
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 January 2023
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1096804
This article is part of the Research TopicMeiosis in Plants: sexual reproduction, genetic variation and crop improvementView all 8 articles
 Zhaoran Tian1†
Zhaoran Tian1† Chengyan Ji1,2†
Chengyan Ji1,2† Zhengqing Xie1
Zhengqing Xie1 Xinjie Shi1,2
Xinjie Shi1,2 Baoming Tian1
Baoming Tian1 Gangqiang Cao1
Gangqiang Cao1 Xiaochun Wei2
Xiaochun Wei2 Yan Yang1
Yan Yang1 Fang Wei1*
Fang Wei1* Gongyao Shi1*
Gongyao Shi1*Trigenomic Brassica allohexaploids (AABBCC, 2n = 6x = 54) have great potential in oilseed breeding and genetic diversity. However, Brassica allohexaploids do not exist naturally, and the underlying mechanism regulating pollen fertility in artificially synthesized Brassica allohexaploids is still unclear. In this study, synthetic Brassica allohexaploids were produced by crossing allotetraploid B. carinata (BBCC, 2n = 4x = 34) and diploid B. rapa (AA, 2n = 2x = 20), followed by chromosome doubling. The results showed that the pollen fertility was significantly reduced and the pollen structures were mostly distorted, but the nursing anther tapetum developed normally in the synthetic Brassica allohexaploids. Furthermore, the data showed that the meiotic events occurred irregularly with uneven chromosome segregation and microspore development appeared mostly abnormal. Transcription analysis showed that the upregulation of genes related to the negative regulation of flower development and the downregulation of genes related to chromosome segregation might play an essential role in reduction of pollen fertility in the Brassica allohexaploids. In conclusion, this study elucidated the related mechanisms affecting pollen fertility during male gametophytic development at the cytological and transcriptomic levels in the newly synthesized Brassica allohexaploids.
Interspecific hybridization and polyploidization are among the dominant driving forces in plant speciation and evolution (Bennett, 2004). Interspecific hybridization can improve crop quality and yield by transferring favorable traits from distant relatives to synthesized hybrids (Snowdon et al., 2000; Peterka et al., 2004). Polyploidization of hybrids (allopolyploidy) can contribute to phenotypic diversity and adaptation to a wider range of environmental conditions (Sattler et al., 2016). Allopolyploids usually form between relatively closely related species with crossability, either within the same genus or at least within the same tribe (Fitzjohn et al., 2007). This means that there are many regions of sequence similarity in common by descents between the two parental genomes in the new allopolyploids. During meiosis, the presence of these homoeologous regions suggests that the cell machinery often have difficulty in correctly identifying homologous chromosomes in the newly formed allopolyploids.
The pairing homoeologous 1 (Ph1) locus in bread wheat has been identified as controlling diploid-like chromosome pairing by preventing homeologous chromosome pairing (HECP) (Griffiths et al., 2006; Bhullar et al., 2014). In addition, some potential quantitative trait loci for the accurate regulation of meiosis in allopolyploids have only been identified in haploid B. napus (Jenczewski et al., 2003) and Arabidopsis suecica (Henry et al., 2014; Lloyd and Bomblies, 2016). However, the mechanism regulating the meiotic stability of newly synthesized Brassica allohexaploids remains unclear. Compared with the established allopolyploids, the newly synthesized allopolyploids have more synaptic multivalents lasting until metaphase I (MI), resulting in chromosome missegregation, aneuploid gametes and compromised fertility (Szadkowski et al., 2010; Xiong et al., 2011).
Not only meiosis can affect pollen fertility, but abnormal microspore development may lead to decreased pollen fertility. Microgametogenesis includes an asymmetric division to form a big vegetative and a small generative nucleus, and the generative nucleus produces two male gametes after a mitotic division (Huang et al., 2021). In addition, the tapetum is the innermost layer of the four sporophytic layers in the anther wall, which can directly affect the development of gametophytes and is of great importance for the development from microspores to pollen grains (Parish and Li, 2010; Du et al., 2019). As the secretory cell layer, the tapetum provides enzymes for the microspores to be released from the tetrads and nutrients for pollen development.
Allopolyploidy leads to transcriptome reprogramming, and the transcriptome changes of allopolyploidy may be an adaptive mechanism to promote the establishment of gene expression programs and stabilize the evolution of species (Pikaard, 2001). The genus Brassica includes six cultivated Brassica species, three diploids (B. rapa, 2n = 20, AA; B. nigra, 2n = 16, BB; B. oleracea, 2n = 18, CC) have evolved into three allotetraploids (B. napus, 2n = 38, AACC; B. carinata, 2n = 34, BBCC; B. juncea, 2n = 36, AABB) through pairwise hybridization and subsequent chromosome doubling in natural conditions, termed as “U’s triangle”, which represents a classical evolutionary process and serves as a useful model for polyploidization and chromosomal evolution (Warwick and Al-Shehbaz, 2006). However, there are no Brassica allohexaploid species (2n = 54, AABBCC) in nature. Numerous attempts have been made to synthesize Brassica allohexaploids through different cross combinations, however, the newly synthesized Brassica allohexaploids tend to have lower pollen fertility than the parents (Chen et al., 2020). A series of broad transcriptome changes were produced in Brassica allohexaploids compared with their parents, and these changes were gernerally related with the plant growth and development, as well as gamete development (Zhao et al., 2013; Ji et al., 2022).
In the present study, comprehensive cytological and transcriptomic analyses were performed to reveal the underlying mechanism related to pollen fertility in newly formed trigenomic Brassica allohexaploids (AABBCC) obtained by hybridization of the allotetraploid B. carinata and the diploid B. rapa, followed by chromosome doubling. This study provides insights into pollen fertility in newly formed Brassica allohexaploids, which will help us in Brassica polyploid breeding and genetic improvements in future.
The materials used in this study were inbred and kept in our laboratory. The synthetic Brassica allohexaploid (AABBCC, 2n = 54) was generated via the crossing between the inbreeding line B. carinata (2n = 34, BBCC, genotype ‘VI047487’) as the maternal parent and B. rapa (2n = 20, AA, genotype ‘JK66-83’) by manual pollination (Yang et al., 2020), following chromosome doubling with colchicine treatment. At 9–12 days after pollination (dap), the immature embryos were rescued on MS agar medium in the growth room. The putatively trigenomic hybrids (ABC, 2n = 27) were treated with colchicine (200 mg/L) for 10 days to double the chromosome numbers on MS agar medium. All the plants were grown in the greenhouse under a 16 h light/8 h dark photoperiod at 22°C.
The pistils were collected from the developing buds, treated with 2 mM 8-hydroxyquinoline at 20°C for 4 h, then washed with distilled water and fixed in Carnot fixed solution for 4 h. Cytogenetic observation was carried out according to previous study (Miyashita et al., 2011). Fresh young leaves of B. rapa, B. carinata and Brassica allohexaploid plants were taken for ploidy detection. Flow cytometry was performed according to the procedure detailed in previous studies (Yin et al., 2020). The peak fluorescence intensity X-Mean is proportional to the cellular DNA content, so the ploidy of the sample was determined according to the peak position of B. rapa and B. carinata. Finally, five hexaploid plants (euploid) were confirmed to have 54 chromosomes, so we selected these five Brassica allohexaploid plants and five randomly selected B. rapa and B. carinata respectively for this study.
Pollen grain stainability was used as an indication of pollen viability, which was determined by staining at least 3000 pollen grains from randomly selected flower buds of each plant in Alexander staining (Solarbio, Beijing, China) (Alexander, 1969).
Anthers with determined stages were first fixed in formalin-acetic acid-alcohol (FAA) under a vacuum for 1 h. After dehydration in a graded ethanol series and diaphaneity in a clearing medium (xylene), the samples were embedded in paraffin (Leica, Weztlar, Germany). Sections (8 μm) were obtained with a Leica Reichert Supernova microtome (Leica, Weztlar, Germany), placed on glass slides, and stained with hematoxylin and eosin (Solarbio, Beijing, China) following manufacturer specifications (Park et al., 1998). The sections were examined with a fluorescence microscope (Olympus BX43, Tokyo, Honshu, Japan), and images were captured by a CCD attached camera DP73 (Olympus, Tokyo, Japan) with CellSens Standard software.
The developing microspores and mature pollens were stained with 4′,6′-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO, USA) solution according to the procedures detailed (Oh et al., 2010). After the whole inflorescences were fixed with Carnot solution, the microspores at different development stages were stained with DAPI solution. Using a fluorescence microscope (Olympus BX43, Tokyo, Honshu, Japan) under either bright field or epifluorescence, microscopy imaging was carried out.
