- 1Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa
- 2Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
- 3Instituto de Biología Molecular y Celular de Plantas, Consejo Superior de Investigaciones Científicas - Universitat Politècnica de València (IBMCP-CSIC-UPV), Valencia, Spain
Avocado is an important agricultural food crop in many countries worldwide. Phytophthora cinnamomi, a hemibiotrophic oomycete, remains one of the most devastating pathogens within the avocado industry, as it is near impossible to eradicate from areas where the pathogen is present. A key aspect to Phytophthora root rot disease management is the use of avocado rootstocks partially resistant to P. cinnamomi, which demonstrates an increased immune response following infection. In plant species, Nucleotide binding-Leucine rich repeat (NLR) proteins form an integral part of pathogen recognition and Effector triggered immune responses (ETI). To date, a comprehensive set of Persea americana NLR genes have yet to be identified, though their discovery is crucial to understanding the molecular mechanisms underlying P. americana-P. cinnamomi interactions. In this study, a total of 161 PaNLR genes were identified in the P. americana West-Indian pure accession genome. These putative resistance genes were characterized using bioinformatic approaches and grouped into 13 distinct PaNLR gene clusters, with phylogenetic analysis revealing high sequence similarity within these clusters. Additionally, PaNLR expression levels were analyzed in both a partially resistant (Dusa®) and a susceptible (R0.12) avocado rootstock infected with P. cinnamomi using an RNA-sequencing approach. The results showed that the partially resistant rootstock has increased expression levels of 84 PaNLRs observed up to 24 h post-inoculation, while the susceptible rootstock only showed increased PaNLR expression during the first 6 h post-inoculation. Results of this study may indicate that the partially resistant avocado rootstock has a stronger, more prolonged ETI response which enables it to suppress P. cinnamomi growth and combat disease caused by this pathogen. Furthermore, the identification of PaNLRs may be used to develop resistant rootstock selection tools, which can be employed in the avocado industry to accelerate rootstock screening programs.
Introduction
Avocados (Persea americana Mill.) are an agriculturally important crop in many countries, including South Africa, Spain, and Mexico (Bulagi et al., 2016; Vargas-Canales et al., 2020). The annual gross production value of avocados in South Africa increased by 14.2% in 2018–2019 to a total of R1.42 billion, when compared to 2017–2018. Phytophthora root rot, caused by the hemibiotrophic oomycete, Phytophthora cinnamomi Rands, remains the largest threat to the avocado industry, in countries where the pathogen is present (Hardham and Blackman, 2018). The pathogen infects the fine feeder roots of avocado trees, leading to decreased water and nutrient transportation between cells (Coffey, 1987). A decline in tree health is observed which ultimately leads to plant death. Phytophthora cinnamomi can survive in soils over long periods of time through the production of chlamydospores and oospores, thus limiting the number of effective control methods for Phytophthora root rot (Dobrowolski et al., 2008; Belisle et al., 2019). Phosphite trunk injections, use of partially resistant rootstocks and organic mulching practices are methods currently employed by the avocado industry to control P. cinnamomi (Giblin et al., 2005). However, research has shown that P. cinnamomi has the potential to become less sensitive toward phosphite trunk injections (Dobrowolski et al., 2008). Continued screening for P. cinnamomi resistant rootstocks is thus of utmost importance and can be accelerated when host-pathogen interactions are understood.
Plant immune responses influence host-pathogen interactions and involve a myriad of proteins which activate complex, multilayered signaling pathways in response to pathogen attack (Dangl and Jones, 2001; Naveed et al., 2020). These can be categorized into two main responses; the Pathogen associated molecular pattern (PAMP) triggered immune response (PTI) and the Effector triggered immune response (ETI; Davis and Hahlbrock, 1987; Jones and Dangl, 2006). The recognition of PAMPs by membrane-bound Pattern recognition receptors (PRRs) activate an innate immune response, which is lower in amplitude when compared to the ETI response, and forms part of the plant’s first line of defense against pathogens (Matzinger, 2007). Pathogens, in turn, produce effector proteins which are secreted into plant cells to interfere with this process. These effector molecules may then be recognized by intracellular proteins, such as Resistance (R) proteins, either directly or indirectly (Monteiro and Nishimura, 2018). Upon effector recognition, R proteins are activated and trigger ETI—a high amplitude, robust immune response. The primary mode of action of ETI is to activate localized cell death caused by the Hypersensitive response (HR), aimed at arresting pathogen growth (Cui et al., 2015).
Resistance proteins are classified into five diverse groups based on protein structure and domains (Bezerra-Neto et al., 2020). The largest group consists of proteins with Nucleotide binding and Leucine rich repeat domains (LLRs), referred to as Nucleotide binding-Leucine rich repeats (NLRs; McDowell and Woffenden, 2003). Other groups include Receptor-like proteins (RLPs), Receptor-like kinases (RLKs), and Transmembrane Coiled-coil proteins (TM-CCs). The NLR group can be further sub-divided into two classes, based on the NLR’s N-terminus domain. The first class has a Coiled-coil (CC) domain, while the second class has a Toll/interleukin-1 receptor (TIR) structure domain. These NLRs are termed CNLs and TNLs, respectively. The CNL class also includes NLR proteins with both a CC domain and a RPW8 domain (resistance to powdery mildew), termed CCR-NLRs or CRNLs (Zhong and Cheng, 2016). CNLs are more abundant in the genomes of tree species when compared to TNLs, although the opposite is seen in Arabidopsis (Neale et al., 2017). Certain angiosperm and conifer species also do not follow this pattern due to TNL duplications, resulting in increased TNL:CNL ratios. Tandem duplications and NLR gene family expansions may have increased fitness levels of tree species that need long-term defense strategies against pathogens (Tobias and Guest, 2014). As a result, these duplicated gene sequences are mostly found in gene clusters within plant genomes (Meyers et al., 2003). Head-to-head NLR genes may express proteins which interact to form homo– or heterodimers, often vital for proper NLR function (Liang et al., 2019). These NLR protein dimers greatly increase the pathogen recognition potential of different NLR protein complexes (Van Wersch and Li, 2019).
A few NLR proteins are constantly expressed at low basal levels which allow plants to “scan” for invading pathogens (Meyers et al., 2002). Most of the genes coding for NLR proteins, however, show differential expression patterns after pathogen attack. This is influenced by the species of pathogen, excreted effector proteins and the plant’s genotype (Christie et al., 2016; Andam et al., 2020). In Eucalyptus grandis challenged by Leptocybe invasa and Chrysoporthe austroafricana, 218 and 343 NLRs were differentially expressed, respectively (Christie et al., 2016). RGA1, a TNL protein in the tree species Salix viminalis, showed higher expression in the resistant host when compared to its susceptible counterpart after Melamspora larici-epitea infection (Martin et al., 2016). Higher RGA1 expression allows for earlier ETI activation which ultimately leads to enhanced disease resistance. The level and timing of NLR expression is crucial, as this ultimately governs whether a plant would be successful in countering pathogen attack (Umadevi and Anandaraj, 2017). Transgenic plants with higher NLR gene expression demonstrated increased resistance to plant pathogens, even when these plants were transformed with non-native NLR genes. Expression of ZmNB25, a NLR first identified in maize, increased the resistance levels of Arabidopsis and rice toward Pseudomonas syringae pv. tomato DC3000 and Bipolaris maydis, respectively (Xu et al., 2018). Understanding how the expression of NLRs change during pathogen infection, and subsequently influence disease resistance, is vital to understanding complex plant-pathogen interactions.
To date, 49 complete putative NLR genes have been identified in avocado using microarray and RNA-sequencing analysis (Van den Berg et al., 2018; Pérez-Torres et al., 2021). In the study done by Pérez-Torres et al. (2021), Hass avocado stems were infected with Fusarium kuroshium, which causes Fusarium dieback disease in avocado. However, only four NLR genes were differentially expressed after F. kuroshium infection. Additionally, only a single avocado NLR gene has been implicated in the defense against P. cinnamomi infection in an avocado rootstock (Van den Berg et al., 2018). This NLR, functionally annotated as RPP13-like protein 4, showed increased expression after P. cinnamomi infection in a partially resistant rootstock. The use of RNA-seq and microarray data to identify NLR genes is limited by the fact that these genes need to be expressed to enable detection and identification. Avocado NLR gene identification has further been hampered by the lack of a high-quality genome assembly. Three avocado genomes, the Mexican landrace cultivar (Persea americana var. drymifolia), the Hass fruiting cultivar (Rendón-Anaya et al., 2019) and the West-Indian pure accession (WI) rootstock (Avocado Genome Consortium, Article in preparation) have only recently been sequenced, providing an opportunity to identify significantly more NLR genes within the avocado genome.
The discovery of NLR genes within the avocado genome could provide novel insight into the interactions of this plant with various pathogens. The current study set out to identify avocado NLR genes using the available genome sequences, and subsequently assess their expression during P. cinnamomi infection of partially resistant and susceptible rootstocks. We identified 161 putative PaNLR genes in the WI rootstock avocado genome, based on amino acid sequences characteristic of conserved NLR domains. Furthermore, we analyzed the expression of the candidate PaNLR genes in a partially resistant and susceptible rootstock following P. cinnamomi inoculation using an RNA-seq approach. We found significantly higher expression levels of 84 PaNLR genes in the partially resistant rootstock when compared to the susceptible rootstock after P. cinnamomi inoculation. This knowledge may benefit future rootstock screening programs aimed at increasing resistance levels toward P. cinnamomi. The PaNLR gene sequences identified in this study serve as an invaluable resource which can be used to pinpoint proteins that play a role in defense responses against other avocado pathogens.
