- 1College of Agronomy, Qingdao Agricultural University, Qingdao, China
- 2State Agricultural Biotechnology Centre, College of Science, Health, Engineering and Education, Murdoch University, Perth, WA, Australia
- 3The Key Laboratory of the Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao, Shandong, China
Introduction: Protein kinases play an important role in plants in response to environmental changes through signal transduction. As a large family of protein kinases, sucrose non-fermenting-1 (SNF1)-related kinases (SnRKs) were found and functionally verified in many plants. Nevertheless, little is known about the SnRK family of Zea mays.
Methods: Evolutionary relationships, chromosome locations, gene structures, conserved motifs, and cis-elements in promoter regions were systematically analyzed. Besides, tissue-specific and stress-induced expression patterns of ZmSnRKs were determined. Finally, functional regulatory networks between ZmSnRKs and other proteins or miRNAs were constructed.
Results and Discussion: In total, 60 SnRK genes located on 10 chromosomes were discovered in maize. ZmSnRKs were classified into three subfamilies (ZmSnRK1, ZmSnRK2, and ZmSnRK3), consisting of 4, 14, and 42 genes, respectively. Gene structure analysis showed that 33 of the 42 ZmSnRK3 genes contained only one exon. Most ZmSnRK genes contained at least one ABRE, MBS, and LTR cis-element and a few ZmSnRK genes had AuxRR-core, P-box, MBSI, and SARE ciselements in their promoter regions. The Ka:Ks ratio of 22 paralogous ZmSnRK gene pairs revealed that the ZmSnRK gene family had experienced a purifying selection. Meanwhile, we analyzed the expression profiles of ZmSnRKs, and they exhibited significant differences in various tissues and abiotic stresses. In addition, A total of eight ZmPP2Cs, which can interact with ZmSnRK proteins, and 46 miRNAs, which can target 24 ZmSnRKs, were identified. Generally, these results provide valuable information for further function verification of ZmSnRKs, and improve our understanding of the role of ZmSnRKs in the climate resilience of maize.
Introduction
Terrestrial plants must cope with environmental changes including drought; excess salt, heat, cold, and toxic metals; and nutrient deficiency (Zhu, 2016). Protein phosphatase and kinase-regulated signal transduction play a vital role in climate resilience in plants (Hunter, 1995; Cohen, 1988). In recent years, several protein kinases have been identified, including receptor protein kinase (RLK) (Tör et al., 2009), mitogen-activated protein kinase (MAPK) (Zhang and Zhang, 2022), calcium-dependent protein kinase (CDPK) (Bender et al., 2018), and sucrose non-fermenting-1 (SNF1)-related kinase (SnRK) (Maszkowska et al., 2021).
SnRKs in plants have a high sequence identity with SNF1 of fungi and AMP-activated protein kinase (AMPK) of mammals (Halford and Hardie, 1998). The first plant SnRK genes were identified in Secale cereale, and showed 48% amino acid homology with SNF1 and AMPK (Alderson et al., 1991). In plants, the SnRK family has expanded and diverged into three subfamilies (SnRK1, SnRK2, and SnRK3) (Hrabak et al., 2003). The N-terminal of three SnRK subfamilies have a conserved domain with serine/threonine protein kinase, while the C-terminal displayed variation. In detail, SnRK1s were most similar to SNF1 and AMPK in terms of structure and function. SnRK2s contain acidic amino acids in the C-terminal domain, either Glu or Asp (Kulik et al., 2011). SnRK3s were widely recognized as CIPKs (CBL-interacting protein kinases) (Tominaga et al., 2010), which have two conserved domains in the C-terminal: NAF and PPI (Masaru et al., 2003; Albrecht et al., 2014).
SnRK1s are evolutionarily conserved energy-sensing protein kinases in plants. When energy supplies become limited, SnRK1s can activate transcriptional regulatory networks and maintain cellular energy homeostasis (Wurzinger et al., 2018; Wang et al., 2021). Therefore, SnRK1s were known as conserved energy sensors. For example, Wang et al. (2021) reported that the α-subunit of OsSnRK1 could phosphorylate JMJ705 to stimulate its demethylase activity, thus targeting and activating a series of low-energy stress-responsive transcription factors. SnRK1 phosphorylates the transcription factor basic leucine zipper63 (bZIP63), which could directly bind the promoter of auxin response factor19 (ARF19) and activate gene expression to mediate lateral root development (Muralidhara et al., 2021). In peach fruit, SnRK1 could phosphorylate sorbitol dehydrogenase and lead to sugar accumulation (Yu et al., 2021). In maize, SnRK1 could coordinate carbon and nitrogen absorption in seeds by phosphorylating E3 ubiquitin ligase ZmRFWD3 (Li et al., 2020).
SnRK2s are much smaller than SnRK1 and are associated with the ABA signal transduction pathway (Cutler et al., 2010; Maszkowska et al., 2021). In rice, a mutation of OsSAPK8, a member of SnRK2, exhibited sensitivity to diverse abiotic stresses at the vegetative stages, such as drought stresses, excess of salt, and low temperature (Zhong et al., 2020). In Arabidopsis, overexpression of AtSnRK2.8 showed higher resistance to drought. AtSnRK2.10 was induced by salt stress and was involved in salt tolerance through increasing photosynthesis and protecting against oxidative damage (Mazur et al., 2021). The activity of SnRK2.4 was significantly increased in roots after 30s of salt exposure, but it disappeared 1h later (Mcloughlin et al., 2012). In rice, SnRK2.6/OST1 was immediately activated in an ABA non-responsive pathway to elevate low temperature adaptation (Ding et al., 2015).
SnRK3s could combine with Ca2+-dependent CBL to regulate ion transport and enhance environmental adaption in plants, especially in abiotic stress. For instance, AtCIPK24 protein could interact with AtCBL4 to phosphorylate and activate plasma membrane H+/Na+ antiporter (SOS1) in a Ca2+-responsive manner in Arabidopsis (Qiu et al., 2002). In rice, the expression of OsCIPK31 was up-regulated under several abiotic stresses during germination and seedling growth (Piao et al., 2010). Ma et al. (2021) indicated that CaCIPK3 positively regulated drought tolerance by the CBL-CIPK network, thus mediating MeJA signaling and the antioxidant defense system. Moreover, the OsCBL1-OsCIPK23 complex could enhance Os-AKT1-mediated K+ uptake (Li et al., 2014).
Maize is the most commonly cultivated food crop globally. However, research concerning the role of SnRK genes is still rare in maize. With the rapid progress of molecular biology and the whole-genome sequencing of maize (Myburg et al., 2014), it becomes possible for us to have a comprehensive understanding of ZmSnRK gene families. Here, 60 candidate ZmSnRK genes were discovered by bioinformatic analysis. Then, evolutionary relationships, chromosome locations, gene structures, conserved motifs, and cis-elements in promoter regions were systematically analyzed. Besides, tissue-specific and stress-induced expression patterns of ZmSnRKs were determined. Finally, functional regulatory networks between ZmSnRKs and other proteins or miRNAs were constructed. All these results offer valuable information for further function verification of ZmSnRKs and help to better comprehend the role of ZmSnRKs in the climate resilience of maize.
Materials and methods
Identification and general characterization analysis of ZmSnRK proteins
ZmSnRK proteins were searched using query probes of AtSnRKs and OsSnRKs by BLASTP. We used Zm-B73 V5.0 database from the MaizeGDB (https://maizegdb.org/) to conduct BLASP. SnRKs in Arabidopsis and rice were extracted from ensemble plants (https://plants.ensembl.org/). Meanwhile, the HMMER 3.0 was utilized to search the ZmSnRKs protein sequence according to the Hidden Markov Model (HMM) files of SnRKs acquired from the Pfam database (http://pfam.xfam.org/). The SMART database was used to reconfirm sequences (Letunic et al., 2012). The molecular weights and isoelectric points of the proteins were analyzed through the ExPASy tool (www.expasy.org/tools/). Subcellular localization was also predicted through WoLF PSORT (https://wolfpsort.hgc.jp/).
Phylogenetic analysis and classification of ZmSnRKs
Multiple homology alignment of 60 ZmSnRK non-redundant amino acid sequences and SnRK protein sequences from Arabidopsis thaliana, Oryza sativa, and Zea mays were conducted by ClustalW with default parameters (Larkin et al., 2007). Then, we used MEGA 7.0 to construct a phylogenetic tree via neighbor-joining (NJ) method (Kumar et al., 2016), with the following parameters: Poisson model, pairwise deletion, and 1,000 bootstrap replications.
Chromosome localization and collinearity analysis of ZmSnRKs
Chromosome localization of ZmSnRKs was conducted using MapChart v. 2.3 Software (Voorrips, 2002). The collinearity of the orthologous SnRK genes between Zea mays and other species, such as Arabidopsis thaliana, Oryza sativa, and Hordeum vulgare, was determined through MCScanX software (Wang et al., 2012). The non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) of ZmSnRKs in the gene collinearity were calculated by using the ParaAT tool (Zhang et al., 2012). The Ka/Ks values were calculated by Calculator 2.0 software (Wang et al., 2010). The gene duplicated time was assessed based on Ks/2λ (Wang et al., 2010), where λ = 1.5 × 10−8.
Analysis of gene structure and motifs of ZmSnRKs
The exon and intron arrangement of ZmSnRK genes was determined via the Gene Structure Display Server (GSDS) tool (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The conserved motif sequences of ZmSnRK proteins were identified using Multiple Expectation Maximization for Motif Elicitation (MEME) online program (http://meme.sdsc.edu/meme/itro.html) (Bailey et al., 2009). InterProScan (www.ebi.ac.uk/Tools/pfa/iprscan/) was utilized to annotate the motifs (Jones et al., 2014). The gene structures and conserved motifs of ZmSnRKs were visualized through TBtools (Chen et al., 2020).
Predication of cis-acting elements in the promoter regions of ZmSnRKs
Promoter sequences (-1500 bp) of ZmSnRK genes were obtained from the maize B73 genome. The cis-elements were determined by PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Magali et al., 2002). We then use TBtools to visualize the cis-elements (Chen et al., 2020) associated with abiotic stress, including abscisic acid response components ABRE, drought response components MBS, auxin response components AuxRR-core, flavonoid biosynthetic genes regulation components MBSI, gibberellin-response components P-box, salicylic acid response components SARE, and low-temperature response compositions LTR.
Modeling of 3D structures of ZmSnRKs
The 3D structures of ZmSnRK proteins were predicted using Swiss-Model (https://swissmodel.expasy.org/interactive/) (Arnold et al., 2006). Then, the quality of 3D protein structure was assessed via SAVES server (http://nihserver.mbi.ucla.edu/SAVES/).
