- 1Institute of Crops and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Landscape and Architecture, Zhejiang A&F University, Hangzhou, China
- 3College of Advanced Agriculture Sciences, Zhejiang A&F University, Hangzhou, China
- 4State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
Because of labor shortages or resource scarcity, direct seeding is the preferred method for rice (Oryza sativa. L) cultivation, and it necessitates direct seeding at the current density. In this study, two density of direct seeding with high and normal density were selected to identify the genes involved in shade-avoidance syndrome. Phenotypic and gene expression analysis showed that densely direct seeding (DDS) causes a set of acclimation responses that either induce shade avoidance or toleration. When compared to normal direct seeding (NDS), plants cultivated by DDS exhibit constitutive shade-avoidance syndrome (SAS), in which the accompanying solar radiation drops rapidly from the middle leaf to the base leaf during flowering. Simulation of shade causes rapid reduction in phytochrome gene expression, changes in the expression of multiple miR156 or miR172 genes and photoperiod-related genes, all of which leads to early flowering and alterations in the plant architecture. Furthermore, DDS causes senescence by downregulating the expression of chloroplast synthesis-related genes throughout almost the entire stage. Our findings revealed that DDS is linked to SAS, which can be employed to breed density-tolerant rice varieties more easily and widely.
1. Introduction
The world’s population is expected to reach 9.3 billion by 2050, and food security is becoming an increasingly serious global issue (Warnasooriya and Brutnell, 2014). Rice is a common staple food (Chen et al., 2022). Direct seeding of rice has become popular in China during the last few decades (Farooq et al., 2011; Zhou et al., 2019). Furthermore, improving rice yields per unit land area by employing an optimal density of direct seeding is a viable option. Density tolerant rice varieties have greater vigor, allowing the possibility of direct sowing and increasing the yield per unit land area. However, densely direct seeding (DDS) can result in competition by neighboring plants, limiting rice production. Shade-avoidance syndrome (SAS) is the name given to this phenomenon in Arabidopsis thaliana. Rice plants change their type to compete with their neighbors for limited resources in order to optimize their life cycle when SAS occurs (Xie et al., 2017). Elucidating the mechanism that regulates SAS in rice can aid scientists in developing new high density-tolerant rice cultivars, making direct-seeding procedures for rice cultivation more popular as a result.
Light is an essential environmental signal for plants (Takanoa et al., 2009). Phytochromes are believed to be the primary mediators of the SAS (Xie et al., 2017). The phytochrome-PHYTOCHROME-INTERACTING FACTORS(PIFs) signaling pathway and the MIR156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) regulatory module regulates phytochrome-mediated SAS responses in Arabidopsis thaliana (Xie et al., 2017). Five PHY genes exist in Arabidopsis thaliana, i.e. PHYA to PHYE, with only PHYB playing a substantial role in SAS (Xie et al., 2017). Differently, rice has only three genes: PHYA, PHYB, and PHYC (Takanoa et al., 2009). Rice PHYA has been reported to govern photomorphogenesis in two separate modes of photoperception (R/FR), while PHYC is involved in the photoperception of FR for deetiolation, according to physiological investigations of PHYA mutants (Takano et al., 2005; Takanoa et al., 2009). Furthermore, under long-day (LD) conditions, both PHYA and PHYB single mutants are believed to lead to early flowering (Ishikawa et al., 2011; Osugi et al., 2011), suggesting that each rice phytochrome plays a unique role in regulating key daylength recognition gene expression. In plants, miR156 works as a master regulator for a variety of biological processes, including SAS (Huijser and Schmid, 2011). The 11 miR156 genes (pre-miR156a to pre-miR156l) are found in the rice genome, and are involved in regulating critical processes such as seed dormancy, seed germination, plant shoot architecture, and grain size (Miao et al., 2019). During vegetative development in plants, miR156 and miR172 act as antagonistic agents (Chuck et al., 2007; Wang et al., 2009; Wu et al., 2009). The miR156-SPL and miR172-APETALA2 (AP2) modules play a crucial role in flowering as critical regulators of the time of the vegetative to reproductive phase transition (Wang et al., 2009; Wu et al., 2009; Cui, 2020).
SAS has been shown to cause early flowering in previous research (Casal, 2012). Rice is a common short day (SD) crop (Chen et al., 2022). It has evolved two flowering pathways: (the Hd1, Ghd7, DTH8, OsPRR37)-Ehd1-Hd3a/RFT1 long day (LD) flowering suppression pathway and the OsGI-Hd1-Hd3a/RFT1 SD flowering-promotion pathway (Chen et al., 2022). The downstream core flowering regulatory genes Hd3a and RICE FLOWERING LOCUS T1 (RFT1) are two florigen genes (Zhao et al., 2015; Chen et al., 2022). In both LD and SD, Ehd1 encodes a B-type response regulator that promotes heading by upregulating the expression of Hd3a and RFT1 (Zhao et al., 2015; Cho et al., 2016). RID1/Ehd2/OsID1, SE5, OsMADS51 and other upstream genes positively regulate Ehd1 (Kim et al., 2007; Wu et al., 2008; Andres et al., 2009), whereas Heading Date 1 (Hd1), Ghd7, COL4, and DTH8/Ghd8/Hd5 negatively regulate it (Yano et al., 2000; Xue et al., 2008; Lee et al., 2010; Wei et al., 2010; Chen et al., 2022). Hd1 has a dual function that suppresses heading in LD and promotes it in SD (Yano et al., 2000). The status of other essential flowering regulatory genes Ghd7, Hd2, and DTH8 are required for Hd1 to reduce heading in LD (Zhang et al., 2017; Zong et al., 2021). SE5 is involved in the biosynthesis of phytochrome chromophore (Andres et al., 2009). By affecting both the Hd1 and Ehd1 flowering pathways, the se5 mutant exhibited early heading under both SD and LD conditions. The AP2 genes, which contain the miR172 target site, act as Ehd1 repressors (Lee and An, 2012; Lee et al., 2014).
The mechanism of SAS in plants is conservative. Significant progress has been made in understanding shade avoidance during the last decade (Casal, 2012; Ciolfi et al., 2013; Xie et al., 2017; Liu et al., 2019; Paulisic et al., 2021). Rice is important for agriculture and the economy. Direct-seeding must be developed in order to increase grain yield while reducing labor and resource costs. We investigated the shade-avoidance response at various times in this study using gene expression under normal direct seeding (NDS) and densely direct seeding (DDS) conditions. Dynamic expression of the genes involved in the phy-miR156s signaling pathway and the photoperiodic flowering pathway were also investigated in this study because miR156s mediates phytochrome-mediated SAS responses. Furthermore, we also studied the dynamic expression of chlorophyll metabolism. Based on these findings, we argue that altering the phy-miR156s signaling pathway is an approach to adapt direct-seeding to a high-density environment.
2. Materials and methods
2.1. Plant materials and growth conditions
The rice cultivars Nipponbare and four mutants (i.e., phyb、OEmiR156b/c、ipa1-2D、Ostb1) were chosen for this study. Seed dormancy was broken by exposing the seeds to 45°C for 3 days, followed by pre-germination then sowing the seedlings in a nursery in a 2 m by 10 m plot. DDS was implemented by planting two plants with no more than a distance of 1 cm between each plant in a row and 1 cm between the rows. NDS was implemented by planting two plants with a distance of 20 cm between each plant in a row and 20 cm between the rows. Both plots received the same fertilizer management.
2.2. Agronomic trait measurements
Once the plants achieved maturity, the primary agronomic traits (i.e., plant height, tiller, leaf length, leaf breadth, spikelet length, the number of primary branches, and the seed-setting rate) were ascertained. Data from three biological replicates were compiled for each trait. For each attribute, data from three biological replicates was compiled.
