
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 December 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1103235
This article is part of the Research TopicGenetic and Epigenetic Regulatory Mechanisms in Higher Plants in Response to Abiotic StressView all 6 articles
Potassium (K) is an essential nutrient for plant physiological processes. Members of the HAK/KUP/KT gene family act as potassium transporters, and the family plays an important role in potassium uptake and utilization in plants. In this study, the TaHAK13 gene was cloned from wheat and its function characterized. Real-time quantitative PCR (RT-qPCR) revealed that TaHAK13 expression was induced by environmental stress and up-regulated under drought (PEG6000), low potassium (LK), and salt (NaCl) stress. GUS staining indicated that TaHAK13 was mainly expressed in the leaf veins, stems, and root tips in Arabidopsis thaliana, and expression varied with developmental stage. TaHAK13 mediated K+ absorption when heterologously expressed in yeast CY162 strains, and its activity was slightly stronger than that of a TaHAK1 positive control. Subcellular localization analysis illustrated that TaHAK13 was located to the plasma membrane. When c(K+) ≤0.01 mM, the root length and fresh weight of TaHAK13 transgenic lines (athak5/TaHAK13, Col/TaHAK13) were significantly higher than those of non-transgenic lines (athak5, Col). Non-invasive micro-test technology (NMT) indicated that the net K influx of the transgenic lines was also higher than that of the non-transgenic lines. This suggests that TaHAK13 promotes K+ absorption, especially in low potassium media. Membrane-based yeast two-hybrid (MbY2H) and luciferase complementation assays (LCA) showed that TaHAK13 interacted with TaNPF5.10 and TaNPF6.3. Our findings have helped to clarify the biological functions of TaHAK13 and established a theoretical framework to dissect its function in wheat.
Potassium ions (K+) are the most abundant cation in plants and are involved in many physiological and biochemical processes, including cell elongation, enzyme activity regulation, osmotic regulation, stomatal movement, photosynthesis, and protein synthesis (Zhang et al., 2012). Potassium also acts as a transporter for photosynthetic substances (from source to sink) and participates in the regulation of osmotic pressure and the plant response to osmotic stress (Gajdanowicz et al., 2011). A moderate increase in potassium application rate is helpful for enhancing plant resistance to abiotic stress (Wang et al., 2021). Potassium is also closely tied to crop quality (Song et al., 2014). The cytoplasmic concentration of K+ for normal growth in living cells is approximately 100 mM (40-200 mM), which is also the optimum concentration for normal enzymatic function. Compared to the higher concentrations of K+ observed in living cells, the concentration of potassium ions in the root-soil interface tends to be at the micromolar level (0.1-1 mM). In most cases, potassium uptake by plants is an active transport process against a concentration gradient (Maathuis, 2009). The absorption of K+ from the external environment into plant cells and its transport within plant tissues are mainly completed by K transporters and K channel proteins. These genes can be divided into five families according to the structure and function of K transporters and channel proteins, including three transporter families (KUP/HAK/KT, HKT, and CPA families) and two ion channel protein families (Shaker and KCO/TPK families). Transporters combine with K+, undergo conformational changes, and then transport K+ across the cell membrane. Channel proteins form water channels through the lipid bilayer that allow K+ to pass through the membrane when the channel is open (Wang et al., 2010).
The KUP/HAK/KT transporter family is present in bacteria, fungi, and plants, and participates in K+ transmembrane transport (Vastermark et al., 2014). Characterizing the physiological functions of the KUP/HAK/KT potassium transporter family in plants has recently been the focus of much research attention. There are 13 proteins belonging to the KUP/HAK/KT family in Arabidopsis (Mäser et al., 2001), 25 in rice (Yang et al., 2009), 5 in barley (Alemán et al., 2011), 27 in corn (Zhang et al., 2012), and 56 in wheat (Cheng et al., 2018). Proteins in the KUP/HAK/KT family are divided into four clusters: cluster I, cluster II, cluster III, and cluster IV. Members of cluster I have been extensively studied, including AtHAK5 (Arabidopsis thaliana), HvHAK1 (Hordeum vulgare L.), and OsHAK1/OsHAK5 (Oryza sativa L.). The HvHAK1 protein is localized in the plasma membrane and primarily expressed in roots; it mediates the high-affinity absorption of potassium ions and is strongly induced by low potassium conditions (Senn et al., 2001). In Arabidopsis thaliana, AtHAK5 is a member of the high affinity K+ absorption system, which is induced by potassium starvation (no K+ supply) and expressed in plant roots (Ahn et al., 2004; Armengaud et al., 2004; Shin and Schachtman, 2004). AtHAK5 maintains a very high level of expression after seven days of potassium starvation (Gierth et al., 2005). Under low potassium stress (< 50 μM K+), athak5 mutant seeds germinate slowly, root elongation is inhibited, and the ability to absorb K+ decreases, indicating that AtHAK5 mediates the absorption of high affinity K+ and participates in the process of seed germination and later growth and development (Rubio et al., 2008; Pyo et al., 2010). The transcription of OsHAK5 increases during potassium starvation and under salt stress; cells accumulate a large amount of K+ (instead of Na+) when expressed in tobacco BY2 cells, suggesting that OsHAK5 is a salt-sensitive high affinity K+ transporter (Horie et al., 2011). In addition, OsHAK1 and OsHAK5 are the two main K+ transporters active in low potassium stress conditions; the transport activity of OsHAK1, unlike that of OsHAK5, is sensitive to Na+ (Okada et al., 2018). The K+ absorption rate and transport capacity of wild type plants is significantly higher than that of mutant plants with OsHAK1 gene knockout. Over-expression of OsHAK1 significantly enhances K+ absorption capacity, suggesting that this protein affects K+ absorption and may simultaneously mediate K+ absorption and transportation by the two K+ absorption systems (Chen et al., 2015; Chen et al., 2019). Finally, the PhaHAK5 protein was identified in a salt-sensitive reed and belongs to cluster IV. Functional analyses in yeast have found it acts as a high affinity potassium transporter, capable of mediating low affinity sodium ion transport in the presence of high Na+ stress (Takahashi et al., 2007).
A total of 56 HAK/KUP/KT family members (TaHAK1-TaHAK25, containing homologous genes) were identified in wheat in a recent phylogenetic analysis (Cheng et al., 2018). The HAK/KUP/KT family includes vital transporter proteins for potassium homeostasis, but very little is known as to the detailed functions of these proteins in plants. In this study, TaHAK13 was investigated in a physiological function analysis to better understand the molecular mechanisms underlying efficient K+ transport in wheat.
Hexaploid wheat (Triticum aestivum L., cv. Yunong 804) seeds were germinated in the dark at 25°C for two days after sterilization; seeds were sprayed daily with ddH2O to keep them moist. After five days, seedlings were transplanted into Hoagland nutrient solution (Table S1), and this solution was changed every three days. Two-week-old wheat seedlings were divided into four groups, with each group containing at least 30 plantlets. The four groups were: a control (Hoagland medium with full K+ concentration of 1mM KCl), a drought treatment (20% PEG6000+Hoagland medium), a low potassium treatment (K2SO4 concentration in Hoagland medium of 0.01 mM), and a salt stress treatment (200 mM NaCl + Hoagland medium). Plantlets were maintained on the medium at 25°C and a 16 h light/8 h dark photoperiod. Seedling roots were sampled at the following time points: 0, 1, 3, 6, 9, 12, and 24 h. Each sample was immediately frozen in liquid nitrogen and stored at -80°C for further analyses.
Tobacco seeds (Nicotiana benthamiana) were sown on nutrient soil and kept in a growth chamber at 25°C and 50-70% relative humidity with a photoperiod of 16 h light/8 h dark for about four weeks; these were used for the assessment of Agrobacterium-mediated transient fluorescent protein fusion expression.
Wild-type (Col-0) and mutant (athak5, SALK_005604) Arabidopsis seeds were surface sterilized with 70% (v/v) ethanol for 7 min and 0.1% (v/v) NaClO for 5 min, then washed with ddH2O three times. Sterilized seeds were stored at 4°C in the dark for three days to promote synchronous germination. Phenotypic analysis of seedlings was carried out under low potassium conditions on Murashige Skoog (MS) medium following protocols described by Pyo et al. (Pyo et al., 2010). For soil culture, plants were pre-cultured in MS basic medium for seven days, then transferred to nutrient soil and cultured in a growth chamber with a photoperiod of 16 h light/8 h dark at 25°C.
Total RNA was extracted from all samples using Trizol (TransGen Biotech) according to the manufacturer’s instructions. First strand cDNA was synthesized using a PrimeScript™ RT reagent Kit with a gDNA Eraser (Takara). The diluted cDNA was amplified using qPCR SYBR Green Master Mix (Yeasen) on a real-time PCR system (Quantstudio™5) following standard protocols. The primer sequences used are listed in Table S2. The qPCR procedures were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 61°C for 1 min, and 72°C for 5 min. Three biological replicates were used in each independent experiment and three independent experiments were performed for each RT-qPCR data analysis. Relative transcript levels were calculated using the 2−ΔΔCt method with the wheat β-actin gene as an internal reference control (Livaka and Schmittgen, 2001).
The coding sequence (CDS) of the TaHAK13 gene (2,412 bp) was amplified from the cDNA isolated from wheat seedlings. Sequence data for TaHAK13 (ID: TraesCS7D02G456900) were obtained from the wheat genome annotation project (Ensembl Plants database). The primer sequence used for amplifying the coding sequence is provided in Table S2. PCR products were cloned into a pESI-Blunt vector using the Zero TOPO-TA Cloning Kit (Yeasen) and then sequenced.
The coding sequence of TaHAK13, which contains SacI and BamHI restriction sites without a stop codon, was amplified and inserted in front of the GFP gene sequence in a 35S-GFP vector. The TaHAK13-GFP fusion vector, under the control of a 35S promoter, was then transformed into Agrobacterium tumefaciens strain GV3101. Positive strains were injected into tobacco leaf epidermal cells using the agroinfiltration method. Before imaging, transformed plants were grown for two days at 22°C with a 16 h light/8 h dark photoperiod. The GFP signal was visualized with a confocal laser-scanning microscope (Carl Zeiss, Germany). The primer sequences used are given in Table S2.
A 2,250 bp fragment was cloned from upstream of the TaHAK13 start codon in wheat genomic DNA using relevant primers (Table S2) with a restriction site. The amplified DNA fragment used a TaHAK13 promoter, replacing the LacZ and CaMV35S promoter regions, and were constructed in a pCAMBIA1304 vector. The constructed plasmid was transformed into Agrobacterium tumefaciens GV3101 to infect Arabidopsis via the flower soaking method. The T2 generation plants were used for GUS histochemical staining. Using previously published methods (Jefferson et al., 1987), the roots, leaves, stems, flowers, pods, and other tissues of both Arabidopsis seedlings and mature plants were stained with a GUS staining kit (Coolaber). The colored part was observed and photographed using a stereomicroscope (Stemi508, Carl Zeiss, Germany).
TaHAK13 was amplified with specific primers (Table S2), digested with BamHI or XbaI restriction endonucleases, and ligated to the yeast expression vector p416. In a functional complementation experiment, p416, TaHAK13-p416, and TaHAK1-p416 were transformed into yeast strains CY162 (MATα, ura3, his3, ade2, trk1△, trk2△::PCK64) (Anderson et al., 1992) and AXT3K (MATα, ena1::HIS3::ena4, nha1::LEU2, nhx1::KanMX4), respectively (Quintero et al., 2000).
