- 1Department of Forest Resources and Environmental Conservation, Virginia Tech, Blacksburg, VA, United States
- 2Institute for Genomics, Biocomputing and Biotechnology, Mississippi State University, Starkville, MS, United States
- 3Department of Forest Ecosystems and Society, Oregon State University, Corvallis, OR, United States
Both the evolution of tree taxa and whole-genome duplication (WGD) have occurred many times during angiosperm evolution. Transcription factors are preferentially retained following WGD suggesting that functional divergence of duplicates could contribute to traits distinctive to the tree growth habit. We used gain- and loss-of-function transgenics, photoperiod treatments, and circannual expression studies in adult trees to study the diversification of three Populus FLOWERING LOCUS D-LIKE (FDL) genes encoding bZIP transcription factors. Expression patterns and transgenic studies indicate that FDL2.2 promotes flowering and that FDL1 and FDL3 function in different vegetative phenophases. Study of dominant repressor FDL versions indicates that the FDL proteins are partially equivalent in their ability to alter shoot growth. Like its paralogs, FDL3 overexpression delays short day-induced growth cessation, but also induces distinct heterochronic shifts in shoot development—more rapid phytomer initiation and coordinated delay in both leaf expansion and the transition to secondary growth in long days, but not in short days. Our results indicate that both regulatory and protein coding sequence variation contributed to diversification of FDL paralogs that has led to a degree of specialization in multiple developmental processes important for trees and their local adaptation.
Introduction
Distinguishing features of trees include large crowns enabled by extensive wood development and protracted flowering-incompetent phases. Central to woody shoot development is the transition from primary growth—the production of phytomers and stem elongation initiated by the shoot apical meristem (SAM) and rib meristem—to secondary growth, which commences with the formation of a vascular cambium that increases girth by producing secondary xylem (Spicer and Groover, 2010; McKim, 2019). In Populus, this transition is synchronized with leaf maturation (Larson and Isebrands, 1971, 1974). Trees inhabit and often dominate temperate and boreal regions because they evolved the ability to become endodormant and orchestrate cellular adaptations that enable above ground meristems and tissues to survive winter freezing temperatures and dehydration stress (Howe et al., 2003; Preston and Sandve, 2013). The tree growth habit has been lost and gained many times throughout angiosperm evolution (Groover, 2005), which has been characterized by whole-genome duplications (WGDs) as well as segmental and tandem duplications (Hanada et al., 2008; Soltis et al., 2015). Although these events are likely to have had a major role in the repeated evolution of trees, empirical evidence for the role of gene duplicate diversification in processes that define the tree life style is sparse.
Flowering time in diverse plants is cued by indicators of seasonal change, with photoperiod and an extended period of chilling temperatures typically major signals (Bernier and Perilleux, 2005). Vegetative phenology of trees is also cued by these signals and study of tree homologs of Arabidopsis flowering time genes provided some of the first evidence for the contribution of gene duplicate diversification to tree developmental processes (reviewed in Ding and Nilsson, 2016; Brunner et al., 2017). Although reproductive phenology is integrated with vegetative phenology in adult trees, their phenophases are not always coincident or controlled by the same environmental cue. Long days (LDs) induce expression of the transcriptional co-factor and florigen FLOWERING LOCUS (FT) in Arabidopsis leaves and FT homologs in diverse plants have conserved functions in the floral transition (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Taoka et al., 2011). In Populus, FT2 is expressed in leaves and rapidly downregulated by short days (SDs), whereas FT1 expression peaks during winter in multiple tissues within winter buds (Böhlenius et al., 2006; Hsu et al., 2011). Changes in cis-regulatory sequences have generally been considered the predominant mechanism for developmental evolution, but increasing evidence supports a role for protein coding changes and both types of sequence changes can be necessary for the evolution of new transcriptional circuits (Lynch and Wagner, 2008; Bartlett, 2020; Britton et al., 2020). Although their divergent seasonal expression patterns could be sufficient for functional diversification of the Populus FT paralogs, their encoded proteins are not fully equivalent. FT1 is much more effective than FT2 at inducing flowering, suggesting that FT1 could mediate the transition of incipient axillary meristems to inflorescence meristems within winter buds and also promote endodormancy release (Hsu et al., 2011; Rinne et al., 2011; Brunner et al., 2014). Conversely, overexpression of either paralog delayed SD-induced growth cessation, but only FT2 expression is consistent with a growth-promoting function (Böhlenius et al., 2006; Hsu et al., 2011).
A conserved mechanism to promote flowering centers on a complex involving FT and the bZIP transcription factor FLOWERING LOCUS D (FD) that activates the related MADS-box genes FRUITFULL (FUL) in the SAM and APETALA1 (AP1) in lateral floral meristems (Schmid et al., 2003; Abe et al., 2005; Wigge et al., 2005; Torti et al., 2012). The FT-FD module has additional effects under certain environmental conditions and different FT and FD homologs appear to have roles in other developmental processes. For example, in SDs, 35S::FT Arabidopsis transgenics have small, curled leaves and this phenotype is dependent on FD (Teper-Bamnolker and Samach, 2005). Both rice OsFD1 and OsFD2 can form a complex with FT homologs; however, only OsFD1 promoted flowering whereas OsFD2 overexpression affected shoot branching and panicle architecture (Taoka et al., 2011; Tsuji et al., 2013; Brambilla et al., 2017).
Transcription factors are preferentially retained after WGDs, which also provide opportunity for a duplicate regulatory module to evolve in concert (Maere et al., 2005; Freeling, 2009; Wu et al., 2020). The Populus genome contains three FD-LIKE (FDL) genes (Supplementary Figure S1). As is the case for FT1 and FT2, FDL1 and FDL2 resulted from the Salicoid WGD, estimated to have occurred ~60 Ma (Tuskan et al., 2006; Rodgers-Melnick et al., 2012). The growth and morphology of 35S::FDL1 poplar transgenics were similar to wild-type (WT) under LDs, but bud set was delayed under SDs (Tylewicz et al., 2015). FDL1 interacts with ABSCISIC ACID-INSENSITIVE3 (ABI3), a bZIP transcription factor with a role in bud formation (Rohde, 2002; Ruttink et al., 2007; Tylewicz et al., 2015). Overexpression of ABI3 or FDL1 upregulates some of the same genes linked to bud development and stress adaptation, suggesting a role for FDL1 in these processes. Two splice variants of FDL2 induced different phenotypes. Transgenics overexpressing FDL2.1 were dwarf, but their SD growth response did not differ from WT (Tylewicz et al., 2015). Under LDs, 35S::FDL2.2 (referred to as FD1 in Parmentier-Line and Coleman, 2016), poplar transgenics flowered precociously and had small leaves and increased branching, but similar to FDL1 overexpression, SD-induced bud set was delayed.
Here, we report that both regulatory and protein coding divergence contribute to the varying degrees of functional diversification among the three FDL genes. Adding new information to previous studies (Tylewicz et al., 2015; Parmentier-Line and Coleman, 2016), we show that only FDL2.2 can induce precocious flowering and its strong upregulation in developing spring inflorescence buds supports a primary role in the floral transition. Furthermore, we show that the vegetative expression of FDL1 and FDL3 peak at opposite seasons, suggesting diversified roles in phenology. FDL3 overexpression showed a novel photoperiod-dependent phenotype. In LDs, FDL3 induced a delay in leaf maturation and the transition to secondary growth and altered the expression of developmentally responsive gibberellin (GA) synthesis and response genes. Expression studies of AP1/FUL homologs suggest that duplicate FT-FD-AP1/FUL modules could play distinct roles in vegetative and reproductive development.
Materials and Methods
Binary Constructs and Plant Transformation
The Populus deltoides FDL2.2 and FDL3 coding regions were amplified with Pfu DNA polymerase (Stratagene) and inserted into pGEM-T Easy vector (Promega). For dominant repression constructs, the 3′ end of coding regions were extended to encode the SRDX repressor domain (rd; Hiratsu et al., 2003) by designing a reverse primer containing the SRDX coding sequence (LDLDLELRLGFS). All primer sequences are provided in Supplementary Table S1. The coding sequences were excised by BamHI/KpnI digestion and cloned into the pBI121 binary vector (BD Biosciences). Vectors were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Populus tremula × Populus alba clone INRA 717-1B4, hereafter referred to as wild type (WT), as previously described (Meilan and Ma, 2006).
Plant Growth Conditions and Measurements
All transgenic and non-transgenic WT plants were propagated in vitro. Rooted plantlets were transferred from tissue culture to soil (Promix B, Canada) and acclimated in a growth chamber. After acclimation, plants were transferred to two-gallon pots and provided with 48 g Osmocote Plus 15-9-12 fertilizer/pot approximately 3 weeks after transfer. LD growth chamber conditions were 16-h light/8-h dark, with light intensity of 100 μ mol m_2 s_1 at plant level, 20°C–22°C, and 65% relative humidity. For SD conditions, the photoperiod was reduced to 8 h by changing the end of day time. In the greenhouse, ambient daylength was extended to 16 h using high pressure sodium lamps. Leaf plastochron index (LPI) was adopted for measurements and collecting samples (Larson and Isebrands, 1971). LPI1 was defined as the first leaf below the shoot apex (SA) with a lamina length of at least 1 cm. The internode (IN) directly beneath the LPI1 leaf was designated as IN1.
Gene Expression
The P. deltoides samples for seasonal gene expression studies and parameters for qRT-PCR were the same as previously described (Hsu et al., 2011), and sampling is summarized in Supplementary Table S4. To study expression in different tissues, we sampled 4-month-old WT plants grown in LD greenhouse conditions. For SA samples, all leaves visible to the naked eye were removed. Axillary buds (ABs) were collected from LPI10 to LPI20. Young leaf (YL) was from LPI2, and nearly mature leaf (ML) was LPI6 (leaf length ~75% of fully expanded leaf size). IN2 is in the primary growth zone, whereas IN6 is transitioning from primary to secondary growth. Phloem (Ph) and xylem (Xy) were scraped from the stem undergoing secondary growth below IN6. We collected non-woody lateral roots. All samples were collected 2 h after the start of the light period. All samples were immediately frozen in liquid nitrogen and stored at −80°C. Samples from three or more trees were pooled for RNA extraction using the RNeasy Plant Mini Kit (Qiagen) and RNase-Free DNase Set (Qiagen) as previously described (Brunner et al., 2004). Each cDNA was synthesized from 2.0 μg total RNA and an oligo (dT) primer using the High Capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer’s protocol. We used the Power SYBR Green PCR Master Mix kit (Applied Biosystems) and the ABI PRISM™ 7500 Real-Time PCR system (Applied Biosystems) for qRT-PCR reactions with three replications per RNA sample. The PCR program was set up to perform an initial incubation at 95°C for 10 min, followed by 95°C for 15 s, and 60°C for 1 min, for a total of 40 cycles. To enable design of gene-specific primers, the more divergent 3′ coding and untranslated region (UTR) were isolated for three aspen AP1/FUL family members using 3′ Rapid Amplification of cDNA Ends (3′ RACE). For LAP1a and LAP1b, full-length aspen cDNA sequences were already available (GenBank accession numbers AF034093 and AF034094). Using P. tremula × P. alba 717-1B4 cDNA as template, 3′ regions were amplified using gene-specific primers and a 3′ RACE adapter primer. The resulting DNA fragment was ligated into a shuttle vector, pCR 2.1 (TA Cloning Kit, Invitrogen, Carlsbad, CA, United States), and sequenced. 3′ RACE sequences and alignment of 3′ regions of AP1/FUL sequences from different Populus species/hybrids and location of primers are shown in Supplementary Figure S2. All primers are listed in Supplementary Table S1. We used an ubiquitin gene (UBQ2) as an internal reference (Mohamed et al., 2010) and normalized the Ct values across plates, determining relative quantities using comparative Ct method (2−ΔΔCt) as previously described (Livak and Schmittgen, 2001).
