
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 February 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.801456
This article is part of the Research TopicThe Brassicaceae — Agri-Horticultural and Environmental Perspectives, Volume IIView all 19 articles
 Huailin Li1,2†
Huailin Li1,2† Kaidi Yu1,2†
Kaidi Yu1,2† Olalekan Amoo1,2
Olalekan Amoo1,2 Yalun Yu1,2
Yalun Yu1,2 Mixia Guo1,2
Mixia Guo1,2 Songyue Deng1,2
Songyue Deng1,2 Mengting Li1,2
Mengting Li1,2 Limin Hu1,2
Limin Hu1,2 Jingzhen Wang1,2
Jingzhen Wang1,2 Chuchuan Fan1,2*
Chuchuan Fan1,2* Yongming Zhou1,2
Yongming Zhou1,2The diversity of petal and leaf color can improve the ornamental value of rapeseed and promote the development of agriculture and tourism. The two copies of carotenoid isomerase gene (BnaCRTISO) in Brassica napus (BnaA09.CRTISO and BnaC08.CRTISO) was edited using the CRISPR/Cas9 system in the present study. The mutation phenotype of creamy white petals and yellowish leaves could be recovered only in targeted mutants of both BnaCRTISO functional copies, indicating that the redundant roles of BnaA09.CRTISO and BnaC08.CRTISO are vital for the regulation of petal and leaf color. The carotenoid content in the petals and leaves of the BnaCRTISO double mutant was significantly reduced. The chalcone content, a vital substance that makes up the yellow color, also decreased significantly in petals. Whereas, the contents of some carotenes (lycopene, α-carotene, γ-carotene) were increased significantly in petals. Further, transcriptome analysis showed that the targeted mutation of BnaCRTISO resulted in the significant down-regulation of important genes BnaPSY and BnaC4H in the carotenoid and flavonoid synthesis pathways, respectively; however, the expression of other genes related to carotenes and xanthophylls synthesis, such as BnaPDS3, BnaZEP, BnaBCH1 and BCH2, was up-regulated. This indicates that the molecular mechanism regulating petal color variation in B. napus is more complicated than those reported in Arabidopsis and other Brassica species. These results provide insight into the molecular mechanisms underlying flower color variation in rapeseed and provides valuable resources for rapeseed breeding.
Flower color is considered one of the major attractants for pollen transmission in nature. Insects can recognize different flower colors through visual signals and transmit targeted pollen between flowers (Cazzonelli, 2011; Nisar et al., 2015). It is generally recognized that flower color is determined by three main pigments: carotenoids, flavonoids, and betalains (Grotewold, 2006). Carotenoids mainly provide orange and red color, flavonoids provide yellow, and betalains provide a few other colors (Cazzonelli, 2011). They are fat-soluble terpenoids synthesized via the isoprene pathway and have been found in various plants and animals (Cunningham and Gantt, 1998). Carotenoids are important pigments determining the color of fruits, vegetables, and flowers (Lv et al., 2015; Zhang et al., 2015; Li X. et al., 2018). They are found mainly in leaf, flower, fruit, and root tissues and play a vital role during plant development, such as protecting the plants against photo-oxidative damage (Holt et al., 2005; Sun et al., 2018). Carotenoids are also the precursors for the biosynthesis of vitamin A, the plant hormones abscisic acid (ABA) and strigolactone (Walter and Strack, 2011).
The biosynthetic pathway of carotenoid in higher plants has been elucidated, and many genes encoding key enzymes involved in this pathway have been successfully cloned. Based on the presence or absence of oxygen in their molecular structure, carotenoids can be divided into two categories, i.e., carotenes and xanthophylls. Carotenes contain carbon and hydrogen in their molecular structure, such as phytoene, lycopene, and α-carotene; xanthophylls contain oxygen, such as lutein, zeaxanthin, neoxanthin (Montserrat et al., 2015). In general, carotenoids in plants use GGPP (Geranylgeranyl pyrophosphate) as the synthetic substrate except for a few plants (Fraser and Bramley, 2004; Matsufuji et al., 2007). In Arabidopsis thaliana, the biosynthesis of carotenoid is regulated by ten enzymes and eleven genes (Patrick et al., 2018).
Carotenoid isomerase (CRTISO) converts the yellow colored prolycopene into the red colored all-trans lycopene in the carotenoid synthesis pathway (Breitenbach and Sandmann, 2005). Previous studies have shown that down-regulation of CRTISO gene expression in citrus can promote the accumulation of β-carotene (Kato et al., 2007). It was also reported that the expression of CRTISO gene could increase carotenoid accumulation in the endosperm of maize seeds (Wurtzel, 2009), implying that the expression of CRTISO gene has a certain correlation with the accumulation of carotenoids in plants. At present, the CRTISO gene has been cloned in tomato (Isaacson, 2002), maize (Wurtzel, 2009), melon (Galpaz et al., 2013), and A. thaliana (Hyoungshin, 2002). But there is no relevant research report on this gene in rapeseed.
Rapeseed is the third-largest oilseed crop worldwide with multi-function application, which are widely used as edible vegetable oil, vegetables, fodder, biofuel and nectar, improvement of saline and alkaline soils (Liu et al., 2020). The distinctive flower color of rapeseed also shows high ornamental value, which has received increased attention in China. The typical flower color of rapeseed is yellow, but there are also reports of some variant flower colors, such as white, yellowish and orange. Numerous genetic analysis of flower color have indicated that this trait is little affected by environmental factors and shows dominant or incompletely dominant inheritance (Quazi, 1988; Zhang et al., 2010; Huang et al., 2014). Up to date, several genes controlling flower color have been reported in B. napus. The white flower color is controlled by a single dominant gene, BnaC3.CCD4, which encodes a carotenoid cleavage dioxygenase and is involved in carotenoid degradation (Zhang et al., 2015; Han et al., 2019). The insertion of a CACTA-like transposable element in BnaC3.CCD4 leads to its loss-of-function and a subsequently enhanced accumulation of carotenoids; thus, results in a petal color transition from white to yellow (Zhang et al., 2015; Han et al., 2019). Gene silencing of two genes, BnaA09.ZEP and BnaC09.ZEP, confers the change in flower color from yellow to orange (Liu et al., 2020). These two genes are homologous to the nuclear-encoded plastid enzyme zeaxanthin epoxidase (AtZEP) and participate in carotenoid biosynthesis. Recently, Zhao et al. (2021) reported that a yellowish-white flower trait is controlled by a single recessive gene, BnaA08.PDS3, which encodes a phytoene desaturase 3 (Zhao et al., 2021). A C-to-T substitution in the coding region of BnaA08.PDS3 results in a premature translation termination and a subsequent decreased carotenoid biosynthesis; thus, changing the flower color from yellow to yellowish-white. Other researchers used cell fusion technology to obtain the fusion plant with white flower trait (47 chromosomes). When the chromosomes were reduced to 38 by backcrossing, the petal color changed from white to yellow (Sakai, 1995). Therefore, until now, the genes and molecular mechanisms regulating flower color in rapeseed have not been fully elucidated.
In recent years, sequence-specific nucleases (SSNs) have been demonstrated to be an amazing tool for improving crops via site-specific genome editing, and CRISPR/Cas9 is considered the most simple and efficient SSN. The CRISPR/Cas9 system has been effectively utilized in rapeseed to produce the targeted mutations for the improvement of numerous agronomic traits (Braatz et al., 2017; Yang et al., 2017, 2018; Hu et al., 2018; Li C. et al., 2018; Zhai et al., 2019; Ahmar et al., 2021).
Hence, we utilized the CRISPR/Cas9 system to generate efficient knockouts of CRTISO homeologs with stable transformation in rapeseed. In the T1, T2, and T3 generations, mutants containing the desired gene modification were obtained by segregation. The transcriptomic analysis and metabolite profiling of BnaCRTISO mutant plants were used in this current study to investigate the molecular mechanisms that regulate the petal color in B. napus. This study provided valuable germplasm resources for the innovation of different petal color varieties in rapeseed and offered a new way to improve polypoid crops.
In this study, B. napus pure line J9707 was used as the donor plants for transformation, and the seeds were obtained from the National Engineering Research Center of Rapeseed, Wuhan, China. The flowers on the primary inflorescence were marked at anthesis, and the petals with different colors were collected for transcriptomic analysis and metabolite profiling.
The binary pYLCRIPSR/Cas9 multiplex genome targeting vector system was utilized for gene editing in this study (Ma et al., 2015). The selection of sequence-specific sgRNAs in the target gene, CRISPR/Cas9 construct assembly, and Agrobacterium tumefaciens-mediated hypocotyl transformation in B. napus were conducted as previously described (Hu et al., 2018). The oligos employed in constructing the sgRNA vectors are listed in Supplementary Table 1. The resulting construct is described in detail in Figure 1B.
