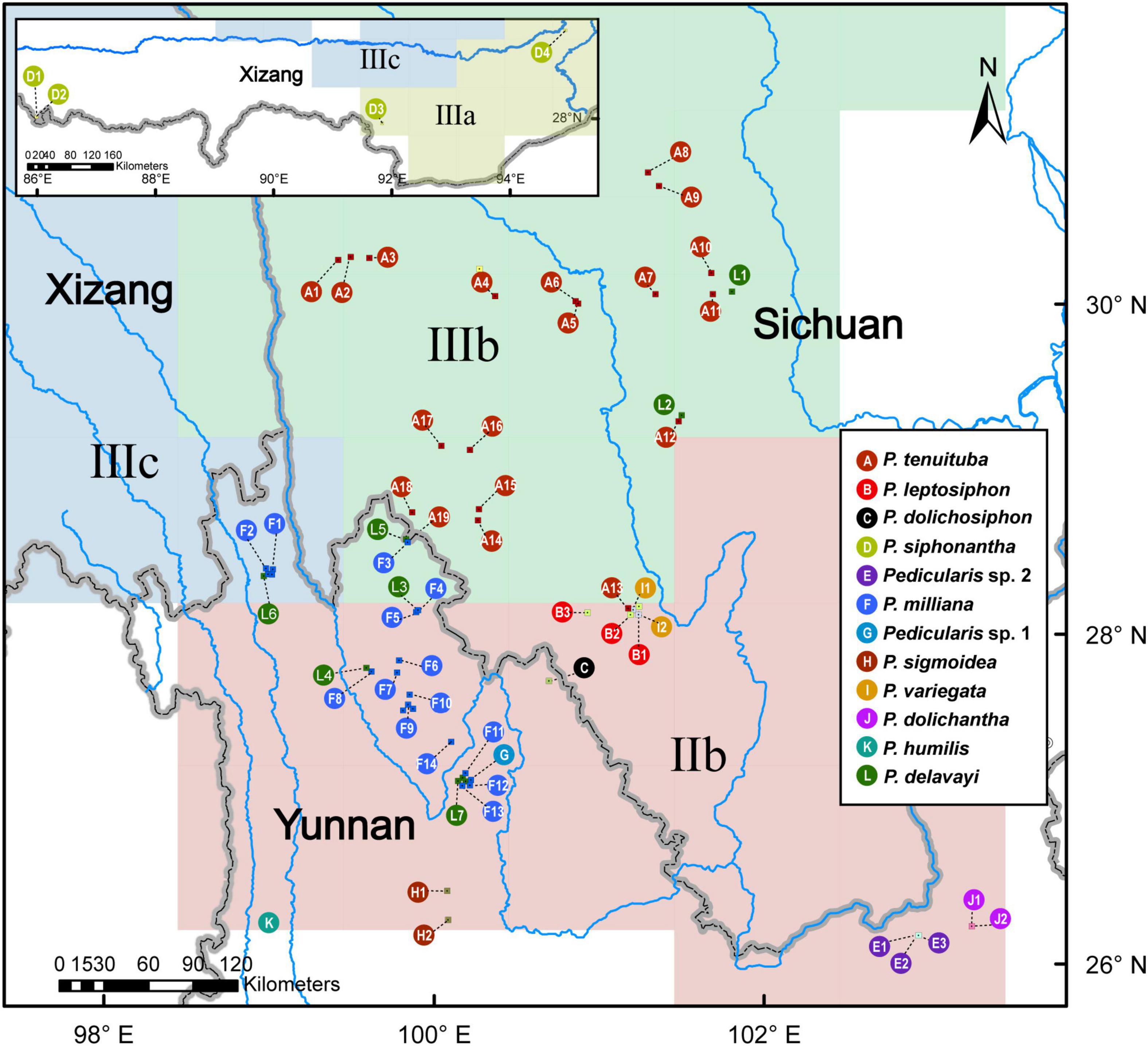
Figure 2. A geographical sampling of the Pedicularis siphonantha complex from the Hengduan Mountains, southwestern China. The colors of circles correspond to different taxa. Numbers and letters indicate populations; these abbreviations are also used in Figures 3, 4, 6. More information regarding collection vouchers can be found in Supplementary Table 1.
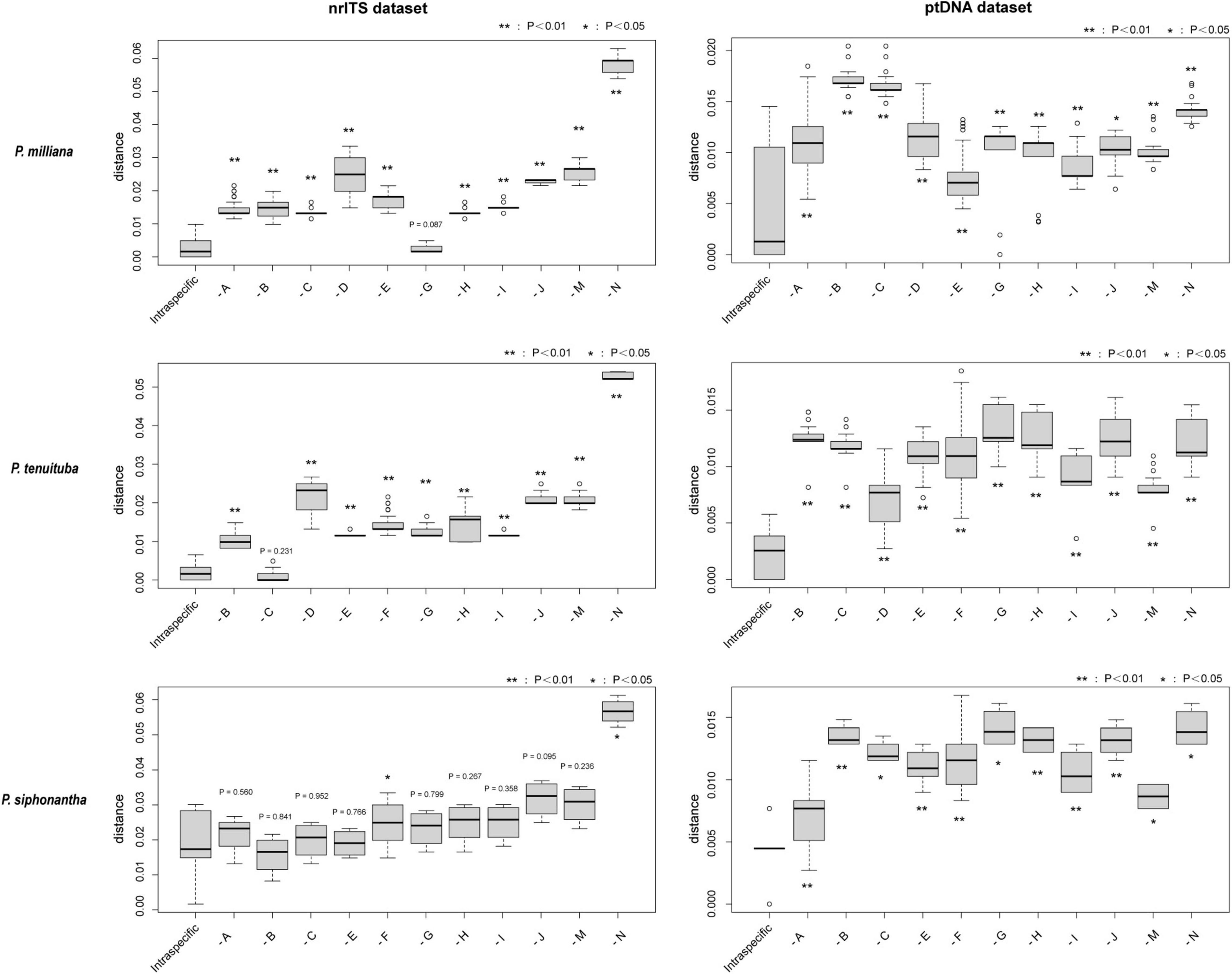
Figure 3. K-S test for intraspecific and interspecific genetic distance. The intraspecific genetic distance of Pedicularis milliana, Pedicularis tenuituba, and Pedicularis siphonantha are estimated. Statistical significance was shown on every group (**P < 0.01, *P < 0.05). -A: to Pedicularis tenuituba; -B: to Pedicularis leptosiphon; -C: to Pedicularis dolichosiphon; -D: to Pedicularis siphonantha; -E: to Pedicularis sp. 2 from Jiaozi Snow Mountain; -F: to Pedicularis milliana; -G: to Pedicularis sp. 1 from Ganheba; -H: to Pedicularis sigmoidea; -I: to Pedicularis variegata; -J: to Pedicularis dolichantha; -M: to Pedicularis amplituba (outgroup); -N: to Pedicularis tahaiensis (outgroup).
