- College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Sucrose non-fermenting-1-related protein kinase 2 (SnRK2) is a class of plant-specific serine/threonine (Ser/Thr) protein kinase that plays an important role in rice stress tolerance, growth and development. However, systematic bioinformatics and expression pattern analysis have not been reported. In the current study, ten OsSnRK2 genes were identified in the rice genome and located on 7 chromosomes, which can be classified into three subfamilies (I, II, and III). Many cis-regulatory elements were identified in the promoter region of OsSnRK2 genes, including hormone response elements, defense and stress responsive elements, indicating that the OsSnRK2 family may play a crucial role in response to hormonal and abiotic stress. Quantitative tissue analysis showed that OsSnRK2 genes expressed in all tissues of rice, but the expression abundance varied from different tissues and showed varietal variability. In addition, expression pattern of OsSnRK2 were analyzed under abiotic stress (salt, drought, salt and drought) and showed obvious difference in diverse abiotic stress. In general, these results provide useful information for understanding the OsSnRK2 gene family and analyzing its functions in rice in response to ABA, salt and drought stress, especially salt-drought combined stress.
Introduction
Plants are exposed to various biotic and abiotic stresses in the natural environment that inhibit normal growth and development. During the evolution, plants have developed complicated and sophisticated mechanisms to cope with various adversity stresses. Protein kinase phosphorylation plays an important role in the signaling pathway, plant growth and development, hormone response and anti-stress by modifying the activity of translated proteins, regulatory enzymes, and various functional proteins (Umezawa et al., 2013; Mao et al., 2020; Sharif et al., 2020; Maszkowska et al., 2021).
Sucrose non-fermenting-1-related protein kinase (SnRK) is a class of serine/threonine (Ser/Thr) protein kinases and widely observed in plants. According to the conservation of the active domain of the kinase, it can be classified into three subfamilies: SnRK1, SnRK2, and SnRK3 (Hrabak et al., 2003). The SnRK2 gene family is a family of plant-specific protein kinases and involved in stress responsive processes. Recent studies have shown that members of the SnRK2 gene family play an important role in stress response by phosphorylation modifications and regulating protein activity and gene expression, thereby participating in the regulation of osmotic stress, stomatal movement through signal transduction (Riichiro et al., 2002; Taishi et al., 2004; Cheng et al., 2017; Fatima et al., 2020; Xu et al., 2020).
As one of important phytohormones, abscisic acid (ABA) is not only involved in plant growth and development but also plays an essential role in plant response to biotic and abiotic stresses (Mahadi et al., 2022). ABA signaling pathway is consisted of ABA receptor, protein phosphatase type 2C and SNF1-related protein kinase (SnRK2) (Li et al., 2000; Taishi et al., 2004; Zhang et al., 2020). In the absence of ABA, PP2C interacts with OsSnRK2 and inhibits its kinase activity, which shuts down the ABA signaling pathway; in the presence of ABA, the PYR/PYL/RCAR receptor protein binds to the hormone to form a complex, which in turn binds to PP2C, relieving its inhibition on SnRK2 and activating the ABA signaling pathway. Therefore, SnRK2 is a key factor with positive regulation of ABA signaling, and the activity of SnRK2 kinase is critical for ABA signaling on/off and abiotic stress response in plants (Fujii et al., 2007; Cheng et al., 2017).
Salt stress and drought stress have become major factors in inhibiting plant growth, development and crop yield. The main risks that the plants suffered from high concentrations of salt in the soil include osmotic stress, ion poisoning, water losing, wilting and metabolic disruption (Kawa et al., 2020; Zelm et al., 2020). In addition, salt stress causes oxidative stress, which destabilizes cell membranes (Munns and Tester, 2008). Drought stress also causes osmotic stress, which leads to the reduction of plant height, leaf area and stomatal conductance, as well as affecting plant root development (Khatun et al., 2021). Numerous studies have reported that SnRK2 is involved in plant responses to salt stress and drought stress. Under salt stress, overexpressing OsSAPK4 reduces the accumulation of Na+ and Cl- in rice leaves than the wild type and promote germination, growth and development (Diedhiou et al., 2008). Among the 11 SnRK2 members in maize, the expression of ZmSnRK2.3 and ZmSnRK2.6 were activated and significantly up-regulated by salt stress (Huai et al., 2008). In recent years, it was found that expression of most VrSnRK2 genes was induced by drought stress, suggesting their potential function in drought stress response; with VrSnRK2.6c being most significantly (12-fold) induced by drought stress (Fatima et al., 2020). Overexpression of TaSnRK2.9 in tobacco enhanced the tolerance of tobacco seedlings and mature plants to drought and salt stress, and increased tobacco survival, seed germination, and root length (Feng et al., 2019). Then, it has been shown that overexpressing MpSnRK2.10 in Malus prunifolia exhibit enhanced drought tolerance in phenotypic appearance associated with drought stress damage, i.e. most of the leaves of transgenic plants appear healthy upon rehydration after drought, while the leaves of wild-type plants show extensive necrosis. Moreover, transgenic plants were observed with less accumulation of ROS and MDA than WT under drought stress (Shao et al., 2018). It has also been confirmed that transgenic plants overexpressing OsSGT1 showed less tolerant to salt stress and that OsSAPK9 can act as a positive regulator for salt stress response and disease resistance by interacting with OsSGT1 (Zhang et al., 2019). All the above results implied that OsSnRK2 family may plays a crucial role in regulating plant response to salt stress and drought stress.
To better understand the OsSnRK2 family members and their responses to abiotic stress, bioinformatics analysis was performed in this study including the physicochemical properties, conserved sequences, gene structures, cis-acting elements, expression profiles, and phylogeny of OsSnRK2. In addition, the expression pattern of OsSnRK2 in different tissues and under different stresses (ABA, salt, drought and salt-drought double) were investigated by qRT-PCR. The results will help to enrich the understanding of the OsSnRK2 gene family and provide a theoretical basis for further studying the function of OsSnRK2 in abiotic stress response of rice.
Materials and methods
Gene identification and chromosome localization
The candidate OsSnRK2 protein sequences were searched for structural domains using the online website SMART: Main page (http://smart.embl-heidelberg.de/) to confirm the presence of the conserved SnRK family domain S_TKc. Based on the OsSnRK2 protein sequences reported in the literature (Yuhko et al., 2004), after collecting the gene information through the online website RiceDate (https://www.ricedata.cn/gene/), TBtools (Chen et al., 2020) was used to chromosome localization maps.
Gene structure and conserved motifs analysis
The protein-coding regions (CDS) and DNA sequences of OsSnRK2 were downloaded in FASTA format from the Whole Genome Data Ensembl website (http://plants.ensembl.org/index.html). The distribution of introns and exons and non-coding regions of the genes were mapped using TBtools. Conserved motifs of the proteins were analyzed online via the MEME website (http://meme-suite.org/) with Motif set to 12 and parameters set to default values, and conserved motifs were mapped using TBtools.
Protein sequence alignment, phylogenetic tree, gene duplication and synteny analysis
Sequence alignment and output of the conserved structural domain of OsSnRK2 proteins were completed by MEGA7 software and geneDoc software and construction of phylogenetic relationships of the OsSnRK2 family, Arabidopsis thaliana and Hordeum vulgare was completed using MEGA7 software. The segmental repeat events of the OsSnRK2 genes were analyzed using TBtools and presented in the form of Circos plots. Also, TBtools was used to do synteny analysis of SnRK2 genes in genomes.
Protein physicochemical properties, subcellular localization, secondary and 3D structure prediction
Protein physicochemical property analysis via ExPASy (https://web.expasy.org/protparam/) online, subcellular localization and nuclear localization were analysis by WoLF (https://wolfpsort.hgc.jp/) and NucPred (https://nucpred.bioinfo.se/uncared/) online respectively. Secondary structure predictive of protein was performed by the SOPMA website (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). Protein 3D structure prediction for OsSnRK2 gene families using the UniProt website (https://www.uniprot.org/).
Protein interaction and cis-acting elements in the promoter region of the OsSnRK2 gene analysis
The STRING (https://cn.string-db.org/) website was used to predict the putative protein-protein interaction networks with Oryza sativa SnRK2 proteins using default settings. Optimising protein-protein interaction networks with Cytoscape (Doncheva et al., 2019).
The sequence of the 2000 bp region upstream of the start codon of the rice OsSnRK2 family gene was downloaded from the whole genome data Ensembl website (http://plants.ensembl.org/index.html) as the promoter sequence. Prediction analysis of cis-acting elements in the promoter region of the gene was performed through the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and promoter cis-acting elements were mapped by TBtools.
Plant growth and stress treatments
The rice material used in this study was japonica rice Nipponbare. The International Rice Research Institute (IRRI) nutrient solution (the pH adjusted to 5.0-5.5 with KOH) was used as the hydroponic medium and changed every 3 days. The seedlings were grown in an artificial climate chamber under light conditions of 14 h at 28°C and 10 h in darkness at 24°C and 70% humidity.
ABA treatment: Rice seedlings of two leaves stage were transferred to a hydroponic medium containing 0 µM, 50 µM and 100 µM ABA respectively and the shoots were taken after 24 h for gene expression analysis.
Salt, drought, and salt-drought combined stress treatments: Rice seedlings grown hydroponically to the two leaves stage were transferred to rice nutrient solutions containing 200 mM NaCl (salt treatment), 13% PEG 6000 (drought treatment) and 200 mM NaCl and 13% PEG 6000 (salt-drought double treatment) for treatment, and shoots were collected 48 h later for gene expression analysis.
Tissue expression and expression profiling of the OsSnRK2 gene
RNA was extracted from roots, stems, leaves, leaf sheath and panicles at the tassel stage, and then analyzed for the expression of OsSnRK2 genes in different tissue using qRT-PCR. The primers used in this assay were listed in Supplementary Table 1.
The CREP database (http://crep.ncpgr.cn/crep-cgi/home.pl) was searched for full fertility expression profile data of the indica rice variety MingChuan63 to obtain the expression of OsSnRK2 genes in five mature organs and 24 different developmental periods, which were clustered hierarchically using TBtools.
RNA extraction and qRT-PCR analysis
Fluorescent quantitative PCR primers were designed by Primer Premier 5 and OsUBQ5 was selected as the internal reference gene. Total RNA was extracted from rice samples using the Total Plant RNA Kit (Tiangen Biochemical Technology Co., Ltd.) and reverse transcribed into cDNA using the AMV First Strand cDNA Synthesis Kit. PCR amplification was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad, USA). The reaction conditions were pre-denaturation at 95 °C for 5 min, denaturation at 94 °C for 35 s, annealing at 63 °C for 40 s, and extension at 72 °C for 20 s. 38 amplification cycles were performed with 4 replicates for each sample and 2-ΔΔCt was used for data processing.
