- 1State Key Laboratory of Crop Stress Biology in Arid Areas and College of Agronomy, Northwest Agriculture and Forestry University, Yangling, Shaanxi, China
- 2Hybrid Rapeseed Research Center of Shaanxi Province, Yangling, Shaanxi, China
MADS-box transcription factors play an important role in regulating floral organ development and participate in environmental responses. To date, the MADS-box gene family has been widely identified in Brassica rapa (B. rapa), Brassica oleracea (B. oleracea), and Brassica napus (B. napus); however, there are no analogous reports in Brassica nigra (B. nigra), Brassica juncea (B. juncea), and Brassica carinata (B. carinata). In this study, a whole-genome survey of the MADS-box gene family was performed for the first time in the triangle of U species, and a total of 1430 MADS-box genes were identified. Based on the phylogenetic relationship and classification of MADS-box genes in Arabidopsis thaliana (A. thaliana), 1430 MADS-box genes were categorized as M-type subfamily (627 genes), further divided into Mα, Mβ, Mγ, and Mδ subclades, and MIKC-type subfamily (803 genes), further classified into 35 subclades. Gene structure and conserved protein motifs of MIKC-type MADS-box exhibit diversity and specificity among different subclades. Comparative analysis of gene duplication events and syngenic gene pairs among different species indicated that polyploidy is beneficial for MIKC-type gene expansion. Analysis of transcriptome data within diverse tissues and stresses in B. napus showed tissue-specific expression of MIKC-type genes and a broad response to various abiotic stresses, particularly dehydration stress. In addition, four representative floral organ mutants (wtl, feml, aglf-2, and aglf-1) in the T0 generation were generated by editing four AGAMOUS (BnaAG) homoeologs in B. napus that enriched the floral organ variant phenotype. In brief, this study provides useful information for investigating the function of MADS-box genes and contributes to revealing the regulatory mechanisms of floral organ development in the genetic improvement of new varieties.
Introduction
Brassica, a genus of Cruciferae, includes many oils, feed, and vegetable crops. Brassica species are the most economically valuable species providing edible oil, industrial oil, vegetables, condiments, and feed necessary for production and life (Cheng et al., 2013). Among them, the significant crops are three diploid plants: Brassica rapa (B. rapa, AA, 2n=20), Brassica oleracea (B. oleracea, CC, 2n=18), and Brassica nigra (B. nigra, BB, 2n=16), and three allotetraploid species: Brassica napus (B. napus, AACC, 2n=38), Brassica juncea (B. juncea, AABB, 2n=36), and Brassica carinata (B. carinata, BBCC, 2n=34), forming the triangle of U model. U’s triangle model, proposed by Nagaharu in 1935, is a typical theory for studying the complexity of evolutionary relationships and polyploidization among brassicas (Nagaharu and Nagaharu, 1935). With the completion and sharing of high-quality and large-scale genomic, resequencing, and comprehensive transcriptome data, a bioinformatics platform was established to explore the gene function of species in U’s triangle. B. rapa was the first U’s triangle species to have its reference genome (Mun et al., 2010; Wang et al., 2011; Cai et al., 2017; Zhang et al., 2018; Zhang et al., 2022), followed by B. oleracea (Kim et al., 2014; Liu et al., 2014; Parkin et al., 2014; Cai et al., 2020). As an important oil crop in the world, B. napus was sequenced in 2014 (Chalhoub et al., 2014), and many genome versions have been published (Sun et al., 2017; Song et al., 2020; Chen et al., 2021), including the winter-type oilseed rape Darmor-bzh, semi-winter oilseed rape Zhongshuang11 (ZS11), and spring-type oilseed rape Westar and so on. Subsequently, the reference genomes of B. juncea (Yang et al., 2016; Paritosh et al., 2021) and B. nigra were published successively (Paritosh et al., 2020; Perumal et al., 2020), and the reference genome of B. carinata was the latest to be reported (Song et al., 2021; Yim et al., 2022). To date, each of the six U’s triangle species has its reference genomes.
MADS-box genes are critical transcription factors (TFs). The term ‘MADS’ originated from four members of MADS family in fungi, plants, and animals: MCM1 in yeast, AGAMOUS in Arabidopsis thaliana (A. thaliana), DEFICIENS in snapdragon, and SERUM RESPONSE FACTOR in human (Jack, 2001). MADS-box genes are characterized by the presence of a conserved DNA-binding domain of approximately 60 amino acids in the N-terminal region of the MADS-box protein, also known as the MADS (M) domain (Yanofsky et al., 1990). MADS-box genes can be classified into Type I (M-type) and Type II (MIKC-type) subclasses according to their evolutionary relationships. Type I generally contains two exons and can further be divided into Mα, Mβ, Mγ, and Mδ subgroups based on the similarities and differences in their exon-intron features (Parenicova et al., 2003), whereas Type II typically consists of 6-8 exons and is divided into two clades, named MIKCC and MIKC*. Except for the highly conserved M domain, MIKC-type TFs also comprise intervening (I), moderately conserved keratin-like (K), and variable C-terminal (C) domains (Egea-Cortines et al., 1999; Henschel et al., 2002). The M domain enables the DNA to bind CArG boxes (Riechmann and Meyerowitz, 1997; Immink et al., 2002); the I and K domains promote the formation of two or more MADS-domain proteins (Fan et al., 1997; Henschel et al., 2002; Yang and Jack, 2004), and the C domain regulates transcriptional activation of the MADS protein (Honma and Goto, 2001). To date, the functions of most MIKC*- and M-type MADS-box genes remain unclear; however, most MIKCC-type genes, playing vital roles in plant growth, development, and response to biotic and abiotic stress, have been elucidated (Kofuji et al., 2003).
Most TFs in the ABCDE model modulate floral organ development and belong to the MIKC-type MADS-box genes (Coen and Meyerowitz, 1991; Pinyopich et al., 2003; Zahn et al., 2006; Silva et al., 2015); examples include FLOWERING LOCUS C (FLC) (Searle et al., 2006), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) (Lee and Lee, 2010), and SHORT VEGETATIVE PHASE (SVP) (Hartmann et al., 2000). In addition, MIKC-type genes are also involved in root (AGL14 and AGL21) (Garay-Arroyo et al., 2013; Yu et al., 2014) and seed development (SHP1/2, FUL) (Ferrandiz et al., 2000; Liljegren et al., 2000). To date, the MADS-box TFs have been systematically investigated in dicotyledon plants, such as tomato (Solanum lycopersicum L.) (Wang et al., 2019), potato (Solanum tuberosum L.) (Shao et al., 2021), B. rapa (Duan et al., 2015), B. oleracea (Sheng et al., 2019), and B. napus (Wu et al., 2018; Zhou et al., 2022), and monocotyledon plants, such as maize (Zea mays L.) (Zhao et al., 2021a), wheat (Triticum aestivum L.) (Ma et al., 2017; Schilling et al., 2020), Foxtail Millet (Setaria italica) (Zhao et al., 2021b), and rice (Oryza sativa) (Arora et al., 2007). However, there have been no reports on the identification of MADS-box genes in three species of U’s triangle: B. nigra, B. juncea, and B. carinata.
In recent years, new results have enriched the functions of genes related to the ABCDE model, including FhAG2, a key male-specific sex determination candidate gene identified using whole-genome sequencing and assembly of two Ficus species, F. microcarpa (monoecious) and F. hispida (functionally dioecious) (Zhang et al., 2020). Owing to polyploidization and gene expansion, the copies of MIKC-type MADS-box genes increased in B. napus compared with that in A. thaliana, limiting the exploration of the function of those genes involved in floral organ development. In this study, the MADS-box gene family in the genome of U’s triangle species was identified for the first time, and gene structure characteristics, conserved domains, gene duplication, synteny events, and expression patterns of MIKC-type MADS-box genes were comprehensively analyzed. A series of bnaag mutants with different phenotypes were obtained using CRISPR/Cas9 technology to edit four BnaAGAMOUS (BnaAG) homologous genes in rapeseed, which offers important insights into the function of different copies of BnaAG.
Materials and methods
Sequence search and identification of MADS-box genes in species of U’s triangle
The genomic sequence, annotated file, and protein sequences of the six Brassica species in U’s triangle were downloaded (Supplementary Table S1), and the longest transcript was used for subsequent analysis. In summary, 105 AtMADS protein sequences downloaded from the TAIR database (https://www.arabidopsis.org/) (Parenicova et al., 2003) were used to perform a BLASTP search of the local protein database of the six U’s triangle species with an E-value < e-10 and identity ≥ 50% (Camacho et al., 2009). The hidden Markov model (HMM) profiles of SRF-TF (PF00319) and K-box (PF01486) were retrieved from the Pfam database (http://pfam-legacy.xfam.org/), and HMMER software (version 3.0) was used to search for homeodomains with an E-value < 1e-5. Subsequently, all MADS-box protein sequences were submitted to the National Center for Biotechnology Information Conserved Domain Database (NCBI-CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Lu et al., 2020) and SMART database (http://smart.embl-heidelberg.de/smart/batch.pl) to examine the integrity of the MADS-box or K-box domains (Letunic and Bork, 2018). The theoretical isoelectric point (pI) and molecular weight (MW) of all candidate MADS-box proteins were predicted using ExPASy (https://www.expasy.org/) (Gasteiger et al., 2003).
Phylogenetic tree, gene structure, and conserved motif analysis
Multiple sequence alignment of MADS-box protein sequences was performed using MAFFT software with the FFT-NS-I method (Katoh et al., 2019). Unrooted and rooted Maximum Likelihood (ML) trees were constructed using FastTree software (Price et al., 2009) and visualized using EvolView (http://www.evolgenius.info/evolview) (Zhang et al., 2012) and iTOL (http://itol.embl.de/) (Letunic and Bork, 2007). The gene structure files of all the MIKC-type genes were extracted using a genome annotation file. The conserved motifs were predicted using the MEME software, with the number of motif sets at 10 (http://meme-suite.org/tools/meme) (Bailey et al., 2009). TBtools was used to draw the exon-intron structure and conserved motif features (Chen et al., 2020).
Gene duplication and genomic synteny analysis
The MIKC-type protein sequences of the U’s triangle species were aligned using BLASTP with an E-value < 1e-10 (Camacho et al., 2009). The tandem and segmental duplication events in each species were analyzed using the MCScanX program (Wang et al., 2012) and visualized using Circos (Krzywinski et al., 2009). Genomic synteny was analyzed using JCVI software and the detailed workflow reference Python version MCscan (https://github.com/tanghaibao/jcvi/wiki/).