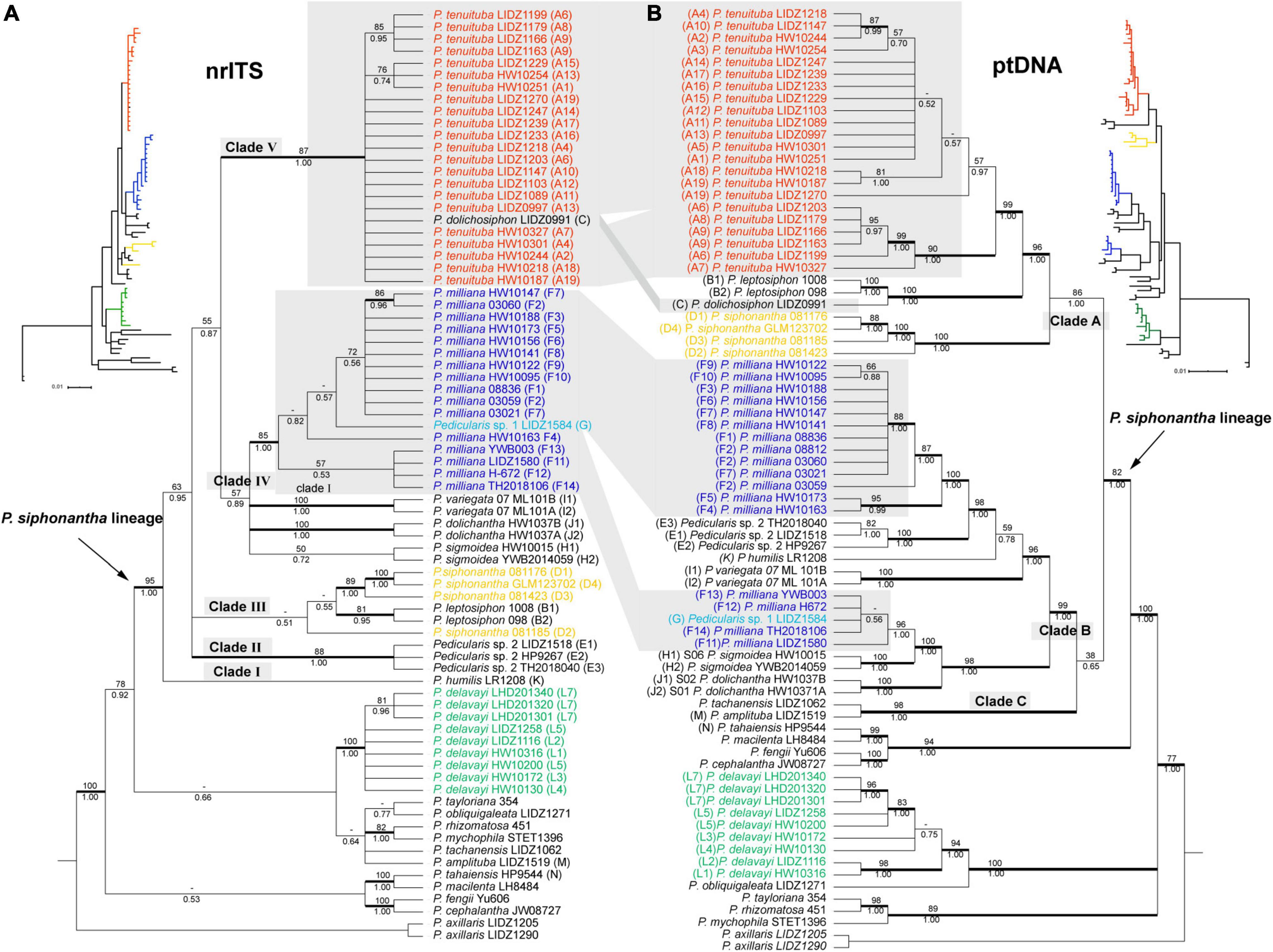
Figure 4. Bayesian Inference (BI) tree comparisons of Pedicularis siphonantha complex inferred from nrITS (A) and the concatenated chloroplast (B) datasets. Phylogenetic incongruence is marked by shadow, Numbers associated with the branches are bootstrap value (BS) and BI posterior probabilities (PP), and thicker lines are indicated as BS ≥ 70 and PP ≥ 0.95. Node with BS < 50 was collapsed.
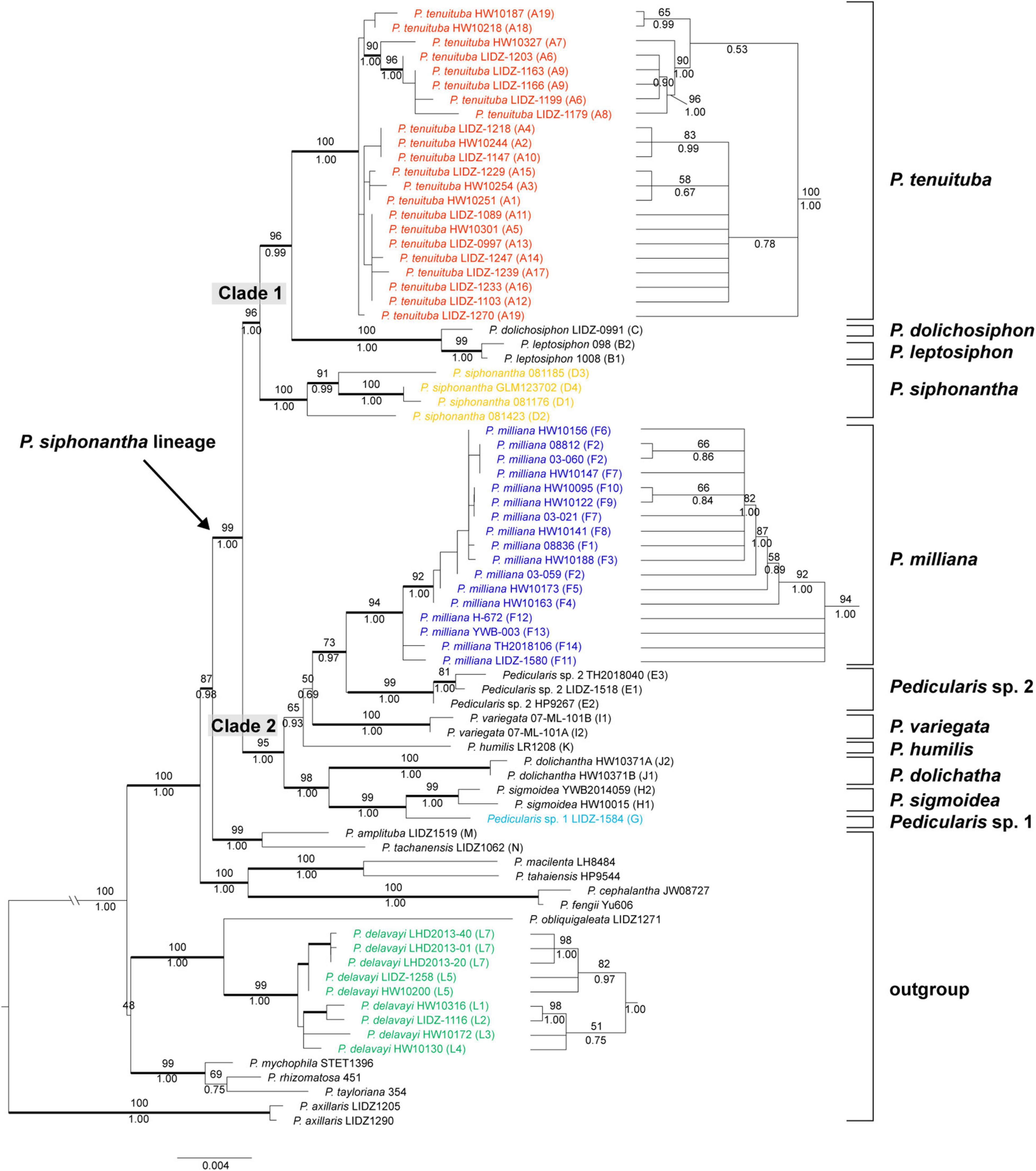
Figure 5. Bayesian inference (BI) tree of the Pedicularis siphonantha complex inferred from the modified nrITS+ptDNA dataset by removing the conflicting sequence nrITS of Pedicularis dolichosiphon (nrITS) and the four ptDNA regions (matK, rbcL, trnH-psbA, and trnL-F) of the samples F11-F14 of Pedicularis milliana in accordance with the topological incongruence between the nrITS and ptDNA phylogenies (see Figure 4 and Table 3). Numbers associated with the branches are ML BS value and BI PP, and thicker lines indicate BS ≥ 70 and PP ≥ 0.95. Node with BS < 50 was collapsed. The topology of the P. milliana clade with short branch lengths appear on the right.