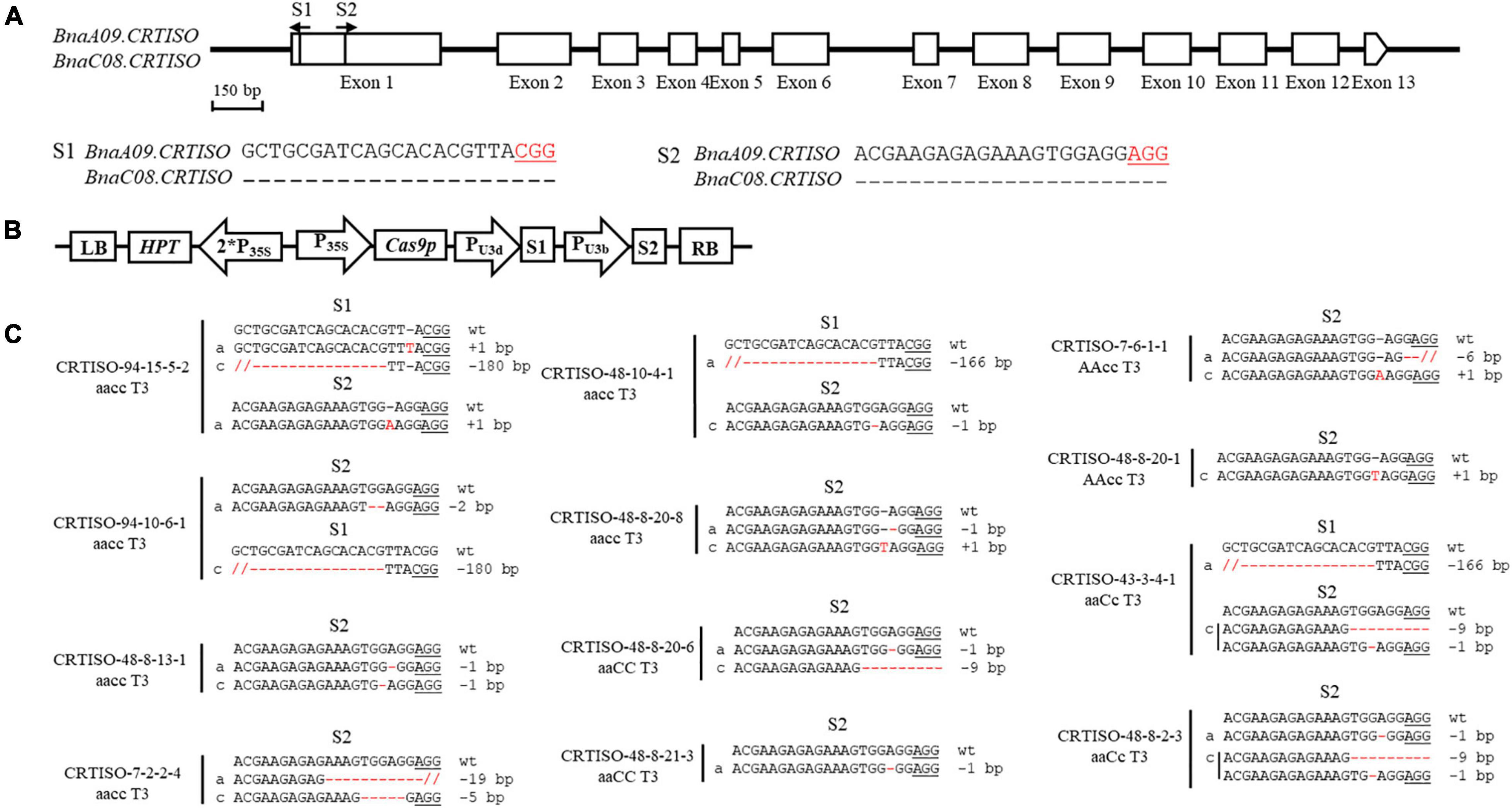
Figure 1. CRISPR/Cas9-induced null mutants of BnaCRTISO in B. napus. (A) The BnaCRTISO gene model includes thirteen exons (box) separated by twelve introns (represented by the solid line). The vertical line in the gene model indicates the target site, and the arrow indicates the sgRNA direction. The target sequences are shown with the protospacer adjacent motif (PAM) underlined. (B) The CRISPR/Cas9 construct houses the following: a hygromycin resistance cassette consisting of the hygromycin phosphotransferase coding sequence driven by the cauliflower mosaic virus 35S promoter; a Cas9 expression cassette comprising the sequence encoding Cas9 driven by 35S promoter; and two sgRNAs (S1-S2) driven by the U3d and U3b promoters from Arabidopsis. (C) Sequences at the sgRNA target sites of BnaCRTISO homozygous mutants in the T3 generation. The PAM is underlined, and nucleotide indels are marked in red, with details labeled at right, “a” and “c” represent the mutated alleles of the target gene on BnaA09.CRTISO and BnaC08.CRTISO, respectively. “aaCC,” “AAcc,” and “aacc” represent homozygous mutations of the target gene in BnaA09.CRTISO, BnaC08.CRTISO and both copies, respectively.
The transgenic plants were screened by hygromycin selection (25 mg/L). Then, the presence of the T-DNA in the construct was assessed by PCR using the specific primer pairs PB-L/PB-R (Supplementary Table 1).
The targeted mutations were determined in transgenic plants using the high-throughput tracking of mutations (Hi-TOM) platform (Zhai et al., 2019). Target-specific and barcoding PCR (two rounds of PCR) were performed to amplify the genomic region encompassing the specific targets of each independent sample, and the resulting PCR products were mixed in equal amounts and purified for next-generation sequencing (the Illumina HiSeq platform at the Novogene Bioinformatics Institute, Beijing, China). The sequencing data was then decoded using a corresponding online tool to track the mutations of the target sites1. The potential off-target sites were identified using CRISPR-P2.02. The primers used to detect targeted and potential off-target mutations are listed in Supplementary Table 1.
Total RNA was prepared using the EasyPure Plant RNA Kit (TransGen Biotech, Beijing, China), and cDNA was synthesized using the Transcript RT Kit (TransGen Biotech). The qRT-PCR was carried out using the TransStart Top Green qPCR SuperMix Kit (TransGen Biotech) on a CFX384 Real-Time System (Bio-Rad). Relative quantification was performed using the comparative cycle threshold method. The relative amount of PCR product that was amplified using the designed primer sets (Supplementary Table 1) was normalized to the reference genes, BnaACT2 and BnaUBC9.
Flower tissues were sampled with three biological replicates. Petals were gently hand dissected from the flower on dry ice, immediately frozen immediately in liquid nitrogen, and stored at −80°C until total RNA extraction.
RNA extraction, cDNA library construction, sequencing, quality control, and read mapping to the reference genome, identification of differentially expressed genes (DEGs), and GO and KEGG pathway enrichment analysis of DEGs were performed using previously established procedure (Shahid et al., 2019; Zhai et al., 2020). Fragments per kilobase of transcript per million mapped reads (FPKM) were calculated as a measure of the level of gene expression. Genes with a false discovery rate (FDR) ≤ 0.05 and an absolute value of log2 fold change ≥ 1 between mutant and wild type (WT) were defined as DEGs. The raw sequence data were deposited in the NCBI Sequence Read Archive (PRJNA749083).
Metabolites were extracted from petals (500 mg dry weight) with three biological replicates and were analyzed using LC-ESI-MS/MS system at the Metware Biotechnology Co., Ltd. (Wuhan, China). Flavonoids were extracted using the same method at the National Engineering Research Center of Rapeseed (Huazhong Agricultural University, Wuhan, China). The sample extraction and metabolic analysis were explicitly done as previously described (Zhai et al., 2020).
For carotenoids analysis, the petals were sampled from double mutants (CRTISO-94-15-5-2, CRTISO-48-8-13-1, CRTISO-7-2-2-4), heterozygous mutant (CRTISO-43-3-4-1, CRTISO-48-8-2-3, CRTISO-48-8-20-5) and WT. Carotenoid pigments extraction and analysis were performed using LC-MS/MS system, as previously described (Lee, 2001; Saladié et al., 2014). Carotenoids were identified based on retention times and absorption spectra as compared to standards. Peak areas were recorded at 286, 348, 473, and 450 nm for phytoene, phytofluene, lycopene, and others, respectively (Xu et al., 2006). The carotenoid levels were quantified using calibration curves prepared with appropriate standards. At least three independent extractions were conducted per sample.
For chlorophyll analysis, the leaves was extracted from double mutants (CRTISO-94-15-5-2, CRTISO-48-8-13-1, CRTISO-7-2-2-4) and WT and quantified using a spectrophotometer, as described previously (Becker, 1994). At least three independent extractions were conducted per sample.
The cDNA sequences of BnaA09.CRTISO and BnaC08.CRTISO without the termination codon were amplified from J9707 using primers CRTISO-15/16 and CRTISO-18/19, respectively (Supplementary Table 1). The amplified cDNA fragments were independently cloned into the pMDC83 vector between the PacI and the AscI site, to generate a C-terminal fusion with GFP under control of the cauliflower mosaic virus 35S promoter. The fused construct was introduced into Nicotiana benthamiana plants by transient Agrobacterium transformation. Samples were observed with a Leica TCSST2 confocal laser microscope (Nikon D760, Tokyo, Japan).
Previous studies revealed that the CRTISO gene is essential for regulating carotenoid content and is highly conserved in many plants (Galpaz et al., 2013; Su et al., 2015; Li et al., 2020; Sun et al., 2020). The carotenoid content of a flower directly determines the color of the petals (Su et al., 2015; Zhang et al., 2015; Liu et al., 2020; Zhao et al., 2021). Thus, CRTISO is one of the ideal candidates for generating different flower colors in rapeseed. There are two CRTISO copies (BnaA09g49740D and BnaC08g44970D, designated as BnaA09.CRTISO and BnaC08.CRTISO, respectively) in B. napus. In order to check for putative mutations in these target genes, we confirmed their genomic DNA and cDNA sequences in the B. napus pure line J9707. Compared to Arabidopsis AtCRTISO, the predicted amino acid sequences of both BnaCRTISO contain an important conserved domain, “the amine oxidase” (Supplementary Figures 1, 2). which meanings BnaA09.CRTISO and BnaC08.CRTISO can produce functional amine oxidase proteins.
BnaA09.CRTISO and BnaC08.CRTISO was 87.94% identical at the nucleotide level and shared 98.64% amino acid identity, suggesting that both genes encode enzymes with similar functions. The sequence alignment of both BnaCRTISO gene copies revealed that polymorphisms distinguished their origins (Supplementary Figure 3).