For in situ hybridization, immature inflorescences were collected from wild Populus trichocarpa trees near Corvallis, OR, United States, and fixed and embedded as previously described (Kelly et al., 1995). Transcripts were detected using antisense riboprobes from the 3′ ends (Supplementary Figure S2) of P. trichocarpa LAP1a (396 bp) and LAP1b (360 bp) cDNAs. Sequences were cloned into pBluscript KS and antisense and sense digoxygenin (DIG) labeled transcripts were produced with T3 and T7 RNA polymerases and DIG RNA Labeling Kit (Roche). Hybridization was done with DIG-labeled T3 and T7 probes (0.5 ng/μl) at 45°C overnight. Hybridized probes were detected by application of Anti-Dig Fab conjugated with alkaline phosphatase (1:1,250 dilution, Roche) and nitroblue tetrazolium/5-bromo-4-chloro-3-indoyl-phosphate.
Microscopic Analysis
For analysis of primary growth and transitional growth internodes, samples were immobilized in 5% agarose and sectioned (60 μm thickness) with a vibratome (Leica VT1200). We sectioned secondary growth internodes with a GSL1-microtome (Swiss Federal Institute for Forest, Snow and Landscape Research WSL, Switzerland). Sections were stained in a drop of the following solution: 1 g phloroglucinol (Sigma-Aldrich) in 100 ml, 95% EtOH, and 16 ml 37% HCL. Procedures for embedding and histology of stem samples and in situ hybridization of developing inflorescences are provided in the section “Materials and Methods.” All images were captured using a Zeiss Axio Imager A1 (Carl Zeiss, Oberkochen, Germany). For more detailed images of stem anatomy (Supplementary Figure S6), samples were fixed and embedded in LR white resin (London Resin Company, Ltd.) as previously described (Grant et al., 2010) and sectioned (2 μm thickness) with glass knives (Leica RM2265). Sections were stained with Toluidine blue/boric acid (0.05% w/v) for 1 min.
Phylogenetic Analysis
FLOWERING LOCUS D family protein sequences (Supplementary Table S2) were aligned using MUSCLE (Edgar, 2004). A maximum-likelihood phylogenetic analysis was performed on the sequence alignment using the JTT + G model, a site coverage cut-off of 90% for alignment gaps/missing data and 100 bootstraps for branch support testing with the program MEGA7 (Kumar et al., 2016). Analysis of AP1/FUL family members (Supplementary Table S3) was the same except that all positions with less than 50% site coverage were eliminated.
Statistics Analyses
Height, leaf length, and new leaf formation data were analyzed in JMP Pro 12 (SAS Institute Inc., 2016–2017), using the Fit model to test the effects of constructs and, when applicable, the events within constructs. We used two-sample t-test to evaluate differences between transgenic and WT means and to test differences between multiple group means, we used the LSMEANS protocol and applied the Tukey–Kramer’s adjustment for all possible pairwise comparisons between group means.
Results
FDL Genes Differ in Reproductive and Seasonal Vegetative Expression
We cloned full-length cDNAs of the three FDL genes from P. deltoides. FDL1 and the FDL2.1 and FDL2.2 splice variants encode proteins nearly identical to those previously reported from P. trichocarpa (Tylewicz et al., 2015; Parmentier-Line and Coleman, 2016) except that the FDL1 reported here contains an additional 39 amino acids at its N-terminus (Supplementary Figure S1). FDL3 shares the conserved C-terminal phosphorylation (T)/SAP motif (Tsuji et al., 2013) and groups with FD in a phylogenetic tree (Supplementary Figure S1).
We studied FDL expression in different vegetative tissues and developmental stages of 4-month-old WT trees grown in a LD greenhouse. Both FDL2 splice variants and FDL3 showed highest expression in shoot apices, whereas FDL1 showed highest expression in a transitional growth internode (Supplementary Figure S3). Study of circannual expression of Populus FT paralogs in different tissues was instrumental in revealing the divergence of FT1 and FT2 functions (Hsu et al., 2011). Hence, we studied expression of the FDLs using these same samples collected from adult P. deltoides growing in Mississippi, United States. Consistent with its ability to induce flowering (Parmentier-Line and Coleman, 2016), FDL2.2 was highly upregulated in newly developing reproductive buds as was the FDL2.1 splice variant (Figures 1B,D). The highest seasonal expression of FDL1 was during late autumn–winter (Figure 1A), consistent with its indicated role in mediating bud maturation and cold adaptation (Tylewicz et al., 2015). FDL3 was more highly expressed in shoot apices during the growing season compared to autumn–winter season and was transiently upregulated in leaves during early autumn at the timepoint when leaf collection shifted from fully expanded leaf (August) to preformed leaf within a terminal bud (September; Figures 1C,E). In shoot apices, the seasonal expression pattern of both FDL3 and FDL2 was opposite of FDL1’s expression pattern (Figures 1E,F).
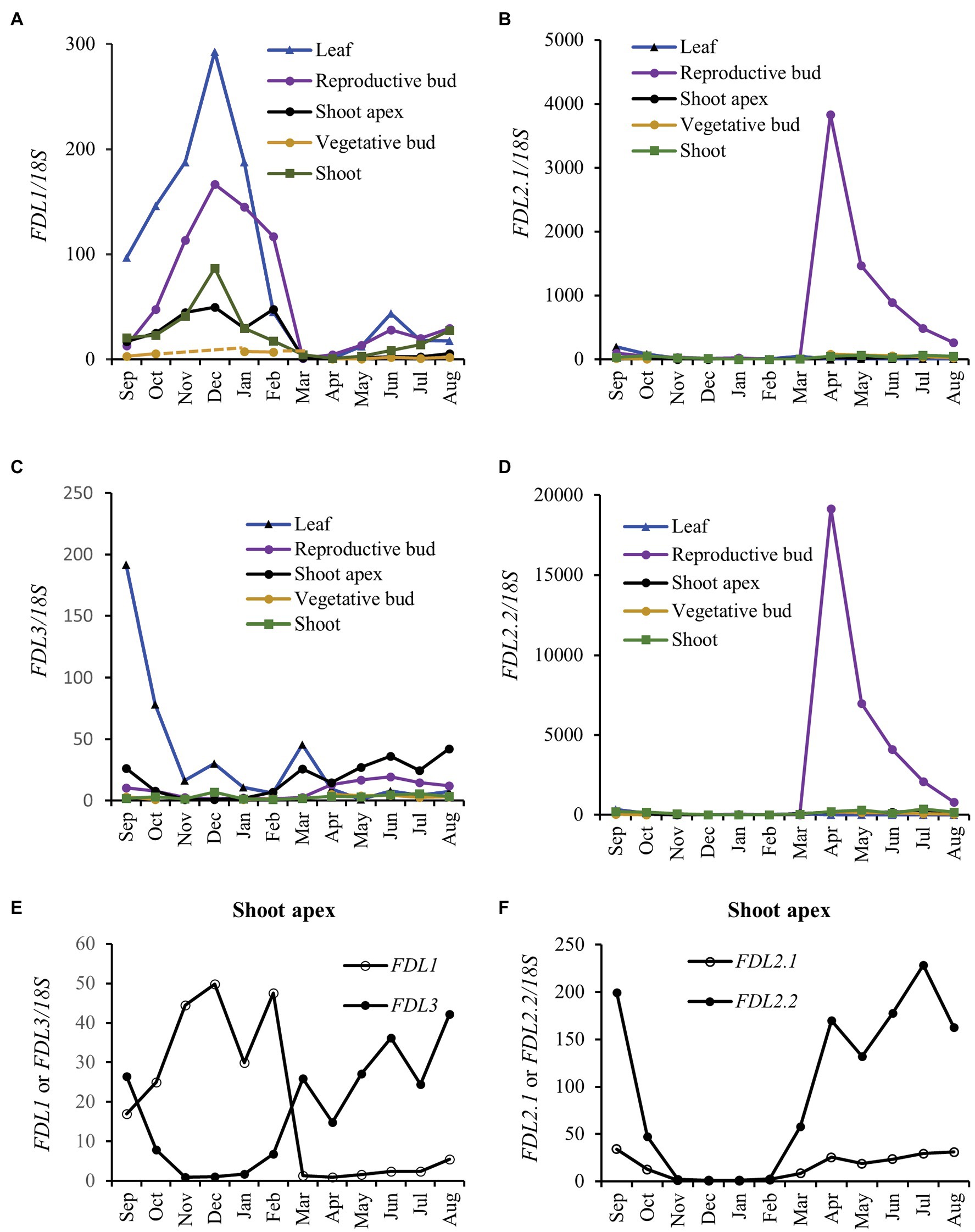
Figure 1. FLOWERING LOCUS D-LIKE (FDL) genes differ in regulation. Relative expression is fold change in transcript levels of (A) FDL1, (B) FDL2.1, (C) FDL3, and (D) FDL2.2 relative to the time point with the lowest expression within a tissue (n = 3, except for shoot apex where the three apices were pooled to provide sufficient sample for analysis). FDL expression was normalized against reference gene 18S rRNA. (E) FDL1 and FDL3 and (F) FDL2.1 and FDL2.2 expression in shoot apices is presented separately to allow comparison of circannual patterns among the different FDLs. Axillary reproductive bud flush began in late February with anthesis reached in March (April sample is newly initiated floral bud). From September to March, preformed leaves and shoots were dissected from terminal buds.
Dominant Repressor Versions of the Poplar FDL Genes Reduce Shoot Elongation to Different Degrees
To compare protein functional equivalency, we first extended each FDL coding sequence to encode the SRDX repressor domain (Hiratsu et al., 2003), and then, each sequence was placed under the control of the 35S promoter and nos terminator. The FDL1rd transgene imposed the most severe effect on shoot development (Figure 2A). Many tiny transgenic shoots, confirmed as positive for the FDL1rd transgene by PCR, regenerated from callus. However, FDL1rd transgenics failed to elongate when sub-cultured on shoot elongation medium and we could not regenerate any rooted plants. Six independent transgenic events of FDL2.1rd were rooted and four events showed short internodes whereas the other two showed WT-like growth in vitro (Supplementary Figure S4A). For transcriptional activators, overexpression of rd-modified and WT proteins is expected to induce opposite phenotypes; however, overexpression of FDL2.1 also reduced shoot growth (Tylewicz et al., 2015). As this suggests that addition of the rd augmented WT FDL2.1 function rather than induced a loss-of-function phenotype, FDL2.1rd transgenics were not studied further. Eleven events of FDL2.2rd were rooted, but only five events showed reduced growth (Supplementary Figure S4A) and the remainder grew similar to WT in vitro. Eight FDL2.2 events, including four showing less growth and four WT-like events were transferred in soil. After growing in the LD greenhouse for 4 months, the FDL2.2rd transgenics were significantly shorter than WT plants (Figure 2D).
