
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 February 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.794171
Evolutionary slowdowns in diversification have been inferred in various plant and animal lineages. Investigation based on diversification models integrated with environmental factors and key characters could provide critical insights into this diversification trend. We evaluate diversification rates in the Cirrhopetalum alliance (Bulbophyllum, Orchidaceae subfam. Epidendroideae) using a time-calibrated phylogeny and assess the role of Crassulacean acid metabolism (CAM) as a hypothesised key innovation promoting the spectacular diversity of orchids, especially those with an epiphytic habit. An explosive early speciation in the Cirrhopetalum alliance is evident, with the origin of CAM providing a short-term advantage under the low atmospheric CO2 concentrations (pCO2) associated with cooling and aridification in the late Miocene. A subsequent slowdown of diversification in the Cirrhopetalum alliance is possibly explained by a failure to keep pace with pCO2 dynamics. We further demonstrate that extinction rates in strong CAM lineages are ten times higher than those of C3 lineages, with CAM not as evolutionarily labile as previously assumed. These results challenge the role of CAM as a “key innovation” in the diversification of epiphytic orchids.
Understanding the dynamics of diversification over space and time and identifying the underlying biotic and abiotic causes of the patterns have been a major focus in evolutionary biology (Ricklefs, 2007). The dynamics of speciation and extinction rates alone, or a combination of both, can lead to various distinctive diversification patterns (Donoghue and Sanderson, 2015; Louca and Pennell, 2020). Theoretically, a slowdown in diversification can be driven by either decreased speciation or increased extinction rates alone, or by the collective effect of both, leading to the “horrible death” of a clade (Donoghue and Sanderson, 2015). A diversification slowdown may be caused by a high net diversification rate early in the history of the clade (cf. the early radiation or the explosive early pattern; Lovette and Bermingham, 1999; Rabosky and Lovette, 2008), followed by a rate decline that is time- or species density-dependent (Moen and Morlon, 2014).
Crassulacean acid metabolism (CAM) represents a striking example of ecological adaptation to CO2-constrained and water-limited environments (Michener and Lajtha, 2007). It is characterised by temporal separation of nocturnal fixation of CO2 and daytime decarboxylation of organic acids to release CO2 that is then refixed by Rubisco in the chloroplasts (Ting, 1985). CAM plants can thereby substantially minimise water loss during the day when evaporative demand is highest and hence use the available water more efficiently than C3 and C4 plants. At least 6% of flowering plants have been determined as having CAM photosynthesis: these taxa are phylogenetically widespread across 343 genera in 35 families (Silvera et al., 2010). CAM photosynthesis is evidently polyphyletic, having arisen independently from multiple C3 ancestors during the Miocene, possibly as a consequence of reduced atmospheric CO2 concentration (Raven and Spicer, 1996). This transition sometimes occurs in parallel with the colonisation of new ecological niches such as increasingly arid habitats, triggering adaptive speciation (Lüttge, 1996). It has been hypothesised that CAM might be a key evolutionary innovation, correlated with extraordinary species diversity in some vascular plant groups, such as Bromeliaceae (Silvestro et al., 2014), Cactaceae (Arakaki et al., 2011), Euphorbiaceae (Horn et al., 2014), and Orchidaceae (Silvera et al., 2009; Givnish et al., 2015).
On the other hand, more detailed studies have indicated that CAM was not an evolutionary driver across the Bromeliaceae as a whole: not all CAM lineages in the family have radiated, and subfamily Tillandsioideae radiated (at least initially) without CAM (Donoghue and Sanderson, 2015). The impact of CAM photosynthesis on diversification in Orchidaceae remains controversial, with conflicting hypotheses proposed. Silvera et al. (2009) suggested that CAM is a drought avoidance mechanism that stimulated the evolution of epiphytism and that epiphytism in turn promoted higher rates of speciation in a wider range of ecological niches. Newly generated data have nevertheless shown that the evolution of CAM and epiphytism in Orchidaceae are so closely associated that it is difficult to disentangle the individual effect of either (Givnish et al., 2015), and Gamisch et al. (2021) demonstrated that the transition from C3 to CAM was not associated with significantly higher diversification rates in Malagasy Bulbophyllum (Orchidaceae subfam. Epidendroideae). Furthermore, caution is necessary before drawing inferences that associate CAM with accelerated diversification rates: extremely high extinction rates, which could have resulted in considerable losses in terms of net diversification, have been demonstrated in CAM lineages compared to species with C3 photosynthesis, although previous authors have argued that the calculation may overestimate the disadvantages of CAM across orchid lineages (Givnish et al., 2015). Importantly, similar patterns have been observed in subfamily Bromelioideae, in which CAM has been associated with a twofold increase in speciation rate but a more than fourfold increase in extinction rate (Silvestro et al., 2014). This phenomenon again highlights the need to re-evaluate the perception of CAM as a putative key innovation.
Epiphytic orchids with species-rich CAM lineages and C3 sister clades represent a good model to investigate diversification dynamics (Givnish et al., 2015; Gamisch et al., 2021). The Cirrhopetalum alliance is a well-supported clade of c. 210 species belonging to the diverse epiphytic genus Bulbophyllum (Hu et al., 2020). A mixture of both CAM and C3 photosynthetic pathways has been reported in Malagasy Bulbophyllum (Gamisch et al., 2021). Here, we use a robust time-calibrated species-level molecular phylogeny of the Cirrhopetalum alliance (Hu et al., 2020), together with data on historic CO2 concentrations (pCO2) and photosynthetic pathways of all species sampled, to: (1) infer the diversification of the Cirrhopetalum alliance; (2) investigate the photosynthesis-dependent diversification integrated with pCO2, specifically focusing on CAM-associated speciation and extinction rates; and (3) re-assess the role of CAM as a possible key innovation in the diversification of epiphytic orchids.
The comparative phylogenetic analyses presented here are based on a 119-accession Bulbophyllum data matrix, plus a further six outgroup taxa from the sister genus Dendrobium, with detailed sampling information provided by Hu et al. (2020). Great effort was made to include samples representative of total morphological, geographical, ecological, and niche diversity of the Cirrhopetalum alliance as a whole. Samples were collected from the field in Hong Kong, Taiwan, South China, Thailand, Laos, the Philippines, Vietnam, and Malaysia, with additional material being incorporated from plants in cultivation at South China Botanical Garden (Guangdong, China), the National Orchid Conservation Center (Shenzhen, China), Kadoorie Farm and Botanic Garden (Hong Kong SAR, China) and Dr. Cecilia Koo Botanic Conservation Center (Taiwan). The resulting 125-accession dataset includes 88 accessions of the Cirrhopetalum alliance, representing 42% of total species diversity, with sampling fraction for the eight major groups ranging from 0.23 to 0.65 (Supplementary Table 1). Although sampling coverage for the “DES-EUB-RHY” group was comparatively low, this group (which includes sect. Desmosanthes) had never previously been considered a member of the Cirrhopetalum alliance, and so this result was unexpected (Hu et al., 2020); coverage for the other groups was generally much higher (Supplementary Table 1). Sequences from two chloroplast (matK and psbA) and two nuclear (ITS and Xdh) regions were used for phylogenetic reconstruction. Since topological incongruence was not observed by visual comparison of phylogenies derived from separate analysis of the individual chloroplast and nuclear datasets (Hu et al., 2020), the cpDNA and nrDNA matrices were concatenated for further Bayesian inference (BI) analyses conducted using MrBayes ver. 3.2.6 (Huelsenbeck and Ronquist, 2001) on CIPRES Science Gateway ver. 3.3 (Miller et al., 2010). Four simultaneous Markov chain Monte Carlo (MCMC) chains (containing three heated chains and one cold chain with a temperature parameter of 0.16) were run for 20 million generations. Convergence of independent runs was examined with the R package AWTY (Nylander et al., 2008) and assessed by checking that the standard deviation of split frequencies was < 0.005. Adequate effective sample sizes (ESS > 200, indicating convergence and confidence) were verified using Tracer ver. 1.6 (Rambaut et al., 2014).