Phylogenetic analysis indicated that all Brassica genes were clustered together with AtCRTISO and CRTISO homologs in other species were clustered on another branch except in tomato (SolCRTISO), suggesting that the CRTISO gene is differentiated among different species (Supplementary Figure 4). BnaA09.CRTISO and BnaC08.CRTISO were closely related to their homologs in B. rapa and B. oleracea, respectively (Supplementary Figure 4), which is in line with their origination from two diploid progenitors.
Analysis of mRNA accumulation patterns of both BnaCRTISO copies based on the public RNA-seq data in yellow-flower rapeseed line showed their expression profiles (Supplementary Figure 5A). In all cases, BnaA09.CRTISO and BnaC08.CRTISO had the highest expression levels in the bud, followed by leaf, and the expression levels of both copies in various tissues were comparable. Thus, we further confirmed that the B. napus genome contains two functional AtCRTISO homologs, BnaA09.CRTISO and BnaC08.CRTISO.
The expression level of both BnaCRTISO copies in J9707 was further examined using qRT-PCR with RNA samples extracted from the petal (Supplementary Figure 5B). Consistent with the public RNA-seq data, BnaC08.CRTISO had a significantly higher expression level than BnaA09.CRTISO in the J9707 petal (Supplementary Figure 5A).
To generate CRISPR/Cas9-induced knockout mutations in the functional copies of BnaCRTISO, two sgRNAs were designed using the CRISPR-P program (Lei et al., 2014). The two sgRNAs [sgRNA1 (S1) and sgRNA2 (S2)] were designed to target the amine oxidase domain, which will induce mutations in the functional domain of the BnaCRTISO gene resulting in the formation of non-functional protein (Figure 1A and Supplementary Figure 3). The designed sgRNAs matched well with both BnaA09.CRTISO and BnaC08.CRTISO copies (Figure 1A). A CRISPR/Cas9 construct containing these two sgRNAs with Cas9 driven by the cauliflower mosaic virus 35S promoter (Figure 1B) was produced based on the CRISPR/Cas9 multiplex genome-editing vector previously described by Yang et al. (2017). The resulting construct was transformed into J9707 using Agrobacterium-mediated transformation. A total of 105 seedlings were regenerated, of which 96 were transgenic positive. And 72 targeted mutants were identified by Sanger DNA sequencing of the PCR products encompassing the target sites, with 37 plants showing a visible knockout phenotype (i.e., creamy white flower; Supplementary Table 2). The overall editing efficiency of the T0 generations is 75.00%, of which the editing efficiency of S1 and S2 were 16.39 and 73.22%, respectively.
To generate stable lines with targeted mutations, 72 independent T0 editing lines of BnaCRTISO were self-pollinated to produce T1, T2, and T3 progenies. The targeted mutations of progenies from these T0 lines were verified by Hi-TOM sequencing analysis of the target sites (Table 1). The results proved that the mutant genotypes could be stably transmitted to the subsequent generations. A total of 10 T3 plants with homozygous mutations in BnaCRTISO were detected, including two BnaA09.CRTISO single mutants, two BnaC08.CRTISO single mutants and six BnaCRTISO double mutants (Table 1 and Figure 1C). These homozygous mutations at the target sites within BnaCRTISO were predicted to cause frameshifts resulting in the production of non-functional proteins (Supplementary Figure 6). As expected, all the double mutants could produce the creamy white flower phenotype (Figure 2B).
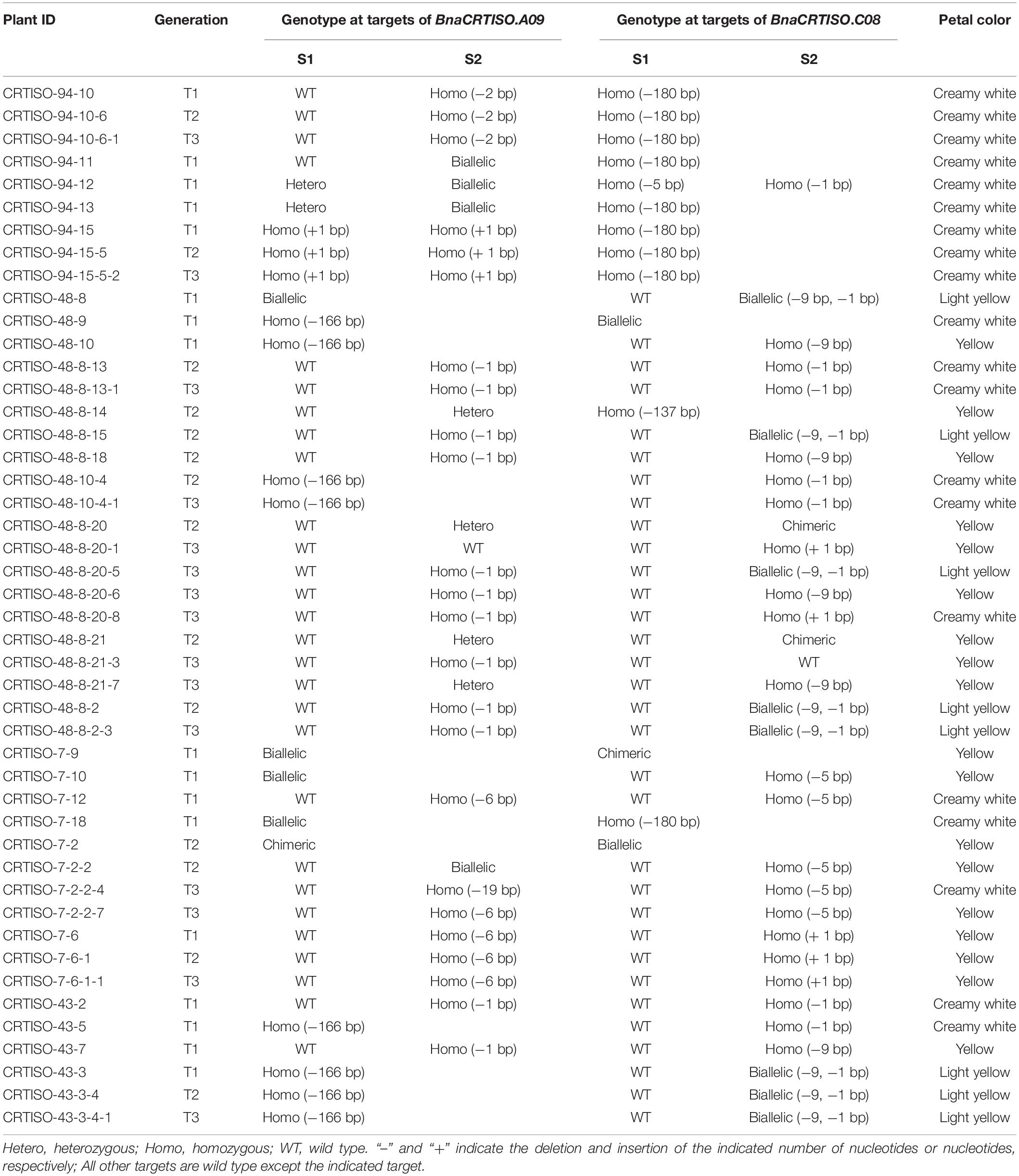
Table 1. Genotypic analysis of BnaCRTISO mutants and their transmission to T1, T2, and T3 generations.