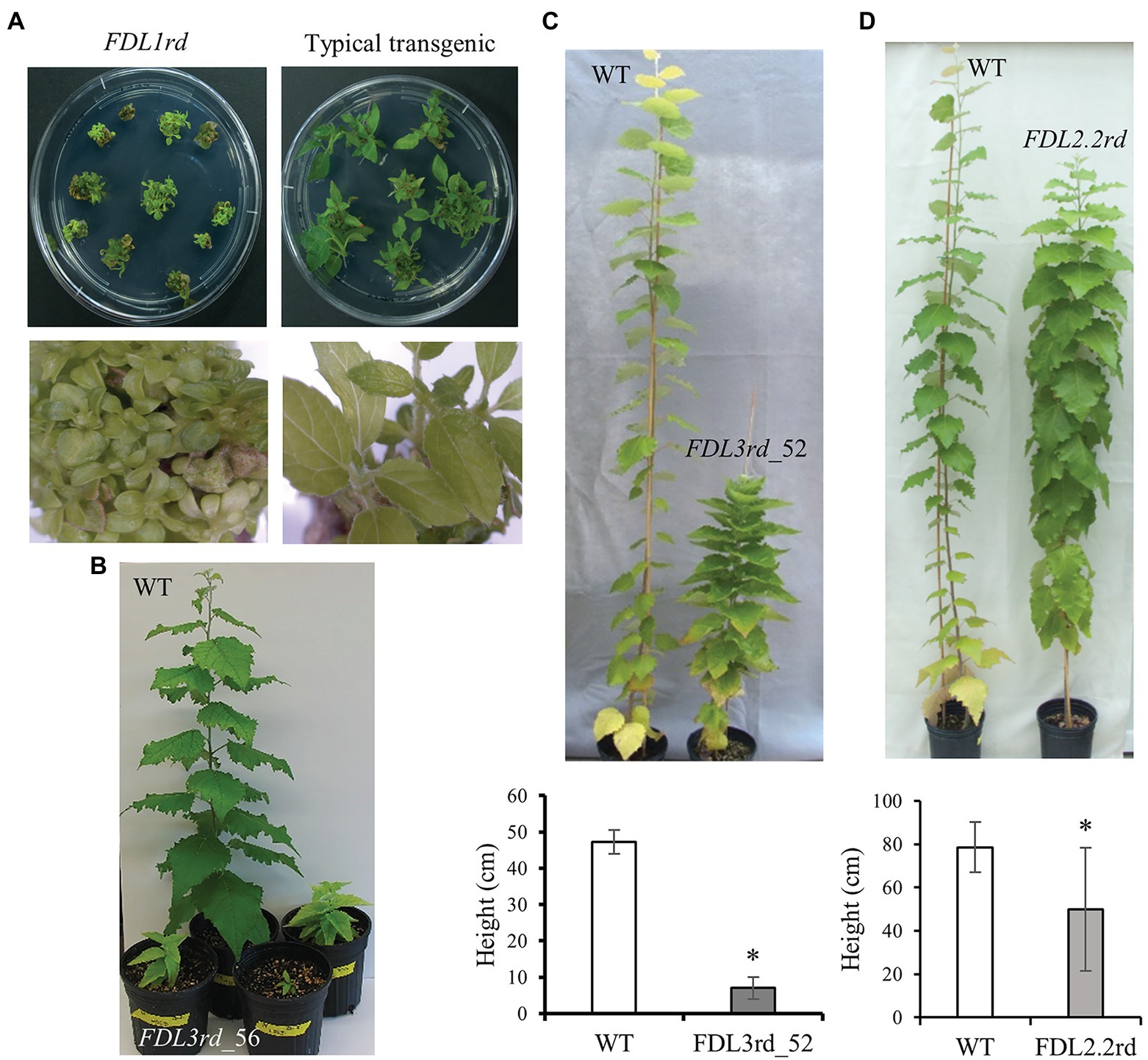
Figure 2. Phenotypic effects of dominant repressor versions of FDL genes. (A) FDL1rd transgenic shoots on shoot elongation medium compared to an unrelated transgenic regenerated at the same time that displays typical shoot elongation. Each clump of shoots corresponds to a single explant that was induced to form callus and then shoots. Bottom photos show shoots from one of the explants in the top photos. (B) FDL3rd_56 transgenics showed reduced shoot growth and set terminal buds within 2 months after potting under LD conditions, whereas WT continued to grow. (C) FDL3rd_52 trees showed reduced shoot elongation. Representative 6-month-old trees are shown and values are means ± SE for two WT and two FDL3rd_52 trees after 3 months of growth in a greenhouse. (D) Representative 6-month-old WT and FDL2.2rd trees and mean heights ± SE after 4 months of growth in a greenhouse. For WT, n = 12; For FDL2.2rd, n = 16 (eight events with two ramets/event). *p < 0.01 compared to WT.
Whereas many FDL3rd transgenic shoots regenerated from callus, rooting was achieved for only five of these, possibly due to severe suppression of shoot elongation by the FDL3rd transgene (Figures 2B, 3A; Supplementary Figure 4B), and nearly all attempts to propagate these rooted shoots in vitro and acclimate FDL3rd transgenics to soil were unsuccessful. Ultimately, we were able to establish only a few ramets of two FDL3rd events in soil. After 2 months in a LD growth chamber, all three ramets of event FDL3rd_56 grew to a height of only 10 cm or less and set terminal buds, as opposed to WT plants which reached 40 cm–50 cm in height and maintained active SAMs (Figure 2B). Two ramets of event FDL3rd_52 survived after transfer to soil and showed reduced height growth compared to WT (Figure 2C). In sum, all of the dominate repressor versions of the different poplar FDL genes reduced shoot elongation, but their effects varied in magnitude with FDL1rd > FDL3rd > FDL2.2rd.

Figure 3. Overexpression of FDL3 affects leaf size and shoot elongation but does not promote flowering. Representative plants showing opposite effects on shoot elongation in vitro of (A) FDL3 dominant repression (FDL3rd) vs. (B) overexpression (FDL3ox). In (A) WT plant is 6-week-old, whereas FDL3rd plant is 10-week-old and in (B), WT and FDL3ox were propagated at the same time and are 4-week-old in the photo. Shoot apices of (C) FDL2.2ox with consecutive axillary inflorescences, (D) WT, and (E) FDL3ox. White arrows in (C) point to axillary inflorescences with multiple female flowers. (F) A premature flowering FDL2.2ox plant with many inflorescences as shown in (C) on both the main shoot and branches. (G) A FDL3ox plant with a few branches, but no flowers. (C–G) All photos are of 6-month-old plants grown at the same time in a greenhouse under a 16-h photoperiod.
FDL2.2 but Not FDL3 Induces Early Flowering Under LDs
We produced transgenics with the 35S promoter directing expression of FDL2.2 or FDL3, designated FDL2.2ox and FDL3ox, respectively. Sixteen independent events each of FDL2.2ox and FDL3ox were regenerated. A previous study showed that FDL2.2 overexpression transgenics had small leaves and flowered in vitro (Parmentier-Line and Coleman, 2016). We observed a similar FDL2.2ox phenotype (Supplementary Figure S4C); however, we only observed in vitro flowering on one FDL2.2ox plant. We consistently observed that, in contrast to FDL3rd plants that were much shorter than WT, FDL3ox shoots elongated faster with longer internodes than WT plants propagated at the same time (Figures 3A,B). Because FDL2.2ox and FDL3ox transgenics had similar small leaf phenotypes in vitro, we directly compared the ability of the two transgenes to induce flowering under LDs. Three to five ramets of five events of FDL2.2ox and five events of FDL3ox were propagated and transferred to soil in parallel with WT plants. The potted plants were grown in the greenhouse under LDs. All transgenics had small leaves as was the case in vitro and also exhibited branching, especially the FDL2.2ox transgenics (Figures 3F,G). Transgenics were also shorter than WT, which was not the case for FDL3ox plants in vitro where sucrose is provided in the medium (Figure 3B).
Within 6 months of growth in the greenhouse, all ramets of all FDL2.2ox events flowered. Transgenics formed consecutive axillary inflorescences and terminal inflorescences also formed on some of the plants (Figure 3C). However, we did not observe flowering on any of the FDL3ox plants (Figure 3E). In addition, two of the FDL3ox events were grown for an additional 10 months with no flowering. Thus, whereas FDL2.2 or FDL3 overexpression induces similar vegetative phenotypes in LDs (Figures 3F,G), they are not equivalent in their ability to induce flowering. This is also consistent with their different expression patterns, particularly the high FDL2.2 expression in newly initiated reproductive buds (Figure 1D). FDL2.2 overexpression has been previously studied (Parmentier-Line and Coleman, 2016); hence, we focused on further characterization of FDL3ox transgenics where vegetative phenotypes could be studied without confounding effects of precious flowering.
Overexpression of FDL3 Accelerates Leaf Initiation but Delays Leaf Expansion and the Transition to Secondary Growth Under LDs
We found that despite their reduced height growth compared to WT, potted FDL3ox transgenics initiated phytomers more rapidly, indicated by the formation of new leaves over time (Figure 4D). In contrast, FDL3ox leaf expansion progressed much more slowly and fully expanded leaf size was reduced (Figure 4). In addition to stalled leaf development, FDL3ox trees had vine-like stems that were not self-supporting (Figure 3G; Supplementary Figure S5A), suggesting defects in secondary vascular tissue development. Under LDs, primary growth of poplar shoots is limited to the leaf development zone and the transition from primary to secondary growth occurs below a leaf that is at least partially mature (Larson and Isebrands, 1971, 1974). Detailed study of leaf and stem development in two FDL3ox events illustrates the coordinate delay in leaf expansion and transition to secondary growth (Figures 5A,B). Vascular development in IN4 and IN6 of FDL3ox (Figures 5B2,B3; Supplementary Figure S6) remained nearly the same as in IN2, with the exception that red-stained lignified cells were present at the position where secondary xylem would normally develop in IN6. Additionally, no phloem fiber bundles were formed in either IN4 or IN6 (Figures 5B2,B3; Supplementary Figure S6), whereas transitional secondary growth in IN4 and secondary growth in IN6 was evident in the stem of WT plants (Figures 5B6,B7; Supplementary Figure S6). Phloem fiber bundles were present in IN10 of FDL3ox, but the secondary xylem still remained in a much less developed state compared to WT (Figures 5B4,B8; Supplementary Figure S6). Secondary xylem was still very poorly developed at IN20 of FDL3ox plants, and while increased xylem development was evident in IN30, it was not present in a continuous ring (Supplementary Figures S5B,5C, S6).
