- Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences (JAAS), Nanjing, China
Magnaporthe oryzae causes rice blast disease and is responsible for major losses in rice production worldwide. Although numerous studies have focused on the interactions between Oryza sativa and M. oryzae, to date, the conserved mechanisms remain in part unclear. In this study, a comparative analysis of transcriptomes of O. sativa L. ssp. japonica cv. ‘Nipponbare’ interacting with three M. oryzae strains (248, 235, and 163) were performed to explore the conserved molecular mechanisms. Differentially expressed genes with similar expression patterns in the interactions between cultivar ‘Nipponbare’ and three M. oryzae strains were defined as Conserved Differentially Expressed Genes (CDEGs). These included 3,647 O. sativa CDEGs and 3,655 M. oryzae CDEGs. Four rice CDEGs (LOC_Os03g19270, LOC_Os07g36600, LOC_Os05g28740, and LOC_Os01g32780) encoding universal stress protein (USP) were induced within 24 h post-inoculation (hpi) by three M. oryzae strains. Meanwhile, overexpression of LOC_Os07g36600 resulted in enhanced rice resistance against M. oryzae. Furthermore, four rice genes coding light-harvesting chlorophyll a/b-binding (LHC) protein (LOC_Os02g52650, LOC_Os09g12540, LOC_Os11g13850, LOC_Os05g22730) were also identified as CDEGs and were induced at 48 hpi, which might contribute to blast resistance through reactive oxygen species (ROS) accumulation. MoCDIP4 is M. oryzae effector inducing rice cell death and were verified that include AA9 CAZy domain (namely GH61 domain). In this study, we found seven MoCDIP4-homologous genes coding proteins with signal peptides and AA9 CAZy domains, which were continuously up-regulated across all infection stages relative to uninoculated control. This study uncovered that genes are required for conserved mechanisms of rice-M. oryzae interaction, which includes rice genes encoding USP proteins and LHC proteins, as well as M. oryzae genes encoding AA9 proteins. This study will help us to understand how O. sativa responds to M. oryzae infections and the molecular mechanisms of M. oryzae pathogenicity.
Introduction
Rice is a staple food that feeds more than half of the world’s population. Rice (Oryzae sativa) blast, the most destructive rice disease worldwide caused by Magnaporthe oryzae (a hemibiotrophic fungal pathogen), can reduce rice yield by 30% (Skamnioti and Gurr, 2009; Dean et al., 2012). Therefore, it is critical to explore the mechanism of rice-M. oryzae interaction and breed for rice cultivars with durable resistance to rice blast.
Approximately 24 h after attaching to the rice leaf surface, M. oryzae forms appressorium, which is an infectious structure that generates enormous turgor pressure and helps M. oryzae to penetrate into the rice cell wall (Howard and Valent, 1996; Ribot et al., 2008). Then, specialized hyphae are produced and expand within rice cells. This process is defined as the biotrophic stage, which lasts for 48 h after adhesion to the leaf surface (Wang et al., 2014). The infection then switches to the necrotrophic phase, during which the rice cells lose viability and disease lesions become evident on the leaf surface.
Plant-pathogen interactions follow the ‘zig-zag’ model, which shows that plants have evolved two main types of innate immunity: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). The PTI system is activated upon direct recognition of PAMPs by two major types of host pattern-recognition receptors (PRRs) (Ausubel, 2005), namely, receptor-like kinases (RLKs) and receptor-like proteins (RLPs) (Boutrot and Zipfel, 2017). The activated PTI signaling subsequently activates downstream targets that result in ROS production, stomatal closure, MAPK activation, and production of defense hormones (Yuan et al., 2021). Pathogens deliver a variety of effectors into the host cells to target PRR complex components, which inhibit the kinase activity of the PRR (Dou and Zhou, 2012). Polymorphic resistance proteins, encoded by plant resistance (R) genes, can directly or indirectly recognize pathogen effectors and activate ETI signaling, which results in enhanced resistance and hypersensitive response (HR) (Cui et al., 2015). Although approximately 100 rice R genes/alleles associated with blast resistance have been identified so far (Ashkani et al., 2015), only 37 race-specific blast R genes have been successfully cloned and most of them encode nucleotide-binding site-leucine-rich-repeat (NBS-LRR) proteins (Li W. et al., 2019). However, the large-scale application of rice cultivars with R genes is limited due to its long-term period of breeding and risk of losing resistance, which resulted from the rapid evolution of M. oryzae (Dean et al., 2005). Except for traditional R genes, defense regulator (DR) genes can regulate blast resistance and received attention due to their partial but durable broad-spectrum blast resistance (Li W. et al., 2019). Take an example, a loss-of-function allele of Pi21, encoding a proline-rich protein, confers broad and durable resistance against M. oryzae (Fukuoka et al., 2009; Liu et al., 2013). Due to conferring broad-spectrum resistance, DR genes might involve in rice response to different distinct strains of M. oryzae. Thus, it is important for DR genes discovery and exploration of their function through dissecting interactions between host plants and different strains of plant pathogens.
In this study, transcriptome sequencing of O. sativa L. ssp. japonica cv. ‘Nipponbare’ (Nip) inoculated with three M. oryzae strains (248, 235, and 162) was used to explore conserved mechanisms of rice-M. oryzae interaction. In our previous work, we inoculated M. oryzae 248, 235, and 162 on 20 rice cultivars. The pathogenicity of three M. oryzae strains are much different (Supplementary Table 1), which suggests M. oryzae 248, 235, and 162 represent different clonal lineages. Based on this, the comparative transcriptomic study was used to dissect the conserved mechanism of rice-M. oryzae interaction. In order to fulfill this, differentially expressed genes (DEGs) displaying similar expression patterns in the three host-pathogen interactions were defined as conserved DEGs (CDEGs). In addition to known plant defense-associated genes (i.e., PR genes, and diterpene biosynthesis genes), we also found that rice genes, encoding universal stress proteins (USPs) and light-harvesting chlorophyll a/b-binding protein (LHC), were induced during the infection stage. Among USP genes mentioned above, OsUSP4 (LOC_Os07g36600) was found that enhance rice resistance to M. oryzae attack. In M. oryzae, seven MoCDIP4-homologous genes encode secreted proteins with a signal peptide and AA9 Carbohydrate-Active enzymes (CAZymes) domain, which were continuously upregulated during the whole interaction stage. Taken together, we suggest that rice USP genes, rice LHC genes, and M. oryzae AA9 genes involve in the conserved mechanism of rice-M. oryzae interaction.
Results
Transcriptome Sequencing and Quality Control
To study common transcriptional changes of different rice-M. oryzae interactions, conidia of three M. oryzae strains (248, 235, and 162) were used as inoculum respectively. The leaf tissues of the inoculated japonica cultivar ‘Nipponbare’ (Nip) were collected at 0, 8, 24, 48, 72, and 96 h post-inoculation (hpi). Nip leaves at 0 hpi and conidia of each M. oryzae strain were defined as control samples (CK). Total RNA was isolated from the samples described above. Low-quality bases or reads were filtered from the raw transcriptome data, and approximately 46 to 63 million pairs of reads from each sample were used in the downstream analysis. All clean reads were mapped to the reference genome of O. sativa L. ssp. japonica cv. ‘Nipponbare’ and M. oryzae 70-15. Approximately 20% of reads unmapped the reference genome and thus were filtered out before expression level calculation. An overview of the mapped statistics is provided in Table 1.
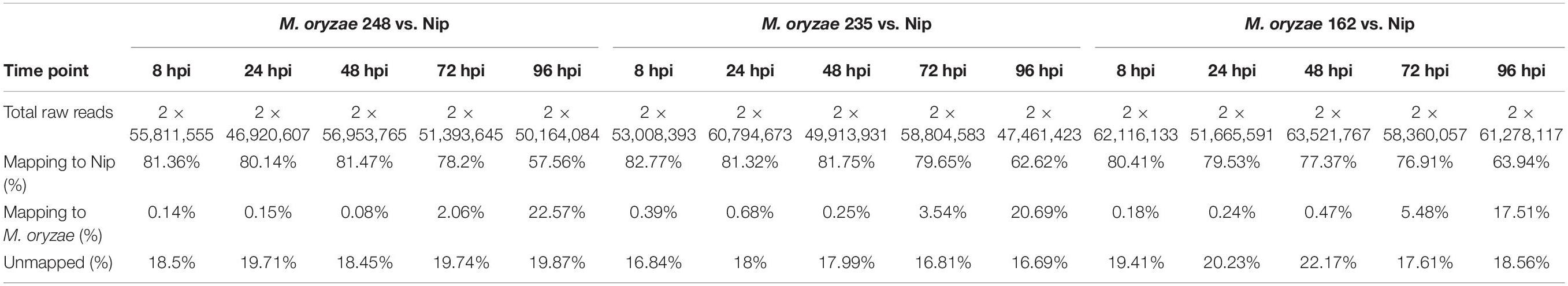
Table 1. Summary of alignment statistics in 15 libraries referring to Oryza sativa L. ssp. japonica genome.
Identification of Conserved Differentially Expressed Genes Related to Rice-M. oryzae Interaction
Differentially expressed genes (DEGs) were identified with adjusted p-values of <0.01 and at least a two-fold change in the normalized (FPKM) expression values. As Supplementary Table 2 shows, 1,913 to 3,612 O. sativa DEGs were identified from the interaction of rice-M. oryzae 248, 1,267 to 4,051 O. sativa DEGs from the interaction of rice-M. oryzae 235, and 2,143 to 4,640 O. sativa DEGs from the interaction of rice-M. oryzae 162. In addition, 4,451 to 5,212, 3,982 to 4,904, and 3,623 to 4,542 DEGs were detected from M. oryzae 248, 235, and 162, respectively.
In order to explore DEGs that may involve in the conserved mechanisms of O. sativa-M. oryzae interaction, we focused on the CDEGs which are DEGs that displayed similar transcriptional changes in the three interactions. As Figure 1 and Supplementary Table 3 displayed, in O. sativa, there were 1,690 CDEGs (880 upregulated and 789 downregulated) at 8 hpi, 2,357 CDEGs (1,071 upregulated and 1,278 downregulated) at 24 hpi, 766 CDEGs (482 upregulated and 273 downregulated) at 48 hpi, 1,606 CDEGs (1,092 upregulated and 513 downregulated) at 72 hpi, and 2,327 CDEGs (1,430 upregulated and 890 downregulated) at 96 hpi. Additionally, in M. oryzae there were 1,394 CDEGs (634 upregulated and 578 downregulated) at 8 hpi, 1,547 CDEGs (857 upregulated and 529 downregulated) at 24 hpi, 1,895 CDEGs (971 upregulated and 716 downregulated) at 48 hpi, 2,428 CDEGs (1,469 upregulated and 888 downregulated) at 72 hpi, and 2,024 CDEGs (1,259 upregulated and 654 downregulated) at 96 hpi. In total, 3,647 and 3,655 CDEGs were identified in O. sativa and M. oryzae, respectively.
