
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 January 2023
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1105988
This article is part of the Research TopicBeyond genetics: modifications of nucleic acid and chromatinView all 6 articles
 Chae Jin Lim1,2†
Chae Jin Lim1,2† Ki Suk Park2†
Ki Suk Park2† Akhtar Ali1,2†
Akhtar Ali1,2† Junghoon Park1,2
Junghoon Park1,2 Seung Min Ryou2
Seung Min Ryou2 Mingzhe Shen3
Mingzhe Shen3 Haris Ali Khan2
Haris Ali Khan2 Zein Eddin Bader2
Zein Eddin Bader2 Shah Zareen2
Shah Zareen2 Min Jae Bae2
Min Jae Bae2 Jong Hyoo Choi2
Jong Hyoo Choi2 Zheng-Yi Xu4
Zheng-Yi Xu4 Jose M. Pardo5
Jose M. Pardo5 Dae-Jin Yun2*
Dae-Jin Yun2*Arabidopsis HOS15/PWR/HDA9 repressor complex, which is similar to the TBL1/NcoR1/HDAC complex in animals, plays a well-known role in epigenetic regulation. PWR and HDA9 have been reported to interact with each other and modulate the flowering time by repressing AGL19 expression, whereas HOS15 and HDA9, together with the photoperiodic evening complex, regulate flowering time through repression of GI transcription. However, the role of the HOS15/PWR/HDA9 core repressor complex as a functional unit in the regulation of flowering time is yet to be explored. In this study, we reported that the loss-of-function hos15-2/pwr/hda9 triple mutant accumulates higher transcript levels of AGL19 and exhibits an early flowering phenotype similar to those of hos15, pwr, and hda9 single mutants. Interestingly, the accumulation of HOS15 in the nucleus was drastically reduced in pwr and hda9 mutants. As a result, HOS15 could not perform its role in histone deacetylation or interaction with H3 in the nucleus. Furthermore, HOS15 is also associated with the same region of the AGL19 promoter known for PWR-HDA9 binding. The acetylation level of the AGL19 promoter was increased in the hos15-2 mutant, similar to the pwr and hda9 mutants. Therefore, our findings reveal that the HOS15/PWR/HDA9 repressor complex deacetylates the promoter region of AGL19, thereby negatively regulating AGL19 transcription, which leads to early flowering in Arabidopsis.
The regulation of flowering time has been well studied in Arabidopsis and involves three major pathways: vernalization, autonomous, and photoperiod pathways. Exposure to cold temperatures in winter is necessary for flowering in spring or summer, and this process is called vernalization (Amasino, 2005). The autonomous or developmental pathway is established by repressing FLOWERING LOCUS C (FLC), a negative regulator of floral integrators, thereby activating the expression of flowering genes, including the mobile ‘florigen’ FLOWERING LOCUS T (FT) (Corbesier and Coupland, 2006). FLC abundance is also repressed by vernalization independently of the autonomous pathway. Lastly, the photoperiod pathway is controlled by light and the circadian clock. The major factors involved in the photoperiod pathway are CONSTANS (CO) and GIGANTEA (GI), that integrate clock and light signals to provide photoperiod-specific induction of FT expression (Jarillo et al., 2008; Song et al., 2014; Shim et al., 2017).
AGAMOUS LIKE 19 (AGL19) was first discovered through phylogenetical identification of conserved MADS-box domain (Alvarez-Buylla et al., 2000). Expression of AGL19 is mainly expressed in roots and is also observed in leaves, seedlings, and flowers. The first function of AGL19 was revealed as a floral activator in the vernalization pathway. AGL19 in this pathway is negatively regulated by POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) subunits and these complexes also target AGL19 chromatin region and affect H3K27me3 enrichment (Schönrock et al., 2006). AGL19 functions not only in flowering related to vernalization but also in non-inductive short-day conditions. To prevent precocious flowering, de-acetylation activity of HISTONE DEACETYLASE 9 (HDA9) regulates the acetylation level of AGL19, which in turn results in repression of FT gene (Kang et al., 2015).
Gene regulation in signaling pathways is determined genetically and epigenetically. Epigenetic regulation of genes is achieved by histone modification, chromatin remodeling, DNA methylation, non-coding RNA, and microRNA (Chinnusamy and Zhu, 2009). Histone modification is a well-known factor involved in epigenetic gene regulation. Histones are modified by acetylation, phosphorylation, ubiquitination, methylation, and sumoylation, which alter the structure of chromatin by either compacting or loosening it (Pfluger and Wagner, 2007; Alaskhar Alhamwe et al., 2018; Zhao et al., 2019). Histone acetylation relaxes chromatin, making it possible for transcription factors to bind to the promoters, thereby activating gene expression. On the contrary, chromatin becomes compacted with histone deacetylation, hindering the interaction of transcription factors with the promoters and repressing the expression of genes (Tamaru, 2010; Park et al., 2018a).
Acetylation and deacetylation of histones are mediated mainly by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. In plants, HDACs are classified into three groups: the RPD3/HDA1 superfamily, HD-tuin family, and SRT (sirtuin) family (Pandey et al., 2002; Hollender and Liu, 2008). Plant HATs are also classified into three groups: GNAT (GCN5- RELATED N-TERMINAL ACETYLTRANSFERASE)-MYST, p300/CREB binding protein (CBP), and TAFII250 related (Pandey et al., 2002). HDACs and HATs in plants regulate numerous plant genes, including those involved in plant development, flowering, and stress responses. For instance, HDA9, an RPD3/HDA1 type histone deacetylase, regulates floral transition in Arabidopsis. HDA9 negatively regulates the flowering-inducer gene AGAMOUS-LIKE 19 (AGL19) by deacetylating its promoter region (Kim et al., 2013; Kim et al., 2016). HDA9 also interacts with the promoters of the flowering genes SOC1 and AGL24 in Brassica juncea (Jiang et al., 2018). In addition, HDA9 is a major component of the evening complex that represses GI (Park et al., 2019).
The Arabidopsis protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15) is a homolog of mammalian TBL1 and TBLR1, which are core components of the SMRT/NCoR1 co-repressor complex (Perissi et al., 2008; Park et al., 2018b). Further analysis revealed that HOS15 is also important for histone deacetylation, supporting the notion that HOS15 functions as a repressor of gene expression (Zhu et al., 2008). Structural analysis showed that HOS15 comprises four major motifs: the LisH motif, WD-40 domain, F-box-like motif, and DWD motif. The LisH motif is involved in protein dimerization (Choi et al., 2008; Joachimiak et al., 2020). The WD-40 motif is important for protein-protein interactions (Xu and Min, 2011). The F-box and DWD motifs are well-known domains for the substrate receptor in the Cullin-base E3 ubiquitin ligase complex. Recent findings suggest that HOS15 acts as a ubiquitin E3 ligase substrate receptor to regulate various signal transduction pathways through targeted protein degradation (Park et al., 2018b; Ali et al., 2019; Shen et al., 2020). HOS15 promotes the degradation of HD2C in low-temperature signaling, regulates drought stress through OST1 (SnRK2.6) degradation, and is responsible for immune responses by targeting NPR1 for degradation (Park et al., 2018b; Ali et al., 2019; Shen et al., 2020). More recently we have shown that HOS15 interacts with POWERDRESS (PWR), known as the SMRT/NCoR1 homolog, and regulates the stability of the HD2C protein under cold stress (Lim et al., 2020). Moreover, it was recently reported that HOS15 in conjunction with PWR-HDA9 regulates leaf senescence (Zareen et al., 2022). HDA9 also forms a complex with PWR and acts as a repressor in various signaling pathways, such as flowering time, leaf senescence, development, thermomorphogenesis, ABA and drought stress response (Yumul et al., 2013; Kang et al., 2015; Chen et al., 2016; Kim et al., 2016; Suzuki et al., 2018; Tasset et al., 2018; Mayer et al., 2019; Ali and Yun, 2020; Khan et al., 2020; Zareen et al., 2022). Overall, these reports suggest the existence of a HOS15-PWR-HDA9 repressor complex which plays an important role in the overall growth, development, and leaf senescence of plants, as well as in overcoming external stimuli.
