
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 January 2023
Sec. Plant Genetics, Epigenetics and Chromosome Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1097534
Spatial distribution and orientation of pods on the main raceme (stem) and branches could affect rapeseed yield. However, genomic regions underlying the pod orientation were not described in Brassica species. Here, we determined the extent of genetic variation in pod orientation, described as the angles of pedicel on raceme (APR) and angles of the pod on pedicel (APP) among 136 rapeseed accessions grown across three environments of the upper, middle and lower Yangtze River in China. The APR ranged from 59° to 109°, while the APP varied from 142° to 178°. Statistical analysis showed that phenotypic variation was due to genotypic (G) and environmental (E) effects. Using the genome-wide association analysis (GWAS) approach, two QTLs for APR (qBnAPR.A02 and qBnAPR.C02) and two for APP (qBnAPP.A05 and qBnAPP.C05), having minor to moderate allelic effects (4.30% to 19.47%) were identified. RNA-seq analysis revealed 606 differentially expressed genes (DEGs) in two rapeseed accessions representing the extreme phenotypes for pod orientation and different alleles at the QTLs of APR. Three DEGs (BnLAZY4.A02, BnSAUR32.A02, and BnSAUR32.C02) were identified as the most likely candidates responsible for variation in pod orientation (APR). This study elucidates the genomic regions and putative candidate genes underlying pod orientation in B. napus.
Rapeseed (Brassica napus L.) is the third largest source of edible vegetable oil, mainly grown for human consumption in various parts of the world, especially in Australia, Canada, China, India, France and Germany. Besides, it is used for biofuel and stock-feed production (Li et al., 2020). Due to the unprecedented growth of the world’s population, increased rapeseed yield is required to meet energy demands. The rapeseed generally bears pods (siliques) on the top of the raceme (primary stem) and side branches (Douglas and Riggs, 2005). After flowering, the rapeseed leaves senescence, falls off, and rely mainly on the green pods for photosynthesis. More than half of the dry matter for seed filling is provided by the pod valves/pericarp (Leng et al., 1992). Therefore, pod orientation plays a vital role in light energy absorption, which is required for photosynthesis and enhanced productivity. Based on the orientation of the pod on the raceme (pod angle), rapeseed can be classified into four types: straight, inclined, flat, and droop (Wu et al., 2007). The straight pod orientation is desired by rapeseed breeders, as they are not prone to ‘shading syndrome’. However, this pod architecture is rare in rapeseed germplasm (Sinhamahapatra et al., 2010). Pods of most cultivated Brassicas tend to be inclined, promoting the plant type to be more compact and disease build-up, especially to Sclerotinia stem rot, caused by the fungus Sclerotinia sclerotiorum.
Plant architecture significantly influences the photosynthetic efficiency and crop harvest index. The growth of plant organs in response to gravity is an essential factor that determines plant morphology. Most plant organs, especially those on mature plants, are displayed at a certain angle to the vertical growth axis and are not parallel to the gravity vector (Lorenzi and Perbal, 1990). Research has shown that the orientation of plant organs is controlled by the antagonistic interaction between the universal mechanism of gravitation and auxin-dependent transport (Roychoudhry et al., 2013).
Although the plant responses to gravity are yet to be fully understood, several key genes involved in the network of gravitational responses were identified in crop plants through genome-wide association studies (GWAS), map-based cloning and mutant studies. For example, in rice, AGPL1, LPA1, ONAC106, PROG1, DWARF3, and TAC1 regulate tillering angle by transmitting gravity signals, suggesting polygenic inheritance (Morita et al., 2006; Okamura et al., 2013; Wu et al., 2013; Sakuraba et al., 2015 (Li et al., 2007; Tan et al., 2008; Sang et al., 2014). In addition, the middle creeping gene LAZY1 also regulates the tillering and leaf angle by changing auxin distribution (Li et al., 2007). Genes such as BRXL4, HSFA2D and OsPIN2 can regulate the expression of the LAZY1 gene (Xu et al., 2005; Chen et al., 2012; Zhang et al., 2018; Li et al., 2019), and the mutation of the SL gene can inhibit the phenotype of lazy1 by inhibiting the biosynthesis of auxin (Xu et al., 2005; Chen et al., 2012; Sang et al., 2014; Zhang et al., 2018; Li et al., 2019). In Arabidopsis thaliana, LAZY1 and TAC1 genes have been shown to regulate the branching angle, similar to rice (de Saint Germain et al., 2013; Waite and Dardick, 2018). In maize, TAC1 can lead to an increase in branching angle (Ku et al., 2011). The genetic loci controlling the branch angle in rapeseed have been studied (Wang et al., 2016). However, the majority of studies focused on the branch angle of crops (Su et al., 2020; Wang et al., 2020; Yang et al., 2020), and few studies have addressed the role of the orientation of reproductive structures responsible for economic yields, such as pods, ears, and glumes (Li et al., 2012). In addition to branch angle, genes related to pod orientation also have been isolated and identified in the model plant Arabidopsis thaliana, tobacco, tomato and cucumber (Hobbie et al., 2000; Li et al., 2001; Venglat et al., 2002; Ha et al., 2007; Khan et al., 2012; Wang et al., 2015; Lin et al., 2016; Sun et al., 2019). The deletion mutation of KNAT1 gene produced downward pods (Venglat et al., 2002). The angle between the pod and raceme of auxin signal mutant axr6 was acute (Hobbie et al., 2000). Generally, the angle between wild-type pod and raceme is about 60°, and the overexpression of the ROP2 gene of Arabidopsis G protein could reach the 90° or above (Li et al., 2001). In contrast, the inhibitory mutants were all smaller than 60° (Li et al., 2001). The pod angle of the actin filament bunching protein genes VILLIN2 and VILLIN3 double mutants vln2vln3 became smaller (van der Honing et al., 2012). The lateral organ boundary genes BOP1 and BOP2 proteins regulated the pod angle through LBD (Ha et al., 2007). The angle between pedicel/fruit and a raceme of spa mutant became smaller (Zhang and Yang, 2012). There are also many studies on the growth direction of Arabidopsis pedicels. Arabidopsis thaliana short pedicel mutant destroyed in KNAT1/BP gene showed decreased pedicel length and downward flowers (Douglas et al., 2002; Venglat et al., 2002). KNAT1/BP negatively regulates KNAT2, KNAT6 and ATH1 to ensure pedicels have a normal upward direction (Ragni et al., 2008; Li et al., 2012). It was found that NtSVP in tobacco, SlAGO7 in tomato and CsUp in cucumber control pedicel and fruit orientation (Wang et al., 2015; Lin et al., 2016; Sun et al., 2019).
GWAS combined with RNA-seq is an effective method to identify key genes which control plant traits (Bai et al., 2022). In this study, the GWAS was employed in a diverse set of rapeseed germplasm, and the associated regions for pod orientation were determined based on linkage disequilibrium (LD). Association between the favourable SNP alleles and pod orientation was assessed by the Mann-Whitney U test to identify SNP markers for marker-assisted selection in the rapeseed breeding programs. We further conducted RNA-seq analysis of pod pedicels of two rapeseed lines with significantly different pod orientations and prioritised candidate genes underlying QTL for pod orientation. Finally, bioinformatics analysis found the protein structure and sequence variation in the population for the candidate genes. Overall, this study aimed to provide a genetic basis of pod orientation in rapeseed so that ideotypes for plant architecture, optimal for increasing photosynthesis efficiency and yields, could be developed.
One hundred and thirty-six rapeseed accessions were planted at three locations in Zunyi City, Guizhou Province (28°N, 107°E), Yangluo City, Hubei Province (30°N, 114°E) and Lu’an City, Anhui Province (32°N, 116°E) (Supplementary Figure 1). These three locations (environments) represented the upper, middle and lower regions of rapeseed cultivation in the Yangtze River valley in China. The experiment was conducted in randomised complete blocks with two replicates across each environment. Each accession was sown in a plot containing three rows with 54 individuals, spaced at 33 cm between rows and 11 cm between plants within each row, with a planting density of 270,000 plants/ha. All experiments were managed according to local field management and cultivation practices
Two quantitative indices related to rapeseed pod orientation were measured: the angle of the pedicel on raceme (APR) and the angle of the pod on pedicel (APP) across 136 rapeseed accessions grown in three environments. We digitally measured the pod-related angles of a GWAS panel in three independent environments. We. At the maturity (BBCH scale 90), five plants of each rapeseed accession were selected from each environment investigation and measured for APR and APP using the image processing method described by Wang et al. (2015). A pod with raceme was cut from each plant and placed on a 20 × 20 cm blackboard for image analysis. A digital camera (DSLR-A350, Sony Inc, Japan), fixed on a tripod, was used to take a picture of the samples from the top, with a focal length of 35 mm. Digital images were downloaded output to the computer to draw the path and measure the angle with the AutoCAD package (https://www.autodesk.com.cn/).
A selected set of 136 rapeseed accessions were genotyped using the Brassica 60K Illumina Infinium@ SNP array (Illumina) as described previously (Liu et al., 2016). After quality control (minor allele frequency>0.05 and missing data<20%) (Liu and Muse, 2005), a total of 21426 SNPs were selected for subsequent GWAS analysis. SNP density map was drawn using the CMplot package (Yin et al., 2021) in R3.3.3 (Team RC, 2013) to gauge the marker coverage across the B. napus genome. Principal component analysis was performed using Tassel v5.0 software (Bradbury et al., 2007). Only the first two principal components were plotted by the ggplot2 package (Wickham, 2011) in R3.3.3 (Team RC, 2013). MEGA-X software (Kumar et al., 2018) was used to draw the phylogenetic tree and show the genetic relationship among GWAS accessions. Both principal component analysis and phylogenetic tree are used to reveal the distribution of population structure. LD (linkage disequilibrium) attenuation was estimated to demonstrate the degree of genetic linkage, and it is drawn by PopLDdecay software (max decay distant = 2500, break = 6000bp, bin1 = 1000, bin2 = 6000) as described previously (Zhang et al., 2019).