Results
Gene identification and chromosome localization analysis of OsSnRK2
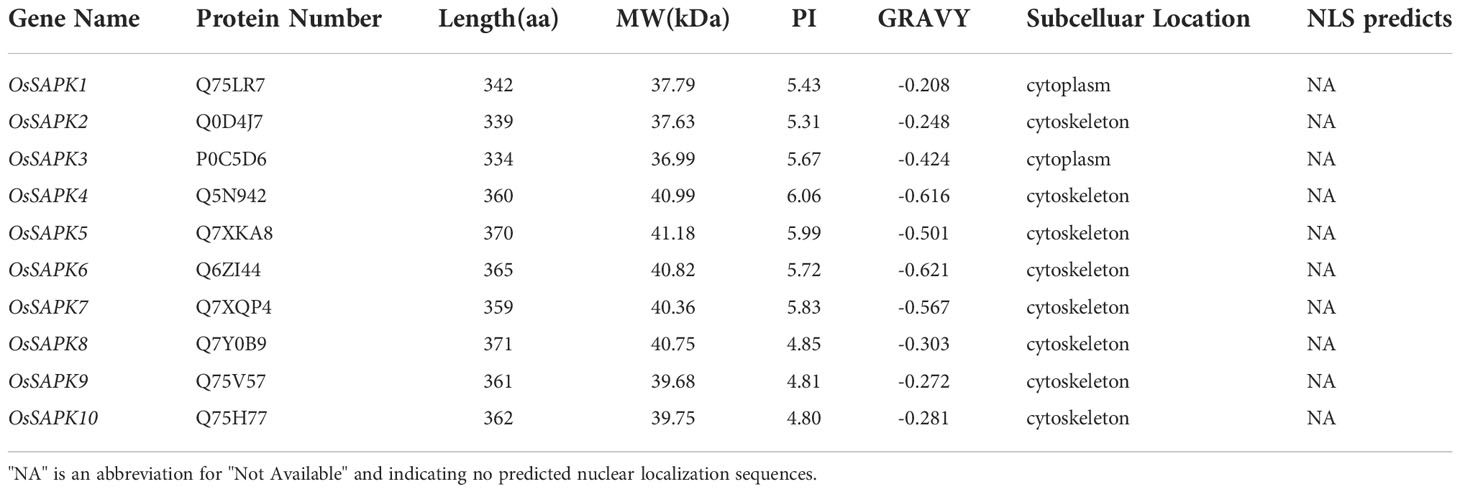
Referring to the OsSAPK1-10 names reported in the published literature (Huai et al., 2008), bioinformatics analysis was performed after collecting gene information via the online website RiceData (https://www.ricedata.cn/gene/). The detailed information of genes was listed in Tables 1, 2. Fox example, the amino acid lengths of the OsSnRK2 family proteins ranged from 334 to 371 aa. Meanwhile, the molecular weight is ranged from 36.99 to 41.18 kDa. The isoelectric points and the hydrophobicity values of OsSnRK2 were ranged from 4.80 to 6.06 and -0.621 to -0.208, respectively. The negative hydrophobicity values means that the OsSnRK2 proteins are all hydrophilic.
Chromosome localization analysis revealed that the 10 genes of OsSnRK2 were distributed on seven chromosomes (Figure 1 and Supplementary Table 2), with OsSAPK1, OsSAPK8, and OsSAPK10 on chromosome 3, OsSAPK5 and OsSAPK7 on chromosome 4, OsSAPK4, OsSAPK6, OsSAPK2, OsSAPK3 and OsSAPK9 are located on chromosomes 1, 2, 7, 10 and 12 respectively.
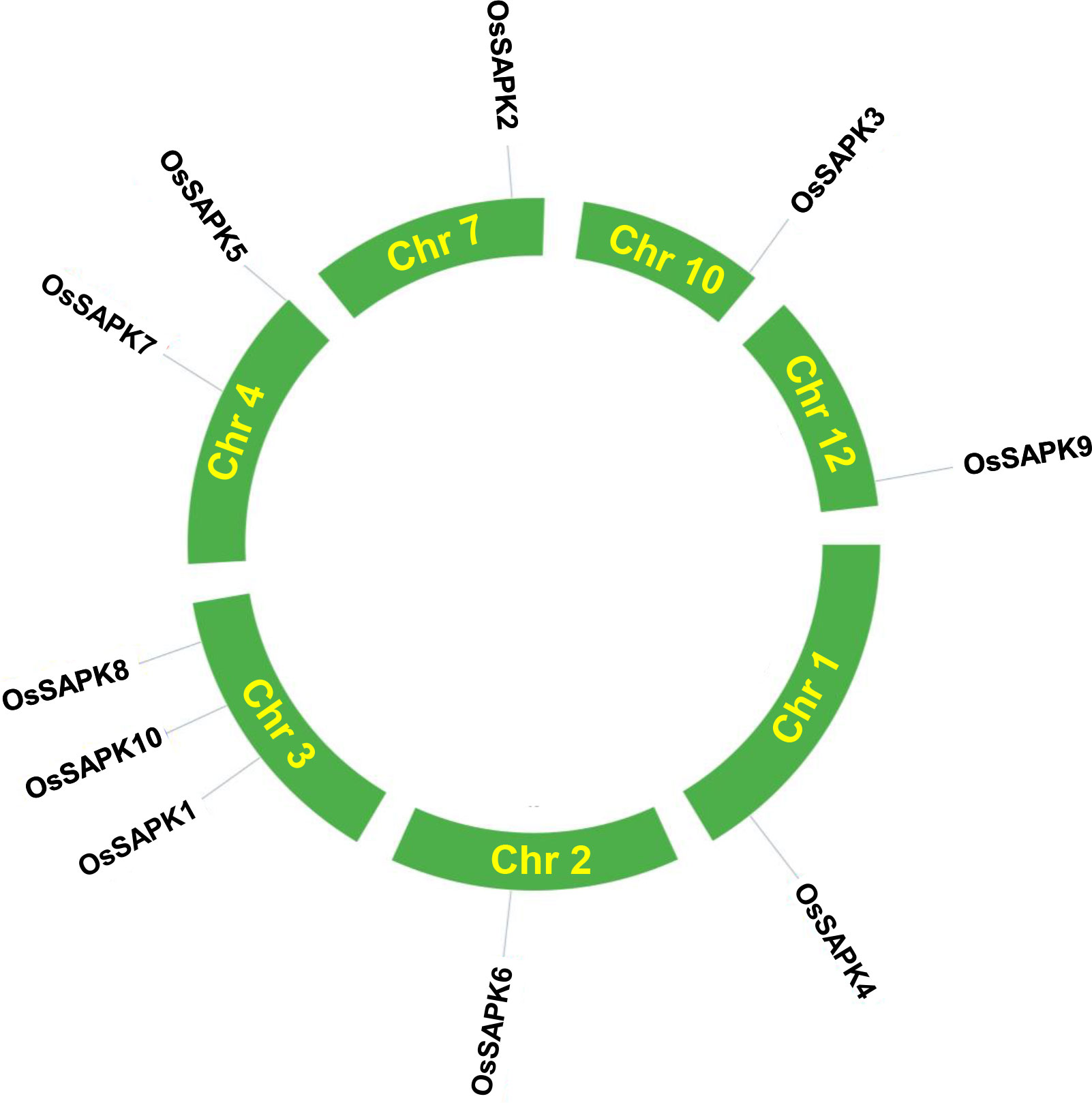
Figure 1 Distribution of OsSnRK2 genes in rice chromosomes. Only show the chromosomes where located the OsSnRK2 gene. The yellow font represents chromosome numbers and Green arc length indicate chromosome size.
Gene structure and protein motif analysis of OsSnRK2
The gene structure analysis of the OsSnRK2 family members showed that the length of the family genes ranged from 1875 (OsSAPK5) to 5459 bp (OsSAPK7). OsSAPK5 and OsSAPK10 had four and seven exons respectively, and the rest of the OsSnRK2 genes all contained nine exons. In addition, the last exon of most OsSnRK2 genes was longer (Figure 2A).
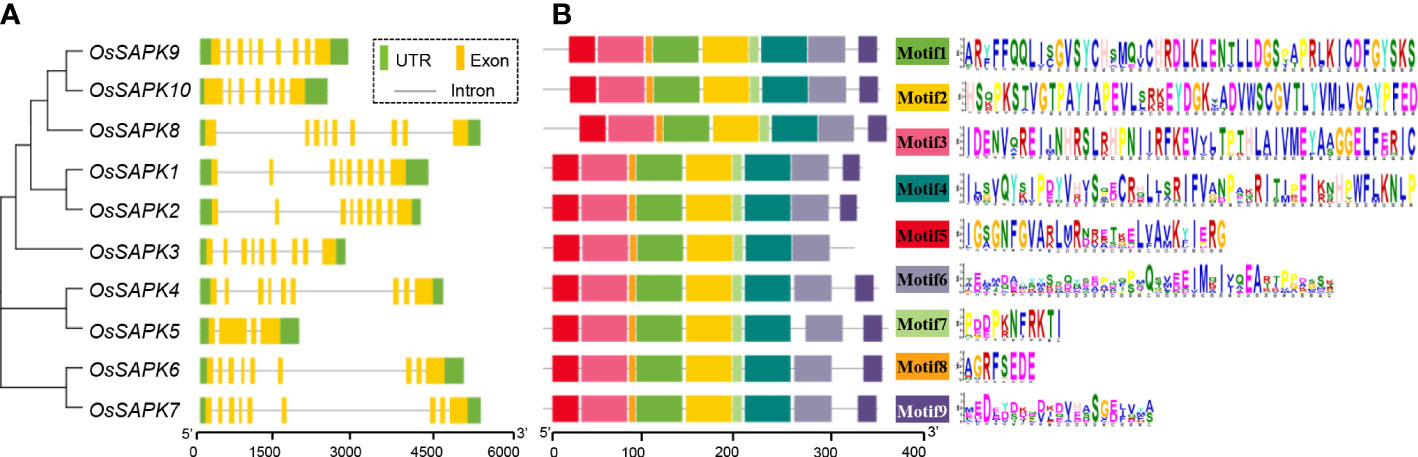
Figure 2 Gene structure and conserved motif analysis of the OsSnRK2 gene. (A) Gene structure analysis of the OsSnRK2. Green boxes, yellow boxes and black lines represent UTR, Exon and Introns respectively. (B) Conserved motif analysis of the OsSnRK2 gene.