Young flowers were harvested and fixed in Carnot fixation solution for 12 h and then transferred to 70% ethanol at 4°C for storage. Meiotic chromosome behavior observations were performed as described previously (Braynen et al., 2017). Meiotic chromosome spreads were prepared as previously described (Zeng et al., 2017). Propidium iodide (PI, Boster Biotechnology, California, USA) solution was applied to stain the prepared slides, and chromosome behavior during meiosis was observed by a fluorescence microscope (Olympus BX43, Tokyo, Honshu, Japan).
For the detection of β-tubulin, procedures from a previous study were adopted, with few modifications (Yang et al., 2019). Fresh inflorescences were collected and fixed immediately in 4% (w/v) paraformaldehyde. The pollen mother cells (PMCs) were first blocked in 5% BSA for 1h. The PMCs were incubated with monoclonal anti-β-tubulin IgG (Sigma-Aldrich, St. Louis, MO, USA) diluted at a 1:100 ratio for 12 h at 4°C in a moist chamber. Then, the PMCs were incubated with a fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA) diluted at a 1:100 ratio for 2 h at 37°C in a dark chamber. Subsequently, a drop of the cell solution was applied to a clean slide after three PBS rinses, and the slide was then stained with 40 g/mL PI. The prepared glass slides were observed and imaged using a Carl Zeiss Confocal Laser Scanning Microscope (LSM 880, Carl Zeiss AG, Oberkochen, Germany).
Genomic DNA from B. nigra was extracted from fresh leaves by the cetyltrimethylammonium bromide (CTAB) method (Springer, 2010). Genomic DNA probes were labeled with Digoxigenin-11-dUTP using a Nick Translation Kit (Roche, Mannheim, Germany). The PMCs were isolated from Brassica allohexaploids during the meiosis stage, fixed in Carnot fixation solution for 12 h, and stored in 70% alcohol. Chromosomes were prepared and GISH was performed by following the previous description (Zhou et al., 2016). The slides were observed under fluorescence microscope (Olympus BX43, Tokyo, Honshu, Japan), and Adobe Photoshop CS6 (SAN Jose, California, USA) was employed for the appropriate adjustment of all the images.
The transcriptome sequencing data of flower buds for Brassica allohexaploids, B. rapa and B. carinata were downloaded from the NCBI Gene Expression Omnibus (GEO) with accession numbers GSE201456, GSE193368 and GSE185639, respectively. The reads were then aligned to the B. rapa genome sequence using HISAT2 v2.1.0, the reference genome and the annotation file were downloaded from B. rapa genome v3.5 sequence (http://Brassicadb.cn, accessed on 23 May 2022) (Kim et al., 2015). Then, the fragments per kilobase per million reads (FPKM) of each gene was calculated to estimate the expression level. DESeq2 was used to perform differential expression analysis on two groups (three biological replicates each), genes with fold change ≥ 2 and false discovery rates (FDR) < 0.05 were designated as DEGs (Love et al., 2014). The Gene Ontology (GO) enrichment analysis was implemented by the hypergeometric test, and GO terms with FDR < 0.05 were considered significantly enriched.
To verify the ploidy of the newly synthesized Brassica allohexaploid plants, we first performed chromosome counting and flow cytometric analysis of these plants. The mitotic cells of immature pistils in the B. rapa, B. carinata and Brassica allohexaploids showing 20, 34 and 54 chromosomes (Supplementary Figures S1A–C). Flow cytometric analysis showed a consistent increase in DNA content in the Brassica allohexaploids, indicating potential allohexaploids (Supplementary Figures S1D–F). Compared with the parental lines, the possible Brassica allohexaploid plants showed vigorous vegetative growth, thickened leaves and larger flowers (Supplementary Figures S1G–O). The morphology of Brassica allohexaploid plants was similar to that of the maternal line, B. carinata, with serrated leaves, yellow flowers and independent vernalization (Supplementary Figures S1J–O).
The pollen fertility of the Brassica allohexaploids was determined by Alexander staining. Compared with both parental lines, the Brassica allohexaploids had lower pollen fertility, with an average pollen fertility rate of 66.1% (n =3106), while the average stainable rate of B. rapa was 95.8% (n = 3959) and that of B. carinata was 98.8% (n = 3795) (Figure 1). These results indicated that we had successfully produced the newly synthetic Brassica allohexaploid plants with reduced pollen fertility.
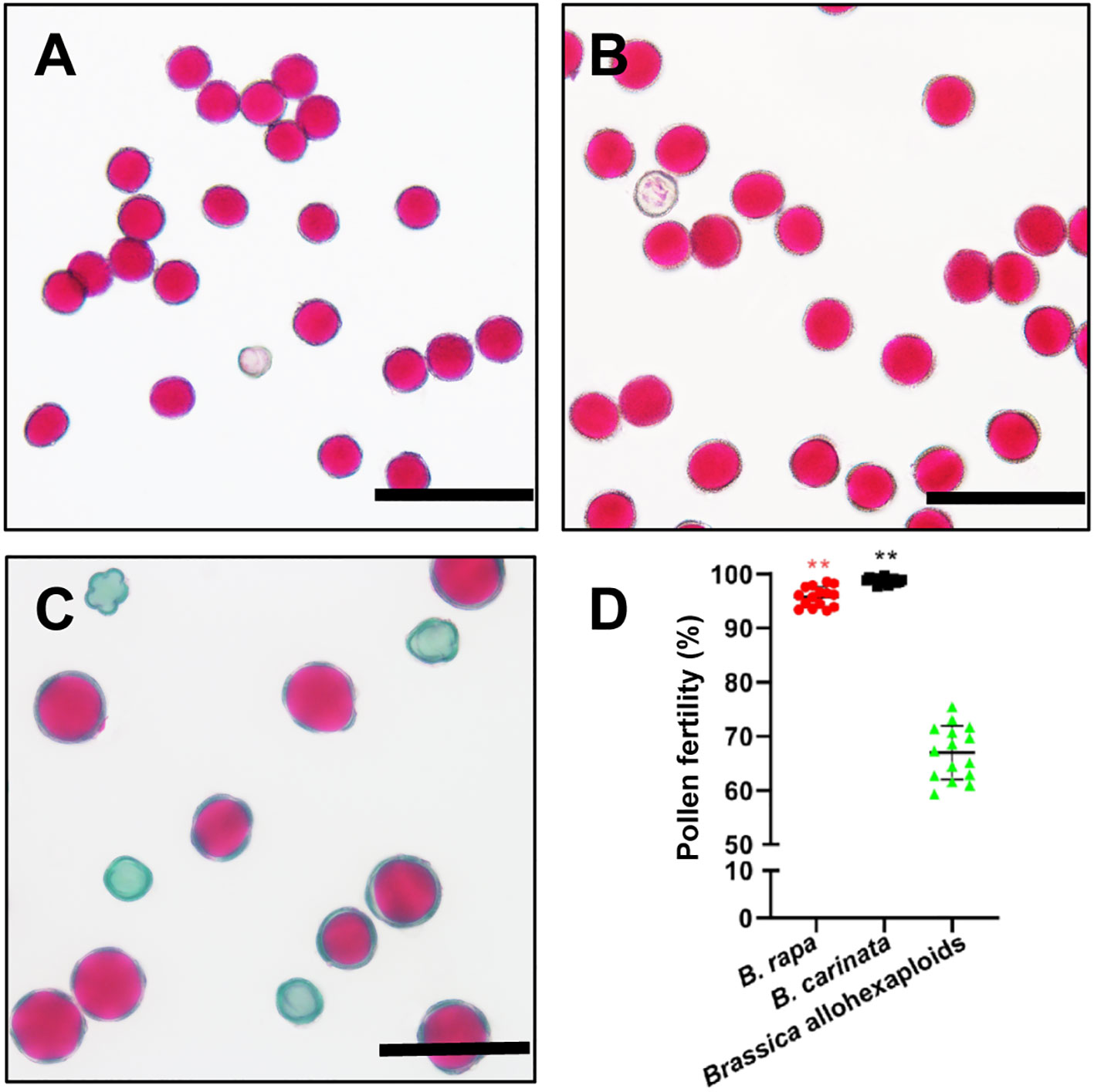
Figure 1 Pollen fertility and morphology observation analyses of B. rapa, B. carinata and Brassica allohexaploids. (A–C) Pollen fertility of B. rapa, B. carinata and Brassica allohexaploids determined by Alexander staining. (D) Statistical analysis of pollen fertility determined by Alexander staining. ** indicates a significant difference at P ≤ 0.01 analyzed by Analysis of Variance (ANOVA).