The yeast complementation analysis was carried out on solid AP-U medium (i.e., an arginine phosphate medium lacking uracil) (Xu et al., 2008), with a supplemental K+ concentration in the range of 0-100 mM. The experimental yeast strains (transformed with p416, TaHAK13-p416, or TaHAK1-p416) were grown overnight in liquid SD-U medium (i.e., a synthetic defined base without uracil) and then transferred to liquid AP-U medium supplemented with different concentrations of K+ (either 1 mM or 100 mM) with the same initial OD600 (~0.001). Once in the AP-U medium, the yeast strains were grown for three days on a shaker at 220 r/min. The OD600 of each strain was measured every 8 h. The experiment was repeated three times.
A K+ depletion experiment was performed according to previously described procedures (Zhang et al., 2018) with minor modifications. Yeast cells (transformed with p416, TaHAK13-p416, or TaHAK1-p416) were grown overnight in liquid SD-U medium at 30°C, and then transferred to AP-U liquid medium for potassium starvation for about 4 h. Cells were suspended in 10 mM MES supplemented with 2% glucose and adjusted to pH 6.0 with Ca(OH)2. At time zero, KCl was added to the culture medium, and samples were collected every 20 min within 2 h.
To measure the K+ content in yeast cells, yeast strains (transformed with p416, TaHAK13-p416, or TaHAK1-p416) were grown in AP-U medium with different K+ concentrations (0-100 mM) at 30°C. Yeast cells were first suspended in pre-cooled sterile water with an OD600 = 0.3, and then repeatedly heated and frozen to break the cells. A flame photometer (FP640) was used to measure the K+ content. The specific operation method was as described by Karabegov (Karabegov, 2011). Three replicates from each sample were tested in total.
The TaHAK13 coding sequence was amplified and used to construct a pCAMBIA1300 vector utilizing a CaMV35S promoter with kanamycin resistance. The resulting plasmid was introduced into Agrobacterium tumefaciens strain GV3101 for transformation into mutant (athak5) and wild-type (Col) Arabidopsis using the floral dip method. Transgenic seedlings (Col/TaHAK13 and athak5/TaHAK13) were confirmed by RT-PCR. Transgenic lines and non-transgenic lines were then planted in MS medium with different K+ concentrations (0, 0.01, 0.1, and 1 mM KCl), and phenotypes (root length and fresh weight) of transgenic and non-transgenic lines measured after ten days.
A non-invasive micro-test technology (NMT) system NMT100-SIM-XY (Younger USA Science and Technology; Xuyue, China) was used to determine the net K+ influx in transgenic Arabidopsis roots, and NMT User Manual 4.1 was referenced for specific operation methods. Arabidopsis seedlings were grown on MS medium for 10 d and then treated with a low potassium solution for 12 h. Before measuring the K+ influx, the ion-selective electrode was calibrated with K+ concentrations of 0.05 mM, 0.1 mM, and 0.5 mM. To take measurements, seedling roots were soaked in a preparatory solution (0.1 mM CaCl2 and 0.3 mM MES, pH 6.0) for 30 min, before transferring to a measuring solution supplied with 0.1 mM KCl or 0.01 mM KCl. The net K+ influx was measured over the course of 8 min under experimental conditions to reduce variability caused by solution fluctuation. Under the microscope, measurement sites were located at 0 μm, 200 μm, 400 μm, and 600 μm away from the root tip, and a microsensor was placed at approximately 150 µm from the root tip to optimize data collection. Flux rates were calculated; note positive values represent efflux, and negative values represent influx. In a separate experiment, measurements were collected from the roots of at least eight Arabidopsis plants, and each plant was measured once.
A membrane-based yeast two-hybrid system was used to screen for proteins interacting with TaHAK13 (Stagljar et al., 1998). The coding sequence of TaHAK13 was introduced into a pBT3-N vector (see Table S2 for primer sequences); the pBT3-N vector was then used as a bait vector, and the bait vector and library plasmid were co-transformed into the yeast strain NMY51 (MATα, his3Δ200, trp1-901, leu2-3, 112, ade2, LYS2::(lexAop)4-HIS3, ura3::(lexAop)8-lacZ, ade2::(lexAop)8-ADE2, GAL4). Proteins interacting with TaHAK13 were identified and then ligated into a pPR3-N vector (Table S2). To assay different protein combinations, the bait vector and prey vector were co-transformed into the yeast NMY51 strain, which was then grown on screening media (SD/-L/-T, SD/-A/-H/-L/-T); proteins were identified using X-Gal after culturing at 30°C for 3 d.
A luciferase complementation assay was used to analyze protein-protein interactions between TaHAK13 and either TaNPF5.10 or TaNPF6.3 (Wang et al., 2020). The coding sequence of TaHAK13 was inserted into a pCAMBIA1300-nLUC vector, and the full CDS of TaNPF5.10 and TaNPF6.3 were separately ligated into pCAMBIA1300-cLUC vectors. The Agrobacterium strain GV3101 carrying these vectors was infiltrated into four-week-old tobacco leaves using the Agrobacterium-mediated transient transformation method (Chen et al., 2008). After three days of infiltration, 1 mM D-fluorescein potassium salt (Yeason) was sprayed on the leaves, which were then kept in the dark for 10 min. The luciferase signal was captured using a plant living image system (Night SHADE LB 985, Berthold, Germany).
Three independent biological repeats were set for each experiment. All data were subjected to analysis of One-way ANOVA according to the model for completely randomized design via SPSS 24.0 software (USA) and represented as mean. Significant differences were calculated based on t-test at P<0.05 level between different treatments.
To quantify TaHAK13 expression under short-term environmental stress, RNA was extracted from the roots of wheat seedlings grown in low potassium, salt, and drought stress conditions. Using RT-qPCR, TaHAK13 expression was then analyzed. Expression was first up-regulated and then down-regulated for all three stresses. In the low potassium (0.01 mM KCl) treatment, TaHAK13 expression reached a maximum (six times that of the control) at 6 h, after then decreasing (Figure 1A). In the salt stress (200 mM NaCl) treatment, TaHAK13 expression showed a similar pattern: up-regulation until the 6 h mark and then a gradual decline (Figure 1B). To determine whether the expression of TaHAK13 was induced by dehydration, 15-day-old plantlets were transferred to a hydroponic solution supplemented with 20% PEG6000. The expression of TaHAK13 was initially up-regulated, reaching a maximum (about ten times the 0 h value) at 3 h post-treatment (Figure 1C). This suggests that TaHAK13 expression was induced by transient abiotic stress (Figure 1 and Figure S1).
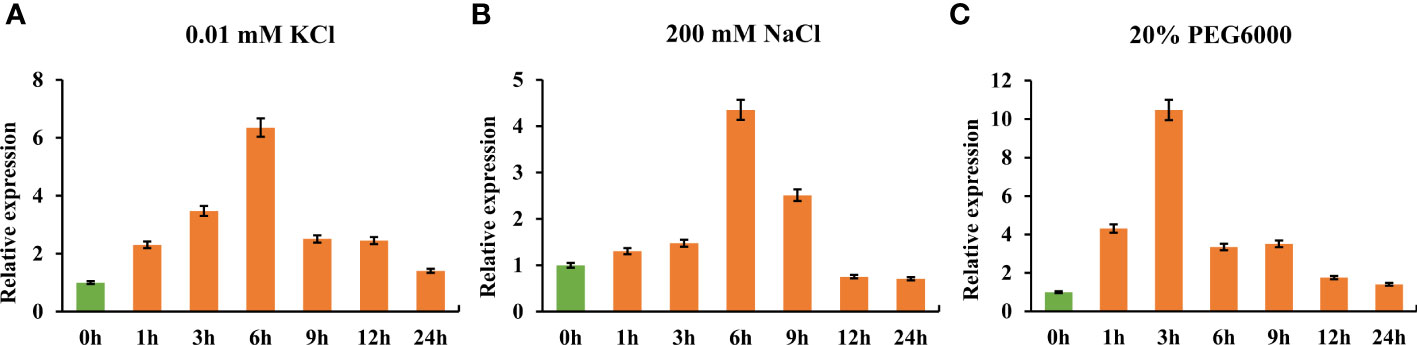
Figure 1 Real-time quantitative PCR expression of TaHAK13 in wheat roots under different stresses. (A) The relative expression of TaHAK13 under low potassium stress (0.01 mM KCl). (B) The relative expression of TaHAK13 under salt stress (200 mM NaCl). (C) The relative expression of TaHAK13 under dehydration stress (20% PEG6000). The relative expression of TaHAK13 at 0 h was taken as a value of one.
The transmembrane domain analysis identified eleven transmembrane structures in the TaHAK13 protein, with the N-terminal of the protein located in the cell membrane and the C-terminal outside the membrane (Figure 2A). The TaHAK13 protein may therefore play an important role in transmembrane transport. To further study the subcellular localization of TaHAK13, a vector containing a TaHAK13-GFP fusion protein was introduced into the epidermal cells of Nicotiana benthamiana leaves via Agrobacterium tumefaciens infection. The subcellular localization of the TaHAK13-GFP fusion protein and 35S-GFP (a control vector) was observed using a laser confocal microscope. The 35S-GFP control was expressed in the nucleus, cell membrane, and cytoplasm, while TaHAK13-GFP was expressed only in the plasma membrane (Figure 2B).
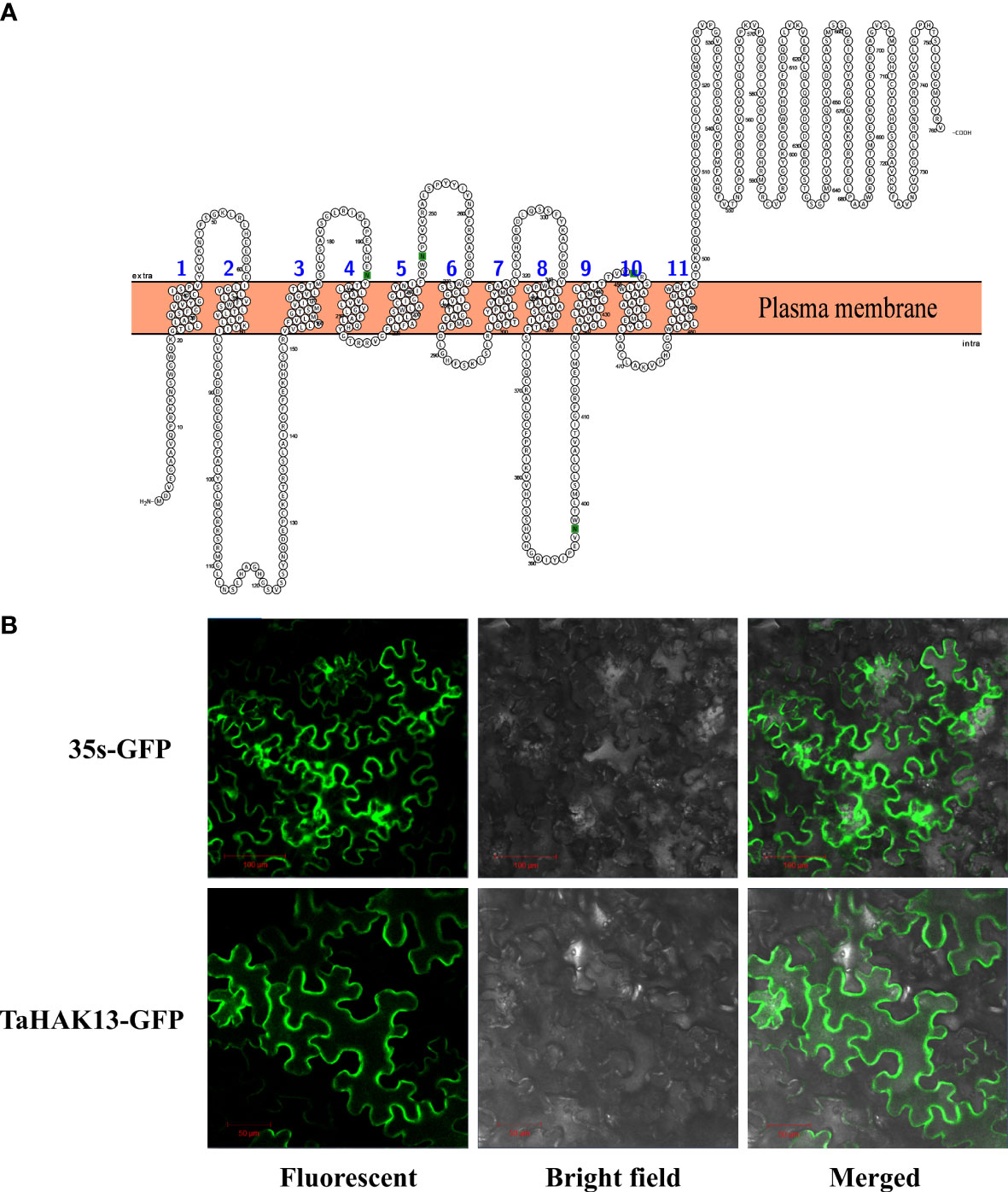
Figure 2 Characteristics of TaHAK13. (A) Predicted transmembrane domains of TaHAK13. (B) Subcellular localization of TaHAK13 in tobacco leaves. 35S-GFP acted as the control. Bar = 50 or 100 μm.