Analysis of gene conserved motifs is useful for exploring the structural composition of proteins. We used MEME online software to analyze the number and distribution of motifs of the OsSnRK2 gene family members, and identified a total of nine motifs, named Motif 1~Motif 9. The results showed that Motif 3 is the most conserved, and the Motif 6 ranked as the second; except for OsSAPK3 which missing the Motif 9 conserved motif, all the other OsSnRK2 genes contain the same number of motifs with the same order arrangement of gene structure (Figure 2B and Supplementary Table 3). The similarities and diversity of these conserved motifs may result from the evolutionary adaptation of the gene functions in OsSnRK2 family.
Protein sequence alignment, phylogenetic relationships, gene duplication and synteny analysis of OsSnRK2
The alignment of amino acid composition of the OsSnRK2 protein sequences showed the presence of a highly conserved S_TKc structural domain in the OsSnRK2 proteins (Figure 3A), which is closely related to their highly conserved function during evolution.
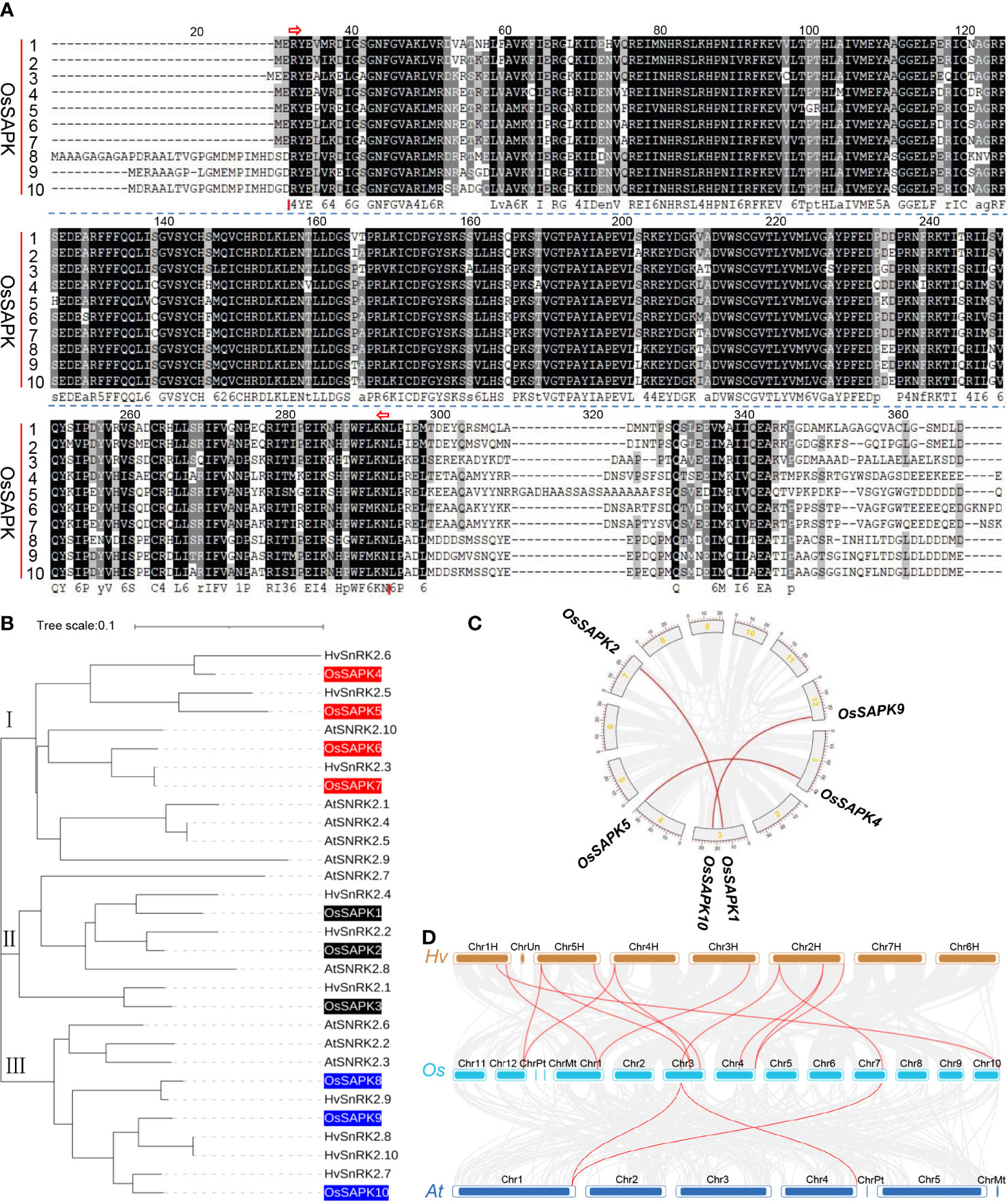
Figure 3 protein sequence alignment, phylogenetic relationships and gene duplication analysis of OsSnRK2. (A) Sequence alignment of the conserved structural domains of the OsSnRK2 gene family, S_TKc structural domain, position 33-292, interval shown by red arrows. (B) Phylogenetic relationships analysis of SnRK2 in Oryza sativa, Arabidopsis thaliana and Hordeum vulgare. (C) Gene segmental duplication analysis of OsSnRK2 in rice. Gray lines indicate all synteny blocks in the rice genome, and the red lines indicate duplicated OsSnRK2 gene pairs. The chromosome number is indicated at the gray arc. The scale at the periphery of the chromosome represents the physical location (Kb). (D) Synteny analysis of SnRK2 genes in the genomes between Hordeum vulgare and Oryza sativa or Arabidopsis thaliana. The gray lines show collinear blocks. The red lines indicate the syntenic gene pairs between Hordeum vulgare and Oryza sativa or Arabidopsis thaliana, respectively.
To further investigate the evolutionary relationships of the OsSnRK2, phylogenetic analysis was performed with amino acid sequences of 10 OsSnRK2 and several important SnRK2 proteins from Arabidopsis thaliana and Hordeum vulgare (Figure 3B). The results showed that the OsSnRK2 proteins could be classified into three subfamilies according to their affinities, with OsSAPK1, OsSAPK2 and OsSAPK3 belonged to subfamily I, OsSAPK4, OsSAPK5, OsSAPK6, OsSAPK7 belonged to subfamily II, and OsSAPK8, OsSAPK9 and OsSAPK10 belonged to subfamily III. The OsSnRK2 family members are closely related to the Arabidopsis thaliana and Hordeum vulgare SnRK2 family members, suggesting that they might have conserved physiological and biochemical functions.
Three segmental duplication events were identified in OsSnRK2 family by the MCScanX function of the software TBtools (Figure 3C) and involving six genes of ten OsSnRK2. All segmental duplication events happened between different chromosomes, but all occur within the same subfamily. The above results demonstrate that a number of OsSnRK2 genes maybe appear in the course of gene duplication, and the segmental duplication events may be responsible for the expansion of SnRK2 genes in rice.
The colinearity of OsSnRK2 gene pairs between Oryza sativa, Hordeum vulgare and Arabidopsis thaliana was compared. The result showed that 2 OsSnRK2 genes exhibited syntenic relationship with AtSnRK2. However, 9 OsSnRK2 genes showed syntenic relationship with HvSnRK2 (Figure 3D and Supplementary Table 4), implying that these genes may play a critical role in evolution of OsSnRK2 family.
Subcellular localization, secondary and 3D structure prediction of OsSnRK2
The predicted subcellular localization results showed that OsSAPK1 and OsSAPK3 proteins were localized in the cytoplasm, the remaining OsSnRK2 proteins were localized in the cytoskeleton. The NucPred scores of the OsSnRK2 family proteins were all below 0.6, indicating no predicted nuclear localization sequences (marked with “NA” in Table 2).
Secondary structures of OsSnRK2 proteins were analyzed by the SOPMA online website (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). All OsSnRK2 proteins were consisted of four secondary structures: alpha helix, extended helix, random coil, and extended strand (Figure 4), and the proportions of alpha helix > random coil > extended strand > extended helix. It is assumed that the alpha helix and random coil are the main conformations of the OsSnRK2 protein. The results of protein 3D structure prediction shows that all OsSnRK2 proteins have similar 3D structure (Figure 5).
