- College of Horticulture, Shenyang Agricultural University, Shenyang, China
Branching is an important agronomic trait determining plant architecture and yield; however, the molecular mechanisms underlying branching in the stalk vegetable, flowering Chinese cabbage, remain unclear. The present study identified two tandem genes responsible for primary rosette branching in flowering Chinese cabbage by GradedPool-Seq (GPS) combined with Kompetitive Allele Specific PCR (KASP) genotyping. A 900 kb candidate region was mapped in the 28.0−28.9 Mb interval of chromosome A07 through whole-genome sequencing of three graded-pool samples from the F2 population derived by crossing the branching and non-branching lines. KASP genotyping narrowed the candidate region to 24.6 kb. Two tandem genes, BraA07g041560.3C and BraA07g041570.3C, homologous to AT1G78440 encoding GA2ox1 oxidase, were identified as the candidate genes. The BraA07g041560.3C sequence was identical between the branching and non-branching lines, but BraA07g041570.3C had a synonymous single nucleotide polymorphic (SNP) mutation in the first exon (290th bp, A to G). In addition, an ERE cis-regulatory element was absent in the promoter of BraA07g041560.3C, and an MYB cis-regulatory element in the promoter of BraA07g041570.3C in the branching line. Gibberellic acid (GA3) treatment decreased the primary rosette branch number in the branching line, indicating the significant role of GA in regulating branching in flowering Chinese cabbage. These results provide valuable information for revealing the regulatory mechanisms of branching and contributing to the breeding programs of developing high-yielding species in flowering Chinese cabbage.
Introduction
Brassica rapa is one of the most important Brassica species with a long cultivation history, has diverse and distinct morphological traits. Flowering Chinese cabbage [Brassica rapa L. ssp. chinensis (L.) Hanelt var. parachinensis (L.H. Bailey)] is that bolts readily (Wang and Kole et al., 2015). It is a stalk vegetable, and the stems with flower buds and leaves are consumed after cooking, especially in southern and central China and southeastern Asian countries. Typically, only one flowering stalk (main stem, without primary rosette branches) can be harvested from the common varieties, while several stalks are produced (12 stalks, primary rosette branches) on the local flowering Chinese cabbage variety named Zengcheng (Niu et al., 2019). Thus, the primary rosette branching trait influences plant architecture and yield in flowering Chinese cabbage. Therefore, identifying genes is important to develop varieties with multiple branches and meet the growing demand for flowering Chinese cabbage.
The activities of meristems, including apical, axillary and inflorescence meristems, basically determine the branching trait (Wang et al., 2018a). In flowering Chinese cabbage, the shoot apical meristem (SAM) turns into an inflorescence meristem that produces flowers directly or flower-bearing shoots after transitioning from the vegetative to the reproductive phase. The primary rosette branches grow from the axillary buds subtended by rosette leaves. Then, the secondary inflorescence branches grow at the axils of the cauline leaves on the elongated internodes of the main inflorescence stem, like that in Arabidopsis thaliana (Wang et al., 2018a; Fichtner et al., 2021). Studies in rice, Arabidopsis, and several other species characterizing the regulatory components of tiller or branch development have improved our understanding of branching (Wang and Li, 2011; Fichtner et al., 2021).
Typically, branching is a quantitative trait controlled by multiple genes and is susceptible to the environment (Ehrenreich et al., 2007; Kebrom et al., 2013) and plant hormones (Wang and Li, 2011). The hormonal control of bud outgrowth is complex and not yet fully understood. Auxin (Morris et al., 2005), cytokinin (Xu et al., 2015), strigolactone (De Jong et al., 2014), gibberellin (Martínez-Bello et al., 2015), abscisic acid (Holalu et al., 2020), and their interactions (Chen et al., 2013; Cao et al., 2017) have been reported to affect branching. Among these, gibberellin significantly influences the growth of branches and main stems. Okada et al. (2020) found that gibberellic acid (GA3) application increased the number of lateral branches on apple trees. Meanwhile, the exogenous spraying of GA3 rescued the dwarf phenotype of the legume’s msd1-2 (multi-seeded1-2) mutant (Li et al., 2021). Researchers have identified a few genes controlling branching in Brassica juncea, Brassica napus, non-heading Chinese cabbage, and purple flowering Chinese cabbage (Li, 2018; Muntha et al., 2019; Li et al., 2020a; Li et al., 2020b). However, branching in flowering Chinese cabbage has not been fully clarified.
Researchers recently proposed a new quantitative trait mapping technique called GradedPool-Seq (GPS) for rapidly mapping the quantitative trait loci (QTL), it can rapidly identify QTLs for complex traits comparing conditional methods (Wang et al., 2019). GPS with high-throughput sequencing scores and assigns the F2 populations derived from a distant cross of parental lines exhibiting contrasting phenotypes into three or more graded groups based on phenotypic values. GPS has been successfully applied to dissect heterotic genes of thousand-grain weight, plant height, heading date, flag leaf angle, and tiller angle in rice. Moreover, the candidate intervals identified by the GPS method is consistent with the mapping interval obtained by the traditional method in rice (Wang et al., 2019).
Therefore, the present study used GPS with Kompetitive Allele Specific PCR (KASP) genotyping to map and identify the candidate genes associated with primary rosette branches in flowering Chinese cabbage. We further analyzed the similarities and variations in the full length and promoters of the candidate genes between the lines with different branching phenotypes. Finally, we investigated the role of GA3 in regulating the development of multiple primary rosette branches in flowering Chinese cabbage. The findings of our study will provide novel insights into the mechanisms of branching and lay a foundation for developing flowering Chinese cabbage cultivars with multiple primary rosette branches.
Materials and methods
Plant materials and growing conditions
‘CX020’ (Branching line), a doubled haploid (DH) line derived by microspore culture from Zengcheng flowering Chinese cabbage [Brassica rapa L. ssp. chinensis (L.) Hanelt var. parachinensis (L.H. Bailey)] with multiple primary rosette branches at harvest, and ‘CX010’ (Non-branching line), a DH line derived from Guangdong flowering Chinese cabbage with one flower-bearing shoot and no primary rosette branch, were used as parents in this study. These DH parents were crossed to generate the F1 and F2 populations for the phenotypic and genetic analyses. These DH parents exhibited stable inheritance after multiple seasons of planting. All the plants (‘CX010’, ‘CX020’, and F2 population) were grown at the Shenyang Agricultural University experimental base (Shenyang, China, 41°82′N, 123°24′E) in 2019. The plants were sown on August 1st, and the number of primary rosette branches was analyzed on September 15th. The primary rosette branches were analyzed on the axillary branches subtended by rosette leaves 5 cm away from the cotyledonary node.