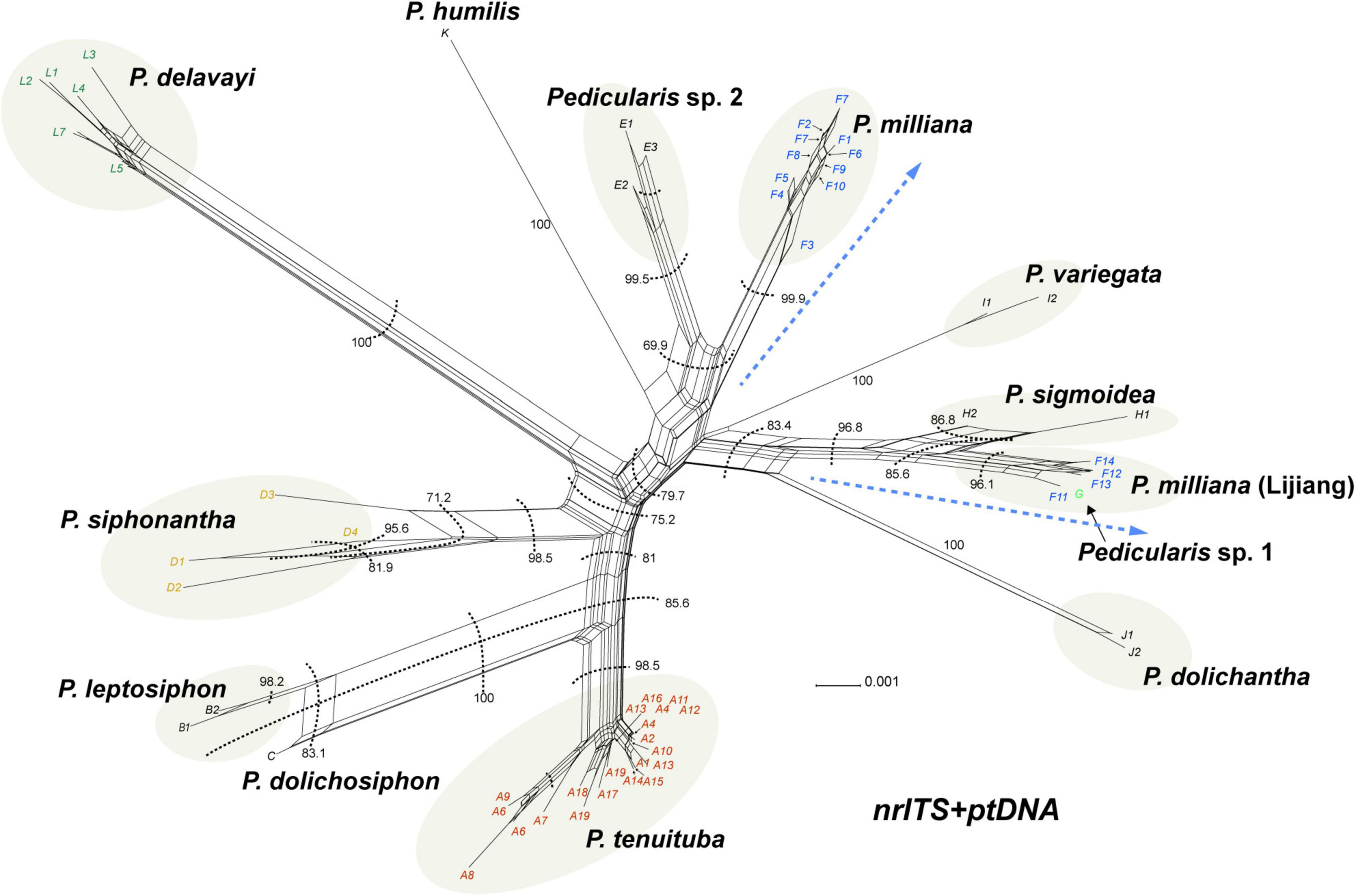
Figure 6. Neighbor-net analysis of Pedicularis siphonantha complex using complete nrITS + ptDNA datasets. Bootstrap support values for clusters are indicated next to the respective cluster delimitation (dashed lines); blue dashed lines indicate the split of Pedicularis milliana.
In this study, we reconstructed a comprehensive phylogeny of the P. siphonantha complex using five DNA loci (nrITS, matK, rbcL, trnH-psbA, and trnL-F) with population-level sampling. Our main goals were to: (1) explore patterns and causes of phylogenetic incongruence between nrITS and plastid DNA datasets in the P. siphonantha complex; (2) revise species delimitations in the P. siphonantha complex; and (3) investigate the causes of species diversification in the P. siphonantha complex.
Materials and Methods
Taxon Sampling
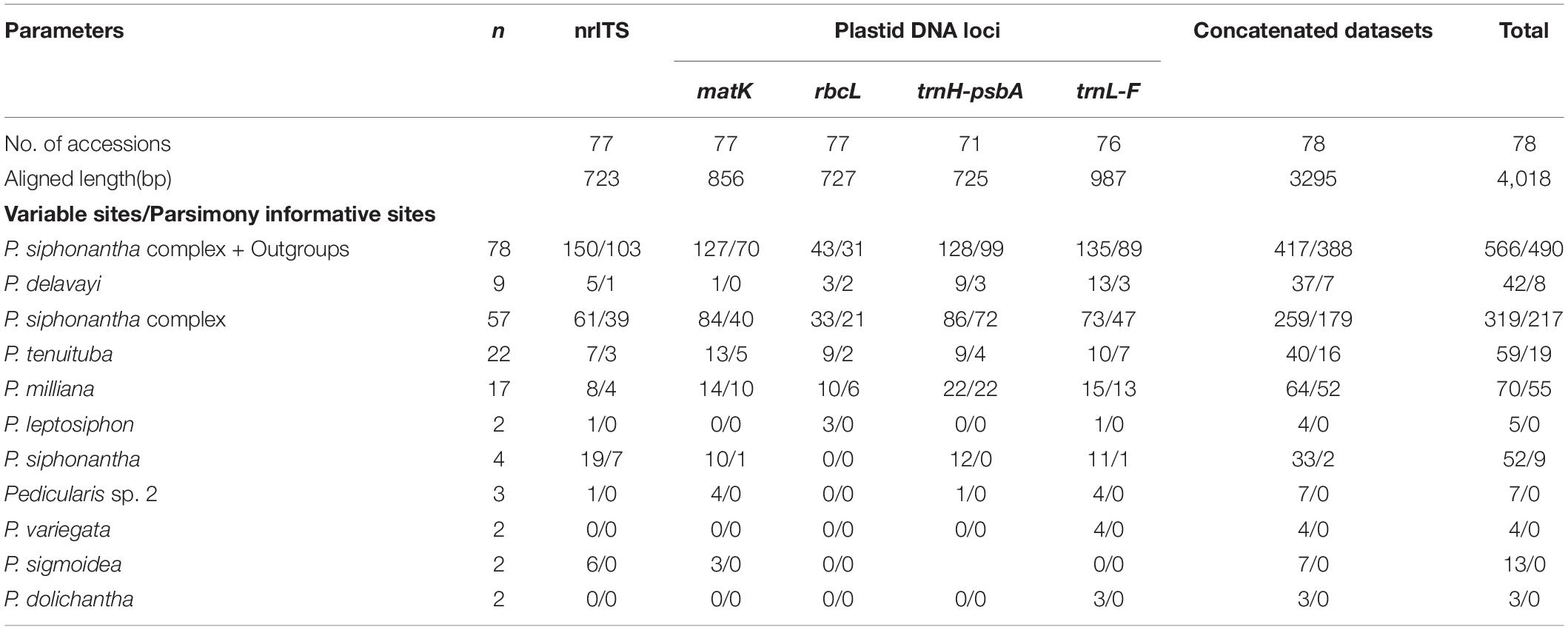
We sampled 78 individuals, mainly from the Hengduan Mountains, as well as the Himalayas and the Yunnan-Guizhou Plateau, representing 11 taxa of the P. siphonantha complex and 12 other Pedicularis species (Supplementary Table 1). The 11 taxa covered nine recognized species of the complex, with the exception of Pedicularis fastigiata Franch., which is known only from the type collection by Orléans H.d’ s.n. (P, barcode P00520823) of the 78 individuals, 50 were newly sampled and sequenced. There were 22 samples (19 populations) of P. tenuituba, widely distributed in western Sichuan, and 17 samples (14 populations) of P. milliana, endemic to the northwestern Yunnan. Three populations of P. milliana (i.e., F11, F12, and F13) were collected from the Yulong Snow Mountain in Lijiang, and population F14 was collected from the Haba Snow Mountain in Shangri-La. In addition, population G (i.e., sample LIDZ1584), collected from Ganheba in the Yulong Snow Mountain represents an unknown taxon, which is similar to P. sigmoidea in the shape of the galea beak but has a smaller corolla. Populations E1–E3, collected from the Jiaozi Snow Mountain, represents another unknown taxon, which is distinguished from P. milliana by its oblate and crested beak. The remaining six taxa of the P. siphonantha complex have narrow distributions, so only a few individuals/populations were included in this study. We included nine samples (seven populations) of P. delavayi from the northwestern Yunnan and western and northern Sichuan, where it overlaps with P. milliana and P. tenuituba. Geographic information for all samples the P. siphonantha complex is shown in Figure 2.
