
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 February 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.807342
 Xiangguang Meng1,2,3
Xiangguang Meng1,2,3 Yuzheng Zhang1,2,3
Yuzheng Zhang1,2,3 Ning Wang1,2,3
Ning Wang1,2,3 Huajie He1,2,3
Huajie He1,2,3 Qiuping Tan1,2,3
Qiuping Tan1,2,3 Binbin Wen1,2,3
Binbin Wen1,2,3 Rui Zhang1,2,3
Rui Zhang1,2,3 Mingyue Sun1,2,3
Mingyue Sun1,2,3 Xuehui Zhao1,2,3
Xuehui Zhao1,2,3 Xiling Fu1,2,3
Xiling Fu1,2,3 Dongmei Li1,2,3
Dongmei Li1,2,3 Wenli Lu1,2,3
Wenli Lu1,2,3 Xiude Chen1,2,3*
Xiude Chen1,2,3* Ling Li1,2,3*
Ling Li1,2,3*
Terpene synthase (TPS) is related to the production of aromatic substances, but there are few studies on the impact of abiotic stress on TPS and its molecular mechanism, especially in peaches. This study found that salt resistance and abscisic acid (ABA) sensitivity of transgenic tomatoes were enhanced by overexpression of PpTPS1. Moreover, it was found that PpTPS1 interacted with and antagonized the expression of the bZIP transcription factor ABA INSENSITIVE 5 (PpABI5), which is thought to play an important role in salt suitability. In addition, PpTCP1, PpTCP13, and PpTCP15 were found to activate the expression of PpTPS1 by yeast one-hybrid (Y1H) and dual-luciferase assays, and they could also be induced by ABA. In summary, PpTPS1 may be involved in the ABA signaling regulatory pathway and play an important role in salt acclimation, providing a new reference gene for the improvement of salt resistance in peaches.
China is a prominent country in the global peach industry, and peach fruit tree cultivation is an important industry for improving the disposable income of the farmers (Deng et al., 2018). Salt damage is an important issue affecting the development of global agriculture in the 21st century, and the saline land area in China is as high as 99 million hm2. As a salt-sensitive, non-saline plant, peach trees can be seriously affected by salt stress in terms of growth, development, fruit yield, and quality of fruit trees (Munns, 2011; Zhu et al., 2018). With bioengineering and gene assembly technology techniques, genetic transformation of salt tolerance genes and molecular biomarker breeding have become important means of improving salt tolerance in plants (Sun et al., 2018). With this process, it has become particularly important to screen excellent, salt-tolerance candidate genes and study their molecular regulatory networks (Nayyeripasand et al., 2021).
Terpene synthases (TPSs) are key enzymes for terpene formation, and use isopentenyl diphosphates of different lengths as substrates. Most plant TPSs have multiple substrates and exhibit catalytic activity, which is an important reason for the rich variety of terpenoids (Chen et al., 2011). TPSs generally have 550–850 amino acid residues, including a C-terminal active structural domain and an N-terminal structural domain (Bohlmann et al., 1998; Chen et al., 2003; Dudareva et al., 2003). Leesburg found that almost all TPSs have a conserved aspartic acid-rich structural domain DDXXD, which plays an important role in the binding of divalent metal ions to substrates (Lesburg et al., 1997). Current studies on TPSs have focused on aroma quality and bio-resistance, and they showed transfer of linalool synthase from fanwort in tomatoes to increase the linalool content in ripe fruits (Lewinsohn et al., 2001); overexpression of geraniol synthase from lemon in tomato was shown to increase the geraniol content (Davidovich-Rikanati et al., 2007); and limonene synthase inhibition in citrus was shown to enhance the resistance of navel orange fruit to Penicillium infestation (Rodriguez et al., 2011, 2014). However, there are few studies on the impact of abiotic stress on TPSs. Mafu et al. (2018) performed oxidative stimulation experiments on maize roots with 1 mM CuSO4 artificially simulating abiotic stress factors and found that dolabralexin-like diterpene accumulated continuously in the roots for 48 h. ZmTPS6 could respond to abscisic acid (ABA) signaling, and overexpression of ZmTPS6 increased drought resistance in maize (Hou et al., 2017; Lee, 2018). The transcription factors FhMYB21 and FhMYC2 of the MYB and MYC families, which are thought to be associated with biotic stress, regulate the expression of FhTPS1 in A. fragrans (Yang et al., 2020). Peach TPS1 was associated with the synthesis of linalool (Liu et al., 2017), so determining whether it is involved in abiotic stresses was of interest.
Abscisic acid can act as a stress hormone (Larosa et al., 1985; Leung and Giraudat, 1998), and under salt stress, ABA can alleviate and mitigate high salt damage to the plant body by inducing a large accumulation of osmolytes in the plant (Shahid et al., 2015; Sharma et al., 2019). It has been demonstrated that ABA alleviates salt stress by regulating target proteins through a series of signal transduction pathways, including ABA receptor recognition, intracellular signaling, and phosphorylation of corresponding transcription factors, to regulate the expression of related genes (Sreenivasulu et al., 2012; Fujii, 2014). ABI5 is a class of bZIP transcription factors downstream of ABA, which can be activated by phosphorylation and involved in stress regulation (Dai et al., 2013). Exogenous application of ABA or stress conditions such as high salt and drought can lead to the induction of ABI5 expression (Lopez-Molina et al., 2001, 2010). Coexpression of Arabidopsis ABI3 and ABI5 in cotton was able to improve cotton resistance to drought stress by regulating reactive oxygen species cluster scavenging and the expression of osmoregulatory marker genes (Mittal et al., 2014). In addition, studies of rice and wheat have also revealed that ABI5 is associated with the plant response to adversity stress (Yang et al., 2010, 2011). ABI5 is involved in the biological process of the plant response to adverse stress and plays an important role in the process of plant adaptation to salt.
In the present study, we overexpressed PpTPS1 in tomatoes and found that ABA sensitivity and resistance were enhanced in transgenic tomatoes. We verified that PpTPS1 interacts with PpABI5 and that PpTCP1, PpTCP13, and PpTCP15 can activate the expression of PpTPS1, and hence we hypothesized that PpTCP1, PpTCP13, PpTCP15, and PpTPS1 play important roles in salt adaptation and their functions are closely related to the ABA signaling pathway.
Beginning in September 2019, samples of the fruit, roots, leaves, flowers, and stems of a 6-year-old “Chun Xue” peach tree were obtained from the Horticulture Experimental Station of Shandong Agricultural University. Tissue samples were stored at −80°C before RNA extraction.
The transgenic tomato variety was “Micro Tom” purchased from Nanjing Fengtai Horticultural Seed (Nanjing, China) Co. Tomato seeds were sterilized, soaked in warm water, germinated on Murashige and Skoog (MS) medium for 3 days, and then used for Agrobacterium infection. The seeds used in the germination rate experiment were treated in the same way. Annual hickory peach seedlings were provided by the National Apple Engineering Center Laboratory. A 3:1:1 mixture of grass charcoal, vermiculite, and perlite was used as a substrate for each tomato material and annual peach seedling planted in a 12-cm-wide square plastic culture bowl. Tomato and annual peach seedlings were cultivated in a growth chamber in which the temperature, relative humidity, photoperiod, and photosynthetic photon flux density were 26°C/20°C, 65%, 14 h/10 h (day/night), 300 μmol m–2 s–1, respectively. The leaves of the annual peach seedlings were sampled in separate periods after spraying with 100 μM ABA, at 4°C, root soaking with 10% PEG, and watering with 150 mM NaCl. Thirty milliliters of 150 mM NaCl solution was applied to the transgenic tomato substrate every afternoon and the wild type (WT) was treated with water for 12 days. The leaves of transgenic tomatoes and the WT were then taken for physiological data analysis.