GradedPool-Seq
Three types of pools (50 plants each), including ‘multiple primary rosette branching (12−15 branches)’, ‘less primary rosette branching (1−3 branches)’ and ‘moderate primary rosette branching (7−8 branches)’ plants were selected via phenotypic analysis from the F2 population, consisting of 1050 individuals, were used for GPS analysis with the two parents. Young and fresh leaves of the parents and the selected F2 individuals were harvested separately for total genomic DNA extraction using the Plant Genomic DNA Kit (Tiangen, Beijing, China), following the manufacturer’s instructions. The quantity and quality of the DNA were ensured using spectrophotometric analysis and 2% agarose gel electrophoresis. The DNA samples were quantified using a Qubit fluorometer and pooled at equimolar concentrations to generate the ‘multiple branching’, ‘less branching’ and ‘moderate branching’ pools separately. Pair-end sequencing (PEN150) on an Illumina Novaseq system (Illumina, USA) was performed following the standard protocol. The sequencing data from each hybrid pool was merged and aligned to the reference genome (http://brassicadb.org/brad/datasets/pub/Genomes/Brassica_rapa/V3.0/) to calculate the depth of each variant. After filtering the variants with low quality and depth using the default parameters, Ridit analysis was performed to calculate the p-value for each variant. The sliding window size was set to 0.2 Mb, the threshold p-value to 10-8, and the candidate regions to peak intervals to reduce background noise and identify the significant variants.
KASP genotyping
KASP was performed on a high-throughput Intelliqube genotyping platform for genotyping of the F2 individuals. DNA was extracted from 150 F2 individuals and the parents using the CTAB method. The DNA concentration and quality were assessed on a BioDropuLite microanalyzer (BioDrop, Britain), and the samples were diluted to a suitable concentration (5−10 ng/μL). KASP primers (Supplementary Table S1) were designed for 19 SNP loci (Supplementary Table S2) using Primer Premier 5.0 (Singh et al., 1998), and KASP assays were conducted in a 384-well plate format on a Hydrocycler (LGC, Middlesex, UK) using the following PCR protocol: 94 °C for 15 min; 94 °C for 20 s, 61 °C for 60 s (-1 °C/cycle, 10 cycles in total), and 94 °C for 20 s; 55 °C for 60 s (26 cycles). The components of the KASP reaction mixture are shown in Supplementary Table S3. The fluorescence signal generated was measured on an IntelliQube (LGC, Middlesex, UK). Finally, based on the KASP genotypic data and phenotypic data of the F2 individuals, the QTL IciMapping software v4.2 (Liu et al., 2019) generated the linkage map to obtain the QTL and further narrow down the candidate interval.
Candidate gene analysis
Gene annotation information of the target region was obtained from the Brassica database (http://brassicadb.org/brad/index.php) and the Arabidopsis database (https://www.arabidopsis.org/). The candidate genes’ full-length sequence and a 2000 bp long promoter sequence were amplified with the specific primers using PCR, and the amplicons were purified using a Gel Extraction Kit (CWBIO, Beijing, China). The purified products were introduced into the pGEM®-T Easy Vector (Promega, USA) and transformed into Top10 competent cells (CWBIO, Beijing, China). The colonies were sequenced at Sangon Biotech (Shanghai, China), and the sequences were aligned using DNAMAN 6.0 (Lynnon Biosoft, Canada). Meanwhile, PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-regulatory elements in the promoter regions of the candidate genes.
Expression analysis of the candidate genes
Total RNA was extracted from the whole roots, rosette stems, tender cauline leaves, flowers of top inflorescences and shoot tips of the parents. The rosette stem of CX020 at five different stages (every ten days), the first sample was taken when CX020 was in the fourth euphylla stage. The RNA was reverse transcribed using the FastKing RT Kit (Tiangen, Beijing). Real-time quantitative reverse transcription PCR (qRT-PCR) was carried out to determine the expression levels of the candidate genes using cDNA as the template with the SYBR Green PCR Master Mix on QuantStudio™ 6 Flex (Applied Biosystems, USA), maintaining three biological replicates per sample. The Actin gene was used as an internal reference control. The primers used for qRT-PCR were as follows: BraA07g041560.3C (F:5′-TGGAGATGATTACTGATGGGTTA-3′; R: 5′-ATTTTCGTGGATGAGAGGGC-3′); BraA07g041570.3C (F:5′-TCCTGGATTTCTGTCCCTTC-3′; R: 5′-ACCCTATGCTTCACGCTTTT-3′); Actin (F: 5′- ATCTACGAGGGTTATGCT-3′; R: 5′-CCACTGAGGACGATGTTT -3′). The relative gene expression levels were calculated following the 2−ΔΔCt method.
Exogenous GA3 and PAC treatment
We predicted that the genes affecting branching might be related to GA3. An experiment was carried out by spraying GA3 (750 mg/L) on the multiple primary rosette branching parent line CX020, and spraying Paclobutrazol (PAC, 0.3×10-3 mg/L on the non-primary rosette branching parent line CX010, using water as a control. The plants were sown on August 1st in green house at Shenyang Agricultural University under long-day conditions and GA3 treatment was carried out on the fourth euphylla stage (August 15th); spraying was carried out every two days until phenotypes of fewer branches appeared. Twenty plants were maintained per treatment, using three biological replicates. The primary rosette branches number was recorded when the branching phenotype is obvious.
Results
Phenotypic characterization of primary rosette branching in flowering Chinese cabbage
During the reproductive growth stage, ‘CX010’ (parent 1, Figure 1A) had only one main stalk but no primary rosette branches on the rosette stem, while ‘CX020’ (parent 2, Figure 1B) had 14 primary rosette branches that contributed to the yield. Moreover, significant differences were observed in the number of primary rosette branches between the non-branching line (‘CX010’) and the multiple branching line (‘CX020’). Phenotypic segregation analysis showed that F1 progeny had a moderate number of primary rosette branches. The number of primary rosette branches in the 1050 F2 plants ranged from 0 to 15 and showed a normal distribution (Figure 1C). These observations indicated the role of QTLs in controlling the number of primary rosette branching in flowering Chinese cabbage.

Figure 1 Phenotype of parent lines CX010, CX020 and distribution of the number of primary rosette branches in flowering Chinese cabbage. (A) Non-primary rosette branching line (‘CX010’). (B) Multiple primary rosette branching line (‘CX020’) Bar = 10 cm. (C) Distribution of the number of primary rosette branches in 1050 individuals of F2 population.
Candidate region for primary rosette branching identified by GPS
The sequences obtained from the three pools (12−15 branches, 7−8 branches, and 1−3 branches) were mapped to the reference genome (http://brassicadb.org/brad/datasets/pub/Genomes/Brassica_rapaV3.0/) to estimate the allelic frequencies. Ridit analysis was implemented with allelic frequencies from three bulks to calculate p-values for each SNP. The background noise complicated the precise localization of the QTLs. Subsequently, the statistical noise-reducing strategy narrowed the interval to about 900 kb (28.0−28.9 Mb; significant peak) on chromosome A07, which was identified as the main QTL controlling the primary rosette branching. A few minor peaks also appeared on the other chromosomes that may be minor QTLs (Figure 2A). However, we further focused on the main QTL.