Molecular Methods
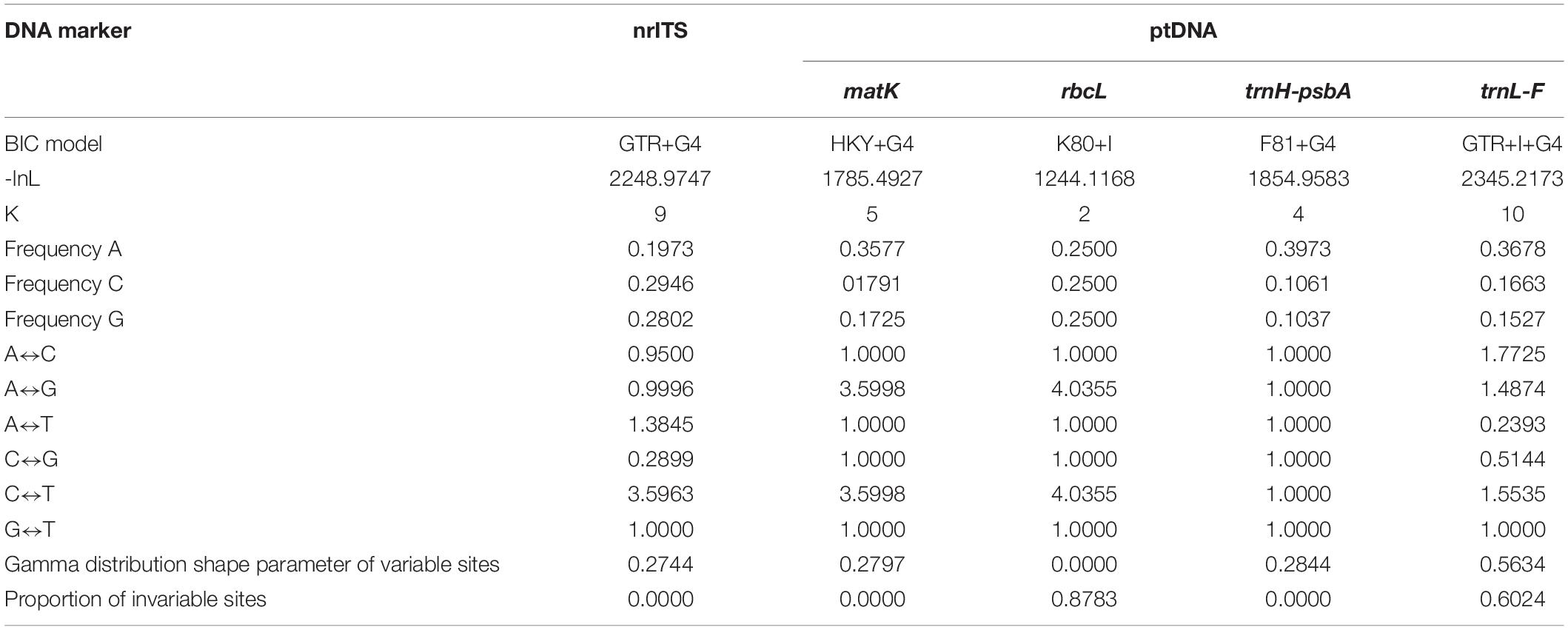
The nrITS and four plastid DNA (ptDNA) markers (matK, rbcL, trnH-psbA, and trnL-F) were amplified and sequenced in this study. Primer information of five DNA markers were presented in previous studies (Yu et al., 2011, 2013, 2018). Genomic DNA of 50 new samples was extracted using a modified CTAB method from silica gel–dried leaves. PCR amplification and sequencing profile followed Yu et al. (2011). Raw sequences were assembled and edited using Geneious 7.1 (Kearse et al., 2012). The nrITS region has multiple copies in the genome. These copies showed evolutionary consistent in the newly sequenced 46 samples, only one sample (P. milliana F2/03-060) had two ambiguous basecalls (i.e., multiple superimposed peaks in chromatograms), and three samples (P. milliana F2/03-059, P. tenuituba A11/HW10187, and P. tenuituba A7/HW10327) had one basecall. The ambiguous site was assigned using IUPAC ambiguity characters. Assembled sequences were aligned using MAFFT 7.4 (Katoh et al., 2019), then adjusted manually using Geneious. Sequence characteristics and Kimura 2-parameter (K-2P) model-based genetic distances among taxa were calculated using MEGA 10.0 (Kumar et al., 2018), and non-parametric two-sample Kolmogorov–Smirnov (K–S) tests of genetic distances between and within species were estimated for widely distributed species P. tenuituba, P. milliana, and P. siphonantha using SPSS 25.0 (IBM, 2017). The nrITS and ptDNA datasets were analyzed separately.
Phylogenetic Analyses
Both Bayesian Inference (BI) and maximum likelihood (ML) were used to reconstruct phylogenetic relationships in the P. siphonantha complex. To explore topological incongruence between nrITS and the concatenated ptDNA phylogenies, the two datasets were analyzed separately. Before concatenating the data sets, we removed sequences that seemed to be the source of conflict. The concatenated datasets (ptDNA and nrITS + ptDNA) were partitioned by gene and the best-fit model was estimated using Modeltest-ng (Darriba et al., 2020) (see Table 1). BI Markov chain Monte Carlo (MCMC) analyses were performed using MrBayes 3.2 (Ronquist et al., 2012) for 2,000,000 generations with two simultaneous runs, each comprising four incrementally heated chains. BI analyses were started with random trees and sampled every 1,000 generations. The first 25% of trees were discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. Posterior probability (PP) values ≥0.95 were considered as well-supported (Alfaro et al., 2003; Kolaczkowski and Thornton, 2007). ML tree search was performed using RAxML version 8.2.12 (Stamatakis, 2014) under GTR + Γ. Node support was evaluated using 1,000 non-parametric bootstrap (BS) replicates. Nodes with BS values ≥70 were considered well-supported (Hillis and Bull, 1993).
To explore patterns of introgression in the complex, we constructed phylogenetic networks of the P. siphonantha complex based on the total dataset by the concatenation of all nrITS and ptDNA sequences by using SplitsTree 4.14.1 (Huson and Bryant, 2006). The Neighbor-net model was performed using the Kimura 2-parameter (K-2P) distance and Ordinary Least Squares Method, with 1,000 BS replicates to estimate split support. Splits with BS ≥ 70 were considered as well-supported.
Topological Conflict Analyses
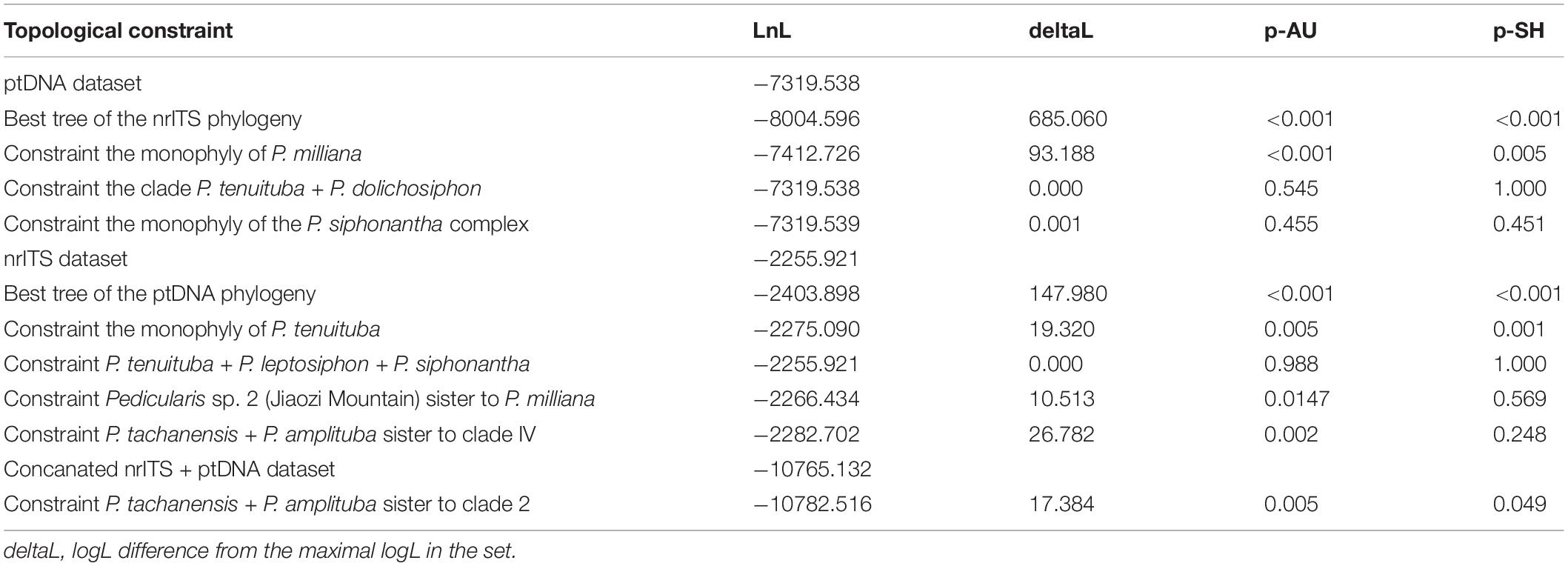
Thresholds of PP ≥ 0.95 and BS ≥ 70 were interpreted as identifying incongruent clades between the nrITS and ptDNA datasets. Based on topological incongruence between the nrITS and ptDNA datasets, the DNA sequence would be considered as heterogeneous one if the phylogenetic cluster was not consistent with the morphological cluster, then the heterogeneous sequence was removed from the concatenated nrITS + ptDNA dataset. Herein, the nrITS sequence of Pedicularis dolichosiphon (Hand.-Mazz.) H. L. Li and the four ptDNA regions (matK, rbcL, trnH-psbA, and trnL-F) of the samples F11–F14 of P. milliana were identified as heterogeneous sequences, so that those sequences were removed from the concatenated nrITS + ptDNA dataset. Then, the concatenated nrITS + ptDNA phylogeny was performed using the same methods as nrITS and the concatenated ptDNA phylogenetic analyses (see above). Additionally, the Shimodaira–Hasegawa (SH) test (Shimodaira and Hasegawa, 1999) and the approximately unbiased (AU) test (Shimodaira, 2002) were used to estimate the degree of topological incongruence among the three datasets (nrITS, ptDNA, and modified nrITS + ptDNA). Constraint trees were constructed in Mesquite version 3.6 (Maddison and Maddison, 2019), and the SH and AU tests were performed using IQ-Tree 1.6 (Lam-Tung et al., 2015).
Phylogeographical Analyses
Ancestral geographical distributions were inferred using BEAST 2.6.3 (Bouckaert et al., 2014). The concatenated nrITS and ptDNA dataset, including all samples of the P. siphonantha complex and two outgroups Pedicularis amplituba H. L. Li and Pedicularis tachanensis Bonati, was imported into BEAUti with “beast-classic package” (Lemey et al., 2009). Samples were assigned to one of four subregions of the Sino-Himalayan flora following Liu et al. (2021). Taking into account the center of endemism identified by Zhang et al. (2016), two samples of Pedicularis siphonantha (D1, D2) collected from Yadong country in the middle Himalaya were assigned to subregion IIIa. The sites model was calibrated using the “bModeltest package” (Bouckaert and Drummond, 2017), the molecular clock model was set to “Relaxed Clock Log Normal,” and the Yule model served as the tree prior. The divergence time of the most recent ancestor between the P. siphonantha complex and the two outgroups was constrained to 9.1 ± 2 Mya. A second calibration point was obtained from the analysis of Yu et al. (2015) (Supplementary Figure 1). MCMC chains were run for 10,000,000 generations, with parameter values and trees sampled every 1,000 generations. Effective sample size (ESS > 200) was assessed using Tracer 1.7 (Rambaut et al., 2018). After discarding 25% of the initial trees as burn-in, the maximum clade credibility (MCC) tree with mean ages and 95% highest posterior density (HPD) intervals on nodes was reconstructed using TreeAnnotator 2.6.3 (Bouckaert et al., 2014).