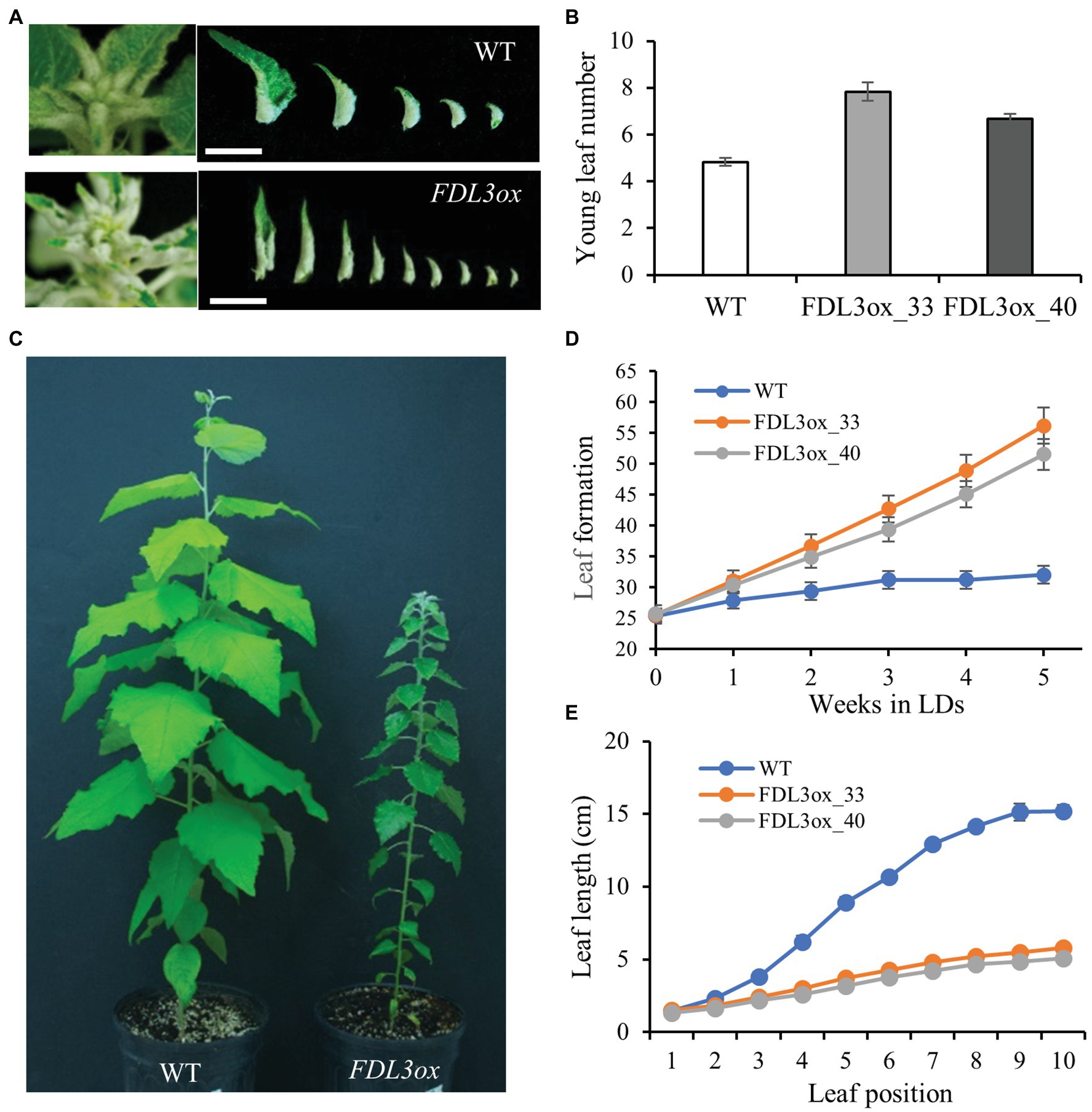
Figure 4. Overexpression of FDL3 accelerates leaf production, but represses leaf growth in long day (LD) conditions. Ramets of two FDL3ox events and WT were grown in a growth chamber under 16-h photoperiods for 2 months. (A) Shoot apices and young rolled leaves of a FDL3ox plant compared with that of a WT plant. Scale bars = 1 cm. (B) Number of young rolled leaves. (C) Representative WT and FDL3ox trees (D) Emergence of new leaves (leaf lamina longer than 1 cm) over time. Leaf number was counted weekly, beginning 3 weeks after transplantation. (E) Progression of leaf length with position on the shoot. Leaf position 1 is the youngest leaf whose lamina is longer than 1 cm. (B,D,E) Means ± SE (n = 6) for two FDL3ox events (33 and 40) and WT.
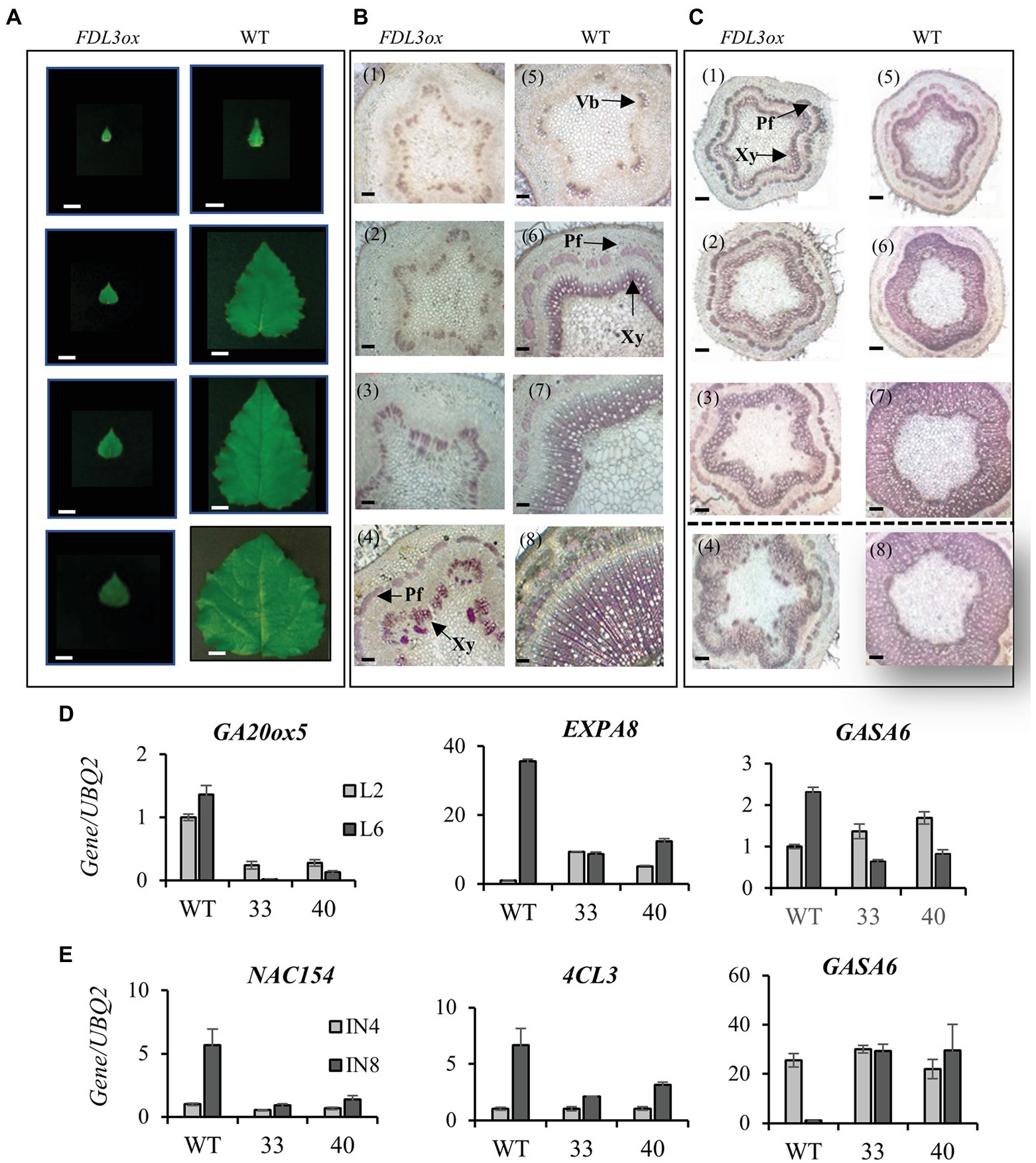
Figure 5. Overexpression of FDL3 synchronously inhibits leaf expansion and the transition to secondary growth in long days (LDs), but secondary growth is restored in short days (SDs). (A–C) Both FDL3ox and WT plants were grown in a LD greenhouse for 6 months and subsequently transferred to a SD growth chamber for 8 weeks. Leaves were counted from top to bottom according to leaf plastochron index (LPI). Internode (IN) number refers to the internode beneath the corresponding LPI. All panels show from top to bottom LPI2, LPI4, LPI6, and LPI10 leaf or corresponding IN. (A) Extremely slow growth of FDL3ox leaves compared to WT in LDs. Scale bars = 2 cm. (B) Severely inhibited secondary growth in FDL3ox plants (B1–4) compared to progressive transition to secondary growth in WT (B5–8) in LDs. (C) Secondary growth in IN2, IN4, and IN6 formed after exposure to SDs (images above dotted line) in FDL3ox (C1–3) and WT (C5–7) plants. Note that FDL3ox INs 4 and 6 (C2,3) now resemble the same INs of WT plants grown in LDs (B6,7). In contrast, IN10 (C4) formed in LDs before SD treatment remained underdeveloped in FDL3ox. After exposure to SDs, WT plants ceased elongation growth, IN2 transitioned to secondary growth (C5) and substantial secondary xylem accumulated in IN4 and IN6 of WT (C6,7). Transverse sections were 60 μm thick, Scale bars = 100 μm. Vb, vascular bundles; Pf, phloem fiber; and Xy, xylem. (D,E) Comparative expression analysis of leaf and stem developmental marker genes in WT and two events of FDL3ox (33 and 40) grown in LDs. Relative expression in LPI2 and LPI6 leaves (D) and in internodes IN4 and IN8 (E). Expression was normalized against reference gene ubiquitin gene (UBQ2).
Little is known about the mechanisms coordinating the timing of leaf maturation and the transition to secondary growth (reviewed in Teixeira et al., 2019); however, GA promotes both leaf expansion and secondary growth (Eriksson et al., 2000). Hence, we studied the expression of GA synthesis and response genes that also show developmentally responsive expression changes. GA20-oxidase 5 (GA20ox5) expression increases as leaves develop, and its expression was reduced in both LPI2 and LPI6 leaves of FDL3ox trees compared to WT (Figure 5D). GA3 application elevates α-EXPANSIN 8 (EXPA8) and GA-STIMULATED ARABIDOPSIS 6 (GASA6) expression in Populus leaves (Xie et al., 2016). In WT, EXPA8 and GASA6 are upregulated in LPI6 compared to LPI2 leaves, but not in FDL3ox plants (Figure 5D). In Arabidopsis, GA promotes GASA6 upregulation and elongation in embryos (Zhong et al., 2015), and in the inflorescence stem, GASA6 expression peaks in regions undergoing maximal elongation (Hall and Ellis, 2013). In Populus stems, GASA6 is most highly expressed in internodes undergoing maximal elongation and downregulated in secondary growth internodes (Dharmawardhana et al., 2010). Whereas GASA6 is downregulated in secondary growth IN8 compared to elongating IN4 in WT, it shows no downregulation in FDL3ox IN8 (Figure 5E). Expression of marker genes for secondary xylem further supports that molecular programs for the transition to secondary growth are not initiating properly in FDL3ox trees. NAC154, a co-ortholog of SECONDARY WALL-ASSOCIATED NAC DOMAIN 2 (Grant et al., 2010), and 4-COUMARATE:COA LIGASE 3 (4CL3; Shi et al., 2010), are several fold higher in IN8 compared to IN4 in WT, but show comparatively low expression in IN8 of FDL3ox trees (Figure 5E).
