- 1Key Laboratory of Horticultural Plant Biology Ministry of Education (MOE), College of Horticulture and Forestry Science, Huazhong Agricultural University, Wuhan, China
- 2Hubei Hongshan Laboratory, Wuhan, China
Sugar transporter proteins (STPs) play vital roles in sugar transport and allocation of carbon sources in plants. However, the evolutionary dynamics of this important gene family and their functions are still largely unknown in citrus, which is the largest fruit crop in the world. In this study, fourteen non−redundant CgSTP family members were identified in pummelo (Citrus grandis). A comprehensive analysis based on the biochemical characteristics, the chromosomal location, the exon–intron structures and the evolutionary relationships demonstrated the conservation and the divergence of CgSTPs. Moreover, CgSTP4, 11, 13, 14 were proofed to be localized in plasma membrane and have glucose transport activity in yeast. The hexose content were significantly increased with the transient overexpression of CgSTP11 and CgSTP14. In addition, antisense repression of CgSTP4 induced the shorter pollen tube length in vitro, implying the potential role of CgSTP4 in pummelo pollen tube growth. Taken together, this work explored a framework for understanding the physiological role of CgSTPs and laid a foundation for future functional studies of these members in citrus species.
Introduction
Soluble sugars such as sucrose, glucose and fructose are the main carbohydrates from photosynthesis of most plants (Rolland et al., 2002). These sugars are the key components of carbon and energy metabolism in plants, providing the skeleton for large molecules such as proteins and nucleic acids (Smeekens and Hellmann, 2014). Besides, sugars can be used as signal transduction molecules regulating various metabolic pathways, biotic and abiotic stress responses, plant growth and development, and regulating the osmotic pressure of plant cells and solutions, thus affecting stomatal opening and closing and other activities (Rolland et al., 2002; Koch, 2004; Rolland et al., 2006; Radchuk et al., 2010; Smeekens and Hellmann, 2014; Huai et al., 2022). In plants, sugars need to pass through membrane several times on its way from the source cells to the sink organs. The cross boundary membrane uptake of sugars is a major event for nutrition in all eukaryotic cells (Tao et al., 2015). The carrier proteins mediating the transmembrane transport of sugars are called sugar transporters. Currently, the vast majority of identified sugar transporters belong to the major facilitator superfamily (MFS), which is usually composed of 12 transmembrane domains known as H+/sugar or Na+/sugar cotransporters (Yan, 2013). According to the different transport substrates, the MFS in plants is mainly divided into disaccharide transporter and monosaccharide transporters (MSTs). Among them, the main disaccharide transporter is sucrose transporters (SUTs) (Chiou and Bush, 1996), also known as sucrose carriers (SUCs), which mediate the transport of sucrose and maltose (Riesmeier et al., 1994; Kuhn and Grof, 2010). The MSTs are composed of a large gene family, which contain seven subfamilies named sugar transporters (STPs) (also known as Hexose transporters, HTs), tonoplast monosaccharide transporters (TMTs), vacuolar glucose transporters (VGTs), plastid glucose transporters (pGlcTs), early response to drought 6-like transporters (ERD6Ls), polyol/monosaccharide transporters (PMTs) and insitol transporters (INTs) (Buttner, 2007; Li et al., 2015a; Fang et al., 2020; Liu et al., 2020). In addition, there is a new class of sugar transporter, which is named as the Sugars Will Eventually be Exported Transporters (SWEETs) (Chen et al., 2010). SWEETs belong to the MtN3-Like membrane protein and have a completely different structure from the MFS sugar transporters (Gautam et al., 2022). These sugar transporters mentioned above have been reported localized in various subcellular locations such as plasma membrane, vacuolar membrane, golgi apparatus membrane, and plastid membrane (Jiu et al., 2018; Fang et al., 2020; Li et al., 2020). The diversity subcellular localization patterns confers a variety of functions for sugar transport proteins. At present, the family of sugar transporters located on the plasma membrane and vacuolar membrane are more studied.
Among the numerous sugar transporter families, the STPs are the most studied monosaccharides transporters, indicating the vital role of STPs in plant development or stress response. STPs are complete membrane proteins with 12 transmembrane domains and are considered to be H+/sugar transporters located on the plasma membrane. So far, the STPs have been identified and studied in many plants, including Arabidopsis thaliana (Buttner, 2010), Manihot esculenta (Liu et al., 2018), Brassica oleracea var. capitata L. (Zhang et al., 2019), Oryza sativa (Toyofuku et al., 2000; Deng et al., 2019), Solanum lycopersicum (Reuscher et al., 2014), Vitis vinifera (Afoufa-Bastien et al., 2010), Fragaria vesca (Jiu et al., 2018; Liu et al., 2020), Pyrus bretschneideri Rehd (Li et al., 2015a), Malus domestica (Wei et al., 2014), Dimocarpus longan Lour (Fang et al., 2020). Based on previous reports, STP members are expressed in different tissues and participate in various metabolic pathways for specific physiological functions. The expression of STPs are also response to hormones, biotic and abiotic stresses, which further significantly affect plant development and stress resistance (Truernit et al., 1996; Buttner et al., 2000; Sade et al., 2013; Murcia et al., 2016; Murcia et al., 2018; Deng et al., 2019; Otori et al., 2019; Paulsen et al., 2019). A total of 14 AtSTPs have been reported in A. thaliana (Buttner, 2010), all of which are localized at the plasma membrane and are responsible for the transport of monosaccharides from the apoplastic space to the cytosol. For example, AtSTP1 is capable of transporting other hexose in addition to fructose, and is involved in the transport of monosaccharides in guard cells (Stadler et al., 2003). AtSTP2 is primarily responsible for absorbing glucose produced by callose degradation during the early stages of pollen maturation (Truernit et al., 1999). AtSTP4, AtSTP6, AtSTP8, AtSTP9, AtSTP10 and AtSTP11 are mainly responsible for the uptake of glucose into the pollen tube of A. thaliana and play a role in the supply of monosaccharides during the growth of the pollen tube (Rottmann et al., 2018). Heterologous expression of the apple hexose transporter gene MdHT2.2 in tomato promotes sucrose, fructose, and glucose accumulation (Wang et al., 2020b) in response to tomato salt tolerance mechanism by balancing cytoplasmic to intercellular ion concentrations and scavenging reactive oxygen species (ROS) (Wang et al., 2020a). In wheat, TaSTP3 is transcriptionally activated by the transcription factor TaWRKY19/61/82 during stripe rust, thereby increasing the sucrose concentration of host cells to guarantee carbon source supply for the fungus (Huai et al., 2022). In apple, MdSTP13a is found to absorb hexose and sucrose simultaneously in the process of sorbitol regulating pollen tube growth to promote the growth of apple pollen tubes (Li et al., 2020). These findings highlight the importance of STPs in plant growth, development, and stress tolerance via sugar transport and carbon source allocation.
The citrus is the largest fruit industry over the world. Studies have shown that the genus citrus originated from three ancestral species: Citrus maxima, Citrus medica, and Citrus reticulate (Wu et al., 2018). It has been suggested that domestication of citrus may have begun with the identification and asexual reproduction of selected, possibly hybrid or mixed individuals. For instance, the cultivation of pummelo (Citrus grandis) is cultivated from the ancestral C. maxima with the introgressions of the other citrus species (Xu et al., 2013; Wu et al., 2014). The fruit quality of citrus is affected by flavor substances, which mainly include soluble sugars, organic acids and volatile compounds, among which the composition and content of soluble sugars is the key factor affecting taste and thus determining fruit flavor quality (Li et al., 2012; Aslam et al., 2019). There are also some reports to support that the relationship between sugar transporters and sugar accumulation in citrus (Zheng et al., 2014). For instance, the soluble sugar-related genes in ‘Rongan’ (RA) and its mutant ‘Huapi’ (HP) kumquat were analyzed, and it was found that high sugar accumulation in HP fruit was associated with up-regulation of SUS, SPS, TST, STP and ERD6L genes (Wei et al., 2021). In addition, artificial thinning can increase the size and sugar content of citrus fruits, affect hormone synthesis and sugar transporter activity, and significantly improve fruit quality (Liu et al., 2022a). However, these studies have only pointed out the potential role of sugar transporters in citrus, but the characteristics and functions of sugar transporters in citrus species are still confused so far.
In this study, the genes encoding STPs in C. grandis genome were identified. The phylogenetic relationships, characteristics, structure, conserved motifs, cis-acting elements and collinearity of CgSTP members were analyzed, revealing the conserved and correlation between homologous and near-homologous genes. Based on the expression pattern and subcellular localization assay, CgSTP4, CgSTP11, CgSTP13 and CgSTP14 were further selected and proofed to have the hexose sugar transport activity. In addition, the function of pollen-specific expression CgSTP4 was explored to be involved in pummelo pollen tube growth via antisense oligonucleotide transformation. Taken together, we identified key candidate CgSTP genes in sugar accumulation, which will be a great scientific significance and potential application for further investigation of the physiological functions of CgSTPs in pummelo or other citrus species.
Materials and methods
Plant materials and growth conditions
The ‘Shatian pummelo’ (Citrus grandis) fruits, leaves, flowers and other tissues were harvested from the Centre of Citrus Plant at Huazhong Agricultural University (Wuhan, China). The pummelo trees were maintained under standard horticultural management and prevention of plant diseases and insect pests. At the popcorn stage, flowers were picked for anther collection, and the anthers were dried in a 28°C oven. The dried anthers and released pollen were collected into 1.5 or 2.0 ml centrifuge tubes sealed with silica gel and stored in -20°C refrigerator for further use. Tobacco plants (Nicotiana benthamiana) were grown in the growth chamber at 23-25°C with 12h light/12 h darkness.
