
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 January 2023
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1092638
This article is part of the Research Topic Aphids as Plant Pests: From Biology to Green Control Technology View all 11 articles
Introduction: Aphids form a stable and mutually beneficial relationship with their primary symbiont Buchnera aphidicola, which play an important role in providing the missing nutrients to the host aphid. Based on the genome sequence of wheat aphid Siotobion miscanthi and its primary symbiont Buchnera that we obtained in our previously study, we identified a metabolic relay gene, ilvA, involved in the isoleucine synthesis pathway between aphids and Buchnera.
Method: In this study, we identified the location and sequence structure of ilvA gene in aphid genome, the expression level in different instars and tissues of aphids, and the effect of reducing ilvA expression on the growth and development of aphids by bioinformatics analysis, quantitative PCR, RNAi and bioassay experiments.
Result: Our study showed that ilvA was expressed at the highest level in the 2nd instar of the aphid, while the expression of this gene was significantly higher in the aphid bacteriocytes than in other tissues. Notably, this gene is localized on the aphid sex chromosome and remains highly conserved and collinearity across different aphid genomes. Knocking down the expression of ilvA reduced the aphid body weight and production. However, the indices of mortality decreased slightly, but were not significantly different, compared to the control.
Discussion: The results show that the relay genes between aphids and their symbionts in the metabolism of essential nutrients have potential roles in the growth and development of aphids, meanwhile, providing target loci and new ideas for RNAi-based aphid green control strategies.
Aphids are important pests that cause significant economic losses in agriculture worldwide. Almost all aphids contain endosymbionts, and one of them, Buchnera aphidicola (hereinafter referred to as Buchnera), which is present in almost all aphids, provides essential nutrients to the host aphid and therefore called primary symbiont (Baumann, 2005). Additionally, aphids have a variety of secondary symbionts in their bodies, and the significance of secondary symbionts in enhancing the adaptation of aphids to adverse environments has been widely reported (De Clerck et al., 2015; Manzano-Marín et al., 2016; Li et al., 2018; Li et al., 2021). In recent years, it is worth noting the growing number of studies have shown that the function of Buchnera not only in providing essential amino acids to the host aphid, but also has potential effects in improving the heat tolerance of aphids (Zhang et al., 2019), revealing the differentiation process (Perreau et al., 2021; Zhang S. et al., 2021) and enhancing their resistance to drugs (Guo et al., 2020). Therefore, exploiting the close and mutually beneficial relationship between aphids and Buchnera may produce new ideas for developing green control strategies of aphids.
In China, the grain aphid Sitobion miscanthi is one of the most prevalent wheat pests and causes substantial economic losses in agriculture (Li et al., 2021). As genome sequencing technologies continuous upgrading and costs decrease, a large number of insect genomic information continues to be deciphered. Based on our previously published genome information of wheat aphid S. miscanthi (LF clone) (Jiang et al., 2019) and its primary symbiont Buchnera (Li et al., 2022), making it more convenient to study the nutrient metabolism interaction network between them. Previously, we used genomic information to identify a key relay gene ilvE, linking Buchnera to aphids in the leucine, isoleucine and valine synthesis pathways. Meanwhile, RNA interference (RNAi) experiment reveals a vital function in three essential amino acid synthesis pathways (Li et al., 2022). However, whether exist other metabolic relay genes are present in the aphid and Buchnera nutrient synthesis chains and can be used as candidate target genes for RNAi is still unknown.
Here, we have mined another key gene ilvA in the aphid-Buchnera relay synthesis of isoleucine pathway through the genomic information obtained in our previous work. Sequence and bioinformatics analysis showed that the ilvA gene was highly conserved in different aphid genomes, while the gene expression profile in different developmental stages and tissues of aphids was clarified by qPCR assay. Subsequently, the effects of ilvA on aphid life parameters were measured by RNAi experiments. Our results indicate that the ilvA gene, which links the aphid and Buchnera amino acid synthesis pathways, has an important effect on aphid weight and offspring, all of which suggest that ilvA gene can be used as candidate target for RNAi against aphids.
The strains of S. miscanthi used in this study was reared on aphid-susceptible wheat seedlings (Triticum aestivum L) in the culture room at 20 ± 1°C with a 75% relative humidity and a light: dark photoperiod of 16: 8 hours. After 10 generations, the aphids were used for the following experiments.
The gene structure and conserved domains were analyzed using NCBI Batch CDD-search, and the results were visualized by TBtools (v 1.09857) (Chen et al., 2020). Conserved motifs of the genes were analyzed by the MEME program with the following parameters: classic mode, with the number of repetitions set to zero or one per sequence and the maximum number of motifs identified set to 6. Meanwhile, the location information of gene on aphid chromosome was obtained by genome annotation file, and the results were visualized by TBtools. We downloaded the chromosome-level genome and annotations of A. pisum (Li et al., 2020), selected the longest representative coding sequences of each gene and translated the nucleotide sequences to amino acid sequences. Then, MCScanX v1.1 (Wang et al., 2012) was used to identify syntenic blocks of genes between A. pisum and the previously published chromosome-level genome of S. miscanthi, and the results were visualized byTBtools.