To determine whether tapetum development was affected in the Brassica allohexaploids, transverse anther sections were further examined by paraffin sectioning at different stages. No obvious morphological difference in the tapetum was observed between the Brassica allohexaploids and the corresponding parents at different anther development stages, and the tapetum, middle layer, endothecium and epidermis all developed normally in the Brassica allohexaploids, as well as in the parental lines (Supplementary Figure S2). During the microsporogenesis stage, PMCs underwent meiosis and formed tetrads. The nursing tapetum began vacuolating and turned into a secretory cell layer, while the middle layer cells became very thin and tended to degenerate (Supplementary Figures S2A, E, I). Subsequently, microspores freely released from the tetrads by callose enzymes produced by the tapetum, which then became more condensed and deeply stained, yet no longer had large vacuoles with hardly visible middle layers (Supplementary Figures S2B, F, J). Microspores gradually increased in size and separated into two unequal daughter cells, while the tapetum was further degraded to provide nutrients and materials for microspore development (Supplementary Figures S2C, G, K). Although the Brassica allohexaploid plants produced some aborted pollen grains at the mature stage, tapetum development and degeneration were normal (Supplementary Figure S2L). These observations suggested that the Brassica allohexaploids exhibited normal tapetum development and tapetum degeneration to provide nutrients and substances for subsequent gametogenesis.
The reduced fertility of mature pollen grains had greatly attracted our interest; thus, we conducted further work to determine whether microspore nucleus underwent abnormal development by observing DAPI-stained spores at different developmental stages. The typical and normal process of microspore development was observed in the parent lines, and some normal microspore development could also be found in the Brassica allohexaploids (Figures 2A–O). Meiosis of PMCs resulted in a tetrad enclosed within a thick callose wall (Figures 2A, F, K). Microspores freely released from the tetrad (Figures 2B, G, L) gradually increased in size and became polarized, with the nucleus displaced to a future germ cell pole (Figures 2C, H, M). Then, a curved cell plate separated the two unequal daughter cells (Figures 2D, I, N). The smaller generative cells, which had nuclei stained intensely with DAPI, underwent pollen mitosis to generate two sperm cells (Figures 2E, J, O). However, some aberrant cell developments were observed at the tetrad stage, with various numbers of nuclei in the Brassica allohexaploids (Figures 2P, Q, U, V). Both B. rapa and B. carinata had 100% normal tetrads composed of four equally sized microspores, but this proportion was reduced to 83.5% at the tetrad stage in the Brassica allohexaploids (Supplementary Figures S3A). Parts of the tetrads (14.7%) were severely abnormal, with highly variable numbers of spores from monad to triad (Figures 2P, Q, U, V). Even relatively normal-looking tetrads in the Brassica allohexaploids gave rise to microspores undergoing defective asymmetric cell division, which might account for the high proportion of abnormal pollen grains (34.6%) with equal nuclei (Figure 2R) and abnormal nuclei (Figure 2W) observed at the early bicellular stage (Supplementary Figure S3B). B. rapa and B. carinata had almost all normal tricellular pollen, while the Brassica allohexaploid plants had 65.5% normal pollen, and the rest showed a mixture of phenotypes, including binucleate and uninucleate pollen grain (Figures 2S, T, X, Y; Supplementary Figure S3C). Taken together, these results clearly showed that the some of microspore development was defective in the synthetic Brassica allohexaploids.
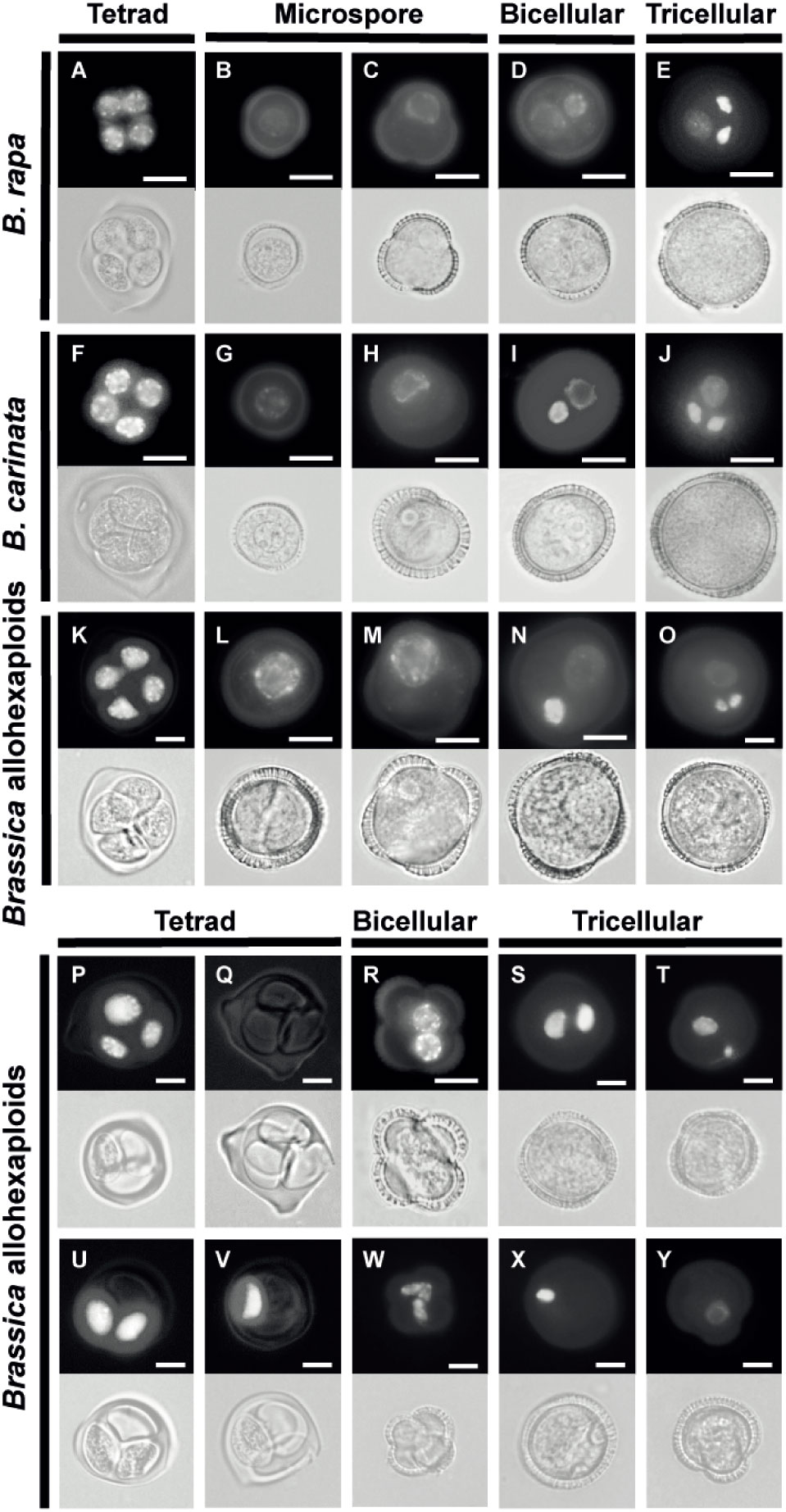
Figure 2 Abnormal pollen development in the synthetic Brassica allohexaploids. Normal pollen development in B. rapa (A–E), B. carinata (F–J) and Brassica allohexaploids (K–O) at different stages. Abnormal pollen development in the Brassica allohexaploids at the tetrad stage (P, Q, U, V), the bicellular stage (R, W) and the tricellular stage (S, T, X, Y). Fluorescent (up) and bright-field images (down) are shown. Bars = 10 μm.
The majority of synthetic polyploids exhibit meiosis aberration, which might lead to abnormalities in pollen development (Tian et al., 2010; Gaebelein et al., 2019). Thus, we assessed chromosome behavior during meiosis in the PMCs of the Brassica allohexaploid plants. Throughout meiosis, some normal chromosome behavior was detected in the Brassica allohexaploids (Figures 3O–U), as was generally observed in B. rapa and B. carinata (Figures 3A–N). These all normal chromosomes were predominantly condensed into bivalents at diakinesis (Figures 3A, H, O), and at metaphase I, the bivalents were orderly aligned along the metaphase plate (Figures 3B, I, P). The homologous chromosomes were evenly separated through anaphase I and telophase I (Figures 3C, D, J, K, Q, R). Then, two groups of condensed sister chromosomes were arranged at the equatorial plate during metaphase II (Figures 3E, L, S). Finally, chromatids separated to each spindle pole at anaphase II and formed tetrads at telophase II (Figures 3F, G, M, N, T, U).