Database searches and identification of STPs in Citrus grandis
In order to identify the STP genes in C. grandis, the whole-genome data of pummelo (Citrus grandis (L.) Osbeck.cv. ‘Wanbaiyou’ v1.0) was downloaded from the Citrus Pan-genome to Breeding Database website (http://citrus.hzau.edu.cn/index.php), which was used to obtain the gene sequences and gene annotations. The Hidden Markov Model (HMM) of the Sugar_tr domain (PF00083) from HMMER (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) was used to search the pummelo protein database at a standard E-value < 1.0 × 10−5 (Finn et al., 2011; Chen et al., 2020; Mistry et al., 2021). A total of 53 hypothetical CgSTP proteins were identified. Furthermore, the conserved domain composition of 14 AtSTP protein sequences were analyzed by CD-search. Then, all the 53 putative protein sequences filtered to submit to the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Finally, 14 STP family members of C. grandis were screened out. They were named CgSTP1 to CgSTP14 based on their relationship with members of the STP family in A. thaliana.
Phylogenetic tree construction and synteny correlation analysis
To explore the evolutionary relationship of CgSTPs between C. grandis and A. thaliana, a phylogenetic tree was constructed by maximum likelihood (ML) method using MEGAX64 software based on the protein sequences of 14 CgSTPs from pummelo and 14 AtSTPs from A. thaliana. The final tree is then beautified through the ITOL website. Among them, the AtSTP protein sequences were downloaded from the The Arabidopsis Information Resource website (https://www.arabidopsis.org/). In order to better understand the conservation of STP genes in evolution, the collinear correlation analysis between species was carried through the MCScanX (Wang et al., 2012) of TBtools software by offering gene annotation and the whole genome sequence of A. thaliana and C. grandis.
Amino acid characteristic and gene structure prediction
The physicochemical properties of the proteins, which included amino acid number (AA), molecular weight (MW), theoretical isoelectric point (PI), and grand average of hydropathy (GRAVY), were obtained in the ExpasyProtParam server (https://web.expasy.org/compute_pi/) (Wilkins et al., 1999). The number of transmembrane domains was predicted by the NovoPro url (https://www.novopro.cn/tools/tmhmm.html). The WoLF PSORT server (https://wolfpsort.hgc.jp/) was applied to predict protein subcellular localization. The gene structure annotation and CDS files of C. grandis were downloaded from the Citrus Pan-genome to Breeding Database website, and the STP genes information was extracted by TBtools using accession number, followed by the Gene Structure Display Server 2.0(http://gsds.gao-lab.org/)to visualize the exon–intron structure of these genes (Hu et al., 2015).
Protein motif and cis-acting elements analysis
To further explore the gene structure of CgSTPs, the MEME Suite web server (https://meme-suite.org/meme/tools/meme) (Bailey and Elkan, 1994; Bailey et al., 2009) was used to predict their protein sequences with a maximum number of motif groups of 12 for conserved motifs, any number of repeats, and an optimal width of the motif ranging from 15 to 60 amino acids. The promoter sequences (2 kb of genomic DNA sequence upstream of the translation initiation codon) of the CgSTP genes were obtained from C. grandis genome files and submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-elements in the promoter (Rombauts et al., 1999; Lescot et al., 2002).
RNA extraction, cDNA synthesis and quantitative real-time analysis
Each 0.2g pummelo tissues, including leaves, flowers and juice sac were snap frozen and ground into fine powder in liquid nitrogen with three independent repetitions. RNA from common tissues was extracted using the Ominplant RNA Kit (Cwbio. Jiangsu, China). And for polyphenol polysaccharide containing tissues, RNA was extracted using the RNAprep Pure Plant Plus Kit (TIANGEN, Wuhan, China) according to the instructions. The concentration and quality of the extracted RNA were confirmed via spectrophotometer and agar gel electrophoresis. Then the EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) reverse transcribed RNA into cDNA. Finally, three technical replicates of qRT-PCR were performed using SYBR Green Supermix kit according to the manual via the Applied biosystem®QuantStudio™ 7 Flex Real-Time PCR System (ABI, Los Angeles, CA, USA), and 2-ΔΔCT method (Udvardi et al., 2008) was used to calculate and analyze the obtained data.
Subcellular localization of CgSTPs
The CDSs of the CgSTP genes were amplified using gene-specific primers without stop codons (Supplementary Table 1). The homologous recombination approach (Rozwadowski et al., 2008) was used to concatenate these target genes with the YFP101 vector, which contained a C-terminal yellow fluorescent protein (YFP) driven by 35S promoter. After the correct comparison between the sequencing results of Tsingke Biological Company (Wuhan, China) and the CDS of the genome through DNAMAN software, the recombinant plasmid was transferred into Agrobacterium tumefaciens GV3101 (Krenek et al., 2015) (Weidi, Wuhan, China). Then, A. tumefaciens containing the recombinant plasmid was injected into tobacco leaves for transient expression (Kato et al., 2002). Finally, the scanning confocal microscope (Leica TCS-SP8, Wetzlar, Germany) was used to image YFP fluorescence with an excitation wavelength of 514 nm and emission at 520-551 nm.
Functional characterization of CgSTPs by heterologous expression in yeast
The CDSs of the target gene were cloned with gene-specific primers containing stop codons (Supplementary Table 1). The yeast expression vector pDR196 was ligated with the gene of interest by homologous recombination, followed by transformation of the recombinant plasmids into the hexose transport deficient yeast strain EBY.VW4000 by lithium acetate method (Soni et al., 1993). The empty vector pDR196 was used as negative control, and the recombinant vector pDR196-AtSTP13 was served as a positive control (Riesmeier et al., 1992; Rottmann et al., 2016; Li et al., 2020). The transformed cells in EBY.VW4000 were pre-incubated in liquid Synthesis Defect (SD)-Ura medium supplemented with 2% maltose (w/v) as the sole carbon source until the OD600 value reached 0.6-0.8 (Wieczorke et al., 1999). Four serial dilutions (10×) of yeast cells were then plated on solid SD-Ura medium containing 2% maltose or glucose as the sole carbon source. The samples were cultured at 30°C for 3 days and then observed and photographed.
Transient expression in tobacco leaves and sugars content analysis
The CDS(s) of CgSTP genes were cloned with gene-specific primers without termination codons (Supplementary Table 1) and inserted into the overexpression vector pK7WG2D with the 35S promoter (Pi et al., 2022). After that, the final recombinant plasmid was transformed into Agrobacterium tumefaciens GV3101, which was further used for injection of tobacco leaves for transient overexpression. The control with empty vector and the recombinant plasmid carrying the target genes (CgSTP(s)-2D) were transformed into the same leaf, as control in the left and CgSTP(s)-2D in the right part of the leaf. Samples were taken on the third day after transformation. The relative gene expression level was determined by RT-PCR assay (the primers were listed in Supplementary Table 1). Then, the content of monosaccharides was measured by GC method as described previously (Liu et al., 2022b). Soluble sugars were extracted in 75% methanol with ribitol (0.12 mg per sample) added as an internal standard and then derivatized sequentially with methoxyamine hydrochloride and N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA). After derivatization, the metabolites were analyzed using a GC equipment (Fuli GC-9720 Pluse, Zhejiang, China) with a HP-FFAP column (30.00 m * 0.32 mm * 0.25 μm) and a 5 m Duraguard column (Agilent Technologies, Palo Alto, CA, USA). Sugar content was quantified based on standard curves generated for each sugar (glucose and fructose) and an internal standard. All results were determined at least three biological replicates.
In vitro pollen tube growth experiments and antisense oligonucleotide transfection
The pollen grains were cultured in liquid germination media (0.02%[w/v]MgSO4, 0.01%[w/v]KNO3, 0.03%[w/v]Ca(NO3)2, 0.01%[w/v]H3B03, 15%[w/v]PEG-4000, 10%[w/v]Sucrose, and liquid NaOH to pH 6.0) (Liang et al., 2017) for pollen tube length assay. Only 1 μl of transfection agent (Biosharp, China) and 1 μl of transfection agent plus 1 μl of oligonucleotide primers (ODN concentration of 100 μM) were added as the control group, and 1 μl of transfection agent plus 1 μl of antisense oligonucleotide primers were added as the experimental group (Meng et al., 2014). Three independent experiments were performed for each treatment, and the same amount of pollen grains were added for germination culture at 25°C in a constant temperature and dark environment. At the 4 h, 6 h and 8 h of culture, the germinated pollen grains were observed and photographed by a type microscope, and then each biological replicate at least 100 germinated pollen grains were measured via image software. Supplementary Table 1 lists the oligonucleotide primer sequences used in this study. For gene expression assay, individual duplicate pollen tubes were collected into a 10 ml centrifuge tube and centrifuged under 12000g at 4°C for 10 min. The samples could be re-suspended once in pure water and centrifuged again to remove the supernatant under the same conditions, after that the samples were snap frozen in liquid nitrogen for RNA extraction and gene expression assay by qRT-PCR.
Result
Genome-wide identification and basic information analysis of STP family genes in C. grandis
Using the Hidden Markov Model (HMM) and the conserved domains of MSF_STP (cd17361) and Sugar_tr (pfam00083) in STPs, a total of 14 STP family members were screened in the C. grandis genome database via the HMMER and Batch CD-Search websites (Table 1; Figure 1 and Supplementary File 1). Following the distant genetic relationship with Arabidopsis STP family genes, the 14 pummelo CgSTPs were named as CgSTP1 - CgSTP14 (Table 1). Furthermore, the phylogenetic tree was constructed based on the protein sequences encoded by CgSTP genes in C. grandis and AtSTP genes in A. thaliana via MEGAX64 (Figure 1B). The results showed that the STP proteins could be classified into four groups (Figure 1B). Among them, tow CgSTPs (CgSTP7 and CgSTP14) together with two AtSTPs from Arabidopsis were attributed to Group I. Group II consisted of four CgSTPs (CgSTP2, CgSTP6, CgSTP8 and CgSTP13), and four AtSTPs. Four CgSTPs (CgSTP3, CgSTP5, CgSTP9 and CgSTP10) clustered with two AtSTPs in Group III. Group IV contained four CgSTPs (CgSTP1, CgSTP4, CgSTP11 and CgSTP12) and six AtSTPs. Furthermore, a total of two sister pairs of CgSTPs were observed in the phylogenetic tree, including CgSTP5 - CgSTP9 and CgSTP1- CgSTP12 (Figure 1B). The result indicated that the STP family members are closely related, which is also a good reference for renaming. Moreover, the position and evolutionary relationships of CgSTP members at the chromosomal level were visualized by TBtools based on the gene structure annotation file (Supplemental Figure 1). Interspecific co-linearity analysis of STP genes revealed that eight pairs of genes are orthologous in two species between C. grandis and A. thaliana, indicating that the STP family was widespread in higher plants and strongly conserved during the evolutionary process.