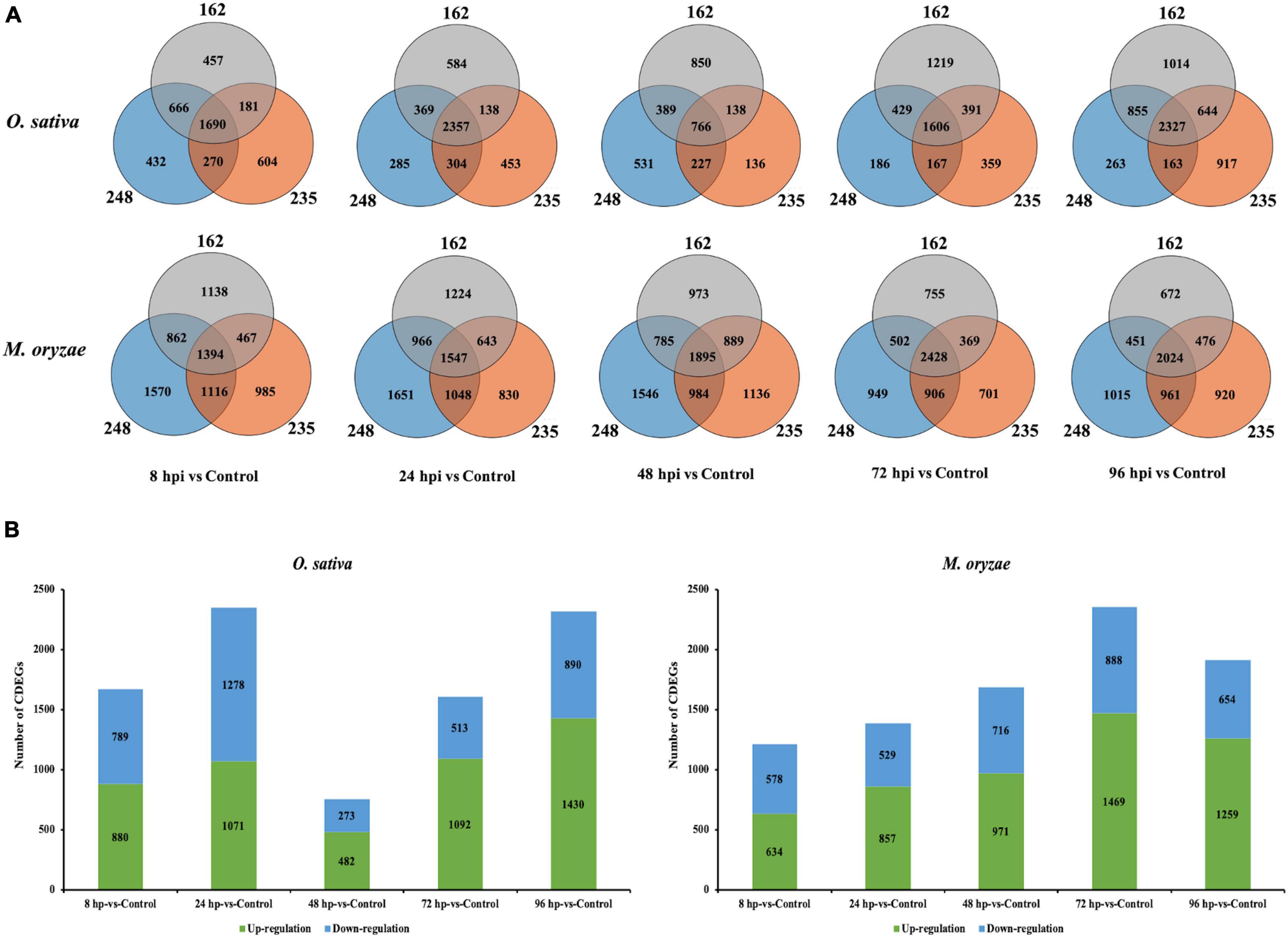
Figure 1. Overview of differentially expressed genes (DEGs) identified in interactions between Nipponbare (Nip) and three strains of Magnaporthe oryzae (248, 235, and 162) at 8, 24, 48, 72, and 96 hpi. (A) Venn diagram displaying overlap between DEGs in interactions between Nipponbare (Nip) and three strains of M. oryzae. OS represent Oryzae sativa L. ssp. japonica cv. Nipponbare and MG represent Magnaporthe oryzae. (B) Expression regulation of conserved DEGs (CDEGs).
Domain annotation of CDEGs identified at every timepoint was performed. The top10 domains of CDEGs at every timepoint were provided in Supplementary Table 4. p450 (PF00067), Pkinase (PF00069), and zf-C3HC4 (PF00097) were found across all timepoints. The CDEGs containing LRR_1 (PF00560), Pkinase_Tyr (PF07714), RRM_1 (PF00076) and Epimerase (PF01370) were specific at early infection stage (8 and 24 hpi). LRR_1 and Pkinase were defined as hallmarks of receptor-like kinases (RLKs) superfamily, which act as important players in rice defense. Take an example, Takai et al. (2008) reported OsFLS2, an RLK protein homologous to flg22, involve in flagellin perception so that promotes rice resistance. We thereby inferred the overrepresented RLKs detected at 8 and 24 hpi may relate to M. oryzae PAMPs perception, such as chitin. The CDEGs containing PTR2 (PF00854), WRKY (PF03106), AMP-binding (PF00501), and Aa_trans (PF01490) were specific at the late infection stage (72 and 96 hpi). Meanwhile, CDEGs specific at 48 hpi were found that contained domain of AP2 (PF00847).
Gene Ontology Enrichment Analysis of the Conserved Differentially Expressed Genes
The results of Gene Ontology (GO) enrichment analysis of the O. sativa CDEGs is provided in Supplementary Table 5. Rice CDEGs, downregulated at 8, 24, 72, and 96 hpi, were enriched in the GO terms ‘photosynthesis, light harvesting in photosystem I,’ ‘photosynthetic electron transport in photosystem I,’ and ‘chlorophyll-binding.’ This indicates that these putative photosynthesis-associated CDEGs may be linked to the reduction in rice green leaf area during infection (Azizi et al., 2015; Sebela et al., 2018). Notably, several rice CDEGs that were upregulated from 48 to 96 hpi were found to be enriched in ‘cinnamic acid biosynthetic process,’ ‘cinnamic acid metabolic process,’ ‘diterpene phytoalexin biosynthetic process,’ and ‘phytoalexin biosynthetic process.’ This finding supports the fact that phytohormones, phytoalexins, and diterpene secondary metabolites play important roles in the rice defense system (Bleecker and Kende, 2000; Li N. et al., 2019). Supplementary Table 6 shows that M. oryzae CDEGs were upregulated from 8 to 96 hpi and enriched in the GO terms ‘carboxylic ester hydrolase activity,’ ‘carbohydrate metabolic process,’ ‘endo-1,4-beta-xylanase activity,’ and ‘endo-1,4-beta-xylanase activity,’ which implies these M. oryzae CDEGs may involve in the decomposition of the rice cell wall.
Pathway Enrichment Analysis of the Conserved Differentially Expressed Genes Identified in O. sativa and M. oryzae
The CDEGs of O. sativa and M. oryzae were mapped against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa, 2002) and pathway enrichment analyses were performed. In O. sativa, we found several CDEGs at 8 to 96 hpi were enriched in photosynthesis-associated pathways, such as ‘photosynthesis - antenna proteins’ and ‘porphyrin and chlorophyll metabolism.’ (Supplementary Table 7). CDEGs at 48 to 96 hpi were enriched in ‘diterpenoid biosynthesis’ and ‘plant-pathogen interaction’ pathways that are linked to the plant defense system.
In this study, MapMan software was used to generate overviews of the biotic stress response and metabolism in rice. We found that more rice CDEGs were upregulated during late infection stages (72 to 96 hpi) compared to early infection stages (8 to 48 hpi), and most of them were assigned terms such as ‘PR-proteins,’ ‘SA,’ ‘JA,’ ‘ethylene,’ ‘WRKY,’ ‘MYB,’ ‘secondary metabolites,’ ‘glutathione-S-transferase,’ and ‘respiratory burst’ (Figure 2). Many CDEGs, assigned to ‘cell wall’ and ‘peroxidases,’ were found to be upregulated from 8 to 24 hpi. Not surprisingly, no CDEGs were detected in ‘R genes.’ We also noticed that many rice CDEGs, downregulating at 8, 24, 72, and 96 hpi but upregulating at 48 hpi, were assigned to the term ‘light reactions’ (Supplementary Figure 1), which is consistent with the result of GO enrichment analysis.
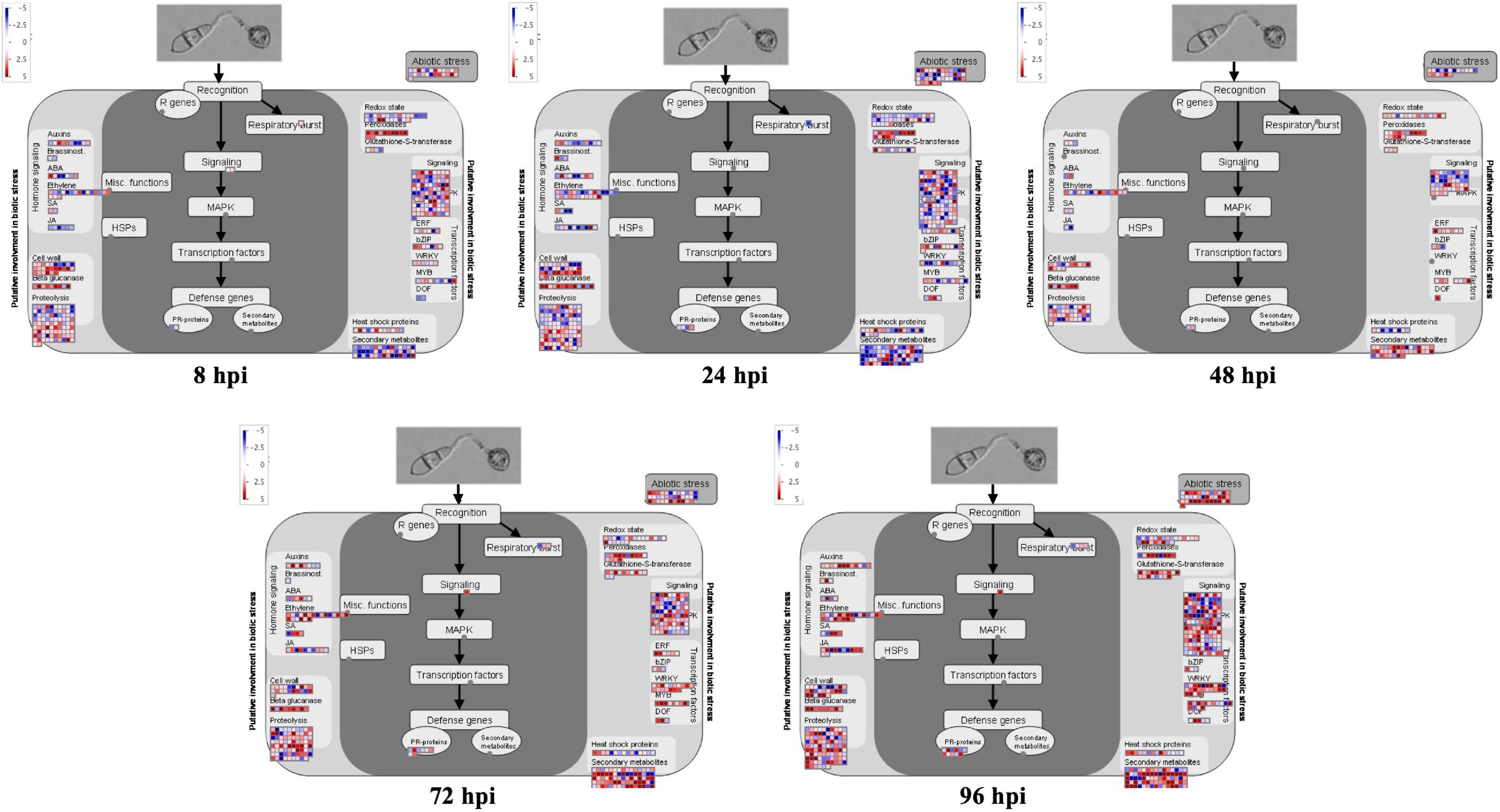
Figure 2. MapMan overviews of biotic stress display transcriptional change at 8, 24, 48, 72, and 96 hpi. CDEGs are significantly upregulated (red) and downregulated (blue) in inoculated leaf samples. Individual genes are represented by small squares. The scale bar displays log2-transformed fold changes.
For M. oryzae, CDEGs at all timepoints were enriched in the pathways ‘biosynthesis of antibiotics,’ ‘nitrogen metabolism,’ and ‘starch and sucrose metabolism’ (Supplementary Table 8). In addition, the pathways of ‘riboflavin metabolism,’ ‘ubiquinone and another terpenoid-quinone biosynthesis,’ ‘phenylalanine metabolism,’ and ‘carbon metabolism’ include CDEGs at early infection stages (8 to 24 hpi). CDEGs at late infection stages (72 to 96 hpi) were enriched in the ‘steroid biosynthesis,’ ‘glycerolipid metabolism,’ ‘pentose and glucuronate interconversions,’ and ‘other glycan degradation’ pathways.
Co-expression Clustering of the Conserved Differentially Expressed Genes Identified in O. sativa and M. oryzae
In order to dissect of the expression pattern of CDEGs identified in O. sativa and M. oryzae, we performed co-expression analysis through the Short Time-series Expression Miner (STEM) toolkit (Ernst and Bar-Joseph, 2006). In brief, 2,869 of the 3,647 rice CDEGs and 2,235 of the 3,655 M. oryzae CDEGs were divided into eight STEM profiles with E-values < 0.01 respectively (Figure 3), which means that CDEGs with similar expression patterns were put into the same STEM profiles.