Expression profiles and interaction networks analysis of ZmSnRKs
The specific expression patterns of SnRKs from various tissues and under abiotic stress in the maize B73 were obtained from qTeller in MaizeGDB (https://qteller.maizegdb.org/) (Opitz et al., 2014; Stelpflug et al., 2016). We used transcript per million (TPM) reads mapped to represent the gene expression values. The genes with |log2 ratio|≥1 were recognized as differentially expressed genes (DEGs). A heatmap was conducted by TBtool to exhibit expression levels in various tissues (log2 (TPM+1))and different abiotic stresses (log2 fold change). Moreover, the protein interactions were predicted by the Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/cgi) (Damian et al., 2021). We used Cytoscape software to create interaction networks (Shannon et al., 2003). A low confidence (0.15) was used as the “minimum required interaction score” when analyzing protein interaction within ZmSnRKs, and high confidence was used as “maximum required interaction score” when analyzing protein interaction between ZmSnRKs and ZmPP2Cs.
Prediction of microRNAs-ZmSnRKs regulatory networks
The microRNA (miRNA)-ZmSnRK regulatory networks were predicted on the basis of the previous methods (Su et al., 2021). In brief, the ZmSnRK-targeting miRNAs were predicted with coding sequences of ZmSnRKs through the psRNATarget server (http://plantgrn.noble.org/psRNATarget/home) under default parameters except for that penalty for G:U pair was 1 and mismatches allowed in seed region was 0. The miRNA-targeted sites were those highly complementary to coding sequences of ZmSnRKs. The interaction networks were created using the Cytoscape V3.9.1 software (https://cytoscape.org/download.html).
Plant growth and treatment
The hydroponic experiment was conducted in a greenhouse at Qingdao Agricultural University, Qingdao, China. Maize inbred line B73 was used in this study. Seeds of maize were treated with 3% H2O2 for 10 min, and were rinsed seven times with distilled water. The seeds were sown in a controlled environment with a day/night temperature of 28/22°C on moist filter papers. After germination, uniform seedlings were transferred to 2 L pots containing 1.5 L of basic nutrient solution (BNS). On the seventh day after transplanting, PEG was added to the containers to form three treatments: BNS (control), BNS plus 10% PEG, and BNS plus 100 mM NaCl. The composition of BNS was (mg L−1) MgSO4·7H2O, 160.21; K2SO4, 130.7; KCl, 7.455; KH2PO4, 34.02; Ca(NO3)2·4H2O, 472.3; Fe(III)-EDTA-Na, 36.7; MnSO4·H2O, 0.17; ZnSO4·7H2O, 0.29; CuSO4·5H2O, 0.025; (NH4)6Mo7O24·4H2O, 0.006; and HBO3, 0.062. The solution pH was adjusted to 5.8–6.0 with NaOH or HCl, as required. Plant leaves and stems were sampled on the seventh day after transplanting. Plant root samples for RNA isolation were collected 24h after treatment. All samples were stored at −80°C for downstream analysis.
Quantitative RT−PCR validation
Quantitative real-time PCR (qRT-PCR) was performed by a QuantStudio3 PCR system (Thermo, USA). First-strand cDNA synthesis was performed using the TOROBlue® qRT Premix with gDNA Eraser 2.0 (TOROIVD, China), followed by qRT-PCR using a TOROGreen® qPCR Master Mix (TOROIVD, China) with ZmActin as a reference. The total PCR volume was 20 μl. The reaction process was 94°C denaturation for 1 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s. Experiments were replicated three times with 2-ΔΔCt relative quantification method. Primers were listed in Table S14.
Statistical analysis
Statistical analyses were performed with a Processing System statistical software package using ANOVA followed by Duncan’s multiple range test.
Results
Identification and evolution of SnRKs in maize
In total, we identified 60 ZmSnRK proteins in the maize inbred line B73 genome. The detailed information in terms of genes and proteins can be seen in Tables S1 and S2. For instance, the amino acid length of 60 ZmSnRKs protein ranges from 215 to 653, and the molecular weight ranges from 24.7 to 72.3 kDa. All ZmSnRKs were predicted to be located in the nucleus, cytoplasm, or mitochondria.
To verify the evolutionary relations of SnRK proteins in angiosperms, we constructed a neighbor-joining tree using 47 rice SnRKs, 39 Arabidopsis SnRKs, and 60 maize SnRKs (Figure 1). The protein sequences in rice and Arabidopsis are listed in Table S3. Hrabak et al., (2003) reported that 39 AtSnRKs were classified into three subfamilies. Here, 60 ZmSnRKs were also clustered into three groups in accordance with phylogenetic analysis. In detail, four proteins containing Pkinase (PF00069), UBA (PF00627), and KA1 (PF02149) domains belong to the ZmSnRK1 subfamily; 14 proteins belong to the ZmSnRK2 subfamily, and the rest of 42 proteins containing Pkinase and NAF (PF03822) domains were clustered into the ZmSnRK3 subfamily (Figure 2A).
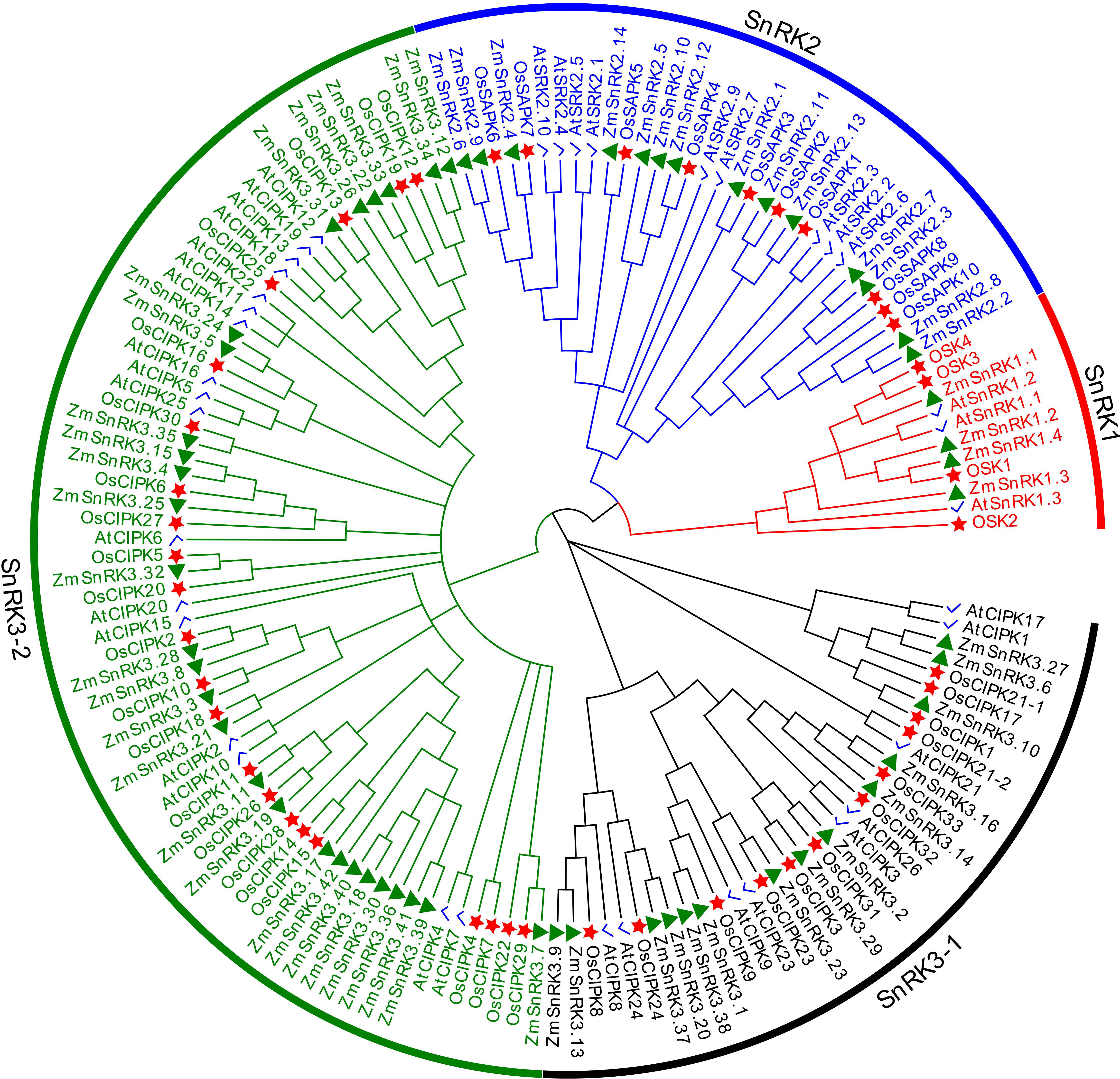
Figure 1 Phylogenetic tree of full-length ZmSnRK, AtSnRK and OsSnRK proteins. The different colored arcs indicate subfamilies of the SnRK proteins. Different colour shapes represent SnRKs from maize (▲), rice (☆), and Arabidopsis (√). The unrooted neighbor-joining phylogenetic tree was constructed using MEGA7 with full-length amino acid sequences and the bootstrap test replicate was set as 1,000 times.
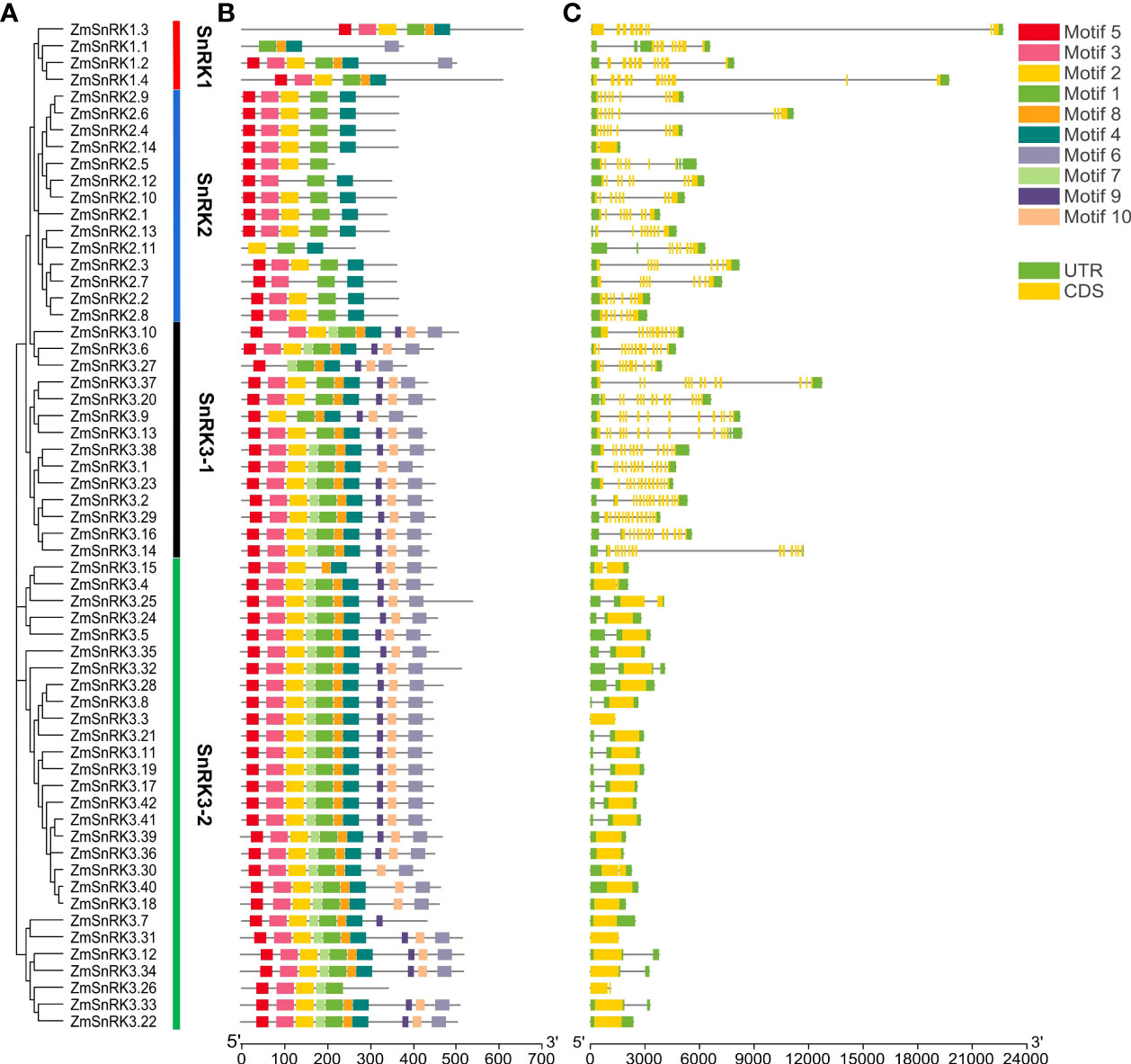
Figure 2 Phylogenetic relationships, architecture of conserved protein motifs and gene structure in SnRK genes from maize. (A) The phylogenetic tree was constructed based on the full-length sequences of maize SnRK proteins using MEGA 7 software. (B) The motif compositions of 60 ZmSnRK proteins. The motifs were identified using the MEME program. Boxes of different colors represent motifs 1 to 10. The length of the amino acid sequences can be estimated by the scale at the bottom. (C) Gene structures of 60 ZmSnRK genes. Yellow boxes represent exons, green boxes represent 5′ or 3′ untranslated regions (UTR), and black lines represent introns. The length of nucleotide sequences of exons/introns/UTRs can be estimated by the scale at the bottom.
