
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 January 2023
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1102196
This article is part of the Research Topic Crop Improvement by Omics and Bioinformatics View all 17 articles
 Jun Zhang1,2†
Jun Zhang1,2† Peng Wu1†
Peng Wu1† Ning Li1†
Ning Li1† Xiaolan Xu1
Xiaolan Xu1 Songxin Wang1
Songxin Wang1 Siyuan Chang1
Siyuan Chang1 Yuping Zhang1
Yuping Zhang1 Xingxing Wang1
Xingxing Wang1 Wangshu Liu2
Wangshu Liu2 Yizan Ma1
Yizan Ma1 Hakim Manghwar1,3
Hakim Manghwar1,3 Xianlong Zhang3
Xianlong Zhang3 Ling Min1*
Ling Min1* Xiaoping Guo1*
Xiaoping Guo1*Although conventional hybrid breeding has paved the way for improving cotton production and other properties, it is undoubtedly time and labor consuming, while the cultivation of male sterile line can fix the problem. Here, we induced male sterile mutants by simultaneously editing three cotton EXCESS MICROSPOROCYTES1 (GhEMS1) genes by CRISPR/Cas9. Notably, the GhEMS1 genes are homologous to AtEMS1 genes, which inhibit the production of middle layer and tapetum cells as well, leading to male sterility in cotton. Interestingly, there are necrosis-like dark spots on the surface of the anthers of GhEMS1s mutants, which is different from AtEMS1 mutant whose anther surface is clean and smooth, suggesting that the function of EMS1 gene has not been uncovered yet. Moreover, we have detected mutations in GhEMS1 genes from T0 to T3 mutant plants, which had necrosis-like dark spots as well, indicating that the mutation of the three GhEMS1 genes could be stably inherited. Dynamic transcriptomes showed plant hormone pathway and anther development genetic network were differential expression in mutant and wild-type anthers. And the lower level of IAA content in the mutant anthers than that in the wild type at four anther developmental stages may be the reason for the male sterility. This study not only facilitates the exploration of the basic research of cotton male sterile lines, but also provides germplasms for accelerating the hybrid breeding in cotton.
Male sterility is an important tool for the utilization of heterosis such as increasing cotton production and quality with less labor and time. For now, artificial emasculation is still the dominant method used for the production of cotton hybrids in China (Yang et al., 2018). However, the cost of hybrid breeding has been increasing year by year due to the shortage of rural labor, resulting in dramatic decreases of production in the planting area of hybrid cotton (Yang et al., 2018; Zheng et al., 2021). In this way, the creation of male sterile lines is a new breakthrough for the acquisition of hybrid seedlings.
Recently, the CRISPR/Cas9 technology has been widely used in gene editing and the plants acquired can be used for hybrid seed production. Due to its precision, simple operation and high efficiency, the CRISPR/Cas9 technology has been applied for a variety of species such as maize, wheat, soybean and rice (Chen et al., 2018; Ma et al., 2019; Okada et al., 2019; Chen et al., 2021). Novel “transgene clean” thermo-sensitive genic male sterility (TGMS) lines have been created on the basis of the induced specific mutations in TMS5 with the CRISPR/Cas9 technology. To test the combinatorial capacity of the obtained new male sterile mutants, the rice TMS5 mutants have crossed with other lines and found that the offspring have better phenotypes and provide higher yields (Zhou et al., 2016). In addition, using the CRISPR/Cas9 technology to target ABORTED MICROSPORES (AMS) congeners in soybeans to produce stable male sterility lines. Furthermore, they have eventually figured out that the editing of GmAMS1 is related to not only the formation of the pollen wall but also the degradation of the tapetum (Chen et al., 2021). Ramadan et al. have successfully generated a wide scale of genotypically and phenotypically mutagenesis using CRISPR/Cas9 mediated pooled sgRNAs assembly, paving the way for creation of cotton male sterile lines (Ramadan et al., 2021).
As shown in the anther and pollen-related gene regulatory network diagram (Wilson and Zhang, 2009), the early anther cell differentiation gene EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) encodes leucine-rich repeat (LRR) receptor kinase which is located on the cell membrane, and the protein is expressed in primary cell wall and tapetum cells. Hence, mutations in EMS1/EXS gene is related to the absence of tapetum in Arabidopsis thaliana, eventually resulting in pollen abortion (Canales et al., 2002). Moreover, the MULTIPLE SPOROCYTE (OsMSP1) gene in rice is homologous to the AtEMS1 genes, which also encodes LRR receptor kinases as well, and the OsMSP1 mutant exhibits a highly similar phenotypes to the Arabidopsis EMS1/EXS mutant (Ken-Ichi Nonomura et al., 2003). Therefore, the EMS1 is an important candidate gene for obtaining male sterile lines. At the same time, we also found that the expression of GhEMS1 was affected in the high temperature sensitive line by high temperature. Here, we have created a complete male sterile line by knocking out the GhEMS1s using CRISPR/Cas9 technology. Through the comparison of pollen fertility and tapetum development of edited male sterile lines, the most suitable mutant was identified. This study not only facilitates the exploration of the basic research of cotton sterile lines, but also provides germplasms for accelerating the hybrid breeding using male sterile lines in cotton.
Jin668, an upland cotton (Gossypium hirsutum L.) line developed by the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University. We have described the transformation system of Jin668 previously (Jin et al., 2006; Li et al., 2019). The wild-type (negative control) and transgenic lines were planted in Wuhan, Hubei under normal farming practices or grown in the greenhouse during the winter in 2018 - 2020. The greenhouses were kept at a temperature of 28–35/20–28°C day/night.
We conducted a genome-wide assessment and chose sgRNA through the CRISPR-P 2.0 (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR) (Liu et al., 2017). The process of vector construction refers to our previous report (Wang et al., 2018). The different vectors were transformed into Agrobacterium GV3101 which then was transformed into cotton Jin668. Refer to published articles for cotton transgene (Jin et al., 2006).
In order to detect the editing efficiency of transgenic lines, the targeted genomic DNA was amplified by PCR with a pair of site-specific primers at the 5’ end with common bridging sequences. Specific steps were similar to previous reports (Liu et al., 2019; Ramadan et al., 2021), and the primers used ware shown in Supplementary Table 1. PCR products were sequenced on Illumina HiSeq platform (Illumina, USA) after recovery. Hi-TOM website (http://www.hi-tom.net/hi-tom/) was used to analyze the sequencing results. In order to detect the off-target situation in the transgene lines editing process, “sgRNAcas9_3.0.5” (Xie et al., 2014) was used to predict off-target sites, following the software default settings. The “extract_targetSeq.pl” script was used in the software package to extract the flanking sequence of the off-target site on the genome. We designed off-target site primers in batches through the “batchprimer3” website (http://batchprimer3.bioinformatics.ucdavis.edu), PCR products were sequenced on Illumina HiSeq platform (Illumina, USA). “CRISPResso2” (Clement et al., 2019) was employed for sequencing results to analyze the off-target sites. The off-target sites and sequences are shown in Supplementary Table 2, 3. The primers for amplification of off-target sites are shown in Supplementary Table 1.
To detect pollen viability, the anther at 0 days post-anthesis (DPA) of WT and mutants was immersed in 2,3,5-Triphenyl tetrazolium chloride (TTC) solution (8 g TTC dissolved in 1 L phosphate buffer) according to a previous report (Min et al., 2013). After being cultured in a 37°C incubator for 30 min, the staining reaction was terminated with 2% (v/v) sulfuric acid solution. Pollen grains were placed on a microscope slide and the Zeiss (Oberkochen, Germany) Axio Scope A1 microscope was used to collect images.
In polyacrylamide gel electrophoresis (PAGE) separations, the 8% non-denaturing polyacrylamide gel (acrylamide: methylene bisacrylamide = 29:1) containing the PCR amplification products were placed in the electrophoresis chamber, and the driving force was set to 60 W. After 1 hour, the sample products were immersed in 0.2% silver nitrate solution for 10 minutes. After that, the products were washed twice with ddH2O and then put them in the chromogenic solution (1.5% sodium hydroxide, 0.4% formaldehyde) for 5 minutes. Finally, protein band patterns could be visualized and subjected to adequate analysis.
