- College of Horticulture, Xinjiang Agricultural University, Xinjiang, China
Coreopsis tinctoria Nutt. (C. tinctoria) has a long history of application and high economic and medicinal value. Flavonoids, the main active components of C. tinctoria, are widely studied in pharmacology and food development. However, the flavonoid biosynthesis pathway in C. tinctoria is unclear. In this study, we comprehensively compared the transcriptomes and metabolite profiles of two colors of C. tinctoria flowers (LS and JS) at different flowering stages. A total of 165 flavonoids (46 flavonoids, 42 flavonols, 22 anthocyanins, 18 chalcones, 12 dihydroflavonols, nine isoflavones, eight dihydroflavonoids, six flavanols, and two tannins) were identified in LS and JS at different flowering stages. Thirty-three metabolites (11 anthocyanins, 11 flavonols, seven flavonoids, two dihydroflavonols, one dihydroflavone, and one chalcone) were found to be statistically significantly different in the LS vs. JS groups. LS flowers accumulated higher levels of 10 anthocyanins (seven cyanidins and three pelargonidins) than JS flowers. Furthermore, candidate genes related to the regulation of flavonoid and anthocyanin synthesis were identified and included 28 structural genes (especially F3H, Cluster-28756.299649, and 3GT, Cluster-28756.230942) in LS and JS, six key differentially expressed transcription factors (especially MYB90a, Cluster-28756.143139) in LS and JS, and 17 other regulators (mainly including transporter proteins and others) in LS. Our results provide valuable information for further studies on the mechanism underlying flavonoid biosynthesis in C. tinctoria.
Introduction
Coreopsis tinctoria Nutt. (C. tinctoria) is an annual herb in the Compositae family, also referred to as the Asteraceae family. In China, Xinjiang is the main production area of C. tinctoria, and the quality of the C. tinctoria produced in Keliyang Township, Hetian, is known to be the best (Jing and Lan, 2012). The main active ingredients of C. tinctoria are flavonoids, and the medicinal value or effects of this herb, such as its effects in lowering hyperlipidemia, hypertension, and hyperglycemia, its antioxidative effects, its anti-inflammatory properties and its effects in preventing cardiovascular diseases, are also due to the extracted flavonoids (Guo et al., 2014, 2015; Du et al., 2018; Zeng and He, 2019; Shen et al., 2021).
Flavonoids are important products of the secondary metabolism of plants, and they have important ecological significance and physiological functions in regulating flower coloring, physiological activities and adaptation to unfavorable environments (Havsteen, 2002). Subclasses of flavonoids include flavones, isoflavones, flavanols, flavanones, flavonols, and anthocyanins. To date, more than 50 flavonoids, such as quercetin, luteolin, catechin, kaempferol, and rutin, have been extracted and identified from C. tinctoria (Guo et al., 2015). Many studies have shown that the total flavonoid content of C. tinctoria is correlated with many factors; the total flavonoid contents exhibit statistically significant differences according to habitats (Sun et al., 2012; Chen et al., 2013), harvesting periods (Gu et al., 2015), agronomic measures (Liang et al., 2016) and determination methods (Ma et al., 2019). For example, Du and Wu (2015) found that the total flavonoid content of C. tinctoria produced on the Hetian Plateau (132.58 μg/mL) was significantly higher than that of C. tinctoria produced on the Dabancheng Plain (99.47 μg/mL), Yang et al. (2016) used the HPLC-DAD (high-performance liquid chromatography with diode-array detection) method to quantitatively analysis of eight compounds in 23 batches, including okanin, and found that the total compound content of C. tinctoria produced in Keliyang Township was the highest. Gu et al. (2015) found that there were significant differences in the total flavonoid content in different growth stages of C. tinctoria, and the total flavonoid content in the bud stage was the highest, in the range of 12.57–13.73%. Although many basic studies have been performed on the use of the total flavonoid content as an index for evaluating the quality of C. tinctoria, a systematic analysis of flavonoid components is still lacking.
At present, the flavonoid synthesis pathway in model plants is relatively well understood. Phenylalanine lyase (PAL), cinnamic acid hydroxylase (C4H), coumadin CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonol synthase (FLS), anthocyanidin synthase (ANS), and other key genes involved in flavonoid synthesis have been isolated and identified from model plants, such as Arabidopsis thaliana (Routaboul et al., 2006). With the development and improvement in omics technology, the mechanism underlying flavonoid synthesis in apple (Henry-Kirk et al., 2012), peanut (Wan et al., 2020), Ziziphus jujuba (Zhang et al., 2020) and other plants has been studied. However, plant flavonoid biosynthesis is specific, and flavonoid biosynthesis and related gene expression differ in different plant varieties, developmental stages, tissues and organs (Wen et al., 2020). Transcription factors (TFs) such as MYB, bHLH, and WD40, play important roles in in regulation of flavonoid biosynthesis. For example, in Arabidopsis thaliana, the MYB12 of R2R3-MYB TF can transcribe and regulate the expression of chalcone synthase and flavonol synthase, and its expression level is closely related to flavonol content. Some MYB proteins form ternary complexes with the basic helix-loop-helix (BHLH) protein Transparent Testa Glabra 1 (TTG1) WD (beta-transducin)-repeat (WDR) subfamily to regulate the expression of genes related to the biosynthesis of anthocyanins and procyanidins (PA) (Pelletier and Shirley, 1996). The study of a large number of key genes and gene expression regulatory factors in the flavonoid synthesis pathway is an important way to understand this process and increase flavonoid contents. However, the studies on the mechanism of flavonoid synthesis from C. tinctoria are unclear.
Coreopsis tinctoria is bright in color, with common ligulate flowers exhibiting reddish-brown color at the base and yellow color at the top (the color ratio of reddish-brown to yellow varies from 0 to 9/10) (Edwin and Hampton, 1971; Science Press, 1979). Coreopsis tinctoria planted in Xinjiang produces two colors of ligulate flowers, namely, ligulate flowers with red–brown/yellow ratios of 2/3–3/4 and entirely yellow ligulate flowers. In this study, the two colors of C. tinctoria are labeled LS (ligulate flowers with red–brown base/yellow top) and JS (entirely yellow ligulate flowers). The accumulation pattern and flavonoid synthesis pathway of LS and JS were studied in different flowering stages, and this study would provide a theoretical basis for identifying and improving the quality and scientific utilization of characteristic resources.
Materials and Methods
Plant Materials and Treatments
The seeds of LS and JS flowers were collected from Keliyang Township, Hetian, China (lat. 37°37′17′′N, long. 78°16′58.80′′E; altitude, 2,196 m), and were potted from March to August 2021 (V peat: V perlite: V vermiculite = 3:1:1) in the solar greenhouse of the Institute of Agricultural Mechanization, Xinjiang Academy of Agricultural Sciences. Eighty pots (pot diameter: 22 cm) were planted for each flower, and each pot contain three plants. Beginning in the seedling stage (30 days after sowing), 1/4–1 standard Hoagland nutrient solution was regularly added (every 7 days; 600 mL per pot) (Supplementary Table 1). The buds and inflorescences of the LS and JS plants were sampled at the bud stage (94 days after sowing, stage 1), the initial flowering stage (101 days after sowing, stage 2), the full flowering stage (116 days after sowing, stage 3) and the final flowering stage (131 days after sowing, stage 4) (Figure 1A). Three biological replicates were collected in each stage of LS and JS plant growth. After collection, the samples were immediately placed into a liquid nitrogen tank at −80°C for storage. The four stages (from stage 1 to stage 4) of LS and JS flowers were labeled as L1, L2, L3, L4 and J1, J2, J3, and J4, respectively.
