- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 2State Key Laboratory of Agrobiotechnology and Ministry of Agriculture Key Laboratory of Plant Pathology, College of Plant Protection, China Agricultural University, Beijing, China
- 3Plant Protection Station, Department of Agriculture Sichuan Province, Chengdu, China
- 4Institute of Plant Protection, Sichuan Academy of Agricultural Sciences, Chengdu, China
Magnaporthe oryzae is the causative agent of rice blast, a devastating disease in rice worldwide. Based on the gene-for-gene paradigm, resistance (R) proteins can recognize their cognate avirulence (AVR) effectors to activate effector-triggered immunity. AVR genes have been demonstrated to evolve rapidly, leading to breakdown of the cognate resistance genes. Therefore, understanding the variation of AVR genes is essential to the deployment of resistant cultivars harboring the cognate R genes. In this study, we analyzed the nucleotide sequence polymorphisms of eight known AVR genes, namely, AVR-Pita1, AVR-Pii, AVR-Pia, AVR-Pik, AVR-Pizt, AVR-Pi9, AVR-Pib, and AVR-Pi54 in a total of 383 isolates from 13 prefectures in the Sichuan Basin. We detected the presence of AVR-Pik, AVR-Pi54, AVR-Pizt, AVR-Pi9, and AVR-Pib in the isolates of all the prefectures, but not AVR-Pita1, AVR-Pii, and AVR-Pia in at least seven prefectures, indicating loss of the three AVRs. We also detected insertions of Pot3, Mg-SINE, and indels in AVR-Pib, solo-LTR of Inago2 in AVR-Pizt, and gene duplications in AVR-Pik. Consistently, the isolates that did not harboring AVR-Pia were virulent to IRBLa-A, the monogenic line containing Pia, and the isolates with variants of AVR-Pib and AVR-Pizt were virulent to IRBLb-B and IRBLzt-t, the monogenic lines harboring Pib and Piz-t, respectively, indicating breakdown of resistance by the loss and variations of the avirulence genes. Therefore, the use of blast resistance genes should be alarmed by the loss and nature variations of avirulence genes in the blast fungal population in the Sichuan Basin.
Introduction
Rice blast disease is one of the most destructive fungal diseases in rice worldwide caused by the filamentous fungal pathogen Magnaporthe oryzae (M. oryzae) (syn. Pyricularia grisea B. C. Couch) (Talbot, 2003; Dean et al., 2012). Rice blast disease causes 10 to 35% yield loss each year, even no yield in some regions with severe epidemics (Dean et al., 2012; Fisher et al., 2012). Developing and application of resistant cultivars are the most effective and environmental-friendly method to control the disease. To date, more than 100 blast resistance (R) genes have been mapped and 38 of them have been cloned (Wang et al., 2017; Zhao et al., 2018; Xie et al., 2019; Liu et al., 2020). According to the gene-for-gene paradigm, cultivars that harbor major blast R genes can prevent the infection of strains carrying the corresponding avirulence (AVR) genes, which are established on the recognitions between R genes and AVR genes (Silue et al., 1992). However, AVR genes are highly variable and the variated AVR protein escape from the recognition mediated by cognate R protein, resulting in breakdown of the resistance function of the R gene (Deng et al., 2017). The blast population can overcome the R gene-mediated resistance in the field due to the heavy selection pressure caused by the monocropping of a single-resistant cultivar for years (Skamnioti and Gurr, 2009; Valent and Khang, 2010). Accordingly, analyzing the variations of AVR genes can evaluate the efficacy of cognate resistance genes (Selisana et al., 2017). Therefore, it is an effective measure to predict R genes efficacy and prevent blast pandemics by monitoring the variation of AVR genes in the field blast fungal population.
To date, more than 40 AVR genes have been identified in the blast fungus and 12 of them (PWL1, PWL2, ACE1, AVR1-CO39, AVR-Pita1, AVR-Pii, AVR-Pia, AVR-Pik, AVR-Pizt, AVR-Pi9, AVR-Pib, and AVR-Pi54) have been cloned (Kang et al., 1995; Sweigard et al., 1995; Farman and Leong, 1998; Orbach et al., 2000; Fudal et al., 2005; Ma et al., 2006; Li et al., 2009; Yoshida et al., 2009; Wu et al., 2015; Zhang et al., 2015; Ray et al., 2016). The avirulent function of AVR genes can be invalidated by different mechanisms, including mutation, genetic recombination, and sexual mating (Noguchi et al., 2006; Tsujimoto Noguchi, 2011). Mutation commonly occurs, such as insertion, point mutation, and deletion. Transposable elements (TEs) can enrich the population diversity through deleting/inactivating genes or horizontal gene transfer (Chuma et al., 2011; Yoshida et al., 2016). Transposon insertion often happens in the promoter or genomic region. For example, Pot3 has been identified as a major reason to generate different variations of AVR-Pita1, AVR-Pib, AVR-Pii, AVR-Pizt, and AVR-Pia (Kang et al., 2001; Yasuda et al., 2006; Li et al., 2009; Yoshida et al., 2009; Singh et al., 2014; Olukayode et al., 2019). Point mutation of AVR-Pita1 and AVR-Pik could generate novel alleles and some of the alleles can escape from the recognition of the cognate R gene (Yoshida et al., 2009; Dai et al., 2010; Kanzaki et al., 2012; Longya et al., 2019; Damchuay et al., 2020). Segmental deletion of coding sequence also results in virulent mutation of AVR-Pita1 and AVR-Pib (Orbach et al., 2000; Zhang et al., 2015). The complete deletion of AVR genes was found in AVR-Pita1, AVR-Pii, and AVR-Pia, generating virulence strains to cultivars harboring their cognate R genes (Yoshida et al., 2009). AVR-Pita1 and AVR-Pii locate adjacently to the telomere of their chromosomes; therefore, loss of chromosome tips is the reason for their frequent spontaneous loss (Yasuda et al., 2006; Khang et al., 2008; Li et al., 2009). Moreover, homologous recombination can cause DNA rearrangement, deletion, translocation, and even horizontal gene transfer between strains. For example, homologous recombination between two repetitive sequences results in the deletion of AVR-Pia (Sone et al., 2013). Parasexual recombination and sexual mating can also result in the genetic exchange of DNA and they are reasons for the pathogenicity variation of rice blast fungus. Therefore, dynamic adaptation and evolution of rice blast fungus could be achieved through these variations of AVR genes in a population, leading to the breakdown of R genes.
The Sichuan Basin is one major base of rice production in southwestern China. There are more than 1.5 million hm2 paddy fields and the production is enough for feeding 100 million people (Wang and Valent, 2009). The climate of the basin is warm and humid, which is greatly conducive to the epidemics of rice blast disease. In the past several decades, three serious rice blast epidemics have been happened with around 10 years interval, since the first epidemic happened in 1984 and 1985, caused by long-term large-scale monocropping of a single-resistant cultivar Shanyou-2 (Lu et al., 2017). Therefore, surveillance of the loss and variations of the AVR genes in the blast fungal population is vital to guarantee rice production in this area.
In this study, we analyzed the distribution and the variation of eight AVR genes, namely, AVR-Pi9, AVR-Pi54, AVR-Pita1, AVR-Pib, AVR-Pia, AVR-Pizt, AVR-Pii, and AVR-Pik, in 383 M. oryzae isolates from 13 prefectures in the Sichuan Basin. We tested the pathogenicity of strains that did not contain an AVR-Pia gene or contain the AVR-Pib and AVR-Pizt variants to the monogenic lines harboring their cognate R genes. Our data indicate that both the loss and variation of an AVR gene in a strain led to breakdown of its cognate R gene. Therefore, the use of Pia, Pib, and Piz-t should be highly alarmed in the Sichuan Basin.