HOS15 and HDA9 have been shown to be important components of the evening complex that regulate GI (Park et al., 2019). In a similar study, the PWR-HDA9 complex was shown to negatively regulate the expression of the AGL19 through histone deacetylation (Kang et al., 2015; Kim et al., 2016). However, the molecular basis of HOS15-PWR-HDA9 complex interference with flowering time is yet to be determined. In this study, we report that HOS15, together with PWR and HDA9, forms a repressor complex and negatively regulates floral transition by inhibiting the expression of AGL19 through histone deacetylation. Furthermore, HDA9 and PWR are required for the nuclear translocation of HOS15 to repress the expression of target genes. Plants lacking the HOS15-HDA9-PWR complex exhibit hyperacetylation of the AGL19 promoter region, which leads to an early flowering phenotype. Our findings demonstrated that HOS15-HDA9-PWR is a repressor complex that negatively regulates floral transition through histone deacetylation.
The Arabidopsis thaliana ecotype Col-0 and mutants pwr-2 (SALK_071811), pwr-3 (SALK_006823), hda9-1 (GK_305G03), hda9-2 (SALK_007123), and hos15-2 (GK_785B10) mutants, and HOS15pro::HOS15-HA/hos15-2 (Park et al., 2018a), HOS15pro::HOS15-FLAG/hos15-2 (Park et al., 2018b) transgenic plants were used in this study. Double (pwr-2 hos15-2, hda9-2 hos15-2, pwr-2 hda9-2) and triple mutants (pwr-2 hda9-2 hos15-2) were generated by crossing. Seeds were surface-sterilized in a solution containing 3% (v/v) sodium hypochlorite solution (Yakuri Pure Chemicals, Kyoto, Japan) for 5 min and then rinsed five times with sterilized water. After stratification at 4°C for 3 days in the dark, the plants were grown on half-strength Murashinge and Skoog (MS) medium or soil at 23°C ± 1°C under 16-h light/8-h-dark conditions with 100 µM m-2s-1 light intensity.
Flowering time analysis of the WT and mutant plants was performed at 23°C ± 1°C in long-day conditions. Flowering time was measured by counting the number of days-to-flowering and the total number of rosette leaves from the primary meristems at bolting.
Total RNA was extracted from two-week-old plants using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase (Sigma-Aldrich). cDNA was synthesized using reverse transcriptase (Toyobo, Osaka, Japan). The SYBR Green PCR Master Mix Kit (Bio-Rad) was used for quantitative PCR using the indicated gene-specific primers. Primer sequences are provided in Supplementary Table 1. Relative expression levels were analyzed using the comparative cycle threshold method (Bio-Rad).
Total protein was isolated from Arabidopsis seedlings using protein extraction buffer containing 150 mM NaCl, 100 mM Tris-Cl (pH 7.5), 0.5% (v/v) NP-40, 1 mM EDTA, 3 mM DTT, and protease inhibitors (1 mM PMSF, 5 μg mL-1 leupeptin, 1 μg mL-1 aprotinin, 1 μg mL-1 pepstatin, 5 μg mL-1 antipain, 5 μg mL-1 chymostatin, 2 mM Na2VO3, 2 mM NaF, and 50 mM MG132). The proteins were separated and detected using SDS-PAGE and immunoblottied with the appropriate antibodies, anti-HOS15 (Park et al., 2018a; Lim et al., 2020), anti-α-tubulin (Sigma-Aldrich), anti-Ac-H3 (Millipore), anti-H3 (Abcam), and anti-FLAG (Sigma-Aldrich) for 2 h at room temperature or overnight at 4°C. Membranes were developed using peroxidase-conjugated secondary anti-rabbit IgG (Santa Cruz Biotechnology) and anti-mouse IgG (Santa Cruz Biotechnology). Western blot membranes were incubated with Clarity™ Western ECL Substrate solution (Bio-Rad), and signals were detected using an imaging system (ChemiDoc™MP, Bio-Rad).
Nuclei were extracted from two-week-old plants using Honda’s buffer (0.4 M sucrose, 2.5% Ficoll 400, 5% dextran T-40, 10 mM MgCl2, 25 mM Tris-Cl (pH 7.5), 10 mM β-mercaptoethanol, 100 mg/mL phenylmethylsulfonyl fluoride, 0.5 mg/mL antipain, and 0.5 mg/mL leupeptin) (Folta and Kaufman, 2006; Park et al., 2018b). The samples were filtered through a 60-µm nylon filter (Millipore) and incubated on ice for 15 min. Triton X-100 was added at a final concentration of 0.5% and the sample was centrifuged at 1500g for 5 min. The supernatants were transferred to new tubes, and cytosolic proteins were used. The pellet was washed with 1 mL Honda’s buffer containing 0.1% Triton X-100. The cells were gently resuspended and centrifuged at 100g for 5 min to pellet the starch and cell debris. The supernatant was transferred to a tube and centrifuged at 1800g for 5 min to pellet the nuclei. The nuclear and cytosolic extracts were suspended in SDS-PAGE loading buffer. Immunoblots were performed using anti-HOS15, anti-α-tubulin (Sigma), and anti-H3 (Abcam) antibodies, and antigen proteins were visualized by chemiluminescence using ECL-detecting reagent (Bio-Rad).
The chromatin immunoprecipitation (ChIP) assay was performed as described by Lim et al. (2020). Nuclear proteins were extracted from two-week-old Arabidopsis plants, and immunoprecipitation was performed using salmon sperm DNA/protein A agarose beads (Upstate Biotechnology) fused to anti-HOS15 and anti-acetylated H3 antibodies (Millipore). Beads treated with anti-rabbit IgG were used as negative controls for each ChIP assay. The immunoprecipitated DNA was purified by phenol-chloroform-isoamyl (PCI) extraction and ethanol precipitation. The precipitated DNA was dissolved in TE buffer and quantified using RT-qPCR. The primers used in the ChIP assays are listed in Supplementary Table 1, and an UBQ10 DNA fragment was used for normalization.