Gene and protein structures in ZmSnRKs
The web server GSDS was used to understand the gene structure of ZmSnRKs, which had 1 to 14 exons unevenly. In detail, SnRK1 subfamily genes contain 8–12 exons. SnRK2 subfamily genes contain 7–9 exons, except ZmSnRK2.2 and ZmSnRK2.14 containing one exon. Interestingly, ZmSnRK3 subfamily genes exhibit a large difference in the number of exons, ranging from 1 to 14 (Figure 2C). For example, half of the genes have only one exon, five genes have two exons, and another gene has 12–14 exons. Hence, ZmSnRK3-1 and ZmSnRK3-2 were classified in terms of the number of exons.
We used MEME to determine protein motifs. Generally, 10 conserved motifs were discovered in ZmSnRKs and named motifs 1–10 (Figure 2B; Table S4). Among them, the motifs ½/3/4/5 belong to phosphokinase domains. Almost all of ZmSnRKs have motifs 1, 2, 3, and 4, except ZmSnRK1.1 with motif 2/3/4, ZmSnRK3.27 and ZmSnRK2.7 with motif 1/3/4, ZmSnRK2.11 with motif 1/2/4, and ZmSnRK3.26 with motif 1/2/3. In addition, motif 6/7/9/10 exists exclusively in ZmSnRK3, except ZmSnRK1.1 and ZmSnRK1.4 containing motif 6. Moreover, motif 8 exists only in ZmSnRK1 and ZmSnRK3, except ZmSnRK3.26. In addition, 3D structures of ZmSnRK proteins were predicted and it showed that different ZmSnRKs own similar 3D structures in the N-terminal but disparate in the C-terminal (Figure 3).
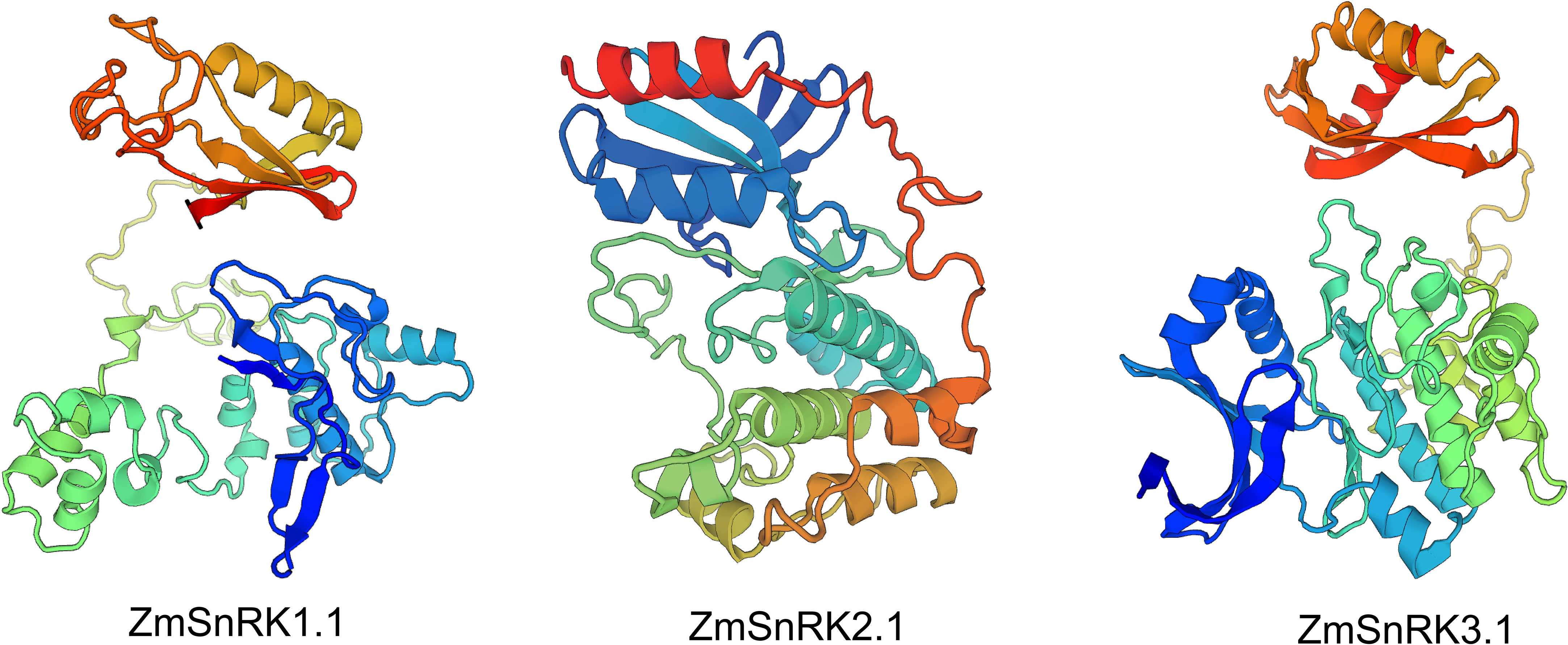
Figure 3 The 3D structure modeling of ZmSnRK proteins. The pymol software was used to create the structural image.
Chromosomal localization and duplication of ZmSnRKs
The chromosome location of ZmSnRK genes was analyzed and 60 ZmSnRKs were mapped to the maize genome (Figure 4). In detail, the gene members of the ZmSnRK1 subfamily were localized in chromosomes 1, 2, 6, and 8. Both ZmSnRK2 and ZmSnRK3 subfamily genes were located in all chromosomes. We used the approach of BLAST and MCScanX to search for gene duplication. Finally, 22 segmental duplication events were detected in ZmSnRKs, and each gene pair was located on a distinct chromosome (Figure 5; Table S5). It demonstrated that most of ZmSnRKs were probably derived from gene duplication, and segmental duplication events could take a dominant role in the expansion of ZmSnRK genes in maize. Moreover, the syntenic relations of the SnRK gene families between maize genome, barley genome, Arabidopsis genome, and rice genome were also determined. The results showed that two SnRK gene pairs were found in maize and Arabidopsis, and four SnRK gene pairs were discovered in maize and rice (Figure 6; Table S6, S7). For instance, AtCIPK15 showed colinearity with ZmSnRK3.11 and ZmSnRK3.17. Furthermore, 24 ZmSnRKs exhibited syntenic relations with SnRKs in barley (Figure 6; Table S8).
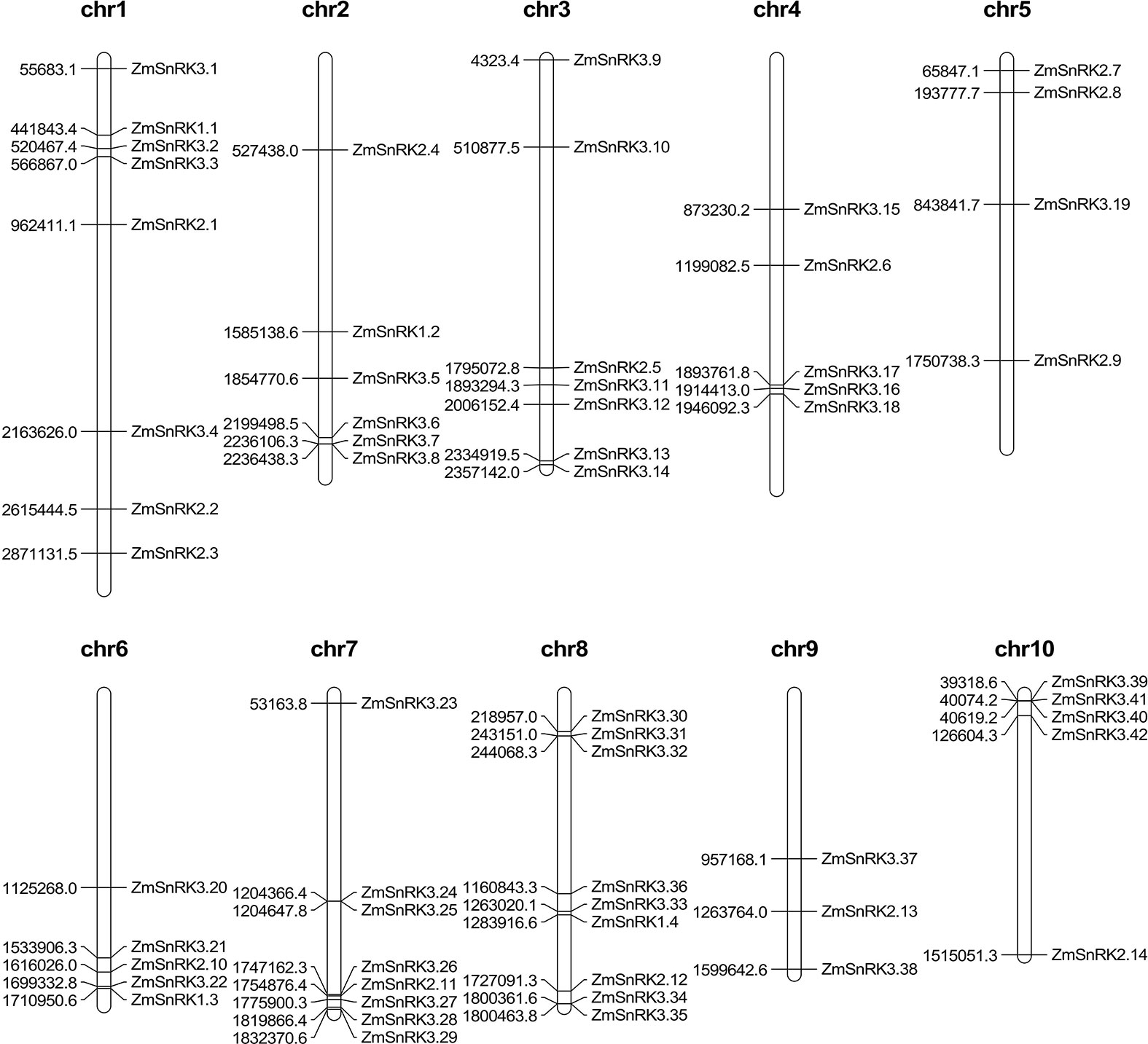
Figure 4 Distribution of ZmSnRK genes in maize chromosomes. The chromosome numbers are indicated at the top of each chromosome image.