Anthers from transgenic lines and wild-type at different developmental stages were immersed in 50% FAA (50% ethanol, 5% propionic acid, and 3.7% formaldehyde) and vacuum infiltrated for 2 h at 4°C, and placed at 4°C for 24 h to fix the tissue. For dehydration, a graded ethanol series (50, 70, 80, 95, and 100%) was used and samples were embedded in paraffin. The embedded tissues were sectioned into 10 μm sections. Anther sections were stained with toluidine blue solution (1%) and the Zeiss (Oberkochen, Germany) Axio Scope A1 microscope was used to collect images. TUNEL detection of apoptosis was performed similar to previous report (Min et al., 2013). Paraffin sections of the anthers were used for TUNEL analysis of the fragmented DNA of apoptotic cells using the DeadEnd™ Fluorometric TUNEL System (G3250, Promega). The analytical wavelengths of fluorescein and propidium iodide were 520 ± 10 nm and 640 ± 10 nm by a confocal microscope (TCS SP2; Leica), respectively.
Anthers from transgenic lines and wild-type were sampled and total RNA was extracted. The library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. Raw data of fastq format were firstly processed through FastQC (Andrianov et al., 2010). Paired-end clean reads were aligned to the G. hirsutum genome using Hisat2 v2.0.5 (Kim et al., 2015). FeatureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene (Liao et al., 2014). And then fragments per kilobase of exon model per million mapped fragments of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1) (Love et al., 2014). Genes with Padj <0.05 and |log2FoldChange| >1 were assigned as differentially expressed. The GO enrichment was performed by the R package ‘clusterProfiler’ (Yu et al., 2012).
Extraction and measurement of the endogenous IAA were as described by Miao et al. (Miao et al., 2019). Three replicates, each of 100 mg of anthers from transgenic lines and wild-type, were sampled at anther developmental stage 6, 7, 9 and 10, mixed with 750 μL of ice-cold 80% methanol containing 2H5-IAA (OlChemlm Ltd, CAS: 76934-78-5, 10 ng ml-1) as internal standard, and shake for 16 hours in the dark at 4°C. After centrifugation at 13,000 rpm for 5 minutes, the supernatant was dried with nitrogen, and the residue was reconstituted in 300 μL of 80% methanol. Finally, the IAA content was measured using an Agilent 4000Q-TRAP HPLC-MS system.
The Arabidopsis EXCESS MICROSPOROCYTES1 (AT5G07280, AtEMS1) controls somatic and reproductive cell development in anthers (Zhao et al., 2002). To determine whether the EMS1 gene participated in the reproductive cell development in cotton, we used AtEMS1 as a query to perform BLASTP searches and identified 11 EMS1 members in G. hirsutum. To get a better understanding of the phylogenetic relationships between EMS1, a phylogenetic tree was constructed based on these 11 G. hirsutum EMS1 and AtEMS1 protein sequences. Clearly, the EMS1s were classified into four branches, the Ghir_A08G010860 (GhEMS1_A08), Ghir_D08G010810 (GhEMS1_D08), and Ghir_A09G018830 (GhEMS1_A09) in the same branch with AtEMS1 (Figure 1A). GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09 have leucine rich repeat N-terminal domain and leucine-rich repeat sequences (Supplementary Figure 1). This result indicated that the three GhEMS1 genes may have similar functions with AtEMS1.
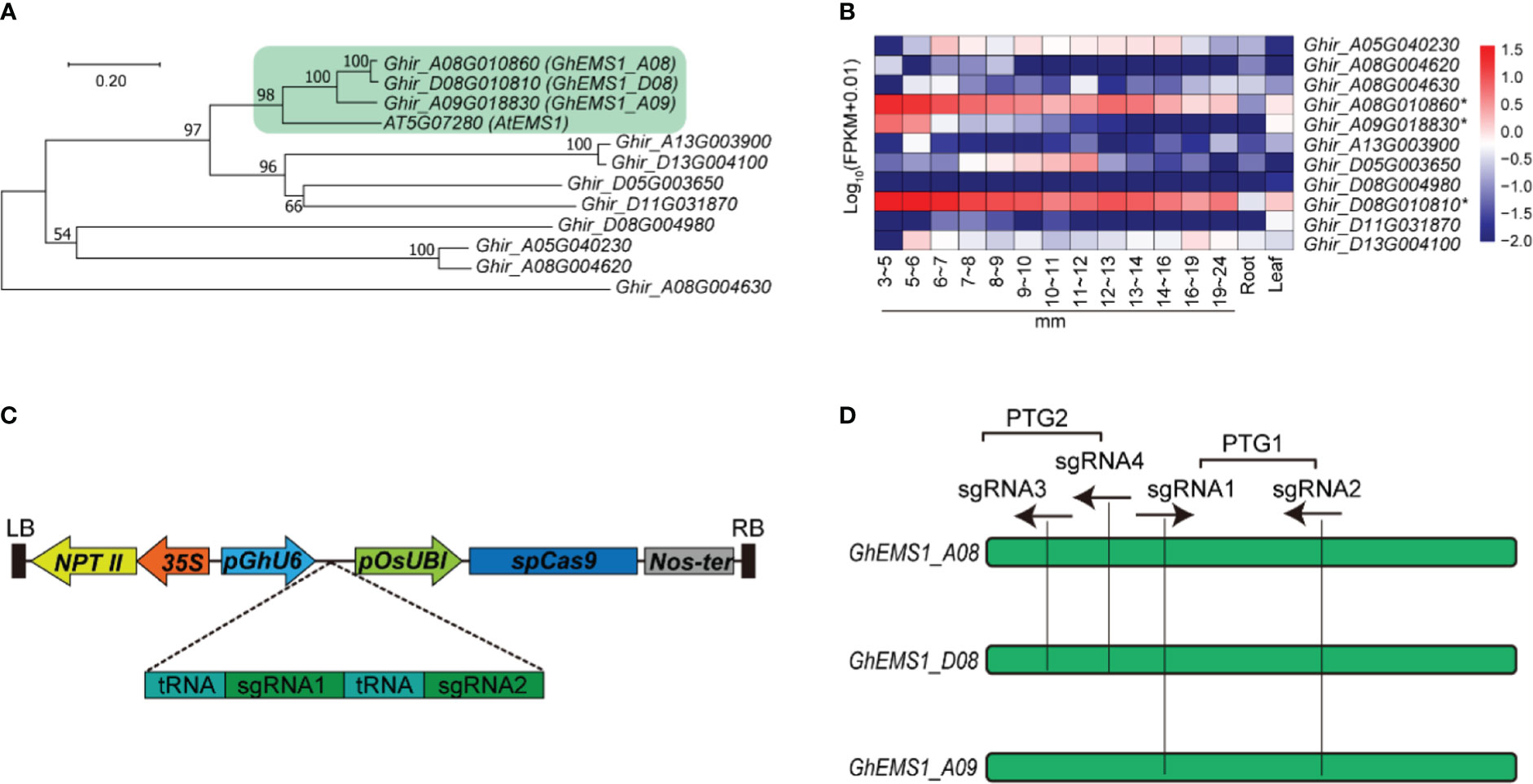
Figure 1 Creation of GhEMS1 genes mutants by using CRISPR/Cas9. (A) A phylogenetic tree for EMS1 genes in Gossypium hirsutum and AtEMS1 was constructed using the neighbor-joining method in MEGA 10.1.8 followed by bootstrapping with 1,000 replicates; (B) Expression profiles of GhEMS1 genes in flower buds of different lengths (3~5 mm, 5~6 mm, 6~7 mm, 7~8 mm, 8~9 mm, 9~10 mm, 10~11 mm, 11~12 mm, 12~13 mm, 13~14 mm, 14~16 mm, 16~19 mm, 19~24 mm)in G. hirsutum H05 determined by transcriptome sequencing (Zhang et al., 2022); (C) Creation of a male-sterile line pool by CRISPR/Cas9. Two sgRNAs were serially connected to each vector to increase the knockout efficiency; (D) Two pairs of sgRNAs on exons were designed and vectors containing polycistronic tRNA–gRNA genes (PTG). Ghir_A08G010860 (GhEMS_A08), Ghir_D08G010810 (GhEMS_D08), and Ghir_A09G018830 (GhEMS_A09) were knocked out by sgRNA1 and sgRNA2 (PTG1). GhEMS_A08 and GhEMS_D08 (PTG2) were knocked out by sgRNA3 and sgRNA4 (PTG2).