Materials and Methods
Putative PaNLR Gene Identification
The P. americana West-Indian pure accession genome was obtained from the Avocado Genome Consortium (Article in preparation). Gene and protein names assigned during genome annotation (Peame105C00g000000) were abbreviated to PC00g000000. The Hass fruiting cultivar (P. americana cv. Hass; GCA_008087245.1) and Mexican rootstock (P. americana var. drymifolia; GCA_008033785.1) genomes (Rendón-Anaya et al., 2019) were obtained from GenBank (NCBI Genbank). Putative Resistance genes were identified and classified using the Resistance Gene Analog (RGA) prediction pipeline, RGAugury1 (Li et al., 2016; downloaded in September 2020). The avocado WI genome, as well as whole genome protein sequences from the WI, Mexican and Hass genomes were used as input with default parameters. The pipeline identifies conserved RGA sequences and domains using five programs: BLAST v. 2.10.1 (Camacho et al., 2009), nCoil2 v. 2.2 (Lupas et al., 1991), InterProScan3 v. 5.52-86.0 (Zdobnov and Apweiler, 2001), Pfam_scan4 v. 1.6 (Finn et al., 2010), and Phobius5 v. 1.01 (Käll et al., 2004). Putative NLR proteins were classified based on the identified domains, namely Nucleotide binding site (NB), Coiled-coil domain (CC), Coiled-coil with RPW8 domain (CCR), Toll/interleukin-1 receptor (TIR), and Leucine rich repeat domain (LRR). Here, N, C, T, and L represent NB, CC, TIR, and LRR domains, respectively. Thus, a protein classified as CNL has a CC, NB, and LRR domain, and a CN protein only has a CC and NB domain. RLKs, RLPs, and TM-CC classifications were annotated if the protein sequences contained a transmembrane domain. After identification and classification, protein functional annotation was done by performing BLASTp analysis in the non-redundant NCBI database. Searches were performed using an expected threshold value of 0.00001, with only the top hit for each candidate NLR gene being considered. If no significant match could be identified, proteins were annotated as Disease resistance-like (DRL) proteins.
PaNLR Gene Cluster Identification
Gene clusters were defined based on appropriate definitions from Meyers et al. (2003), Kohler et al. (2008), and Christie et al. (2016). A gene cluster was defined as: a genomic region which contained three or more NLR genes, with less than nine other genes between adjacent NLR genes, and with two adjacent NLR genes being less than 250 kb apart. The WI genome general feature format (GFF) file was used to indicate the distance and number of neighboring genes between NLR genes. The position of NLR genes were visualized using CViT6 v. 1.3 (Cannon and Cannon, 2011).
Phylogenetic Analysis
Phylogenetic analysis was used to assess whether PaNLR genes from the same gene cluster have high sequence similarity. Phylogenetic analysis included 161 P. americana NB-domain protein sequences, 10 complete protein sequences from Cinnamomum micranthum f. kanehirae (RWR97694.1, RWR95032.1, RWR91786.1, RWR92004.1, RWR93015.1, RWR98067.1, RWR88343.1, RWR88103.1, RWR87020.1, and RWR85657.1; Chaw et al., 2019) and one complete protein sequence from Solanum bulbocastanum (Q7XBQ9.1; Song et al., 2003). This S. bulbocastanum sequence was used since no RGA2 sequences were identified in C. micranthum f. kanehirae (Chaw et al., 2019). Sequence alignment was performed using ClustalW v2.1 with default parameters in MEGA X (Thompson et al., 1994; Kumar et al., 2018). A maximum likelihood phylogenetic tree was produced using the Jones-Taylor-Thornton substitution model and 1,000 bootstrap replications.
Plant Inoculation and RNA Sequencing
PaNLR expression data were obtained by dual RNA-sequencing of P. americana inoculated with P. cinnamomi. Roots from partially resistant (Dusa®) and susceptible (R0.12) rootstocks were inoculated by dipping in P. cinnamomi (isolate GKB4) zoospore suspension with a concentration of 1.4 × 105 zoospores/ml. Thereafter, plantlets were replanted in a mixture of vermiculite and perlite (1:1 ratio) and roots were harvested after 6, 12, 24, and 120 h post-inoculation (hpi). Three biological replicates from three independent plants were harvested at each time point. For control samples, three plantlets per rootstock were mock-inoculated using sterile water and root samples were harvested at 24 hpi.
Root samples were flash frozen using liquid N2 and stored at −70°C. The samples were then powdered using an IKA® Tube Mill (IKA®, Staufen, DUE). Modified CTAB extractions were performed to extract total RNA (Chang et al., 1993). RNA extractions were purified using a Qiagen RNeasy clean up kit (Qiagen, Valecia, California, United States) following DNase I treatment (Fermentas Life Sciences, Hanover, United States). An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) was used to measure RNA purity and quality. Samples were stored at −70°C before being sent to Novogene (Novogene Corporation Inc., Chula Vista, California, United States) for paired-end (250–300 bp insert cDNA library) sequencing using Illumina Hiseq 2500 with PE150 mode.
Expression Analysis
Dual RNA-sequencing data was analyzed by the Avocado Research Program and used during this study (Article in preparation). In short, RNA-seq reads were trimmed and low-quality bases were removed using Trimmomatic v. 0.39 (Bolger et al., 2014). FASTQC v. 0.11.9 was used to confirm read quality and the resultant reports were summarized using MultiQC (Ewels et al., 2016). RNA-seq reads were aligned to the P. americana WI genome using HISAT v. 2.0.6 (Kim et al., 2015). Gene level transcript abundance was quantified using featureCounts v. 2.0.1 (Liao et al., 2014) during initial expression screens within RNA-seq libraries across all time-points (6, 12, 24, and 120 hpi) using the mock-inoculated or susceptible rootstock libraries as a reference. DESeq2 (Love et al., 2014) was used for the normalization and analysis of counts. Library data points for transcripts with fewer than 10 reads were removed, and transcripts without any read data overall were omitted from further analyses. Quantification data for PaNLR genes were extracted using R studio v. 1.4.1106 (RStudio Team, 2020) and gene IDs previously identified by RGAugury. Expression level differences were analyzed using two approaches: (1) comparing the expression of candidate PaNLR genes 6, 12, 24, and 120 hpi in both the susceptible and partially resistant rootstock to that of their respective mock-inoculated samples, and (2) comparing the expression of candidate PaNLRs in the partially resistant rootstock to the expression in the susceptible avocado rootstock (mock-inoculated, 6, 12, 24, and 120 hpi). PaNLR genes were considered to be up- or downregulated when the Log2 Fold Change (Log2FC) value for each gene was ≥1 or ≤−1, respectively. False discovery rate adjusted values of p ≤ 0.05 generated as part of the DeSeq2 package were used to indicate statistical significance. Heatmaps and dendrograms depicting expression level differences (Log2FC) were generated using the Pheatmap package v. 1.0.12 (Kolde, 2012) in R studio v. 1.4.1106 (RStudio Team, 2020). To assess whether NLR genes within a gene cluster were co-expressed, NLR expression data was analyzed using Clust7 v. 1.12.0 (Abu-Jamous and Kelly, 2018).
Results
Putative PaNLR Genes Identified in the Avocado Genome
The RGAugury pipeline identified 259 putative PaNLR genes within the WI rootstock genome (Figure 1), while no PaNLR genes could be identified within the Mexican and Hass genomes. NLR gene sequences which did not include a LRR domain sequence were removed from further analysis and considered as incomplete PaNLR genes. This resulted in 161 PaNLR sequences which were classified as complete PaNLR genes. Of these genes, 102 were classified as CNLs, two as CRNLs, 56 as NLs and one as TNL, based on the domains present within their predicted amino acid sequences.
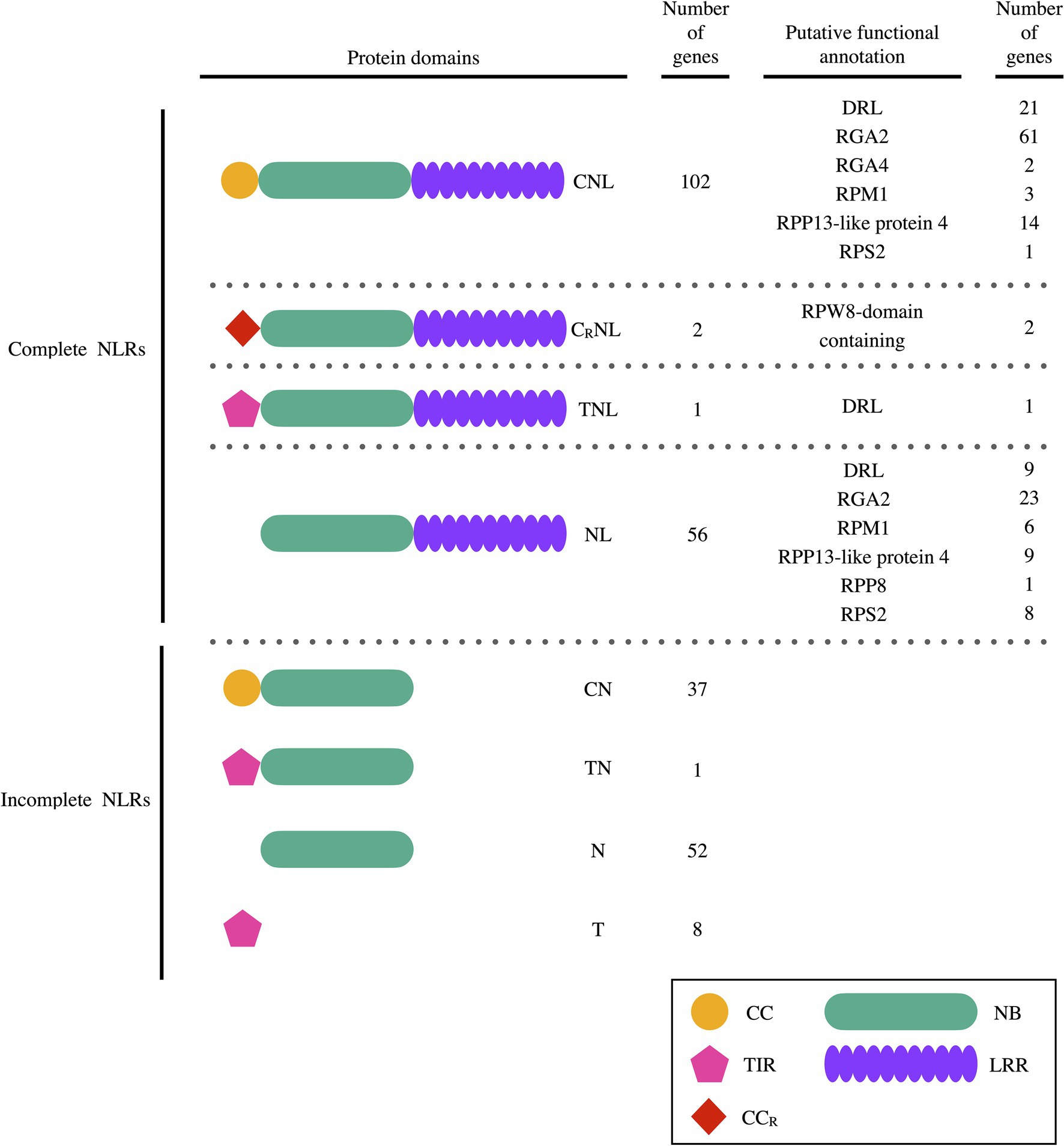
Figure 1. The number of PaNLR genes identified in the West-Indian pure accession Persea americana genome and the set of protein domains each gene encodes for. Putative Nucleotide binding-Leucine rich repeat (NLR) protein functional annotations predicted using BLASTp analysis are also listed (C/CC, coiled-coil domain; CR/CCR, coiled-coil RPW8 domain; DRL, disease resistance-like protein; L/LRR, leucine rich repeat domain; N/NB, nucleotide binding domain; and T/TIR, toll/interleukin-1 receptor domain).