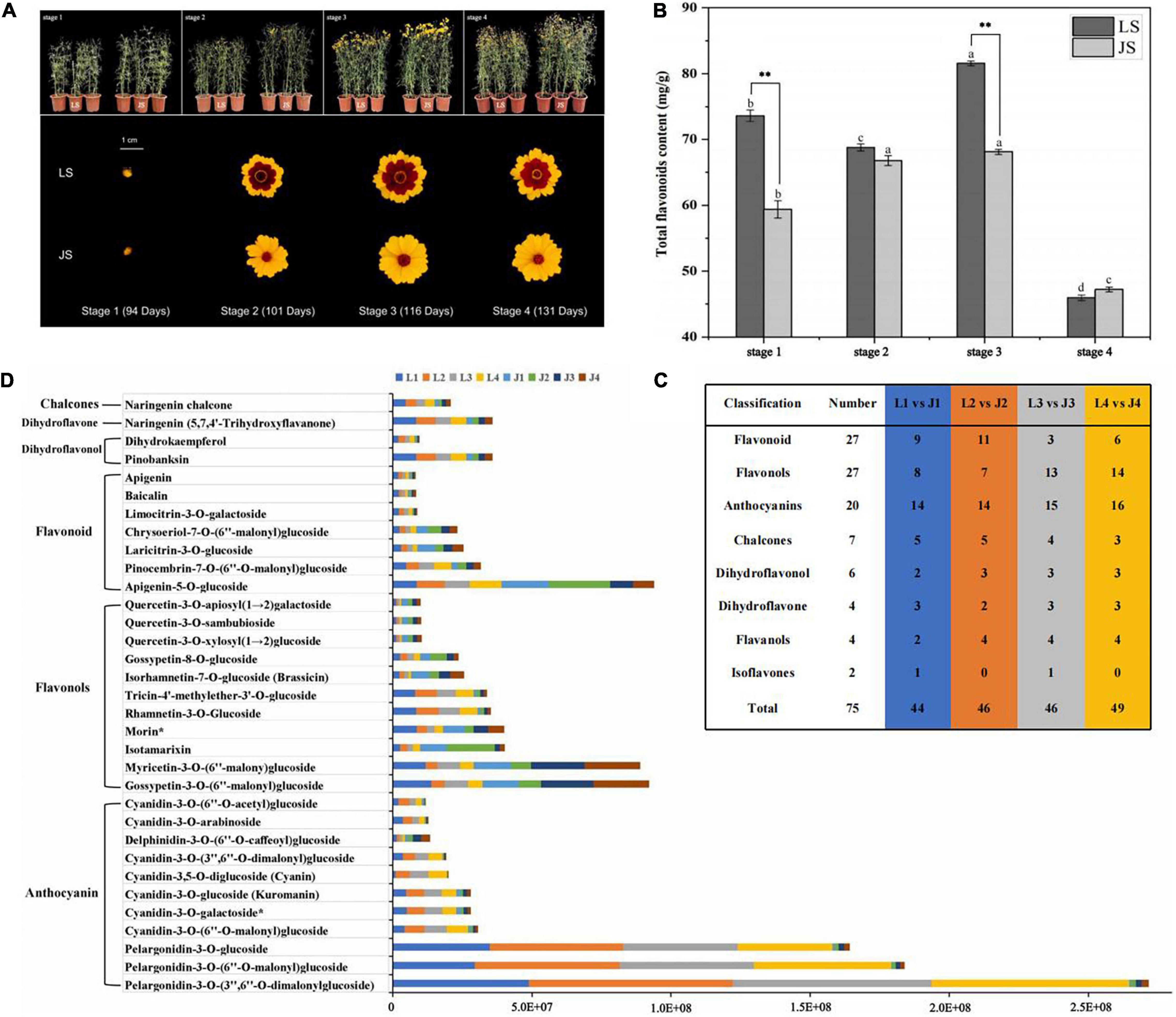
Figure 1. Sampling and analysis of flavonoids metabolites. (A) Morphology of four stages of two colors of C. tinctoria. (B) Total flavonoid content of LS and JS at four stages. (C) Statistics on the types of DAFs among different comparison groups. (D) The relative content of 33 differential accumulation of flavonoids (DAFs) accumulated at different stages in LS and JS.
Measurement of Total Flavonoid Content
The total flavonoid content was determined by spectrophotometry, with specific reference to the methods of Zhang (2013). Samples without rutin were used as the blank control, and the absorbance of rutin was measured at a wavelength of 510 nm; a standard curve was drawn with concentration x as the abscissa and absorbance y as the ordinate. The regression equation obtained was y = 23.707x − 0.7563 (R2 = 0.9959). The absorbance was measured at a wavelength of 510 nm in parallel three times, and the total flavonoid content was calculated.
Qualitative and Quantitative Analysis of Flavonoid Metabolites
The method used to identify flavonoid metabolites was similar to that described by Xiao et al. (2021). First, the freeze-dried C. tinctoria buds and inflorescence were ground to a powder with a grinding instrument (MM 400, Retsch) (30 Hz, 1.5 min), and 100 mg of powder was extracted overnight with 1.0 mL of 70% aqueous methanol at 4°C; during this step, the extraction rate was improved by three eddies. After centrifugation for 10 min at 10,000 g, the sample was filtered, and the supernatant was filtered through a microporous membrane (0.22 μm pore size ANPEL, Shanghai, China).1 The sample for LC–MS/MS (liquid chromatography with tandem mass spectrometry) analysis was kept in a sample injection vial.
Metabolite data acquisition was conducted with a UPLC (Shim-pack UFLC SHIMADZU CBM30A)2 (ultrafast liquid chromatography) instrument coupled to an MS/MS (Applied Biosystems 4500 QTRAP)3 instrument. Chromatographic separation was carried out on a Waters ACQUITY UPLC HSS T3 system (Shim-pack UFLC SHIMADZU CBM30A, see text footnote 2), which was equipped with a C18 column (1.8 μm × 2.1 mm × 100 mm), at 40°C. The injection volume was 5 μL at a flow rate of 0.4 mL/min. The organic phase was acetonitrile (with 0.04% acetic acid), and the mobile phase was ultrapure water (with 0.04% acetic acid). The elution gradient was as follows: 0 min, 95:5 water/acetonitrile (v/v); 11.0 min, 5:95 water/acetonitrile; 12.0 min, 5:95 water/acetonitrile; 12.1 min, 95:5 water/acetonitrile; and 15.0 min, 95:5 water acetonitrile. The metabolites in the samples were analyzed both qualitatively and quantitatively by using MS data, Analyst 1.6.3 software, and metabolite information from the public MetWare database (MWDB, Wuhan MetWare, China).
Based on the results of principal component analysis (PCA), the metabolites with | log2 Fold Change (FC)| ≥ 1 and variable importance in project (VIP) value ≥ 1 in the OPLS-DA (orthogonal projections to latent structures-discriminant analysis) model were considered to be statistically significant different among samples (Saccenti et al., 2013).
RNA Extraction, Library Establishment, and Sequencing
The total RNA was extracted from 24 samples of C. tinctoria by the TRIzol (Invitrogen) method, and the quality of the RNA samples was assessed by agarose gel electrophoresis with a Qubit 2.0 Fluorometer (Life Technologies, CA, United States) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) to ensure that qualified samples were used for transcriptome sequencing. The cDNA library was established by a TruSeq™ RNA sample preparation kit (Illumina, San Diego, CA, United States). mRNA with a polyA tail was enriched with oligo (dT) (deoxythymine) magnetic beads, and the mRNA molecules were broken into short fragments with the addition of fragmentation buffer. Using the short fragment mRNA as the template, first-strand cDNA was synthesized by six-base random primers (random hexamers). Buffer, dNTPs (deoxynucleotide triphosphates; dUTP (2′-deoxyuridine, 5′-triphosphate), dATP (deoxyadenosine triphosphate), dGTP (deoxyguanosine triphosphate), and dCTP (deoxycytidine triphosphate) and DNA polymerase I were then added to synthesize double-stranded cDNA.
The double-stranded cDNA was then purified by AMPure XP beads. The double-stranded cDNA structure had a sticky end that was filled until blunt via the addition of End Repair Mix, and an A base was added to the 30 end to overlap with a T base overhang. The fragment size was then selected by AMPure XP beads, and the final cDNA library was enriched by PCR (polymerase chain reaction) as described in the instructions. After cDNA enrichment by PCR, a Qubit 2.0 instrument was used for preliminary quantification, and an Agilent 2100 instrument was used to detect the insert size of the library. The effective concentration of the library was subsequently determined by Q-PCR (quantitative polymerase chain reaction) (effective concentration of the library > 2 nM), and the Illumina HiSeq platform was used for sequencing (Wuhan Metware).
De novo Assembly and Functional Annotation
The raw data of 24 samples were obtained by de novo transcriptome sequencing, and the raw data underwent filtering and quality control before reassembly. To obtain clean reads, an eliminated adapter was used to remove the adapter reads, reads containing more than 10% unknown nucleotides and low-quality reads (reads containing more than 50% bases with Q ≤ 20 reads). There is no reference genome for C. tinctoria, and the clean reads were spliced by Trinity4 to obtain the transcript sequence, which was used as a reference sequence for subsequent analyses. Hierarchical clustering of transcripts was performed by comparing the read number and expression pattern of transcripts using Corset.5 Hierarchical clustering was conducted with Corset to ultimately obtain the longest cluster sequence as a follow-up analysis on unigenes.
Then, we used BLAST software to compare the unigene sequences with seven public databases, including the NCBI non-redundant protein (NR),6 TrEMBL (a computer annotated supplement to SwissProt), Pfam,7 SwissProt,8 Kyoto Encyclopedia of Genes and Genomes (KEGG),9 Gene Ontology (GO)10 and euKaryotic Ortholog Groups (KOG)11 databases. The RNA-Seq data set was uploaded to NCBI12 (accession no. PRJNA698724).
Quantitative Gene Expression and Differentially Expressed Genes Analysis
The clean reads of 24 samples were mapped back to a reference sequence using Bowtie 2,13 and the fragments per kilobase of transcript per million fragments mapped (FPKM) method was used to normalize the expression of the mapped reads. The read count of genes was determined using feature counts. DESeq2 was used to analyze the difference in the read count data. The multiple hypothesis test correction of the P value (probability) was carried out using the Benjamini–Hochberg method, and the false discovery rate (FDR) was determined. Genes that satisfied the conditions of | log2 FC| ≥ 1 and FDR < 0.05 were considered to be differentially expressed (Love et al., 2014).
Annotation and Analysis of Transcription Factors
Transcription factor prediction was performed using iTAK software, which integrates the PlnTFDB and PlantTFDB databases and uses the well-defined TF family and rules in the database. TFs were identified through hmmscan comparison, TFs related to flavonoid synthesis were identified through a reference review. The selected TFs with DAFs/DAAs Spearson’s correlation (ρ) values > 0.80 or <−0.80 were selected to draw the interaction network diagram.