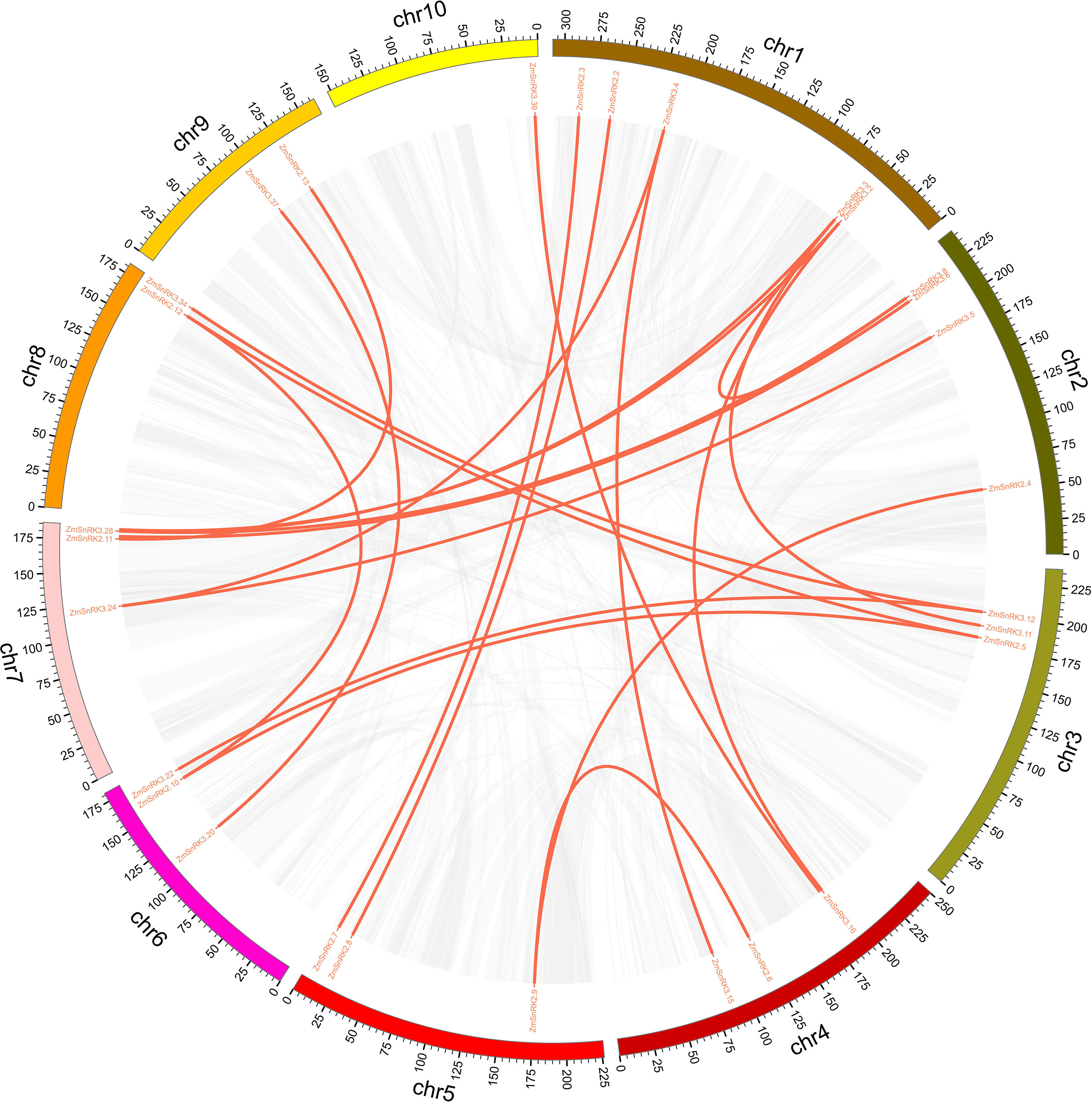
Figure 5 The synteny analysis of ZmSnRK family in maize. Gray lines indicate all synteny blocks in the maize genome. The genes linked by red lines represent homologues.
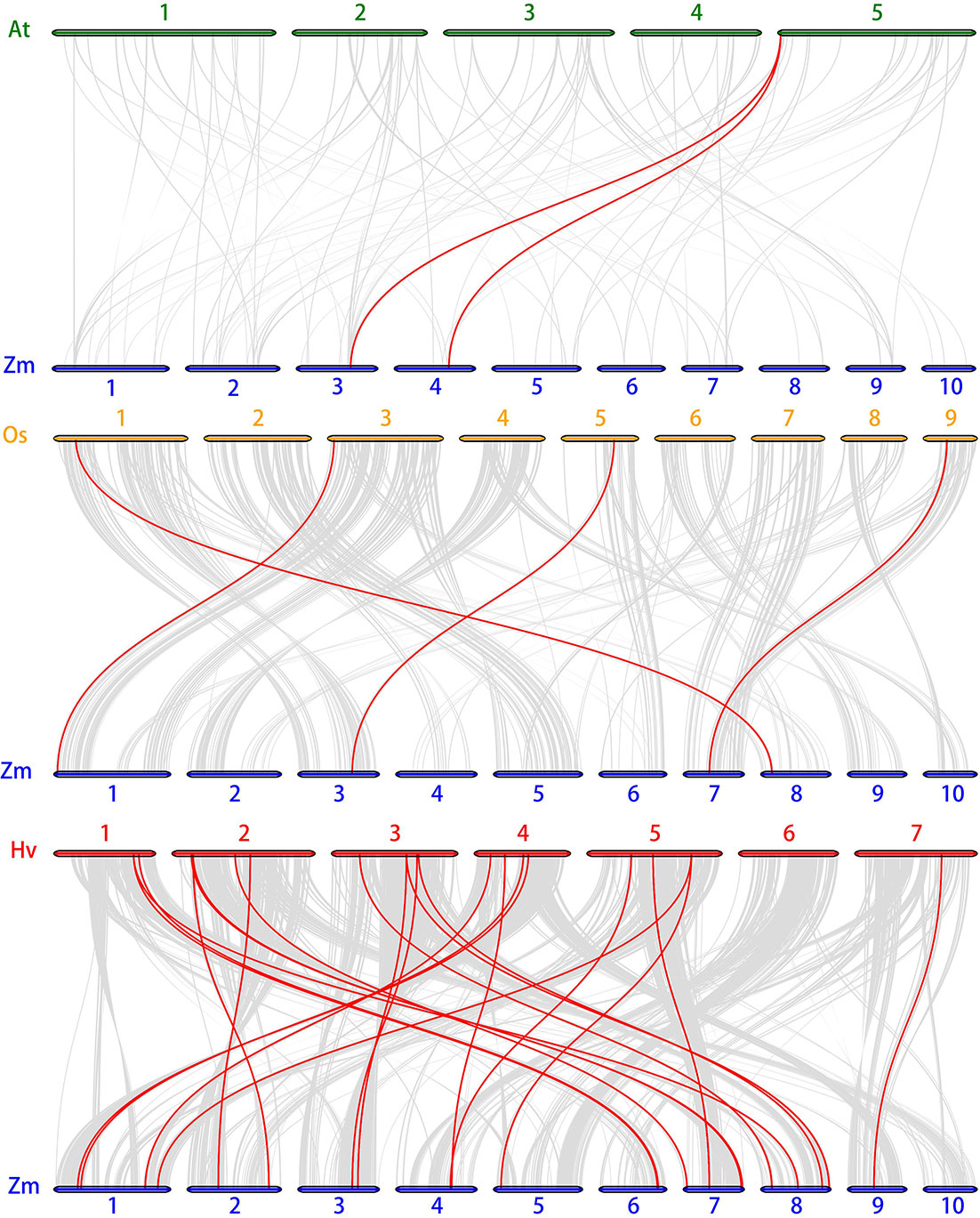
Figure 6 Synteny analysis of SnRK genes between maize, Arabidopsis, rice and barley. Gray lines: all collinear blocks within maize and other plant genomes. Red lines: the synteny of SnRK gene pairs. The specie names with the prefixes, Hv, Zm, At, and Os indicate barley, maize, Arabidopsis, and rice, respectively.
To understand the evolutionary constraints acting on paralogous SnRK family genes, Ks value, Ka value, Ka : Ks ratio, and divergence time were calculated (Table S5). Most of the segmental duplication of ZmSnRK gene pairs displayed Ka/Ks ratios less than 1, except ZmSnRK3.3/ZmSnRK3.8 and ZmSnRK3.6/ZmSnRK3.27 with 2.69 and 1.02, respectively. The average value of ZmSnRK3 (Ka/Ks = 0.56) was higher than ZmSnRK2 gene pairs (Ka/Ks = 0.13). Divergence time was therefore estimated to occur between 4.304 Mya and 68.812 Mya ago. The above results implied that ZmSnRK gene families were likely to have suffered strong purifying selective pressure in the course of evolution.
Cis-elements analysis in the promoters of ZmSnRKs
Promoter sequences of all ZmSnRK genes were obtained from the maize B73 genome database. A total of 60 ZmSnRKs were assessed to analyze cis-elements, including ABRE, AuxRR-core, MBS, MBSI, P-box, SARE, LTR, involving in ABA, auxin, drought-inducibility, flavonoid biosynthetic genes regulation, gibberellin, salicylic acid and low temperature response (Figure 7; Table S9). In general, 50 ZmSnRKs genes (90%) owned ABRE cis-elements, 22 ZmSnRK genes (36.7%) carried MBS cis-elements, and 28 ZmSnRK genes (46.7%) had LTR cis-elements. Six ZmSnRK genes had AuxRR-core elements, and 12 ZmSnRK genes had P-box. Meanwhile, only ZmSnRK2.4 and ZmSnRK2.12 had MBSI and SARE elements, respectively.