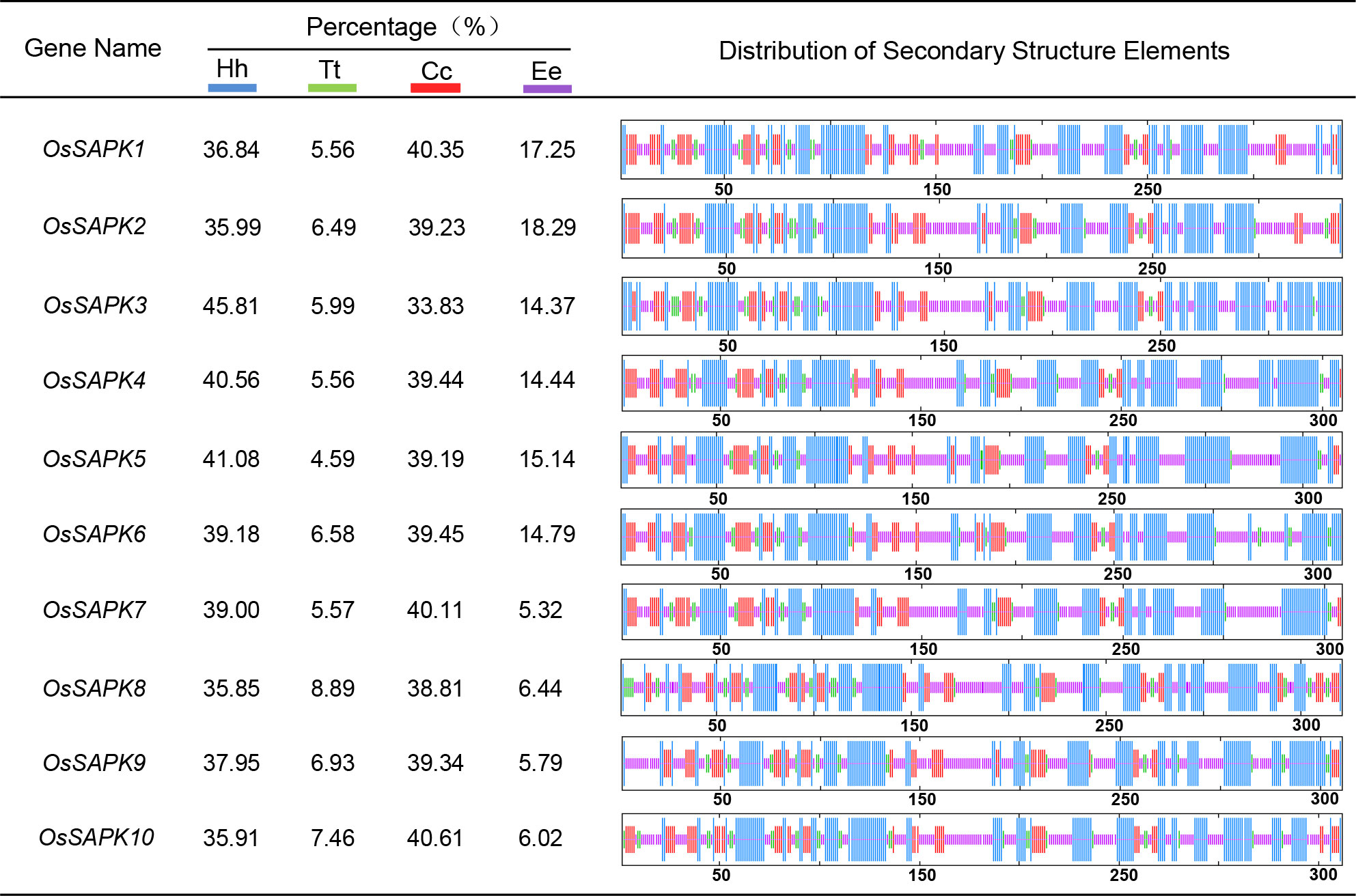
Figure 4 Secondary structure analysis of OsSnRK2 proteins. The blue color represents alpha helix (Hh), the green color represents extended helix (Tt), the yellow color represents random coil (Cc) and the red color represents extended strand (Ee).
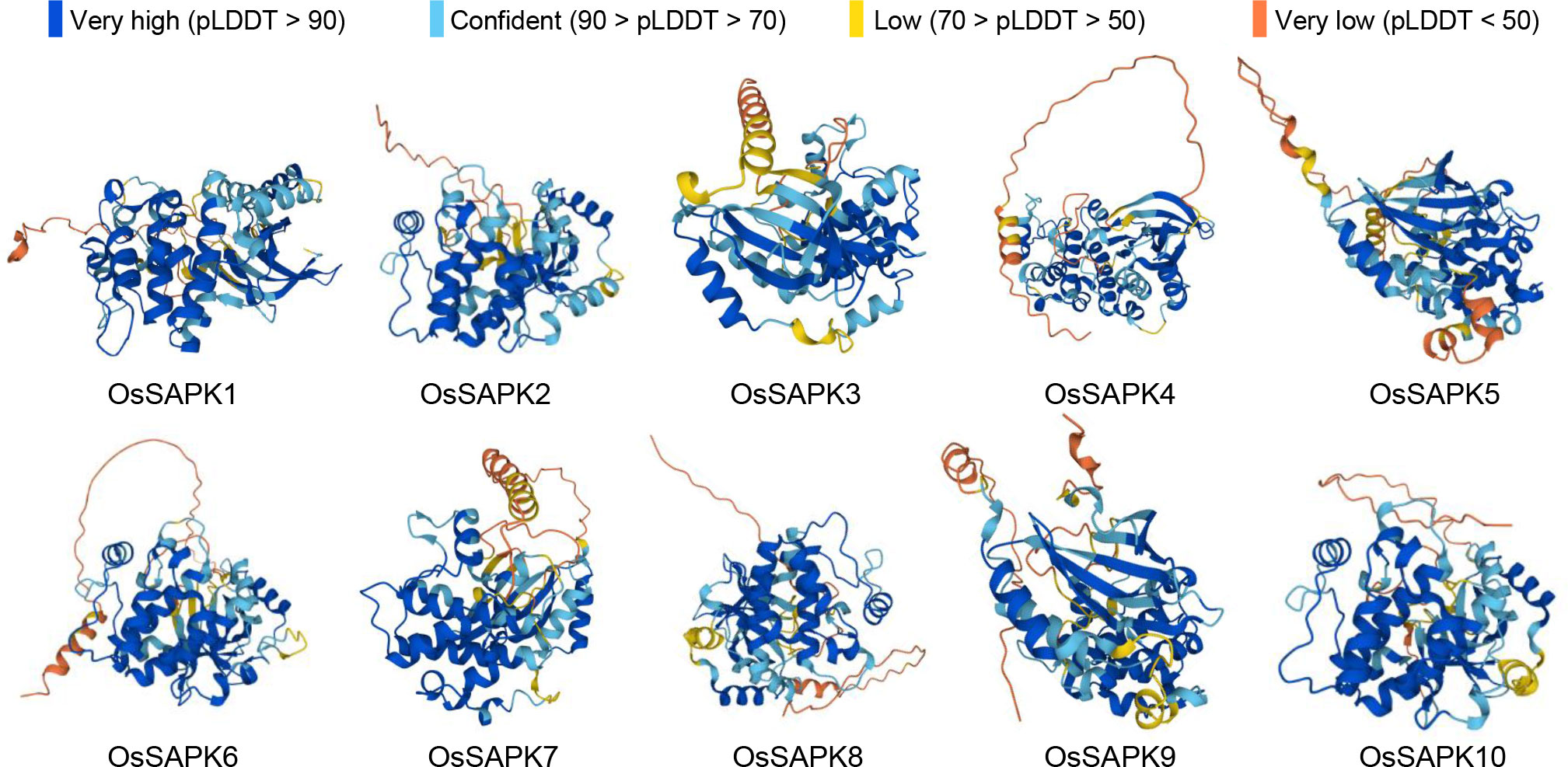
Figure 5 Protein 3D structure prediction model of OsSnRK2 gene families. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100.
Protein-protein interaction analysis of OsSnRK2 proteins
To identify potential interacting proteins with the OsSnRK2 proteins, a protein - protein interaction (PPI) network was generated with the STRING database (Figure 6). Notably, several proteins belonging to OsBZIP and OsPP2C proteins interacted with OsSAPK1, OsSAPK2, OsSAPK3, OsSAPK5, OsSAPK6, OsSAPK7, OsSAPK9, OsSAPK10, suggested their regulatory role in ABA signaling. Furthermore, there are other potential interacting proteins for OsSnRK2 proteins. Among them, OsDOS (a CCCH-Type Zinc Finger Protein) and OsDjC28 (belongs to heat shock protein DnaJ), are both showing tight relationships with different OsSnRK2 proteins, respectively.

Figure 6 Protein-protein interaction network assembly of OsSnRK2 proteins. The area and color of the circles represent the number of interacting proteins, the larger area and darker color of the circles indicate more interacting proteins. The dashed lines represent protein interaction score ≤ 0.5, and solid lines means protein interaction score > 0.5.
Cis-elements analysis in OsSnRK2 promoters
To explore the underlying function of the OsSnRK2 genes, Plant-CARE was adopted for the analysis of cis-elements in their promoter region. The sequence of 2000 bp upstream of the OsSnRK2 gene start codon was downloaded from the whole genome data Ensembl website and submitted for the cis-elements assay. The promoter sequences of the OsSnRK2 gene contained a number of light-responsive cis-acting elements (e.g. AE-box, Box4, G-Box, GT1-motif, sp1), ABA-responsive cis-acting elements (ABRE), stress-responsive cis-acting elements (DRE, LTR, MYB, MYC, TC-rich repeats), salicylic acid response-related elements (TCA-element), MeJA-related elements (CGTCA-motif, TGACG-motif) and meristematic tissue expression elements (CAT-box) (Figure 7 and Supplementary Table 5). This indicates that OsSnRK2 genes may be by exogenous factors such as light, hormones, stress and involved in the stress response.
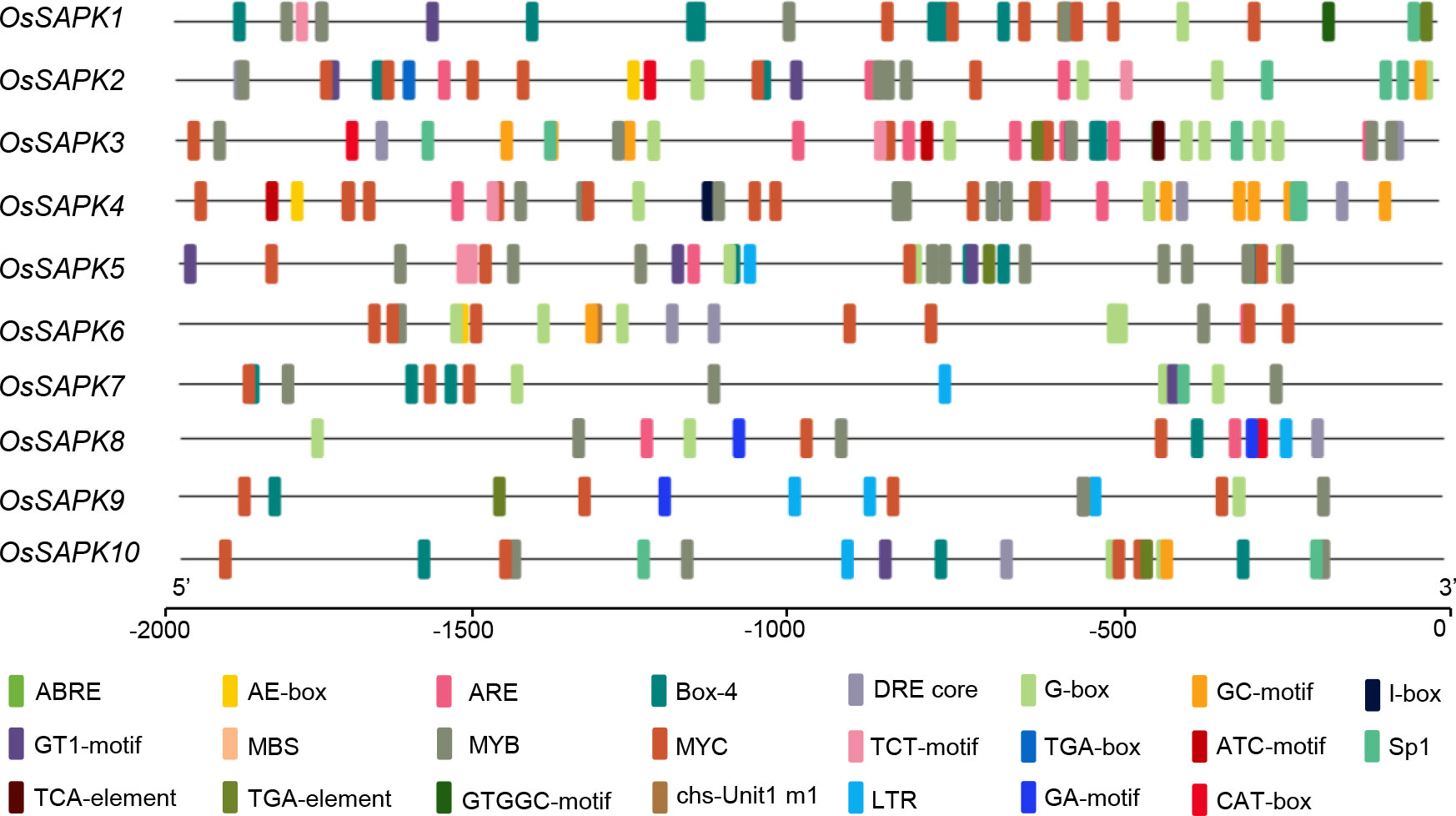
Figure 7 Predicted cis-acting elements in OsSnRK2 promoter. Promoter sequences (−2 Kb) of 10 OsSnRK2 were analyzed by PlantCARE. Different cis-elements are represented by different colors.