Considering that the ilvA gene is localized on the aphid sex chromosome, we collected newly emerged winged and wingless adult aphids for transcriptome analysis in order to understand the gene expression on the aphid autosomes and sex chromosome. Total RNA from the winged and wingless aphids were extracted with the total RNA extraction regent kit (Tianmo, Beijing, China) following the manufacturer’s instructions. The quality of the RNA samples was evaluated on a 1% (w/v) agarose gel by electrophoresis and quantified by a Nanodrop 20000 spectrophotometer (DNovix, Washington, DC, United States). And enrichment of mRNA with polyA tails by Oligo (dT) magnetic beads. The obtained mRNA was then randomly interrupted with divalent cations in NEB Fragmentation Buffer, and the library was built for the following Illumina sequencing. In order to ensure the quality and reliability of data analysis, the raw reads were filtered by removing reads with adapters, reads with unidentifiable base information (noted as N) or the low-quality reads (reads with Qphred <= 20 with more than 50% of the entire read length in number of bases). Then, fast and accurate comparison of clean reads with our published reference genome (Jiang et al., 2019) using HISAT2 software (Kim et al., 2015). The differentially expressed genes (DEGs) between the winged and wingless aphids were analyzed. Our sequence data have been deposited in the National Center for Biotechnology information’s Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA908645).
ilvA was amplified by PCR from S. miscanthi cDNA with the specific primers listed in Table 1. The nucleotide sequence of the ilvA gene from S. miscanthi in this paper has been deposited in GenBank under accession number OQ093134. To quantify the ilvA transcript levels in different tissues and different developmental stages of aphids, qRT-PCR was performed with the specific primers listed in Table 1. The expression of the ilvA gene was normalized to the expression of the aphid housekeeping gene NADH (Zhang S. Y. et al., 2021). The amplification efficiency amplified with primers was 100.5 and 99.0% for ilvA and NADH. All treatments had three biological replicates, and each replicate consisted of three technical replicates.
The molecules of dsRNA targeting S. miscanthi ilvA (dsilvA) and the gene sequence of the green fluorescent protein (dsGFP), used as negative control, were synthetized according to the specific primers listed in Table 1. Meanwhile, the dsRNA and control were diluted to 500 ng/µl in an artificial diet (20% sucrose), and pure aphid artificial diet was used as the blank control. A total of 500 newly born winged adult S. miscanthi were picked from fresh wheat plants. After starvation for 2 hours, 15 active S. miscanthi were transferred into each feeding devices, and three replicate tubes were set up. After silencing, S. miscanthi individuals were collected at different time after treatment with dsRNA. Then, the surviving aphids were counted to calculated mortality. Additionally, the surviving aphids were used to detect RNAi efficiency by qPCR.
In addition, six aphids per 3 pairs were picked from the artificial device to perform the weight measurement. The average of these 3 pairs of weight values was calculated as one biological replicate per time point. Six biological replicates were examined. Moreover, the number of aphid production at different times of treatment was also counted. Six biological replicates were examined.
Differences in gene expression level at different time points of RNAi experiment were tested by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test using SPSS version 23.0 software (IBM, Armonk, NY, United States).
Based on our previously reported genome information of the S. miscanthi and its primary symbiont Buchnera (Jiang et al., 2019; Li et al., 2022), we found that the Buchnera genome contains almost all the key genes in the essential amino acid synthesis pathway, however, upstream of the aphid essential amino acid isoleucine synthesis pathway, a threonine dehydratase gene named ilvA, which has the function of hydrolyzing threonine to 2-oxybutanoate, is missing from the Buchnera genome, but present in the aphid genome (Figure 1).
To verify the accuracy of genome sequencing and understand the function of the ilvA gene, we cloned the ilvA gene using specific primers (Table 1). The full-length ilvA gene (1272 bp) was obtained by PCR amplification, with GenBank accession number OK431491. The ilvA gene is localized on S. miscanthi chromosome 8 (SmChr_8), encoding 423 amino acids with a deduced MW of 45.4 kDa and possessing a threonine dehydratase structural domain (Figure 2). Interestingly, S. miscanthi chromosome 8 is derived from a sex chromosome split that is thought to be highly homozygous and conserved in different aphid genomes (sex chromosome splitting due to chromosome splicing problems during the pre-sequencing process cannot be excluded). Additionally, synteny analysis between the S. miscanthi and pea aphid (A. pisum) genomes revealed that ilvA is also highly conserved in terms of gene location (Figure 2A). Phylogenetic analysis showed that ilvA gene sequences in different insects clustered into different branches, implying that the gene is highly conserved in different insects. Domain structure and motif analysis showed that ilvA genes are highly conserved in Hemiptera, especially in aphids (Figures 3, 4).
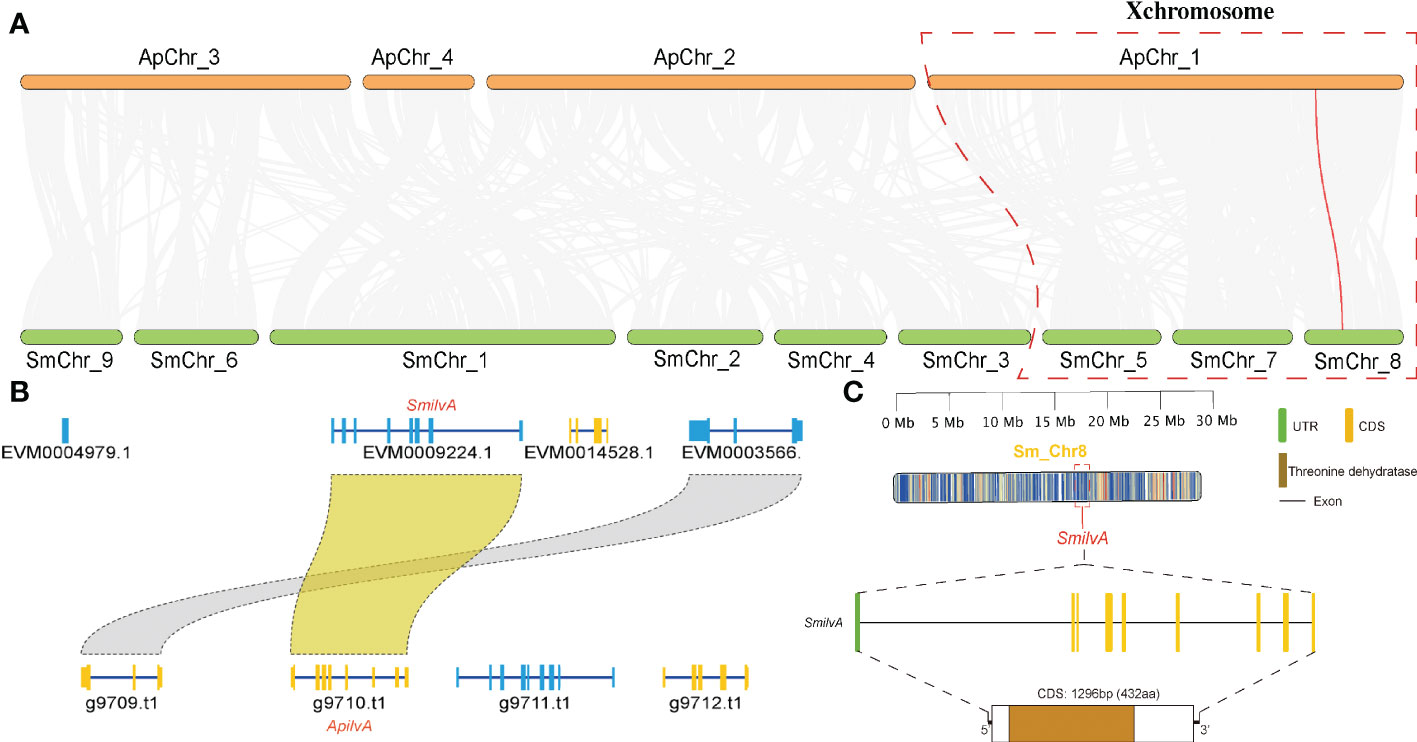
Figure 2 Sequence characteristics, structure and genome collinearity analysis. (A) Genome collinearity analysis of wheat aphid S. miscanthi and pea aphid A. pisum. Red lines represent the ilvA gene. (B) Colinearity analysis of ilvA gene in different aphid genomes. (C) Gene structure analysis of ilvA..