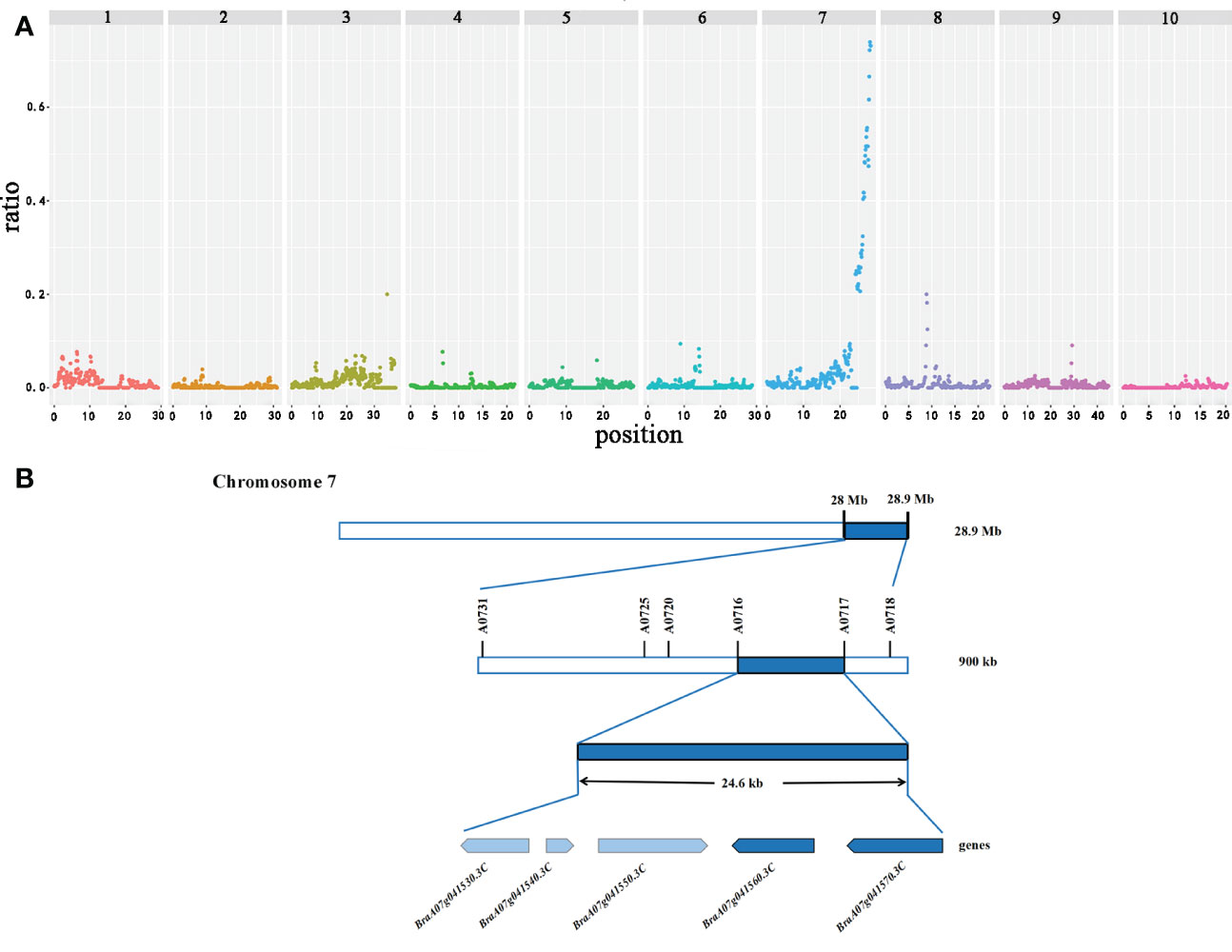
Figure 2 Candidate interval and genes identified by GPS on chromosome A07 in flowering Chinese cabbage. (A) The X-axis value is set at a midpoint at each defined genomic interval, and the Y-axis value corresponds to the ratio. (B) Candidate genes identified in the targeted interval.
Further mapping of the primary rosette branching gene
A total of 178 genes were identified in the candidate region based on the gene information in the Brassica database (Supplementary Table S4). Then, to locate the candidate genes controlling the primary rosette branching in this region, we developed 18 KASP markers according to SNPs in this region. The genotype of 150 F2 individuals and two parents was detected. Only one QTL was discovered associated with the number of primary rosette branches in the F2 based on KASP, which accounted for 20.81% of the phenotypic variance. Markers A0716 and A0717, located at the two sides of the candidate gene, were the most closely associated with the candidate gene, and the physical distance between these markers was 24.6 kb, which contained five genes (Figure 2B).
Identification of the candidate genes related to primary rosette branching
Then, all the candidate genes in the candidate region were analyzed based on the Brassica database to identify the candidate genes that influence primary rosette branching. To further identify the candidate genes that influence primary rosette branching, five annotated genes in the candidate region were analyzed (Table 1). Among these, two tandem genes, BraA07g041560.3C and BraA07g041570.3C, were found homologous to the Arabidopsis thaliana AT1G78440, which encodes gibberellin 2-oxidase that acts on C19 gibberellins. In rice, GA2oxs influence the number of tillers (Lo et al., 2008). Therefore, we hypothesized that these two tandem genes might be associated with the number of primary rosette branches in flowering Chinese cabbage.
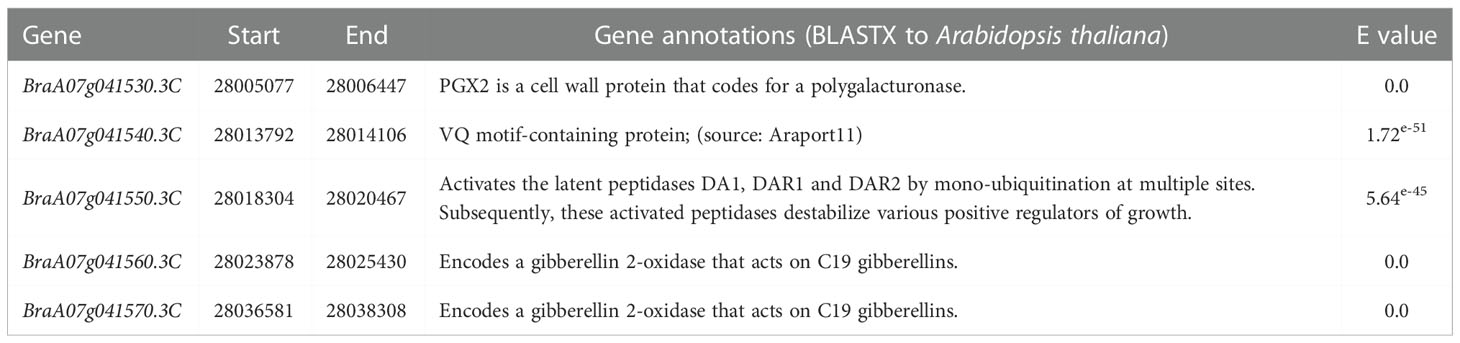
Table 1 Annotation of the genes within the mapped region on chromosome A07 in flowering Chinese cabbage.
Candidate gene cloning and sequence analysis
These two full-length genes (BraA07g041560.3C and BraA07g041570.3C) were cloned to analyze the variances between the parents (Supplementary Figures S1, S2). Sequence analysis revealed that the full-length sequence of BraA07g041560.3C was identical in the parents, whereas BraA07g041570.3C had a synonymous SNP mutation from A to G at the 290th position (Supplementary Figure S2). The promoter sequence of BraA07g041560.3C had a 31 bp deletion in CX020 (Supplementary Figure S3). There were many differences of the promoter sequence in BraA07g041560.3C between CX020 and CX010 that resulted in the absence of an ERE cis-regulatory element and the position changes of many cis-acting elements in CX020 (Supplementary Figure S3; Figure 3A). Compared to CX010, CX020 had an SNP in the promoter of BraA07g041570.3C, 103 bp upstream of the translation initiation site, resulting in the absence of an MYB cis-regulatory element (Supplementary Figure S4; Figure 3B).