The expression specificity of TaHAK13 in different tissues (roots, stems, leaves, spikes, and grains) was analyzed using RNA-Seq data from the wheat expression database (Figure 3). The average TaHAK13 expression was highest in roots, followed by spikes and grains, and relatively low in stems; the lowest values were seen in leaves.
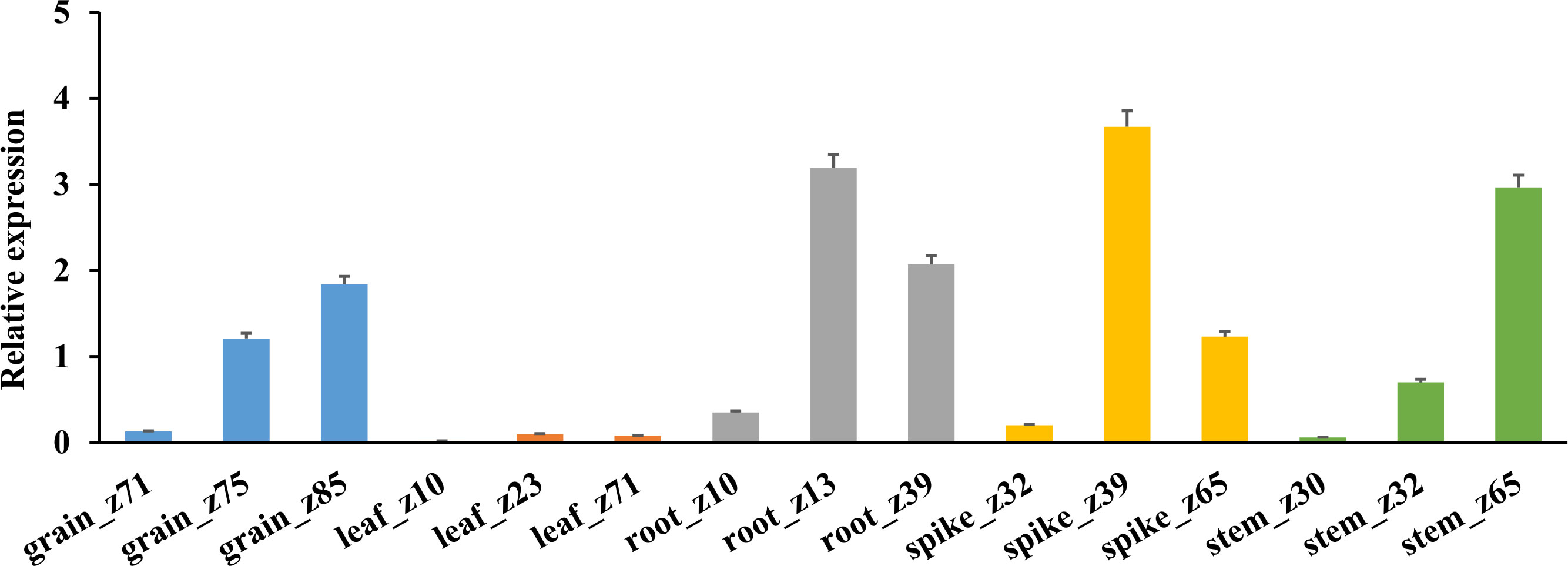
Figure 3 TaHAK13 expression in different tissues. An RNA-seq analysis of wheat expression data was used to assess the expression specificity of TaHAK13 in different tissues.
To study TaHAK13 expression in greater detail, the 2,250 bp fragment upstream of the gene was cloned and used as a promoter. The promoter region of the TaHAK13 gene contained a TATA-box, CAAT-box, stress response elements (e.g., an MYB binding site involved in drought resistance), cis-acting elements (involved in defense and stress responses), and a WRKY-binding W-box. Thus, the TaHAK13 gene promoter is regulated by many factors. In addition, a key element (the Root motif TAPOX1) required for root specific expression was also found in the promoter, indicating that TaHAK13 is highly expressed in roots (Table S3).
Transgenic plants harboring a TaHAK13 promoter-GUS fusion vector were used to investigate specific tissue expression patterns. Strong signals were detected in whole Arabidopsis plantlets in a GUS staining assay (Figure 4). The GUS gene (controlled by a TaHAK13 promoter) was mainly expressed in the veins (Figures 4A, B), vascular bundle tissue of the embryonic axis (Figure 4C), taproot (Figure 4D), and lateral root apex (Figure 4E) of transgenic Arabidopsis seedlings. The highest TaHAK13 specificity was observed in roots. To explore whether expression was sensitive to developmental stage, GUS staining was also carried out in the roots, leaves, stems, flowers, and pods of mature Arabidopsis plants. The veins (Figure 4O), roots (Figure 4F), and stems (Figure 4M) of transgenic mature Arabidopsis plants were deeply stained. Interestingly, as the depth of root penetration increased, the staining became more pronounced (Figures 4G–K), consistent with early TaHAK13 expression in roots and root tips at the seedling stage. Epidermal hairs (Figure 4N) and flowers (Figure 4P) were only stained lightly. Overall, the TaHAK13 promoter predominantly drove GUS expression in mature roots and the vascular tissues of transgenic Arabidopsis plants, and the level of root-specific expression was influenced by developmental stage.
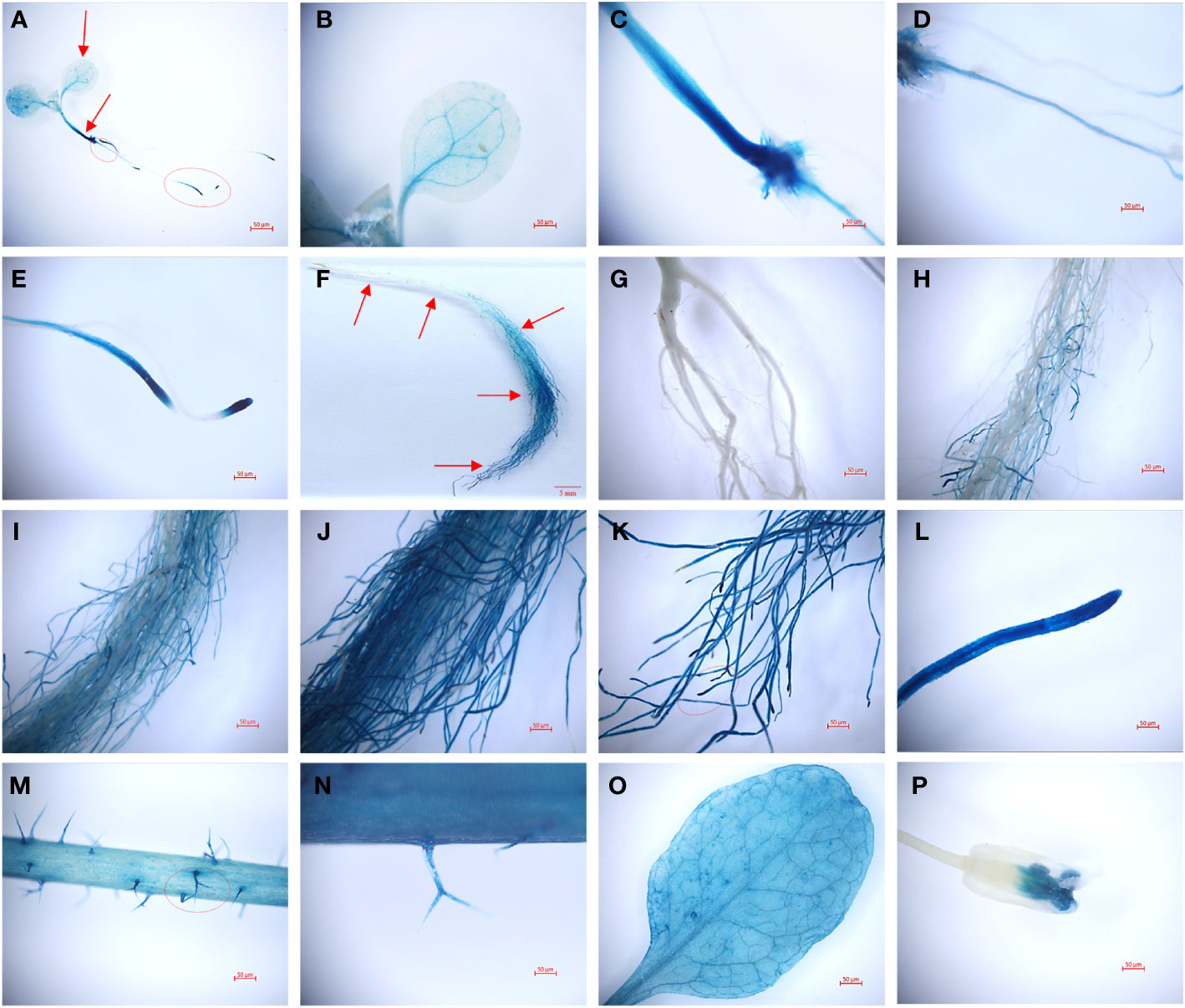
Figure 4 GUS histochemical staining with a TaHAK13 promoter in transgenic Arabidopsis thaliana. (A-E) Zoomed out image of a transgenic Arabidopsis seedling (A) and its leaf (B), stem (C), taproot (D), and lateral root apex (E). (F-P) Zoomed out image of a root (F) and enlarged images of the indicated root positions (marked with red arrows) (G-L) in mature transgenic Arabidopsis plants; also pictured are the stem (M), stem surface (N), leaf (O), and flower (P).
