- Department of Life Sciences, School of Agriculture, Meiji University, Kawasaki, Japan
Phosphorus (P) is an essential nutrient for plant growth and plants use inorganic phosphate (Pi) as their P source, but its bioavailable form, orthophosphate, is often limited in soils. Hence, plants have several mechanisms for adaptation to Pi starvation. One of the most common response strategies is “Pi recycling” in which catabolic enzymes degrade intracellular constituents, such as phosphoesters, nucleic acids and glycerophospholipids to salvage Pi. Recently, several other intracellular degradation systems have been discovered that salvage Pi from organelles. Also, one of sphingolipids has recently been identified as a degradation target for Pi recycling. So, in this mini-review we summarize the current state of knowledge, including research findings, about the targets and degradation processes for Pi recycling under Pi starvation, in order to further our knowledge of the whole mechanism of Pi recycling.
Introduction
Plants take up inorganic phosphate (Pi; PO43-) and use it as their phosphorus (P) source. In the plant, the Pi is metabolized into organic phosphate, and it is also used for post-translational protein modification to regulate protein activity. Thus, Pi is essential for proper plant growth. Since Pi makes easily complex with metal ions such as aluminum and iron ions in soils and thus render it unavailable to plants, however, soils with low orthophosphate, which is an available form of Pi, are widespread throughout the world (Raghothama, 1999; Lynch, 2011). So, to overcome Pi starvation stress, plants have various systems to respond to Pi starvation. One of the Pi starvation responses is “Pi recycling”, which is a complex subject due to the existence of many intracellular components that contain Pi and the many degradation systems that can salvage Pi from them (Figure 1). Therefore, this mini-review aims to summarize our current understanding of Pi recycling in plant cells under Pi starvation and highlight areas that require further study to develop a better understanding of the overall Pi recycling process.
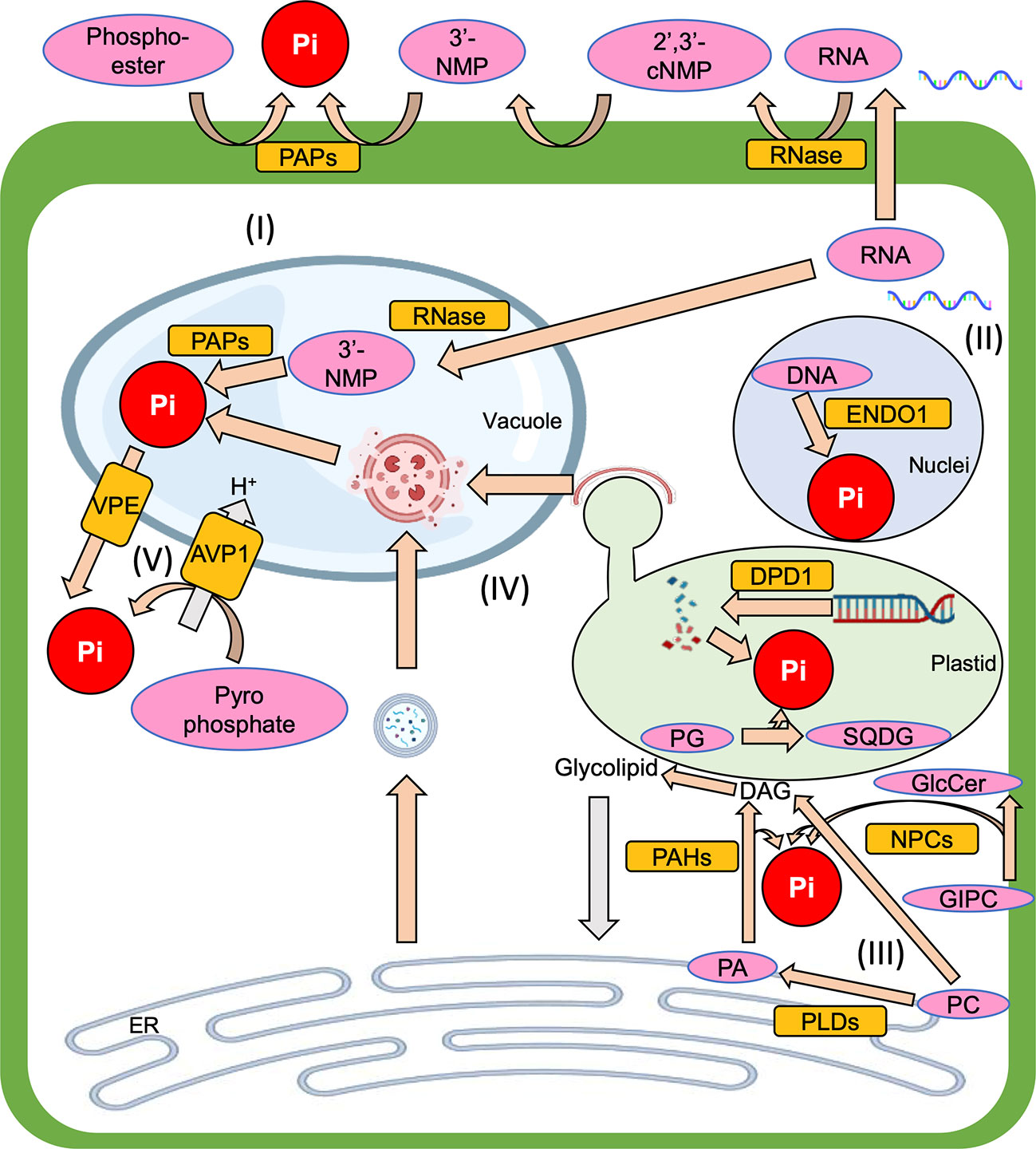
Figure 1 The overview of Pi recycling systems which are introduced in this mini-review, (I) Phosphoester degradation, (II) Nucleic acid degradation, (III) Membrane lipid remodeling, (IV) Autophagy, (V) Vacuolar transporter. Pi, inorganic phosphate; PAP, purple acid phosphatase; RNase, Ribonuclease; 2’,3’-cNMP, 2’,3’-cyclic nucleoside monophosphate intermediate; 3’-NMP, 3’-nucleoside monophosphate; ENDO1, endonuclease 1; DPD1, defective in pollen organelle DNA degradation 1; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol; GIPC, glycosylinositolphosphorylceramide; GlcCer, glucosylceramide; ER, endoplasmic reticulum; PC, phosphatidylcholine; PA, phosphatidic acid; DAG, diacylglycerol; PLC, phospholipase C; NPC, non-specific PLC; PLD, phospholipase D; PAH, phosphatidic acid phosphohydrolase; VPE, vacuolar pi efflux transporter; AVP1, arabidopsis vacuolar pyrophosphatase 1.