To investigate the function of the HOS15-PWR-HDA9 complex in regulating the flowering time, we analyzed flowering phenotypes of the loss-of-function hos15-2 mutant and three complementation lines expressing the construct HOS15pro::HOS15-HA in the hos15-2 background (Park et al., 2018a). As expected, hos15-2 mutant plants exhibited early flowering (Park et al., 2019), while the flowering time of complementation lines was similar to that of WT plants (Supplemental Figures 1A–C). Using RNA-seq analysis data, we previously showed that the expression level of the AGL19 was upregulated in hos15-2 mutants (Park et al., 2019), which could promote flowering time (Kang et al., 2015; Park et al., 2019). To confirm this, we analyzed the transcript level of AGL19 using RT-qPCR, which was significantly increased in the hos15-2 mutant. In contrast, the complementation lines possessed a similar level of AGL19 transcription to that of the WT plants (Supplemental Figure 1D; Supplemental Figure 2). These results suggest that the early flowering phenotype of hos15-2 might be due to the upregulation of AGL19.
The transcript level of AGL19 has previously been reported to be upregulated in pwr and hda9 knockout mutants, and as a result, these mutants show early flowering phenotypes (Kang et al., 2015; Kim et al., 2016). Since HOS15 knockout plants also show early flowering and the upregulation of AGL19 transcript levels (Supplemental Figure 1), we assumed that HOS15 might work together with PWR-HDA9 in the same repressor complex, thereby negatively regulating flowering time by repressing AGL19 transcription. To test our hypothesis, we generated double and triple (pwr hos15, hda9 pwr, hda9 hos15, hda9 hos15 pwr) mutant plants and tested their flowering phenotypes. As expected, double and triple knockout mutants of the HOS15-PWR-HAD9 complex showed an early flowering phenotype statistically similar to single mutants of HOS15, PWR, and HDA9, signifying that these mutations were not additive (Figure 1A–C). In addition to flowering phenotypes, the transcript level of AGL19 was upregulated in single, double, and triple mutant plants compared to that in WT plants (Figure 1D). Furthermore, double and triple mutants of HOS15, PWR, and HDA9 exhibited dwarf phenotypes, such as small plant size and short and blunt-end siliques, which are common phenotypes of hos15-2, pwr, and hda9 single mutants (Supplemental Figures 3 and 4) (Kang et al., 2015; Kim et al., 2016; Mayer et al., 2019; Lim et al., 2020). Overall, these findings indicate that the HOS15-PWR-HDA9-complex negatively regulates flowering time by inhibiting AGL19 expression.
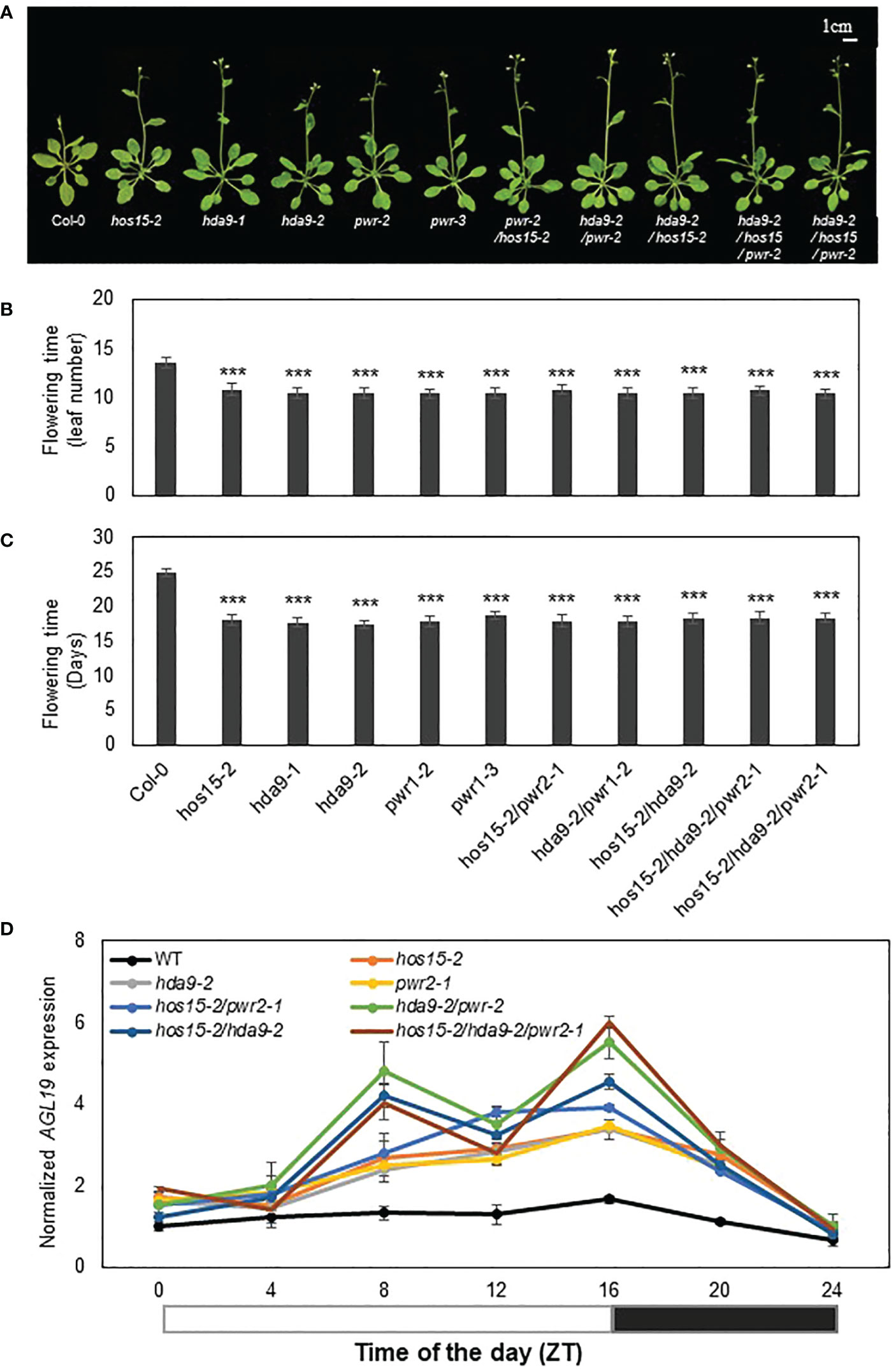
Figure 1 Involvement of HOS15, HDA9, and PWR in the same flowering pathway. (A–C) Flowering time of wild type (Col-0), hos15-2, hda9, pwr-2, as well as double and triple mutant plants under long-day conditions (16 h light/8 h dark). The number of rosette leaves at bolting (B) and days to flowering (C) were counted from (A). Data represent means ± SD (n = 19). Significant difference was determined by a Student’s t-test (***p<0.001). (D) The transcript level of AGL19 in two-week-old Col-0, hos15-2, hda9, pwr, as well as double and triple mutant Arabidopsis plants. Samples were harvested according to Zeitgeber time (ZT) and were analyzed by RT-qPCR and normalized with ACTIN2. Data represent means ± SD from three biological replicates with three technical repeats each (n = 3). The black and white bar represents night and daylight periods, respectively.