Expression profiles of MIKC-type MADS-box genes in B. napus
RNA-seq data for six tissues (roots, stems, leaves, flowers, seeds, and siliques) and four abiotic stressors (dehydration, salt, ABA, and cold) in B. napus were downloaded from the National Genomics Data Center (NGDC) (project ID: CRA001775) (https://ngdc.cncb.ac.cn/?lang=en). The transcriptome data of embryos and seed coats at different developmental stages and two different white powder-resistant rapeseed lines after inoculation with powdery mildew were downloaded from the NCBI database (project ID: PRJNA641876 and PRJNA778657). All transcriptome data were mapped to the Darmor-bzh genome using HISAT2 (Kim et al., 2015). The Transcripts Per Million (TPM) values were calculated using the FeatureCounts R package, and a heatmap of expression level was plotted using TBtools software (Liao et al., 2014; Chen et al., 2020).
The sgRNA design, vector construction, and hypocotyl genetic transformation in B. napus
First, four BnaAG genes with the highest homology to A. thaliana (AT4G18960) were selected for designing single guide RNA (sgRNA). The sgRNA1 was designed to target the MADS-box and sgRNA2 to target the K-box, and then a multiplex genome editing tool CRISPR/Cas9 with two sgRNAs was used to mutate BnaAG genes (Xing et al., 2014). Finally, the successfully constructed recombinant plasmid (pHSE401-BnaAG) was transformed into Agrobacterium tumefaciens strain GV3101, followed by genetic transformation of 19YB437 (recipient material) using a detailed transgenic operation method described by Bhalla and Singh (Bhalla and Singh, 2008). Primers used to construct the vectors are listed in Supplementary Table S12.
Identification of positive mutants and characterization of floral organ phenotype
All positive tissue culture plantlets were transplanted into humus soil and grown under a 16 h light/8 h dark photoperiod at 22°C until they reached the five-leaf stage. After four weeks of vernalization, the seedlings were transplanted into a normal environment for growth. The variant phenotypes of floral organs were observed and recorded; DNA was extracted from leaf tissues of each type of flower organ mutant, and the fragments near the two target sequences were amplified using PCR with specific primers (Supplementary Table S12). The PCR products and positive colonies were sequenced, and the editing events of each mutant were analyzed according to the results of Sanger sequencing.
Results
MADS-box genes in the six U’s triangle species
In total, 1430 MADS-box genes were identified in the genome of the six U’s triangle species. To uncover the kinship relationships of MADS-box genes among A. thaliana and U’s triangle species, the ML method was used to construct the phylogenetic tree based on the alignment of 105 MADSs in A. thaliana and 1430 MADS-box proteins in U’s triangle species (Figure 1A; Supplementary Table S2). The 1,535 MADS-box genes were divided into M-type and MIKC-type subfamilies; among them, the MIKC subfamily had the highest number of members (842), followed by the Mα (287), Mβ (143), Mγ (164), and Mδ (99) subfamilies (Figure 1B; Supplementary Table S3). The three diploid Brassica species B. rapa (AA), B. nigra (BB), and B. oleracea (CC) in the U’s triangle have undergone a whole genome triplication event (WGT) (Wang et al., 2011; Woodhouse et al., 2014) resulting in the expansion of MADS-box genes compared with A. thaliana; however, the gene numbers were significantly different. For example, 159, 184, and 180 MADS-box genes were identified in B. rapa, B. nigra, and B. oleracea genomes, respectively. In addition, although allopolyploidy facilitated the expansion of MADS-box genes, the total number of MADS-box genes identified in allotetraploid species differed from the total number of genes in its two progenitor species. For example, 240 MADS-box genes were identified in the genome of B. juncea, which was significantly less than the sum of genes in its progenitor species B. rapa (159) and B. nigra (184), and the number of MADS-box genes in B. carinata genome (335) was slightly lower than the sum of genes in its progenitor species B. nigra (184) and B. oleracea (180). The 1430 MADS-box genes were renamed based on the gene information about their distribution on chromosomes, and the protein length, molecular weight (MW), and isoelectric points (pI) of each MADS protein were analyzed synthetically to understand the basic information more comprehensively (Supplementary Table S3).
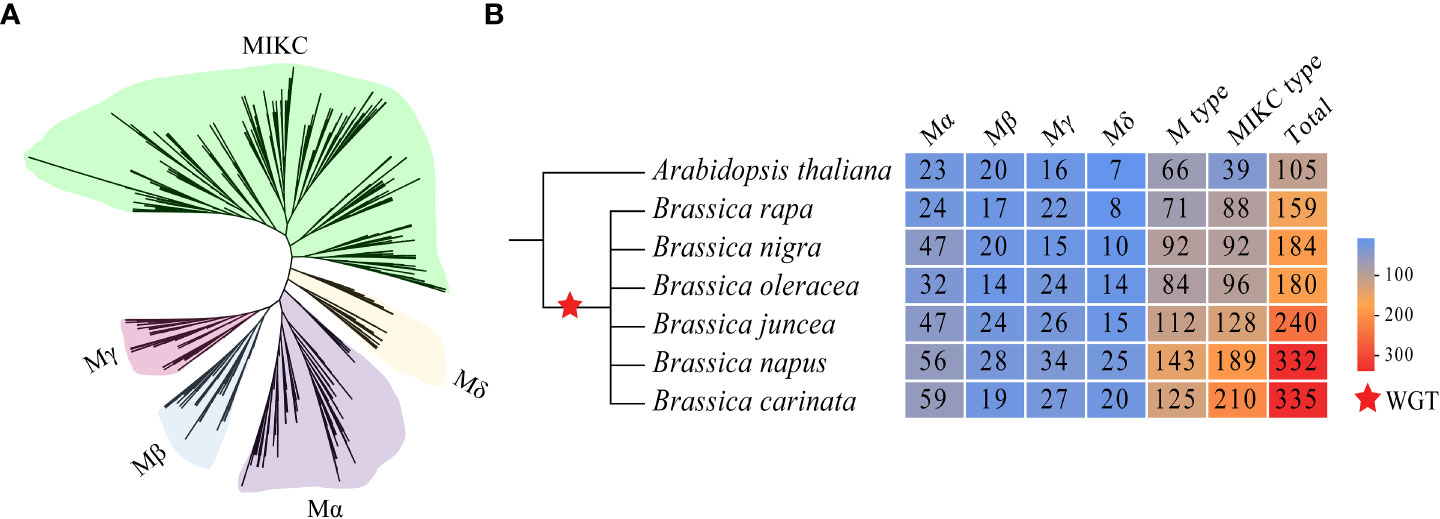
Figure 1 Evolutionary tree of MADS-box proteins in the six U’s triangle species and A thaliana and the number of subfamily members. (A) The phylogenetic tree of 1535 MADS proteins was constructed using the maximum likelihood method, and different colors represent different subfamilies. (B) The red star represents that the genome has undergone a genome-wide triplication event (WGT).
Phylogenetic tree of MIKC-type MADS-box genes
MIKC-type MADS-box genes are numerous, and their classification in plants is complicated. The ML phylogenetic tree was constructed for 39 MIKC-type genes in A. thaliana and 803 in the U’s triangle species (Figure 2). Subsequently, 842 MIKC-type genes were divided into 35 subclades according to classification criteria and nomenclature in A. thaliana. Among them, 32 subclades contained MIKC-type genes from each species, and these subclades were renamed referring to the corresponding name in A. thaliana. The other three subclades (AGL72-like, AGL18-like, and FLC-like) did not contain corresponding homologous genes in A. thaliana; therefore, their renaming was based on the name of the branch closest to their evolutionary relationships (Figure 2). AGL72-like subclades had the highest number of genes, which may be another sister group of AGL72 subclades. In addition, most subclades had a clear evolutionary classification, whereas the SEP subclade had the highest number of genes and a complicated branch topology, implying that SEP subclades may have undergone multiple duplication events in the U’s triangle species.
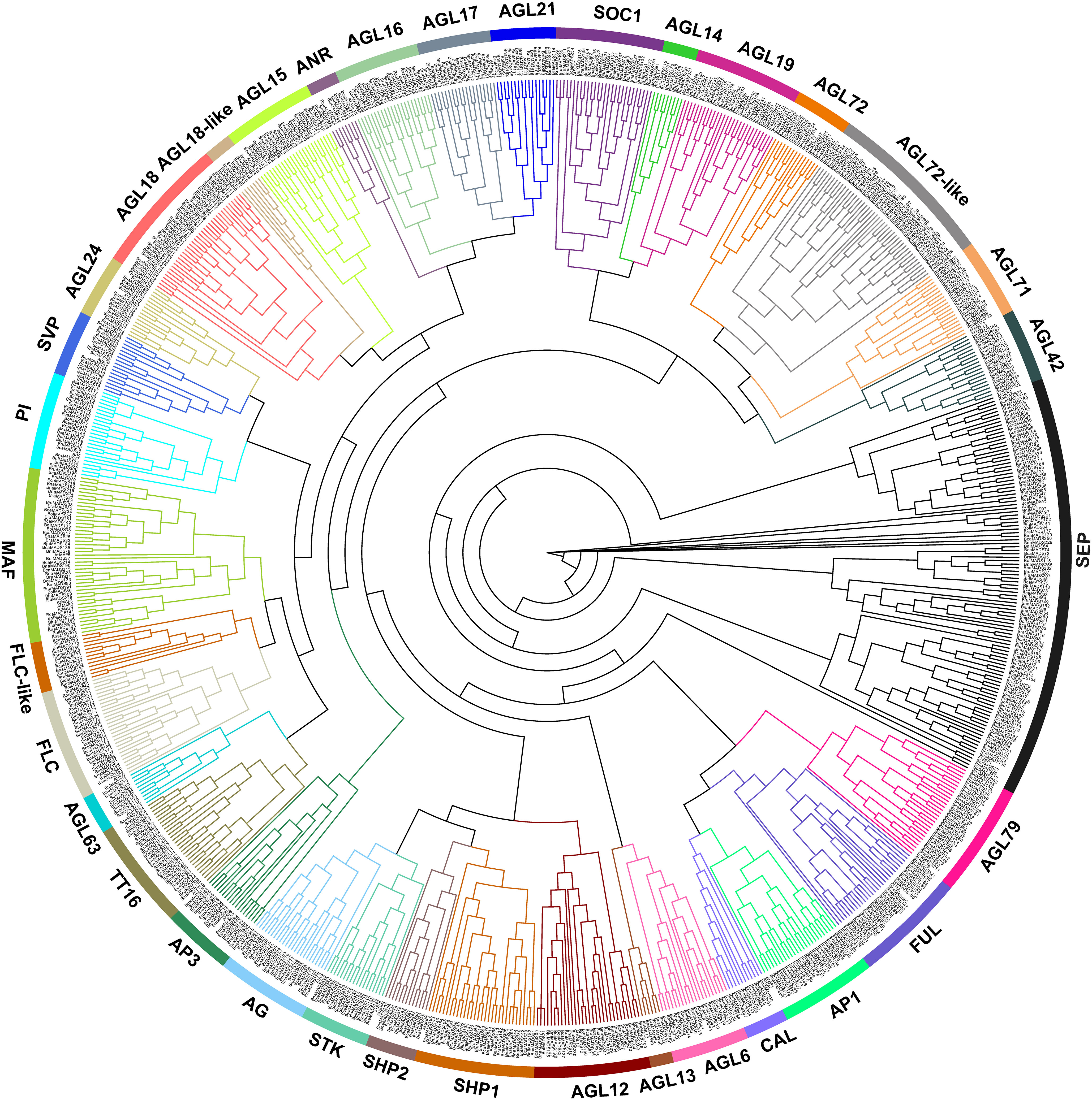
Figure 2 Neighbor-joining (NJ) tree of MIKC-type MADS-box genes identified in the six U’s triangle species; the genes were divided into 35 subgroups. Different colors represent different subclusters.