To further identify the biological function of the EMS1 genes involved in cotton-specific developmental processes, we have summarized the expression of EMS1 genes in different organs/tissues (including roots, leaves, anthers in different length buds) of G. hirsutum. As shown in Figure 1B, most of GhEMS1 genes have expressed in anthers, and Ghir_A08G010860 (GhEMS1_A08), Ghir_D08G010810 (GhEMS1_D08), and Ghir_A09G018830 (GhEMS1_A09) were all predominantly expressed in early-stage anthers (stage 4/5, bud length: 3~5 mm) and their expression gradually decreased with anthers development, implying that these genes may play crucial roles in identification of reproductive cell development in early stage anthers.
Due to combination of two sgRNAs with tRNA can improve the transcription and knockout efficiency (Xie et al., 2015; Wang et al., 2018). Thus, two sgRNAs targeting the same GhEMS1 genes were combined by overlap extension PCR and then ligated to the expression vector pRGEB32-GhU6.7, to produce polycistronic tRNA-gRNA genes PTG1 and PTG2 vectors (Figure 1C). The PTG1 contained sgRNA1 and sgRNA2, which was designed to knock out GhEMS1_A08, GhEMS1_D08, and GhEMS1_A09 (Figure 1D). The PTG2 contained sgRNA3 and sgRNA4, which was designed to knock out GhEMS1_A08 and GhEMS1_D08 at the same time (Figure 1D). The prepared vectors were transformed the cotton by Agrobacterium (GV3101) (Supplementary Figure 2). We obtained 3 sterile plants (KO1~3) for PTG1, and 2 sterile plants (KO4~5) for PTG2 (Figure 2A). Five independent transformation plants showed different degrees of necrosis-like dark spots on the surface of anther and have different pollen viability (Figures 2A-C). KO1-KO3, showed obvious necrosis-like dark spots on the surface of the anthers (Figures 2A, B). There was no pollen in the anthers of KO1 and KO2, and only a few shriveled pollen grains in the KO3 anthers (Figure 2C). KO4 and KO5 showed a few anthers with dark spots, and the pollen quantity and viability were lower than the wild type (WT) but higher than those of KO1- KO3 (Figures 2A-C).
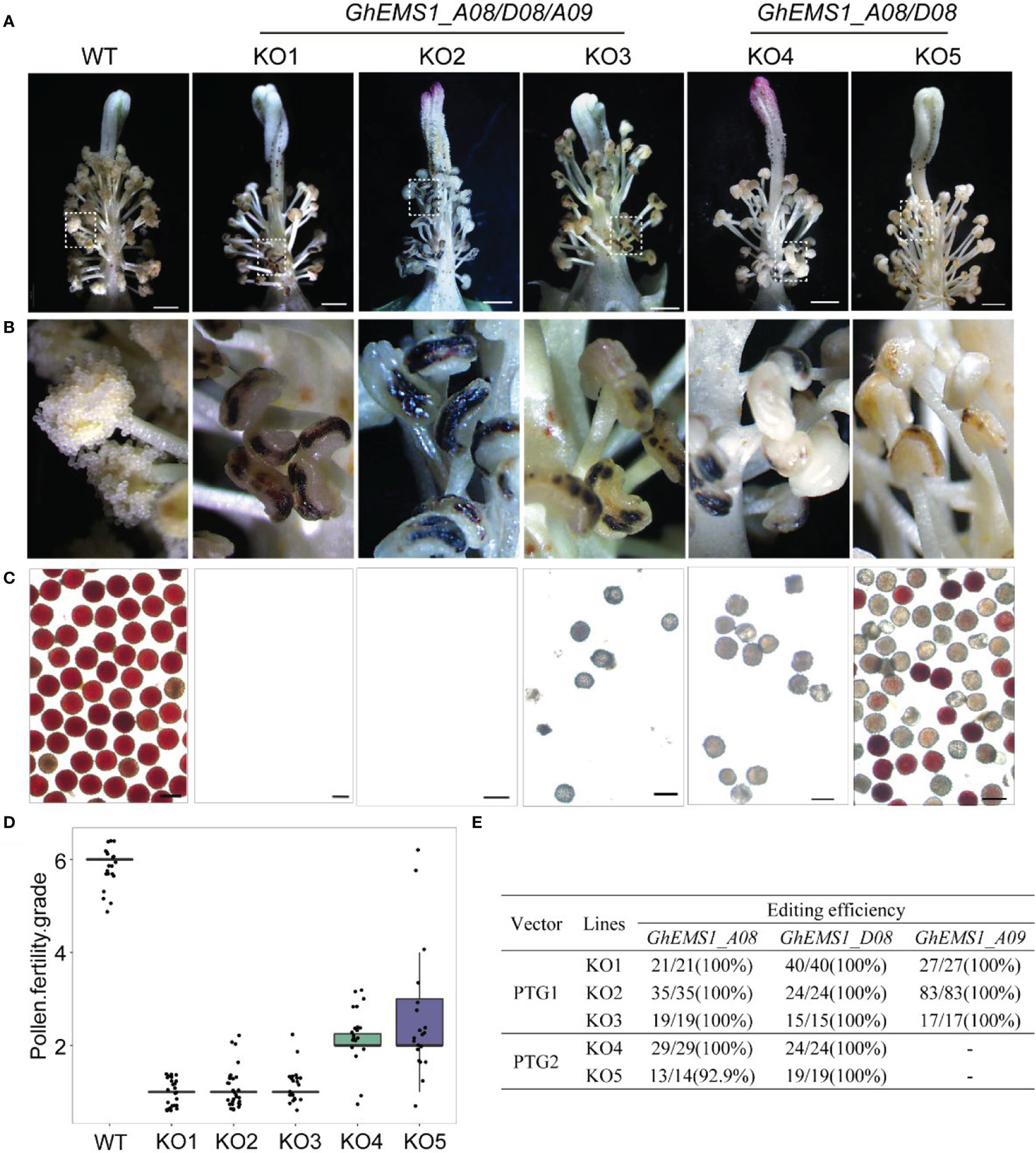
Figure 2 Phenotypes of GhEMS1s mutants. (A) The phenotypes of the WT and KO1 - KO5 transgenic lines, three genes Ghir_A08G010860 (GhEMS_A08), Ghir_D08G010810 (GhEMS_D08), and Ghir_A09G018830 (GhEMS_A09) of PTG1 in KO1- KO3 sterile plants. GhEMS_A08 and GhEMS_D08 (PTG2) were knocked out in KO4 and KO5 plants. Scale bars: 2 mm; (B) Partial enlarged view of anther in picture (a); (C) TTC (2,3,5-triphenyl tetrazolium chloride) was used to detect the pollen viability. No pollen was observed in the anthers of KO1 and KO2 plants. Scale bars: 100 µm. (D) Statistics for pollen fertility in the GhEMS1 mutants. During the phenotypic investigation, we divided plant fertility into six levels: Grade 1 indicates that anthers with necrosis-like dark spots and all anthers without pollen; Grade 2 indicates that < 25% of the anthers have a few inactive pollen grains without dehiscence; Grade 3, 4, and 5 indicate that 25%, 50%, and 75% of the anthers spread pollen, respectively; Grade 6 indicates that all anthers dehiscence and release active pollen. The phenotype of every flower was recorded, and the fertility of different plants was counted. (E) Analysis of gene editing efficiency in different GhEMS mutant lines by Sanger sequencing.