The budding yeast S. cerevisiae has been shown to be an excellent model for studying ion transport and ion homeostasis (Mao et al., 2022). Mutant strains lacking their own ion transport systems serve as an efficient tool for the molecular study of higher eukaryote transporters via their expression in yeast cells (Xu et al., 2008). TaHAK13 was inserted into the yeast strain CY162 to explore the K+ sensitivity of the TaHAK13 transporter. CY162 is defective in high-affinity potassium uptake and cannot grow on low K+ (≤1 mM) AP plates. A yeast complementation experiment was conducted on solid AP-U media with different K+ levels (0, 1, 2, 10, or 100 mM KCl). The p416-TaHAK1 recombinant plasmid was transferred into CY162 for use as a positive control, while the p416 vector was transferred into CY162 to create a negative control. All test strains (including those with transgenes or the empty vector) grew uniformly on AP plates with 2, 10, or 100 mM KCl (Figure 5A). With only 1 mM KCl, the yeast strain transformed with TaHAK13 showed similar growth to the positive control (TaHAK1); growth was poor for the empty vector transformant (negative control) (Figure 5A). Therefore, TaHAK13 can restore growth in CY162 on low K+ media and has K+ transport ability.
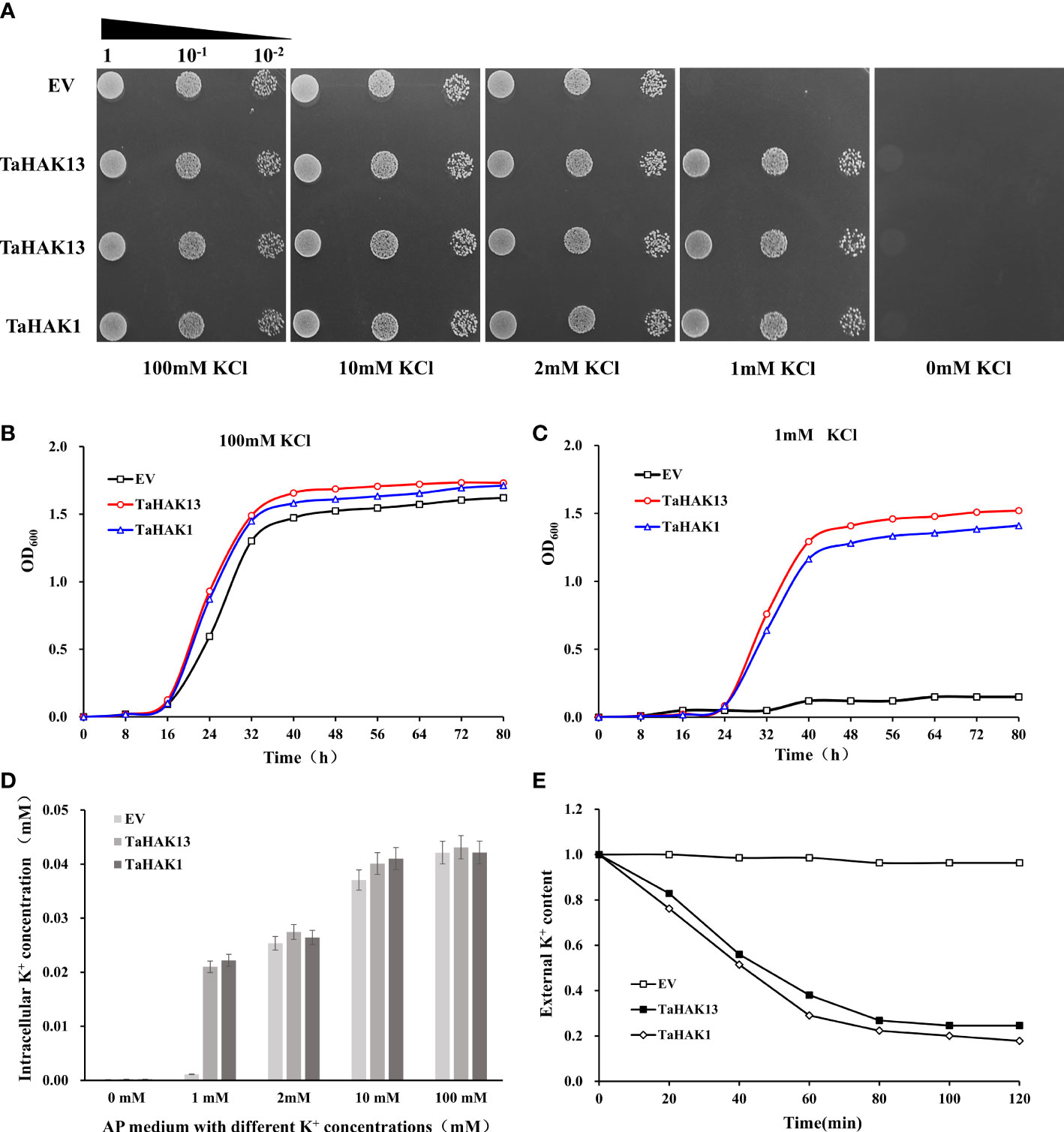
Figure 5 TaHAK13 complementarity analysis in a Trk1 and Trk2 K+ uptake system deficient yeast strain (A) Growth of mutant CY162 in AP-U solid media with different concentrations of K+. CY162 was transformed with the empty vector p416 (EV), TaHAK13-p416 (TaHAK13), or TaHAK1-p416 (TaHAK1). After serial dilution, each strain was added to an agar plate for culture. (B) Growth curves for CY162 transformed with the empty vector, TaHAK13, or TaHAK1 in AP-U liquid medium supplemented with 1mM K+ or 100 mM K+. (C) Determination of the K+ content in CY162 transformed with the empty vector p416, TaHAK13, or TaHAK1 in solid AP-U media with various concentrations of K+. (D) K+ depletion experiment in the presence of 1 mM K+ in AP-U medium. CY162 (transformed with p416, TaHAK13, or TaHAK1) was subjected to K+ starvation for 4 h before beginning the experiment. The K+ content in the buffer was measured at intervals of 2 h.
As the RT-qPCR results revealed that TaHAK13 can respond to salt stress, both the empty vector p416 and TaHAK13-p416 were transferred into the yeast strain AXT3K respectively. AXT3K does not possess any of the major endogenous sodium transporters essential for salt tolerance and is incapable of growing on AP plates with NaCl concentrations greater than 10 mM. Yeast drop experiments on AP media with different salt concentrations (0, 10, 20, 30, and 50 mM NaCl) were carried out. The growth of AXT3K transformed with TaHAK13-p416 and p416 was similar and neither survived at higher NaCl concentrations (Figure S2), indicating that TaHAK13 does not transport Na+.
Patterns of yeast cell growth in AP-U liquid media with different concentrations of K+ further confirmed the growth restoration ability of the TaHAK13 transformant. At 100 mM KCl, the transgenic yeast strain CY162 containing TaHAK13 had the same growth rate as that of the transgenic yeast strain TaHAK1 (positive control) and empty vector (negative control). At 1 mM KCl, TaHAK13 and TaHAK1 transformants grew normally, but the yeast strain transformed with an empty vector exhibited little growth (Figure 5B).
To further characterize the relationship between CY162 growth and K+ absorption, the K+ content of yeast strains cultured with various K+ concentrations was measured. Yeast expressing TaHAK13, TaHAK1 (positive control), or an empty vector (negative control) maintained a stable K+ intracellular concentration when 100, 10, or 2 mM of external K+ was added to the AP liquid medium. However, under K+ stress (1 mM K+ AP medium), only yeast expressing TaHAK13 or TaHAK1 showed effective K+ absorption; the empty vector transformant showed growth deficits as the K+ content declined (Figure 5C). Meanwhile, CY162 transformed with TaHAK13 or TaHAK1 depleted the available K+ in the culture medium (1 mM), but no such depletion was observed by the empty vector strain (Figure 5D). In conclusion, TaHAK13 was regulated by intracellular K+ and strictly controlled the intracellular K+ content to maintain ion balance.
The Arabidopsis thaliana mutant athak5 is sensitive to low potassium, and its primary root length is shorter than that of wild-type plants (Gierth et al., 2005; Pyo et al., 2010). To further characterize the function of TaHAK13 in plants, it was amplified and expressed in the Arabidopsis mutant athak5 and the wild-type (Col). Under normal potassium conditions (0.1 and 1 mM KCl), the root length and fresh weight of the complementary lines (athak5/TaHAK13) were not significantly different from those of the mutant lines (athak5), but when growing under low K conditions (0 and 0.01 mM KCl), the athak5 mutant showed serious growth defects. The expression of TaHAK13 rescued the sensitive phenotype of athak5 (Figures 6A, B, S5), and the root length and fresh weight of the two transgenic lines were significantly higher than those of the athak5 mutant (Figures 6C, D). Similar patterns were seen for the over-expression lines (Col/TaHAK13) and wild-type lines (Col) (Figures 7A, B). Under normal potassium conditions (0.1 and 1 mM KCl), the root length and fresh weight of Col/TaHAK13 transgenic lines did not differ from those of the Col non-transgenic lines, while under low K conditions (0 and 0.01 mM KCl), the expression of TaHAK13 increased plant tolerance of low potassium (Figures 7C, D). These results further confirm that TaHAK13 acts as a high affinity potassium transporter that mediates K+ uptake in plants under low potassium conditions.