Materials and Methods
Fungal Isolates and Culture
We collected rice blast-infected leaf samples from rice blast hotspots in 13 nurseries of the Sichuan Basin, including DZ, YS, TJ, YA, QW, ST, QS, ZZ, PX, NX, NB, AZ, and JY (Figure 1A). Then, single-spore isolation method was used to establish single-spore isolates following a previous report (Jia, 2009). A total of 383 single-spore isolates were obtained from the infected leaves of the susceptible variety Lijiangxin Tuan Heigu (LTH). Isolates were cultured on PDA medium (200 g/l potato, 20 g/l dextrose, and 13 g/l agar) in a growth chamber at 25°C in a 12-h light/12-h dark photoperiod for 7 days and were stored at −20°C on filter paper.
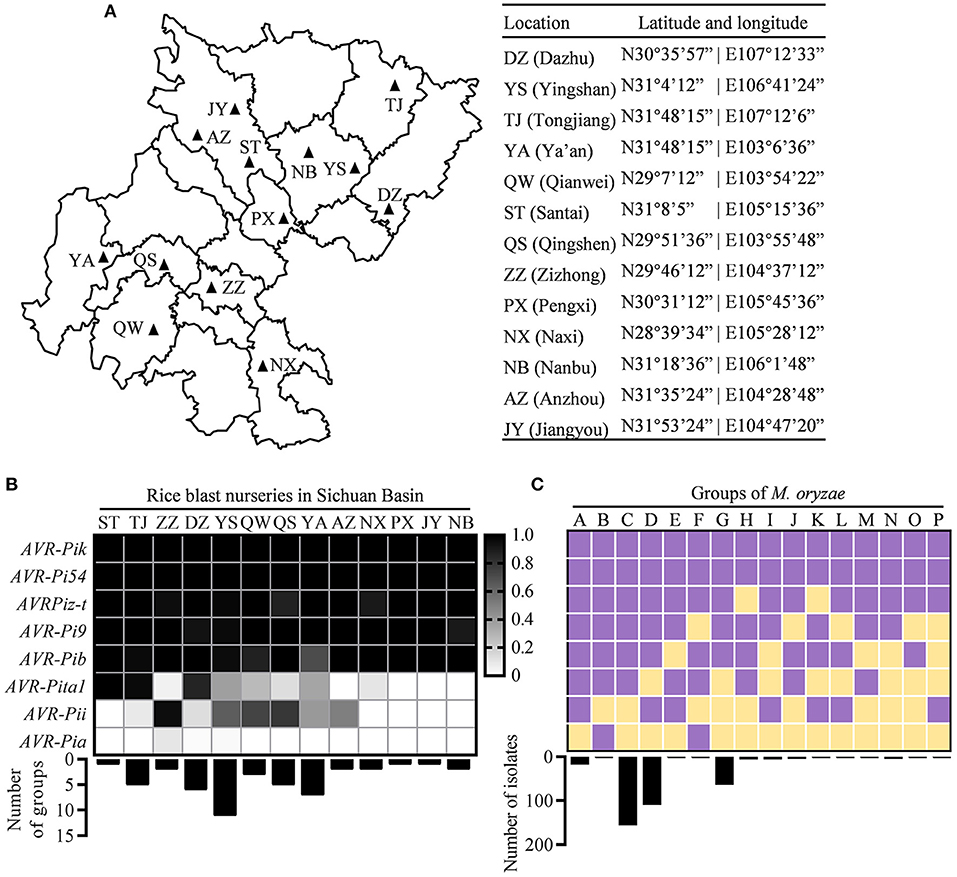
Figure 1. Distribution of avirulence (AVR) genes in the Sichuan Basin. (A) Location of rice blast nurseries in the Sichuan Basin. (B) The distribution of eight AVR genes in the indicated nurseries. The presence of each AVR gene was indicated by shaded box (shadow key) in the 383 field isolates from 13 prefectures (top) where the strains formed different number of groups (bottom). (C) The classification of Magnaporthe oryzae (M. oryzae) strains based on the presence of AVR genes. Upper panel shows the presence/absence of each AVR gene and lower panel shows the number of isolates in each group. Purple, presence; yellow, absence.
Deoxyribonucleic Acid Preparation, PCR Amplification, and Sequencing
The fungal isolates were cultured on PDA medium in plates covered with cellophane membrane at 25°C in a 12-h light/12-h dark photoperiod for 7 days. Then, mycelia were harvested and frozen in liquid nitrogen for grinding to fine powder. Total genomic DNA was extracted via cetyltrimethylammonium bromide (CTAB) method [10 mM Tris-HCl, pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA), 100 mM NaCl, 2% sodium dodecyl sulfate (SDS)] for all the fungal isolates (Saghai-Maroof et al., 1984). PCR was performed with gene-specific primers (Supplementary Table 1) for amplifying eight avirulence genes, namely, AVR-Pib, AVR-Pik, AVR-Pia, AVR-Pi9, AVR-Pi54, AVR-Pita1, AVR-Pizt, and AVR-Pii from isolated DNA samples. Each PCR reaction contains 2.5 μl of 10X EasyTaq buffer, 0.25 μl of 5 U/μl EasyTaq DNA polymerase (TransGen Biotech Corporation Ltd., Beijing, China), 0.5 μl of 10 mM deoxynucleotide triphosphates (dNTPs), 0.5 μl of each 10 μM primer, 1 μl of 20–50 ng/μl fungal genomic DNA, and 19.5 μl of distilled water. Amplifications were performed in the Bio-Rad Thermal Cycler (C1000, Bio-Rad Laboratories, Life Science Research, California, USA) with the following PCR program: 95°C for 3 min, 30 cycles at 95°C for 30 s, 52 to 58°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 8 min. PCR products were examined by electrophoresis in 1% agarose gel using 120 V for 30 min and the gel was stained with GelRed Nucleic Acid Stain (Biotium, USA). The sizes of PCR amplicons were estimated by Trans2000 DNA Ladder (TransGen Biotech Corporation Ltd., Beijing, China). The PCR amplicons were sequenced by TsingKe Biotech Corporation Ltd. (Beijing, China). The obtained sequences were aligned with the sequences of each AVR gene to detect natural variations.
Plant Growth and Pathogenicity Assay
Rice blast fungal isolates on PDA medium were cultured on OTA media (50 g/l oatmeal, 200 ml/l tomato juice, and 15 g/l agar) at 25°C with 12-h light followed by 12-h dark treatment for producing mycelium. 7 days later, the mycelia were scraped with a cell spreader and the plates were exposed to continuous light at 28°C for 3 to 5 days to promote sporulation. Then, the conidia were washed with 2 ml double-distilled water (ddH2O) per petri dish and the suspension was adjusted the concentration to 1 × 105 conidia/ml.
The indicated lines and susceptible control LTH were used for pathogenicity assays in laboratory. The rice seeds were immersed in water for 2 days at 37°C in darkness for germination and then grown in the greenhouse with 28/24 ± 1°C day/night temperature, 70% relative humidity, and 14-h/10-h light/dark period. At three-leaf to five-leaf seedling stage, the second youngest leaves were detached and placed on the ddH2O with 6-benzylaminopurinehydrochloride (6-BA) in square petri dish after punch wounded. Then, the punch-wounded leaves were drop inoculated with 10 μl of spore suspension at the wound sites as a previous report (Kong et al., 2012). The inoculated leaves in petri dish were incubated in dark at 26°C in the greenhouse with 90% relative humidity for 24 h (Fang et al., 2018). Lesion size was examined at 4 to 7 days after inoculation to assess the pathogenicity of isolates.
Data Analysis
The DNA sequences were assembled and aligned with DNAMAN9 (Lynnon Biosoft) and the reference sequences of AVR genes were obtained from the National Center for Biotechnology Information (NCBI) with GenBank accessions given in Supplementary Table 1.