SDs Restore Leaf Expansion and Secondary Growth of FDL3ox Plants
After 3 weeks of exposure to SDs, height growth and formation of new leaves stopped in WT plants, whereas FDL3ox plants showed no sign of growth cessation (Figures 6B,C). After 5 weeks in SDs, WT plants had formed brown apical buds, while shoot apices of FDL3ox plants remained active; FDL3ox plants eventually formed apical buds after 10 weeks in SDs (Figure 6A).
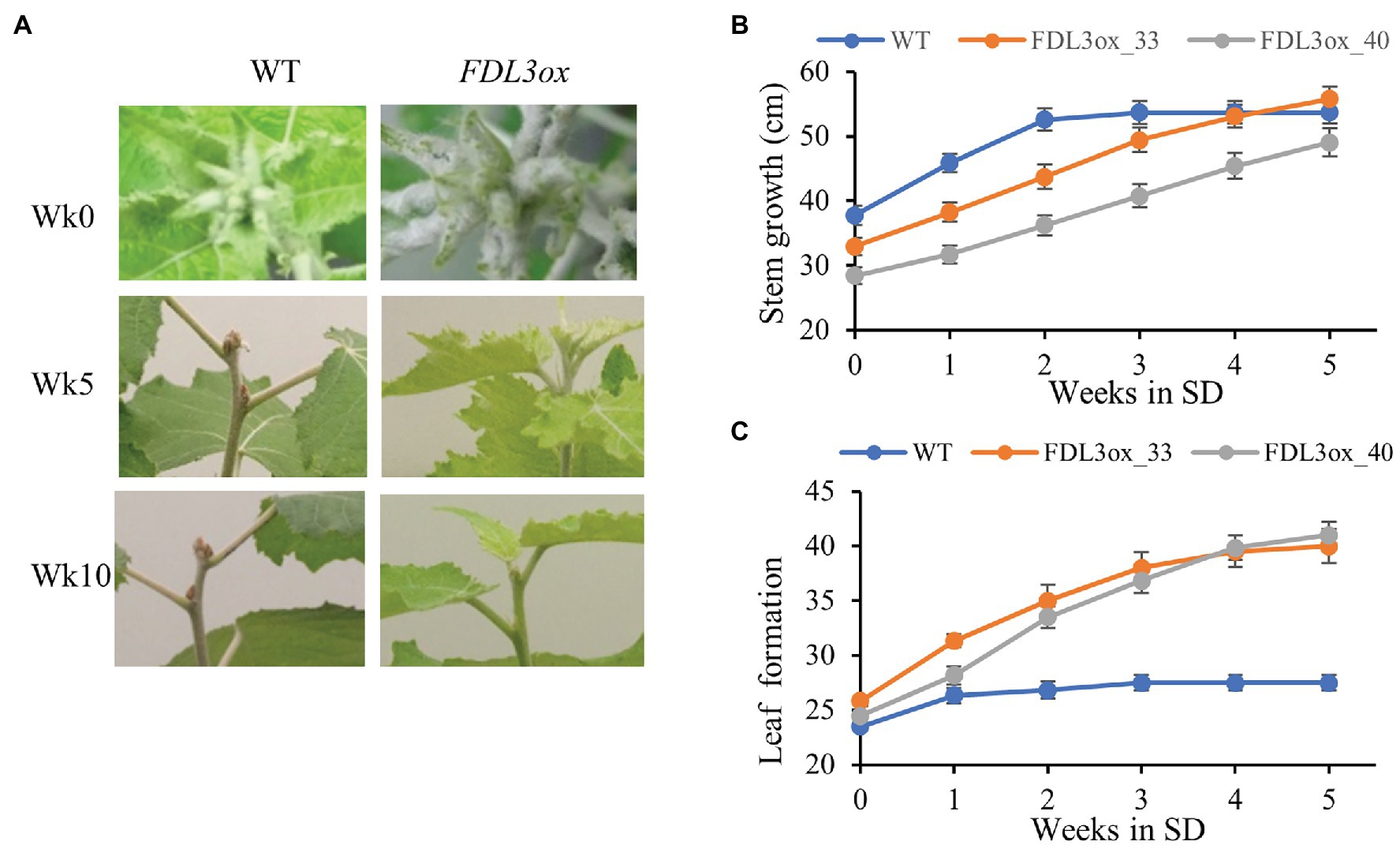
Figure 6. Overexpression of FDL3 delays growth cessation and bud set in short days. Plants of WT and two FDL3ox events (33 and 40) were grown in long days for 2 months before exposure to SDs. (A) Apical bud development of FDL3ox plants compared to WT after 5 and 10 weeks in SD (Week 5 and Week 10). In Week 5, WT plants had formed buds. In contrast, FDL3ox plants maintained actively growing apex. By Week 10, FDL3ox plants formed buds. (B,C) Cumulative stem growth (B) and leaf formation (C) were measured weekly during the first 5 weeks in SDs. (B) Plant height and (C) leaf numbers are means ± SE (n = 6).
Intriguingly, FDL3ox plants not only continued to grow in SDs, but also the development of leaves formed after transfer to SDs was similar to that of leaves of WT grown in LDs (Figures 7A–C). In WT plants, leaves directly below the forming apical bud and formed under LDs continued to expand after transfer to SDs (Figures 7A,B). In contrast, small FDL3ox leaves formed during LD treatment did not increase in size during SD treatment. Strikingly, FDL3ox leaves that formed after transfer from LDs to SDs expanded rapidly, exceeding the length of those produced under LDs by ~2-fold. The return of SD-treated FDL3ox plants to LDs again led to the production of the small leaves like those produced during the first LD treatment (Figures 7D,E). We confirmed that the effect of FDL3ox overexpression on leaf development is dependent on photoperiod alone and not location (i.e., moving plants from greenhouse to growth chamber) by growing plants entirely in growth chambers with only photoperiod altered. Within 4 weeks in SDs, leaf expansion and height of FDL3ox were the same as WT plants that had started to form apical buds (Supplementary Figures S7B,C). In contrast, FDL3ox plants grown in LD conditions were shorter with small leaves (Supplementary Figures S7A,C).
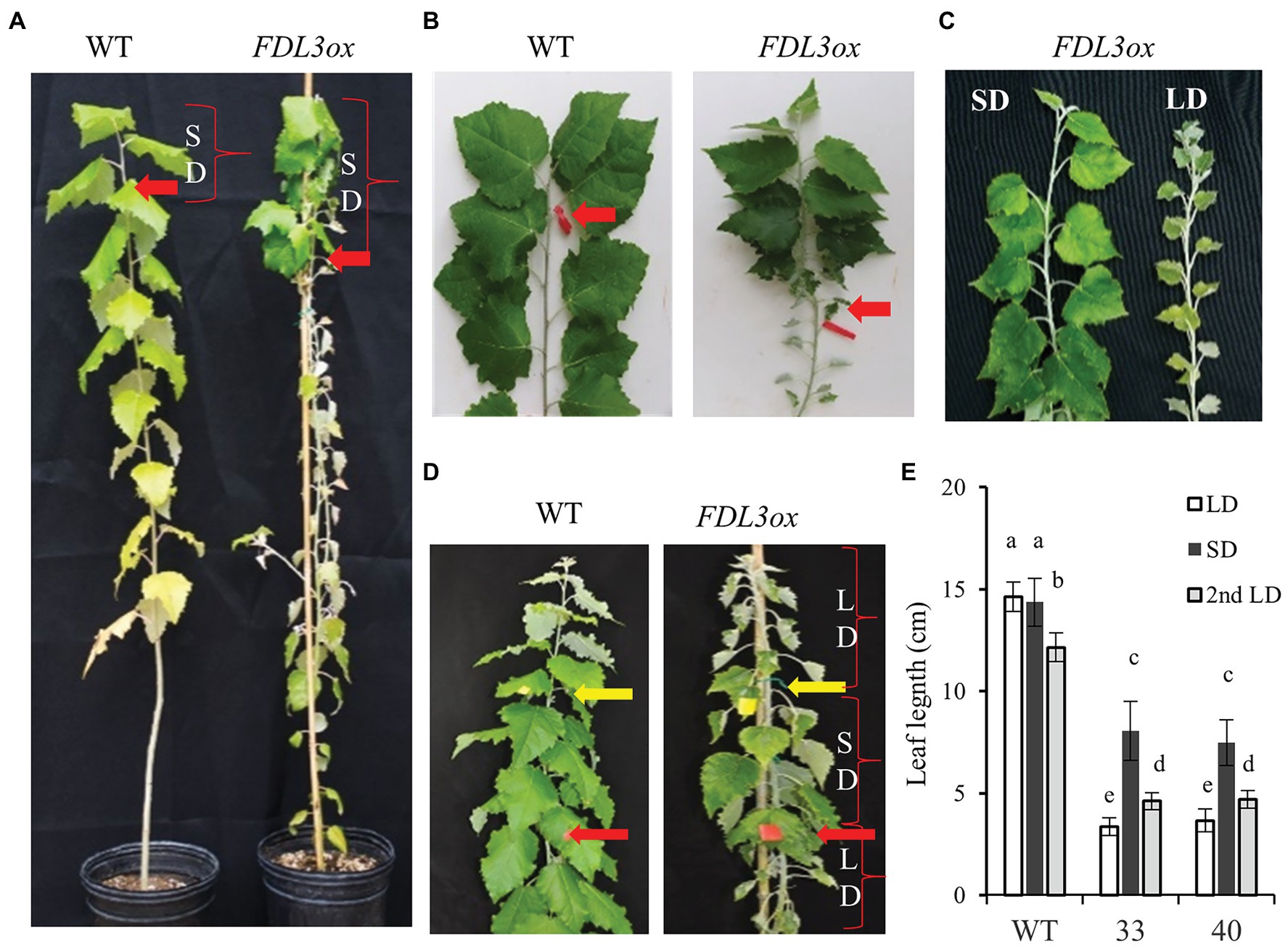
Figure 7. FDL3ox trees resume leaf development in short days. (A,B) Both FDL3ox and WT plants were grown in a long day (LD) greenhouse for 6 months and then were transferred into a SD growth chamber for 8 weeks. FDL3ox leaves formed after transfer to SDs (above the red arrows) showed leaf development similar to actively growing WT plants, in contrast to underdeveloped leaves formed on FD3ox plants in LDs (below the red arrows). Plants (A) and shoots (B) were imaged after 8 weeks exposure to SDs. (C) Shoots from ramets of the same FDL3ox event grown 8 weeks in SDs or LDs. (D,E) The changes in leaf expansion size of FDL3ox plants followed the changes of photoperiod duration. (D) FDL3ox and WT plants were grown for 2 months in LDs (below the red arrows), followed by 4 weeks of SDs (between red arrows and yellow arrows), and then 3 weeks of LDs (above the yellow arrows). (E) Fully expanded leaf length of WT and FDL3ox plants formed in SDs and LDs. Six fully expanded leaves were measured for each plant. Leaf length is mean ± SE (n = 4); different letters indicated significant differences, p < 0.0001, Tukey–Kramer’s test.