Tissue expression and expression profile analysis of OsSnRK2
The expression patterns of the OsSnRK2 gene were investigated in different tissues (root, stem, leaf, sheath and panicle) of rice (Nipponbare) (Figure 8A). It was found that OsSAPK2 and OsSAPK7 were most abundantly expressed in the root and expressed at the lowest level in the panicle; OsSPK3, OsSAPKA4, and OsSAPK9 were the most highly expressed in the stem, OsSAPK3, OsSAPK4 were the least expressed in panicle and OsSAPK9 was the least expressed in leaf sheath; OsSAPK1, OsSAPK5, OsSAPK8, and OsSAPK10 were the most highly expressed in leaves, OsSAPK1, OsSAPK5 and OsSAPK8 were the least expressed in panicle and OsSAPK10 was the least expressed in stems. These results indicated that the expression pattern of the ten OsSnRK2 genes were tissue-specific, suggesting that OsSnRK2 might play distinct roles in different tissue in growth and stress response of rice.
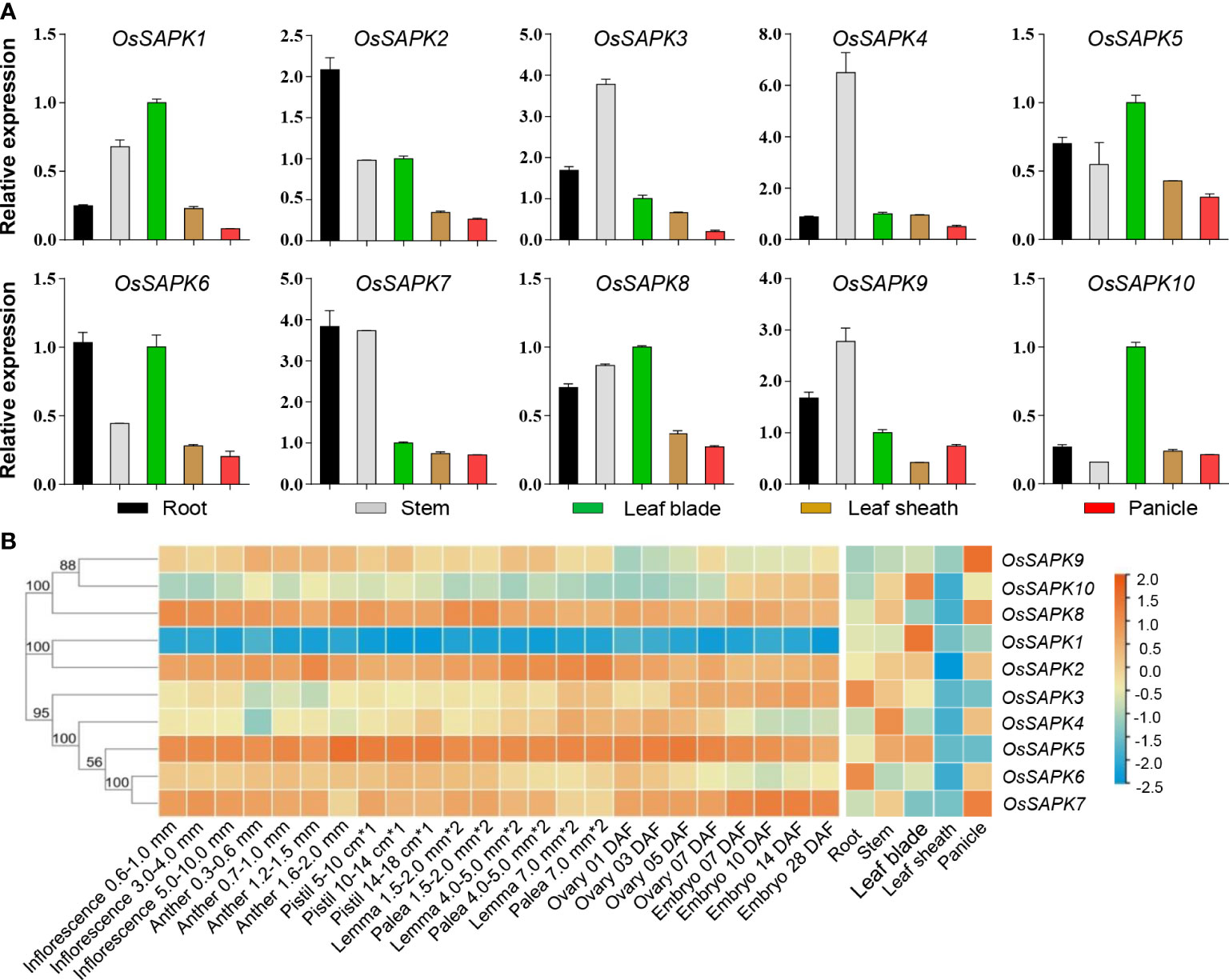
Figure 8 Expression analysis of the OsSnRK2 in different tissues. (A) Expression analysis of OsSnRK2 in root, stem, leaf blade, leaf sheath and panicle by qRT-PCR. OsUBQ5 was used for normalization. The data represent means ± standard deviation (n = 3). (B) Expression profiles of OsSnRK2 in an indica rice variety Minghui 63 obtained from CREP database. The color scale represents relative expression levels from low (blue) to high (red).
The expression file of the OsSnRK2 genes of Indica rice Mingchu 63 obtained from the CREP database (https://ricexpro.dna.affrc.go.jp/data-set.html) was hierarchically clustered in root, stem, leaf blade, leaf sheath, and panicle (Figure 8B and Supplementary Table 6), and the results showed that the expression level of OsSAPK1 and OsSAPK10 were highest in leaves, OsSAPK2 was highest in stems and OsSAPK8 was lowest in panicles, and these were consistent with the expression pattern of the OsSnRK2 genes that we showed in the japonica rice variety Nipponbare. However, the expression pattern of OsSAPK7 and OsSAPK9 obtained from the CREP database in Mingchu 63 differed dramatically from that in Nipponbare, with OsSAPK7 and OsSAPK9 in Mingchu 63 having the highest expression in the panicle. Taken together, the results indicate that there is some variability in the tissue expression of OsSnRK2 among different rice varieties.
Meanwhile, the expression of the OsSnRK2 family genes of Indica rice Mingchu 63 obtained from the CREP database was hierarchically clustered at 24 different developmental stages (Figure 8B and Supplementary Table 6). On the basis of the expression patterns of the OsSnRK2 genes in the reproductive period, they could be classified into three groups. Group 1 had low expression in all periods and contained OsSAPK1. Group 2 include OsSAPK2, OsSAPK5, OsSAPK7, and OsSAPK8, and they were expressed at relatively high level in most periods. The rest of OsSnRK2 genes were belong to the Group 3 and had moderate expression during all the periods. These results implied that OsSnRK2 showed different expression level in different developmental periods.
Expression patterns of OsSnRK2 under ABA and abiotic stress
To investigate the response of the OsSnRK2 genes to ABA, the expression level of OsSnRK2 genes in rice seedlings after 24 h treatment with different concentrations of ABA (0 µM, 50 µM, 100 µM) was analyzed (Figure 9). The results showed that the expression of OsSAPK3 and OsSAPK8 remained unchanged under different concentrations of ABA applications; the expression trends of OsSAPK2, OsSAPK4, OsSAPK7, and OsSAPK9 were significantly lower than that of the control with 50 µM ABA treatment. The expression of OsSAPK2, OsSAPK4, OsSAPK7, and OsSAPK9 was 0.002-fold, 0.31-fold, 0.37-fold, and 0.51-fold lower than the control with 50 µM ABA treatment, respectively; the expression of OsSAPK1 and OsSAPK6 tended to increase and then decrease with increasing concentration of ABA treatment. The expression levels of OsSAPK1 and OsSAPK6 in 50 µM ABA treatment were 1.66 and 4.32 times higher than the control group, respectively. The expression levels of OsSAPK5 and OsSAPK10 were enhanced as the ABA concentration increased, and the levels of OsSAPK5 and OsSAPK10 in 100 µM ABA treatment were 1.85 and 1.77 times higher than the control group, respectively.