Divergence times were estimated using the Bayesian relaxed molecular clock model with uncorrelated log-normal rates in BEAST ver. 2.4.4 (Bouckaert et al., 2014). The birth–death (BD) prior on node ages was selected against the Yule prior after model testing by Bayes factors (BF > 100) based on preliminary runs as implemented in Tracer. Two calibration points were applied to date the phylogeny. Firstly, a normal distribution with mean 30.17 Ma (million years ago) (standard deviation, SD: 3.48 Ma; 95% highest posterior density, HPD: 36.99–23.3 Ma) was assigned to the root of the phylogeny based on the median stem node age of the Bulbophyllum-Dendrobium clade [adopted from Gamisch et al. (2015) and based on robust estimation with several calibration schemes as sensitive analyses]. Secondly, a lognormal distribution with a minimum stem age (23.2 Ma) was used for a Dendrobium winikaphyllum macrofossil from the early Miocene (23–20 Ma) of New Zealand that was assigned to the node basal to the Australian-New Zealand Dendrobium clade (Conran et al., 2009). We conducted five independent MCMC runs of 50 million generations with sampling every 5,000 generations. After evaluating convergence with the R package AWTY and effective sample sizes in Tracer (ESS > 200), we combined the five independent runs with LogCombiner ver. 2.1.3 (Rambaut and Drummond, 2014), setting the burn-in to 25% of the initial samples of each run. We further used TreeAnnotator ver. 2.1.3 (Bouckaert et al., 2014) to compute the maximum clade credibility (MCC) tree for downstream comparative study.
Downstream analyses were based on either the 88-accession time-calibrated MCC tree of the Cirrhopetalum alliance (Dataset 1) or, where possible, 100 and 1,000 randomly selected post-burn-in trees from BEAST, chosen with R package APE ver. 3.4 (Paradis et al., 2004) (Dataset 2 and Dataset 3, respectively). Deviations in diversification across clades from a constant BD model and potential rate shifts in speciation and extinction were detected using three complementary approaches: LASER ver. 2.4 (Rabosky, 2006), Diversity-Dependent Diversification (DDD) ver. 3.5 (Etienne et al., 2012), and Bayesian analysis of macro-evolutionary mixtures (BAMM) ver. 2.5 (Rabosky, 2014; Table 1).
Firstly, the γ statistic test for departure from a constant rate (CR) pure birth model (Pybus and Harvey, 2000) was conducted with both CR test and Monte Carlo CR (MCCR) test as implemented in LASER. A γ value ≤ 1.645 (p < 0.05) was interpreted as supporting the rejection of the pure-birth model under a one-tailed test, providing support for the alternative hypothesis of a slowdown in diversification as predicted with an explosive early pattern. 1,000 post-burn-in pruned random trees (Dataset 3) were used in the CR test, taking account of phylogenetic uncertainty. To account for incomplete sampling in the MCCR test (Cusimano et al., 2012), the taxon sampling fraction of the Cirrhopetalum alliance (frac = 88/210) was applied in tree simulation with the “sim.bd.taxa.age” function in R package TREESIM ver. 2.3 (Stadler, 2011). Speciation and extinction rates used for tree simulation were obtained by fitting the CR BD model to the empirical data (Dataset 1). Simulated trees were then randomly pruned to the actual sample size (n = 88). The null distribution of γ values based on the simulated 1,000 trees against the empirical γ value based on Dataset 1 was compared using the MCCR test statistic.
Three DD diversification models (DD1: linear dependence in speciation rate; DD2: exponential dependence in speciation rate; and DD3: linear dependence in extinction rate) were tested against the simpler CR BD DD model based on Dataset 1, simulated 100 times under CR and each DD model with the R package DDD. The settings for different conditions followed Etienne et al. (2016).
Three TD diversification models (Supplementary Table 2) specifically developed to test the explosive early speciation pattern were applied to further disentangle speciation-extinction dynamics through time. A similar Maximum likelihood (ML) method as implemented in LASER was used for model comparison with the time-dependent CR BD model.
Thirdly, BAMM was used to infer net diversification, speciation and extinction rates across the Cirrhopetalum alliance based on Dataset 1. BAMM explores a vast number of candidate models of lineage diversification using reversible jump Markov chain Monte Carlo (rjMCMC). To account for incomplete taxon sampling, data on the non-random incomplete sampling of the Cirrhopetalum alliance were incorporated following BAMM protocols by calculating the proportion of species sampled per group within the Cirrhopetalum alliance and estimating the backbone sampling as the overall proportion of groups sampled in this study (Supplementary Table 1). The priors were generated based on Dataset 1 using the “setBAMMpriors” function in R package BAMMTOOLS ver. 2.1.6 (Rabosky et al., 2014). The estimation was conducted over 2 × 107 generations, sampling every 2,000 generations and discarding the first 20% of the sampled data as burn-in. The convergence and confidence for each run was subsequently checked using BAMMTOOLS, calculating the ESS for the likelihood. The “plotRateThroughTime” function in BAMMTOOLS was used to plot speciation, extinction and net diversification rates.
Finally, lineage-through-time (LTT) plots were generated using APE.
We used the environment-dependent diversification model (Morlon et al., 2016) to test whether past pCO2 might have impacted diversification of the Cirrhopetalum alliance. Specifically, four palaeoenvironment-dependent diversification models with different combinations of constraint on speciation (or birth) and extinction (or death) rates (Table 2) were fitted and compared to relevant TD diversification models, implemented in R package RPANDA ver. 1.3 (Morlon et al., 2016) using Dataset 2. We extracted palaeoclimatic pCO2 data covering the past 20 million years (Figure 1A; Foster et al., 2017). An ML approach was used to estimate speciation and extinction, taking incomplete taxon sampling into account.
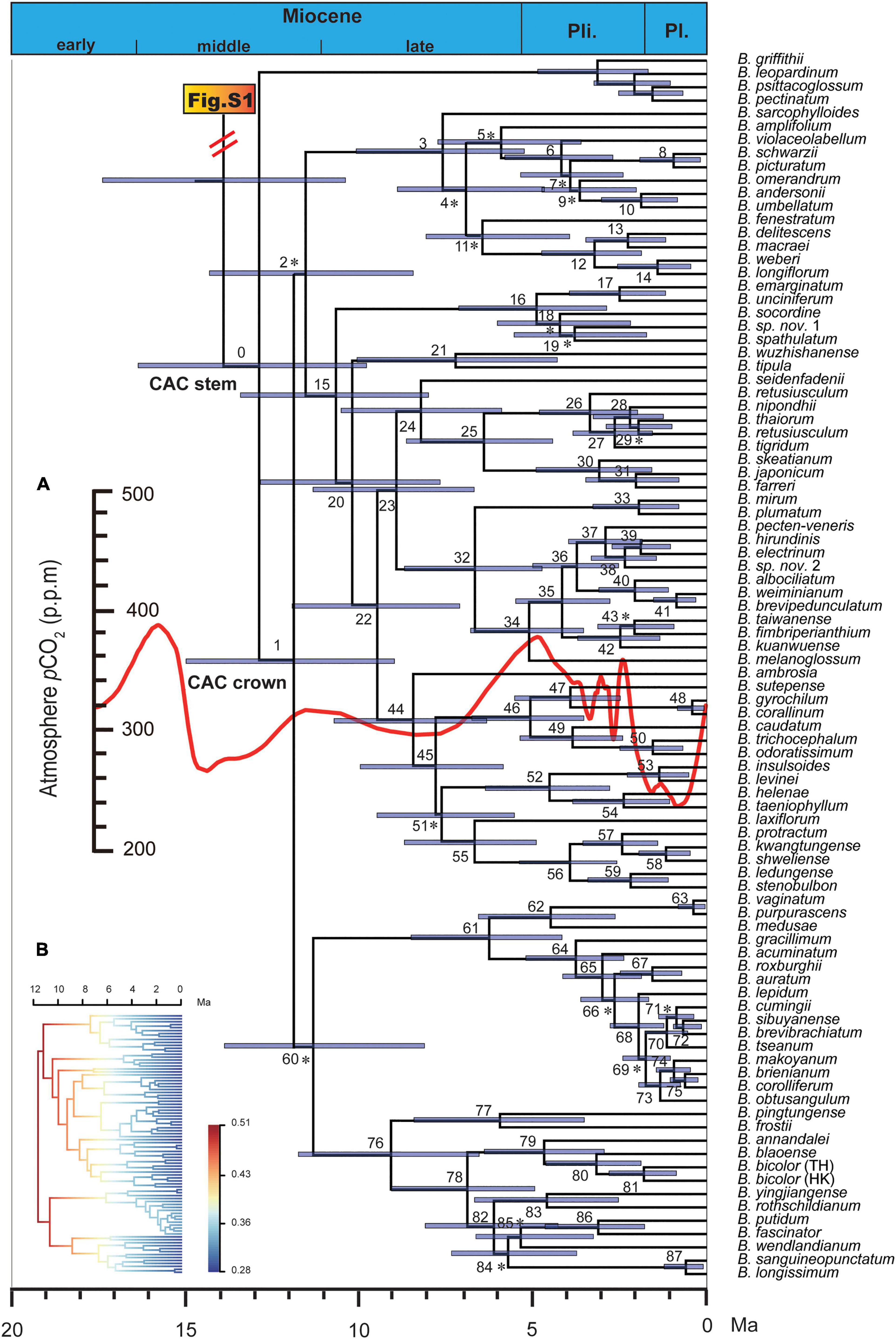
Figure 1. Maximum clade credibility chronogram of the Cirrhopetalum alliance clade (CAC) and its sister group. Asterisk indicates nodes with posterior clade probabilities (PP) < 0.95; otherwise, PP ≥ 0.95. Node numbers correspond with those in Supplementary Table 4. The inserted curve (A) represents estimated mean atmospheric CO2 concentration change based on palaeoclimatic pCO2 data extracted from Foster et al. (2017); the inserted phylorate plot (B) corresponds to the topology of CAC, showing net species diversification rate (lineage/Myr; cool colours = slow, warm = fast). B., Bulbophyllum; Ma, million years ago; Pl., Pleistocene; Pli., Pliocene.
