
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 March 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.765213
This article is part of the Research Topic Interactions Between Biochemical Pathways Producing Plant Colors and Scents View all 19 articles
As an important member of the MYB transcription factor (TF) family, the MYB-related TFs play multiple roles in regulating the synthesis of secondary metabolites and developmental processes, as well as in response to numerous biotic and abiotic stressors in plants. However, little is known regarding their roles in regulating the formation of floral volatile organic compounds (VOCs). In this study, we conducted a genome-wide analysis of MYB-related proteins in sweet osmanthus; 212 OfMYB-related TFs were divided into three distinct subgroups based on the phylogenetic analysis. Additionally, we found that the expansion of the OfMYB-related genes occurred primarily through segmental duplication events, and purifying selection occurred in all duplicated gene pairs. RNA-seq data revealed that the OfMYB-related genes were widely expressed in different organs of sweet osmanthus, and some showed flower organ/development stage-preferential expression patterns. Here, three OfMYB-related genes (OfMYB1R70/114/201), which were expressed nuclearly in floral organs, were found to be significantly involved in regulating the synthesis of floral VOCs. Only, OfMYB1R201 had transcriptional activity, thus implying that this gene participates in regulating the expression of VOC synthesis related genes. Remarkably, the transient expression results suggested that OfMYB1R70, OfMYB1R114, and OfMYB1R201 are involved in the regulation of VOC synthesis; OfMYB1R114 and OfMYB1R70 are involved in accelerating β-ionone formation. In contrast, OfMYB1R201 decreases the synthesis of β-ionone. Our results deepen our knowledge of the functions of MYB-related TFs and provide critical candidate genes for the floral aroma breeding of sweet osmanthus in the future.
MYB-related transcription factors (TFs), named for their highly conserved 52 amino acid MYB domain, are predominant and critical members of the MYB TF family in plants (Riechmann and Ratcliffe, 2000; Dubos et al., 2010). MYB TFs can be generally divided into four subfamilies based on the number of MYB domains present: 1R-MYB (MYB-related), R2R3-MYB, 3R(R1R2R3)-MYB, and 4R-MYB proteins (Rosinski and Atchley, 1998). The 1R-MYB are collectively called MYB-related proteins and include proteins with a single or a partial MYB repeat (Dubos et al., 2010). Recently, many studies performing systematic identification and genome-wide characterization of the MYB family have reported genes in various plants, such as Petunia hybrida, potato (Solanum tuberosum), peach (Amygdalus persica), and Chinese jujube (Ziziphus jujuba) (Zhang et al., 2018; Ji et al., 2019; Liu et al., 2020; Chen et al., 2021). The MYB-related and R2R3-MYB subfamily contain the highest quantity of genes within the MYB family (Liu et al., 2020). To date, R2R3-MYB has been comprehensively characterized and reported to have diverse functions (Nesi et al., 2001; Koes et al., 2005; Ambawat et al., 2013), particularly in enhancing volatile aroma production, promoting anthocyanin accumulation, and responding to abiotic stresses. In contrast, several MYB-related proteins control diverse physiological processes, particularly the regulation of the anthocyanin biosynthesis pathway and the amelioration of multiple abiotic stresses. The first MYB-related TF identified in plants was in potato (MYBSt1) (Baranowskij et al., 1994), and since then, the MYB family has gradually gained more attention. However, to date, the function of MYB-related genes in regulating the synthesis of floral aroma remains largely unknown.
According to the most recent classification based on the highly conserved motif, MYB-related TFs are divided into three subgroups: the CPC-like subgroup of R3-type single-MYB proteins consisting of CPC (CAPRICE)-like, TRY (TRIPTYCHON)-like, ETC (Enhancer of TRY and CPC)-like and MYBL2; the CCA1-like/R-R subgroup, including the CCA1 (Circadian Clock Associated1)-like, TBP (Telomeric DNA-binding Protein)-like, RAD (RADIALIS)-like, TRF-like, LHY (Late Elongated Hypocotyl), and RVE (Early Phytochrome Responsive1)-like proteins; and the GARP subgroup, which is further classified into the LUX, MYB_CC, KANADI, ARR, HHO, GLK (GOLDEN2-LIKE), and GARP-like subfamilies. The MYB-related genes were subsequently confirmed to play fundamental roles as transcriptional regulators, associated repressors, circadian clock, and telomeric repeat-binding proteins in various biological processes. AtMYBD, a member of the CCA1-like/R-R subgroup in Arabidopsis thaliana, regulates anthocyanin biosynthesis in a circadian clock-related manner (Nguyen and Lee, 2016). SlTRY, an R3 MYB member of the CPC-like subgroup, has been confirmed to be involved in anthocyanin biosynthesis and is considered a core regulator of root-hair development and the plant trichome (Rumi et al., 2013). The grape TF AQUILO, a member of the GARP subgroup, increases cold tolerance by promoting the accumulation of raffinose family oligosaccharides (Sun et al., 2018). In recent years, MYB-related TFs have been found to be involved in regulating anthocyanin biosynthesis in various pathways and products. The proteins MdLUX and MdPCL-like, containing a single MYB-like repeat, promote anthocyanin accumulation through DNA hypomethylation in apple (Malus domestica) (Li et al., 2021). The functions of MYB-related proteins in metabolite biosynthesis have been characterized in various land plants, for instance, Arabidopsis, soybean (Glycine max), grapevine (Vitis vinifera), lily (Lilium spp.), and tiger lily (Lilium lancifolium) (Matsui et al., 2008; Niu et al., 2016; Sakai et al., 2019; Yong et al., 2019).
Sweet Osmanthus (Osmanthus fragrans) is an evergreen woody flowering member of the Oleaceae. Because of its pleasing floral fragrance and multitudinous flowers, sweet osmanthus is widely cultivated in China as a common landscape tree species (Huang et al., 2015). Sweet osmanthus is relevant to several industries, including tea, food, and perfume production (Li et al., 2020a). The floral fragrance of sweet osmanthus has been comprehensively studied in terms of compound identification and classification. Studies have identified that β-ionone is an essential substance that strongly affects the floral fragrance of sweet osmanthus, and CCD4 is a vital enzyme regulating the formation of β-ionone (Han et al., 2016). Previous studies have confirmed that OfCCD4 increases the production of β-ionone, based on the existence of β-carotene, and both OfWRKY3 and OfERF61 are positive regulators of the OfCCD4 gene and enable the synthesis of β-ionone in petals of sweet osmanthus (Han et al., 2016, 2019). According to data on the sweet osmanthus genome, we found that the OfCCD4 promoter region contains numerous cis-acting regulatory elements, thus indicating that OfCCD4 might be regulated by other TFs (Yang et al., 2018). However, functional identification of related TFs and examination of the transcriptional regulation mechanism of floral volatile organic compounds (VOCs) have been limited. Therefore, studies on the MYB-related TFs’ regulation of enhancing floral VOC production are needed to improve understanding of the mechanism of VOC production and the genetic quality of sweet osmanthus.
Here, we conducted a comprehensive analysis of sweet osmanthus’ MYB-related TF family members, including their potential roles in regulating the formation of floral VOCs. In this study, 212 MYB-related members were identified from the genome of sweet osmanthus, and their chromosomal locations, conserved motifs, and expression patterns were analyzed and visualized. Additionally, we selected three candidate MYB-related genes that had flower organ/development stage-preferential expression patterns and then performed detailed gas chromatography-mass spectrometry (GC-MS) analysis of floral VOCs between transient expression plants and empty vector plants.
Sweet osmanthus genomic sequences were acquired from the genome database for O. fragrans. The sequence of the MYB DNA binding domain (DBD) was downloaded from the Pfam database (http://pfam.xfam.org/, accession PF00249) (Finn et al., 2016), and HMMER software (version 3.0) with a default E-value. The NCBI Batch Web CD-Search Tool (Marchler-Bauer and Bryant, 2004) was further used to assess the numbers of DBDs in the sequences and retain the genes with one DBD. Finally, the putative MYB-related genes, according to the number of DBDs and the position information extracted from the General Feature Format (GFF) files, were identified and renamed according to their locations on the chromosomes. The online tool ExPASy was used to verify the physicochemical characteristics of these MYB-related protein sequences, such as the theoretical isoelectric point (pI) and molecular weight.
A total of 132 AtMYB-related protein sequences1 and 212 putative OfMYB-related proteins were selected to construct a phylogenetic tree through neighbor-joining method with a bootstrap score of 1,000 and p-distance parameter (MEGA7.0). The complete amino acid sequences of OfMYB-related proteins were used to obtain the DBDs and validated in the Pfam database. The alignment and visualization of 212 OfMYB-related domains and subsequent protein analysis were accomplished with ClustalX.
The OfMYB1R gene structures and coding sequences (CDSs) were derived from the sweet osmanthus genomic GFF3 file. TBtools software was used to visualize the gene structures, CDS boundaries, and intron distribution (Chen et al., 2020). The MEME Suite webserver was used to screen the conserved motifs in OfMYB-related TFs, identifying a maximum of 20 motifs2 (Bailey et al., 2009).
The chromosomes corresponding to MYB-related genes were screened and mapped with MG2C.3 The Multiple Collinearity Scan toolkit (MCscanX) was used to analyze the tandem and segmental duplication pattern of each MYB-related gene.4 Ks (synonymous) and Ka (non-synonymous) substitution ratios of gene pairs were evaluated with TBtools software (Chen et al., 2020).