HDA9 negatively regulates AGL19 expression by associating with the promoter region of AGL19 and promoting histone deacetylation (Kang et al., 2015). Therefore, we tested whether HOS15 is associated with the promoter region of AGL19 to modulate chromatin status. A chromatin immunoprecipitation (ChIP) assay showed that HOS15 was indeed associated with the E region of the AGL19 promoter, in a region already known for HDA9 association (Figure 2; Kang et al., 2015). These data suggest that HOS15 binds to the promoter of AGL19 to co-regulate the histone acetylation status of AGL19 chromatin.
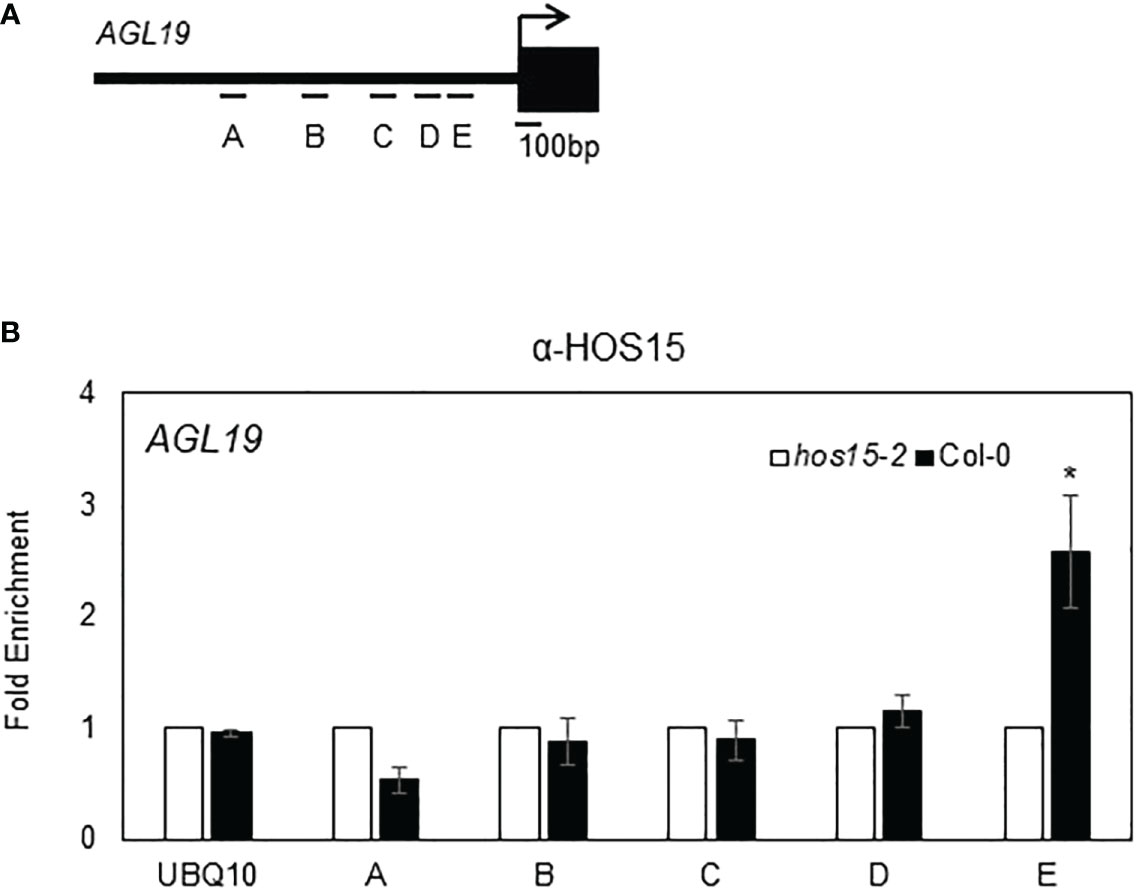
Figure 2 Association of HOS15 with the AGL19 promoter region. (A) Amplicon regions (A–E) in the AGL19 promoter used for ChIP-qPCR. The arrow indicates the transcriptional start site. (B) The ChIP assays were performed using the anti-HOS15 antibody on samples from two-week-old wild-type (Col-0) and hos15-2 mutant plants. The amount of DNA in the immunoprecipitated complex was determined by RT-qPCR using primers specific to the different regions (A–E) of the AGL19 promoter and is presented as the fold enrichment after normalization with the corresponding input and compared with hos15-2 mutant plants. Data represent means ± SD from three biological replicates with three technical repeats each (n = 3). Significant difference was determined by a Student’s t-test (* p<0.05).
HOS15 forms a complex with PWR and HDA9 (Park et al., 2018b; Suzuki et al., 2018; Mayer et al., 2019). Furthermore, HOS15, PWR, and HDA9 were predominantly localized in the nucleus (Zhu et al., 2008; Kang et al., 2015; Lim et al., 2020), and HDA9 accumulation was reduced in the nuclei of pwr and hos15 mutant plants (Chen et al., 2016; Mayer et al., 2019). Based on these precedents, we investigated whether the accumulation of HOS15 in nuclei also required PWR-HDA9. A nuclear-cytoplasm fractionation analysis using pwr and hda9 mutant plants showed that HOS15 nuclear accumulation was greatly decreased in pwr and hda9 mutants as compared with WT (Figure 3). Notably, compared with WT plants, HOS15 transcript levels were not changed in pwr and hda9 mutants; in contrast, the total protein amount was decreased in these mutants (Supplemental Figure 5). HOS15 also interacts with other RPD3-type class 1 histone deacetylases, such as HDA6 and HDA19 (Park et al., 2018a; Park et al., 2018b). However, unlike hda9, hda6, and hda19 mutants did not exhibit early flowering; instead, they exhibited partial late-flowering phenotypes (Supplemental Figures 6A, B). Furthermore, HOS15 total protein remained unchanged in hda6 and hda19 mutant plants compared with that in WT plants (Supplemental Figure 6C). Overall, these results suggest that HOS15 forms a complex with PWR-HDA9 in the nucleus and regulates floral transition by repressing AGL19 expression.