Putative protein functional annotation of the 161 complete PaNLR candidates were assigned using BLASTp. In total, 31 sequences were assigned as DRL proteins. More than 52% of sequences were putatively identified as RGA2 proteins (Figure 1). Other sequence identifications included RGA4, RPM1, RPP13-like protein 4, RPP8, RPS2, and RPW8-domain containing type proteins.
The RGAugury pipeline also identified RLP, RLK, and TM-CC proteins using WI whole genome protein sequences. These protein sequences were separated from NLR sequences if a transmembrane domain sequence was identified. In total, 106 RLP sequences, 889 RLK sequences and 189 TM-CC sequences were identified.
PaNLR Gene Clusters Identified in the WI Genome
PaNLR gene clusters were identified based on neighboring PaNLR genes being less than 250 kb apart, and having less than three non-NLR genes between them. In total, 13 PaNLR gene clusters were identified, accounting for 74 (45.9%) of the complete PaNLR gene sequences (Figure 2). Thirteen PaNLR genes were mapped to unanchored chromosomes and were thus excluded from the cluster analysis. Chromosome 2 had four gene clusters (the largest set of clusters on any of the chromosomes) and also contained the largest gene cluster (consisting of nine PaNLR sequences). No gene clusters were identified on chromosomes 4, 5, 8, 9, 10, and 12. Eight of the gene clusters contained sequences which encode RGA2 proteins, with the gene clusters occurring on chromosomes 6 and 7 lacking PaNLR genes encoding RGA2 proteins (Table 1).
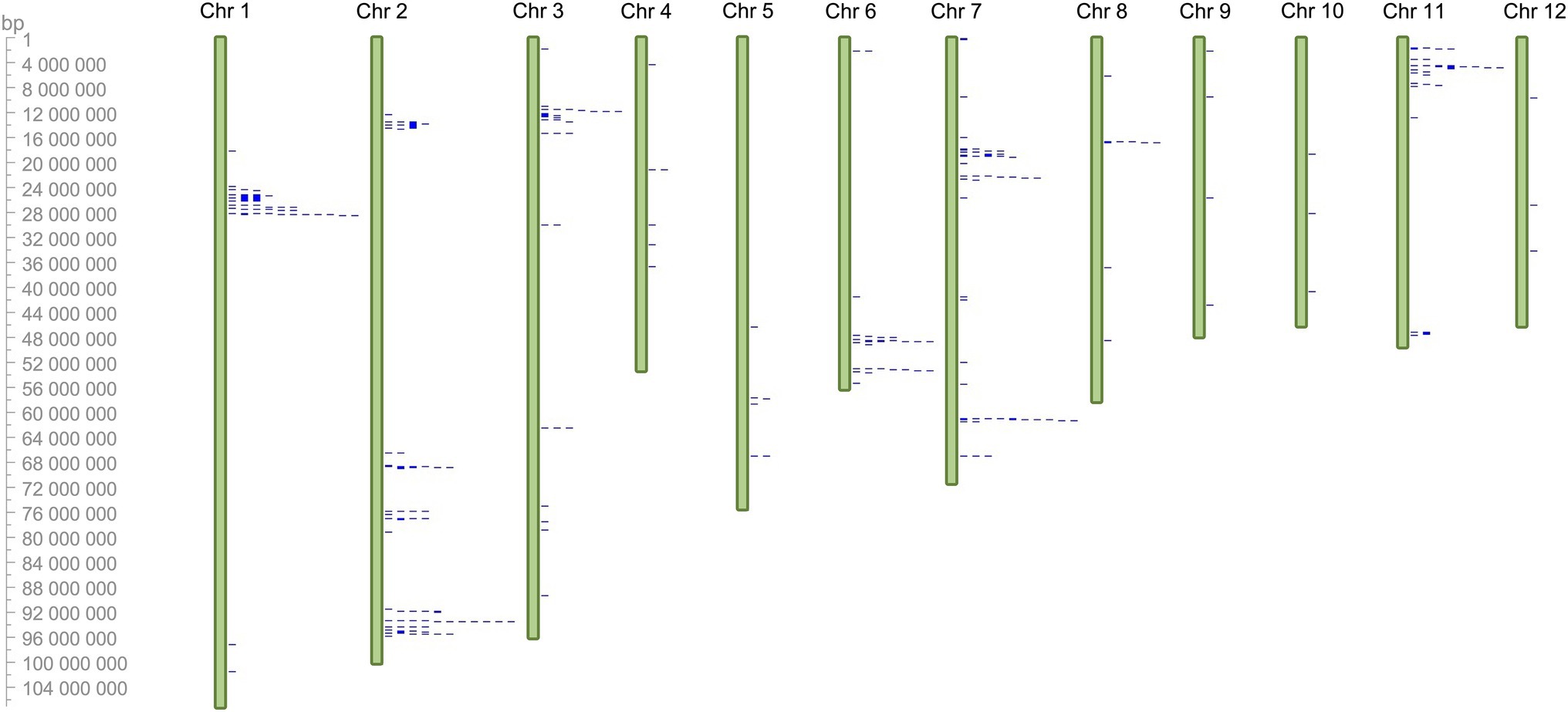
Figure 2. Chromosomal location of 148 putative PaNLR genes identified within the Persea americana West-Indian pure accession genome (represented by blue marks). The genes were mapped to 12 chromosomes (green bars) using CViT. Chromosome 0 was excluded from the analysis as it is not representative of a true chromosome, thus 13 PaNLR genes could not be mapped to chromosomes 1–12 and are not shown in the figure.
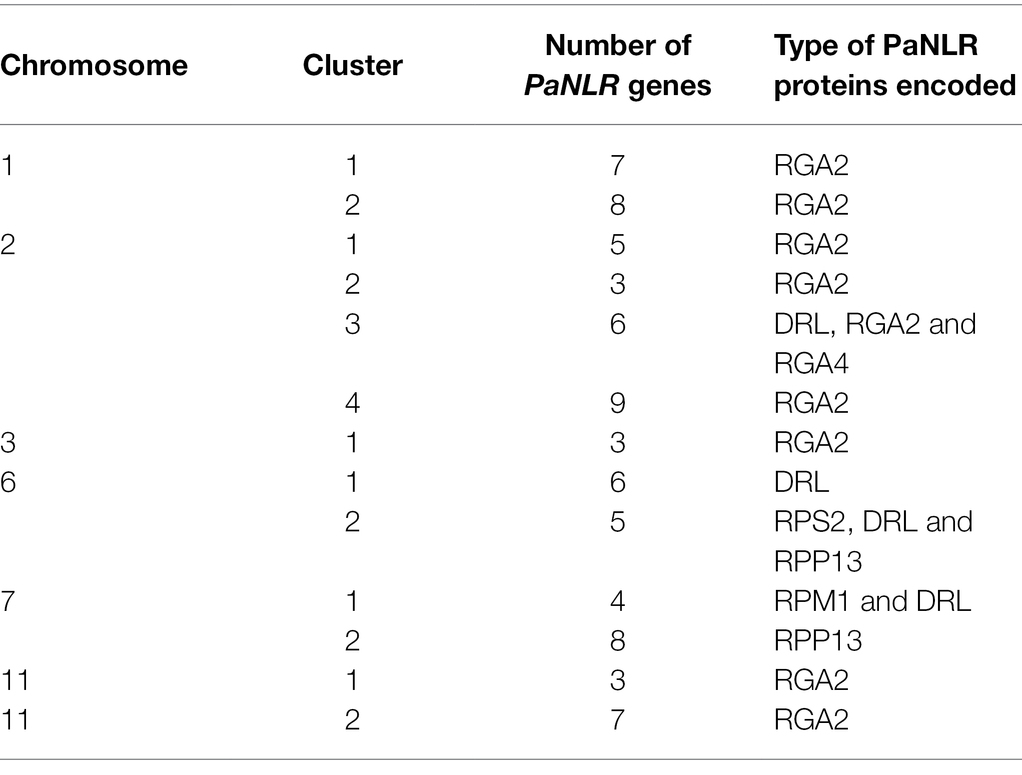
Table 1. Types of resistance genes found within PaNLR gene clusters on different chromosomes within the genome of Persea americana (West-Indian pure accession).