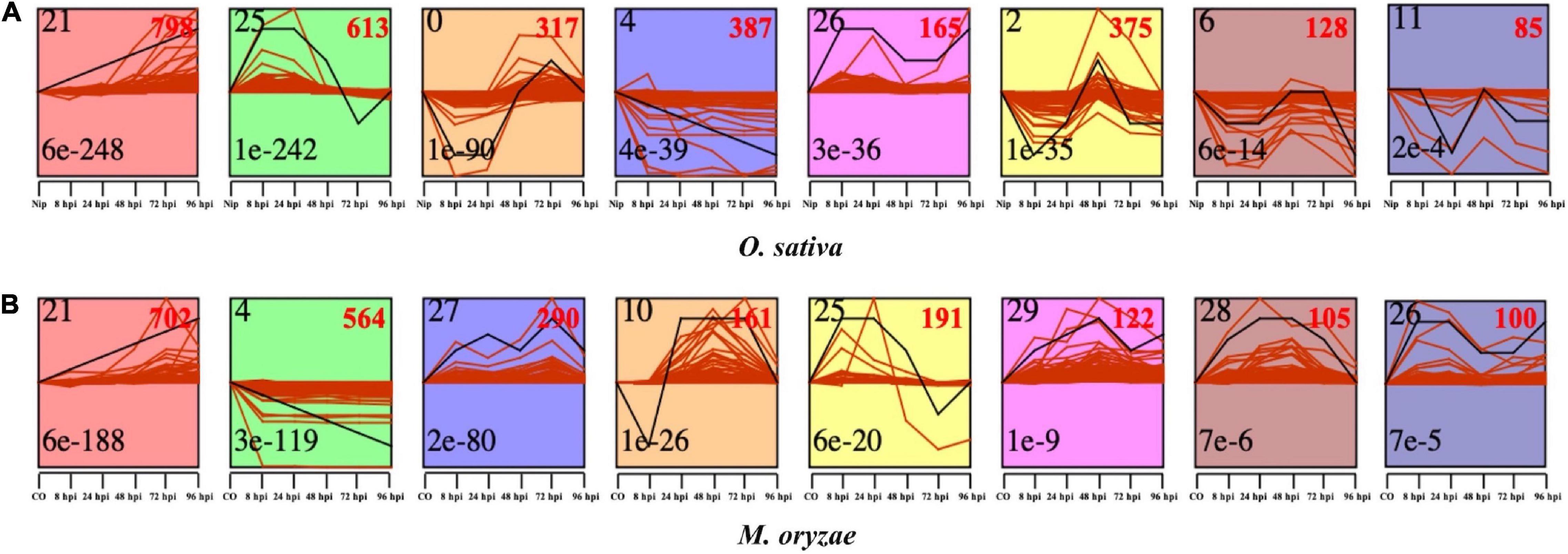
Figure 3. Short Time-series Expression Miner (STEM) toolkit identified the temporal expression profiles of CDEGs in O. sativa (A) and M. oryzae (B) with a p-value < 0.05. The number on the top left corner of each profile box was Profile ID assigned by STEM, the number on the bottom left represents the adjusted p-value, and the number on the top right corner represents the CDEGs count of each cluster.
Rice CDEGs used for co-expression analysis were provided in Supplementary Table 9. Profiles 25 and 26 consists of CDEGs that were specifically upregulated during the early interaction stages (8 to 24 hpi); profiles 0, 2, and 6 contain CDEGs that were specifically upregulated during the late interaction stages (48 to 96 hpi); continuously upregulated rice CDEGs were put into profile 21, and CDEGs in profile 4 showed decreasing expression trends during all infection stages. The result of pathway enrichment analysis for each rice co-expression profile was provided in Supplementary Table 10. For profiles of early upregulation (profiles 25 and 26), enriched pathways mainly include ‘Carbon metabolism,’ ‘Valine, leucine and isoleucine degradation,’ ‘Propanoate metabolism,’ and so on. For profiles of late upregulation (profile 0, 2, and 6), pathways of ‘Photosynthesis - antenna proteins,’ ‘Photosynthesis’ and ‘Carbon fixation in photosynthetic organisms’ were enriched. For profiles of continuous upregulation (profile 21), pathways of ‘Biosynthesis of secondary metabolites,’ ‘Diterpenoid biosynthesis,’ and ‘Flavonoid biosynthesis’ were enriched.
For M. oryzae, as Supplementary Table 11 displayed, profiles 21, 27, and 29 consisted of continuously upregulated CDEGs; profiles 25 and 26 included CDEGs that were specifically upregulated during the early infection stages; profile 10 contained CDEGs specifically upregulated during the late infection stages; CDEGs in profile 4 showed continuously decreasing expression levels from 8 to 96 hpi. The result of pathway enrichment analysis for each M. oryzae co-expression profile was provided in Supplementary Table 12. For profiles of early upregulation (profile 25 and 26), M. oryzae CDEGs in these profiles mainly enriched in the pathway of ‘Biosynthesis of secondary metabolites.’ For profiles of late upregulation (profile 10), enriched pathways mainly include the pathways of ‘Metabolic pathways.’ For profiles of continuous upregulation (profile 21, 27, and 29), pathways of ‘Biosynthesis of secondary metabolites,’ ‘Starch and sucrose metabolism,’ ‘Galactose metabolism’ and ‘Glycolysis/Gluconeogenesis’ were enriched.
Conserved Differentially Expressed Genes of Pathogenesis-Related Genes Are M. oryzae-Responsive
Pathogenesis-related (PR) genes are essential components of PAMP-triggered immunity (Duran-Flores and Heil, 2016). Here, we collected 1,074 rice PR genes retrieved from Zhang et al. (2016). The 112 O. sativa PR genes were divided into thirteen subfamilies (PR1, PR2, PR3, PR4, PR5, PR6, PR8, PR9, PR-10, PR-12, PR-14, PR-15, and PR-16) (Supplementary Table 13). As Supplementary Figure 2A shows, PR genes in profile 21 were found across diverse PR subfamilies except for PR-12. PR genes in profiles 25 and 26 mainly belong to PR8 and PR9 subfamilies, which have putative activities of chitinase and lignin-forming peroxidases (Irigoyen et al., 2020). Based on their expression profiles, nine PR8 genes and seven PR9 genes were specifically upregulated during the early infection stage (Supplementary Figure 2B). PR proteins in profiles 0, 2, and 6 are concentrated in the PR-14 subfamily. Six PR-14 genes in profiles 2 and 6 were specifically upregulated at 48, 72, and 96 hpi, which demonstrates the importance of O. sativa PR-14 proteins during the late host-pathogen interaction stages.
Universal Stress Proteins Might Respond to M. oryzae Infection
Plant phytohormones, such as salicylic acid (SA), jasmonate acid (JA), and ethylene (ET), are essential regulators of the plant defense system, which activate the appropriate and effective responses to pathogen infection. Using MapMan analysis, we found six rice CDEGs (LOC_Os01g32780, LOC_Os03g19270, LOC_Os05g28740, LOC_Os12g36630, LOC_Os07g36600, and LOC_Os01g19820), annotated as universal stress proteins (USPs) and clustered in profiles 25 or 26 (Table 2), were assigned to the branch pathways of ‘hormone metabolism and ‘ethylene induced regulated-responsive-activated’ (BinCode 17.5.3). Figure 4 shows the six USP-coding genes were specifically upregulated from 8 to 24 hpi. the qRT-PCR result revealed that transcripts of LOC_Os03g19270, LOC_Os07g36600, LOC_Os05g28740, and LOC_Os01g32780 were highly abundant during the early interaction stage (8 and 24 hpi), with almost 15 to 1200-fold compared to the control sample (Figure 5A), which suggest that LOC_Os03g19270, LOC_Os07g36600, LOC_Os05g28740, and LOC_Os01g32780 might involve in the conserved mechanism of responding to M. oryzae attack.
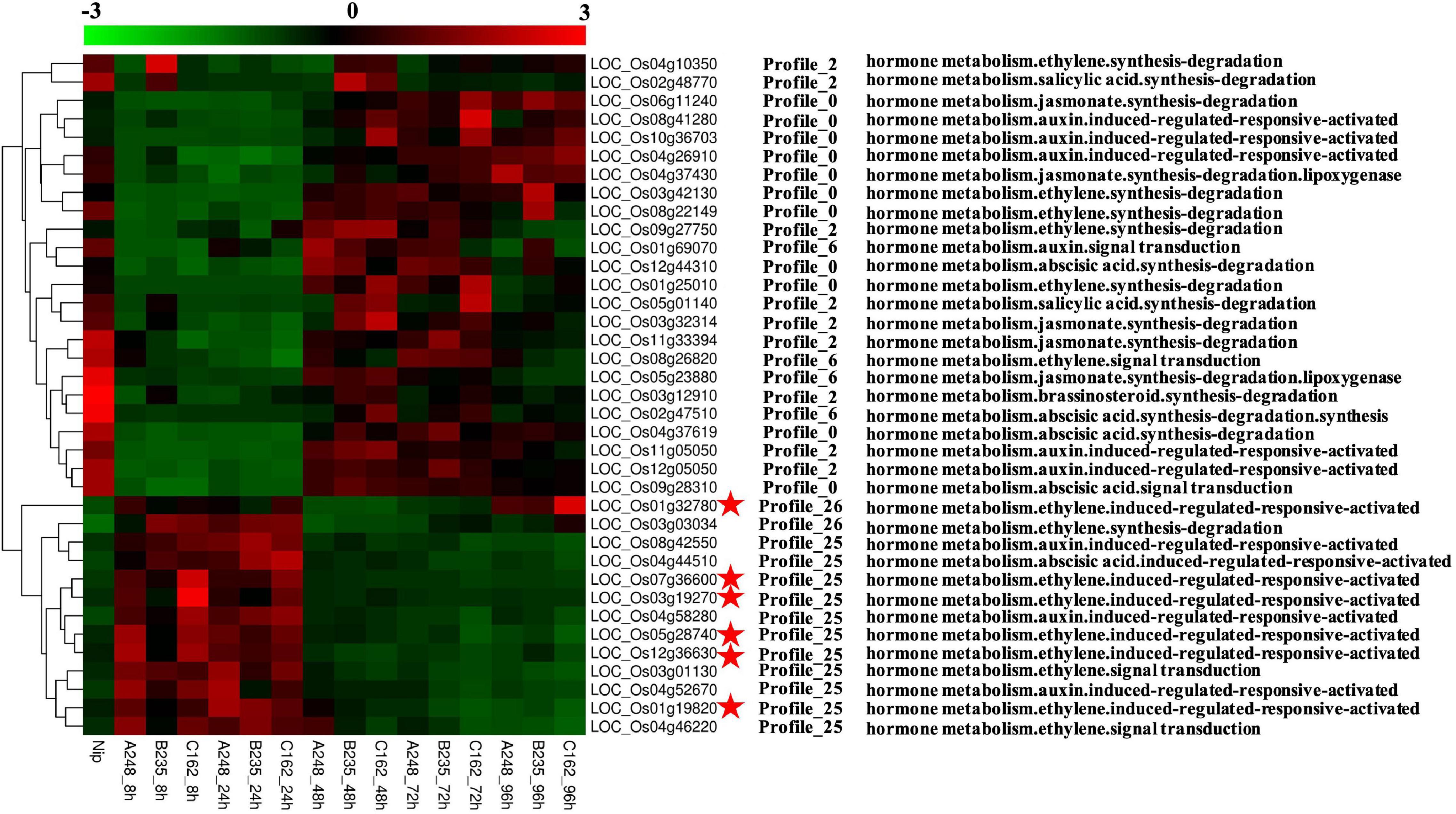
Figure 4. Overview of O. sativa CDEGs assigned to hormone metabolism or hormone signaling based on the annotation of MapMan functional categories. Heat maps display expression patterns of these O. sativa CDEGs. The middle part of the figure shows these O. sativa CDEGs belong to Profile 25, 26, 0, 2, and 6 according to co-expression clustering. The right side of the figure displays MapMan annotation of these O. sativa CDEGs. Among these O. sativa CDEGs, encoding genes of universal stress proteins (USPs), induced by ethylene according to MapMan annotation, were marked by red asterisks.