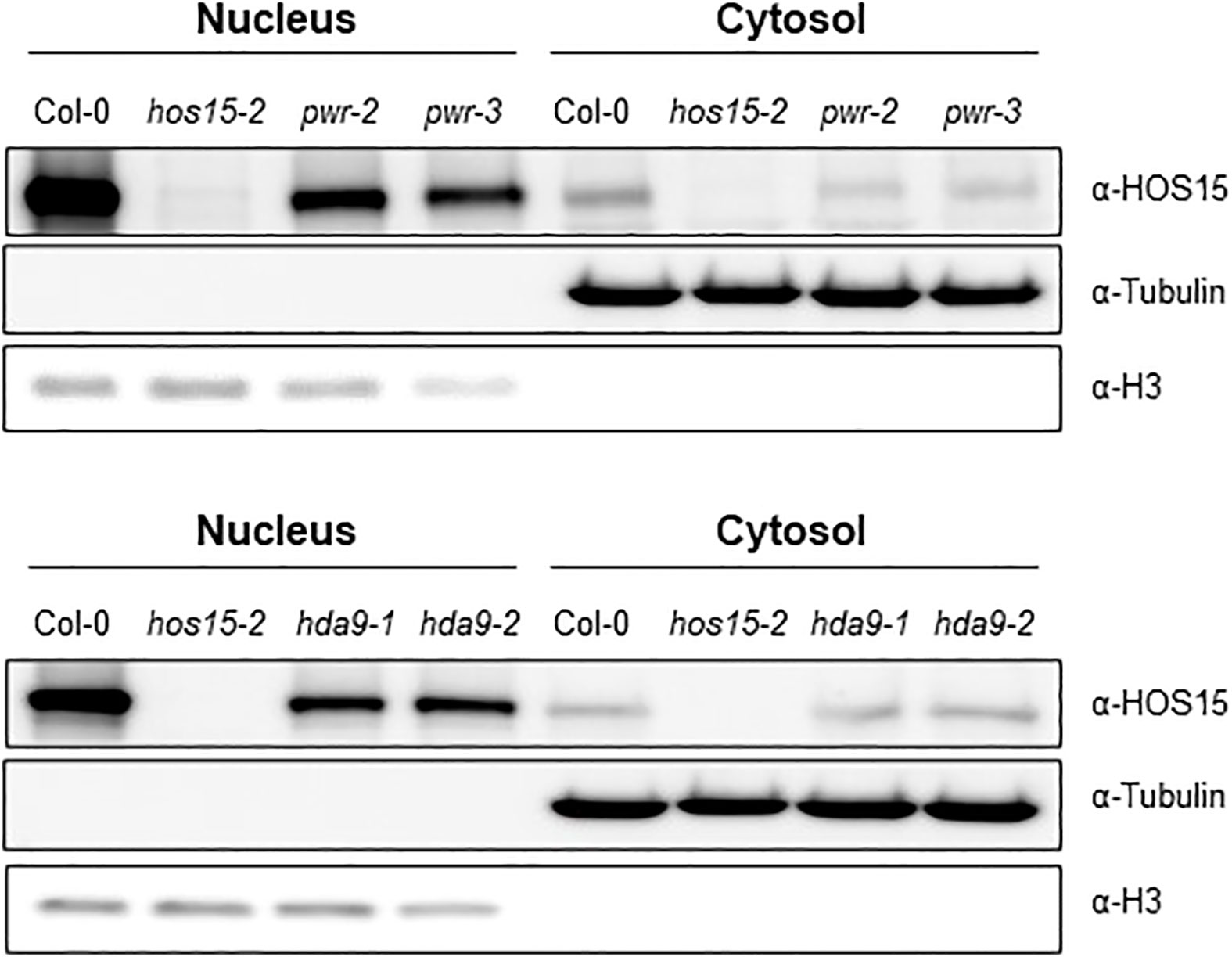
Figure 3 Decreased HOS15 nuclear accumulation in pwr and hda9 mutants. Immunoblotting of HOS15 protein in two-week-old plants of Col-0, hos15-2, hda9 and pwr mutants. Total protein was extracted and separated into cytoplasmic (cytosol) and nuclear (nucleus) fractions and immunoblotted with an anti-HOS15 antibody. α-Tubulin and histone H3 served as loading controls for the cytosol and nucleus, respectively.
Previous reports have revealed that HOS15, PWR, and HDA9 repress target gene expression by regulating histone H3 acetylation status (Kim et al., 2016; Mayer et al., 2019; Park et al., 2019). To investigate whether HOS15 interacts with H3, we performed a co-IP assay using two independent HOS15pro::HOS15-Flag/hos15-2 complementation plant lines (Park et al., 2018b). As shown in Figure 4A, HOS15 interacted with H3 in vivo. As HOS15 stability in the nucleus is dependent on PWR-HDA9, we tested HOS15 and H3 interactions in pwr and hda9 mutants. Notably, the interaction of HOS15 with H3 was drastically reduced in pwr and hda9 mutants compared with that in the WT plants (Figure 4B). In addition, total histone acetylation levels were increased in hos15, pwr and hda9 single, double, and triple mutant plants compared with WT plants, and the increment was similar between single and higher order mutants (Figure 5A) (Zareen et al., 2022). In line with this, we next tested the acetylation status of AGL19 chromatin in hos15, pwr and hda9 single and hos15/pwr/hda9 triple mutant plants using a ChIP assay with acetylated H3 (AcH3) antibody. H3 acetylation of AGL19 chromatin was higher in hos15-2 mutant plants than in the WT plants (Figure 5B, Supplemental Figure 7). Consistent with the notion that HOS15 functions together with the PWR-HDA9 complex, no significant difference in the increase in H3 acetylation level was observed between these single and triple mutant plants (Figure 5B). Taken together, these findings suggest that HOS15, PWR, and HDA9 work together in the same repressor complex and deacetylate AGL19 chromatin region, thereby negatively regulating AGL19 expression.
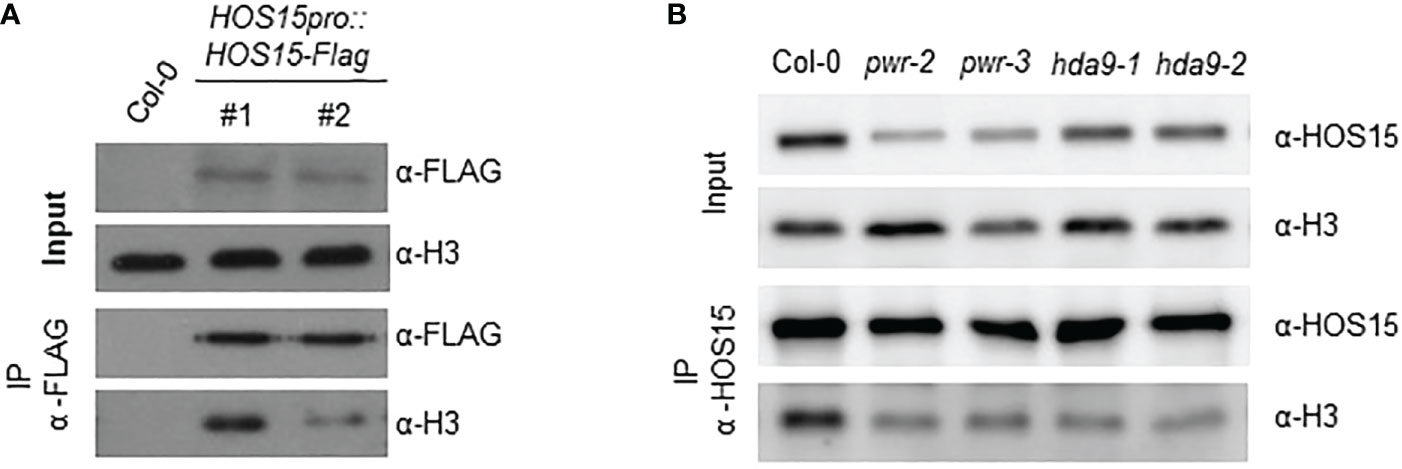
Figure 4 Interaction of HOS15 with histone H3. (A) Co-IP assay using two-week-old Arabidopsis Col-0 and HOS15pro:HOS15-Flag/hos15-2 complementation lines. Protein extracts (Input) were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-H3 antibodies (B) Co-IP assay using two-week-old Arabidopsis Col-0 hos15-2, hda9 and pwr mutant plants. Protein extracts (Input) were immunoprecipitated with anti-HOS15 antibody and immunoblotted with anti-HOS15 and anti-H3 antibodies.