Weighted Gene Co-expression Network Analysis
The overlapping differentially expressed genes (DEGs) (2,140 out of 7,483 DEGs) and differential anthocyanins accumulation (DAAs) were selected for co-expression network analysis via weighted gene co-expression network analysis (WGCNA). The content of overlapping DAAs in LS was used as a trait for WGCNA, and modules were obtained through WGCNA with default settings. Furthermore, the correlation coefficients between the hub genes in the module were determined. The hub genes with DAAs Pearson’s correlation coefficient (| PCC| > 0.80) values were selected to draw the interaction network diagram.
Validation of RNA-Seq Data by Quantitative Real Time-PCR
The nine DEGs related to flavonoid biosynthesis identified in the RNA-seq data were selected for validation by qRT–PCR (real-time quantitative reverse transcription). The total RNA of three biological repeat sequences was extracted from flower buds and inflorescences by TRIzol reagent (Invitrogen, Shanghai, China) for qRT–PCR analysis. The RNA of each sample was reverse transcribed into cDNA by the PrimeScript fragments RT reagent Kit with gDNA Eraser (Perfect Real Time). A Bio-Rad CFX Connect fluorescence quantitative PCR detection system was used. The amplification cycle procedure was as follows: 95°C for 3 min, 95°C for 10 s, 55°C for 20 s, 72°C for 20 s, 75°C for 5 s, repeated for 40 cycles. Primers targeting the reference gene GAPDH were designed according to the conserved region in a GAPDH gene sequence alignment of Helianthus annuus. The 2–ΔΔCT method was used to calculate relative gene expression, and the error bars represent the mean ± standard deviation (SD). The primer information is shown in the attached Supplementary Table 2.
Data Analysis
Excel 2010 was used for data processing, SPSS Statistics 23.0 software was used for analysis of variance, and Origin 2018 software was used for generating the figures. The R software (R version 3.6.1) stats package was used to drawing the correlation network diagram.
Results
Metabolic Differences Between the Two Colors of Coreopsis tinctoria
To explore the mechanism underlying flavonoid metabolism in C. tinctoria, the total flavonoid contents of LS and JS were measured. Among the four flowering stages of LS, the total flavonoid content was significantly higher in stage 3 (81.60 mg/g) than in the other stages. This was followed by the flavonoid contents of LS in stage 1, stage 2, and stage 4, and the total flavonoid contents during these stages were 73.65, 68.78, and 45.98 mg/g, respectively. The total flavonoid content of JS first increased and then decreased with of the progression of flowering stage. The total flavonoid content of JS was highest at stage 3, 68.11 mg/g. This was followed by the contents of JS in stage 1, stage 2, and stage 4, and the total flavonoid contents during these stages were 59.38, 66.79, and 47.24 mg/g, respectively. The differences in the total flavonoid contents of LS in stage 1 and stage 3 from those of JS in the same stages were statistically significant, i.e., the total flavonoid contents in LS were 24 and 20% higher than those in JS, respectively. The results showed the dynamic difference in total flavonoid content of LS and JS in the four different stages, especially in stage 1 and stage 3 (Figure 1B).
To further analyze the differences in flavonoid metabolites in C. tinctoria (LS and JS) at different stages, the flavonoid metabolites were detected by UHPLC–ESI–MS/MS. The total ion flow map (TIC) obtained from QC samples indicates the sum of all ion strengths at different time points (Supplementary Figure 1A). In the detection multipeak map (XIC), the chromatographic peaks of different colors correspond to different flavonoid metabolites (Supplementary Figure 1B). The superposition of the total ion flow map (TIC map) of the QC samples shows that the response intensity and retention time of each peak strongly overlapped, indicating that the data were reliable (Supplementary Figure 1C). A total of 165 flavonoids were identified in the buds and inflorescences of LS and JS at different flowering stages (Supplementary Table 3), and these flavonoids included 46 flavonoids, 42 flavonols, 22 anthocyanins, 18 chalcones, 12 dihydroflavonols, nine isoflavones, eight dihydroflavonoids, six flavanols, and two tannins (Supplementary Figure 2). The relative contents of flavonoids were analyzed by hierarchical cluster thermography (HCA), and there were significant differences in the patterns of flavonoid accumulation in LS and JS at different stages. The more relative contents of flavonoids accumulated in LS and JS in stage 1, and more flavonoids accumulated during the flowering stages of LS than at that of JS (Supplementary Figure 3). Principal component analysis (PCA) showed that the L1 vs. J1, L2 vs. J2, L3 vs. J3, and L4 vs. J4 groups were clearly separated, and principal components PC1 (38.15%) and PC2 (23.00%) showed a degree of change of 61.50% (Supplementary Figure 4), indicating the difference between the LS and JS groups. The PCA results were consistent with the HCA results, which showed the repeatability and reliability of the data.
Based on the data obtained from the metabolome, 44, 46, 46, and 49 DAFs were identified between the L1 vs. J1, L2 vs. J2, L3 vs. J3, and L4 vs. J4 groups, respectively (Figure 1C). In addition, 33 DAFs (E05 and above and | log2 FC| > 1) were identified in LS and JS at different stages. Compared to JS, there were 20 DAFs (10 anthocyanins, four flavonoids, two flavonols, two dihydroflavonols, one dihydroflavone, and one chalcone) that significantly accumulated in LS at one or more stages. Compared to LS, 13 DAFs (nine flavonols, three flavonoids, and one anthocyanin) accumulated in JS at one or more stages. Seventeen DAFs (10 anthocyanins, two flavonols, two dihydroflavonols, one flavonoid, one dihydroflavone, and one chalcone) significantly accumulated in LS and JS during the four stages (Figure 1D).
The DAFs with the largest fold changes among the different stages were mainly enriched in anthocyanins in LS. Pelargonidin-3-O-(3′′,6′′-O-dimalonylglucoside) (log2FC = −6.25 to −4.82), pelargonidin-3-O-(6′′-O-malonyl) glucoside (log2FC = −5.96 to −4.86), pelargonidin-3-O-glucoside (log2FC = −5.61 to −4.09), cyanidin-3,5-O-diglucoside (Cyanin) (log2FC = −5.49 to −3.45), cyanidin-3-O-(3′′,6′′-O-dimalonyl) glucoside (log2FC = −3.71 to −3.16), cyanidin-3-O-(6′′-O-acetyl)glucoside (log2FC = −3.65 to −1.56), cyanidin-3-O-arabinoside (log2 FC = −4.11 to −2.38), and cyanidin-3-O-(6′′-O-malonyl)glucoside (log2FC = −3.11 to −2.31) substantially accumulated in LS during four stages. In JS, flavonols obviously accumulated in a few stages, among which quercetin-3-O-xylosyl(1→2)glucoside, quercetin-3-O-apiosyl(1→2)galactoside and quercetin-3-O-sambubioside accumulated in stage 3, and the Log2FC values were 1.45, 1.45, and 1.41, respectively. In addition, delphinidin-3-O-(6-O-caffeoyl) glucoside accumulated in JS in stage 3 and stage 4, changing by a multiple of 1.42–1.72 (log2FC value) (Supplementary Table 4). These DAFs might be the key metabolites that influence the changes in flavonoids in LS and JS. And these significantly accumulated anthocyanins might affect the coloration of LS and JS.
Transcriptome Sequencing, Assembly, and Statistics
To further study the difference in the mechanism underlying flavonoid synthesis between LS and JS, de novo transcriptome sequencing was performed on 24 samples of buds and inflorescences of C. tinctoria. For each sample, 42,768,396–53,410,008 raw reads were obtained by high-throughput sequencing, and after splices and low-quality sequences were excluded, 40,703,558–51,523,378 clean reads were obtained. Q30 bases accounted for greater than 91.79% of the total, and the average GC content was 43.90% (Supplementary Table 5). After assembly with Trinity software, 463,574 transcripts with an average length of 786, 447,813 unigenes with an average length of 804 bp, N50 lengths of 1,049 and 1,061 bp, and 23,017 transcripts and unigenes with sequence lengths greater than or equal to 2,000 bp were identified (Supplementary Table 6). Comparing a total of 447,813 unigenes with information from seven public databases (NR, Trembl, Swiss-Prot, Pfam, KOG, GO, and KEGG), it was found that the comparison rate of all unigenes in at least one database was as high as 60.40% (Supplementary Table 7). In summary, the RNA-seq data were reliable and had high quality, and these data could be used for further analysis.
Differentially Expressed Genes Analysis of Expression and KEGG Enrichment
First, Pearson correlation analysis was carried out on 24 samples, and the results showed that the repeatability of the same group of samples was good, and there were significant differences in correlation between different groups (Supplementary Figure 5). The DEGs were analyzed by DESeq2 software, and the selected DEGs (| log2FC| ≥ 1 and FDR < 0.05) were then compared and analyzed. There were 38,126, 41,086, 39,820, and 37,213 DEGs in the L1 vs. J1, L2 vs. J2, L3 vs. J3, and L4 vs. J4 groups, respectively, of which 24,477, 27,890, 26,274, and 24,880 were upregulated and 13,649, 13,196, 13,546, and 12,333 were downregulated, respectively. Compared to those expressed by LS, the number of upregulated genes expressed by JS in stage 2 and stage 3 was the highest, and the number of downregulated genes expressed by JS in stage 1 and stage 3 was the highest. Stage 3 was the critical period of LS and JS (Figure 2A). The Venn diagram results showed that 7,458 DEGs were co-expressed among the four comparison groups, which indicated that these DEGs might perform key functions in flavonoid regulation in LS and JS (Figure 2B).