The general linear model (GLM) was constructed using genotype, environment, and phenotype data. The ANOVA was carried out to determine whether the genotypic effect, environmental effect and genotype×environment interaction effect exist. The multi-environment mixed linear model (MLM) in Tassel5.0 software (Bradbury et al., 2007) was used to analyse the association between phenotype and genotype across three environments (Zunyi, Yangluo and Lu’an). The kinship relationship (K matrix) was added to the model to eliminate false positives caused by the genetic relationship. In this study, the p-value indicates whether an SNP was related to the corresponding trait, and r2 shows the phenotypic variation explained by the marker. The standard of significant sites obtained by GWAS is P<0.0001. The Manhattan and QQ plots were drawn using the qqman package (Turner, 2014) in R3.3.3 (Team RC, 2013).
All genes in the candidate region were annotated in the NR (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/) and Swiss-port databases (https://www.expasy.org/resources/uniprotkb-swiss-prot) using blast software (Korf et al., 2003). The function of homologous genes in A. thaliana was screened and checked on the TAIR website (https://www.arabidopsis.org/index.jsp).
The QTL alleles that positively affected the smaller APR and APP were referred to as “favourable alleles”. In contrast, the alleles that cause the larger angles are referred to as “unfavourable alleles”. We selected the most significant SNP in each candidate region and selected favourable alleles for APR and APP. Because the phenotypes corresponding to marker genotypes did not conform to the normal distribution, we used a nonparametric test, the Mann-Whitney U test, to analyse the differences. Box plots were drawn by the ggplot2 software package (Wickham, 2011) in R3.3.3.
The pedicels of mature pods of rapeseed accessions: line 9319 (APR: inclined rapeseed, 59°, the APP is 170°) and line ZS11 (the APR of flat rapeseed, 90°, APP 170°) were collected for RNA sequencing (Figure 1). Both accessions were planted in triplicates under the controlled environment room maintained at 25°C light and dark regimes (16/8 h) till sample collection. RNA libraries were constructed using NEB Next® Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. Libraries were sequenced on Illumina HiSeq 4000 platform, resulting in 150 bp paired-end reads. The quality control of RNA-seq reads was carried out by fastqc (Andrews, 2010) software and summarised by MultiQC (Ewels et al., 2016). For RNA degradation, the transcript integrity index (TIN) was checked using RSeQC following the method described by Wang et al. (2012). After quality control and filtration, the transcriptome sequencing Reads were compared to the reference genome assembly of ZS11 (version 20200127) using hierarchical indexing for spliced alignment transcripts (HISAT) version 2.1.0 (Kim et al., 2015). StringTie software v1.3.6 (Pertea et al., 2015) was used to assemble new transcripts. RSEM v1.2.31 was used to quantify the gene expression level. The accuracy and sensitivity of mutation detection using GATK Best Practices (O'Connor and Van Auwera, 2020) were similar to those using SAMtools mpileup (Li et al., 2009). Considering the rapidity and convenience of mutation detection by SAMtools mpileup, sample mutation detection was adopted by SAMtools mpileup based on the comparison data between HISAT2 and genome, and the results were stored in VCF format.
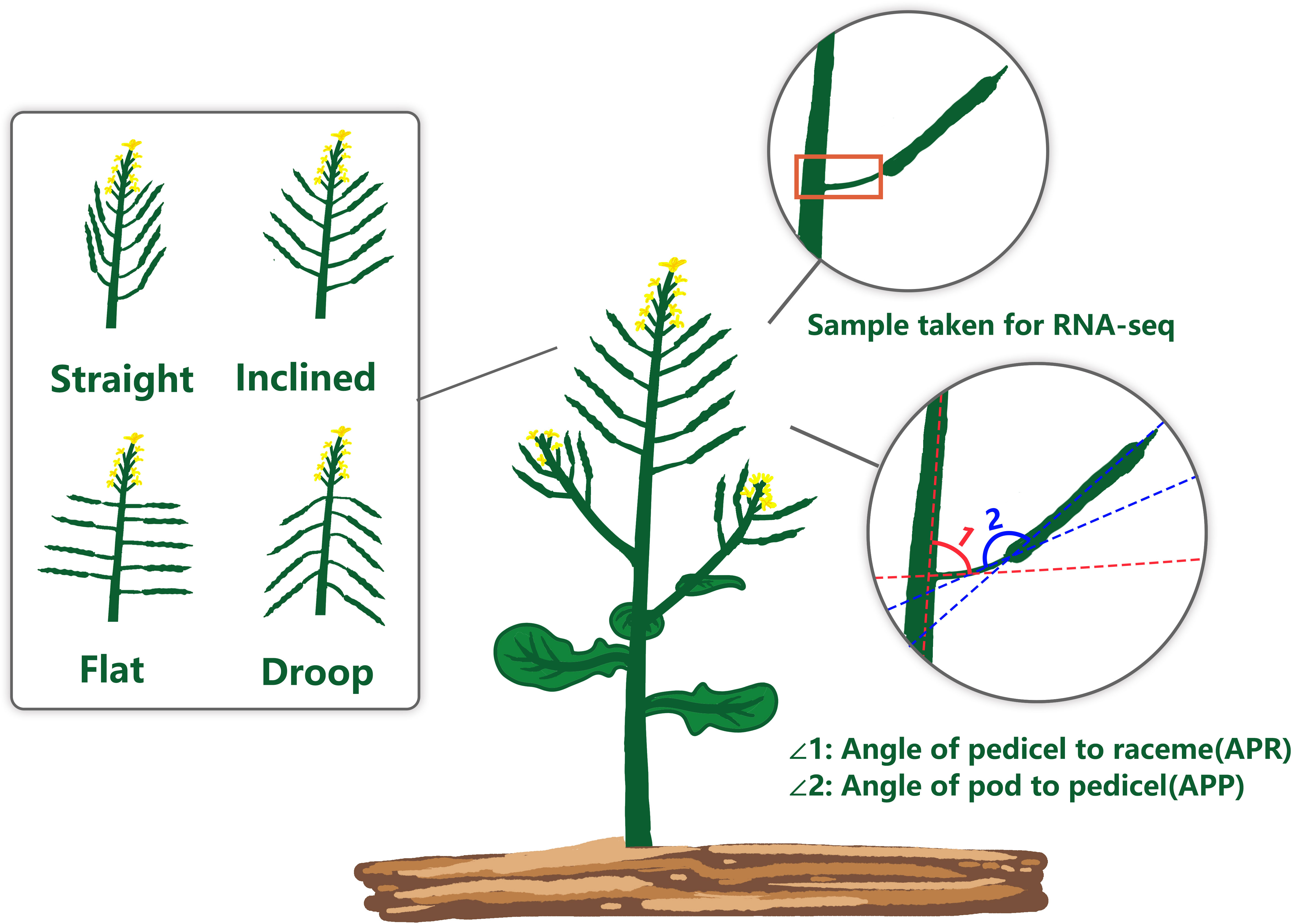
Figure 1 Schematic diagram of the rapeseed pod orientation showing two indices of pod orientation; APR, Angle of pedicel to raceme; and APP, Angle of the pod to the pedicel.
We downloaded candidate protein sequences involved in the pod orientation of different species from the NCBI website (https://www.ncbi.nlm.nih.gov/) and established a local protein database. Seven genes of IGT families in Arabidopsis thaliana were compared with the local protein database by BLAST (Ye et al., 2006). Mafft software (Katoh et al., 2002) was used to align the IGT family genes, and finally, the evolutionary tree was constructed using FastTree software (Price et al., 2009). SAUR gene family had hidden Markov model (PF02519) on the Pfam website (http://pfam.xfam.org/), so we used Hmmer (Finn et al., 2011) software to identify the SAUR gene family, and also used Mafft software to align the SAUR gene family, and then used FastTree software to construct the phylogenetic tree. The NWK format file obtained from tree building is used in R to draw the unrooted phylogenetic tree by TreeAndLeaf (Cardoso et al., 2022) package. Different species were depicted as different colours of evolutionary leaves, and the genetic relationship with rapeseed was used as the size of evolutionary leaves. The distance of genetic relationship was referred to the Taxonomy Common Tree of NCBI (https://www.ncbi.nlm.nih.gov/).
Alphafold2 (Jumper et al., 2021) software can predict the protein structure with 90% accuracy. The protein structure was predicted by alphafold2 software, and the rank_0 model with the best prediction score was selected from the rank_0 to 4 models of the predicted results to draw the protein structure. ChimeraX software (Pettersen et al., 2021) was used to draw protein structure, calculate B-factor value and map to protein structure.
The Pi value of SNP diversity was calculated from 289 rapeseed core accessions collected worldwide (Xuan et al., 2020). The original data of the Next-generation Sequencing was downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/SRA/SRP15312). The sequences were compared to the reference genome of ZS11 (Chen et al., 2021) by Sentieon software (Freed et al., 2017), and the GVCF file was used for detection variation among 289 accessions. GATK software was used to identify GVCF variant files as SNP and InDel VCF files (O'Connor and Van Auwera, 2020). According to the distribution of each parameter in the population, GATK software was used to control the data quality of the variant file, and the filter parameter used by the SNP file is QD < 2.0; FS > 50; MQ < 20; MQ Rank Sum > -12.5; Read Pos Rank Sum > -8.0. Vcftools software (Danecek et al., 2011) was used to calculate diversity. A 100-bp sliding window with a 25-bp step size was used to calculate nucleotide diversity. Using R language, the nucleotide diversity of the gene and its upstream and downstream 2Kb regions was fitted by the loess method, with a span of 0.3.
To determine the extent of natural variation in pod orientation, as APR and APP (Figure 1), 136 rapeseed accessions were grown in the field across three different environments. Statistical analysis showed that the variation in APR was due to genotypic (G) and environmental effects (E), but no G × E interaction existed. However, the effects of G, E and G × E were significant for the APP (Table 1). Broad sense heritability (H2) was calculated, and it ranged from 49.65% (APR) to 76.65% (APP). The frequency distribution of the APP and APR showed continuous variation (Figure 2), suggesting that the pod angle-related characteristics were inherited quantitatively.