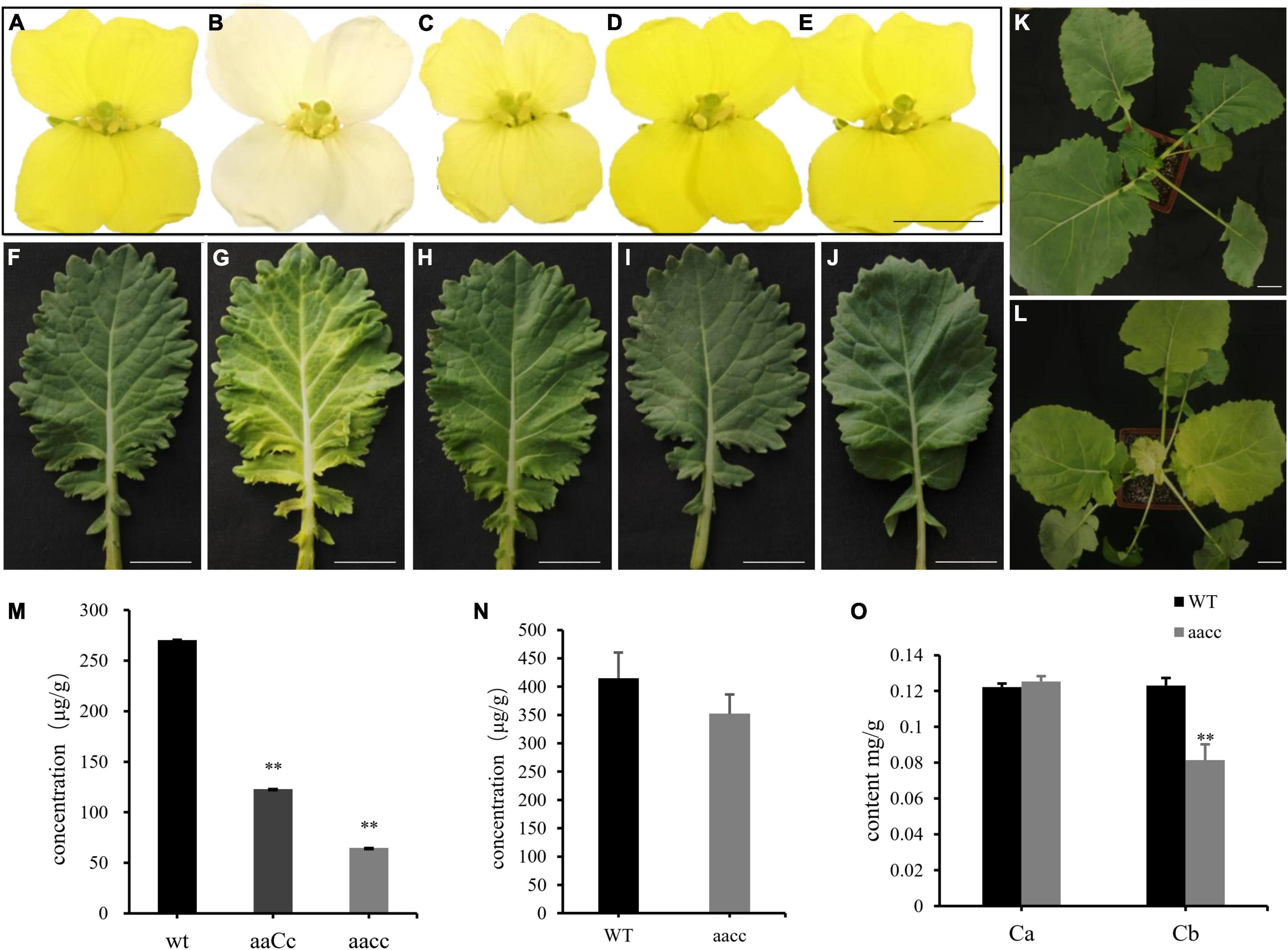
Figure 2. Mutations in the BnaCRTISO gene affected the color of petals and leaves. (A–J) The phenotype of flower (At the first day after flowering) and leaf in WT, aacc, aaCc, AAcc, aaCC, respectively. “aaCC,” “AAcc,” and “aacc” represent mutations of the target gene in BnaA09.CRTISO, BnaC08.CRTISO and both copies, respectively. (K) WT seeding. (L) aacc seeding. Bars, 1 cm. (M) The total carotenoid contents (μg/g fresh weight) of petals in aacc, aaCc, WT, respectively. (N) The total carotenoid contents of leaves in aacc, WT, respectively. (O) The content of chlorophyll a, b in the leaves of WT and aacc, respectively. (M–O) Data are presented as means ± SE (n ≥ 3); t-test was used for statistical analysis between the mutant and its WT (**P > 0.01).
The single mutants showed a similar yellow flower phenotype as WT (Figures 2A,D,E), while the heterozygous mutants (aaCc) showed a lighter yellow flower phenotype (Figure 2C). Thus, both copies of the BnaCRTISO gene function redundantly in regulating flower color. In addition, the inner leaves of the double mutants were also more yellowish than those of the WT, single mutants, and heterozygous mutants (Figures 2F–L), indicating that the BnaCRTISO gene also functions in the leaves.
To ascertain whether off-targeting occurred in the present study, we searched the B. napus genome for putative off-target sites with high homology to S1 and S2 according to the CRISPR-P program (Lei et al., 2014). A total of seven putative off-target sites were identified for both sgRNAs (Supplementary Table 10), and no off-target editing was detected in T0 and T1 by gene-specific primers amplification and Sanger sequencing (Supplementary Table 10). This result shows that the off-target effect is negligible when the specificity of each sgRNA is fully considered based on the genome sequence. Thus, the CRISPR/Cas9 system has a high specificity for targeted mutagenesis in B. napus.
In order to explore the subcellular localization of BnaA09.CRTISO and BnaC08.CRTISO, GFP was fused to the C terminal of each gene and transiently expressed in tobacco leaves. The green fluorescence signal overlaps closely with the chloroplast red autofluorescence signal observed by confocal microscopy (Figure 3). Thus, BnaCRTISO was predicted to be a chloroplast-localized protein, which was consistent with previous reports that carotenoids is synthesized and stored in plastids (Lange and Ghassemian, 2005).
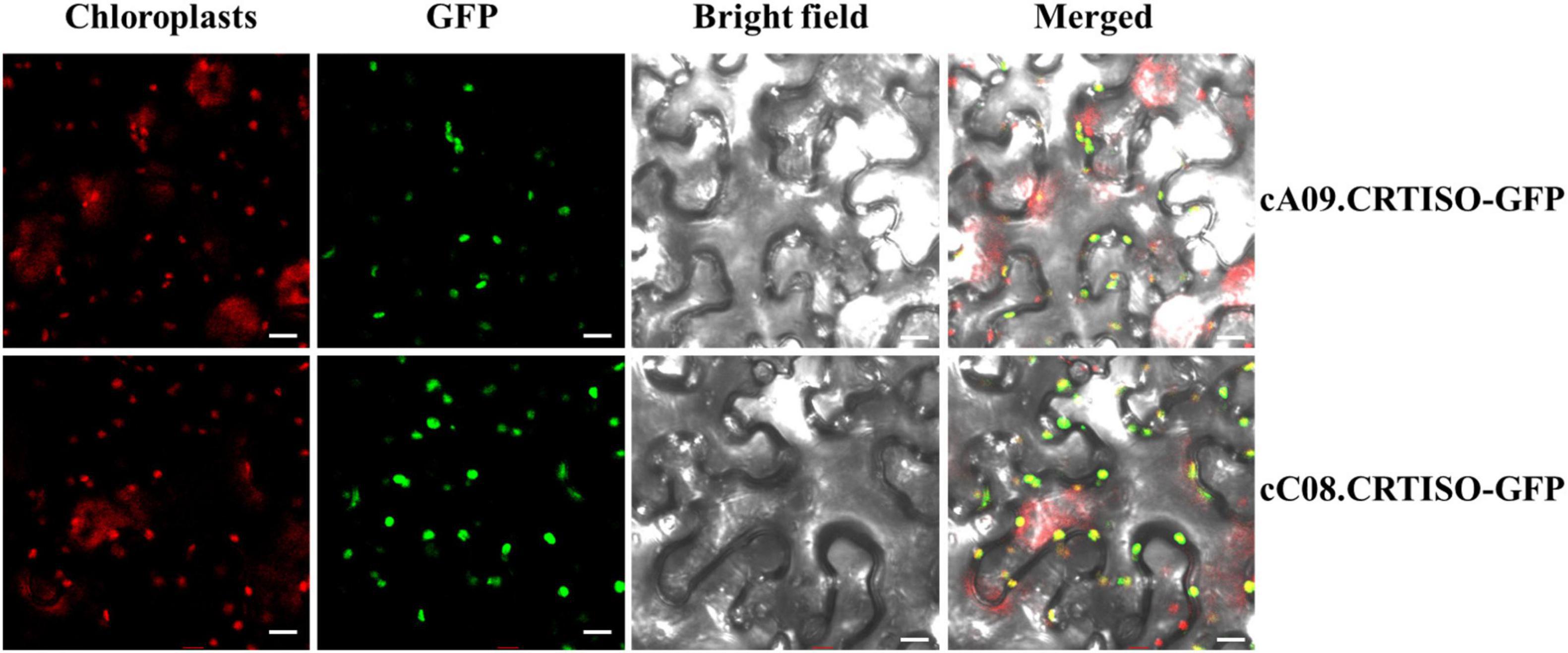
Figure 3. Subcellular localization of BnaA09.CRTISO and BnaC08.CRTISO in tobacco leaves. The fusion proteins generated using the coding sequence (cA09.CRTISO-GFP and cC08.CRTISO-GFP) were independently introduced into tobacco leaves. Scale bar = 10 μm.
The petals from flowers during anthesis were collected to compare the expression profiles between BnaCRTISO double mutants (CRTISO-94-15-5-2, CRTISO-48-8-13-1, CRTISO-7-2-2-4) and corresponding WT to investigate the transcriptome changes underlying the petal color variation (Supplementary Table 3). In subsequent analysis, a total of 41,186 genes were expressed in the developing petals during the same period. In the double mutants and WT, the pearson correlation coefficient between any two of the three biological repeats is very high (R = 0.93–0.98), indicating that the transcriptome sequencing data used in this study is highly reliable (Supplementary Figure 7).
Comparing the transcript abundance in these petals, it was observed that there were 2,058 DEGs between the BnaCRTISO double mutants and its corresponding WT at the same period (Supplementary Table 4). Overall, 953 genes were up-regulated in the double mutant petals, while 1,105 genes were down-regulated, which may be related to the changes observed in petal color (Supplementary Figure 8). The GO and KEGG enrichment analysis of these identified DEGs showed that the metabolic processes of carotenoid and flavonoid were significantly enriched among the down-regulated DEGs in mutants relative to WT (Supplementary Figures 9–11 and Supplementary Tables 5–7).
As the targeted mutated gene, BnaC08.CRTISO and BnaA09.CRTISO were down-regulated by almost eight and three times, respectively. The expression levels of most carotenoid biosynthesis-related genes were significantly up-regulated, such as BnaPDS3, BnaZDS, BnaCYP97A3, BnaBCH1, BnaBCH1, BnaZEP; whereas, five copies of BnaPSY were significantly down-regulated. BnaPSY, BnaPDS3, BnaZDS, and BnaCRTISO are involved in carotene synthesis; BnaCYP97A3, BnaBCH1 BnaBCH1, and BnaZEP are related to xanthophylls synthesis. PSY, encoding a phytoene synthase, acts like a faucet and plays a significant role at the initial stage in the carotenoid biosynthesis pathway. Moreover, some genes in the carotenoid degradation pathway were also significantly down-regulated, including BnaCCD4, BnaCCD8, BnaNCED2, and BnaNCED3 (Figure 4 and Supplementary Table 7).