Results
Presence of Avirulence Genes in Field Isolates From the Sichuan Basin
We isolated the single spores from diseased leaves of susceptible variety LTH in 13 nurseries of the Sichuan Basin (Figure 1A). Finally, 383 isolates were obtained and subjected to investigate the variations of AVR genes. We analyzed the nucleotide sequence polymorphisms of eight known AVRs, namely, AVR-Pi9, AVR-Pi54, AVR-Pita1, AVR-Pib, AVR-Pia, AVR-Pizt, AVR-Pii, and AVR-Pik. First, we investigated their presence based on PCR amplification and classified the 383 isolates into the 16 race groups based on the presence of AVR genes. We found that five AVR genes, namely, AVR-Pik, AVR-Pi54, AVR-Pizt, AVR-Pi9, and AVR-Pib, were present in all the nurseries and the isolates containing the five AVR genes formed the major race groups of M. oryzae in the Sichuan Basin (Figures 1B,C). Nevertheless, AVR-Pita1 was present in the isolates from ST, TJ, and DZ, but almost (the present ratio < 0.1) or completely absent in five rice blast nurseries, including ZZ, AZ, PX, JY, and NB. AVR-Pii was present in the isolates of ZZ, but almost or completely absent in six rice blast nurseries, namely, ST, TJ, NX, PX, JY, and NB (Figure 1B). AVR-Pia was only present in a few isolates obtained from ZZ, DZ, and YS.
We speculated that the isolates that did not contain AVR-Pia may become virulent to the monogenic IRBL line carrying cognate R gene Pia. We inoculated these isolates to IRBLa-A. As our expectation, IRBLa-A showed susceptibility to all the isolates (Figure 2), indicating that loss of AVR-Pia breaks down the resistance of Pia. Similarly, those strains that did not contain AVR-Pita1, AVR-Pii, and AVR-Pia may breakdown their cognate R genes.
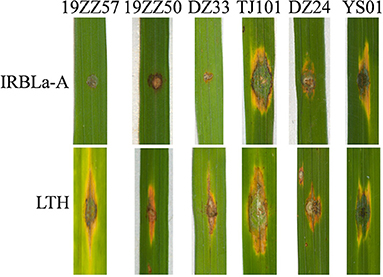
Figure 2. Loss of AVR-Pia broke down the resistance of Pia in rice. The representative blast disease phenotypes displayed on the leaves of susceptible line Lijiangxin Tuan Heigu (LTH) upon inoculation with the indicated strains. IRBLa-A is the monogenic line harboring the blast resistance gene Pia. AVR-Pia is present in the three isolates, namely, 19ZZ57, 19ZZ50, and DZ33, but not in the others, TJ101, DZ24, and YS01. The representative figures were taken at 5 days postinoculation (dpi).
We classified the race group of M. oryzae based on their harbored AVR genes. The 383 isolates were divided into the 16 race groups (Figure 1C). The groups C and D are the two largest groups consisting of 156 and 110 isolates, respectively. The groups G and A consist of 64 and 18 isolates, respectively. The rest groups each consist of fewer than 10 isolates. To estimate the diversity of M. oryzae in each prefecture, we calculated the number of the race groups in each nursery (Figure 1B). There are 11 groups existed in YS, suggesting that it has the most abundant diversity of M. oryzae. The rest nurseries have one to seven groups. Among them, the seven groups were identified in YA, while only the one and two groups were identified in ST and ZZ, respectively. There are similar amounts of detected isolates in these nurseries, such as 20 in YA, 19 in ST, and 19 in ZZ (Figure 1A). These data indicate that the diversity of M. oryzae shows distinct geographic feature in the Sichuan Basin.
Sequence Diversity and Distribution of AVR-Pib
Although AVR-Pib has high presence in most isolates (Figure 1B), its cognate Pib gene has been lost resistance in the Sichuan Basin (Zhang et al., 2017). Therefore, we investigated the sequence diversity of AVR-Pib. The genomic sequence covering the coding sequence (CDS) region was amplified via PCR using AVR-Pib-specific primers (Figure 3A; Supplementary Table 1). The PCR products can be divided into four different amplification patterns (APs) to denote no amplicon (AP0) and different sizes of amplicons (AP1-PA3) (Figures 3A,B). These APs include 10 variations that are designed haplotype 1 (H1) to haplotype 10 (H10) (Figure 3C). AP1 is the biggest PCR amplicon with over 2,000 bps, which was detected in 235 isolates and is larger than the expected size of AVR-Pib (GeneBank: KM887844.1), indicating that there is an insertion. Consistently, sequencing analysis identified an 1,861 bp insertion that shared 99.84% nucleotide sequence identity to the transposon Pot3 (GeneBank: AF333034.1) and flanked with two base pairs (5′-TA-3′) target site duplication (TSD). The insertion locates at four novel sites in 5′-untranslated region (5′-UTR) of AVR-Pib, including at −54, −231, −235, and −241 bp upstream of the start codon, which belong to H2, H3, H4, and H5 (Figure 3C). We developed an internal primer pairs, AvrPibF2/AvrPib, which could distinguish H2 from H3, H4, and H5 (Figures 3A–C). AP2 was detected in seven isolates from YA with near 1,000 bp, which defined as H1. Sequence alignment revealed that AP2 had a 482-bp insertion at the position of −85 bp from the start codon of AVR-Pib. This insertion showed 97.1% nucleotide sequence identity to the non-LTR retrotransposon Mg-SINE (GeneBank: U35313.1) and was flanked with a 12-bp sequence (CTTTTGCTTCGA) (Figure 3C). AP3 matches with the expected size of AVR-Pib (Figure 3B). Sequencing analysis identified several indels in the CDS and 5′-UTR of AVR-Pib (Figure 3C). A deletion occurs at positions 37 and 38 of the CDS region (H6) and several insertions occur in the 5′-UTR, including insertion of AAC at −124, C at −216 or −220, TAACT or TAAGT at −231, and TAACGT at −235 (H7-H9) (Figure 3C). Moreover, the sequencing chromatogram uncovered that some AP3 has double peaks, which decoded into two types of sequences with insertions at −231, −216, and −124 (H10). One sequence has insertion of TAACGT at −231, C at −216, and AAC at −124 and another sequence has insertion of TAACT at −231, C at −216, and AAC at −124 (Figure 3C). Besides, we failed to obtain amplicons from 15 isolates, which were from YS (5 isolates), TJ (2 isolates), YA (6 isolates), and QW (2 isolates) (Supplementary Table 2). These data indicate that the diverse ways were employed by AVR-Pib for escaping from R protein detection, such as insertion of Pot3, Mg-SINE, indels, and gene loss.