Gene structure and conserved motifs of MIKC-type MADS-box genes
The exon-intron structural characteristics of MIKC-type genes varied remarkably in different subclades; however, the number and arrangement of exon-introns were similar in the same subclade (Figure S1). Most of the genes in the AG and AP3 subclades comprised seven exons and six introns (Figures 3A, D), AP1 and CAL subclades possessed the closest evolutionary relationship, and the majority of MIKC-type genes in AP1 and CAL subgroups generally contained eight exons and seven introns (Figure 3B); the PI subclade (Figure 3E) had six exons, and the SEP subclade had eight exons (Figure 3C).
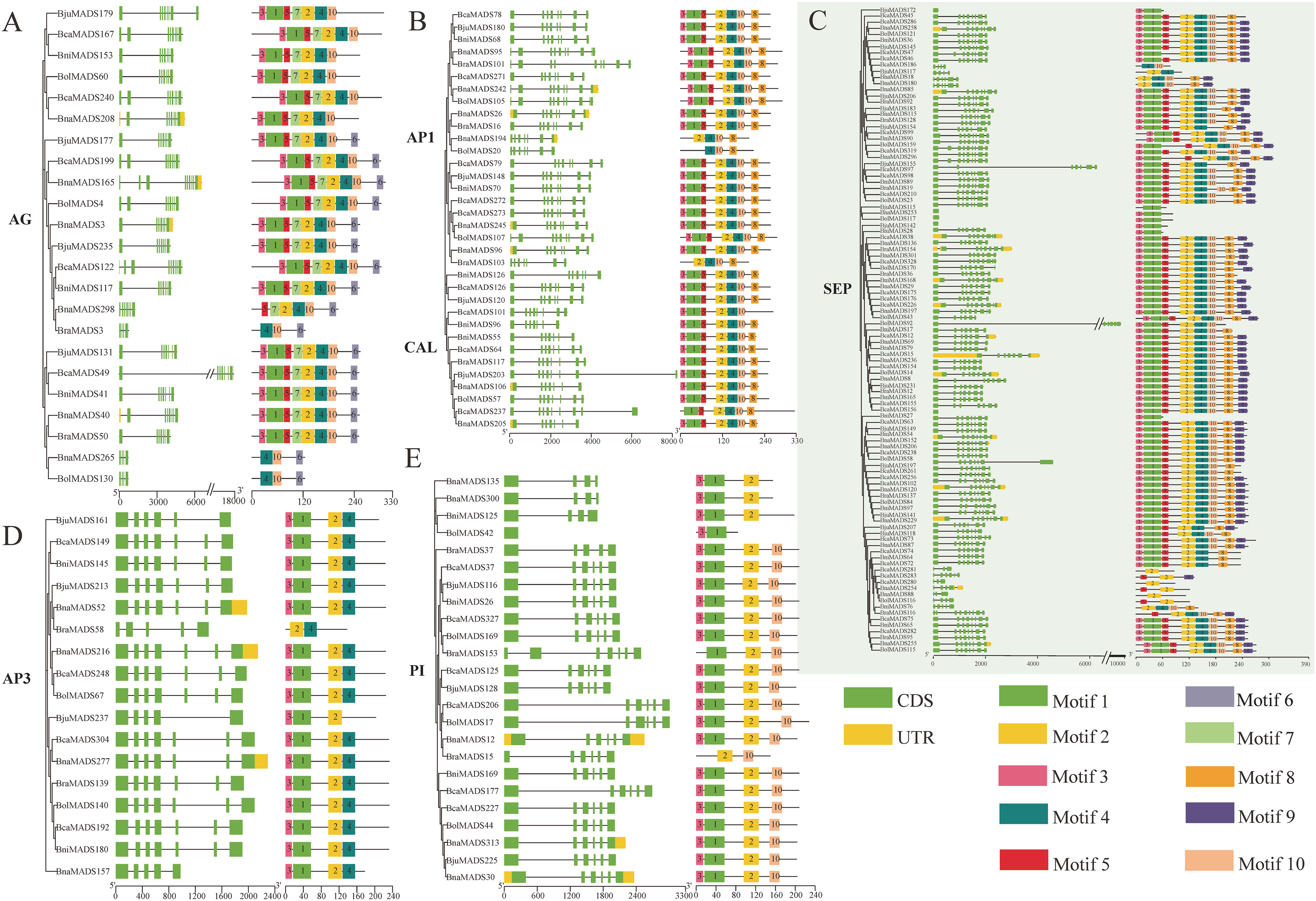
Figure 3 Gene structure and conserved motifs of six MIKC-type subclades [AG (A), AP1 and CAL (B), SEP (C), AP3 (D), and PI (E)]. The green boxes, yellow boxes, and block lines represent the exons, UTR, and introns, respectively. Ten different colored boxes represent ten distinct motifs.
Ten conserved motifs were observed in 803 MIKC-type protein sequences (Supplementary Table S4). Motif 1 and motif 3 existed in most of the MADS-box domains, and motif 2, comparatively conserved in the K-box domain, may be responsible for the conservation of MIKC-type proteins among different subclades. However, other motifs were specifically distributed in different subclades and determined the specificity of the MIKC-type subclades (Figure 3; Figure S1). For example, comparing the conserved motifs of the AG, AP1, and CAL subclades (Figures 3A, B), motif 8 was only found in the AP1 and CAL subclades, whereas motifs 6 and 7 were only found in the AG subclade. Furthermore, motif 4 and motif 10 were distinctively distributed in the AP3 and PI subclades (Figures 3D, E), respectively, determining the specificity of these two subclades. The gene structure and conserved motifs of the other subclades are shown in Figure S1.
Gene duplication and synteny of MIKC-type MADS-box genes
BLAST and MCScanX software were used to analyze the gene duplication events of MIKC-type genes in the U’s triangle species. A mass of tandem and segmental duplication events were detected in the genomes of B. rapa (3/69) (Figure 4A), B. nigra (4/75) (Figure 4B), B. oleracea (8/66) (Figure 4C), B. juncea (2/137) (Figure 4D), B. napus (4/194) (Figure 4E), and B. carinata (8/281) (Figure 4F). The duplication event numbers of MIKC-type genes in three diploid basic species were nearly the same. However, there was a significant difference among the three allotetraploids, particularly in segmental duplication events (Supplementary Tables S5, 6), suggesting that MIKC-type genes have different evolutionary processes in the three allotetraploid species. Furthermore, the subgenome distribution of segmental duplication events in the genomes of three tetraploid Brassica species was analyzed. In B. juncea, 88 segmental duplication events occurred across the AA and BB subgenomes, 22 across the AA subgenome, and 27 across the BB subgenome (Figure 4D). In B. napus, 125 segmental duplication events occurred across the AA/CC subgenome, 41 across the AA subgenome, and 28 across the CC subgenome (Figure 4E). In B. carinata, 159 segmental duplication events occurred across the BB/CC subgenome, 85 across the BB subgenome, and 37 across the CC subgenome (Figure 4F). These results indicate that segmental duplication events among the three tetraploid Brassica species mainly occurred across the different subgenomes.