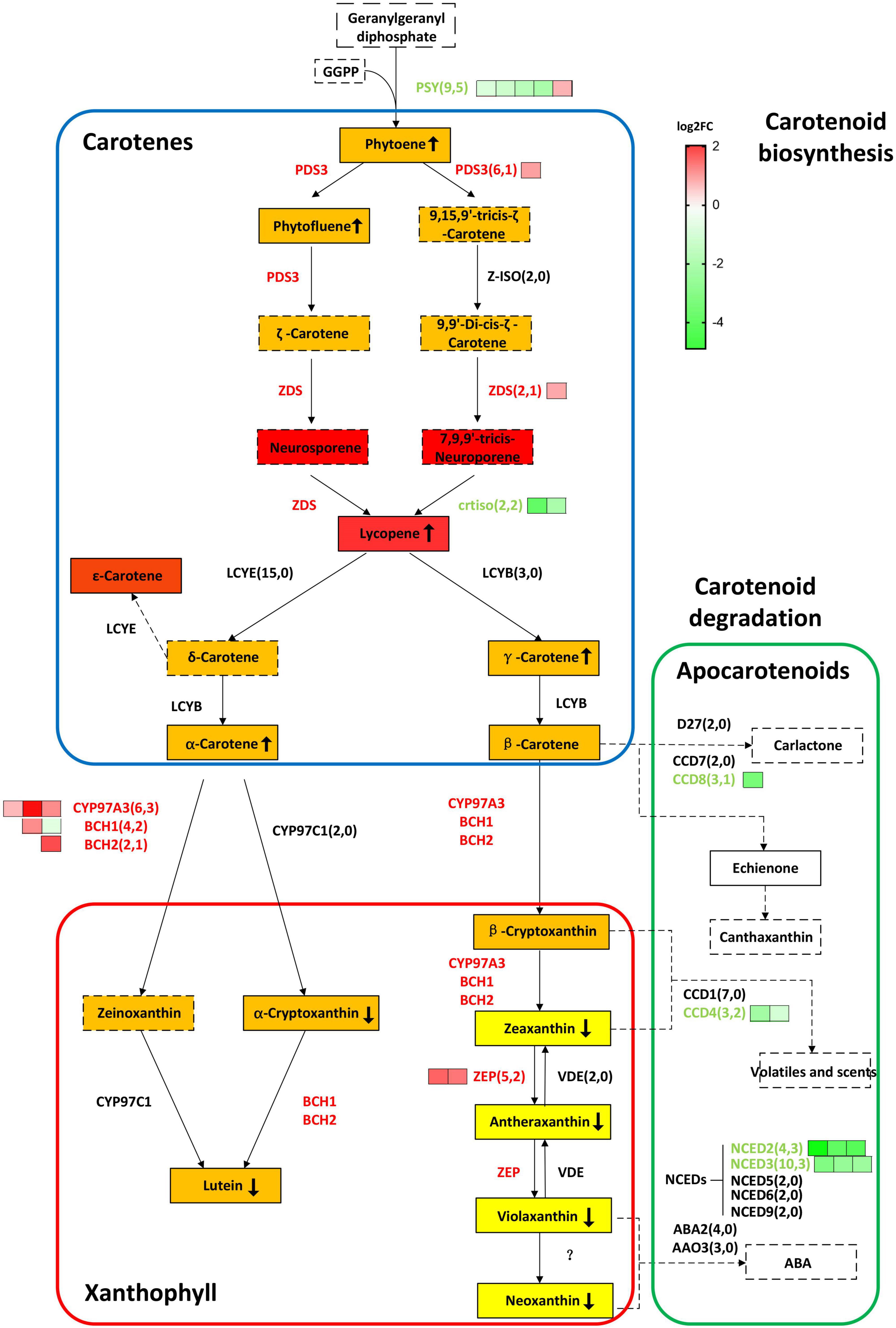
Figure 4. BnaCRTISO regulates genes related to carotenes and xanthophyll in rapeseed. In this study, the gene copy numbers from the rapeseed genome and DEGs are listed in parentheses. The log2 fold change (crtiso/WT) of DEGs of different copies of the same gene is represented by green (down-regulated) or red (up-regulated) squares arranged from left to right. Gene names are represented in capital letters, and corresponding mutants are represented in small letters. Genes that are up-regulated and down-regulated are represented in red and blue, respectively. The background color of the box indicates the color of the substance. The detected carotenoids in present study were indicated in the solid box, and undetected carotenoids in present study were indicated in the dotted box. The up or down arrow in the box indicates increase or decrease of the corresponding substance content, respectively. PSY, PHYTOENE SYNTHASE; PDS3, 15-cis-zeta-carotene isomerase; Z-ISO, 15-cis-zeta-carotene isomerase; ZDS, zeta-carotene desaturase; CRTISO, carotenoid isomerase; LCYE, lycopene beta/epsilon cyclase protein; LCYB, lycopene cyclase; BCH1, beta-hydroxylase 1; BCH2, beta-carotene hydroxylase 2; CYP97A3, cytochrome P450, family 97, subfamily A, polypeptide 3; CYP97C1, cytochrome P450 superfamily protein; ZEP, zeaxanthin epoxidase; VDE, non-photochemical quenching 1; NCED5,nine-cis-epoxycarotenoid dioxygenase 5; NCED3, nine-cis-epoxycarotenoid dioxygenase 3; NCED6, nine-cis-epoxycarotenoid dioxygenase 6; NCED9, nine-cis-epoxycarotenoid dioxygenase 9; NCED2, nine-cis-epoxycarotenoid dioxygenase 2; CCD4, nine-cis-epoxycarotenoid dioxygenase 4; ABA2, NAD(P)-binding Rossmann-fold superfamily protein; AAO3, abscisic aldehyde oxidase 3; CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2; CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4; CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3; CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1; D27, beta-carotene isomerase D27-like protein; CCD7, carotenoid cleavage dioxygenase 7; CCD8, carotenoid cleavage dioxygenase 8.
To verify the reliability of the RNA-seq data, 29 DEGs in the petals were selected for qRT-PCR verification analysis. These DEGs include 13 genes involved in the carotenoid synthesis, five genes involved in the flavonoid metabolism, and 11 randomly selected genes. Linear regression analysis showed that the correlation coefficient between the transcript levels assessed by the two analytic systems was very high (R = 0.812; Supplementary Figure 12), which further confirmed the reliability of the RNA-seq data.
Together, these results indicated the importance of the BnaCRTISO gene in the metabolic pathway of carotenoid. This also further illustrates the complex regulatory mechanisms of the BnaCRTISO gene in carotenoid synthesis of rapeseed.
To assess the impact of targeted mutation of BnaCRTISO on the carotenoid metabolic pathway, double (aacc: CRTISO-94-15-5-2, CRTISO-48-8-13-1, CRTISO-7-2-2-4) and heterozygous (aaCc: CRTISO-43-3-4-1, CRTISO-48-8-2-3, CRTISO-48-8-20-5) mutant T3 lines were grown in the field with their WT control. The total carotenoid content of petal and leaf samples were analyzed using spectrophotometer. It showed that the total carotenoid content of petals were significantly decreased in aacc mutants compared with aaCc and WT (aacc < aaCc < WT) (Figure 2M). The similar trend was observed in the total carotenoid content of leaves (Figure 2N). The two types of chlorophyll (chlorophyll a and chlorophyll b) concentration were further measured in leaves. And the chlorophyll b concentration of aacc mutants were significantly decreased relative to WT; whereas, no difference in chlorophyll a concentrations were observed between aacc mutants and WT (Figure 2O). This result indicated that BnaCRTISO may be involved in the anabolism of chlorophyll b in leaves.
Furthermore, carotenoid were analyzed using an LC-MS/MS system. Almost all carotene and xanthophyll contents showed a significant difference among aacc, aaCc, and WT. The carotene content of aacc was significantly increased when compared to the aaCc mutant and WT (aacc > aaCc > WT); while the xanthophyll content was significantly decreased (aacc < aaCc < WT) (Figure 5 and Supplementary Table 8). (E/Z)-phytoene, phytofluene, and lycopene are the main components of carotene. In this study, no lycopene was detected in the petals of WT; unexpectedly, the lycopene contents of the aacc and aaCc mutant were significantly increased (aacc > aaCc > WT; Supplementary Table 8). It should be noted that lycopene produced by the BnaCRTISO enzyme catalyzes prolycopene conversion in the carotenoid pathway. This result indicates the occurrence of another pathway for lycopene biosynthesis or degradation in the BnaCRTISO mutant, which converts prolycopene into all-trans lycopene through a non-enzymatic reaction under light illumination (Tuan et al., 2011; Su et al., 2015). In the xanthophyll metabolic pathway, the contents of α-cryptoxanthin (synthetic lutein precursor substance), β-cryptoxanthin (zeaxanthin precursor substance), lutein, zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin were all significantly decreased in aacc mutants (Figure 5 and Supplementary Table 8). These changes of xanthophylls content agreed well with the phenotypic variations in these materials: the higher the xanthophylls content, the yellower the petals.