High PaNLR Sequences Similarity Within NLR Gene Clusters
Phylogenetic analysis was performed to infer evolutionary relatedness between the 161 identified putative PaNLR genes using NB-domain protein sequences (Figure 3). Complete NLR protein sequences from C. micranthum f. kanehirae and S. bulbocastanum were included during analysis. The analysis revealed that PaNLR genes from the same NLR gene cluster grouped together within a clade, indicating high sequence similarity within clusters and possible gene duplication events. Most PaNLRs did not form a clade with C. micranthum f. kanehirae NLRs, indicating high diversification of P. americana NLRs after these two species diverged, especially RGA2 type PaNLRs.
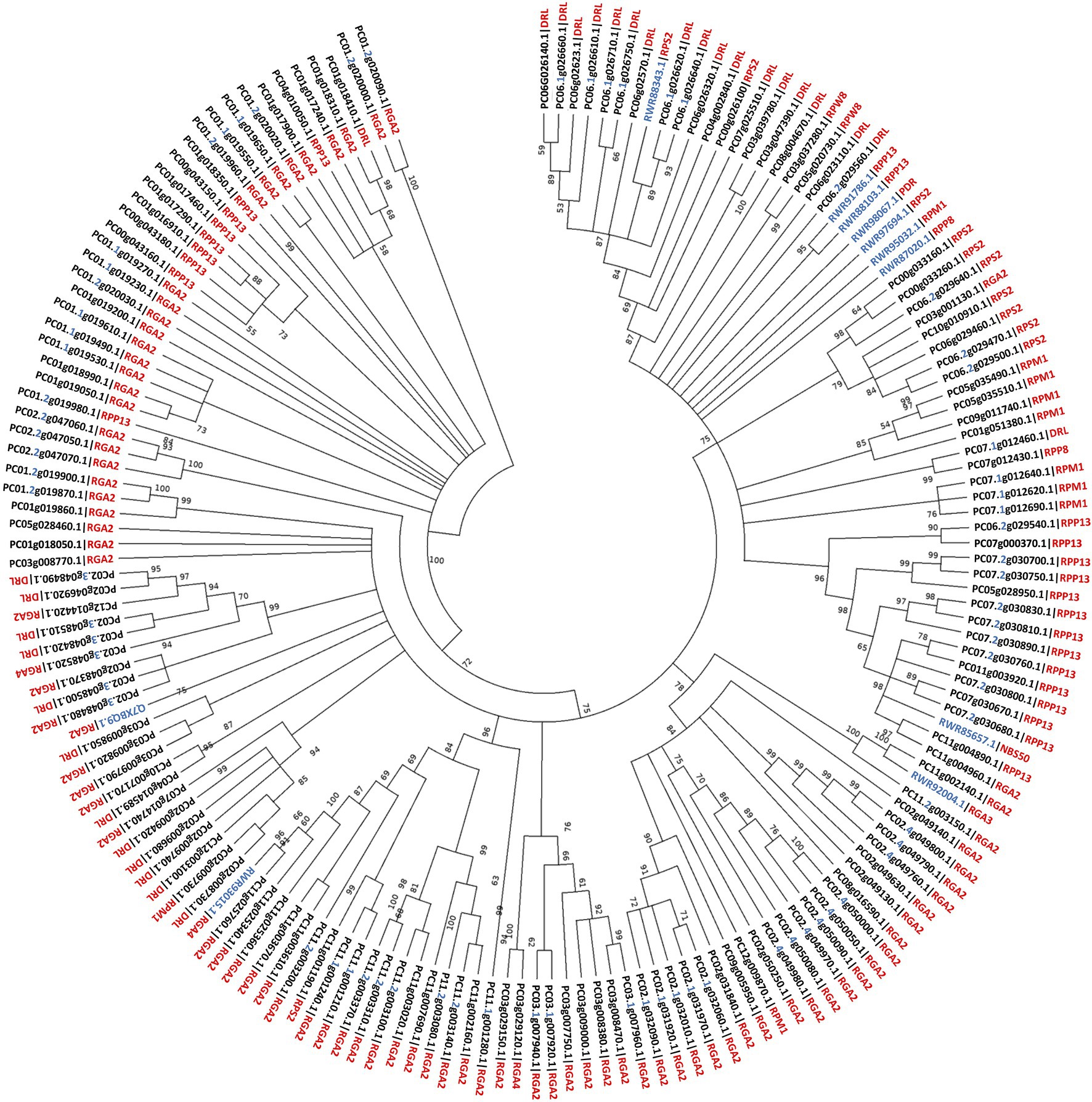
Figure 3. Phylogenetic relationship of 161 Persea americana (West-Indian pure accession) Nucleotide binding domains from putative PaNLR genes. Evolutionary history was inferred using the Maximum likelihood method and JTT matrix-based model following ClustalW alignment. A total of 1,000 bootstrap replicates were performed, with bootstrap values over 50 being shown above branch points. NB-domain protein sequences of Persea americana (PC) with complete NLR sequences from Cinnamomum micranthum f. kanehirae (RWR) and Solanum bulbocastanum (Q) were used during the analysis. Persea americana identification numbers include the gene cluster number, where appropriate (in blue) and protein type (in red). Unidentified PaNLR protein types were termed Disease resistance-like (DRL) proteins. Sequences from other species also include protein type (NBS50, NBS-LRR disease resistance protein NBS50; PDR, disease resistance-like protein isoform X1).
PaNLR Expression Following Phytophthora cinnamomi Inoculation
Expression analysis was performed using dual RNA-sequencing data obtained from partially resistant and susceptible avocado rootstocks inoculated with P. cinnamomi zoospores. In total, 145 of the 161 identified complete PaNLR genes were expressed in the roots of both rootstocks, across all timepoints. A clear difference in PaNLR expression was observed between the two rootstocks in response to P. cinnamomi inoculation. In the partially resistant rootstocks (Dusa®), a total of 84 PaNLR genes showed a significant (p ≤ 0.05) change in expression level during at least one timepoint after P. cinnamomi inoculation, when compared to mock-inoculated samples (Figure 4). However, only 74 PaNLRs showed a significant (p ≤ 0.05) change in expression in the susceptible rootstocks (R0.12) after inoculation, when compared to mock-inoculated samples. The number of PaNLR genes with expression level differences in response to P. cinnamomi inoculation differed most notably between the two rootstocks at 12 and 24 hpi (Table 2). Only six PaNLR genes were differentially expressed in R0.12 at 12 and 24 hpi, compared to 74 PaNLR in Dusa®. PaNLR genes within a cluster were shown not to be co-expressed based on the Clust analysis.
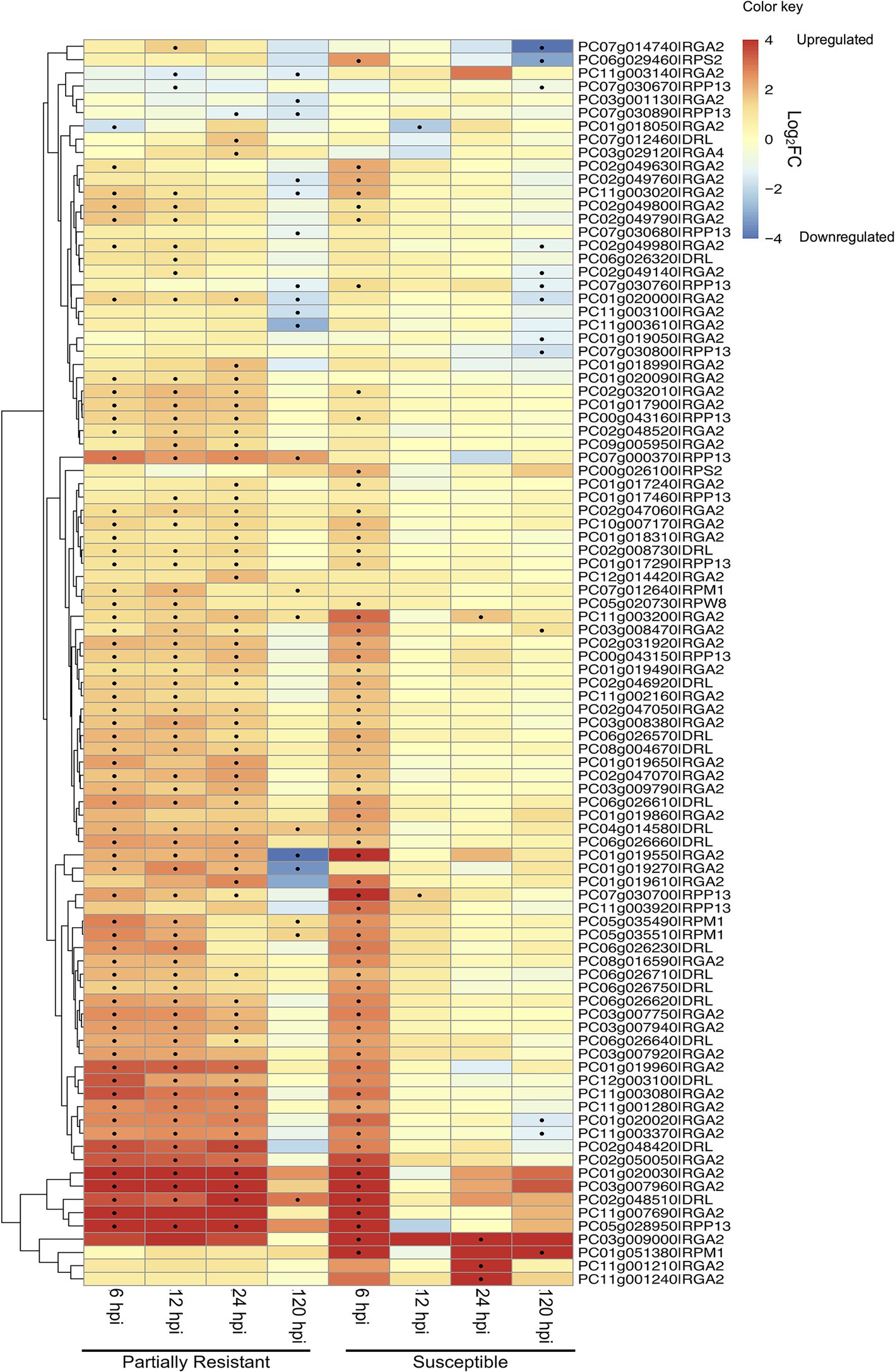
Figure 4. Heatmap and dendrogram showing the expression (as Log2 Fold Change) of 94 PaNLR genes following Phytophthora cinnamomi inoculation of a partially resistant (Dusa®) and susceptible (R0.12) avocado rootstock. Dots indicate a significant change (p ≤ 0.05 and |Log2FC| ≥ 1) in expression level when compared to mock-inoculated samples (hpi, hours post-inoculation).