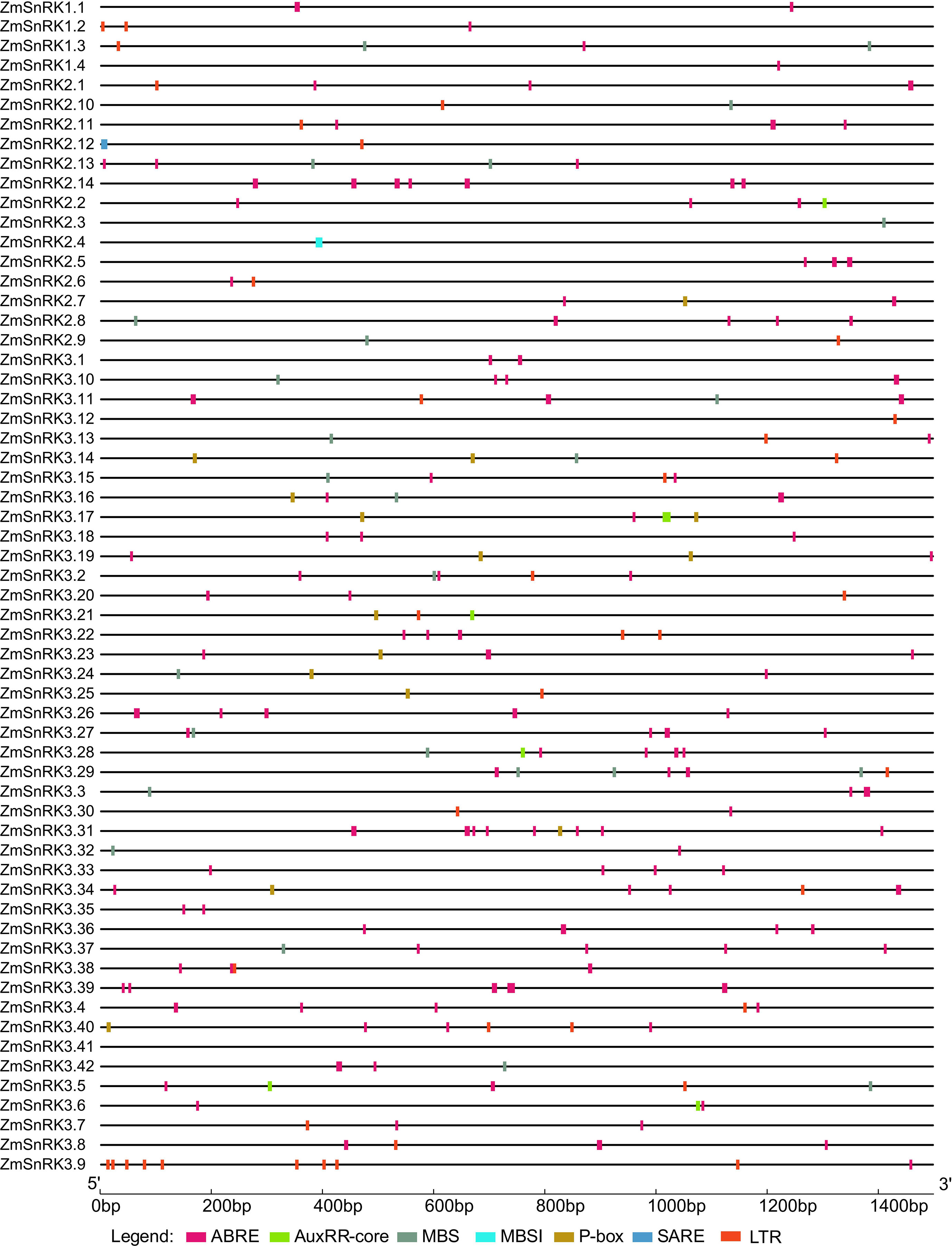
Figure 7 Predicted cis-regulatory elements in ZmSnRK promoters. Promoter sequences (about 1.5 kb) of 60 ZmSnRK genes were analyzed by PlantCARE. The upstream length to the translation starting site can be inferred according to the scale at the bottom.
Tissue-specific expression patterns of ZmSnRKs
We compared tissue-specific expression patterns of 60 ZmSnRK genes in maize B73. In terms of different expression patterns, 60 ZmSnRK genes were classified into three groups (Figure 8; Table S10). Group 1 contains three genes (ZmSnRK3.26/3.34/3.40), and they are not expressed in all analyzed tissues. Group 2 includes 12 genes, which were expressed only in specific tissues. For instance, ZmSnRK3.1 was expressed only in stem and leaves, and did not display expression in other tissues. Group 3 contains 45 genes expressed in all analyzed tissues. Group 3 could be classified into three subgroups. There were 14 ZmSnRKs belonging to subgroup 1 with high expression patterns (log2TPM+1>2) in all analyzed tissues, including three ZmSnRK1s, five ZmSnRK2s, and six ZmSnRK3s. Subgroup 2 contained only ZmSnRK3.23 with log2TPM+1<0.5 in all analyzed tissues. The remaining 30 genes were assigned to subgroup 3.
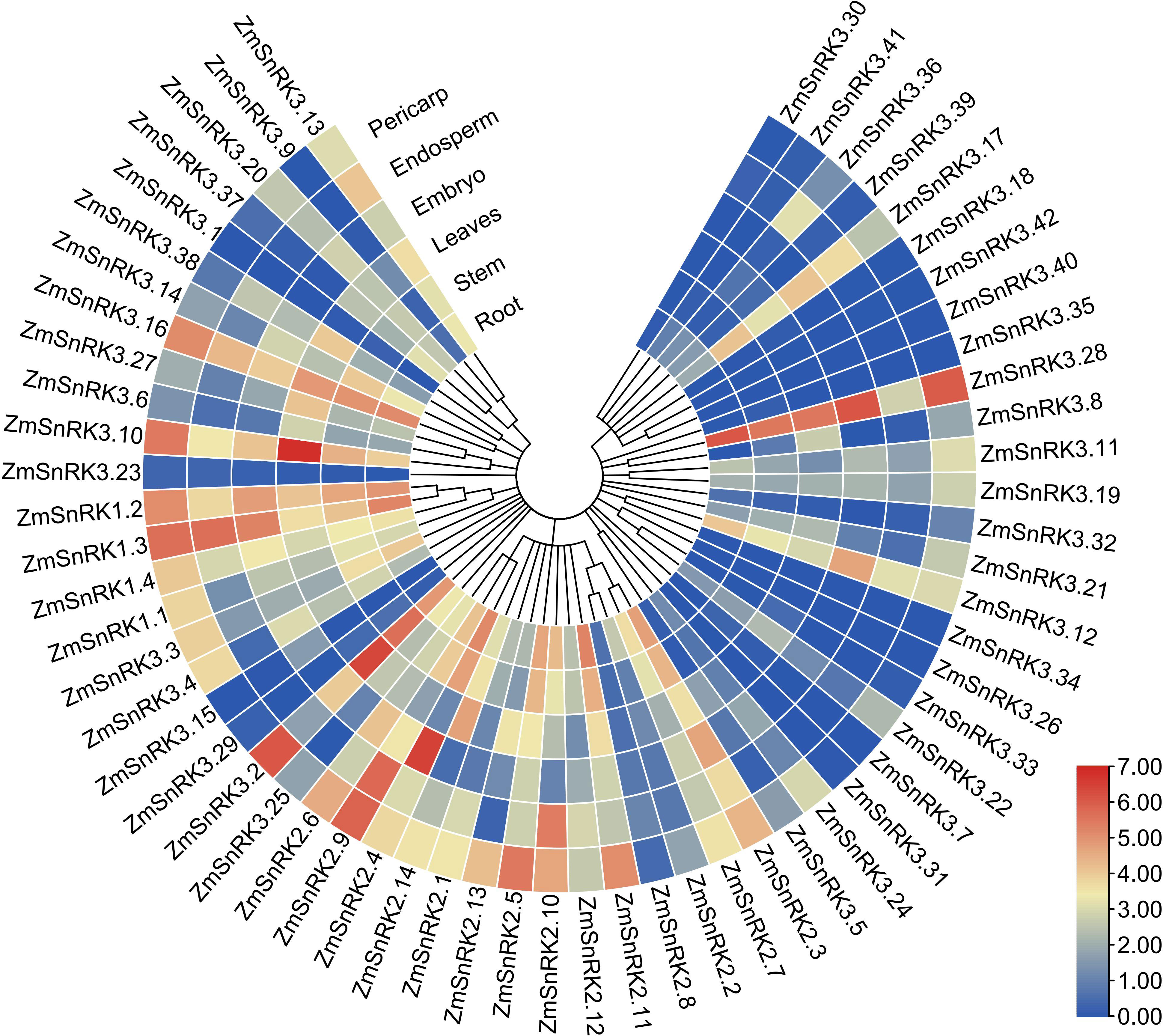
Figure 8 Expression profiles of the ZmSnRK genes in different tissues. The color scale represents expression data with row scale. Blue: low expression; red: high expression.
Expression patterns of ZmSnRKs under diverse abiotic stresses
We collected and analyzed the transcriptome data of maize SnRK genes under drought, salt, high temperature, low temperature, and ultraviolet light. In general, the expression of several ZmSnRK genes showed significant alterations under abiotic stresses (Figure 9; Table S11). For example, ZmSnRK2.13 exhibited up-regulation after drought stress. The expression pattern of ZmSnRK2.2 increased under cold treatment. ZmSnRK3.1 was down-regulated under cold and salt stress. On the contrary, some ZmSnRKs were not induced by any abiotic stresses listed in this study. For instance, 11 ZmSnRK genes showed almost no expression changes under all analyzed stresses, such as ZmSnRK1.4, ZmSnRK2.3, and ZmSnRK3.13. In addition, many genes showed opposite expression profiles in response to different abiotic stress. For example, ZmSnRK2.2 displayed expression inhibition in UV treatment, but expression enhancement in cold stress. In addition, several ZmSnRK genes were chosen for qRT-PCR to verify the reliability of transcriptome data, and the results were uniform to the sequencing data (Figure S2, S3).