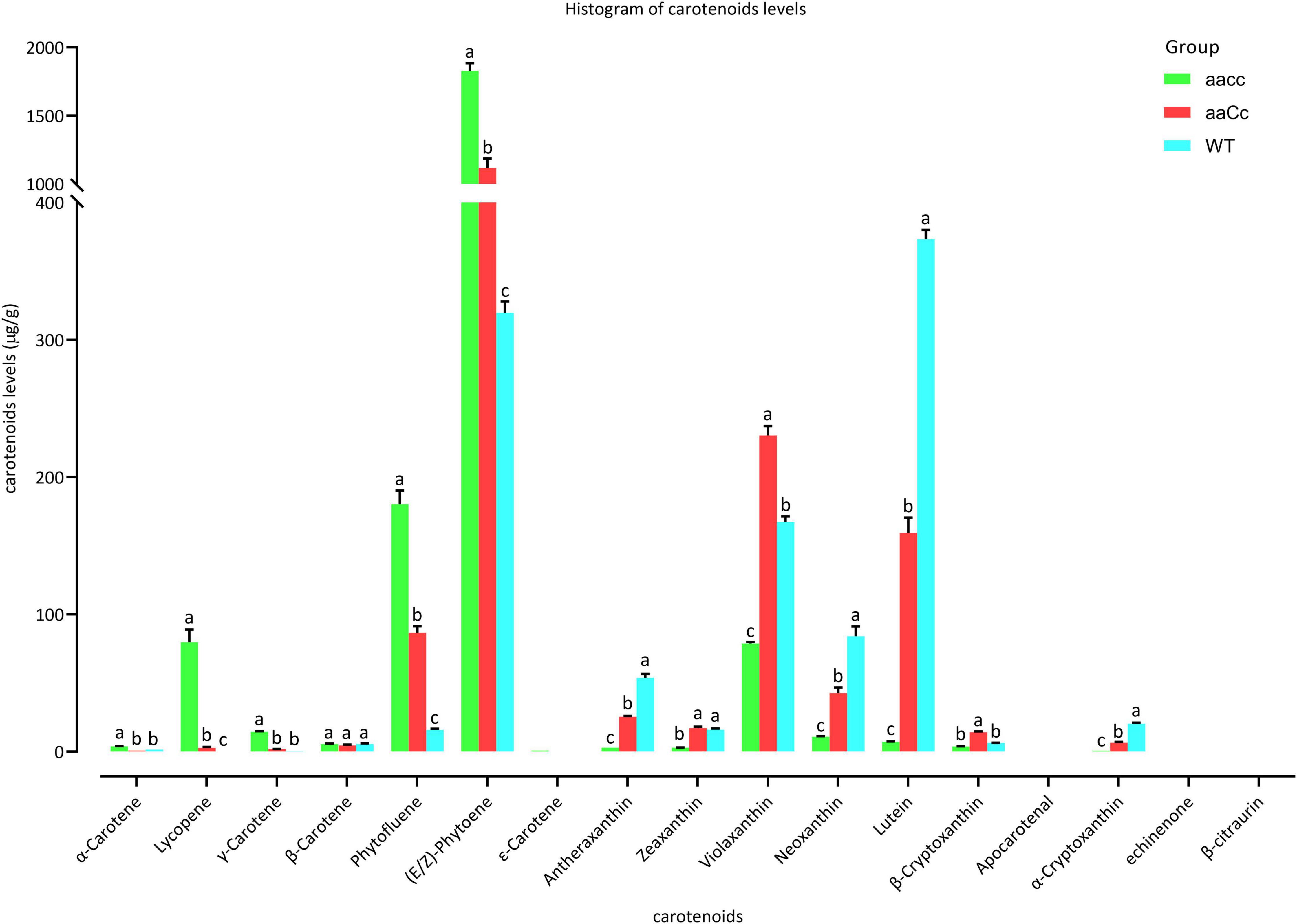
Figure 5. The different carotenoids levels in the petals of mutants and WT. aacc, homozygous double mutants; aaCc, the heterozygous mutants with a lighter yellow flower phenotype; WT, wild type; LSD-test was used for statistical analysis between the aacc and WT (Lower case letter indicates the significant difference at 0.05 level).
The flavonoid metabolic profiling of petal samples from the double-mutant (CRTISO-94-15-5-2, CRTISO-48-8-13-1, CRTISO-7-2-2-4) and its WT was analyzed using LC-ESI-MS/MS system. Most of the identified flavonoid metabolites show a significant difference between the double-mutant and WT petals (Supplementary Table 8). Among them, naringenin chalcone, the main flavonoid metabolites that constitutes yellow petals, were significantly decreased in the mutant. It is consitent with the transcriptomic analysis of the mutant petals, which indicates that the expression of naringenin chalcone metabolic genes (BnaC4H, BnaTT4, BnaTT7, BnaPAL2, Bna4CL3) were decreased in the double mutant (Figure 6, Supplementary Figure 12, and Supplementary Table 9). The content of other anthocyanins (apigenin and luteolin) in the mutants also decreased significantly (Figure 6 and Supplementary Table 9). Thus, these results are in line with the phenotypes and the transcriptomic analysis of the mutant petal.
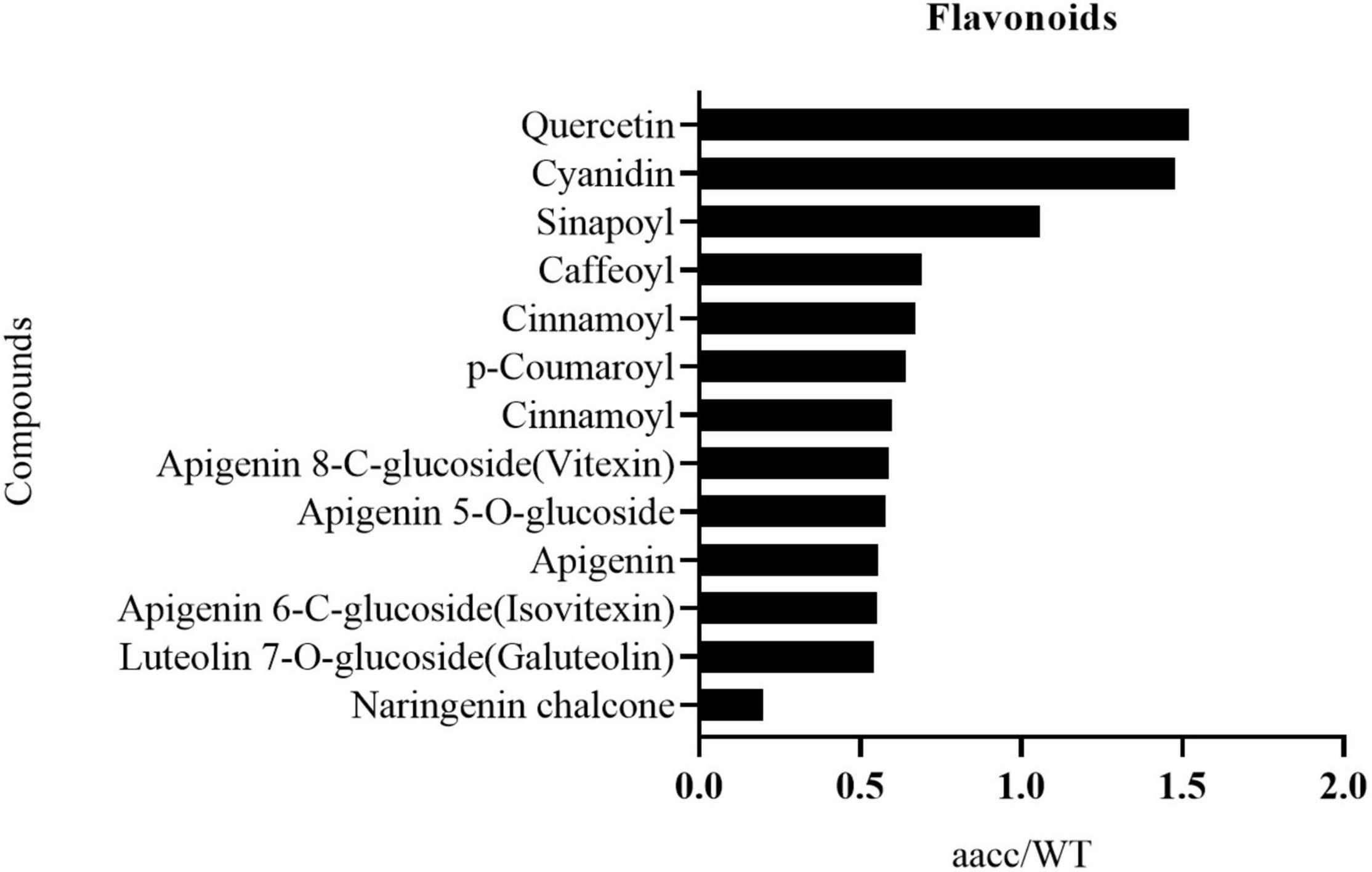
Figure 6. The identified flavonoid metabolites in petals with significant difference between the BnaCRTISO double mutant (aacc) and WT. The level of most of these metabolites decreased in mutant petals. And they are listed in the order of decreased fold changes (aacc/WT) from top to down.
Although flower color in B. napus is a very important trait and has attracted much interest since 1929 (Pearson, 1929), the underlying molecular mechanisms have not been well characterized. The white flower is an ornamental trait and ideal phenotypic marker in assisted breeding. Studies has also indicated that the white flower trait of B. napus is closely linked with high erucic acid (Liu et al., 2004). Therefore, it is significant to develop phenotypic markers and improve the ornamental value of low erucic acid rapeseed. To this end, an effective approach is needed to produce targeted mutations in these well-conserved CRTISO homologs in B. napus. The newly developed CRISPR/Cas9 technology provides a powerful approach for creating novel allelic variations. Thus far, it has been successfully utilized to modify several important agronomic traits (multilocular silique, plant height, architecture, pod shatters resistance, etc.) in rapeseed by generating specific gene knockout (Yang et al., 2017, 2018; Li C. et al., 2018; Zhai et al., 2019).