2.3. Small RNA blot analysis
Total RNA was isolated from rice leaves using the TRIzol reagent (Invitrogen); 10 µg of total RNA were fractionated on 17% polyacrylamide gels under denaturing conditions (i.e., 7 M urea). Blots were hybridized using digoxigenin end-labeled LNA oligonucleotides (Exiqon, Vedbaek, Denmark) complementary to the miR156 or miR172 sequences. The assay was conducted as previously described (Guo et al., 2016). The following DNA oligonucleotides were used: miR156 probe:5’-GTGCTCACTCTCTTCTGTCA-3’, miR172 probe: 5’-GCAGCACCATCAAGATTCAC - 3’, U6 probe: 5’- TGTATCGTTCCAATTTTATCGGATGT - 3’.
2.4. RNA Extraction and RT-PCR
Total RNA was extracted from rice leaves using the TRIzol reagent (Invitrogen), and cDNA was made using 1 µL of RNA and ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO), as directed by the manufacturer. Each cDNA replicate was subjected to qPCR, and all samples were processed in duplicate. A Power SYBR Green PCR Master Mix kit was used for the quantitative real-time PCR investigations (Applied Biosystems). PCR conditions were 95 °C for 10 minutes, 40 cycles of 95 °C for 15 seconds, 60 °C for 1 minute, and a final melt curve step from 60 °C to 95 °C. The average expression ratios were obtained from the equation 2-△△CT, where △△CT represents △CT (gene of interest in stage evaluated)-△CT (gene of interest at control stage), according to the protocol reported by Cui (Cui et.al., 2020).
2.5. Radiation intensity measurement
At noontime, a spectrograph (Spectrometer MAYA 2000, 830 Douglas Ave., Dunedin, FL, USA) was used to measure the absolute visible light radiation intensity at the flag leaf, the middle leaf and the base leaf from NDS and DDS plants during the flowering period. Three biological replicates were used.
2.6. Chlorophyll content measurement
The content of chlorophyll was determined using Xu’s technique (Xu et al., 2017). Cut leaves (0.2 g fresh weight) were soaked in 10 mL ethanol for 48 h in the dark. The remaining plant debris was removed by centrifugation. A UV-VIS spectrophotometer (Shimadzu, UV-2600, Japan) was used to analyze the supernatants at 663, 645, and 470 nm, respectively.
2.7. Statistical analysis
Based on three biological replicates, the data were expressed as the mean values with the standard deviation (SD). Student’s unpaired t test and Duncan’s multiple range test were used to determine statistical significance. Probability values of 5% were considered statistically significant; a single asterisk (*) and a double asterisk (**) denote statistical significance at the levels of 5% and 1%, respectively. The methodologies used in this study for the statistical analysis of the gene relative expression levels and the agronomic traits are all described above.
3. Results
3.1. Phenotypic and agronomic traits analysis
Shade-avoidance responses have previously been shown to include increased leaf angles to the horizontal, reduced branching, reduced leaf blade area, and early flowering (Casal, 2012). DDS, on the other hand, has consistently shown shade-avoidance traits throughout the life of the plant. Tiller number, plant height, flag leaf length and width, panicle length, grain number per panicle, seed-setting rate, and primary and secondary branch length and width were all measured. DDS plants have no or very little tiller during their lives (approximately only one tiller in total) (Figures 1A, D). In comparison to NDS plants, DDS plants had just 64% flag leaf length and 69% flag leaf width on average (Figures 1B-D). Furthermore, DDS plants exhibited early flowering and a short panicle length (~22% shorter than NDS), resulting in a significant reduction in grain number per panicle (~70% of NDS), primary branch (~92% of NDS), and secondary branch (~47% percent of NDS). However, the agronomic parameters (i.e., plant height and the seed-setting rate) did not differ substantially between DDS and NDS plants (Figure 1) (Figures 1B-D). Additionally, DDS plants displayed senescence. These observations indicate that DDS plants exhibited typical shade-avoidance traits in both the vegetative and adult stages.
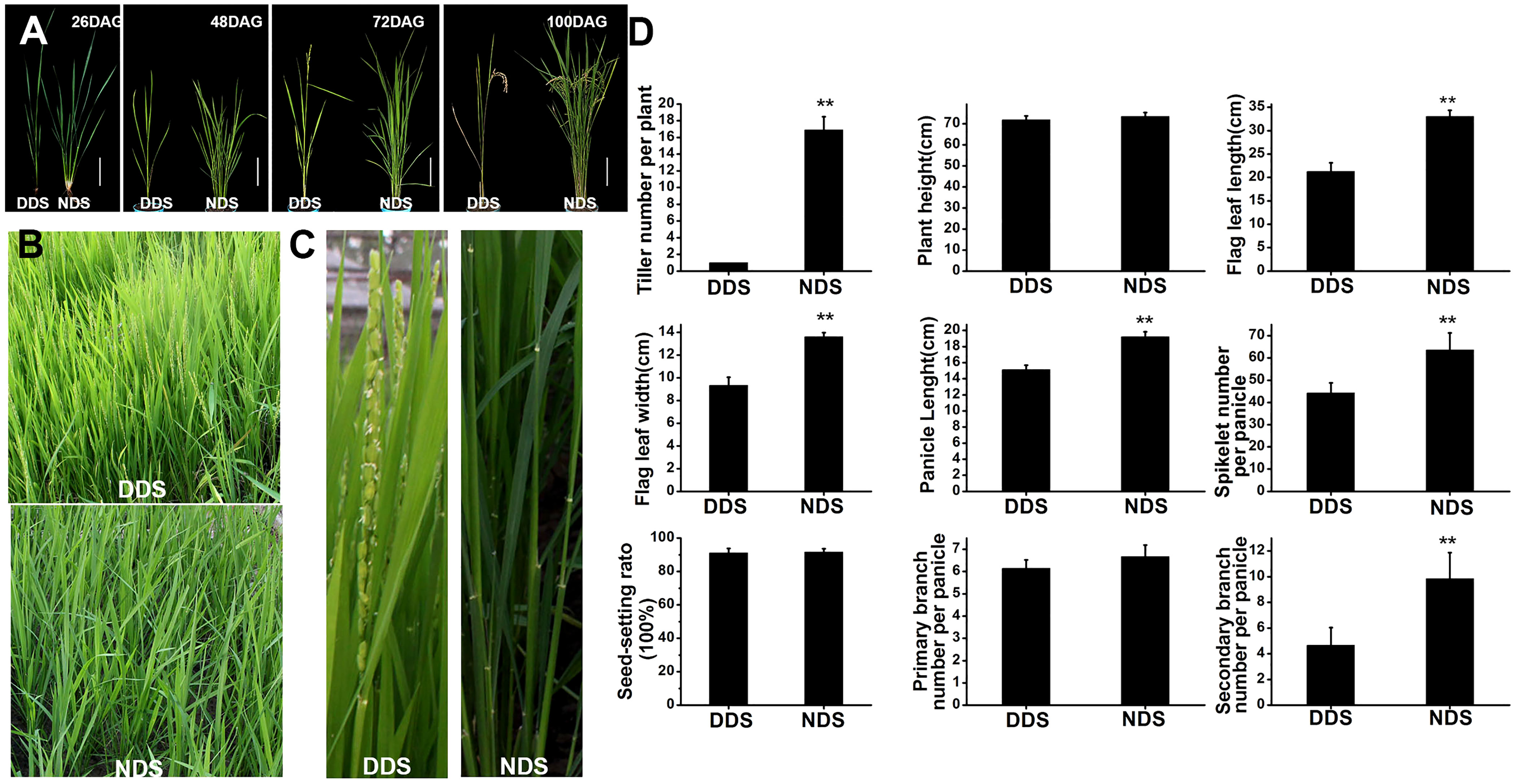
Figure 1 DDS plants exhibit constitutive SAS. (A) Comparison of the tiller at different stages under DDS or NDS conditions. 26, 48, 72, 100 DAG treatment were photographed. Bar = 15 cm. (B, C) Earlier heading dates and yellow leaves were observed in DDS at 72 DAG. (D) Agronomic traits in these plants. Data represent the mean ± SD of three biological replicates (Student’s t test: **P ≤ 0.01).