The SD-mediated restoration of leaf expansion in FDL3ox (Figure 7B), prompted us to examine secondary growth of the stem segments formed under SD conditions. After 8 weeks in SD conditions, substantial secondary xylem and phloem fiber bundles were present in IN2 of WT plants, which set bud before the internode was collected (Figure 5C5). In contrast to the poor secondary growth of FDL3ox plants under LD conditions (Figures 5B1–4), a closed circle of secondary xylem and phloem fiber bundles were present in all three internodes of FDL3ox plants formed under SD (Figures 5C1–3). Similar to leaves of FDL3ox plants formed under LDs and their failure to expand and mature under SDs (Figures 7A,B), the secondary xylem of IN10 formed in FDL3ox plants grown under LDs remained poorly developed in SDs (Figure 5C4).
FDL3ox and Daylength Alter FT2 and AP1/FUL Expression
The rapid change in FDL3ox leaf development following the shift from LD to SD conditions (Figure 7) prompted us to study changes in expression of FT2 and possible transcriptional targets of FDL3. In rice, FT-FD complexes form in leaves as well as in SAM and the small leaf phenotype of 35S::FT Arabidopsis under SDs requires FD and ectopic expression of FUL (Teper-Bamnolker and Samach, 2005; Brambilla et al., 2017). In source leaves, FT2 is rapidly downregulated in response to SDs (Böhlenius et al., 2006; Hsu et al., 2011; Supplementary Figure S11) and studies suggest that LIKE-AP1a (LAP1a) acts downstream of a FT-FDL1 complex (Azeez et al., 2014; Tylewicz et al., 2015). Moreover, Populus AP1/FUL family members were upregulated in 35S:FDL2.2 transgenics (Parmentier-Line and Coleman, 2016), and inducible FT1 or FT2 expression upregulated FUL expression (Hsu et al., 2011).
The P. trichocarpa genome contains five members of the AP1/FUL family (Supplementary Figure S8A). In FDL3ox transgenics, only LAP1a, LAP1b, and FUL were upregulated under LDs and their expression levels were correlated (R2 ≥ 0.79) with expression level of the FDL3ox transgene (Supplementary Figure S8). In WT trees under LDs, FT2 was dramatically upregulated as leaves near full expansion (LPI6), but leaf stage had comparatively little effect on FDL3 expression (Supplementary Figure S9). Compared to WT plants, FT2 expression in LPI6 leaf was 4–6-fold higher in FDL3ox transgenics in LDs (Figure 8A). In WT plants under LDs, LAP1a, b transcripts were low to barely detectable in shoot apices and LPI6 leaf, whereas FUL was relatively highly expressed and downregulated in multiple tissues in response to SDs (Supplementary Figure S10). However, all three genes were upregulated in LPI6 leaf and shoot apices of FDL3ox trees under LDs (Figures 8B–D, Supplementary Figure S12). In SDs, expression of FT2 and the AP1/FUL homologs was reduced in both WT and FDL3ox leaf and/or shoot apex (Figures 8A–D, Supplementary Figure S11). These results suggest that under LDs, FDL3ox transgenics could have elevated levels of a FT2-FDL3 complex in leaves as well as shoot apices that activates AP1/FUL homologs but that under SDs, reduced FT2 levels limit complex formation and AP1/FUL expression.
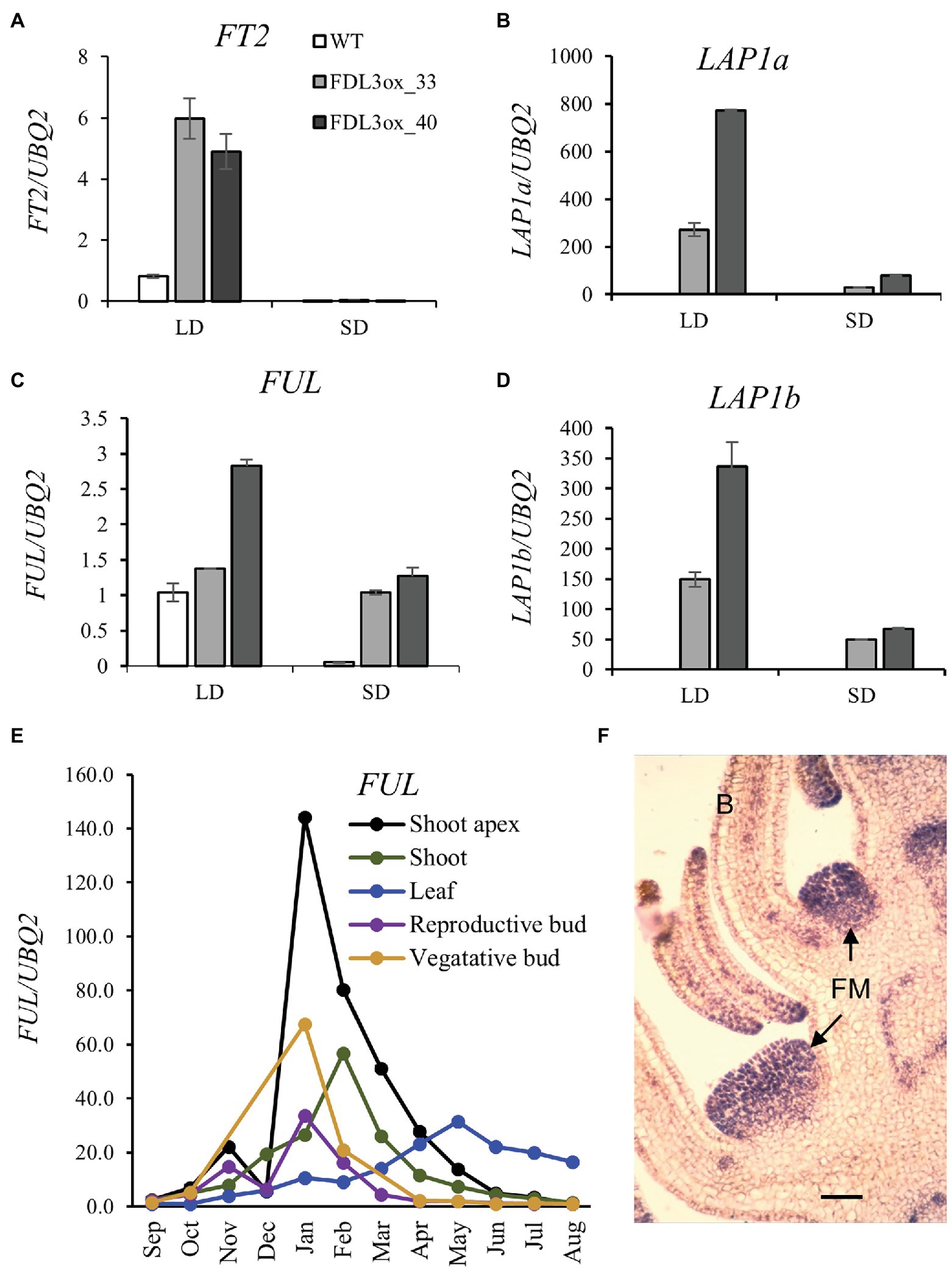
Figure 8. FDL3ox and daylength alter expression of FT2 and three AP1/FUL homologs with diverse vegetative and reproductive expression patterns. (A–D) Fully expanded leaves were collected from WT and two independent events of FDL3ox plants grown for 2 months in LDs, followed by 3 weeks in SDs. Relative fold changes in transcript levels of FT2 (A), LAP1a (B), FUL (C), and LAP1b (D). Expression of the other two members of the AP1/FUL family (Supplementary Figure 8), MADS14 and MADS28, was not detectable in either LDs or SDs. The expression was normalized against reference gene UBQ2. (E) Seasonal expression pattern of FUL in adult Populus deltoides. Relative expression is fold change in transcript levels relative to the time point with the lowest expression within a tissue (n = 3 biological replicates except that three technical replicates were assayed from a pool of three shoot apices). (F) In situ hybridization showing LAP1b expression in initiating floral meristems of an immature male Populus trichocarpa inflorescence. FM, floral meristem; B, bract. Scale bar = 100 μm. Additional LAP1a and LAP1b in situ hybridizations are provided in Supplementary Figure S12.
To elucidate possible endogenous roles and diversification of the three AP1/FUL homologs, we studied their expression patterns. Because FUL was expressed in various tissues of juvenile trees (Supplementary Figure S10), we studied its circannual expression in adult P. deltoides. FUL was expressed in all tissues, being most highly upregulated in shoot apices in winter with expression declining to lowest levels after shoots had set terminal buds in summer (Figure 8E). In juvenile samples, expression of the AP1 co-orthologs (LAP1a and LAP1b) was low compared to FUL expression (Supplementary Figure S10), and transcriptome profiling of poplar floral bud development indicated a role for LAP1 in flowering (Chen et al., 2018). Thus, we studied the spatial expression patterns of the LAP1 paralogs in developing reproductive buds. In situ hybridization of early developmental stages showed strong expression in initiating floral meristems (Figure 8F; Supplementary Figure S12), consistent with the conserved role of AP1 orthologs in specifying floral meristem identity (Pabon-Mora et al., 2013).
Discussion
Both Regulatory and Protein Variation Likely Contribute to the Functional Diversification of FDL Paralogs
The different tissue and seasonal expressional patterns of poplar FDL genes indicate that regulatory diversification of FDLs is linked to distinct roles in flowering and vegetative phenology. FDL2 is distinct in that it is the only FDL showing predominately reproductive expression (Figure 1). Both splice variants are upregulated in initiating axillary inflorescence buds, consistent with a canonical function in the control of the floral transition. However, only FDL2.2 induces early onset of flowering (Figure 3; Tylewicz et al., 2015; Parmentier-Line and Coleman, 2016). FDL1 expression was highest in late fall and winter (Figures 1A,E), consistent with one of its previously proposed functions, cold adaptation (Tylewicz et al., 2015). Conversely, FDL3 is expressed in shoot apices during the growing season (Figures 1C,E) and FDL3ox plants initiated new leaves at a faster rate than WT under LDs (Figure 4D), suggesting that FDL3 might be the paralog with a primary role in promoting apical growth.