Results
Matrix Characteristics
Matrix characteristics of nrITS, four plastid DNA, and the concatenated ptDNA datasets are shown in Table 2. Similar to previous studies (Yu et al., 2011, 2013, 2015, 2018), these five loci included an adequate numbers of variable sites and parsimony-informative sites for subsequent phylogenetic analyses in the P. siphonantha complex. The nrITS dataset is the most informative, followed by two plastid intergenic spacer datasets (trnH-psbA and trnL-F). The two protein-coding genes (rbcL and matK) are less informative than the three spacer datasets. Sequences of P. siphonantha and P. milliana show the highest variation at the species level for both nrITS and ptDNA datasets.
Genetic Distance Estimation
Comparisons of genetic distances within and between species using the K–S test are shown in Figure 3. The intraspecific genetic distances among P. milliana sequences were significantly smaller (P < 0.05) than the distance between P. milliana and any other taxon, with one exception; nrITS sequences of P. milliana and Pedicularis sp. 1 did not differ significantly. The same pattern holds for P. tenuituba, with the exception being a non-significant distance with nrITS sequences P. dolichosiphon. While the ptDNA distances within P. siphonantha were significantly less than the distance between P. siphonantha and ptDNA of any other taxon, distances of nrITS sequences within P. siphonantha were significantly smaller than the distance between P. milliana and Pedicularis tahaiensis Bonati (P < 0.05).
Phylogenetic Analyses of the nrITS Dataset
Maximum likelihood and BI analyses obtained identical topologies for the nrITS dataset (Figure 4A). The P. siphonantha complex was recovered as monophyletic (BS/PP = 95/1.00), and P. delavayi was not included in that clade. Within the P. siphonantha complex, five major clades were recovered. Clade I had only Pedicularis humilis Bonati, and it was moderately supported as sister to the remaining four clades (BS/PP = 63/0.95). The relationships among Clades II–V were not well-resolved. Clade II consisted of three individuals of Pedicularis sp. 2, and clade III included two species, Pedicularis leptosiphon H. L. Li and P. siphonantha, with P. leptosiphon nested within P. siphonantha, although with weak support. Clades IV and V were weakly supported as sister lineages (BS/PP = 55/0.87). Clade IV consisted of five species, Pedicularis dolichantha Bonati, P. milliana, P. sigmoidea, Pedicularis variegata H. L. Li, and Pedicularis sp. 1 (G). In this clade, Pedicularis sp. 1 (G) was nested within P. milliana. Clade V consisted of P. dolichosiphon nested within 22 samples of P. tenuituba.
Phylogenetic Analyses of the ptDNA Dataset
Maximum likelihood and BI obtained identical topologies from the ptDNA dataset (Figure 4B). Unlike the nrITS phylogenies, the P. siphonantha complex was not recovered as monophyletic, including P. amplituba + P. tachanensis not considered here to be members of the complex. The core species of the P. siphonantha complex were split into two major clades (i.e., Clades A and B + C), with Clade C (P. amplituba and P. tachanensis) weakly supported as sister to the remaining members of Clade B (BS/PP = 38/0.65). Clade A included four species, P. dolichosiphon, P. leptosiphon, P. siphonantha, and P. tenuituba, which were strongly supported as monophyletic (BS/PP ≥ 99/1.00). Within Clade A, P. siphonantha was sister to the remaining three species with P. dolichosiphon and P. leptosiphon forming a clade sister to P. tenuituba.
Clade B contained seven species, which were split into two subclades. One subclade included P. dolichantha, P. sigmoidea, Pedicularis sp. 1, and four samples of P. milliana from the Yunlong Snow Mountain and the Haba Snow Mountain. In this subclade, P. dolichantha was sister to the remaining species, with P. sigmoidea as the sister to a clade including Pedicularis sp. 1 nested within P. milliana. The other subclade included P. humilis, P. variegata, Pedicularis sp. 2, and 13 samples of P. milliana, which were all supported as monophyletic. Within this subclade, P. variegata was sister to the remaining species, with P. humulis arising as sister to a clade including Pedicularis sp. 2 + P. milliana.
Topological Conflicts Between the nrITS and ptDNA Phylogenies
NrITS and ptDNA phylogenies are incongruent in the placement of P. milliana, P. tenuituba, and their relatives. In the nrITS phylogeny, 17 samples of P. milliana were only monophyletic if they included Pedicularis sp. 1 (BS/BP = 85/1.00). In the ptDNA phylogeny, they were separated into two distant clades, i.e., four samples (F11–F14) formed a clade by including Pedicularis sp. 1 (BS/BP = 100/1.00) as sister to P. sigmoidea, and the remaining 13 samples were monophyletic (BS/BP = 96/1.00) as sister to Pedicularis sp. 2. Twenty-four samples of P. tenuituba are strongly supported as monophyletic (BS/BP = 99/1.00) in the ptDNA phylogeny, but they are made paraphyletic by the inclusion of P. dolichosiphon in the nrITS phylogeny. Therefore, the four ptDNA regions of P. milliana samples F11–F14 and the nrITS sequence of P. dolichosiphon were identified as heterogeneous sequences. In addition, the nrITS dataset supports monophyly of the P. siphonantha complex (BS/BP = 95/100), but the ptDNA dataset does not, by including two short-tubular species P. amplituba and P. tachanensis.
Results of the AU and SH test for alternative hypotheses are summarized in Table 3. If phylogeny is constrained by the ptDNA dataset, SH and AU tests rejected (P < 0.01) the best tree topology of the nrITS dataset and monophyly of P. milliana. These tests failed to reject the monophyly of P. tenuituba + P. dolichosiphon and monophyly of the P. siphonantha complex. Meanwhile, when constrained by the nrITS dataset, SH and AU tests failed to reject (P > 0.05) the monophyly of P. tenuituba + P. leptosiphon + P. siphonantha, and the monophyly of P. milliana + Pedicularis sp. 2, but rejected the best tree topology of the ptDNA dataset, and the short tubular P. tachanensis + P. amplituba clade sister to the Clade B. Moreover, for the modified nrITS + ptDNA dataset, the null hypothesis that short tubular P. tachanensis + P. amplituba is sister to Clade 2 was rejected (P < 0.05).
Phylogenetic Analyses of the Concatenated nrITS + ptDNA Dataset
After removing conflicting sequences, P. dolichosiphon (nrITS) and P. milliana (ptDNA regions of F11–F14), ML and BI analyses produced nearly the same topology (Figure 5). Within the strongly monophyletic (BS/BP = 99/1.00) P. siphonantha complex, there were two major clades. Clade 1 (BS/BP = 96/1.00) included P. dolichosiphon, P. leptosiphon, P. siphonantha, and P. tenuituba. In this clade, P. siphonantha was sister to the remaining taxa, with the monophyletic P. tenuituba (BS/BP = 100/1.00) sister to a clade including P. dolichosiphon + P. leptosiphon (BS/BP = 100/1.00). Clade 2 (BS/BP = 95/1.00) included six taxa forming two subclades. In the larger subclade, P. milliana was monophyletic (BS/BP = 94/1.00), and sister to Pedicularis sp. 2 (BS/BP = 99/1.00). A clade including P. milliana + Pedicularis sp. 2 + P. variegata was sister to P. humilis (BS/BP = 65/0.93). In the other subclade, P. dolichantha was sister to P. sigmoidea + Pedicularis sp. 1 (BS/BP = 99/1.00).
Phylogenetic Network of the Pedicularis siphonantha Complex
The phylogenetic network of the concatenated nrITS and ptDNA dataset showed P. milliana split into two clusters (Figure 6), identical to those recovered in the ptDNA topology. Four samples from the Yulong Snow Mountain and the Haba Snow Mountain were nested with Pedicularis sp. 1 as sister to P. sigmoidea (BS = 96.8), and the other 13 samples were monophyletic as sister to Pedicularis sp. 2. In addition, P. dolichosiphon was resolved as either sister to P. leptosiphon (BS = 100) or P. tenuituba (BS = 85.6).
Phylogeographical Analyses of the Pedicularis siphonantha Complex
Phylogeographical analysis indicated that the most common ancestor of the P. siphonantha complex diverged from other Pedicularis in the late Miocene (6.04Mya–10.38Mya), south of the Hengduan Mountains (IIb), which harbors nine of eleven species/taxa of this complex (Figure 7). Species diversification of this complex mainly occurred in the Pliocene (2.48Mya–5.3Mya). After the initial divergence of Clades 1 and 2, members of Clade 1 migrated to the north and west, with Clade 2 diversifying in situ. In the Clade 1, P. siphonantha diverged and diversified in the Himalayan region (IIIa), P. leptosiphon and P. dolichosiphon diverged in situ (IIb), and P. tenuituba diverged north of the Hengduan Mountains. In Clade 2, seven species/taxa diverged in situ (IIb), with some populations of P. milliana migrating northward to IIIb and IIIc.