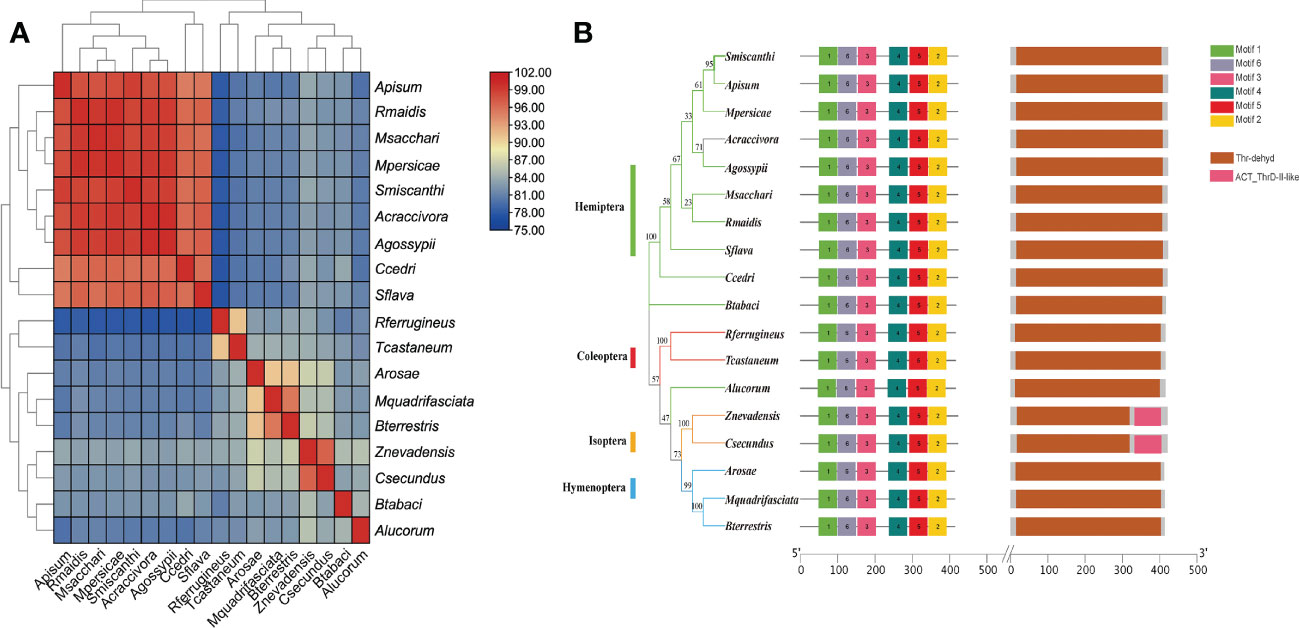
Figure 4 Sequence similarity, motif and domain analysis of ilvA gene in different insects (A) Sequence homology heat map analysis of ilvA gene in different insects. (B) Analysis of motifs and domains in ilvA gene sequence.
Considering the specific location of ilvA on the sex chromosome of aphids, and sexually mature aphids are rarely found in S. miscanthi, therefore, we sequenced the transcriptomes of winged and wingless adult aphids to investigate the differences in gene expression patterns on the autosomes and sex chromosomes of S. miscanthi. Surprisingly, the expression of genes on autosomes was significantly higher than that on sex chromosomes in both winged and wingless adult aphids (Figure 5A). To investigate the selective pressure on genes with expression levels in winged and wingless adult aphids, we estimated Ka/Ks value for paralogous genes within the autosome and sex chromosome of S. miscanthi, meanwhile, we also estimated Ka/Ks value for single-copy orthologous genes for a pair of related aphid species (S. miscanthi/A. pisum). The results showed that the selection pressure on sex chromosomes was significantly higher than that on autosomes, whether it was paralogous genes on aphid chromosomes (p < 0.0056, Kruskal-Wallis rank sum test) or orthologous genes on different aphid chromosomes (p < 0.0001) (Figures 5B, C). In addition, the Ka/Ks values of ilvA and our previously reported ilvE gene in different aphids were very low, 0.166 and 0.087, respectively (Figure 5D), suggesting that these genes are under relaxed purifying selection and the function is stable in aphid genome.