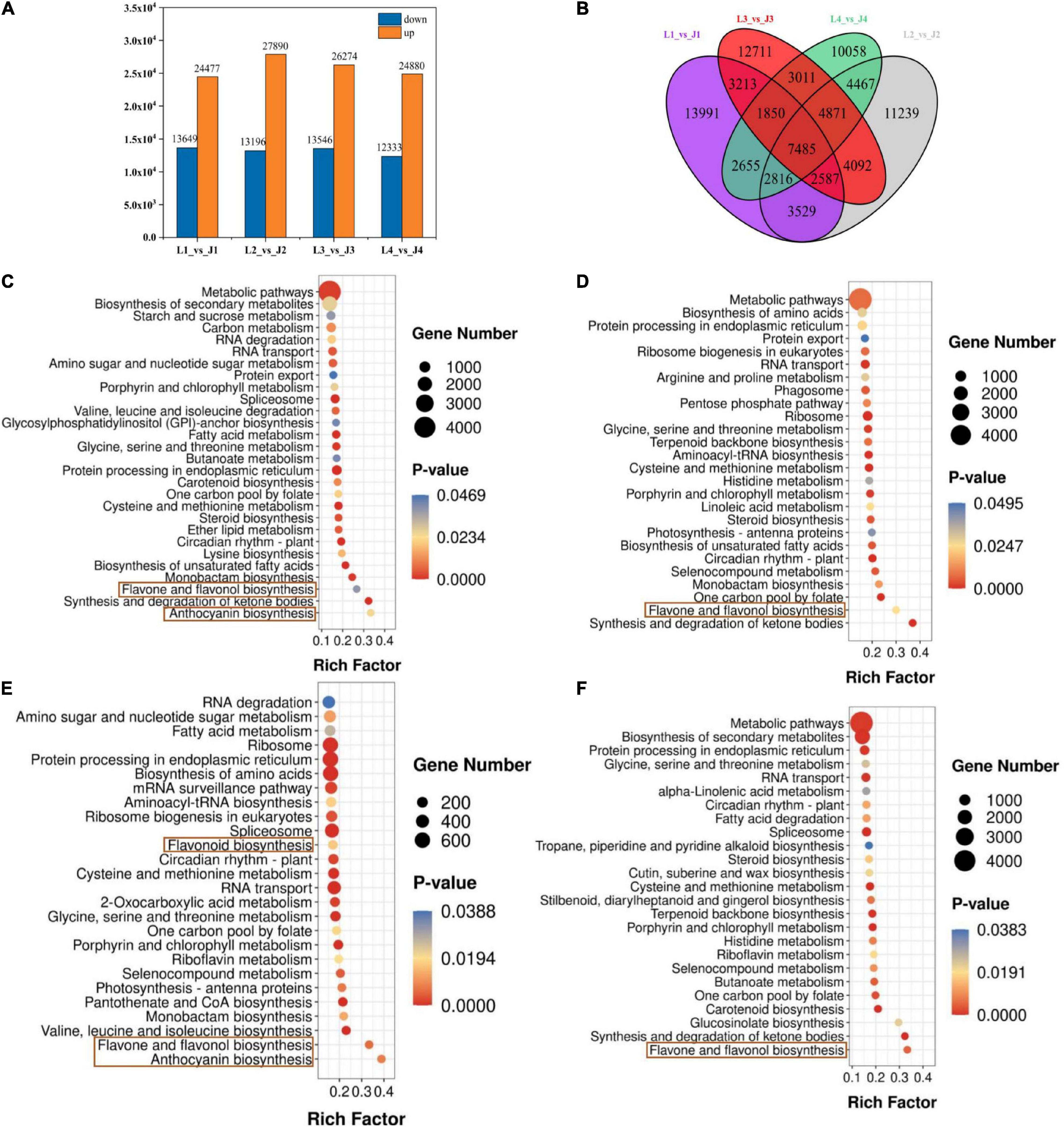
Figure 2. Differentially expressed genes (DEGs) in LS and JS. (A) Statistics of DEGs in different comparison groups of LS and JS. Red indicates upregulated DEGs. Green indicates downregulated DEGs. (B) Venn diagram of DEGs. (C–F) DEGs enrichment KEGG scatter plot in four stages of LS and JS comparison groups. (C) L1 vs. J1 group. (D) L2 vs. J2 group. (E) L3 vs. J3 group. (F) L4 vs. J4 group.
The DEGs identified in the LS vs. JS comparison groups were involved in a total of 137 KEGG metabolic pathways, and 28, 26, 26, and 25 significantly enriched metabolic pathways were identified in the L1 vs. J1, L2 vs. J2, L3 vs. J3, and L4 vs. J4 groups, respectively (Supplementary Table 8). Among them, flavone and flavonol biosynthesis (ko00944) was significantly enriched among the four comparative groups. Anthocyanin biosynthesis (ko00942) was significantly enriched in the L1 vs. J1 and L3 vs. J3 comparison groups, and flavonoid biosynthesis (ko00941) was significantly enriched in the L3 vs. J3 comparison groups. It is suggested that flavonoid biosynthesis-related pathways play an important role in different stages of LS and JS development (Figures 2C–F).
Analysis of Differentially Expressed Genes Related to Flavonoid and Anthocyanin Biosynthesis
Based on KEGG enrichment analysis and gene functional annotation, 61 DEGs (at least one stage FPKM > 10) were shown to be significantly enriched in pathways related to flavonoid biosynthesis. The 61 DEGs included 29 DEGs (19 HCT genes, eight PGT1 genes, one C3′H gene, one CCoAOMT gene) involved upstream of flavonoid biosynthesis and 32 DEGs (two C4H genes, two CHS genes, five CHI genes, six F3H genes, nine F3′H genes, three DFR genes, two 3GT genes, two LDOX genes, and one UF3RT gene) involved in flavonoid biosynthesis and anthocyanin biosynthesis (Supplementary Table 9). Furthermore, the Spearman correlation between the expression level of 61 DEGs and the total flavonoid content and the relative content of 33 DAFs were analyzed. The results showed that there was a positive correlation between the expression of four DEGs (one HCT gene, Cluster-28756.178701; one 3GT gene, Cluster-28756.220920; two LDOX genes, Cluster-28756.215472, Cluster-28756.227869) and total flavonoid content, whereas the expression of two DEGs (one C4H gene Cluster-28756.151433; one F3H gene, Cluster-28756.279662) had a negative correlation with total flavonoid content (P < 0.05; Figure 3A). In addition, the expression levels of 28 genes (seven HCT genes, four PGT1 genes, one C3′H gene, one C4H gene, four F3H genes, five F3′H genes, three DFR genes, one LDOX gene, and two 3GT genes) were significantly associated with the levels of 25 DAFs (10 anthocyanins; six flavonols; five flavonoids; two dihydroflavonol; one chalcones; and one dihydroflavone). The expression levels of the F3H gene (Cluster-28756.299649) and 3GT gene (Cluster-28756.230942) were significantly positively correlated with the levels of 7 types of anthocyanins (pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside (Kuromanin), cyanidin-3-O-galactoside, pelargonidin-3-O-(6′′-O-malonyl)glucoside, cyanidin-3-O-(6′′-O-malonyl)glucoside, pelargonidin-3-O-(3′′,6′′-O-dimalonylglucoside), cyanidin-3-O-(3′′,6′′-O-dimalonyl)glucoside). On the other hand, the expression of these two DEGs were significantly negatively correlated with the levels of three types of flavonols [quercetin-3-O-apiosyl(1→2)galactoside, quercetin-3-O-sambubioside, quercetin-3-O-xylosyl(1→2)glucoside]. The expression levels of these two DEGs in LS in the four stages were significantly higher than those in JS, especially in stage 3, suggesting that the two DEGs may play an essential role in anthocyanin accumulation in LS. Otherwise, the expression of HCT gene (Cluster-28756.81340), PGT1 gene (Cluster-28756.230443), and F3H gene (Cluster-28756.165750) were significantly negatively correlated with the level of naringenin chalcone. The expression of the PGT1 gene (Cluster-28756.230443) and F3H gene (Cluster-28756.165750) were significantly negatively correlated with the levels of three anthocyanins [cyanidin-3-O-arabinoside, pelargonidin-3-O-(6′′-O-malonyl)glucoside, and pelargonidin-3-O-(3′′,6′′-O-dimalonylglucoside)] and 1 flavonoid [pinocembrin-7-O-(6′′-O-malonyl)glucoside]. The expression levels of the HCT gene (Cluster-28756.81340) and the PGT1 gene (Cluster-28756.230443) were significantly negatively correlated with the levels of two flavonols (rhamnetin-3-O-glucoside and tricin-4′-methyl ether-3′-O-glucoside) and one dihydroflavonol (dihydrokaempferol). These three genes were only highly expressed in JS in the four stages but not in LS in the four stages, indicating that the three DEGs might negatively regulate flavonoid synthesis in JS (Figures 3B,C).