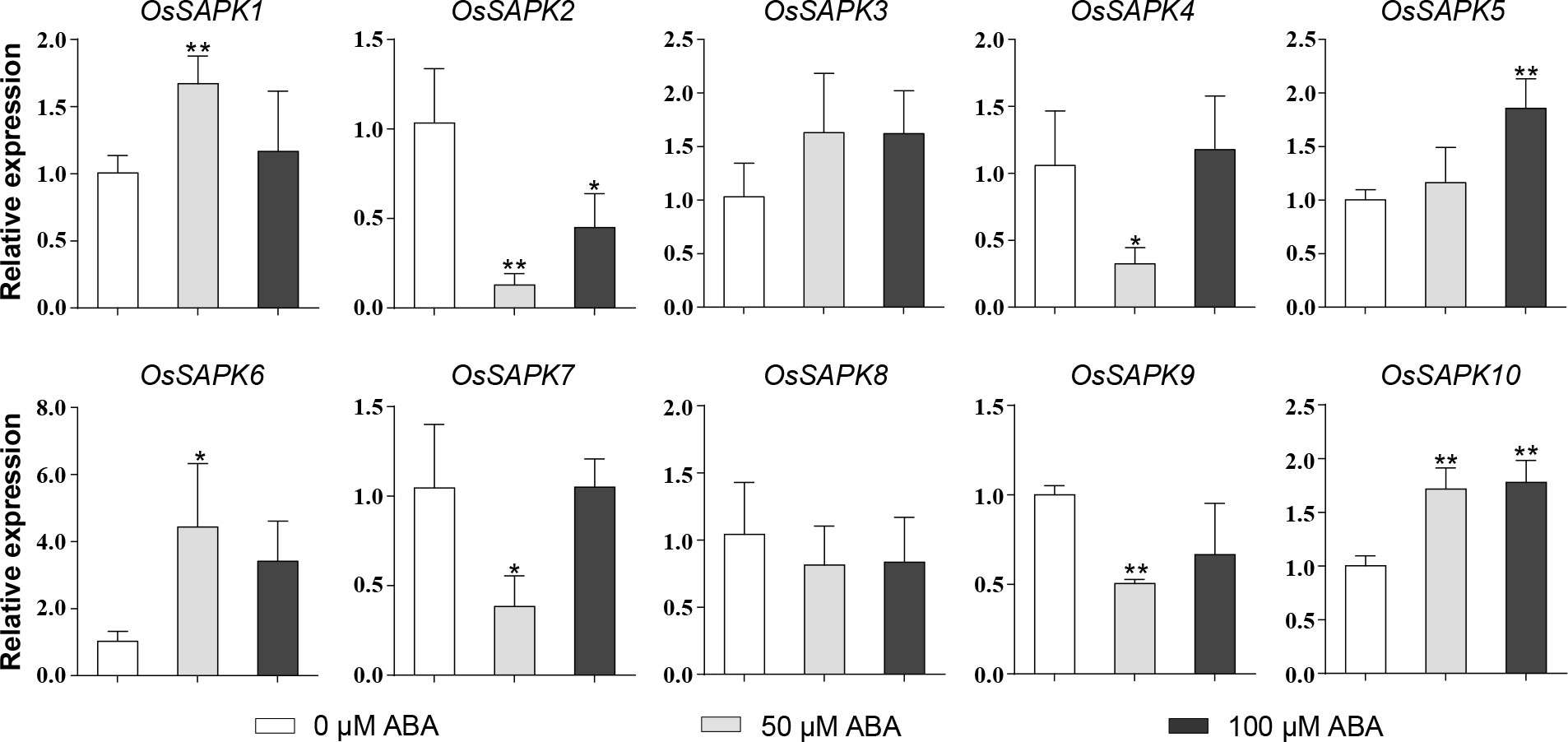
Figure 9 Expression analysis of OsSnRK2 under different ABA concentration treatments. OsUBQ5 was used for normalization. Total RNA was extracted from shoots of 14-day-old rice seedlings that were treatment or not. The data bars indicated Mean ± SD of three replicates. Asterisks represents significant differences from 0 µM ABA treatment (*p < 0.05; **p < 0.01) by Student’s t test.
To investigate the gene expression of OsSnRK2 gene under salt and drought stress, the transcription level of the OsSnRK2 gene in rice seedlings under three different treatments of 200 mM NaCl (salt treatment), 13% PEG 6000 (drought treatment) and 200 mM NaCl combined with 13% PEG 6000 (salt-drought double treatment) was compared using qRT-PCR (Figure 10). Under salt stress, the expression level of OsSAPK1, OsSAPK2, OsSAPK4, OsSAPK6, OsSAPK7 and OsSAPK8 were dramatically increased. In contrast, OsSAPK3 and OsSAPK5 were significantly decreased. However, the expression of other OsSnRK2s were not altered by salt stress. Under drought stress, OsSAPK10 was observed significantly up-regulated, which was 2.65-fold higher than the control. Some of the OsSnRK2 genes were inhibited by drought stress, with OsSAPK2 and OsSAPK4 being 0.40 and 0.48 times than the control, respectively. The expression of OsSAPK1 and OsSAPK6 was significantly up-regulated than the other genes under the salt-drought double stress, with 13.96 and 3.75 times higher than the control, respectively. The above results demonstrate the potential important role of the OsSnRK2 family in response to abiotic stress.
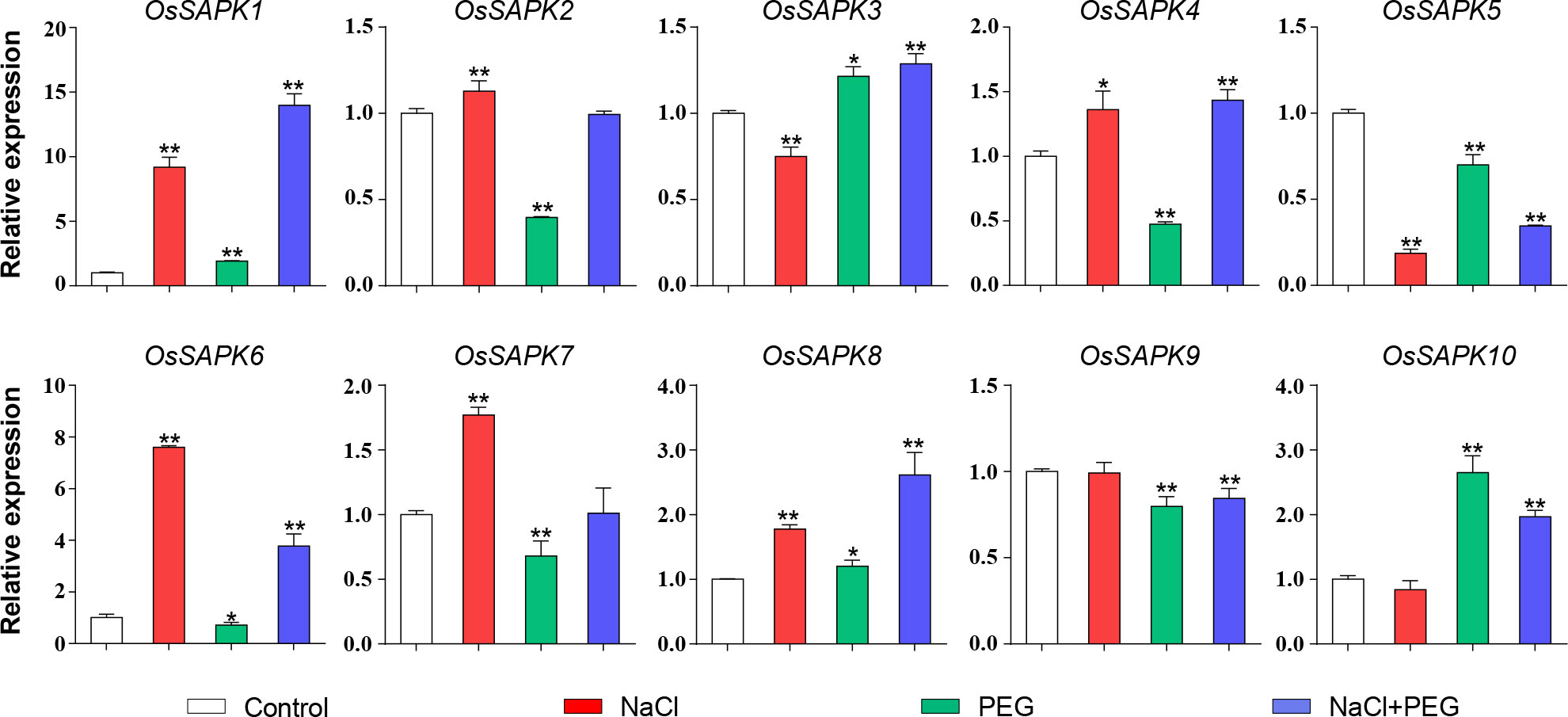
Figure 10 Expression analysis of OsSnRK2 under salt, drought and salt-drought double stress. OsUBQ5 was used for normalization. Total RNA was extracted from shoots of 14-day-old rice seedlings that were treatment or not. The data bars indicated Mean ± SD of three replicates. Asterisks represents significant differences from Control (*p < 0.05; **p < 0.01) by Student’s t test.
Discussion
Detailed characterization and evolution of SnRK2 in rice
SnRK2 genes were found only in the plant kingdom and play a crucial role in plant growth, stress response, and have been identified in a variety of plants, such as Arabidopsis thaliana, oat, Hevea brasiliensis (Cheng et al., 2017; Shao et al., 2018; Wang et al., 2019), etc. Researchers has also identified OsSnRK2 in rice, but not systematically. In the current study, ten OsSnRK2 gene (named OsSAPK1-OsSAPK9) sequences were obtained from website RiceDate for detailed bioinformatics analysis. Ten OsSnRK2 genes were unevenly distributed on seven chromosomes (Figure 1). The same subfamily genes often share similar intron-exon organization and motif (Zhang et al., 2022), For example, most SnRK2 genes have 9 exons in Nicotiana tabacum (Li et al., 2022). In this study, we found that majority of OsSnRK2 genes have 9 exons and the same motif arranged in the same order (Figure 2). The similarity shown above implied the functional redundancy among the members of OsSnRK2 gene family. However, there were differences in the structure of individual OsSnRK2 compared with genes of the same subclade. For example, the number of exons of OsSAPK5 is significantly less than other members in the same subfamily, which only has 5 exons, and OsSAPK3 has 8 motif, but others member have 9 motif. It is speculated that OsSnRK2 members may have new functions by increased or decreased introns in the process of evolution to adapt to environmental changes.