Carbon isotope ratio [δ13C (‰)] was used as a proxy indicator of photosynthetic pathways (Ehleringer and Osmond, 1989). Leaf carbon isotope ratios were determined using small fragments (≤ 3 mg) of dried leaf tissues for all samples of the Cirrhopetalum alliance included in this study (Supplementary Table 3). Duplicate samples of the same species from different sources (i.e., samples collected from the wild vs. samples collected from plants maintained in cultivation with regular watering) were included for comparison where available. Measurements were conducted with an isotope ratio mass spectrometer (SIRMS, CF mode) at the Stable Isotope Laboratory in the School of Biological Sciences, The University of Hong Kong.
Signature δ13C values for typical C3 plants range from −33.0 to −22.1‰, whereas values for obligate CAM plants typically range from −18.0 to −12.0‰ (Ehleringer and Osmond, 1989). Species with δ13C values ranging from −22.0 to −18.1‰ are usually interpreted as C3-CAM intermediates (Bone et al., 2015). Photosynthetic pathways were coded to generate binary character state data for downstream comparative phylogenetic studies according to the following two schemes: Scheme 1 (S1, defined by cut-off δ13C values as −18‰), in which C3 and C3-CAM intermediates were coded as “0” and obligate CAM as “1”; and Scheme 2 (S2, defined by cut-off δ13C values as −22‰), in which C3 was coded as “0” and C3-CAM intermediates and obligate CAM were collectively coded as “1” (Supplementary Table 3). Considering the relative uncertainty and the facultative features of C3-CAM intermediates, this study mainly focuses on strong CAM with discussion based on S1.
To test whether CAM and C3 photosynthetic pathways are associated with decreased diversification and the dynamics of speciation and extinction, we applied diverse models implemented in BiSSE (binary-state speciation and extinction; Maddison et al., 2007) and HiSSE (hidden state speciation and extinction; Beaulieu and O’Meara, 2016).
The BiSSE model was implemented in the R package DIVERSITREE ver. 0.9–10 (FitzJohn, 2012). ML optimisation was employed in BiSSE to estimate state-dependent rates of speciation (i.e., λ0, λ1) and extinction (μ0, μ1), as well as rates of transition from C3 to CAM (q01) and vice versa (q10), based on the full model (with all rates free). Nine alternative diversification models, with speciation and/or extinction and/or transition rates constrained, were then compared against the full model. These nine models were: three models in which these parameters were constrained to be equal (i.e., λ0 = λ1; μ0 = μ1; q01 = q10); and six models in which each had one parameter fixed to zero (i.e., λ0, λ1, μ0, μ1, q01 or q10 = 0) (Table 3). The model with the lowest AIC was preferred and considered as strongly supported with ΔAIC ≥ 2 compared to other alternative models (Burnham and Anderson, 2002).
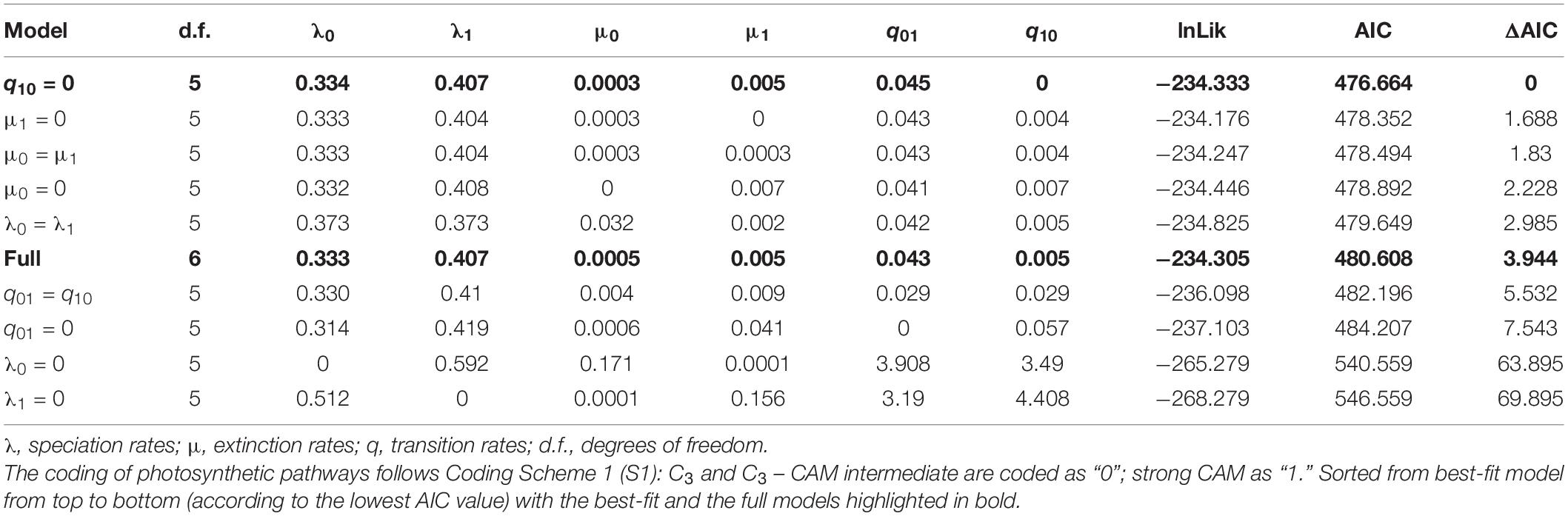
Table 3. The fit of BiSSE models of photosynthetic pathway evolution in the Cirrhopetalum alliance based on Dataset 3.
Exercising caution for type I errors arising in BiSSE (i.e., incorrectly finding neutral traits correlated with higher diversification rates; Rabosky and Huang, 2015), we further applied the HiSSE model to test the hypothesis that shifts in photosynthetic pathways could be correlated with hidden state speciation and extinction (Beaulieu and O’Meara, 2016). The testing of HiSSE models was implemented in the R package HISSE ver. 2.1.6 (Beaulieu and O’Meara, 2016), based on Dataset 1 and Coding Scheme S1. We tested the same 24 models proposed by Beaulieu and O’Meara (2016), together with a HiSSE model with all possible rates varying independently (following Landis et al., 2018). These 25 models included four models corresponding to the BiSSE analysis with a variety of constrained parameters; 17 HiSSE models that assumed a hidden state associated with observed character states with a variety of turn-over rates, extinction rates, and transition rates constrained; and four trait-independent models. Model selection was based on the lowest AIC and ΔAIC scores as indicated above.
Because sampling fractions cannot be specified based on the proportion of C3 and CAM species in the tree, we were unable to correct for non-random and incomplete sampling in both the BiSSE and HiSSE analyses (as recommended by FitzJohn et al., 2009). The results are therefore discussed with caution since the C3/CAM phenotype cannot be predicted without complete carbon isotopic data or physiological measurements of living plants representing all unsampled species.
We implemented the BiSSE likelihood function in R package DIVERSITREE to reconstruct the ancestral photosynthetic pathway. Reconstructions were conducted under the best-fitting BiSSE model based on Dataset 3 and two Coding Schemes (S1 and S2).
More than 80% of the clades in the time-calibrated phylogeny of the Cirrhopetalum alliance clade (CAC) were strongly supported (Figure 1 and Supplementary Table 4). The median crown age of the Dendrobium-Bulbophyllum clade was estimated as 31.62 Ma (HPD: 37.6–26.2 Ma; Supplementary Figure 1), broadly consistent with Xiang et al. (2016): 30.3 Ma (HPD: 34–26.9 Ma). The crown age of the Bulbophyllum clade was estimated as 16.7 Ma (HPD: 21.2–12.4 Ma; Supplementary Figure 1), largely congruent with Gamisch et al. (2015): 15.7 Ma (HPD: 22.5–9.8 Ma). The stem and crown ages of the CAC were reconstructed as 12.8 Ma (HPD: 16.3–9.8 Ma) and 11.8 Ma (HPD: 14.9–9 Ma), respectively.
