- 1Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China
- 2University of the Chinese Academy of Sciences, Beijing, China
- 3Bright Agricultural Development (Group) Co., Ltd., Shanghai, China
- 4National Key Laboratory of Plant Molecular Genetics, Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China
- 5Institute of Advanced Biotechnology and School of Life Sciences, Southern University of Science and Technology, Shenzhen, China
Protochlorophyllide oxidoreductase (POR) plays a key role in catalyzing the light-dependent reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), and thus promotes the transit from etiolated seedlings to green plants. In this study, by exploring ethyl methanesulfonate (EMS)-mediated mutagenesis in Chenopodium quinoa NL-6 variety, we identified a mutant nl6-35 that displays faded green leaf and reduced chlorophyll (Chl) and carotenoid contents. Bulk segregant analysis (BSA) revealed that a mutation in CqPORB gene is genetically associated with the faded green leaf of the nl6-35 mutant. Further study indicates that the nl6-35 mutant exhibits abnormal grana stacks and compromised conversion of Pchlide to Chlide upon illumination, suggesting the important role of CqPORB in producing photoactive Pchlide. Totally three CqPOR isoforms, including CqPORA, CqPORA-like, and CqPORB are identified in NL-6 variety. Transcriptional analysis shows that the expression of all these three CqPOR isoforms is regulated in light- and development-dependent manners, and in mature quinoa plants only CqPORB isoform is predominantly expressed. Subcellular localization analysis indicates that CqPORB is exclusively localized in chloroplast. Together, our study elucidates the important role of CqPORB in the regulation of Chl biosynthesis and chloroplast development in quinoa.
Introduction
Chenopodium quinoa Willd. (quinoa) is an annual pseudocereal crop that belongs to Amaranthaceae family, and this family also encompasses other economically important species such as spinach (Spinacia olereaceae L.) and sugar beet (Beta vulgaris L.) (Mizuno et al., 2020). Quinoa originated from the Andean region of South America and is now cultivated worldwide, especially since the declaration of the year 2013 as the International Year of Quinoa by the United Nations Food and Agricultural Organization (FAO) (Mizuno et al., 2020). Compared with the major staple cereal crops, such as rice, wheat, oat, and maize, that lack enough lysine for the daily requirement of human being, quinoa seeds contain a more balanced amino acid profile, particularly for the rich of lysine, histidine, and methionine (Dakhili et al., 2019). In addition, quinoa is rich in protein, starch, lipid, a variety of vitamins, minerals, and some health-beneficial phytochemicals (Graf et al., 2016; Cai and Gao, 2020). Due to its exceptional nutritional value, quinoa is considered as a “perfect and strategic food” by FAO (Dakhili et al., 2019), and it was selected as desirable food for astronauts in space by the National Aeronautics and Space Administration (NASA) of the United States (Yasui et al., 2016). In addition to its high nutritional value, quinoa is regarded as a halophyte that tolerates multiple abiotic stresses, including high salinity, drought, and low temperature, which makes it a promising crop to grow in marginal areas (Hariadi et al., 2011; Böhm et al., 2018). In recent years, three quinoa reference genomes from the accessions of Kd, QQ74, and Real have been independently sequenced and assembled (Yasui et al., 2016; Jarvis et al., 2017; Zou et al., 2017), which opens a door to study the agronomic traits of quinoa in a genome-wide and molecular levels.
Although quinoa was domesticated over 7,000 years ago in the Andean region of South America (Dillehay et al., 2007), the average yield of quinoa is still relatively low and highly dependent on climate and growth conditions. Therefore, yield improvement is currently an urgent issue for quinoa breeding to meet the increasing of global demand for quinoa. Photosynthesis is an important biological process that enables plants to convert sunlight energy to carbohydrates, and thus promotes plant growth and yield (Eberhard et al., 2008). Engineering of photosynthesis machinery is a feasible and useful strategy to improve crop yield, which has been reported in several studies (Long et al., 2015; Chen et al., 2020). Chloroplast is the cellular organelle in plants that serves as the site for photosynthesis, and chlorophyll (Chl) biosynthesis occurred in chloroplast is essential for plants to execute photosynthesis. Chl is the most abundant pigment in land plants and algae that is indispensable for light absorption and energy transfer during photosynthesis (Allen et al., 2011; Wang and Grimm, 2021). Chl biosynthesis has been studied for decades, and it has been known that enzymes, including magnesium chelatase (MgCh), Mg-protoporphyrin IX methyltransferase (MgMT), Mg-protoporphyrin IX monomethyl ester cyclase (MgCy), protochlorophyllide oxidoreductase (POR), divinylchlorophyllide reductase (DVR), and Chl synthase, are involved in this process (Tanaka and Tanaka, 2007). Chl biosynthesis is tightly controlled in response to environmental light signal. In darkness, Chl biosynthesis is halted at the step of protochlorophyllide (Pchlide), which is the precursor of chlorophyllide (Chlide), while upon illumination, Pchlide is converted to Chlide, and subsequently Chl is synthesized (Reinbothe et al., 2010). The conversion of Pchlide to Chlide is considered as a key step for de-etiolation in plants (Masuda et al., 2003). Studies have shown that POR proteins are required for catalyzing the light-dependent reduction of Pchlide to Chlide in plants (Reinbothe et al., 1999; Sakuraba et al., 2013). In the dark, POR and its substrate Pchlide are assembled into prolamellar bodies (PLBs) in etioplasts, and upon light exposure, photon is absorbed by the bound pigment and POR catalyzes the reduction of the C17-C18 double bond of the D-ring of Pchlide, leading to the production of Chlide (Heyes and Hunter, 2005; Blomqvist et al., 2008).
PORs are highly conserved proteins that are widely identified in angiosperms (Reinbothe et al., 2010). In Arabidopsis, there are three POR isoforms (AtPORA, AtPORB, and AtPORC) that display distinct developmental and light-regulated expression patterns (Armstrong et al., 1995; Masuda et al., 2003).AtPORA primarily presents in etiolated seedlings and its transcript level is rapidly decreased upon illumination (Armstrong et al., 1995). AtPORB is expressed in both etiolated seedlings and light-grown plants, while AtPORC is rarely detected in etiolated seedlings but is increased after several hours of light exposure (Masuda et al., 2003). Both AtPORB and AtPORC are required for bulk Chl synthesis throughout whole plant development in Arabidopsis (Paddock et al., 2010; Paddock et al., 2012). Under normal light conditions, neither AtporB nor AtporC single mutant exhibits obvious phenotypes, but AtporB AtporC double mutant displays seedling-lethal xantha phenotype at the cotyledon stage, suggesting that AtPORB and AtPORC function redundantly in the regulation of light-dependent Chl biosynthesis in Arabidopsis (Frick et al., 2003). In rice, there are only two POR isoforms, namely OsPORA and OsPORB (Kwon et al., 2017). The expression patterns of these two OsPOR genes are reminiscent of Arabidopsis PORA and PORB genes. OsPORA gene is mainly expressed in the dark during early leaf development, while OsPORB is expressed throughout the whole leaf growth stage independent of light signal. faded green leaf (fgl) mutant, which is disrupted in OsproB gene, displays faded green leaf, which is caused by the reduced Chl and carotenoid contents (Sakuraba et al., 2013). In addition, necrotic lesions occurring from the top area of older leaf blades are detected in the fgl mutant, which are largely caused by the increased accumulation of non-photoactive Pchlide and subsequently increased singlet oxygen upon illumination (Sakuraba et al., 2013). The association of Pchlide accumulation with singlet oxygen production has been well-elucidated in flu mutant in Arabidopsis. flu mutation leads to increased overaccumulation of Pchlide in darkness, which in turn results in a rapid elevation of singlet oxygen in chloroplasts upon illumination, and finally triggers cell death (Op Den Camp et al., 2003; Kim et al., 2012). In addition to Arabidopsis and rice, the function of POR genes has also been characterized in many other plant species, including barley (Hordeum vulgare L.) (Holtorf et al., 1995), wheat (Triticum aestivum) (Teakle and Griffiths, 1993), maize (Zea mays) (Selstam et al., 2002), tobacco (Nicotiana tabacum L.) (Selstam et al., 2002), and cabbage (Brassica oleracea cv. Capitata) (Nie et al., 2021).
In this study, we performed genetic screening for the mutants with reduced Chl content in quinoa, and a mutant with faded green leaf was identified. Based on bulk segregant analysis (BSA), we found that the faded green leaf phenotype was caused by a mutation in CqPORB gene. Further study revealed that disruption of CqPORB gene resulted in abnormal grana stacks and reduced conversion rate of photoactive Pchlide to Chlide upon light exposure. Expression patterns of CqPOR genes in response to light and in different developmental stages were also analyzed in this study.
Materials and methods
Plant materials and growth conditions
The quinoa variety NL-6 and Longli-4 were obtained from Prof. Heng Zhang and Prof. Pengshan Zhao, respectively. Quinoa seeds were directly sown in soil and kept at a growth chamber at 28°C under a light cycle of 12 h light/12 h dark. The phenotype of plants was observed and photographed after growth for indicated days. Arabidopsis Columbia (Col-0) ecotype was used for the transformation of 35S::CqPORB-GFP. Arabidopsis seeds were sterilized and sown on half Murashige and Skoog (MS) solid medium and kept at 4°C for 3 days before being transferred to an incubator at 22°C with a long-day cycle (16 h light/8h dark).
