
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Plant Sci. , 19 December 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1088938
This article is a correction to:
Identification and Validation of Quantitative Trait Loci for Wheat Dwarf Virus Resistance in Wheat (Triticum spp.)
 Anne-Kathrin Pfrieme1‡
Anne-Kathrin Pfrieme1‡ Britta Ruckwied1†‡
Britta Ruckwied1†‡ Antje Habekuß1
Antje Habekuß1 Torsten Will1
Torsten Will1 Andreas Stahl1*
Andreas Stahl1* Klaus Pillen2
Klaus Pillen2 Frank Ordon3
Frank Ordon3A Corrigendum on
Identification and validation of quantitative trait loci for Wheat Dwarf Virus resistance in wheat (Triticum spp.)
by Pfrieme A-K, Ruckwied B, Habekuß A, Will T, Stahl A, Pillen K and Ordon F (2022) 13:828639. doi: 10.3389/fpls.2022.828639
In the published article, there was an error in Table 4. Table 4 represents the P values of Supplementary Table S7. In Table S7, false P-values and means were reported for the populations FixFa, FixS, FixRe, and RexFi. After their correction, the total number of significant differences decreased. However, there are no differences in the QTL that show significance. In conclusion, 14 of the 35 QTL that were identified by GWAS are shown to be significant. The overall statement remains unchanged. Table 4 is mentioned in Results “Validation of Quantitative trait loci”. For this reason, an adjustment is also made in the text. The corrected Table 4 and its caption appears below.
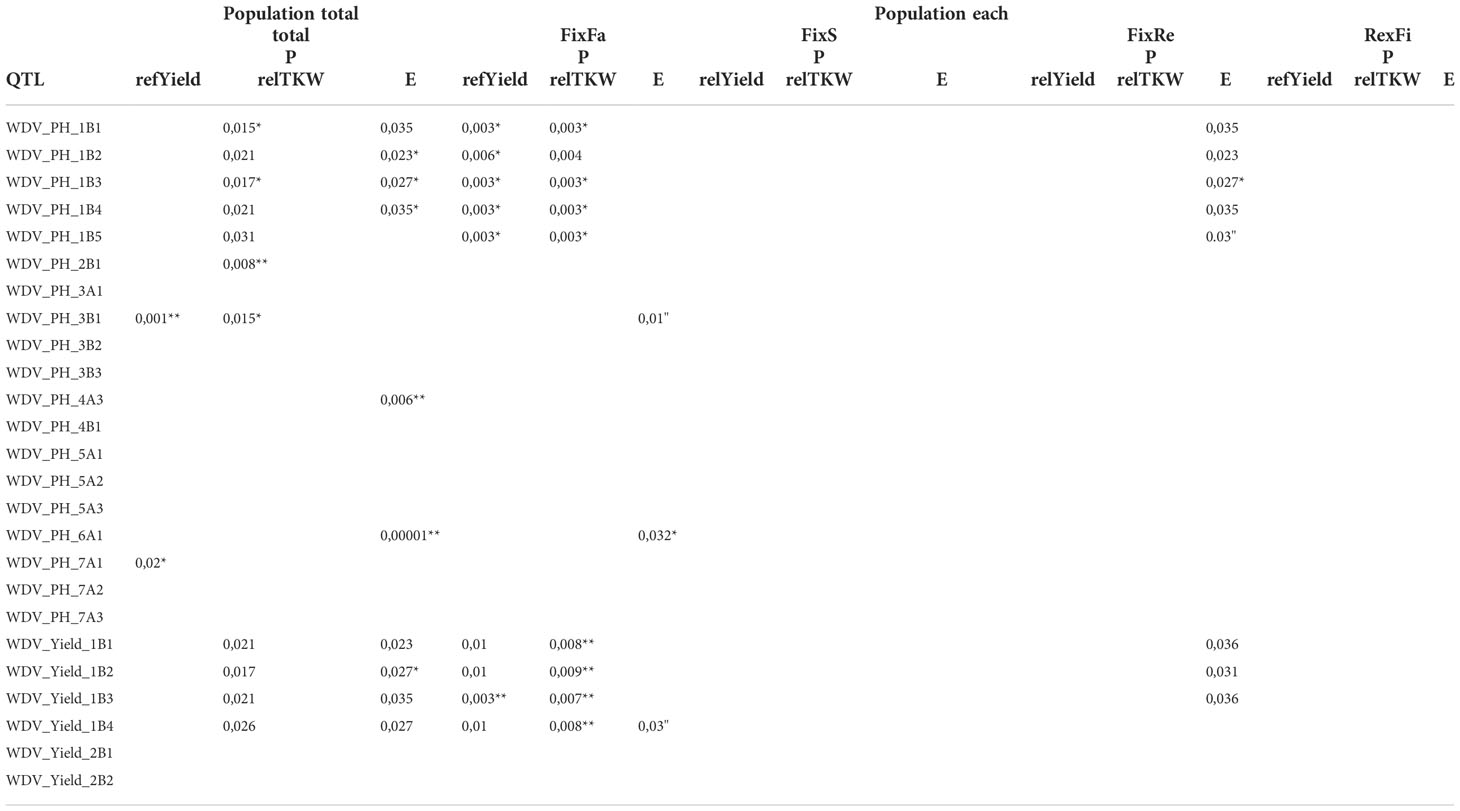
Table 4 Validations of the identified QTL using a t-Test, *, ** indicate significant differences between allele means at 5% and 1% level, respectively.