Both the CR and MCCR tests rejected the pure-birth model under a one-tailed test with γ values < −1.645 [−3.36 (p = 0.001) and −2.43 (p = 0.003), respectively], providing support for the alternative hypothesis of a slowdown in diversification. All three DD diversification models were rejected with p values greatly exceeding 0.05, supporting the alternative diversity-independent model (DD1: p = 0.3, power of test = 0.25; DD2: p = 0.93, power of test = 0.43; DD3: p = 0.92, power of test = 0.39). In the TD model test, SPVAR was selected as the best-fitting model (Supplementary Table 2). The LTT plots for the Cirrhopetalum alliance derived from Dataset 1 and Dataset 3 further suggest a slowdown of lineage accumulation over time, with evidence of recent intensification, while BAMM revealed a slowdown in net diversification and speciation rates across the Cirrhopetalum alliance (Figure 1B and Supplementary Figure 2). The overall speciation rate (λ) of the Cirrhopetalum alliance calculated under the Yule model was 0.49 lineages/Myr (million years).
In the palaeoenvironment-dependent diversification test, the pCO2-dependent model with an exponentially variable speciation and constant extinction was selected as the best-fit model (Table 2). Following the best-fit model, the initial speciation rate of the Cirrhopetalum alliance was estimated as 0.4012 lineages/Myr, with a subsequent drop to −0.0004 lineages/Myr being positively correlated to a fall in atmospheric pCO2 with alpha < 0.
Results of various diversification model tests are summarised in Table 1.
Results of the fit BiSSE and HiSSE models are summarised in Table 3 and Supplementary Tables 5, 6. In the BiSSE full model (in which all rates are free) based on Coding Scheme S1 (Table 3 and Supplementary Figure 3), speciation rates were higher in CAM lineages (CAM: λ1 = 0.407; C3: λ0 = 0.333), whereas CAM-associated extinction rates were significantly (10 times) higher than that in C3 lineages. The rate of transition from C3 to strong CAM (q01) was 0.043 but near zero for the reverse (q10 = 0.005). When compared to the full BiSSE model, the overall best-fit was the model in which the transition rate q10 (from strong CAM to C3) was set to zero (Table 3). Joint analyses based on Coding Scheme S2 identified the same model (q01 = 0) as the best-fitting model (Supplementary Table 5). We note that the other three models (μ0 = 0; μ1 = 0; and μ0 = μ1) also received considerable support (ΔAIC < 2) and revealed an extremely low q10 value and asymmetric transitions between C3 and CAM (Table 3 and Supplementary Table 5).
The fit of 25 diversification models in the HiSSE framework indicated that the BiSSE model with equal extinction rates (ε0 = ε1) was the best-supported model (i.e., BiSSE models performed better than the HiSSE or the CID models; Supplementary Table 6). Although a more complex HiSSE mode (i.e., τ0A = τ1A = τ0B, ε’s equal, q0B1B = 0, q1B0B = 0, all other q’s equal) received considerable support (ΔAIC = 0.227; Supplementary Table 6), the simpler BiSSE model is preferred (Beaulieu and O’Meara, 2016). The HiSSE results suggest that photosynthetic pathways explain most of the diversification heterogeneity in the Cirrhopetalum alliance. The following discussion therefore mainly focuses on the results based on more comprehensive BiSSE model comparison (Table 3 and Supplementary Table 5).
Following the best-fit BiSSE model with q01 set to zero, ancestral photosynthetic pathway reconstruction based on Coding Scheme S1 revealed all CAM lineages to be derived from C3 ancestors and that the pathway evolved independently on at least nine occasions (Table 4 and Figure 2). Strong CAM was found to be established across all lineages in Clade 60 following initial split from C3 ancestors, and also latterly in Clade 35. Similar patterns were found when analyses were conducted based on the second-best model (results not shown).
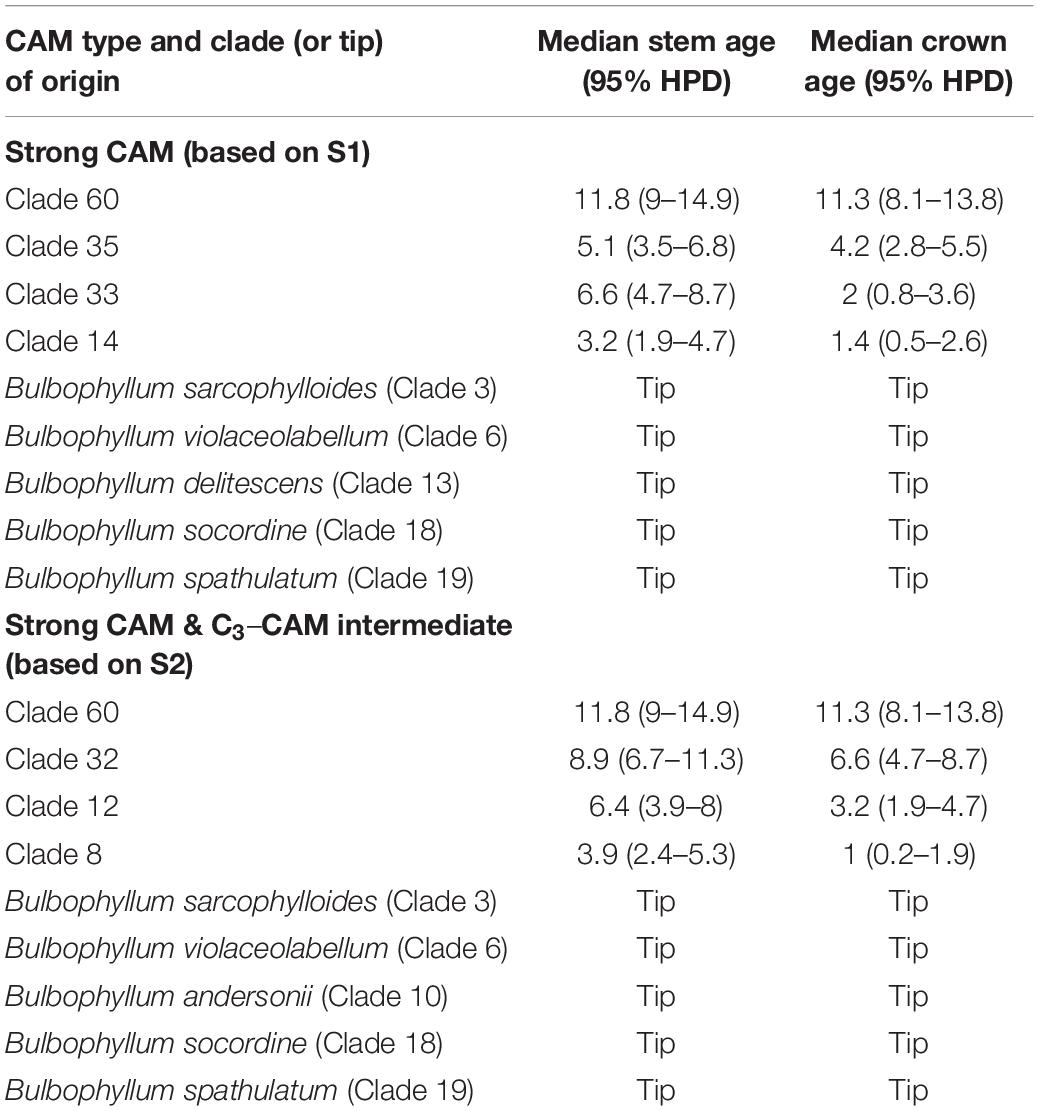
Table 4. Divergence times for the origin of CAM within the Cirrhopetalum alliance clade based on Coding Schemes S1 and S2.