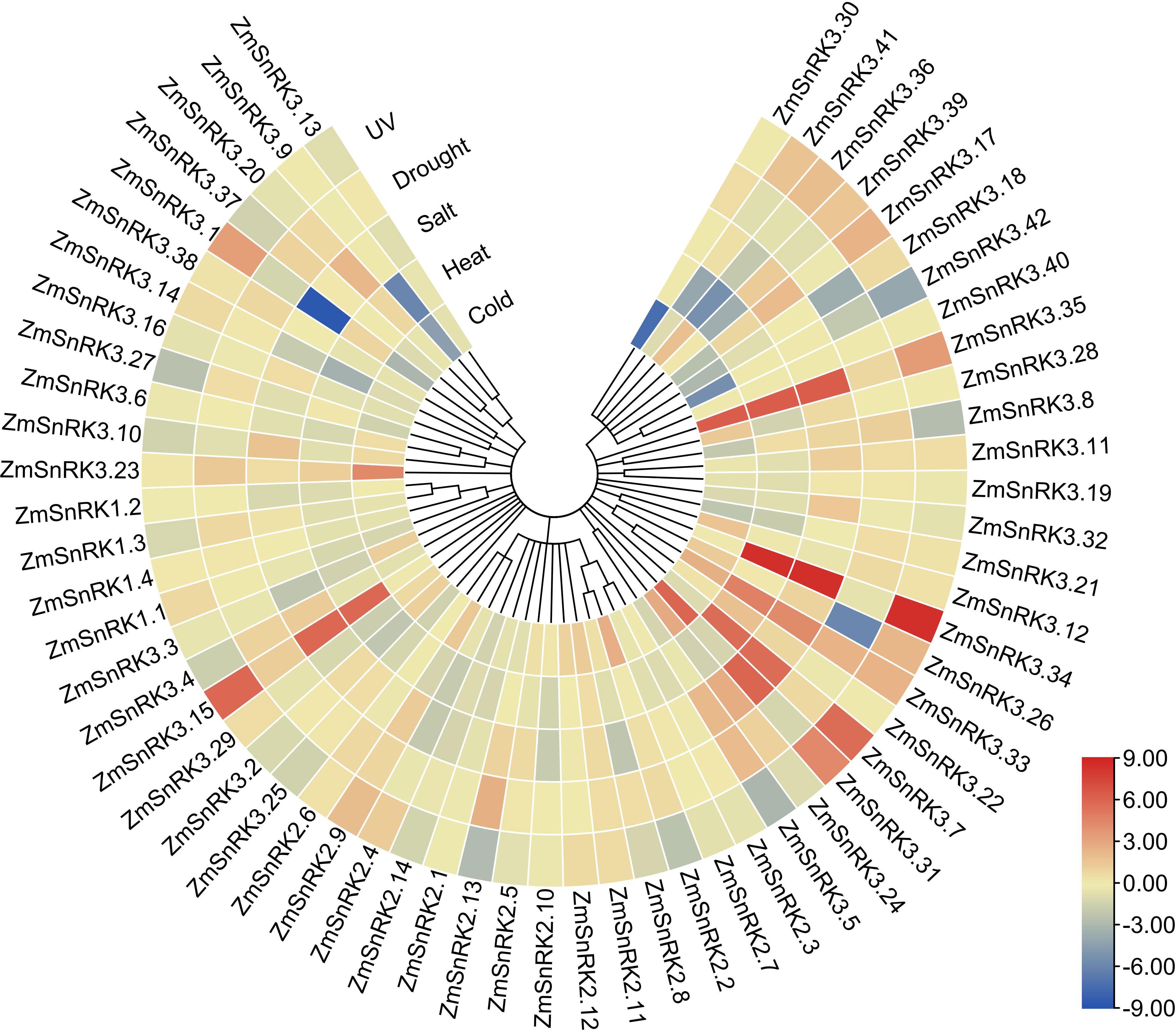
Figure 9 Expression profiles of the ZmSnRK genes under different abiotic stresses. Expression data were the ratio to control values. The color scale represents expression levels from upregulation (red) to downregulation (blue).
Function and regulatory network of ZmSnRKs
Protein phosphatases PP2C could dephosphorylate and inactivate plant SnRKs, the process of which prevents SnRKs from targeting ABA-dependent genes and ion channels. Hence, we used the web server of STRING to predict the protein–protein interaction (PPI) between ZmSnRKs and ZmPP2Cs to comprehend the regulatory networks of ZmSnRKs (Figure 10; Table S12). Here, eight ZmPP2Cs that could interact with ZmSnRK proteins were found. In detail, ZmPP2C65 and ZmPP2C67 could bind six ZmSnRK proteins; ZmPP2C51, ZmPP2C68, and ZmPP2C69 could bind four ZmSnRK proteins; ZmPP2C4 and ZmPP2C71 could bind three ZmSnRK proteins; PP2C17 could dephosphorylate ZmSnRK2.11 specifically. Moreover, there are also interactions within ZmSnRK proteins. For instance, ZmSnRK1.1 could bind with ZmSnRK2.14 and ZmSnRK3.12 (Figure S1).
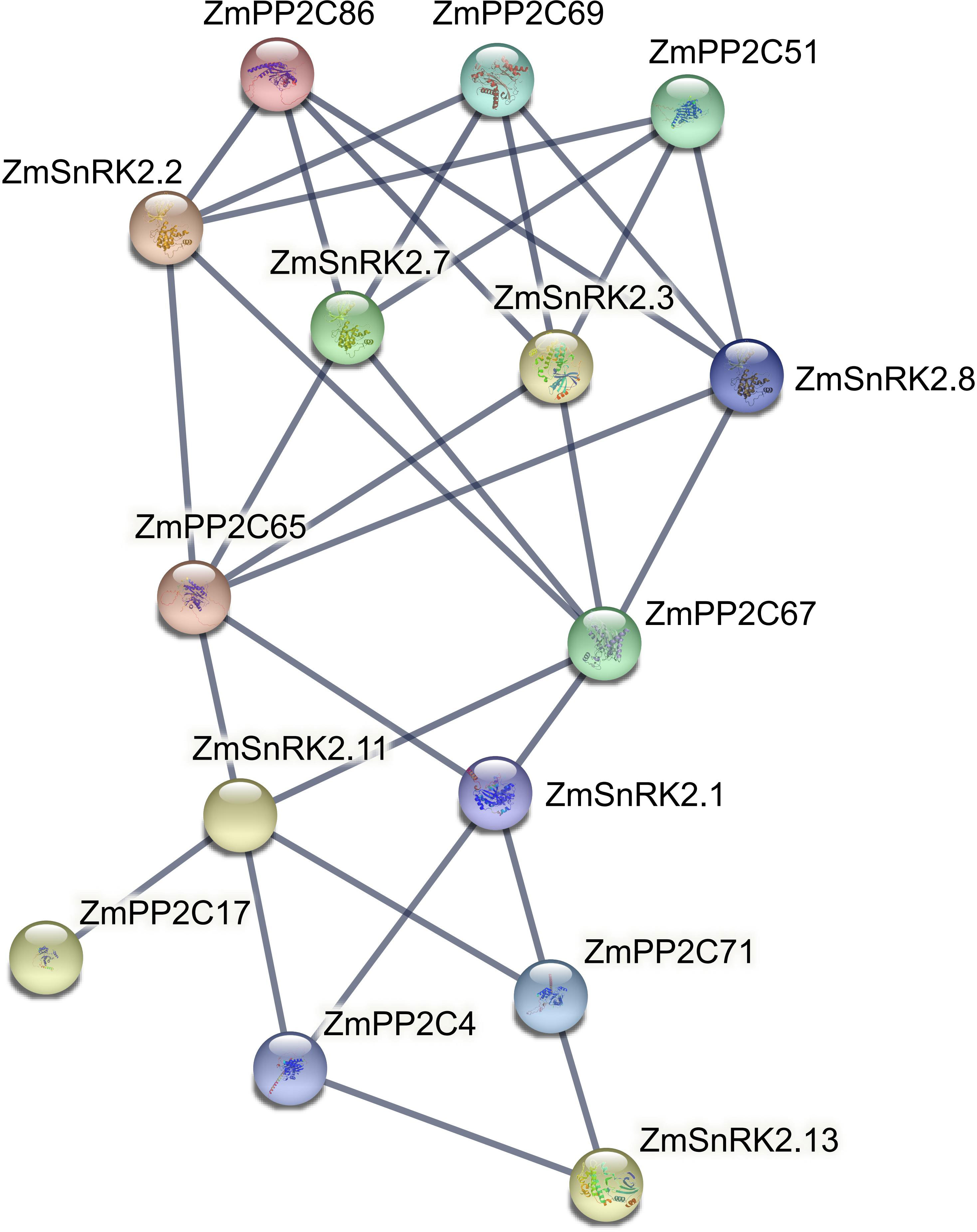
Figure 10 Interaction networks between ZmSnRK and ZmPP2C proteins. Information on ZmPP2Cs was shown in Table S12.
We constructed the miRNA-mRNA regulatory network between 321 published miRNA in maize and 60 ZmSnRKs. A total of 46 putative miRNAs were identified to have a potential capacity to target and regulate 24 ZmSnRKs (Figure 11; Table S13). The miRNAs of ZmSnRKs could be grouped into 11 networks: group 1 involving two ZmSnRKs (ZmSnRK3.1 and ZmSnRK3.38), group 2 involving seven ZmSnRKs (ZmSnRK 2.2/2.4/2.6/2.8 and ZmSnRK 3.26/3.33), group 3 involving four ZmSnRKs (ZmSnRK 2.11/2.13 and ZmSnRK 3.6/3.20), group 5 involving ZmSnRK3.2 and ZmSnRK3.28, and group 7 involving ZmSnRK 3.30/3.40/3.42, group 4/6/8/9/10/11 involving ZmSnRK 3.27/1.2/3.19/3.31/3.17, respectively.
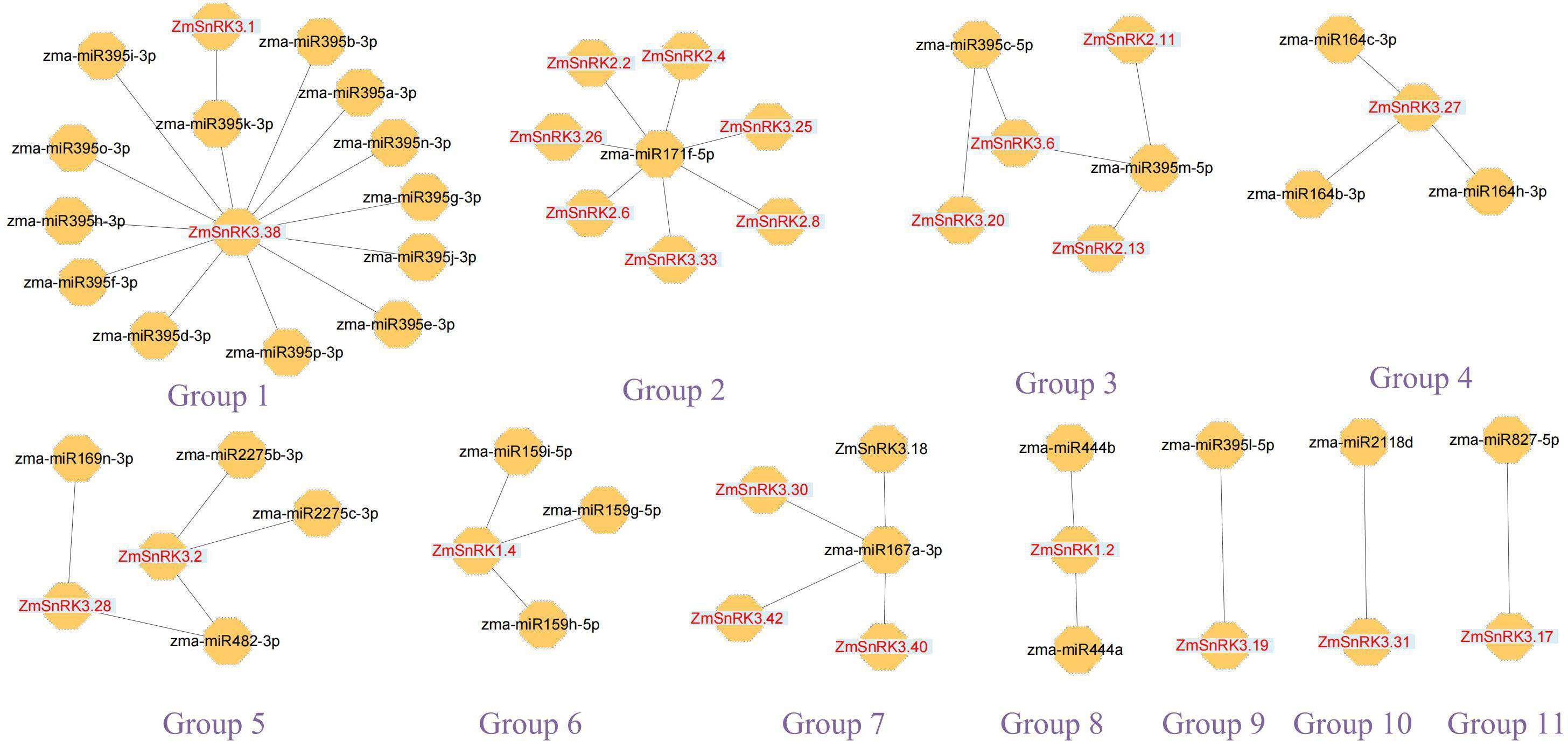
Figure 11 Interaction networks between miRNAs and miRNAs-acted ZmSnRKs. Information on miRNAs and miRNAs-acted ZmSnRKs was shown in Table S13.