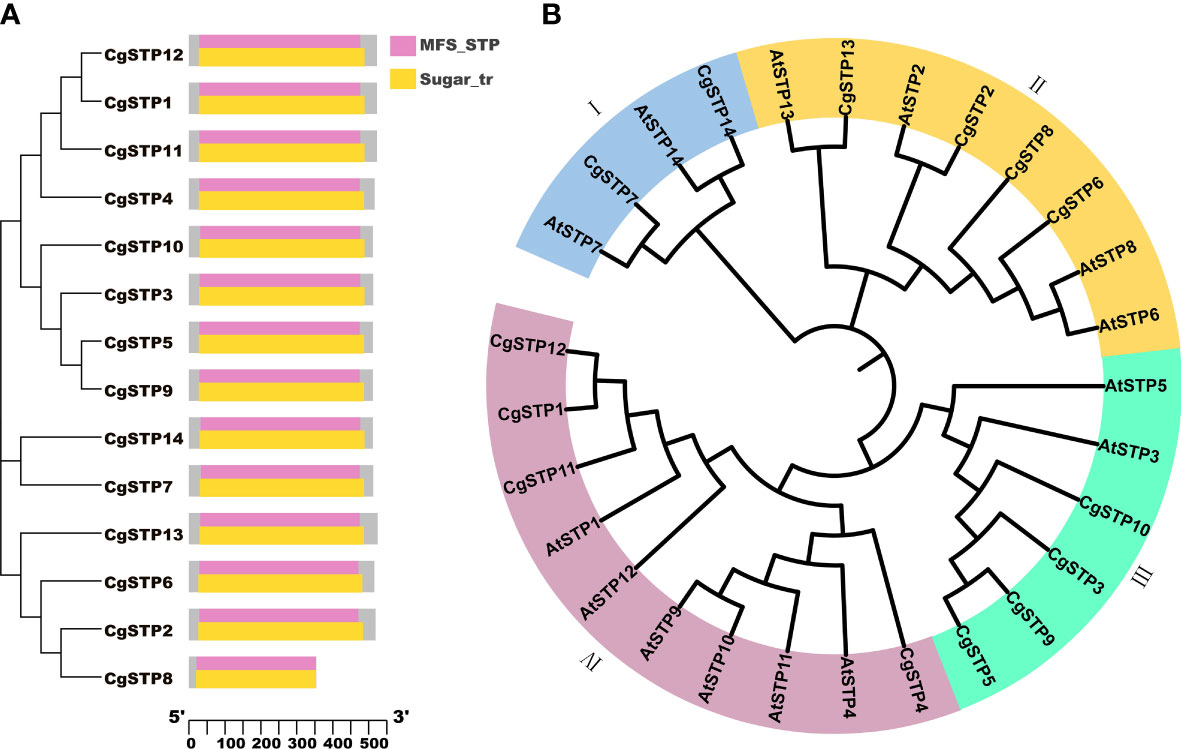
Figure 1 Phylogenetic analysis of STP members from C grandis and A thaliana, and conserved domain analysis of CgSTPs. (A) The conserved domain analysis was performed through the NCBI Batch CD-search tool. (B) A total of 14 STP protein sequences from C grandis and 14 STP protein sequences from A thaliana were aligned by ClustalW method. The MEGAX program was applied to construct the phylogenetic tree by the ML method in the default parameters. The beautification of the tree was carried out on the iTOL website.
According to the genomic information of C. grandis chromosomes, the distribution of 14 STP genes on the chromosome was analyzed. It was found that all CgSTP members were unevenly distributed on seven chromosomes, with the most abundantly distributed 7 genes on chromosome 9 (Table 1 and Supplemental Figure 1). Protein sequence characteristics, including number of amino acids (AA), molecular weight (Mw), theoretical isoelectric point, grand average of hydropathicity (GRAVY), transmembrane domain (TMD) and subcellular localization prediction, were also analyzed. As shown in Table 1, the average STP protein length was 504 aa, with the longest 524 aa from CgSTP13 and the shortest 353 aa from CgSTP8. Accordingly, the molecular weight of these proteins ranged from 38.7 kDa (CgSTP8) to 57.9 kDa (CgSTP11). The theoretical isoelectric points ranged from 8.86 (CgSTP11) to 9.56 (CgSTP9), which were all weakly basic. According to the relevant principle of grand average of hydropathicity (GRAVY), CgSTP1, CgSTP7, CgSTP10, CgSTP11, CgSTP12 and CgSTP13 were defined as amphoteric proteins with the value between -0.5 to 0.5, while the other proteins were hydrophobic proteins with more positive value (Table 1). More importantly, the number of CgSTPs transmembrane domains was presented from 8 to 12 based on the prediction by the NovoPro website. The vast majority of CgSTP members had 12 transmembrane domains, which was consistent with the characteristics of the MFS superfamily (Yan, 2013). In addition, the subcellular localization prediction of the protein sequences of CgSTP genes was performed via the WoLF PSORT (Table 1). Most of the CgSTP proteins are predicted to be plasma membrane proteins, but some are vacuolar membrane proteins, including CgSTP3, CgSTP5, CgSTP8 and CgSTP10, which are different from the localization of STP proteins in other species. These results indicate that CgSTP proteins may play different roles in sugar transport relying on their protein characterization and subcellular localization.
Gene structure, conserved motifs and promoter cis-acting elements analysis of the CgSTPs
To understand the structure of CgSTP genes, exons, introns, and untranslated regions were analyzed via GSDS2.0 and TBtools. It was found that CgSTP1, CgSTP2, CgSTP3, CgSTP4, CgSTP6, CgSTP8, and CgSTP12 didn’t have upstream and downstream UTRs. While CgSTP5 didn’t have upstream UTR, and the other six members all contained upstream and downstream UTRs (Figure 2B). Besides, all members contained exons and introns, but differed in specific numbers and locations. The exon number of the CgSTP genes ranged from 2 to 5 (Figure 2B). CgSTP13 gene had five exons, whereas CgSTP12 only had two exons. The different of gene structures might contribute to the functional diversity of closely related STP genes. Moreover, 12 putative conserved motifs were predicted through MEME analysis in most CgSTP proteins, and all these motifs were arranged in a stable order in the protein sequence (motif 2, motif 4, motif 10, motif 3, motif 7, motif 5, motif 6, motif 9, motif 12, motif 1, motif 8, and motif 11) (Figure 2A). The length of the conserved motifs ranges from 16 to 50 amino acids, and these 12 motifs are contained in all CgSTP proteins, except for CgSTP8 only has 8 motifs (Figure 2A; Supplementary Table 2). The high uniformity of these conserved motifs fully reflected the relatively conserved function of CgSTP proteins in the evolutionary process.
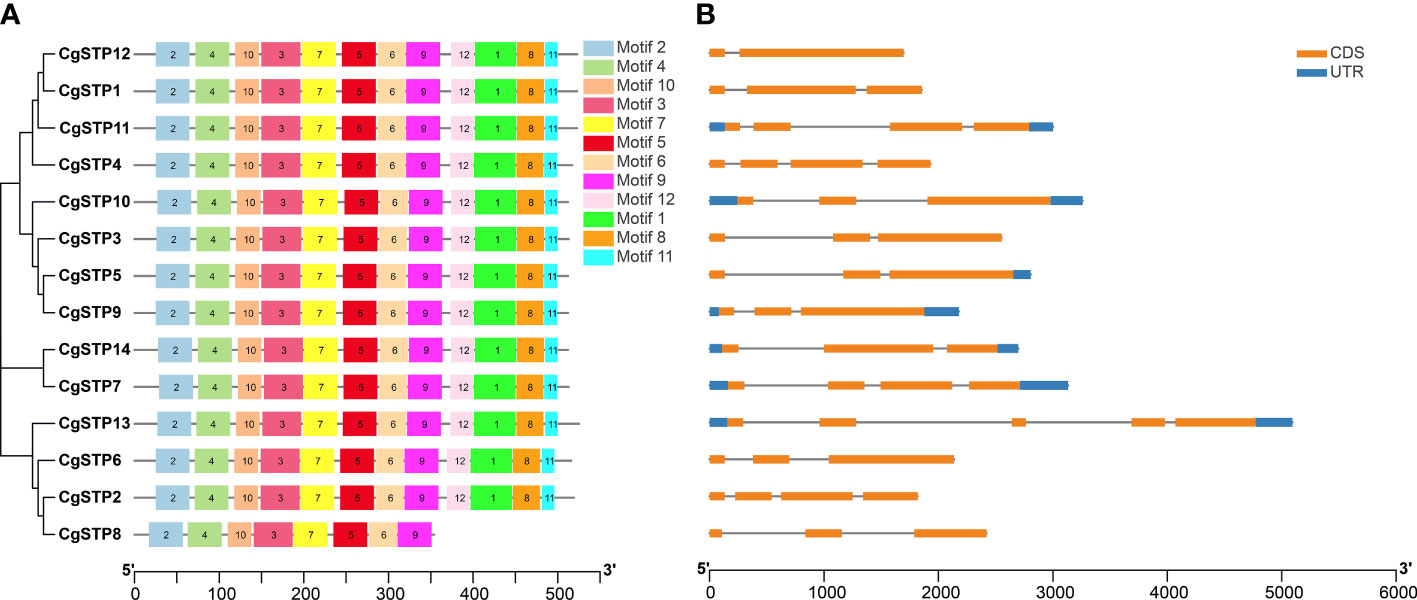
Figure 2 Motif, gene structure and phylogenetic relationship analysis of CgSTP genes. (A) The motif analysis was performed on the MEME. Twelve motifs were identified. The detail of motif sequence information was presented in Supplemental Table 2. (B) The gene exon–intron structure analysis. The blue color was the untranslated region, while the orange was the coding sequence. The line without color referred to introns. The neighbor-joining phylogenetic tree of the CgSTP genes sequences was constructed using 1000 bootstrap replicates by MEGAX64.