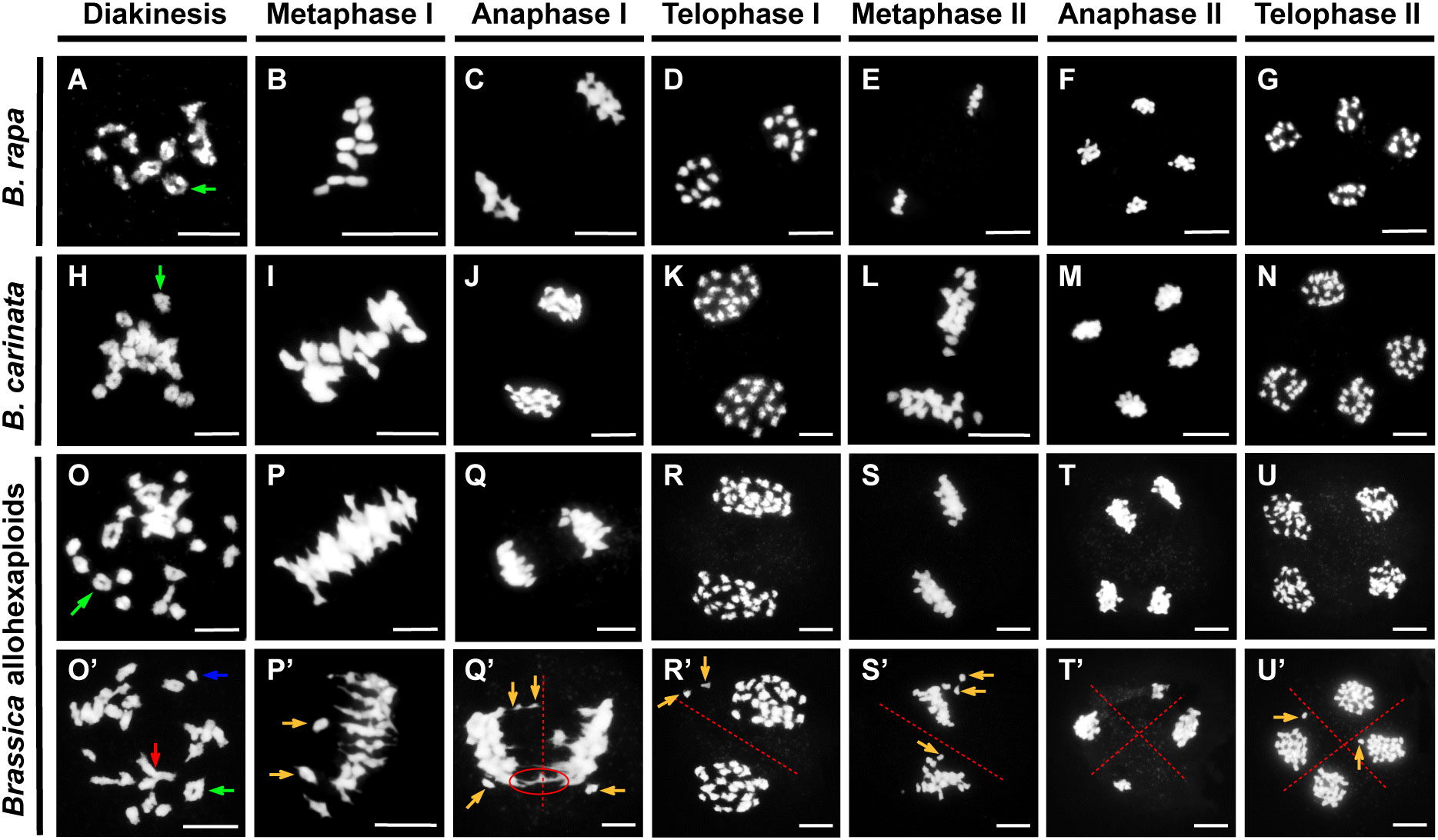
Figure 3 Meiotic chromosome behavior in B. rapa, B. carinata and Brassica allohexaploids. Normal meiosis processes in B. rapa (A–G), B. carinata (H–N) and Brassica allohexaploids (O–U). (O’–U’) Abnormal chromosome behavior of pollen mother cells (PMCs) in the Brassica allohexaploid plants. The green arrow indicates a normal bivalent; the blue arrow indicates a univalent; the red arrow indicates a multivalent; and the brown arrow indicates a lagging chromosome. The chromosome bridge is indicated with a red ellipse. Bars = 10 µm.
Normal chromosome behavior could be observed in the Brassica allohexaploid plants, and bivalents were detected with high frequency, but univalents and multivalents were also frequently observed in PMCs from the Brassica allohexaploids at diakinesis (Figures 3O, O’). At metaphase I, 85.6% of PMCs carried certain chromosomes detached from the equatorial plate with lagging chromosomes (Figures 3P’; Supplementary Figure S4). Subsequently, meiotic chromosomes were segregated, and chromosome bridges and unequal segregation events occurred in 72.4% of PMCs at anaphase I and in 36.9% of PMCs at anaphase II (Figures 3Q’, R’; Supplementary Figure S4). At metaphase II, 86.5% PMCs were observed with lagging chromosomes, which finally led to unbalanced gametes with unequal chromosome numbers in each microspore after telophase II (Figures 3S’, U’; Supplementary Figure S4). These observations suggested that the development of PMCs had severe defects in the Brassica allohexaploids.
In order to observe the microtubule dynamics of the newly synthesized Brassica allohexaploids during meiosis, the microtubule dynamics with separating chromosomes during meiosis were studied by β-tubulin immunostaining. Balanced chromosome segregation and complete meiotic cytokinesis were observed in B. rapa and B. carinata (Figure 4). During pachytene and diakinesis, condensed chromosomes and some microtubules appeared as intense foci around the perinuclear zone (Figures 4A, B, A’, B’). At metaphase I, microtubules were organized into the spindle structure and then attached to the kinetochores, forming a typical bipolar fusiform configuration at the metaphase plate (Figures 4C, C’). Then, the spindle pulled each group of homologous chromosomes to the polar side of the cell at anaphase I (Figures 4D, D’). During meiosis II, two pairs of spindles were constructed at metaphase II (Figures 4E, E’) and ensured the accurate disjunction of sister chromatids at anaphase II (Figures 4F, F’), consequently contributing to tetrad formation, where radial microtubule arrays were normally generated surrounding the nuclei at telophase II (Figures 4G, G’).
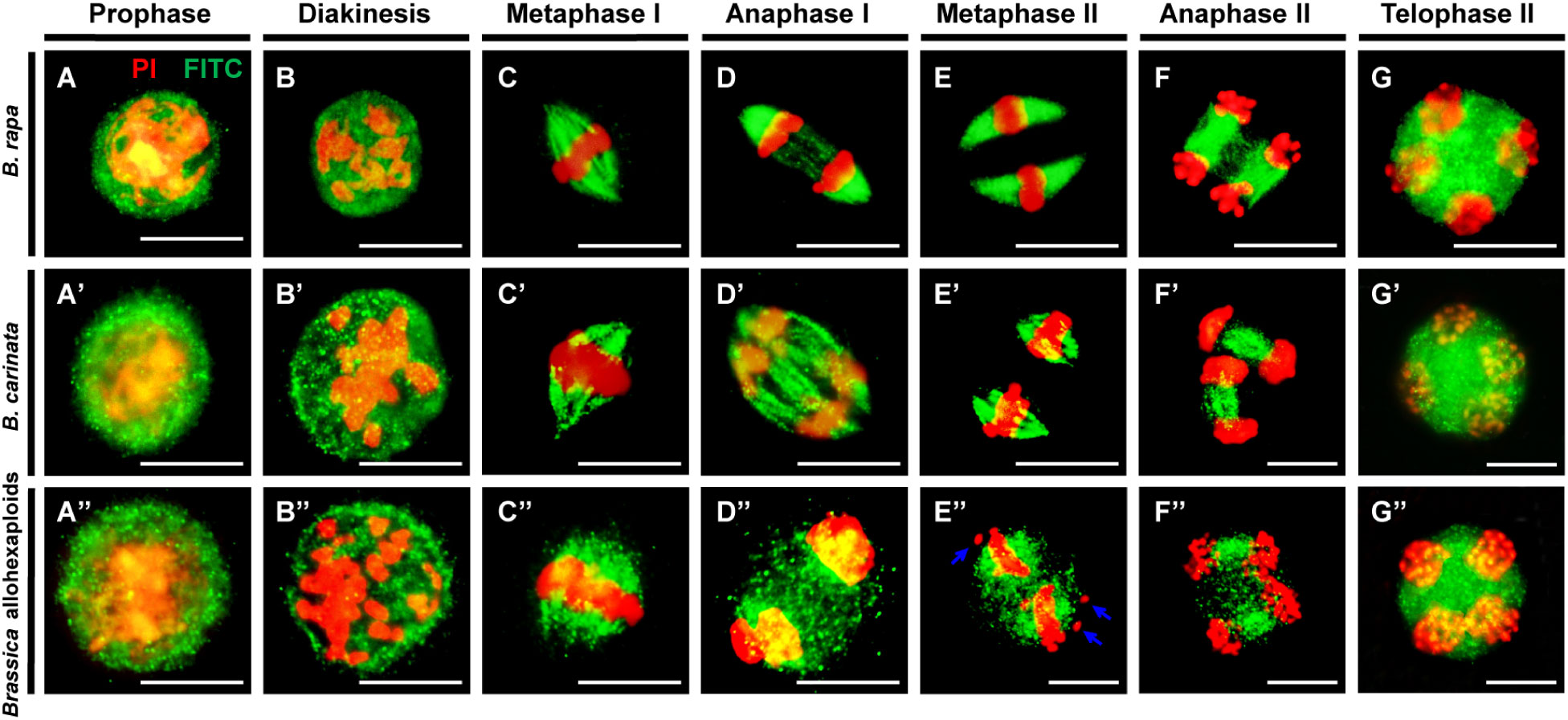
Figure 4 Spindle organization during meiosis in B. rapa, B. carinata and Brassica allohexaploids. (A–G) Normal spindle organization and chromosome behavior observed in pollen mother cells (PMCs) of B rapa. (A’–G’) Normal spindle organization and chromosome behavior observed in PMCs of B carinata. (A”–G”) Abnormal spindle organization and chromosome behavior observed in PMCs of the Brassica allohexaploids. Microtubules and chromosomes are colored in green and red, respectively. Blue arrows indicate lagging chromosomes. Bars = 10 µm.