Phosphoester degradation
Pi starvation-induced acid phosphatases PURPLE ACID PHOSPHATASEs (PAPs), which have phosphoester hydrolase activity, have been purified and characterized in many plant species, such as Arabidopsis thaliana (Li et al., 2002), Solanum lycopersicum (Bozzo et al., 2002; Bozzo et al., 2006; Srivastava et al., 2020), Nicotiana banthaminana (Lung et al., 2008), Lupinus albus (Ozawa et al., 1995; Li and Tadano, 1996; Miller et al., 2001; Aslam et al., 2022). It was reported that the expressions of some PAP genes are upregulated by Pi starvation in A. thaliana and S. lycopersicum (del Pozo et al., 1999; Li et al., 2002; Wang et al., 2011; Srivastava et al., 2020). Of these, AtPAP10, 12, 17, and 26 are secreted acid phosphatases (Veljanovski et al., 2006; Hurley et al., 2010; Tran et al., 2010; Wang et al., 2014; O’Gallagher et al., 2022), whereas, AtPAP12, 15, and 16 are known to be major intracellular acid phosphatases in A. thaliana, because intracellular acid phosphatase activity in pap12, pap15, and pap26 is less than that in wild-type (Wang et al., 2014). Additionally, since AtPAP15 is not expressed in the root hairs or epidermal cells, it is unlikely to be secreted into the root exudates (Kuang et al., 2009). Recently, it was also reported that under Pi starvation AtPAP17 localizes in the lytic vacuole, in addition to extracellular spaces (O’Gallagher et al., 2022). AtPAP17 and 26 are bifunctional enzymes, both phosphatase and peroxidase (del Pozo et al., 1999; Veljanovski et al., 2006). Since it is known that Pi starvation enhances accumulation of reactive oxygen species (ROS), which leads to programmed cell death (Van Camp et al., 1998; Overmyer et al., 2000; Tyburski et al., 2009; Yoshitake et al., 2022), that proteins may contribute to plant growth not only through the resupply of Pi from intracellular components but also because increase ROS degradation under Pi starvation. It is known that a phosphohydrolase for inositol pyrophosphates (PP-InsPs), one of the phosphoesters, has a role in Pi starvation response, but it is unclear whether this enzyme is involved in Pi recycling (Gaugler et al., 2022).
Nucleic acid degradation
Pi can be salvaged from degradation of nucleic acids under Pi starvation. Nucleic acids contain Pi, and most nucleic acid in plant cells is ribosomal RNA (rRNA) (Veneklaas et al., 2012). They are broken down by Ribonucleases (RNases). The T2 family of RNases are highly conserved among viruses, bacteria, fungi, animals, and plants (Deshpande and Shankar, 2002), and can be grouped into three subclasses (Igic and Kohn, 2001). Expression of class I RNases is induced by stresses such as wounding, drought, and salinity (Hillwig et al., 2008; Bustos et al., 2010; MacIntosh et al., 2010), and also by Pi starvation (Bariola et al., 1994; Köck et al., 1995; Dodds et al., 1996; Bariola et al., 1999). These facts, plus the following facts lead us to hypothesize that these RNases degrade apoplastic RNA and salvage Pi from apoplast under Pi starvation. First, most class I RNases are secreted to out of the cell (Köck et al., 1995; Bariola et al., 1999; Smith et al., 2018). Also, Arabidopsis apoplastic fluid contains small RNAs and long noncoding RNAs, including circular RNAs (Zand Karimi et al., 2022). Furthermore, exogenous addition of RNA in media recovers growth defect of primary roots under Pi starvation (Chen et al., 2000). It is also thought that carnivorous plants, Drosera aldelae and Nepenthes ventricose, salvage Pi from the degradation of prey RNA by RNase (Okabe et al., 2005; Stephenson and Hogan, 2006).
Furthermore, it is thought that class I RNase can salvage intracellular Pi during Pi starvation. For example, in S. lycopersicum, RNase LX is a Pi starvation-induced intracellular RNase (Köck et al., 1995), which is retained in endoplasmic reticulum (ER) by C-terminal tetrapeptide HDEF (Löffler et al., 1993; Lehmann et al., 2001). Class II RNase also appear to be involved in response to Pi starvation. For example, Arabidopsis class II RNase, RNS2, is localized in the vacuole and degrade vacuolar RNA during development (Hillwig et al., 2011; Floyd et al., 2017). In addition, because the expressions of class II RNase genes are upregulated by Pi starvation, it is considered that these RNases have not only a housekeeping role in rRNA degradation but also a role in response to Pi starvation (Taylor et al., 1993; Liang et al., 2002; MacIntosh et al., 2010; Hillwig et al., 2011; Floyd et al., 2017).
Recently, it has been reported that AtRNSs can cleavage transfer RNAs (tRNAs) and that RNS1 is responsible for the accumulation of specific tRNA fragments (glycine and aspartic acid) in leaves under Pi starvation (Megel et al., 2019). However, it is still unclear whether the generation of tRNA fragments is involved in Pi recycling.
Pi is also released from 3’-nucleoside monophosphate (3’-NMP) by PAP (Abel et al., 2000). 3’-NMP can be generated from 2’,3’-cyclic nucleoside monophosphate intermediate (2’,3’-cNMP), which is generated from degradation of RNAs, by a side reaction of class I and II RNases (Nürnberger et al., 1990). However, the catabolic enzyme of 2’,3’-cNMP, a Pi starvation-induced cyclic nucleotide phosphodiesterase, was discovered in cultured L. esculentum cells, and it had higher activity of 2’,3’-cNMP degradation was higher than RNases (Abel et al., 2000). Thus, it appears that Pi salvaging from cyclic nucleotides is mainly mediated by this cyclic nucleotide phosphodiesterase pathway.