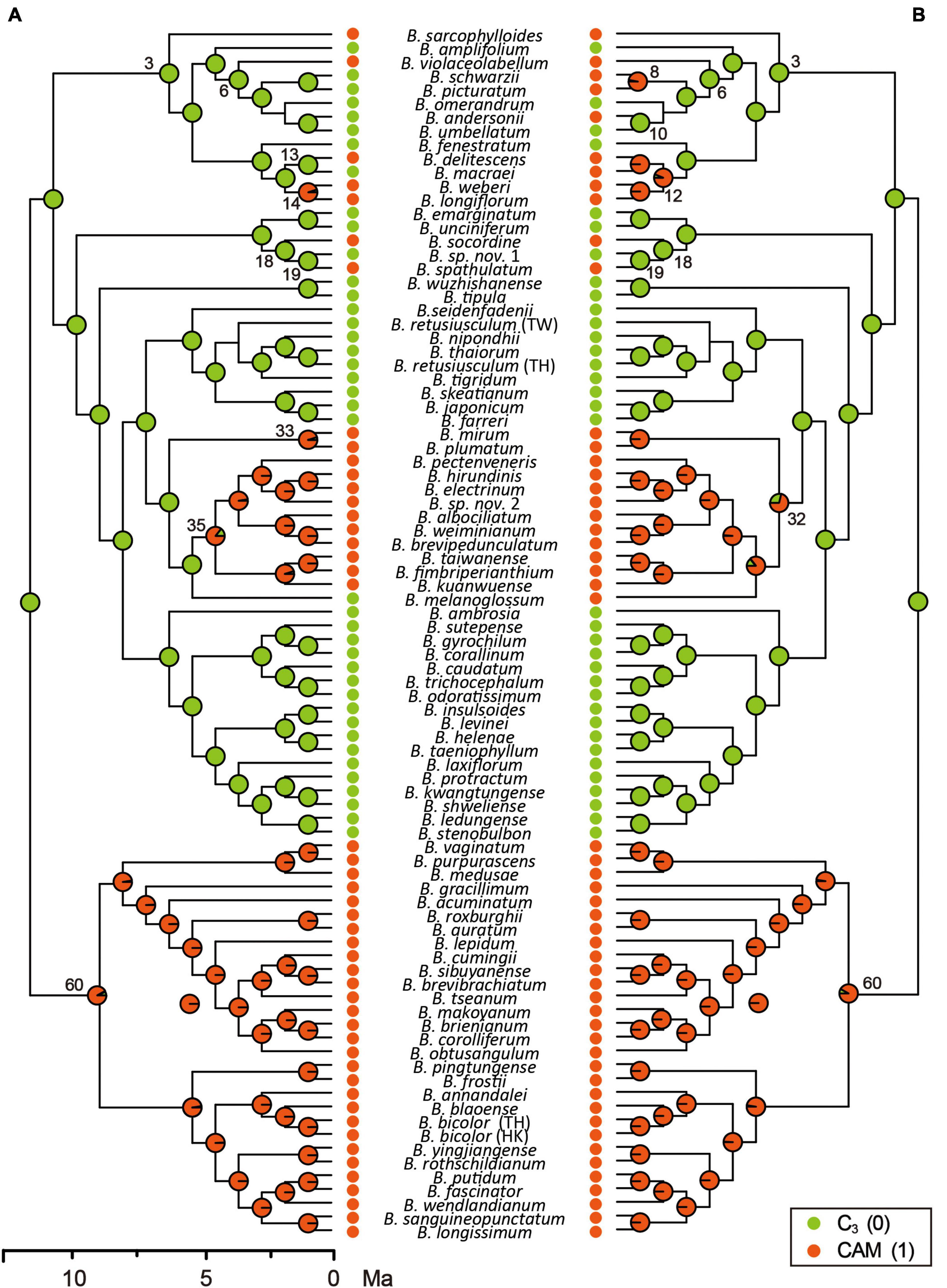
Figure 2. Reconstruction of the evolution of photosynthetic pathways in the Cirrhopetalum alliance clade, based on two coding schemes of photosynthetic modes described in the main text. Panel (A) corresponds to Coding Scheme S1 and panel (B) corresponds to Coding Scheme S2. Pie charts at internal nodes show relative probabilities assigned to each ancestral state. Clade numbers correlate with those shown in Figure 1 and Supplementary Table 4. Ma, million years ago.
C3 photosynthesis is reconstructed here as the ancestral photosynthetic pathway in the Cirrhopetalum alliance, with the divergence of CAM lineages estimated at c. 11.8 Ma. Strong CAM is inferred to have evolved independently at least nine times in four clades within the alliance, with CAM fixed in Clade 60 at c. 11.3 Ma and in Clade 35 at c. 4.2 Ma. Furthermore, there are several examples of CAM evolving at tips in other clades in the phylogeny (Table 4 and Figure 2), leading to a characteristic “twiggy” phylogenetic pattern (Schwander and Crespi, 2009).
It has been suggested that the striking global climatic changes in the mid-Miocene (c. 12–6 Ma)—with sharply decreased atmospheric CO2 concentrations that subsequently coincided with cooling and aridification in the late Miocene—may have created novel ecological opportunities in terrestrial ecosystems across all continents (Kürschner et al., 2008). The extensive aridification under low CO2 conditions in the mid- to late-Miocene is likely to have promoted selection for increased drought tolerance in many plant lineages across the world, including succulents such as Cactaceae (Arakaki et al., 2011), Aizoaceae (Valente et al., 2014), and Agavoideae (Heyduk et al., 2016). There is also evidence that the origin of alternative photosynthetic pathways (i.e., CAM and C4) is correlated to the expansion of arid ecosystems in the Miocene (Horn et al., 2014; Spriggs et al., 2014). The origin and fixation of the CAM photosynthetic pathway in the Cirrhopetalum alliance as revealed in the present study is likely to represent another example of such an adaptation. The combination of global atmospheric changes in the mid-Miocene and the final extension of the uplift of the Himalayas and Qinghai-Tibetan Plateau during the late Miocene and Pliocene is likely to have further intensified aridification in Central Asia, leading to the expansion of modern arid ecosystems (Miao et al., 2012). These changes have been linked to rapid radiations in many plant lineages in Asia (Wen et al., 2014). It is also worth noting that past monsoon activity has been implicated in playing a crucial role in diversification of Malagasy Bulbophyllum (Gamisch et al., 2021), as well as in the slowdown in diversification of Primulina (Gesneriaceae) in southern China (Kong et al., 2017). The impact of other environmental and climatic factors (i.e., global temperature changes and reginal monsoons) should be thoroughly investigated in future studies.
Multiple diversification model tests based on ML and Bayesian methods consistently revealed an explosive early speciation (Table 1) at the root of the Cirrhopetalum alliance during the late Miocene (Figure 1B), coinciding with the initial divergence of CAM lineages (Figure 2). This process mirrors the concept of a “confluence,” referring to the sequential occurrence of a suite of traits (innovations and synnovations), environmental changes and geographic movement along the branches of a phylogenetic tree (Donoghue and Sanderson, 2015). The coincidence and joint impact of ecological opportunity and the innovation of CAM could provide at least a partial explanation for the early explosion in the alliance. This initial burst of species diversity, however, was shown in the BAMM analysis to be followed by a gradual decline in diversification and speciation rate (Supplementary Figure 2).
Several recent studies have revealed strong evidence for a diversification slowdown (Phillimore and Price, 2008; Morlon et al., 2010; Moen and Morlon, 2014; but see Cusimano and Renner, 2010), with a meta-analysis of 289 phylogenies demonstrating that nearly 63% of clades spanning a variety of plant and animal taxa have experienced a slowdown (Morlon et al., 2010). Caution is nevertheless necessary as biases associated with our relatively small dataset plus incomplete and non-random sampling could give rise to ambiguous or unfounded evolutionary histories (Cusimano and Renner, 2010). In addition, the potential for infinite diversification scenarios based on extant phylogenetic trees has been linked to over-interpretation of certain diversification patterns (Louca and Pennell, 2020; Morlon et al., 2020). The development of analytical tools for comparative phylogenetic studies, however, has greatly improved our power to detect extinction and hence avoid this bias (Condamine et al., 2013; Moen and Morlon, 2014) and others, such as incompletely or non-randomly sampled phylogenies (Cusimano et al., 2012). In the present study, we integrated multiple diversification model tests based on both Bayesian and ML methods to derive a consensus in potential diversification histories (Table 1). To reduce certain highlighted biases, we integrated simulated data into our empirical tree to calculate the null model and perform DDD testing, and sampling fraction was applied in BAMM. Most importantly, we emphasise that alternative explanations such as environment-dependent models should be considered (Moen and Morlon, 2014) in understanding diversification patterns. Indeed, the palaeoenvironment-dependent diversification model testing suggests that the observed diversification slowdown in the Cirrhopetalum alliance could be explained by a long-term response to environmental changes in pCO2 (Table 2): exponential speciation in the alliance is positively correlated with a fall in atmospheric CO2 until the late Miocene, followed by a gradual increase in CO2. Even so, while pCO2 appears to be correlated with the initial radiation linked to the innovation of CAM photosynthesis, it remains unclear how character and environmental changes connect mechanistically to the dynamics of diversification (Donoghue and Sanderson, 2015). A variety of additional environmental factors, including temperature, monsoons, aridity, light intensity, growth season temperature, and precipitation, may influence the competitiveness of different photosynthetic strategies. In this regard, it is also worth noting that Hu et al. (2020) revealed several unexpected evolutionary transitions within the Cirrhopetalum alliance, with sect. Desmosanthes shown to be nested within this group and a variety of floral characters (pertaining to the petals, sepals, and lip) receiving moderate to strong phylogenetic signal, despite the lack of any correlation between floral characters and diversification. Future research with broader taxa sampling could provide more comprehensive insights into the interplay between niche and floral evolution in this hyper-diverse genus as a whole.