There were also errors in Supplementary Tables S7 and S8. In Table S7, false P-values and means were reported for the populations FixFa, FixS, FixRe, and RexFi. After their correction, the total number of significances has decreased. However, there are no differences in the number of QTL that show a significant difference. In the conclusion, 14 of the 35 GWAS QTL are shown to be significant. The statement remains unchanged.
In Supplementary Table S8, the determined values of the simple and multiple regression analysis are given. An error occurred in the simple regression analysis of FixS and FixRe (values of FixFa were accidently given in both columns). However, the correction only affects this table, as it is not described in the results section. The values for the multiple regression analysis are correct.
The description of the validated QTL had to be changed because they referred to the error in Table 4. A correction has been made in Results “Validation of Quantitative trait loci”. This sentence previously stated:
“One QTL for relYield and ten for the extinction showed significant effects to varying degrees across all populations. The most significant effects were observed for TKW with 11 significant QTL (Table 4 and Supplementary Table S7). Looking at the significances at the population level, most were observed for the FixRe population. In particular, four QTL for relPH and four QTL for relYield on chromosome 1B (WDV_PH_1B1, WDV_PH_1B2, WDV_PH_1B3, WDV_PH_1B4, WDV_Yield_1B1, WDV_Yield_1B2, WDV_Yield_1B3, WDV_Yield_1B4) showed highly significant and consistent effects in all populations. The effects of WDV_PH_7A1 for the trait relYield and those of WDV_Yield_1B2- 1B4 were only validated in two populations. An effect of WDV_PH_4A3, WDV_PH_6A1, WDV_Yield_1B1, WDV_Yield_1B3 and WDV_Yield_1B4 was only confirmed in the FixRe population.”
The corrected sentence appears below:
“Two QTL for relYield and ten for the extinction value (relative virus titer) showed significant effects to varying degrees across all populations. The most significant effects were observed for TKW with 11 significant QTL (Table 4 and Supplementary Table S7). Looking at significant differences at the population level, most were observed for the FixFa population. In particular, one QTL for relYield on chromosome 1B (WDV_Yield_1B1) showed highly significant and consistent effects in all populations. The contributions of WDV_PH_1B1-1B5 and WDV_Yield_1B1-WDV_Yield_1B4 were significant in three populations. The effects of WDV_PH_7A1 were only validated in two populations. An effect of WDV_PH_3B1 and WDV_PH_6A1 was only confirmed in the FixFa population.”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Wheat dwarf virus (WDV), genome-wide association study (GWAS), wheat (Triticum aestivum), quantitative trait loci (QTL), resistance breeding, Psammotettix alienus
Citation: Pfrieme A-K, Ruckwied B, Habekuß A, Will T, Stahl A, Pillen K and Ordon F (2022) Corrigendum: Identification and validation of Quantitative Trait Loci for Wheat dwarf virus resistance in wheat (Triticum spp.). Front. Plant Sci. 13:1088938. doi: 10.3389/fpls.2022.1088938
Received: 03 November 2022; Accepted: 18 November 2022;
Published: 19 December 2022.
Edited and Reviewed by:
Denise Tieman, University of Florida, Gainesville, United StatesCopyright © 2022 Pfrieme, Ruckwied, Habekuß, Will, Stahl, Pillen and Ordon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Stahl, YW5kcmVhcy5zdGFobEBqdWxpdXMta3VlaG4uZGU=
†Present address: Britta Ruckwied, Gene Bank Department, Resources Genetics and Reproduction, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany
‡These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.