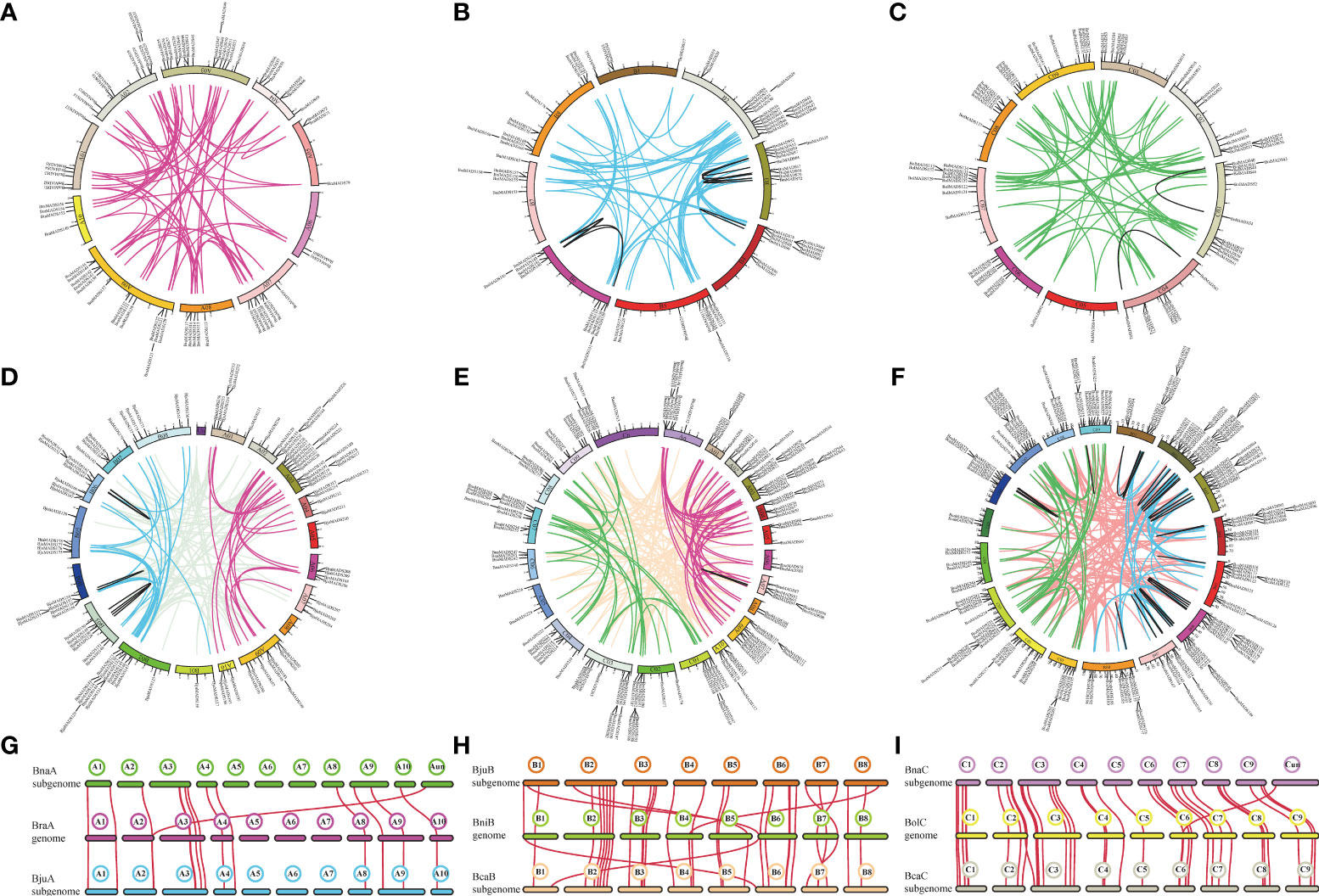
Figure 4 Duplication events and synteny of MIKC-type MADS-box genes. (A-F) Chromosome location information and segmental duplication of MIKC-type MADS-box genes. (A–F) indicate the segmental duplication events occurring within the genomes of B. rapa, B. nigra, B. oleracea, B. juncea, B. napus, and B. carinata, respectively. Colored lines represent replication events identified within different subgenomes, and black lines represent duplication gene pairs that occur on the same chromosome. (G-I) Genomic synteny between progenitor Brassica species B. rapa, B. nigra, and B. oleracea) and the allotetraploid species (B. carinata, B. juncea, and B. napus).
To elucidate the evolutionary process of MIKC-type genes, synteny between three allotetraploid species (B. carinata, B. juncea, and B. napus) and its progenitor Brassica species (B. rapa, B. nigra, and B. oleracea) was performed (Supplementary Table S7). A total of 15 gene pairs were identified between B. rapa and the A subgenome of B. napus and B. juncea (Figure 4G). Among them, BnaMADS148 (BnaAnng06990D) may be located on chrA02 of B. napus because its syntenic gene pairs were all located on chrA02. In addition, 32 gene pairs were detected across the B. nigra genome and B subgenomes of B. juncea and B. carinata (Figure 4H), of which 28 gene pairs were shared on the same chromosome among the B genome of progenitor species and B subgenome of its hybridized allotetraploid species, whereas the other four were shared on different chromosomes. Additionally, 37 MIKC-type gene pairs were identified across the B. oleracea genome and C subgenomes of B. napus and B. carinata (Figure 4I). Overall, the number of syntenic gene pairs in subgenome A was significantly lower than that in subgenome B or C.
Expression profile analysis of MIKC-type BnMADS-box genes in various tissues of B. napus
B. napus is an important oil crop in the U’s triangle, and its abundant and reliable transcriptome data provide a basis for exploring the expression patterns of MIKC-type genes. Therefore, transcriptome data of various tissues, embryos, and seed coats at different developmental stages were downloaded from the public RNA-seq database to analyze the expression characteristics of MIKC-type genes. A total of 63 MIKC-type BnaMADS-box genes were screened from the expression matrix of different tissues (Supplementary Table S8), which were classified into five subgroups according to their expression patterns (Figure 5A). Subgroup I genes were highly expressed in stems and leaves, and some genes were also highly expressed in siliques and other tissues; however, all genes were expressed at low levels in the seed. Subgroup II genes were mainly expressed in flowers, seeds, and siliques, but their expression was low in roots. The subgroup III genes were root-specific. Nine genes, including especially BnaMADS37, BnaMADS111, BnaMADS155, and BnaMADS189, in subgroup IV were highly expressed in seeds. The genes in subgroup V, including BnaMADS12, BnaMADS30, BnaMADS300, and BnaMADS313, were only highly expressed in the flowers. Overall, the expression patterns of the 63 BnMADS-box genes were tissue-specific, suggesting that these genes may have vital biological functions in tissues during specific developmental processes.
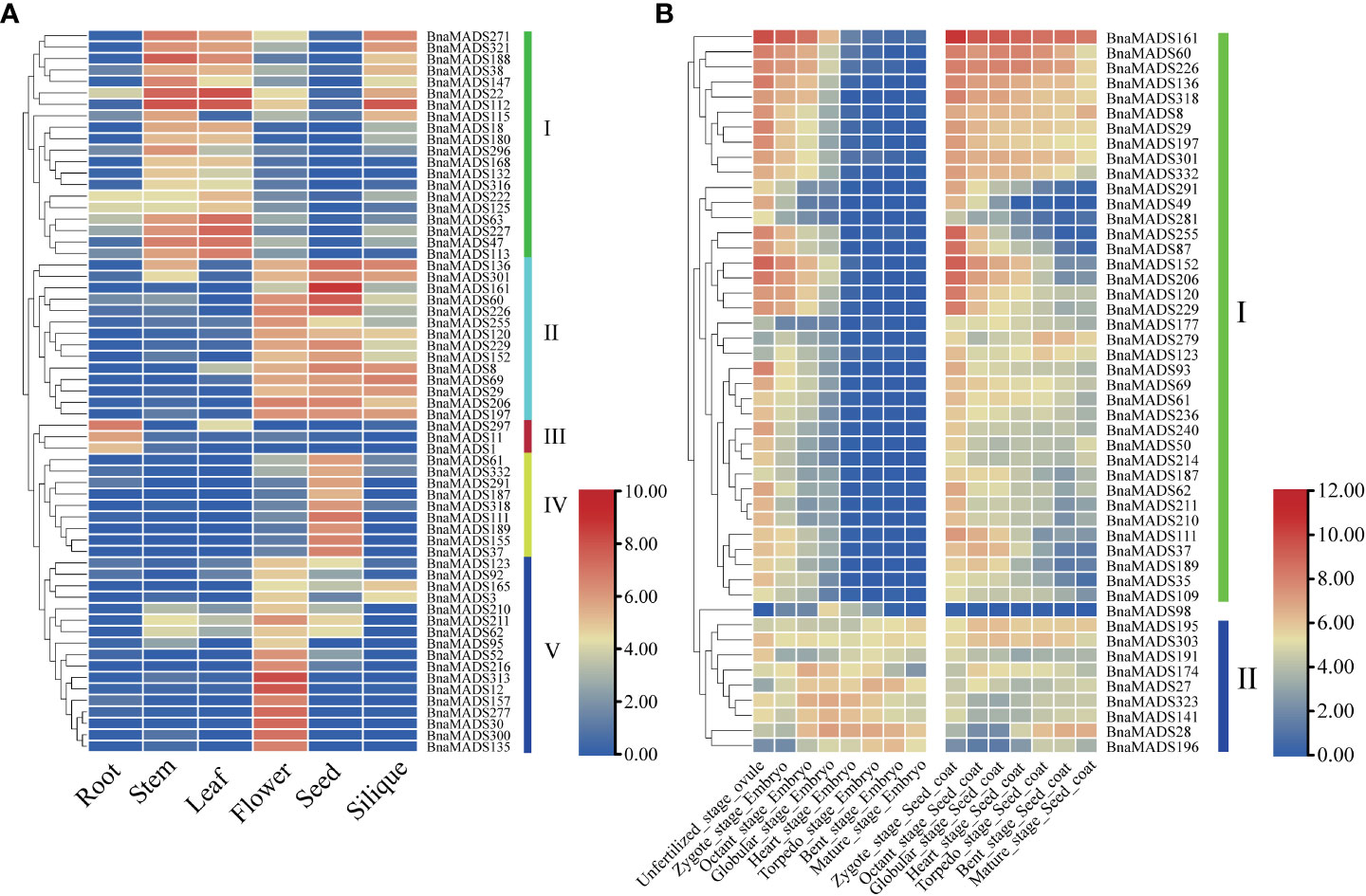
Figure 5 Expression patterns of MIKC-type MADS-box genes in different tissues and developmental stages. (A) Heatmap of TPM values in six different tissues (root, stem, leaf, flower, seed, and silique). (B) The gene expression trends during different unfertilized ovule, embryo, and seed coat stages.
To explore the expression patterns of MIKC-type BnaMADS-box genes at different developmental stages of the embryo and seed coat, the expression patterns of 48 BnaMADS-box genes were divided into two major subgroups (Figure 5B; Supplementary Table S9). Subgroup I genes were mainly expressed during the unfertilized ovule stage and first three periods of embryonic development (zygote, octant, and globular stages), whereas subgroup II genes were generally expressed during the embryo and seed coat development stages, especially in the middle and late stages of embryo development, such as BnaMADS27 was highly expressed in torpedo and bent stage embryos, BnaMADS141 and BnaMADS323 were highly expressed in octant, globular, and heart stage embryos. In addition, BnaMADS98 was highly expressed in globular stage embryo, whereas it was less expressed during seed coat development. Therefore, analyzing the expression pattern of MIKC-type genes in B. napus provided a guide for exploring the biological functions of MIKC-type genes in various tissues during the entire developmental process.