Discussion
Protein kinases and phosphatases can recognize and transduce stress signals to diverse cellular compartments. SNF1-related kinases (SnRKs) universally exist in all eukaryotes, especially in plants. To date, the SnRK family has been identified in a number of plant species, including Arabidopsis thaliana (Hrabak et al., 2003), Oryza sativa (Kobayashi et al., 2004), Brassica napus (Zhu et al., 2020), and Brachypodium distachyon (Wang et al., 2015). However, the report concerning whole-genome analysis of the ZmSnRK genes has not yet been published in maize. In the current study, 60 ZmSnRKs have been found in the Zea mays genome, which were considered as ZmSnRK1 to ZmSnRK60 based on their chromosome distribution (Figures 1, 4). Previously, SnRK genes were classified into three subfamilies based on different domains in C-terminus in Arabidopsis and rice (Kolukisaoglu et al., 2004). In the maize genome, 60 ZmSnRK genes were also classified into three subfamilies, including 4 ZmSnRK1s, 14 ZmSnRK2s, and 42 ZmSnRK3s (Figures 1, 2; Table S2). Then, ZmSnRK genes family was systematically investigated, including evolutionary relationships, conserved motifs, subcellular localization, chromosome location, gene divergence time, promoter elements analysis, and expression patterns.
The plant SnRK1 subfamily is the key regulator of energy homeostasis (Crepin and Rolland, 2019). SnRK1 controls the activity of many enzymes and the transcription activity of several genes (Baena-González et al., 2007; Nukarinen et al., 2016), such as bZIP, ethylene insensitive3 (EIN) and indeterminate domain (IDD) (Jeong et al., 2015; Zhai et al., 2017; Muralidhara et al., 2021). For example, SnRK1 delayed senescence through phosphorylating the EIN3 transcription factor and led to its destabilization (Zhai et al., 2017). SnRK1 strongly phosphorylated bZIP63 and enhanced its dimer formation, either homodimerization or heterodimerization with other bZIPs, thus contributing to DNA binding (Mair et al., 2015). Meanwhile, Jeong et al. (2015) reported that SnRK1 was responsible for regulating various developmental processes in plants, such as hypocotyl elongation or blooming (Simon et al., 2018). In the current study, ZmSnRK1 has four members and exhibited a high expression pattern in all tissues, but showed no alteration upon abiotic stress (Figures 1, 8, 9; Figure S2, S3; Table S2, S10, S11). Moreover, ZmSnRK1.3 and ZmSnRK1.4 were predicted to be located in mitochondria (Table S1). ZmSnRK1.1 and ZmSnRK1.2 were likely to be located in the nucleus and cytoplasm but with 0.657 and 0.568 reliable-index if located in mitochondria (data not shown). It suggests ZmSnRK1 may also be crucial in plant development as an energy sensor.
Plant SnRK2s, a key component of ABA signaling pathways, are regulated by ABA receptors (PYR/PYL/RCAR) and protein phosphatase 2Cs (PP2CAs). In this study, eight ZmPP2Cs were predicted to interact with seven ZmSnRK2s (Figure 10). It suggests that ZmPP2Cs could also dephosphorylate ZmSnRKs in maize. The SnRK2 subfamily in plants is extremely responsible for responding to abiotic stress, such as drought (Umezawa et al., 2004; Barajas-Lopez et al., 2018; Hasan et al., 2021) and salt (Zhong et al., 2020). When plants are subjected to drought stresses, they generate more ABA, thus leading to defensive stress responses to activate many SnRK2s via ABA-dependent or ABA-independent pathways (Hasan et al., 2022). Here, the expression level of ZmSnRK2.13 was significantly increased after drought stress (Figure 9; Figure S2; Table S11), which was likely to be dephodphrylated by ZmPP2C4 and ZmPP2C71 (Figure 10). Moreover, the promoter of ZmSnRK2.13 had three ABRE cis-elements and one MBS cis-element, which was associated with ABA responsiveness and drought stress (Figure 7; Table S9). One segmental duplication event was detected between ZmSnRK2.11 and ZmSnRK2.13 (Figure 5; Table S5), but drought exposure does not change the gene expression in ZmSnRK2.11 (Figure 9; Table S11), indicating that ZmSnRK2.13 was positively selected when subjected to drought stress even though it can be cleaved by the same miRNA (Figure 11; Table S13). Belda-Palazón et al. (2022) reported that SnRK2 could interact with SnRK1 and consequently cause subcellular localization changes of the SnRK1 α-subunit, the process of which are vital for repressing the target of rapamycin (TOR) and inhibiting root growth in an ABA-dependent manner. In this study, ZmSnRK2.13 could also bind to ZmSnRK1.1 and ZmSnRK2.8 (Figure S1). It suggested that SnRK1 could interact with SnRK2 and jointly participate in plant growth in maize. In rice, OsSAPK8 (orthologous to ZmSnRK2.3 and ZmSnRK2.7) acts as a positive regulator when plants are subjected to cold, drought, and salt stress (Zhong et al., 2020). Meanwhile, Frazier et al. (2011) reported that miR395 was up-regulated under drought stress. The current study showed that miR395 could target and cut the mRNA of ZmSnRK2.3 (Figure 11; Table S13). ZmSnRK2.3 and ZmSnRK2.7, as a duplicated gene pair, do not respond to abiotic stress (Figures 5, 9; Table S5, S11), but showed high expression in all tested tissues (Figure 8; Table S10), indicating different mechanism in diverse species. Interestingly, ZmSnRK2.4 have only MBSI cis-element, which is involved in flavonoid biosynthesis (Figure 7; Figure S2; Table S9), and its expression level was repressed under salt stress (Figure 9; Table S11). Watkins et al. (2017) found that flavonoids are involved in plant development through affecting auxin, ethylene, and ABA signaling. HSFB2b and GmMYB173 activated a number of flavonoid biosynthesis-related genes and promoted flavonoid accumulation, thus positively regulating salt tolerance (Pi et al., 2018; Bian et al., 2020). This indicates that ZmSnRK2.4 may also be involved in flavonoid biosynthesis and regulate plant growth under salt stress.
The SnRK3 subfamily was widely recognized as CBL-interacting protein kinases (CIPKs). In plants, CBL-CIPK signaling network regulates multiple stimuli or signals (Ma et al., 2020). For example, Qiu et al. (2002) reported that AtCIPK24 could interact with AtCBL4 and activate the H+/Na+ antiporter. In the current study, ZmSnRK3.33 showed a high expression level under five abiotic stresses (Figure 9; Table S11), which can be regulated by zma-miR171. Besides, ZmSnRK2.2, ZmSnRK2.4, ZmSnRK2.6, and ZmSnRK2.8 were also cut by zma-miR171 (Figure 11; Table S13). These indicate that ZmSnRK3 and ZmSnRK2 may have a similar function, at least in one aspect. In soybean, GmCIPK2 acts as a positive regulator in response to drought stress in an ABA-dependent manner (Xu et al., 2021). Here, ZmSnRK3.26 showed down-regulation with 5.9-fold changes after drought stress (Figure 9; Table S11), the promoter of which has five ABRE elements (Figure 7; Table S9). Hence, ZmSnRK3 could also respond to abiotic stresses through an ABA-dependent pathway.
In short, this research carried out a comprehensive analysis of the SnRK gene family in Zea mays. With the above results, we deduced that ZmSnRKs might play a pivotal role in the long-term climate resilience of maize. Numerous functional verification work is still necessary for comprehending the biological functions of ZmSnRKs in future research.
Conclusion
SnRK genes play important roles in signaling pathways, including responses to abiotic stresses in plants. In the present study, genome-wide identification and characterization of SnRK genes were conducted in Zea mays. A total of 60 ZmSnRK genes were characterized and divided into three subfamilies. Phylogenetic and synteny analysis of SnRK genes among Arabidopsis, rice, and maize provide valuable clues for the evolutionary characteristics of the ZmSnRK genes. Moreover, the cis-acting elements, gene expression, and regulatory network of ZmSnRK families were also determined. These results provide insights into the functional differences, evolutionary relationships, and expression profiles of SnRKs in Zea mays.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XF and WL conceived and designed the research. XF, QM, QY, JZ, DX, and XD performed the experiments and data analyses. WL, XF, and QM wrote the article. LG and WM revised the manuscript. All authors read and approved the final article.