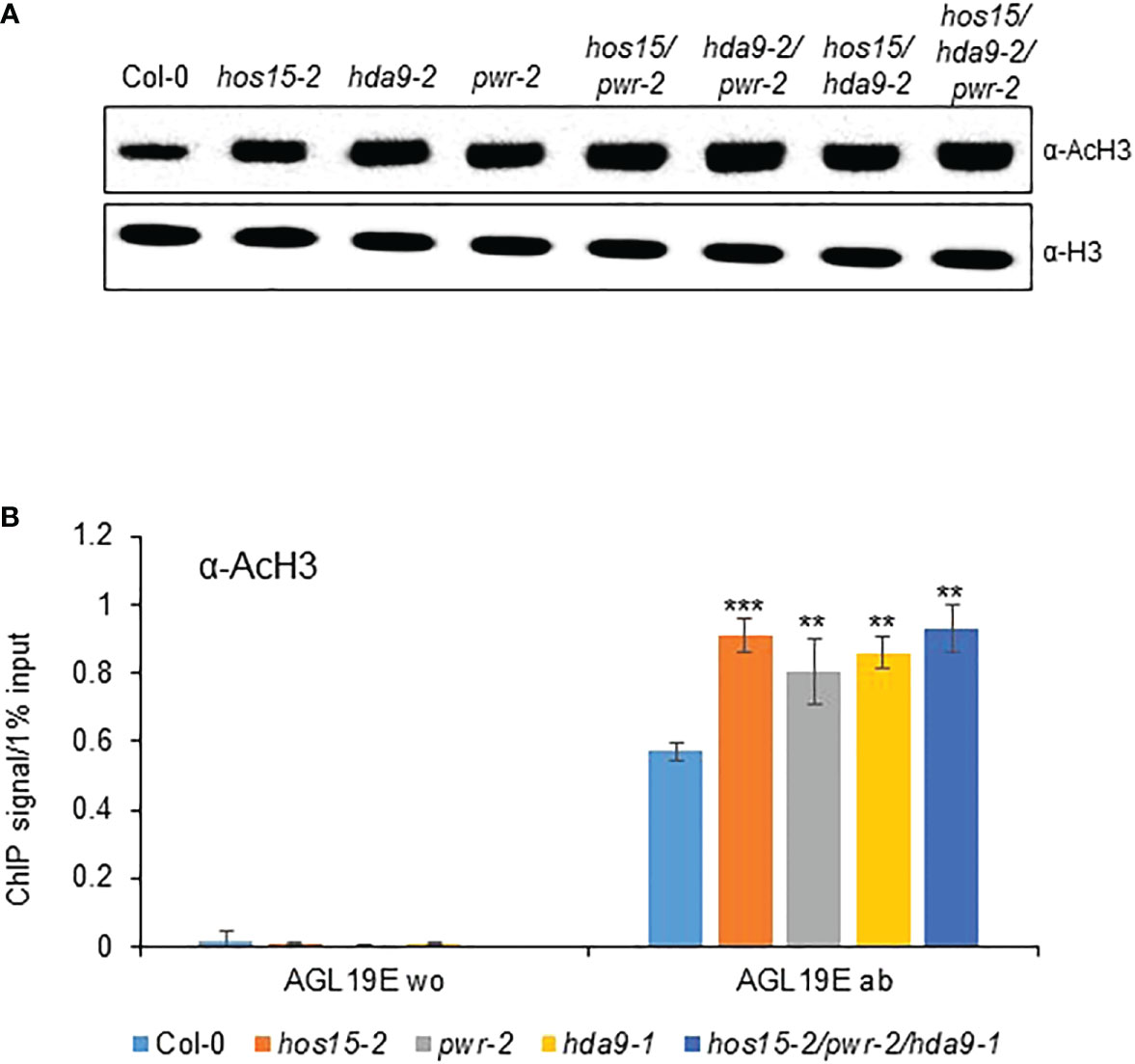
Figure 5 Regulation of AGL19 histone acetylation by PWR-HOS15-HDA9 complex. (A) Total proteins were extracted from two-week-old Col-0, hos15-2, hda9, pwr, as well as double and triple mutant plants and immunoblotted with anti-AcH3 antibody. Antibodies against histone H3 were used as the loading control. (B) Chromatin complexes from Col-0, hos15, pwr-2, hda9-1, and hos15-2 pwr-2 hda9-1 triple mutant plants were immunoprecipitated with anti-AcH3 (ab). A control reaction was processed in parallel with rabbit IgG only (w/o). ChIP and input-DNA samples were quantified by real-time qPCR using primers specific to the different regions (A–E) of the AGL19 promoter region. Data represent means ± SD from three biological replicates with three technical repeats each (n = 3). Significant difference was determined by a Student’s t-test (**p<0.01, ***p<0.01).
Epigenetic regulation plays an essential role in various signaling pathways in plants. Our study revealed the role of the HOS15-PWR-HDA9 co-repressor complex in the epigenetic regulation of flowering time through the expression of AGL19, a flowering-regulating gene (Kim et al., 2013; Kang et al., 2015). Although the regulatory pathways leading to flowering in Arabidopsis have been well documented, there is still room for further investigation to uncover the complete mechanisms of floral transitions. Using genetic interactions, proteomics, and chromatin immunoprecipitation assays, we found that the HOS15-PWR-HDA9 co-repressor complex inhibits AGL19 expression through histone modification of AGL19 chromatin thereby negatively regulating flowering time in Arabidopsis (Figure 6). Loss-of-function single, double, or triple mutants of the HOS15-HDA9-PWR complex resulted in early flowering phenotypes (Figure 1). Furthermore, the expression of AGL19 was strongly upregulated in hos15, hda9 and pwr mutants, which highlights the involvement of HOS15-HDA9-PWR complex in floral transition. The diurnal expression pattern of AGL19 was altered in the triple and the double mutants hos15 hda9 and pwr hda9 with a decline at ZT12 (Figure 1). The reason for this behavior is unknown, but the HOS15/PWR/HDA9 complex is likely to impact the expression of other circadian and photoperiodic genes whose altered expression may, in turn, impinge on the diurnal rhythm of AGL19 transcripts enhancing the oscillations (Brambilla and Fornara, 2017).