DNA also can be source of Pi. The ENDONUCLEASE 1 (ENDO1) gene encodes a type I nuclease and its expression in Petunia is induced by Pi starvation (Pérez-Amador et al., 2000; Jones et al., 2021). Under Pi starvation, plastid DNA is degraded by DEFECTIVE IN POLLEN ORGANELLE DNA DEGRADATION 1 (DPD1), exonuclease. However, Pi remobilization from old leaves to younger leaves under Pi starvation is suppressed in the dpd1 mutant (Takami et al., 2018), suggesting that DPD1 may be required for Pi recycling. However, it is unclear how the metabolites of plastid DNA are transported out of plastids.
Membrane lipid remodeling
The major components of the biological membrane are glycerolipids, sphingolipids, and sterols (Harayama and Riezman, 2018). The glycerolipids containing Pi in the polar heads are called phospholipids, whereas, those containing carbohydrates are called glycolipids. Under Pi starvation, phospholipids are degraded and the resultant diacylglycerols (DAGs), which are precursors of glycolipids, are delivered to the plastid. The glycolipids are transferred to extraplastidic membranes such as the ER membrane, plasma membrane, and mitochondria membrane to maintain membrane structures (Härtel and Benning, 2000; Dörmann and Benning, 2002; Jouhet et al., 2004; Nakamura, 2013). This process is called “membrane lipid remodeling”. In this process, phosphatidylglycerol (PG) which is an anionic phospholipid, and phosphatidylcholine (PC) which forms a lipid bilayer, are replaced respectively by sulfoquinovosyldiacylglycerol (SQDG) and digalactosyldiacylglycerol (DGDG) (Härtel and Benning, 2000; Dörmann and Benning, 2002; Yu et al., 2002). PLASTID LIPASE 1 (PLIP1) and its paralogues (PLIP2, 3) degrade PG, but they produce fatty acids and lyso-PG rather than Pi (Wang et al., 2017; Wang et al., 2018). So, for full understanding of the Pi recycling system from PG, it is still necessary to identify the PG catabolic enzymes which produce Pi from the polar head of PG. SQDG is synthesized by SQDG SYNTHASE 2 (SQD2) from DAG and UDP-sulfoquinovose (Yu et al., 2002), with the UDP-sufoquinovose being generated by SQD1 (Sanda et al., 2001). The expression of both SQD1 and 2 are upregulated by Pi limitation (Essigmann et al., 1998; Yu et al., 2002). SQD2 also synthesizes glucuronosyldiacylglycerol (GlcADG) and sqd2 mutant showed more severe growth defect phenotypes than sqd1 mutant under Pi starvation (Okazaki et al., 2013). Therefore, it appears that GlcADG has an important role in Pi starvation tolerance. However, it is not clear what lipids are replaced by GlcADG under Pi starvation.
There are two PC degradation pathways during membrane lipid remodeling. One pathway generates DAG from PC directly by phospholipase C (PLC), while the other generates DAG by phosphatidic acid phosphatase from phosphatidic acid (PA) which is produced from PC by phospholipase D (PLD). In A. thaliana, the expression of two NON-SPECIFIC PLC (NPC4, 5) genes is upregulated by Pi starvation. Since NPC4 and NPC5 localize to the plasma membrane and cytoplasm respectively, it is thought that the localization for NPC4 and NPC5 degradation of PC is in the plasma membrane and ER membrane, respectively (Nakamura et al., 2005; Gaude et al., 2008). PHOSPHATE STARVATION-INDUCED GENE 2 (PS2) and its homologue, PHOSPHOETHANOLAMINE/PHOSPHOCHOLINE PHOSPHATASE 1 (PECP1), salvage Pi from phosphocholine (Angkawijaya and Nakamura, 2017; Hanchi et al., 2018; Angkawijaya et al., 2019).
A. thaliana has 12 PLD genes, of which two AtPLDζ (PLDζ1 and PLDζ2) gene expression is upregulated by Pi starvation. Furthermore, pldζ1 pldζ2 mutant has a defect in membrane lipid remodeling (Zhang et al., 2005; Li et al., 2006a; Li et al., 2006b; Su et al., 2018). PHOSPHATIDIC ACID PHOSPHOHYDROLASE 1, 2 (PAH1, PAH2) are soluble phosphatidic acid phosphatases and are localized in the cytoplasm (Nakamura et al., 2009). Under Pi starvation, the pah1 pah2 double mutant shows growth defect phenotypes and its membrane lipid remodeling is suppressed, indicating that PAHs are important in the Pi recycling from phospholipids (Nakamura et al., 2009). It has also been found that while the growth phenotype of pah1 and pah2 single mutants was similar to that of wild-type, the pah1 pah2 double mutant showed abnormal phenotypes, such as stutter rosette leaves, shorter siliques, and phospholipid accumulation, even under Pi-sufficient conditions (Eastmond et al., 2010). Therefore, it appears that PAH1 and PAH2 are functionally redundant. The more moderate phenotype of pldζ1 pldζ2 double mutant compared with the pah1 pah2 suggests that other PLDs provide PA to PAH for membrane lipid remodeling (Li et al., 2006b; Nakamura et al., 2009). After DAGs are generated by NPC or PAH, they are delivered to plastids by SECOND LptD-FAMILY PROTEIN (LPTD1) under Pi starvation (Hsueh et al., 2017). Then, the DAGs are converted into glycolipids on the plastid envelope membrane. MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 1 (MGD1), Type-A monogalactosyldiacylglycerol (MGDG) synthase, localizes on the inner envelope membrane of plastids and works in photosynthetic tissues even under Pi sufficient conditions (Awai et al., 2001; Kobayashi et al., 2007). In contrast, Type-B MGD (MGD2, 3) localizes on the outer envelope membranes of plastids, and the expression of these genes is strongly activated by Pi starvation (Awai et al., 2001; Kobayashi et al., 2004). MGDG, which is produced by Type-B MGD, is the substrate of DGDG SYNTHASE (DGD) that is transported to extraplastidic membranes to replace PC (Kelly et al., 2003). However, it is unclear how DGDG is delivered to extraplastidic membranes.
Recently, it has been reported that under Pi starvation the Pi-containing sphingolipid glycosylinositolphosphorylceramide (GIPC) is degraded by NPC4 whereas the amount of glucosylceramide (GlcCer), a non-Pi-containing sphingolipid, is increased (Yang et al., 2021). This finding suggests that GIPC is a Pi store and is replaced by GlcCer or metabolites from GlcCer, to maintain membrane functions. Therefore, both glycerophospholipids and phosphosphingolipids could facilitate Pi storage under Pi starvation.