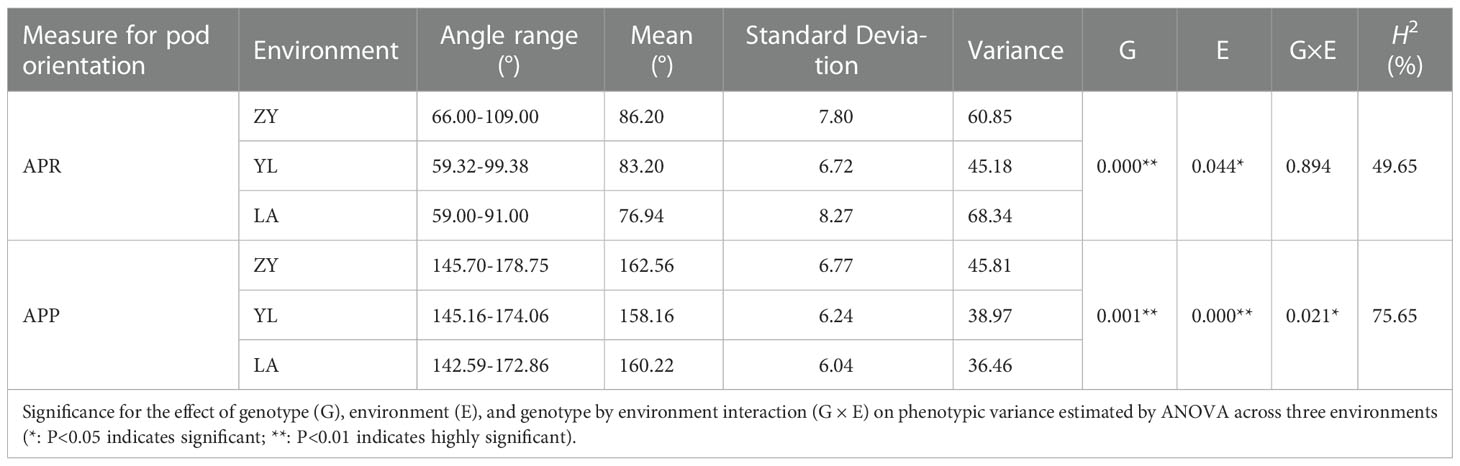
Table 1 Summary statistics on the phenotypic variation of pod orientation measured as Angle of the Pedicle to Raceme (APR) and Angle of the Pod to Pedicel (APP) in the 136 rapeseed accessions grown across three environments: ZY: Zunyi; YL: Yangluo, and LA: Lu’an.
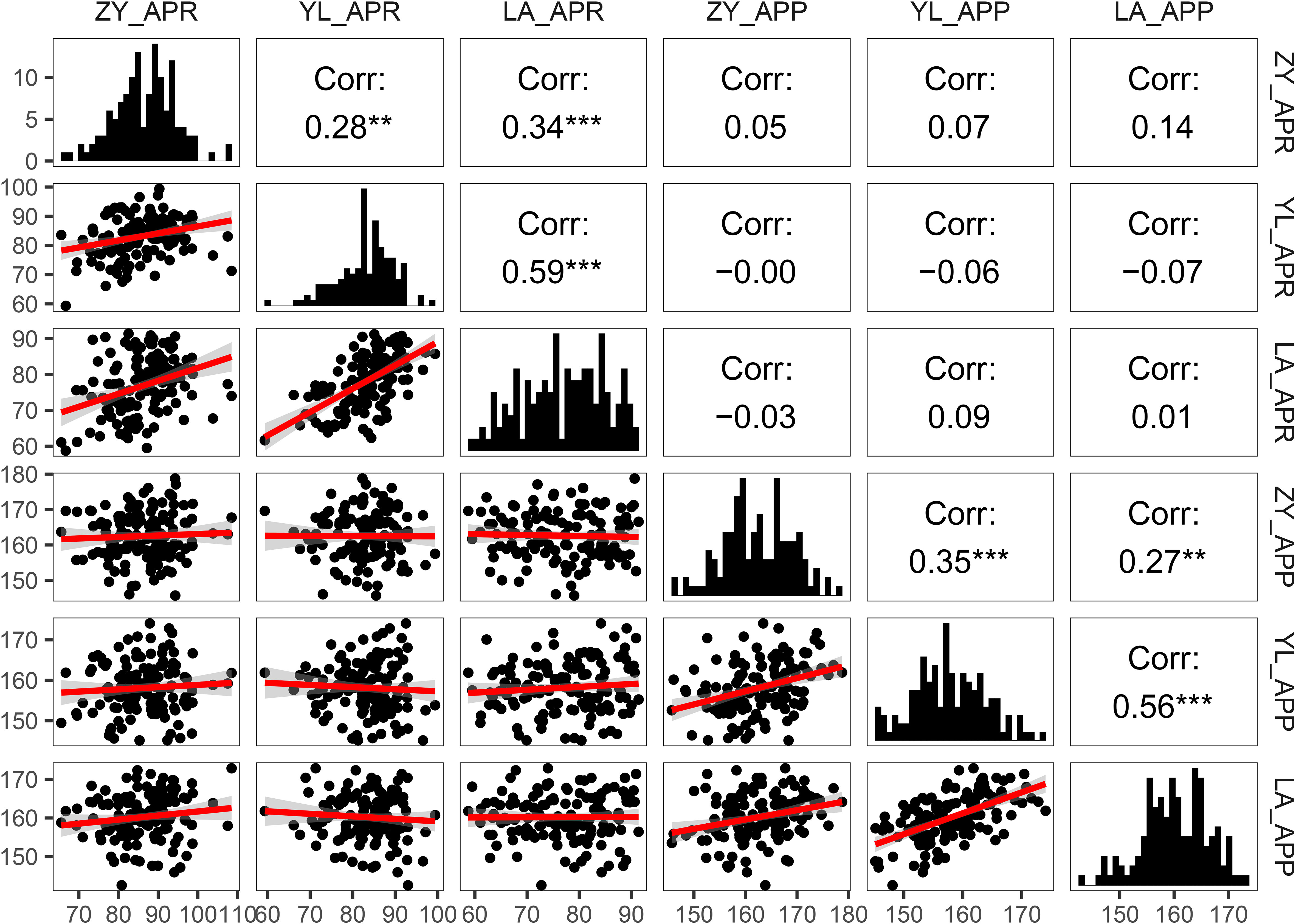
Figure 2 Pearson correlations and frequency distribution for pod orientation traits (Angle of the pedicle to raceme: APR and angle of the pod to pedicel: APP) among 136 rapeseed accession grown across three environments (ZY: Zunyi, YL: Yangluo, LA: Lu’an). The lower left part of the picture is the scatter diagram among the environments; the red line shows simple linear regression of variables. The diagonal corner is the frequency distribution diagram of the environments. The upper right corner is the correlation between the environments. **: P<0.01 indicates highly significant correlation; ***: P<0.001 indicates extremely significant correlation.
Genetic variation for pod orientation among 136 accessions of rapeseed is presented (Figure S3). The APR varied from 66° to 109°, 59.32° to 99.38°, and 59° to 91° with the mean values of 86.20 ± 7.80°, 83.20 ± 6.72° and 76.94 ± 8.27° in the ZY, YL and LA environments, respectively. Most accessions (80%) had APR ranging from 73° to 89°. The Australian cultivar, Rivette and F459, had a small APR angle (APR<70°), while Cibrabra and OG3186 had a large APR angle (APR>90°). The Australian variety, Rivette, had the minimum APR (59.32°), while the European variety, Nilla, had the maximum APR (109°). The APP varied from 142.59° to 178.75° across environments (Table 1). The 80% germplasm accessions had APP varying from 153° to 167°. Rapeseed accessions: BLN3342 and R11 had small APP angles (APP<150°), whereas OG3190 and P10 had a large APP angle (APP>170°).
We ranked rapeseed accession based on their angles to identify accessions of interest for rapeseed breeding. In this study, we defined the pod orientation as the sum of APR and APP less than 200° as straight type; greater than 200° and less than 250° were inclined types; more than 250° and less than 300° degrees were flat type, and greater than 300° was droop type (Figure 2). We classified pod orientation based on their angle into discrete categories: straight type with the APR of ~ 0° and the APP of ~ 180°; the inclined type having an APR of ~ 45° and the APP of ~ 180°; the flat type with APR of ~ 90°, and the APP of ~ 180°; droop type which had APR and APP of ~ 180° and 180°, respectively, Most accessions had inclined-type pods while none had straight and drooped types pods (Supplementary Figure 4).
Pearson correlation analysis between angle-related traits (APR and APP) across different environments showed significant positive correlations (r = 0.27 to 0.59, Figure 2). These results suggested that at least part of the genetic control of variation in angle-related traits was due to the environments (Zunyi, Yangluo and Lu’an). There was no relationship between APR and APP across three environments (r = -0.06 to 0.05), which indicated that different genetic factors might control variation for both measures of pod orientation.
To uncover the genetic basis of pod orientation, the significance of the associations between phenotypes and 21,426 genome-wide SNPs markers (Liu et al., 2016) was evaluated using the linear mixed (Q+K) model. There were 8,878 polymorphic SNPs (41.44%) in the A sub-genome and 12,548 (58.56%) in the C sub-genome, covering 238.13 Mb and 405.53 Mb sub-genomes, respectively, among the 136 rapeseed accessions (Supplementary Table 1). SNPs were evenly distributed across the A sub-genome and C sub-genomes (Figure 3A). The average LD decay of sub-genome A was smaller than that of sub-genome C. Under the threshold of R2 = 0.5, the LD decay of the whole population was about 147 kb, and under the threshold of R2 = 0.2, it was about 2.1 Mb (Supplementary Table 2, Figure 3B).