3.2. Absolute visible light radiation intensity declined under DDS
DDS permits plants to compete for limited resources with their neighbors (Ciolfi et al., 2013). We measured the radiation intensity at various leaves to determine the absolute visible light radiation intensity (i.e., the flag leaf, middle leaf and the base leaf). The radiation intensity at the flag leaf did not differ significantly between the DDS and the NDS plants at the flowering stage (Figure 2), however, the radiation intensity at the intermediate leaf and the base leaf was dramatically reduced in the DDS plants (Figure 2B). These results suggest that the normal R:FR ratio conditions have been altered, which simulated shade under DDS conditions.
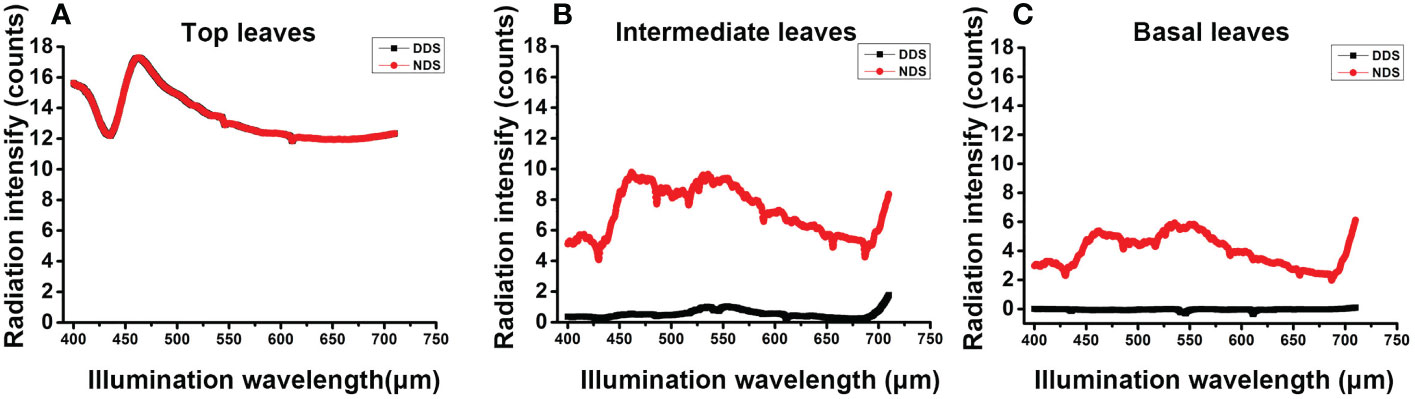
Figure 2 Radiation intensity declined under DDS. (A) The radiation intensity showed no difference at the top leaves at 72 DAG. (B) The radiation intensity was reduced in the DDS plants in intermediate leaves at 72 DAG. (C) The radiation intensity was reduced in the DDS basal leaves at 72 DAG. Data were measure for at least three biological replicates.
3.3. The transcript levels of PHYA, PHYB, PHYC decreased under DDS conditions
Phytochromes, which mediate SAS in plants, primarily respond to red (R)/far-red light (FR). We discovered that at 40, 48, 56, and 64 DAG under DDS conditions, the expression of the rice phytochrome genes PHYA, PHYB, and PHYC was down-regulated, indicating that the DDS plants had high overshadow. Only few differences were identified at 24 and 32 DAG, when both DDS and NDS plants were in the juvenile-to-adult stage (Figure 3). The DDS plants became a little flattened around 100 DAG, which slowed down the neighbors’ competition. We thus concluded that red (R)/far-red light (FR) of PHYA/B/C were largely weakened under DDS conditions.
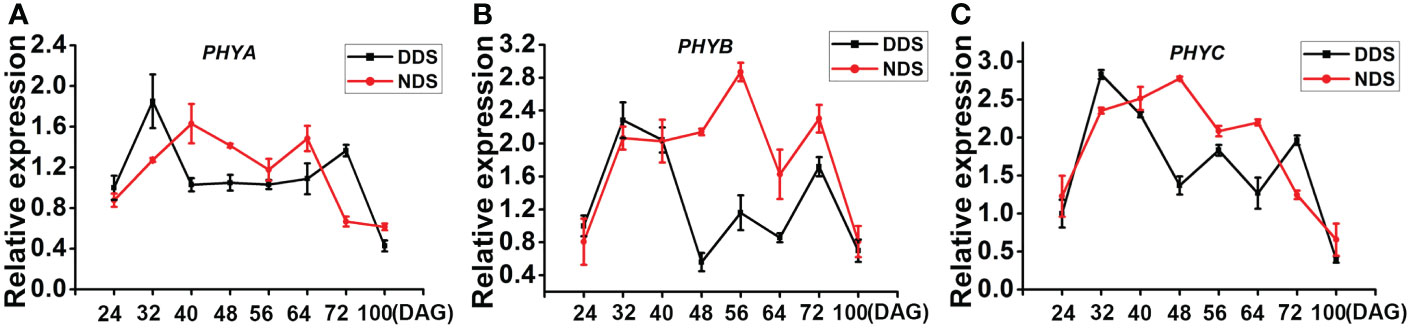
Figure 3 The transcript levels of PHYA, PHYB, PHYC decreased under DDS conditions. (A-C) The qRT-PCR analysis of temporal expression in the whole plants PHYA, PHYB, PHYC between DDS and NDS at 24、32、40、56、64、72、100 DAG. Error bars represent mean ± SD (n = 3) where the different letters indicate statistical differences according to Duncan’s multiple range test (p ≤ 0.05).
3.4. The transcript levels of miR156s and miR172s increase under DDS
In plants, miR156 is a conserved regulator of developmental time. Overexpression of rice miR156B/C resulted in more tillers, shorter and narrower leaves, a longer duration of the expression of juvenile vegetative features, and later flowering (Cui 2020). We looked at the expression of leaves under both conditions to see if miR156s played a role in the response to DDS. We employed a northern blot to detect the level under both conditions to measure the total level of miR156s in vivo and evaluate the dynamic expression of miR156s. At 24 DAG, 32 DAG, 40 DAG, 48 DAG, 56 DAG, 64 DAG, 72 DAG, and 100 DAG, RNA samples were collected from the leaves of entire plants. Under DDS conditions, we saw a higher concentration of miR156 transcripts throughout the stage than under NDS conditions (Figure 4). On the other hand, SAS caused miR156 to be downregulated in Arabidopsis. Th miR156 transcript level dropped at first and later increased under both conditions. A quantitative reverse transcriptase RT-PCR experiment showed that the primary transcripts of miR156D and miR156H were clearly enhanced under DDS conditions (Figure 4B). During vegetative development in plants, miR156 and miR172 act as antagonistic agents (Chuck et al., 2007; Wang et al., 2009; Wu et al., 2009), so we looked at the levels of miR172s. In this study, the expression of miR172s steadily decreased in the DDS plants during shoot growth and maturation, while it gradually declined and then sharply decreased in the NDS plants at 100DAG (Figure 4A). DDS plants produced 10.2 leaves on average during their entire life cycle, 3-4 leaves less than the NDS plants (Figure 4). When compared to NDS, DDS heading dates were 4 days earlier. These findings reveal that under DDS conditions, both miR156 and miR172 were misregulated, resulting in early flowering, reduced organ size, and narrow leaves, and the DDS plants were more sensitive compared with the NDS plants.