Ethyl methanesulfonate (EMS) mutagenesis in quinoa
For EMS treatment, a set of healthy seeds of quinoa NL-6 variety were presoaked in distilled water at 4°C for 8 h, and then the seeds were immersed in 1.0% EMS solution (v/v) (Sigma-Aldrich, M0880) and incubated at 4°C for 12 h. After treatment, a volume of 10% sodium thiosulphate (w/v) solution was added to the EMS solution, and the quinoa seeds were gently stirred and stand for 10 min before they were washed five times with distilled water to neutralize and remove EMS. The mutated seeds were sown in field for the collection of progeny seeds.
Measurement of photosynthetic pigments
For Chl and carotenoid measurement, the leaves of four quinoa seedlings grown in the conditions of 28°C/26°C day/night temperatures and a 12 h/12 h light (216 µmol m−2·s−1)/dark cycle were sampled. Total pigments were extracted using 95% ethanol. The contents of Chl a, Chl b and carotenoid were measured using a NanoDrop™ 2000C UV-vis spectrophotometer (Thermo Scientific) as described previously (Lichtenthaler, 1987).
Total Pchlide and Chlide were calculated following the method described previously (Kwon et al., 2017). 20 seedlings were grown in darkness for 5 days or further exposed to white light for 10 s, 30 s, and 5 min. The seedlings were homogenized in 500 μL of ice-cold 80% acetone and incubated for 8 h in darkness (4°C). After spinning for 5 min, 150 μL of the supernatant was transferred to a 96-well plate (black, LABSELECT, 33113) that is made of polypropylene. Fluorescence was detected at an excitation wavelength of 440 nm, and emissions were collected from top between 600 nm and 720 nm at room temperature using a Thermo Scientific Varioskan Flash Plate Reader.
Quinoa protoplast assay
The quinoa seeds were sown directly in soil and grown in a light incubator at 22°C (light)/20°C (dark) in long-day conditions (16 h light/8 h dark). The first or second pair of the true leaves of quinoa seedlings after growth for 15-20 days were collected using adhesive tape and dissociated in a dissociation solution (1.5% Cellulase R10, 0.4% Macerozyme R10, 0.5 M Mannitol, 20 mM KCl, 20 mM MES (pH 5.7), 10 mM CaCl2, and 0.1% BSA). The dissociation was carried out on a shaker with a speed of 50 rpm for 2.5 h under darkness at 22°C. The protoplasts were filtered through a Filter Mesh 100 and the filtered cells were centrifuged at 100 g for 3 min. The supernatant was removed and the cells were washed with 5-10 mL of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES (pH 5.7), and 0.1% (w/v) glucose) for three time with gentle shake. After washing, appropriate amount of MMg solution (0.4 M Mannitol, 15 mM MgCl2, and 4 mM MES (pH 5.7)) was added to adjust the cell concentration to about 2 × 105 cells/mL. Subsequently, 100 μL protoplasts, 5 μL CqPORB-GFP plasmid (800-1000 ng/μL), and 105 μL 40% (w/v) PEG (0.2 M mannitol and 100 mM Ca(NO3)2) were slowly mixed, and the mixture was incubated at room temperature for 15 min. The transformation was terminated by adding 500 μL W5 solution. After centrifugation at 100 g for 3 min, the supernatant was discarded and protoplasts were washed twice with W5 solution. The protoplasts were transferred to cell culture plate and incubated at 22°C under low light conditions (5 µmol·m−2·s−1) for 36 h. Then protoplasts were collected and total Chl was extracted under light conditions, and Chl was measured according to the protocol described above.
Transmission electron microscopy (TEM) analysis
The first true leaves of NL-6 and nl6-35 harvested from 10-day-old seedlings grown in low-light conditions were used for the detection of granum in chloroplast. The samples were immersed in TEM fixative (G1102, Servicebio, China) containing 2.5% glutaraldehyde overnight at 4°C, and rinsed with 0.1 M sodium phosphate buffer (pH 7.4), and post-fixed with a 1% (w/v) aqueous solution of OsO4 at room temperature for 7 h. The materials were then dehydrated in a gradient ethanol series, and embedded in EMBed 812 resin (90529-77-4, SPI, USA). Tissues were processed into ultrathin sections, and stained with a 2% (w/v) uranium acetate and subsequently with 2.6% (w/v) lead citrate solution. The chloroplast was observed using a TEM device (HT7800, HITACHI, Japan).
BSA-seq analysis of mutations in nl6-35
For BSA-seq analysis, nl6-35 was backcrossed with its parent NL-6. In F2 population, 80 plants with faded green leaf were collected and pooled together for total DNA isolation using the DNeasy Plant Maxi kit (Qiagen). The library construction, sequencing, and bioinformatic analysis were all conducted by Personalbio (Shanghai, China). 150 bp pair-end sequencing libraries were generated and sequenced with an Illumina NovaSeq 6000 platform (Illumina lnc., San Diego, CA, USA). The clean reads that were filtered from raw reads were aligned to the reference genome of NL-6 using BWA (Li and Durbin, 2009). Picard 1.107 tools were used to convert alignment files to bam files and remove duplicated reads. GATK was applied to conduct local realignment around Indels and single-nucleotide polymorphism (SNP) calling (Zhu et al., 2014). Annovar program (Wang et al., 2010) was used to annotate all the SNPs based on the GFF3 files of the reference genome of NL-6. The SNP-index (Takagi et al., 2013) was calculated for all SNPs to identify candidate regions associated with the mutant trait, and the SNPs with a read depth ≥ 8 and SNP-index ≥ 0.3 were retained, and the average SNP-index across a 1-Mb genomic interval was measured individually using a 100-kb sliding-window approach and then plotted against all the chromosomes of quinoa genome. Finally, the SNPs with SNP-index = 1 were chosen for the manual analysis of candidate genes. To verify the SNP in CqPORB gene in nl6-35 mutant, genomic sequence of CqPORB gene was amplified and sequenced using traditional Sanger sequencing. The gene-specific primers used for sequencing are listed in Supplementary Table 1.
Molecular marker-associated analysis of CqPORB gene mutation in nl6-35
The identified SNP in CqPORB gene was used to design Kompetitive Allele Specific PCR (KASP) marker. The developed marker was used for the linkage analysis of the F2 population plants derived from a hybridization between nl6-35 and its parent NL-6. Totally 93 plants with mutant phenotype and 95 plants with wild type phenotype were selected for KASP analysis. Besides, the mutant-type plants from the F2 progenies of Longli-4 × nl6-35 crossing population were also selected for linkage analysis using the gene-specific KASP marker in CqPORB gene.
Protein sequence alignment and phylogenetic analysis
Homologs of CqPORB were obtained by a Blastp search of GenomeNet (www.genome.jp), Plant GDB (www.plantgdb.org), and The Rice Annotation Project Database. Multiple protein sequences were aligned by ClustalW and conserved regions were visualized by CLC sequence viewer 8. The phylogenetic tree was constructed by iqtree with the best-fit model and 1,000 replicates of ultrafast bootstrap (Minh et al., 2020). The resulting tree files were submitted to iTOL (Letunic and Bork, 2021) for modification.
Gene expression analysis
To analyze the expression pattern of CqPORs in response to light, surface sterilized seeds were sowed on ½ MS solid medium and incubated at 22°C for 3 days, and then etiolated seedlings were collected after exposure to white light for 0, 3, and 6 h. For the analysis of CqPOR gene expression in different developmental stages, the first true leaves of 10-day-old seedlings, leaves of early flowering stage (30 d), and late flowering stage (50 d) were sampled. Three independent replicates were performed. Total RNAs of all these samples were extracted by using EastepTM Super Total RNA Extraction Kit (Promega, United States). cDNAs were synthesized using HiScript III RT SuperMix for qPCR (+gDNA wiper) according to the manufacturer’s instructions (Vazyme, Nanjing, China). qRT-PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) and Bio-Rad CFX connect real time detection system (BIO-RAD, United States). Primers used for qRT-PCR are listed in Supplementary Table 1. The transcript levels of all three CqPOR genes in four different tissues were obtained from previous RNA-seq data (Jiang et al., 2022). Gene expression patterns of CqPOR genes in different tissues were visualized in heat map using TBtools software package (Minh et al., 2020).
Plasmid construction and plant transformation
CqPORB was amplified using the cDNA template generated from the leaves of NL-6 using gene-specific primers (Supplementary Table 1). CqPORB was constructed into plant expression binary vector pCAMBIA1305, which carries a GFP reporter gene driven by the cauliflower mosaic virus (CaMV) 35S promoter. The recombinant plasmid CqPORB-GFP was introduced into the competent cells of Agrobacterium tumefaciens GV3101 strain, which was further used for transformation into Nicotiana benthamiana and Arabidopsis thaliana (Columbia). For transient expression in N. benthamiana, GV3101 strain harboring CqPORB-GFP was infiltrated into leaves according to the protocol described previously (Liu et al., 2010). Transformation of Arabidopsis plants was performed by using floral dip method (Clough and Bent, 1998). To detect the subcellular localization of CqPORB, GFP fluorescence was detected by Leica confocal laser scanning microscope SP8 with 488 nm excitation light and 500-550 nm emission light.