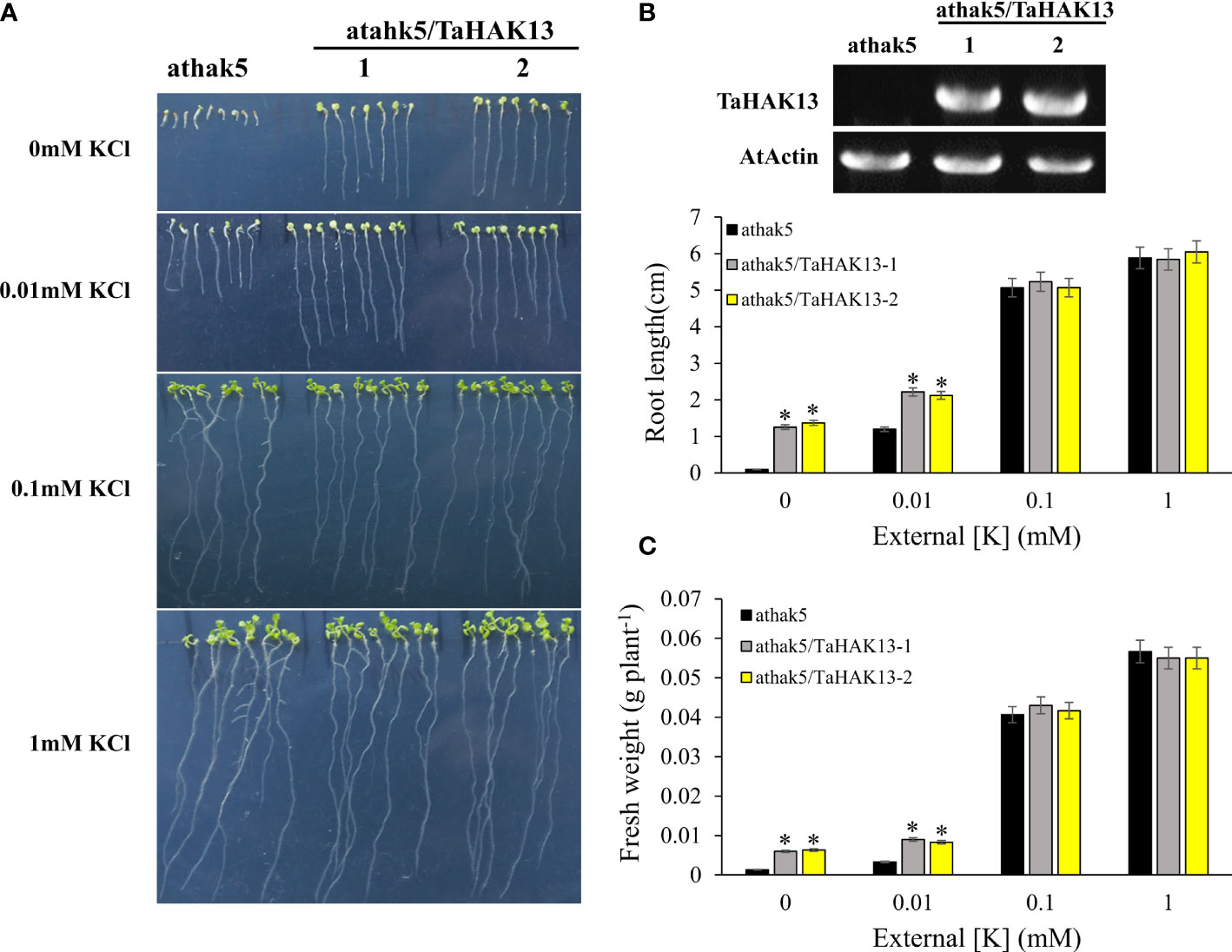
Figure 6 The expression of TaHAK13 rescued the sensitive phenotype of Arabidopsis mutant athak5 under low potassium condition. (A) The athak5 mutant and TaHAK13 transgenic lines (athak5/TaHAK13) were grown on MS media with different K+ concentrations for 10 d. (B) The expression of TaHAK13 in athak5 and the transgenic lines (athak5/TaHAK13) was analyzed using RT-PCR. (C) The root length in each bar represents the average root length of 20 seedlings from three independent experiments. (D) The plant fresh weight in each bar represents the average fresh weight of 20 seedlings from three independent experiments. The Student t-test (*p<0.05) was used to analyze the statistical significance.
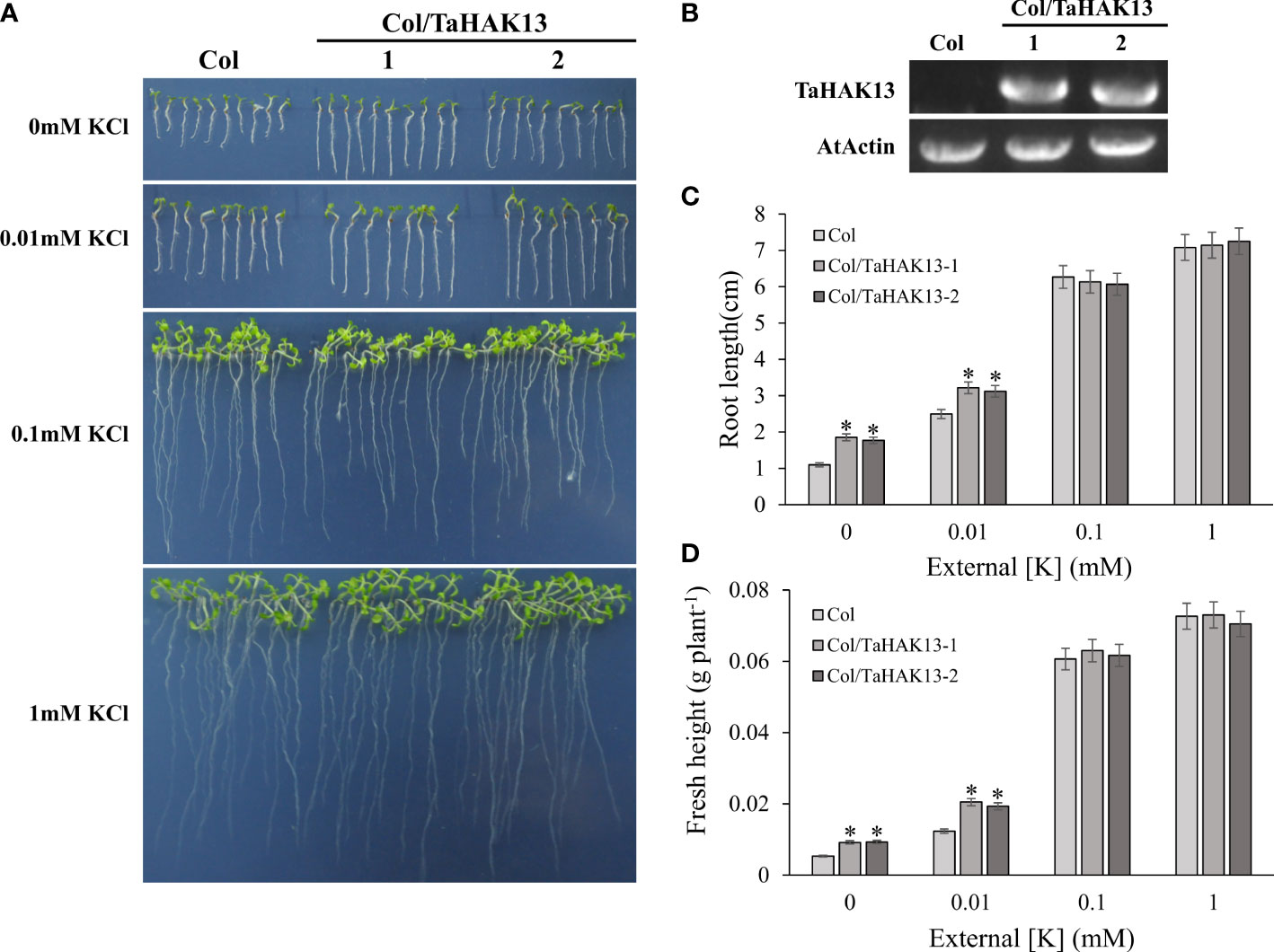
Figure 7 Functional verification of TaHAK13 in wild-type Arabidopsis thaliana. (A) Wild-type (Col) and transgenic lines (Col/TaHAK13) were grown on MS media with different K+ concentrations for 10 d. (B) The expression of TaHAK13 in Col and transgenic Arabidopsis thaliana was quantified via RT-PCR. (C) The root length of the plant in each bar represents the average root length of 20 seedlings from three independent experiments. (D) The fresh weight of plants in each bar represents the average fresh weight of 20 seedlings from three independent experiments. The Student t-test (*p<0.05) was used to analyze the statistical significance.
To determine whether TaHAK13 is needed for potassium acquisition in Arabidopsis roots in low potassium environments, the net K influx in seedling primary roots was measured using non-invasive micro-test technology (NMT) (Figure S6). The net K influx was then compared between the TaHAK13 complementary lines (athak5/TaHAK13) and overexpression lines (Col/TaHAK13) and their respective wild-types. In eight minutes of measurement, no differences were detected between the athak5/TaHAK13 lines and the athak5 mutant when seedlings were supplied with 0.1 mM K+ over the measurement period (Figures 8A, B). However, when the K+ concentration supplied was 0.01 mM, the net K influx was larger in athak5/TaHAK13 lines versus the athak5 mutant over the six minutes measurement (Figure 8C). On average, TaHAK13 expression in the athak5 mutant increased the net K influx about 3.5 times (Figure 8D). A similar pattern was seen for the Col/TaHAK13 lines versus wild-type. When 0.01 mM K+ was supplied, the wild-type had a much lower net K influx, only about 50% of that observed for the Col/TaHAK13 lines (Figures 8G, H). Increasing the K+ concentration from 0.01 to 0.1 mM dramatically augmented the net K influx in both Col/TaHAK13 lines and wild-type; over time, the K influx in the wild-type largely caught up, finishing only about 5% less than the Col/TaHAK13 influx rate (Figures 8E, F). Therefore, TaHAK13 is directly involved in the acquisition of root K+, especially in low potassium conditions.
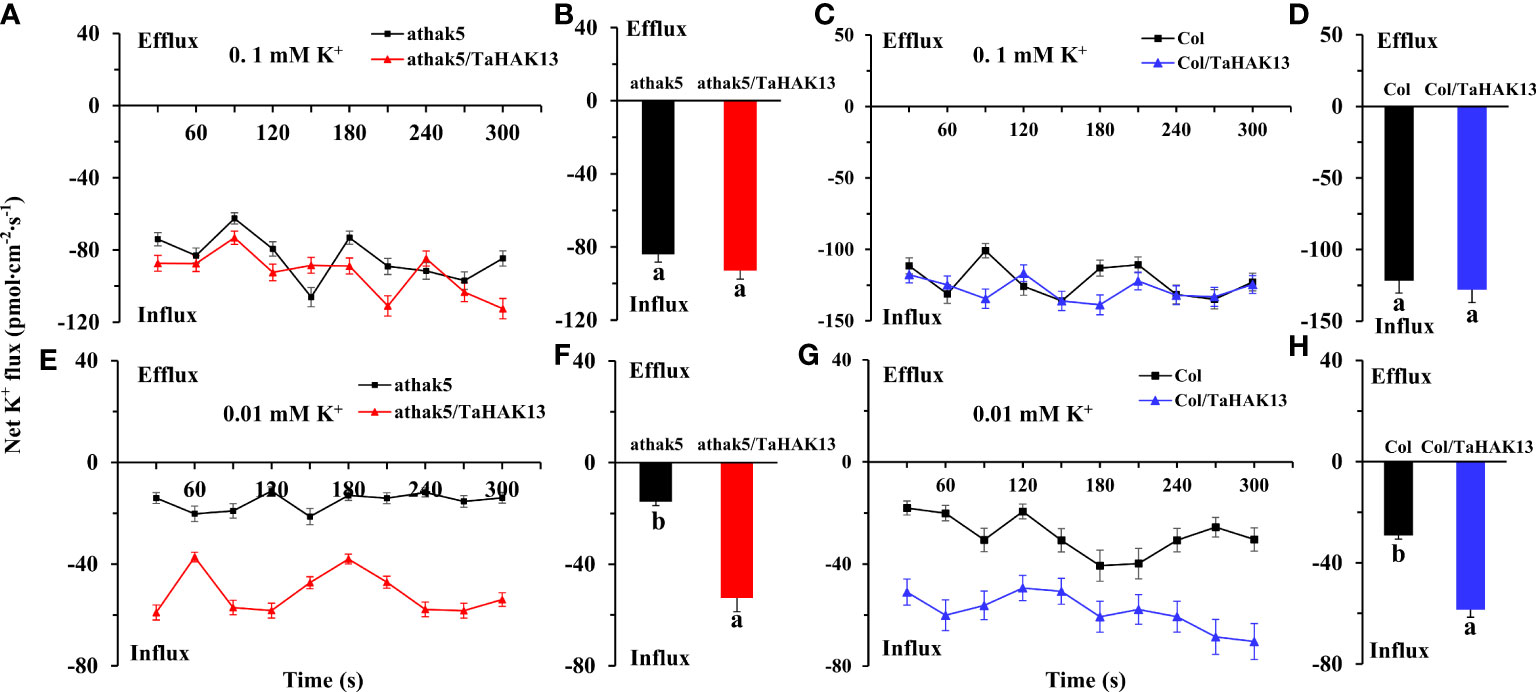
Figure 8 Effect of TaHAK13 expression on the net K influx in primary root meristem supplied with different concentrations of (K) (A, C) The net K influx of Arabidopsis thaliana mutant athak5 and its complementary line (athak5/TaHAK13) over 8 min (see ‘Materials and methods’). (B, D) Mean net K influx of athak5 and athak5/TaHAK13 when 0.1 mM K+ (A) or 0.01 mM K+ (C) was supplied; average values were taken over the whole eight minutes of influx data. (E, G) The net K influx of wild-type Arabidopsis thaliana (Col) and its overexpression line (Col/TaHAK13) over 8 min. (F, H) Mean net K influx of Col and Col/TaHAK13 when 0.1 mM K+ (A) or 0.01 mM K+ (C) was supplied; average values were taken over the whole eight minutes of influx data. Significant differences between transgenic lines and their respective non-transgenic lines are indicated by different letters.