Total RNA of sweet osmanthus was extracted from the bud-pedicel (S1), bud-eye (S2), primary blooms (S3), full bloom (S4), and post-bloom flower (S5) stages. Three biological samples were collected and processed with the RNAprep Pure Plant Kit (Tiangen, Beijing, China). After removing the gDNA from the total RNA, cDNA was synthesized with SuperMix (Transgen, Beijing, China) under the following conditions: 42°C for 15 min and 85°C for 5 s (Yue et al., 2021). The qRT-PCR primers for OfMYB-related genes were designed in Primer Premier 5.0. The expression of OfACTIN, OfRAN, and OfRBP2 genes served as average references for normalization of the expression levels in different flower stages (Zhang et al., 2015) (Supplementary Table 1). Each qRT-PCR assay was performed on three biological samples and three technical replicates. Data of comparative threshold cycle (Ct) values were used to calculate the relative gene expression levels (Yue et al., 2021).
To construct 35S:OfMYBMYB1R70/114/201:GFP, the Super 1,300 vector containing the CDS of OfMYB-relared70/114/201, without the stop codon was used. SmaI and KpnI restriction sites were utilized. Agrobacterium tumefaciens (GV3101) containing 35S:OfMYB1R70/114/201:GFP and negative control vectors were infiltrated into tobacco (Nicotiana benthamiana) leaves for the analysis of subcellular localization. The tobacco plants were grown for 35 days in a growth chamber with 15/9 h light/dark (144 μmol⋅m–2⋅s–1) and 26 ± 2°C. After 3 days of growth, green fluorescent protein (GFP) fluorescence signals were confirmed in the infiltrated plants via DAPI staining with a LSM710 microscope (Zeiss, Germany).
Additionally, through the NdeI and EcoRI restriction sites, the CDS of OfMYB-relared70/114/201, lacking the termination codon, were transformed into the pGBKT7 vector. Then the Saccharomyces cerevisiae yeast strain AH109 (WeidiBio, Shanghai, China) was used to obtain the four vectors (three types of pGBKT7-OfMYB1Rs and one negative control). Finally, transformed yeast with overexpression vectors were grown in selective defined SD/-Trp, SD/-Trp-Ade, and SD/-Trp-Ade + X-α-gal media with incubation at 30°C for 3 days in the dark.
To explore the regulatory function of OfMYB1R70/114/201 genes in volatile metabolic components, we selected tobacco leaves to analyze floral VOCs by transient expression. The method to determine the transient expression of candidate genes in Nicotiana benthamiana leaves has been successfully used to explore the functions of genes involved in VOC synthesis, such as OfTPS1/2 (Zeng et al., 2016), AtTPS10/14 (Ginglinger et al., 2013), and MpGPS.SSU (Yin et al., 2017). GV3101 containing 35S:OfMYB1R70/114/201:GFP and empty vectors were used to infiltrate 35-day-old tobacco leaves. Six biological replicates of each OfMYB1R sample were injected after 48 h, and the empty vectors were injected in four biological replicates. After growth for 2 days (light/dark: 15/9 h, 144 μmol⋅m–2⋅s–1), headspace solid-phase microextraction (SPME), a precise method for measuring aroma, was used to collect the floral VOCs. In every 4-ml SPME vial, 0.4 g samples of fresh leaves were added to the bottom, and 1 μl of 100000-fold diluted ethyl caprate was added to the middle (Shanghai, Macklin Inc., China) (Yang et al., 2018). After exposure at the middle of the capped vials for 35 min at 42°C, the SPME fibers of DB-5 MS were injected into the heated syringe port for 3 min, and desorption was performed at 250°C (Bellefonte, Supelco Inc., United States). Subsequently, the desorbed floral VOCs were identified with the Trace DSQ GC-MS device (Ji, 2021), and the N-alkanes were used to calculate the RI value of the volatile compounds for comparison with the RIL value within DB-5MS (30 × 0.25 × 0.25) to confirm the aroma compound detection5 (Ji, 2020, 2021). In the software SIMCA 13.0 (Umetrics AB, Umea, Sweden), divergence in the metabolite levels of plants with transient expression was detected with partial least squares discriminant analysis (PCA-X).
To validate the empty vector and 35S:OfMYB1R70/114/201:GFP expression in transgenic tobacco plants, we snap-froze the leaves of 35-day-old tobacco with transient expression 48 h after injection of the vector (light/dark: 15/9 h, 144 μmol⋅m–2⋅s–1) in liquid nitrogen. According to the RNAprep Pure Plant Kit (Tiangen, Beijing, China) cDNA transcribed from total RNA isolated from the leaves of transgenic plants and empty vector plants was used as the template (Ding et al., 2019). Each OfMYB1R assay was performed on six biological samples, and empty vector plants were performed on four biological replicates. The primers of OfMYB1Rs for semi-quantitative RT-PCR (RT-PCR) were the same primers used in the qRT-PCR analysis. The gene NbL25 was used as a reference to normalize the expression levels in tobacco (Supplementary Table 1). The primers for NbCCD4s and NbLCYB, which are homologs of OfCCD4 and OfLCYB, were selected to verify the participation of candidate genes in regulating floral aroma synthesis (Supplementary Table 1).
Utilizing the MYB DBD HMM profile (Pfam ID PF00249), MYB and MYB-like domains for the entire sweet osmanthus genome were screened. According to the results, verified by Pfam and NCBI-CDD analysis, a total of 212 non-redundant OfMYB1R protein sequences were identified, a number greater than that for most species (Table 1). Based on the chromosomal position of the MYB-related genes, these genes were successively renamed OfMYB1R1 to OfMYB1R212. Basic information on OfMYB-related genes, such as gene ID, renamed name, chromosomal location, pI, and molecular weight, can be found in Supplementary Table 2. The protein sequences are provided in Supplementary Table 3. The total length of the OfMYB-related protein sequences ranged from 82 to 1,816 amino acids, the molecular weights ranged from 9.21 to 199.69 kDa, and the predicted pI values were 4.49–10.1.
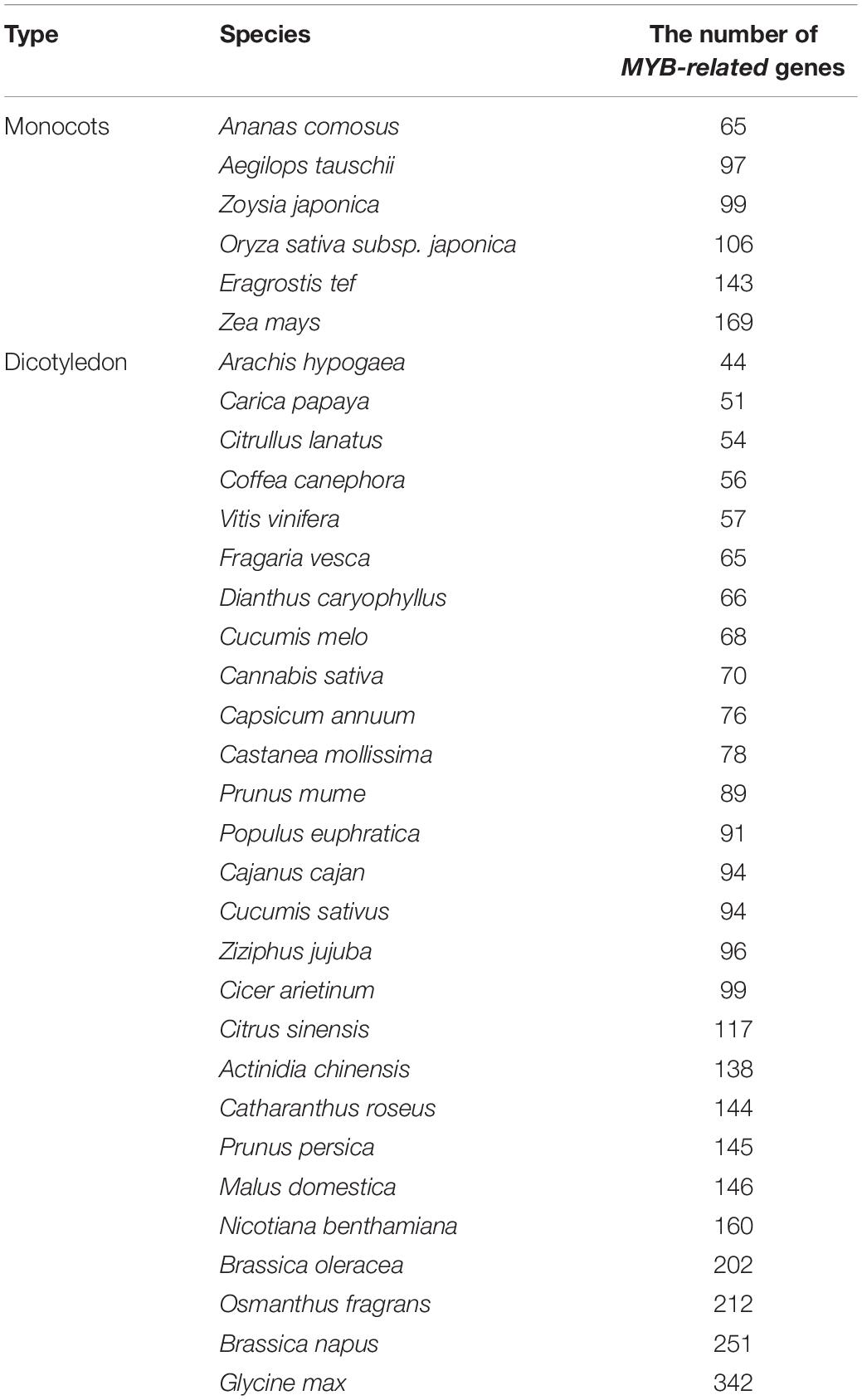
Table 1. Comparison of the MYB-related gene family size between Osmanthus fragrans and other species.