This study shows the successful utilization of CRISPR/Cas9 for targeted mutations of the BnaCRTISO gene in rapeseed with an editing efficiency of 75.00%, which is higher than those reported in previous research (14.4–45.0%) (Yang et al., 2017; Zhai et al., 2019; Ahmar et al., 2021). The editing efficiency at the S2 target site (73.22%) is higher than S1 (16.39%), indicating that the efficiency of the sgRNA promoter U3b is higher than that of U3d as our previous reports (Yang et al., 2017; Zhai et al., 2019; Ahmar et al., 2021). In this study, the targeted mutations of the two functional copies of BnaCRTISO in rapeseed generated the creamy white petal phenotype (Supplementary Table 2 and Figure 2), indicating that the BnaCRTISO gene is essential for flower color regulation and highly conservative in Brassica plants. The phenotypic analysis shows that the flower color of the double mutants is creamy white, while the BnaA09.CRTISO and BnaC08.CRTISO single mutants showed a comparable phenotype to that of the WT. Hence, both copies of the BnaCRTISO gene have functional redundancy in regulating flower color formation, and their contribution is equal. In addition, the heterozygous mutant (aaCc) identified in this study had a biallelic mutation (−9, −1 bp; e.g., CRTISO-8-15) on the BnaC08.CRTISO with “aa” genotype, and the plant shows a slight change in petal color (Figure 2B). While the double allelic mutation (−6, −19 bp; e.g., CRTISO-7-2-2) on BnaA09.CRTISO with “cc” genotype shows no change in petal color (Table 1). A homozygous −9 bp mutation on BnaC08.CRTISO with “aa” genotype (CRTISO-48-10, CRTISO-48-8-18, CRTISO-48-8-20-6, CRTISO-48-8-21-7, CRTISO-43-7) also shows no change in petal color (Table 1). Thus, these results indicated that different mutation types have different effects on BnaCRTISO enzyme activity.
Flavonoids and carotenoids are the pivotal pigments for the formation of most flower colors (Zhang et al., 2015). It has been reported that BnaC3.CCD4 and CmCCD4a contribute to the white color formation in the petals of B. napus and chrysanthemum, respectively, by degrading carotenoids into colorless compounds (Ohmiya et al., 2006; Zhang et al., 2015). Recently, Zhao et al. (2021) identified a mutation in a PDS3 gene in carotenoid biosynthesis pathway causes yellowish-white petals in rapeseed (Zhao et al., 2021). To date, no gene regulating the white flower traits other than the CCD4 and PDS3 gene has been reported in rapeseed.
In Arabidopsis, the crtiso mutant exhibiting partial inhibition of lutein biosynthesis in light-grown tissue and the accumulation of poly-cis-carotene precursors in dark-grown tissue with cotyledons colors changing from yellow to orange (Hyoungshin, 2002). The loss of function of the BrCRTISO gene confers orange color to the inner leaves and induces changes in flower color from yellow to orange in Chinese cabbage (B. rapa) (Su et al., 2015). Recently, the targeted editing of BoaCRTISO changed the leaf color from green to yellow, with the significant reduction of the concentrations of carotenoids and chlorophylls in Chinese kale (Sun et al., 2020). The crtiso mutant in tomato accumulates prolycopene instead of all-trans-lycopene, which could result in the production of orange fruit (Kato et al., 2007). It shows that the CRTISO gene in different plants can effectively change the carotenoid content in the flower, leaf, or fruit. In rapeseed, the cotyledon color of the BnaCRTISO double mutant showed a similar color change as that in B. rapa under the dark treatment (Supplementary Figure 13). Additionally, the petal color of the BnaCRTISO mutant changed from yellow to creamy white, which is different from that of B. rapa and Arabidopsis. It implies that the function of CRTISO in allotetraploid B. napus has become more complex in the course of evolution, and more information about this gene involved in the carotenoid pathway is yet to be ascertained.
PSY is the first key enzyme in the synthesis pathway, and its expression is regulated by feedback from upstream and downstream genes and metabolites (Yonekura-Sakakibara et al., 2019). The BnaPSY gene is also an important gene encoding the rate-limiting enzyme in the carotenoid synthesis pathway (Zhou et al., 2015). Down-regulation of the BnaPSY gene leads to a decrease in the total carotenoids content in the petals of the BnaCRTISO double mutants. Moreover, the xanthophyll content also decreased drastically in the BnaCRTISO mutants, and there are almost no xanthophylls in the petals of the double mutants. In contrast, the content of lycopene synthesized by the catalysis of BnaCRTISO protein increased significantly in the BnaCRTISO mutant. This result is consistent with the changes in the carotenoids content measured in the leaves of Arabidopsis CRTISO mutants (Isaacson, 2002). The expression of key genes BnaPDS3 and BnaZDS in the carotenoid synthesis pathway was also significantly up-regulated. This shows that the steady down-regulation of BnaCRTISO resulted in the up-regulation of BnaPDS3 and BnaZDS, which promote the accumulation of prolycopene and the mass synthesis of lycopene via a non-enzymatic pathway (Figure 4).
The expression levels of several xanthophyll synthetic-related genes (BnaCYP97A3, BnaBCH1, BnaBCH2, BnaZEP) in the mutants are up-regulated. Compared with the slight change in the synthetic substrate content, the content of the downstream substances (α-cryptoxanthin, lutein, zeaxanthin, antheraxanthin, violaxanthin, neoxanthin) are decreased significantly (Figure 4 and Supplementary Table 8). Interestingly, the expression of genes that inhibit ABA hormone signal transduction, such as BnaHAI1, BnaABI1, and BnaHAI2, are significantly down-regulated (Supplementary Table 7). In contrast, the expression of the ABA hormone signaling receptor BnaPYL4 is significantly up-regulated (Supplementary Table 7). It shows that the ABA hormone signal transduction process is accelerated in the mutant. Previoly reports indicated that ABA is synthesized from xanthophylls, e.g., zeaxanthin, violaxanthin and neoxanthin (Li and Walton, 1990; Parry et al., 1990). Thus, the low content of xanthophylls in the BnaCRTISO mutant is probably due to its degradation.
In the past, researchers created various types of light-colored flowers in different plants by changing the expression of the CHS gene to generate either white or light white flowers (Wang et al., 2018). In this study, the expression of multiple copies of the BnaCHS gene, which is the upstream gene regulating the flavonoid metabolism, was significantly down-regulated in the BnaCRTISO mutant resulting in the closure of the entire flavonoid synthesis pathway and a sharp decrease in the flavonoids content in petal (Supplementary Table 7). BnaF3′H is a key gene regulating for anthocyanin synthesis in the flavonoid synthesis pathway (Dubos et al., 2010; He et al., 2013), and its expression in the mutant is also greatly down-regulated. Collectively, these results indicate that the BnaCRTISO is an important gene involved in both the carotenoid and flavonoid pathways.
At present, there is no direct correlation between the carotenoid pathway and flavonoid synthesis pathway in plants (Grotewold, 2006; Cazzonelli and Pogson, 2010). It is known that they jointly regulate the color of flowers through the accumulation and mixing of pigments to produce their colored substances. In this study, a creamy white flower mutant was obtained by the mutation of the BnaCRTISO gene. Through metabolome and transcriptome analysis, we observed that the loss of BnaCRTISO gene function affects not only the expression of related genes in the carotenoid pathway, but also the expression of key genes involved in the flavonoid synthesis pathway. This reveals that some unknown mechanism of interactions exists between the carotenoid and flavonoid pathway in B. napus that are worthy of further study.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YZ and CF conceived the study and designed the experiments. LH performed the experiments. KY performed the bioinformatic analysis and wrote the manuscript. YY, ML, MG, and SD helped in the material sampling. LH, OA, and CF helped in the revision of this manuscript. CF supervised the study. All authors contributed to the article and approved the submitted version.
The study was financially supported by the National Natural Science Foundation of China (31671279; 31971976; 31371240), and Fundamental Research Funds for the Central Universities (2662015PY172).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.801456/full#supplementary-material
Ahmar, S., Zhai, Y., Huang, H., Yu, K., Hafeez, U. K. M., Shahid, M., et al. (2021). Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop J. doi: 10.1016/j.cj.2021.03.023
Becker, EW (ed.) (1994). Microalgae Biotechnology and Microbiology. Cambridge: Cambridge University Press, 56–62.
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., and Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935. doi: 10.1104/pp.17.00426
Breitenbach, J., and Sandmann, G. (2005). zeta-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220, 785–793. doi: 10.1007/s00425-004-1395-2
Cazzonelli, C. I. (2011). Carotenoids in nature: insights from plants and beyond. Funct. Plant Biol. 38, 833–847. doi: 10.1071/FP11192
Cazzonelli, C. I., and Pogson, B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trend Plant Sci. 15, 266–274. doi: 10.1016/j.tplants.2010.02.003
Cunningham, F. X., and Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 557–583. doi: 10.1146/annurev.arplant.49.1.557
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). Myb transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581.
Fraser, P. D., and Bramley, P. M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43, 228–265. doi: 10.1016/j.plipres.2003.10.002
Galpaz, N., Burger, Y., Lavee, T., Tzuri, G., Sherman, A., Melamed, T., et al. (2013). Genetic and chemical characterization of an EMS induced mutation in Cucumis melo CRTISO gene. Arch. Biochem. Biophys. 539, 117–125. doi: 10.1016/j.abb.2013.08.006
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57, 761–780. doi: 10.1146/annurev.arplant.57.032905.105248
Han, F., Cui, H., Zhang, B., Liu, X., Yang, L., Zhuang, M., et al. (2019). Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B. oleracea. BMC Genomics 20:242. doi: 10.1186/s12864-019-5596-2
He, H., Ke, H., Keting, H., Qiaoyan, X., and Silan, D. (2013). Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS One 8:e74395. doi: 10.1371/journal.pone.0074395
Holt, N. E., Zigmantas, D., Valkunas, L., Li, X. P., Niyogi, K. K., and Fleming, G. R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436. doi: 10.1126/science.1105833
Hu, L., Zhang, H., Yang, Q., Meng, Q., Han, S., Nwafor, C. C., et al. (2018). Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor. Appl. Genet. 131, 1–10. doi: 10.1007/s00122-018-3184-5
Huang, Z., Ban, Y., Bao, R., Zhang, X., Xu, A., and Ding, J. (2014). Inheritance and gene mapping of the white flower in Brassica napus L. N. Z. J. Crop Hortic. Sci. 42, 111–117.