In contrast, the Brassica allohexaploids showed alterations in some microtubule dynamics. During the early and later stages of meiosis, the Brassica allohexaploids exhibited a microtubule distribution pattern similar to that in B. rapa and B. carinata (Figures 4A”, B”). However, there were some irregular spindles with punctate foci of signals indicating a failure of the typical bipolar spindle structure (Figure 4C”). Notably, some chromosomes were not attached to the meiotic spindles in the Brassica allohexaploids (Figure 4E”). Some microtubule fibers were more scattered at anaphase I, anaphase II and telophase II, which could not guarantee the accomplishment of successful and accurate chromosome segregation in the Brassica allohexaploids (Figures 4D”, F”, G”). The abnormal microtubules were counted as 6.61%, 27.84% and 61.52% in B. rapa, B. carinata and Brassica allohexaploids, respectively (Supplementary Figure S5). These results demonstrated that impaired bipolar spindle during meiosis may contribute to low pollen fertility in the newly synthesized Brassica allohexaploids.
Chromosomes must first recognize their homologous partners and then pair with them during early meiotic prophase I to ensure accurate chromosome segregation (Xiong et al., 2021). Hence, chromosome segregation defects generally exist in plants with chromosome pairing problems. To reveal the interactions between the three genomes resulting from hybridization and genome doubling in the Brassica allohexaploid plants, GISH analysis was performed with the B genome probes from the B genome of B. nigra.
Homologous chromosomes were closely aligned at the pachytene in both B. rapa and B. carinata, indicating complete synapsis and pairing (Figures 5A, B). Although almost all of the chromosomes could complete synapsis and pairing in the Brassica allohexaploids, the existence of unpaired chromosomes in most PMCs of the Brassica allohexaploids suggested the occurrence of incomplete pairing and synapsis (Figure 5C). Meanwhile, GISH results showed that all of the chromosomes of the B genome were cohesively placed on the equatorial plate at metaphase I (Figures 6A, G, M) and metaphase II (Figures 6D, J, P), while lagging chromosomes were stained red without the FITC label, indicating that the lagging chromosomes were from the A and C genomes rather than the B genome. Moreover, the lagging chromosomes and chromosome bridges also existed without B genome-labeled chromosomes at anaphase I (Figures 6B, H, N) and anaphase II (Figures 6E, K, Q), demonstrating no B genome chromosome loss in the Brassica allohexaploids. In addition, the chromosome sets of the B genome were regularly distributed in dyad PMCs, with eight chromosomes in each pole at telophase I (Figures 6C, I, O), and could be equally segregated into tetrads at telophase II (Figures 6F, L, R). Therefore, these results indicated that B genome was less prone to missegregation compared to A and C genome during meiosis in the Brassica allohexaploids.
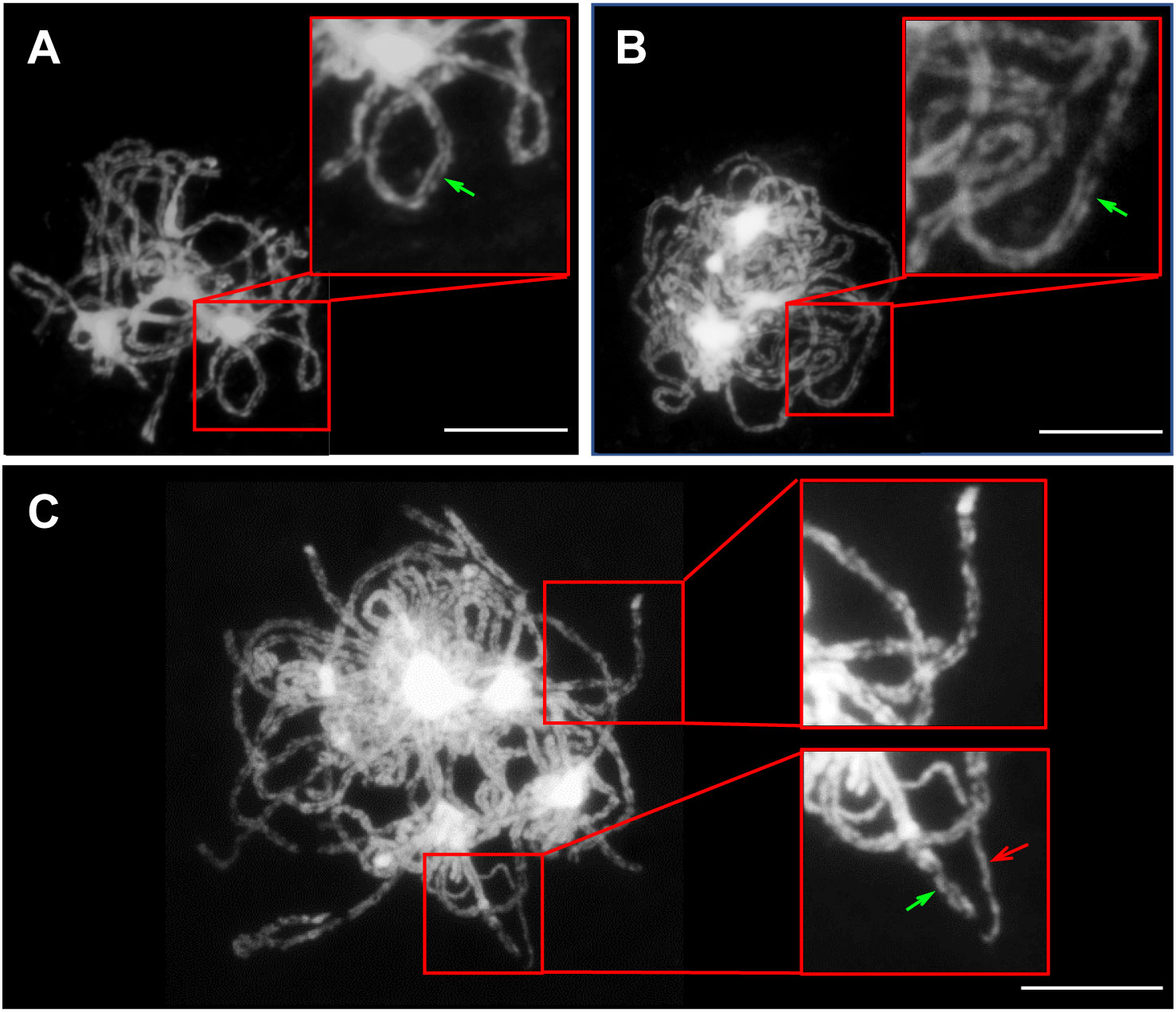
Figure 5 Pairing and synapsis of homologous or homoeologous chromosomes at pachytene. Fully synapsed and paired chromosomes in B. rapa (A) and B. carinata (B); in contrast, partially synapsed and pairing chromosomes at pachytene in the Brassica allohexaploids (C). Green arrows indicate normal synapsed and paired chromosomes, while red arrows indicate the non-synapsed and unpaired chromosomes. Bars = 10 µm.