Funding
This work was supported by the National Natural Science Foundation of China (grant No. 32001449; 32101660) and the Natural Science Foundation of Shandong Province (grant No. ZR2021QC052).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1087839/full#supplementary-material
References
Albrecht, V., Ritz, O., Linder, S., Harter, K., Kudla, J. (2014). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. doi: 10.1093/emboj/20.5.1051
Alderson, A., Sabelli, P. A., Dickinson, J. R., Cole, D., Richardson, M., Kreis, M., et al. (1991). Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl. Acad. Sci. U.S.A. 88, 8602–8605. doi: 10.1073/pnas.88.19.8602
Arnold, K., Bordoli, L., Kopp, J., Schwede, T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Baena-González, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 23, 938–942. doi: 10.1038/nature06069
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Barajas-Lopez, J. D., Moreno, J. R., Gamez-Arjona, F. M., Pardo, J. M., Punkkinen, M., Zhu, J. K., et al. (2018). Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 93, 107–118. doi: 10.1111/tpj.13761
Belda-Palazón, B., Costa, M., Beeckman., T., Rolland, F., Baena-González, E. (2022). ABA represses TOR and root meristem activity through nuclear exit of the SnRK1 kinase. Proc. Natl. Acad. Sci. U.S.A. 119, e2204862119. doi: 10.1073/pnas.2204862119
Bender, K. W., Zielinski, R. E., Huber, S. C. (2018). Revisiting paradigms of Ca2+ signaling protein kinase regulation in plants. Biochem. J. 475, 207–223. doi: 10.1042/BCJ20170022
Bian, X. H., Li, W., Niu, C. F., Wei, W., Hu, Y., Han, J. Q., et al. (2020). A class b heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 225 (1), 268–283. doi: 10.1111/nph.16104
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cohen, P. (1988). Review lecture: Protein phosphorylation and hormone action. Proc. R. Soc Lond. 234, 115–44. doi: 10.1098/rspb.1988.0040
Crepin, N., Rolland, F. (2019). SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 51, 29–36. doi: 10.1016/j.pbi.2019.03.006
Cutler, S. R., Rodriguez, P. L., Finklestein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Ann. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Damian, S., Gable, A. L., Nastou, K. C., David, L., Rebecca, K., Sampo, P., et al. (2021). The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, 605–612. doi: 10.1093/nar/gkab835
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., Yang, S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in arabidopsis. Dev. Cell. 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Frazier, T. P., Sun, G., Burklew, C. E., Zhang, B. (2011). Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 49, 159–165. doi: 10.1007/s12033-011-9387-5
Halford, N. G., Hardie, D. G. (1998). SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Molec Biol. 37, 735–748. doi: 1023/A:1006024231305
Hasan, M. M., Gong, L., Nie, Z. F., Feng, X., Ahammed, G. J., Fang, X. W. (2021). ABA-induced stomatal movements in vascular plants during dehydration versus rehydration. Environ. Exp. Bot. 186, 104436. doi: 10.1016/j.envexpbot.2021.104436
Hasan, M. M., Liu, X. D., Waseem, M., Guang-Qian, Y, Alabdallah, N. M., Jahan, M. S., Fang, X. W. (2022). ABA activated SnRK2 kinases: an emerging role in plant growth and physiology. Plant Signal Behav. 17, 2071024. doi: 10.1080/15592324.2022.2071024
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hunter, T. (1995). Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. doi: 10.1016/0092-8674(95)90405-0
Jeong, E. Y., Seo, P. J., Woo, J. C., Park, C. M. (2015). AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in arabidopsis. BMC Plant Biol. 15, 110. doi: 10.1186/s12870-015-0503-8
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kobayashi, Y., Yamamoto, S., Minami, H., Kagaya, Y., Hattori, T. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 16, 1163–1177. doi: 10.1105/tpc.019943
Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., Kudla, J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kulik, A., Wawer, I., Krzywi ´nska, E., Bucholc, M., Dobrowolska, G. (2011). SnRK2 protein kinases–key regulators of plant response to abiotic stresses. Omics 15, 859. doi: 10.1089/omi.2011.0091
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–29348. doi: 10.1093/bioinformatics/btm404
Letunic, I., Doerks, T., Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 302–305. doi: 10.1093/nar/gkr931
Li, J., Long, Y., Qi, G. N., Li, J., Xu, Z. J., Wu, W. H., et al. (2014). The os-AKT1 channel is critical for k+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell. 26, 3387–3402. doi: 10.1105/tpc.114.123455
Li, C., Qi, W., Liang, Z., Yang, X., Ma, Z., Song, R. (2020). A SnRK1-ZmRFWD3-Opaque2 signaling axis regulates diurnal nitrogen accumulation in maize seeds. Plant Cell. 32, 2823–2841. doi: 10.1105/tpc.20.00352
Magali, L., Patrice, D., Gert, T., Kathleen, M., Yves, M., Yves, V. D. P., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Mair, A., Pedrotti, L., Wurzinger, B., Anrather, D., Simeunovic, A., Weiste, C., et al. (2015). SnRK1-triggered switch of bZIP63 dimerization mediates the lowenergy response in plants. eLife 4, e05828. doi: 10.7554/eLife.05828.043
Ma, X., Li, Y., Gai, W. X., Li, C., Gong, Z. H. (2021). The CaCIPK3 gene positively regulates drought tolerance in pepper. Hortic. Res. 8, 216. doi: 10.1038/s41438-021-00651-7
Ma, X., Li, Q. H., Yu, Y. N., Qiao, Y. M., Haq., S. U., Gong, Z. H. (2020). The CBL-CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 215668. doi: 10.3390/ijms21165668
Masaru, O., Yan, G., Ursula, H., Jian-Kang, Z. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100, 11771–11776. doi: 10.1073/pnas.2034853100
Maszkowska, J., Szymańska, K. P., Kasztelan, A., Krzywińska, E., Sztatelman, O., Dobrowolska, G. (2021). The multifaceted regulation of SnRK2 kinases. Cell 102180. doi: 10.3390/cells10092180
Mazur, R., Maszkowska, J., Anielska-Mazur, A., Garstka, M., Polkowska-Kowalczyk, L., Czajkowska, A., et al. (2021). The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 187, 2785–2802. doi: 10.1093/plphys/kiab438
Mcloughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Laurière, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architectureduring salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Muralidhara, P., Weiste, C., Collani, S., Krischke, M., Kreisz, P., Draken, J., et al. (2021). Perturbations in plant energy homeostasis prime lateral root initiation via SnRK1-bZIP63-ARF19 signaling. Proc. Natl. Acad. Sci. U.S.A. 118, e2106961118. doi: 10.1073/pnas.2106961118
Myburg, A. A., Dario, G., Tuskan, G. A., Uffe, H., Hayes, R. D., Jane, G., et al. (2014). The genome of eucalyptus grandis. Nature 510, 356–362. doi: 10.1038/nature13308
Nukarinen, E., Nägele, T., Pedrotti, L., Wurzinger, B., Mair, A., Landgraf, R., et al. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6, 31697. doi: 10.1038/srep31697
Opitz, N., Paschold, A., Marcon, C., Malik, W. A., Lanz, C., Piepho, H. P., et al. (2014). Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genomics 15, 741. doi: 10.1186/1471-2164-15-741
Piao, H. L., Xuan, Y. H., Park, S. H., Je, B. I., Park, S. J., Park, S. H., et al. (2010). OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cell. 30, 19–27. doi: 10.1007/s10059-010-0084-1
Pi, E., Zhu, C., Fan, W., Huang, Y., Qu, L., Li, Y., et al. (2018). Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell Proteomics. 17, 1209–1224. doi: 10.1074/mcp.RA117.000417
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Shannon, P., Markeil, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Simon, N. M. L., Kusakina, J., Fernández-López, Á., Chembath, A., Belbin, F. E., Dodd, A. N. (2018). The energy-signaling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol. 176, 1299–1310. doi: 10.1104/pp.17.01395
Stelpflug, S. C., Sekhon, R. S., Vaillancourt, B., Hirsch, C. N., Buell, C. R., de Leon, N., et al. (2016). An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome. 9. doi: 10.3835/plantgenome2015.04.0025
Su, W., Raza, A., Zeng, L., Gao, A., Lv, Y., Ding, X., et al. (2021). Genome-wide analysis and expression patterns of lipid phospholipid phospholipase gene family in Brassica napus l. BMC Genomics 22, 548. doi: 10.1186/s12864-021-07862-1
Tominaga, M., Harada, A., Kinoshita, T., Shimazaki, K. (2010). Biochemical characterization of calcineurin b-like-interacting protein kinase in vicia guard cells. Plant Cell Physiol. 51, 408–421. doi: 10.1093/pcp/pcq006
Tör, M., Lotze, M. T., Holton, N. (2009). Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654. doi: 10.1093/jxb/erp233
Umezawa, T., Yoshida, R., Maruyama, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (2004). SRK2C, a SNF1-relatedprotein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 17306–17311. doi: 10.1073/pnas.0407758101
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, 49. doi: 10.1093/nar/gkr1293
Wang, L., Hu, W., Sun, J., Liang, X., Yang, X., Wei, S., et al. (2015). Genome-wide analysis of SnRK gene family in brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 237, 33–45. doi: 10.1016/j.plantsci.2015.05.008
Wang, W., Lu, Y., Li, J., Zhang, X., Hu, F., Zhao, Y., et al. (2021). SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis. Plant Cell. 33, 3721–3742. doi: 10.1093/plcell/koab224
Wang, D., Zhang, Z., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Watkins, J. M., Chapman, J. M., Muday, G. K. (2017). Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol. 175, 1807–1825. doi: 10.1104/pp.17.01010
Wurzinger, B., Nukarinen, E., Nagele, T., Weckwerth, W., Teige, M. (2018). The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 176, 1085–1094. doi: 10.1104/pp.17.01404
Xu, M., Li, H., Liu, Z., Wang, X. H., Xu, P., Dai, S. J., et al. (2021). The soybean CBL-interacting protein kinase, GmCIPK2, positively regulates drought tolerance and ABA signaling. Plant Physiol. Biochem. 167, 980–989. doi: 10.1016/j.plaphy.2021.09.026
Yu, W., Peng, F., Wang, W., Liang, J., Xiao, Y., Yuan, X. (2021). SnRK1 phosphorylation of SDH positively regulates sorbitol metabolism and promotes sugar accumulation in peach fruit. Tree Physiol. 41, 1077–1086. doi: 10.1093/treephys/tpaa163
Zhai, Z., Liu, H., Shanklin, J. (2017). Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell. 29, 871–889. doi: 10.1105/tpc.17.00019
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool 665 for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419, 779–781. doi: 10.1016/j.bbrc.2012.02.101
Zhang, M., Zhang, S. (2022). Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64, 301–341. doi: 10.1111/jipb.13215
Zhong, R., Wang, Y., Gai, R., Xi, D., Mao, C., Ming, F. (2020). Rice SnRK protein kinase OsSAPK8 acts as a positive regulator in abiotic stress responses. Plant Sci. 292, 110373. doi: 10.1016/j.plantsci.2019.110373
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: maize (Zea mays), SnRK, genome-wide, expression patterns, climate-resilience
Citation: Feng X, Meng Q, Zeng J, Yu Q, Xu D, Dai X, Ge L, Ma W and Liu W (2022) Genome-wide identification of sucrose non-fermenting-1-related protein kinase genes in maize and their responses to abiotic stresses. Front. Plant Sci. 13:1087839. doi: 10.3389/fpls.2022.1087839
Received: 02 November 2022; Accepted: 05 December 2022;
Published: 22 December 2022.
Edited by:
Mehanathan Muthamilarasan, University of Hyderabad, IndiaReviewed by:
Roshan Singh, National Institute of Plant Genome Research (NIPGR), IndiaPooja Choudhary, Jaypee Institute of Information Technology, India
Sumi Rana, University of Hyderabad, India
Copyright © 2022 Feng, Meng, Zeng, Yu, Xu, Dai, Ge, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxing Liu, bGl1d3hAcWF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xue Feng
Xue Feng Quan Meng1†
Quan Meng1† Jianbin Zeng
Jianbin Zeng Dengan Xu
Dengan Xu Wujun Ma
Wujun Ma Wenxing Liu
Wenxing Liu