Hyoungshin, P. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14, 321–332. doi: 10.1105/tpc.010302
Isaacson, T. (2002). Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14, 333–342. doi: 10.1105/tpc.010303
Kato, M., Matsumoto, H., Ikoma, Y., Kuniga, T., Nakajima, N., and Yoshida, T. (2007). Accumulation of carotenoids and expression of carotenoid biosynthetic genes and carotenoid cleavage dioxygenase genes during fruit maturation in the juice sacs of ‘Tamami,’ ‘Kiyomi’ Tangor, and ‘Wilking’ Mandarin. Engei Gakkai Zasshi 76, 103–111. doi: 10.2503/jjshs.76.103
Lange, B. M., and Ghassemian, M. (2005). Comprehensive post-genomic data analysis approaches integrating biochemical pathway maps. Phytochemistry 66, 413–451. doi: 10.1016/j.phytochem.2004.12.020
Lee, H. S. (2001). Characterization of carotenoids in juice of red navel orange (Cara Cara). J. Agric. Food Chem. 49, 2563–2568. doi: 10.1021/jf001313g
Lei, Y., Lu, L., Liu, H., Li, S., Xing, F., and Chen, L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496. doi: 10.1093/mp/ssu044
Li, C., Hao, M., Wang, W., Wang, H., Fan, C., and Chu, W. (2018). An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 9:442. doi: 10.3389/fpls.2018.00442
Li, C., Ji, J., Wang, G., Li, Z., Wang, Y., and Fan, Y. (2020). Over-Expression of LcPDS, LcZDS, and LcCRTISO, genes from wolfberry for carotenoid biosynthesis, enhanced carotenoid accumulation, and salt tolerance in tobacco. Front. Plant Sci. 11:119. doi: 10.3389/fpls.2020.00119
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Li, Y., and Walton, D. (1990). Violaxanthin is an abscisic acid precursor in water-stressed dark-grown bean leaves. Plant Physiol. 92, 551–559. doi: 10.1104/pp.92.3.551
Liu, X., Tu, J., Chen, B., and Fu, T. (2004). Identification of the linkage relationship between the flower colour and the content of erucic acid in the resynthesized Brassica napus L. Acta Genet. Sin. 31, 357–362. doi: 10.1046/j.1439-0523.2002.00711.x
Liu, Y., Ye, S., Yuan, G., Ma, X., Heng, S., Yi, B., et al. (2020). Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 104, 932–949. doi: 10.1111/tpj.14970
Lv, P., Li, N., Liu, H., Gu, H., and Zhao, W. (2015). Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars. Food Chem. 174, 52–59. doi: 10.1016/j.foodchem.2014.11.022
Ma, X., Zhang, Q., and Zhu, Q. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. ScienceDirect Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Matsufuji, H., Ishikawa, K., Nunomura, O., Chino, M., and Taketa, M. (2007). Anti-oxidant content of different coloured sweet peppers, white, green, yellow, orange and red (Capsicum annuum L.). Int. J. Food Sci. Technol. 42, 1482–1488.
Montserrat, R. B., Inés, G., Carlos, V., Maria, B., and Rosa, M. E. (2015). Carotenoids from haloarchaea and their potential in biotechnology. Mar. Drugs 13, 5508–5532. doi: 10.3390/md13095508
Nisar, N., Li, L., Shan, L., Khin, N. C., and Pogson, B. (2015). Carotenoid metabolism in plants. Mol. Plant 8, 68–82.
Ohmiya, A., Kishimoto, S., Aida, R., Yoshioka, S., and Sumitomo, K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in Chrysanthemum petals. Plant Physiol. 142, 1193–1201. doi: 10.1104/pp.106.087130
Parry, A., Babiano, M., and Horgan, R. (1990). The role of cis-carotenoids in abscisic acid biosynthesis. Planta 182, 118–128. doi: 10.1007/BF00239993
Patrick, S., Marta, R. F., Ian, C. C., Florian, W., and Ralf, W. (2018). Establishment of an Arabidopsis callus system to study the interrelations of biosynthesis, degradation and accumulation of carotenoids. PLoS One 13:e0192158. doi: 10.1371/journal.pone.0192158
Quazi, M. H. (1988). Interspecific hybrids between Brassica napus L. and B. oleracea L. developed by embryo culture. Theor. Appl. Genet. 75, 309–318.
Sakai, T., Iwabuchi, M., Kohno-Murase, J., Liu, H. J., and Imamura, J. (1995). “Transfer of radish CMS-restorer gene into Brassica napus by intergeneric protoplast fusion,” in Proceedings of the 9th International Rapeseed Congress, Cambridge, 3–5.
Saladié, M., Wright, L. P., Garcia-Mas, J., Rodriguez-Concepcion, M., and Phillips, M. A. (2014). The 2-C-methylerythritol 4-phosphate pathway in melon is regulated by specialized isoforms for the first and last steps. J. Exp. Bot. 65, 5077–5092. doi: 10.1093/jxb/eru275
Shahid, M., Cai, G., Zu, F., Zhao, Q., Qasim, M., Hong, Y., et al. (2019). Comparative transcriptome analysis of developing seeds and silique wall reveals dynamic transcription networks for effective oil production in Brassica napus L. Int. J. Mol. Sci. 20:1982. doi: 10.3390/ijms20081982
Su, T., Yu, S., Zhang, J., Yu, Y., Zhang, D., and Zhao, X. (2015). Loss of function of the carotenoid isomerase gene BrCRTISO confers orange color to the inner leaves of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 33, 648–659. doi: 10.1007/s11105-014-0779-0
Sun, B., Jiang, M., Zheng, H., Jian, Y., and Tang, H. (2020). Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 7:161. doi: 10.1038/s41438-020-00379-w
Sun, T., Yuan, H., Cao, H., Yazdani, M., Tadmor, Y., and Li, L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11, 58–74. doi: 10.1016/j.molp.2017.09.010
Tuan, P. A., Kim, J. K., Park, N. I., Lee, S. Y., and Park, S. U. (2011). Carotenoid content and expression of phytoene synthase and phytoene desaturase genes in bitter melon (Momordica charantia). Food Chem. 126, 1686–1692. doi: 10.1016/j.foodchem.2010.12.058
Walter, M. H., and Strack, D. (2011). Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 28, 663–692. doi: 10.1039/c0np00036a
Wang, Z., Yu, Q., Shen, W., El Mohtar, M., Zhao, X., and Gmitter, F. G. (2018). Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 18:189. doi: 10.1186/s12870-018-1418-y
Wurtzel, V. E. T. (2009). Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol. 150, 562–572. doi: 10.1104/pp.109.137042
Xu, C.-J., Fraser, P. D., Wang, W.-J., and Bramley, P. M. (2006). Differences in the carotenoid content of ordinary citrus and lycopeneaccumulating mutants. J. Agric. Food Chem. 54, 5474–5481. doi: 10.1021/jf060702t
Yang, H., Wu, J., Tang, T., Liu, K., and Dai, C. (2017). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7:4877. doi: 10.1038/s41598-017-07871-9
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, M. H. U., et al. (2018). Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. doi: 10.1111/pbi.12872
Yonekura-Sakakibara, K., Higashi, Y., and Nakabayashi, R. (2019). The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 10:943. doi: 10.3389/fpls.2019.00943
Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., and Fan, C. (2019). CRISPR/Cas9-mediated genome editing reveals differences in the contribution of indehiscent homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. doi: 10.1007/s00122-019-03341-0
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhang, B., Liu, C., Wang, Y., Yao, X., Wang, F., Wu, J., et al. (2015). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206, 1513–1526. doi: 10.1111/nph.13335
Zhang, B., Lu, C. M., Kakihara, F., and Kato, M. (2010). Effect of genome composition and cytoplasm on petal colour in resynthesized amphidiploids and sesquidiploids derived from crosses between Brassica rapa and Brassica oleracea. Plant Breed. 121, 297–300. doi: 10.1046/j.1439-0523.2002.722295.x
Zhao, C., Safdar, L. B., Xie, M., Shi, M., Liu, S., Dong, Z., et al. (2021). Mutation of the phytoene desaturase 3 gene causes yellowish-white petals in Brassica napus. Crop J. 9, 1124–1134.
Keywords: Brassica napus, flower color, BnaCRTISO, gene editing, carotenoid
Citation: Li H, Yu K, Amoo O, Yu Y, Guo M, Deng S, Li M, Hu L, Wang J, Fan C and Zhou Y (2022) Site-Directed Mutagenesis of the Carotenoid Isomerase Gene BnaCRTISO Alters the Color of Petals and Leaves in Brassica napus L. Front. Plant Sci. 13:801456. doi: 10.3389/fpls.2022.801456
Received: 25 October 2021; Accepted: 03 January 2022;
Published: 10 February 2022.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Fuyou Fu, Agriculture and Agri-Food Canada (AAFC), CanadaCopyright © 2022 Li, Yu, Amoo, Yu, Guo, Deng, Li, Hu, Wang, Fan and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuchuan Fan, ZmFuY2h1Y2h1YW5AbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.