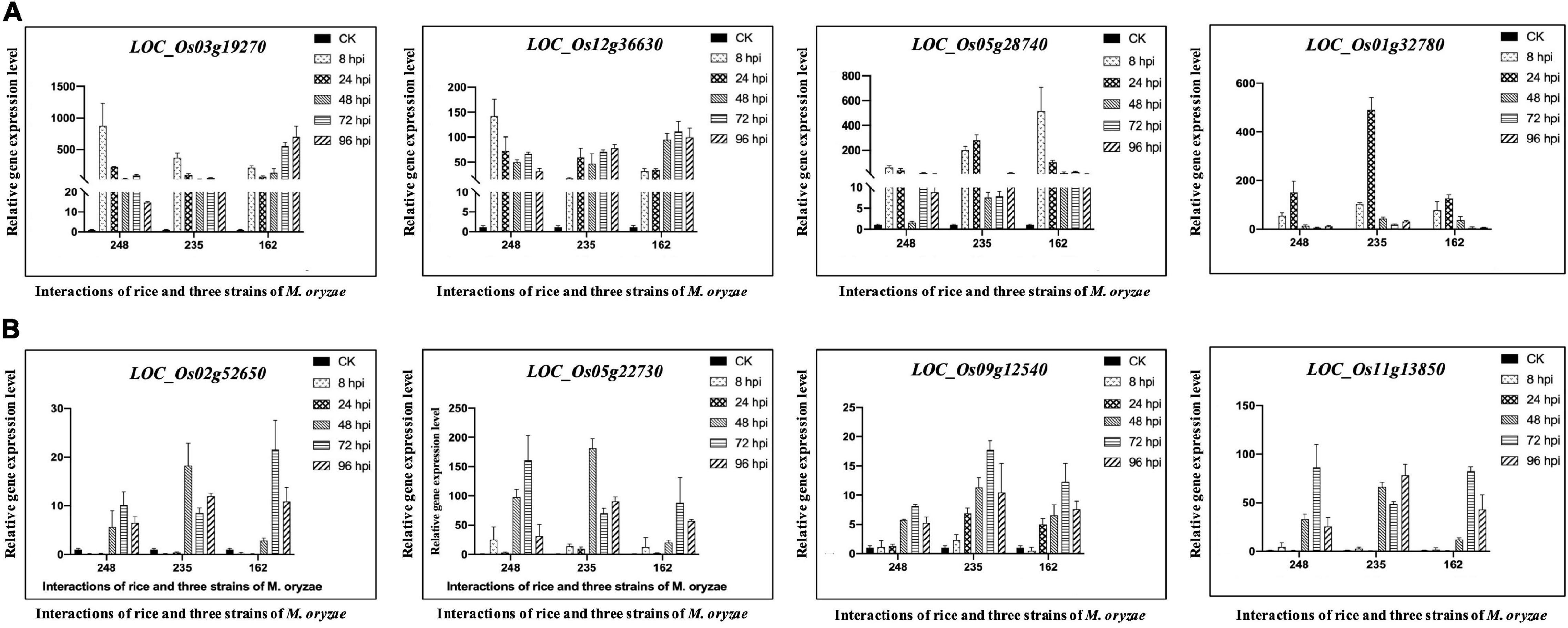
Figure 5. Quantitative Real-Time PCR verification of expression of selected coding genes of Light-harvesting chlorophyll a/b-binding proteins and universal stress proteins in O. sativa L. ssp. japonica cv. Nipponbare (Nip) during interactions with Magnaporthe oryzae 248, 235, and 162. Un-incubated Nip plant was used as the control sample and normalized to 1.0. The actin genes of Nip (LOC_Os03g50885) were used as internal reference genes. Data displayed as the average of three biological replicates and error bars indicate standard deviations. 8, 24, 48, 72, and 96 hpi represent hours post-incubation with three M. oryzae strains. (A) Quantitative Real-Time PCR verification for Light-harvesting chlorophyll a/b-binding proteins coding genes in Nip. (B) Quantitative Real-Time PCR verification for universal stress protein-coding genes in Nip.
OsUSP4 (LOC_Os07g36600) Is Involved in Blast Resistance
According to the analysis above, we found four USPs may involve in the conserved mechanism of response to M. oryzae. To investigate this, overexpression transgenic line of three rice USPs coding genes (OsUSP3: LOC_Os03g19270; OsUSP4: LOC_Os07g36600; OsUSP5: LOC_Os05g28740) were produced via PXQ vector by transforming PXQ:OsUSPs into ZH11 cultivar (wild type, Zhonghua 11), and were designed as OsUSP3OX, OsUSP4OX, and OsUSP5OX respectively. A total of seventy-six independent transgenic T1 lines were obtained (thirty-two for OsUSP3OX, fourteen for OsUSP4OX, thirty for OsUSP5OX). Based on the qRT-PCR analysis, it is notable that OsUSP3, OsUSP4, and OsUSP5 showed significantly higher expression levels compare to that in ZH11 (Figure 6C). Thus, overexpression transgenic lines of OsUSP3OX (PXQ3-14, PXQ3-19, PXQ3-28), OsUSP4OX (PXQ4-1, PXQ4-6, PXQ4-14), and OsUSP5OX (PXQ5-17, PXQ5-18, PXQ5-19, 5-25) were selected for inoculation assays. Selected overexpression transgenic lines were grown in a greenhouse for 2 weeks and were inoculated with M. oryzae isolates Guy11. Seven-days after inoculation, the diseased leaf area of PXQ4-1, PXQ4-6 and PXQ4-14 are significantly smaller than that of ZH11 (Figures 6A,B). However, there were no significant differences between the lesion area of ZH11 and overexpression transgenic lines of OsUSP3OX and OsUSP5OX. Moreover, ZH11 and three overexpression transgenic lines of OsUSP4 were also inoculated by M. oryzae 248, 235, and 162. As Supplementary Figure 3 showed, the diseased leaf ZH11 were 21.11, 25.52, and 12.32% after being inoculated by M. oryzae 248, 235, and 162, which are significantly larger than that of three overexpression transgenic lines of OsUSP4. This result suggests that OsUSP4 (LOC_Os07g36600) might be blast pathogen-responsive and slow disease response to rice blast.
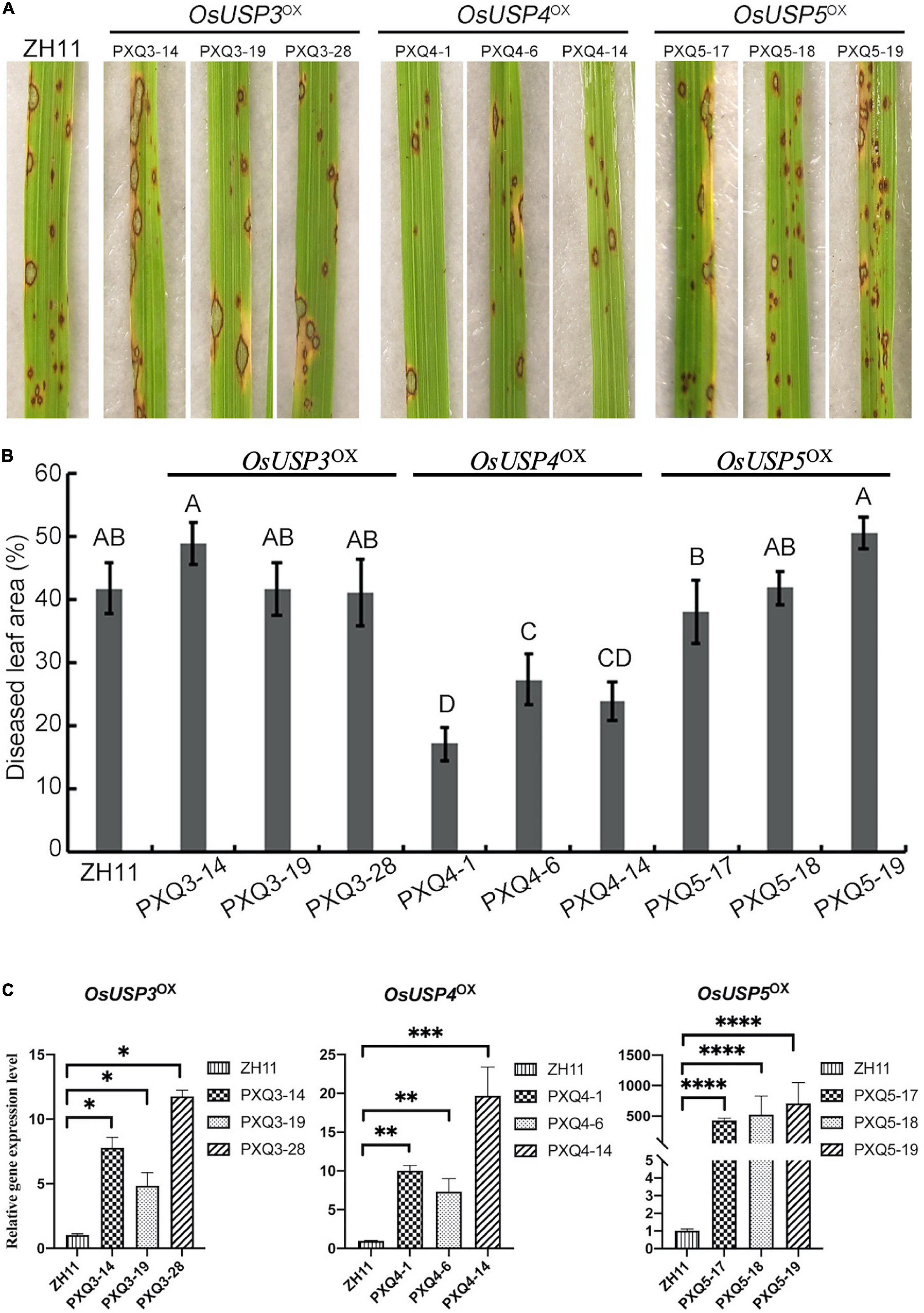
Figure 6. Disease reactions of ZH11, OsUSP3OX, OsUSP4OX, OsUSP5OX leaves incubated by Guy11. (A) Photographs showing disease reaction of indicated rice lines and races: wild-type (ZH11); overexpression transgenic line of OsUSP3OX (PXQ3-14, PXQ3-19, PXQ3-28), OsUSP4OX (PXQ4-1, PXQ4-6, and PXQ4-14), and OsUSP5OX (PXQ5-17, PXQ5-18, PXQ5-19). (B) Disease lesion area was assessed by Image J. Lesions were photographed and measured or scored at 6 days post-inoculation by isolation of Guy11. A, B, C, and D represent the significant difference (one-way ANOVA test, P < 0.01). (C) qRT-PCR was used for expression-level evaluation of universal stress proteins (USPs) coding genes in ZH11 and corresponding transgenic lines: OsUSP3 in PXQ3-14, PXQ3-19, and PXQ3-28; OsUSP4 in PXQ4-1, PXQ4-6 and PXQ4-14; OsUSP5 in PXQ5-17, PXQ5-18, and PXQ5-19. T-test Asterisks denote significant differences compared to ZH11 plants (Student t-test with two-sided and three replicates, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Light-Harvesting Chlorophyll a/b-Binding Protein Is Associated With Response to M. oryzae
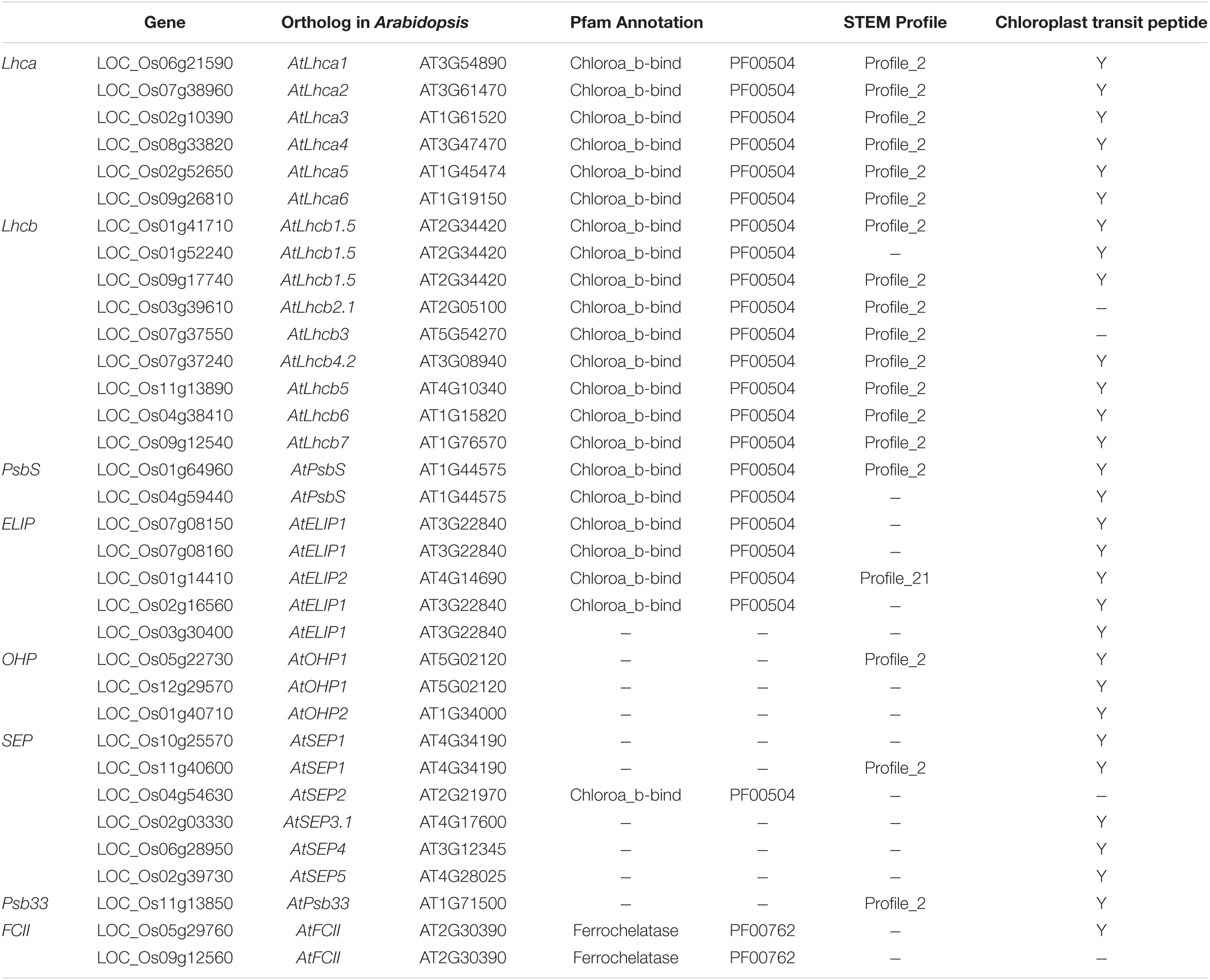
Based on previous KEGG and MapMan analysis, we found that photosynthesis-associated pathways are important during the rice-M. oryzae interaction and rice genes involved in these pathways encode light-harvesting chlorophyll a/b-binding protein (LHC). LHC superfamily consists of eight subfamilies: Lhca, Lhcb, PsbS (photosystem II subunit S), FCII (ferrochelatase II), OHP (one-helix protein), SEP (stress-enhanced protein), ELIP (early light-induced protein), and Psb33 (photosystem II protein 33) (Klimmek et al., 2006; Engelken et al., 2010; Zou and Yang, 2019).