Autophagy
(Macro)autophagy is an evolutionally conserved process for degradation of protein and/or organelles in eukaryotes. In this process, targets are engulfed by an isolation membrane, forming an autophagosome. Then, the autophagosomes are delivered to the vacuole, where the targets are degraded. Plant autophagy is induced by nutrient starvation, such as carbon, nitrogen, and zinc deficiency (Yoshimoto et al., 2004; Izumi et al., 2010; Shinozaki et al., 2020). Pi starvation also induces autophagy in tobacco BY-2 cells and A. thaliana (Tasaki et al., 2014; Naumann et al., 2019; Yoshitake et al., 2022). Also, experiments using an ER stress sensor mutant have shown that Pi starvation-mediated autophagy is regulated by ER stress response (Naumann et al., 2019). ER stress induces ER-phagy, a type of autophagy that degrades ER specificity (Liu et al., 2012). Recently, it has also been reported that ER stress is induced by oxidative lipid accumulation and that ER-phagy contributes to Pi recycling in leaf cells (Yoshitake et al., 2022). Furthermore, the timing of inducing the ER-phagy mediated Pi recycling system is earlier than that of PHOSPHATE STARVATION RESPONSE 1 transcription factor regulated Pi starvation responses, such as membrane lipid remodeling (Pant et al., 2015), suggesting that plants have two phases in their Pi starvation response, to effectively adapt to natural fluctuations in Pi starvation.
Another kind of autophagy, chlorophagy, which involves specific chloroplast degradation by autophagy, also contributes to Pi recycling. For example, under Pi starvation, excess supply of nitrate induces Rubisco-containing body (RCB)-mediated chlorophagy, in which the chloroplast is partially degraded. This chlorophagy is induced by a reduction in the carbon/nitrogen ratio and is conducive to Pi recycling (Yoshitake et al., 2021). Therefore, in addition to the Pi recycling system that is induced by sole Pi starvation, plants seem to have another one which is induced by complex nutrient status.
Ribophagy is the process of ribosome degradation by autophagy, and it has been discovered in plants as well as in yeast (Kraft et al., 2008). RNA-containing granules have been shown to be incorporated into vacuoles of Zea mays primary root meristem (Niki et al., 2014). Also, a study on A. thaliana reported that the number of autophagosomes containing ribosomes was increased by defective of RNS2 (Floyd et al., 2015). Therefore, autophagy may play a role in RNA and ribosome degradation, although it is unclear whether this contributes to Pi recycling.
Vacuolar transporter
The vacuole is a compartment for degradation of intracellular components and salvaging nutrients. Therefore, knowledge of the Pi transporters on the vacuolar membrane (tonoplast) is important for understanding the system of Pi homeostasis and Pi recycling. In yeast, the VACUOLAR TRANSPORTER CHAPERONE complex (Vtc1-4) transports Pi from the cytoplasm into the vacuole and generates polyphosphates, a linear chain of anywhere between three to thousands of Pi units (Secco et al., 2012b). Polyphosphate can be hydrolased to Pi by PHOSPHATE METABOLISM 5 (Phm5) in the vacuole and it is transported from the vacuole to the cytoplasm by the vacuolar Pi transporter Pho91 (Ogawa et al., 2000; Hürlimann et al., 2007). Vtc2-4 and Pho91 have SIG1/Pho81/XPR1 (SPX) domain, and it is known that plants have Pi transporters containing the SPX domain, SPX-MAJOR FACILITATOR SUPERFAMILY (SPX-MFS) family proteins (Secco et al., 2012a; Secco et al., 2012b). In A. thaliana, there are three SPX-MFS proteins, PHOSPHATE TRANSPORTER 5 (PHT5;1, PHT5;2, and PHT5;3) (Liu et al., 2016). The expression of PHT5;1/VPT1 is induced by high Pi conditions in order to maintain the Pi concentration in the cytoplasm (Liu et al., 2015). There are also three SPX-MFS (OsSPX-MFS1-3) proteins in Oryza sativa (Wang et al., 2015). OsSPX-MFS3 transports Pi from the vacuolar lumen to the cytoplasm, whereas OsSPX-MFS1 and 2 import Pi to the vacuole (Wang et al., 2015; Liu et al., 2016; Guo et al., 2022). Other VACUOLAR Pi EFFLUX TRANSPORTERs (VPEs) have been identified in O. sativa, and phylogenic analysis has revealed that these VPE proteins evolved from an ancient plasma membrane glycerol-3-phosphate transporter during terrestrial plant evolution (Xu et al., 2019). The expression of VPE genes is induced by Pi starvation when they control Pi homeostasis in plant cells (Ramaiah et al., 2011; Xu et al., 2019). Some transporter proteins on the tonoplast generate Pi. For example, it has been shown that the transcription and translation of ARABIDOPSIS VACUOLAR PYROPHOSPHATASE 1 (AVP1), type I H+-pyrophosphatase, are enhanced by Pi starvation (Yang et al., 2007), and that vacuolar pyrophosphatase generates two Pis from one pyrophosphate during transportation of H+ into the vacuole (Zhen et al., 1994; Maeshima, 2000). Furthermore, overexpression of AVP1 improved plant growth under Pi starvation in A. thaliana, S. lycopersicum, and O. sativa (Yang et al., 2007; Heuer et al., 2016), and AVP1 overexpressors have been shown to exhibit enhanced rhizosphere acidification under Pi starvation which enhanced Pi uptake (Yang et al., 2007). However, it is still unclear whether this pyrophosphatase activity relates to Pi recycling.
Conclusion
Pi recycling systems are the plant response mechanism to Pi starvation. Therefore, it is important to increase our understanding of this important process. So, we examine the catabolic pathways producing Pi from intracellular components under Pi starvation in this mini-review. We have shown that there are various plant responses to Pi starvation. For example, while Pi starvation induced the expression of ENDO1 in Petunia, this did not occur in A. thaliana grown on Pi-depleted media for 14 days (Pérez-Amador et al., 2000; Jones et al., 2021). Also, recent studies using A. thaliana showed that ER-phagy only contributes to Pi recycling in the early phase of Pi starvation (2-3 days) and that RCB-mediated chlorophagy salvages Pi under high nitrate/Pi starvation conditions (Yoshitake et al., 2021; Yoshitake et al., 2022). Hence, AtENDO1 may be involved in Pi recycling in early phase of Pi starvation or under certain conditions.