Furthermore, a three-month fertility assay was performed on WT and the five transgenic plants. We divided plant fertility into six levels: Grade 1 indicates that anthers with necrosis-like dark spots and all anthers without pollen; Grade 2 indicates that < 25% of the anthers have a few inactive pollen grains without dehiscence with a few anthers have dark spots; Grade 3, 4, and 5 indicate that 25%, 50%, and 75% of the anthers spread pollen, respectively; Grade 6 indicates that all anthers dehiscence and release active pollen. The phenotype of every flower was recorded, and the fertility of different plants was counted. The results showed that the fertility of the WT plants was relatively stable at grade 6, with pollen viability higher than 99.5% and anthers normal dehiscence and no dark spots; KO1- KO3 were below grade 2, with no pollen or very few inactive pollen grains, and dark spots on the anther surface (Figures 2C, D). KO1 was the most stable, with 100% anthers of all flowers having necrosis-like dark spots on the anther surface, and no pollen (Figure 2D). The fertility of KO4 and KO5 was above grade 2 and below grade 6, with a few anthers have dark spots (Figures 2B-D).
To check the mutation at the selected target site in KO1-KO5 lines, the sanger sequencing was performed. We found that three genes, GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09, all were 100% edited in KO1-KO3 plants (Figure 2E), and the deletion length was in the range of 2 to 29 bp (Supplementary Figure S3). In the KO4 and KO5 plants, GhEMS1_A08 and GhEMS1_D08 were successfully edited with 100% editing efficiency, and no editing in the GhEMS1_A09 (Figure 2E and Supplementary Figure S3B). Moreover, the expression level of GhEMS1_A09 was lower in the early stage anthers, compared with the expression of GhEMS1_A08 and GhEMS1_D08 in the same stage anthers, and the expression level of GhEMS1_A09 decreased earlier (Figure 1B). In all, the male fertility of KO4 and KO5 was better than that of KO1-KO3 (Figure 2C), indicating that the three GhEMS1 genes were essential for male fertility and played crucial roles in male fertility. In addition, 38 potential off-target genes of the two sgRNAs in KO1 (complete male sterility plant) were analyzed, and no off-target effects were found in KO1 (Supplementary Tables 2, 3), this result suggested the formation of male sterility of KO1 only caused by the mutations of GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09 genes.
Whether the sterile phenotype can be stably inherited to the offspring is related to the successful application of sterile mutant to cross breeding. To test the genetic stability of the necrosis-like dark spots as a marker of sterility, we applied WT pollen to the stigma of sterile KO1 plants. Then, the target site fragments in PTG1 of GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09 in WT, KO1, T1 generation (KO1×WT) were amplified by PCR, and polyacrylamide gel electrophoresis (PAGE) was used to identify the editing. The results showed that the T1 generation had the same fragment with T0 generation, but due to WT pollination, some new editing types were generated, such as in T1-3, for which two sgRNAs generated new editing types (Supplementary Figure 3C). The individual sgRNAs showed different efficiencies in the detection of PAGE, indicating that it is necessary to connect two sgRNAs in tandem with one vector (Supplementary Figure 3C). In addition, the necrosis-like dark spots on the anthers could also be observed from the T0 to T3 (Figures 2A, B and 3A), T3 plants (n=64) had 35.9% (23/64) complete necrosis anthers (Figure 3A). Gene editing types were identified by Hi-TOM (Liu et al., 2019), which revealed that the T3 generation had the same editing type as the T0 generation, such as the mutations (-1 bp, -5 bp, -6 bp) at the sgRNA1 target site (Figures 4A, C, E, G), and mutations (-2 bp, -20 bp, -1 bp) at the sgRNA2 target site (Figures 4B, D, F, H). However, due to WT pollination and the retention of Cas9, some new editing types have also been generated, such as the mutation (+1 bp) at the sgRNA1 target site (Figure 4A). Moreover, the pollen quantity and viability of T3 plants were analyzed, the results were consistent with the gene editing efficiency and the number of necrosis-like dark spots on the anthers in these plants, suggested the GhEMS1 mutants displayed the genetic stability of the necrosis-like dark spots as a marker of male sterility.

Figure 3 The sterile phenotype of surface necrosis of T3. (A) wild type, KO1 T3 plants, and enlarged images. (B) Changes in the anthers with different degrees of necrosis-like spots. Compared with those of the WT, yellow spots appeared on the anthers of sterile KO1 plants at stage 7, these spots gradually deepened at stage 10 and stage 14 to form necrosis-like dark spots eventually. KO1, GhEMS 3-genes simultaneous mutant. The white arrows indicate necrotic spots.
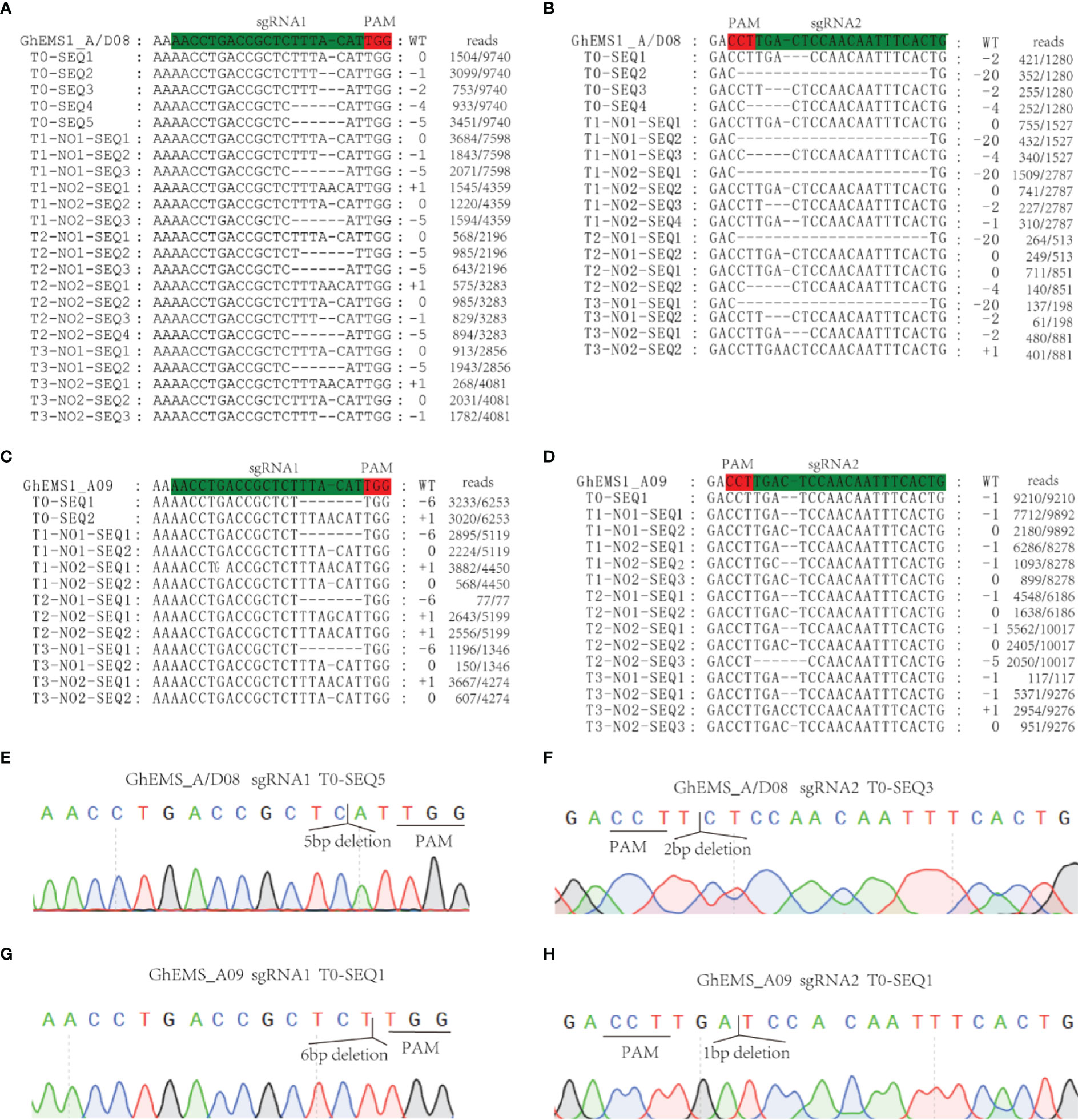
Figure 4 KO1 editing types were inherited in T0 to T3. (A, B) GhEMS_A/D08 were edited in sgRNA1 and sgRNA2, respectively. (C, D) GhEMS_A09 were edited in sgRNA1 and sgRNA2, respectively. (E) Sequencing peak photos of GhEMS_A/D08 with 5 bp deletions at the sgRNA1 site. (F) Sequencing peak photos of GhEMS_A/D08 with 2 bp deletions at the sgRNA2 site. (G) Sequencing peak photos of GhEMS_A09 with 6 bp deletions at the sgRNA1 site. (H) Sequencing peak photos of GhEMS_A09 with 1 bp deletions at the sgRNA2 site.