Results
nl6-35 mutant exhibits reduced Chl
To investigate the regulatory mechanism of Chl biosynthesis in quinoa, we performed EMS-mediated mutagenesis for quinoa NL-6 variety and screened for mutants that displayed a reduced Chl content. NL-6 is a coastal germplasm that originates from lowland Chilean and exhibits relatively stable agronomic traits (Benlhabib et al., 2016). Here we report one of the mutants, designed nl6-35, that showed faded green leaf when grown in both greenhouse and field (Figure 1A, Supplementary Figures 1A, B). Specifically, we noted that at cotyledon stage, there was no significant difference between the wild type NL-6 and nl6-35 (Supplementary Figure 1C), but the leaves of nl6-35 became faded green after growth for approximately 14 days under white light (Figure 1A). Accompanying the faded green leaf, the phenotype of curled leaf was also observed in the nl6-35 mutant, but not in the wild type NL-6 (Figure 1B). Genetic analysis indicated that the phenotypes of faded green leaf and curled leaf were tightly associated, suggesting that they are likely caused by a same mutation in the nl6-35 mutant. Quantification of Chl content revealed that Chl a and Chl b, and total Chl contents were significantly reduced in the nl6-35 mutant (Figure 1C), but the ratio of Chl a/Chl b was slightly increased in the nl6-35 mutant (Figure 1D). In addition, the carotenoid content was also reduced by approximately 15.6% in the nl6-35 (Figure 1E). These results suggested that the faded green leaf in nl6-35 was likely caused by the reduced Chl and carotenoid contents.
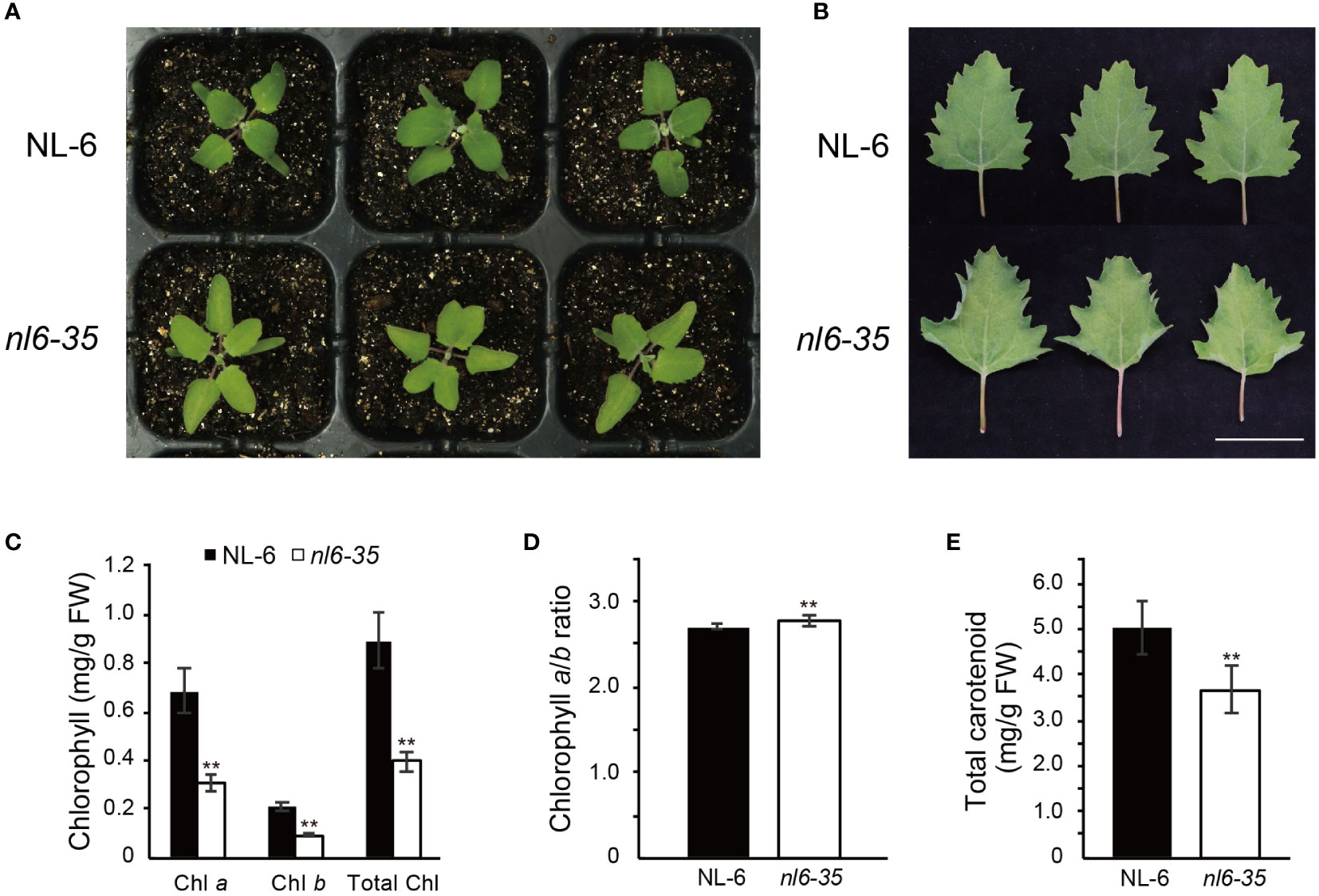
Figure 1 Identification of quinoa mutant nl6-35 that exhibits faded green leaf and reduced Chl. (A) Phenotype of wild type (NL-6) and nl6-35 plants grown on soils for 14 days under a light cycle of 12 h light/12 h dark. (B) Phenotype of the leaves of 30-day-old plants grown under a light cycle of 12 h light/12 h dark. Scale bar, 5 cm. (C) Measurement of Chl a, Chl b, and total Chl in wild type (NL-6) and nl6-35 plants. Values are the means ± SD of six biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test). (D) The ratio of Chl a/Chl b in wild type (NL-6) and nl6-35 plants. Values are the means ± SD of six biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test). (E) Quantification of total carotenoid content in each genotype. Values are the means ± SD of six biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test).
Identification of mutations in nl6-35 by using BSA−seq
To identify the mutated gene in nl6-35 that is responsible for the phenotype of faded green leaf, nl6-35 was backcrossed with its parent NL-6. The F1 generation showed a wild type phenotype and F2 population exhibited a segregation of 3:1 (Table 1), indicating that nl6-35 is a recessive mutation controlled by a single gene. To perform BSA-seq analysis, 80 plants of F2 population with faded green leaf were collected and mixed for whole genome sequencing. After filtering the raw reads, a total of 324,781,844 high quality reads were obtained, and 99.94% of the reads were mapped to NL-6 reference genome (1.39 GB), with an average of 34× reads coverage. Totally 11,960 SNPs were identified after removing the ones with low coverage (<8 reads). Because EMS mutagenesis most frequently causes C to T or G to A change in the genome, so we searched for these two types of nucleotide acid changes and finally obtained 3,013 SNPs with C to T or G to A change (Figure 2A, Supplementary Figure 2, Supplementary Table 2). Among these SNPs, 35 were identified with a mutation rate of 100%, and 15 of them were located on chromosome (chr)17 (Supplementary Table 3), suggesting that the objective gene is most likely located on chr17. Analysis of these 15 SNPs showed that 14 SNPs were located either in intronic or intergenic regions (Supplementary Table 3), while one SNP (G2258A) was identified in the fourth exon region of CqNL-6_047471, resulting in an amino acid change of Leu279 to Phe (Figure 2B). CqNL-6_047471 gene was annotated as a light-dependent protochlorophyllide reductase, which is homologous to POR genes reported in other plant species. It is worth noting that the faded green leaf of the nl6-35 mutant is similar to the phenotype reported in OsporB mutant in rice (Kwon et al., 2017), suggesting that CqNL-6_047471 gene mutation is responsible for the phenotype of nl6-35 mutant in quinoa. Because CqNL-6_047471 gene mutation resulted in a similar phenotype as the OsporB mutant in rice, we named CqNL-6_047471 as CqPORB.
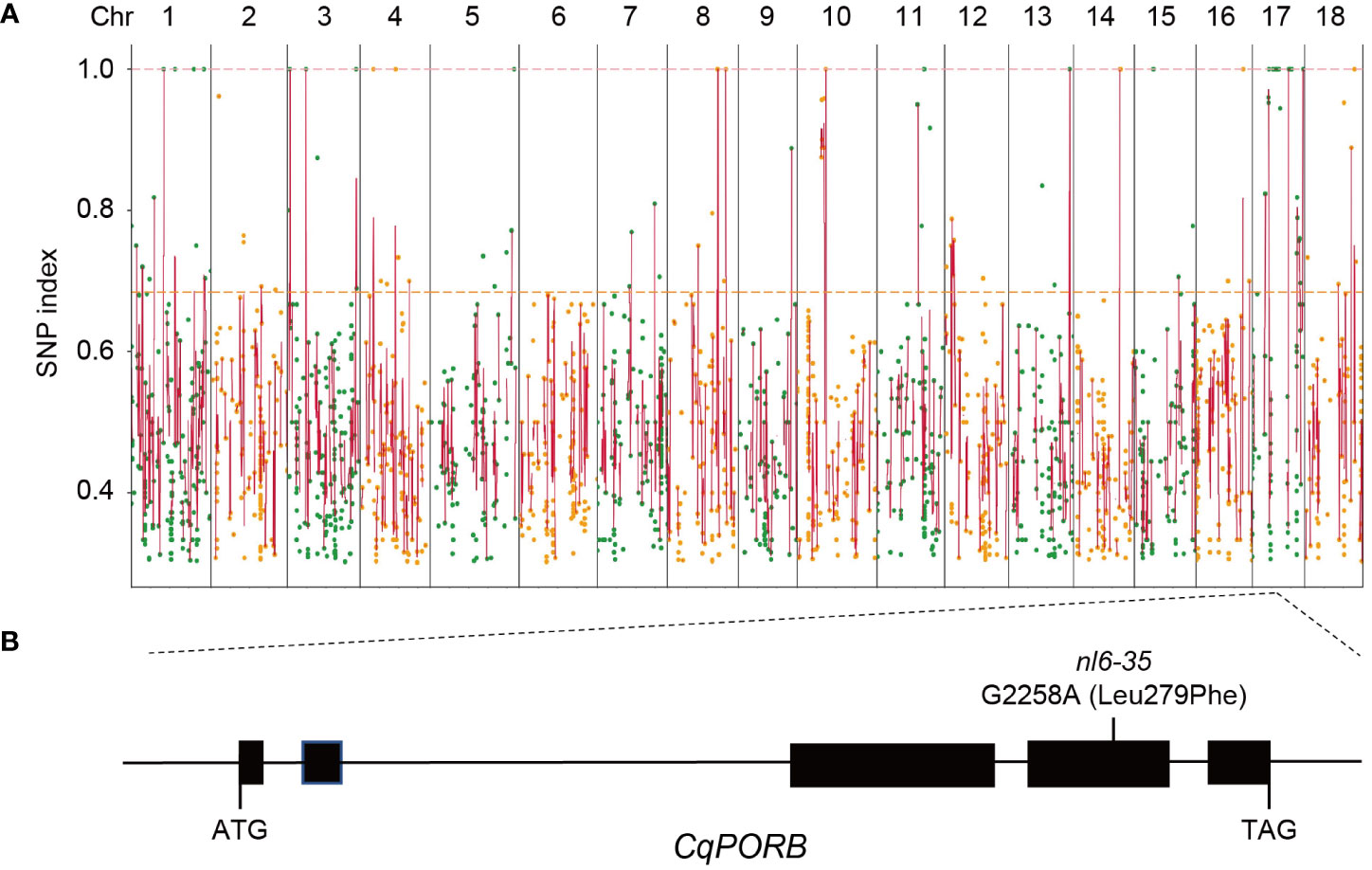
Figure 2 Identification of the mutated genes in nl6-35 using BSA-seq analysis. (A) Identification of SNPs in nl6-35 using the F2 hybridization population of the nl6-35 mutant with its parent NL-6. Round dots indicate all the SNPs with a C to T or G to A change in the nl6-35 mutant. X axis indicates the distribution of SNPs on all 18 chromosomes in quinoa. Y axis indicates the SNP index of each SNP. (B) Diagram illustrating the genomic structure of CqPORB gene. The G2258A mutation (Leu279phe) in nl6-35 mutant is shown.