In yeast, the VTC complex is required for microautophagy, another type of autophagy in which the tonoplast invaginates and directly engulfs the target (Uttenweiler et al., 2007). But, while microautophagy is known to be induced by photodamage, sucrose starvation, and ammonium stress (Nakamura and Izumi, 2018; Goto-Yamada et al., 2019; Robert et al., 2021), it is still unclear whether Pi starvation induces microautophagy. Also, it is possible that other intracellular degradation systems could be involved in Pi recycling, in addition to microautophagy.
Therefore, in conclusion, further analysis is still needed to fully understand the whole Pi recycling system. This includes more research under various Pi starvation conditions, and studies to establish the extent to which other degradation systems are involved in Pi recycling.
Author contributions
YY and KY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Grant-in Aid for Scientific Research (B) [20H03281]; the Grant-in-Aid for Scientific Research on Innovative Areas [19H05713]; the Joint Usage/Research Center, Institute of Plant Science and Resources, Okayama University [R439].
Acknowledgments
We would like to thank Iain McTaggart for critical reading and English proofreading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abel, S., Nürnberger, T., Ahnert, V., Krauss, G. J., Glund, K. (2000). Induction of an extracellular cyclic nucleotide phosphodiesterase as an accessory ribonucleolytic activity during phosphate starvation of cultured tomato cells. Plant Physiol. 122, 543–552. doi: 10.1104/pp.122.2.543
Angkawijaya, A. E., Nakamura, Y. (2017). Arabidopsis PECP1 and PS2 are phosphate starvation-inducible phosphocholine phosphatases. Biochem. Bioph. Res. Co. 494, 397–401. doi: 10.1016/j.bbrc.2017.09.094
Angkawijaya, A. E., Ngo, A. H., Nguyen, V. C., Gunawan, F., Nakamura, Y. (2019). Expression profiles of 2 phosphate starvation-inducible phosphocholine/phosphoethanolamine phosphatases, PECP1 and PS2, in Arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00662
Aslam, M. M., Pueyo, J. J., Pang, J., Yang, J., Chen, W., Chen, H., et al. (2022). Root acid phosphatases and rhizobacteria synergistically enhance white lupin and rice phosphorus acquisition. Plant Physiol. 190, 2449–2465. doi: 10.1093/plphys/kiac418
Awai, K., Maréchal, E., Block, M. A., Brun, D., Masuda, T., Shimada, H., et al. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. P. Natl. Acad. Sci. U. S. A. 98, 10960–10965. doi: 10.1073/pnas.181331498
Bariola, P. A., Howard, C. J., Taylor, C. B., Verburg, M. T., Jaglan, V. D., Green, P. J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6, 673–685. doi: 10.1046/j.1365-313X.1994.6050673.x
Bariola, P. A., MacIntosh, G. C., Green, P. J. (1999). Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol. 119, 331–342. doi: 10.1104/pp.119.1.331
Bozzo, G. G., Dunn, E. L., Plaxton, W. C. (2006). Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ. 29, 303–313. doi: 10.1111/j.1365-3040.2005.01422.x
Bozzo, G. G., Raghothama, K. G., Plaxton, W. C. (2002). Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur. J. Biochem. 269, 6278–6286. doi: 10.1046/j.1432-1033.2002.03347.x
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Pérez-Pérez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet. 6, e1001102. doi: 10.1371/journal.pgen.1001102
Chen, D. L., Delatorre, C. A., Bakker, A., Abel, S. (2000). Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211, 13–22. doi: 10.1007/s004250000271
del Pozo, J. C., Allona, I., Rubio, V., Leyva, A., de la Pena, A., Aragoncillo, C., et al. (1999). A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 19, 579–589. doi: 10.1046/j.1365-313X.1999.00562.x
Deshpande, R. A., Shankar, V. (2002). Ribonucleases from T2 family. Crit. Rev. Microbiol. 28, 79–122. doi: 10.1080/1040-840291046704
Dodds, P. N., Clarke, A. E., Newbigin, E. (1996). Molecular characterisation of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol. Biol. 31, 227–238. doi: 10.1007/BF00021786
Dörmann, P., Benning, C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7, 112–118. doi: 10.1016/S1360-1385(01)02216-6
Eastmond, P. J., Quettier, A. L., Kroon, J. T. M., Craddock, C., Adams, N., Slabas, A. R. (2010). Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22, 2796–2811. doi: 10.1105/tpc.109.071423
Essigmann, B., Güler, S., Narang, R. A., Linke, D., Benning, C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. P. Natl. Acad. Sci. U. S. A. 95, 1950–1955. doi: 10.1073/pnas.95.4.1950
Floyd, B. E., Morriss, S. C., MacIntosh, G. C., Bassham, D. C. (2015). Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11, 2199–2212. doi: 10.1080/15548627.2015.1106664
Floyd, B. E., Mugume, Y., Morriss, S. C., MacIntosh, G. C., Bassham, D. C. (2017). Localization of RNS2 ribonuclease to the vacuole is required for its role in cellular homeostasis. Planta 245, 779–792. doi: 10.1007/s00425-016-2644-x
Gaude, N., Nakamura, Y., Scheible, W. R., Ohta, H., Dörmann, P. (2008). Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56, 28–39. doi: 10.1111/j.1365-313X.2008.03582.x
Gaugler, P., Schneider, R., Liu, G., Qiu, D., Weber, J., Schmid, J., et al. (2022). Arabidopsis PFA-DSP-Type phosphohydrolases target specific inositol pyrophosphate messengers. Biochemistry 61, 1213–1227. doi: 10.1021/acs.biochem.2c00145
Goto-Yamada, S., Oikawa, K., Bizan, J., Shigenobu, S., Yamaguchi, K., Mano, S., et al. (2019). Sucrose starvation induces microautophagy in plant root cells. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01604
Guo, R., Zhang, Q., Ying, Y., Liao, W., Liu, Y., Whelan, J., et al. (2022). Functional characterization of the three Oryza sativa SPX-MFS proteins in maintaining phosphate homoeostasis. Plant Cell Environ. doi: 10.1111/pce.14414