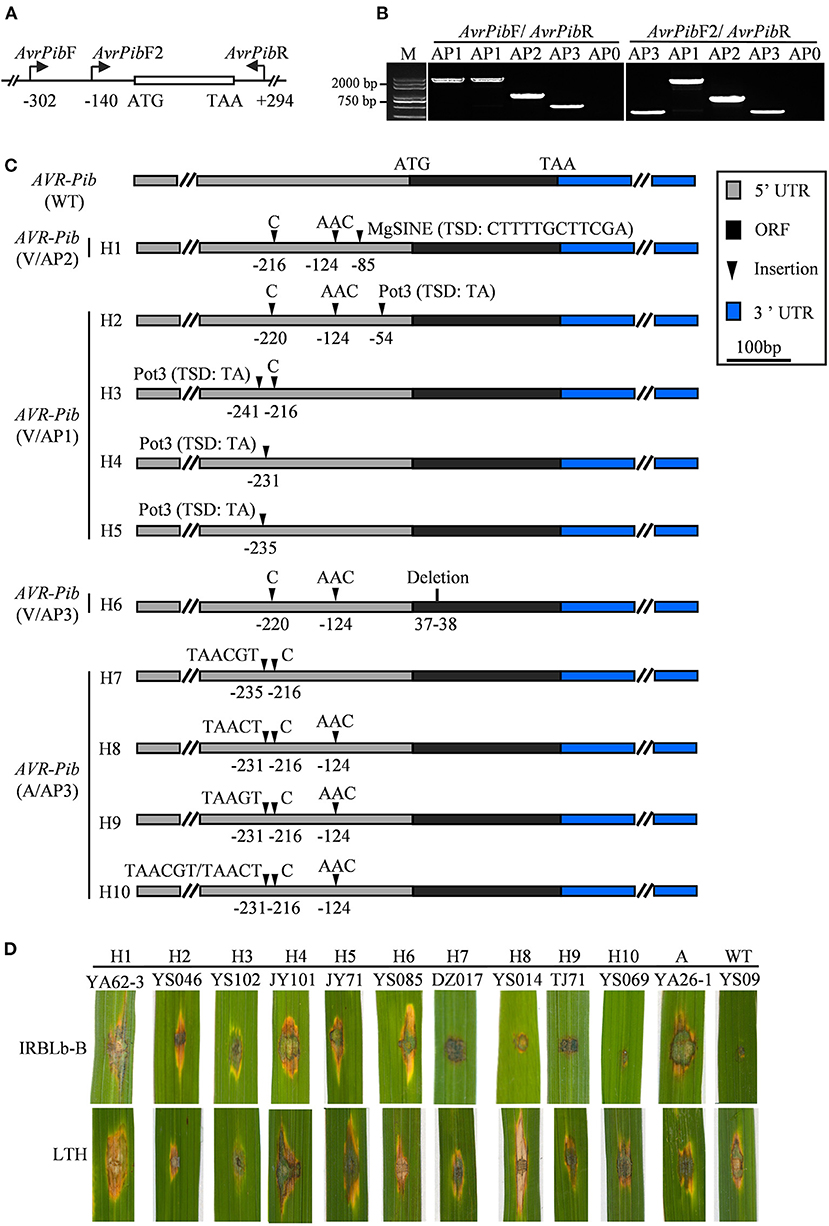
Figure 3. AVR-Pib variants and their pathogenicity to IRBLb-B. (A) Graph illustrates the location of the primer pairs AvrPibF/AvrPibR and AvrPibF2/AvrPibR used to examine the variation in AVR-Pib locus. (B) Amplification patterns of AVR-Pib. Four amplification patterns (AP0-AP3) were classified by the size of the fragments, which amplified from different isolates by indicated primers. AP0 indicates no amplicon. AP1, AP2, and AP3 indicate different sizes of amplicons. M, marker. (C) Characterization of allelic variations at AVR-Pib. H1 to H10, ten haplotypes of AVR-Pib contain different variations. (D) Pathogenicity of the indicated isolates on the monogenic line IRBLb-B carrying Pib. A, absence of AVR-Pib in the indicated strain.
Then, we tested whether the isolates harboring AVR-Pib variations were virulent to IRBLb-B containing cognate Pi-b. To this end, we performed pathogenicity assays and found that isolates with short insertions are avirulent to IRBLb-B, including H7, H8, H9, and H10 (Figures 3C,D), indicating that the short sequence insertions in 5′-UTR may not impact the function of AVR-Pib. In contrast, the isolates virulent to IRBLb-B carry the other four types of variations, including Mg-SINE insertion (H1), Pot3 insertion (H2-H5), deletion in CDS (H3), and absence of AVR-Pib (Figures 3C,D), indicating that these variations caused function loss of AVR-Pib to breakdown of the Pib resistance.
To further investigate the perniciousness of invalid AVR-Pib in the Sichuan Basin, we calculated the ratios of AVR-Pib variants in all the isolates (Figure 4). Our data indicate that 26.63% isolates contain the valid AVR-Pib, i.e., wild type (WT) and short sequence insertions in 5′-UTR of AVR-Pib. The invalid AVR-Pib exists in 73.37% isolates, consisting with the conclusion that its cognate Pib gene has been lost resistance in the Sichuan Basin (Zhang et al., 2017). There are four variations of invalid AVR-Pib, Pot3 insertion, 2 bp deletion in CDS (CDS 37–38Del), absence, and Mg-SINE insertion, which, respectively, exist in 61.36, 6.27, 3.91, and 1.83% isolates, suggesting that Pot3 insertion is the major way to invalidate AVR-Pib. In addition, the isolates carrying Pot3 insertion extensively exist in 11 nurseries, except YA and QW, and preponderantly in seven nurseries, namely, QS, ZZ, AZ, JY, ST, PX, and DZ. Variants with 2 bp deletion in CDS and Mg-SINE insertion of AVR-Pib were only detected in part of isolates from YS and YA, respectively. Some isolates lost AVR-Pib gene in YS, YA, TJ, and QW. These varieties of invalid AVR-Pib were carried by most isolates in YS and YA. However, the valid AVR-Pib existed in over half of isolates in QW, NB, NX, and TJ (Figure 4), hinting that the Pib gene may still confer resistance in these four regions, especially in QW and NX.
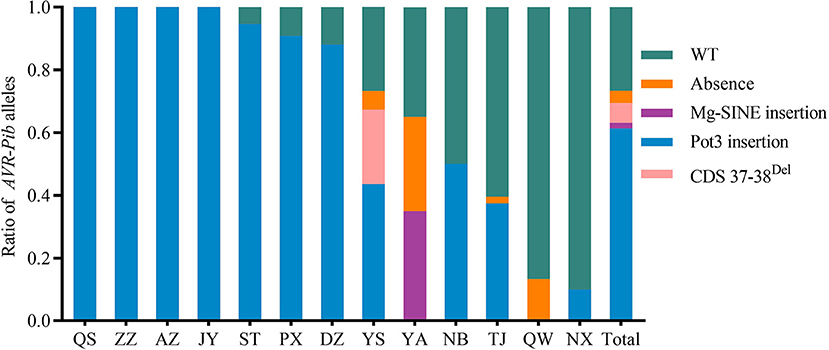
Figure 4. Distribution of isolates carrying variants of invalid AVR-Pib in 13 nurseries of the Sichuan Basin. Four variants, namely, absence, Mg-SINE insertion, Pot3 insertion, and coding sequence (CDS) 37-38Del, which caused function loss of AVR-Pib. Wild type (WT) denotes isolates have similar amplification size of AVR-Pib WT. Although some of variants may exist short insertions in the promoter of AVR-Pib, it is still avirulence to the monogenic line IRBLb-B carrying Pib.
Variations in AVR-Pizt and AVR-Pita1
Previously, insertions were reported in the 5′-UTR of AVR-Pizt, resulting in the function loss of AVR-Pizt and breakdown of the Pizt resistance (Wang et al., 2020). The fragment was amplified via PCR with primers AvrPiztF and AvrPiztR (Figure 5A). There are two sizes of the amplicons obtained in 97.91% of the isolates, but failed in a few isolates from YS, QS, ZZ, and NX (Figure 5B; Supplementary Table 2). Sequencing analysis indicated that there is an insertion of 201 bp at −181 bp upstream of the start codon of AVR-Pizt (Figure 5A). The insertion shared 96% sequence identity to solo-LTR of retrotransposon Inago2 (GeneBank: AB334125.1). The insertion was detected in 59 isolates obtained from the nurseries of YS and DZ (Supplementary Table 2). According to the pathogenicity assays, three representative isolates, namely, YS046, YS125, and DZ25, harboring the insertion are virulent to the monogenic rice blast resistance line IRBLzt-T carrying Pizt (Figure 5C), indicating that the insertion disrupts the avirulent function of AVR-Pizt, leading to breakdown of the Pizt-mediated resistance in the Sichuan Basin.