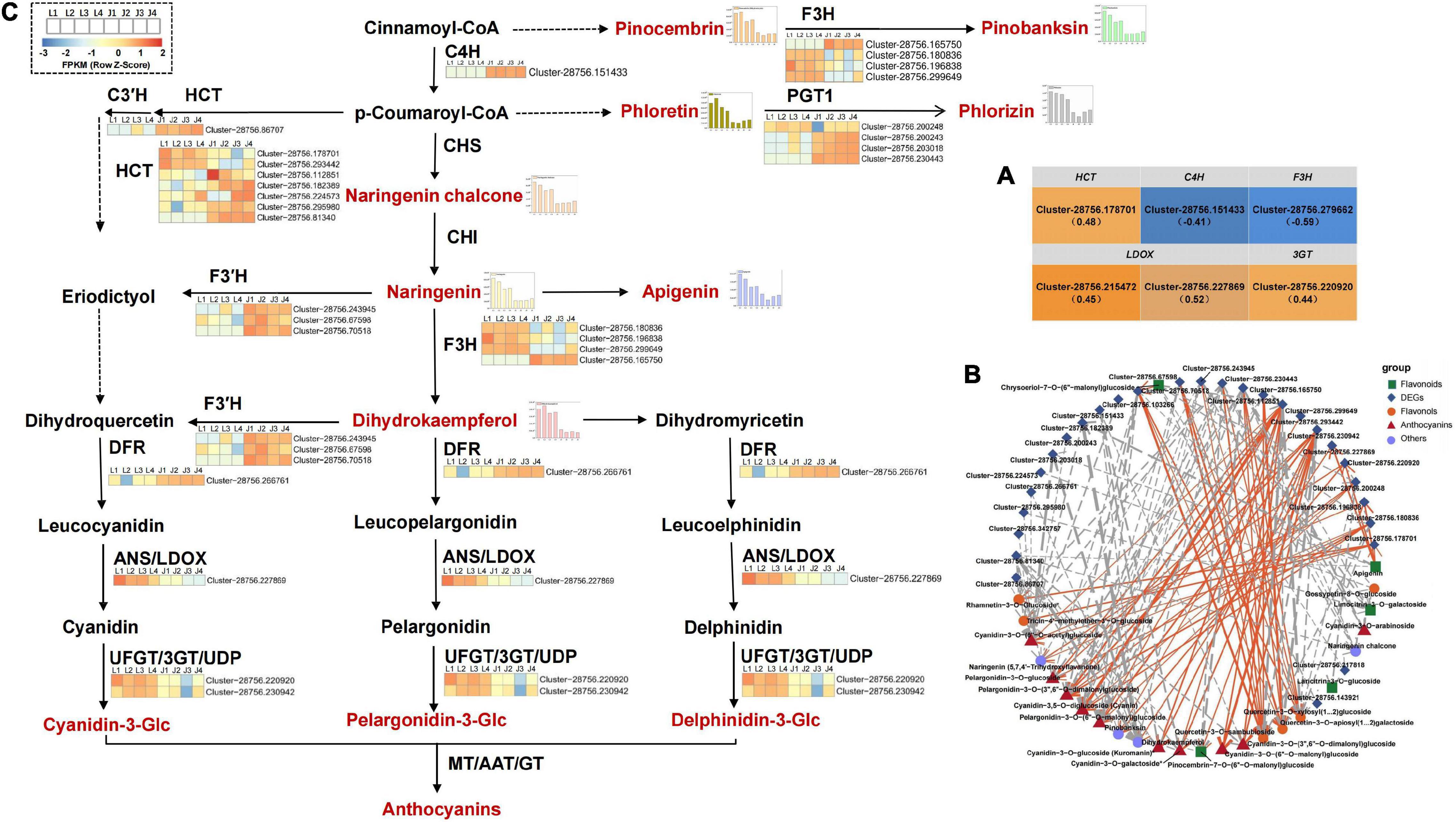
Figure 3. DEGs analysis of flavonoid and anthocyanin synthesis in C. tinctoria. (A) Six DEGs correlated with total flavonoids content. The brackets indicated the correlation coefficient ρ. (B) There was a significant correlation between 28 DEGs and 25 DAFs (P < 0.01, —ρ— > 0.80). The solid line indicates the positive correlation, the dotted line indicates the negative correlation, and the thickness of the line indicates the correlation strength. (C) The 28 DEGs (in B) involved in flavonoid and anthocyanin biosynthesis pathway of LS and JS. C3′H, coumaroylquinate 3′-monooxygenase; C4H, trans-cinnamate 4-monooxygenase; HCT, shikimate O-hydroxycinnamoyltransferase; PGT1, phlorizin synthase; F3H, naringenin 3-dioxygenase; F3′H, flavonoid 3′-monooxygenase; DFR, bifunctional dihydroflavonol 4-reductase; LDOX, leucoanthocyanidin dioxygenase; 3GT, anthocyanidin 3-O-glucosyltransferase.
Quantitative Real Time-PCR
To verify the accuracy of the RNA-seq data, nine DEGs involved in the flavonoid metabolic pathway were selected, and their expression was verified by qRT–PCR. The results showed that the trends of qRT–PCR and transcriptome expression were basically consistent with those of gene expression, indicating that the measured transcriptome data were effective and reliable (Supplementary Figure 6).
Analysis of Transcription Factor-Mediated Regulation of Flavonoid and Anthocyanin Biosynthesis
The flavonoid biosynthesis pathway is mainly regulated by external factors and TFs, and the TFs involved in the regulation include MYB, bHLH (MYC), and WD40 family (Zhang et al., 2014). In this study, many kinds of TFs were identified in LS and JS in the four stages. Among them, the largest number of TF families (the top 10 categories) was identified in the L1 vs. J1, L2 vs. J2, L3 vs. J3, and L4 vs. J4 groups, and these TF families included MYB, C3H, C2H2, NAC, AP2/ERF-ERF, GRAS, bHLH, bZIP, and so on. The number of MYB (including MYB-related) gene families was the highest among the four comparison groups (Supplementary Figure 7). The eight MYB and five bHLH TFs (at least one stage FPKM > 10) related to flavonoid biosynthesis were screened. The expression levels of these TFs were different in LS and JS in one or more stages (Niu et al., 2016; Supplementary Figure 8). There was a significant correlation between the expression of six TFs and the levels of six cyanidins. The expression levels of the MYB90a, MYB90b, and MYB111c TFs all had significant positive correlations with the levels of one or more cyanidins. However, the expression levels of MYB111a, MYC2b, and MYC2c all had significant negative correlations with the levels of one or more cyanidins. Among them, the expression of MYB90a (Cluster-28756.143139) was significantly positively correlated with the levels of five types of cyanidins [cyanidin-3-O-glucoside (kuromanin), cyanidin-3-O-galactoside, cyanidin-3-O-(6′′-O-acetyl)glucoside, cyanidin-3,5-O-diglucoside (cyanin), and cyanidin-3-O-(3′′,6′′-O-dimalonyl)glucosidec], while there was a significant negative correlation between the expression of MYC2b and the levels of these five types of cyanidins (Supplementary Figure 9). MYB90a was only highly expressed in LS in the four stages, indicating that the MYB90a TF might positively regulate anthocyanin biosynthesis (cyanidins) in LS. In contrast, the expression of MYC2b was only observed in JS during the four stages, indicating that the MYC2b TF might negatively regulate anthocyanin biosynthesis (cyanidins) in JS.
Identification of Anthocyanin Synthesis-Related Differentially Expressed Genes in LS by Weighted Gene Co-expression Network Analysis
To investigate the gene regulatory network of anthocyanin accumulation in LS, WGCNA was conducted using the FPKM values of the overlapping DEGs (2,140 out of 7,483 DEGs, FPKM > 1 at least in one stage in LS) and DAFs (10 accumulated anthocyanins in LS) as the source data. A total of 24 DEG modules were identified in the cluster dendrogram (Figure 4A), four of which MEgreen, MEdarkgreen, MEmagenta, and MEroyablue were highly correlated with the contents of more types of anthocyanins (Figure 4B). To explore the expression patterns of the DEGs in these modules, heatmaps were generated using the FPKM values of the DEGs in the four modules. The heatmap results of the four modules showed that most DEGs were expressed at higher levels in the flowering stages (stages 2–4) (Supplementary Figure 10). Moreover, the interaction network diagram was constructed between the hub genes (top 20 DEGs with WGCNA weights of each module) in the MEgreen (weight > 3.44), MEdarkgreen (weight > 0.65), MEmagenta (weight > 3.29), and MEroyablue (weight > 2.04) modules and the anthocyanin content. In the modules, 17 DEGs were selected to construct the regulatory network, including five enzyme genes, eight protein-related genes, two flower-related genes, and two genes with unknown functions (Supplementary Table 10). These genes both positively regulated one or more types of DAAs (cyanidin-3,5-O-diglucoside (cyanin), cyanidin-3-O-(3′′,6′′-O-dimalonyl)glucoside, cyanidin-3-O-(6′′-O-malonyl)glucoside, cyanidin-3-O-galactoside, cyanidin-3-O-glucoside (Kuromanin), pelargonidin-3-O-(3′′,6′′-O-dimalonyl)glucoside, pelargonidin-3-O-(6′′-O-malonyl)glucoside, and pelargonidin-3-O-glucosidec). As shown in Figure 4C, cyanidin-3-O-(6′′-O-malonyl)glucoside was at the core of the network diagram, and it was positively regulated by eight genes. Then, pelargonidin-3-O-(6′′-O-malonyl)glucoside, pelargonidin-3-O-(3′′,6′′-O-dimalonyl)glucoside, and cyanidin-3-O-(3′′,6′′-O-dimalonyl)glucoside were positively regulated by five genes. This finding indicated the main contribution of these four DAAs to LS coloration, and the important regulatory role of these 17 genes in LS anthocyanin synthesis was hypothesized.