Hanchi, M., Thibaud, M. C., Légeret, B., Kuwata, K., Pochon, N., Beisson, F., et al. (2018). The phosphate fast-responsive genes PECP1 and PPsPase1 affect phosphocholine and phosphoethanolamine content. Plant Physiol. 176, 2943–2962. doi: 10.1104/pp.17.01246
Harayama, T., Riezman, H. (2018). Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Bio. 19, 281–296. doi: 10.1038/nrm.2017.138
Härtel, H., Benning, C. (2000). Can digalactosyldiacylglycerol substitute for phosphatidylcholine upon phosphate deprivation in leaves and roots of Arabidopsis? Biochem. Soc T. 28, 729–732. doi: 10.1042/bst0280729
Heuer, S., Gaxiola, R., Schilling, R., Herrera-Estrella, L., López-Arredondo, D., Matthias, W., et al. (2016). Improving phosphorus use efficiency: a complex trait with emerging opportunities. Plant J. 90, 868–885. doi: 10.1111/tpj.13423
Hillwig, M. S., Contento, A. L., Meyer, A., Ebany, D., Bassham, D. C., MacIntosh, G. C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. P. Natl. Acad. Sci. U. S. A. 108, 1093–1098. doi: 10.1073/pnas.1009809108
Hillwig, M. S., LeBrasseur, N. D., Green, P. J., MacIntosh, G. C. (2008). Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Mol. Genet. Genomics 280, 249–261. doi: 10.1007/s00438-008-0360-3
Hsueh, Y. C., Ehmann, C., Flinner, N., Ladig, R., Schleiff, E. (2017). The plastid outer membrane localized LPTD1 is important for glycerolipid remodelling under phosphate starvation. Plant Cell Environ. 40, 1643–1657. doi: 10.1111/pce.12973
Hurley, B. A., Tran, H. T., Marty, N. J., Park, J., Snedden, W. A., Mullen, R. T., et al. (2010). The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 153, 1112–1122. doi: 10.1104/pp.110.153270
Hürlimann, H. C., Stadler-Waibel, M., Werner, T. P., Freimoser, F. M. (2007). Pho91 is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell. 18, 4201–4689. doi: 10.1091/mbc.e07-05-0457
Igic, B., Kohn, J. R. (2001). Evolutionary relationships among self-incompatibility RNases. P. Natl. Acad. Sci. U. S. A. 98, 13167–13171. doi: 10.1073/pnas.231386798
Izumi, M., Wada, S., Makino, A., Ishida, H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154, 1196–1209. doi: 10.1104/pp.110.158519
Jones, M., Bai, S., Lin, Y., Chapin, L. (2021). The senescence-associated endonuclease, PhENDO1, is upregulated by ethylene and phosphorus deficiency in Petunia. Horticulturae 7, 46. doi: 10.3390/horticulturae7030046
Jouhet, J., Maréchal, E., Baldan, B., Bligny, R., Joyard, J., Block, M. A. (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167, 863–874. doi: 10.1083/jcb.200407022
Kelly, A. A., Froehlich, J. E., Dörmann, P. (2003). Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15, 2694–2706. doi: 10.1105/tpc.016675
Kobayashi, K., Awai, K., Takamiya, K., Ohta, H. (2004). Arabidopsis type B monogalactosyldiacylglycerol synthase genes are expressed during pollen tube growth and induced by phosphate starvation. Plant Physiol. 134, 640–648. doi: 10.1104/pp.103.032656
Kobayashi, K., Kondo, M., Fukuda, H., Nishimura, M., Ohta, H. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. P. Natl. Acad. Sci. U. S. A. 104, 17216–17221. doi: 10.1073/pnas.0704680104
Köck, M., Löffler, A., Abel, S., Glund, K. (1995). cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol. Biol. 27, 477–485. doi: 10.1007/BF00019315
Kraft, C., Deplazes, A., Sohrmann, M., Peter, M. (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610. doi: 10.1038/ncb1723
Kuang, R., Chan, K. H., Yeung, E., Lim, B. L. (2009). Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 151, 199–209. doi: 10.1104/pp.109.143180
Lehmann, K., Hause, B., Altmann, D., Köck, M. (2001). Tomato ribonuclease LX with the functional endoplasmic reticulum retention motif HDEF is expressed during programmed cell death processes, including xylem differentiation, germination, and senescence. Plant Physiol. 127, 436–449. doi: 10.1104/pp.010362
Liang, L., Lai, Z., Ma, W., Zhang, Y., Xue, Y. (2002). AhSL28, a senescence- and phosphate starvation-induced S-like RNase gene in Antirrhinum. BBA-Gene Struct. Expr. 1579, 64–71. doi: 10.1016/S0167-4781(02)00507-9
Li, M., Qin, C., Welti, R., Wang, X. (2006a). Double knockouts of phospholipases D ζ 1 and D ζ 2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140, 761–770. doi: 10.1104/pp.105.070995
Li, M., Tadano, T. (1996). Comparison of characteristics of acid phosphatases secreted from roots of lupin and tomato. Soil Sci. Plant Nutr. 42, 753–763. doi: 10.1080/00380768.1996.10416623
Liu, Y., Burgos, J. S., Deng, Y., Srivastava, R., Howell, S. H., Bassham, D. C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 24, 4635–4651. doi: 10.1105/tpc.112.101535
Liu, T. Y., Huang, T. K., Yang, S. Y., Hong, Y. T., Huang, S. M., Wang, F. N., et al. (2016). Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 7, 11095. doi: 10.1038/ncomms11095
Liu, J., Yang, L., Luan, M., Wang, Y., Zhang, C., Zhang, B., et al. (2015). A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. P. Natl. Acad. Sci. U. S. A. 112, E6571–E6578. doi: 10.1073/pnas.1514598112
Li, M., Welti, R., Wang, X. (2006b). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761. doi: 10.1104/pp.106.085647
Li, D., Zhu, H., Liu, K., Liu, X., Leggewie, G., Udvardi, M., et al. (2002). Purple acid phosphatases of Arabidopsis thaliana: Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 277, 27772–27781. doi: 10.1074/jbc.M204183200
Löffler, A., Glund, K., Irie, M. (1993). Amino acid sequence of an intracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur. J. Biochem. 214, 627–633. doi: 10.1111/j.1432-1033.1993.tb17962.x