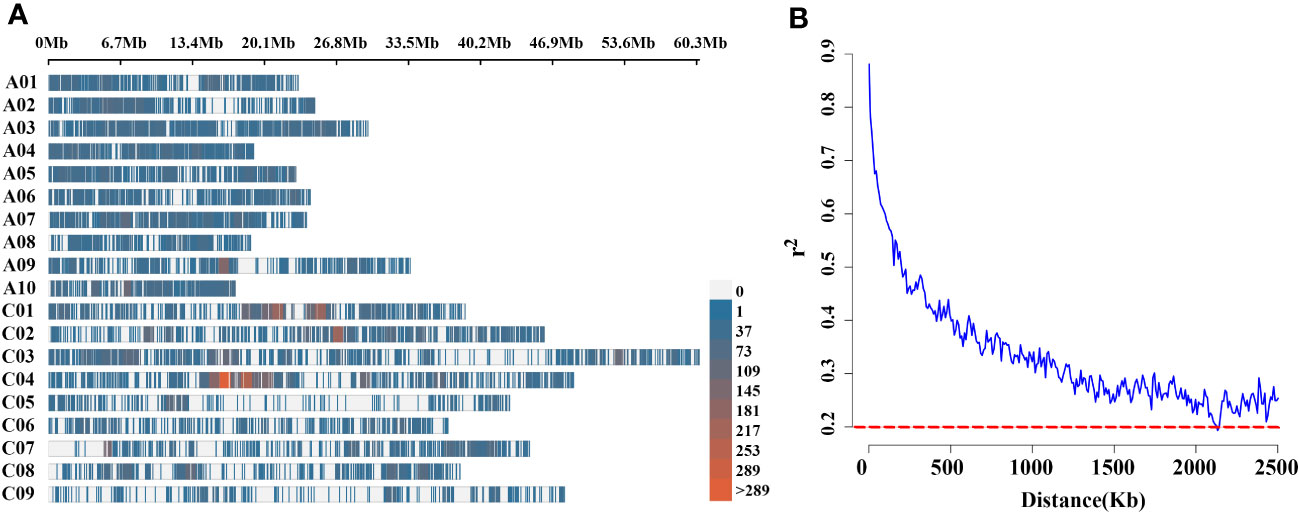
Figure 3 (A) High-density physical map of 136 rapeseed accessions, revealed with Brassica 60K Illumina Infinium@ SNP array. Different colours indicate the SNPs contained in the 1Mb genomic region. The red indicates SNPs rich region, the blue area indicates low-density SNP regions and the gray area indicates the absence of SNP markers; (B) LD decay of Brassica genome of 136 natural population using 60K Illumina Infinium@ SNP array. The dotted line in red colour shows that r2 is 0.2. X-axis: physical distance is shown in Kb.
Manhattan and Quantile-Quantile (Q-Q) plots showed significant genetic associations for APP and APR across environments (Figure 4). Using the -log10(P) ≥4 as threshold LOD, we identified four QTL, designated as qBnAPR.A02, qBnAPP.A05, qBnAPR.C02, and qBnAPP.C05 on chromosomes A02, A05, C02, and C05, respectively (Table 2, Table S3). Interestingly, qBnAPR.A02 and qBnAPR.C02 were located in the homologous regions on the group 2 chromosomes (A02/C02), while qBnAPR.A05 and qBnAPP.C05 were mapped to homologous regions on A05/C05 chromosomes (Supplementary Figure 5). SNP markers accounted for 4.50% (APR) to 16.78% (APP) of the phenotypic variance (Table 2). In addition to four significant QTL, two minor QTL (one for APR on the A08 and one APP on chromosome A06) were also detected (original data not shown). However, these QTL had low LOD scores (LOD>3, but <threshold LOD 4). Two false QTL (one for APR on the A03 chromosome and one APP on chromosome A10) were excluded because only one SNP was significant at these loci.
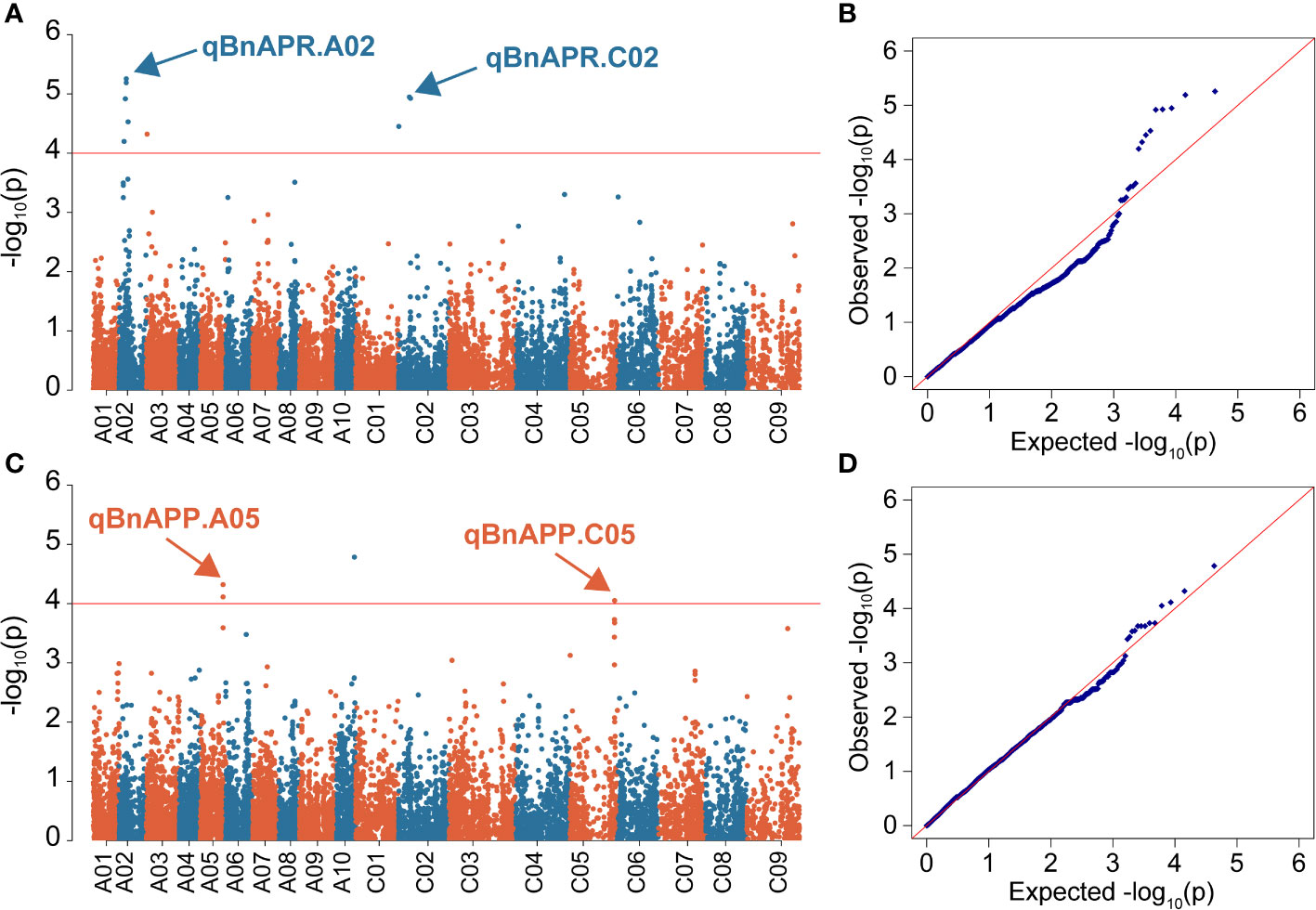
Figure 4 Manhattan and quantile-quantile (QQ) plots showing associations between SNPs and two pod orientation traits in 136 rapeseed accessions. GWAS was conducted by the mixed linear model (MLM) in Tassel v5.0. (A) Manhattan plot for the angle of pedicel to raceme (APR) across three environments; (B) Q-Q plot for the APR across three environments; (C) Manhattan plot for the angle of the pod to pedicel (APP); (D) Q-Q plot for APP. The horizontal red line represents the genome-wide significant threshold (p=1.0 × 10-4) For clarity, only SNPs with a −log10P-value > = 4 are shown. SNPs are plotted according to their physical positions on the ZS11 reference genome.
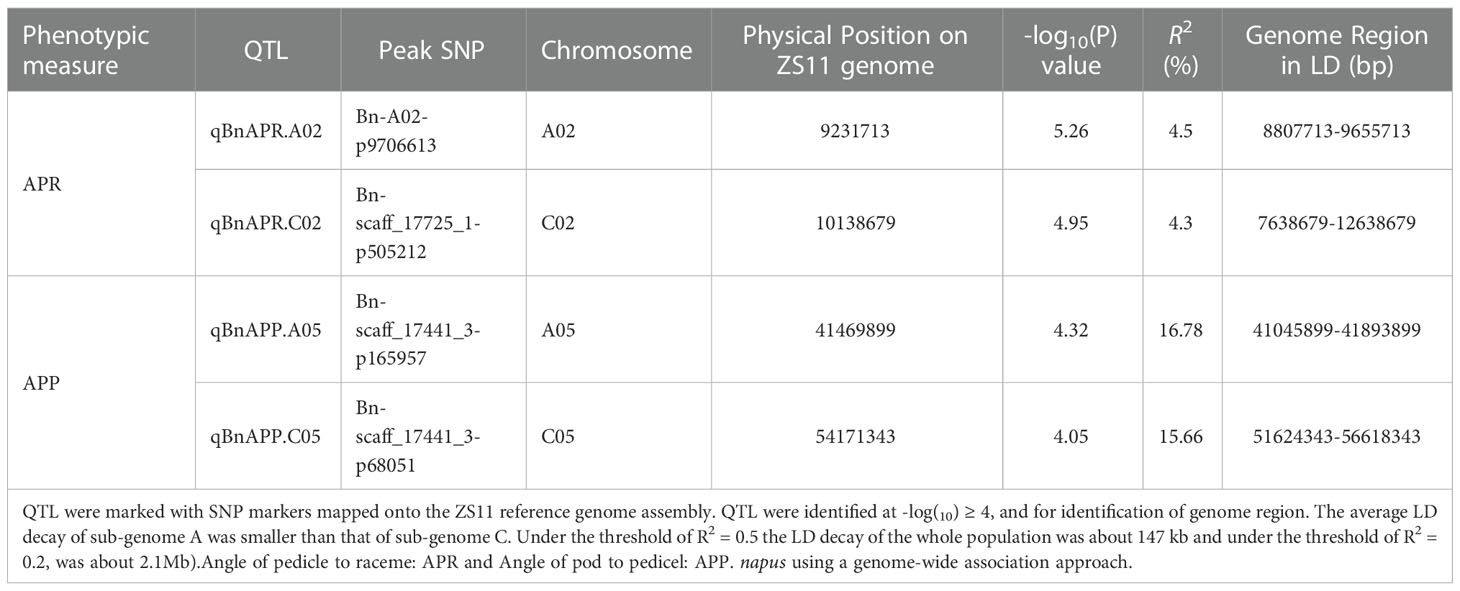
Table 2 QTL for pod orientation (Angle of the Pedicle to Raceme: APR and Angle of the Pod to Pedicel: APP) identified in a panel of 136 accessions of B.
We observed that phenotypes corresponding to different genotypes did not show normal distribution. Therefore, the Mann-Whitney U test (Supplementary Table 4) was applied to determine whether the phenotypes (different measures of pod orientation) correspond to marker alleles. The AA SNP allele of Bn-A02-p9706613 decreased the APR compared to GA and GG. Similar results were observed with the AA allele of Bn-scaff_17725_1-p505212, which showed significant differentiation from large pod angle accessions with GA or GG alleles. AA alleles at Bn-scaff_17441_3-p165957 and Bn-scaff_17441_1-p1146459 also showed an association with reduced APP compared to GA and GG alleles (Figure 5).