Cis-elements are important molecular switches involved in the regulation of gene transcription during plant growth and development or abiotic stress response. To detect regulatory factors and predict cis-elements of CgSTPs, each of the 2 kb promoter region (upstream of the start codon of the gene) of 13 CgSTP genes was retrieved from the C. grandis genome sequence. But for the CgSTP2 gene, the identified sequence was shorter than 2 kb (1047bp), since the presence of another gene located less than 2 kb upstream of CgSTP2 start codon site (Supplementary File 2). Finally, 14 cis-elements in the promoter region of CgSTP genes were predicted by PlantCARE database (Supplementary Figure 2A). A heat map was further constructed to show the frequency of different cis-elements (Supplementary Figure 2B). These predicted cis-elements respond to different phytohormones (Gibberellin, Abscisic acid, Auxin, Methyl jasmonate, Salicylic acid) and various environmental factors, as well as abiotic and biotic stresses. Among them, hormone-related elements were widely presented in the promoter region of CgSTPs, so it was suspected that hormones play a potential role in regulating the expression of CgSTP genes. Besides, the anaerobic induction regulator is presented in the promoters of all genes except CgSTP2, which suggested that these genes might be induced by anaerobic conditions, such as waterlogging stress. In addition, some the other gene-specific cis-elements were also identified in the promoter region of the CgSTP genes, like the MYB binding site involved in drought-inducibility, low-temperature responsiveness, and the cis-acting element involved in defense and stress responsiveness, which provided the possibility to regulate gene expression and respond to various conditions of CgSTPS. Overall, cis-acting element analysis will provide a good reference for further studies on transcriptional regulation of CgSTP members.
Expression profiles of CgSTP Genes in different tissues
The spatiotemporal expression is a critical aspect in determining gene function. Accordingly, three different tissues of leaf, flower and juice sac of ‘Shatian pummelo’ were collected to perform the tissue expression pattern analysis of STP family members via qRT-PCR assay (Figure 3). The results showed that CgSTPs were widely expressed in leaf, flower and juice sac. Among them, CgSTP7, CgSTP9, CgSTP11, CgSTP13 and CgSTP14 had a higher expression level in two or three different tissues (Figure 3). For instance, CgSTP13 was highly expressed in leaf and flower tissues (Figures 3A, B), while CgSTP7 was highly expressed in leaf and juice sac tissues (Figures 3A, C). CgSTP9, CgSTP11 and CgSTP14 were highly expressed in leaf, flower and juice sac (Figure 3), implying that these members should have a universal functions in different tissues. What’s more, the tissue-limited expression pattern was also presented that CgSTP4 and CgSTP6 were specifically expressed in flower (Figure 3B). For some other members were expressed at relatively low levels in all three tissue, such as CgSTP1, CgSTP2, CgSTP8 and CgSTP10, which might be expressed at specific developmental stage or in response to certain stimuli such as abiotic and biotic stresses. Taken together, these expression patterns in different tissues suggested that CgSTPs should have distinct physiology functions throughout the plant.
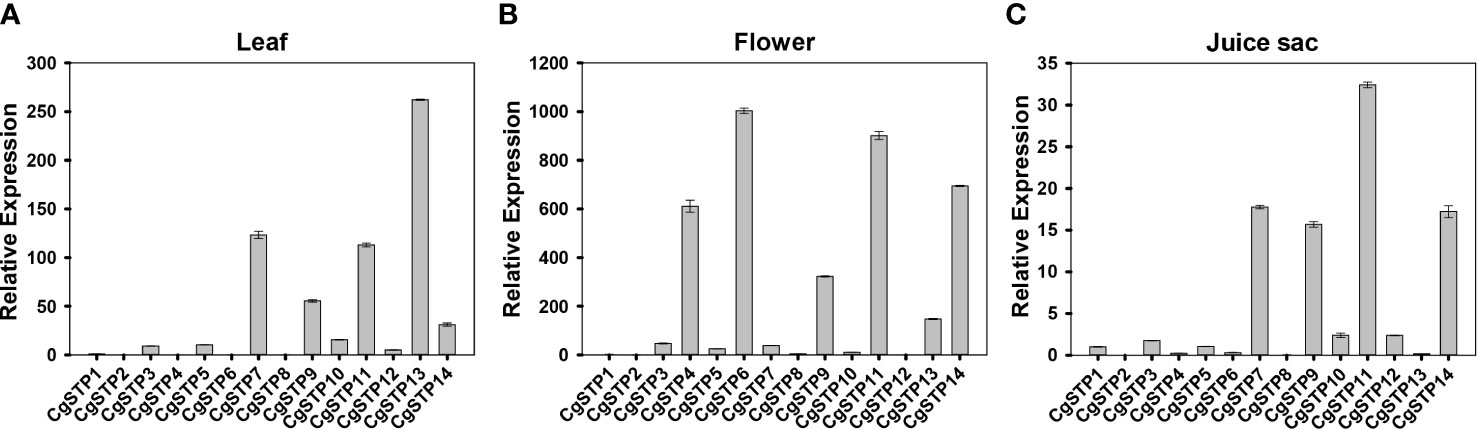
Figure 3 Relative expression patterns of CgSTPs in different tissues. (A) Leaf. (B) Flower. (C) Juice sac. The relative expression of all CgSTP members was determined by qRT-PCR assay. The CgActin was defined as the reference and the CgSTP1’s expression level was normalized as 1. Data are mean ± SE (n=3).
Functional characterization of CgSTPs by heterologous expression in yeast
To investigate the transport function of CgSTPs, the subcellular localization was first identified. According to the expression level, the 6 higher expressed genes (CgSTP4, CgSTP7, CgSTP9, CgSTP11, CgSTP13, CgSTP14) were selected for further analysis. The cell membrane subcellular localization of CgSTPs were observed by co-expressing the YFP fusion proteins and the plasma membrane marker (PM marker) in tobacco leaves (Figure 4), which is consistent with the prediction results of the WoLF PSORT (Table 1). Based on previous reports, STPs had hexose sugar (like glucose and fructose) transport activity (Buttner et al., 2000; McCurdy et al., 2010; Liu et al., 2018; Kong et al., 2022). To characterize the transport properties of the CgSTP proteins, the monosaccharide uptake incompetent yeast mutant strain EBY.VW4000 (only grow on maltose medium) was applied for sugar uptake assay (Wieczorke et al., 1999). For that, the yeast expression vector pDR196 containing CgSTP4, CgSTP7, CgSTP9, CgSTP11, CgSTP13, CgSTP14 were respectively transformed into the EBY.VW4000, and the empty vector and AtSTP13 were used as negative and hexose-uptake positive control in yeast growth assay. Glucose was added as sole carbon source to detect the sugar transport activity of CgSTPs. All yeast cells grew well on the synthesis deficient (SD-Ura) medium containing 2% maltose (Figure 5), indicating the successfully expression of vector in yeast. As expected, negative control yeast cells transformed with empty vector pDR196 did not grow on glucose medium, while positive control yeast cells transformed with CgSTP13-pDR196 grew normally on glucose medium (Figure 5). For yeast cells carrying CgSTP4, CgSTP11, CgSTP13, and CgSTP14 could be grown on SD medium containing glucose, suggesting that these four CgSTP proteins could have glucose transport activity. In addition, the other two CgSTP members (CgSTP7 and CgSTP9) were unable to recovery yeast growth on glucose medium (Figure 5), implying that these two CgSTP members didn’t have or had a weak hexose transport activity. The subcellular localization and the verification of the monosaccharide transport activity of CgSTPs would provide a foundation for further physiological function research.