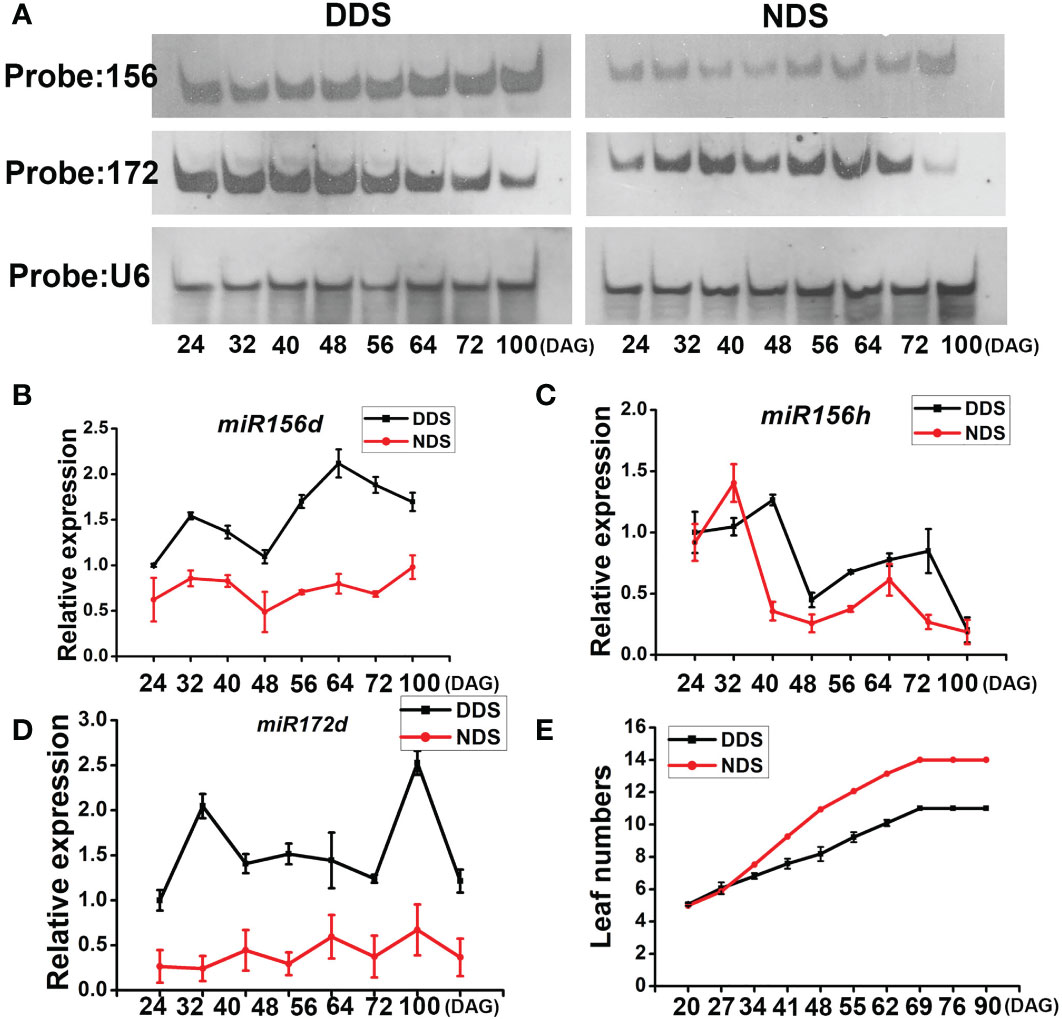
Figure 4 Ectopic expression of miR156s and miR172s in DDS plants. (A) Northern blot of miR156s and miR172s expression levels in whole plants at either the adult or juvenile stages between DDS and NDS. The RNA samples in each lane were extracted from whole plants. U6 was used as a loading control. (B, C) A Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of pri-miR156d/h expression between DDS and NDS at 24、32、40、56、64、72、100 DAG. (D) A qRT-PCR analysis of temporal expression of pri-miR172d between DDS and NDS at 24、32、40、56、64、72、100 DAG. (E) A shorter plastochron and fewer leaves were observed for DDS compared to NDS. Error bars represent mean ± SD (n = 3) where the different letters indicate statistical differences by Duncan’s multiple range test (p<0.05).
3.5. DDS show early flowering
Rice is a typical SD plant with a strong connection to phytochromes. Compared to NDS, the DDS heading dates were 4 days ahead (Figure 5). Since the expression of PHYA, PHYB, and PHYC decreases after 40 DAG, this study focused on the flowering regulatory genes to study phytochrome-related responses. Under DDS conditions, the expression of SE5 decreased from 48 DAG to 64 DAG (Figure 5). The downstream genes in the flowering pathway, RFT1 and Hd3a, showed identical expression patterns under both LD and SD conditions. In this study, RFT1 and Hd3a expression remained low for LD, but was dramatically up-regulated for SD under both DDS and NDS conditions (Figure 5C). In the LD state, however, RFT1 expression was higher for DDS than NDS (Figure 5). Despite the fact that Hd3a expression did not differ considerably, there was a modest upregulation at 48 DAG (Figure 5). Early heading date 1 (Ehd1) integrates many upstream regulatory signals to control the expression of the two florigen genes Hd3a and RFT1. For DDS, Ehd1 showed much higher expression than NDS at 24 and 32 DAG, but only small differences at subsequent stages (Figure 5). Heading date 1 (Hd1) is a key gene in rice for flowering regulation; both DDS and NDS showed comparable tendencies throughout the stage, with the exception of overexpression at 24 and 32 DAG (Figure 5). In LD, Ghd7 acts as a heading repressor. Ghd7 showed an opposite expression trend after 40 DAG (Figure 5). Hd5 expression was equivalent for DDS and NDS, but higher for DDS at 32-64 DAG (Figure 5). SNB and IDS1 are two AP2 transcription factors that function downstream of miR172. At 32 to 64 DAG, both SNB and IDS1 expression were increased (Figure 5I). These observations indicate that a gradual alteration of the expression of miR156 and miR172 results in changes in developmental phase transitions, leading early flowering under DDS conditions.
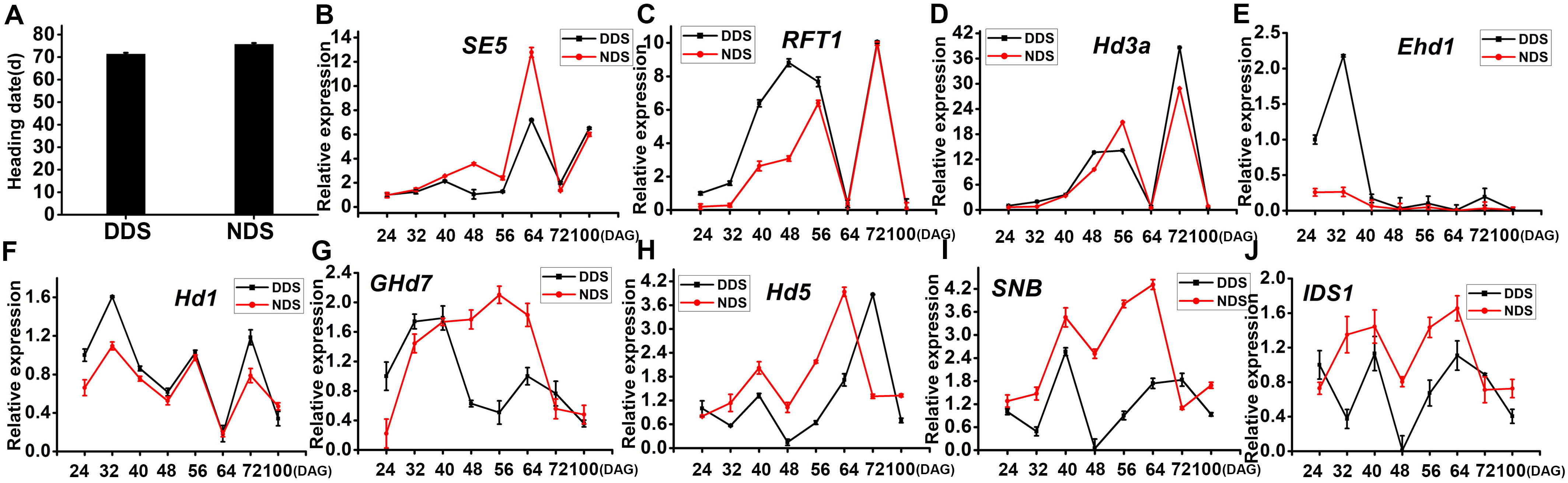
Figure 5 DDS showed early flowering and HD related genes. (A) The heading dates of NDS plants were delayed by 4 d, compared with the DDS. (B-J) qRT-PCR analysis of temporal expression in the whole plant of SE5, RFT1, Hd3a, Ehd1, Hd1, Ghd7, Hd5, SNB, IDS1 between DDS and NDS at 24、32、40、56、64、72、100 DAG. Error bars represent mean ± SD (n = 3) where the different letters indicate statistical differences by Duncan’s multiple range test (p ≤ 0.05).