The necrosis-like dark spots on the mature anthers could be inherited, but when and how the necrosis-like dark spots appeared on the anther surface? To explore the formation period of necrosis-like dark spots, we obtained the stage 6 to stage 14 anthers of KO1. At stage 7, yellow spots appeared on the anthers of sterile phenotype plants, which gradually deepened at stage 10 and stage 14 to form necrosis-like dark spots eventually (Figure 3B). Therefore, the dark spots on the surface of the anthers started earlier, which can help us to screen sterile plants at the early stage.
To observe the microspore development of KO1, anther tissue cross-sections were made. At stage 6, the WT exhibited four complete anther cell layers (from outside to inside: epidermis, endothecium, middle layer, and tapetum layer) and microsporocytes. However, the sterile mutant anthers lacked middle layer and tapetum cells (Figures 5A, C). Furthermore, in the WT anthers, the microsporocytes completed nuclear division at stage 6 (Figure 5A), tetrads formed at stage 7 (Figure 5A), microspores were released from tetrads at stage 8 (Figure 5A), and then microspores developed into mature pollen during stage 9 to stage 11 with the tapetum development and degeneration (Figure 5A). However, the mutants could complete nuclear division but not cytoplasmic division, causing the enlarged microsporocytes and undetached microsporocytes at stages 7 to 9 (Figure 5A). At stage 10, the microsporocytes of mutants started to degrade, and they were completely degraded at stage 11 (Figure 5A). What’s more, the degree of DNA fragmentation in the anthers was detected by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling). In the WT, there were a few yellow fluorescence signals in the tapetum layer, a few microspores at stage 9, and the yellow fluorescence signals were observed enhanced at stage 10. However, no fluorescence signals were observed in the stage 9 anthers of the mutant, and only faint fluorescence signals were observed appearing in the degrading microsporocytes in the stage 10 anther locules in the mutant (Figures 5B, C). This result indicated that there was no normally developed pollen in the chamber of the mutant and pollen deformity was caused by the absence of tapetum formation and the PCD process of microspore mother cells.
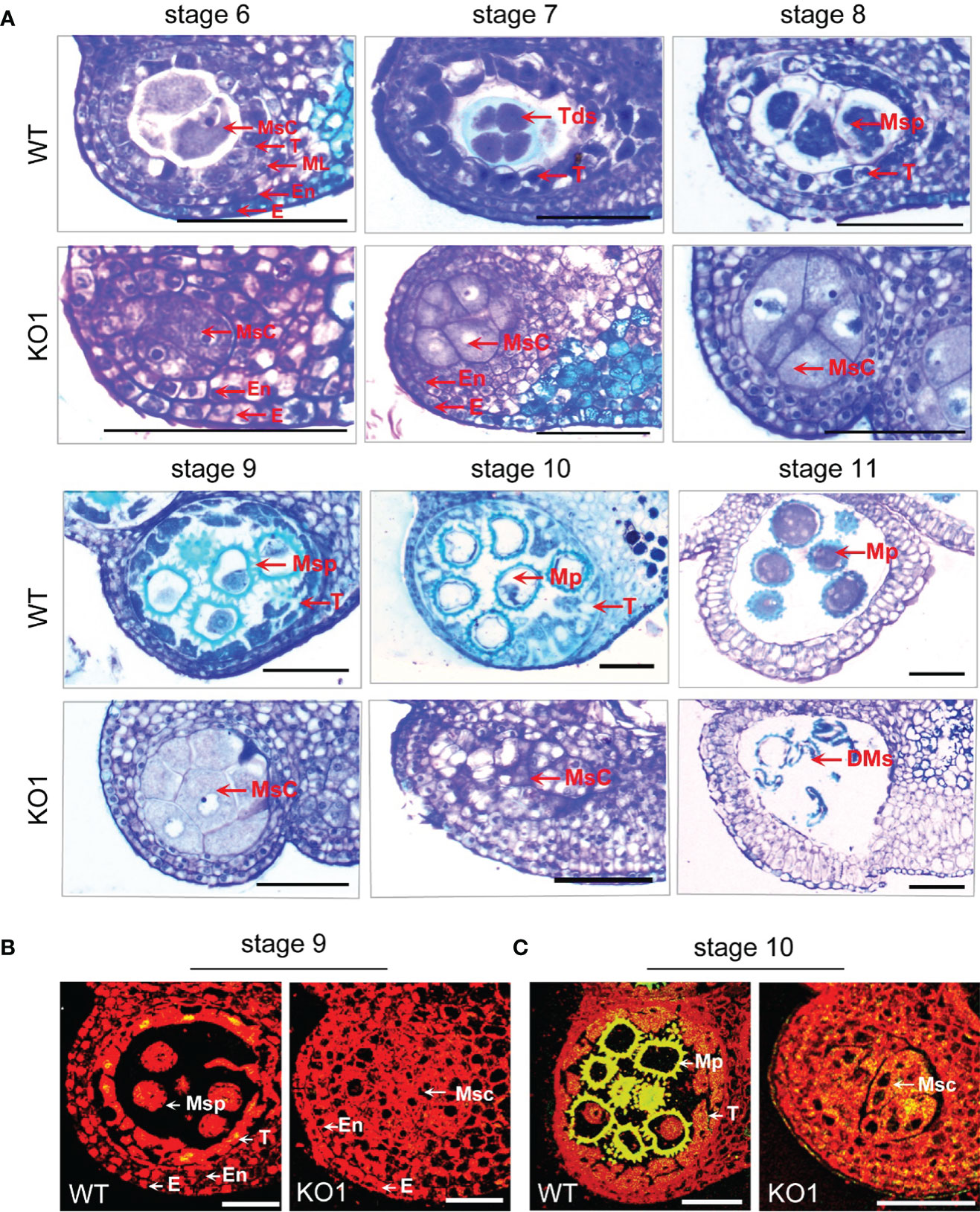
Figure 5 Comparison of the histological characteristics of the WT and KO1 sterile line T3 anthers. (A) Stage 6-11 histological characteristics of the WT and KO1, Scale bars: 100 µm (B, C) Analysis of DNA damage in anthers of WT and KO1 male-sterile plants. The degree of DNA fragmentation of anthers was detected by TUNEL. Scale bars: 100 µm. DMs: degenerated microspores; E, epidermis; En, endothecium; Msc, microsporocyte; Mp, mature pollen; ML, middle layer; Msp, microspore; T, tapetum; Tds, tetrads; WT, wild type.