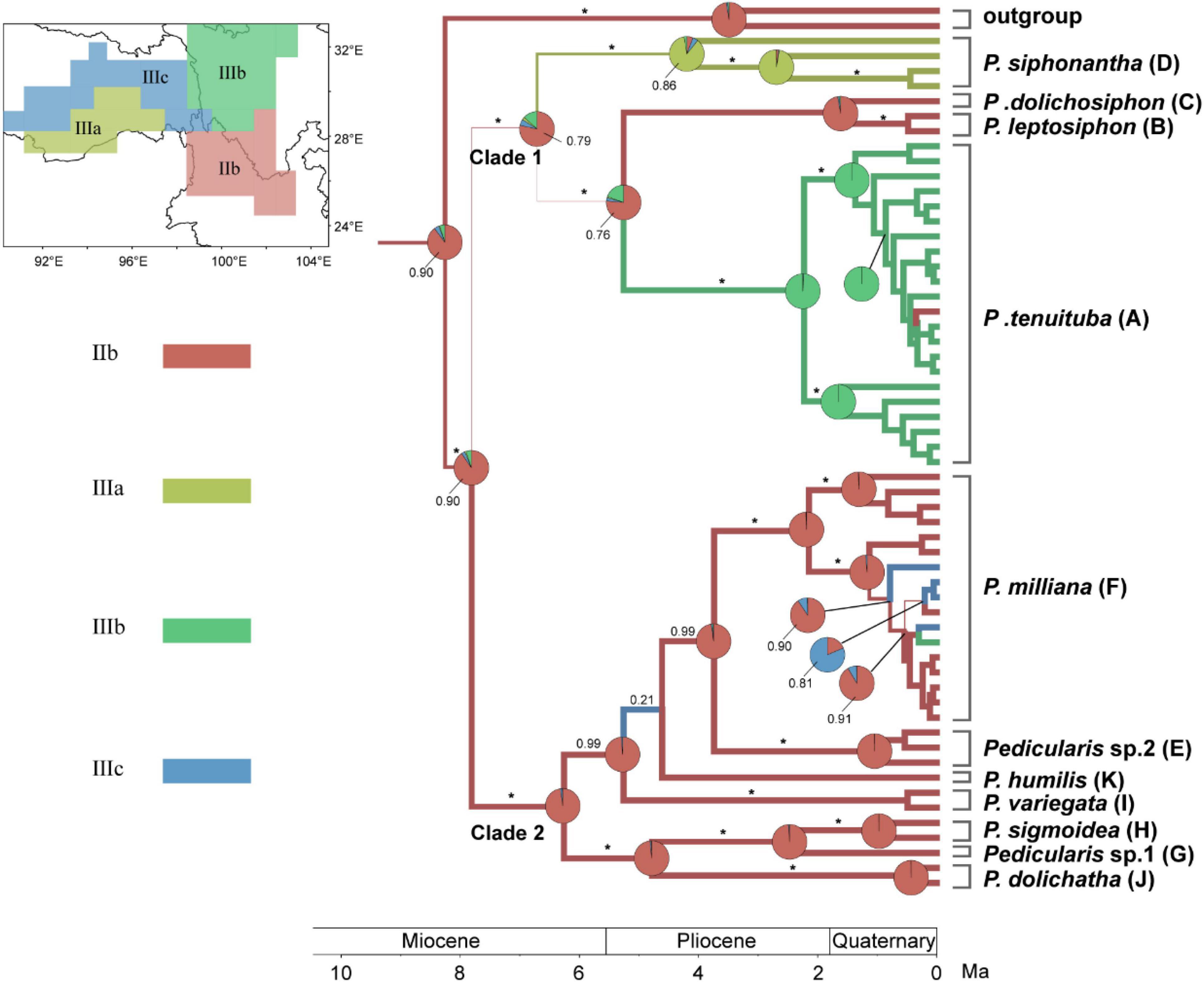
Figure 7. Bayesian phylogeographical reconstruction of ancestral area of the Pedicularis siphonantha complex in the Sino-Himalayan region using the modified nrITS + ptDNA dataset (see above). Pie charts and thickness of branch represent the marginal probabilities for potential ancestral areas with each subregion (Liu et al., 2021), represented by a different color. BI PPs are above branches (* = 1.00).
Discussion
Topological Incongruence Between the nrITS and ptDNA Phylogenies
Topological incongruence between nuclear/nrITS and ptDNA phylogenies has been reported in many taxa (Rieseberg et al., 1996; Buckley et al., 2006; Stegemann et al., 2012; Yi et al., 2015; Stull et al., 2020; Ye et al., 2021). In this study, incongruences between the nrITS and ptDNA datasets were found among species within the P. siphonantha complex and in the sister relationship between the P. siphonantha complex and P. amplituba + P. tachanensis in phylogenetic analyses, as well as the estimation of genetic distance. The incongruence could be caused by convergent sequence evolution, incomplete lineage sorting, hybridization/introgression, horizontal gene transfer, and gene duplication/loss (Rokas et al., 2003; Degnan and Rosenberg, 2009). Phylogenetic network analyses suggested that introgression between P. milliana and P. sigmoidea and between P. leptosiphon and P. tunuituba is the most plausible explanation for discordance. However, incomplete lineage sorting, convergent sequence evolution, or others cannot be ruled out (Joly et al., 2009).
Introgression might have been common within recently derived species complexes when their distributions overlap (Acosta and Premoli, 2010; Liu et al., 2017; Liu et al., 2020). Pedicularis spp. are outcrossed and exclusively pollinated by bumblebees, and pollinator-mediated interspecific gene flow may cause hybridization and introgression among Pedicularis species in the same community (Hong and Li, 2005; Yang et al., 2007; Eaton et al., 2012). In the case of Pedicularis sect. Cyathophora, Yu et al. (2013) have documented that the plastid genome of P. cyathophylloides was likely captured from an ancestor of P. cyathophylla in the West Sichuan. Similarly, our results show that paraphyletic P. milliana populations were associated with distinct geographical ranges, suggesting that either genetic divergence occurred between two clusters due to allopatry and/or a plastid genome capture event. We, therefore, propose an ancient hybridization event between the ancestors of P. milliana (♀) and P. sigmoidea/Pedicularis sp. 1 (♂) in the Yulong Snow Mountain. In this scenario, high-altitude (≥3,800 m) hybrids (♀) backcrossed with ancestors of P. milliana (♀), and low-altitude (<3,800 m) hybrids became established as species at lower altitudes. Therefore, morphological consistency was found among high-altitude populations of P. milliana, while low altitude Pedicularis sp. 1 diverged from P. milliana in the shape of the beak and low lip of corolla. Sample C of P. dolichosiphon might also be the result of introgression between P. tenuituba (♀) and P. leptosiphon (♂), but greater population-level sampling is required to investigate this fully. In addition, more genomic evidence from organelle and nuclear genomes were needed to test these speculations.
Phylogenetic Species Delimitation in the Pedicularis siphonantha Complex
Traditionally, the P. siphonantha complex together with other long-tubular species belonged to Ser. Longiflorae (Li, 1949; Tsoong, 1956). Phylogenetic analyses showed that Ser. Longiflorae is polyphyletic, but the P. siphonantha complex was monophyletic (Yu et al., 2015, 2018). In this study, ptDNA phylogenies rejected the monophyly of the P. siphonantha complex by including two short-tubular species, P. amplituba and P. tachanensis. The AU and SH tests also could not reject the inclusion of P. tachanensis + P. amplituba in the P. siphonantha complex using the nrITS dataset or the monophyly of the P. siphonantha complex using the ptDNA dataset. Moreover, phylogenies of the concatenated nrITS + ptDNA dataset strongly supported the monophyly of the P. siphonantha complex (BS/BP = 99/1.00). Therefore, the monophyly of the P. siphonantha complex should be accepted, but more nuclear genes and more robust phylogeny should be applied for evaluating this complex in the future.
Although introgression may confound phylogenetic species delimitation of P. dolichosiphon and Pedicularis sp. 1, species delimitations of the remaining nine species are well-resolved, which is consistent with morphological identification. For Pedicularis sp. 2, the corolla beak shape and lower-lip lobes are quite different than its sister species, P. milliana. Moreover, Pedicularis sp. 2 occurs on the Jiaozi Snow Mountain. Morphological, molecular, and biogeographic evidence all support Pedicularis sp. 2 to be a new species. It is worth noting that relatively high intraspecies genetic and phenotypic variations suggest P. siphonantha in the Himalayas needs further investigations [also reviewed by Yu et al. (2018)]. The phylogenetic position of P. humilis is still not well-resolved; however, it is an isolated species of the Gaoligong Mountain, perhaps the result of allopatric speciation.
Traditionally, morphological character similarity was the main evidence for assessing species relationships, but this criterion might be not suitable in the P. siphonantha complex. Because floral characters are labile in Pedicularis, morphologically similar species might be only distantly related. For example, phylogenetic analyses showed that P. milliana was clustered with morphologically different species including P. variegata, P. humilis, P. dolichantha, and P. sigmoidea, rather than the morphologically similar species P. siphonantha, contra previous placements [e.g., Li (1949); Tsoong (1963), and Yang et al. (1998)]. Moreover, the long-tubular species P. delavayi (excluded from the P. siphonantha complex) was clustered with short-tubular species Pedicularis obiquigaleata W. B. Yu and H. Wang. Understanding taxonomic affinities within the P. siphonantha complex requires morphological, geographical, and molecular evidence.