The dominant repressor version of each of the FDL genes reduced shoot elongation in LDs to markedly different degrees (Figures 2, 3A; Supplementary Figure S4), indicating that the proteins share partial functional equivalency. In contrast, growth was extended in SDs by overexpression of FDL1, FDL2.2, or FDL3 (Figure 6; Tylewicz et al., 2015; Parmentier-Line and Coleman, 2016), but detection of any differences in degree of this SD phenotype was not possible as each FDL transgenic was studied in a different lab. However, transgenics overexpressing WT versions also supported partial functional equivalency of the three FDL proteins because these transgenics differed in regard to other phenotypic effects. FDL1 overexpression did not induce obvious changes in growth and development under LDs (Tylewicz et al., 2015), whereas overexpression of either FDL2.2 or FDL3 altered vegetative phenotypes, but only FDL2.2 induced flowering (Parmentier-Line and Coleman, 2016; Figure 3).
The only difference between the proteins encoded by the two FDL2 splice variants (FDL2.1 and FDL2.2) is an additional 29 amino acids within the FDL2.1 bZIP domain (Supplementary Figure S1CS). FDL2.2 promoted dramatic changes in vegetative phenotype and precocious flowering, while FDL2.1 overexpression inhibited growth (Tylewicz et al., 2015), yielding a similar albeit weaker version of the growth suppression resulting from FDL2.1rd overexpression (Supplementary Figure S4). Perhaps the 29 additional amino acids in FDL2.1 interfere with transcriptional activation of FDL2 target genes, weakly mimicking the FDL2.1rd phenotype. That the seasonal fluctuations of FDL2.1 and FDL2.2 expression in the shoot apex and tissue-level expression patterns of these variants are similar (Figure 1; Supplementary Figure S3) also points to the possibility that FDL2.1 acts as a regulator of FDL2.2.
With knowledge of the varied differences among the FDL-induced phenotypes, comparison of protein sequences can suggest domains to target for future analysis to determine the sequence variation responsible for protein functional divergence. For example, both FDL2.2ox and FDL3ox plants had small leaves and vine-like stems in LDs, but FDL3ox did not induce flowering (Figure 3). Tsuji et al. (2013) proposed that the LSL motif (Supplementary Figure S1C) is important for flowering as it was present in most eudicot FDs, but among diverse Poaceae FDs, it was limited to the subgroup containing flowering-promoting FDs. In FDL3, the first leucine of the LSL motif is replaced with serine, suggesting a candidate mutation for the absence of flowering-promoting activity.
The FD, FT, and AP1/FUL families have conserved roles in flowering, but studies in diverse angiosperms have also shown different patterns of gene duplication/loss and subsequent functional evolution (Litt and Irish, 2003; Abe et al., 2005; Wigge et al., 2005; Hsu et al., 2011; Taoka et al., 2011; Tsuji et al., 2013; Tylewicz et al., 2015). Whereas the two Populus FTs derive from the Salicoid WGD, the FD and AP1/FUL families are more complex, with Salicoid duplicates retained for only some members (FDL1/FDL2 and LAP1a/LAP1b; Rodgers-Melnick et al., 2012). Similar to the FT paralogs, FDL1 and FDL2 show highly divergent expression patterns (Figure 1; Hsu et al., 2011). Whereas it remains to be determined if this allowed FT and FDL duplicates to evolve in concert, the overlap in seasonal peak expression (winter) for FT1 and FDL1 (Figure 1A; Hsu et al., 2011) and that their proteins interact (Tylewicz et al., 2015) suggest this as a possibility. FT1, FDL2.2., and FT2, albeit less effectively, induce precocious flowering when overexpressed (Figure 3; Böhlenius et al., 2006; Hsu et al., 2006, 2011; Parmentier-Line and Coleman, 2016). In contrast, overexpression of FDL1 or LAP1a did not induce early flowering (Azeez et al., 2014; Tylewicz et al., 2015), but insufficiency does not preclude a role in flowering. FT1 is upregulated in winter vegetative buds, and detailed study of reproductive development indicated that inflorescence buds subsequently (i.e., shortly after bud flush) develop in the axils of late preformed leaves (Yuceer et al., 2003; Hsu et al., 2011). Although FT2 is predominately expressed in source leaves, it is also expressed coincident with FDL2.2 in developing spring inflorescence buds (Figure 1D; Hsu et al., 2011) and LAP1a and LAP1b are expressed in initiating floral meristems (Figure 8F, Supplementary Figure S12). We previously posited that both FT1 and FT2 could have roles in flowering but act at different stages—FT1 might promote the transition of incipient axillary meristems within winter buds to inflorescence meristems, while FT2 might promote floral meristem initiation within the developing inflorescence (Brunner et al., 2014). Perhaps FDL1 and FDL2 could have similarly diversified roles in flowering as well as the suggested diversification of three FDLs in vegetative development. Gene editing can potentially clarify their gene-specific functions; however, given the difficulties in generating plants with FDL1rd or FDL3rd transgenes, this might require mutations that reduce or alter only specific sequences/functions rather than knock out gene activity. Use of an FT-mediated early flowering poplar system (Azeez and Busov, 2019) could help delineate roles in flowering. Moreover, FDL2 is potentially a candidate for CRISPR-mediated manipulation to prevent flowering and mitigate gene flow from plantations. For forest trees, not only both male and female reproductive sterility but prevention of flower formation is desired for biosafety (Fritsche et al., 2018).
FDL3 Overexpression Induces Heterochronic Shifts in Shoot Ontogeny Depending on Photoperiod
In addition to a faster rate of phytomer initiation in LDs, leaf expansion and the transition to secondary growth were greatly delayed in FDL3ox shoots (Figures 4, 5; Supplementary Figures S5, S6). The formation of a vascular cambium introduces another sink; thus, the occurrence of this transition at a distance from an active SAM and below a leaf that is at least partially mature reflects sink-source relationships (Larson and Isebrands, 1971, 1974). Moreover, the direction of carbon transport changes as leaves develop (Dickson and Larson, 1981; Isebrands and Nelson, 1983). In general, transitional leaves transport carbon upward to younger leaves and SAM, recently mature leaves transport in both directions, and older leaves transport carbon to the lower stem and roots. The delayed transition to secondary growth in FDL3ox plants (Figure 5B; Supplementary Figure S6) might be a direct consequence of stalled leaf development preventing production and transport of sufficient sugar to initiate and support secondary growth. A more active SAM (i.e., more rapid phytomer initiation; Figure 4D) and hence increased SAM sink strength in FDL3ox plants might further limit sugar import for leaf expansion and concurrently, reduce carbon availability for cambium formation. FT2 upregulation is strongly correlated with leaf maturation (Supplementary Figure S9B). In Arabidopsis, the signaling sugar trehalose-6-phosphate (T6P) increases FT expression in leaves to promote florigen (Wahl et al., 2013). Thus, it will be interesting to determine if the T6P pathway acts to induce FT2 expression in source leaves, linking FT2 signaling to leaf sugar status and export direction to apical and cambial meristems.
Overexpression of GA20ox in poplar increased leaf size, height and diameter in LDs and delayed SD-induced growth cessation but not FT2 downregulation, suggesting that FT2 and GA act in parallel growth-promoting pathways (Eriksson et al., 2000, 2015). In FDL3ox trees, reduced GA20ox5 expression in leaves and that GA-responsive genes GASA6 and EXPA8 were not upregulated suggests that reduced GA levels and signaling contributed to reduced leaf expansion (Figure 5D). In tobacco, GA signaling from maturing leaves is important for both shoot elongation and radial growth (Dayan et al., 2012) and GA can act systemically to delay SD-induced growth cessation in poplar (Miskolczi et al., 2019). However, the FDL3ox phenotype does not support that reduced GA levels in leaves reduces apical growth as FDL3ox trees initiate phytomers at a faster rate and SD-induced growth cessation is delayed (Figures 4D, 6). Moreover, FDL3ox plants grew faster with longer internodes than WT in vitro on sugar-containing media, the opposite of FDL3rd plants (Figures 3A,B). This suggests that the reduced height growth of potted FDL3ox plants (Figures 3G, 4C) could be a secondary effect of small transgenic leaves providing less photosynthate than WT leaves.
Gibberellin is synthesized in apices and signaling might be predominately local as grafting onto GA-overexpressing rootstocks was less effective at delaying SD-induced growth cessation in WT scions than grafting onto FT-overexpressing rootstocks (Miskolczi et al., 2019). Expression of stem elongation marker gene GASA6 was similarly high in WT and FDL3ox IN4, but unlike in WT, showed no decrease in IN8 of FDL3ox trees, consistent with their protracted primary growth phase (Figure 5E). Although GA promotes wood formation, GASA6 expression in FDL3ox stems suggests that a localized attenuation of GA-promoted stem elongation might be necessary for internodes to transition to secondary growth.
A striking phenotype of FDL3ox trees was the rapid change in leaf and stem development with photoperiod (Figure 7), indicating that effect of FDL3 on shoot ontogeny depends on genes whose activity is controlled by daylength. In LDs, FT2, LAP1, and FUL expression is elevated in FDL3ox trees, but as in WT, all are downregulated by SDs (Figure 8; Supplementary Figure S11). However, in SDs, LAP1 and FUL expression is still higher in FDL3ox plants compared to WT. This suggests that FDL3 overexpression could increase the level of a FDL3-FT2 complex in leaf as well as SAM under LDs, elevating the expression of downstream targets to promote SAM activity, but limiting leaf expansion and the transition to secondary growth. Under SDs, perhaps reduced LAP1/FUL expression is sufficient to maintain SAM activity, but not to delay leaf development and the transition to secondary growth. However, elevated LAP1a expression is not sufficient to explain the FDL3ox phenotype as it did not alter leaf development (Azeez et al., 2014). Various FT and FD homologs have been shown to interact with other proteins (Mimida et al., 2011; Tsuji et al., 2013; Tylewicz et al., 2015; Jung et al., 2016; Brambilla et al., 2017; Li et al., 2019); thus, the effect of FDL3 on leaf development and secondary growth could be independent of FT2. Heterochronic mutants have helped reveal genetic mechanisms controlling seed maturation and vegetative phase change (reviewed in Buendia-Monreal and Gillmor, 2018). Thus, further study of FDL3ox transgenics could provide an inroad into understanding the genetic pathways that link leaf and stem ontogenies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material .