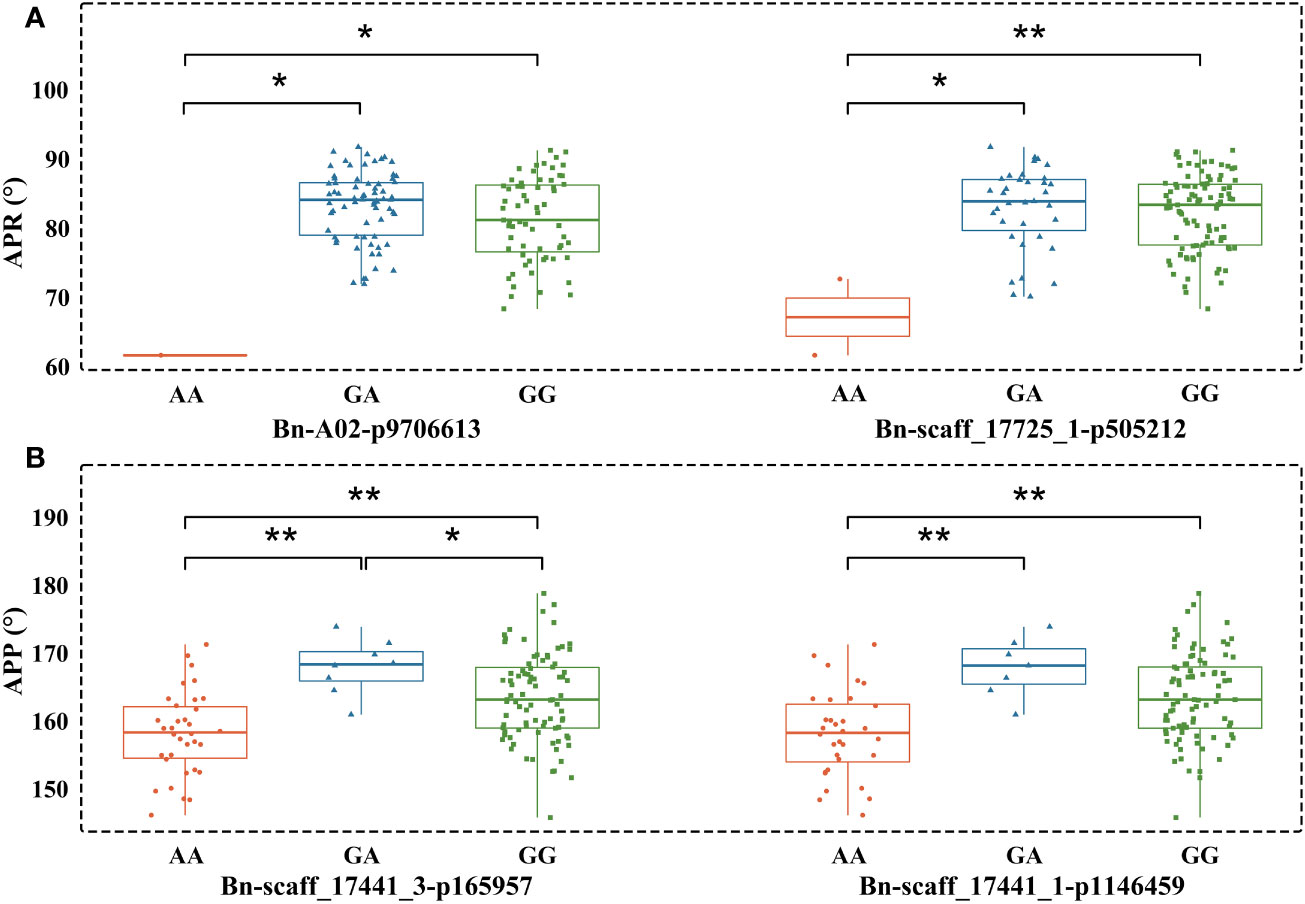
Figure 5 Boxplots showing the association between alleles of two peak SNP markers and pod orientation. (A) Angle of the pod on raceme (APR) and corresponding SNP alleles; (B) Angle of the pod on pedicel (APP) and corresponding alleles. All statistical associations are marked with *(at P<0.05) and with ** (at P<0.01).
We selected two accessions that had extreme phenotypes: line 9319 (APR: inclined rapeseed, 59°, the APP is 170°) and line ZS11 (the flat rapeseed, APR of 90°, APP 170°) (Figure 6A). The DEGs were analysed in both lines with extreme phenotypes by RNA-seq approach (Supplementary Table 5). Through RNA seq analysis, 128 million reads were generated. The data of each library ranged from 5.8 Gb to 8.2 Gb, and the Q30 base calling was ~95%. 85% of the reads were mapped to the reference ZS11 Genome.
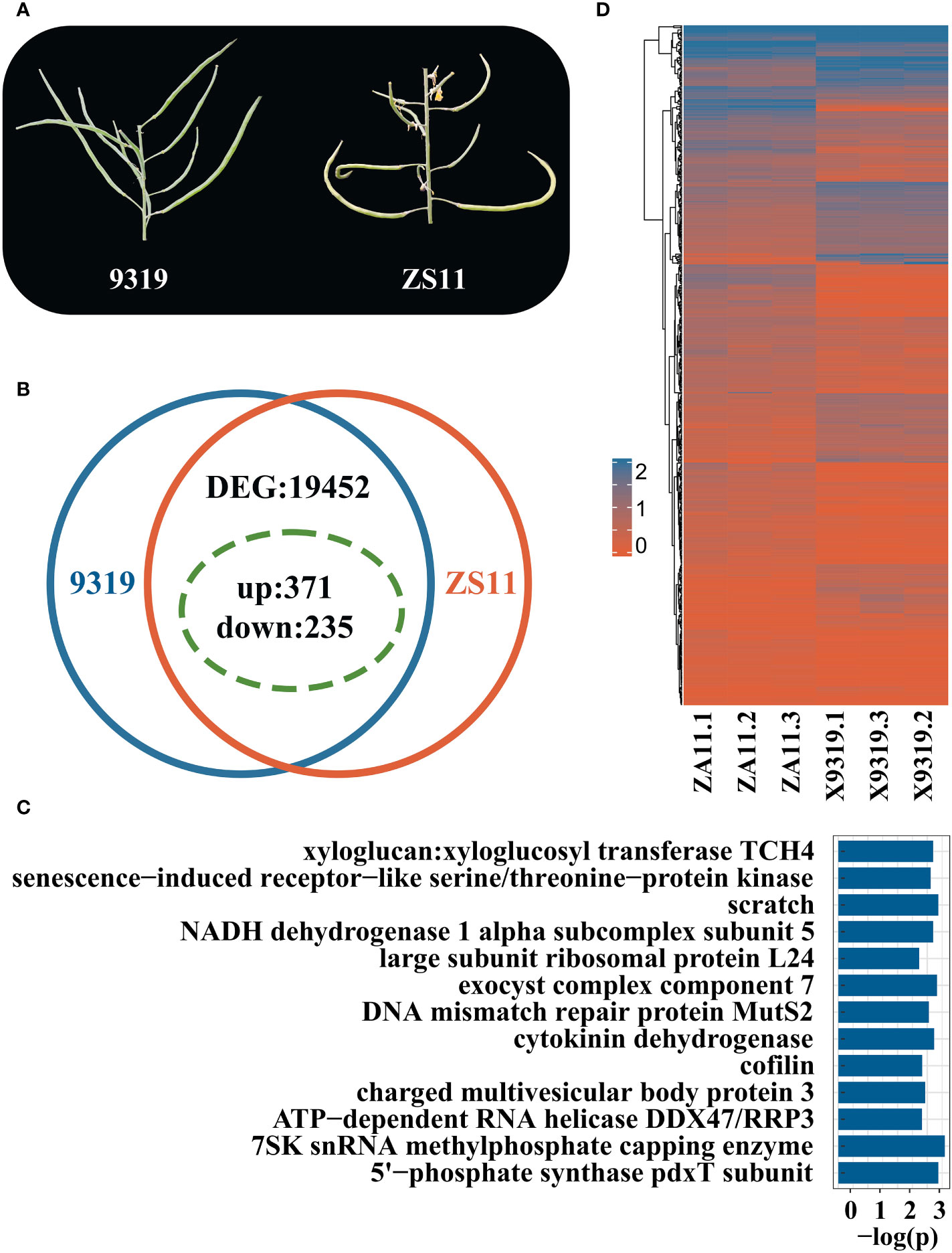
Figure 6 RNA-seq analysis of rapeseed lines with contrasting pod orientation phenotypes. (A) Phenotypic difference of pod orientation between 9319 line (Angle of the pedicle to raceme (inclined type, 59°, the angle of the pod to pedicel is 170°) and ZS11 line (Angle of pedicel to raceme, flat type, 90°; Angle of the pod to pedicel:170°); (B) Statistics summary of differentially expressed genes (DEGs) between 9319 and ZS11. The green dotted circle indicates the DEGs in QTL; (C) Enriched KEGG pathways of candidate DEG between 9319 and ZS11; (D) Heat map of candidate genes of DEG expression, the expression levels were transformed by log10(x+1), 0 represents that the gene was not expressed, and 2 represents that the expression level was high.
A total of 19,452 DEGs were identified between 9319 and ZS11 accessions, including 12,205 up-regulated genes and 7,247 down-regulated genes (Figures 6B, C). Of them, 606 DEGs were identified, underlying the QTL regions for pod orientation. Among 606 candidate genes, 235 DEGs were down-regulated, and 371 were up-regulated (Figure 6B). We detected 292 DEGs underlying qBnAPR.A02 and 314 DEGs underlying the qBnAPR.C02 (Supplementary Table 6).