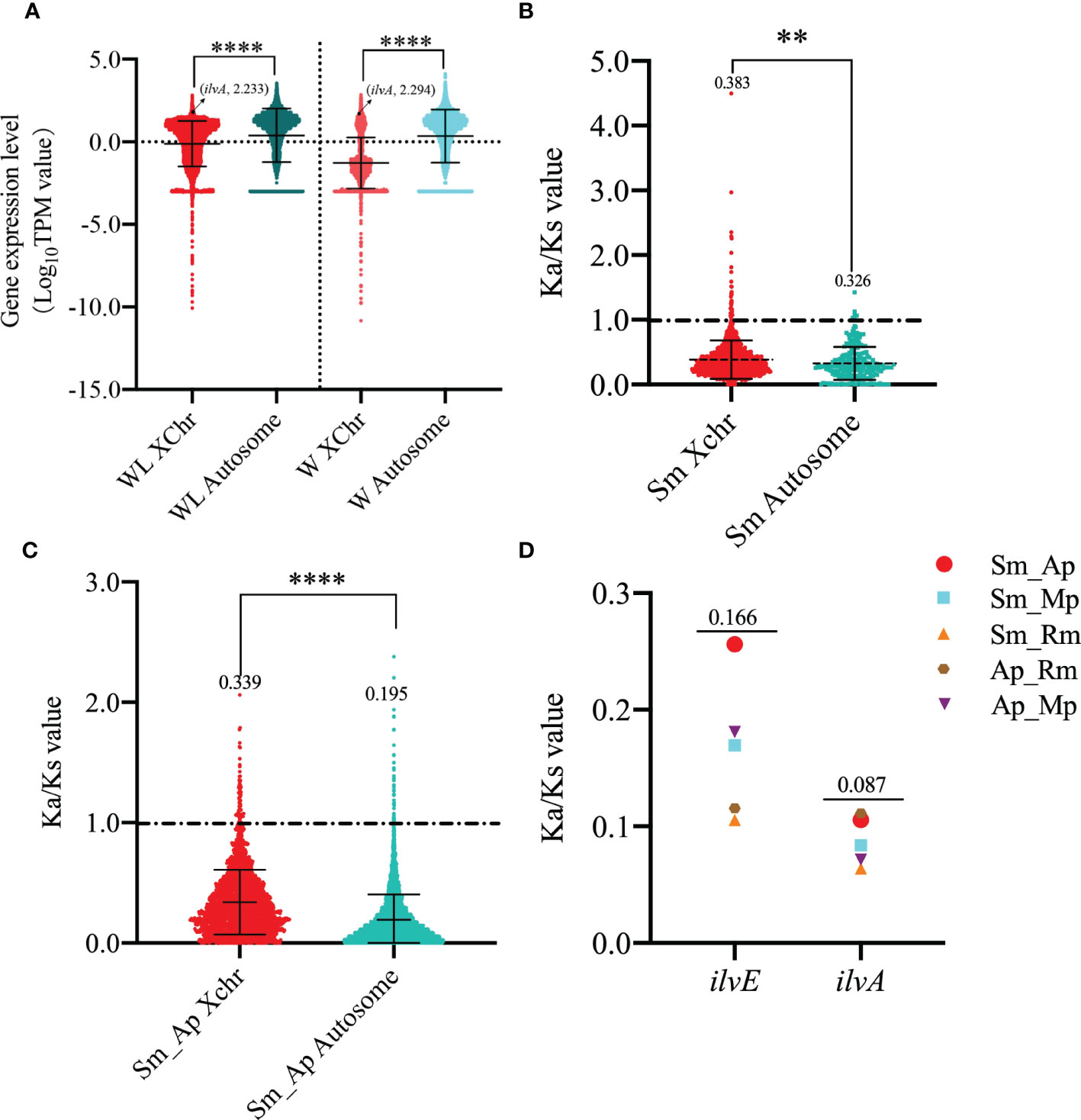
Figure 5 Analysis of gene expression levels and selection pressure on different type of chromosomes in winged and wingless adult aphids (A) Analysis of gene expression on different chromosome of winged and wingless adult aphids. (B) Analysis of selection pressure of different chromosome paralogous genes in S. miscanthi. (C) Analysis of selection pressure of different chromosome orthologous genes in S. miscanthi and A. pisum. (D) Selection pressure analysis of ilvA and ilvE orthologous genes in different aphid genomes. The “**” and “****” indicates significant differences based on the Mann-Whitney U test for two sample comparison at P < 0.001 and P < 0.00001, respectively.
The expression profile of ilvA at different tissues and developmental stages was examined using real-time PCR. Interestingly, the ilvA gene was expressed in all instars of aphids, with the highest expression in the 2nd instar and the lowest in the 1st instar (Figure 6A). In order to further reveal the expression specificity of ilvA gene in different tissues of aphids, we dissected the head, thorax, abdomen, gut, cornicle and bacteriocytes of aphids and performed the qPCR experiment. Unexpectedly, the results revealed that the expression of the ilvA gene was significantly higher in the bacteriocytes of aphids than in the other tissues (Figure 6B).