To explore the molecular mechanisms of anther abortion in GhEMS1 mutants, we compared the transcriptomes of WT and Ghems1 anthers at four developmental stages (stages 6, 7, 9, 10). A total of 7,172 genes were found to be differentially expressed between Ghems1 and WT at four anther developmental stages (Supplementary Table 4). Of these differentially expressed genes (DEGs), 2,070 (28.86%) were up-regulated and 5,102 (71.14%) were down-regulated (|log2(fold change)|≥1 and padj < 0.05) in Ghems1 (Figure 6A). Compared with WT, Ghems1 had more up-regulated genes than down-regulated genes at stages 6 and 7 (Figure 6A). However, at stages 9 and 10, Ghems1 had more down-regulated genes than up-regulated genes (Figure 6A). Of these genes, 239, 1,250, 551 and 631 genes were unique at stages 6, 7, 9 and 10, respectively, and 23 genes showed differential expression in all four developmental stages (Supplementary Table 5 and Figure 6B). These 23 genes included three GhEMS1 genes that have been edited, and the expression of three GhEMS1 genes was downregulated in all four stage mutant anthers (Figure 6E). The expression level of GhEMS_D08 was higher than that of GhEMS_A08/A09, and gradually decreased following the anther development. GO enrichment analysis, “peroxidase activity” was highly enriched in stage 6 and 7 anthers of mutants, and “monooxygenase activity” is highly enriched in stage 6, 7, 9, which is related to peroxide in GO enrichment analysis (Supplementary Figure 4 and Table 6). So, we detected the content of peroxide at four developmental stages of Ghems1 and WT anthers (Figure 6G). The results showed that compared with the WT, the mutants had lower peroxide content in stage 6 and 7, but higher peroxide content in stage 9 and 10. Black spots began to appear in stage 7 and appeared in large numbers in stage 9, might be responsible for the appearance of black spots on the surface of anthers. In mutant stage 6 down-regulated genes, “activation of MAPKK activity” and “MAP kinase kinase kinase activity” were enriched. In stage 6, 9 down-regulated genes, “pollen exine formation” was enriched, while in stage 9 and 10 DEGs was enriched in “plant- type cell wall modification”, possibly related to pollen formation (Supplementary Figure 4 and Table 6).
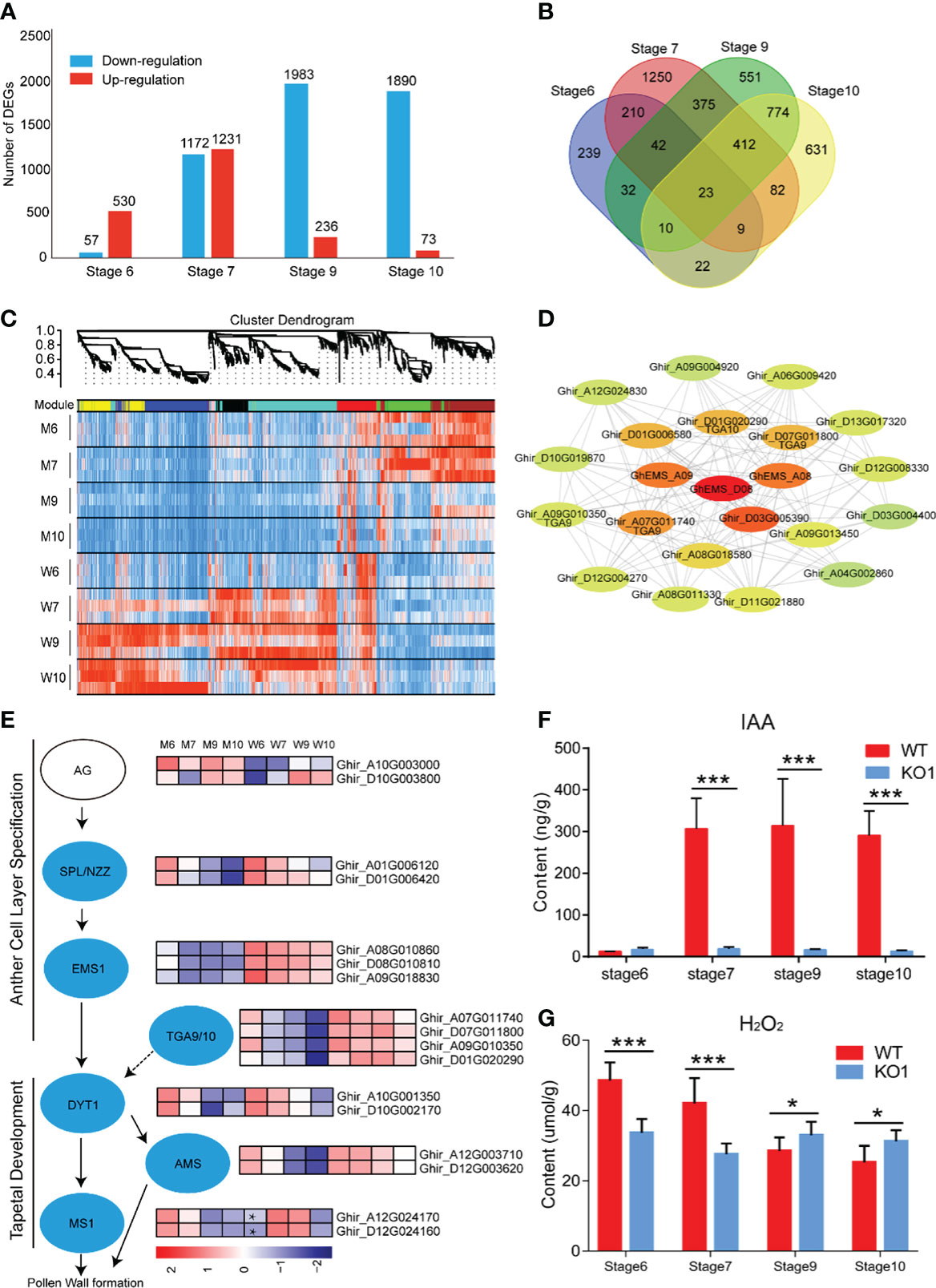
Figure 6 Transcriptome analysis of four developmental stages. (A) Difference gene statistics of wild type and mutant in four periods; (B) Intersection of differential genes at different stages; (C) WGCNA co-expression module, a total of 8 different modules; (D) Analysis of the co-expression network of the red module. The shades of color represent correlations; (E) Gene expression of the regulatory network of pollen development; (F) IAA content at four developmental stages; (G) H2O2 content at four developmental stages. W, wild type; M, mutant; AG, AGAMOUS; AMS, ABORTED MICROSPORE; DYT1, DYSFUNCTIONAL TAPETUM 1; EMS1, EXCESS MICROSPOROCYTES 1; MS1, MALE STERILITY 1; NZZ/SPL, SPOROCYTELESS/NOZZLE. Data are presented as means ± SE from five biologically independent experiments. Asterisks indicate statistically significant differences (***, P <0.001; *, P <0.05); by Student’s t-test.
To generate co-expression networks for all DEGs and biological samples, the weighted gene co-expression network analysis was performed. A total of 8 gene modules were identified (Figure 6C). EMS1 genes were distributed in the red module. We extracted the red module genes that may be related to GhEMS1 genes to do a further co-expression network analysis and showed that there are four leucine-zipper transcription factors TGACG9/10 (TGA9/10, Ghir_D01G020290, Ghir_A07G011740, Ghir_D07G011800 and Ghir_A09G010350) genes, one tetrapeptide alpha-pyrone reductase 1 (TKPR1, Ghir_D03G005390), and one indole-3-acetic acid-amido synthetase (GH3.6, Ghir_D01G006580), were associated with GhEMS1. TGA9 and TGA10 are expressed throughout early anther primordia, and mutations in TGA9 and TGA10 lead to male sterility and differential defects in abaxial (Murmu et al., 2010). In the genetic framework for control of anthers development (Wilson and Zhang, 2009), the expression trends of SPL/NZZ, EMS1, TGA9/10, DYT1 and AMS were the same, but MS1 in the stage 6 anthers of mutant was higher than WT (Figure 6E). Previously reported that TGA9/10 was located downstream of SPL/NZZ and upstream or in parallel with DYT1 in the genetic hierarchy that controls anther development (Murmu et al., 2010), similar to EMS1. Interestingly, we also found that there was co-expression of TGA9/10 and EMS1 (Figure 6D), it was suggested that TGA9/10 and EMS1 may interact with each other to regulate anther development.