RNA from the samples was extracted using the RNA Pure Plant Kit (DNase I) (CW0559) and first strand cDNA was obtained using the SuperRT cDNA Synthesis Kit (CW0741) for qRT-PCR. An Ultra SYBR Mixture Kit (CW0957) was used for qRT-PCR analysis, and the primers are listed in Supplementary Table 1. PpActin (Prupe.6G163400) is an internal reference gene of peach (Zhao et al., 2020), and SlAction (Solyc11g005330) is the internal reference gene of tomato (Hu et al., 2021). These kits were purchased from Kangwei Century Biotechnology (Jiangsu) Co., and primer synthesis was performed by Biotech Bioengineering (Shanghai) Co.
Primers (5′- ATGGCATTGTTTTCTATGGCCA-3′; 5′- CACAGATTCAGTTTCATACAGCATCG-3′) were designed by searching the NCBI database using the cDNA sequence Prupe.4G030400 (PpTPS1). The amplification reaction was performed using the cDNA of “Chun Xue” peach as the template, and the amplified products were sequenced and compared with the NCBI database. PpTPS1 was cloned into the plant expression vector PBI121-GFP using SmaI single enzyme digestion (Supplementary Figure 1). The recombinant plasmid was transformed into GV3101 Agrobacterium using the freeze-thaw method, and the transformed Agrobacterium was then used to infect the “Micro Tom” tomato explants (Jian et al., 2019). After induction of germination, rooting, and identification, overexpressed transgenic tomatoes were obtained.
Tomato leaves were taken after treatment to measure the relative water content, relative electrical measurements, and MDA contents, and the analysis was performed as described previously by Roger (2001). The proline, soluble protein, and soluble sugar contents were measured in the treated tomato leaves, according to Zhuang et al. (2019).
NCBI was used to design specific primers for PpABI5 (5′-GAGAGTGGTGAATGGGGGTG-3′; 5′-ACCTTTTCCACCGGTCCATC-3′), ligate the amplified PpABI5 fragment with the TRV2 plasmid using restriction enzymes EcoRI and BamHI, and then transfer it into the Agrobacterium GV3101. The PpABI5-TRV2 and TRV2 Agrobacterium liquid were mixed with the Agrobacterium liquid of TRV1 in a 1:1 ratio, and they were injected into the green ripe peach fruit, followed by vacuum treatment for 10 min and dark treatment for 1 day. The sample was then left under normal lighting condition for 3 days. The pulp was used as a sample for qRT-PCR analysis (Zhang et al., 2015).
The peach yeast two-hybrid library was established. Then PpTPS1-pGBKT7 vector was constructed and transferred it into Y2H and cultured on -Trp, -Trp/X-α-gal medium at 30°C to verify the autonomous activation activity. Afterward, the library was initially screened for interacting genes, and the screened PpABI5 (Prupe.7G112200.1) was inserted into the pGADT7 vector (Zhang et al., 2021). The Matchmaker Yeast Two-Hybrid System User Manual was followed to perform mutual verification again.
PpABI5 was cloned into a pGEX-6P-1 vector containing a GST tag, and PpTPS1 was cloned into a pET32a vector containing a His-tag. The vectors were transformed into BL21 DE3 Escherichia coli, and 1 mM isopropyl β-D-thiogalactoside (IPTG) was used to induce the expression of the fusion protein in the bacteria on a shaker at 37°C (Zhao et al., 2020). After the induced PpTPS1-His fusion protein and PpABI5-GST protein were purified by dialysis, the protein was mixed according to the combination of PpTPS-His/GST and PpTPS-His/PpABI5-GST, purified by a GST purification column, and then subjected to Western blot analysis. The GST/His-tagged Protein Purification Kit (P2262/P2229), His/GST Tag Mouse Monoclonal Antibody (AF5060/AF5063), and goat anti-mouse IgG (A0216) used in the experiment were purchased from Shanghai Biyuntian Biotechnology Co.
The PpABI5-pSPYNE and PpTPS1-pSPYCE recombinant plasmids were constructed and transformed into Agrobacterium tumefaciens GV3101. The two bacterial solutions were mixed at a 1:1 ratio to transfect onion epidermal cells for 30 min, and PpABI5-pSPYNE + pSPYCE and PpTPS1-pSPYCE + pSPYNE, 1:1 mixed bacterial liquid, were used as the two groups of controls (Chen et al., 2018). After 48 h of culture in MS medium, the cells were observed with a laser-scanning confocal microscope (LSM880).
PpTPS1100–400 (primers: 5′-AAAGGCAGAGCCATGCTTAG-3′; 5′-TAAAGAGACCCTCGCGGGCT-3′) was amplified using 5′−100 to −400 bp genomic DNA of the 2-kb upstream promoter of PpTPS1 as a template. FastDigest BstBI was used to linearize the plasmid, transform it into yeast one-hybrid (Y1H) Gold, and detect the lowest concentration of AbA that inhibits the growth of bait yeast strains on Ura medium at 30°C. Next, the lowest concentration of AbA was used for the Y1H screening library (Wang et al., 2020). The screening results were sequenced and compared with the sequences in NCBI.
PpTCP1, PpTCP13, and PpTCP15 were inserted into the pGreenII0029 62-SK vector, and the promoter fragment PpTPS1100–400 was inserted into the pGreenII0800-Luc vector (Walter et al., 2004). The vectors were then transformed into A. tumefaciens strain GV3101 pSoup. A mixture of four Agrobacterium strains was injected on the back of tobacco leaves: PpTCP1/PpTCP13/PpTCP15-62SK + PpTPS1-Luc, PpTCP1/PpTCP13/PpTCP15-62SK + Luc, and 62SK + PpTPS1-Luc. After 2–3 days, 1 mM D-luciferin and sodium salt were applied to the backs of the tobacco leaves, and an in vivo imaging system was used to detect fluorescence.
Biological triplicates were performed for each sample, and the data are expressed as the mean ± standard error (SE). Two-way ANOVA was performed using SPSS for Windows 24 when applicable. The qRT-PCR data were analyzed by Duncan’s test. A significance level of P < 0.05 was used.
The leaves, flowers, stems, roots, fruit, and seeds of 6-year-old “Chun Xue” peach trees were used as samples to perform qRT-PCR analysis to study the expression patterns of PpTPS1 in different tissues of peaches. The results showed PpTPS1 had the highest expression in fruit, followed by leaves, and the lowest expression was observed in flowers, indicating that PpTPS1 may be widely involved in a variety of physiological activities in plants (Figure 1A).