We collected known LHC genes from Umate (2010) and obtained 34 rice LHC genes following the method of Zhao et al. (2020) (Figure 7A). Among them, 19 rice LHC genes were identified as rice CDEGs and most of them were arranged into STEM Profile_2, which include six Lhca genes, eight Lhcb genes, one PsbS gene (LOC_Os01g64960), one OHP gene (LOC_Os05g22730), one SEP gene (LOC_Os11g40600) and one Psb33 gene (LOC_Os01g64960) (Table 3). Combined with the expression profile, it is obvious that these rice LHC genes were induced at 48 hpi (Figure 7B), which include LHCB5 (LOC_Os11g13890). Liu et al. (2019) reported LHCB5 phosphorylation was activated by M. oryzae Guy11 and contributes to blast resistance through ROS accumulation. Through verification of qRT-PCR assays, transcription of LOC_Os02g52650 (Lhca subfamily), LOC_Os09g12540 (Lhcb subfamily), LOC_Os11g13850 (Psb33 subfamily), and LOC_Os05g22730 (OHP subfamily) obviously increased with 5- to 150-fold expression level since 48 hpi (Figure 5B). Taken together, we speculate that LOC_Os02g52650, LOC_Os09g12540, LOC_Os11g13850, and LOC_Os05g22730 might involve blast resistance with a similar function to LHCB5.
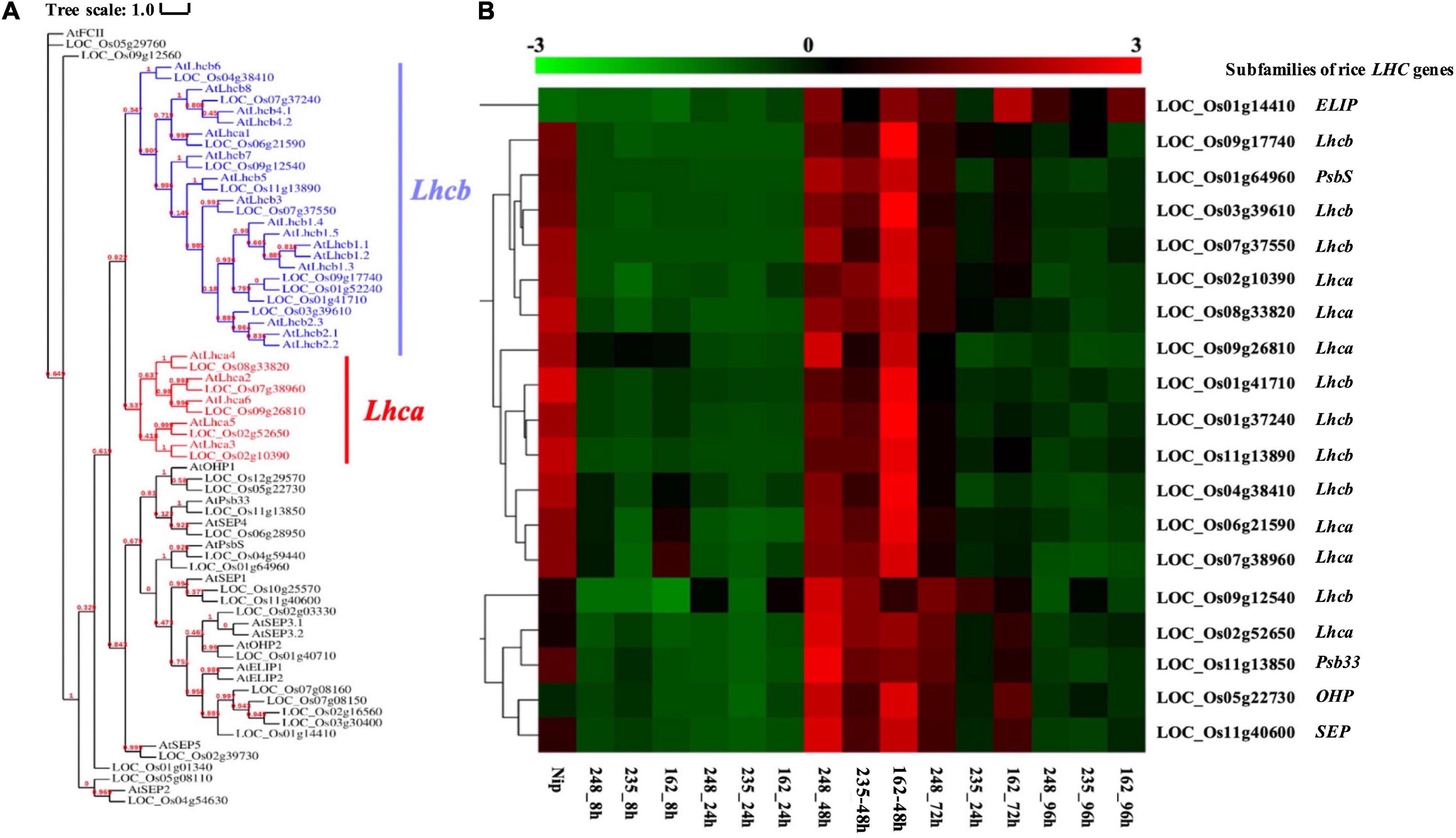
Figure 7. Analysis of O. sativa light-harvesting chlorophyll a/b-binding proteins (LHCs) superfamily. (A) Phylogenetic tree of LHCs proteins identified in O. sativa and Arabidopsis thaliana Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). Subfamilies of Lhcb and Lhca were marked in blue and red color. (B) RNAseq Expression level of Lhcb and Lhca genes during interacting with M. oryzae. The LHCs subfamily and their homologous genes in Arabidopsis thaliana were listed on the right side of the figure.
Diterpene Phytoalexins Are Related to the Conserved Mechanism of the Rice Response to M. oryzae Infection
Some continuously upregulated rice CDEGs were found to be enriched in secondary metabolite biosynthesis pathways according to the MapMan analysis. Thus, antiSMASH1 was used to predict secondary metabolite biosynthesis clusters in rice using the default parameters.
A total of 40 O. sativa secondary metabolite biosynthesis clusters were identified (Supplementary Figure 4A), which consist of 14 saccharide clusters, six lignan clusters, five polyketide clusters, and five terpene clusters. Fifteen rice CDEGs were identified as being associated with terpene biosynthesis, which obviously exceeds the number of CDEGs in other secondary metabolite clusters (Supplementary Table 14). These terpene biosynthesis-associated CDEGs were distributed in clusters c12 and c16 (Supplementary Table 14). Five of these CDEGs (i.e., LOC_Os02g36020, LOC_Os02g36140, LOC_Os02g36210, LOC_Os04g09900, and LOC_Os04g10060) mapped to the ‘diterpenoid biosynthesis’ pathway and encode core terpene biosynthesis enzymes (Supplementary Figures 4B,C). Notably, the four CDEGs coding core terpene biosynthesis enzymes were arranged into profile 21 (Supplementary Table 14), which suggests continuously increasing their expression level during whole rice-M. oryzae interaction stage. Furthermore, the activity prediction of the four core terpene biosynthesis enzymes was also performed. LOC_Os02g36210 encode ent-copalyl diphosphate synthase (EC: 5.5.1.13). LOC_Os02g36140 are predicted to function as ent-cassa-12,15-diene synthase (EC: 4.2.3.28). LOC_Os04g09900 have an enzymatic activity as syn-copalyl-diphosphate synthase (EC: 5.5.1.14). LOC_Os04g10060 was annotated as syn-pimara-7,15-diene synthase (EC: 4.2.3.35).
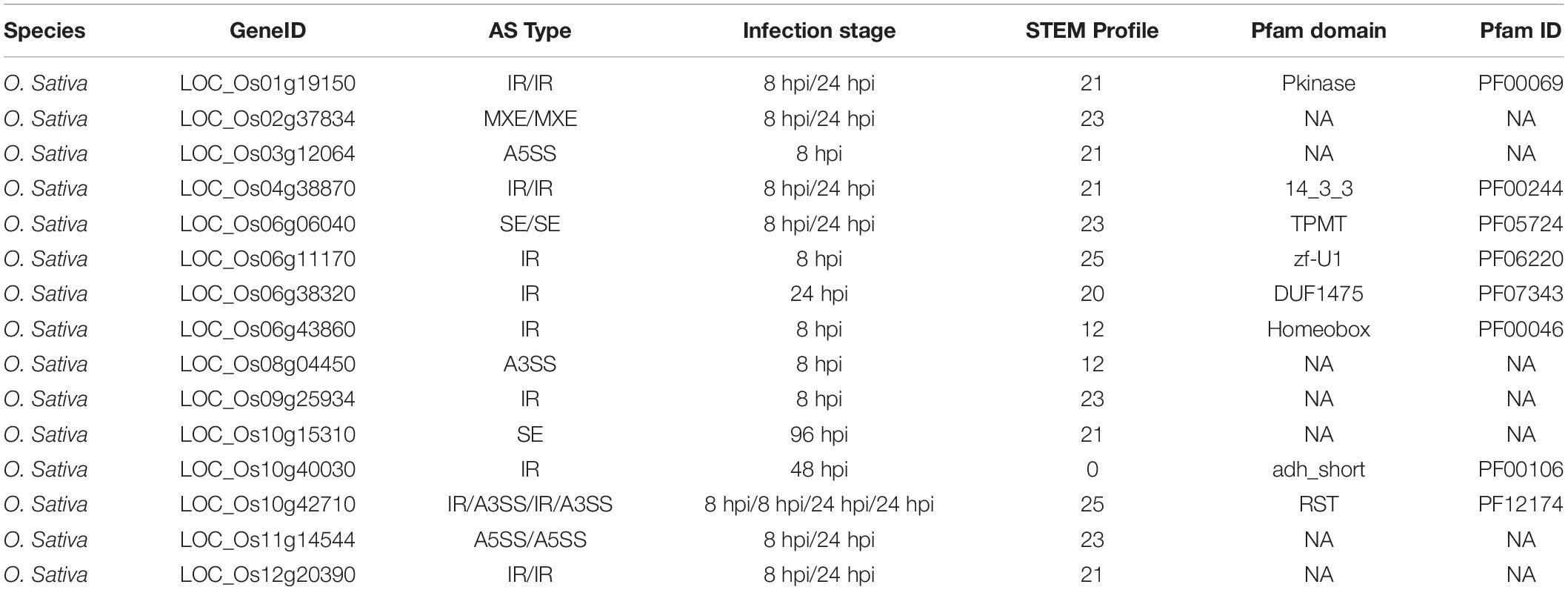
Alternative Splicing of O. sativa RCD1-SRO-TAF4 (RST) Gene Associated With Response to M. oryzae
Putative alternative splicing (AS) events that occurred in CDEGs of O. sativa and M. oryzae were identified via rMATs software (false discovery rate (FDR) cutoff of ≤0.05 and ΔPSI of ≥10%). There are no AS events predicted among M. oryzae CDEGs. In contrast, a total of 24 AS events, distributed across 15 O. sativa CDEGs, were detected (Table 4). Thirteen AS events were intron retention (IR), which was followed by alternative 3′ sites (A3SS, three AS events), alternative 5′ sites (A5SS, three events), exon-skipping (SE, three events), and mutually exclusive exons (MXE, two events). Overall, almost all AS events occurred at 8 or 24 hpi of rice-M. oryzae interaction. These AS events-occurred O. sativa CDEGs encode proteins containing diverse domains such as Pkinase (PF00069), 14_3_3 (PF00244), TPMT (PF05724), zf-U1 (PF06220), Homeobox (PF00046), adh_short (PF00106), and RST (PF12174). Seven of these CDEGs were annotated as “unknown function.” Interestingly, LOC_Os10g42710 (OsSRO1a), with an RST domain, was found to suppress Xanthomonas oryzae pv oryzae (Xoo) infection through the OsMYC2-mediated JA signaling pathway (Kashihara et al., 2020). There are seven transcripts of LOC_Os10g42710 (LOC_Os10g42710.1 to LOC_Os10g42710.7) that were detected based on in silico prediction (Supplementary Figure 5A). LOC_Os10g42710.2 contain complete RST domain and are major transcripts with the highest expression value at 8 hpi of rice respond to three M. oryzae strains (Supplementary Figure 5B). An intron retention event occurred at 8 and 24 hpi, which result in the gene model of LOC_Os10g42710 being switched from other transcripts to LOC_Os10g42710.2. Taken together, LOC_Os10g42710.2, containing complete RST domain, might be associated with regulation of JA-induced resistance compared to other transcripts.