Crassulacean acid metabolism photosynthesis has often been interpreted as a highly labile photosynthetic pathway (Dodd et al., 2002; Winter et al., 2008), with some individuals of a species exhibiting facultative CAM, in which the degree of CAM expression greatly varies due to different developmental stages (e.g., juvenile vs. mature individuals; Winter et al., 2008) or environmental conditions (e.g., induced drought, salinity, high light intensity, low temperature, or anoxic conditions; Cushman and Borland, 2002). Empirical studies based on well-characterised facultative CAM plants, however, show that once CAM is established, it might not revert to a C3 mode, even after alleviation of the adverse environmental conditions which gave rise to it, as demonstrated in Clusia minor (Winter et al., 2008) and Mesembryanthemum crystallinum (Winter and Holtum, 2007), for example, though we note the opposite to be the case in Clusia pratensis (Winter et al., 2008). Similarly, significant CAM expression was detected in Cirrhopetalum alliance species based on carbon isotope ratios (δ13C), indicating consistent regulation of CAM, both in wild plants and in those under cultivation with regular watering (present study; multiple parallel δ13C data shown in Supplementary Table 3), even though CAM is energetically costly to produce and maintain as compared with C3 (Shameer et al., 2018). It is noticeable that neither favourable nor unfavourable conditions are capable of changing CAM to C3, presumably because the ontogenetic programme of the plant does not allow such modification (Griffiths et al., 2002; Winter et al., 2008). Restricted reversion to C3 might therefore be explained in part by the constitutive pre-set processes of development and growth that feature in CAM plants (Osmond, 2007), involving a series of developmental constraints (Heyduk et al., 2016).
From a phylogenetic perspective, the switch from C3 to obligate CAM has been suggested to be relatively rare to moderate at the species level, as reflected by few speciation events: e.g., three times over 6–10 Myr in Asparagaceae subfam. Agavoideae (Heyduk et al., 2016); three times over 7.36 Myr in Malagasy Bulbophyllum Gamisch et al. (2021); four times over 11.5 Myr in Orchidaceae subtribe Eulophiinae (Bone et al., 2015); 10 times over 7 Myr in Bromeliaceae subfam. Bromelioideae (Silvestro et al., 2014); 20 times over 15 Myr in Euphorbia (Euphorbiaceae) (Horn et al., 2014); 10 times among the Orchidaceae (Silvera et al., 2009); and nine times over 11.8 Myr in the Cirrhopetalum alliance in the present study (Table 4, based on S1). The reversal of CAM to C3, however, appears to be extremely rare as revealed by a strong asymmetry of transition rates between the two states. Comparison of 25 diversification models with HiSSE suggests that photosynthetic pathways explain most of the diversification heterogeneity in the Cirrhopetalum alliance; more comprehensive BiSSE models indicate a trend toward the acquisition of CAM in the Cirrhopetalum alliance, but with a near-zero transition rate for the reversion from CAM to C3 (Table 3). Similar asymmetrical transitions between CAM (or C4) and C3 were also found in different plant families (Horn et al., 2014; Silvestro et al., 2014; Bone et al., 2015; Heyduk et al., 2016), indicating lower flexibility and adaptive potential in CAM lineages compared to counterpart C3 lineages.
Key evolutionary innovations can be defined as morphologically or physiologically adaptive changes in individual traits that are causally linked to an increased diversification rate, either by increasing speciation rates or by decreasing extinction rates (Hunter, 1998; Ng and Smith, 2014). In contrast, extensive morphological, physiological, and ecological specialisation observed in adaptive traits may provide a short-term selective advantage, but in the long term reduce the capability of a species to adapt and survive (i.e., low adaptive potential) in changing environments, representing a substantial risk of future extinction (i.e., evolutionary “dead-ends”; Wright et al., 2013). Several traits have been suggested as likely causes of evolutionary dead-ends, including self-fertilisation in flowering plants (Igic and Busch, 2013), sexual dimorphism in ostracods (Martins et al., 2018), and sociality in spiders (Agnarsson et al., 2006), although support for some hypotheses of this nature remains ambiguous (Gamisch et al., 2015).
A recent study focusing on the Malagasy Bulbophyllum (Gamisch et al., 2021) indicated that CAM may be selectively advantageous even in habitats with high rainfall and suggested CAM as an evolutionary “gateway” trait that widened the ecological niche of the genus in Madagascar, albeit without significant effect on species diversity and diversification rates. We argue that CAM photosynthesis, in certain lineages, might represent another example of an evolutionary dead-end, thereby challenging the prevailing “key innovation” hypothesis of CAM in tropical orchids (e.g., Gravendeel et al., 2004; Givnish et al., 2015). Comprehensive empirical studies have implicated a phenomenon of CAM-associated high extinction rates in Bromeliaceae (Silvestro et al., 2014), Euphorbiaceae (Horn et al., 2014), and Orchidaceae (Givnish et al., 2015). In the present study, we demonstrate that the origin of CAM might have triggered an “early explosion” of speciation in the Cirrhopetalum alliance, concordant with the decline of atmospheric pCO2 during the late Miocene, with CAM initially providing a short-term evolutionary advantage, but with the alliance subsequently experiencing a gradual decrease in speciation and net diversification. Our study further reveals estimated extinction rates in CAM lineages that are ten times higher than those in C3 lineages, as inferred from the best-fit BiSSE model (Table 3), whereas CAM was not significantly linked to a higher speciation rate according to the full model, the best-fit and the second best-fit models in BiSSE (Table 3 and Supplementary Figure 3). The increased extinction rate is typically predicted to result in a “tippy” or “twiggy” phylogenetic pattern, in which the derived traits are distributed across the tips of the phylogenetic trees (Schwander and Crespi, 2009); this is observed in the Cirrhopetalum alliance (Figure 2) and in other CAM or C4 lineages, such as Bromeliaceae subfam. Bromelioideae (Silvestro et al., 2014) and Euphorbia (Euphorbiaceae) (Horn et al., 2014). The repeated origin and extinction of CAM suggests a conflict between the short-term benefits and long-term costs of producing and maintaining CAM. Although the benefits of CAM may initially outweigh the costs of maintaining C3 (hence leading to the multiple, independent evolution of CAM), in the longer term, CAM—as a more expensive strategy with low adaptive potential—may result in higher extinction. The significant negative influence on net diversification arising from a dramatically increased extinction rate has long been neglected, since research has primarily focused on the adaptive advantages of CAM: estimation of extinction rate is associated with considerable uncertainty (Silvestro et al., 2014).
An evolutionary dead-end can be alleviated if the possibility of reversal to the ancestral state is relaxed (Goldberg and Igić, 2008). Alternative photosynthetic strategies with greater flexibility and adaptive potential would be predicted to cope better with the challenges presented by a changing environment. Nevertheless, it is worth noting that facultative CAM induced or up-regulated by environmental factors has not yet been demonstrated unequivocally for any species in Orchidaceae (Winter and Holtum, 2014). Despite recent progress in genomics, proteomics, metabolomics, and computational modelling of the dynamic network that regulates CAM (De Paoli et al., 2014; Zhang et al., 2016; Yin et al., 2018), the genomic features and regulatory mechanisms of CAM have yet to be fully elucidated (Yang et al., 2015). Future studies should therefore focus on exploring the underlying genomic, metabolomic and ecological mechanisms of CAM-associated high extinction rates in diverse plant lineages, in order to evaluate how the potential evolutionary dead-end presented by the impacts of a changing environment might be evaded.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
A-QH, SWG, and RMKS conceptualised the research and drafted the manuscript. A-QH performed the laboratory experiments, data collection, and analyses. All authors discussed the results, reviewed the article, and approved the final article.