Figure 3 The cis-regulatory elements analysis of BraA07g041560.3C and BraA07g041570.3C promoters in parents CX010 and CX020. (A) Analysis of BraA07g041560.3C gene promoters. (B) Analysis of BraA07g041570.3C gene promoters.
Expression analysis of candidate genes
Further, qRT-PCR was used to detect the relative expression levels of BraA07g041560.3C and BraA07g041570.3C in CX010 and CX020. The relative expression level of BraA07g041560.3C in the stem and flower of CX020 was significantly higher than that in CX010, while that in the leaf and shoot tip of CX020 was markedly lower than that in CX010 (Figure 4A). The relative expression level of BraA07g041570.3C in CX020 was substantially higher in root, stem, flower, and shoot tip than that of CX010 but significantly lower in the leaf (Figure 4B). We further analyzed the differences in the expression levels of BraA07g041560.3C and BraA07g041570.3C in CX020 stem at different stages. The analysis revealed that the BraA07g041560.3C expression was the highest at the second stage (Figure 5A). Meanwhile, the BraA07g041570.3C expression level at the last stage was significantly different from those at the other stages (Figure 5B).
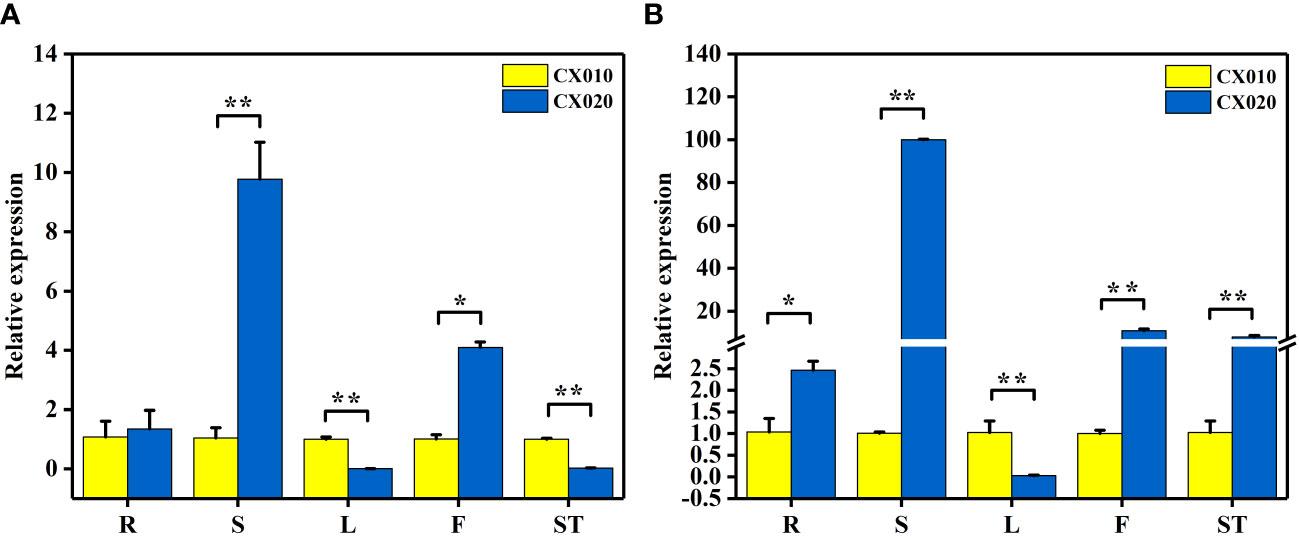
Figure 4 Expression levels of BraA07g041560.3C and BraA07g041570.3C in different tissues of CX010 and CX020 plants based on qRT-PCR. (A) The expression level of BraA07g041560.3C. (B) The expression level of BraA07g041570.3C. R, whole roots; S, rosette stems; L, tender cauline leaves; F, flowers of top inflorescences; ST, shoot tips. The data shown are the means of three replicates (± SD). * and ** indicate significant differences in expression levels at P < 0.05 and P < 0.01, respectively (Student’s t-test).
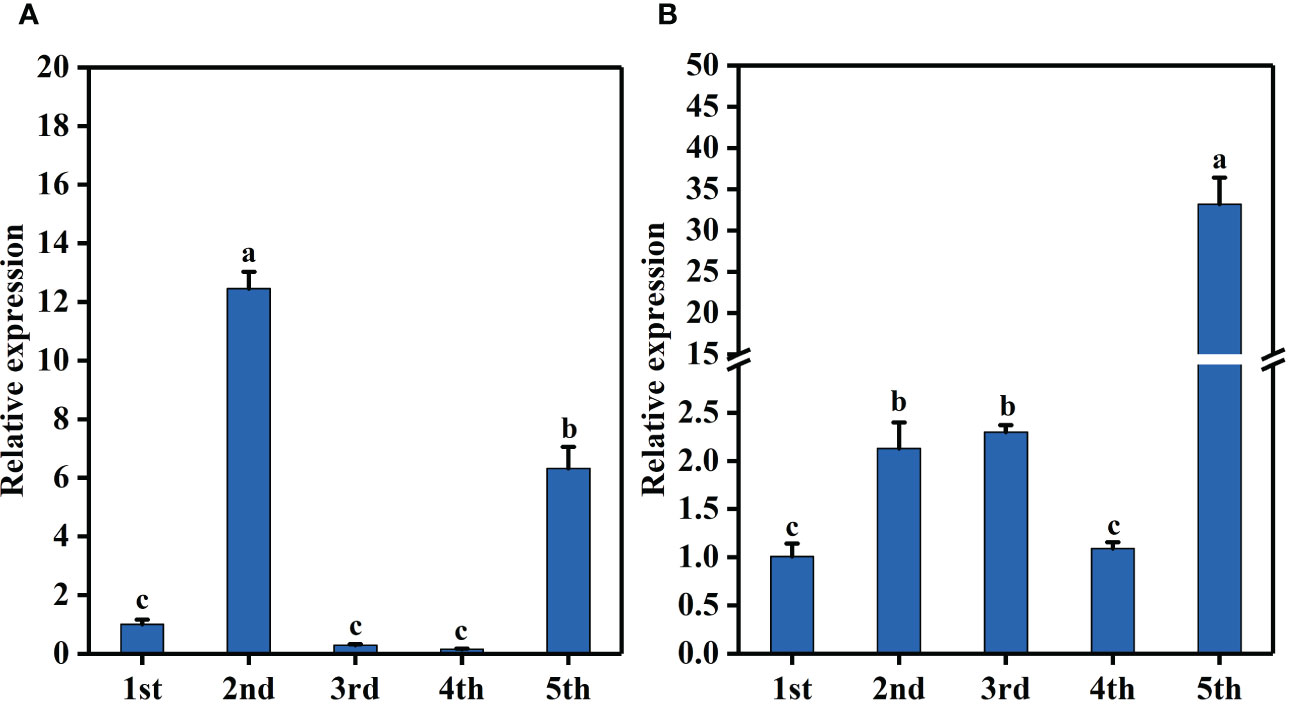
Figure 5 Expression levels of BraA07g041560.3C and BraA07g041570.3C in CX020 rosette stem at different stages based on qRT-PCR. (A) The expression level of BraA07g041560.3C. (B) The expression level of BraA07g041570.3C. The data shown are the means of three replicates (± SD). The different lowercase letters above the means are significantly different at P=0.05 level.