Expression profiles of MIKC-type BnMADS-box genes under biotic and abiotic stressors in B. napus
To determine the mechanism of MIKC-type genes responding to biotic and abiotic stress, transcriptome data for dehydration, salt, ABA, and cold stress were downloaded from the public RNA-seq database (Supplementary Table S10). Based on gene expression trends under abiotic stress, the 38 MIKC-type genes were divided into three categories: subgroups I, II, and III (Figure 6A). Subgroup I genes were expressed under various types of abiotic stresses. BnaMADS227, BnaMADS286, and BnaMADS297 were significantly upregulated after dehydration, salt, ABA, and cold treatment, whereas BnaMADS104 and BnaMADS311 were more susceptible to dehydration, salt, and ABA stress. BnaMADS222 was downregulated at 1 h and 8 h of dehydration stress, indicating that BnaMADS222 was more sensitive to dehydration stress. A small subset of the genes in subgroup II was sensitive to abiotic stress. BnaMADS2 and BnaMADS164 were induced by dehydration and cold treatment; BnaMADS38 and BnaMADS302 were upregulated under dehydration and salt stress, and BnaMADS56 was significantly downregulated after dehydration stress. Subgroup III genes had low expression under abiotic treatments; however, individual genes were upregulated after stress treatment. For example, BnaMADS123 and BnaMADS63 were significantly highly expressed after 24 h of cold treatment, and BnaMADS35 was upregulated after 24 h of ABA treatment. The rapeseed leaf transcriptome data of resistant and susceptible lines after inoculation with powdery mildew were downloaded for further analysis (Figure 6B; Supplementary Table S11). A total of 21 MIKC-type genes responded to powdery mildew stress, of which 16 genes, including BnaMADS113, BnaMADS118, and BnaMADS321, showed significantly higher expression in resistant strains than in sensitive strains. Furthermore, BnaMADS2, BnaMADS11, BnaMADS164, BnaMADS190, and BnaMADS215 showed higher expression in susceptible strains than in resistant strains.
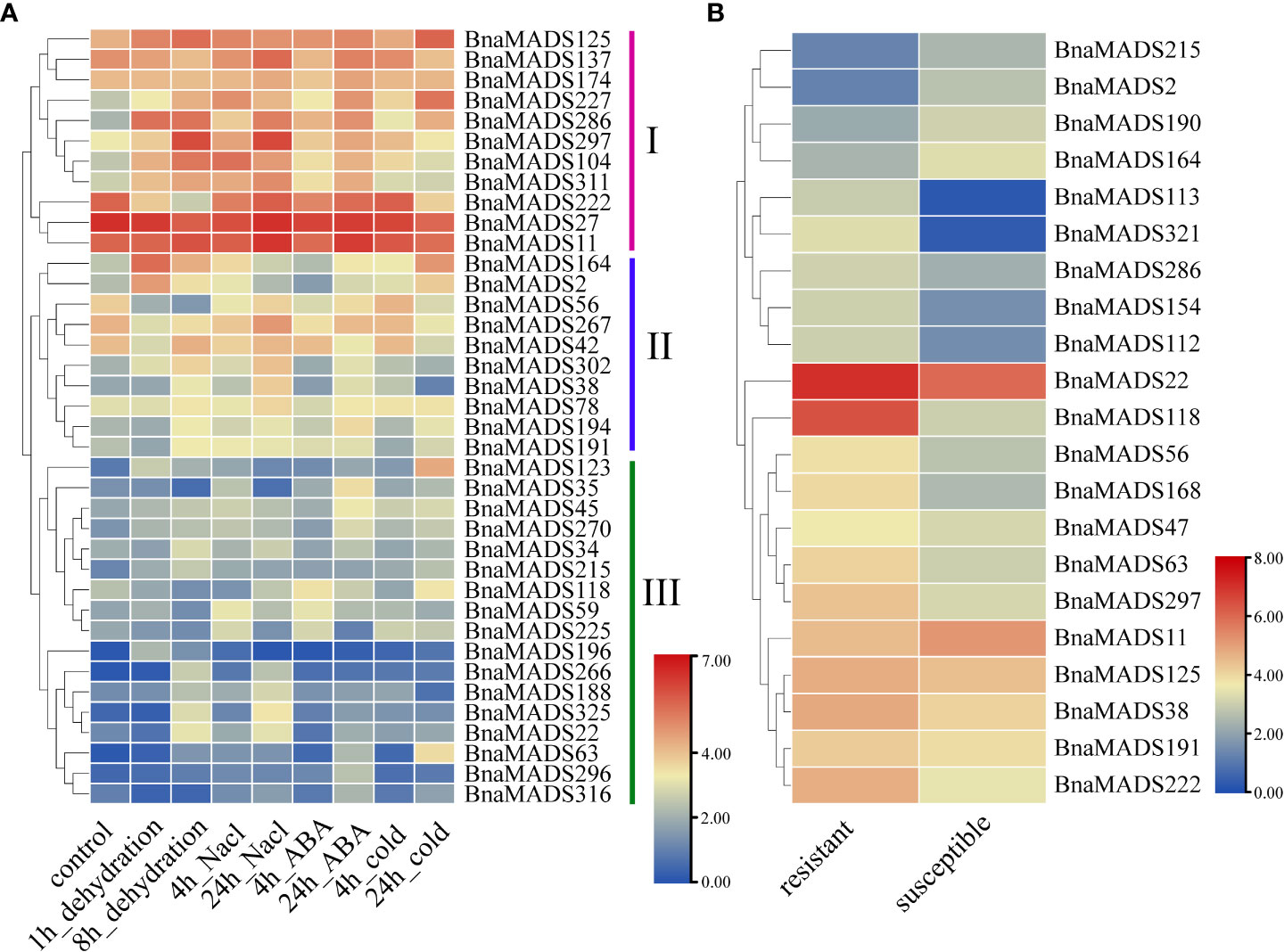
Figure 6 Expression of MIKC-type MADS-box genes in response to biotic and abiotic stresses. (A) Expression profiles of genes in rapeseed leaves under four different abiotic stressors (dehydration, salt, ABA, and cold). (B) The leaves expression patterns of two different white powder-resistant rapeseed lines after inoculation with powdery mildew.
Sequence analysis of BnaAG and vector construction
To further verify the contribution of BnaAG genes to floral organ development, four highly homologous BnaAGs, BnaA01g09760D, BnaA03g43820D, BnaC01g11460D, BnaC03g62970D (also named BnaAG.A01, BnaAG.A03, BnaAG.C01, and BnaAG.C03, respectively), located on chromosomes A01, A03, C01, and C03, respectively, were isolated by Blastp with AtAG. Phylogenetic tree analysis showed that BnaAG.A01 and BnaAG.C01 had the closest relationships with AtAG (Figure 7A). The observation of gene structure features showed that most of the BnaAG genes contained seven exons, whereas BnaAG.C01 was composed of nine exons but had no effect on its conserved domain at the gene structure level (Figures 7B). Based on multiple alignments of BnaAG homologous genes, CRISPR/Cas9 vectors were constructed to target the conserved regions of four BnaAG homoeologs. In brief, the consensus sequence on the first exon of BnaAG.A01, BnaAG.A03, BnaAG.C03, and the third exon of BnaAG.C01 were targeted by sgRNA1. SgRNA2 targeted the consensus sequence on the third exon of BnaAG.A01, BnaAG.A03, BnaAG.C03, and the fifth exon of BnaAG.C01 (Figures 7C). Based on these results, sgRNA1 was located on the MADS-box domain and sgRNA2 on the K-box domain.
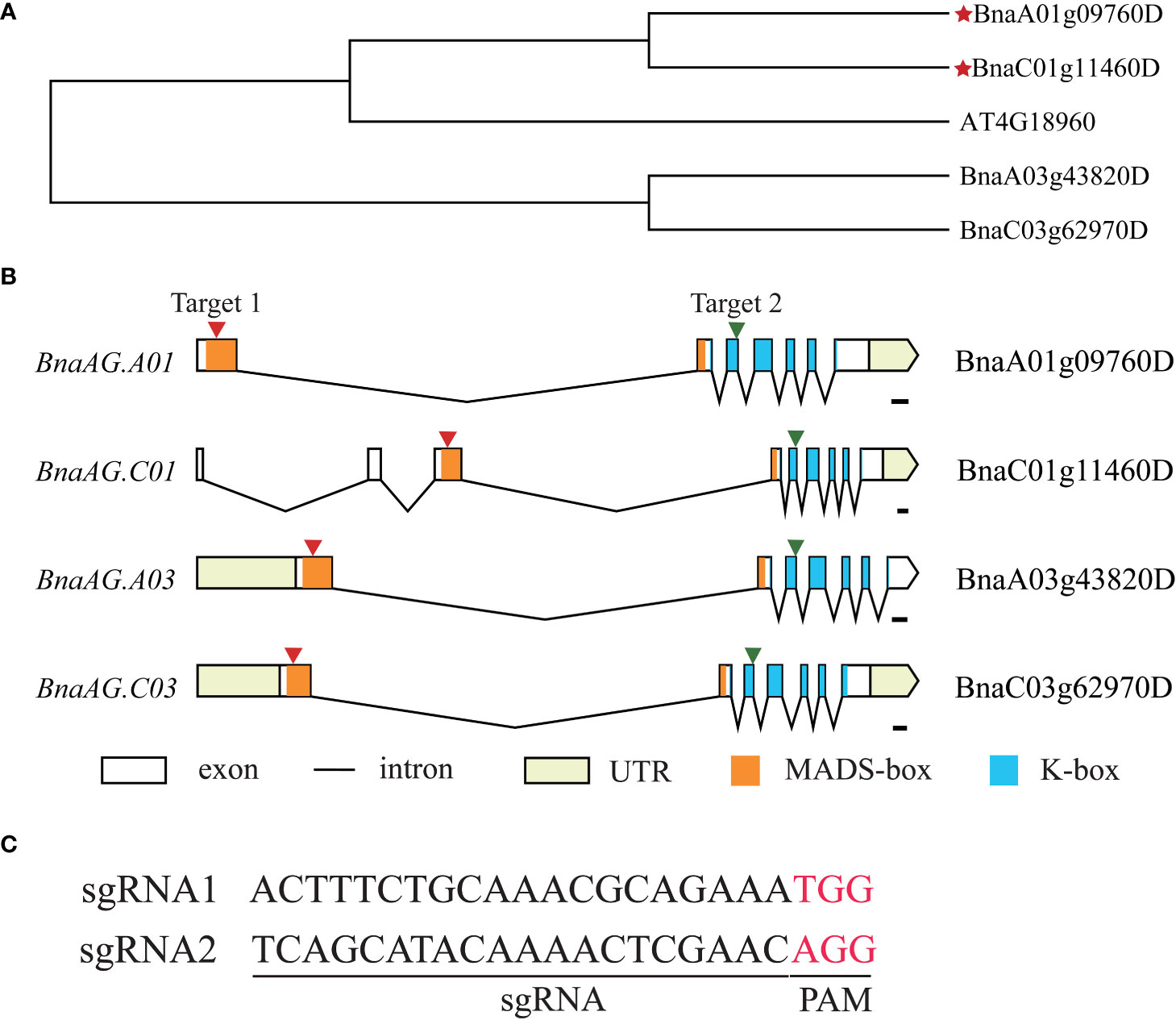
Figure 7 Construction of vectors for targeting BnaAG homologous genes. (A) NJ tree of BnaAG homologous genes in B napus and A thaliana. The red stars represent the BnaAG homologs of B napus closely related to the AG gene in A thaliana. (B) Gene structure, conserved domain, and distribution of sgRNA1 and sgRNA2 targeting four homologous genes of BnaAG. The black line represents introns, and the black boxes represent the exon. The red arrows indicate the location of sgRNA targets on the exon, and the orange and blue boxes represent MADS-box and K-box domains, respectively. (C) Sequence information of two sgRNA, PAM is indicated in red.