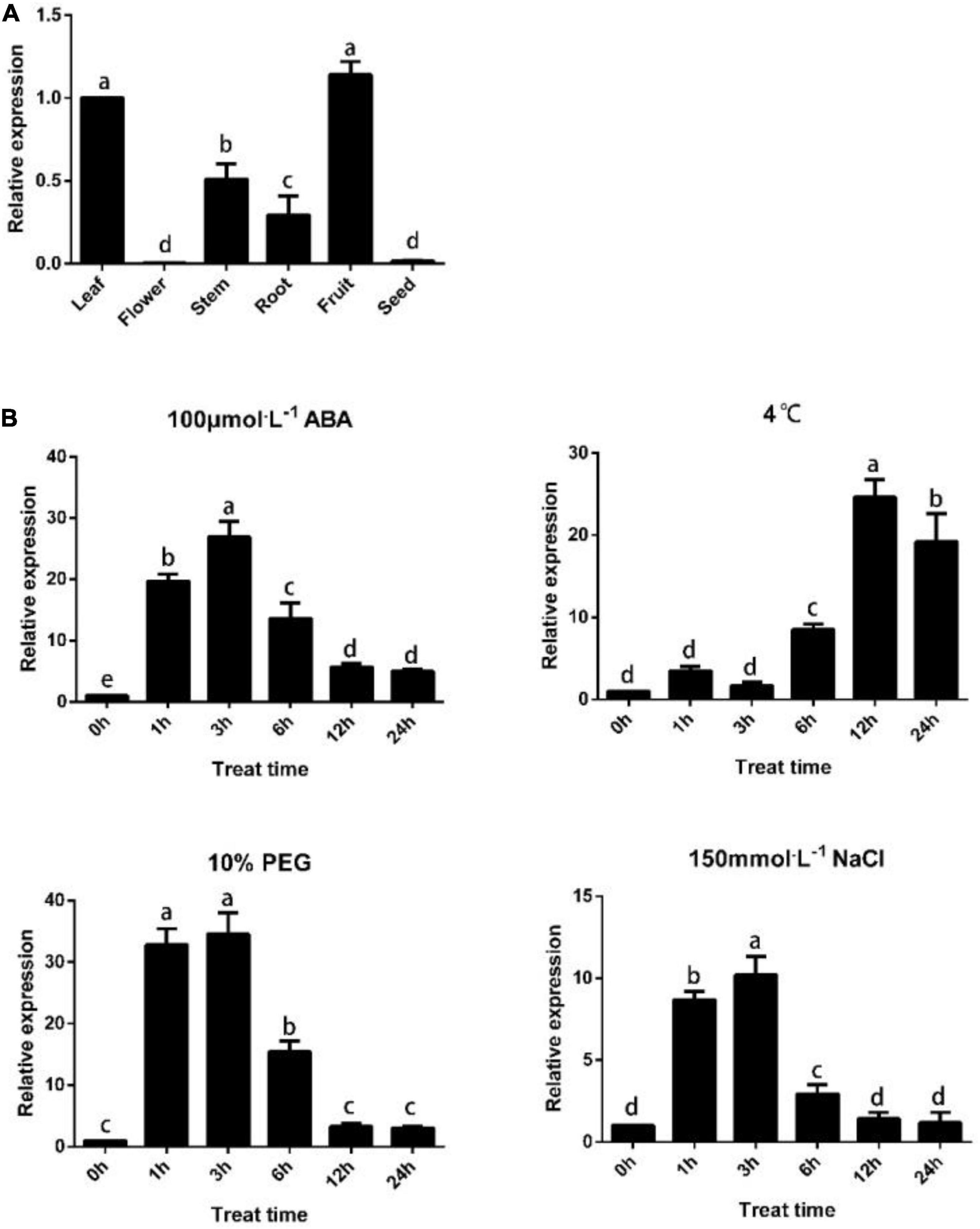
Figure 1. PpTPS1 tissue specificity analysis and expression pattern analysis under abiotic stress. (A) PpTPS1 tissue specificity analysis. (B) Annual peach leaves treated with 100 μM ABA, 4°C, 10% PEG and 150 mM NaCl for 0, 1, 3, 6, 12, and 24 h were used as samples to analyze the expression of the PpTPS1 model. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
Most TPSs genes contain abundant promoter cis-elements related to abiotic stress, but no further research has been conducted. To initially explore whether PpTPS1 is induced by abiotic stress, we analyzed its expression pattern under exogenous ABA, low-temperature, salt, and drought treatments. The results showed that the target gene was significantly induced under the treatment of 100 μM ABA, 4°C, 10% PEG, and 150 mM NaCl. Interestingly, the expression pattern of PpTPS1 under NaCl and PEG treatment was similar to that under ABA treatment, and its expression level increased rapidly at first, reached a maximum at 3 h and then decreased (Figure 1B). This result indicated that PpTPS1 could play a role in most abiotic stresses.
The amplification products of “Chun Xue” were sequenced and compared with the NCBI, and the sequences were consistent with PpTPS1 (Genbank ID: OL855977). The results of RT-PCR (Supplementary Figure 2) and qRT-PCR analyses, showed that PpTPS1 was successfully overexpressed in tomatoes, and three overexpressing tomato lines were obtained (Figure 2A). We treated transgenic and WT “Micro TOM” tomatoes that were grown for 5 weeks and grew consistently with 150 mmol⋅L–1 NaCl solution for 12 days, and the control group was treated with water. As shown in Figure 2B, the leaves of WT tomatoes treated with NaCl showed curling, yellowing, and necrosis, while the transgenic tomatoes grew better, and the higher the expression of PpTPS1 was, the better the plants grew; the control group exhibited no significant difference. We measured the relative water content of transgenic and WT tomatoes after salt treatment, and the results demonstrated that the relative water content of transgenic tomatoes was significantly higher than that of WT tomatoes. The relative electrical and malondialdehyde (MDA) content measurements showed that WT tomatoes suffered significantly more damage than transgenic tomatoes, and the trend was consistent with the expression level (Figure 2C). This result suggests that salt treatment may cause a relatively large disruption of the cellular structure of WT tomatoes compared with transgenic tomatoes, disrupting the ionic balance, but the exact mechanism needs further experimental verification.
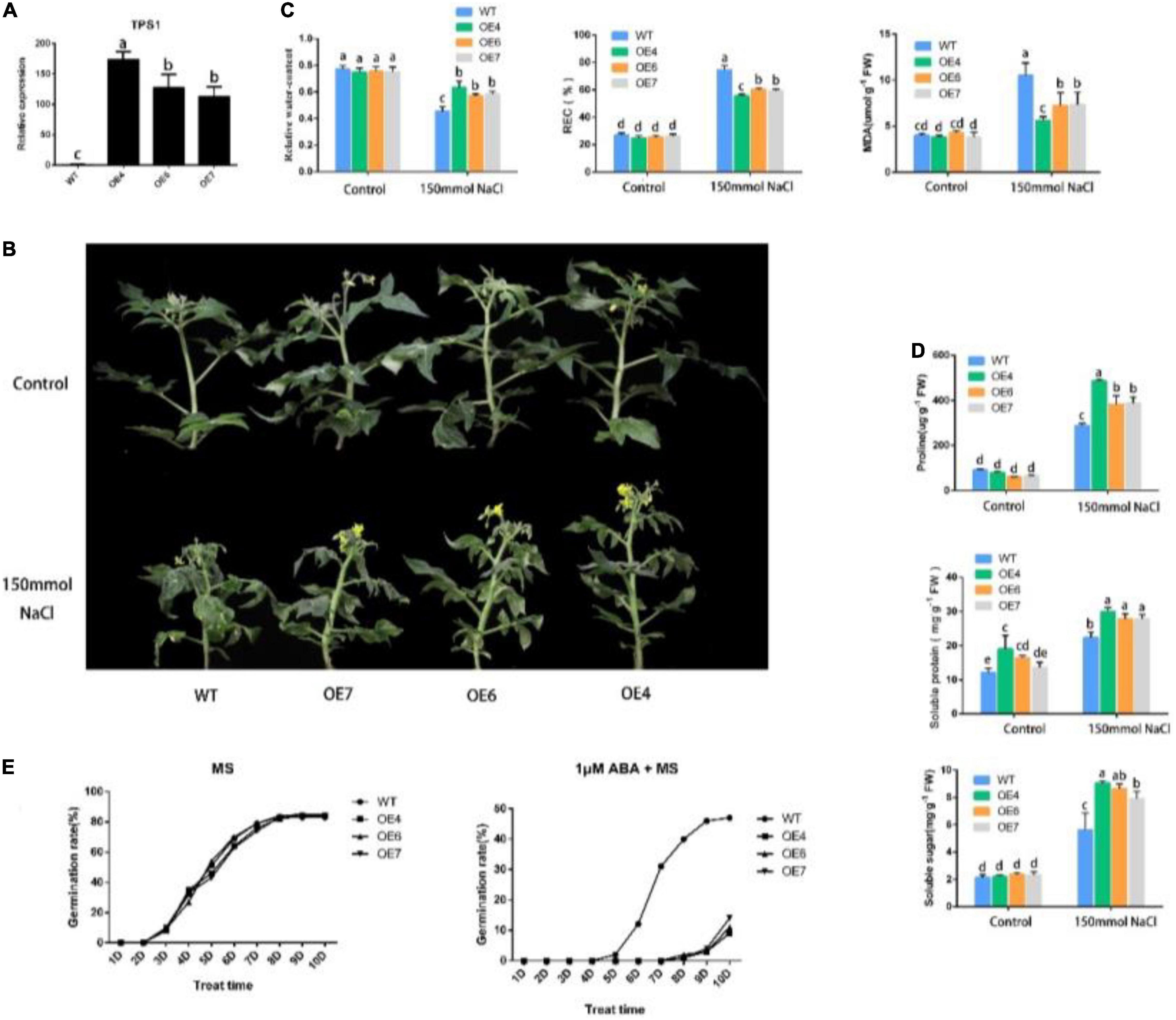
Figure 2. Functional identification of PpTPS1. (A) qRT-PCR analysis of PpTPS1 transgenic tomato lines under normal condition. Transgenic tomato lines under normal condition. (B) Growth status of WT and transgenic tomatoes treated with NaCl for 12 days. (B,C) The leaves of WT and transgenic tomatoes treated with NaCl for 12 days were taken to measure the relative electrical, and MDA contents Growth status of WT and transgenic tomatoes treated with NaCl for 12 days. (C) Analysis of the relative electrical measurement and MDA content and relative water content of WT and transgenic tomatoes treated with NaCl for 12 days. (E) Analysis of the seed germination rate of WT, OE4, OE6, OE7 on MS and MS + 1 μM ABA media for 10 days. Each line contained 100 seeds. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
Content of osmotic adjustment substances can be used as an evaluation index to measure salt tolerance. Numerous studies have shown that plants can increase their resistance to abiotic stress by increasing the content of osmotic adjustment substances. To preliminarily explore the mechanism of PpTPS1 transgenic tomatoes in response to salt stress, we measured the content of its osmotic adjustment substances. The results showed that the proline, soluble protein, and soluble sugar contents of transgenic tomatoes after NaCl treatment were significantly higher than those of the WT tomatoes, especially OE4 (Figure 2D). Therefore, we speculate that the increased content of osmotic regulators may be one reason transgenic tomatoes have better salt resistance.