Phenotypic features after GA3 and PAC treatment
GA3 treatment significantly decreased the number of primary rosette branches (Figures 6A, B). On the contrary, after PAC treatment on the non-branching line CX010, the number of primary rosette branches were significantly increased (Figures 6C, D). The number of rosette branches significantly decreased from 10 to 0 after GA3 treatment (Figure 7A), while the number of rosette branches significantly increased from 0 to 8 after PAC treatment (Figure 7B).

Figure 6 Phenotypes of CX020 and CX010 after treatments with exogenous GA3 and PAC. (A, B) The CK and exogenous GA3 treatment of CX020. Bar = 8 cm. (C, D) The CK and exogenous PAC treatment of CX010. Bar = 5 cm.
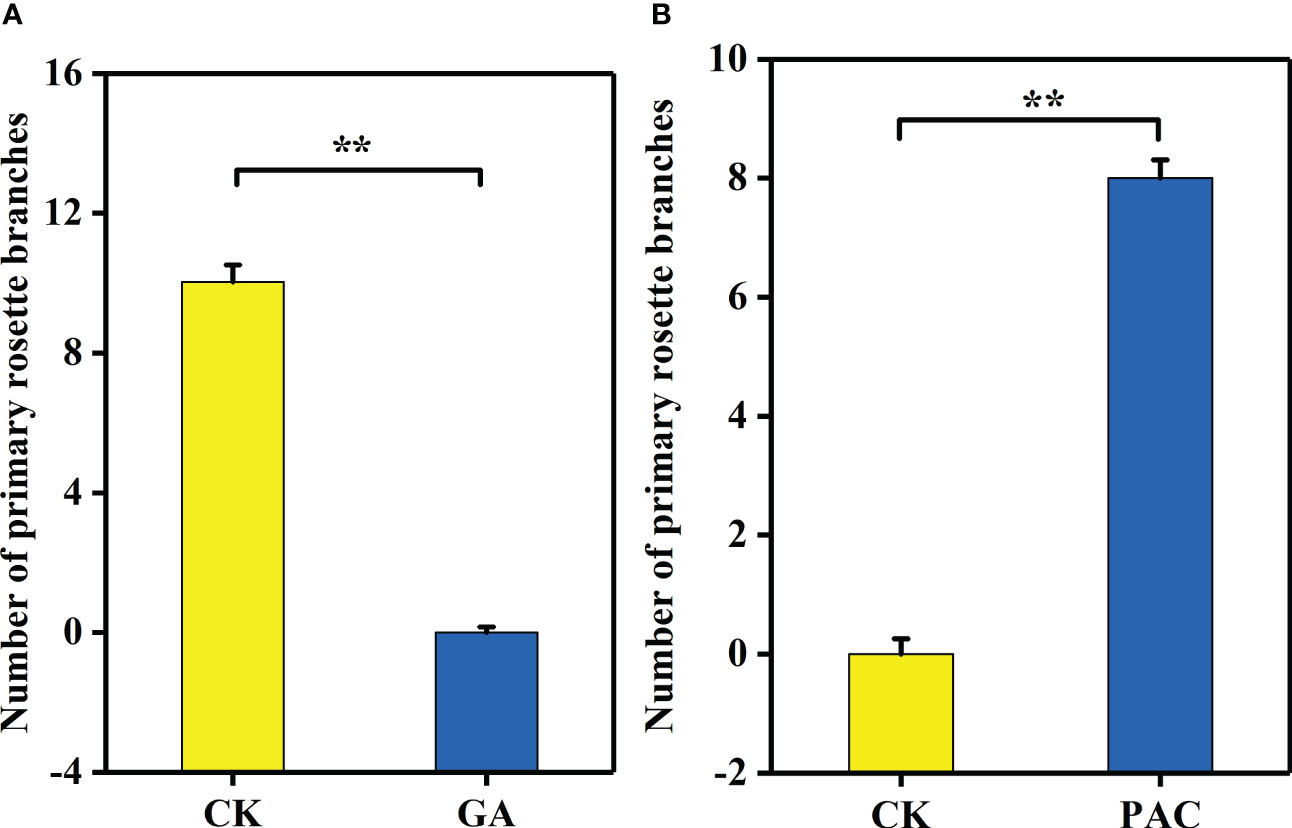
Figure 7 The number of primary rosette branches of CX020 and CX010 treated with exogenous GA3 and PAC. (A) The CK and exogenous GA3 treatment of CX020. (B) The CK and exogenous PAC treatment of CX010. The data shown are the means of three replicates (± SD). ** indicate significant differences in expression levels at P < 0.01 (Student’s t-test).
Discussion
Branching is an important trait that determines plant architecture, directly influences yield, and is closely related to environmental adaptation (Teichmann and Muhr, 2015; Mathan et al., 2016). The primary rosette branches that contribute to yield in flowering Chinese cabbage are different from the rosette branches in non-heading Chinese cabbage and tillers in cereal crops. In non-heading Chinese cabbage, basal branches arise from axillary meristems in the leaf axils subtended by rosette leaves during the vegetative stage (Cao et al., 2016). Then the inflorescence branches grow out from the axillary meristems. In cereal crops, tillers arise from non-elongated internodes at the base of the parent shoot during the vegetative growth phase and survive even if the primary shoot dies because tillers generally produce adventitious roots (Kebrom et al., 2013). While, the branches in flowering Chinese cabbage develop only after transitioning to the reproductive phase, the stage at which tiller development ceases in cereal crops, such as wheat, barley, and rice. Studies have identified genes related to branching in Brassica crops. Li et al. predicted BnaA09.ELP6 controls effective primary cauline branching (arising from the main stem) in Brassica napus (Li et al., 2020a). Meanwhile, shoot branching in non-heading Chinese cabbage (Brassica rapa ssp. chinensis Makino) is controlled by BrSB9.1 (Bra007056), which is homologous to MOC1 that controls tillering in rice (Li et al., 2020b). Bra004212, the homolog of TCP1, was identified as the candidate gene for tillering in purple flowering Chinese cabbage (Li, 2018). PAT1 (Phytochrome A signal transduction 1), which belongs to the GRAS transcription factor family, negatively regulates branching in leafy Brassica juncea (Muntha et al., 2019). However, the genes controlling primary rosette branching in Chinese cabbage differ from these reported species. The present study the first time identified for two tandem genes, BraA07g041560.3C and BraA07g041570.3C, which are homologous to AT1G78440 encoding GA2ox1 oxidase, as potential candidate genes responsible for primary rosette branching.