Validation of the association of CqPORB mutation with nl6-35 mutant phenotype
Currently genetic transformation in quinoa is still technically limited, so we explored other strategies to verify the association of CqPORB gene mutation with the faded green leaf of nl6-35. In the F2 hybridization population of nl6-35 with NL-6, we selected 93 plants that showed faded green leaf phenotype and 95 plants that showed wild type phenotype for the analysis of G2258A mutation in CqPORB gene. To perform genotyping in a high-throughput way, KASP (Kompetitive Allele Specific PCR) molecular marker that targeted this mutation site was designed. KASP analysis showed that in all 93 plants with mutant phenotype, the nucleotide acid G2258 was mutated to A, while in all 95 plants with wild type phenotype, G2258 site was either wild type or heterozygous, with a ratio of 27:68 (close to 1:2), verifying that CqPORB gene mutation is genetically associated with the mutant phenotype of nl6-35 mutant.
In addition, we also crossed nl6-35 mutant with another quinoa variety Longli-4 that originates from Bolivia. In F2 generation, the segregation of the faded green leaf phenotype is approximately 3:1 (Table 1), corroborating that the mutant phenotype in nl6-35 was controlled by a recessive mutation. Similarly, 180 plants with faded green phenotype were selected for KASP analysis. The result showed that all these plants with mutant-type phenotype carried CqPORB allele that were derived from nl6-35 but not from Longli-4 variety, which further supported that CqPORB gene mutation is associated with the faded green leaf of nl6-35 mutant.
To further demonstrate that the reduced chlorophyll content in nl6-35 mutant was caused by the CqPORB gene mutation, we generated protoplasts from both wild type NL-6 and nl6-35 and transformed wild type CqPORB to the protoplasts of nl6-35. In protoplast, total Chl content in the nl6-35 mutant was much lower than that of the NL-6. However, transformation of CqPORB gene, but not the empty vector, to the protoplasts of nl6-35 significantly increased Chl content (Supplementary Figure 3), suggesting that CqPORB gene mutation is responsible for the reduced Chl content in the nl6-35 mutant.
nl6-35 shows abnormal chloroplast development and reduced conversion of Pchlide to Chlide upon illumination
Studies have shown that disruption of POR genes affects the development of thylakoids in both Arabidopsis and rice (Paddock et al., 2010; Paddock et al., 2012; Sakuraba et al., 2013). Here we took a close view of chloroplast in nl6-35 by using transmission electron microscopy. In the wild type NL-6, numerous grana stacks were observed in the chloroplasts of light-grown plants. In the nl6-35 mutant, however, only unstacked thylakoids were observed (Figure 3A), indicating that CqPORB is crucial for grana development in quinoa.
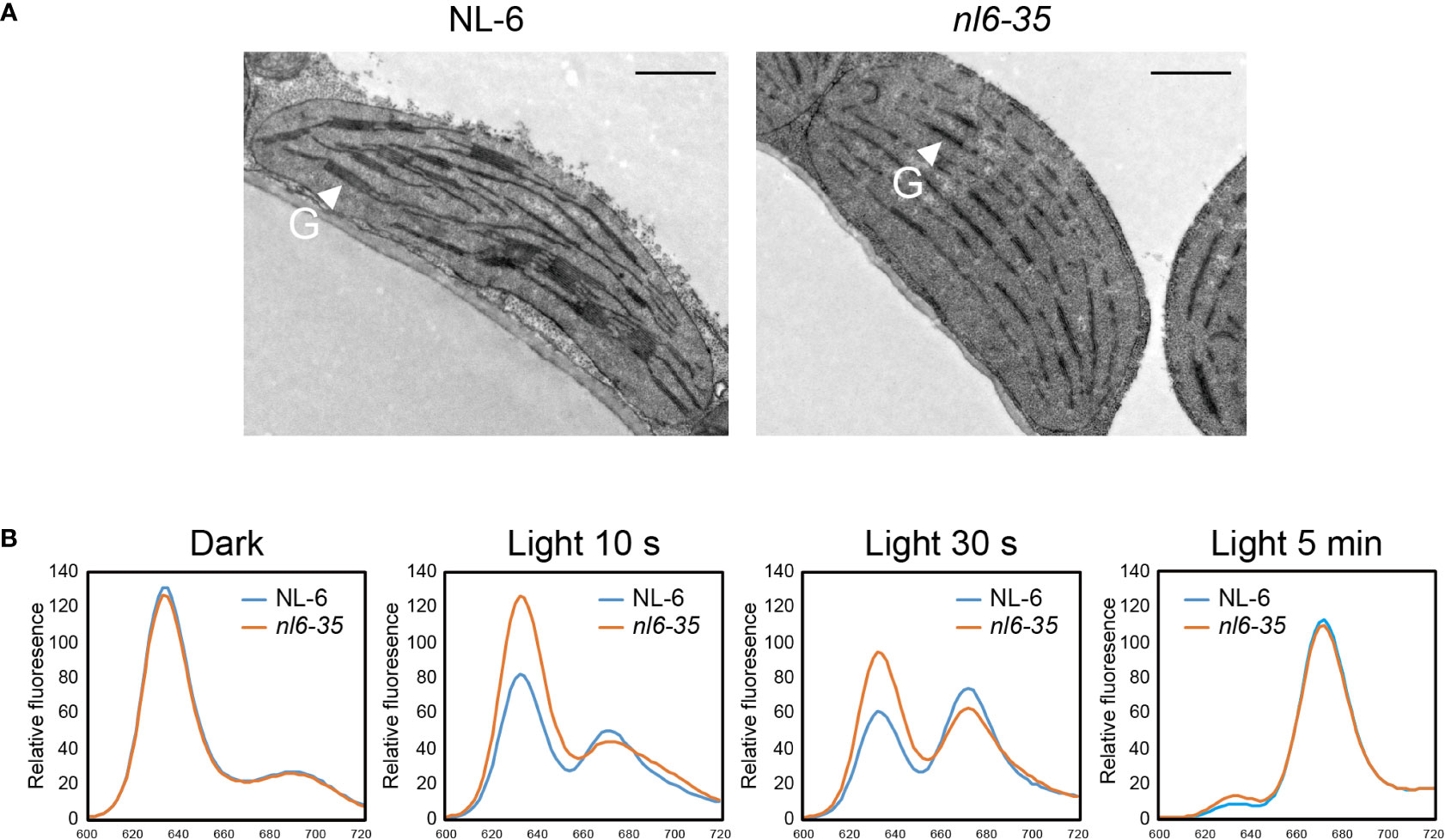
Figure 3 Transmission electron microscopy (TEM) analysis of plastids and measurement of Pchlide in nl6-35 mutant. (A) 10-day-old wild type (NL-6) and nl6-35 leaves grown under long-day conditions (16 h light/8 h dark) were collected for the analysis of grana in chloroplast. G, grana stacks. Scale bar, 1.0 μm. (B) Analysis of the conversion of Pchlide to Chlide upon illumination. Pigments were extracted from five-day-old etiolated seedlings in the dark or after being exposure to light for 10 s, 30 s, and 5 min. Total Pchlide was calculated from the fluorescence emission at 634 nm, while Chlide was calculated from the fluorescence emission at 672 nm.