Lung, S. C., Leung, A., Kuang, R., Wang, Y., Leung, P., Lim, B.-L. (2008). Phytase activity in tobacco (Nicotiana tabacum) root exudates is exhibited by a purple acid phosphatase. Phytochem. 69, 365–373. doi: 10.1016/j.phytochem.2007.06.036
Lynch, J. P. (2011). Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156, 1041–1049. doi: 10.1104/pp.111.175414
MacIntosh, G. C., Hillwig, M. S., Meyer, A., Flagel, L. (2010). RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol. Genet. Genomics 283, 381–396. doi: 10.1007/s00438-010-0524-9
Maeshima, M. (2000). Vacuolar H+-pyrophosphatase. BBA-Biomembranes. 1465, 37–51. doi: 10.1016/S0005-2736(00)00130-9
Megel, C., Hummel, G., Lalande, S., Ubrig, E., Cognat, V., Morelle, G., et al. (2019). Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 47, 941–952. doi: 10.1093/nar/gky1156
Miller, S. S., Liu, J., Allan, D. L., Menzhuber, C. J., Fedorova, M., Vance, C. P. (2001). Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol. 127, 594–606. doi: 10.1104/pp.010097
Nakamura, Y. (2013). Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 52, 43–50. doi: 10.1016/j.plipres.2012.07.002
Nakamura, Y., Awai, K., Masuda, T., Yoshioka, Y., Takamiya, K., Ohta, H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280, 7469–7476. doi: 10.1074/jbc.M408799200
Nakamura, S., Izumi, M. (2018). Regulation of chlorophagy during photoinhibition and senescence: lessons from mitophagy. Plant Cell Physiol. 59, 1135–1143. doi: 10.1093/pcp/pcy096
Nakamura, Y., Koizumi, R., Shui, G., Shimojima, M., Wenk, M. R., Ito, T., et al. (2009). Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. P. Natl. Acad. Sci. U. S. A. 106, 20978–20983. doi: 10.1073/pnas.0907173106
Naumann, C., Müller, J., Sakhonwasee, S., Wieghaus, A., Hause, G., Heisters, M., et al. (2019). The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol. 179, 460–476. doi: 10.1104/pp.18.01379
Niki, T., Saito, S., Gladish, D. K. (2014). Granular bodies in root primary meristem cells of Zea mays L. var. Cuscoensis K. (Poaceae) that enter young vacuoles by invagination: A novel ribophagy mechanism. Protoplasma. 251, 1141–1149. doi: 10.1007/s00709-014-0622-3
Nürnberger, T., Abel, S., Jost, W., Glund, K. (1990). Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 92, 970–976. doi: 10.1104/pp.92.4.970
O’Gallagher, B., Ghahremani, M., Stigter, K., Walker, E. J. L., Pyc, M., Liu, A. Y., et al. (2022). Arabidopsis PAP17 is a dual-localized purple acid phosphatase up-regulated during phosphate deprivation, senescence, and oxidative stress. J. Exp. Bot. 73, 382–399. doi: 10.1093/jxb/erab409
Ogawa, N., DeRisi, J., Brown, P. O. (2000). New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell. 11, 4051–4411. doi: 10.1091/mbc.11.12.4309
Okabe, T., Iwakiri, Y., Mori, H., Ogawa, T., Ohyama, T. (2005). An S-like ribonuclease gene is used to generate a trap-leaf enzyme in the carnivorous plant Drosera adelae. FEBS Lett. 579, 5729–5733. doi: 10.1016/j.febslet.2005.09.043
Okazaki, Y., Otsuki, H., Narisawa, T., Kobayashi, M., Sawai, S., Kamide, Y., et al. (2013). A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 4, 1510. doi: 10.1038/ncomms2512
Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., Jr., et al. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 12, 1849–1862. doi: 10.1105/tpc.12.10.1849
Ozawa, K., Osaki, M., Matsui, H., Honma, M., Tadano, T. (1995). Purification and properties of acid phosphatase secreted from lupin roots under phosphorus-deficiency conditions. Soil Sci. Plant Nutr. 41, 461–469. doi: 10.1080/00380768.1995.10419608
Pant, B. D., Burgos, A., Pant, P., Cuadros-Inostroza, A., Willmitzer, L., Scheible, W. R. (2015). The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 66, 1908–1918. doi: 10.1093/jxb/eru535
Pérez-Amador, M. A., Abler, M. L., De Rocher, E. J., Thompson, D. M., van Hoof, A., LeBrasseur, N. D., et al. (2000). Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 122, 169–180. doi: 10.1104/pp.122.1.169
Raghothama, K. G. (1999). Phosphate acquisition. Annu. Rev. Plant Phys. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Ramaiah, M., Jain, A., Baldwin, J. C., Karthikeyan, A. S., Raghothama, K. G. (2011). Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol. 157, 279–291. doi: 10.1104/pp.111.178541
Robert, G., Yagyu, M., Koizumi, T., Naya, L., Masclaux-Daubresse, C., Yoshimoto, K. (2021). Ammonium stress increases microautophagic activity while impairing macroautophagic flux in Arabidopsis roots. Plant J. 105, 1083–1097. doi: 10.1111/tpj.15091
Sanda, S., Leustek, T., Theisen, M. J., Garavito, R. M., Benning, C. (2001). Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 276, 3941–3946. doi: 10.1074/jbc.M008200200
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012a). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Secco, D., Wang, C., Shou, H., Whelan, J. (2012b). Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586, 289–295. doi: 10.1016/j.febslet.2012.01.036
Shinozaki, D., Merkulova, E. A., Naya, L., Horie, T., Kanno, Y., Seo, M., et al. (2020). Autophagy increases zinc bioavailability to avoid light-mediated reactive oxygen species production under zinc deficiency. Plant Physiol. 182, 1284–1296. doi: 10.1104/pp.19.01522
Smith, A. P., Fontenot, E. B., Zahraeifard, S., DiTusa, S. F. (2018). Molecular components that drive phosphorus-remobilisation during leaf senescence. Annu. Plant Rev. 48, 159–186. doi: 10.1002/9781119312994.apr0521
Srivastava, R., Akash, Parida, A. P., Chauhan, P. K., Kumar, R. (2020). Identification, structure analysis, and transcript profiling of purple acid phosphatases under Pi deficiency in tomato (Solanum lycopersicum L.) and its wild relatives. Int. J. Biol. Macromol. 165, 2253–2266. doi: 10.1016/j.ijbiomac.2020.10.080