3.6. Relative expression of genes related to chloroplast development in DDS
Rice exhibited early flowering and senescence under DDS conditions. Chlorophyll metabolism and chloroplast formation are linked to senescence, so we looked at the transcript abundance of genes involved in chlorophyll synthesis and chloroplast development for both DDS and NDS. Initially, we determined the levels of the Chl a, Chl b, and carotenoid (Car) pigments for DDS and NDS flag leaves. We discovered that the content of Chl a and Chl b decreased dramatically (by 24% and 41%, respectively, compared to NDS) (Figure 6). Except for OsCAO, genes involved in chlorophyll synthesis (i.e., LchP2, OsCHLH, OsDVR, OsHEMA, OsPORA, OsPORB, and OsYGL) were considerably down-regulated under DDS compared to NDS conditions (Figures 6B–I). These results suggest that chloroplast biogenesis and development were seriously affected by DDS.
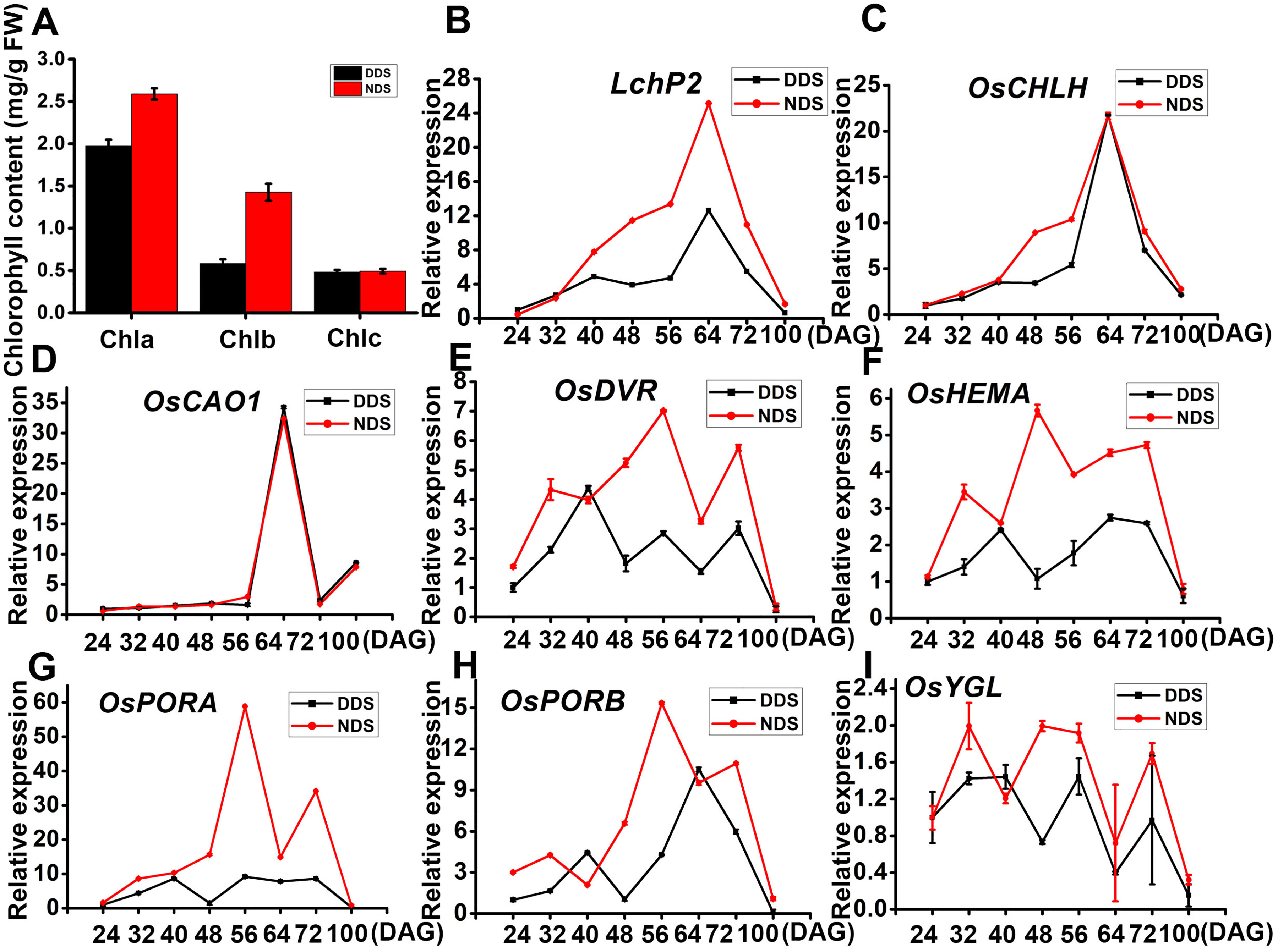
Figure 6 Relative expression of genes related to chloroplast development in leaves from DDS and NDS. (A) Content of photosynthetic pigments in flag leaves from DDS and NDS plants. (B-I) A qRT-PCR analysis of temporal expression in the whole plant with genes associated with chlorophyll synthesis (i.e., LchP2, OsCHLH, OsCAO1,OsDVR, OsHEMA, OsPORA, OsPORB, and OsYGL) between DDS and NDS plants at 24、32、40、56、64、72、100 DAG. Error bars represent mean ± SD (n = 3) where the different letters indicate statistical differences according to Duncan’s multiple range test (p ≤ 0.05).
3.7. The expression of several SPLs were repressed in DDS plants
SPL genes are regulated by miR156 and play a role in a variety of developmental processes. In Arabidopsis thaliana, active SPL genes control various morphological changes linked to shade avoidance responses. In DDS plants at 70 DAG, the expression of OsSPL3, OsSPL13, and OsSPL14 decreased (Figures 7A–C). Comparing DDS to NDS, the potential yield was lower for DDS plants (Figure 7). In Arabidopsis, a link between gibberellin-mediated and miR156/SPL module-mediated flowering pathways has been established (Yu et al., 2012).In order to check whether DELLA protein SLR1(slender rice 1) was regulated by miR156/SPL genes, we analyzed SLR. The expression of SLR was decreased from 32 to 56 DAG (Figure 8). Taken together, these results suggest a link between of OsSPLs and SLR in DDS leading to fewer tillers.