To explore whether PpTPS1 responds to ABA, we analyzed the seed germination rate of PpTPS1-overexpressing tomatoes under ABA treatment. The results showed that there was no significant difference between the seed germination rate of WT and transgenic tomatoes on a normal MS medium. On 1 μM ABA + MS medium, the seed germination rate of transgenic tomatoes was significantly lower than that of the WT (Figure 2E), and it was opposite to the expression trend. This result shows that overexpression of PpTPS1 can increase the sensitivity of plants to ABA.
After testing for autonomous activation activity, we found PpTPS1 had no autonomous activation activity (Figure 3A). We used PpTPS1-BD as the “bait” to screen its interacting proteins in the library. From the preliminary screening results (Supplementary Table 2), we found that PpABI5 may interact with PpTPS1, and previous studies of ABI5 have shown that it is involved in salt stress. We cloned the full-length PpABI5, constructed the PpABI5-AD vector, and cotransformed Y2H competent yeast with PpTPS1-BD. The results showed that PpTPS1 can interact with PpABI5 (Figure 3B). To further verify the interaction between PpTPS1 and PpABI5, we conducted in vitro protein interaction verification (pull-down) and bimolecular fluorescence complementation (BiFC) analysis (Figures 3C,D), and the results supported the above conclusions.
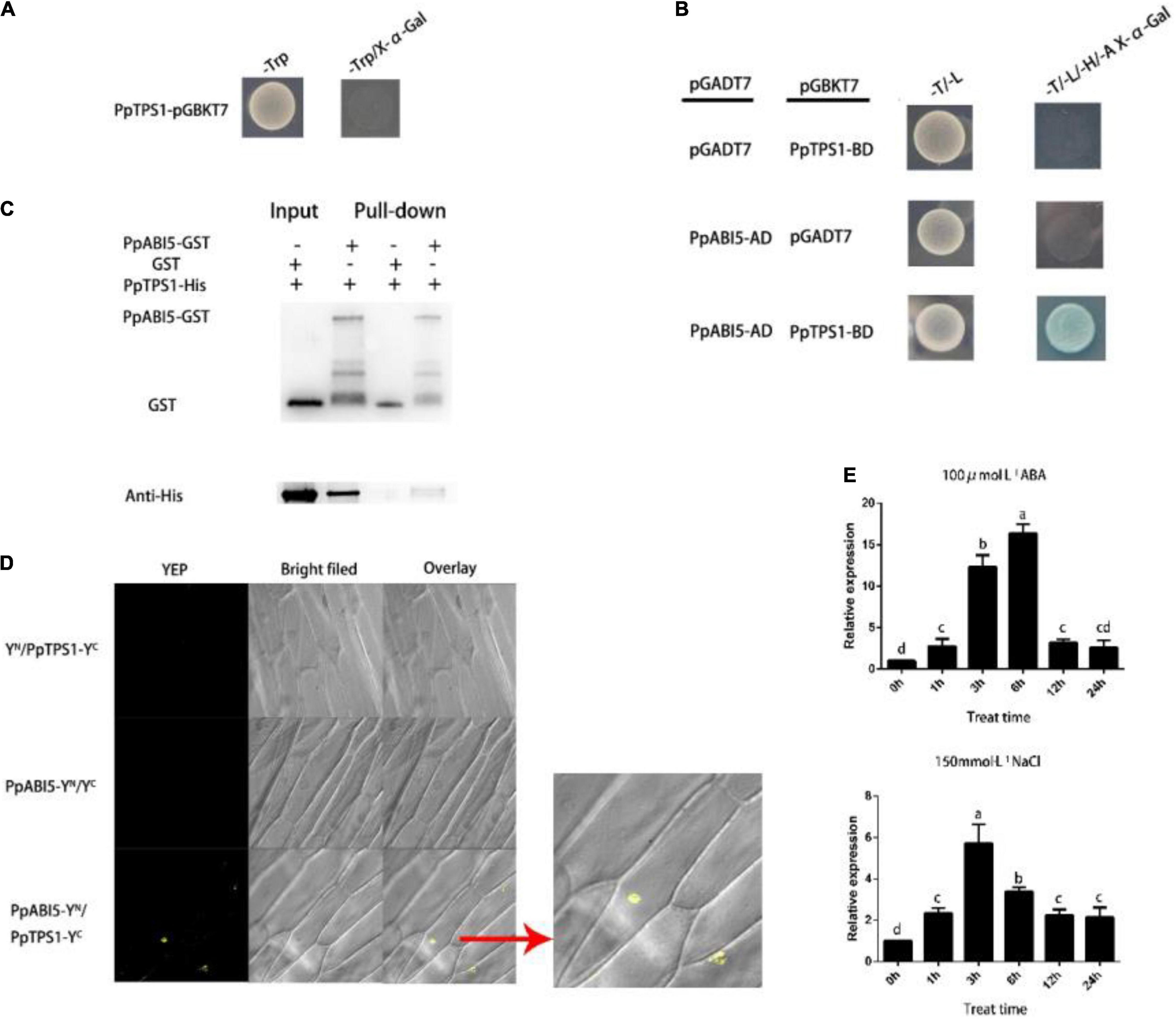
Figure 3. PpABI5 interacts with PpTPS1 and responds to salt stress. (A) PpTPS1 has no autonomous activation activity. (B) Verification of the interaction between PpTPS1 and PpABI5 by a yeast two-hybrid (Y2H) experiment. Yeast containing PpTPS1-BD and PpABI5-AD can grow on DDO (SD/-Leu/-Trp), and they can also grow and turn blue on QDO (SD-Leu/-Trp/-His/-Ade/X-α-Gal). (C) GST pull-down analysis verified the interaction of PpTPS1 and PpABI5. GST, PpTPS1-His, and PpABI5-GST proteins were all purified once before the experiment. (D) Bimolecular fluorescence complementation (BiFC) verification of the interaction between PpTPS1 and PpABI5. (E) Annual peach leaves treated with 100 μM ABA and 150 mM NaCl for 0, 1, 3, 6, 12, and 24 h were used as samples to analyze the expression of the PpABI5 model. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
To verify whether PpABI5 responded to salt stress, we analyzed the expression pattern of PpABI5 after exogenous ABA and NaCl treatment. The results showed that PpABI5 can be induced by salt stress, which is consistent with our hypothesis and the results of previous studies (Figure 3E).