Traditional QTL mapping using genetic map construction by traditional markers and phenotyping is a reliable approach to isolate genes or QTLs associated with agronomic traits (Li et al., 2020b). Hundreds of SSR (Simple Sequence Repeat), InDels, or SLAF (Specific-Locus Fragment)markers have been developed to construct a primary genetic linkage map based on the population of the F2 generation, doubled haploid (DH) or recombinant inbred line (RIL) and locate QTL according to the phenotype (Liu et al., 2019). Usually, to narrow down the region and screen for a few candidate genes, a near-isogenic line (NIL) population is needed. However, the process of developing the NIL population is time-consuming and labor-intensive. GPS is a quick and efficient method to ascertain the genomic regions that harbor QTL for complex quantitative traits (Wang et al., 2019). It accelerates gene mapping by sequencing the graded pools; here, only the F2 population is needed. Bulked-segregant analysis (BSA) also rapidly and effectively locates genes by constructing segregating F2 populations from parents with significant phenotypic differences and selecting individuals with extreme traits to build a DNA pool for sequencing (Giovannoni et al., 1991; Michelmore et al., 1991; Takagi et al., 2013; Zou et al., 2016). However, the GPS has a higher resolution (~400 kb in rice) (Wang et al., 2019) than BSA (3 Mb) (Wang et al., 2018b; Liu et al., 2019; Yang et al., 2021). GPS identified a QTL controlling fruit size in melons, and traditional QTL mapping validated the results (Lian et al., 2021). Liu et al. (2020) successfully identified two candidate intervals controlling extremely late flowering in rice by GPS. In this work, one major candidate region associated with primary rosette branching was finally located in a 900 kb region on chromosome A07 based on three graded pools according to the primary rosette branching number of the F2 population in flowering Chinese cabbage, with 178 genes. To our knowledge, this is the first report on the genetic control of primary rosette branching in flowering Chinese cabbage using GPS.
KASP genotyping based on uniplex SNP is a novel approach to fine map genes using F2 individuals combined with their phenotype data (Xu et al., 2018; Cheng et al., 2021). Liu et al. (2019) narrowed the candidate region from 3.29 Mb to 790 kb by QTL analysis using KASP markers with 147 F2 individuals in tomatoes and identified the Cf-10 gene (Cladosporium fulvum). Lei et al. (2020) narrowed the genome interval from 4.17 Mb to 222 kb by 26 KASP markers genotyping with 199 individuals randomly selected from F2:3 and identified a major QTL and candidate gene for salt tolerance in rice. In this study, we genotyped 150 F2 individuals using 18 KASP markers and narrowed down the candidate region from 900 kb to 24.6 kb, with five genes.
GA2 oxidases are dioxygenases encoded by multiple genes and the key enzymes involved in gibberellin metabolism (Hedden and Phillips, 2000). GA2 oxidase typically transforms the bioactive GA1 and GA4 into the inactive catabolic metabolites GA8 and GA34, respectively, reducing the activity of GA and maintaining the balance between bioactive and inactive GA (Claeys et al., 2014). Studies have demonstrated the role of GA2ox1 in regulating rosette branching in some crops. Higher expression levels of GA2ox genes have been correlated to low concentrations of bioactive GAs (Schomburg et al., 2003; Dijkstra et al., 2008; Zhou et al., 2011). Meanwhile, turfgrass (Paspalum notatum Flugge) overexpressing AtGA2ox1 had significantly lower levels of active GA but more tillers than wild-type plants (Agharkar et al., 2007). Overexpression of the OsGA2ox in rice increased tiller number (Lo et al., 2008), consistent with an increased number of tillers observed with the overexpression of PvGA2ox5 and PvGA2ox9 in switchgrass (Panicum virgatum L.) (Wuddineh et al., 2014). On the other hand, silencing of five GA2ox genes in tomatoes significantly increased GA4 content and inhibited lateral branches (Martínez-Bello et al., 2015). Therefore, we speculated that the two tandem genes, BraA07g041560.3C and BraA07g041570.3C, homologous to the Arabidopsis thaliana gene (AT1G78440) encoding a gibberellin 2-oxidase, found in the candidate region might be associated with primary rosette branching in flowering Chinese cabbage. While in the candicate region, the other three genes were not found to be related to branching. BraA07g041530.3C was the homologs gene of AtPGX2, which has been demonstrated regulating root hair development in response to phospho-starved (Zhang et al., 2022). BraA07g041540.3C was the homologs gene of AtVQ10, which has been shown to interact with WRKY33 to affect plant sizes at mature stages in Arabidopsis (Cheng et al., 2012). BraA07g041550.3C was found to be homologous to Arabidopsis At1g78420 (DA2), which encoded RING-type protein with E3 ubiquitin ligase activity and regulated seed size by restricting cell proliferation in the maternal integuments of developing seeds (Xia et al., 2013).
Typically, variations in the promoter regions may lead to changes in gene expression levels (Mito et al., 1996). The expression levels of these two genes were significantly higher in rosette stems of the branching line than in the non-branching line. Detailed analysis revealed differences in the promoter sequences between the parents. The promoter of the BraA07g041560.3C gene missed an ERE cis-regulatory element in branching line CX020, with a difference in the location of a TATA-box and the cis-regulatory element. However, there is no evidence for the role of the ERE cis-regulatory element in the formation of branching. Meanwhile, the promoter sequence of the BraA07g041570.3C gene lacked an MYB cis-regulatory element in CX020. Studies presented associated several genes belonging to the MYB family (MYB2, MYB37, and MYB181) with axillary meristem development and branching in Arabidopsis (Guo and Gan, 2011; Keller et al., 2006; Yang et al., 2018). AtMYB2 protein represses the formation of axillary meristems in response to salt and drought stresses (Jia et al., 2020). Meanwhile, the overexpression of GmMYB181 in Arabidopsis altered the plant architecture, increased lateral branches, and reduced plant height (Yang et al., 2018). Therefore, we hypothesized that the MYB gene control primary rosette branching in flowering Chinese cabbage.
GAs are a large group of diterpenoid natural products characterized by tetracyclic 6-5-6-5 ring derived from ent-gibberellane (MacMillan and Takahashi, 1968; Peters, 2010). The biosynthesis of GA is a complex multi-step process requiring a variety of functional enzymes to catalyze the different intermediates (Wei et al., 2019). GA normally inhibits shoot branching, and plants overexpressing GA catabolic genes and GA-deficient mutants exhibit more shoot branching comparing to the wild-type (Silverstone et al., 1997; Agharkar et al., 2007; Lo et al., 2008). GA regulates internode elongation in rice, where the bioactive GA probably prevents from reaching the nodes below the shoot apex and inhibits internode elongation during the vegetative phase (Sakamoto et al., 2001).In this work, exogenous GA3 application significantly reduced the rosette branches of flowering Chinese cabbage.
Conclusions
The present study identified two tandem genes, BraA07g041560.3C and BraA07g041560.3C, homologous to AT1G78440 encoding GA2ox1 oxidase, as the candidates genes related to primary rosette branching in flowering Chinese cabbage. We detected differences in cis-regulatory elements in the promoter sequences of BraA07g041560.3C and BraA07g041570.3C between the branching and non-branching lines, which indicated the role of genes in regulating branching. These results provide valuable information for revealing the branching regulatory mechanisms in flowering Chinese cabbage. Further studies should investigate and conform the possible function and the promoter activity of BraA07g041560.3C and BraA07g041570.3C in flowering Chinese cabbage.