Table 2. Number of PaNLR genes expressed in two avocado rootstocks in response to Phytophthora cinnamomi inoculation at different timepoints post-inoculation, when compared to mock-inoculated rootstocks (hpi, hours post-inoculation).
PC03g007960|RGA2 was the most upregulated PaNLR gene in Dusa®, with a Log2FC value of 8.02 (p < 0.01) at 12 hpi (Figure 4). This gene was also upregulated in Dusa® at both 6 (Log2FC = 7.2; p < 0.01) and 24 hpi (Log2FC = 7.8; p < 0.01), while only being upregulated in R0.12 at 6 hpi (Log2FC = 7.8; p < 0.01). PC03g009000|RGA2 was the most upregulated PaNLR gene in R0.12 at 6 hpi with the largest Log2FC value of 8.46 (p < 0.01) of all samples. This gene did not show any significant changes in expression in any of the samples collected from Dusa®. Furthermore, PC11g001210|RGA2 and PC11g001240|RGA2 were upregulated in R0.12 at 24 hpi (Log2FC = 7.6; p < 0.01 and Log2FC = 5.1; p < 0.05, respectively), but did not show any significant change in expression in Dusa®, at any time point.
When PaNLR gene expression was compared between the two rootstocks, with susceptible rootstock (R0.12) set as the reference, results indicated that PaNLR gene expression was higher in the partially resistant rootstocks (Dusa®), overall (Table 3). This was evident at the 12 and 24 hpi time points especially, with up to 74 PaNLR genes having higher expression (p ≤ 0.05) in Dusa® at 12 hpi (Figure 5). PC11g001210|RGA2 and PC11g001240|RGA2 were two PaNLR genes that were expressed at significantly higher levels in Dusa® when compared to R0.12, in all samples collected including mock-inoculated roots, even though both PaNLRs were significantly upregulated at 24 hpi in R0.12 when compared to mock-inoculated samples (Figure 4). The Log2FC values for both PC11g001210|RGA2 and PC11g001240|RGA2 were larger than 8.5 (p < 0.01) in all samples except at 24 hpi, where the Log2FC values decreased to 3.8 and 6.2 (p < 0.01), respectively (Figure 5). The PaNLR gene with the highest expression level in R0.12 when compared with Dusa®, was PC02g009680|DRL. However, this PaNLR was only expressed at significantly higher levels in R0.12 at 6 hpi (Log2FC = −7.6; p < 0.01), with no significant difference in expression levels being observed at any other time point.

Table 3. Number of PaNLR genes with a significantly higher expression in either the partially resistant (Dusa®) or susceptible (R0.12) avocado rootstock before and following Phytophthora cinnamomi inoculation (hpi, hours post-inoculation).
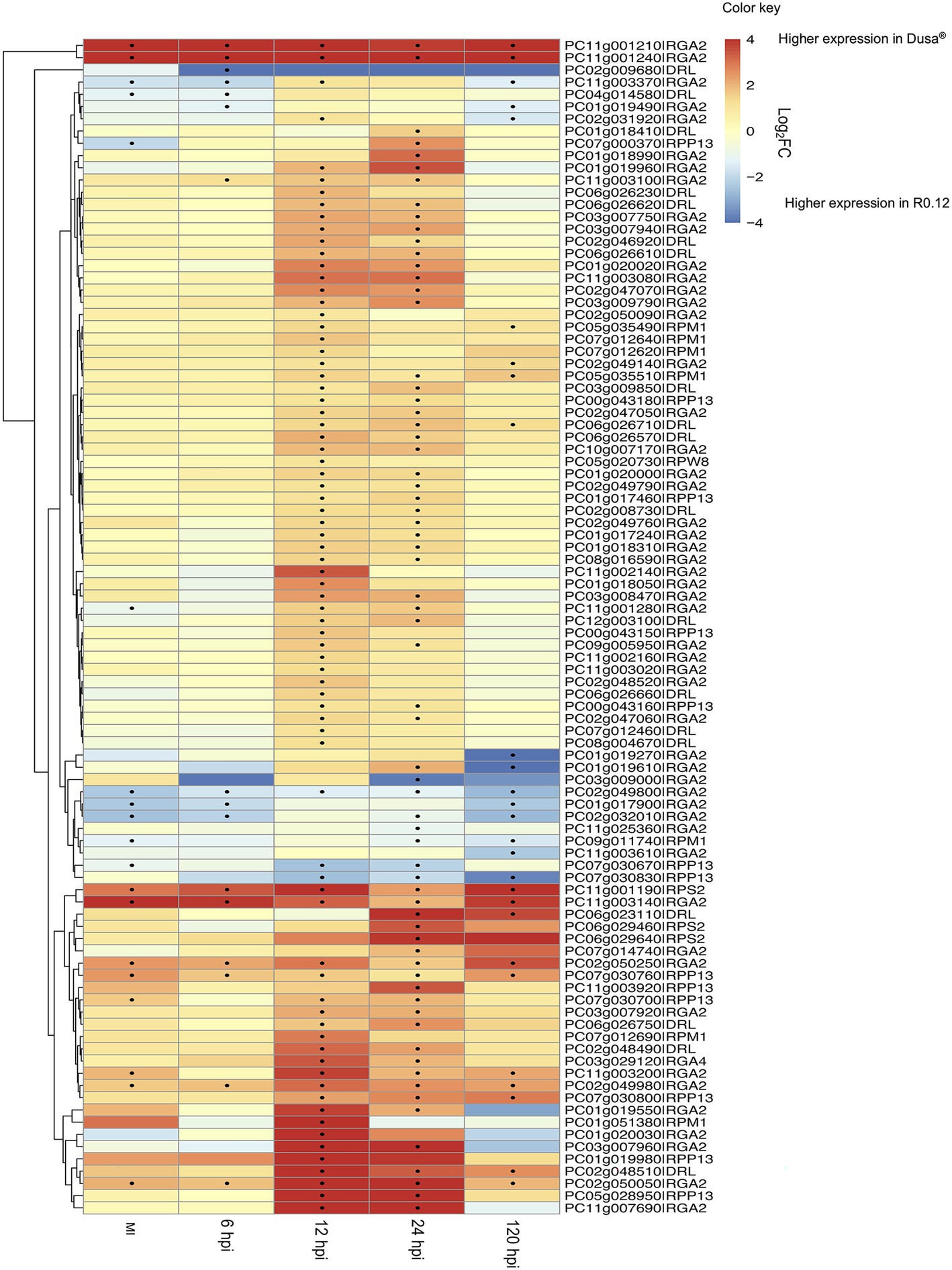
Figure 5. Heatmap and dendrogram showing PaNLR expression levels in a partially resistant avocado rootstock (Dusa®) mock-inoculated (MI) and following Phytophthora cinnamomi inoculation (hpi, hours post-inoculation) using a susceptible rootstock (R0.12) as the reference. A positive Log2FC indicates higher expression in the partially resistant rootstock, while a negative Log2FC indicates higher expression in the susceptible rootstock. Dots indicate a significant difference (p ≤ 0.05 and |Log2FC| ≥ 1) in expression between the two rootstocks.
Discussion
Nucleotide binding-Leucine rich repeat proteins play a crucial role in plant immune responses by recognizing effector molecules produced by invading pathogens. Following effector recognition, NLR proteins activate ETI through complex signaling pathways, which leads to pathogen resistance (Monteiro and Nishimura, 2018). NLR proteins have been studied extensively in many other crops including S. bulbocastanum, Zea mays, Oryza sativa, and Triticum monococcum (Collins et al., 1998; Mago et al., 1999; Bozkurt et al., 2007; Lokossou et al., 2010). The results of these studies ultimately led to the breeding of crops with increased resistance toward various pathogens (Farnham and Baulcombe, 2006; Wang et al., 2019). Thus far, a comprehensive set of P. americana NLR genes have not been identified, and moreover, P. americana NLR gene expression has never been studied during P. cinnamomi infection. Thus, a large knowledge gap remains in understanding ETI activation during P. cinnamomi infection in avocado rootstocks (Van den Berg et al., 2018). The knowledge of avocado NLR functionality is vital to understanding resistance toward P. cinnamomi in avocado rootstocks and can be used by the avocado industry for molecular breeding purposes.
Using the P. americana WI genome, we identified 161 putative complete PaNLR gene sequences. No PaNLR gene sequences could be identified in the Mexican and Hass genome assemblies, since these genomes are highly fragmented (Rendón-Anaya et al., 2019; Talavera et al., 2019). Of the 161 complete PaNLR sequences, 102 were classified as CNL proteins, two as CRNL proteins, 56 as NL proteins, and one as a TNL protein (Figure 1). The 56 gene sequences encoding NL proteins were found to be expressed during P. cinnamomi infection, indicating that these proteins may play a role in this host-pathogen interaction, even though they lack CC and TIR domains. NLR sequences lacking these motifs are also expressed in other plant species, including E. grandis, Malus x domestica, and Vitis vinifera, further suggesting that these NLR proteins are still functional (Arya et al., 2014; Christie et al., 2016; Goyal et al., 2020). A higher CNL:TNL ratio was also observed in E. grandis, M. x domestica, and V. vinifera woody species, making it unsurprising to observe a higher CNL:TNL ratio in P. americana. However, it was not expected that only one TNL sequence would be identified. This could be a result of the genome assembly and annotation programs used not identifying full length gene sequences, thus producing truncated protein sequences as a result. BLASTp analysis was performed on the entire set of P. americana protein sequences and no additional PaNLR sequences could be identified. Furthermore, an independent study found only one TNL gene being expressed in Hass avocado stems during F. kuroshium infection (Pérez-Torres et al., 2021). This validated that no TNL motifs were missed during PaNLR identification using the RGAugury program and WI genome.