KEGG enrichment analysis of 66 genes showed that the different candidate genes involved 309 pathways (Supplementary Table 7) and enriched for 13 pathways, including K00279 (Figure 6D). Among them, cytokinin dehydrogenase and xylloglucan: xylloglucosyl transferase TCH4 pathways are related to plant growth and development. Cytokinin dehydrogenase can promote the differentiation and growth of buds (Zhang et al., 2008). Xyloglucan: xyloglucosyl transferase TH4 plays an important role in the formation and reconstruction of cross-linking in xyloglucan and participates in the plant cell wall modification process (Jan et al., 2004). Annotations revealed 7 genes among 606 candidate genes related to auxin: BnaC02G0079400ZS, BnaC02G0175200ZS, BnaC02G0118700ZS, BnaC02G0102400ZS, BnaC02G0050100ZS, BnaC02G0166500ZS, and BnaA02G0138300ZS.
We focused on three interesting DEGs which underlie QTL for pod orientation: the first two are homologous genes related to auxin synthesis. BnaA02G0138300ZS and BnaC02G0175200ZS, designated as BnSAUR32.A02 and BnSAUR32.C02, respectively. The third gene was BnaA02G0200300ZS, which was related to root angle growth in Arabidopsis thaliana (Yang et al., 2020). BnaA02G0200300ZS was designated as BnLAZY4.A02. BnSAUR32.A02 and BnSAUR32.C02 genes are homologues that map in the proximity 1604 kb and 654 kb upstream of the peak SNP of the APR trait (Table 3). The BnSAUR32.A02 is a homologue of SAUR32 (AT5G53590) encoding SAUR-like auxin-responsive protein in Arabidopsis. BnSAUR32.C02 and AT5G53590 had poor homology and are SAUR32-like genes.
The BnLAZY4.A02 gene belongs to the IGT gene family, with 167 IGT family genes in 13 species. To investigate the relationship of BnLAZY4.A02 that was expressed in the pedicel of line 9319 but not in the pedicel of line ZS11, with other species, we carried out cluster analysis using neighbour-joining approach (Figure 7A). BnLAZY4.A02 was clustered on the same branch as Chinese cabbage and cabbage, which is consistent with the genetic lineage of Brassica species. There were three helical structures in the BnLAZY4.A02 protein with high B-factor values, which may regulate upstream and downstream genes (Figure 7B).
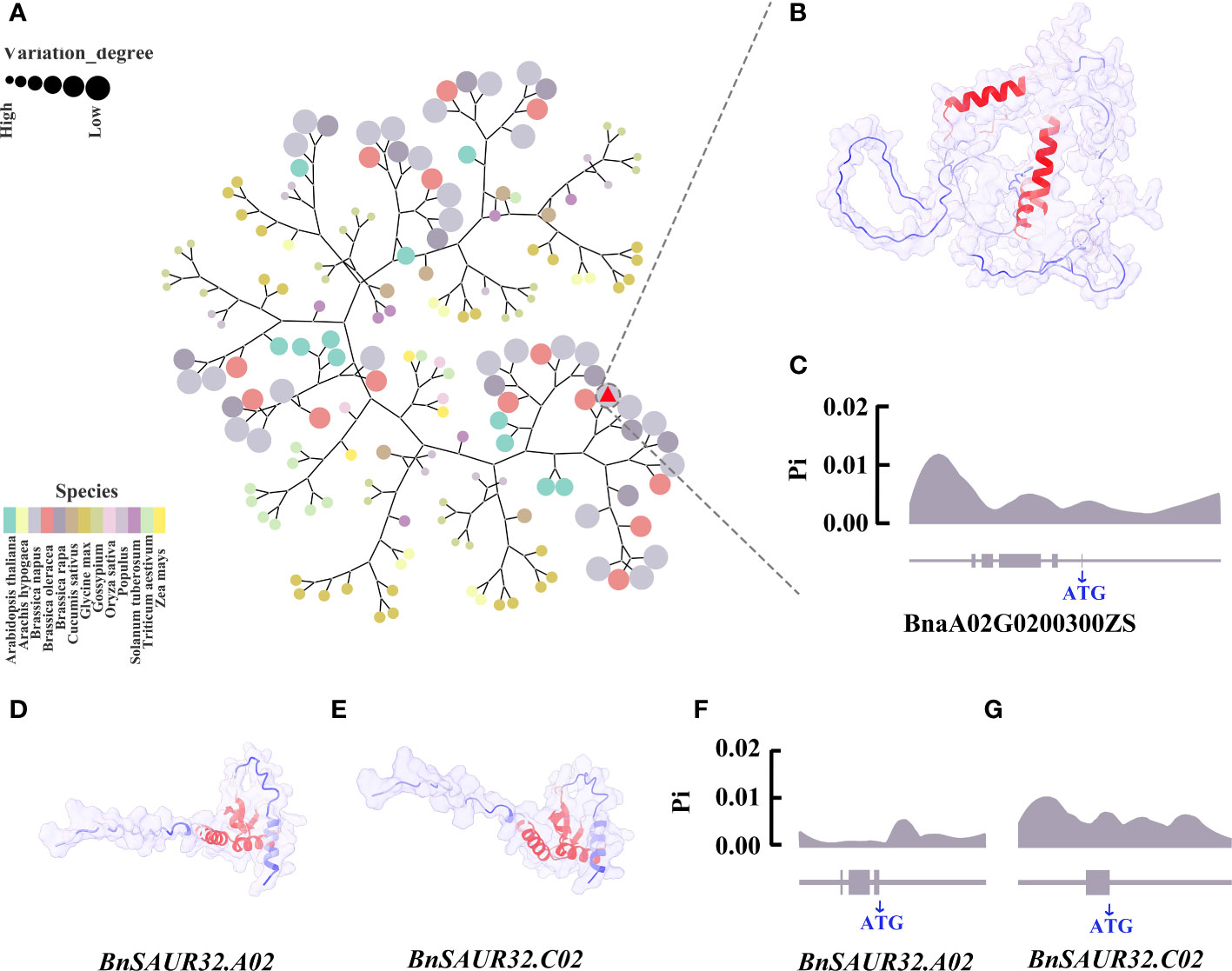
Figure 7 Bioinformatics analysis of three key candidate genes (A) Phylogenetic tree of IGT gene family in 13 species, including Arabidopsis thaliana, Arachis hypogaea, Brassica oleracea, Brassica napus, Brassica rapa, Cucumis sativus, Glycine max, Gossypium, Oryza sativa, Populus, Solanum tuberosum, Triticum aestivum and Zea mays. Different colours represent different species, the size of the circle represents the genetic relationship with rapeseed, and the larger the circle, the closer it is to rapeseed. Red triangle marked LAZY4. (B) Prediction of BnLAZY4.A02 protein structure using Alphafold2. The blue the protein colour, the lower the B-factor, and the red the protein colour, the higher the B-factor. (C) The nucleotide diversity distribution of BnLAZY4.A02 in 289 core germplasm collections; (D) Protein structure map predicted by BnSAUR32.A02 gene. The bluer the protein colour, the lower the B-factor, and the red the protein colour, the higher the B-factor; (E) Protein structure map predicted by BnSAUR32.C02 gene; (F) Distribution of nucleotide diversity of BnSAUR32.A02 gene in 289 core germplasm collections; (G) Distribution of nucleotide diversity of BnSAUR32.C02 gene in 289 core germplasm collections.
To gain insights into nucleotide diversity in candidate genes involved in pod orientation, we utilised public genomic sequence data of 289 rapeseed core collections (Xuan et al., 2020). Results showed that the nucleotide diversity peak appeared downstream of the BnLAZY4.A02 gene (Figure 7C), which could regulate the expression of this gene and hence, responsible for genetic variation for pod orientation in rapeseed. Further research is required to substantiate this proposed hypothesis. BnSAUR32.A02 and BnSAUR32.C02 genes belong to the SAUR gene family. We identified 1,857 SAUR family genes by gene family analysis in 13 species, including Arabidopsis thaliana, Arachis hypogaea, Brassica oleracea, Brassica napus, Brassica rapa, Cucumis sativus, Glycine max, Gossypium, Oryza sativa, Popular, Solanum tuberosum, Triticum aestivum and Zea mays (Supplementary Figure 6). The protein structures of BnSAUR32.A02 and BnSAUR32.C02 genes were very similar (Figures 7D, E). The active region of the protein was concentrated in the centre of the protein. In the core collection of 289 rapeseed accessions, the nucleotide of BnSAUR32.A02 and BnSAUR32.C02 genes had almost no variation, and its Pi value was low, 0.0001 (The critical value of nucleotide diversity is 0.005.) (Figures 7F, G). However, there was considerable nucleotide diversity (Pi value: 0.0087, Pi>0.005), in the upstream region of BnSAUR32.A02, which could affect the gene expression.
Crop architecture is an important physiological and agronomic trait that could drive rapeseed yield. The branch angle determines the overall plant shape, while the pod angle contributes to the photosynthesis rate. The ideal plant architecture can make the best use of resources such as light, water and nutrients, achieving the maximum yield and possibly the best quality. In recent years, crop architecture has become a vital breeding aim of genetic improvement programs. Investigating pod orientation via GWAS expands the research in two ways. First, this study mined the natural variation in pod orientation traits in a diverse GWAS panel of accessions rather than two parents, used in most biparental QTL mapping studies. Secondly, we identified four QTL and candidate genes associated with pod orientation.
Earlier studies investigated genetic variation for pod orientation using qualitative scores: straight, inclined, flat and droop (Wu et al., 2007). However, these assessments are subjective and less accurate for genomic analysis. In this study, we followed a more accurate technique, employing image processing to assess genetic variation in pod orientation (APP and APR). This approach avoided errors in the manual angle measurement under field conditions. Moreover, we used quantitative scores to classify pod orientation. We only found accessions with flat and inclined types in our GWAS panel. Further research is required to screen national and international diverse accessions for straight types desired by the rapeseed improvement programs. Moderate correlations across some environments suggest that pod orientation traits can be selected in rapeseed breeding and are suitable for selection in the early generation.
There were statistically significant G×E interactions for both APP. APR across environments. Therefore, we employed the multi-environment-based MLM model, thus excluding the impact of the environment on the genotype-phenotype association, as opposed to the approach followed by (Raman et al., 2022). We identified four significant QTL regions that accounted for a small proportion of variation (4.5 to 17%). These results suggest that variation in pod orientation is polygenic and influenced by the environment. In this study, we did not identify QTL associated with QTL × Environment interaction.