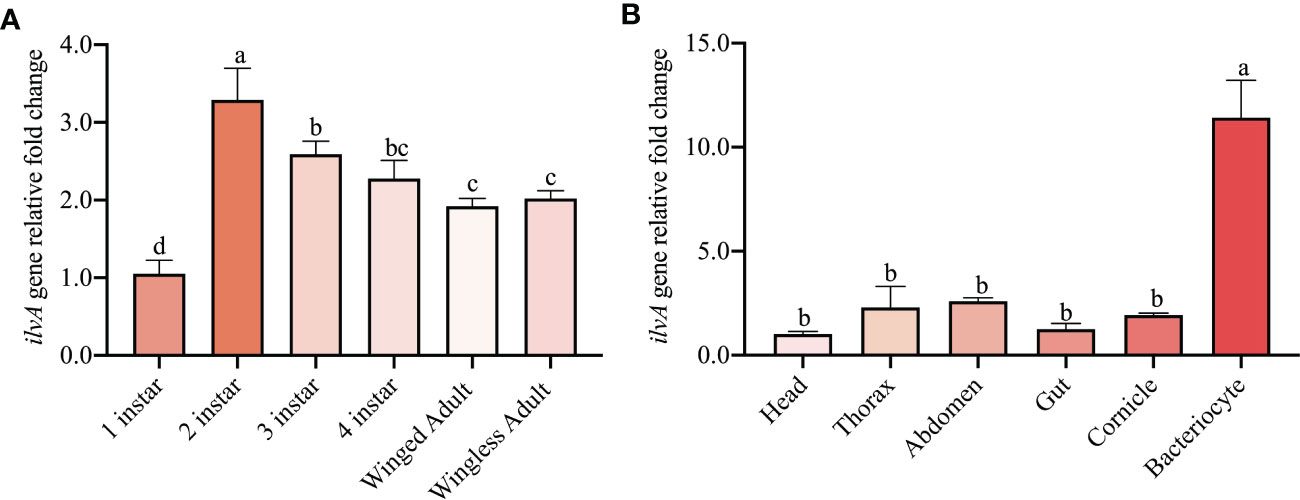
Figure 6 Expression of ilvA genes at different tissues and developmental stages in S. miscanthi (A) Expression level of ilvA gene in aphids at different developmental stages. (B) Expression level of ilvA gene in different tissues of aphid. Different letters above the bars indicate significant differences at P < 0.05.
To further verify the potential function of ilvA in aphid development, we synthesized dsRNA in vitro for RNA interference experiments. As shown in Figure 7A, the expression level of ilvA gene decreased by 46.3% and 54.3%, respectively, after 72 h and 96 h of RNAi treatment (Figure 7A). The results showed that the synthesized dsRNA fragments could effectively interfere with the expression of ilvA gene. Bioassays showed that compared with feeding dsGFP and sucrose control (CK), feeding dsilvA for 24 and 48 hs had no significant effect on aphid body weight and aphid production, but decreased significantly at 72 and 96 hs (Figures 7C, D). Additionally, the indices of mortality decreased slightly, but were not significantly different in all time points, compared to the control (Figure 7B). All these results indicate that interference with the ilvA gene has a negative effect on the growth and development of aphids.
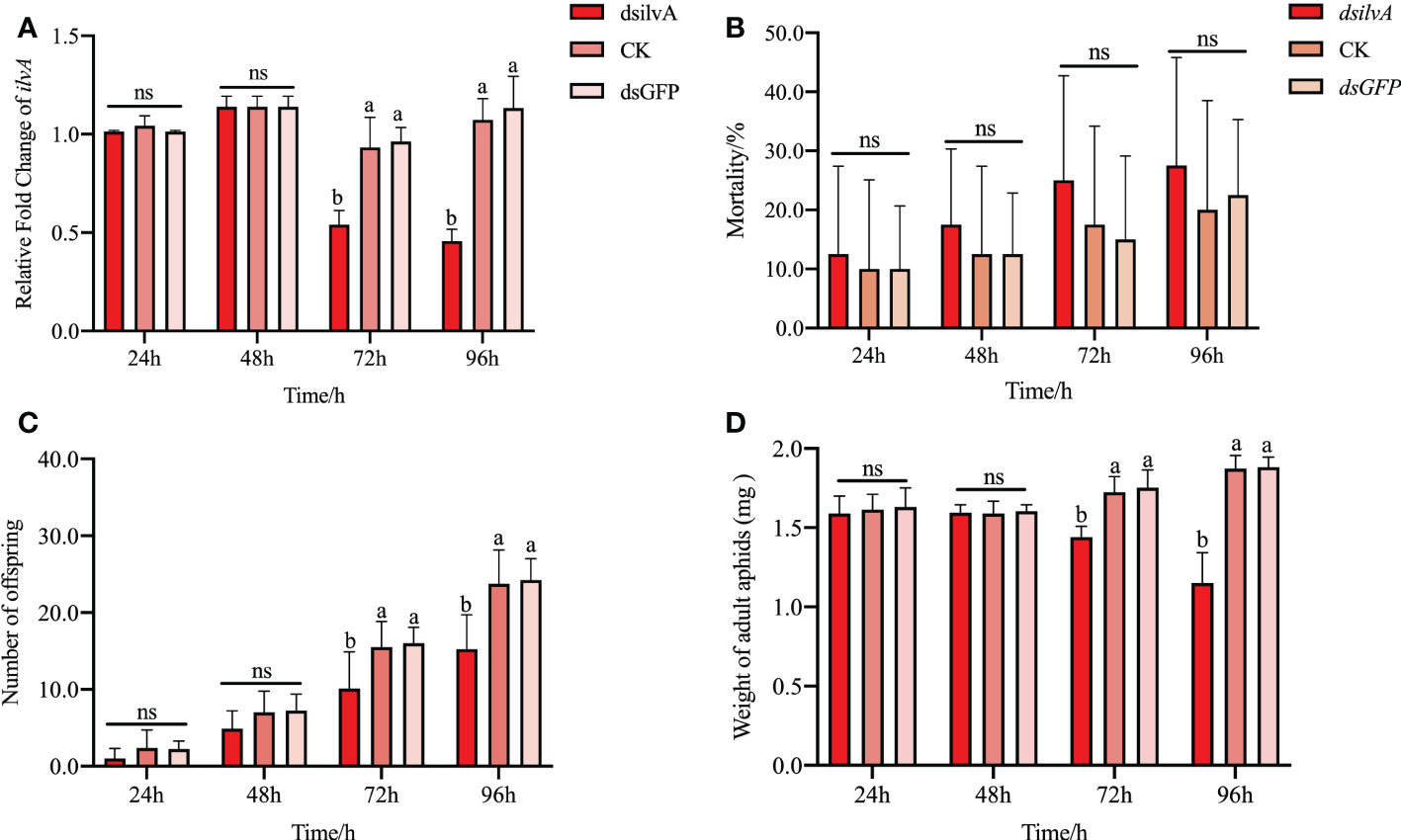
Figure 7 Effect of RNA interference with ilvA gene on life parameters of S. miscanthi (A) Effect of feeding dsRNA at different times on the expression of ilvA. (B) Effect of interfering with ilvA at different times on the aphid mortality. (C) Effect of interfering with ilvA on offspring of S. miscanthi. (D) Effect of interfering with ilvA on aphid body weight of S. miscanthi. Different letters above the bars indicate significant differences at P < 0.05, while ns indicates no significant difference.