Putative protein functional annotation revealed that more than 50% of the identified PaNLR genes encode RGA2-like proteins (Figure 1). This type of NLR protein was first identified in S. bulbocastanum and is encoded for by Rpi-blb1 (Van der Vossen et al., 2003). RGA2 proteins elicit an immune response and confer resistance toward Phytophthora infestans in potato and tomato plants, after recognizing ipiO RxLR proteins (Champouret et al., 2009). Recently, two P. cinnamomi RxLR proteins with high sequence similarity to P. infestans ipiO RxLRs were identified by Joubert et al. (2021). One of these RxLRs, PcinRxLR34a, was significantly upregulated in P. cinnamomi during infection of the susceptible rootstock R0.12, when compared to expression in mycelia. This suggests that this RxLR plays a role during pathogen infection. Future research should focus on identifying whether P. americana RGA2 proteins recognize these P. cinnamomi RxLRs.
RPP13-like protein 4 type proteins were the second largest group of PaNLR proteins identified in P. americana. RPP13-like protein 4 and RPP8 has been shown to confer resistance toward Peronospora parasitica and Hyaloperonospora arabidopsidis, respectively, in Arabidopsis thaliana (Bittner-Eddy et al., 1999; Mohr et al., 2010). Peronospora parasitica, H. arabidopsidis, and P. cinnamomi are oomycetes, suggesting that these pathogens may express Avirulence (Avr) proteins with similar structure and function (Cooke et al., 2000). This indicates that RPP13-like protein 4 and RPP8 in avocado may recognize P. cinnamomi effectors and play a role in rootstock resistance toward P. cinnamomi. The same assumption can be made regarding RPS2, which confers partial resistance toward Phytophthora sojae, a close relative to P. cinnamomi, in Glycine max (Mideros et al., 2007).
RPM1-like NLR proteins were also identified in avocado. Homologs of RPM1-like NLR genes in A. thaliana are responsible for recognizing P. syringae effectors during infection (Boyes et al., 1998). Pseudomonas syringae has been isolated from avocados; however, no symptoms of infection were observed (Scortichini et al., 2003). This might also explain why so few (5.6% of NLRs) RPM1-like genes were identified in avocado. Furthermore, since P. syringae infection does not present a threat to the avocado industry, NLR genes which confer resistance toward this pathogen would likely be of limited use in avocado screening programs. Lastly, two PaNLRs were annotated as RGA4-like proteins; in O. sativa, RGA4 proteins form heterodimers with RGA5 proteins, which recognize Magnaporthe oryzae infection (Césari et al., 2013). RGA5 proteins act as a receptor for M. oryzae Avr proteins and as a repressor of RGA4. Once RGA5 recognizes Avr proteins, RGA4 is released and activates cell death responses. Thus, in the absence of RGA5 proteins, RGA4 activates cell death in an Avr-independent manner (Césari et al., 2014). Since no P. americana proteins were identified as RGA5 proteins, it remains unclear whether the RGA4 proteins would respond to P. cinnamomi Avr proteins in avocado.
Gene cluster analysis was performed to identify possible duplication events of P. americana NLRs (Meyers et al., 2003). If NLR genes within a cluster were shown to be functionally important for rootstock resistance, NLR gene clusters can be targeted during molecular screening strategies. In total, 13 PaNLR gene clusters were identified in the P. americana genome (Table 1). Of these, four clusters were identified on chromosome 2 with one containing nine PaNLR gene sequences. No clusters were observed on chromosomes 4, 5, 8, 9, 10, and 12 (Figure 1). Eight clusters only contained RGA2 protein sequences, indicating that these genes may have originated from gene duplication events as described by Meyers et al. (1998) and López et al. (2003). Retained NLRs following duplication indicate functional relevance, suggesting that these RGA2 NLRs may play an important role in avocado defense responses. In Phaseolus vulgaris, RGA2 gene clusters were identified as Quantitative trait loci (QTL), which confer resistance toward Colletotrichum lagenarium (López et al., 2003). Further investigation focusing on functional significance will help identify whether the PaNLR gene clusters in P. americana can be used as QTL molecular markers during rootstock breeding programs. Ultimately, these clusters serve as a reservoir for NLR diversity since duplicated genes are free to mutate, which may lead to novel NLRs being able to recognize novel effector proteins from pathogens (Innes et al., 2008).
Phylogenetic analysis revealed high similarity between PaNLRs within gene clusters, further indicating that PaNLR gene clusters may have originated from gene duplication events (Shao et al., 2014). During phylogenetic tree construction, 161 PaNLR Nucleotide binding domain protein sequences were used together with protein sequences from C. micranthum f. kanehirae and S. bulbocastanum (Song et al., 2003; Chaw et al., 2019). Sequences from C. micranthum f. kanehirae were used since this species is the closest relative to P. americana (both species form part of the Lauraceae family) in which NLRs have been identified (Wu et al., 2017). A RGA2 sequence from S. bulbocastanum was also included, since no RGA2 proteins were identified in C. micranthum f. kanehirae (Chaw et al., 2019). Phylogenetic analysis revealed that NB domain sequences within a PaNLR gene cluster grouped together, indicating high sequence similarity within these clusters (Figure 2). Moreover, few PaNLRs formed a clade with NLR sequences from C. micranthum f. kanehirae, indicating large NLR diversification within P. americana species. These observations might be the result of different pathogens shaping the PaNLR arsenal during the coevolutionary arms race between hosts and pathogens (Anderson et al., 2010).
Once putative PaNLR genes were identified in the WI genome, their expression was analyzed using dual transcriptomic data from partially resistant (Dusa®) and susceptible (R0.12) rootstocks inoculated with P. cinnamomi. Of the 161 PaNLRs identified in this study, 16 PaNLRs were not expressed in either rootstock at any timepoint. Many NLRs have tissue-specific expression levels in other plants, making these results unsurprising (Munch et al., 2018). Since this study investigated PaNLR expression in root tissues, it is expected that these 16 PaNLRs might play a role in recognizing pathogens which infect other avocado tissues. Interestingly, PaNLR genes within a gene cluster did not show similar expression patterns after P. cinnamomi inoculation. This was also observed in E. grandis when infected with C. austroafricana and L. invasa. The authors attributed this to expressed NLR genes being functionally relevant, and not the result of being located within active transcription zones by coincidence (Christie et al., 2016). Thus, we can hypothesize that PaNLRs in gene clusters being expressed following P. cinnamomi inoculation, do indeed have functional significance in activating defense responses against the invading pathogen.
During the first 6 h of infection, more than 60 PaNLR genes showed a significant increase in expression in either rootstock, with a similar pattern of expression activation for 54 of the same PaNLR genes in both rootstocks (Figure 4). This indicates that both rootstocks have similar responses with regards to PaNLR expression during the first 6 h of P. cinnamomi infection. PaNLR genes with the largest increase in expression at 6 hpi, were mainly RGA2 type proteins (PC01g020030, PC03g007960, and PC11g007690). RGA2 proteins activate the HR, and higher RGA2 transcript levels were associated with increased P. infestans resistance in S. bulbocastanum (Bradeen et al., 2009). This upregulation of RGA2 in both avocado rootstocks would likely result in a strong HR, which may limit P. cinnamomi growth.
PaNLR gene expression levels in Dusa® was higher when compared to R0.12, at both 12 and 24 hpi. Very few PaNLR genes showed differential expression patterns at 12 and 24 hpi in R0.12, which might indicate a decrease in ETI activation compared to Dusa®. Thus, the expression analysis revealed that Dusa® rootstocks overall have a stronger, more prolonged response to P. cinnamomi inoculation when compared to R0.12 rootstocks. NLR expression in susceptible varieties of S. viminalis, C. arietinum L. and Brassica oleracea do not show such stark differences in the expression when compared to resistant varieties, when infected with Melampsora larici-epitea, Ascochyta rabiei, and Fusarium oxysporum f. sp. conglutinans, respectively (Martin et al., 2016; Andam et al., 2020; Liu et al., 2020). It was thus expected that a greater portion of PaNLRs would show increased expression in R0.12 at these timepoints. These results might be due to either the pathogen interfering with PaNLR expression, or the pathogen suppressing host responses in R0.12. For example, W boxes, which are cis-regulatory elements recognized by WRKY transcriptions factors, are often overrepresented in plant defense-related gene promoters including NLR promoter sequences (Mohr et al., 2010). In A. thaliana, WRKY expression was downregulated by Avr3a-type effectors from Phytophthora parasitica (Li et al., 2019). This would subsequently lead to decreased NLR expression. It would be interesting to see whether P. cinnamomi uses similar tactics to influence ETI in P. americana. Thus, investigating which cis-regulatory elements are shared between PaNLR genes would be of interest in future research. Moreover, P. cinnamomi RxLRs were shown to have increased expression levels at 12 and 24 hpi in R0.12 (Joubert et al., 2021). Since some RxLRs suppress programed cell death, P. cinnamomi RxLRs could influence PaNLR expression and contribute to the results observed for R0.12 (Dalio et al., 2018). This data will help understand which PaNLR proteins might be important for recognizing P. cinnamomi effectors during infection and limiting P. cinnamomi growth. However, it must be noted that further studies, including protein–protein interaction studies, are needed to concretely state which individual PaNLR proteins recognize P. cinnamomi effectors.