Additionally, 132 AtMYB-related proteins were analogously identified from the A. thaliana genome (Dubos et al., 2010; Liu et al., 2020). The phylogenetic and evolutionary relationships between the 344 AtMYB- and OfMYB-related proteins were analyzed (Figure 1). In accordance with the classification of the MYB-related proteins of Arabidopsis, 212 OfMYB-related proteins were divided into three subgroups: CPC-like, GARP-like (MYB_CC, ARR, HHO, LUX, KANADI, and GRAP-related, considered an unknown subfamily), and a heterogeneous subgroup (consisting of CCA1-like/R-R, TRF-like, TBP-like, RAD-like, and atypical MYB-related proteins).
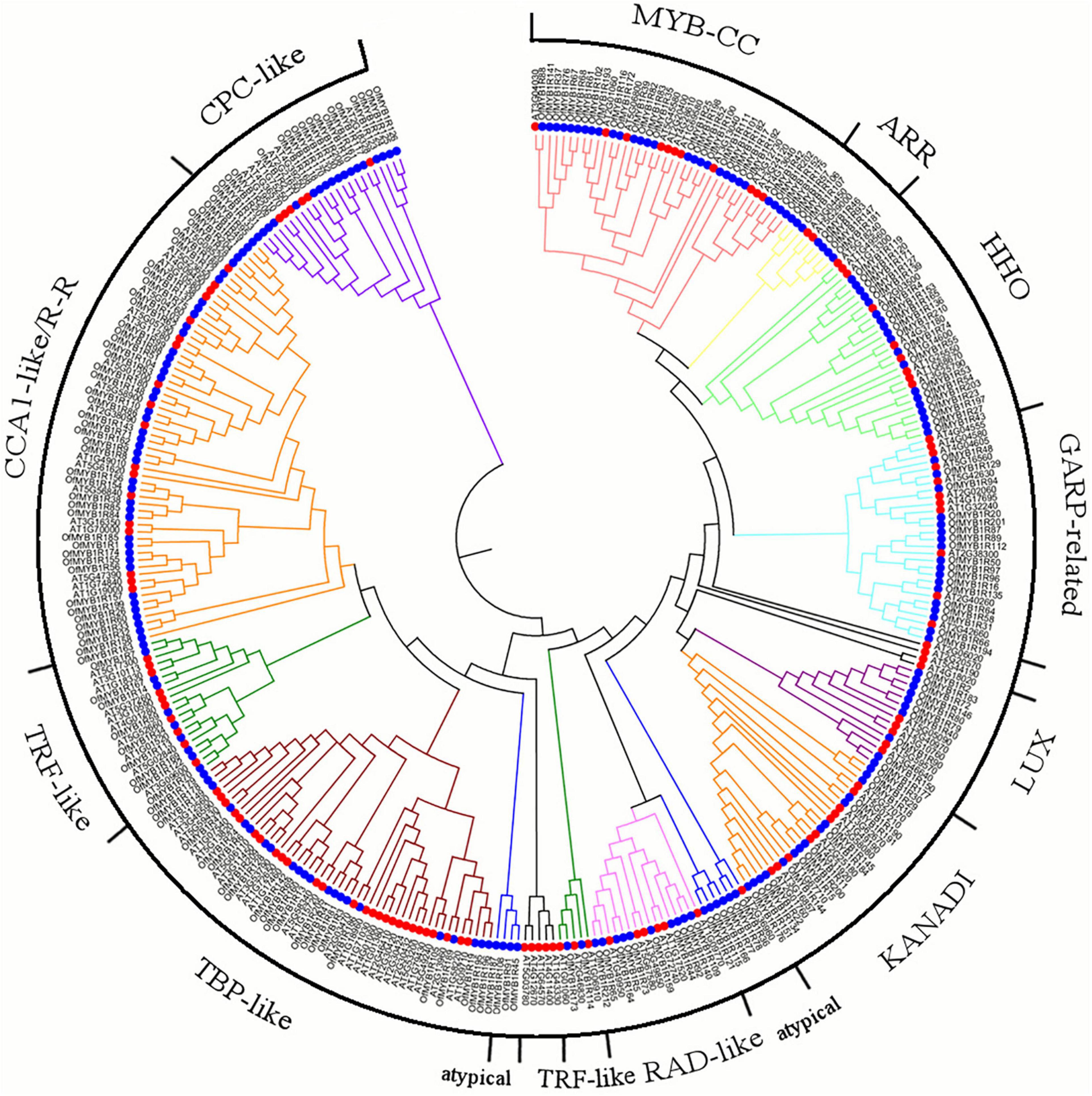
Figure 1. Phylogenetic relationship between the 212 O. fragrans (blue circles) and 132 Arabidopsis (red circles) MYB-related proteins, illustrated by a maximum likelihood (ML) tree. Alignment was performed with the ClustalW program, and these proteins were imported to MEGA 7.0 to construct the phylogenetic tree with 1,000 bootstrap iterations. MYB1Rs represents OfMYB-related proteins.
The three subgroups, comprising 212 OfMYB-related proteins, were further classified into 12 categories according to highly conserved motifs. The GARP-like subgroup was divided into six clades, whereas the CCA1-like/R-R subgroup consisted of five clades (Figure 2). Moreover, 22 OfMYB-related proteins in the TBP-like subgroup were split into three major clades whose sequence alignments implied high similarity within each clade, thus indicating that these genes may have similar functions.
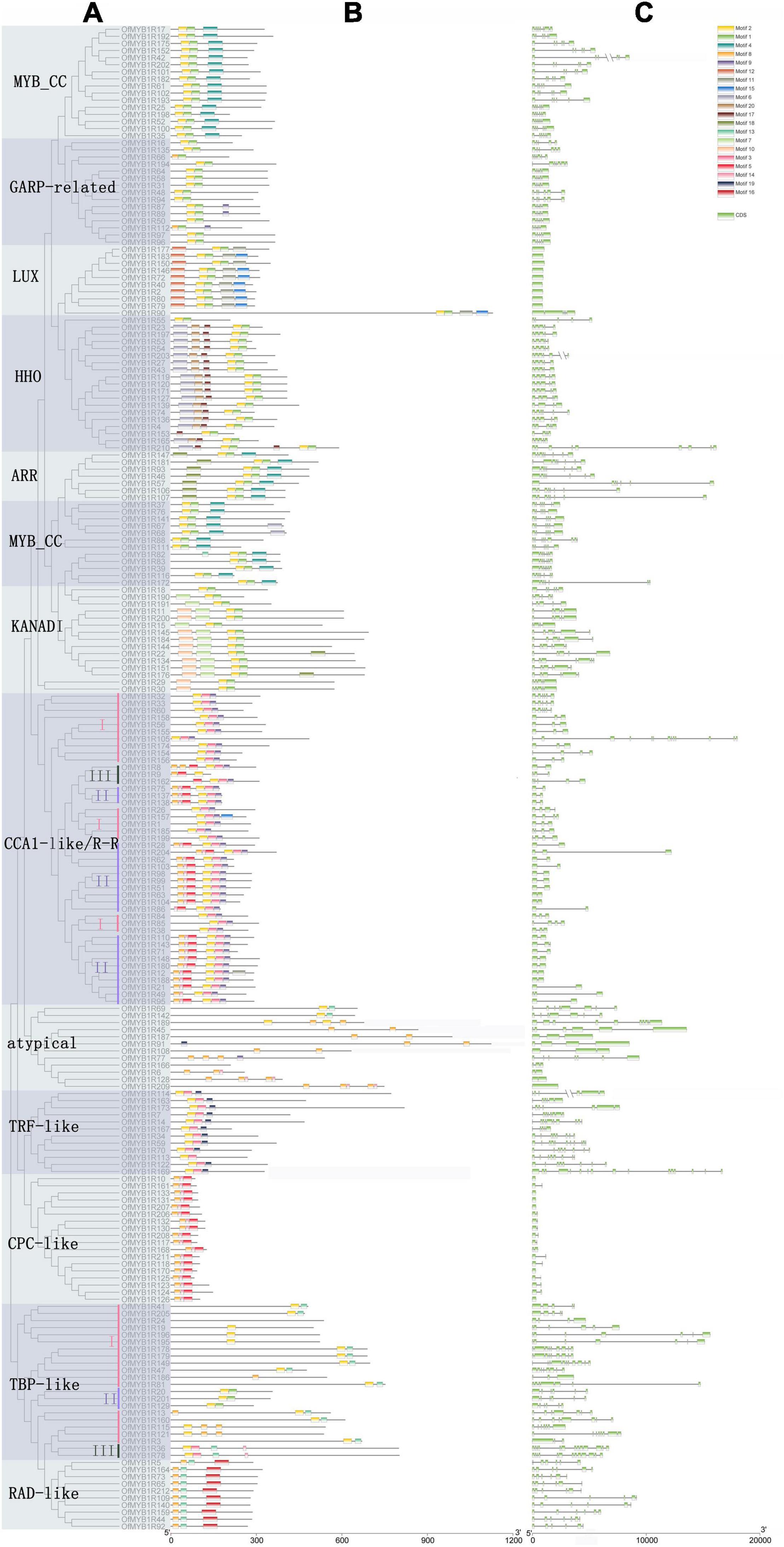
Figure 2. Phylogenetic relationships of 212 OfMYB-related proteins, visualized by a ML tree (A). Conserved motifs of OfMYB-related proteins (B). Exon/intron structures of OfMYB-related genes. Exon(s) and intron(s) are shown with green boxes and gray lines, respectively (C). The 20 conserved motifs in the OfMYB-related TFs were predicted by the MEME Suite web server. I–III indicate clades I–III.