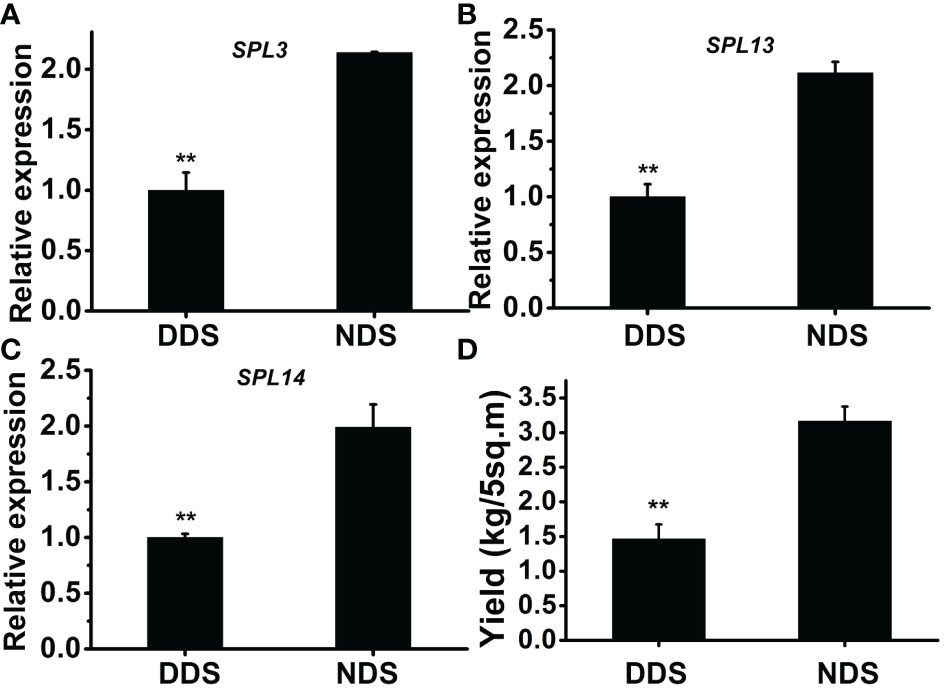
Figure 7 The theoretical yield was reduced in DDS plants. (A-C) A qRT-PCR analysis of temporal expression in the whole plant containing SPL3, SPL13, SPL14 genes. (D) The theoretical yields were estimated. Data represent the mean ± SD of three biological replicates (Student’s t test: **P ≤ 0.01).
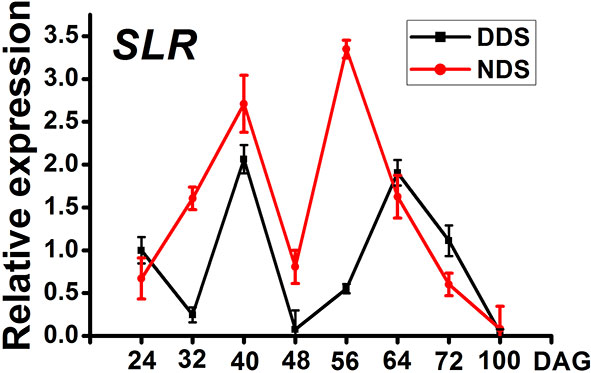
Figure 8 A qRT-PCR analysis of the temporal expression in the whole plants containing SLR between DDS and NDS at 24、32、40、56、64、72、100 DAG. Error bars represent mean ± SD (n = 3) where the different letters indicate statistical differences by Duncan’s multiple range test (p ≤ 0.05).
4. Discussion
The phytochrome-miR156 pathway regulates plant responses to SAS, allowing for rapid reconfiguration of the plant body while avoiding an unfavorable reaction to low R/FR (Xie et al., 2017). As a result, it has been widely assumed that plants have evolved the SAS during crop domestication and genetic improvement. Deciphering the mechanism that modulates SAS in DDS would thus aid scientists in the development of new density-tolerant cultivars for improved agronomic performance.
DDS causes a plant architecture phenotype in Arabidopsis similar to SAS (Xie et al., 2017). The majority of thorough SAS investigations have been in Arabidopsis thaliana (Casal, 2012; Xie et al., 2017; Liu et al., 2019; Paulisic et al., 2021). In response to simulated shade, a collection of signaling integrators between light and other signals has been shown. During SAS, inactivated phyB causes a rapid buildup of PIF proteins that bind directly to the promoters of numerous miR156 genes, preventing the miR156s from inhibiting their target SPL genes. In this study, the dynamic expression of genes involved in the phytochromes-miR156s/miR172s or other signaling pathways were investigated using DDS. Fewer tillers, decreases in leaf length, width, grain number per panicle, and early flowering are all classic shade-avoidance responses induced by high density. These responses may have a favorable or negative impact on rice yield. The lower sunlight intensity in middle and base leaves under DDS conditions repressed the expression of rice phytochrome genes PHYA, PHYB, and PHYC, particularly PHYB. Phytochrome genes are thought to be entirely responsible for the perception of red and far-red light. The genes miR156 and miR172 are antagonistic toward vegetative development in plants. SAS inhibited the expression of miR156s in Arabidopsis. On the other hand, miR156s/miR172s were highly expressed under DDS conditions, mimicking the typical phenotype of OEmiR156 (i.e., short and narrow leaves) (Cui et al., 2020). The difference in the expression of miR156s/miR172s between Arabidopsis and rice may due to treatment differences. In Arabidopsis, only the EOD-FR treatment was adopted to mimic shady conditions (Xie et al., 2017). However, under DDS conditions, rice plants might compete with their neighbors not only for light but also for the space necessary to complete their life cycle. Our findings suggest that DDS may have other effects in addition to SAS. The gene miR156 regulates a variety of morphological changes linked with shade-avoidance responses by acting as a master upstream regulator of numerous genes such as SPLs. The decrease of yield is caused by the downregulation of SPL3, SPL13, and SPL14 under DDS conditions, which causes a variety of morphological abnormalities. In Arabidopsis, a link between gibberellin-mediated and miR156/SPL module-mediated flowering pathways has been established (Yu et al., 2012). DELLAs interact with multiple flowering activators and repressors, resulting in late flowering phenotypes (Feng et al., 2008; Yu et al., 2012). Unlike Arabidopsis, which contains five DELLA genes, rice has only one, i.e. SLR1. Similar to the situation in Arabidopsis, SLR is also involved in the Ehd1-Hd3a/RFT1 flowering pathway, resulting in late flowering (Zhang et al., 2022). Furthermore, the rice DELLA protein SLR1 interacts with MOC1 to regulate the tiller number (Liao et al., 2019). In this study, the decreased expression of SLR under DDS implies that GA is also involved in miR156/SPL module-mediated flowering pathways (Figure 8).
In rice, miR172 acts as a significant regulator of developmental changes. Plants with high levels of miR172d have been found to activate the flowering promotion genes Ehd1 and Hd3a. The expression of Hd3a has been found to be briefly up-regulated at 48 DAG, when developmental changes associated with DDS occur, resulting in flowering four days early. The up-regulation of miR172s under DDS conditions causes the expression of SNB and IDS1 to drop for practically the entire experimental duration. Furthermore, when compared to NDS, SE5, GHD7, and HD5 all showed lower expression under DDS conditions at 48 to 56 DAG, indicating that loss-of-function mutants for these genes flower early. At 48 to 56 DAG, HD1 and RFT1 had the reverse expression pattern. DDS may switch genes on or off by indirectly affecting miR156 and miR172 expression, according to these findings. Due to the adaptability of rice cultivars to different geographical locations and cropping seasons, great emphasis should be paid to uncovering the genetic and molecular mechanism of rice photoperiodic blooming under DDS conditions. Because there are so many predominant alleles, such as HD1, Ehd1, and others (Doi et al., 2004; Leng et al., 2020) in different locations, the degree of SAS resulting from DDS may vary. The flowering of four mutants (i.e., phyb、OEmiR156b/c、ipa1-2D、Ostb1) was different for WT(NPB) under DDS and NDS conditions (Figure 9), indicating these alleles may retard SAS. Future studies may discover density-tolerant alleles in flowering-suppression or flowering-promotion pathways. Chloroplast dysfunction causes yellow leaves in DDS plants at 70 DAG, due to a decreased pigment content and the down-regulation of genes associated with chlorophyll metabolism and chloroplast development as a result of DDS.