Data availability statement
The data presented in the study are deposited in the SRA repository of the National Center for Biotechnology Information, accession number PRJNA908111 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA908111.
Ethics statement
The authors note that this research was performed and reported in accordance with ethical standards of scientific conduct.
Author contributions
YZ and HF designed the experiments. JG and JL conducted the experiments and wrote the manuscript. JG, JL, and QY performed the data analysis. YZ revised the manuscript. All authors reviewed and approved this manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (Grant No. 31972404) and Science Study Foundation of Liaoning (LJKZ0640) and China Agriculture research system (CARS-23).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1083528/full#supplementary-material
Supplementary Figure 1 | The full-length of gene BraA07g041560.3C.
Supplementary Figure 2 | The full-length of gene BraA07g041570.3C.
Supplementary Figure 3 | The promoter sequence of BraA07g041560.3C.
Supplementary Figure 4 | The promoter sequence of BraA07g041570.3C.
Supplementary Table 1 | KASP primer sequence used in this study.
Supplementary Table 2 | SNP site information.
Supplementary Table 3 | The components of the KASP reaction mixture.
Supplementary Table 4 | The 178 genes identified in the candidate region based on the gene information in the Brassica database.
References
Agharkar, M., Lomba, P., Altpeter, F., Zhang, H. N., Kenworthy, K., Lange, T. (2007). Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 5, 791–801. doi: 10.1111/j.1467-7652.2007.00284.x
Cao, X. W., Cui, H. M., Yao, Y., Xiong, A. S., Hou, X. L., Li, Y. (2017). Effects of endogenous hormones on variation of shoot branching in a variety of non-heading Chinese cabbage and related gene expression. J. Plant Biol. 60, 343–351. doi: 10.1007/s12374-016-0124-2
Cao, X. W., Cui, H. M., Li, J. (2016). Heritability and gene effects for tiller number and leaf number in non-heading Chinese cabbage using joint segregation analysis. Scientia Horticulturae 203, 199–206. doi: 10.1016/j.scienta.2016.03.018
Cheng, Z. K., Liu, Z. G., Xu, Y. C., Ma, L. L., Chen, J. Y., Gou, J. Q., et al. (2021). Fine mapping and identification of the candidate gene BFS for fruit shape in wax gourd (Benincasa hispida). Theor. Appl. Genet. 134, 3983–3995. doi: 10.1007/s00122-021-03942-8
Cheng, Y., Zhou, Y., Yang, Y. (2012). Structural and functional analysis of VQ motif-containing proteins in arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. (Rockville) 159 (2), 810. doi: 10.1104/pp.112.196816
Chen, X. L., Zhou, X. Y., Xi, L., Li, J. X., Zhao, R. Y., Ma, N., et al. (2013). Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema x grandiflora cv. jinba). PLoS One 8, e61717. doi: 10.1371/journal.pone.0061717
Claeys, H., De Bodt, S., Inzé, D. (2014). Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 19, 231–239. doi: 10.1016/j.tplants.2013.10.001
De Jong, M., George, G., Ongaro, V., Williamson, L., Willetts, B., Ljung, K., et al. (2014). Auxin and strigolactone signaling are required for modulation of Arabidopsis shoot branching by nitrogen supply. Plant Physiol. 166, 384–395. doi: 10.1104/pp.114.242388
Dijkstra, C., Adams, E., Bhattacharya, A., Page, A. F., Anthony, P., Kouurmptli, S., et al. (2008). Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Rep. 27, 463–470. doi: 10.1007/s00299-007-0471-z
Ehrenreich, I. M., Stafford, P. A., Purugganan, M. D. (2007). The genetic architecture of shoot branching in Arabidopsis thaliana: A comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics 176, 1223–1236. doi: 10.1534/genetics.107.071928
Fichtner, F., Barbier, F. F., Annunziata, M. G., Feil, R., Olas, J. J., Mueller-Roeber, B., et al. (2021). Regulation of shoot branching in Arabidopsis by trehalose 6-phosphate. New Phytol. 229, 2135–2151. doi: 10.1111/nph.17006
Giovannoni, J. J., Wing, R. A., Ganal, M. W., Tanksley, S. D. (1991). Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Res. 19, 6553–6558. doi: 10.1093/nar/19.23.6553
Guo, Y. F., Gan, S. S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156, 1612–1619. doi: 10.1104/pp.111.177022
Hedden, P., Phillips, A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. doi: 10.1016/S1360-1385(00)01790-8
Holalu, S. V., Reddy, S. K., Blackman, B. K., Finlayson, S. A. (2020). Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant,Cell Environ. 43, 2224–2238. doi: 10.1111/pce.13824
Jia, T., Zhang, K., Li, F., Huang, Y. F., Fan, M. M., Huang, T., et al. (2020). The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep 39, 1755–1765. doi: 10.1007/s00299-020-02602-3
Kebrom, T. H., Spielmeyer, W., Finnegan, E. J. (2013). Grasses provide new insights into regulation of shoot branching. Trends Plant Sci. 18, 41–48. doi: 10.1016/j.tplants.2012.07.001
Keller, T., Abbott, J., Moritz, T., Doerner, P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611. doi: 10.1105/tpc.105.038588
Lei, L., Zheng, H. L., Bi, Y. L., Yang, L. M., Liu, H. L., Wang, J. G., et al. (2020). Identification of a major QTL and candidate gene analysis of salt tolerance at the bud burst stage in rice (Oryza sativa l.) using QTL-seq and RNA-seq. Rice 13, 22. doi: 10.1186/s12284-020-00416-1
Li, Y. X. (2018). Genetic analysis and QTL mapping of tillering in purple caitai (Wuhan(HB: Huazhong Agricultural University).