Aphids and their primary symbiont Buchnera aphidicola have formed a long-term and stable symbiotic relationship. At the same time, they are considered to be typical cases in studying the coevolution relationship between insects and endosymbionts (Clark et al., 2000). Numerous studies have shown that Buchnera provides aphids with essential nutrients that are missing from the phloem sap of feeding host plants (Shigenobu et al., 2000). There is persuasive experimental evidence that the Buchnera genome provides aphids with almost all of the key genes in the essential amino acid synthesis pathway in the model insect pea aphid A. pisum, however, little is known about the consistency of this tight nutrient supply chain model in other aphids. Given that almost all aphids contain Buchnera, which plays a vital role in aphids, therefore, taking Buchnera as the starting point, developing a strategy to break the stable nutrient supply chain between them may become an effective new idea for green prevention and control of aphids. At present, RNA interference technology is widely used in insect gene function verification, is considered to be a new direction for the development of green biological pesticides (Bautista et al., 2009; Belles, 2010; Wuriyanghan et al., 2011; Liu et al., 2020). However, RNA interference technology cannot be effectively implemented in prokaryotes, resulting in direct silencing of aphid primary symbiont Buchnera gene is difficult to achieve. Therefore, it has become a new research direction to excavate and identify the relay genes between host aphid and Buchnera in the synthesis pathway of essential nutrients and using them as RNA interference target sites. It is gratifying that the availability of the genomes of wheat aphid S. miscanthi and its primary symbiont Buchnera makes this work feasible.
It has been known for decades that Buchnera lacks some genes encoding essential amino acid biosynthesis enzymes and compensates via the host pea aphid A. pisum (Shigenobu et al., 2000; Wilson et al., 2010). Meanwhile, in our previous study, we found that a metabolic relay gene ilvE, a branched-chain amino acid transferase gene required for the final step in the synthesis of the three essential amino acids valine, leucine and isoleucine, was absent in the Buchnera genome but was present in the aphid genome (Li et al., 2022). The result was consistent with the previous studies on pea aphid genome (Wilson et al., 2010). In addition, RNAi of the ilvE gene significantly increased aphid mortality. Therefore, nutrient synthesis relay genes between aphids and Buchnera can be used as candidate targets for RNA interference. However, whether there are other metabolic relay genes in the aphid genome and have potential effect on the aphid development is still rarely reported.
In this study, combined with our previously reported genome data, we identified a threonine dehydratase gene called ilvA in the upstream process of isoleucine synthesis, which is absent in the Buchnera genome but exists in the aphid genome, and the function of the gene is to hydrolyze threonine to intermediate product 2-Oxobutanoate (Figure 1). Therefore, to further verify the function of the gene, we cloned the ilvA gene and obtained a complete CDS sequence. Bioinformatics analysis showed that the gene was highly conserved in sequence and location in different aphids (Figures 3, 4). Meanwhile, it is worth noting that this gene is located on the sex chromosome of aphids (Figure 2A). In recent years, with the continuous advancement of genome sequencing technology, more and more aphid genomes have been resolved (Chen et al., 2019; Roberto et al., 2020; Zhang S. et al., 2021). Related studies have reported that in different aphid genomes, sex chromosomes are highly conserved, and the selection pressure of genes on sex chromosomes of A. pisum is significantly higher than that of autosomes (Li et al., 2020). Moreover, the average expression level of genes on sex chromosomes of pea aphid was significantly lower than that on autosomes, which may be the reason for the significant increase of selection pressure (Jaquiery et al., 2018). In our study, we performed the transcriptome analysis of winged and wingless adult aphids and determined that the expression levels of genes on sex chromosomes were significantly lower than those on autosomes, the results are consistent with previous studies. Moreover, the selective pressure on sex chromosomes was also significantly higher than that on autosomes, whether it was paralogous gene pairs within chromosomes or orthologous gene pairs between different aphids (Figure 5). However, the expression level of ilvA and ilvE gene is much higher than the average value of other genes on sex chromosomes (Supplementary Table S1). Meanwhile, ilvA and ilvE are under low selection pressure in different aphids, which means that the function of this kind of gene in aphid is stable. In general, a lower level of gene expression may imply less important to phenotypes (Nabholz et al., 2013), and previous study implicate that the aphid sex chromosome as a less preferred location for highly expressed genes (Li et al., 2020). However, in this study, we found that although the expression level of genes on sex chromosomes is low overall, there are still some highly expressed genes, and the gene has a potential role in the synthesis of some important nutrients. This result also means that the aphid sex chromosome is still a mysterious region worthy of further study.
To investigate the expression level of the ilvA gene in aphids, we next determined the expression pattern of the ilvA in different developmental stages and tissues of S. miscanthi by qPCR. Our results showed that the expression of the ilvA gene was significantly higher in the 2rd instars of aphids than in the other instars and exhibited lowest expression level in 1st instars (Figure 3). Surprisingly, the ilvA gene was highly expressed in the bacteriocytes of aphids, where Buchnera shelters. Interestingly, the specific expression location of this gene is consistent with another synthetic relay gene ilvE (Li et al., 2022). Previous reports suggest that Buchnera plays an important role in maintaining amino acid synthesis and supply homeostasis in aphids (Wilson et al., 2010). Therefore, with the growth and development of aphids, the increasing titer of Buchnera may have a potential role in regulating the expression of ilvA and ilvE. Moreover, considering the specificity of the expression location of ilvA and ilvE genes, we hypothesized that the closer distance to Buchnera may be more convenient for the synthesis and transport of essential amino acids regulated by aphids between bacteriocytes and hemocoel.