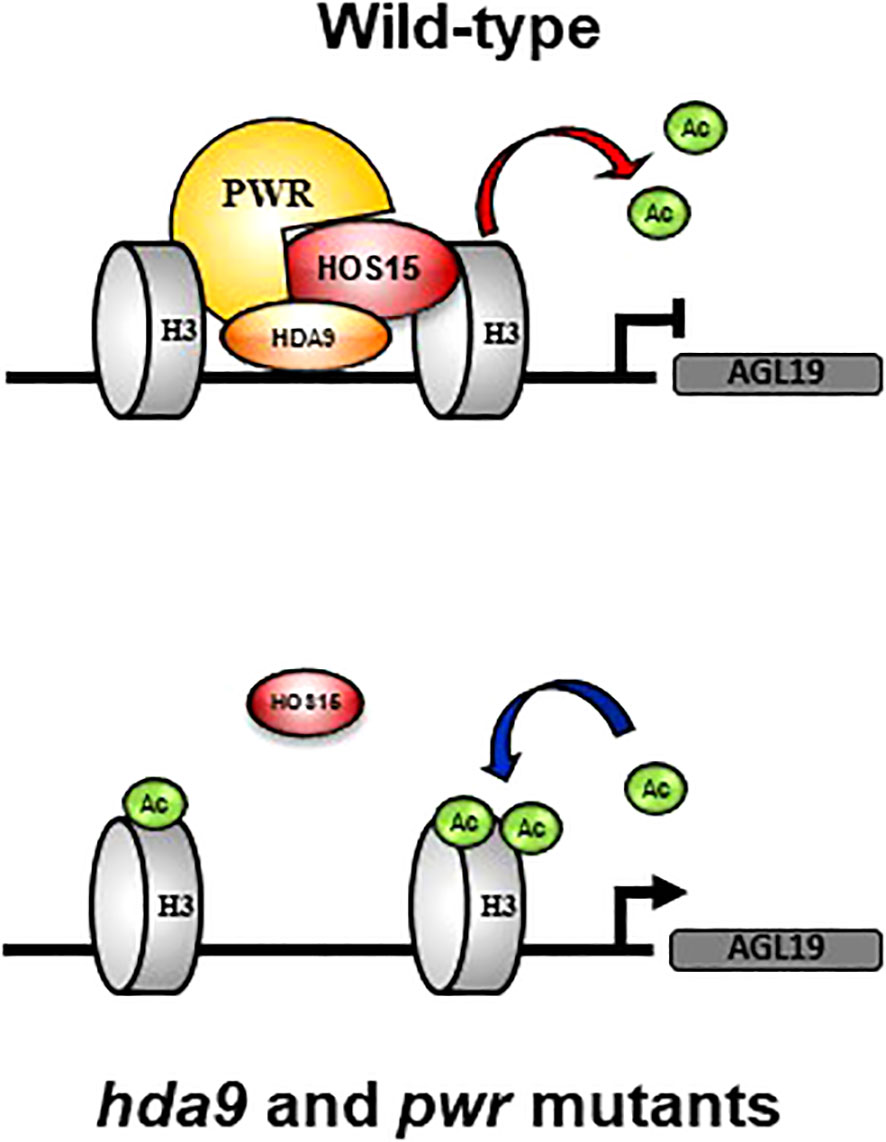
Figure 6 Working model for HOS15-PWR-HDA9 complex in flowering. Under natural conditions (wild-type), the HOS15-PWR-HDA9 complex negatively regulates AGL19 gene expression through histone deacetylation. In the absence of PWR and HDA9, which interact with HOS15 to form a complex, the accumulation of HOS15 in the nucleus is reduced. Disruption of the HOS15-PWR-HDA9 repressor complex results in enhanced histone acetylation and relaxed chromatin. AGL19 gene expression increases compared to that in the wild type, and flowering is promoted.
Previously we have shown that HDA9 and HOS15 regulate flowering time by interfering with GI transcription (Park et al., 2019). We have also shown that PWR is required for HOS15-mediated HD2C degradation upon cold stress (Lim et al., 2020). These findings suggest that HDA9 and PWR are required for functional HOS15. To confirm this, we analyzed HOS15 accumulation in hda9 and pwr mutants both in nucleus and cytosol. As shown in Figure 3, HOS15 accumulation in the nucleus was greatly reduced in hda9 and pwr mutants compared with WT. By contrast, HOS15 accumulation in the cytosol was almost similar among all tested lines (Figure 3), suggesting that HDA9 and PWR are required for nuclear accumulation of HOS15. Furthermore, HOS15 interaction with H3 was impaired in hda9 and pwr mutants (Figure 4) which further strengthened the notion that functional HDA9 and PWR are critical for HOS15’s nuclear activity.
Recent reports have shown that HOS15-HDA9-PWR is a co-repressor complex which modulates histone acetylation status thereby regulating physiological processes including senescence (Mayer et al., 2019; Zareen et al., 2022). Consistent with previous report, acetylated H3 was accumulated abundantly in hos15, hda9 and pwr single, double, and triple mutants. In addition to this, we have also found that HOS15-HDA9-PWR complex regulates flowering time through regulation of AGL19 chromatin (Figure 5). Using chromatin immunoprecipitation assay, we found that AGL19 chromatin was hyperacetylated in loss of function mutants of HOS15-HDA9-PWR complex (Figure 5), highlighting the importance of this complex in floral transition via AGL19 regulation. In the WT plants, the HOS15-PWR-HDA9 co-repressor complex negatively regulated the expression of AGL19 by promoting histone deacetylation at the AGL19 promoter, which compacts chromatin. In the absence of PWR and HDA9, the HOS15 protein was unstable in the nucleus. The removal of either one of the components of the repressor complex prevented histone deacetylation, thereby relaxing AGL19 chromatin, the increase in the expression of AGL19 and accelerating the flowering time (Figure 6). This study revealed that HOS15 together with HDA9 and PWR control flowering time by repressing AGL19 through increased histone deacetylation.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
CL, KP, AA, and D-JY conceived and designed the research. D-JY supervised the experiments. CL, KP, MJ, and SR performed the experiments. MS, JP, HK, ZB, SZ, MB, JC and Z-YX provided technical assistance and advice to CL and KP CJL, KSP, AA, JMP, and D-JY wrote the manuscrip. All authors contributed to the article and approved the submitted version.