The full-length TaHAK13 sequence was inserted into a pBT3-N vector for use as a bait vector to screen for protein interactions in wheat. Self-activation and toxicity tests revealed that the bait vector was normally expressed in a yeast system, was non-toxic to yeast, and had no self-activation, so it was used to subsequent screening experiments (Figure S7). After screening, five genes were identified; these genes were involved in many aspects of plant disease resistance, including signal transduction, stress resistance, and nutritional stress resistance (Table S4). The above five cDNA sequences were inserted into a prey vector (pPR3-N); the bait and prey vectors were then transformed into the yeast strain NMY51. NMY51 was grown on SD/-Leu/-Trp (DDO) and SD/-His/-Leu/-Trp/-Ade (QDO) medium for 3 d, and then X-Gal was used for identification. Except for the negative control, the positive control and the verified transformation solution both grew normally and turned blue on the QDO medium (Figure 9A). Interestingly, the genes TaNPF5.10 and TaNPF6.3 belong to the NPT/PTR family of wheat, which plays an important role in the absorption, transport, and distribution of nitrate in plant cells, tissues, and organs. Therefore, a luciferase complementation assay (LCA) was carried out to assess protein interactions. Only the co-transformed areas of TaHAK13 and TaNPF5.10 or TaNPF6.3 emitted fluorescence. The transformed empty vector (nLUC + cLUC) and other combinations (TaHAK13-nLUC + cLUC, nLUC + TaNPF5.10-cLUC, and nLUC + TaNPF6.3-cLUC) did not emit fluorescence (Figures 9B, C), suggesting that TaHAK13 interacts with both TaNPF5.10 and TaNPF6.3.
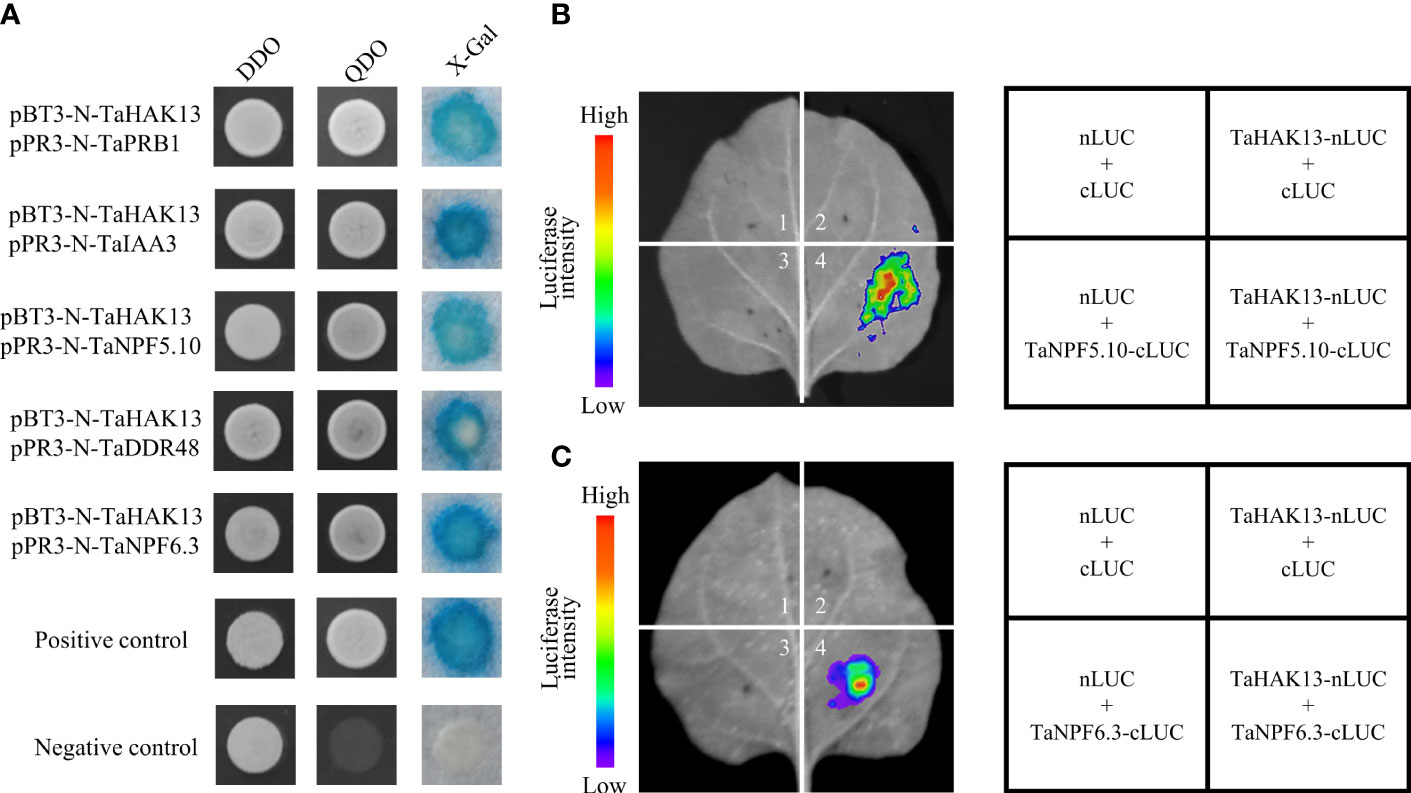
Figure 9 The interaction of TaHAK13 with TaNPF5.10 and TaNPF6.3. (A) The interaction of TaHAK13 with screened proteins was verified using a split-ubiquitin yeast two-hybrid system; different protein combinations were detected on appropriate screening media (SD/-L-T, SD/-A-/H/-L/-T, SD/-A/-H/-L/-T+20μg/mL X-Gal). (B) LCA of the interaction between TaHAK13 and TaNPF5.10. (C) LCA of TaHAK13 and TaNPF6.3. As shown in the graphics, various combinations were used to co-transform tobacco leaves. Luciferase signals were captured by a NightSHADE LB 985 plant in vivo imaging system.
Soil K+ concentrations are often low for plant growth and development, meaning that plants often experience low potassium stress (Wang and Wu, 2009). The KUP/HAK/KT family of potassium transporters constitutes the primary system for K+ uptake in plants under low-K+ concentrations. Many high affinity potassium transporter genes have been identified to date, such as AtHAK5 (Arabidopsis thaliana), GhHAK5 (Gossypium hirsutum), HvHAK1 (Hordeum vulgare), LeHAK5 (Lycopersicon esculentum), and OsHAK1 (Oryza sativa) (Santa-María et al., 1997; Bañuelos et al., 2002; Wang et al., 2002; Gierth et al., 2005; Chao et al., 2018). Studying these genes can provide a guide to the molecular mechanisms underlying K+ transport. In this study, a wheat gene that is homologous to AetHAK13 in Aegilops was identified and researched (Figures S3, S4).
To explore how TaHAK13 functions in plants, the TaHAK13 promoter was cloned from the hexaploid common wheat variety Yunong 804. The promoter sequence was found to contain a TATA-box and CAAT-box, two key components responsible for initiating and regulating transcription in eukaryotes (Luo et al., 2015). The TaHAK13 promoter also contained many photo-responsive elements, such as an ACE, G-box, GT1-motif, and others. Both the G-box (Heng et al., 2019) and GT1-motif (Zhao et al., 2012) are essential for genes to respond to light signals. In addition, the TaHAK13 promoter also contained a MYB binding site and a WRKY-binding W-box, so TaHAK13 expression is likely also regulated by upstream transcription factors (Table S3). Previous research has shown that HAK family genes, such as OsHAK21 (Shen et al., 2015), AtKUP7 (Han et al., 2016), SiHAK10 (Liu et al., 2019), and SiHAK20 (Wang et al., 2020), are mainly expressed in roots. The tissue-specificity of TaHAK13 was examined, and expression was found to be mainly focused in plant veins, stems, and root tips at the seedling stage; in mature plants TaHAK13 was highly expressed in the roots, suggesting that expression varied with developmental stage. Expression was higher in mature plants versus seedlings, consistent with the characteristics of most high affinity potassium transporters (Figure 4). Most HAK family genes belonging to cluster I are induced by low potassium stress, such as OsHAK5 (Yang et al., 2014), AtHAK5 (Nieves-Cordones et al., 2019), and OsHAK16 (Feng et al., 2019). GUS staining was stronger in low potassium conditions versus normal conditions, indicating higher gene expression level. In addition, OsHAK8 (part of cluster II) is also induced by low potassium stress (Wang et al., 2021). Therefore, the expression of TaHAK13 in plants seems to be affected by low potassium stress.
In rice, OsHAK1 improves the growth of yeast at KCl concentrations of 0.05-1 mM. Furthermore, for any K concentration, yeast expressing OsHAK1 is also more tolerant to salt stress (Chen et al., 2015). Compared to yeast transformed with an empty pYES2 vector, yeast strains expressing OsHAK16 can tolerate up to 200 mM NaCl (Feng et al., 2019). In addition, OsHAK16 expression improves the growth of the low potassium-sensitive yeast mutant R5421 at 0.1, 1, and 10 mM K+ supply rates. Collectively, these results suggest that OsHAK16 can enhance K+ absorption in yeast cells (Feng et al., 2019). In this study, TaHAK13, TaHAK1, and an empty vector were inserted into the yeast strain CY162. The CY162 strain transformed with an empty vector (p416) did not grow normally on low potassium medium. However, the addition of TaHAK1 or TaHAK13 restored the defective phenotype of CY162; these strains grew normally on AP medium with 1 mM KCl, suggesting that TaHAK13 plays an important function in K+ absorption (Figure 5). In previous studies, WΔ3 yeast cells expressing CaHAK1 can survive on the media with K+ concentrations of less than 1 μM, but this ability is inhibited by micromolar concentrations of (Martínez-Cordero et al., 2004). Under low potassium stress, yeast strains transformed with SiHAK1 show higher growth rates than positive controls (Zhang et al., 2018). In addition, the expression of HbHAK1 in yeast strain CY162 promotes K+ absorption when potassium levels are extremely low, and reduces sodium toxicity to support yeast cell survival under high salt stress (Zhang et al., 2020). Here, TaHAK13, TaSOS1, and an empty vector were transferred into the mutant AXT3K, which has high salt sensitivity. The TaHAK13 mutant exhibited poor growth under high sodium conditions, as did that with an empty vector; only the positive control (TaSOS1) grew well. Therefore, TaHAK13 does not transport Na+. Presumably, the main function of TaHAK13 is as a K+ transporter that shows Na+ sensitivity (Figure S2).