In Chl biosynthesis pathway, POR is an essential enzyme that catalyzes the reduction of Pchlide to Chlide in a light-dependent way (Tanaka and Tanaka, 2007). POR-bound Pchlide is termed ‘photoactive’ and be able to converted to Chlide instantaneously in the presence of light, whereas Pchlide not bound by POR is termed ‘non-photoactive’ and has been indicated as a factor causing photooxidative damage (Paddock et al., 2010). Here, we measured the content of total Pchlide and Chlide in NL-6 and nl6-35 before and after light exposure for 10 s, 30s, and 5 min. Under dark conditions, the total Pchlide content was comparable between wild type NL-6 and nl6-35. However, after illumination, the photoactive Pchlide was reduced in a higher speed in NL-6 than in nl6-35 mutant, while Chlide was increased in a higher rate in NL-6 than in nl6-35 mutant, suggesting that CqPORB is important for the conversion of Pchlide to Chlide (Figure 3B).
Characterization of POR genes in quinoa
The genomic length of CqPORB annotated in NL-6 genome was 2780 nucleotide acids, and this gene harbors five exons and encodes a protein of 395 amino acids. CqPORB shares high sequence similarities with its homologs in other plants species, including Arabidopsis thaliana At5g54190/PORA (77.5%), At4g27440/PORB (76.6%) and At1g03630/PORC (76.6%); Oryzae sativa Os04g0678700/PORA (70.3%) and Os10g0496900/PORB (72.8%); Physcomitrella patens Ppls146_112V6.1 (78.2%) and Ppls108_171V6.1 (68.2%); Selaginella moellendorffii SMpep_g142334 (71.0%); and Chlamydomonas reinhardtii Cre01.g015350.t1.1 (65.8%) (Supplementary Figure 4). This result suggested that POR proteins are conserved from algae to higher plants.
Alignment of CqPORB against the protein database of NL-6 genome revealed that there are two additional isoforms, CqNL-6_007045 and CqNL-6_022507, which are located on Chr4 and Chr12, respectively, based on the annotation of NL-6 genome. Quinoa is an allotetraploid plant that consists of A sub-genome and B sub-genome that originated from diploid Chenopodium pallidicaule and diploid Chenopodium suecicum, respectively (Jarvis et al., 2017). Based on synteny analysis of quinoa genome, it has been shown that Chr4 and Chr12 are the colinear chromosomes in quinoa that originated from the A and B sub-genomes of its progenitors (Jarvis et al., 2017). These clues suggest that CqNL-6_007045 and CqNL-6_022507 are evolutionarily the same isoform, and thereby they were designated as CqPORA (CqNL-6_007045) and CqPORA-like (CqNL-6_022507), respectively.
Interestingly, we only identified one copy of CqPORB on Chr17 in NL-6 variety that originated from A sub-genome, indicating that the copy of CqPORB might be lost in the B sub-genome. To understand whether this is also the case in other quinoa varieties, we aligned CqPORB protein sequence with the protein database of the sequenced QQ74 variety (Jarvis et al., 2017). Unexpectedly, there are four CqPOR isoforms in QQ74 variety, and the two copies of CqPORA and CqPORB were identified in the A sub-genome and B sub-genome, respectively (Supplementary Table 4). These results indicated that the CqPORB copy was only lost in the NL-6, but not in QQ74. To understand whether CqPORB-like gene was truly lost in the genome of NL-6 or it was caused by the incomplete genome sequencing. The collinearity of the CqPORB-like gene and its flanking genes between QQ74 and NL-6 varieties was analyzed. Totally 20 genes, including CqPORB-like gene, were selected in the QQ74 genome, and among them 9 genes were also identified in the collinear region of NL-6 genome, but CqPORB-like gene and its neighboring genes was completely lost in the NL-6 genome (Figure 4A). To further exclude the possibility that the loss of CqPORB-like gene in NL-6 was caused by incomplete sequencing, primers that matched to both CqPORB and CqPORB-like genes were used for gene amplification using the genomic DNAs isolated from NL-6, Longli-4 and Real varieties, and PCR products were sequenced by sanger sequencing. The result showed that only one isoform was amplified in the NL-6 variety, but two isoforms were amplified in both Longli-4 and Real varieties (Figure 4B), verifying that CqPORB-like gene was lost in the NL-6 genome.
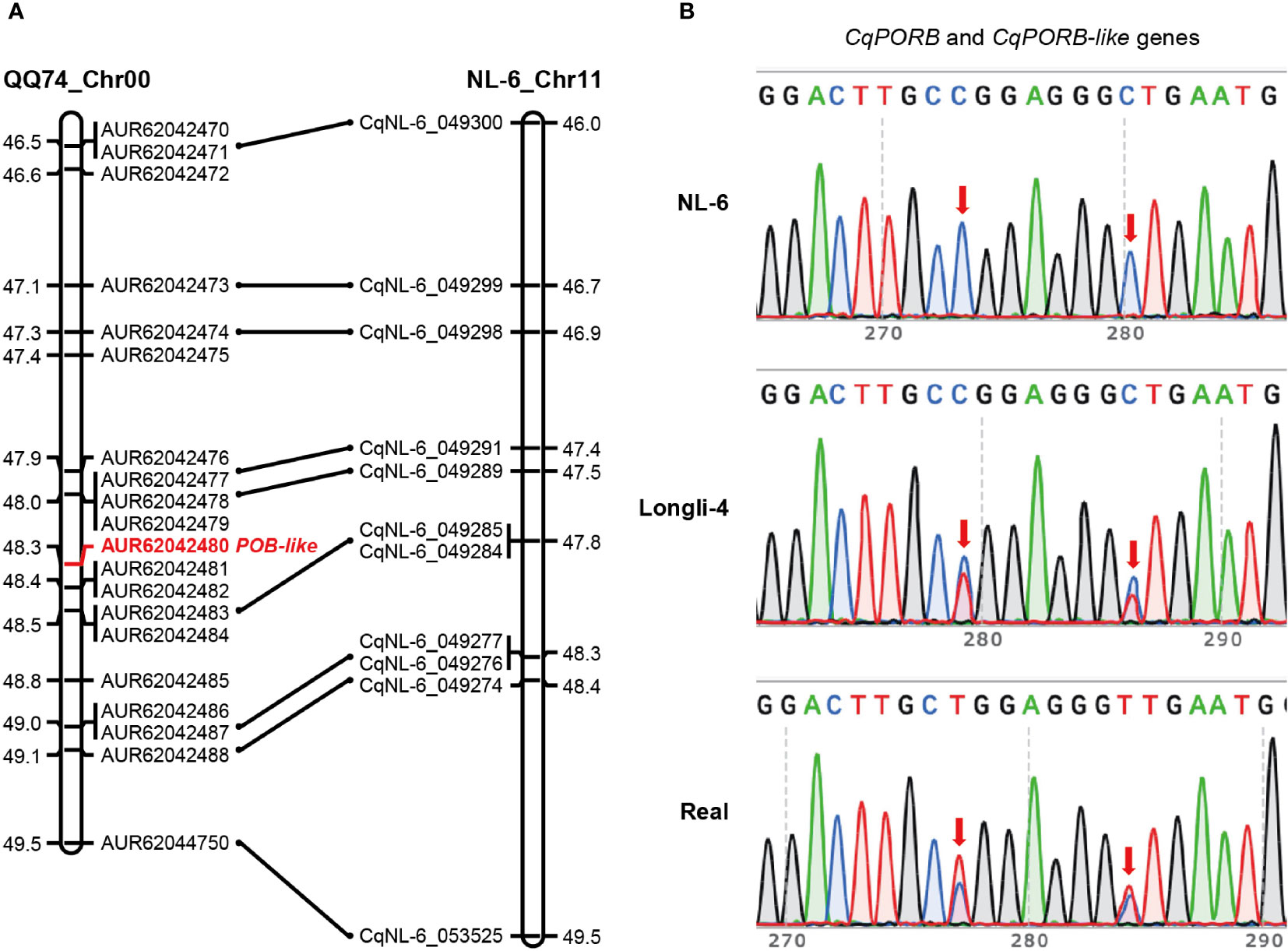
Figure 4 Collinearity of the CqPORB-like gene-located region between QQ74 and NL-6 varieties. (A) The CqPORB-like gene (AUR62042480) and its flanking genes in QQ74 variety were selected to align with the collinear chromosomal region in NL-6 variety. Note that CqPORB-like gene is lost in the NL-6 genome. (B) PCR amplification of CqPORB and CqPORB-like genes using a pair of primers matched to both genes in NL-6, Longli-4, and Real varieties. The PCR products were sequenced by sanger sequencing. Arrows indicate the variations between CqPORB and CqPORB-like genes.
Gene expression analysis of CqPORs in quinoa
In a previous study, we have performed RNA-seq analysis for four different tissues, including leaf, root, stamen, and pistil in NL-6 variety (Jiang et al., 2022). Using these transcriptomic data, we found that all the three CqPOR genes were highly expressed in the leaves, but with a much less level in pistil, and they were almost not detected in root and stamen (Figure 5A). This gene expression pattern is consistent with the role of POR genes in the regulation of Chl biosynthesis in photosynthetic tissues. Moreover, the transcriptomic data showed that CqPORA and CqPORA-like genes rather than CqPORB gene exhibited a more similar gene expression pattern (Figure 5A).