To further study the function of ilvA in aphid, the RNAi experiment was performed. Unlike the ilvE gene, the indices of mortality decreased slightly, but were not significantly different, after feeding dsilvA 72 and 96 hs (Figure 7B). However, the weight and production of aphids decreased significantly (Figures 7C, D). Recent studies have shown that common ancestor of Hymenoptera lose all key genes in the valine, leucine and isoleucine synthesis pathway, but parasitoids are able to control related pathways in their host insect, providing them with missing essential nutrients (Ye et al., 2022). Compared with the commensalism relationship between parasitoids and host insects, aphids form a long-term stable mutualistic relationship with their primary symbiont Buchnera. The relay synthesis process in the nutrient synthesis pathway is important evidence of coevolution. Therefore, the destruction of synthetic relay chain may be the reason for the negative impact on the growth and development of S. miscanthi after knocking down the expression of ilvA and ilvE. However, there are also relevant study showed that the Buchnera protein HisC could functionally replace the missing ilvE, catalyzing the terminal reaction in these pathways (Shigenobu et al., 2000). It is worth noting that the ilvA gene is the upstream gene of the pathway, which is different from the downstream gene ilvE in the synthesis pathway. Therefore, whether reducing the expression of ilvA will cause functional complementation or expression response of some genes in the downstream pathway is still needs further exploration. At the same time, reduced expression of the ilvE gene significantly inhibited Leu, Ile and Val production in aphids (Li et al., 2022). Although in this study, we did not quantify isoleucine production in aphids after ilvA interference, but as a result of our previous studies, we speculated that the decrease in aphid growth fitness may be potentially associated with amino acid production, meanwhile, the decrease in the production of single one essential amino acid may also have a much smaller negative effect on aphids than the three essential amino acids. Such speculation also provides an explanation for why silencing the ilvA gene does not result in a significant increase in mortality.
Moreover, secondary symbiont in insect also has potential functions in compensating for the lack of amino acids (Gómez-Valero et al., 2004; Ju et al., 2017). Whether secondary symbiont rescue the negative effects on aphid growth and development caused by RNA interference remains unknown. It is worth noting that ilvE and ilvA are highly conserved in different aphids, so whether the RNAi targeting them has broad consistency is worthy of further study, which will also provide a theoretical basis for screening broad-spectrum RNAi target sites.
Numerous reports have indicated that almost all insects contain various types of endosymbionts (Baumann, 2005). However, so far, a large number of insect endosymbionts have not been cultured in vitro, making the study of their function dependent on host insects (Moran and Mira, 2001). Moreover, RNAi technology is not available for prokaryotic endosymbiont-associated genes, making it difficult to realize the strategy of using symbionts to control insects, necessitating new ideas. In this study, based on our previous studies, we identified another highly conserved isoleucine synthesis pathway upstream gene ilvA that is absent in the Buchenra genome but is present in the aphid genome, which plays an essential role in influencing the body weight and reproduction of aphids. With the growing popularity of genome sequencing, an increasing number of data resources on the genomes of insects and their endosymbionts have been deciphered. Our study may provide a future direction for targeting important junctions in the endosymbiont-insect metabolic relay process to control agricultural pests.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA908645.
QL, JF and JC conceived and designed the experiments. QL and YC performed the experiments. QL analyzed the data. QL and YC wrote the paper. All of the authors read and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
The authors are thankful for the financial support provided by the National Natural Science Foundation of China (32001900), the China Postdoctoral Science Foundation (2020M680786), the National Key R & D Plan of China (2017YFD0201700), the Earmarked fund for China Agriculture Research System (CARS-22) and China’s Donation to the CABI Development Fund (IVM10051).
We would like to thank Ms. Yanxia Liu for aphid rearing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1092638/full#supplementary-material
Baumann, P. (2005). Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. doi: 10.1146/annurev.micro.59.030804.121041
Bautista, M. A. M., Miyata, T., Miura, K., Tanaka, T. (2009). RNA Interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem. Mol. Biol. 39, 38–46. doi: 10.1016/j.ibmb.2008.09.005
Belles, X. (2010). Beyond drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 55, 111–128. doi: 10.1146/annurev-ento-112408
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Xia, R. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Shakir, S., Bigham, M., Richter, A., Fei, Z. J., Jander, G. (2019). Genome sequence of the corn leaf aphid (Rhopalosiphum maidis Fitch). Gigascience. 8, 1–12. doi: 10.1093/gigascience/giz033
Clark, M. A., Moran, N. A, Baumann, P., Wernegreen, J. J. (2000). Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution. 54, 517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x