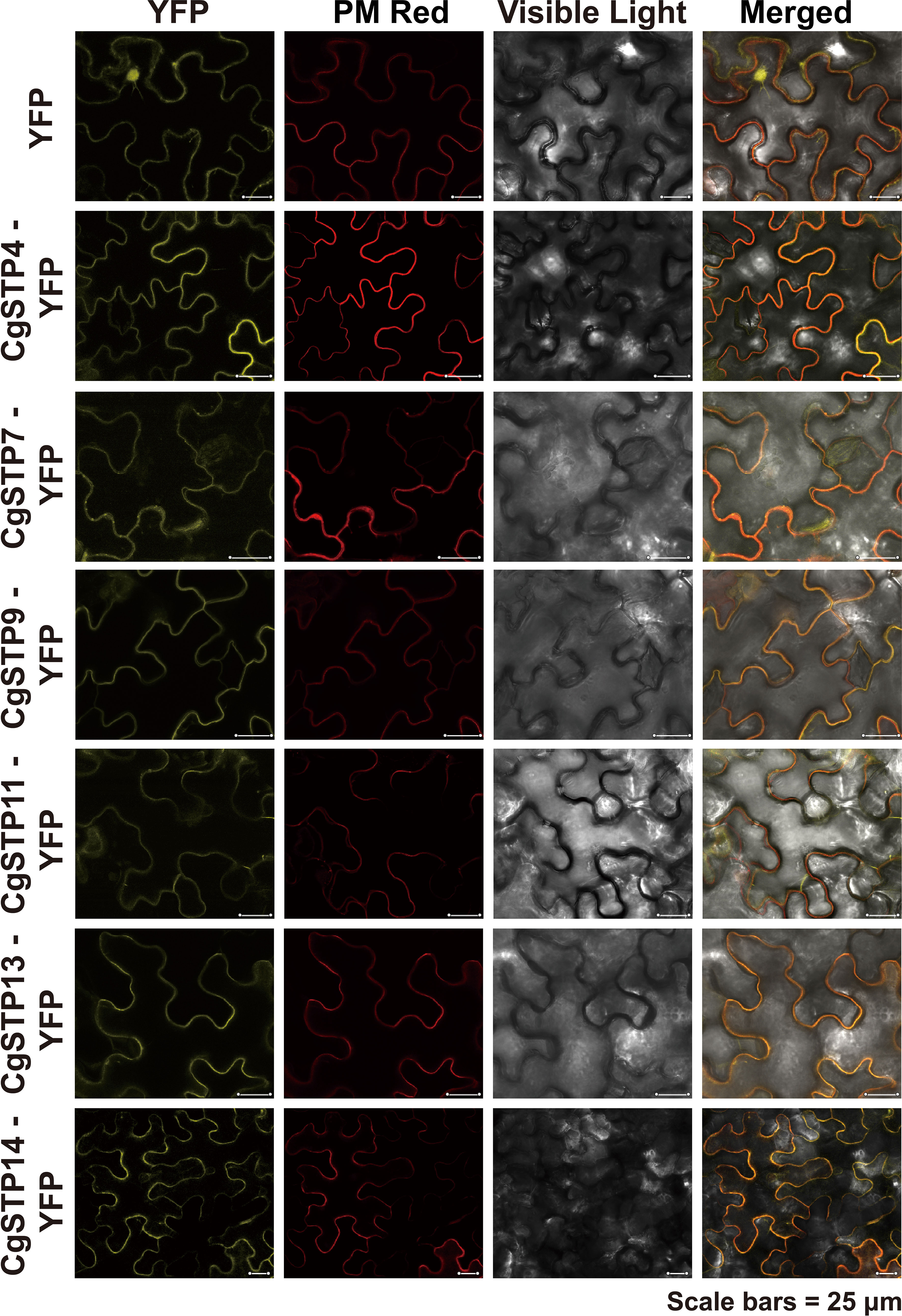
Figure 4 Subcellular Localization of CgSTPs. Tobacco leaves expressing 35S:YFP or 35S:CgSTP(s)-YFP, in colocalization with plasma membrane Deep Red marker, were visualized with a confocal laser microscope. Scale bars = 25 μm.
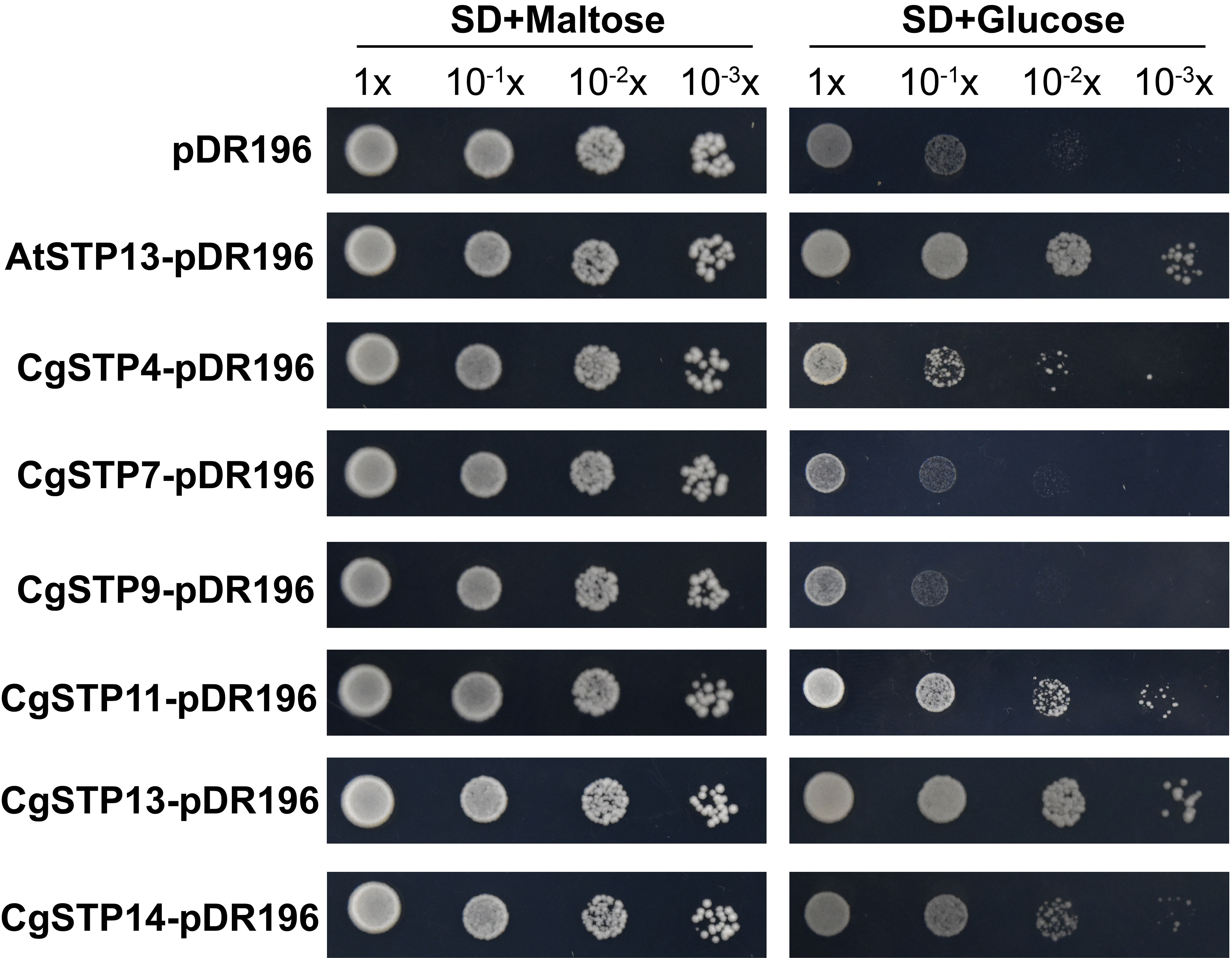
Figure 5 The sugar uptake assay of CgSTPs in Yeast (S. cerevisiae) Strain EBY.VW4000. Yeast growth assay of EBY.VW4000 transformed with pDR196 empty vector, AtSTP13-pDR196 or CgSTPs-pDR196 alone on 2% (w/v) maltose or glucose SD-Ura medium.
Validation of sugar accumulation by transient overexpression of CgSTPs
To verify and analyze the sugar accumulation function of CgSTPs, the genes that have been confirmed to have transport activity and highly expression level in fruit or leaf tissues were selected. The overexpression vectors of pK7WG2D-CgSTP11, CgSTP13, CgSTP14 were constructed under the 35S promoter. After that, the final expression vectors containing CgSTP11/13/14, and the empty vector control were transformed into tobacco leaves for transient overexpression (Supplementary Figure 3). In consideration of the hexose transport activity, the main monosaccharides’ (glucose and fructose) content were determined by GC-FID method (Li et al., 2015b). As shown in Figure 6, the glucose and fructose contents with the overexpression of target CgSTPs were increased in comparison with the control. For CgSTP11 and CgSTP14-OE samples, both glucose and fructose had a significantly higher level, implying that they did have the function of accumulating monosaccharides in plant. Overall, the transient overexpression assay provide the important clues for further more detail research of CgSTPs in citrus species.
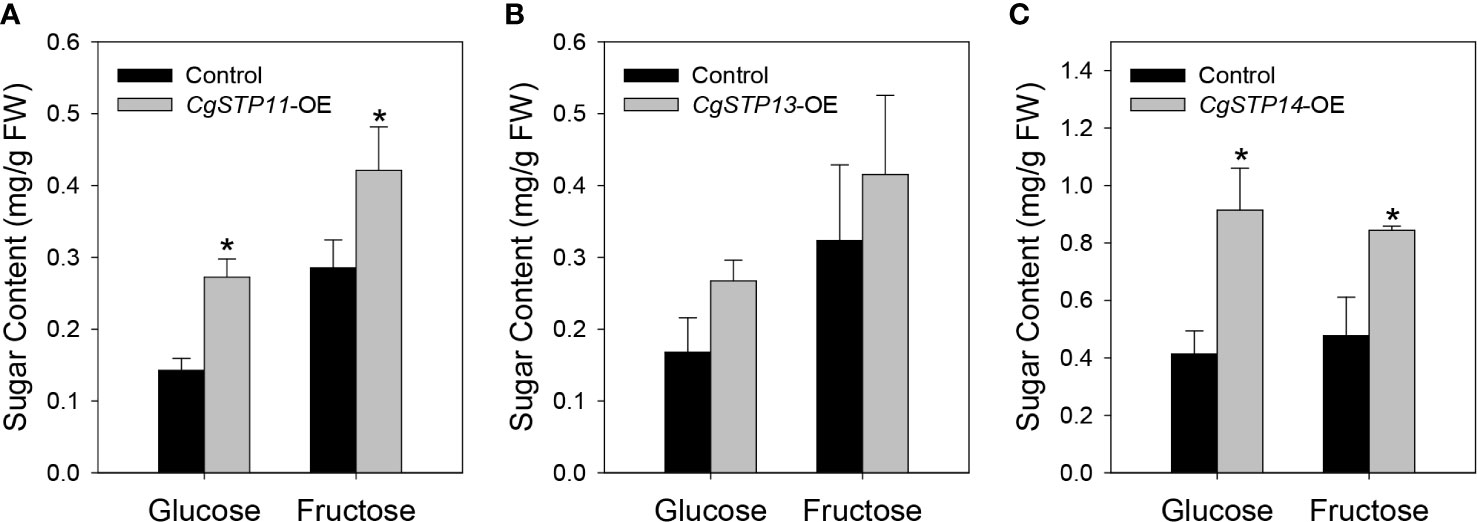
Figure 6 Determination of monosaccharide content in CgSTPs transient overexpression samples. The content of glucose and fructose in CgSTP11 (A), CgSTP13 (B), CgSTP14 (C) transient overexpression tobacco leaves. Data are mean ± SE (n=3).