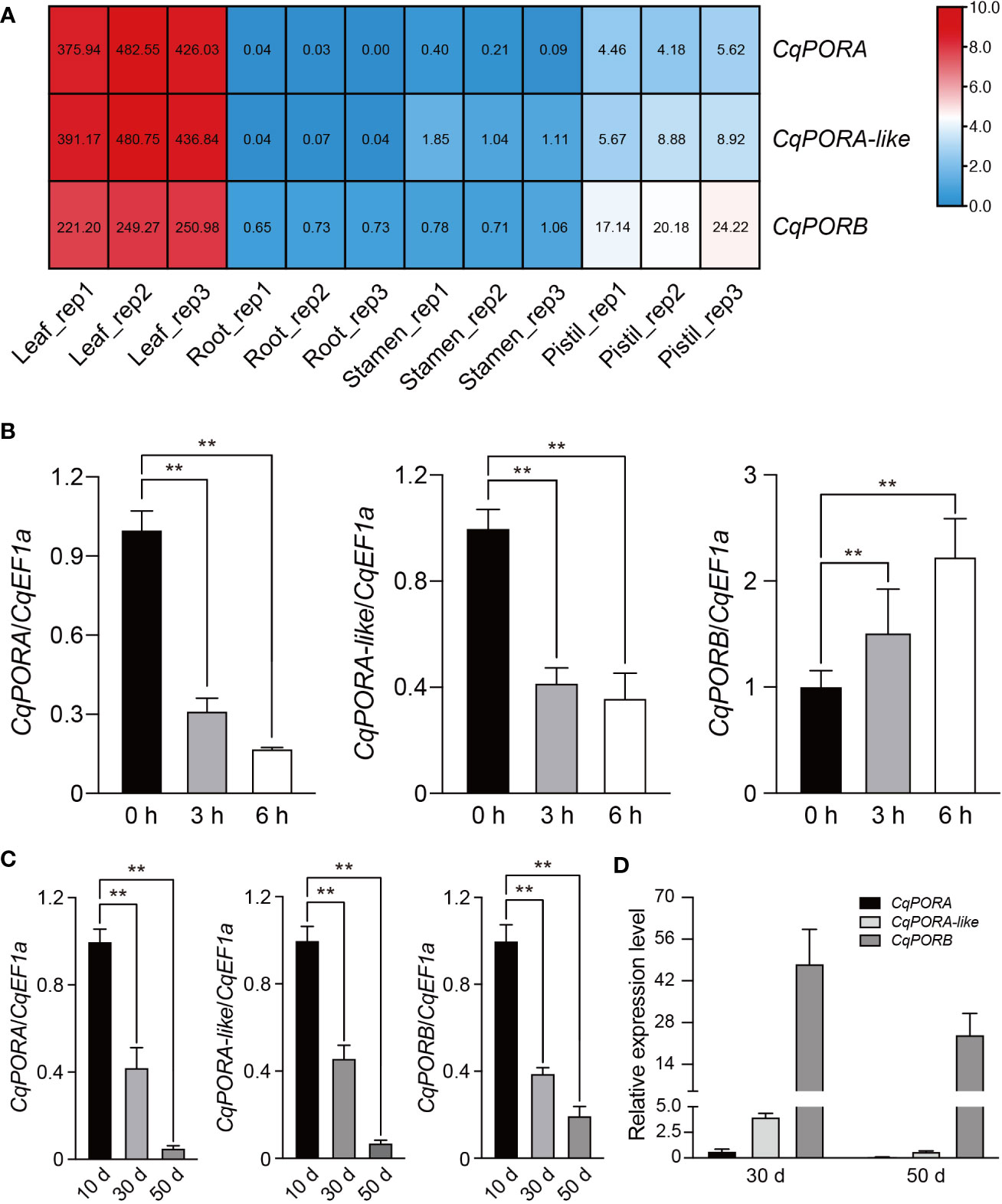
Figure 5 Expression patterns of POR genes in quinoa. (A) RNA-seq assay was performed for the leaf, root, stamen, and pistil tissues of NL-6 variety, and three independent replicates were conducted for each tissue. Different colors in the heatmap indicate the relative expression level of each CqPOR gene in different tissues based on log2-transformed FPKM. The FPKM values of CqPOR genes in four tissues are shown. (B) qRT-PCR analysis of the expression level of each CqPOR gene in etiolated seedlings after being exposed to white light for 0, 3, and 6 h. CqEF1a was used as an internal control. The transcript level of each CqPOR gene at 0 h was set as 1. Values are the means ± SD of three biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test). (C) qRT-PCR-analysis of the expression level of each CqPOR gene in NL-6 after growth on soils for 10, 30, and 50 days. CqEF1a was used as an internal control. The transcript level of each CqPOR gene at 10 d was set as 1. Values are the means ± SD of three biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test). (D) Comparison of the relative expression levels of the three CqPOR genes in NL-6 after growth on soils for 30 and 50 days. Values are the means ± SD of three biological replicates.
It has been known that the expression of POR genes is altered upon illumination (Armstrong et al., 1995). Here, we tested the expression of these three CqPOR genes in NL-6 etiolated seedlings during the transit from dark to white light. qRT-PCR analysis showed that both CqPORA and CqPORA-like genes were down-regulated, while CqPORB gene was up-regulated after exposure to light (Figure 5B). The similar expression pattern of CqPORA and CqPORA-like genes in response to light further supported that they are evolutionarily the same isoform. The different expression pattern between CqPORA and CqPORB genes indicated that these two genes may exhibit a distinct function in the regulation of Chl synthesis in response to ambient light signal. We also analyzed the expression of these three POR genes in different developmental stages in quinoa, and found that after growth on soils for 10, 30, and 50 d, the expression levels of all these three POR genes were gradually decreased, but the CqPORB gene was reduced in a slower rate than the CqPORA genes (Figure 5C). At later developmental stages, we noted that the transcript level of CqPORB was much higher than the CqPORA and CqPORA-like genes (Figure 5D), indicating that CqPORB plays a predominant role in Chl biosynthesis and chloroplast development in mature plants.
CqPORB is localized in chloroplast
To study the subcellular localization of CqPORB in quinoa, we infiltrated Agrobacterium strain harboring 35S::CqPORB-GFP plasmid into quinoa leaves according to the protocol described previously (Xiao et al., 2022). However, after transformation for 48 h, no GFP fluorescence signal was detected in quinoa leaves, suggesting that the efficiency of transient gene expression in quinoa leaves is still very low. We then transformed 35S::CqPORB-GFP plasmid to tobacco leaves, and found that CqPORB was exclusively localized in chloroplast (Figure 6A). Besides, 35S::CqPORB-GFP was transformed into Arabidopsis to obtain stable transgenic plants, and in these transgenic plants, GFP fluorescence was also clearly observed in chloroplast (Figure 6B).
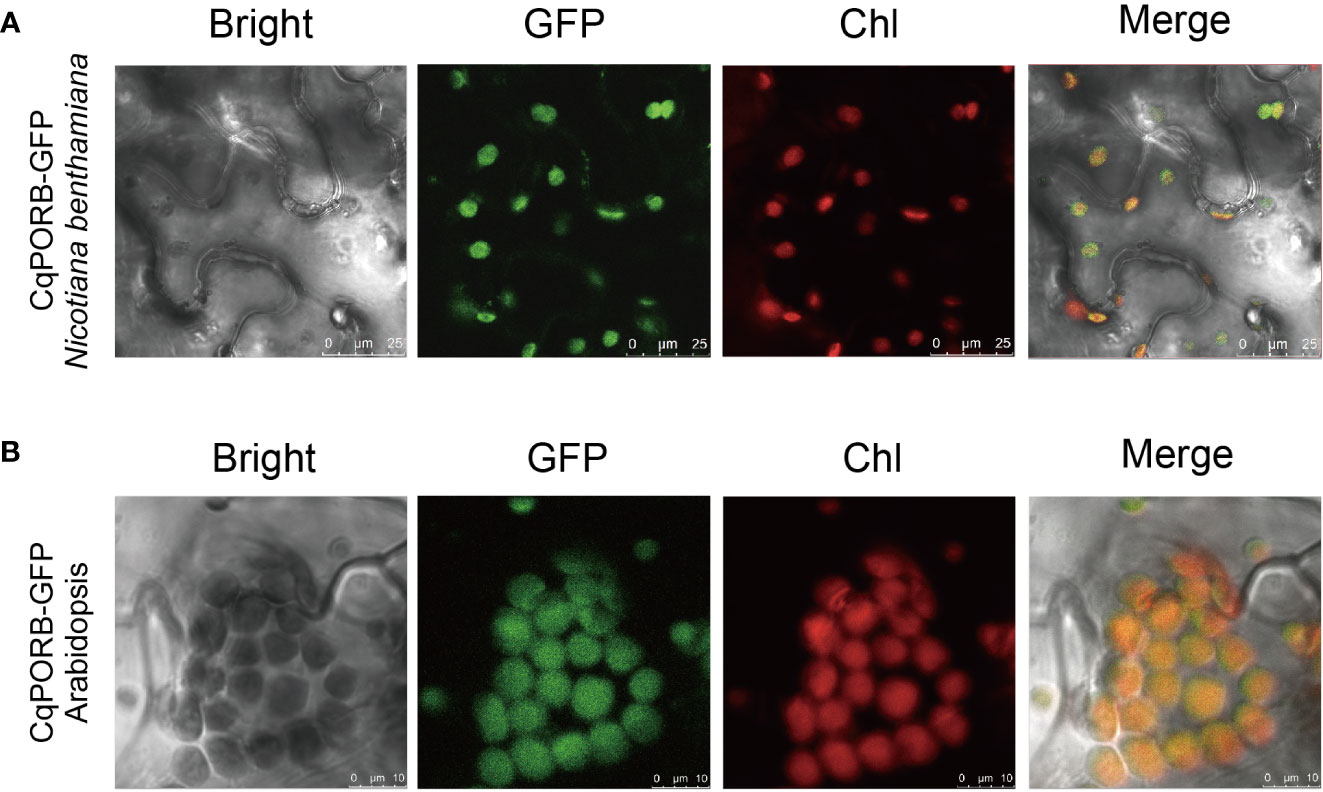
Figure 6 Analysis of the subcellular localization of CqPORB. (A) CqPORB-GFP was transiently expressed in Nicotiana benthamiana, and fluorescence was detected using confocal laser scanning microscopy. Scale bar, 25 µm. (B) The subcellular localization of CqPORB was analyzed in Arabidopsis transgenic plants expressing CqPORB-GFP. Fluorescence was detected using confocal laser scanning microscopy. Scale bar, 10 µm.