To identify favourable alleles for pod architecture, we sought an association between SNP marker alleles (Figure 5) and phenotypes. The flat type of pods occupies considerable space, is more prone to shading, limiting the photosynthetic area and is not suitable for high-density cultivation. While the straight or inclined type pods can maximise the function of light energy and space utilisation and have more advantages. Based on the above assumption, we defined the alleles that promote pod orientation to straight or inclined as the favourable allele. The AA marker genotype is a favourable allele for short pod angle on peak SNPs of four significant loci. qAPR.A02 delimited with Bn-A02-p9706613 marker and qAPP.A05 delimited with Bn-scaff_17441_3-p165957 marker cumulatively contributed 21.28% variation in pod orientation. GG alleles detected with SNPs markers (Bn-A02-p9706613 and Bn-scaff_17725_1-p505212) are in the smallest inclined type materials. Two SNPs, Bn-scaff_17441_3-p165957 and Bn-scaff_17441_3-p68051, are AA genotypes, and not all four loci are favourable alleles of pod orientation. On this basis, the germplasms of inclined type with GG alleles of qBnAPP.A05 and qBnAPP.C05 might be improved to AA alleles obtain straight type.
We followed two approaches to identify the candidate gene underlying QTL for pod orientation. Firstly, we examined genomic regions in LD (upstream and downstream 424 Kb for the A subgenome and 2.5Mb for the C subgenome). Secondly, we identified DEG underlying QTL regions. We also searched Arabidopsis thaliana priori candidate genes whose role in branch and inflorescence angle has been confirmed.
We prioritised three candidate genes: BnLAZY4.A02, BnSAUR32.A02 and BnSAUR32.C02, underlying pod orientation QTL (Table 2). BnLAZY4.A02 was expressed in the pod pedicel of 9319 but not in the pod pedicel of ZS11. The BnLAZY4.A02 gene belongs to the IGT family, which plays a role in the gravity signals of several plant species (Waite and Dardick, 2021). LAZY homologues have been shown to regulate shoot branching angles in Arabidopsis, rice, maize and several perennial tree crops (Roychoudhry and Kepinski, 2015). In Arabidopsis thaliana, recent studies have shown that increasing LAZY4 expression in pifq mutants can partially save the interrupted hypocotyl gravity phenotype of dark growing IFQ but cannot save the development of endoderm amyloplast (Yang et al., 2020). During the growth in Arabidopsis, amyloplast in columella cell sank due to gravity, and LAZY protein regulated auxin transport in a polarised way after feeling this change. The polar transport of auxin leads to anti-gravity growth (Furutani et al., 2020). Therefore, the mechanism of LAZY4 regulating pod angle may be the same. In a recent study on the effect of jasmonic acid on the lateral root orientation of Arabidopsis thaliana, the transcription activation of LAZY4 mediated by a jasmonic acid-related gene MYC2 also indicates that the LAZY4 gene is related to plant growth angle (Sharma et al., 2022).
Genetic variation in pod orientation QTL on A02 and C02 (Table 2) could be conditioned by small auxin up-regulated RNA genes (SAURs) homologues: BnSAUR32.A02 and BnSAUR32.C02. SAURs are implicated in regulating dynamic and adaptive growth in Arabidopsis and sunflower (Atamian et al., 2016; Bemer et al., 2017; Stortenbeker and Bemer, 2019). Auxin and other upstream factors can also regulate the expression of the SAURs gene (Favero et al., 2017; van Mourik et al., 2017; Hu et al., 2018). SAURs can interact with PP2C.D phosphatases (Spartz et al., 2014) to induce plant growth by regulating cell wall acidification. SAURs can also act independently of auxin. Zhou et al. identified 98 SAUR gene families in apple, and screened 25 QTLs for regulating root growth angle. Seven SAURs were selected as candidate genes for regulating root growth angle through protein-protein interaction network (Zhou et al., 2022). In the leaf angle of sorghum, a strong candidate gene SAUR36 was identified by RNA-seq, and its expression may increase the leaf angle (Natukunda et al., 2022). It is worth noting that the variation of the promoter region of BnSAUR32.A02 could also regulate genetic variation in pod orientation underlying QTL on A02 and C02 chromosomes.
In conclusion, we determined genetic variation in pod orientation in diverse rapeseed accessions. Utilising GWAS, we identified four significant QTL for pod orientation on A02, A05, C02, and C05 chromosomes in diverse rapeseed accessions. In addition, we performed RNA-seq analysis and prioritised three candidate genes: differentially expressed between inclined and flat pods and localised within the QTL associated with APP and APR. Thus, our GWAS and transcriptome studies delineated three putative candidate genes underlying pod orientation in B. napus. Overall, the results of this study provided the genetic tools and resources for understanding the molecular basis of variation in pod orientation of rapeseed. These findings may facilitate the development of and cultivation of rapeseed ideotypes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/?lang=en, CRA007893.
JL designed and supervised the study. WW and JL collected the samples. QH provided the germplasm. YY performed genetic, and transcriptome data analyses. YY, HR and JL discussed results and prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of China (U19A2029), the Central Public-interest Scientific Institution Basal Research Fund (No. 161017202203 and 1610172020001) and an open project (KF2020007) of the Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, P. R. China. We thank the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences for computational and experimental support. We also thank Grains Research and Development Corporation for supporting collaborative research carried out at NSW DPI and OCRI, Wuhan, under the DAN00208 project.
The handling editor CL declared a shared affiliation with the authors YY, WW, QH, JL at the time of review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1097534/full#supplementary-material
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data (Babraham Institute, Cambridge, United Kingdom: Babraham Bioinformatics).
Atamian, H. S., Creux, N. M., Brown, E. A., Garner, A. G., Blackman, B. K., Harmer, S. L. (2016). Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 353, 587–590. doi: 10.1126/science.aaf9793
Bai, S., Hong, J., Su, S., Li, Z., Wang, W., Shi, J., et al. (2022). Genetic basis underlying tiller angle in rice (Oryza sativa l.) by genome-wide association study. Plant Cell Rep. 41, 1707–1720. doi: 10.1007/s00299-022-02873-y
Bemer, M., van Mourik, H., Muiño, J. M., Ferrándiz, C., Kaufmann, K., Angenent, G. C. (2017). FRUITFULL controls SAUR10 expression and regulates Arabidopss growth and architecture. J. Exp. Bot. 68, 3391–3403. doi: 10.1093/jxb/erx184
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cardoso, M. A., Rizzardi, L. E., Kume, L. W., Groeneveld, C. S., Trefflich, S., Morais, D. A., et al. (2022). TreeAndLeaf: an R/Bioconductor package for graphs and trees with focus on the leaves. Bioinformatics 38, 1463–1464. doi: 10.1093/bioinformatics/btab819
Chen, X., Tong, C., Zhang, X., Song, A., Hu, M., Dong, W., et al. (2021). A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 19, 615–630. doi: 10.1111/pbi.13493
Chen, Y., Fan, X., Song, W., Zhang, Y., Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
de Saint Germain, A., Bonhomme, S., Boyer, F.-D., Rameau, C. (2013). Novel insights into strigolactone distribution and signalling. Curr. Opin. Plant Biol. 16, 583–589. doi: 10.1016/j.pbi.2013.06.007
Douglas, S. J., Riggs, C. D. (2005). Pedicel development in Arabidopsis thaliana: contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Dev. Biol. 284, 451–463. doi: 10.1016/j.ydbio.2005.06.011
Douglas, S. J., Chuck, G., Dengler, R. E., Pelecanda, L., Riggs, C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14, 547–558. doi: 10.1105/tpc.010391
Ewels, P., Magnusson, M., Lundin, S., Käller, M. (2016). MultiQC: summarise analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Favero, D. S., Le, K. N., Neff, M. M. (2017). Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-# 3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J. 89, 1133–1145. doi: 10.1111/tpj.13451
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Freed, D., Aldana, R., Weber, J. A., Edwards, J. S. (2017). The sentieon genomics tools-a fast and accurate solution to variant calling from next-generation sequence data. BioRxiv, 115717. doi: 10.1101/115717
Furutani, M., Hirano, Y., Nishimura, T., Nakamura, M., Taniguchi, M., Suzuki, K., et al. (2020). Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 11 (1), 1–13. doi: 10.1038/s41467-019-13729-7
Ha, C. M., Jun, J. H., Nam, H. G., Fletcher, J. C. (2007). BLADE-ON-PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19, 1809–1825. doi: 10.1105/tpc.107.051938
Hobbie, L., McGovern, M., Hurwitz, L. R., Pierro, A., Liu, N. Y., Bandyopadhyay, A., et al. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127, 23–32. doi: 10.1242/dev.127.1.23
Hu, W., Yan, H., Luo, S., Pan, F., Wang, Y., Xiang, Y., et al. (2018). Genome-wide analysis of poplar SAUR gene family and expression profiles under cold, polyethylene glycol and indole-3-acetic acid treatments. J. Plant Physiol. 128, 50–65. doi: 10.1016/j.plaphy.2018.04.021
Jan, A., Yang, G., Nakamura, H., Ichikawa, H., Kitano, H., Matsuoka, M., et al. (2004). Characterisation of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 136, 3670–3681. doi: 10.1104/2Fpp.104.052274
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Katoh, K., Misawa, K., Kuma, K. I., Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Khan, M., Xu, M., Murmu, J., Tabb, P., Liu, Y., Storey, K., et al. (2012). Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 158, 946–960. doi: 10.1104/pp.111.188573
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/2Fmolbev%2Fmsy096
Ku, L., Wei, X., Zhang, S., Zhang, J., Guo, S., Chen, Y. (2011). Cloning and characterisation of a putative TAC1 ortholog associated with leaf angle in maise (Zea mays l.). PloS One 6, e20621. doi: 10.1371/journal.pone.0020621
Leng, S. H., Zhu, G. G., Deng, X. L. (1992). Studies on the sources of the dry matter in the seed of rapeseed. Acta Agronomica Sin 18(4), 250–257.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, H., Shen, J.-J., Zheng, Z.-L., Lin, Y., Yang, Z. J. P. P. (2001). The rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126, 670–684. doi: 10.1104/pp.126.2.670
Li, L., Chen, B., Yan, G., Gao, G., Xu, K., Xie, T., et al. (2020). Proposed Strategies and current progress of research and utilization of oilseed rape germplasm in China. J. Plant Genet. Resour. 21, 1–19.