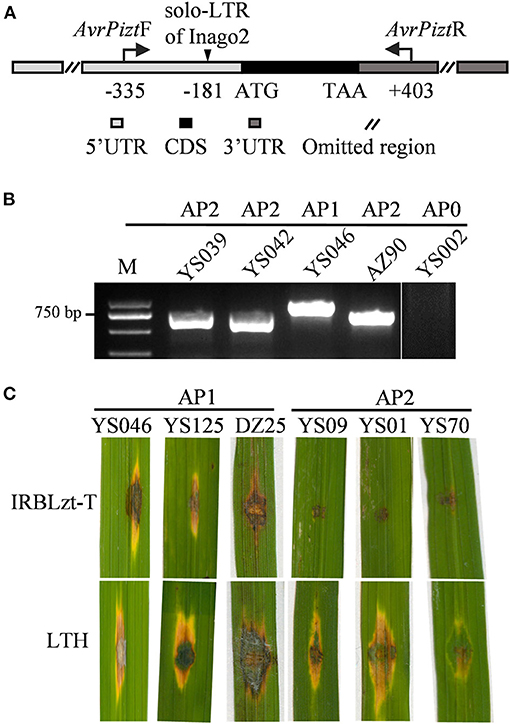
Figure 5. AVR-Pizt variants and their pathogenicity to IRBLzt-T. (A) Schematic diagram of AVR-Pizt and the insert site of a solo-LTR of Inago2. (B) Amplification patterns of AVR-Pizt from the indicated isolates. The primers AvrPiztF/AvrPiztR were used to distinguish three alleles. AP0 indicates no amplicon. AP1, AP2, and AP3 indicate different sizes of amplicons. (C) Pathogenicity of representative strains on the monogenic line IRBLzt-T carrying Piz-t. AP1 strains have an insertion of solo-LTR in AVR-Pizt gene and AP2 strains have WT allele of AVR-Pizt gene.
AVR-Pita1 locates close to the telomere region on the third chromosome. Some literatures reported that AVR-Pita1 is easy to be lost or exists as different avirulent haplotypes (Zhou et al., 2007; Singh et al., 2014; Damchuay et al., 2020). In this study, we used gene-specific primers to amplify fragments containing the whole CDS region of AVR-Pita1 (Supplementary Table 1). The expected size fragment was successfully amplified from 191 (49.9%) isolates. Analyzing the sequence of fragment, we identified five variations for AVR-Pita1 (Supplementary Table 3). All these variations had been reported to be still avirulent (Li et al., 2014; Damchuay et al., 2020). However, over half of isolates (192, 50.1%) failed to produce an amplificon, indicating that AVR-Pita1 is losing and going to breakdown the resistance function of Pita in the Sichuan Basin.
Haplotype Identification and Distribution of AVR-Pik Alleles
To investigate the natural variation of AVR-Pik in the field isolates of the Sichuan Basin, we analyzed the sequence of CDS region of AVR-Pik. The AVR-Pik gene was successfully amplified from all the isolates, indicating that AVR-Pik exists widely in the isolates from the Sichuan Basin. Sequencing analysis identified five AVR-Pik alleles, namely, AVR-PikA, AVR-PikB, AVR-PikC, AVR-PikD, and AVR-PikE (Figure 6A). Among them, single AVR-PikA, AVR-PikB, AVR-PikC, or AVR-PikD was characterized in less than 10% isolates. Especially, AVR-PikC is present only in 0.26% isolates. In contrast, AVR-PikE is abundant and exists in 35.25% isolates. The rest isolates (53.00%) were characterized as harboring two AVR-Pik alleles, namely, AVR-PikD + AVR-PikA and AVR-PikD + AVR-PikE, which were detected in 42.56 and 10.44% isolates, respectively (Figure 6B). Therefore, AVR-PikE and AVR-PikD + AVR-PikA are dominant AVR-Pik alleles in the blast fungal populations in the Sichuan Basin. In addition, we identified a new deletion of CTTT, which happened at −42 to −39 upstream of the start codon of AVR-PikD in the AVR-PikD + AVR-PikE harbored isolates (Figure 6A).
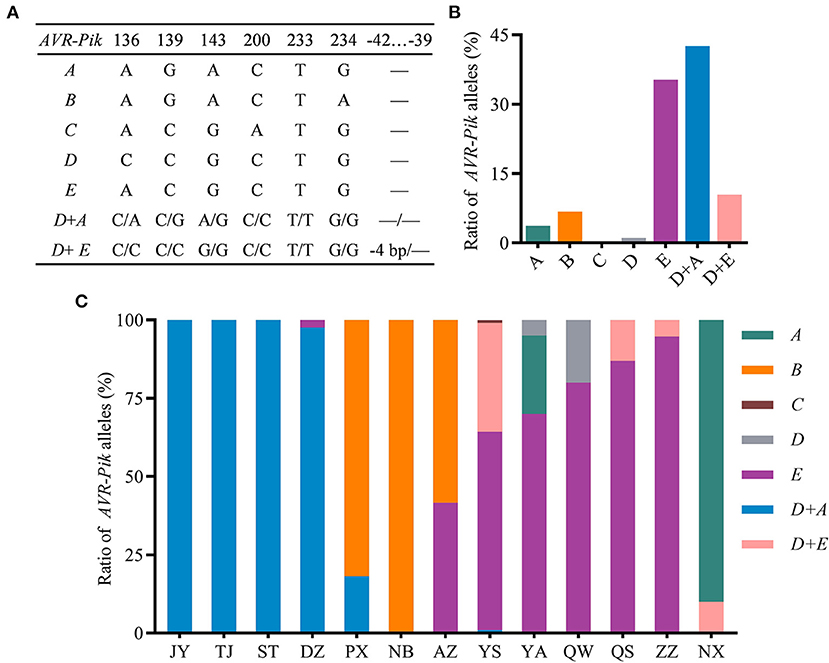
Figure 6. The variation and distribution of AVR-Pik alleles in the Sichuan Basin. (A) AVR-Pik alleles characterized in the isolates from the Sichuan Basin. −4bp, CTTT is deleted at −42 to −39 of the promoter of AVR-PikD. —, no change compared with WT. (B) Ratio of isolates harboring different AVR-Pik alleles. (C) Ratio of AVR-Pik alleles in the indicated 13 prefectures in the Sichuan Basin.
Meanwhile, we analyzed the distribution of different AVR-Pik alleles in the 13 nurseries (Figure 6C). Single AVR-PikA was found in isolates from YA (25%) and NX (90%). AVR-PikB was detected in isolates from NB (100%), PX (81.82%), and AZ (58.33%). AVR-PikC was detected only in one isolate from YS. Single AVR-PikD was detected in YA (5%) and QW (20%). Single AVR-PikE was detected in five nurseries that accounted for 63.37, 70, 80, 86.96, and 94.74% of the isolates from YS, YA, QW, QS, and ZZ, respectively. Single AVR-PikE was also detected in two isolates from DZ and five isolates from AZ. Double alleles of AVR-PikD + AVR-PikA were detected in DZ (97.62%), YS (0.99%), TJ (100%), ST (100%), PX (18.18%), and JY (100%), whereas another double alleles of AVR-PikD + AVR-PikE were found in YS (34.65%), QS (13.04%), ZZ (5.26%), and NX (10%). Taken together, AVR-PikA was the dominant allele of AVR-Pik in the nursery of NX. AVR-PikB was dominant in the PX, NB, and AZ. AVR-PikE was dominant in the YS, YA, QW, QS, and ZZ. AVR-PikD + AVR-PikA were dominant in the JY, TJ, ST, and DZ. These data indicate that AVR-Pik alleles existed in all the isolates with distinct geographical feature and, thus, cultivars carrying Pikm can be exploited in the Sichuan Basin.
Mating Type of Magnaporthe oryzae in the Sichuan Basin
Specific primers for MAT1-1 and MAT1-2 (Supplementary Table 1) were used to investigate the mating type composition of isolates in the M. oryzae population. In total, 171 isolates are MAT1-1 and 174 isolates are MAT1-2 (Figure 7A), indicating that the distribution of MAT1-1 and MAT1-2 is relatively balanced in the Sichuan Basin. In addition, we also identified hermaphroditic type in 38 isolates, which have both the mating types simultaneously. Single mating type was found in the isolates from 6 of 13 nurseries, namely, ST, PX, NB, JY, QS, and ZZ. Two mating types were detected in the rest seven nurseries and the hermaphroditic isolates were found in TJ, DZ, and YA (Figure 7B). Even though the sexual status is hardly observed under natural conditions, sexual reproduction potential is still present in regions where the isolates have both the mating types, hinting that the parasexual recombination may be an important way to promote variation of AVR genes in the Sichuan Basin.