Magnaporthe oryzae Effectors Related to the Conserved Infection Mechanism
During the period of host colonization, M. oryzae secretes a set of effectors that disturb plant immune systems (van der Does and Rep, 2007; Oliva et al., 2010). In this study, 46 known M. oryzae effectors were retrieved from the study of Gómez Luciano et al. (2019), and 16 of these verified effectors were assigned into six STEM profiles based on the co-expression analysis (Supplementary Table 15). As Figure 8A showed, most known effector genes grouped in profile 10 (i.e., AvrPi9, BAS162, BAS3, BAS1, BAS2, BAS4, and AVR-Pia), which was specifically upregulated from 24 to 72 hpi (Figure 8B). SPD9, MoCDIP4, SPD2, and MoCDIP1 were upregulated throughout the whole infection process and were therefore classified as profile 21. Profile 25 includes MoCDIP2 and MoHEG16, which supports the idea that MoCDIP2 and MoHEG16 may play roles during early infection stages (from 8 to 24 hpi). Moreover, Avr-Pi54, MoHEG13, and BAS113 belong to profiles 4, 28, and 29, respectively.
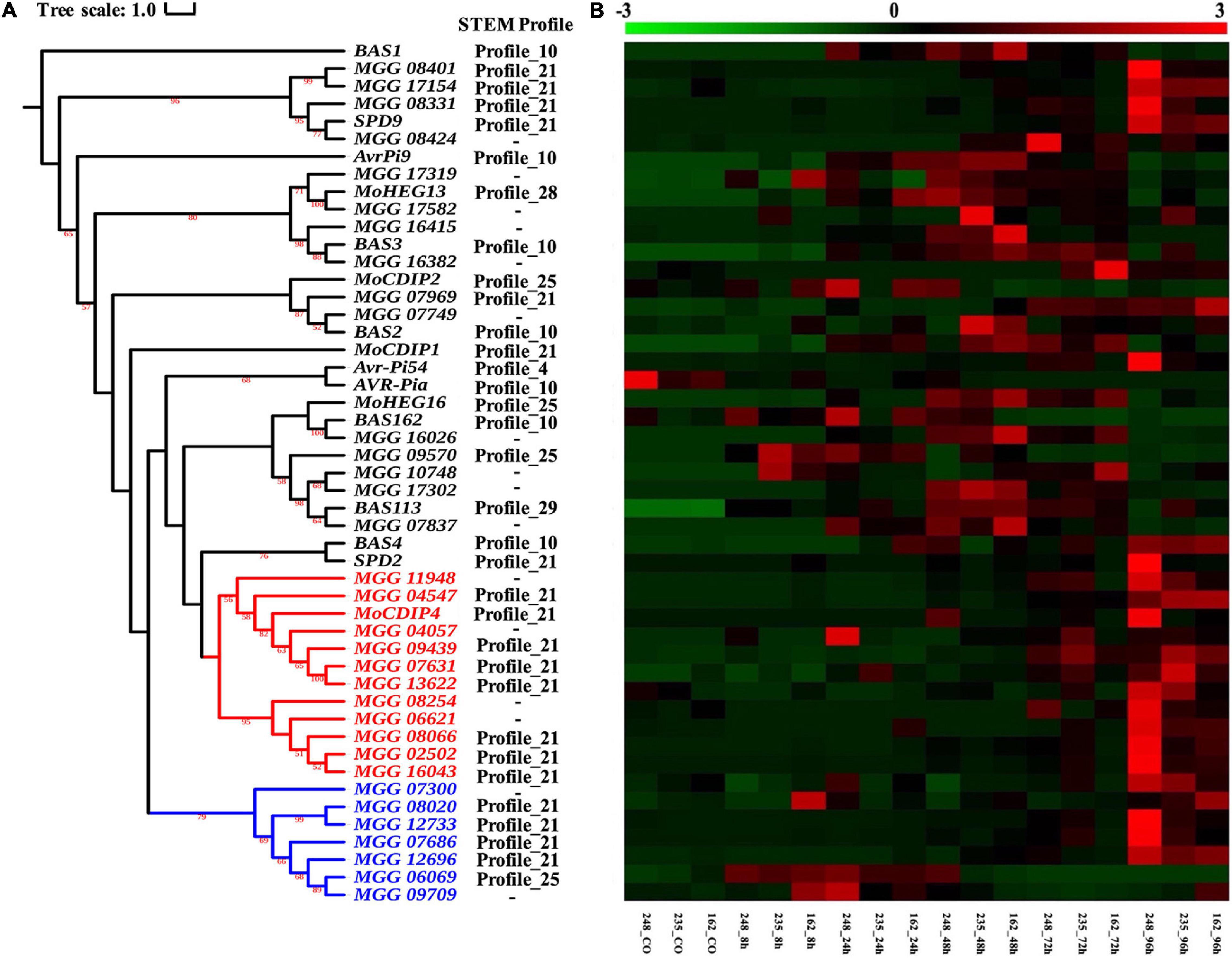
Figure 8. Analysis of verified and potential effectors. (A) Phylogenetic tree of 45 verified effectors in M. oryzae and their homology genes. The maximum likelihood tree with 1,000 bootstraps was displayed (values displayed per branch). Red and blue branches represent the MoCDIP4 effector and its homolog genes. (B) Expression pattern of verified and potential effectors which followed the topologic order of phylogeny tree.
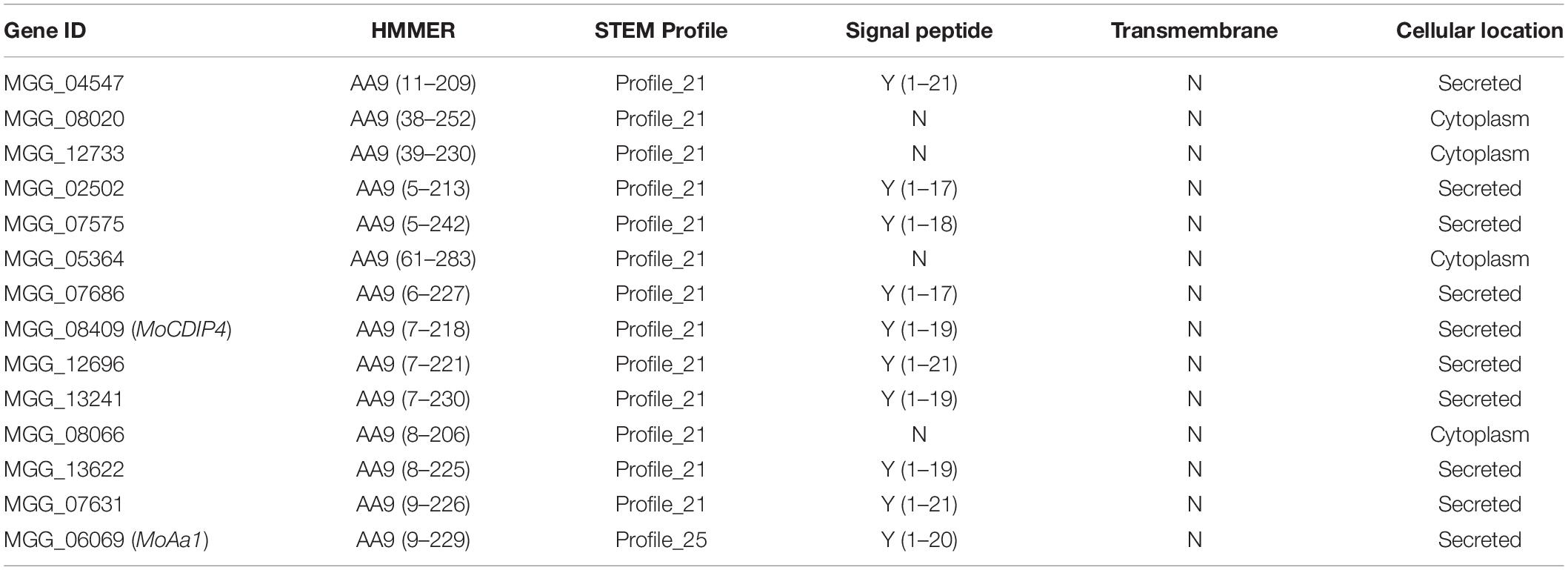
For searching more putative effector genes in the CDEGs, the BLASTP toolkit (E-value threshold: 1E-15) and MCL software (inflation threshold: 4.0) were used to cluster M. oryzae genes based on their protein sequences. As Supplementary Table 15 showed, SPD9, MoCDIP2, MoCDIP4, BAS113, BAS162, MoHEG13, BAS3, BAS2, and BAS4 were found to have between 1 and 18 homologous genes. There are 18 homologous genes of effector MoCDIP4 (MGG_08409), which is more than that of other effectors. 11 out of MoCDIP4-homologous genes were assigned to profile 21 (as does MoCDIP4 itself), which showed continuous upregulation during the whole interaction stage between and three strains of M. oryzae (Figures 3B, 8B). Figure 8A displayed MoCDIP4 and its homologous genes are concentrated in two distantly related clades, which suggests different functions. MGG_11948, MGG_04547, MGG_04057, MGG_09439, MGG_07631, MGG_13622, MGG_08245, MGG_06621, MGG_08066, MGG_02502 and MGG_16043 share same phylogenic clade and STEM profile with MoCDIP4 (red branches in Figure 8A), which infers their similar function to MoCDIP4. MoCDIP4-homologous genes in distantly related clade (MGG_07300, MGG_08020, MGG_12733, MGG_07686, MGG_12696, MGG_06069, and MGG_09709) were marked by blue color in Figure 8A and displayed their continuous upregulation expression level and potential different function with MoCDIP4.
Carbohydrate-Active Enzymes Associated With Pathogenicity
Phytopathogenic fungi are known to produce cell wall degrading enzymes (CWDEs) to breach the plant cell wall, which is the most important physical barrier during plant-pathogen interactions. Therefore, we predicted CAZymes in M. oryzae using the dbCAN web server2 (Yin et al., 2012), HMMER (Finn et al., 2011), DIAMOND (Buchfink et al., 2015), and Hotpep (Busk et al., 2017). We identified 399 CAZymes-coding genes and 164 of them were identified as CDEGs. Most of the CDEGs related to CAZymes belonged to the subfamilies of AA9, GH3, GH10, GH2, and GH31 (Supplementary Figure 6A and Table 5). Based on signal peptide prediction, we found ten secreted AA9 proteins (Supplementary Figure 6B), which include MGG_04547, MGG_02502, MGG_07575, MGG_07686, MGG_12696, MGG_13241, MGG_13622, MGG_07631 MoCDIP4 (MGG_08409) and MoAa1 (MGG_06069). Except for MGG_13241 and MGG_07575, seven of them are homologous to MoCDIP4 (the sequence similarities between them and MoCDIP4 are range from 29.2 to 42.4%) (Supplementary Figure 6C). Taken together, the AA9 CAZy subfamily may play an important role in conserved mechanisms during M. oryzae attacking rice.
Discussion
The O. sativa-M. oryzae interactions are critically important due to the huge threat M. oryzae infection poses to rice yield. However, the conserved molecular mechanisms underlying the O. sativa-M. oryzae interactions are unclear. To describe this, we compared transcriptome data from O. sativa L. ssp. japonica cv. ‘Nipponbare’ interacted with three M. oryzae strains (248, 235, and 162).
In conclusion, we have identified the potential roles played by members of several rice gene families in host resistance to M. oryzae. For example, the PR-8 and PR-9 genes were specifically upregulated during the initial infection stages, but PR-14 genes showed the opposite expression trend. Although the function of PR genes in blast resistance was well-known, we revealed that PR8, PR9, and PR-14 subfamilies are an important component of conserved mechanisms of the rice response to M. oryzae infection. Moreover, the expression pattern of core terpene biosynthesis enzymes revealed that increasing production plays a certain role in the conserved mechanism of rice-M. oryzae interaction, which consists of previous publications. Shimura et al. (2007) reported that LOC_Os02g36210 (OsCyc2) and LOC_Os02g36140 (OsDTC1) encode major enzymes in the biosynthesis of the diterpene phytoalexins phytocassane A-E, and LOC_Os04g09900 (OsCyc1) and LOC_Os04g10060 (OsKS4) are responsible for the production of Momilactones A and B. In addition, regarding cytochrome P450, the expression of genes, such as LOC_Os04g10160 (CYP99A2) and LOC_Os04g09920 (CYP99A3), is induced by the chitin oligosaccharide elicitor of pathogens (Shimura et al., 2007).