CRISPR/Cas9 mediated mutation in BnaAG homoeologs and enriched floral organ mutant phenotypes
After the genetic transformation of the rapeseed hypocotyl mediated by Agrobacterium tumefaciens, more than 40 T0 generation hygromycin B-positive plants were obtained, of which 20 lines consisted of four concentric wheels and showed no difference from the wild-type (WT), whereas the other lines showed a disorganized floral structure. Therefore, T0 generation lines can be classified into four types based on their floral organ mutation phenotypes. To verify the mutation type, plants of all phenotypes were screened using Sanger sequencing, and the editing efficiency of sgRNA1 was found to be significantly lower than that of sgRNA2 (Figures S2, 3). Specifically, compared to wild type (WT) (Figures 8A–C), there were no phenotypic differences found in wt-like (wtl) (Figures 8D–F). Sequencing results showed that no editing events existed in target 1, and all BnaAG homologs had a single base inserted except BnaAG.A01 in target 2 (Figure 8S). The female flower-like (feml) mutant (Figures 8G–I), which led to the conversion of stamens into petals and expansion of carpels, generated low seed-setting and distorted siliques after cross-pollination (Figure 8P). The sequencing results showed that no editing events happened except for single base insertion in BnaAG.C01 of target 1. BnaAG.C01, BnaAG.C03, and BnaAG.A03 had a single base insertion in target 2, whereas BnaAG.A01 had a deletion of three bases (CGA) in target 2 that did not cause frameshift mutation (Figure S4). The next mutant, like ag-2 mutant in A. thaliana, gave rise to the following phenotype: six stamens replaced by six petals, and the carpel replaced by a new flower with an elongated pedicel, in which stamens and carpels were replaced by petals (also called agamous-like flower-2 (aglf-2) (Yanofsky et al., 1990; Tsutsui and Higashiyama, 2017) (Figures 8J–L, Q, R). Target 1 of the BnAG homologous gene was not edited; however, insertion or deletion occurred in all BnAG homologous genes in target 2, resulting in a frameshift mutation. The phenotype of the last mutant was similar to the ag-1 mutant in A. thaliana (Yanofsky et al., 1990); its outermost whorl had four normal sepals, while its third whorl stamens and fourth whorl carpel were all replaced by a multitude of petals. The resulting flower mutant, agamous-like flower-1 (aglf-1), contained more than 85 petals (Figures 8M–O), in which BnaAG.A01, BnaAG.C01, and BnaAG.A03 with base insertion or deletion in target 2 caused a frameshift mutation, whereas BnaAG.C03 had a single base insertion in both the target squences, resulting in a frameshift mutation.
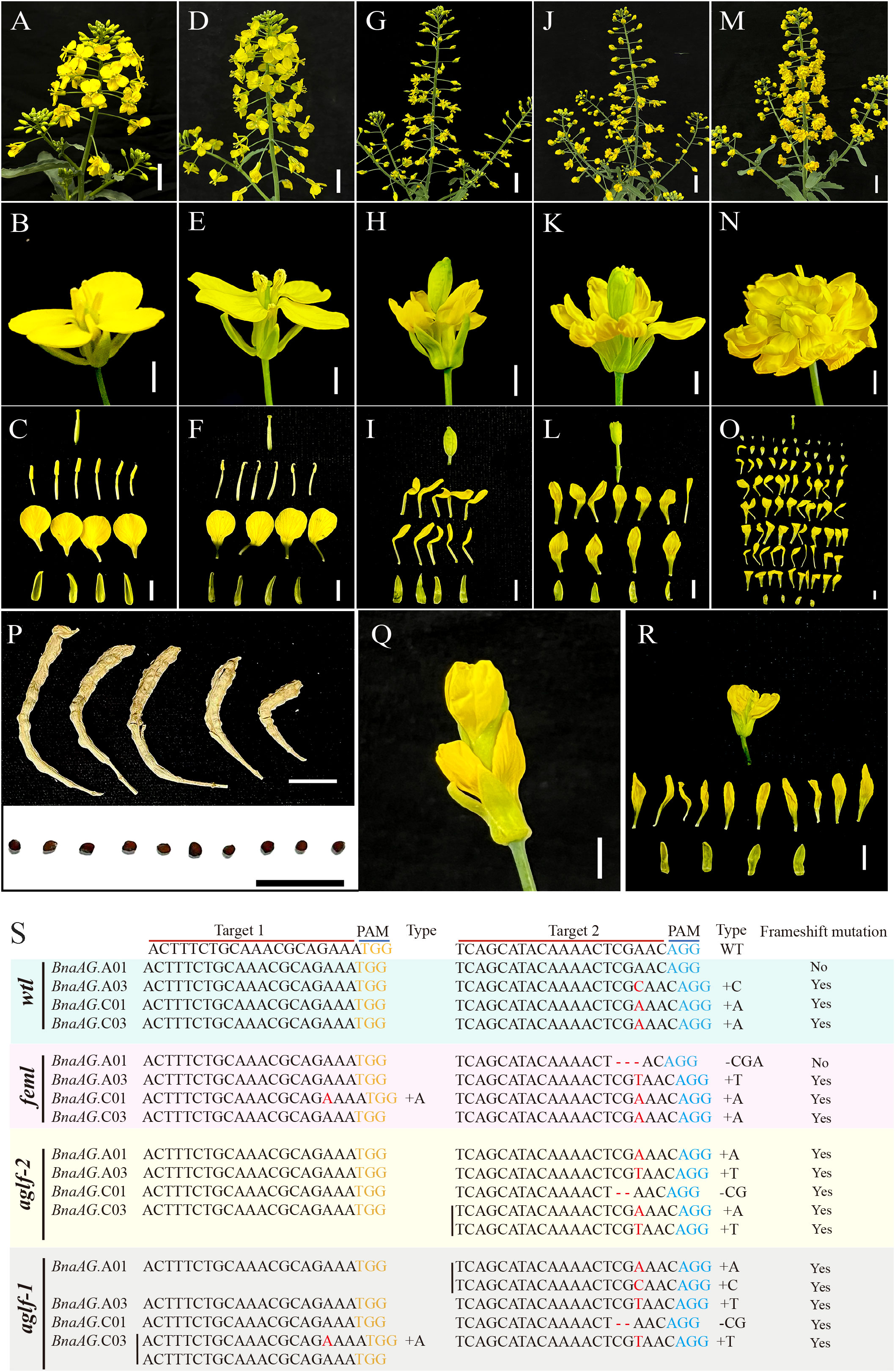
Figure 8 Phenotype and target sequence validation of the T0 generation of the gene-edited BnaAG genes. mInflorescences of WT (A), wtl (D), feml (G), aglf-2 (J), and aglf-1 (M). Scale bars, 3 cm. The flowers of WT (B), wtl (E), feml (H), aglf-2 (K), and aglf-1 (N) plants. Scale bars, 0.5 cm. Components from the anatomizing flowers of WT (C), wtl (F), feml (I), aglf-2 (L), and aglf-1 (O). Scale bars, 0.5 cm. (P) Mature siliques and seeds of feml lines after cross-pollination. Scale bars, 1 cm. (Q, R) Flower (Q) and components from the anatomizing flower (R) of aglf-2 during the flowering stage. (S) Validation of target sequences for various mutant phenotypes. Yellow and blue fonts represent the PAM of target sequences 1 and 2, respectively. Red ‘-’ means deletions, and red font represents single base insertions. Yes or no indicates that the BnaAG gene underwent a frameshift mutation.
Discussion
The U’s triangle model represents the six globally important Brassica species and explains the genetic relationships among them. The B. carinata reference genome was published in 2021, marking the start of functional genomics. It also promoted the exploration of genetic evolution and molecular design breeding in the U’s triangle species (Song et al., 2021; Yim et al., 2022). The MADS-box gene family is an important transcription factor that functions in floral homeostasis, morphogenesis, angiosperm reproductive development, and vegetative development (Okada and Shimura, 1994; Becker et al., 2000; Alvarez-Buylla et al., 2019).
The MIKC-type MADS-box genes were significantly expanded in the genome of six U’s triangle species
Previous studies showed that a total of 160, 91, and 307 MADS-box genes have been identified in B. rapa, B. oleracea, and B. napus genomes, respectively. However, there was no related research regarding MADS-box genes in B. nigra, B. juncea, and B. carinata genomes (Duan et al., 2015; Wu et al., 2018; Sheng et al., 2019). With the disclosure of the genome information of the remaining three Brassica species (B. nigra, B. juncea, and B. carinata), this study was the first to perform the systematic identification and classification of MADS-box genes in U’s triangle species. A total of 159 MADS-box genes were identified in the B. rapa genome, which is consistent with previous results (Duan et al., 2015). However, 180 MADS-box genes were investigated in the B. oleracea genome, which was approximately twice the number previously reported (Sheng et al., 2019), and 332 MADS-box genes were identified in the B. napus genome, which was 25 more than originally reported (Wu et al., 2018) (Figure 1B). The differences in the number of members may be confined to the reference genome quality of previously assembled Brassica species. The MADS-box genes can be divided into M-type and MIKC-type according to their phylogenetic relationships. The gene number ratio of the two types was 1.69 (66:39) in A. thaliana, 0.8 (71:88) in B rapa, 1.0 (92:92) in B. nigra, 0.87 (84:96) in B. oleracea, 0.87 (112:128) in B. juncea, 0.75 (143:189) in B. napus, and 0.59 (125:210) in B. carinata, indicating that the ratio of M-type and MIKC-type genes decreased gradually compared with that of A. thaliana. Moreover, the genome of the three diploid U’s triangle species has undergone genome-wide triploid events during the evolutionary process (Wang et al., 2011; Woodhouse et al., 2014), leading to a significantly higher ratio of MIKC-type genes in A. thaliana and three diploid species (A. thaliana:B. rapa:B. nigra:B. oleracea = 39:88:92:96) than that of M-type genes (A. thaliana:B. rapa:B. nigra:B. oleracea = 66:71:92:84). The function of M-type genes in plants is poorly understood, whereas MIKC-type genes are implicated in plant growth and development and environmental adaptation (Kofuji et al., 2003; Yang et al., 2016; Guo et al., 2021). Therefore, the significant expansion of MIKC-type genes in the U’s triangle species may be an adaptive evolution.