Stephenson, P., Hogan, J. (2006). Cloning and characterization of a ribonuclease, a cysteine proteinase, and an aspartic proteinase from pitchers of the carnivorous plant Nepenthes ventricosa Blanco. Int. J. Plant Sci. 167, 239–248. doi: 10.1086/499284
Su, Y., Li, M., Guo, L., Wang, X. (2018). Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J. 94, 315–326. doi: 10.1111/tpj.13858
Takami, T., Ohnishi, N., Kurita, Y., Iwamura, S., Ohnishi, M., Kusaba, M., et al. (2018). Organelle DNA degradation contributes to the efficient use of phosphate in seed plants. Nat. Plants 4, 1044–1055. doi: 10.1038/s41477-018-0291-x
Tasaki, M., Asatsuma, S., Matsuoka, K. (2014). Monitoring protein turnover during phosphate starvation-dependent autophagic degradation using a photoconvertible fluorescent protein aggregate in tobacco BY-2 cells. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00172
Taylor, C. B., Bariola, P. A., delCardayré, S. B., Raines, R. T., Green, P. J. (1993). RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. P. Natl. Acad. Sci. U. S. A. 90, 5118–5122. doi: 10.1073/pnas.90.11.5118
Tran, H. T., Qian, W., Hurley, B. A., She, Y. M., Wang, D., Plaxton, W. C. (2010). Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 33, 1789–1803. doi: 10.1111/j.1365-3040.2010.02184.x
Tyburski, J., Dunajska, K., Tretyn, A. (2009). Reactive oxygen species localization in roots of Arabidopsis thaliana seedlings grown under phosphate deficiency. Plant Growth Regul. 59, 27–36. doi: 10.1007/s10725-009-9385-9
Uttenweiler, A., Schwarz, H., Neumann, H., Mayer, A. (2007). The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell. 18, 166–175. doi: 10.1091/mbc.E06-08-0664
Van Camp, W., Van Montagu, M., Inzé, D. (1998). H2O2 and NO: Redox signals in disease resistance. Trends Plant Sci. 3, 330–334. doi: 10.1016/S1360-1385(98)01297-7
Veljanovski, V., Vanderbeld, B., Knowles, V. L., Snedden, W. A., Plaxton, W. C. (2006). Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 142, 1282–1293. doi: 10.1104/pp.106.087171
Veneklaas, E. J., Lambers, H., Bragg, J., Finnegan, P. M., Lovelock, C. E., Plaxton, W. C., et al. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195, 306–320. doi: 10.1111/j.1469-8137.2012.04190.x
Wang, K., Froehlich, J. E., Zienkiewicz, A., Hersh, H. L., Benning, C. (2017). A plastid phosphatidylglycerol lipase contributes to the export of acyl groups from plastids for seed oil biosynthesis. Plant Cell. 29, 1678–1696. doi: 10.1105/tpc.17.00397
Wang, K., Guo, Q., Froehlich, J. E., Hersh, H. L., Zienkiewicz, A., Howe, G. A., et al. (2018). Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell. 30, 1006–1022. doi: 10.1105/tpc.18.00250
Wang, L., Li, Z., Qian, W., Guo, W., Guo, X., Huang, L., et al. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 157, 1283–1299. doi: 10.1104/pp.111.183723
Wang, L., Lu, S., Zhang, Y., Li, Z., Du, X., Liu, D. (2014). Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 56, 299–314. doi: 10.1111/jipb.12184
Wang, C., Yue, W., Ying, Y., Wang, S., Secco, D., Liu, Y., et al. (2015). OsSPX-MFS3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 169, 2822–2831. doi: 10.1104/pp.15.01005
Xu, L., Zhao, H., Wan, R., Liu, Y., Xu, Z., Tian, W., et al. (2019). Identification of vacuolar phosphate efflux transporters in land plants. Nat. Plants. 5, 84–94. doi: 10.1038/s41477-018-0334-3
Yang, H., Knapp, J., Koirala, P., Rajagopal, D., Peer, W. A., Silbart, L. K., et al. (2007). Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol. J. 5, 735–745. doi: 10.1111/j.1467-7652.2007.00281.x
Yang, B., Li, M., Phillips, A., Li, L., Ali, U., Li, Q., et al. (2021). Nonspecific phospholipase C4 hydrolyzes phosphosphingolipids and sustains plant root growth during phosphate deficiency. Plant Cell. 33, 766–780. doi: 10.1093/plcell/koaa054
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshitake, Y., Nakamura, S., Shinozaki, D., Izumi, M., Yoshimoto, K., Ohta, H., et al. (2021). RCB-mediated chlorophagy caused by oversupply of nitrogen suppresses phosphate-starvation stress in plants. Plant Physiol. 185, 318–330. doi: 10.1093/plphys/kiaa030
Yoshitake, Y., Shinozaki, D., Yoshimoto, K. (2022). Autophagy triggered by iron-mediated ER stress is an important stress response to the early phase of Pi starvation in plants. Plant J 110, 1370–1381. doi: 10.1111/tpj.15743
Yu, B., Xu, C., Benning, C. (2002). Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. P. Natl. Acad. Sci. U. S. A. 99, 5732–5737. doi: 10.1073/pnas.082696499
Zand Karimi, H., Baldrich, P., Rutter, B. D., Borniego, L., Zajt, K. K., Meyers, B. C., et al. (2022). Arabidopsis apoplastic fluid contains sRNA- and circular RNA–protein complexes that are located outside extracellular vesicles. Plant Cell. 34, 1863–1881. doi: 10.1093/plcell/koac043
Zhang, W., Yu, L., Zhang, Y., Wang, X. (2005). Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. BBA – Mol. Cell Biol. L. 1736, 1–9. doi: 10.1016/j.bbalip.2005.07.004
Keywords: phosphate recycling, phosphatase, nuclease, membrane lipid remodeling, autophagy, vacuolar transporter
Citation: Yoshitake Y and Yoshimoto K (2023) Intracellular phosphate recycling systems for survival during phosphate starvation in plants. Front. Plant Sci. 13:1088211. doi: 10.3389/fpls.2022.1088211
Received: 03 November 2022; Accepted: 23 December 2022;
Published: 17 January 2023.
Edited by:
Rahul Kumar, University of Hyderabad, IndiaReviewed by:
Ahmed H. El-Sappah, Zagazig University, EgyptAdwaita Prasad Parida, University of Delhi, India
Copyright © 2023 Yoshitake and Yoshimoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kohki Yoshimoto, a29oa2lfeW9zaGltb3RvQG1laWppLmFjLmpw
 Yushi Yoshitake
Yushi Yoshitake Kohki Yoshimoto
Kohki Yoshimoto