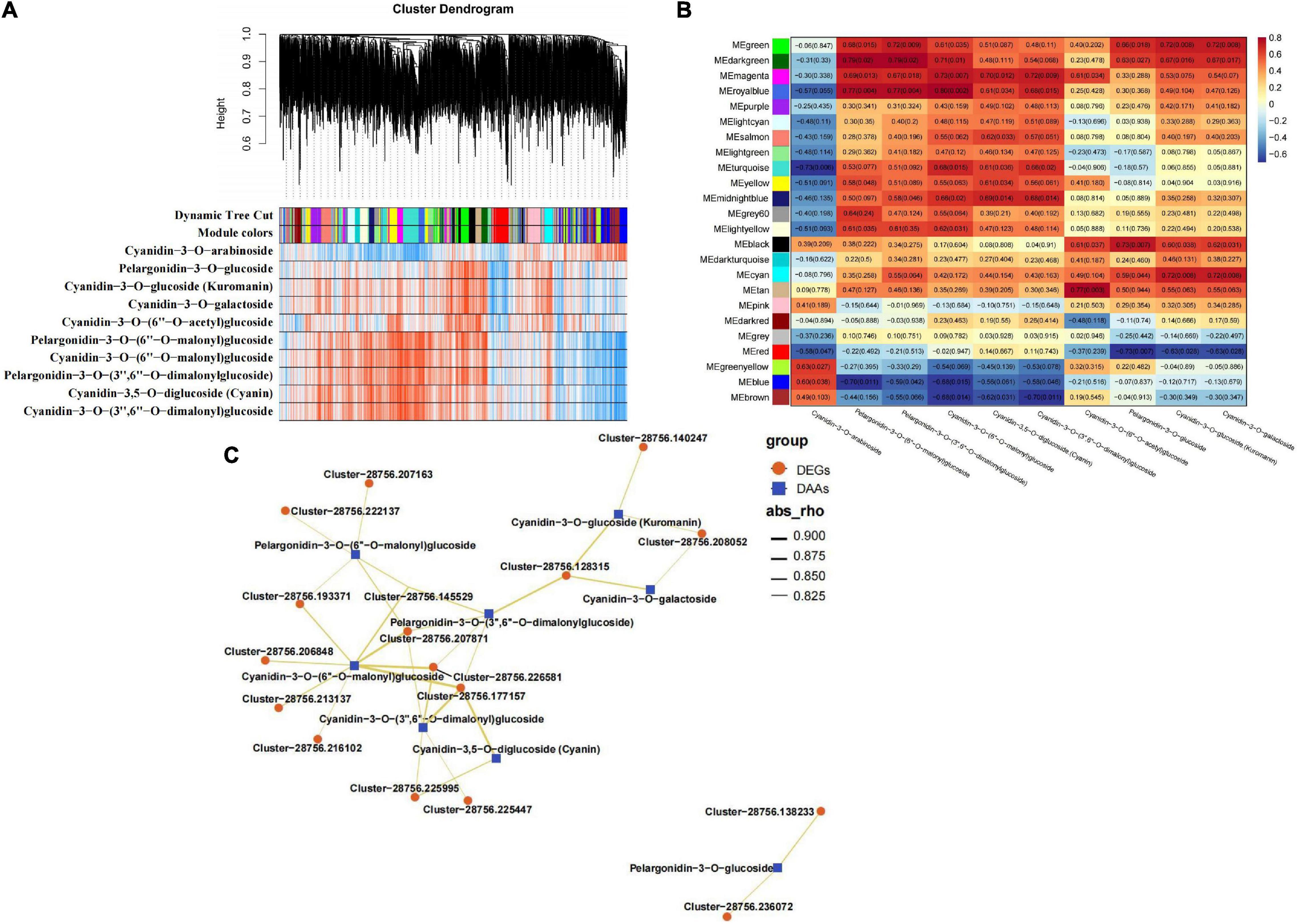
Figure 4. WGCNA results of 2140 DEGs (with 10 DAAs —PCC— > 0.80). (A) Hierarchical clustering tree (cluster dendrogram) results showed 24 expression modules, labeled with different colors. (B) Module-anthocyanin relationship analysis. The value inside each box represents Pearson’s correlation coefficient between the module with anthocyanin, and the number in each parentheses represents p-value. The color scale on the right represents the degree of correlation between modules and anthocyanins and the red represent high correlation. (C) The interaction network diagram between DEGs and 10 DAAs. Red triangles represent DEGs and blue triangles represent DAAs.
Discussion
Identification of Flavonoid Metabolites in Coreopsis tinctoria
Flavonoids contribute in important ways to the medicinal effects of C. tinctoria, and studies on the identification of flavonoids in C. tinctoria are mostly focused on LS (Wang et al., 2015; Shen et al., 2021). However, there are few studies on JS, an important flower color type of C. tinctoria. In this study, the total flavonoid content in LS and JS during four flowering stages was determined. The results showed that there were significant changes in the total flavonoid content in the same or different stages of the two colors of C. tinctoria (LS and JS). In both LS and JS, the total flavonoid content was highest in stage 3, 81.6 and 68.11 mg/g, respectively. Then, 165 flavonoid metabolites in LS and JS during the four stages were identified by LC–MS/MS; these metabolites included 46 flavones, 42 flavonols, 22 anthocyanins, 18 chalcones, 12 dihydroflavonols, and nine isoflavones. The results showed that flavones, flavonols and anthocyanins were the main types of flavonoids in the flowering stages of C. tinctoria. Thirty-three DAFs that exhibited statistically meaningful changes were identified in the LS vs. JS groups, and these DAFs included 11 anthocyanins, 11 flavonols, seven flavonoids, two dihydroflavonols, one chalcone, and one dihydroflavone. There were significant differences in the patterns of accumulation of these DAFs in LS and JS during different stages. The degree of anthocyanin accumulation in LS was higher than that in JS; otherwise, the degree of flavonoid accumulation in JS was higher than that in LS. Compared with the limitations of previous studies that determined the types of flavonoids in C. tinctoria, the results of this study greatly enrich our knowledge of the types and components of flavonoids in C. tinctoria.
Anthocyanin Differences in Coreopsis tinctoria
Anthocyanin is the main chromogenic substance in flavonoids, and cyanidin, pelargonidin and delphinidin are common anthocyanins (Wu et al., 2018) in plants. Usually, cyanidin has a purplish red color, delphinidin has a blue–purple color, and pelargonidin has a brick-red color (Yu et al., 2019). Early on, Shimokoriyama indicated that the red–brown component of the ligulate flower of LS was cyanidin-3-O-glucoside (Shimokoriyama, 1957). In this study, we screened 10 anthocyanins that exhibited significantly different accumulation patterns in LS at the four stages, including seven types of cyanidins [cyanidin-3-O-(6′′-O-acetyl)glucoside, cyanidin-3-O-arabinoside, cyanidin-3-O-glucoside (kuromanin), cyanidin- 3-O-galactoside, cyanidin-3-O-(3′′,6′′-O-dimalonyl)glucoside, cyanidin-3-O-(6′′-O-malonyl)glucoside, and cyanidin-3,5-O-diglucoside (cyanin)] and three types of pelargonidins [pelargonidin-3-O-(3′′,6′′-O-dimalonylglucoside), pelargon idin-3-O-(6′′-O-malonyl) glucoside, and pelargonidin-3-O-glucoside]. The composition and content of anthocyanins greatly affect flower color. Jiang et al. (2020) and Wang F. et al. (2021) observed anthocyanins, such as cyanidin-3-O-glucoside and cyanidin-3,5-O-diglucoside (cyanin), from purple Salvia miltiorrhiza and purple Ficus carica Linn., respectively. Fu et al. (2021) found that cyanidin-3-O-(6′′-O-malonyl)glucoside was the main contributor to the color of Camellia japonica. The results of these studies were similar to those of this study, and these DAAs may be the main reason for the difference in flower color between LS and JS.