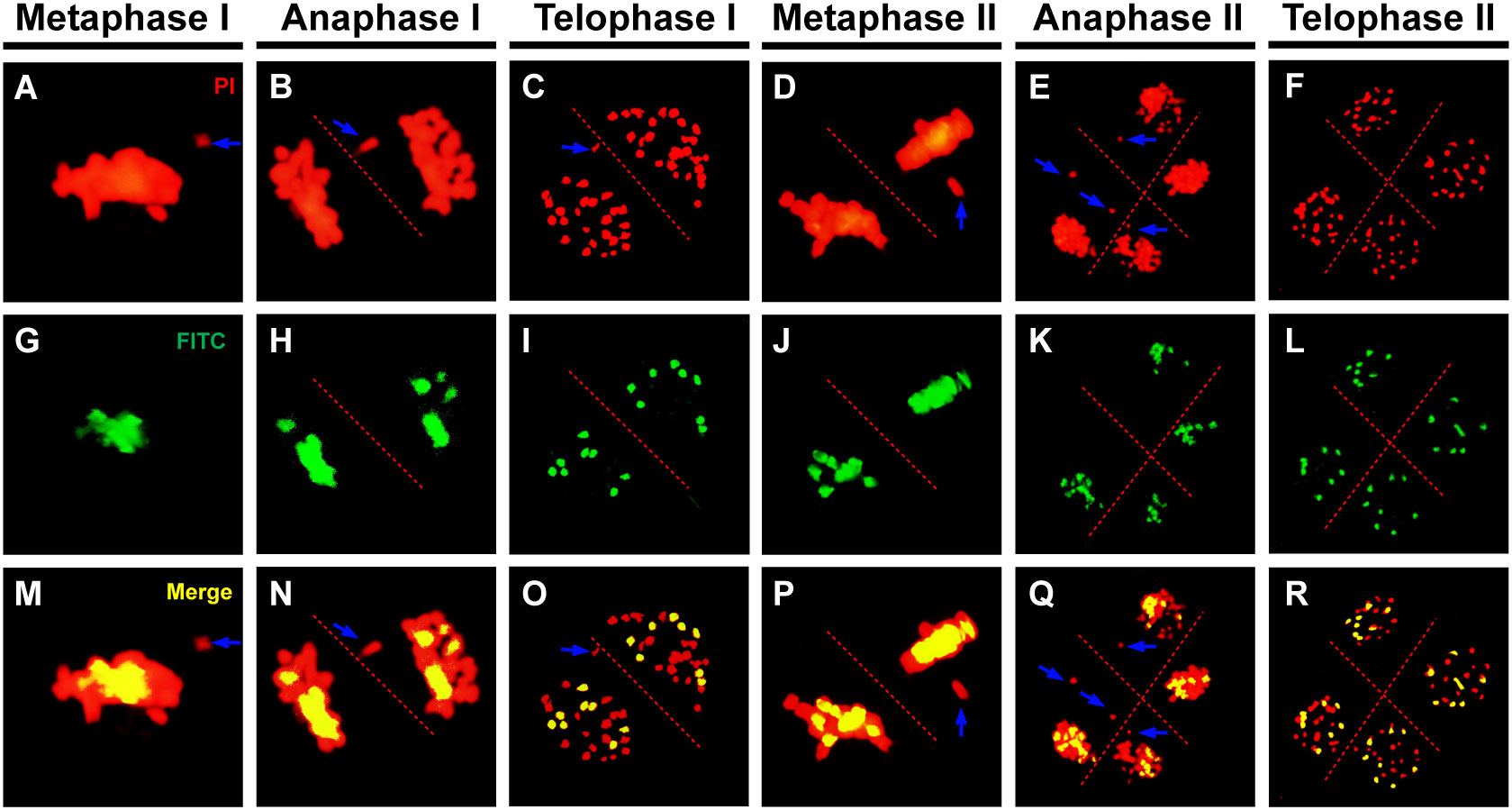
Figure 6 Genomic in situ hybridization (GISH) analysis of the Brassica allohexaploids during meiosis. (A–F) Chromosomes stained with propidium iodide (PI, red). (G–L) B genome chromosomes labeled by fluorescein isothiocyanate (FITC, green). (M–R) Merged overlay of the two signals (yellow). Blue arrows indicate lagging chromosomes from the A or C genome. Bars = 10 µm.
To determine the gene expression changes of floral buds in the synthesized Brassica allohexaploids, transcriptome analysis of the Brassica allohexaploids and their parents was performed using RNA-Seq. In the flower buds of the Brassica allohexaploids, 8249 genes were upregulated compared with B. rapa and 8150 genes were upregulated compared with B. carinata (Figure 7A). Venn diagram showed that there were 3463 genes upregulated in the flower buds of the Brassica allohexaploids compared to their parents (Figure 7A). The flower buds of the Brassica allohexaploids had 7013 downregulated genes compared with B. rapa and 4214 downregulated genes compared with B. carinata (Figure 7B). The 1349 genes were found to be downregulated in the flower buds of the Brassica allohexaploids as compared to their parents (Figure 7B). These findings demonstrated that the Brassica allohexaploids had more upregulated genes than downregulated genes compared with their parents. To explore the functional differences of DEGs among the flower buds between the Brassica allohexaploids and their parents, we focused on the significantly enriched GO Biological Process (BP) terms. In the flower buds of the Brassica allohexaploids, genes upregulated compared to their parents were mainly significantly enriched in microtubule-based movements (GO:0007018), DNA repair (GO:0006281), negative regulation of flower development (GO:0009910), double-strand break repair via homologous recombination (GO:0000724) and synapsis (GO:0007129), while genes downregulated compared to their parents were more enriched in postreplication repair (GO:0006301), fructose metabolic process (GO:0006000), response to abscisic acid (GO:0009737), chromosome segregation (GO:0007059) and ubiquitin-dependent protein catabolic process (GO:0006511)(Figure 7C).
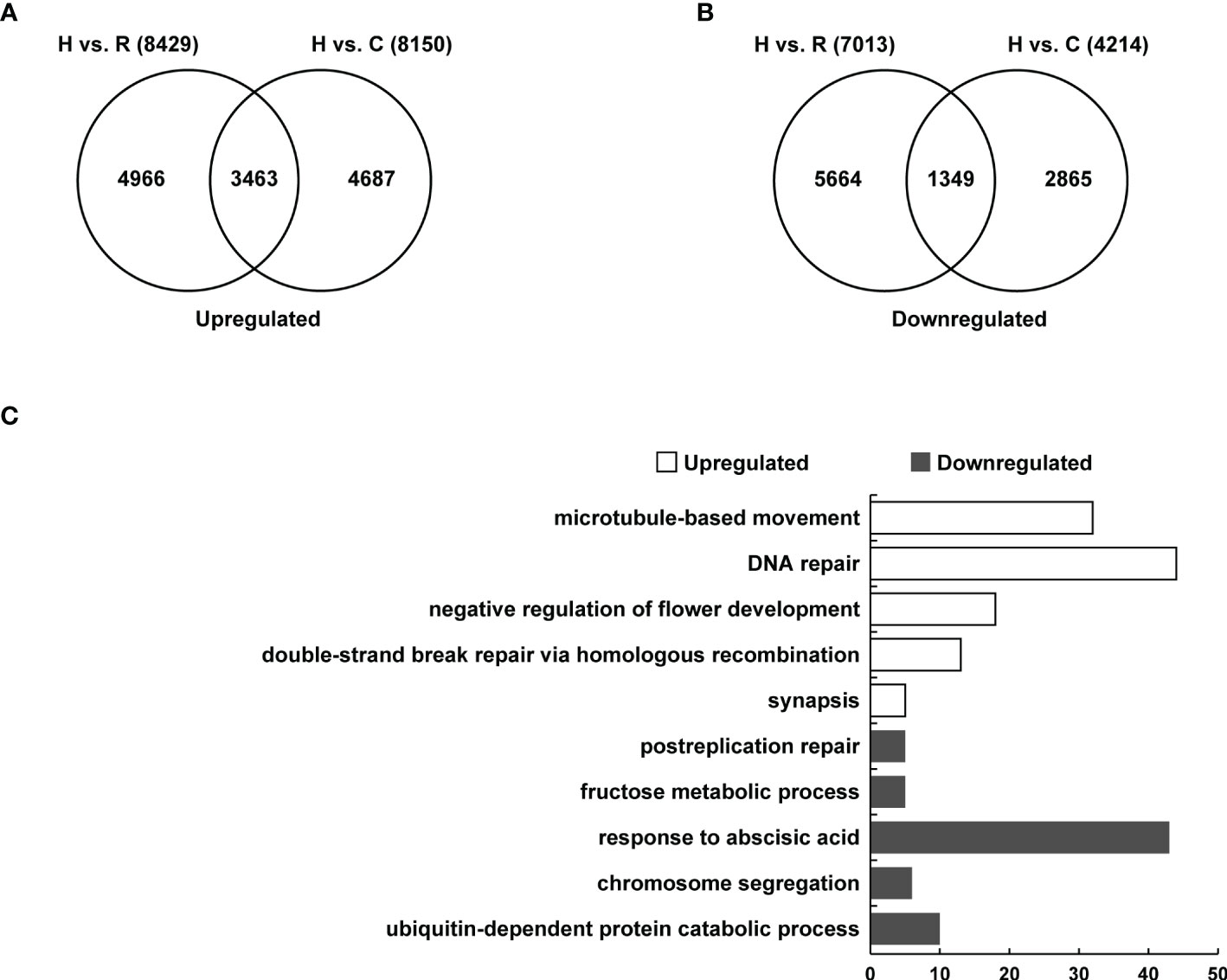
Figure 7 Transcriptome analysis of flower buds at meiosis in the Brassica allohexaploids. (A) Venn diagram showing the overlap of upregulated genes between Brassica allohexaploids vs. B. rapa and Brassica allohexaploids vs. B. carinata. (B) Venn diagram showing the overlap of downregulated genes between Brassica allohexaploids vs. B. rapa and Brassica allohexaploids vs. B. carinata. (C) GO classification of the overlapping genes upregulated and downregulated between Brassica allohexaploids vs. B. rapa and Brassica allohexaploids vs. B. carinata.