Author Contributions
XS, C-YH, and AB designed experiments, performed research, and analyzed results. CM produced overexpression transgenics used in this study. XS and AB drafted the paper. C-YH and CM revised and provided comments on the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the US Department of Energy Office of Science, Office of Biological and Environmental Research, Grant No. DE-SC0012574 and the USDA National Institute of Food and Agriculture, McIntire Stennis project 1025004.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Cetin Yuceer for his role in initiating this study, Takeshi Fujino for isolating aspen 3′ RACE clones and Eric Beers for helpful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.805101/full#supplementary-material
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Azeez, A., and Busov, V. B. (2019). Improved heat FT induction leads to earlier and more prolific flowering in Poplar. J. Bot. Res. 1, 15–18. doi: 10.30564/jrb.v1i2.1013
Azeez, A., Miskolczi, P., Tylewicz, S., and Bhalerao, R. P. (2014). A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr. Biol. 24, 717–724. doi: 10.1016/j.cub.2014.02.037
Bartlett, M. (2020). Looking back to look forward: protein-protein interactions and the evolution of development. New Phytol. 225, 1127–1133. doi: 10.1111/nph.16179
Bernier, G., and Perilleux, C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3, 3–16. doi: 10.1111/j.1467-7652.2004.00114.x
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Brambilla, V., Martignago, D., Goretti, D., Cerise, M., Somssich, M., de Rosa, M., et al. (2017). Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 29, 2801–2816. doi: 10.1105/tpc.17.00645
Britton, C. S., Sorrells, T. R., and Johnson, A. D. (2020). Protein-coding changes preceded cis-regulatory gains in a newly evolved transcription circuit. Science 367, 96–100. doi: 10.1126/science.aax5217
Brunner, A. M., Evans, L. M., Hsu, C.-Y., and Sheng, X. (2014). Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front. Plant Sci. 5:732. doi: 10.3389/fpls.2014.00732
Brunner, A. M., Varkonyi-Gasic, E., and Jones, R. C. (2017). “Phase change and phenology in trees,” in Comparative and Evolutionary Genomics of Angiosperm Trees. eds. A. Groover and Q. Cronk (Cham, Switzerland: Springer), 227–274.
Brunner, A. M., Yakovlev, I. A., and Strauss, S. H. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4:14. doi: 10.1186/1471-2229-4-14
Buendia-Monreal, M., and Gillmor, C. S. (2018). The times they are A-Changin': Heterochrony in plant development and evolution. Front. Plant Sci. 9:1349. doi: 10.3389/fpls.2018.01349
Chen, Z., Rao, P., Yang, X., Su, X., Zhao, T., Gao, K., et al. (2018). A Global view of transcriptome dynamics during male floral bud development in Populus tomentosa. Sci. Rep. 8:722. doi: 10.1038/s41598-017-18084-5
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Dayan, J., Voronin, N., Gong, F., Sun, T. P., Hedden, P., Fromm, H., et al. (2012). Leaf-induced gibberellin signaling is essential for internode elongation, cambial activity, and fiber differentiation in tobacco stems. Plant Cell 24, 66–79. doi: 10.1105/tpc.111.093096
Dharmawardhana, P., Brunner, A. M., and Strauss, S. H. (2010). Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11:150. doi: 10.1186/1471-2164-11-150
Dickson, R. E., and Larson, P. R. (1981). 14C fixation, metabolic labeling patterns, and translocation profiles during leaf development in Populus deltoides. Planta 152, 461–470. doi: 10.1007/bf00385364
Ding, J., and Nilsson, O. (2016). Molecular regulation of phenology in trees-because the seasons they are a-changin. Curr. Opin. Plant Biol. 29, 73–79. doi: 10.1016/j.pbi.2015.11.007
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Eriksson, M. E., Hoffman, D., Kaduk, M., Mauriat, M., and Moritz, T. (2015). Transgenic hybrid aspen trees with increased gibberellin (GA) concentrations suggest that GA acts in parallel with FLOWERING LOCUS T2 to control shoot elongation. New Phytol. 205, 1288–1295. doi: 10.1111/nph.13144
Eriksson, M. E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Fritsche, S., Klocko, A. L., Boron, A., Brunner, A. M., and Thorlby, G. (2018). Strategies for engineering reproductive sterility in plantation forests. Front. Plant Sci. 9:1671. doi: 10.3389/fpls.2018.01671
Grant, E. H., Fujino, T., Beers, E. P., and Brunner, A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337–352. doi: 10.1007/s00425-010-1181-2
Groover, A. T. (2005). What genes make a tree a tree? Trends Plant Sci. 10, 210–214. doi: 10.1016/j.tplants.2005.03.001
Hall, H., and Ellis, B. (2013). Transcriptional programming during cell wall maturation in the expanding Arabidopsis stem. BMC Plant Biol. 13:14. doi: 10.1186/1471-2229-13-14
Hanada, K., Zou, C., Lehti-Shiu, M. D., Shinozaki, K., and Shiu, S. H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003. doi: 10.1104/pp.108.122457
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313X.2003.01759.x
Howe, G. T., Aitken, S. N., Neale, D. B., Jermstad, K. D., Wheeler, N. C., and Chen, T. H. H. (2003). From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can. J. Bot. 81, 1247–1266. doi: 10.1139/b03-141
Hsu, C. Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U. S. A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Hsu, C. Y., Liu, Y., Luthe, D. S., and Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18, 1846–1861. doi: 10.1105/tpc.106.041038
Isebrands, J. G., and Nelson, N. D. (1983). Distribution of [14C]-labeled photosynthates within intensively cultured Populus clones during the establishment year. Physiol. Plant. 59, 9–18. doi: 10.1111/j.1399-3054.1983.tb06563.x
Jung, J. H., Lee, H. J., Ryu, J. Y., and Park, C. M. (2016). SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 9, 1647–1659. doi: 10.1016/j.molp.2016.10.014
Kelly, A. J., Bonnlander, M. B., and Meeks-Wagner, D. R. (1995). NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell 7, 225–234. doi: 10.1105/tpc.7.2.225
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larson, P. R., and Isebrands, J. G. (1971). The plastochron index as applied to developmental studies in cottonwood. Can. J. For. Res. 1, 1–11. doi: 10.1139/x71-001
Larson, P. R., and Isebrands, J. G. (1974). Anatomy of the primary-secondary transition zone in stems of Populus deltoides. Wood Sci. Technol. 8, 11–26. doi: 10.1007/bf00350638
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis class II TCP transcription factors integrate with the FT-FD module to control flowering. Plant Physiol. 181, 97–111. doi: 10.1104/pp.19.00252
Litt, A., and Irish, V. F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165, 821–833. doi: 10.1093/genetics/165.2.821
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lynch, V. J., and Wagner, G. P. (2008). Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
McKim, S. M. (2019). How plants grow up. J. Integr. Plant Biol. 61, 257–277. doi: 10.1111/jipb.12786
Meilan, R., and Ma, C. (2006). Poplar (Populus spp.). Methods Mol. Biol. 344, 143–151. doi: 10.1385/1-59745-131-2:143
Mimida, N., Kidou, S., Iwanami, H., Moriya, S., Abe, K., Voogd, C., et al. (2011). Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiol. 31, 555–566. doi: 10.1093/treephys/tpr028
Miskolczi, P., Singh, R. K., Tylewicz, S., Azeez, A., Maurya, J. P., Tarkowska, D., et al. (2019). Long-range mobile signals mediate seasonal control of shoot growth. Proc. Natl. Acad. Sci. U. S. A. 116, 10852–10857. doi: 10.1073/pnas.1902199116
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Pabon-Mora, N., Hidalgo, O., Gleissberg, S., and Litt, A. (2013). Assessing duplication and loss of APETALA1/FRUITFULL homologs in Ranunculales. Front. Plant Sci. 4:358. doi: 10.3389/fpls.2013.00358
Parmentier-Line, C. M., and Coleman, G. D. (2016). Constitutive expression of the poplar FD-like basic leucine zipper transcription factor alters growth and bud development. Plant Biotechnol. J. 14, 260–270. doi: 10.1111/pbi.12380
Preston, J. C., and Sandve, S. R. (2013). Adaptation to seasonality and the winter freeze. Front. Plant Sci. 4:167. doi: 10.3389/fpls.2013.00167
Rinne, P. L., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Rodgers-Melnick, E., Mane, S. P., Dharmawardhana, P., Slavov, G. T., Crasta, O. R., Strauss, S. H., et al. (2012). Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22, 95–105. doi: 10.1101/gr.125146.111
Rohde, A. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14, 1885–1901. doi: 10.1105/tpc.003186
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. doi: 10.1105/tpc.107.052811
Schmid, M., Uhlenhaut, N. H., Godard, F., Demar, M., Bressan, R., Weigel, D., et al. (2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. doi: 10.1242/dev.00842
Shi, R., Sun, Y. H., Li, Q., Heber, S., Sederoff, R., and Chiang, V. L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51, 144–163. doi: 10.1093/pcp/pcp175
Soltis, P. S., Marchant, D. B., Van de Peer, Y., and Soltis, D. E. (2015). Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125. doi: 10.1016/j.gde.2015.11.003
Spicer, R., and Groover, A. (2010). Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592. doi: 10.1111/j.1469-8137.2010.03236.x
Taoka, K.-I., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Teixeira, R. T., Sheng, X., and Brunner, A. M. (2019). “Activity of the shoot apical and cambial meristems: coordination and responses to environmental signals,” in Molecular Physiology and Biotechnology of Trees. ed. F. M. Cánovas (London, United Kingdom: Elsevier), 185–199.
Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675. doi: 10.1105/tpc.105.035766
Torti, S., Fornara, F., Vincent, C., Andres, F., Nordstrom, K., Gobel, U., et al. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24, 444–462. doi: 10.1105/tpc.111.092791
Tsuji, H., Nakamura, H., Taoka, K., and Shimamoto, K. (2013). Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 54, 385–397. doi: 10.1093/pcp/pct005
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Tylewicz, S., Tsuji, H., Miskolczi, P., Petterle, A., Azeez, A., Jonsson, K., et al. (2015). Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. U. S. A. 112, 3140–3145. doi: 10.1073/pnas.1423440112
Wahl, V., Ponnu, J., Schlereth, A., Arrivault, S., Langenecker, T., Franke, A., et al. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707. doi: 10.1126/science.1230406
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Wu, S., Han, B., and Jiao, Y. (2020). Genetic contribution of Paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13, 59–71. doi: 10.1016/j.molp.2019.10.012
Xie, J., Tian, J., Du, Q., Chen, J., Li, Y., Yang, X., et al. (2016). Association genetics and transcriptome analysis reveal a gibberellin-responsive pathway involved in regulating photosynthesis. J. Exp. Bot. 67, 3325–3338. doi: 10.1093/jxb/erw151
Yuceer, C., Land, S. B., Kubiske, M. E., and Harkess, R. L. (2003). Shoot morphogenesis associated with flowering in Populus deltoides (Salicaceae). Am. J. Bot. 90, 196–206. doi: 10.3732/ajb.90.2.196
Keywords: FLOWERING LOCUS D, FT, FRUITFULL, gene duplication, heterochrony, leaf development, phenology, secondary growth
Citation: Sheng X, Hsu C-Y, Ma C and Brunner AM (2022) Functional Diversification of Populus FLOWERING LOCUS D-LIKE3 Transcription Factor and Two Paralogs in Shoot Ontogeny, Flowering, and Vegetative Phenology. Front. Plant Sci. 13:805101. doi: 10.3389/fpls.2022.805101
Edited by:
Hisayo Yamane, Kyoto University, JapanReviewed by:
Ove Nilsson, Umeå University, SwedenErika Varkonyi-Gasic, The New Zealand Institute for Plant and Food Research Ltd., New Zealand
Nobuhiro Kotoda, Saga University, Japan
Copyright © 2022 Sheng, Hsu, Ma and Brunner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy M. Brunner, YWJydW5uZXJAdnQuZWR1
 Xiaoyan Sheng
Xiaoyan Sheng Chuan-Yu Hsu2
Chuan-Yu Hsu2 Amy M. Brunner
Amy M. Brunner