Because of the interaction between PpTPS1 and PpABI5, we were curious about the expression of SlABI5 in PpTPS1-overexpressing tomatoes. The qRT-PCR results showed that the expression of SlABI5 in transgenic tomatoes was significantly lower than that in WT tomatoes (Figure 4A). Then we constructed the PpABI5-TRV2 vector and successfully silenced PpABI5 in peach fruit (Figure 4B). The qRT-PCR results showed that the expression of PpTPS1 in peach fruit was significantly increased after PpABI5 was silenced compared with the control (Figure 4C). This result indicated that there is an antagonistic relationship between PpTPS1 and PpABI5.
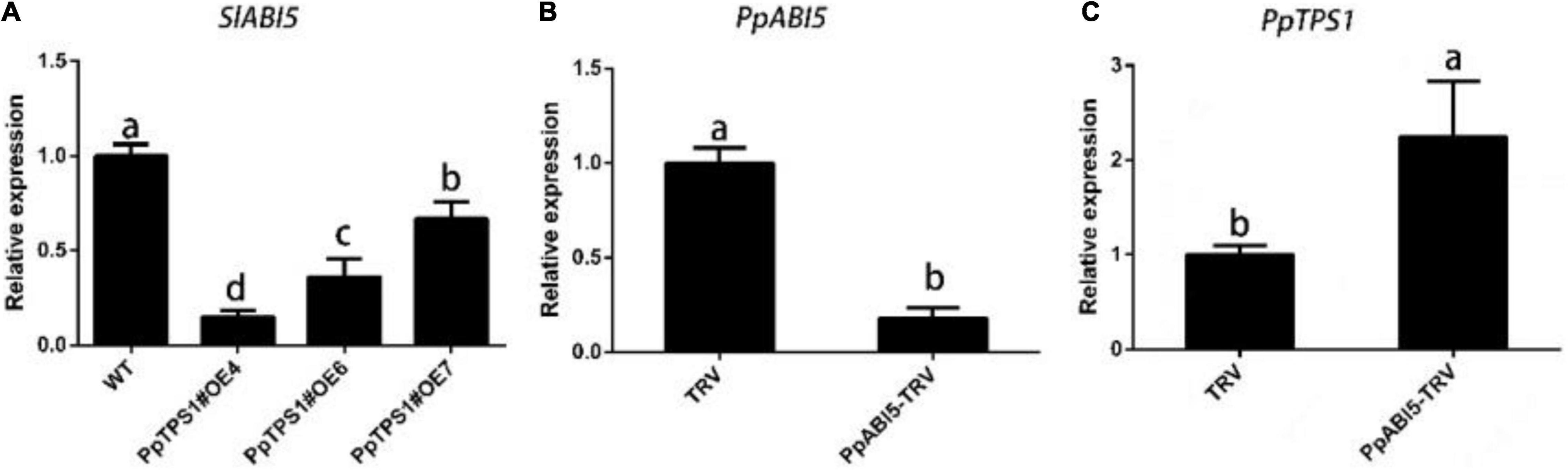
Figure 4. Analysis of the antagonistic relationship between PpTPS1 and PpABI5. (A) Analysis of the expression of the PpABI5 homologous gene SlABI5 in three transgenic tomato lines, OE4, OE6, and OE7. (B) The expression level of PpABI5 after 4 days of transient silencing of PpABI5 in peach fruit by the VIGS system using TRV as a vector. (C) Analysis of PpTPS1 expression after transient silencing of PpABI5. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
To explore the regulatory relationship of PpTPS1 upstream genes, we analyzed the 2-kb promoter sequence upstream of ATG, the translation start site of the PpTPS1 gene, and found that there were multiple TCP-binding motifs, G (T/C) GGNCCCAC motifs, in the 118–378 bp fragment of the promoter (Cubas, 2002; Kosugi and Ohashi, 2002; Li et al., 2005). Previous studies revealed that signal transmission between TCP transcription factors and ABA played an important role in salt stress (Tatematsu et al., 2008). The experiments in the present study showed that the use of 200 ng mL–1 AbA can inhibit the growth of the PpTPS1100–400-pAbAi “bait” yeast strain (Figure 5A). A Y1H screen library was performed using PpTPS1100–400-pAbAi as bait, and the initial screening results contained three TCP family genes: PpTCP1 (Prupe.3G240200.1), PpTCP13 (Prupe.3G252600.1), and PpTCP15 (Prupe.5G191200.1) (Supplementary Table 3). Further verification revealed that the PpTPS1100–400-pAbAi “bait” yeast transferred into PpTCP1-AD, PpTCP13-AD, and PpTCP15-AD plasmids grew normally on SD/-Ura-Leu/300 ng⋅mL–1 AbA, indicating that PpTCP1, PpTCP13, and PpTCP15 could bind to the PpTPS1 promoter (Figure 5B).
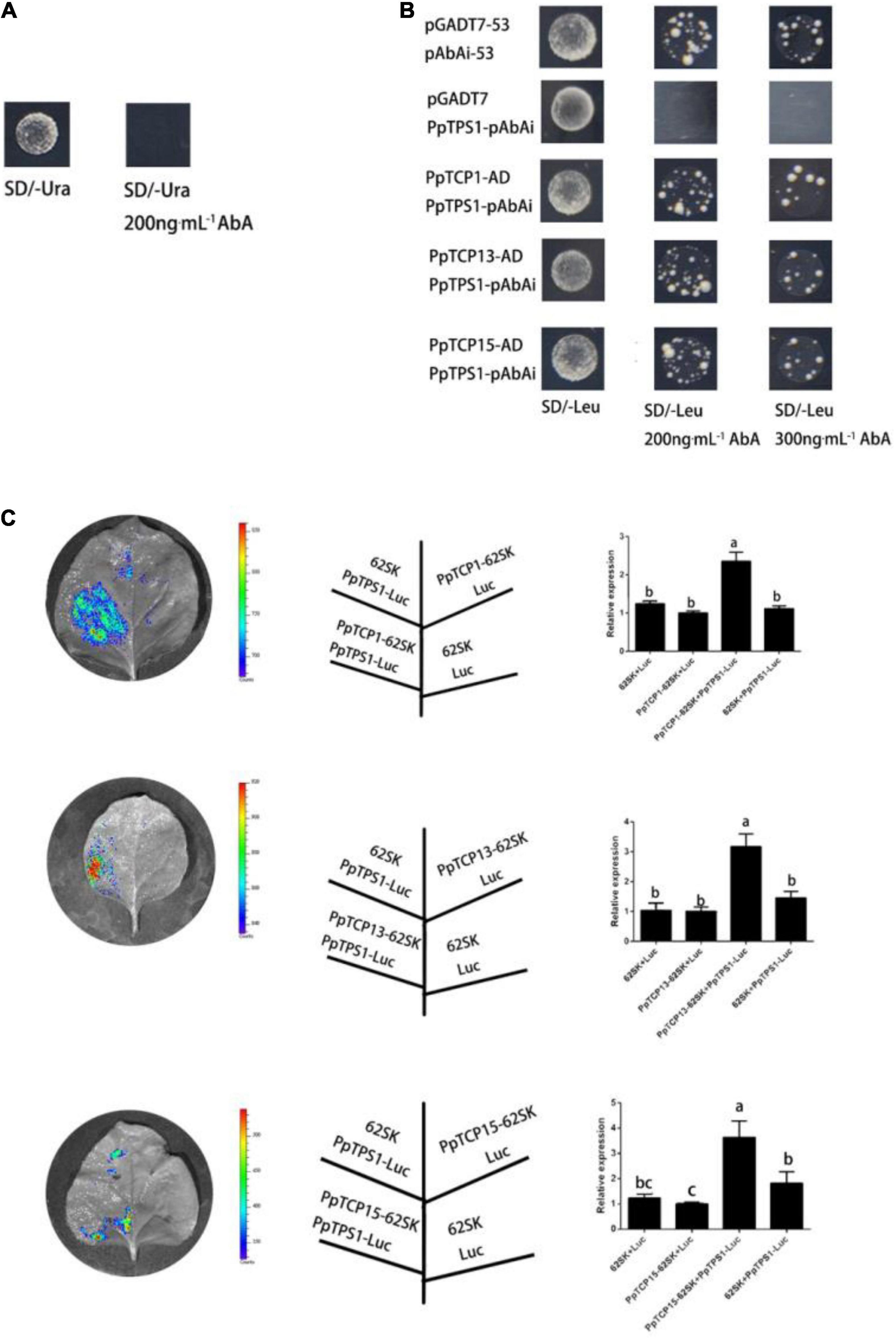
Figure 5. PpTCP1, PpTCP13, and PpTCP15 can activate the expression of PpTPS1. (A) The PpTPS1-pAbAi “bait” strain was inhibited by 200 ng⋅mL–1 AbA. (B) The PpTCP1-pGADT7, PpTCP13-pGADT7, and PpTCP15-pGADT7 plasmids were constructed, transformed into Y1H medium containing PpTPS1100–400-pAbAi, and cultured in Leu medium at 30°C in the dark for 3 days. The grown yeast was then diluted with 0.9% NaCl solution (OD600 = 0.002), spotted on SD/-Leu at concentrations of 200 and 300 ng⋅mL–1 AbA, and cultured in the dark at 30°C for 3–5 days. (C) A tobacco dual-luciferase experiment verified that PpTCP1, PpTCP13, and PpTCP15 could activate the expression of PpTPS1. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
To explore how PpTCP1, PpTCP13, and PpTC15 regulate the expression of PpTPS1, we performed a dual-luciferase assay. The results showed that the fluorescence intensity of the regions where PpTPS1100–400-Luc coexpressed with PpTCP1-62SK, PpTCP13-62SK, and PpTCP15-62SK increased significantly (Figure 5C), indicating that PpTCP1, PpTCP13, and PpTCP15 could promote the expression of PpTPS1.
We analyzed the 1.5 kb promoter region upstream of PpTCP1, PpTCP13, and PpTCP15, and the results showed that these three genes all contained the cis-acting element ABRE, which is involved in the ABA response (Table 1). We then performed qRT-PCR analysis on peach seedlings treated with ABA. As shown in Figure 6, ABA significantly induced the expression of PpTCP1, PpTCP13, and PpTCP15, but the expression patterns of the three were different. We speculated that these genes could play different roles in the process of ABA signaling.
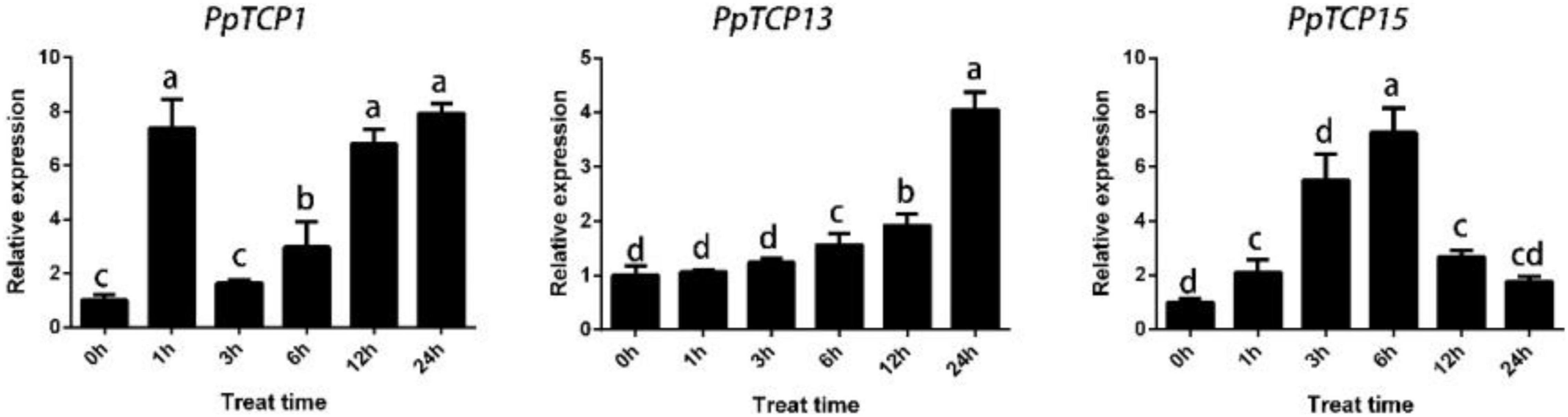