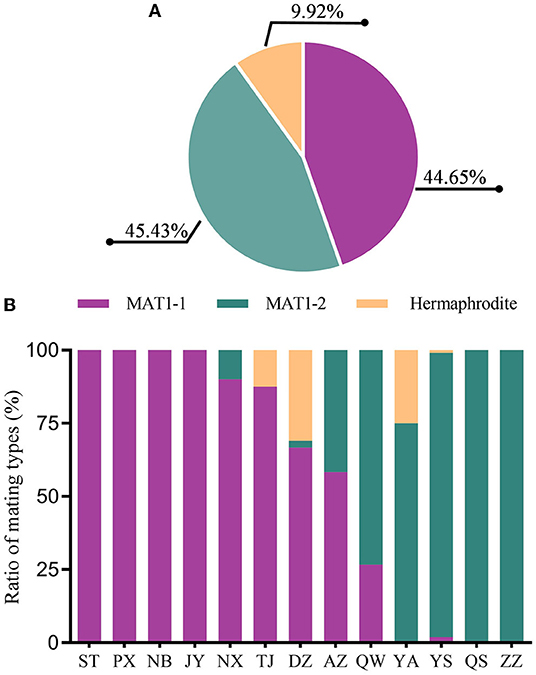
Figure 7. The composition and distribution of MAT1-1 and MAT1-2 isolates in the M. oryzae population in the Sichuan Basin. (A) The composition of MAT1-1 and MAT1-2 in the isolates. (B) The distribution of isolates with different mating types in the Sichuan Basin.
Discussion
The dominant population of M. oryzae displays distinct geographic feature, which results from the different environments and rice varieties determined by local preferences. As the diversity of rice varieties has abundant, the geographic feature is increasingly apparent in population of M. oryzae. Therefore, monitoring the presence and variation of AVR genes is an efficient way to crack the geographic code in M. oryzae population (Selisana et al., 2017). Usually, commercial cultivars or monogenetic resistance lines were subjected for monitoring. It can select resistance cultivars or clear the valid R genes for variety deployment, but cannot reflect the population of M. oryzae accurately for prediction of rice blast epidemic. In this study, we collected disease samples from the susceptible rice accession LTH that does not have any known resistance genes, to establish single spore isolates, which are more accurate to represent the populations of M. oryzae in paddy fields.
We obtained 383 M. oryzae isolates from LTH in 13 nurseries located in the Sichuan Basin. According to the gene-for-gene paradigm, activation of resistance depends on the recognition between R and AVR protein. Loss and variations in AVRs can result in breakdown of the cognate R genes. To speculate the valid R genes in different regions of the Sichuan Basin, we analyzed eight AVR genes, namely, AVR-Pi9, AVR-Pi54, AVR-Pita1, AVR-Pib, AVR-Pia, AVR-Pizt, AVR-Pii, and AVR-Pik, in the 383 isolates. We found that the AVR genes of M. oryzae employed different mechanisms, such as transposon insertion, short sequence insertion, nucleotide deletion and substitution, gene duplication, as well as gene loss (Figures 1, 3, 5, 6; Supplementary Tables 2, 3), to avoid perception mediated by host R genes for pathogenesis in the Sichuan Basin.
The resistance function of Pib seems completely invalidated due to the high frequency of loss and sequence variations in AVR-Pib in the Sichuan Basin. According to previous reports, IRBLb-B carrying Pib is susceptible to more than 60% of M. oryzae strains isolated from Cambodia and Hunan province in China (Fukuta et al., 2014; Xing et al., 2017). IRBLb-B also shows susceptible to all the 248 field isolates from Bohol of Philippines because of the prevailing insertion of Pot3 in AVR-Pib (Olukayode et al., 2019). AVR-Pib seems undergone different genetic events, including transposable elements insertion, segmental deletion, complete absence, and point mutation in 300 isolates collected from the southern, northeastern, and far northeastern regions of China (Zhang et al., 2015). These reports indicated that AVR-Pib showed diverse variations in the nature, causing loss of avirulence. In this study, we detected AVR-Pib in 96.08% of the isolates from the Sichuan Basin. However, AVR-Pib demonstrated abundant variations in different regions of the Sichuan Basin, including effective variations that cause the loss of the avirulent function in AVR-Pib. Among these variations, the main variation came from the Pot3 insertions in the 5′-UTR of AVR-Pib, which led to breakdown of the Pib resistance (Figure 3). Besides, we also identified some novel sites of Pot3 and Mg-SINE insertion, which generated different virulent haplotypes of AVR-Pib (Figure 3).
AVR-Pik is the most complicated AVR gene locus among the eight AVRs. To date, six AVR-Pik alleles have been identified based on polymorphic amino acids at five sites, namely, AVR-PikA, AVR-PikB, AVR-PikC, AVR-PikD, AVR-PikE, and AVR-PikF (Yoshida et al., 2009; Longya et al., 2019). Among them, AVR-PikD was the most popular allele and it is most likely the ancestral allele, from which the AVR-PikE, A, C, and B alleles were derived (Yoshida et al., 2009; Kanzaki et al., 2012). AVR-Pik alleles can be specifically recognized by different Pik genes to trigger immune responses. The ancestral allele AVR-PikD can be recognized by five Pik alleles (Pik, Pi-km, Pi-kh, Pi-kp, and Pi-ks). AVR-PikE is recognized by three Pik alleles (Pik, Pi-km, and Pi-kh), while AVR-PikA is recognized by Pi-km and Pi-kh. However, all the Pik alleles cannot recognize AVR-PikB, AVR-PikC, and AVR-PikF (Yoshida et al., 2009; Kanzaki et al., 2012; Wu et al., 2014; Zhai et al., 2014; Wang et al., 2017). AVR-PikD, AVR-PikA, AVR-PikC, and AVR-PikE exist commonly worldwide, but AVR-PikB was previously identified only in a Japan isolate and the Chinese isolate Zhong-10-8-14 and AVR-PikF is a novel emerging allele reported in Thailand (Yoshida et al., 2009; Kanzaki et al., 2012; Cao et al., 2017; Longya et al., 2019). Here, we did not detect AVR-PikF in the Sichuan Basin. We identified five AVR-Pik (A-E) alleles and found that some isolates harboring two copies of AVR-Pik alleles containing AVR-PikD + AVR-PikA or AVR-PikD + AVR-PikE (Figure 6; Supplementary Table 2). AVR-PikD + AVR-PikA were previously reported presumably generated from gene duplication (Wang et al., 2017), AVR-PikD + AVR-PikE might be a novel gene duplication event occurred to the blast fungal population in the Sichuan Basin. As one of the main fungal adaption and evolution mechanisms, gene duplication can lead to the emergence of novel virulent alleles of AVR genes (Chuma et al., 2011). Isolates harboring two copies of AVR-Pik are likely to be virulent through losing AVR-PikD. Furthermore, virulent alleles AVR-PikB and AVR-PikC have already occurred in three nurseries (PX, NB, and AZ) and one prefecture (YS) in the Sichuan Basin, respectively (Figure 6C). Isolates harboring different AVR-Pik alleles seem gradually become aggressive in the Sichuan Basin.