Discussion
Quinoa is an allotetraploid plant that harbors two similar genomes (Jarvis et al., 2017), so genetic study of a specific gene function in quinoa is a challenging work. However, using EMS-mediated mutagenesis of NL-6 variety, we identified some mutants with altered phenotypes, including early flowering, dwarfism, yellow leaves, and branched plants, suggesting that some phenotypes of quinoa are control by a single gene derived from A sub-genome or B sub-genome of quinoa or the biological functions of two gene copies originated from progenitors diverged during evolution. Based on BSA-seq analysis, 3,013 SNPs that were derived from C to T or G to A change were identified in nl6-35 mutant, and a region containing candidate mutated genes was clearly detected, indicating that EMS-mediated mutagenesis is effective in quinoa, and whole genome sequencing-based mapping can be efficiently used to identify mutated genes in quinoa.
In this study, we report that mutation of CqporB gene alone leads to faded green leaf phenotype, which is mainly caused by the reduced Chl and carotenoid contents. Homolog analysis showed that only one copy of CqPORB that locates in A sub-genome was identified in NL-6, implying that CqPORB copy in B sub-genome was lost during evolution, and this could be one of the reasons that we could identify CqporB mutant based on EMS mutagenesis-mediated approach. It has been reported that, compared to other quinoa varieties, NL-6 variety exhibits yellow panicles and relatively reduced biomass (Benlhabib et al., 2016), but the molecular mechanisms underlying these phenotypes are still unknown. Based on our data, we can speculate that these phenotypes may be caused by the existence of only one copy of CqPORB in the genome of NL-6 variety. In future, the reasons underlying the loss of CqPORB-like gene in NL-6 but not in other quinoa varieties worth further investigation.
The function of PORs has been studied in many different plant species, including Arabidopsis, rice, wheat, and barley (Teakle and Griffiths, 1993; Armstrong et al., 1995; Holtorf et al., 1995; Sakuraba et al., 2013). The number of POR genes vary in different plant species. In Arabidopsis, there are three POR genes, AtPORA, AtPORB, and AtPORC, and these three genes exhibit distinct expression patterns in response to light signal (Armstrong et al., 1995). In rice, only two POR genes, OsPORA and OsPORB, were identified, and their expression levels were both altered after illumination, despite in an opposite way (Sakuraba et al., 2013). Similar to rice, there are two POR isoforms in quinoa, and these two isoforms also exhibited an opposite response to light, with CqPORA down-regulated and CqPORB up-regulated after light exposure, suggesting that these two POR genes are conserved in terms of gene expression pattern in different plant species. Our result showed that nl6-35 mutant, in which CqPORB gene is mutated, did not exhibit obvious phenotypes in young seedling stage compared with the wild type, but all mutant plants display faded green leaf after growth for approximately 14 days. This phenomenon can be explained by the fact that both CqPORA and CqPORB genes are expressed in young seedings, but in mature plants, only CqPORB gene is predominately expressed. In line with the role of POR proteins in the biosynthesis of Chl, CqPORB is exclusively localized in the chloroplast, and RNA-seq data showed that all three CqPOR genes in NL-6 variety were highly expressed in leaves, but much less or not expressed in root, stamen, and pistil.
CqPORB gene encodes a NADPH:protochlorophyllide oxidoreductase that is required for the photo-reduction of Pchlide to Chlide (Heyes and Hunter, 2005). Recently, the structure-based mechanism of the reduction of Pchlide to Chlide upon illumination has been addressed. Plant POR is able to bind to Pchlide, NADPH, and lipids to forms helical filaments around lipid bilayer tubes, which facilitates the target of its product, Chlide, to the membrane for chlorophyll biosynthesis (Nguyen et al., 2021). Our result showed that the conversion of photoactive Pchlide to Chlide was disrupted in CqporB mutant in quinoa, which is similar to that observed in OsporB mutant in rice (Sakuraba et al., 2013), suggesting that PORB proteins in different plant species exhibit a similar function. Our data also showed that CqporB gene mutation resulted in abnormal grana stacks, which is similar to that of AtporB AtporC double mutant in Arabidopsis and that of OsporB mutant in rice (Paddock et al., 2010; Sakuraba et al., 2013), demonstrating that POR proteins play a structural role in the formation of grana stacks in chloroplast. In rice, it has been shown that disruption of CqporB gene results in necrotic lesions in leaves, which is largely caused by the increased accumulation of non-photoactive Pchlide and increased production of singlet oxygen (Sakuraba et al., 2013). In future, the production of singlet oxygen in CqporB mutant upon illumination is worth investigating. In addition, the role of FLU protein in the regulation of Pchlide accumulation and singlet oxygen production in quinoa needs to be further explored.
Quinoa has been regarded as a nutritional crop to contribute to global food security, and the demand for quinoa is expected to be rapidly increased in near future (Bazile et al., 2016). Because of the importance of quinoa as nutritional food, governments and companies in many countries have put increasing efforts to expand quinoa cultivation in non-native areas and improve the yield of quinoa. Due to some limited agronomic traits, large-scale cultivation of quinoa in non-native areas is still limited (Mizuno et al., 2020), so breeding of quinoa varieties with improved agronomic traits that adapt to diverse climate and soil conditions becomes increasingly urgent. Taking advantage of the high-quality quinoa reference genome and diverse quinoa varieties, molecular marker-assisted breeding will be an effective way to cultivate quinoa with improved agronomic traits. Photosynthesis is often explored as a target for the crop yield improvement (Long et al., 2015; Walter and Kromdijk, 2022), so identification of genes involved in photosynthesis in quinoa will provide valuable genetic loci for yield improvement of this pseudocereal crop.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI, GSE198572.
Author contributions
CL, MR, and CZ contributed to conception and design of the study. CL, MR, JL, and XW performed the most of experiments. QW and HZ conducted protein subcellular localization analysis. QZ, JY, and FY provided quinoa materials and assisted the growth of quinoa. HZ and J-KZ participated in scientific discussions. CZ wrote the manuscript with contributions from CL and MR. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Shanghai Agriculture Applied Technology Development Program, China (Grant No. X20200101).
Acknowledgments
We would like to thank Dr. Peng Wang for the assistance of Pchlide content analysis and Prof. Pengshan Zhao for providing quinoa Longli-4 variety.
Conflict of interest
Authors QZ, JY, and FY are employed by Bright Agricultural Development (Group) Co., Ltd, Shanghai, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1083438/full#supplementary-material
Supplementary Figure 1 | The growth phenotype of nl6-35 mutant. (A) Phenotype of wild type (NL-6) and nl6-35 plants grown in a growth chamber for 40 days. Scale bar, 10 cm. (B) Phenotype of wild type (NL-6) and nl6-35 plants grown in field for 80 days.(C) Cotyledons of wild type (NL-6) and nl6-35 seedlings grown on soils for 7 days. Scale bar, 1 cm.
Supplementary Figure 2 | Identification of SNPs in nl6-35 mutant using BSA-seq. The F2 plants of the crossing population of nl6-35 and NL-6 were collected for whole-genome sequencing. All SNPs with C to T or G to A change were selected and distributed on the chromosomes of quinoa.
Supplementary Figure 3 | Transformation of CqPORB gene rescues the reduced Chl content in nl6-35 mutant using protoplast system. Protoplasts from wild type NL-6 and nl6-35 were generated, and CqPORB gene was transformed into the protoplasts of nl6-35 before total Chl was measured. The protoplasts transformed with empty vector were used as control. Values are the means ± SD of three biological replicates. Asterisks indicate statistically significant differences (**p < 0.01, Student’s t-test).
Supplementary Figure 4 | Alignment of POR homologs in different plant species. (A) Alignment of POR proteins in Chenopodium quinoa, Arabidopsis thaliana, Oryzae sativa, Physcomitrella patens, Selaginella moellendorffii, and Chlamydomonas reinhardtii. (B) Phylogenetic analysis of PORs in different plant species. Phylogenetic tree was generated by using IQ-TREE2 program.