The largest subgroup was GARP-like, which contained 94 OfMYB-related proteins, comprising MYB_CC, LUX, HHO, KANADI, ARR, and GARP-related, with diversely conserved motifs and CDS patterns (Figure 2); however, the GLK subfamilies did not have BLAST results. The GARP-like subgroup was categorized by the conserved motif Wx3LHx2Fx6L(G)Gx5PK/Sx5Mx4L and the DNA-binding helix 3 domain, harboring the SH(A/L)QK(F/Y) sequence, which is commonly found in diverse plant genomes (Antje et al., 2011). Apart from the common conserved motifs, several unique motifs were detected in different subfamilies, such as the LHEQLE motif in MYB_CC, which contained 28 members and was the largest clade. The smallest clade belonged to ARR, with seven proteins. The second-largest subgroup, with three W residues, consisted of the CCA1-like/R-R, TBP-like, TRF-like, RAD-like, and an atypical MYB-related clade. Notably, the TBP-like subgroup was classified into three distinct clades based on remarkably conserved motifs, and the most common conserved motif of TBP-like was LKDKW(R/K) (N/T) (Supplementary Figure 1). The smallest subgroup, containing only 44 OfMYB-related proteins, was CCA1-like/R-R, an R3-type MYB TF, which had a highly conserved SHAQK(Y/F) (F/Y) motif in the MYB domain third helix—the distinctive characteristic in this subgroup (Supplementary Figure 1).
The 212 OfMYB-related genes were located on all of the 23 uneven sweet osmanthus chromosomes (Figure 3). Remarkably, chromosome (Chr) 03 contained the largest number of OfMYB-related genes (17), followed by Chr08 and Chr11 (each with 15 genes), and then Chr01 (14 genes). In contrast, only two genes were mapped to Chr23 and Chr06. The pattern of OfMYB1Rs followed a random distribution (sig < 0.05) (Supplementary Figure 2).
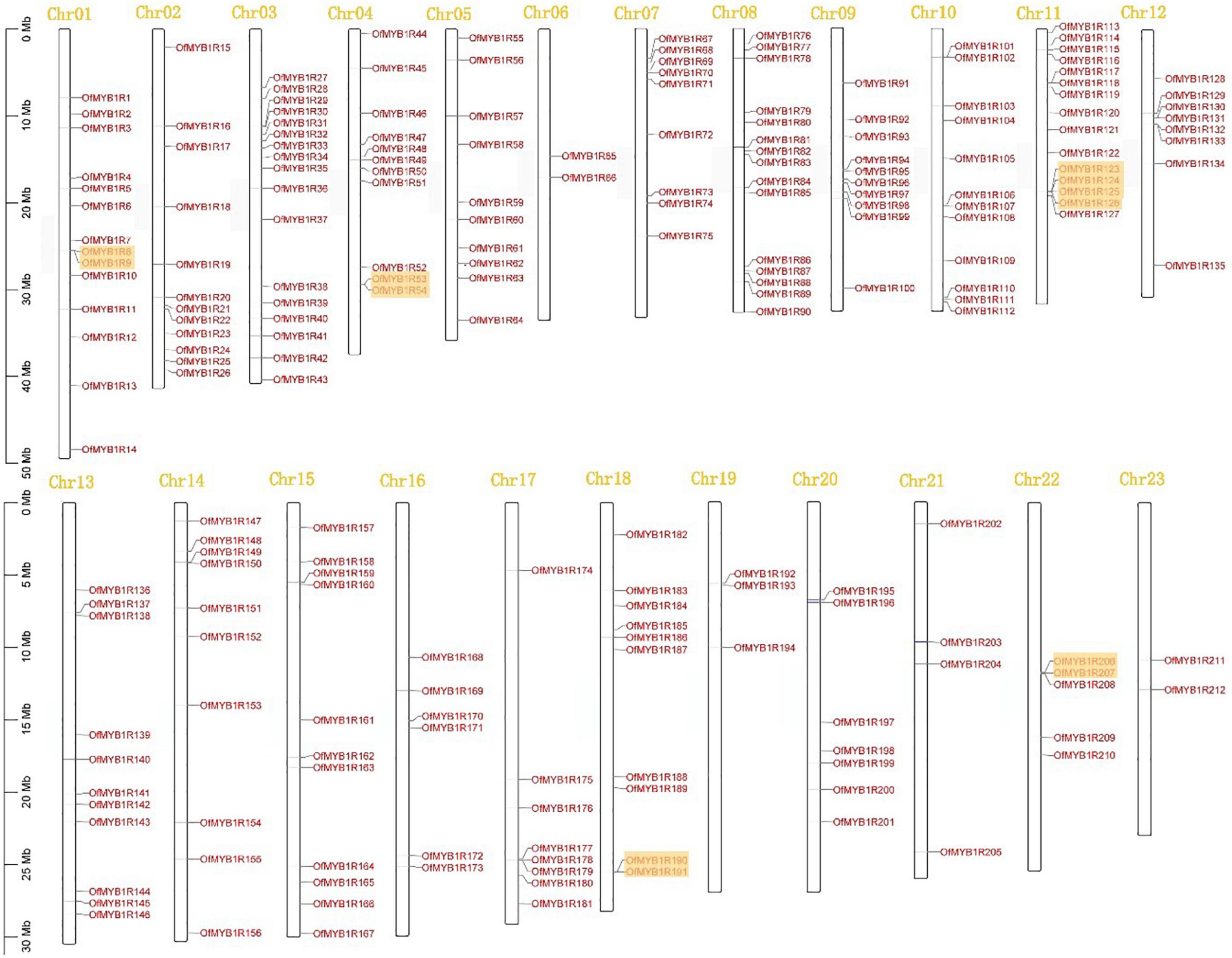
Figure 3. Chromosomal locations of 212 OfMYB-related genes, mapped on the basis of the GFF files of O. fragrans. The numbers at the top represent each chromosome. Orange boxes represent clusters of tandemly duplicated genes.
Gene duplication plays a crucial role in driving genetic diversity and is common in plant genomes. This research identified seven tandemly duplicated pairs of 12 OfMYB-related genes and 68 segmentally duplicated out of the 212 OfMYB-related genes. Tandemly duplicated gene pairs were found only in chromosomes 01, 04, 11, 18, and 22. Chr11 had three pairs of tandemly duplicated genes continuously, and segmentally duplicated genes were found on all chromosomes (Figure 3). The tandemly duplicated cluster on Chr11 provides interesting insight into the expansion pattern of sweet osmanthus expansion genes in this large gene family. Moreover, 68 segmental duplication events were distributed across all chromosomes in the synteny blocks (Supplementary Figure 3). Every tandemly duplicated OfMYB-related gene pair had a Ka/Ks value less than 1, which evolved in purifying selection; in particular, duplicated gene pairs of OfMYB1R124 (Chr11) VS. OfMYB1R125(Chr11) and OfMYB1R125 (Chr11) VS. OfMYB1R126(Chr11) were significantly different from 1 (Supplementary Table 4). Based on these results, we concluded that some OfMYB-related genes originated from gene duplication events, whereas the evolution of the OfMYB-related genes was influenced by segmental duplication.
The expression patterns of OfMYB-related genes in different organs and flower developmental stages were analyzed with RNA-sequencing (Supplementary Table 5). Four sweet osmanthus organs were sequenced and used to acquire the reads per kilobase per million mapped reads (RPKM) values of 194 OfMYB-related genes, whereas the values of the remaining 17 genes were not found. Eight OfMYB-related genes were observed in a single stage: three genes were observed only in the root or primary-bloom stage, two genes (OfMYB1R74 and OfMYB1R156) were observed only in the stem stage. The remaining 186 OfMYB-related genes showed different expression patterns, and 142 were present in all samples. Based on the of the conserved motif analysis results, 194 expressed OfMYB-related genes were classified into three clear subgroups. Most members of the CCA1-like/R-R subgroup had high expression in leaves as well as the primary- and full-bloom stage of flowers, whereas a wide range of CPC subgroup genes were expressed mainly in the root, stem, and flower fading (S5) stages. The members of the GARP subgroup showed particularly high expression in the root (Figure 4). Remarkably, in the CCA1-like/R-R subgroup, three OfMYB-related genes (OfMYB1R70, OfMYB1R114, and OfMYB1R201) had the highest expression in the flower organ/development stage, thus indicating a preferential expression pattern.