Key Structural Genes Responsible for Flavonoid Synthesis in Coreopsis tinctoria
At present, the flavonoid synthesis pathway has been clearly studied in Arabidopsis thaliana and other model plants (Kleindt et al., 2010; Martens et al., 2010). The biosynthesis pathway of flavonoids in most plants is relatively conserved, but the transcription levels of structural genes (PAL, 4CL, C4H, CHS, CHI, F3H, etc.) involved in the flavonoid biosynthesis pathway are different in different plants or in the same plants in different developmental stages and tissues, thus affecting the regulation of flavonoid synthesis (Chen et al., 2021). The anthocyanin synthesis pathway is a branch of the flavonoid biosynthesis pathway, and related studies have focused on different plants with color formation, such as sweet potato (He et al., 2020), radish (Park et al., 2011), and passion fruit (Qiu et al., 2020). In this study, C4H (1), F3H (4), F3′H (5), DFR (3), LDOX (1), and 3GT (2) structural genes that were directly involved in the flavonoid biosynthesis pathway and anthocyanin biosynthesis pathway were identified in C. tinctoria (LS and JS), which was consistent with the results of related studies on Rhododendron (Nakatsuka et al., 2008) and Chinese herbaceous peony (Zhao et al., 2012). Considering the difference in anthocyanin contents between LS and JS in different stages, two genes (the F3H gene, Cluster-28756.299649, and the 3GT gene, Cluster-28756.230942) whose expression was significantly positively correlated with the levels of more types of anthocyanins were identified. F3H is the key enzyme in the flavonoid metabolic pathway, and it can catalyze naringin to produce dihydroflavonol (Castellarin et al., 2007). Dihydroflavonol is the starting substrate of anthocyanin synthesis, and it produces anthocyanins after catalysis by a series of key enzymes. Finally, colored anthocyanins are catalyzed by anthocyanidin 3-O-glucosyltransferases (UFGT/3GT) to combine with glucoside to produce colored anthocyanins. These two DEGs were specifically expressed by LS, especially in stage 3, which was similar to the results obtained in relevant studies of grapes by Mohammad et al. (2011). These two genes promoted the accumulation of anthocyanins in LS. We hypothesize that these two genes may play key roles in anthocyanin accumulation in C. tinctoria. In addition, the expression levels of seven number of HCT genes upstream of flavonoid synthesis were significantly different in LS and JS, which is similar to the screening results of Wang R. et al. (2021) in safflower. Correlation analysis with DAFs showed that the expression levels of three number of HCT genes (Cluster-28756.81340, Cluster-28756.178701, and Cluster-28756.112851) were statistically significantly correlated with the levels of a variety of flavones, flavonols, and anthocyanins, indicating that the HCT gene promotes the accumulation of flavonoids.
Transcription Factors and Other Candidate Genes Related to Anthocyanin Synthesis
Plant flavonoid biosynthesis is affected not only by the expression levels of structural genes but also by the levels of MYB, bHLH, and WD40 TFs and their MYB–bHLH–WD40 (MBW) complex (Hichri et al., 2011; Liu et al., 2021). This study found that the number of MYB TFs (including MYB-related TFs) in C. tinctoria in different stages was the highest, and some bHLH and bZIP TFs were also identified, but no WD40 TFs were identified, indicating that MYB was the main TF category that regulates the synthesis of flavonoids in C. tinctoria. MYB TFs can be divided into 1R-MYB (1R-MYB and MYB-related), R2R3-MYB, 3R-MYB, and 4R-MYB (Niu et al., 2016) according to the number of highly conserved DNA-binding domain repeats they contain. At present, most relevant studies focus on the R2R3-MYB category, such as apple MdMYB23 (An et al., 2018), strawberry MYB10 (Castillejo et al., 2020) and grape VvMYB5a (Deluc et al., 2006), which regulate anthocyanins and flavonols. In this study, the expression of MYB90a (Cluster-28756.143139) was associated with the levels of five cyanidins and was highly expressed in LS. Bac-Molenaar et al. (2015) found that MYB90 was the key TF with phenotypes ranging from green to completely red/purple rosettes in Arabidopsis thaliana, and MYB90 could regulate flower color of Arabidopsis thaliana (Stracke et al., 2001; Niu et al., 2016), and it is speculated that MYB90 may be involved in the color accumulation of LS (Reddish-brown).
The formation of anthocyanins depends on glycosylation, hydroxylation, acylation, and methoxylation to maintain stability, and this process is controlled by some transporters and other proteins (Fu et al., 2021). In this study, key genes involved in the color formation of LS were identified by WGCNA, and 17 genes exhibiting the highest correlation were selected to construct a network diagram. The results showed that these eight anthocyanins were regulated by 2–8 genes. In WGCNA, one CHY1 gene (Cluster-28756.128315), two elf-5A genes (Cluster-28756.207163; Cluster-28756.207871), one LOC107827976 gene (Cluster-28756.222137), two guaA genes (Cluster-28756.216102; Cluster-28756.177157), one LOS1 gene (Cluster-28756.206848), two UTP7 genes (Cluster-28756.225447; Cluster-28756.225995), one EMB2768 gene (Cluster-28756.145529), one L195_g025361 (Cluster-28756.193371), and one TOPP6 gene (Cluster-28756.226581) were shown to target one, one, four, two, one, three, one, one, one, four, two, and three anthocyanins, respectively. These results indicate that these 17 genes might play important roles in color formation in LS, but the specific functions need to be further studied.
Conclusion
In summary, we comprehensively compared the transcriptomes and metabolite profiles of two colors of C. tinctoria flowers (LS and JS) at different developmental stages. The flavonoid metabolites (anthocyanins) and DEGs of C. tinctoria were revealed, and a few candidate genes related to the main DAFs/DAAs were identified. Our results will provide new information about the synthesis of secondary metabolites, the regulation of flower color of C. tinctoria, and the functional identification of key genes of interest.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, bioproject/PRJNA698724.
Author Contributions
HJ conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and tables, authored or reviewed drafts of the manuscript, and approved the final draft. ZL performed the experiments, analyzed the data, and approved the final draft. XJ collected the samples and approved the final draft. YQ conceived and designed the experiments, authored or reviewed drafts of the manuscript, and approved the final draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Xinjiang Autonomous Region Graduate Research and Innovation Project (XJ2020G138); Xinjiang Uygur Autonomous Region Key Discipline Funding Project (2016-10758-3); and National Natural Science Foundation of China (31360319).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.810422/full#supplementary-material
Footnotes
- ^ http://www.anpel.com.cn/
- ^ http://www.shimadzu.com.cn/
- ^ http://www.appliedbiosystems.com.cn/
- ^ http://trinityrnaseq.sourceforge.net/
- ^ https://code.google.com/p/corset-project/
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://xfam.org/
- ^ http://www.expasy.ch/sprot
- ^ http://www.genome.jp/kegg
- ^ http://www.geneontology.org
- ^ http://www.ncbi.nlm.nih.gov/COG/
- ^ https://www.ncbi.nlm.nih.gov/bioproject/PRJNA698724
- ^ http://bowtie-bio.sourceforge.net
References
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
Bac-Molenaar, J. A., Fradin, E. F., Rienstra, J. A., Vreugdenhil, D., and Keurentjes, J. J. B. (2015). GWA Mapping of Anthocyanin Accumulation Reveals Balancing Selection of MYB90 in Arabidopsis thaliana. PLoS One 10:e0143212. doi: 10.1371/journal.pone.0143212
Castellarin, S. D., Peterlunger, E., Sivilotti, P., Degan, M., Pfeiffer, A., and Gaspero, G. (2007). Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 30, 1381–1399. doi: 10.1111/j.1365-3040.2007.01716.x
Castillejo, C., Waurich, V., Wagner, H., Ramos, R., Oiza, N., Munoz, P., et al. (2020). Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) Fruit. Plant Cell 32, 3723–3749. doi: 10.1105/tpc.20.00474
Chen, C., Zhou, G., Chen, J., Liu, X., Lu, X., Chen, H., et al. (2021). Integrated metabolome and transcriptome analysis unveils novel pathway involved in the formation of yellow peel in cucumber. Int. J. Mol. Sci. 22:1494. doi: 10.3390/ijms22031494
Chen, G., Pa, L. D., Zhu, J., Fan, C. Z., Wu, M. Y., and Li, X. J. (2013). Effect of site condition on quality of Kunlun snow chrysanthemum. Mod. Chin. Med. 15, 1060–1063. doi: 10.13313/j.issn.1673-4890.2013.12.003
Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511. doi: 10.1104/pp.105.067231
Du, D., Yao, L. B., and Zhang, R. (2018). Protective effects of flavonoids from Coreopsis tinctoria Nutt. on experimental acute pancreatitis via Nrf-2/ARE-mediated antioxidant pathways. J. Ethnopharmacol. 224, 261–272. doi: 10.1016/j.jep.2018.06.003
Du, J., and Wu, Z. H. (2015). Using HPLC to determine the Content of chlorogenic acid and rutin in Coreopsis tinctoria Nutt. from different areas. J. Henan Univ. Technol. 2, 71–74. doi: 10.16433/j.cnki.issn1673-2383.2015.02.037
Edwin, B. S., and Hampton, M. P. (1971). Biosystematic study of Coreopsis tinctoria and C. Cardaminefolia (Compostitae). Brittonia 23, 161–170.
Fu, M. Y., Yang, X., Zheng, J. R., Wang, L., Yang, X. Y., Tu, Y., et al. (2021). Unraveling the regulatory mechanism of color diversity in camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 12:685136. doi: 10.3389/fpls.2021.685136
Gu, N. N., Qin, Y., Liang, H. B., and Chen, A. X. (2015). Analysis of the effects of different harvest periods on the quality of Coreopsis tinctoria Nutt. LingShizhen Med. Medica Res. 26, 2247–2250. doi: 10.3969/j.issn.1008-0805.2015.09.078
Guo, L. M., Zhang, P., Zhang, Q., and Li, S. M. (2014). Recent advances in the analysis, extraction, characterization and biological activities of chemical substances of Kunlun chrysanthemum (Coreopsis tinctoria). Food Sci. 35, 298–303. doi: 10.7506/spkx1002-6630-201407058
Guo, L., Zhang, W., Li, S., and Ho, C. T. (2015). Chemical and nutraceutical properties of Coreopsis tinctoria. J. Funct. Foods 13, 11–20. doi: 10.1016/j.jff.2014.11.011
Havsteen, B. H. (2002). The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96, 67–202. doi: 10.1016/S0163-7258(02)00298-X
He, L., Liu, X., Liu, S., Zhang, J., Zhang, Y., Sun, Y., et al. (2020). Transcriptomic and targeted metabolomic analysis identifies genes and metabolites involved in anthocyanin accumulation in tuberous roots of sweetpotato (Ipomoea batatas L.). Plant Physiol. Biochem. 156, 323–332. doi: 10.1016/j.plaphy.2020.09.021
Henry-Kirk, R. A., McGhie, T. K., Andre, C. M., Hellens, R. P., and Allan, A. C. (2012). Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. J. Exp. Bot. 63, 5437–5450. doi: 10.1093/jxb/ers193
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Jiang, T., Zhang, M., Wen, C., Xie, X., Tian, W., Wen, S., et al. (2020). Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. flowers. BMC Plant Biol. 20:349. doi: 10.1186/s12870-020-02553-7
Jing, Y. X., and Lan, W. (2012). Determination of total flavonoids in Kunlun snow chrysanthemum from different producing areas. Xinjiang J. Tradit. Chin. Med. 30, 62–64.