Arabidopsis thaliana mutants (athak5) showed serious growth defects when grown on MS medium without additional K+. The expression of short awn barley HbHAK1, millet SiHAK1, corn ZmHAK5, or rice OsHAK21 restored growth to similar levels as the wild-type under low potassium conditions (Shen et al., 2015; Zhang et al., 2018; Qin et al., 2019., Zhang et al., 2020). Loss of function of KUP7 and KUP9 in Arabidopsis thaliana results in short roots and yellow leaves on low potassium medium; the K+ absorption rate and potassium content in xylem sap also decreases (Han et al., 2016; Zhang et al., 2020). To assess whether TaHAK13 has a similar function in plants, TaHAK13 was introduced into the mutant athak5. Under low potassium conditions (0 mM KCl and 0.01 mM KCl), athak5 had short roots and yellow leaves, while the TaHAK13 transgenic strain did not show similar deficits: root length and fresh weight were significantly higher than in athak5 (Figure 6 and S5). These results indicate that TaHAK13 mediated the absorption and transportation of K+. In addition, the expression of some HAK/KUP/KT family genes can enhance plant salt tolerance. For example, oshak16 knockout lines show reduced K+ absorption and a lower K+/Na+ ratio, while OsHAK16 overexpression lines have higher K+ absorption and greater root-to-shoot transport, thus improving salt tolerance (Feng et al., 2019). Similarly, wild-type and zmhak4 knockout mutants do not differ under normal conditions, but under salt stress (100 mM NaCl), zmhak4 knockout mutants are about 15% smaller than wild-type controls, suggesting that ZmHAK4 promotes salt tolerance by maintaining a steady state (of Na+ and K+) and constant K+/Na+ ratio (Zhang et al., 2019). Here, the expression of TaHAK13 in Arabidopsis thaliana may improve the salt tolerance of plants.
The DUAL membrane system is a yeast two-hybrid system of membrane proteins mediated by split-ubiquitin. It provides a method of in vivo protein analysis different from the conventional yeast two-hybrid system, which makes it possible to analyze interactions among membrane proteins (Johnsson and Varshavsky, 1994). Here, TaHAK13 was inserted into a pBT3-N bait vector, and a wheat cDNA library was screened for proteins interacting with the protein encoded by this gene. Among the proteins identified, TaNPF5.10 and TaNPF6.3 had the strongest interactions with TaHAK13 (Figure 9), so we speculate that this interaction may co-regulate the absorption of K+ and by plants. In Arabidopsis thaliana, the nrt1.1 knockout mutant showed poor K+ absorption and root-shoot distribution, as well as growth stagnation when K+ is restricted. These K+ absorption-related interactions depend on H+ consumption mechanisms related to NRT1.1-mediated H+/ co-metabolism (Fang et al., 2020). The interactions between K+ and different N forms is realized by NRT1.5 modulation of root-derived ethylene signals that regulate K+ transport from root to shoot. upregulates the transcription activity of ET-insensitive 3 (EIN3) but inhibits the expression of NRT1.5. The addition of strongly inhibits the activity of EIN3, while upregulating the expression of AtNRT1.5 and increasing the K+ concentration (Chen et al., 2021). Plants can sense the ratio of K+/ in the soil, adjusting the K+/ transport ratio between roots and shoots to maintain a balance of these ions in their tissues. Transcription factor MYB59 aids in this process by regulating the transcription of AtNRT1.5/AtNPF7.3 in response to low potassium stress (Du et al., 2019). Studies have reported positive interactions between potassium and nitrogen in wheat, i.e., high potassium can alleviate ammonium nitrogen stress, improving growth by promoting the absorption of nutrients and the production of assimilation products (Guo et al., 2019). In this study, two genes were identified interacting with TaHAK13 using a MbY2H and a luciferase complementation assay (LCA). TaNPF5.10 and TaNPF6.3 belong to the NPT/PTR family in wheat; this family plays an important role in the absorption, transport, and distribution of nitrate nitrogen in plant cells, tissues, and organs. In addition, all three proteins were localized in the cell membrane and had a transmembrane domain (Figures S8, S9); therefore, TaHAK13 most likely interacts with TaNPF5.10 and TaNPF6.3 within the plasma membrane.
TaHAK13 is a member of cluster II that is widely distributed in plant tissues, including in roots, stems, leaves, and flowers. The members of cluster II not only participate in K+ absorption, but also play a role in growth regulation. Isotope 32P labeling has been used to analyze the expression of AtKUP1, AtKUP2, AtKUP3, and AtKUP4 in different tissues under normal potassium supply; all four genes were expressed in roots, leaves, and flowers, without tissue expression specificity (Kim et al., 1998). The HAK protein of cluster II plays a variety of physiological roles in plants, such as promoting K+ absorption, maintaining intracellular K+ concentrations, and participating in cell expansion and growth. It indirectly affects the transmembrane transport of root auxin by regulating the intracellular H+ content, thus initiating the development of root hairs and the geotropism of roots (Rodrίguez-Navarro, 2000). Therefore, TaHAK13 may also participate in K+ absorption and cell expansion and growth, but further research is required to verify this hypothesis.
In this study, the TaHAK13 gene was cloned from wheat and its function characterized. RT-qPCR showed that TaHAK13 expression was up-regulated under drought, low potassium, and salt stress. GUS staining indicated that TaHAK13 was mainly expressed in the leaf veins, stems, and root apex in Arabidopsis thaliana, and expression varied with developmental stage. The subcellular localization analysis illustrated that TaHAK13 was located to the cytoplasmic membrane. In yeast and Arabidopsis, the overexpression of TaHAK13 improved their ability to absorb K+ under low potassium condition, but did not have the ability to transport Na+. Membrane-based yeast two-hybrid (MbY2H) and luciferase complementation assays (LCA) showed that TaHAK13 interacted with TaNPF5.10 and TaNPF6.3 proteins. Overall, our study revealed the role of TaHAK13 in plants and the mechanism of low potassium tolerance in plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YR and HX conceived and designed the experiments. YR, XC, WD, YD, YZ, and BL performed the experiments. XC and TL analyzed the data. YR wrote the manuscript. HX revised the article. All authors read and approved the submitted version.
This work was funded by Henan Provincial Science and Technology Research Project (221100110300) and Natural Science Foundation of Henan Province (202300410217). These funding bodies neither influenced the selected experiments of this study nor their design.
We thank Professor Sheng Luan and Dr Renjie Tang from University of California, Berkeley for kindly offering the yeast strain CY162.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1103235/full#supplementary-material
Supplementary Figure 1 | Real-time quantitative PCR expression of TaHAK13 in wheat roots under normal conditions.
Supplementary Figure 2 | Salt sensitivity of transgenic TaHAK13 yeast. The mutant AXT3K grew on AP-U solid media with different concentrations of NaCl. AXT3K strain was transformed into empty vector (EV) or TaHAK13 and positive control TaSOS1. Each strain was diluted continuously and dropped on agar plate for culture.
Supplementary Figure 3 | Amino acid sequence alignment of HAK13-like K+ transporters from different plant species. The amino acid sequences of nine K+ transporters were aligned using DNAMAN software. These transporters included TaHAK13 from wheat, AtKUP9, AtKUP10 and AtKUP13 from Arabidopsis, AetHAK13 from Aegilops, HvHAK13 from barley, OsHAK13 from rice, PvHAK13 from switchgrass, SbHAK13 from sorghum, BdHAK13 from brachypodium distachyon, PhaHAK13 from Panicum hallii, SiHAK13 from millet, SvHAK13 from Green bristlegrass.
Supplementary Figure 4 | Phylogenetic analysis of HAK13-like K+ transporters. phylogenetic tree was constructed by MEGA5.0 software using the neighbor-joining method, and statistical support for the nodes was assessed by bootstrap analysis (1000 replicates).
Supplementary Figure 5 | Growth condition of athak5 mutant and TaHAK13 transgenic lines under normal and low potassium conditions. (A) athak5 mutant and TaHAK13 transgenic lines grew on MS medium and low potassium media for ten days. (B) The plant survival rate of athak5 mutant and TaHAK13 transgenic lines on MS medium and low potassium media.
Supplementary Figure 6 | Measurement chart of net K+ flux in Arabidopsis roots. On the left side, the root zone of Arabidopsis, and on the right side, the flow sensor (3-5 μm) is used to test the root zone of Arabidopsis (150 μm from the root tip), and each planet is measured for eight minutes.
Supplementary Figure 7 | MbY2H screening using TaHAK13 as a bait. (A) Schematic diagram of MbY2H screening. The cDNA library developed using the total RNAs of low potassium stressed wheat roots was screened using TaHAK13-pBT3-N as bait. (B) Self-activation and toxicity tests of bait vector TaHAK13-pBT3-N. (C) PCR analysis of positive clones. PCR amplification was performed for the positive clones using the primers pPR3-N-F and pPR3-N-R (Table S2).
Supplementary Figure 8 | Predicted transmembrane domains of TaNPF5.10 and TaNPF6.3.
Supplementary Figure 9 | Subcellular localization of TaNPF5.10 and TaNPF6.3 in tobacco leaves. 35S-GFP acted as the control.
Supplementary Table 1 | Improved Hogland nutrient solution. Note: (1) Low potassium stress: the final concentration of K+ was 1 × 10-5mol·L-1, and K2SO4 was used to add K+. (2) Salt stress: NaCl with final concentration of 200 mM was added to the above nutrient solution. (3) Drought stress (20% PEG6000): PEG6000 with a final concentration of 20% was added to the above nutrient solution.
Supplementary Table 2 | Primer sequences used in this study.
Supplementary Table 3 | Analysis of cis-elements in the promoters of TaHAK13.
Supplementary Table 4 | Functional analysis of TaHAK13 interacting protein.