Li, P., Wang, Y., Qian, Q., Fu, Z., Wang, M., Zeng, D., et al. (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. doi: 10.1038/cr.2007.38
Li, Y., Pi, L., Huang, H., Xu, L. (2012). ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. J. Exp. Bot. 63, 1423–1433. doi: 10.1093/jxb/err376
Li, Z., Liang, Y., Yuan, Y., Wang, L., Meng, X., Xiong, G., et al. (2019). OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localisation. Mol. Plant 12, 1143–1156. doi: 10.1016/j.molp.2019.05.014
Lin, D., Xiang, Y., Xian, Z., Li, Z. G. (2016). Ectopic expression of SlAGO7 alters leaf pattern and inflorescence architecture and increases fruit yield in tomato. Physiologia plantarum 157, 490–506. doi: 10.1111/ppl.12425
Liu, K., Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Liu, J., Wang, W., Mei, D., Wang, H., Fu, L., Liu, D., et al. (2016). Characterising variation of branch angle and genome-wide association mapping in rapeseed (Brassica napus L.). Front. Plant Sci 7, 21. doi: 10.3389/fpls.2016.00021
Lorenzi, G., Perbal, G. (1990). Root growth and statocyte polarity in lentil seedling roots grown in microgravity or on a slowly rotating clinostat. Physiologia Plantarum 78, 532–537. doi: 10.1111/j.1399-3054.1990.tb05238.x
Morita, M. T., Sakaguchi, K., Si, K., Taira, K., Kato, T., Nakamura, M., et al. (2006). A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 47, 619–628. doi: 10.1111/j.1365-313X.2006.02807.x
Natukunda, M. I., Mantilla-Perez, M. B., Graham, M. A., Michelle, A. G., Peng, L., Maria, G. S. (2022). Dissection of canopy layer-specific genetic control of leaf angle in sorghum bicolor by RNA sequencing. BMC Genomics 23, 95. doi: 10.1186/s12864-021-08251-4
O'Connor, B., Van Auwera, G. (2020). Genomics in the cloud: using docker, gatk, and WDL in Terra (CA, USA: O'Reilly Media Sebastopol).
Okamura, M., Hirose, T., Hashida, Y., Yamagishi, T., Ohsugi, R., Aoki, N. (2013). Starch reduction in rice stems due to a lack of OsAGPL1 or OsAPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture. Funct. Plant Biol. 40, 1137–1146. doi: 10.1071/FP13105
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2021). UCSF ChimeraX: Structure visualisation for researchers, educators, and developers. Protein Sci. 30, 70–82. doi: 10.1002/pro.3943
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Ragni, L., Belles-Boix, E., Pautot, V. (2008). Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20, 888–900. doi: 10.1105/tpc.108.058230
Raman, H., Raman, R., Pirathiban, R., Mcvittie, B., Sharma, N., Liu, S., et al. (2022). Multi-environment QTL analysis delineates a major locus associated with homoeologous exchanges for water-use efficiency and seed yield in allopolyploid brassica napus. Plant Cell Environ. 45, 12019–12036. doi: 10.1111/pce.14337
Roychoudhry, S., Del Bianco, M., Kieffer, M., Kepinski, S. (2013). Auxin controls gravitropic setpoint angle in higher plant lateral branches. Curr. Biol. 23, 1497–1504. doi: 10.1016/j.cub.2013.06.034
Roychoudhry, S., Kepinski, S. (2015). Shoot and root branch growth angle control–the wonderfulness of lateralness. Curr. Opin. Plant Biol. 23, 124–131. doi: 10.1016/j.pbi.2014.12.004
Sakuraba, Y., Piao, W., Lim, J.-H., Han, S.-H., Kim, Y.-S., An, G., et al. (2015). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339. doi: 10.1093/pcp/pcv144
Sang, D., Chen, D., Liu, G., Liang, Y., Huang, L., Meng, X., et al. (2014). Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. 111, 11199–11204. doi: 10.1073/pnas.1411859111
Sharma, M., Sharma, M., Jamsheer, K. M., Laxmi, A. (2022). Jasmonic acid coordinates with light, glucose and auxin signalling in regulating branching angle of arabidopsis lateral roots. Plant Cell Environ. 45, 1554–1572. doi: 10.1111/pce.14290
Sinhamahapatra, S., Raman, R. B., Roy, S., Roy, U. S., Raut, N. M., Ashok, K. V. (2010). Breeding for an ideal plant type in yellow sarson (Brassica rapa l. yellow sarson). Electronic J. Plant Breed. 1, 689–694.
Spartz, A. K., Ren, H., Park, M. Y., Grandt, K. N., Lee, S. H., Murphy, A. S., et al. (2014). SAUR inhibition of PP2C-d phosphatases activates plasma membrane h+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26, 2129–2142. doi: 10.1105/tpc.114.126037
Stortenbeker, N., Bemer, M. (2019). The SAUR gene family: the plant's toolbox for adaptation of growth and development. J. Exp. Bot. 70, 17–27. doi: 10.1093/jxb/ery332
Su, S.-H., Keith, M. A., Masson, P. H. (2020). Gravity signaling in flowering plant roots. Plants 9, 1290. doi: 10.3390/plants9101290
Sun, J., Xiao, T., Nie, J., Chen, Y., Lv, D., Pan, M., et al. (2019). Mapping and identification of CsUp, a gene encoding an auxilin-like protein, as a putative candidate gene for the upward-pedicel mutation (up) in cucumber. BMC Plant Biol. 19, 1–12. doi: 10.1186/s12870-019-1772-4
Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197
R Core Team (2013). R: A language and environment for statistical computing. Vienna, Austria. Available at: http://www.R-project.org/.
Turner, S. D. (2014). Qqman: an r package for visualising GWAS results using QQ and manhattan plots. Biorxiv, 005165. doi: 10.1101/005165
van der Honing, H. S., Kieft, H., Emons, A. M. C., Ketelaar, T. (2012). Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 158, 1426–1438. doi: 10.1104/pp.111.192385
van Mourik, H., van Dijk, A. D., Stortenbeker, N., Angenent, G. C., Bemer, M. (2017). Divergent regulation of Arabidopsis SAUR genes: a focus on the SAUR10-clade. BMC Plant Biol. 17, 1–14. doi: 10.1186/s12870-017-1210-4
Venglat, S., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., et al. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. 99, 4730–4735. doi: 10.1073/pnas.072626099
Waite, J. M., Dardick, C. (2018). TILLER ANGLE CONTROL 1 modulates plant architecture in response to photosynthetic signals. J. Exp. Bot. 69, 4935–4944. doi: 10.1073/pnas.072626099
Wang, H., Cheng, H., Wang, W., Liu, J., Hao, M., Mei, D., et al. (2016). Identification of BnaYUCCA6 as a candidate gene for branch angle in brassica napus by QTL-seq. Sci. Rep. 6, 38493. doi: 10.1038/srep38493
Wang, L., Wang, S., Li, W. (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. doi: 10.1093/bioinformatics/bts356
Wang, W. X., Hu, Q., Mei, D. S., Li, Y. C., Wang, H., Fu, L., et al. (2015). Evaluation branch and pod angle measurement based on digital images from Brassica napus l. (in Chinese). Chin. J. Oil Crop Sci. 37, 566. doi: 10.7505/j.issn.1007-9084.2015.04.020
Wang, X., Yu, R., Wang, J., Lin, Z., Han, X., Deng, Z., et al. (2020). The asymmetric expression of SAUR genes mediated by ARF7/19 promotes the gravitropism and phototropism of plant hypocotyls. Cell Rep. 31, 107529. doi: 10.1016/j.celrep.2020.107529
Wickham, H. (2011). Wiley interdisciplinary reviews: Computational statistics. ggplot2. 3, 180–185. doi: 10.1002/wics.147
Wu, X. M., Chen, B. Y., Lu, G. Y. (2007). Description specification and data standard of rapeseed germplasm resources (in Chinese) (Wuhan, Hubei: China Agricultural Publishing House).
Wu, X., Tang, D., Li, M., Wang, K., Cheng, Z. (2013). Loose plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 161, 317–329. doi: 10.1104/pp.112.208496
Xuan, L., Yan, T., Lu, L., Zhao, X., Wu, D., Hua, S., et al. (2020). Genome-wide association study reveals new genes involved in leaf trichome formation in polyploid oilseed rape (Brassica napus l.). Plant Cell Environ. 43, 675–691. doi: 10.1111/pce.13694
Xu, M., Zhu, L., Shou, H., Wu, P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Yang, P., Wen, Q., Yu, R., Han, X., Deng, X. W., Chen, H. (2020). Light modulates the gravitropic responses through organ-specific PIFs and HY5 regulation of LAZY4 expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 18840–18848. doi: 10.1073/pnas.2005871117
Ye, J., McGinnis, S., Madden, T. L. (2006). BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34, W6–W9. doi: 10.1093/nar/gkl164
Yin, L., Zhang, H., Tang, Z., Xu, J., Yin, D., Zhang, Z., et al. (2021). rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinf. 19, 619–628. doi: 10.1016/j.gpb.2020.10.007
Zhang, C., Dong, S.-S., Xu, J.-Y., He, W.-M., Yang, T.-L. (2019). PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788. doi: 10.1093/bioinformatics/bty875
Zhang, F., Guo, D. P., Lin, M. L., Yan, M. M. (2008). Physiological, biochemical and molecular properties of cytokinin Oxidase/Dehydrogenase (in Chinese). Plant Physiol. Commun. 44, 797–803. doi: 10.1111/j.1399-3054.1994.tb00668.x
Zhang, S. C., Yang, C. W. (2012). Identification and function analysis of the silique angle gene SPA in Arabidopsis (in Chinese). J. South China Normal Univ. 44, 107–111. doi: 10.6054/j.jscnun.2012.06.023
Zhang, N., Yu, H., Yu, H., Cai, Y., Huang, L., Xu, C., et al. (2018). A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 30, 1461–1475. doi: 10.1105/tpc.18.00063
Keywords: rapeseed, pod orientation, GWAS, RNA-seq, candidate genes
Citation: Yang Y, Wang W, Hu Q, Raman H and Liu J (2023) Genome-wide association and RNA-seq analyses identify loci for pod orientation in rapeseed (Brassica napus). Front. Plant Sci. 13:1097534. doi: 10.3389/fpls.2022.1097534
Received: 14 November 2022; Accepted: 21 December 2022;
Published: 13 January 2023.
Edited by:
Chengzhen Liang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Mingxun Chen, Northwest A&F University, ChinaCopyright © 2023 Yang, Wang, Hu, Raman and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Liu, bGl1amlhMDJAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.