Allopatric Speciation in the Pedicularis siphonantha Complex
Species diversification of Pedicularis in the Mountains of Southwest China is thought to be associated with the uplift of the Qinghai-Tibet Plateau and the establishment of the Asian monsoon climatic cycle (Ding et al., 2020). This rapid diversification resulted in dramatic variation in the form and shape of the corolla (Eaton et al., 2012), though the reasons for several independent transitions of the corolla tube from short to long remain unknown (Huang and Fenster, 2007; Yu et al., 2015, 2018; Huang et al., 2016). Macior and his colleagues (Macior, 1990; Macior and Tang, 1997; Macior et al., 2001) proposed that long-tubular corollas might have some adaptative advantages in the alpine meadow by extending the reproductive organs to attract bumblebee pollinators. The P. siphonantha complex, having long-tubular corollas, could be a good model for investigating species diversification in the Mountains of Southwest China. Phylogeographical analysis suggested that ancestors of the P. siphonantha complex originated from south of the Hengduan Mountains in the late Miocene, then rapidly expanded westward to the Himalayas, northward of the Hengduan Mountains, and eastward to the Yunnan-Guizhou plateau. Intense orogeny of southern Hengduan Mountains during the late Miocene and Pliocene (Lai et al., 2007; Wang et al., 2012; Favre et al., 2015; Zhang et al., 2017; Farnsworth et al., 2019) likely contributed to environmental heterogeneity driving rapid species divergence in the P. siphonantha complex. Therefore, P. sigmoidea, P. dolichantha, and Pedicularis sp. 2 are restricted to the margin of the Hengduan Mountains, while P. milliana and P. tenuituba in the heartland of the Hengduan Mountains, as well as P. siphonantha in the Himalayas, are widely distributed in the vast contiguous alpine meadows of those ranges.
Phylogeny of the modified nrITS + ptDNA dataset resolved the P. siphonantha complex as two major clades. In Clade 1, P. siphonantha is distributed in the western Himalaya (the type specimen was collected from Nepal), rather than widely distributed in the eastern Himalaya, Sichuan, and Yunnan, as described in the Flora of China (Yang et al., 1998); P. leptosiphon and P. dolichosiphon are restricted to Ninglang, Yunnan, and Muli, Sichuan; and P. tenuituba is widely distributed in western Sichuan, and partly overlaps with P. leptosiphon in southwestern Sichuan. Geographic separation may have driven P. siphonantha to diverge from the remaining species in this clade. Moreover, the carmine speckles on the lower lobes of P. tenuituba may appear distinct from P. leptosiphon to bumblebee pollinators where they both occur. It is also worth noting that P. tenuituba mainly grows in humid grasslands, but P. leptosiphon prefers to grow in the sandy, dry meadows. Niche specification and difference in pollinators may mediate reproductive isolation between P. tenuituba and P. leptosiphon. However, occasional hybridization might have been responsible for producing the suspected hybrid P. dolichosiphon.
The mountainous terrain of the home range of the P. siphonantha complex likely maintains genetic isolation among geographically isolated species. For example, P. dolichantha and its sister species P. sigmoidea are distributed in isolated mountains near Huize and Eryuan, Yunnan, while species closely related to them, P. humilis and P. variegata, occur in the Gaoligong Mountains and southwest Yunnan and Muli, Sichuan, respectively. P. milliana is widely distributed in northwestern Yunnan, and its sister Pedicularis sp. 2 (Jiaozi Snow Mountain) is only found in the Jiaozi Snow Mountain, Dongchuan, Yunnan. Of the seven species in this clade, P. milliana and Pedicularis sp. 1 co-occur in the Yulong Snow Mountain and P. sigmoidea has been collected from the south margin of the distribution range of P. milliana in Heqing, Eryuan, and Dali. Therefore, geographical isolation likely drove species divergence in this complex, with the exception that introgression between P. milliana and P. sigmoidea may have produced the suspected hybrid Pedicularis sp. 1 and the plastome capture of P. milliana from P. sigmoidea in Lijiang and south Shangri-La, northwest Yunnan.
Conclusion
Overall, phylogenetic analyses of five DNA loci (nrITS, matK, rbcL, trnH-psbA, and trnL-F) clarify species delimitation within the P. siphonantha complex. Differences in geographical distribution and altitude can be important supplementary indicators to identify species of the P. siphonantha complex despite the lack of diagnostic morphological characters in herbarium specimens. The P. siphonantha complex likely originated from allopatric speciation. The origin of P. milliana and Pedicularis sp. 1 in the Lijiang region was plausibly due to an ancestral hybridization event. The morphological, molecular, and biogeographic evidence support taxonomic recognition of Pedicularis sp. 2. To better understand the evolution of the P. siphonantha complex, further studies of phenotype and environmental factors are needed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
W-BY, HW, and D-ZL conceived the study. W-BY, HW, J-BY, and RC collected the data. RL and W-BY analyzed the data. RL, W-BY, CR, and D-ZL interpreted the results. All authors wrote and revised the article and approved the final version of the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31870196 and 32071670), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000), and the Ten Thousand Talents Program of Yunnan for Top-notch Young Talents.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer C-LX declared a shared affiliation, with no collaboration, with several of the authors HW, J-BY, and D-ZL to the handling editor at the time of the review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Jie Cai, LiNa Dong, Lian-Ming Gao, Hua-Jie He, Wei Jiang, Rong Li, Bin Liu, En-De Liu, Jie Liu, Min-Lu Liu, Lu Lu, Yang Luo, Hui Tang, Chun-Lei Xiang, Ji-Dong Ya, Qiu-Lin Yang, Xiu-Long Yang, and Shu-Dong Zhang for their help in the field work and/or providing plant samples, and to Jing Yang and Zhi-Rong Zhang for their help and suggestions in the lab work, and to the physical support from the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, and the HPC Platform of the Public Technology Service Center, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.811206/full#supplementary-material
Supplementary Figure 1 | Maximum clade credibility tree of Pedicularis from BEAST divergence time analysis. The secondary calibration of the Orobanchaceae crown was constrained to 56 ± 10 Mya which was obtained from http://timetree.org/. The estimated age of nodes is presented above the branch. Node bars represent the 95% highest posterior density (HPD) interval.
References
Acosta, M. C., and Premoli, A. C. (2010). Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenet. Evol. 54, 235–242. doi: 10.1016/j.ympev.2009.08.008
Alfaro, M. E., Zoller, S., and Lutzoni, F. (2003). Bayes or bootstrap? a simulation study comparing the performance of bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 20, 255–266. doi: 10.1093/molbev/msg028
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/j.tree.2006.11.004
Bouckaert, R. R., and Drummond, A. J. (2017). bModelTest: bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 17:42. doi: 10.1186/s12862-017-0890-6
Bouckaert, R., Heled, J., Kuehnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comp. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Boufford, D. E. (2014). Biodiversity hotspot: china’s hengduan mountains. Arnoldia (Jamaica Plain) 72, 24–35.
Buckley, T. R., Cordeiro, M., Marshall, D. C., and Simon, C. (2006). Differentiating between hypotheses of lineage sorting and introgression in New Zealand alpine cicadas (Maoricicada Dugdale). Syst. Biol. 55, 411–425. doi: 10.1080/10635150600697283
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi: 10.1093/molbev/msz189
Degnan, J. H., and Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. doi: 10.1016/j.tree.2009.01.009
Ding, W.-N., Ree, R. H., Spicer, R. A., and Xing, Y.-W. (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578–581. doi: 10.1126/science.abb4484
Don, D., Hamilton, F., and Wallich, N. (1825). Prodromus Florae Nepalensis: Sive Enumeratio Vegetabilium Quae In Itinere Per Nepaliam Proprie Dictam Et Regiones Conterminas, Ann. 1802-1803. Londini: J. Gale.
Eaton, D. A. R., Fenster, C. B., Hereford, J., Huang, S.-Q., and Ree, R. H. (2012). Floral diversity and community structure in Pedicularis (Orobanchaceae). Ecology 93, S182–S194. doi: 10.1890/11-0501.1
Farnsworth, A., Lunt, D. J., Robinson, S. A., Valdes, P. J., Roberts, W. H. G., Clift, P. D., et al. (2019). Past East Asian monsoon evolution controlled by paleogeography, not CO2. Sci. Adv. 5:eaax1697. doi: 10.1126/sciadv.aax1697
Favre, A., Michalak, I., Chen, C. H., Wang, J. C., Pringle, J. S., Matuszak, S., et al. (2016). Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J. Biogeogr. 43, 1967–1978. doi: 10.1111/jbi.12840
Favre, A., Paeckert, M., Pauls, S. U., Jaehnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the qinghai-tibetan plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253. doi: 10.1111/brv.12107
Hebert, P. D. N., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hebert, P. D. N., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Roy. Soc. Lond. B. Biol. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Hollingsworth, P. M., Li, D. Z., Van Der Bank, M., and Twyford, A. D. (2016). Telling plant species apart with DNA: from barcodes to genomes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 371:20150338. doi: 10.1098/rstb.2015.0338
Hong, W., and Li, D. Z. (2005). Pollination biology of four Pedicularis species (Scrophulariaceae) in northwestern Yunnan, China. Ann. Mo. Bot. Gard. 92, 127–138.
Hoorn, C., Mosbrugger, V., Mulch, A., and Antonelli, A. (2013). Biodiversity from mountain building. Nat. Geosci. 6, 154–154. doi: 10.1038/ngeo1742
Huang, S.-Q., and Fenster, C. B. (2007). Absence of long-proboscid pollinators for long-corolla-tubed Himalayan Pedicularis species: implications for the evolution of corolla length. Int. J. Plant Sci. 168, 325–331. doi: 10.1086/510209
Huang, S.-Q., Wang, X.-P., and Sun, S.-G. (2016). Are long corolla tubes in Pedicularis driven by pollinator selection? J. Integr. Plant Biol. 58, 698–700. doi: 10.1111/jipb.12460
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Joly, S., Mclenachan, P. A., and Lockhart, P. J. (2009). A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Nat. 174, E54–E70. doi: 10.1086/600082
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kolaczkowski, B., and Thornton, J. W. (2007). Effects of branch length uncertainty on bayesian posterior probabilities for phylogenetic hypotheses. Mol. Biol. Evol. 24, 2108–2118. doi: 10.1093/molbev/msm141
Kress, W. J. (2017). Plant DNA barcodes: applications today and in the future. J. Syst. Evol. 55, 291–307. doi: 10.1111/jse.12254
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai, Q., Ding, L., Wang, H., Yue, Y., and Cai, F. (2007). Constraining the stepwise migration of the eastern Tibetan Plateau margin by apatite fission track thermochronology. Sci. China Earth Sci. 50, 172–183. doi: 10.1007/s11430-007-2048-7
Lam-Tung, N., Schmidt, H. A., Von Haeseler, A., and Bui Quang, M. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Lemey, P., Rambaut, A., Drummond, A. J., and Suchard, M. A. (2009). Bayesian phylogeography finds its roots. PLoS Comp. Biol. 5:e1000520. doi: 10.1371/journal.pcbi.1000520
Li, H.-L. (1948). A revision of the genus Pedicularis in China. part I. Proc. Acad. Nat. Sci. Phila. 100, 205–378.