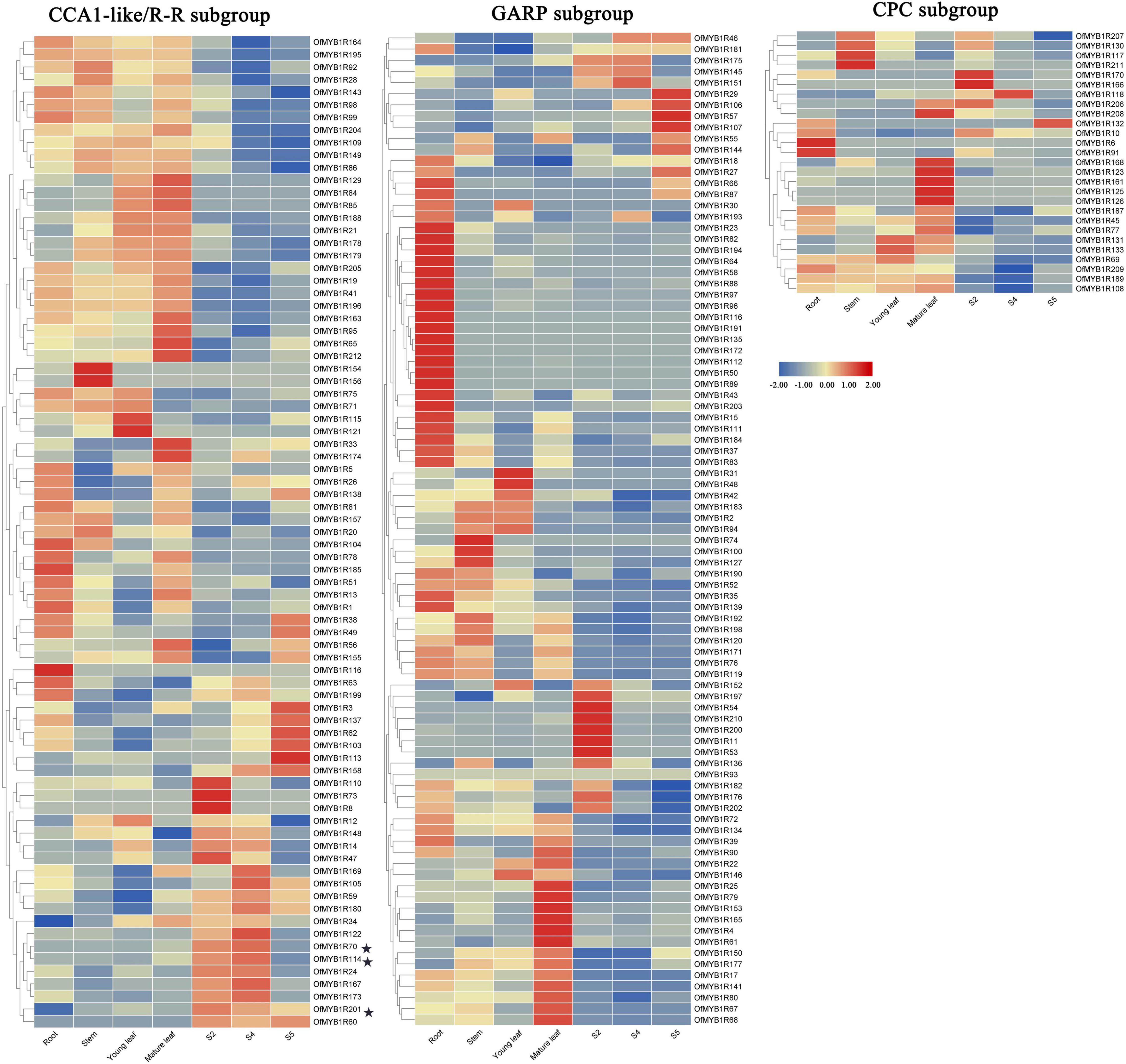
Figure 4. Expression analysis of 194 OfMYB-related genes in seven organs. The RPKM values were log2 transformed. S2 represents the bud-eye stage, S4 the represents full blooming stage, and S5 represents the flower fading stage. Genes with no transcriptome data are not shown. The candidate genes in the biosynthesis of floral aroma are marked by asterisks.
According to the expression patterns and RPKM values of OfMYB-related genes (Figure 4), three OfMYB-related genes (OfMYB1R70/114/201) of the CCA1-like/R-R subgroup were further selected for validation experiments in five flowering stages. The results were reflected those of the RNA-seq experiment, thus verifying the accuracy of the RNA-seq data. The expression of OfMYB1R70 was similar to that of OfMYB1R114, which was generally up-regulated during flowering (S1 and S3). Meanwhile, S3 had the lowest expression, and other processes were down-regulated. Interestingly, the expression of OfMYB1R70 in S3–S5 was similar to that of OfMYB1R114, but the expression of OfMYB1R70/114 was considerably elevated in stages S3–S4, whereas the expression of OfMYB1R201 increased slightly in the same stages (Figure 5). For reliable normalization, we selected three reference genes, ACTIN, RAN, and RBP2, for the qRT-PCR analysis. The expression levels of OfMYB1Rs were obtained from the average of three reference genes (Figure 5), and the independent expression results of ACTIN, RAN, and RBP2 are shown in Supplementary Figure 4.
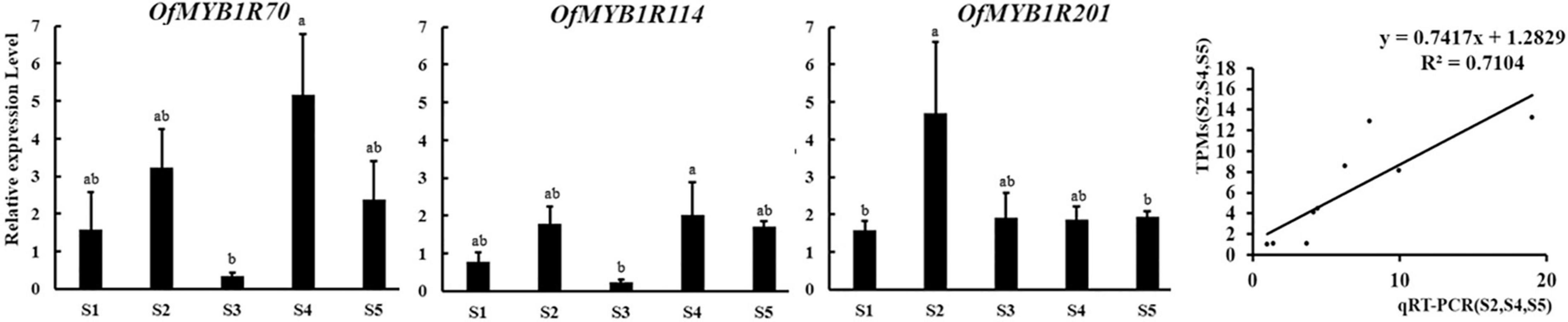
Figure 5. Expression patterns of three candidate OfMYB-related genes using the average of three reference genes (ACTIN, RAN, and RBP2) in different stages of sweet osmanthus flowers. Three biological and technological replicate measurements were performed for each stage, and the data are shown as the mean ± SD (n = 3) at the top of each bar. The small letters above the error bars indicate significant differences obtained in SPSS 25 with Tukey’s test at P < 0.05. S1–S5 represent the bud-pedicel stage, bud-eye stage, primary blooming, full blooming, and flower fading stage, respectively, on the abscissa.
To thoroughly examine the potential function of OfMYB-related genes in transcriptional regulation, we transformed tobacco leaves to assess the subcellular localization. The GFP fluorescence of the p35S:OfMYB1R70:GFP fusion protein was intense in the nucleus, as confirmed by DAPI staining (Supplementary Figure 5). The fluorescence of p35S:OfMYB1R114:GFP, like that of the control GFP protein lacking OfMYB-related genes, was dispersed in the cytoplasm and nucleus (Supplementary Figure 5). In contrast, the fluorescence of the p35S:OfMYB1R70/201:GFP fusion protein was detected in the nucleus and was dispersed in the plasma membrane. To further identify the transcriptional activation functions of the OfMYB-related genes, we cloned the CDSs of selected OfMYB-related genes into the yeast expression vector pGBKT7. The negative control pGBKT7 and OfMYB-related genes showed comparable growth on SD/-Trp medium with serial dilutions (Figure 6). Nevertheless, except for pGBKT7–OfMYB1R201, no yeast cells containing pDEST-GBKT7-OfMYB1R plasmids survived and turned blue on SD/-Trp-Ade medium with X-α-Gal.
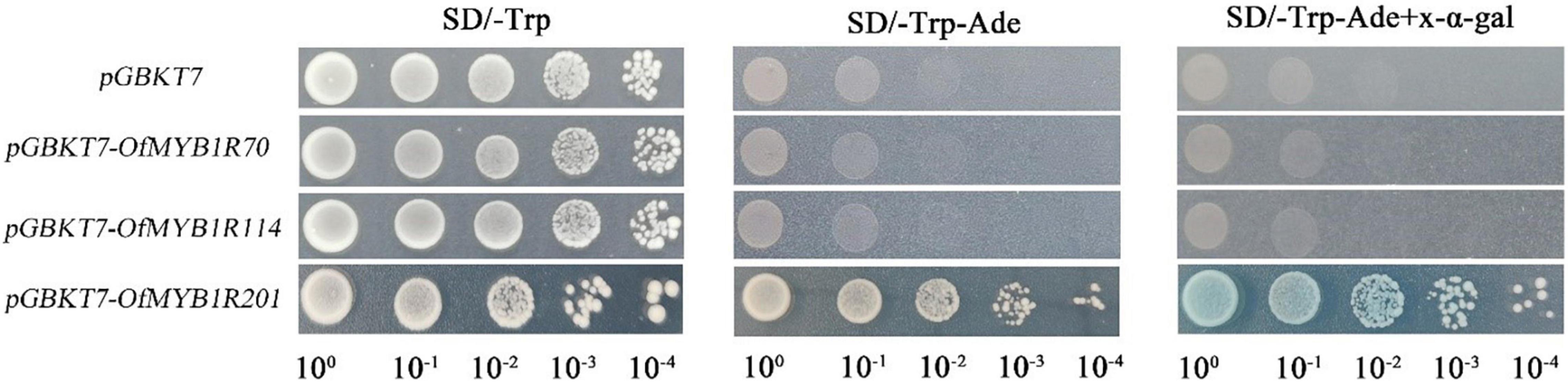
Figure 6. Transcriptional activation analysis of candidate OfMYB1Rs. Growth of transformants containing pGBKT7 and pGBKT7-OfMYB1R70/114/201 on SD/-Trp medium with serial dilutions. Only pGBKT7-OfMYB1R201 survived and turned blue on SD/-Trp-Ade medium in the presence of X-A-gal.