This research was funded by the Ph.D. student fellowship support to A-QH from Kadoorie Farm and Botanic Garden (KFBG). A-QH was also funded by a Sainsbury Orchid Fellowship from the Royal Botanic Gardens, Kew.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank J. L. Zhang, L. Simpson, F. L. Condamine, D. Silvestro, O. A. Peérez-Escobar, and B. Xue for their insightful comments and assistance in data analysis. We are grateful to two reviewers and editor SW for comments that helped to improve the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.794171/full#supplementary-material
Agnarsson, I., Avilés, L., Coddington, J. A., and Maddison, W. P. (2006). Sociality in Theridiid spiders: repeated origins of an evolutionary dead end. Evolution 60, 2342–2351. doi: 10.1111/j.0014-3820.2006.tb01869.x
Arakaki, M., Christin, P.-A., Nyffeler, R., Lendel, A., Eggli, U., Ogburn, R. M., et al. (2011). Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc. Natl. Acad. Sci. U.S.A. 108, 8379–8384. doi: 10.1073/pnas.1100628108
Beaulieu, J. M., and O’Meara, B. C. (2016). Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601. doi: 10.1093/sysbio/syw022
Bone, R. E., Smith, J. A. C., Arrigo, N., and Buerki, S. (2015). A macro-ecological perspective on crassulacean acid metabolism (CAM) photosynthesis evolution in Afro-Madagascan drylands: eulophiinae orchids as a case study. New Phytol. 208, 469–481. doi: 10.1111/nph.13572
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2, a software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537I
Burnham, K. P., and Anderson, D. R. (2002). Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd Edn. New York, NY: Springer-Verlag.
Condamine, F. L., Rolland, J., and Morlon, H. (2013). Macroevolutionary perspectives to environmental change. Ecol. Lett. 16, 72–85. doi: 10.1111/ele.12062
Conran, J. G., Bannister, J. M., and Lee, D. E. (2009). Earliest orchid macrofossils: early miocene Dendrobium and Earina (Orchidaceae: epidendroideae) from New Zealand. Am. J. Bot. 96, 466–474. doi: 10.3732/ajb.0800269
Cushman, J. C., and Borland, A. M. (2002). Induction of crassulacean acid metabolism by water limitation. Plant Cell Environ. 25, 295–310. doi: 10.1046/j.0016-8025.2001.00760.x
Cusimano, N., and Renner, S. S. (2010). Slowdowns in diversification rates from real phylogenies may not be real. Syst. Biol. 59, 458–464. doi: 10.1093/sysbio/syq032
Cusimano, N., Stadler, T., and Renner, S. S. (2012). A new method for handling missing species in diversification analysis applicable to randomly or nonrandomly sampled phylogenies. Syst. Biol. 61, 785–792. doi: 10.1093/sysbio/sys031
De Paoli, H. C., Borland, A. M., Tuskan, G. A., Cushman, J. C., and Yang, X. (2014). Synthetic biology as it relates to CAM photosynthesis: challenges and opportunities. J. Exp. Bot. 65, 3381–3393. doi: 10.1093/jxb/eru038
Dodd, A. N., Borland, A. M., Haslam, R. P., Griffiths, H., and Maxwell, K. (2002). Crassulacean acid metabolism: plastic, fantastic. J. Exp. Bot. 53, 569–580. doi: 10.1093/jexbot/53.369.569
Donoghue, J. M., and Sanderson, J. M. (2015). Confluence, synnovation, and depauperons in plant diversification. New Phytol. 207, 260–274. doi: 10.1111/nph.13367
Ehleringer, J. R., and Osmond, C. B. (1989). “Stable isotopes,” in Plant Physiological Ecology: Field Methods and Instrumentation, eds R. W. Pearcy, J. R. Ehleringer, H. A. Mooney, and P. W. Rundel (London: Chapman and Hall), 281–290.
Etienne, R. S., Haegeman, B., Stadler, T., Aze, T., Pearson, P. N., Purvis, A., et al. (2012). Diversity-dependence brings molecular phylogenies closer to agreement with the fossil record. Proc. Royal Soc. B 279, 1300–1309. doi: 10.1098/rspb.2011.1439
Etienne, R. S., Pigot, A. L., and Phillimore, A. B. (2016). How reliably can we infer diversity-dependent diversification from phylogenies? Methods Ecol. Evol. 7, 1092–1099. doi: 10.1093/sysbio/syq052
FitzJohn, R. G. (2012). Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092. doi: 10.1111/j.2041-210x.2012.00234.x
FitzJohn, R. G., Maddison, W. P., and Otto, S. P. (2009). Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. doi: 10.1093/sysbio/syp067
Foster, G., Royer, D., and Lunt, D. (2017). Future climate forcing potentially without precedent in the last 420 million years. Nat. Commun. 8:14845. doi: 10.1038/ncomms14845
Gamisch, A., Fischer, G. A., and Comes, H. P. (2015). Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum) in light of the hypothesis of selfing as an evolutionary dead end. BMC Evol. Biol. 15:192. doi: 10.1186/s12862-015-0471-5
Gamisch, A., Winter, K., Fischer, G. A., and Comes, H. P. (2021). Evolution of crassulacean acid metabolism (CAM) as an escape from ecological niche conservatism in malagasy Bulbophyllum (Orchidaceae). New Phytol. 231, 1236–1248. doi: 10.1111/nph.17437
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Royal Soc. B 282:20151553. doi: 10.1098/rspb.2015.1553
Goldberg, E. E., and Igić, B. (2008). On phylogenetic tests of irreversible evolution. Evolution 62, 2727–2741. doi: 10.1111/j.1558-5646.2008.00505.x
Gravendeel, B., Smithson, A., Slik, F. J. W., and Schuiteman, A. (2004). Epiphytism and pollinator specialization: drivers for orchid diversity? Philos. Trans. R. Soc. B 359, 1523–1535.
Griffiths, H., Helliker, B., Roberts, A., Haslam, R. P., Girnus, J., Robe, W. E., et al. (2002). Regulation of Rubisco activity in crassulacean acid metabolism plants: better late than never. Funct. Plant Biol. 29, 689–696.
Heyduk, K., McKain, M. R., Lalani, F., and Leebens-Mack, J. (2016). Evolution of a CAM anatomy predates the origins of crassulacean acid metabolism in the Agavoideae (Asparagaceae). Mol. Phylogenet. Evol. 105, 102–113. doi: 10.1016/j.ympev.2016.08.018
Horn, J. W., Xi, Z., Riina, R., Peirson, J. A., Yang, Y., Dorsey, B. L., et al. (2014). Evolutionary bursts in Euphorbia (Euphorbiaceae) are linked with photosynthetic pathway. Evolution 68, 3485–3504. doi: 10.1111/evo.12534
Hu, A.-Q., Gale, S. W., Liu, Z.-J., Suddee, S., Hsu, T.-C., Fischer, G. A., et al. (2020). Molecular phylogenetics and floral evolution of the Cirrhopetalum alliance (Bulbophyllum, Orchidaceae): evolutionary transitions and phylogenetic signal variation. Mol. Phylogenet. Evol. 143:106689. doi: 10.1016/j.ympev.2019.106689
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hunter, J. P. (1998). Key innovations and the ecology of macroevolution. Trends Ecol. Evol. 13, 31–36. doi: 10.1016/s0169-5347(97)01273-1
Igic, B., and Busch, J. W. (2013). Is self-fertilization an evolutionary dead end? New Phytol. 198, 386–397.
Kong, H., Condamine, F. L., Chen, J., Pan, B., Möller, M., et al. (2017). Both temperature fluctuations and East Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol Ecol. 26, 6414–6429. doi: 10.1111/mec.14367
Kürschner, W. M., Kvacěk, Z., and Dilcher, D. L. (2008). The impact of Miocene atmospheric carbon dioxide fluctuations on climate and the evolution of terrestrial ecosystems. Proc. Natl Acad. Sci. U.S.A 105, 449–453. doi: 10.1073/pnas.0708588105
Landis, J. B., Bell, C. D., Hernandez, M., Zenil-Ferguson, R., McCarthy, E. W., Soltis, D. E., et al. (2018). Evolution of floral traits and impact of reproductive mode on diversification in the phlox family (Polemoniaceae). Molec. Phylogen. Evol. 127, 878–890. doi: 10.1016/j.ympev.2018.06.035
Louca, S., and Pennell, M. W. (2020). Extant timetrees are consistent with a myriad of diversification histories. Nature 580, 502–505. doi: 10.1038/s41586-020-2176-1
Lovette, I. J., and Bermingham, E. (1999). Explosive speciation in the New World Dendroica warblers. Proc. Royal Soc. B 266, 1629–1636.