The tissue-specific expression patterns of MIKC-type BnaMADS-box genes
Public transcriptome data helps effectively identify valuable genes. In this study, a large number of tissue-specific MIKC-type BnaMADS-box genes were discovered in various tissues and during embryo development processes in B napus, of which BnaMADS1 and BnaMADS297 were highly expressed in roots (Figure 5A). AtAGL14 (XAL2, AT4G11880), a homologous gene of BnaMADS297 in A. thaliana, which binds to the CArG box (a cis-regulatory element of PIN1 and PIN4), regulates root elongation by upregulating the abundance of PIN protein (Garay-Arroyo et al., 2013). AGL21 (AT4G37940), a homolog of BnaMADS1, mainly regulates lateral root development, especially during lateral root growth under low-nitrogen conditions (Yu et al., 2014). BnaMADS111 is specifically expressed in seeds and highly expressed during the early stage of seed coat development (zygote, octant, and globular stage) (Figure 5B); its homolog AGL32 (TT16, AT5G23260) modulates the development of the sub-epidermal integument cell layer in A. thaliana seeds (Coen et al., 2017). BnMADS12 and BnMADS313 were specifically highly expressed only in flowers, and their homologous gene PI in A. thaliana acts as a floral homeotic gene mainly involved in the regulation of petal and stamen development (Goto and Meyerowitz, 1994). BnaMADS28 is highly expressed during the later stages of embryo and seed coat development, and its homologous gene AGL15 (AT5G13790) facilitates somatic embryogenesis in A. thaliana (Paul et al., 2022). BnaMADS161 is highly expressed during the early stage of embryo development and throughout the seed coat development process, and its homologous gene AGL11 (STK, AT4G09960) in Arabidopsis is involved in seed and silique development (Di Marzo et al., 2022).
MIKC-type BnaMADS-box genes are widely involved in drought stress in B. napus
Plant responses to biotic and abiotic stresses reflect resilience to external challenges. Plants usually respond to drought stress by flowering early to complete their life cycle, a phenomenon called drought escape (Waadt et al., 2022). Transcriptome data analysis showed that some BnaMADS-box genes involved in flowering time are also highly expressed under drought stress, possibly regulating drought escape. For example, AGL20 (SOC1, AT2G45660), a homologous gene of BnaMADS227 in A. thaliana, is upregulated by ABF3 and ABF4 under drought stress, thus promoting flowering (Hwang et al., 2019). BnaMADS227 is not only upregulated under drought stress but also induced by salt, ABA, and cold stress, indicating that BnaMADS227 participates in drought response and other abiotic stresses. In addition, BnaMADS2 and BnaMADS164 also respond to various abiotic stresses, such as dehydration and cold stress, and homologous AGL16 (AT3G57230) negatively regulates drought stress through stomatal density and movement in A. thaliana (Zhao et al., 2020). Thus, we speculated that these two genes may be involved in drought or cold stress. The expression of some genes, such as BnaMADS118, BnaMADS113, and BnaMADS321, was high in resistant strains and low in susceptible strains; however, the homologs of these genes in Arabidopsis are not reportedly involved in biotic stress. Exploring the mechanism of these genes in powdery disease can enrich the function of MIKC-type BnaMADS-box genes.
The BnaAG homolog genes may be functionally redundant
Polyploid plants enriched the floral organ phenotype due to the complex gene copy number but interfered with the systematic study of floral organ development. AG is responsible for stamen identity, carpel identity, and floral meristem determination (Theissen et al., 2000; Dreni and Kater, 2014). In this study, CRISPR/Cas9 technology was used to knock-out four BnaAG genes, and four lines with significant phenotypic differences in the T0 generation were obtained: wtl, feml, aglf-2, and aglf-1 (Figure 8; Figure S3). The BnaAG.A01 was not edited in wtl, suggesting that the presence of only one copy of BnaAG can maintain the normal development of floral organs. However, the amino acid mutation of BnaAG.A01 did not cause a termination mutation, and the frameshift mutation occurred on the other three homologous genes, resulting in the feml phenotype. In addition, termination mutations of four BnaAG homologs in aglf-2 and aglf-1 resulted in phenotypes similar to those in Arabidopsis (Yanofsky et al., 1990). These results confirmed that BnaAG genes were involved in the development of third and fourth whorls in flowers, suggesting that there may be functional redundancy among the four BnaAG genes.
BnaAG may be a sex-identification marker gene in B. napus
AG is responsible for the floral organ sex determination of plants (Zhang et al., 2020; Wang et al., 2021). The flowers of B. napus are monoecious, and the creation and application of male sterile materials in rapeseed can greatly enrich the utilization of heterosis. The feml phenotype, similar to the female flower, was found in the T0 generation after editing BnaAG. Although the carpel was deformed, a few seeds could be obtained by cross-pollination, implying that the feml was a female flower without stamens. Thus, BnaAG may be a sex-determining marker gene in B. napus.
Conclusions
In conclusion, 1430 MADS-box genes were identified in the six U’s triangle species and classified into M-type and MIKC-type based on their evolutionary relationships. Gene structure, conserved motifs, duplication events, and synteny between the subgenomes of different MIKC-type genes were analyzed. The expression profiles of different tissues revealed that the MIKC-type genes exhibited tissue-specific expression. Transcriptome analysis of genes induced by abiotic and biotic stress showed that MIKC-type MADS-box genes responded to external stimuli. Moreover, four types of floral organ mutants were created by editing BnaAG homologous genes, which revealed the mechanisms by which BnaAG regulates floral organ development in rapeseed. Our study provides novel insights into the regulation of floral organ development in Brassica species and may help develop new genetic varieties.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SWH designed the experiments. MS wrote the manuscript. MS and YFZ executed the experiments. QJ and SHH analyzed the data. RA and NC prepared the figures. JM and YTZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Shaanxi Creative Talents Promotion Plan-Technological Innovation Team (2020TD-051), the Scientific Research and Sharing Platform Construction Project of Shaanxi Province (2021PT-036), the Key Research and Development Program of Henan Province (221111110200), the Key Research and Development Program of Yangling Seed Industry Innovation Center (Ylzy-yc-2021-02), and the National Key Research and Development Program of China (2016YFD0101900).
Acknowledgments
We are thankful to Qijun Chen (China Agricultural University) for providing the pHSE401 vector.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1115513/full#supplementary-material
Supplementary Figure 1 | Gene structure and conserved motifs of MIKC-type subclades except for AG, AP1, CAL, AP3, PI, and SEP subclades.
Supplementary Figure 2 | Editing efficiency of sgRNA-1 and sgRNA-2.
Supplementary Figure 3 | Multiple sequence alignment of BnaAG.A01 in WT and feml.
References
Alvarez-Buylla, E. R., Garcia-Ponce, B., Sanchez, M. P., Espinosa-Soto, C., Garcia-Gomez, M. L., Pineyro-Nelson, A., et al. (2019). MADS-box genes underground becoming mainstream: plant root developmental mechanisms. New Phytol. 223, 1143–1158. doi: 10.1111/nph.15793
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8, 242. doi: 10.1186/1471-2164-8-242
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Becker, A., Winter, K. U., Meyer, B., Saedler, H., Theissen, G. (2000). MADS-box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17, 1425–1434. doi: 10.1093/oxfordjournals.molbev.a026243
Bhalla, P. L., Singh, M. B. (2008). Agrobacterium-mediated transformation of brassica napus and brassica oleracea. Nat. Protoc. 3, 181–189. doi: 10.1038/nprot.2007.527
Cai, C., Wang, X., Liu, B., Wu, J., Liang, J., Cui, Y., et al. (2017). Brassica rapa genome 2.0: A reference upgrade through sequence re-assembly and gene re-annotation. Mol. Plant 10, 649–651. doi: 10.1016/j.molp.2016.11.008
Cai, X., Wu, J., Liang, J., Lin, R., Zhang, K., Cheng, F., et al. (2020). Improved brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theor. Appl. Genet. 133, 3187–3199. doi: 10.1007/s00122-020-03664-3
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, F., Mandakova, T., Wu, J., Xie, Q., Lysak, M. A., Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Chen, X., Tong, C., Zhang, X., Song, A., Hu, M., Dong, W., et al. (2021). A high-quality brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 19, 615–630. doi: 10.1111/pbi.13493
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Coen, O., Fiume, E., Xu, W., De Vos, D., Lu, J., Pechoux, C., et al. (2017). Developmental patterning of the sub-epidermal integument cell layer in arabidopsis seeds. Development 144, 1490–1497. doi: 10.1242/dev.146274
Di Marzo, M., Viana, V. E., Banfi, C., Cassina, V., Corti, R., Herrera-Ubaldo, H., et al. (2022). Cell wall modifications by alpha-XYLOSIDASE1 are required for control of seed and fruit size in arabidopsis. J. Exp. Bot. 73, 1499–1515. doi: 10.1093/jxb/erab514
Dreni, L., Kater, M. M. (2014). MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol. 201, 717–732. doi: 10.1111/nph.12555
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2015). Genome-wide analysis of the MADS-box gene family in brassica rapa (Chinese cabbage). Mol. Genet. Genomics 290, 239–255. doi: 10.1007/s00438-014-0912-7
Egea-Cortines, M., Saedler, H., Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in antirrhinum majus. EMBO J. 18, 5370–5379. doi: 10.1093/emboj/18.19.5370
Fan, H. Y., Hu, Y., Tudor, M., Ma, H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12, 999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x
Ferrandiz, C., Liljegren, S. J., Yanofsky, M. F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during arabidopsis fruit development. Science 289, 436–438. doi: 10.1126/science.289.5478.436
Garay-Arroyo, A., Ortiz-Moreno, E., de la Paz Sanchez, M., Murphy, A. S., Garcia-Ponce, B., Marsch-Martinez, N., et al. (2013). The MADS transcription factor XAL2/AGL14 modulates auxin transport during arabidopsis root development by regulating PIN expression. EMBO J. 32, 2884–2895. doi: 10.1038/emboj.2013.216
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Goto, K., Meyerowitz, E. M. (1994). Function and regulation of the arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Guo, N., Wang, S., Gao, L., Liu, Y., Wang, X., Lai, E., et al. (2021). Genome sequencing sheds light on the contribution of structural variants to brassica oleracea diversification. BMC Biol. 19, 93. doi: 10.1186/s12915-021-01031-2
Hartmann, U., Hohmann, S., Nettesheim, K., Wisman, E., Saedler, H., Huijser, P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Munster, T., Theissen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss physcomitrella patens. Mol. Biol. Evol. 19, 801–814. doi: 10.1093/oxfordjournals.molbev.a004137
Honma, T., Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Immink, R. G., Gadella, T. W., Jr., Ferrario, S., Busscher, M., Angenent, G. C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. U S A. 99, 2416–2421. doi: 10.1073/pnas.042677699
Jack, T. (2001). Plant development going MADS. Plant Mol. Biol. 46, 515–520. doi: 10.1023/a:1010689126632
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, H. A., Lim, C. J., Kim, S., Choe, J. K., Jo, S. H., Baek, N., et al. (2014). High-throughput sequencing and de novo assembly of brassica oleracea var. capitata l. for transcriptome analysis. PLoS One 9, e92087. doi: 10.1371/journal.pone.0092087
Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., et al. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20, 1963–1977. doi: 10.1093/molbev/msg216
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Letunic, I., Bork, P. (2007). Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. doi: 10.1038/ncomms4930
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ma, J., Yang, Y., Luo, W., Yang, C., Ding, P., Liu, Y., et al. (2017). Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum l.). PLoS One 12, e0181443. doi: 10.1371/journal.pone.0181443
Mun, J. H., Kwon, S. J., Seol, Y. J., Kim, J. A., Jin, M., Kim, J. S., et al. (2010). Sequence and structure of brassica rapa chromosome A3. Genome Biol. 11, R94. doi: 10.1186/gb-2010-11-9-r94
Nagaharu, U., Nagaharu, N. J. J. J. B. (1935). Genome analysis in brassica with special reference to the experimental formation of b. napus and peculiar mode of fertilization. Jpn J. Bot. 7, 389–452.