Figure 6. Expression patterns of PpTCP1, PpTCP13, and PpTCP15 after ABA treatment. One-year-old peaches were sprayed with 100 μM ABA, and samples were taken at 0, 1, 3, 6, 12, and 24 h for qRT-PCR analysis. Biological triplicates were performed for each sample. Different letters represent significant differences according to ANOVA and Duncan’s test (P < 0.05).
Plants perceive biotic and abiotic stress signals and initiate a series of hormonal signaling pathways that regulate the expression of downstream defense genes to generate relevant defense responses. Among these hormones, ABA plays an important role in the defense against abiotic stresses such as drought, high salt, cold, high temperature, and mechanical damage (Zhang et al., 2006; Lata and Prasad, 2011). Many studies have shown that an increase in ABA content plays an indispensable role in improving the salt tolerance of plants. The specific role of ABA is to relieve osmotic stress and ion stress caused by excessive salt, maintain water balance, and protect the integrity of the cell membrane structure. The accumulation of osmotic regulators, such as proline, soluble protein, and soluble sugar, can alleviate the damage caused by abiotic stress and improve plant tolerance. Under salt stress, ABA can induce a large accumulation of proline, an osmotic regulator in plants, and increase the activity of related protective enzymes (Espasandin et al., 2014). Salt stress-related genes can regulate osmotic substances by participating in ABA signal transduction to achieve their related functions. GhMYB73 can positively regulate ABA signaling, increase the proline content, and enhance salt tolerance in transgenic cotton (Zhao et al., 2019). WRKY family transcription factors such as GhWEKY17, GhWRKY41, ZmWRKY33, and DoWRY5 can regulate the ABA metabolic pathway, participate in ABA signal transmission, and, thus, participate in the regulation of osmotic stress (Li et al., 2013; Yan et al., 2014; Chu et al., 2016). In this study, we found that PpTPS1 can be induced by exogenous ABA, and the seed germination rate experiment of transgenic tomato showed that PpTPS1 enhanced the sensitivity of plants to ABA (Figure 2E). The relative water content, relative electrical measurement, and MDA content of transgenic plants after salt stress treatment were lower than those of WT plants, while the content of osmoregulatory substances was significantly higher than that of WT plants. Therefore, we speculate that PpTPS1 increases plant salt resistance by participating in the ABA signaling pathway and, thus, increasing the accumulation of osmotic substances. ABI5 is involved in the biological process of the plant’s response to adverse stress. Plants with OsABI5 overexpression exhibited faster expansion loss, yellowing, and growth inhibition under salt stress, while OsABI5-antisense plants were salt tolerant (Zou et al., 2007). Transgenic tobacco ZmABI5 showed reduced chlorophyll content, proline accumulation, and antioxidant enzyme activity under drought, salt, heat, and cold stresses (Yan et al., 2012). ABI5 also plays an important role in the feedback regulation of ABA core signaling and ABA biosynthesis. In Arabidopsis, Wang found that ABI1 ABI2, encoding a PP2C phosphatase that negatively regulates ABA signaling, is directly activated by ABI5 (Wang et al., 2019). In barley HvSnRK2.1, HvPP2C4 and the key ABA biosynthetic enzyme HvNCED1 are activated in the hvabi5 synapse (Zhu et al., 2020). We verified that PpTPS1 interacted with PpABI5 by Y2H, GST pull-down, and BiFC assays, and consistent with expectations, PpABI5 expression was also induced by ABA and salt stress. A qRT-PCR analysis of transgenic tomato and peach TRV samples revealed that PpTPS1 and PpABI5 antagonized each other (Figures 3, 4). Therefore, we speculated that PpABI5 may play a negative regulatory role during salt stress in peach, and PpTPS1 can be positively involved in ABA signaling by ABA activation, while PpTPS1 can enhance ABA biosynthesis and signaling to enhance salt resistance of plants by antagonizing PpABI5 expression. However, further experimental evidence is still required.