Morphological analysis showed that pollen fertility was 66.1%, on average, in Brassica allohexaploids, which was significantly reduced compared to that in the parental lines (Figure 1). Tapetum development in the corresponding Brassica allohexaploids was normal during anther development (Supplementary Figure S2). This meant that tapetum development became normal and could support the normal anther development of Brassica allohexaploid plants during early cytological diploidization. However, the development of microspore nucleus was partially abnormal in the Brassica allohexaploids. The percentage of abnormal microspore development increased from the tetrad stage to the tricellular stage (from 16.5% to about 34%; Supplementary Figure S3) in Brassica allohexaploids, indicating partially abnormal microspore development in nascent allohexaploids. Collectively, in addition to the typically reported meiotic defects in polyploids, these observations indicate the existence of defective microspore development in Brassica allohexaploids, which might be another result of meiotic defective or genetic exchange between different genomes or even a nucleo-cytoplasmic interaction (Comai, 2005; Geng et al., 2013; Yang et al., 2016). These results suggest that microspore abnormalities may affect the fertility of newly synthesized Brassica allohexaploids. The gene expression patterns synthesized Brassica allohexaploids and their parents were compared in this study, to further reveal the molecular mechanisms affecting pollen development. Enrichment of upregulated genes in the negative regulation of flower development may affect pollen fertility.
Meiosis is the key biological process that underpins sexual reproduction (Osman et al., 2011). Thus, successful and accurate chromosome segregation during meiosis is significant for genetic stability during sexual reproduction, which might be a crucial challenge in polyploids consisting of more than two sets of chromosomes (Tian et al., 2010; Mwathi et al., 2017). For Brassica allotriploids, severely unstable meiosis occurred with variations in chromosome behavior, such as univalents and multivalents at diakinesis, lagging chromosomes at metaphase, anaphase and telophase, unequal segregation and chromosome bridges at anaphase in all PMCs (Yang et al., 2020). For the corresponding Brassica allohexaploids, stable chromosomal behavior was observed in some PMCs (Figures 3O–U). However, unstable meiosis still occurred in Brassica allohexaploid plants (Figures 3O’–U’), putatively resulting from unstable chromosome pairing and genetic recombination between homoeologous and non-homologous regions during early cytological diploidization in Brassica allohexaploids (Tian et al., 2010; Geng et al., 2013; Gupta et al., 2016; Zhou et al., 2016). The downregulation of ASK1 suggested that chromosome segregation may have been shown to perform important roles in pollen fertility in the Brassica allohexaploids (Zhao et al., 2006). Moreover, the bipolar spindle is essentially required for chromosome movement and segregation during meiosis, and precise chromosome segregation is accomplished by the proper attachment of chromosomes to spindle microtubules via the kinetochore (Duro and Marston, 2015; Severson et al., 2016). This study demonstrated that no significant reduction of microtubule fibers occurred during meiosis in Brassica allohexaploids; however, incomplete spindle organization (Figure 4) could not provide enough force for accurate chromosome movements, thereby causing meiosis abnormalities (Hotta et al., 2012). Genes for chromosome segregation were downregulated may lead to reduced pollen fertility.
In the present work, we investigated the complete cytological process during male gamete formation in Brassica trigenomic allohexaploids via a cross between natural allotetraploid B. carinata and diploid B. rapa followed with chromosome doubling. In the newly synthesized Brassica allohexaploids, anther tapetum developed normally, while the microspore development was defective. In addition, chromosome behavior was mostly irregular, and the bipolar spindle during meiosis was partially abnormal in the Brassica allohexaploids. At the transcriptional level, the upregulation of genes related to the negative regulation of flower development and the downregulation of genes related to chromosome segregation may influence pollen fertility in the Brassica allohexaploids. Taken together, our results provide detailed cytological and transcriptomic insights into pollen development in the newly synthesized Brassica allohexaploids, which would be considered as a useful germplasm for Brassica polyploid breeding.
The data presented in the study are deposited in the NCBI, accession number GSE201456, GSE193368 and GSE185639.
FW, GS and XW conceived, designed and instructed the study. BT, GC and YY bred the plant materials. ZX and XS assisted with material identification. XS, ZT and CJ performed the main experiments and data analysis, and wrote the manuscript. ZT, CJ and FW amended the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by Henan Provincial Natural Science Foundation of China (No. 202300410366), and the Program for Science & Technology Innovation Talents in Universities of Henan Province (No. 19HASTIT014), and Youth Innovation Project of Key discipline of Zhengzhou University (No. XKZDQN202002), and the Fostering Project for Basic Research of Zhengzhou University (No. JC21310015).
We would like to express our thanks to the reviewers for their useful comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1096804/full#supplementary-material
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Bennett, M. D. (2004). Perspectives on polyploidy in plants - ancient and neo. Biol. J. Linn. Soc. 82, 411–423. doi: 10.1111/j.1095-8312.2004.00328.x
Bhullar, R., Nagarajan, R., Bennypaul, H., Sidhu, G. K., Sidhu, G., Rustgi, S., et al. (2014). Silencing of a metaphase I-specific gene results in a phenotype similar to that of the pairing homeologous 1 (Ph1) gene mutations. Proc. Natl. Acad. Sci. United States America 111, 14187–14192. doi: 10.1073/pnas.1416241111
Braynen, J., Yang, Y., Wei, F., Cao, G. Q., Shi, G. Y., Tian, B. M., et al. (2017). Transcriptome analysis of floral buds deciphered an irregular course of meiosis in polyploid brassica rapa. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00768
Chen, F. F., Zhu, Z. Y., Tong, L., Guo, X. C., Xu, S. L., Chen, J. H., et al. (2020). Production of allohexaploid brassica hybrid between tuber mustard (Brassica juncea l. var. crassicaulis Chen &Yang) and Chinese kale (Brassica oleracea var. alboglabra bailey). Sci. Hortic. 270:109412. doi: 10.1016/j.scienta.2020.109412
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Duro, E., Marston, A. L. (2015). From equator to pole: Splitting chromosomes in mitosis and meiosis. Genes Dev. 29, 109–122. doi: 10.1101/gad.255554.114
Du, K., Xiao, Y. Y., Liu, Q. E., Wu, X. Y., Jiang, J. J., Wu, J., et al. (2019). Abnormal tapetum development and energy metabolism associated with sterility in SaNa-1A CMS of brassica napus l. Plant Cell Rep. 38, 545–558. doi: 10.1007/s00299-019-02385-2
Fitzjohn, R. G., Armstrong, T. T., Newstrom-Lloyd, L. E., Wilton, A. D., Cochrane, M. (2007). Hybridisation within brassica and allied genera: Evaluation of potential for transgene escape. Euphytica 158, 209–230. doi: 10.1007/s10681-007-9444-0
Gaebelein, R., Alnajar, D., Koopmann, B., Mason, A. S. (2019). Hybrids between brassica napus and b. nigra show frequent pairing between the b and A/C genomes and resistance to blackleg. Chromosome Res. 27, 221–236. doi: 10.1007/s10577-019-09612-2