A previous study, also done on R0.12 and Dusa® rootstocks, showed that R0.12 had significantly higher P. cinnamomi pathogen loads when compared to Dusa®, at all tested time-points (Engelbrecht et al., 2013). The increased PaNLR expression in Dusa®, especially RGA2 PaNLRs, at 12 and 24 hpi is likely to increase the amplitude of ETI activation and the HR, assuming successful P. cinnamomi Avr detection. Studies have shown that overexpression of NLR genes leads to higher levels of resistance and subsequent decreased disease symptoms. In Nicotiana benthamiana plants, overexpression of the Vitis amurensis NLR gene, VaRGA1, resulted in increased resistance toward P. parasitica (Li et al., 2017). Two RGA2 NLRs (PC11g001210 and PC11g001240) showed much higher expression in Dusa® when compared to R0.12, in all samples (Figure 5). As described earlier, the RxLR in P. cinnamomi with high similarity to a RGA2 protein counterpart, ipiO1, showed increased expression in R0.12 at 12 hpi, when compared to mycelia control samples (Joubert et al., 2021). Since R0.12 RGA2 proteins are not upregulated at this timepoint, it may suggest that fewer of these RxLR effectors are recognized, resulting in a compromised HR. However, high expression of RGA2 NLRs in Dusa® at all timepoints could result in increased ETI and might lead to decreased pathogen growth rates and/or decreased zoospore germination. However, in R0.12, pathogen load could be higher due to decreased ETI. These differences might be why Dusa® is able to survive P. cinnamomi attack for longer periods of time and show less disease symptoms.
This study is the first to identify and classify putative PaNLR genes using the P. americana WI genome. Phylogenetic analysis revealed that many PaNLRs found within NLR gene clusters may have originated from gene duplication events. Up to 94 PaNLR genes showed expression differences in response to P. cinnamomi attack, indicating a possible role in P. cinnamomi recognition and ETI activation. Furthermore, PaNLRs showed sustained, increased expression in a partially resistant rootstock (Dusa®) after inoculation, which could explain how this rootstock is able to suppress P. cinnamomi growth. This research paves the way toward understanding P. americana-P. cinnamomi interactions on a molecular level. Future studies should focus on investigating protein–protein interactions between PaNLRs and P. cinnamomi Avr proteins, and how P. cinnamomi is able to suppress PaNLR expression in R0.12 rootstocks. Furthermore, future studies should also include functionally characterizing the identified PaNLRs and investigating their role in defense responses against other P. americana pathogens. However, the lack of an efficient transformation system for P. americana greatly limits functional studies, and the results of this study highlights the need for the development of an improved system.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA675400.
Author Contributions
AF analyzed all data and drafted the manuscript. RB performed the early analysis of RNA sequencing data and data curation. AB completed the assembly of the WI genome. JE designed and performed the experiments. VS and NB provided supervision of the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Project funding was generously provided by the Hans Merensky Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors appreciatively acknowledge the Avocado Genome Consortium for providing the Persea americana WI rootstock genome. We would also like to thank Westfalia Fruit for plant material and the Hans Merensky Foundation for project funding.
Footnotes
1. ^https://bitbucket.org/yaanlpc/rgaugury/
3. ^http://www.ebi.ac.uk/interpro/about/-interproscan/
References
Abu-Jamous, B., and Kelly, S. (2018). Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol. 19:172. doi: 10.1186/s13059-018-1536-8
Andam, A., Azizi, A., Majdi, M., and Abdolahzadeh, J. (2020). Comparative expression profile of some putative resistance genes of chickpea genotypes in response to ascomycete fungus, Ascochyta rabiei (Pass.) Labr. Braz. J. Bot. 43, 123–130. doi: 10.1007/s40415-020-00576-w
Anderson, J. P., Gleason, C. A., Foley, R. C., Thrall, P. H., Burdon, J. B., and Singh, K. B. (2010). Plants versus pathogens: an evolutionary arms race. Funct. Plant Biol. 37, 499–512. doi: 10.1071/FP09304
Arya, P., Kumar, G., Acharya, V., and Singh, A. K. (2014). Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS One 9:e107987. doi: 10.1371/journal.pone.0107987
Belisle, R. J., Hao, W., McKee, B., Arpaia, M. L., Manosalva, P., and Adaskaveg, J. E. (2019). New oomycota fungicides with activity against Phytophthora cinnamomi and their potential use for managing avocado root rot in California. Plant Dis. 103, 2024–2032. doi: 10.1094/PDIS-09-18-1698-RE
Bezerra-Neto, J. P., Araújo, F. C., Ferreira-Neto, J. R. C., Silva, R. L. O., Borges, A. N. C., Matos, M. K. S., et al. (2020). “NBS-LRR genes—plant health sentinels: structure, roles, evolution and biotechnological applications,” in Applied Plant Biotechnology for Improving Resistance to Biotic Stress. eds. P. Poltronieri and Y. Hong (Amsterdam, Netherlands: Academic Press), 63–120.
Bittner-Eddy, P., Can, C., Gunn, N., Pinel, M., Tör, M., Crute, I., et al. (1999). Genetic and physical mapping of the RPP13 locus, in Arabidopsis, responsible for specific recognition of several Peronospora parasitica (downy mildew) isolates. Mol. Plant-Microbe Interact. 12, 792–802. doi: 10.1094/MPMI.1999.12.9.792
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyes, D. C., Nam, J., and Dangl, J. L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. U. S. A. 95, 15849–15854. doi: 10.1073/pnas.95.26.15849
Bozkurt, O., Hakki, E. E., and Akkaya, M. S. (2007). Isolation and sequence analysis of wheat NBS-LRR type disease resistance gene analogs using degenerate PCR primers. Biochem. Genet. 45, 469–486. doi: 10.1007/s10528-007-9089-7
Bradeen, J. M., Iorizzo, M., Mollov, D. S., Raasch, J., Kramer, L. C., Millett, B. P., et al. (2009). Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol. Plant-Microbe Interact. 22, 437–446. doi: 10.1094/mpmi-22-4-0437
Bulagi, M., Hlongwane, J., and Belete, A. (2016). Analyzing the linkage between agricultural exports and agriculture’s share of gross domestic products in South Africa. J. Agric. Stud. 4:142. doi: 10.5296/jas.v4i1.8918
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cannon, E. K. S., and Cannon, S. B. (2011). Chromosome visualization tool: a whole genome viewer. Int. J. Plant Genomics 2011:373875. doi: 10.1155/2011/373875
Césari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. doi: 10.15252/embj.201487923
Césari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Champouret, N., Bouwmeester, K., Rietman, H., van der Lee, T., Maliepaard, C., Heupink, A., et al. (2009). Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant-Microbe Interact. 22, 1535–1545. doi: 10.1094/MPMI-22-12-1535
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 11, 113–116. doi: 10.1007/BF02670468
Chaw, S. M., Liu, Y. C., Wu, Y. W., Wang, H. Y., Lin, C. Y. I., Wu, C.-S., et al. (2019). Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 5, 63–73. doi: 10.1038/s41477-018-0337-0
Christie, N., Tobias, P. A., Naidoo, S., and Külheim, C. (2016). The Eucalyptus grandis NBS-LRR gene family: physical clustering and expression hotspots. Front. Plant Sci. 6:1238. doi: 10.3389/fpls.2015.01238
Coffey, D. M. (1987). Phytophthora root rot of avocado: an integrated approach to control in California. Plant Dis. 71, 1046–1053.
Collins, N. C., Webb, C. A., Seah, S., Ellis, J. G., Hulbert, S. H., and Pryor, A. (1998). The isolation and mapping of disease resistance gene analogs in maize. Mol. Plant-Microbe Interact. 11, 968–978. doi: 10.1094/MPMI.1998.11.10.968
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., and Brasier, C. M. (2000). A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. doi: 10.1006/fgbi.2000.1202
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dalio, R. J. D., Maximo, H. J., Oliveira, T. S., Dias, R. O., Breton, M. C., Felizatti, H., et al. (2018). Phytophthora parasitica effector PpRxLR2 suppresses Nicotiana benthamiana immunity. Mol. Plant-Microbe Interact. 31, 481–493. doi: 10.1094/MPMI-07-17-0158-FI
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Davis, K. R., and Hahlbrock, K. (1987). Induction of defense responses in cultured parsley cells by plant cell wall fragments. Plant Physiol. 84, 1286–1290. doi: 10.1104/pp.84.4.1286
Dobrowolski, M. P., Shearer, B. L., Colquhoun, I. J., O’Brien, P. A., and Hardy, G. E. S. (2008). Selection for decreased sensitivity to phosphite in Phytophthora cinnamomi with prolonged use of fungicide. Plant Pathol. 57, 928–936. doi: 10.1111/j.1365-3059.2008.01883.x
Engelbrecht, J., Duong, T. A., and Van den Berg, N. (2013). Development of a nested quantitative real-time PCR for detecting Phytophthora cinnamomi in Persea americana rootstocks. Plant Dis. 97, 1012–1017. doi: 10.1094/PDIS-11-12-1007-RE
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Farnham, G., and Baulcombe, D. C. (2006). Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. U. S. A. 103, 18828–18833. doi: 10.1073/pnas.0605777103
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Giblin, F., Pegg, K., Willingham, S., Anderson, J., Coates, L., Cooke, T., et al. (2005). “Phytophthora revisited,” in Proceedings of the New Zealand and Australia Avocado Grower’s Conference; September 20–22, 2005; (Tauranga: New Zealand Avocado Growers Association).
Goyal, N., Bhatia, G., Sharma, S., Garewal, N., Upadhyay, A., Upadhyay, S. K., et al. (2020). Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 112, 312–322. doi: 10.1016/j.ygeno.2019.02.011
Hardham, A. R., and Blackman, L. M. (2018). Phytophthora cinnamomi. Mol. Plant Pathol. 19, 260–285. doi: 10.1111/mpp.12568
Innes, R. W., Ameline-Torregrosa, C., Ashfield, T., Cannon, E., Cannon, S. B., Chacko, B., et al. (2008). Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148, 1740–1759. doi: 10.1104/pp.108.127902
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joubert, M., Backer, R., Engelbrecht, J., and Van den Berg, N. (2021). Expression of several Phytophthora cinnamomi putative RxLRs provides evidence for virulence roles in avocado. PLoS One 16:e0254645. doi: 10.1371/journal.pone.0254645
Käll, L., Krogh, A., and Sonnhammer, E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. doi: 10.1016/j.jmb.2004.03.016
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kohler, A., Rinaldi, C., Duplessis, S., Baucher, M., Geelen, D., Duchaussoy, F., et al. (2008). Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 66, 619–636. doi: 10.1007/s11103-008-9293-9
Kolde, R. (2012). Pheatmap: Pretty Heatmaps. R Package Version, 2 Edn, Vol. 1. Available at: https://cran.r-project.org/web/packages/pheatmap/ (Accessed August 2020).