To investigate the volatile metabolic components, we obtained the GC-MS total ion current chromatograms of 22 tobacco samples from four types of plants with transient expression. Approximately 23 volatile compounds were identified in the plants with transient expression, including esters, phenolics, ketones, alkanes, and monoterpenes (Supplementary Table 6). PCA-X plots, generated by comparing three types of plants with transient expression and empty vector (NC), showed significant metabolic differences (Figure 7). Notably, according to two principal components, tobacco leaves infiltrated with 35S:OfMYB1R70/114/201:GFP differed from those with control vectors in the production of volatile aromatic compounds. OfMYB1R70/114 and NC clustered separately in the PCA-X. OfMYB1R201 was slightly scattered in the PCA (Figure 7), thus implying a role in regulating the production of volatile aromatic compounds. Among the three types of plants with transient expression, the plants with transient OfMYB1R114 expression had a greater ability to regulate aromatics synthesis than the other two types. The comprehensive regulation pattern of OfMYB1R114 was greater than OfMYB1R70, in view of the number of differentially present substances satisfying VIP > 1 and P < 0.05.
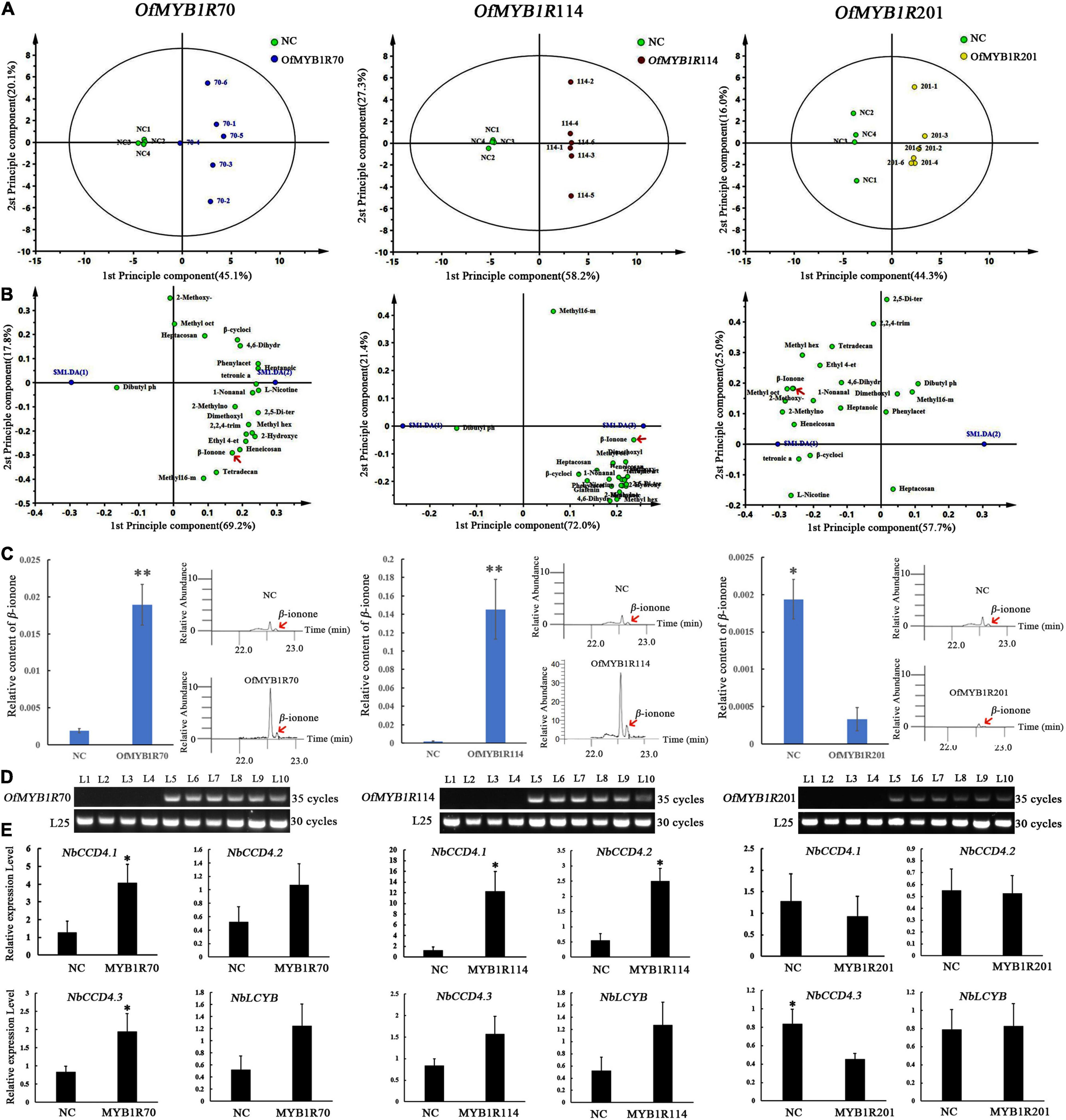
Figure 7. Score scatter plot of the leaf GC-MS profiles of three types plants with transient expression of different MYB1R transcription factors in in tobacco, determined with PCA-X analysis in SIMCA13.0 (A). Loading scatter plot analysis of floral VOCs in plants after infiltration of 35S:OfMYBMYB1R70/114/201:GFP and control vectors. The red arrow marks the floral aroma in plants with transient expression (B). Floral composition analysis of β-ionone after infiltration with 35S:OfMYBMYB1R70/114/201:GFP and control vectors (C). RT-PCR to explore the regulation of gene transcription OfMYBMYB1R70/114/201 levels. A fragment of the NbL25 gene was amplified as an internal control. L1–4 represent different plants infiltrated with empty vectors, and L5–10 represent different plants infiltrated with 35S:OfMYB1R:GFP vectors (D). qRT-PCR analysis of the NbCCD4 transcript levels in control and transiently transformed tobacco leaves (E). *P < 0.05, **P < 0.01.
Interestingly, further analysis revealed that OfMYB1R114 regulates the synthesis of six esters, two ketones, one monoterpene, one alkane, and one aldehyde, which showed significant discrepancies in the GC-MS analysis of the aroma content with VIP > 1 and SPSS with P < 0.05. The substances that slightly changed included floral VOCs such as β-ionone, nonanal, and benzeneacetaldehyde (Supplementary Table 6). Notably, OfMYB1R114 mainly altered the production of nonanal, which is common in rose and sweet orange, and caused a clear difference in β-ionone. Interestingly, in agreement with the results of correlation analysis, OfMYB1R114 positively regulated the production of β-cyclocitral. OfMYB1R70 is predominantly involved in the synthesizing of four esters and two ketones, and the most striking change in floral fragrance compounds was observed for β-ionone, a significant food flavor. In contrast, OfMYB1R201 showed significantly negative regulation of four esters, one ketone, one aldehyde, and one naphthenic compound, and the production of floral VOCs (nonanal and β-ionone) was comparatively low. Simultaneously, the results of RT-PCR demonstrated that OfMYB1R70/114/201 were transiently expressed in the plants. OfMYB1R70/114/201 was detectable in 35S:OfMYB1R:GFP plants with transient expression, whereas OfMYB1R70/114/201 expression was undetectable in transformed empty vector plants. According to the qRT-PCR results, NbCCD4.1 was significantly up-regulated in plants with transient expression of OfMYB1R70 and OfMYB1R114, and NbCCD4.2 was markedly elevated in the plants with transient expression of OfMYB1R114. Interestingly, NbCCD4.3 was significantly elevated in the plants with transient expression of OfMYB1R70 and dramatically diminished in the plants with transient expression of OfMYB1R201 (Figure 7). These results indicated that the transient overexpression of OfMYB1R70 activated the expression of NbCCD4.1 and NbCCD4.2; OfMYB1R114 might have activated NbCCD4.1 and NbCCD4.3; and OfMYB1R201 repressed the transcription of NbCCD4.3 in tobacco plants, thus leading to the changes in β-ionone content in the plants with transient expression of the three OfMYB1Rs.
To date, MYB TFs, mainly comprising MYB-related and R2R3 MYB, modulate the transcription levels of numerous essential genes in secondary metabolism biosynthetic pathways, thereby regulating the production of diverse floral VOCs (Jian et al., 2019). Several MYB members have been identified as a conserved module regulating terpene synthesis. MsMYB, a novel negative regulator in Mentha spicata associated with the promoter region of geranyl diphosphate synthase (GPPS), decreases the production of monoterpenes by inhibiting the activity of GPPS (Reddy et al., 2017). Recently, the function of R2R3MYBs in floral aroma formation in Hedychium coronarium has been extensively reported (Abbas et al., 2021). However, further studies of MYB-related proteins, particularly the close relationship between the function of MYB-related TFs and floral aroma synthesis, remain unclear.
The number of OfMYB-related genes was more significant than that observed in other plants reported over the recent years (Table 1; Jin et al., 2017), such as 109 genes in peach (Zhang et al., 2018). In our study, we identified 212 MYB-related genes in O. fragrans, a value close to the number of R2R3-MYB genes in O. fragrans (243 genes) (Li et al., 2020a), thus representing the largest subfamily of MYB genes, which play critical roles in various processes of growth, development, and responses to abiotic or biotic stress. While high, the number of R2R3-MYB subfamily members did not exceed the number of 1R-MYB subfamily members. The amount of 1R-MYBs in Ganoderma was twice the number of R2R3-MYBs. Moreover, previous studies have performed genome-wide analyses of MYB-related proteins from 16 species of flowering plants, moss, Selaginella, and algae (Du et al., 2013). In contrast to the detailed studies on the R2R3-MYB subfamily, little attention has been paid to the MYB-related (1R-MYB) subfamily, which has been reported only in a few common crops in recent years: potato, sunflower (Helianthus annuus), radish (Raphanus sativus), and oil palm (Elaeis guineensis) (Li et al., 2020b; Liu et al., 2020; Zhou et al., 2020; Muleke et al., 2021).