Li, H.-L. (1949). A revision of the genus Pedicularis in China. part II. Proc. Acad. Nat. Sci. Phila. 101, 1–378.
Liu, B.-B., Campbell, C. S., Hong, D.-Y., and Wen, J. (2020). Phylogenetic relationships and chloroplast capture in the Amelanchier-Malacomeles-Peraphyllum clade (Maleae, Rosaceae): Evidence from chloroplast genome and nuclear ribosomal DNA data using genome skimming. Mol. Phylogenet. Evol. 147:106784. doi: 10.1016/j.ympev.2020.106784
Liu, M.-L., Yu, W.-B., and Wang, H. (2013). Rapid identification of plant species and iFlora application of DNA barcoding in a large temperate genus Pedicularis (Orobanchaceae). Plant Divers. 35, 707–714. doi: 10.7677/ynzwyj201313168
Liu, X., Wang, Z., Shao, W., Ye, Z., and Zhang, J. (2017). Phylogenetic and taxonomic status analyses of the Abaso section from multiple nuclear genes and plastid fragments reveal new insights into the North America origin of Populus (Salicaceae). Front. Plant Sci. 7:2022. doi: 10.3389/fpls.2016.02022
Liu, Y., Ye, J.-F., Hu, H.-H., Peng, D.-X., Zhao, L.-N., Lu, L.-M., et al. (2021). Influence of elevation on bioregionalisation: a case study of the Sino-Himalayan flora. J. Biogeogr. 48, 2578–2587. doi: 10.1111/jbi.14222
Macior, L. W. (1990). Pollination ecology of Pedicularis punctata Decne. (Scrophulariaceae) in the Kashmir Himalaya. Plant Species Biol. 5, 215–223. doi: 10.1111/j.1442-1984.1990.tb00181.x
Macior, L. W., and Tang, Y. (1997). A preliminary study of the pollination ecology of Pedicularis in the Chinese Himalaya. Plant Species Biol. 12, 1–7. doi: 10.1111/j.1442-1984.1997.tb00150.x
Macior, L. W., Tang, Y., and Zhang, J.-C. (2001). Reproductive biology of Pedicularis (Scrophulariaceae) in the Sichuan Himalaya. Plant Species Biol. 16, 83–89. doi: 10.1046/j.1442-1984.2001.00048.x
Maddison, W. P., and Maddison, D. R. (2019). Mesquite: A Modular System For Evolutionary Analysis. V3.7.0 [Online]. Available online at: http://www.mesquiteproject.org/ [Accessed August 10, 2020].
Maximowicz, C. J. (1888). Diagnoses plantarum novarum Asiaticarum. Bull. Acad. Sci. St Petersburg 32, 427–629.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ree, R. H. (2005). Phylogeny and the evolution of floral diversity in Pedicularis (Orobanchaceae). Int. J. Plant Sci. 166, 595–613. doi: 10.1086/430191
Rieseberg, L. H., Whitton, J., and Linder, C. R. (1996). Molecular marker incongruence in plant hybrid zones and phylogenetic trees. Acta Bot. Neerl. 45, 243–262. doi: 10.1111/j.1438-8677.1996.tb00515.x
Rokas, A., Williams, B. L., King, N., and Carroll, S. B. (2003). Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425, 798–804. doi: 10.1038/nature02053
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508.
Shimodaira, H., and Hasegawa, M. (1999). Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stegemann, S., Keuthe, M., Greiner, S., and Bock, R. (2012). Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. U.S.A. 109, 2434–2438. doi: 10.1073/pnas.1114076109
Struck, T. H., Feder, J. L., Bendiksby, M., Birkeland, S., Cerca, J., Gusarov, V. I., et al. (2018). Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 33, 153–163. doi: 10.1016/j.tree.2017.11.007
Stull, G. W., Soltis, P. S., Soltis, D. E., Gitzendanner, M. A., and Smith, S. A. (2020). Nuclear phylogenomic analyses of asterids conflict with plastome trees and support novel relationships among major lineages. Am. J. Bot. 107, 790–805. doi: 10.1002/ajb2.1468
Tsoong, P.-C. (1963). “Scrophulariaceae (Pars II),” in Flora Reipublicae Popularis Sinacae, Vol. 68, eds S.-S. Chien and W.-Y. Chun (Beijing: Science Press), 61–378.
Wang, E., Kirby, E., Furlong, K. P., Van Soest, M., Xu, G., Shi, X., et al. (2012). Two-phase growth of high topography in eastern Tibet during the Cenozoic. Nat. Geosci. 5, 640–645. doi: 10.1038/ngeo1538
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yang, C.-F., Gituru, R. W., and Guo, Y.-H. (2007). Reproductive isolation of two sympatric louseworts, Pedicularis rhinanthoides and Pedicularis longiflora (Orobanchaceae): how does the same pollinator type avoid interspecific pollen transfer? Biol. J. Linn. Soc. 90, 37–48. doi: 10.1111/j.1095-8312.2007.00709.x
Yang, F. S., Qin, A. L., Li, Y. F., and Wang, X. Q. (2012). Great genetic differentiation among populations of Meconopsis integrifolia and its implication for plant speciation in the Qinghai-Tibetan Plateau. PLoS One 7:e37196. doi: 10.1371/journal.pone.0037196
Yang, H.-B., Holmgren, N. H., and Mill, R. R. (1998). “Pedicularis Linn,” in Flora of China, Vol. 18, eds Z.-Y. Wu and P. H. Raven (St. Louis, MI: Missouri Botanical Garden Press & Science Press), 97–209. doi: 10.1016/j.biopha.2017.10.133
Ye, X. Y., Ma, P. F., Guo, C., and Li, D. Z. (2021). Phylogenomics of Fargesia and Yushania reveals a history of reticulate evolution. J. Syst. Evol. 59, 1183–1197. doi: 10.1111/jse.12719
Yi, T.-S., Jin, G.-H., and Wen, J. (2015). Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian – New World disjunct plant genus Osmorhiza (Apiaceae). Mol. Phylogenet. Evol. 85, 10–21. doi: 10.1016/j.ympev.2014.09.028
Yu, W. B., Huang, P. H., Li, D. Z., and Wang, H. (2013). Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS One 8:e74828. doi: 10.1371/journal.pone.0074828
Yu, W. B., Huang, P.-H., Ree, R. H., Liu, M.-L., Li, D.-Z., and Wang, H. (2011). DNA barcoding of Pedicularis L. (Orobanchaceae): evaluating four universal barcode loci in a large and hemiparasitic genus. J. Syst. Evol. 49, 425–437. doi: 10.1111/j.1759-6831.2011.00154.x
Yu, W. B., Liu, M. L., Wang, H., Mill, R. R., Ree, R. H., Yang, J. B., et al. (2015). Towards a comprehensive phylogeny of the large temperate genus Pedicularis (Orobanchaceae), with an emphasis on species from the Himalaya-Hengduan Mountains. BMC Plant Biol. 15:176. doi: 10.1186/s12870-015-0547-9
Yu, W. B., Wang, H., Liu, M. L., Grabovskaya-Borodina, A. E., and Li, D. Z. (2018). Phylogenetic approaches resolve taxonomical confusion in Pedicularis (Orobanchaceae): reinstatement of Pedicularis delavayi and discovering a new species Pedicularis milliana. PLoS One 13:e0200372. doi: 10.1371/journal.pone.0200372
Zha, H.-G., Milne, R. I., and Sun, H. (2010). Asymmetric hybridization in Rhododendron agastum: a hybrid taxon comprising mainly F(1)s in Yunnan, China. Ann. Bot. 105, 89–100. doi: 10.1093/aob/mcp267
Zhang, D.-C., Ye, J.-X., and Sun, H. (2016). Quantitative approaches to identify floristic units and centres of species endemism in the Qinghai-Tibetan Plateau, south-western China. J. Biogeogr. 43, 2465–2476. doi: 10.1111/jbi.12819
Keywords: Pedicularis siphonantha complex, phylogenetic delimitation, speciation, mountains of Southwest China, the Hengduan Mountains
Citation: Liu R, Wang H, Yang J-B, Corlett RT, Randle CP, Li D-Z and Yu W-B (2022) Cryptic Species Diversification of the Pedicularis siphonantha Complex (Orobanchaceae) in the Mountains of Southwest China Since the Pliocene. Front. Plant Sci. 13:811206. doi: 10.3389/fpls.2022.811206
Received: 08 November 2021; Accepted: 21 February 2022;
Published: 24 March 2022.
Edited by:
Susann Wicke, Humboldt University of Berlin, GermanyReviewed by:
Richard Ree, Field Museum of Natural History, United StatesChun-Lei Xiang, Kunming Institute of Botany (CAS), China
Alex Twyford, University of Edinburgh, United Kingdom
Copyright © 2022 Liu, Wang, Yang, Corlett, Randle, Li and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Bin Yu, yuwenbin@xtbg.ac.cn
 Rong Liu
Rong Liu