Parasexual recombination and sexual mating can result in the occurrence of complex rice blast races and arising of pathogenic variants (Noguchi et al., 2006; Wang and Valent, 2009). Sexual cycles can be observed on culture medium and are rarely found in the nature, but the sexual recombination can also occur in some limited regions in south Asia (Saleh et al., 2012). Moreover, hermaphroditic strains identified in some restricted areas lead to the assumption that sexual reproduction could occur in the field (Wang and Valent, 2009). Hermaphroditic isolates are rare and previous studies showed that hermaphroditic strains could be found in Thailand, India, Philippines, and Yunnan province of China (Hayashi et al., 1997; Kumar et al., 1999; Mekwatanakarn et al., 1999). In this study, hermaphroditic strains are found in four regions, three of them locate in the northeastern of the Sichuan Basin and isolates collected in five regions have both the mating types (Figure 7). These findings suggested that sexual reproduction may occur in some limited regions in the Sichuan Basin, which may generate complex blast fungus races and change the population structure through DNA exchange. Intriguingly, the most abundance of AVR-Pib alleles was identified in the nurseries YS, YA, and TJ (Figure 4), where they have hermaphroditic strains (Figure 7). But, two virulence AVR-Pik alleles, namely, AVR-PikB and AVR-PikC, escaped from recognition by Pik alleles found in the nurseries PX, NB, AZ, YA, and QW, respectively (Figure 6C), where they have single, couple, or hermaphroditic of the two mating types. It is interesting to further dissect the relationship between parasexual recombination or sexual mating and mutation, absence, or duplication of AVR genes. Furthermore, the distribution of either AVR genes or their variations has distinct geographical features (Figures 4, 6C). It may be caused by different selection pressure conferred by rice variety diversity in various regions of the Sichuan Basin. To further monitoring the dynamics of the AVR gene variations, we need to collect more isolates and perform further analysis in the future. Meanwhile, the pan-genome analysis on isolates from different years may help to understand the molecular mechanism of the rice blast epidemic in the Sichuan Basin.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Y-YH and W-MW conceived the experiment. Z-JH, X-YL, HF, S-XZ, YX, X-XL, CL, R-MZ, MP, Y-PJ, X-HH, G-BL, J-HZ, HW, and D-QL carried out the experiment. W-SZ, C-HF, Z-XZ, Yu-LP, FH, and Yo-LP managed the blast nurseries. Z-JH, J-WZ, JF, YL, and MH analyzed the data. Z-JH wrote the manuscript. Y-YH and W-MW edited the manuscript. All authors contributed to the article and approved the submitted version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (U19A2033 to W-MW) and the Department of Science and Technology of Sichuan Province (2020YJ0332 to W-MW and 2021YJ0304 to YL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Yi-Fu Wang and Jin-Feng Chen for their help with the isolation of single spore strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.788876/full#supplementary-material
References
Cao, J., Yu, Y., Huang, J., Liu, R., Chen, Y., Li, S., et al. (2017). Genome re-sequencing analysis uncovers pathogenecity-related genes undergoing positive selection in Magnaporthe oryzae. Sci. China Life Sci. 60, 880–890. doi: 10.1007/s11427-017-9076-4
Chuma, I., Isobe, C., Hotta, Y., Ibaragi, K., Futamata, N., Kusaba, M., et al. (2011). Multiple translocation of the Avr-pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and Related Species. PLOS Pathogens 7, e1002147. doi: 10.1371/journal.ppat.1002147
Dai, Y., Jia, Y., Correll, J., Wang, X., and Wang, Y. (2010). Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae. Fungal Genet. Biol. 47, 973–980. doi: 10.1016/j.fgb.2010.08.003
Damchuay, K., Longya, A., Sriwongchai, T., Songkumarn, P., Parinthawong, N., Darwell, K., et al. (2020). High nucleotide sequence variation of avirulent gene, AVR-Pita1, in Thai rice blast fungus population. J. Genetic. 99, 8. doi: 10.1007/s12041-020-01197-8
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Fang, W. W., Liu, C. C., Zhang, H. W., Xu, H., Zhou, S., Fang, K. X., et al. (2018). Selection of differential isolates of Magnaporthe oryzae for postulation of blast resistance genes. Phytopathology 108, 878–884. doi: 10.1094/PHYTO-09-17-0333-R
Farman, M. L., and Leong, S. A. (1998). Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: discrepancy between the physical and genetic maps. Genetics 150, 1049–1058. doi: 10.1093/genetics/150.3.1049
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Fudal, I., Böhnert, H. U., Tharreau, D., and Lebrun, M.-H. (2005). Transposition of MINE, a composite retrotransposon, in the avirulence gene ACE1 of the rice blast fungus Magnaporthe grisea. Fungal Genetic. Biol. 42, 761–772. doi: 10.1016/j.fgb.2005.05.001
Fukuta, Y., Koga, I., Ung, T., Sathya, K., Kawasaki-Tanaka, A., Koide, Y., et al. (2014). Pathogenicity of rice blast (Pyricularia oryzae Cavara) isolates from Cambodia. Japan Agricult. Res. Q. JARQ 48, 155–166. doi: 10.6090/jarq,0.48.155
Hayashi, N., Li, C., Li, J., Iwano, M., Naito, H., Yoshino, R., et al. (1997). Distribution of fertile Magnaporthe grisea fungus pathogenic to rice in Yunnan Province, China. Japanese J. Phytopathol. 63, 316–323. doi: 10.3186/jjphytopath.63.316
Jia, Y. (2009). A user-friendly method to isolate and single spore the fungi magnaporthe oryzae and magnaporthe grisea obtained from diseased field samples. Plant Health Progr. 10, 25. doi: 10.1094/PHP-2009-1215-01-BR
Kang, S., Lebrun, M. H., Farrall, L., and Valent, B. (2001). Gain of Virulence Caused by Insertion of a Pot3 Transposon in a Magnaporthe grisea Avirulence Gene. Mol. Plant-Microbe Interact. 14, 671–674. doi: 10.1094/MPMI.2001.14.5.671
Kang, S., Sweigard, J. A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8, 939–948. doi: 10.1094/MPMI-8-0939
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Khang, C. H., Park, S.-Y., Lee, Y.-H., Valent, B., and Kang, S. (2008). Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant-Microbe Interact. 21, 658–670. doi: 10.1094/MPMI-21-5-0658
Kong, L. A., Yang, J., Li, G. T., Qi, L. L., Zhang, Y. J., Wang, C. F., et al. (2012). Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8, e1002526. doi: 10.1371/journal.ppat.1002526
Kumar, J., Nelson, R. J., and Zeigler, R. S. (1999). Population structure and dynamics of Magnaporthe grisea in the Indian himalayas. Genetics 152, 971–984. doi: 10.1093/genetics/152.3.971
Li, J., Lu, L., Jia, Y., and Li, C. (2014). Effectiveness and durability of the rice pi-ta gene in Yunnan province of China. Phytopathology 104, 762–768. doi: 10.1094/PHYTO-11-13-0302-R
Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., et al. (2009). The Magnaporthe oryzae Avirulence Gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 22, 411–420. doi: 10.1094/MPMI-22-4-0411
Liu, M. H., Kang, H., Xu, Y., Peng, Y., Wang, D., Gao, L., et al. (2020). Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice. Plant Biotechnol. J. 18, 1376–1383. doi: 10.1111/pbi.13300
Longya, A., Chaipanya, C., Franceschetti, M., Maidment, J. H. R., Banfield, M. J., and Jantasuriyarat, C. (2019). Gene duplication and mutation in the emergence of a novel aggressive allele of the AVR-Pik effector in the rice blast fungus. Mol. Plant Microbe. Interact. 32, 740–749. doi: 10.1094/MPMI-09-18-0245-R
Lu, D., He, F., Gong, X., Wang, J., Fu, R., Han, Y., et al. (2017). Review and reflection on rice variety certification for identifying rice blast resistance in sichuan for 30 years. Chinese Agricult. Sci. Bull. 33, 150–154.