This paper was supported by Konkuk University in 2022.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1105988/full#supplementary-material
Alaskhar Alhamwe, B., Khalaila, R., Wolf, J., von Bulow, V., Harb, H., Alhamdan, F., et al. (2018). Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 14, 39. doi: 10.1186/s13223-018-0259-4
Ali, A., Kim, J. K., Jan, M., Khan, HA, Khan, IU, Shen, M, et al. (2019). Rheostatic control of ABA signaling through HOS15-mediated OST1 degradation. Mol. Plant 12, 1447–1462. doi: 10.1016/j.molp.2019.08.005
Ali, A., Yun, D. J. (2020). Arabidopsis HOS15 is a multifunctional protein that negatively regulate ABA-signaling and drought stress. Plant Biotechnol. Rep. 14, 163–167. doi: 10.1007/s11816-020-00600-1
Alvarez-Buylla, E. R., Liljegren, S. J., Pelaz, S., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. doi: 10.1046/j.1365-313x.2000.00891.x
Amasino, R. M. (2005). Vernalization and flowering time. Curr. Opin. Biotechnol. 16, 154–158. doi: 10.1016/j.copbio.2005.02.004
Brambilla, V., Fornara, F. (2017). Y flowering? regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim. Biophys. Acta 1860, 655–660. doi: 10.1016/j.bbagrm.2016.10.009
Chen, X., Lu, L., Mayer, K. S., Scalf, M, Qian, S, Lomax, A, et al. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in arabidopsis. Elife 5, e17214. doi: 10.7554/eLife.17214
Chinnusamy, V., Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Choi, H.-K., Choi, K.-C., Kang, H.-B., Kim, HC, Lee, YH, Haam, S, et al. (2008). Function of multiple lis-homology domain/WD-40 repeat-containing proteins in feed-forward transcriptional repression by silencing mediator for retinoic and thyroid receptor/nuclear receptor corepressor complexes. Mol. Endocrinol. 22, 1093–1104. doi: 10.1210/me.2007-0396
Corbesier, L., Coupland, G. (2006). The quest for florigen: a review of recent progress. J. Exp. Bot. 57, 3395–3403. doi: 10.1093/jxb/erl095
Folta, K. M., Kaufman, L. S. (2006). Isolation of arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat. Protoc. 1, 3094–3100. doi: 10.1038/nprot.2006.471
Hollender, C., Liu, Z. (2008). Histone deacetylase genes in arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Jarillo, J. A., Del Olmo, I., Gómez-Zambrano, A., Lázaro, A., López-González, L., Miguel, E., et al. (2008). Review. photoperiodic control of flowering time. Spanish J. Agric. Res. 6, 221–244. doi: 10.5424/sjar/200806S1-391
Jiang, W., Wei, D., Zhou, W., Wang, Z, Tang, Q. (2018). HDA9 interacts with the promoters of SOC1 and AGL24 involved in flowering time control in brassica juncea. Biochem. Biophys. Res. Commun. 499, 519–523. doi: 10.1016/j.bbrc.2018.03.180
Joachimiak, E., Waclawek, E., Niziolek, M., Osinka, A., Fabczak, H., Gaertig, J., et al. (2020). The LisH domain-containing n-terminal fragment is important for the localization, dimerization, and stability of Katnal2 in tetrahymena. Cells 9, 1–19. doi: 10.3390/cells9020292
Kang, M. J., Jin, H. S., Noh, Y. S., Noh, B. (2015). Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in arabidopsis. New Phytol. 206, 281–294. doi: 10.1111/nph.13161
Khan, I. U., Ali, A., Khan, H. A., Baek, D, Park, J, Lim, CJ, et al. (2020). PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in arabidopsis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00623
Kim, W., Latrasse, D., Servet, C., Zhou, D. X. (2013). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398. doi: 10.1016/j.bbrc.2012.11.102
Kim, Y. J., Wang, R., Gao, L., Li, D, Xu, C, Mang, H, et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863. doi: 10.1073/pnas.1618618114
Lim, C. J., Park, J., Shen, M., Park, HJ, Cheong, MS, Park, KS, et al. (2020). The histone-modifying complex PWR/HOS15/HD2C epigenetically regulates cold tolerance1[OPEN]. Plant Physiol. 184, 1097–1111. doi: 10.1104/pp.20.00439
Mayer, K. S., Chen, X., Sanders, D., Chen, J, Jiang, J, Nguyen, P, et al. (2019). Hda9-pwr-hos15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 180, 342–355. doi: 10.1104/pp.18.01156
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Park, H. J., Baek, D., Cha, J. Y., Liao, X, Kang, SH, McClung, CR, et al. (2019). Hos15 interacts with the histone deacetylase hda9 and the evening complex to epigenetically regulate the floral activator gigantea. Plant Cell 31, 37–51. doi: 10.1105/tpc.18.00721
Park, J., Lim, C. J., Khan, IU, Jan, M, Khan, HA, Park, HJ, et al. (2018b). Identification and molecular characterization of HOS15-interacting protein in arabidopsis thaliana. J. Plant Biol. 61, 336–345. doi: 10.1007/s12374-018-0313-2
Park, J., Lim, C. J., Shen, M., Park, HJ, Cha, JY, Iniesto, E, et al. (2018a). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. U.S.A. 115 (23), E5400–E5409. doi: 10.1073/pnas.1721241115
Perissi, V., Scafoglio, C., Zhang, J., Ohgi, K. A., Rose, D. W., Glass, C. K., et al. (2008). TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol. Cell. 29, 755–766. doi: 10.1016/j.molcel.2008.01.020
Pfluger, J., Wagner, D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. doi: 10.1016/j.pbi.2007.07.013
Schönrock, N., Bouveret, R., Leroy, O., Borghi, L., Köhler, C., Gruissem, W., et al. (2006). Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20 (12), 1667–1678. doi: 10.1101/gad.377206
Shen, M., Lim, C. J., Park, J., Kim, J. E., Baek, D., Nam, J., et al. (2020). HOS15 is a transcriptional corepressor of NPR1-mediated gene activation of plant immunity. Proc. Natl. Acad. Sci. U.S.A. 117, 30805–30815. doi: 10.1073/pnas.2016049117
Shim, J. S., Kubota, A., Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in arabidopsis: constans is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., MacCoss, M. J., et al. (2014). Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Suzuki, M., Shinozuka, N., Hirakata, T., Nakata, M. T., Demura, T., Tsukaya, H., et al. (2018). Oligocellula1/high expression of osmotically responsive genes15 promotes cell proliferation with histone deacetylase9 and powerdress during leaf development in arabidopsis thaliana. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00580
Tamaru, H. (2010). Confining euchromatin/heterochromatin territory: Jumonji crosses the line. Genes Dev. 24, 1465–1478. doi: 10.1101/gad.1941010
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in arabidopsis thaliana. PloS Genet. 14 (3), e1007280. doi: 10.1371/journal.pgen.1007280
Xu, C., Min, J. (2011). Structure and function of WD40 domain proteins. Protein Cell 2, 202–214. doi: 10.1007/s13238-011-1018-1
Yumul, R. E., Kim, Y. J., Liu, X., et al. (2013). POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PloS Genet. 9 (1), e1003218. doi: 10.1371/journal.pgen.1003218
Zareen, S., Ali, A., Lim, C. J., Khan, H. A., Park, J. H., Xu, Z. Y., et al. (2022). The transcriptional corepressor HOS15 mediates dark-induced leaf senescence in arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.828264
Zhao, T., Zhan, Z., Jiang, D. (2019). Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genomics 46, 467–476. doi: 10.1016/j.jgg.2019.09.005
Keywords: Arabidopsis, flowering time, chromatin remodeling, histone acetylation and deacetylation, HOS15, HDA9, PWR, AGL19
Citation: Lim CJ, Park KS, Ali A, Park J, Ryou SM, Shen M, Khan HA, Bader ZE, Zareen S, Bae MJ, Choi JH, Xu Z-Y, Pardo JM and Yun D-J (2023) Negative regulation of floral transition in Arabidopsis by HOS15-PWR-HDA9 complex. Front. Plant Sci. 13:1105988. doi: 10.3389/fpls.2022.1105988
Received: 23 November 2022; Accepted: 16 December 2022;
Published: 06 January 2023.
Edited by:
Jungnam Cho, Center for Excellence in Molecular Plant Sciences (CAS), ChinaCopyright © 2023 Lim, Park, Ali, Park, Ryou, Shen, Khan, Bader, Zareen, Bae, Choi, Xu, Pardo and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dae-Jin Yun, ZGp5dW5Aa29ua3VrLmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.