In the co-expression network, we found TKPR1 which co-expressed with EMS1 (Figure 6D), TKPR1 involved in the biosynthesis of hydroxylated tetraketide compounds that serve as sporopollenin precursors (the main constituents of exine) was essential for pollen wall development (Tang et al., 2009; Grienenberger et al., 2010). And GO enrichment analysis shows that “pollen exine formation” was highly enriched in stage 6 and 9 anthers of mutants. To confirm whether the synthesis of sporopollenin precursors in the mutants had been affected, we measured the autofluorescence intensity of sporopollenin of Ghems1 and WT anthers at stages 6, 7, 9, 10 by microscopy with UV light illumination (Supplementary Figure 5) (Ma et al., 2022). In the mutant, the autofluorescence of sporopollenin was not detected in the four stages, but in WT, the autofluorescence of sporopollenin was detected in the 9 and 10 stages. This showed that the synthesis of sporopollenin was affected in the mutant.
Phytohormones play an important role in the regulation of anther development. In the co-expression network, we found GH3.6 which co-expressed with EMS1 (Figure 6D), GH3.6 catalyzes the synthesis of indole-3-acetic acid (IAA)-amino acid conjugates, providing a mechanism for the plant to cope with the presence of excess auxin (Staswick et al., 2005). Compared with WT, the expression of GH3.6 in the mutant gradually decreased (Supplementary Figure 6). So, we detected the dynamic changes of auxin (IAA) in anthers of Ghems1s male sterile line (Figure 6F). During the four anther developmental stages, the free IAA content of male sterile plants KO1 changed little with the development of anther and was at a lower level. However, the free IAA content of WT increased greatly between the stage 6 and stage 7 of anthers development, and maintained a high level after that. The lower IAA content in the stage 7, 9, 10 anthers of Ghems1s mutants may be closely related to the occurrence of male sterility.
From the above results, we found that after the editing of both GhEMS1_A08 and GhEMS1_D08, there were yellow spots on the surface of the anthers and a few fertile pollen grains in the chamber. Moreover, the results of simultaneously editing three GhEMS1 genes showed that there were necrosis-like dark spots on the surface of the anthers, which contained completely aborted pollen grains, thus the necrosis-like dark spots can serve as a marker of completely male-sterile cotton lines with GhEMS1s mutants, and can help breeders to screen sterile plants at the early anther stage.
In recent years, CRISPR/Cas9 technology has played an important role in the creation of sterile materials. The sterile genes cloned in model plants Arabidopsis and rice have potential applications in cotton. AtEMS1, OsMSP1 encode LRR receptor kinases, and the mutants have the same phenotype, including the production of a large number of microspore mother cells, no tapetum layer and middle layer. Compared with Arabidopsis thaliana, cotton GhEMS1 mutant anther surface has obvious necrotic phenotype, and the molecular mechanism needs to be further studied.
Previous reports have shown that the young microspore stage and flowering stage in rice are very sensitive to high temperature (HT) stress, and HT stress destroys the function of tapetum during microspore formation and leads to poor anther dehiscence (Endo et al., 2009). It was worth noting that the expression of GhEMS1 genes at the tetrad stage were differently respond to HT in the HT-tolerant and -sensitive lines, could provide a theoretical basis for the study of male sterility of cotton caused by high temperature (Supplementary Figure 7).
Morphological trait markers have broad application prospects in cotton production. Among them, pigment glands, okra leaf shape, and virescently traits were more commonly studied (Zhu et al., 2008; Ma et al., 2013; Ma et al., 2016). The virescently marker is linked to sterility gene and can be used to identify sterile lines at the early seedling stage (Ma et al., 2013). For now, there has been no marker found on the anther associated with fertility. In our study, the necrosis-like dark spots appeared on the anthers in the early stage, which could be screened during early bud periods, reducing waste of resources and allowing for hybrid breeding.
Upland cotton is a polyploid species with a larger genome (2.5 Gb), so most genes have multiple copies and high sequence similarity due to the polyploidization of At and Dt sub-genomes, which makes cotton gene engineering very difficult (Wang et al., 2019). In this study, many EMS1-like genes were aligned in the upland cotton genome through the amino acid sequence of the Arabidopsis AtEMS1 gene. The three genes of GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09 may have functional redundancy because they are in one branch and the mutant is completely male sterile by knocking out GhEMS1_A08, GhEMS1_D08 and GhEMS1_A09 at the same time, but the two gene mutants show partial infertility by knocking out GhEMS1_A08, GhEMS1_D08. Multiple genes control male sterility, which makes it difficult to find EMS genes using map-based cloning. Completely sterile plants cannot produce tapetum and intermediate layers, confirming that cotton GhEMS1 is a key gene regulating cotton anther and microspore development. Thus, to establish a cotton hybrid system, the GhEMS1 mutant can be used as the male sterile line, and the negative sterile plants (Cas9-free) can be selected and crossed with other cultivars to create excellent hybrids. The positive sterile plants (with Cas9) can be crossed with transgenic acceptors or cultivars to breed sterile lines (Supplementary Figure 8).
The effect of IAA on plant male sterility was often reported. The reduction of the express of IAA related gene in wheat was associated with the occurrence of male-sterility (Su et al., 2019). At the third stage, the contents of IAA, GA3 and ZR in CMS were remarkably lower than its maintainer of Chinese Cabbage (Liu et al., 2014). When IAA was depleted, the vascular bundles develop abnormally, and the passage of water and nutrients into the drug compartment was blocked, resulting in abnormal microspore development and pollen abortion (Liu et al., 2014). ABA and IAA are involved in PCD of microsporocytes during meiosis in Petunia hybrida L. (Kovaleva et al., 2018). Sugar and IAA may be the key regulators of cotton anther response to high temperature stress (Min et al., 2014). In this study, we found that among the genes co-expressed by EMS1 through the co-expression network, there was an auxin synthesis related gene GH3.6. At the same time, there was a significant difference in IAA content between the male sterile mutant and the wild type during the 6-10 stage of anthers development, which is very likely to be the cause of male sterility. We also tested other endogenous hormones, but found no regular changes. As to whether the male sterility can be restored by spraying IAA, further study is needed.
In summary, we created a new male-sterile cotton line, which may further promote the utilization of the genic male-sterile lines and the development of cotton hybrid breeding. In addition, the cotton line with three mutated EMS1 homolog genes provides the basis for the study of cotton anther tapetum and microspore development.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA827503.