References
Allen, J. F., de Paula, W. B. M., Puthiyaveetil, S., Nield, J. (2011). A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 16, 645–655. doi: 10.1016/j.tplants.2011.10.004
Armstrong, G. A., Runge, S., Frick, G., Sperling, U., Apel, K. (1995). Identification of NADPH:Protochlorophyllide oxidoreductases a and b: A branched pathway for light-dependent chlorophyll biosynthesis in arabidopsis thaliana. Plant Physiol. 108, 1505–1517. doi: 10.1104/pp.108.4.1505
Bazile, D., Jacobsen, S. E., Verniau, A. (2016). The global expansion of quinoa: trends and limits. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00622
Benlhabib, O., Boujartani, N., Maughan, P. J., Jacobsen, S. E., Jellen, E. N. (2016). Elevated genetic diversity in an F2:6 population of quinoa (chenopodium quinoa) developed through an inter-ecotype cross. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01222
Blomqvist, L. A., Ryberg, M., Sundqvist, C. (2008). Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth. Res. 96, 37–50. doi: 10.1007/s11120-007-9281-y
Böhm, J., Messerer, M., Müller, H. M., Scholz-Starke, J., Gradogna, A., Scherzer, S., et al. (2018). Understanding the molecular basis of salt sequestration in epidermal bladder cells of chenopodium quinoa. Curr. Biol. 28, 3075–3085. doi: 10.1016/j.cub.2018.08.004
Cai, Z. Q., Gao, Q. (2020). Comparative physiological and biochemical mechanisms of salt tolerance in five contrasting highland quinoa cultivars. BMC Plant Biol. 20, 70. doi: 10.1186/s12870-020-2279-8
Chen, J. H., Chen, S. T., He, N. Y., Wang, Q. L., Zhao, Y., Gao, W., et al. (2020). Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants 6, 570–580. doi: 10.1038/s41477-020-0629-z
Clough, S. J., Bent, A. F. (1998). Floral dip: A simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dakhili, S., Abdolalizadeh, L., Hosseini, S. M., Shojaee-Aliabadi, S., Mirmoghtadaie, L. (2019). Quinoa protein: Composition, structure and functional properties. Food Chem. 299, 125–161. doi: 10.1016/j.foodchem.2019.125161
Dillehay, T. D., Rossen, J., Andres, T. C., Williams, D. E. (2007). Preceramic adoption of peanut, squash, and cotton in northern Peru. Science 316, 1890–1893. doi: 10.1126/science.1141395
Eberhard, S., Finazzi, G., Wollman, F. A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515. doi: 10.1146/annurev.genet.42.110807.091452
Frick, G., Su, Q., Apel, K., Armstrong, G. A. (2003). An arabidopsis porB porC double mutant lacking light-dependent NADPH: Protochlorophyllide oxidoreductases b and c is highly chlorophyll-deficient and developmentally arrested. Plant J. 35, 141–153. doi: 10.1046/j.1365-313X.2003.01798.x
Graf, B. L., Rojas Silva, P., Rojo, L. E., Delatorre Herrera, J., Baldeón, M. E. (2016). Innovations in health value and functional food development of quinoa (Chenopodium quinoa willd.). Compr. Rev. Food Sci. Food Saf. 14, 431–445. doi: 10.1111/1541-4337.12135
Hariadi, Y., Marandon, K., Tian, Y., Jacobsen, S. E., Shabala, S. (2011). Ionic and osmotic relations in quinoa (Chenopodium quinoa willd.) plants grown at various salinity levels. J. Exp. Bot. 62, 185–193. doi: 10.1093/jxb/erq257
Heyes, D. J., Hunter, C. N. (2005). Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem. Sci. 30, 642–649. doi: 10.1016/j.tibs.2005.09.001
Holtorf, H., Reinbothe, S., Reinbothe, C., Bereza, B., Apel, K. (1995). Two routes of chlorophyllide synthesis that are differentially regulated by light in barley (Hordeum vulgare l.). Proc. Natl. Acad. Sci. U. S. A. 92, 3254–3258. doi: 10.1073/pnas.92.8.3254
Jarvis, D. E., Ho, Y. S., Lightfoot, D. J., Schmöckel, S. M., Li, B., Borm, T. J. A., et al. (2017). The genome of chenopodium quinoa. Nature 542, 307–312. doi: 10.1038/nature21370
Jiang, W., Li, C., Li, L., Li, Y., Wang, Z., Yu, F., et al. (2022). Genome-wide analysis of CqCrRLK1L and CqRALF gene families in chenopodium quinoa and their roles in salt stress response. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.918594
Kim, C., Meskauskiene, R., Zhang, S., Lee, K. P., Ashok, M. L., Blajecka, K., et al. (2012). Chloroplasts of arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039. doi: 10.1105/tpc.112.100479
Kwon, C. T., Kim, S. H., Song, G., Kim, D., Paek, N. C. (2017). Two NADPH: protochlorophyllide oxidoreductase (POR) isoforms play distinct roles in environmental adaptation in rice. Rice 10, 1. doi: 10.1186/s12284-016-0141-2
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, 293–296. doi: 10.1093/nar/gkab301
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, L., Zhang, Y., Tang, S., Zhao, Q., Zhang, Z., Zhang, H., et al. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in nicotiana benthamiana. Plant J. 61, 893–903. doi: 10.1111/j.1365-313X.2009.04109.x
Long, S. P., Marshall-Colon, A., Zhu, X. G. (2015). Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. doi: 10.1016/j.cell.2015.03.019
Masuda, T., Fusada, N., Oosawa, N., Takamatsu, K., Yamamoto, Y. Y., Ohto, M., et al. (2003). Functional analysis of isoforms of NADPH: Protochlorophyllide oxidoreductase (POR) In arabidopsis thaliana. Plant Cell Physiol. 44, 963–974. doi: 10.1093/pcp/pcg128
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mizuno, N., Toyoshima, M., Fujita, M., Fukuda, S., Kobayashi, Y., Ueno, M., et al. (2020). The genotype-dependent phenotypic landscape of quinoa in salt tolerance and key growth traits. DNA Res. 27, dsaa022. doi: 10.1093/dnares/dsaa022
Nguyen, H. C., Melo, A. A., Kruk, J., Frost, A., Gabruk, M. (2021). Photocatalytic LPOR forms helical lattices that shape membranes for chlorophyll synthesis. Nat. Plants 7, 437–444. doi: 10.1038/s41477-021-00885-2
Nie, L., Zheng, Y., Zhang, L., Wu, Y., Zhu, S., Hou, J., et al. (2021). Characterization and transcriptomic analysis of a novel yellow-green leaf wucai (Brassica campestris l.) germplasm. BMC Genomics 22, 258. doi: 10.1186/s12864-021-07573-7
Op Den Camp, R. G. L., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. Plant Cell 15, 2320–2332. doi: 10.1105/tpc.014662
Paddock, T., Lima, D., Mason, M. E., Apel, K., Armstrong, G. A. (2012). Arabidopsis light-dependent protochlorophyllide oxidoreductase a (PORA) is essential for normal plant growth and development. Plant Mol. Biol. 78, 447–460. doi: 10.1007/s11103-012-9873-6
Paddock, T. N., Mason, M. E., Lima, D. F., Armstrong, G. A. (2010). Arabidopsis protochlorophyllide oxidoreductase a (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 72, 445–457. doi: 10.1007/s11103-009-9582-y
Reinbothe, C., Bakkouri, M., Buhr, F., Muraki, N., Nomata, J., Kurisu, G., et al. (2010). Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–624. doi: 10.1016/j.tplants.2010.07.002
Reinbothe, C., Lebedev, N., Reinbothe, S. (1999). A protochlorophyllide light-harvesting complex involved in de-etiolation of higher plants. Nature 397, 80–84. doi: 10.1038/16283
Sakuraba, Y., Rahman, M. L., Cho, S. H., Kim, Y. S., Koh, H. J., Yoo, S. C., et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase b and is essential for chlorophyll synthesis under high light conditions. Plant J. 74, 122–133. doi: 10.1111/tpj.12110
Selstam, E., Schelin, J., Brain, T., Williams, W. P. (2002). The effects of low pH on the properties of protochlorophyllide oxidoreductase and the organization of prolamellar bodies of maize (Zea mays). Eur. J. Biochem. 269, 2336–2346. doi: 10.1046/j.1432-1033.2002.02897.x
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Tanaka, R., Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448
Teakle, G. R., Griffiths, W. T. (1993). Cloning, characterization and import studies on protochlorophyllide reductase from wheat (Triticum aestivum). Biochem. J. 296, 225–230. doi: 10.1042/bj2960225
Walter, J., Kromdijk, J. (2022). Here comes the sun: How optimization of photosynthetic light reactions can boost crop yields. J. Integr. Plant Biol. 64, 564–591. doi: 10.1111/jipb.13206
Wang, P., Grimm, B. (2021). Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. 26, 484–495. doi: 10.1016/j.tplants.2020.12.005
Wang, K., Li, M., Hakonarson, H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Xiao, X., Meng, F., Satheesh, V., Xi, Y., Lei, M. (2022). An agrobacterium-mediated transient expression method contributes to functional analysis of a transcription factor and potential application of gene editing in chenopodium quinoa. Plant Cell Rep. 41, 1975–1985. doi: 10.1007/s00299-022-02902-w
Yasui, Y., Hirakawa, H., Oikawa, T., Toyoshima, M., Matsuzaki, C., Ueno, M., et al. (2016). Draft genome sequence of an inbred line of chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res. 23, 535–546. doi: 10.1093/dnares/dsw037
Zhu, P., He, L., Li, Y., Huang, W., Xi, F., Lin, L., et al. (2014). OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PloS One 9, e97507. doi: 10.1371/journal.pone.0097507
Keywords: protochlorophyllide oxidoreductase (POR), quinoa (Chenopodium quinoa Willd), protochlorophyllide, chlorophyll, grana stacks
Citation: Li C, Ran M, Liu J, Wang X, Wu Q, Zhang Q, Yang J, Yi F, Zhang H, Zhu J-K and Zhao C (2022) Functional analysis of CqPORB in the regulation of chlorophyll biosynthesis in Chenopodium quinoa. Front. Plant Sci. 13:1083438. doi: 10.3389/fpls.2022.1083438
Received: 29 October 2022; Accepted: 29 November 2022;
Published: 12 December 2022.
Edited by:
Benoît Castandet, Université Paris Cité, FranceReviewed by:
Juan B. Arellano, Spanish National Research Council (CSIC), SpainXueyun Hu, Yangzhou University, China
Copyright © 2022 Li, Ran, Liu, Wang, Wu, Zhang, Yang, Yi, Zhang, Zhu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunzhao Zhao, Y3p6aGFvQGNlbXBzLmFjLmNu
†These authors have contributed equally to this work
 Chao Li1†
Chao Li1† Chunzhao Zhao
Chunzhao Zhao