Ahn, S. J., Shin, R., Schachtman, D. P. (2004). Expression of KT/KUP genes in arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134, 1135–1145. doi: 10.1104/pp.103.034660
Alemán, F., Nieves-Conlones, M., Martínez, V., Rubio, F. (2011). Root K+ acquisition in plants: The arabidopsis thaliana model. Plant Cell Physiol. 52, 1603–1612. doi: 10.1093/pcp/pcr096
Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., Gaber, R. F. (1992). Functional expression of a probable arabidopsis thaliana potassium channel in saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 89, 3736–3740. doi: 10.1073/pnas.89.9.3736
Armengaud, P., Breitling, R., Amtmann, A. (2004). The potassium-dependent transcriptome of arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136, 2556–2576. doi: 10.1104/pp.104.046482
Bañuelos, M. A., Garciadeblas, B., Cubero, B., Rodríguez-Navarro, A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130, 784–795. doi: 10.1104/pp.007781
Chao, M., Wen, Q., Zhang, Z., Hu, G., Zhang, J., Wang, G., et al. (2018). Sequence characteristics and expression analysis of potassium transporter gene GhHAK5 in upland cotton (Gossypium hirsutum L.). Acta Agron. Sin. 44, 236–244. doi: 10.3724/SP.J.1006.2018.00236
Cheng, X., Liu, X., Mao, W., Zhang, X., Chen, S., Zhan, K., et al. (2018). Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L). Int. J. Mol. Sci. 19, 3969. doi: 10.3390/ijms19123969
Chen, G., Hu, J., Lian, J., Zhang, Y., Zhu, L., Zeng, D., et al. (2019). Functional characterization of OsHAK1 promoter in response to osmotic/drought stress by deletion analysis in transgenic rice. Plant Growth Regul. 88, 241–251. doi: 10.1007/s10725-019-00504-3
Chen, G., Hu, Q., Luo, L., Yang, T., Zhang, S., Hu, Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. doi: 10.1111/pce.12585
Chen, H., Zhang, Q., Wang, X., Zhang, J., Ismail, A. M., Zhang, Z. (2021). Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell Environ. 44, 3806–3818. doi: 10.1111/pce.14182
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 323–324. doi: 10.1104/pp.107.111740
Du, X., Wang, F., Li, H., Jing, S., Yu, M., Li, J., et al. (2019). The transcription factor MYB59 regulates K+/ translocation in the arabidopsis response to low k+ stress. Plant Cell. 31, 699–714. doi: 10.1105/tpc.18.00674
Fang, X., Liu, X., Zhu, Y., Ye, J., Jin, C. (2020). The K+ and interaction mediated by NITRATE TRANSPORTER 1.1 ensures better plant growth under K+-limiting conditions. Plant Physiol. 184, 1900–1916. doi: 10.1104/pp.20.01229
Feng, H., Tang, Q., Cai, J., Xu, B., Xu, G., Yu, L. (2019). Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 250, 549–561. doi: 10.1007/s00425-019-03194-3
Gajdanowicz, P., Michard, E., Sandmann, M., Rocha, M., Corrêa, L. G. G., Ramírez-Aguilar, S. J., et al. (2011). Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc. Natl. Acad. Sci. U. S. A. 108, 864–869. doi: 10.1073/pnas.1009777108
Gierth, M., Mäser, P., Schroeder, J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 k+ channel contribution to K+ uptake kinetics in arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Guo, J., Jia, Y., Chen, H., Zhang, L., Yang, J., Zhang, J., et al. (2019). Growth, photosynthesis, and nutrient uptake in wheat are affected by differences in nitrogen levels and forms and potassium supply. Sci. Rep. 9, 1248. doi: 10.1038/s41598-018-37838-3
Han, M., Wu, W., Wu, W., Wang, Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in arabidopsis root under K+-limited conditions. Mol. Plant 9, 437–446. doi: 10.1016/j.molp.2016.01.012
Heng, Y., Lin, F., Jiang, Y., Ding, M., Yan, T., Lan, H., et al. (2019). B-box containing proteins BBX30 and BBX31, acting downstream of hy5, negatively regulate photomorphogenesis in arabidopsis. Plant Physiol. 180, 497–508. doi: 10.1104/pp.18.01244
Horie, T., Sugawara, M., Okada, T., Taira, K., Kaothien-Nakayama, P., Katsuhara, M., et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 111, 346–356. doi: 10.1016/j.jbiosc.2010.10.014
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Johnsson, N., Varshavsky, A. (1994). Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91, 10340–10344. doi: 10.1016/s0076-6879(05)99050-2
Karabegov, M. (2011). Flame photometers. basic parameters and metrological support. Meas. Tech. 54, 735–742. doi: 10.1007/s11018-011-9796-7
Kim, E. J., Kwak, J. M., Uozumi, N., Schroeder, J. I. (1998). AtKUP1: an arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell. 10, 51–62. doi: 10.2307/3870628
Liu, J., Liu, J., Liu, J., Cui, M., Huang, Y., Tian, Y., et al. (2019). The potassium transporter SiHAK10 is involved in mycorrhizal potassium uptake. Plant Physiol. 180, 465–479. doi: 10.1104/pp.18.01533
Livaka, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Y., Li, J., Zhang, S., Du, K. (2015). Research progress on plant promoter. North. Hortic. 22, 186–189. doi: 10.11937/bfyy.201522048
Maathuis, F. J. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12, 250–258. doi: 10.1016/j.pbi.2009.04.003
Mao, P., Run, Y., Wang, H., Han, C., Zhang, L., Zhan, K., et al. (2022). Genome-wide identification and functional characterization of the chloride channel TaCLC gene family in wheat (Triticum aestivum l.). Front. Genet. 13. doi: 10.3389/fgene.2022.846795
Martínez-Cordero, M. A., Martínez, V., Rubio, F. (2004). Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol. Biol. 56, 413–421. doi: 10.1007/s11103-004-3845-4
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646
Nieves-Cordones, M., Lara, A., Ródenas, R., Amo, J., Rivero, R. M., Martínez, V., et al. (2019). Modulation of K+ translocation by AKT1 and AtHAK5 in arabidopsis plants. Plant Cell Environ. 42, 2357–2371. doi: 10.1111/pce.13573
Okada, T., Yamane, S., Yamaguchi, M., Kato, K., Shinmyo, A., Tsunemitsu, Y., et al. (2018). Characterization of rice KT/HAK/KUP potassium transporters and K+ uptake by HAK1 from oryza sativa. Plant Biotechnol. 35, 101–111. doi: 10.5511/plantbiotechnology.18.0308a
Pyo, Y. J., Gierth, M., Schroeder, J. I., Cho, M. H. (2010). High-affinity K+ transport in arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 153, 863–875. doi: 10.1104/pp.110.154369
Qin, Y., Wu, W., Wang, Y. (2019). ZmHAK5 and ZmHAK1 function in K+ uptake and distribution in maize under low k+ conditions. J. Integr. Plant Biol. 61, 691–705. doi: 10.1111/jipb.12756
Quintero, F. J., Blatt, M. R., Pardo, J. M. (2000). Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 471, 224–228. doi: 10.1016/s0014-5793(00)01412-5
Rodríguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469, 1–30. doi: 10.1016/s0304-4157(99)00013-1
Rubio, F., Nieves-Cordones, M., Alemán, F., Martínez, V. (2008). Relative contribution of AtHAK5 and AtAKT1 to k+ uptake in the high-affinity range of concentrations. Physiol. Plant 134, 598–608. doi: 10.1111/j.1399-3054.2008.01168.x
Santa-María, G. E., Rubio, F., Dubcovsky, J. (1997). A rodríguez-Navarro, the HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 9, 2281–2289. doi: 10.1105/tpc.9.12.2281
Senn, M. E., Rubio, F., Bañuelos, M. A., Rodríguez-Navarro, A. (2001). Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J. Biol. Chem. 276, 44563–44569. doi: 10.1074/jbc.m108129200
Shen, Y., Shen, L., Shen, Z., Jing, W., Ge, H., Zhao, J., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 38, 2766–2779. doi: 10.1111/pce.12586
Shin, R., Schachtman, D. P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. U. S. A. 101, 8827–8832. doi: 10.1073/pnas.0401707101
Song, Y., Dong, L., Jin, Y., Shi, S., Zhang, L., Liu, C., et al. (2014). Subcellular localization and expression analysis of nicotiana sylvestris KUP/HAK/KT family K+ transporter gene NsHAK11. Sci. Agric. Sin. 47, 1058–1071. doi: 10.3864/j.issn.0578-1752.2014.06.003
Stagljar, I., Korostensky, C., Johnsson, N., Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 95, 5187–5192. doi: 10.1073/pnas.95.9.5187
Takahashi, R., Nishio, T., Ichizen, N., Takano, T. (2007). High-affinity k+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows na+ permeability under NaCl stress. Plant Cell Rep. 26, 1673–1679. doi: 10.1007/s00299-007-0364-1
Vastermark, A., Wollwage, S., Houle, M. E., Rio, R., Saier, M. H., Jr. (2014). Expansion of the APC superfamily of secondary carriers. Proteins: Struct. Funct. Bioinf. 82, 2797–2811. doi: 10.1002/prot.24643
Wang, Y., Chen, Y., Wu, W. (2021). Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52. doi: 10.1111/jipb.13053
Wang, Y., Garvin, D. F., Kochian, L. V. (2002). Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots, evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 130, 1361–1370. doi: 10.1104/pp.008854
Wang, Y., He, L., Li, H., Xu, J., Wu, W. (2010). Potassium channel α-subunit AtKC1 negatively regulates AKT1-mediated k+ uptake in arabidopsis roots under low-k+ stress. Cell Res. 20, 826–837. doi: 10.1038/cr.2010.74
Wang, Z., Hong, Y., Zhu, G., Li, Y., Niu, Q., Yao, J., et al. (2020). Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J. 39, e103256. doi: 10.15252/embj.2019103256
Wang, X., Li, J., Pan, Y., Cai, D., Mao, D., Chen, L., et al. (2021). Rice potassium transporter OsHAK8 mediates K+ uptake and translocation in response to low K+ stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.730002
Wang, Y., Wu, W. (2009). Molecular genetic mechanism of high efficient potassium uptake in plants. Chin. Bull. Bot. 44, 27–36. doi: 10.3969/j.issn.1674-3466.2009.01.003
Wang, F., Zhang, N., Guo, Y., Gong, B., Li, J. (2020). Split nano luciferase complementation for probing protein-protein interactions in plant cells. J. Integr. Plant Biol. 62, 1065–1079. doi: 10.1111/jipb.12891
Xu, H., Jiang, X., Zhan, K., Cheng, X., Chen, X., Pardo, J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473, 8–15. doi: 10.1016/j.abb.2008.02.018
Yang, Z., Gao, Q., Sun, C., Li, W., Gu, S., Xu, C. (2009). Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa l.). J. Genet. Genomics 36, 161–172. doi: 10.1016/s1673-8527(08)60103-4
Yang, T., Zhang, S., Hu, Y., Wu, F., Hu, Q., Chen, G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166, 945–959. doi: 10.1104/pp.114.246520
Zhang, M., Huang, P., Ji, Y., Wang, S., Wang, S., Li, Z., et al. (2020). KUP9 maintains root meristem activity by regulating K+ and auxin homeostasis in response to low K. EMBO Rep. 21, e50164. doi: 10.15252/embr.202050164
Zhang, M., Liang, X., Wang, L., Cao, Y., Song, W., Shi, J., et al. (2019). A HAK family na+ transporter confers natural variation of salt tolerance in maize. Nat. plants. 5, 1297–1308. doi: 10.1038/s41477-019-0565-y
Zhang, H., Xiao, W., Yu, W., Jiang, Y., Li, R. (2020). Halophytic hordeum brevisubulatum HbHAK1 facilitates potassium retention and contributes to salt tolerance. Int. J. Mol. Sci. 21, 5292. doi: 10.3390/ijms21155292
Zhang, H., Xiao, W., Yu, W., Yao, L., Li, L., Wei, J., et al. (2018). Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low k+ or salt stress. Plant Cell Rep. 37, 1533–1546. doi: 10.1007/s00299-018-2325-2
Zhang, Z., Zhang, J., Chen, Y., Li, R., Wang, H., Wei, J. (2012). Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 39, 8465–8473. doi: 10.1007/s11033-012-1700-2
Keywords: Wheat (Triticum aestivum L.), TaHAK13, Low potassium stress, function characterization, interaction protein
Citation: Run Y, Cheng X, Dou W, Dong Y, Zhang Y, Li B, Liu T and Xu H (2022) Wheat potassium transporter TaHAK13 mediates K+ absorption and maintains potassium homeostasis under low potassium stress. Front. Plant Sci. 13:1103235. doi: 10.3389/fpls.2022.1103235
Received: 20 November 2022; Accepted: 09 December 2022;
Published: 23 December 2022.
Edited by:
Mokhtar Guerfel, Shaqra University, Saudi ArabiaReviewed by:
Yibing Hu, Nanjing Agricultural University, ChinaCopyright © 2022 Run, Cheng, Dou, Dong, Zhang, Li, Liu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haixia Xu, aGF1eGh4QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.