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, P., Quan, X., Jia, G., Xiao, J., Cloutier, S., and You, F. M. (2016). RGAugury: a pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics 17:852. doi: 10.1186/s12864-016-3197-x
Li, T., Wang, Q., Feng, R., Li, L., Ding, L., Fan, G., et al. (2019). Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. New Phytol. doi: 10.1111/nph.16139 [Epub ahead of print].
Li, X., Zhang, Y., Yin, L., and Lu, J. (2017). Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 254, 957–969. doi: 10.1007/s00709-016-1005-8
Liang, W., van Wersch, S., Tong, M., and Li, X. (2019). TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol. 221, 2054–2066. doi: 10.1111/nph.15534
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, X., Zhao, C., Yang, L., Zhuang, M., Zhang, Y., Wang, Y., et al. (2020). A time-resolved dual transcriptome analysis reveals the molecular regulating network underlying the compatible/incompatible interactions between cabbage (Brassica oleracea) and Fusarium oxysporum f. sp. conglutinans. Plant Soil 448, 455–478. doi: 10.1007/s11104-020-04437-z
Lokossou, A., Rietman, H., Wang, M., Krenek, P., Schoot, H., Henken, B., et al. (2010). Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Mol. Plant-Microbe Interact. 23, 1206–1216. doi: 10.1094/MPMI-23-9-1206
López, C. E., Acosta, I. F., Jara, C., Pedraza, F., Gaitán-Solís, E., Gallego, G., et al. (2003). Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology 93, 88–95. doi: 10.1094/phyto.2003.93.1.88
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. doi: 10.1126/science.252.5009.1162
Mago, R., Nair, S., and Mohan, M. (1999). Resistance gene analogues from rice: cloning, sequencing and mapping. Theor. Appl. Genet. 99, 50–57. doi: 10.1007/s001220051207
Martin, T., Rönnberg-Wästljung, A.-C., Stenlid, J., and Samils, B. (2016). Identification of a differentially expressed TIR-NBS-LRR gene in a major QTL associated to leaf rust resistance in Salix. PLoS One 11:e0168776. doi: 10.1371/journal.pone.0168776
Matzinger, P. (2007). Friendly and dangerous signals: is the tissue in control? Nat. Immunol. 8, 11–13. doi: 10.1038/ni0107-11
McDowell, J. M., and Woffenden, B. J. (2003). Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 21, 178–183. doi: 10.1016/s0167-7799(03)00053-2
Meyers, B. C., Chin, D. B., Shen, K. A., Sivaramakrishnan, S., Lavelle, D. O., Zhang, Z., et al. (1998). The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10, 1817–1832. doi: 10.1105/tpc.10.11.1817
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Meyers, B. C., Morgante, M., and Michelmore, R. W. (2002). TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 32, 77–92. doi: 10.1046/j.1365-313x.2002.01404.x
Mideros, S., Nita, M., and Dorrance, A. E. (2007). Characterization of components of partial resistance, Rps2, and root resistance to Phytophthora sojae in soybean. Phytopathology 97, 655–662. doi: 10.1094/PHYTO-97-5-0655
Mohr, T. J., Mammarella, N. D., Hoff, T., Woffenden, B. J., Jelesko, J. G., and McDowell, J. M. (2010). The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant-Microbe Interact. 23, 1303–1315. doi: 10.1094/MPMI-01-10-0022
Monteiro, F., and Nishimura, M. T. (2018). Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 56, 243–267. doi: 10.1146/annurev-phyto-080417-045817
Munch, D., Gupta, V., Bachmann, A., Busch, W., Kelly, S., Mun, T., et al. (2018). The Brassicaceae family displays divergent, shoot-skewed NLR resistance gene expression. Plant Physiol. 176, 1598–1609. doi: 10.1104/pp.17.01606
Naveed, Z. A., Wei, X., Chen, J., Mubeen, H., and Ali, G. S. (2020). The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 11:593905. doi: 10.3389/fpls.2020.593905
Neale, D. B., Martínez-García, P. J., De La Torre, A. R., Montanari, S., and Wei, X. X. (2017). Novel insights into tree biology and genome evolution as revealed through genomics. Annu. Rev. Plant Biol. 68, 457–483. doi: 10.1146/annurev-arplant-042916-041049
Pérez-Torres, C. A., Ibarra-Laclette, E., Hernández-Domínguez, E. E., Rodríguez-Haas, B., Pérez-Lira, A. J., Villafán, E., et al. (2021). Molecular evidence of the avocado defense response to Fusarium kuroshium infection: a deep transcriptome analysis using RNA-Seq. PeerJ 9:e11215. doi: 10.7717/peerj.11215
Rendón-Anaya, M., Ibarra-Laclette, E., Méndez-Bravo, A., Lan, T., Zheng, C., Carretero-Paulet, L., et al. (2019). The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. U. S. A. 116, 17081–17089. doi: 10.1073/pnas.1822129116
RStudio Team (2020). RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. Available at: http://www.rstudio.com/
Scortichini, M., Marchesi, U., Dettori, M. T., and Rossi, M. P. (2003). Genetic diversity, presence of the syrB gene, host preference and virulence of Pseudomonas syringae pv. syringae strains from woody and herbaceous host plants. Plant Pathol. 52, 277–286. doi: 10.1046/j.1365-3059.2003.00860.x
Shao, Z. Q., Zhang, Y. M., Hang, Y. Y., Xue, J. Y., Zhou, G. C., Wu, P., et al. (2014). Long-term evolution of nucleotide-binding site-Leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol. 166, 217–234. doi: 10.1104/pp.114.243626
Song, J., Bradeen, J. M., Naess, S. K., Raasch, J. A., Wielgus, S. M., Haberlach, G. T., et al. (2003). Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. U. S. A. 100, 9128–9133. doi: 10.1073/pnas.1533501100
Talavera, A., Soorni, A., Bombarely, A., Matas, A. J., and Hormaza, J. I. (2019). Genome-wide SNP discovery and genomic characterization in avocado (Persea americana Mill.). Sci. Rep. 9:20137. doi: 10.1038/s41598-019-56526-4
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tobias, P. A., and Guest, D. I. (2014). Tree immunity: growing old without antibodies. Trends Plant Sci. 19, 367–370. doi: 10.1016/j.tplants.2014.01.011
Umadevi, P., and Anandaraj, M. (2017). Genotype specific host resistance for Phytophthora in black pepper (Piper nigrum L.). Physiol. Mol. Plant Pathol. 100, 237–241. doi: 10.1016/j.pmpp.2017.10.011
Van den Berg, N., Mahomed, W., Olivier, N. A., Swart, V., and Crampton, B. G. (2018). Transcriptome analysis of an incompatible Persea americana-Phytophthora cinnamomi interaction reveals the involvement of SA- and JA-pathways in a successful defense response. PLoS One 13:e0205705. doi: 10.1371/journal.pone.0205705
Van Der Vossen, E., Sikkema, A., Hekkert, B. T. L., Gros, J., Stevens, P., Muskens, M., et al. (2003). An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. doi: 10.1046/j.1365-313X.2003.01934.x
Van Wersch, S., and Li, X. (2019). Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 24, 688–699. doi: 10.1016/j.tplants.2019.05.005
Vargas-Canales, J. M., Carbajal-Flores, G., Bustamante-Lara, T. I., Camacho-Vera, J. H., Fresnedo-Ramírez, J., Palacios-Rangel, M. I., et al. (2020). Impact of the market on the specialization and competitiveness of avocado production in Mexico. Int. J. Fruit Sci. 20, S1942–S1958. doi: 10.1080/15538362.2020.1837711
Wang, L., Zhao, L., Zhang, X., Zhang, Q., Jia, Y., Wang, G., et al. (2019). Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. U. S. A. 116, 18479–18487. doi: 10.1073/pnas.1910229116
Wu, C. C., Chu, F. H., Ho, C. K., Sung, C. H., and Chang, S. H. (2017). Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung 71, 189–197. doi: 10.1515/hf-2016-0133
Xu, Y., Liu, F., Zhu, S., and Li, X. (2018). The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and Arabidopsis. Front. Plant Sci. 9:1033. doi: 10.3389/fpls.2018.01033
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Keywords: NLR, avocado (Persea americana Mill.), Phytophthora, NB-LRR, resistance gene, NLR expression, Phytophthora root rot
Citation: Fick A, Swart V, Backer R, Bombarely A, Engelbrecht J and van den Berg N (2022) Partially Resistant Avocado Rootstock Dusa® Shows Prolonged Upregulation of Nucleotide Binding-Leucine Rich Repeat Genes in Response to Phytophthora cinnamomi Infection. Front. Plant Sci. 13:793644. doi: 10.3389/fpls.2022.793644
Edited by:
Maria Raffaella Ercolano, University of Naples Federico II, ItalyReviewed by:
Vittorio Nicolis, Stellenbosch University, South AfricaEdgar Huitema, University of Dundee, United Kingdom
Copyright © 2022 Fick, Swart, Backer, Bombarely, Engelbrecht and van den Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alicia Fick, YWxpY2lhLmZpY2tAdXAuYWMuemE=
 Alicia Fick
Alicia Fick Velushka Swart
Velushka Swart Robert Backer
Robert Backer Aureliano Bombarely
Aureliano Bombarely Juanita Engelbrecht
Juanita Engelbrecht Noëlani van den Berg
Noëlani van den Berg