Ma, J. H., Wang, L., Feng, S. J., Lin, F., Xiao, Y., and Pan, Q. H. (2006). Identification and fine mapping of AvrPi15, a novel avirulence gene of Magnaporthe grisea. Theor. Appl. Genet. 113, 875–883. doi: 10.1007/s00122-006-0347-6
Mekwatanakarn, P., Kositratana, W., Phromraksa, T., and Zeigler, R. S. (1999). Sexually fertile Magnaporthe grisea rice pathogens in Thailand. Plant Dis. 83, 939–943. doi: 10.1094/PDIS.1999.83.10.939
Noguchi, M. T., Yasuda, N., and Fujita, Y. (2006). Evidence of genetic exchange by parasexual recombination and genetic analysis of pathogenicity and mating type of parasexual recombinants in rice blast fungus, Magnaporthe oryzae. Phytopathology® 96, 746–750. doi: 10.1094/PHYTO-96-0746
Olukayode, T., Quime, B., Shen, Y. C., Yanoria, M. J., Zhang, S., Yang, J., et al. (2019). Dynamic insertion of pot3 in Avrpib prevailing in a field rice blast population in the philippines led to the high virulence frequency against the resistance Gene Pib in Rice. Phytopathology 109, 870–877. doi: 10.1094/PHYTO-06-18-0198-R
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., and Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. The Plant Cell 12, 2019–2032. doi: 10.1105/tpc.12.11.2019
Ray, S., Singh, P. K., Gupta, D. K., Mahato, A. K., Sarkar, C., Rathour, R., et al. (2016). Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 7, 1140. doi: 10.3389/fpls.2016.01140
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceed. Nat. Acad. Sci. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Saleh, D., Xu, P., Shen, Y., Li, C., Adreit, H., Milazzo, J., et al. (2012). Sex at the origin: an Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol. Ecol. 21, 1330–1344. doi: 10.1111/j.1365-294X.2012.05469.x
Selisana, S. M., Yanoria, M. J., Quime, B., Chaipanya, C., Lu, G., Opulencia, R., et al. (2017). Avirulence (AVR) gene-based diagnosis complements existing pathogen surveillance tools for effective deployment of resistance (r) genes against rice blast disease. Phytopathology 107, 711–720. doi: 10.1094/PHYTO-12-16-0451-R
Silue, D., Notteghem, J. L., and Tharreau, D. (1992). Evidence of a gene-for-gene relationship in the oryza sativa-magnaporthe grisea pathosystem. Phytopathology 82, 577–580. doi: 10.1094/Phyto-82-577
Singh, P. K., Thakur, S., Rathour, R., Variar, M., Prashanthi, S. K., Singh, A. K., et al. (2014). Transposon-based high sequence diversity in Avr-Pita alleles increases the potential for pathogenicity of Magnaporthe oryzae populations. Funct. Integr. Genom. 14, 419–429. doi: 10.1007/s10142-014-0369-0
Skamnioti, P., and Gurr, S. J. (2009). Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150. doi: 10.1016/j.tibtech.2008.12.002
Sone, T., Takeuchi, S., Miki, S., Satoh, Y., Ohtsuka, K., Abe, A., et al. (2013). Homologous recombination causes the spontaneous deletion of AVR-Pia in Magnaporthe oryzae. FEMS Microbiol. Lett. 339, 102–109. doi: 10.1111/1574-6968.12058
Sweigard, J. A., Carroll, A. M., Kang, S., Farrall, L., Chumley, F. G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7, 1221–1233. doi: 10.1105/tpc.7.8.1221
Talbot, N. J. (2003). On the Trail of a Cereal Killer: Exploring the Biology of Magnaporthe grisea. Ann. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Tsujimoto Noguchi, M. (2011). Parasexual Recombination in Magnaporthe oryzae. Japan Agricult. Res. Q.: JARQ 45, 39–45. doi: 10.6090/jarq.45.39
Valent, B., and Khang, C. H. (2010). Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 13, 434–441. doi: 10.1016/j.pbi.2010.04.012
Wang, B.-h., Ebbole, D. J., and Wang, Z.-h. (2017). The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integrat. Agricult. 16, 2746–2760. doi: 10.1016/S2095-3119(17)61746-5
Wang, Q., Li, J., Lu, L., He, C., and Li, C. (2020). Novel variation and evolution of AvrPiz-t of Magnaporthe oryzae in field isolates. Front. Genet. 11, 746. doi: 10.3389/fgene.2020.00746
Wang, X., and Valent, B. (2009). Advances in genetics, genomics and control of rice blast disease: springer science and business. Media. 21, 9. doi: 10.1007/978-1-4020-9500-9
Wu, J., Kou, Y., Bao, J., Li, Y., Tang, M., Zhu, X., et al. (2015). Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 206, 1463–1475. doi: 10.1111/nph.13310
Wu, W., Wang, L., Zhang, S., Li, Z., Zhang, Y., Lin, F., et al. (2014). Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant. Microbe. Interact. 27, 759–769. doi: 10.1094/MPMI-02-14-0046-R
Xie, Z., Yan, B., Shou, J., Tang, J., Wang, X., Zhai, K., et al. (2019). A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180308. doi: 10.1098/rstb.2018.0308
Xing, J., Jia, Y., Peng, Z., Shi, Y., He, Q., Shu, F., et al. (2017). Characterization of molecular identity and pathogenicity of rice blast fungus in Hunan Province of China. Plant Dis. 101, 557–561. doi: 10.1094/PDIS-03-16-0288-RE
Yasuda, N., Tsujimoto Noguchi, M., and Fujita, Y. (2006). Partial mapping of avirulence genes AVR-Pii and AVR-Pia in the rice blast fungus Magnaporthe oryzae. Canad. J. Plant Pathol. 28, 494–498. doi: 10.1080/07060660609507325
Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., et al. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591. doi: 10.1105/tpc.109.066324
Yoshida, K., Saunders, D. G. O., Mitsuoka, C., Natsume, S., Kosugi, S., Saitoh, H., et al. (2016). Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genom. 17, 370. doi: 10.1186/s12864-016-2690-6
Zhai, C., Zhang, Y., Yao, N., Lin, F., Liu, Z., Dong, Z., et al. (2014). Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS One 9, e98067. doi: 10.1371/journal.pone.0098067
Zhang, S., Wang, L., Wu, W., He, L., Yang, X., and Pan, Q. (2015). Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 5, 11642. doi: 10.1038/srep11642
Zhang, S., Zhong, X., Qiao, G., Shen, L., Zhou, T., and Peng, Y. (2017). Difference in Virulence of Magnaporthe oryzae from Sichuan, Chongqing and Guizhou. Southwest China J. Agricult. Sci. 30, 359–365. doi: 10.16213/j.cnki.scjas.2017.2.020
Zhao, H., Wang, X., Jia, Y., Minkenberg, B., Wheatley, M., Fan, J., et al. (2018). The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 4. doi: 10.1038/s41467-018-04369-4
Keywords: rice blast, Magnaporthe oryzae, avirulence gene, variation, monogenetic lines, resistance gene
Citation: Hu Z-J, Huang Y-Y, Lin X-Y, Feng H, Zhou S-X, Xie Y, Liu X-X, Liu C, Zhao R-M, Zhao W-S, Feng C-H, Pu M, Ji Y-P, Hu X-H, Li G-B, Zhao J-H, Zhao Z-X, Wang H, Zhang J-W, Fan J, Li Y, Peng Y-L, He M, Li D-Q, Huang F, Peng Y-L and Wang W-M (2022) Loss and Natural Variations of Blast Fungal Avirulence Genes Breakdown Rice Resistance Genes in the Sichuan Basin of China. Front. Plant Sci. 13:788876. doi: 10.3389/fpls.2022.788876
Received: 03 October 2021; Accepted: 10 March 2022;
Published: 12 April 2022.
Edited by:
Corina Vlot, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Muxing Liu, Nanjing Agricultural University, ChinaChatchawan Jantasuriyarat, Kasetsart University, Thailand
Devanna BN, National Rice Research Institute (ICAR), India
Copyright © 2022 Hu, Huang, Lin, Feng, Zhou, Xie, Liu, Liu, Zhao, Zhao, Feng, Pu, Ji, Hu, Li, Zhao, Zhao, Wang, Zhang, Fan, Li, Peng, He, Li, Huang, Peng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ming Wang, ajMxNndlbm1pbmd3YW5nQHNpY2F1LmVkdS5jbg==; Yan-Yan Huang, aDE5ODV5eUAxNjMuY29t
 Zi-Jin Hu1
Zi-Jin Hu1 Yan-Yan Huang
Yan-Yan Huang Wen-Sheng Zhao
Wen-Sheng Zhao Xiao-Hong Hu
Xiao-Hong Hu Guo-Bang Li
Guo-Bang Li Zhi-Xue Zhao
Zhi-Xue Zhao He Wang
He Wang Jing Fan
Jing Fan Yan Li
Yan Li Min He
Min He Wen-Ming Wang
Wen-Ming Wang