Kleindt, C. K., Stracke, R., Mehrtens, F., and Weisshaar, B. (2010). Expression analysis of flavonoid biosynthesis genes during Arabidopsis thaliana silique and seed development with a primary focus on the proanthocyanidin biosynthetic pathway. BMC Res. Notes 3:255. doi: 10.1186/1756-0500-3-255
Liang, H. B., Qin, Y., Jia, S. N., and Chen, A. X. (2016). Effects of phosphate fertilizer application on the growth and quality of Coreopsis tinctoria Nutt. Jiangsu Agric. Sci. 44, 248–250. doi: 10.15889/j.issn.1002-1302.2016.10.068
Liu, K. Y., Wang, M. L., Xin, H. B., Zhang, H., Cong, R. C., and Huang, D. Z. (2021). Anthocyanin biosynthesis and regulate mechanisms in plants: a Review. Chin. Agric. Sci. Bull. 37, 41–51.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, M. D., Ma, M. Q., and Zhao, X. T. (2019). The main active ingredient and its function and detection method of Coreopsis tinctoria Nutt. Bot. Res. 8, 286–292. doi: 10.12677/br.2019.83037
Martens, S., Preuß, A., and Matern, U. (2010). Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71, 1040–1049. doi: 10.1016/j.phytochem.2010.04.016
Mohammad, B. A., Susanne, H., Chen, S. W., Wang, Y. C., Oliver, Y., Laszlo, G. K., et al. (2011). Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol. 11:7. doi: 10.1186/1471-2229-11-7
Nakatsuka, A., Mizuta, D., Kii, Y., Miyajima, I., and Kobayashi, N. (2008). Isolation and expression analysis of flavonoid biosynthesis genes in evergreen azalea. Sci. Hortic. 118, 314–320. doi: 10.1016/j.scienta.2008.06.016
Niu, Y. L., Jiang, X. M., and Xu, X. Y. (2016). Reaserch advances on transcription factor MYB gene family in plant. Mol. Plant Breed. 14, 2050–2059. doi: 10.13271/j.mpb.014.002050
Park, N. I., Xu, H., Li, X., Jang, I. H., Park, S., Ahn, G. H., et al. (2011). Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus). J. Agric. Food Chem. 11, 491–496. doi: 10.1007/s10142-011-0229-0
Pelletier, M. K., and Shirley, B. W. (1996). Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Plant Physiol. 11, 339–345. doi: 10.1104/pp.111.1.339
Qiu, W., Su, W., Cai, Z., Dong, L., Li, C., Xin, M., et al. (2020). Combined analysis of transcriptome and metabolome reveals the potential mechanism of coloration and fruit quality in yellow and purple passiflora edulis sims. J. Agric. Food Chem. 68, 12096–12106. doi: 10.1021/acs.jafc.0c03619
Routaboul, J. M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Einhorn, J., et al. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224, 96–107. doi: 10.1007/s00425-005-0197-5
Saccenti, E., Hoefsloot, H. C. J., Smilde, A. K., Westerhuis, J. A., and Hendriks, M. M. W. B. (2013). Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 10, 361–374. doi: 10.1007/s11306-013-0598-6
Shen, J., Hu, M., Tan, W., Ding, J., Jiang, B., Xu, L., et al. (2021). Traditional uses, phytochemistry, pharmacology, and toxicology of Coreopsis tinctoria Nutt.: a review. J. Ethnopharmacol. 269:113690. doi: 10.1016/j.jep.2020.113690
Shimokoriyama, M. (1957). Anthochlor pigments of Coreopsis tinctoria. J. Am. Chem. Soc. 79, 214–220. doi: 10.1021/ja01558a057
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, Z. Z., Abuduklimu, B., Zhang, Y. L., and Simayil, A. (2012). Quantitate Chemical Composition and Chemical Pattern Recognition in Coriopsis ticntoria from Different Areas. Chin. J. Exp. Tradit. Med. Formulae 18, 174–177.
Wan, L., Lei, Y., Yan, L., Liu, Y., Pandey, M. K., Wan, X., et al. (2020). Transcriptome and metabolome reveal redirection of flavonoids in a white testa peanut mutant. BMC Plant Biol. 20:161. doi: 10.1186/s12870-020-02383-7
Wang, F., Ji, G. S., Xu, Z. B., Feng, B., Zhou, Q., Fan, X. L., et al. (2021). Metabolomics and transcriptomics provide insights into anthocyanin biosynthesis in the developing grains of purple wheat (Triticum aestivum L.). J. Agric. Food Chem. 69, 11171–11184. doi: 10.1021/acs.jafc.1c01719
Wang, R., Ren, C., Dong, S., Chen, C., Xian, B., Wu, Q., et al. (2021). Integrated metabolomics and transcriptome analysis of flavonoid biosynthesis in safflower (Carthamus tinctorius L.) with different colors. Front. Plant Sci. 12:712038. doi: 10.3389/fpls.2021.712038
Wang, W., Chen, W., Yang, Y., Liu, T., Yang, H., and Xin, Z. (2015). New phenolic compounds from Coreopsis tinctoria Nutt. and their antioxidant and angiotensin i-converting enzyme inhibitory activities. J. Agric. Food Chem. 63, 200–207. doi: 10.1021/jf504289g
Wen, W., Alseekh, S., and Fernie, A. R. (2020). Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol. 55, 100–108. doi: 10.1016/j.pbi.2020.04.004
Wu, X. X., Zhang, A. D., Zhu, Z. W., Yao, J., Zha, D. S., and Li, X. (2018). Research on plants anthocyanin biosynthesis metabolic pathway and its regular factors. Acta Agric. Shanghai 34, 127–132. doi: 10.1007/s00425-014-2131-1
Xiao, L., Cao, S., Shang, X., Xie, X., Zeng, W., Lu, L., et al. (2021). Metabolomic and transcriptomic profiling reveals distinct nutritional properties of cassavas with different flesh colors. Food Chem. Mol. Sci. 2:100016. doi: 10.1016/j.fochms.2021.100016
Yang, Y., Sun, X., Liu, J., Kang, L., Chen, S., Ma, B., et al. (2016). Quantitative and qualitative analysis of flavonoids and phenolic acids in snow chrysanthemum (Coreopsis tinctoria Nutt.) by HPLC-DAD and UPLC-ESI-QTOF-MS. Molecules 21:1307. doi: 10.3390/molecules21101307
Yu, K., Jun, J. H., Duan, C., and Dixon, R. A. (2019). VvLAR1 and VvLAR2 are bifunctional enzymes for proanthocyanidin biosynthesis in grapevine. Plant Physiol. 180, 1362–1374. doi: 10.1104/pp.19.00447
Zeng, X. M., and He, X. Y. (2019). Research progress on chemical composition and efficacy of Coreopsis tinctoria. Sci. Technol. Food Ind. 13, 335–339.
Zhang, J. (2013). Determination of total flavonoids in artificial and wild Coreopsis tinctoria. Xinjiang J. Tradit. Chin. Med. 31, 49–50.
Zhang, Q., Wang, L., Liu, Z., Zhao, Z., Zhao, J., Wang, Z., et al. (2020). Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chem. 312:125903. doi: 10.1016/j.foodchem.2019.125903
Zhang, X., Song, J. Y., Hu, Y. L., Xu, J., Xu, Z. C., Ji, A. J., et al. (2014). Research progress of the regulation on active compound biosynthesis by the bHLH transcription factors in plants. Acta Pharm. Sin. 49, 435–442.
Keywords: Coreopsis tinctoria Nutt., metabonomic, transcriptomic, flavonoid biosynthesis, anthocyanins, candidate genes
Citation: Jiang H, Li Z, Jiang X and Qin Y (2022) Comparison of Metabolome and Transcriptome of Flavonoid Biosynthesis in Two Colors of Coreopsis tinctoria Nutt. Front. Plant Sci. 13:810422. doi: 10.3389/fpls.2022.810422
Received: 06 November 2021; Accepted: 14 February 2022;
Published: 09 March 2022.
Edited by:
Yanran Li, University of California, Riverside, United StatesReviewed by:
Xiujuan Qi, Zhengzhou Fruit Research Institute (CAAS), ChinaJiang Chen, Chengdu University of Traditional Chinese Medicine, China
Copyright © 2022 Jiang, Li, Jiang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Qin, eGpuZHFpbnlvbmdAMTI2LmNvbQ==
 Hong Jiang
Hong Jiang Zhiyuan Li
Zhiyuan Li