Multiple sequence alignment of OsSnRK2 proteins showed that the N-terminus of this family was highly conserved protein kinase catalytic domain (S_TKc structural) and contained similar motif composition, which has also been reported in pepper (Capsicum annuum L.) (Wu et al., 2020; Figure 3). In the analysis of gene duplication and collinearity of OsSnRK2, we identified three pairs of segmental duplication events in ten genes, which speculated that segmental duplication may be the main way of SnRK2 gene expansion in rice. A pair of duplication events, OsSAPK1 and OsSAPK2, have been confirmed to act synergistically and become positive regulators of salt tolerance in rice (Lou et al., 2018). Moreover, the more syntenic gene pairs between Hordeum vulgare and Oryza sativa than between Oryza sativa and Arabidopsis thaliana, illustrating the greater affinity between Hordeum vulgare and Oryza sativa, which was further demonstrated by the results of phylogenetic relationships analysis (Figure 3).
OsSnRK2 genes play important roles in ABA pathway
The results of protein-protein interaction analysis show that the OsSnRK2 proteins are closely related to the OsBZIP and OsPP2C proteins (Figure 6). Among the OsBZIP proteins, OsBZIP23 and OsBZIP46 are positive regulators in ABA-mediated drought resistance and are involved in ABA signaling (Yoshida et al., 2010; Guo et al., 2013; Vanitha et al., 2022). In addition, among the OsPP2C protein family, there are key players in ABA signal transduction, including OsPP2C50 and OsPP2C53, which act by negatively regulating ABA responses (Rodriguez et al., 1998; Merlot et al., 2001; Ma et al., 2009; Li et al., 2015; Zhao et al., 2021). These proteins have been shown to interact with some of the OsSnRK2 family proteins, suggesting that the OsSnRK2 family proteins are widely involved in the ABA signaling pathway.
Cis-acting elements are important switches in the regulation of gene transcription. Analysis of cis-acting elements of OsSnRK2 promoter showed that there were enriched in cis-elements for the responses to ABA, such as ABRE, AER, etc. (Figure 7). This results are similar to those in pepper (Wu et al., 2020), Brassica napus (Yoo et al., 2016), Ammopiptanthus nanus (Tang et al., 2021). Further analysis revealed that 90% of OsSnRK2 genes had ABRE, an ABA-related regulatory element (Kazuo and Kazuko, 2013), suggesting that the potential involvement of OsSnRK2 family genes in ABA response. For example, mutations in ABA-activated SNF1-associated protein kinase 2 (SnRK2s) – SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3 in Arabidopsis thaliana cause up-regulation of ABA repressor gene expression and down-regulation of ABA activator gene expression, resulting in severe growth defects during seed development, such as loss of dormancy function (Kazuo et al., 2009).
Protein interaction and cis-acting elements analysis of OsSnRK2 indicate that this family likely involved in ABA signaling transduction pathway, which was further confirmed by the expression analysis of OsSnRK2 gene under ABA treatment. Most OsSnRK2 genes expression were significantly altered by exogenous ABA, some were up-regulated (OsSAPK1, 6, 5, 10) and some were down-regulated (OsSAPK2, 4, 7, 9), indicating that OsSnRK2 genes play a dual role in ABA response and plant growth (Figure 9). Mahadi et al. demonstrate this point and proposed that OsSnRK2 can promote plant growth under normal conditions and while inhibiting plant growth in the absence of ABA (Mahadi et al., 2022).
OsSnRK2 genes play important roles in salt and drought stress responses
In the natural environment, plants are subjected to various abiotic stresses, such as drought, salinity low temperature, etc. As protein kinases, OsSnRK2 can phosphorylate or dephosphorylate the interacting proteins and involved in signal transduction, which is essential for plants to sense and adapt to various stress (Lou et al., 2017; Wang J et al., 2020; Wang Y et al., 2020; Li et al., 2021; Jia et al., 2022). In current study, we identified kinds of potential interacting factor for OsSnRK2 proteins including heat shock protein DnaJ and bZIP transcription factors that play a crucial role in salt and drought stress, and indicating OsSnRK2 involved in stress response (Figure 6). In addition, a large number of cis-acting elements were found in the SnRK2 genes promoters, which is important in response to different hormones and abiotic stresses in wheat, potato, cotton (Zhang et al., 2016; Bai et al., 2017; Liu et al., 2017). Similar results were also verified in rice. The promoters of almost all OsSnRK2 contain drought and salt stress response cis-acting elements, such as ABRE, G-box, and DRE, indicated that OsSnRK2 family play a crucial role in the process of rice adaptive stress.
Previous studies have demonstrated that SnRK2 genes are involved in a various of abiotic stresses responses (Ding et al., 2015; Soma et al., 2017; Tan et al., 2018). AtSnRK2.4 and AtSnRK2.10 are rapidly and transiently activated and regulate ROS homeostasis involved salt stress in Arabidopsis (Szymańska et al., 2019). The expression of TaSnRK2 genes was induced by drought and salt, and overexpression of TaSnRK2.4 in Arabidopsis significantly increased salt tolerance (Zhang et al., 2016). Under salt and PEG treatment, five GhSnRK2 genes expression were found notably upregulated in cotton (Gossypium hirsutum) (Liu et al., 2017). In Populus trichocarpa, heterologously overexpression of PtSnRK2.5 and PtSnRK2.7 genes could enhance Arabidopsis salt stress tolerance (Song et al., 2016). In rice, several SnRK2 genes have also been identified to be involved in stress responses. For example, overexpression of OsSAPK4, OsSAPK6 or OsSAPK7 in rice could increase salt tolerance (Diedhiou et al., 2008; Nam et al., 2012; Zeng et al., 2021), and overexpression OsSAPK2 could improve grain yield by regulating nitrogen utilization under reproductive drought stress (RDS) (Lou et al., 2020). However, there is no systematic gene expression analysis under abiotic stress, especially under salt-drought combined stress, though they can coexist in the agroecosystem. In our results, almost all OsSnRK2 members responded to salt and drought treatment simultaneously, except OsSAPK9 and OsSAPK10 which only responded to drought stress. Some OsSnRK2s showed opposite responses to salt and drought, but some are the same. For example, OsSAPK1 and OsSAPK8 expression were up-regulated by salt and drought simultaneously, and OsSAPK2, OsSAPK4, OsSAPK6 and OsSAPK7 were up-regulated by salt and down-regulated by drought stress (Figure 10). Under salt and drought stress, most OsSnRK2 genes, including OsSAPK1, OsSAPK3, OsSAPK4, OsSAPK6, OsSAPK8 and OsSAPK10, were up-regulated and two genes (OsSAPK5/9) were significantly down-regulate than control(Figure 10). The gene expressions of OsSAPK1 and OsSAPK8 under salt-drought combined stress were significantly higher than those under single stress (salt or drought). In addition, OsSAPK1 and OsSAPK8 were highly induced by salt and drought simultaneously, which suggested that OsSAPK1 and OsSAPK8 may play an important positive regulatory role in response to salt-drought combined stress, and overexpression of OsSAPK1 or OsSAPK8 in rice may significantly improve the salt and drought tolerance and increase rice yield. The above results showed that OsSnRK2 genes exhibit different expression profiles in response to salt and drought, which indicated that OsSnRK2 genes play different role, and may be have different mechanisms in response to salt and drought stress.
Conclusion
Here, we studied the SnRK2 gene family in rice by bioinformatics method. The results revealed the characteristics of the OsSnRK2 gene in terms of physicochemical properties, phylogenetic relationships, structural domain distribution, chromosomal localization, motif composition, intron-exon structure, tissue expression, etc. In addition, the tissue-specific expression and response of the OsSnRK2 gene to ABA, salt, drought, and salt-drought double stress were analyzed using qRT-PCR. The results showed that OsSnRK2 genes were expressed in rice roots, stems, leaves, leaf sheaths and panicles, but the expression levels varied from different tissues and varieties. The expression level of most OsSnRK2 genes were induced by ABA, salt, drought, and salt-drought double stress, indicating that OsSnRK2 genes may be extensively involved in stress response. In summary, this study adopted bioinformatics way and qRT-PCR to unveil the physicochemical properties, tissue expression patterns, and responses of the OsSnRK2 gene to different stresses, expanding the understanding of SnRK2 gene family in rice and providing a reference for further investigation of their functions in response to ABA treatment and different abiotic stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
DX, QT, and TY conceived and designed the research; TY, QC, LK, WM, XQZ, YF, XZ, and QT performed the experiments and data analyses. DX supervised the experiments and gave advice on laboratory work. TY and QT wrote the manuscript. DX, QT, and WM revised the manuscript. All authors read and approved the final article.