Antisense repression of CgSTP4 reduces pollen tube growth
According to the previous report, sugar transport proteins were essential for pollen tube fast growth (Cheng et al., 2015; Rottmann et al., 2018; Li et al., 2020). In this study, it was found that CgSTP4 had the specifically high expression level in flower (Figure 3). Therefore, the expression profile of CgSTP4 in flower and various tissues of flower, including pollen, pollen tube, receptor, style, filament and petal of C. grandis, were further detected by qRT-PCR. The results revealed that CgSTP4 was highly expressed in pollen grains and pollen tubes (Figure 7A). Given the importance of sugar absorption for pollen tube growth, we explored the potential role of CgSTP4 in pollen tubes via the antisense oligonucleotide transfection assay (Meng et al., 2014; Meng et al., 2018). Compared with control and sense oligonucleotide transfection, the expression of CgSTP4 can be significantly inhibited by as-ODN treatment during the germination of pollen in ‘Shatian pummelo’ (Figure 7B). As expected, the pollen tube length was shorter with the lower expression of CgSTP4 (Figure 7C), indicating that CgSTP4 plays an important potential role in the sugar uptake for the growth of pollen tubes in C. grandis.
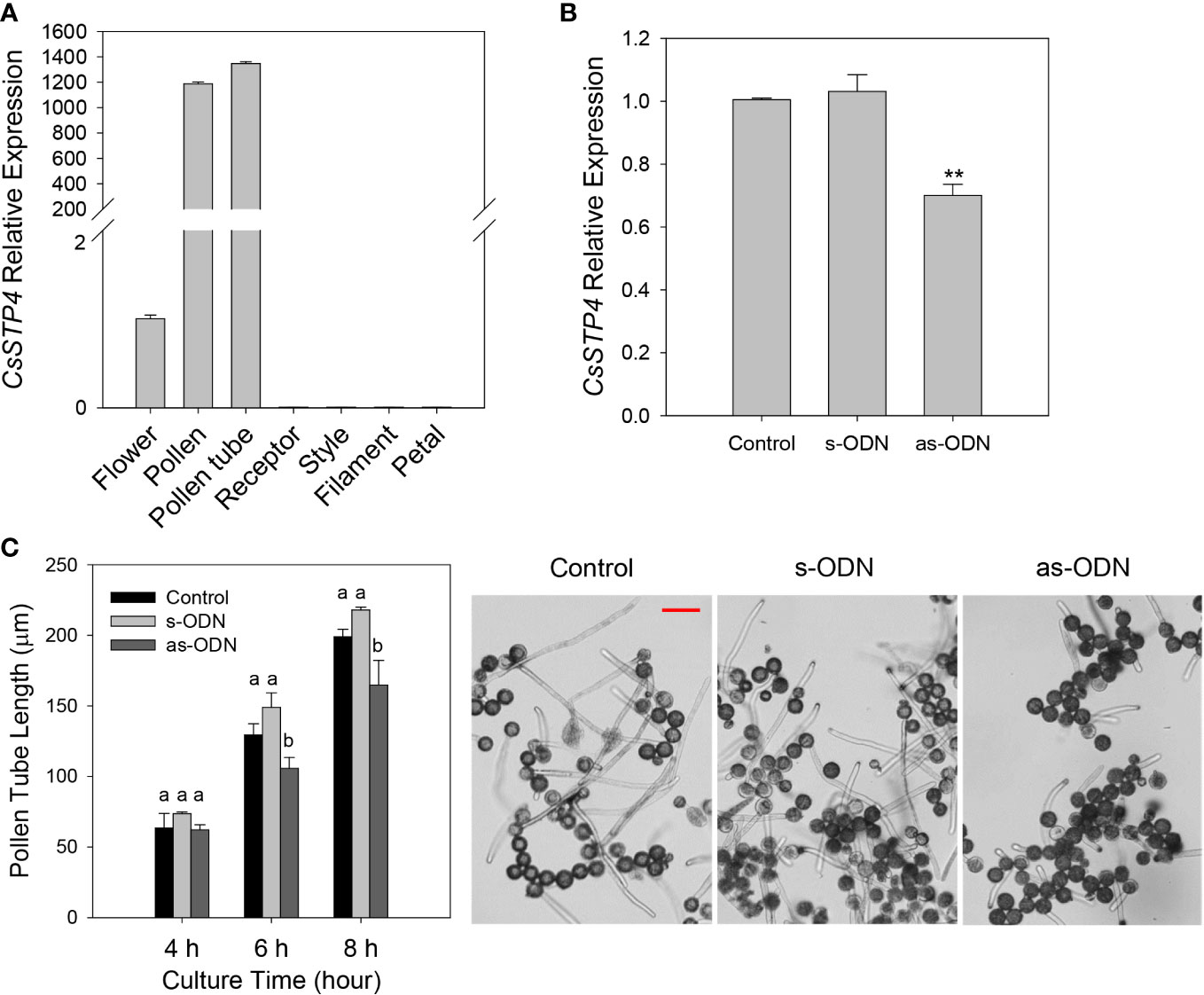
Figure 7 The pollen tube growth was inhibited with the lower expression of CgSTP4. (A) The expression of CgSTP4 in different tissues of flower was determined by qRT-PCR. The CgActin was defined as the reference and the CgSTP4’s expression level in flower tissue was normalized as 1. (B) Expression levels of CgSTP4 in pollen tubes transfected with antisense or sense oligonucleotide of CgSTP4 (s-CgSTP4, as-CgSTP4), or transfection agent alone (Control). The CgActin was defined as the reference and the CgSTP4’s expression level in control condition was normalized as 1. Data are mean ± SE (n=3). * Represents significant difference in comparison with control using Student’s t test at p < 0.05. (C) Pollen tube growth assay after transfection with as-CgSTP4, s-CgSTP4, or transfection agent alone (Control). Scale bar = 50 μm. Data are mean ± SE (n=3). Letters (A–B) indicate significant differences at each sampling point (p < 0.05) using Duncan’s multiple range test (MRT) after ANOVA.
Discussion
Sugar transport proteins (STPs or HTs) are mainly involved in the absorption and transport of hexose in plants. They have been reported to play a key role in plant response to biotic or abiotic stresses, growth and development (Fotopoulos et al., 2003; Buttner, 2010; Slewinski, 2011). Moreover, the sugar accumulation in plant, especially in fruit tissues, is closely related to STPs. So far, there has been no systematic study on the STPs, including their expression profile, localization, physiological functions in citrus, which is the most productive fruit in the world. The acquisition of genome sequence of citrus species provides a good opportunity to identify STP family members. Here, a total of 14 CgSTPs were identified by BLAST search and HMMER analysis of C. grandis genome. All selected STP proteins contained MSF_STP (cd17361) and Sugar_tr (pfam00083) conserved domains (Figure 1), which belong to the STP family of the MFS (Yan, 2013). The amino acid number of CgSTPs is between 353 aa (CgSTP8) and 524 aa (CgSTP13) (Table 1), which is similar to the other reported STP families in Arabidopsis (Buttner, 2010), tomato (Reuscher et al., 2014), strawberry (Jiu et al., 2018; Liu et al., 2020) and pear (Li et al., 2015a), indicating the relatively stable of STPs during the whole evolutionary process. Besides, this study was the first to comprehensively analyze the gene structure, conserved motifs and cis-acting elements of CgSTP family. The expression pattern and function analysis of CgSTP gene were further studied.
Phylogenetic analysis presented that CgSTP proteins were classified into four groups in combination with AtSTPs in each group (Figure 1). The result of collinear correlation revealed eight pairs of homologous genes between pummelo and Arabidopsis (Figure S1), indicating potential similarities in evolutionary relationship and functions between the two plants. This will provide a useful reference for the future functional research of CgSTPs. It has been reported that most STP proteins have 12 transmembrane domains (TMD1-TMD12), including N-domain (TMD1-TMD6) and C-domain (TMD7-TMD12) (Hirai et al., 2002; Yan, 2013). In this study, 10 of the 14 CgSTP proteins contained the all of 12 TMDs, while CgSTP2 had 11 TMDs and 2 CgSTP had 10 TMDs (Table 1). However, CgSTP8 carries only 8 TMDs (Table 1), alone with a missing sequence in the middle of the protein. These results suggested that loss of N-terminal or C-terminal regions may have occurred in some CgSTP members during evolution. Consistent with this, similar STP protein structures have been observed in cassava (Liu et al., 2018), tomato (Reuscher et al., 2014), and grapevine (Afoufa-Bastien et al., 2010). As the transmembrane transports, most of the CgSTP were predicted to be localized in the plasma membrane, while a few be localized the vacuolar membrane based on the WoLF PSORT database (Table 1). Subcellular localization assay indicated that CgSTP4, CgSTP7, CgSTP9, CgSTP11, CgSTP13 and CgSTP14 were located to the plasma membrane (Figure 4), which was consistent with the predicted results. Accordingly, the most plasma membrane-localized members were proofed to have hexose sugar transport activity via the yeast mutant recovery growth assay (Figure 5).
In addition, the conserved motif analysis was carried out to further reveal the relationship between the various members and the potential roles of CgSTPs. Interestingly, the result demonstrated that CgSTPs contained all motif sequences except CgSTP8 (Figure 2A). This is similar to the characterization in Arabidopsis (Buttner, 2010) and cassava (Liu et al., 2018). The conservation and divergence of motifs in STP proteins may lead to functional similarities or differences among members of different STP families. We also analyzed the gene structure of CgSTPs and found that most CgSTP genes had three exons and two introns (Figure 2B). The variation in the number of exons and introns of the STP gene ranged from two to five, consistent with the other plant species, such as Arabidopsis (Buttner, 2010), cabbage (Zhang et al., 2019), grapevine (Afoufa-Bastien et al., 2010) and pear (Li et al., 2015a). The diversity of STP gene structures may be due to the structural differentiation formed by the insertion or deletion of exons and introns (Xu et al., 2012). Moreover, cis-acting element analysis was performed on the promoter sequence of CgSTPs (Figure S2). The predicted results indicated that CgSTPs may be involved in plant hormone (GA, ABA, IAA, MeJA and SA) or abiotic stress responses.