As an important part of the MYB family, MYB-related genes were initially classified into five subgroups: the CCA1-like, CPC-like, TBP-like, I-box-binding-like, R-R-type (Zhang et al., 2018). However, after detailed studies on the GARP family, MYB-related genes were divided into three subgroups: CCA1-like/R-R, CPC-like, and GARP-like (Liu et al., 2020). After extensive further studies, a merger occurred in the Circadian Clock Associated 1-like and R-R-type, equivalent to the CCA1-like/R-R subgroups in this study; the results are consistent with those of the most recent studies (Liu et al., 2020). In previous work, CCA1-like proteins have been found to be closely associated with R-R type MYB repeats, thus implying that R-R-type gene loss events may directly lead to CCA1-like proteins. In this classification, 44 members of the CCA1-like/R-R group contained the conserved motif SHAQK (motif 2 in Supplementary Figure 1) in the MYB repeat, in agreement with previous studies in Arabidopsis (Ji et al., 2019). The CAPR-like subgroups initially comprised maize G2-like and Arabidopsis ARR-B, which were newly derived in angiosperms. In agreement with previous research, the GARP-like subgroups were classified into six clades in this identification (Liu et al., 2020).
As a typical MYB member, the CPC-like subgroup has been reported to be involved in the basic function of regulating root-hair development and the plant trichome. It has been confirmed to regulate the synthesis of secondary metabolites in recent years. For instance, AtCPC, characterized as a repressor of anthocyanin production, has provided novel insight into the transcriptional regulation of flavonoid biosynthesis. SlTRY and SlGL3, MYB-related homologs of tomato, also influence synthesis of anthocyanin (Jian et al., 2019). In contrast, AtMYBL2, an R3-MYB protein, has been found to restrain flavonoid synthesis. Interestingly, MYBL2 inhibits flavonoid biosynthesis and trichome development (Matsui et al., 2008). LlMYB3, a MYB related homolog from tiger lily, positively regulates the stress tolerance of plants and is involved in the anthocyanin biosynthesis pathway, thereby regulating stress tolerance (Yong et al., 2019).
In this expression analysis, OfMYB1R70, OfMYB1R114, and OfMYB1R201, which were preferentially expressed in floral organs, had higher RPKM values than other OfMYB1Rs, particularly OfMYB1R70 and OfMYB1R114, which had the top two RPKM values. Moreover, subcellular localization prediction indicated that they were mainly located in the nucleus, thus suggesting that these candidate genes may play important roles in regulating floral aroma synthesis during the bloom-stage of sweet osmanthus; therefore, they were selected for follow-up experiments. However, no prior studies have focused on the functions of MYB-related TFs in floral aroma biosynthesis. The relationship with the synthesis of floral VOCs has been reported only for 2RMYB (Reddy et al., 2017), although 2RMYB and MYB-related TFs jointly compose the main family in plants and are involved in many biological processes, particularly secondary metabolism, and responses to environmental factors, in divergent ways (Jian et al., 2019; Zhao et al., 2019).
Transcription factors have been reported to regulate the synthesis of floral VOCs by directly binding the promoter regions of target enzyme genes and controlling their expression levels. In H. coronarium, HcMYB genes directly bind the promoters of key structural volatile synthesis genes and regulate the biosynthesis of terpenoids and benzenoids (Abbas et al., 2021). In O. fragrans, OfERF61 up-regulates the expression of OfCCD4 and influences the synthesis of β-ionone (Han et al., 2019). In petunia, PhMYB4 controls the production of floral volatile benzenoid/phenylpropanoid by suppressing the downstream gene transcription (Thomas et al., 2011). PhERF6 binds the promoters of EOBI protein, which influences floral scent, thus negatively regulating the production of volatile compounds (Liu et al., 2017). In spearmint, MsMYB represses the expression of the GPPS gene, thus modifying secondary terpene metabolism (Reddy et al., 2017). In this study, we found that OfMYB1R201 is nuclearly located with self-activation activity its transient overexpression decreases the synthesis of β-ionone (Figure 7) and other VOCs in tobacco leaves (Supplementary Figure 6). The expression of NbCCD4.3 was significantly diminished in transformed plants with transient expression of the OfMYB1R201 vector (Figure 7). These results indicated that OfMYB1R201 could participate in negatively regulating VOC synthesis.
Remarkably, OfMYB1R70 and OfMYB1R114 were found to have nuclear localization (Supplementary Figure 5), and their expression during flower development was highly correlated (Figure 4). The transient transformation results showed that OfMYB1R70 and OfMYB1R114 were both involved in the regulation of aroma component synthesis, as confirmed by the markedly greater production of β-ionone and other emitted metabolites than that in the empty vector plants (Supplementary Figure 6). The expression of NbCCD4.1 and NbCCD4.3 was elevated in plants with transient expression of the OfMYB1R114 vector, and the expression of NbCCD4.1 and NbCCD4.2 was markedly elevated in plants with transient expression of the OfMYB1R70 vector (Figure 7). However, we found that OfMYB1R70 and OfMYB1R114 had no transcriptional activity, thus indicating that they do not directly activate the transcription of their downstream genes. Previous studies have reported that TFs interact with other proteins in regulating the synthesis of secondary metabolites by the MBW complex (Koes et al., 2005). For example, FhMYBx, containing a bHLH-binding motif in the R3 domains, suppresses the formation of the MYB-bHLH-WD40 (MBW) complex via an association with bHLH proteins, thus down-regulating anthocyanin metabolism (Li et al., 2020c). SmelMYBL1, an R3 MYB repressor in eggplant, acts as an inhibitor of the MBW complex by competing with MYB activators (SmelANT1 and SmelAN2) for the bHLH binding site involved in anthocyanin biosynthesis (Andrea et al., 2020). Hence, we concluded that OfMYB1R70/114/201 regulates aroma component formation by interacting with other TFs or acting as activators in competing pathways that alter the availability of precursors. The molecular mechanism of OfMYB1R70/114/201 in regulating the aroma metabolite synthesis will be further explored in following studies.
In this study, to provide novel insight into the regulation of floral aroma by all MYB-related genes in sweet osmanthus, we comprehensively identified 212 putative MYB-related members by genome-wide analysis. We categorized them into three subgroups, including 12 categories related to 132 MYB-related members of A. thaliana in the phylogenetic tree. Additionally, the information on conserved motifs in genes was visualized in figures, and the expression patterns indicated that three candidate OfMYB1Rs might be associated with the development and growth of flowers. Notably, members of the CC1-like/R-R subgroup in sweet osmanthus, OfMYB1R70/114/201, localized primarily in the nucleus. Remarkably, transient expression studies demonstrated that all those genes were associated with the regulation of floral VOC synthesis, and OfMYB1R114 had an apparent ability to positively regulate the synthesis of floral VOCs, such as β-ionone and nonanal, whereas OfMYB1R201, as a TF, negatively regulated floral VOCs. Together, our findings provide insight into OfMYB-related genes, explaining their roles in plants as regulators of the production of volatile aroma compounds. Our future efforts will focus on modulating the functions of OfMYB1R70/114/201 in regulating the production of floral aroma and investigating the roles of upstream regulatory factors.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YY and XnY conceived the original idea. XnY and XW performed the experimental work. XnY and WD contributed to the data analysis. XnY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant nos. 31870695 and 32071828) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Hannah Thomas for critical editing of the manuscript. We also thank Xiaoyue Ji, a lab master at the Advanced Analysis Testing Center (AATC), Nanjing Forestry University, China, for assistance in the detection of the plants with transient expression.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.765213/full#supplementary-material
Supplementary Figure 1 | Sequence logos of the MYB-related domains of subgroups.
Supplementary Figure 2 | The classification and frequency of each subgroup on each chromosome.
Supplementary Figure 3 | Tandemly and segmentally duplicated genes visualized in the O. fragrans genome.
Supplementary Figure 4 | The expression patterns of three candidate OfMYB1Rs, on the basis of three reference genes (ACTIN, RAN, and RBP2).
Supplementary Figure 5 | Subcellular localization of candidate OfMYB-related proteins.
Supplementary Figure 6 | Floral composition analysis after infiltration with 35S:OfMYBMYB1R70/114/201:GFP and control vectors (VIP > 1 and P < 0.05).
Supplementary Figure 7 | OPLS-DA plots generated from comparisons among three types of plants with transient expression and negative control vector plants with transient expression (NC).
Supplementary Figure 8 | The process of transient expression in N. benthamiana.