XG, XZ, LM and JZ designed the studies. JZ, XX, NL, PW, SW, YZ, XW, SC, YM performed the experiments and data analysis. WL, and HM improved the grammar of the manuscript. JZ, PW, XG and LM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by funding from the National Key Research and Development Program of China (2022YFD1200800), Fundamental Research Funds for the Central Universities (2662019PY073), the National Natural Science Foundation of China (32072024), and the Fundamental Research Funds for the Central Universities (2021ZKPY019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1102196/full#supplementary-material
Andrianov, V., Borisjuk, N., Pogrebnyak, N., Brinker, A., Dixon, J., Spitsin, S., et al. (2010). Tobacco as a production platform for biofuel: Overexpression of arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8 (3), 277–287. doi: 10.1111/j.1467-7652.2009.00458.x
Canales, C., Bhatt, A. M., Scott, R., Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in arabidopsis. Curr. Biol. 12 (20), 1718–1727. doi: 10.1016/S0960-9822(02)01151-X
Chen, R., Xu, Q., Liu, Y., Zhang, J., Ren, D., Wang, G., et al. (2018). Generation of transgene-free maize Male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01180
Chen, X., Yang, S., Zhang, Y., Zhu, X., Yang, X., Zhang, C., et al. (2021). Generation of male-sterile soybean lines with the CRISPR/Cas9 system. Crop J. 9 (6), 1270–1277. doi: 10.1016/j.cj.2021.05.003
Clement, K., Rees, H., Canver, M. C., Gehrke, J. M., Farouni, R., Hsu, J. Y., et al. (2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37 (3), 224–226. doi: 10.1038/s41587-019-0032-3
Endo, M., Tsuchiya, T., Hamada, K., Kawamura, S., Yano, K., Ohshima, M., et al. (2009). High temperatures cause Male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 50 (11), 1911–1922. doi: 10.1093/pcp/pcp135
Grienenberger, E., Kim, S. S., Lallemand, B., Geoffroy, P., Heintz, D., Souza, C. D. A., et al. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22 (12), 4067–4083. doi: 10.1105/tpc.110.080036
Jin, S., Liang, S., Zhang, X., Nie, Y., Guo, X. (2006). An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum l.). Plant Cell Tissue Organ Cult. 85 (2), 181–185. doi: 10.1007/s11240-005-9068-9
Ken-Ichi Nonomura, K. M., Eiguchi, M., Suzuki, T., Miyao, A., Hirochika, H., Kurata, N. (2003). The MSP1 gene is necessary to restrict the number of cells entering into Male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15 (8), 1728–1739. doi: 10.1105/tpc.012401
Kim, D., Landmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/Nmeth.3317
Kovaleva, L. V., Voronkov, A. S., Zakharova, E. V., Andreev, I. M. (2018). ABA and IAA control microsporogenesis in petunia hybrida l. Protoplasma 255 (3), 751–759. doi: 10.1007/s00709-017-1185-x
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi: 10.1093/bioinformatics/btt656
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., Chen, L. L. (2017). CRISPR-p 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10 (3), 530–532. doi: 10.1016/j.molp.2017.01.003
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., et al. (2019). Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62 (1), 1–7. doi: 10.1007/s11427-018-9402-9
Liu, H., Wu, K., Yang, M., Zhou, X., Zhao, Y. (2014). Variation of soluble sugar, starch and plant hormones contents in sesame dominant genic male sterile line during bud development. Oil Crop Sci. 36 (2), 175–180. doi: 10.7505/j.issn.1007-9084.2014.02.006
Li, J., Wang, M., Li, Y., Zhang, Q., Lindsey, K., Daniell, H., et al. (2019). Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnol. J. 17 (2), 435–450. doi: 10.1111/pbi.12988
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
Ma, K., Han, J., Hao, Y., Yang, Z., Chen, J., Liu, Y. G., et al. (2019). An effective strategy to establish a male sterility mutant mini-library by CRISPR/Cas9-mediated knockout of anther-specific genes in rice. J. Genet. Genomics 46 (5), 273–275. doi: 10.1016/j.jgg.2019.03.005
Ma, D., Hu, Y., Yang, C., Liu, B., Fang, L., Wan, Q., et al. (2016). Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7, 10456. doi: 10.1038/ncomms10456
Ma, J., Wei, H., Liu, J., Song, M., Pang, C., Wang, L., et al. (2013). Selection and characterization of a novel photoperiod-sensitive male sterile line in upland cotton. J. Integr. Plant Biol. 55 (7), 608–618. doi: 10.1111/jipb.12067
Ma, H., Wu, Y., Lv, R., Chi, H., Zhao, Y., Li, Y., et al. (2022). Cytochrome P450 mono-oxygenase CYP703A2 plays a central role in sporopollenin formation and ms5ms6 fertility in cotton. J. Integr. Plant Biol. 64, 2009–2025. doi: 10.1111/jipb.13340
Miao, Y. H., Xu, L., He, X., Zhang, L., Shaban, M., Zhang, X. L., et al. (2019). Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. Plant J. 98 (2), 329–345. doi: 10.1111/tpj.14222
Min, L., Li, Y., Hu, Q., Zhu, L., Gao, W., Wu, Y., et al. (2014). Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164 (3), 1293–1308. doi: 10.1104/pp.113.232314
Min, L., Zhu, L. F., Tu, L. L., Deng, F. L., Yuan, D. J., Zhang, X. L. (2013). Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75 (5), 823–835. doi: 10.1111/tpj.12245
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154 (3), 1492–1504. doi: 10.1104/pp.110.159111
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17 (10), 1905–1913. doi: 10.1111/pbi.13106
Ramadan, M., Alariqi, M., Ma, Y., Li, Y., Liu, Z., Zhang, R., et al. (2021). Efficient CRISPR/Cas9 mediated pooled-sgRNAs assembly accelerates targeting multiple genes related to male sterility in cotton. Plant Methods 17 (1), 16. doi: 10.1186/s13007-021-00712-x
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M., Maldonado, M. C., et al. (2005). Characterization of an arabidopsis enzyme family that conjugates amino acids to indole-3-Acetic acid. Plant Cell 17 (2), 616–627. doi: 10.1105/tpc.104.026690
Su, Q., Yang, J., Fu, Q. Y., Jia, F. Y., Li, S. P., Li, Y., et al. (2019). Profiling of indole metabolic pathway in thermo-sensitive bainong male sterile line in wheat (Triticum aestivum l.). Physiol. Mol. Biol. Plants 25 (1), 263–275. doi: 10.1007/s12298-018-0626-0
Tang, L. K., Chu, H., Yip, W. K., Yeung, E. C., Lo, C. (2009). An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in arabidopsis. New Phytol. 181 (3), 576–587. doi: 10.1111/j.1469-8137.2008.02692.x
Wang, M., Tu, L., Yuan, D., Shen, C., Li, J., Liu, F., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, gossypium hirsutum and gossypium barbadense. Nat. Genet. 51 (2), 224–229. doi: 10.1038/s41588-018-0282-x
Wang, P., Zhang, J., Sun, L., Ma, Y., Zhang, X. (2018). High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16 (1), 137–150. doi: 10.1111/pbi.12755
Wilson, Z. A., Zhang, D. B. (2009). From arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60 (5), 1479–1492. doi: 10.1093/jxb/erp095
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112 (11), 3570–3575. doi: 10.1073/pnas.1420294112
Xie, S., Shen, B., Zhang, C., Huang, X., Zhang, Y. (2014). sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS One 9 (6), e100448. doi: 10.1371/journal.pone.0100448
Yang, L., Wu, Y., Zhang, M., Zhang, J., Stewart, J. M., Xing, C., et al. (2018). Transcriptome, cytological and biochemical analysis of cytoplasmic male sterility and maintainer line in CMS-D8 cotton. Plant Mol. Biol. 97 (6), 537–551. doi: 10.1007/s11103-018-0757-2
Yu, G. C., Wang, L. G., Han, Y. Y., He, Q. Y. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Zhang, R., Zhou, L. L., Li, Y. L., Ma, H. H., Li, Y. W., Ma, Y. Z., et al. (2022). Rapid identification of pollen- and anther-specific genes in response to high-temperature stress based on transcriptome profiling analysis in cotton. Int. J. Mol. Sci. 23 (6), 3378. doi: 10.3390/ijms23063378
Zhao, D. Z., Wang, G. F., Speal, B., Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the arabidopsis anther. Genes Dev. 16 (15), 2021–2031. doi: 10.1101/gad.997902
Zheng, H., Wang, R., Jiang, Q., Zhang, D., Mu, R., Xu, Y., et al. (2021). Identification and functional analysis of a pollen fertility-associated gene GhGLP4 of gossypium hirsutum l. Theor. Appl. Genet. 134 (10), 3237–3247. doi: 10.1007/s00122-021-03888-x
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive genic Male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6, 37395. doi: 10.1038/srep37395
Keywords: cotton, GhEMS1s, CRISPR/Cas9, male-sterile line, necrosis-like dark spots
Citation: Zhang J, Wu P, Li N, Xu X, Wang S, Chang S, Zhang Y, Wang X, Liu W, Ma Y, Manghwar H, Zhang X, Min L and Guo X (2023) A male-sterile mutant with necrosis-like dark spots on anthers was generated in cotton. Front. Plant Sci. 13:1102196. doi: 10.3389/fpls.2022.1102196
Received: 18 November 2022; Accepted: 19 December 2022;
Published: 09 January 2023.
Edited by:
Zhichao Wu, National Institutes of Health (NIH), United StatesReviewed by:
Rabarijaona Romer, University of Antananarivo, MadagascarCopyright © 2023 Zhang, Wu, Li, Xu, Wang, Chang, Zhang, Wang, Liu, Ma, Manghwar, Zhang, Min and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoping Guo, eHBndW9AbWFpbC5oemF1LmVkdS5jbg==; Ling Min, bGluZ21pbkBtYWlsLmh6YXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.