During plant growth and development, sugar accumulation and distribution has proved to be closely linked to the STP members. The most physiological functions also depend on gene expression pattern. In this study, the expression of CgSTPs was investigated by qRT-PCR, and it was found that CgSTPs had a various expression level in different pummelo tissues (Figure 3), which was consistent with the AtSTPs in Arabidopsis (Buttner, 2010). For example, AtSTP1 is mainly expressed in the root for the hexose absorption from the extracellular (Otori et al., 2019). AtSTP4, AtSTP6, AtSTP8, AtSTP9, AtSTP10 and AtSTP11 are highly expressed in pollen and are mainly responsible for pollen tube growth via regulating glucose uptake (Rottmann et al., 2018). In here, CgSTPs also showed tissue-specific expression, such as CgSTP4 was specifically expressed in pollen and pollen tubes (Figure 7). The further pollen tube growth experiment proofed that CgSTP4 was involved in regulating the pummelo pollen tube length in vitro (Figure 7). Given the important of sugar content for the fruit flavor quality, the CgSTP11 and CgSTP14, which were highly expressed in juice sac tissue, were selected for sugar accumulation assay. The results shown that the hexose was dramatically increased with the overexpression of these two genes, implying the potential functions in improvement of the citrus fruit sweet quality. Except for these members, there are some other CgSTP genes that had lower expression level in detected tissues, like CgSTP1, CgSTP2, or CgSTP8. The more work was required in future to decipher the speculation if they were induced by special development stage or environmental conditions based on cis-element assay in their promoter sequence. In summary, the identification, expression pattern analysis, biological and physiological function assays of CgSTPs explored the functional CgSTP members in sugar accumulation and pollen tube growth, and also paved a way for further elucidating the more functions and regulatory mechanisms of sugar transport proteins in citrus species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CL and J-HL designed the experiments. WX performed the experiments with assistance from ZL, ZZ, SZ and ML. WX, DG, J-HL, and CL analyzed and discussed the results. WX and CL finalized writing and revision of the manuscript. All authors have read and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key Research and Development Program of China (2019YFD1000100), Hubei Hongshan Laboratory (2021hszd016), and the Huazhong Agricultural University (start-up funding to CL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1106219/full#supplementary-material
References
Afoufa-Bastien, D., Medici, A., Jeauffre, J., Coutos-Thevenot, P., Lemoine, R., Atanassova, R., et al. (2010). The vitis vinifera sugar transporter gene family: Phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 10, 245. doi: 10.1186/1471-2229-10-245
Aslam, M. M., Deng, L., Wang, X. B., Wang, Y., Pan, L., Liu, H., et al. (2019). Expression patterns of genes involved in sugar metabolism and accumulation during peach fruit development and ripening. Sci. Hortic. 257, 108633. doi: 10.3390/ijms17122112
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Buttner, M. (2007). The monosaccharide transporter(-like) gene family in arabidopsis. FEBS Lett. 581, 2318–2324. doi: 10.1016/j.febslet.2007.03.016
Buttner, M. (2010). The arabidopsis sugar transporter (AtSTP) family: An update. Plant Biol. 12, 35–41. doi: 10.1111/j.1438-8677.2010.00383.x
Buttner, M., Truernit, E., Baier, K., Scholz-Starke, J., Sontheim, M., Lauterbach, C., et al. (2000). AtSTP3, a green leaf-specific, low affinity monosaccharide-h+ symporter of arabidopsis thaliana. Plant Cell Environ. 23, 175–184. doi: 10.1046/j.1365-3040.2000.00538.x
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, J. T., Wang, Z. Y., Yao, F. Z., Gao, L. H., Ma, S., Sui, X. L., et al. (2015). Down-regulating CsHT1, a cucumber pollen-specific hexose transporter, inhibits pollen germination, tube growth, and seed development. Plant Physiol. 168, 635–63+. doi: 10.1104/pp.15.00290
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–U199. doi: 10.1038/nature09606
Chiou, T. J., Bush, D. R. (1996). Molecular cloning, immunochemical localization to the vacuole, and expression in transgenic yeast and tobacco of a putative sugar transporter from sugar beet. Plant Physiol. 110, 511–520. doi: 10.1104/pp.110.2.511
Deng, X. L., An, B. G., Zhong, H., Yang, J., Kong, W. L., Li, Y. S. (2019). A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 10, 239. doi: 10.3390/genes10030239
Fang, T., Peng, Y., Rao, Y., Li, S. H., Zeng, L. H. (2020). Genome-wide identification and expression analysis of sugar transporter (ST) gene family in longan (Dimocarpus longan l.). Plants-Basel 9, 342. doi: 10.3390/plants9030342
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fotopoulos, V., Gilbert, M. J., Pittman, J. K., Marvier, A. C., Buchanan, A. J., Sauer, N., et al. (2003). The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atbetafruct1, are induced in arabidopsis during infection with the fungal biotroph erysiphe cichoracearum. Plant Physiol. 132, 821–829. doi: 10.1104/pp.103.021428
Gautam, T., Dutta, M., Jaiswal, V., Zinta, G., Gahlaut, V., Kumar, S. (2022). Emerging roles of SWEET sugar transporters in plant development and abiotic stress responses. Cells 11, 1303. doi: 10.3390/cells11081303
Hirai, T., Heymann, J. A., Shi, D., Sarker, R., Maloney, P. C., Subramaniam, S. (2002). Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9, 597–600. doi: 10.1038/nsb821
Huai, B. Y., Yuan, P., Ma, X. X., Zhang, X. R., Jiang, L. H., Zheng, P. J., et al. (2022). Sugar transporter TaSTP3 activation by TaWRKY19/61/82 enhances stripe rust susceptibility in wheat. New Phytol. 236, 266–282. doi: 10.1111/nph.18331
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiu, S. T., Haider, M. S., Kurjogi, M. M., Zhang, K. K., Zhu, X. D., Fang, J. G. (2018). Genome-wide characterization and expression analysis of sugar transporter family genes in woodland strawberry. Plant Genome 11(3). doi: 10.3835/plantgenome2017.11.0103
Kato, N., Pontier, D., Lam, E. (2002). Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Physiol. 129, 931–942. doi: 10.1104/pp.005496
Koch, K. (2004). Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246. doi: 10.1016/j.pbi.2004.03.014
Kong, L. Y., Li, X. N., Zhan, Z. X., Piao, Z. Y. (2022). Sugar transporters in plasmodiophora brassicae: Genome-wide identification and functional verification. Int. J. Mol. Sci. 23, 5264. doi: 10.3390/ijms23095264
Krenek, P., Samajova, O., Luptovciak, I., Doskocilova, A., Kornis, G., Samaj, J. (2015). Transient plant transformation mediated by agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 33, 1024–1042. doi: 10.1016/j.biotechadv.2015.03.012
Kuhn, C., Grof, C. P. (2010). Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 13, 288–298. doi: 10.1016/j.pbi.2010.02.001
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liang, M., Yang, W., Su, S. Y., Fu, L. L., Yi, H. L., Chen, C. W., et al. (2017). Genome-wide identification and functional analysis of s-RNase involved in the self-incompatibility of citrus. Mol. Genet. Genomics 292, 325–341. doi: 10.1007/s00438-016-1279-8
Li, M. J., Feng, F. J., Cheng, L. L. (2012). Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PloS One 7 (3), e33055. doi: 10.1371/journal.pone.0033055
Li, S. J., Liu, X. J., Xie, X. L., Sun, C. D., Grierson, D., Yin, X. R., et al. (2015b). CrMYB73, a PH-like gene, contributes to citric acid accumulation in citrus fruit. Sci. Hortic. 197, 212–217. doi: 10.1016/j.scienta.2015.09.037
Li, C. L., Meng, D., Pineros, M. A., Mao, Y. X., Dandekar, A. M., Cheng, L. L., et al. (2020). A sugar transporter takes up both hexose and sucrose for sorbitol-modulated In vitro pollen tube growth in apple. Plant Cell 32, 449–469. doi: 10.1105/tpc.19.00638
Liu, Q., Dang, H. J., Chen, Z. J., Wu, J. Z., Chen, Y. H., Chen, S. B., et al. (2018). Genome-wide identification, expression, and functional analysis of the sugar transporter gene family in cassava (Manihot esculenta). Int. J. Mol. Sci. 19 (4), 987. doi: 10.3390/ijms19040987
Liu, H. T., Ji, Y., Liu, Y., Tian, S. H., Gao, Q. H., Zou, X. H., et al. (2020). The sugar transporter system of strawberry: Genome-wide identification and expression correlation with fruit soluble sugar-related traits in aFragaria x ananassagermplasm collection. Horticult. Res. 7, 132. doi: 10.1038/s41438-020-00359-0
Liu, C., Li, D. X., Huang, X. B., Zhang, F. Q., Xie, Z. Z., Zhang, H. Y., et al. (2022a). Manual thinning increases fruit size and sugar content of citrus reticulata blanco and affects hormone synthesis and sugar transporter activity. J. Integr. Agric. 21, 725–735. doi: 10.1016/S2095-3119(20)63502-x
Liu, S. C., Liu, X. C., Gou, B. R., Wang, D. L., Liu, C. R., Sun, J., et al. (2022b). The interaction between CitMYB52 and CitbHLH2 negatively regulates citrate accumulation by activating CitALMT in citrus fruit. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.848869
Li, J. M., Zheng, D. M., Li, L. T., Qiao, X., Wei, S. W., Bai, B., et al. (2015a). Genome-wide function, evolutionary characterization and expression analysis of sugar transporter family genes in pear (Pyrus bretschneideri rehd). Plant Cell Physiol. 56, 1721–1737. doi: 10.1093/pcp/pcv090