Lüttge, U. (1996). “Clusia: plasticity and diversity in a genus of C3/CAM intermediate tropical trees,” in Crassulacean Acid Metabolism: Biochemistry, Ecophysiology and Evolution, eds K. Winter and J. A. C. Smith (Berlin: Springer-Verlag), 296–311. doi: 10.1007/978-3-642-79060-7_20
Maddison, W. P., Midford, P. E., and Otto, S. P. (2007). Estimating a binary character’s effect on speciation and extinction. Syst. Biol. 56, 701–710. doi: 10.1080/10635150701607033
Martins, M. J. F., Puckett, T. M., Lockwood, R., Swaddle, J. P., and Hunt, G. (2018). High male sexual investment as a driver of extinction in fossil ostracods. Nature 556, 366–369. doi: 10.1038/s41586-018-0020-7
Miao, Y., Wu, F., Warny, S., Fang, X., Lu, H., Fu, H., et al. (2012). Miocene fire intensification linked to continuous aridification on the Tibetan Plateau. Geology 47, 303–307. doi: 10.1130/g45720.1
Michener, R., and Lajtha, K. (2007). “Sources of variation in the stable isotopic composition of plants,” in Stable Isotopes in Ecology and Environmental Science, 2nd Edn, eds K. Lajtha and R. Michener (Malden, MA: Blackwell), 22–34. doi: 10.1002/9780470691854.ch2
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Paper Presented at Proceedings of the Gateway Computing Environments Workshop (GCE) (New Orleans, LA: IEEE), 1–8
Moen, D., and Morlon, H. (2014). Why does diversification slow down? Trends Ecol. Evol. 29, 190–197. doi: 10.1016/j.tree.2014.01.010
Morlon, H., Hartig, F., and Robin, S. (2020). Prior hypotheses or regularization allow inference of diversification histories from extant timetrees. bioRxiv [preprint]. Available online at: https://doi.org/10.1101/2020.07.03.185074 [accessed on Jul 4, 2020].
Morlon, H., Lewitus, E., Condamine, F. L., Manceau, M., Clavel, J., and Drury, J. (2016). RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol. 7, 589–597. doi: 10.1111/2041-210x.12526
Morlon, H., Potts, M. D., and Plotkin, J. B. (2010). Inferring the dynamics of diversification: a coalescent approach. PLoS Biol. 8:e1000493. doi: 10.1371/journal.pbio.1000493
Ng, J., and Smith, S. D. (2014). How traits shape trees: new approaches for detecting character state-dependent lineage diversification. J. Evol. Biol. 27, 2035–2045. doi: 10.1111/jeb.12460
Nylander, J. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583.
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Phillimore, A. B., and Price, T. D. (2008). Density-dependent cladogenesis in birds. PLoS Biol. 6:e71. doi: 10.1371/journal.pbio.0060071
Pybus, O. G., and Harvey, P. H. (2000). Testing macro-evolutionary models using incomplete molecular phylogenies. Proc. Royal Soc. B 267, 2267–2272. doi: 10.1098/rspb.2000.1278
Rabosky, D. L. (2006). LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol. Bioinform. 2, 247–250.
Rabosky, D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9:e89543. doi: 10.1371/journal.pone.0089543
Rabosky, D. L., Grundler, M., Anderson, C., Title, P., Shi, J. J., Brown, J. W., et al. (2014). BAMM tools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707. doi: 10.1111/2041-210x.12199
Rabosky, D. L., and Huang, H. (2015). A robust semi-parametric test for detecting trait-dependent diversification. Syst. Biol. 65, 181–193. doi: 10.1093/sysbio/syv066
Rabosky, D. L., and Lovette, I. J. (2008). Density-dependent diversification in North American wood warblers. Proc. Royal Soc. B 275, 2363–2371. doi: 10.1098/rspb.2008.0630
Rambaut, A., and Drummond, A. J. (2014). LogCombiner ver. 2.1.3. Edinburgh, UK: Institute of Evolutionary Biology
Rambaut, A., Suchard, M. A., Xie, D., and Drummond, A. J. (2014). Tracer ver. 1.6. Available online at: http://beast.bio.ed.ac.uk/Tracer
Raven, J. A., and Spicer, R. A. (1996). “The evolution of Crassulacean acid metabolism,” in Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution, eds K. Winter and J. A. C. Smith (Berlin: Springer-Verlag), 360–385.
Ricklefs, R. E. (2007). Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 22, 601–610. doi: 10.1016/j.tree.2007.06.013
Schwander, T., and Crespi, B. J. (2009). Twigs on the tree of life? Neutral and selective models for integrating macroevolutionary patterns with microevolutionary processes in the analysis of asexuality. Mol. Ecol. 18, 28–42. doi: 10.1111/j.1365-294X.2008.03992.x
Shameer, S., Baghalian, K., Cheung, C. Y. M., Ratcliffe, R. G., and Sweetlove, L. J. (2018). Computational analysis of the productivity potential of CAM. Nat. Plants 4, 165–171. doi: 10.1038/s41477-018-0112-2
Silvera, K., Neubig, K. M., Whitten, W. M., Williams, N. H., Winter, K., and Cushman, J. C. (2010). Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 37, 995–1010. doi: 10.1071/fp10084
Silvera, K., Santiago, L. S., Cushman, J. C., and Winter, K. (2009). Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 149, 1838–1847. doi: 10.1104/pp.108.132555
Silvestro, D., Zizka, G., and Schulte, K. (2014). Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae). Evolution 68, 163–175. doi: 10.1111/evo.12236
Spriggs, E. L., Christin, P. A., and Edwards, E. J. (2014). C4 photosynthesis promoted species diversification during the Miocene grassland expansion. PLoS One 9:e97723. doi: 10.1371/journal.pone.0105923
Stadler, T. (2011). Simulating trees on a fixed number of extant species. Syst. Biol. 60, 676–684. doi: 10.1093/sysbio/syr029
Valente, L. M., Britton, A. W., Powell, M. P., Papadopulos, A. S., Burgoyne, P. M., and Savolainen, V. (2014). Correlates of hyperdiversity in southern African ice plants Aizoaceae. Bot. J. Linn. Soc. 174, 110–129. doi: 10.1111/boj.12117
Wen, J., Zhang, J.-Q., Nie, Z.-L., Zhong, Y., and Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet. 5:24575120. doi: 10.3389/fgene.2014.00004
Winter, K., Garcia, M., and Holtum, J. A. M. (2008). On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchoe and Opuntia. J. Exp. Bot. 59, 1829–1840. doi: 10.1093/jxb/ern080
Winter, K., and Holtum, J. A. M. (2007). Environment or development? Lifetime net CO2 exchange and control of the expression of crassulacean acid metabolism in Mesembryanthemum crystallinum. Plant Physiol. 143, 98–107. doi: 10.1104/pp.106.088922
Winter, K., and Holtum, J. A. M. (2014). Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 65, 3425–3441. doi: 10.1093/jxb/eru063
Wright, S. I., Kalisz, S., and Slotte, T. (2013). Evolutionary consequences of self-fertilization in plants. Proc. Royal Soc. B 280:20130133. doi: 10.1098/rspb.2013.0133
Xiang, X.-G., Mi, X.-C., Zhou, H.-L., Li, J.-W., Chung, S.-W., Li, D.-Z., et al. (2016). Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr. 43, 1310–1323.
Yang, X., Cushman, J. C., Borland, A. M., Edwards, E. J., Wullschleger, S. D., Tuskan, G. A., et al. (2015). A roadmap for research on Crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 207, 491–504. doi: 10.1111/nph.13393
Yin, H., Guo, H.-B., Weston, D. J., Borland, A. M., Ranjan, P., Abraham, P. E., et al. (2018). Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave. BMC Genom. 19:588. doi: 10.1186/s12864-018-4964-7
Keywords: CAM, diversification slowdown, evolutionary dead-end, key innovation, pCO2
Citation: Hu A-Q, Gale SW, Liu Z-J, Fischer GA and Saunders RMK (2022) Diversification Slowdown in the Cirrhopetalum Alliance (Bulbophyllum, Orchidaceae): Insights From the Evolutionary Dynamics of Crassulacean Acid Metabolism. Front. Plant Sci. 13:794171. doi: 10.3389/fpls.2022.794171
Received: 13 October 2021; Accepted: 10 January 2022;
Published: 03 February 2022.
Edited by:
Susann Wicke, Humboldt University of Berlin, GermanyReviewed by:
Jianquan Liu, Lanzhou University, ChinaCopyright © 2022 Hu, Gale, Liu, Fischer and Saunders. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephan W. Gale, c3RlcGhhbmdhbGVAa2ZiZy5vcmc=; Richard M. K. Saunders, c2F1bmRlcnNAaGt1Lmhr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.