De Clerck, C., Fujiwara, A., Joncour, P., Leonard, S., Felix, M. L., Francis, F., et al. (2015). A metagenomic approach from aphid’s hemolymph sheds light on the potential roles of co-existing endosymbionts. Microbiome. 3, 63. doi: 10.1186/s40168-015-0130-5
Gómez-Valero, L., Soriano-Navarro, M., Perez-Brocal, V., Heddi, A., Moya, A., Garcia-Verdugo, J. M., et al. (2004). Coexistence of wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 186, 6626–6633. doi: 10.1128/JB.186.19.6626-6633.2004
Guo, S. K., Gong, Y. J., Chen, J. C., Shi, P., Cao, L. J., Yang, Q., et al. (2020). Increased density of endosymbiotic Buchnera related to pesticide resistance in yellow morph of melon aphid. J. Pest Sci. 93, 1281–1294. doi: 10.1007/s10340-020-01248-0
Jaquiery, J., Peccoud, J., Ouisse, T., Legeai, F., Prunier-Leterme, N., Gouin, A., et al. (2018). Disentangling the causes for faster-X evolution in aphids. Genome. Biol. Evol. 10, 507–520. doi: 10.1093/gbe/evy015
Jiang, X., Zhang, Q., Qin, Y. G., Yin, H., Zhang, S. Y., Li, Q., et al. (2019). A chromosome-level draft genome of the grain aphid Sitobion miscanthi. GigaScience. 8, 1–8. doi: 10.1093/gigascience/giz101
Ju, J. F., Hoffmann, A. A., Zhang, Y. K., Duan, X. Z., Guo, Y., Gong, J. T., et al. (2017). Wolbachia-induced loss of male fertility is likely related to branch chain amino acid biosynthesis and ilvE in Laodelphax striatellus. Insect. Biochem. Mol. Biol. 85, 11–20. doi: 10.1016/j.ibmb.2017.04.002
Li, Q., Fan, J., Cheng, Y., Hou, M. L., Chen, J. L. (2022). ilvE as a potential RNAi target to inhibit amino acid synthesis to control the wheat aphid Sitobion miscanthi. Entomol. Gen. doi: 10.1127/entomologia/2022/1528
Li, Q., Fan, J., Sun, J. X., Wang, M. Q., Chen, J. L. (2018). Effect of the secondary symbiont Hamiltonella defensa on fitness and relative abundance of Buchnera aphidicola of wheat aphid, Sitobion miscanthi. Front. Microbiol. 9, 582. doi: 10.3389/fmicb.2018.00582
Li, Q., Sun, J. X., Qin, Y. G., Fan, J., Zhang, Y., Tan, X. L., et al. (2021). Reduced insecticide sensitivity of the wheat aphid Sitobion miscanthi after infection by the secondary bacterial symbiont Hamiltonella defensa. Pest Manag Sci. 77, 1936–1944. doi: 10.1002/ps.6221
Li, Y. Y., Zhang, B., Moran, N. A. (2020). The aphid X chromosome is a dangerous place for functionally important genes: diverse evolution of hemipteran genomes based on chromosome-level assemblies. Mol. Biol. Evol. 8, 2357–2368. doi: 10.1093/molbev/msaa095
Liu, S. S., Jaouannet, M., Dempsey, D. M. A., Christine, J. I., Coustau, C., Kogel, K. H. (2020). RNA-Based technologies for insect control in plant production. Biotechnol. Adv. 39, 107463. doi: 10.1016/j.biotechadv.2019.107463
Manzano-Marín, A., Simon, J. C., Latorre, A. (2016). Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia symbiotic consortia between the distantly related lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol. Evol. 8, 1440–1458. doi: 10.1093/gbe/evw085
Moran, N. A., Mira, A. (2001). The process of genome shrinkage in the obligate symbiont, Buchnera aphidicola. Genome. Biol. 2, research0054.1–research 0054.12. doi: 10.1186/gb-2001-2-12-research0054
Nabholz, B., Ellegren, H., Wolf, J. (2013). High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol. Biol. Evol. 30, 272–284. doi: 10.1093/molbev/mss238
Perreau, J., Zhang, B., Maeda, G. P., Kirkpatrick, M., Moran, N. A. (2021). Strong within-host selection in a maternally inherited obligate symbiont: Buchnera and aphids. Proc. Nati. Acad. Sci. U. S. A. 118, 1–8. doi: 10.1073/pnas.2102467118
Roberto, B., Archana, S., Cindayniah, J. G., Felicidad, F. F., Sam, T. M., Glen, P., et al. (2020). A chromosome-level genome assembly of the woolly apple aphid, Eriosoma lanigerum hausmann (Hemiptera: Aphididae). Mol. Ecol. Resour. 21, 316–326. doi: 10.1111/1755-0998.13258
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., Ishikawa, H. (2000). Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp aps. Nature. 407, 81–86. doi: 10.1038/35024074
Wang, Y. P., Tang, H. B., DeBarry, J. D., Tan, X., Li, J. P., Wang, X. Y., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wilson, A. C.C., Ashton, P. D., Calevro, F., Charles, F., Colella, S., Febvay, G., et al. (2010). Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Nucleic. Acids Res. 19, 249–258. doi: 10.1111/j.1365-2583.2009.00942.x
Wuriyanghan, H., Rosa, C., Falk, B. W. (2011). Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PloS One 6, e27736. doi: 10.1371/journal.pone.0027736
Ye, X., Xiong, S. J., Teng, Z. W., Yang, Y., Wang, J. L., Yu, K. L., et al. (2022). Genome of the parasitoid wasp Cotesia chilonis sheds light on amino acid resource exploitation. BMC Biology. 20, 118. doi: 10.1186/s12915-022-01313-3
Zhang, S., Gao, X. K., Wang, L., Jiang, W. L., Su, H. H., Jing, T. X., et al. (2021). Chromosome-level genome assemblies of two cotton-melon aphid Aphis gossypii biotypes unveil mechanisms of host adaption. Mol. Ecol. Resour. 00, 1–15. doi: 10.1111/1755-0998.13521
Zhang, B., Leonard, S. P., Li, Y. Y., Moran, N. A. (2019). Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proc. Nati. Acad. Sci. U. S. A. 116, 24712–24718. doi: 10.1073/pnas.1915307116
Keywords: Sitobin miscanthi, Buchnera aphidicola, ilvA, metabolic relay, RNAi
Citation: Li Q, Cheng Y, Fan J and Chen JL (2023) Metabolic relay gene of aphid and primary symbiont as RNAi target loci for aphid control. Front. Plant Sci. 13:1092638. doi: 10.3389/fpls.2022.1092638
Received: 10 November 2022; Accepted: 23 December 2022;
Published: 18 January 2023.
Edited by:
Huipeng Pan, South China Agricultural University, ChinaReviewed by:
Zezhong Yang, Tianjin Academy of Agricultural Sciences, ChinaCopyright © 2023 Li, Cheng, Fan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian Chen, Y2hlbmp1bGlhbkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.