T encode plant-specific transcription factors containing a bHLH motif that can bind to DNA or produce protein–protein interactions. Previous studies have revealed that TCPs can participate in ABA regulatory pathways and play an important role in abiotic stresses. In Arabidopsis, DOF6 is an ABA biosynthesis gene, and AtTCP14 can interact with its target DOF6 to inhibit ABA biosynthesis (Tatematsu et al., 2008). Under stress, rice OsTCP19 causes significant upregulation of the ABA biosynthesis gene ABI4; overexpression of OsTCP19 decreases water loss and reactive oxygen species (ROS) in the plant, thereby improving plant salinity tolerance (Mukhopadhyay and Tyagi, 2015); OsPCF6 and OsTCP21 improve plant tolerance to cold stress by altering ROS scavenging capacity Zhou et al. (2013); and (Wang et al., 2014) found that miR319-targeted AtTCP14 improves plant tolerance during drought and salt stress. The upstream region of the PpTPS1 promoter contains multiple TCP binding sites G (T/C) GGNCCCAC motifs, and PpTCP1, PpTCP13, and PpTCP15 can activate the expression of PpTPS1, as verified by Y1H and dual-luciferase assays (Figure 5). Promoter analysis revealed that all three genes contained ABRE cis-elements (Table 1). A qRT-PCR analysis revealed that they could all be activated by ABA, and analysis of their expression patterns in response to ABA with PpTPS1 revealed that PpTCP1 was significantly expressed in the early stages of ABA treatment, similar to the expression patterns of PpTPS1 in response to ABA, drought, and salt stress. PpTCP13 was expressed at the late stage of ABA treatment, while PpTCP15 had two expression peaks, presumably regulating the PpTPS1 and ABA signaling pathways in different ways.
In summary, PpTCP1, PpTCP13, and PpTCP15 were induced by ABA to activate PpTPS1 expression, while PpTPS1 was able to interact with PpABI5 to participate in the ABA regulatory pathway, and it is speculated that PpTPS1 could play a role in the regulation of salt acclimation in plants.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
LL, XC, and XM designed the study. XM, NW, BW, RZ, and HH performed the experiments and analyzed the data. XM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was partially supported by the National Key Research and Development Plan (2018YFD1000104); the National Natural Science Foundation of China (31872041); Natural Science Foundation of Shandong Provincial (ZR2018MC023); Shandong Province Agricultural Good Seed Project grant (Nos. 2020LZGC007 and 2020LZGC00702); and Shandong Provincial Fruit Industry Technology System – Cultivation and Soil Fertilization Post (SDAIT-06-04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank Fu Xiling, Li Dongmei, and Xiao Wei for providing reagents, materials, and analysis tools.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.807342/full#supplementary-material
Bohlmann, J., Meyer-Gauen, G., and Croteau, R. (1998). Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. U S A. 95, 4126–4133. doi: 10.1073/pnas.95.8.4126
Chen, F., Tholl, D., Bohlmann, J., and Pichersky, E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Chen, F., Tholl, D., D’Auria, J. C., Farooq, A., Pichersky, E., and Gershenzon, J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell. 15, 481–494. doi: 10.1105/tpc.007989
Chen, M., Liu, X., Jiang, S., Wen, B., Yang, C., Xiao, W., et al. (2018). Transcriptomic and functional analyses reveal that PpGLK1 regulates chloroplast development in peach (Prunus persica). Front. Plant Sci. 9:34. doi: 10.3389/fpls.2018.00034
Chu, X., Chen, W., Chen, X., Lu, W., Han, L., Wang, X., et al. (2016). Correction: the cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic nicotiana benthamiana. PLoS One 11:e0157026. doi: 10.1371/journal.pone.0157026
Cubas, P. (2002). Role of TCP genes in the evolution of morphological characters in angiosperms. Systemat. Associat. Special 65, 247–266. doi: 10.1201/9781420024982.ch13
Dai, M., Xue, Q., McCray, T., Margavage, K., Chen, F., Lee, J. H., et al. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell. 25, 517–534. doi: 10.1105/tpc.112.105767
Davidovich-Rikanati, R., Sitrit, Y., Tadmor, Y., Iijima, Y., Bilenko, N., Bar, E., et al. (2007). Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat. Biotechnol. 25, 899–901. doi: 10.1038/nbt1312
Deng, X. X., Shuang, H., Hao, Y. J., Xu, Q., Han, M. Y., Zhang, S. B., et al. (2018). A review of the development of fruit tree discipline in 100 years. J. Agron. 8, 24–34.
Dudareva, N., Martin, D., Kish, C. M., Kolosova, N., Gorenstein, N., Faldt, J., et al. (2003). (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell. 15, 1227–1241. doi: 10.1105/tpc.011015
Espasandin, F. D., Maiale, S. J., Calzadilla, P., Ruiz, O. A., and Sansberro, P. A. (2014). Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem. 76, 29–35. doi: 10.1016/j.plaphy.2013.12.018
Fujii, H. (2014). Abscisic Acid Implication in Plant Growth and Stress Responses. Phytohormones: A Window to Metabolism. Signal. Biotechnol. Applicat. 2014, 37–54. doi: 10.1007/978-1-4939-0491-4_2
Hou, J. Y., Fu, J. Y., Zhao, Y., Qiu, Y., Li, C. C., and Wang, Q. (2017). Analysis of differential expression of terpene synthase genes in maize roots in response to drought and hormone treatments. Maize Sci. 2017, 63–67. doi: 10.13597/j.cnki.maize.science.20170510
Hu, T., Wang, S., Wang, Q., Xu, X., Wang, Q., and Zhan, X. (2021). A tomato dynein light chain gene SlLC6D is a negative regulator of chilling stress. Plant Sci. 303:110753. doi: 10.1016/j.plantsci.2020.110753
Jian, W., Cao, H., Yuan, S., Liu, Y., Lu, J., Lu, W., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6:22. doi: 10.1038/s41438-018-0098-y
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Larosa, P. C., Handa, A. K., Hasegawa, P. M., and Bressan, R. A. (1985). Abscisic acid accelerates adaptation of cultured tobacco cells to salt. Plant Physiol. 79, 138–142. doi: 10.1104/pp.79.1.138
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, S. Y. (2018). Functional and expression regulation studies of the maize terpene synthase gene TPS6. Ph. D. thesis. Beijing: Chinese Academy of Agricultural Sciences.