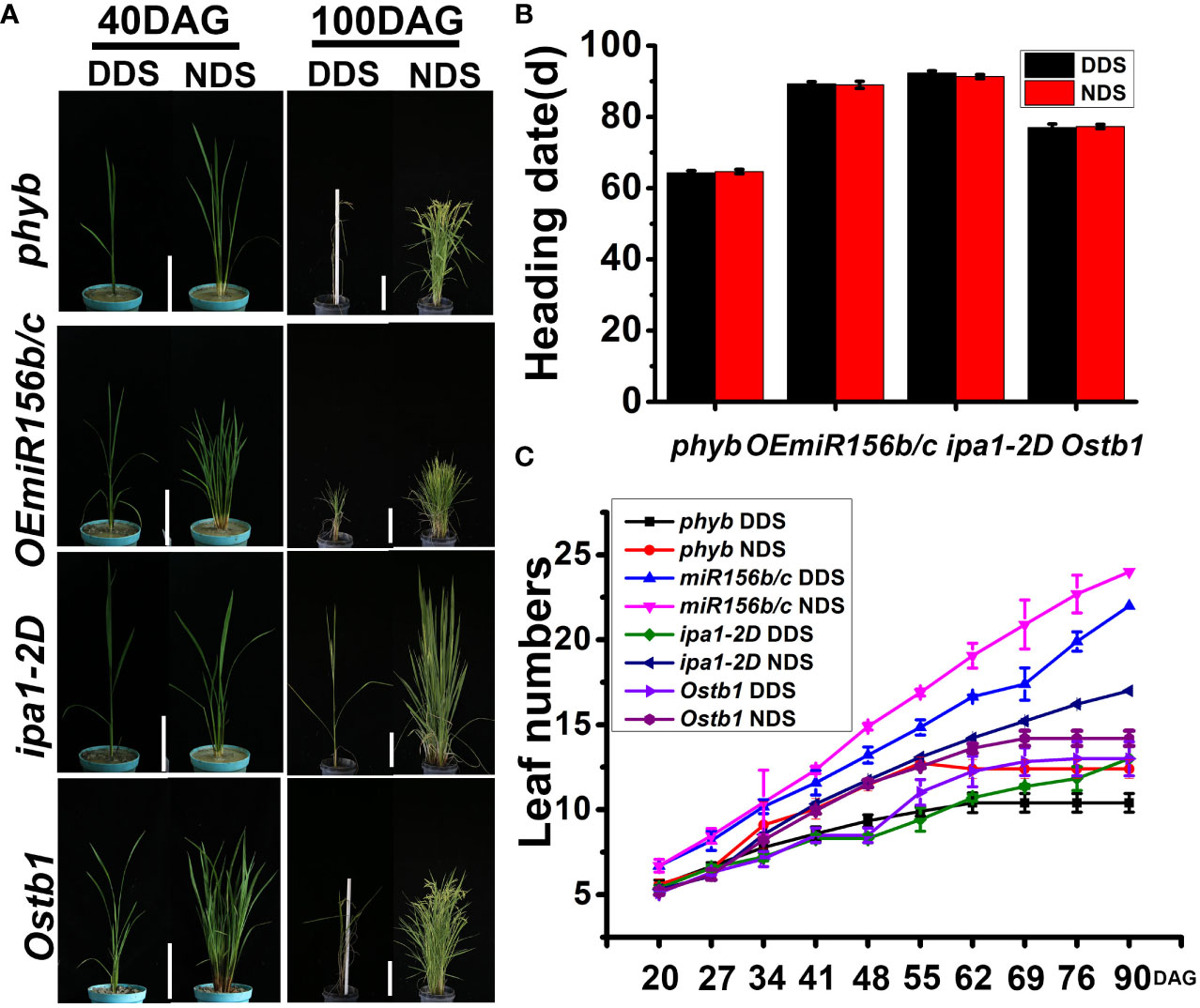
Figure 9 (A) A comparison of four mutants (i.e., phyb、OEmiR156b/c、ipa1-2D、Ostb1) of the tiller at different stages under DDS or NDS conditions. 40, 100 DAG treatment were photographed. Bar = 15 cm (left) Bar = 20 cm (right). (B) The same heading dates for four mutants (i.e., phyb、OEmiR156b/c、ipa1-2D、Ostb1) were observed under DDS conditions. (C) A fewer leaves were observed under DDS compared to NDS conditions in the four mutants (i.e., phyb、OEmiR156b/c、ipa1-2D、Ostb1). Error bars represent mean ± SD (n = 3) and different letters indicate significant differences as indicated by Duncan’s multiple range test (p ≤ 0.05).
The yield of direct seeding is determined by the efficiency with which the plants use radiation and the capacity with which they intercept light. Plants display a degree of mutual shading under a high planting density (i.e., under DDS conditions). According to increasing evidence, the SAS regulatory module is highly conserved in plants, and many genes involved in the SAS pathway may have been selected or discarded during rice domestication or breeding for better plant architecture and agronomic performance. A reduction of SAS could allow for higher plant productivity at higher densities or higher yield at current densities. This will be accomplished through the selection of natural variants as well as the generation of SAS mutations involving the genes mentioned above via genome editing. Therefore, further research should be focused on the effect of DDS on the SAS genes mentioned above.
5. Conclusion
We compared the responses of rice plants to NDS and DDS in this study. We proposed a putative DDS model based on our findings (Figure 10). In this model, DDS plants exhibit a constitutive SAS, in which solar radiation drops rapidly from the flag leaf to the base leaf, lowering phytochrome gene expression and changing the expression of multiple miR156 or miR172 genes. Because miR156 is a conservative regulator of developmental timing in plants, a mutation in the miR156 or miR172 gene causes the expression of photoperiod-related genes to be misregulated, resulting in early flowering. Furthermore, DDS causes senescence by downregulating the expression of chloroplast synthesis-related genes throughout almost the entire stage. As a result, we hypothesized that predominant alleles (such as HD1, Ehd1, and so on) in different geographical regions may exhibit different degrees of SAS under DDS. In the future, density tolerant alleles could be found for the SAS-suppression or SAS-promotion pathways.
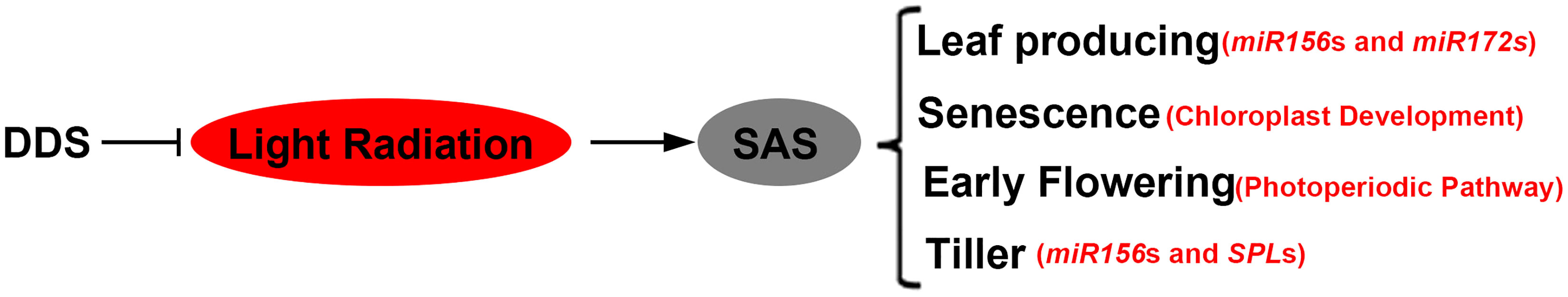
Figure 10 Simplified schematic model depicting the shade-avoidance response in DDS. Arrow: activate; Bar: repress.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
YTC, LBG conceived the project and designed the research. YTC, MHZ, JS, HHF, JYW and JJW performed the qRT-PCR assay. YTC, MHZ, LBG performed miRNA northern blotting. JS, HHF, XZX and JJW performed phenotype observations. YTC wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ22C130007, National Natural Science Foundation of China under Grant No. 32101791 and Key Laboratory of Digital Dry Land Crops of Zhejiang Province under Grant No.2022E10012.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer FZ declared a shared affiliation with the author LG to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1105882/full#supplementary-material
References
Andres, F., Galbraith, D. W., Talon, M., Domingo, G. (2009). Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. 151, 681–690. doi: 10.1104/pp.109.139097
Casal, J. J. (2012). Shade avoidance, Arabidopsis Book, 10:e0157. doi: 10.1199/tab.0157. Epub 2012 Jan 19.