Okada, K., Shimura, Y. (1994). Genetic analyses of signalling in flower development using arabidopsis. Plant Mol. Biol. 26, 1357–1377. doi: 10.1007/BF00016480
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Paritosh, K., Pradhan, A. K., Pental, D. (2020). A highly contiguous genome assembly of brassica nigra (BB) and revised nomenclature for the pseudochromosomes. BMC Genomics 21, 887. doi: 10.1186/s12864-020-07271-w
Paritosh, K., Yadava, S. K., Singh, P., Bhayana, L., Mukhopadhyay, A., Gupta, V., et al. (2021). A chromosome-scale assembly of allotetraploid brassica juncea (AABB) elucidates comparative architecture of the a and b genomes. Plant Biotechnol. J. 19, 602–614. doi: 10.1111/pbi.13492
Parkin, I. A., Koh, C., Tang, H., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid brassica oleracea. Genome Biol. 15, R77. doi: 10.1186/gb-2014-15-6-r77
Paul, P., Joshi, S., Tian, R., Diogo Junior, R., Chakrabarti, M., Perry, S. E. (2022). The MADS-domain factor AGAMOUS-Like18 promotes somatic embryogenesis. Plant Physiol. 188, 1617–1631. doi: 10.1093/plphys/kiab553
Perumal, S., Koh, C. S., Jin, L., Buchwaldt, M., Higgins, E. E., Zheng, C., et al. (2020). A high-contiguity brassica nigra genome localizes active centromeres and defines the ancestral brassica genome. Nat. Plants 6, 929–941. doi: 10.1038/s41477-020-0735-y
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Riechmann, J. L., Meyerowitz, E. M. (1997). Determination of floral organ identity by arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 8, 1243–1259. doi: 10.1091/mbc.8.7.1243
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S., Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1111/nph.16122
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Shao, Z., He, M., Zeng, Z., Chen, Y., Hanna, A. D., Zhu, H. (2021). Genome-wide identification and expression analysis of the MADS-box gene family in sweet potato [Ipomoea batatas (L.) lam]. Front. Genet. 12(1664-8021 (1664-8021 (Print). doi: 10.3389/fgene.2021.750137
Sheng, X. G., Zhao, Z. Q., Wang, J. S., Yu, H. F., Shen, Y. S., Zeng, X. Y., et al. (2019). Genome wide analysis of MADS-box gene family in brassica oleracea reveals conservation and variation in flower development. BMC Plant Biol. 19, 106. doi: 10.1186/s12870-019-1717-y
Silva, C. S., Puranik, S., Round, A., Brennich, M., Jourdain, A., Parcy, F., et al. (2015). Evolution of the plant reproduction master regulators LFY and the MADS transcription factors: The role of protein structure in the evolutionary development of the flower. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01193
Song, J. M., Guan, Z., Hu, J., Guo, C., Yang, Z., Wang, S., et al. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Song, X., Wei, Y., Xiao, D., Gong, K., Sun, P., Ren, Y., et al. (2021). Brassica carinata genome characterization clarifies u's triangle model of evolution and polyploidy in brassica. Plant Physiol. 186, 388–406. doi: 10.1093/plphys/kiab048
Sun, F. M., Fan, G. Y., Hu, Q., Zhou, Y. M., Guan, M., Tong, C. B., et al. (2017). The high-quality genome of brassica napus cultivar 'ZS11' reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468. doi: 10.1111/tpj.13669
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Tsutsui, H., Higashiyama, T. (2017). pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in arabidopsis thaliana. Plant Cell Physiol. 58, 46–56. doi: 10.1093/pcp/pcw191
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
Wang, X., Song, M., Flaishman, M. A., Chen, S., Ma, H. (2021). AGAMOUS gene as a new sex-identification marker in fig (Ficus carica l.) is more efficient than RAN1. Front. Plant Sci. 12(1664-462X (1664-462X (Print). doi: 10.3389/fpls.2021.755358
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wang, Y., Zhang, J., Hu, Z., Guo, X., Tian, S., Chen, G. (2019). Genome-wide analysis of the MADS-box transcription factor family in solanum lycopersicum. Int. J. Mol. Sci. 20, 2961. doi: 10.3390/ijms20122961
Woodhouse, M. R., Cheng, F., Pires, J. C., Lisch, D., Freeling, M., Wang, X. (2014). Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. U.S.A. 111, 5283–5288. doi: 10.1073/pnas.1402475111
Wu, Y., Ke, Y., Wen, J., Guo, P., Ran, F., Wang, M., et al. (2018). Evolution and expression analyses of the MADS-box gene family in brassica napus. PLoS One 13, e0200762. doi: 10.1371/journal.pone.0200762
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. doi: 10.1186/s12870-014-0327-y
Yang, Y., Jack, T. (2004). Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 55, 45–59. doi: 10.1007/s11103-004-0416-7
Yang, J., Liu, D., Wang, X., Ji, C., Cheng, F., Liu, B., et al. (2016). The genome sequence of allopolyploid brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232. doi: 10.1038/ng.3657
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., Meyerowitz, E. M. (1990). The protein encoded by the arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yim, W. C., Swain, M. L., Ma, D., An, H., Bird, K. A., Curdie, D. D., et al. (2022). The final piece of the triangle of U: Evolution of the tetraploid brassica carinata genome. Plant Cell. 34, 4143–4172. doi: 10.1093/plcell/koac249
Yu, L. H., Miao, Z. Q., Qi, G. F., Wu, J., Cai, X. T., Mao, J. L., et al. (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Zahn, L. M., Feng, B. M., Ma, H. (2006). Beyond the ABC-model: Regulation of floral homeotic genes. Adv. Botanical Research: Incorporating Adv. Plant Pathol. 44, 163–207. doi: 10.1016/S0065-2296(06)44004-0
Zhang, L., Cai, X., Wu, J., Liu, M., Grob, S., Cheng, F., et al. (2018). Improved brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5(2662-6810 (2662-6810 (Print), 50. doi: 10.1038/s41438-018-0071-9
Zhang, H., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40 (Web Server issue), W569–W572. doi: 10.1093/nar/gks576
Zhang, Z., Guo, J., Cai, X., Li, Y., Xi, X., Lin, R., et al. (2022). Improved reference genome annotation of brassica rapa by pacific biosciences RNA sequencing. Front. Plant Sci. 13(1664-462X (1664-462X (Print). doi: 10.3389/fpls.2022.841618
Zhang, X., Wang, G., Zhang, S., Chen, S., Wang, Y., Wen, P., et al. (2020). Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell 183, 875–889.e817. doi: 10.1016/j.cell.2020.09.043
Zhao, D., Chen, Z., Xu, L., Zhang, L., Zou, Q. (2021a). Genome-wide analysis of the MADS-box gene family in maize: Gene structure, evolution, and relationships. Genes (Basel) 12, 1956. doi: 10.3390/genes12121956
Zhao, P. X., Miao, Z. Q., Zhang, J., Chen, S. Y., Liu, Q. Q., Xiang, C. B. (2020). Arabidopsis MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 71, 6092–6106. doi: 10.1093/jxb/eraa303
Zhao, W., Zhang, L. L., Xu, Z. S., Fu, L., Pang, H. X., Ma, Y. Z., et al. (2021b). Genome-wide analysis of MADS-box genes in foxtail millet (Setaria italica l.) and functional assessment of the role of SiMADS51 in the drought stress response. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.659474
Keywords: Brassica, U’s triangle, MADS-box genes, floral organ development, AGAMOUS
Citation: Song M, Zhang Y, Jia Q, Huang S, An R, Chen N, Zhu Y, Mu J and Hu S (2023) Systematic analysis of MADS-box gene family in the U’s triangle species and targeted mutagenesis of BnaAG homologs to explore its role in floral organ identity in Brassica napus. Front. Plant Sci. 13:1115513. doi: 10.3389/fpls.2022.1115513
Received: 04 December 2022; Accepted: 23 December 2022;
Published: 12 January 2023.
Edited by:
Helin Tan, Nanjing Agricultural University, ChinaCopyright © 2023 Song, Zhang, Jia, Huang, An, Chen, Zhu, Mu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengwu Hu, c3dodTgzMjUxQG53c3VhZi5lZHUuY24=; Jianxin Mu, anhtc3h5Y0AxNjMuY29t
†These authors contributed equally to this work
 Min Song
Min Song Yanfeng Zhang
Yanfeng Zhang Qingli Jia2
Qingli Jia2 Shuhua Huang
Shuhua Huang Yantao Zhu
Yantao Zhu Jianxin Mu
Jianxin Mu