Geng, X. X., Chen, S., Astarini, I. A., Yan, G. J., Tian, E., Meng, J., et al. (2013). Doubled haploids of novel trigenomic brassica derived from various interspecific crosses. Plant Cell Tissue Organ Cult. 113, 501–511. doi: 10.1007/s11240-013-0292-4
Griffiths, S., Sharp, R., Foote, T. N., Bertin, I., Wanous, M., Reader, S., et al. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752. doi: 10.1038/nature04434
Gupta, M., Atri, C., Agarwal, N., Banga, S. S. (2016). Development and molecular-genetic characterization of a stable brassica allohexaploid. Theor. Appl. Genet. 129, 2085–2100. doi: 10.1007/s00122-016-2759-2
Henry, I. M., Dilkes, B. P., Tyagi, A., Gao, J., Christensen, B., Comai, L. (2014). The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of arabidopsis. Plant Cell 26, 181–194. doi: 10.1105/tpc.113.120626
Hotta, T., Kong, Z. S., Ho, C. M. K., Zeng, C. J. T., Horio, T., Fong, S., et al. (2012). Characterization of the arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell 24, 1494–1509. doi: 10.1105/tpc.112.096610
Huang, J. Y., Dong, J., Qu, L. J. (2021). From birth to function: Male gametophyte development in flowering plants. Curr. Opin. Plant Biol. 10 (10), 3687–99. doi: 10.1016/j.pbi.2021.102118
Jenczewski, E., Eber, F., Grimaud, A., Huet, S., Lucas, M. O., Monod, H., et al. (2003). PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164, 645–653. doi: 10.1093/genetics/164.2.645
Ji, C., Tian, Z., Liu, Y., Shi, G., Tian, B., Chen, W., et al. (2022). Transcriptome profiling identifies candidate genes contributing to Male and female gamete development in synthetic brassica allohexaploids. Plants (Basel) 11 (12), 1556. doi: 10.3390/plants11121556
Kim, D., Landmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–U121. doi: 10.1038/nmeth.3317
Lloyd, A., Bomblies, K. (2016). Meiosis in autopolyploid and allopolyploid arabidopsis. Curr. Opin. Plant Biol. 30, 116–122. doi: 10.1016/j.pbi.2016.02.004
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Miyashita, T., Araki, H., Hoshino, Y. (2011). Ploidy distribution and DNA content variations of lonicera caerulea (caprifoliaceae) in Japan. J. Plant Res. 124, 1–9. doi: 10.1007/s10265-010-0341-6
Mwathi, M. W., Gupta, M., Atri, C., Banga, S. S., Batley, J., Mason, A. S. (2017). Segregation for fertility and meiotic stability in novel brassica allohexaploids. Theor. Appl. Genet. 130, 767–776. doi: 10.1007/s00122-016-2850-8
Oh, S. A., Park, K. S., Twell, D., Park, S. K. (2010). The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 64, 839–850. doi: 10.1111/j.1365-313X.2010.04374.x
Osman, K., Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Franklin, F. C. H. (2011). Pathways to meiotic recombination in arabidopsis thaliana. New Phytol. 190, 523–544. doi: 10.1111/j.1469-8137.2011.03665.x
Parish, R. W., Li, S. F. (2010). Death of a tapetum: A programme of developmental altruism. Plant Sci. 178, 73–89. doi: 10.1016/j.plantsci.2009.11.001
Park, S. K., Howden, R., Twell, D. (1998). The arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125, 3789–3799. doi: 10.1242/dev.125.19.3789
Peterka, H., Budahn, H., Schrader, O., Ahne, R., Schutze, W. (2004). Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor. Appl. Genet. 109, 30–41. doi: 10.1007/s00122-004-1611-2
Pikaard, C. S. (2001). Genomic change and gene silencing in polyploids. Trends Genet. 17, 675–677. doi: 10.1016/S0168-9525(01)02545-8
Sattler, M. C., Carvalho, C. R., Clarindo, W. R. (2016). The polyploidy and its key role in plant breeding. Planta 243, 281–296. doi: 10.1007/s00425-015-2450-x
Severson, A. F., Von Dassow, G., Bowerman, B. (2016). Oocyte meiotic spindle assembly and function. Essays Dev. Biol. Pt A 116, 65. doi: 10.1016/bs.ctdb.2015.11.031
Snowdon, R. J., Winter, H., Diestel, A., Sacristan, M. D. (2000). Development and characterisation of brassica napus-sinapis arvensis addition lines exhibiting resistance to leptosphaeria maculans. Theor. Appl. Genet. 101, 1008–1014. doi: 10.1007/s001220051574
Springer, N. M. (2010). Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB. Cold Spring Harb. Protoc. 2010 (11), pdb.prot5515. doi: 10.1101/pdb.prot5515
Szadkowski, E., Eber, F., Huteau, V., Lode, M., Huneau, C., Belcram, H., et al. (2010). The first meiosis of resynthesized brassica napus, a genome blender. New Phytol. 186, 102–112. doi: 10.1111/j.1469-8137.2010.03182.x
Tian, E. T., Jiang, Y. F., Chen, L. L., Zou, J., Liu, F., Meng, J. L. (2010). Synthesis of a brassica trigenomic allohexaploid (B. carinata x b. rapa) de novo and its stability in subsequent generations. Theor. Appl. Genet. 121, 1431–1440. doi: 10.1007/s00122-010-1399-1
Warwick, S. I., Al-Shehbaz, I. A. (2006). Brassicaceae: Chromosome number index and database on CD-rom. Plant Syst. Evol. 259, 237–248. doi: 10.1007/s00606-006-0421-1
Xiong, Z. Y., Gaeta, R. T., Edger, P. P., Cao, Y., Zhao, K. L., Zhang, S. Q., et al. (2021). Chromosome inheritance and meiotic stability in allopolyploid brassica napus. G3-Genes Genomes Genet. 11 (2), jkaa011. doi: 10.1093/g3journal/jkaa011
Xiong, Z., Gaeta, R. T., Pires, J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid brassica napus. Proc. Natl. Acad. Sci. U.S.A. 108, 7908–7913. doi: 10.1073/pnas.1014138108
Yang, S., Chen, S., Geng, X. X., Yan, G., Li, Z. Y., Meng, J. L., et al. (2016). The first genetic map of a synthesized allohexaploid brassica with a, b and c genomes based on simple sequence repeat markers. Theor. Appl. Genet. 129, 689–701. doi: 10.1007/s00122-015-2657-z
Yang, Y., Wei, F., Braynen, J., Wei, X., Tian, B., Shi, G., et al. (2019). Cytological and proteomic analyses of floral buds reveal an altered atlas of meiosis in autopolyploid brassica rapa. Cell Biosci. 9, 49. doi: 10.1186/s13578-019-0313-z
Yang, Y., Yan, G., Li, Z. S., Yuan, J. C., Wei, X. C., Wei, F., et al. (2020). Cytological atlas at meiosis reveals insights into pollen fertility in synthetic brassica allotriploids between allotetraploid b. carinata and diploid b. rapa. Plant Physiol. Biochem. 148, 237–245. doi: 10.1016/j.plaphy.2020.01.003
Yin, L., Zhu, Z., Luo, X., Huang, L., Li, Y., Mason, A. S., et al. (2020). Genome-wide duplication of allotetraploid brassica napus produces novel characteristics and extensive ploidy variation in self-pollinated progeny. G3 (Bethesda) 10, 3687–3699. doi: 10.1534/g3.120.401493
Zeng, X. H., Yan, X. H., Yuan, R., Li, K. Q., Wu, Y. H., Liu, F., et al. (2017). Identification and analysis of MS5(d): A gene that affects double-strand break (DSB) repair during meiosis I in brassica napus microsporocytes. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01966
Zhao, D., Yang, X., Quan, L., Timofejeva, L., Rigel, N. W., Ma, H., et al. (2006). ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in arabidopsis. Plant Mol. Biol. 62, 99–110. doi: 10.1007/s11103-006-9006-1
Zhao, Q., Zou, J., Meng, J. L., Mei, S. Y., Wang, J. B. (2013). Tracing the transcriptomic changes in synthetic trigenomic allohexaploids of brassica using an RNA-seq approach. PloS One 8 (7), e68883. doi: 10.1371/journal.pone.0068883
Keywords: anther, Brassica allohexaploid, chromosome segregation, pollen fertility, tapetum, meiosis, microspore development, mitosis
Citation: Tian Z, Ji C, Xie Z, Shi X, Tian B, Cao G, Wei X, Yang Y, Wei F and Shi G (2023) Integrated cytological and transcriptomic analysis reveals insights into pollen fertility in newly synthetic Brassica allohexaploids. Front. Plant Sci. 13:1096804. doi: 10.3389/fpls.2022.1096804
Received: 12 November 2022; Accepted: 21 December 2022;
Published: 13 January 2023.
Edited by:
Changbin Chen, Arizona State University, United StatesReviewed by:
Yingxiang Wang, Fudan University, ChinaCopyright © 2023 Tian, Ji, Xie, Shi, Tian, Cao, Wei, Yang, Wei and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Wei, ZmFuZ3dlaUB6enUuZWR1LmNu; Gongyao Shi, c2hpZ3lAenp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.