Chen, R. Z., Deng, Y. W., Ding, Y. G., Guo, J. X., Qiu, J., Wang, B., et al. (2022). Rice functional genomics: decades’ efforts and roads ahead. Sci. China Life Sci. 65, 33–92. doi: 10.1007/s11427-021-2024-0
Cho, L. H., Yoon, J., Pasriga, R., An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Chuck, G., Cigan, A. M., Saeteurn, K., Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544–549. doi: 10.1038/ng2001
Ciolfi, A., Sessa, G., Sassi, M., Possenti, M., Salvucci, S., Carabelli, M., et al. (2013). Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 163, 331–353. doi: 10.1104/pp.113.22154
Cui, Y.T., Cheng, J. F., Ruan, S., Qi, P.P., Liu, W., Bian, H. W., et al. (2020). The heterochronic gene Oryza sativa LIKE HETEROCHROMATIN PROTEIN 1 modulates miR156b/c/i/e levels. J Integr Plant Biol. 62, 1839–1852. doi: 10.1111/jipb.12991
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a b-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Farooq, M., Siddique, K. H. M., Rehman, H., Aziz, T., Lee, D. J., Wahid, A. (2011). Rice direct seeding: Experiences, challenges and opportunities. Soil Tillage Res. 111, 87–98. doi: 10.1016/j.still.2010.10.008
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Guo, F., Han, N., Xie, Y., Fang, K., Yang, Y. N., Zhu, M. Y., et al. (2016). The miR393a/target module regulates seed germination and seedling establishment under submergence in rice (Oryza sativa l.). Plant Cell Environ. 39, 2288–2302. doi: 10.1111/pce.12781
Huijser, P., Schmid, M. (2011). The control of developmental phase transitions in plants. Development 138, 4117–4129. doi: 10.1242/dev.063511
Ishikawa, R., Aoki, M., Kurotani, K., Yokoi, S., Shinomura, T., Takano, M., et al. (2011). Phytochrome b regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285, 461–470. doi: 10.1007/s00438-011-0621-4
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Lee, D.-Y., An, G. (2012). Two AP2 family genes, supernumerary bract (SNB) and osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69, 445–461. doi: 10.1111/j.1365-313X.2011.04804.x
Lee, Y. S., Jeong, D. H., Lee, D. Y., Yi, J., Ryu, C.-H., Kim, S. L., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30. doi: 10.1111/j.1365-313X.2010.04226.x
Lee, Y. S., Lee, D. Y., Cho, L., An, G. (2014). Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7, 31. doi: 10.1186/s12284-014-0031-4
Leng, Y. J., Gao, Y. H., Chen, L., Yang, Y. L., Huang, L. C., Dai, L. P., et al. (2020). Using heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol. J. 18, 119–128. doi: 10.1111/pbi.13177
Liao, Z. G., Yu, H., Duan, J. B., Yuan, K., Yu, C. J., Meng, X. B., et al. (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10, 2738. doi: 10.1038/s41467-019-10667-2
Liu, Y., Wei, H. B., Ma, M. D., Li, Q. Q., Kong, D. X., Sun, J., et al. (2019). Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell. 31, 2089–2106. doi: 10.1105/tpc.18.00991
Miao, C., Wang, Z., Zhang, L., Yao, J. J., Hua, K. J., Liu, X. J., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10, 3822. doi: 10.1038/s41467-019-11830-5
Osugi, A., Itoh, H., Ikeda-Kawakatsu, K., Takano, M., Izawa, T. (2011). Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 157, 1128–1137. doi: 10.1104/pp.111.181792
Paulisic, S., Qin, W., Arora Veraszto, H., Then, C., Alary, B., Nogue, F., et al. (2021). Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 40, e104273. doi: 10.15252/embj.2019104273
Takanoa, M., Inagaki, N., Xie, X. Z., Kiyota, S., Baba-Kasai, A., Tanabata, T., et al. (2009). Phytochromes are the sole photoreceptorsd for perceiving red/far-red light in rice. PNAS 106, 14705–14710. doi: 10.1073/pnas.0907378106
Takano, M., Inagaki, N., Xie, X. Z., Yuzurihara, N., Hihara, F., Ishizuka, T., et al. (2005). Distinct and cooperative functions of phytochromes a, b, and c in the control of deetiolation and flowering in rice. Plant Cell. 17, 3311–3325. doi: 10.1105/tpc.105.035899
Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Warnasooriya, S. N., Brutnell, T. P. (2014). Enhancing the productivity of grasses under high-density planting by engineering light responses: from model systems to feedstocks. J. Exp. Bot. 65, 2825–2834. doi: 10.1093/jxb/eru221
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S. H., Yu, C. Y., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, C. Y., You, C. J., Li, C. S., Long, T., Chen, G. X., Byrne, M. E., et al. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. PNAS 105, 12915–12920. doi: 10.1073/pnas.0806019105
Xie, Y., Liu, Y., Wang, H., Ma, X. J., Wang, B. B., Wu, G. X., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in arabidopsis. Nat. Commun. 8, 348. doi: 10.1038/s41467-017-00404-y
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Xu, J., Wang, L., Zhou, M. Y., Zeng, D. L., Hu, J., Zhu, L., et al. (2017). Narrow albino leaf 1 is allelic to CHR729, regulates leaf morphogenesis and development by affecting auxin metabolism in rice. Plant Growth Regulation 82, 175–186. doi: 10.1007/s10725-017-0249-4
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell. 12, 2473–2483. doi: 10.2307/3871242
Yu, S., Galvão, V. C., Zhang Y, C., Horrer, D., Zhang, T. Q., Hao, Y. H., et al. (2012). Gibberellin regulates the arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 24, 3320–3332. doi: 10.1105/tpc.112.101014
Zhang, S., Deng, L., Cheng, R., Hu, J., Wu, C. Y. (2022). RID1 sets rice heading date by balancing its binding with SLR1 and SDG722. J. Integr. Plant Biol. 64 (1), 149–165. doi: 10.1111/jipb.13196
Zhang, Z. Y., Hu, W., Shen, G. J., Liu, H. Y., Hu, Y., Zhou, X. C., et al. (2017). Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci. Rep. 7, 5388. doi: 10.1038/s41598-017-05873-1
Zhao, J., Chen, H., Ren, D., Tang, H. W., Qiu, R., Feng, J. L., et al. (2015). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa.L). New Phytol. 208, 936–948. doi: 10.1111/nph.13503
Zhou, W., Guo, Z., Chen, J., Jiang, J., Hui, D. F., Wang, X., et al. (2019). Direct seeding for rice production increased soil erosion and phosphorus runoff losses in subtropical China. Sci. Total Environ. 695, 133845. doi: 10.1016/j.scitotenv.1019.133845
Keywords: oryza sativa. L, densely direct seeding, phytochrome, miR156, miR172, early flowering
Citation: Cui Y, Zhu M, Song J, Fan H, Xu X, Wu J, Guo L and Wang J (2023) Expression dynamics of phytochrome genes for the shade-avoidance response in densely direct-seeding rice. Front. Plant Sci. 13:1105882. doi: 10.3389/fpls.2022.1105882
Received: 23 November 2022; Accepted: 30 December 2022;
Published: 18 January 2023.
Edited by:
Xuke Lu, Institute of Cotton Research (CAAS), ChinaReviewed by:
Fengli Zhao, China National Rice Research Institute, ChinaJian-Hong Xu, Zhejiang University, China
Tiankang Wang, Hunan Hybrid Rice Research Center, China
Copyright © 2023 Cui, Zhu, Song, Fan, Xu, Wu, Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Wang, d2FuZ2pqNDE5N0AxNjMuY29t; Longbiao Guo, Z3VvbG9uZ2JpYW9AY2Fhcy5jbg==
 Yongtao Cui
Yongtao Cui Minhua Zhu2
Minhua Zhu2 Longbiao Guo
Longbiao Guo