Although the pathways of SA and JA are essential for rice resistance, we noticed the ethylene biosynthesis pathway also might play an important role in the conserved mechanism of rice-M. oryzae interaction. Ethylene biosynthesis is induced in response to abiotic and biotic stresses (Bleecker and Kende, 2000; Li N. et al., 2019). For example, Sauter et al. (2002) found that ethylene can regulate the expression of OsUSP1 (one rice USP gene) during adaptation to submergence stress. This suggests that some USP genes are induced by ethylene and are associated with tolerance to several abiotic stresses, such as drought and cold (Loukehaich et al., 2012; Melencion et al., 2017). Furthermore, ERF-like transcription factors, the components of the ethylene pathway, were reported that activate the expression of genes involved in various aspects of the systemic induced defense responses (Broekaert et al., 2006). Remarkably, in this study, we found four rice USP genes (LOC_Os03g19270, LOC_Os07g36600, LOC_Os05g28740, and LOC_Os01g32780) are associated with a pathway of ‘ethylene induced regulated-responsive-activated’ and specifically upregulated at 8 and 24 hpi by three strains M. oryzae. We therefore speculate the four rice USPs genes might be induced by ethylene. We next obtained overexpression transgenic rice line of three USP genes above (OsUSP3: LOC_Os03g19270; OsUSP4: LOC_Os07g36600; OsUSP5: LOC_Os01g32780). The transgenic line of OsUSP4 was found to have a milder disease progression compared to ZH11 (wild type) despite there being no difference in inoculation phenotype between ZH11 and the transgenic lines of OsUSP3 and OsUSP5 (Figure 6). Gou et al. (2020) recently found that MfUSP1, one of the USP genes in Medicago falcata, regulates the antioxidant defense system to maintain ROS homeostasis, which limits the growth of plant pathogens. Combined with this, we thereby suggest that induction of OsUSP4 (LOC_Os07g36600) may involve the conserved mechanism of response to M. oryzae infection and confer partial blast resistance.
Due to the massive amount of energy required during the induction of the plant defense system (Swarbrick et al., 2006), there is an increased demand for photosynthesis, the major pathway that provides required carbon sources during plant-pathogen interactions. However, photosynthesis-related genes receive little attention in the previous rice-M. oryzae transcriptome studies. Herein, we noticed that the Lha and Lhb genes, two subfamilies that encode light-harvesting chlorophyll a/b-binding proteins (LHCs), were downregulated during early infection stages, which was not the expected result. A similar phenomenon was also observed by Barriuso et al. (2008) and Ishiga et al. (2009). One possible explanation that has been proposed is that the reduced photosynthesis limits carbon source availability, which can be obtained by pathogens, or that downregulating photosynthesis can protect the plant cell against oxidative damage (Blokhina et al., 2003; Bolton, 2009). In our study, we found that four photosynthesis-associated genes (LOC_Os02g52650: Lhca subfamily; LOC_Os09g12540: Lhcb subfamily; LOC_Os11g13850: Psb33 subfamily; LOC_Os05g22730: OHP subfamily) were induced after 48 hpi. Combined with previous research (Liu et al., 2019), the hypothesis was proposed that the four photosynthesis-related genes in rice might promote ROS generation so that contribute to rice blast resistance, which is activated by infection of three strains M. oryzae. However, the detailed mechanism of this will be focused on in future studies.
The study of Chen et al. (2013) initially reported MoCDIP4, containing AA9 domain (namely GH61 domain), that induces light-dependent cell death in Nicotiana benthamiana and light-independent cell death in rice, which may facilitate the colonization of M. oryzae. This suggests that light harvesting in plants may be associated with MoCDIP4-induced cell death and even disease progression of M. oryzae. Here we found that 11 MoCDIP4-homologous genes showed a trend of continuously upregulated expression following inoculation with M. oryzae (Figure 8) and seven of them contain a signal peptide and AA9 CAZyme domain (Supplementary Figure 6). Among them, MGG_13622, MGG_07631, MGG_04547, and MGG_02502 share the same phylogenic clade and own higher sequence similarity with MoCDIP4, which suggest their similar function to MoCDIP4. However, MGG_07686, MGG_12696, and MGG_06069 locate in different phylogenic clades with MoCDIP4 and own lower sequence similarities with MoCDIP4, which suggests their potential different function with MoCDIP4. Notably, MoAa91 (MGG_06069) was found to promote appressorium formation and suppress the chitin-induced plant immune response by competing with the immune receptor chitin elicitor-binding protein precursor (CEBiP) (Li et al., 2020). Taken together, the induction of MoCDIP4-homologous genes revealed the important role of the AA9 subfamily in rice blast pathogenicity.
Overall, this study will contribute to our understanding of the conserved molecular mechanisms of rice-M. oryzae host-pathogen interaction. Rice USP genes, rice LHC genes, and M. oryzae AA9 genes might be required for the conserved mechanism of rice-M. oryzae interaction. We also verified that OsUSP4 (LOC_Os07g36600) involve in rice resistance to M. oryzae attack. This study will deepen our understanding of rice-M. oryzae and broad ideas for further studies.
Materials and Methods
Plant Materials, Fungal Materials, and Growth Conditions
Wild-type (O. sativa L. ssp. japonica cv. Nipponbare (Nip) and Zhonghua 11) and overexpression transgenic lines of rice USPs coding genes (OsUSP3OX, OsUSP4OX, OsUSP5OX) were used in this study. All rice seeds were rinsed twice with demineralized water and germinated for 3 days at 28°C on sterilized wet filter paper. Germinated seeds were placed in a disposable plastic cup and grown in a greenhouse for 2 weeks (16/8 h light/dark cycle, 28 ± 2°C, and 75% humidity). Three M. oryzae strains of 248, 235, and 162 were selected from strains that we collected from the disease nursery of two municipal rice breeding institutes (Jintan: 31°40′20″N, 119°21′34″E; Ganyu: 34°54′10″N, 118°59′32″E).
Inoculation Assays
Conidia of Magnaporthe oryzae strains 248, 235, 162, and Guy11 were used for inoculation assays. Two-week-old Nip rice plants were used for inoculation with M. oryzae strains 248, 235, and 162 (all three strains are compatible). Concentration of conidia suspensions were adjusted to 5 × 105 spores/mL with.2% (w/v) gelatin solution. Then, 5 ml conidia suspensions were sprayed onto leaves of inoculated plants, which were kept in a dark chamber at 85% humidity and 28°C for the first 24 h. After 24 hpi, fungal-inoculated rice seedlings were moved to a growth chamber with the same conditions as that of the greenhouse. Leaves at 8, 24, 48, 72, and 96 hpi were harvested for transcriptome sequencing and qRT-PCR assays. Two-week-old rice plants of ZH11, OsUSP3OX, OsUSP4OX, and OsUSP5OX were used for inoculation with M. oryzae Guy11 according to the method above. Leaves were harvested for disease severity assessment at 6 days after inoculation.
RNA Isolation and Illumina Sequencing
Leaves of un-incubated Nip plants were used as control samples and leaves of incubated Nip plants at 8, 24, 48, 72, and 96 hpi were used as treatment samples. Total RNA was isolated from control and treatment samples using Qiagen RNAeasy Mini kit (Qiagen Inc., Valencia, CA, United States) according to the manufacturer’s protocol. Isolated RNA was analyzed for its quality by gel electrophoresis and quantified by spectrophotometer (Nano-Drop 2000, Thermo Fisher Scientific, Wilmington, DE, United States). RNA integrity number (RIN) was calculated by using Agilent 2100 Bioanalyzer (Agilent Technologies, Thermo Fisher Scientific Inc., Waltham, MA, United States) RNA samples with RIN greater than or equal to 7 were used for library and cDNA preparation. The fragment library for RNA sequencing was prepared using Illumina True-Seq RNA Library Prep Kit (San Diego, CA, United States) according to the manufacturer’s protocol. Illumina HiSeq 2000 platform was used to generate large amounts of sequencing data performing paired-end sequencing runs using 1 g of high-quality total RNA (RIN > 7) to obtain 150 bp sequence length reads. The RNA sequencing data are deposited at the SRA website, accession numbers SRP324816 and SRP324897.
Generation of OsUSP3OX, OsUSP4OX, and OsUSP5OX Transgenic Lines
The coding sequences of OsUSP3 (LOC_Os03g19270), OsUSP4 (LOC_Os07g36600), and OsUSP5 (LOC_Os01g32780) were amplified using cDNA isolated from 2-week-old ZH11 as PCR templates. The amplified coding sequences were cloned into the rice transformation PXQ vector. The final construct PXQ::OsUSP3, PXQ::OsUSP4, and PXQ::OsUSP5 were transformed into ZH11 by Agrobacterium (strain EHA105)-mediated co-cultivation. Transgenic plants were selected on growth media containing hygromycin (40 mg/L).
Normalization of Expression Levels and Detection of Differentially Expressed Genes
FastQC3 was used to assess read quality. Reads with contaminant primer/adapters and long stretches of poor-quality bases were removed. Clean reads were mapped to the reference genomes of rice and M. oryzae, respectively (rice: MSU Rice Genome Annotation Project Release 74; M. oryzae: M. oryzae 70-15 v3.05) using TopHat version 2.1.16 with default parameters (Trapnell et al., 2009). Calculation of raw read count and normalization to fragments per kilobase per million (FPKM) were performed using Cufflinks version 2.2.1 (Trapnell et al., 2012). Due to no replicate of each RNA-Seq sample, we applied the R-package DEseq7 to detect DEGs, following the parameters of Anders and Huber (2010). Genes with a combination of FDR≦0.01 and the absolute value of log2 (fold change) ≧1 were regarded as DEGs.
Additional Bioinformatic Methods
The multiple alignment analysis was performed with MUSCLE version 3.8.318 (Edgar, 2004). The maximum-likelihood phylogeny trees were constructed with IQ-TREE version 1.6.129 (Nguyen et al., 2015) with 1,000 bootstrap values. OmicShare tools10 was used for enrichment analysis of GO and KEGG pathways. Prediction of secondary metabolites in rice and M. oryzae were performed with antismash version 511 with default settings. The TBtools kit12 (Chen et al., 2018) was used to visualize the genome location of rice CDEGs associated with terpene biosynthesis.
Validation of Gene Expression Using Quantitative qRT-PCR
Using Qiagen RNAeasy Mini kit (Qiagen Inc., Valencia, CA, United States), RNA isolation was performed, and cDNA synthesis was carried out using the Superscript IV Reverse transcriptase cDNA synthesis kit (TB Green ® Premix Ex Taq™ II)13 using 2 ug template RNA. All cDNA samples were diluted to 20 ng––1 prior to qRT-PCR. The gene expression levels were evaluated using qRT-PCR (Bio-Rad Real-Time PCR cycler using SYBG as the fluorescent dye)14. The actin gene of rice (LOC_Os03g50885) was used as an internal reference gene. Primers were designed in Primer315 and the NCBI BLASTN web platform16 was used to check the specificity of the sequences for the genes in question, with the low complexity filter turned off. The internal reference genes list above were used to normalize the expression levels of selected candidates.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA739552 (rice-rice blast interaction data) and PRJNA739674 (conidia and mycelium of M. oryzae which were used as control case for M. oryzae).
Author Contributions
YnL, DL, and ZQ planned and designed the research. DL and ZQ performed the experiments. DL drafted this manuscript. YD, JY, MY, RZ, HC, XP, TS, and JQ participate in isolation of Magnaporthe oryzae 248, 235, and 162. YnL, YuL, and ZC supervised the manuscript, whole research and provided guidance. All authors had access to the final manuscript and approved the submission of the article.
Funding
This work was supported by funding to YnL from the National Natural Science Foundation of China (Grant/Award Number: 31861143011). This work also received funding from Jiangsu Agriculture Science and Technology Innovation Fund [Grant/Award Number: CX19(1008)] and The Revitalization Foundation of Seed Industry of Jiangsu [Grant/Award Number: JBGS(2021)005].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.723356/full#supplementary-material
Supplementary Figure 1 | MapMan overviews of metabolism display transcriptional change at 8, 24, 48, 72, and 96 hpi. CDEGs significantly upregulated (red) and downregulated (blue) inoculated leaf samples relative to the control sample are illustrated. Individual genes are represented by small squares. The scale bar displays log2-transformed fold changes.
Supplementary Figure 2 | Analysis of O. sativa PR genes relates to CDEGs. (A) Sankey plot depicting association of PR subfamilies and STEM Profiles. (B) RNAseq Expression profiles of O. sativa PR genes assigned into STEM Profile 25, 26, 0, 2, and 6 of co-expression.
Supplementary Figure 3 | Diseased reactions of ZH11, OsUSP3OX, OsUSP4OX, OsUSP5OX leaves incubated by M. oryzae 248, 235, and 162. (A) Photographs showing disease reaction of indicated rice lines and races: wild-type (ZH11); overexpression transgenic line of OsUSP4OX (PXQ4-1, PXQ4-6, PXQ4-14). (B) Disease lesion area was assessed by Image J. Lesions were photographed and measured or scored at 6 days post-inoculation by isolation of M. oryzae 248, 235, and 162. A, B, and C represent the significant differences (one-way ANOVA test, P < 0.01).