Funding
This work is supported by the Natural Science Foundation of Zhejiang province in China (LY21C130007; LY20C140003; LY19C130001), Hangzhou Scientific and Technological Major Project (202203A01) and Hangzhou Scientific and Technological Project (20201203B107).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1088281/full#supplementary-material
References
Bai, J., Mao, J., Yang, H., Khan, A., Fan, A., Liu, S., et al. (2017). Sucrose non-ferment 1 related protein kinase 2 (SnRK2) genes could mediate the stress responses in potato (Solanum tuberosum l.). BMC Genet. 18, 41. doi: 10.1186/s12863-017-0506-6
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, C., Wang, Z., Ren, Z., Zhi, L., Yao, B., Su, C., et al. (2017). SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PloS Genet. 13, e1006947. doi: 10.1371/journal.pgen.1006947
Diedhiou, C. J., Popova, O. V., Dietz, K. J., Golldack, D. (2008). The SNF1-type serine-threonine protein kinase SAPK4 regulates stressresponsive gene expression in rice. BMC Plant Bio. 8, 49. doi: 10.1186/1471-2229-8-49
Ding, S., Zhang, B., Qin, F. (2015). Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell. 27, 3228–3244. doi: 10.1105/tpc.15.00321
Doncheva, N. T., Morris, J. H., Gorodkin, J., Jensen, L. J. (2019). Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 18, 623–632. doi: 10.1021/acs.jproteome.8b00702
Fatima, A., Khan, M. J., Awan, H. M., Akhtar, M. N., Bibi, N., Sughra, K., et al. (2020). Genome-wide identification and expression analysis of SnRK2 gene family in mungbean (Vigna radiata) in response to drought stress. Crop Pasture Sci. 71, 469–476. doi: 10.1071/CP19392
Feng, J., Wang, L., Wang, L., Wu, Y., Luo, Q., Zhang, Y., et al. (2019). TaSnRK2.9, a sucrose non-fermenting 1-related protein kinase gene, positively regulates plant response to drought and salt stress in Transgenic tobacco. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.02003
Fujii, H., Verslues, P., Zhu, J.-K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Guo, C., Ge, X., Ma, H. (2013). The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 82, 239–253. doi: 10.1007/s11103-013-0057-9
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132 (2), 666–80. doi: 10.1104/pp.102.011999
Huai, J., Wang, M., He, J., Zheng, J., Dong, Z., Lv, H., et al. (2008). Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 27, 1861–1868. doi: 10.1007/s00299-008-0608-8
Jia, M., Meng, X., Song, X., Zhang, D., Kou, L., Zhang, J., et al. (2022). Chilling-induced phosphorylation of IPA1 by OsSAPK6 activates chilling tolerance responses in rice. Cell Discovery 8, 71. doi: 10.1038/s41421-022-00413-2
Kawa, D., Meyer, A. J., Dekker, H. L., Abd-El-Haliem, A. M., Gevaert, K., Slijke, E. V. D., et al. (2020). SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 182, 361–377. doi: 10.1104/pp.19.00818
Kazuo, N., Kazuko, Y.-S. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Kazuo, N., Yasunari, F., Norihito, K., Takeshi, K., Taishi, U., Satoshi, K., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Khatun, M., Sarkar, S., Era, F. M., Mominul Islam, A. K. M., Anwar, P., Fahad, H., et al. (2021). Drought stress in Grain legumes: Effects, tolerance mechanisms and management. Agronomy. 11, 2374. doi: 10.3390/AGRONOMY11122374
Li, C., Shen, H., Wang, T., Wang, X. (2015). ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol. 56, 2396–2408. doi: 10.1093/pcp/pcv154
Li, J., Song, J., Li, C., Ma, J., Liu, J., Zhu, X., et al. (2022). Genome-wide identification and expression profile analysis of the SnRK2 gene family in Nicotiana tabacum. Biochem. Genet. 60, 1511–1526. doi: 10.1007/s10528-021-10170-8
Liu, Z., Ge, X., Yang, Z., Zhang, C., Zhao, G., Chen, E., et al. (2017). Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum l.). BMC Genet. 18 (1), 54. doi: 10.1186/s12863-017-0517-3
Li, J., Wang, X., Watson, M. B., Assmann, S. M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 287, 300–303. doi: 10.1126/science.287.5451.300
Li, X., Yu, B., Wu, Q., Min, Q., Zeng, R., Xie, Z., et al. (2021). OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PloS Genet. 17, e1009699. doi: 10.1371/journal.pgen.1009699
Lou, D., Chen, Z., Yu, D., Yang, X. (2020). SAPK2 contributes to rice yield by modulating nitrogen metabolic processes under reproductive stage drought stress. Rice. 13, 35. doi: 10.1186/s12284-020-00395-3
Lou, D., Wang, H., Liang, G., Yu, D. (2017). OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00993
Lou, D., Wang, H., Yu, D. (2018). The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 18, 203. doi: 10.1186/s12870-018-1408-0
Mahadi, H. M., Xudong, L., Muhammed, W., Yao, G., Alabdallah, N. M., Shah, J. M., et al. (2022). ABA activated SnRK2 kinases: An emerging role in plant growth and physiology. Plant Signal. Behav. 17, 2071024. doi: 10.1080/15592324.2022.2071024.
Mao, X., Li, Y., Rehman, S. U., Miao, L., Zhang, Y., Chen, X., et al. (2020). The sucrose non-fermenting 1-related protein kinase 2 (SnRK2) genes are multifaceted players in plant growth, development and response to environmental stimuli. Plant Cell Physiol. 61, 225–242. doi: 10.1093/pcp/pcz230
Maszkowska, J., Szymańska, K. P., Kasztelan, A., Krzywińska, E., Sztatelman, O., Dobrowolska, G. (2021). The multifaceted regulation of SnRK2 kinases. Cells. 10, 2180. doi: 10.3390/cells10092180
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D. L., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 324, 1064–1068. doi: 10.1126/science.1172408
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nam, M. H., Huh, S. M., Kim, K. M., Park, W. J., Seo, J. B., Cho, K., et al. (2012). Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci. 10, 25. doi: 10.1186/1477-5956-10-25
Riichiro, Y., Tokunori, H., Kazuya, I., Tsuyoshi, M., Fuminori, T., Jose, A., et al. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. doi: 10.1093/pcp/pcf188
Rodriguez, P. L., Benning, G., Grill, E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421, 185–190. doi: 10.1016/s0014-5793(97)01558-5
Shao, Y., Zhang, X., Nocker, S. V., Gong, X., Ma, F. (2018). Overexpression of a protein kinase gene MpSnRK2.10 from Malus prunifolia confers tolerance to drought stress in transgenic Arabidopsis thaliana and apple. Gene. 692, 26–34. doi: 10.1016/j.gene.2018.12.070
Sharif, R., Xie, C., Wang, J., Cao, Z., Zhang, H., Chen, P., et al. (2020). Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus l. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 158, 502–520. doi: 10.1016/j.ijbiomac.2020.04.124
Soma, F., Mogami, J., Yoshida, T., Abekura, M., Takahashi, F., Kidokoro, S., et al. (2017). ABA unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants. 3, 16204. doi: 10.1038/nplants.2016.204
Song, X., Yu, X., Hori, C., Demura, T., Ohtani, M., Zhuge, Q. (2016). Heterologous overexpression of poplar SnRK2 genes enhanced salt stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00612
Szymańska, K. P., Polkowska-Kowalczyk, L., Lichocka, M., Maszkowska, J., Dobrowolska, G. (2019). SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int. J. Mol. Sci. 20, 143. doi: 10.3390/ijms20010143
Taishi, U., Riichiro, Y., Kyonoshin, M., Kazuko, Y.-S., Kazuo, S. (2004). SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. P Natl. Acad. Sci. U.S.A. 101, 17306–17311. doi: 10.1073/pnas.0407758101
Tang, Y., Lu, F., Feng, W., Liu, Y., Cao, Y., Li, W., et al. (2021). Genome-wide identification and expression analyses of AnSnRK2 gene family under osmotic stress in Ammopiptanthus nanus. Plants. 10, 882. doi: 10.3390/PLANTS10050882
Tan, W., Zhang, D., Zhou, H., Zheng, T., Yin, Y., Lin, H. (2018). Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PloS Genet. 14, e1007336. doi: 10.1371/journal.pgen.1007336
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8. doi: 10.1126/scisignal.2003509
Vanitha, P., Preethi, V., Akshay, U., Akashata, D., Sujitha, D., Ashwin, V., et al. (2022). Novel small molecules targeting bZIP23 TF improve stomatal conductance and photosynthesis under mild drought stress by regulating ABA. FEBS J. 289, 6058–6077. doi: 10.1111/febs.16461
Wang, Y., Hou, Y., Qiu, J., Wang, H., Wang, S., Tang, L., et al. (2020). Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 228, 1336–1353. doi: 10.1111/nph.16774
Wang, J., Ren, Y., Liu, X., Luo, S., Zhang, X., Liu, X., et al. (2020). Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 14, 315–329. doi: 10.1016/j.molp.2020.11.022
Wang, X., Wu, Q., Yuan, H. (2019). Cloning, prokaryotic expression and purification of SnRK2.6 in Hevea brasiliensis. MPB. 17, 2828–2834. doi: 10.13271/j.mpb.017.002828
Wu, Z., Cheng, J., Hu, F., Qin, C., Xu, X., Hu, K. (2020). The SnRK2 family in pepper (Capsicum annuum l.): genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genom. 42, 1117–1130. doi: 10.1007/s13258-020-00968-y
Xu, P., Zhang, X., Su, H., Liu, X., Wang, Y., Hong, G. (2020). Genome-wide analysis of PYL-PP2C-SnRK2s family in Camellia sinensis. Bioengineered. 11, 103–115. doi: 10.1080/21655979.2019.1710932
Yoo, M.-J., Ma, T., Zhu, N., Liu, L., Harmon, A. C., Wang, Q., et al. (2016). Genome-wide identification and homeolog-specific expression analysis of the SnRK2 genes in Brassica napus guard cells. Plant Mol. Biol. 91, 211–227. doi: 10.1007/s11103-016-0456-9
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yuhko, K., Shuhei, Y., Hideyuki, M., Yasuaki, K., Tsukaho, H. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 16, 1163–1177. doi: 10.1105/tpc.019943
Zelm, E. V., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Zeng, D., Wang, C., Xie, J., Zhang, F., Lu, J., Shi, X. (2021). Stress-activated protein kinase OsSAPK7 regulates salt-stress tolerance by modulating diverse stress-defensive responses in rice. Rice Sci. 28, 547–594. doi: 10.1016/j.rsci.2021.09.003
Zhang, Z., Ali, S., Zhang, T., Wang, W., Xie, L. (2020). Identification, evolutionary and expression analysis of PYL-PP2C-SnRK2s gene families in soybean. Plants. 9, 1356. doi: 10.3390/plants9101356
Zhang, J., Liao, J., Ling, Q., Xi, Y., Qian, Y. (2022). Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 23, 125–125. doi: 10.1186/s12864-022-08345-7
Zhang, H., Li, W., Mao, X., Jing, R., Jia, H. (2016). Differential activation of the wheat SnRK2 family by abiotic stresses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00420
Zhang, F., Zeng, D., Huang, L., Shi, Y., Chen, T., Zhang, F., et al. (2019). Stress-activated protein kinase OsSAPK9 regulates tolerance to salt stress and resistance to bacterial blight in rice. Rice. 12, 80. doi: 10.1186/s12284-019-0338-2
Keywords: rice, OsSnRK2, genome−wide, abiotic stress, expression patterns
Citation: Yu T, Cen Q, Kang L, Mou W, Zhang X, Fang Y, Zhang X, Tian Q and Xue D (2022) Identification and expression pattern analysis of the OsSnRK2 gene family in rice. Front. Plant Sci. 13:1088281. doi: 10.3389/fpls.2022.1088281
Received: 03 November 2022; Accepted: 23 November 2022;
Published: 13 December 2022.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Chenchen Zhao, University of Tasmania, AustraliaShengchun Xu, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2022 Yu, Cen, Kang, Mou, Zhang, Fang, Zhang, Tian and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quanxiang Tian, cXVhbnhpYW5nQGh6bnUuZWR1LmNu; Dawei Xue, ZHd4dWVAaHpudS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Tongyuan Yu
Tongyuan Yu Qiwen Cen†
Qiwen Cen† Lihua Kang
Lihua Kang Yunxia Fang
Yunxia Fang Xian Zhang
Xian Zhang Quanxiang Tian
Quanxiang Tian Dawei Xue
Dawei Xue