Abbas, F., Ke, Y. G., Zhou, Y. W., Yu, Y. Y., Waseem, M., Ashraf, U., et al. (2021). Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12:623742. doi: 10.3389/FPLS.2021.623742
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Andrea, M., Francesco, E. F., Sergio, I., Alessandra, G., Maria, M. A., Cinzia, C., et al. (2020). Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena L.). PLoS One 15:e0232986. doi: 10.1371/journal.pone.0232986
Antje, F., Katja, M., Braun, E. L., and Erich, G. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Bailey, T. L. Bodén, M., Buske, F. A., Frith, M. C., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. doi: 10.1093/nar/gkp335
Baranowskij, N., Frohberg, C., Prat, S., and Willmitzer, L. (1994). A novel DNA binding protein with homology to MYB oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G. Q., He, W. Z., Guo, X. X., and Pan, J. S. (2021). Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int. J. Mol. Sci. 22:4838. doi: 10.3390/IJMS22094838
Ding, W. J., Ouyang, Q. X., Li, Y. L., Shi, T. T., Li, L., Yang, X. L., et al. (2019). Genome-wide investigation of WRKY transcription factors in sweet osmanthus and their potential regulation of aroma synthesis. Tree Physiol. 40, 557–572. doi: 10.1093/treephys/tpz129
Du, H., Wang, Y. B., Xie, Y., Liang, Z., Jiang, S. J., Zhang, S. S., et al. (2013). Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 20, 437–448. doi: 10.1093/dnares/dst021
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L., et al. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., and Mitchell, A. L. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Ginglinger, J., Boachon, B., Höfer, R., Paetz, C., Köllner, T., Miesch, L., et al. (2013). Gene co-expression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25, 4640–4657. doi: 10.1105/tpc.113.117382
Han, Y. J., Wang, H. Y., Wang, X. D., Li, K., Dong, M. F., Li, Y., et al. (2019). Mechanism of floral scent production in Osmanthus fragrans and the production and regulation of its key floral constituents, β-ionone and linalool. Hortic. Res. 6, 1359–1380. doi: 10.1038/s41438-019-0189-4
Han, Y. J., Wu, M., Cao, L. Y., Yuan, W. J., Dong, M. F., Wang, X. H., et al. (2016). Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol. Biol. 91, 485–496. doi: 10.1007/s11103-016-0483-6
Huang, B., Chen, H. Q., and Shao, L. Q. (2015). The ethanol extract of Osmanthus fragrans attenuates Porphyromonas gingivalis lipopolysaccharide-stimulated inflammatory effect through the nuclear factor erythroid 2-related factor-mediated antioxidant signalling pathway. Arch. Oral Biol. 60, 1030–1038. doi: 10.1016/j.archoralbio.2015.02.026
Ji, Q., Wang, D. W., Zhou, J., Xu, Y. L., Shen, B. G., and Zhou, F. (2019). Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ 7:e6353. doi: 10.7717/peerj.6353
Ji, X. Y. (2020). Comparative analysis of volatile organic compounds and bioactive compounds in typical coniferous and broad-leaved tree species. J. Essent Oil Bearing Plants 23, 1105–1117. doi: 10.1080/0972060X.2020.1854128
Ji, X. Y. (2021). Comparative investigation of volatile components and bioactive compounds in beers by multivariate analysis. Flavour Fragr. J. 36, 374–383. doi: 10.1002/FFJ.3649
Jian, W., Cao, H. H., Yuan, S., Liu, Y. D., Lu, J. F., Lu, W. J., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6:22. doi: 10.1038/s41438-018-0098-y
Jin, J. P., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/J.TPLANTS.2005.03.002
Li, H. Y., Yue, Y. Z., Ding, W. J., Chen, G. W., Li, L., Li, Y. L., et al. (2020a). Genome-wide identification, classification, and expression profiling reveals R2R3-MYB transcription factors related to monoterpenoid biosynthesis in Osmanthus fragrans. Genes 11:353. doi: 10.3390/genes11040353
Li, J. J., Liu, H., Yang, C., Wang, J., Yan, G. J., Si, P., et al. (2020b). Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus L. Indust. Crops and Prod. 143:111924. indcrop.2019.111924 doi: 10.1016/j
Li, Y. Q., Shan, X. T., Gao, R. F., Han, T. T., Zhang, J., Wang, Y. N., et al. (2020c). MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 3:396. doi: 10.1038/s42003-020-01134-6
Li, W. F., Ning, G. X., Zuo, C. W., Chu, M. Y., Yang, S. J., Ma, Z. H., et al. (2021). MYB_SH[AL]QKY[RF] transcription factors MdLUX and MdPCL-like promote anthocyanin accumulation through DNA hypomethylation and MdF3H activation in apple. Tree Physiol. 41, 836–848. doi: 10.1093/TREEPHYS/TPAA156
Liu, F., Xiao, Z. N., Yang, L., Chen, Q., Shao, L., Liu, J. X., et al. (2017). PhERF6, interacting with EOBI, negatively regulates fragrance biosynthesis in petunia flowers. New Phytol. 215, 1490–1502. doi: 10.1111/nph.14675
Liu, Y. H., Zeng, Y. T., Li, Y. M., Liu, Z., Lin, W. K., Espley, R. V., et al. (2020). Genomic survey and gene expression analysis of the MYB-related transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 164, 2450–2464. doi: 10.1016/j.ijbiomac.2020.08.062
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Muleke, E. M., Wang, Y., Zhang, W., Xu, L., Ying, J., Karanja, B. K., et al. (2021). Genome-wide identification and expression profiling of MYB transcription factor genes in radish (Raphanus sativus L.). J. Integr. Agric. 20, 120–131. doi: 10.1016/S2095-3119(20)63308-1
Nesi, N., Jond, C., Debeaujon, I., and Caboche, M. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2311–2322. doi: 10.1105/TPC.010098
Nguyen, N. H., and Lee, H. (2016). MYB-related transcription factors function as regulators of the circadian clock and anthocyanin biosynthesis in Arabidopsis. Plant Signal. Behav. 11:e1139278. doi: 10.1080/15592324.2016.1139278
Niu, T. Q., Gao, Z. D., Zhang, P. F., Zhang, X. J., Gao, M. Y., Ji, W., et al. (2016). MYBA2 gene involved in anthocyanin and flavonol biosynthesis pathways in grapevine. Genet. Mol. Res. 15:gmr15048922. doi: 10.4238/gmr15048922
Reddy, V. A., Wang, Q., Dhar, N., Kumar, N., Venkatesh, P., Rajan, C., et al. (2017). Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 15, 1105–1119. doi: 10.1111/pbi.12701
Riechmann, J., and Ratcliffe, O. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. doi: 10.1016/S1369-5266(00)00107-2
Rosinski, J. A., and Atchley, W. R. (1998). Molecular evolution of the MYB Family of transcription factors: evidence for polyphyletic origin. J. Mol. Evol. 46, 74–83. doi: 10.1007/PL00006285
Rumi, T. W., Yuka, N., and Takuji, W. (2013). Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8:e24575. doi: 10.4161/psb.24575
Sakai, M., Yamagishi, M., and Matsuyama, K. (2019). Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep. 38, 609–622. doi: 10.1007/s00299-019-02391-4
Sun, X. M., Matus, J. T., Wong, D., Wang, Z. M., Chai, F. M., Zhang, L. L., et al. (2018). The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 69, 1749–1764. doi: 10.1093/jxb/ery020
Thomas, A. C., Joo, Y. K., Ashlyn, E. W., Laura, A. L., Kyle, C. S., Robert, C. S., et al. (2011). PhMYB4 fine-tunes the floral volatile signature of Petunia×hybrida through PhC4H. J. Exp. Bot. 62, 1133–1143. doi: 10.1093/jxb/erq342
Yang, X. L., Li, H. Y., Yue, Y. Z., Ding, W. J., Chen, X., Shi, T. T., et al. (2018). Transcriptomics analysis of the candidate genes related to aroma formation in Osmanthus fragrans. Molecules 23:1604. doi: 10.3390/molecules23071604
Yin, J. L., Wong, W. S., Jang, I. C., and Chua, N. H. (2017). Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 213, 1133–1144. doi: 10.1111/nph.14280
Yong, Y. B., Zhang, Y., and Lyu, Y. N. (2019). A MYB-related transcription factor from Lilium lancifolium L. (LlMYB3) is involved in anthocyanin biosynthesis pathway and enhances multiple abiotic stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 20:3195. doi: 10.3390/ijms20133195
Yue, Y. Z., Du, J. H., Li, Y., Thomas, H. R., Frank, M. H., Wang, L. G., et al. (2021). Insight into the petunia Dof transcription factor family reveals a new regulator of male-sterility. Indust. Crops Prod. 161:113196. doi: 10.1016/J.INDCROP.2020.113196
Zeng, X. L., Liu, C., Zheng, R. R., Cai, X., Luo, J., Zou, J. J., et al. (2016). Emission and accumulation of monoterpene and the key terpene synthase (TPS) associated with monoterpene biosynthesis in Osmanthus fragrans Lour. Front. Plant Sci. 6:1232. doi: 10.3389/fpls.2015.01232
Zhang, C. H., Ma, R. J., Xu, J. L., Yan, J., Guo, L., Song, J., et al. (2018). Genome-wide identification and classification of MYB superfamily genes in peach. PLoS One 13:e0199192. doi: 10.1371/journal.pone.0199192
Zhang, C., Fu, J. X., Wang, Y. G., Bao, Z. Y., and Zhao, H. B. (2015). Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of sweet osmanthus (Osmanthus fragrans Lour.). PLoS One 10:e0136355. doi: 10.1371/journal.pone.0136355
Zhao, P. C., Hou, S. L., Guo, X. F., Jia, J. T., Yang, W. G., Liu, Z. J., et al. (2019). A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 19:564. doi: 10.1186/s12870-019-2159-2
Keywords: sweet osmanthus, MYB-related, transcription factor, GC-MS, floral aroma
Citation: Yan X, Ding W, Wu X, Wang L, Yang X and Yue Y (2022) Insights Into the MYB-Related Transcription Factors Involved in Regulating Floral Aroma Synthesis in Sweet Osmanthus. Front. Plant Sci. 13:765213. doi: 10.3389/fpls.2022.765213
Received: 26 August 2021; Accepted: 03 January 2022;
Published: 09 March 2022.
Edited by:
Xiumin Fu, South China Botanical Garden (CAS), ChinaReviewed by:
Dengwei Jue, Chongqing University of Arts and Sciences, ChinaCopyright © 2022 Yan, Ding, Wu, Wang, Yang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiulian Yang, eGx5QG5qZnUuZWR1LmNu; Yuanzheng Yue, eXVleXVhbnpoZW5nQG5qZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.