McCurdy, D. W., Dibley, S., Cahyanegara, R., Martin, A., Patrick, J. W. (2010). Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 3, 1049–1063. doi: 10.1093/mp/ssq050
Meng, D., Gu, Z., Li, W., Wang, A., Yuan, H., Yang, Q., et al. (2014). Apple MdABCF assists in the transportation of s-RNase into pollen tubes. Plant J. 78, 990–1002. doi: 10.1111/tpj.12524
Meng, D., He, M., Bai, Y., Xu, H., Dandekar, A. M., Fei, Z., et al. (2018). Decreased sorbitol synthesis leads to abnormal stamen development and reduced pollen tube growth via an MYB transcription factor, MdMYB39L, in apple (Malus domestica). New Phytol. 217, 641–656. doi: 10.1111/nph.14824
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Murcia, G., Pontin, M., Piccoli, P. (2018). Role of ABA and gibberellin A(3) on gene expression pattern of sugar transporters and invertases in vitis vinifera cv. Malbec during berry ripening. Plant Growth Regul. 84, 275–283. doi: 10.1007/s10725-017-0338-4
Murcia, G., Pontin, M., Reinoso, H., Baraldi, R., Bertazza, G., Gomez-Talquenca, S., et al. (2016). ABA and GA(3) increase carbon allocation in different organs of grapevine plants by inducing accumulation of non-structural carbohydrates in leaves, enhancement of phloem area and expression of sugar transporters. Physiol. Plant. 156, 323–337. doi: 10.1111/ppl.12390
Otori, K., Tanabe, N., Tamoi, M., Shigeoka, S. (2019). Sugar transporter protein 1 (STP1) contributes to regulation of the genes involved in shoot branching via carbon partitioning in arabidopsis. Biosci. Biotechnol. Biochem. 83, 472–481. doi: 10.1080/09168451.2018.1550355
Paulsen, P. A., Custodio, T. F., Pedersen, B. P. (2019). Crystal structure of the plant symporter STP10 illuminates sugar uptake mechanism in monosaccharide transporter superfamily. Nat. Commun. 10 (1), 407. doi: 10.1038/s41467-018-08176-9
Pi, M., Zhong, R., Hu, S., Cai, Z., Plunkert, M., Zhang, W., et al. (2022). A GT-1 and PKc domain-containing transcription regulator SIMPLE LEAF1 controls compound leaf development in woodland strawberry. New Phytol. doi: 10.1111/nph.18589
Radchuk, R., Emery, R. J., Weier, D., Vigeolas, H., Geigenberger, P., Lunn, J. E., et al. (2010). Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 61, 324–338. doi: 10.1111/j.1365-313X.2009.04057.x
Reuscher, S., Akiyama, M., Yasuda, T., Makino, H., Aoki, K., Shibata, D., et al. (2014). The sugar transporter inventory of tomato: Genome-wide identification and expression analysis. Plant Cell Physiol. 55, 1123–1141. doi: 10.1093/pcp/pcu052
Riesmeier, J. W., Willmitzer, L., Frommer, W. B. (1992). Isolation and characterization of a sucrose carrier cdna from spinach by functional expression in yeast. EMBO J. 11, 4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x
Riesmeier, J. W., Willmitzer, L., Frommer, W. B. (1994). Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13, 1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x
Rolland, F., Baena-Gonzalez, E., Sheen, J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Rolland, F., Moore, B., Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 Suppl, S185–S205. doi: 10.1105/tpc.010455
Rombauts, S., Dehais, P., Van Montagu, M., Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rottmann, T., Klebl, F., Schneider, S., Kischka, D., Ruscher, D., Sauer, N., et al. (2018). Sugar transporter STP7 specificity for l-arabinose and d-xylose contrasts with the typical hexose transporters STP8 and STP12. Plant Physiol. 176, 2330–2350. doi: 10.1104/pp.17.01493
Rottmann, T., Zierer, W., Subert, C., Sauer, N., Stadler, R. (2016). STP10 encodes a high-affinity monosaccharide transporter and is induced under low-glucose conditions in pollen tubes of arabidopsis. J. Exp. Bot. 67, 2387–2399. doi: 10.1093/jxb/erw048
Rozwadowski, K., Yang, W., Kagale, S. (2008). Homologous recombination-mediated cloning and manipulation of genomic DNA regions using gateway and recombineering systems. BMC Biotechnol. 8, 88. doi: 10.1186/1472-6750-8-88
Sade, D., Brotman, Y., Eybishtz, A., Cuadros-Inostroza, A., Fernie, A. R., Willmitzer, L., et al. (2013). Involvement of the hexose transporter gene LeHT1 and of sugars in resistance of tomato to tomato yellow leaf curl virus. Mol. Plant 6, 1707–1710. doi: 10.1093/mp/sst036
Slewinski, T. L. (2011). Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 4, 641–662. doi: 10.1093/mp/ssr051
Smeekens, S., Hellmann, H. A. (2014). Sugar sensing and signaling in plants. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00113
Soni, R., Carmichael, J. P., Murray, J. A. (1993). Parameters affecting lithium acetate-mediated transformation of saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet. 24, 455–459. doi: 10.1007/bf00351857
Stadler, R., Buttner, M., Ache, P., Hedrich, R., Ivashikina, N., Melzer, M., et al. (2003). Diurnal and light-regulated expression of AtSTP1 in guard cells of arabidopsis. Plant Physiol. 133, 528–537. doi: 10.1104/pp.103.024240
Tao, Y., Cheung, L. S., Li, S., Eom, J. S., Chen, L. Q., Xu, Y., et al. (2015). Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527, 259–263. doi: 10.1038/nature15391
Toyofuku, K., Kasahara, M., Yamaguchi, J. (2000). Characterization and expression of monosaccharide transporters (OsMSTs) in rice. Plant Cell Physiol. 41, 940–947. doi: 10.1093/pcp/pcd016
Truernit, E., Schmid, J., Epple, P., Illig, J., Sauer, N. (1996). The sink-specific and stress-regulated arabidopsis STP4 gene: Enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8, 2169–2182. doi: 10.1105/tpc.8.12.2169
Truernit, E., Stadler, R., Baier, K., Sauer, N. (1999). A male gametophyte-specific monosaccharide transporter in arabidopsis. Plant J. 17, 191–201. doi: 10.1046/j.1365-313x.1999.00372.x
Udvardi, M. K., Czechowski, T., Scheible, W. R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737. doi: 10.1105/tpc.108.061143
Wang, Z. Y., Liang, Y. H., Jin, Y. R., Tong, X. L., Wei, X. Y., Ma, F. W., et al. (2020a). Ectopic expression of apple hexose transporter MdHT2.2 reduced the salt tolerance of tomato seedlings with decreased ROS-scavenging ability. Plant Physiol. Biochem. 156, 504–513. doi: 10.1016/j.plaphy.2020.10.001
Wang, Y. P., Tang, H. B., Debarry, J. D., Tan, X., Li, J. P., Wang, X. Y., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Z. Y., Wei, X. Y., Yang, J. J., Li, H. X., Ma, B. Q., Zhang, K. K., et al. (2020b). Heterologous expression of the apple hexose transporter MdHT2.2 altered sugar concentration with increasing cell wall invertase activity in tomato fruit. Plant Biotechnol. J. 18, 540–552. doi: 10.1111/pbi.13222
Wei, X. Y., Liu, F. L., Chen, C., Ma, F. W., Li, M. J. (2014). The malus domestica sugar transporter gene family: identifications based on genome and expression prof ling related to the accumulation of fruit sugars. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00569
Wei, Q. J., Ma, Q. L., Zhou, G. F., Liu, X., Ma, Z. Z., Gu, Q. Q. (2021). Identification of genes associated with soluble sugar and organic acid accumulation in 'Huapi' kumquat (Fortunella crassifolia swingle) via transcriptome analysis. J. Sci. Food Agric. 101, 4321–4331. doi: 10.1002/jsfa.11072
Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C. P., Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in saccharomyces cerevisiae. FEBS Lett. 464, 123–128. doi: 10.1016/s0014-5793(99)01698-1
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wu, G. A., Prochnik, S., Jenkins, J., Salse, J., Hellsten, U., Murat, F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. doi: 10.1038/nbt.2906
Wu, G. A., Terol, J., Ibanez, V., Lopez-Garcia, A., Perez-Roman, E., Borreda, C., et al. (2018). Genomics of the origin and evolution of citrus. Nature 554, 311–316. doi: 10.1038/nature25447
Xu, Q., Chen, L. L., Ruan, X., Chen, D., Zhu, A., Chen, C., et al. (2013). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66. doi: 10.1038/ng.2472
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yan, N. (2013). Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38, 151–159. doi: 10.1016/j.tibs.2013.01.003
Zhang, W., Wang, S. Y., Yu, F. W., Tang, J., Yu, L., Wang, H., et al. (2019). Genome-wide identification and expression profiling of sugar transporter protein (STP) family genes in cabbage (Brassica oleracea var. capitata l.) reveals their involvement in clubroot disease responses. Genes 10 (1), 71. doi: 10.3390/genes10010071
Keywords: Citrus grandis, sugar transporter protein, gene expression, sugar accumulation, pollen tube growth
Citation: Xu W, Liu Z, Zhao Z, Zhang S, Li M, Guo D, Liu J-H and Li C (2023) The functional analysis of sugar transporter proteins in sugar accumulation and pollen tube growth in pummelo (Citrus grandis). Front. Plant Sci. 13:1106219. doi: 10.3389/fpls.2022.1106219
Received: 23 November 2022; Accepted: 12 December 2022;
Published: 04 January 2023.
Edited by:
Chao Li, Northwest A&F University, ChinaReviewed by:
Junhong Zhang, Zhejiang Agriculture and Forestry University, ChinaMengxia Zhang, Cornell University, United States
Copyright © 2023 Xu, Liu, Zhao, Zhang, Li, Guo, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunlong Li, Y2wyNDQ0QG1haWwuaHphdS5lZHUuY24=
 Weiwei Xu1
Weiwei Xu1 Zeqi Zhao
Zeqi Zhao Ji-Hong Liu
Ji-Hong Liu Chunlong Li
Chunlong Li