Supplementary Figure 4 | Analysis of O. sativa CDEGs associated with secondary metabolites biosynthesis. (A) Distribution of CDEGs in different secondary metabolites biosynthesis clusters predicted by antiSMASH. (B) Two terpene biosynthesis clusters include CDEGs involved in terpene biosynthesis. (C) Visualization based on KEGG pathway annotation of terpene synthase relates to CDEGs.
Supplementary Figure 5 | Alternative splicing analysis of one RCD1-SRO-TAF4 (RST) protein (LOC_Os10g42710). (A) The expression value of seven transcripts of LOC_Os10g42710 at control sample and 8 hpi. (B) Structure and Sashimi plot of LOC_Os10g42710.1 and LOC_Os10g42710.2 transcripts. The plot of domain coordinate was displayed above the transcript structure plot.
Supplementary Figure 6 | Analysis of M. oryzae Carbohydrate-Active enzyme (CAZyme) relates to pathogenicity. (A) The proportion of each CAZyme subfamilies. (B) The count of secreted proteins is assigned to the CAZyme subfamily of AA9, GH10, GH3, CE5, and GH7. (C) The expression level of MoCDIP4 and its seven homologous genes coding proteins with a signal peptide and AA9 CAZy domain.
Abbreviations
STEM, Short Time-series Expression Miner.
Footnotes
- ^ https://antismash.secondarymetabolites.org
- ^ http://bcb.unl.edu/dbCAN2/
- ^ https://anaconda.org/bioconda/fastqc
- ^ http://rice.uga.edu/
- ^ https://www.ncbi.nlm.nih.gov/genome/?term=magnaporthe.+oryzae
- ^ https://ccb.jhu.edu/software/tophat/downloads/
- ^ http://bioinfo.au.tsinghua.edu.cn/software/degseq
- ^ https://anaconda.org/etetoolkit/muscle
- ^ http://www.iqtree.org/
- ^ http://www.omicshare.com/tools
- ^ http://plantismash.secondarymetabolites.org/
- ^ https://github.com/CJ-Chen/TBtools/releases
- ^ https://www.takarabio.com/products/real-time-pcr/real-time-pcr-kits/qpcr-with-tb-green-detection/tb-green-premix-ex-taq-ii-(tli-rnase-h-plus)
- ^ https://www.bio-rad.com/en-us/category/real-time-pcr-systems?ID=059db09c-88a4-44ad-99f8-78635d8d54db&WT_mc_id=200911029047&WT_srch=1&WT_knsh_id=d7b619e0-b912-446a-9ab2-ff44bb3e61ac&gclid=CjwKCAiAg6yRBhBNEiwAeVyL0Lok9TxVYu6kejVBO7Bt6JWY0uu9FLcZGpT5aVHFGM3t9PcovawBChoCrUsQAvD_BwE
- ^ https://bioinfo.ut.ee/primer3-0.4.0/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Nat. Prec. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Ashkani, S., Yusop, M. R., Shabanimofrad, M., Azady, A., Ghasemzadeh, A., Azizi, P., et al. (2015). Allele Mining Strategies: principles and Utilisation for Blast Resistance Genes in Rice (Oryza sativa L.). Curr. Issues Mol. Biol. 17, 57–73. doi: 10.21775/cimb.017.057
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. doi: 10.1038/ni1253
Azizi, P., Rafii, M. Y., Mahmood, M., Abdullah, S. N., Hanafi, M. M., Nejat, N., et al. (2015). Differential Gene Expression Reflects Morphological Characteristics and Physiological Processes in Rice Immunity against Blast Pathogen Magnaporthe oryzae. PLoS One 10:e0126188. doi: 10.1371/journal.pone.0126188
Barriuso, J., Solano, B. R., and Gutierrez Manero, F. J. (2008). Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus sp. on Arabidopsis thaliana. Phytopathology 98, 666–672. doi: 10.1094/PHYTO-98-6-0666
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Blokhina, O., Virolainen, E., and Fagerstedt, K. V. (2003). Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91, 179–194. doi: 10.1093/aob/mcf118
Bolton, M. D. (2009). Primary metabolism and plant defense—fuel for the fire. Mol. Plant Microbe Interact. 22, 487–497. doi: 10.1094/MPMI-22-5-0487
Boutrot, F., and Zipfel, C. (2017). Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Broekaert, W. F., Delauré, S. L., De Bolle, M. F., and Cammue, B. P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. doi: 10.1146/annurev.phyto.44.070505.143440
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.15496/publikation-1176
Busk, P. K., Pilgaard, B., Lezyk, M. J., Meyer, A. S., and Lange, L. (2017). Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics 18:214. doi: 10.1186/s12859-017-1625-9
Chen, C., Chen, H., He, Y., and Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv [Preprint]. doi: 10.1101/289660
Chen, S., Songkumarn, P., Venu, R., Gowda, M., Bellizzi, M., Hu, J., et al. (2013). Identification and characterization of in planta–expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant Microbe Interact. 26, 191–202. doi: 10.1094/MPMI-05-12-0117-R
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Dou, D., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Duran-Flores, D., and Heil, M. (2016). Sources of specificity in plant damaged-self recognition. Curr. Opin. Plant Biol. 32, 77–87. doi: 10.1016/j.pbi.2016.06.019
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Engelken, J., Brinkmann, H., and Adamska, I. (2010). Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 10:233. doi: 10.1186/1471-2148-10-233
Ernst, J., and Bar-Joseph, Z. (2006). STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Gómez Luciano, L. B., Tsai, I. J., Chuma, I., Tosa, Y., Chen, Y.-H., Li, J.-Y., et al. (2019). Blast fungal genomes show frequent chromosomal changes, gene gains and losses, and effector gene turnover. Mol. Biol. Evol. 36, 1148–1161. doi: 10.1093/molbev/msz045
Gou, L., Zhuo, C., Lu, S., and Guo, Z. (2020). A Universal Stress Protein from Medicago falcata (MfUSP1) confers multiple stress tolerance by regulating antioxidant defense and proline accumulation. Environ. Exp. Bot. 178:104168. doi: 10.1016/j.envexpbot.2020.104168
Howard, R. J., and Valent, B. (1996). Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. doi: 10.1146/annurev.micro.50.1.491
Irigoyen, M. L., Garceau, D. C., Bohorquez-Chaux, A., Lopez-Lavalle, L. A. B., Perez-Fons, L., Fraser, P. D., et al. (2020). Genome-wide analyses of cassava Pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid. BMC Genomics 21:93. doi: 10.1186/s12864-019-6443-1
Ishiga, Y., Uppalapati, S. R., Ishiga, T., Elavarthi, S., Martin, B., and Bender, C. L. (2009). The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 181, 147–160. doi: 10.1111/j.1469-8137.2008.02639.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kashihara, K., Onohata, T., Yariuchi, R., Tanaka, S., Akimitsu, K., and Gomi, K. (2020). The overexpression of OsSRO1a, which encodes an OsNINJA1- and OsMYC2-interacting protein, negatively affects OsMYC2-mediated jasmonate signaling in rice. Plant Cell Rep. 39, 489–500. doi: 10.1007/s00299-019-02504-z
Klimmek, F., Sjodin, A., Noutsos, C., Leister, D., and Jansson, S. (2006). Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140, 793–804. doi: 10.1104/pp.105.073304
Li, N., Han, X., Feng, D., Yuan, D., and Huang, L.-J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20:671. doi: 10.3390/ijms20030671
Li, W., Chern, M., Yin, J., Wang, J., and Chen, X. (2019). Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 50, 114–120. doi: 10.1016/j.pbi.2019.03.015
Li, Y., Liu, X., Liu, M., Wang, Y., Zou, Y., You, Y., et al. (2020). Magnaporthe oryzae Auxiliary Activity Protein MoAa91 Functions as Chitin-Binding Protein To Induce Appressorium Formation on Artificial Inductive Surfaces and Suppress Plant Immunity. mBio 11:e03304. doi: 10.1128/mBio.03304-19
Liu, M., Zhang, S., Hu, J., Sun, W., Padilla, J., He, Y., et al. (2019). Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 17572–17577. doi: 10.1073/pnas.1905123116
Liu, W. D., Liu, J. L., Ning, Y. S., Ding, B., Wang, X. L., Wang, Z. L., et al. (2013). Recent Progress in Understanding PAMP- and Effector-Triggered Immunity against the Rice Blast Fungus Magnaporthe oryzae. Mol. Plant 6, 605–620. doi: 10.1093/mp/sst015
Loukehaich, R., Wang, T., Ouyang, B., Ziaf, K., Li, H., Zhang, J., et al. (2012). SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 63, 5593–5606. doi: 10.1093/jxb/ers220
Melencion, S. M. B., Chi, Y. H., Pham, T. T., Paeng, S. K., Wi, S. D., Lee, C., et al. (2017). RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants. Int. J. Mol. Sci. 18:2546. doi: 10.3390/ijms18122546
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oliva, R., Win, J., Raffaele, S., Boutemy, L., Bozkurt, T. O., Chaparro-Garcia, A., et al. (2010). Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 12, 705–715. doi: 10.1111/j.1462-5822.2010.01471.x
Ribot, C., Hirsch, J., Balzergue, S., Tharreau, D., Nottéghem, J.-L., Lebrun, M.-H., et al. (2008). Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 165, 114–124. doi: 10.1016/j.jplph.2007.06.013
Sauter, M., Rzewuski, G., Marwedel, T., and Lorbiecke, R. (2002). The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331. doi: 10.1093/jxb/erf096
Sebela, D., Quinones, C., Cruz, C. V., Ona, I., Olejnickova, J., and Jagadish, K. S. V. (2018). Chlorophyll Fluorescence and Reflectance-Based Non-Invasive Quantification of Blast, Bacterial Blight and Drought Stresses in Rice. Plant Cell Physiol. 59, 30–43. doi: 10.1093/pcp/pcx144
Shimura, K., Okada, A., Okada, K., Jikumaru, Y., Ko, K. W., Toyomasu, T., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018. doi: 10.1074/jbc.M703344200
Skamnioti, P., and Gurr, S. J. (2009). Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150. doi: 10.1016/j.tibtech.2008.12.002
Swarbrick, P. J., Schulze-Lefert, P., and Scholes, J. D. (2006). Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 29, 1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x
Takai, R., Isogai, A., Takayama, S., and Che, F. S. (2008). Analysis of Flagellin Perception Mediated by flg22 Receptor OsFLS2 in Rice. Mol. Plant Microbe Interact. 21, 1635–1642. doi: 10.1094/MPMI-21-12-1635
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Umate, P. (2010). Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal. Behav. 5, 1537–1542. doi: 10.4161/psb.5.12.13410
van der Does, H. C., and Rep, M. (2007). Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Mol. Plant Microbe Interact. 20, 1175–1182. doi: 10.1094/MPMI-20-10-1175
Wang, Y., Kwon, S. J., Wu, J., Choi, J., Lee, Y. H., Agrawal, G. K., et al. (2014). Transcriptome Analysis of Early Responsive Genes in Rice during Magnaporthe oryzae Infection. Plant Pathol. J. 30, 343–354. doi: 10.5423/PPJ.OA.06.2014.0055
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. doi: 10.1093/nar/gks479
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zhang, Y., Zhao, J., Li, Y., Yuan, Z., He, H., Yang, H., et al. (2016). Transcriptome Analysis Highlights Defense and Signaling Pathways Mediated by Rice pi21 Gene with Partial Resistance to Magnaporthe oryzae. Front. Plant Sci. 7:1834. doi: 10.3389/fpls.2016.01834
Zhao, Y., Kong, H., Guo, Y., and Zou, Z. (2020). Light-harvesting chlorophyll a/b-binding protein-coding genes in jatropha and the comparison with castor, cassava and arabidopsis. PeerJ 8:e8465. doi: 10.7717/peerj.8465
Keywords: rice-Magnaporthe oryzae interaction, comparative transcriptome, universal stress proteins, light-harvesting chlorophyll a/b-binding proteins, AA9 proteins
Citation: Liang D, Qi Z, Du Y, Yu J, Yu M, Zhang R, Cao H, Pan X, Qiao J, Song T, Liu Y, Chen Z and Liu Y (2022) Identification of Differentially Expressed Genes Reveal Conserved Mechanisms in the Rice-Magnaporthe oryzae Interaction. Front. Plant Sci. 13:723356. doi: 10.3389/fpls.2022.723356
Received: 10 June 2021; Accepted: 18 January 2022;
Published: 05 April 2022.
Edited by:
Guotian Li, Huazhong Agricultural University, ChinaReviewed by:
Qinhu Wang, Northwest A&F University, ChinaXiaofei Liang, Northwest A&F University, China
Copyright © 2022 Liang, Qi, Du, Yu, Yu, Zhang, Cao, Pan, Qiao, Song, Liu, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Liu, MTk5NzAwMTNAamFhcy5hYy5jbg==
 Dong Liang
Dong Liang Zhongqiang Qi
Zhongqiang Qi Junjie Yu
Junjie Yu