Lian, Q., Fu, Q. S., Xu, Y. Y., Hu, Z. C., Zheng, J., Zhang, A. A., et al. (2021). QTLs and candidate genes analyses for fruit size under domestication and differentiation in melon (Cucumis melo l.) based on high resolution maps. BMC Plant Biol. 21, 126. doi: 10.1186/s12870-021-02904-y
Li, B., Gao, J., Chen, J., Wang, Z., Shen, W., Yi, B., et al. (2020a). Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor. Appl. Genet. 133, 771–783. doi: 10.1007/s00122-019-03506-x
Li, W., Ma, Q., Yin, P., Wen, J., Pei, Y., Niu, L., et al. (2021). The GA 20-oxidase encoding gene MSD1 controls the main stem elongation in medicago truncatula. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.709625
Li, P., Su, T., Zhang, B., Li, P., Xin, X., Yue, X., et al. (2020b). Identification and fine mapping of qSB.A09, a major QTL that controls shoot branching in brassica rapa ssp. chinensis makino. Theor. Appl. Genet. 133, 1055–1068. doi: 10.1007/s00122-020-03531-1
Liu, J., Gong, J. Y., Wei, X., Yang, S. H., Huang, X. H., Li, C., et al. (2020). Dominance complementation of Hd1 and Ghd8 contributes to extremely late flowering in two rice hybrids. Mol. Breed. 40, 76. doi: 10.1007/s11032-020-01162-4
Liu, G., Zhao, T., You, X. Q., Jiang, J. B., Li, J. F., Xu, X. Y. (2019). Molecular mapping of the Cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato (Solanum lycopersicum). BMC Plant Biol. 19, 15. doi: 10.1186/s12870-018-1616-7
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618. doi: 10.1105/tpc.108.060913
MacMillan, J., Takahashi, N. (1968). Proposed procedure for the allocation of trivial names to the gibberellins. Nature 217, 170–171. doi: 10.1038/217170a0
Martínez-Bello, L., Moritz, T., López-Díaz, I. (2015). Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 66, 5897–5910. doi: 10.1093/jxb/erv300
Mathan, J., Bhattacharya, J., Ranjan, A. (2016). Enhancing crop yield by optimizing plant developmental features. Development 143, 3283–3294. doi: 10.1242/dev.134072
Michelmore, R. W., Paran, I., Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Mito, N., Wimmers, L., Bennett, A. (1996). Sugar regulates mRNA abundance of h+-ATPase gene family members in tomato. Plant Physiol. 112, 1229–1236. doi: 10.1104/pp.112.3.1229
Morris, S. E., Cox, M. C., Ross, J. J., Krisantini, S., Beveridge, C. A. (2005). Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 138, 1665–1672. doi: 10.1104/pp.104.058743
Muntha, S. T., Zhang, L., Zhou, Y., Zhao, X., Hu, Z., Yang, J., et al. (2019). Phytochrome a signal transduction 1 and CONSTANS-LIKE 13 coordinately orchestrate shoot branching and flowering in leafy Brassica juncea. Plant Biotechnol. J. 17, 1333–1343. doi: 10.1111/pbi.13057
Niu, L. J., Shi, F. Y., Feng, H., Zhang, Y. (2019). Efficient doubled haploid production in microspore culture of zengcheng flowering Chinese cabbage (Brassica campestris L. ssp. Chinensis [L.] makino var. utilis tsen et Lee). Scientia Hortic. 245, 57–64. doi: 10.1016/j.scienta.2018.09.076
Okada, K., Wada, M., Takebayashi, Y., Kojima, M., Sakakibara, H., Nakayasu, M., et al. (2020). Columnar growth phenotype in apple results from gibberellin deficiency by ectopic expression of a dioxygenase gene. Tree Physiol. 40, 1205–1216. doi: 10.1093/treephys/tpaa049
Peters, R. J. (2010). Two rings in them all: The labdane-related diterpenoids. Natural Product Rep. 27, 1521–1530. doi: 10.1039/C0NP00019A
Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., et al. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125, 1508–1516. doi: 10.1104/pp.125.3.1508
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A., Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. doi: 10.1105/tpc.005975
Silverstone, A. L., Mak PiuYing, A., Casamitjana Martinez, E., Sun, T. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 (3), 1087–1099. doi: 10.1038/s41598-017-10823-y
Singh, V. K., Mangalam, A. K., Dwivedi, S., Naik, S. (1998). Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques 24, 318–319. doi: 10.2144/98242pf02
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Teichmann, T., Muhr, M. (2015). Shaping plant architecture. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00233
Wang, X. W., Kole, C. (2015). The Brassica rapa Genome: Economic/Academic Importance of Brassica rapa (New York: Springer-Verlag Berlin Heidelberg), 1–2. doi: 10.1007/978-3-662-47901-8
Wang, Y., Li, J. Y. (2011). Branching in rice. Curr. Opin. Plant Biol. 14, 94–99. doi: 10.1016/j.pbi.2010.11.002
Wang, N., Liu, Z. Y., Zhang, Y., Li, C. Y., Feng, H. (2018b). Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theor. Appl. Genet. 131, 673–684. doi: 10.1007/s00122-017-3028-8
Wang, B., Smith, S. M., Li, J. (2018a). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wang, C., Tang, S., Zhan, Q., Hou, Q., Zhao, Y., Zhao, Q., et al. (2019). Dissecting a heterotic gene through GradedPool-seq mapping informs a rice-improvement strategy. Nat. Communication 10 (1), 2982. doi: 10.1038/s41467-019-11017-y
Wei, C. H., Zhu, C. Y., Yang, L. P., Zhao, W., Ma, R. X., Li, H., et al. (2019). A point mutation resulting in a 13 bp deletion in the coding sequence of Cldf leads to a GA-deficient dwarf phenotype in watermelon. Horticulture Res. 6, 132. doi: 10.1038/s41438-019-0213-8
Wuddineh, W., Mazarei, M., Zhang, J. Y., Poovaiah, C., Mann, D., Ziebell, A., et al. (2014). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum l.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 13, 636–647. doi: 10.1111/pbi.12287
Xia, T., Li, Na., Dumenil, J., Li, J., Kamenski, A., Bevan, M. W., et al. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in arabidopsis. Plant Cell 25, 3347–3359. doi: 10.1105/tpc.113.115063
Xu, X., Ji, J., Xu, Q., Qi, X., Weng, Y., Chen, X. (2018). The major-effect quantitative trait locus CsARN6.1 encodes an AAA ATPase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 93, 917–930. doi: 10.1111/tpj.13819
Xu, J., Zha, M., Li, Y., Ding, Y., Chen, L., Ding, C., et al. (2015). The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa l.). Plant Cell Rep. 34, 1647–1662. doi: 10.1007/s00299-015-1815-8
Yang, S. J., Tian, X. X., Wang, Z. Y., Wei, X. C., Zhao, Y. Y., Su, H. N., et al. (2021). Fine mapping and candidate gene identification of a white flower gene BrWF3 in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.646222
Yang, H., Xue, Q., Zhang, Z. Z., Du, J. Y., Yu, D. Y., Huang, F. (2018). GmMYB181, a soybean R2R3-MYB protein, increases branch number in transgenic Arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01027
Zhang, Q., Deng, A. W., Xiang, M. (2022). The root hair development of pectin polygalacturonase PGX2 activation tagging line in response to phosphate deficiency. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.862171
Zhou, B., Peng, D., Lin, J., Huang, X., Peng, W., He, R., et al. (2011). Heterologous expression of a gibberellin 2-oxidase gene from Arabidopsis thaliana enhanced the photo-synthesis capacity in Brassica napus l. J. Plant Biol. 54, 23–32. doi: 10.1007/s12374-010-9139-2
Keywords: flowering Chinese cabbage, primary rosette branches, gene identification, tandem genes, GA2ox1 oxidase
Citation: Guan J, Li J, Yao Q, Liu Z, Feng H and Zhang Y (2022) Identification of two tandem genes associated with primary rosette branching in flowering Chinese cabbage. Front. Plant Sci. 13:1083528. doi: 10.3389/fpls.2022.1083528
Received: 29 October 2022; Accepted: 01 December 2022;
Published: 19 December 2022.
Edited by:
Xiangshu Dong, Yunnan University, ChinaReviewed by:
Tongbing Su, Beijing Academy of Agricultural and Forestry Sciences, ChinaXiaoming Song, North China University of Science and Technology, China
Copyright © 2022 Guan, Li, Yao, Liu, Feng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Zhang, emhhbmd5dW41MTFAc3lhdS5lZHUuY24=
†These authors have contributed equally to this work
 Jian Guan
Jian Guan Jinyan Li
Jinyan Li Qingyu Yao
Qingyu Yao Zhiyong Liu
Zhiyong Liu Hui Feng
Hui Feng Yun Zhang
Yun Zhang