Lesburg, C. A., Zhai, G., Cane, D. E., and Christianson, D. W. (1997). Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science 277, 1820–1824. doi: 10.1126/science.277.5333.1820
Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:199. doi: 10.1146/annurev.arplant.49.1.199
Lewinsohn, E., Schalechet, F., Wilkinson, J., Matsui, K., Tadmor, Y., Nam, K. H., et al. (2001). Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127, 1256–1265. doi: 10.2307/4280182
Li, C., Potuschak, T., Colon-Carmona, A., Gutierrez, R., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U S A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, H., Gao, Y., Xu, H., Dai, Y., Deng, D., and Chen, J. (2013). ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 70, 207–216. doi: 10.1007/s10725-013-9792-9
Liu, H., Cao, X., Liu, X., Xin, R., Wang, J., Gao, J., et al. (2017). UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 40, 2261–2275. doi: 10.1111/pce.13029
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lopez-Molina, L., Seébastien, M., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2010). ABI5 acts downstream of ABI3 to execute an aba-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Mafu, S., Ding, Y., Murphy, K. M., Yaacoobi, O., Addison, J. B., Wang, Q., et al. (2018). Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in Maize. Plant Physiol. 176, 2677–2690. doi: 10.1104/pp.17.01351
Mittal, A., Gampala, S. S., Ritchie, G. L., Payton, P., Burke, J. J., and Rock, C. D. (2014). Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 12, 578–589. doi: 10.1111/pbi.12162
Mukhopadhyay, P., and Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5:9998. doi: 10.1038/srep12381
Munns, R. (2011). In Plant Responses to Drought and Salinity Stress - Developments in a Post-Genomic Era. Florida, FL: Academic Press, 1–32. doi: 10.1016/B978-0-12-387692-8.00001-1
Nayyeripasand, L., Garoosi, G. A., and Ahmadikhah, A. (2021). Genome-Wide association study (GWAS) to identify salt-tolerance QTLs carrying novel candidate genes in rice during early vegetative stage. Rice 14:9. doi: 10.1186/S12284-020-00433-0
Rodriguez, A., San Andres, V., Cervera, M., Redondo, A., Alquezar, B., Shimada, T., et al. (2011). Terpene down-regulation in orange reveals the role of fruit aromas in mediating interactions with insect herbivores and pathogens. Plant Physiol. 156, 793–802. doi: 10.1104/PP.111.176545
Rodriguez, A., Shimada, T., Cervera, M., Alquezar, B., Gadea, J., Gomez-Cadenas, A., et al. (2014). Terpene down-regulation triggers defense responses in transgenic orange leading to resistance against fungal pathogens. Plant Physiol. 164, 321–339. doi: 10.1104/PP.113.224279
Roger, M. R. (2001). Handbook of plant ecophysiology techniques. London: Kluwer Academic Press, 207–212.
Shahid, M. A., Balal, R. M., Pervez, M. A., Abbas, T., Aqeel, M. A., Riaz, A., et al. (2015). Exogenous 24-epibrassinolide elevates the salt tolerance potential of pea (Pisum sativum L.) by improving osmotic adjustment capacity and leaf water relations. J. Plant Nutr. 38, 1050–1072. doi: 10.1080/01904167.2014.988354
Sharma, A., Shahzad, B., Kumar, V., Kohli, S. K., Sidhu, G. P. S., Bali, A. S., et al. (2019). Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomol 9:285. doi: 10.3390/biom9070285
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., and Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. doi: 10.1016/j.gene.2012.06.076
Sun, Z., Li, H., Zhang, Y., Li, Z., Ke, H., Wu, L., et al. (2018). Identification of SNPs and candidate genes associated with salt tolerance at the seedling stage in cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:1011. doi: 10.3389/fpls.2018.01011
Tatematsu, K., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wang, Q., Xu, G., Zhao, X., Zhang, Z., Wang, X., Liu, X., et al. (2020). Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 71, 1585–1597. doi: 10.1093/jxb/erz516
Wang, S. T., Sun, X. L., Hoshino, Y., Yu, Y., Jia, B., Sun, Z. W., et al. (2014). MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L). PLoS One 9:e91357. doi: 10.1371/journal.pone.0091357
Wang, X., Guo, C., Peng, J., Li, C., Wan, F., Zhang, S., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221, 341–355. doi: 10.1111/nph.15345
Yan, F., Deng, W., Wang, X., Yang, C., and Li, Z. (2012). Maize (Zea mays L.) homologue of ABA-insensitive (ABI) 5 gene plays a negative regulatory role in abiotic stresses response. Plant Growth Regul. 68, 383–393. doi: 10.1007/s10725-012-9727-x
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yang, H., Zhang, H., Mao, X., Wang, C., and Jing, R. (2010). Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One 5:16041. doi: 10.1371/journal.pone.0016041
Yang, Y., Yu, X., Song, L., and An, C. (2011). ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol. 156, 873–883. doi: 10.1104/pp.111.175950
Yang, Z., Li, Y., Gao, F., Jin, W., Li, S., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71, 4140–4158. doi: 10.1093/jxb/eraa184
Zhang, J., Jia, W., Yang, J., and Ismail, A. M. (2006). Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 97, 111–119. doi: 10.1016/j.fcr.2005.08.018
Zhang, L., Zhu, L. X., Chuan, X. U., Cui, C. M., Sheng, H. Y., Rui, L. I., et al. (2015). The effect of silencing chalcone synthase on anthocyanin metabolism in peach. Acta Hortic. Sin. 2015:554. doi: 10.16420/j.issn.0513-353x.2014-0554
Zhang, X., Shen, H., Wen, B., Li, S., and Li, L. (2021). BTB-TAZ domain protein PpBT3 modulates peach bud endodormancy by interacting with PpDAM5. Plant Sci. 310:110956. doi: 10.1016/j.plantsci.2021.110956
Zhao, X., Han, X., Wang, Q., Wang, X., Chen, X., Li, L., et al. (2020). EARLY BUD BREAK 1 triggers bud break in peach trees by regulating hormone metabolism, the cell cycle, and cell wall modifications. J. Exp. Bot. 71, 3512–3523. doi: 10.1093/jxb/eraa119
Zhao, Y., Yang, Z., Ding, Y., Liu, L., Han, X., Zhan, J., et al. (2019). Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 286, 28–36. doi: 10.1016/j.plantsci.2019.05.021
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.4161/psb.28700
Zhu, J. F., Cui, Z. R., Wu, C. H., Deng, C., Chen, J. H., et al. (2018). Research progress and prospect of saline land greening in China. World For. Res. 2018, 70–75. doi: 10.13348/j.cnki.sjlyyj.2018.0034.y
Zhu, T., Li, L., Feng, L., and Ren, A. M. (2020). StABI5 involved in the regulation of chloroplast development and photosynthesis in potato. Int. J. Mol. Sci. 21:21031068. doi: 10.3390/ijms21031068
Zhuang, K., Kong, F., Zhang, S., Meng, C., Yang, M., Liu, Z., et al. (2019). Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. New Phytol. 221, 1998–2012. doi: 10.1111/nph.15532
Keywords: peach, PpTPS1, ABA, salt stress, PpABI5
Citation: Meng X, Zhang Y, Wang N, He H, Tan Q, Wen B, Zhang R, Sun M, Zhao X, Fu X, Li D, Lu W, Chen X and Li L (2022) Prunus persica Terpene Synthase PpTPS1 Interacts with PpABI5 to Enhance Salt Resistance in Transgenic Tomatoes. Front. Plant Sci. 13:807342. doi: 10.3389/fpls.2022.807342
Received: 02 November 2021; Accepted: 11 January 2022;
Published: 23 February 2022.
Edited by:
Abidur Rahman, Iwate University, JapanCopyright © 2022 Meng, Zhang, Wang, He, Tan, Wen, Zhang, Sun, Zhao, Fu, Li, Lu, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiude Chen, Y2hlbnhpdWRlQDE2My5jb20=; Ling Li, bGlsaW5nMjE3QHNkYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.